Abstract
The kidney is vulnerable to hypoxia-induced injury. One of the mechanisms underlying this phenomenon is cell apoptosis triggered by hypoxia-inducible factor-1- α (HIF-1α) activation. MicroRNA-210 (miR-210) is known to be induced by HIF-1α and can regulate various pathological processes, but its role in hypoxic kidney injury remains unclear. Here, in both rat systemic hypoxia and local kidney hypoxia models, we found miR-210 levels were upregulated significantly in injured kidney, especially in renal tubular cells. A similar increase was observed in hypoxia-treated human renal tubular HK-2 cells. We also verified that miR-210 can directly suppress HIF-1α expression by targeting the 3′ untranslated region of HIF-1α mRNA in HK-2 cells in severe hypoxia. Accordingly, miR-210 overexpression caused significant inhibition of the HIF-1α pathway and attenuated apoptosis caused by hypoxia, while miR-210 knockdown exerted the opposite effect. Taken together, our findings verify that miR-210 is involved in the molecular response in hypoxic kidney lesions in vivo and attenuates hypoxia-induced renal tubular cell apoptosis by targeting HIF-1α directly and suppressing HIF-1α pathway activation in vitro.
INTRODUCTION
Oxygen is a vital microenvironmental substrate for sustaining tissue homeostasis. Many physiological and pathological conditions cause insufficiency in oxygen availability, or hypoxia, which can eventually lead to apoptosis if the hypoxia is severe (1,2). Despite constituting merely 0.4%–0.5% of a person’s total body weight, the kidney is responsible for almost 7% of the body’s oxygen consumption (3). The high sensitivity of the kidney makes it prone to hypoxic injury and even renal tubular damage and diffuse kidney lesions (4). In the clinic, many diseases induce systemic hypoxia, such as high-altitude disease, postoperative hypoxemia, adult respiratory distress syndrome and shock. Systemic hypoxia drives multiorgan damage, especially kidney injury (5–7). Another kind of kidney hypoxic injury is caused by local hypoxia, in which oxygen sufficiency usually originates from a decreased renal or intrarenal blood supply (8). Both kinds of kidney hypoxic lesion can lead to acute kidney injury (AKI), which is a common cause of in-hospital mortality (9). As for the mechanism of hypoxic kidney injury, the excessive and sustaining activation of hypoxia-inducible factor-1-α (HIF-1α) was deemed a critical event (10–12).
Hypoxia-inducible factors (HIFs) are the key transcriptional factors in the cell to induce hypoxia response. Three isoforms of HIF are found in mammals: HIF-1α, HIF-2α and HIF-3α. Only HIF-1α is found in all cell types, and it acts as a core regulator (1). In addition to transcribing adaptive genes, HIF-1α also transcribes some pro-apoptotic genes, such as Bcl-2 interacting protein 3 (BNIP3) and Bcl-2 interacting protein 3-like (NIX) (13), and stabilizes the tumor suppressor p53 (14) to induce apoptosis. Therefore, HIF-1α has been reported as a risk factor for renal disease (15). HIF-1α is controlled by a series of complex and elaborate mechanisms because of its abovementioned vital function. Several studies have reported an interesting resolution pattern of HIF-1α, which showed that HIF-1α is not continuous, but undergoes a transient increase followed by a decline under hypoxic conditions (16,17). But canonical oxygen-dependent protein degradation mediated by prolyl hydroxylase domain-containing enzymes (PHDs) cannot competently explain this phenomenon (18). Other regulatory mechanisms of HIF-1α resolution, such as microRNA (miRNA)-mediated regulation, have attracted interest in recent years (16).
miRNAs are small non-coding RNAs that were discovered in recent decades. They regulate target genes primarily by silencing mRNA at the post-transcriptional level through interacting with mRNA untranslated regions (UTRs). Hypoxia regulates a cluster of specific miRNAs, which are termed hypoxamiRs (19). microRNA-210 (miR-210) is known as the master hypoxamiR, as it is ubiquitously expressed in a wide range of cell types, is induced by HIF-1α and regulates a variety of responses to cope with hypoxia-induced stress (20,21). However, to date, the role of miR-210 in hypoxic renal injury is unknown. Moreover, the relationship between miR-210 and HIF-1α has never been studied in the kidney before. A small number of studies, the results of which were controversial, focused on the impact of miR-210 on HIF-1α. One study showed that miR-210 regulates HIF-1α via indirect positive feedback (22), whereas another group observed that miR-210 regulates HIF-1α via negative feedback in differentiated T cells (23).
In this study, we established a systemic hypoxia rat model using a hypobaric chamber (7500 m, 7 h) and a local kidney hypoxia rat model using a microvascular clamp to induce hypoxic injury. We observed that both hypoxic conditions led to kidney lesions accompanied by obvious HIF-1α pathway activation. Simultaneously, miR-210 was also found to be significantly upregulated in both kinds of kidney lesions and connected to injury severity. Using the human renal tubular epithelial HK-2 cell line, we proved that miR-210 protects against apoptosis induced by severe hypoxia. Subsequently, we verified that HIF-1α is a target gene of miR-210 in HK-2 cells, and HIF-1α activation can thus be suppressed by miR-210 to attenuate hypoxia-induced apoptosis. Collectively, these results reveal that increased miR-210 expression in hypoxia-induced kidney lesions attenuates cell injury and apoptosis by targeting HIF-1α in severe hypoxia.
MATERIALS AND METHODS
Animal Maintenance, Systemic Hypoxia and Local Kidney Hypoxia Models
Six-to-eight-week-old male Sprague–Dawley rats with body weights of 180–220 g were housed in a pathogen-free and temperature-controlled room (22°C ± 2°C) with a 12 h light and 12 h dark cycle. Food and water were available ad libitum. For the systemic hypoxia model, the rats chosen randomly for the experimental group were put in a hypobaric chamber (Fenglei) to mimic an elevated altitude of 7500 m (9.0% O2) within 10 min, and the rats in the control group were placed in the corresponding normoxia condition. An acute kidney injury model was used to induce local kidney hypoxia. In brief, Sprague-Dawley rats were anesthetized with sodium pentobarbital (30 mg/kg intraperitoneally; Sigma) under aseptic conditions. The bilateral renal arteries were then exposed and occluded using a microvascular nontraumatic bulldog clamp via a flank incision. Following warm ischemia of 60 min, the rats were euthanized and kidney tissue were removed for further study. The sham group rats were only anesthetized and their kidneys were exposed under the same conditions, without clamping. All animal experimental procedures complied fully with the regulations of the Institute of Basic Medical Sciences.
Cell Culture, Hypoxia and Transfection
HK-2 cells were cultured in Dulbecco’s modified Eagle’s medium/Ham’s Nutrient Mixture F-12 (Gibco) supplemented with 10% (v/v) fetal bovine serum (Gibco), 25 U/mL penicillin (Hyclone) and 25 μg/mL streptomycin (Hyclone) at 37°C in a humidified incubator containing 5% (v/v) CO2. The hypoxic environment of cultured cells was realized in a hypoxia glove box (Coy), in which nitrogen gas was infused to obtain an oxygen concentration of 0.3% and to maintain a CO2 concentration of 5% at a temperature of 37°C. Cells were transfected with 100 nM miR-210 mimic, inhibitor, HIF-1α siRNA and negative control (Ribo) and 1ug plasmids using TurboFect Reagent (Thermo), according to the manufacturer’s protocols.
RNA Extraction and Real-Time Polymerase Chain Reaction
Total RNA was extracted from tissue, cell, serum and medium using RNAiso plus (Takara), according to the manufacturer’s instructions. Reverse transcription (RT) was performed using an RT kit (Takara) with miRNA-specific primers (Ribo) or oligo dT (Takara), respectively. pri-miR-210 reverse transcriptional primer: 5′-CACAGATCAGCCGCTGTCAC-3′. Real-time RNA quantification was conducted on an ABI StepOne Plus Detection System (Applied Biosystems) using Power SYBR Green PCR Master Mix (Applied Biosystems) and specific primers. The primer sequences are shown in Supplementary Table S1. The miR-210 primer, RNU6 primer and miR-39 primer were purchased from Ribo. Spike-in miR-39 miRNA (Ribo) was used for quantification of extracellular miR-210 levels.
