Abstract
Genome-wide studies of aging have identified subsets of genes that show age-related changes in expression. Although the types of genes that are age-regulated vary among different tissues and organisms, some patterns emerge from these large data sets. First, aging is associated with a broad induction of stress response pathways, although the specific genes and pathways involved differ depending on cell type and species. In contrast, a wide variety of functional classes of genes are downregulated with age, often including tissue-specific genes. Whereas the upregulation of age-regulated genes is likely to be governed by stress-responsive transcription factors, questions remain as to why particular genes are susceptible to age-related transcriptional decline. Here, we discuss recent findings showing that splicing is misregulated with age. While defects in splicing could lead to changes in protein isoform levels, they could also impact gene expression through nonsense-mediated decay of intron-retained transcripts. The discovery that splicing is misregulated with age suggests that other aspects of gene expression, such as transcription elongation, termination and polyadenylation, must also be considered as potential mechanisms for age-related changes in transcript levels. Moreover, the considerable variation between genome-wide aging expression studies indicates that there is a critical need to analyze the transcriptional signatures of aging in single cell types rather than whole tissues. Since age-associated decreases in gene expression could contribute to a progressive decline in cellular function, understanding the mechanisms that determine the aging transcriptome provides a potential target to extend healthy cellular lifespan.
Introduction
Aging is associated with increased mortality, progressive physiological decline, and increased risk of human pathologies such as cancer, heart disease and neurodegenerative disease [1]. The progressive decline in physiological function of an organism is generally referred to as senescence [2], while the term cellular senescence specifically refers to the proliferative arrest observed in cells grown in culture after a finite number of divisions, also known as the Hayflick limit [3]. The rate and progression of senescence is influenced both by the chronological age of the organism and by genetic and environmental factors. Dynamic changes in gene expression occur during aging, and are influenced by environmental stimuli and genetic factors. The transcriptome of a cell reflects both transcription and RNA processing events such as splicing and polyadenylation. Here, we broadly define transcriptional signatures of aging as the set of processed transcripts that are differentially expressed during chronological aging following completion of development.
The molecular changes that occur during senescence have been categorized into nine hallmarks of aging [1]. One such hallmark of aging is depletion of stem cell reserves, resulting in part from cellular senescence due to telomere attrition [1, 4]. Other hallmarks of aging include genomic instability, mitochondrial dysfunction, epigenetic alterations, altered intracellular communication, deregulated nutrient sensing and loss of proteostasis [1]. These molecular hallmarks of aging both impact, and are influenced by, transcriptional changes. The transcriptional signatures of aging have been identified for a number of species in different cell types and tissues, with remarkably little overlap [5–8]. While individually these studies have identified potential biomarkers for aging, they also raise the question as to the long-term effect of cumulative changes in expression of multiple genes within a cell: Are these transcriptional changes protective or detrimental? Identifying the mechanisms that lead to age-associated transcriptional changes could provide potential targets for therapies to delay the onset of age-associated diseases by enhancing protective responses and suppressing detrimental changes. However, the low correlation in transcriptional signatures of aging observed in different studies provides a challenge to identifying such mechanisms.
There are different models for aging that have implications for the potential mechanisms that could lead to age-associated transcriptional changes [9]. Evolutionary theories of aging for species that reproduce repeatedly throughout their lifespan seek to explain longevity in terms of natural selection on the level of the organism rather than the cell. These aging theories can be broadly categorized as programmed or passive [9]. The concept of aging as a genetically programmed trait, framed in evolutionary terms, is based on the idea that aging is beneficial for the species as a whole [10]. Removing older individuals could benefit the population by preventing overcrowding and increasing the rate of evolution [9]. While this model is widely disputed [9], it is clear that aging can be regulated since mutations in genes such as daf-2 or daf-16, encoding the IGF-1 receptor and FOXO respectively, can extend lifespan in worms (Caenorhabditis elegans) [11]. Moreover, there are clear correlations between genetic loci, such as APOE, FOXO3, 5q33.3 and ACE, and longevity [11–20]. While these genes clearly modulate lifespan, their association with longevity does not necessarily imply that lifespan itself is under selection pressure. Arguing against this idea is the observation that the force of natural selection decreases with age [21]. Since animals are vulnerable to predation or disease, an animal’s potential to produce future offspring declines with age, resulting in decreasing natural selection pressure with chronological age [21]. This decreasing natural selection pressure during aging forms the basis of the three major passive theories of aging that each seek to explain aging at the organismal level.
The passive theories for aging center around the concept that aging could have evolved by sacrificing late survival for early reproduction [22]. In these models, diverting resources towards growth and reproduction enhances fitness at the cost of maintaining cellular function [2, 22, 23]. These theories suggest that tissues within an organism, and individual cells within those tissues, age because they lack either the resources or mechanisms to fully maintain long-term cellular function. The first of these theories, the disposable soma theory, states that an organism balances growth and reproduction with preventing cellular damage by diverting resources towards growth and reproduction, thus enhancing fitness at the cost of maintaining cellular function [22, 23]. The second antagonistic pleiotropy theory proposes that some genes that are important at the early stages of the life of an organism reduce fitness later during life [2]. The third mutation accumulation theory proposes that age-associated deleterious mutations would be weakly selected against, resulting in their accumulation in a species over evolutionary time [21]. All three of these theories describe how aging could have evolved by sacrificing late survival for early reproduction [22]. These concepts fit with the idea that aging is a balance between damage and repair [11, 24], with organisms distributing energy expenditure between maintaining function and producing offspring. Genetic or environmental factors that shift this balance can alter aging.
In this review, we compare aging transcriptome studies to obtain insight into the mechanisms that could be involved in age-associated changes in gene expression. We focus on those studies examining chronological aging, rather than longevity per se, in healthy wild-type animals. We identify common classes of genes that are misregulated with age across different species, tissues and cell types. We highlight new studies describing a pervasive role for splicing and RNA processing in aging. Additionally, we describe potential mechanisms, consistent with the passive theories of aging, to describe how specific aspects of transcription and RNA processing could be particularly vulnerable to aging.
Global versus local changes in gene expression
The failure to maintain proper cellular homeostasis with age might be expected to result in a global deregulation of gene expression. An early study implicating gene expression changes as a major component of aging found a decrease in the amount of processed mRNA in the cytoplasm of aged rat liver cells, consistent with a global decrease in gene expression [25]. However, later studies in a variety of cell types and organisms indicated that relatively few genes show age-related changes in transcript levels. In contrast to a global deregulation of gene expression with age, only 4% of genes show age-related changes in expression in either human brain or kidney tissue, or in monkey (Macaca mulatta) skeletal muscle [26–29]. Moreover, fewer than 2% of genes show altered expression profiles with age in human skin, achilles tendon, blood leucocytes and retina, or rodent brain, skeletal muscle, liver and heart tissue [6, 30–39]. Further, no more than 4% of mouse genes have age-associated changes in expression in either photoreceptor neurons or microglia [40, 41]. The small number of genes with age-associated changes in expression is not limited to mammals; only 3% of genes in fruit fly (Drosophila melanogaster) heart tissue show altered expression profiles during normal aging [42].
