In nonphotosynthetic glandular trichomes, terpenoid metabolism is powered by oxidative phosphorylation, fermentation and a unique ferredoxin/ferredoxin-NADP+ reductase pair.
Abstract
The commercially important essential oils of peppermint (Mentha × piperita) and its relatives in the mint family (Lamiaceae) are accumulated in specialized anatomical structures called glandular trichomes (GTs). A genome-scale stoichiometric model of secretory phase metabolism in peppermint GTs was constructed based on current biochemical and physiological knowledge. Fluxes through the network were predicted based on metabolomic and transcriptomic data. Using simulated reaction deletions, this model predicted that two processes, the regeneration of ATP and ferredoxin (in its reduced form), exert substantial control over flux toward monoterpenes. Follow-up biochemical assays with isolated GTs indicated that oxidative phosphorylation and ethanolic fermentation were active and that cooperation to provide ATP depended on the concentration of the carbon source. We also report that GTs with high flux toward monoterpenes express, at very high levels, genes coding for a unique pair of ferredoxin and ferredoxin-NADP+ reductase isoforms. This study provides, to our knowledge, the first evidence of how bioenergetic processes determine flux through monoterpene biosynthesis in GTs.
Plant secondary (or specialized) metabolites are frequently synthesized and accumulated in specialized tissues or cell types. Anatomical structures that harbor specialized metabolic pathways can take many forms and vary widely across the plant lineages. Some examples are resin ducts, secretory cavities, glandular trichomes (GTs), laticifers, and gum ducts (Fahn, 1988). GTs are particularly widespread across the angiosperm lineages and accumulate, among others, the specialized metabolites responsible for the psychoactive effects of cannabis (Cannabis sativa), the bitter flavor of hops (Humulus lupulus), the insect-repellant principles of tomato (Solanum lycopersicum) leaves, the antimalarial activity of sweet wormwood (Artemisia annua), and the essential oils of members of the Lamiaceae (Lange and Turner, 2013).
Peppermint (Mentha × piperita) and its close relatives have a long history as model systems for furthering our understanding of GT structure and function. Pioneering studies in the late 1980s and early 1990s reported on methods for purifying secretory cells of GTs away from other leaf cell types and established that the entire biosynthetic pathway from imported sugars to the monoterpene end products is confined to these specialized cell types (Gershenzon et al., 1992; McCaskill et al., 1992; McCaskill and Croteau, 1995). In the mid to late 1990s, these protocols were modified to allow the development of an EST library, the sequencing of which revealed the stunning metabolic specialization of mint GTs (Lange et al., 2000). These data sets also enabled the cloning and characterization of genes encoding the entire complement of enzymes responsible for p-menthane monoterpenoid biosynthesis in peppermint (Lange et al., 1998; Burke et al., 1999; Lange and Croteau, 1999a, 1999b; Ringer et al., 2003, 2005; Davis et al., 2005). Kinetic mathematical models facilitated the discovery of feedback control, GT developmental patterns, and epigenetic processes as critical determinants of metabolic outcomes (Rios-Estepa et al., 2008, 2010; Lange and Rios-Estepa, 2014; Ahkami et al., 2015). Protocols for the Agrobacterium tumefaciens-mediated transformation of mint species had been developed in the late 1990s (Berry et al., 1996; Caissard et al., 1996; Diemer et al., 1998; Niu et al., 1998) and were later employed, supported in part by mathematical modeling, to successfully modulate monoterpenoid essential composition and yield in transgenic peppermint plants (Mahmoud and Croteau, 2001; Mahmoud et al., 2004; Lange et al., 2011, 2013). The draft genome sequence and genetic map of Mentha longifolia, a diploid mint related to the ancestors of present-day peppermint and spearmint (Mentha × spicata; which are both polyploid), are resources that now also enable advanced breeding approaches for cultivar improvement (Vining et al., 2017).
The nonphotosynthetic GTs of peppermint (Werker et al., 1985), which fill up with monoterpenoid essential oil within only 1 d (Turner et al., 2000a, 2000b), sustain an exceptionally high flux through central carbon metabolism and the monoterpene biosynthetic pathway. Therefore, a better understanding of how flux is regulated in these cells will have a tremendous impact on the development of novel metabolic engineering and molecular breeding strategies toward mint cultivar improvement. One promising approach involves modeling metabolism more globally. Genome-scale stoichiometric models have been produced for various model plant systems, including leaf cells of Arabidopsis (Arabidopsis thaliana; Poolman et al., 2009; de Oliveira Dal’Molin et al., 2010a; Mintz-Oron et al., 2012; Cheung et al., 2013; Arnold and Nikoloski, 2014), maize (Zea mays; de Oliveira Dal’Molin et al., 2010b; Saha et al., 2011), rice (Oryza sativa; Poolman et al., 2013), and embryos of rapeseed (Brassica napus; Hay and Schwender, 2011). Building on these preexisting models, we developed a stoichiometric model reflecting metabolism in mint GTs by integrating newer transcriptome data sets with the wealth of knowledge on this subject in the published literature. We then employed a series of simulations, based on flux balance analysis (Fell and Small, 1986; Savinell and Palsson, 1992), to predict reactions with critical functions in monoterpene formation under extraordinarily high flux. Follow-up experimental testing revealed shared, carbon source-dependent roles for oxidative phosphorylation and ethanolic fermentation in ATP regeneration and the occurrence of a unique pair of ferredoxin (Fd) and ferredoxin-NADP+ reductase (FNR) in heterotrophic peppermint GTs.
RESULTS
Reconstruction of Metabolism in Peppermint GTs
To automate various processes relevant to building metabolic models, we developed a software package called YASMEnv. Because of space constraints, the capabilities cannot be outlined in the main body of this article, and extensive details are provided in Supplemental Methods and Data File S1. Further instructions and the entire code are available freely at https://bitbucket.org/seanrjohnson/yasmenv.
To assemble the reactions required for central carbon metabolism in peppermint GTs, we surveyed the literature for a particularly well-curated metabolic reconstruction and selected the Arabidopsis Core Model (Arnold and Nikoloski, 2014) to serve as the foundation for our efforts. Additional metabolic reactions were extracted from the AraCyc and MetaCyc databases (Mueller et al., 2003; Caspi et al., 2016). A process termed instantiation (Latendresse et al., 2012) was employed, using an implementation of the relevant algorithm in YASMEnv, to ensure that only mass-balanced reactions were included in the model. Each reaction was associated with a subcellular compartment based on the records in the Arabidopsis Core Model, AraCyc, and MetaCyc, and these associations were confirmed by searches against the TargetP and SUBAcon databases (Emanuelsson et al., 2000; Hooper et al., 2014). Sequences of transcripts represented in recently acquired transcriptome data sets for peppermint GTs (Ahkami et al., 2015) were compared, based on global identity (Myers and Miller, 1988; Tian and Skolnick, 2003), with those associated with reactions in the Arabidopsis Core Model, AraCyc, and MetaCyc. The first iteration of the menpiGT metabolic reconstruction consisted of all reactions of the Arabidopsis Core Model and reactions from AraCyc and MetaCyc with at least one associated peppermint transcript (Supplemental Methods and Data File S1).
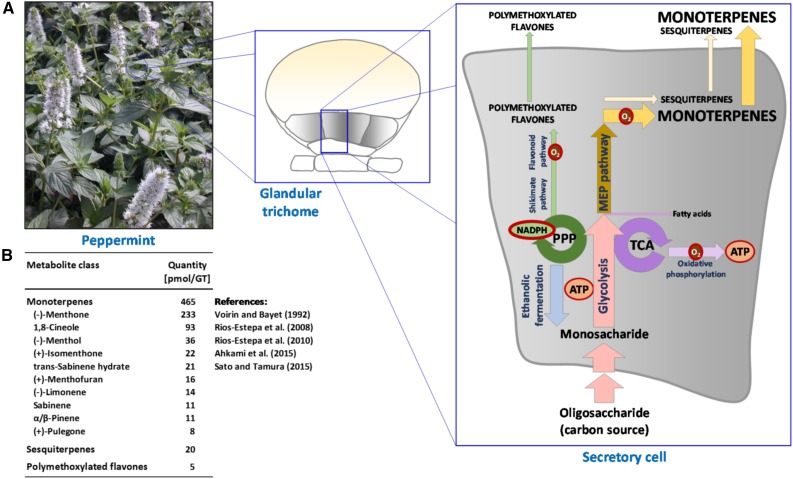
The model was refined in five additional stages: (1) completion of pathways to known GT metabolites, (2) removal of thermodynamically infeasible loops, (3) removal of reactions retained from the Arabidopsis Core Model that are not supported by peppermint transcripts, (4) removal of reactions that cannot carry flux because they are not connected to the remainder of the network, and (5) modifications to the model based on the experimental data presented in this article (Supplemental Methods and Data File S1). The final model (termed menpiGT_2015) contains 624 reactions, accounting for 757 expressed transcripts. The menpiGT_2015 model is a reflection of the astounding specialization of secretory cells within peppermint GTs during secretory phase (Fig. 1A). An imported oligosaccharide (raffinose or Suc) is converted, through central carbon metabolism and then dedicated pathways, primarily into monoterpenes (94%–95% [v/v] of essential oil), sesquiterpenes (4%), and polymethoxylated flavones (1%–2%; Fig. 1B), while genes involved in other pathways that commonly carry significant flux, such as amino acid conversion into proteins, sugar nucleotide interconversions into cell wall macromolecules, and fatty acid/lipid biosynthesis, are expressed only at fairly low levels (Supplemental Table S1).
Figure 1.
Reconstruction of metabolism in peppermint GTs. A, Leaf surfaces contain abundant GTs. Eight secretory cells (gray shading), arranged in a disc, are responsible for the biosynthesis of monoterpenoid essential oils, which are then secreted into a preformed cavity. The major pathways operating in peppermint GTs, based on transcriptomic and metabolomic data, are depicted. PPP, Pentose phosphate pathway; TCA, tricarboxylic acid cycle. B, Monoterpenes, sesquiterpenes, and polymethoxylated flavones are the most significant biomass outputs of GTs at secretory stage, and the values in this table were the basis for flux balance analysis, using the menpiGT_2015 metabolic reconstruction, to determine the distribution of carbon flow through the network of reactions.
Knockout Simulations Suggest Essential Roles for ATP Regeneration and Nonphotosynthetic Fd Reduction
To test the utility of menpiGT_2015 for identifying processes required for maintaining high flux toward monoterpenes, we employed a process called simulated reaction deletions (Edwards and Palsson, 2000). Reactions were removed from menpi_2015 one at a time, and the impact on monoterpene formation was evaluated (Supplemental Table S2). Thirty-five reactions were found to be essential for producing (−)-menthol from an imported carbon source (raffinose; Table I). The requirements for catabolism of raffinose (the primary carbon source considered in the model), the exchange of small molecules across membranes (balancing C and O budgets), and the reactions involved directly in the biosynthesis of precursors and end products of the monoterpene pathway were expected outcomes of these in silico predictions. Interestingly, the regeneration of nucleotide triphosphates (in particular CTP and ATP) and the recycling of reduced Fd also were returned as essential reactions in the simulated knockout study (Table I). These reactions are linked to the bioenergetics of GTs, which were poorly understood at the outset of our study; therefore, we designed experiments to further test how these processes may affect monoterpene formation.
