Highlights
-
•
At least five distinct species of Bartonella infect R. rattus rodents and their ectoparasites in Madagascar.
-
•
Infection dynamics for zoonotic B. elizabethae are consistent with an SIS or SIR model in rodent hosts.
-
•
Infection dynamics for non-zoonotic B. phoceensis & rattimassiliensis are consistent with an SI model in rodent hosts.
-
•
Transmission of B. elizabethae and B. phoceensis & rattimassiliensis, respectiviely, are affected by S. fonquerniei & X. cheopsis fleas and Polyplax spp. lice.
-
•
S. fonquerniei/X. cheopsis fleas may vector B. elizabethae and Polyplax spp. lice vector B. phoceensis/rattimassiliensis.
Keywords: Bartonella spp., Rattus rattus, Synopsyllus fonquerniei, Madagascar, Force of infection
Abstract
Bartonella spp. are erythrocytic bacteria transmitted via arthropod vectors, which infect a broad range of vertebrate hosts, including humans. We investigated transmission dynamics and host-parasite-vector relationships for potentially zoonotic Bartonella spp. in invasive Rattus rattus hosts and associated arthropod ectoparasites in Madagascar. We identified five distinct species of Bartonella (B. elizabethae 1, B. elizabethae 2, B. phoceensis 1, B. rattimassiliensis 1, and B. tribocorum 1) infecting R. rattus rodents and their ectoparasites. We fit standard epidemiological models to species-specific age-prevalence data for the four Bartonella spp. with sufficient data, thus quantifying age-structured force of infection. Known zoonotic agents, B. elizabethae 1 and 2, were best described by models exhibiting high forces of infection in early age class individuals and allowing for recovery from infection, while B. phoceensis 1 and B. rattimassiliensis 1 were best fit by models of lifelong infection without recovery and substantially lower forces of infection. Nested sequences of B. elizabethae 1 and 2 were recovered from rodent hosts and their Synopsyllus fonquerniei and Xenopsylla cheopsis fleas, with a particularly high prevalence in the outdoor-dwelling, highland-endemic S. fonquerniei. These findings expand on force of infection analyses to elucidate the ecological niche of the zoonotic Bartonella elizabethae complex in Madagascar, hinting at a potential vector role for S. fonquerniei. Our analyses underscore the uniqueness of such ecologies for Bartonella species, which pose a variable range of potential zoonotic threats.
1. Introduction
Bartonella spp. are facultative, intracellular Gram-negative bacteria, which infect the endothelial cells and erythrocytes of a broad diversity of vertebrate hosts. Bartonella spp. have been documented infecting a wide range of mammals (rodents, bats, carnivores, ungulates, and humans, among others; Chomel et al., 2009, Kosoy, 2010), as well as some birds and reptiles (Mascarelli et al., 2014, Valentine et al., 2007). At least twelve known species or subspecies of Bartonella bacteria are now recognized zoonotic agents (Vayssier-Taussat et al., 2009, Veikkolainen et al., 2014), and two others, B. bacilliformis and B. quintana (respectively, the causative agents for Carrion’s disease and trench fever) can maintain transmission in human hosts (Vayssier-Taussat et al., 2009). Zoonotic Bartonella spp. typically cause fever, lymphadenopathy, skin lesions, cardiopathy, endocarditis, or neuroretinitis in humans (Breitschwerdt and Kordick, 2000, Kosoy et al., 2003), as exemplified by Bartonella henselae, the causative agent in Cat Scratch Disease (Welch et al., 1992). Reservoir hosts typically do not experience substantial pathology upon infection, instead hosting the pathogen in an as-of-yet-undefined primary niche, from which bacteria are seeded into the blood stream to establish long-term erythrocytic infections (Dehio, 2001). Among Bartonella spp., B. bacilliformis (previously identified only in humans and sandfly vectors) is an exception in causing widespread and life-threatening hemolytic anemia in its primary host (Ihler, 1996).
Five of the recognized zoonotic Bartonella spp. (B. elizabethae, B. vinsonii subsp. arupensis, B. grahamii, B. washoensis, and B. tribocorum) are associated with rodent reservoirs (Buffet et al., 2013, Kandelaki et al., 2016, Kosoy et al., 2010, Vayssier-Taussat et al., 2016), and others are likely to be described in future research (Veikkolainen et al., 2014). In previous studies, Bartonella spp. prevalence declined with increasing rodent weight (indicative of increasing age or health), suggestive of immune clearance mechanisms (Kosoy et al., 2004b, Morway et al., 2008). One study demonstrated antibody reactivity to Bartonella spp. antigens in rodent serum samples (Kosoy et al., 2004a, Kosoy et al., 2004b) but, because Bartonella spp. were pooled in weight trend analyses, species-specific infection dynamics could not be resolved (Kosoy et al., 2004a, Kosoy et al., 2004b). In non-rodent hosts (i.e. cats and humans), antibody-mediated immune responses to persistent infections appear to be common (Brouqui et al., 2005, Kabeya et al., 2006, Pearce et al., 2006).
Because infection dynamics often vary with the age structure of the host population, reporting of apparent prevalence, the proportion of individual hosts testing positive for a disease of interest, can belie a disease’s foothold in a host community (Heisey et al., 2006). In contrast, age-class specific reporting of prevalence data (or, minimally, weight-class grouping of rodent-borne Bartonella spp.; Kosoy et al., 2004b, Morway et al., 2008) can be used to estimate the age-specific force of infection (FOI), or the per capita rate at which susceptible hosts become infected (Muench, 1959). Age-specific FOI can then help identify age-classes most critical in driving population-level patterns of transmission for a given infection and elucidate the influence of specific cohorts on the magnitude and timing of epidemics (Long et al., 2010). In the case of zoonotic infections (i.e. rodent-borne Bartonella spp.), the ‘spillover force of infection,’ or the force of infection from animals to humans, is a product of its constituent factors: infection dynamics within the reservoir animal population, animal-human contact rates, and the probability of viable human infection given such a contact (Keeling and Gilligan, 2000, Lloyd-Smith et al., 2009).
We investigated the first of these three factors—infection dynamics within the reservoir host—for rodent-borne Bartonella spp. in central Madagascar. In particular, we describe the diversity, distribution, and age-structured transmission dynamics of Bartonella spp. infecting peridomestic Rattus rattus and their associated arthropod ectoparasites (fleas, lice, mites, and ticks). Vector ecology has been largely understudied in Bartonella systems (Chomel et al., 2009), though it is thought that most Bartonella spp. are transmitted between hosts by diverse hematophagous arthropods (Billeter et al., 2008), ranging from sandflies (Battisttini, 1931) to lice (Swift, 1920) to fleas (Bown et al., 2004, Chomel et al., 1996), among others. The geographical extent of a vector-borne pathogen is determined by the combined range limits of the host, vector, and pathogen (Reisen, 2010). We explored vector dependency and range limits for potentially zoonotic rodent-borne Bartonella spp. in Madagascar.
Previous work has reported B. quintana in human lice (Sangaré et al., 2014) and novel genotypes of Bartonella in bats and ectoparasite bat flies (Brook et al., 2015, Wilkinson et al., 2016) in Madagascar. To our knowledge, our study represents the first published record of Bartonella spp. infecting rodents and their arthropods in Madagascar. Because invasive rodents live in close proximity, often sympatry, with humans in Madagascar (Rahelinirina et al., 2010), the records we report here are important for understanding zoonotic risk. With this analysis, we specifically aimed to: (1) identify Bartonella spp. infecting peridomestic rats in Madagascar, including genotypes previously known to be zoonotic, (2) model the age-prevalence of specific rodent-borne Bartonella genotypes to develop our understanding of the influence of age-structured force of infection on their transmission, and (3) construct a Bartonella spp. phylogeny from both rodent hosts and associated ectoparasites to elucidate possible vector relationships in our system.
