A plant intracellular immune receptor mediates broad-spectrum resistance by recognizing a small conserved PAMP-like region within the pathogen protein.
Abstract
Plants use both cell surface-resident pattern recognition receptors (PRRs) and intracellular nucleotide binding leucine-rich repeat (NLR) receptors to detect various pathogens. Plant PRRs typically recognize conserved pathogen-associated molecular patterns (PAMPs) to provide broad-spectrum resistance. By contrast, plant NLRs generally detect pathogen strain-specific effectors and confer race-specific resistance. Here, we demonstrate that the tomato (Solanum lycopersicum) NLR Sw-5b confers broad-spectrum resistance against American-type tospoviruses by recognizing a conserved 21-amino acid peptide region within viral movement protein NSm (NSm21). Sw-5b NB-ARC-LRR domains directly associate with NSm21 in vitro and in planta. Domain swap, site-directed mutagenesis and structure modeling analyses identified four polymorphic sites in the Sw-5b LRR domain that are critical for the recognition of NSm21. Furthermore, recognition of NSm21 by Sw-5b likely disturbs the residues adjacent to R927 in the LRR domain to weaken the intramolecular interaction between LRR and NB-ARC domains, thus translating recognition of NSm21 into activation of Sw-5b. Natural variation analysis of Sw-5b homologs from wild tomato species of South America revealed that the four polymorphic sites in the Sw-5b LRR domain were positively selected during evolution and are all necessary to confer resistance to tospovirus. The results described here provide a new example of a plant NLR mediating broad-spectrum resistance through recognition of a small conserved PAMP-like region within the pathogen effector.
INTRODUCTION
Plant and animal immune systems rely on membrane-resident pattern recognition receptors (PRRs) and intracellular nucleotide binding oligomerization domain-like receptors (NLRs) to recognize and prevent pathogen invasion (Couto and Zipfel, 2016; Jones et al., 2016). Plant PRRs typically include receptor kinases that recognize conserved pathogen-associated molecular patterns (PAMPs), such as flagellin or chitin, to provide broad-spectrum pathogen resistance (Zipfel, 2008; Couto and Zipfel, 2016). By contrast, plant NLRs generally recognize specific pathogen effectors and confer race-specific resistance (Caplan et al., 2008; Jones et al., 2016). Plant NLRs form the largest class of intracellular immune receptors ranging from 34 in papaya (Carica papaya) to 737 in apple (Malus domestica; Jones et al., 2016). Although animal NLRs provide broad-spectrum resistance by recognizing conserved PAMPs, it remains unknown if any plant NLRs can recognize small conserved PAMP-like structures derived from pathogens to impart broad-spectrum resistance.
A subclass of N-terminal coil-coil (CC) domain-containing NLRs (CC-NLRs) including Mi-1.2 and its homologs Hero and Sw-5b from the Solanaceae family is known to exhibit broad-spectrum resistance to several pathogens or pests. Tomato (Solanum lycopersicum) Mi-1.2 confers resistance to multiple root knot nematodes, Meloidogyne spp (Milligan et al., 1998; Vos et al., 1998), Macrosiphum euphorbiae aphid (Rossi et al., 1998; Vos et al., 1998), Bemisia tabaci whitefly (Nombela et al., 2003), and Bactericerca cockerelli psyllids (Casteel et al., 2006). In tomato, the Hero gene confers broad-spectrum resistance to all pathotypes of the nematodes Globodera rostochiensis and G. pallida (Ernst et al., 2002), whereas the Sw-5b gene confers resistance to several tomato-infecting tospoviruses (Brommonschenkel et al., 2000). How this subclass of plant NLRs provides broad-spectrum resistance to pathogens and/or pests remains largely unknown.
In addition to the N-terminal CC domain, the CC-NLRs contain a central nucleotide-binding, Apaf-1, R-protein, and CED-4 (NB-ARC) domain (van der Biezen and Jones, 1998) and the C-terminal leucine-rich repeat (LRR) domain. The NB-ARC domain can bind ADP to maintain the autoinhibited or “off” state or bind ATP to switch to the activated or “on” state (Tameling et al., 2006; Williams et al., 2011). It is hypothesized that NLR recognition of pathogen effectors directly or indirectly (Caplan et al., 2008; Jones et al., 2016) leads to changes in conformation of NLRs from the autoinhibited state to the activated state (Takken and Tameling, 2009). How this recognition event leads to an activation of NLR is poorly understood.
Tospoviruses are severe plant pathogens with a very broad host range, infecting more than 1000 plant species belonging to 80 different families (Goldbach and Peters, 1996). Tospoviruses cause an estimated annual crop loss of one billion U.S. dollars and are considered one of the most devastating plant viruses worldwide (Goldbach and Peters, 1996; Kormelink et al., 2011; Scholthof et al., 2011). Tomato Sw-5b CC-NLR is an effective dominant resistance gene that confers broad-spectrum resistance to tomato-infecting tospoviruses including Tomato spotted wilt virus (TSWV), Groundnut ring spot virus (GRSV), and Tomato chlorotic spot virus (TCSV) (Brommonschenkel et al., 2000; Spassova et al., 2001). The Sw-5b NLR recognizes TSWV movement protein NSm (López et al., 2011; Hallwass et al., 2014; Peiró et al., 2014), which is required for cell-to-cell movement and systemic infection of tospoviruses (Kormelink et al., 1994; Li et al., 2009; Feng et al., 2016). The cognate effector recognized by Mi-1.2 or Hero is currently unknown. The Sw-5b NLR and tospovirus NSm are an excellent model system to examine the molecular basis of NLR-mediated broad-spectrum resistance.
Here, we report that Sw-5b confers broad-spectrum resistance to multiple tospoviruses through interacting with a conserved 21-amino acid region in the NSm (NSm21). Using biochemical, molecular, and structure modeling-based approaches, we show that Sw-5b NB-ARC-LRR directly associates with NSm21. We identified four polymorphic sites and an adjacent arginine residue at position 927 (R927) in the LRR domain that are critical for translating recognition of NSm21 into activation of Sw-5b. Our results illustrate a new example of a plant NLR that confers broad-spectrum immunity through recognition of a small conserved pathogen protein-derived peptide region similar to PAMP. These findings provide molecular insights into the mechanistic basis of pathogen recognition and activation of NLRs that confers broad-spectrum resistance.
RESULTS
Sw-5b Confers Broad-Spectrum Resistance to American-Type Tospoviruses
Previous studies showed that tomato Sw-5b confers broad-spectrum resistance to tospoviruses including TSWV, GRSV, and TCSV (Boiteux and Giordano, 1993; Brommonschenkel et al., 2000). Because of the rapid increase in the characterization of new members in the Tospovirus genus in recent years, currently the genus includes more than 20 recognized and tentative species. These species are primarily classified as American and Euro/Asian groups (Supplemental Figure 1A and Supplemental Data Set 1) based on the amino acid sequences of nucleocapsid protein (>10% divergence is recognized as a new tospovirus species according to the criteria set by the International Committee on Taxonomy of Viruses). This prompted us to investigate if Sw-5b can confer resistance to members in both American- and Euro/Asian-type groups. We selected TSWV and Impatiens necrotic spot virus (INSV) to represent the American type tospoviruses and Tomato zonate spot virus (TZSV) and Hippeastrum chlorotic ringspot virus (HCRV) to represent the Euro/Asian type (Supplemental Figure 1A). These viruses were inoculated individually onto a previously characterized transgenic Nicotiana benthamiana plants expressing tomato Sw-5b (Nb:Sw-5b) (Chen et al., 2016). Nb:Sw-5b plants showed a hypersensitive cell death response (HR) in the TSWV or INSV inoculated leaves by 1 to 2 d postinoculation (Supplemental Figure 1B) and no systemic symptoms (Supplemental Figures 1C and 1D). However, Nb:Sw-5b plants inoculated with TZSV or HCRV showed no detectable HR in the inoculated leaves (Supplemental Figure 1B) but showed systemic symptoms (Supplemental Figures 1C and 1D). These results, combined with a previous report that Sw-5b confers resistance to GRSV and TCSV (Boiteux and Giordano, 1993; Brommonschenkel et al., 2000), suggest that Sw-5b confers broad-spectrum resistance to American-type but not to Euro/Asian-type tospoviruses.
Sw-5b Induces HR to American-Type Tospoviruses by Recognizing NSm Protein
Tomato Sw-5b induces HR through recognition of the movement protein NSm of TSWV (López et al., 2011; Hallwass et al., 2014; Peiró et al., 2014). To understand the molecular basis of susceptibility in Sw-5b plants to Euro/Asian-type tospoviruses, we cloned NSm from American-type tosposviruses (TSWV and INSV) and Euro/Asian-type tospoviruses (TZSV and HCRV) and fused them individually with a 3xHA tag at the C terminus. These NSms were coexpressed with full-length genomic Sw-5b (p2300S-Sw-5b) (Chen et al., 2016) in N. benthamiana leaves by agroinfiltration. Leaf areas coinfiltrated with TSWV-NSm or INSV-NSm with p2300S-Sw-5b showed cell death by 36 to 48 h postinfiltration (hpi) (Supplemental Figure 1E). By contrast, the leaf areas coinfiltrated with TZSV-NSm or HCRV-NSm with p2300S-Sw-5b did not show any visible cell death (Supplemental Figure 1E). Immunoblot assay with HA antibody showed that all NSm proteins were expressed in the infiltrated leaf areas (Supplemental Figure 1F). These results indicate that Sw-5b recognizes NSm from the analyzed American-type tospoviruses, but not those from the Euro/Asian-type tospoviruses.
A 21-Amino Acid Region in TSWV-NSm Is Sufficient to Induce Sw-5b-Mediated Cell Death
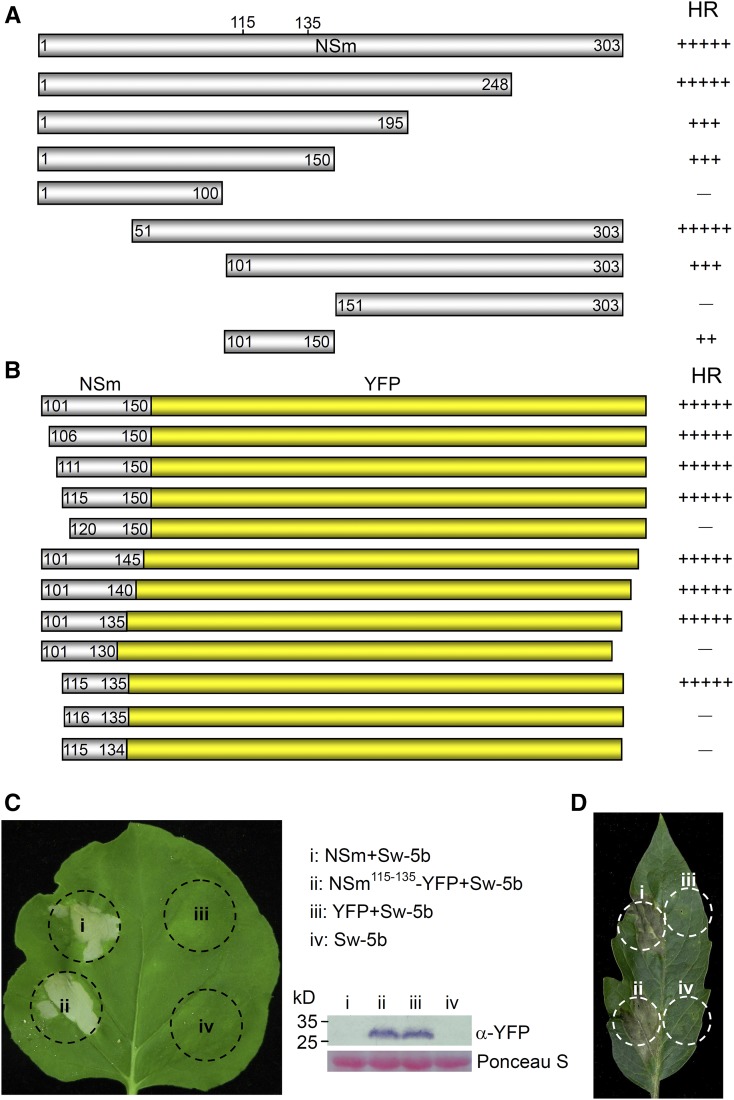
To determine the region in the NSm from American-type tospovirus that is recognized by Sw-5b, we generated a series of TSWV-NSm deletion mutants (Figure 1A). Coinfiltration of individual NSm deletion mutants with p2300S-Sw-5b into N. benthamiana leaves showed that amino acid residues 101 to 150 in TSWV-NSm are sufficient to induce moderate cell death (Figure 1A; the cell death scale is shown in Supplemental Figure 2). Because immunoblotting with NSm antibodies failed to detect TSWV-NSm101–150 (Supplemental Figure 3A), we reasoned that the observed moderate cell death could be due to lower accumulation of NSm101–150. Therefore, we fused yellow fluorescence protein (YFP) to the C terminus of TSWV-NSm101–150 (TSWV-NSm101–150-YFP). We observed significant accumulation of TSWV-NSm101–150-YFP in immunoblot analyses (Supplemental Figure 3B). Furthermore, coexpression of TSWV-NSm101–150-YFP with Sw-5b in N. benthamiana leaves induced an extensive cell death, similar to that induced by the full-length TSWV-NSm (Figure 1B).
Figure 1.
A 21-Amino Acid Peptide Region in NSm Is Sufficient to Induce Sw-5b-Mediated Cell Death.
(A) Analyses of NSm deletion mutants for the induction of cell death in the presence of Sw-5b. The truncated mutants of NSm with their remaining amino acids are shown below the full-length 303-amino acid NSm. The severity of the cell death induced by each construct is shown on the right side. (−) to (+++++) indicate the severity of cell death on a scale of 0 to 5, as shown in Supplemental Figure 2.