Table I. Reactions returned as being essential for (−)-menthol production from raffinose, as determined by simulated reaction deletions.
| Reaction Description | Pathway | Compartment |
|---|---|---|
| 1-Deoxy-d-xylulose 5-phosphate synthase | Methylerythritol phosphate (MEP) pathway | Plastid |
| 1-Deoxy-d-xylulose 5-phosphate reductoisomerase | MEP pathway | Plastid |
| 4-Diphosphocytidyl-2C-methyl-d-erythritol synthase | MEP pathway | Plastid |
| 4-Diphosphocytidyl-2C-methyl-d-erythritol kinase | MEP pathway | Plastid |
| 2C-Methyl-d-erythritol 2,4-cyclodiphosphate synthase | MEP pathway | Plastid |
| 4-Hydroxy-3-methylbut-2-en-1-yl diphosphate synthase | MEP pathway | Plastid |
| 4-Hydroxy-3-methylbut-2-en-1-yl diphosphate reductase | MEP pathway | Plastid |
| Geranyl diphosphate synthase | Prenyl elongation | Plastid |
| (−)-Limonene 3-hydroxylase | Monoterpene modifications | Cytosol |
| (−)-trans-Isopiperitenol dehydrogenase | Monoterpene modifications | Mitochondrion |
| (−)-trans-Isopiperitenone reductase | Monoterpene modifications | Cytosol |
| (+)-cis-Isopulegone isomerase | Monoterpene modifications | Cytosol |
| (+)-Pulegone reductase | Monoterpene modifications | Cytosol |
| (−)-Menthone:(−)-menthol reductase | Monoterpene modifications | Cytosol |
| (−)-Limonene transport | Monoterpene modifications | Plastid to cytosol (ER) |
| (−)-trans-Isopiperitenol transport | Monoterpene modifications | Cytosol to mitochondrion |
| (−)-trans-Isopiperitenone transport | Monoterpene modifications | Mitochondrion to cytosol |
| (−)-Menthol export | Monoterpene modifications | Cytosol to oil cavity |
| Proton diffusion | O2/CO2/H2O/H diffusion | Mitochondrion to cytosol |
| CO2 diffusion | O2/CO2/H2O/H diffusion | Cytosol to plastid |
| Water diffusion | O2/CO2/H2O/H diffusion | Cytosol to mitochondrion |
| Oxygen diffusion | O2/CO2/H2O/H diffusion | Cytosol to mitochondrion |
| Oxygen exchange | O2/CO2/H2O/H diffusion | External to cytosol |
| CO2 exchange | O2/CO2/H2O/H diffusion | Cytosol to external |
| Raffinose import | Raffinose catabolism | External to cytosol |
| UTP:Gal-1-phosphate uridylyltransferase | Raffinose catabolism | Cytosol |
| UDP:Gal 4-epimerase | Raffinose catabolism | Cytosol |
| Raffinose galactosidase | Raffinose catabolism | Cytosol |
| Galactokinase | Raffinose catabolism | Cytosol |
| CDP kinase | CTP regeneration | Plastid |
| CMP kinase | CTP regeneration | Plastid |
| Inorganic phosphatase | CTP regeneration | Plastid |
| Respiratory complex I | Oxidative phosphorylation | Mitochondrion |
| ATP synthase | Oxidative phosphorylation | Mitochondrion |
| Fd-NADP+ reductase | Fd reduction | Plastid |
Sensitivity of Monoterpene Production to the Inhibition of Oxidative Phosphorylation Depends on the Concentration of Carbon Source
Peppermint GTs are nonphotosynthetic structures (Werker et al., 1985), and one important question pertains to the regeneration of ATP to power their highly active metabolism. In heterotrophic animal cells, oxidative phosphorylation in mitochondria has a critical role in supplying ATP; thus, one would hypothesize that this process also might provide energy equivalents in nonphotosynthetic GTs. Indeed, the genes coding for the enzyme complexes of the oxidative phosphorylation pathway were expressed at fairly high levels in peppermint GTs (Fig. 2A), indicating that this pathway of ATP regeneration is likely operational. Isolated GTs are capable of synthesizing monoterpenes from externally supplied sugar precursors (Gershenzon et al., 1992; McCaskill et al., 1992; McCaskill and Croteau, 1995) and, thus, would seem to be an excellent model system in which to study this process. However, previous feeding experiments employed complex media that contained various enzyme cofactors, including ATP; thus, it was important to test if ATP could be omitted from the incubation buffer. In experiments that evaluated various combinations of cofactors, we determined that the presence of NADP+ and ADP is sufficient to support monoterpene production in isolated peppermint GTs (Supplemental Methods and Data File S2).
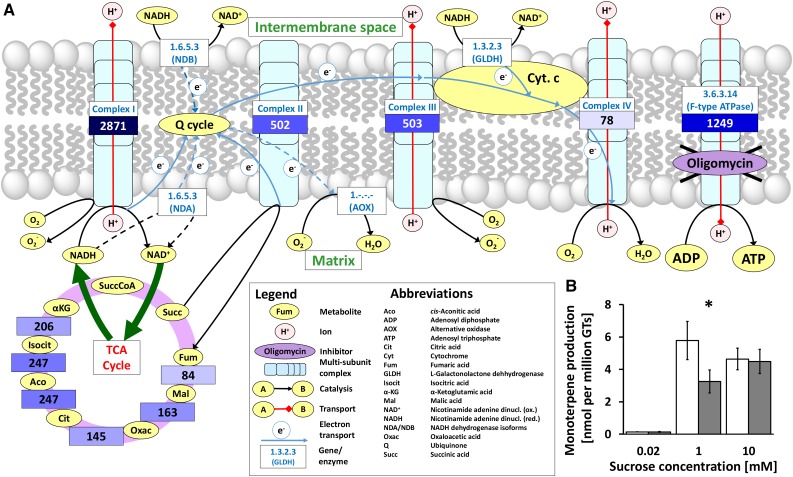
Figure 2.
Relevance of oxidative phosphorylation for monoterpene formation in peppermint GTs. A, Mapping of gene expression patterns (values are transcripts per kilobase million [TPM] in secretory phase GTs) related to enzyme complexes involved in oxidative phosphorylation. TCA, Tricarboxylic acid. B, Monoterpene production from Suc by isolated secretory cells. White and gray bars indicate values obtained with controls and oligomycin-treated cell clusters, respectively. Values are means ± sd (n = 3). The asterisk indicates P < 0.05 for the difference between control and treated samples, based on a two-tailed Student’s t test.
Two additional experimental considerations need to be mentioned. (1) An alternative pathway for regenerating ATP, independent of oxidative phosphorylation, would be through the activity of adenylate kinase (direct phosphorylation of proffered ADP), and to disable this potentially cooperative reaction for ATP formation, the incubation buffer was supplemented with AMP, a known inhibitor of the enzyme (Atkinson, 1968). (2) Although raffinose is the primary transport oligosaccharide in many Lamiaceae (and likely imported into GTs; Büchi et al., 1998; Olennikov and Tankhaeva, 2007), there is no commercial source for the radiolabeled tracer; therefore, we employed [U-14C]Suc in our labeling experiments (Suc is formed during raffinose catabolism).
Isolated GTs were incubated in media containing three different Suc concentrations and subjected to oligomycin (omitted in controls), which inhibits the ATP synthase component of the enzyme complexes performing oxidative phosphorylation (Huijing and Slater, 1961; Penefsky, 1985). At the end of the experimental period, radiolabeled monoterpenes were extracted into hexane:ether (1:1, v/v) and quantified by liquid scintillation counting. Based on data obtained in control experiments (no oligomycin), the maximum incorporation of label from [U-14C]Suc into monoterpenes occurs at 1 mm Suc (5.8 nmol per million gland cells), with a slight decrease at 10 mm (Fig. 2B). Oligomycin had no effect on monoterpene accumulation at 0.02 mm Suc, when the monoterpene yield (and likely the ATP demand) was very low. At 1 mm Suc, the addition of oligomycin resulted in a significant 2-fold decrease in the quantity of monoterpenes produced (from 5.8 to 3.2 nmol per million gland cells). Surprisingly, at 10 mm Suc (the highest concentration tested), oligomycin had no impact on monoterpene production (4.5 nmol per million gland cells). These results could not be explained with the reactions included in version 5 of menpiGT_2015 and indicated that alternative mechanisms for ATP regeneration, particularly at high sugar concentrations, needed to be considered.
One such possibility would be the coupling of ATP recycling to fermentative steps, such as those occurring in microbes and plants under oxygen-limiting conditions. To the best of our knowledge, fermentative reactions have not been included in any metabolic reconstruction for plants; therefore, we added the relevant reactions manually to menpiGT_2015 (version 6). In addition, transcriptome data obtained with isolated peppermint GTs at secretory stage (Ahkami et al., 2015) were incorporated using a modified SPOT algorithm (Kim et al., 2016; details are outlined in Supplemental Methods and Data File S1). Allowed carbon sources were oligosaccharides, ammonia, oxygen, hydrogen sulfide, sulfate, inorganic phosphate, water, and carbon dioxide. The experimentally derived biomass export reactions, as presented in Figure 1B, were disabled and, therefore, became values to be predicted by the model based on gene expression patterns. Allowed outputs were monoterpenes, sesquiterpenes, polymethoxylated flavones, ethanol, lactate, cellulose, amino acids, water, and carbon dioxide (Supplemental Table S3). Interestingly, our simulations predicted a significant formation of fermentative reaction products (lactate and ethanol; Fig. 3B); thus, we embarked on experiments, described in the following paragraph, to test the hypothesis that fermentation plays a role in energizing peppermint GTs.
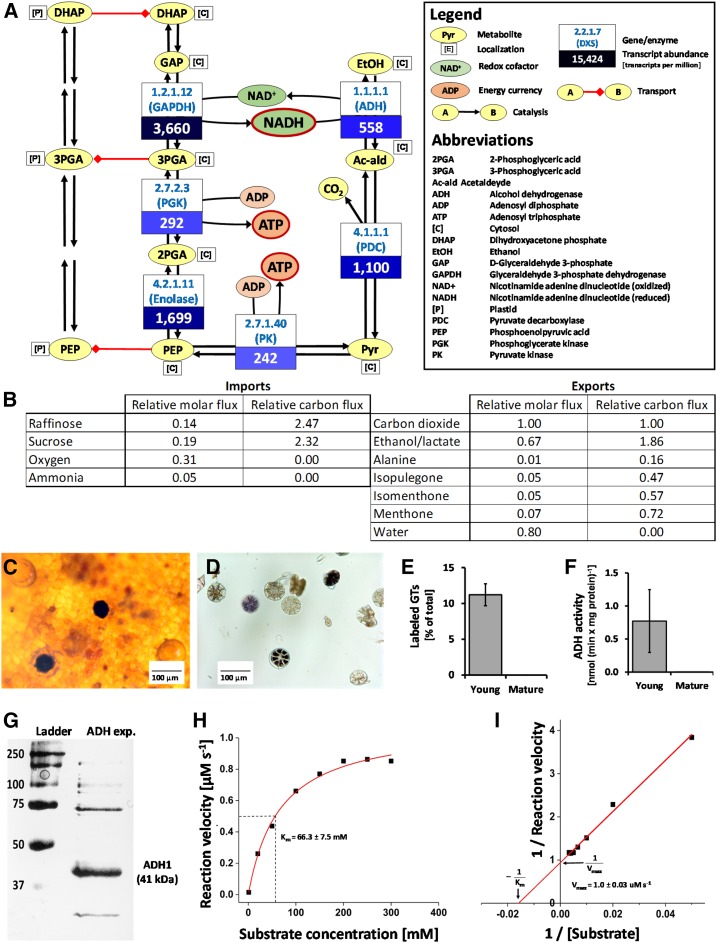
Figure 3.
