Abstract
Background
There is now increasing evidence that asthma and atopy originate in part in utero, with disease risk being associated with the altered epigenetic regulation of genes.
Objective and Methods
To determine the relationship between variations in DNA methylation at birth and the development of allergic disease, we examined the methylation status of CpG loci within the promoter regions of Th1/2 lineage commitment genes (GATA3, IL-4, IL-4R, STAT4 and TBET) in umbilical cord DNA at birth in a cohort of infants from the Southampton Women’s Survey (n=696) who were later assessed for asthma, atopic eczema and atopy.
Results
We found that higher methylation of GATA3 CpGs -2211/-2209 at birth was associated with a reduced risk of asthma at ages 3 (median ratio [median methylation in asthma group/median methylation in non-asthma group] =0.74, p=0.006) and 6 (median ratio 0.90, p=0.048) years. Furthermore, we demonstrated that the GATA3 CpG loci associated with later risk of asthma lie within a NF-κB binding site, and that methylation here blocks transcription factor binding to the GATA3 promoter in the Human Jurkat T cell line. Associations between umbilical cord methylation of CpG loci within IL-4R with atopic eczema at 12 months (median ratio 1.02, p=0.028), and TBET with atopy (median ratio 0.98, p=0.017) at 6-7 years of age were also observed.
Conclusions and Clinical Relevance
Our findings provides further evidence of a developmental contribution to the risk of later allergic disorders, and suggests that involvement of epigenetic mechanisms in childhood asthma is already demonstrable at birth.
Keywords: Methylation, Th1/Th2, GATA3, TBET, biomarkers
1. Introduction
Asthma, atopic eczema and other allergic conditions are complex disorders associated with a variety of environmental factors. They are among some of the most prevalent chronic conditions affecting children in economically developed nations and are becoming increasingly prevalent in developing countries (1–3). Allergic asthma is characterized by Th2-type inflammation which results in bronchial hyperresponsiveness, airway obstruction and tissue remodeling4,5. Effective type-2 immune responses are generated by type-2 helper CD4+ T cells (Th2) as well as type-2 innate effector cells. Naïve T cells differentiate into Th1, Th2, Th17 or inducible Tregs, depending upon the cytokine environment(4–6). IFN-γ and IL-12 induce the differentiation of naïve T cells into Th1 cells, while IL-4 promotes Th2 differentiation. IFNγ acting through STAT1 promotes transcription of T-bet, a Th1-specific T box transcription factor, while IL-12, via STAT 3/4 maintain TBET expression. TBET initiates Th1 lineage commitment from naive T cells through the chromatin remodelling of the IFNγ locus, upregulation of IL-12R and by repressing Th2 related genes(7, 8). In contrast, IL-4 acts via STAT6 to activate the transcription factor GATA3, which is critical for Th2 differentiation9. In Th2 cells, GATA3 inhibits IFN-γ expression, and activates the expression of the Th2 related cytokines IL-4, IL-5 and IL-13(9, 10). DNA methylation plays a key role in the maintenance of these different T cell lineages, with gene specific methylation being well documented within IL-4, IFNγ and GATA3 in the Th cell lineage where these genes are transcriptionally silenced(11).
Differences in the immune response are detectable at birth, suggesting that in utero environmental exposures have the capacity to modify the set point of the immune system at birth(12). The differentiation of T cells plays an important role in preventing fetal rejection during pregnancy, resulting in a skewing to T helper (Th)2 immune responses and suppression of Th1-mediated immunity. This bias against proinflammatory cytokines leaves the newborn susceptible to microbial infection(13) and therefore development of optimal regulation of gene expression is required to obtain a balanced postnatal immune response. Immaturity in Th1 as well as Treg activity may increase the risk of an inappropriate persistence of a Th2 allergy-prone immune phenotype(14). Consistent with a Th1/Th2 imbalance being associated with an increased asthma risk, a significant increase in GATA3 expression has been found in the airways of asthmatics compared with those of controls(15). Several single nucleotide polymorphisms (SNP’s) in the GATA3 promoter and exons of GATA3 have been associated with atopic eczema and asthma-related phenotypes including high IgE(16, 17).
There is growing evidence that childhood asthma and atopy together with a number of other chronic diseases may originate at least in part in utero(18). Epidemiological studies have found associations between prenatal nutritional, allergenic and toxicant exposures and an increased susceptibility to atopic disorders(19, 20). Early life environmental exposure may alter disease risk through epigenetic processes, such as DNA methylation, which induce heritable changes in gene expression without a change in gene sequence(21). For example, maternal exposure to polycyclic aromatic hydrocarbons was associated with the increased methylation of the CpG island of acyl-coA synthetase long chain family member 3 (ACDL3) and the development of asthma symptoms in children under 5 years of age(22). A meta-analysis across 13 cohorts has also reported an association between maternal smoking and newborn blood DNA methylation at over 6000 CpG sites (23), while Gruzieva,O et al., (24) has reported an association between NO2 exposure during pregnancy and altered methylation of 3 CpG sites within mitochrondria-related genes in the cord blood of newborn infants. Furthermore, DNA methylation of the interleukin-4 receptor gene has been associated with an increased incidence of asthma at age 18 years(25), although whether this methylation change was induced by an early life environmental exposure is not known.
Although DNA methylation patterns are often tissue specific, a number of studies have shown inter-tissue methylation correlations(26, 27). Thus, in human fetal tissues at birth, the targeted measurement of the methylation of genes involved in determining allergic outcome, such as those involved in the Th1/Th2 differentiation pathway, may provide an indicator of environmental influences affecting early development and may permit the identification of individuals at increased risk of asthma, atopic eczema and atopy in childhood. In this study, to determine the relationship between variations in DNA methylation at birth and the development of allergic disease, we measured the methylation status of CpG loci within highly conserved regulatory regions of genes involved in Th1/2 lineage commitment (GATA3, IL-4, IL-4R, STAT4, and TBET) in umbilical cord DNA obtained at birth from a cohort of children who were later assessed for asthma, atopic eczema and atopy.