(B) Fine mapping of the region in NSm required for the induction of cell death. The NSm truncated mutants shown were fused individually to the N terminus of YFP, and transiently coexpressed with Sw-5b in N. benthamiana leaves. The strength of the cell death induced by each construct is shown on the right side. (−) to (+++++) indicate the severity of cell death on a scale of 0 to 5, as shown in Supplemental Figure 2.
(C) and (D) Cell death induced by the full-length NSm or NSm115–135-YFP in the presence of Sw-5b in wild-type N. benthamiana plants (C) and in tomato (D). Cell death in the infiltrated leaves was photographed at 4 d postinoculation. Expression of NSm115–135–YFP and YFP in the infiltrated leaves was confirmed by immunoblot assay using YFP antibody (C). The sizes of proteins in kilodaltons are shown to the left of the blot.
To further define the region within TSWV-NSm101–150 that is required for Sw-5b recognition, we generated sequential deletions of one to five amino acids starting from the N or C terminus of TSWV-NSm101–150. Coinfiltration of individual deletion mutants with p2300S-Sw-5b in leaves of N. benthamiana or transgenic Nb::Sw-5b demonstrated that only the 21-amino acid residues from 115 to 135 of TSWV-NSm are sufficient to induce extensive cell death (Figures 1B and C; Supplemental Figures 3B and 3C). In addition, this 21-amino acid region alone was sufficient to induce cell death when coexpressed with Sw-5b in tomato leaves (Figure 1D). Further single amino acid deletions at either the N or C terminus of the 21-amino acid peptide region abolished their ability to induce cell death (Figure 1B) even though these deletions are expressed at the same level as the 21-amino acid peptide fused to YFP (Supplemental Figure 3B). Therefore, TSWV-NSm residues 115 to 135 (hereafter referred to as NSm21) are sufficient to induce Sw-5b-mediated cell death.
The NSm21 Region in American-Type Tospoviruses Is Highly Conserved and Sufficient to Induce Sw-5b-Mediated Cell Death
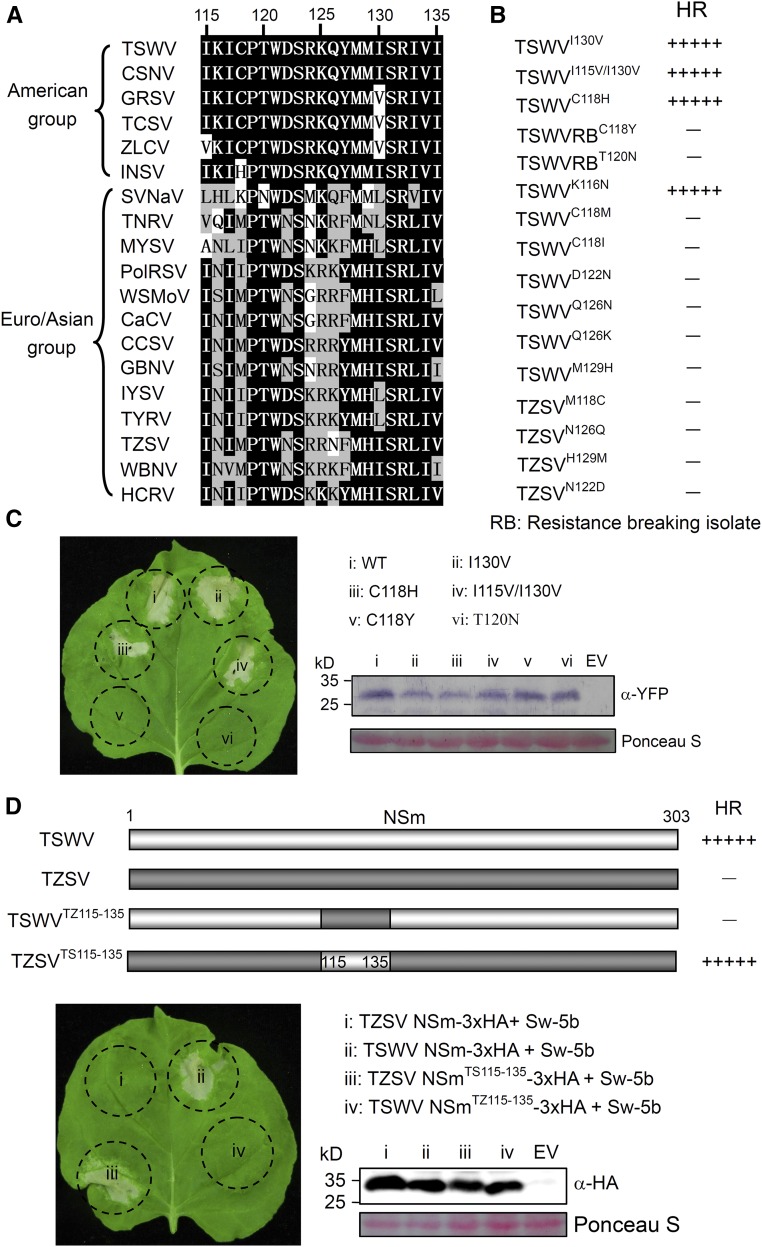
Sequence alignment using ClustalW showed that the NSm21 region from different American-type tospoviruses is highly conserved except for I130V in GRSV and TCSV, I115V/I130V in ZLCV, and C118H in INSV (Figure 2A). Introduction of I115V, I130V, I115V/I130V, or C118H into TSWV-NSm showed that these changes have no effect on the induction of cell death in the presence of Sw-5b (Figures 2B and 2C).
Figure 2.
NSm21 from American-Type but Not Euro/Asian-Type Tospoviruses Functions as an Active Inducer of Sw-5b-Mediated Cell Death.
(A) Alignment of the 21-amino acid region in NSm (NSm21) from American- and Euro/Asian-type tospoviruses by ClustalW.
(B) Cell death induction by various NSm21 mutants carrying specific amino acid substitutions. (−) to (+++++) indicate the severity of cell death on a scale of 0 to 5, as shown in Supplemental Figure 2.
(C) C118Y or T120N substitutions in NSm21-YFP abolish its ability to induce cell death. The leaf was photographed at 5 d postinfiltration. Expression of fusion proteins was confirmed by immunoblot analysis using YFP antibody. The sizes of proteins in kilodaltons are shown to the left of the blot.
(D) Replacement of the 21-amino acid region of TZSV NSm with TSWV induces cell death. TZSV NSm carrying TSWV NSm21 strongly induced cell death. TSWV NSm with TZSV NSm21 failed to induce cell death. Expression of chimeric NSm was confirmed by immunoblot assays using HA-specific antibody. The sizes of proteins in kilodaltons are shown to the left of the blot.
Two natural TSWV-NSm variants (NSmC118Y and NSmT120N) were shown to overcome the Sw-5b-mediated resistance in tomato (López et al., 2011). Interestingly, these two sites are located within the NSm21 region (Figure 2A). To test the effect of these two sites, we introduced these mutations in NSm21 fused to YFP at the C terminus (NSm21-YFP) and then coinfiltrated these constructs individually with p2300S-Sw-5b into N. benthamiana leaves. In contrast to the TSWV-NSm21 control, coinfiltration of TSWV-NSm21(C118Y)-YFP or TSWV-NSm21(T120N)-YFP with Sw-5b failed to induce cell death (Figure 2C).
Comparison of the NSm21 region from the American-type tospoviruses with that from the Euro/Asian type revealed that over nine residues in this 21-amino acid region differ between the two geographic types (Figure 2A). To test whether the NSm21 from an American-type tospovirus is sufficient to convert a Euro/Asian tospovirus NSm into an Sw-5b eliciting effector, we replaced amino acid residues 115 to 135 in TSWV-NSm with those from TZSV-NSm (TSWVTZ115-135) and in TZSV-NSm with those from TSWV-NSm (TZSVTS115-135). All wild-type NSm and NSm chimeric constructions were tagged with 3xHA at the C terminus to facilitate detection through immunoblot analyses. Coinfiltration of these chimeric constructs individually with p2300S-Sw-5b into N. benthamiana leaves showed that TZSVTS115-135 induced cell death, whereas TSWVTZ115-135 did not (Figure 2D). Immunoblot analysis confirmed that the chimeric NSm mutants were produced at similar amounts (Figure 2D). These results indicated that the 21-amino acid conserved region in the NSm of the American-type tospovirus is sufficient to convert Euro/Asian-type NSm into a form that elicits Sw-5b-mediated cell death.
To further map the key residues in the 21-amino acid peptide region responsible for cell death induction, we generated K116N, C118M, C118I, D122N, Q126N, Q126K, and M129H (all from TZSV-NSm) individually in TSWV-NSm. Immunoblot analyses indicated that all these mutants are expressed (Supplemental Figure 4). Interestingly, among these mutants, only TSWVK116N induced cell death (Figure 2B). Similarly, when the M118C, N122D, N126Q, and H129M mutation (identified in TSWV-NSm) were individually introduced into the corresponding site in TZSV-NSm, none of them induced cell death (Figure 2B; Supplemental Figure 4). However, introduction of TSWV-NSm residues into all six sites (M118C, N122D, R125K, N126Q, H129M, and LIV133-135IVI) of TZSV-NSm successfully triggered cell death (Supplemental Figure 5). These results indicate that a highly conserved 21-amino acid region of NSm from American tospoviruses could induce Sw-5b-mediated cell death. In addition, the eight residues in NSm at positions 118, 122, 125, 126, 129, and 133 to 135 are critical for Sw-5b-mediated cell death induction.
Sw-5b Interacts with NSm21
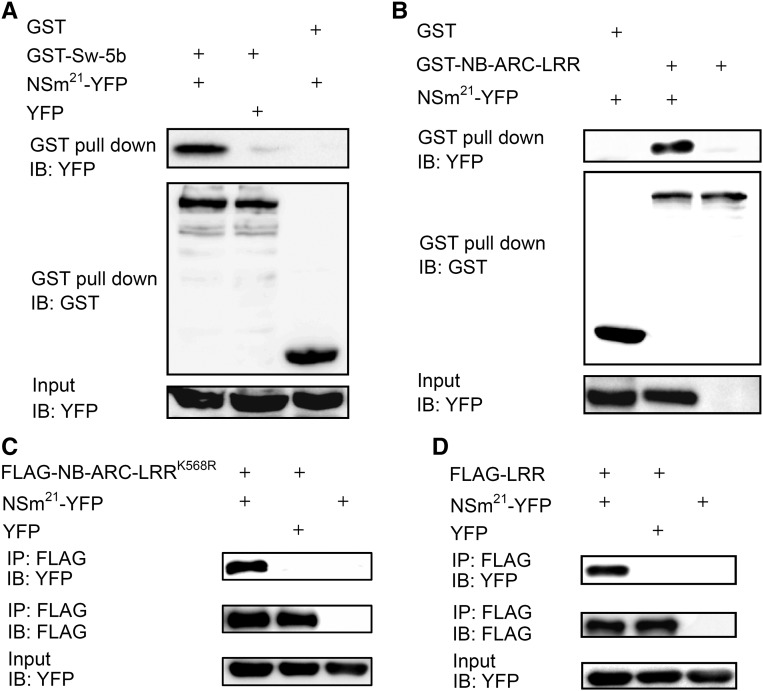
To determine if Sw-5b interacts with the conserved 21-amino acid peptide region of TSWV-NSm, we coexpressed Sw-5b and TSWV-NSm21-YFP in N. benthamiana by agroinfiltration. Because coexpression of Sw-5b with NSm21-YFP caused very strong cell death, it was difficult to perform a coimmunoprecipitation (co-IP) assay. Therefore, we expressed Sw-5b fused to GST at the N terminus in Escherichia coli. Although the expression level was low, we were able to detect GST-Sw-5b fusion protein by immunoblot analyses (Figure 3A, middle panel). We performed a GST pull-down assay by mixing GST-Sw-5b with protein extracts from N. benthamiana leaf tissue expressing NSm21-YFP. GST-Sw-5b was able to pull down a substantial amount of NSm21-YFP compared with the YFP control (Figure 3A). GST alone did not pull down NSm21-YFP (Figure 3A). These results indicate that Sw-5b physically associates with the conserved 21-amino acid region of the TSWV-NSm effector.
Figure 3.
Sw-5b Interacts with NSm21 in Vitro and in Planta.
(A) GST pull-down assay of the interaction between the full-length Sw-5b and NSm21-YFP. GST-Sw-5b or GST was expressed and purified from E. coli. NSm21-YFP was expressed in N. benthamiana plants and harvested at 24 h postagroinfiltration. The purified GST-Sw-5b or GST was mixed with plant extracts of NSm21-YFP and pulled down with GST beads. GST-Sw-5b pulled down a significant amount of NSm21-YFP compared with the YFP control. GST alone failed to pull down NSm21-YFP.
(B) Sw-5b NB-ARC-LRR and NSm21-YFP interact in an in vitro GST pull-down assay. Purified GST-Sw-5b-NB-ARC-LRR expressed in E. coli pulled down purified NSm21-YFP-6xhis. NSm21-YFP-6xhis was not pulled down with GST alone.
(C) Sw-5b NB-ARC-LRRK568R associates with NSm21-YFP in planta. Sw-5b P-loop mutant NB-ARC-LRRK568R fused to FLAG tag coimmunoprecipitated NSm21-YFP when coexpressed in N. benthamiana leaves but not with YFP alone.
(D) Sw-5b LRR associates with NSm21-YFP in planta. Sw-5b LRR fused to FLAG tag coimmunoprecipitated with NSm21-YFP when coexpressed in N. benthamiana leaves but not with YFP alone.
IB, immunoblot with specific antibody; IP, immunoprecipitation with specific antibody.