Studies that have identified larger numbers of age-regulated genes can be largely explained by the statistical power of the study, cell cycle changes, or by tissue heterogeneity. For example, studies with large statistical power such as a meta-analysis of approximately 1500 human blood samples identified 12.5% of expressed genes as being differentially expressed with age [8]. In addition, changes in cell cycle profiles in old yeast cells and aged mouse hematopoietic stem cells largely account for the transcriptional changes observed in these aging cells [43–46]. Tissue heterogeneity provides a particular challenge in interpreting transcriptome data from aging cells. This becomes apparent when comparing the large number of fruit fly genes (23 – 33%) identified as being age-regulated in whole flies or large body parts, with the much smaller number of genes (3%) that change in tissues such as the heart [42, 47–49]. Moreover, there are considerable differences in the number of genes that are age-regulated in different mouse tissues ranging from 0% in liver to 1.5% of the mouse genome in thymus and eye [7]. Further, the macular and peripheral neural retina, the cortex and medulla sections of the kidney and the hippocampus, entorhinal cortex, superior-frontal and post central gyrus of the brain all show different aging transcriptional signatures in humans [27, 28, 33]. The inherent heterogeneity in aging tissues, which will consist of a mixture of senescent cells, growth arrested but growth competent cells, and post-mitotic cells, is a major limitation for identifying transcriptional signatures of aging. However, it is clear that within a single cell type or tissue, only a small number of genes show transcriptional changes with age, arguing against the idea of global, widespread misregulation of gene expression during aging.
Heterogeneity in gene expression with age
Since aging is influenced both by chronological age and environmental factors, different organisms within a population will age at different rates. Likewise, different cell types within a tissue could age differently. Further, individual cells within a given cell type could also age differently. Very few studies have examined gene expression changes in single cells with age, but the limited available data on single gene targets by qPCR analysis support an increase in cellular variability in gene expression with age within a given tissue. For example, mice cardiomyocytes show a significant increase in gene expression heterogeneity relative to younger cells [50]. High levels of variability in gene expression were also observed in heart tissue from fruit flies, although this variability did not differ between young and old flies [42]. However, single cell RNA-seq data from old mouse hematopoietic stem cells showed that differing transcriptome profiles of cells reflected their cell cycle status, accounting for some of the heterogeneity in the population [51]. Recent advances in single cell RNA-sequencing techniques [52] provide an exciting opportunity to simultaneously compare age-related gene expression changes across multiple cell types and individual cells within a given tissue.
More data support the idea that there is variability in changes in gene expression across different organisms within a population with age. Middle aged individuals in a population show older or younger transcriptional signatures, depending on their genetic background and environmental exposure. For example, microarray studies of gene expression in the human frontal cortex from post-mortem samples from different individuals indicates that there is considerable variation in the gene expression profile from middle aged individuals [26]. Whereas the gene expression profiles of very old and young individuals highly correlate with other individuals in those groups, individuals within the middle aged group did not resemble each other, and instead correlated more strongly with either the young or old group [26]. Additionally, there are greater transcriptional changes between middle age and young or old samples in some human tissues, as compared with young versus old [32]. Importantly, the transcriptional signature of aging within blood from individual human patients is significantly associated with aging-associated phenotypes such as blood pressure, waist-hip ratio and smoking [8]. These data suggest that gene expression is likely to be heavily influenced by environmental or genetic factors, in addition to time itself, both in single cells within a tissue and in different individuals within a population.
What types of genes show age-associated changes in expression?
Despite the fact that only a small fraction of the genome changes with age, genes involved in a number of stress response pathways are reproducibly upregulated with age across multiple species, tissues and cell types. For example, increased expression of genes involved in stress response and oxidative damage is observed in aging human brain, retina, skin and fibroblasts (Table 1) [26, 27, 30, 32, 53]. Similarly, genes involved in stress response and inflammation show increased expression with age in rodent brain, kidney, liver, muscle, and pancreatic cells, and in fruit flies [34, 37, 39, 54]. In addition, genes that function in DNA repair are upregulated with age in both worms and fruit flies [55]. Moreover, upregulation of genes involved in MAPK signaling and the immune response is also observed in fruit flies and mice and humans [8, 36, 41, 56–58]. An increase in age-associated expression of stress response and immune response pathways is reproducibly observed in aging studies; meta-analysis of 27 data sets from mice, rats and humans identifies 56 genes consistently overexpressed with age that are enriched for immune response pathways [5]. Further, genes encoding apolipoprotein D, which protects against oxidative stress, are consistently upregulated with age in humans, monkeys, mice and flies [59–61]. Together, these studies indicate that aging is associated with an induction of general cellular stress response pathways.
Table 1.
Functional classes of genes that change with age in different organisms and tissues in the respective studies. Organisms are grouped by row, and tissue types are broadly grouped into columns. Pathways that increase with age are shown in red, and pathways that decrease with age are shown in blue. References are indicated in brackets.
| Human | Brain | Eyes and Retinas | Kidney | T-cells and Blood | Skin, Fibroblasts, Achilles Tendon |
| Stress response [26, 27] DNA repair [26, 27] Inflammation [27] MAPK [56] Synaptic Transmission [26, 27] Axon Guidance [56] Calcium Signaling [56] Protein Synthesis [27] |
Stress Response [30] Immune Response [30, 33] Cell metabolism [33] Metabolic processes [33] |
Cellular Matrix [28] Cellular Polarity [28] Insulin-like Factors [28] Heatshock [28] |
T-cell Activation [57] Cell Differentiation [57] RNA Processing [6] |
Cellular/Oxidative Damage [32, 53] Metabolic Processes [32] Cell Cycle [31, 53] Extracellular Matrix [31] |
|
| Rodents | Brain | Retinas, Photoreceptor cells and Microglia | Kidney and Liver | Skeletal and Cardiac Muscle | Pancreatic, Antibody Secreting, Stem Cells |
| Stress Response [34] Inflammation [34] Inflammation [35] Protein Turnover [34] Immune response [35] Heat Shock [35] |
Retinoic Acid Receptor [40] Insulin Metabolism [40] Immune Response [41, 58] |
Stress response [39] Inflammation [39] Apoptosis [39] Cell Cycle [39] Liver Functions [39] |
Inflammation [37] MAPK [36] Extracellular Matrix [65] Protein Synthesis [65] Inflammation [37] Signal Transduction [37] |
Inflammation [54] Apoptosis [54, 104] Glucose metabolism [104] Cell Cycle [45, 54] |
|
| Fruit Flies | Whole Organism | Head | Heart | ||
| Stress Response [47] Immune Response [47] Calcium Binding [48] Heat Shock [48] |
Synaptic Transmission[49] | Extracellular matrix [42] DNA repair [42] Potassium Ion Transport [42] ATPase Activity [42] |
|||
| Worm | Whole Organism | Muscle | |||
| Heat Shock [105] Insulin-like [105] |
Translation [106] Mitochondrial Matrix [106] |
||||
Intriguingly, one of the key regulators of longevity, FOXO, encodes a transcription factor that is required for induction of a variety of cellular pathways, including stress response [62, 63]. Thus, the upregulation of stress response pathways might play a protective role in aging cells [24]. Consistent with the passive models of aging, an upregulation in expression of genes with age is likely to reflect a response to, rather than being a driver of, the cellular decline associated with aging. For example, an increase in oxidative stress in aging cells could induce the increased DNA binding shown by the transcription factor NF-κb in aging mouse cardiac muscle [64]. Notably, NF-kb is also implicated in the age-related transcriptional changes observed in human skin [32]. In addition, since environmental stresses could influence the timing and level of upregulation of stress response genes, inducible upregulation of these genes could explain much of the heterogeneity observed both between individual cells in a tissue, and between different individuals in a population.