Relevance of fermentation for monoterpene formation in peppermint GTs. A, Mapping of gene expression patterns (values are TPM in secretory phase GTs) in the context of coupling the NADH-to-NAD+ conversion catalyzed by ADH to the ATP-generating steps of glycolysis. B, The menpiGT_2015 model predicts fermentative reactions to carry significant flux during the secretory phase of GTs. C, ADH activity staining darkens GTs in secretory phase on intact leaves. D, ADH activity staining darkens isolated secretory cells. E, Percentage of secretory cells labeled by ADH activity staining correlates with the proportion of GTs at secretory phase. F, ADH activity in GTs isolated from young and mature leaves. Values are means ± sd (n = 3). G, Purification of recombinant peppermint ADH1 from E. coli. H, Michaelis-Menten plot for the dependence of reaction velocity of peppermint ADH1 on the substrate concentration. H, Lineweaver-Burk plot to determine the Km and Vmax values for peppermint ADH1 graphically.
Fermentation Is Active in Peppermint GTs
In fermentative processes, NADH and pyruvate formed in glycolysis are converted into NAD+ and an organic molecule (most often ethanol or lactic acid), which is coupled to reactions of the bottom half of glycolysis that regenerate ATP (Fig. 3A). Based on transcriptome data (Ahkami et al., 2015), genes involved in both glycolysis (glyceraldehyde 3-phosphate dehydrogenase, phosphoglycerate kinase, enolase, and pyruvate kinase) and fermentation (pyruvate decarboxylase and alcohol dehydrogenase [ADH]) are expressed at high (in some cases very high) levels in peppermint GTs (Fig. 3A). When isolated GTs were supplied with appropriate reactants, significant ADH activity (assayed with NAD+, NBT, and ethanol) was detected in about 12% of gland cell clusters (observed as dark staining; Fig. 3, C–E), which corresponds well with the proportion of GTs in secretory phase on young leaves (Turner et al., 2000a, 2000b). Enzyme assays performed with crude protein extracts indicated a fairly high specific ADH activity in GTs isolated from young leaves (0.8 nmol min−1 mg−1 protein) but no measurable activity in GTs obtained from mature leaves (where no monoterpene biosynthesis occurs; Fig. 3F). These data support the hypothesis that fermentation plays an important role in energizing nonphotosynthetic peppermint GTs and justify the inclusion of these reactions in version 6 of the menpiGT_2015 model.
Given that monoterpenol dehydrogenases are among the most highly expressed genes in secretory phase GTs (Lange et al., 2000; Ahkami et al., 2015), one may postulate that the ADH activity we observed is a nonspecific side activity of these enzymes. To test this hypotheses, we first performed a BLASTx search with the well-characterized ADH1 sequence from potato (Solanum tuberosum; Matton et al., 1990) against the peppermint GT transcriptome data set. This search indicated the presence of two putative orthologs (at roughly 88% identity for the translated peptide sequence) that were expressed at high levels in peppermint GTs (accession nos. MF579445 and MF579446; Supplemental Fig. S1). The cDNAs corresponding to the peppermint ADH candidates were cloned by PCR with gene-specific primers and ligated into an appropriate expression vector. Each vector carrying a peppermint candidate cDNA was transformed into Escherichia coli, the expression of the transgene was induced, and the recombinant protein was purified. The sizes of the proteins encoded by MF579445 and MF579446 were both roughly 41 kD (Fig. 3G). One ADH-like candidate (recombinant protein produced from MF579445) turned out to be inactive in our assays, while the other (produced from MF579446; henceforth referred to as ADH1) had the properties of a typical ADH [Km(ethanol) = 53.9 mm, kcat = 603.5 s−1; Fig. 3, H and I]. These results confirm the presence of a specific ADH activity, which is independent of monoterpenol dehydrogenases, in nonphotosynthetic peppermint GTs.
Mint GTs Contain a Unique Fd/FNR Complex
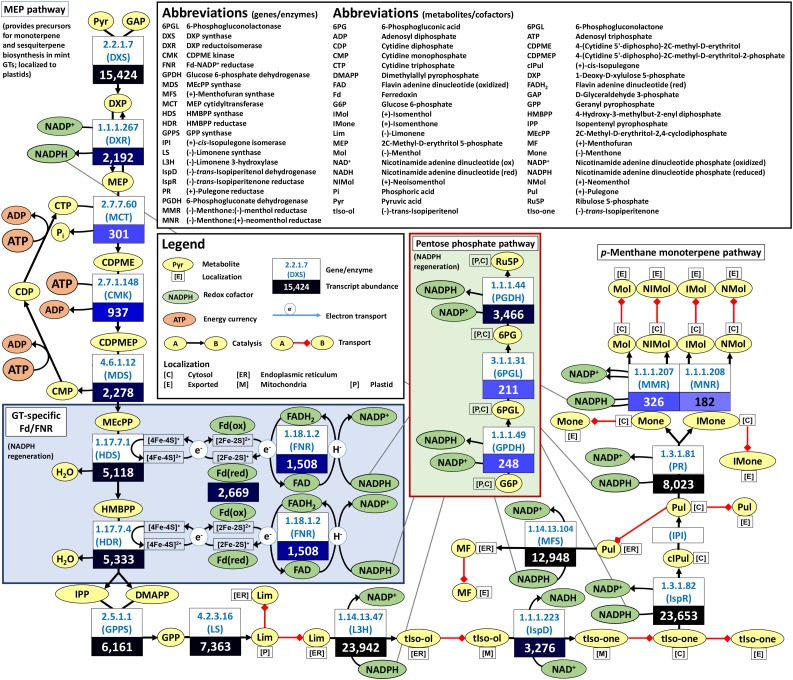
The precursors for monoterpene and sesquiterpene biosynthesis in peppermint GTs are provided exclusively by the MEP pathway (McCaskill and Croteau, 1995; Eisenreich et al., 1997; Lange et al., 2001). The genes involved in the MEP pathway and downstream monoterpene-specific reactions are expressed at very high levels in peppermint GTs (Fig. 4). The electron transfer reactions of reductive steps of the MEP pathway, catalyzed by 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate synthase (HDS) and 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate reductase (HDR), employ Fd and FNR (Seemann et al., 2006; Rekittke et al., 2013). This cofactor requirement is in accordance with our modeling prediction that Fd recycling is essential for monoterpene biosynthesis (Table I). The nonphotosynthetic isoforms of Fd and FNR have redox potentials that are significantly different from those in photosynthetic cells (Aliverti et al., 2001); thus, electrons can flow in a different direction from NADPH to FNR, then from FNR to Fd, and finally from Fd to the iron-sulfur clusters of HDS and HDR (Fig. 4). Thus far, nonphotosynthetic Fd and FNR isoforms have only been characterized from roots (Hanke et al., 2005; Aliverti et al., 2008; Onda et al., 2000); therefore, it was of interest to evaluate which isoforms are present in nonphotosynthetic GTs.
Figure 4.
Relevance of Fd and FNR for reductive steps in the biosynthesis of monoterpene precursors in peppermint GTs (values are TPM in secretory phase GTs).
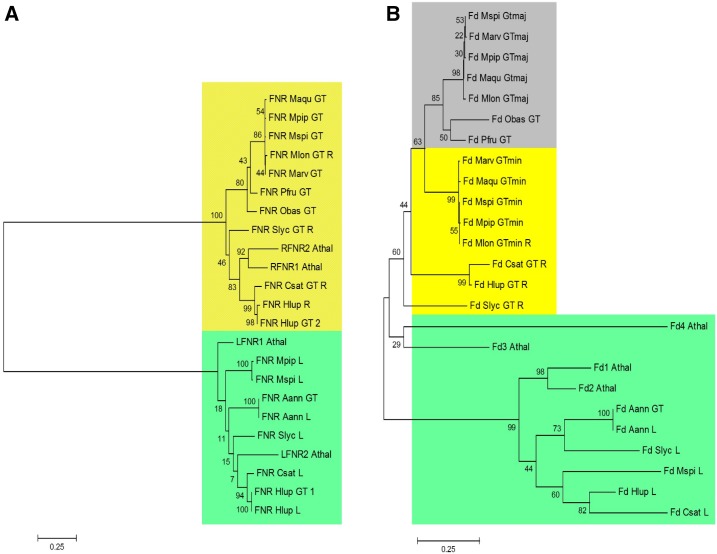
Public databases were surveyed for data sets covering leaf, root, and GT transcriptomes of angiosperms. Phylogenetic trees were then generated based on alignments of nucleotide sequences of Fd and FNR isoforms (Supplemental Table S4). FNRs expressed in GTs (in some cases more than one isoform) separated into two different clades: those expressed in nonphotosynthetic GTs (cannabis, peppermint, and basil [Ocimum basilicum]) were identical to the root isoform or clustered with root isoforms, whereas those expressed in photosynthetic GTs (tomato and sweet wormwood) clustered with leaf isoforms (Fig. 5A). Generally speaking, similar patterns were observed with GT-expressed isoforms of Fd (separation of leaf from root/GT isoforms). However, there was one significant difference: the major isoforms expressed in nonphotosynthetic GTs (particularly in the mint family) formed a subclade that was distinguishable from that of root isoforms (Fig. 5B).
Figure 5.
A unique Fd/FNR pair occurs in monoterpene-accumulating GTs. A, Maximum likelihood tree of FNR isoforms. B, Maximum likelihood tree of Fd isoforms. Comparisons employ nucleotide sequences corresponding to the mature proteins (excluding plastidial targeting sequences). Scale bars represent the number of substitutions per site. Species abbreviations are as follows: Aann (Artemisia annua), Athal (Arabidopsis thaliana), Csat (Cannabis sativa), Hlup (Humulus lupulus), Maqu (Mentha acquatica), Marv (Mentha arvensis), Mlon (Mentha longifolia), Mpip (Mentha × piperita), Mspi (Mentha × spicata), Obas (Ocimum basilicum), Pfru (Perilla frutescens), Slyc (Solanum lycopersicum). Other abbreviations are as follows: L, leaf isoform; maj, major GT isoform; min, minor GT isoform; R, root isoform. Colors indicate isoforms present in leaves (green), roots (gray), GTs (yellow), and both roots and GTs (yellow/gray).
Did GT-specific Fd isoforms evolve in mint? The hybrid hexaploid nature of peppermint makes it difficult to answer this question due to the potential for the presence of many Fd isoforms in genomes retained from three different ancestral species. However, the recently completed draft genome assembly for M. longifolia, a diploid species related to one of the ancestors of present-day peppermint (Vining et al., 2017), allowed us to examine all Fd isoforms. Organ- and tissue-specific transcriptome data for M. longifolia were recently deposited in public archives (root and GT transcriptomes with National Center for Biotechnology Information [NCBI] short read archive accession nos. SRP083887 and SRR3623199, respectively). The sequence of the minor Fd isoform of M. longifolia GTs (average expression of 289 TPM) was identical to that of the root isoform, whereas the major isoform (average expression of 2,586 TPM) was distinct from the root isoform (Fig. 5B). The major and minor isoforms from genus Mentha GTs differed in the mature peptide sequence at 12 positions (Fig. 6A). To ensure that these findings were not due to assembly errors from short reads, we PCR amplified and sequenced cDNAs corresponding to both Fd isoforms (KY748231 [GT major] and KY748232 [GT minor/root]) and thereby confirmed the unique sequence of the major GT isoform. The major peppermint GT-specific isoform of Fd was essentially identical to the major isoform of other mint species, and the same sequence conservation across species was observed for the minor GT/root isoform (Fig. 5B). Mapping of the Fd isoforms to the M. longifolia genome revealed that they map to different contigs: the major Fd isoform of GTs was mapped to contig 19,491 (refers to genome assembly with National Center for Biotechnology Information BioProject accession no. PRLNA310613), whereas the minor GT isoform mapped to contig 5,531, indicating that these genes likely have separate origins.