The Sw-5b NB-ARC-LRR Region Directly Associates with NSm21 in Vitro and in Planta
We recently showed that the Sw-5b NB-ARC-LRR region lacking N terminus and CC domains was autoinhibited in the absence of NSm but activated in the presence of NSm (Chen et al., 2016). To determine if the Sw-5b NB-ARC-LRR region could induce cell death in the presence of NSm21, we coexpressed nontagged or N-terminally FLAG-tagged NB-ARC-LRR and NSm21-YFP in N. benthamiana leaves. The coexpression of NB-ARC-LRR (or FLAG-NB-ARC-LRR) with NSm21-YFP led to a strong cell death response in the infiltrated leaves but not with YFP alone (Supplemental Figures 6A and 6B). To determine whether Sw-5b NB-ARC-LRR could directly interact with NSm21, GST-NB-ARC-LRR fusion protein and NSm21-YFP-(His)6 were expressed in E. coli. Two purified proteins were mixed at a 1:1 ratio and pulled down using the GST beads. In contrast to GST alone, GST-NB-ARC-LRR directly pulled down NSm21-YFP (Figure 3B).
To confirm these results, FLAG-NB-ARC-LRR and NSm21-YFP were coexpressed in N. benthamiana leaves. However, our repeated attempts showed that FLAG-NB-ARC-LRR and NSm21-YFP were rapidly degraded upon onset of cell death, which made it difficult to perform co-IP assays. Previous reports have shown that mutation in the P-loop of Arabidopsis RPP1 NLR and tobacco N NLR abolished their ability to induce cell death, but not their ability to associate with cognate effectors (Krasileva et al., 2010; Padmanabhan et al., 2013). We showed recently that K568R mutation in the P-loop of Sw-5b NB-ARC-LRR abolished its ability to induce HR (Chen et al., 2016). Therefore, we tested if Sw-5b FLAG-NB-ARC-LRRK568R could associate with NSm21 in planta. Coexpression of FLAG-NB-ARC-LRRK568R with NSm21-YFP in N. benthamiana leaves did not induce HR (Supplemental Figure 6C) but FLAG-NB-ARC-LRRK568R coimmunoprecipitated with NSm21-YFP but not with YFP alone (Figure 3C). These findings confirmed that Sw-5b NB-ARC-LRR interacts with NSm21 in planta.
Four Polymorphic Sites in the Sw-5b LRR Domain Are Critical for NSm21 Recognition
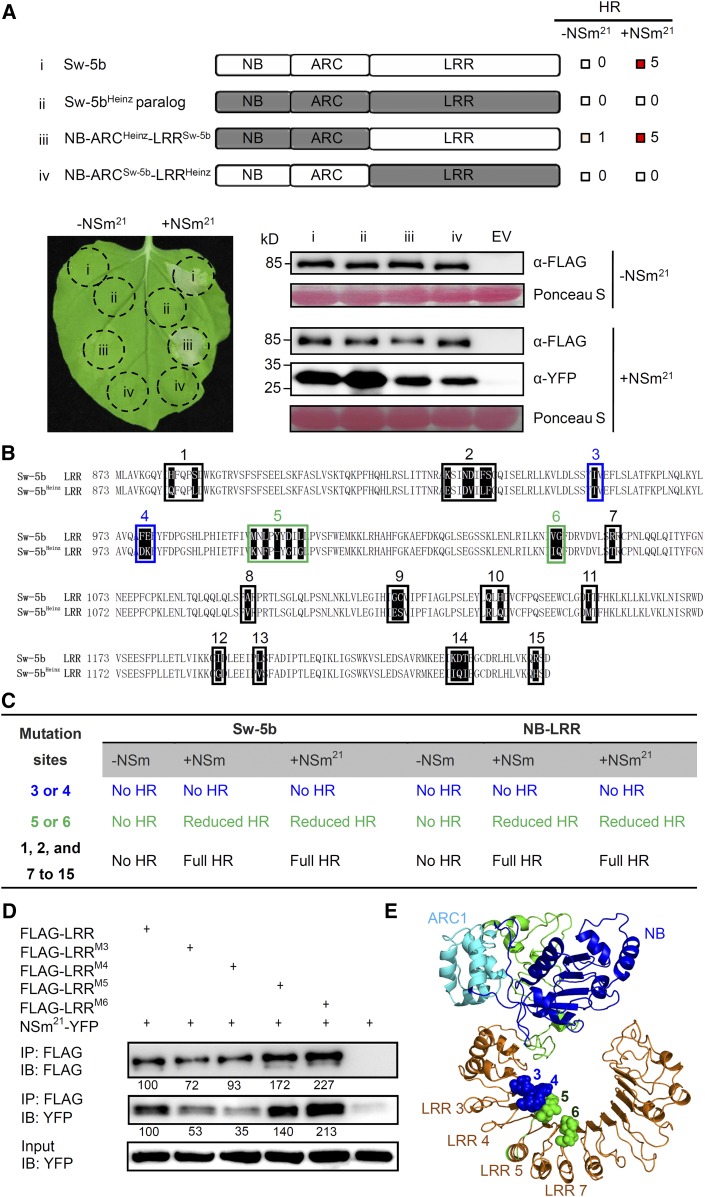
The tomato cultivar Heinz1706 is susceptible to TSWV infection. We analyzed the Heinz1706 genome sequence (Tomato Genome Consortium, 2012) and identified the Sw-5b paralog (Sw-5bHeinz). We cloned Sw-5bHeinz NB-ARC-LRR and coexpressed this construct with NSm21-YFP in N benthamiana leaves. As expected, it failed to induce cell death, suggesting that Sw-5bHeinz is not able to recognize NSm21 (Figure 4A). Comparative analyses of protein sequences of Sw-5b and Sw-5bHeinz showed very few polymorphic residues (Supplemental Figure 7). To determine the region responsible for recognition of NSm21-YFP and induction of cell death, we generated NB-ARC and LRR domain swaps (Figure 4A). Replacement of the LRR domain of Sw-5bHeinz with that of Sw-5b (NB-ARCHeinz-LRRSw-5b) was sufficient to induce cell death in the presence of NSm21-YFP (Figure 4A). Interestingly, this chimeric protein also induced weak cell death in the absence of NSm21-YFP (Figure 4A; the cell death scale is shown in Supplemental Figure 2). Replacement of the LRR domain of Sw-5b with Heinz LRR (NB-ARCSw-5b-LRRHeinz) was unable to induce cell death in the presence of NSm21-YFP (Figure 4A). These results suggest that the Sw-5b LRR domain is responsible for the recognition of NSm21.
Figure 4.
Determination of the NSm21 Recognition Sites in Sw-5b.
(A) Expression of FLAG-tagged Sw-5b (i) and chimeric NB-ARCHeinz-LRRSw-5b (iii) induces cell death when coexpressed with NSm21 in N. benthamiana plants. Sw-5bHeinz (ii) and chimeric NB-ARCSw-5b-LRRHeinz (iv) failed to induce cell death when coexpressed with NSm21. Expression of individual proteins was confirmed by immunoblot assay using FLAG and NSm antibodies.
(B) Alignment of the LRR sequence of Sw-5b and Sw-5bHeinz. Boxed regions indicate 15 different polymorphic sites/region between Sw-5b and Sw-5bHeinz.
(C) Each polymorphic site in Sw-5b LRR was replaced with the cognate site found in the Heinz paralog in full-length Sw-5b and Sw-5b-NB-ARC-LRR and expressed alone or coexpressed with NSm or NSm21 in N. benthamiana plants. Normal cell death, reduced cell death, or no cell death response was recorded 5 d after expression. The polymorphic sites 3 and 4 are shown in blue; the polymorphic sites 5 and 6 are shown in green; the polymorphic sites 1, 2, and 7 to 15 are shown in black.
(D) Sw-5b FLAG-LRR, FLAG-LRRM3, FLAG-LRRM4, FLAG-LRRM5, or FLAG-LRRM6 immunoprecipitate NSm21-YFP when coexpressed in N. benthamiana leaves. The amount of NSm21-YFP, FLAG-LRR and mutants precipitated were quantified using ImageQuant software. The relative fold of mutant in each lane was compared with immunoprecipitated wild type. Similar results were obtained from more than three biological repeats. IP, immunoprecipitation with specific antibody; IB, immunoblot with specific antibody.
(E) Mapping of four polymorphic sites 3, 4, 5, and 6 on the three-dimensional homology structure model of Sw-5b-NB-ARC-LRR. I954 at polymorphic site 3, and F977 and E978 at polymorphic site 4 are in a blue sphere. M996 (highlighted with sphere), L998, Y1000, D1002, and L1004 at polymorphic sites 3 are shown in green. V1047 and G1048 at polymorphic site 4 are shown in a green sphere.
To further map the critical residues in Sw-5b LRR responsible for NSm21 recognition, we generated 15 substitution mutants in the Sw-5b LRR domain with those identified in the Heinz LRR domain (Figure 4B). Coexpression of individual substitution mutants with NSm21-YFP in N. benthamiana leaves showed that Sw-5b LRR with substitutions at the polymorphic sites 1, 2, or 7 through 15 were still able to induce strong cell death similar to that induced by the wild-type Sw-5b or Sw-5b NB-ARC-LRR (Figures 4B and 4C). Interestingly, substitution at polymorphic site 3 (M3) or 4 (M4) abolished the cell death and substitution at polymorphic site 5 (M5) or 6 (M6) resulted in reduced cell death in the presence of NSm21-YFP (Figures 4B and 4C; Supplemental Figure 8A). Time-course ion leakage analyses showed that the conductivity in tissue infiltrated with M4 was reduced to the basal level and M3 was reduced to 34 to 62% at 24 to 48 hpi compared with the wild type (Supplemental Figure 8B). At 24 to 42 hpi, the conductivity in tissue infiltrated with M5 was reduced to 12 to 38% of tissue infiltrated with the wild type and that of tissue infiltrated with M6 was reduced to 11 to 39% (Supplemental Figure 8B). Immunoblot analysis indicated that the abolished or reduced cell death caused by the M3, M4, M5, and M6 mutants is not due to a reduction of protein accumulation or stability of NSm21-YFP (Supplemental Figure 8C).
To elucidate the role of Sw-5b LRR polymorphic sites 3 to 6 in recognizing NSm21, we generated FLAG-tagged wild-type and mutant versions of LRR. We coexpressed these mutants with NSm21-YFP in N. benthamiana leaves and performed a co-IP assay. FLAG-LRRWT did pull down NSm21-YFP efficiently but not YFP (Figures 3D and 4D). Because the loading of the four polymorphic LRR mutants was always not equal to that of the LRRWT control in three independent experiments, we normalized the amount of NSm21-YFP to the amount of LRR protein pulled down. This analysis indicated that FLAG-LRRM3, FLAG-LRRM4, FLAG-LRRM5, and FLAG-LRRM6 mutants pulled down 26, 62, 19, and 6% less NSm21-YFP compared with FLAG-LRRWT (Figure 4D).
To further understand the role of polymorphic sites in LRR, we analyzed these sites in the three-dimensional Sw-5b NB-ARC-LRR homology structure model that we reported previously (Chen et al., 2016). Analysis of the Sw-5b LRR sequence revealed that it has 15 LRRs and polymorphic sites 3, 4, 5, and 6 are in the LRR 3, 4, 5, and 7, respectively (Supplemental Figure 9A; Figure 4E). Although these four sites are in four different LRRs, they cluster together on the three-dimensional homology structure (Figure 4E) and are all surface-exposed (Supplemental Figure 9B). These findings suggest that these residues might play important roles in the recognition of the NSm21.
