SUMMARY
AMP-activated protein kinase (AMPK) is a master sensor and regulator of cellular energy status. Upon metabolic stress, AMPK suppresses anabolic and promotes catabolic processes to regain energy homeostasis. Cancer cells can occasionally suppress the growth restrictive AMPK pathway by mutation of an upstream regulatory kinase. Here, we describe a widespread mechanism to suppress AMPK through its ubiquitination and degradation by the cancer-specific MAGE-A3/6-TRIM28 ubiquitin ligase. MAGE-A3 and MAGE-A6 are highly similar proteins normally expressed only in the male germline, but frequently re-activated in human cancers. MAGE-A3/6 are necessary for cancer cell viability and sufficient to drive tumorigenic properties of non-cancerous cells. Screening for targets of MAGE-A3/6-TRIM28 revealed that it ubiquitinates and degrades AMPKα1. This leads to inhibition of autophagy, activation of mTOR signaling, and hypersensitization to AMPK agonists, such as metformin. These findings elucidate a germline mechanism commonly hijacked in cancer to suppress AMPK.
INTRODUCTION
Cells must coordinate multiple metabolic processes in order to balance their energy usage with nutrient availability. One prominent way that this balance is accomplished is through the activity of the AMP-activated protein kinase (AMPK). AMPK is a hetero-trimeric kinase comprised of catalytic α and regulatory β and γ subunits that is regulated by the cellular concentrations of ATP, ADP, and AMP (Hardie et al., 2012b). When cellular levels of ATP fall and ADP/AMP rise, ATP that is bound to the γ subunit is replaced by ADP and/or AMP, resulting in activation of the catalytic kinase subunit (Landgraf et al., 2013; Suter et al., 2006). Once activated, AMPK generally opposes anabolic energy-consuming pathways while promoting catabolic ATP-generating pathways. For example, AMPK inhibits ACC1 and mTOR to block fatty acid and protein synthesis, respectively, while at the same time it promotes autophagy via multiple pathways involving mTOR, ULK1 and VPS34 (Egan et al., 2011; Gwinn et al., 2008; Hardie et al., 2012b; Kim et al., 2013; Kim et al., 2011). In addition to changes in energy levels, upstream kinases such as LKB1/STK11 and CaMKK regulate AMPK activity by phosphorylation of its activation loop at T172 (Hawley et al., 2003; Hawley et al., 2005; Shaw et al., 2004; Woods et al., 2005).
Although AMPK may in some cases promote late-stage tumor growth (Laderoute et al., 2014), multiple lines of evidence suggest AMPK has critical tumor suppressor activities in both humans and experimental models, including mice (Hardie and Alessi, 2013; Shackelford and Shaw, 2009). For example, knockout of AMPKα1 in the mouse accelerates development of c-Myc-driven lymphomas (Faubert et al., 2013). AMPK’s role in suppressing tumor initiation and progression is multifaceted, including growth suppression by inhibiting synthesis of cellular macromolecules (Hardie et al., 2012b), particularly through downregulating the mTOR signaling pathway (Gwinn et al., 2008; Inoki et al., 2003), and promoting cell cycle arrest through stabilizing p53 and cyclin-dependent kinase inhibitors (Imamura et al., 2001; Jones et al., 2005; Liang et al., 2007). Additionally, AMPK can oppose the Warburg effect in favor of oxidative phosphorylation through up-regulating oxidative enzymes and promoting mitochondrial biogenesis (Canto et al., 2009; Winder et al., 2000). Furthermore, AMPK has recently been shown to inhibit epithelial-to-messenchymal transition (EMT) by modulating the Akt-MDM2-Foxo3 signaling axis (Chou et al., 2014).
Given the importance of metabolic control and AMPK’s role as master sensor and regulator of cellular energy, it is not surprising that this signaling axis is de-regulated in a variety of disease states, including cancer (Hardie and Alessi, 2013; Shackelford et al., 2009). For example, in approximately 20% of lung adenocarcinomas and cervical cancers, signaling through this axis is reduced by loss of function mutation or deletion of Lkb1/Stk11 (Matsumoto et al., 2007; Sanchez-Cespedes et al., 2002; Wingo et al., 2009). Additionally, AMPK levels have been shown to be reduced in some cases of hepatocellular carcinomas and B-RAF V600E can downregulate AMPK signaling through inhibition of Lkb1/Stk11 in melanomas (Esteve-Puig et al., 2009; Lee et al., 2012; Zheng et al., 2009; Zheng et al., 2013). From these multiple lines of converging evidence on AMPK’s critical role in tumor suppression, there is great interest in the utilization of compounds that stimulate AMPK activity, such as metformin, in the prevention and treatment of cancer and many clinical trials are ongoing (Hadad et al., 2011; Hardie et al., 2012a; Niraula et al., 2012; Pernicova and Korbonits, 2014).
Melanoma antigen (MAGE) genes are conserved in all eukaryotes, encode for proteins with a common MAGE homology domain, and have rapidly expanded to comprise almost 50 unique genes in humans (Chomez et al., 2001; Feng et al., 2011). Approximately two-thirds of human MAGEs are considered cancer-testis antigens because they are normally restricted to expression in the testis, but are aberrantly re-expressed in cancer and have antigenic properties (Simpson et al., 2005). The functional significance of MAGEs in tumors is not well understood, but accumulating evidence supports their importance. For example, knockdown of MAGE-A3/6 impairs tumor growth in mice, whereas expression of MAGE-A3 in MAGE-negative cells drives tumor growth and metastasis in vivo (Liu et al., 2008; Yang et al., 2007). Importantly, we recently showed that a defining characteristic of MAGE proteins is their ability to bind and potentiate the activity of specific E3 ubiquitin ligases (Doyle et al., 2010). For example, MAGE-L2 binds to the TRIM27 ubiquitin ligase and promotes ubiquitination of the WASH actin assembly complex to facilitate endosomal protein recycling (Hao et al., 2013).
Here, we present evidence for a regulatory axis engaged in cancer cells that downregulates AMPK through ubiquitination and degradation of AMPKα1 by the normally testis-restricted MAGE-A3/6-TRIM28 E3 ubiquitin ligase complex activated in cancer. These findings identify a widespread mechanism for downregulating AMPK signaling during tumorigenesis and elucidate an unanticipated mechanism of action for oncogenic MAGE cancer-testis antigens.
RESULTS
MAGE-A3 and MAGE-A6 are physiologically restricted to expression in the testis, but are aberrantly expressed in cancer
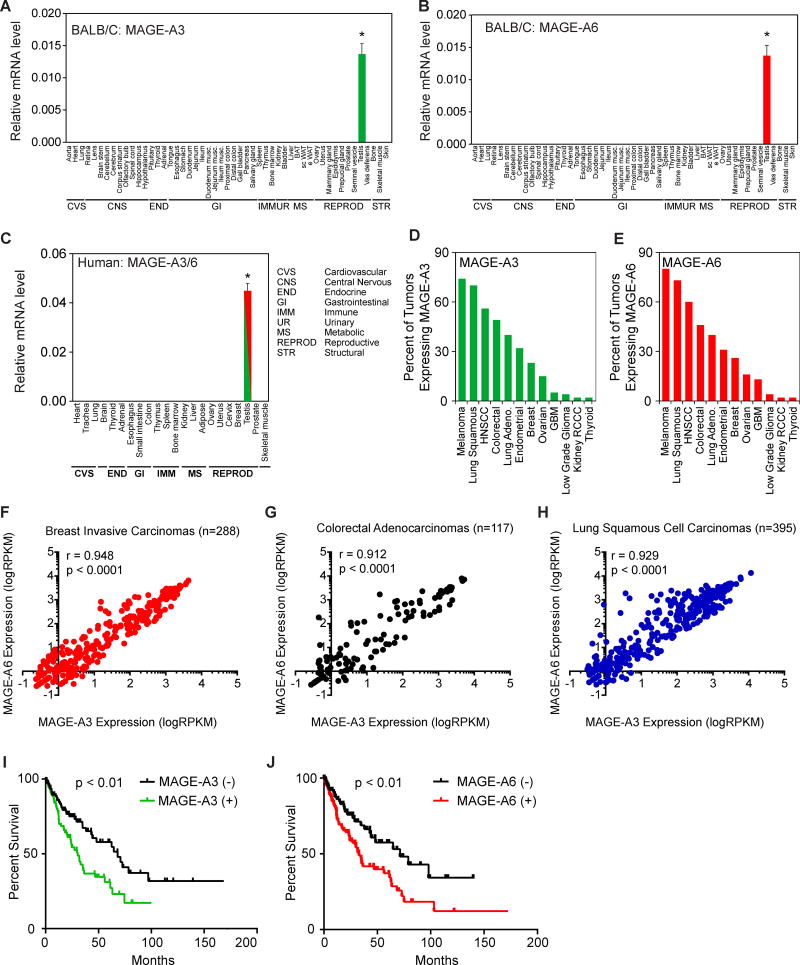
MAGE-A3 and MAGE-A6 are highly similar, neighboring genes located on the X chromosome that encode proteins with 96% identity (Figure S1A–B). Given their homology and functional redundancy (see below) we refer to these genes as simply MAGE-A3/6 herein. To thoroughly examine the expression pattern of MAGE-A3/6, we analyzed their expression by RT-QPCR in >50 mouse tissues from two strains of mice (C57BL/6 and BALB/C). Consistent with previous findings (De Plaen et al., 1994), mouse MAGE-A3/6 were completely restricted to expression in the testis with no detectable expression in any other tissue (Figures 1A–B, S1C–D). We extended these analyses to a panel of >20 human tissues and found that human MAGE-A3/6 are similarly restricted to expression only in the human testis (Figure 1C). Like other cancer-testis antigen genes, MAGE-A3/6 have been reported to be aberrantly expressed in tumors (Jang et al., 2001; Shantha Kumara et al., 2012). Our analysis of a variety of different tumors types from patients revealed that MAGE-A3/6 are commonly expressed in many cancer types, including breast invasive carcinomas (25%), colon adenocarcinomas (50%), and lung squamous cell carcinomas (75%; Figure 1D–E). Additionally, expression of MAGE-A3 and MAGE-A6 was significantly correlated in breast invasive carcinomas, colon adenocarcinomas, and lung squamous cell carcinomas (Figure 1F–H). However, expression of MAGE-A3 was not significantly correlated with expression of unrelated MAGE-A11 or MAGE-B2 (Figure S1E–F), suggesting that MAGE-A3 and MAGE-A6 expression is selectively coordinated. Furthermore, to determine whether MAGE-A3/6 expression correlates with patient outcome, we analyzed whether expression of MAGE-A3/6 correlates with overall survival. Indeed, patients with lung squamous cell carcinomas expressing MAGE-A3 or MAGE-A6 have a significant decrease in overall survival time (Figure 1I–J). Patients with tumors expressing MAGE-A3 had a >50% reduced overall survival time compared to patients with MAGE-A3-negative tumors (30 versus 69 months, respectively; 2.0 hazard ratio; Supplemental Table 1). Similarly, patients with tumors expressing MAGE-A6 had a >50% reduced survival time (33 vs 71 months; hazard ratio of 1.9; Supplemental Table 1). Together, these results suggest MAGE-A3/6 are physiologically restricted to the testis in both humans and mice, but are frequently found in a wide variety of cancer types and their expression correlates with poor patient prognosis.