Figure 6.
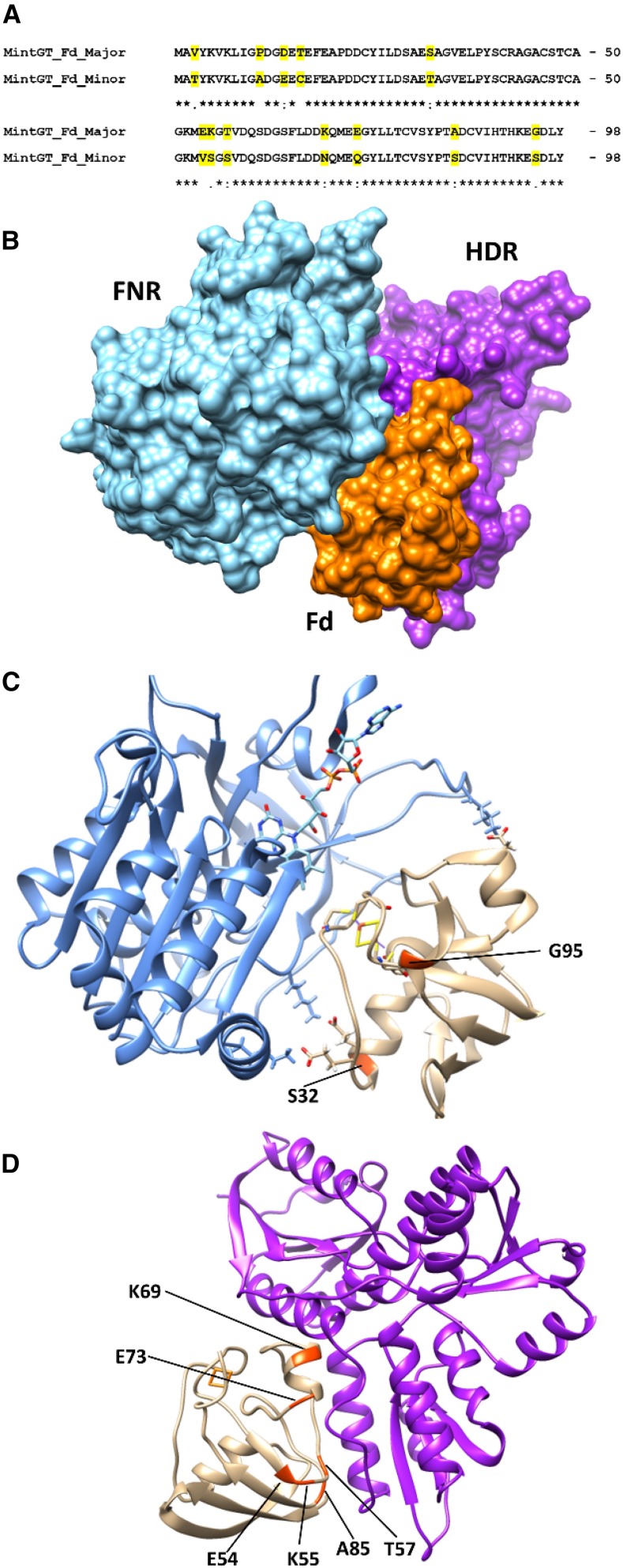
Protein-protein interactions in the ternary FNR-Fd-HDR complex of peppermint GTs. A, Alignment of mature peptide sequences of Fd isoforms present in peppermint GTs. Residues that differ between the major and minor isoform are highlighted in yellow. B, Docking conformation of the FNR-Fd-HDR ternary complex. C, Ribbon diagram of the FNR-Fd interaction. Residues that differ between the major and minor Fd isoforms of GTs are highlighted in red. D, Ribbon diagram of the Fd-HNR interaction. Residues that differ between the major and minor Fd isoforms of GTs are highlighted in red.
We also obtained transcriptome data for GTs isolated from Perilla frutescens (short read archive accession no. SRR4106089) and included previously published GT-specific cDNA sequences from basil (Gang et al., 2001) in our phylogenetic analysis, which expands the scope of sequence comparisons to other Lamiaceae with high monoterpenoid essential oil content. The GT-specific Fd isoforms of these species clustered with the major isoform of mint species, indicating that nonphotosynthetic GTs with high flux toward monoterpenes may harbor unique Fd isoforms across the mint family.
To assess the potential implications of a GT-specific Fd isoform in peppermint, we modeled the structures of Fd (GT major isoform), FNR (GT/root isoform), and HDR based on published data for closely related isoforms from plants (Fig. 6). Because of the more speculative nature of this analysis, we are providing the details in “Discussion.”
DISCUSSION
menpiGT_2015 Is the First Genome-Scale Reconstruction of Specialized Metabolism in GTs
Large-scale mathematical models of plant metabolism were initially focused on species for which genome sequences were available (Arabidopsis, rice, maize, and other model organisms), but with the advent of cost-competitive technologies for obtaining transcriptome sequences, metabolic reconstructions coupled with flux balance analysis also have been performed for nonmodel plants (Hay and Schwender, 2011). Most of these models were generated to represent reactions in a major organ, and only a few accurately reflect metabolism specific to a certain tissue (de Oliveira Dal’Molin et al., 2010b; Hay and Schwender, 2011; Pilalis et al., 2011; Mintz-Oron et al., 2012). To our knowledge, the menpiGT_2015 model is the first attempt to gather data from various sources (literature, transcriptomics, metabolomics, and biochemical assays) and capture metabolism in nonphotosynthetic GTs. We decided to start with a well-curated, compartmentalized, metabolic reconstruction recently generated for a model plant (Arabidopsis Core Model; Arnold and Nikoloski, 2014) and then incorporated additional information from Web-based databases focusing on metabolism (AraCyc and MetaCyc) to produce a consensus set of reactions. The YASMEnv toolbox, which we developed specifically to automate various steps in the generation of metabolic models, was invaluable in these efforts, and we hope that others will take advantage of the freely available scripts (https://bitbucket.org/seanrjohnson/yasmenv). Further model development steps included eliminating thermodynamically infeasible loops, ensuring that all reactions are mass balanced, and associating reactions with transcripts (adding reactions unique to mint GTs and eliminating reactions not supported by mint transcripts). These adjustments reduced the total number of reactions in mepiGT_2015 to 624 (from an initial 2,639 reactions; Supplemental Methods and Data File S1) and are reflective of the remarkable specialization of mint GTs.
Since mint GT cells are nonphotosynthetic, an imported carbon source (likely raffinose; McCaskill et al., 1992; Büchi et al., 1998; Olennikov and Tankhaeva, 2007) is converted, via central carbon metabolism, to precursors for terpenoid biosynthesis (which correspond to more than 95% of products accumulated in essential oil). Importantly, precursors for both C10-derived monoterpenes and C15-derived sesquiterpenes are provided exclusively by the plastidial MEP pathway (McCaskill and Croteau, 1995; Eisenreich et al., 1997; Lange et al., 2001), which is reflected in the extremely high expression levels of the relevant genes (whereas expression levels of genes of the cytosolic/peroxisomal mevalonate pathway are low or undetectable; Supplemental Table S1). The pathway toward methoxylated flavones is clearly active at the transcriptional level, with biosynthetic genes being expressed at appreciable levels (Supplemental Table S1), but the quantities of these specialized metabolites in mint GTs are generally very low (1%–2% of the essential oil, with no significant occurrence in other cell types; Voirin and Bayet, 1992; Sato and Tamura, 2015).
Transcripts related to cellular maintenance reactions (e.g. protein synthesis and degradation, nucleotide biosynthesis, and fatty acid/lipid biosynthesis) are detectable in published mint data sets (Ahkami et al., 2015), but their abundance is very low compared with those involved in terpenoid and polymethoxylated flavone biosynthesis. Some genes involved in nucleotide interconversion reactions are expressed at high levels (e.g. adenosine kinase at 2,122 TPM and adenylate kinase at 788 TPM), but these are linked to the regeneration of ATP, which is needed to support the highly active biosynthetic pathways of GTs (the MEP pathway alone requires six ATP molecules to generate one molecule of geranyl diphosphate [a monoterpene precursor]). The conversions from 1-deoxy-d-xylulose 5-phosphate to one molecule of (−)-menthol require 10 molecules of NADPH, and the high expression levels of genes involved in the oxidative branch of the pentose phosphate pathway (coding for Glc-6-P dehydrogenase, 6-phosphogluconolactonase, and 6-phosphogluconate dehydrogenase; Fig. 4), which support NADPH regeneration (Kruger and von Schaewen, 2003), are in accordance with the expected demands. In summary, menpiGT_2015 captures the specialization of mint GTs at secretory stage and allows us to test initial assumptions about metabolism computationally, which can then be assessed further experimentally.
Iterative Mathematical Modeling and Experimental Testing Provide Insights into the Processes That Power Metabolism in Nonphotosynthetic GTs
Reaction knockout simulations with menpiGT_2015 (version 5) were performed to determine which reactions are essential for (−)-menthol production (Table I; Supplemental Table S2). Surprisingly, of the seven pathways or processes identified by this simulation as being essential, only two, the MEP pathway and the monoterpene biosynthetic pathway, were strongly supported by prior experimental evidence (Lange, 2015). Three other essential processes, Suc/raffinose import and metabolism (carbon source), CTP regeneration (required to provide CTP as reactant in the MEP pathway), and exchange of small molecules across membranes (trivial), did not appear to warrant further exploration at this time. The remaining two essential processes, oxidative phosphorylation and regeneration of reduced Fd, had not been studied at all in GTs and, therefore, were deemed worthy of experimental testing.
Feeding experiments with isolated GTs were employed to assess the contribution of oxidative phosphorylation to the formation of organic soluble products (primarily monoterpenes) from 14C-labeled Suc. One of the surprises in our experimental data was the finding that monoterpene production in the presence of the ATP synthase inhibitor oligomycin was dependent on the concentration of Suc. We observed inhibitory effects at 1 mm Suc but not at 0.02 or 10 mm Suc (Fig. 3). The lack of inhibition at 0.02 mm Suc may be due to a low overall metabolic rate (measured as monoterpene production per unit of time) at such a low Suc concentration. At 1 mm Suc, oligomycin treatment reduced monoterpene production to about half of that observed in the absence of oligomycin, and one might interpret this as incomplete inhibition of ATP synthase. However, this would not explain why there was no effect of the oligomycin treatment on monoterpene accumulation at 10 mm Suc. An alternative explanation was the involvement of processes other than oxidative phosphorylation in regenerating ATP. Adenylate kinase was inhibited by the addition of AMP in our assays (Atkinson, 1968) and, therefore, was unlikely to be a significant contributor to ATP formation, leaving fermentation as the only remaining candidate for providing energy equivalents. After adding fermentative reactions to our model and incorporating gene expression patterns acquired previously (Ahkami et al., 2015), simulations predicted that fermentation products (lactate and ethanol) should indeed accumulate in peppermint GTs. Follow-up biochemical experiments provided evidence that peppermint GTs contain highly active ADH1, the signature enzyme of fermentation (Fig. 3), thereby confirming the model predictions. It is important to note that this enzyme is clearly distinct from monoterpenol dehydrogenases, which were demonstrated previously to not possess an ADH side activity (Kjonaas et al., 1985; Gershenzon et al., 1989).