2. Methods
2.1. Southampton Women’s Survey
In the Southampton Women’s Survey (SWS), Southampton, UK, we studied a prospective cohort of women aged 20-34 years. Women who are not pregnant were recruited; those who subsequently conceived were followed through pregnancy and their offspring through infancy and childhood (28). The umbilical cord was frozen at birth and stored at -70°C.
Outcome assessments were as follows: Research nurses examined the child’s skin, looking for evidence of visible atopic eczema at ages 12 months and 6-7 years. Mothers were also asked whether the child had dry skin or an itchy skin condition since birth. Case definition of atopic eczema was based on the UK Working Party diagnostic criteria for the definition of atopic eczema(29); we omitted a history of asthma or hay fever as a criterion at 12 months because the children were too young to have developed these disorders in infancy. At 3 and 6-7 years, the research nurses asked the mothers whether a doctor had ever diagnosed “asthma” in their child (International Study of Asthma and Allergy in Childhood questionnaire). A positive response was considered evidence of doctor-diagnosed asthma. Atopy was defined using skin prick testing as a response of ≥ 3 mm to cat, dog, house dust mite (Dermatophagoides pteronyssinus), grass pollens, egg and milk allergens (Hollister-Stier, Spokane, WA) at age 3 years. Tree pollen was added (ALK Abelló Hørsholm, Denmark) at 6-7 years. Valid readings were those with appropriate positive and negative control responses. Cohort characteristics are shown in Table 1.
Table 1.
Table showing cohort characteristics of the participants in the smaller and extended sample sets.
| Characteristic | % or mean (standard deviation) for smaller sample (n=366) | % or mean (standard deviation) for extended sample (n=696) |
|---|---|---|
| Maternal age at child's birth | 30.5 (3.6) | 30.8 (3.6) |
| Maternal pre-pregnancy BMI | 25.3 (4.4) | 25.2 (4.4) |
| Maternal smoking during pregnancy | 12.9% | 12.3% |
| Maternal atopy | 43.6% | 42.9% |
| Female child | 47.0% | 47.3% |
| Birthweight in grams | 3463 (489) | 3500 (501) |
| Ever breastfed | 86.6% | 85.6% |
| Atopic eczema at 12 months | 12.6% (cases=46) | 11.3% (cases=76) |
| Doctor diagnosed asthma at 3 yrs | 5.8% (cases=21) | 5.9% (cases=34) |
| Atopy at 3 yrs | 16.0% (cases=48) | 16.3% (cases=76) |
| Atopic eczema at 6-7 yrs | 16.8% (cases=53) | 16.3% (cases=102) |
| Doctor diagnosed asthma at 6-7 yrs | 12.9% (cases=47) | 14.3% (cases=86) |
| Atopy at 6-7 yrs | 22.6% (cases=60) | 24.0% (cases=104) |
2.2. Ethics statement
The study was conducted according to the guidelines in the Declaration of Helsinki, and the Southampton and South West Hampshire Research Ethics Committee approved all procedures (276/97, 307/97, 089/99, and 06/Q1702/104). Written informed consent was obtained from all participating women and by parents or guardians with parental responsibility on behalf of their children.
2.3. Methylation analysis
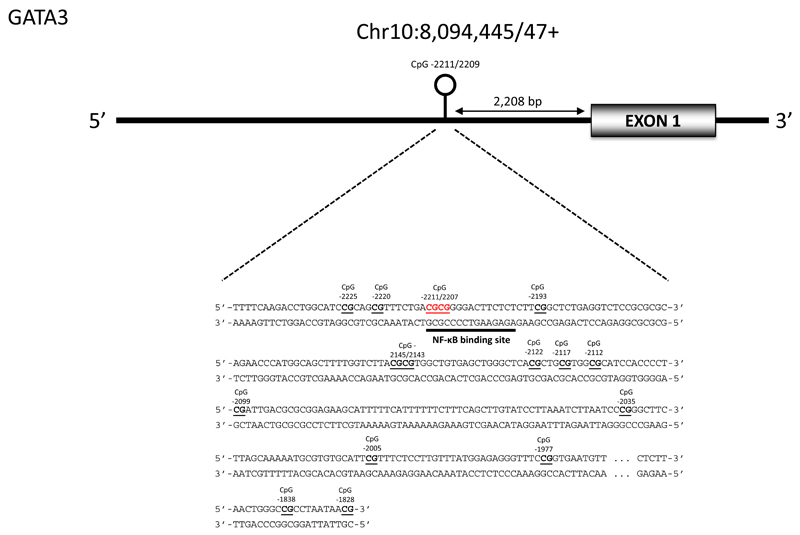
The methylation of CpG loci within GATA binding protein 3 (GATA3), Interleukin-4 (IL-4), Interleukin 4 receptor (IL-4R), signal transducer and activator of transcription 4 (STAT4), and T-box transcription factor (TBET); genes that are known to play key roles in maintaining Th1/Th2 balance, were analysed in umbilical cord DNA from SWS participants. Sequencing assays were designed to cover the key regulatory regions within these genes that had been shown previously to be differentially methylated during T cell differentiation(11, 30) or associated with asthma incidence(25) (Figure 1A, Supplementary Figure 1, Supplementary Table 1). Sodium bisulfite pyrosequencing was used to measure DNA methylation in IL-4, IL-4R, STAT4 and TBET but because of the sequence context of the CpGs within GATA3, Seqeunom EpiTYPER MassARRAY analysis was used to analyse the methylation status of GATA3. The location of the regions analysed and sequence of primers used are shown in Supplementary Table 2 (Pyrosequencing amplicons) and Supplementary Table 3 (GATA3 Sequenom amplicon), together with the methylation ranges for IL-4, IL-4R, TBET and STAT4 (Supplementary Table 4) and for GATA3 (Supplementary Table 5). The methods used for pyrosequencing and Sequenom analysis are described in the Supplementary Information. Information on co-methylation of sites is shown using a plot similar to a coMET plot (31) in Supplementary Figure 2.