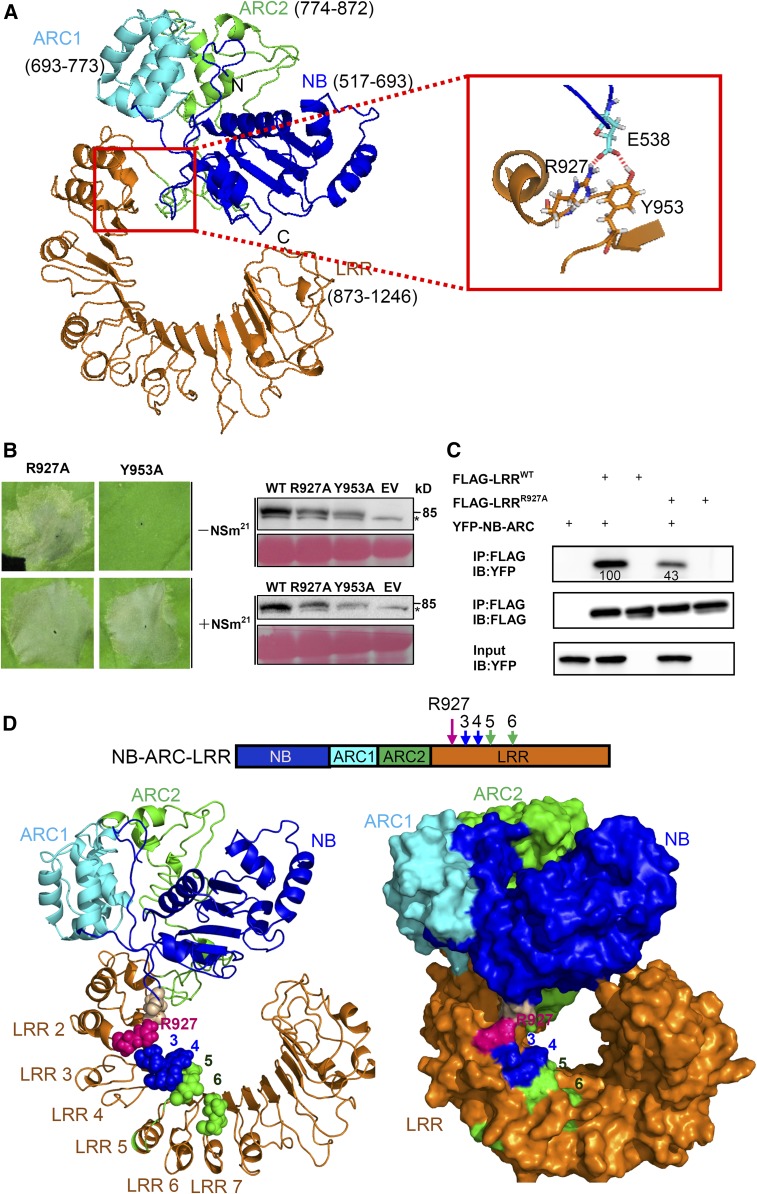
R927, Which Resides Adjacent to Four Polymorphic Sites in the LRR Domain, Is Required to Keep Sw-5b in an Autoinhibited State
We showed previously that the Sw-5b LRR domain retains the NB-ARC domain in an autoinhibited state and that deletion of LRR autoactivates the NB-ARC domain, leading to cell death (Chen et al., 2016). To understand how the LRR interacts with the NB-ARC domain to keep Sw-5b in an autoinhibited state, we searched for the potential interaction sites between LRR and NB-ARC domains in the predicted Sw-5b NB-ARC-LRR structure model (Figure 5A). This analysis showed that the N terminus of the LRR and NB-ARC are located in close proximity (Figure 5A, boxed region). Furthermore, R927 and Y953 in the LRR could potentially interact with E538 in a loop-like structure formed by the hhGRExE motif in NB (Figure 5A, enlarged box). To test these predictions, we introduced the R927A and Y953A mutation individually into NB-ARC-LRR. Transient expression of these mutants individually in N. benthamiana leaves showed that the R927A mutant but not Y953A, constitutively induced cell death in the absence of NSm21 (Figure 5B). To investigate the effect of the R927A mutation on NB-ARC and LRR interactions, we coexpressed FLAG-tagged wild-type LRR (FLAG-LRRWT) or FLAG-LRRR927A with YFP-NB-ARC in N. benthamiana leaves followed by co-IP. The R927A mutation strongly reduced the interaction between LRR and NB-ARC compared with FLAG-LRRWT (Figure 5C). To determine whether the autoactivity of R927A required a functional nucleotide binding/hydrolysis or the mutant LRR had adopted a permanent conformational change, we generated the K568R mutation in the P-loop of FLAG-NB-ARC-LRRR927A (FLAG-NB-ARC-LRRK568RandR927A). The FLAG-NB-ARC-LRRK568RandR927A mutant was unable to induce cell death in the absence or presence of NSm21-YFP (Supplemental Figure 10). These results suggest that nucleotide binding is still required for autoactivation of NB-ARC-LRRR927A.
Figure 5.
The R927 Residue Adjacent to Four Polymorphic Sites Keeps Sw-5b in an Autoinhibited State.
(A) Putative interaction interface between the LRR and NB-ARC domain on the three-dimensional homology structure of Sw-5b NB-ARC-LRR is shown in the red box. NB (blue), ARC1 (cyan), ARC2 (green), LRR (amino acid residue 873–1246; orange) are subdomains of Sw-5b NB-ARC-LRR. The enlarged red boxed on the right shows that amino acid R927 and Y953 in the LRR and E538 in the NB can form a putative contact site.
(B) The R927A (YFP-NB-ARC-LRRR927) but not the Y953A (YFP-NB-ARC-LRRY953) mutant results in cell death in the absence of NSm21 in leaves of N. benthamiana plants. Expression of R927A and Y953A induces cell death in the presence of NSm21. The right panels show the accumulation of proteins using an anti-YFP antibody. The sizes of proteins in kilodaltons are shown to the right of the blot. Asterisk represents the nonspecific band. Ponceau S staining was used to show protein loading.
(C) The effect of the R927A mutant on the physical interaction between the LRR and NB-ARC domains. Wild-type Sw-5b LRR fused to FLAG tag (FLAG-LRRWT) or R927A mutant (FLAG-LRRR927A) was used to immunoprecipitate YFP-NB-ARC. The amount of YFP-NB-ARC precipitated by the wild type or R927A was quantified by ImageQuant software. IP, immunoprecipitation with specific antibody; IB, immunoblot with specific antibody.
(D) Location of R927 residue and the polymorphic sites 3, 4, 5, and 6 on the three-dimensional homology structure of Sw-5b NB-ARC-LRR. The upper panel shows NB-ARC-LRR with the indicated position of the R927 residue and the four polymorphic sites. The ribbon and surface homology structure model of Sw-5b NB-ARC-LRR are shown in the left and right, respectively. The R927 residue is shown in a purple sphere, while the four polymorphic sites are shown as in Figure 4E.
The R927 residue is located on LRR2 of Sw-5b and is upstream of the four polymorphic sites important for NSm21 recognition (Figure 5D; Supplemental Figure 9A). When we mapped the R927 residue and the four polymorphic sites together onto the three-dimensional structure model of Sw-5b NB-ARC-LRR, R927 was located next to the four polymorphic sites (Figure 5D).
Mutual Interaction between Four Polymorphic Sites and the R927 Residue on Sw-5b LRR
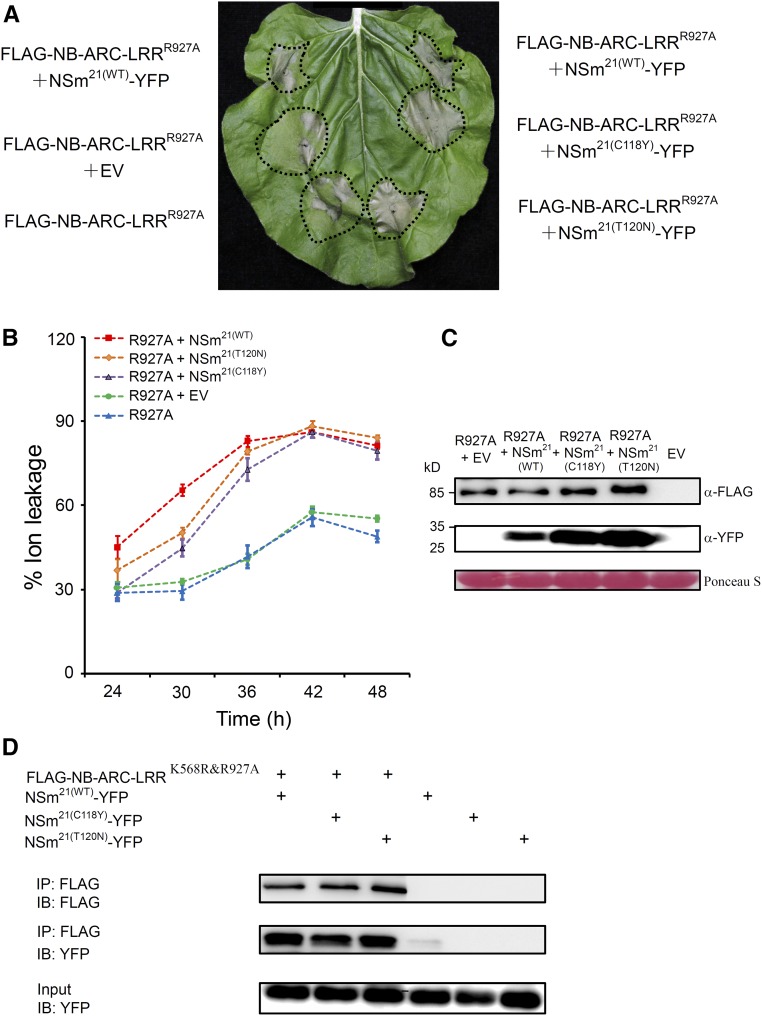
Three-dimensional structure modeling indicated that residue R927 and the four polymorphic sites were clustered together and might mutually interact. To test the potential interaction between the R927 residue and NSm21, we coexpressed noneliciting NSm effector mutants, NSm21(C118Y) or NSm21(T120N), with Sw-5b FLAG-NB-ARC-LRRR927A in N. benthamiana leaves. We used the 8th and 9th leaves (down from the apex) of 12-week-old N. benthamiana plants for this assay to reduce the autoactivity of Sw-5b FLAG-NB-ARC-LRRR927A, as older leaves tend to show less extensive HR cell death (Bendahmane et al., 2002). In this assay, Sw-5b FLAG-NB-ARC-LRRR927A induced moderate autoactivated cell death (Figure 6A). Surprisingly, the addition of NSm21(C118Y) or NSm21(T120N) induced much more extensive cell death with Sw-5b FLAG-NB-ARC-LRRR927A than was caused by Sw-5b FLAG-NB-ARC-LRRR927A alone (Figures 6A and 6C; Supplemental Table 1). A time-course ion leakage assay confirmed that the conductivity of tissue coexpressing FLAG-NB-ARC-LRRR927A and NSm21(C118Y) or NSm21(T120N) was 12 to 38% higher than that in FLAG-NB-ARC-LRRR927A alone or when coexpressed with the empty vector at 30 to 48 hpi (Figure 6B). To examine the association of NSm21(C118Y) and NSm21(T120N) with Sw-5b FLAG-NB-ARC-LRRR927A, we coexpressed NSm21(C118Y)-YFP or NSm21(T120N)-YFP with FLAG-NB-ARC-LRRK568RandR927A in N. benthamiana leaves followed by co-IP. The results showed that both NSm21(C118Y) and NSm21(T120N) associate with Sw-5b FLAG-NB-ARC-LRRK568RandR927A (Figure 6D). We also examined the interaction of noneliciting NSm21(C118Y) and NSm21(T120N) mutants with wild-type Sw-5b FLAG-NB-ARC-LRRK568R. Both mutants associate with Sw-5b FLAG-NB-LRRK568R, similar to wild-type NSm21-YFP (Supplemental Figure 11). Collectively, these results suggest that these two noneliciting NSm21 mutants were able to enhance cell death induced by Sw-5b NB-ARC-LRRR927A.
Figure 6.
Nonelicitor NSm21(C118Y) and NSm21(T120N) Could Enhance Cell Death Induced by Sw-5b NB-ARC-LRRR927A.
(A) Cell death assay of NSm21(C118Y) and NSm21(T120N) with Sw-5b FLAG-NB-ARC-LRRR927A in the leaves of 12-week-old N. benthamiana plants. The 8th leaf on a 12-week-old N. benthamiana plant was coinfiltrated with Sw-5b FLAG-NB-ARC-LRRR927A and NSm21-YFP, FLAG-NB-ARC-LRRR927A and NSm21(C118Y)-YFP or FLAG-NB-ARC-LRRR927A and NSm21(T120N)-YFP. Areas infiltrated with FLAG-NB-ARC-LRRR927A alone or with both FLAG-NB-ARC-LRRR927A and the empty expression vector were used as controls. The infiltrated leaf was photographed at 5 d postinoculation. The infiltrated area for each treatment is marked with dotted line.
(B) Time course of ion leakage caused by expression of Sw-5b FLAG-NB-ARC-LRRR927A (R927A), FLAG-NB-ARC-LRRR927A + empty vector (R927A+EV), FLAG-NB-ARC-LRRR927A + NSm21(WT)-YFP [R927A+NSm21(WT)], FLAG-NB-ARC-LRRR927A + NSm21(C118Y)-YFP [R927A+NSm21(C118Y)], and FLAG-NB-ARC-LRRR927A + NSm21(T120N)-YFP [R927A+NSm21(T120N)] in the leaves of 12-week-old N. benthamiana plants are shown.
(C) Immunoblot detection of FLAG-NB-ARC-LRRR927A and the wild-type and mutant NSm21 proteins using a FLAG-specific and an YFP-specific antibody, respectively. The sizes of proteins in kilodaltons are shown to the left of the blot. Ponceau S staining was used as a protein loading control.
(D) FLAG-NB-ARC-LRR K568RandR927A was coexpressed with NSm21(WT)-YFP, NSm21(C118Y)-YFP, or NSm21(T120N)-YFP in N. benthamiana leaves followed by co-IP assays using a FLAG-specific and a YFP-specific antibody. Leaf tissues expressing NSm21(WT)-YFP, NSm21(C118Y)-YFP, or NSm21(T120N)-YFP were used as a negative control. IP, immunoprecipitation with specific antibody; IB, immunoblot with specific antibody.
To further determine the possible interaction between four polymorphic sites and the R927 residue on the LRR, we tested whether mutation of the four polymorphic sites would affect R927A-mediated autoactivation. To this end, we mutagenized each polymorphic site in Sw-5b according to its cognate sites in Sw-5bHeinz. Mutations introduced into the polymorphic site 3, 4, or 5 severely reduced the autoactivity of Sw-5b NB-ARC-LRRR927A (Supplemental Table 2 and Supplemental Figure 12). However, mutation at polymorphic site 6 did not affect the autoactivity (Supplemental Table 2). Further analysis showed that introduction of mutations into any two polymorphic sites simultaneously or to all four polymorphic sites in Sw-5b NB-ARC-LRRR927A affected the autoactivity (Supplemental Table 2). These results indicate that all four polymorphic sites in Sw-5b LRR contribute to keep R927 at a proper configuration for activation. To investigate whether these polymorphic sites also play roles in other autoactivation sites, we mutagenized polymorphic sites 3, 4, and 5 individually in the two previously reported autoactivation mutants, Sw-5b NB-ARC-LRRD642E (mutation at the NB subdomain) and Sw-5b NB-ARC-LRRD857V (mutation at the ARC2 subdomain) (Chen et al., 2016). The mutations at three polymorphic sites in Sw-5b NB-ARC-LRRD642E or Sw-5b NB-ARC-LRRD857V did not alter their autoactivation (Supplemental Table 2 and Supplemental Figure 12). These results indicate that these polymorphic sites specifically function in the R927-mediated autoactivation. Collectively, we conclude that four polymorphic sites and R927 in the Sw-5b LRR likely functionally interact.
