Abstract
Anthracnose caused by Colletotrichum species is a serious disease of more than 30 plant genera. Several Colletotrichum species have been reported to infect chili in different countries. Although China is the largest chili-producing country, little is known about the species that have been infecting chili locally. Therefore, we collected samples of diseased chili from 29 provinces of China, from which 1285 strains were isolated. The morphological characters of all strains were observed and compared, and multi-locus phylogenetic analyses (ITS, ACT, CAL, CHS-1, GAPDH, TUB2, and HIS3) were performed on selected representative strains. Fifteen Colletotrichum species were identified, with C. fioriniae, C. fructicola, C. gloeosporioides, C. scovillei, and C. truncatum being prevalent. Three new species, C. conoides, C. grossum, and C. liaoningense, were recognised and described in this paper. Colletotrichum aenigma, C. cliviae, C. endophytica, C. hymenocallidis, C. incanum, C. karstii, and C. viniferum were reported for the first time from chili. Pathogenicity of all species isolated from chili was confirmed, except for C. endophytica. The current study improves the understanding of species causing anthracnose on chili and provides useful information for the effective control of the disease in China.
Keywords: DNA phylogeny, multi-gene analysis, plant pathogen, systematics
INTRODUCTION
Chili (Capsicum spp.) is an important vegetable crop worldwide. China maintains the largest planted area of chili, producing more than 28 M tons per year for domestic consumption and export (Li et al. 2009). One of the most destructive diseases restricting chili production is anthracnose, caused by Colletotrichum spp. (Bailey & Jeger 1992, Poonpolgul & Kumphai 2007, Than et al. 2008), resulting in up to 40 % yield loss in China (Lin et al. 2004).
Colletotrichum species can infect more than 30 plant genera (Perfect et al. 1999, Dean et al. 2012, Farr & Rossman 2016). More than 10 Colletotrichum species have been reported from chili, with different distributions among countries (Than et al. 2008, Liao et al. 2012, Kanto et al. 2014, Sharma et al. 2014, Diao et al. 2015). For example, anthracnose on chili is caused by C. coccodes, C. fructicola, C. siamense, and C. truncatum in India (Sharma & Shenoy 2014); by C. acutatum, C. coccodes, and C. gloeosporioides in the USA; by C. acutatum, C. dematium, C. gloeosporioides, and C. truncatum in Australia; by C. acutatum, C. coccodes, C. dematium, C. gloeosporioides, and C. panacicola in Korea (Than et al. 2008); and by C. acutatum, C. gloeosporioides, C. truncatum, and C. coccodes in China (Shin et al. 1999, Liao et al. 2012). Most of these reports, however, were based on morphology and ITS sequences or a combination of ITS and TUB2 sequences, which have been shown to be insufficient in distinguishing closely related taxa in several species complexes. In addition, these records were mostly based on a small sampling from restricted areas, and, thus, may underestimate the species diversity.
The current study aimed to investigate the Colletotrichum species causing anthracnose on chili in China, by employing large-scale sampling and isolation, and via morphological characterisation and multi-locus phylogeny of the obtained strains.
MATERIALS AND METHODS
Sample collection and isolation
From 2008 to 2014, fruits and leaves of chili (Capsicum spp.) with anthracnose symptoms were collected from 50 locations in 29 provinces of China (Fig. 1). In each location, a hierarchical sampling method was used as previously described (Kohli et al. 1995). Five fields were chosen at each sampling location, and 25 chili fruits and also leaves in some cases were collected from each field along a diagonal transect. Colletotrichum species were isolated as described by Cai et al. (2009). All isolates were grown at 28 °C for further study. Type specimens of new species from this study were deposited in the Mycological Herbarium, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China (HMAS), and ex-type living cultures were deposited in the China General Microbiological Culture Collection Centre (CGMCC), Beijing, China.
Fig. 1.
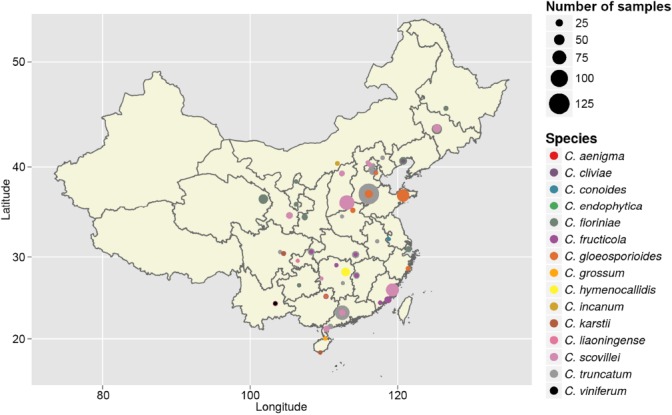
Map showing locations in China where chili was sampled for Colletotrichum species. Each coloured circle represents one species by preliminary identification, and the size of the circle indicates the number of isolates collected from that location.
Morphological characterisation
Mycelial plugs (5 mm) were transferred from the edge of actively growing cultures to fresh potato dextrose agar (PDA, 1.5 %, Difco) plates. Cultures were kept at 28 °C with a 12/12 h fluorescent light/dark cycle. The morphological characters for all isolates, including colony and conidial characteristics, were observed. Microscopic characters were examined with the Carl Zeiss Imager A2 microscope after 1 mo of cultivation. Among the 1 285 obtained isolates (Table 1), 121 representative isolates were selected for further multilocus phylogenetic analyses based on geographical location, morphology (colony shape and colour and characteristics of aerial mycelia and conidia), and ITS sequences. Different morphological types were selected from each location, and the number of representative isolates selected depended on the number of isolates with different morphologies. Furthermore, if the number of isolates with high morphological and ITS sequence similarities was less than 10 in one location, then one isolate was randomly selected as a representative. If the number was more than 10, on the other hand, then an additional isolate (one from each of the 10 isolates) was selected for multi-locus phylogenetic analyses. The length and width of 30 conidia for each isolate were measured in lactic acid, and mean values calculated. The formation of appressoria was induced as described by Cai et al. (2009).
Table 1.
A list of all Colletotrichum isolates collected from chili in China based on preliminary identification.