Figure 1. MAGE-A3 and MAGE-A6 are normally restricted to expression in the testis, but are aberrantly expressed in cancer and predict poor patient prognosis.
(A–B) RT-QPCR analysis (n=3) of the normalized expression of mouse MAGE-A3 (A) and MAGE-A6 (B) in the indicated tissues from BALB/C mice.
(C) RT-QPCR analysis (n=3) of the normalized expression of human MAGE-A3/6 (one primer set detects both) in the indicated human tissues.
(D–E) Percent of patient tumors expressing MAGE-A3 (D) and MAGE-A6 (E) is shown.
(F–H) MAGE-A3 and MAGE-A6 are co-expressed in breast invasive carcinomas (F), colorectal adenocarcinomas (G), and lung squamous cell carcinomas (H).
(I–J) Expression of MAGE-A3 (C) or MAGE-A6 (D) in patients with lung squamous carcinomas correlates with poor overall survival.
Data are represented as the mean +SD. Asterisks indicates p<0.05. See also Figure S1 and Table S1.
MAGE-A3/6 are required for cancer cell viability and function as oncogenes
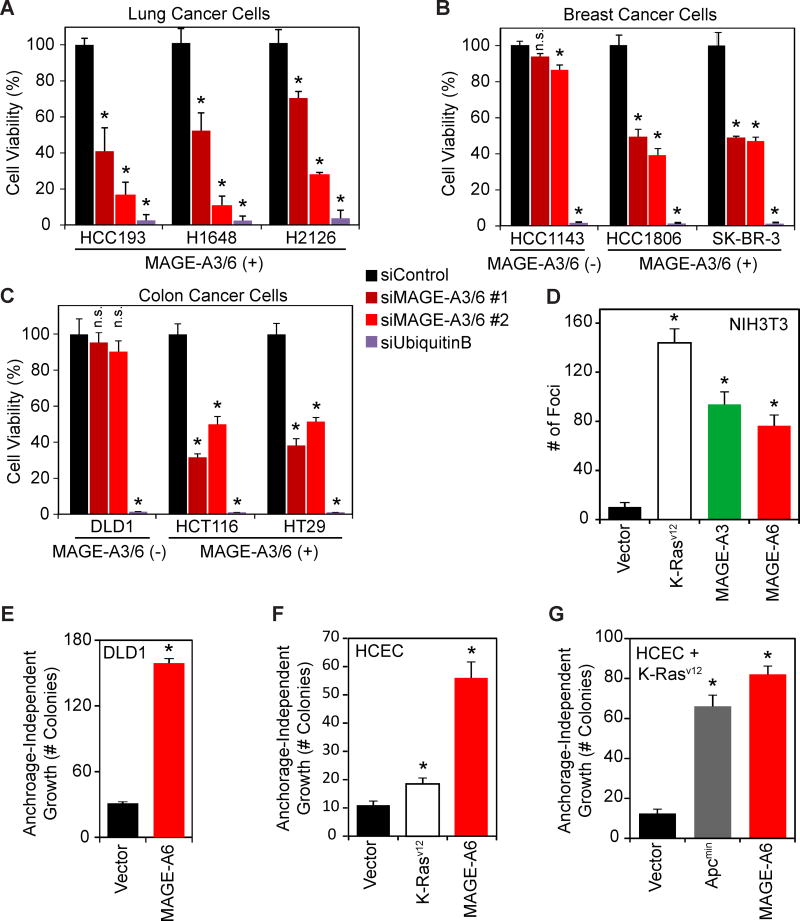
MAGE-A3/6 could be “passenger” genes that have little functional role or significance in tumorigenesis and are simply biomarkers. Alternatively, MAGE-A3/6 may be oncogenic “driver” genes that are involved in promoting tumor initiation and/or progression. To determine if MAGE-A3/6 have important functional roles in cancer cells, we examined whether patient-derived breast, colon, and lung cancer cells require the expression of MAGE-A3/6 for viability. Indeed, knockdown of MAGE-A3/6 using two independent siRNAs (Figure S2A–B) in multiple lung (HCC193, H1648, and H2126), breast (HCC1806 and SK-BR-3), and colon (HCT116 and HT29) cancer cells resulted in a significant decrease in cell viability and clonogenic survival (Figures 2A–C and S2C–D). Importantly, these effects are likely on-target, because MAGE-A3/6 siRNAs do not significantly alter the viability of MAGE-A3/6-negative cells (HCC1143 and DLD1), even though the cytotoxic siRNA targeting Ubiquitin B was just as lethal in all cell lines (Figure 2A–C). These results suggest that upon expression of MAGE-A3/6, cells become dependent on their expression for viability, similar to other reports of “oncogene addiction” (Weinstein, 2002).
Figure 2. MAGE-A3/6 are potent oncogenes whose expression is necessary for viability of cancer cells and is sufficient to transform cells.
(A–C) MAGE-A3/6 depletion reduces viability of MAGE-A3/6 expressing tumor cell lines. Lung (A), breast (B), and colon (C) cancer cells were treated with siControl, two distinct siMAGE-A3/6, or cytotoxic siUbiquitinB for a transfection control. Cell viability was measured by MTT assay.
(D) MAGE-A3 and MAGE-A6 have oncogenic activity. NIH3T3 fibroblasts were transfected with MAGE-A3, MAGE-A6, or mutant K-Rasv12 as a positive-control and foci formation was assayed. Foci were stained with crystal violet and counted.
(E) MAGE-A6 promotes anchorage-independent growth of DLD1 colon cancer cells. MAGE-A-negative DLD1 cells stably expressing vector or MAGE-A6 were assayed for anchorage-independent growth in soft agar colony formation assays.
(F–G) MAGE-A6 promotes anchorage-independent growth of non-transformed, immortalized human colonic epithelial cells (HCECs) without (F) or with (G) expression of mutant K-Rasv12. The indicated HCEC cells were assayed for anchorage-independent growth in soft agar colony formation assays.