2. Materials and methods
2.1. Field sampling
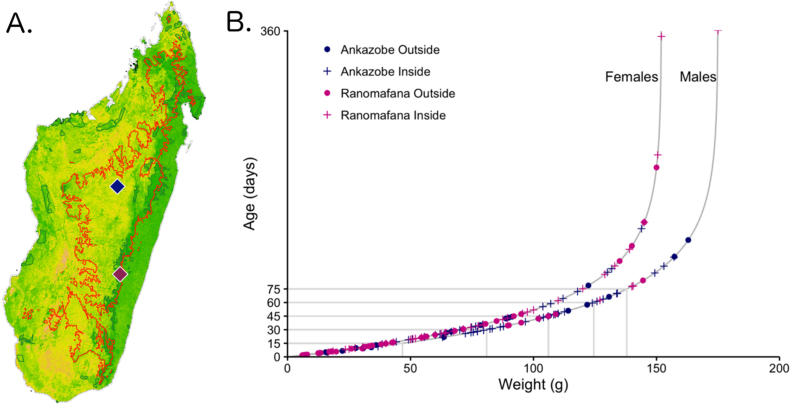
In total, 158 R. rattus were trapped and lethally sampled in two different regions in central Madagascar: in peridomestic communities surrounding Ambohitantely Special Reserve, District of Ankazobe, in July 2013 and in both forest and human communities surrounding Ranomafana National Park in June 2014 (Fig. 1A). Kidney tissue samples were collected from all dispatched animals, frozen on site in liquid nitrogen, then transported to −80 °C freezers at the Institut Pasteur of Madagascar, Antananarivo, to await export. Rats in the District of Ankazobe were also flea-combed, and all visible ectoparasites were collected, stored in ethanol, and additionally exported. Of particular note to the analyses presented in this manuscript, the District of Ankazobe is situated firmly on Madagascar’s High Plateau, within the range limits of the endemic Synopsyllus fonquerniei rat flea (Rahelinirina et al., 2010), while Ranomafana National Park is located on the peripheral, subtropical boundary of this region. Further details of our field sampling methodology are elucidated in Text S1 (Gerber et al., 2010, Vallan, 2000).
Fig. 1.
A. Vegetation map of Madagascar (green = forest; yellow = grassland; orange = desert) with 800 m elevation range limit of S. fonquerniei, the endemic Malagasy rat flea, highlighted in red. Ankazobe (navy) and Ranomafana (fuschia) sites are marked as diamonds. B. Age-for-weight relationships for captured R. rattus as estimated from the von Bertalanffy equation (Ricker, 1979): W(t) = W(1 − e−k(t−t0)) in which W represents the highest weight class rodent in the data subset (males: 175 g, females: 152 g). Data from Ankazobe sites are depicted in navy (outside sampling sites = circles, inside sampling sites = pluses) and from Ranomafana in fuchsia.
2.2. Ethics statement
All field studies were carried out under permit authorization from Madagascar National Parks and the Madagascar Ministry for Water and Forests (research permits: N0162/13/MEF/SG/DGF/DCB.SAP/SCB and N0130/13/MEF/SG/DGF/DCB.SAP/SCB; export permit N0149N-EA08/MG14) and in strict accordance with guidelines posted by the American Veterinary Medical Association. All field protocols employed were pre-approved by the Princeton University Institutional Animal Care and Use Committee (IACUC protocol # 1926, July 2013; #1989, June 2014), and every effort was made to minimize discomfort to animals.
2.3. Molecular analysis
Frozen kidney tissue samples from all lethally sampled rats were transported to the Centers for Disease Control and Prevention (CDC), Fort Collins, CO. Briefly, DNA was extracted using Qiagen QIAamp tissue kits (QIAGEN, Valencia, CA, USA) according to the manufacturer’s instructions, and DNA extractions were examined for Bartonella spp. by conventional PCR targeting NADH dehydrogenase gamma subunit (nuoG) gene, which is optimal for tissue studies (Bai et al., 2015).
Ectoparasite samples collected from R. rattus in the District of Ankazobe were processed at the University at Buffalo (Buffalo, NY, USA), following the same procedures outlined for host tissues. DNA was extracted from a subset of samples, representing at least of one of each arthropod type recovered from an individual rat host (for a total of 24 Synopsyllus fonquerniei, 8 Xenopsylla cheopsis, 12 Echidnophaga gallinacea, 2 Polyplax sp. lice, 6 Haemaphysalis sp. ticks, and 6 mesostigmatid mites). Extractions were amplified for Bartonella spp. DNA by conventional PCR targeting the nuoG gene. Ectoparasite voucher specimens were slide-mounted and identified using available taxonomic keys.
The obtained nuoG sequences from both rodents and arthropod ectoparasites were clustered to the putative species level by comparing similarity with Bartonella spp. sequences previously deposited in GenBank (Table S1), including species identified in rats of the genus Rattus. Further details of our molecular processing, including primer sequences, are delineated in Text S1.
2.4. Quantitative analysis
All quantitative analysis was conducted in R v. 3.1 for MacIntosh (R Foundation for Statistical Computing, Vienna, Austria). Analyses followed two major themes: (1) Building on techniques from epidemiology, we first modeled age-prevalence for each of the four main species of rodent-borne Bartonella highlighted in our molecular analyses, allowing for age-specific variation in the force of infection and accounting for deviations across sampling sites; and (2) Focusing on the subset of our dataset for which ectoparasite samples were available, we statistically described the distribution and prevalence patterns of arthropod ectoparasites of R. rattus rodents in Madagascar, highlighting phylogenetic relationships across the host-parasite-vector continuum that could underpin observed patterns of infection in the host community. We here summarize our data analysis procedures in brief; the full details of our methodology are elucidated in Text S2.
2.4.1. Force of infection in age-prevalence models
Explicit age determination of wild-caught R. rattus is virtually impossible; thus, we instead estimated age from the rodents’ weights using the von Bertalanffy equation (Ricker, 1979): W(t) = W(1 − e−k(t−t0)) where W is the mass of the largest individual in the dataset and k is a species-specific constant. In our model, k = 0.0207 for R. rattus from the literature (Rajagopalan, 1970), though see Text S3 for consideration of alternative values for k. The von Bertalanffy approximation is believed to provide one of the most reliable estimates of age for the Rodentia order (Griffiths and Brook, 2005, Zullinger et al., 1984). We calculated rat age (in days) separately for male and female individuals and also classed rodents into biologically-relevant 15-day age bins (i.e. infant: 0–15 days; juvenile: 16–30 days; adult: 30+ days), though see Text S3 for evaluation of the sensitivity of our results to variation in age bin duration (Fig. 1B; Yoon et al., 2014).
Using these estimates for rodent age, we followed Long et al. (2010) and Pomeroy et al. (2015); to model age-prevalence for each of the four Bartonella spp. and quantify the age-specific force of infection (FOI, λ), or the per capita rate at which susceptible hosts become infected. In the case of a non-immunizing persistent infection, hosts are born susceptible, and the total time of infection exposure increases with age, resulting in higher prevalence in later age classes because individuals of a given age (a) will have experienced a cumulative FOI throughout their lifetime. Such data will be best fit by a Susceptible-Infected (SI) model. Similar methods can be applied to estimate FOI for transient, fully immunizing infections from age-seroprevalence data; in such cases, seroprevalence will also increase with age, and corresponding data (not available in this study) can be fit by a Susceptible-Infected-Recovered (SIR) model. By contrast, non-immunizing, transient infections should demonstrate an age-prevalence profile that first increases in early age classes, reaches a peak, then declines to some lower endemic prevalence in later age classes. Older individuals will have had greater opportunity to progress through the compartments of a corresponding Susceptible-Infected-Susceptible (SIS) model, resulting in the return of many late life individuals to the susceptible class.
We used likelihood ratio tests (LRTs) to separately select amongst SI and SIS models of varying numbers of age bins for each of the four Bartonella spp. in our age-prevalence dataset. We employed partial profile likelihood to compute confidence intervals on each age-specific FOI estimate in the final, best fit model (Bolker, 2008). We compared each best fit model with an alternative model allowing for deviations in the age-specific FOI across the four sites investigated in this study (Ankazobe Outside, Ankazobe Inside, Ranomafana Outside, and Ranomafana Inside). Details of all model constructions, including relevant R-code, and comparison of fits for various model forms are outlined in Text S2, Tables S2 and S3.
This framework makes several assumptions: that rodents experience no heterogeneity in infection exposure, that the sensitivity of our PCR assay is perfect, and, in the case of the SI models, that infection is lifelong and infection-induced mortality negligible. Because our data are derived from two cross-sectional sampling events during the same month of two different years, we also ignore the possibility of seasonal variations in the age-specific force of infection.
2.4.2. Host-parasite-vector relationships
Focusing on the subset of our data for which ectoparasite information was available (District of Ankazobe), we next sought to understand host-parasite-vector relationships that might elucidate the force of infection trends outlined above. Using representative nuoG sequences obtained from both host and ectoparasite PCR assays (see Molecular analysis, above), we constructed a maximum likelihood phylogeny highlighting nested relationships between Bartonella spp. genotypes recovered from rats and their arthropods. Phylogenetic alignment was performed using Brucella abortus as an outgroup and the default parameters in MUSCLE (Edgar, 2004). We used jModelTest 2 to identify TIM2 + I + G as the best fit model for the data (Darriba et al., 2012) and evaluated phylogenetic relationships using a GTR + G + I model with 1000 bootstrap replicates and data partitioned by codon in RAxML (Stamatakis, 2006).