The next question was if the consideration of fermentation could explain the data obtained in the experiments in which oxidative phosphorylation was inhibited. More specifically, why did we observe a significant reduction of monoterpene production in isolated GTs treated with oligomycin at 1 mm, but not at 10 mm, Suc concentration? In all plant ADH enzymes characterized thus far, the Km value is always in the millimolar range (Mayne and Lea, 1984; Tong and Lin, 1988; Osterman et al., 1993; Hadizadeh and Keyhani, 2004). We calculated a Km of 66.3 mm for peppermint ADH1, which is consistent with these earlier studies. Therefore, we hypothesize that acetaldehyde does not reach an intracellular concentration that is sufficiently high to drive ADH activity toward ethanolic fermentation at lower Suc concentrations. Under those conditions, ATP regeneration does not rely on fermentation and is dependent primarily on oxidative phosphorylation (thus, the effect of the ATP synthase inhibitor on monoterpene production at 1 mm Suc). The situation would be different at high Suc concentrations, when intracellular levels of acetaldehyde can be high enough for ADH to operate in the direction of ethanol production, which complements or maybe even substitutes for oxidative phosphorylation (thus, no effect of the ATP synthase inhibitor on monoterpene production at 10 mm Suc). Based on measurements in various plants, the intracellular concentration of oligosaccharides can be up to 100 mm (Nadwodnik and Lohaus, 2008), indicating that the concentration of acetaldehyde in mint GTs could be sufficiently high to support ethanolic fermentation.
Peppermint GTs synthesize oxygenated monoterpenes at high flux (each individual gland fills within about 24 h after reaching secretory stage; Turner et al., 2000a, 2000b), which places significant demands on the availability of molecular oxygen for biosynthetic cytochrome P450-dependent monooxygenases (Lupien et al., 1999; Bertea et al., 2001). Mint GTs are covered by a thick waxy cuticle (Turner et al., 2000a, 2000b), which may prohibit a direct diffusion of atmospheric oxygen into the cells responsible for monoterpenoid essential oil biosynthesis. Together, these factors may contribute to oxygen-limiting conditions in GTs, which would favor fermentation as a mechanism for ATP regeneration (Fig. 4).
Occurrence of GT-Specific Fd/FNR Isoforms in Monoterpene-Accumulating Plants
The regeneration of reduced Fd was identified in this study as an essential process by computational knockout simulations (Table I). This is a logical consequence of the fact that Fd is a cofactor for electron transfer reactions toward two reductive enzymes of the MEP pathway (Seemann et al., 2006; Rekittke et al., 2013) and flavone hydroxylases (Berim et al., 2014), which are involved in the formation of the major products of mint GTs (monoterpenes and methoxylated flavones, respectively). In earlier versions of menpiGT_2015, Fd served as a substrate for FNR, which is the direction of electron flow in photosynthetic cells. With the removal of PSI from the model, due to the lack of support from gene expression data, the direction of FNR had to be reversed in order to maintain the capacity for monoterpene production, which is the direction of electron flow suggested to occur in nonphotosynthetic root cells (Aliverti et al., 2001).
A phylogenetic analysis indicated that the most highly expressed FNR isoform of mint GTs clustered with the nonphotosynthetic isoforms of other plants, which was an expected outcome. Surprisingly, however, two different Fd isoforms were found to be expressed in GTs of the genus Mentha. The minor isoform (lower expression levels) of M. piperita and M. longifolia was identical to the isoform most highly expressed in roots of the same species. The major isoform (10-fold higher expression than the minor isoform) was unique to GTs. The major isoform also was conserved across several members of the Lamiaceae. This begs the questions of whether there is a functional consequence to this sequence diversity. One possibility would be that the interaction between the major Fd isoform of GTs and reductive enzymes of the MEP pathway, HDS and HDR, facilitates high flux through these steps and the remainder of the monoterpene biosynthetic pathway.
What are the potential implications of a novel Fd isoform specifically expressed in nonphotosynthetic GTs of the genus Mentha (and possibly across the Lamiaceae)? Based on a molecular model of ternary peppermint FNR-Fd-HDR complexes, we asked if there were differences among the sequences of peppermint Fd isoforms (Fig. 6A) that had the potential to affect the interaction(s) with FNR and/or HDR (Fig. 6B). Prior crystal structure data of the electron transfer complex between Fd and FNR provided evidence for a close proximity of the [2Fe-2S] cluster of Fd and the FAD prosthetic group of FNR (Kurisu et al., 2001). The authors suggested that pairs of charged residues form intermolecular salt bridges, which was in accordance with prior biochemical data (for review, see the context of plant Fd-FNR interactions by Aliverti et al., 2008). Two residues implicated previously in the interaction of Fd and FNR were different in peppermint isoforms (Shinohara et al., 2017): Ser-32 and Gly-95 in the GT major isoform corresponded to Thr-32 and Ser-95, respectively, in the minor isoform (numbering based on the mature polypeptide, excluding the plastidial targeting sequence, of the major isoform of peppermint; Fig. 6C).
Crystal structures have been resolved for both HDS and HDR, but only one study modeled the binding with Fd. Rekittke et al. (2013) employed docking calculations to select the energetically most favorable complexes between HDR and the cognate Fd from Plasmodium falciparum. The authors concluded that Fd was most likely to attach to a shallow well formed by the N terminus, helix α1B, and the loops near strands β4A and β4B. Our FNR-Fd-HDR docking simulations indicated that one of the lowest energy ternary complexes had a molecular surface that was very similar to that suggested previously (Rekittke et al., 2013). Interestingly, six residues purportedly involved in the Fd-HDR interaction differed between the major and minor Fd isoforms of peppermint GTs (Glu-54, Lys-55, Thr-57, Lys-69, Glu-73, and Ala-85 in the major isoform corresponded to Val-54, Ser-55, Ser-57, Asn-69, Gln-73, and Ser-85 in the minor isoform, respectively; Fig. 6D).
We hypothesize that the differences between the major and minor Fd isoforms in GTs may affect the stability of the FNR-Fd-HDR complex, which, in turn, may impact flux through the biosynthetic step catalyzed by HDR. The situation would be different in photosynthetic GTs characterized thus far. In type VI GTs of Solanum habrochaites, the accumulation of terpenoids occurs over a longer period of time, while in peppermint GT, filling concludes within a matter of days (Turner et al., 2000a, 2000b). Terpenoids in S. habrochaites GTs are derived from both the mevalonate and MEP pathways (Bergau et al., 2015; Balcke et al., 2017), whereas in peppermint, terpenoids of GTs are produced solely via the MEP pathway and, therefore, the reactions catalyzed by HDS and HDR (McCaskill and Croteau, 1995; Eisenreich et al., 1997; Lange et al., 2001). It is tempting to speculate that the major GT-specific Fd isoform of peppermint (and possibly other Lamiaceae) might have evolved to facilitate the particularly high flux through the reactions catalyzed by HDS and HDR en route to monoterpenoid end products.
MATERIALS AND METHODS
Simulations Using the menpiGT_2015 Model
The step-wise development of a metabolic reconstruction and its data-driven iterations are described in Supplemental Methods and Data File S1. To simulate gene knockouts, upper and lower bounds were set such that CO2 and raffinose were the only permitted carbon sources in the network, oxygen uptake was permitted, but NH4 import was disabled. The upper and lower flux bounds for menthol export were set to 1, and the objective function was set to minimize raffinose import. A reference minimum for raffinose import was calculated with no internal reactions disabled. Upper and lower flux bounds for each reaction were then set to 0, one at a time, and raffinose import was computed.
The SPOT algorithm was implemented according to Kim et al. (2016), with the addition of Boolean constraints to prevent reversible reactions from being counted twice in the objective function (referred to as Boolean constrained SPOT). Prior to running Boolean constrained SPOT, boundary fluxes were modified to disable the experimentally derived biomass export reaction and to allow the optional import and export of small molecules. Allowed carbon sources were Suc, Glc, Fru, or raffinose. Additional allowed imports were ammonia, oxygen, hydrogen sulfide, sulfate, inorganic phosphate, water, and carbon dioxide. Allowed exports were monoterpenes, sesquiterpenes, polymethoxylated flavones, ethanol, lactate, cellulose, amino acids, water, and carbon dioxide. Optimization problems were solved with Gurobi Optimizer (Gurobi Optimization), accessed through YASMEnv, a modeling tool we developed specifically for this study (Supplemental Methods and Data File S1).
Chemicals and Biochemicals
Chemicals were purchased from Sigma-Aldrich, unless noted otherwise. Biochemical reagents were sourced from Thermo Fisher Scientific, unless noted otherwise. [U-14C]Suc (432 mCi mol−1 in 90% aqueous ethanol) was obtained from MP Biomedicals.
GT Isolation
GTs were isolated based on the protocol developed by Gershenzon et al. (1992), with modifications. Roughly 20 g of leaves was harvested into a plastic bag on ice. Depending on the purpose of the experiments, young leaves (less than half expanded) and/or mature leaves (fully expanded) were harvested when the first flower buds emerged. Leaves were weighed and transferred into a bead beater chamber containing 100 g of glass beads (0.5 mm diameter) and 300 mL of isolation buffer (250 mm sorbitol, 10 mm KCl, 2 mm MgCl2, 25 mm 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid, 1% [w/v] polyvinylpyrrolidone [average mass of 40,000 g mol−1], and 0.6% [w/v] methyl cellulose; adjusted to pH 7.4). Bead beating was performed as three 1-min pulses, with 1 min of cooling on ice between pulses. The contents of the bead beater chamber were poured onto a nylon mesh (350 µm) and rinsed with wash buffer (200 mm sorbitol, 50 mm KCl, 25 mm 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid, 5 mm MgCl, 1 mm ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid, 0.5 mm Na2HPO4, and 0.1 mm NaP2O7; adjusted to pH 7.4). These wash steps were repeated with nylon mesh of two sizes (first 105 µm, then 20 µm). Secretory cells of GTs were retained on 20-µm mesh and carefully transferred into a tube containing 1 mL of wash buffer. To remove residual monoterpenes, the mixture was centrifuged for 5 min at 2,000g (at 4°C), secretory cells were resuspended in 1 mL of wash buffer, and cell clusters were counted using a hemacytometer.
Conversion of Radiolabeled Suc to Monoterpenes by Isolated Secretory Cells
Feeding experiments were performed based on previously developed protocols (McCaskill et al., 1992; McCaskill and Croteau, 1995), with modifications. In a 2-mL screw-cap glass vial, about 80,000 isolated secretory cell octets were suspended in reaction buffer containing [U-14C]Suc (0.5 µCi per assay), unlabeled Suc (0.02, 1, or 10 mm), 2 mm AMP (to inhibit adenylate kinase), 0.5 mm ADP, and 0.1 mm NADP+, which was then overlaid with 500 µL of hexane:ether (1:1, v/v) and sealed with a Teflon-lined cap. The mixture was incubated for 4 h at 23°C on an orbital shaker set to 200 rpm. Combinations of other cofactors and inhibitors were tested as described in Supplemental Methods and Data File S2. At the end of the experimental period, vials were vortexed (30 s at highest setting) and centrifuged for 2 min at 20,000g. A 250-µL aliquot of the organic phase was transferred to 3 mL of EcoScint (National Diagnostics) and counted with a Tri-Carb 2100 TR scintillation counter (PerkinElmer).
