ABSTRACT
Here we assess histone modification, chromatin remodeling, and DNA methylation processes that coordinately control the expression of the bone master transcription factor Sp7 (osterix) during mesenchymal lineage commitment in mammalian cells. We find that Sp7 gene silencing is mediated by DNA methyltransferase1/3 (DNMT1/3)-, histone deacetylase 1/2/4 (HDAC1/2/4)-, Setdb1/Suv39h1-, and Ezh1/2-containing complexes. In contrast, Sp7 gene activation involves changes in histone modifications, accompanied by decreased nucleosome enrichment and DNA demethylation mediated by SWI/SNF- and Tet1/Tet2-containing complexes, respectively. Inhibition of DNA methylation triggers changes in the histone modification profile and chromatin-remodeling events leading to Sp7 gene expression. Tet1/Tet2 silencing prevents Sp7 expression during osteoblast differentiation as it impairs DNA demethylation and alters the recruitment of histone methylase (COMPASS)-, histone demethylase (Jmjd2a/Jmjd3)-, and SWI/SNF-containing complexes to the Sp7 promoter. The dissection of these interconnected epigenetic mechanisms that govern Sp7 gene activation reveals a hierarchical process where regulatory components mediating DNA demethylation play a leading role.
KEYWORDS: DNA demethylation, epigenetics, osteoblast differentiation, Sp7, Tet enzymes
INTRODUCTION
The Sp7 (osterix) gene codes for a transcription factor (TF) that is expressed in a highly restricted manner in mesenchymal cells that are differentiating to osteoblasts and chondrocytes (1), where it recognizes a GC-rich sequence at regulatory regions of target bone-related genes (2). Sp7 expression is required for bone formation: homozygous Sp7 knockout mice die shortly after birth, exhibiting severe skeletal abnormalities, including a drastic reduction in bone mineralization due to impaired osteoblast differentiation (1). Preosteoblastic cells isolated from Sp7-null mouse embryos reveal that the expression level of the Runx2 TF, the master regulator of osteogenic lineage commitment, remains unaltered, indicating that Sp7 functions downstream of Runx2 (3, 4). Sp7 also exhibits an essential role in adult bone homeostasis (5). Targeted depletion of the Sp7 gene in mice results in bone abnormalities due to the inhibition of osteoblast differentiation, altered cartilage accumulation, and impaired function of osteocytes (6). Similarly, mutations at the human Sp7 gene have been found to be associated with bone abnormalities (7), and different single nucleotide polymorphisms at the Sp7 locus are associated with alterations in bone mineral density (8, 9).
Sp7 promoter activity is upregulated by transcriptional pathways mediated by mitogen-activated protein kinase (MAPK) effectors (10) and the Runx2 and Dlx5 TFs (11, 12) as well as in response to parathyroid hormone (13), ascorbic acid (14), and vitamin D3 (15, 16). Incubation of mesenchymal cells with the osteoblast differentiation-promoting factor bone morphogenetic protein 2 (BMP2) results in enhanced Sp7 mRNA expression (1). This BMP2-dependent effect is mediated by the Dlx5 TF, which binds to the Sp7 promoter and recruits the coactivator p300, an event that is modulated by the p38 (11) and phosphatidylinositol 3 (PI3)-kinase/Akt (17) signaling pathways. Recent studies also indicated the role of specific epigenetic modifiers in Sp7 gene expression, which mediate changes in histone posttranslational modifications (PTMs) at the Sp7 promoter. Thus, Pbx1-mediated repression of the Sp7 gene in bone marrow stem cells involves decreased histone acetylation due to the reduced binding of the p300 acetyltransferase and increased interactions of histone deacetylase 1 (Hdac1) and Hdac2 (18). The H3K27me3 demethylases Jmjd3 and Utx have been shown to mediate the reduction of this repressive mark that accompanies Sp7 gene transcription during osteoblast differentiation and bone formation (19–21). It has also been reported that treatment of C2C12 promyoblastic cells with 5-azacytidine (5AzaC), a DNA methyltransferase (Dnmt) inhibitor, results in the activation of Sp7 gene expression (22, 23). It was found, however, that most 5AzaC-treated cells (>90%) were differentiated to the myogenic phenotype due to elevated expression of muscle-related master TFs (e.g., myogenic differentiation 1 [MyoD]) in most of the cells and to the remaining levels of CpG methylation at the Sp7 promoter. Together, these results indicate that successful osteogenic commitment requires a significant reduction in CpG methylation and changes in histone PTMs at the Sp7 promoter that drive sufficient Sp7 expression. Therefore, there is a necessity to understand what the main components of these mechanisms are and how they are coordinated during osteogenic differentiation.
One principal pathway leading to DNA demethylation in mammals is initiated by the conversion from 5-methyl-CpG (5mC) to 5-hydroxymethyl-CpG (5hmC) mediated by the Ten-Eleven Transformation (Tet) family of dioxygenases (24, 25). Importantly, both Tet1 and Tet2 enzymes have been found to be required during lineage commitment of mouse embryonic stem cells (25–27) and to be critical components during mouse development (27, 28). Tet proteins have also been reported to function as scaffolding proteins, mediating the formation of multienzymatic regulatory complexes with several other epigenetic modifiers, including SWI/SNF chromatin remodelers, histone methylases, and demethylases (29–31).
We report that a key regulatory component of the mechanisms leading to Sp7 expression during osteoblast lineage commitment is the Tet1/Tet2-mediated DNA demethylation of the Sp7 promoter. This process is coordinated with nucleosome remodeling, “erasing” of the H3K9me3/H3K27me3 repressive marks, and “writing” of the H3Ac/H3K4me3 transcription-associated marks.
RESULTS
Histone marks and epigenetic mediators that control Sp7 gene expression.
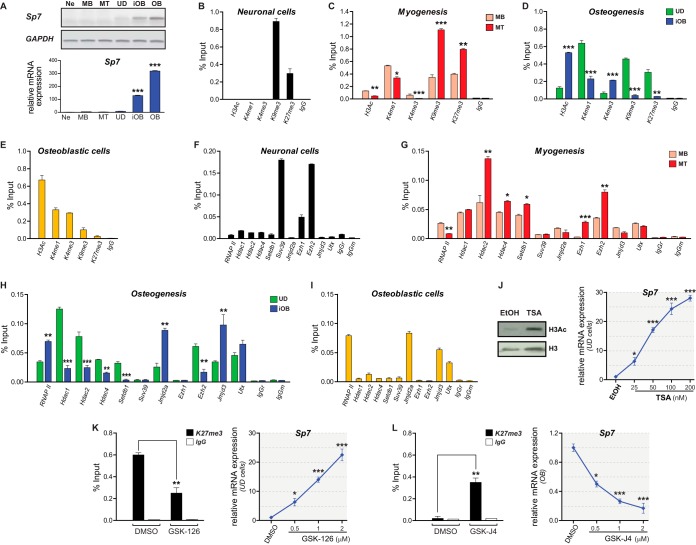
To assess the epigenetic mechanisms controlling the transcriptional activation and silencing of the Sp7 bone master regulator gene, we determined by chromatin immunoprecipitation (ChIP) analyses whether specific histone PTMs and candidate chromatin modifiers (writers and erasers) bind to the proximal Sp7 promoter in different mouse-derived cell systems that express or do not express this gene (Fig. 1A). Nonosteoblastic cells, including N2a neurons (Ne), C2C12 myoblasts (MB), and myotubes (MT), as well as uncommitted mesenchymal multipotent C3H cells (UD), do not express Sp7 mRNA. In contrast, UD cells induced to differentiate to osteoblasts by treatment with BMP2 (iOB) and MC3T3 osteoblasts (OB) express elevated levels of Sp7 mRNA (Fig. 1A). As an additional control, it was shown that Ne and MT cells express their respective phenotypic gene markers (see Fig. S1 in the supplemental material).
FIG 1.
Sp7 gene expression is associated with an epigenetic signature at the Sp7 promoter. (A) The Sp7 gene is selectively expressed in osteoblastic cells. Sp7 mRNA levels were determined by RT-PCR. (Top) Conventional PCR; (bottom) qPCR. Relative mRNA expression values were established by normalization against the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA level. (B to E) Enrichment of histone posttranslational modifications at the Sp7 gene promoter determined by ChIP of samples from N2a neuronal (Ne) cells (B), C2C12 myoblasts (MB) and C2C12 myoblasts differentiated to myotube (MT) cells (C), C3H uncommitted multipotent cells (UD) and C3H cells differentiated to osteoblasts (iOB) in response to BMP2 (D), and MC3T3 osteoblastic (OB) cells (E). (F to I) Enrichments of RNA polymerase II (RNAPII) and chromatin-associated regulators at the Sp7 promoter determined by ChIP of samples from the cell types described above. (J) Effect of TSA-dependent chromatin hyperacetylation on Sp7 mRNA expression in UD cells. (Left) Western blotting using anti-H3Ac and anti-H3 antibodies; (right) RT-qPCR analysis of Sp7 mRNA expression. EtOH, ethanol. (K) Effect of GSK-126-dependent inhibition of Ezh2-mediated H3K27 methylation on Sp7 mRNA expression in UD cells. (Left) ChIP to evaluate H3K27me3 enrichment at the Sp7 promoter; (right) Sp7 mRNA expression levels measured by RT-qPCR. DMSO, dimethyl sulfoxide. (L) Effect of GSK-J4-dependent inhibition of H3K27me3 demethylases on Sp7 mRNA expression in OB cells. (Left) H3K27me3 enrichment at the Sp7 promoter measured by ChIP; (right) Sp7 mRNA expression levels measured by RT-qPCR. ChIP results are expressed as a percentage of the input. Data represent the means of results from at least three independent experiments ± standard errors of the means. Student's t test was performed to establish statistical significance. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Ne cells exhibit enrichment in the H3K9me3/H3K27me3 marks at the Sp7 promoter and an absence of PTMs associated with active or poised promoters (H3Ac/H3K4me1/H3K4me3) (Fig. 1B). MB cells show reduced, but detectable, levels of H3Ac and enrichment of H3K4me1/H3K9me3/H3K27me3 marks (Fig. 1C). Interestingly, MT cells show further enriched levels of H3K9me3/H3K27me3 together with a reduced enrichment of H3Ac/H3K4me3 (Fig. 1C), indicating that myogenic differentiation proceeds with the progressive deposition of repressive PTMs at the Sp7 promoter. When UD cells are differentiated to osteoblasts (iOB), H3Ac/H3K4me3 marks are significantly enriched at the Sp7 promoter (Fig. 1D), concomitant with its transcriptional activation. These enrichments are accompanied by decreased levels of H3K9me3/H3K27me3 marks (Fig. 1D), whereas H3K4me1 exhibits only a partial reduction. This epigenetic signature is equivalent to that found at the Sp7 promoter in OB cells (Fig. 1E), indicating that they represent a pattern strongly associated with Sp7 gene transcription in osteogenic cells.
It was next determined that in Ne cells, this promoter is enriched in the Suv39H1 and Ezh2 methyltransferases, which have been shown to mediate the deposition of the H3K9me3 and H3K27me3 marks, respectively (Fig. 1F). Reduced, but significant, binding of Ezh1 was also detected (Fig. 1F), suggesting that the interaction of a PRC2 complex containing Ezh1 and/or Ezh2 can contribute to maintaining both H3K27me3 levels and transcriptional repression at the Sp7 promoter in these cells. Binding of additional epigenetic modifiers, including Hdac1/2/4, Setdb1, Jmjd2a, Jmjd3, and Utx, as well as interactions of RNA polymerase II (RNAPII) were not detected at this promoter in neuronal cells (Fig. 1F). In contrast, we found that Hdac1/2/4, Setdb1, and Ezh2 are present at the Sp7 promoter in MB cells (Fig. 1G). Importantly, these cells show reduced, although detectable, levels of the RNAPII, Jmjd3, and Utx proteins at this promoter (Fig. 1G). Following myogenic differentiation, MT cells exhibit further enriched levels of the Hdac2/4, Setdb1, and Ezh2 proteins at the Sp7 promoter, concomitant with the release of RNAPII and binding of Ezh1 (Fig. 1G). Jmjd3 and Utx remain poorly associated at this region in MT cells (Fig. 1G). Together, these results indicate that Sp7 gene repression in promyoblastic cells is reflected by an epigenetic signature on the Sp7 promoter that is further enforced as the cells engage terminal myogenesis.
