Abstract
This study is the first to comprehensively characterize m6A patterns in the Arabidopsis chloroplast and mitochondria transcriptomes based on our open accessible data deposited in NCBI's Gene Expression Omnibus with GEO Series accession number of GSE72706. Over 86% of the transcripts were methylated by m6A in the two organelles. Over 550 and 350 m6A sites were mapped, with ~5.6 to ~5.8 and ~4.6 to ~4.9 m6A sites per transcript, to the chloroplast and mitochondria genome, respectively. The overall m6A methylation extent in the two organelles was greatly higher than that in the nucleus. The m6A motif sequences in the transcriptome of two organelles were similar to the nuclear motifs, suggesting that selection of the m6A motifs for RNA methylation was conserved between the nucleus and organelle transcriptomes. The m6A patterns of rRNAs and tRNAs in the organelle were similar to those in the nucleus. However, the m6A patterns in coding RNAs were distinct between the nucleus and the organelle, suggesting that that regulation of the m6A methylation patterns may be different between the nuclei and the organelles. The extensively methylated transcripts in the two organelles were mainly associated with rRNA, ribosomal proteins, photosystem reaction proteins, tRNA, NADH dehydrogenase and redox. On average, 64% and 79% of the transcripts in the two organelles showed differential m6A methylation across three organs of the leaves, flowers and roots. The m6A methylation extent in the chloroplast was higher than that in the mitochondria. This study provides deep insights into the m6A methylation topology and differentiation in the plant organelle transcriptomes.
Introduction
Chemical modifications have been found ubiquitously distributing in RNAs of the living species[1–10]. Among those, N6-methyladenosine (m6A) has been found prevalently distributing in nuclear mRNA, rRNA, tRNA, and some snRNA of eukaryotes[3,5,7,10–16]. The m6A topology was found highly conserved in the eukaryote transcritptome[17,18]. For example, most m6A sites enriched near the stop codon or 3'untranslated regions (UTR) in the nuclear mRNAs of the higher living species[17–20]. RNA m6A modification in the nuclear RNAs was responsible for certain important metabolisms, e.g. RNA splicing, RNA export[21], RNA stability[17,18], control of the circadian clock[22], regulation of gene expression[23,24], decision of cell differential fate[10,15,25] and regulation of RNA-protein interaction[26]. Silencing the m6A methyltransferase significantly influences gene expression and alternative splicing patterns, resulting in initiation of the p53signaling pathway and apoptosis [17]. m6A modification is selectively recognized by binding proteins to affect the translation status and lifetime of mRNA [18]. Specific inhibition of m6A methylation by silencing of the m6A methylase Mettl3 is sufficient to elicit circadian period elongation and RNA processing delay [22]. Increased m6A methylation promotes there programming of mouse embryonic fibroblasts(MEFs) to pluripotent stem cells; in contrast, a reduced m6A level impedes reprogramming [25]. The methylation and demethylation of m6A is contemporarily and precisely regulated to be balanced due to stimuli as to maintain an appropriate metabolism in the cell[24]. Defect in m6A methylation or demethylation will result in severe physiological consequences[27], e.g. abnormal reproductive development[28,29], obesity[30], or cancer[10–15,31,32] in mammals.
Most of the mysteries concerning m6A RNA methylation were derived from mammals aforementioned. However, some phenomena associated with m6A RNA methylation were discovered in plants, which adds our knowledge in this area. Plant mRNA contains m6A methylation level similar to that in animal cells[28,33,34]. N6-methyladenosine mRNA methylase is essential for embryonic development in Arabidopsis thaliana[28]. Inactivation of the Arabidopsis mRNA adenosine methylase (MTA) results in failure of the developing embryo to arrest at the globular stage [28]. mRNAs in the arrested seeds contain deficient m6A methylation [28]. A 90% reduction of m6A levels during later growth stages gives rise to plants with altered growth patterns and reduced apical dominance [19]. The flowers of the mutant plants show defects in their floral organ number, size, and identity [19]. MTA expression is highly associated with dividing tissues, particularly reproductive organs, shoot meristems, and emerging lateral roots [28]. Over 85% of the modified transcripts show high m6A methylation extent compared to their transcript level in Arabidopsis thaliana [35]. Highly m6A methylated transcripts are mainly associated with transporters, stress responses, redox, regulation factors, and some non-coding RNAs [35]. m6A may be another important contributor to organ differentiation in Arabidopsis and rice [34,35]. Most of the transposable element transcripts retain a fragmented form with a relatively low transcript level and high m6A methylation in the cells, which is suitable for roles of the transposable elements[35]. Therefore, m6A RNA methylation also plays important roles connecting critical metabolisms in plants.
High efficiency and specific binding ability of the m6A antibody provides a useful tool for a transcriptome-wide analysis of the m6A patterns in several species [17,18,21,30,33,34]. The successful experiments are fulfilled through use of RNA immunoprecipitation (RIP) and m6A-seq [17,18,21,30,33,34]. RIP experiment in the m6A-seq study was aimed to enrich RNAs containing m6A through use of m6A antibody to the fragmented RNA pool [17]. The enriched m6A RNA pool is used for RNA-seq, called ‘m6A-seq’[17,18,21,30,33,34].
Chloroplast and mitochondria are two important organelles mainly for photosynthesis and respiration in plants, respectively[36,37]. Amyloplast was derived from chloroplast and evolved mainly for food storage in plant organs, e.g, in roots, fruits and seeds[38]. m6A modification was recently found in the chloroplast transcritptome[34]. But information of m6A modification in mitochondria is unclear. In addition, m6A methylation differences of RNAs between organelles and nucleus have not been well assayed. The nuclear m6A RNA methylation was deeply surveyed in previous studies [17,18,21,30,33,34]. However, little is known of this modification in the organelles. This study aimed to (i) comprehensively characterize m6A distributing patterns in the Arabidopsis chloroplast/amyloplast and mitochondria transcriptomes, (ii) analyze relationship between the transcript level and the m6A modification extent in the Arabidopsis chloroplast/amyloplast and mitochondria, and (iii) characterize differential patterns of the m6A methylation across leaves, flowers and roots in the Arabidopsis chloroplast/amyloplast and mitochondria. This is the first study to comprehensively analyze the m6A distributing and differential patterns across organs in the plant organelles.
Methods
Ethics statement
All plant materials used in this study are freely available to all researchers without any protection for the intellectual property right. This research meets all applicable requirements for the ethics of experimentation and research integrity from all five institutes that provide support to this study.
Plant growth conditions and treatments
Wild Columbia ecotype (‘Col-0’) of Arabidopsis thaliana was used in this research as we in our recent publication [35]. When the plants were fully flowered (five weeks after seed germination), the materials of flowers, rosette leaves and roots were separately collected, promptly frozen in liquid nitrogen and stored at -80°C until use.
RNA extraction
The modified cetyl trimethylammonium bromide (CTAB) method was used for RNA isolation as described in our previous study[35]. LiCl solution (8.0 M) was used to precipitate RNA. The purified RNA pellet was stored at -80°C until use.
RNA fragmentation and RNA immunoprecipitation (RIP)
The purified total RNA was fragmented into ~100-nucleotide-long usingthe ZnCl2 buffer (10mM ZnCl2 and 10mM Tris-HCl, pH 7) according to the protocol developed by Dominissini et al. [17]. The fragmented RNA was prepared for RIP and m6A-seq.
The m6A-specific binding antibody (Merck Millipore, Billerica, MA, USA) was used for the RIP experiments according to the protocol of Dominissini et al.’s [17]. Ethanol and glycogen were used to precipitate the pulled-down RNA by the m6A antibody. The m6A RNA pellet was cleaned using 80% ethanol and then resuspended into 15 μl dd-H2O for m6A-seq, high-performance liquid chromatography (HPLC) and mass spectrometry (MS) analysis.
RNA-seq, m6A-seq and input RNA-seq
To perform RNA sequencing from numerous RNAs including coding RNAs without polyA in the organelles, the Ribo-Zero rRNA Removal Kit (Madison, WI, USA) was used to remove rRNA (actually rRNA can not be completely removed). High throughput m6A-seq, RNA-seq and input RNA-seq were performed on HiSeq 2000 (Illumina Inc, San Diego, CA) at Purdue University Genomics Core Facility (http://www.genomics.purdue.edu/services/core.shtml). All RNA sequencing of three samples of leaves, flowers and roots was performed at the same batch on the same sequencer.
Alignment of reads and visualization of m6A peaks
All RNA sequencing data sets were mapped to the Arabidopsis genome (TAIR10) using TopHat2 under a parameter of ‘-b2-fast’[39]. The potential PCR duplicates were removed by a parameter of ‘rmdup’ rooted in SAM tools [40]. The fragment numbers for each transcript were estimated using the feature Counts with a parameter of “-p”[41].
The distributing patterns of the mapped m6A-seq data were visualized using free software, Integrative Genomics Viewer (IGV2.3, Boston, MA, USA) [42]. IGV is used for high-performance visualization of interactive exploration of large, integrated genomic datasets [42]. The tool is able to present extent of m6A methylation, sequencing depth, sequencing fragment alignment, and gene ID of the sequencing data [42].
Because of an extensively low non-specific immunoprecipitation rate (< 1%) in this study[35], all the mapped reads in the m6A-seq were assumed to be derived from specific immunoprecipitation of the RNA fragments containing m6A modification. Thus, an estimation of m6A peak number of a m6A modified transcript was estimated by this formula: total mapped length covered by m6A fragments within the transcript/150, considered that library construction for m6A-seq was created from a m6A RNA pool with an average RNA length of ~106 nucleotides (S1 Fig) and average coverage of a peak in m6A-seq data was ~150-nucleotide long in this study as visualized by IGV 2.3 [35].
