Abstract
Retroviral integrases (IN) catalyze the integration of the reverse-transcribed viral DNA into the host genome, an essential process leading to virus replication. For Moloney murine leukemia virus (M-MuLV) IN, the limited solubility of the recombinant protein has restricted the development of biophysical and structural analyses. Herein, recombinant M-MuLV IN proteins, either full length or two nonoverlapping domain constructs, were purified under non-denaturing conditions from solubilized bacterial extracts by Ni2+-NTA resins. Additionally, WT IN was further purified by heparin chromatography. All of the purified proteins were shown to be active and stable. WT M-MuLV IN chromatographed with a peak corresponding with a dimer by gel filtration chromatography. In contrast, the single point mutant C209A IN migrated predominantly as a tetramer. For both proteins, fractions in equilibrium between dimers and tetramers were competent to assemble concerted two-end integrations and yielded a unique strand-transfer profile in the presence of a 28-mer U5 oligonucleotide substrate, indicative of a distinct conformation within the synaptic complex. This specific target-site selection was not observed with a shorter 20-mer U5 substrate. These studies provide the foundation for biophysical and structural analysis on M-MuLV IN and the mechanism of retroviral integration.
Introduction
An essential step of the retroviral life cycle is the integration of the reverse-transcribed viral DNA into the host genome. This two-step process is carried out by the viral enzyme integrase (IN), encoded by the pol gene. First, two deoxynucleotides are removed from both 3′ termini of the viral long terminal repeats (LTRs), exposing the conserved CA at the recessed 3′ ends (3′-processing) (Brown et al., 1989; Fujiwara and Mizuuchi, 1988; Katzman et al., 1989; Roth et al., 1989). The 3′ ends of the viral DNA are then joined to 5′ staggered sites in the target DNA in a concerted transesterification reaction (strand transfer) (Craigie et al., 1990; Engelman et al., 1991; Katz et al., 1990). Integration is completed by repair of the 5′ overhang of the viral LTR and the single-strand gaps flanking the integrated viral DNA, presumably by host enzymes. Repair of the single-strand gaps creates the hallmark duplication of the target DNA sequence flanking the retroviral integration site.
Sequence comparison and mutational analysis have identified three functional domains in the retroviral IN proteins. The N-terminal domain contains a conserved His-His/Cys-Cys (HHCC) region (Johnson et al., 1986), which can coordinate a zinc ion in an ordered conformation with increased α-helical content (Burke et al., 1992; McEuen et al., 1992; Yang et al., 1999a). For Moloney murine leukemia virus (M-MuLV), the HHCC is essential for IN function in vivo and optimal integration in vitro (Jonsson et al., 1996; Roth et al., 1990). Biophysical and biochemical analyses showed that the HHCC domain is involved in protein multimerization (Lee et al., 1997; Yang et al., 1999a; Zheng et al., 1996) and it is required for the formation of stable IN–LTR complexes (Donzella et al., 1996; Engelman et al., 1993; VanDenEnt et al., 1999; Vink et al., 1994). For M-MuLV IN, we have previously shown that the isolated HHCC domain forms dimers in solution (Yang et al., 1999a). Preincubation of the isolated M-MuLV HHCC domain with the LTR substrate increases the efficiency of two-end concerted integration products, in a complementation assay with a core/C-terminus IN construct (Yang and Roth, 2001).
The central core domain of IN is characterized by a conserved D-D(35)-E triad motif. Mutation of the conserved aspartic or glutamic acid residues results in loss of all catalytic activities, indicating a role for these three residues in active site function (Drelich et al., 1992; Engelman and Craigie, 1992; Kulkosky et al., 1992; Leavitt et al., 1993). Solution of crystal structures of isolated catalytic core domains of human immunodeficiency virus type 1 (HIV-1) and avian sarcoma virus (ASV) IN revealed that the catalytic domains of retroviral INs fold with similar topology (Bujacz et al., 1995; Dyda et al., 1994). Recently, structures of the core plus the C-terminus domains of HIV-1 IN (Chen et al., 2000a) and HIV-1 core with the N-terminal HHCC domains (Wang et al., 2001) have been solved. In these models, the catalytic triad is on the outer faces of the dimer.
The C-terminal region is less conserved and has been demonstrated to possess nonspecific DNA-binding activity (Khan et al., 1991; Mumm and Grandgenett, 1991; Vink et al., 1993; Woerner and Marcus-Sekura, 1993). The NMR structure of the DNA-binding domain of HIV-1 IN appears as a saddle-shaped dimer (Lodi et al., 1995), and the two-domain structure with the catalytic core shows flexibility of the C-terminus domain with respect to the catalytic core (Chen et al., 2000a, 2000b).
Although the independently folding domains of IN have been delineated and some of their structures solved, the manner in which these domains interact to form the catalytically active protein is still not understood. Evidence that integrase is a multimeric enzyme has been obtained through direct physical measurement of purified IN, cross-linking experiments (Gao et al., 2001; Heuer and Brown, 1998), and in vitro complementation analyses (Engelman et al., 1993; Jonsson et al., 1996; vanGent et al., 1993; Yang et al., 1999a; Yang and Roth, 2001). Purified recombinant INs can exist in a dynamic equilibrium that includes monomers, dimers, tetramers, and higher-order oligomers (Bao et al., 2003; Jones et al., 1992; Sherman and Fyfe, 1990; Vincent et al., 1993). Sedimentation analysis of ASV IN along with activity assays has revealed that the enzyme is active as a multimer, minimally a dimer which assembles to a tetramer in the presence of DNA (Bao et al., 2003; Jones et al., 1992). For M-MuLV integrase, the limited solubility of the recombinant protein has impeded structural analyses and detailed studies of subunit composition.
Multiple strategies have been developed to purify retro-viral INs due to their limited solubility. The expression and purification of isolated IN regions have facilitated the biochemical and biophysical characterization of isolated domains. Full-length, recombinant M-MuLV IN proteins required extraction from insoluble fractions under harsh denaturing conditions followed by slow refolding schemes of renaturation (Jonsson et al., 1993). Structural studies of HIV-1, Rous sarcoma virus (RSV), and simian immunodeficiency virus (SIV) INs have been greatly advanced through the mutational analysis of hydrophobic residues for improved solubility. For each, one point mutation improved the solubility and facilitated structural studies, including F185K for HIV-1 (Jenkins et al., 1996), F199K for RSV (Hyde et al., 1999), and F185H for SIV (Li et al., 1999).
In this study, recombinant tagged M-MuLV integrase proteins were purified under nondenaturing conditions from solubilized bacterial extracts. All of the purified IN proteins were shown to be active and stable in solution. Size fractionation of wild-type (WT) and a single point mutant C209A identified multimerization states competent to assemble concerted two-end integrations. These studies provide the foundation for structural analysis on M-MuLV integrase and retroviral integration.
Results
M-MuLV IN constructs
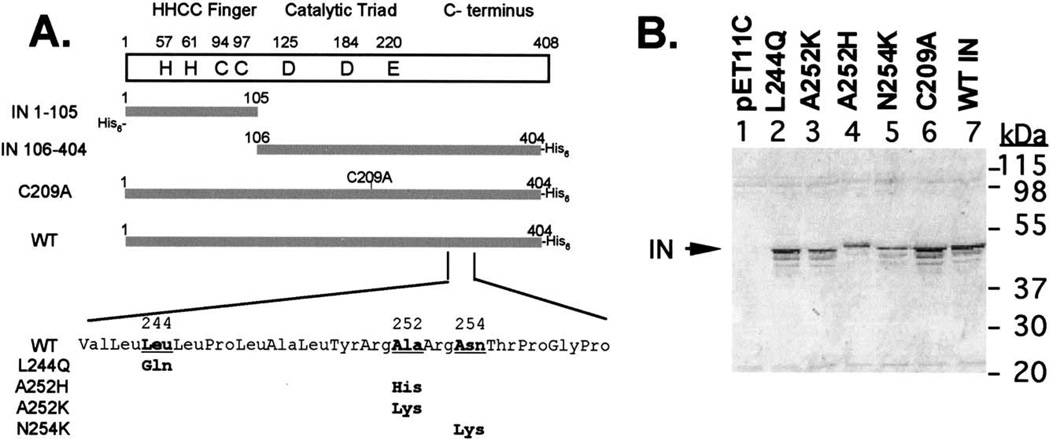
A schematic representation of the M-MuLV IN and mutants utilized in this study is shown in Fig. 1A. Previous work in our laboratory has shown functional complementation between two nonoverlapping IN domains, consisting of the HHCC domain (IN 1–105) and the core/C-terminal region (IN 106–404) (Yang et al., 1999a; Yang and Roth, 2001). Recombinant, purified C209A IN is active using in vitro oligonucleotide-based assays (Jonsson et al., 1996); however, in vivo, passage of virions bearing IN C209A are extremely delayed (Donzella et al., 1998). Modification of Cys209 with N-ethylmaleimide (NEM) blocked IN function and complementation with IN 1–105. C209A IN was insensitive to modification with NEM (Donzella et al., 1998) and resulted in the loss of complementation with IN 1–105 (Jonsson et al., 1996). M-MuLV C209A IN was included in these studies due to the implication of the central cysteine residue in IN multimerization.
Fig. 1.
(A) Illustration of IN proteins used in this study. The name of each construct is indicated to the left of each protein. (B) Solubility of the point mutants generated on the full-length M-MuLV IN protein; 0.005% of the soluble fraction (1 mL) for each mutant protein was loaded on SDS–PAGE and analyzed by Western blotting with anti-IN antibody (Tanese et al., 1986). Migration of molecular weight markers are indicated.
Studies of the HIV-1 (Jenkins et al., 1996), RSV (Hyde et al., 1999), and SIV (Li et al., 1999) IN proteins have been greatly advanced through the mutational analysis of hydrophobic residues for improved solubility. The Genthreader program (Jones, 1999) was applied to align and thread the M-MuLV IN against a known IN structure for steric analysis. By this approach, A252 within M-MuLV IN was identified as the equivalent to HIV-1 IN F185, and N254 was found to be equivalent to RSV IN F199 (data not shown). These residues were targeted for mutagenesis (Fig. 1A): Ala252 was replaced by His and Lys; N254 was mutated to Lys. Additionally, a third substitution was generated within a region of high hydrophobicity to replace Leu244 to Gln.