We next determined whether the above-described epigenetic modulators are also associated with the Sp7 promoter in UD cells and during osteogenesis-dependent Sp7 gene activation. UD cells exhibit binding of RNAPII, Hdac1/2/4, Setdb1, Jmjd2a, Ezh2, Jmjd3, and Utx at the Sp7 promoter (Fig. 1H). Osteogenic differentiation (iOB) resulted in significant enrichments of RNAPII, Jmjd2a, and Jmjd3 at this region and reduced interactions of Hdac1/2/4, Setdb1, and Ezh2 (Fig. 1H). Utx binding remained unaltered (Fig. 1H). This signature is similar to that found at this region in OB cells (Fig. 1I), indicating that changes in the recruitment of epigenetic modulators can result in an epigenetic profile that promotes Sp7 gene transcription during osteogenesis.
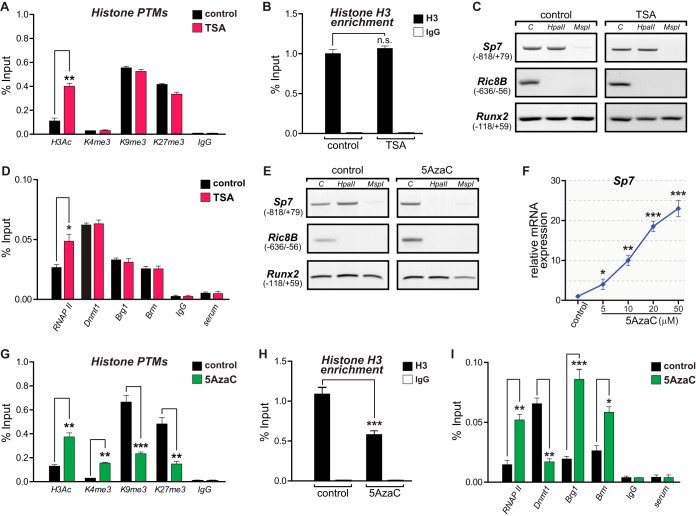
To further assess whether the presence of these epigenetic components is associated with Sp7 gene expression, we evaluated the effect of drugs that selectively inhibit some of the key enzymes found at the Sp7 promoter in UD cells. We first incubated cells with increasing concentrations of trichostatin A (TSA), a paninhibitor of HDAC activity (32, 33), including Hdac1/2/4. TSA treatment resulted in increased H3Ac protein levels (Fig. 1J, left) and a dose-dependent induction of Sp7 mRNA expression (Fig. 1J, right), accompanied by enhanced H3Ac enrichment at the Sp7 promoter (see Fig. 4A). Treatment of UD cells with the Ezh2 inhibitor GSK-126 (34) resulted in a dose-dependent induction of Sp7 mRNA expression (Fig. 1K, right) that was paralleled by a marked reduction in H3K27me3 levels at the Sp7 promoter (Fig. 1K, left). Together, these results confirm a critical role of HDACs and Ezh2 in maintaining Sp7 gene silencing in uncommitted mesenchymal cells. On the other hand, OB cells incubated with the demethylase (Jmjd3 and Utx) inhibitor GSK-J4 (35) exhibited H3K27me3 enrichment at the Sp7 promoter (Fig. 1L, left) and a dose-dependent inhibition of Sp7 gene transcription (Fig. 1L, right). Hence, enhanced H3K27me3 levels at the Sp7 promoter repress Sp7 gene expression in osteoblasts, and Jmjd3/Utx activity contributes to maintaining reduced levels of this mark at this promoter and thereby contributes to stimulating Sp7 gene transcription.
FIG 4.
Coordinated changes in epigenetic parameters accompany transcriptional activation of the Sp7 gene during osteoblast differentiation. UD cells were grown in the presence of the HDAC inhibitor trichostatin A (TSA) (A to D) or the Dnmt inhibitor 5-azacytidine (5AzaC) (E to I) to induce chromatin hyperacetylation or DNA demethylation, respectively. (A and B) H3Ac, H3K4me3, H3K9me3, and H3K27me3 levels (A) as well as histone H3 enrichment at the Sp7 promoter (B) were determined by ChIP of chromatin samples from cells treated with TSA (50 nM) or the vehicle (ethanol). (C) Genomic DNAs from TSA-treated and control cells were analyzed for changes in the 5mC status at the Sp7 promoter by assessing cleavage by HpaII and MspI, as described in the legend to Fig. 2. (D) Enrichment of RNAPII, Dnmt1, Brg1, and Brm at the Sp7 gene promoter in chromatin samples from TSA-treated and control cells measured by ChIP. (E) DNA demethylation at the Sp7 promoter in UD cells treated with 5AzaC (10 μM) and the vehicle (dimethyl sulfoxide). (F) 5AzaC-dependent demethylation of the Sp7 promoter and Sp7 expression. mRNA levels were determined as described in the legend to Fig. 1. (G) 5AzaC-dependent demethylation of the Sp7 promoter and enrichment of H3Ac, H3K4me3, H3K9me3, and H3K27me3 marks at this sequence. (H) Decreased histone H3 enrichment at the Sp7 promoter as a consequence of 5AzaC-dependent demethylation. (I) Changes in enrichment of the RNAPII, Dnmt1, Brg1, and Brm proteins at the Sp7 promoter as a consequence of 5AzaC-dependent demethylation. Enrichments were determined by ChIP as described in the legend to Fig. 1. Data represent means ± standard errors of the means (n ≥ 3). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001 (as determined by Student's t test).
Active DNA demethylation accompanies Sp7 gene activation during osteoblast differentiation.
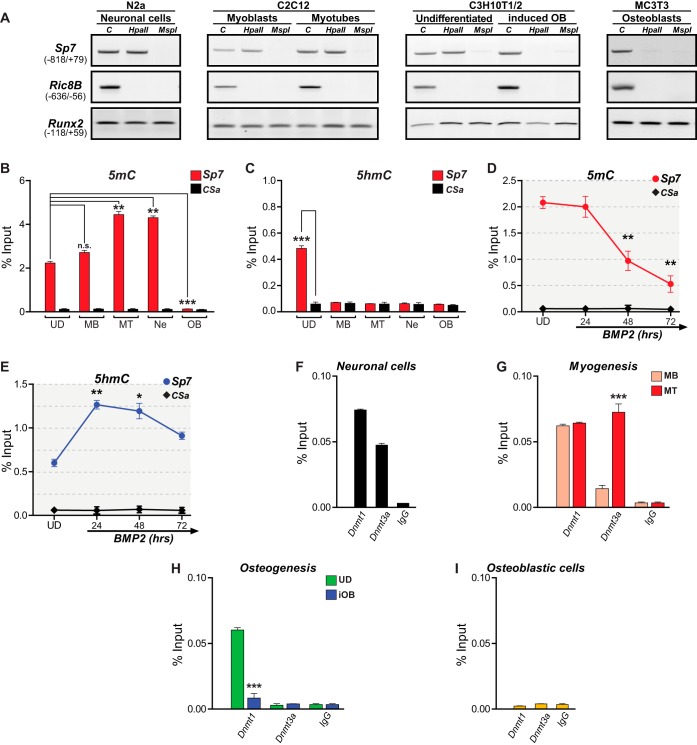
DNA methylation accompanies Sp7 gene repression in nonosteoblastic cells (23). We confirmed this result in our cell models by first evaluating cleavage susceptibility at the Sp7 promoter to the isoschizomers HpaII and MspI (two sites within the promoter sequence under analysis), which are sensitive and insensitive to methylation, respectively (36). As necessary controls in all the experiments performed, we evaluated a region of the proximal Runx2 gene P1 promoter that does not contain MspI/HpaII cleavage sites and a region of the Ric-8B promoter that contains an unmethylated CpG island (37). As shown in Fig. 2A, the Sp7 promoter DNA is methylated in Ne, MB, MT, and UD cells. Interestingly, this pattern is also detected in human embryonic and mesenchymal stem cells (see Fig. S2 in the supplemental material) that do not express Sp7, indicating that this repressive mark is conserved and can be established early during development. In contrast, both OB and iOB cells exhibit demethylated DNA at their transcriptionally active Sp7 promoter (Fig. 2A). These results indicate that DNA demethylation at the Sp7 promoter accompanies the transcription of this gene during osteogenesis.
FIG 2.
Sp7 gene expression is directly associated with the Sp7 promoter DNA methylation/hydroxymethylation status. (A, B, and D) DNA methylation (5mC) at the Sp7 gene promoter in samples obtained from the indicated cell types was assessed by evaluating susceptibility to cleavage by the restriction endonucleases HpaII (sensitive to 5mC) and MspII (insensitive to 5mC) (A) as well as by MedIP analysis (B and D). (C and E) DNA hydroxymethylation (5hmC) was also determined by MedIP. In panel A, Ric-8B is a nonmethylated CpG-rich promoter sequence, and Runx2 is a low-CpG-containing promoter sequence that does not contain MspI/HpaII cleavage sites. In all MedIP analyses, CSa is a well-established unmethylated DNA sequence used as a control; MedIP values are expressed as a percentage of the input. (F to I) Enrichment of Dnmt1 and Dnmt3a at the Sp7 gene promoter determined by ChIP, using antibodies and chromatin samples isolated from Ne (F), MB and MT (G), UD and iOB (H), and OB (I) cells. ChIP values are expressed as a percentage of the input. See the legend to Fig. 1 for an explanation of abbreviations and symbols. Data represent means ± standard errors of the means (n ≥ 3). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; n.s., not significant (as determined by Student's t test).
To quantify these changes in DNA methylation, we performed methylated DNA immunoprecipitation (MedIP) assays (see Materials and Methods). Elevated 5mC at the Sp7 promoter was detected in cells that do not express Sp7, whereas an absence of significant levels of this modification was determined in OB cells. Importantly, DNA hypermethylation at the Sp7 promoter accompanies myogenic differentiation, reaching values that are comparable to those found in neuronal and adipogenic cells (Fig. 2B and Fig. S3). In recent years, DNA demethylation has been linked to the activity of Tet hydroxylases (38, 39). Therefore, MedIP analyses were performed to determine 5hmC levels at the Sp7 promoter in the different cell types under study (Fig. 2C). Significant 5hmC levels were detected only in UD cells (Fig. 2C). Moreover, it was determined that 5mC and 5hmC levels at the Sp7 promoter change during osteoblast differentiation (Fig. 2D and E): a reduced level of 5mC is detected after 48 h of BMP2 treatment (Fig. 2D) in parallel with a robust increase in the level of 5hmC (Fig. 2E). These results strongly argue in favor of a role of 5hmC (and therefore of Tet enzymes) in Sp7 gene transcription activation during osteogenesis.
Using ChIP, we then confirmed that Dnmts are bound to the Sp7 promoter in cells that are not expressing Sp7 and that this interaction is decreased as the cells differentiate to osteoblasts. As shown in Fig. 2F, maintenance and de novo Dnmts, the Dnmt1 and Dnmt3a proteins, respectively, are bound to the Sp7 promoter in Ne cells. Also, MB cells show both proteins bound to the Sp7 promoter, and in MT cells, the Dnmt3a interaction is significantly increased (Fig. 2G). In contrast, reduced Dnmt1 enrichment and an absence of Dnmt3a at the Sp7 promoter are found during osteoblast differentiation (UD to iOB) (Fig. 2H). This decreased Dnmt1/Dnmt3a binding pattern is similar to that detected in OB cells (Fig. 2I), indicating that DNA demethylation that accompanies the transcription of the Sp7 gene during osteogenesis involves the release of Dnmts from the Sp7 promoter.
