Abstract
Obesity and the metabolic disorders that constitute metabolic syndrome are a primary cause of morbidity and mortality in the world. Nonetheless, the changes in the proteins and the underlying molecular pathways involved in the relevant pathogenesis are poorly understood.
In this study a proteomic analysis of the visceral adipose tissue isolated from metabolically healthy and unhealthy obese patients was used to identify presence of altered pathway(s) leading to metabolic dysfunction. Samples were obtained from 18 obese patients undergoing bariatric surgery and were subdivided into two groups based on the presence or absence of comorbidities as defined by the International Diabetes Federation. Two dimensional difference in-gel electrophoresis coupled with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry was carried out. A total of 28 proteins were identified with a statistically significant difference in abundance and a 1.5-fold change (ANOVA, p ≤ 0.05) between the groups. 11 proteins showed increased abundance while 17 proteins were decreased in the metabolically unhealthy obese compared to the healthy obese. The differentially expressed proteins belonged broadly to three functional categories: (i) protein and lipid metabolism (ii) cytoskeleton and (iii) regulation of other metabolic processes. Network analysis by Ingenuity pathway analysis identified the NFκB, IRK/MAPK and PKC as the nodes with the highest connections within the connectivity map. The top network pathway identified in our protein data set related to cellular movement, hematological system development and function, and immune cell trafficking. The VAT proteome between the two groups differed substantially between the groups which could potentially be the reason for metabolic dysfunction.
Keywords: healthy obese, metabolic syndrome, obesity, proteomics, visceral adipose tissue, unhealthy obese
INTRODUCTION
The rising prevalence of obesity worldwide due to not only an increasingly sedentary lifestyle and consumption of sugary beverages but also the rapidly changing socioeconomic status is known to increasingly predispose individuals to comorbid conditions associated with obesity (Ng et al., 2014). Obesity contributes to a large proportion of this risk, because it plays a central role in the development of insulin resistance, impaired glucose tolerance, dyslipidemia, vascular diseases such as hypertension, and even cancers. These factors, when present simultaneously, are defined as metabolic syndrome (MetS) and are known to increase the risk of cardiovascular disease (Reaven, 2011). Recent evidence has shown that obesity does not always lead to the development of these adverse metabolic and cardiovascular conditions and a sub-group of the obese individuals exist that are at a lower risk of these complications (Alfadda, 2014; van Vliet-Ostaptchouk et al., 2014). Sub-phenotypes of obesity have been identified based on the presence or absence of these metabolic disorders and obese patients are categorized as either metabolically healthy obese (MHO) or metabolically unhealthy obese (MUHO) (Bluher, 2014; Fabbrini et al., 2015; Gurnell et al., 2003; Kramer et al., 2013) according to the different definitions of the MetS.
Although there are discrepancies among studies on the MetS definition used, broadly, the MHO group is classified as obese individuals with a body mass index (BMI) of > 30 and the absence of any comorbidities, whereas the MUHO subgroup is characterized by a BMI of > 30 along with 3 or more components of MetS, of which, elevated triglyceride levels and increased waist circumference are factors best known to increase the risk of cardiovascular disease in MUHO (Pataky et al., 2010). The heterogeneity noted in these metabolic disturbances in both the groups, irrespective of their BMI, has been related to relative differences in the distribution of fat in the different depots rather than its total volume.
Adipose tissue which plays a central role in body weight homeostasis, inflammation, and development of insulin resistance is also known to be markedly different between the two groups. Adipocytes in MHO group are smaller, more hyperplastic with preserved insulin sensitivity while they are hypertrophic in the MUHO with dysmetabolic phenotype and increased synthesis and release of a wide range of inflammatory adipokines. Visceral adipose tissue (VAT) MUHO is known to have a higher rate of dysfunction, characterized by an imbalance in secretion of pro- and anti-inflammatory adipokines and cytokines, dysregulation of lipid storage and release, decreased insulin-stimulated glucose uptake and mitochondrial dysfunction, presence of different immune cell population and is known to be the causative factor of both obesity and MetS (Alfadda., 2014; Badoud et al., 2015).
The exact molecular mechanisms of adipose tissue dysfunction within the obese population and the manner or time of transition from healthy to unhealthy phenotype remain unclear. It is still not known whether this transition is age dependent or triggered by environmental factors (Pataky et al., 2010). Research into the MHO and MUHO phenotypes may give some clues to the differences between these phenotypes and point to the underlying pathophysiology. The BioSHaRE-EU consortium recently shared the same thoughts reflecting the need to study the metabolic dysregulation in obesity (van Vliet-Ostaptchouk et al., 2014).
Proteomic technology has served as a powerful tool for identifying key components of the adipose tissue proteome (Peinado et al., 2012). Proteomic (Murri et al., 2013) and metabolomic (Bohm et al., 2014) studies on adipose tissue and adipocytes have identified markers that may predict insulin resistance, body weight and glucose intolerance in the obese. A recent study by Gomez-Serrano et al used the iTRAQ labelling and LC-MS technique to compare between VAT proteome from obese individuals with established diabetes on treatment and those without diabetes along with its influence on age and gender (Gomez-Serrano et al., 2016). Previously, Doumatey et al. (2016) shot gun label free proteomics approach and compared the serum proteome of the MHO with the MUHO in African American cohort. Our aim in the present study was to compare the VAT proteome in two distinct obesity phenotypes, the MHO and the MUHO, by using a complimentary proteomics 2D-DIGE mass spectrometry approach to elucidate the changes in the tissue proteome that may increase the risk of metabolic alterations in obese individuals. Identification of the differences in the proteome between these distinct obesity phenotypes may provide insights into the understanding of the development of metabolic dysregulation in obesity, along with the identification of VAT related functional proteins that may serve as biomarkers for obesity related metabolic derangements.
MATERIALS AND METHODS
Ethical approval and consent to participate
Prior to implementation, all procedures and protocols were reviewed and approved by the Institutional Review Board, College of Medicine; King Saud University No 08/1960/E. Written informed consent to use the samples for research was obtained from all the participants. This study was conducted at the Obesity Research Center, College of Medicine, King Saud University, Riyadh, Saudi Arabia.
Study design and patient selection
A group of 18 obese patients (9 women and 9 men) aged between 24 and 40 years were selected from a group of Saudi patients undergoing bariatric surgery at the King Khalid University Hospital in Riyadh, Kingdom of Saudi Arabia. Prior to the operation, all patients provided medical history and underwent a physical examination. Weight (in kilograms) was measured in light clothing and without shoes to the nearest 0.1 kg. Height was measured using a stadiometer, and BMI was calculated. The subjects’ BMIs ranged from 32.9 to 71.9 kg/m2. Body fat analyzer (Gima Spa, Italy) was used to calculate the fat percentage and lean mass. The patients were subdivided into two groups based on the presence or absence of comorbidities as defined by the International Diabetes Federation (IDF). The Subjects were classified as metabolically healthy if they had not met any of the IDF components for MetS. The components included in the criteria are fasting triglycerides level ≥ 1.7 mmol/L; high density lipoprotein (HDL) cholesterol level < 1.04 mmol/L in men or < 1.3 mmol/L in women or administration of lipid-lowering medication; systolic/diastolic blood pressure ≥ 130/85 mmHg or administration of antihypertensive medication; and fasting glucose level ≥ 5.56 mmol/L or administration of antidiabetic medication. Accordingly, 9 patients were considered to be MHO and 9 were considered to be MUHO and had at least 3 of the components of MetS. The exclusion criteria included the presence of acute inflammation, infection, or malignancy.