Characterization of M-MuLV IN and mutants from the soluble fraction
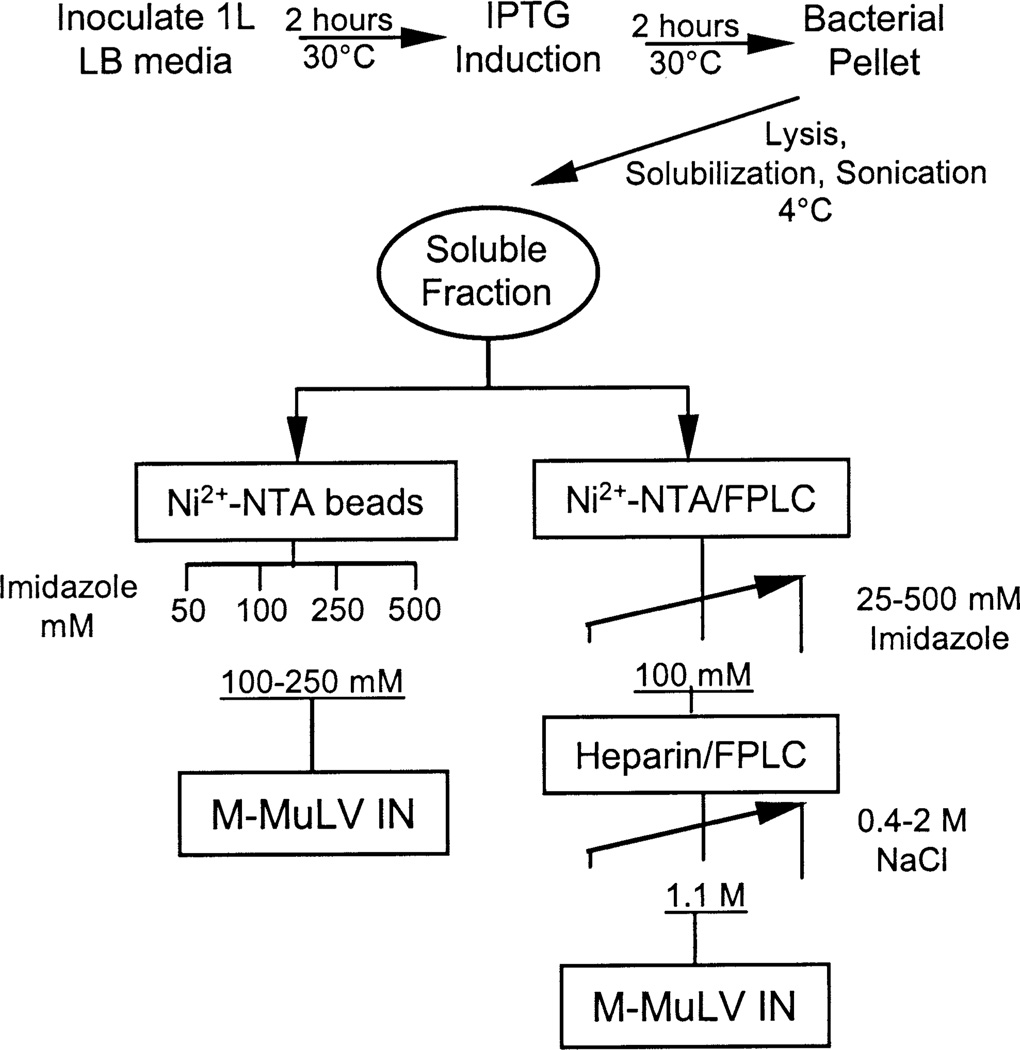
Fig. 2 shows a schematic of the expression and purification procedure utilized to obtain purified WT M-MuLV IN from solubilized bacterial extracts. A similar approach has been recently used to express and purify HTLV-2 IN (Pieper and Jonsson, 2002) and the Sin Nombre virus N protein (Jonsson et al., 2001), a member of the hantavirus genera, family Bunyaviridae. Overnight bacterial cultures were diluted and induced with IPTG for 2 h at 30°C. Bacterial pellets were resuspended in a solubilization buffer containing 300 mM NaCl and 10 mM of the zwitterionic Chaps detergent. Bacterial lysis was triggered at 4°C by lysozyme, Dounce homogenization, and extensive but controlled sonication cycles at 4°C. Sonication in the presence of Chaps and high salt released IN proteins from cellular nucleic acid into a soluble fraction. The soluble fraction was clarified by centrifugation and analyzed for solubility or subjected to further purification as discussed below.
Fig. 2.
Purification scheme. Major steps in order to obtain soluble fraction are shown. Below, the two procedures to obtain purified M-MuLV IN proteins. Left: Proteins bound to Ni2+-NTA beads are eluted with the indicated solutions of imidazole. Right: Proteins bound to Ni2+-NTA column/FPLC system are eluted with an imidazole gradient running between the indicated concentrations. Pooled proteins are then loaded into a heparin column/FPLC system and eluted with a NaCl gradient running between the indicated concentrations.
Analysis of the solubility of WT and mutant M-MuLV IN
Four mutations, A252H, A242K, N254K, and L244Q, were introduced into M-MuLV IN within the region homologous to that which resulted in increased solubility for the HIV-1 (Jenkins et al., 1996), RSV (Hyde et al., 1999), and SIV (Li et al., 1999) IN proteins (Fig. 1A). The solubility of these four point-mutant proteins and C209A IN were compared to that of the WT IN protein in the context of the current solubilization conditions. Cultures were induced with IPTG and lysed, and soluble fractions were obtained under conditions similar to those for purifying the wild-type protein (Fig. 2). A representative Western blot of SDS– PAGE of the soluble fractions from the different mutants and the parental WT IN is shown in Fig. 1B. The 45-kDa protein corresponding to M-MuLV IN was visible in the soluble fraction and induced in the presence of IPTG (compare control pET11C (lane 1) to WT IN (lane 7)). The 45-kDa protein detected in the L244Q, A252H, A252K, and N254L IN mutants was comparable to the WT IN (lanes 2–5). Interestingly, the migration of IN A252H was altered compared to the related IN A252K substitution (compare lane 4 to lane 3). A small increase in the level of 45-kDa IN was detected in the soluble fraction of C209A IN when compared with that of the WT IN protein (compare lanes 6 and 7). This was further analyzed in a large-scale purification of the proteins (see below). Western blot analysis identified two additional faster migrating species. These are predicted to be N-terminal truncations, as they did not bind to Ni+2 NTA affinity columns (see below).
Purification of M-MuLV IN and subdomains from the soluble fraction
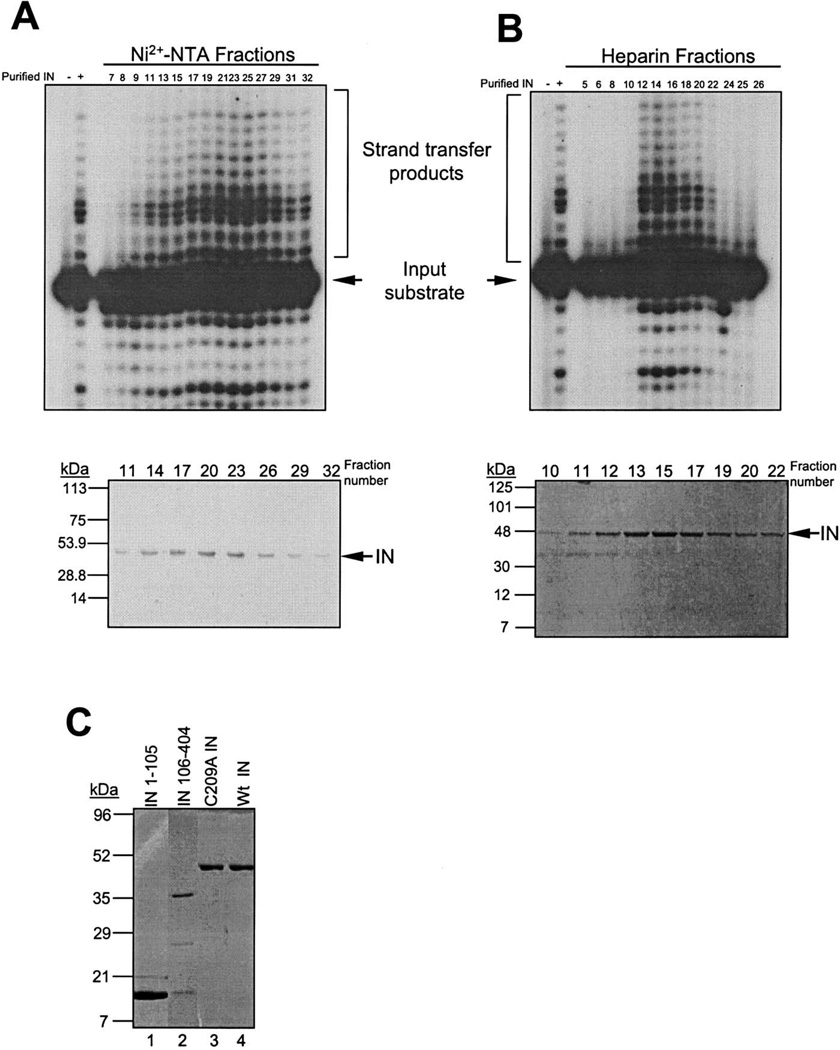
The soluble fraction was isolated after clarification by centrifugation and was subjected to two different purification schemes, one utilizing batch purification and the other FPLC column chromatography (Fig. 2). In order to obtain highly purified full-length IN protein, the soluble fraction was applied to a Ni2+-NTA resin column using a FPLC system (Fig. 2, right). After washing the column, proteins were eluted with a linear imidazole gradient. Fractions were subjected to SDS-PAGE and assayed for strand-transfer activity. This assay uses a 5′-labeled oligonucleotide substrate mimicking the U5 LTR terminus, which lacks the two terminal bases removed during 3′-processing, thus exposing the 5′-CA-3′ end, as illustrated in Fig. 3A. A representative profile of activity of the eluted fractions from the Ni2+-NTA column is shown in Fig. 4A. The predominant protein detected within the peak of integrase activity (fractions 21–25) was the 45-kDa IN product, visualized by Coomassie staining (Fig. 4A, lower panel). The peak of strand-transfer activity corresponded with 100 mM imidazole. Although the bulk of eluted IN protein shows a significant level of purification, pooled fractions were subjected to further purification through a FPLC–heparin resin. Bound proteins were then subjected to elution by a linear gradient of NaCl (0.4–2 M, Fig. 2) and fractions were assayed by strand-transfer activity and SDS–PAGE of the fractions. A representative column profile of WT IN activity is shown in Fig. 4B. There is a sharper peak of strand-transfer activity (i.e., fractions 12–18, upper panel) at 1.1 M NaCl which correlates with the elution of the protein as visualized by Coomassie staining (i.e., fractions 12–19, Fig. 4B, bottom panel). Furthermore, a smaller protein contaminant eluted prior to the 45-kDa IN protein peak. The active WT IN fractions were pooled giving a yield of 0.83 mg of highly purified active protein per liter of induced bacterial culture.
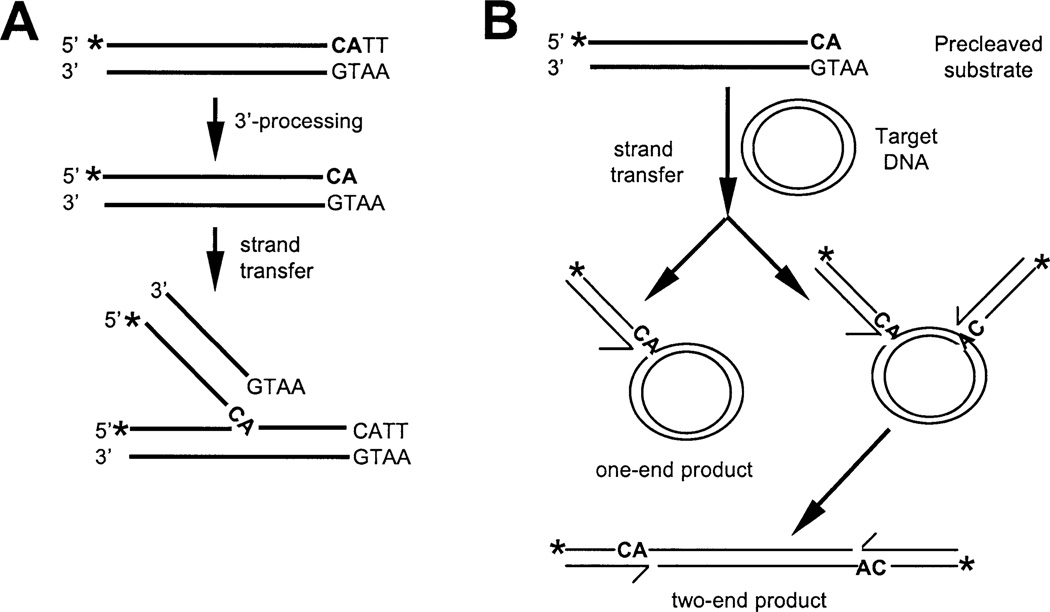
Fig. 3.