Figure 1.
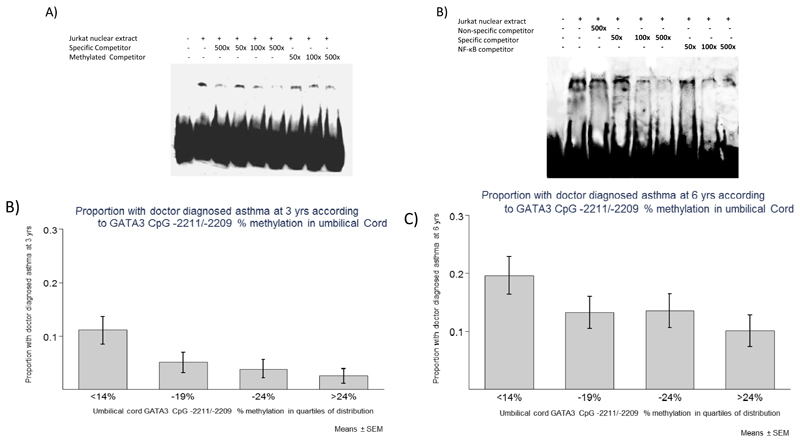
A) Schematic diagram showing the promoter region of GATA3 and the location of the CpG sites measured. B/C) Methylation of GATA3 CpGs -2211/-2209 in umbilical cord at birth are associated with doctor diagnosed asthma incidence at 3 (B) and 6-7(C) years of age. Methylation has been divided into 4 equal groups according to rank; means and standard error means are plotted for each group.
2.6. Statistical analysis
Statistical analysis was carried out using Stata™ 12 (StataCorp, Texas). Distributions of methylation percentages at CpG sites were investigated (Supplementary Tables 4, 5). CpG sites were excluded from analysis if the median methylation was not ≥ 5% or if the difference between the 5th and 95th percentiles was ≤10%. All CpG sites fulfilled these criteria. The distributions of methylation measurements for many of the CpG’s were skewed, so non-parametric tests were used. Associations between cord CpG methylation and asthma and allergy outcomes (doctor diagnosed asthma at 3 and 6-7 years, atopic eczema at 6-7 years and atopy at 6-7 years) were investigated. The difference between the central location (i.e. median) of each group (allergic/non-allergic) was tested using Mann-Whitney tests and the results presented as the median ratio (median CpG methylation in allergic group/median CpG methylation in non-allergic group) with the corresponding Mann-Whitney p value. Where the median ratio is less than 1 this indicates a protective effect of higher CpG methylation on the allergic outcome. When CpG sites were significantly associated with asthma and allergy phenotypes, logistic regression was used to derive odds ratios after adjusting for appropriate covariates (child’s sex, maternal smoking during pregnancy and a batch effect for methylation analysis). The odds ratio describes the multiplicative change in odds for a 1% increase in methylation, all other variables in the model remaining constant. An odds ratio of less than 1 indicates a protective effect of higher CpG methylation on the allergic outcome. Where adjusted logistic regressions indicated a significant association between CpG methylation and allergic outcome an additional analysis was undertaken to determine the influence of outliers on the regression coefficients using Pregibon’s dbeta measure (32).P values are presented uncorrected for multiple testing. A p value of <0.05 was considered significant.
2.7. Electrophoretic mobility shift assays
To examine transcription factor binding at the CpG sites of interest we undertook electrophoretic mobility shift assays (see Supplementary Information for Methods).
3. Results
3.1. Cohort characteristics
The DNA methylation of GATA3 was measured in the umbilical cord of 696 children from the Southampton Women’s Survey (SWS) cohort (28). Due to limited resources and DNA availability, the DNA methylation of the other candidate genes was measured in the umbilical cord of 366 children, a subset of the 696 children. In the subset of 366 offspring, 13% of the mothers smoked during pregnancy, mean maternal age at the child’s birth was 30.5 years and pre-pregnancy body mass index was 25.3 kg/m2 (Table 1). 47% of the infants were female; mean birth weight was 3463 g. 46 infants (13%) fulfilled the criteria for atopic eczema in infancy, and 53 children (15%) at age 6-7 years. At age 3 years, the parents of 21 participants (6%) reported that a doctor had diagnosed asthma in their child; the number of diagnoses of asthma increased to 47 (13%) at age 6-7 years. Supplementary Table 10 shows further information on the children in whom the methylation status of IL4-R and IL4, TBET and STAT4 was measured. The cohort characteristics for all samples sets were very similar to the main cohort. Maternal smoking was slightly lower in prevalence in the IL4-R and IL4, TBET and STAT4 sample sets, as was a doctor diagnosis of asthma at age 3 years. The prevalences of atopy at 3 years and atopic eczema at 6-7 years were slightly higher in the IL4-R sample set, but these differences were not significant (p=0.23, p=0.73, p=0.43 and p=0.41 respectively).
