Abstract
Muga (Antheraea assamensis) is an economically important silkmoth endemic to the states of Assam and Meghalaya in India and is the producer of the strongest known commercial silk. However, there is a scarcity of genomic and proteomic data for understanding the organism at a molecular level. Our present study is on decoding the complete mitochondrial genome (mitogenome) of A. assamensis using next generation sequencing technology and comparing it with other available lepidopteran mitogenomes. Mitogenome of A. assamensis is an AT rich circular molecule of 15,272 bp (A+T content ~80.2%). It contains 37 genes comprising of 13 protein coding genes (PCGs), 22 tRNA and 2 rRNA genes along with a 328 bp long control region. Its typical tRNAMet-tRNAIle-tRNAGln arrangement differed from ancestral insects (tRNAIle-tRNAGln-tRNAMet). Two PCGs cox1 and cox2 were found to have CGA and GTG as start codons, respectively as reported in some lepidopterans. Interestingly, nad4l gene showed higher transversion mutations at intra-species than inter-species level. All PCGs evolved under strong purifying selection with highest evolutionary rates observed for atp8 gene while lowest for cox1 gene. We observed the typical clover-leaf shaped secondary structures of tRNAs with a few exceptions in case of tRNASer1 and tRNATyr where stable DHU and TΨC loop were absent. A significant number of mismatches (35) were found to spread over 19 tRNA structures. The control region of mitogenome contained a six bp (CTTAGA/G) deletion atypical of other Antheraea species and lacked tandem repeats. Phylogenetic position of A. assamensis was consistent with the traditional taxonomic classification of Saturniidae. The complete annotated mitogenome is available in GenBank (Accession No. KU379695). To the best of our knowledge, this is the first report on complete mitogenome of A. assamensis.
Introduction
Mitochondria are known to have descended from α-proteobacterium endosymbionts and have retained numerous bacterial features [1]. Apart from being the powerhouse of the cell, they are involved in various cellular processes like fatty acid metabolism, apoptosis and aging [2]. These functions are carried out by many nuclear-encoded genes along with extra-nuclear genes that include protein-coding genes (PCGs), transfer RNA (tRNA) and ribosomal rRNA (rRNA) genes and are present in the mitogenome which is mostly circular and self-replicative. It also consists of several non-coding regions with the longest being AT-rich control region comprising of several conserved regions and repeats. Although small in size, the mitogenome governs maternal inheritance and has several unique features like faster evolution rate, low or absence of homologous recombination, evolutionary conserved gene products and richness in genetic polymorphism. This makes it a potential marker for barcoding, phylogeographic and phylogenetic studies [3]. It plays a potential role in molecular evolutionary studies by elucidating evolutionary models and substitution patterns that vary timely and across sequences. Compared to individual genes, whole mitogenomes are more informative phylogenetic models due to its multiple genome level features like gene position, content, secondary structures of RNA and control region.
Over the past few decades, animal mitogenomes, particularly insects (~80% of the sequenced arthropods), have been widely studied for comparative genomics and molecular systematics [3–4]. More than 250 Lepidopteran insect mitogenomes have been sequenced till now using Sanger sequencing and next generation sequencing (NGS) technologies as found in GenBank (https://www.ncbi.nlm.nih.gov/). The mitogenome ranges from 14–16 kilo basepairs (kb) in majority of the lepidopterans and consists of 37 genes (13 PCGs, 2 rRNA genes, 22 tRNA genes) along with a control region. The PCGs encode 2 subunits of ATPase (ATP6 and ATP8), 3 subunits of cytochrome c oxidase (COI, COII and COIII), 1 subunit of cytochrome b (CYTB) and 7 subunits of NADH dehydrogenase (ND1, ND2, ND3, ND4, ND4L, ND5 and ND6). These proteins are responsible for oxidative phosphorylation (OXPHOS) as they form essential mitochondrial membrane-associated protein complex systems [3]. The control region plays a role in the replication and transcription of mitogenome. The utilities and potential of mitochondrial PCGs (cox1 and cox2) as barcode markers have been well demonstrated in the order Lepidoptera [5]. The comparative mitogenome analysis also elucidated sequence divergence patterns among domesticated and non-domesticated lepidopteran mitogenomes [6–7]. Although frequency of mitogenome sequencing of lepidopterans has increased, the evolutionary relationships among many family members of the same order have been hardly investigated [8–9].
Saturniidae is the most diverse family of wild silkmoths which include giant moths, royal moths and emperor moths. Most of these silkmoths are unexplored and might have potential significance in sericulture [10]. Till date, mitogenome data of eleven species have been sequenced and made available in GenBank. However, numerous individual gene sequences of various wild species are also available whose complete mitogenome sequencing has not been done. Antheraea assamensis, muga silkmoth is a semi-domesticated and one of the economically important moths of the Saturniidae family. It is endemic to erstwhile undivided Assam and its adjoining hilly areas located in the North-Eastern part of India. It is a multivoltine and polyphagous moth that primarily thrives on two main host plants Persea bombycina and Litsea monopetala [11]. Its silk has wide applications in textile industry and has great potential as a biomaterial due to its unique biophysical properties like golden lustre, tenacity and absorption of UV radiation [12]. Like other Antheraea species, it produces reelable silk which is the most expensive silks of the world. However, its semi-domesticated nature and extraction of fibroin directly from cocoon fibers limit its extensive rearing and prospects of global applicability as a biomaterial. The whole genome of A. assamensis is not yet available. However, de novo transcriptome data from our laboratory is available in GenBank (Accession Number- SRX1293136, SRX1293137 and SRX1293138) [13].
In the present study, we report the whole mitogenome sequence of A. assamensis using NGS and comparative analysis of its sequences and genome architectures with that of the other lepidopterans. The comparative study was based on several characteristics such as genome arrangement, PCGs, tRNAs, rRNAs, nucleotide composition, codon usage, evolutionary rates, gene divergence, conserved regions in control region, etc. Furthermore, phylogenetic trees inferred using datasets like nucleotide sequences of 13 PCGs and 13 PCGs+2 rRNAs were analyzed to elucidate the relationships among lepidopteran insects. This study will facilitate a better understanding of the comparative and evolutionary biology of A. assamensis with the other lepidopteran insects.
