Abstract
Infections have been a major cause of disease throughout the history of humans on earth. With the introduction of antibiotics, it was thought that infections had been conquered. However, bacteria have been able to develop resistance to antibiotics at an exponentially increasing rate. The growing threat from multi-drug resistant organisms calls for intensive action to prevent the emergence of totally resistant and untreatable infections. Novel, non-invasive, non-antibiotic strategies are needed that act more efficiently and faster than current antibiotics. One promising alternative is antimicrobial photodynamic inactivation (APDI), an approach that produces reactive oxygen species when dyes and light are combined. So far, it has been questionable if bacteria can develop resistance against APDI. This review paper gives an overview of recent studies concerning the susceptibility of bacteria towards oxidative stress, and suggests possible mechanisms of the development of APDI-resistance that should at least be addressed. Some ways to potentiate APDI and also to overcome future resistance are suggested.
Keywords: Antimicrobial photodynamic inactivation, oxidative stress, oxidative stress response, resistance to APDI, sub-lethal APDI
1. Introduction
Microorganisms have existed on the earth for more than 3.8 billion years and exhibit the greatest genetic and metabolic diversity of any known life form. They are an important component of the biosphere and serve a vital role in the maintenance of ecosystems. In order to survive, they have evolved mechanisms that enable them to respond to selective pressure exerted by a range of different environments and competitive challenges. Humans have continuously exposed pathogenic microbial populations to antibiotics, antiseptics, and other antimicrobial agents, in attempts to control infectious disease. These microorganisms have then responded by developing a variety of resistance mechanisms to escape this offensive against their survival. Currently, antimicrobial resistance among bacteria, viruses, parasites, and other disease-causing organisms is a serious worldwide threat to management of infectious disease.
Antibiotics were discovered in the middle of the 20th century, and almost immediately reduced the threat of infectious bacterial diseases, which had devastated the human population for centuries. But, surprisingly soon after the discovery of penicillin in 1940, a number of treatment failures and isolation of some bacterial strains such as staphylococci which were no longer sensitive to penicillin started to occur. This marked the beginning of the era of antibiotic resistance [1].
Resistance can be categorized into two mechanisms.
Intrinsic or natural resistance whereby microorganisms naturally either do not possess the appropriate target sites for the specific drug (and therefore the drug does not affect them), or they naturally have only low permeability to those agents, because of the differences in the chemical nature of the drug and the microbial membrane structures. This is particularly relevant for those antibiotics that require entry into the microbial cell in order to effect their action.
Acquired resistance whereby a naturally susceptible microorganism develops mechanisms that prevent it from being affected by the drug.
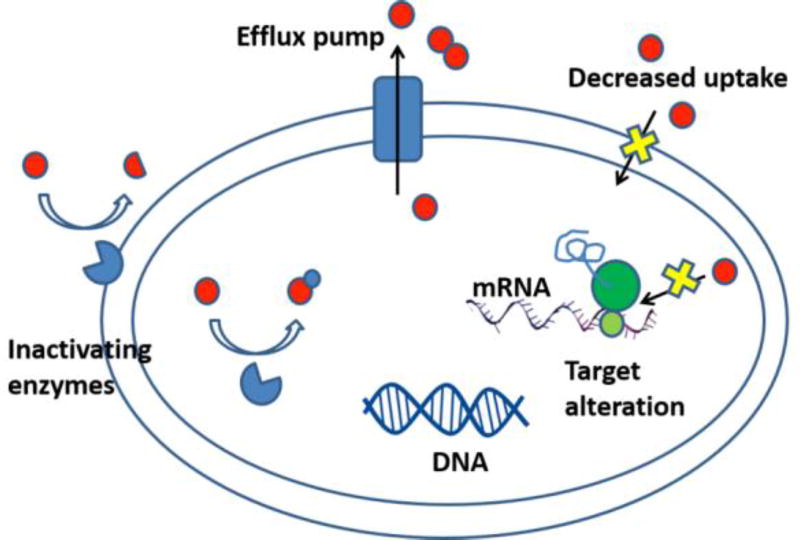
Acquired resistance mechanisms can occur through various ways (summarized in Figure 1) [2].
-
-
the presence of an enzyme that inactivates the antimicrobial agent
-
-
a mutation in the antimicrobial target of the agent, which reduces the binding of the antimicrobial agent
-
-
post-transcriptional or post-translational modification of the antimicrobial target of the agent, which reduces binding of the antimicrobial agent
-
-
reduced uptake of the antimicrobial agent into the cell
-
-
active efflux of the antimicrobial agent out of the cell
Figure 1.
Antibiotic resistance mechanisms in bacteria
Resistance elements can be acquired by transmission of free (naked) DNA from one bacterial species to another bacterial species (horizontal gene transfer). Genes responsible for antibiotic resistance in one species of bacteria can be transferred to another species of bacteria through various mechanisms such as F-pilus generation as well as via bacteriophages [3].
Although antimicrobial resistance is a naturally occurring biological phenomenon, it is often enhanced as a consequence of the adaptation forced upon microbes by continuous or repeated exposure to antimicrobial agents used to prevent or treat infections in humans or livestock, and the widespread routine use of disinfectants in farms, hospitals, and households [1]. It is now accepted that excessive antimicrobial use is the single most important factor responsible for increased antimicrobial resistance [4, 5]
In 2015 the O’Neill report garnered much international attention when it delivered the alarming forecast that by 2050 (if nothing were done to stem the growth of multi-drug-resistant bacteria) there would have been 300 million premature deaths that would have cost the world economy $100 trillion [6].
The current worldwide increase in drug-resistant bacteria and the simultaneous decline in efforts by both academic laboratories and pharmaceutical companies directed towards the discovery of new antibacterial agents to combat resistant strains now poses a serious threat to the treatment of life-threatening infections. Therefore, it is necessary to develop novel noninvasive and non-toxic antimicrobial strategies that act more efficiently and faster than the current antibiotics, and to which pathogens will not easily develop resistance [7]. One promising alternative to current antibiotics is antimicrobial photodynamic inactivation (APDI).
2 Antimicrobial photodynamic inactivation
APDI is defined as the application of a non-toxic dye known as a photosensitizer (PS), which can be photo-activated with light of the appropriate wavelength in the presence of oxygen to generate cytotoxic reactive oxygen species (ROS) such as singlet oxygen and/or free radicals) [8–10].
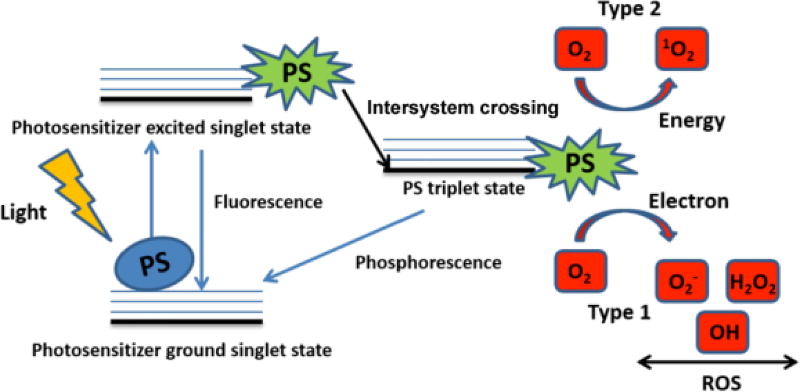
The process is initiated when a ground state PS (S0) absorbs light of an appropriate wavelength and is converted into an electronic excited singlet state (S1). Many PS molecules in this shortlived state can decay back to the ground state with the emission of light (fluorescence) or heat (internal conversion) [11], but some can also be transformed by intersystem crossing into a much longer-lived excited triplet state (T1, T3). The excited triplet state PS can do one of two things; it can react with molecular oxygen by energy transfer generating singlet oxygen (a process which is termed a type II reaction) or else it can undergo an electron transfer reaction to form PS radical ions which in turn react with oxygen to produce cytotoxic species such as superoxide, hydrogen peroxide, and/or hydroxyl radicals (which is termed a type I reaction) (Figure 2). The singlet oxygen or other reactive oxygen species (ROS) can cause damage to bacterial cells or other microbes through several mechanisms. These include oxidation of membrane lipids and amino acids in proteins, cross-linking of proteins and oxidative damage to nucleic acids with the subsequent disturbance of the normal functioning of the pathogen [12, 13].
Figure 2.
Principals of Antimicrobial Photodynamic Inactivation shown in a Jablonski Diagram
It has been proposed that the ROS generated by light-activated PS can trigger microbial killing, and cell damage via three mechanisms: (a) damage of the cell membrane (or virus envelope); (b) inactivation of essential enzymes and proteins; and/or (c) damage to DNA [9].
The APDI-induced photo-damage to microbial constituents can result in considerable morphological and functional changes in the microbial cells themselves. Functional damage results from loss of enzyme activities, protein oxidation and formation of protein-protein cross-links, and inhibition of cellular metabolic processes (e.g. DNA synthesis, glucose transport, etc). Morphological alterations include alteration of the mesosome structure. Direct damage to the cell membrane leads to leakage of cellular contents following inactivation of the membrane transport system [14].
There is a very wide variation in the cellular structure and organization among different classes of microbial cells. These structural variations influence the interaction of exogenous PS with different cellular components, and can also affect the effectiveness and the mechanism of action of the APDI with different pathogens. Differences in the cell walls of Gram-positive and Gram-negative bacteria play an important role in the susceptibility of bacteria to APDI. Grampositive bacteria have a thick and porous peptidoglycan layers that surround a cytoplasmic membrane, while Gram-negative bacteria possess an outer membrane, surrounding a thinner peptidoglycan layer, inside which is the cytoplasmic membrane [15, 16]. To perform efficient APDI, the PS needs to penetrate (or at least bind to) the cell wall of the bacteria and end up in the plasma membrane or in the cytoplasm; however, the membrane barriers of the bacterial cell limit the simple diffusion of PS into the bacterial cytosol [17, 18]. Therefore, APDI of Grampositive bacteria is definitely much easier to accomplish than that of Gram-negative bacteria. The cell walls of fungal cells have a structure that is intermediate in permeability between Gram-positive and Gram-negative bacteria. The outer part is a moderately porous layer of β-glucan and mannan polysaccharides.
3. Oxidative stress
Microbial life first evolved in a world without oxygen and was rich with reduced iron. By three billion years ago, microbial life forms shared basic biochemical mechanisms and a common metabolic system, which persist even today. The subsequent oxygenation of the atmosphere by photosynthetic organisms created a disaster for primitive life: oxygen is a reactive chemical species, and organisms had to develop strategies to defend themselves against it [19].
Molecular oxygen (O2) is small and nonpolar, and it diffuses across typical biological membranes as quickly as it does through water [20]. Therefore, even the most active cells cannot consume through respiration rapidly enough to reduce the intracellular O2 concentration considerably below the concentration immediately outside the cell. O2 reacts poorly with cellular biomolecules, however its reactivity derives from the metabolic formation of ROS [21], which results from the uncontrolled addition of consecutive electrons to O2, instead of the normal controlled addition to form water. This uncontrolled addition generates superoxide (O2•−), hydrogen peroxide (H2O2), the hydroxyl radical (•OH) [22]. When the balance between ROS and their scavenger systems is disturbed, ROS accumulate within the cells leading to a condition called oxidative stress [23].
Some microorganisms escape oxidative stress by living in anaerobic microhabitats; all others must deal with the consequences of intracellular O2. Different examinations show that the ability to do so varies widely: obligate anaerobes cannot tolerate oxygen at all, microaerophilic organisms require a relatively low-micromolar O2 concentration, while aerobes have adapted to grow in air-saturated fluids. However, almost all of these microorganisms suffer poor growth, elevated mutagenesis or even death when they are exposed to O2 levels that exceed those of their native habitats [23].