Detection of Cell Apoptosis by Flow Cytometry
The HK-2 cells were trypsinized, collected and then labeled using an Annexin V-PI apoptosis kit (Dijindo). The cells were incubated with fluorescein isothiocyanate (FITC)-labeled Annexin V and propidium iodide (PI) solution in the dark for 15 min. Flow cytometry was conducted using FACS Calibur FL1 (530 nm) bandpass filters (Becton Dickinson), and the data were analyzed using CellQuest software (BD).
Western Blotting and Analysis
HK-2 cells were lysed using radioimmunoprecipitation assay (RIPA) buffer (Pierce) supplemented with protease inhibitor cocktail (Roche). The protein samples were subjected to routine sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane and incubated with primary antibodies overnight. The primary antibodies against HIF-1α (H1α 67), FAS (A-20) and β-actin (I-19), were purchased from Santa Cruz Biotechnology. Antibodies against NIX and BNIP3 (EPR4034) were purchased from Abcam. Antibody against p53 was purchased from Cell Signaling Technology. Antibody against HIF-2α was purchased from Novus Biologicals. Antibody against glucose transporter-1 (GLUT-1) was purchased from Proteintech Group. A horseradish peroxidase–conjugated secondary antibody (Santa Cruz Biotechnology) and enhanced chemiluminescence (Pierce) were applied for protein detection. The relative expression of the protein bands was quantified by densitometric scanning using Image Pro Plus (Media Cybernetics) and standardized for densitometric analysis to β-actin levels.
Kidney Periodic Acid-Schiff Staining and TdT-Mediated dUTP Nick-End Labeling Assay
Kidney tissues were fixed with 10% neutral buffered formalin for 48 h and then embedded in paraffin and cut into 3–5 μm sections using routine methods. The kidney tissue samples were stained with periodic acid–Schiff (PAS) (Sigma), and histologic structures were examined under a light microscope. Kidney TdT-mediated dUTP Nick-end Labeling (TUNEL) assay was performed using an In Situ Cell Death Detection Kit (Roche), according to the manufacturer’s instructions. Three images per kidney (eight kidneys per group) were acquired, and the positive cells were counted individually.
Luciferase Reporter Assay
The predicted seed sequence of miR-210 in the 3′UTR of human HIF-1α mRNA (length: 220 bp) and the corresponding mutant sequence (length: 220 bp) were constructed into modified pGL3-Luc control luciferase reporter plasmids (Promega). These plasmids were co-transfected into HK-2 cells with a miR-210 mimic or a negative control, as indicated. After 48 h of transfection, the cells were lysed with passive lysis buffer (Promega). Luciferase activity was measured with a dual luciferase assay system (Promega), in accordance with the manufacturer’s protocol.
Primary Glomerular and Tubular Cell Isolation
The kidneys of rats were removed, then cortical tissue was separated from the medulla and minced into a paste. The cortical paste was pressed through a 106 μm metal sieve to collect large tubular fragments and suspended in phosphate-buffered saline. This suspension was then triturated and poured over a 75 μm sieve. The glomeruli were trapped on the sieve, and the tubules were filtered through the sieve. Isolated glomerular and tubular cells were used for detection of RNA and protein expression.
Statistical Analysis
Data are shown as the mean ± standard deviation (SD) and were analyzed using Student t test, except in special cases. P < 0.05 was considered statistically significant.
All supplementary materials are available online at www.molmed.org.
RESULTS
Systemic Hypoxia and Local Kidney Hypoxia Led to Rat Renal Lesions Accompanied by HIF-1α Activation
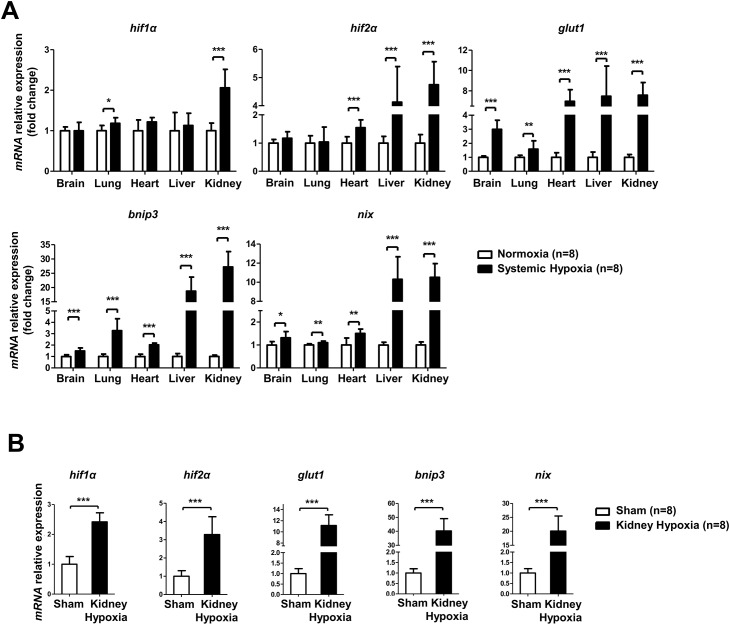
There is no evidence of HIF-1α pathway activity change in vital organs in systemic hypoxia. In our study, we used a hypobaric hypoxic rat model (7500 m for 7 h) to investigate changes in HIF-1α pathway activity in the brain, lung, heart, liver and kidney. As shown in Figure 1A, our analysis demonstrates that among these hypoxic organs, the rat kidney was found to have the most significantly increased HIF-1α mRNA expression (more than twofold). Moreover, the expression levels of HIF-1α pro-apoptotic target genes in kidney, such as BNIP3 and NIX, were upregulated 10-fold compared to baseline (Figure 1A). A similar and more significant activation of the HIF-1α pathway was also observed in the local kidney hypoxia rat model when treated with the acute kidney injury operation (Figure 1B).
Figure 1.
Systemic hypoxia and local kidney hypoxia induced HIF-1α activation in a rat model. Six-to-eight-week-old male Sprague-Dawley rats were randomly divided into normoxia (n = 8), systemic hypoxia (7500 m, 7 h; n = 8), sham (n = 8) and local kidney hypoxia (ischemia, bilateral, 60 min; n = 8) groups. (A) Changes in HIF-1α pathway genes (HIF-1α, GLUT-1, BNIP3 and NIX) and HIF-2α mRNA expression in vital organs (brain, lung, heart, liver and kidney) of normoxia and systemic hypoxia groups. (B) Changes in HIF-1α, HIF-2α, GLUT-1, BNIP3 and NIX mRNA expression in kidney of sham and local kidney hypoxia groups. All mRNA levels were assessed by real-time polymerase chain reaction and normalized to β-actin. Data are shown as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001.