| Species | Location | Host tissue | Year | Number of isolates | Longitude | Latitude |
|---|---|---|---|---|---|---|
| C. aenigma | Yangliuqing, Tianjin | Fruit | 2012 | 1 | 39.4 | 117.01 |
| C. cliviae | Xingcheng, Liaoning | Fruit | 2012 | 1 | 40.63 | 120.74 |
| C.conoides | Nanjing, Jiangsu | Fruit | 2010 | 3 | 32.06 | 118.79 |
| C. endophytica | Mile, Yunnan | Fruit | 2011 | 1 | 24.41 | 103.41 |
| C. fioriniae | Fengxiang, Shanxi | Fruit | 2011 | 17 | 34.55 | 107.4 |
| Changchun | Fruit | 2011 | 47 | 43.81 | 125.32 | |
| Xining, Qinghai | Fruit | 2011 | 42 | 36.61 | 101.78 | |
| Sanya, Hainan | Fruit | 2012 | 2 | 18.25 | 109.51 | |
| Guiyang, Guizhou | Fruit | 2012 | 3 | 26.64 | 106.63 | |
| Xingcheng, Liaoning | Fruit | 2012 | 29 | 40.63 | 120.74 | |
| Yinchuan, Ningxia | Fruit | 2012 | 4 | 38.48 | 106.23 | |
| Guyuan, Ningxia | Fruit | 2012 | 8 | 36.01 | 106.24 | |
| Fengxian, Shanghai | Fruit | 2012 | 16 | 30.91 | 121.47 | |
| Harbin, Heilongjiang | Fruit | 2012 | 9 | 45.8 | 126.53 | |
| C. fructicola | Fuzhou, Fujian | Fruit | 2011 | 2 | 26.07 | 119.29 |
| Guilin, Guangxi | Fruit | 2011 | 10 | 25.27 | 110.29 | |
| Hengyang, Hunan | Fruit | 2012 | 4 | 29.03 | 111.69 | |
| Changsha, Hunan | Fruit | 2012 | 20 | 28.23 | 112.94 | |
| Laiyang, Shandong | Fruit | 2011 | 32 | 36.99 | 120.74 | |
| Wucheng, Shandong | Fruit | 2012 | 15 | 37.16 | 116.08 | |
| Zhangzhou, Fujian | Fruit | 2008 | 5 | 24.51 | 117.64 | |
| Quanzhou, Fujian | Fruit | 2009 | 23 | 24.87 | 118.67 | |
| Fengxiang, Shanxi | Fruit | 2011 | 5 | 34.55 | 107.4 | |
| Xinxiang, Henan | Fruit | 2011 | 5 | 35.3 | 113.93 | |
| Yichun, Jiangxi | Fruit | 2011 | 6 | 27.81 | 114.41 | |
| Jianyang, Sichuan | Fruit | 2011 | 8 | 30.41 | 104.55 | |
| Mile, Yunnan | Fruit | 2011 | 10 | 24.41 | 103.41 | |
| Yangliuqing, Tianjin | Fruit | 2012 | 4 | 39.4 | 117.01 | |
| Wuhan, Hubei | Fruit | 2012 | 4 | 30.28 | 114.29 | |
| Xingcheng, Liaoning | Fruit | 2012 | 4 | 40.63 | 120.74 | |
| Shizhu, Chongqing | Fruit | 2013 | 4 | 30.6 | 108.29 | |
| C. gloeosporioides | Guilin, Guangxi | Fruit | 2011 | 1 | 25.27 | 110.29 |
| Qingyuan, Guangdong | Fruit | 2013 | 21 | 23.28 | 112.48 | |
| Qingyuan, Guangdong | Fruit | 2014 | 1 | 23.28 | 112.48 | |
| Laiyang, Shandong | Fruit | 2011 | 64 | 36.99 | 120.74 | |
| Wucheng, Shandong | Fruit | 2011 | 30 | 37.16 | 116.08 | |
| Fengxiang, Shanxi | Fruit | 2011 | 10 | 34.55 | 107.4 | |
| Xinxiang, Henan | Fruit | 2011 | 10 | 35.3 | 113.93 | |
| Taizhou, Zhejiang | Fruit | 2011 | 8 | 28.65 | 121.42 | |
| Changsha, Hunan | Fruit | 2012 | 20 | 28.23 | 112.94 | |
| Mile, Yunnan | Fruit | 2011 | 5 | 24.41 | 103.41 | |
| Jianyang, Sichuan | Fruit | 2011 | 4 | 30.41 | 104.55 | |
| Guyuan, Ningxia | Fruit | 2012 | 2 | 36.01 | 106.24 | |
| Wuqing, Tianjin | Fruit | 2012 | 1 | 39.38 | 117.04 | |
| Xingcheng, Liaoning | Fruit | 2012 | 4 | 40.63 | 120.74 | |
| C. grossum | Haikou, Hainan | Fruit | 2011 | 3 | 20.04 | 110.19 |
| C. hymenocallidis | Changsha, Hunan | Fruit | 2012 | 35 | 28.23 | 112.94 |
| C. incanum | Helingeer, Inner Mongolia | Fruit | 2012 | 6 | 40.37 | 111.82 |
| C. karstii | Sanya, Hainan | Fruit | 2012 | 3 | 18.25 | 109.51 |
| Jianyang, Sichuan | Fruit | 2011 | 5 | 30.41 | 104.55 | |
| Mile, Yunnan | Fruit | 2011 | 1 | 24.41 | 103.41 | |
| C. liaoningense | Xingcheng, Liaoning | Fruit | 2012 | 11 | 40.63 | 120.74 |
| Shapingba, Chongqing | Fruit | 2012 | 1 | 29.54 | 106.46 | |
| C. scoville | Yanqing, Beijing | Fruit | 2011 | 4 | 40.45 | 115.97 |
| Changping, Beijing | Fruit | 2013 | 8 | 40.22 | 116.23 | |
| Fuzhou, Fujian | Fruit | 2011 | 68 | 26.07 | 119.29 | |
| Gangu, Gansu | Fruit | 2012 | 21 | 34.73 | 105.33 | |
| Jida, Jilin | Fruit | 2013 | 33 | 43.88 | 125.31 | |
| Changzhi, Shanxi | Fruit | 2011 | 84 | 36.19 | 113.11 | |
| Shuozhou, Shanxi | Fruit | 2012 | 14 | 39.33 | 112.43 | |
| Sanya, Hainan | Fruit | 2012 | 1 | 18.25 | 109.51 | |
| Zhijiang, Hunan | Fruit | 2011 | 1 | 27.44 | 109.68 | |
| Zhanjiang, Guangdong | Fruit | 2011 | 18 | 21.27 | 110.35 | |
| Qingyuan, Guangdong | Fruit | 2012 | 20 | 23.28 | 112.48 | |
| C. truncatum | Qingyuan, Guangdong | Fruit | 2013 | 80 | 23.28 | 112.48 |
| Qingyuan, Guangdong | Fruit | 2014 | 10 | 23.28 | 112.48 | |
| Maoming, Guangdong | Fruit | 2013 | 13 | 21.55 | 110.88 | |
| Yichun, Jiangxi | Leave | 2011 | 20 | 27.81 | 114.41 | |
| Shizhu, Chongqing | Fruit | 2013 | 23 | 30.6 | 108.29 | |
| Wuhan, Hubei | Fruit | 2013 | 25 | 30.28 | 114.29 | |
| Fengxiang, Shanxi | Fruit | 2011 | 12 | 34.55 | 107.4 | |
| Wucheng, Shandong | Fruit | 2011 | 125 | 37.16 | 116.08 | |
| Laiyang, Shandong | Fruit | 2011 | 10 | 36.99 | 120.74 | |
| Yangliuqing, Tianjin | Fruit | 2012 | 11 | 39.4 | 117.01 | |
| Langfang, Hebei | Fruit | 2011 | 20 | 39.52 | 116.61 | |
| Chengde, Hebei | Fruit | 2013 | 5 | 40.95 | 117.96 | |
| Daxing, Beijing | Fruit | 2011 | 9 | 39.73 | 116.34 | |
| Shunyi, Beijing | Fruit | 2011 | 10 | 40.13 | 116.65 | |
| Xingcheng, Liaoning | Fruit | 2012 | 16 | 40.63 | 120.74 | |
| Changchun, Jilin | Fruit | 2012 | 7 | 43.71 | 125.54 | |
| Chengdu, Sichuan | Fruit | 2011 | 3 | 30.57 | 104.07 | |
| Hefei, Anhui | Fruit | 2011 | 2 | 31.82 | 117.23 | |
| Fuzhou, Fujian | Fruit | 2011 | 2 | 26.07 | 119.29 | |
| Luoyang, Henan | Fruit | 2011 | 1 | 34.62 | 112.45 | |
| Xinxiang, Henan | Fruit | 2011 | 2 | 35.3 | 113.93 | |
| Changsha, Hunan | Fruit | 2012 | 1 | 28.23 | 112.94 | |
| Hengyang, Hunan | Fruit | 2012 | 1 | 26.89 | 112.57 | |
| Changde, Hunan | Fruit | 2012 | 3 | 29.03 | 111.69 | |
| Mile, Yunnan | Fruit | 2011 | 4 | 24.41 | 103.41 | |
| Zhanjiang, Guangdong | Fruit | 2011 | 5 | 21.27 | 110.36 | |
| Xining, Qinghai | Fruit | 2011 | 1 | 36.61 | 101.78 | |
| Shuozhou, Shanxi | Fruit | 2012 | 1 | 39.33 | 112.43 | |
| C. viniferum | Mile, Yunnan | Fruit | 2011 | 1 | 24.41 | 103.41 |
| Total | 1285 |
DNA extraction, PCR amplification, and sequencing
Genomic DNA was extracted from 121 representative isolates as previously described (Murray & Thompson 1980, Diao et al. 2015). The following loci were amplified with the indicated primers: the internal transcribed spacer regions and intervening 5.8S nrRNA gene (ITS) with primers ITS4/ITS5 (White et al. 1990); partial sequences of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) with primers GDF1/GDR1 (Templeton et al. 1992); actin gene (ACT) with primers ACT512F/ACT783R (Carbone & Kohn 1999); beta-tubulin (TUB2) with primers T1/Bt2b (Glass & Donaldson 1995, O’Donnell & Cigelnik 1997); calmodulin (CAL) with primers CL1/CL2A (O’Donnell et al. 2000); chitin synthase 1 (CHS-1) with primers CHS-79F/CHS-345R (Carbone & Kohn 1999); and histone3 (HIS3) with primers CYLH3F/CYLH3R (Crous et al. 2004b). PCR reactions were performed as described by Damm et al. (2009). DNA sequencing was conducted by Sunbiotech, Beijing, China with a 3730 DNA Analyzer (Applied Biosystems, USA). The sequences obtained from forward and reverse primers were used to obtain consensus sequences with DNAMAN v. 6.0 (Lynnon Biosoft, USA). Sequences were aligned using MAFFT v. 6 (Katoh & Toh 2010).