Aerobic organisms use O2 for respiration or oxidation of nutrients to obtain energy. Reactive by-products of oxygen, such as superoxide anion radical (O2•−), hydrogen peroxide (H2O2), and the highly reactive hydroxyl radicals (·OH), are generated continuously in cells grown aerobically [24].
Environmental agents such as ionizing radiation, UV radiation, or numerous compounds that generate intracellular O2•−, (redox-cycling agents such as menadione) can also cause oxidative stress, which arises when the concentration of ROS increases to a level that exceeds the cell’s defense capacity [24]. High temperatures can result in high oxidative stress, leading to damage to proteins, DNA double-strand breaks and cell death [25, 26]. Moreover antioxidant enzymes function poorly at high temperatures. In addition, cold temperatures can cause oxidative stress; cells of the Antarctic bacterium Pseudomonas fluorescens, grown at 4°C, suffer an increasing amount of free radicals and show enhanced activity of two antioxidant enzymes [27]. Some immune cells (neutrophils and macrophages) use the NADPH oxidase enzyme, activated upon invasion by pathogenic bacteria, as a weapon to kill microbes during phagocytosis [24].
The biological targets for ROS are DNA, RNA, proteins and lipids. Much of the damage is caused by hydroxyl radicals generated from H2O2 via the Fenton reaction, which requires iron (or another divalent metal ion, such as copper) and a source of reducing equivalents (possibly NADH) to regenerate the metal. Moreover direct one-electron of H2O2 can give •OH. Lipids are major targets for damage caused during oxidative stress. Free radicals can attack directly polyunsaturated fatty acids in membranes and initiate lipid peroxidation. The initial effect of lipid peroxidation is a decrease in membrane fluidity, which changes the membrane properties and can significantly disrupt membrane-bound proteins. This effect acts as an amplifier, more radicals are formed, and polyunsaturated fatty acids are degraded to a variety of products. Some of these products, such as the aldehydes (malondialdehyde and 4-hydroynonenal), are also very reactive and can damage molecules such as proteins [28]. Unlike reactive free radicals, aldehydes are rather long lived and can diffuse from the site of their origin and reach and attack targets which are distant from the initial free-radical event, acting as “toxic second messengers” of the complex lipid peroxidation chain reactions. DNA is also a main target; ROS attack both the base and the sugar moieties producing single- and double-strand breaks in the backbone, adducts of base and sugar groups, and cross-links to other molecules, lesions that block DNA replication [29, 30]. Several different kinds of oxidative damage to proteins have been documented [31, 32], including oxidation of sulfhydryl groups, oxidation of methionine to a sulfoxide, introduction of carbonyl groups, oxidative modification of amino acid residues close to metal-binding sites (via metal-catalyzed oxidation), addition of aldehydes to amino groups, modification of prosthetic groups or metal clusters, protein-protein cross-linking and peptide fragmentation. All these modifications are harmful to the cell, since they lead to a loss of function of membranes and proteins, and block DNA replication or cause mutations [24].
Singlet oxygen is the single most important ROS involved in APDI of microbial cells and the question arises of whether there are any antioxidant defense mechanisms devoted to protection against 1O2? Certainly in green plants in which 1O2 is routinely produced during photosynthesis (especially at high light intensities) by energy transfer from triplet excited chlorophylls in the chloroplasts, sophisticated defense mechanisms have evolved [33]. Carotenoids are considered to be the main 1O2 quenchers in chloroplasts, and light stress can induce the oxidation of the carotenoid β-carotene in Arabidopsis plants, leading to the accumulation of oxidation products, such as β-cyclocitral. This compound was found to induce changes in the expression of a large set of genes, but had little effect on the expression of gene markers related to protection against H2O2 [34].
3.1 Oxidative stress sensing and response in bacteria
An effective defense response against oxidative stress is a required item in the basic survival kit of all aerobic organisms, as well as those anaerobes that exist in environments subject to transient exposures to oxygen [22].
When ROS levels exceed safe limits, bacteria have the ability to organize an inducible response, resulting in increased expression of ROS detoxification enzymes along with additional protective systems that repair oxidative damage, protect susceptible enzymes from inactivation, and control the levels of free Fe2+ [35].
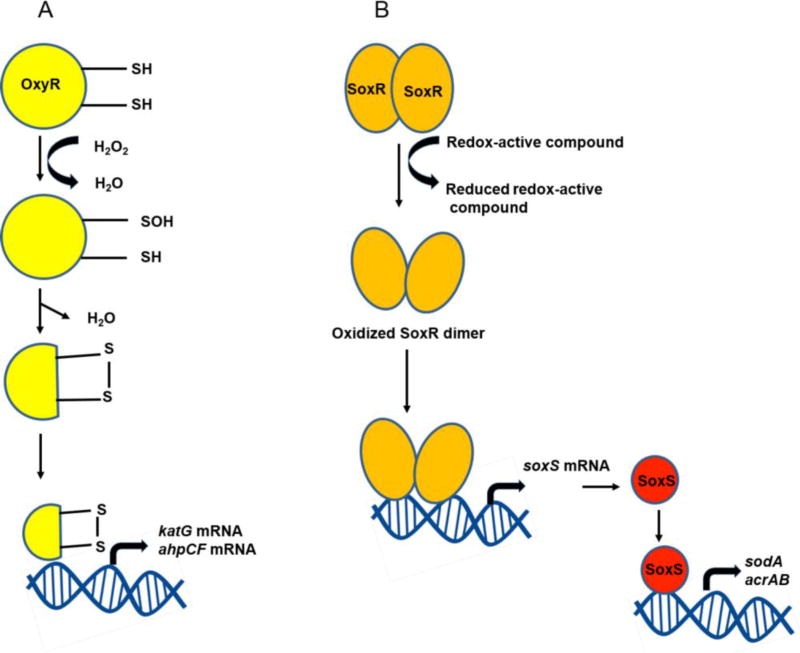
The regulation of the expression of the genes involved in the bacterial defense response to oxidative stress is complex, and often under the control of regulators that can directly sense the levels of specific ROS and activate or de-repress transcription of their target genes. Three wellstudied examples of these transcriptional regulators are the peroxide responsive regulators OxyR, PerR, and OhrR [36]. OxyR and PerR are primarily sensors of H2O2, while OhrR senses organic peroxides (ROOH) and sodium hypochlorite (NaOCl). OxyR and OhrR sense oxidants by means of the reversible oxidation of specific cysteine residues in their structure. In contrast, PerR senses H2O2 via the Fe-catalyzed oxidation of histidine residues. These transcription regulators also affect complex biological phenomena, such as biofilm formation, the evasion of host immune responses, and antibiotic resistance via the direct regulation of specific proteins [37].
Oxidized OxyR (Figure 3A) binds to promoters co-operatively with the RNA polymerase in E. coli [38], and positively regulates a group of peroxide stress defense genes, such as ahpCF, dps, and katG, whose expression is also induced by H2O2 [39]. PerR (peroxide resistance regulator) is the major regulator of peroxide stress defense in Gram-positive bacteria, such as Bacillus subtilis, Staphylococcus aureus, and Streptococcus pyogenes, and in some Gram-negative bacteria, including Campylobacter jejuni and Helicobacter hepaticus [40–42]. As a member of the Fur (ferric uptake regulator) family of metallo-regulators, B. subtilis PerR senses the intracellular Fe/Mn ratio and requires metal ions, including Zn2+ as a structural component, and Mn2+ and Fe2+ as regulatory ions [43]. While Fe2+ mediates PerR regulation of peroxide defense genes, such as katA, ahpCF, and mrgA (a homolog of dps), the negative auto-regulation of perR involves Mn2+ in B. subtilis. Although B. subtilis PerR regulates peroxide defense genes, perR transcription is not affected by H2O2 [44]. Instead, conformational changes in the PerR protein caused by H2O2 stress contribute to the regulatory function of PerR. Oxidation of one of two histidine residues (i.e., H37 and H91) by H2O2 in B. subtilis PerR results in the dissociation of Fe2+ from PerR, and the de-metallated PerR can no longer bind to DNA, so these conformational changes in PerR induce gene expression [45].
Figure 3.
Activation of redox-sensitive transcriptional regulators in Escherichia coli.
Another well-characterized regulatory system of oxidative stress defense is SoxRS, which is dedicated to the regulation of superoxide defense in E. coli and Salmonella [46]. SoxR (superoxide response regulator) was first identified as a genetic locus that positively regulates protein expression after exposure to superoxide-generating agents, such as paraquat [47, 48]. Redox-cycling compounds that directly generate the superoxide anion activate SoxR [49], and the activated SoxR stimulates expression of SoxS, which subsequently induces oxidative stress defense genes (Figure 3B) [50].
3.2 Defense mechanisms to oxidative stress
The emergence of oxygen in the atmosphere led to the development of defense mechanisms that either maintained the concentration of the O2-derived radicals within acceptable levels, or else repaired oxidative damage. Iron plays a significant role in biology (transport, storage and activation of molecular oxygen, reduction of ribonucleotides, activation and decomposition of peroxides, and electron transport) and Fe2+ is required for the growth of almost all living cells. Due to its potential damaging effects, in bacteria, iron solubilization and metabolism is strictly regulated at two levels: (i) uptake into the cell by specific membrane-bound receptors; and (ii) storage inside the cell by two proteins, bacterioferritin (very similar to the eukaryotic ferritin) but presenting ferroxidase activity. Some molecules are constitutively present and help to maintain an intracellular reducing environment, or to scavenge chemically reactive ROS. These molecules comprise pools of non-enzymatic antioxidants such as NADPH and NADH, β-carotene, ascorbic acid, α-tocopherol, and glutathione (GSH). GSH, present at high concentrations, maintains a strong reducing environment inside the cell, and its reduced form is maintained by glutathione reductase using NADPH as a source of reducing power. In addition, specific enzymes decrease the steady-state levels of reactive oxygen. Two superoxide dismutases (SOD), which convert O2•− to H2O2 and O2, have been described in Escherichia coli: an iron-containing enzyme, whose expression is modulated by intracellular iron levels [51], and a manganese containing SOD, the predominant enzyme expressed during aerobic growth, whose expression is transcriptionally regulated by at least six control systems [52]. A third SOD activity with properties similar to eukaryotic CuZn-SOD, has been found in the E. coli periplasmic space [53]. In E. coli, H2O2 is removed by two catalases (converting it to H2O and O2): hydroperoxidase I (HPI), which is present during aerobic growth and transcriptionally controlled at different levels [54], and hydroperoxidase II (HPII), which is induced during the stationary phase [55]. Glutathione peroxidase and DT-diaphorase are also ROS scavenging enzymes. Secondary defenses include DNA-repair systems, and proteolytic and lipolytic enzymes. DNA repair enzymes include endonuclease IV, which is induced by oxidative stress, and exonuclease III, which is induced in the stationary phase and in starving cells. Both enzymes act on duplex DNA by removing DNA 3' termini [56]. Prokaryotic cells contain catalysts able to directly repair some covalent modifications to the primary structure of proteins. One of the most frequent repair modifications is the reduction of oxidized disulfide bonds: (i) thioredoxin reductase transfers electrons from NADPH to thioredoxin via a flavin carrier, (ii) glutaredoxin is also able to reduce disulfide bonds, but using GSH as an electron donor and, (iii) protein disulfide isomerase facilitates disulfide exchange reactions with large inactive protein substrates, besides having chaperone activity. Oxidation of methionine to methionine sulfoxide can be repaired by methionine sulfoxide reductase. Recent experimental data described that surface-exposed methionine residues surrounding the entrance to the active site were preferentially oxidized without loss of catalytic activity, and suggested that methionine residues could function as a “last-chance” antioxidant detection system for proteins [57].
3.3 Oxidative stress induced by APDI
The most relevant molecules produced by APDI that can induce oxidative stress in bacteria are singlet oxygen, hydroxyl radicals, superoxide anions, and hydrogen peroxide. How much and in what quantity these different ROS are generated depends on the chemical origin of the given PS and its microenvironment [58].