Materials and methods
Sample processing, DNA sequencing and assembly
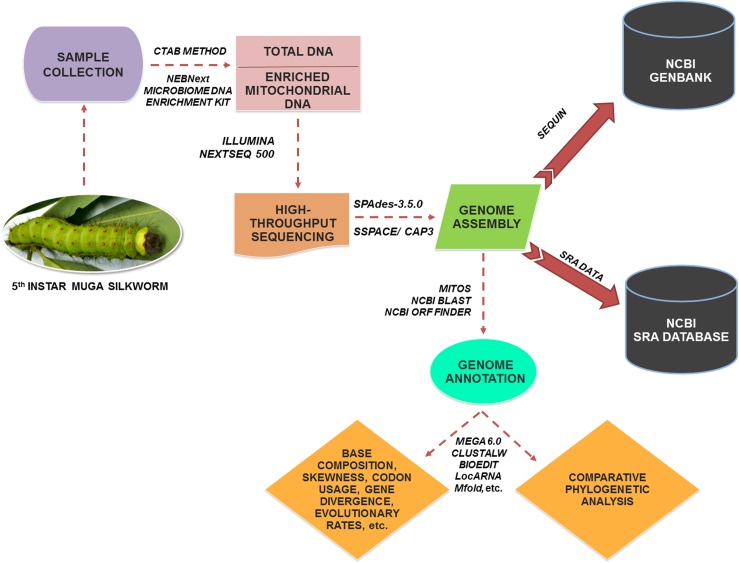
The larvae of A. assamensis were reared on Persea bombycina or Som (a primary host plant) in the experimental field of Central Muga Eri Research and Training Institute, Lahdoigarh, Jorhat, Assam, India following recommended package of practices from brushing till attainment of maturity [14]. The effective rate of rearing of silkworms referred as the number of mature larvae collected from the total brushed larvae was found to be 61.63%. Other rearing parameters like body weight of mature larvae (11.84 g), cocoon weight (6.26 g) and shell weight (0.55 g) were found to be suitable for experimental work. The fifth instar larval stage of A. assamensis was used for the present study (Sample ID: CMERI-Aa-001). The larvae were sterilized by washing with 70% ethanol before being processed for mitogenome studies. The larvae were stored in 95% absolute ethanol at -80°C for future use. The total DNA was extracted using CTAB (Cetyl trimethylammonium bromide) based buffer and silica column by the service provider (Genotypic Technology Pvt. Ltd. Bengaluru, India). Subsequently, mitochondrial DNA was enriched from total DNA extracted once the integrity, quantity and purity of extracted DNA was confirmed by agarose gel electrophoresis, light absorbance and fluorescence spectroscopy. The complete overview of sequencing and analysis of mitochondrial genome of A. assamensis is represented in Fig 1.
Fig 1. A pictorial overview of the methodologies used for sequencing and analysis of A. assamensis mitogenome.
Briefly, the preferential enrichment of mitochondrial DNA was carried out with NEBNext microbiome DNA enrichment kit (New England Biolabs, USA) which selectively removes CpG-methylated eukaryotic nuclear DNA. The enriched mitochondrial DNA was subjected to DNA sequencing at Genotypic Technology’s Genomics facility where the enriched DNA was acoustically sheared to 300–500 bp using a specially designed Covaris microTube (for focused ultra-sonication). The fragmented mitochondrial DNA was cleaned up using HighPrep beads (MagBio Genomics, Inc, Gaithersburg, Maryland) and subjected to end-repair, A-tailing and ligation with multiplex adaptors using the NEXTFlex DNA Sequencing kit (Catalogue # 5140–02, Bioo Scientific). The ligated DNA was cleaned with HighPrep beads and subjected to amplification via PCR as follows: initial denaturation at 98˚C for 2 min; 10 cycles of denaturation at 98˚C for 30 sec, annealing at 65˚C for 30 sec and extension at 72˚C for 60 sec; and a final extension at 72˚C for 4 min) using the primers provided by NEXTFlex DNA Sequencing kit. Finally, the PCR product was purified with HighPrep beads, quantified and fragment range was assessed using Qubit fluorometer and the Agilent D1000 Tape (Agilent Technologies), respectively prior to sequencing. The sequencing was carried out in Illumina NextSeq 500 sequencer (Illumina Inc, Sandiego, USA) through 2 × 150 bp paired-end chemistry. The raw paired end data obtained was de-multiplexed using Bcl2fastQ, assessed with FastQC tool [15] to remove low quality bases (Q<30) and preprocessed using ABLT-Scripts (Genotypic technology, Bangalore India). The SPAdes-3.5.0 was used for sequence assembly followed by the filling of gaps. Finally, scaffolding of assembled contigs and clustering were carried out with SSPACE and CAP3 programs, respectively [16–17].
Genome annotation, visualization and comparative analysis
The assembled scaffolds were annotated using MITOS web-server to predict the location of protein coding regions/genes, tRNAs, rRNAs and secondary structures of tRNAs. MITOS is a widely used webserver for the annotation of metazoan mitochondrial genomes due to its advanced annotation methodology [18]. The location of PCGs, tRNAs and rRNAs was further confirmed using tools like NCBI (National Center for Biotechnology Information)–BLAST, BioEdit and Clustal Omega by comparing the sequences of A. assamensis with the respective sequences published for other lepidopteran insects [19–20]. The boundaries of PCGs i.e. initiation and termination codons were determined using NCBI ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) by specifying the invertebrate mitochondrial genetic code. Further, the number of overlapping or spacer regions between the genes were visualized and calculated manually. The control region was validated by comparing with the available sequences in GenBank and the tandem repeats in this region were searched through Tandem Repeat Finder [20–21]. Then, the whole mito-map was constructed and visualized using Blast Ring Image Generator (BRIG) tool [22]. Finally, the complete annotated file of mitochondrial genome was prepared using NCBI Sequin tool (http://www.ncbi.nlm.nih.gov/Sequin/) and the sequin file along with SRA data were submitted to NCBI GenBank.