ADH Activity Assays
Intact GTs on leaves and isolated secretory cells were stained for ADH activity by a method originally developed for viability staining of pollen grains (Freeling, 1976; Stinson and Mascarenhas, 1985). For whole-leaf staining, leaves were harvested, submerged in phosphate buffer (100 mm, pH 7.3), and maintained at −20°C for 3 h. Leaves were allowed to thaw at room temperature and submerged in staining solution containing sodium phosphate (90 mm, pH 7.3), NBT chloride (0.27 mm), NAD+ (0.9 mm), and 10% (v/v) ethanol. The leaves were rocked gently in the staining solution for 4 h at 23°C, washed with water, and destained with 50% aqueous methanol and then 90% aqueous methanol (30 min for each wash step). Isolated secretory cells were suspended in sodium phosphate (90 mm, pH 7.3), NBT chloride (0.27 mm), NAD+ (0.27 mm), phenazine methosulfate (0.027 mm), and 10% (v/v) ethanol. Cells were rocked gently in the staining solution for 4 h at 23°C and washed with 90% methanol for 45 min and then sodium phosphate buffer (100 mm, pH 7.3) for 5 min. The secretory cell suspension was transferred to a hemacytometer, and cell discs were counted using an DMLB fluorescence microscope (Leica Microsystems). Images were taken with a model 3.2.0 camera (SPOT Imaging).
For specific activity measurements, isolated secretory cells (obtained as described above) were suspended in lysis buffer containing Tris (50 mm, pH 8), sorbitol (10%, v/v), E-65 proteinase inhibitor (0.04 mm), pepstatin A (0.02 mg mL−1), 1,10-phenanthroline (20 mm), and 4-(2-aminoethyl)benzenesulfonyl fluoride (4 mm). Cells were disrupted in a glass homogenizer and the lysate was centrifuged at 15,000g for 10 min (4°C). The supernatant was filtered through a 0.22-µm filter and desalted using a Bio-Gel P6 column (Bio-Rad). The concentration of total protein was determined (Bradford, 1976), and ADH activity was measured using the method of Mayer and Arnold (2002) with modifications (an aliquot of the crude protein extract was boiled for 5 min to serve as a negative control). The protein extract (15 µg of protein per microtiter plate well for young leaves; 3 µg per well for mature leaves) was diluted with assay buffer to finally contain Tris (50 mm, pH 8), NBT chloride (0.18 mm), NAD+ (0.18 mm), phenazine methosulfate (0.018 mm), and gelatin (0.08%, w/v). The reaction was started by the addition of 0.2 m ethanol and absorption at 580 nm (formation of NADH) measured over a period of 4 h (µ-Quant; BioTek Instruments). The slope of the absorption curve was compared with that of a standard curve generated by adding known quantities of NADH directly to the assay buffer (no cells). The specific activity was obtained by subtracting the absorbance determined in assays performed without adding substrate.
Cloning and Characterization of ADH from Peppermint
The sequence of the well-characterized ADH1 from potato (Solanum tuberosum; P14673; Matton et al., 1990) was used to query our peppermint (Mentha × piperita) transcriptome assembly. Two contigs that contained genes with high identity (∼88% at the peptide sequence level) and the corresponding cDNAs were amplified using Phusion DNA polymerase (New England Biolabs) with an annealing temperature of 60°C. The primer sequences were 5′-TGGCTGATCAAACCAAACCAG-3′ (ADH-like gene; forward 1), 5′-TATGGCTGATCAAACCAAACCAG-3′ (ADH-like gene; forward 2), 5′-GATCCTCAGAACTTGATAATAACCTTGAC-3′ (ADH-like gene; reverse 1), 5′-CTCAGAACTTGATAATAACCTTGAC-3′ (ADH-like gene; reverse 2), 5′-TGTCGACTACTCATGGTCAAG-3′ (ADH1 gene; forward 1), 5′-TATGTCGACTACTCATGGTCAA-3′ (ADH1 gene; forward 2), 5′-GATCCTTACATTTTAACAACACAACGAAG-3′ (ADH1 gene; reverse 1), and 5′-CTTACATTTTAACAACACAACGAAG-3′ (ADH1 gene; reverse 2). The target vector (pSBET; Schenk et al., 1995) was digested with NdeI and BamHI and then mixed with PCR amplicons in a sticky-end ligation reaction. The identity of constructs was confirmed by sequencing. Constructs were transformed into Escherichia coli BL21 cells for recombinant protein production.
A single colony was picked and grown in Luria-Bertani solution (5 mL) containing 50 µg mL−1 kanamycin for 12 h. The mixture was added to a larger Erlenmeyer flask containing 150 mL of Luria-Bertani medium, and cells were grown at 37°C to an optical density of 0.8. Transgene expression was induced by the addition of 0.1 mm isopropyl β-d-1-thiogalactopyranoside, and the flask was incubated at 16°C for 16 h. The turbid broth was centrifuged in a Sorvall RC3C centrifuge (Thermo Fisher Scientific) at 5,000g for 10 min. Precipitated cells were suspended in 50 mm 3-morpholino-2-hydroxypropanesulfonic acid buffer (containing 10% [v/v] glycerol and 1 mm DTT) and sonicated for three cycles of 15 s each on ice. The resulting suspension was centrifuged in a model 4515 D microfuge (Eppendorf) at 13,000g for 30 min. The supernatant was collected and loaded onto hydroxyapatite (Sigma-Aldrich) in batch mode and incubated for 1 h. After washing with 3-morpholino-2-hydroxypropanesulfonic acid buffer, the sample was eluted with 200 mm phosphate buffer. To assess the purity of the preparation, 15 μL of the eluent was processed by SDS-PAGE and stained using Coomassie Brilliant Blue. Following destaining using a 20:7:73 (v/v/v) mixture of methanol:acetic acid:water, protein bands were detected by imaging (Alpha Innotech). Activity assays contained 10 µm purified enzyme and varying concentrations of ethanol. The conversion of NAD to NADH was used to monitor the progress of the reaction at 340 nm (Synergy H1 Hybrid Reader; BioTek). The Origin 2017 software package (OriginLab) was employed to fit data and deduce kinetic constants.
FNR and Fd Sequence Comparisons
Mentha longifolia, Mentha arvensis, and Perilla frutescens mRNA was purified from isolated GT tissue as described above. For M. longifolia and P. frutescens, strand-specific paired-end libraries were prepared and sequenced using the HiSeq3000 platform (Illumina). For M. arvensis, single-end libraries were sequenced by the HiSeq2000 platform (Illumina). Sequences for other species were obtained from the literature or from publicly accessible databases (Supplemental Table S3). In some cases, the literature data included information on gene expression levels. In other cases, we assembled contigs and determined relative expression levels. To generate contigs, reads were trimmed using Skewer (Jiang et al., 2014) and assembled with either Trinity (Grabherr et al., 2011; in the case of Illumina reads) or MIRA (Chevreux et al., 2004; in the case of 454 reads). Expression levels were estimated as average read coverage in the case of MIRA assemblies or calculated using Bowtie (Langmead et al., 2009) and RSEM (Li et al., 2010) in the case of Trinity assemblies. Sequences were aligned with Clustal Omega (Sievers et al., 2011; 5′ end corresponding to the plastidial targeting sequence removed) and maximum likelihood trees generated using MEGA7 (Kumar et al., 2016) with 1,000 iterations of bootstrapping.
Cloning of GT-Specific Isoforms of Peppermint Fd and FNR
GTs were isolated, RNA was purified, and first-strand cDNA was generated as described previously (Ahkami et al., 2015). cDNAs were amplified using Phusion DNA polymerase (New England Biolabs) with an annealing temperature of 60°C. The primer sequences were 5′-AGTGTACATATGATCAGAAAGTTCAGAAACTC-3′ (Fd, major GT isoform, forward), 5′-TCTATGGGATCCCACGATTTCGATTGTTCATCCC-3′ (Fd, major GT isoform, reverse), 5′-AGTGTACATATGGCAACTCCGTCTAAGAAGC-3′ (Fd, minor GT isoform, forward), 5′-TCTATGGGATCCTTCCCTCATATCTAACAACAAGC-3′ (Fd, minor GT isoform, reverse), 5′-AGTGTACATATGAGCATCGTTATTTTCCGAC-3′ (FNR, GT/root isoform, forward), 5′-TCTATGGGATCCGGGGTCTGTTTTAATAATTGC-3′ (FNR, GT/root isoform, reverse), 5′-AGTGTACATATGACTGCTGCTGTAAGTGCCGCA-3′ (FNR, leaf isoform, forward), and 5′-TCTATGGGATCCTCAGTACACTTCCACATTCCAC-3′ (FNR, leaf isoform, reverse). Following digestion with NdeI and BamHI, amplicons were ligated into the pGEM-T Easy vector (Promega), and the identity of constructs was confirmed by sequencing.
Simulating Protein-Protein Interactions in the Peppermint GT FNR-Fd-HDR Complex
The sequences of peppermint Fd (minor GT/root isoform), Fd (major GT isoform), FNR (GT/root isoform), and HDR were submitted to the I-TASSER server for protein structure prediction (Roy et al., 2010; Yang et al., 2015). A structural representation in Protein Data Bank format was returned and employed in structural alignments using the Chimera molecular visualization program (Pettersen et al., 2004). Protein Data Bank files of the recently published crystal structures of the root isoforms of maize Fd and FNR (Shinohara et al., 2017) were uploaded into Chimera. A structural alignment was then performed by overlaying the corresponding peppermint Fd and FNR isoforms. Using the Swap function in Chimera, the sequences of the maize Fd and FNR isoforms were changed manually to reflect the residues that are present in the appropriate peppermint Fd and FNR isoforms. The structures of the peppermint Fd and FNR isoforms obtained by this approach (excluding iron-sulfur clusters and ligands) were energy minimized in Chimera, and the iron-sulfur clusters and ligands were subsequently reintroduced by docking. The same method was employed to produce a peppermint HDR structure (following alignment to the published Plasmodium falciparum HDR structure; Rekittke et al., 2013). The structure files for peppermint Fd, FNR, and HDR were submitted to the GRAMM-X server (Tovchigrechko and Vasker, 2006) to assess interactions in a three-protein complex (Fd-FNR as receptor and HDR as ligand). The GRAMM-X algorithm generated multiple docked confirmations. The conformation that most closely maintained the amino acid distances suggested previously to contribute to protein-protein interaction surfaces (Rekittke et al., 2013; Shinohara et al., 2017) was selected for structure-function studies.