Discernment of m6A topological patterns
The consensus m6A motif sequences were figured out by Luo et al.’s protocol [33]. The typical m6A patterns of different types of RNAs were captured by screenshot from IGV2.3 visualization of the m6A mapping results after normalization.
m6A methylation extent versus transcript level
Sequencing depth in RNA-seq was normalized using the algorithm of Fragments Per Kilobase of Transcript Per Million Fragments Mapped (FPKM = Counts of mapped fragments × 109) / (Length of transcript × Total count of the mapped fragments))[43]. While the sequencing depth in m6A-seq was normalized using a modified FPKM (MFPKM = Counts of mapped fragments × 109) / (Total mapped length covered by m6A fragments within the transcript × Total count of the mapped m6A fragments))[35].
The m6A methylation extent of a transcript were categorized into three groupings based on comparison of MFPKM of the transcript in the m6A-seq with FPKM of the same transcript in the RNA-seq using χ2 test:(1) the m6A methylation extent ‘equivalent’ to the transcript level (‘equivalent’, ratio of FPKM to MFPKM fits 1:1 (p < 0.05)), (2) the methylation extent higher than the transcript level (‘Hi’, ratio of FPKM to MFPKM < 1 (p < 0.05)), and (3) the methylation extent lower than the transcript level (‘Low’, ratio of FPKM to MFPKM > 1 (p < 0.05))[35].
Differential transcript level and differential m6A methylation among plant organs
RNA-seq data was normalized by FPKM as described above. χ2 tests were used to estimate whether FPKM was significantly different between two organs using R 3.1 (http://cran.r-project.org/bin/windows/base/). The transcripts with a fold change in FPKM > 2.0 or < 0.5, and FDR < 0.02 were considered differentially expressed between two organs[17].
To minimize influence of the transcript level on estimation of differentiation of the m6A extent, m6A-seq data was normalized by a specific algorithm, NFPKM (NFPKM = MFPKM in m6A-seq/LOG (FPKM in RNA-seq, 2)). χ2 tests were also used to estimate whether NFPKM of a m6A modified transcript was significantly different between two organs using R 3.1. The transcripts with a fold change of NFPKM > 2.0 or < 0.5, and FDR < 0.005 were considered differentially methylated between two organs[17,35].
qRT-PCR
Quantitative real-time PCR (qRT–PCR) was performed to assess relative abundance (RA)of m6A RNA in the RIP samples. All purified RNA templates were transferred into cDNA using Quanta qScript™ cDNA Synthesis Kits (Quanta BioSciences, Inc, Gaithersburg, MD, USA). Six genes were randomly chosen for this test (S1 Table). qRT-PCR primers were designed to span exon-exon junctions in order to eliminate potential amplification of the genomic DNA. qRT-PCR was performed on C1000 Thermal Cycler (Bio-RAD) using SYBR Green SuperMix buffer (Bio-RAD) and 300 ng total cDNA template for amplification. Because the qRT-PCR amplicon spanned an exon-exon junction with a length of 80–150 bp (Fig 1a), cDNA of the Actin2 gene was used for housekeeping gene and was used for normalization of total RNA in qRT-PCR. The relative abundance of m6A RNA in the qRT-PCR amplicons was estimated using this algorithm: RA = 100 × 2-ΔC. Expected abundance (EA) of m6A RNA in the m6A-seq data set was estimated by this algorithm: EA = 100× (the mapped m6A RNA reads of the test gene in m6A-seq and in the cDNA region for qRT-PCR test/the mapped RNA reads of the Actin2 gene in RNA-seq and in the cDNA region for qRT-PCR test). Consistency between the AR and ER results among three organs was compared (S2 Fig). The correlation coefficient between the average RA and the average EA was calculated using SPSS 13.0 (SPSS, USA).
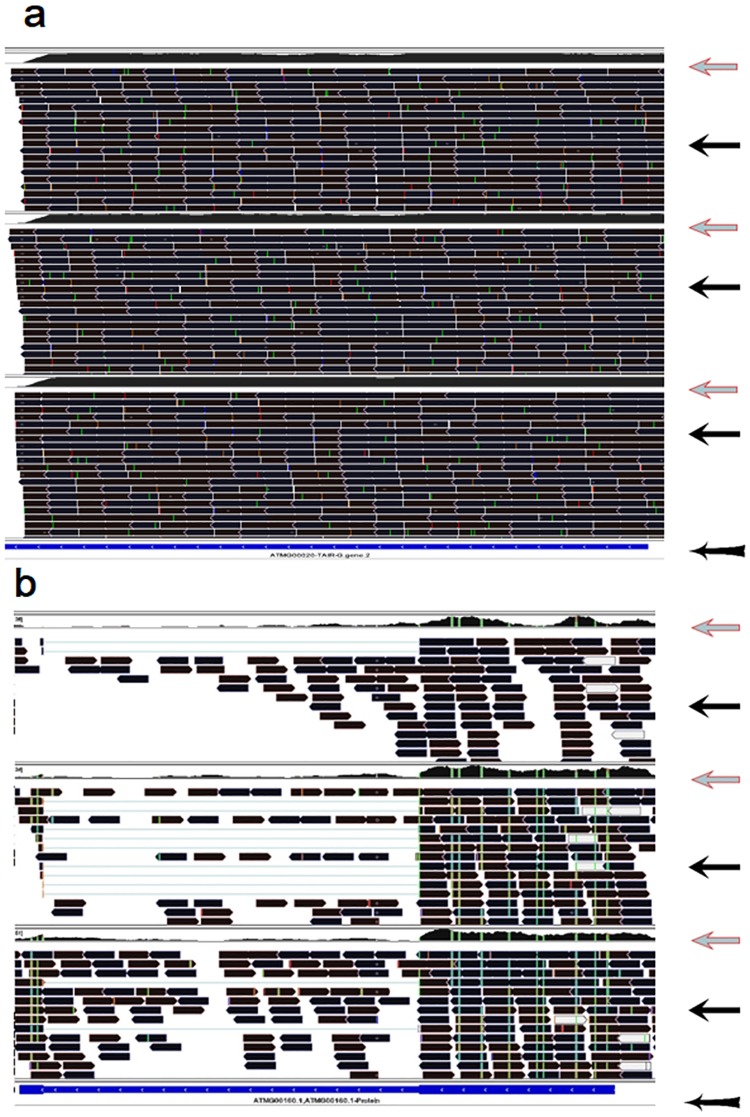
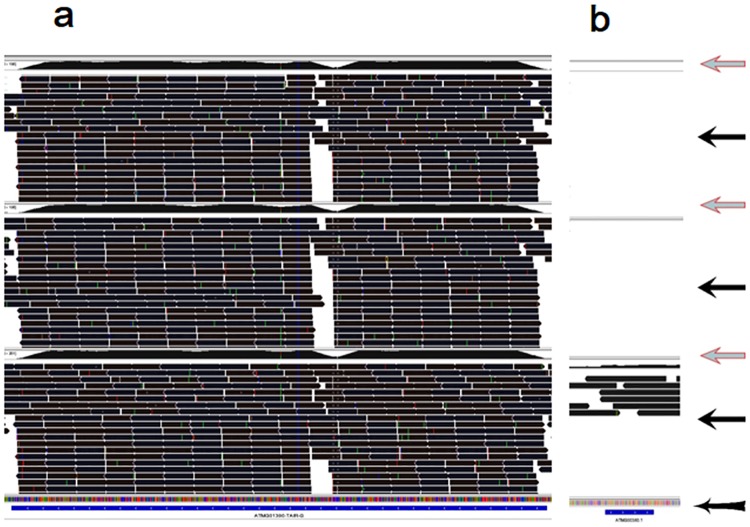
Fig 1. Screenshots from the IGV visualized program present two typical types of m6A topologies in the coding RNAs in the Arabidopsis mitochondria.
Extent of m6A methylation, sequencing depth, sequencing fragment alignment, and gene ID of the sequencing data can be clearly visualized by the IGV program [42]. The area in the screenshot indicated by the arrow, “Red leftwards arrow”, presents m6A methylation extent across the transcript. The area in the screenshot indicated by the arrow, “Black leftwards arrow”, presents sequencing fragment alignment across the transcript. The area in the screenshot indicated by the arrow, “Black leftwards arrow with tail”, presents gene ID information including gene ID, sequence reading direction, the intron and exon regions. (a) Type 1 (representative gene, ‘ATMG00920’, expressed for ‘a hypothetical protein’), the whole transcript without intron was highly methylated by m6A; (b) Type 2 (representative gene, ‘ATMG00160’, expressed for ‘cytochrome oxidase 2’), the exon was highly methylated but the intron was not methylated by m6A. Trace files of three organs, leaves (the upper), flowers (in the middle) and roots (the lower) were presented within a screenshot.