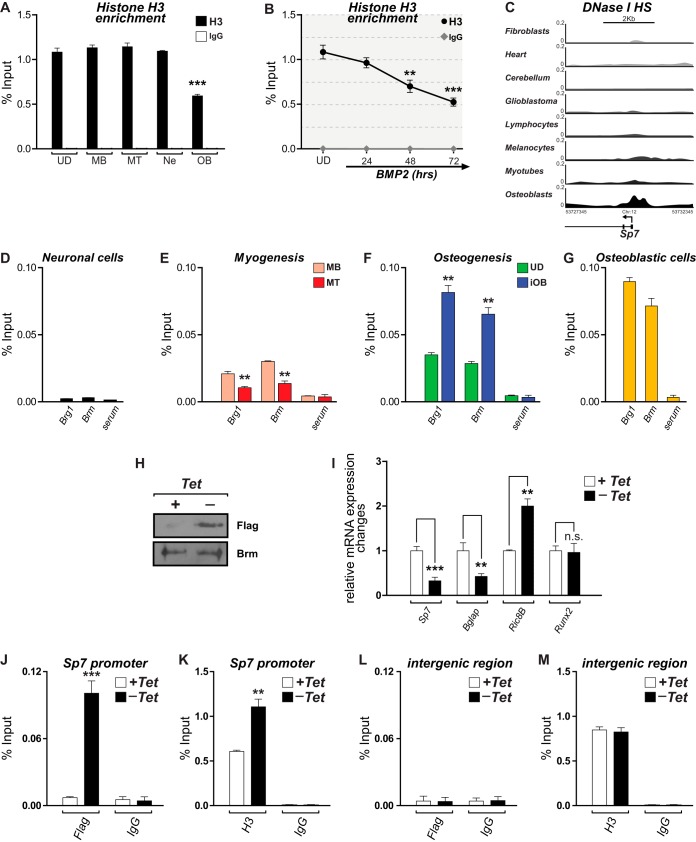
Nucleosome content modulates the accessibility to regulatory regions as well as the presence of specific epigenetic signatures, including histone PTMs and DNA methylation (40–42). Therefore, we evaluated whether changes in nucleosomal histone H3 accompany the above-described epigenetic profile at the Sp7 promoter during Sp7 gene activation. As shown in Fig. 3A, the promoter of this gene exhibits high-level and comparable histone H3 enrichment among cells that are not expressing Sp7. In contrast, OB cells show a significant reduction (about 50%) in histone H3 binding (Fig. 3A). Moreover, the histone H3 reduction is progressive during osteoblast differentiation (Fig. 3B), indicating that the transcriptional activation of this gene during osteogenesis involves a chromatin-remodeling event that likely includes the removal and/or displacement of nucleosomes at regulatory regions that are proximal to the transcriptional start site (TSS). This conclusion is further supported by data available at the ENCODE Project Consortium (43) website (accessed through the UCSC genome browser [http://genome.ucsc.edu/]) (44), where OB cells exhibit enhanced DNase I hypersensitivity at the Sp7 promoter, in contrast to several other nonosseous cell types (Fig. 3C).
FIG 3.
Reduced histone H3 enrichment and SWI/SNF-dependent chromatin remodeling at the Sp7 promoter accompany Sp7 expression in osteoblasts. (A and B) Histone H3 enrichment at the proximal promoter region of the Sp7 gene determined by ChIP of chromatin samples obtained from the indicated cell types (A) as well as from UD cells incubated with BMP2 for up to 72 h (B). See the legend to Fig. 1 for an explanation of cell types and symbols. (C) DNase I hypersensitivity (DNase I HS) profiles at the Sp7 gene promoter in different human cell types obtained from the ENCODE public database. Profiles are shown as the density of DNase I cleavage signals. (D to G) ChIP analyses to detect binding of the SWI/SNF catalytic subunits Brg1 and Brm to the proximal promoter region of the Sp7 gene in the different cell types. (H) Western blot analyses showing the expression of the Flag-tagged Brm mutant protein (Brm-NTP) in the ROSBrmTA osteoblastic cell line grown in the presence (+) or absence (−) of tetracycline (Tet) for 3 days. Blots were revealed by using specific anti-Flag (top) or anti-Brm (bottom) antibodies. (I) Sp7, Bglap, Ric-8B, and Runx2 mRNA expression levels in ROSBrmTA cells grown in the presence and absence of tetracycline. mRNA levels were measured by RT-qPCR and are shown as values relative to the expression level of each gene in the presence of tetracycline, normalized to the level of Gapdh. (J and K) Binding of the Flag-tagged mutant Brm protein (J) and enrichment of histone H3 at the Sp7 promoter (K) were determined by ChIP of samples from cells grown in the presence or absence of tetracycline. (L and M) Evaluation of an unrelated genomic region, devoid of known regulatory sequences (96), as a control. ChIP values and statistical significance were determined as described in the legend of Fig. 1. Data represent means ± standard errors of the means (n ≥ 3). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001 (as determined by Student's t test).
Changes in nucleosome distribution are strongly associated with the activity of ATP-dependent chromatin remodelers (42). Hence, we next determined whether complexes from the SWI/SNF family of chromatin remodelers are associated with the Sp7 promoter in the different cell systems under study. ChIP assays were performed by using antibodies against the Brg1 and Brm catalytic subunits of the SWI/SNF complexes (45). As shown in Fig. 3D, these proteins are not bound to the Sp7 promoter in neuronal cells. Similarly, the enrichment of Brg1 and Brm on this promoter is significantly decreased during myogenic differentiation (Fig. 3E). In contrast, both proteins are significantly enriched at the Sp7 promoter in iOB cells (Fig. 3F), reaching values that are comparable to those found in OB cells (Fig. 3G). To establish a more direct relationship between SWI/SNF activity and Sp7 gene transcription, we took advantage of a previously developed osteoblastic cell line (ROSBrmTA) that conditionally expresses (tetracycline-inducible Tet-off system) a Flag-tagged Brm mutant protein that lacks ATP hydrolysis activity (46, 47). This Brm protein is competent for assembling nonfunctional SWI/SNF complexes (46, 48) that interact with target promoters and inhibit chromatin-remodeling events (37, 46). As shown in Fig. 3H, ROSBrmTA cells grown in the absence of tetracycline for 4 days expressed significant levels of the mutant Flag-tagged Brm protein. This expression is coupled to an important decrease in the level of Sp7 mRNA (Fig. 3I), confirming that SWI/SNF activity is required for Sp7 expression in osteoblasts. In agreement with data from previous reports, the expression of the mutant Brm protein results in decreased Bglap and increased Ric-8B mRNA expression levels (Fig. 3I) (37, 46) and does not affect the expression of the Runx2-p57 gene (47). Importantly, we confirmed by ChIP that this mutant Flag-tagged Brm protein binds to the Sp7 promoter in these cells (Fig. 3J) and that this interaction results in a significant increase in histone H3 enrichment (Fig. 3K), which parallels Sp7 gene transcriptional inhibition. The interaction and the subsequent functional effect of the mutant Brm protein at the Sp7 promoter are selective, as the mutant Brm protein neither binds to a nonrelated intergenic sequence (Fig. 3L) nor alters histone H3 enrichment at this region (Fig. 3M). Together, these results confirm that an SWI/SNF-dependent nucleosome-remodeling event accompanies the transcriptional activity of the Sp7 promoter in osteoblasts.
DNA demethylation is a primary event during transcriptional activation of the Sp7 gene.
As chromatin acetylation and DNA demethylation are critical components of the molecular mechanisms that mediate Sp7 gene transcription during osteoblast differentiation, it became relevant to address whether there is a hierarchical relationship between both epigenetic mechanisms. This was assessed by using the well-described effects of TSA and 5AzaC, inhibitors of HDAC and DNMT enzymes, respectively (49, 50). As shown in Fig. 1J, incubation of UD cells with TSA results in a dose-dependent increase of the Sp7 mRNA expression level. As described in previous reports (51), this Sp7 gene activation is accompanied by an increase in the H3Ac level at the Sp7 promoter (Fig. 4A). However, TSA-treated cells neither show an enrichment of the active mark H3K4me3 nor exhibit a relevant decrease of the levels of the repressive marks H3K9me3 and H3K27me3 (although the latter shows a slight reduction) at this region (Fig. 4A). In addition, histone H3 enrichment (Fig. 4B) and DNA methylation (Fig. 4C) at the Sp7 promoter do not change following TSA treatment. It was also found that although RNAPII enrichment is significantly enhanced at the Sp7 promoter by TSA treatment, binding of Dnmt1, Brg1, and Brm is not altered (Fig. 4D). These results indicate that although TSA-dependent upregulation of Sp7 expression in UD cells is accompanied by increased binding of H3Ac and RNAPII to the Sp7 promoter, this gene activation does not involve the other two epigenetic hallmarks of this process identified during osteogenesis, DNA demethylation and nucleosome remodeling. When UD cells were grown in the presence of 5AzaC, a reduction of CpG methylation at the Sp7 promoter (Fig. 4E) was accompanied by the dose-dependent transcriptional activation of the Sp7 gene (Fig. 4F). These changes were paralleled by H3Ac/H3K4me3 enrichment and H3K9me3/H3K27me3 reduction at the Sp7 promoter (Fig. 4G), thus matching the profile previously found for the transcriptionally active Sp7 gene (Fig. 1). Moreover, this active epigenetic signature is accompanied by reductions in both histone H3 enrichment (Fig. 4H) and Dnmt1 binding (Fig. 4I), together with increased interactions of RNAPII and the SWI/SNF subunits Brg1 and Brm (Fig. 4I). Together, these results indicate that DNA demethylation represents a critical primary molecular event for the transcriptional activation of the Sp7 gene during osteoblast differentiation.
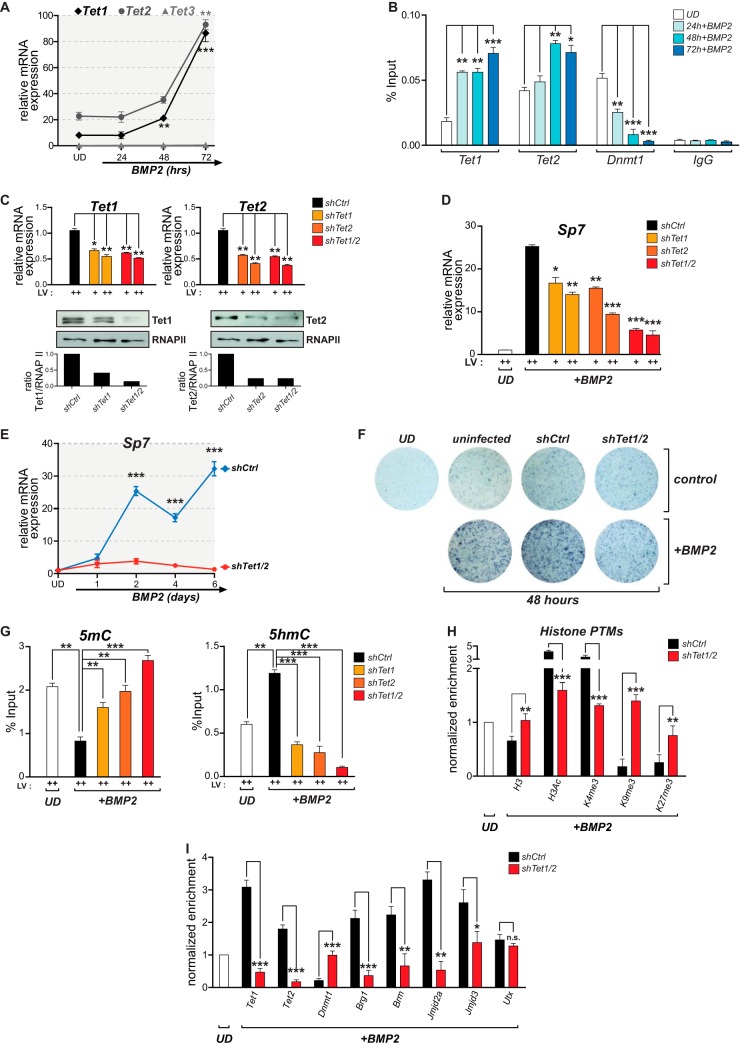
Tet-mediated 5hmC at the Sp7 promoter is required for sustained Sp7 gene transcription during osteoblast differentiation.