3.2. Association between umbilical cord DNA methylation at birth and doctor diagnosed asthma at age 3 years and 6-7 years
The methylation of 35 CpG loci in the regulatory regions of Th1/Th2 candidate genes was measured at birth in umbilical cord in relation to doctor diagnosed asthma at age 3 and 6-7 years (Supplementary Table 7). The CpG sites measured in this study were selected as they were located within known regulatory regions which had previously been reported to be important for the expression of the gene during Th1/Th2 differentiation(11, 30) or in the case of IL-4R, previously associated with asthma incidence(25)(Supplementary Figure 1 and Supplementary Table 1). For all CpGs examined, the presence of SNPs was excluded by direct sequencing. For GATA3, 16 CpGs loci, located 2.5 kb upstream of the proximal promoter were measured (Figure 1A). This region has been shown to be highly conserved amongst species(33) and contains several Notch and GATA3 binding sites which have been shown to be critical for GATA3 induction during Th2 differentiation(33, 34). We found that higher methylation of umbilical cord GATA3 CpGs -2211/-2209bp were associated with a decreased risk of asthma at age 3 years (cases=31, controls=500, median ratio 0.74, p=0.006) and at 6-7 years (cases=79, controls=475, median ratio 0.90, p=0.048) (Table 2). As we found significant associations between asthma at 3 and 6-7 years and GATA3 CpGs, we then used logistic regression to allow adjustment for appropriate covariates. Logistic regression with asthma at age 3 years as the outcome, GATA3 CpGs -2211/-2209bp as the predictor, and adjusting for child’s sex, maternal smoking during pregnancy and a batch effect for methylation analysis, showed an odds ratio for asthma of 0.940 (95%CI 0.895 to 0.987, cases=31, controls=498, p=0.012). Logistic regression with asthma at age 6-7 years adjusting for the same covariates, showed an odds ratio for asthma of 0.976 (95% CI 0.949 to 1.003, cases=78, controls=474 p=0.077).. There was also an association between higher methylation of IL-4R CpG +28239 (Supplementary Figure 1C) (0.98, cases=15, controls=85, p=0.029) and a lower incidence of asthma at 6-7 years, although this association was not seen at 3 years (Table 2, Supplementary Table 7) and was lost at age 6-7 years after adjusting for the covariates listed above.
Table 2.
Significant associations between the methylation of specific CpG sites in umbilical cord at birth with doctor diagnosed asthma at ages 3 and 6-7 years (* denotes p<0.05).
| CpG | Genomic coordinates | Median ratio at age 3 years (yes/no)(N cases/ N controls) |
P value |
|---|---|---|---|
| GATA3 CpG -2211/-2209 | chr10:8134451,53+ | 0.737 (31/500) | 0.006* |
| Median ratio at age 6-7 years (yes/no)(N cases/ N controls) |
|||
| GATA3 CpG -2211/-2209 | chr10:8134451,53+ | 0.895 (79/475) | 0.048* |
| IL-4R CpG +28239 | chr16:27260974+ | 0.98 (15/85) | 0.029* |
3.3. Association between DNA methylation at birth and childhood atopic eczema
Higher methylation of GATA3 CpGs -2145/-2143bp were associated with a higher incidence of atopic eczema at 12 months (median ratio 1.20, cases=67, controls=575, p=0.05), but this association was attenuated after adjusting for child’s sex, maternal smoking during pregnancy and a batch effect for methylation analysis (cases=67, controls=573, p=0.083). There was no association between the methylation of these CpGs and atopic eczema at age 6-7 years (Table 3, Supplementary Table 8). An association was observed between the methylation of IL-4R CpG +28239 and atopic eczema at age 12 months (median ratio 1.02, cases=13, controls=87, p=0.028), and there were borderline associations with methylation of IL-4R CpG +28269 bp (1.03, cases=13, controls=81, p=0.059) and STAT4 CpG -229 (Supplementary Figure 1B) (0.981, cases=19, controls=154, p=0.067). At 6-7 years, higher methylation of IL-4 CpG -9664bp (Supplementary Figure 1A) was, associated with an increased risk of atopic eczema (1.02, cases=15, controls=104, p=0.0498) (Table 3, Supplementary Table 8).
Table 3.
Significant associations found between the methylation of specific CpG sites in umbilical cord at birth with atopic eczema at 12 months of age (* denotes p<0.05).
| CpG | Genomic coordinate | Median ratio at age 12 months (yes/no) (N cases/ N controls) | P value |
|---|---|---|---|
| GATA3 CpG -2145/-2143 | chr10:8134517,19+ | 1.20 (67/575) | 0.054 |
| IL-4R CpG +28239 | chr16:27260974+ | 1.02 (13/87) | 0.028* |
| IL-4R CpG +28269 | chr16:27261004+ | 1.03 (13/81) | 0.059 |
| STAT4 CpG -229 | chr2:191724460- | 0.981 (19/154) | 0.067 |
| Median ratio at age 6-7 years (yes/no) | |||
| IL-4 CpG -9664 | chr5:132027913+ | 1.02 (15/104) | 0.0498* |
3.4. Association between DNA methylation at birth and atopy at 6-7 years
Higher methylation of TBET CpG -18424 (Supplementary Figure 1D), which is located within the upstream enhancer of TBET(35), was associated with a decreased risk of atopy at 6-7 years (0.98, cases=37, controls=104, p=0.017) (Table 4, Supplementary Table 9). Logistic regression with atopy at 6-7 years as the outcome, TBET CpG -18424 as the predictor, and adjusting for child’s sex, maternal smoking during pregnancy and a batch effect for methylation analysis, showed an odds ratio for atopy of 0.883 (95% CI 0.781 to 0.997, cases=35, controls=103, p=0.045). Methylation at the other CpG sites measured was not associated with atopy at age 6-7 years.
Table 4.
Significant associations were found between the methylation of specific CpG sites in umbilical cord at birth with atopy at 6-7 years of age (* denotes p<0.05).
| CpG | Genomic coordinate | Median ratio at age 6-7 years (yes/no) (N cases/ N controls) | P value |
|---|---|---|---|
| TBET CpG -18424 | chr17:43147185+ | 0.98 (37/104) | 0.017* |
3.5. Functional significance of altered GATA3 CpG3 and 4 methylation
To determine the functional consequences of CpG -2211 and -2209 methylation within the GATA3 promoter, we examined transcription factor binding to the promoter region of GATA3 using electrophoretic mobility shift assays on nuclear extracts from the human T cell line Jurkat. We found one specific protein complex bound to a labelled oligonucleotide containing CpGs -2211/-2209. Binding to this probe was competed out by the addition of an excess of unlabelled specific competitor but not with an excess of a nonspecific competitor (Figure 2A). Addition of an unlabelled oligonucleotide containing methylated cytosines at CpG -2211/-2209 also failed to compete out binding to the labelled probe (Figure 2A), suggesting that CpG -2211/-2209 methylation blocks transcription factor binding to this region of GATA3.