The APDI-induced damage to bacteria is specific in terms of the localization of a given PS. That means, when a PS is only attached to the surface of a bacterium, oxidative damage of proteins and fatty acids appears only at the site of localization due to the high reactivity, short lifetime and limited diffusion of the generated ROS [59, 60].
4. Can resistance develop after sub-lethal APDI?
It is known that repeated usage of antimicrobial agents at low (sub-lethal) concentrations may lead to the development of bacterial population that are more resistant (or at least more tolerant) to higher concentration of these agents [61].
If APDI was used in the treatment of infections and the PS would reach the target site at only sub-lethal concentrations, and was therefore activated by light producing sub-lethal of ROS, any microorganism viable at the site of infection would be exposed to doses of APDI that would not result in total cell death, i.e., sub-lethal doses of APDI (sAPDI), exposing survivors to ROS stress. Since aPDI can kill all microbial cells if the dose is high enough, increased tolerance would be manifested by the need for much higher concentrations of PS and/or higher doses of light. It is clear that ROS can damage DNA. Such stress also leads to increased mutational events, which could lead to selection for survival of more resistant or less susceptible strains [62].
The most frequently employed method of testing for a microbe’s ability to become resistant to a particular agent is by subjecting the said microbe to routine continual exposure to a particular agent. Giuliani et al. analyzed whether the Gram-positive bacteria Staphylococcus aureus, and the Gram-negative bacteria Pseudomonas aeruginosa could become resistant to APDI via repeated exposure to tetracationic PS Zn(II) phthalocyanine derivative in concert with 30 J/cm2 of 600–700 nm light. It was shown that after 20 consecutive APDI treatments at PS concentrations corresponding to the previously determined minimum inhibitory concentrations (MIC), S. aureus, and P. aeruginosa were all incapable of developing resistance to APDI. However, when the 20 exposures were repeated without light, their results showed that the MIC of Zn (II) phthalocyanine derivative for S. aureus in the dark did increase. This shows that S. aureus may be able to develop some ability to protect itself against the dark toxic effect of a PS, perhaps by up-regulating efflux pumps or altering its membrane structure [63].
In order to assess the possible development of tolerance to antimicrobial blue light alone in P. aeruginosa, 10 repeated cycles of sub-lethal inactivation of bacteria in vitro, followed by bacterial re-growth, were carried out by Amin et al [64]. This tolerance study showed no evidence of the development of tolerance by P. aeruginosa to blue light after 10 consecutive cycles of sub-lethal inactivation. Their finding was in agreement with that obtained before when an Acinetobacter baumannii strain was treated with blue light for 10 consecutive cycles of sub-lethal inactivation [65].
Tavares et al. also attempted to model APDI resistance in bioluminescent E. coli and Vibrio fischeri by repeated exposure to 5,10,15-tris(1-methylpyridinium-4-yl)-20-(pentafluorophenyl)-porphyrin triiodide (TriPy+-Me-PF) excited with broad-band white light. After an initial exposure to APDI conditions in suspension (4 log10 reduction in cell viability), cells were plated, re-grown overnight, and repeatedly exposed to APDI 10 times thereafter using the aforementioned protocol. Their results demonstrated that neither E. coli nor V. fischeri could develop resistance to APDI [66].
Cassidy et al. exposed S. aureus and P. aeruginosa to the PS meso-tetra (N-methyl-4-pyridyl) porphine tetra tosylate (TMP) and methylene blue (MB) for 72 h in an effort to “habituate” cells. It was shown that sub-lethal APDI did not decrease the susceptibility to commonly employed antibiotics. Similarly, habituation with sub-lethal APDI did not reduce susceptibility of P. aeruginosa isolates to APDI protocols previously determined as lethal (eradication). A reduction in susceptibility to APDI following habituation was apparent for two S. aureus isolates with MB and for 1 S. aureus isolate with TMP as the photosesnitizer [67].
Pourhajibagher et al. studied the effects of APDI with toluidine blue O (TBO) and light emitting diode irradiation, on virulence features and expression profiling of genes encoding potential virulence factors in a colistin-resistant extensively-drug resistant (XDR) clinical isolate of A. baumannii (CR-XDR-AB) and in A. baumannii ATCC 19606 by studying the cells surviving APDI. Their results showed that the sAPDI could lead to modulation of the virulence of A. baumanii strains in surviving cells in planktonic growth mode by suppressing the expression of the genes (csuE, epsA, and abaI) associated with biofilm formation, but not blsA (the gene corresponding to attenuation in biofilm formation) at the transcriptional level. Following sAPDI, there was a significant up-regulation of blsA in CR-XDR-AB, but no up-regulation in the 19606 strain. Insignificant changes in the gene encoding bacterial heat shock protein (dnaK) and DNA repair gene (recA) in surviving cells demonstrated that sAPDI did not induce the typical cell stress fund with other antibacterial treatments, and there was no observable DNA damage when applied to A. baumanii strains in the planktonic growth mode. CR-XDR-AB cells surviving sAPDI showed a reduction of cell metabolic activity, increase in outer membrane permeability, and inhibition of efflux pump systems. sAPDI reduced the minimum inhibitory concentrations of the most tested antimicrobials by ≥2-fold in CR-XDR-AB strain [68].
Pourhajibagher et al. also evaluated whether Enterococcus faecalis cells exposed to indocyanine green (ICG), TBO, and MB-sAPDI exhibited changes in metabolic activity and biofilm formation. The anti-metabolic and anti-biofilm potential at sub-lethal APDI doses mediated by ICG, TBO, and MB against E. faecalis was analyzed using the XTT reduction assay, crystal violet assay, and scanning electron microscopy. Their results showed that higher doses of sAPDI adversely affected biofilm formation ability and metabolic activity. ICG-, TBO-, and MB-APDI at a maximum sub-lethal dose clearly reduced the formation of biofilm up to 42.8%, 22.6%, and 19.5%, respectively. ICG-, TBO-, and MB-sAPDI showed an obvious reduction in bacterial metabolic activity by 98%, 94%, and 82%, respectively. ICG-APDI showed a stronger inhibitory effect on biofilm formation in E. faecalis than MB- and TBO-APDI at sub-lethal levels. Interestingly, a gradual increase in metabolic activity and biofilm formation were observed upon exposure to a lower dose of sAPDI [69].
Kashef et al. study aimed to determine the effect of sAPDI on the antibiotic susceptibility of clinical S. aureus isolates. Forty clinical S. aureus isolates were exposed to sAPDI with TBO and MB. After exposure, susceptibility of surviving organisms to a range of antibiotics was determined and compared with the susceptibility of an untreated control. It was observed that the effect of sAPDI on antibiotic susceptibility patterns of isolates was strain-dependent. Thirty-five (85.4%) isolates were resistant to oxacillin but after exposure to sub-lethal TBO-APDI, only 13 (31.7%) remained resistant to this antibiotic (p < 0.001). In contrast, only 9 (22%) isolates were resistant to erythromycin, however, sub-lethal TBO-APDI increased resistance to this antimicrobial agent (61%), (p < 0.001). Exposure to sub-lethal TBO-APDI also increased resistance to amoxicillin-clavulanate. Exposure to sub-lethal MB-APDI decreased resistant to oxacillin and piperacillin. A reduction in susceptibility to amikacin and erythromycin following sAPDI was apparent with MB [70].
It is very important to emphasize one severe limitation of these studies. APDI is designed to be a relatively brief clinical procedure, as opposed to antibiotics, which are applied for relatively prolonged periods. Accordingly, each of these aforementioned studies exposed microbial samples to APDI conditions (light and PS) for brief periods of time, then removed these conditions and allowed the remaining bacteria to re-grow. This approach to APDI resistance, fails to address a serious problem: when microbes acquire resistance to antimicrobial agents, they must continuously grow in the presence of low-levels of antimicrobial agents. This is best exemplified by the emergence of antibiotic-resistance microbial isolates from the widespread employment of antibiotics in livestock feedstuffs [71].
The approach of chemical antimicrobials is quite opposite to that of APDI. APDI is inherently designed to be a brief process in which excess PS is washed away and after delivery of light of the appropriate wavelength and desired fluence, light is removed from the region exposed to APDI. Thus, a more realistic way to establish a possible increased tolerance or resistance to APDI would be by allowing microbial samples to grow continuously under sAPDI conditions; i.e. exposed to defined sub-lethal levels of PS and light. This also, has obvious limitations in that APDI is not designed to be an extended procedure; however, such a protocol could be clinically rationalized by considering extended microbe-PS interactions; antimicrobial PSs are generally cationic molecules such that there is sufficient interaction between the PS and the anionic cell envelope of Gram-positive and Gram-negative bacteria. If a patient were to receive an APDI treatment, removal of all excess PS after light exposure would prove entirely impossible. This then follows that as the patient goes about his or her daily life, exposure to ambient light sources may be unavoidable, thus creating quasi-sub-lethal PDI stress.
5 Mechanisms determining bacterial susceptibility to APDI
Studies have focused on bacterial protection from radical-type oxygen species such as superoxide and hydrogen peroxide. However, the mechanism by which bacteria respond to APDI-mediated stresses remains relatively unknown [72].
5.1 Protective pigments
In recent years, many reports about the efficacy of APDI against P. aeruginosa have been published, and it has become clear that P. aeruginosa is particularly tolerant to photodynamically induced oxidative stress [73–75]. The ability of P. aeruginosa to produce pigments, combined with the ability to elicit an oxidative stress response [76], may thus contribute to survival to APDI-induced oxidative stress.
Orlandi et al. [77] investigated the role of pigments in tolerance of P. aeruginosa to APDI-induced oxidative stress. To this end, differentially pigmented transposon mutants of P. aeruginosa PAO1 were isolated and submitted to APDI with two different PS, a phenothiazinium dye, TBO, that acts mainly via a type I mechanism, and a porphyrin dye, 5,10,15,20-tetrakis-(1-methyl-4-pyridyl)-21H,23-porphine tetra-p-tosylate (TMPyP), that acts mainly via a type II mechanism. In general, in the presence of pigments a higher tolerance to APDI-induced photo-oxidative stress was observed. Hyper-production of pyomelanin makes the cells much more tolerant to stress caused by either radicals or singlet oxygen generated by different PS upon photo-activation. Phenazines, pigments characteristic of P. aeruginosa (pyocyanin and phenazine-1-carboxylic acid) produced in different amounts depending on the culture conditions, are able to counteract both types of APDI-elicited ROS. Hyper-production of pyoverdin, caused by a mutation in a quorum-sensing gene, made P. aeruginosa more tolerant to a PS that generates mainly singlet oxygen, although in this case the observed tolerance to photo-oxidative stress cannot be completely attributed to the presence of the pigment.
Carotenoids are naturally occurring terpenoid pigments, formed from isoprene residues making up a polyene chain of conjugated double bonds. These pigments are responsible for the wide variety of orange-red colors seen in nature, and absorb light in the wavelength range of 300– 600 nm [78].
For many years the role of carotenoids as accessory pigments in photosynthesis was thought to be the only function in photosynthetic organisms. However, Sistrom et al. [79] suggested that carotenoids might be acting as protective agents against photodynamic action in bacteria. They studied the wild type and an isogenic mutant of Rhodopseudomonas spheroides (“blue-green”) that did not produce colored carotenoids but instead accumulated the colorless carotenoid precursor, phytoene. These workers found that when the blue-green mutant was grown in the presence of light and air, growth stopped in the light, chlorophyll was destroyed and the cells were killed. On the other hand, the wild type, with its normal level of carotenoid pigments, is not injured by growth in light and air; in the presence of air, chlorophyll synthesis stops and the organism grows by aerobic metabolism. It should be noted that the deletion of the carotenoid pigments did not markedly affect the ability of the blue-green mutant to grow photosynthetically in the presence of light, but in an atmosphere of nitrogen (bacterial photosynthesis is anaerobic). Only when both light and air were present did the lack of carotenoids lead to photosensitization. The authors were able to show that it was the bacteriochlorophyll that was responsible for the lethal photosensitization of the blue-green mutant and that both oxygen and light were necessary for the destructive reaction to occur (photodynamic action). They concluded that the carotenoid pigments functioned as protective agents against this photodynamic killing and that the bacteriochlorophyll was the endogenous photosensitizer. These findings that carotenoids could protect against chlorophyll photosensitization, have been confirmed in other photosynthetic bacteria and algae [80].
