ABSTRACT
In Vibrio species, AphB is essential to activate virulence cascades by sensing low-pH and anaerobiosis signals; however, its regulon remains largely unknown. Here, AphB is found to be a key virulence regulator in Vibrio alginolyticus, a pathogen for marine animals and humans. Chromatin immunoprecipitation followed by high-throughput DNA sequencing (ChIP-seq) enabled the detection of 20 loci in the V. alginolyticus genome that contained AphB-binding peaks. An AphB-specific binding consensus was confirmed by electrophoretic mobility shift assays (EMSAs), and the regulation of genes flanking such binding sites was demonstrated using quantitative real-time PCR analysis. AphB binds directly to its own promoter and positively controls its own expression in later growth stages. AphB also activates the expression of the exotoxin Asp by binding directly to the promoter regions of asp and the master quorum-sensing (QS) regulator luxR. DNase I footprinting analysis uncovered distinct AphB-binding sites (BBS) in these promoters. Furthermore, a BBS in the luxR promoter region overlaps that of LuxR-binding site I, which mediates the positive control of luxR promoter activity by AphB. This study provides new insights into the AphB regulon and reveals the mechanisms underlying AphB regulation of physiological adaptation and QS-controlled virulence in V. alginolyticus.
IMPORTANCE In this work, AphB is determined to play essential roles in the expression of genes associated with QS, physiology, and virulence in V. alginolyticus, a pathogen for marine animals and humans. AphB was found to bind directly to 20 genes and control their expression by a 17-bp consensus binding sequence. Among the 20 genes, the aphB gene itself was identified to be positively autoregulated, and AphB also positively controlled asp and luxR expression. Taken together, these findings improve our understanding of the roles of AphB in controlling physiological adaptation and QS-controlled virulence gene expression.
KEYWORDS: AphB, ChIP-seq, binding motif, QS, Vibrio alginolyticus
INTRODUCTION
Bacterial pathogens leverage a plethora of signaling pathways to sense and to respond to environmental and host cues to optimize their virulence expression and physiological adaptation for survival. The elucidation of regulatory cascades and their interaction networks will pinpoint the signals required to activate bacterial virulence gene expression and help to illuminate how pathogens intertwine various pathways to orchestrate the expression of genes associated with virulence and physiological adaptation. Furthermore, bacterial pathogens evolve capabilities to take advantage of some nondetrimental signals (e.g., temperature fluctuations or anoxic conditions), integrating them into preset regulatory pathways (e.g., quorum-sensing [QS] systems) to render magnified signaling outputs (1). Living in both aquatic environments and human intestines with fluctuating cues, some Vibrio species must have been endowed with such sophisticated regulatory capabilities. In the notorious human pathogen Vibrio cholerae, QS signaling has been proved to be essential for inhibition of the expression of the cholera toxin and toxin-coregulated pilus (2). Furthermore, the diverse environmental inputs that are present in the intestine, including pH, bicarbonate (3), reduced oxygen (4), bile (5), and unsaturated fatty acids (6), as well as temperature (7), integrate into the QS circuit and modulate the virulence gene expression by various mechanisms, tightly connecting virulence regulation and physiological adaptation and thus representing an exquisite regulatory system among bacterial pathogens.
AphB, a LysR family protein, represents one of the key regulators that transmit the aforementioned signals for optimal virulence output in V. cholerae (4, 8). AphB cooperates with AphA, the low-cell-density master QS regulator (MQSR), to activate tcpPH expression and the subsequent expression cascade of virulence genes. AphB forms a tetramer with a ligand-binding pocket to respond to two known signals, i.e., low pH and anaerobiosis (9). The N-terminal DNA-binding domain folds into a typical winged helix-turn-helix motif to activate gene expression upon the conformational transition brought about by a thiol-based switch, i.e., cysteine 235, in an anoxic environment (4, 9). Thus, the interplay of AphA and AphB couple, at the critical levels, the signaling of QS and oxygen tension for the exquisite regulation of virulence gene expression in V. cholerae. Moreover, recent investigations revealed that AphB cooperates with OhrR to jump-start tcpP and ohrA transcription, respectively, with their distinct thiol-based switches for fine-tuned pathogenesis and responses to anoxic conditions (10, 11). In addition to its established essential roles in the activation of tcpP and toxR to control virulence expression, AphB has been revealed to play critical roles in the regulation of genes associated with acid tolerance and anaerobic responses in both V. cholerae and Vibrio vulnificus (12, 13). AphB binds to the promoters of cadC to regulate its expression and that of four other genes to counteract external acidification in these bacteria. Notably, a microarray screen predicted that 489 genes are potentially regulated by AphB in V. vulnificus, suggesting AphB as a global regulator (14). Thorough investigation of this regulon will further illustrate the mechanisms underlying these regulatory functions.
Vibrio alginolyticus is an opportunistic pathogen that infects a great diversity of sea animals and causes intraintestinal and extraintestinal diseases or wound infections in humans (15). Known factors associated with the virulence of V. alginolyticus include extracellular proteases, e.g., Asp (16). These factors and other virulence-related genes are closely regulated by the LuxR-LuxO-centered QS regulatory system in the bacterium (17, 18). We determined previously that AphA and LuxR, the high-cell-density MQSR, inhibited and activated, respectively, the expression of QS-controlled virulence gene expression in the bacterium (19, 20). LuxR also regulates itself by binding the promoter in two regions, known as RBSI and RBSII (19). LuxR binds RBSI to activate luxR expression and interacts with RBSII to repress its expression. AphA negatively regulates luxR expression by directly binding to the promoter at the AphA-binding site (ABS). In this study, we further explored whether AphB regulates virulence factors and how it associates with the QS regulatory pathway. Chromatin immunoprecipitation (ChIP) followed by high-throughput DNA sequencing (ChIP-seq) technology following MEME analysis (21) allowed us to identify novel gene targets and the consensus sequence that is directly bound by AphB. The data presented here indicate that AphB modulates QS, demonstrating another layer of QS control to regulate the pathogenesis of Vibrio species.
RESULTS
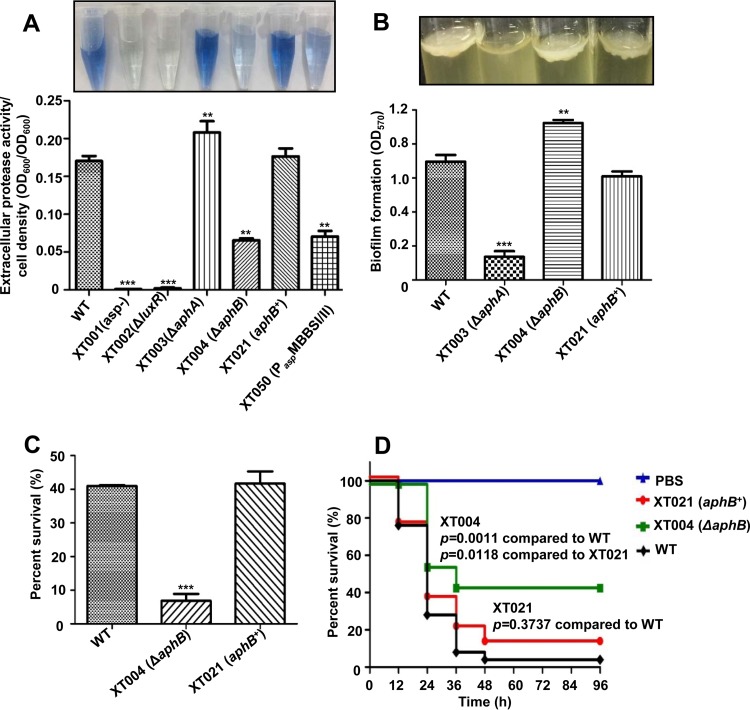
Essential roles of AphB in ECP production, biofilm formation, acid resistance, and in vivo survival of zebra fish.