Figure 2.
Methylation at CpGs -2211/-2209 within a NFκB binding site blocks binding to the GATA3 promoter. Nuclear extract from the human Jurkat T cell line was incubated with a labelled probe covering CpGs -2211/-2209 and A) an increasing concentration of unmethylated or methylated competitor or B) with an increasing concentration an oligonucleotide containing the consensus NFκB binding site.
As CpGs -2211/-2209 lie within a putative NF-κB site in the GATA3 promoter, we investigated whether binding to this region could be competed out by the addition of an excess of unlabelled oligonucleotide containing the NF-κB binding consensus binding site. We found that the addition of an excess of an unlabelled oligonucleotide containing the NF-κB consensus binding sequence very effectively competed out binding (Figure 2B), supporting the proposition that NF-κB binds to this region of the GATA3 promoter.
4. Discussion
We report that the methylation of specific CpG loci associated with genes involved in Th1/Th2 balance at birth were associated with a later risk of allergic disease. Specifically, we found that higher methylation of CpGs -2211/-2209 located upstream of the transcription start site of the GATA3 gene in umbilical cord tissue at birth were associated with a lower incidence of asthma in childhood. GATA3, which is a member of the GATA family of zinc-finger transcription factors, plays a critical role in Th1/Th2 balance, as it is required for Th2 cell differentiation, maintenance and cytokine production(30). The regulation of GATA3 expression is crucial during development, as over expression of GATA3 can disrupt Th1/Th2 balance. Naive Th cells express a low level of GATA3, levels of GATA3 then increase during Th2 differentiation but are down-regulated during Th1 differentiation(30). Transgenic mice over-expressing GATA3 in thymocytes and T cells develop thymic lymphomas and spontaneous formation of committed Th2 cells(36, 37) while ectopic expression in developing Th1 cells interferes with the methyl-CpG binding domain protein-2 and promotes histone hyperacetylation on the IL-4 locus(38). Moreover, mice over-expressing GATA3 develop exacerbated allergen-induced airway inflammation and airway remodeling(39, 40). Elevated expression of GATA3 is also found in the airways of individuals with asthma and single nucleotide polymorphisms within GATA3 are associated with an increased incidence of asthma and other allergic phenotypes(16, 17). GATA3 transcription is regulated by IL-4, STAT6, Notch and itself during T cell differentiation(34, 41–43), although GATA3 has also been shown to be regulated by a variety of other factors including USF, ZEB, E2A, HEB and NF-κB(44, 45). Interestingly, the GATA3 CpGs -2211/-2209, the sites most strongly associated with later asthma in our data lie within a putative NF-κB binding site. Consistent with NF-κB binding to this region, oligonucleotides containing the NF-κB consensus binding sequence effectively competed out binding to this region, suggesting that NF-κB binds to this region within the GATA3 promoter across the CpG loci associated with childhood asthma. Methylation of the cytosines at CpGs -2211/-2209 led to a decrease in protein binding to the sequence, suggesting that differential methylation at this sequence may interfere with NF-κB binding, resulting in downstream effects on GATA3 expression.
The association between GATA CpG methylation at -2211/-2209 and asthma at 3 years of age remained after adjustment for maternal smoking, batch effect for methylation analysis and child’s sex, while the association at 6-7 years weakened slightly after adjustment for these variables. . Our analyses also found an association between the methylation of specific CpGs within IL-4R in cord tissue at birth and the incidence of atopic eczema in later childhood. Methylation of IL-4R CpG +28180 was previously reported by Soto-Ramírez et al.(25) to be positively associated with asthma incidence in the peripheral blood of 18 year old adolescents. However, in our study there was no association between CpG+28180 and asthma incidence found, instead we found a positive association between umbilical cord IL-4R CpG +28269 methylation, the neighbouring CpG site and incidence of atopic eczema at 12 months. Such differences between the two studies may reflect tissue specific differences in DNA methylation, as we studied methylation in umbilical cord as opposed to in peripheral blood, or differing predominant “asthma” phenotypes in children which we studied, compared to the adolescents in the previous study. Likewise differences have been seen between early and late onset atopic dermatitis, in an Asian mother–offspring cohort, GUSTO (Growing Up in Singapore Towards healthy Outcomes) early onset atopic dermatitis was found to be mainly associated with familial factors, while late onset atopic dermatitis was associated with consumption of antibiotics or probiotics(46).
We also identified an association between TBET CpG methylation and atopy at age 6-7 years, which remained significant after adjustment for confounders. Further studies will be required to determine the robustness of such associations; nonetheless, but our study suggests that the early life environment may modulate the methylation of key Th1/Th2 genes affecting the later risk of asthma, atopic dermatitis and atopy.