In contrast to the age upregulated genes, there is considerably more variety in the types of genes that are downregulated with age. Several studies have identified age-associated downregulation of genes involved in metabolism, mitochondrial function, protein synthesis, processing, transport and turnover [8, 27, 30, 34, 38, 65, 66], consistent with the loss of proteostasis and mitochondrial dysfunction identified as hallmarks of aging [1]. Intriquingly, genes associated with mitochondrial function are also implicated in longevity [55, 67, 68]. There are similarities in the aging transcriptome of related tissues in mice, suggesting that the age-related changes in gene expression are likely to be dependent on cell type [7]. Notably, in tissues such as the brain or heart, the genes that are downregulated with age tend to be associated with more tissue-specific functions [59, 65] (Table 1). For example, those genes that are downregulated with age in the human brain are enriched for neuron-specific functions such as synaptic transmission, axon guidance, and calcium signaling [26, 27, 56]. Moreover, genes associated with cardiac function, such as desmoglein-2 and dynein, which are necessary for epithelial and myocardial cell-cell junctions and muscle contractions respectively, are downregulated with age in mouse heart [65]. Additionally, there is a decline in expression of some genes in liver that are required for xenobiotic metabolism, the breakdown of toxic of foreign compounds in the body [39]. Although in general there is more variability in the types of genes downregulated with age, there is a consistent decrease in expression of genes involved in splicing and mRNA processing in rodents and humans [6, 35, 69–73].
We note that the interpretation of these studies is complicated by the previously discussed fact that most tissues are composed of a variety of different cell types. Thus, transcriptional signatures of aging reflect both changes in the proportion of each cell type in the tissue and cell cycle status. For example, the brain comprises mitotic glial cells and post-mitotic neurons, each of which has its own aging transcriptome [71]. In addition, underlying differences in sample preparation and handling, RNA isolation (polyA versus total), statistical power, and analysis approaches can all impact the types of genes identified. In particular, transcriptome analysis from human tissue samples can be challenging due to sample collection limitations, especially for post-mortem tissues. Identifying transcriptional signatures of aging in single cell types, and in fact in single cells if possible, will be critical to separate out changes in individual cells from alterations in the composition of the tissue itself.
What causes the age-related transcriptional decline of some genes?
The increased expression of stress response genes, including those that respond to DNA damage, indicates that aging cells experience higher levels of genomic damage. Indeed, accumulation of genomic DNA damage is a consistent feature of the aging cell [74]. Early studies recognized that accumulation of genomic damage could result in transcriptional defects over the life span of the organism [75]. However, whereas UV-induced DNA damage results in transcriptional silencing of a large proportion of the genome [76, 77], aging is associated with the decreased expression of only a small fraction of genes. Although the level of DNA damage in aging tissues is much lower than UV-irradiated cultured cells, the question remains as to why specific genes would be particularly vulnerable to DNA damage? One possible explanation is provided by the observation that the promoters of a subset of genes, that show age-related decreases in expression in the human brain, are more vulnerable to oxidative DNA damage in cultured neurons [26]. Thus, differences in the susceptibility of particular regions of the genome to DNA damage could account for the specificity of changes in gene expression. Another possibility is that some transcription factors might be more susceptible to oxidative damage, thereby resulting in downregulation of their target genes [78, 79]. If these transcription factors include terminal selectors, whose continual expression is required to maintain cellular identity [80], then decreased levels of these transcription factors would result in decreased expression of their tissue-specific target genes. Additionally, the expression or activity of specific transcriptional repressors could increase with age, leading to downregulation of their target genes [79]. For example, the neuron-restrictive silencer factor (REST/NRSF) is upregulated in the aging brain, where it plays a neuroprotective role [81]. Although transcription factor binding to promoter or enhancer elements in DNA is necessary for the initial stages of the transcription cycle, it is not sufficient for proper gene expression. Following recruitment of RNA polymerase II to promoters, subsequent events occur that include transcription elongation, termination, and co-transcriptional splicing and polyadenylation. All of these events contribute to proper gene expression. RNA processing events occur co-transcriptionally and are intimately associated with the process of transcription itself [82].
Notably, recent studies suggest that RNA processing, in particular splicing, plays an important role in the aging transcriptome [6]. Decreased expression of splicing factors has been observed in both mouse muscle, and human blood and brain [6, 69–71]. In fact, as many as a third of splicing factors exhibit altered expression with age in human blood cells [70], in which 6 out of 7 age-regulated gene ontology pathways are related to mRNA processing [6]. Further, spliceosomal genes show age-associated changes in expression in brain tissue from the African turquoise killifish (Nothobranchius furzeri) [83]. Moreover, levels of splicing factor expression correlate with lifespan in both humans and mice, suggesting that proper splicing might be critical for maintaining proper cellular function [6, 69, 71].
Are there age-associated defects in splicing?
The observed decreased expression of splicing factors with age does in fact correlate strongly with significant changes in alternative splicing in rodent brain, skin, muscle, bone, thymus, adipose tissue, spleen and muscle, and in human blood and brain [6, 35, 69–73] (Table 2). In human brain, 1174 exons are significantly differentially expressed with age [71]. Further in mouse skin, skeletal muscle, bone, thymus and white adipose tissue, between 0.3% – 3.2% of transcripts show changes in alternative splicing with age [73]. The types of age-related changes in alternative splicing, such as exon skipping or inclusion, vary from gene to gene. Although most changes in alternative splicing are observed during development, 30% of all alternative splicing changes that occur in an organism happen during aging [72, 84]. These data suggest that there are substantial age-associated changes in splicing, and that these changes in splicing could be caused by the age-associated decrease in expression of splicing factors (Table 2). Notably, the genes that show differences in splicing with age in the human brain include those with neuronal-specific functions such as synaptic transmission [71]. Similarly, genes involved in collagen production and post translational modification show age-associated changes in splice isoform abundance in human achilles tendon [31]. This suggests that at least some of the same types of tissue-specific genes that show transcriptional decline with age are also more vulnerable to changes in splicing with age.