For understanding evolutionary relationships of A. assamensis with various lepidopterans as listed in Table 1, the sequences of whole mitogenome, coding regions, tRNAs, rRNAs and control region were retrieved from the NCBI GenBank database and a comparative analysis was performed.
Table 1. List of species considered for comparative mitogenome study.
(Highlighted ones are the organisms of Bombycoidea superfamily used for comparative mitogenome analysis with respect to A. assamensis).
| Superfamily | Family | Species | Size (bp) | Accession No. | References |
|---|---|---|---|---|---|
| Bombycoidea | Saturniidae | Antheraea assamensis | 15,272 | KU379695 | This study |
| Attacus atlas | 15,282 | KF006326 | [23] | ||
| Samia cynthia ricini | 15,384 | JN215366 | [24] | ||
| Antheraea pernyi | 15,575 | AY242996 | [25] | ||
| Antheraea yamamai | 15,338 | EU726630 | [26] | ||
| Antheraea frithi | 15,338 | NC_027071 | GenBank | ||
| Actias selene | 15,236 | NC_018133 | [27] | ||
| Saturnia boisduvalii | 15,360 | NC_010613 | [28] | ||
| Eriogyna pyretorum | 15,327 | FJ685653 | [29] | ||
| Bombycidae | Bombyx mandarina | 15,682 | AY301620 | [30] | |
| Bombyx mori | 15,656 | AB070264 | [31] | ||
| Rondotia menciana | 15,301 | KC881286 | [32] | ||
| Sphingidae | Manduca sexta | 15,516 | EU286785 | [33] | |
| Sphinx morio | 15,299 | KC470083 | [34] | ||
| Geometroidea | Geometridae | Biston panterinaria | 15,517 | NC_020004 | [35] |
| Phthonandria atrilineata | 15,499 | NC_010522 | [36] | ||
| Cossoidea | Cossidae | Eogystia hippophaecolus | 15,431 | KC831443 | [37] |
| Papilionoidea | Riodinidae | Abisara fylloides | 15,301 | HQ259069 | [38] |
| Lycaenidae | Spindasis takanonis | 15,349 | HQ184266 | [39] | |
| Protantigius superans | 15,248 | HQ184265 | [39] | ||
| Papilionidae | Papilio maraho | 16,094 | FJ810212 | [40] | |
| Parnassius bremeri | 15,389 | NC_014053 | [41] | ||
| Nymphalidae | Danaus plexippus | 15,314 | NC_021452 | GenBank | |
| Eumenis autonoe | 15,489 | GQ868707 | [42] | ||
| Noctuoidea | Lymantriidae | Gynaephora menyuanensis | 15,770 | KC185412 | [43] |
| Lymantria dispar | 15,569 | NC_012893 | GenBank | ||
| Yponomeutoidea | Plutellidae | Plutella xylostella | 16,014 | KM023645 | [44] |
| Tortricoidea | Tortricidae | Adoxophyes honmai | 15,680 | DQ073916 | [45] |
| Cydia pomonella | 15,253 | JX407107 | [46] | ||
| Pyraloidea | Crambidae | Ostrinia furnacalis | 14,536 | NC_003368 | [47] |
| Ostrinia nubilalis | 14,535 | NC_003367 | [47] | ||
| Diptera | Drosophila melanogaster | 19,517 | NC_001709 | [48] | |
| (Outgroup) | Drosophila yakuba | 16,019 | NC_001322 | [49] | |
| Anopheles gambiae | 15,363 | NC_002084 | [50] |
Comparative mitogenome analysis
Comparative mitogenome analysis was carried out in order to find out similarities and differences of A. assamensis with other insects in terms of characteristics like genome size, organization, structure, gene content and rearrangement. The sequence homology of A. assamensis mitogenome with selected insects was also performed using Clustal Omega [20].
Comparative analysis among protein coding genes (PCGs)
The PCGs of A. assamensis were compared with selected organisms based on their number, length, initiation and termination codons. The sequence similarity among the PCGs was determined by aligning the sequences of A. assamensis with selected Bombycoids using Clustal Omega. MEGA 6.0 tool was used for determining the gene-by-gene divergences in 13 PCGs in terms of phylogenetically informative sites, conserved sites as well as variable sites [51]. The same tool was used for estimating nucleotide substitution and evolutionary rates among the genes in terms of transitions (ts), transversions (tv), rate of synonymous substitutions (Ks), rate of non-synonymous substitutions (Ka) and their ratios. Further, the correlation between GC content and Ka/Ks ratio was studied in order to predict the effect of GC content on evolutionary rates of PCGs.
Comparative analysis among transfer RNAs (tRNAs)
The length, arrangement, secondary structures and variation in the structures were studied among the selected organisms. Homologous sites in their secondary structures were identified by aligning the tRNA sequences using LocARNA tool and the type/number of mismatches were calculated [52].
Comparative analysis among ribosomal RNAs (rRNAs)
The number, type, location and length of rRNAs in A. assamensis were compared with the selected organisms in order to study the conservation pattern. In addition, sequence homology of rRNAs of A. assamensis with the selected organisms was determined and gene-by-gene divergences among the rRNAs were studied using MEGA 6.0 tool [51].
Comparative analysis among control region (A+T rich region)
The control region of A. assamensis was compared with selected organisms based on the length and location. Multiple sequence alignment of control region was then carried out using Clustal Omega and their conserved regions, repeats and indels were visualized using BioEdit tool [19].
Comparative analysis among overlapping sequence (OS) and intergenic spacer (IGS) regions
The OS and IGS regions of A. assamensis mitogenome were compared with selected organisms (Table 1) in terms of number, length and location. The sequence homology of OS and IGS was determined through sequence alignment using Clustal Omega and BioEdit tools to find conserved motifs.