(A) Integration assays based on oligonucleotide 5′-labeled substrates. 3′-processing and strand-transfer reactions are indicated. (B) Concerted two-end integration assay. Target DNA and the two major types of reaction products are shown. The third product (not indicated) is identical to that shown in (A).
Fig. 4.
Activity column profiles from Ni2+-NTA/FPLC (A) or from heparin column/FPLC system (B). Fractions were assayed for strand-transfer activity (upper panels) and SDS-PAGE and Coomassie staining (lower panels). (C) M-MuLV IN proteins purified using batch elution. Lane 1, IN 1–105; lane 2, IN 106–404; lane 3, C209A IN; lane 4, WT IN.
Rapid purification of multiple constructs was obtained using batch purification on Ni2+-NTA. Fig. 4C summarizes the purified fractions of IN 1–105, IN 106–404, C209A IN, and WT IN. Fractions were bound in batch to Ni2+-NTA and eluted at 50, 100, 250, and 500 mM imidazole steps. Fractions were pooled based on purity across the individual steps. For IN 1–105 (Fig. 4C, lane 1), C209A IN (Fig. 4C, lane 3), and WT IN (Fig. 4C, lane 4), fractions were pooled from the 50 and 100 mM imidazole steps. For IN 106–404, the fractions were pooled from the 50–250 mM steps (Fig. 4C, lane 2). Two additional proteins of Mr 26,000 and 15,000 are detected in the IN 106–404 preparation. The 15-kDa protein was recognized by anti-IN antibodies on Western blot (data not shown) and therefore represents a low level of breakdown product. Starting from 1 L of induced cultures, this purification scheme yield 3 mg of IN 1–105, 1.65 mg of IN 106–404, 0.3 mg of C209A IN, and 0.15 mg of WT IN. These fractions were suitable for the biochemical studies on the M-MuLV IN proteins presented below.
Catalytic properties of the purified IN proteins
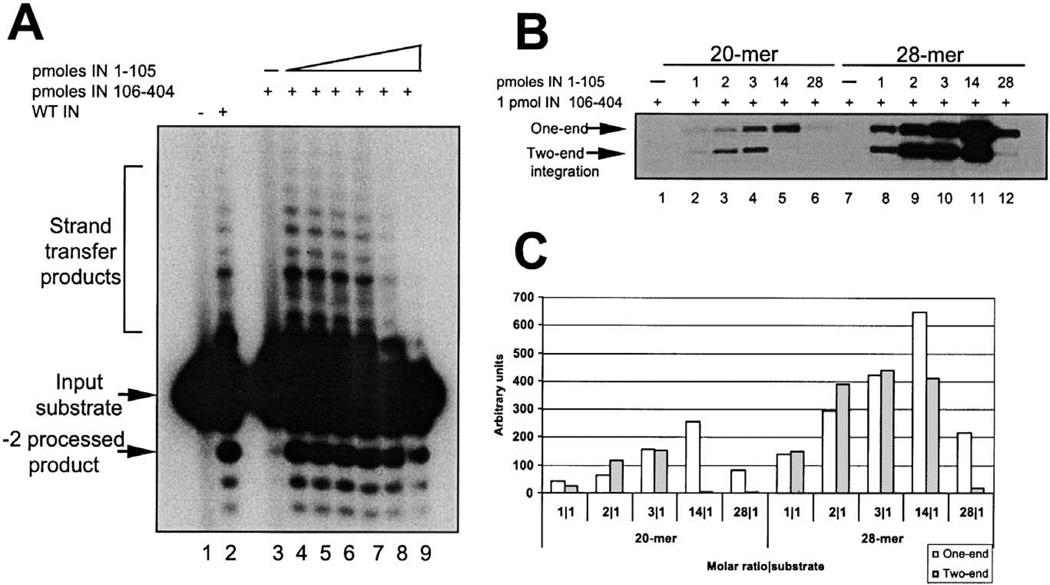
Functional complementation between two nonoverlapping fragments of M-MuLV IN, the HHCC N-terminal domain (IN 1–105), and the core/C-terminal region (IN 106–404) is observed in vitro (Yang et al., 1999a; Yang and Roth, 2001). The purified IN proteins used for those studies were extracted from the insoluble fractions followed by slow steps of renaturation (Jonsson et al., 1993). To examine the activity of the protein subdomains purified from the soluble fraction, the activity of the nonoverlapping domains was determined in a complementation assay for the 3′-processing reaction. In this assay, a 20-mer 5′-labeled blunt-ended oligonucleotide mimicking the terminal U5 at the LTR is used as the substrate for the reaction (Fig. 3A). Upon resolution of the products on denaturing electrophoresis, both a −2 processed product migrating larger than the input substrate and heterogeneous sized strand-transfer products migrating larger than the input substrate can be visualized. Fig. 5A shows a complementation assay in which the catalytic subunit of IN (IN 106–404) was held constant with a titration of the N-terminal domain (IN 1–105) in the reaction. No products were detected in the absence of enzyme (Fig. 5A, lane 1) or in the presence of IN 106–404 alone (Fig. 5A, lane 3). Upon addition of the HHCC domain (for a molar ratio IN 1–105:IN 106–404 of 2.3), both the 3′-processing and the strand-transfer products can be visualized (Fig. 5A, lane 4). Higher levels of the HHCC domain in the reaction inhibited catalysis (lanes 5–9). Optimal activity was observed at 10 mM MnCl2 and 100 mM of KCl. A low level of strand-transfer activity was observed in 1 mM of MgCl2 (data not shown).
Fig. 5.
Complementation assays of purified IN 1–105 and IN 106–404. (A) 3′-processing and strand-transfer reactions using 0.5 pmol of a blunt-end 20-mer-labeled oligonucleotide-based assay. IN 106–404 (1 pmol) was incubated alone (lane 3), or in the presence of IN 1–105 at 2.3 pmol (lane 4), 4.6 pmol (lane 5), 9.2 pmol (lane 6), 13.8 pmol (lane 7), 27 pmol (lane 8), and 55 pmol (lane 9). Products were resolved by 16% sequencing gels. Lanes 1 and 2 correspond to reactions in the absence of presence of 5 pmol of purified WT IN protein, respectively. (B) Concerted two-end integration assay; 1 pmol of IN 106–404 IN was assayed with increasing amounts of IN 1–105 using a 20-mer (lanes 1–7) or with a 28-mer (lanes 8–14) donor-labeled substrate and a target DNA. Reactions products were resolved by agarose gels and the radioactive bands quantified as shown in (C).
The catalytic activity of the nonoverlapping IN fragments was also assayed for concerted two-end integration (Yang and Roth, 2001). In this assay, the 5′-labeled precleaved oligonucleotide substrate is used as the donor DNA and a circular plasmid is the target DNA for integration (Fig. 3B). Two different length U5 oligonucleotide mimics were assayed: a 20-mer and a 28-mer (Fig. 5B). The IN 1–105 domain was titrated against 1 pmol of IN 106–404. Reaction products were resolved by agarose gels (Fig. 5B) and quantified (Fig. 5C). The efficiency of both one-end and two-end integration reactions is reproducibly increased four- to fivefold in the presence of the 28 versus the 20-mer substrate (compare panels 20-mer and 28-mer, Fig. 5C). This result suggests that longer substrates promote a more efficient assembly of a catalytic competent complex and/or further stabilize the interactions between the domains. This is of interest, as the region between positions 20 and 28 is outside of the conserved 13-bp inverted repeat sequence at the termini of the LTR.
Despite the efficiency differences between the 20-mer and the 28-mer U5 LTR substrate, the single-end and the two-end products were similar with respect to the ratios of IN 1–105 to IN 106–404. Preferential synthesis of two-end integration products was observed at an IN 1–105:IN 106–404 ratio of 2:1 (lanes 3 and 9, Fig. 5B). At a molar ratio IN 1–105:IN 106–404 of 3:1, the overall yield of products increased; however, the ratio of one-end:two-end integration was near stoichiometric (Fig. 5C). The two-end integration products were more sensitive to the presence of excess of IN 1–105 zinc-binding domain (Fig. 5B, lanes 5 and 11). The presence of a large excess of IN 1–105 (28-fold molar excess over IN 106–404, Fig. 5B, lanes 6 and 12) was inhibitory to the two-end integration reaction.
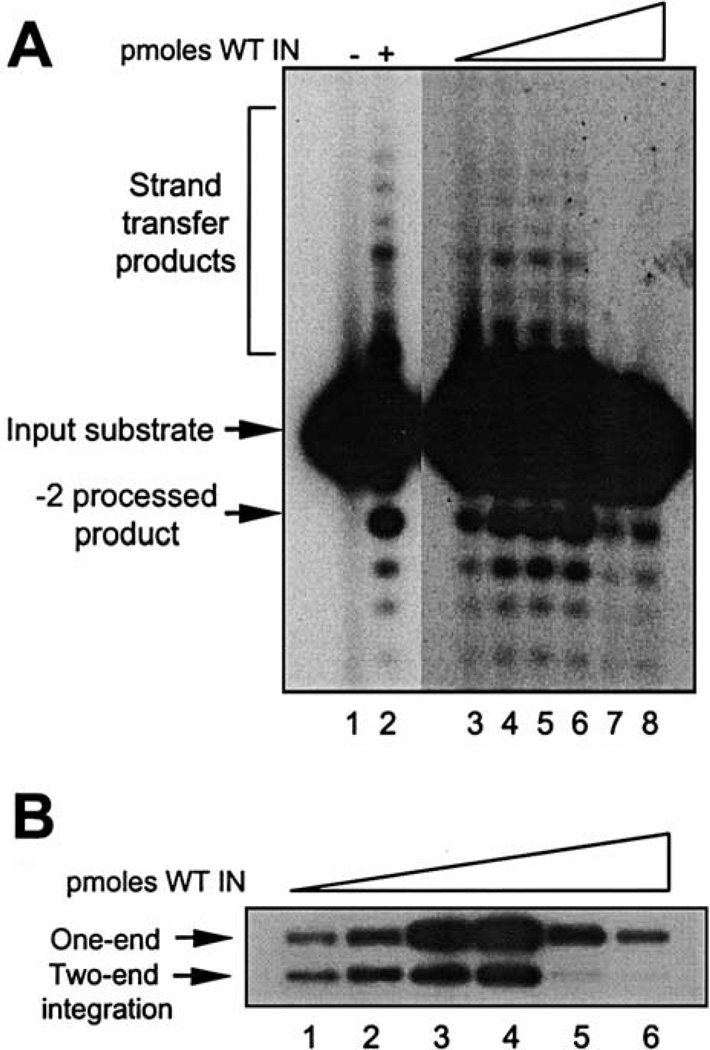
The catalytic activities of the full-length, wild-type IN protein purified under the current scheme were also analyzed. Fig. 6A shows a titration of WT IN (1.4 to 10 pmol) in the presence of 0.5 pmol of a blunt-ended 20-mer oligonucleotide substrate. Maximal 3′-processing was observed with 5 pmol of IN enzyme (lane 6), and increasing the amount of enzyme to 7.6 pmol or higher yielded inhibition of both 3′-processing and strand-transfer reactions (Fig. 6A, lanes 7 and 8).