Table 2.
Age associated changes in splicing defined as changes in expression of splicing factors, changes in isoform usage, or increases in intron retention. Organisms are grouped by row, and separated into tissue types. Age-associated changes are indicated by arrows, ↓ or ↑ signifying decrease or increase with age respectively, change denoted by Δ. No symbol indicates that the indicated process was not examined in the study.
| Age-associated changes in: | |||||
|---|---|---|---|---|---|
| Tissues Tested | References | Expression of splicing factors | Isoform usage | Intron Retention | |
| Human | Blood | Harries, 2011 | ↓ | Δ | |
| Lee, 2016 | ↓ | Δ | |||
| Brain | Tollervey, 2011 | Δ | |||
| Mazin, 2013 | ↓ | Δ | ↑ | ||
| Endothelial and Fibroblasts | Holly 2013 | ↓ | Δ | ||
| Rodents | Brain | Wood, 2013 | Δ | ||
| Skin, Muscle, Bone, Thymus, and Adipose | Rodríguez, 2016 | Δ | |||
| Spleen and Muscle | Lee, 2016 | ↓ | Δ | ↑ | |
Could an age-associated defect in splicing provide an explanation as to why specific genes show decreased transcript levels with age in particular tissues? Although defects in splicing can change alternative splice isoform usage, they can also result in intron retention or frameshifts leading to nonsense-mediated decay of the improperly spliced transcript [85, 86]. Data suggests that intron retention increases with age, and that there is a corresponding increase in the expression of genes required for nonsense-mediated decay during aging [6, 72, 87]. Is it possible that some genes are more likely to become improperly spliced with age? Indeed, higher frequencies of intron retention correlate with short intron length, high gene expression level, weak splice sites, and density of regulatory elements [88]. This suggests that some genes could be relatively more susceptible to intron retention, and that this could be influenced by splicing regulatory proteins whose expression differs between cell types. Additionally, some of the classes of tissue-specific genes that show both age-associated decreases in transcript levels and changes in alternative splicing, such as genes involved in synaptic transmission in neurons, have a large number of exons and possible alternative transcripts. It is plausible that defects in splicing could disproportionately affect the expression of these heavily spliced transcripts as compared to transcripts that contain fewer exons. Splicing occurs co-transcriptionally, and splice site selection is influenced by transcription rate [82, 89]. Thus, the splice isoform changes observed in old cells could also reflect changes in transcription rate at those genes. In addition, the interplay between these aspects of gene expression is complicated by the fact that defects in splicing could indirectly affect transcription. For example, age-associated changes in splicing of the transcription factor STAT1 could influence transcription of its target genes [6].
Intriguingly, the changes in splicing observed during normal aging might also provide insight into disease states since many human pathologies are associated with misregulation of splicing [90]. For example, about 20% of the genes that change with respect to alternative splicing in normal aging, also show similar changes in splicing in a premature aging disease model [73]. Further, the changes in splicing observed during normal aging in the human brain closely mimic the changes observed in Alzheimer’s disease [71]. In addition, mutations in splicing factor genes, which result in extensive changes in splicing patterns, occur frequently in adult myeloid malignancies [90]. Changes in alternative splice isoform usage can also directly cause human disease. For instance, mutations in Lamina A that activate a cryptic splice site within exon 11 have been implicated in Hutchinson-Gilford progeria syndrome [91]. Thus, understanding the interplay between splicing and transcription in the aging transcriptome that occurs with normal aging, might also provide insight into the similar transcriptome changes associated with human disease.
Could other aspects of gene expression be vulnerable to aging?
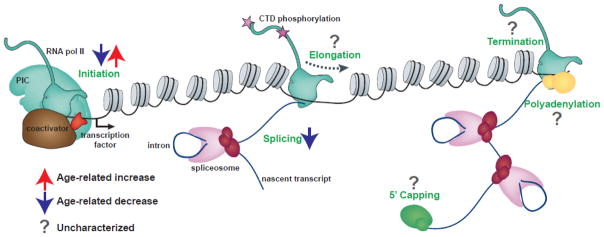
Gene expression at the level of mRNA production involves a coordinated series of events beginning with recruitment of RNA polymerase II and ending with export of the processed mRNA to the cytoplasm for translation [82]. In this review, we have focused on new studies that suggest that RNA processing, and in particular splicing, might be deregulated with age. Indeed, as discussed previously, genes involved in splicing and post-transcriptional processing are downregulated with age [6]. However other aspects of transcription could also be vulnerable to aging (Figure 1). Following its recruitment to promoters, RNA polymerase II often transcribes a short distance into the gene and then pauses, in a process termed promoter proximal pausing [92]. The release of RNA polymerase II from this promoter proximal pause site is highly regulated for many developmentally important genes, including those that are tissue-specifically expressed [93]. In addition, productive transcription requires the successful passage of the elongating RNA polymerase II throughout the gene body [94]. RNA splicing and processing are tightly coupled with transcription, and are modulated by transcription rate and chromatin structure [82]. Little attention has focused on transcription elongation in aging studies, but techniques such as global nuclear run on-sequencing (GRO-seq), precision nuclear run on-sequencing (PRO-seq) and native elongating transcript sequencing (NET-seq) that measure distribution of the actively transcribing RNA polymerase II could provide insight into if and how the mechanism of transcription itself alters with age [95–97]. In addition, there are hints that alternative polyadenylation is important for stem cell function, and that polyadenylation activity decreases in aging neurons [98, 99]. Moreover, all aspects of gene expression including transcription elongation and RNA processing occur in the context of chromatin, and multiple studies have identified changes both in histone modifications and chromatin organization with age [100–102].
Figure 1.
Co-transcriptional RNA processing could be misregulated with age. Processes that have been shown to be up or downregulated with age are indicated by the red and blue arrows respectively. Processes that could be misregulated with age, but have not yet been examined in sufficient detail are indicated by grey question marks. CTD, carboxy-terminal domain of RNA polymerase II, PIC, pre-initiation complex; CTD; RNA pol II, RNA polymerase II.
In this review, we have focused on how transcription and splicing become defective with age, however these are just the beginning steps in the process of gene expression. Protein synthesis and post-translational modifications have also been shown to be disrupted with age [103]. Understanding the aging transcriptional and proteomic signatures within cell types will be a challenging, but necessary next step, to determining if and how the cellular decline associated with age can be delayed.