We next used logistic regression to quantify the extent to which Bartonella spp. infection in the host rat predicted infection in the associated arthropod ectoparasite. We chose this model form over the more intuitive inverse (i.e. vector predicting infection in host) due to uncertainty over the extent to which we completely sampled the entire vector community on a given host. Because we may have missed collection of some arthropods on some hosts, we cannot accurately assess the extent to which ectoparasite infections predict host infections. We are confident in the validity of our host sampling and PCR-assay (though, see Discussion for consideration of missed mixed infections), and therefore, chose to instead evaluate host infection status as a predictor for infection status in arthropods. Because B. phoceensis 1 and B. rattimassiliensis 1 were each recovered from one ectoparasite only, we restricted our analysis to B. elizabethae 1 and 2 only. Using the glmer function in the lme4 package in R (Bates et al., 2015), we related the occurrence of B. elizabethae 1 and 2 in ectoparasite arthropods to two predictors: parasite type (5 levels: S. fonquerniei, X. cheopsis, E. gallinacea fleas, and Haemaphysalis spp. ticks) and infection status in the rat host (5 levels: B. elizabethae 1-positive, B. elizabethae 2-positive, B. elizabethae 1-positive, B. phoceensis 1-positive, B. rattimassiliensis 1-positive, and negative), with a random effect on rat identity to allow for recovery of multiple arthropods from the same rat host.
We also quantified host-ectoparasite interactions using a permutation test to examine whether the proportion of concordant Bartonella spp. infections within each host-ectoparasite pair (i.e. separately for each ectoparasite type) was greater than expected under the null hypothesis that the Bartonella spp. infections of a host and its ectoparasite are independent (details in Text S4). We computed a one-tailed p-value as the proportion of permutated datasets that yielded a proportion of concordance more extreme than the corresponding statistic as calculated in the unpermuted, raw data.
Finally, we explored the question of whether Bartonella species ranges might be limited by habitat restrictions in the vector population, with a focus on the arthropods for which sufficient data were available: the three fleas (S. fonquerniei, X. cheopsis, and E. gallinacea). We employed logistic regression to compare the occurrence of each flea species in turn against the presence of the other two flea species on the same rat host, as well as sampling site (two levels: Ankazobe Outside, Ankazobe Inside).
3. Results
A total of 158 R. rattus rats were sampled in this study, 93 of which (58.9%) were positive for some species of Bartonella, which we identified as belonging to five distinct genotypes (# positive; prevalence): B. elizabethae 1 (22; 13.9%), B. elizabethae 2 (6; 3.8%), B. phoceensis 1 (40; 25.3%), B. rattimassiliensis 1 (21; 13.3%), B. tribocorum 1 (1; 0.01%). Because only one positive sample was recovered for B. tribocorum 1, the remainder of our quantiative analyses report results for the first four species of rodent-borne Bartonella only.
3.1. Force of infection in age-prevalence models
We depict rodent age estimates from each of four sampling sites in Fig. 1B, along with biologically relevant 15-day developmental age classes, by which we binned piece-wise estimates for the age-structured force of infection. Table S2 shows the full set of age-specific SI and SIS model comparisons for all four Bartonella spp. genotypes, and Text S2 describes the analytical process in detail, including relevant R-code. Text S3 and Table S3 provide a detailed analysis of the sensitivity of our results to variations in the age-for-weight estimates and the duration of each age bin.
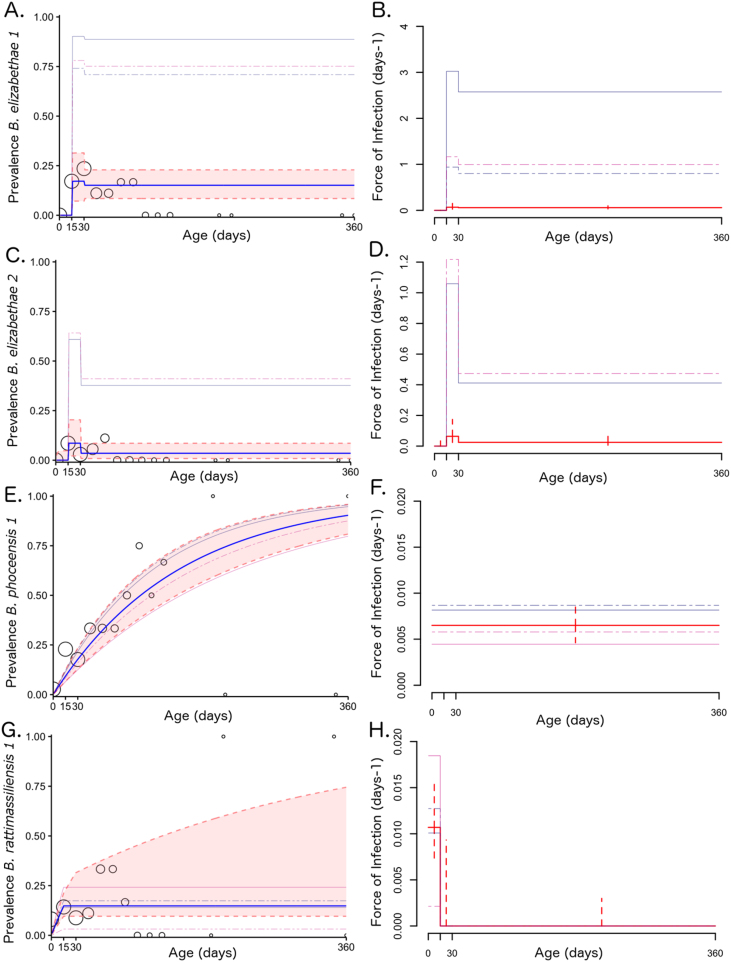
A three-age class SIS model offered the best fit for B. elizabethae 1 and 2 genotypes, while an SI model with constant FOI best represented the B. phoceensis 1 data. Like B. phoceensis 1, the B. rattimassiliensis 1 data were also best described by an SI model (i.e. persistent infection), though inclusion of age structure (three classes) here significantly improved the model’s ability to recover patterns in the data. Models for B. elizabethae 1 and 2 demonstrated substantially higher estimates for FOI in the juvenile age class (15–30 day) than in both the infant (0–15 days) and post-juvenile (30+) classes, while the B. rattimassiliensis 1 model exhibited its highest FOI in the infant class. For these three Bartonella species, young rodents appear to represent the age cohort supporting the majority of transmission, while B. phoceensis 1-infected rats experience a constant FOI across a lifespan (Fig. 2).
Fig. 2.
Age-prevalence and force of infection (respectively) for sampled Rattus rattus infected with B. elizabethae 1 (A,B), B. elizabethae 2 (C,D), B. phoceensis 1 (E,F), and B. rattimassiliensis 1 (G,H). In the age-prevalence charts (A,C,E,G), open circles signify age-stratified prevalence from the data binned over 15-day intervals, and circle size correlates to sample size within each bin. The blue line represents the expected proportion infected in each age class from the best fit model for each species (SIS with 3 age classes for B. elizabethae 1 and 2; SI with a constant force of infection for B. phoceensis 1; and SI with 3 age classes for B. rattimassiliensis 1). Pink shading encompasses the 95% confidence interval as determined via partial profile likelihood, and faint background lines depict predicted prevalence from the more relaxed version of the model allowing for deviations in age-specific FOI by sampling site (navy solid = Ankazobe Outside, navy dashed = Ankazobe Inside; fuchsia solid = Ranomafana Outside; fuchsia solid = Ranomfana Inside). Confidence intervals for site-specific FOIs are listed in Table S4 (not shown in figure for ease of viewing). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To explore the possibility of geographic range limits for our various Bartonella spp. subtypes, we compared the best fit model for each Bartonella genotype with a more relaxed form of the same model, which allowed for site-specific deviations in each age-class-specific estimate of the FOI (Table 1; Fig. 2). In all cases, site-specific deviations led to a significantly better fit. Because some sampling sites had zero positive cases of a particular genotype (i.e. there was no B. elizabethae 1 recovered from Ranomafana Outside sites and no B. elizabethae 2 recovered from Ankazobe Inside or Ranomafana Outside sites), these models were fit with only the subset of data from sites for which the pathogen was present. For this reason, we present the results for both model forms. It is possible that our more restrictive models (which do not allow for site-specific deviations in λ) violate the assumption of no heterogeneity in infection exposure if certain sampling sites are outside the habitat range for the parasite or the vector; in this case, our summary models likely under-predict the true FOI for B. elizabethae 1 and 2. If this assumption is not violated, then it is also possible that our more relaxed models allowing for site-specific deviations in λ over-predict the true FOI for B. elizabethae 1 and 2 because data from low prevalence sites (zeroes in our sampling) have been excluded. More extensive field sampling of both rat hosts and ectoparasite vectors will be critical to resolving this uncertainty in the future.