Fig. 6.
Integration assays of purified M-MuLV IN. (A) 3′-processing and strand-transfer reactions in an oligonucleotide-based assay; 0.5 pmol of blunt-end-labeled 20-mer substrate was incubated with increasing amounts of purified IN: 1.26 pmol (lane 3), 2.52 pmol (lane 4), 3.8 pmol (lane 5), 5 pmol (lane 6), 7.6 pmol (lane 7), and 10 pmol (lane 8). Reaction products were resolved on 16% sequencing gels, dried, and exposed. (B) Concerted two-end integration; 1 pmol of a 28-mer-labeled substrate was incubated with increasing amounts of purified WT IN: 0.63 pmol (lane 1), 1.26 pmol (lane 2), 2.52 pmol (lane 3), 5 pmol (lane 4), 7.6 pmol (lane 5), and 10 pmol (lane 6) and with the target DNA for 1 h at 37°C. Reaction products were resolved by 1% agarose gels and the dried gels exposed to films.
The full-length IN enzyme was also tested in the concerted two-end integration assay (Fig. 6B). The WT IN was titrated for strand transfer into a plasmid target DNA using a 5′-labeled precleaved oligonucleotide substrate, and the products were analyzed by agarose gels. The yield of one-end and two-end products increased with respect to IN protein (lanes 1–3), with maximal products detected at 5 pmol enzyme in the presence of 1 pmol of 28-mer U5 LTR substrate (lane 4). Concentrations of enzyme over this amount inhibited both the one-end and the two-end strand-transfer products (lanes 5 and 6). Although the one-end integrant remains the preferred product, the yield of two-end concerted integrants is improved utilizing the protein purified from the soluble fraction.
Characterization of soluble M-MuLV IN by size-exclusion chromatography
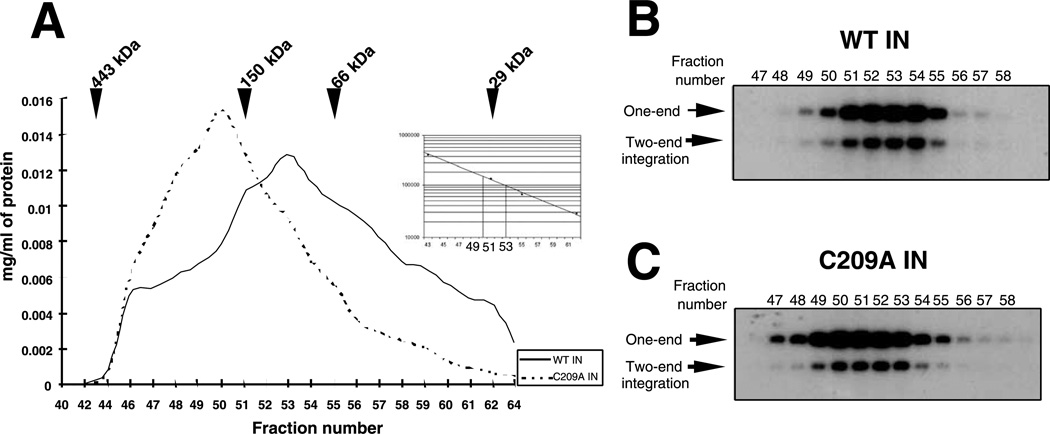
To determine the multimerization state of M-MuLV IN, the WT and C209A IN were further characterized by sizeexclusion chromatography. Within M-MuLV IN, there are three Cys residues; two within the zinc-binding domain and the third, C209, within the catalytic core. Previous studies have shown that IN 106–404 bearing C209A was unable to complement IN 1–105, indicating that the mutation affected the multimerization of IN (Jonsson et al., 1996). IN C209A was therefore analyzed by size fractionation. The soluble fraction of WT IN and C209A (Fig. 2) was purified by Ni2+-NTA affinity columns, eluted with step concentrations of imidazole, and pooled. WT IN and C209A IN were further concentrated and applied into a FPLC S200 sizeexclusion column. The elution profiles are shown in Fig. 7A. The elution position of the globular protein markers is indicated. No proteins were found in the void volume of the column (data not shown). WT IN protein eluted in a broad protein profile with a major peak between 150- and 66-kDa protein markers. From the standard curve, the calculated molecular mass of the predominant WT IN species was 95 kDa, corresponding to a dimer of WT IN. C209A IN protein eluted in a more limited profile with a major peak between 443- and 150-kDa protein markers, corresponding to a calculated molecular mass of 165 kDa, consistent with a tetramer of C209A IN.
Fig. 7.
Chromatographic resolution of M-MuLV integrase proteins. (A) Elution profile of WT IN and C209A IN on S200 gel filtration chromatography. The relative position of migration of the globular molecular weight markers is indicated. The volume of each fraction was 275 µL. Inset: Migration is plotted vs the molecular weight of the markers. Dotted lines show molecular weight of fractions 50 and 53. The profile shows only fractions containing proteins. Aliquots (1 µL) of the fractions of WT IN (B) or C209A IN (C) were assayed in the concerted two-end integration assay, using a 28-mer 5′-labeled oligonucleotide donor substrate and target plasmid DNA as acceptor.
Fractions from the S200 column were further analyzed for integration activity. Equivalent aliquots of the fractions containing IN proteins were incubated with a precleaved 5′-labeled 28-mer oligonucleotide substrate and strand-transfer products into a plasmid DNA target were analyzed by agarose gels (Yang and Roth, 2001). Fig. 7B shows the profile of concerted strand-transfer activity from the fractions containing the WT IN protein. Although further assembly of protein and DNA occurs under the assay conditions, analysis across the S200 column identifies IN multimerization states capable of functional assembly. The peak of concerted strand-transfer activity is located between fractions 52 and 54 with a maximum yield obtained from fraction 53. Low strand-transfer activity was detected when the input IN into the assay consisted of fractions expected to contain tetramers (Fig. 6B, fraction 50) or those expected to contain monomers of WT IN (Fig. 7B, fraction 59) (data not shown). A profile of concerted strand-transfer activity of the fractions containing the C209A IN protein is shown in Fig. 7C. The peak of strand-transfer activity is concentrated between fractions 49 and 53 with a maximum yield obtained from fraction 50, consistent with the structure of the input C209 IN of a tetramer. C209A IN fractions predicted to contain IN dimers maintained significant levels of activity for this assay (Fig. 7C, fraction 53). Fractions expected to be monomers are not active (fraction 59, data not shown).
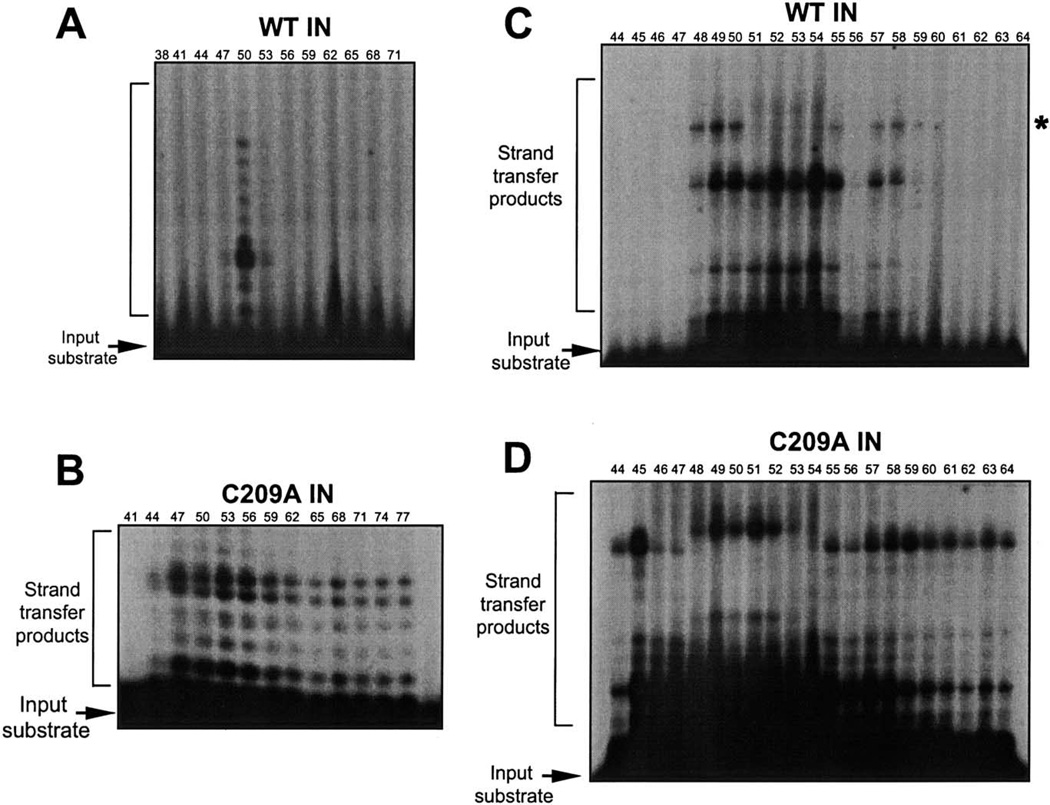
Fractions resolved by exclusion chromatography were further analyzed in the oligonucleotide-based assays (Fig. 3A). Initial assays utilized a 20-mer precleaved substrate (Fig. 8A), and limiting amounts of the fraction (1 µl) were assayed for strand transfer. The profile of activity from fractions containing WT IN protein is shown in Fig. 8A. A single peak of strand-transfer activity at fraction 50 was detected under these conditions. These results suggest that under limiting enzyme, preassembled tetramers are active for strand transfer, under conditions where preassembled dimers present at higher concentrations (fraction 53) are not active.
Fig. 8.
(A, B) Strand-transfer activity of the S200 chromatography fractions using a 20-mer oligonucleotide substrate. (A) Activity profile of WT IN assayed at 1 µL across the fractions. (B) Activity profile of C209A IN assayed at 2 µL across the fractions. Reaction products were resolved by 16% sequencing gels. (C, D) Strand-transfer activity of the exclusion chromatography fractions using a 28-mer oligonucleotide substrate. WT IN was assayed using aliquots (2 µL) of the fractions (C) or C209A IN (C). Reaction products were resolved by 16% sequencing gels.
