Abstract
Chromophobe renal cell carcinoma (ChRCC) is characterized by major changes in chromosomal copy number (CN). No model is available to precisely elucidate the molecular drivers of this tumor type. HNF1B is a master regulator of gene expression. Here we report that the transcription factor HNF1B is downregulated in the majority of ChRCC and that the magnitude of HNF1B loss is unique to ChRCC. We also observed a strong correlation between reduced HNF1B expression and aneuploidy in ChRCC patients. In murine embryonic fibroblasts or ACHN cells, HNF1B deficiency reduced expression of the spindle checkpoint proteins MAD2L1 and BUB1B, and the cell cycle checkpoint proteins RB1 and p27. Further, it altered the chromatin accessibility of Mad2l1, Bub1b and Rb1 genes and triggered aneuploidy development. Analysis of the TCGA database revealed TP53 mutations in 33% of ChRCC where HNF1B expression was repressed. In clinical specimens, combining HNF1B loss with TP53 mutation produced an association with poor patient prognosis. In cells, combining HNF1B loss and TP53 mutation increased cell proliferation and aneuploidy. Our results show how HNF1B loss leads to abnormal mitotic protein regulation and induction of aneuploidy. We propose that coordinate loss of HNF1B and TP53 may enhance cellular survival and confer an aggressive phenotype in ChRCC.
Keywords: Chromophobe renal cell carcinoma (ChRCC), HNF1B, TP53, spindle assembly checkpoint (SAC), Chromosomal instability (CIN)
Introduction
ChRCC was first described in 1985 by Thoenes and colleagues (1) and accounts for approximately 5 percent of renal malignancies (2). Based on immunohistochemical stains, DNA methylation analysis and mRNA expression patterns, ChRCC is postulated to arise from the distal convoluted tubule of the nephron (3) (4). ChRCC is characterized by loss of entire copies of chromosomes 1, 2, 6, 10, 13, and 17, with other chromosomes lost at a lower frequency (4). Focal chromosomal loss is not seen in ChRCC (5). This pattern of aneuploidy distinguishes ChRCC from other renal tumors (6) (7), and the constellation of chromosomal copy number (CN) abnormalities in ChRCC suggests a profound defect exists in the mitotic regulation of the cellular chromosomal CN.
Aneuploidy, is frequently observed in malignancies. The presence of aneuploidy may be causally related to, but is conceptually distinct from chromosomal instability (CIN), which describes the “rate” of karyotypic change (8). Factors that induce CIN, and ultimately aneuploidy, include spindle assembly checkpoint defects and chromosome cohesion defects (8) (9). An assessment of the genomic defects in ChRCC genomic databases, including The Cancer Genome Atlas (TCGA), does not point to an obvious driver of aneuploidy (4). Patients with Birt-Hogg-Dube (BHD) syndrome, an autosomal dominant disorder driven by germline mutations in folliculin (FLCN), have a significant probability of developing ChRCC (10). However, FLCN mutations are rarely found in sporadic ChRCC (11) (12) (4), nor is folliculin known to be associated with maintenance of chromosomal CN fidelity. To date, no mutation has been consistently linked with sporadic ChRCC, and the driver of aneuploidy is still missing (13)(14)(15)(16)(17,18). HNF1B encodes the tissue-restricted transcription factor HNF1B (TCF2, vHNF1, LFB3). HNF1B was recently shown to be a master regulator of gene expression via maintaining active transcription from its target promoters, or counterbalancing the silencing effect induced by mitotic chromatin condensation (13) (14). The Gene Atlas database reports that HNF1B mRNA is expressed at a high level in kidney and pancreatic islets (15). In mice, Hnf1b is expressed in early embryogenesis, and homozygous Hnf1b deletion is lethal at embryonic day 6 due to defective visceral endoderm differentiation (16). At a later stage, HNF1B is expressed in kidney, pancreas, liver, genital tract, gut and the hindbrain (17). During kidney development, HNF1B is expressed throughout the entire kidney, but predominantly in the distal nephron and collecting duct (18) (19). HNF1B mutations represent the most common known monogenic cause of developmental kidney disease (20). HNF1B mutations in humans were reported to cause multi-system diseases such as renal defects (19), maturity onset diabetes of the young type 5 (MODY5) (21) and genitourinary tract abnormalities (22).
In 2004 a somatic HNF1B mutation was first reported in a patient with ChRCC (23). Following that, Lebrun et al demonstrated a biallelic inactivation of HNF1B in a second ChRCC patient that arose from the combination of a germline mutation and a somatic gene deletion (24). During the same year, another report identified biallelic inactivation of HNF1B in two out of 12 patients with ChRCC (25). More recently, downregulation of HNF1B mRNA and protein was reported in an additional cohort of patients with ChRCC (26) (27), suggesting deficiency of HNF1B expression might play a significant role in the pathogenesis of ChRCC.
With the publication of The Cancer Genome Atlas (TCGA) ChRCC project, multiparametric data from 66 ChRCC tumor samples along with paired 25 normal tissues became available (4). This dataset allows researchers to perform further in silico hypothesis generating analyses. We have taken advantage of this resource to guide the development of our mechanistic model of ChRCC, and to test the hypothesis that decreased HNF1B expression is a driver of ChRCC development.
Materials and Methods
Cell culture, immunoblotting and materials
ACHN cell line was obtained from the ATCC in 2015. Cells were validated in March 2015 and March 2017 by STR DNA fingerprinting using the Promega 16 High Sensitivity STR Kit (Catalog # DC2100). The STR profiles were compared to online search databases (DSMZ/ATCC/JCRB/RIKEN) of 2455 known profiles; along with the MD Anderson Characterized Cell Line Core (CCLC) database of 2556 know profiles. The STR profiles matched known DNA fingerprints or were unique. Length of time between thawing and use was less than two weeks, and fewer than 10 passages. Mycoplasma contamination was tested using DAPI staining monthly, and was confirmed in March 2017 using Lonza MycoAlert Kit. The MEFs were isolated from E12.5–D13.5 day-pregnant mice as described previously (28), and in all experiments fibroblast derived from siblings were compared. Only the first three generations of MEF cells were used for experiments and mycoplasma test was not performed. Hnf1bflox/flox mice were a kind gift from Drs. Pontoglio and Dr. Igarashi. Immortalized and primary MEF cells were grown in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA), 100 U/ml penicillin, 100 μg/ml streptomycin and in humidified, 37°C chambers with 5% CO2. The primers for PCR were from integrated DNA Technologies (Coralville, IA). To determine protein concentration, cell protein was extracted using 1% Triton buffer after the treatment. The amount of protein was adjusted to the same concentration, transferred to nitrocellulose membrane, incubated with primary and secondary antibodies, The following sources provided antibodies: Hnf1b for western (MyBioSource, San Diego, CA), p53 for human cells, p21, acetyl-α-tubulin, Mad1l1, Bub1b, Rb1, p16 for murine cells and secondary antibodies (Santa Cruz Biotechnology); p53 for murine cells, p21 for human cells, p27, Mad2l1, and p-S10-Histon 3 (Cell Signaling Technology, Beverly, MA); β-gal senescence staining kit was purchased from Cell Signaling Technology, Beverly, MA.
Cell cycle analysis
For cell cycle profile analysis, trypsinized MEFs were fixed with 70% ethanol, followed by washing with phosphate-buffered saline and incubated in Propidium Iodide (PI)/RNase Staining Solution (Cell Signaling Technology, Beverly, MA) 30 min, and cells were subjected to cell cycle analysis by Flow cytometry.