Blood collection and biochemical assays
Blood samples were taken under fasting conditions at the time of the operation. Plasma was separated immediately by centrifugation, and aliquots were frozen at −80°C for subsequent analysis. Biochemical investigations were carried out on all the patients’ samples prior to the operation. The levels of serum glucose, triglycerides, total cholesterol, and HDL cholesterol were determined using a Dimension Xpand Plus integrated clinical chemistry autoanalyzer (Siemens Healthcare Diagnostics, USA). The serum levels of low-density lipoprotein (LDL) cholesterol were calculated using Friedewald’s equation. The plasma insulin concentration was determined by electro-chemiluminescence on a Cobas e411 immunoanalyzer (Roche Diagnostics, USA). Insulin resistance was determined using “homeostasis model assessment of insulin resistance” (HOMA-IR), which was calculated according to the following equation: HOMA-IR = fasting plasma glucose (mmol/L) × fasting plasma insulin (mIU/L) ÷ 22.5. The percentage of glycosylated hemoglobin (HbA1c) was measured using a turbidimetric inhibition immunoassay on a Dimension Xpand Plus autoanalyzer (Siemens Healthcare Diagnostics).
Tissue collection and protein extraction
VAT (approximately 100 mg) was excised from the greater omentum of the abdominal adipose tissue by our collaborating surgeon. All tissues were immediately snap-frozen in liquid nitrogen and stored at −80°C until analysis. At the time of analysis, tissue samples were thawed and washed 3 times with PBS to remove any traces of blood and other contaminants before homogenization. Proteins were extracted from VAT by using a T25 digital ULTRA TURRAX homogenizer (IKA, Germany) directly in lysis buffer (0.5 ml, pH 8.8, 30 mM Tris-HCl, 7 M urea, 2 M thiourea, 2% CHAPS, and a 1× protease inhibitor mix) on ice. The suspension was shaken for 1 h at room temperature and then sonicated (Microsonicator, Qsonica Sonicators, USA; 30% pulse, two intervals of 1 min each, separated by a 1 min gap). 50mM dithiothreitol (DTT) was then added and the protein extracts were centrifuged (20,000 × g, 1 h, 4°C) according to the procedure developed by our group (Alfadda et al., 2013; Benabdelkamel et al., 2015). The pellet was removed and the solubilized protein in the supernatant was precipitated using the 2D Clean-up Kit according to the manufacturer’s protocol (GE Healthcare, USA).
Protein labeling with cyanine dyes
The protein pellets were solubilized in labeling buffer (7 M urea, 2 M thiourea, 30 mM Tris-HCl, 4% CHAPS, pH 8.5). Insoluble material was pelleted by centrifugation (12,000 × g, room temperature, 5 min), protein concentrations were determined in triplicate using the 2D-Quantkit (GE Healthcare, USA), and pH of the samples was adjusted to 8.5 using NaOH (100 mM). Proteins were labeled (400 pmol of CyDye™ DIGE Fluor dyes, GE Healthcare, UK) in 1 μl of DMF and then mixed with a sample containing 50 μg of protein. The samples were incubated on ice for 30 min in the dark. The labeling reaction was terminated by adding 1 μl of 10 mM lysine. Each sample was covalently labeled with a fluorophore, either Cy3 or Cy5. A pool of equal amounts of protein from each sample in the experiment was labeled with Cy2 and served as the internal standard. (See Supplementary Table S1).
Two-dimensional (2D) electrophoresis, image scanning and preparative gel analysis
First dimension analytical gel electrophoresis was performed as follows. Nine Immobiline Dry Strips (24 cm, pH 3–11; GE Healthcare, Sweden) were passively re-hydrated (30 V, 12 h). This was followed by isoelectric focusing using an Ettan IPGphor IEF unit (GE Healthcare, Sweden). Focusing was performed at 20°C, at 50 μA per strip. A stepwise protocol was used for the first-dimension IEF. After the first dimension, the strips were equilibrated and separated on 12.5% (SDS-PAGE) gels using an Ettan Dalt Six device (GE Healthcare, Sweden). The gels were scanned with a Typhoon 9400 scanner (GE Healthcare) using appropriate wavelengths and filters for Cy2, Cy3 and Cy5 dyes. Preparative gels were prepared using total protein (1 mg) obtained from a pool of equal protein amounts of the 18 VAT samples. This was denatured in lysis buffer and mixed with a rehydration buffer. Gels were stained with Colloidal Coomassie Blue as described previously (Alfadda et al., 2013; Benabdelkamel et al., 2015).
Protein Identification by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (MS)
Coomassie stained gel spots were excised manually, then washed and digested according to the previously described protocol (Benabdelkamel et al., 2015). In brief, destaining was carried out in steps, first by adding 50 mM NH4HCO3 and 50% CH3CN, then by adding 100% CH3CN. The gel pieces were then dried by vacuum centrifugation (40°C, 5 min), rehydrated, and digested (60 minutes, 10 μl of ice-cold trypsin solution, 20 ng sequencing grade modified porcine trypsin (Promega, USA) in 25 mM NH4HCO3 pH 8.0 at 4°C). Digestion continued overnight, at 37°C. The digests were transferred to a 0.5 ml tube and the peptides were extracted by the addition of 30% CH3CN containing 0.1% CF3COOH. Next, the samples were concentrated to approximately 10 μl by vacuum centrifugation (Eppendorf, USA) and stored at −80°C until use (Shevchenko et al., 1996). An aliquot of the digestion solution (1 μl) was spotted onto a MALDI target (384 anchorchip MTP 800 μm Anchorchip; Bruker Daltonik, Germany) together with 0.5 μl of a matrix (10 mg α-cyano-4-hydroxycinnamic acid (CHCA) in 1 μl of 30% CH3CN and 0.1% aqueous CF3COOH) and then left to dry (at room temperature) before MS analysis. Protein identification was assigned by peptide mass fingerprinting (PMFs) using an ultrafleXtreme time-of-flight (TOF) mass spectrometer (Bruker Daltonics, Germany) and confirmed by MS/MS analysis of at least three peptides in each sample using LIFT spectra performed by MALDI-TOF-MS/MS. The peptide mass fingerprints were processed using the flexAnalysis™ software (version 2.4, Bruker Daltonics, Germany). The MS data were interpreted by using BioTools v.3.2 (Bruker Daltonics, Germany), together with the Mascot search algorithm (version 2.0.04 updated 10/06/2016; Matrix Science Ltd., UK). Mascot parameters as follows: fixed cysteine modification with propionamide, variable modification due to methionine oxidation, one missed cleavage site (i.e., in case of incomplete trypsin hydrolysis) and a mass tolerance of 100 ppm. Identified proteins were accepted as correct if they showed a Mascot score greater than 56 and p value of ≤ 0.05. Not all spots of interest could be identified because some proteins were of low abundance and did not yield sufficiently intense mass fingerprints; other spots were mixtures of multiple proteins.
DIGE image acquisition and pathway analysis
Differential in-gel electrophoresis (DIGE) images were analyzed using the Progenesis Same Spots v.3.3 software (Nonlinear Dynamics Ltd., UK). The gel images were first aligned together and prominent spots were used to manually assign 60 vectors to the digitized images within each gel. The automatic vector tool was next used to add additional vectors (500 vectors in total), which were manually revised and edited for correction if necessary. These vectors were used to warp and align gel images with a reference image of one internal standard across and within each gel. Gel groups were defined according to the experimental design, and normalized volume of the spots was used to identify statistically significant differences. The software calculated the normalized volume of each spot on each gel from the Cy3 (or Cy5) to Cy2 spot volume ratio. The software performs log transformation of the spot volumes to generate normally distributed data. Log normalized volume was used to quantify differential expression. Independent direct comparisons were made between MUHO and MHO, and fold differences and p - values were calculated using one-way ANOVA. All spots were pre-filtered and manually checked before applying the statistical criteria (ANOVA test, p ≤ 0.05 and fold difference ≥ 1.5). The normalized volume of spots, instead of spot intensities, was used in statistical processing. Only those spots that fulfilled the above mentioned statistical criteria were submitted for MS analysis.