Strand-transfer activity with S200 fractions was further tested using a longer 28-mer precleaved substrate and the products were visualized by denaturing electrophoresis (Figs. 8C and D). Interestingly, two different patterns of target-site utilization were observed for the strand-transfer products. For the WT IN S200 fractions, there is a loss of a high-molecular-weight strand-transfer product from fractions 51 to 54 (Fig. 8C). This corresponds to fractions whose input IN was capable of concerted two-end integration with an exogenous plasmid target DNA (Fig. 7B). The large strand-transfer products result from integration at the end of the target DNA. These results indicate that the assembly of an active synaptic complex on a large target DNA within these fractions is distinct. The generation of different target sites was previously detected in complementation assays using IN 106–404 and IN 1–105. A limited range of target sites were selected in the presence of a zinc-coordinated HHCC region versus a HHCC domain renatured in the presence of EDTA (Yang et al., 1999a). The increase in stringency was interpreted to reflect the tightness of the assembled complexes in presence of Zn2+.
Similarly, two different patterns of strand-transfer products can be visualized when aliquots of fractions containing C209A IN mutant are assayed utilizing the 28-mer U5 LTR oligonucleotide (Fig. 8D). A shift to higher-molecular-weight strand-transfer products is observed between fractions 48 and 52, whose input IN is in the range of dimers and tetramers. These fractions directly correlate with those active for integration into a large exogenous DNA substrate (Fig. 7C). Fractions 44 to 47 (including multimers greater than tetramers) plus fractions 55 to 64 (including monomers) yield smaller sized strand-transfer products. Although similar fractions were positive for strand-transfer activity using a 20-mer substrate (Fig. 8B), no difference in the target-site specificity was observed. The result using the 28-mer was surprising. The generation of a different target site may represent a distinct conformation of the DNA within an active synaptic complex.
Discussion
In this study, we have purified active M-MuLV integrase proteins under nondenaturing conditions. Recombinant M-MuLV IN was previously purified under harsh denaturing conditions involving highly concentrated solutions of either guanidine chloride or urea to solubilize the proteins from inclusion bodies (Jonsson et al., 1993), causing protein denaturation. Slow renaturation by dialysis is necessary after purification, in order to refold the protein into an active conformation. In the current study, IN was solubilized immediately after bacterial lysis, by performing a series of controlled cycles of sonication at 4°C. The strong DNA-binding activity of retroviral IN has been previously reported (Roth et al., 1988). After shearing the DNA, solubilized IN can be directly bound to nickel-affinity resins, eluted with imidazole solutions, and further purified by heparin columns. A similar scheme of lysis was previously used to purify GST-tagged M-MuLV IN by using glutathione–agarose resins (Dotan et al., 1995). In that report, for consistent activity between different preparations, a phosphocellulose column was necessary to remove an inhibitor (Dotan et al., 1995). Using the purification scheme described herein, individual fractions eluted with imidazole could be directly monitored for IN activity. A similar scheme of lysis and solubilization was used to purify an active two-domain derivative of RSV IN (Yang et al., 1999b), feline immunodeficiency virus (FIV) IN (Shibagaki et al., 1997), HTLV-2 IN (Pieper and Jonsson, 2002), and Sin Nombre virus N protein (Jonsson et al., 2001). Other alternative approaches to purify retroviral INs include solubilization from insoluble fractions with 1 M NaCl solutions for HIV-1 IN (Sherman and Fyfe, 1990) and RSV PrA IN (McCord et al., 1998).
Retroviral INs have been modified by site-directed mutagenesis in order to improve their solubility. In an HIV-1 IN study, 29 point mutants were tested in which hydrophobic residues were substituted with Lys or Ala residues (Jenkins et al., 1995). The single point mutation of F185K within the catalytic core improved the solubility of the HIV-1 polypeptide. A single point mutation of F199K for the RSV two-domain integrase (Hyde et al., 1999) and F185H for SIV IN was also found to improve solubility and the ability to obtain diffraction quality crystals (Li et al., 1999). Both of these residues are located at the dimer interface of the catalytic core domain. A similar approach was developed with an N-terminal truncated construct of M-MuLV RT, to modify potentially solvent exposed hydrophobic residues. Of the substituted enzymes, V433K and L435K were substantially more soluble in the absence of detergent than the parental construct and crystals of the N-terminal truncated M-MuLV L435K RT complexed with a duplex of DNA oligonucleotides have been obtained (Das and Georgiadis, 2001). In the present study, we have targeted four positions at the C terminus within the catalytic domain of M-MuLV IN for mutagenesis. Substituted amino acids were found to be structurally equivalent to those previously altered in HIV-1, RSV, and SIV IN that had improved the solubility of the polypeptides. Additionally, we used the previously generated M-MuLV IN point mutant, within the catalytic triad C209A IN, which was known to be active in vitro for 3′-processing, strand-transfer, and disintegration reactions (Donzella et al., 1998; Jonsson et al., 1996). Among the mutants tested, C209A IN yielded twofold increase in the soluble fraction with respect to the parental WT IN. Using the expression and purification schemes presented herein, active and highly purified C209A IN is under intense research in order to obtain diffraction quality crystallization conditions as a model for the full-length M-MuLV IN structural analysis.
There are several signs that the proteins purified from the soluble fractions are greatly improved over renatured preparations in solubility and activity. Previously, the renatured protein did not chromatograph on DEAE, heparin butyl sepharose, or sephadex 75 as a homogenous peak (Jonsson and Roth, unpublished results). On many resins, including Superose 12 columns, the protein never eluted. From the results presented on Fig. 4B, the native protein fractionated on the FPLC– heparin column as a symmetric peak. The stability of both WT IN and C209A IN allowed the fractionation and activity profiles using size-exclusion chromatography. Previously purified preparations of IN 106–404 recovered from the insoluble fraction were sensitive to high DTT concentrations for full complementation activity with IN 1–105 (Donzella et al., 1996; Jonsson et al., 1996), suggesting improper refolding and low stability in solution. The activities of IN 106–404 varied widely between different preparations and frequently yielded inactive protein. With the purification conditions developed herein, the ability to obtain active IN 106–404 has been highly reproducible.
The irreproducibility of the proteins purified from the insoluble fraction makes direct comparison difficult with the current soluble preparations. However, published data on previously optimized proteins purified from the insoluble fractions indicate that the soluble IN 1–105, IN 106–404, and WT IN require, under standard reaction conditions, a lower protein:protein as well as protein:DNA ratio for 3′-end processing and strand-transfer reactions compared with the renatured proteins. Previously a 10-fold molar excess of IN 1–105 to IN 106–404 was required for optimal strand-transfer and 3′-processing (Yang et al., 1999a), whereas a 2.3:1 ratio was optimal for the soluble proteins. Similarly, concerted two-end integration previously required an optimal molar ratio of IN 1–105: IN 106–404 of 8:1 (Yang and Roth, 2001), whereas with the soluble protein, a 2:1 ratio yielded predominantly two-end concerted integration products. Using the soluble wild-type IN, the optimal IN:DNA ratio was 10:1. Using the same assay, M-MuLV IN protein purified from insoluble fractions followed by slow steps of renaturation required a 20:1 ratio of IN:DNA for optimal integration (Jonsson et al., 1996). Similarly, optimal two-end integration using a 28-mer LTR was obtained with the soluble IN at an IN:LTR ratio of 5:1, whereas previously the maximal integration into an exogenous substrate required a 20:1 IN:LTR ratio, with the predominant product being the one-end integrant (Yang and Roth, 2001).
In an infected cell, the two ends of the viral DNA must be integrated into the target host DNA in a concerted reaction, and multiple lines of evidence suggest that integrase functions as a multimer to act on both DNA ends. As previously determined, in solution, both RSV and HIV-1 IN existed as a mixture of monomers, dimers, and tetramers in equilibrium (Jones et al., 1992; Vincent et al., 1993). Multimerization is stimulated by Zn2+ (Lee et al., 1997), Mn2+ (Wolfe et al., 1996), and high-molecular-mass complexes were detected in the presence of fluorescent oligonucleotides and Mg2+ (Vercammen et al., 2002). Protein cross-linking experiments have also shown that IN exists as a multimer (Heuer and Brown, 1997, 1998). Furthermore, multimerization during catalysis is implied by studies indicating complementation between HIV-1 IN mutants (Engelman et al., 1993; vanGent et al., 1993) and functional tetramers were deduced from kinetic studies of disintegration of ASV IN (Bao et al., 2003). The structure of a full-length retroviral IN is not yet known. However, the central catalytic core domain and all two-domain fragments spanning the core plus either the C terminus (Chen et al., 2000a; Yang et al., 2000) or the N terminus (Wang et al., 2001) from different retroviruses form dimers in the crystal structures. Similarly, the structure of the Tn5 transposase synaptic complex consists of a dimer subunit (Davies et al., 2000). Alternatively, tetramers have been isolated from cells expressing HIV-1 IN (Cherepanov et al., 2003), also detected under reducing conditions when HIV-1 IN was analyzed by fluorescence anisotropy decay measurements (Deprez et al., 2000) and by a Zn2+-dependent interaction that requires the zinc finger motif in the amino-terminal domain (Lee et al., 1997). Previously, biochemical characterization of M-MuLV IN was restricted due to the limited solubility of the recombinant protein. In the present study, the multimerization state of M-MuLV IN was functionally analyzed. WT IN was shown to migrate predominantly as a dimer on size-exclusion chromatography, consistent with other retroviral IN systems. Functional analysis indicated that fractions in equilibrium between dimers and tetramers are capable of assembly and catalysis of two viral LTRs into an exogenous target DNA. No differences were observed in the target-site selection of the strand-transfer products when a 20-mer U5-end substrate was assayed across the gel filtration column. Significant differences in the target-site selection were observed when the column was assayed with a 28-mer U5 substrate for strand transfer. With both 20-mer and 28-mer U5 substrates, multimers larger than tetramers and smaller than dimers were shown to be active as well. However, the dimers and tetramers could be differentiated from the other multimers in an assay by a distinct strand-transfer profile and target-site selection, indicative of a different assembly pathway. Similar results were obtained when fractions containing C209A IN were assayed. Although C209A IN migrated predominantly as a tetramer, the peaks of activity were also mainly concentrated on fractions expected to contain dimers–tetramers. These results indicate that multimerization mediated by protein–protein interactions is different than functional multimerization during catalysis, in the presence of a DNA substrate.
The first step in the integration pathway involves the assembly of a stable complex between IN and the viral DNA donor substrate. HIV-1 IN assembled on immobilized substrates was shown to be sufficient to catalyze strand transfer when the target DNA was added subsequent to assembly (Wolfe et al., 1996). Studies using the HIV-1 IN strand-transfer inhibitors diketo acids (DKAs) indicated that the target DNA requires a configuration of the enzyme defined only after the assembly of the specific IN complex (Espeseth et al., 2000). The results presented in Fig. 8 suggest that upon assembly on identical 28-mer oligonucleotide DNA substrates, the target DNA can be positioned within the active complex in at least two distinct configurations, as revealed by their distinct target DNA site selection. Using the same donor 28-mer U5 DNA substrate but a large plasmid DNA (Fig. 7B), we demonstrated that the same fraction which migrated on the S200 as dimers through tetramers were able to perform concerted integration, which is indicative of a functional and coordinated nucleoprotein assembly. Previous results have indicated that plasmid target DNA can access the target binding site and promote the functional and coordinated assembly of the multimer, stabilizing the subunits, in a concerted integration assay (Yang and Roth, 2001). Fractions other than dimers–tetramers were not correctly stabilized by the plasmid target DNA since no strand transfer on the exogenous plasmid could be detected. Substrate coordination features were also analyzed with M-MuLV disintegration reactions (Donzella et al., 1996). Mispairing of the crossbone single-strand region of the substrate affected the intermolecular, but not the intramolecular disintegration reactions, suggesting differential assembly for cis or trans half-site modes of substrate binding (Donzella et al., 1996). It is not known whether these different configurations are ultimately defined by protein–protein and/or DNA–protein interactions. As proposed by Sinha et al. (2002), within the population only a minority of the HIV-1 IN subunits will correctly interact to form a nucleoprotein complex competent for concerted integration. Most of the interactions will lead to one-end integration events.