ShRNA
Lentiviral mediated shRNA was performed using shRNA lentiviral (pLKO.1-puro) plasmids (Sigma). The oligonucleotides containing the TRC Lentiviral Mouse Hnf1b shRNA target sequence that were used are: sequence #1, 5′ - AAACCCAGGGCTGCCTTGGAAAAG - 3′ and sequence #2, 5′ - AAACCCAGGGCTGCCTTGGAAAAG - 3′. One 100 mm dish of 293T cells (Invitrogen) was co-transfected with 3 μg of the pLKO.1-puro plasmids plus lentivirus packaging mix (Sigma) using Lipofectamine 2000. The medium were changed approximately 16 h after transfection, and the cells were cultured an additional 48–72 h. The media were then collected, centrifuged at 3000 rpm for 5 min, and filtered through a 0.45 μM filter. Experimental cells were incubated with the virus-containing medium overnight in 6-well plates, the media were changed, and cells were incubated for 24 h. Cells were transferred to 100 mm dishes and infected cells were selected by incubation in puromycin (4 μg/ml) for about 10 days.
Immunofluorescence
The MEFs grown on coverslips were fixed in 4% paraformaldehyde for 20 min at 37°C, permeabilized by 0.1% Triton X-100, washed in phosphate-buffered saline, followed by 3% BSA blocking for one hour at room temperature and incubated with primary antibodies for overnight at 4°C. Primary antibodies were used as follows: acetyl-a-tubulin antibody (Santa Cruz Biotechnology, Santa Cruz, CA) at 1:3000 dilution. After washes, coverslips were incubated with fluorescein isothiocyanate- conjugated anti-mouse secondary antibodies (Jackson ImmunoResearch, West Grove, PA, USA; at 1:1000 dilution). The DNA was visualized with DAPI (Sigma) and the coverslips were mounted on slides with Vectashield mounting solution (Vector, Burlingame, CA, USA) and sealed. Staining was analyzed using a Nikon confocal A1 microscope (Melville, NY).
Adenovirus infection
Cells were passaged and placed into 6-well plates, after 6 hours, removed medium and replaced with serum free medium, 500MOI adenovirus-RGD-CMV-GFP or Adenovirus-RGD- CMV-Cre-GFP (Vectorbiolab Inc.) were added to infect for 2 hours, followed by culture in 10% serum contained medium for 48 hours or 72 hours, and then proceeded to experimental determination.
RT-PCR
For RT-PCR analysis, RNA was isolated using RNeasy Mini Kit (Qiagen) according to manufacturer’s protocol. The RNA was quantified using a Nano-drop spectrophotometer. The Reverse Transcription-PCR reaction was performed using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Real Time-PCR was performed to quantify mRNA by using the Fast SYBR Green Master (Applied Biosystems) in the reaction with the following primers
Hnf1b, Forward 5′-CCCCTCACCATCAGCCAAG-3′, Reverse 5′-GGTTCTGAGATTGCTGGGGATT-3′
Rb1, Forward 5′-TGCATCTTTATCGCAGCAGTT-3′, Reverse 5′-GTTCACACGTCCGTTCTA ATTTG-3′
Mad1l1, 5′-AGC CGT CTG GAA CAG GAA AAG-3′, Reverse 5′-GGGTTAAGGCTCATGTGTAGCA-3′
Mad2l1, Forward 5′-GTGGCCGAGTTTTTCTCATTTG-3′, Reverse 5′-AGGTGAGTCCATATT TCTGCACT-3′
Bub1b, Forward 5′-GAGGCGAGTGAAGCCATGT-3′, Reverse 5′-TCCAGAGTAAAAGCGGATTTCAG-3′
Cdkn1b, Forward 5′-TCAAACGTGAGAGTGTCTAACG-3′, Reverse 5′-CCGGGCCGAAGAGATTTCTG-3′
Gapdh, Forward 5′-AGGTCGGTGTGAACGGATTTG-3′, Reverse 5′-GGGGTCGTTGATGGCAACA-3′
Chromatin Immunoprecipitation
Chromatin from P3 primary MEF cells was prepared according to Millipore Magna ChIP™ A/G protocol. Chromatin were sonicated by Bioruptor in Nuclear lysis buffer with protease inhibitor for 30 second and paused for 30 second, 10 cycles. We then incubated sheared cross-linked chromatin with antibody to Hnf1b (Santa Cruz, sc22840), antibody to histone H3 (Abcam, ab1791), antibody to histone H3K9me3 (Abcam, Ab8898), antibody to histone H3K9ac (ab4441), or IgG as control. Purified DNA was used to do real time PCR. Primer sequences are listed as following:
| Rb1 | Forward 5′-AATGTGTTCCGTAAAAGTTTCTAGT-3′ | Reverse 5′-TACACTTTCAGCTTTATGGGTTGG-3′ |
| Mad1l1 | Forward 5′-ATTTAGACTCACACTGCCGC-3′ | Reverse 5′-AGGGATGCCCCTGAATGTCT-3′ |
| Mad2l1 | Forward 5′-GAGGGAATCCCCACCTCTTT-3′ | Reverse 5′-AGCAAAACACAGAATGGCGAA-3′ |
| Bub1b | Forward 5′-GCTTAGGCTACCTCCCTTTTT-3′ | Reverse 5′-GAGACAGGGACTGGGAGAGA-3′ |
| Rb1 −0.5kb | Forward 5′-TAAGATCTGTCCCGCTCGGA-3′ | Reverse 5′-ACAAGGGAAGATGGGCGATG-3′ |
| Rb1 0.7kb | Forward 5′-ATGCAACAGTTGGGAGCCTT-3′ | Reverse 5′-AGGCTTTTCTTGGCCTCTCC-3′ |
| Mad2l1 −0.5kb | Forward 5′-CTGCACAGTACGGACCTTGT-3′, | Reverse 5′-AATGAGCAAGCGCAGAGTG-3′ |
| Mad2l1 0.5kb | Forward 5′-TCAGCGTGGCATTTATCCGT-3′, | Reverse 5′-ATGAGCTCGGGGTCAGTAGT-3′ |
| Bub1b −0.5kb | Forward 5′-GGTGTTAAGACCTGGGGCTC-3′ | Reverse 5′-TCTTCTGGGTTGCTCCGAAC-3′ |
| Bub1b 0kb | Forward 5′-AATTTGAAACTTGGCGGCCC-3′ | Reverse 5′-TCAACCTTCAGCTGGCTCTG-3′ |
Statistical analysis
Level 3 TCGA data was downloaded from the FIREHOSE database (http://gdac.broadinstitute.org/) including gene expression and GISTIC CN data. Student’s t-test or analysis of variance (ANOVA) was used to assess association between a continuous variable and a categorical variable. T-test was used for paired data. Fisher’s exact test was used to assess association between two categorical variables. Log-rank test was used to assess survival difference between different patient groups.
Results
Significant downregulation of HNF1B occurs in ChRCC but not in other forms of RCC
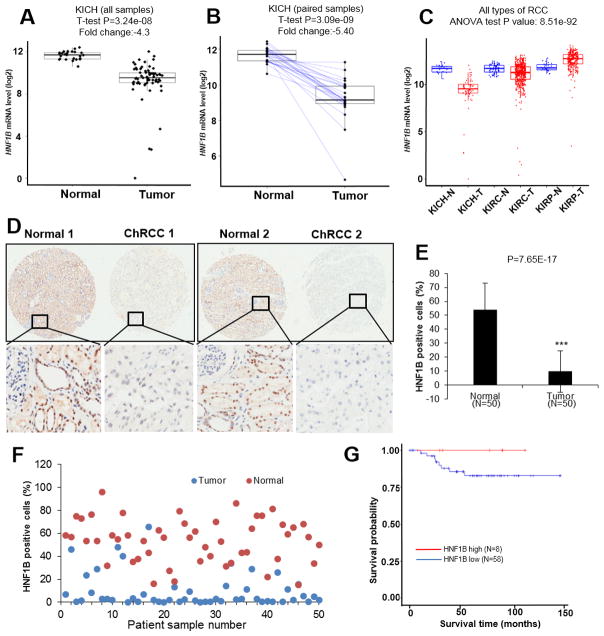
HNF1B underexpression and biallelic inactivation were repeatedly observed in small case reports and case series of ChRCC (23) (24) (25) (26) (27) We queried the ChRCC TCGA database, which includes 66 samples with RNAseq and mutation data, all of which have clinical information available. When we compared HNF1B mRNA levels in normal (N=25) and in ChRCC (N=66) specimens, we observed a significant reduction in mRNA levels (fold change= − 4.3 or 23% of normal, P=3.24e-8, Fig. 1A) in the ChRCC group. We further compared expression of HNF1B mRNA in paired normal (N=25) and ChRCC (N=25) samples, and found an even greater difference in HNF1B (−5.4 or 18% of normal, P=3.09e-09, Fig. 1B). Although reduced HNF1B levels were frequently observed in ChRCC, the expression levels in other forms of RCC has not been described. We assessed TCGA datasets for clear cell RCC (ccRCC) and papillary RCC (pRCC) and discovered that significant and consistent HNF1B downregulation did not occur in ccRCC or in pRCC (Fig. 1C). Furthermore, HNF1B protein levels of 50 paired tumors and uninvolved control samples in a ChRCC tissue microarray (TMA) were evaluated by immunohistochemistry (IHC). Two representative paired normal versus tumor samples are shown in Fig. 1D. HNF1B was highly expressed in nuclear tubule cells in normal kidney tissue. HNF1B levels were dramatically decreased in the majority of ChRCC samples. Average (Fig. 1E) and individual HNF1B positive cell percentages (Fig. 1F) were quantified. In order to assess the prognostic impact of HNF1B expression in ChRCC, we divided ChRCC patients into two groups based on their HNF1B mRNA levels (29). We compared overall survival between these two groups. Patients in the HNF1B high group (N=8) showed 100% survival, while those in the HNF1B low (N=58) group showed a 75% survival probability (Fig. 1G). Taken together, TCGA data analyses indicate decreased HNF1B expression may play a significant role in ChRCC development, and potentially in the development of a lethal phenotype.