Table 1.
Comparisons of best fit FOI model per Bartonella spp. genotype w/& w/out sampling site deviations in λ.
| Bartonella spp. | Model Form | Optimized Parameter Estimates |
neg. log-likeli-hood | LRT† | p-val†† | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Force of Infection |
Site-Specific Deviations to λ |
σ | ||||||||||
| λ (0–15) | λ (16–30) | λ (30+) | Ank. Out | Ank. In | Rano. Out | Rano.In | ||||||
| B. elizabethae 1 | SIS | 1.75−23 | 5.93e-02 | 5.10e-02 | – | – | – | – | 0.286 | 52.56 | – | – |
| SIS (w/λ site deviations) |
4.91e-35¶ | 4.83e-01¶ | 4.11e-01¶ | 6.26 | 1.95 | – | 2.42 | 6.58 | 43.98 | 17.16 | 0.00003*** | |
| B. elizabethae 2 | SIS | 4.26e-09 | 6.38e-02 | 2.46e-02 | – | – | – | – | 0.680 | 23.25 | – | – |
| SIS (w/λ site deviations) |
4.91e-35¶ | 4.38e-01¶ | 1.70e-01¶ | 2.42 | – | – | 2.78 | 6.62 | 19.89 | 6.726 | 0.00950*** | |
| B. phoceensis 1 | SI (null) | (constant FOI) 0.0065 | – | – | – | – | – | 75.28 | – | – | ||
| SI (null) (w/λ site deviations) |
(constant FOI) 0.0067¶ | 1.22 | 1.29 | 0.662 | 0.862 | – | 73.91 | 2.742 | 0.09774* | |||
| B. rattimassiliensis 1 | SI | 1.07e-02 | 1.17e-12 | 2.71e-11 | – | – | – | – | – | 60.84 | – | – |
| SI (w/λ site deviations) |
1.48e-02¶ | 7.20e-13¶ | 8.30e-12¶ | 0.681 | 0.860 | 1.247 | 0.145 | – | 57.27 | 7.136 | 0.00756*** | |
| SIS | (constant FOI) 0.011 | – | – | – | – | 0.050 | 60.66 | – | – | |||
| SIS (w/λ site deviations) |
(constant FOI) 0.515¶ | 2.04 | 2.28 | 3.64 | 0.306 | 6.47 | 57.46 | 6.394 | 0.01145** | |||
†Likelihood ratio test ††associated p-value from a chi-squared distribution comparing the negative binomial log-likelihood of the more restrictive to the less restrictive model via the following equation: 2*(ll(m2)-ll(m1)) where m1 = more restrictive, no site deviation model and m2 = less restrictive, site deviation-permitting model. ¶These values indicate mean lambda values across subset of sites for which each model fit is tabulated; site-specific deviations are added to mean to reproduce values visualized in Fig. 2. *Statistical significance by p-value standard <0.1*, <0.05**, <0.01***.
3.2. Host-parasite-vector relationships
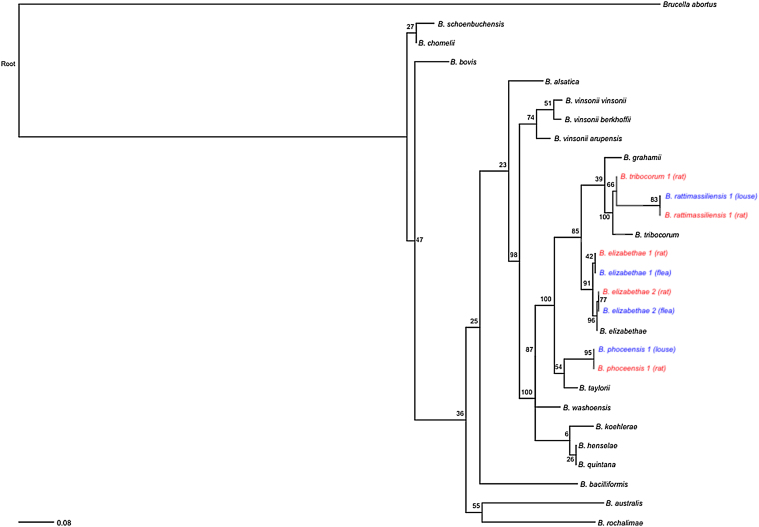
Fig. 3 represents a maximum likelihood phylogeny of representative Bartonella spp. isolates recovered from nuoG sequencing of both host and arthropod ectoparasite samples. B. elizabethae 1 and 2 sequences recovered from S. fonquerniei and X. cheopsis fleas and B. phoceensis 1 and B. rattimassiliensis 1 sequences recovered from Polyplax spp. lice nest with the same respective genotypes recovered from rodent hosts. That said, the ecological landscape of these interactions was complex and revealed considerable discordance between sequences obtained from several host-ectoparasite pairs, a pattern which has been reported in host-ectoparasite relationships for Bartonella spp. elsewhere (Abbot et al., 2007, Brinkerhoff et al., 2010, Tsai et al., 2010). Indeed, neither arthropod type nor host rat sequence significantly correlated with arthropod infection status in our logistic regression model (Table S5). However, our permutation tests (Text S4) demonstrated that host-ectoparasite concordance in Bartonella spp. for S. fonquerniei occurred more frequently than would be expected by chance (p = 0.007), suggestive of transmission between host-ectoparasite pairs. All cases of rat-S. fonquerniei concordancy were for Bartonella elizabethae genotypes (6/8 for B. elizabethae 1 and 2/8 for B. elizabethae 2). By contrast, no host-ectoparasite concordance was observed for X. cheopsis (p = 1).
Fig. 3.
Maximum likelihood phylogeny of representative Bartonella genotypes obtained from nuoG gene sequencing of rodent (red) and arthropod ectoparasite (blue) samples in our dataset (outgroup: Brucella abortus) (RAxML, GTR + G + I model, partitioned by codon position, with 1000 bootstrap replicates) (Stamatakis, 2006). All bootstrap values are shown on corresponding nodes. Branch lengths are scaled by nucleotide substitutions per site. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 2 summarizes the bulk of our findings across the host-parasite-vector-continuum. Most S. fonquerniei fleas recovered from B. elizabethae 1 or B. elizabethae 2-positive rats were PCR- positive for the same Bartonella spp. identified in the rodent host. By contrast, none of the S. fonquerniei fleas recovered from B. phoceensis 1-positive rats were PCR-positive for the Bartonella spp. recovered from their hosts; notably, all of those fleas were PCR-positive for different Bartonella spp. than their host rat (most with B. elizabethae 1 and one with B. elizabethae 2). No X. cheopsis recovered from B. phoceensis 1-positive rats were PCR-positive for the host rat Bartonella spp., but one tested positive for B. elizabethae 1. No fleas of any species recovered from B. rattimassiliensis 1 positive-rats were positive for any sequence of Bartonella.
Table 2.
Host-parasite-vector relationships from the Ankazobe subset of the data.
| Sequence | Prevalence in Rattus rattus [# positive/# tested (%)] |
Ectoparasites on positive rats [# positive/# tested (%)] |
|||
|---|---|---|---|---|---|
| Synopsyllus fonquerniei | Xenopsylla cheopis | Echidnophaga gallinacea | Other | ||
| B. elizabethae 1 | 19/76 (25) | 6/9 (66.7) | – | 0/2 (0) | 1/3 (33.3)c |
| B. elizabethae 2 | 4/76 (5.3) | 2/2 (100) | – | – | – |
| B. phoceensis 1 | 25/76 (32.9) | 0/9a (0) | 0/2b | 0/3 (0) | 1/4 (25)d |
| B. rattimassiliensis 1 | 11/76 (14.5) | 0/2 (0) | 0/1 (0) | 0/2 (0) | 1/2d |
Eight of these 9 were positive for B. elizabethae 1 and 1 for B. elizabethae 2 in spite of host infection.
One of these two was positive for B. elizabethae 1 in spite of host infection.