The quantitative data were then imported into IPA software (Ingenuity® Systems, http://www.ingenuity.com). This software helps to determine the functions and pathways most strongly associated with a protein list by overlaying experimental expression data on networks constructed from published interactions.
Immunoblotting
To independently confirm the findings of the 2D-DIGE experiments, proteins with statistically significant differential abundance were selected and examined by immunoblotting. Antibodies against peptidyl prolyl isomerase A (PPIA), urokinase plasminogen activator receptor (UPAR), plasminogen activator inhibitor-1 (PAI-1), glycerol phosphate dehydrogenase (GPD), β-actin (ACTB), and translocase of the outer membrane 20 (TOM20) were used for immunoblotting. An equal amount of protein from each sample (50 μg) was separated by 1-dimensional discontinuous slab gel electrophoresis (12% SDS-polyacrylamide gel). Proteins were electrotransferred to an Immobilon-P, polyvinylidene difluoride transfer membrane (PVDF, Millipore, USA) using a mini trans-blot electrotransfer cell (BioRad, USA). Following transfer, the membrane was stained with Ponceau-S to confirm the transfer efficiency. The membrane was then blocked in 5% fat free milk (FFM) in Tris-buffered saline (TBS) (10 mM Tris HCl, 150 mM NaCl, 1 h, room temperature) and rinsed with 3 changes of TBS-T (TBS and 0.1% Tween 20) buffer. Samples were then incubated with the specified primary antibodies in blocking buffer. Blots were incubated with the appropriate IgG-HRP-conjugated secondary antibody, and the immunoreactive bands detected by enhanced chemiluminescence (ECL, Thermo Fisher Scientific, USA) were visualized by scanning with Fluorchem Q (Cell Biosciences, USA), and digitalized using the image analysis software Alpha View Q 3.0 (Cell Biosciences).
Statistical analysis
The results on the biochemical parameters in the MHO and MUHO groups are presented as means ± SD, and significant differences between the mean values were assessed using Student’s t-test. All statistical analyses were conducted in the Graph Pad Prism software, version 5.0 for Windows (GraphPad Software, USA).
RESULTS
Anthropometric and biochemical data
Both groups included in the study were obese (BMI > 30 kg/m2) and matched for their age and BMI. There were no significant differences noted between the two groups in relation to their body fat percentage, blood pressure, fasting glucose levels, lipids, liver enzymes and serum creatinine levels. The MUHO group was found to have higher insulin and HOMA-IR levels as compared to the MHO group (Table 1).
Table 1.
Clinical and biochemical characteristics of the study participants
| MHO | MUHO | p value | |
|---|---|---|---|
| N | 9 | 9 | |
| Age (years) | 31.3 ± 6.5 | 32 ± 8.8 | 0.86 |
| Weight (kg) | 128.2 ± 37.3 | 131.2 ± 26.5 | 0.85 |
| BMI (kg/m2) | 48.1 ± 13.18 | 48.66 ± 8.8 | 0.92 |
| Fat percentage | 52.2 ± 15.7 | 55.8 ± 11.3 | 0.61 |
| SBP (mm/Hg) | 122.6 ± 12.4 | 129 ± 5.7 | 0.20 |
| DBP (mm/Hg) | 74.3 ± 7.9 | 72.9 ± 4.9 | 0.67 |
| Fasting glucose (mmol/L) | 7.0 ± 2.4 | 9.6 ± 4.1 | 0.17 |
| Insulin (mIU/ml) | 15.0 ± 3.9 | 24.4 ± 10.9 | 0.05 |
| HOMA -IR | 4.6 ± 2.2 | 10.7 ± 7.5 | 0.05 |
| Total cholesterol (mmol/L) | 4.4 ± 1.0 | 4.4 ± 0.6 | 0.87 |
| Triglycerides (mmol/L) | 1.4 ± 0.8 | 2.0 ± 1.6 | 0.34 |
| HDL cholesterol (mmol/L) | 1.0 ± 0.3 | 0.8 ± 0.1 | 0.11 |
| LDL cholesterol (mmol/L) | 2.7 ± 0.8 | 2.7 ± 0.7 | 0.93 |
| Aspartate transaminase (IU/L) | 25.6 ± 13.9 | 25.8 ± 13.8 | 0.97 |
| Alanine transaminase (IU/L) | 41.0 ± 19.8 | 44.1 ± 28.0 | 0.80 |
| Alkaline phosphatase (IU/L) | 77.3 ± 24.0 | 92.9 ± 15.9 | 0.17 |
| Creatinine (mmol/L) | 72.5 ± 13.1 | 76.1 ± 23.3 | 0.71 |
Data are shown as means ± SD.
Identification of differentially abundant protein spots in MHO and MUHO by 2D-DIGE analysis
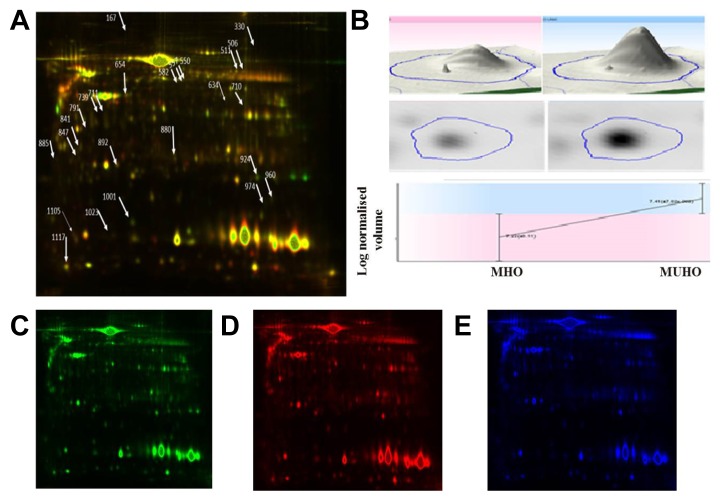
2D-DIGE was used to determine the individual changes in the VAT proteome from 18 samples: i.e., MHO (n = 9), and MUHO (n = 9). Figure 1 shows a representative 2D-DIGE gel from the set of pairwise comparisons that were made between the two groups. The arrows indicate the differentially abundant spots that were either decreased or increased between the groups. The spot patterns were reproducible across the nine gels making the alignment and further analysis possible. Cy2-labeling (internal standard) was used to allow for normalization across the complete set of gels and for quantitative differential analysis of the protein levels. Significant changes in protein abundance levels were based on an ANOVA test (i.e., p ≤ 0.05 and fold-change ≥ 1.5). Progenesis statistical software analysis detected a total of 40 protein spots showing significant increase or decrease in expression between MHO and MUHO.
Fig. 1.
A 2D-DIGE image comparing the VAT proteome of MHO and MUHO patients.