Results
Extent of m6A methylation in the Arabidopsis chloroplast/amyloplast and mitochondria
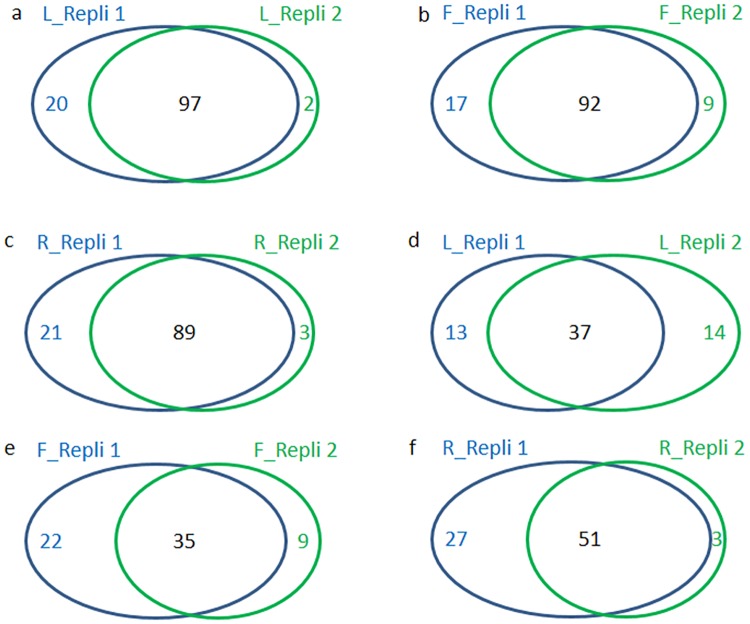
This study shared the same data sets with our recent publication for characterization of m6A methylation in the Arabidopsis nucleus [35]. Six samples from three organs of Arabidopsis leaves, flowers and roots were used for m6A-seq, six samples for RNA-seq, and six samples for input RNA-seq (total fragmented RNA without RIP experiment as the control for m6A-seq), respectively, with two replicates for each RNA sequencing (S2 Table). In m6A-seq, agreement proportion between two replicates was 82%, 78% and 79% from the leaf, flower and root chloroplast/amyloplast samples, respectively (Fig 2a–2c). While in the mitochondria, the agreement proportion was 74%, 70% and 77% from the leaf, flower and root samples, respectively (Fig 2d–2f).
Fig 2. Number of the overlapped m6A transcripts in the two m6A-seq replicates.
(a) in the leaf chloroplast; (b) in the flower chloroplast; (c) in the root amyloplast; (d)in the leaf mitochondria; (e) in the flower mitochondria; and (f) in the root mitochondria.
In total, 133 and 146 genes have been so far discovered in the Arabidopsis chloroplast/amyloplast and mitochondria genome, respectively (Table 1)[36,44]. In this study, we found that 79–80% and 34–64% of the genes were transcribed in the chloroplast/amyloplast and mitochondria transcriptome, respectively (Table 1). This indicated that proportion of the transcribed genes in the chloroplast/amyloplast was close to that of the nuclear genes (p< 0.01)[35]. However, proportion of the transcribed genes in the mitochondria was significantly lower than that of the nuclear genes or the genes from the chloroplast/amyloplast (p< 10−4) (Table 1)[44].
Table 1. Proportion of the transcribed genes methylated by m6A in the chloroplast/amyloplast and mitochondria.
| Transcribed genes | Chloroplast/ amyloplast | Mitochondria | ||||
|---|---|---|---|---|---|---|
| Leaf | Flower | Root | Leaf | Flower | Root | |
| Replicate 1 | ||||||
| Total number of the genes | 133 | 133 | 133 | 146 | 146 | 146 |
| Number of the transcribed genes | 117 | 109 | 114 | 62 | 63 | 94 |
| The transcribed genes methylated by m6A | 117 | 109 | 110 | 51 | 57 | 78 |
| Proportion of the transcribed genes (%) | 88 | 82 | 86 | 42 | 43 | 64 |
| Proportion of the methylated genes (%) | 100 | 100 | 96 | 82 | 90 | 83 |
| Replicate 2 | ||||||
| Number of the transcribed genes | 99 | 101 | 92 | 56 | 49 | 56 |
| The transcribed genes methylated by m6A | 99 | 101 | 92 | 51 | 44 | 54 |
| Proportion of the transcribed genes (%) | 74 | 76 | 72 | 38 | 34 | 38 |
| Proportion of the methylated genes (%) | 100 | 100 | 100 | 91 | 90 | 96 |
We found that98–100%and 86–90% of the transcribed genes were chemically modified by m6A in the chloroplast/amyloplast and mitochondria transcriptome, respectively (Table 1). These two proportions were significantly higher than that (~73%) in the nuclear transcriptome (p< 10−4)[35]. The results also indicated that proportion of the m6A methylated genes in the chloroplast/amyloplast was higher than that in the mitochondria (p< 0.001) (Table 1).
On average, around 620 m6A sites from the leaves, ~580 sites from the flowers, and ~570 sites from the roots were successfully mapped to the Arabidopsis chloroplast genome with an estimation of ~5.6 to ~5.8 m6A sites per transcript (S3 Table). About 280 m6A sites from the leaves, ~340 sites from the flowers, and ~400 sites from the roots were successfully mapped to the Arabidopsis mitochondria genome with an estimation of ~4.6 to ~4.9 m6A sites per transcript (S4 Table). Therefore, the total m6A sites and the number of m6A sites per transcript in the chloroplast transcriptome were significantly higher than those in the mitochondria transcriptome(p< 0.01). In addition, the number of m6A sites per transcript in the two organelles was greatly higher than that in the nuclear transcritptome[33,35].
m6A topology in the Arabidopsis chloroplast/amyloplast and mitochondria
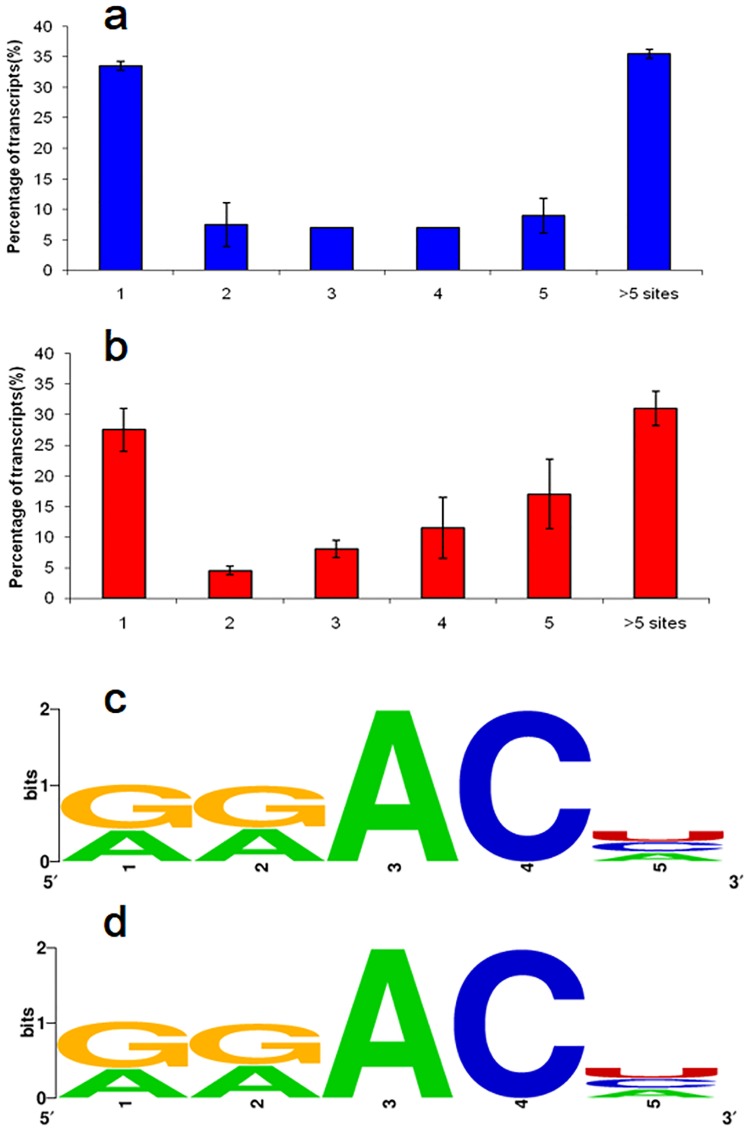
Over 27% of the methylated transcripts was covered by one m6A site in the two organelles (Fig 3a and 3b, details in S5 and S6 Tables). Over 31% contained six or more sites in the two organelles (Fig 3a and 3b, details in S5 and S6 Tables).
Fig 3. Features of m6A methylation in two organelles.
(a) Proportion of the transcribed methylated genes containing different m6A sites in the chloroplast/amyloplast transcriptome; (b) proportion of the transcribed methylated genes containing different m6A sites in the mitochondria transcriptome; (c) the most common consensus motif (RRm6ACH) in the m6A peaks in the chloroplast/amyloplast transcriptome; and (d) the most common consensus motif (RRm6ACH) in the m6A peaks the mitochondria transcriptome.
The consensus sequence of m6A modification has been identified as ‘RRm6ACH’ in the nuclear transcritptome of mammals and plants, where R is A/G and H is A/C/U[17,21,35, 45]. Our data showed that over 65% and 67% of the RIP fragments in m6A-seq contained the consensus sequence of ‘RRm6ACH’ in the Arabidopsis chloroplast and mitochondria, respectively (Fig 3c and 3d). Two mostly observed motifs were GGm6ACC (10.3%) and GGm6ACU (10.7%) in the chloroplast transcriptome(Fig 3c). And GGm6ACA (10.4%) and GGm6ACU (11.0%) were the mostly detected motifs in the mitochondria transcriptome(Fig 3d). Thus the m6A motifs were conserved between the chloroplast and mitochondria transcriptome. Our observation also suggested that the m6A motifs in the two organelles were similar to those in the plant nuclear transcritptome[33,35].