Data (n≥3) are represented as the mean +SD. Asterisks indicates p<0.05. See also Figure S2
To determine whether MAGE-A3/6 are indeed oncogenic driver genes, we analyzed their activity in several classical assays. First, expression of either MAGE-A3 or MAGE-A6 significantly stimulated foci formation of NIH3T3 cells (Figure 2D). Furthermore, MAGE-A6 promoted other hallmarks of cancer, such as anchorage-independent growth of the MAGE-A3/6-negative DLD1 colon cancer cells (Figure 2E). Finally, to more stringently assay the oncogenic activity of MAGE-A3/6, we determined the ability of MAGE-A6 to promote tumorigenic phenotypes in non-transformed, human colonic epithelial cells (HCECs) derived from normal colon biopsies and immortalized with CDK4 and hTERT (Roig et al., 2010). Remarkably, stable expression of MAGE-A6 alone was sufficient to drive transformation of these cells, resulting in robust anchorage-independent growth in soft agar (Figure 2F). In this setting, the oncogenic activity of MAGE-A6 was even more robust than expression of the bona fide K-Rasv12 oncogene (Figure 2F). Additionally, even in the context of mutant K-Rasv12 expression, MAGE-A6 was still competent to drive anchorage-independent growth of HCEC cells to a similar degree as the Apcmin oncogene (Figure 2G). Consistent with these findings, MAGE-A3/6 drives increased tumor growth and metastasis in an orthotopic xenograft mouse model of thyroid cancer (Liu et al., 2008). Collectively, these findings suggest that MAGE-A3/6 are potent driver oncogenes that have therapeutic potential.
The MAGE-A3/6-TRIM28 E3 ubiquitin ligase ubiquitinates and degrades AMPKα1
Previously, we reported that MAGE proteins, including MAGE-A3/6, form complexes with specific E3 ubiquitin ligases to regulate ubiquitination (Doyle et al., 2010; Hao et al., 2013). MAGE-A3/6 specifically bind to the TRIM28 E3 ubiquitin ligase, also known as KAP1 (Doyle et al., 2010). We and others have previously shown that MAGE-A proteins can inhibit the critical p53 tumor suppressor, including via MAGE-A-TRIM28-induced ubiquitination and proteasome-mediated degradation of p53 (Doyle et al., 2010; Marcar et al., 2010; Monte et al., 2006; Wang et al., 2005; Yang et al., 2007). However, several of the cell lines dependent on MAGE-A3/6 for viability (Figure 2A–C) are p53 null (HCC1806 and H1648) or mutant (HCC193, SK-BR-3, and HT-29). Additionally, MAGE-A3/6 stimulated anchorage-independent growth in p53 mutant DLD1 colon cancer cells (Figure 2E). Furthermore, expression of MAGE-A3/6 did not inversely correlate with p53 mutational status (χ2=0.17; p=0.98). Therefore, the MAGE-A3/6-TRIM28 E3 ubiquitin ligase may have additional targets relevant to its function in cancer cells.
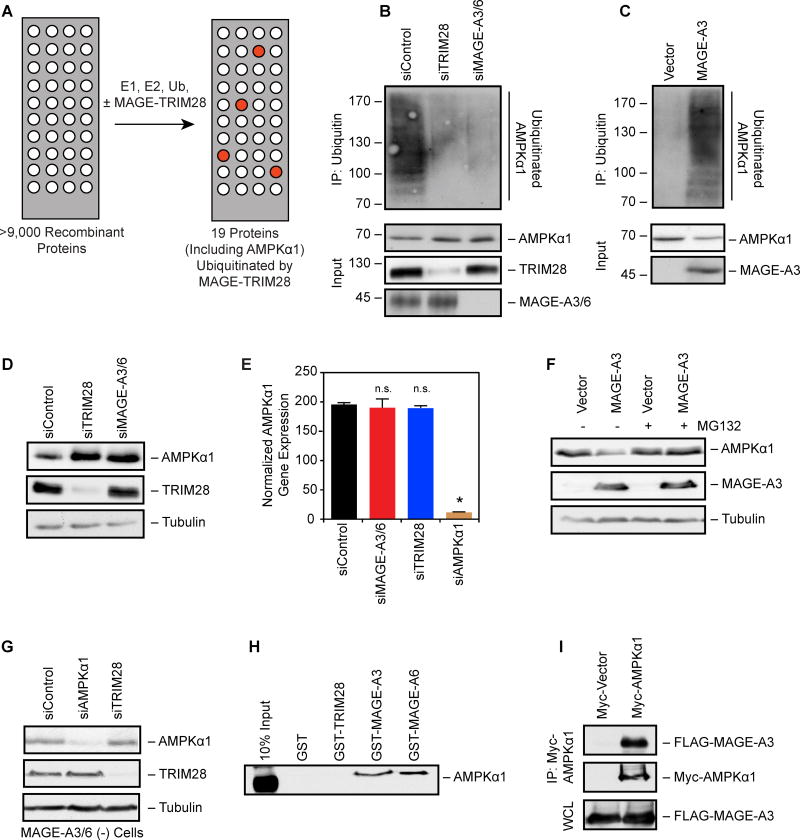
To identify additional targets of the MAGE-TRIM28 E3 ubiquitin ligase, we screened for its direct substrates by performing in vitro ubiquitination reactions on protein microarrays containing >9,000 SF9-purified, recombinant proteins (Figure 3A). This screen yielded 19 potential MAGE-TRIM28 substrates (Supplemental Table 2) whose ubiquitination were significantly (p<0.05) increased by MAGE-TRIM28. To validate the results of the screen, candidates were tested for regulation by MAGE-A3/6-TRIM28, with AMPKα1 being the most consistent and robust target of MAGE-A3/6-TRIM28 (Figure 3B–D). Multiple siRNAs targeting MAGE-A3/6 or TRIM28 decreased ubiquitination of AMPKα1 (Figures 3B and S2F). Expression of MAGE-A3 in MAGE-A3/6-negative cells promoted ubiquitination of AMPKα1 (Figure 3C). Furthermore, knockdown of MAGE-A3/6 or TRIM28 resulted in increased AMPKα1 protein levels (Figure 3D), without affecting mRNA levels (Figure 3E). Additional subunits of the AMPK holoenzyme complex, such as AMPKβ1 and AMPKγ1, were correspondingly elevated (Figure S2G). In contrast, expression of MAGE-A3 in MAGE-A3/6-negative cells decreased AMPKα1 protein levels (Figure 3F), without affecting mRNA levels (Figure S2E). Notably, AMPKα1 protein levels could be rescued by addition of the MG132 proteasome inhibitor (Figure 3F), suggesting that MAGE-A3/6-TRIM28 ubiquitination of AMPKα1 leads to its degradation by the proteasome.
Figure 3. MAGE-A3/6-TRIM28 E3 ubiquitin ligase ubiquitinates and degrades AMPKα1.
(A) Schematic of in vitro screen for MAGE-TRIM28 ubiquitination substrates using protein arrays.
(B) AMPKα1 ubiquitination requires MAGE-A3/6-TRIM28. HeLa (MAGE-A3/6-positive) were treated with the indicated siRNAs for 24 hrs before transfection with Myc-tagged ubiquitin for 48 hrs before anti-Myc immunoprecipitation (IP) and immunoblotting was performed (n=3).
(C) Expression of MAGE-A3 promotes AMPKα1 ubiquitination. MAGE-A3/6-negative HEK293 cells stably expressing FLAG-MAGE-A3 were transfected with Myc-ubiquitin 48 hrs before anti-Myc IP and immunoblotting was performed (n=3).
(D) Knockdown of MAGE-A3/6-TRIM28 increases AMPKα1 protein levels. MAGE-A3/6-positive cells were treated with the indicated siRNAs for 72 hrs and then blotted for the indicated proteins (n > 3).
(E) Knockdown of MAGE-A3/6-TRIM28 does not affect AMPKα1 mRNA levels. MAGE-A3/6-positive cells were treated with the indicated siRNAs for 72 hrs and then AMPKα1 mRNA levels were determined by RT-QPCR (n=3). Data are represented as the mean + SD. Asterisks indicates p<0.05.
(F) MAGE-A3 promotes proteasome-dependent AMPKα1 degradation. MAGE-A3/6-negative cells expressing vector of MAGE-A3 were treated with 5 µM MG132 for 4 hrs before immunoblotting (n=3).
(G) TRIM28-mediated AMPKα1 degradation requires MAGE-A3/6. MAGE-A3/6-negative HEK293 cells were transfected with the indicated siRNA for 72 hrs before cell lysates were immnoblotted (n>3).
(H) GST pulldown assays reveal AMPKα1 directly binds to MAGE-A3 and MAGE-A6, but not TRIM28 (22–418) or GST (n=3).
(I) HeLa cells expressing FLAG-MAGE-A3 or FLAG-vector along with Myc-AMPKα1 were subjected to anti-Myc IP and immunoblotting. WCL represents whole cell lysate. Data representative of multiple experiments (n=2). See also Table S2.
To determine if MAGE-A3/6 enhances degradation of AMPKα1 by TRIM28, as is the case for p53 (Doyle et al., 2010), or if MAGE-A3/6 is required for specifying AMPKα1 degradation by TRIM28, we examined whether TRIM28 regulates AMPKα1 levels in MAGE-A3/6-negative cells. Knockdown of TRIM28 in MAGE-A3/6-negative cells had no effect on AMPKα1 levels (Figure 3G), suggesting that MAGE-A3/6 is required for AMPKα1 degradation by TRIM28. Consistent with this notion, the AMPKα1β1γ1 holoenzyme complex (Figure S2H) and specifically AMPKα1 bound to recombinant GST-MAGE-A3 and GST-MAGE-A6, but not GST-TRIM28 in vitro (Figure 3H). Additionally, overexpressed and endogenous AMPKα1 co-immunoprecipated with MAGE-A3 from cells (Figures 3I and S2I). These results suggest that MAGE-A3/6 specifies ubiquitination of AMPKα1 by the TRIM28 ubiquitin ligase.