AphB belongs to the LysR transcription factor family and has been revealed to be essential in pathogenesis and physiological fitness in V. cholerae and V. vulnificus (12, 14). We identified the 876-bp aphB gene in the genome of V. alginolyticus EPGS. The V. alginolyticus AphB protein displays high levels of similarity to its homologs from other Vibrio species, with 99% (Vibrio harveyi), 96% (Vibrio parahaemolyticus), 89% (V. vulnificus), and 81% (V. cholerae) identity. We created an in-frame deletion mutant of aphB, XT004 (ΔaphB), to characterize the roles of aphB in V. alginolyticus (Table 1). The growth of XT004 was similar to that of the wild-type (wt) strain in Luria-Bertani (LB) medium with 3% NaCl (LBS medium) (data not shown). Asp is the key exotoxin in V. alginolyticus (19). Asp activity in extracellular proteins (ECPs), as specifically determined by hide powder azure (HPA) digestion assays, was significantly decreased in the XT004 (ΔaphB) strain, compared to the wt strain (Fig. 1A). In the complementation strain XT021 (aphB+), extracellular Asp activity was restored to the wt level. This result revealed that AphB positively regulated Asp expression or secretion in V. alginolyticus. Biofilm formation was significantly increased in XT004 (ΔaphB), compared to the wt strain, the complementation strain XT021 (aphB+), and strain XT003 (ΔaphA), which is impaired in biofilm formation (20) (Fig. 1B). When challenged for 60 min in 10 mM sodium citrate buffer (pH 5.8) supplemented with 3.0% NaCl (SCBN), the survival of the wt strain decreased to 40%, while that of XT004 (ΔaphB) was dramatically decreased to 7% (Fig. 1C). The XT021 (aphB+) strain restored the acid tolerance to a level similar to that of the wt strain. The data showed that AphB is essential in responding to low pH in V. alginolyticus.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| E. coli | ||
| DH5α λpir | pir lysogen Δ(ara-leu) araD Δ(lacX74) phoA20 thi-1 rpoB argE(Am) recA1 | 40 |
| SM10 λpir | thi thir leu tonA lecY supE recA::RP4-2-Tc::Mu Kmr (pirR6K) | 41 |
| BL21(DE3) | Host strain for protein expression | Novagen |
| V. alginolyticus | ||
| EPGS | Wild-type pathogenic isolate from fish; Ampr | 25 |
| XT001 | EPGS asp−; Cmr | 16 |
| XT002 | EPGS ΔluxR | 16 |
| XT003 | EPGS ΔaphA | 20 |
| XT004 | EPGS ΔaphB | This study |
| XT005 | EPGS ΔaphA ΔaphB | This study |
| XT020 | ΔaphB/pBAD33-aphB-flag; Cmr | This study |
| XT021 | aphB+, ΔaphB/pBAD33-aphB-flag2; Cmr | This study |
| XT022 | ΔaphB/pBAD33-aphB; Cmr | This study |
| XT023 | ΔaphB/pBAD33; Cmr | This study |
| XT024 | EPGS/pBAD33-Flag; Cmr | This study |
| XT025 | EPGS/pDM8-Pasp-luxAB; Cmr | This study |
| XT026 | ΔaphB/pDM8-Pasp-luxAB; Cmr | This study |
| XT027 | EPGS/pDM8-PaspMBSI/II-luxAB; Cmr | This study |
| XT028 | EPGS/pDM8-PluxR-luxAB; Cmr | This study |
| XT029 | ΔaphB/pDM8-PluxR-luxAB; Cmr | This study |
| XT030 | ΔluxR/pDM8-PluxR-luxAB; Cmr | This study |
| XT031 | ΔaphA/pDM8-PluxR-luxAB; Cmr | This study |
| XT032 | ΔaphA ΔaphB/pDM8-PluxR-luxAB; Cmr | This study |
| XT034 | EPGS/pDM8-PaphB-luxAB; Cmr | This study |
| XT035 | ΔaphB/pDM8-PaphB-luxAB; Cmr | This study |
| XT036 | EPGS/pDM8-PluxRΔBBS-luxAB; Cmr | This study |
| XT037 | EPGS/pDM8-PluxRΔABS-luxAB; Cmr | This study |
| XT038 | EPGS/pDM8-PluxRΔRBSI-luxAB; Cmr | This study |
| XT040 | EPGS/pDM8-PluxRMBBS-luxAB; Cmr | This study |
| XT041 | EPGS/pDM8-PluxRMRBSI-luxAB; Cmr | This study |
| XT042 | EPGS/pDM8-PluxRMRBSPI-luxAB; Cmr | This study |
| XT043 | EPGS/pDM8-PluxRΔABSMBBS-luxAB; Cmr | This study |
| XT057 | BL21/pET28a-aphA; Kmr | 20 |
| XT058 | BL21/pET28a-luxR; Kmr | 16 |
| XT046 | BL21/pET28a-aphB; Kmr | This study |
| XT050 | PaspMBBSI/II, Pasp with scrambled AphB binding sites I/II | This study |
| XT051 | EPGS/pDM8-PaspMBBSI-luxAB; Cmr | This study |
| XT052 | EPGS/pDM8-PaspMBBSII-luxAB; Cmr | This study |
| XT053 | ΔaphB/pDM8-PaspMBBSI-luxAB; Cmr | This study |
| XT054 | ΔaphB/pDM8-PaspMBBSII-luxAB; Cmr | This study |
| XT055 | ΔaphB/pDM8-PaspMBBSI/II-luxAB; Cmr | This study |
| Plasmids | ||
| pDM4 | Suicide vector, pir dependent, R6K, SacBR; Cmr | 42 |
| pBAD33-mob | pBAD33, mob+ allowing for conjugation; Cmr | 20 |
| pDM8 | pSup202 containing promoterless lacZ; Cmr | 43 |
| pET28a | Expression vector; Kmr | Novagen |
| pDM4::aphA | pDM4 with aphA fragment deletion of 4 to 573 bp | 20 |
| pDM4-aphB | pDM4 carrying aphB with deletion of 4 to 873 bp | This study |
| pBAD33-aphB-flag | pBAD33-derived aphB-Flag expression plasmid | This study |
| pBAD33-aphB-flag2 | pBAD33 derivative expressing aphB-Flag by PaphB | This study |
| pBAD33-flag | pBAD33 derivative expressing Flag alone | This study |
| pBAD33-aphB | pBAD33-derived aphB expression plasmid | This study |
| pET28a-aphB | pET28a with aphB ORF | This study |
| pET28a-aphA | pET28a with aphA ORF | 20 |
| pET28a-luxR | pET28a with luxR ORF | 18 |
| pDM8-Pasp-luxAB | pDM8 with Pasp-luxAB | This study |
| pDM8-PaspMBBSI/II-luxAB | pDM8 with mutated AphB boxes of Pasp-luxAB | This study |
| pDM8-PaspMBBSI-luxAB | pDM8 with mutated AphB box I of Pasp-luxAB | This study |
| pDM8-PaspMBBSII-luxAB | pDM8 with mutated AphB box II of Pasp-luxAB | This study |
| pDM8-PluxR-luxAB | pDM8 with PluxR-luxAB | This study |
| pDM8-PaphB-luxAB | pDM8 with PaphB-luxAB | This study |
| pDM8-PluxRΔRBSI-luxAB | pDM8-PluxR-luxAB with LuxRI box deleted | This study |
| pDM8-PluxRΔABS-luxAB | pDM8-PluxR-luxAB with AphA box deleted | This study |
| pDM8-PluxRΔBBS-luxAB | pDM8-PluxR-luxAB with AphB box deleted | This study |
| pDM8-PluxRMBBS-luxAB | pDM8-PluxR-luxAB with AphB box mutated | This study |
| pDM8-PluxRMRBSI-luxAB | pDM8-PluxR-luxAB with LuxRI box mutated | This study |
| pDM8-PluxRMRBSPI-luxAB | pDM8-PluxR-luxAB with LuxRI box partially mutated | This study |
| pDM8-PluxRΔABSMBBS-luxAB | pDM8-PluxR-luxAB with AphB box mutated and AphA box deleted | This study |
Kmr, kanamycin resistant; Ampr, ampicillin resistant; Cmr, chloramphenicol resistant.
FIG 1.
Essential roles of AphB in various phenotypes. (A) The extracellular Asp production assay was based on HPA digestion. All strains were cultured in LBS medium for 9 h at 30°C and then each of the supernatants was used for HPA tests. The data were normalized with respect to cell densities and are shown as the mean ± standard error of the mean (SEM) (n = 3). **, P < 0.01; ***, P < 0.001, Student's t test, relative to the wt strain. (B) Biofilm formation in glass tubes was quantitatively assayed with crystal violet staining of the biofilm cells. The absorbance (OD570) of the crystal violet that was released from the biofilm cells was measured and normalized with respect to the OD600 values of the cultures. **, P < 0.01; ***, P < 0.001, Student's t test, relative to the wt strain. (C) Cultures grown in LBS medium were subjected to the acid challenge at pH 5.8. The survival rate was calculated by using the values that were determined immediately after inoculation as those of 100% survival. The results are shown as the mean ± standard deviation (SD) (n = 3). ***, P < 0.001, Student's t test, relative to the wt strain. (D) Groups of 30 fish were infected with the indicated strains. The numbers of fish that survived were recorded for 4 days. The P values were calculated using a Kaplan-Meier survival analysis with a log rank test, with Prism software.
To further evaluate the influence of aphB on the virulence of V. alginolyticus, adult zebra fish (n = 30) were challenged by intramuscular (i.m.) injection with 5 × 106 CFU/fish of the wt EPGS strain, the XT004 (ΔaphB) strain, or the XT021 (aphB+) strain. The deaths of fish challenged with XT004 occurred later than those of fish challenged with the wt strain or the complementation strain. The survival rate for fish challenged with XT004 (∼42%) was significantly higher than the rates for fish challenged with the wt strain (∼8%) or XT021 (∼15%) (Fig. 1D). The significant virulence attenuation that resulted from aphB abrogation suggested that AphB is involved in the virulence expression in V. alginolyticus. Taken together, these data indicate that AphB plays a plethora of roles in the physiological adaptation and virulence expression in V. alginolyticus.
Genome-wide identification of AphB-binding regions by ChIP-seq.
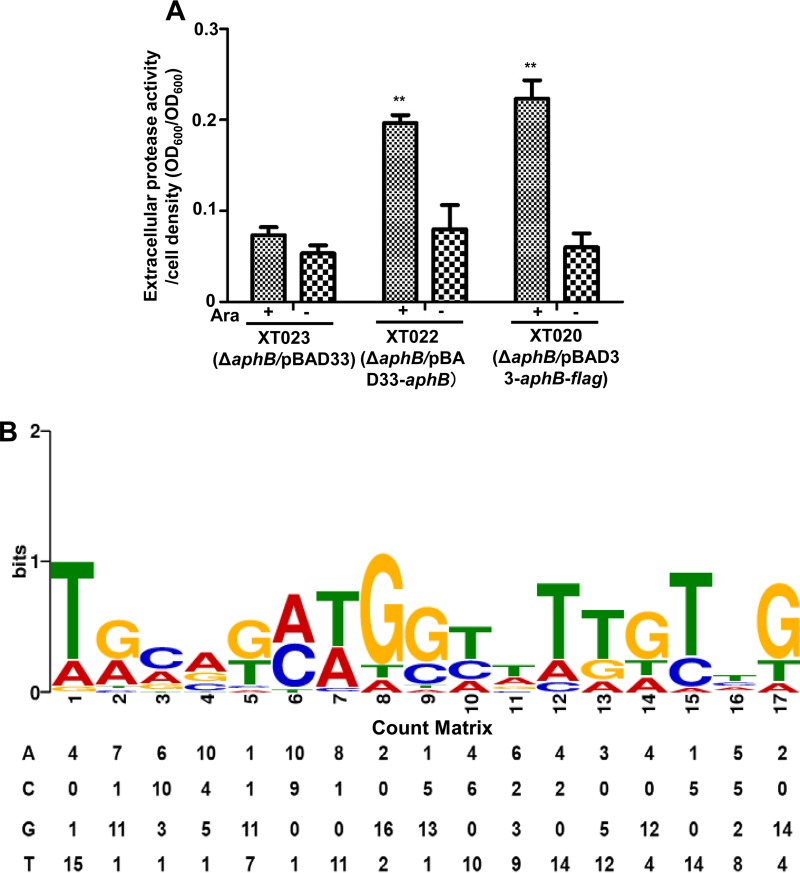
We were intrigued by the mechanisms underlying the control of multiple phenotypes by AphB. ChIP-seq was used to investigate the AphB-binding regions on the chromosomes of V. alginolyticus strain XT020 expressing Flag-tagged AphB driven by pBAD (ΔaphB/pBAD33-aphB-flag). The AphB-Flag fusion protein was expressed in XT020 and behaved similarly to wt AphB expressed in XT022 (ΔaphB/pBAD33-aphB) in vivo, as determined by assaying their activities in the activation of Asp production (Fig. 1A and 2A).
FIG 2.
ChIP-seq analysis of in vivo binding sites of AphB on V. alginolyticus chromosomes. (A) The function of Flag-tagged AphB (AphB-Flag) remained intact in V. alginolyticus. AphB-Flag or AphB expression driven by pBAD was induced by l-arabinose (Ara), and extracellular Asp production was assayed in the indicated strains with (+) or without (−) the addition of Ara. **, P < 0.01, Student's t test, relative to results for the absence of arabinose. (B) The significant motif that resulted from the ChIP-seq binding sequences was identified. The height of each letter represents the relative frequency of each base at each position in the consensus sequence. The count matrix for the binding sequences is also shown.