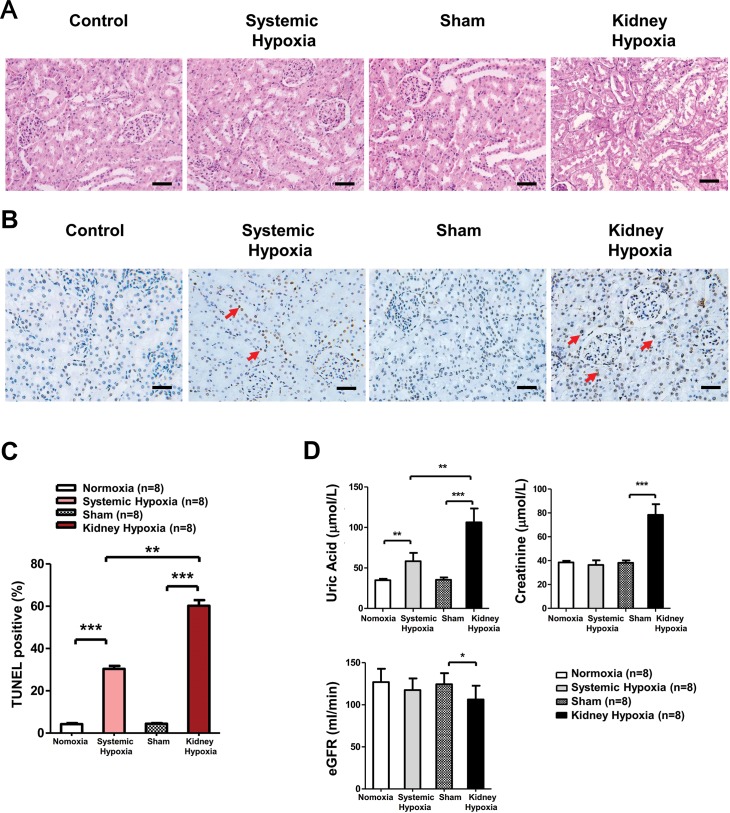
We used PAS stain and TUNEL assays to evaluate renal histopathological changes caused by hypoxia. Obvious tubular injury was observed by PAS staining in the local kidney hypoxia group (Figure 2A), while remarkable TUNEL-positive apoptotic renal cells were found in both groups (Figure 2B, C). It is worth noting that these apoptotic cells were predominantly localized in the renal tubule, in which HIF-1α is primarily expressed (24). Impaired histostructure usually comes with abnormal biochemical indicators. With regard to renal biochemical indicators, we found serum uric acid (UA), an early marker of renal injury, significantly elevated in both groups (25), while serum creatinine (Cr) and evaluated glomerular filtration rates (eGFRs), which were deemed more serious indicators, were significantly worse, predominantly in the local kidney hypoxia group (Figure 2D). Taken together, these data demonstrate the effectiveness of our hypoxia injury model and suggest that local kidney hypoxia resulted in severe renal lesion, and systemic hypoxia can also lead to moderate and early renal injury. Moreover, these results reveal that both systemic and local kidney hypoxia treatments lead to HIF-1α pathway activation accompanied by kidney lesion formation.
Figure 2.
Systemic hypoxia and local kidney hypoxia induced rat kidney lesion. (A) Representative kidney PAS staining of normoxia (n = 8), systemic hypoxia (n = 8), sham (n = 8) and local kidney hypoxia (n = 8) groups. (B) Representative kidney TUNEL staining. (C) TUNEL positive cell quantification in the kidney. The numbers of positive TUNEL stained cells were counted in eight high-power fields. Scale bar = 50 μm. The red arrows indicate typical apoptotic cells. (D) Measurement of rat serum UA, Cr, eGFR levels (n = 8). Data are shown as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001.
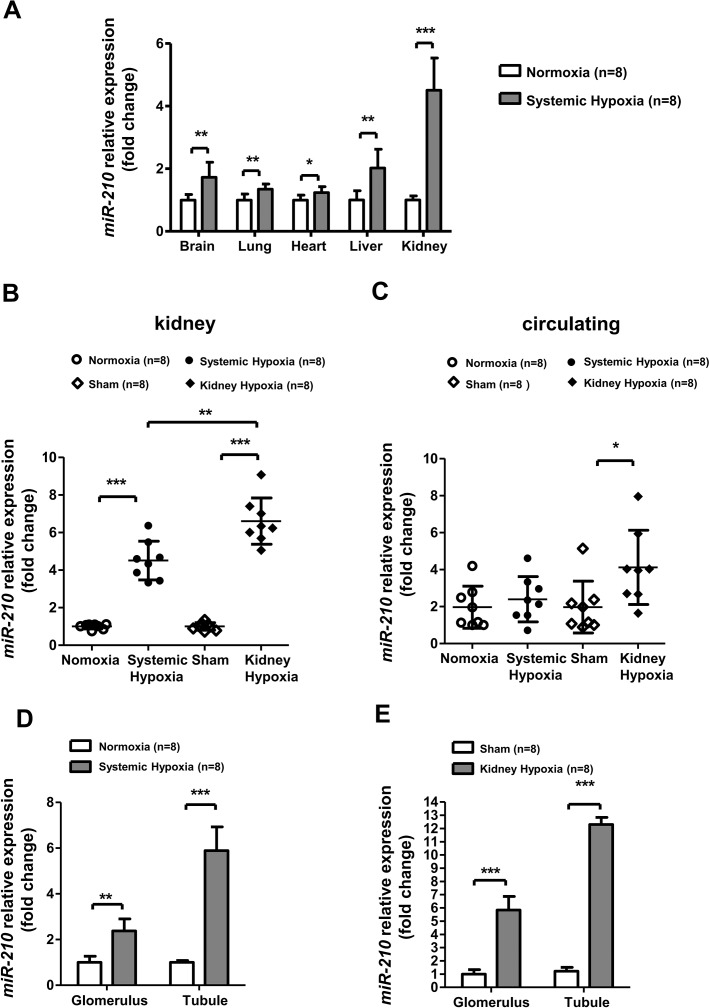
Levels of miR-210 Significantly Increased in Rat Kidney and Circulation after Systemic or Local Kidney Hypoxia
Except for the abovementioned changes in histostructure, biochemical indicator and HIF-1α activation, we also examined the expression level of miR-210 in the two hypoxia treatment groups, which was reported to be induced by HIF-1α predominantly (21). Notably, for systemic hypoxia, the most significantly upregulated miR-210 expression was found in the rat kidney among our screened vital organs (Figure 3A). For local kidney hypoxia, we observed increased miR-210 levels not only in kidney tissue (Figure 3B), but also in peripheral circulation (Figure 3C). To address the expression profile of miR-210 in the kidney in detail, we investigated miR-210 expression in primary isolated renal tubular and glomerular cells. The results demonstrate that, compared with glomerulus, miR-210 expression increased more significantly in renal tubule after systemic (Figure 3D) and local (Figure 3E) hypoxia treatments where hypoxia-induced apoptosis was predominantly present, as mentioned above.
Figure 3.
miR-210 increased significantly in rat kidney after systemic and local kidney hypoxia, especially in tubular cells. miR-210, another target of HIF-1α, was measured in normoxia (n = 8), systemic hypoxia (7500 m, 7 h; n = 8), sham (n = 8) and local kidney hypoxia (ischemia, bilateral, 60 min; n = 8) groups. (A) miR-210 expression in the vital organs (brain, lung, heart, liver and kidney) was measured after systemic hypoxia. (B) miR-210 expression in (B) kidney and (C) circulation were measured in the four groups. miR-210 expression in isolated primary tubular cells and glomerular cells from renal cortex after (D) systemic hypoxia and (E) local hypoxia. miR-210 expression in isolated primary tubular cells and glomerular cells from renal cortex after systemic hypoxia. miR-210 levels were measured by real-time PCR. To measure circulating miR-210 level, miR-39 was used for the spike-in. Renal miR-210 levels were normalized to RNU6, and circulating miR-210 levels were normalized to miR-39. Data are shown as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001.