The one positive “other” for B. elizabethae 1 was a mesostigmatid mite.
The positive ‘others’ for B. phoceensis 1 and B. rattimassiliensis 1 were Polyplax spp. lice.
Though not the focus of our sampling, we opportunistically collected two Polyplax spp. lice from a B. phoceensis 1 positive rat and a B. rattimassiliensis 1 positive rat. Both lice tested positive for the Bartonella genotype with which their host rats were infected. Table S6 reports raw prevalence of each Bartonella spp. genotype recovered from the arthropod ectoparasites assayed in our study. In addition to those arthropods already highlighted, we also sequenced several E. gallinacea fleas, Haemaphysalis spp. ticks, and mesostigmatid mites; of these, only one mite tested positive for one Bartonella genotype (B. elizabethae 1), suggesting a relatively minor role for these additional arthropods in transmission of the four Bartonella genotypes explored in this analysis.
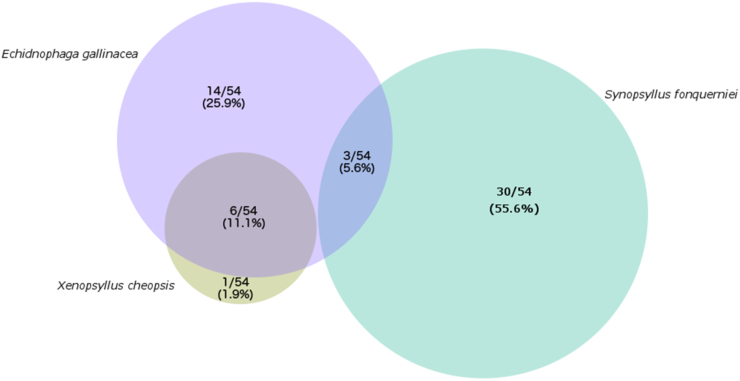
Infestation with S. fonquerniei was significantly, negatively correlated with infestation with both X. cheopsis and E. gallinacea; in fact, no individual rats were co-infected with S. fonquerniei and X. cheopsis, though E. gallinacea co-infested with both other flea types (Fig. 4; Table 3). Sampling site proved to be a statistically significant predictor of flea infestation for all flea types, with “outside” captures positively correlated with infestation with S. fonquerniei and negatively correlated with infestation with X. cheopsis and E. gallinacea; X. cheopsis was never recovered from an outside sampling site. These findings are in congruence with previous reports of flea distributions in Madagascar (Duplantier et al., 2005). The relative absence of S. fonquerniei in outdoor localities and X. cheopsis in indoor localities in the District of Ankazobe underlines the assumptions in our FOI analyses: if a certain ectoparasite serves as the exclusive vector for one Bartonella spp. genotype, then its absence in certain habitats could indicate that rodent infection exposure is heterogenous by habitat type, thus validating the omission of sites lacking in data for a given pathogen in our model fits.
Fig. 4.
Venn Diagram of flea co-infestation for 54 R. rattus rats from our dataset, for which ectoparasites were isolated and identified. Numbers within each cell give the raw number and corresponding percent (%) of these 54 rats found infested with each combination of ectoparasites. Circle size scales with percentage.
Table 3.
Predictors of infestation by flea type on R. rattus, from GLM.
| Flea | Predictor | Slope | Lower CI† | Upper CI† | p-val |
|---|---|---|---|---|---|
| S. fonquerniei | X. cheopsis | –never recovered together– | |||
| E. gallinacea | −0.896 | −2.197 | 0.4061 | 0.1775 | |
| Outside site | 1.515 | 0.1732 | 2.857 | 0.0269** | |
| X. cheopsis | S. fonquerniei | –never recovered together– | |||
| E. gallinacea | 0.613 | −1.816 | 3.042 | 0.621 | |
| Outside site | –never recovered outside– | ||||
| E. gallinacea | S. fonquerniei | −0.896 | −2.197 | 0.4059 | 0.1775 |
| X. cheopsis | 1.267 | −1.031 | 3.565 | 0.2799 | |
| Outside site | −2.285 | −3.555 | −1.016 | 0.0004 *** | |
†Confidence intervals were computed for the slope of each interaction as slope ±1.96*standard error. * Indicates statistical significance if predictor removed from model, p-value <0.05**, <0.01***
4. Discussion
We here report the first account of Bartonella spp. infection in rodent hosts and associated arthropod ectoparasites in Madagascar. Our molecular analyses identified five distinct species of Bartonella (B. elizabethae 1, B. elizabethae 2, B. phoceensis 1, B. rattimassiliensis 1, and B. tribocorum 1) infecting R. rattus rats and/or associated arthropod ectoparasites in two disparate ecosystems of central Madagascar. We first explored the transmission dynamics of each of four Bartonella species for which there were sufficient data; age-structured SIS models best recovered patterns in the age-prevalence data exhibited by rats infected with B. elizabethae 1 and 2, while SI models provided a better fit to the data for B. phoceensis 1 and B. rattimassiliensis 1. We then investigated host-parasite-vector relationships in our data, recovering nested sequences of all four Bartonella spp. in rat hosts and associated ectoparasites, with B. elizabethae 1 and 2 found in S. fonquerniei and X. cheopsis fleas and B. phoceensis 1 and B. rattimassiliensis 1 in Polyplax spp. lice. These patterns are suggestive of a role for these arthropods in the transmission of respective Bartonella spp. In particular, concordant pairings of B. elizabethae 1 and 2 genotypes in host-ectoparasite pairs occurred more frequently than expected by chance for S. fonquerniei fleas, underlining the likely importance of these arthropods in the transmission dynamics of the B. elizabethae complex. We demonstrated significant habitat restrictions on the range of S. fonquerniei, which could limit the range of the B. elizabethae, a known zoonotic agent, in Madagascar.
For zoonotic pathogens, the spillover force of infection is influenced by pathogen dynamics in the reservoir host, the animal-human contact rate, and the susceptibility of the spillover host (i.e. humans) to infection (Lloyd-Smith et al., 2009). Though explicit models of animal-to-human pathogen spillover are rare in the scientific literature, the few that do exist present scenarios in which spillover events are driven by epizootic peaks of infection in the reservoir population, concomitant with large-scale shedding of infectious material into the environment (Keeling and Gilligan, 2000, Sauvage et al., 2007). Consistent with these model scenarios, we found that zoonotic Bartonella species demonstrated higher forces of infection than genotypes not known to pose zoonotic risks. Indeed, B. elizabethae 1 and 2 in juvenile rats demonstrated forces of infection some ten orders of magnitude higher than anything recovered for B. rattimassiliensis 1 or B. phoceensis 1, thus offering a potential avenue for zoonotic transmission. Additionally, higher forces of infection are generally associated with more virulent, highly replicating pathogens, likely to cause pathogen-induced mortality and/or place greater evolutionary pressure on hosts to develop resistance; such theories are also consistent with our finding that a transmission model incorporating infection recovery (indicative of immune defense) provided the best fit to the B. elizabethae 1 and 2 data. Intriguingly, our SIS model for B. elizabethae 1 and 2 did not completely recapitulate the data trend of prevalence declining to zero in highest age class rodents (instead, leveling out to an equilibrium prevalence), suggesting that a Susceptible-Infected-Recovered (SIR) model form, by which pathogen clearance is immunizing, or an SI model incorporating pathogen-induced mortality might be more appropriate. Careful genotype-specific reporting of age-structured seroprevalence patterns against particular Bartonella spp. antigens would enable these inferences; previous attempts to investigate the serological landscape of rat-borne Bartonella spp. have yielded conflicting results (Kosoy et al., 2004a, Kosoy et al., 2004b, Kosoy et al., 1997).
Our site-specific estimates for age-specific FOI were also substantially higher for B. elizabethae 1 and 2 than our cumulative estimates in models not allowing for location-specific deviations because sites in which the pathogen was not detected were excluded while fitting the deviation-permitting model. We are left with the question of whether our cumulative model under-predicts the force of infection for these Bartonella spp. by including susceptible hosts outside the habitat range of the pathogen or whether our site-deviation model over-predicts the force of infection by excluding data from low prevalence sites.