Figure 1. HNF1B expression is reduced in ChRCC patients.
HNF1B mRNA (log2) level in normal (N=25) and ChRCC (N=66) samples from TCGA database (A). HNF1B mRNA levels of paired samples from normal (N=25) and ChRCC (N=25) compared by T-test (B). HNF1B mRNA (log2) levels in ChRCC, ccRCC and pRCC and their normal controls (C) (KICH, Kidney Chromophobe, N=normal, T=tumor; KIRC, Kidney clear cell carcinoma, KIPA, Kidney papillary cell carcinoma). 50 paired ChRCC tumor and normal TMA tissues analyzed by IHC for HNF1B staining (D). Averages of HNF1B percentage positivity in normal and tumor cells (E). Scatter plot of percentage HNF1B staining positivity in 50 matched tumor/normal pairs (F). ChRCC patient survival in HNF1B high and HNF1B low groups (G).
Loss of HNF1B but not of chromosome 17 correlated with aneuploidy in ChRCC
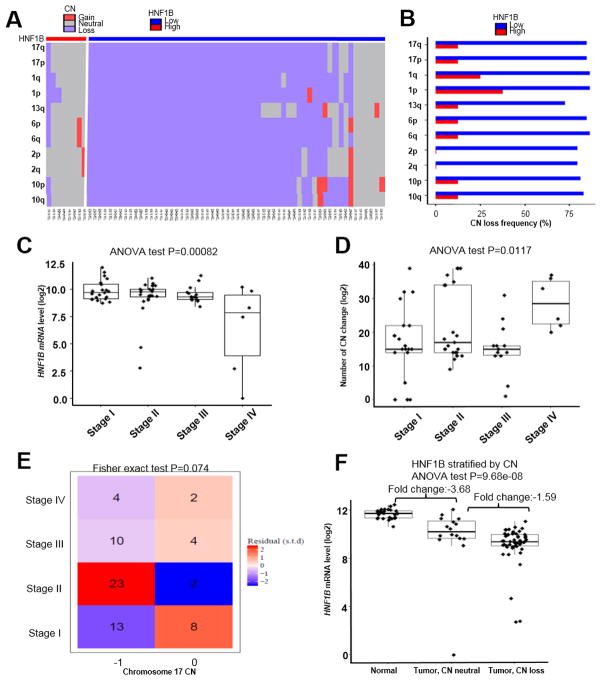
To determine whether HNF1B deficiency correlates with broad chromosome CN alterations or aneuploidy in ChRCC, we analyzed CN status of the HNF1B high and low groups in TCGA ChRCC patient database. The majority of the low HNF1B group showed CN loss in chromosomes 1, 2, 6, 10, 13 and 17 while most patients in the HNF1B high group were CN neutral (Fig. 2A). CN loss frequency, which is defined by percentage of patients who have any CN loss, was significantly higher (>=75%) in the HNF1B low group than that in the HNF1B high group (Fig. 2B). We divided ChRCC patients into 4 stages based on their clinical parameters and found that stage IV tumors showed an even greater drop in HNF1B levels (P=0.00082) (Fig. 2C). On the other hand, CN change was also increased in stage IV patients (Fig. 2D), further implicating decreased HNF1B in the evolution of aneuploidy in ChRCC. Since HNF1B is located on chromosome 17, one copy of which is frequently lost in ChRCC (7), we performed a correlation analysis of chromosome 17 loss with stage, and found no significant correlation (Fig. 2E). To determine whether expression of other genes on chromosome 17 is similarly decreased in ChRCC, we assessed FLCN mRNA levels in TCGA ChRCC database. Germline mutations in FLCN are often found in familial ChRCC as part of the Birt-Hogg Dube syndrome, which is linked to the development of familial ChRCC. Surprisingly, FLCN mRNA levels are not significantly altered (Fold change=1.17) in ChRCC when compared to normal control tissue (Supplementary Fig. S1A). FLCN mutations were not present in ChRCC TCGA samples. In addition, FLCN expression was not differentially suppressed in ChRCC when compared to other RCC histotypes (Supplementary Fig. S1B), suggesting the importance of HNF1B but not FLCN loss in sporadic ChRCC. Furthermore, we then assessed HNF1B levels in chromosome 17 CN neutral ChRCC patients (N=16). Interestingly, HNF1B was already greatly decreased in that group compared with normal tissue (N=25), with only a relatively minor further decrease in HNF1B in the group with chromosome 17 CN loss (N=50) (Fig. 2F), further supporting the hypothesis that HNF1B repression is a driver for CN loss. In addition, this result indicated HNF1B loss alone is necessary but may not be sufficient for CN loss.
Figure 2. Loss of HNF1B correlates with aneuploidy in ChRCC patients and not with chromosome 17 loss.
Copy number (CN) alteration of frequently lost chromosomes (1, 2, 6, 10, 13, 17) in HNF1B high (N=8) and HNF1B low (N=58) groups. Hierarchical clustering constructed with Euclidean distance and Ward’s linkage. One row represents one frequently lost chromosome arm, and one column represents one patient (A). Whole chromosome CN loss frequencies in HNF1B high and HNF1B low groups (B). HNF1B mRNA level comparisons among stage I–IV patients by ANOVA (C). Total number of chromosome CN change in stage I–IV patients by ANOVA (D). Correlation between patient stage and chromosome 17 CN loss via Fisher exact test (E). HNF1B mRNA levels in normal controls (N=25), tumor CN neutral (N=16) and tumor CN abnormal (N=50) groups by ANOVA (F).