Tet enzymes can mediate DNA demethylation through their DNA hydroxylase activity (39, 52). Therefore, we evaluated whether these enzymes contribute to Sp7 gene activity during osteoblast differentiation. As shown in Fig. 5A, Tet1 and Tet2, but not Tet3, are expressed in UD cells, and this expression is enhanced during BMP2-induced osteoblast differentiation. The Tet1 and Tet2 proteins bind to the Sp7 promoter in UD cells, an interaction that is increased following treatment with BMP2 and that is concomitant with Dnmt1 dissociation from this promoter (Fig. 5B). Knockdown of Tet1 and/or Tet2 expression in UD cells was next assessed by using specific short hairpin RNAs (shRNAs) that significantly reduce the expression of each mRNA and protein (Fig. 5C). The independent knockdown of either Tet1 or Tet2 results in a significant inhibition (about 50%) of Sp7 gene transcription in iOB cells (Fig. 5D). Moreover, the double knockdown of Tet1/2 proteins (Fig. 5C) produces an additive decrease in the Sp7 mRNA expression level (about 80%) (Fig. 5D). This effect is maintained beyond 6 days of BMP2 treatment (Fig. 5E) and prevents the normal progression of the osteoblast differentiation program, as reflected by the markedly reduced alkaline phosphatase activity at the extracellular matrix of treated cells (Fig. 5F) (53). These results indicate that Tet1 and Tet2 are required for Sp7 gene expression and likely for the expression of downstream osteoblast-related genes during osteogenic lineage commitment. Additionally, these results suggest that Tet1 and Tet2 functions are, at least partially, complementary at the Sp7 promoter. Tet1 or Tet2 knockdown results in the retention of 5mC at the Sp7 promoter in BMP2-treated cells (Fig. 5G, left), whereas the double knockdown of Tet1/2 produces higher 5mC levels at this promoter that are comparable to those detected in UD cells (Fig. 5G, left). The increased 5mC level is accompanied by a significant reduction in the level of 5hmC at the Sp7 promoter (Fig. 5G, right), further indicating that this 5hmC mark is mediated by Tet1 and/or Tet2.
FIG 5.
Tet-dependent DNA demethylation of the Sp7 gene promoter and Sp7 expression during osteoblast differentiation. (A) Tet1 and Tet2, but not Tet3, are expressed during BMP2-induced osteogenic differentiation. The Tet mRNA expression levels were assessed by RT-qPCR using specific primers against each gene. mRNA values were normalized and are presented as described in the legend to Fig. 1. (B) Binding of the Tet1, Tet2, and Dnmt1 proteins to the Sp7 gene promoter during osteoblast differentiation determined by ChIP using specific antibodies. (C) Knockdown of Tet and Tet2 expression in UD cells. Lentiviral particles (LV) encoding shRNAs against Tet1 (shTet1) or Tet2 (shTet2) were used to transduce UD cells separately or in combination (shTet1/2). Tet1 and Tet2 mRNA expression levels (top) were determined by RT-qPCR. Tet1 and Tet2 protein levels were confirmed by Western blotting. RNAPII protein levels were assessed to confirm equal protein loading and to discard nonspecific effects. Relative quantification of the levels of Tet1 and Tet2 proteins (shown below each representative Western blot) was assessed by using ImageJ software. An unrelated nonspecific shRNA (shCtrl) was used as a control. (D) Effects of Tet1, Tert2, and Tet1/2 knockdowns on the expression of Sp7 mRNA during osteoblast differentiation. (E) Effect of Tet1/2 knockdown on the long-term expression of the Sp7 gene in differentiating osteoblasts. Days of differentiation are indicated below the graph. (F) Cell culture staining to reveal alkaline phosphatase activity in UD cells transduced with shCtrl and shTet1/2 lentiviruses and subsequently grown in the presence or absence of BMP2 for 48 h. The degree of blue staining reflects the level of alkaline phosphatase activity associated with the extracellular matrix of the cells. (G) Effects of Tet1, Tet2, and Tet1/2 knockdowns on the presence of 5mC (left) and 5hmC (right) at the Sp7 promoter of iOB cells. 5mC and 5hmC were assessed by MedIP as described in the legend to Fig. 2. (H and I) Effect of Tet1/2 knockdown on the presence of posttranslationally modified histones (H) and epigenetic modifiers (I) at the Sp7 promoter in iOB cells. Enrichments were assessed as described in the legend to Fig. 1. In panels H and I, enrichment values are expressed as fold changes relative to the value for UD cells (set as 1). Data represent means ± standard errors of the means (n ≥ 3). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001 (as determined by Student's t test).
To address whether alterations in 5mC/5hmC levels at the Sp7 promoter in Tet1/2-depleted cells also affect nucleosomal histone enrichment and the histone PTM signature described above, ChIP assays were performed. Tet1/2-depleted cells exhibit elevated histone H3 binding at the Sp7 promoter upon BMP2 stimulation (Fig. 5H), reaching enrichment values that are comparable to those found in UD cells. The levels of the H3Ac and H3K4me3 marks are reduced when Tet1/2 expression is silenced, and the repressive marks H3K9me3/H3K27me3 are enhanced at the Sp7 promoter in these Tet1/2-deficient iOB cells (Fig. 5H). The binding of the epigenetic regulators previously found to be associated with Sp7 gene regulation was also evaluated under these experimental conditions. As expected, the interaction of Tet1 and Tet2 with the Sp7 promoter was significantly reduced in these cells (Fig. 5I). Similarly, the enrichment of Brg1, Brm, Jmjd2a, and Jmjd3 was markedly decreased (Fig. 5I). In contrast, the binding of Utx was found to be unaltered (Fig. 5I), indicating that the interaction of Utx with the Sp7 promoter is Tet independent. Dnmt1 binding was retained at the Sp7 promoter in Tet1/2-depleted cells treated with BMP2, with values that were comparable to those detected in UD cells (Fig. 5I). Together, these results indicate that Tet-dependent DNA demethylation of the Sp7 promoter is required for the transcriptional activation of this gene during osteoblast differentiation as well as for the binding of most of the regulatory machinery at the Sp7 promoter that mediates the writing and erasing of the epigenetic signature that accompanies Sp7 expression during osteogenesis.
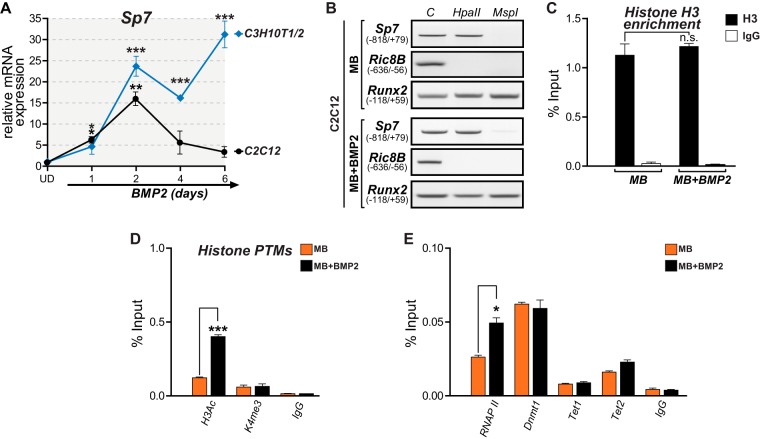
Previous reports indicated that myoblastic C2C12 cells represent a model to study osteogenic differentiation, as BMP2 stimulation results in the expression of numerous bone-related genes and the downregulation of muscle-related genes (54). However, this BMP2-dependent induction of the osteogenic program is often found to be associated with the inconsistent expression of late bone phenotypic markers (53, 55). Here we determined that treatment of these MB cells with BMP2 results in a significant increase in the Sp7 mRNA expression level that peaks after 48 h of incubation (Fig. 6A). Nevertheless, the Sp7 mRNA level in BMP2-treated cells decreases nearly to basal levels after 96 and 144 h of incubation (Fig. 6A). Interestingly, this transient Sp7 mRNA expression is not found associated with decreased DNA methylation (Fig. 6B), reduced histone H3 enrichment (Fig. 6C), or increased H3K4me3 levels (Fig. 6D) at the Sp7 promoter. Instead, this transiently elevated Sp7 mRNA expression level in BMP2-treated MB cells is accompanied by increased H3Ac and RNAPII binding to the Sp7 promoter (Fig. 6D and E, respectively). This pattern is similar to that found during Sp7 activation induced by enhanced histone acetylation using the HDAC inhibitor TSA (Fig. 4A to D). It was also found that Tet2 is the main Tet family member associated with the Sp7 promoter in BMP2-treated MB cells (Fig. 6E), although this level of interaction is significantly lower than that seen in OB cells (Fig. 5B). Together, these results suggest that although C2C12 cells are capable of engaging osteogenic lineage commitment by expressing Sp7 (as well as several other osteoblast-related genes [22]), this expression is only transient and does not involve the establishment of an epigenetic signature at the Sp7 promoter that resembles that found in osteoblastic cells. These data also confirm that Sp7 gene transcription in nonosteoblastic mesenchymal cells can be induced by artificially enhancing histone acetylation at the Sp7 gene promoter using the HDAC inhibitor TSA.
FIG 6.
DNA demethylation at the Sp7 promoter accompanies long-term expression of the Sp7 gene. (A) BMP2 treatment results in transient and stable Sp7 gene expression in MB and UD mesenchymal cells, respectively. Sp7 mRNA levels were determined by RT-qPCR as explained in the legend to Fig. 1 and are presented as values relative to the values for the corresponding untreated cells. (B) Treatment of MB cells with BMP2 for 48 h does not induce DNA demethylation at the Sp7 promoter. (C) Histone H3 enrichment at the Sp7 promoter is not reduced in MB cells treated with BMP2 (48 h). (D) MB cells incubated with BMP2 exhibit increased enrichment of H3Ac but not of H3K4me3 at the Sp7 promoter. (E) Enrichment of RNAPII, Dnmt1, and Tet2 (but not Tet1) at the Sp7 gene promoter in MB cells treated with BMP2 (48 h). Data represent means ± standard errors of the means (n ≥ 3). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001 (as determined by Student's t test).
Coupling of DNA demethylation, H3K4 methylation, and H3K9me3 demethylation activities during activation of the Sp7 gene.
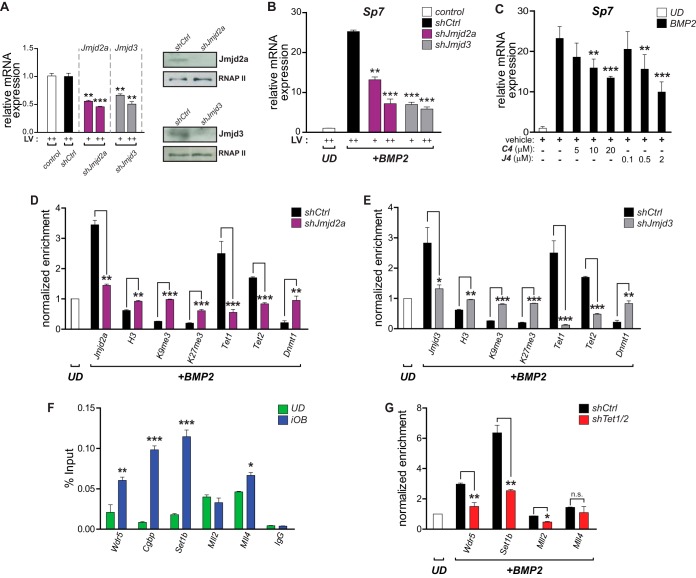
The demethylases Jmjd2a and Jmjd3 bind to the Sp7 promoter during the osteoblast differentiation-linked activation of this gene (Fig. 1H). In addition, shRNA-mediated knockdown of Jmjd2a or Jmjd3 expression (Fig. 7A) inhibits Sp7 mRNA expression during BMP2-induced osteoblast differentiation (Fig. 7B). The effectiveness of these knockdowns was also confirmed by ChIP (Fig. 7D and E); a decrease in the Jmjd2a or Jmjd3 expression level is reflected by the reduced enrichment of these two proteins at the Sp7 promoter and by the corresponding increase in the levels of the H3K9me3 and H3K27me3 marks. Interestingly, an increase in the levels of these two repressive marks is also produced when each demethylase is downregulated independently (Fig. 7D and E), suggesting that the erasing of these histone PTMs at the Sp7 promoter may occur through a coordinated mechanism. The independent depletion of Jmjd2a and Jmjd3 also results in an enrichment of histone H3 at the Sp7 promoter (Fig. 7D and E), in agreement with the idea that the downregulation of Sp7 expression is accompanied by nucleosome enrichment at the Sp7 promoter. The contribution of Jmjd3 and Jmjd2a activities to Sp7 gene expression during osteogenic differentiation was also addressed by determining the effect of the drugs GSK-J4 and C4, selective inhibitors of Jmjd3 and Jmjd2a, respectively (35, 56). As shown in Fig. 7C, the BMP2-mediated upregulation of Sp7 mRNA expression is inhibited, in a dose-dependent manner, by coincubation with either GSK-J4 (Fig. 7C, left) or C4 (Fig. 7C, right). Accordingly, the binding of Tet1 and Tet2 to the Sp7 promoter is significantly impaired in iOB cells where the Jmjd2a and Jmjd3 proteins have been knockdown (Fig. 7D and E, respectively). These results further support the idea that a coordinated mechanism is operating at the Sp7 promoter, involving the recruitment of activities that both demethylate DNA and erase the H3K9me3 and H3K27me3 repressive marks. In agreement with this, Dnmt1 enrichment at this promoter was found to be significantly enhanced in Jmjd2a- and Jmjd3-depleted iOB cells (Fig. 7D and E, respectively), supporting the concept that a mutually exclusive association of DNA methylation and DNA demethylation regulatory machineries is directly related to the H3K9me3 and H3K27me3 levels on the Sp7 promoter.