Perspectives and Conclusions
During aging, only a small proportion of transcripts change with age in terms of either transcript levels or splice isoform usage (Figure 2). This suggests that certain genes are susceptible to age-related changes in gene expression. Multiple studies suggest that aging is associated with an induction in the expression of a broad range of stress response genes, and a decline in the expression of specific subsets of genes, which differ between tissues. The transcriptional decline of specific genes that is observed with age is of interest, since decreased levels of the encoded proteins could contribute to the age-associated decline in cellular function. Thus, changes in gene expression could be both a consequence, and cause, of the progressive decline in function that defines aging. Unrepaired cellular damage, such as oxidative damage to proteins or DNA, triggers the induction of stress response genes. Increases in levels of transcriptional repressors could directly contribute to the decline in expression of other genes. However, the same unrepaired cellular damage that increases gene expression could also directly reduce expression of other genes if specific transcription factors or promoters are damaged. Decreased expression of genes that encode spliceosomal or transcriptional regulators could further decrease gene expression. For example, genes that are highly expressed and heavily spliced might be particularly vulnerable to defects in splicing, leading to intron retention and/or changes in splice isoform usage. Intron retention and/or frameshifts could result in nonsense-mediated decay, further reducing transcript levels. Together, changes in the transcriptome that occur with age could initiate a downward spiral towards progressive cellular decline. However, the heterogeneity in transcriptional signatures of aging between individuals [8, 24] suggests that this downward spiral can be slowed by environmental and genetic factors. Thus, understanding the mechanisms involved in determining the aging transcriptome could provide therapeutic targets to slow aging and delay the onset of age-related disease.
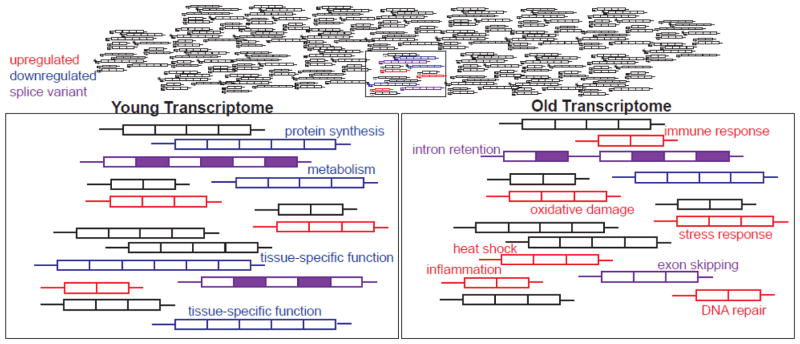
Figure 2.
An overview of age-associated transcriptome changes. Only 2 – 3 % of the whole genome (upper panel) shows age-associated changes in transcript levels. These changes between young and old cells are depicted in the two lower panels. Transcripts are shown with exons depicted as boxes, and introns and untranslated regions as lines. Transcripts that increase or decrease with age are shown in red and blue respectively, versus unchanged transcripts in black. Potential splice variants are shown in purple that include intron retention (third transcript from top) and exon skipping (bottom right).
Highlights.
A relatively small percentage of an organism’s transcriptome changes with age
A broad group of stress-responsive genes increase with age
A variety of genes decrease with age depending on tissue and cell type
Splice isoform usage and intron retention change with age
Acknowledgments
We thank the National Eye Institute of the National Institutes of Health for their support under Award Number R01EY024905 to V.W.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Williams GC. Pleiotropy, Natural-Selection, and the Evolution of Senescence. Evolution. 1957;11:398–411. [Google Scholar]
- 3.Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 4.Sahin E, Depinho RA. Linking functional decline of telomeres, mitochondria and stem cells during ageing. Nature. 2010;464:520–8. doi: 10.1038/nature08982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Magalhães JP, Curado J, Church GM. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics. 2009;25:875–81. doi: 10.1093/bioinformatics/btp073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harries LW, Hernandez D, Henley W, Wood AR, Holly AC, Bradley-Smith RM, et al. Human aging is characterized by focused changes in gene expression and deregulation of alternative splicing. Aging Cell. 2011;10:868–78. doi: 10.1111/j.1474-9726.2011.00726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zahn JM, Poosala S, Owen AB, Ingram DK, Lustig A, Carter A, et al. AGEMAP: a gene expression database for aging in mice. PLoS Genet. 2007;3:e201. doi: 10.1371/journal.pgen.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peters MJ, Joehanes R, Pilling LC, Schurmann C, Conneely KN, Powell J, et al. The transcriptional landscape of age in human peripheral blood. Nat Commun. 2015;6:8570. doi: 10.1038/ncomms9570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kowald A, Kirkwood TB. Can aging be programmed? A critical literature review. Aging Cell. 2016 doi: 10.1111/acel.12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldsmith TC. On the programmed/non-programmed aging controversy. Biochemistry Biokhimiia. 2012;77:729–32. doi: 10.1134/S000629791207005X. [DOI] [PubMed] [Google Scholar]
- 11.Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–12. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 12.Anselmi CV, Malovini A, Roncarati R, Novelli V, Villa F, Condorelli G, et al. Association of the FOXO3A locus with extreme longevity in a southern Italian centenarian study. Rejuvenation Res. 2009;12:95–104. doi: 10.1089/rej.2008.0827. [DOI] [PubMed] [Google Scholar]
- 13.Broer L, Buchman AS, Deelen J, Evans DS, Faul JD, Lunetta KL, et al. GWAS of longevity in CHARGE consortium confirms APOE and FOXO3 candidacy. J Gerontol A Biol Sci Med Sci. 2015;70:110–8. doi: 10.1093/gerona/glu166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nebel A, Kleindorp R, Caliebe A, Nothnagel M, Blanche H, Junge O, et al. A genome-wide association study confirms APOE as the major gene influencing survival in long-lived individuals. Mechanisms of Ageing and Development. 2011;132:324–30. doi: 10.1016/j.mad.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 15.Schachter F, Fauredelanef L, Guenot F, Rouger H, Froguel P, Lesueurginot L, et al. Genetic Associations with Human Longevity at the Apoe and Ace Loci. Nature Genetics. 1994;6:29–32. doi: 10.1038/ng0194-29. [DOI] [PubMed] [Google Scholar]