ChIP-seq assays identified 20 enriched loci harboring AphB-binding peaks (Table 2), which were enriched >4-fold (P < 0.001) more than those in the control samples from the XT024 strain expressing the Flag tag (EPGS/pBAD33-Flag) (Table 1). These 20 loci are located across the genome, specifically in upstream regions (4 genes), overlap start regions (4 genes), overlap end regions (2 genes), and within coding regions (10 genes), suggesting that AphB is a global transcriptional regulator in V. alginolyticus (Table 2). Using the MEME suite, a 17-bp AphB consensus sequence (TGCAGN7TGTTG) was identified (Fig. 2B). The count matrix is shown to portray the alignment of AphB-binding sites (BBS) and represents the relative frequencies of each base at different positions in the consensus sequence (Fig. 2B).
TABLE 2.
AphB ChIP-seq list
| Gene identification | Annotationa | AphB box-like sequence | Locationb | Average fold enrichment | Presence in other Vibrio speciesc | P |
|---|---|---|---|---|---|---|
| EPGS_01005 | Transcriptional regulator (aphB) | TCGGTAATTTAATGTGT | UP | 12.3 | Y | <0.001 |
| EPGS_03654 | Methionine synthase I (metH) | TGATGCTGCTTAAATAG | IN | 5.86 | N | <0.001 |
| EPGS_03653 | Na+/H+ antiporter | TACCGCAGGTTCAATGG | IN | 5.46 | Y | <0.001 |
| EPGS_04302 | Cadaverine/lysine antiporter (cadB) | TACAGCTGCATTGGTTG | UP | 5.2 | N | <0.001 |
| EPGS_01006 | CheY-like receiver | GGCCTCAGGTCTTGCTG | IN | 4.58 | Y | <0.001 |
| EPGS_04385 | Adenylate cyclase (cyaA) | AAAGTATGGTTTTGTAT | OS | 5.36 | Y | <0.001 |
| EPGS_04384 | Porphobilinogen deaminase (hemC) | AACAGTAAGCATTTCTT | OE | 5.28 | N | <0.001 |
| EPGS_04454 | UDP-N-acetylmuramate dehydrogenase (murB) | TGAGTCTGGTATTGTCA | IN | 5.18 | N | <0.001 |
| EPGS_01393 | Capsular polysaccharide biosynthesis protein | TGCGGCCGGTTTTATTG | IN | 4.46 | N | <0.001 |
| EPGS_01394 | UDP-glucose pyrophosphorylase | TGAAGCCGGTATGGATG | IN | 4.92 | N | <0.001 |
| EPGS_10421 | LPS heptosyltransferase | TGCAGAAGGCGTTGCCG | OS | 5.08 | N | <0.001 |
| EPGS_01420 | Glycosyltransferase involved in LPS biosynthesis | TGAAGATGGTACGTTTG | OE | 5.56 | N | <0.001 |
| EPGS_01419 | Lauroyl/myristoyl acyltransferase | TGCAGAAGGCGTTGCCG | IN | 4.98 | N | <0.001 |
| EPGS_01418 | Glyceromanno-heptose 6-epimerase (gmhD) | TGGCGCTGGCATGATCG | IN | 4.44 | N | <0.001 |
| EPGS_01532 | Methylglyoxal synthase | AGCGGATGGTTATGTTG | IN | 4.64 | N | <0.001 |
| EPGS_01531 | Hypothetical protein | AGCATCAGCCTTATTAG | IN | 5.46 | N | <0.001 |
| EPGS_01530 | Ferritin | TAAATAAGGAGTGGCAT | OS | 5 | N | <0.001 |
| EPGS_01529 | Hypothetical protein | TGGATATGGCTTTGTAG | OS | 4.68 | N | <0.001 |
| EPGS_00555 | Asp | TTAACATTCACTTTTTG | UP | 6.6 | N | <0.001 |
| EPGS_01835 | LuxR | TATCAATAATAATGACA | UP | 5.8 | N | <0.001 |
The genes discussed in the text are in bold. LPS, lipopolysaccharide.
The location of the identified binding site, relative to the gene, is indicated. UP, upstream of the start codon; OS, overlapping the start codon; IN, inside the gene coding sequence; OE, overlapping the end codon.
Multiple roles of AphB in the binding and expression of various genes.
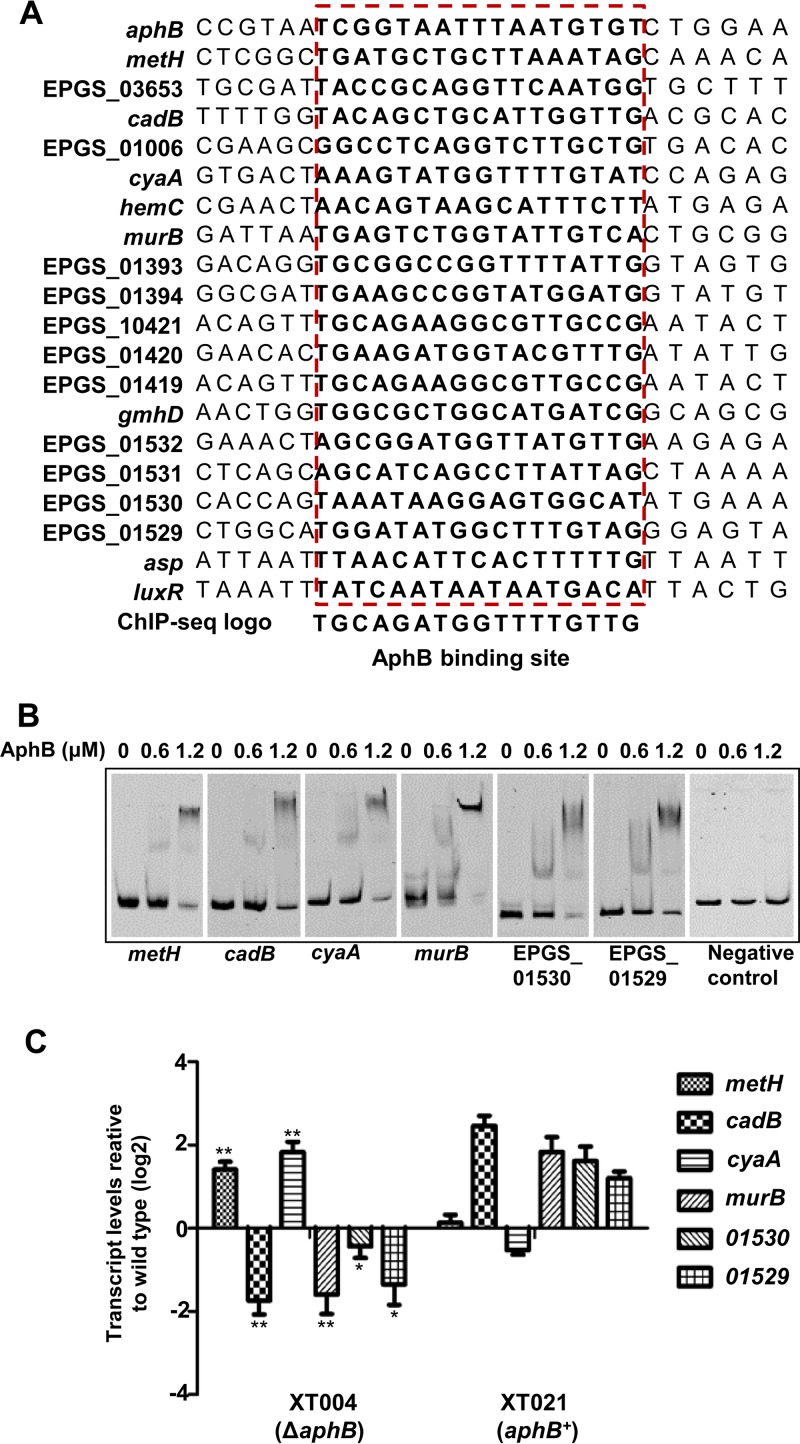
Using the MEME suite, the BBS in the 20 ChIP-seq-identified genes could be aligned (Fig. 3A). We further analyzed the ChIP-seq data by choosing six genes of interest in the list (i.e., metH, cadB, cyaA, murB, EPGS_01529, and EPGS_01530) (Table 2). The electrophoretic mobility shift assay (EMSA) results indicated that AphB bound efficiently to all probes, in a concentration-dependent manner, in the presence of high concentrations (10-fold excess of the probe DNA) of a nonspecific competitor, whereas the negative control (gyrB) remained unbound even at the highest protein concentration (Fig. 3B). We further performed quantitative real-time PCR (qRT-PCR) with the wt, XT004 (ΔaphB), and XT021 (aphB+) strains, to test the expression of the genes. The transcription of metH and cyaA was significantly increased but that of cadB, murB, EPGS_01529, and EPGS_01530 decreased in the XT004 (ΔaphB) strain (Fig. 3C); furthermore, the expression of all of the genes was restored in the overexpression strains. Taken together, these results confirmed the ChIP-seq data and suggested multiple novel roles of AphB in the various processes of V. alginolyticus.
FIG 3.
Validation of ChIP-seq results in vitro and in vivo. (A) Alignment of the ChIP-enriched regions of the 20 genes that contain an AphB-binding box. The conserved consensus motif is boxed. (B) AphB binding specifically to the selected target regions, as revealed by the EMSA. The promoter regions of metH, cadB, cyaA, murB, EPGS_01530, and EPGS_01529 were chosen, and the PCR products containing the indicated fragment were added to the reaction mixtures with different concentrations of AphB protein and a 10-fold excess of the nonspecific competitor poly(dI-dC). The negative control (the gyrB gene fragment) showed no binding to AphB. (C) qRT-PCR analysis of the transcripts of the selected genes. The wt, XT004 (ΔaphB), and XT020 (aphB+) strains were cultured in LBS medium for 4 h, and mRNA transcripts were detected by qRT-PCR. The gyrB gene was selected as a control. The results are displayed as the mean ± SD (n = 3). *, P < 0.05; **, P < 0.01, Student's t test, relative to the wt strain.
AphB repression of its own expression in later growth stages.