Comparative analysis with respect to nucleotide composition, skewness and codon usage
The nucleotide composition of sequences of whole mitochondrial genome, concatenated and individual PCGs, tRNAs, rRNAs, spacers and control region was calculated using MEGA 6.0 software [51]. The base composition skewness was also calculated for all the regions of mitogenome using the formula (Eq 1 and Eq 2) described by Junqueira et al., 2004 [53].
| (1) |
| (2) |
Where A, T, G and C denote the frequencies of respective bases.
The codon usage and relative synonymous codon usage (RSCU) values of PCGs were determined using MEGA 6.0.
Comparative phylogenetic analysis
About 34 organisms representing 13 different families of 8 super families within the order Lepidoptera were considered for phylogenetic analysis along with three organisms belonging to the order Diptera as outgroups (Table 1). The phylogenetic relationship of A. assamensis was elucidated using two criteria’s: (i) concatenated nucleotide sequences of 13 PCGs and (ii) concatenated nucleotide sequences of 13 PCGs and 2 rRNAs.
The nucleotide sequences of each PCG were translated via amino acid alignment using MAFFT algorithm in TranslatorX server which were then back translated into nucleotide alignment with GBlocks [54]. The individual nucleotide sequences obtained from GBlocks and rRNA sequences were then concatenated for further phylogenetic analysis. Substitution model optimization for each dataset was performed in jModelTest 2.1.7 [55–56]. The GTR+G+I model was selected as the best substitution model for both the datasets to conduct maximum likelihood (ML) and bayesian inference (BI) analysis. Phylogenetic trees for both the datasets were inferred using maximum likelihood (ML) method implemented in RaxmlGUI v1.3 with 1000 bootstrap replicates [57]. The BI analysis was conducted using Markov chain Monte Carlo (MCMC) method in MrBayes v3.2.6 for 1,150,000 and 1,320,000 generations for PCG and PCG+rRNA datasets, respectively [58–59]. Trees were sampled every 1000 generations and the first 25% were discarded as burn-in. The stationary was considered to be reached when the average standard deviation of split frequency reached below 0.01. Other parameters like effective sample size (ESS) and potential scale reduction factor (PSRF) were evaluated for stationary using Tracer v1.6 [60]. The consensus phylogenetic trees obtained for both the datasets were visualized using iToL v3.6.1 tool [61].
Results and discussion
Sample processing, DNA sequencing and assembly
Total DNA extracted from fifth instar larvae of A. assamensis was found to be optimal (1363.23 ng/μl concentration by NanoDrop spectrophotometer and 234.36 ng/μl concentration by Qubit fluorometer) for mitochondrial DNA enrichment. The mitochondrial DNA was enriched from total DNA for library preparation. The library profile showed that size of mitogenome fragments were in the range of 180 to 880 bp. However, total insert size distribution was from 300 to 1000 bp as it involved ~120 bp adapters along with mitogenome fragments. In addition to the proper distribution of fragments, their concentrations (~27.2 ng/μl) were also found to be adequate for sequencing (S1 Fig). The sequencing resulted in 32,12,599 total number of raw reads, out of which around 28,69,153 high quality reads were processed for scaffold preparation. These reads were used for mitogenome scaffold preparation through de novo assembly of sequenced contigs using SPAdes-3.5.0, SSPACE and CAP3 program. Finally, a scaffold of 15,272 bp length was obtained which represents the whole mitogenome sequence of A. assamensis.
Genome annotation, visualization and comparative analysis
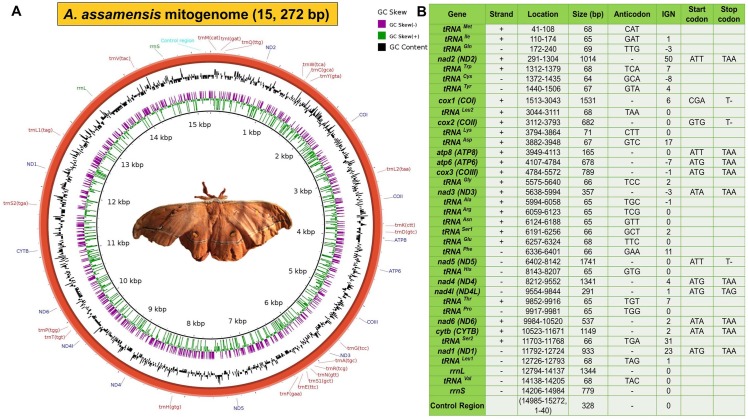
The annotated mitochondrial genome of A. assamensis appeared to be a closed circular structure comprising of 37 genes (13 PCGs, 22 tRNAs and 2 rRNAs) along with a non-coding control region over its 15,272 bp length. 23 genes were found to be encoded by the majority strand or J-strand (+) and the remaining 14 genes by the minority strand or N-strand (-). The whole mito-map constructed using BRIG along with its characteristics like gene location and arrangement are shown in Fig 2. The full annotated mitogenome sequence and SRA data of A. assamensis submitted to NCBI GenBank are available under the accession numbers KU379695 and SRR3948351, respectively.
Fig 2. A. assamensis mitochondrial genome.
(A) Circular map (B) Characteristic features of the mitogenome. tRNAs are represented as trn followed by the IUPAC-IUB single letter amino acid codes e.g. trnM denote tRNAMet. IGN represents (+) values as intergenic nucleotides and (-) values as overlapping regions.
Comparative mitogenome analysis
In order to study the conservation pattern, features like genome size, gene content, genome organization, structure, rearrangement and sequence of A. assamensis mitogenome were compared with other lepidopterans belonging to various levels of taxonomic classification (Table 1).
We found that the length of A. assamensis mitogenome was smaller than that of other sequenced Bombycoids and just bigger than that of Actias selene (15,236 bp) and A. artemis (15,243 bp) [27, 62]. Notably, it lies within the characteristic size of most of the insects (14 to 20 kb). In insects, mitogenome size variation may be attributed to variation in non-coding regions especially the control region that shows great differences in length as well as pattern. It collectively leads to higher degree of gene rearrangements. In contrast, PCGs are quite stable in the mitogenome [23–24]. Smaller mitogenome size in A. assamensis may be favored to reduce accumulation of slightly deleterious mutations during its evolution.