Phylogenetic analysis
All sequences of the 121 representative isolates were blasted in GenBank. Sequences with high similarities were selected and included in the analyses (Table 2a, 2b, 2c). Concatenated analyses of ITS, GAPDH, CHS-1, HIS3, ACT, and TUB2 were conducted for the C. acutatum species complex and Colletotrichum species with curved conidia, while ITS, GAPDH, CAL, ACT, CHS-1, and TUB2 were concatenated for the analysis of the C. gloeosporioides species complex and other species. Maximum parsimony (MP) analyses based on the combined datasets were conducted using PAUP v. 4.0b10 (Swofford 2002). Phylogenetic trees were generated using the heuristic search option with TBR branch swapping and 1 000 random sequence additions. Maxtrees were unlimited, with branches of zero length collapsed, and all multiple parsimonious trees were saved. Clade stability was assessed using a bootstrap analysis with 1 000 replicates. Afterward, tree length (TL), consistency index (CI), retention index (RI), rescaled consistency index (RC), and homoplasy index (HI) were calculated. Bayesian inference (BI) was used to reconstruct the phylogenetic tree using MrBayes v. 3.2.1 (Ronquist & Huelsenbeck 2003). Best-fit models of nucleotide substitution were selected using MrModelTest v. 2.3 (Nylander 2004). Two analyses of four MCMC chains were run from random trees for 1 000 000 generations, and trees were sampled every 100 generations resulting in 20 000 total trees. The first 25 % of the trees were discarded as the burn-in phase of each analysis, and the remaining trees were used to calculate posterior probabilities (Cai et al. 2006, Liu et al. 2012, 2013). An additional Maximum likelihood (ML) analysis was implemented in the C. gloeosporioides species complex using the CIPRES Science Gateway v. 3.3 (www.phylo.org), and the RAxML-HPC BlackBox was selected with default parameters. Sequences derived in this study were deposited in GenBank (Table 2), and the concatenated alignments were deposited in TreeBASE (http://treebase.org/treebase-web/home.html; study S17061), and the descriptions and nomenclature in MycoBank (Crous et al. 2004a).
Table 2a.
Strains used for the phylogenetic analysis of the Colletotrichum gloeosporioides species complex and other species with details about host, location, and GenBank accession numbers.
* = Ex-type culture. Strains studied in this paper are in bold font.
Table 2b.
Strains used for the phylogenetic analysis of the Colletotrichum acutatum species complex with details about host, location, and GenBank accession numbers.
* = Ex-type culture. Strains studied in this paper are in bold font.
Table 2c.
Strains used for the phylogenetic analysis of Colletotrichum species with curved conidia with details on host, location, and GenBank accession numbers.
* = Ex-type culture. Strains studied in this paper are in bold
Genealogical concordance phylogenetic species recognition analysis
New species and their most closely related neighbours were analysed using the Genealogical Concordance Phylogenetic Species Recognition (GCPSR) model with a pairwise homoplasy index (PHI) test as described by Quaedvlieg et al. (2014). The PHI tests were performed in SplitsTree4 (Huson 1998, Huson & Bryant 2006) to determine the recombination level within phylogenetically closely related species using a six-locus concatenated dataset (ACT, CAL, CHS, GAPDH, ITS, and TUB2) for C. conoides, C. grossum, and C. liaoningense and their respective related species. A pairwise homoplasy index below a 0.05 threshold (Φw < 0.05) indicated the presence of significant recombination in the dataset. The relationship between closely related species was visualised by constructing a split graph.
Pathogenicity assay
Seven chili cultivars, Capsicum annuum cv. Chaotianjiao, Denglongjiao, Sanyingjiao, Zidantou, C. frutescens cv. Shuangla, Xiaomila and C. chinense cv. Huangdijiao, were inoculated with representative strains of 15 Colletotrichum species respectively (Table 3, 4). Pathogenicity tests were conducted on chili following the methods described by Montri et al. (2009) and Mongkolporn et al. (2010). Healthy, ripe red and green chili fruits were surface sterilised in 1 % NaClO for 5 min separately, washed twice with sterile-distilled water, and air dried on sterile filter paper. Each fruit was inoculated with 1 μL of a conidial suspension (1 × 106 conidial/mL), which was injected onto the non-wounded fruit surface using a microsyringe (Shanghai, China). Control fruits were treated with 1 μL of distilled water. Each isolate was inoculated to five replicate fruits. The inoculated fruits were incubated in a moist chamber at 28 °C and were examined for symptoms daily for 9 d. The virulence and pathotypes were evaluated as described by Montri et al. (2009). The experiment was conducted twice.
Table 3.
Anthracnose severity scores on a 0–9 scale and pathotypes of 15 Colletotrichum species isolates at the ripe red fruit stage of seven chili cultivars.
| Isolate |
Capsicum annuum |
C. frutescens |
C. chinense |
Mean | Pathotype | ||||
|---|---|---|---|---|---|---|---|---|---|
| Chaotianjiao | Denglongjiao | Sanyingjiao | Zidantou | Shuanla | Xiaomila | Huangdijiao | |||
| C. aenigma CAUG26 | 5 | 5 | 7 | 5 | 5 | 7 | 5 | 6 | PC1-R |
| C. cliviae CAUOS5 | 7 | 7 | 5 | 7 | 7 | 5 | 0 | 5 | PC2-R |
| C. conoides CAUG17 | 7 | 7 | 7 | 7 | 7 | 5 | 5 | 6 | PC1-R |
| C. endophytica CAUG28 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | PC3-R |
| C. fioriniae CAUT34 | 7 | 7 | 9 | 9 | 9 | 9 | 7 | 8 | PC1-R |
| C. fructicola CAUG1 | 7 | 7 | 9 | 9 | 9 | 9 | 7 | 8 | PC1-R |
| C. gloeosporioides CAUG2 | 7 | 7 | 9 | 9 | 7 | 7 | 5 | 7 | PC1-R |
| C. grossum CAUG7 | 7 | 7 | 7 | 7 | 5 | 5 | 7 | 6 | PC1-R |
| C. hymenocallidis CAUG9 | 5 | 7 | 9 | 9 | 7 | 7 | 0 | 6 | PC2-R |
| C. incanum CAUT34 | 5 | 7 | 7 | 5 | 7 | 5 | 5 | 6 | PC1-R |
| C. karstii CAUOS1 | 7 | 5 | 7 | 7 | 9 | 9 | 7 | 7 | PC1-R |
| C. liaoningense CAUOS2 | 9 | 7 | 9 | 9 | 5 | 9 | 5 | 8 | PC1-R |
| C. scovillei CAUA1 | 7 | 9 | 9 | 9 | 9 | 7 | 9 | 8 | PC1-R |
| C. truncatum CAUT1 | 7 | 7 | 9 | 7 | 9 | 9 | 7 | 8 | PC1-R |
| C. viniferum CAUG27 | 5 | 7 | 9 | 9 | 9 | 9 | 5 | 8 | PC1-R |
| Mean | 6 | 6 | 7 | 7 | 7 | 7 | 5 | 6 | – |
Table 4.