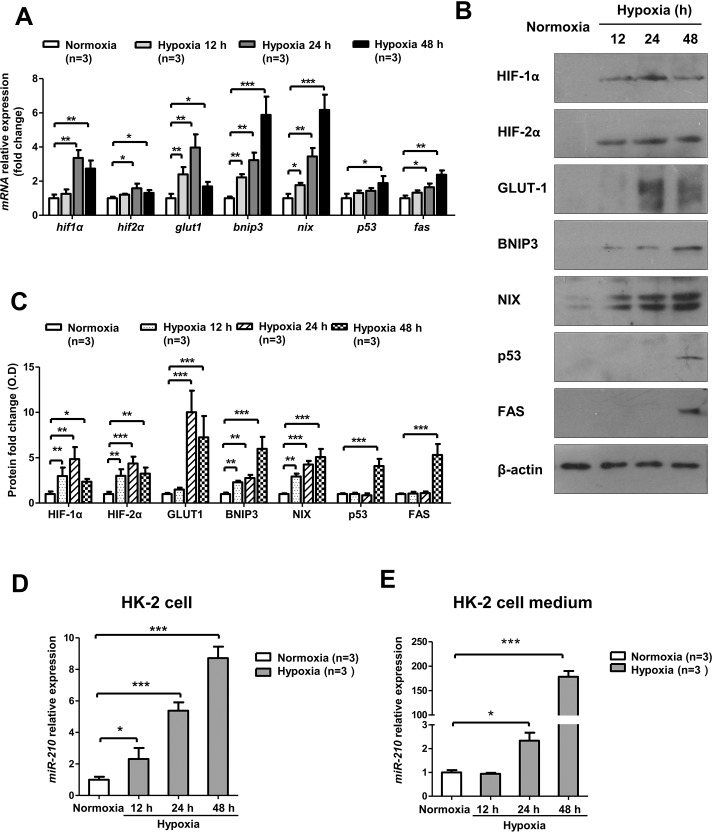
Severe Hypoxia-Induced HIF-1α Pathway Activation and Apoptotic Gene Expression Accompanied by miR-210 Elevation in Human Renal Tubular HK-2 Cells
As most of the apoptotic cells were localized in the renal tubules, where miR-210 was remarkably upregulated in vivo, we conducted an in depth study in vitro involving human renal tubular epithelial HK-2 cells.
HIF-1α pathway activation was also observed in HK-2 cells after severe hypoxia (0.3% O2). Interestingly, HIF-1α mRNA levels were increased in the early phase of hypoxia and later decreased, though the hypoxic condition remained unchanged. Similar findings were observed regarding GLUT-1 and HIF-2α mRNA levels. However, the mRNA levels of BNIP3 and NIX, the pro-apoptotic target genes of HIF-1α, gradually increased (Figure 4A). Regarding protein expression, these genes displayed changes that were similar to those in their mRNA expression patterns (Figures 4B, C). In addition, the mRNA levels of some cell apoptotic genes, such as p53 and FAS, were also increased after hypoxia (Figure 4A), and similar changes in their protein expression were detected (Figures 4B, C). Moreover, we found that HK-2 cells exposed to hypoxia experienced a significant increase in cellular (Figure 4D) and extracellular (Figure 4E) miR-210 expression compared with normoxia, a finding consistent with our results in vivo. To examine which step of miR-210 biogenesis might be modulated by hypoxia, we measured pri-miR-210 after hypoxia and found that it was apparently upregulated after hypoxia, which indicates that miR-210 was regulated at the transcriptional level in hypoxia (Figure S2).
Figure 4.
HIF-1α activation and apoptotic gene expression were observed after severe hypoxia in HK-2 cells, with simultaneous miR-210 upregulation. HK-2 cells were cultured in normoxia (21% O2) or hypoxia (0.3% O2) and harvested at 12 h, 24 h and 48 h. (A) mRNA analysis and (B) protein analysis of HIF-1α pathway genes (HIF-1α, GLUT-1, BNIP3 and NIX), HIF-2α and the apoptosis-associated genes p53 and FAS. (C) Optical density quantitative analysis of Western blots in (B). (D) Real-time PCR analysis of miR-210 levels in HK-2 cells. (E) Real-time PCR analysis of miR-210 levels in HK-2 cell medium. All mRNA expression levels were normalized to β-actin (n = 3). Cellular miR-210 levels were normalized to RNU6 (n = 3), and extracellular miR-210 levels were normalized to miR-39 (n = 3). The Western blotting relative expressions are compared with β-actin (n = 3). Data are shown as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001.
In summary, severe hypoxia induced HIF-1α and apoptosis pathway activation in renal tubular cells and increased miR-210 expression in vitro.
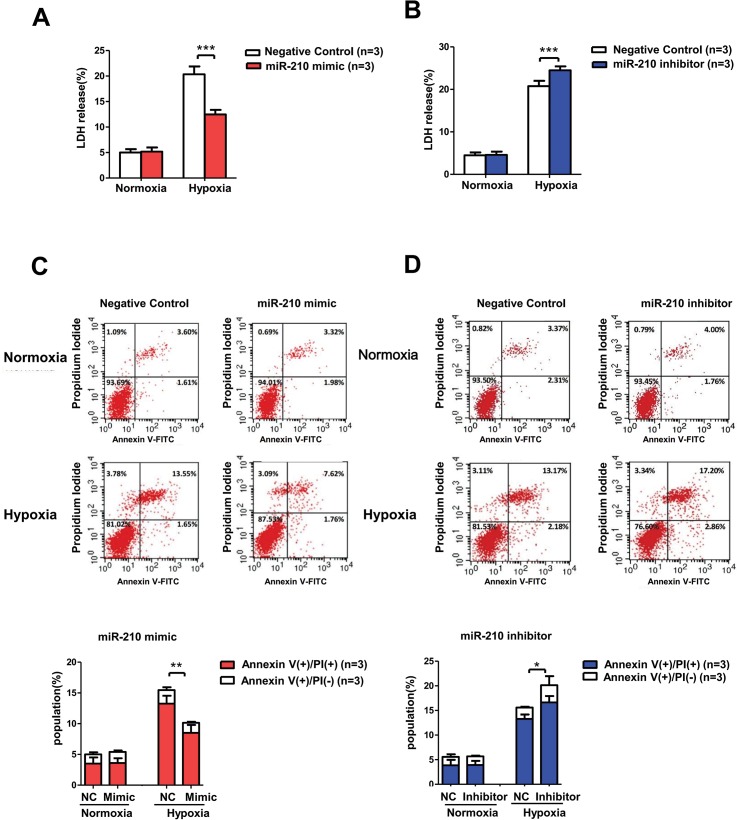
miR-210 Alleviated Hypoxia-Induced Cell Injury and Apoptosis in HK-2 Cells
Based on our findings that miR-210 levels increased in the renal tubular cells after hypoxia both in vivo and in vitro, we subsequently explored the pathophysiological role of miR-210 using miR-210 overexpression and knockdown techniques. Cellular miR-210 level was notably increased ( >2000-fold) when transfected with mimic, while it decreased to 10% after being transfected with inhibitor (Figure S1). Lactose dehydrogenase (LDH) assay was performed to assess cell injury. The results of the LDH assay demonstrate that severe hypoxia (0.3% O2) damaged HK-2 cells, as LDH release increased by approximately fourfold after 48 h of treatment (Figure 5A, NC group). This increase could be partially blocked by the miR-210 mimic (Figure 5A) and aggravated by the miR-210 inhibitor (Figure 5B). Moreover, we employed annexin V/PI double-staining flow cytometry assay to measure cell apoptosis. Consistent with the LDH assay results, the flow cytometry data showed that the total apoptosis rate of the miR-210 mimic group was significantly lower than that of the NC group after hypoxia treatment (Figure 5C), while the total apoptosis rate of the miR-210 inhibitor group, particularly the late apoptosis rate, was augmented compared with that of the NC group (Figure 5D). These results suggest that miR-210 plays a protective role in renal tubular cells by attenuating hypoxic injury and apoptosis in severe hypoxia.
Figure 5.
miR-210 overexpression reduced hypoxic cell injury and apoptosis in HK-2 cells, while miR-210 knockdown exerted the opposite effect. We transfected miR-210 mimic and inhibitor into HK-2 cells and conducted LDH and annexin V/PI assays after normoxia (21% O2) or hypoxia (0.3% O2) treatment for 48 h. LDH release was evaluated in cell medium after (A) miR-210 overexpression or (B) miR-210 knockdown in HK-2 cells treated with hypoxia. Cell apoptosis was analyzed in the (C) miR-210 mimic group and (D) miR-210 inhibitor group using annexin V/PI staining flow cytometry assay. Units of the Y and X axes are fluorescence intensity. Annexin V (+) PI (+), annexin V (+) PI (−), annexin V (−) PI (+) and annexin V (−) PI (−) indicated late apoptotic, early apoptotic, necrotic and living cells, respectively. All data are repeated from three independent experiments (n = 3). Data are shown as mean ± SD. NC, negative control. *P < 0.05; **P < 0.01; ***P < 0.001.