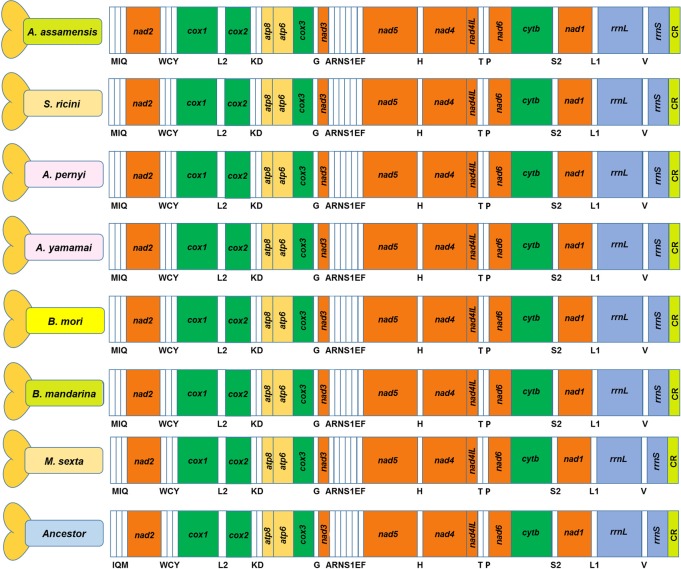
The existence of a typical gene content i.e. 37 genes and a control region was evident in A. assamensis mitogenome as observed in other sequenced mitogenomes of lepidopteran insects. The arrangement of mitochondrial genes was found to be in the order identical to the selected Bombycoid insects (Fig 3). Three typical rearrangements were observed in the mitogenome of A. assamensis similar to most of the other lepidopteran insects [35, 37, 44, 45] as shown in Fig 3. These were (i) tRNAMet-Ile-Gln (trnM/trnI/trnQ or MIQ) cluster, (ii) tRNALys-Asp (K-D) cluster and (iii) tRNAAla-Arg-Asn-Ser1-Glu-Phe (ARNS1EF) cluster. The typical MIQ cluster rearrangement located between the control region and nad2 gene differed from the ancestral pattern tRNAIle-Gln-Met (trnI/trnQ/trnM or IQM) as reported in two ghost moths (Fig 3) belonging to Hepialidae family—a non-ditrysian lineage (Lepidoptera: Exoporia: Hepialoidea) [63–64]. This suggests that A. assamensis along with other lepidopteran insects have evolved with a typical gene arrangement after splitting from its stem lineage during the course of time. A typical ARNS1EF arrangement is found in most of the lepidopteran insects. However, some rearrangement in this cluster has been reported in few lepidopteran insects different from A. assamensis [65–66]. Further, sequence similarity performed among selected Bombycoids showed that A. assamensis shared highest similarity with Antheraea species (A. yamamai—92.1%, A. pernyi—91.7%) followed by the other Bombycoids like S. ricini (88.9%), M. sexta (85.5%), B. mori (83.4%) and B. mandarina (82.9%) indicating a significant level of homology among Bombycoidea superfamily of Lepidoptera.
Fig 3. Mitogenome arrangement of A. assamensis with respect to other selected species.
Ancestor here denotes Thitarodes renzhiensis.
Comparative analysis among protein coding genes (PCGs)
The mitogenome of A. assamensis comprised of a total of 13 PCGs which spanned 11,208 bp constituting around 73.38% of the total mitogenome. The total size of PCGs and their individual sizes were found to be similar to the other Bombycoid species (S1 Table). The PCGs were distributed over both the strands of double stranded mitogenome, 9 genes in the J-strand and 4 genes in the N-strand.
The ATN sequence was found to be the start codon for all the genes in A. assamensis similar to other Bombycoid insects [24, 25, 26, 31, 33]. These codons include ATT (nad2, atp8, nad5), ATA (nad3, nad6, cytb) and ATG (atp6, cox3, nad4, nad4l, nad1) (Fig 2B). Interestingly, start codons of cox1 and cox2 genes in A. assamensis were found to deviate from the canonical ATN initiation codon pattern. Similarly, A. pernyi [25], A. yamamai [26], S. boisduvalii [28], B. mandarina [30], B. mori [31] and M. sexta [33] were also reported to diverge from this pattern. In addition to the gene sequence similarity analysis, comparative amino acid sequence alignment of the reported lepidopteran insects confirmed CGA and GTG as the initiation codons for both cox1 and cox2 genes of A. assamensis, respectively. CGA is also reported as the start codon for cox1 gene in some of the members of Lepidoptera such as A. atlas [23], E. pyretorum [29], M. sexta [33], A. honmai [45], Maruca vitrata [67] and dipteran insect such as Drosophila melanogaster [68]. On the other hand, GTG is the start codon for cox2 gene in S. ricini [24], S. boisduvalii [28] and E. pyretorum [29].
Further, three genes cox1, cox2 and nad5 showed incomplete termination codon (only T) instead of canonical termination codon pattern TAA or TAG (Fig 2B). This is similar to A. pernyi which also shows incomplete termination codon in cox1, cox2 and nad5 genes [25]. The existence of incomplete stop codon has been observed as a common phenomenon of mitochondrial genes of metazoans like lepidopterans in order to minimize IGS and OS [25, 26, 31, 33, 49]. Several studies reveal that it could be a recognition site for an endonuclease that truncates the polycistronic pre-mRNA. These incomplete codons are rectified by the post-transcriptional polyadenylation to yield functional stop codon with TAA termini [33].
The PCGs of A. assamensis were also compared with seven different organisms belonging to various levels of taxonomic classification in Insecta class at nucleotide level and corresponding amino acid level in order to determine the conserved, variable and phylogenetically informative sites (S2 Table). Various species of Antheraea showed 84–90% conserved sites at nucleotide level while at amino acid level it was comparatively higher (86–98%). This might be due to synonymous substitution where a non-beneficial mutation is purified by same amino acid encoding codons. However, atp8 gene exhibited lower values (76.8%) at amino acid level which may suggest that mutation in the gene was needed to help the organism in its evolution. Various comparative analyses were further carried out for the organisms of different genus, family, superfamily and order. The conserved sites were found to decrease with higher hierarchical organisms while variable and phylogenetically informative sites increased in number. Among all PCGs, cox1 and cox2 genes showed maximal conservation both at nucleotide and amino acid levels across the organisms of various classification levels. Similarly, M. sexta and P. maraho were also reported to exhibit higher conservation for cox1 and cox2 genes [33, 40].