MHO and MUHO samples were labeled with Cy3 and Cy5, respectively, while the protein reference pool (internal standard) was labeled with Cy2. VAT proteins were focused on linear pH 3–11, IPG strips (24 cm), and were then separated by 12.5% polyacrylamide gels. (A) Cy3 and Cy5 channel overlap image. Differentially expressed protein spots identified by MALDI-TOF are marked on the gels by arrows. (B) An example of protein spot (# 960, Peptidylprolyl Isomerase A) analysis showing differential expression of proteins in 2D and 3D. Individual 2D-DIGE gel image of VAT samples labeled with Cy3 (C), Cy5 (D), and Cy2 (E).
All differentially abundant protein spots were selected for excision and identification by MALDI-TOF MS. PMFs of 28 from the 40 spots were successfully matched to entries in the SWISS-PROT database by Mascot with high confidence. In some cases, variants of the same protein were found at several locations on the gel. Table 2 lists the 28 protein spots identified as differentially abundant between the groups. A total of 11 protein spots were up regulated while 17 were down regulated in the MUHO in comparison to the MHO group.
Table 2.
Differentially abundant proteins between MUHO vs. MHO in VAT. MALDI-TOF mass spectrometry was used to identify the protein spots cut from the 2D-DIGE gels
| Spot no.a | Accession no.b | Protein name | MASCOT ID | Pic | MWd | Cov% | Scoree | p Value (ANOVA) | Ratio MUHO vs. MHOf |
|---|---|---|---|---|---|---|---|---|---|
| 710 | Q6S8J3 | POTE ankyrin domain family member E | POTEE | 5.83 | 122882 | 17 | 60 | 0.006 | 1.6 |
| 847 | Q9H254 | Spectrin beta chain, non-erythrocytic 4 | SPTN4 | 5.72 | 290006 | 10 | 61 | 0.011 | 2.22 |
| 841 | P52565 | Rho GDP-dissociation inhibitor 1 | GDIR1 | 5.02 | 23193 | 15 | 72 | 0.018 | 1.6 |
| 1001 | P02766 | Transthyretin | TTHY | 5.52 | 15877 | 24 | 95 | 0.038 | 1.96 |
| 880 | P04792 | Heat shock protein beta-1 | HSP1 | 5.98 | 22826 | 51 | 102 | 0.029 | 1.85 |
| 960 | P62937 | Peptidyl-prolyl cis-trans isomerase A | PPIA | 7.68 | 18001 | 27 | 73 | 0.021 | 2.22 |
| 924 | Q03405 | Urokinase plasminogen activator surface receptor | UPAR | 6.20 | 38605 | 74 | 79 | 0.039 | 1.61 |
| 582 | P05121 | Plasminogen activator inhibitor 1 | PAI1 | 6.68 | 46088 | 28 | 58 | 0.009 | 1.56 |
| 974 | P30043 | Flavin reductase (NADPH) | BLVRB | 7.13 | 22219 | 28 | 57 | 0.039 | 1.61 |
| 1117 | A8K979 | ERI1 exoribonuclease 2 | ERI2 | 9.12 | 78549 | 19 | 57 | 0.008 | 2.32 |
| 974 | A8MWP6 | Uncharacterized protein ENSP00000382042 | YQ019 | 5.66 | 18362 | 34 | 60 | 0.016 | 2.17 |
| 791 | P08758 | Annexin A5 | ANXA5 | 4.94 | 35971 | 49 | 108 | 0.008 | − 2.24 |
| 634 | P63261 | Actin, cytoplasmic 2 | ACTG | 5.31 | 42108 | 45 | 88 | 0.003 | − 1.82 |
| 654 | P63261 | Actin, cytoplasmic 2 | ACTG | 5.31 | 42108 | 34 | 74 | 0.003 | − 1.82 |
| 330 | P60709 | Actin, cytoplasmic 1 | ACTB | 5.89 | 41710 | 10 | 128 | 0.004 | − 1.89 |
| 711 | P60709 | Actin, cytoplasmic 1 | ACTB | 5.29 | 42052 | 43 | 82 | 0.029 | − 1.73 |
| 1105 | P09382 | Galectin-1 | LEG1 | 5.34 | 15048 | 66 | 72 | 0.05 | − 1.6 |
| 721 | P21695 | Glycerol-3-phosphate dehydrogenase [NAD(+)], cytoplasmic | GPDA | 5.81 | 38171 | 38 | 65 | 0.029 | − 1.73 |
| 739 | P21695 | Glycerol-3-phosphate dehydrogenase [NAD(+)], cytoplasmic | GPDA | 581 | 38171 | 42 | 91 | 0.05 | − 1.37 |
| 892 | P02647 | Apolipoprotein | APOA1 | 5.56 | 30759 | 13 | 62 | 0.011 | − 1.91 |
| 167 | P12109 | Collagen alpha-1(VI) chain | CO6A1 | 5.26 | 108462 | 20 | 67 | 0.023 | − 1.57 |
| 550 | Q13228 | Selenium-binding protein 1 | SBP1 | 5.93 | 52928 | 27 | 67 | 0.008 | − 1.74 |
| 506 | P04040 | Catalase | CATA | 6.90 | 59947 | 35 | 65 | 0.036 | − 1.49 |
| 511 | P04040 | Catalase | CATA | 6.90 | 59947 | 36 | 88 | 0.012 | − 1.46 |
| 1023 | Q6UXN7 | TOMM20-like protein 1 | TO20L | 8.87 | 17973 | 60 | 63 | 0.04 | − 1.55 |
| 557 | Q5VTR2 | E3 ubiquitin-protein ligase BRE1A | BRE1A | 5.73 | 114220 | 20 | 66 | 0.0295 | − 3 |
| 885 | Q8TDE3 | Ribonuclease 8 | RNA58 | 8.69 | 17600 | 75 | 60 | 0.006 | − 1.53 |
| 330 | Q9Y651 | Transcription factor SOX-21 | SOX21 | 9.74 | 28676 | 26 | 60 | 0.004 | − 1.89 |
Spot numbers correspond to those included in the 2D-image
Protein accession number for SWISSPROT Database
Theoretical isoelectric point
Theoretical molecular weight
MASCOT score
Protein expression between MUHO vs. MHO( positive values indicate an increase in the protein abundance in MUHO; negative values indicate a decrease in the protein abundance in MUHO)
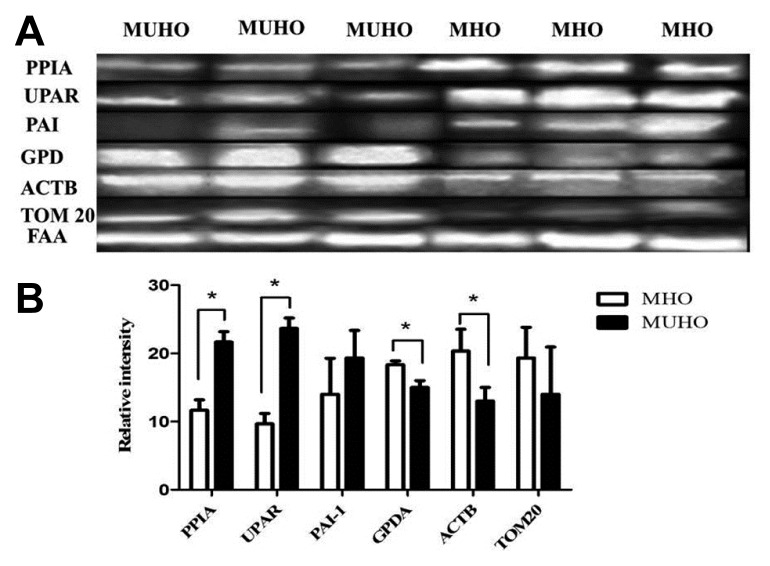
Confirmation of changes in selected proteins by immunoblotting
Several of the differences in protein levels identified between MHO and MUHO in adipose tissue by 2D-DIGE were confirmed by immunoblot analysis of the selected proteins targeted for confirmation which were PPIA, UPAR, PAI-1, GPD, ACTB and TOM20 (Fig. 2). The immunoblots confirmed that these proteins showed fold changes that were similar to those obtained from the 2D-DIGE experiment in the adipose tissue isolated from MHO and MUHO groups. Immunoblot data were normalized to the housekeeping protein fumaryl aceto acetase (FAA).