Loss of Hnf1b induced polyploidy followed by cell death and chromosome instability in proliferating MEF cells
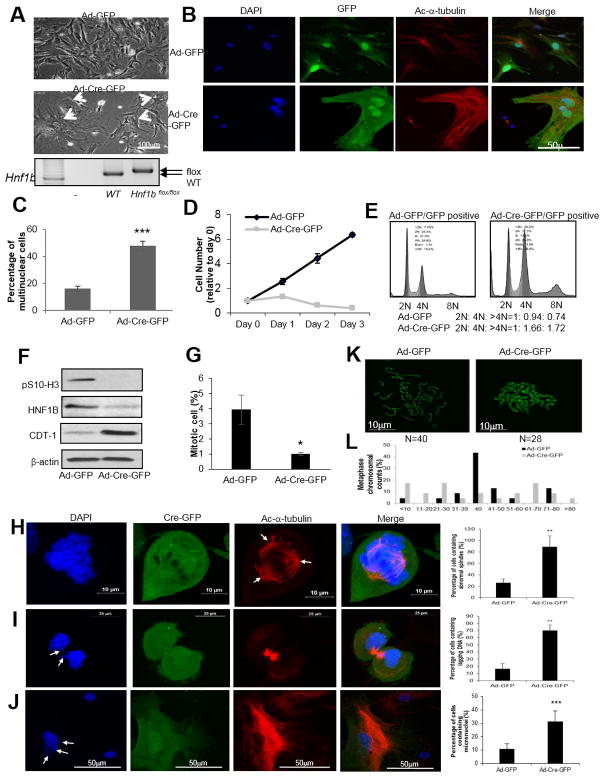
We then obtained Hnf1bflox/flox mice to better understand the impact of HNF1B loss on cellular phenotypes (30). Mouse genotypes were confirmed by DNA gel as shown in Fig. 3A, lower panel. After infection with Ad-Cre-GFP for 3 days, primary (P3) Hnf1bflox/flox MEFs displayed a significant increase in polynuclear cells (Fig. 3A arrows and Fig. 3B–3C) compared with Ad-GFP infected cells. Interestingly, we found Hnf1b deficient cells ceased to proliferate (Fig. 3D). In addition, we used spontaneously immortalized MEFs (28) which have similar proliferating rates as P1–P3 primary MEFs, and then knocked down Hnf1b by using shRNA as a parallel system to assess the effect of downregulation of HNF1B on cell cycle and cell death. We observed the morphology of Hnf1b knockdown cells was consistently altered, and it showed a higher percentage of multinuclear cells compared with mock cells that were infected with scrambled shRNA (Supplementary Fig. S2A–S2B). To better understand the effect of Hnf1b loss on cell cycle regulation, the cell cycle profile was examined in GFP positive early passage (P1–P3) Hnf1bflox/flox MEFs after infection with Ad-GFP or Ad-Cre-GFP. GFP-positive cells exhibited dramatically higher 4N and above 4N percentages in Ad-Cre-GFP than Ad-GFP infected cells (Fig. 3E). Immortalized Hnf1b knockdown MEFs showed a comparable profile, with an increased population of cells in 4N phase and above 4N than mock cells (Supplementary Fig. S2C). The ratio of cells containing DNA content shown as 2N: 4N: >4N further suggested the substantial elevation of 4N and above 4N cell populations in response to Hnf1b depletion. Serine 10 phosphorylated histone H3 was measured to examine the mitotic index. Its level was largely abolished in Hnf1b deficient cells after Ad-Cre-GFP infection in primary P3 MEFs (Fig. 3F) and shRNA knockdown (Supplementary Fig. S2D) in immortalized MEFs, indicating the increase of 4N was not due to mitotic arrest. After knockdown of Hnf1b, immortalized MEFs were prone to create abnormal mitotic spindles when compared to mock cells (Supplementary Fig. S2E–S2F). To further illustrate the cell phase of 4N and above 4N cell accumulation, the G1 marker CDT-1 (31) was immunoblotted and found to be greatly enhanced in Hnf1b depleted MEFs (Fig. 3F), suggesting those 4N and above 4N cells were arrested in G1 phase instead of G2 or M. This conclusion was further demonstrated by a remarkable reduction of the mitotic ratio in Hnf1b depleted cells (Fig. 3G). Live images are also shown in Supplementary Fig. S3A and S3B to further illustrate the mitotic process. Quantified time of nuclear envelope breakdown (NEB) to anaphase was compared between mock and Hnf1b depleted cells, and no difference was observed (Supplementary Fig. S3C), further confirming that Hnf1b loss did not arrest cells in mitosis. Thus, the above results demonstrate that Hnf1b deficiency induced polynuclearity with very low viability in proliferating cells is the result of G1 arrest rather than G2 or mitotic arrest.
Figure 3. Loss of Hnf1b induced chromosomal instability (CIN) and aneuploidy in MEFs.
Hnf1b flox/flox P2 MEFs infected with Ad-Cre-GFP or Ad-GFP for 2 h in serum free media, followed by complete media culture for 72 h. Cell morphologies captured after 3-day infection. Arrowheads point to polynuclear cells (A). DAPI, GFP and acetyl-α-tubulin staining (B). Percentages of polyploid cells in infected cells (C). Cell survival curves (D). Cell cycle profiles recorded by fluorescence assisted cell sorting (E). Phospho-S10-Histone H3, CDT-1 and HNF1B protein levels (F). Percentage of mitotic cells in Ad-GFP and Ad-Cre-GFP infected cells (G). Ad-Cre-GFP infected P3 Hnf1b flox/flox MEFs were immunostained by DAPI and acetylated-α-tubulin. (H) Abnormal spindle (arrows), (I) anaphase lagging chromosome (arrows), and (J) micronuclei (arrows) are shown in Ad-Cre-GFP infected cells, along with corresponding quantification results. Chromosome numbers were counted by metaphase spread method in Ad-GFP and Ad-Cre-GFP cells. 50 mitotic cells in each group were randomly selected and counted. Representative images are shown in (K) and quantification results are shown in (L). Data are expressed as mean±SD, N=3, *P<0.05; **P<0.01; ***P<0.001.
Genomic instability and aneuploidy are the hallmarks of tumor initiation. To evaluate the impact of Hnf1b loss on genomic stability, chromosome characteristics were examined. In Ad-Cre-GFP infected primary P1–P3 MEFs, cells were more likely to develop spindle abnormalities (Fig. 3H arrow), anaphase DNA lagging (Fig. 3I arrow) and micronuclei (Fig. 3J arrow). Quantification is shown in each corresponding panel. Moreover, we analyzed chromosome number per metaphase to further define chromosome instability. The results demonstrated that more than 90 percent of Ad-Cre-GFP infected cells displayed aneuploidy, with both losses and gains, while more than 40 percent of Ad-GFP cells maintained a diploid genome (Fig. 3K–3L).
Hnf1b deficiency reduced spindle checkpoint protein (MAD2L1, BUB1B) and cell cycle checkpoint protein (RB1 and p27) expression, and altered chromatin access of Mad2l1, Bub1b and Rb1 genes
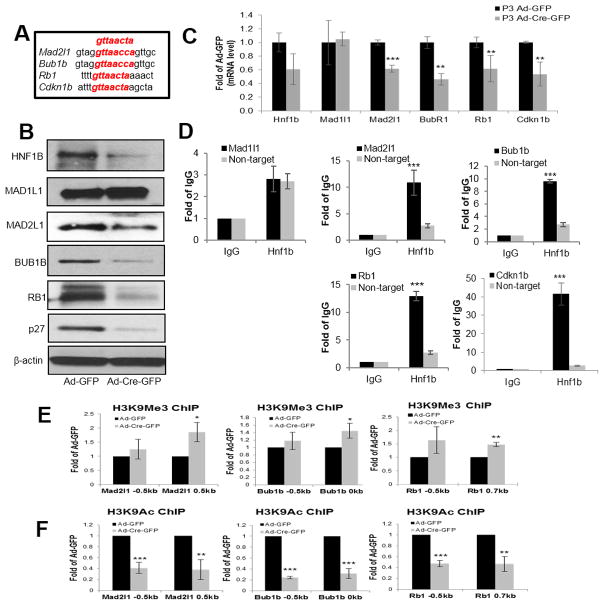
Recent publications showed that HNF1B may be a master regulator of chromatin accessibility, and modulation of HNF1B may regulate a number of genes simultaneously (13) (14). Mutations of various checkpoint proteins, including spindle assembly complex proteins and cell cycle checkpoint proteins, are rare in human malignancies, but aberrant expression of these proteins can induce aneuploidy (9) (32). Animal models of SAC deficiency show the development of tumorigenesis in a tissue and second-hit context dependent manner, and are associated with significant aneuploidy (33). Therefore, we hypothesized that spindle checkpoint proteins and cell cycle checkpoint protein expression levels are altered after Hnf1b depletion. The DNA consensus sequence that murine HNF1B binds to is “gttaacta”. Based on a predicted substrate score created by Jaspar software analysis, SACs Mad1l1, Mad2l1, Bub1b and cell cycle checkpoints Rb1 and Cdkn1b were good candidates (Supplementary Fig. S4A–S4B) and predicted highest score sequences of each substrates were aligned (Fig. 4A). We found that in both primary and immortalized MEFs, depletion of Hnf1b resulted in MAD2L1, BUB1B, RB1 and p27 protein down-regulation (Fig. 4B and Supplementary Fig. S4C), but not MAD1L1 downregulation. We showed that after thymidine treatment, loss of Hnf1b induced the same results. (Supplementary Fig. S4D). We then found mRNA levels of checkpoints Mad2l1, Bub1b, Rb1 and Cdkn1b were significantly down regulated after Hnf1b loss (Fig. 4C). To validate our hypothesis that HNF1B directly regulates these genes, we performed a chromatin immunoprecipitation (ChIP) assay of HNF1B, followed by qRT-PCR against predicted binding site primers of substrates, using immunoglobulin IgG ChIP as a control. We also used a non-targeted primer as an additional control in parallel with predicted binding site primers. ChIP results confirmed our previous observations: Mad2l1, Bub1b, Rb1 and Cdkn1b all had significantly stronger binding affinity than both IgG and non-target primers, whereas Mad1l1 did not (Fig. 4D). HNF1B has been shown to reopen chromatin and facilitate transcription of target genes after mitotic silencing (13) (14), We performed ChIP of repressing (H3K9me3) (Fig. 4E) or activating (H3K9ac) (Fig. 4F) histone H3 modifications of Mad2l1, Bub1b and Rb1 at various locations relative to transcription start sites in Ad-GFP and Ad-Cre-GFP infected primary P3 MEF cells. For Cdkn1b, a different mechanism was proposed (data not shown). As we expected, these genes underwent silencing upon Hnf1b depletion. The acetylation of H3K9 was significantly decreased, and trimethylation of H3K9 was increased, indicating the activating chromatin modification on the target gene was lost after Hnf1b loss. Taken together, our results suggest HNF1B affected the chromatin accessibility of these checkpoint genes in proliferating cells, thereby altering their gene expression levels.