Most S. aureus strains produce an orange membrane-bound carotenoid pigment known as staphyloxanthin. It is a typical secondary metabolite. It is not necessary for the growth and reproduction of S. aureus but might serve a role in survival in infected hosts and in avoiding the immune system [81, 82]. Liu et al. showed that a S. aureus mutant with disrupted carotenoid biosynthesis was more susceptible to oxidant killing, indicating that carotenoids could act as a virulence factor [83]. Carotenoids could reduce the penetration of singlet oxygen by decreasing membrane fluidity [84].
5.2 Capsular polysaccharide
Due to the short diffusion distance of APDI-generated ROS, its efficiency to kill bacteria depends on the degree to which the PS binds to bacteria, and whether the PS can penetrate to a sensitive intracellular site. Capsular polysaccharide could either increase or decrease the binding of the PS to the bacterial cell and, independently of binding, could also act as a barrier to penetration of the PS into the interior of the microorganism, where the generation of ROS would be more likely to lead to cell death. Gad et al. used two isogenic pairs of wild-type and transposon mutants deficient in capsular polysaccharide and slime production generated from strains of Staphylococcus epidermidis and S. aureus to examine the effects of extracellular slime on susceptibility to APDI mediated by two cationic PS (a polylysine-chlorin e6 conjugate, pL-ce6, and MB) and an anionic molecule, free ce6, and subsequent exposure to 665-nm light. Free ce6 gave more killing of mutant strains than wild type, despite the latter taking up more PS. The cationic pL-ce6 and MB gave similar uptakes and killing despite a 50-fold difference in incubation concentration. Differences in susceptibility between strains observed with free ce6 largely disappeared with the cationic compounds despite significant differences in uptake. These data suggested that slime production can be a problem against APDI for gram-positive bacteria but that this problem can be overcome by using cationic PS [85].
5.3 Efflux pumps
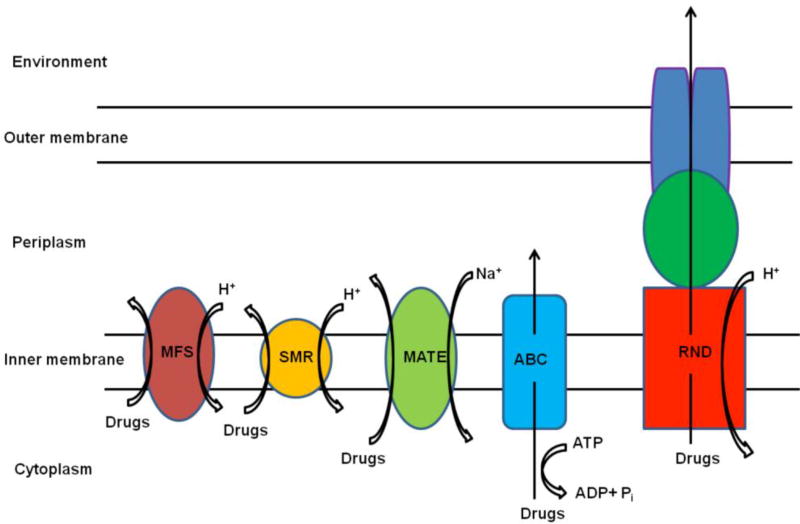
One possible mechanism determining the efficiency of APDI of bacteria might be its dependence on multi-drug resistant efflux pumps [86]. Efflux pumps (summarized in Figure 4) are transport proteins involved in the extrusion of toxic substrates from within cells into the external environment [87]. Such pumps may be specific for one substrate or may transport a range of structurally dissimilar compounds.
Figure 4.
Schematic illustration of the main types of bacterial drug efflux pumps (MFS: major facilitator superfamily, SMR: small multi drug resistance, MATE: multi drug and toxic compound extrusion, ABC: ATP-binding cassette family, and RND: resistance-nodulation-division family)
Phenothiazinium dyes have long been established as nontoxic and clinically useful compounds both for staining living tissues [88] and for some pharmacological indications [89, 90]. This consideration together with their ready availability has been important in their selection as antimicrobial PSs for the few clinical indications in which APDI is carried out. Tegos and Hamblin demonstrated for the first time that phenothiazinium-based PSs were substrates of multi-drug resistant efflux pumps in bacteria. This discovery raised the possibility of combining the phenothiazinium dyes with one of range of possible efflux pump inhibitors [86].
Efflux pump inhibitors (EPI) had a dramatic effect in potentiating the killing effect of APDI with phenothiazinium dyes [91]. Killing of S. aureus mediated by TBO and red light was greatly increased by coincubation with known inhibitors of the major facilitator pump NorA (diphenyl urea INF271, reserpine, 5'-methoxyhydnocarpin, and the polyacylated neohesperidoside, ADH7). The potentiation effect was greatest in the case of mutants that overexpressed NorA and least in NorA null cells. Addition of the EPI before TBO, had a bigger effect than addition of the EPI after TBO. Cellular uptake of TBO was increased by EPI. EPI increased APDI mediated by other phenothiazinium dyes, such as methylene blue and dimethylmethylene blue, but not that mediated by non-phenothiazinium PS, such as Rose Bengal and benzoporphyrin derivative. Killing of Gram-negative P. aeruginosa mediated by TBO and light was also potentiated by the resistance nodulation division pump (MexAB-OprM) inhibitor phenylalanine-arginine betanaphthylamide but to a lesser extent than for S. aureus. Kishen et al. [92] evaluated the ability of MB and rose bengal (RB) to inactivate biofilms of E. faecalis. The role of a specific microbial EPI, verapamil hydrochloride in the MB-APDI of E. faecalis biofilms was also investigated. The results showed that E. faecalis biofilms exhibited significantly higher resistance to APDI when compared with E. faecalis in suspension. APDI with cationic MB produced superior inactivation of E. faecalis strains in a biofilm along with significant destruction of biofilm structure when compared with anionic RB. The ability to inactivate biofilm bacteria was further enhanced when the EPI was used with MB. These experiments demonstrated the advantage of MB combined with an EPI to inactivate biofilm bacteria and disrupt biofilm structure.
Grinholc et al. [93] investigated whether the uptake and activity of the multi-drug resistance pumps might influence their previously observed variations [94] among the clinical strains to protoporphyrin-derived, amphipilic protoporphyrin diarginate mediated APDI. Using a new set of four selected methicillin-resistant (MRSA) and methicillin-susceptible (MSSA) clinical strains as well as ATCC S. aureus, they confirmed that the bactericidal effect of the APDI was straindependent as it showed a wide variation ranging from 0 to 5 log10-unit reduction in viable counts. However, neither variation in levels of PS uptake nor the pharmacological inhibition of efflux pump explained such a phenomenon.
5.4 Antibiotic resistance mechanisms
Another study by Grinholc group [95] aimed to provide relevant evidence as to whether the response of MRSA strains to APDI differs significantly from that of MSSA, and whether it results from antimicrobial resistance mechanisms or susceptibility to routinely used antimicrobials. S. aureus response to protoporphyrin IX (PPIX)-mediated photo-inactivation was studied for 424 MRSA/MSSA isolates. VITEK 2 Advanced Expert System was used to detect antimicrobial resistance mechanisms and strains’ susceptibility to antibiotic therapy. Their results demonstrated that MRSA were significantly more resistant to photo-inactivation than MSSA strains; however, the difference observed did not result from antimicrobial susceptibility or resistance mechanisms. Moreover, it was determined that the ability to form biofilms in vitro did not explain the observed differences between MRSA and MSSA strains.
Tang et al [96] compared APDI with pL-ce6 and TBO in clinical MRSA and ESBL producing E. coli, together with their corresponding ATCC reference strains. pL-ce6 at 8 µM, 30 J/cm2 gave 5 logs killing for ESBL-producing E. coli and E. coli (ATCC 25922); 4 log killing for MRSA, and 3 log killing for S. aureus (ATCC 25923). TBO at 80 µM, 30 J/cm2 only gave 3 logs killing of MRSA and 2 log killing of S. aureus (ATCC 25923). TBO (400 µM, 30 J/cm2 induced equal killing for both E. coli strains The MRSA isolate responded better than reference S. aureus to both PS.
5.5 Anti-oxidant enzymes
To understand if the antioxidant enzyme status may be involved in the S. aureus response to APDI, Nakonieczna et al. [97] checked survival rates of the isogenic sod mutant and wild-type strains of S. aureus and compared the effect of SOD in the response to APDI at both the protein as well as gene expression levels. Superoxide dismutase forms the first line of bacterial defense against oxidative stress, therefore it was expected that a correlation might exist between the SOD status in the cell and the response to APDI. The effectiveness of APDI towards S. aureus and its SOD isogenic mutants deprived of either of the two SOD activities, namely SodA or SodM or both of them showed similar results, regardless of the SOD status cultured in TSB medium. On the contrary, in the special CL medium (without Mn2+ ions) the double SodAM knockout mutant was highly susceptible to APDI. Among 8 clinical isolates of S. aureus analyzed (4 MRSA and 4 MSSA), strains, highly resistant and strains highly susceptible to APDI were identified. They observed that SOD activity, as well as sodA and sodM transcript level increased after protoporphyrin IX-based APDI, but only in APDI-sensitive strains. They confirmed that porphyrin-based photo-killing efficacy was a strain-dependent phenomenon. They showed that oxidative stress sensitivity caused by the lack of both SOD enzymes can be relieved in the presence of Mn2+ ions and partially in the presence of Fe2+ ions. The fact that SOD activity increased was observed only in APDI-susceptible cells emphasizes that this is probably not a direct factor affecting S. aureus vulnerability to porphyrin based APDI.
5.6 Agr status
As a global transcription regulator, the accessory gene regulator (Agr) plays a central role in gene regulation in S. aureus [98]. Activation of the Agr system down-regulates genes associated with bacterial adhesion and up-regulates genes for exoprotein production [99]. Park et al. [100] wanted to gain insight into the mechanism of bacterial response to APDI by exploring the genetic response of S. aureus to APDI combined with the photosensitizer Ce6 and laser light. It was shown that Agr was involved in S. aureus response to oxidative stress induced by APDI. Transcriptional profiling revealed that sAPDI induced a general stress response and also activated Agr-dependent gene regulation. Moreover, mutant S. aureus lacking Agr function showed hyper-susceptibility to two different APDI regimens with higher energy densities, which led to the hypothesis that the function of the Agr was necessary for protection of S. aureus from APDI-mediated oxidative stress.
S. aureus expresses a large variety of virulence factors that are accurately regulated by a complex network of different transcriptional factors (e.g.: mgrA, ơB) and several two-component systems (e.g.: sae) [101, 102]. The sae system up-regulates a wide variety of virulence factors including proteins that link bacteria to the extracellular matrix of the host, like the adhesins Eap or FnBPs [103]. Moreover, both agr and sae systems have been found to be involved in the response to oxidative stress [104, 105]. Gándara et al. [106] studied the differential susceptibility to TBO-APDI of S. aureus mutants in different key regulators of virulence factors involved in planktonic lifestyles, and which have been related (to some extent) to oxidative stress. They showed that the two-component system sae impaired the response to TBO-APDI through a mechanism not related to the Eap adhesin. They also showed that the agr locus does not seem to be related to TBO-APDI sensitivity. However, by employing S. aureus mutants lacking the agr system, it was shown that agr induced a certain degree of resistance against APDI employing protoporphyrin diarginate and chlorin e6 as PS [100, 107]. It is therefore important to consider that different PS elicit photo-damage by different mechanisms, and this may be related to the different strain responses to APDI.