Fig. 2.
Confirmation of the proteomic data using immunoblot analysis of selected proteins, identified by 2D-DIGE analysis.
(A) The results obtained by immunoblotting were similar to the results obtained by 2D-DIGE. (B) Graphical representation of the relative intensity values of normalized protein bands for MHO and MUHO. The data are presented as histograms of mean ± SD.
The most significantly related functions extracted from this overlapping network included, cellular movement (12 proteins, P = 6.02E-03 − 5.37E-08), free radical scavenging (7 proteins, P = 5.03E-03 − 1.68E-06), carbohydrate metabolism (6 proteins, P = 6.03E-03 − 2.90E-06), lipid metabolism (7 proteins, P = 6.03E-03 − 2.90E-06), and small molecule biochemistry (9 proteins, P = 6.03E-03 − 2.90E-06; Table S2). The number of proteins participating in an identified function was compared with the total number of proteins known to be involved in that particular function in the Ingenuity Knowledge Base, and it was used to calculate the statistical significance. The most interesting canonical pathways included death receptor signaling (P =1.05E-04, 3.3% overlap), coagulation system (P = 5.64E-04, 5.7% overlap), acute phase response signaling (P = 6.26E-04, 1.8% overlap), Rho GDI signaling (P = 6.70E-04, 1.7% overlap), and NRF2-mediated oxidative stress response (P =7.52E-04, 1.7% overlap). Details of the canonical pathways identified in this study are summarized in Supplementary Table S2.
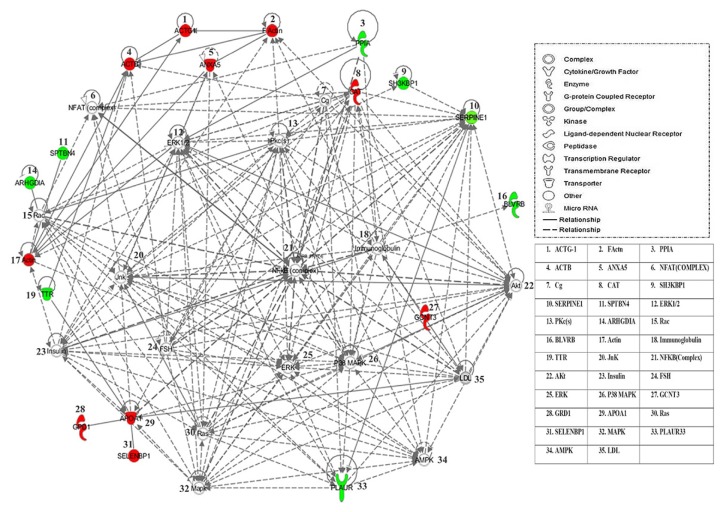
Protein - protein interaction networks
The biological roles of the differentially abundant proteins identified in MUHO (compared to MHO) were studied using the IPA software. To generate a network pathway of significance, the software computes a score based on the best fit of the input data set of proteins and a list of biological functions stored in its knowledge database. The generated network is preferentially enriched for proteins with specific and extensive interactions, the interacting proteins being represented as nodes and their biological relation as lines. In our data, two interaction networks were identified for the proteins exhibiting differential expression in MUHO (Supplementary Table S2). The highest scoring network (Fig. 3), score 40, incorporated 17 out of the 24 focus molecules and related them to pathways involved in cellular movement, hematological system development and function, immune cell trafficking, and cellular growth and proliferation pathways. The second network, score 9, included 5 focus molecules related to pathways associated with protein trafficking, cancer, and organismal injury and abnormalities pathways.
Fig. 3.
Schematic representation of the most significant IPA network from differentially regulated proteins in the MHO compared to those from the MUHO group.
IPA analysis indicated a functional interaction of networks with the highest score of 40 related to the cellular movement, hematological system development and function, and immune cell trafficking, revealing NFκB, ERK1/2, IL-13 and P38 MAPK as central nodes that are deregulated in MUHO. Nodes in green and red correspond to the up and down-regulated proteins in MUHO vs. MHO, respectively. Uncoloured nodes are proposed by IPA and indicate potential targets functionally coordinated with the differentially expressed proteins. Solid lines indicate direct molecular interactions, and dashed lines represent indirect relations.
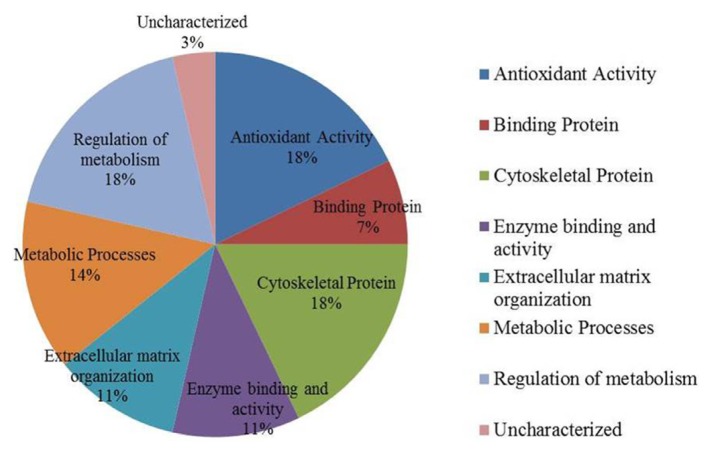
Classification of key proteins based on function
To gain a better understanding of the molecular function and biological implications of the 28 differentially abundant proteins identified in the data set between the MUHO and MHO, functional annotations were carried out with Gene ontology (GO) terms using UniprotKb. The dominant functional categories highlighted were for cytoskeletal proteins (18%), regulators of metabolic processes (18%), protein with antioxidant activity (18%) followed by those proteins that served as part of metabolic processes (14%), extracellular matrix organisation (11%), as enzymes (11%), binding proteins (7%) and others (3%). Figure 4 depicts graphically the percentage of involvement of each of these proteins within the biologically functional categories as a function of their total number for our set of identified proteins.
Fig. 4.
Comparative depiction of the differentially abundant identified proteins categorized into groups according to their function based on the GO terms using UniprotKB.
The representative pie diagram shows the (%) of involvement for different functional categories of the proteins between MHO and MUHO group.
DISCUSSION
Obesity is characterized by a chronic low grade inflammatory state which results in insulin resistance within the insulin sensitive target tissues (liver, muscle and adipose tissue). The strong link between body fat mass and insulin resistance was established in both humans and mice through studies done using the lipodystrophy disease model - a condition which recapitulates several features of MetS. It suggests that an optimal range for adipose tissue mass exists, which, when breached on either side of the spectrum results in insulin resistance (Gurnell, 2003). Visceral adiposity is a major determinant of the metabolic health and links obesity with metabolic disease. A higher percentage of VAT [which has an increased capacity for synthesis and release of adipokines (Ouchi et al., 2011)] compared to SAT was shown to be independently associated with development of insulin resistance and the metabolic deterioration from MHO to MUHO phenotype (Hwang et al., 2015).