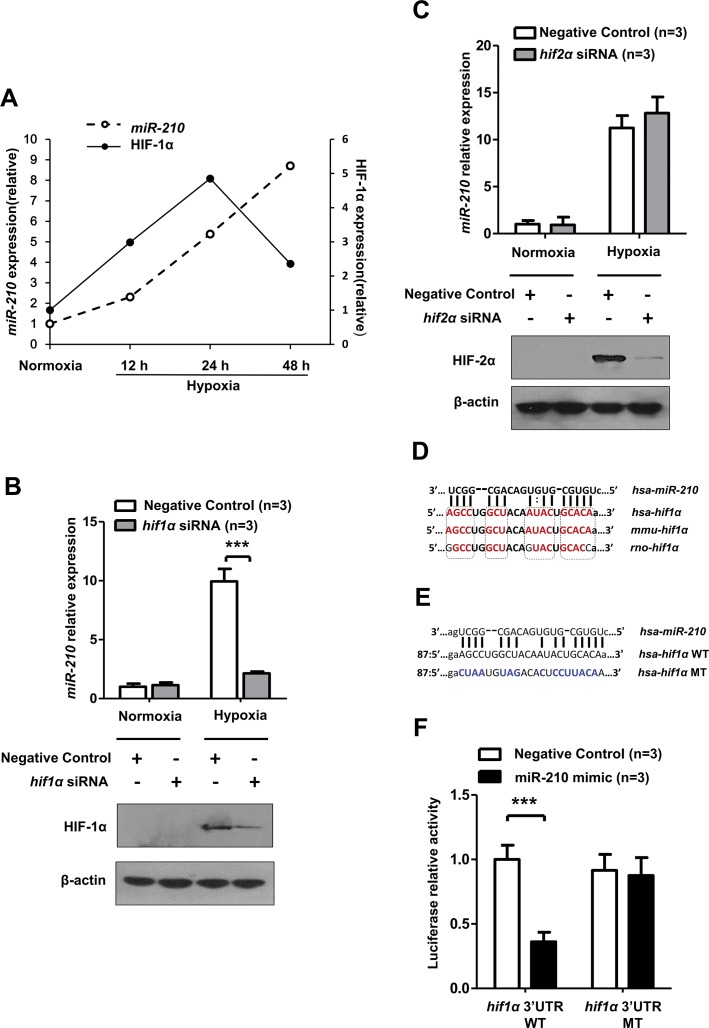
miR-210 Targeted 3′UTR of HIF-1α mRNA Directly in Hypoxia
We noted that HIF-1α protein expression began to decrease in late hypoxia in conjunction with miR-210 accumulation, suggesting that miR-210 may negatively regulate HIF-1α when hypoxia stress is aggravated (Figure 6A). The correlation between miR-210 and HIF-1α in hypoxic kidney lesion has not been studied before. In addition, although miR-210 is a known target of HIF-1α, no consensus has been reached regarding how miR-210 affects HIF-1α, as controversial results in different cell types and under different conditions have been obtained by multiple research groups (22,23).
Figure 6.
miR-210 targeted 3′UTR of HIF-1α mRNA directly. (A) The relevance of miR-210 and HIF-1α protein expression in hypoxia-treated HK-2 cells (0.3% O2). The densitometry results for relative HIF-1α protein expression were normalized to β-actin. Detection of miR-210 expression after (B) HIF-2α and (C) HIF-1α siRNA transfection and in HK-2 cells. miR-210 expression was normalized to RNU6. The predicted binding site of miR-210-3p in the 3′UTR of human HIF-1α mRNA was found at the microRNA.org website (www.microRNA.org). (D) The potential binding site of miR-210-3p in the 3′UTR of HIF-1α mRNA is highly conserved among the selected mammals. The high conserved bases are shown in red. (E) The mutant-type (MT) design of the miR-210-3p binding sequence in the 3′UTR of human HIF-1α mRNA. WT, wild-type. (F) Luciferase reporter assays were conducted to determine the impact of miR-210-3p mimic on the activity of the 3′UTR of the human WT HIF-1α or the 3′UTR of the MT HIF-1α luciferase plasmid in HK-2 cells. Luciferase activity data were obtained after normalizing the firefly luciferase value to the renilla luciferase value. All quantitative results are from three independent experiments and are shown as mean ± SD. The Western blot data are representative results from three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
After knockdown of HIF-1α, miR-210 upregulation in hypoxia-treated HK-2 cells was significantly inhibited, which indicates that HIF-1α is necessary for miR-210 induction in renal tubular epithelial cells (Figure 6B). HIF-2α knockdown had no such impact on miR-210 (Figure 6C) and other gene (Supplementary Figure S3) expressions. Subsequently, we successfully found the seed sequence of miR-210-3p in the 3′UTR of human HIF-1α mRNA via using the Memorial Sloan-Kettering Cancer Center miRNA database (www.microrna.org/). This sequence is highly conserved in several mammals, as reported by other groups (23) (Figure 6D). We inserted this seed sequence (wild-type) and the corresponding mutated sequence (mutant-type) into a luciferase reporter vector to examine whether miR-210 binds the 3′UTR of human HIF-1α mRNA directly in HK-2 cells (Figure 6E). As shown in Figure 6F, the miR-210 mimic sharply decreased the activity of the wild-type 3′UTR of HIF-1α mRNA but had no significant impact on the mutant-type 3′UTR.
These results indicate that, in renal tubular cells, HIF-1α is necessary for miR-210 induction, and miR-210 can target the 3′UTR of HIF-1α mRNA reversely, which implies that miR-210 may contribute to the prevention of excessive accumulation of HIF-1α in the abovementioned hypoxic renal injury.
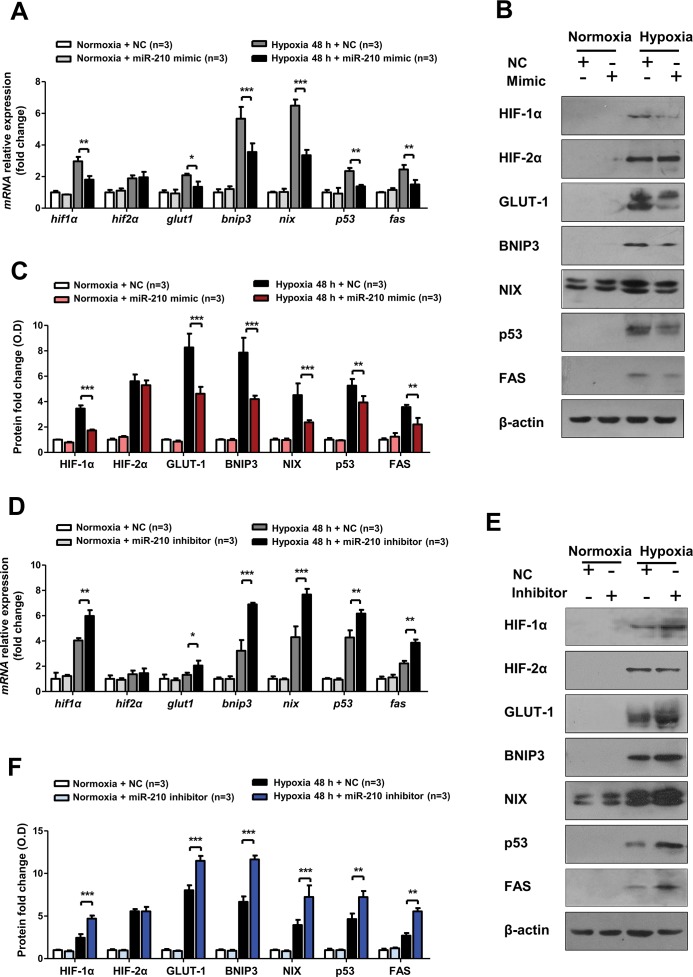
miR-210 Attenuated Hypoxic Apoptosis by Suppressing HIF-1α Activation
Previous studies have shown that HIF-1α activation is a risk factor for kidney disease (10,26,27) and that activated HIF-1α may transcribe some pro-apoptotic genes, such as BNIP3 and NIX, or stabilize p53, leading to apoptosis. We explored how the HIF-1α and apoptosis pathways are altered by miR-210 overexpression or knockdown, since miR-210 targeted 3′UTR of HIF-1α mRNA directly.