Natural Variation at the Four Polymorphic Sites and R927 in Sw-5b Homologs from Wild Tomato Species
American-type tospoviruses originated in South America (de Avila et al., 1990; Bezerra et al., 1999; Pozzer et al., 1999; Williams et al., 2001). Interestingly, various tomato species also originated from South America (Tomato Genome Consortium, 2012; Lin et al., 2014). The Sw-5b gene was derived from wild tomato species Solanum peruvianum in South America and later introduced into cultivated tomato (Stevens et al., 1992). The same geographic distribution of American-type tospoviruses and Sw-5b suggest that Sw-5b might have evolved in tomato to resist tospoviruses. To investigate the natural variation at four polymorphic sites and the R927 in Sw-5b, we obtained 27 accessions representing all 17 wild tomato species from South America. We cloned and sequenced Sw-5b homologs from 25 out of 27 accessions because seeds of two accessions did not germinate (Table 1). Interestingly, R927 is conserved in all sequenced Sw-5 homologs. Sw-5b homologs with all four polymorphic sites conserved were able to induce cell death when coexpressed with NSm or NSm21 (Table 1; Supplemental Figure 13) and also conferred resistance to TSWV infection assayed by ELISA (Table 1). By contrast, homologs with changes in polymorphic site 3 alone with other sites conserved were not able to induce cell death or confer resistance to TSWV. Furthermore, changes in 4, 5, and 6 sites and conservation of site 3 are also not sufficient to induce cell death (Table 1; Supplemental Figure 13). These findings suggest that the four polymorphic sites in the Sw-5b LRR domain were positively selected during evolution and these polymorphic residues in all four distinct sites are necessary to confer resistance to TSWV.
Table 1. Natural Variation Analysis of R927 and the Four Polymorphic Sites (3, 4, 5, and 6) in the Sw-5 LRR Domain of Wild Tomato Species.
| Accessions | Taxon | HRa | ELISAb | R927 | 3 (I) | 4 (FE) | 5 (MNLPYYDIL) | 6 (VG) |
|---|---|---|---|---|---|---|---|---|
| LA0370 | S. peruvianum | + | − | √ | √ | √ | √ | √ |
| LA1407 | S. cheesmaniae | + | − | √ | √ | √ | √ | √ |
| LA1722 | S. corneliomulleri | + | − | √ | √ | √ | √ | √ |
| LA1365 | S. huaylasense | + | − | √ | √ | √ | √ | √ |
| LA2809 | S. huaylasense | + | − | √ | √ | √ | √ | √ |
| LA1350 | S. arcanum | + | − | √ | √ | √ (FK) | √ | √ |
| LA0103 | S. corneliomulleri | − | − | √ | S | √ (FK) | √ | √ |
| LA2581 | S. peruvianum | − | + | √ | S | √ (FK) | √ | √ |
| LA4445 | S. peruvianum | − | − | √ | SM | √ (FK) | √ | √ |
| LA0716 | S. pennellii | − | n | √ | √ | YK | KNFPYYDIV | VE |
| LA0751 | S. pennellii | − | + | √ | √ | YK | KNFPYYDIV | VE |
| LA0927 | S. cheesmaniae | − | n | √ | T | DK | KNFP-YGIG | IQ |
| LA0747 | S. galapagense | − | + | √ | T | DK | KNFP-YGIG | IQ |
| LA1204 | S. lycopersicum | − | + | √ | T | DK | KNFP-YGIG | IQ |
| LA0446 | S. peruvianum | − | + | √ | T | DK | KNFP-YGIG | IQ |
| LA0411 | S. pimpinellifolium | − | + | √ | T | DK | KNFP-YGIG | IQ |
| LA1331 | S. corneliomulleri | − | + | √ | N | DK | KNFP-YNIG | IG |
| LA2113 | S. neorickii | − | n | √ | T | DK | KNLP-YGIV | IQ |
| LA1777 | S. habrochaites | − | + | √ | T | DK | KNLP-YGTG | IG |
| LA1358 | S. huaylasense | − | − | √ | T | DE | KNYP-YRMG | IG |
| LA1930 | S.chilense | − | + | √ | N | DK | KNFP-YNIGL | IG |
| LA0445 | S. peruvianum | − | − | √ | √ | YK | KNFPYYDIV | √ |
| LA1028 | S. chmielewskii | − | − | Truncated | ||||
| LA2951 | S. lycopersicoides | − | + | Truncated | ||||
| LA1974 | S. sitiens | − | n | No PCR product |
Twenty-seven accessions representing all 17 wild tomato species from South America were obtained from TGRC, while two accessions of tomato seeds, S. ochranthum LA2160 and S. juglandifolium LA3322, did not germinate and were not used in further analysis. The “√” refers to the conserved site. As Sw-5b with the FK site variation is still functionally resistant to TSWV, we still mark this site as a functionally conserved variation. The “n” represents “not tested.”
Sw-5b homolog is coexpressed with NSm for the HR assay.
The presence of virus in the systemic leaves of wild tomato species inoculated with TSWV was assayed by ELISA.
DISCUSSION
Sw-5b Recognizes a Conserved NSm21 Epitope to Confer Broad-Spectrum Resistance to American-Type Tospoviruses
Most animal NLRs are known to recognize conserved PAMPs to provide broad-spectrum resistance (Maekawa et al., 2011; Jones et al., 2016). By contrast, plant intracellular NLRs are generally known to detect pathogen strain-specific effectors and confer race-specific resistance (Caplan et al., 2008; Jones et al., 2016). In this study, using tomato Sw-5b NLR and the TSWV model system, we demonstrate that a plant NLR Sw-5b confers broad-spectrum resistance to various American-type tospoviruses by recognizing a conserved 21-amino acid peptide region in the viral movement protein NSm. Our findings provide a new example of a plant NLR that is somewhat similar to a mammalian NLR by recognizing a small conserved peptide of phylogenetically related pathogens.
Tobacco N NLR has been shown to confer broad-spectrum resistance to most strains of Tobamovirus except the Ob strain (Tobias et al., 1982; Csillery et al., 1983). The 50-kD helicase domain (p50) of the replicase from the Tobacco mosaic virus (TMV)-U1 was identified as the elicitor of the N-mediated HR (Abbink et al., 1998; Erickson et al., 1999). Although p50 of TMV replicase is recognized by N NLR, attempts to identify the small region within p50 that is recognized by N NLR has not been successful (Erickson et al., 1999; Abbink et al., 2001). Domain swap experiments between p50 of the N eliciting TMV-U1 strain and N noneliciting TMV-Ob strain indicated that four different regions within p50 are required for N recognition (Abbink et al., 2001). Similar to N, our findings described here show that Sw-5b NLR confers broad-spectrum resistance to many American-type tospoviruses through recognition of movement protein NSm. However, unlike TMV p50, we were able to identify and show that a highly conserved 21-amino acid peptide region of NSm (amino acid residues 115–135) among American-type tospoviruses is sufficient to induce Sw-5b-mediated HR.
A conserved 22-amino acid peptide within bacterial flagellin (flg22) and 18-amino acid peptide within bacterial elongation factor Tu (elf18) are known to act as PAMPs that are recognized by plant PRRs FLS2 and EFR, respectively (Felix et al., 1999; Gómez-Gómez and Boller, 2000; Zipfel et al., 2006). A 23-amino acid Arabidopsis endogenous peptide (AtPep1) is also known to function as a damage-associated molecular pattern that is recognized by PEPR1 and PEPR2 receptors (Yamaguchi et al., 2006, 2010). The highly conserved NSm21 peptide region among American-type Tospovirus that is sufficient to trigger Sw-5b-mediated HR represents like a PAMP. Although PAMPs are generally conserved among microbes, some PAMPs are only narrowly conserved (Brunner et al., 2002; Pruitt et al., 2015). Pep-13, a surface-exposed peptide region of a calcium-dependent cell wall transglutaminase (TGase), is conserved only among oomycete Phytophthora species, but not among oomycete Phythium, Albugo, Peronospora, and Plasmopara species (Nürnberger et al., 1994; Brunner et al., 2002). Similarly, a sulfated, 21-amino acid RaxX peptide (RaxX21-sY) recognized by rice (Oryza sativa) PRR Xa21 is conserved among Xanthomonas but not present in other bacteria (Pruitt et al., 2015).
We found that NSm21 from Euro/Asian-type tospoviruses fails to trigger Sw-5b-mediated HR. The sequence of the NSm21 region contains over nine different residues between two geographic types of tospoviruses. This indicates that the NSm21 peptide region from Euro/Asian-type tospoviruses “escapes” recognition by the Sw-5b NLR. In a similar manner, it has been shown that some PAMPs also “escape” recognition by a PRR by evolving the recognized sequence (Felix et al., 1999; Sun et al., 2006; Wang et al., 2015). Flg22 peptide from plant-associated bacteria Agrobacterium tumefaciens and Rhizobium fail to trigger FLS2-mediated immunity. Comparison of the flg22 sequence from Pseudomonas syringae (QRLSTGSRINSAKDDAAGLQIA) with Agrobacterium (ARVSSGLRVGDASDNAAYWSIA) and Rhizobium (AHVSSGLRVGQAADNAAYWSIA) indicates that these flg22 sequences are significantly different (Felix et al., 1999). Furthermore, flagellin polymorphism within a species has also been demonstrated for Xanthomonas campestris pv campestris (Xcc) (Sun et al., 2006). Phylogenetic analysis of a set of Xcc strains with diverse origin showed that the Xcc flagellin product falls into two distinct groups and the flagellin from only one group triggers plant defense. Further studies identified that a Val43Asp polymorphism within the flg22 peptide region determines the eliciting or noneliciting nature of Xcc flagellins (Sun et al., 2006). Not only Xcc can evade Arabidopsis FLS2-mediated defenses, but Xanthomonas oryzae pv oryzae and Xanthomonas oryzae pv oryzicola can also evade the rice FLS2-mediated perception of bacterial flagellins because of the changes in the flg22 sequence (Wang et al., 2015).
Close homologs of Sw-5b, such as Mi-1.2 and Hero NLRs, have also been reported to provide a broad-spectrum resistance (Boiteux and Giordano, 1993; Milligan et al., 1998; Rossi et al., 1998; Vos et al., 1998; Brommonschenkel et al., 2000; Ernst et al., 2002; Nombela et al., 2003; Casteel et al., 2006). However, the molecular basis for such recognition is currently unknown. Our findings described here should facilitate mechanistic studies on broad-spectrum resistance mediated by NLRs. Interestingly, tomato Mi-1.2 NLR confers resistance to four unrelated pests, i.e., nematode, aphid, whitefly, and psyllid (Milligan et al., 1998; Rossi et al., 1998; Vos et al., 1998; Nombela et al., 2003; Casteel et al., 2006). It will be interesting to determine if there is a conserved NSm21-like epitope in these pest effectors that Mi-1.2 has evolved to recognize and provide broad-spectrum resistance.
Sw-5b NB-ARC-LRR Region Directly Interacts with NSm21
PRRs are known to detect PAMPs through direct interaction (Zipfel, 2008; Sun et al., 2013; Couto and Zipfel, 2016). Direct interaction between plant NLRs and pathogen effectors have been reported. For example, rice Pi-ta NLR has been shown to interact with the Magnaporthe grisea fungal effector AvrPita by far-western assay (Jia et al., 2000). Interaction between flax M or L NLR and its cognate effectors has been shown using a yeast two-hybrid assay (Dodds et al., 2001, 2006; Bernoux et al., 2016). Indirect interaction between effector and intracellular NLRs through an intermediary host factor has also been shown (Caplan et al., 2008; Collier and Moffett, 2009; Jones et al., 2016). For example, NLR proteins RPM1 and RPS2 detect pathogen effectors through monitoring the state of the RIN4 that is targeted for phosphorylation by the pathogen effectors AvrB or AvrRpm1 (Chung et al., 2011) or targeted for the cleavage by the bacterial effector AvrRpt2 (Mackey et al., 2002; Axtell and Staskawicz, 2003). RPS5 monitors the state of protein kinase PBS1 that is targeted for proteolytic cleavage by pathogen effector AvrPphB (Shao et al., 2003). These NLRs typically recognize the activity of the effectors rather than their structures or sequences. However, some NLRs that directly interact with effectors could also detect the activity of effectors. The recently reported RPS4 and RRS1 pair for integrated decoy model detect the activity of bacterial effector that target WRKY. Both AvrRPS4 and PopP2 effectors interact with RRS1 WRKY domains (Le Roux et al., 2015; Sarris et al., 2015), and direct interaction between RRS1 WRKY and PopP2 has also been observed (Le Roux et al., 2015). The rice NLR pairs RGA4/RGA5 and Pikp-1/Pikp-2 may also belong to this category. RGA5 carries a C-terminal RATX1 or HMA domain that may act as a decoy. This domain directly interacts with effectors AVRPia and AVR1-CO39 from M. oryzae (Cesari et al., 2013). The HMA domain that is integrated between the CC and the NB-ARC domain of Pikp-1 directly interacts with a different effector, Avr-Pik (Maqbool et al., 2015).
Here, we demonstrated interaction between tomato Sw-5b NLR and TSWV NSm21. We show that Sw-5b NB-ARC-LRR directly interacts with viral NSm21 epitope in vitro. Furthermore, Sw-5b NB-ARC-LRR also interacts with this epitope in vivo and it is sufficient to induce cell death. Because NB-ARC-LRR is a core region of Sw-5b that is regulated by the CC and N-terminal domain (Chen et al., 2016; De Oliveira et al., 2016), we hypothesize that the full-length Sw-5b also directly interacts with NSm21. Thus, our Sw-5b and NSm21 interaction shown here adds another novel way of recognizing effector by direct interaction. This is different from those above described NLRs that recognize effector activity. It is possible that the rice Pi-ta, flax M, or L NLRs in the direct interaction model may also recognize the structures or sequences of corresponding effectors or may even recognize a small conserved peptide region of effectors. Our findings that Sw-5b recognizes 21-amino acid peptide sequences within NSm provide a novel example of recognition of small peptide by NLR that is similar to PRR-mediated recognition of PAMPs.