FIG 7.
Coupling of DNA demethylation with H3K4 methylation/H3K9me3 demethylation at the Sp7 promoter during osteoblast differentiation. (A) Demethylases Jmjd2a and Jmjd3 were knocked down in UD cells by infecting the cells with lentiviruses coding for specific shRNAs (shJmjd2a and shJmjd3, respectively). Decreased expression levels of both demethylases were confirmed by RT-qPCR (mRNAs) (left) and Western blotting (proteins) (right). Infection with an unrelated random shRNA (shCtrl) was used as a control. Normalizations were carried out as described in the legend to Fig. 5. (B) Knockdown of Jmjd2a and Jmjd3 impairs Sp7 gene expression during osteoblast differentiation. Infected UD cells were treated with BMP2 for 48 h, and mRNA levels (Sp7 and Gapdh) were determined by RT-qPCR. (C) Treatment of differentiating (iOB) cells with the demethylase inhibitor GSK-J4 (selective for Jmjd3) or C4 (selective for Jmjd2a) inhibits Sp7 mRNA expression in a dose-dependent manner. (D and E) Knockdowns of Jmjd2a (D) and Jmjd3 (E) decrease their presence at the Sp7 promoter and affect the enrichment (measured by ChIP) of histone H3, H3K9me3, H3K27me3, Tet1, Tet2, and Dnmt1 at this promoter sequence. Normalized enrichments are shown as fold changes relative to the values for UD cells. (F) Binding (determined by ChIP) of key components of COMPASS and COMPASS-like complexes to the Sp7 promoter during osteoblast lineage commitment. Enrichment values are shown as a percentage of the input. (G) Tet1/2 knockdown impairs the interaction (determined by ChIP) of the COMPASS complex subunits Wdr5 and Set1b with the Sp7 promoter in iOB cells. In contrast, only minor effects on the binding of Mll2- and Mll4-containing complexes are observed. Results are shown as fold changes relative to the value for UD cells (set as 1). Data represent means ± standard errors of the means (n ≥ 3). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001 (as determined by Student's t test).
Previous studies demonstrated the contribution of Tet enzymes to the recruitment of H3K4 methylase-containing COMPASS complexes to target sequences (57). As shown in Fig. 7F, we confirmed that Wdr5, a critical component of COMPASS and COMPASS-like complexes (58, 59), binds to the Sp7 promoter in iOB cells. This interaction of Wdr5 is accompanied by the binding of the Set1b methyltransferase and Cgbp, both of which are components of the Set1/COMPASS complex (59). Also, the COMPASS-like methyltransferases Mll2 and Mll4 were found to interact with this promoter in iOB cells (Fig. 7F). Importantly, the Wdr5, Cgbp, and Set1b proteins were only poorly bound to the Sp7 promoter in UD cells, indicating that the interaction of Set1/COMPASS accompanies H3K4me3 enrichment and the expression of the Sp7 gene during osteoblast differentiation. On the other hand, Mll2 and Mll4 were found to be enriched at the Sp7 promoter in samples from UD cells (Fig. 7F), suggesting that their function at this locus precedes Sp7 gene transcription. The association of Wdr5 and Set1b with the Sp7 promoter was found to be dependent on Tet1/2 expression and binding to this regulatory region (Fig. 7G). In contrast, the binding of Mll4 to the Sp7 promoter was not significantly impaired by Tet1/2 depletion (Fig. 7G). Together, these results indicate that the enrichment of Tet1/2 proteins at the Sp7 promoter during osteogenic differentiation is required for the interaction of COMPASS and COMPASS-like complexes that favor Sp7 gene expression.
DISCUSSION
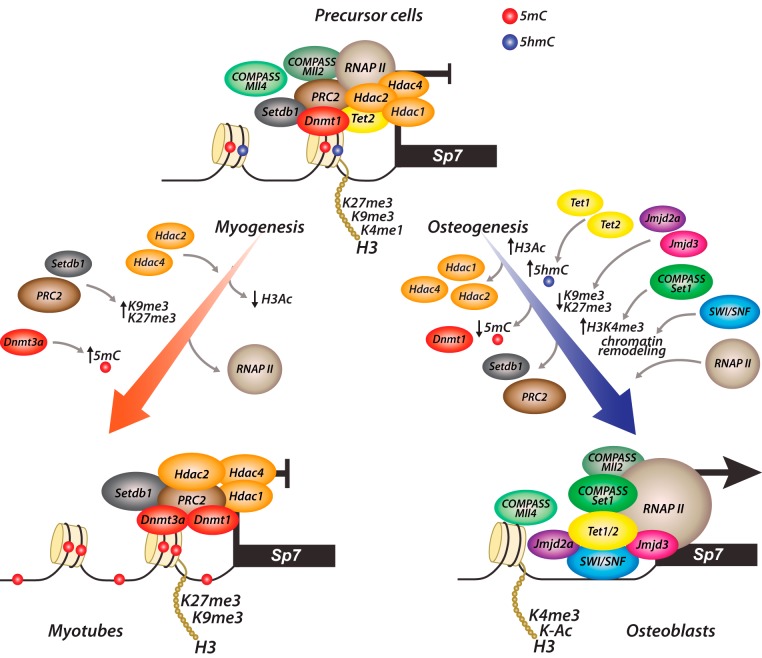
Our results demonstrate that DNA methylation is a critical component within the mechanisms that maintain the stable repression of the Sp7 gene in nonosteoblastic and preosteogenic cells. We determined that Tet-mediated DNA demethylation not only accompanies the expression of this bone master regulator during osteoblast differentiation but also is directly linked to the establishment of an epigenetic histone mark profile at the Sp7 promoter that supports Sp7 gene transcription during BMP2-induced osteogenesis (Fig. 8). Accumulating evidence demonstrates that Tet-mediated 5hmC in mammalian genomes can represent a principal component during the active DNA demethylation process that leads to transcriptional activation (60–66). Moreover, our data indicate that the binding of both the Tet1 and Tet2 proteins to the Sp7 promoter is critical for the recruitment of enzymatic machineries that mediate both the writing and erasing that generate the epigenetic signature that accompanies Sp7 gene transcription during osteogenesis (Fig. 8). In agreement with previously reported results (29, 30), our data suggest that the Tet1 and Tet2 proteins play a role not only during the transcriptional activation of the Sp7 gene as 5mC hydroxylases but also by contributing to scaffold regulatory proteins that mediate epigenetic control that favors transcription.
FIG 8.
Proposed model for the epigenetic control of Sp7 gene expression during osteoblast lineage commitment. Key regulatory events and their cognate regulatory complexes are schematically represented. Enrichments or reductions of epigenetic hallmarks at the Sp7 gene promoter are indicated by arrows. Relative enrichments of the regulatory complexes are represented by the sizes of the spheres.
The necessity of DNA demethylation for a stable Sp7 gene expression profile was further demonstrated by using promyoblastic cells. We found that BMP2-mediated transient Sp7 expression in these cells is mainly correlated with enhanced H3Ac levels and increased binding of RNAPII at Sp7 promoter sequences. Other parameters of the epigenetic signature associated with active and stable Sp7 gene expression during osteoblast differentiation, including decreased binding of Dnmt1 and histone H3 together with increased enrichment of H3K4me3 and Tet1/2 (Fig. 8), were not identified at the Sp7 promoter in myoblastic cells treated with BMP2. These results raise significant concern about using BMP2-treated C2C12 cells as a model to address epigenetic mechanisms controlling the expression of this bone master regulator. Additionally, it was recently shown that the inability to activate Sp7 expression following incubation with well-established osteoblast induction medium represents a critical limitation for using mesenchymal stem cells derived from human umbilical cords as a source of pro-osteogenic cells (67). Therefore, these results, together with results of our analyses using the HDAC inhibitor TSA in uncommitted mesenchymal cells, indicate that the activation of Sp7 gene transcription can be induced in nonosteogenic cells independent of DNA demethylation and nucleosome remodeling at the Sp7 promoter although concomitantly with increased histone acetylation and enhanced RNAPII binding. Nevertheless, the stable Sp7 gene transcription observed during BMP2-induced osteoblast differentiation involves Tet-dependent DNA demethylation and SWI/SNF-associated nucleosome remodeling at the Sp7 promoter.
Our results indicate that what can be identified as independent epigenetic mechanisms are also operating in a coordinated manner at the Sp7 promoter to mediate DNA demethylation, nucleosome remodeling, and erasing of the H3K9me3 and H3K27me3 repressive marks during osteogenic differentiation. Hence, we identify an interdependent association of the demethylases Jmjd2a/Jmjd3 and the Tet1/2 proteins with the Sp7 promoter. It is important to note that these interdependent interactions are not due to unrelated variations in the expression levels of the genes encoding these enzymes that are not directly targeted by our shRNA-mediated strategy (data not shown). Previous studies also demonstrated a strong correlation between DNA methylation and the presence of the H3K9me3 repressive mark (68–72). In contrast, currently, there is not clear evidence supporting a direct relationship between DNA methylation and H3K27me3 levels (73). Whether the H3K27me3 enrichment that accompanies the decreased binding of Tet1/Tet2 to the Sp7 promoter results only from the reduced interaction of a Jmjd3-containing complex will require further clarification, as our analyses show that the binding of the other H3K27me3 demethylase, Utx, to this promoter remains unaltered in Tet1/2-depleted cells. Hence, these results raise the possibility that Utx activity is inhibited under these experimental conditions by an unknown mechanism. These relevant aspects are currently being investigated by our team. Similarly, future analyses will need to confirm the putative dynamic formation at the Sp7 promoter of high-molecular-weight complexes, including these histone demethylases and other proteins mediating epigenetic activities identified in this study (e.g., Tet proteins). In line with this possibility, it was reported previously that Jmjd3 forms a complex with Wdr5 and Brg1, key components of COMPASS and SWI/SNF complexes, respectively (74, 75), and that Tet proteins can interact with these same complexes (29, 57).
Our results demonstrate that the binding of the Tet1 and Tet2 proteins to the Sp7 promoter during osteogenic differentiation is necessary for the interaction of H3K4-methylating complexes. This result is in agreement with data from previous reports supporting an inverse relationship between DNA methylation and H3K4me3 enrichment (76). Interestingly, it was recently reported that H3K4me3-dependent reexpression of epigenetically silenced genes requires a significant reduction of DNA methylation levels at the TSS of these genes (77), supporting the idea that a hierarchically organized network of epigenetic mechanisms is operating at regulatory regions of numerous genes in mammalian cells. Our results show that the stable and robust expression of the Sp7 gene during osteoblast differentiation is associated with H3K4me3 enrichment, the release of Dnmt1, and reduced levels of 5mC at the Sp7 promoter. Also, DNA methylation inhibition in our mesenchymal cells was accompanied by H3K4me3 enrichment at the Sp7 promoter. We identified Mll2-, Mll4-, and Set1b-containing complexes at the Sp7 promoter, suggesting a functional overlap of these H3K4-methylating complexes. Although the physiological relevance of these results and their mechanistic implications are presently unclear, our results are in agreement with data from previous reports indicating that genes can be recognized for more than one H3K4 methylase complex (78–80). A recent study provides strong evidence for a mechanism that, based on the selective sumoylation of COMPASS and COMPASS-like complex subunits, supports a model where H3K4me3-mediated epigenetic control is achieved through the dynamic formation of selective H3K4-methylating complexes at target genes (80). Interestingly, we also find that the binding of Mll proteins to the Sp7 promoter precedes transcriptional activation. Recent reports indicate that Mll3/4-containing complexes can have a role during gene repression by mediating the deposit and maintenance of the H3K4me1 mark at target promoters (79, 81). Although it remains to be determined whether these methylases have similar repressive roles at the Sp7 gene in uncommitted mesenchymal cells, it is encouraging that their binding to this promoter is accompanied by elevated H3K4me1 levels. Current experiments by our team are specifically addressing this question.