Inhibition of AMPK by MAGE-A3/6-TRIM28 impacts cellular metabolic processes
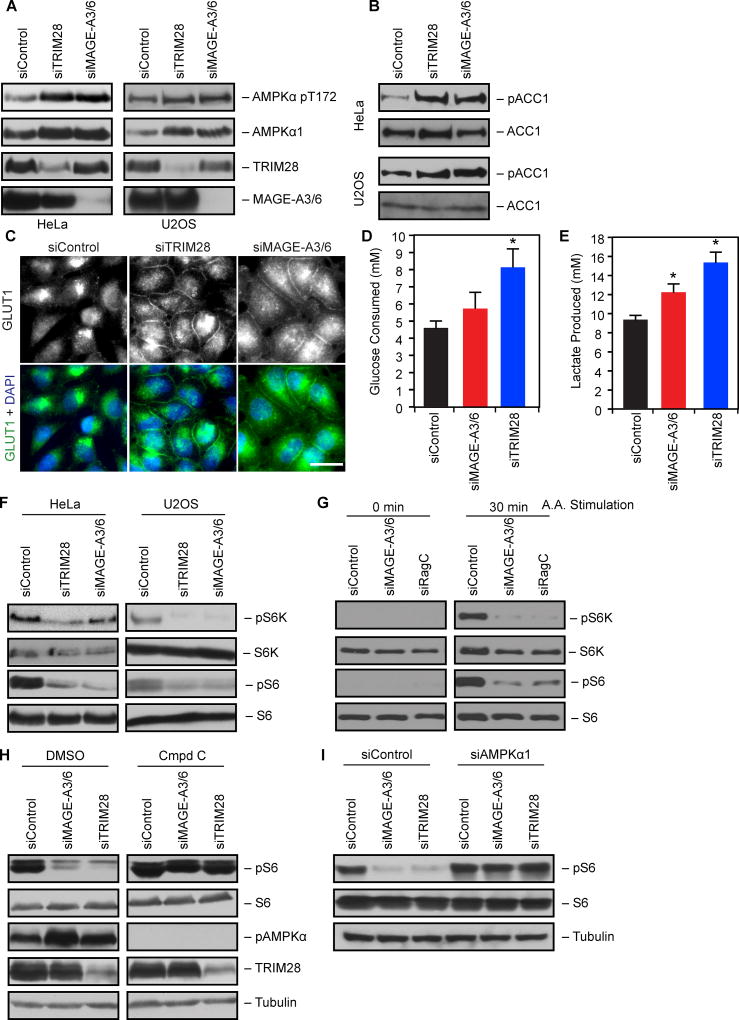
Next, we determined whether modulation of AMPK protein levels by MAGE-A3/6-TRIM28 had a functional impact on AMPK activity and the cellular metabolic processes it controls. Knockdown of MAGE-A3/6 or TRIM28 increased both total and active (phosphorylated, pT172) AMPKα1 (Figure 4A). Furthermore, the knockdown of MAGE-A3/6 or TRIM28 resulted in increased phosphorylation of ACC1 (Figure 4B), a prototypical target of AMPK (Carling et al., 1987). Although AMPK generally antagonizes the Warburg effect (Faubert et al., 2013), acute stimulation of AMPK can promote glycolysis through a variety of activities, including plasma membrane localization of the GLUT1 glucose transporter and increased glucose consumption (Barnes et al., 2002; Hardie et al., 2012b). Knockdown of MAGE-A3/6 or TRIM28 resulted in increased plasma membrane localized GLUT1 (Figure 4C). Furthermore, knockdown of TRIM28 increased glucose consumption (Figure 4D) and correspondingly increased lactate production (Figure 4E) in MAGE-A3/6-positive cells. These results suggest that MAGE-A3/6-TRIM28 has a functional impact on cellular metabolism.
Figure 4. MAGE-A3/6-TRIM28 ubiquitin ligase regulates AMPK controlled metabolic processes.
(A–B) MAGE-A3/6-TRIM28 knockdown increases phospho-AMPK (A) and phospho-ACC1 (B) signaling. HeLa or U2OS cells were treated with the indicated siRNAs for 72 hrs before cell lysates immunoblotted (n≥3).
(C) MAGE-A3/6-TRIM28 knockdown increases Glut1 plasma membrane localization. HeLa cells were treated with the indicated siRNAs for 72 hrs before immunostaining for Glut1 (n=3). Scale bar represent 20 µm.
(D–E) MAGE-A3/6-TRIM28 knockdown alters glucose metabolism. HeLa cells were treated with MAGE-A3/6 or TRIM28 siRNA for 72 hrs and then fed with fresh media. After 6 hrs, media was collected and glucose (D) and lactate (E) levels in media were analyzed via nova analyzer. Data (n=3) represent mean +SD.
(F) MAGE-A3/6-TRIM28 is required for mTOR signaling. HeLa or U2OS cells were treated with the indicated siRNAs for 72 hrs before immunoblotting (n=3).
(G) MAGE-A3/6-TRIM28 is required for amino acid-induced mTOR activity. HeLa cells were treated with siControl, siMAGE-A3/6, or siRagC (positive control) siRNAs for 72 hrs before 6 hr starvation in EBSS (0 min) or starvation followed by 30 min amino acid stimulation (n=3).
(H–I) Inhibition of AMPK reverses mTOR inhibition by MAGE-A3/6 or TRIM28 knockdown. Cells were transfected with the indicated siRNAs for 72 hrs before collection and immunoblotting (H) or treatment for 4 hrs with 10 µM Compound C or vehicle (DMSO) (I) (n ≥ 2).
Asterisks indicates p<0.05.
In addition to regulating cellular glucose metabolism, AMPK is well documented to inhibit anabolic processes, such as mTOR-dependent protein synthesis, to conserve energy in the context of reduced ATP levels (Gwinn et al., 2008; Inoki et al., 2003). Therefore, we knocked down MAGE-A3/6 or TRIM28 and followed mTOR activity by examining phosphorylation of p70 ribosomal S6 kinase and ribosomal S6 protein. Upon knockdown of MAGE-A3/6 or TRIM28, mTOR signaling was severely inhibited and phosphorylation of both p70 ribosomal S6 kinase and ribosomal S6 protein were reduced (Figure 4F). Similarly, amino acid-induced mTOR activity was significantly reduced upon depletion of MAGE-A3/6 (Figure 4G). Importantly, reduction in basal mTOR activity was rescued by treatment with the AMPK inhibitor, compound c (Figure 4H) or co-depeltion of AMPKα1 (Figure 4I). Together, these results suggest that the MAGE-A3/6-TRIM28 ubiquitin ligase is functionally important for maintenance of mTOR activity, likely through inhibition of AMPK.