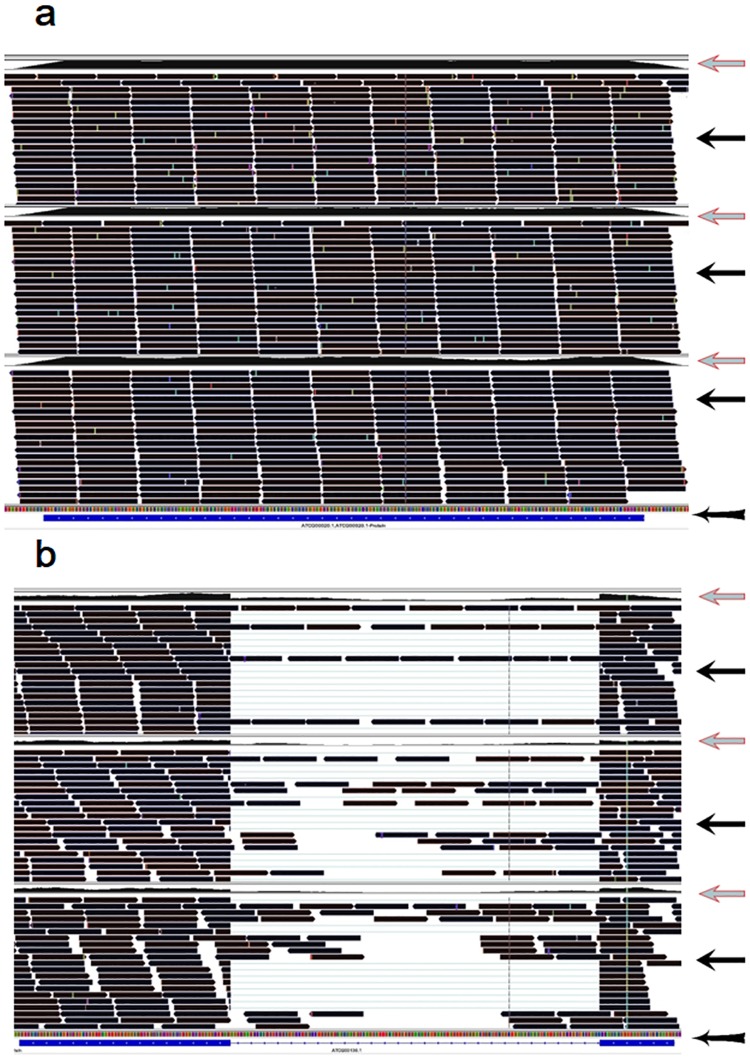
Most of the modified transcripts had similar m6A patterns between the two organelles (Figs 1 and 4–6). Two typical m6A patterns were found in the coding RNAs in the two organelles (Figs 1 and 4): (1) the whole coding RNA without intron was highly methylated by m6A (Figs 1a and 4a), and (2) the exon of the transcript was highly methylated but the intron was much less methylated by m6A (Figs 1b and 4b). All rRNAs were extensively methylated by m6A in the two organelles (Figs 5a and 6a). Most of tRNAs, with or without intron, were highly methylated by m6A in the Arabidopsis chloroplast/amyloplast (Fig 5b and 5c).
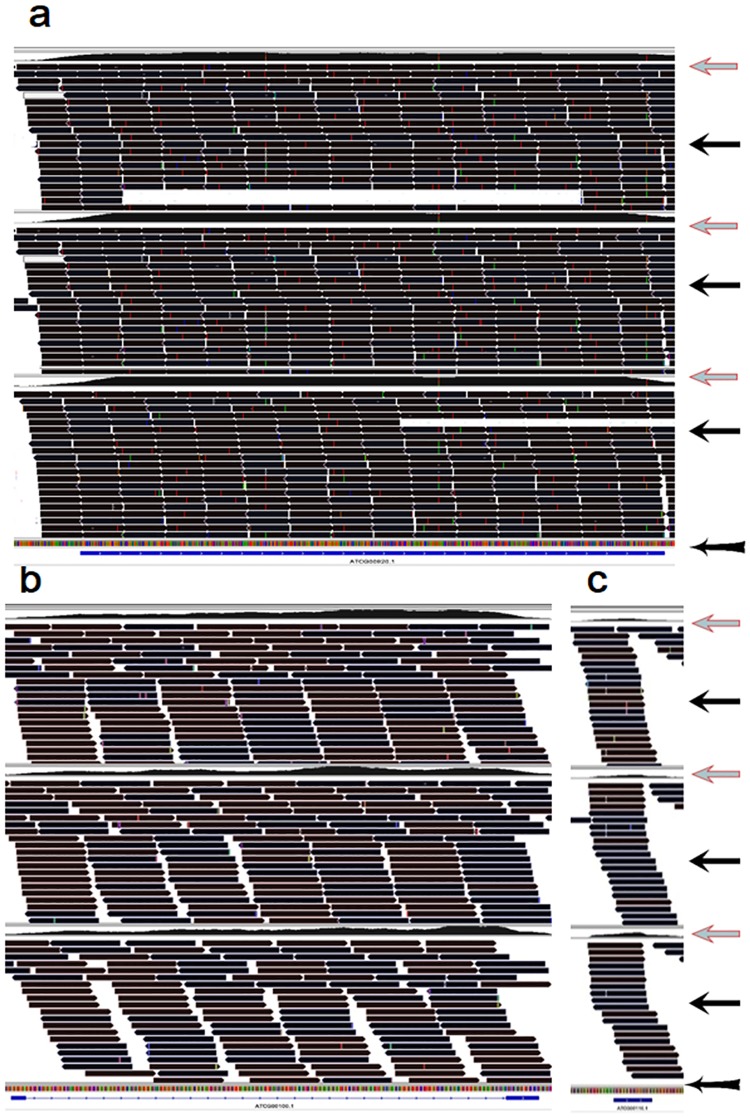
Fig 4. Screenshots from the IGV visualized program present two typical types of m6A topologies in the coding RNAs in the Arabidopsis chloroplast/amyloplast.
Extent of m6A methylation, sequencing depth, sequencing fragment alignment, and gene ID of the sequencing data can be clearly visualized by the IGV program [42]. The area in the screenshot indicated by the arrow, “Red leftwards arrow”, presents m6A methylation extent across the transcript. The area in the screenshot indicated by the arrow, “Black leftwards arrow”, presents sequencing fragment alignment across the transcript. The area in the screenshot indicated by the arrow, “Black leftwards arrow with tail”, presents gene ID information including gene ID, sequence reading direction, the intron and exon regions. (a) Type 1 (representative gene, ‘ATCG00020’, expressed for ‘photosystem II reaction center protein A’), the whole transcript without intron was highly methylated by m6A; and (b) Type 2 (representative gene, ‘ATCG00130’, expressed for ‘ATPase, F0 complex, subunit B/B’), the exon was highly methylated but the intron was less methylated by m6A. Trace files of three organs, leaves (the upper), flowers (in the middle) and roots (the lower) were presented within a screenshot.
Fig 6. Screenshots from the IGV visualized program present m6A topologies in rRNA and tRNAs in the Arabidopsis mitochondria.
Extent of m6A methylation, sequencing depth, sequencing fragment alignment, and gene ID of the sequencing data can be clearly visualized by the IGV program [42]. The area in the screenshot indicated by the arrow, “Red leftwards arrow”, presents m6A methylation extent across the transcript. The area in the screenshot indicated by the arrow, “Black leftwards arrow”, presents sequencing fragment alignment across the transcript. The area in the screenshot indicated by the arrow, “Black leftwards arrow with tail”, presents gene ID information including gene ID, sequence reading direction, the intron and exon regions. (a) The whole rRNA was highly methylated by m6A, representative rRNA, ‘ATMG01390’; (b) The whole tRNA was slightly methylated by m6A, representative tRNA, ‘ATMG00380’, expressed for tRNA-Asn. The Trace files of three organs, leaves (the upper), flowers (in the middle) and roots (the lower) were presented within a screenshot.
Fig 5. Screenshots from the IGV visualized program present m6A topologies in rRNA and tRNAs in the Arabidopsis chloroplast/amyloplast.
Extent of m6A methylation, sequencing depth, sequencing fragment alignment, and gene ID of the sequencing data can be clearly visualized by the IGV program [42]. The area in the screenshot indicated by the arrow, “Red leftwards arrow”, presents m6A methylation extent across the transcript. The area in the screenshot indicated by the arrow, “Black leftwards arrow”, presents sequencing fragment alignment across the transcript. The area in the screenshot indicated by the arrow, “Black leftwards arrow with tail”, presents gene ID information including gene ID, sequence reading direction, the intron and exon regions. (a) The whole rRNA was highly methylated by m6A, representative rRNA, ‘ATCG00920’; (b) The whole tRNA with intron was highly methylated, representative tRNA, ‘ATCG00100’; and (c) The whole tRNA without intron was highly methylated, representative tRNA, ‘ATCG00110’. The Trace files of three organs, leaves (the upper), flowers (in the middle) and roots (the lower) were presented within a screenshot.
m6A methylation extent versus gene transcript level in the two organelles
To compare m6A methylation extent in m6A-seq with gene transcript level in RNA-seq in Arabidopsis, our previous study categorized the m6A methylation extent into three groupings based on comparison of ‘the modified fragments per kilobase of transcript per million fragments mapped of the transcript in m6A-seq (MFPKM)’ with ‘the fragments per kilobase of transcript per million fragments mapped of the counterpart in the RNA-seq (FPKM)’using χ2 test[35]. This study also applied this method for analysis of extent of m6A methylation in the transcriptome of two organelles.
The chloroplast/amyloplast showed a different m6A methylation extent among three organs. On average, 79% and 52% of the methylated transcripts in the leaf and flower chloroplast showed a high m6A modification level, while 5% and 15% of the m6A modified transcripts had a low modification extent, respectively(Table 2). However, 33% of the methylated transcripts in the root amyloplast showed a high m6A modification level, and 40% had a low m6A modification level (Table 2).