- 16.Soerensen M, Dato S, Christensen K, McGue M, Stevnsner T, Bohr VA, et al. Replication of an association of variation in the FOXO3A gene with human longevity using both case-control and longitudinal data. Aging Cell. 2010;9:1010–7. doi: 10.1111/j.1474-9726.2010.00627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilicox BJ, Donlon TA, He Q, Chen R, Grove JS, Yano K, et al. FOXO3A genotype is strongly associated with human longevity. P Natl Acad Sci USA. 2008;105:13987–92. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sebastiani P, Solovieff N, Dewan AT, Walsh KM, Puca A, Hartley SW, et al. Genetic signatures of exceptional longevity in humans. PLoS One. 2012;7:e29848. doi: 10.1371/journal.pone.0029848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shadyab AH, LaCroix AZ. Genetic factors associated with longevity: a review of recent findings. Ageing Res Rev. 2015;19:1–7. doi: 10.1016/j.arr.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 20.Walter S, Atzmon G, Demerath EW, Garcia ME, Kaplan RC, Kumari M, et al. A genome-wide association study of aging. Neurobiol Aging. 2011;32:2109, e15–28. doi: 10.1016/j.neurobiolaging.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Medawar PB. In: An unsolved problem of biology. Lewis HK, editor. London: Published for the college; 1952. [Google Scholar]
- 22.Kirkwood TB, Rose MR. Evolution of senescence: late survival sacrificed for reproduction. Philos Trans R Soc Lond B Biol Sci. 1991;332:15–24. doi: 10.1098/rstb.1991.0028. [DOI] [PubMed] [Google Scholar]
- 23.Kirkwood TB. Evolution of ageing. Nature. 1977;270:301–4. doi: 10.1038/270301a0. [DOI] [PubMed] [Google Scholar]
- 24.Haigis MC, Yankner BA. The aging stress response. Mol Cell. 2010;40:333–44. doi: 10.1016/j.molcel.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yannarell A, Schumm DE, Webb TE. Age-dependence of nuclear RNA processing. Mech Ageing Dev. 1977;6:259–64. doi: 10.1016/0047-6374(77)90026-4. [DOI] [PubMed] [Google Scholar]
- 26.Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, et al. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–91. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 27.Berchtold NC, Cribbs DH, Coleman PD, Rogers J, Head E, Kim R, et al. Gene expression changes in the course of normal brain aging are sexually dimorphic. Proc Natl Acad Sci U S A. 2008;105:15605–10. doi: 10.1073/pnas.0806883105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodwell GE, Sonu R, Zahn JM, Lund J, Wilhelmy J, Wang L, et al. A transcriptional profile of aging in the human kidney. PLoS Biol. 2004;2:e427. doi: 10.1371/journal.pbio.0020427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kayo T, Allison DB, Weindruch R, Prolla TA. Influences of aging and caloric restriction on the transcriptional profile of skeletal muscle from rhesus monkeys. Proc Natl Acad Sci U S A. 2001;98:5093–8. doi: 10.1073/pnas.081061898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshida S, Yashar BM, Hiriyanna S, Swaroop A. Microarray analysis of gene expression in the aging human retina. Invest Ophthalmol Vis Sci. 2002;43:2554–60. [PubMed] [Google Scholar]
- 31.Peffers MJ, Fang Y, Cheung K, Wei TK, Clegg PD, Birch HL. Transcriptome analysis of ageing in uninjured human Achilles tendon. Arthritis Res Ther. 2015;17:33. doi: 10.1186/s13075-015-0544-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haustead DJ, Stevenson A, Saxena V, Marriage F, Firth M, Silla R, et al. Transcriptome analysis of human ageing in male skin shows mid-life period of variability and central role of NF-κB. Sci Rep. 2016;6:26846. doi: 10.1038/srep26846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cai H, Fields MA, Hoshino R, Priore LV. Effects of aging and anatomic location on gene expression in human retina. Front Aging Neurosci. 2012;4:8. doi: 10.3389/fnagi.2012.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee CK, Weindruch R, Prolla TA. Gene-expression profile of the ageing brain in mice. Nat Genet. 2000;25:294–7. doi: 10.1038/77046. [DOI] [PubMed] [Google Scholar]
- 35.Wood SH, Craig T, Li Y, Merry B, de Magalhães JP. Whole transcriptome sequencing of the aging rat brain reveals dynamic RNA changes in the dark matter of the genome. Age (Dordr) 2013;35:763–76. doi: 10.1007/s11357-012-9410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edwards MG, Sarkar D, Klopp R, Morrow JD, Weindruch R, Prolla TA. Age-related impairment of the transcriptional responses to oxidative stress in the mouse heart. Physiol Genomics. 2003;13:119–27. doi: 10.1152/physiolgenomics.00172.2002. [DOI] [PubMed] [Google Scholar]
- 37.Bronikowski AM, Carter PA, Morgan TJ, Garland T, Ung N, Pugh TD, et al. Lifelong voluntary exercise in the mouse prevents age-related alterations in gene expression in the heart. Physiol Genomics. 2003;12:129–38. doi: 10.1152/physiolgenomics.00082.2002. [DOI] [PubMed] [Google Scholar]
- 38.Lee CK, Klopp RG, Weindruch R, Prolla TA. Gene expression profile of aging and its retardation by caloric restriction. Science. 1999;285:1390–3. doi: 10.1126/science.285.5432.1390. [DOI] [PubMed] [Google Scholar]
- 39.Cao SX, Dhahbi JM, Mote PL, Spindler SR. Genomic profiling of short- and long-term caloric restriction effects in the liver of aging mice. Proc Natl Acad Sci U S A. 2001;98:10630–5. doi: 10.1073/pnas.191313598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parapuram SK, Cojocaru RI, Chang JR, Khanna R, Brooks M, Othman M, et al. Distinct signature of altered homeostasis in aging rod photoreceptors: implications for retinal diseases. PLoS One. 2010;5:e13885. doi: 10.1371/journal.pone.0013885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma W, Cojocaru R, Gotoh N, Gieser L, Villasmil R, Cogliati T, et al. Gene expression changes in aging retinal microglia: relationship to microglial support functions and regulation of activation. Neurobiol Aging. 2013;34:2310–21. doi: 10.1016/j.neurobiolaging.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cannon L, Zambon AC, Cammarato A, Zhang Z, Vogler G, Munoz M, et al. Expression patterns of cardiac aging in Drosophila. Aging Cell. 2017;16:82–92. doi: 10.1111/acel.12559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lesur I, Campbell JL. The transcriptome of prematurely aging yeast cells is similar to that of telomerase-deficient cells. Mol Biol Cell. 2004;15:1297–312. doi: 10.1091/mbc.E03-10-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laun P, Ramachandran L, Jarolim S, Herker E, Liang P, Wang J, et al. A comparison of the aging and apoptotic transcriptome of Saccharomyces cerevisiae. FEMS Yeast Res. 2005;5:1261–72. doi: 10.1016/j.femsyr.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 45.Kowalczyk MS, Tirosh I, Heckl D, Rao TN, Dixit A, Haas BJ, et al. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 2015;25:1860–72. doi: 10.1101/gr.192237.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rossi DJ, Bryder D, Zahn JM, Ahlenius H, Sonu R, Wagers AJ, et al. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc Natl Acad Sci U S A. 2005;102:9194–9. doi: 10.1073/pnas.0503280102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pletcher SD, Macdonald SJ, Marguerie R, Certa U, Stearns SC, Goldstein DB, et al. Genome-wide transcript profiles in aging and calorically restricted Drosophila melanogaster. Curr Biol. 2002;12:712–23. doi: 10.1016/s0960-9822(02)00808-4. [DOI] [PubMed] [Google Scholar]