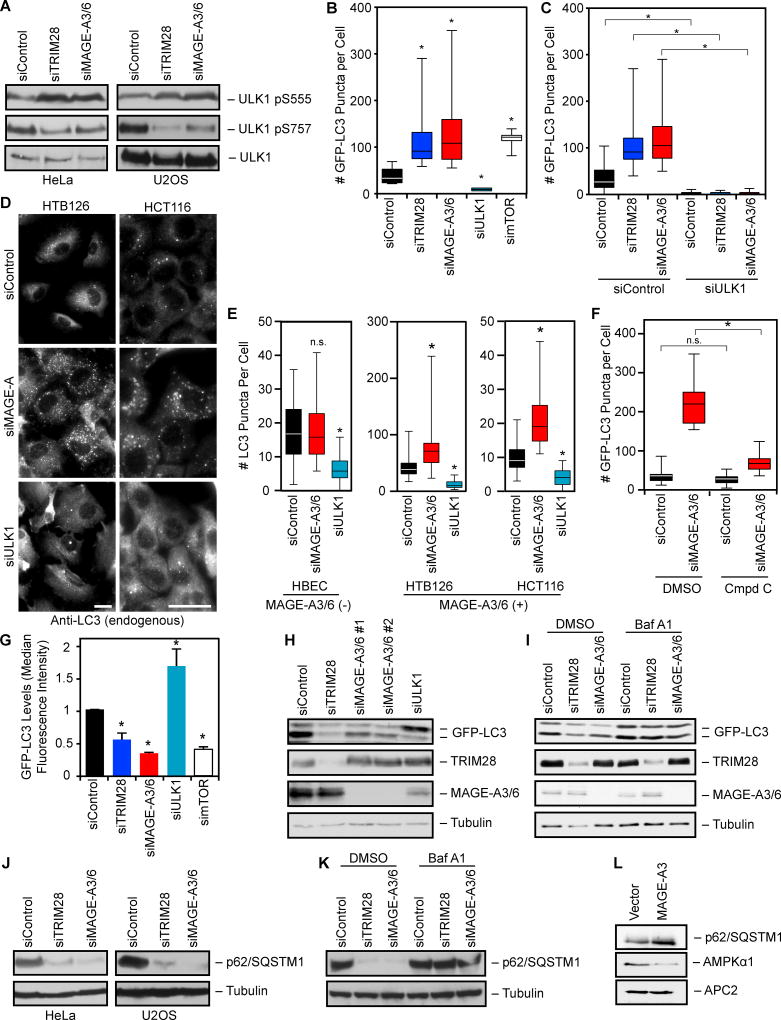
MAGE-A3/6-TRIM28 ubiquitin ligase inhibits autophagy
Because MAGE-A3/6-TRIM28 regulates both AMPK and mTOR activities and both of these signaling pathways converge to opposingly modulate autophagy (Egan et al., 2011; Kim et al., 2011), we examined whether MAGE-A3/6-TRIM28 influences autophagy. One mechanism by which AMPK and mTOR regulate autophagy is through phosphorylation of the proximal ULK1 kinase required for autophagosome formation. AMPK phosphorylation of ULK1 S555 promotes ULK1 activity and autophagy, whereas mTOR phosphorylation of ULK1 S757 inhibits ULK1 activity and autophagy (Egan et al., 2011; Kim et al., 2011). Knockdown of MAGE-A3/6 or TRIM28 upregulated ULK1 S555 phosphorylation (AMPK site) and downregulated ULK1 S757 phosphorylation (mTOR site) (Figure 5A). Changes in ULK1 phosphorylation by MAGE-A3/6 or TRIM28 knockdown were accompanied by the expected increase in GFP-LC3 puncta, a marker of autophagy (Figures 5B and S3A–C). The increased GFP-LC3 puncta in MAGE-A3/6 and TRIM28 depleted cells was blocked by co-depletion of ULK1 (Figure 5C). The magnitude of increased GFP-LC3 puncta upon MAGE-A3/6 or TRIM28 knockdown was similar to knockdown of mTOR, an established potent inhibitor of autophagy (Figure 5B). To complement our results using cells stably expressing GFP-LC3, we examined the number of endogenous LC3 puncta upon knockdown of MAGE-A3/6. Similarly to GFP-LC3, siRNAs targeting MAGE-A3/6 induced the accumulation of endogenous LC3 puncta in MAGE-A3/6-positive cells, but had no effect in MAGE-A3/6-negative cells (Figure 5D–E). Importantly, short-term inhibition of AMPK with compound c attenuated MAGE-A3/6-RNAi-induced GFP-LC3 puncta formation (Figure 5F).
Figure 5. MAGE-A3/6-TRIM28 ubiquitin ligase inhibits autophagy.
(A) HeLa or U2OS cells were transfected with siRNA for 72 hrs before immunoblotting (ULK1 pS555 AMPK target site; ULK1 pS757 mTOR target site) (n>3).
(B) U2OS cells stably expressing GFP-LC3 were treated with the indicated siRNAs for 72 hrs before imaging and quantitation of GFP-LC3 puncta (n=3). Box plots are mean and quartiles.
(C) U2OS cells stably expressing GFP-LC3 were treated with the indicated siRNAs for 72 hrs before imaging (n=3). Box plots represent mean and quartiles of n>50 cells.
(D–E) Seventy-two hrs after transfection cells were stained for endogenous LC3 (D). Scale bars represent 20 µm. Box plots of number of LC3 puncta per cell (E). Box plots are mean and quartiles of n>50 cells.
(F) U2OS cells were transfected with the indicated siRNA for 72 hrs before treatment for 4 hrs with 10 µM Compound C or vehicle (DMSO). Box plots represent mean and quartiles of n>50 cells.
(G) U2OS GFP-LC3 cells were transfected with the indicated siRNA for 72 hrs. Median GFP fluorescent intensity +SD as determined by flow cytometry is shown (n=3).
(H) Cells stably expressing GFP-LC3 were treated with siRNAs for 72 hrs before immunoblotting (n>3).
(I) Knockdown of MAGE-A3/6 or TRIM28 increases autophagic flux. GFP-LC3 cells were transfected with the indicated siRNA for 72 hrs. Cells were treated with Bafilomycin A for 4 hrs before cell lysates were immunoblotted (n=3).
(J–K) Knockdown of MAGE-A3/6 or TRIM28 increases p62 consumption. HeLa or U2OS cells were treated with the indicated siRNA for 72 hrs. Cells lysates were collected (J) or cells were treated with DMSO or Bafilomycin A for 4 hrs before cell lysates were collected (K) and the indicated proteins were detected by immunoblotting (n ≥ 2).
(L) MAGE-A3/6-negative HBEC cells were stably transfected with Myc-MAGE-A3 and cell lysates were immunoblotted (n=2).
Asterisks indicates p<0.05. See also Figure S3.
Since an increase in LC3 puncta may represent either a block in autophagosome fusion with lysosomes or an increase in autophagy, we measured the consumption (levels) of GFP-LC3 by flow cytometry. We observed a significant decrease in GFP-LC3 fluorescence upon knockdown of MAGE-A3/6 or TRIM28 and this was again similar to the degree of GFP-LC3 consumption upon mTOR depletion (Figures 5G and S3D). These results were further confirmed by western blotting where knockdown of MAGE-A3/6 or TRIM28 promoted a marked decrease in GFP-LC3 protein levels (Figure 5H). These changes in GFP-LC3 were not due to alterations in GFP-LC3 mRNA levels (Figure S3E). To determine if the decrease in LC3 was the consequence of increased autophagic flux, we treated cells depleted of MAGE-A3/6 or TRIM28 with bafilomycin A1 to prevent acidification of lysosomes and degradation of proteins by autophagy. Short-term treatment of cells with bafilomycin A1 blocked consumption of GFP-LC3 by knockdown of MAGE-A3/6 or TRIM28 (Figure 5I). Finally, we examined the levels of autophagy in MAGE-A3/6 or TRIM28 depleted cells by an independent measure, consumption of the p62/SQSTM1 autophagy adaptor. Similarly to LC3, endogenous p62/ SQSTM1 was consumed upon knockdown of MAGE-A3/6 or TRIM28 (Figure 5J) and this could be rescued by bafilomycin A1 (Figure 5K). Furthermore, the ability of MAGE-A3/6 to inhibit autophagy was also confirmed by expression of MAGE-A3 in normal, non-transformed cells that typically are negative for MAGE-A3/6. MAGE-A3 expression induced the degradation of AMPKα1 and the accumulation of p62/ SQSTM1 (Figure 5L), consistent with reduced autophagy in these cells. Collectively, these results suggest that MAGE-A3/6-TRIM28 inhibits autophagy and that depletion of MAGE-A3/6 or TRIM28 dramatically increases autophagic flux.
MAGE-A3/6 regulation of AMPKα1 is relevant in human tumors
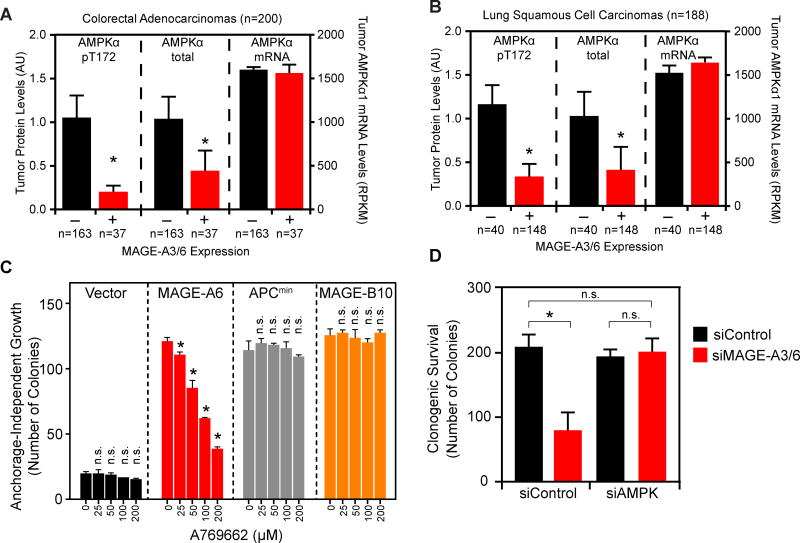
Our results suggest that the oncogenic MAGE-A3/6-TRIM28 ubiquitin ligase regulates several cellular metabolic regulatory pathways through ubiquitination and degradation of AMPKα1. To determine the relevance of these findings to human tumors, we examined whether MAGE-A3/6 expression inversely correlated with AMPK activity and protein levels in patient tumor samples. Indeed, breast invasive carcinoma (Figure S4A), colon adenocarcinoma (Figure 6A), and lung squamous cell carcinoma (Figure 6B) tumors expressing MAGE-A3/6 had significantly reduced total and active (phospho-T172) AMPKα protein levels. This reduction was not a consequence of decreased AMPKα1 mRNAs in these tumors (Figures 6A–B and S4A). Consistent with these findings, the phosphorylated form of AMPK is downregulated in high proportion of cases of breast cancer (Hadad et al., 2009). In addition, MAGE-A3/6 expression in tumors correlated with reduced downstream AMPK signaling, such as increased markers of mTOR activity (Figure S4B).