Table 2. Three groupings of the m6A methlylation extent compared to the transcript level in the chloroplast/amyloplast transcriptome of three organs in Arabidopsis.
| Plant organs | High | Low | Equivalent | |||
|---|---|---|---|---|---|---|
| No. of transcribed genes | Proportion(%) | No. of transcribed genes | Proportion(%) | No. of transcribed genes | Proportion(%) | |
| Leaves_1 | 95 | 82 | 7 | 6 | 15 | 13 |
| Leaves_2 | 75 | 76 | 4 | 4 | 20 | 19 |
| Average | 79 | 5 | 16 | |||
| Flowers_1 | 54 | 50 | 26 | 24 | 29 | 26 |
| Flowers_2 | 54 | 53 | 5 | 5 | 42 | 42 |
| Average | 52 | 15 | 34 | |||
| Roots_1 | 47 | 43 | 52 | 47 | 11 | 10 |
| Roots_2 | 21 | 23 | 29 | 32 | 42 | 45 |
| Average | 33 | 40 | 28 | |||
‘High’, ‘Low’ and ‘equivalent’ were categorized by comparison of the m6A-seq depth (MFPKM, the methlylation extent of m6A) of each transcript with that in the RNA-seq (FPKM, the transcript level). ‘High’ or ‘Low’ referred to as a relatively high or low m6A methlylation extent compared with its transcript level based on χ2 test (p< 0.05); ‘equivalent’, suggested that the m6A methlylation depth was relatively ‘equivalent’ to the transcript level (ratio of MFPKM to FPKM fits 1:1) based on χ2 test (p< 0.05).‘_1’ and ‘_2’ represent two replicates.
The mitochondria showed a similar m6A methylation trend in three organs of the leaf, flower and root. Proportion of three groupings representing m6A methylation level in the mitochondria had non-significant differences among three organs (p> 0.6) (Table 3). On average, 85–89% of the methylated transcripts had a high modification level, while 4–6% of the modified transcripts presented a low modification extent in the three organs. In addition, proportion of the transcripts showing a high methylation extent in the mitochondria was higher than that in the chloroplast/amyloplast (Tables 2 and 3).
Table 3. Three groupings of the m6A methlylation extent compared to the transcript level in the mitochondria transcriptome of three organs in Arabidopsis.
| Plant organs | High | Low | Equivalent | |||
|---|---|---|---|---|---|---|
| No. of transcribed genes | Proportion(%) | No. of transcribed genes | Proportion(%) | No. of transcribed genes | Proportion(%) | |
| Leaves_1 | 40 | 80 | 4 | 8 | 6 | 12 |
| Leaves_2 | 46 | 90 | 2 | 4 | 3 | 6 |
| Average | 85 | 6 | 9 | |||
| Flowers_1 | 52 | 91 | 2 | 4 | 3 | 5 |
| Flowers_2 | 38 | 86 | 2 | 5 | 4 | 9 |
| Average | 89 | 4 | 7 | |||
| Roots_1 | 69 | 89 | 5 | 6 | 4 | 5 |
| Roots_2 | 48 | 89 | 2 | 4 | 4 | 7 |
| Average | 89 | 5 | 6 | |||
‘High’, ‘Low’ and ‘equivalent’ were categorized by comparison of the m6A-seq depth (MFPKM, the methlylation extent of m6A) of each transcript with that in the RNA-seq (FPKM, the transcript level). ‘High’ or ‘Low’ referred to as a relatively high or low m6A methlylation extent compared with its transcript level based on χ2 test (p< 0.05); ‘equivalent’, suggested that the m6A methlylation depth was relatively ‘equivalent’ to the transcript level (ratio of MFPKM to FPKM fits 1:1) based on χ2 test (p< 0.05).‘_1’ and ‘_2’ represent two replicates.
To better understand relationship between the m6A methylation extent in m6A-seq and the transcript level in RNA-seq in the chloroplast/amyloplast and mitochondria, the transcript level was categorized into three groupings: high, moderate and low as the method we previously used[35]. And each category contained one-third of the m6A modified transcripts from the highest to the lowest FPKM in RNA-seq as we described in our previous study[35].
In the both leaf and flower chloroplasts, comparison of ratio of average MFPKM in m6A-seq to average FPKM in RNA-seq between three groupings using t-test (Table 4) showed that most of the highly expressed transcripts were relatively less modified by m6A, and most transcripts with a low expression level were more likely modified by m6A (p<0.05). The moderately expressed transcripts tended to be moderately methylated in the leaf and flower chloroplasts (p< 0.05). However, the root amyloplast presented this methylation feature that the moderately expressed transcripts were more likely to be methylated, and those expressed at the two extremes were less methylated by m6A (Table 4). Intriguingly, the mitochondria transcripts in all three organs presented this feature: most of the highly expressed transcripts were relatively less methylated by m6A, and most transcripts with a low expression level were more likely modified by m6A (p< 0.05). The moderately expressed transcripts tended to be moderately methylated by m6A in the mitochondria (p< 0.05) (Table 5). Therefore, in most cases, features of the methylation extent versus the transcript level in the two organelles were similar to those recently found in the nuclear transcripts in Arabidopsis[35].
Table 4. Relationship between the m6A methlylation extent and the transcript level in the chloroplast/amyloplast transcriptome.
| Plant organs | High | Moderate | Low | ||||||
|---|---|---|---|---|---|---|---|---|---|
| MFPKM | FPKM | Ratio | MFPKM | FPKM | Ratio | MFPKM | FPKM | Ratio | |
| Leaves_1 | 82964.4 | 32491.2 | 2.6 | 6002.6 | 1990.8 | 3.0 | 558.6 | 230.3 | 2.4 |
| Leaves_2 | 32128.9 | 33644.2 | 1.0 | 12436.3 | 5137.6 | 2.4 | 5792.8 | 1060.7 | 5.5 |
| Average | 1.8 | 2.7 | 4.0 | ||||||
| Flowers_1 | 151788.5 | 53586.9 | 2.8 | 45698.8 | 6592.2 | 6.9 | 6448.6 | 896.4 | 7.2 |
| Flowers_2 | 30304.4 | 28351.9 | 1.1 | 12931.0 | 7610.2 | 1.7 | 7183.2 | 2068.3 | 3.5 |
| Average | 1.9 | 4.3 | 5.4 | ||||||
| Roots_1 | 113745.4 | 37647.1 | 3.0 | 36667.2 | 7074.2 | 5.2 | 4762.5 | 1708.5 | 2.8 |
| Roots_2 | 26555.5 | 31256.0 | 0.8 | 14101.1 | 14295.3 | 1.0 | 10214.8 | 5962.2 | 1.7 |
| Average | 1.9 | 3.1 | 2.3 | ||||||
‘High’, ‘Moderate’, and ‘Low’ refers to as three groupings of the transcript levels from the highest to the lowest FPKM in RNA-seq. Each grouping included one-third numbers of the m6A modified transcripts. t-test on ratio of the average MFPKM in m6A-seq to the average FPKM in RNA-seq in each grouping showed significantly different (p< 0.05) ratios between three groupings. ‘_1’ and ‘_2’ represent two replicates.
Table 5. Relationship between the m6A methlylation extent and the transcript level in the mitochondria transcriptome.
| Plant organs | High | Moderate | Low | ||||||
|---|---|---|---|---|---|---|---|---|---|
| MFPKM | FPKM | Ratio | MFPKM | FPKM | Ratio | MFPKM | FPKM | Ratio | |
| Leaves_1 | 23180.3 | 22058.9 | 1.1 | 9544.0 | 2080.8 | 4.6 | 4633.4 | 403.9 | 11.5 |
| Leaves_2 | 45144.4 | 39388.0 | 1.1 | 3678.0 | 1657.7 | 2.2 | 4282.0 | 373.6 | 11.5 |
| Average | 1.1 | 3.4 | 11.5 | ||||||
| Flowers_1 | 39204.6 | 41593.0 | 1.0 | 6451.7 | 1122.6 | 5.7 | 9909.9 | 291.0 | 34.1 |
| Flowers_2 | 44625.7 | 32068.5 | 1.4 | 4087.3 | 719.3 | 5.7 | 2412.2 | 178.6 | 13.5 |
| Average | 1.2 | 5.7 | 23.9 | ||||||
| Roots_1 | 186592.6 | 25723.2 | 7.3 | 7697.9 | 587.1 | 13.1 | 4160.2 | 138.3 | 30.1 |
| Roots_2 | 41081.8 | 38538.0 | 1.1 | 6729.9 | 667.1 | 10.1 | 1869.4 | 117.9 | 15.9 |
| Average | 4.4 | 12.6 | 23.0 | ||||||
‘High’, ‘Moderate’, and ‘Low’ refers to as three groupings of the transcript levels from the highest to the lowest FPKM in RNA-seq. Each grouping included one-third numbers of the m6A modified transcripts. t-test on ratio of the average MFPKM in m6A-seq to the average FPKM in RNA-seq in each grouping showed significantly different (p< 0.05) ratios between three groupings. ‘_1’ and ‘_2’ represent two replicates.
The transcripts extensively methylated by m6A in organelles
We found that ~15%, 6% and 8% of the modified transcripts were extensively methylated by m6A in the chloroplast/amyloplast of leaves, flowers and roots, respectively (with a ratio of MFPKM in the m6A-seq to FPKM in the RNA-seq ≥ 5, False discovery rate (FDR)<10−12, and the cleaned read number per transcript ≥ 20) (Tables 6 and 7). In total, 20 transcripts were found extensively methylated by m6A in the chloroplast/amyloplast of the three Arabidopsis organs (Table 6). These transcripts extensively modified by m6A were mainly associated with chloroplast-encoded ribosomal RNA, ribosomal proteins, photosystem reaction proteins or tRNA (Table 6).