Four Polymorphic Sites within the LRR Region of Sw-5b NLR Are Required for the Recognition of the NSm21 Epitope
Analysis of Arabidopsis FLS2 through alanine scanning mutagenesis and phylogenetic studies using orthologs from related species has shown that the flg22 binding sites are on the solvent-exposed regions of the LRR domain (Dunning et al., 2007). The crystal structure of FLS2 and its coreceptor BAK1 ectodomains complexed with flg22 peptide revealed that a conserved and a nonconserved site from the inner surface of the FLS2 associate with the C- and N-terminal segment of flg22, respectively (Sun et al., 2013). Our domain swap and site-directed mutagenesis along with co-IP experiments identified only four polymorphic sites on the Sw-5b LRR domain that are important for NSm21 binding and recognition. These four polymorphic recognition sites are surface exposed and clustered together along the inner surface of LRR 3-7 in the three-dimensional structure model. We speculate that these surfaces exposed polymorphic sites could facilitate the specific binding of NSm21. Further structural studies will be required to reveal the precise binding-interfaces of Sw-5b LRR and NSm21.
Binding of NSm21 to LRR Might Alter the Critical R927 Site to Activate Sw-5b
Plant NLRs have been hypothesized to function as molecular switches (Lukasik and Takken, 2009; Takken and Tameling, 2009). NLRs are in an autoinhibited state in the absence of pathogen and switch to an activated state upon perception of pathogen effector (Lukasik and Takken, 2009; Takken and Tameling, 2009). However, how NLR recognition of effector leads to an activated state remains unknown. Interestingly, our structure model-based functional analysis identified R927 in Sw-5b LRR2 as a residue critical for keeping the NB-ARC domain in an autoinhibited state. The R927A substitution resulted in an autoactivation of Sw-5b. However, introduction of the K568R P-loop mutation abolished the autoactivation. Therefore, R927A still requires functional ATP binding or hydrolysis for its autoactivation. It has been proposed that activation and conformation change of plant NLRs require ATP binding or hydrolysis (Takken and Tameling, 2009). Consistent with this, we found that the Sw-5b R927A mutant weakened the interaction between the NB-ARC and LRR domains. We hypothesize that this weakened intramolecular interaction might either promote ATP binding or reduce ATP hydrolysis leading to the autoactivation of this mutant.
How does NSm21 binding relieve R927 to activate Sw-5b? Our structural model showed that the four polymorphic sites important for NSm21 recognition are located next to the R927 residue. Consistently, mutational analysis revealed that these four polymorphic sites are specifically required to keep the R927 site in a sensitive configuration for autoactivation but are not required for autoactivation of the NB subdomain (D624E) or ARC2 subdomain (D857V) mutants. Strikingly, NSm21(C118Y) or NSm21(T120N) mutants that fail to induce cell death in the presence of wild-type Sw-5b could enhance cell death induced by the R927A autoactive mutant. We hypothesize that NSm21(C118Y) or NSm21(T120N) mutants may not be able to alter the residues adjacent to the R927 site to weaken the interaction between NB-ARC and LRR. Consequently, they fail to induce cell death in the wild-type Sw-5b background. On the other hand, if interaction between NB-ARC and LRR is already weakened, as is the case in R927A mutant, binding of NSm21(C118Y) and NSm21(T120N) mutants may further disrupt the interaction between NB-ARC and LRR, leading to more extensive cell death.
A single amino acid deletion at the N or C terminus of NSm115-135 eliminated the ability to trigger Sw-5b-mediated HR (Figure 1B). Similarly, any single amino acid mutation at residues 118, 120, 122, 126, and 129 on the NSm21 peptide region completely inhibited HR induction (Figure 2B). Co-IP analysis showed that nonelicitor mutant NSm21(C118Y) or NSm21(T120N) interacts with Sw-5b NB-ARC-LRR as well as the wild-type NSm21. These results suggest that the interaction between NSm and Sw-5b alone may not be sufficient to induce cell death. Our result is consistent with the findings that the tobacco N NLR could associate with both the elicitor p50-U1 and nonelicitor p50-Ob; however, only p50-U1 induces cell death (Padmanabhan et al., 2013). Furthermore, the rice intracellular NLR Pik interacts with mutant AVR-PikE or AVR-PikA, but fails to induce cell death (Maqbool et al., 2015). Pathogen recognition and NLR activation might be two independent steps in the activation of defense. The observations that four polymorphic sites and the R927 residue localized next to each other on the three-dimensional structure model also support the notion that the NSm21 recognition sites and competent R927 activation site might play independent roles but their roles are closely interrelated and mutually dependent.
Our natural variation analyses of Sw-5b homologs from wild tomato species originating from South America revealed that the R927 residue is conserved, whereas the four polymorphic sites are positively selected and might have evolved in a stepwise manner. Although R927 is present in all Sw-5b homologs of wild tomato species, site-directed mutagenesis analyses revealed that positive selection at four polymorphic sites is required for maintaining the R927 residue at a competent state for activation. The positive selection of the four polymorphic sites in Sw-5b may have created two important functions. First, it created a dedicated recognition site for viral NSm21; second, it created a competent configuration at the R927 site that is poised for activation. The evolutionary selection of NSm21 recognition sites together with the findings that NSm21 recognition sites and a competent R927 activation site are mutually dependent on each provide insights into how Sw-5b NLR evolved to recognize the viral NSm protein and how this NLR is activated upon perception of viral invasion.
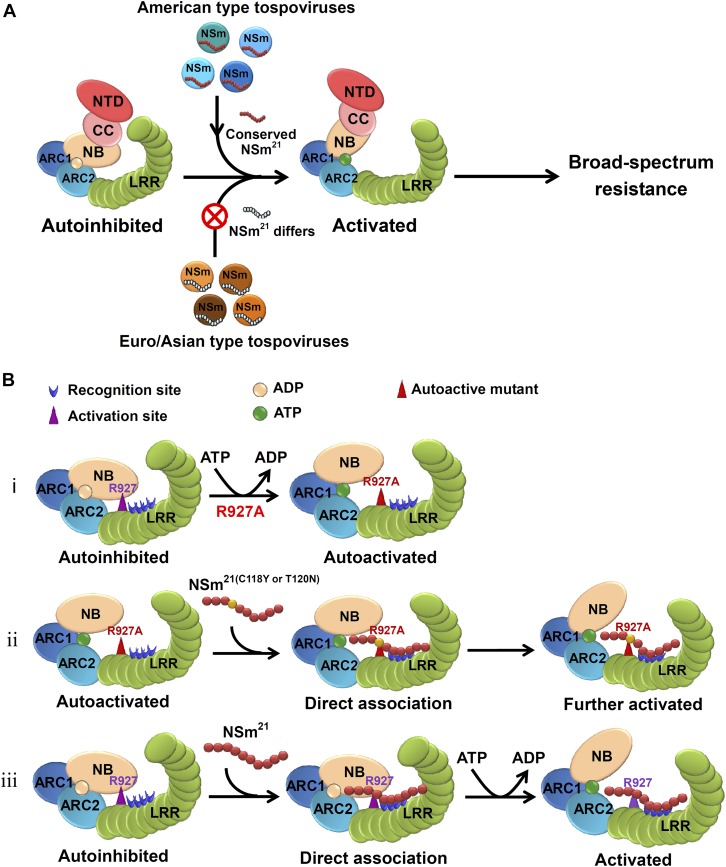
On the basis of our findings described here, we propose a model for Sw-5b-mediated broad-spectrum resistance to American-type tospoviruses (Figures 7A and 7B). A 21-amino acid epitope in the NSm is highly conserved among American-type tospoviruses. Recognition of this conserved NSm21 epitope by four polymorphic sites on the LRR modulates the adjacent R927 residue to release the interaction between LRR and NB-ARC domains leading to the activation of Sw-5b. NSm21 of Euro/Asian-type tospoviruses differs greatly from that of American-type tospoviruses, and Sw-5b cannot be activated by Euro/Asian-type tospoviruses. This model illustrates an important mechanism that translates pathogen recognition into activation of Sw-5b NB-ARC-LRR. In addition to NB-ARC-LRR, we have shown recently that the CC and N-terminal domain are involved in multilayered autoregulation of Sw-5b and this regulation is required for Sw-5b to confer resistance (Chen et al., 2016). We hypothesize that, upon recognition of NSm21, the multilayered inhibitory effects of the CC and N-terminal domain are relieved from NB-ARC-LRR and Sw-5b NLR is then switched from “autoinhibited” or “off” state to “activated” or “on” state to mediate broad-spectrum immunity against American-type tospoviruses. Our findings and new model may have broad implications for the mechanistic studies of pathogen recognition and activation for other plant NLR receptors to mediate their immunity.
Figure 7.
Models for Viral Effector Recognition and Activation of Tomato NLR Sw-5b to Mediate Broad-Spectrum Resistance to American Tospoviruses.
(A) Sw-5b-mediated broad-spectrum resistance to American-type tospoviruses. Tomato NLR receptor Sw-5b confers a broad-spectrum resistance to American-type tospoviruses but not Euro-Asian-type tospoviruses through the recognition of a conserved epitope in viral movement protein NSm (NSm21).
(B) NSm21 recognition and activation of Sw-5b NB-ARC-LRR. i, R927 residue is important for LRR to interact with and to lock NB-ARC in an autoinhibited state. Substitution of R927 with an alanine residue results in autoactivation of Sw-5b NB-ARC-LRR. ii, The nonelicitor NSm21(C118Y) and NSm21(T120N) enhance the HR induced by Sw-5b NB-ARC-LRRR927A. iii, The R927 residue and the four polymorphic sites 3, 4, 5, and 6 on Sw-5b LRR are located next to each other. Recognition of NSm21 by four polymorphic sites on the Sw-5b LRR domain modulates the R927 site to weaken the intramolecular interaction between NB-ARC and LRR, thus translating recognition of NSm21 peptide into activation of Sw-5b NB-ARC-LRR.
METHODS
Plasmid Construction
Plasmids p2300S-Sw-5b, p2300S-Sw-5b NB-ARC-LRR, and p2300S-NSm were described previously (Chen et al., 2016). All other Sw-5b, Sw-5bHeinz, and NSm derivatives were cloned downstream of the 35S promoter in the pCambia2300S binary vector as described previously (Feng et al., 2013). A 1xFLAG or 3xHA tag was fused to the N terminus of Sw-5b, Sw-5bHeinz NB-ARC-LRR, NSm, and other derivatives for immunoblotting or coimmunoprecipitation assays. YFP fusion constructs and other site-directed mutants were generated by two-step overlap PCR as described (Chen et al., 2016). Full-length Sw-5 homolog genes were amplified from genomic DNA isolated from collected wild tomato species and were cloned individually into a plant expression vector p2300S. Three independent clones representing each wild tomato species were sequenced from both ends. Primers and details of constructs are listed in Supplemental Data Set 2.
Transient Expression, Virus Inoculation, and Plant Growth
Agrobacterium tumefaciens-mediated transient expression was performed as previously described (Chen et al., 2016). Agrobacterium strains GV3101 and EHA105 were used for transient expression assays in Nicotiana benthamiana and tomato (Solanum lycopersicum) leaves, respectively. TSWV, INSV, TZSV, and HCRV were isolated from local diseased plants in Yunnan province and propagated on N. benthamiana plants. For virus inoculation, crude saps of N. benthamiana leaves infected with a specific virus isolate were rub-inoculated onto leaves of Sw-5b transgenic N. benthamiana plants, as described previously by our group (Chen et al., 2016). Tomato cultivar Xianke #1 was used for the cell death assay. Seeds of wild-type S. arcanum, S. cheesmaniae, S. chilense, S. chmielewskii, S. corneliomulleri, S. galapagense, S. habrochaites, S. huaylasense, S. lycopersicum, S. neorickii, S. pennellii, S. peruvianum, S. pimpinellifolium, and S. sitiens plants were from the Tomato Genetic Resource Center, University of California, Davis, and were grown inside a growth chamber. Crude saps from virus-infected N. benthamiana leaves were inoculated onto the leaves of 6-week-old Nb:Sw-5b transgenic N. benthamiana plants or 3-week-old wild tomato seedlings to determine their resistance to TSWV. All agroinfiltrated or virus-inoculated plants were grown inside a growth chamber with white fluorescent light (∼150 μmol/m2/s) and set at 23°C/25°C (day/night) and 16-h-light/8-h-dark light cycles.
Immunoblot and Immunoprecipitation Assay
Immunoblot and immunoprecipitation assays were performed as described (Chen et al., 2016). Briefly, N. benthamiana leaves agroinfiltrated with NSm, NSm21, Sw-5b NB-ARC-LRR, or their mutant derivatives were harvested at 20 to 24 hpi. Agroinfiltrated leaf tissues were ground in prechilled extraction buffer (10% [v/v] glycerol, 25 mM Tris-HCl, pH 7.5, 1 mM EDTA, 150 mM NaCl, 10 mM dithiothreitol, 2% [w/v] polyvinylpolypyrrolidone, 0.5% [v/v] Triton X-100, and 1× protease inhibitors cocktail) followed by 30 min centrifugation at 18,000g at 4°C. The supernatant (1 mL) was mixed with 25 μL anti-FLAG (M2; Sigma-Aldrich; catalog no. F2426) agarose beads and then pelleted by 5 min centrifugation at 1000g at 4°C. For the immunoblot assay, supernatants (30 μL each) were heated at 95°C for 5 min and separated by SDS-PAGE. The resulting blots were probed using an anti-FLAG-HRP (1:5000; Sigma-Aldrich; catalog no. A8592), anti-His (1:5000; Sigma-Aldrich; catalog no. SAB2702218), anti-YFP (1:5000; Sigma-Aldrich; catalog no. SAB4301138), or anti-GST antibody (1:8000; Sigma-Aldrich; catalog no. SAB1305539) followed by HRP-conjugated goat anti-rabbit (1:10,000; Sigma-Aldrich; catalog no. A6154)/mouse antibody (1:10,000; Sigma-Aldrich; catalog no. A4416) and developed using the ECL Substrate Kit as described (Thermo Scientific). Alternatively, the blots were probed with primary antibody and detected by AP-conjugated goat anti-rabbit (1:10,000; Sigma-Aldrich; A3687)/mouse IgG (1:10,000; Sigma-Aldrich; catalog no. A0418) and 5-bromo-4-chloro-3-indolylphosphate/nitroblue tetrazolium staining (Sangon Biotech).