- 48.Zou S, Meadows S, Sharp L, Jan LY, Jan YN. Genome-wide study of aging and oxidative stress response in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2000;97:13726–31. doi: 10.1073/pnas.260496697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Girardot F, Lasbleiz C, Monnier V, Tricoire H. Specific age-related signatures in Drosophila body parts transcriptome. BMC Genomics. 2006;7:69. doi: 10.1186/1471-2164-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bahar R, Hartmann CH, Rodriguez KA, Denny AD, Busuttil RA, Dollé ME, et al. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature. 2006;441:1011–4. doi: 10.1038/nature04844. [DOI] [PubMed] [Google Scholar]
- 51.Kowalczyk MS, Tirosh I, Heck D, Rao TN, Dixit A, Haas BJ, et al. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Research. 2015;25:1860–72. doi: 10.1101/gr.192237.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–14. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marthandan S, Baumgart M, Priebe S, Groth M, Schaer J, Kaether C, et al. Conserved Senescence Associated Genes and Pathways in Primary Human Fibroblasts Detected by RNA-Seq. PLoS One. 2016;11:e0154531. doi: 10.1371/journal.pone.0154531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xin Y, Okamoto H, Kim J, Ni M, Adler C, Cavino K, et al. Single-Cell RNAseq Reveals That Pancreatic β-Cells From Very Old Male Mice Have a Young Gene Signature. Endocrinology. 2016;157:3431–8. doi: 10.1210/en.2016-1235. [DOI] [PubMed] [Google Scholar]
- 55.McCarroll SA, Murphy CT, Zou S, Pletcher SD, Chin CS, Jan YN, et al. Comparing genomic expression patterns across species identifies shared transcriptional profile in aging. Nat Genet. 2004;36:197–204. doi: 10.1038/ng1291. [DOI] [PubMed] [Google Scholar]
- 56.Lipinski MM, Zheng B, Lu T, Yan Z, Py BF, Ng A, et al. Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer’s disease. Proc Natl Acad Sci U S A. 2010;107:14164–9. doi: 10.1073/pnas.1009485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lazuardi L, Herndler-Brandstetter D, Brunner S, Laschober GT, Lepperdinger G, Grubeck-Loebenstein B. Microarray analysis reveals similarity between CD8+CD28− T cells from young and elderly persons, but not of CD8+CD28+ T cells. Biogerontology. 2009;10:191–202. doi: 10.1007/s10522-008-9167-1. [DOI] [PubMed] [Google Scholar]
- 58.Carter TA, Greenhall JA, Yoshida S, Fuchs S, Helton R, Swaroop A, et al. Mechanisms of aging in senescence-accelerated mice. Genome Biol. 2005;6:R48. doi: 10.1186/gb-2005-6-6-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Loerch PM, Lu T, Dakin KA, Vann JM, Isaacs A, Geula C, et al. Evolution of the aging brain transcriptome and synaptic regulation. PLoS One. 2008;3:e3329. doi: 10.1371/journal.pone.0003329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Walker DW, Muffat J, Rundel C, Benzer S. Overexpression of a Drosophila homolog of apolipoprotein D leads to increased stress resistance and extended lifespan. Curr Biol. 2006;16:674–9. doi: 10.1016/j.cub.2006.01.057. [DOI] [PubMed] [Google Scholar]
- 61.Sanchez D, López-Arias B, Torroja L, Canal I, Wang X, Bastiani MJ, et al. Loss of glial lazarillo, a homolog of apolipoprotein D, reduces lifespan and stress resistance in Drosophila. Curr Biol. 2006;16:680–6. doi: 10.1016/j.cub.2006.03.024. [DOI] [PubMed] [Google Scholar]
- 62.Martins R, Lithgow GJ, Link W. Long live FOXO: unraveling the role of FOXO proteins in aging and longevity. Aging Cell. 2016;15:196–207. doi: 10.1111/acel.12427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Greer EL, Brunet A. FOXO transcription factors in ageing and cancer. Acta Physiol (Oxf) 2008;192:19–28. doi: 10.1111/j.1748-1716.2007.01780.x. [DOI] [PubMed] [Google Scholar]
- 64.Helenius M, Hanninen M, Lehtinen SK, Salminen A. Aging-induced up-regulation of nuclear binding activities of oxidative stress responsive NF-kB transcription factor in mouse cardiac muscle. Journal of molecular and cellular cardiology. 1996;28:487–98. doi: 10.1006/jmcc.1996.0045. [DOI] [PubMed] [Google Scholar]
- 65.Lee CK, Allison DB, Brand J, Weindruch R, Prolla TA. Transcriptional profiles associated with aging and middle age-onset caloric restriction in mouse hearts. Proc Natl Acad Sci U S A. 2002;99:14988–93. doi: 10.1073/pnas.232308999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Edwards MG, Sarkar D, Klopp R, Morrow JD, Weindruch R, Prolla TA. Impairment of the transcriptional responses to oxidative stress in the heart of aged C57BL/6 mice. Ann N Y Acad Sci. 2004;1019:85–95. doi: 10.1196/annals.1297.017. [DOI] [PubMed] [Google Scholar]
- 67.Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, et al. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497:451. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Landis G, Shen J, Tower J. Gene expression changes in response to aging compared to heat stress, oxidative stress and ionizing radiation in Drosophila melanogaster. Aging-Us. 2012;4:768–89. doi: 10.18632/aging.100499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee BP, Pilling LC, Emond F, Flurkey K, Harrison DE, Yuan R, et al. Changes in the expression of splicing factor transcripts and variations in alternative splicing are associated with lifespan in mice and humans. Aging Cell. 2016;15:903–13. doi: 10.1111/acel.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Holly AC, Melzer D, Pilling LC, Fellows AC, Tanaka T, Ferrucci L, et al. Changes in splicing factor expression are associated with advancing age in man. Mech Ageing Dev. 2013;134:356–66. doi: 10.1016/j.mad.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tollervey JR, Wang Z, Hortobágyi T, Witten JT, Zarnack K, Kayikci M, et al. Analysis of alternative splicing associated with aging and neurodegeneration in the human brain. Genome Res. 2011;21:1572–82. doi: 10.1101/gr.122226.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mazin P, Xiong J, Liu X, Yan Z, Zhang X, Li M, et al. Widespread splicing changes in human brain development and aging. Mol Syst Biol. 2013;9:633. doi: 10.1038/msb.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rodríguez SA, Grochová D, McKenna T, Borate B, Trivedi NS, Erdos MR, et al. Global genome splicing analysis reveals an increased number of alternatively spliced genes with aging. Aging Cell. 2016;15:267–78. doi: 10.1111/acel.12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moskalev AA, Shaposhnikov MV, Plyusnina EN, Zhavoronkov A, Budovsky A, Yanai H, et al. The role of DNA damage and repair in aging through the prism of Koch-like criteria. Ageing Res Rev. 2013;12:661–84. doi: 10.1016/j.arr.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 75.Gensler HL, Bernstein H. DNA damage as the primary cause of aging. Q Rev Biol. 1981;56:279–303. doi: 10.1086/412317. [DOI] [PubMed] [Google Scholar]