The strengths of the study include the relatively large sample size for GATA3 CpG methylation and prospective collection of data. Moreover, atopic eczema was ascertained by a standardised algorithm incorporating history and examination, while atopy was assessed objectively by skin prick testing. We also report an association between the methylation of CpG sites within the promoter region of GATA3 and asthma and show that these CpG sites lie within an NFkB binding site suggesting functional relevance. While further replication of this finding and the other associations observed is required in a larger number of subjects it does suggest that the methylation status of Th1/Th2 genes at birth may have utility as indicators of later allergic disease risk. There are however some limitations to this study. Firstly, the self-reported nature of some of the information, raising the possibility of recall and information bias. Also, although the sample size in which GATA3 methylation was measured was relatively large (n=696 in the extended sample), the sample size for the analysis of IL-4, STAT4, TBET and IL-4R methylation, due to limited DNA availability, was smaller and together with the low prevalence of some allergic phenotypes (5.9% for asthma at 3 years) makes it therefore more challenging to detect associations. Secondly we have analysed methylation in umbilical cord samples and differences in cellular heterogeneity within the cord may contribute to the differences seen in methylation between individuals. Although, cord tissue is also a good surrogate for tissues of mesodermal origin shown by hierarchical clustering analysis of cord with 25 primary tissues/cells from the Epigenome Roadmap project, we do not have Infinium450K data or other genome wide data on the SWS umbilical cord samples, precluding a Houseman correction (47) or reference free surrogate variable analysis (48). Notwithstanding the above, none of the top 500 leukocyte related CpGs from Houseman et al. reside within the regions analysed here and umbilical cord tissue is predominantly made up of mesenchyme derived cells, and less heterogeneous than cord blood. Nevertheless, even if these methylation changes do reflect differences in cellular heterogeneity, our data do suggest altered DNA methylation at these sites may provide a valuable means of identifying individuals at risk of later allergic disease.
A third limitation was that we measured the methylation of 35 CpG sites/groups, raising the likelihood that some of the weaker associations identified could represent chance findings. Although the associations between GATA3 CpG -2211/-2209 and childhood asthma do not survive a strict Bonferroni or 5% FDR correction it is unlikely that these results are false positives. The CpG sites were chosen using a prior hypothesis that Th2 lineage determination genes were likely to be associated with allergic outcomes. In addition some of the CpG sites are highly correlated as illustrated by the coMET plot (31) in Supplementary Figure 2. The allergic outcomes are also significantly associated with each other (p<0.0001) for outcomes at age 6-7 years, Bonferroni relies on the assumption that each of the tests are independent (49). However, the main purpose of a Bonferroni correction is to guard against false positives and in this study we sought to functionally validate the association between GATA3 methylation and asthma by examining the significance of altered methylation of the CpG sites associated with asthma risk in human T cells. Our finding that methylation at these CpGs blocked the binding of the transcription factor NfKb to the promoter region of GATA3 points away from the association of GATA3 methylation with asthma being a false positive finding.
A further limitation inherent in the Sequenom technique, which we used to measure the methylation of CpGs within GATA3, is that it cannot distinguish some closely neighbouring CpG sites with the mass, hence their reporting as groups (such as CpG -2211/-2209), but because of the sequence content of this region, designing amplicons to use on the pyrosequencer was not feasible. Finally, we did not have information on serum IgE concentrations or on rigorously diagnosed food allergy.
5. Conclusions
In conclusion, we have demonstrated that higher methylation of GATA3 CpGs -2211/-2209 at birth was associated with a reduced risk of asthma at ages 3 and 6-7 years. The associations identified by our study provide further evidence of a developmental contribution to later risk of allergic disorders and may in time lead to new biomarkers aiding the diagnosis and treatment of infants at risk of developing these chronic and potentially debilitating diseases.
Supplementary Material
Acknowledgements
We thank the EpiGen Operational Management Group for their project management. This work was supported by funding from the Medical Research Council, Nutricia Research, British Heart Foundation, Arthritis Research UK, National Osteoporosis Society, International Osteoporosis Foundation, Cohen Trust. KMG is supported by the National Institute for Health Research through the NIHR Southampton Biomedical Research Centre and by the European Union's Seventh Framework Programme (FP7/2007-2013), project EarlyNutrition under grant agreement n°289346.
Footnotes
Competing Interests
KMG has received reimbursement for speaking at conferences sponsored by companies selling nutritional and pharmaceutical products. The research groups involved in this work are part of an academic consortium that has received research funding from Abbott Nutrition, Nestec and Danone.
Authors Contributions
KL, KMG, GB and AS conceived and designed the study. SB and HI carried out the statistical analysis. SN, PC, EG, EA, RCH, RM and TB performed the laboratory work and analysis. SB, KL, KMG and SE wrote the manuscript in conjunction with GB, CC, HI and EvB.
References