Table 6. The transcripts presenting extensive high m6A methylation in the Arabidopsis chloroplast/amyloplast.
| Organs | Gene ID | Gene functions |
|---|---|---|
| Leaves | ATCG01160, ATCG00970, ATCG01210, ATCG01180, ATCG00950, ATCG00920 | Chloroplast-encoded ribosomal RNA[54] |
| ATCG00640, ATCG00650 | ribosomal proteins[55,56] | |
| ATCG00400 | tRNA[44] | |
| ATCG00550, ATCG00510 | photosystem reaction proteins[57] | |
| ATCG00140 | ATP synthase subunit C family protein[58] | |
| ATCG01130 | Ycf1 protein[59] | |
| ATCG01010 | NADH-Ubiquinone oxidoreductase[57] | |
| ATCG01040 | Cytochrome C assembly protein[57] | |
| ATCG00660 | PETG[55–57] | |
| Flowers | ATCG01180, ATCG01210, ATCG00950, ATCG00920 | Chloroplast-encoded ribosomal RNA[54] |
| ATCG00550 | photosystem reaction proteins[57] | |
| ATCG00140 | ATP synthase subunit C family protein[58] | |
| Roots | ATCG01180, ATCG00950, ATCG01210, ATCG00920 | Chloroplast-encoded ribosomal RNA[60] |
| ATCG01310, ATCG00110 | ribosomal proteins[57,60] | |
| ATCG00390 | tRNA[44] | |
| ATCG00890 | NADH-Ubiquinone/plastoquinone (complex I) protein |
Table 7. The transcripts presenting extensive high m6A methylation in the Arabidopsis mitochondria.
| Organs | Gene ID | Gene functions |
|---|---|---|
| Leaves | ATMG01380, ATMG00020, ATMG01390 | mitochondrial ribosomal RNA[54] |
| ATMG00030, ATMG01200, ATMG01130 | Proteins of unknown functions | |
| ATMG00650, ATMG00285 | NADH dehydrogenase subunits[61] | |
| ATMG00160 | Cytochrome oxidase 2 | |
| ATMG00280 | Ribulose bisphosphate carboxylase large chain | |
| Flowers | ATMG01380, ATMG00020, ATMG01390 | mitochondrial ribosomal RNA |
| ATMG00510, ATMG00650, ATMG00060, ATMG00070, ATMG00580 | NADH dehydrogenase subunits | |
| ATMG00640 | hydrogen ion transporting ATP synthases | |
| ATMG01130, ATMG00030, ATMG00660, ATMG00690 | Proteins of unknown functions | |
| ATMG00730 | cytochrome c oxidase subunit | |
| ATMG00080 | ribosomal protein | |
| ATMG01360 | cytochrome oxidase | |
| ATMG01170 | ATPase | |
| Roots | ATMG01380 | mitochondrial ribosomal RNA |
| ATMG00285, ATMG00510, ATMG00580, ATMG00513, ATMG01120, ATMG01320, ATMG00270, ATMG00650 | NADH dehydrogenase subunits | |
| ATMG00900, ATMG00830, ATMG00516, ATMG00180, ATMG00960 | cytochrome C biogenesis | |
| ATMG00980, ATMG00210, ATMG00080 | Ribosomal proteins | |
| ATMG00570 | Sec-independent periplasmic protein translocase | |
| ATMG00220 | apocytochrome b | |
| ATMG01190, ATMG00640 | ATP synthase | |
| ATMG00060, ATMG01020, ATMG01130, ATMG00660, ATMG01320 | Proteins of unknown functions | |
| ATMG00590 | Cytochrome b/b6 protein | |
| ATMG00560 | Nucleic acid-binding, OB-fold-like protein | |
| ATMG00640 | hydrogen ion transporting ATP synthases | |
| ATMG01170 | ATPase |
And ~20%, 34% and 45% of the modified transcripts were extensively methylated by m6A in the mitochondria of leaves, flowers and roots, respectively. In total, 38 transcripts were discovered extensively methylated by m6A in the mitochondria of the three Arabidopsis organs (Table 7). These transcripts extensively modified by m6A were mainly related with mitochondria-encoded ribosomal RNA, ribosomal proteins, NADH dehydrogenase subunits, protein for redox, or proteins of unknown functions (Table 7).
Differential m6A methylation across organs in the transcriptomes of two organelles
As we described in our previous study[35], we applied an algorithm ‘MFPKM in m6A-seq divided by LOG (FPKM in RNA-seq, 2) (NFPKM)’to each transcript to estimate differential m6A methylation among three organs of leaves, flowers and roots (see details in the Methods section of this paper). Two fold change and chi-square were applied for estimation of differential m6A methylation and differential gene transcript level between two organs[35].
On average, 72% of the transcripts in the chloroplast/amyloplast presented differential transcript level between two organs(fold change of FPKM between two organs > 2 or < 0.5, and FDR< 0.05). However, 64%of the modified transcripts in the chloroplast/amyloplast showed differential methylation between two organs (fold change of NFPKM between two organs > 2 or < 0.5, and FDR< 0.05) (Table 8). A paired analysis indicated that proportion of transcripts in the chloroplast/amyloplast showing differential transcript level across organs was higher than that showing differential m6A methylation extent among the three Arabidopsis organs (p< 0.003). On average, 69% of the transcripts in the mitochondria presented differential transcript level (fold change of FPKM between two organs > 2 or < 0.5, and FDR< 0.05). However, 79%of the m6A transcripts in the mitochondria showed differential methylation between two organs (fold change of NFPKM between two organs > 2 or < 0.5, and FDR< 0.05) (Table 9). Proportion of the transcripts in the mitochondria showing differential transcript level was significantly lower than that showing the differential m6A methylation extent among the three Arabidopsis organs (p< 0.05). The comparison also showed that two organelles in the leaves exhibited the highest extent of m6A methylation among three organs followed by that in the flower organelles. And the transcripts in the root organelles were less likely methylated among three organs (Table 8).
Table 8. The transcripts presenting differential transcript level and differential m6A methylation in the chloroplast/amyloplast among three organs in Arabidopsis (fold change >2 or <0.5, FDR < 0.02).
| Differential level | Leaves vs Flowers | Leaves vs Roots | Flowers vs Roots | |||
|---|---|---|---|---|---|---|
| Hi-leaves | Hi-flowers | Hi-leaves | Hi-roots | Hi-flowers | Hi-root | |
| Differential transcript level | ||||||
| Replicate 1 | ||||||
| No. of transcripts | 71 | 13 | 20 | 71 | 28 | 39 |
| Proportion (%) | 62 | 11 | 17 | 60 | 25 | 35 |
| Total (%) | 73 | 77 | 60 | |||
| Replicate 2 | ||||||
| No. of transcripts | 59 | 17 | 20 | 59 | 21 | 61 |
| Proportion (%) | 52 | 15 | 20 | 59 | 20 | 58 |
| Total (%) | 67 | 79 | 78 | |||
| Differential m6A extent | ||||||
| Replicate 1 | ||||||
| No. of transcripts | 60 | 13 | 47 | 41 | 45 | 21 |
| Proportion (%) | 52 | 11 | 39 | 34 | 40 | 19 |
| Total (%) | 63 | 73 | 59 | |||
| Replicate 2 | ||||||
| No. of transcripts | 34 | 30 | 36 | 24 | 30 | 23 |
| Proportion (%) | 36 | 32 | 40 | 27 | 33 | 25 |
| Total (%) | 68 | 67 | 58 | |||
Table 9. The transcripts presenting differential transcript level and differential m6A methylation in the mitochondria among three organs in Arabidopsis (fold change >2 or <0.5, FDR < 0.02).
| Differential level | Leaves vs Flowers | Leaves vs Roots | Flowers vs Roots | |||
|---|---|---|---|---|---|---|
| Hi-leaves | Hi-flowers | Hi-leaves | Hi-roots | Hi-flowers | Hi-root | |
| Differential transcript level | ||||||
| Replicate 1 | ||||||
| No. of transcripts | 29 | 8 | 32 | 8 | 15 | 15 |
| Proportion (%) | 57 | 16 | 53 | 13 | 29 | 29 |
| Total (%) | 73 | 66 | 58 | |||
| Replicate 2 | ||||||
| No. of transcripts | 27 | 8 | 32 | 3 | 28 | 0 |
| Proportion (%) | 61 | 18 | 67 | 6 | 64 | 0 |
| Total (%) | 79 | 73 | 64 | |||
| Differential m6A extent | ||||||
| Replicate 1 | ||||||
| No. of transcripts | 42 | 1 | 41 | 0 | 24 | 16 |
| Proportion (%) | 93 | 2 | 89 | 0 | 44 | 29 |
| Total (%) | 95 | 89 | 73 | |||
| Replicate 2 | ||||||
| No. of transcripts | 20 | 14 | 29 | 5 | 23 | 5 |
| Proportion (%) | 44 | 31 | 63 | 11 | 55 | 12 |
| Total (%) | 75 | 74 | 67 | |||
Six genes were randomly chosen for validation of our analysis of m6A differential methylation in the two organelles(S1 Table). Because the amplicons of qRT-PCR cover a short span in the transcriptome, 50 to 150 bp[33], two flanks of the amplicon containing one m6A peak in IGV program and showing differential m6A methylation were chosen to design primers (S1 Table). The correlation coefficient between the qRT-PCR and the RIP-seq results was significant (r = 0.9294, n = 18 genes, and p< 10−5), indicating that our qRT-PCR data were consistent with the data estimated by m6A-seq and RNA-seq using the IGV program (S2 Fig).
Discussion
Similarities and differences of m6A methylation between nucleus and organelle transcriptomes
MFPKM in m6A-seq represents the methylation level of the transcripts[35]. The average m6A MFPKM in the two organelles(Tables 4 and 5) was extensively (more than a hundred times) higher than that in the nucleus[35]. In addition, proportion of the m6A modified transcripts (over 90%) in the two organelles (Table 1) was also significantly higher than that (~73%) in the nucleus in Arabidopsis[35]. Therefore, the overall m6A methylation extent in the two organelles was greatly higher than that in the nucleus.
m6A motifs were similar between the nucleus and organelle transcriptome (Fig 3c and 3d), suggesting that recognition of motif for m6A methylation was conserved between the nucleus and the two organelles. Because the genes responsible for RNA m6A methylation have not been detected in the organelles, the enzymes of these genes may be expressed from nucleus and transported to the organelles form6A methylation therein.
m6A patterns in rRNAs were also similar between the nucleus and the organelles. For example, the whole rRNA transcripts were highly methylated by m6A in the both nucleus and organelles (Figs 5a and 6a)[35]. m6A patterns in tRNAs in the chloroplast/amyloplast were also similar to those in the nuclear transcripts (Fig 5b and 5c)[35]. However, m6A patterns in the coding RNAs were apparently different between nucleus and organelle transcripts. A dominant m6A peak nears top codon or in the 3′UTR was observed in most of the nuclear mRNA[14,17,18,23,35]. However, this dominant m6A peak was not observed in the coding RNAs in the two organelles (Figs 1 and 4). While most of the coding RNAs were highly methylated with numerous m6A peaks evenly distributing in the transcript exons including stop codon or 3′UTR though the extensively lower m6A peaks were observed in the introns of the coding RNAs in the two organelles(Figs 1 and 4). This suggested that regulation of the m6A methylation patterns may be somewhat different between the nuclei and the organelles.