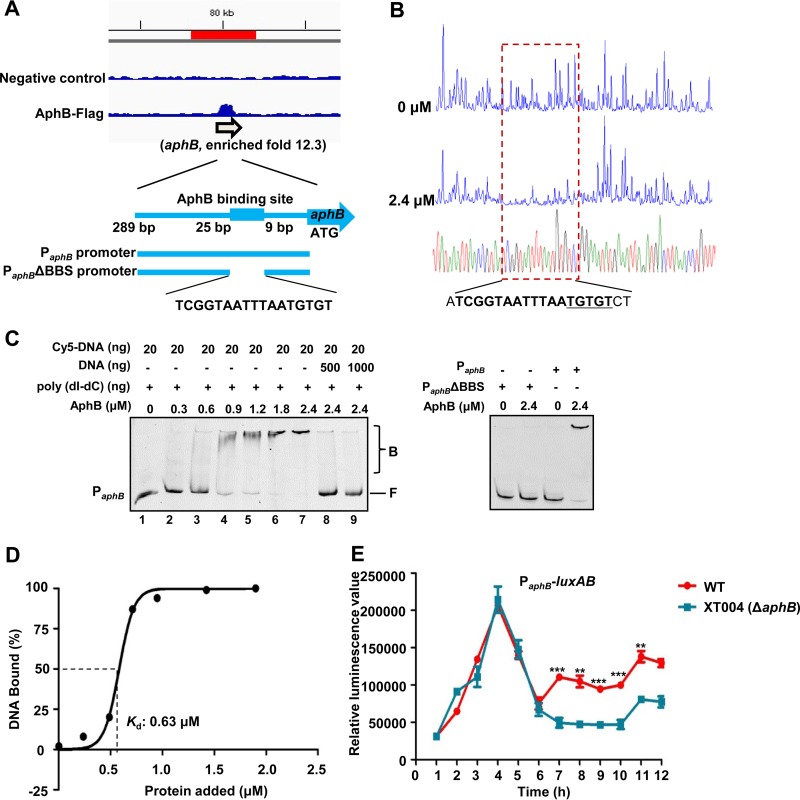
AphB has been established to bind the aphB promoter and autorepress its own expression in V. cholerae (8). Among the 20 enriched loci identified in the ChIP-seq data (Table 2), aphB represented one of the highest peaks (12.3-fold) in the V. alginolyticus genome bound by AphB (Fig. 4A). To map the binding site, we performed DNase I footprinting analysis of the two strands of the promoter region of aphB (Fig. 4B). The result showed that AphB protects 5′-ATCGGTAATTTAATGTGTCT-3′, a region ranging from position −9 to position −25, relative to its ATG start codon (Fig. 4A and B). As expected, the AphB protein bound directly to the aphB promoter region, in a concentration-dependent manner, in the EMSA in the presence of high concentrations (10-fold) of nonspecific poly(dI-dC) competitor (Fig. 4C, lanes 1 to 7) and could be titrated out by adding the unlabeled aphB probe (Fig. 4C, lanes 8 and 9). The absence of the BBS in PaphB abolished the capacity of AphB to bind to its own promoter region (Fig. 4A and C). The Kd (dissociation constant) value for AphB binding to aphB was calculated to be 0.63 μM (Fig. 4D). Given these results, we next determined whether aphB expression regulated itself in vivo. We compared the expression levels of PaphB-luxAB (Table 1) in the context of the wt and XT004 (ΔaphB) strains. At 1 to 6 h, there was no apparent difference in luminescence levels in the wt and XT004 strains. During later hours of incubation, the luminescence in XT004 was significantly lower than that in the wt strain (Fig. 4E). These results showed that AphB binds directly to its own promoter to positively regulate the expression of aphB in V. alginolyticus at later growth stages.
FIG 4.
AphB binding directly to its own promoter for self-activation. (A) AphB binding to its own promoter region, as illustrated by the peak (enriched 12.3-fold) identified in the ChIP-seq experiment. The AphB-binding site and the probes used for the EMSA studies shown in panel C are illustrated. (B) DNase I footprinting analysis of AphB binding to the aphB promoter. Electropherograms, after DNase I digestion, of the aphB promoter probe (200 ng) incubated with 0 or 2.4 μM AphB are shown. The imperfect dyad of the binding site (TGTGT) is underlined. (C) EMSA of the aphB promoter region (PaphB) (left) and PaphBΔBBS (right) with purified AphB. Twenty nanograms of each Cy5-labeled probe was added to the EMSA reaction mixtures. A 10-fold excess of nonspecific competitor DNA [poly(dI-dC)] was added to the EMSA reaction mixtures. A 25- or 50-fold excess of unlabeled specific competitor DNA was also included (left, lanes 8 and 9). AphB-bound (labeled B) or unbound (labeled F) DNA is shown. (D) Plot showing the affinity of AphB binding to its own promoter. The densitometric intensities of bound DNA fragments are plotted against the AphB concentrations. The concentration of AphB causing half-maximal binding (Kd) is shown. A representative plot is shown (n = 3). (E) PaphB-luxAB transcriptional analysis in wt and XT004 (ΔaphB) strains. The wt and XT004 strains carrying the PaphB-luxAB reporter plasmid were cultured in LBS medium for 12 h and assayed for luminescence every 1 h. The results are shown as the mean ± SD (n = 3). **, P < 0.01; ***, P < 0.005, Student's t test, relative to the wt strain.
AphB activation of asp expression by direct binding to its promoter.
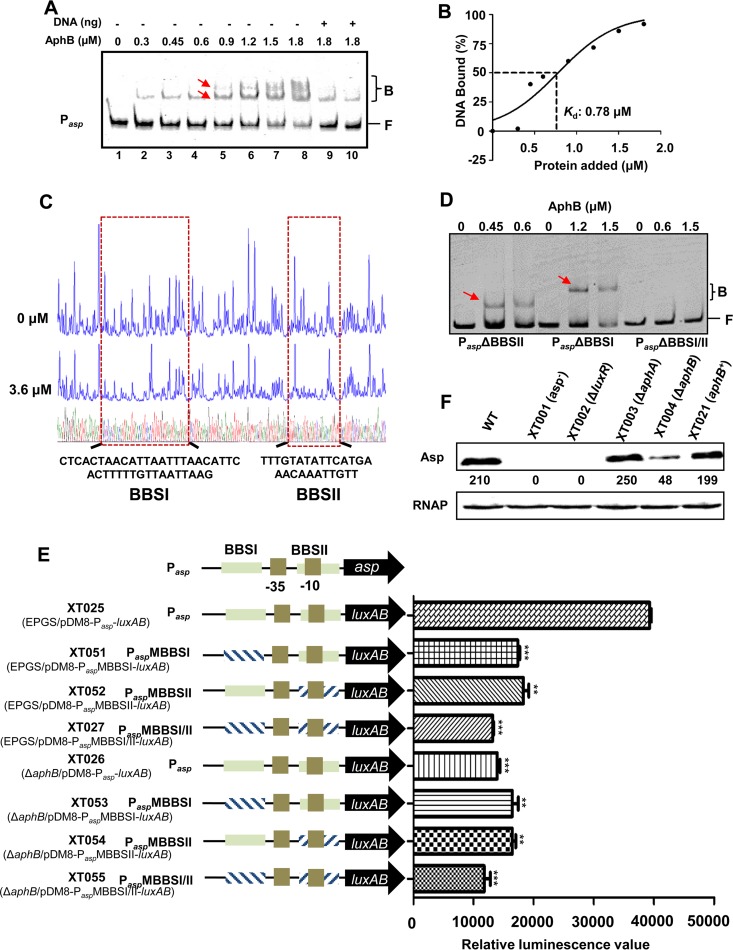
ChIP-seq data indicated that asp was also bound by AphB and enriched 6.6-fold (Table 2). An EMSA was carried out with increasing amounts of AphB with a 302-bp asp promoter probe. As shown in Fig. 5A, lanes 1 to 8, the addition of AphB resulted in a concentration-dependent ladder of two bands with retarded movement, suggesting that two binding sites with different affinities for AphB could be present within the asp regulatory region. The binding of AphB was also specific, because of the presence of excess poly(dI-dC). Furthermore, the unlabeled 302-bp DNA sequence competed for the binding of AphB in a dose-dependent manner (Fig. 5A, lanes 9 and 10), confirming that AphB binds specifically to the probe, with a Kd value of 0.78 μM (Fig. 5B).
FIG 5.
AphB binding directly to the asp promoter to regulate its expression. (A) EMSA of the asp promoter with purified AphB. Twenty nanograms of each Cy5-labeled asp probe was added to the EMSA reaction mixtures. A 25- to 50-fold excess of unlabeled specific DNA (+) (lanes 9 and 10) and a 10-fold excess of nonspecific competitor DNA [poly(dI-dC)] (lanes 1 to 10) were included. The arrows indicate the two binding sites for the AphB protein (BBSI and BBSII) in the asp promoter. (B) Plot showing the affinity of AphB for the asp promoter. (C) DNase I footprinting analysis of BBSI and BBSII in the asp promoter. (D) The EMSA with the asp promoter variants and AphB. Variants of the promoter region of asp lacking BBSI (PaspΔBBSI), BBSII (PaspΔBBSII), or both binding sites (PaspΔBBSI/II) were used. A 10-fold excess of nonspecific competitor DNA [poly(dI-dC)] and 20 ng of each Cy5-labeled probe were added to the EMSA reaction mixtures. Arrows indicate the specific bands relative to BBSI and BBSII. B, bound bands; F, free or unbound bands. (E) Promoter activities of Pasp and its variants fused to luxAB and assayed in wt or ΔaphB strains. The fluorescence data are shown as the mean ± SEM from three experiments. **, P < 0.01; ***, P < 0.005, Student's t test, relative to the wt strain. (F) Western blot analysis of Asp production, performed using an Asp-specific antiserum. The optical density values counted with Gel-Pro analyzer software (Media Cybernetics) are shown below the bands. RNAP was used as a control for blot loading.
To determine whether there were two specific BBS in the asp promoter, a DNase I footprinting assay was performed with the 302-bp DNA probe used in the EMSA studies described above. A comparison of the electropherograms with and without AphB revealed two specific AphB-binding sites, i.e., a 29-bp motif (bp −252 to −211, relative to the asp start codon) (BBSI) and a 26-bp motif (bp −136 to −110) (BBSII) (Fig. 5C), consistent with the EMSA results. Subsequently, we repeated the EMSA using three truncated asp promoter probes, without the 29-bp motif (PaspΔBBSI), without the 26-bp motif (PaspΔBBSII), or without both of them (PaspΔBBSI/II). While AphB bound to the former two probes, each of which maintained one specific AphB-binding site, AphB lost its capacity to bind to the probe that lacked both binding sites (Fig. 5D).
To further test how Asp expression is controlled by AphB, we constructed strain XT050 (EPGS/PaspMBBSI/II), which featured the asp promoter carrying scrambled AphB-binding sites I/II in the genome, and we assayed the Asp activities in the ECPs of the strain. The data indicated that deletion of both aphB and the AphB-binding sites resulted in significantly decreased Asp production (Fig. 1A). Then, we generated a luxAB transcriptional reporter by placing luxAB under the control of the asp promoter or its variants. Compared to Pasp-luxAB in the wt strain (XT025), Pasp-luxAB in the ΔaphB context (XT026) and PaspMBBSI/II-luxAB in the wt strain (XT027) exhibited significantly reduced (by ∼3-fold) bioluminescence (Fig. 5E). The bioluminescence in the wt and ΔaphB strains with PaspMBBSI-luxAB (XT051 and XT053, respectively) or PaspMBBSII-luxAB (XT052 and XT054, respectively) was also markedly decreased (by ∼2-fold). PaspMBBSI/II-luxAB expression in the ΔaphB strain (XT055) showed further attenuated bioluminescence, demonstrating that AphB is essential for asp transcription. A Western blot assay with an Asp-specific antibody demonstrated that the Asp production in XT004 (ΔaphB) was less than that in the wt strain (Fig. 5F). Taken together, these results indicated that AphB plays essential roles in modulating, possibly at the transcriptional level, the production of the exotoxin Asp in V. alginolyticus.