- 1.Vercelli D. Advances in asthma and allergy genetics in 2007. The Journal of allergy and clinical immunology. 2008;122(2):267–71. doi: 10.1016/j.jaci.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 2.Lemanske RF, Jr, Busse WW. Asthma: clinical expression and molecular mechanisms. The Journal of allergy and clinical immunology. 2010;125(2 Suppl 2):S95–102. doi: 10.1016/j.jaci.2009.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eder W, Ege MJ, von Mutius E. The asthma epidemic. The New England journal of medicine. 2006;355(21):2226–35. doi: 10.1056/NEJMra054308. [DOI] [PubMed] [Google Scholar]
- 4.Seder RA, Paul WE. Acquisition of lymphokine-producing phenotype by CD4+ T cells. Annual review of immunology. 1994;12:635–73. doi: 10.1146/annurev.iy.12.040194.003223. [DOI] [PubMed] [Google Scholar]
- 5.Reiner SL, Seder RA. T helper cell differentiation in immune response. Current opinion in immunology. 1995;7(3):360–6. doi: 10.1016/0952-7915(95)80111-1. [DOI] [PubMed] [Google Scholar]
- 6.Murphy KM, Ouyang W, Farrar JD, Yang J, Ranganath S, Asnagli H, et al. Signaling and transcription in T helper development. Annual review of immunology. 2000;18:451–94. doi: 10.1146/annurev.immunol.18.1.451. [DOI] [PubMed] [Google Scholar]
- 7.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100(6):655–69. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 8.Mullen AC, High FA, Hutchins AS, Lee HW, Villarino AV, Livingston DM, et al. Role of T-bet in commitment of TH1 cells before IL-12-dependent selection. Science. 2001;292(5523):1907–10. doi: 10.1126/science.1059835. [DOI] [PubMed] [Google Scholar]
- 9.Zhou M, Ouyang W. The function role of GATA-3 in Th1 and Th2 differentiation. Immunologic research. 2003;28(1):25–37. doi: 10.1385/IR:28:1:25. [DOI] [PubMed] [Google Scholar]
- 10.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89(4):587–96. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 11.Janson PC, Winerdal ME, Winqvist O. At the crossroads of T helper lineage commitment-Epigenetics points the way. Biochimica et biophysica acta. 2009;1790(9):906–19. doi: 10.1016/j.bbagen.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 12.Martino DJ, Prescott SL. Silent mysteries: epigenetic paradigms could hold the key to conquering the epidemic of allergy and immune disease. Allergy. 2010;65(1):7–15. doi: 10.1111/j.1398-9995.2009.02186.x. [DOI] [PubMed] [Google Scholar]
- 13.Levy O. Innate immunity of the newborn: basic mechanisms and clinical correlates. Nature reviews Immunology. 2007;7(5):379–90. doi: 10.1038/nri2075. [DOI] [PubMed] [Google Scholar]
- 14.Nauta AJ, Ben Amor K, Knol J, Garssen J, van der Beek EM. Relevance of pre- and postnatal nutrition to development and interplay between the microbiota and metabolic and immune systems. The American journal of clinical nutrition. 2013;98(2):586S–93S. doi: 10.3945/ajcn.112.039644. [DOI] [PubMed] [Google Scholar]
- 15.Nakamura Y, Ghaffar O, Olivenstein R, Taha RA, Soussi-Gounni A, Zhang DH, et al. Gene expression of the GATA-3 transcription factor is increased in atopic asthma. The Journal of allergy and clinical immunology. 1999;103(2 Pt 1):215–22. doi: 10.1016/s0091-6749(99)70493-8. [DOI] [PubMed] [Google Scholar]
- 16.Arshad SH, Karmaus W, Kurukulaaratchy R, Sadeghnejad A, Huebner M, Ewart S. Polymorphisms in the interleukin 13 and GATA binding protein 3 genes and the development of eczema during childhood. The British journal of dermatology. 2008;158(6):1315–22. doi: 10.1111/j.1365-2133.2008.08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pykalainen M, Kinos R, Valkonen S, Rydman P, Kilpelainen M, Laitinen LA, et al. Association analysis of common variants of STAT6, GATA3, and STAT4 to asthma and high serum IgE phenotypes. The Journal of allergy and clinical immunology. 2005;115(1):80–7. doi: 10.1016/j.jaci.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 18.Pike KC, Hanson MA, Godfrey KM. Developmental mismatch: consequences for later cardiorespiratory health. BJOG. 2008;115(2):149–57. doi: 10.1111/j.1471-0528.2007.01603.x. [DOI] [PubMed] [Google Scholar]
- 19.Hsu JT, Missmer SA, Young MC, Correia KF, Twarog FJ, Coughlin IB, et al. Prenatal food allergen exposures and odds of childhood peanut, tree nut or sesame seed sensitization. Annals of allergy, asthma & immunology : official publication of the American College of Allergy, Asthma, & Immunology. 2013;111(5):391–6. doi: 10.1016/j.anai.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 20.Ferrante G, Antona R, Malizia V, Montalbano L, Corsello G, La Grutta S. Smoke exposure as a risk factor for asthma in childhood: a review of current evidence. Allergy and asthma proceedings : the official journal of regional and state allergy societies. 2014;35(6):454–61. doi: 10.2500/aap.2014.35.3789. [DOI] [PubMed] [Google Scholar]
- 21.Prescott S, Saffery R. The role of epigenetic dysregulation in the epidemic of allergic disease. Clinical epigenetics. 2011;2(2):223–32. doi: 10.1007/s13148-011-0028-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perera F, Tang WY, Herbstman J, Tang D, Levin L, Miller R, et al. Relation of DNA methylation of 5'-CpG island of ACSL3 to transplacental exposure to airborne polycyclic aromatic hydrocarbons and childhood asthma. PloS one. 2009;4(2):e4488. doi: 10.1371/journal.pone.0004488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joubert BR, Felix JF, Yousefi P, Bakulski KM, Just AC, Breton C, et al. DNA Methylation in Newborns and Maternal Smoking in Pregnancy: Genome-wide Consortium Meta-analysis. American journal of human genetics. 2016;98(4):680–96. doi: 10.1016/j.ajhg.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gruzieva O, Xu CJ, Breton CV, Annesi-Maesano I, Anto JM, Auffray C, et al. Epigenome-Wide Meta-Analysis of Methylation in Children Related to Prenatal NO2 Air Pollution Exposure. Environmental health perspectives. 2017;125(1):104–10. doi: 10.1289/EHP36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soto-Ramirez N, Arshad SH, Holloway JW, Zhang H, Schauberger E, Ewart S, et al. The interaction of genetic variants and DNA methylation of the interleukin-4 receptor gene increase the risk of asthma at age 18 years. Clinical epigenetics. 2013;5(1):1. doi: 10.1186/1868-7083-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Talens RP, Boomsma DI, Tobi EW, Kremer D, Jukema JW, Willemsen G, et al. Variation, patterns, and temporal stability of DNA methylation: considerations for epigenetic epidemiology. The FASEB Journal. 2010;24(9):3135–44. doi: 10.1096/fj.09-150490. [DOI] [PubMed] [Google Scholar]