Protein Expression, Purification, and in Vitro GST Pull-Down
Constructs pGEX-2TK-Sw-5b, pGEX-2TK-NB-ARC-LRR, and pET28-NSm21-YFP-6xhis were individually transformed into Escherichia coli strain Rosetta (DE3). To express the recombinant proteins, 10 mL of overnight culture was transferred to a 1-liter flask and incubated at 37°C till OD600 reached 0.5 to 0.6 in a shaker incubator. Protein expression was induced with 0.1 mM isopropyl β-d-thiogalactopyranoside overnight at 18°C. GST-fused recombinant proteins were purified using Glutathione Sepharose beads as instructed by the manufacturer (Qiagen). The purification of 6xHis-tag recombinant proteins was described (Li et al., 2015). GST pull-down assays were performed using a previously reported protocol (Cui et al., 2010).
Cell Death Assay and Ion Leakage Measurement
Development of cell death in leaves was monitored through daily observation in the infiltrated leaves and through measurement of ion leakage using leaf discs of 5 mm in diameter harvested from the infiltrated leaves as described (Mittler et al., 1999). Briefly, five leaf discs were floated on 10 mL of double distilled water at room temperature for 3 h. After incubation, the solution without leaf discs was measured and referred to as value A. The leaf discs were then returned to the solution and treated at 95°C for 25 min. Then, the solution was measured and referred to as value B. The conductivity was expressed as percent ion leakage: (value A/value B) × 100.
Sequence Alignment, Phylogenetic Tree Construction, and Homology Structure Modeling
Sequences of Sw-5b (AY007366.1) and Sw-5bHeinz paralog (XM_004247995.3) were aligned using ClustalW (www.ebi.ac.uk). The phylogenetic tree based on the deduced amino acid sequence of nucleocapsid protein was constructed using the MEGA 5.01 software package (Tamura et al., 2011). The deduced amino acid sequences of NSm and NSm21 were analyzed by multiple sequence alignment using the ClustalW method in the BioEdit program (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). Homology structure modeling of Sw-5b NB-ARC-LRR was conducted as described (Chen et al., 2016). The homology models were then presented using the PyMOL Molecular Graphics System, version 1.5.0.4.
Accession Numbers
The nucleotide sequences of nucleocapsid (N) and movement protein NSm from different tospoviruses were obtained from the GenBank database: GQ478668 (Alstroemeria necrotic streak virus, AlNSV), NC_008301 (capsicum chlorosis virus, CaCV), AY867502 (calla lily chlorotic spot virus, CCSV), AF067068 (chrysanthemum stem necrosis virus, CSNV), L12048 (groundnut ringspot virus, GRSV), KU356853 (Hippeastrum chlorotic ringspot virus, HCRV), X66972 (impatiens necrotic spot virus, INSV), AF001387 (iris yellow spot virus, IYSV), EU275149 (melon severe mosaic virus, MeSMV), AB038343 (melon yellow spot virus, MYSV), U27809 (peanut bud necrosis virus, PBNV), AF080526 (peanut chlorotic fan-spot virus, PCSV), KF383956 (pepper chlorotic spot virus, PCSV), EF445397 (poligonum ringspot virus, PolRSV), AF013994 (peanut yellow spot virus, PYSV), HQ728387 (soybean vein necrosis-associated virus, SVNaV), AF282982 (tomato chlorotic spot virus, TCSV), FJ489600 (tomato necrotic ringspot virus, TNRV), AF020660 (tomato spotted wilt virus, TSWV), AY686718 (tomato yellow ring virus, TYRV), EF552433 (tomato zonate spot virus, TZSV), EU249351 (watermelon bud necrosis virus, WBNV), NC_003843 (watermelon silver mottle virus, WSMoV), and AF067069 (zucchini lethal chlorosis virus, ZLCV) for N gene; HQ728387 (soybean vein necrosis virus, SVNV); KT004454 (CCSV), NC_008303 (CaCV), AF213675 (CSNV), AF213673 (GRSV), JX833565 (HCRV), NC_003616 (INSV), AF214014 (IYSV), AB061773 (MYSV), U42555 (GBNV), EU271753 (PolRSV), HQ728386 (SVNaV), AF213674 (TCSV), FJ947152 (TNRV), AF208498 (TSWV), JN560177 (TYRV), NC_010490 (TZSV), FJ694963 (WBNV), NC_003841 (WSMoV), and AF213676 (ZLCV) for NSm genes.Full-length nucleotide sequences representing the Sw-5 gene of 24 wild tomato species have been deposited in GenBank under accession numbers S. peruvianum LA4445, KY006940; S. huaylasense LA2809, KY006941; S. peruvianum LA2581, KY006942; S. neorickii LA2113, KY006943; S. chilense LA1930, KY006944; S. habrochaites LA1777, KY006945; S. corneliomulleri LA1722, KY006946; S. huaylasense LA1365, KY006947; S. huaylasense LA1358, KY006948; S. arcanum LA1350, KY006949; S. corneliomulleri LA1331, KY006950; S. lycopersicum LA1204, KY006951; S. cheesmaniae LA0927, KY006952; S. pennellii LA0751, KY006953; S. galapagense LA0747, KY006954; S. pennellii LA0716, KY006955; S. peruvianum LA0446, KY006956; S. peruvianum LA0445, KY006957; S. pimpinellifolium LA0411, KY006958; S. peruvianum LA0370, KY006959; S. corneliomulleri LA0103, KY006960; and S. cheesmaniae LA1407, KY006961.
Supplemental Data
Supplemental Figure 1. Sw-5b confers broad-spectrum resistance to American-type tospoviruses but not Euro/Asian-type tospoviruses.
Supplemental Figure 2. HR scale used to determine the extent of cell death.
Supplemental Figure 3. Immunoblot analysis and cell death induction by NSm deletion mutants in N. benthamiana and transgenic plants expressing Sw-5b (Nb::Sw-5b).
Supplemental Figure 4. Immunoblot analyses of TSWV and TZSV NSm mutants transiently expressed in N. benthamiana.
Supplemental Figure 5. Mapping the critical residue(s) in NSm21 required for cell death induction.
Supplemental Figure 6. Full-length Sw-5b and Sw-5b-NB-ARC-LRR recognize NSm21 to induce cell death.
Supplemental Figure 7. Alignment of NB-ARC-LRR sequence of Sw-5b and Sw-5bHeinz.
Supplemental Figure 8. Cell death, ion leakage, and immunoblot analysis of wild-type Sw-5b NB-ARC-LRR and substitution polymorphic mutants.
Supplemental Figure 9. Localization of R927 residue and the four polymorphic sites in the amino acid sequence and the three-dimensional structure model of Sw-5b NB-ARC-LRR.
Supplemental Figure 10. A functional P-loop is required for the autoactivation of NB-ARC-LRRR927A.
Supplemental Figure 11. Nonelicitor NSm21 mutants associate with NB-ARC-LRRK568R.
Supplemental Figure 12. Immunoblot analysis of Sw-5b NB-ARC-LRRR927A with mutations at the four polymorphic sites.
Supplemental Figure 13. Immunoblot analysis of YFP tagged Sw-5b homologs from tomato wild species transiently expressed in N. benthamiana.
Supplemental Table 1. Measuring the amount of collapse area as a percentage of the infiltrated area for Sw-5b NB-ARC-LRRR927A alone or with NSm21, NSm21(C118Y), and NSm21(T120N).
Supplemental Table 2. Requirement of the four polymorphic sites on autoactivation activity of Sw-5b NB-ARC-LRRR927A.
Supplemental Data Set 1. Alignments used to generate the phylogeny presented in Supplemental Figure 1A.
Supplemental Data Set 2. List of primers used in this study.
Acknowledgments
We thank Xin Shun Ding (The Samuel Roberts Noble Foundation) for critical reading of the manuscript. This work was supported by the National Natural Science Foundation of China (31630062 and 31471746 to X.T.), by the National Program on Key Basic Research Project of China (973 Program, 2014CB138400 to X.T.), by the Youth Talent Support Program of China and Distinguished Professor of Jiangsu Province to X.T., by the Special Fund for Agro-Scientific Research in the Public Interest (201303028 to X.T.), and by National Science Foundation grants (NSF-IOS-1354434 and NSF-IOS-1339185 to S.P.D.-K.). The materials of tomato wild species were developed by and/or obtained from the UC Davis/C.M. Rick Tomato Genetics Resource Center and maintained by the Department of Plant Sciences, University of California, Davis.
AUTHOR CONTRIBUTIONS
M.Z., L.J., and X.T. designed the research. M.Z., L.J., B.B., W.Z., X.C., J.L., Z.C., B. W., C.W., and Q.W. performed the research. M.Z., L.J., W. Z., Q.S., and S.P.D.-K. analyzed the data. Y. L. contributed to discussions. M.Z., S.P.D.-K., and X.T. wrote the article.
Glossary
- PRR
pattern recognition receptor
- PAMP
pathogen-associated molecular pattern
- TSWV
Tomato spotted wilt virus
- GRSV
Groundnut ring spot virus
- TCSV
Tomato chlorotic spot virus
- HCRV
Hippeastrum chlorotic ringspot virus
- INSV
Impatiens necrotic spot virus
- TZSV
Tomato zonate spot virus
- HR
hypersensitive cell death response
- hpi
hours postinfiltration
- co-IP
coimmunoprecipitation
- TMV
Tobacco mosaic virus
References
- Abbink T.E., de Vogel J., Bol J.F., Linthorst H.J. (2001). Induction of a hypersensitive response by chimeric helicase sequences of tobamoviruses U1 and Ob in N-carrying tobacco. Mol. Plant Microbe Interact. 14: 1086–1095. [DOI] [PubMed] [Google Scholar]
- Abbink T.E.M., Tjernberg P.A., Bol J.F., Linthorst H.J.M. (1998). Tobacco mosaic virus helicase domain induces necrosis in N gene-carrying tobacco in the absence of virus replication. Mol. Plant Microbe Interact. 11: 1242–1246. [Google Scholar]
- Axtell M.J., Staskawicz B.J. (2003). Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112: 369–377. [DOI] [PubMed] [Google Scholar]
- Bendahmane A., Farnham G., Moffett P., Baulcombe D.C. (2002). Constitutive gain-of-function mutants in a nucleotide binding site-leucine rich repeat protein encoded at the Rx locus of potato. Plant J. 32: 195–204. [DOI] [PubMed] [Google Scholar]
- Bernoux M., Burdett H., Williams S.J., Zhang X., Chen C., Newell K., Lawrence G.J., Kobe B., Ellis J.G., Anderson P.A., Dodds P.N. (2016). Comparative analysis of the flax immune receptors L6 and L7 suggests an equilibrium-based switch activation model. Plant Cell 28: 146–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezerra I.C., de O Resende R., Pozzer L., Nagata T., Kormelink R., De Ávila A.C. (1999). Increase of tospoviral diversity in Brazil with the identification of two new tospovirus species, one from chrysanthemum and one from zucchini. Phytopathology 89: 823–830. [DOI] [PubMed] [Google Scholar]
- Boiteux L.S., Giordano L.D. (1993). Genetic-basis of resistance against 2 tospovirus species in tomato (Lycopersicon esculentum). Euphytica 71: 151–154. [Google Scholar]
- Brommonschenkel S.H., Frary A., Frary A., Tanksley S.D. (2000). The broad-spectrum tospovirus resistance gene Sw-5 of tomato is a homolog of the root-knot nematode resistance gene Mi. Mol. Plant Microbe Interact. 13: 1130–1138. [DOI] [PubMed] [Google Scholar]
- Brunner F., Rosahl S., Lee J., Rudd J.J., Geiler C., Kauppinen S., Rasmussen G., Scheel D., Nürnberger T. (2002). Pep-13, a plant defense-inducing pathogen-associated pattern from Phytophthora transglutaminases. EMBO J. 21: 6681–6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan J., Padmanabhan M., Dinesh-Kumar S.P. (2008). Plant NB-LRR immune receptors: from recognition to transcriptional reprogramming. Cell Host Microbe 3: 126–135. [DOI] [PubMed] [Google Scholar]
- Casteel C.L., Walling L.L., Paine T.D. (2006). Behavior and biology of the tomato psyllid, Bactericerca cockerelli, in response to the Mi-1.2 gene. Entomol. Exp. Appl. 121: 67–72. [Google Scholar]
- Cesari S., et al. (2013). The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 25: 1463–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Zhu M., Jiang L., Zhao W., Li J., Wu J., Li C., Bai B., Lu G., Chen H., Moffett P., Tao X. (2016). A multilayered regulatory mechanism for the autoinhibition and activation of a plant CC-NB-LRR resistance protein with an extra N-terminal domain. New Phytol. 212: 161–175. [DOI] [PubMed] [Google Scholar]
- Chung E.H., da Cunha L., Wu A.J., Gao Z., Cherkis K., Afzal A.J., Mackey D., Dangl J.L. (2011). Specific threonine phosphorylation of a host target by two unrelated type III effectors activates a host innate immune receptor in plants. Cell Host Microbe 9: 125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier S.M., Moffett P. (2009). NB-LRRs work a “bait and switch” on pathogens. Trends Plant Sci. 14: 521–529. [DOI] [PubMed] [Google Scholar]
- Couto D., Zipfel C. (2016). Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 16: 537–552. [DOI] [PubMed] [Google Scholar]
- Csillery G., Tobias I., Rusko J. (1983). A new pepper strain of tomato mosaic virus. Acta. Phytopathol. Acad. Sci. Hung. 18: 195–200. [Google Scholar]
- Cui H., Wang Y., Xue L., Chu J., Yan C., Fu J., Chen M., Innes R.W., Zhou J.M. (2010). Pseudomonas syringae effector protein AvrB perturbs Arabidopsis hormone signaling by activating MAP kinase 4. Cell Host Microbe 7: 164–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Avila A.C., Huguenot C., Resende Rde.O., Kitajima E.W., Goldbach R.W., Peters D. (1990). Serological differentiation of 20 isolates of tomato spotted wilt virus. J. Gen. Virol. 71: 2801–2807. [DOI] [PubMed] [Google Scholar]