Our subsequent descriptive analyses build on our FOI analyses to offer evidence for the first hypothesis, supporting the role of S. fonquerniei, and, to a lesser extent, X. cheopsis, in the transmission of B. elizabethae 1 and 2 among Malagasy R. rattus rats. The ranges of vector-borne parasites are limited by the ranges of their hosts and vectors, and ectoparasite infestations in R. rattus examined in our study showed distinctive signatures of habitat limitations and mutual exclusion. S. fonquerniei was recovered predominantly from outside sampling sites and never in concert with X. cheopsis, which was found exclusively on rats trapped inside households. If S. fonquerniei plays an essential role in the transmission cycle of B. elizabethae, then we might expect rats recovered from indoor sampling sites to experience a lower force of infection for B. elizabethae genotypes than rats trapped outside; broadly, this is the pattern exhibited in our data. Previous work demonstrates that S. fonquerniei is limited in geographic extent to outdoor localities above an 800 m elevation gradient in Madagascar, a factor believed to be important in predicting the range limits for another zoonotic bacterial pathogen for which S. fonquerniei serves as a vector: Yersinia pestis, the causative agent in bubonic plague.
Though ectoparasite samples were not collected from the Ranomafana region, we hypothesize that sites in this region are located at the elevation limit (∼900 m) of the highland distribution (800 m-restricted) of S. fonquerniei. It is possible that S. fonquerniei fleas and the pathogens which they support are consequently less abundant in this locale. Future studies including more categorically lowland sites will be critical to evaluating whether B. elizabethae 1 and 2 can be independently maintained among R. rattus rodent hosts and X. cheopsis arthropods in Madagascar, or whether, as seems to be the case for plague, these Bartonella subtypes require the contributions of S. fonquerniei to their transmission cycle for persistence. Additionally, if future serological work confirms that a model incorporating parasite-induced mortality better recapitulates the dynamics of B. elizabethae infections in rodent hosts, then a strategy of independent pathogen maintenance in arthropod vectors (i.e. S. fonquerniei) may be necessary to explain Bartonella spp. persistence. Recent work posits a role for Bartonella spp. as intracellular endosymbionts of their arthropod vectors (similar to Wolbachia spp.) (Tsai et al., 2011); the diversity of Bartonella genotypes, independent of the rodent host population, exhibited amongst potential arthropod vectors in Madagascar is compatible with such a hypothesis.
Our work also recovered nested sequences of B. phoceensis 1 and B. rattimassiliensis 1 in Polyplax spp. lice and associated rodents, suggestive of a role for lice in the transmission of these other Bartonella species. This interaction is inconclusive due to sampling size; however, B. phoceensis and B. rattimassiliensis sequences have been recovered from louse ectoparasites of sampled rodents before (Reeves et al., 2006), while previous attempts to isolate these sequences from the fleas of infected rats have also failed (Gundi et al., 2004). Reviews of the literature thus posit a role for lice as vectors of B. phoceensis and B. rattimassiliensis (Billeter et al., 2008), a hypothesis which our data support.
Our study highlights the power of age-structured pathogen prevalence data, by which age-specific patterns in the force of infection can be inferred. We estimated rodent age from mass via the von Bertalanffy equation (Ricker, 1979), though future studies could incorporate more precise measurements of age. In particular, in lethally sampled mammals, including Rattus spp. rats, dried eye lens weight has been demonstrated to correlate closely with animal age (since eye lenses grow continuously throughout a mammal’s life) (Hardy et al., 1983) and could offer a more rigorous approximation of rodent age for future analyses. Additionally, future field studies should include a skeletal measure of rodent size (akin to the routine forearm measurement for bats) so that mass-per-body size relationships can be inferred. We have used rodent weight (mass) to estimate age; however, animals will vary substantially in physiological condition within a population and across a season. Among adult (75+ day) rats in particular, skeletal growth tends to cease (Yoon et al., 2014), and weight for body size may offer a better reflection of the nutritional condition of the rodent host, which can play a key role in immune mechanisms of pathogen clearance and control (Dowell, 2001).
Finally, we report the results of PCR amplification and sequence alignment of one genotype of Bartonella spp. for each positive rodent host and arthropod vector. Mixed infections of multiple Bartonella spp. in the same host and/or vector have been reported in the past (Abbot et al., 2007, Gurfield et al., 2001, La Scola et al., 2002, Rolain et al., 2003), and it is possible that our molecular analyses have preferentially amplified certain Bartonella genotypes over others and missed identification of coinfected individuals. In the future, more diverse sampling of multiple host tissues and/or red blood cell could help deter preferential amplification of certain genotypes. If, as exhibited in our data, more diverse tissue sampling continues to recover only one Bartonella spp. per rodent host, then such patterns could indicate that simultaneous infections with the multiple species of Bartonella present in this system are often fatal for their hosts. The extent to which coexistence of these multiple Bartonella spp. is mediated by inter-species competition versus landscape-level niche partitioning among parasite species with different ranges (subject to the range limits of their respective vectors) remains a question for further study.
5. Conclusions
Our analyses shed light on the transmission dynamics and genotype-specific host-parasite-vector associations for Bartonella spp. infections in Rattus spp. hosts and associated arthropod ectoparasites in Madagascar. We distinguish between the infection dynamics of the B. elizabethae complex, a known zoonotic pathogen, which appears to maintain a transient presence in its rodent hosts, and those exhibited by B. phoceensis and B. rattimassiliensis, which appear to operate as persistent infections. Many ecological studies have reported data from diverse Bartonella species as a single composite Bartonella spp. in the past (e.g. Bai et al., 2008, Janecek et al., 2012, Morway et al., 2008, Young et al., 2014). While such a practice may provide heightened statistical power, our findings suggest that this method will not reveal genotype-specific ecological associations and infection dynamics for diverse pathogens that span a broad range of host virulence strategies and differ substantially in their potential for zoonotic transmission.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
CEB, APD, CJEM, MYK, and KD devised the study. CEB, EOY, and HCR collected field samples. YB and MYK performed molecular analyses of rodent tissue samples. HS and KD performed molecular analyses and species identifications of arthropod ectoparasites. CEB, CJM, EOY, and APD carried out the statistical analyses of the data. CEB, CJEM, APD, MYK, and KD wrote the manuscript, which all other authors read and edited.
Acknowledgements
CEB gratefully acknowledges funding from the National Science Foundation Graduate Research Fellowship Program, the National Geographic Society Young Explorers Grant Program (YEG #9269-13) and Princeton University’s Centre for Health and Well-being. The authors acknowledge the CDC’s Global Diseases Detection Program (YB and MYK), a Wellcome Trust Sustaining Health Grant (106866/Z/15/Z; CJEM), the Bill and Melinda Gates Foundation (CJEM), and a National Science Foundation Division of Environmental Biology grant (1050793; KD) for additional financial support. We thank Evaline Cheng, Dr. Summer Arrigo-Nelson, Dr. Jean-Michel Héraud, and the Virology Unit at Institut Pasteur de Madagascar (IPM) for field support and the Madacascar Institut pour la Conservation des Ecosystèmes Tropicaux (MICET) for help with permits and logistics. We gratefully acknowledge Dr. Steve Bellan for helpful comments on the manuscript.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.epidem.2017.03.004.
Contributor Information
Cara E. Brook, Email: caraeb@princeton.edu, cbrook04@gmail.com.
Ying Bai, Email: bby5@cdc.gov.
Emily O. Yu, Email: eoyu00@gmail.com.
Hafaliana C. Ranaivoson, Email: rchristian@pasteur.mg.
Haewon Shin, Email: haewonsh@buffalo.edu.
Andrew P. Dobson, Email: dobber@princeton.edu.
C. Jessica E. Metcalf, Email: cmetcalf@princeton.edu.
Michael Y. Kosoy, Email: mck3@cdc.gov.