5.7 Heat shock proteins
Heat shock proteins (HSPs) are indicative of stressful conditions that may affect cell viability. They are a group of ubiquitous chaperone proteins responsible for the refolding, repair and recycling of damaged proteins, and stabilization of lipid membranes during cellular stress [108– 110]. In microbial cells, the heat shock proteome has best been characterized in E. coli, and it has been found that two major HSP families—GroEL / GroES and DnaK/ DnaJ / GrpE—are mainly responsible for protection against stress in both Gram-negative and Gram-positive bacteria [111–114]. Up-regulation of HSPs during oxidative, antibiotic, osmotic and acid stress is associated with resistance to these stresses, and up-regulation of HSPs prior to subsequent stress enables bacterial cells to acquire “tolerance” to that particular stress [114].
To date, only two studies have looked at the bacterial heat shock response to APDI. The first of these studies was performed by Bolean et al. [115]. They tested whether the expression of GroEL by Streptococcus mutans was enhanced after APDI with rose bengal. Higher HSP expression was detected in bacteria after APDI treatment as compared with light alone or PS alone. The expression of HSP after APDI was similar to that induced by osmotic stress. No DNA degradation was observed after APDI of S. mutans. They concluded that APDI may cause effects similar to those of other stressful conditions in S. mutans, and cell death induced by this treatment reflects the inability of the bacteria to protect itself sufficiently against the lethal effects of APDI with rose bengal.
St Denis et al. [116] investigated whether APDI induced protective responses such as HSPs in bacteria. Using TBO at sub-lethal APDI conditions, a sevenfold increase in bacterial HSP GroEL and a three-fold increase in HSP DnaK were observed in E. coli post APDI. Pretreatment with 50°C heat for 30 min reduced APDI killing in both E. coli and in Enterococcus faecalis. Inhibition of the highly conserved chaperone DnaK using a small molecule benzylidene lactam HSP inhibitor potentiated (but not significantly) the effect of APDI in E. faecalis; however, this effect was not observed in E. coli presumably because inhibitor could not gain access due to the Gram-negative permeability barrier. They concluded that induction of HSPs may be a mechanism whereby bacteria could become tolerant to APDI and called for the need for further study in the application of dual APDI-HSP-inhibition therapies.
6. Biofilm resistance to APDI
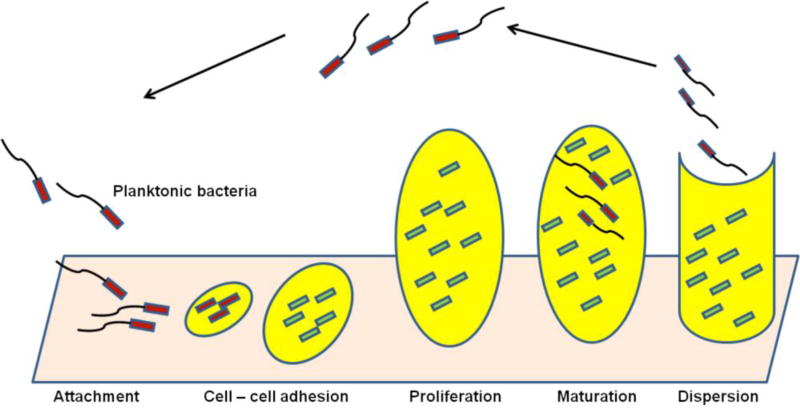
A structured consortium of microbial cells (mostly bacteria or fungi) attached onto a living or inert surface is formed by the cells sticking to each other where they are surrounded by the self-produced extracellular polymeric matrix (composed of polysaccharides, proteins, lipids, and extracellular DNA) and is known as a biofilm (Figure 5). The formation of biofilm is considered an adaptation of microbes to hostile environments [117, 118]. Bacteria that live in a biofilm community possess several advantages, including structural stability, firm adherence to biotic or abiotic surfaces, increased virulence, and resistance to both antimicrobial therapy and the host immune response [18, 119, 120].
Figure 5.
Stages of biofilm development
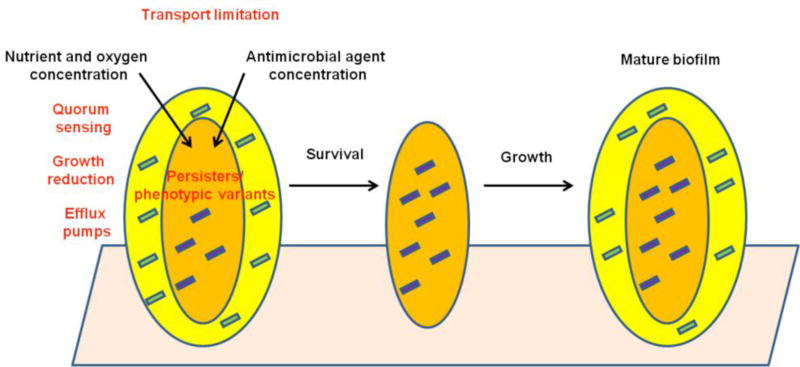
The principal hypotheses to explain reduced antibiotic susceptibility of bacteria in biofilms are the possibility of slow or incomplete penetration of the antibiotic into the biofilm (due to biofilm matrix components proving a mechanical barrier to diffusion), modification of microbial enzymes and efflux pumps, an altered chemical microenvironment within the biofilm (low oxygen concentration and slow growth), a unique, and highly protected, phenotypic state (persister cells), and high cell density due to quorum sensing (Figure 6) [121, 122].
Figure 6.
Mechanisms of reduced antimicrobial susceptibility of bacteria in biofilm (1-Antimicrobial agents kill susceptible outer cells, 2- Inner persister cells survive, 3- Entire biofilm regenerates)
It has been shown that bacteria growing as biofilms are less susceptible to APDI compared with their equivalent planktonic forms. The dye concentration and the light dose needed for the photo-inactivation of biofilms are considerably higher than those required to inactivate planktonic bacterial suspensions [123–125]. In fact, cells growing in biofilms differ from their planktonic counterparts in a number of aspects, such as the cell wall structure and composition, rate of growth, and presence of polysaccharide intercellular adhesin (PIA), which may block both the uptake of the PS and the penetration of light, and thereby reduce the photosensitizing efficiency [126, 127]. Generally, longer pre-incubation times (up to 24 h), higher concentrations (up to 25 times) and longer light exposure times (up to 30 min) are required to reach a phototoxicity of 3 log10 steps against biofilm growing bacteria compared to their planktonic counterparts [58]. To date no specific regulatory process is known for biofilm tolerance to APDI.
6.1 Can Nanotechnology Potentiate APDI of Biofilm?
The use of nanotechnology is an attractive approach to improving the effectiveness of APDI against resistant microbial cells [128–133] and to improve PS delivery to biofilms [134–136]. This goal may be achieved in different ways, for instance by enhancing the delivery of PS to microorganisms (encapsulating the PS in nanoparticles) [137] or by increasing the 1O2 yield of the PS (by covalently binding the PS to the surface of the nanoparticles [138], or simply by mixing the nanoparticles and PS together). In some cases the nanoparticles themselves (for instance TiO2, fullerenes or quantum dots) have been shown to act as PS themselves and are capable of photodynamically inactivating the microorganisms [139].
Shrestha and Kishen assessed the antibacterial effect of a novel PS (Rose Bengal functionalized chitosan nanoparticles [CSRBnp]) to eliminate bacteria in the presence of various root canal constituents that are known to inhibit the antibacterial efficacy of root canal disinfectants. Chitosan is an attractive material from which to make antimicrobial nanoparticles as it is a naturally occurring polymer, is biodegradable and biocompatible, and moreover it posseses an intrinsic cationic charge [140]. The antibacterial effect of CSRBnp was tested on planktonic E. faecalis with or without pretreatment by using different inhibiting agents such as dentin, dentin-matrix, pulp tissue, bacterial lipopolysaccharides, and bovine serum albumin (BSA). Pulp and BSA inhibited the antibacterial effect of CSRBnp (without photoactivation) significantly (P < 0.05) even after 24 hours of interaction. CSRBnp completely eliminated the bacteria after 24 hours of interaction after photodynamic therapy [132].
Chitosan (CS) nanoparticles were also used in a study by Chen et al. to investigate their ability to potentiate the activity of erythrosine (ER) against bacteria and yeast. CS nanoparticles loaded with ER were prepared by an ionic gelation method and tested for their APDI efficacy against planktonic cells and biofilms of S. mutans, P. aeruginosa and Candida albicans. Significant phototoxicity was observed when the cells were exposed to light irradiation after treatment with free ER or ER/CS nanoparticles. The antimicrobial activity of ER/CS nanoparticles was significantly higher than ER in free form [129].
Pagonis et al. studied the in vitro effects of poly(lactic-co-glycolic acid) (PLGA) nanoparticles loaded with MB and light against E. faecalis (ATCC 29212). E. faecalis species were sensitized in planktonic phase and in experimentally infected root canals of human extracted teeth with MBloaded nanoparticles followed by exposure to red light at 665 nm. The synergism of light and MB-loaded nanoparticles led to approximately 2 and 1 log10 reduction of colony-forming units (CFUs) in planktonic phase and root canals, respectively. In both cases, mean log10 CFU levels were significantly lower than controls and MB-loaded nanoparticles without light [141].
Conjugating PSs to the surface of gold nanoparticles (GNPs) is a modern approach to increase APDI efficacy. Darabpour et al. aimed to determine whether GNPs could enhance the efficiency of MB-APDI of MRSA in a mature biofilm. Positively charged MB was immobilized onto the negatively-charged GNPs through electrostatic interaction. Four day-old biofilms of MRSA were treated with MB-conjugated GNPs and subsequently exposed to red light. MB-conjugated GNPs showed significant photo-inactivation against 4-day-old biofilm of MRSA (>5 log10 CFU reduction) while free MB-APDI resulted in less than 1 log10 CFU reduction [142].
6.2 Potentiation of APDI by Inorganic Salts
We have discovered that APDI can be potentiated (up to 6 logs of extra killing) by addition of simple inorganic salts, depending on the PS structure and photochemical mechanism. The most powerful and versatile salt is potassium iodide [143], but potassium bromide [143], sodium thiocyanate [144], sodium azide [145] and even sodium nitrite have also shown potentiation. The mechanism of potentiation with iodide is believed to be singlet oxygen addition to iodide to form iodine radicals, hydrogen peroxide and molecular iodine [146]. Another mechanism involves two-electron oxidation of iodide/bromide taking place in titanium dioxide photocatalysis to form hypohalites [147, 148]. A third mechanism involves a oneelectron oxidation of azide anion to form azide radical [145].
The addition of iodide has been shown to improve the performance of aPDT in vivo using several animal models of localized infection. Potentiation of MB-APDT using red light, by addition of iodide was shown in 3rd degree burn wound in mice infected with bioluminescent MRSA [143]. Another study used a cationic fullerene (LC16) with a deca-quaternary chain and a second attached chain of ten tertiary amino groups excited by either white or UVA light [149]. A mouse model of a partial thickness skin abrasion infected with bioluminescent A. baumannii gave an increased rate of loss of bioluminescence signal when iodide (10 mM) was combined with LC16 and then illuminated with either UVA or with white light. Finally addition of iodide was tested in a mouse model of oral candidiasis infection mediated by MB and red light [150]. Although we have not yet specifically published a paper looking at the effect of iodide on potentiating APDI destruction of bacterial biofilms, preliminary data have shown that KI makes a surprisingly large difference to the number of logs of killing. Studies are ongoing.