The study of nucleotide variation pattern (transition-transversion ratio) among the PCGs of A. assamensis with respect to various members (A. pernyi, A. yamamai, S. ricini, M. sexta, B. mori and B. mandarina) of Bombycoidea superfamily revealed that the transition-transversion ratio decreased with distant organisms in comparison with closer ones across all the PCGs [69]. For example, ratio for atp8 gene varied from 1.7 to 0.1 when A. assamensis was compared with its close relative A. pernyi (same genus) to distant organism M. sexta (different family). However in case of nad4l gene, a lower ratio was observed for A. pernyi (0.24) than distant ones. Further comparative analysis of all the sequenced species of Antheraea genus was carried out in order to find out variation in nad4l gene within the genus. The nad4l gene of A. assamensis showed lower ratio across the species whereas higher ratio was observed in comparison with other species. Interestingly, it exhibited higher ratio with S. ricini which belongs to different genus of same family. This apparently depicts that nad4l gene is a decisive marker for evolution to form A. assamensis within Antheraea genus. Further, we may hypothesize that A. assamensis might have acquired gene from S. ricini during its evolution as both the organism are found in Northeast India and share the same evolutionary space [70].
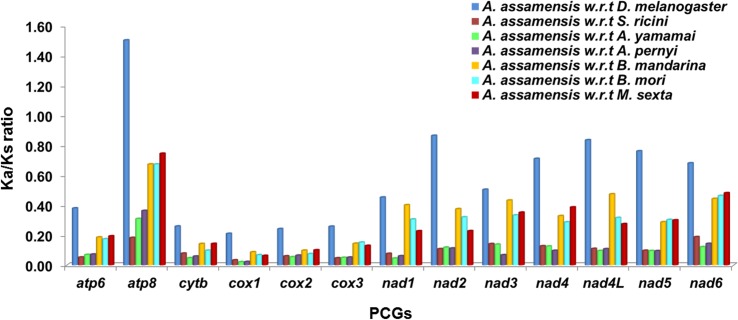
Different evolutionary rates among the genes reflect different functional constraints affecting the mitogenome that varies among species. Therefore, in order to determine the evolutionary rates in A. assamensis, the PCGs were compared with that of seven different organisms belonging to various hierarchy levels in Insecta class. The average rate of synonymous substitutions (Ks) and the average rate of non-synonymous substitutions (Ka) along with their ratios (Ka/Ks) were calculated for all 13 PCGs. The ratio Ka/Ks less than, greater than and equal to 1 indicates that genes are under negative (purifying) selection, positive (adaptative) selection and neutral evolution, respectively [71]. Among 13 PCGs, atp8 gene encoding ATPase subunit 8 exhibited 1.5 ratio with reference to that of D. melanogaster indicating positive/ relaxed selection acting on this gene (Fig 4). This indicates that mutation in atp8 of A. assamensis was due to a necessity to re-organize its structure. These changes may be due to the requirement of energy for the production and secretion of silk fibers which are not observed in D. melanogaster. It depicts that functional change in an organism needs commitment mutation in responsible genes in order to support survival of the fittest. Notably, the ratio for remaining PCGs of A. assamensis with all organisms was found to be less than one which suggests that mutation was against the requirement and hence mutation was replaced by synonymous nucleotides. This suggests that all the protein coding genes evolved under strong purifying (negative) selection as is evident from the number of conserved and variable sites. The Ka/Ks ratio in atp8 with reference to B. mandarina, B. mori and M. sexta was found to be significantly higher. These organisms belong to different family with respect to A. assamensis and hence significant variation in atp8 gene can be expected due to certain changes in the mitogenome kinetics. As S. ricini, A. yamamai, A. pernyi belong to the same family of A. assamensis, significant Ka/Ks ratio was not observed across the PCGs. Overall comparison of PCGs exhibited that mutation in atp8 gene was essential in insects to yield several strains with different kinetics as other PCGs showed lower ratios. The lowest evolutionary rates were observed for cox1 gene indicating that it is least susceptible to variation in protein sequence and hence shows potential as a barcode for evolutionary studies in silkworms. In addition, other genes like cox2, cox3, cytb and nad genes with slightly low evolutionary rates next to cox1 can also serve as barcode markers. Furthermore, the effect of GC content on Ka/Ks ratio was studied and we found a significant negative correlation between them (S2 Fig). This indicates that change in GC content may result in the variation in nucleotide substitution pattern or evolutionary pattern among the PCGs.
Fig 4. Evolutionary rates (Ka/Ks) of individual PCG of A. assamensis with selected species.
w.r.t. here denotes with reference to-.
Comparative analysis among transfer RNAs (tRNAs)
Twenty two tRNA genes with a total length of 1465 bp were found to be present in the mitogenome of A. assamensis. 14 tRNA genes were encoded by the J strand while 8 tRNA genes were encoded by the N-strand (Fig 2B). The individual length of each gene varied from 64 to 71 bp as reported for other lepidopterans (S1 Table).