Figure 4. Hnf1b deficiency reduced spindle checkpoint (MAD2L1, BUB1B) and cell cycle checkpoint (RB1, and p27) expression at protein, mRNA levels and affected chromatin access of Mad2l1, Bub1b and Rb1 genes in proliferating MEFs.
Consensus DNA sequence of HNF1B binding aligned with predicted highest score sequences of each substrate (A). Protein levels of HNF1B, MAD1L1, MAD2L1, BUB1B, RB1, p27 and b-actin examined by immunoblotting after Ad-GFP and Ad-Cre-GFP infection for 72 h in P3 primary Hnf1bflox/flox MEFs (B). Relative mRNA levels normalized to Gapdh and determined by qRT-PCR after Ad-GFP and Ad-Cre-GFP infection for 48 h (C). ChIP of HNF1B or IgG against Mad1l1, Mad2l1, Bub1b, Rb1 and Cdkn1b primers based on best postulated binding scores as well as non-targeted primers (D). Chromatin immunoprecipitation of repressing (H3K9me3) (E) and activating (H3K9ac) (F) histone H3 modifications of Mad2l1, Bub1b and Rb1 at two different locations close to transcriptional start site. Data are expressed as mean±SD, N=3, *P<0.05; **P<0.01; ***P<0.001.
Coordinate loss of Bub1b and Rb1 recapitulated polynuclearity and larger cell size seen with Hnf1b depletion
To determine whether loss of these proteins was responsible for the phenotype generated by Hnf1b loss, we knocked down Rb1, p27, Mad2l1 and Bub1b individually and in combination in primary P3 MEFs. Among these combinations, we found that combined loss of Rb1 and Bub1b recapitulated the polynuclear phenotype observed after Hnf1b loss. Similar to Hnf1b loss, knockdown of Bub1b and Rb1 together enlarged the cell size (Supplementary Fig. S5A). Knockdown of Rb1 alone, knockdown of Mad2l1 and Rb1, and knockdown of Rb1, p27, Mad2l1 and Bub1b together induced polynuclearity but to a lesser extent (Supplementary Fig. S5A–S5B). Cell death, another phenotype induced by Hnf1b loss, was also observed in these knockdown combinations in primary MEFs (Supplementary Fig. S5C). The corresponding protein levels are shown in Supplementary Fig. S5D. These data indicate that loss of Rb1 may be sufficient for inducing chromosome instability. Loss of Rb1 and Bub1b together even more closely mimicked the phenotype we observed after Hnf1b loss.
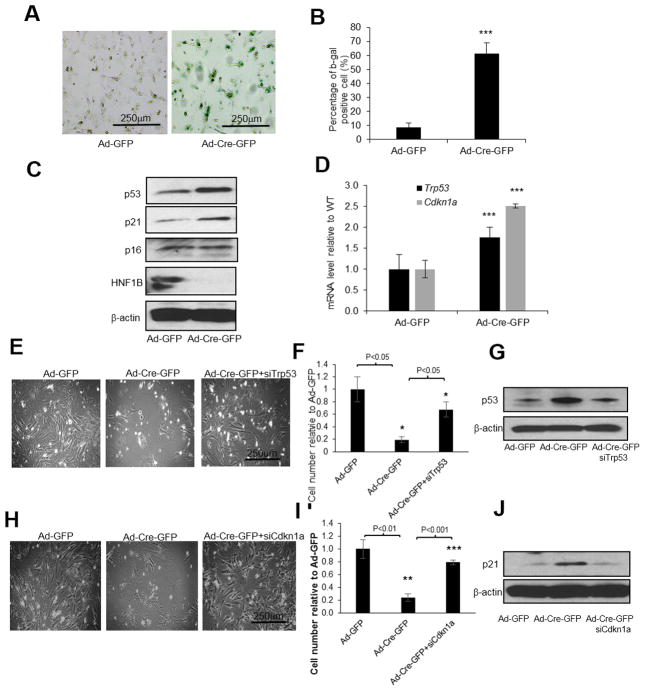
Loss of HNF1B induced p53/p21 pathway activation and cell death
We have so far shown that loss of Hnf1b in proliferating MEFs can result in chromosomal copy number change, followed by cell death. It has been reported that if nontransformed mammalian cells develop polyploidy, tetraploid checkpoint proteins, including p53 and Rb, are activated (34), followed by G1 arrest, induction of senescence and cell death (35). To determine whether Hnf1b loss induced senescence in MEFs, we stained our cells with beta-galactosidase, a senescence marker. As we expected, the intensity of beta galactosidase was dramatically enhanced in Hnf1b null cells (Fig. 5A–5B). As expected, cell death via apoptosis was also induced by Hnf1b loss (data not shown). p53 and p21 protein levels and Trp53 and Cdkn1a mRNA levels were dramatically increased in response to Hnf1b loss (Fig. 5C–5D). In addition, we used siRNA to transiently knock down either Trp53 or Cdkn1a, followed by Cre activation to deplete Hnf1b. We found that cell death was dramatically rescued with either Trp53 (Fig. 5E–5G) or Cdkn1a knockdowns (Fig. 5H–5J), providing evidence that cell death induced by Hnf1b depletion is dependent on tetraploid checkpoint p53/p21 activation.
Figure 5. Inactivation of p53/p21 pathway rescued cell death induced by Hnf1b loss in MEFs.
Primary Hnf1b flox/flox P3 MEF cells were infected by Ad-Cre-GFP. After 48 h, b-gal senescence assay was performed (A) and quantification results are shown in (B). Protein levels of p53, p21, p16 as well as HNF1B were measured by immunoblotting (C). mRNA levels of Trp53 and Cdkn1a were determined by RT-PCR (D). Primary P3 MEFs were transiently transfected with either siRNA against Trp53 or Cdkn1a. After 24 h, cells were infected with Ad-GFP or Ad-Cre-GFP for an additional 48 h. Cell density images, quantification and corresponding protein levels by immunoblotting after Trp53 knockdown (E–G) or Cdkn1a knockdown (H–J) are shown. Data are expressed as mean±SD, N=3, *P<0.05; **P<0.01; ***P<0.001.