7 Conclusion
APDI is an exciting new approach for the selective inactivation and eradication of microbial pathogens. As antibiotic resistance becomes an even greater issue, APDI will likely be introduced more widely into the clinical setting, however, it is crucial that APDI efficacy be practically evaluated.
The studies mentioned in this work have attempted to model and evaluate possible bacterial resistance mechanisms to APDI. While antibiotics generally work on a “key-and-lock” principle, with each drug having a single target in the bacterial metabolism, APDI is very different. APDI produces ROS that can damage a host of microbial biomolecules, most of which can be lethal, rather than just inhibiting growth. Therefore the chances that microbes can develoip tolerance or resistance to APDI must be considered highly unlikely. Although it seems that there is no universal agreement that can be reached at this time, one thing is certain: more evaluation is needed. The scientific community should be cautious and not get too excited over numerous in vitro studies reporting the killing of a wide variety of species. Nevertheless, in vivo studies of APDT continue to show promise for carefully chosen types of infection [151, 152].
Acknowledgments
MRH was funded by US NIH R01AI050875 and R21AI121700. Nasim Kashef was supported by University of Tehran.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Walsh C. Molecular mechanisms that confer antibacterial drug resistance. Nature. 2000;406(6797):775–81. doi: 10.1038/35021219. [DOI] [PubMed] [Google Scholar]
- 2.Fluit AC, Visser MR, Schmitz FJ. Molecular detection of antimicrobial resistance. Clin Microbiol Rev. 2001;14(4):836–71. doi: 10.1128/CMR.14.4.836-871.2001. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74(3):417–33. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aarestrup FM, et al. Effect of abolishment of the use of antimicrobial agents for growth promotion on occurrence of antimicrobial resistance in fecal enterococci from food animals in Denmark. Antimicrob Agents Chemother. 2001;45(7):2054–9. doi: 10.1128/AAC.45.7.2054-2059.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byarugaba DK. A view on antimicrobial resistance in developing countries and responsible risk factors. Int J Antimicrob Agents. 2004;24(2):105–10. doi: 10.1016/j.ijantimicag.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 6.O'Neill J. Tackling a global health crisis:initial steps. The Review on Antimicrobial Resistance Chaired by Jim O’Neill. 2015 [Google Scholar]
- 7.Taylor PW, Stapleton PD, Paul Luzio J. New ways to treat bacterial infections. Drug Discov Today. 2002;7(21):1086–91. doi: 10.1016/s1359-6446(02)02498-4. [DOI] [PubMed] [Google Scholar]
- 8.Hamblin MR. Antimicrobial photodynamic inactivation: a bright new technique to kill resistant microbes. Curr Opin Microbiol. 2016;33:67–73. doi: 10.1016/j.mib.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamblin MR, Hasan T. Photodynamic therapy: a new antimicrobial approach to infectious disease? Photochem Photobiol Sci. 2004;3(5):436–50. doi: 10.1039/b311900a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang L, Dai T, Hamblin MR. Antimicrobial photodynamic inactivation and photodynamic therapy for infections. Methods Mol Biol. 2010;635:155–73. doi: 10.1007/978-1-60761-697-9_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green TJ, et al. Phosphorimeters for analysis of decay profiles and real time monitoring of exponential decay and oxygen concentrations. Anal Biochem. 1988;174(1):73–9. doi: 10.1016/0003-2697(88)90520-9. [DOI] [PubMed] [Google Scholar]
- 12.Ochsner M. Photophysical and photobiological processes in the photodynamic therapy of tumours. J Photochem Photobiol B. 1997;39(1):1–18. doi: 10.1016/s1011-1344(96)07428-3. [DOI] [PubMed] [Google Scholar]
- 13.Luksiene Z. Photodynamic therapy: mechanism of action and ways to improve the efficiency of treatment. Medicina (Kaunas) 2003;39(12):1137–50. [PubMed] [Google Scholar]
- 14.Jori G, et al. Photodynamic therapy in the treatment of microbial infections: basic principles and perspective applications. Lasers Surg Med. 2006;38(5):468–81. doi: 10.1002/lsm.20361. [DOI] [PubMed] [Google Scholar]
- 15.Bourre L, et al. Effective photoinactivation of Gram-positive and Gram-negative bacterial strains using an HIV-1 Tat peptide-porphyrin conjugate. Photochem Photobiol Sci. 2010;9(12):1613–20. doi: 10.1039/c0pp00146e. [DOI] [PubMed] [Google Scholar]
- 16.Sperandio FF, Huang YY, Hamblin MR. Antimicrobial Photodynamic Therapy To Kill Gram-Negative Bacteria. Recent Pat Antiinfect Drug Discov. 2013 doi: 10.2174/1574891x113089990012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bertoloni G, et al. Photosensitizing activity of water- and lipid-soluble phthalocyanines on prokaryotic and eukaryotic microbial cells. Microbios. 1992;71(286):33–46. [PubMed] [Google Scholar]
- 18.Maisch T, et al. Antibacterial photodynamic therapy in dermatology. Photochem Photobiol Sci. 2004;3(10):907–17. doi: 10.1039/b407622b. [DOI] [PubMed] [Google Scholar]
- 19.Anbar AD. Oceans. Elements and evolution. Science. 2008;322(5907):1481–3. doi: 10.1126/science.1163100. [DOI] [PubMed] [Google Scholar]
- 20.Ligeza A, et al. Oxygen permeability of thylakoid membranes: electron paramagnetic resonance spin labeling study. Biochim Biophys Acta. 1998;1365(3):453–63. doi: 10.1016/s0005-2728(98)00098-x. [DOI] [PubMed] [Google Scholar]
- 21.Gerschman R, et al. Oxygen poisoning and x-irradiation: a mechanism in common. Science. 1954;119(3097):623–6. doi: 10.1126/science.119.3097.623. [DOI] [PubMed] [Google Scholar]
- 22.Imlay JA. Pathways of oxidative damage. Annu Rev Microbiol. 2003;57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- 23.Imlay JA. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol. 2013;11(7):443–54. doi: 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cabiscol E, Tamarit J, Ros J. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int Microbiol. 2000;3(1):3–8. [PubMed] [Google Scholar]
- 25.Chen J, et al. Oxidative stress at high temperatures in Lactococcus lactis due to an insufficient supply of Riboflavin. Appl Environ Microbiol. 2013;79(19):6140–7. doi: 10.1128/AEM.01953-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murata M, et al. Molecular strategy for survival at a critical high temperature in Eschierichia coli. PLoS One. 2011;6(6):e20063. doi: 10.1371/journal.pone.0020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chattopadhyay MK, et al. Increase in oxidative stress at low temperature in an antarctic bacterium. Curr Microbiol. 2011;62(2):544–6. doi: 10.1007/s00284-010-9742-y. [DOI] [PubMed] [Google Scholar]
- 28.Humphries KM, Szweda LI. Selective inactivation of alpha-ketoglutarate dehydrogenase and pyruvate dehydrogenase: reaction of lipoic acid with 4-hydroxy-2-nonenal. Biochemistry. 1998;37(45):15835–41. doi: 10.1021/bi981512h. [DOI] [PubMed] [Google Scholar]
- 29.Sies H. Damage to plasmid DNA by singlet oxygen and its protection. Mutat Res. 1993;299(3–4):183–91. doi: 10.1016/0165-1218(93)90095-u. [DOI] [PubMed] [Google Scholar]
- 30.Sies H, Menck CF. Singlet oxygen induced DNA damage. Mutat Res. 1992;275(3–6):367–75. doi: 10.1016/0921-8734(92)90039-r. [DOI] [PubMed] [Google Scholar]
- 31.Fucci L, et al. Inactivation of key metabolic enzymes by mixed-function oxidation reactions: possible implication in protein turnover and ageing. Proc Natl Acad Sci U S A. 1983;80(6):1521–5. doi: 10.1073/pnas.80.6.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stadtman ER. Metal ion-catalyzed oxidation of proteins: biochemical mechanism and biological consequences. Free Radic Biol Med. 1990;9(4):315–25. doi: 10.1016/0891-5849(90)90006-5. [DOI] [PubMed] [Google Scholar]
- 33.Ramel F, et al. Chemical quenching of singlet oxygen by carotenoids in plants. Plant Physiol. 2012;158(3):1267–78. doi: 10.1104/pp.111.182394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramel F, et al. Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc Natl Acad Sci U S A. 2012;109(14):5535–40. doi: 10.1073/pnas.1115982109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pomposiello PJ, Demple B. Global adjustment of microbial physiology during free radical stress. Adv Microb Physiol. 2002;46:319–41. doi: 10.1016/s0065-2911(02)46007-9. [DOI] [PubMed] [Google Scholar]
- 36.Hillion M, Antelmann H. Thiol-based redox switches in prokaryotes. Biol Chem. 2015;396(5):415–44. doi: 10.1515/hsz-2015-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dubbs JM, Mongkolsuk S. Peroxide-sensing transcriptional regulators in bacteria. J Bacteriol. 2012;194(20):5495–503. doi: 10.1128/JB.00304-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tao K, Fujita N, Ishihama A. Involvement of the RNA polymerase alpha subunit C-terminal region in co-operative interaction and transcriptional activation with OxyR protein. Mol Microbiol. 1993;7(6):859–64. doi: 10.1111/j.1365-2958.1993.tb01176.x. [DOI] [PubMed] [Google Scholar]
- 39.Zheng M, et al. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J Bacteriol. 2001;183(15):4562–70. doi: 10.1128/JB.183.15.4562-4570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Belzer C, et al. PerR controls peroxide- and iron-responsive expression of oxidative stress defense genes in Helicobacter hepaticus. Eur J Microbiol Immunol (Bp) 2011;1(3):215–22. doi: 10.1556/EuJMI.1.2011.3.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mongkolsuk S, Helmann JD. Regulation of inducible peroxide stress responses. Mol Microbiol. 2002;45(1):9–15. doi: 10.1046/j.1365-2958.2002.03015.x. [DOI] [PubMed] [Google Scholar]
- 42.van Vliet AH, et al. Campylobacter jejuni contains two fur homologs: characterization of iron-responsive regulation of peroxide stress defense genes by the PerR repressor. J Bacteriol. 1999;181(20):6371–6. doi: 10.1128/jb.181.20.6371-6376.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Helmann JD. Specificity of metal sensing: iron and manganese homeostasis in Bacillus subtilis. J Biol Chem. 2014;289(41):28112–20. doi: 10.1074/jbc.R114.587071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fuangthong M, et al. Regulation of the Bacillus subtilis fur and perR genes by PerR: not all members of the PerR regulon are peroxide inducible. J Bacteriol. 2002;184(12):3276–86. doi: 10.1128/JB.184.12.3276-3286.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee JW, Helmann JD. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature. 2006;440(7082):363–7. doi: 10.1038/nature04537. [DOI] [PubMed] [Google Scholar]
- 46.Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem. 2008;77:755–76. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Greenberg JT, et al. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc Natl Acad Sci U S A. 1990;87(16):6181–5. doi: 10.1073/pnas.87.16.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsaneva IR, Weiss B. soxR, a locus governing a superoxide response regulon in Escherichia coli K-12. J Bacteriol. 1990;172(8):4197–205. doi: 10.1128/jb.172.8.4197-4205.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gu M, Imlay JA. The SoxRS response of Escherichia coli is directly activated by redox-cycling drugs rather than by superoxide. Mol Microbiol. 2011;79(5):1136–50. doi: 10.1111/j.1365-2958.2010.07520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pomposiello PJ, Bennik MH, Demple B. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J Bacteriol. 2001;183(13):3890–902. doi: 10.1128/JB.183.13.3890-3902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Niederhoffer EC, et al. Control of Escherichia coli superoxide dismutase (sodA and sodB) genes by the ferric uptake regulation (fur) locus. J Bacteriol. 1990;172(4):1930–8. doi: 10.1128/jb.172.4.1930-1938.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Compan I, Touati D. Interaction of six global transcription regulators in expression of manganese superoxide dismutase in Escherichia coli K-12. J Bacteriol. 1993;175(6):1687–96. doi: 10.1128/jb.175.6.1687-1696.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benov LT, Fridovich I. Escherichia coli expresses a copper- and zinc-containing superoxide dismutase. J Biol Chem. 1994;269(41):25310–4. [PubMed] [Google Scholar]