Anthracnose severity scores on a 0–9 scale and pathotypes of 15 Colletotrichum species isolates at the mature green fruit stage of seven chili cultivars.
| Isolate |
Capsicum annuum |
C. frutescens |
C. chinense |
Mean | Pathotype | ||||
|---|---|---|---|---|---|---|---|---|---|
| Chaotianjiao | Denglongjiao | Sanyingjiao | Zidantou | Shuanla | Xiaomila | Huangdijiao | |||
| C. aenigma CAUG26 | 5 | 5 | 5 | 5 | 0 | 0 | 0 | 3 | PC1-G |
| C. cliviae CAUOS5 | 7 | 9 | 5 | 7 | 7 | 5 | 7 | 7 | PC2-G |
| C. conoides CAUG17 | 7 | 5 | 0 | 5 | 7 | 5 | 0 | 4 | PC3-G |
| C.endophyticaCAUG28 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | PC4-G |
| C. fioriniae CAUT34 | 9 | 9 | 7 | 7 | 9 | 9 | 7 | 8 | PC2-G |
| C. fructicola CAUG1 | 7 | 9 | 9 | 5 | 0 | 7 | 7 | 6 | PC5-G |
| C.gloeosporioidesCAUG2 | 5 | 9 | 7 | 5 | 0 | 7 | 7 | 6 | PC5-G |
| C. grossum CAUG7 | 3 | 3 | 0 | 5 | 5 | 5 | 0 | 3 | PC3-G |
| C.hymenocallidis CAUG9 | 5 | 5 | 5 | 5 | 7 | 7 | 7 | 6 | PC2-G |
| C. incanum CAUT34 | 5 | 5 | 5 | 3 | 7 | 5 | 5 | 5 | PC2-G |
| C. karstii CAUOS1 | 5 | 7 | 7 | 7 | 0 | 0 | 0 | 4 | PC1-G |
| C.liaoningense CAUOS2 | 5 | 5 | 9 | 7 | 5 | 3 | 7 | 6 | PC2-G |
| C. scovillei CAUA1 | 7 | 7 | 7 | 7 | 9 | 9 | 7 | 7 | PC2-G |
| C. truncatum CAUT1 | 7 | 5 | 5 | 7 | 7 | 7 | 5 | 6 | PC2-G |
| C. viniferum CAUG27 | 5 | 5 | 5 | 7 | 9 | 9 | 7 | 7 | PC2-G |
| Mean | 5 | 6 | 5 | 5 | 5 | 5 | 4 | 5 | – |
RESULTS
Disease survey and strain isolation
Symptoms of anthracnose were circular or angular sunken lesions on chili fruits and irregularly shaped brown spots with dark brown edges on leaves. A total of 1 285 isolates of Colletotrichum spp. were obtained from 29 provinces in China (Fig. 1, Table 1). Attempts were made to collect samples from multiple locations in Tibet and Xinjiang provinces for several years but failed to locate symptomatic plants. All strains were isolated from fruits except those from Jiangxi province, where serious damage was found on chili leaves rather than on fruits.
Group assessment
Based on megablast searches in GenBank using ITS sequences and the colony morphologies on PDA, all strains were assigned to four groups, i.e., those that produce cylindrical conidia with round ends were assigned to the C. gloeosporioides species complex; those that produce acute ends or ± cylindrical conidia with only one acute end were assigned to the C. acutatum species complex; those that produce dark setae and curved conidia were assigned to the Colletotrichum species with curved conidia; and the remaining strains were assigned to a fourth group. Among the 121 isolates, 31 belonged to the C. gloeosporioides complex; 48 belonged to the C. acutatum complex; 34 belonged to the Colletotrichum species with curved conidia, and eight belonged to the fourth group (Damm et al. 2012a, b, 2013, 2014, Weir et al. 2012, Crouch 2014).
Multi-locus phylogenetic analyses
The 121 representative isolates from chili were subjected to multi-locus phylogenetic analyses (Table 2a, 2b, 2c). The trees generated from the Bayesian and RaxML analyses were essentially similar to that from the MP analysis (Fig. 2) and are therefore not shown. In Fig. 2, the 31 isolates in the C. gloeosporioides complex clustered in eight clades, eight with C. fructicola, 13 with C. gloeosporioides, and four with C. aenigma, C. endophytica, C. hymenocallidis, and C. viniferum, respectively. In addition, two distinct lineages, which clustered distantly from any known species in the complex, were recognised as new species and herein described as C. conoides and C. grossum (Fig. 2). In Fig. 3, the isolates of the C. acutatum complex clustered in two clades, 31 with C. scovillei and 17 with C. fioriniae. In the Colletotrichum species with curved conidia, 33 isolates clustered with C. truncatum, and one clustered with C. incanum (Fig. 4). The remaining isolates were assigned to C. cliviae and C. karstii. A new lineage belonging to the fourth group, distinct from all known species, is herein described as a new species, C. liaoningense (Fig. 6).
Fig. 2.
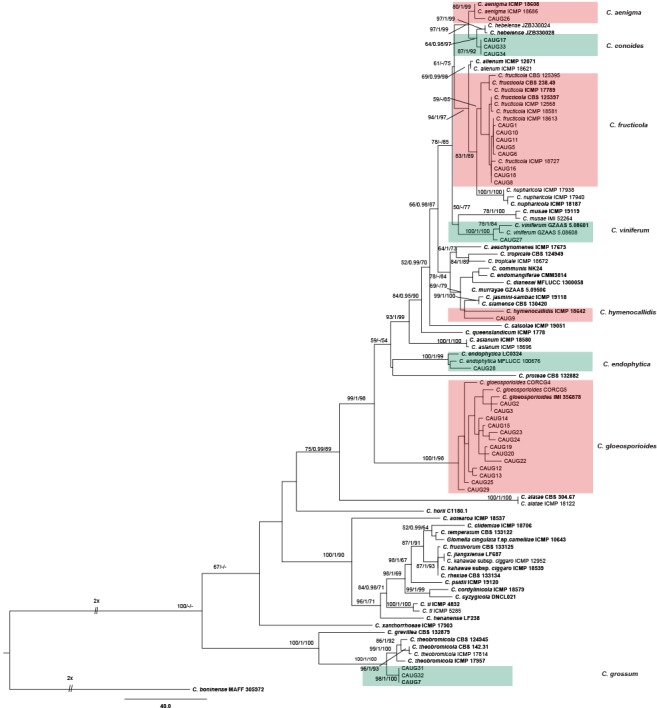
Maximum parsimony tree of isolates in the Colletotrichum gloeosporioides species complex obtained from a heuristic search of combined ACT, CAL, CHS-1, GAPDH, ITS, and TUB2 gene sequences. Colletotrichum boninense was used as the outgroup. Bootstrap support values ≥ 50 %, Bayesian posterior probability values ≥ 0.95 and RAxML bootstrap support values (ML ≥ 50 %) are shown at the nodes. Tree length = 1665, CI = 0.672, RI = 0.889, RC = 0.597, HI = 0.328. Ex-type strains are emphasised in bold.
Fig. 3.

Maximum parsimony tree of isolates in the Colletotrichum acutatum species complex obtained from a heuristic search of combined ACT, CHS-1, GAPDH, HIS3, ITS, and TUB2 gene sequences. Colletotrichum gloeosporioides was used as the outgroup. Bootstrap support values ≥ 50 % and Bayesian posterior probability values ≥ 0.95 are shown at the nodes. Tree length = 943, CI = 0.757, RI = 0.912, RC = 0.691, HI = 0.243. Ex-type strains are emphasised in bold.
Fig. 4.
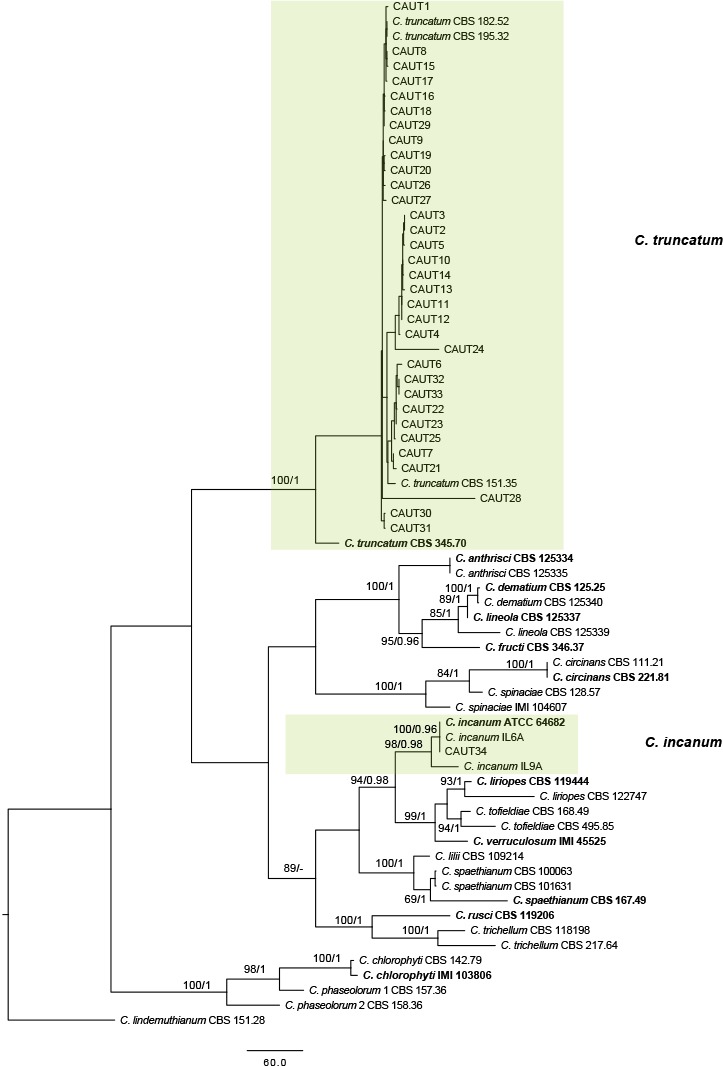
Maximum parsimony tree of Colletotrichum species with curved conidia obtained from a heuristic search of combined ACT, CHS-1, GAPDH, HIS3, ITS, and TUB2 gene sequences. Colletotrichum lindemuthianum was used as the outgroup. Bootstrap support values ≥ 50 % and Bayesian posterior probability values ≥ 0.95 are shown at the nodes. Tree length = 2853, CI = 0.467, RI = 0.859, RC = 0.401, HI = 0.533. Ex-type strains are emphasised in bold.