The MUHO phenotype in turn is a major cause of morbidity and mortality due to its associated micro- and macro-vascular complications which fuel the atherogenic drive leading to cerebrovascular and ischemic heart disease. Not only the adipocyte as a whole, but also dysfunction of the adipocyte organelles namely the endoplasmic reticulum, mitochondria, and lipid droplet (LD) also affect the differentiation or dedifferentiation programs and result in systemic metabolic alterations and insulin resistance (Rodriguez et al., 2007). Additional risk factors such as elevated plasma levels of fibrinogen and PAI-1, and microalbuminuria are also known to contribute to the MUHO phenotype. Previous proteomic study by Gomez-Serrano et al. (2016) compared the VAT from established T2DM obese and non-diabetic obese patients using iTRAQ technology followed with LC-MS analysis. They studied a cohort of patients who had been treated with medication for 2 years that is in contrast to our study population who had impaired fasting glucose level and were not on medication.
In the present study we carried out a proteomic analysis of whole VAT homogenates in the obese with and without metabolic syndrome to elucidate the differences in protein abundance between the two conditions. These differences may contribute to the understanding of differences in the overall metabolism observed between healthy and unhealthy obesity in individuals matched for BMI and age, based on the presence of Mets components as defined by IDF.
Proteins with increased abundance in MUHO as compared to MHO
In our study, the protein spots relating to several unique proteins namely; POTE ankyrin domain family member E (POTEE), Spectrin beta chain non-erythrocytic 4 (SPTN4), Rho GDP dissociation inhibitor 1 (GDIR1), Transthyretin (TTHY), Heat shock protein β1 (HSP1), Peptidyl-prolyl cis-trans isomerase A (PPIA), Urokinase plasminogen activator surface receptor (UPAR/PLAUR), Plasminogen activator inhibitor 1 (PAI-1), Biliverdin B (BLVRB), ERI1 exoribonuclease 2 (ERI2) and the uncharacterized protein (YQ019) were significantly increased in abundance in the VAT proteome of the MUHO compared to the MHO group. Our findings are similar to Gomez-Serrano et al. (2016) who also identified an increase in SPTN4, GD1R1, TTHY and ANXA5 in the obese diabetic group.
The significant increase in the abundance of POTEE ankyrin, GD1R1 and the non-erythroid spectrin cytoskeletal proteins in the MUHO found in our dataset is similar to Hodges et al., who identified these in the LDs proteome (Hodges et al., 2010). Like adipocytes, LDs once considered as inert fat containing globules are now known to be metabolically active (Olofsson et al., 2009) regulating not just lipid but also other major metabolic pathways. In obesity, the enlarged size and hypertrophy of adipocytes is mainly attributed to the enlargement of the unilocular LD, its increased lipid storage capacity and resistance to hormone-stimulated lipolysis, a dysfunction of which may contribute to development of insulin resistance and MetS. The finding of an increase in spot abundance of these proteins may indicate a heightened LD metabolic activity within the VAT of MUHO. Similarly, TTHY a secretory protein that circulates both independently and bound to retinol binding protein 4 (RBP4), was found to be significantly increased in the MUHO. Increased TTHY has been shown to induce insulin resistance. On the other hand, decreasing the levels of TTHY was shown to improve glucose homeostasis and has been suggested as a potential therapeutic approach for the treatment of T2DM (Zemany et al., 2015).
The significant increase in the abundance of spots related to PAI-1 and UPAR, proteins belonging to the plasminogen activation system, indicate the increased involvement of this pathway in the MUHO group compared to the MHO. Both PAI-1 and UPAR are involved in the regulation of cellular motility, migration and adhesion through their binding to extracellular matrix (ECM) components. Elevated PAI-1 and UPAR levels lead to changes in ECM and to the development of inflammation, insulin resistance, and athero-thrombotic complications (Smith et al., 2010). Juhan-Vague et al have shown that an increase in PAI-1 levels are strongly associated with BMI, visceral fat, blood pressure, plasma levels of insulin or pro-insulin, triglycerides, small dense LDL particles, free fatty acids, and HDL cholesterol (Juhan-Vague et al., 2003).
The MUHO group also demonstrated an increase in the abundance of spots relating to HSP1, a chaperone protein known to mitigate cytokine-induced islet apoptosis and improve insulin signaling in blood monocytes from obese subjects (Oliva et al., 2013). Another interesting protein significantly increased in abundance is BLVRB, a flavin reductase, which converts biliverdin IXβ to bilirubin Ixβ and the latter in turn increases insulin sensitivity (O’Brien et al., 2015) as probable defense mechanisms against the ensuing insulin resistance. The increase in the BLBVRB protein in the MUHO group has not been previously reported.
Proteins with decreased abundance in MUHO compared to MHO group
A significant decrease in abundance of unique proteins namely; Annexin A5 (ANXA5), Actin cytoplasmic 2 (ACTG), Actin cytoplasmic 1 (ACTB), Galectin-1 (LEG1), Glycerol-3-phosphate dehydrogenase [NAD(+)] cytoplasmic (GPDA), Apolipoprotein (APOA1), Collagen alpha-1 (VI) chain (CO6A10), Selenium-binding protein 1 (SBP1), Catalase (CATA), TOMM20-like protein 1 (TOM20/TO20L), E3 ubiquitin-protein ligase BRE1A (BRE1A), Ribonuclease 8 (RNA58), and Transcription factor SOX-21 (SOX21) were noted in the VAT proteome of the MUHO compared to the MHO group.
Four of the identified proteins (ANXA5, ACTG, ACTB, and LEG1) are cytoskeletal proteins that play a major role in the modulation of the ECM and in determining the plasticity of the adipose tissue. Adipose tissue has great capacity to remodel its ECM, expand and increase its storage capacity in the state of over nutrition. Gomez-Serrano et al. in their study have also demonstrated a similar decrease in the ECM proteins in the diabetic and aging obese individuals (Gomez-Serrano et al., 2016). ANXA5, in particular, is involved in signal transduction and has been implicated as inhibitor of the pro inflammatory-protein kinase C pathway (Rothhut et al., 1995; Sato et al., 2000) which results in reduced local vascular and systemic inflammation, reduce vascular remodeling, and improved vascular function (Wen et al., 1999). Olivia et al., in their study in women with gestational diabetes, demonstrated a positive correlation between significantly lowered ANXA5 levels and increased in inflammation (Oliva et al., 2013). Actins B and G, that were significantly decreased in the VAT of MUHO group compared to the MHO group, are also known to participate in the expansion capability of the ECM (Alfadda et al., 2013; van Vliet-Ostaptchouk et al., 2014). The function and role of ANXA5, ACTG, ACTB, and LEG1 have been shown in our previous proteomic studies (Alfadda et al., 2013; Benabdelkamel et al., 2015).
In the present study, we detected a decrease in the abundance of CO6A1and galectin 1 in the MUHO. Collagen 6, the most abundant matrix protein in the differentiated mature adipocytes of the adipose tissue (Mori et al., 2014), is known to be dysfunctional in obesity and contributes to the development of the MetS (Lackey et al., 2014). Its increased expression in the MHO group indicates that VAT in this group retains its plasticity and expansion capacity to allow for additional storage of triglycerides and protects against the development of metabolic dysfunction. Similarly Galectin 1 regulates cellular functions (He et al., 2006) and may be involved in increasing the expansion capacity of the adipocytes (Marsich et al., 2008). These data suggest a decrease in the expansion capacity of VAT in the MUHO, which could lead to a more dysregulated metabolic profile and development of insulin resistance.