Figure 6. Regulation of AMPK by MAGE-A3/6-TRIM28 is relevant to human tumors.
(A–B) TCGA data were analyzed for MAGE-A3/6 mRNA levels and total and active (pT172) AMPKα protein levels. Data are mean +SE with number (n) of tumors indicated. Asterisks indicates p<0.01.
(C) Anchorage-independent growth assays of the indicated HCEC cells were performed in presence of the indicated concentrations of A769662. Number of colonies >100 µm were counted (n=3). Data are mean +SD. Asterisks indicates p<0.05.
(D) Colony formation assays were performed in HeLa cells treated with the indicated siRNAs. Data are mean +SD, n=3. Asterisks indicates p<0.05. See also Figure S4 and S5.
Finally, AMPK agonists are of significant interest in treatment and prevention of cancer (Hardie et al., 2012a). Thus, we determined whether AMPK agonists could reverse the phenotypes of MAGE-A3/6 driven anchorage-independent growth and cancer cell viability. The AMPK activating compounds, aminoimidazole carboxamide ribonucleotide (AICAR) and metformin, suppressed the ability of MAGE-A6 to promote anchorage-independent growth of normal HCEC cells and DLD1 colon cancer cells in a dose-dependent manner (Figure S4D–F). Importantly, these effects were specific to MAGE-A6 expressing cells as AICAR and metformin minimally affected Apcmin or MAGE-B10 driven anchorage-independent growth of HCEC cells (Figure S4D–E). Since the cellular effects of both AICAR and metformin extend beyond just activation of AMPK, including affecting mitochondrial respiration (Hardie et al., 2012a), we also examined whether a direct allosteric activator of AMPK, A769662 (Cool et al., 2006; Landgraf et al., 2013), or genetic manipulation of AMPKα1 could alter phenotypes associated with MAGE-A3/6. MAGE-A6-induced, but not Apcmin- or MAGE-B10-induced anchorage-independent growth of HCEC and DLD1 cells was significantly impaired by A769662 in a dose-dependent manner (Figures 6C and S4G). Furthermore, co-depletion of AMPKα1 rescued MAGE-A3/6-RNAi-induced decrease in cell viability (Figure 6D). Taken together, these results suggest that regulation of AMPK by MAGE-A3/6 is relevant to human tumors and pharmacological agonists of AMPK may have therapeutic potential in MAGE-A3/6-positive tumors.
DISCUSSION
AMPK senses and responds to the energy status of cells to regulate multiple metabolic processes and limit energy expenditure. Significant effort has been directed towards understanding the role and dysregulation of AMPK in cancer. One known mechanism of reducing AMPK activity in cancer is mutation/deletion of its upstream regulatory kinase Lkb1/Stk11. However, this is a rare event in most tumor types other than lung adenocarcinomas and cervical cancers (Wingo et al., 2009). In this study, we demonstrate that the MAGE-A3/6-TRIM28 E3 ligase complex ubiquitinates and degrades AMPKα1. Thus, the prominent activation of MAGE-A3/6 expression in many cancer types may represent an alternative mechanism for downregulating the AMPK signaling pathway (Figure S5). Consistent with this, expression of MAGE-A3/6 and mutation of Lkb1/Stk11 are rarely found in the same lung adenocarcinoma tumors (Figure S4C, p<0.01).
MAGE-A3/6 are normally exclusively expressed in the testis, but are frequently turned on in many tumor types, including colon, lung and breast tumors (Figure 1). In combination with previous studies, our findings suggest that activation of MAGE-A3/6 in cancer cells is not a by-product, passenger event during cellular transformation and tumorigenesis, but rather MAGE-A3/6 are driver genes that support multiple phenotypes associated with tumors, including metabolic dysregulation. We propose that one critical oncogenic function of MAGE-A3/6 is downregulation of AMPK and alteration of cellular metabolism in cancer cells. Strikingly, this mode of AMPK regulation does not occur in normal somatic cells that do not express MAGE-A3/6, but only occurs upon reactivation of the testicular MAGE-A3/6 program in cancer cells.
Although AMPK coordinates many different actions in the cell, one key process it controls is autophagy. While the role of autophagy in the progression of cancer is multifaceted, loss of autophagy has been implicated in the initiation of tumorigenesis (Choi et al., 2013; Wei et al., 2013; White, 2012). Our results suggest that aberrant activation of MAGE-A3/6 in tumors may provide a unique mechanism for inhibition of tumor-suppressive autophagy during tumor initiation. Interestingly, MAGE-A3/6 expression is undetectable in never-smokers, but is aberrantly found in the lungs of smokers before they have any clinical signs of disease (Jang et al., 2001). Thus, MAGE-A3/6 expression may occur early during tumor initiation and could be one mechanism to downregulate autophagy during this stage. Identification of the factors that regulate MAGE-A3/6 expression in adult tissues may provide insights into understanding events leading to tumor initiation. One major regulatory mechanism controlling expression of MAGE cancer-testis antigens is promoter CpG methylation in normal somatic cells (Simpson et al., 2005). However, simple demethylation of MAGEs is not sufficient to drive expression (Weber et al., 1994). The identification of additional transcriptional regulators will be of utmost importance.
Our findings of the association of MAGE-A3/6 expression with AMPK degradation in human tumors has important and potentially immediate implications on the utilization of AMPK activating compounds, such as metformin and A769662, that are vigorously being pursued in the prevention and treatment of cancer (Quinn et al., 2013). While AMPK activating drugs are currently in clinical trials for treatment of a variety of tumor types, the early results thus far have been mixed with no apparent explanation (Quinn et al., 2013). We propose that MAGE-A3/6 expression status may be a useful enrollment biomarker to select patients with the greatest potential response to AMPK agonists. Additionally, since MAGE-A3/6 expression increases signaling through the mTOR pathway, the use of currently approved mTOR inhibitors may be effective in the future treatment of MAGE-A3/6-driven tumors.
Little is known about the physiological role of MAGE-A3/6 in the testis. Our findings on the molecular and cellular functions of MAGE-A3/6-TRIM28 in cancer provide intriguing insights into their normal physiological function during spermatogenesis. Interestingly, germ cells in the testis switch their carbon energy sources as they differentiate from spermatogonia stem cells to maturing haploid spermatids (Nakamura et al., 1984). We propose that MAGE-A3/6 may function to protect maturing spermatocytes from energy stress by dampening AMPK activation. Also, MAGE-A3/6 might enable developmental stage-dependent activation of anabolic pathways required for normal spermatogenesis, such as lipid and protein synthesis. Furthermore, developing spermatocytes also express an unusual splice variant of LKB1 with a different C-terminal region, which is required for spermiogenesis (Towler et al., 2008). Consistent with these ideas, we have found that mouse MAGE-A genes are highly expressed in pre-pachytene spermatocytes (data not shown) where these regulator events are occurring and previous studies have shown that testis-specific knockout of TRIM28 blocks spermatocyte differentiation, resulting in testis degeneration (Weber et al., 2002).
In summary, our findings illuminate a previously unrecognized, widespread regulation of AMPK during tumorigenesis by a testis-specific ubiquitin ligase, provide an unprecedented molecular mechanism by which MAGE cancer-testis antigens drive tumorigenesis, and have important implications to maximizing the clinical utility of AMPK-directed chemotherapies.
EXPERIMENTAL PROCEDURES
Cell culture and transfections
Cells were cultured under standard conditions and transfected according to manufacturer’s recommendation. Detailed descriptions of cell culture conditions, transfection procedures, siRNA sequences, and antibodies are described in the Extended Experimental Procedures.
RNA preparation and RT-QPCR
Preparation of RNA from tissues and cells and RT-QPCR analysis was performed by standard molecular biology techniques and described in the Extended Experimental Procedures. All procedures and use of mice were approved by the Institutional Animal Care and Use Committee of UT Southwestern Medical Center.
Colony formation and anchorage-independent growth soft agar assays
For colony formation assays on plastic, cells were transfected for 72 hrs with siRNAs and then re-plated at single cell density. After three weeks, cells were fixed and stained with crystal violet (0.05% (w/v)) and counted. For anchorage-independent growth soft agar growth assays, cells were suspended in 0.375% Noble agar (Difco) supplemented with regular growth medium and overlaid on 0.5% Noble agar. Cells were allowed to grow for 2–4 weeks before colonies ≥ 100 µm in size were counted.
Immunofluorescence and microscopy
Immunofluorescence was performed essentially as described previously (Hao et al., 2013) and in Extended Experimental Procedures.
In vitro ubiquitination screen
ProtoArray containing >9,000 GST-tagged recombinant proteins purified from SF9 insect cells was purchased from Invitrogen. In vitro ubiquitination on the slide was performed according to manufacture instructions with minor modifications described in Extended Experimental Procedures.