Fig. 6.
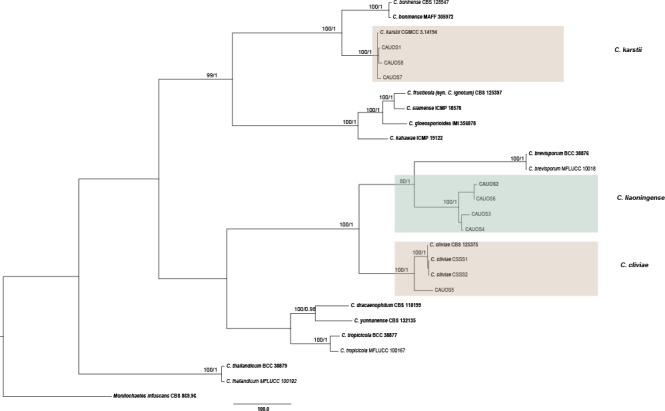
Maximum parsimony tree of isolates of Colletotrichum species in the fourth group obtained from a heuristic search of combined ACT, CAL, CHS-1, GAPDH, ITS, and TUB2 gene sequences. Monilochaetes infuscans was used as the outgroup. Bootstrap support values ≥ 50 % and Bayesian posterior probability values ≥ 0.95 are shown at the nodes. Tree length = 2913, CI = 0.717, RI = 0.870, RC = 0.624, HI = 0.283. Ex-type strains are emphasised in bold.
Pathogenicity
All tested isolates except that of C. endophytica were pathogenic to most of the detached ripe red chili fruits (Table 3). All Capsicum annuum and Ca. frutescens cultivars were susceptible to all tested Colletotrichum species (except C. endophytica), with disease scores from 5 to 9. Capsicum chinense was susceptible to most of the Colletotrichum species, except C. cliviae CAUOS5, C. endophytica CAUG28, and C. hymenocallidis CAUG9. Capsicum annuum and Ca. frutescens were the most susceptible, with average scores of 7. Three pathotypes (PC1-R, PC2-R, and PC3-R) were identified (Table 3) based on three differential reactions of tested strains with Capsicum chinense cv. Huangdijiao, Ca. annuum and Ca. frutescens. Host reactions of the mature green fruit were similar to those of the ripe fruit (Table 4). Similar to the ripe fruit, all three cultivars of the mature green fruit of Capsicum annuum, Zidantou, Denglongjiao, and Sanyingjiao were susceptible to all tested Colletotrichum species, except C. endophytica, with average scores from 5 to 6. Most of the Capsicum frutescens cultivars and Ca. chinense cv. Huangdijiao at the green fruit stage were susceptible to most isolates, except C. aenigma CAUG26, C. conoides CAUG17, C. gloeosporioides CAUG2, C. grossum CAUG7, C. fructicola CAUG1 and C. karstii CAUOS1. Five pathotypes were identified based on the differential reactions with Xiaomila, Shuangla, and Huangdijiao (Table 4). All of the pathogenic isolates formed sunken, brown to dark lesions on the fruits. No symptoms developed on the negative controls.
Prevalence of Colletotrichum species
To determine the prevalence of the Colletotrichum species associated with chili in China, the sample locations and the number of isolates were assessed for each species. Isolates with highly similar morphology and ITS sequences to those of the ex-type of C. truncatum appear to be most common (N = 422), representing 33 % of all isolates, and presenting in 56 % of all sampling locations (Fig. 5). All 34 isolates chosen from this group for multi-locus phylogenetic analysis were confirmed to be C. truncatum (Fig. 4). It therefore appears that C. truncatum is the most prevalent species of Colletotrichum on chili in China. The next most prevalent species included C. scovillei, C. gloeosporioides, C. fioriniae, and C. fructicola, which accounted for 21, 14, 14, and 13 % of all the isolates, respectively. The remaining species were detected in less than 3 % of the sampling locations.
Fig. 5.

Prevalence of Colletotrichum species on chili in China based on preliminary identifications. a. The percentage of isolates represented by the indicated Colletotrichum species on chili; b. number of sampling locations where the seven most prevalent species were isolated.
Taxonomy
Based on the morphology and the multi-locus phylogeny, the 121 isolates were assigned to 15 species. Seven species (C.aenigma, C. cliviae, C. endophytica, C. hymenocallidis, C. incanum, C. karstii, and C. viniferum) were reported from chili for the first time. Three other species (C. fioriniae, C. fructicola, and C. scovillei) were reported for the first time in China, and a further three species newly described.
Colletotrichum conoides Y.Z. Diao, C. Zhang, L. Cai & X.L. Liu, sp. nov. — MycoBank MB812003; Fig. 7
Fig. 7.
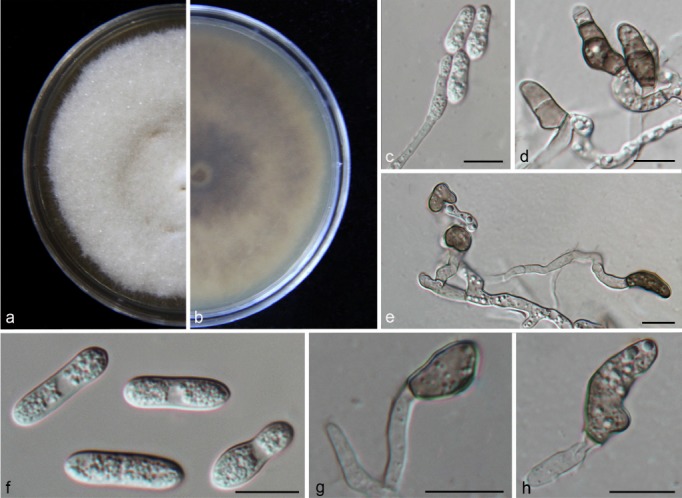
Colletotrichum conoides (CAUG17). a–b. Colonies on PDA above and below; c. conidiophores; d–e, g–h: appressoria; f. conidia. — Scale bars: c–h = 10 μm.
Etymology. Referring to the host variety (Capsicum annuum var. conoides) from which the fungus was first collected.
Colonies on PDA attaining 53–55 mm diam in 4 d at 28 °C; aerial mycelia greyish white; reverse light grey to medium grey with white margin. Chlamydospores not observed. Vegetative hyphae hyaline, smooth-walled, septate, branched. Conidiomata and setae not observed. Conidiophores formed directly on aerial mycelium, hyaline, aseptate. Conidiogenous cells hyaline, cylindrical to clavate, 22–30 × 3.5–5 μm, opening 2.5–3 μm. Conidia hyaline, aseptate, smooth-walled, cylindrical to clavate, both ends obtusely rounded, contents granular and mostly equally distributed, 13–17.5 × 5–6.5 μm (av. = 15.9 × 5.9 μm), L/W ratio = 2.7. Appressoria single or in small groups, medium to dark brown, aseptate, mostly ellipsoidal to irregular in outline, and crenate or deeply lobed at margin, 4–11.5 × 6–10.5 μm (av. = 8.35 × 7.1 μm), L/W ratio = 1.2. Sexual morph not observed after 8 wk.