MATERIALS AND METHODS
Chromatin immunoprecipitation.
ChIP analyses were performed with cross-linked chromatin samples as described previously (37, 82), with the following considerations. Cell cultures were incubated for 10 min with 1× phosphate-buffered saline (PBS) containing 1% formaldehyde, with gentle agitation at room temperature, after which a 10× glycine solution (1.25 M) was added to stop the cross-linking reaction. Cross-linked cells were washed with 1× PBS three times, resuspended in 5 volumes of cell lysis buffer (CLB) (50 mM HEPES [pH 7.8], 3 mM MgCl2, 20 mM KCl, 0.1% NP-40, and proteinase inhibitors), and homogenized with a Dounce homogenizer. The cell extracts were collected by centrifugation at 3,000 × g for 5 min, resuspended in 500 μl of sonication buffer (50 mM HEPES [pH 7.9], 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% deoxycholate acid, 0.1% SDS, and proteinase inhibitors), and then incubated for 10 min on ice. Chromatin was sheared in a Bioruptor water bath sonicator (Diagenode Inc., NJ) to obtain fragments of 200 to 500 bp. Extracts were sonicated at high power for three pulses of 5 min each and centrifuged at 16,000 × g for 15 min at 4°C. Supernatants were collected, aliquoted, frozen in liquid nitrogen, and stored at −80°C; one aliquot was used for A260 measurements. The fragmented chromatin size was confirmed by electrophoretic analysis. For immunoprecipitation (IP), 25 μg of cross-linked chromatin extracts was diluted in sonication buffer and precleared by incubation with 2 μg of normal IgG and 50 μl of protein A/G-agarose beads (Santa Cruz Biotechnology, Dallas, TX) for 1 h at 4°C. Chromatins were first centrifuged at 4,000 × g for 5 min to collect the supernatant and then incubated with specific antibodies overnight at 4°C (see Table S1 in the supplemental material for the list of antibodies used). The immunocomplexes were recovered by the addition of protein A- or G-agarose beads, followed by incubation for 1 h at 4°C. Immunoprecipitated complexes were washed once with sonication buffer, twice with LiCl buffer (100 mM Tris-HCl [pH 8.0], 500 mM LiCl, 1% NP-40, and 1% deoxycholic acid), and once with Tris-EDTA (TE) buffer (pH 8.0). Finally, the immunocomplexes were eluted in elution buffer (50 mM NaHCO3 and 1% SDS) and incubated for 15 min at 65°C. Samples were centrifuged at 10,000 × g for 5 min, and supernatants were collected for incubation overnight at 65°C, to reverse cross-linking in the presence of RNase I (10 μg/ml). After incubation with proteinase K (100 μg/ml) for 2 h at 50°C, the DNAs were recovered from samples by phenol-chloroform extraction followed by ethanol precipitation using glycogen (20 μg/ml) as a precipitation carrier. Immunoprecipitated DNA was quantified by quantitative PCR (qPCR) analysis, determining enrichment levels as a percentage of the amount of input material. In some cases, values are shown as “normalized enrichment.” This corresponds to the enrichment values determined for each specific immunoprecipitation that are then expressed relative to the corresponding values determined in undifferentiated C3H10T1/2 cells. Primers used are listed in Table S2 in the supplemental material.
Reverse transcription-qPCR (RT-qPCR) analysis.
Total RNA was extracted with TRIzol (Thermo Fisher, Waltham, MA), according to the manufacturer's protocol, using 2 μg of RNA for reverse transcription. qPCR was performed by using Brilliant II SYBR green qPCR master mix (Agilent Technologies, Santa Clara, CA) in a Stratagene Mx3000P thermal cycler (Agilent Technologies). All primer concentrations and melting temperatures for the qPCRs were previously standardized for approximately 100% efficiency. Sequences of primers used are listed in Table S2 in the supplemental material.
Cell culture.
Each cell line used was purchased from the American Type Culture Collection (ATCC) and was cultured according to the corresponding standard indications. C3H10T1/2 cells (ATCC CCL-226) were cultured at low confluence in Dulbecco's modified Eagle's medium (DMEM) (catalog number 11965-092; Gibco, MA) supplemented with 10% fetal bovine serum (FBS) and 50 U/ml penicillin-streptomycin. C2C12 cells (ATCC CRL-1772) were cultured at low confluence in DMEM–F-12 medium (catalog number 12500-062; Gibco) supplemented with 10% FBS and 50 U/ml penicillin-streptomycin. N2a cells (ATCC CCL-131) were cultured in DMEM supplemented with 10% FBS and 50 U/ml penicillin-streptomycin. MC3T3 cells (ATCC CRL-2593) were cultured in alpha-minimal essential medium (alpha-MEM) (catalog number A10490-01; Gibco) with 10% FBS and 50 U/ml of penicillin-streptomycin. For osteoblast differentiation experiments, C3H10T1/2 cells were stimulated according to previously described protocols (83). A total of 150,000 cells were seeded per 100-mm dish, maintained for 3 days until they reached 80% confluence, and then changed to fresh medium supplemented with 300 ng/ml of BMP2 (catalog number 4577; BioVision, CA). Treated and control cells were cultured for 48 h or as indicated in each figure. BMP2 treatments of C2C12 cells were performed similarly, and samples were collected at the indicated times. For myogenic differentiation, C2C12 cells were treated according to previously described protocols (84). A total of 150,000 cells were seeded per 100-mm dish, maintained for 2 days until they reached 80% confluence, and then changed to DMEM–F-12 medium containing 10% horse serum. This differentiation medium was replaced every 2 days. Myotubes were collected after 4 days of differentiation. Neuronal N2a cells were grown until they reached 90% confluence, after which medium was replaced with DMEM containing 1% FBS to arrest proliferation and thus promote the acquisition of the characteristic neuronal phenotype (85). Cells were collected 4 days later, replacing the medium every 2 days. Preosteoblastic MC3T3 cells were seeded at low confluence (150,000 cells per 100-mm dish), cultured until they reached confluence, and then differentiated for 5 days in alpha-MEM containing ascorbic acid (50 ng/ml) to acquire a mature osteoblastic phenotype (37).
Knockdown of gene expression.
Gene silencing experiments were performed with validated lentiviral particles carrying control or gene-specific shRNAs (86) obtained from Sigma-Aldrich (MO). Infection protocols were carried out according to standard methods, as previously reported (87), in the presence of 10 μg/ml of Polybrene. Cells were infected at low confluence levels (∼40%) and cultured for the indicated times. The efficiency of knockdown was finally checked by RT-qPCR and Western blot analysis.
Methylated DNA immunoprecipitation analysis.
MedIP analysis was performed as described previously (88, 89). We extracted genomic DNA from cultured cell samples by overnight proteinase K treatment in lysis buffer (20 mM Tris [pH 8.0], 4 mM EDTA, 20 mM NaCl, and 1% SDS), phenol-chloroform extraction, ethanol precipitation, and RNase digestion. Genomic DNA was then sonicated to produce fragments of ∼400 bp. Four micrograms of fragmented genomic DNA was used for each immunoprecipitation reaction mixture. Genomic DNA was then denatured for 10 min at 95°C and immunoprecipitated overnight at 4°C with 10 μl of anti-5-methylcytidine (Eurogentec) or anti-5-hydroxymethylcitydine (Active-Motif) antibodies in a final volume of 500 ml of IP buffer (10 mM sodium phosphate [pH 7.0], 140 mM NaCl, 0.05% Triton X-100). The mixture was incubated with 50 μl of protein A-agarose beads for 2 h at 4°C and washed twice with 700 μl of IP buffer. The beads with were then treated with 7 μl of proteinase K (10 mg/ml) in 250 μl of digestion buffer (50 mM Tris [pH 8.0], 10 mM EDTA, and 0.5% SDS) for 3 h at 50°C. Immunoprecipitated DNA was recovered by phenol-chloroform extraction followed by ethanol precipitation using glycogen as the precipitation carrier. Purified DNA was then evaluated by qPCR analysis, defining the enrichment levels as a percentage of the input material. Primers used for qPCR analysis, including those used to evaluate negative and positive control genomic sequences (CSa and intracisternal A particle [IAP], respectively), are listed in Table S2 in the supplemental material.
DNA methylation analyses using restriction endonucleases.
Purified genomic DNA was obtained from the different cellular samples after overnight proteinase K treatment in lysis buffer (20 mM Tris [pH 8.0], 4 mM EDTA, 20 mM NaCl, and 1% SDS), phenol-chloroform extraction, ethanol precipitation, and RNase digestion. Five micrograms of genomic DNA was cleaved overnight with 20 U of the HpaII (methylation-sensitive) or MspI (methylation-insensitive) enzyme (New England BioLabs, Ipswich, MA) in the corresponding buffers at 37°C (90). With this approach, two HpaII/MspI sites (positions −313 and −114) were analyzed at the proximal Sp7 promoter. For the analysis of each digested DNA, 200 ng was used as the template for PCRs. Amplified products were then electrophoresed in 1% agarose gels. Primer sequences used to amplify the sequences associated with the Sp7, Runx2-P1, and Ric-8B promoters are listed in Table S2 in the supplemental material.
Tet-off system to generate SWI/SNF-deficient osteoblastic cells.
To determine whether Sp7 expression requires the activity of SWI/SNF chromatin-remodeling complexes, we used the ROSBrmTA osteoblastic cell line that expresses an ATPase-deficient Flag-tagged Brm mutant protein under the control of the tetracycline-inducible promoter system (37, 46, 47). ROSBrmTA cells were maintained in F-12 medium (5% FBS, 14 mM NaHCO3, 800 μM CaCl2, and 28 mM HEPES) supplemented with the antibiotics hygromycin (50 μg/ml) and Geneticin (100 μg/ml) for selective pressure and tetracycline (10 μg/ml) to control inducible expression by the Tet-off system. The mutant Brm protein is expressed in these cells after growth in medium lacking tetracycline for 3 days. Cells were then collected and processed for Western blotting, RT-qPCR, and ChIP analyses.
Drugs to inhibit chromatin-modifying enzymes.
All drugs used in this study were previously described (33–35, 91, 92). 5-Azacytidine (catalog number A2385; Sigma-Aldrich) inhibits DNA methyltransferases, trichostatin A (catalog number T8552; Sigma-Aldrich) inhibits a wide range of histone deacetylases (HDACs), GSK-126 (catalog number 2282; BioVision) inhibits the Ezh2 methyltransferase, and GSK-J4 (catalog number SML0701; Sigma-Aldrich) inhibits both the Utx and Jmjd3 histone demethylases.
Analyses of nuclear proteins by Western blotting.
Nuclear extracts were prepared according to standard methods (93). Purified histones were obtained by using a standard acid extraction method (94), using Slide-A-Lyzer dialysis cassettes with a molecular mass cutoff of 3.5 kDa. In both cases, protein levels were quantified by Bradford's assay, using bovine serum albumin as a standard (95).
For Western blot analyses, 10 or 5 μg of total nuclear extracts or purified histones, respectively, was subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and then transferred to a nitrocellulose membrane. Immunoblotting was performed with secondary antibodies conjugated to horseradish peroxide (HRP) and enhanced chemiluminescence solutions (Perkin-Elmer, Waltham, MA). Primary antibodies used are listed in Table S1 in the supplemental material. Six percent and 12% acrylamide gels were used to resolve proteins of high and low molecular weights, using RNAPII or TFIIB levels to control for protein loading.