- De Oliveira A.S., Koolhaas I., Boiteux L.S., Caldararu O.F., Petrescu A.-J., Resende R.O., Kormelink R. (2016). Cell death triggering and effector recognition by Sw-5 SD-CNL proteins from resistant and susceptible tomato isolines to Tomato spotted wilt virus. Mol. Plant Pathol. 17: 1442–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds P.N., Lawrence G.J., Ellis J.G. (2001). Six amino acid changes confined to the leucine-rich repeat beta-strand/beta-turn motif determine the difference between the P and P2 rust resistance specificities in flax. Plant Cell 13: 163–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds P.N., Lawrence G.J., Catanzariti A.M., Teh T., Wang C.I., Ayliffe M.A., Kobe B., Ellis J.G. (2006). Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. USA 103: 8888–8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning F.M., Sun W., Jansen K.L., Helft L., Bent A.F. (2007). Identification and mutational analysis of Arabidopsis FLS2 leucine-rich repeat domain residues that contribute to flagellin perception. Plant Cell 19: 3297–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson F.L., Holzberg S., Calderon-Urrea A., Handley V., Axtell M., Corr C., Baker B. (1999). The helicase domain of the TMV replicase proteins induces the N-mediated defence response in tobacco. Plant J. 18: 67–75. [DOI] [PubMed] [Google Scholar]
- Ernst K., Kumar A., Kriseleit D., Kloos D.U., Phillips M.S., Ganal M.W. (2002). The broad-spectrum potato cyst nematode resistance gene (Hero) from tomato is the only member of a large gene family of NBS-LRR genes with an unusual amino acid repeat in the LRR region. Plant J. 31: 127–136. [DOI] [PubMed] [Google Scholar]
- Feng Z., Chen X., Bao Y., Dong J., Zhang Z., Tao X. (2013). Nucleocapsid of Tomato spotted wilt tospovirus forms mobile particles that traffic on an actin/endoplasmic reticulum network driven by myosin XI-K. New Phytol. 200: 1212–1224. [DOI] [PubMed] [Google Scholar]
- Feng Z., Xue F., Xu M., Chen X., Zhao W., Garcia-Murria M.J., Mingarro I., Liu Y., Huang Y., Jiang L., Zhu M., Tao X. (2016). The ER-membrane transport system is critical for intercellular trafficking of the NSm movement protein and tomato spotted wilt tospovirus. PLoS Pathog. 12: e1005443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix G., Duran J.D., Volko S., Boller T. (1999). Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18: 265–276. [DOI] [PubMed] [Google Scholar]
- Goldbach R., Peters D. (1996). Molecular and biological aspects of Tospoviruses. In The Bunyaviridae, Elliott R.M., ed (New York: Plenum Press; ), pp. 129–157. [Google Scholar]
- Gómez-Gómez L., Boller T. (2000). FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5: 1003–1011. [DOI] [PubMed] [Google Scholar]
- Hallwass M., de Oliveira A.S., de Campos Dianese E., Lohuis D., Boiteux L.S., Inoue-Nagata A.K., Resende R.O., Kormelink R. (2014). The Tomato spotted wilt virus cell-to-cell movement protein (NSM ) triggers a hypersensitive response in Sw-5-containing resistant tomato lines and in Nicotiana benthamiana transformed with the functional Sw-5b resistance gene copy. Mol. Plant Pathol. 15: 871–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y., McAdams S.A., Bryan G.T., Hershey H.P., Valent B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19: 4004–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D., Vance R.E., Dangl J.L. (2016). Intracellular innate immune surveillance devices in plants and animals. Science 354: aaf6395. [DOI] [PubMed] [Google Scholar]
- Kormelink R., Storms M., Van Lent J., Peters D., Goldbach R. (1994). Expression and subcellular location of the NSM protein of tomato spotted wilt virus (TSWV), a putative viral movement protein. Virology 200: 56–65. [DOI] [PubMed] [Google Scholar]
- Kormelink R., Garcia M.L., Goodin M., Sasaya T., Haenni A.L. (2011). Negative-strand RNA viruses: the plant-infecting counterparts. Virus Res. 162: 184–202. [DOI] [PubMed] [Google Scholar]
- Krasileva K.V., Dahlbeck D., Staskawicz B.J. (2010). Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. Plant Cell 22: 2444–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roux C., et al. (2015). A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell 161: 1074–1088. [DOI] [PubMed] [Google Scholar]
- Li J., Feng Z., Wu J., Huang Y., Lu G., Zhu M., Wang B., Mao X., Tao X. (2015). Structure and function analysis of nucleocapsid protein of tomato spotted wilt virus interacting with RNA using homology modeling. J. Biol. Chem. 290: 3950–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Lewandowski D.J., Hilf M.E., Adkins S. (2009). Identification of domains of the Tomato spotted wilt virus NSm protein involved in tubule formation, movement and symptomatology. Virology 390: 110–121. [DOI] [PubMed] [Google Scholar]
- Lin T., et al. (2014). Genomic analyses provide insights into the history of tomato breeding. Nat. Genet. 46: 1220–1226. [DOI] [PubMed] [Google Scholar]
- López C., Aramburu J., Galipienso L., Soler S., Nuez F., Rubio L. (2011). Evolutionary analysis of tomato Sw-5 resistance-breaking isolates of Tomato spotted wilt virus. J. Gen. Virol. 92: 210–215. [DOI] [PubMed] [Google Scholar]
- Lukasik E., Takken F.L. (2009). STANDing strong, resistance proteins instigators of plant defence. Curr. Opin. Plant Biol. 12: 427–436. [DOI] [PubMed] [Google Scholar]
- Mackey D., Holt B.F. III, Wiig A., Dangl J.L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108: 743–754. [DOI] [PubMed] [Google Scholar]
- Maekawa T., Kufer T.A., Schulze-Lefert P. (2011). NLR functions in plant and animal immune systems: so far and yet so close. Nat. Immunol. 12: 817–826. [DOI] [PubMed] [Google Scholar]
- Maqbool A., Saitoh H., Franceschetti M., Stevenson C.E., Uemura A., Kanzaki H., Kamoun S., Terauchi R., Banfield M.J. (2015). Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor. eLife 4: e08709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan S.B., Bodeau J., Yaghoobi J., Kaloshian I., Zabel P., Williamson V.M. (1998). The root knot nematode resistance gene Mi from tomato is a member of the leucine zipper, nucleotide binding, leucine-rich repeat family of plant genes. Plant Cell 10: 1307–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R., Herr E.H., Orvar B.L., van Camp W., Willekens H., Inzé D., Ellis B.E. (1999). Transgenic tobacco plants with reduced capability to detoxify reactive oxygen intermediates are hyperresponsive to pathogen infection. Proc. Natl. Acad. Sci. USA 96: 14165–14170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nombela G., Williamson V.M., Muñiz M. (2003). The root-knot nematode resistance gene Mi-1.2 of tomato is responsible for resistance against the whitefly Bemisia tabaci. Mol. Plant Microbe Interact. 16: 645–649. [DOI] [PubMed] [Google Scholar]
- Nürnberger T., Nennstiel D., Jabs T., Sacks W.R., Hahlbrock K., Scheel D. (1994). High affinity binding of a fungal oligopeptide elicitor to parsley plasmamembranes triggers multiple defense response. Cell 78: 449–460. [DOI] [PubMed] [Google Scholar]
- Padmanabhan M.S., Ma S., Burch-Smith T.M., Czymmek K., Huijser P., Dinesh-Kumar S.P. (2013). Novel positive regulatory role for the SPL6 transcription factor in the N TIR-NB-LRR receptor-mediated plant innate immunity. PLoS Pathog. 9: e1003235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiró A., Cañizares M.C., Rubio L., López C., Moriones E., Aramburu J., Sánchez-Navarro J. (2014). The movement protein (NSm) of Tomato spotted wilt virus is the avirulence determinant in the tomato Sw-5 gene-based resistance. Mol. Plant Pathol. 15: 802–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzer L., Bezerra I.C., Kormelink R., Prins M., Peters D., Resende R.O., de Ávila A.C. (1999). Characterization of a tospovirus isolate of iris yellow spot virus associated with a disease in onion fields in brazil. Plant Dis. 83: 345–350. [DOI] [PubMed] [Google Scholar]
- Pruitt R.N., et al. (2015). The rice immune receptor XA21 recognizes a tyrosine-sulfated protein from a Gram-negative bacterium. Sci. Adv. 1: e1500245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi M., Goggin F.L., Milligan S.B., Kaloshian I., Ullman D.E., Williamson V.M. (1998). The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proc. Natl. Acad. Sci. USA 95: 9750–9754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarris P.F., et al. (2015). A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell 161: 1089–1100. [DOI] [PubMed] [Google Scholar]
- Scholthof K.B., Adkins S., Czosnek H., Palukaitis P., Jacquot E., Hohn T., Hohn B., Saunders K., Candresse T., Ahlquist P., Hemenway C., Foster G.D. (2011). Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 12: 938–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao F., Golstein C., Ade J., Stoutemyer M., Dixon J.E., Innes R.W. (2003). Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 301: 1230–1233. [DOI] [PubMed] [Google Scholar]
- Spassova M.I., Prins T.W., Folkertsma R.T., Klein-Lankhorst R.M., Hille J., Goldbach R.W., Prins M. (2001). The tomato gene Sw5 is a member of the coiled coil, nucleotide binding, leucine-rich repeat class of plant resistance genes and confers resistance to TSWV in tobacco. Mol. Breed. 7: 151–161. [Google Scholar]
- Stevens M.R., Scott S.J., Gergerich R.C. (1992). Inheritance of a gene for resistance to Tomato spotted wilt virus (TSWV) from Lycopersicon peruvianum Mill. Euphytica 59: 9–17. [Google Scholar]
- Sun W., Dunning F.M., Pfund C., Weingarten R., Bent A.F. (2006). Within-species flagellin polymorphism in Xanthomonas campestris pvcampestris and its impact on elicitation of Arabidopsis FLAGELLIN SENSING2-dependent defenses. Plant Cell 18: 764–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Li L., Macho A.P., Han Z., Hu Z., Zipfel C., Zhou J.M., Chai J. (2013). Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 342: 624–628. [DOI] [PubMed] [Google Scholar]
- Takken F.L.W., Tameling W.I.L. (2009). To nibble at plant resistance proteins. Science 324: 744–746. [DOI] [PubMed] [Google Scholar]
- Tameling W.I., Vossen J.H., Albrecht M., Lengauer T., Berden J.A., Haring M.A., Cornelissen B.J., Takken F.L. (2006). Mutations in the NB-ARC domain of I-2 that impair ATP hydrolysis cause autoactivation. Plant Physiol. 140: 1233–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobias I., Rast A.T.B., Maat D.Z. (1982). Tobamoviruses of pepper, eggplant and tobacco: comparative host reactions and serological relationships. Neth. J. Plant Pathol. 88: 257–268. [Google Scholar]
- Tomato Genome Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Biezen E.A., Jones J.D. (1998). The NB-ARC domain: a novel signalling motif shared by plant resistance gene products and regulators of cell death in animals. Curr. Biol. 8: R226–R227. [DOI] [PubMed] [Google Scholar]
- Vos P., et al. (1998). The tomato Mi-1 gene confers resistance to both root-knot nematodes and potato aphids. Nat. Biotechnol. 16: 1365–1369. [DOI] [PubMed] [Google Scholar]
- Wang S., Sun Z., Wang H., Liu L., Lu F., Yang J., Zhang M., Zhang S., Guo Z., Bent A.F., Sun W. (2015). Rice OsFLS2-mediated perception of bacterial flagellins is evaded by Xanthomonas oryzae pvs. oryzae and oryzicola. Mol. Plant 8: 1024–1037. [DOI] [PubMed] [Google Scholar]
- Williams L.V., López Lambertini P.M., Shohara K., Biderbost E.B. (2001). Occurrence and geographical distribution of Tospovirus species infecting tomato crops in Argentina. Plant Dis. 85: 1227–1229. [DOI] [PubMed] [Google Scholar]
- Williams S.J., Sornaraj P., deCourcy-Ireland E., Menz R.I., Kobe B., Ellis J.G., Dodds P.N., Anderson P.A. (2011). An autoactive mutant of the M flax rust resistance protein has a preference for binding ATP, whereas wild-type M protein binds ADP. Mol. Plant Microbe Interact. 24: 897–906. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y., Pearce G., Ryan C.A. (2006). The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proc. Natl. Acad. Sci. USA 103: 10104–10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Huffaker A., Bryan A.C., Tax F.E., Ryan C.A. (2010). PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 22: 508–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C. (2008). Pattern-recognition receptors in plant innate immunity. Curr. Opin. Immunol. 20: 10–16. [DOI] [PubMed] [Google Scholar]
- Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J.D.G., Boller T., Felix G. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760. [DOI] [PubMed] [Google Scholar]