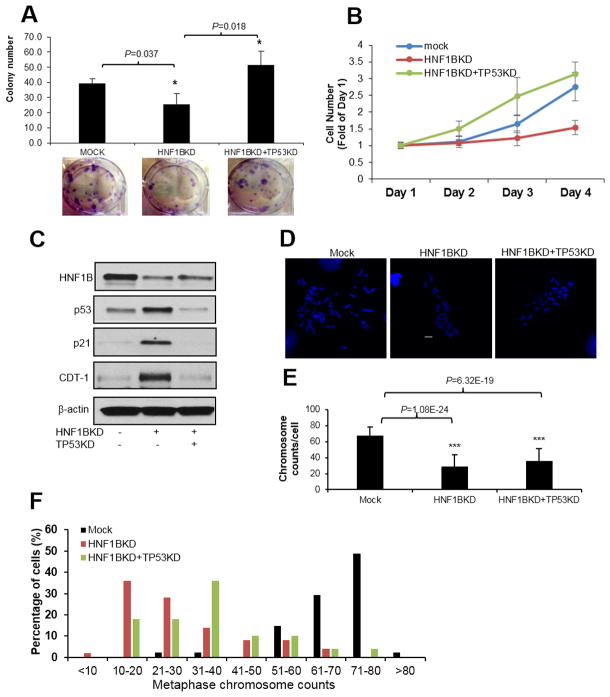
Loss of TP53 in ACHN cells rescued HNF1B loss induced cell death but not aneuploidy
In addition to MEFs, we used the TP53 wildtype human renal cell carcinoma cell line ACHN to examine the effect of TP53 loss in the presence of HNF1B loss on cell survival and aneuploidy. As expected, knockdown of HNF1B dramatically increased cell death. Knockdown of TP53 rescued HNF1B loss induced cell death, as shown by colony formation assays (Fig. 6A) and cell survival assays (Fig. 6B) while TP53 knockdown alone showed a slight increase in cell survival (data not shown). To validate that cell death induced by HNF1B loss in ACHN cells was via activation of the p53 pathway and induction of G1 arrest, protein levels of p53, p21 and CDT-1 were assayed by immunoblotting in these cell lines. Results confirmed the p53/p21 tetraploid checkpoint pathway was activated after HNF1B loss and cells were arrested in G1 phase. Knockdown of TP53 eliminated the induction of p21 and released cells from G1 arrest induced by HNF1B loss (Fig. 6C). We then assessed chromosome number alterations after manipulation of these two genes. HNF1B knockdown significantly reduced chromosome number compared with mock ACHN cells, but TP53 knockdown was not able to rescue HNF1B loss induced aneuploidy (Fig. 6D–6F). TP53 knockdown alone did not affect chromosome number (data not shown).
Figure 6. Loss of TP53 in ACHN cells rescued HNF1B loss induced cell death but not aneuploidy.
HNF1B or both HNF1B and TP53 were knocked down in ACHN cells. Cell proliferation was measured by either colony formation (A) or cell survival assay (B). HNF1B, p53, p21 and CDT-1 levels in ACHN cells were measured by immunoblotting (C). Chromosome number in mock, HNF1BKD and HNF1BKD+TP53KD cells was counted by metaphase spread assay. Images (D) and quantification results (E) are shown. Chromosome count distribution quantification results are shown in (F). 50 mitotic cells in each cell line were randomly counted. Data are expressed as mean±SD, N=3, *P<0.05; **P<0.01; ***P<0.001
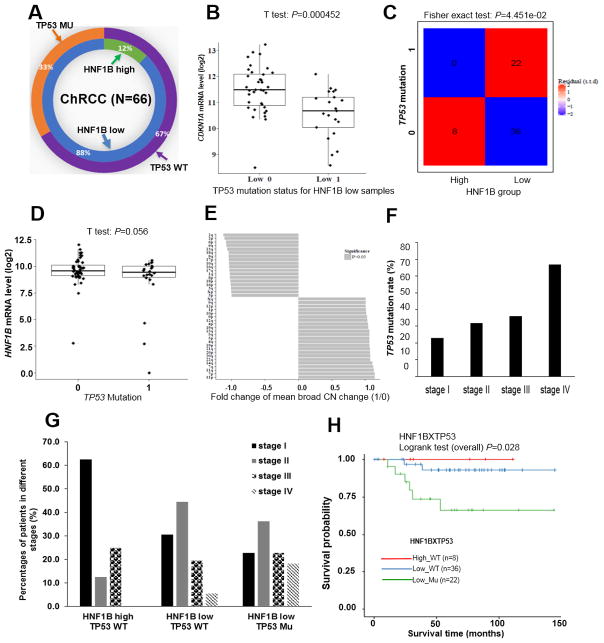
Biallelic inactivation of TP53 facilitates the development of ChRCC in HNF1B low expressing patients
TP53 is one of the most common mutated genes in cancer (36). For unclear reasons, it is rarely mutated in ccRCC (6). However, in sporadic ChRCC, approximately 33% of patients harbor TP53 mutations (7). All of these 33% TP53 mutated patient samples are found in the HNF1B low expression group (Fig. 7A). Furthermore, we analyzed CDKN1A mRNA levels in TP53 mutant samples (N=22) in comparison with TP53 wild type samples (N=36) of these HNF1B low expression patients. As expected, CDKN1A mRNA level was significantly dysregulated by TP53 mutation (Fig. 7B). The TP53 gene is also located on chromosome 17, one copy of which is frequently lost in ChRCC. Similar to our findings with FLCN, overall TP53 mRNA expression in ChRCC tumors is maintained at about 79.1% of normal levels (data not shown). To determine whether a correlation exists between low HNF1B expression and TP53 mutations, we performed a Fisher exact test and found a positive correlation (P=0.045) (Fig. 7C). In the TP53 mutated group, there was no difference in HNF1B mRNA levels compared with the TP53 wildtype group (Fig. 7D), suggesting that a decrease in HNF1B might precede the TP53 mutation. Although TP53 was reported to limit aneuploidy induced proliferation and tumorigenesis (37) (38), its inactivation is not causally related to aneuploidy (39). To determine whether TP53 status is associated with aneuploidy in ChRCC, we examined the correlation between TP53 status and CN alterations. There was no significant correlation found between any single chromosome CN change and TP53 mutational status (Fig. 7E). We then determined the relationship between TP53 status and tumor stage, and found TP53 mutation rates increased with increasing stage (Fig. 7F). Furthermore, we divided patients into three groups according to HNF1B expression level and TP53 mutation status. Results showed that in the HNF1B high-TP53 wild type group, about 60% patients had stage I disease, and no patients had stage IV disease. In the HNF1B low-TP53 wild type group, a larger number of patients had stage II and stage III disease, and about 5.1% patients had stage IV disease In the HNF1B low-TP53 mutant group, fully 18.2% of the patients had stage IV disease (Fig. 7G) and a significantly lower survival probability (Fig. 7H).
Figure 7. Biallelic inactivation of TP53 is associated with low HNF1B expression and a lethal phenotype in ChRCC patients.
HNF1B expression and TP53 mutation status in 66 ChRCC patients from TCGA database. WT: wild type. MU: mutant (A). CDKN1A mRNA levels in WT TP53/HNF1B low and mutant TP53/HNF1B low samples (B). Association between HNF1B expression level and TP53 mutation status using Fisher’s exact test (C). HNF1B mRNA levels in TP53 wildtype (N=44) and mutated (N=22) samples using T-test (D). Correlation between TP53 mutation and chromosome CN change (E). TP53 mutation rate in stage (I–IV) patients (F). ChRCC patients were divided into three groups according to HNF1B expression level and TP53 mutation status. Percentage of stage distribution in each group is shown (G). Survival probability curves from patients in using log rank test (H).