Katharina Dittmar, Email: kd52@buffalo.edu.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Abbot P., Aviles A.E., Eller L., Durden L.A. Mixed infections, cryptic diversity, and vector-borne pathogens: evidence from Polygenis fleas and Bartonella species. Appl. Environ. Microbiol. 2007;73:6045–6052. doi: 10.1128/AEM.00228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Kosoy M.Y., Ray C., Brinkerhoff R.J., Collinge S.K. Temporal and spatial patterns of Bartonella infection in black-tailed prairie dogs (Cynomys ludovicianus) Microb. Ecol. 2008;56:373–382. doi: 10.1007/s00248-007-9355-6. [DOI] [PubMed] [Google Scholar]
- Bai Y., Rizzo M.F., Alvarez D., Moran D., Peruski L.F., Kosoy M. Coexistence of Bartonella henselae and B. clarridgeiae in populations of cats and their fleas in Guatemala. J. Vector Ecol. 2015;40:327–332. doi: 10.1111/jvec.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D., Mächler M., Bolker B.M., Walker S.C. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2015;67:1–48. [Google Scholar]
- Battisttini T. Verruga peruana: Sa transmission par le phlébotome. Rev. Sud-Americaine Med. Chir. 1931;2:719–724. [Google Scholar]
- Billeter S.A., Levy M.G., Chomel B.B., Breitschwerdt E.B. Vector transmission of Bartonella species with emphasis on the potential for tick transmission. Med. Vet. Entomol. 2008;22:1–15. doi: 10.1111/j.1365-2915.2008.00713.x. [DOI] [PubMed] [Google Scholar]
- Bolker B. Princeton University Press; 2008. Ecological Models and Data in R. [Google Scholar]
- Bown K.J., Bennett M., Begon M. Flea-borne Bartonella grahamii and Bartonella taylorii in bank voles. Emerg. Infect. Dis. 2004;10:6–9. doi: 10.3201/eid1004.030455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitschwerdt E.B., Kordick D.L. Bartonella infection in animals: carriership, reservoir potential, pathogenicity, and zoonotic potential for human infection. Clin. Microbiol. Rev. 2000;13:428–438. doi: 10.1128/cmr.13.3.428-438.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkerhoff R.J., Kabeya H., Inoue K., Bai Y., Maruyama S. Detection of multiple Bartonella species in digestive and reproductive tissues of fleas collected from sympatric mammals. ISME J. 2010;4:955–958. doi: 10.1038/ismej.2010.22. [DOI] [PubMed] [Google Scholar]
- Brook C.E., Bai Y., Dobson A.P., Osikowicz L.M., Ranaivoson H.C., Zhu Q., Kosoy M.Y. Bartonella spp. in fruit bats and blood-feeding ectoparasites in Madagascar. PLoS Negl. Trop. Dis. 2015;10:e0003532. doi: 10.1371/journal.pntd.0003532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouqui P., Stein A., Dupont H.T., Gallian P., Badiaga S., Rolain J.M., Mege J.L., Scola B., La Berbis P., Raoult D. Ectoparasitism and vector-borne diseases in 930 homeless people from Marseilles. Medicine (Baltimore) 2005;84:61–68. doi: 10.1097/01.md.0000152373.07500.6e. [DOI] [PubMed] [Google Scholar]
- Buffet J.-P., Kosoy M., Vayssier-Taussat M. Natural history of Bartonella-infecting rodents in light of new knowledge on genomics, diversity and evolution. Future Microbiol. 2013;8:1117–1128. doi: 10.2217/fmb.13.77. [DOI] [PubMed] [Google Scholar]
- Chomel B.B., Kasten R.W., Chi B., Yamamoto K., Gurfield A.N., Abbott R.C., Pedersen N.C., Koehler E., Chomel B.B., Kasten R.W., Floyd-hawkins K.I.M., Chi B., Yamamoto K., Roberts-wilson J., Gurfield A.N., Abbott R.C., Pedersen N.C., Koehler J.E. Experimental transmission of Bartonella henselae by the cat flea. J. Clin. Microbiol. 1996;34:1952–1956. doi: 10.1128/jcm.34.8.1952-1956.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomel B.B., Boulouis H.-J., Breitschwerdt E.B., Kasten R.W., Vayssier-Taussat M., Birtles R.J., Koehler J.E., Dehio C. Ecological fitness and strategies of adaptation of Bartonella species to their hosts and vectors. Vet. Res. 2009;40:29. doi: 10.1051/vetres/2009011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D., Taboada G.L., Doallo R., Posada D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehio C. Bartonella interactions with endothelial cells and erythrocytes. Trends Microbiol. 2001;9:279–285. doi: 10.1016/s0966-842x(01)02047-9. [DOI] [PubMed] [Google Scholar]
- Dowell S.F. Seasonal variations in host suceptibility and cycles of certain infectious diseases. Emerg. Infect. Dis. 2001;7:369–374. doi: 10.3201/eid0703.010301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duplantier J.-M., Duchemin J.-B., Chanteau S., Carniel E. From the recent lessons of the Malagasy foci towards a global understanding of the factors involved in plague reemergence. Vet. Res. 2005;36:437–453. doi: 10.1051/vetres:2005007. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber B., Karpanty S.M., Crawford C., Kotschwar M., Randrianantenaina J. An assessment of carnivore relative abundance and density in the eastern rainforests of Madagascar using remotely-triggered camera traps. Oryx. 2010;44:219. [Google Scholar]
- Griffiths A.D., Brook B.W. Body size and growth in tropical small mammals: examining variation using non-linear mixed effects models. J. Zool. 2005;267:211–220. [Google Scholar]
- Gundi V.A.K.B., Davoust B., Khamis A., Raoult D., La Scola B. Isolation of Bartonella rattimassiliensis sp. nov. and Bartonella phoceensis sp. nov. from European Rattus norvegicus. J. Clin. Microbiol. 2004;42:3816–3818. doi: 10.1128/JCM.42.8.3816-3818.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurfield A.N., Boulouis H.J., Chomel B.B., Kasten R.W., Heller R., Bouillin C., Gandoin C., Thibault D., Chang C.C., Barrat F., Piemont Y. Epidemiology of Bartonella infection in domestic cats in France. Vet. Microbiol. 2001;80:185–198. doi: 10.1016/s0378-1135(01)00304-2. [DOI] [PubMed] [Google Scholar]
- Hardy A.R., Quy R.J., Huson L.W. Estimation of age in the Norway rat (Rattus norvegicus Berkenhout) from the weight of the eyelens. J. Appl. Ecol. 1983;20:97–102. [Google Scholar]
- Heisey D.M., Joly D.O., Messier F. The fitting of general force-of-infection models to wildlife disease prevalence data. Ecology. 2006;87:2356–2365. doi: 10.1890/0012-9658(2006)87[2356:tfogfm]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Ihler G.M. Bartonella bacilliformis: dangerous pathogen slowly emerging from deep background. FEMS Microbiol. Lett. 1996;144:1–11. doi: 10.1111/j.1574-6968.1996.tb08501.x. [DOI] [PubMed] [Google Scholar]
- Janecek E., Mietze A., Goethe R., Schneider T., Strube C. Bartonella spp. infection rate and B. grahamii in ticks. Emerg. Infect. Dis. 2012;18:1689–1690. doi: 10.3201/eid1810.120390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya H., Sase M., Yamashita M., Maruyama S. Predominant T Helper 2 immune responses against Bartonella henselae in naturally infected cats. Microbiol. Immunol. 2006;50:171–178. doi: 10.1111/j.1348-0421.2006.tb03783.x. [DOI] [PubMed] [Google Scholar]
- Kandelaki G., Malania L., Bai Y., Chakvetadze N., Katsitadze G., Imnadze P., Nelson C., Harrus S. Human lymphadenopathy caused by ratborne Bartonella, Tbilisi, Georgia. Emerg. Infect. Dis. 2016;22:1–3. doi: 10.3201/eid2203.151823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling M.J., Gilligan C.A. Metapopulation dynamics of bubonic plague. Nature. 2000;407:903–906. doi: 10.1038/35038073. [DOI] [PubMed] [Google Scholar]
- Kosoy M.Y., Regnery R.L., Tzianabos T., Marston E.L., Jones D.C., Green D., Maupin G., Olson J.G., Childs A.E. Distribution, diversity, and host specificity of Bartonella in rodents from the southeastern United States. Am. J. Trop. Med. Hyg. 1997;57:578–588. doi: 10.4269/ajtmh.1997.57.578. [DOI] [PubMed] [Google Scholar]
- Kosoy M., Murray M., Gilmore R.D., Bai Y., Gage K.L. Bartonella strains from ground squirrels are identical to Bartonella washoensis isolated from a human patient. J. Clin. Microbiol. 2003;41:645–650. doi: 10.1128/JCM.41.2.645-650.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosoy M., Mande E., Green D., Marston E., Jones D., Childs J. Prospective studies of Bartonella of rodents Part II. Diverse infections in a single rodent community. Vector Borne Zoonotic Dis. 2004;4:296–305. doi: 10.1089/vbz.2004.4.296. [DOI] [PubMed] [Google Scholar]