As shown in Figures 7A, B and C, the transfected miR-210 mimic had suppressive effects on both HIF-1α mRNA and protein expression in HK-2 cells in hypoxia. miR-210 overexpression significantly inhibited the target genes of HIF-1α, such as GLUT-1, BNIP3 and NIX, at the mRNA and protein levels after 48 h of hypoxia. Additionally, the miR-210 mimic blocked expression of the pivotal apoptotic genes p53 and FAS at both the mRNA and protein levels (Figures 7A–C). In contrast, ectopic expression of the miR-210 inhibitor promoted HIF-1α mRNA and protein expression, as well as expression of the abovementioned HIF-1α target genes and p53 and FAS (Figures 7D–F). However, there was no obvious change in HIF-2α expression when miR-210 was overexpressed or knocked down.
Figure 7.
miR-210 overexpression suppressed HIF-1α pathway activation in HK-2 cells after hypoxia, while miR-210 knockdown displayed the reverse effect. (A)Real-time PCR and (B) representative Western blotting were employed to detect the mRNA and protein expression levels of the HIF-1α pathway genes (HIF-1α, GLUT-1, BNIP3 and NIX), HIF-2α and the pivotal apoptotic genes (p53 and FAS) in HK-2 cells transfected with a miR-210 mimic and negative control under normoxia (21% O2) or hypoxia (0.3% O2) conditions for 48 h. (C) Optical density quantitative analysis of Western blots in (B). Regarding miR-210 inhibitor transfection, the mRNA expression levels of the abovementioned genes are shown in (D), and the corresponding protein expression are shown in (E). The optical density quantitative analysis of Western blots in (F). All quantitative results are from three independent experiments. All mRNA expression levels and Western blotting relative expressions were compared with β-actin (n = 3). Data are shown as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001.
In summary, these findings show that, in hypoxic renal tubular cell injury, HIF-1α is a target gene of miR-210, and miR-210 can attenuate cell apoptosis by blocking HIF-1α pathway activation.
DISCUSSION
AKI is a syndrome characterized by rapid deterioration of kidney function within several hours to several days, is a common disease in critically ill patients and is often diagnosed in the context of other acute illnesses (28). Among the primary causes of AKI are renal hypoxia and energy supply failure after kidney ischemia. Expanding our understanding of the hypoxic pathophysiology of AKI will provide useful therapeutic methods in the clinic. In kidneys, miRNAs play an indispensable role in maintaining renal homeostasis and are involved in various kidney diseases (29).
Many studies have demonstrated that miR-210 is the most consistently upregulated miRNA induced by hypoxia (20,30) and HIF-1α (21,22,31). Indeed, miR-210 was highly increased in both kidney (32) and serum (33) in clear cell renal cell carcinoma patients, which is associated with a mutation of von Hippel-Lindau–induced HIF-1α accumulation. But its expression in hypoxic kidneys is unknown. In our work, we observed HIF-1α pathway activation, apoptosis formation and robust miR-210 upregulation simultaneously occurring in the rat kidney after hypoxia. In recent years, circulating miRNAs were found (34) and regarded as novel biomarkers for some diseases, and our study observed that serum miR-210 was increased only in the local hypoxia group. The reason for the serum miR-210 remaining unchanged in systemic hypoxia may partly be because the kidney hypoxia was mild and early in this situation. Similar results were observed in HK-2 cells in hypoxia. We also found that the expression levels of pro-apoptotic target genes of HIF-1α, such as BNIP3 and NIX, were remarkably increased. Using TUNEL assay, we observed that the numbers of apoptotic cells increased notably in the kidney, especially in the renal tubular region, where HIF-1α is predominantly expressed (24). Compared with other vital organs, the kidney, especially the renal tubule, expresses high levels of miR-210. In summary, we found that miR-210 and HIF-1α are closely related with respect to their expression and localization in hypoxic kidney lesions in vivo. We also observed elevated serum Cr, UA levels and declined eGFR in hypoxia-treated rats, which is further support of the harmfulness of hypoxia.
Although it is known that miR-210 can be induced directly by HIF-1α (21,22,31), no consensus has been reached regarding how miR-210 impacts HIF-1α. In HK-2 cells, we observed increases in HIF-1α and miR-210 expression in the early phase of hypoxia, similar to our results in vivo. Interestingly, in sustained hypoxia, HIF-1α mRNA and protein expression tended to decrease after 24 h, whereas miR-210 expression continued to increase. HIF-1α protein stability is primarily negatively regulated by PHD-dependent hydroxylation in normoxia, which results in ubiquitination and proteasome degradation (35). In hypoxia, PHD activity is inhibited, leading to increased HIF-1α protein levels (36). However, this mechanism does not sufficiently explain our observation of resolution of HIF-1α expression in prolonged hypoxia, which has also been reported by others (17,37). miRNA-mediated regulation has become a new theory illustrating the resolution of HIF-1α (16). Several studies have demonstrated that HIF-1α can be regulated by microRNAs, including miR-210 (23,38–42). However, one group reported that miR-210 negatively regulated HIF-1α in mouse T cells (23), while another group found that miR-210 overexpression indirectly resulted in HIF-1α accumulation and created a positive feedback loop in hypoxia (22). In our study, we found a potential miR-210 binding sequence in the 3′UTR of human HIF-1α mRNA. Furthermore, using a luciferase reporter assay, we verified that HIF-1α was a target gene of miR-210 in HK-2 cells. Based on the collective findings, we concluded that a negative feedback loop involving miR-210 and HIF-1α exists in hypoxic human kidney, as well as the mouse immune system. Though HIF-2α was reported to have overlapping functions with HIF-1α, in our study, knockdown of HIF-2α had no obvious effect on miR-210 and other genes, which further supports that HIF-1α was the dominant regulator for miR-210 induction (31,43). More importantly, we found that miR-210 contributes to HIF-1α resolution in prolonged hypoxia.