Discussion
CIN and aneuploidy are two of the hallmarks of cancer (8). Nonetheless, patterns of aneuploidy vary significantly between RCC histologies. ChRCC is characterized by losses in chromosomes 1, 2, 6, 10 and 17. These losses are present early, and remain relatively stable throughout stages I–III, suggesting they are linked to the acquisition of a ChRCC phenotype In cell line experiments, the induction of aneuploidy is frequently associated with decreased cellular viability (Fig. 3D) (8). Thus, although a mitotic defect exists in ChRCC that creates a pattern of widespread and specific aneuploidy, this aneuploidy of itself does not drive a lethal tumor phenotype. Recent data from TCGA suggest that ChRCC tumor viability may be enabled by the CN loss of other checkpoints including CDKN1A, which will allow ChRCC cells to survive despite its inherently unfavorable aneuploid state (7).
The underlying drivers of CIN and aneuploidy in ChRCC have remained elusive to date. Our findings suggest that attenuated HNF1B expression may be responsible for CIN and induction of the aneuploid phenotype in ChRCC. Our data show that decreased levels of HNF1B exist even in tumors that possess both copies of chromosome 17, and that HNF1B levels do not follow CN stoichiometry (Fig. 2F). These data lend credence to the hypothesis that loss of HNF1B expression is a bona fide early event, and an initiator of ChRCC.
Our cell line experiments, using both knockout and knockdown of Hnf1b in MEFs, provide strong evidence that the HNF1B protein acts as a regulator of SAC gene expression. The SAC is the major gatekeeper of intact mitotic spindle function and its role in the maintenance of chromosome stability has been demonstrated in numerous animal models including Caenorhabditis elegans, Drosophila, and mice (40) (41) (42). Mutations of various checkpoint proteins, including spindle assembly checkpoint and cell cycle checkpoints, are well-studied possible causes of CIN (32). Biallelic mutation of Bub1b, another central SAC protein, was reported to cause constitutional aneuploidy and cancer predisposition (43). Inactivating one allele of Mad2 was shown to induce aneuploidy and tumorigenesis in mice (44) (42). Therefore, loss of a transcription factor that controls expression of several SAC genes is likely to induce CIN. While we clearly see the development of CIN, with wholesale decrease in chromosomal number in some cells, as well as increase in others (Fig. 3L), the pattern of CN change we observe in our MEF model does not exactly recapitulate those seen in ChRCC. However, in human ACHN cells, HNF1B loss induced a dramatic reduction in the overall chromosomal copy number, which matches the broad pattern seen in ChRCC, suggesting that species and cell line specific differences in programming determine which CN changes are permissible and viable.
In our current study, loss of HNF1B induced G1 arrest in both MEFs (Fig. 3, Supplementary Fig. S3) and human ACHN cells (Fig. 6). We found that senescence was greatly elevated in Hnf1b deficient MEFs. p53 is a well-known tetraploid checkpoint (35). As expected, we found that p53 and p21 were activated in our cells. Furthermore, inactivation of p53/p21 by either siRNA knockdown of Trp53 or Cdkn1a in MEFs or knockdown of TP53 in ACHN cells was sufficient to rescue the senescence and cell death induced by HNF1B depletion. TCGA analysis provides similar observations. Biallelic inactivation of TP53 was found in 22 out of 66 (33%) specimens. Each of these 22 specimen expressed low HNF1B, suggesting low expression of HNF1B antedates TP53 mutation, and the addition of mutated TP53 is a key driver of a lethal ChRCC phenotype. The absence of universal TP53 loss in the remaining ChRCC specimens suggests there are other factors besides TP53 loss that provide a permissive environment for cellular survival and the maintenance of a nonlethal ChRCC phenotype.
Taken together, our results demonstrate that decrease or loss of HNF1B expression leads to SAC, RB1 and p27 repression and creates chromosome instability. The additional loss of Trp53/Cdkn1a enables these cells to evade senescence and death. In vivo experiments in ACHN cells further validate the significant roles of combined HNF1B and TP53 loss in generating CN abnormalities and tumor growth. TCGA data analyses confirmed patients with loss of HNF1B together with TP53 inactivation have a worse prognosis. Our aggregate findings provide evidence that decreased expression of HNF1B may be a key driver of ChRCC development, and the additional loss of TP53 may engender a lethal phenotype (Supplementary Fig. S6). These findings provide a foundation for the development of relevant animal models, which will ultimately lead to a better understanding of ChRCC pathogenesis and permit the development of therapies that specifically target ChRCC biology.
Supplementary Material
Acknowledgments
We gratefully acknowledge the excellent technical support from Microscope Core Facility and Flow Cytometry core at the Texas A&M University Institute of Biosciences and Technology
Financial support:
E. Jonasch received philanthropic support from The George A. Robinson Foundation, Catherine and Dewey Stringer, Judy Stringer, the Monteleone Family Foundation and the Cure Fur Cancer Foundation. This work was also supported by the NIH/NCI Core Grant under award number P30CA016672-39 (E. Jonasch) and used the University of Texas MD Anderson Cancer Center shRNA Core Facility.
References
- 1.Thoenes W, Storkel S, Rumpelt HJ. Human chromophobe cell renal carcinoma. Virchows Archiv B, Cell pathology including molecular pathology. 1985;48:207–17. doi: 10.1007/BF02890129. [DOI] [PubMed] [Google Scholar]
- 2.Storkel S, Eble JN, Adlakha K, Amin M, Blute ML, Bostwick DG, et al. Classification of renal cell carcinoma: Workgroup No. 1. Union Internationale Contre le Cancer (UICC) and the American Joint Committee on Cancer (AJCC) Cancer. 1997;80:987–9. doi: 10.1002/(sici)1097-0142(19970901)80:5<987::aid-cncr24>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 3.Prasad SR, Narra VR, Shah R, Humphrey PA, Jagirdar J, Catena JR, et al. Segmental disorders of the nephron: histopathological and imaging perspective. The British journal of radiology. 2007;80:593–602. doi: 10.1259/bjr/20129205. [DOI] [PubMed] [Google Scholar]
- 4.Davis CF, Ricketts CJ, Wang M, Yang L, Cherniack AD, Shen H, et al. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer cell. 2014;26:319–30. doi: 10.1016/j.ccr.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Speicher MR, Schoell B, du Manoir S, Schrock E, Ried T, Cremer T, et al. Specific loss of chromosomes 1, 2, 6, 10, 13, 17, and 21 in chromophobe renal cell carcinomas revealed by comparative genomic hybridization. The American journal of pathology. 1994;145:356–64. [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–9. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research N. Linehan WM, Spellman PT, Ricketts CJ, Creighton CJ, Fei SS, et al. Comprehensive Molecular Characterization of Papillary Renal-Cell Carcinoma. The New England journal of medicine. 2016;374:135–45. doi: 10.1056/NEJMoa1505917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nature reviews Genetics. 2012;13:189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- 9.Varetti G, Pellman D, Gordon DJ. Aurea mediocritas: the importance of a balanced genome. Cold Spring Harbor perspectives in biology. 2014;6:a015842. doi: 10.1101/cshperspect.a015842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nickerson ML, Warren MB, Toro JR, Matrosova V, Glenn G, Turner ML, et al. Mutations in a novel gene lead to kidney tumors, lung wall defects, and benign tumors of the hair follicle in patients with the Birt-Hogg-Dube syndrome. Cancer cell. 2002;2:157–64. doi: 10.1016/s1535-6108(02)00104-6. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt LS, Warren MB, Nickerson ML, Weirich G, Matrosova V, Toro JR, et al. Birt-Hogg-Dube syndrome, a genodermatosis associated with spontaneous pneumothorax and kidney neoplasia, maps to chromosome 17p11. 2. American journal of human genetics. 2001;69:876–82. doi: 10.1086/323744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagy A, Zoubakov D, Stupar Z, Kovacs G. Lack of mutation of the folliculin gene in sporadic chromophobe renal cell carcinoma and renal oncocytoma. International journal of cancer Journal international du cancer. 2004;109:472–5. doi: 10.1002/ijc.11694. [DOI] [PubMed] [Google Scholar]