Glucose consumption and lactate measurements
Twenty-four hrs after plating, cells were transfected with siRNA. Seventy-two hrs after siRNA transfections, cells were changed into fresh media for 6 hrs. Media was collected and analyzed using Nova Analyzer to quantitate amount of glucose and lactate in the media.
Recombinant protein purification and in vitro binding assays
Recombinant proteins were produced using standard procedures described in the Extended Experimental Procedures. In vitro binding assays were performed as described previously (Doyle et al., 2010; Hao et al., 2013) and specified in the Extended Experimental Procedures.
Assessment of mRNA/protein expression levels in human tumors and statistical analysis
mRNA levels, survival data, and mutational status were determined using the cancer genome atlas. Tumor protein expression levels were determined previously by reverse phase protein arrays performed on tumors with matching RNA-seq data (Cancer Genome Atlas Research, 2014).
Supplementary Material
Acknowledgments
We thank members of the Potts lab for helpful discussions and critical reading of the manuscript. We also thank Drs. Ralph Deberardinis, Beth Levine, John Minna, Jerry Shay, and Hongtao Yu for guidance and critical reagents. This work was supported by Michael L. Rosenberg Scholar in Medical Research fund (PRP), CPRIT R1117 (PRP), DOD Discovery Award W81XWH-12-1-0446 (PRP), and WELCH Foundation I-1821 (PRP).
Footnotes
AUTHOR CONTRIBUTIONS
C.T.P., S.R., M.B.P., M.A.W., and P.R.P. contributed to experimental design. C.T.P., S.R., K.F.T., J.L.W., M.B.P., Y.O., and P.R.P. performed experiments, data analysis, and figure composition. C.T.P. and P.R.P. wrote the manuscript. S.R., K.F.T., J.L.W., M.B.P., and P.R.P. proofed the manuscript.
References
- Barnes K, Ingram JC, Porras OH, Barros LF, Hudson ER, Fryer LG, Foufelle F, Carling D, Hardie DG, Baldwin SA. Activation of GLUT1 by metabolic and osmotic stress: potential involvement of AMP-activated protein kinase (AMPK) Journal of cell science. 2002;115:2433–2442. doi: 10.1242/jcs.115.11.2433. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research, N. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carling D, Zammit VA, Hardie DG. A common bicyclic protein kinase cascade inactivates the regulatory enzymes of fatty acid and cholesterol biosynthesis. FEBS letters. 1987;223:217–222. doi: 10.1016/0014-5793(87)80292-2. [DOI] [PubMed] [Google Scholar]
- Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- Chomez P, De Backer O, Bertrand M, De Plaen E, Boon T, Lucas S. An overview of the MAGE gene family with the identification of all human members of the family. Cancer Res. 2001;61:5544–5551. [PubMed] [Google Scholar]
- Chou CC, Lee KH, Lai IL, Wang D, Mo X, Kulp SK, Shapiro CL, Chen CS. AMPK Reverses the Mesenchymal Phenotype of Cancer Cells by Targeting the Akt-MDM2-Foxo3a Signaling Axis. Cancer Res. 2014;74:4783–4795. doi: 10.1158/0008-5472.CAN-14-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cool B, Zinker B, Chiou W, Kifle L, Cao N, Perham M, Dickinson R, Adler A, Gagne G, Iyengar R, et al. Identification and characterization of a small molecule AMPK activator that treats key components of type 2 diabetes and the metabolic syndrome. Cell metabolism. 2006;3:403–416. doi: 10.1016/j.cmet.2006.05.005. [DOI] [PubMed] [Google Scholar]
- De Plaen E, Arden K, Traversari C, Gaforio JJ, Szikora JP, De Smet C, Brasseur F, van der Bruggen P, Lethe B, Lurquin C, et al. Structure, chromosomal localization, and expression of 12 genes of the MAGE family. Immunogenetics. 1994;40:360–369. doi: 10.1007/BF01246677. [DOI] [PubMed] [Google Scholar]
- Doyle JM, Gao J, Wang J, Yang M, Potts PR. MAGE-RING protein complexes comprise a family of E3 ubiquitin ligases. Molecular cell. 2010;39:963–974. doi: 10.1016/j.molcel.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve-Puig R, Canals F, Colome N, Merlino G, Recio JA. Uncoupling of the LKB1-AMPKalpha energy sensor pathway by growth factors and oncogenic BRAF. PloS one. 2009;4:e4771. doi: 10.1371/journal.pone.0004771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faubert B, Boily G, Izreig S, Griss T, Samborska B, Dong Z, Dupuy F, Chambers C, Fuerth BJ, Viollet B, et al. AMPK is a negative regulator of the Warburg effect and suppresses tumor growth in vivo. Cell metabolism. 2013;17:113–124. doi: 10.1016/j.cmet.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Gao J, Yang M. When MAGE meets RING: insights into biological functions of MAGE proteins. Protein Cell. 2011;2:7–12. doi: 10.1007/s13238-011-1002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Molecular cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadad S, Iwamoto T, Jordan L, Purdie C, Bray S, Baker L, Jellema G, Deharo S, Hardie DG, Pusztai L, et al. Evidence for biological effects of metformin in operable breast cancer: a pre-operative, window-of-opportunity, randomized trial. Breast cancer research and treatment. 2011;128:783–794. doi: 10.1007/s10549-011-1612-1. [DOI] [PubMed] [Google Scholar]
- Hadad SM, Baker L, Quinlan PR, Robertson KE, Bray SE, Thomson G, Kellock D, Jordan LB, Purdie CA, Hardie DG, et al. Histological evaluation of AMPK signalling in primary breast cancer. BMC cancer. 2009;9:307. doi: 10.1186/1471-2407-9-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao YH, Doyle JM, Ramanathan S, Gomez TS, Jia D, Xu M, Chen ZJ, Billadeau DD, Rosen MK, Potts PR. Regulation of WASH-Dependent Actin Polymerization and Protein Trafficking by Ubiquitination. Cell. 2013;152:1051–1064. doi: 10.1016/j.cell.2013.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG, Alessi DR. LKB1 and AMPK and the cancer-metabolism link - ten years after. BMC Biol. 2013;11:36. doi: 10.1186/1741-7007-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG, Ross FA, Hawley SA. AMP-activated protein kinase: a target for drugs both ancient and modern. Chem Biol. 2012a;19:1222–1236. doi: 10.1016/j.chembiol.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nature reviews Molecular cell biology. 2012b;13:251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley SA, Boudeau J, Reid JL, Mustard KJ, Udd L, Makela TP, Alessi DR, Hardie DG. Complexes between the LKB1 tumor suppressor, STRAD alpha/beta and MO25 alpha/beta are upstream kinases in the AMP-activated protein kinase cascade. J Biol. 2003;2:28. doi: 10.1186/1475-4924-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley SA, Pan DA, Mustard KJ, Ross L, Bain J, Edelman AM, Frenguelli BG, Hardie DG. Calmodulin-dependent protein kinase kinase-beta is an alternative upstream kinase for AMP-activated protein kinase. Cell metabolism. 2005;2:9–19. doi: 10.1016/j.cmet.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Imamura K, Ogura T, Kishimoto A, Kaminishi M, Esumi H. Cell cycle regulation via p53 phosphorylation by a 5'-AMP activated protein kinase activator, 5-aminoimidazole- 4-carboxamide-1-beta-D-ribofuranoside, in a human hepatocellular carcinoma cell line. Biochemical and biophysical research communications. 2001;287:562–567. doi: 10.1006/bbrc.2001.5627. [DOI] [PubMed] [Google Scholar]
- Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- Jang SJ, Soria JC, Wang L, Hassan KA, Morice RC, Walsh GL, Hong WK, Mao L. Activation of melanoma antigen tumor antigens occurs early in lung carcinogenesis. Cancer Res. 2001;61:7959–7963. [PubMed] [Google Scholar]
- Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, Birnbaum MJ, Thompson CB. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Molecular cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- Kim J, Kim YC, Fang C, Russell RC, Kim JH, Fan W, Liu R, Zhong Q, Guan KL. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell. 2013;152:290–303. doi: 10.1016/j.cell.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nature cell biology. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laderoute KR, Calaoagan JM, Chao WR, Dinh D, Denko N, Duellman S, Kalra J, Liu X, Papandreou I, Sambucetti L, et al. 5'-AMP-activated Protein Kinase (AMPK) Supports the Growth of Aggressive Experimental Human Breast Cancer Tumors. The Journal of biological chemistry. 2014;289:22850–22864. doi: 10.1074/jbc.M114.576371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf RR, Goswami D, Rajamohan F, Harris MS, Calabrese MF, Hoth LR, Magyar R, Pascal BD, Chalmers MJ, Busby SA, et al. Activation of AMP-activated protein kinase revealed by hydrogen/deuterium exchange mass spectrometry. Structure. 2013;21:1942–1953. doi: 10.1016/j.str.2013.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CW, Wong LL, Tse EY, Liu HF, Leong VY, Lee JM, Hardie DG, Ng IO, Ching YP. AMPK promotes p53 acetylation via phosphorylation and inactivation of SIRT1 in liver cancer cells. Cancer Res. 2012;72:4394–4404. doi: 10.1158/0008-5472.CAN-12-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Shao SH, Xu ZX, Hennessy B, Ding Z, Larrea M, Kondo S, Dumont DJ, Gutterman JU, Walker CL, et al. The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nature cell biology. 2007;9:218–224. doi: 10.1038/ncb1537. [DOI] [PubMed] [Google Scholar]