The secondary structures of tRNAs of A. assamensis exhibited a typical clover-leaf structure similar to other lepidopteran species (S3 and S4 Figs). This depicts that arrangement of all the tRNA genes is mostly conserved among insects. However, some variation in the structures of A. assamensis was observed such as aminoacyl acceptor stem of tRNAMet, TΨC loop of tRNAIle, tRNACys, tRNATyr, tRNAArg, tRNAAsn, tRNAGlu, tRNAPhe, tRNATrp, tRNASer2, dihydrouridine (DHU) arm of tRNASer1, etc. The lack of stable DHU arm in tRNASer1 and TΨC loop in tRNATyr has also been reported in lepidopterans such as S. ricini, A. yamamai, A. pernyi, B. mori, B. mandarina, M. sexta, E. pyretorum, A. fylloides, B. panterinaria, E. autonoe, etc. [24, 25, 26, 29, 30, 31, 33, 35, 38, 42]. These structural variations might be attributed to variation in the number of base pairs responsible for the formation of aminoacyl acceptor (AA) stem, DHU arm, TΨC arm and anticodon (AC) stem-loop (S5 and S6 Figs). Among various structural parts of tRNAs, AA stem of A. assamensis was found to be more consistent in size across all the tRNAs and in other lepidopterans as observed from comparative analysis. AC stem, AC loop and DHU stem also showed consistency in the number of base pairs across lepidopterans. However, the consistency of these stems and loops varied with the type of tRNAs. In addition, anticodons of tRNAs were found to be identical to the selected Bombycoids (S3 Table) and majority of other lepidopteran insects [26, 28, 30, 35]. On the contrary, TѰC stem, TѰC loop and DHU loop exhibited randomness in the number of base pairs across the organisms and tRNAs. For instance, TѰC loop was absent in tRNATyr of A. assamensis while tRNATrp of A. assamensis and tRNATyr of B. mori had 7 bp and 5 bp loops, respectively. The consistency in the number of base pairs of AA stem, AC stem, AC loop and DHU stem was probably to avoid dysfunctioning of tRNAs actively participating in protein synthesis and thus critical for the survival in the evolutionary milieu.
We identified a total of 35 mismatched base pairs in tRNAs of A. assamensis scattered over the AA stem, TΨC stem, AC and DHU regions of 19 tRNAs. 28 mismatches were observed in G-U combinations while 6 and 1 mismatches were found to be in U-U and G-A combinations, respectively (S3 Table). These mismatches lead to changes in tRNA structures; for instance, two U-U mismatches in tRNASer2 resulted in the formation of an extra loop in AC stem.
From the comparative analysis, we found that the number of total mismatches was higher in A. assamensis (35 No.s) than the selected Bombycoids like S. ricini (32 No.s), B. mori (29 No.s), M. sexta (25 No.s), etc. Numerically, this mismatch was higher than many Saturniids like E. pyretorum (24 No.s), S. cynthia (28 No.s), etc. Similarly, mismatches such as G-U, U-U, G-A, A-A, A-C and C-U have been previously reported for many lepidopterans like E. autonoe (26 No.s), A. fylloides (27 No.s), etc. [38, 42]. Among various mismatches in A. assamensis, G-U mismatches were found be the highest (28 No.s) than the selected Bombycoids and other lepidopterans e.g. A. atlas (25 No.s) and E. autonoe (20 No.s). The details of mismatches found in various stem-loop structures of different tRNAs of Bombycoid members are compared in S3 Table. The non-canonical pairing might occur due to the insertion/deletion of nucleotides or nucleotide substitution of bases within the tRNA genes as a form of ancient insertional editing. The presence of high number of mismatches indicates higher nucleotide substitution in the tRNA sequences of A. assamensis as compared to the selected Bombycoids which may result in genome structure and sequence evolution. These mismatches are likely to be corrected by RNA editing mechanism as observed in other arthropods [72].
Comparative analysis among ribosomal RNAs (rRNAs)
A. assamensis comprised of two genes for rRNA coded by (–) strand of the mitogenome. rrnL (lrRNA) is located between tRNALeu1 and tRNAVal genes, whereas rrnS (srRNA) is accommodated between tRNAVal and control region as reported for other lepidopterans. The lengths of rrnL and rrnS were 1344 bp and 779 bp, respectively (S1 Table) and thus similar to many reported lepidopteran insects [38, 43]. The sequence homology of two rRNA genes of A. assamensis with the selected Bombycoids revealed that the sequence identity of rrnS gene (84 to 95%) was higher than that of rrnL (80 to 89%). The divergence study in rRNA genes (rrnL and rrnS) also exhibited low variability sites which suggest that these are highly conserved sites and may have potential application in molecular systematics. This may be due to the natural effort to preserve the structure and function of rRNA as protein synthesis depends on it.
Comparative analysis among control region (A+T rich region)
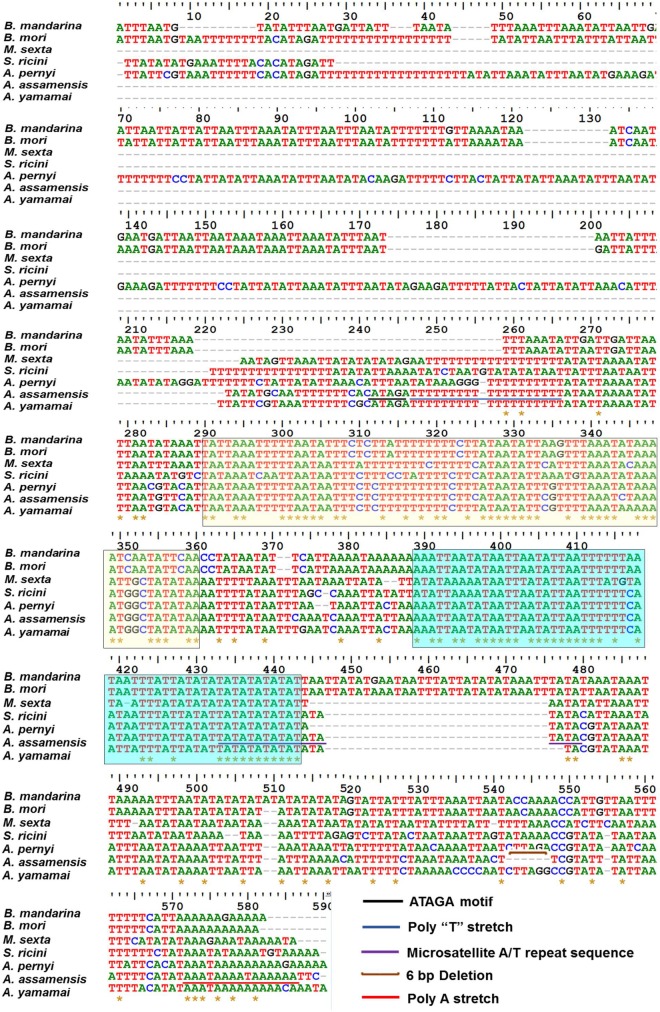