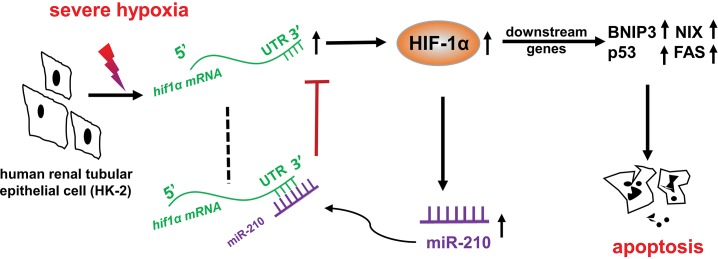
Based on the above result that HIF-1α is a target gene of miR-210 in HK-2 cells, we used overexpression or knockdown techniques to investigate the function of miR-210 in hypoxic HK-2 cells and the underlying mechanism. Using LDH and flow cytometry assays, we found that miR-210 mimic can protect HK-2 cells from hypoxic injury and apoptosis. In contrast, miR-210 inhibitor exerts adverse effects on hypoxic HK-2 cells. Many studies have shown that HIF-1α activation is a risk factor for cell survival in hypoxia (44,45) and that HIF-1α inhibition may be used as a therapeutic strategy for treating kidney damage (10). Previous studies revealed that activated HIF-1α can damage cells by inducing BNIP3 and NIX (46–48) or by stabilizing the tumor suppressor p53 (14). We observed that in late hypoxia, though HIF-1α began to decline, the apoptosis-inducing factor BNIP3, a target gene of HIF-1α, also known as the target gene of miR-210 (49), was strengthened. This might be because p53 (50) activation induced a BNIP3 increase in late hypoxia. Except that miR-210 could suppress BNIP3 via HIF-1α, it has a direct suppressive effect on BNIP3. However, our results show that the extent of BNIP3 downregulation was similar to other hypoxia-related genes after miR-210 mimic or inhibitor transfection, implying that BNIP3 was mostly regulated in a HIF-1-dependent way, supported by a previous study (47). We found that the levels of p53 and FAS were both decreased after miR-210 overexpression. In contrast, miR-210 knockdown resulted in HIF-1α and apoptosis pathway activation. Though miRNA mimic is chemically synthesized, these results suggest that miR-210 possesses the capacity to exert renoprotective effects by suppressing HIF-1α pathway activation. In summary, we found that miR-210 expression increases significantly in the kidney in hypoxia-induced injury and has the potential to alleviate hypoxic renal tubular apoptosis by directly targeting HIF-1α (Figure 8), Further work will be required to determine the consequence of miR- 210 overexpression through genetic approaches.
Figure 8.
Schematic diagram of the negative feedback loop involving miR-210 and HIF-1α in renal tubular cells under severe hypoxic conditions.
miR-210 was proven to have beneficial effect on ischemic diseases such as acute peripheral ischemia (51), cerebral ischemia (52) and ischemic heart disease (53). It has been reported that plasma miR-210 was upregulated (54) and serum miR-210 was decreased (55) in AKI patients. However, the biologic function of miR-210 in kidney ischemia remains unclear. Our results were obtained immediately after renal ischemia in vivo, which is normally prior to the manifestation of clinical AKI. And the study in vitro showed for the first time that increased miR-210 has a renoprotective function in kidney hypoxic injury. miRNAs are regarded as sensitive biomarkers in different diseases and at different scales. Our findings imply that miR-210 has the possibility to be a promising biomarker and potential therapeutic target for early detection and treatment of hypoxic kidney injury.
CONCLUSION
This study reveals that both HIF-1α and its target miR-210 are significantly upregulated in hypoxic kidney injury in vivo and in vitro. Increased miR-210 reversely targets HIF-1α directly to suppress its expression in renal tubular cells. This negative feedback loop contributes to HIF-1α resolution in severe hypoxia and further inhibits the downstream pro-apoptotic gene expression of HIF-1α, thereby attenuating hypoxic kidney damage. Considering the complexity and poor outcome of AKI, our results provide an expanded understanding of AKI mechanism and points to a potential role for miR-210 in developing novel diagnosis and intervention of hypoxic kidney injury in the future.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the grants from the Natural Science Foundation of China (Nos. 81430044 and 31271205) and the National Basic Research Program of China (Nos. 2011CB910800 and 2012CB518200).
Footnotes
Online address: http://www.molmed.org
DISCLOSURE
The authors declare they have no competing interests as defined by Molecular Medicine, or other interests that might be perceived to influence the results and discussion reported in this paper.
Cite this article as: Liu L-L, et al. (2017) miR-210 protects renal cell against hypoxia-induced apoptosis by targeting HIF-1 alpha. Mol. Med. 23:258–71.
REFERENCES
- 1.Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valtin H. Renal Function: Mechanisms Preserving Fluid and Solute Balance in Health. Boston: Little Brown; 1983. [Google Scholar]
- 4.Legrand M, Mik EG, Johannes T, Payen D, Ince C. Renal hypoxia and dysoxia after reperfusion of the ischemic kidney. Mol Med. 2008;14:502–16. doi: 10.2119/2008-00006.Legrand. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horkan CM, et al. The association of acute kidney injury in the critically ill and postdischarge outcomes: a cohort study. Crit Care Med. 2015;43:354–64. doi: 10.1097/CCM.0000000000000706. [DOI] [PubMed] [Google Scholar]
- 6.Masewu A, et al. Acute kidney injury is a powerful independent predictor of mortality in critically ill patients: a multicenter prospective cohort study from Kinshasa, the Democratic Republic of Congo. BMC Nephrol. 2016;17:118. doi: 10.1186/s12882-016-0333-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Semenza GL, et al. Hypoxia, HIF-1, and the pathophysiology of common human diseases. Adv Exp Med Biol. 2000;475:123–30. doi: 10.1007/0-306-46825-5_12. [DOI] [PubMed] [Google Scholar]
- 8.Thadhani R, Pascual M, Bonventre JV. Acute renal failure. N Engl J Med. 1996;334:1448–60. doi: 10.1056/NEJM199605303342207. [DOI] [PubMed] [Google Scholar]
- 9.Vaidya VS, Ferguson MA, Bonventre JV. Biomarkers of acute kidney injury. Annu Rev Pharmacol Toxicol. 2008;48:463–93. doi: 10.1146/annurev.pharmtox.48.113006.094615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu Q, et al. Silencing of hypoxia-inducible factor-1alpha gene attenuated angiotensin II-induced renal injury in Sprague-Dawley rats. Hypertension. 2011;58:657–64. doi: 10.1161/HYPERTENSIONAHA.111.177626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nangaku M, Eckardt KU. Hypoxia and the HIF system in kidney disease. J Mol Med (Berl) 2007;85:1325–30. doi: 10.1007/s00109-007-0278-y. [DOI] [PubMed] [Google Scholar]
- 12.Kimura K, et al. Stable expression of HIF-1alpha in tubular epithelial cells promotes interstitial fibrosis. Am J Physiol Renal Physiol. 2008;295:F1023–9. doi: 10.1152/ajprenal.90209.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Althaus J, et al. Expression of the gene encoding the pro-apoptotic BNIP3 protein and stimulation of hypoxia-inducible factor-1alpha (HIF-1alpha) protein following focal cerebral ischemia in rats. Neurochem Int. 2006;48:687–95. doi: 10.1016/j.neuint.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 14.An WG, et al. Stabilization of wild-type p53 by hypoxia-inducible factor 1alpha. Nature. 1998;392:405–8. doi: 10.1038/32925. [DOI] [PubMed] [Google Scholar]
- 15.Mao S, Huang S. The signaling pathway of hypoxia inducible factor and its role in renal diseases. J Recept Signal Transduct Res. 2013;33:344–8. doi: 10.3109/10799893.2013.830130. [DOI] [PubMed] [Google Scholar]
- 16.Bruning U, et al. MicroRNA-155 promotes resolution of hypoxia-inducible factor 1alpha activity during prolonged hypoxia. Mol Cell Biol. 2011;31:4087–96. doi: 10.1128/MCB.01276-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chamboredon S, et al. Hypoxia-inducible factor-1alpha mRNA: a new target for destabilization by tristetraprolin in endothelial cells. Mol Biol Cell. 2011;22:3366–78. doi: 10.1091/mbc.E10-07-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henze AT, et al. Prolyl hydroxylases 2 and 3 act in gliomas as protective negative feedback regulators of hypoxia-inducible factors. Cancer Res. 2010;70:357–66. doi: 10.1158/0008-5472.CAN-09-1876. [DOI] [PubMed] [Google Scholar]