- 13.Verdeguer F, Le Corre S, Fischer E, Callens C, Garbay S, Doyen A, et al. A mitotic transcriptional switch in polycystic kidney disease. Nature medicine. 2010;16:106–10. doi: 10.1038/nm.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Massa F, Garbay S, Bouvier R, Sugitani Y, Noda T, Gubler MC, et al. Hepatocyte nuclear factor 1beta controls nephron tubular development. Development. 2013;140:886–96. doi: 10.1242/dev.086546. [DOI] [PubMed] [Google Scholar]
- 15.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–7. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barbacci E, Reber M, Ott MO, Breillat C, Huetz F, Cereghini S. Variant hepatocyte nuclear factor 1 is required for visceral endoderm specification. Development. 1999;126:4795–805. doi: 10.1242/dev.126.21.4795. [DOI] [PubMed] [Google Scholar]
- 17.Cereghini S, Ott MO, Power S, Maury M. Expression patterns of vHNF1 and HNF1 homeoproteins in early postimplantation embryos suggest distinct and sequential developmental roles. Development. 1992;116:783–97. doi: 10.1242/dev.116.3.783. [DOI] [PubMed] [Google Scholar]
- 18.Bingham C, Hattersley AT. Renal cysts and diabetes syndrome resulting from mutations in hepatocyte nuclear factor-1beta. Nephrology, dialysis, transplantation : official publication of the European Dialysis and Transplant Association - European Renal Association. 2004;19:2703–8. doi: 10.1093/ndt/gfh348. [DOI] [PubMed] [Google Scholar]
- 19.Gresh L, Fischer E, Reimann A, Tanguy M, Garbay S, Shao X, et al. A transcriptional network in polycystic kidney disease. The EMBO journal. 2004;23:1657–68. doi: 10.1038/sj.emboj.7600160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clissold RL, Hamilton AJ, Hattersley AT, Ellard S, Bingham C. HNF1B-associated renal and extra-renal disease-an expanding clinical spectrum. Nature reviews Nephrology. 2015;11:102–12. doi: 10.1038/nrneph.2014.232. [DOI] [PubMed] [Google Scholar]
- 21.Horikawa Y, Iwasaki N, Hara M, Furuta H, Hinokio Y, Cockburn BN, et al. Mutation in hepatocyte nuclear factor-1 beta gene (TCF2) associated with MODY. Nat Genet. 1997;17:384–5. doi: 10.1038/ng1297-384. [DOI] [PubMed] [Google Scholar]
- 22.Ryffel GU. Mutations in the human genes encoding the transcription factors of the hepatocyte nuclear factor (HNF)1 and HNF4 families: functional and pathological consequences. J Mol Endocrinol. 2001;27:11–29. doi: 10.1677/jme.0.0270011. [DOI] [PubMed] [Google Scholar]
- 23.Bellanne-Chantelot C, Chauveau D, Gautier JF, Dubois-Laforgue D, Clauin S, Beaufils S, et al. Clinical spectrum associated with hepatocyte nuclear factor-1beta mutations. Annals of internal medicine. 2004;140:510–7. doi: 10.7326/0003-4819-140-7-200404060-00009. [DOI] [PubMed] [Google Scholar]
- 24.Lebrun G, Vasiliu V, Bellanne-Chantelot C, Bensman A, Ulinski T, Chretien Y, et al. Cystic kidney disease, chromophobe renal cell carcinoma and TCF2 (HNF1 beta) mutations. Nature clinical practice Nephrology. 2005;1:115–9. doi: 10.1038/ncpneph0054. [DOI] [PubMed] [Google Scholar]
- 25.Rebouissou S, Vasiliu V, Thomas C, Bellanne-Chantelot C, Bui H, Chretien Y, et al. Germline hepatocyte nuclear factor 1alpha and 1beta mutations in renal cell carcinomas. Human molecular genetics. 2005;14:603–14. doi: 10.1093/hmg/ddi057. [DOI] [PubMed] [Google Scholar]
- 26.Buchner A, Castro M, Hennig A, Popp T, Assmann G, Stief CG, et al. Downregulation of HNF-1B in renal cell carcinoma is associated with tumor progression and poor prognosis. Urology. 2010;76:507e6–11. doi: 10.1016/j.urology.2010.03.042. [DOI] [PubMed] [Google Scholar]
- 27.Wang CC, Mao TL, Yang WC, Jeng YM. Underexpression of hepatocyte nuclear factor-1beta in chromophobe renal cell carcinoma. Histopathology. 2013;62:589–94. doi: 10.1111/his.12026. [DOI] [PubMed] [Google Scholar]
- 28.Xu J. Preparation, culture, and immortalization of mouse embryonic fibroblasts. Curr Protoc Mol Biol. 2005;Chapter 28(Unit 28):1. doi: 10.1002/0471142727.mb2801s70. [DOI] [PubMed] [Google Scholar]
- 29.Tong P, Chen Y, Su X, Coombes KR. SIBER: systematic identification of bimodally expressed genes using RNAseq data. Bioinformatics. 2013;29:605–13. doi: 10.1093/bioinformatics/bts713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coffinier C, Gresh L, Fiette L, Tronche F, Schutz G, Babinet C, et al. Bile system morphogenesis defects and liver dysfunction upon targeted deletion of HNF1beta. Development. 2002;129:1829–38. doi: 10.1242/dev.129.8.1829. [DOI] [PubMed] [Google Scholar]
- 31.Wohlschlegel JA, Dwyer BT, Dhar SK, Cvetic C, Walter JC, Dutta A. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science. 2000;290:2309–12. doi: 10.1126/science.290.5500.2309. [DOI] [PubMed] [Google Scholar]
- 32.Colombo R, Moll J. Destabilizing aneuploidy by targeting cell cycle and mitotic checkpoint proteins in cancer cells. Curr Drug Targets. 2010;11:1325–35. doi: 10.2174/1389450111007011325. [DOI] [PubMed] [Google Scholar]
- 33.Baker DJ, Jin F, Jeganathan KB, van Deursen JM. Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer cell. 2009;16:475–86. doi: 10.1016/j.ccr.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borel F, Lohez OD, Lacroix FB, Margolis RL. Multiple centrosomes arise from tetraploidy checkpoint failure and mitotic centrosome clusters in p53 and RB pocket protein-compromised cells. Proc Natl Acad Sci U S A. 2002;99:9819–24. doi: 10.1073/pnas.152205299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Campisi J, d’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nature reviews Molecular cell biology. 2007;8:729–40. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 36.Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harbor perspectives in biology. 2010;2:a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li M, Fang X, Baker DJ, Guo L, Gao X, Wei Z, et al. The ATM-p53 pathway suppresses aneuploidy-induced tumorigenesis. Proc Natl Acad Sci U S A. 2010;107:14188–93. doi: 10.1073/pnas.1005960107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson SL, Compton DA. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol. 2010;188:369–81. doi: 10.1083/jcb.200905057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bunz F, Fauth C, Speicher MR, Dutriaux A, Sedivy JM, Kinzler KW, et al. Targeted inactivation of p53 in human cells does not result in aneuploidy. Cancer Res. 2002;62:1129–33. [PubMed] [Google Scholar]
- 40.Baker DJ, Jeganathan KB, Cameron JD, Thompson M, Juneja S, Kopecka A, et al. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet. 2004;36:744–9. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- 41.Kitagawa R, Rose AM. Components of the spindle-assembly checkpoint are essential in Caenorhabditis elegans. Nature cell biology. 1999;1:514–21. doi: 10.1038/70309. [DOI] [PubMed] [Google Scholar]
- 42.Michel LS, Liberal V, Chatterjee A, Kirchwegger R, Pasche B, Gerald W, et al. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409:355–9. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- 43.Hanks S, Coleman K, Reid S, Plaja A, Firth H, Fitzpatrick D, et al. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat Genet. 2004;36:1159–61. doi: 10.1038/ng1449. [DOI] [PubMed] [Google Scholar]
- 44.Dobles M, Liberal V, Scott ML, Benezra R, Sorger PK. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell. 2000;101:635–45. doi: 10.1016/s0092-8674(00)80875-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.