AphB action on luxR expression by direct binding to its promoter.
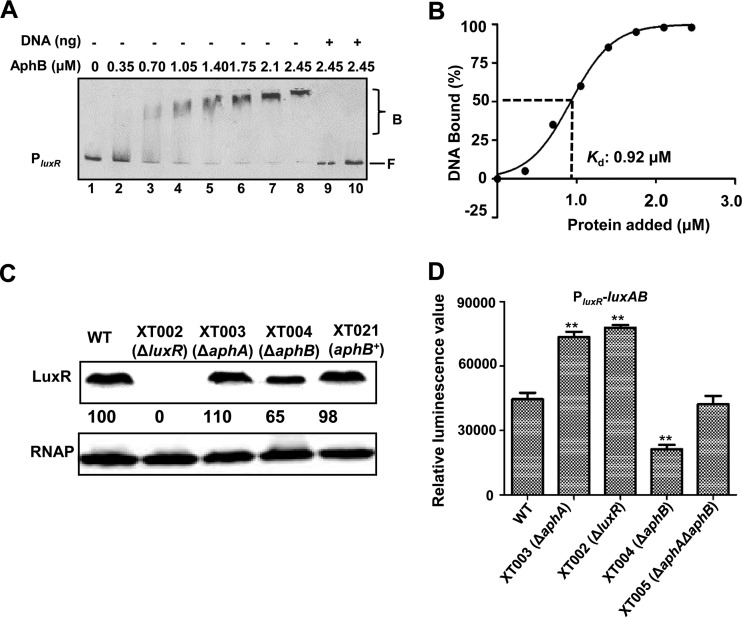
It was intriguing to find a luxR-related region included as one of the ChIP-seq hits (Table 2). To further test the interaction between AphB and luxR, a 286-bp Cy5-labeled DNA fragment containing the luxR regulatory region was incubated with increasing amounts of AphB and then subjected to electrophoresis. The addition of AphB resulted in the bands of the fragment moving more slowly (Fig. 6A, lanes 1 to 8). The binding of AphB to the luxR promoter was also specific, because the assays were performed in the presence of excess poly(dI-dC) as a nonspecific competitor, and unlabeled 286-bp DNA competed for the binding of AphB (Fig. 6A, lanes 9 and 10). The Kd value for AphB binding to the luxR promoter was calculated to be 0.92 μM (Fig. 6B).
FIG 6.
Direct AphB regulation of luxR expression. (A) EMSA of the promoter fragment of luxR with purified AphB. Twenty nanograms of each Cy5-labeled probe was added to the EMSA reaction mixtures. A 25- to 50-fold excess of unlabeled specific DNA and a 10-fold excess of nonspecific competitor DNA [poly(dI-dC)] (lanes 9 and 10) were used. B, bound bands; F, free or unbound bands. (B) Plot showing the affinity of AphB for the luxR promoter (n = 3). (C) Western blot analysis of the cellular pellets, performed using a LuxR-specific antiserum. RNAP was used as a control for blot loading. (D) Promoter activity of luxR fused to luxAB in the wt strain and different mutant strains. The fluorescence levels were assayed in the strains harboring the PluxR-luxAB reporter plasmid. Data are shown as the mean ± SEM (n = 3). **, P < 0.01, Student's t test, relative to the wt strain or other strains.
Western blotting with the LuxR antibody indicated that AphB abrogation resulted in a notably lower level of LuxR, compared to that found in the wt strain and the complementation strains in the stationary growth phase (9 h) (Fig. 6C). Furthermore, a luxR transcriptional fusion reporter exhibited a significantly lower level of luxR transcription in the XT004 (ΔaphB) strain than in the wt strain (Fig. 6D). Collectively, these results indicated that AphB binds directly to the luxR promoter region to control its expression in V. alginolyticus.
Mapping and characterization of an AphB-binding site in the luxR promoter region.
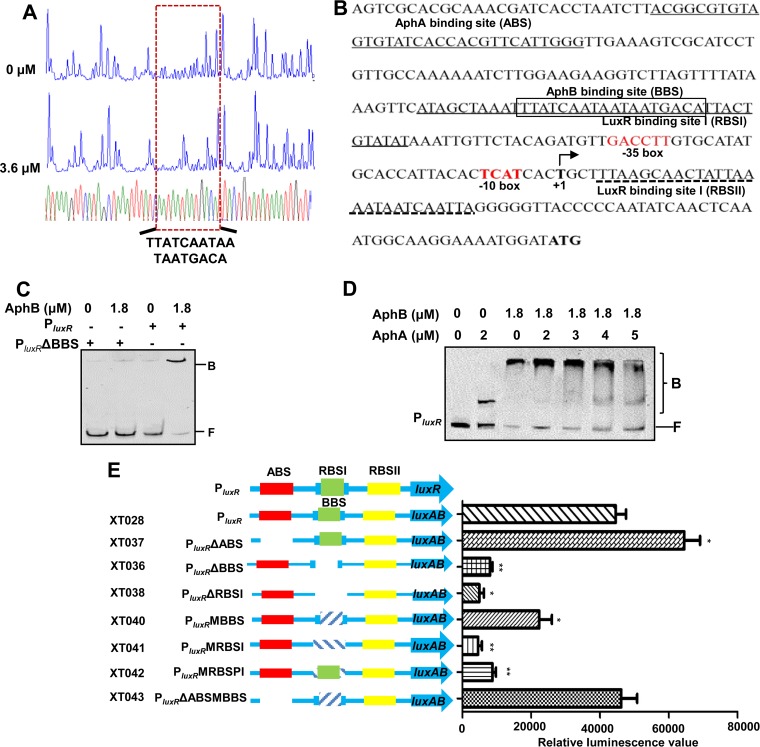
A DNase I footprint assay was performed on both strands of a 286-bp DNA fragment containing the promoter region of luxR (Fig. 7A). An 18-bp fragment (5′-TTATCAATAATAATGACA-3′) was found to be the putative binding box of AphB. The BBS overlaps with the LuxR-binding site I (RBSI) (Fig. 7B) (19), and the deletion of the BBS abolished the capacity for AphB binding to the luxR promoter (Fig. 7C).
FIG 7.
AphB regulation of luxR expression without the cooperation of AphA. (A) DNase I footprinting analysis of the BBS in the luxR promoter. Electropherograms, after digestion with DNase I, of the luxR promoter probe (200 ng) incubated with 0 or 3.6 μM AphB are shown. (B) Nucleotide sequence of the luxR promoter. LuxR-binding sites I and II (RBSI and RBSII), the ABS, and the BBS are shown. The BBS region that is protected by AphB is boxed. The transcription start site (TSS) is labeled +1. (C and D) EMSAs of the intact luxR promoter (PluxR) or that lacking the AphB binding box (PluxRΔBBS) with purified AphB alone (C) or that of PluxR with AphA-AphB mixtures (D). A 10-fold excess of nonspecific competitor DNA [poly(dI-dC)] and 20 ng of each Cy5-labeled probe were added to the EMSA reaction mixtures. B, bound bands; F, free or unbound bands. (E) Promoter activities of luxR and its variants fused to luxAB. The fluorescence levels were assayed for each of the strains. The data are shown as the mean ± SEM (n = 3). *, P < 0.05; **, P < 0.01, Student's t test, relative to the wt luxR promoter.
AphA and LuxR are established repressors of luxR expression, and the deletion of AphA and LuxR resulted in elevated luxR promoter activity (Fig. 6D) (19, 20). We further tested whether AphB modulates the expression of luxR in an AphA-dependent manner. We constructed the double mutant strain XT005 (ΔaphA ΔaphB) and investigated the PluxR activities in the strain. In comparison with the ΔaphA strain, the deletion of aphB in the ΔaphA strain markedly downregulated luxR promoter activity; however, the activity was still at a level significantly higher than that found in the XT004 (ΔaphB) strain, suggesting that AphB might independently modulate the inhibitory effects of AphA on luxR expression. EMSA studies indicated that, when both AphA and AphB were included in the PluxR binding reaction, AphA competed for the probe bound by AphB as the AphA concentration increased. The result suggested that AphA and AphB can simultaneously bind to the luxR promoter region and there might be no cooperative interplay in their binding to the region (Fig. 7D).
We next characterized the roles of the binding sites of AphA, AphB, and LuxR in the luxR promoter region in luxR expression (Fig. 7E). As expected, the luxR promoter activity was significantly reduced with the deletion (PluxRΔBBS [XT036]) or mutation (PluxRMBBS [XT040]) of BBS, and an exaggerated decrease in the luxR activity was observed upon the deletion (PluxRΔRBSI [XT038]) or mutation (PluxRMRBSI [XT041]) of RBSI, demonstrating that these cis-acting elements and AphB were essential for activation of the luxR promoter. Moreover, the nucleotide substitution in the RBSI with the intact BBS (PluxRMRBSPI [XT042]) reduced the luxR promoter activity to a level lower than that of PluxRMBBS (XT040) but higher than that of PluxRMRBSI (XT041). Furthermore, the activity of the luxR promoter with elimination of the ABS (PluxRΔABS [XT037]) and mutation of the BBS (PluxRΔABSMBBS [XT043]) was similar to that of the wt promoter. Collectively, these data support the idea that luxR expression is subjected to a sophisticated regulation network in which AphB, LuxR, and AphA interact.
DISCUSSION
Quorum sensing is a bacterial cell-to-cell communication process that modifies collective bacterial behaviors. As a global regulatory network, this process directly controls the expression of ∼110 genes, which modulate the expression of ∼625 genes in V. harveyi (22). The research on V. cholerae indicates that multiple-level regulatory networks are required to optimize the spatiotemporal production and activity levels of HapR and AphA to avoid fitness costs (23). Furthermore, this exquisite regulatory network seems to be well deployed in bacterial pathogens to couple and to relay various environmental stimuli other than cell density signals (i.e., autoinducers) for both proper physiological adaptation and virulence expression. In the zoonotic pathogen V. alginolyticus, the QS system, which is analogous to that in V. harveyi, has been characterized; this system includes the central regulator LuxR (16–19, 24, 25). Another MQSR, AphA, was recently determined to control the expression of more than 40 genes, including luxR in the low-cell-density growth stage (20). Here, we characterized the aphB gene of V. alginolyticus and found the targets of AphB by the ChIP-seq method. An AphB-dependent binding motif was identified and many novel regulation targets of AphB were revealed in V. alginolyticus. Especially, we found that AphB controls virulence through the direct regulation of asp and luxR expression. The analysis also demonstrated the essential roles of AphB in acid tolerance and biofilm formation in V. alginolyticus. Given that V. cholerae AphB uses thiol-based switches (C76, C94, and C235) to control its virulence cascade (4, 10) and these cysteine residues are conserved in AphB in V. alginolyticus, our data suggested that AphB may also mediate the QS output by sensing anaerobic conditions or other cues (e.g., acid) in V. alginolyticus.