Specimen examined. CHINA, Jiangsu Province, Nanjing City, on fruits of Capsicum annuum var. conoides, Sept. 2010, Y.Z. Diao (holotype HMAS 246481, ex-type living culture CGMCC 3.17615 = CAUG17 = LC6226); ibid., NJ26, living culture CAUG33; ibid., NJ27, living culture CAUG34.
Notes — Colletotrichum conoides is phylogenetically most closely related to C. hebeiense (Fig. 2). Sequence data from ITS and CHS-1 could not separate the two species, but they can be distinguished by GAPDH (12 bp), ACT (4 bp), or TUB (3 bp). The two species also differ in the following characteristics: the granules are uniformly distributed in the conidia of C. conoides but mostly present at the polar ends in the conidia of C. hebeiense; most appressoria of C. conoides are ovoid ellipsoidal with crenate or deeply lobed margin, while those of C. hebeiense are clavate to subglobose; conidia of C. conoides are slightly larger than those of C. hebeiense (13–17.5 × 5–6.5 μm vs 11.6–15.3 × 4.47–6.88 μm). In addition, C. conoides was described from Capsicum annuum var. conoides, while C. hebeiense was described from Vitis vinifera (Yan et al. 2015). A PHI test revealed no significant recombination event between C. conoides and C. hebeiense (Fig. 8).
Fig. 8.
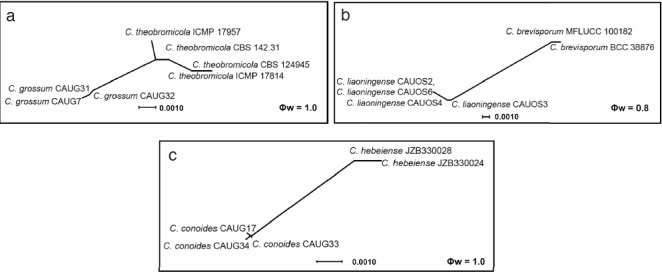
The results of the pairwise homoplasy index (PHI) test of closely related species using both LogDet transformation and splits decomposition. PHI test results (Φw) < 0.05 indicate significant recombination within the dataset.
Colletotrichum grossum Y.Z. Diao, C. Zhang, L. Cai & X.L. Liu, sp. nov. — MycoBank MB812006; Fig. 9
Fig. 9.
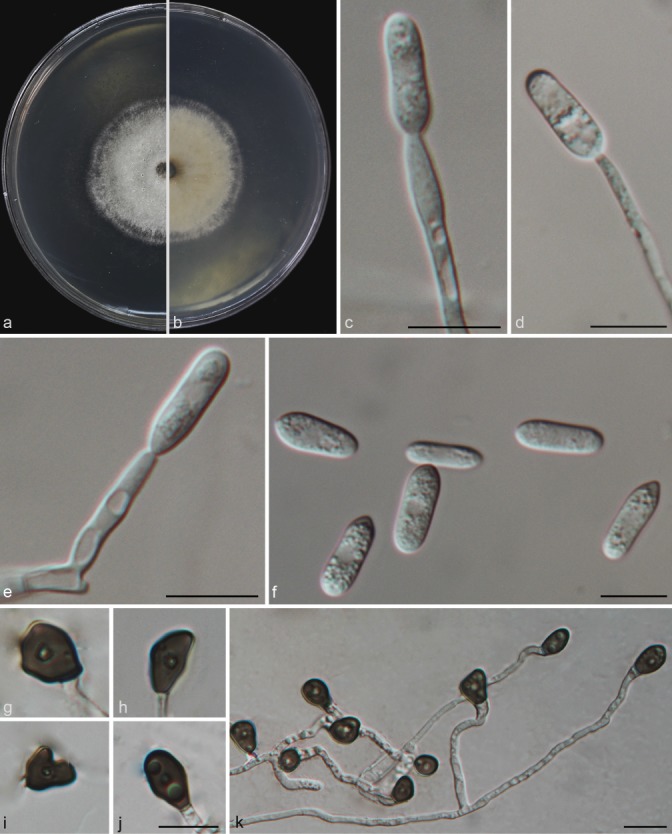
Colletotrichum grossum (CAUG7). a–b. Colonies on PDA above and below; c–e: conidiophores; f. conidia; g–k: appressoria. — Scale bars: c–f, j–k = 10 μm (j applied to g–j).
Etymology. Referring to the host variety (Capsicum annuum var. grossum) from which the fungus was first collected.
Colonies on PDA attaining 49–52 mm diam in 4 d at 28 °C; aerial mycelia white, reverse light grey with white margin. Chlamydospores not observed. Vegetative hyphae hyaline, smooth-walled, septate, branched. Conidiomata and setae not observed. Conidiophores formed directly on aerial mycelium, hyaline, aseptate. Conidiogenous cells hyaline, cylindrical to clavate, 22–32 × 3–3.5 μm, opening 2–2.5 μm. Conidia hyaline, aseptate, smooth-walled, cylindrical to clavate, both ends rounded or one end acute, contents granular and mostly present at the polar ends, 14.5–20.5 × 5–7.5 μm (av = 16.8 × 6.3 μm), L/W ratio = 2.7. Appressoria single, medium brown, aseptate, mostly ovoid or ellipsoidal to irregular in outline, and crenate in margin. 5.5–11.5 × 4–10.5 μm (av = 8.65 × 6.1 μm), L/W ratio = 1.4. Sexual morph not observed after 8 wk.
Specimen examined. CHINA, Hainan Province, Haikou city, on chili fruits (Capsicum annuum var. grossum), Oct. 2010, Y.Z Diao (holotype HMAS 246480, ex-type living culture CGMCC3.17614 = CAUG7 = LC6227); ibid., HN2, living culture CAUG31; ibid., HN3, living culture CAUG32.
Notes — Colletotrichum grossum is phylogenetically most closely related to C. theobromicola (Fig. 2). The sequence data of ITS and CAL do not separate the two species, but they can be distinguished by GAPDH (3 bp), ACT (5 bp), and TUB (8 bp). In morphology, C. grossum differs from C. theobromicola by having wider conidia (14.5–20.5 × 5–7.5 μm vs 14.5–18.7 × 4.5–5.5 μm) and colonies that are flat white rather than black as in C. theobromicola (Rojas et al. 2010). A PHI test revealed no significant recombination event between C. grossum and C. theobromicola (Fig. 8).
Colletotrichum liaoningense Y.Z. Diao, C. Zhang, L. Cai & X.L. Liu, sp. nov. — MycoBank MB812007; Fig. 10
Fig. 10.
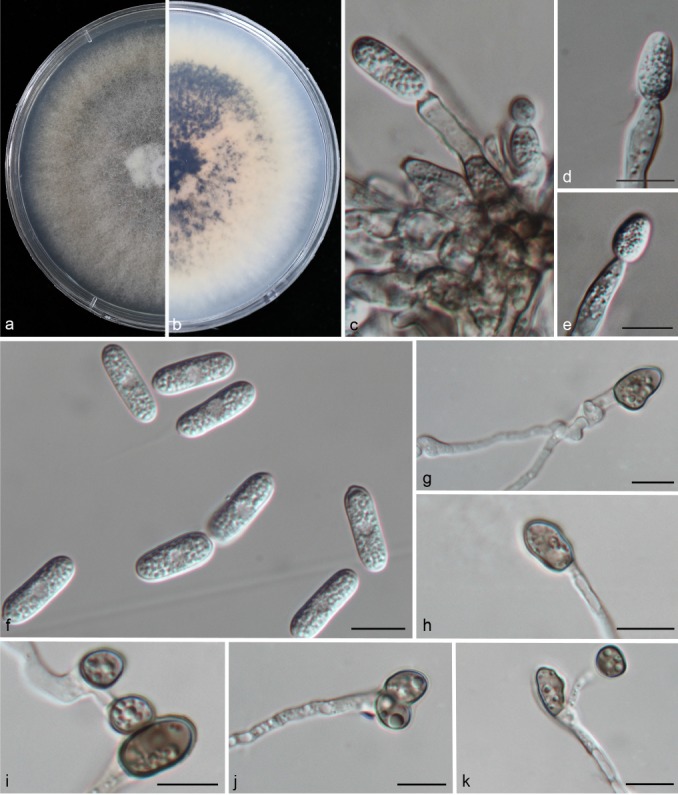
Colletotrichum liaoningense (CAUOS2). a–b. Colonies on PDA above and below; c–e: conidiophores; f. conidia; g–k: appressoria. — Scale bars: d–k = 10 μm (d applies to c–d).
Etymology. Referring to the province in China where the fungus was first collected.