This study also demonstrated that both of the transcript level and the m6A methylation extent in the transcriptome of two organelles(Tables 8 and 9) showed a higher differential ratio than that in the nuclear transcritptome[35].
Similarities and differences of m6A methylation between chloroplast/amyloplast and mitochondria
m6A patterns in the coding RNAs and rRNAs were similar between two organelles in Arabidopsis(Figs 1 and 4–6), suggesting an alike machinery for m6A methylation between the chloroplast/amyloplast and mitochondria. However, the m6A patterns in tRNAs were distinct between two organelles. For example, most of tRNAs were highly methylated by m6A in the chloroplast/amyloplast (Fig 5c). However, only few tRNAs was detected to be methylated by m6A in the mitochondria (Fig 6b). This may be due to too low transcript level of tRNAs for detection of their m6A methylation because very few tRNAs with an extremely low transcript level were also observed in the RNA-seq data in the mitochondria.
The average m6A MFPKM in the chloroplast/amyloplast (Table 4) was significantly higher than that in the mitochondria (Table 5) (p<0.001). In addition, proportion of the m6A modified transcripts (nearly 100%) in the chloroplast/amyloplast was also significantly higher than that (over 90%) in the mitochondria (Table 1, p< 0.05). Therefore, the overall m6A methylation extent in the chloroplast/amyloplast was higher than that in the mitochondria.
Potential functions of m6A methylation in the transcriptome of two organelles
m6A methylation in the nucleus acts as a signal for transport of RNA from the nucleus to the cytoplasm[17,21]. The dysfunction of m6A methylation may result in a failure of RNA transport from the nucleus to the cytoplasm, or RNA degradation[21]. Nuclear mRNAs translated into proteins located in mitochondria or chloroplast were also found highly methylated by m6A in our previous study[35]. The overall m6A methylation extent in the two organelles was found in this study (Tables 1, 4 and 5) extensively higher than that in the nuclear transcripts[35]. However, the biological functions responsible for this phenomenon need further investigation.
In the both chloroplast and mitochondria, introns were much less methylated than exons in the coding RNAs (Figs 1b and 4b). This phenomenon was similar to that in the nuclear mRNAs[35], suggesting that m6A methylation in the two organelles may also be responsible for RNA splicing[17,35]. Mitochondria confer a role of regulation of cellular proliferation and differentiation[46]. m6A in the nuclear transcripts is also related to regulation of differentiation and fate of the stem cells[10,47]. However, effects of m6A methylation in the organelles on the cellular proliferation and differentiation need further investigation.
Expressions of some genes were mutually regulated by each other between the organelle and the nucleus. Whether and how m6A methylation in the nucleus regulates gene expression in the two organelles, or vice versa, is unclear. Gene silencing of METTL3, a gene responsible for m6A modification, can result in an arrest of embryo development at the globular stage in Arabidopsis[28]. The male-infertility line plays an important role in crop breeding[48–51]. Some infertility phenomena are caused by organelle dysfunctions or interactions between the organelle and nucleus genes[52,53]. Nevertheless, an investigation whether m6A methylation in the organelles affects fertility and development of the reproductive organ may provide insights in our future breeding programs.
High productivity in crops is highly related to relative high photosynthesis and low respiration in plants[36,37]. Chloroplast will be switched into amyloplast in mature seeds, fruits or tubes, and mainly used for food storage in plants[37]. The transcripts expressed for photosystem reaction proteins, NADH dehydrogenase subunits, and protein for redox, were extensively methylated by m6A in the two organelles (Tables 6 and 7). m6A methylation has been found to have function in regulation of gene expression[23,24]. Further studies in investigation of molecular functions of m6A methylation in the chloroplast/amyloplast and mitochondria may facilitate our better control of metabolisms in these two organelles, thus to potentially increase crop productivity to ensure the global food and energy security in the future.
Conclusions
To our knowledge, this is the first study for comprehensive and transcriptome-wide characterization of RNA m6A patterns, relationship between m6A methylation extent and gene transcript level, and differential features of m6A methylation across leaves, flowers and roots in the chloroplast/amyloplast and mitochondria.
Over 600 and 400 m6A sites were successfully mapped to the Arabidopsis chloroplast/amyloplast and mitochondrial genomes, respectively, with an estimation of ~4.6 to ~5.8 m6A sites per m6A transcript, in the two organelles. Over 86% of the transcripts were chemically modified by m6A in the two organelle transcriptomes. Around two thirds of the m6A sites in the transcripts in the two organelles contained motifs, ‘RRm6ACH’, which were similar to that in the nuclear transcripts. The average MFPKM of m6A-seq in the chloroplast/amyloplast and mitochondria was over a hundred times higher than that in the nucleus. In most cases, the m6A methylation extent was relatively high compared to the transcript level in the two organelles (p< 0.05). The m6A extensively methylated transcripts in the two organelles were mainly associated with ribosomal RNA, ribosomal proteins, photosystem reaction proteins, NADH dehydrogenase subunits, protein for redo or tRNA. The m6A patterns in rRNAs were similar between the nucleus and organelle transcripts, i.e, the whole rRNAs were highly methylated by m6A. A dominant m6A peak enriched near stop codon or at 3'UTR in most of the nuclear mRNAs was not observed in the coding RNAs in the chloroplast/amyloplast and mitochondria. On average, 64% and 79% of the transcripts showed differential m6A methylation across three organs in the chloroplast/amyloplast and mitochondria, respectively. Intriguingly, the overall m6A methylation extent in the chloroplast/amyloplast was greatly higher than that in the mitochondria.
Supporting information
(A) RNA quality of the total RNA for the RNA-seq sample was high with RIN over 8.5. (B) RNA fragmentation for the m6A-seq samples was consistent in the RIP experiments, with an average length of 106 nt.
(PDF)
(A) RA for ‘ATCG00360’. (B) EA for ‘ATCG00360’. (C) RA for ‘ATCG0083’. (D) EA for ‘ATCG0083’. (E) RA for ‘ATCG00890’. (F) EA for ‘ATCG00890’. (G) RA for ‘ATMG00510’. (H) EA for ‘ATMG00510’. (I) RA for ‘ATMG00513’. (J) RA for ‘ATMG00513’. (K) RA for ‘ATMG00580’. (L) EA for ‘ATMG00580’.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability
All data are available from the GEO database (accession number GSE72706).