We were intrigued that luxR expression was modulated by AphB. With EMSA and DNase I footprinting studies, we could pinpoint the binding site in the luxR promoter region to which AphB binds directly (Fig. 6A and 7A). Interestingly, the AphB-binding sequence in the luxR promoter region is not highly similar to the consensus sequence identified by ChIP-seq (Fig. 3A and 7A), and the binding affinity is relatively low (Kd = 0.92 μM), compared to that of aphB promoter (Fig. 4D). We speculate that other factors, perhaps including AphA (Fig. 7D), in a bacterial cell influence its affinity for the luxR promoter region. These analyses further demonstrated that a sophisticated regulatory network exists to control luxR expression. Future work will shed light on the detailed molecular mechanisms of AphB interactions with other factors, such as the recently established σE-based RNA polymerase (RNAP) interaction, in the luxR promoter region (19).
Our data suggested that V. alginolyticus AphB might adopt different regulatory roles in its own expression, compared to its V. cholerae homolog (8). In V. cholerae, AphB autorepresses aphB promoter expression; in V. alginolyticus, however, aphB expression appeared to be positively regulated by AphB at later growth stages (Fig. 4E). The promoter structures of PaphB in V. alginolyticus and V. cholerae are similar, and the BBS are mapped between the putative transcriptional start site and the start codon (ATG) of PaphB in these two bacteria (Fig. 4A) (8). In V. cholerae, the BBS consensus sequence (TGCAAN7TGTCA) has been identified (8). In V. alginolyticus, we could not find the perfect AphB binding box but we found the imperfect dyad of the binding site (TGTGT) (Fig. 4B). This distinction in the BBS consensus sequences in PaphB in these two bacteria might account for the different roles of AphB in the regulation of aphB expression in the bacteria. The mechanism of how AphB modulates the expression of aphB remains to be elucidated. The different autoregulation mechanisms in different Vibrio species may also indicate that their different lifestyles have shaped much of their strategies to cope with acidic or anaerobic conditions.
In V. vulnificus, 489 genes across the two chromosomes are regulated by AphB (14). Furthermore, 18 genes are activated by AphB in V. cholerae, including cadB, cadA, cadC, and a Na+/H+ antiporter, and 3 genes are repressed, including a bactoferritin-associated ferredoxin (bfd). Among the 20 detected peaks in the ChIP-seq data, 5 loci of V. alginolyticus were also revealed to be regulated in V. cholerae (12), including EPGS_03653 (Na+/H+ and K+/H+ antiporter), cadB, cyaA, hemC, and aphB (Table 2). The overlapping lists of genes supported the reliability of our ChIP-seq analysis. Moreover, our ChIP-seq data also suggested the multifunctional roles of AphB in different bacterial contexts. Specifically, AphB may not only mediate multiple environmental signals to the QS cascade but also be involved in the regulation of non-QS processes in different bacterial contexts.
The MEME program allows an AphB-binding consensus sequence (TGCAGN7TGTTG) to be found from the ChIP-seq enriched peaks. We made an alignment with several sequences that are bound by AphB in V. cholerae and V. vulnificus (13, 26, 27). The results indicated that the AphB-binding sites in V. alginolyticus are slightly different from those of tcpP (TGCAAN7TTGCA), toxR (TGCAAN7ATGGA), and aphB (TGCAAN7TGTCA) in V. cholerae (8, 27) and are highly divergent from other established binding motifs, such as those of cadC I (TTAAAN7ACTTA) and cadC II (TACGTN7GGCTA) in V. vulnificus (13). These results may further show that the binding sites of AphB vary in different bacteria and in the genes participating in various processes. The mechanism that would explain why the essential A neighboring N7 in V. cholerae (8) could be replaced by G in the V. alginolyticus AphB-binding box is not yet known. Further bioinformatics analysis of these consensus sequences has identified many more potential targets that are bound by AphB in V. alginolyticus, which might allow for a detailed examination of the binding mechanism of AphB in various bacteria.
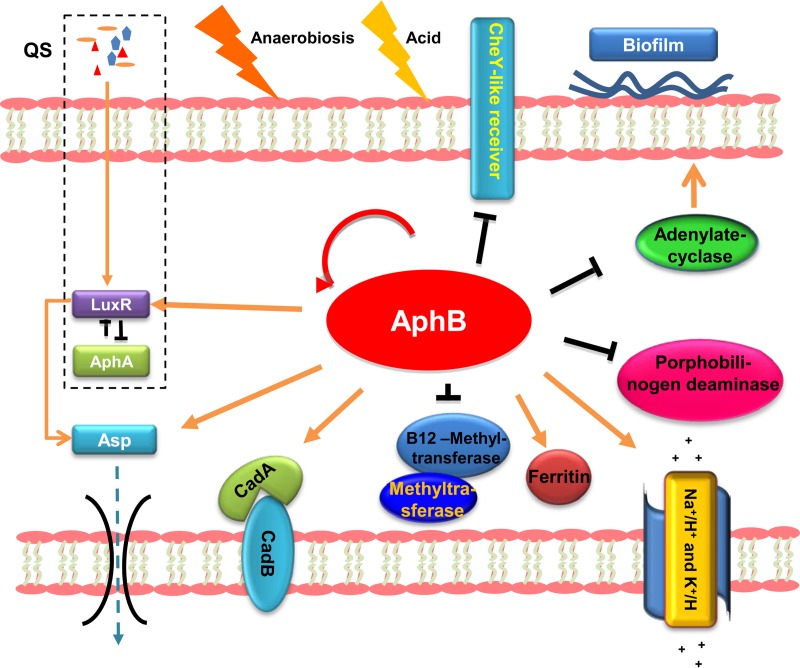
We analyzed the cellular pathways of the products of the genes in which an AphB-binding motif was present (Fig. 8). AphB-binding targets include several transcription factors such as aphB, which is self-activated (Fig. 8). The product of EPGS_03654 (metH) is a vitamin B12 methyltransferase, which plays an important role in the biosynthesis of methionine (28). In addition, the ChIP-seq data (Table 2) also indicated that AphB could regulate heme biosynthesis (29), sodium potassium pumps, and amino acid metabolism (Fig. 8). In V. vulnificus and V. cholerae, AphB regulates CadC, which in turn activates cadBA in response to low pH (12, 13). In V. alginolyticus, AphB could directly regulate cadBA (Fig. 3B and C), which encodes proteins that catalyze the decarboxylation of lysine to cadaverine and its excretion. Thus, our data indicated that AphB is involved in acid responses with different mechanisms among the Vibrio species. The cyaA gene, encoding adenylate cyclase, was negatively regulated by AphB (Fig. 3). The cAMP-cAMP receptor protein (CRP) regulatory network globally controls processes of virulence and physiology (30–32), and cyaA has been proved to be essential for biofilm formation (20, 33) in various bacteria. We thus speculate that increased cyaA expression in the aphB deletion strain XT004 might contribute to enhanced biofilm formation in the strain (Fig. 1B). Furthermore, expression of the exotoxin Asp was regulated by AphB via direct binding to the asp promoter region (Fig. 5), and this regulatory circuit was strengthened by AphB modulation of the QS cascade (Fig. 6 and 7). All of these alterations in the expression of the AphB regulon, including established or currently unknown virulence factors (Fig. 8), might contribute to the attenuated virulence in fish upon deletion of AphB in V. alginolyticus (Fig. 1D).
FIG 8.
Schematic summarization of the regulatory roles of AphB in V. alginolyticus. Kobas 2.0 (38) revealed AphB-involved pathways, and their respective cellular locations are illustrated with arrows (activation) or bar-ended lines (repression) and are discussed in the text. The QS signaling circuit is simplified and boxed.
In summary, the results presented in this work characterized the roles of AphB in global regulatory networks in V. alginolyticus. The ChIP-seq results facilitated identification of the binding motif in the AphB regulon and offer valuable information for the future study of other processes and targets regulated by AphB. Moreover, this study may improve the understanding of how AphB mediates multiple environmental signals to the QS system so that Vibrio species can adapt to different environments and reprogram their virulence expression accordingly.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. The V. alginolyticus strains were grown at 30°C in LBS medium. Escherichia coli strains were grown at 37°C in LB medium. Antibiotics and inducers were used at the following concentrations: carbenicillin, 100 μg/ml; chloramphenicol, 25 μg/ml; kanamycin, 100 μg/ml; l-arabinose, 0.04% (wt/vol); isopropyl-β-d-1-thiogalactopyranoside (IPTG), 1 mM.
Construction of V. alginolyticus mutant strains.
Construction of the aphB mutant strain XT004 (ΔaphB) with the suicide plasmid pDM4 followed a previously reported strategy (34). PCRs were performed to amplify upstream (586-bp) and downstream (369-bp) fragments of aphB by using primers aphB 1/aphB 2 and aphB 3/aphB 4, respectively (see Table S1 in the supplemental material). The two PCR products were cloned into XbaI-digested pDM4 by isothermal assembly and transformed into E. coli DH5α λpir cells, as described previously (35). After being sequenced, the resulting plasmids were transformed into E. coli SM10 λpir and then mated into EPGS by conjugation. Double-crossover processes were selected sequentially on LBS medium containing carbenicillin and chloramphenicol and then on LBS medium with 12% (wt/vol) sucrose. Creation of the targeted mutants was confirmed by PCR using the primer pair of aphB out-F and aphB out-R (Table S1) to sequence the deleted region. Construction of the other mutants was accomplished in the same manner.
To construct the aphA/aphB double-deletion strain XT005 (ΔaphA ΔaphB), the pDM4::aphA plasmid (20) was mated from SM10 λpir into XT004 (ΔaphB) for double-crossover screening. To construct the aphB-flag fusion, PCR was performed with the primer pair of aphB-Flag-F and aphB-Flag-R1 (Table S1) to amplify the DNA sequence upstream of the termination codon of aphB (TAA), and the Flag DNA fragment (20) was inserted in front of TAA. Then, the PCR product was inserted into the plasmid pBAD33-mob (Table S1) (20) under the control of the arabinose-induced pBAD promoter (for pBAD33-aphB-flag) or the native aphB promoter (for pBAD33-aphB-flag2). The resultant plasmids were introduced into XT004 (ΔaphB) by conjugation.
The construction of strain XT050 with the asp promoter (Pasp) carrying scrambled AphB-binding sites I and II (BBSI/II), PaspMBBSI/II, in the chromosome also followed the double-crossover screening strategy with the suicide plasmid pDM4. PaspMBBSI/II was amplified with primer pairs of PaspMBBSI-P1/PaspMBBSI-P2 and PaspMBBSII-P3/PaspMBBSII-P4 (Table S1) by overlap PCR. The DNA fragment was cloned into pDM4 by isothermal assembly and then introduced into wt V. alginolyticus for subsequent homologous recombination in the asp locus.