Alkaline phosphatase activity assays.
C3H10T1/2 cell monolayers were washed twice with Dulbecco's PBS (without Ca2+/Mg2+), fixed with a 1% formaldehyde solution during 5 min, washed again with Dulbecco's PBS (without Ca2+/Mg2+) plus 0.05% Tween 20, and finally rinsed in staining solution (0.1 M Tris-HCl [pH 9.5], 0.1 M NaCl, 0.05 M MgCl2, 0.1% Tween 20) containing NBT (Nitro Blue Tetrazolium)-BCIP [5-bromo-4-chloro-3-indolylphosphate] reagent (catalog number 11681451001; Roche, Mannheim, Germany) with incubation at room temperature for 30 min in the dark (87). Staining corresponds to the formation of a blue precipitate that was captured by using conventional photo cameras.
Supplementary Material
ACKNOWLEDGMENTS
We thank Brigitte van Zundert for helpful discussions and critical evaluation of the manuscript.
This work was supported by grants from FONDECYT (1130706; Fondo Nacional de Desarrollo Cientifico y Tecnologico) and FONDAP (15090007; Fondo de Financiamiento de Centros de Investigacion en Areas Prioritarias). H.S. was supported by CONICYT PhD fellowship (21100548).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00177-17.
REFERENCES
- 1.Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B. 2002. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 108:17–29. doi: 10.1016/S0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 2.Zhang C, Cho K, Huang Y, Lyons JP, Zhou X, Sinha K, McCrea PD, de Crombrugghe B. 2008. Inhibition of Wnt signaling by the osteoblast-specific transcription factor Osterix. Proc Natl Acad Sci U S A 105:6936–6941. doi: 10.1073/pnas.0710831105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Long F. 2011. Building strong bones: molecular regulation of the osteoblast lineage. Nat Rev Mol Cell Biol 13:27–38. doi: 10.1038/nrm3254. [DOI] [PubMed] [Google Scholar]
- 4.Sinha KM, Zhou X. 2013. Genetic and molecular control of osterix in skeletal formation. J Cell Biochem 114:975–984. doi: 10.1002/jcb.24439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang C. 2010. Transcriptional regulation of bone formation by the osteoblast-specific transcription factor Osx. J Orthop Surg Res 5:37. doi: 10.1186/1749-799X-5-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou X, Zhang Z, Feng JQ, Dusevich VM, Sinha K, Zhang H, Darnay BG, de Crombrugghe B. 2010. Multiple functions of Osterix are required for bone growth and homeostasis in postnatal mice. Proc Natl Acad Sci U S A 107:12919–12924. doi: 10.1073/pnas.0912855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lapunzina P, Aglan M, Temtamy S, Caparros-Martin JA, Valencia M, Leton R, Martinez-Glez V, Elhossini R, Amr K, Vilaboa N, Ruiz-Perez VL. 2010. Identification of a frameshift mutation in Osterix in a patient with recessive osteogenesis imperfecta. Am J Hum Genet 87:110–114. doi: 10.1016/j.ajhg.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Timpson NJ, Tobias JH, Richards JB, Soranzo N, Duncan EL, Sims AM, Whittaker P, Kumanduri V, Zhai G, Glaser B, Eisman J, Jones G, Nicholson G, Prince R, Seeman E, Spector TD, Brown MA, Peltonen L, Smith GD, Deloukas P, Evans DM. 2009. Common variants in the region around Osterix are associated with bone mineral density and growth in childhood. Hum Mol Genet 18:1510–1517. doi: 10.1093/hmg/ddp052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao J, Bradfield JP, Li M, Zhang H, Mentch FD, Wang K, Sleiman PM, Kim CE, Glessner JT, Frackelton EC, Chiavacci RM, Berkowitz RI, Zemel BS, Hakonarson H, Grant SF. 2011. BMD-associated variation at the Osterix locus is correlated with childhood obesity in females. Obesity (Silver Spring) 19:1311–1314. doi: 10.1038/oby.2010.324. [DOI] [PubMed] [Google Scholar]
- 10.Lu X, Gilbert L, He X, Rubin J, Nanes MS. 2006. Transcriptional regulation of the osterix (Osx, Sp7) promoter by tumor necrosis factor identifies disparate effects of mitogen-activated protein kinase and NF kappa B pathways. J Biol Chem 281:6297–6306. doi: 10.1074/jbc.M507804200. [DOI] [PubMed] [Google Scholar]
- 11.Ulsamer A, Ortuno MJ, Ruiz S, Susperregui AR, Osses N, Rosa JL, Ventura F. 2008. BMP-2 induces Osterix expression through up-regulation of Dlx5 and its phosphorylation by p38. J Biol Chem 283:3816–3826. doi: 10.1074/jbc.M704724200. [DOI] [PubMed] [Google Scholar]
- 12.Nishio Y, Dong Y, Paris M, O'Keefe RJ, Schwarz EM, Drissi H. 2006. Runx2-mediated regulation of the zinc finger Osterix/Sp7 gene. Gene 372:62–70. doi: 10.1016/j.gene.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 13.Barbuto R, Mitchell J. 2013. Regulation of the osterix (Osx, Sp7) promoter by osterix and its inhibition by parathyroid hormone. J Mol Endocrinol 51:99–108. doi: 10.1530/JME-12-0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xing W, Singgih A, Kapoor A, Alarcon CM, Baylink DJ, Mohan S. 2007. Nuclear factor-E2-related factor-1 mediates ascorbic acid induction of osterix expression via interaction with antioxidant-responsive element in bone cells. J Biol Chem 282:22052–22061. doi: 10.1074/jbc.M702614200. [DOI] [PubMed] [Google Scholar]
- 15.Meyer MB, Benkusky NA, Lee CH, Pike JW. 2014. Genomic determinants of gene regulation by 1,25-dihydroxyvitamin D3 during osteoblast-lineage cell differentiation. J Biol Chem 289:19539–19554. doi: 10.1074/jbc.M114.578104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maehata Y, Takamizawa S, Ozawa S, Kato Y, Sato S, Kubota E, Hata R. 2006. Both direct and collagen-mediated signals are required for active vitamin D3-elicited differentiation of human osteoblastic cells: roles of osterix, an osteoblast-related transcription factor. Matrix Biol 25:47–58. doi: 10.1016/j.matbio.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Mandal CC, Drissi H, Choudhury GG, Ghosh-Choudhury N. 2010. Integration of phosphatidylinositol 3-kinase, Akt kinase, and Smad signaling pathway in BMP-2-induced osterix expression. Calcif Tissue Int 87:533–540. doi: 10.1007/s00223-010-9419-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gordon JA, Hassan MQ, Koss M, Montecino M, Selleri L, van Wijnen AJ, Stein JL, Stein GS, Lian JB. 2011. Epigenetic regulation of early osteogenesis and mineralized tissue formation by a HOXA10-PBX1-associated complex. Cells Tissues Organs 194:146–150. doi: 10.1159/000324790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang D, Okamura H, Teramachi J, Haneji T. 2015. Histone demethylase Utx regulates differentiation and mineralization in osteoblasts. J Cell Biochem 116:2628–2636. doi: 10.1002/jcb.25210. [DOI] [PubMed] [Google Scholar]
- 20.Yang D, Okamura H, Nakashima Y, Haneji T. 2013. Histone demethylase Jmjd3 regulates osteoblast differentiation via transcription factors Runx2 and osterix. J Biol Chem 288:33530–33541. doi: 10.1074/jbc.M113.497040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang F, Xu L, Xu L, Xu Q, Karsenty G, Chen CD. 2015. Histone demethylase JMJD3 is required for osteoblast differentiation in mice. Sci Rep 5:13418. doi: 10.1038/srep13418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hupkes M, van Someren EP, Middelkamp SH, Piek E, van Zoelen EJ, Dechering KJ. 2011. DNA methylation restricts spontaneous multi-lineage differentiation of mesenchymal progenitor cells, but is stable during growth factor-induced terminal differentiation. Biochim Biophys Acta 1813:839–849. doi: 10.1016/j.bbamcr.2011.01.022. [DOI] [PubMed] [Google Scholar]
- 23.Lee JY, Lee YM, Kim MJ, Choi JY, Park EK, Kim SY, Lee SP, Yang JS, Kim DS. 2006. Methylation of the mouse DIx5 and Osx gene promoters regulates cell type-specific gene expression. Mol Cells 22:182–188. [PubMed] [Google Scholar]
- 24.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. 2010. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. 2011. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 27.Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, Lahesmaa R, Orkin SH, Rodig SJ, Daley GQ, Rao A. 2011. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dawlaty MM, Breiling A, Le T, Barrasa MI, Raddatz G, Gao Q, Powell BE, Cheng AW, Faull KF, Lyko F, Jaenisch R. 2014. Loss of Tet enzymes compromises proper differentiation of embryonic stem cells. Dev Cell 29:102–111. doi: 10.1016/j.devcel.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, Weng Z, Rando OJ, Fazzio TG. 2011. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell 147:1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neri F, Incarnato D, Krepelova A, Rapelli S, Pagnani A, Zecchina R, Parlato C, Oliviero S. 2013. Genome-wide analysis identifies a functional association of Tet1 and Polycomb repressive complex 2 in mouse embryonic stem cells. Genome Biol 14:R91. doi: 10.1186/gb-2013-14-8-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. 2011. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshida M, Kijima M, Akita M, Beppu T. 1990. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem 265:17174–17179. [PubMed] [Google Scholar]
- 33.Woan KV, Sahakian E, Sotomayor EM, Seto E, Villagra A. 2012. Modulation of antigen-presenting cells by HDAC inhibitors: implications in autoimmunity and cancer. Immunol Cell Biol 90:55–65. doi: 10.1038/icb.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A III, Diaz E, LaFrance LV, Mellinger M, Duquenne C, Tian X, Kruger RG, McHugh CF, Brandt M, Miller WH, Dhanak D, Verma SK, Tummino PJ, Creasy CL. 2012. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 35.Kruidenier L, Chung CW, Cheng Z, Liddle J, Che K, Joberty G, Bantscheff M, Bountra C, Bridges A, Diallo H, Eberhard D, Hutchinson S, Jones E, Katso R, Leveridge M, Mander PK, Mosley J, Ramirez-Molina C, Rowland P, Schofield CJ, Sheppard RJ, Smith JE, Swales C, Tanner R, Thomas P, Tumber A, Drewes G, Oppermann U, Patel DJ, Lee K, Wilson DM. 2012. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature 488:404–408. doi: 10.1038/nature11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vardimon L, Kressmann A, Cedar H, Maechler M, Doerfler W. 1982. Expression of a cloned adenovirus gene is inhibited by in vitro methylation. Proc Natl Acad Sci U S A 79:1073–1077. doi: 10.1073/pnas.79.4.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grandy R, Sepulveda H, Aguilar R, Pihan P, Henriquez B, Olate J, Montecino M. 2011. The Ric-8B gene is highly expressed in proliferating preosteoblastic cells and downregulated during osteoblast differentiation in a SWI/SNF- and C/EBPbeta-mediated manner. Mol Cell Biol 31:2997–3008. doi: 10.1128/MCB.05096-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, Surani MA. 2013. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science 339:448–452. doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kohli RM, Zhang Y. 2013. TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502:472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee JY, Lee TH. 2012. Effects of histone acetylation and CpG methylation on the structure of nucleosomes. Biochim Biophys Acta 1824:974–982. doi: 10.1016/j.bbapap.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee JY, Lee J, Yue H, Lee TH. 2015. Dynamics of nucleosome assembly and effects of DNA methylation. J Biol Chem 290:4291–4303. doi: 10.1074/jbc.M114.619213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clapier CR, Cairns BR. 2009. The biology of chromatin remodeling complexes. Annu Rev Biochem 78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 43.ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. 2002. The human genome browser at UCSC. Genome Res 12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bartholomew B. 2014. Regulating the chromatin landscape: structural and mechanistic perspectives. Annu Rev Biochem 83:671–696. doi: 10.1146/annurev-biochem-051810-093157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Villagra A, Cruzat F, Carvallo L, Paredes R, Olate J, van Wijnen AJ, Stein GS, Lian JB, Stein JL, Imbalzano AN, Montecino M. 2006. Chromatin remodeling and transcriptional activity of the bone-specific osteocalcin gene require CCAAT/enhancer-binding protein beta-dependent recruitment of SWI/SNF activity. J Biol Chem 281:22695–22706. doi: 10.1074/jbc.M511640200. [DOI] [PubMed] [Google Scholar]