- 76.Mayne LV, Lehmann AR. Failure of RNA synthesis to recover after UV irradiation: an early defect in cells from individuals with Cockayne’s syndrome and xeroderma pigmentosum. Cancer Res. 1982;42:1473–8. [PubMed] [Google Scholar]
- 77.Rockx DA, Mason R, van Hoffen A, Barton MC, Citterio E, Bregman DB, et al. UV-induced inhibition of transcription involves repression of transcription initiation and phosphorylation of RNA polymerase II. Proc Natl Acad Sci U S A. 2000;97:10503–8. doi: 10.1073/pnas.180169797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Berlett BS, Stadtman ER. Protein oxidation in aging, disease, and oxidative stress. J Biol Chem. 1997;272:20313–6. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- 79.Roy AK. Transcription factors and aging. Mol Med. 1997;3:496–504. [PMC free article] [PubMed] [Google Scholar]
- 80.Hobert O. Regulatory logic of neuronal diversity: terminal selector genes and selector motifs. Proc Natl Acad Sci U S A. 2008;105:20067–71. doi: 10.1073/pnas.0806070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu T, Aron L, Zullo J, Pan Y, Kim H, Chen Y, et al. REST and stress resistance in ageing and Alzheimer’s disease. Nature. 2014;507:448–54. doi: 10.1038/nature13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bentley DL. Coupling mRNA processing with transcription in time and space. Nat Rev Genet. 2014;15:163–75. doi: 10.1038/nrg3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Baumgart M, Groth M, Priebe S, Savino A, Testa G, Dix A, et al. RNA-seq of the aging brain in the short-lived fish N. furzeri - conserved pathways and novel genes associated with neurogenesis. Aging Cell. 2014;13:965–74. doi: 10.1111/acel.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Somel M, Guo S, Fu N, Yan Z, Hu HY, Xu Y, et al. MicroRNA, mRNA, and protein expression link development and aging in human and macaque brain. Genome Res. 2010;20:1207–18. doi: 10.1101/gr.106849.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Losson R, Lacroute F. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc Natl Acad Sci U S A. 1979;76:5134–7. doi: 10.1073/pnas.76.10.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Maquat LE, Kinniburgh AJ, Rachmilewitz EA, Ross J. Unstable beta-globin mRNA in mRNA-deficient beta o thalassemia. Cell. 1981;27:543–53. doi: 10.1016/0092-8674(81)90396-2. [DOI] [PubMed] [Google Scholar]
- 87.Blanco FJ, Bernabeu C. Alternative splicing factor or splicing factor-2 plays a key role in intron retention of the endoglin gene during endothelial senescence. Aging Cell. 2011;10:896–907. doi: 10.1111/j.1474-9726.2011.00727.x. [DOI] [PubMed] [Google Scholar]
- 88.Sakabe NJ, de Souza SJ. Sequence features responsible for intron retention in human. BMC Genomics. 2007;8:59. doi: 10.1186/1471-2164-8-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, et al. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell. 2003;12:525–32. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 90.Scotti MM, Swanson MS. RNA mis-splicing in disease. Nat Rev Genet. 2016;17:19–32. doi: 10.1038/nrg.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Eriksson M, Brown WT, Gordon LB, Glynn MW, Singer J, Scott L, et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–8. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nechaev S, Adelman K. Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation. Biochim Biophys Acta. 2011;1809:34–45. doi: 10.1016/j.bbagrm.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Levine M. Paused RNA polymerase II as a developmental checkpoint. Cell. 2011;145:502–11. doi: 10.1016/j.cell.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jonkers I, Lis JT. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol. 2015;16:167–77. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–73. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–8. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kwak H, Fuda NJ, Core LJ, Lis JT. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science. 2013;339:950–3. doi: 10.1126/science.1229386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mueller AA, Cheung TH, Rando TA. All’s well that ends well: alternative polyadenylation and its implications for stem cell biology. Curr Opin Cell Biol. 2013;25:222–32. doi: 10.1016/j.ceb.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lindholm DB. Decreased transcription of neuronal polyadenylated RNA during senescence in nuclei from rat brain cortex. J Neurochem. 1986;47:1503–6. doi: 10.1111/j.1471-4159.1986.tb00785.x. [DOI] [PubMed] [Google Scholar]
- 100.Hu Z, Chen K, Xia Z, Chavez M, Pal S, Seol JH, et al. Nucleosome loss leads to global transcriptional up-regulation and genomic instability during yeast aging. Genes Dev. 2014;28:396–408. doi: 10.1101/gad.233221.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pal S, Tyler JK. Epigenetics and aging. Sci Adv. 2016;2:e1600584. doi: 10.1126/sciadv.1600584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Feser J, Truong D, Das C, Carson JJ, Kieft J, Harkness T, et al. Elevated histone expression promotes life span extension. Mol Cell. 2010;39:724–35. doi: 10.1016/j.molcel.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rattan SI, Derventzi A, Clark BF. Protein synthesis, posttranslational modifications, and aging. Ann N Y Acad Sci. 1992;663:48–62. doi: 10.1111/j.1749-6632.1992.tb38648.x. [DOI] [PubMed] [Google Scholar]
- 104.Kannan S, Dawany N, Kurupati R, Showe LC, Ertl HC. Age-related changes in the transcriptome of antibody-secreting cells. Oncotarget. 2016;7:13340–53. doi: 10.18632/oncotarget.7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lund J, Tedesco P, Duke K, Wang J, Kim SK, Johnson TE. Transcriptional profile of aging in C. elegans. Curr Biol. 2002;12:1566–73. doi: 10.1016/s0960-9822(02)01146-6. [DOI] [PubMed] [Google Scholar]
- 106.Ma X, Zhan G, Sleumer MC, Chen S, Liu W, Zhang MQ, et al. Analysis of C. elegans muscle transcriptome using trans-splicing-based RNA tagging (SRT) Nucleic Acids Res. 2016;44:e156. doi: 10.1093/nar/gkw734. [DOI] [PMC free article] [PubMed] [Google Scholar]