- 54.Finn GJ, Condon S. Regulation of catalase synthesis in Salmonella typhimurium. J Bacteriol. 1975;123(2):570–9. doi: 10.1128/jb.123.2.570-579.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.von Ossowski I, et al. Nucleotide sequence of Escherichia coli katE, which encodes catalase HPII. J Bacteriol. 1991;173(2):514–20. doi: 10.1128/jb.173.2.514-520.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Demple B, Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu Rev Biochem. 1994;63:915–48. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 57.Levine RL, et al. Methionine residues as endogenous antioxidants in proteins. Proc Natl Acad Sci U S A. 1996;93(26):15036–40. doi: 10.1073/pnas.93.26.15036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maisch T. Resistance in antimicrobial photodynamic inactivation of bacteria. Photochem Photobiol Sci. 2015;14(8):1518–26. doi: 10.1039/c5pp00037h. [DOI] [PubMed] [Google Scholar]
- 59.Sabbahi S, et al. The role of reactive oxygen species in Staphylococcus aureus photoinactivation by methylene blue. Water Sci Technol. 2008;58(5):1047–54. doi: 10.2166/wst.2008.471. [DOI] [PubMed] [Google Scholar]
- 60.Huang L, et al. Type I and Type II mechanisms of antimicrobial photodynamic therapy: an in vitro study on gram-negative and gram-positive bacteria. Lasers Surg Med. 2012;44(6):490–9. doi: 10.1002/lsm.22045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McMahon MA, et al. Changes in antibiotic susceptibility in staphylococci habituated to sub-lethal concentrations of tea tree oil (Melaleuca alternifolia) Lett Appl Microbiol. 2008;47(4):263–8. doi: 10.1111/j.1472-765X.2008.02420.x. [DOI] [PubMed] [Google Scholar]
- 62.Moody CS, Hassan HM. Mutagenicity of oxygen free radicals. Proc Natl Acad Sci U S A. 1982;79(9):2855–9. doi: 10.1073/pnas.79.9.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Giuliani F, et al. In vitro resistance selection studies of RLP068/Cl, a new Zn(II) phthalocyanine suitable for antimicrobial photodynamic therapy. Antimicrob Agents Chemother. 2010;54(2):637–42. doi: 10.1128/AAC.00603-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Amin RM, et al. Antimicrobial blue light inactivation of Pseudomonas aeruginosa by photoexcitation of endogenous porphyrins: In vitro and in vivo studies. Lasers Surg Med. 2016;48(5):562–8. doi: 10.1002/lsm.22474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y, et al. Antimicrobial blue light therapy for multidrug-resistant Acinetobacter baumannii infection in a mouse burn model: implications for prophylaxis and treatment of combat-related wound infections. J Infect Dis. 2014;209(12):1963–71. doi: 10.1093/infdis/jit842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tavares A, et al. Antimicrobial photodynamic therapy: study of bacterial recovery viability and potential development of resistance after treatment. Mar Drugs. 2010;8(1):91–105. doi: 10.3390/md8010091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cassidy CM, Donnelly RF, Tunney MM. Effect of sub-lethal challenge with Photodynamic Antimicrobial Chemotherapy (PACT) on the antibiotic susceptibility of clinical bacterial isolates. J Photochem Photobiol B. 2010;99(1):62–6. doi: 10.1016/j.jphotobiol.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 68.Pourhajibagher M, et al. Modulation of virulence in Acinetobacter baumannii cells surviving photodynamic treatment with toluidine blue. Photodiagnosis Photodyn Ther. 2016;15:202–12. doi: 10.1016/j.pdpdt.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 69.Pourhajibagher M, et al. Sub-lethal doses of photodynamic therapy affect biofilm formation ability and metabolic activity of Enterococcus faecalis. Photodiagnosis Photodyn Ther. 2016;15:159–66. doi: 10.1016/j.pdpdt.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 70.Kashef N, Akbarizare M, Kamrava SK. Effect of sub-lethal photodynamic inactivation on the antibiotic susceptibility and biofilm formation of clinical Staphylococcus aureus isolates. Photodiagnosis Photodyn Ther. 2013;10(4):368–73. doi: 10.1016/j.pdpdt.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 71.Holmberg SD, et al. Drug-resistant Salmonella from animals fed antimicrobials. N Engl J Med. 1984;311(10):617–22. doi: 10.1056/NEJM198409063111001. [DOI] [PubMed] [Google Scholar]
- 72.Ziegelhoffer EC, Donohue TJ. Bacterial responses to photo-oxidative stress. Nat Rev Microbiol. 2009;7(12):856–63. doi: 10.1038/nrmicro2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Philippova TO, et al. The antimicrobial properties of new synthetic porphyrins. Journal of Porphyrins and Phthalocyanines. 2003;7(11):755–760. [Google Scholar]
- 74.Tegos GP, et al. Protease-stable polycationic photosensitizer conjugates between polyethyleneimine and chlorin(e6) for broad-spectrum antimicrobial photoinactivation. Antimicrob Agents Chemother. 2006;50(4):1402–10. doi: 10.1128/AAC.50.4.1402-1410.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kashef N, Borghei YS, Djavid GE. Photodynamic effect of hypericin on the microorganisms and primary human fibroblasts. Photodiagnosis Photodyn Ther. 2013;10(2):150–5. doi: 10.1016/j.pdpdt.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 76.Ochsner UA, et al. Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF. J Bacteriol. 2000;182(16):4533–44. doi: 10.1128/jb.182.16.4533-4544.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Orlandi VT, et al. Pigments influence the tolerance of Pseudomonas aeruginosa PAO1 to photodynamically induced oxidative stress. Microbiology. 2015;161(12):2298–309. doi: 10.1099/mic.0.000193. [DOI] [PubMed] [Google Scholar]
- 78.Sandmann G. Carotenoid biosynthesis in microorganisms and plants. Eur J Biochem. 1994;223(1):7–24. doi: 10.1111/j.1432-1033.1994.tb18961.x. [DOI] [PubMed] [Google Scholar]
- 79.Sistrom WR, Griffiths M, Stanier RY. The biology of photosynthetic bacterium which lacks colored carotenoids. J Cell Comp Physiol. 1956;48(3):473–515. doi: 10.1002/jcp.1030480309. [DOI] [PubMed] [Google Scholar]
- 80.Anderson SM, Krinsky NI. Protective action of carotenoid pigments against photodynamic damage to liposomes. Photochem Photobiol. 1973;18(5):403–8. doi: 10.1111/j.1751-1097.1973.tb06440.x. [DOI] [PubMed] [Google Scholar]
- 81.Pelz A, et al. Structure and biosynthesis of staphyloxanthin from Staphylococcus aureus. J Biol Chem. 2005;280(37):32493–8. doi: 10.1074/jbc.M505070200. [DOI] [PubMed] [Google Scholar]
- 82.Clauditz A, et al. Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infect Immun. 2006;74(8):4950–3. doi: 10.1128/IAI.00204-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu GY, et al. Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J Exp Med. 2005;202(2):209–15. doi: 10.1084/jem.20050846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Subczynski WK, Markowska E, Sielewiesiuk J. Effect of polar carotenoids on the oxygen diffusion-concentration product in lipid bilayers. An EPR spin label study. Biochim Biophys Acta. 1991;1068(1):68–72. doi: 10.1016/0005-2736(91)90061-c. [DOI] [PubMed] [Google Scholar]
- 85.Gad F, et al. Effects of growth phase and extracellular slime on photodynamic inactivation of gram-positive pathogenic bacteria. Antimicrob Agents Chemother. 2004;48(6):2173–8. doi: 10.1128/AAC.48.6.2173-2178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tegos GP, Hamblin MR. Phenothiazinium antimicrobial photosensitizers are substrates of bacterial multidrug resistance pumps. Antimicrob Agents Chemother. 2006;50(1):196–203. doi: 10.1128/AAC.50.1.196-203.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Van Bambeke F, et al. Antibiotic efflux pumps in prokaryotic cells: occurrence, impact on resistance and strategies for the future of antimicrobial therapy. J Antimicrob Chemother. 2003;51(5):1055–65. doi: 10.1093/jac/dkg224. [DOI] [PubMed] [Google Scholar]
- 88.Czarnecki DJ, Feider HK, Splittgerber GF. Toluidine blue dye as a breast localization marker. AJR Am J Roentgenol. 1989;153(2):261–3. doi: 10.2214/ajr.153.2.261. [DOI] [PubMed] [Google Scholar]
- 89.Clifton J, 2nd, Leikin JB. Methylene blue. Am J Ther. 2003;10(4):289–91. doi: 10.1097/00045391-200307000-00009. [DOI] [PubMed] [Google Scholar]
- 90.Schirmer RH, et al. Methylene blue as an antimalarial agent. Redox Rep. 2003;8(5):272–5. doi: 10.1179/135100003225002899. [DOI] [PubMed] [Google Scholar]
- 91.Tegos GP, et al. Inhibitors of bacterial multidrug efflux pumps potentiate antimicrobial photoinactivation. Antimicrob Agents Chemother. 2008;52(9):3202–9. doi: 10.1128/AAC.00006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kishen A, et al. Efflux pump inhibitor potentiates antimicrobial photodynamic inactivation of Enterococcus faecalis biofilm. Photochem Photobiol. 2010;86(6):1343–9. doi: 10.1111/j.1751-1097.2010.00792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Grinholc M, et al. Evaluation of the role of the pharmacological inhibition of Staphylococcus aureus multidrug resistance pumps and the variable levels of the uptake of the sensitizer in the strain-dependent response of Staphylococcus aureus to PPArg(2)-based photodynamic inactivation. Photochem Photobiol. 2010;86(5):1118–26. doi: 10.1111/j.1751-1097.2010.00772.x. [DOI] [PubMed] [Google Scholar]
- 94.Grinholc M, et al. Bactericidal effect of photodynamic inactivation against methicillin-resistant and methicillin-susceptible Staphylococcus aureus is strain-dependent. J Photochem Photobiol B. 2008;90(1):57–63. doi: 10.1016/j.jphotobiol.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 95.Grinholc M, et al. Multiresistant strains are as susceptible to photodynamic inactivation as their naive counterparts: protoporphyrin IX-mediated photoinactivation reveals differences between methicillin-resistant and methicillin-sensitive Staphylococcus aureus strains. Photomed Laser Surg. 2014;32(3):121–9. doi: 10.1089/pho.2013.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tang HM, Hamblin MR, Yow CM. A comparative in vitro photoinactivation study of clinical isolates of multidrug-resistant pathogens. J Infect Chemother. 2007;13(2):87–91. doi: 10.1007/s10156-006-0501-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nakonieczna J, et al. Superoxide dismutase is upregulated in Staphylococcus aureus following protoporphyrin-mediated photodynamic inactivation and does not directly influence the response to photodynamic treatment. BMC Microbiol. 2010;10:323. doi: 10.1186/1471-2180-10-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Novick RP. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol. 2003;48(6):1429–49. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- 99.Dunman PM, et al. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J Bacteriol. 2001;183(24):7341–53. doi: 10.1128/JB.183.24.7341-7353.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Park HJ, et al. Agr function is upregulated by photodynamic therapy for Staphylococcus aureus and is related to resistance to photodynamic therapy. Microbiol Immunol. 2013;57(8):547–52. doi: 10.1111/1348-0421.12070. [DOI] [PubMed] [Google Scholar]