- 19.Leung AK, Sharp PA. MicroRNA functions in stress responses. Mol Cell. 2010;40:205–15. doi: 10.1016/j.molcel.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan YC, Banerjee J, Choi SY, Sen CK. miR-210: the master hypoxamir. Microcirculation. 2012;19:215–23. doi: 10.1111/j.1549-8719.2011.00154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang X, et al. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol Cell. 2009;35:856–67. doi: 10.1016/j.molcel.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly TJ, Souza AL, Clish CB, Puigserver P. A hypoxia-induced positive feedback loop promotes hypoxia-inducible factor 1alpha stability through miR-210 suppression of glycerol-3-phosphate dehydrogenase 1-like. Mol Cell Biol. 2011;31:2696–706. doi: 10.1128/MCB.01242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H, et al. Negative regulation of Hif1a expression and TH17 differentiation by the hypoxia-regulated microRNA miR-210. Nat Immunol. 2014;15:393–401. doi: 10.1038/ni.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosenberger C, et al. Expression of hypoxia-inducible factor-1alpha and -2alpha in hypoxic and ischemic rat kidneys. J Am Soc Nephrol. 2002;13:1721–32. doi: 10.1097/01.asn.0000017223.49823.2a. [DOI] [PubMed] [Google Scholar]
- 25.Patschan D, Patschan S, Gobe GG, Chintala S, Goligorsky MS. Uric acid heralds ischemic tissue injury to mobilize endothelial progenitor cells. J Am Soc Nephrol. 2007;18:1516–24. doi: 10.1681/ASN.2006070759. [DOI] [PubMed] [Google Scholar]
- 26.Kapitsinou PP, et al. Preischemic targeting of HIF prolyl hydroxylation inhibits fibrosis associated with acute kidney injury. Am J Physiol Renal Physiol. 2012;302:F1172–9. doi: 10.1152/ajprenal.00667.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malhotra R, Tyson DW, Rosevear HM, Brosius FC 3rd. Hypoxia-inducible factor-1alpha is a critical mediator of hypoxia induced apoptosis in cardiac H9c2 and kidney epithelial HK-2 cells. BMC Cardiovasc Disord. 2008;8:9. doi: 10.1186/1471-2261-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singbartl K, Joannidis M. Short-term effects of acute kidney injury. Crit Care Clin. 2015;31:751–62. doi: 10.1016/j.ccc.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 29.Bhatt K, Mi QS, Dong Z. microRNAs in kidneys: biogenesis, regulation, and pathophysiological roles. Am J Physiol Renal Physiol. 2011;300:F602–10. doi: 10.1152/ajprenal.00727.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giannakakis A, et al. miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biol Ther. 2008;7:255–64. doi: 10.4161/cbt.7.2.5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakada C, et al. Overexpression of miR-210, a downstream target of HIF1alpha, causes centrosome amplification in renal carcinoma cells. J Pathol. 2011;224:280–8. doi: 10.1002/path.2860. [DOI] [PubMed] [Google Scholar]
- 32.Neal CS, Michael MZ, Rawlings LH, Van der Hoek MB, Gleadle JM. The VHL-dependent regulation of microRNAs in renal cancer. BMC Med. 2010;8:64. doi: 10.1186/1741-7015-8-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao A, Li G, Peoc’h M, Genin C, Gigante M. Serum miR-210 as a novel biomarker for molecular diagnosis of clear cell renal cell carcinoma. Exp Mol Pathol. 2013;94:115–20. doi: 10.1016/j.yexmp.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 34.Chen X, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 35.Semenza GL. Oxygen sensing, hypoxia-inducible factors, and disease pathophysiology. Annu Rev Pathol. 2014;9:47–71. doi: 10.1146/annurev-pathol-012513-104720. [DOI] [PubMed] [Google Scholar]
- 36.Epstein AC, et al. C elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 37.Uchida T, et al. Prolonged hypoxia differentially regulates hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha expression in lung epithelial cells: implication of natural antisense HIF-1alpha. J Biol Chem. 2004;279:14871–8. doi: 10.1074/jbc.M400461200. [DOI] [PubMed] [Google Scholar]
- 38.Cascio S, et al. miR-20b modulates VEGF expression by targeting HIF-1 alpha and STAT3 in MCF-7 breast cancer cells. J Cell Physiol. 2010;224:242–9. doi: 10.1002/jcp.22126. [DOI] [PubMed] [Google Scholar]
- 39.Cha ST, et al. MicroRNA-519c suppresses hypoxia-inducible factor-1alpha expression and tumor angiogenesis. Cancer Res. 2010;70:2675–85. doi: 10.1158/0008-5472.CAN-09-2448. [DOI] [PubMed] [Google Scholar]
- 40.Lei Z, et al. Regulation of HIF-1alpha and VEGF by miR-20b tunes tumor cells to adapt to the alteration of oxygen concentration. PLoS One. 2009;4:e7629. doi: 10.1371/journal.pone.0007629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rane S, et al. Downregulation of miR-199a depresses hypoxia-inducible factor-1alpha and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ Res. 2009;104:879–86. doi: 10.1161/CIRCRESAHA.108.193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taguchi A, et al. Identification of hypoxia-inducible factor-1 alpha as a novel target for miR-17–92 microRNA cluster. Cancer Res. 2008;68:5540–5. doi: 10.1158/0008-5472.CAN-07-6460. [DOI] [PubMed] [Google Scholar]
- 43.Camps C, et al. hsa-miR-210 Is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res. 2008;14:1340–8. doi: 10.1158/1078-0432.CCR-07-1755. [DOI] [PubMed] [Google Scholar]
- 44.Carmeliet P, et al. Role of HIF-1alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature. 1998;394:485–90. doi: 10.1038/28867. [DOI] [PubMed] [Google Scholar]
- 45.Krick S, et al. Role of hypoxia-inducible factor-1alpha in hypoxia-induced apoptosis of primary alveolar epithelial type II cells. Am J Respir Cell Mol Biol. 2005;32:395–403. doi: 10.1165/rcmb.2004-0314OC. [DOI] [PubMed] [Google Scholar]
- 46.Greijer AE, van der Wall E. The role of hypoxia inducible factor 1 (HIF-1) in hypoxia induced apoptosis. J Clin Pathol. 2004;57:1009–14. doi: 10.1136/jcp.2003.015032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sowter HM, Ratcliffe PJ, Watson P, Greenberg AH, Harris AL. HIF-1-dependent regulation of hypoxic induction of the cell death factors BNIP3 and NIX in human tumors. Cancer Res. 2001;61:6669–73. [PubMed] [Google Scholar]
- 48.Bruick RK. Expression of the gene encoding the proapoptotic Nip3 protein is induced by hypoxia. Proc Natl Acad Sci USA. 2000;97:9082–7. doi: 10.1073/pnas.97.16.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang F, et al. miR-210 suppresses BNIP3 to protect against the apoptosis of neural progenitor cells. Stem Cell Res. 2013;11:657–67. doi: 10.1016/j.scr.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 50.Wang EY, et al. p53 mediates autophagy and cell death by a mechanism contingent on Bnip3. Hypertension. 2013;62:70–7. doi: 10.1161/HYPERTENSIONAHA.113.01028. [DOI] [PubMed] [Google Scholar]
- 51.Zaccagnini G, et al. Hypoxia-induced miR-210 modulates tissue response to acute peripheral ischemia. Antioxid Redox Signal. 2014;21:1177–88. doi: 10.1089/ars.2013.5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zeng LL, et al. Lentivirus-mediated overexpression of microRNA-210 improves long-term outcomes after focal cerebral ischemia in mice. CNS Neurosci Ther. 2016;22:961–9. doi: 10.1111/cns.12589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu S, et al. MicroRNA-210 as a novel therapy for treatment of ischemic heart disease. Circulation. 2010;122:S124–31. doi: 10.1161/CIRCULATIONAHA.109.928424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lorenzen JM, et al. Circulating miR-210 predicts survival in critically ill patients with acute kidney injury. Clin J Am Soc Nephrol. 2011;6:1540–6. doi: 10.2215/CJN.00430111. [DOI] [PubMed] [Google Scholar]
- 55.Aguado-Fraile E, et al. A pilot study identifying a set of microRNAs as precise diagnostic biomarkers of acute kidney injury. PLoS One. 2015;10:e0127175. doi: 10.1371/journal.pone.0127175. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.