The analysis of complementation assays of IN 1–105 with IN 106–404 M-MuLV IN nonoverlapping constructs indicated a more efficient assembly of multimeric complexes (four- to fivefold) when using 28-mer versus 20-mer oligonucleotide substrates mimicking the viral terminal U5 in a concerted integration reaction (Fig. 5B). Interestingly, positions 20 to 28 are outside of the conserved 13-bp inverted repeat sequences at the termini of the M-MuLV LTR. For HIV-1 IN, the extension of 13 nucleotides on the 15 terminal bases IN recognition sequence enhanced 3′-processing of the termini in an oligonucleotide-based assay (Vink et al., 1991). Reconstitution systems have also used purified IN and linear retrovirus-like LTRs to recapitulate concerted two-end integration reactions (about 500-bp-long) for HIV-1 (Hindmarsh et al., 1999) and RSV (McCord et al., 1998). For RSV, as well as in vivo for HIV-1 (Brown et al., 1999) and M-MuLV (Murphy and Goff, 1992), deletions or modifications of internal sequences (up to 12 nucleotides from the end) have major effects in avian LTRs to promote two-end integration in vitro (Chiu and Grand-genett, 2000). The role of either the sequence or the structure of nucleotides outside of the terminal sequences is not clear. A 22-bp deletion starting at position 12 from the M-MuLV LTR end has little effect on IN-associated properties in the preintegration complexes (Wei et al., 1998). For SIV IN, two-end integration events showed an increase in efficiency with an increase in the length of the donor DNA from 134 to 433 bp (Goodarzi et al., 1999). For HIV-1, positions 17–20 were found to be required for concerted integration (Brin and Leis, 2002). However, preliminary results indicate that position 20 of the M-MuLV U5 substrate tolerates any nucleotide when assayed in the complementation assay for concerted integration (data not shown). More studies are necessary to define the role of the outside sequences in recognition by retroviral integrases.
Material and methods
Materials
Crude [γ-32P]ATP (7000 Ci/mmol) was purchased from ICN. T4 polynucleotide kinase, T4 DNA ligase, Vent DNA polymerase and restriction enzymes were obtained from New England Biolabs. Ni2+-nitrilotriacetic acid agarose (Ni2+-NTA) was purchased from Qiagen. Ni2+-NTA Su-perflow (5 mL) and HiTrap heparin HP (5 ml) were purchased from Amersham Biosciences.
Oligonucleotides
DNA oligonucleotides were prepared by the University of Medicine and Dentistry of New Jersey, DNA Synthesis Facility and purified by electrophoresis on 20% denaturing polyacrylamide gels (Jonsson et al., 1993). Oligonucleotides used in this study were labeled with [γ-32P]ATP by a kinase reaction as previously described (Jonsson et al., 1993). Oligonucleotide 5319 (28-mer) 5′-GACTACCCGT-CAGCGGGGGTCTTTCATT-3′ and its complementary strand 5321 5′-AATGAAAGACCCCCGCTGACGGGTA-GTC-3′ or oligonucleotide 2783 (20-mer) 5′-GTCAG-CGGGGGTCTTTCATT-3′ and its complementary strand 2785 5′-AATGAAAGACCCCCGCTGAC-3′ were used as the blunt-end U5 end substrate for the 3′-processing reactions. Oligonucleotide 5320 (26-mer) 5′-GACTACCCGT-CAGCGGGGGTCTTTCA-3′ and its complementary strand 5321, or oligonucleotide 2784 (18-mer) 5′-GTCAGCG-GGGGTCTTTCA-3′ and its complementary oligonucleotide 2785 mimicking a precleaved U5 end, were used as the substrate for strand-transfer and two-end integration reactions.
Generation of IN point mutants
Point mutations in M-MuLV IN were generated by overlapping PCR. For the oligonucleotides shown below, underlined positions indicate the codon changed. For individual PCR reactions, the first fragment was obtained with a common 5′ primer 5′-GATCACCGGTTTTCATTACACAGT-GACTGATA-3′ (100 pmoles) and with 5′-CAGGGCTA-AGGGGAGTTGGAGCACCCAGTCTCT-3′ (for L244Q, 100 pmol), 5 ‘-GCCCGGCGTGTTGCGCTTTCGGTACA-GGGCTA-3′ (for A252K, 100 pmol), 5′-GCCCGGCGT-GTTGCGGTGTCGGTACAGGGCTA-3′ (for A252H, 100 pmol) or 5′ -ATGGGGGCCCGGCGTCTTGCGGGCTGC-GTACAG-3′ (for N254K, 100 pmol) on the template pET-HTWT (Jonsson et al., 1993) and Vent DNA polymerase. Cycle conditions were as follows: 2 min at 95°C, 35 cycles of 30 s at 95°C, 30 s at 63°C and 90 s at 68°C, and a final incubation of 10 min at 68°C. A 750-bp fragment in each case was gel-isolated. The second PCR fragment was obtained with 5′ -AGAGACTGGGTGCTCCAACTCCCCT-TAGCCCTG-3′ (for L244Q, 100 pmol), 5′-TAGCCCTG- TACCGAAAGCGCAACACGCCGGGC-3′ (for A252K, 100 pmol), 5′ -TAGCCCTGTACCGACACCGCAACACG-CCGGGC-3′ (for A252H, 100 pmol), or 5′-CTGTACCG-AGCCCGCAAGACGCCGGGCCCCCAT-3′ (for N254K, 100 pmol), each one incubated with a common 3′ primer 5′-GGTGGATCCAGTACTGACCCCTCTG-3′ (100 pmol) using template pET-HTWT and Vent DNA polymerase as described above. A 500-bp fragment in each case was gel-isolated. For overlapping PCR reactions, equimolar amounts of the 750 bases fragment and 500 bases fragment for each mutant were incubated with the 5′ and 3′ common primers (100 pmol each) and Vent DNA polymerase. Cycle conditions were as follows: 2 min at 95°C, 35 cycles of 30 s at 95°C, 30 s at 53°C, 3 min at 68°C, and a final incubation of 10 min at 68°C. The 1.2-kb product fragments were digested with SacII and BamHI (contained within 3′ primer). For each construct, an 890-bp fragments was gel-isolated and exchanged into pET-HTWT. Introduction of the single point mutations were verified by sequencing with the 3′ common primer at the Core DNA Facility, UMDNJ/ RWJMS.
Expression and purification of M-MuLV integrase
Previously reported expression vectors for recombinant M-MuLV IN (pET-HTWT wild type) (Jonsson et al., 1993), IN C209A (Jonsson et al., 1993), IN 106–404 (Jonsson et al., 1993), and IN 1–105 (Yang et al., 1999a) containing a hexahistidine tag were expressed in Escherichia coli BL21 (DE3) (Novagen). A similar procedure has been recently reported for HTLV-2 IN (Pieper and Jonsson, 2002). For each construct, 100 mL of Luria broth medium (LB) containing 200 µg/mL Carbenicillin (LB-Carb200) was inoculated and grown overnight at 37°C with constant shaking. Cultures were then centrifuged, resuspended into 20 mL of LB-Carb200, and used to inoculate 1 L of LB-Carb200 in a 2-L flask. Cultures were grown for 2 h at 30°C, induced with IPTG (0.256 mM) for 2 h at 30°C, and resuspended into 40 mL of ice-cold solubilization buffer (50 mM sodium phosphate buffer, pH 8.0, 300 mM NaCl, 10 mM imidazole, 10 mM Chaps (3-[(3-cholamidopropyl)dimethylammonio]-1-propane sulfonate), 1 protease EDTA-free tablet (Boehringer Mannheim) per 50 milliliters of buffer, at 4°C. One hundred milligrams of lysozyme was added, dounced until mixed, and kept at 4°C for 15 min. Samples were then sonicated for 15 s and placed on ice for 1 min. Sonication was repeated until samples appeared clear (not viscous) and then they were centrifuged at 16,000 rpm in a Sorvall SS34 rotor for 1 h at 4°C. Supernatants containing soluble IN proteins were subjected to purification either by batch purification or using columns on FPLC systems.
(i) Batch purification
Solubilized extracts (40 mL) were mixed with 1 mL of Ni2+-NTA preequilibrated in solubilization buffer, and incubated overnight at 4°C with rocking. Decanted resin was washed with 100 mL of washing buffer (50 mM sodium phosphate, pH 8.0, 1 M NaCl, 25 mM imidazole, 10 mM Chaps, 10 mM 2-mercaptoethanol, 20% glycerol). Step elution (1-mL fractions) was performed with 5 mL each of washing buffer containing 50, 100, 250, 500 mM imidazole. Elution of the specific IN proteins and subdomains varied between 50 and 250 mM imidazole, as indicated. IN proteins or subdomains after one round of batch purification were active and suitable for biochemical studies. Fractions were pooled and dialyzed against 20 mM Hepes, pH 7.4, 1.5 mM DTT, 400 mM KCl, 2 mM Chaps, 50% glycerol, and 10 µM ZnCl2 and stored at −80°C.
(ii) FPLC-based purification
Five milliliters of Ni2+-NTA column-FPLC was sequentially equilibrated with 7 column volumes of buffer A (50 mM sodium phosphate, pH 8.0, 1 M NaCl, 10 mM Chaps, 5 mM 2-mercaptoethanol, and 10% glycerol) with 10 mM imidazole, with 7 column volumes of buffer A containing 25 mM of imidazole, and with 7 column volumes of buffer A with 10 mM imidazole. Solubilized extracts containing WT full-length IN were filtered through 0.45-µm filters and applied to the column. Bound proteins were washed with 8 column volumes of buffer A containing 25 mM imidazole or until baseline and then eluted with 9.4 column volumes of a 10 to 500 mM imidazole gradient in buffer A, collecting 2.5-mL fractions. Aliquots of the fractions were analyzed by SDS–PAGE and for strand-transfer activity in an oligonucleotide-based assay (see below). Selected fractions were pooled and diluted to 0.4 M NaCl with buffer C (50 mM Hepes pH 7.4, 1 mM DTT, 2 mM Chaps, 10% glycerol). The heparin column–FPLC (5 mL) was sequentially equilibrated with 4 column volumes of buffer C containing 0.4 M NaCl, with 2 column volumes of buffer C containing 2 M NaCl and, with 5 0.4 M NaCl, with 2 column volumes of buffer C containing 2 M NaCl and, with 5 the Ni2+-NTA column were loaded onto the heparin column and eluted with 7 column volumes of a 0.4 to 2 M NaCl gradient in buffer C (2.5-mL fractions). Aliquots of the fractions were analyzed by SDS-PAGE and for strand-transfer activity in an oligonucleotide-based assay (see below).