Funding Statement
This work was funded by the 2012 Shaanxi Province Fund for Returnees Scientists from Foreign Study (A289021201), China Scholarship Council Project (22861057), Initiation Grant to Dr. Yizhen Wan as the Emeritus Professor by Jiangsu University of Science and Technology, USA NIH grants (R01GM070795 and R01GM059138) to Kai Tang, and National Science Foundation of China (31101166) to Dayong Zhang. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Finkel D, and Groner Y. Methylations of adenosine residues (m6A) in pre-mRNA are important for formation of late simian virus 40 mRNAs. Virology.1983; 131:409–425. doi: 10.1016/0042-6822(83)90508-1 [DOI] [PubMed] [Google Scholar]
- 2.Kane SE, and Beemon K. Precise localization of m6A in Rous sarcoma virus RNA reveals clustering of methylation sites: implications for RNA processing. Mol Cell Biol. 1985; 5: 2298–2306. doi: 10.1128/MCB.5.9.2298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matera AG, Terns RM and Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev. 2007; 8: 209–220. doi: 10.1038/nrm2124 [DOI] [PubMed] [Google Scholar]
- 4.Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, et al. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res.2011; 39: D195–201. doi: 10.1093/nar/gkq1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Globisch D, Pearson D, Hienzsch A, Bruckl T, Wagner M, Thoma I, et al. Systems-based analysis of modified tRNA bases. AngewChemIntEd Engl. 2011; 50: 9739–9742. doi: 10.1002/anie.201103229 [DOI] [PubMed] [Google Scholar]
- 6.He C. Grand challenge commentary: RNA epigenetics? Nat Chem Biol. 2010;6:863–865. doi: 10.1038/nchembio.482 [DOI] [PubMed] [Google Scholar]
- 7.Pan T. N6-methyl-adenosine modification in messenger and long non-coding RNA. Trends Biochem Sci. 2013; 38: 204–209. doi: 10.1016/j.tibs.2012.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee M, Kim B and Kim VN. Emerging roles of RNA modification: m(6)A and U-tail. Cell. 2014;158: 980–987. doi: 10.1016/j.cell.2014.08.005 [DOI] [PubMed] [Google Scholar]
- 9.Bodi Z, Bottley A, Archer N, May ST and Fray RG. Yeast m6A methylated mRNAs are enriched on translating ribosomes during meiosis, and under rapamycin treatment. PLoS One. 2015;10: e0132090 doi: 10.1371/journal.pone.0132090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen T, Hao YJ, Zhang Y, Li MM, Wang M, Han W, et al. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell. 2015; 16: 289–301. doi: 10.1016/j.stem.2015.01.016 [DOI] [PubMed] [Google Scholar]
- 11.Liu J, and Jia G. Methylation modifications in eukaryotic messenger RNA. J Genet Genomics. 2014;41: 21–33. doi: 10.1016/j.jgg.2013.10.002 [DOI] [PubMed] [Google Scholar]
- 12.Desrosiers R, Friderici K and Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci USA. 1974;71: 3971–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei CM, Gershowitz A and Moss B. Methylated nucleotides block 5' terminus of HeLa cell messenger RNA. Cell. 1975; 4:379–386. doi: 10.1016/0092-8674(75)90158-0 [DOI] [PubMed] [Google Scholar]
- 14.Bodi Z, Button JD, Grierson D and Fray RG. Yeast targets for mRNA methylation. Nucleic Acids Res. 2010; 38: 5327–5335. doi: 10.1093/nar/gkq266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell.2014; 15: 707–719. doi: 10.1016/j.stem.2014.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grosjean H. Fine-tuning of RNA functions by modification and editing. 2005; Springer-Verlag. [Google Scholar]
- 17.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012; 485: 201–206. doi: 10.1038/nature11112 [DOI] [PubMed] [Google Scholar]
- 18.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE and Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell.2012;149: 1635–1646. doi: 10.1016/j.cell.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bodi Z, Zhong S, Mehra S, Song J, Graham N, Li H, et al. Adenosine methylation in Arabidopsis mRNA is associated with the 3' End and reduced levels cause developmental defects. Front Plant Sci. 2012; 3: 48 doi: 10.3389/fpls.2012.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwartz S, Agarwala SD, Mumbach MR, Jovanovic M, Mertins P, Shishkin A, et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell.2013; 155: 1409–1421. doi: 10.1016/j.cell.2013.10.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature.2014; 505:117–120. doi: 10.1038/nature12730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell.2013;155:793–806. doi: 10.1016/j.cell.2013.10.026 [DOI] [PubMed] [Google Scholar]
- 23.Meyer KD and Jaffrey SR. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol. 2014; 15: 313–326. doi: 10.1038/nrm3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu Y, Dominissini D, Rechavi G and He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet. 2014; 5: 293–306. doi: 10.1038/nrg3724 [DOI] [PubMed] [Google Scholar]
- 25.Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science.2015; 347: 1002–1006. doi: 10.1126/science.1261417 [DOI] [PubMed] [Google Scholar]
- 26.Liu N, Dai Q, Zheng G, He C, Parisien Mand Pan T. N6-methyladenosine-dependent RNA structural switches regulate RNA–protein interactions. Nature.2015; 518:560–564. doi: 10.1038/nature14234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niu Y, Zhao X, Wu YS, Li MM, Wang XJ and Yang YG. N6-methyl-adenosine (m6A) in RNA: an old modification with a novel epigenetic function. Genomics Proteomics Bioinformatics.2013; 11: 8–17. doi: 10.1016/j.gpb.2012.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong S, Li H, Bodi Z, Button J, Vespa L, Herzog M, et al. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell.2008; 20:1278–1288. doi: 10.1105/tpc.108.058883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011; 7:885–887. doi: 10.1038/nchembio.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu N, Parisien M, Dai Q, Zheng G, He C and Pan T. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA.2013; 19:1848–1856. doi: 10.1261/rna.041178.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res.2014; 24: 177–189. doi: 10.1038/cr.2014.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo GZ, MacQueen A, Zheng G, Duan H, Dore LC, Lu Z, et al. Unique features of the m6Amethylome in Arabidopsis thaliana. Nat Commun. 2014; 5: 5630 doi: 10.1038/ncomms6630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Wang X, Li C, Hu S, Yu J and Song S. Transcriptome-wide N(6)-methyladenosine profiling of rice callus and leaf reveals the presence of tissue-specific competitors involved in selective mRNA modification. RNA Biol. 2014;11:1180–1188. doi: 10.4161/rna.36281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wan Y, Tang K, Zhang D, Xie S, Zhu X, Wang Z, et al. Transcriptome-wide high-throughput deep m(6)A-seq reveals unique differential m(6)A methylation patterns between three organs in Arabidopsis thaliana. Genome Biol. 2015; 16: 272 doi: 10.1186/s13059-015-0839-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henze K and Martin W. Evolutionary biology: essence of mitochondria. Nature. 2003; 426: 127–128. doi: 10.1038/426127a [DOI] [PubMed] [Google Scholar]
- 37.Nakayama T and Archibald JM. Evolving a photosynthetic organelle. BMC Biol. 2012; 10: 35 doi: 10.1186/1741-7007-10-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wise R. The Diversity of Plastid Form and Function. 2007; Springer; 23, 3–26. 39. [Google Scholar]
- 39.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R and Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013; 14: R36 doi: 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAM tools. Bioinformatics.2009; 25: 2078–2079. doi: 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liao Y, Smyth GK and Shi W. Feature Counts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30: 923–930. doi: 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- 42.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G and Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011; 29: 24–26. doi: 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010; 28: 511–515. doi: 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sato S, Nakamura Y, Kaneko T, Asamizu E and Tabata S. Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Res. 1999; 6: 283–290. doi: 10.1093/dnares/6.5.283 [DOI] [PubMed] [Google Scholar]
- 45.Csepany T, Lin A, Baldick CJ Jr and Beemon K. Sequence specificity of mRNA N6-adenosine methyltransferase. J Biol Chem. 1990; 265: 20117–20122. [PubMed] [Google Scholar]
- 46.Weinberg F and Chandel NS. Mitochondrial metabolism and cancer. Ann N Y Acad Sci. 2009; 1177: 66–73. doi: 10.1111/j.1749-6632.2009.05039.x [DOI] [PubMed] [Google Scholar]
- 47.Zhao BS and He C. Fate by RNA methylation: m6A steers stem cell pluripotency. Genome Biol. 2015; 16: 43 doi: 10.1186/s13059-015-0609-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bongaarts J and Potter RG. Fertility, biology, and behavior: an analysis of the proximate determinants. 1983; Academic Press, New York. [Google Scholar]
- 49.Stover J. Revising the proximate determinants of fertility framework: what have we learned in the past 20 years? Stud Fam Plann. 1998; 29: 255–267. doi: 10.2307/172272 [PubMed] [Google Scholar]
- 50.Richards A.J. (1986).Plant breeding systems. London: George Allen &Unwin. [Google Scholar]
- 51.Rao MK, Devi KU and Arunilati A. Applications of genic male sterility in plant breeding. Plant Breeding.1990;105:1–25. doi: 10.1111/j.1439-0523.1990.tb00447.x [Google Scholar]
- 52.Couvet D, Bonnemaison F and Gouyon PH. The maintenance of females among hermaphrodites: the importance of nuclear-cytoplasmic interactions. Heredity.1986; 57: 325–30. doi: 10.1038/hdy.1986.130 [Google Scholar]
- 53.De Haan AA, Luyten RMJM, Bakx-Schotman TJMT and Van Damme JMM. The dynamics of gynodioecy in Plantagolanceolata L. I. Frequencies of male-steriles and their cytoplasmic male sterility types. Heredity.1997;79: 453–62. doi: 10.1038/hdy.1997.184 [Google Scholar]
- 54.Spremulli L.Protein synthesis, assembly, and degradation In: Biochemistry and Molecular Biology of Plant. 2000; American Society of Plant Biologists. [Google Scholar]
- 55.Zybailov B, Rutschow H, Friso G, Rudella A, Emanuelsson O, Sun Q, et al. Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS One. 2008;3: e1994 doi: 10.1371/journal.pone.0001994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bruley C, Dupierris V, Salvi D, Rolland N and Ferro M. AT_CHLORO: A chloroplast protein database dedicated to sub-plastidial localization. Front Plant Sci. 2012; 3: 205 doi: 10.3389/fpls.2012.00205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, et al. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005; 37: 501–506. doi: 10.1038/ng1543 [DOI] [PubMed] [Google Scholar]
- 58.Peltier JB, Ytterberg AJ, Sun Q and van Wijk KJ. New functions of the thylakoid membrane proteome of Arabidopsis thaliana revealed by a simple, fast, and versatile fractionation strategy. J Biol Chem. 2004; 279: 49367–49383. doi: 10.1074/jbc.M406763200 [DOI] [PubMed] [Google Scholar]
- 59.Kikuchi S, Bedard J, Hirano M, Hirabayashi Y, Oishi M, Imai M, et al. Uncovering the protein translocon at the chloroplast inner envelope membrane. Science.2013; 339: 571–574. doi: 10.1126/science.1229262 [DOI] [PubMed] [Google Scholar]
- 60.Mitra SK, Gantt JA, Ruby JF, Clouse SD and Goshe MB. Membrane proteomic analysis of Arabidopsis thaliana using alternative solubilization techniques. J Proteome Res. 2007; 6: 1933–1950. doi: 10.1021/pr060525b [DOI] [PubMed] [Google Scholar]
- 61.Sugiura M and Takeda Y. Respiration and Photorespiration. In: Biochemistry & Molecular Biology of Plants. 2005; Amer Soc Plant Biol.pp.676. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) RNA quality of the total RNA for the RNA-seq sample was high with RIN over 8.5. (B) RNA fragmentation for the m6A-seq samples was consistent in the RIP experiments, with an average length of 106 nt.
(PDF)
(A) RA for ‘ATCG00360’. (B) EA for ‘ATCG00360’. (C) RA for ‘ATCG0083’. (D) EA for ‘ATCG0083’. (E) RA for ‘ATCG00890’. (F) EA for ‘ATCG00890’. (G) RA for ‘ATMG00510’. (H) EA for ‘ATMG00510’. (I) RA for ‘ATMG00513’. (J) RA for ‘ATMG00513’. (K) RA for ‘ATMG00580’. (L) EA for ‘ATMG00580’.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
All data are available from the GEO database (accession number GSE72706).