Construction of promoter-luxAB plasmids and bioluminescence assays.
The plasmid pDM8 (Table 1) was used to construct promoter-luxAB fusions. PaphB-luxAB (20) was amplified with the primers pDM8-PaphB-luxAB-P1, pDM8-PaphB-luxAB-P2, pDM8-PaphB-luxAB-P3, and luxAB-pDM8-P4 (Table S1) via overlap PCR and then was cloned into the SmaI site in pDM8 by isothermal assembly (35). The Pasp-luxAB and PluxR-luxAB fragments were generated by overlap PCR and then cloned into pDM8 with similar strategies. All of these luxAB reporter plasmids were introduced into the wt and XT004 (ΔaphB) strains (Table 1).
For bioluminescence assays, the reporter strains were cultured overnight, diluted to 6 × 106 CFU/ml in fresh LBS medium, and then incubated at 30°C, with shaking at 200 rpm. A 200-μl aliquot was sampled every 1 h for 12 h and, after the addition of 1% decanal, the bioluminescence was measured in an Orion II bioluminescence reader (Berthold Detection Systems, Pforzheim, Germany), as described previously (36). Meanwhile, the bacterial growth was also monitored by measuring the optical density at 600 nm (OD600) (Biotek, Winooski, VT).
Quantitative real-time PCR.
The overnight cultures were inoculated into fresh LBS medium and incubated for an additional 4 or 9 h. One microgram of RNA was used to generate cDNA (Toyobo, Tsuruga, Japan) using random primers (TaKaRa, Dalian, China). The cDNA was then amplified with FastStart Universal SYBR green master (Tiangen, Beijing, China) with specific primer pairs (Table S1) in a 7500 real-time system (Applied Biosystems, Foster City, CA). We performed three independent experiments (each in triplicate) and normalized the transcript levels to those of gyrB in each of the samples by the ΔΔCT method.
Purification of AphB.
The aphB open reading frame (ORF) sequence was amplified by PCR using the primers aphBpET28a-F and aphBpET28a-R (Table S1) and then was subcloned into pET28a to produce AphB with a His6 tag at the carboxyl terminus (His6-AphB). The resulting plasmid was then transformed into E. coli BL21(DE3). This expression strain was grown to an OD600 value of 0.8 to 1.0 and treated with 1 mM IPTG at 25°C for 14 to 15 h to induce AphB expression. Finally, the protein was purified using nickel affinity chromatography.
Electrophoretic mobility shift assays.
For the EMSA studies, different Cy5-labeled promoter regions of luxR, asp, aphB, metH, cadB, cyaA, murB, EPGS_01530, and EPGS_01529 were amplified using specific primers (Table S1). The fragments were incubated for 30 min at 25°C in reaction mixtures with various concentrations of purified His6-AphB in 20 μl of the binding buffer (250 mM NaCl, 1 mM dithiothreitol, 20 mM HEPES [pH 9.0]). The samples were then loaded onto a 6% polyacrylamide gel, and electrophoresis was performed at 100 V for 130 min, on ice. The gels were then scanned with an FLA 9500 scanner (GE Healthcare, Uppsala, Sweden).
ChIP-seq analysis.
ChIP-seq was performed following the protocol that we established previously (19, 20). The pBAD33-aphB-flag plasmid, encoding AphB-Flag, was transferred to XT004 (ΔaphB) cells for ChIP assays. In addition, the pBAD33-flag plasmid, encoding the Flag tag alone, was transferred to XT004 (ΔaphB) as a negative control. The two resulting strains (XT020 and XT022) were cultured overnight in LBS medium and then diluted (1:1,000) into 50 ml of fresh medium with the addition of 0.04% l-arabinose. After 4 h of growth at 30°C with shaking, 1% formaldehyde was added to the cultures for 10 min at 30°C, with shaking, to form cross-links among protein and DNA fragments. The cross-linking process was stopped by the addition of 1.25 M glycine. Following washing with phosphate-buffered saline (PBS), the bacteria were harvested and then resuspended in 25 ml of SDS lysis buffer (20). The bacteria were broken and the DNA was sonicated into 100- to 500-bp fragments, followed by immunoprecipitation experiments with anti-Flag beads (Sigma-Aldrich). After 12 h of incubation, the beads were washed twice with a low-salt wash buffer (20), twice with a high-salt wash buffer (20), twice with a LiCl wash buffer (20), and twice with Tris-HCl–EDTA (TE) buffer. The beads and input samples were resuspended in 200 μl of elution buffer (20). After centrifugation, 8 μl of 5 M NaCl was added to all samples, which were then incubated at 65°C for 12 h to resolve the DNA-protein cross-links. One microliter of RNase was added to each tube following incubation at 37°C for 4 h. After that, the mixture was added to each tube with 8 μl of 1 M Tris-HCl, 4 μl of 0.5 M EDTA, and 1 μl of proteinase K and incubated at 45°C for 2 h.
The DNA was purified by phenol-chloroform extraction and was used for library construction with the VAHTS Turbo DNA library prep kit (Vazyme, Nanjing, China). We quantitated each library and sequenced the libraries using a MiSeq sequencer (Illumina, San Diego, CA). The ChIP-seq reads were mapped to the V. alginolyticus EPGS genome. The enriched peaks were then identified using MACS software (37). Finally, the AphB-binding motif was generated by MEME (21). We also used Kobas 2.0 to analyze the relevant KEGG pathway and to illustrate the functions of the enriched genes (38).
DNase I footprinting analysis.
The dye/primer-based DNase I footprinting assay was performed as described previously (19). Briefly, the promoter region of aphB was PCR amplified with Pfu DNA polymerase using PaphB-F and PaphB-R primers (carrying 6-carboxyfluorescein [6-FAM] at the 5′ end) (Table S1). The FAM-labeled probes were purified and quantified with a NanoDrop 2000C spectrophotometer (Thermo Fisher Inc.). For each assay, 400 ng of probes was incubated with different amounts of AphB in a total volume of 40 μl. After incubation for 30 min at 25°C, 10 μl of solution containing approximately 0.015 units of DNase I (Promega, Madison, WI) and 100 nmol of freshly prepared CaCl2 was added, and the solution was further incubated for 1 min at 25°C. The reaction was stopped by adding 140 μl of DNase I stop solution (containing 200 mM unbuffered sodium acetate, 30 mM EDTA, and 0.15% SDS). The samples were extracted with phenol-chloroform and precipitated with ethanol, and the pellets were dissolved in 10 μl of Milli-Q water. Approximately 2 μl of digested DNA was added to 7.9 μl of HiDi formamide (Applied Biosystems) and 0.1 μl of GeneScan 500 LIZ size standards (Applied Biosystems). The samples were analyzed with a 3730 DNA analyzer, using the G5 dye set and running an altered default genotyping module that increased the injection time to 30 s and the injection voltage to 3 kV. The results were analyzed with GeneMapper 4.0 (Applied Biosystems).
Extracellular Asp activity assay.
We used HPA digestion to detect the Asp activity in ECPs, as described previously (34). The supernatants from the 9-h culture were filtered with 0.22-μm filters (Millipore) and mixed with 1 ml of PBS (pH 7.0) and 10 mg of HPA (Sigma-Aldrich, Shanghai, China). After the mixture had been incubated for 2 h at 37°C with shaking at 200 rpm, 1 ml of 10% trichloroacetic acid (TCA) was added to stop the reaction. The mixture was then centrifuged at 8,000 × g for 3 min, and the absorbance of the supernatants was measured at 600 nm.
Western blot assay.
The bacterial cell pellets or concentrated ECP were suspended in 1 volume of PBS to normalize the culture densities based on OD600 values. Then, 15 μl of each sample was resolved in a 12% denaturing polyacrylamide gel before being transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore, Bedford, MA). The membrane was then blocked with 10% skim milk powder solution for 2 h and incubated overnight at 4°C with a 1:2,500 dilution of polyclonal rabbit anti-LuxR or anti-Asp antiserum (GL Peptide Ltd., Shanghai, China). After three washes with PBS with 500 μl of Tween 20 (PBST), the blots were incubated for 1.5 h with a 1:2,000 dilution of horseradish peroxidase-conjugated goat anti-rabbit IgG antiserum (Santa Cruz Biotechnology, Santa Cruz, CA). Finally, the immunoreactive protein bands were visualized with ECL reagent (Thermo Fisher Scientific Inc., Waltham, MA).
Acid challenges.
The acid tolerance was assessed in SCBN and filter sterilized (13). The cultures of V. alginolyticus were grown at 30°C, with shaking, to OD600 values of 0.8 to 1.0 and inoculated into SCBN to reach a final concentration of ∼105 CFU/ml. The cultures were incubated for 60 min at 30°C with shaking at 100 rpm. Samples were withdrawn immediately after the inoculation or at an indicated time and were plate counted in triplicate on LBS agar.
Biofilm formation assay.
The wt, XT004 (ΔaphB), XT021 (aphB+), and other strains cultured overnight were diluted to OD600 values of 1.0. Fifty microliters of the cultures were inoculated into 5 ml of LBS medium in glass tubes and grown for 48 h at 30°C without shaking. After being stained with 2% crystal violet for 20 min, the biofilms were dissolved in 33% (vol/vol) acetic acid, and the solution was measured at 570 nm.
Infection of fish.
The wt, XT004 (ΔaphB), and XT021 (aphB+) strains were grown in fresh LBS medium for 9 h. Zebra fish were injected i.m. with PBS or the indicated strain at a dose of 5 × 106 CFU/fish, as described previously (39). Thirty fish were injected with PBS or one of the strains, and three parallel experiments were conducted. All animal experiments presented in this study were approved by the Animal Care Committee of the East China University of Science and Technology (protocol 2006272). The Experimental Animal Care and Use Guidelines from the Ministry of Science and Technology of China (publication MOST-2011-02) were strictly adhered to.
Accession number(s).
The ChIP-seq data sets have been deposited in GenBank under accession number SRP111492.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China (grants 41376128, 31372560, and 30301059), the Ministry of Agriculture of China (grants CARS-50 and nyhyzx-201303047), and the Shanghai Pujiang Program (grant 16PJD018).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00252-17.