Size-exclusion chromatography
Solubilized extracts of WT IN and C209 IN (corresponding to 2 L of culture) were bound in batch to 2 mL of Ni2+-NTA resin and step-eluted with 5 mL of washing buffer containing 100 and 250 mM imidazole and dialyzed at 4°C for 6–8 h against 50 mM Hepes, pH 8.0, 1 M NaCl, 1.5 mM DTT, 2 mM Chaps, 10 µM ZnCl2, and 10% glycerol. Proteins were concentrated to 250 µL by centricon columns (YM-10: 10,000 molecular weight cut-off); 0.5 mg of each IN protein (250 µL) was applied to a S200 10/30 gel filtration column (Pharmacia) preequilibrated with 50 mM Hepes, pH 8.0, 400 mM KCl, 1.5 mM DTT, 2 mM Chaps, and 10% glycerol; 0.275-mL fractions were collected. The column was standardized using gel filtration molecular weight markers (Sigma): thyroglobulin (669 kDa), apofer-ritin (443 kDa), alcohol dehydrogenase (150 kDa), bovine serum albumin (66 kDa), and carbonic anhydrase (29 kDa).
Integration assays
3′-processing and strand-transfer reactions contained 0.5 pmol of labeled substrate and reaction buffer containing 20 mM morpholineethanesulfonic acid (MES, pH 6.2), 100 mM KCl, 10 mM MnCl2, 10 mM DTT, and 10% glycerol, unless otherwise indicated (Jonsson et al., 1993). The reaction conditions (35 µL) for two-end integration reactions (Yang and Roth, 2001) were the same as those for 3′-processing and strand-transfer reactions except that 200 mM KCl was added in the presence of 1 pmol of U5 substrate and 1.2 µg of target plasmid DNA. Oligonucleotide substrates were labeled at the 5′ end by T4 polynucleotide kinase and mixed with the complementary strand at a ratio of 1:2 (labeled oligonucleotide vs complementary strand) in 100 mM NaCl. The oligonucleotides were annealed by heating for 3 min at 95°C and then cooled to room temperature. Complementation assays were performed by preincu-bating the IN 1–105 with the U5 substrate for 5 min on ice. IN 106–404 was added, followed by incubation at 37°C for 5 min. When assaying the full-length IN protein, the U5 LTR substrate was preincubated with aliquots of the protein for 5 min on ice followed by addition of target DNA plasmid and KCl and the reactions were incubated at 37°C for 2 h. Reactions were stopped by the addition of 10 mM EDTA (pH 8.0)-0.5% SDS and 100 µg/mL of proteinase K at 37°C for 1 h. Reaction products were visualized by electrophoresis on a 1% agarose gel and autoradiography of dried gels.
Western blot
Western blots were performed using anti-MuLV IN antibody (rabbit 4, bleed 5) (Tanese et al., 1986).
Acknowledgments
This work was supported by Grants RO1 CA76545 and RO1 CA90174, awarded to M.J.R. We thank Carlos Rivera for technical assistance.
References
- Bao KK, Wang H, Miller JK, Erie DA, Skalka AM, Wong I. Functional oligomeric state of avian sarcoma virus integrase. J. Biol. Chem. 2003;278:1323–1327. doi: 10.1074/jbc.C200550200. [DOI] [PubMed] [Google Scholar]
- Brin E, Leis J. HIV-1 integrase interaction with U3 and U5 terminal sequences in vitro defined using substrates with random sequences. J. Biol. Chem. 2002;277:18357–18364. doi: 10.1074/jbc.M201354200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown H, Chen H, Engelman A. Structure-based mutagenesis of the human immunodeficiency virus type 1 DNA attachment site: effects on integration and cDNA synthesis. J. Virol. 1999;73:9011–9020. doi: 10.1128/jvi.73.11.9011-9020.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PO, Bowerman B, Varmus HE, Bishop JM. Retroviral integration: structure of the initial covalent product and its precursor, and a role for the viral IN protein. Proc. Natl. Acad. Sci. USA. 1989;86:2525–2529. doi: 10.1073/pnas.86.8.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujacz G, Jaskolski M, Alexandratos J, Wlodawer A, Merkel G, Katz RA, Skalka AM. High-resolution structure of the catalytic domain of avian sarcoma virus integrase. J. Mol. Biol. 1995;253:333–346. doi: 10.1006/jmbi.1995.0556. [DOI] [PubMed] [Google Scholar]
- Burke CJ, Sanyal G, Bruner MW, Ryan JA, LaFemina RL, Rob-bins HL, Zeft AS, Middaugh CR, Cordingly MG. Structural implications of spectroscopic characterization of a putative zinc finger peptide from HIV-1 integrase. J. Biol. Chem. 1992;267:9639–9644. [PubMed] [Google Scholar]
- Chen JC-H, Krucinski J, Miercke LJW, Finer-Moore JS, Tang AH, Leavitt AD, Stroud RM. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding. Proc. Natl. Acad. Sci. USA. 2000a;97:8233–8238. doi: 10.1073/pnas.150220297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Yan Y, Munshi S, Li Y, Zugay-Murphy J, Xu B, Witmer M, Felock P, Wolfe A, Sardana V, Emini EA, Hazuda D, Kuo LC. X-ray structure of simian immunodeficiency virus inte-grase containing the core and C-terminal domain (residues 50–293)—an initial glance of the viral DNA-binding platform. J. Mol. Biol. 2000b;296:521–533. doi: 10.1006/jmbi.1999.3451. [DOI] [PubMed] [Google Scholar]
- Cherepanov P, Maertens G, Proost P, Devreese B, Beeumen JV, Engelborghs Y, Clercq ED, Debyser Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- Chiu R, Grandgenett DP. Avian retrovirus DNA internal attachment site requirements for full-site integration in vitro. J. Virol. 2000;74:8292–8298. doi: 10.1128/jvi.74.18.8292-8298.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigie R, Fujiwara T, Bushman F. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990;62:829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- Das D, Georgiadis MM. A directed approach to improving the solubility of Moloney murine leukemia virus reverse transcriptase. Protein Sci. 2001;10:1936–1941. doi: 10.1110/ps.16301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies DR, Goryshin IY, Reznikoff WS, Rayment I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science. 2000;289:77–85. doi: 10.1126/science.289.5476.77. [DOI] [PubMed] [Google Scholar]
- Deprez E, Tauc P, Leh H, Mouscadet J-F, Auclair C, Bronchon J-C. Olligomeric states of the HIV-1 integrase as measured by time-resolbed fluorescence anisotropy. Biochemistry. 2000;39:9275–9284. doi: 10.1021/bi000397j. [DOI] [PubMed] [Google Scholar]
- Donzella GA, Jonsson CB, Roth MJ. Coordinated disintegration reactions mediated by Moloney murine leukemia virus integrase. J. Virol. 1996;70:3909–3921. doi: 10.1128/jvi.70.6.3909-3921.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donzella GA, Leon O, Roth MJ. Implications of a central cysteine residue and the HHCC domain of Moloney murine leukemia virus integrase protein in functional multimerization. J. Virol. 1998;72:1691–1698. doi: 10.1128/jvi.72.2.1691-1698.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotan I, Scottoline BP, Heuer TS, Brown PO. Characterization of recombinant murine leukemia virus integrase. J. Virol. 1995;69:456–468. doi: 10.1128/jvi.69.1.456-468.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drelich M, Wilhelm R, Mous J. Identification of amino acid residues critical for endonuclease and integration activities of HIV-1 IN protein in vitro. Virology. 1992;188:459–468. doi: 10.1016/0042-6822(92)90499-f. [DOI] [PubMed] [Google Scholar]
- Dyda F, Hickman AB, Jenkins TM, Engelman A, Craigie R, Da-vies DR. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- Engelman A, Bushman FD, Craigie R. Identification of discrete functional domains of HIV-1 integrase and their organization within an active multimeric complex. EMBO J. 1993;12:3269–3275. doi: 10.1002/j.1460-2075.1993.tb05996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman A, Craigie R. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J. Virol. 1992;66:6361–6369. doi: 10.1128/jvi.66.11.6361-6369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman A, Mizuuchi K, Craigie R. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell. 1991;67:1211–1221. doi: 10.1016/0092-8674(91)90297-c. [DOI] [PubMed] [Google Scholar]
- Espeseth AS, Felock P, Wolfe A, Witmer M, Grobler J, Anthony N, Egbertson M, Melamed JY, Young S, Hamill T, Cole JL, Hazuda DJ. HIV-1 integrase inhibitors that compete with the target DNA substrate define a unique strand transfer conformation for integrase. Proc. Natl. Acad. Sci. USA. 2000;97:11244–11249. doi: 10.1073/pnas.200139397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Mizuuchi K. Retroviral DNA integration: structure of an integration intermediate. Cell. 1988;54:497–504. doi: 10.1016/0092-8674(88)90071-2. [DOI] [PubMed] [Google Scholar]
- Gao K, Butler SL, Bushman F. Human immunodeficiency virus type 1 integrase: arrangement of protein domains in acitve cDNA complexes. EMBO J. 2001;20:3565–3576. doi: 10.1093/emboj/20.13.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi G, Pursley M, Felock P, Witmer M, Hazuda D, Brack-mann K, Grandgenet D. Efficiency and fidelity of full-site integration reactions using recombinant simian immunodeficiency virus integrase. J. Virol. 1999;73:8104–8111. doi: 10.1128/jvi.73.10.8104-8111.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer TS, Brown PO. Mapping features of HIV-1 integrase near selected sites on viral and target DNA molecules in an active enzyme-DNA complex by photo-cross-linking. Biochemistry. 1997;36:10655–10665. doi: 10.1021/bi970782h. [DOI] [PubMed] [Google Scholar]
- Heuer TS, Brown PO. Photo-cross-linking studies suggest a model for the architecture of an active human immunodeficiency virus type 1 integrase-DNA complex. Biochemistry. 1998;37:6667–6678. doi: 10.1021/bi972949c. [DOI] [PubMed] [Google Scholar]
- Hindmarsh P, Ridky T, Reeves R, Andrake M, Skalka AM, Leis J. HMG protein family members stimulate human immunodeficiency virus type 1 and avian sarcoma virus concerted DNA integration in vitro. J. Virol. 1999;73:2994–3003. doi: 10.1128/jvi.73.4.2994-3003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde CC, Bushman FD, Mueser TC, Yang Z-N. Crystal structure of an active two-domain derivative of rous sarcoma virus integrase. J. Mol. Biol. 1999;296:535–538. doi: 10.1006/jmbi.1999.3463. [DOI] [PubMed] [Google Scholar]
- Jenkins T, Kickman A, Dyda F, Ghirlando R, Davies D, Craigie R. Catalytic domain of HIV-1 integrase: identification of a soluble mutant by systematic replacement of hydrophobic residues. Proc. Natl. Acad. Sci. USA. 1995;92:6057–6061. doi: 10.1073/pnas.92.13.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins TM, Engelman A, Ghirlando R, Craigie R. A soluble active mutant of HIV-1 integrase. J. Biol. Chem. 1996;271:7712–7718. doi: 10.1074/jbc.271.13.7712. [DOI] [PubMed] [Google Scholar]