- 101.Janzon L, Arvidson S. The role of the delta-lysin gene (hld) in the regulation of virulence genes by the accessory gene regulator (agr) in Staphylococcus aureus. EMBO J. 1990;9(5):1391–9. doi: 10.1002/j.1460-2075.1990.tb08254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Novick RP, et al. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12(10):3967–75. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Giraudo AT, et al. Studies on the expression of regulatory locus sae in Staphylococcus aureus. Curr Microbiol. 2003;46(4):246–50. doi: 10.1007/s00284-002-3853-z. [DOI] [PubMed] [Google Scholar]
- 104.Baba T, et al. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J Bacteriol. 2008;190(1):300–10. doi: 10.1128/JB.01000-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Herbert S, et al. Repair of global regulators in Staphylococcus aureus 8325 and comparative analysis with other clinical isolates. Infect Immun. 2010;78(6):2877–89. doi: 10.1128/IAI.00088-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gandara L, et al. Sae regulator factor impairs the response to photodynamic inactivation mediated by Toluidine blue in Staphylococcus aureus. Photodiagnosis Photodyn Ther. 2016;16:136–141. doi: 10.1016/j.pdpdt.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 107.Grinholc M, et al. The connection between agr and SCCmec elements of Staphylococcus aureus strains and their response to photodynamic inactivation. Photomed Laser Surg. 2011;29(6):413–9. doi: 10.1089/pho.2010.2854. [DOI] [PubMed] [Google Scholar]
- 108.Horvath I, et al. Membrane-associated stress proteins: more than simply chaperones. Biochim Biophys Acta. 2008;1778(7–8):1653–64. doi: 10.1016/j.bbamem.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 109.Schlesinger MJ. Heat shock proteins. J Biol Chem. 1990;265(21):12111–4. [PubMed] [Google Scholar]
- 110.Torok Z, et al. Evidence for a lipochaperonin: association of active protein-folding GroESL oligomers with lipids can stabilize membranes under heat shock conditions. Proc Natl Acad Sci U S A. 1997;94(6):2192–7. doi: 10.1073/pnas.94.6.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Arsene F, Tomoyasu T, Bukau B. The heat shock response of Escherichia coli. Int J Food Microbiol. 2000;55(1–3):3–9. doi: 10.1016/s0168-1605(00)00206-3. [DOI] [PubMed] [Google Scholar]
- 112.Fredriksson A, et al. Defense against protein carbonylation by DnaK/DnaJ and proteases of the heat shock regulon. J Bacteriol. 2005;187(12):4207–13. doi: 10.1128/JB.187.12.4207-4213.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Laport MS, et al. Transcriptional analysis of the groE and dnaK heat-shock operons of Enterococcus faecalis. Res Microbiol. 2004;155(4):252–8. doi: 10.1016/j.resmic.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 114.Singh VK, et al. Role for dnaK locus in tolerance of multiple stresses in Staphylococcus aureus. Microbiology. 2007;153(Pt 9):3162–73. doi: 10.1099/mic.0.2007/009506-0. [DOI] [PubMed] [Google Scholar]
- 115.Bolean M, et al. Photodynamic therapy with rose bengal induces GroEL expression in Streptococcus mutans. Photomed Laser Surg. 2010;28(Suppl 1):S79–84. doi: 10.1089/pho.2009.2635. [DOI] [PubMed] [Google Scholar]
- 116.St Denis TG, et al. Analysis of the bacterial heat shock response to photodynamic therapymediated oxidative stress. Photochem Photobiol. 2011;87(3):707–13. doi: 10.1111/j.1751-1097.2011.00902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.de la Fuente-Nunez C, et al. Bacterial biofilm development as a multicellular adaptation: antibiotic resistance and new therapeutic strategies. Curr Opin Microbiol. 2013;16(5):580–9. doi: 10.1016/j.mib.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 118.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2(2):95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 119.Resch A, et al. Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Appl Environ Microbiol. 2005;71(5):2663–76. doi: 10.1128/AEM.71.5.2663-2676.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ehrlich GD, Arciola CR. From Koch's postulates to biofilm theory. The lesson of Bill Costerton. Int J Artif Organs. 2012;35(10):695–9. doi: 10.5301/ijao.5000169. [DOI] [PubMed] [Google Scholar]
- 121.Hoiby N, et al. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents. 2010;35(4):322–32. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 122.Stewart PS, Costerton JW. Antibiotic resistance of bacteria in biofilms. Lancet. 2001;358(9276):135–8. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 123.Kashef N, Karami S, Djavid GE. Phototoxic effect of hypericin alone and in combination with acetylcysteine on Staphylococcus aureus biofilms. Photodiagnosis Photodyn Ther. 2015;12(2):186–92. doi: 10.1016/j.pdpdt.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 124.Demidova TN, Hamblin MR. Effect of cell-photosensitizer binding and cell density on microbial photoinactivation. Antimicrob. Agents Chemother. 2005;49(6):2329–2335. doi: 10.1128/AAC.49.6.2329-2335.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Usacheva MN, Teichert MC, Biel MA. Comparison of the methylene blue and toluidine blue photobactericidal efficacy against gram-positive and gram-negative microorganisms. Lasers Surg Med. 2001;29(2):165–73. doi: 10.1002/lsm.1105. [DOI] [PubMed] [Google Scholar]
- 126.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284(5418):1318–22. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 127.Zanin IC, et al. Photosensitization of in vitro biofilms by toluidine blue O combined with a lightemitting diode. Eur J Oral Sci. 2006;114(1):64–9. doi: 10.1111/j.1600-0722.2006.00263.x. [DOI] [PubMed] [Google Scholar]
- 128.Aoshima H, et al. Antimicrobial activity of fullerenes and their hydroxylated derivatives. Biocontrol Sci. 2009;14(2):69–72. doi: 10.4265/bio.14.69. [DOI] [PubMed] [Google Scholar]
- 129.Chen CP, Chen CT, Tsai T. Chitosan nanoparticles for antimicrobial photodynamic inactivation: characterization and in vitro investigation. Photochem Photobiol. 2012;88(3):570–6. doi: 10.1111/j.1751-1097.2012.01101.x. [DOI] [PubMed] [Google Scholar]
- 130.Nafee N, et al. Antibiotic-free nanotherapeutics: hypericin nanoparticles thereof for improved in vitro and in vivo antimicrobial photodynamic therapy and wound healing. Int J Pharm. 2013;454(1):249–58. doi: 10.1016/j.ijpharm.2013.06.067. [DOI] [PubMed] [Google Scholar]
- 131.Shrestha A, Kishen A. The effect of tissue inhibitors on the antibacterial activity of chitosan nanoparticles and photodynamic therapy. J Endod. 2012;38(9):1275–8. doi: 10.1016/j.joen.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 132.Shrestha A, Kishen A. Antibacterial efficacy of photosensitizer functionalized biopolymeric nanoparticles in the presence of tissue inhibitors in root canal. J Endod. 2014;40(4):566–70. doi: 10.1016/j.joen.2013.09.013. [DOI] [PubMed] [Google Scholar]
- 133.Tawfik AA, Alsharnoubi J, Morsy M. Photodynamic antibacterial enhanced effect of methylene blue-gold nanoparticles conjugate on Staphylococcal aureus isolated from impetigo lesions in vitro study. Photodiagnosis Photodyn Ther. 2015;12(2):215–20. doi: 10.1016/j.pdpdt.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 134.Darabpour E, Kashef N, Mashayekhan S. Chitosan nanoparticles enhance the efficiency of methylene blue-mediated antimicrobial photodynamic inactivation of bacterial biofilms: An in vitro study. Photodiagnosis Photodyn Ther. 2016;14:211–7. doi: 10.1016/j.pdpdt.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 135.Misba L, Kulshrestha S, Khan AU. Antibiofilm action of a toluidine blue O-silver nanoparticle conjugate on Streptococcus mutans: a mechanism of type I photodynamic therapy. Biofouling. 2016;32(3):313–28. doi: 10.1080/08927014.2016.1141899. [DOI] [PubMed] [Google Scholar]
- 136.Sherwani MA, et al. Gold Nanoparticle-Photosensitizer Conjugate Based Photodynamic Inactivation of Biofilm Producing Cells: Potential for Treatment of C. albicans Infection in BALB/c Mice. PLoS One. 2015;10(7):e0131684. doi: 10.1371/journal.pone.0131684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sadasivam M, et al. Self-assembled liposomal nanoparticles in photodynamic therapy. Eur J Nanomed. 2013;5(3) doi: 10.1515/ejnm-2013-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chen RJ, et al. Formation of gold decorated porphyrin nanoparticles and evaluation of their photothermal and photodynamic activity. Mater Sci Eng C Mater Biol Appl. 2016;63:678–85. doi: 10.1016/j.msec.2016.03.034. [DOI] [PubMed] [Google Scholar]
- 139.Huang YY, et al. Can nanotechnology potentiate photodynamic therapy? Nanotechnol Rev. 2012;1(2):111–146. doi: 10.1515/ntrev-2011-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Dai T, et al. Chitosan preparations for wounds and burns: antimicrobial and wound-healing effects. Expert Rev Anti-Infect Ther. 2011;9(7):857–79. doi: 10.1586/eri.11.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Pagonis TC, et al. Nanoparticle-based endodontic antimicrobial photodynamic therapy. J Endod. 2010;36(2):322–8. doi: 10.1016/j.joen.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Darabpour E, et al. Fast and effective photodynamic inactivation of 4-day-old biofilm of methicillin-resistant Staphylococcus aureus using methylene blue-conjugated gold nanoparticles. Journal of Drug Delivery Science and Technology. 2017;37:134–140. [Google Scholar]
- 143.Vecchio D, et al. Bacterial photodynamic inactivation mediated by methylene blue and red light is enhanced by synergistic effect of potassium iodide. Antimicrob Agents Chemother. 2015;59(9):5203–12. doi: 10.1128/AAC.00019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.St Denis TG, et al. Thiocyanate potentiates antimicrobial photodynamic therapy: In situ generation of the sulfur trioxide radical anion by singlet oxygen. Free Radic Biol Med. 2013;65C:800–810. doi: 10.1016/j.freeradbiomed.2013.08.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Huang L, et al. Paradoxical potentiation of methylene blue-mediated antimicrobial photodynamic inactivation by sodium azide: role of ambient oxygen and azide radicals. Free Radic Biol Med. 2012;53(11):2062–71. doi: 10.1016/j.freeradbiomed.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Huang L, et al. Potassium iodide potentiates broad-spectrum antimicrobial photodynamic inactivation using Photofrin. ACS Infect Dis. 2017 doi: 10.1021/acsinfecdis.7b00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Huang YY, et al. Broad-Spectrum Antimicrobial Effects of Photocatalysis Using Titanium Dioxide Nanoparticles Are Strongly Potentiated by Addition of Potassium Iodide. Antimicrob Agents Chemother. 2016;60(9):5445–53. doi: 10.1128/AAC.00980-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wu X, et al. Broad-spectrum antimicrobial photocatalysis mediated by titanium dioxide and UVA is potentiated by addition of bromide ion via formation of hypobromite. Free Radic Biol Med. 2016;95:74–81. doi: 10.1016/j.freeradbiomed.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhang Y, et al. Potentiation of antimicrobial photodynamic inactivation mediated by a cationic fullerene by added iodide: in vitro and in vivo studies. Nanomedicine (Lond) 2015;10(4):603–14. doi: 10.2217/nnm.14.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Freire F, et al. Photodynamic therapy of oral Candida infection in a mouse model. J Photochem Photobiol B. 2016;159:161–168. doi: 10.1016/j.jphotobiol.2016.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kharkwal GB, et al. Photodynamic therapy for infections: Clinical applications. Lasers Surg Med. 2011;43(7):755–67. doi: 10.1002/lsm.21080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Dai T, Huang YY, Hamblin MR. Photodynamic therapy for localized infections-State of the art. Photodiagnosis Photodyn Ther. 2009;6(3–4):170–188. doi: 10.1016/j.pdpdt.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]