- 27.Murphy SK, Huang Z, Hoyo C. Differentially methylated regions of imprinted genes in prenatal, perinatal and postnatal human tissues. PloS one. 2012;7(7):e40924. doi: 10.1371/journal.pone.0040924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inskip HM, Godfrey KM, Robinson SM, Law CM, Barker DJ, Cooper C, et al. Cohort profile: The Southampton Women's Survey. International journal of epidemiology. 2006;35(1):42–8. doi: 10.1093/ije/dyi202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams HC, Burney PG, Pembroke AC, Hay RJ. The U.K. Working Party's Diagnostic Criteria for Atopic Dermatitis. III. Independent hospital validation. The British journal of dermatology. 1994;131(3):406–16. doi: 10.1111/j.1365-2133.1994.tb08532.x. [DOI] [PubMed] [Google Scholar]
- 30.Scheinman EJ, Avni O. Transcriptional regulation of GATA3 in T helper cells by the integrated activities of transcription factors downstream of the interleukin-4 receptor and T cell receptor. The Journal of biological chemistry. 2009;284(5):3037–48. doi: 10.1074/jbc.M807302200. [DOI] [PubMed] [Google Scholar]
- 31.Martin TC, Yet I, Tsai PC, Bell JT. coMET: visualisation of regional epigenome-wide association scan results and DNA co-methylation patterns. BMC bioinformatics. 2015;16:131. doi: 10.1186/s12859-015-0568-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pregibon D. Logistic regression diagnostics Annals of Statistics. 1981;9:705–24. [Google Scholar]
- 33.Huber JP, Gonzales-van Horn SR, Roybal KT, Gill MA, Farrar JD. IFN-alpha suppresses GATA3 transcription from a distal exon and promotes H3K27 trimethylation of the CNS-1 enhancer in human Th2 cells. Journal of immunology. 2014;192(12):5687–94. doi: 10.4049/jimmunol.1301908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fang TC, Yashiro-Ohtani Y, Del Bianco C, Knoblock DM, Blacklow SC, Pear WS. Notch directly regulates Gata3 expression during T helper 2 cell differentiation. Immunity. 2007;27(1):100–10. doi: 10.1016/j.immuni.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Y, Ochando JC, Bromberg JS, Ding Y. Identification of a distant T-bet enhancer responsive to IL-12/Stat4 and IFNgamma/Stat1 signals. Blood. 2007;110(7):2494–500. doi: 10.1182/blood-2006-11-058271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nawijn MC, Dingjan GM, Ferreira R, Lambrecht BN, Karis A, Grosveld F, et al. Enforced expression of GATA-3 in transgenic mice inhibits Th1 differentiation and induces the formation of a T1/ST2-expressing Th2-committed T cell compartment in vivo. Journal of immunology. 2001;167(2):724–32. doi: 10.4049/jimmunol.167.2.724. [DOI] [PubMed] [Google Scholar]
- 37.Nawijn MC, Ferreira R, Dingjan GM, Kahre O, Drabek D, Karis A, et al. Enforced expression of GATA-3 during T cell development inhibits maturation of CD8 single-positive cells and induces thymic lymphoma in transgenic mice. Journal of immunology. 2001;167(2):715–23. doi: 10.4049/jimmunol.167.2.715. [DOI] [PubMed] [Google Scholar]
- 38.Lee HJ, Takemoto N, Kurata H, Kamogawa Y, Miyatake S, O'Garra A, et al. GATA-3 induces T helper cell type 2 (Th2) cytokine expression and chromatin remodeling in committed Th1 cells. The Journal of experimental medicine. 2000;192(1):105–15. doi: 10.1084/jem.192.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kiwamoto T, Ishii Y, Morishima Y, Yoh K, Kikuchi N, Haraguchi N, et al. Blockade of cysteinyl leukotriene-1 receptors suppresses airway remodelling in mice overexpressing GATA-3. Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology. 2011;41(1):116–28. doi: 10.1111/j.1365-2222.2010.03571.x. [DOI] [PubMed] [Google Scholar]
- 40.Kiwamoto T, Ishii Y, Morishima Y, Yoh K, Maeda A, Ishizaki K, et al. Transcription factors T-bet and GATA-3 regulate development of airway remodeling. American journal of respiratory and critical care medicine. 2006;174(2):142–51. doi: 10.1164/rccm.200601-079OC. [DOI] [PubMed] [Google Scholar]
- 41.Kurata H, Lee HJ, O'Garra A, Arai N. Ectopic expression of activated Stat6 induces the expression of Th2-specific cytokines and transcription factors in developing Th1 cells. Immunity. 1999;11(6):677–88. doi: 10.1016/s1074-7613(00)80142-9. [DOI] [PubMed] [Google Scholar]
- 42.Ouyang W, Lohning M, Gao Z, Assenmacher M, Ranganath S, Radbruch A, et al. Stat6-independent GATA-3 autoactivation directs IL-4-independent Th2 development and commitment. Immunity. 2000;12(1):27–37. doi: 10.1016/s1074-7613(00)80156-9. [DOI] [PubMed] [Google Scholar]
- 43.Zhu J, Guo L, Watson CJ, Hu-Li J, Paul WE. Stat6 is necessary and sufficient for IL-4's role in Th2 differentiation and cell expansion. Journal of immunology. 2001;166(12):7276–81. doi: 10.4049/jimmunol.166.12.7276. [DOI] [PubMed] [Google Scholar]
- 44.Gregoire JM, Romeo PH. T-cell expression of the human GATA-3 gene is regulated by a non-lineage-specific silencer. The Journal of biological chemistry. 1999;274(10):6567–78. doi: 10.1074/jbc.274.10.6567. [DOI] [PubMed] [Google Scholar]
- 45.Das J, Chen CH, Yang L, Cohn L, Ray P, Ray A. A critical role for NF-kappa B in GATA3 expression and TH2 differentiation in allergic airway inflammation. Nature immunology. 2001;2(1):45–50. doi: 10.1038/83158. [DOI] [PubMed] [Google Scholar]
- 46.Loo EX, Shek LP, Goh A, Teoh OH, Chan YH, Soh SE, et al. Atopic Dermatitis in Early Life: Evidence for at Least Three Phenotypes? Results from the GUSTO Study. International archives of allergy and immunology. 2015;166(4):273–9. doi: 10.1159/000381342. [DOI] [PubMed] [Google Scholar]
- 47.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28(6):882–3. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goeman JJ, Solari A. Multiple hypothesis testing in genomics. Stat Med. 2014;33(11):1946–78. doi: 10.1002/sim.6082. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.