- Johnson MS, McClure MA, Feng DF, Gray J, Doolittle RF. Computer analysis of retroviral pol genes: assignment of enzymatic functions to specific sequences and homologies with nonviral enzymes. Proc. Natl. Acad. Sci. USA. 1986;83:7648–7652. doi: 10.1073/pnas.83.20.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. GenTHREADER: an efficient and reliable protein fold recognition method for genomic sequences. J. Mol. Biol. 1999;287:797–815. doi: 10.1006/jmbi.1999.2583. [DOI] [PubMed] [Google Scholar]
- Jones KS, Coleman J, Merkel GW, Laue TM, Skalka AM. Retroviral integrase functions as a multimer and can turn over catalyt-ically. J. Biol. Chem. 1992;287:16037–16040. [PubMed] [Google Scholar]
- Jonsson C, Gallegos J, Fero P, Severson W, Su X, Schmaljohn C. Purification and characterization of the Sin Nombre virus nu-cleocapsid protein expressed in Escherichia coli. Protein Expr. Purif. 2001;23:134–141. doi: 10.1006/prep.2001.1489. [DOI] [PubMed] [Google Scholar]
- Jonsson CB, Donzella GA, Gaucan E, Smith CM, Roth MJ. Functional domains of Moloney murine leukemia virus integrase defined by mutation and complementation analysis. J. Virol. 1996;70:4585–4597. doi: 10.1128/jvi.70.7.4585-4597.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson CB, Donzella GA, Roth MJ. Characterization of the forward and reverse integration reactions of the Moloney murine leukemia virus integrase protein purified from Escherichia coli. J. Biol. Chem. 1993;268:1462–1469. [PubMed] [Google Scholar]
- Katz RA, Merkel G, Kulkosky J, Leis J, Skalka AM. The avian retroviral IN protein is both necessary and sufficient for integra-tive recombination in vitro. Cell. 1990;63:87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- Katzman M, Katz R, Skalka AM, Leis J. The avian retroviral integration protein cleaves the terminal sequences of linear DNA at the in vivo sites of integration. J. Virol. 1989;63:5319–5327. doi: 10.1128/jvi.63.12.5319-5327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan E, Mack JP, Katz RA, Kulkosky J, Skalka AM. Retroviral integrase domains: DNA binding and the recognition of LTR sequences. Nucleic Acids Res. 1991;19:851–860. doi: 10.1093/nar/19.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkosky J, Jones KS, Katz RA, Mack JPG, Skalka AM. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol. Cell. Biol. 1992;12:2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavitt AD, Shiue L, Varmus HE. Site-directed mutagenesis of HIV-1 Integrase demonstrates differential effects on integrase function in vitro. J. Biol. Chem. 1993;268:2113–2119. [PubMed] [Google Scholar]
- Lee SP, Xiao J, Knutson JR, Lewis MS, Han MK. Zn2+ promotes the self-association of human immunodeficiency virus type-1 integrase in vitro. Biochemistry. 1997;36:173–180. doi: 10.1021/bi961849o. [DOI] [PubMed] [Google Scholar]
- Li Y, Yan Y, Zugay-Murphy J, Xu B, Cole JL, Witmer M, Felock P, Wolfe A, Hazuda D, Sardana MK, Chen Z, Kuo LC, Sar-dana VV. Purification, solution properties and crystallization of SIV integrase containing a continuous core and C-terminal domain. Acta Crystallogr. D Biol. Crystallogr. 1999;55:1906–1910. doi: 10.1107/s0907444999009610. [DOI] [PubMed] [Google Scholar]
- Lodi PL, Ernst JA, Kuszewski J, Hickman AB, Engelman A, Craigie R, Clore GM, Grononborn AM. Solution structure of the DNA binding domain of HIV-1 integrase. Biochemistry. 1995;34:9826–9833. doi: 10.1021/bi00031a002. [DOI] [PubMed] [Google Scholar]
- McCord M, Stahl S, Mueser T, Hyde CC, Vora A, Grandgenett D. Purification of recombinant Rous sarcoma virus integrase possessing physical and catalytic properties similar to virion-derived integrase. Protein Expr. Purif. 1998;14:167–177. doi: 10.1006/prep.1998.0954. [DOI] [PubMed] [Google Scholar]
- McEuen AR, Edward B, Koepke DA, Ball AE, Jennings BA, Wol-stenholme AJ, Danson MJ, Hough DW. Zinc binding by retroviral integrase. Biochem. Biophys. Res. Commun. 1992;189:813–818. doi: 10.1016/0006-291x(92)92275-3. [DOI] [PubMed] [Google Scholar]
- Mumm SR, Grandgenett DP. Defining nucleic acid-binding properties of avian retrovirus integrase by deletion analysis. J. Virol. 1991;65:1160–1167. doi: 10.1128/jvi.65.3.1160-1167.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy JE, Goff SP. A mutation at one end of Moloney murine leukemia virus DNA blocks cleavage of both ends by the viral integrase in vivo. J. Virol. 1992;66:5092–5095. doi: 10.1128/jvi.66.8.5092-5095.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piefer A, Jonsson CB. A comparative study of the human T-cell leukemia virus type 2 integrase expressed in and purified from Escherichia coli and Pichia pastoris. Protein Exp. Purif. 2002;25:291–299. doi: 10.1016/s1046-5928(02)00011-6. [DOI] [PubMed] [Google Scholar]
- Roth MJ, Schwartzberg P, Tanese N, Goff SP. Analysis of mutations in the integration function of Moloney murine leukemia virus: effects on DNA binding and cutting. J. Virol. 1990;64:4709–4717. doi: 10.1128/jvi.64.10.4709-4717.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth MJ, Schwartzberg PL, Goff SP. Structure of the termini of DNA intermediates in the integration of retroviral DNA: dependence of IN function and terminal DNA sequence. Cell. 1989;58:47–54. doi: 10.1016/0092-8674(89)90401-7. [DOI] [PubMed] [Google Scholar]
- Roth MJ, Tanese N, Goff SP. Gene product of Moloney murine leukemia virus required for proviral integration is a DNA-binding protein. J. Mol. Biol. 1988;203:131–139. doi: 10.1016/0022-2836(88)90097-6. [DOI] [PubMed] [Google Scholar]
- Sherman PA, Fyfe JA. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleavage activity. Proc. Natl. Acad. Sci. USA. 1990;87:5119–5123. doi: 10.1073/pnas.87.13.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibagaki Y, Holmes ML, Appa RS, Chow SA. Characterization of Feline immunodeficiency virus integrase and analysis of functional domains. Virology. 1997;230:1–10. doi: 10.1006/viro.1997.8466. [DOI] [PubMed] [Google Scholar]
- Sinha S, Pursley MH, Grandgenett DP. Efficient concerted integration by recombinant human immunodeficiency virus type 1 integrase without cellular or viral cofactors. J. Virol. 2002;76:3105–3113. doi: 10.1128/JVI.76.7.3105-3113.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanese N, Roth MJ, Goff SP. Analysis of retroviral pol gene products with antisera raised against fusion proteins produced in Escherichia coli. J. Virol. 1986;59:328–340. doi: 10.1128/jvi.59.2.328-340.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDenEnt FMI, Vos A, Plasterk RHA. Dissecting the role of the N-terminal domain of human immunodeficiency virus integrase by trans-complementation analysis. J. Virol. 1999;73:3176–3183. doi: 10.1128/jvi.73.4.3176-3183.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- vanGent DC, Vink C, Groeneger AAMO, Plasterk RHA. Complementation between HIV integrase proteins mutated in different domains. EMBO J. 1993;12:3261–3267. doi: 10.1002/j.1460-2075.1993.tb05995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercammen J, Maertens G, Gerard M, Clercq ED, Debyser Z, Engelborghs Y. DNA-induced polymerization of HIV-1 inte-grase analyzed with fluorescence fluctuation spectroscopy. J. Biol. Chem. 2002;277:38405–38052. doi: 10.1074/jbc.M205842200. [DOI] [PubMed] [Google Scholar]
- Vincent KA, Ellison V, Chow SA, Brown PO. Characterization of human immunodeficiency virus type 1 integrase expressed in Escherichia coli and analysis of variants with amino-terminal mutations. J. Virol. 1993;67:425–437. doi: 10.1128/jvi.67.1.425-437.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink C, Groeneger AAMO, Plasterk RHA. Identification of the catalytic and DNA-binding region of the human immunodeficiency virus type 1 integrase protein. Nucleic. Acids. Res. 1993;21:1419–1425. doi: 10.1093/nar/21.6.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink C, Lutzke RA, Plasterk RHA. Formation of a stable complex between the human immunodeficiency virus integrase protein and viral DNA ends. Nucleic Acids Res. 1994;22:4103–4110. doi: 10.1093/nar/22.20.4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink C, VanGent DC, Elgersma Y, Plasterk RHA. Human immunodeficiency virus integrase protein requires a subterminal position of its viral DNA recognition sequence for efficient cleavage. J. Virol. 1991;65:4636–4644. doi: 10.1128/jvi.65.9.4636-4644.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J-Y, Ling H, Yang W, Craigie R. Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. EMBO J. 2001;20:7333–7343. doi: 10.1093/emboj/20.24.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S-Q, Mizuuchi K, Craigie R. Footprint on the viral DNA ends in Moloney murine leukemia virus preintegration complexes reflect a specific association with integrase. Proc. Natl. Acad. Sci. USA. 1998;95:10535–10540. doi: 10.1073/pnas.95.18.10535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woerner AM, Marcus-Sekura CJ. Characterization of a DNA binding domain in the C-terminus of HIV-1 integrase by deletion mutagenesis. Nucleic Acids Res. 1993;21:3507–3511. doi: 10.1093/nar/21.15.3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AL, Felock PJ, Hastings JC, Blau CU, Hazuda DJ. The role of manganese in promoting multimerization and assembly of human immunodeficiency virus type 1 integrase as a catalytically active complex on immobilized long terminal repeat substrates. J. Virol. 1996;70:1424–1432. doi: 10.1128/jvi.70.3.1424-1432.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Leon O, Greenfield NJ, Roth MJ. Functional interactions of the HHCC domain of Moloney murine leukemia virus integrase revealed by non-overlapping complementation and zinc dependent dimerization. J. Virol. 1999a;73:1809–1817. doi: 10.1128/jvi.73.3.1809-1817.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Roth MJ. Assembly and catalysis of concerted two-end integration events by Moloney murine leukemia virus integrase. J. Virol. 2001;75:9561–9570. doi: 10.1128/JVI.75.20.9561-9570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z-N, Mueser T, Bushman F, Hyde CC. Crystal structure of an active two-domain derivative of Rous sarcoma virus inte-grase. J. Mol. Biol. 1999b;296:535–548. doi: 10.1006/jmbi.1999.3463. [DOI] [PubMed] [Google Scholar]
- Yang ZN, Mueser TC, Hyde FDBCC. Crystal structure of an active two-domain derivative of Rous sarcoma virus integrase. J. Mol. Biol. 2000;296:535–548. doi: 10.1006/jmbi.1999.3463. [DOI] [PubMed] [Google Scholar]
- Zheng R, Jenkins TM, Craigie R. Zinc folds: the N-terminal domain of HIV-1 integrase, promotes multimerization, and enhances catalytic activity. Proc. Natl. Acad. Sci. USA. 1996;93:13659–13664. doi: 10.1073/pnas.93.24.13659. [DOI] [PMC free article] [PubMed] [Google Scholar]