Accession Numbers
Accession numbers are as follows: M. piperita Fd cDNA clone (major GT isoform), GenBank KY748231; M. piperita Fd cDNA clone (minor GT/root isoform), GenBank KY748232; M. piperita FNR cDNA clone (GT/root isoform), GenBank KY748233; M. piperita FNR cDNA clone (leaf isoform), GenBank KY748234; M. piperita HDR cDNA sequence, GenBank KY888887; M. piperita ADH-like cDNA sequence, GenBank MF579445; M. piperita ADH1 cDNA sequence, GenBank MF579446; P. frutescens GT transcriptome, SRR4106089; M. arvensis GT transcriptome, SRR1271560 (young leaves) and SRR1271559 (mature leaves); M. longifolia (CMEN585 accession) GT transcriptome, SRR3623199.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Alignment of alcohol dehydrogenase sequences.
Supplemental Table S1. Reactions included in various iterations of the menpiGT_2015 model.
Supplemental Table S2. Predicted outcomes of simulated reaction deletions.
Supplemental Table S3. Predicted reaction outcomes of the menpiGT_2015 model using Boolean constrained SPOT and gene expression patterns to estimate flux distribution.
Supplemental Table S4. Sequences and relative expression levels of genes coding for FNR and Fd isoforms across angiosperms that harbor GTs.
Supplemental Methods and Data File S1. Reconstructing mint GT metabolism (all iterations of the menpiGT_2015 model).
Supplemental Methods and Data File S2. Establishing experimental conditions to test the effects of in vivo ATP synthase inhibition on monoterpene formation in isolated peppermint GTs.
Acknowledgments
We thank Susan Vogtman for tending plants, Kelly Vining (Oregon State University) for sharing data prior to publication, and Jeremy Jewell, Amber Parrish, and Glenn Turner for helpful discussions and technical assistance.
Glossary
- GT
glandular trichome
- MEP
methylerythritol phosphate
- TPM
transcripts per kilobase million
Footnotes
This research was supported by grants from the DOE Office of Secretary of Energy (grant no. DE-SC0001553 to B.M.L.) and the HHS National Institutes of Health training grant program (grant no. T32GM083864 to S.R.J.).
Articles can be viewed without a subscription.
References
- Ahkami A, Johnson SR, Srividya N, Lange BM (2015) Multiple levels of regulation determine monoterpenoid essential oil compositional variation in the mint family. Mol Plant 8: 188–191 [DOI] [PubMed] [Google Scholar]
- Aliverti A, Faber R, Finnerty CM, Ferioli C, Pandini V, Negri A, Karplus PA, Zanetti G (2001) Biochemical and crystallographic characterization of ferredoxin-NADP(+) reductase from nonphotosynthetic tissues. Biochemistry 40: 14501–14508 [DOI] [PubMed] [Google Scholar]
- Aliverti A, Pandini V, Pennati A, de Rosa M, Zanetti G (2008) Structural and functional diversity of ferredoxin-NADP(+) reductases. Arch Biochem Biophys 474: 283–291 [DOI] [PubMed] [Google Scholar]
- Arnold A, Nikoloski Z (2014) Bottom-up metabolic reconstruction of Arabidopsis and its application to determining the metabolic costs of enzyme production. Plant Physiol 165: 1380–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson DE. (1968) The energy charge of the adenylate pool as a regulatory parameter: interaction with feedback modifiers. Biochemistry 7: 4030–4034 [DOI] [PubMed] [Google Scholar]
- Balcke GU, Bennewitz S, Bergau N, Athmer B, Henning A, Majovsky P, Jiménez-Gómez JM, Hoehenwarter W, Tissier A (2017) Multi-omics of tomato glandular trichomes reveals distinct features of central carbon metabolism supporting high productivity of specialized metabolites. Plant Cell 29: 960–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergau N, Bennewitz S, Syrowatka F, Hause G, Tissier A (2015) The development of type VI glandular trichomes in the cultivated tomato Solanum lycopersicum and a related wild species S. habrochaites. BMC Plant Biol 15: 289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berim A, Park JJ, Gang DR (2014) Unexpected roles for ancient proteins: flavone 8-hydroxylase in sweet basil trichomes is a Rieske-type, PAO-family oxygenase. Plant J 80: 385–395 [DOI] [PubMed] [Google Scholar]
- Berry C, Eck JMV, Kitto SL, Smigocki A (1996) Agrobacterium-mediated transformation of commercial mints. Plant Cell Tissue Organ Cult 44: 177–181 [Google Scholar]
- Bertea CM, Schalk M, Karp F, Maffei M, Croteau R (2001) Demonstration that menthofuran synthase of mint (Mentha) is a cytochrome P450 monooxygenase: cloning, functional expression, and characterization of the responsible gene. Arch Biochem Biophys 390: 279–286 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Büchi R, Bachmann M, Keller F (1998) Carbohydrate metabolism in source leaves of sweet basil (Ocimum basilicum L.), a starch-storing and stachyose-translocating labiate. J Plant Physiol 153: 308–315 [Google Scholar]
- Burke CC, Wildung MR, Croteau R (1999) Geranyl diphosphate synthase: cloning, expression, and characterization of this prenyltransferase as a heterodimer. Proc Natl Acad Sci USA 96: 13062–13067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caissard JC, Faure O, Jullien F, Colson M, Perrin A (1996) Direct regeneration in vitro and transient GUS expression in Mentha x piperita. Plant Cell Rep 16: 67–70 [DOI] [PubMed] [Google Scholar]
- Caspi R, Billington R, Ferrer L, Foerster H, Fulcher CA, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Mueller LA, et al. (2016) The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 44: D471–D480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung CYM, Williams TCR, Poolman MG, Fell DA, Ratcliffe RG, Sweetlove LJ (2013) A method for accounting for maintenance costs in flux balance analysis improves the prediction of plant cell metabolic phenotypes under stress conditions. Plant J 75: 1050–1061 [DOI] [PubMed] [Google Scholar]
- Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Müller WEG, Wetter T, Suhai S (2004) Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res 14: 1147–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis EM, Ringer KL, McConkey ME, Croteau R (2005) Monoterpene metabolism: cloning, expression, and characterization of menthone reductases from peppermint. Plant Physiol 137: 873–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Dal’Molin CG, Quek LE, Palfreyman RW, Brumbley SM, Nielsen LK (2010a) AraGEM, a genome-scale reconstruction of the primary metabolic network in Arabidopsis. Plant Physiol 152: 579–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Dal’Molin CG, Quek LE, Palfreyman RW, Brumbley SM, Nielsen LK (2010b) C4GEM, a genome-scale metabolic model to study C4 plant metabolism. Plant Physiol 154: 1871–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diemer F, Jullien F, Faure O, Moja S, Colson M, Matthys-Rochon E, Caissard JC (1998) High efficiency transformation of peppermint (Mentha × piperita L.) with Agrobacterium tumefaciens. Plant Sci 136: 101–108 [Google Scholar]
- Edwards JS, Palsson BO (2000) The Escherichia coli MG1655 in silico metabolic genotype: its definition, characteristics, and capabilities. Proc Natl Acad Sci USA 97: 5528–5533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenreich W, Sagner S, Zenk MH, Bacher A (1997) Monoterpenoid essential oils are not of mevalonoid origin. Tetrahedron Lett 38: 3889–3892 [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Fahn A. (1988) Tansley Review No. 14. Secretory tissues in vascular plants. New Phytol 108: 229–257 [DOI] [PubMed] [Google Scholar]
- Fell DA, Small JR (1986) Fat synthesis in adipose tissue: an examination of stoichiometric constraints. Biochem J 238: 781–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M. (1976) Intragenic recombination in maize: pollen analysis methods and the effect of parental adh1 isoalleles. Genetics 83: 701–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gang DR, Wang J, Dudareva N, Nam KH, Simon JE, Lewinsohn E, Pichersky E (2001) An investigation of the storage and biosynthesis of phenylpropenes in sweet basil. Plant Physiol 125: 539–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershenzon J, Maffei M, Croteau R (1989) Biochemical and histochemical localization of monoterpene biosynthesis in the glandular trichomes of spearmint (Mentha spicata). Plant Physiol 89: 1351–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershenzon J, McCaskill D, Rajaonarivony JIM, Mihaliak C, Karp F, Croteau R (1992) Isolation of secretory cells from plant glandular trichomes and their use in biosynthetic studies of monoterpenes and other gland products. Anal Biochem 200: 130–138 [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadizadeh M, Keyhani E (2004) Detection and kinetic properties of alcohol dehydrogenase in dormant corm of Crocus sativus L. Acta Hortic 650: 127–139 [Google Scholar]
- Hanke GT, Okutani S, Satomi Y, Takao T, Suzuki A, Hase T (2005) Multiple iso-proteins of FNR in Arabidopsis: evidence for different contributions to chloroplast function and nitrogen assimilation. Plant Cell Environ 28: 1146–1157 [Google Scholar]
- Hay J, Schwender J (2011) Metabolic network reconstruction and flux variability analysis of storage synthesis in developing oilseed rape (Brassica napus L.) embryos. Plant J 67: 526–541 [DOI] [PubMed] [Google Scholar]
- Hooper CM, Tanz SK, Castleden IR, Vacher MA, Small ID, Millar AH (2014) SUBAcon: a consensus algorithm for unifying the subcellular localization data of the Arabidopsis proteome. Bioinformatics 30: 3356–3364 [DOI] [PubMed] [Google Scholar]
- Huijing F, Slater EC (1961) The use of oligomycin as an inhibitor of oxidative phosphorylation. J Biochem 49: 493–501 [DOI] [PubMed] [Google Scholar]
- Jiang H, Lei R, Ding SW, Zhu S (2014) Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics 15: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MK, Lane A, Kelley JJ, Lun DS (2016) E-Flux2 and SPOT: validated methods for inferring intracellular metabolic flux distributions from transcriptomic data. PLoS ONE 11: e0157101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjonaas RB, Venkatachalam KV, Croteau R (1985) Metabolism of monoterpenes: oxidation of isopiperitenol to isopiperitenone, and subsequent isomerization to piperitenone by soluble enzyme preparations from peppermint (Mentha piperita) leaves. Arch Biochem Biophys 238: 49–60 [DOI] [PubMed] [Google Scholar]
- Kruger NJ, von Schaewen A (2003) The oxidative pentose phosphate pathway: structure and organisation. Curr Opin Plant Biol 6: 236–246 [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurisu G, Kusunoki M, Katoh E, Yamazaki T, Teshima K, Onda Y, Kimata-Ariga Y, Hase T (2001) Structure of the electron transfer complex between ferredoxin and ferredoxin-NADP(+) reductase. Nat Struct Biol 8: 117–121 [DOI] [PubMed] [Google Scholar]
- Lange BM. (2015) Biosynthesis and biotechnology of high-value p-menthane monoterpenes, including menthol, carvone, and limonene. Adv Biochem Eng Biotechnol 148: 319–353 [DOI] [PubMed] [Google Scholar]
- Lange BM, Croteau R (1999a) Isoprenoid biosynthesis via a mevalonate-independent pathway in plants: cloning and heterologous expression of 1-deoxy-D-xylulose-5-phosphate reductoisomerase from peppermint. Arch Biochem Biophys 365: 170–174 [DOI] [PubMed] [Google Scholar]