- Kosoy M., Mandel E., Green D., Marston E., Childs J. Prospective studies of Bartonella of rodents: part I. Demographic and temporal patterns in population dynamics. Vector Borne Zoonotic Dis. 2004;4:285–295. doi: 10.1089/vbz.2004.4.285. [DOI] [PubMed] [Google Scholar]
- Kosoy M., Bai Y., Sheff K., Morway C., Baggett H., Maloney S.A., Boonmar S., Bhengsri S., Dowell S.F., Sitdhirasdr A., Lerdthusnee K., Richardson J., Peruski L.F. Identification of Bartonella infections in febrile human patients from Thailand and their potential animal reservoirs. Am. J. Trop. Med. Hyg. 2010;82:1140–1145. doi: 10.4269/ajtmh.2010.09-0778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosoy M.Y. Ecological associations between bacteria of the genus Bartonella and mammals. Biol. Bull. 2010;37:716–724. [Google Scholar]
- La Scola B., Liang Z., Zeaiter Z., Houpikian P., Grimont P.A.D., Raoult D. Genotypic characteristics of two serotypes of Bartonella henselae. J. Clin. Microbiol. 2002;40:2002–2008. doi: 10.1128/JCM.40.6.2002-2008.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Smith J.O., George D., Pepin K.M., Pitzer V.E., Pulliam J.R.C., Dobson A.P., Hudson P.J., Grenfell B.T. Epidemic dynamics at the human-animal interface. Science (80-.) 2009;326:1362–1367. doi: 10.1126/science.1177345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long G.H., Sinha D., Read A.F., Pritt S., Kline B., Harvill E.T., Hudson P.J., BjØrnstad O.N. Identifying the age cohort responsible for transmission in a natural outbreak of Bordetella bronchiseptica. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascarelli P.E., McQuillan M., Harms C.A., Harms R.V., Breitshwerdt E.B. Bartonella henselae and B. koehlerae DNA in birds. Emerg. Infect. Dis. 2014;20:6–9. doi: 10.3201/eid2003.130563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morway C., Kosoy M., Eisen R., Montenieri J., Sheff K., Pamela J., Powers N., Reynolds P.J. A longitudinal study of Bartonella infection in populations of woodrats and their fleas. J. Vector Ecol. 2008;33:353–364. doi: 10.3376/1081-1710-33.2.353. [DOI] [PubMed] [Google Scholar]
- Muench H. Harvard University Press; Boston, MA, USA: 1959. Catalytic Models in Epidemiology. [Google Scholar]
- Pearce L.K., Radecki S.V., Brewer M., Lappin M.R. Prevalence of Bartonella henselae antibodies in serum of cats with and without clinical signs of central nervous system disease. J. Feline Med. Surg. 2006;8:315–320. doi: 10.1016/j.jfms.2006.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeroy L.W., Bjornstad O.N., Kim H., Jumbo S.D., Abdoulkadiri S., Garabed R. Serotype-specific transmission and waning immunity of endemic foot-and-mouth disease virus in Cameroon. PLoS One. 2015;10:1–16. doi: 10.1371/journal.pone.0136642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahelinirina S., Duplantier J.M., Ratovonjato J., Ramilijaona O., Ratsimba M., Rahalison L. Study on the movement of Rattus rattus and evaluation of the plague dispersion in Madagascar. Vector Borne Zoonotic Dis. 2010;10:77–84. doi: 10.1089/vbz.2009.0019. [DOI] [PubMed] [Google Scholar]
- Rajagopalan P.K. Breeding behaviour and development of Rattus rattus wroughtoni Hinton 1919 (Rodentia: Muridae) in the laboratory. J. Bombay Nat. Hist. Soc. 1970;67:552–558. [Google Scholar]
- Reeves W.K., Szumlas D.E., Moriarity J.R., Loftis A.D., Abbassy M.M., Helmy I.M., Dasch G.a. Louse-borne bacterial pathogens in lice (Phthiraptera) of rodents and cattle from Egypt. J. Parasitol. 2006;92:313–318. doi: 10.1645/GE-717R.1. [DOI] [PubMed] [Google Scholar]
- Reisen W.K. Landscape epidemiology of vector-borne diseases. Annu. Rev. Entomol. 2010;55:461–483. doi: 10.1146/annurev-ento-112408-085419. [DOI] [PubMed] [Google Scholar]
- Ricker W.E. Growth rates and models. Fish Physiol. 1979;11:677–743. [Google Scholar]
- Rolain J.M., Franc M., Davoust B., Raoult D. Molecular detection of Bartonella quintana, B., koehlerae, B., henselae, B., clarridgeiae, Rickettsia felis, and Wolbachia pipientis in cat fleas, France. Emerg. Infect. Dis. 2003;9:0–4. doi: 10.3201/eid0903.020278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangaré A.K., Boutellis A., Drali R., Socolovschi C., Barker S.C., Diatta G., Rogier C., Olive M.-M., Doumbo O.K., Raoult D. Detection of Bartonella quintana in African body and head lice. Am. J. Trop. Med. Hyg. 2014;91:294–301. doi: 10.4269/ajtmh.13-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvage F., Langlais M., Pontier D. Predicting the emergence of human hantavirus disease using a combination of viral dynamics and rodent demographic patterns. Epidemiol. Infect. 2007;135:46–56. doi: 10.1017/S0950268806006595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Swift H. Trench fever. Arch. Intern. Med. 1920;36:76–98. [Google Scholar]
- Tsai Y.-L., Chuang S.-T., Chang C.-C., Kass P.H., Chomel B.B. Bartonella species in small mammals and their ectoparasites in Taiwan. Am. J. Trop. Med. Hyg. 2010;83:917–923. doi: 10.4269/ajtmh.2010.10-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y.L., Chang C.C., Chuang S.T., Chomel B.B. Bartonella species and their ectoparasites: selective host adaptation or strain selection between the vector and the mammalian host? Comp. Immunol. Microbiol. Infect. Dis. 2011;34:299–314. doi: 10.1016/j.cimid.2011.04.005. [DOI] [PubMed] [Google Scholar]
- Valentine K.H., Harms C.A., Cadenas M.B., Birkenheuer A.J., Marr H.S., Braun-McNeil J., Maggi R.G., Breitshwerdt E.B. Bartonella DNA in loggerhead sea turtles. Emerg. Infect. Dis. 2007;13:6–7. doi: 10.3201/eid1306.061551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallan D. Influence of forest fragmentation on amphibian diversity in the nature reserve of Ambohitantely, highland Madagascar. Biol. Conserv. 2000;96:31–43. [Google Scholar]
- Vayssier-Taussat M., Le Rhun D., Bonnet S., Cotté V. Insights in Bartonella host specificity. Ann. N. Y. Acad. Sci. 2009;1166:127–132. doi: 10.1111/j.1749-6632.2009.04531.x. [DOI] [PubMed] [Google Scholar]
- Vayssier-Taussat M., Moutailler S., Féménia F., Raymond P., Croce O., Scola B., La Fournier P., Raoult D. Identification of novel zoonotic activity of Bartonella spp., France. Emerg. Infect. Dis. 2016;22:457–462. doi: 10.3201/eid2203.150269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veikkolainen V., Vesterinen E.J., Lilley T.M., Pulliainen A.T. Bats as reservoir hosts of human bacterial pathogen, Bartonella mayotimonensis. Emerg. Infect. Dis. 2014;20 doi: 10.3201/eid2006.130956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch D.F., Pickett D.A., Slater L.N., Steigerwalt A.G., Brenner D.J. Rochalimaea henselae sp. nov. a cause of septicemia, bacillary angiomatosis, and parenchymal bacillary peliosis. J. Clin. Microbiol. 1992;30:275–280. doi: 10.1128/jcm.30.2.275-280.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson D.A., Duron O., Cordonin C., Gomard Y., Ramasindrazana B., Mavingui P., Goodman S.M., Tortosa P. The bacteriome of bat flies (Nycteribiidae) from the Malagasy region: a community shaped by host ecology, bacterial transmission mode, and host-vector specificity. Appl. Environ. Microbiol. 2016 doi: 10.1128/AEM.03505-15. (AEM. 03505-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon B.H., Kim C.J., Romero R., Jun J.K., Park K.H., Choi S.T., Chi J.G., Sengupta P. The laboratory rat: relating its age with human’s. Am. J. Obstet. Gynecol. 2014;4:624–630. [Google Scholar]
- Young H.S., Dirzo R., Helgen K.M., McCauley D.J., Billeter S.a., Kosoy M.Y., Osikowicz L.M., Salkeld D.J., Young T.P., Dittmar K. Declines in large wildlife increase landscape-level prevalence of rodent-borne disease in Africa. Proc. Natl. Acad. Sci. 2014;1–6 doi: 10.1073/pnas.1404958111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zullinger E.M., Ricklefs R.E., Redford K.H., Mace G.M. Fitting sigmoidal equations to mammalian growth curves. J. Mammal. 1984;65:607–636. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.