Colonies on PDA attaining 48–51 mm diam in 4 d at 28 °C; aerial mycelia light grey, reverse medium to dark brown with white margin. Chlamydospores not observed. Vegetative hyphae hyaline, smooth-walled, septate, branched. Conidiomata acervular. Setae medium grey, smooth-walled to verruculose, 3–6-septate, 46–68 μm long, base cylindrical, conical, or slightly inflated, 4.5–6.5 μm diam at the widest part, tip rounded. Conidiophores formed directly on aerial mycelium, hyaline, aseptate. Conidiogenous cells hyaline, cylindrical to clavate, 27–30 × 3.5–4.5 μm, opening 2–4 μm. Conidia cylindrical to clavate, both ends rounded or one end acute, contents granular and mostly present at the polar ends, hyaline, aseptate, smooth-walled, 14–18.5 μm × 5–7.5 μm (av. = 16.3 × 6.1 μm), L/W ratio = 2.7. Appressoria single, medium to dark brown, aseptate, mostly ellipsoidal to irregular in outline, and crenate at margin, 3.5–5 × 2.5–4.5 μm (av. = 4.1 × 2.9 μm), L/W ratio = 1.4. Sexual morph not observed after 8 wk.
Specimen examined. CHINA, Xingcheng city, Liaoning Province on chili fruits (Capsicum annuum var. conoides), Oct. 2012, Y.Z. Diao (holotype HMAS 246479, ex-type living culture CGMCC3.17616 = CAUOS2 = LC6228); ibid., LN3, living culture CAUOS3; ibid., LN4, living culture CAUOS4; ibid., LN6, living culture CAUOS6.
Notes — Colletotrichum liaoningense is phylogenetically most closely related to C. brevisporum (Fig. 6). The sequence data from ITS and ACT could not separate the two species; however, they can be distinguished from each other via GAPDH (10 bp) or TUB (12 bp). The granules are equally distributed in the conidia of C. liaoningense but mostly present at the polar ends in conidia of C. brevisporum. The appressoria of C. liaoningense are smaller than those of C. brevisporum (3.5–5 × 2.5–4.5 μm vs 10–13 × 8–11 μm) (Noireung et al. 2012). A PHI test revealed no significant recombination event between C. liaoningense and C. brevisporum (Fig. 8).
DISCUSSION
Colletotrichum truncatum, the most frequently isolated species in this study, has been reported from more than 460 plant species (Farr & Rossman 2016). This taxon has also been shown to cause serious damage to chili production in Australia, China, India, Thailand, and other countries (Poonpolgul & Kumphai 2007, Than et al. 2008, Sharma et al. 2014, Diao et al. 2015). In China, C. truncatum has been reported from tomato, dragon fruit, pumpkin, and other crops (Chai et al. 2014, Cheng et al. 2014, Diao et al. 2014, Guo et al. 2014). Geographic populations of C. truncatum in China exhibit significant genetic differentiation and recombination abilities, which can probably be attributed to the prevalence of this species (Diao et al. 2015).
Colletotrichum gloeosporioides has been reported to infect chili in Australia, China, India, Korea, Thailand, the USA, and other countries (Shin et al. 1999, Kim et al. 2008, Than et al. 2008). However, a recent study revealed this taxon to be a species complex comprising many morphologically similar taxa (Weir et al. 2012). Therefore, this new classification system necessitates a re-investigation of species in the C. gloeosporioides species complex on chili, as species in this complex exhibit biological and physiological differences. In the current study, C. gloeosporioides s.str. and C. fructicola were revealed to be most prevalent in this complex, representing 47 % and 42 % of the isolates, respectively (Fig. 2). Colletotrichum fructicola was originally isolated from coffee berries (Prihastuti et al. 2009), and has since been found on a wide range of host plants (Weir et al. 2012). However, this is the first report of C. fructicola infecting chili. In previous studies, C. gloeosporioides s.str. was shown to be an uncommon pathogen on chili and other fruits in the tropics (Phoulivong et al. 2010). Additionally, we failed to isolate C. gloeosporioides s.str. from chili in the tropical regions of China, e.g. Hainan, south of Guangdong, and Yunnan provinces (Table 1), which suggested a significant effect of climate on the distribution of these pathogens. Pathogenicity of all obtained species from chili in this study was confirmed by inoculation tests, except for that of C. endophytica. Colletotrichum endophytica, which was originally reported as an endophytic fungus in tropical grasses (Manamgoda et al. 2013), did not show pathogenicity to any chili cultivars in our test, further underlining the possible endophytic nature of this species.
Colletotrichum acutatum is a commonly reported species, and causes anthracnose on numerous plants worldwide (Damm et al. 2012a). It was originally described from Carica papaya, Capsicum frutescens, and Delphinium ajacis in Australia (Simmonds 1965), but has subsequently been reported to infect chili in almost all pepper-growing countries, such as Australia, China, India, Korea, New Zealand, Thailand, and the USA (Than et al. 2008). Like C. gloeosporioides, C. acutatum has also been shown to represent a species complex (Damm et al. 2012a). Interestingly, C. acutatum s.str. was not found on chili in China (Fig. 3). Only C. scovillei and C. fioriniae were identified from this complex (Fig. 3).
No Colletotrichum species were detected on chili in Tibet and Xinjiang, despite the fact that several field trips have been made to these provinces, and attempts have been made for to isolate these fungi. The failure to detect Colletotrichum species from these regions might be explained by the high latitude, small growing area, dry climate, and high day/night variation in temperature. Colletotrichum fructicola and C. truncatum were isolated from leaves in the Jiangxi province, and were also found from fruits in other sampling regions. In previous studies, these two species were primarily isolated from fruits from various plants (Poonpolgul & Kumphai 2007, Than et al. 2008, Alaniz et al. 2015, Diao et al. 2015).
In summary, the current study represents the hitherto most intensive investigation of Colletotrichum species on chili in China, which revealed 15 species, with the dominant species being C. fioriniae, C. fructicola, C. gloeosporioides, C. scovillei, and C. truncatum. The information provided here could prove useful for the control of anthracnose on chili, as well as for the screening of new chili cultivars against anthracnose.
Acknowledgments
This work was supported by the Special Fund for Agro-scientific Research in the Public Interest of China (No. 201303023), and also partially supported by the National High Technology Research and Development Program of China (2012CB111401).