Besides the ECM proteins, we found a significant decrease in abundance of the spots related to GPDH (an enzyme known to regulate lipid metabolism) in the MUHO group compared to the MHO group. Adipocytes lack a glycerol kinase; therefore, GPDH produced during glycolysis serves as the main precursor for lipid biosynthesis and metabolism, triacylglycerol synthesis, and is crucial for the maintenance of the NAD+/NADH potential and redox state (Xu et al., 2011). The decrease in abundance of GPDH in the MUHO group indicates a reduced capacity of the unhealthy VAT to synthesize triglycerides while this capacity is retained in the MHO group. An interesting finding in our study is the identification of a decrease in abundance of TOM20 in the MUHO group in comparison to the MHO group. TOM20 is an outer mitochondrial membrane protein that is involved in the import of nuclear encoded proteins into the mitochondria along with the uptake of the UCP1 (Scarpulla, 2008; Schleiff et al., 1997; 2000).
Additionally, a decrease in the abundance of the antioxidant enzymes, namely catalase and selenium binding protein 1 (SBP1) was noted in the MUHO group in comparison to the MHO group. Catalase is known to counter the production of reactive oxygen species and takes part in the peroxisomal β-oxidation of fatty acids. SBP1on the other hand is the main binding protein for antioxidant enzymes including glutathione peroxidase that catalyzes the reduction of lipid hydroperoxide and hydrogen peroxide levels in adipose tissue. The comparative decrease in the expression of these enzymes in the MUHO group indicates a decrease in the antioxidant capacity of the VAT that can in turn lead to metabolic dysregulation (Musgrove, 1990).
The findings of our proteomic analysis of VAT from healthy and unhealthy obese subjects indicates differential expression of proteins representing highly interlinked complex pathways linked to insulin dysregulation. The set of proteins upregulated in the VAT of MHO shows enrichment in cytoskeletal and antioxidant proteins along with those involved in mitochondrial import and transcriptional activity, while the protein set upregulated in MUHO were enriched in those proteins that increased the metabolic dysfunction in the ECM, mitochondria and LD. The network analysis of all the proteins revealed that they interacted strongly with the central nodes of the network which related to the IRK/MAPK/ERK/Akt pathway of insulin signaling, along with the involvement of pro inflammatory signaling pathways-NFκB, Jun N-terminal kinases (JNKs), and PKc. Altered insulin signaling, which can be considered either a dysfunctional or a protective response, may be the main differentiating factor between MUHO and MHO phenotypes. It is well known that activation of the above mentioned protein kinases induces inflammation, which has a broad role in driving the pathogenesis of systemic insulin resistance. Given that BMI was matched between the groups, the only observed difference was an increase in insulin levels, essentially indicating that insulin levels, irrespective of BMI, contribute to the development of metabolic dysfunction.
Similarly the existence of different phenotypes of obesity indicate that there exist individual differences in the metabolic response to the increasing weight as well as within the stages of development of obesity. The body’s response to this continuum of changes shifts between a reactive to protective or compensatory mechanism and the point where one process replaces the other and leads to metabolic complications remains unclear. This is not only due to the dynamic nature of the human body but also due to the individuals adaptations with his environment.
CONCLUSION
There is heterogeneity in the metabolic status among the obese individuals matched for both age and BMI; this heterogeneity places them into distinct sub-groups. Using a proteomic approach, we found that MHO expresses proteins that improve the expansion capacity of adipose tissue along with enhancing its antioxidant activity. The MUHO group on the other hand showed an increase in proteins that reflected a greater alteration in the mitochondrial and LD metabolic activity. This alteration along with a limited capacity for expansion of the ECM could make them more predisposed to these adverse metabolic consequences along with an increase in the risk of cardiovascular disease.
Supplementary data
ACKNOWLEDGMENTS
The authors would like to extend their sincere appreciation to the Deanship of Scientific Research at King Saud University for funding this research through the Research Group Project no. RGP-334.
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Alfadda A.A. Circulating adipokines in healthy versus unhealthy overweight and obese subjects. Int J Endocrinol. 2014;2014:170434. doi: 10.1155/2014/170434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfadda A.A., Benabdelkamel H., Masood A., Moustafa A., Sallam R., Bassas A., Duncan M. Proteomic analysis of mature adipocytes from obese patients in relation to aging. Exp Gerontol. 2013;48:1196–1203. doi: 10.1016/j.exger.2013.07.008. [DOI] [PubMed] [Google Scholar]
- Badoud F., Perreault M., Zulyniak M.A., Mutch D.M. Molecular insights into the role of white adipose tissue in metabolically unhealthy normal weight and metabolically healthy obese individuals. FASEB J. 2015;29:748–758. doi: 10.1096/fj.14-263913. [DOI] [PubMed] [Google Scholar]
- Benabdelkamel H., Masood A., Almidani G.M., Alsadhan A.A., Bassas A.F., Duncan M.W., Alfadda A.A. Mature adipocyte proteome reveals differentially altered protein abundances between lean, overweight and morbidly obese human subjects. Mol Cell Endocrinol. 2015;401:142–154. doi: 10.1016/j.mce.2014.11.021. [DOI] [PubMed] [Google Scholar]
- Bluher M. Are metabolically healthy obese individuals really healthy? Eur. J Endocrinol. 2014;171:R209–219. doi: 10.1530/EJE-14-0540. [DOI] [PubMed] [Google Scholar]
- Bohm A., Halama A., Meile T., Zdichavsky M., Lehmann R., Weigert C., Fritsche A., Stefan N., Konigsrainer A., Haring H.U., et al. Metabolic signatures of cultured human adipocytes from metabolically healthy versus unhealthy obese individuals. PLoS One. 2014;9:e93148. doi: 10.1371/journal.pone.0093148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doumatey A.P., Zhou J., Zhou M., Prieto D., Rotimi C.N., Adeyemo A. Proinflammatory and lipid biomarkers mediate metabolically healthy obesity: A proteomics study. Obesity (Silver Spring) 2016;24:1257–1265. doi: 10.1002/oby.21482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbrini E., Yoshino J., Yoshino M., Magkos F., Tiemann Luecking C., Samovski D., Fraterrigo G., Okunade A.L., Patterson B.W., Klein S. Metabolically normal obese people are protected from adverse effects following weight gain. J Clin Invest. 2015;125:787–795. doi: 10.1172/JCI78425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Serrano M., Camafeita E., Garcia-Santos E., Lopez J.A., Rubio M.A., Sanchez-Pernaute A., Torres A., Vazquez J., Peral B. Proteome-wide alterations on adipose tissue from obese patients as age-, diabetes- and gender-specific hallmarks. Sci Rep. 2016;6:25756. doi: 10.1038/srep25756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurnell M., Savage D.B., Chatterjee V.K., O’Rahilly S. The metabolic syndrome: peroxisome proliferator-activated receptor gamma and its therapeutic modulation. J Clin Endocrinol Metabol. 2003;88:2412–2421. doi: 10.1210/jc.2003-030435. [DOI] [PubMed] [Google Scholar]