- Lange BM, Croteau R (1999b) Isopentenyl diphosphate biosynthesis via a mevalonate-independent pathway: isopentenyl monophosphate kinase catalyzes the terminal enzymatic step. Proc Natl Acad Sci USA 96: 13714–13719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange BM, Ketchum REB, Croteau RB (2001) Isoprenoid biosynthesis: metabolite profiling of peppermint oil gland secretory cells and application to herbicide target analysis. Plant Physiol 127: 305–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange BM, Lange I, Turner GW, Herron BK (2013) Utility of aromatic plants for the biotechnological production of sustainable chemical and pharmaceutical feedstocks. Med Aromat Plants 2: 5 [Google Scholar]
- Lange BM, Mahmoud SS, Wildung MR, Turner GW, Davis EM, Lange I, Baker RC, Boydston RA, Croteau RB (2011) Improving peppermint essential oil yield and composition by metabolic engineering. Proc Natl Acad Sci USA 108: 16944–16949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange BM, Rios-Estepa R (2014) Kinetic modeling of plant metabolism and its predictive power: peppermint essential oil biosynthesis as an example. Methods Mol Biol 1083: 287–311 [DOI] [PubMed] [Google Scholar]
- Lange BM, Turner GW (2013) Terpenoid biosynthesis in trichomes: current status and future opportunities. Plant Biotechnol J 11: 2–22 [DOI] [PubMed] [Google Scholar]
- Lange BM, Wildung MR, McCaskill D, Croteau R (1998) A family of transketolases that directs isoprenoid biosynthesis via a mevalonate-independent pathway. Proc Natl Acad Sci USA 95: 2100–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange BM, Wildung MR, Stauber EJ, Sanchez C, Pouchnik D, Croteau R (2000) Probing essential oil biosynthesis and secretion by functional evaluation of expressed sequence tags from mint glandular trichomes. Proc Natl Acad Sci USA 97: 2934–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latendresse M, Krummenacker M, Trupp M, Karp PD (2012) Construction and completion of flux balance models from pathway databases. Bioinformatics 28: 388–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Ruotti V, Stewart RM, Thomson JA, Dewey CN (2010) RNA-seq gene expression estimation with read mapping uncertainty. Bioinformatics 26: 493–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien S, Karp F, Wildung M, Croteau R (1999) Regiospecific cytochrome P450 limonene hydroxylases from mint (Mentha) species: cDNA isolation, characterization, and functional expression of (−)-4S-limonene-3-hydroxylase and (−)-4S-limonene-6-hydroxylase. Arch Biochem Biophys 368: 181–192 [DOI] [PubMed] [Google Scholar]
- Mahmoud SS, Croteau RB (2001) Metabolic engineering of essential oil yield and composition in mint by altering expression of deoxyxylulose phosphate reductoisomerase and menthofuran synthase. Proc Natl Acad Sci USA 98: 8915–8920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud SS, Williams M, Croteau R (2004) Cosuppression of limonene-3-hydroxylase in peppermint promotes accumulation of limonene in the essential oil. Phytochemistry 65: 547–554 [DOI] [PubMed] [Google Scholar]
- Matton DP, Constabel P, Brisson N (1990) Alcohol dehydrogenase gene expression in potato following elicitor and stress treatment. Plant Mol Biol 14: 775–783 [DOI] [PubMed] [Google Scholar]
- Mayer KM, Arnold FH (2002) A colorimetric assay to quantify dehydrogenase activity in crude cell lysates. J Biomol Screen 7: 135–140 [DOI] [PubMed] [Google Scholar]
- Mayne RG, Lea PJ (1984) Alcohol dehydrogenase in Hordeum vulgare: changes in isoenzyme levels under hypoxia. Plant Sci Lett 37: 73–78 [Google Scholar]
- McCaskill D, Croteau R (1995) Monoterpene and sesquiterpene biosynthesis in glandular trichomes of peppermint (Mentha x piperita) rely exclusively on plastid-derived isopentenyl diphosphate. Planta 197: 49–56 [Google Scholar]
- McCaskill D, Gershenzon J, Croteau R (1992) Morphology and monoterpene biosynthetic capabilities of secretory cell clusters isolated from glandular trichomes of peppermint (Mentha piperita L.). Planta 187: 445–454 [DOI] [PubMed] [Google Scholar]
- Mintz-Oron S, Meir S, Malitsky S, Ruppin E, Aharoni A, Shlomi T (2012) Reconstruction of Arabidopsis metabolic network models accounting for subcellular compartmentalization and tissue-specificity. Proc Natl Acad Sci USA 109: 339–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller LA, Zhang P, Rhee SY (2003) AraCyc: a biochemical pathway database for Arabidopsis. Plant Physiol 132: 453–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers EW, Miller W (1988) Optimal alignments in linear space. Comput Appl Biosci 4: 11–17 [DOI] [PubMed] [Google Scholar]
- Nadwodnik J, Lohaus G (2008) Subcellular concentrations of sugar alcohols and sugars in relation to phloem translocation in Plantago major, Plantago maritima, Prunus persica, and Apium graveolens. Planta 227: 1079–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu X, Lin K, Hasegawa PM, Bressan RA, Weller SC (1998) Transgenic peppermint (Mentha × piperita L.) plants obtained by cocultivation with Agrobacterium tumefaciens. Plant Cell Rep 17: 165–171 [DOI] [PubMed] [Google Scholar]
- Olennikov DN, Tankhaeva LM (2007) Lamiaceae carbohydrates. II. Water-soluble polysaccharides from Mentha x piperita. Chem Nat Compd 43: 648–651 [Google Scholar]
- Onda Y, Matsumura T, Kimata-Ariga Y, Sakakibara H, Sugiyama T, Hase T (2000) Differential interaction of maize root ferredoxin:NADP(+) oxidoreductase with photosynthetic and non-photosynthetic ferredoxin isoproteins. Plant Physiol 123: 1037–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterman JC, Chiang Y, Markwell J (1993) Characterization of mutation-induced changes in the maize (Zea mays L.) ADH1-1S1108 alcohol dehydrogenase. Biochem Genet 31: 497–506 [DOI] [PubMed] [Google Scholar]
- Penefsky HS. (1985) Mechanism of inhibition of mitochondrial adenosine triphosphatase by dicyclohexylcarbodiimide and oligomycin: relationship to ATP synthesis. Proc Natl Acad Sci USA 82: 1589–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Pilalis E, Chatziioannou A, Thomasset B, Kolisis F (2011) An in silico compartmentalized metabolic model of Brassica napus enables the systemic study of regulatory aspects of plant central metabolism. Biotechnol Bioeng 108: 1673–1682 [DOI] [PubMed] [Google Scholar]
- Poolman MG, Kundu S, Shaw R, Fell DA (2013) Responses to light intensity in a genome-scale model of rice metabolism. Plant Physiol 162: 1060–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poolman MG, Miguet L, Sweetlove LJ, Fell DA (2009) A genome-scale metabolic model of Arabidopsis and some of its properties. Plant Physiol 151: 1570–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekittke I, Olkhova E, Wiesner J, Demmer U, Warkentin E, Jomaa H, Ermler U (2013) Structure of the (E)-4-hydroxy-3-methyl-but-2-enyl-diphosphate reductase from Plasmodium falciparum. FEBS Lett 587: 3968–3972 [DOI] [PubMed] [Google Scholar]
- Ringer KL, Davis EM, Croteau R (2005) Monoterpene metabolism: cloning, expression, and characterization of (−)-isopiperitenol/(−)-carveol dehydrogenase of peppermint and spearmint. Plant Physiol 137: 863–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringer KL, McConkey ME, Davis EM, Rushing GW, Croteau R (2003) Monoterpene double-bond reductases of the (−)-menthol biosynthetic pathway: isolation and characterization of cDNAs encoding (−)-isopiperitenone reductase and (+)-pulegone reductase of peppermint. Arch Biochem Biophys 418: 80–92 [DOI] [PubMed] [Google Scholar]
- Rios-Estepa R, Lange I, Lee JM, Lange BM (2010) Mathematical modeling-guided evaluation of biochemical, developmental, environmental, and genotypic determinants of essential oil composition and yield in peppermint leaves. Plant Physiol 152: 2105–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios-Estepa R, Turner GW, Lee JM, Croteau RB, Lange BM (2008) A systems biology approach identifies the biochemical mechanisms regulating monoterpenoid essential oil composition in peppermint. Proc Natl Acad Sci USA 105: 2818–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5: 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha R, Suthers PF, Maranas CD (2011) Zea mays iRS1563: a comprehensive genome-scale metabolic reconstruction of maize metabolism. PLoS ONE 6: e21784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A, Tamura H (2015) High antiallergic activity of 5,6,4′-trihydroxy-7,8,3′-trimethoxyflavone and 5,6-dihydroxy-7,8,3′,4′-tetramethoxyflavone from eau de cologne mint (Mentha × piperita citrata). Fitoterapia 102: 74–83 [DOI] [PubMed] [Google Scholar]
- Savinell JM, Palsson BO (1992) Network analysis of intermediary metabolism using linear optimization. I. Development of mathematical formalism. J Theor Biol 154: 421–454 [DOI] [PubMed] [Google Scholar]
- Schenk PM, Baumann S, Mattes R, Steinbiss HH (1995) Improved high-level expression system for eukaryotic genes in Escherichia coli using T7 RNA polymerase and rare ArgtRNAs. Biotechniques 19: 196–198, 200 [PubMed] [Google Scholar]
- Seemann M, Tse Sum Bui B, Wolff M, Miginiac-Maslow M, Rohmer M (2006) Isoprenoid biosynthesis in plant chloroplasts via the MEP pathway: direct thylakoid/ferredoxin-dependent photoreduction of GcpE/IspG. FEBS Lett 580: 1547–1552 [DOI] [PubMed] [Google Scholar]
- Shinohara F, Kurisu G, Hanke G, Bowsher C, Hase T, Kimata-Ariga Y (2017) Structural basis for the isotype-specific interactions of ferredoxin and ferredoxin:NADP(+) oxidoreductase: an evolutionary switch between photosynthetic and heterotrophic assimilation. Photosynth Res (in press) [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinson J, Mascarenhas JP (1985) Onset of alcohol dehydrogenase synthesis during microsporogenesis in maize. Plant Physiol 77: 222–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian W, Skolnick J (2003) How well is enzyme function conserved as a function of pairwise sequence identity? J Mol Biol 333: 863–882 [DOI] [PubMed] [Google Scholar]
- Tong WF, Lin SW (1988) Purification and biochemical properties of rice alcohol-dehydrogenase. Bot Bull Acad Sin 29: 245–253 [Google Scholar]
- Tovchigrechko A, Vakser IA (2006) GRAMM-X public web server for protein-protein docking. Nucleic Acids Res 34: W310–W314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner GW, Gershenzon J, Croteau RB (2000a) Distribution of peltate glandular trichomes on developing leaves of peppermint. Plant Physiol 124: 655–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner GW, Gershenzon J, Croteau RB (2000b) Development of peltate glandular trichomes of peppermint. Plant Physiol 124: 665–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vining KJ, Johnson SR, Ahkami A, Lange I, Parrish AN, Trapp SC, Croteau RB, Straub SCK, Pandelova I, Lange BM (2017) Draft genome sequence of Mentha longifolia and development of resources for mint cultivar improvement. Mol Plant 10: 323–339 [DOI] [PubMed] [Google Scholar]
- Voirin B, Bayet C (1992) Developmental variations in leaf flavonoid aglycones of Mentha x piperita. Phytochemistry 31: 2299–2304 [Google Scholar]
- Werker E, Ravid U, Putievsky E (1985) Structure of glandular hairs and identification of the main components of their secreted material in some species of the Labiatae. Isr J Bot 34: 31–45 [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y (2015) The I-TASSER suite: protein structure and function prediction. Nat Methods 12: 7–8 [DOI] [PMC free article] [PubMed] [Google Scholar]