- Liu W, Cheng S, Asa SL, Ezzat S. The melanoma-associated antigen A3 mediates fibronectin-controlled cancer progression and metastasis. Cancer Res. 2008;68:8104–8112. doi: 10.1158/0008-5472.CAN-08-2132. [DOI] [PubMed] [Google Scholar]
- Marcar L, Maclaine NJ, Hupp TR, Meek DW. Mage-A cancer/testis antigens inhibit p53 function by blocking its interaction with chromatin. Cancer Res. 2010;70:10362–10370. doi: 10.1158/0008-5472.CAN-10-1341. [DOI] [PubMed] [Google Scholar]
- Matsumoto S, Iwakawa R, Takahashi K, Kohno T, Nakanishi Y, Matsuno Y, Suzuki K, Nakamoto M, Shimizu E, Minna JD, et al. Prevalence and specificity of LKB1 genetic alterations in lung cancers. Oncogene. 2007;26:5911–5918. doi: 10.1038/sj.onc.1210418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte M, Simonatto M, Peche LY, Bublik DR, Gobessi S, Pierotti MA, Rodolfo M, Schneider C. MAGE-A tumor antigens target p53 transactivation function through histone deacetylase recruitment and confer resistance to chemotherapeutic agents. Proc Natl Acad Sci U S A. 2006;103:11160–11165. doi: 10.1073/pnas.0510834103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Okinaga S, Arai K. Metabolism of round spermatids: evidence that lactate is preferred substrate. Am J Physiol. 1984;247:E234–242. doi: 10.1152/ajpendo.1984.247.2.E234. [DOI] [PubMed] [Google Scholar]
- Niraula S, Dowling RJ, Ennis M, Chang MC, Done SJ, Hood N, Escallon J, Leong WL, McCready DR, Reedijk M, et al. Metformin in early breast cancer: a prospective window of opportunity neoadjuvant study. Breast cancer research and treatment. 2012;135:821–830. doi: 10.1007/s10549-012-2223-1. [DOI] [PubMed] [Google Scholar]
- Pernicova I, Korbonits M. Metformin--mode of action and clinical implications for diabetes and cancer. Nature reviews Endocrinology. 2014;10:143–156. doi: 10.1038/nrendo.2013.256. [DOI] [PubMed] [Google Scholar]
- Quinn BJ, Kitagawa H, Memmott RM, Gills JJ, Dennis PA. Repositioning metformin for cancer prevention and treatment. Trends Endocrinol Metab. 2013;24:469–480. doi: 10.1016/j.tem.2013.05.004. [DOI] [PubMed] [Google Scholar]
- Roig AI, Eskiocak U, Hight SK, Kim SB, Delgado O, Souza RF, Spechler SJ, Wright WE, Shay JW. Immortalized epithelial cells derived from human colon biopsies express stem cell markers and differentiate in vitro. Gastroenterology. 2010;138:1012–1021. e1011–1015. doi: 10.1053/j.gastro.2009.11.052. [DOI] [PubMed] [Google Scholar]
- Sanchez-Cespedes M, Parrella P, Esteller M, Nomoto S, Trink B, Engles JM, Westra WH, Herman JG, Sidransky D. Inactivation of LKB1/STK11 is a common event in adenocarcinomas of the lung. Cancer Res. 2002;62:3659–3662. [PubMed] [Google Scholar]
- Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nature reviews Cancer. 2009;9:563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelford DB, Vasquez DS, Corbeil J, Wu S, Leblanc M, Wu CL, Vera DR, Shaw RJ. mTOR and HIF-1alpha-mediated tumor metabolism in an LKB1 mouse model of Peutz-Jeghers syndrome. Proc Natl Acad Sci U S A. 2009;106:11137–11142. doi: 10.1073/pnas.0900465106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shantha Kumara HM, Grieco MJ, Caballero OL, Su T, Ahmed A, Ritter E, Gnjatic S, Cekic V, Old LJ, Simpson AJ, et al. MAGE-A3 is highly expressed in a subset of colorectal cancer patients. Cancer Immun. 2012;12:16. [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Kosmatka M, Bardeesy N, Hurley RL, Witters LA, DePinho RA, Cantley LC. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc Natl Acad Sci U S A. 2004;101:3329–3335. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nature reviews Cancer. 2005;5:615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- Suter M, Riek U, Tuerk R, Schlattner U, Wallimann T, Neumann D. Dissecting the role of 5'-AMP for allosteric stimulation, activation, and deactivation of AMP-activated protein kinase. The Journal of biological chemistry. 2006;281:32207–32216. doi: 10.1074/jbc.M606357200. [DOI] [PubMed] [Google Scholar]
- Towler MC, Fogarty S, Hawley SA, Pan DA, Martin DM, Morrice NA, McCarthy A, Galardo MN, Meroni SB, Cigorraga SB, et al. A novel short splice variant of the tumour suppressor LKB1 is required for spermiogenesis. The Biochemical journal. 2008;416:1–14. doi: 10.1042/BJ20081447. [DOI] [PubMed] [Google Scholar]
- Wang C, Ivanov A, Chen L, Fredericks WJ, Seto E, Rauscher FJ, 3rd, Chen J. MDM2 interaction with nuclear corepressor KAP1 contributes to p53 inactivation. The EMBO journal. 2005;24:3279–3290. doi: 10.1038/sj.emboj.7600791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J, Salgaller M, Samid D, Johnson B, Herlyn M, Lassam N, Treisman J, Rosenberg SA. Expression of the MAGE-1 tumor antigen is up-regulated by the demethylating agent 5-aza-2'-deoxycytidine. Cancer Res. 1994;54:1766–1771. [PubMed] [Google Scholar]
- Weber P, Cammas F, Gerard C, Metzger D, Chambon P, Losson R, Mark M. Germ cell expression of the transcriptional co-repressor TIF1beta is required for the maintenance of spermatogenesis in the mouse. Development. 2002;129:2329–2337. doi: 10.1242/dev.129.10.2329. [DOI] [PubMed] [Google Scholar]
- Wei Y, Zou Z, Becker N, Anderson M, Sumpter R, Xiao G, Kinch L, Koduru P, Christudass CS, Veltri RW, et al. EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell. 2013;154:1269–1284. doi: 10.1016/j.cell.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein IB. Cancer. Addiction to oncogenes--the Achilles heal of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- White E. Deconvoluting the context-dependent role for autophagy in cancer. Nature reviews Cancer. 2012;12:401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winder WW, Holmes BF, Rubink DS, Jensen EB, Chen M, Holloszy JO. Activation of AMP-activated protein kinase increases mitochondrial enzymes in skeletal muscle. Journal of applied physiology. 2000;88:2219–2226. doi: 10.1152/jappl.2000.88.6.2219. [DOI] [PubMed] [Google Scholar]
- Wingo SN, Gallardo TD, Akbay EA, Liang MC, Contreras CM, Boren T, Shimamura T, Miller DS, Sharpless NE, Bardeesy N, et al. Somatic LKB1 mutations promote cervical cancer progression. PloS one. 2009;4:e5137. doi: 10.1371/journal.pone.0005137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A, Dickerson K, Heath R, Hong SP, Momcilovic M, Johnstone SR, Carlson M, Carling D. Ca2+/calmodulin-dependent protein kinase kinase-beta acts upstream of AMP-activated protein kinase in mammalian cells. Cell metabolism. 2005;2:21–33. doi: 10.1016/j.cmet.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Yang B, O'Herrin SM, Wu J, Reagan-Shaw S, Ma Y, Bhat KM, Gravekamp C, Setaluri V, Peters N, Hoffmann FM, et al. MAGE-A, mMage-b, and MAGE-C proteins form complexes with KAP1 and suppress p53-dependent apoptosis in MAGE-positive cell lines. Cancer Res. 2007;67:9954–9962. doi: 10.1158/0008-5472.CAN-07-1478. [DOI] [PubMed] [Google Scholar]
- Zheng B, Jeong JH, Asara JM, Yuan YY, Granter SR, Chin L, Cantley LC. Oncogenic B-RAF negatively regulates the tumor suppressor LKB1 to promote melanoma cell proliferation. Molecular cell. 2009;33:237–247. doi: 10.1016/j.molcel.2008.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Yang W, Wu F, Wang C, Yu L, Tang L, Qiu B, Li Y, Guo L, Wu M, et al. Prognostic significance of AMPK activation and therapeutic effects of metformin in hepatocellular carcinoma. Clin Cancer Res. 2013;19:5372–5380. doi: 10.1158/1078-0432.CCR-13-0203. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.