- He J., Baum L.G. Galectin interactions with extracellular matrix and effects on cellular function. Methods Enzymol. 2006;417:247–256. doi: 10.1016/S0076-6879(06)17017-2. [DOI] [PubMed] [Google Scholar]
- Hodges B.D., Wu C.C. Proteomic insights into an expanded cellular role for cytoplasmic lipid droplets. J Lipid Res. 2010;51:262–273. doi: 10.1194/jlr.R003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang Y.C., Hayashi T., Fujimoto W.Y., Kahn S.E., Leonetti D.L., McNeely M.J., Boyko E.J. Visceral abdominal fat accumulation predicts the conversion of metabolically healthy obese subjects to an unhealthy phenotype. Int J Obes. 2015;39:1365–1370. doi: 10.1038/ijo.2015.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhan-Vague I., Alessi M.C., Mavri A., Morange P.E. Plasminogen activator inhibitor-1, inflammation, obesity, insulin resistance and vascular risk. J Thrombosis Haemostasis. 2003;1:1575–1579. doi: 10.1046/j.1538-7836.2003.00279.x. [DOI] [PubMed] [Google Scholar]
- Kramer C.K., Zinman B., Retnakaran R. Are metabolically healthy overweight and obesity benign conditions?: a systematic review and meta-analysis. Ann Int Med. 2013;159:758–769. doi: 10.7326/0003-4819-159-11-201312030-00008. [DOI] [PubMed] [Google Scholar]
- Lackey D.E., Burk D.H., Ali M.R., Mostaedi R., Smith W.H., Park J., Scherer P.E., Seay S.A., McCoin C.S., Bonaldo P., et al. Contributions of adipose tissue architectural and tensile properties toward defining healthy and unhealthy obesity. Am J Physiol Endocrinol Metabol. 2014;306:E233–246. doi: 10.1152/ajpendo.00476.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsich E., Mozetic P., Ortolani F., Contin M., Marchini M., Vetere A., Pacor S., Semeraro S., Vittur F., Paoletti S. Galectin-1 in cartilage: expression, influence on chondrocyte growth and interaction with ECM components. Matrix Biol. 2008;27:513–525. doi: 10.1016/j.matbio.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Mori S., Kiuchi S., Ouchi A., Hase T., Murase T. Characteristic expression of extracellular matrix in subcutaneous adipose tissue development and adipogenesis; comparison with visceral adipose tissue. Int J Biol Sci. 2014;10:825–833. doi: 10.7150/ijbs.8672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murri M., Insenser M., Bernal-Lopez M.R., Perez-Martinez P., Escobar-Morreale H.F., Tinahones F.J. Proteomic analysis of visceral adipose tissue in pre-obese patients with type 2 diabetes. Mol Cell Endocrinol. 2013;376:99–106. doi: 10.1016/j.mce.2013.06.010. [DOI] [PubMed] [Google Scholar]
- Musgrove J.P. Family and community health in a new medical school. Med Education. 1990;24:124–128. doi: 10.1111/j.1365-2923.1990.tb02510.x. [DOI] [PubMed] [Google Scholar]
- Ng M., Fleming T., Robinson M., Thomson B., Graetz N., Margono C., Mullany E.C., Biryukov S., Abbafati C., Abera S.F., et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2014;384:766–781. doi: 10.1016/S0140-6736(14)60460-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien L., Hosick P.A., John K., Stec D.E., Hinds T.D., Jr Biliverdin reductase isozymes in metabolism. Trends Endocrinol Metabol. 2015;26:212–220. doi: 10.1016/j.tem.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva K., Barker G., Rice G.E., Bailey M.J., Lappas M. 2D-DIGE to identify proteins associated with gestational diabetes in omental adipose tissue. J Endocrinol. 2013;218:165–178. doi: 10.1530/JOE-13-0010. [DOI] [PubMed] [Google Scholar]
- Olofsson S.O., Bostrom P., Andersson L., Rutberg M., Perman J., Boren J. Lipid droplets as dynamic organelles connecting storage and efflux of lipids. Biochim Biophys Acta. 2009;1791:448–458. doi: 10.1016/j.bbalip.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Ouchi N., Parker J.L., Lugus J.J., Walsh K. Adipokines in inflammation and metabolic disease. Nat Rev Immunol. 2011;11:85–97. doi: 10.1038/nri2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pataky Z., Bobbioni-Harsch E., Golay A. Open questions about metabolically normal obesity. Int J Obes. 2010;34(Suppl 2):S18–23. doi: 10.1038/ijo.2010.235. [DOI] [PubMed] [Google Scholar]
- Peinado J.R., Pardo M., de la Rosa O., Malagon M.M. Proteomic characterization of adipose tissue constituents, a necessary step for understanding adipose tissue complexity. Proteomics. 2012;12:607–620. doi: 10.1002/pmic.201100355. [DOI] [PubMed] [Google Scholar]
- Reaven G.M. The metabolic syndrome: time to get off the merry-go-round? J. Int Med. 2011;269:127–136. doi: 10.1111/j.1365-2796.2010.02325.x. [DOI] [PubMed] [Google Scholar]
- Rothhut B., Dubois T., Feliers D., Russo-Marie F., Oudinet J.P. Inhibitory effect of annexin V on protein kinase C activity in mesangial cell lysates. Eur J Biochem. 1995;232:865–872. [PubMed] [Google Scholar]
- Sato H., Ogata H., De Luca L.M. Annexin V inhibits the 12-O-tetradecanoylphorbol-13-acetate-induced activation of Ras/extracellular signal-regulated kinase (ERK). signaling pathway upstream of Shc in MCF-7 cells. Oncogene. 2000;19:2904–2912. doi: 10.1038/sj.onc.1203615. [DOI] [PubMed] [Google Scholar]
- Scarpulla R.C. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- Schleiff E., McBride H. The central matrix loop drives import of uncoupling protein 1 into mitochondria. J Cell Sci. 2000;113(Pt 12):2267–2272. doi: 10.1242/jcs.113.12.2267. [DOI] [PubMed] [Google Scholar]
- Schleiff E., Shore G.C., Goping I.S. Human mitochondrial import receptor, Tom20p. Use of glutathione to reveal specific interactions between Tom20-glutathione S-transferase and mitochondrial precursor proteins. FEBS Lett. 1997;404:314–318. doi: 10.1016/s0014-5793(97)00145-2. [DOI] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Smith H.W., Marshall C.J. Regulation of cell signalling by uPAR. Nat Rev Mol Cell Biol. 2010;11:23–36. doi: 10.1038/nrm2821. [DOI] [PubMed] [Google Scholar]
- van Vliet-Ostaptchouk J.V., Nuotio M.L., Slagter S.N., Doiron D., Fischer K., Foco L., Gaye A., Gogele M., Heier M., Hiekkalinna T., et al. The prevalence of metabolic syndrome and metabolically healthy obesity in Europe: a collaborative analysis of ten large cohort studies. BMC Endocr Disord. 2014;14:9. doi: 10.1186/1472-6823-14-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y., Edelman J.L., Kang T., Sachs G. Lipocortin V may function as a signaling protein for vascular endothelial growth factor receptor-2/Flk–1. Biochem Biophys Res Commun. 1999;258:713–721. doi: 10.1006/bbrc.1999.0678. [DOI] [PubMed] [Google Scholar]
- Xu S.P., Mao X.Y., Ren F.Z., Che H.L. Attenuating effect of casein glycomacropeptide on proliferation, differentiation, and lipid accumulation of in vitro Sprague-Dawley rat preadipocytes. J Dairy Sci. 2011;94:676–683. doi: 10.3168/jds.2010-3827. [DOI] [PubMed] [Google Scholar]
- Zemany L., Bhanot S., Peroni O.D., Murray S.F., Moraes-Vieira P.M., Castoldi A., Manchem P., Guo S., Monia B.P., Kahn B.B. Transthyretin antisense oligonucleotides lower circulating RBP4 levels and improve insulin sensitivity in obese mice. Diabetes. 2015;64:1603–1614. doi: 10.2337/db14-0970. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.