- 47.Cruzat F, Henriquez B, Villagra A, Hepp M, Lian JB, van Wijnen AJ, Stein JL, Imbalzano AN, Stein GS, Montecino M. 2009. SWI/SNF-independent nuclease hypersensitivity and an increased level of histone acetylation at the P1 promoter accompany active transcription of the bone master gene Runx2. Biochemistry 48:7287–7295. doi: 10.1021/bi9004792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de la Serna IL, Carlson KA, Imbalzano AN. 2001. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat Genet 27:187–190. doi: 10.1038/84826. [DOI] [PubMed] [Google Scholar]
- 49.Jones PA, Taylor SM. 1980. Cellular differentiation, cytidine analogs and DNA methylation. Cell 20:85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- 50.Monneret C. 2005. Histone deacetylase inhibitors. Eur J Med Chem 40:1–13. doi: 10.1016/j.ejmech.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 51.Lee HW, Suh JH, Kim AY, Lee YS, Park SY, Kim JB. 2006. Histone deacetylase 1-mediated histone modification regulates osteoblast differentiation. Mol Endocrinol 20:2432–2443. doi: 10.1210/me.2006-0061. [DOI] [PubMed] [Google Scholar]
- 52.Zhao H, Chen T. 2013. Tet family of 5-methylcytosine dioxygenases in mammalian development. J Hum Genet 58:421–427. doi: 10.1038/jhg.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee MH, Javed A, Kim HJ, Shin HI, Gutierrez S, Choi JY, Rosen V, Stein JL, van Wijnen AJ, Stein GS, Lian JB, Ryoo HM. 1999. Transient upregulation of CBFA1 in response to bone morphogenetic protein-2 and transforming growth factor beta1 in C2C12 myogenic cells coincides with suppression of the myogenic phenotype but is not sufficient for osteoblast differentiation. J Cell Biochem 73:114–125. [PubMed] [Google Scholar]
- 54.Katagiri T, Yamaguchi A, Komaki M, Abe E, Takahashi N, Ikeda T, Rosen V, Wozney JM, Fujisawa-Sehara A, Suda T. 1994. Bone morphogenetic protein-2 converts the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. J Cell Biol 127:1755–1766. doi: 10.1083/jcb.127.6.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hayashi M, Maeda S, Aburatani H, Kitamura K, Miyoshi H, Miyazono K, Imamura T. 2008. Pitx2 prevents osteoblastic transdifferentiation of myoblasts by bone morphogenetic proteins. J Biol Chem 283:565–571. doi: 10.1074/jbc.M708154200. [DOI] [PubMed] [Google Scholar]
- 56.Chu CH, Wang LY, Hsu KC, Chen CC, Cheng HH, Wang SM, Wu CM, Chen TJ, Li LT, Liu R, Hung CL, Yang JM, Kung HJ, Wang WC. 2014. KDM4B as a target for prostate cancer: structural analysis and selective inhibition by a novel inhibitor. J Med Chem 57:5975–5985. doi: 10.1021/jm500249n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deplus R, Delatte B, Schwinn MK, Defrance M, Mendez J, Murphy N, Dawson MA, Volkmar M, Putmans P, Calonne E, Shih AH, Levine RL, Bernard O, Mercher T, Solary E, Urh M, Daniels DL, Fuks F. 2013. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J 32:645–655. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Steward MM, Lee JS, O'Donovan A, Wyatt M, Bernstein BE, Shilatifard A. 2006. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat Struct Mol Biol 13:852–854. doi: 10.1038/nsmb1131. [DOI] [PubMed] [Google Scholar]
- 59.Shilatifard A. 2012. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem 81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Valinluck V, Sowers LC. 2007. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res 67:946–950. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- 61.Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, Zhang X, Cheng X. 2012. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res 40:4841–4849. doi: 10.1093/nar/gks155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ji D, Lin K, Song J, Wang Y. 2014. Effects of Tet-induced oxidation products of 5-methylcytosine on Dnmt1- and DNMT3a-mediated cytosine methylation. Mol Biosyst 10:1749–1752. doi: 10.1039/c4mb00150h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. 2011. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X, Dai Q, Song CX, Zhang K, He C, Xu GL. 2011. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maiti A, Drohat AC. 2011. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J Biol Chem 286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nabel CS, Jia H, Ye Y, Shen L, Goldschmidt HL, Stivers JT, Zhang Y, Kohli RM. 2012. AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation. Nat Chem Biol 8:751–758. doi: 10.1038/nchembio.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sepulveda H, Aguilar R, Prieto CP, Bustos F, Aedo S, Lattus J, van Zundert B, Palma V, Montecino M. 28 March 2017. Epigenetic signatures at the RUNX2-P1 and Sp7 gene promoters control osteogenic lineage commitment of umbilical cord-derived mesenchymal stem cells. J Cell Physiol doi: 10.1002/jcp.25627. [DOI] [PubMed] [Google Scholar]
- 68.Nguyen CT, Weisenberger DJ, Velicescu M, Gonzales FA, Lin JC, Liang G, Jones PA. 2002. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2′-deoxycytidine. Cancer Res 62:6456–6461. [PubMed] [Google Scholar]
- 69.Hashimoto H, Vertino PM, Cheng X. 2010. Molecular coupling of DNA methylation and histone methylation. Epigenomics 2:657–669. doi: 10.2217/epi.10.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kurimoto K, Yabuta Y, Hayashi K, Ohta H, Kiyonari H, Mitani T, Moritoki Y, Kohri K, Kimura H, Yamamoto T, Katou Y, Shirahige K, Saitou M. 2015. Quantitative dynamics of chromatin remodeling during germ cell specification from mouse embryonic stem cells. Cell Stem Cell 16:517–532. doi: 10.1016/j.stem.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 71.Sharif J, Endo TA, Nakayama M, Karimi MM, Shimada M, Katsuyama K, Goyal P, Brind'Amour J, Sun MA, Sun Z, Ishikura T, Mizutani-Koseki Y, Ohara O, Shinkai Y, Nakanishi M, Xie H, Lorincz MC, Koseki H. 2016. Activation of endogenous retroviruses in Dnmt1(−/−) ESCs involves disruption of SETDB1-mediated repression by NP95 binding to hemimethylated DNA. Cell Stem Cell 19:81–94. doi: 10.1016/j.stem.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 72.Zhang T, Termanis A, Ozkan B, Bao XX, Culley J, de Lima Alves F, Rappsilber J, Ramsahoye B, Stancheva I. 2016. G9a/GLP complex maintains imprinted DNA methylation in embryonic stem cells. Cell Rep 15:77–85. doi: 10.1016/j.celrep.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Kin Sung KW, Rigoutsos I, Loring J, Wei CL. 2010. Dynamic changes in the human methylome during differentiation. Genome Res 20:320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. 2007. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 75.Miller SA, Mohn SE, Weinmann AS. 2010. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol Cell 40:594–605. doi: 10.1016/j.molcel.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cano-Rodriguez D, Gjaltema RA, Jilderda LJ, Jellema P, Dokter-Fokkens J, Ruiters MH, Rots MG. 2016. Writing of H3K4Me3 overcomes epigenetic silencing in a sustained but context-dependent manner. Nat Commun 7:12284. doi: 10.1038/ncomms12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang P, Lin C, Smith ER, Guo H, Sanderson BW, Wu M, Gogol M, Alexander T, Seidel C, Wiedemann LM, Ge K, Krumlauf R, Shilatifard A. 2009. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol Cell Biol 29:6074–6085. doi: 10.1128/MCB.00924-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cheng J, Blum R, Bowman C, Hu D, Shilatifard A, Shen S, Dynlacht BD. 2014. A role for H3K4 monomethylation in gene repression and partitioning of chromatin readers. Mol Cell 53:979–992. doi: 10.1016/j.molcel.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nayak A, Viale-Bouroncle S, Morsczeck C, Muller S. 2014. The SUMO-specific isopeptidase SENP3 regulates MLL1/MLL2 methyltransferase complexes and controls osteogenic differentiation. Mol Cell 55:47–58. doi: 10.1016/j.molcel.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 81.Hu D, Gao X, Morgan MA, Herz HM, Smith ER, Shilatifard A. 2013. The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol Cell Biol 33:4745–4754. doi: 10.1128/MCB.01181-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Soutoglou E, Talianidis I. 2002. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science 295:1901–1904. doi: 10.1126/science.1068356. [DOI] [PubMed] [Google Scholar]
- 83.Date T, Doiguchi Y, Nobuta M, Shindo H. 2004. Bone morphogenetic protein-2 induces differentiation of multipotent C3H10T1/2 cells into osteoblasts, chondrocytes, and adipocytes in vivo and in vitro. J Orthop Sci 9:503–508. doi: 10.1007/s00776-004-0815-2. [DOI] [PubMed] [Google Scholar]
- 84.Mancini A, Sirabella D, Zhang W, Yamazaki H, Shirao T, Krauss RS. 2011. Regulation of myotube formation by the actin-binding factor drebrin. Skelet Muscle 1:36. doi: 10.1186/2044-5040-1-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang YP, Wang ZF, Zhang YC, Tian Q, Wang JZ. 2004. Effect of amyloid peptides on serum withdrawal-induced cell differentiation and cell viability. Cell Res 14:467–472. doi: 10.1038/sj.cr.7290249. [DOI] [PubMed] [Google Scholar]
- 86.Davidson BL, Harper SQ. 2005. Viral delivery of recombinant short hairpin RNAs. Methods Enzymol 392:145–173. doi: 10.1016/S0076-6879(04)92009-5. [DOI] [PubMed] [Google Scholar]
- 87.Rojas A, Aguilar R, Henriquez B, Lian JB, Stein JL, Stein GS, van Wijnen AJ, van Zundert B, Allende ML, Montecino M. 9 October 2015. Epigenetic control of the bone-master Runx2 gene during osteoblast-lineage commitment by the histone demethylase JARID1B/KDM5B. J Biol Chem doi: 10.1074/jbc.M115.657825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. 2005. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet 37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 89.Mohn F, Weber M, Schubeler D, Roloff TC. 2009. Methylated DNA immunoprecipitation (MeDIP). Methods Mol Biol 507:55–64. doi: 10.1007/978-1-59745-522-0_5. [DOI] [PubMed] [Google Scholar]
- 90.Zuo T, Tycko B, Liu TM, Lin JJ, Huang TH. 2009. Methods in DNA methylation profiling. Epigenomics 1:331–345. doi: 10.2217/epi.09.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Muller A, Florek M. 2010. 5-Azacytidine/azacitidine. Recent Results Cancer Res 184:159–170. doi: 10.1007/978-3-642-01222-8_11. [DOI] [PubMed] [Google Scholar]
- 92.Helin K, Dhanak D. 2013. Chromatin proteins and modifications as drug targets. Nature 502:480–488. doi: 10.1038/nature12751. [DOI] [PubMed] [Google Scholar]
- 93.Schreiber E, Harshman K, Kemler I, Malipiero U, Schaffner W, Fontana A. 1990. Astrocytes and glioblastoma cells express novel octamer-DNA binding proteins distinct from the ubiquitous Oct-1 and B cell type Oct-2 proteins. Nucleic Acids Res 18:5495–5503. doi: 10.1093/nar/18.18.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shechter D, Dormann HL, Allis CD, Hake SB. 2007. Extraction, purification and analysis of histones. Nat Protoc 2:1445–1457. doi: 10.1038/nprot.2007.202. [DOI] [PubMed] [Google Scholar]
- 95.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 96.Aguilar R, Grandy R, Meza D, Sepulveda H, Pihan P, van Wijnen AJ, Lian JB, Stein GS, Stein JL, Montecino M. 2014. A functional N-terminal domain in C/EBPbeta-LAP* is required for interacting with SWI/SNF and to repress Ric-8B gene transcription in osteoblasts. J Cell Physiol 229:1521–1528. doi: 10.1002/jcp.24595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.