REFERENCES
- Alaniz S, Hernández L, Mondino P. 2015. Colletotrichum fructicola is the dominant and one of the most aggressive species causing bitter rot of apple in Uruguay. Tropical Plant Pathology 40: 265–274. [Google Scholar]
- Bailey JA, Jeger MJ. 1992. Colletotrichum: biology, pathology and control. CAB International, Wallingford. [Google Scholar]
- Cai L, Hyde KD, Taylor PWJ, et al. 2009. A polyphasic approach for studying Colletotrichum. Fungal Diversity 39: 183–204. [Google Scholar]
- Cai L, Jeewon R, Hyde KD. 2006. Phylogenetic investigations of Sordariaceae based on multiple gene sequences and morphology. Mycological Research 110: 137–150. [DOI] [PubMed] [Google Scholar]
- Carbone I, Kohn LM. 1999. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553–556. [Google Scholar]
- Chai AL, Zhao YJ, Shi YX, et al. 2014. Identification of Colletotrichum capsici (Syd.) Butler causing anthracnose on pumpkin in China. Canadian Journal of Plant Pathology 36: 121–124. [Google Scholar]
- Cheng BP, Huang YH, Peng AT, et al. 2014. First report of leaf and fruit spot of Citrus reticulata Blanco cv. Nian Ju caused by Colletotrichum truncatum in China. Plant Disease 98: 422. [DOI] [PubMed] [Google Scholar]
- Crouch JA. 2014. Colletotrichum caudatum s.l. is a species complex. IMA Fungus 5: 17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Gams W, Stalpers JA, et al. 2004a. MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19–22. [Google Scholar]
- Crous PW, Groenewald JZ, Risède JM, et al. 2004b. Calonectria species and their Cylindrocladium anamorphs: species with sphaeropedunculate vesicles. Studies in Mycology 50: 415–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm U, Cannon PF, Liu F, et al. 2013. The Colletotrichum orbiculare species complex: important pathogens of field crops and weeds. Fungal Diversity 61: 29–59. [Google Scholar]
- Damm U, Cannon PF, Woudenberg JHC, et al. 2012a. The Colletotrichum acutatum species complex. Studies in Mycology 73: 37–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm U, Cannon PF, Woudenberg JHC, et al. 2012b. The Colletotrichum boninense species complex. Studies in Mycology 73: 1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm U, O’Connell RJ, Groenewald JZ, et al. 2014. The Colletotrichum destructivum species complex – hemibiotrophic pathogens of forage and field crops. Studies in Mycology 79: 49–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm U, Woudenberg JHC, Cannon PF, et al. 2009. Colletotrichum species with curved conidia from herbaceous hosts. Fungal Diversity 39: 45–87. [Google Scholar]
- Dean R, Van Kan JA, Pretorius ZA, et al. 2012. The Top 10 fungal pathogens in molecular plant pathology. Molecular Plant Pathology 13: 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao YZ, Zhang C, Lin D, et al. 2014. First report of Colletotrichum truncatum causing anthracnose of tomato in China. Plant Disease 98: 687. [DOI] [PubMed] [Google Scholar]
- Diao YZ, Zhang C, Xu JP, et al. 2015. Genetic differentiation and recombination among geographic populations of the fungal pathogen Colletotrichum truncatum from chili peppers in China. Evolutionary Applications 8: 108–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr DF, Rossman AY. 2016. Fungal Databases, Systematic Mycology and Microbiology Laboratory, ARS, USDA; Retrieved February 6, 2016, from http://nt.ars-grin.gov/fungaldatabases/. [Google Scholar]
- Glass NL, Donaldson GC. 1995. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61: 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo LW, Wu YX, Ho HH, et al. 2014. First report of dragon fruit (Hylocereus undatus) anthracnose caused by Colletotrichum truncatum in China. Journal of Phytopathology 162: 272–275. [Google Scholar]
- Huson DH. 1998. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14: 68–73. [DOI] [PubMed] [Google Scholar]
- Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution 23: 254–267. [DOI] [PubMed] [Google Scholar]
- Kanto T, Uematsu S, Tsukamoto T, et al. 2014. Anthracnose of sweet pepper caused by Colletotrichum scovillei in Japan. Journal of General Plant Pathology 80: 73–78. [Google Scholar]
- Katoh K, Toh H. 2010. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 26: 1899–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JT, Park SY, Choi W, et al. 2008. Characterization of Colletotrichum isolates causing anthracnose of pepper in Korea. The Plant Pathology Journal 24: 17–23. [Google Scholar]
- Kohli Y, Brunner LJ, Yoell H, et al. 1995. Clonal dispersal and spatial mixing in populations of the plant pathogenic fungus, Sclerotinia sclerotiorum. Molecular Ecology 4: 69–77. [Google Scholar]
- Li Q, Han YZ, Zhang GC. 2009. Status and development trends of hot pepper industry home and abroad. Hubei Agricultural Science 9: 2278–2281. [Google Scholar]
- Liao CY, Chen MY, Chen YK, et al. 2012. Characterization of three Colletotrichum acutatum isolates from Capsicum spp. European Journal of Plant Pathology 133: 599–608. [Google Scholar]
- Lin Q, Lv Z, Huang R, et al. 2004. Screening of pepper germplasm for resistance to TMV, CMV, phytophthora blight and anthracnose. Southwest China Journal of Agricultural Sciences 18: 108–110. [Google Scholar]
- Liu F, Damm U, Cai L, et al. 2013. Species of the Colletotrichum gloeosporioides complex associated with anthracnose diseases of Proteaceae. Fungal Diversity 61: 89–105. [Google Scholar]
- Liu JK, Phookamsak R, Doilom M, et al. 2012. Towards a natural classification of Botryosphaeriales. Fungal Diversity 57: 149–210. [Google Scholar]
- Manamgoda DS, Udayanga D, Cai L, et al. 2013. Endophytic Colletotrichum from tropical grasses with a new species C. endophytica. Fungal Diversity 61: 107–115. [Google Scholar]
- Mongkolporn O, Montri P, Supakaew T, et al. 2010. Differential reactions on mature green and ripe chili fruit infected by three Colletotrichum spp. Plant Disease 94: 306–310. [DOI] [PubMed] [Google Scholar]
- Montri P, Taylor PWJ, Mongkolporn O. 2009. Pathotypes of Colletotrichum capsici, the causal agent of chili anthracnose, in Thailand. Plant Disease 93: 17–20. [DOI] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research 8: 4321–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noireung P, Phoulivong S, Liu F, et al. 2012. Novel species of Colletotrichum revealed by morphology and molecular analysis. Cryptogamie Mycologie 33: 347–362. [Google Scholar]
- Nylander JAA. 2004. MrModelTest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University. [Google Scholar]
- O’Donnell K, Cigelnik E. 1997. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7: 103–116. [DOI] [PubMed] [Google Scholar]
- O’Donnell K, Nirenberg HI, Aoki T, et al. 2000. A multigene phylogeny of the Gibberella fujikuroi species complex: Detection of additional phylogenetically distinct species. Mycoscience 41: 61–78. [Google Scholar]
- Perfect SE, Hughes HB, O’Connell RJ, et al. 1999. Colletotrichum: a model genus for studies on pathology and fungal – plant interactions. Fungal Genetics and Biology 27: 186–198. [DOI] [PubMed] [Google Scholar]
- Phoulivong S, Cai L, Chen H, et al. 2010. Colletotrichum gloeosporioides is not a common pathogen on tropical fruits. Fungal Diversity 44: 33–43. [Google Scholar]
- Poonpolgul S, Kumphai S. 2007. Chilli pepper anthracnose in Thailand. In: The First International Symposium on Chili Anthracnose: 23 Convention Center, Seoul National University, Korea. [Google Scholar]
- Prihastuti H, Cai L, Chen H, et al. 2009. Characterization of Colletotrichum species associated with coffee berries in northern Thailand. Fungal Diversity 39: 89–109. [Google Scholar]
- Quaedvlieg W, Binder M, Groenewald JZ, et al. 2014. Introducing the Consolidated Species Concept to resolve species in the Teratosphaeriaceae. Persoonia 33: 1–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas EI, Rehner SA, Samuels GJ, et al. 2010. Colletotrichum gloeosporioides s.l. associated with Theobroma cacao and other plants in Panama: multilocus phylogenies distinguish host-associated pathogens from asymptomatic endophytes. Mycologia 102: 1318–1338. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Sharma G, Kumar PA, Damodara SB. 2014. Infra-specific diversity of Colletotrichum truncatum associated with chilli anthracnose in India based on microsatellite marker analysis. Archives of Phytopathology and Plant Protection 47: 2509–2523. [Google Scholar]
- Sharma G, Shenoy BD. 2014. Colletotrichum fructicola and C. siamense are involved in chilli anthracnose in India. Archives of Phytopathology and Plant Protection 47: 1179–1194. [Google Scholar]
- Shin H, Xu T, Zhang C, et al. 1999. The comparative study of capsicum anthracnose pathogens from Korea with that of China. Journal of Zhejiang University (Agriculture and Life Sciences) 26: 629–634. [Google Scholar]
- Simmonds JH. 1965. A study of the species of Colletotrichum causing ripe fruit rots in Queensland. Queensland Journal of Agricultural and Animal Science 22: 437–459. [Google Scholar]
- Swofford D. 2002. PAUP 4.0 b10: Phylogenetic analysis using parsimony (*and other methods). Computer programme. Sinauer Associates, Sunderland, MA, USA. [Google Scholar]
- Templeton MD, Rikkerink EHA, Solon SL, et al. 1992. Cloning and molecular characterization of the glyceraldehyde-3-phosphate dehydrogenase-encoding gene and cDNA from the plant pathogenic fungus Glomerella cingulata. Gene 122: 225–230. [DOI] [PubMed] [Google Scholar]
- Than PP, Prihastuti H, Phoulivong S, et al. 2008. Chilli anthracnose disease caused by Colletotrichum species. Journal of Zhejiang University Science B 9: 764–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Johnston PR, Damm U. 2012. The Colletotrichum gloeosporioides species complex. Studies in Mycology 73: 115–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee J, et al. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, et al. (eds), PCR protocols: a guide to methods and applications: 315–322. Academic Press, San Diego, California, USA. [Google Scholar]
- Yan JY, Jayawardena M, Goonasekara ID, et al. 2015. Diverse species of Colletotrichum associated with grapevine anthracnose in China. Fungal Diversity 71: 233–246. [Google Scholar]