REFERENCES
- 1.Kramer R, Jung K. 2010. Bacterial signaling. Wiley-VCH, Weinheim, Germany. [Google Scholar]
- 2.Matson JS, Withey JH, DiRita VJ. 2007. Regulatory networks controlling Vibrio cholerae virulence gene expression. Infect Immun 75:5542–5549. doi: 10.1128/IAI.01094-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abuaita BH, Withey JH. 2009. Bicarbonate induces Vibrio cholerae virulence gene expression by enhancing ToxT activity. Infect Immun 77:4111–4120. doi: 10.1128/IAI.00409-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Z, Yang M, Peterfreund GL, Tsou AM, Selamoglu N, Daldal F, Zhong Z, Kan B, Zhu J. 2011. Vibrio cholerae anaerobic induction of virulence gene expression is controlled by thiol-based switches of virulence regulator AphB. Proc Natl Acad Sci U S A 108:810–815. doi: 10.1073/pnas.1014640108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang M, Liu Z, Hughes C, Stern AM, Wang H, Zhong Z, Kan B, Fenical W, Zhu J. 2013. Bile salt-induced intermolecular disulfide bond formation activates Vibrio cholerae virulence. Proc Natl Acad Sci U S A 110:2348–2353. doi: 10.1073/pnas.1218039110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lowden MJ, Skorupski K, Pellegrini M, Chiorazzo MG, Taylor RK, Kull FJ. 2010. Structure of Vibrio cholerae ToxT reveals a mechanism for fatty acid regulation of virulence genes. Proc Natl Acad Sci U S A 107:2860–2865. doi: 10.1073/pnas.0915021107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weber GG, Kortmann J, Narberhaus F, Klose KE. 2014. RNA thermometer controls temperature-dependent virulence factor expression in Vibrio cholerae. Proc Natl Acad Sci U S A 111:14241–14246. doi: 10.1073/pnas.1411570111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kovacikova G, Skorupski K. 2002. Binding site requirements of the virulence gene regulator AphB: differential affinities for the Vibrio cholerae classical and El Tor tcpPH promoters. Mol Microbiol 44:533–547. doi: 10.1046/j.1365-2958.2002.02914.x. [DOI] [PubMed] [Google Scholar]
- 9.Taylor JL, de Silva RS, Kovacikova G, Lin W, Taylor RK, Skorupski K, Kull FJ. 2012. The crystal structure of AphB, a virulence gene activator from Vibrio cholerae, reveals residues that influence its response to oxygen and pH. Mol Microbiol 83:457–470. doi: 10.1111/j.1365-2958.2011.07919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Z, Wang H, Zhou Z, Sheng Y, Naseer N, Kan B, Zhu J. 2016. Thiol-based switch mechanism of virulence regulator AphB modulates oxidative stress response in Vibrio cholerae. Mol Microbiol 102:939–949. doi: 10.1111/mmi.13524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Z, Wang H, Zhou Z, Naseer N, Xiang F, Kan B, Goulian M, Zhu J. 2016. Differential thiol-based switches jump-start Vibrio cholerae pathogenesis. Cell Rep 14:347–354. doi: 10.1016/j.celrep.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kovacikova G, Lin W, Skorupski K. 2010. The LysR-type virulence activator AphB regulates the expression of genes in Vibrio cholerae in response to low pH and anaerobiosis. J Bacteriol 192:4181–4191. doi: 10.1128/JB.00193-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhee JE, Jeong HG, Lee JH, Choi SH. 2006. AphB influences acid tolerance of Vibrio vulnificus by activating expression of the positive regulator CadC. J Bacteriol 188:6490–6497. doi: 10.1128/JB.00533-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeong HG, Choi SH. 2008. Evidence that AphB, essential for the virulence of Vibrio vulnificus, is a global regulator. J Bacteriol 190:3768–3773. doi: 10.1128/JB.00058-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruwandeepika HAD, Jayaweera TSP, Bhowmick PP, Karunasagar I, Bossier P, Defoirdt T. 2012. Pathogenesis, virulence factors and virulence regulation of vibrios belonging to the Harveyi clade. Rev Aquac 4:59–74. doi: 10.1111/j.1753-5131.2012.01061.x. [DOI] [Google Scholar]
- 16.Rui HP, Liu Q, Wang QY, Ma Y, Liu H, Shi CB, Zhang YX. 2009. Role of alkaline serine protease, Asp, in Vibrio alginolyticus virulence and regulation of its expression by LuxO-LuxR regulatory system. J Microbiol Biotechnol 19:431–438. [DOI] [PubMed] [Google Scholar]
- 17.Wang QY, Liu Q, Ma Y, Rui HP, Zhang YX. 2007. LuxO controls extracellular protease, haemolytic activities and siderophore production in fish pathogen Vibrio alginolyticus. J Appl Microbiol 103:1525–1534. doi: 10.1111/j.1365-2672.2007.03380.x. [DOI] [PubMed] [Google Scholar]
- 18.Rui HP, Liu Q, Ma Y, Wang QY, Zhang YX. 2008. Roles of LuxR in regulating extracellular alkaline serine protease A, extracellular polysaccharide and mobility of Vibrio alginolyticus. FEMS Microbiol Lett 285:155–162. doi: 10.1111/j.1574-6968.2008.01185.x. [DOI] [PubMed] [Google Scholar]
- 19.Gu D, Guo M, Yang MJ, Zhang YX, Zhou XH, Wang QY. 2016. A σE-mediated temperature gauge controls a switch from LuxR-mediated virulence gene expression to thermal stress adaptation in Vibrio alginolyticus. PLoS Pathog 12:e1005645. doi: 10.1371/journal.ppat.1005645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu D, Liu H, Yang Z, Zhang YX, Wang QY. 2016. Chromatin immunoprecipitation sequencing technology reveals global regulatory roles of low-cell-density quorum-sensing regulator AphA in the pathogen Vibrio alginolyticus. J Bacteriol 198:2985–2999. doi: 10.1128/JB.00520-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Machanick P, Bailey TL. 2011. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Kessel JC, Ulrich LE, Zhulin IB, Bassler BL. 2013. Analysis of activator and repressor functions reveals the requirements for transcriptional control by LuxR, the master regulator of quorum sensing in Vibrio harveyi. mBio 4:e00378-13. doi: 10.1128/mBio.00378-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rutherford ST, van Kessel JC, Shao Y, Bassler BL. 2011. AphA and LuxR/HapR reciprocally control quorum sensing in vibrios. Genes Dev 25:397–408. doi: 10.1101/gad.2015011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang QY, Liu Q, Ma Y, Zhou L, Zhang YX. 2007. Isolation, sequencing and characterization of cluster genes involved in the biosynthesis and utilization of the siderophore of marine fish pathogen Vibrio alginolyticus. Arch Microbiol 188:433–439. doi: 10.1007/s00203-007-0261-6. [DOI] [PubMed] [Google Scholar]
- 25.Liu H, Wang QY, Liu Q, Cao XD, Shi C, Zhang YX. 2011. Roles of Hfq in the stress adaptation and virulence in fish pathogen Vibrio alginolyticus and its potential application as a target for live attenuated vaccine. Appl Microbiol Biotechnol 91:353–364. doi: 10.1007/s00253-011-3286-3. [DOI] [PubMed] [Google Scholar]
- 26.Kovacikova G, Skorupski K. 2001. Overlapping binding sites for the virulence gene regulators AphA, AphB and cAMP-CRP at the Vibrio cholerae tcpPH promoter. Mol Microbiol 41:393–407. doi: 10.1046/j.1365-2958.2001.02518.x. [DOI] [PubMed] [Google Scholar]
- 27.Xu X, Stern AM, Liu Z, Kan B, Zhu J. 2010. Virulence regulator AphB enhances toxR transcription in Vibrio cholerae. BMC Microbiol 10:3. doi: 10.1186/1471-2180-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Byerly KA, Urbanowski ML, Stauffer GV. 1991. The metR binding site in the Salmonella typhimurium metH gene: DNA sequence constraints on activation. J Bacteriol 173:3547–3553. doi: 10.1128/jb.173.11.3547-3553.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee MJ, Kim HJ, Lee JY, Kwon AS, Jun SY, Kang SH, Kim P. 2013. Effect of gene amplifications in porphyrin pathway on heme biosynthesis in a recombinant Escherichia coli. J Microbiol Biotechnol 23:668–673. doi: 10.4014/jmb.1302.02022. [DOI] [PubMed] [Google Scholar]
- 30.Botsford JL, Harman JG. 1992. Cyclic AMP in prokaryotes. Microbiol Rev 56:100–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Görke B, Stülke J. 2008. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol 6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- 32.McDonough KA, Rodriguez A. 2011. The myriad roles of cyclic AMP in microbial pathogens: from signal to sword. Nat Rev Microbiol 10:27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jackson DW, Simecka JW, Romeo T. 2002. Catabolite repression of Escherichia coli biofilm formation. J Bacteriol 184:3406–3410. doi: 10.1128/JB.184.12.3406-3410.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheng LL, Lv YZ, Liu Q, Wang QY, Zhang YX. 2013. Connecting type VI secretion, quorum sensing, and c-di-GMP production in fish pathogen Vibrio alginolyticus through phosphatase PppA. Vet Microbiol 162:652–662. doi: 10.1016/j.vetmic.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 35.Gibson DG, Young L, Chuang R, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 36.Yin KY, Wang QY, Xiao JF, Zhang YX. 2017. Comparative proteomic analysis unravels a role for EsrB in the regulation of reactive oxygen species stress responses in Edwardsiella piscicida. FEMS Microbiol Lett 364:fnw269. doi: 10.1093/femsle/fnw269. [DOI] [PubMed] [Google Scholar]
- 37.Feng J, Liu T, Qin B, Zhang Y, Liu XS. 2012. Identifying ChIP-seq enrichment using MACS. Nat Protoc 7:1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L. 2011. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 39:W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lv YZ, Xiao JF, Liu Q, Wu HZ, Zhang YX, Wang QY. 2012. Systematic mutation analysis of two-component signal transduction systems reveals EsrA-EsrB and PhoP-PhoQ as the major virulence regulators in Edwardsiella tarda. Vet Microbiol 157:190–199. doi: 10.1016/j.vetmic.2011.12.018. [DOI] [PubMed] [Google Scholar]
- 40.Wang QY, Millet YA, Chao MC, Sasabe J, Davis BM, Waldor MK. 2015. A genome-wide screen reveals that the Vibrio cholerae phosphoenolpyruvate phosphotransferase system modulates virulence gene expression. Infect Immun 83:3381–3395. doi: 10.1128/IAI.00411-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang WL, Wang SX, Yu FG, Zhang LJ, Qi GM, Liu YQ, Gao SY, Kan B. 2003. Construction and evaluation of a safe, live, oral Vibrio cholerae vaccine candidate, IEM108. Infect Immun 71:5498–5504. doi: 10.1128/IAI.71.10.5498-5504.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang SY, Lauritz J, Jass J, Milton DL. 2002. A ToxR homolog from Vibrio anguillarum serotype O1 regulates its own production, bile resistance, and biofilm formation. J Bacteriol 184:1630–1639. doi: 10.1128/JB.184.6.1630-1639.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Croxatto A, Chalker VJ, Lauritz J, Jass J, Hardman A, Williams P, Cámara M, Milton DL. 2002. VanT, a homologue of Vibrio harveyi LuxR, regulates serine, metalloprotease, pigment, and biofilm production in Vibrio anguillarum. J Bacteriol 184:1617–1629. doi: 10.1128/JB.184.6.1617-1629.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.