The control region is the longest non-coding region of A. assamensis mitogenome and with a length of 328 bp, is similar to other Bombycoids. This region is located between rrnS and MIQ cluster. The comparative sequence homology of control region displayed higher similarity with its close relatives such as A. yamamai (89.2%) and A. pernyi (82.7%) while showed lower similarity with distant organisms such as B. mori (~68%). It was found that two sequence blocks of more than 50 bp in length displayed 96–100% similarity with other Antheraea species while S. ricini (78–96%), M. sexta (68–88%) and B. mori (73–80%) showed low similarity. Additionally, some conserved structures and repeats were observed in the control region of A. assamensis (Fig 5). These were ‘ATAGA’ motif, poly T stretch (T)19, microsatellite (TA)9 repeat sequence, poly-A region and a 6 bp deletion. ‘ATAGA’ motif and (T)19 are known to be important for gene regulation and serve as a recognition site for replication initiation of minor or light strand. The poly A tail has been proposed to be required for RNA maturation and serve as a sequence for controlling transcription or replication initiation in insects [73]. The microsatellites (TA)9 also known as simple sequence repeats (SSR) are proposed to be used as molecular markers due to their abundance and highly polymorphic nature [24]. A microsatellite (AT)n element is a well conserved site and can be used for evolutionary and conservation genetic studies [25, 26, 74]. The control region of A. assamensis lacked tandem repeat elements similar to other completely sequenced Antheraea species except A. pernyi which harbored six 38 bp tandem repeats. The significance of these tandem repeats is unclear and needs further taxon sampling and mitogenome characterization. Presence of highly conserved yet distinct consensus sequences in the control region thus shows its high potential as a phylogenetic marker at lower taxonomic levels.
Fig 5. Multiple sequence alignment of control region of A. assamensis mitogenome showing conserved structural elements.
Yellow and green highlighted blocks represent 1st and 2nd conserved blocks, respectively.
Comparative analysis among overlapping sequence (OS) and intergenic spacer (IGS) regions
The overlapping sequences (OS) and intergenic spacers (IGS) are commonly found in the mitogenome of lepidopterans. These regions may vary in length and location from species to species during their evolution. In case of A. assamensis, six OS and seventeen IGS were found to be present in the mitogenome. The size of OS and IGS were in the range of 1–8 bp (cumulative length of 23 bp) and 1–50 bp (cumulative length of 171 bp), respectively. The presence of OS, IGS and their corresponding lengths were similar to many of the reported lepidopterans [23, 27, 32, 43, 45] as given in S4 Table. A characteristic conserved overlapping junction (ATGATAA) was observed in between atp8 and atp6 genes as reported for most of the lepidopterans [38, 42, 45].
The spacer between the genes tRNAGln and nad2 (tRNAGln—nad2 spacer) was found to be the largest (50 bp) in A. assamensis but slightly shorter than that observed in A. yamamai (53 bp), S. cynthia (54 bp), etc. Similarly, it has been detected as the largest spacer in many other lepidopterans like A. atlas, A. selene, S. boisduvali, E. pyretorum, P. bremeri and A. honmai [23, 27, 28, 29, 41, 45]. Further, the spacers cytb-tRNASer2 (31 bp), tRNASer2-nad1 (23 bp), tRNALys- tRNAAsp (17 bp), etc. were also found to exist in A. assamensis mitogenome (Fig 2B). The spacer cytb-tRNASer2 may serve as a binding site for the mtTERM, which is a transcription termination peptide [24, 26, 33]. A conserved motif ‘ATACTAA’ was found in the spacer tRNASer2-nad1 on the comparison of A. assamensis with selected Bombycoid species. This spacer is implicated in mitochondrial transcription termination where the ‘ATACTAA’ motif is essential as a recognition site for mtTERM [24, 26, 33].
The spacers in A. assamensis mitogenome also exhibited sequence homology with their adjacent genes as observed in many of the lepidopteran insects [24–26]. For instance, spacer tRNAGln-nad2 showed 68% similarity with its adjacent nad2 gene. This might be because of the partial duplication of nad2 gene in order to provide another origin of replication [26, 28, 33]. This also suggests that A. assamensis may have undergone rapid sequence divergence attributed to the non-coding nature of IGS as seen in other organisms [26]. Apart from adjacent genes, IGS also showed sequence homology with their distant genes in A. assamensis. For example, spacer tRNALys-tRNAAsp showed homology with rrnS (82%), nad5 (76%) and atp8 (82%) genes. Similarly, the same spacer in S. ricini has also been reported to exhibit homology with rrnS (78%) and nad5 (72%) genes [24]. This might be attributed to gene duplication and degeneration.
Comparative analysis with respect to nucleotide composition, skewness and codon usage
The nucleotide composition in mitogenome of A. assamensis was analyzed in terms of specific nucleotide content, AT and GC skewness. Whole mitogenome nucleotide composition revealed 80.2% AT content (40.8% T + 39.4% A) and remaining 19.8% GC content (12.1% C + 7.7% G). Similar to A. assamensis, many lepidopterans insects also showed enhanced AT content [37, 39]. This AT content was found to vary with different regions of the mitogenome. The AT content was maximal (~91.2%) in the control region followed by rRNAs genes (84.3%), tRNAs genes (80.8%) and PCGs (78%). The individual PCGs also exhibited variation in the corresponding AT content. For instance, atp8, nad4l and nad6 genes had the highest AT content while cox1 and cox3 genes had lowest values. This variation pattern was also detected in the other lepidopterans like E. pyretorum, P. atrilineata, G. menyuanensis, etc. [29, 36, 43]. A close analysis of the third codon position in all 13 PCGs of A. assamensis exhibited AT biasness which was 93% than the first (73%) and second (70%) codon positions similar to the other Bombycoids [24, 25, 29]. This may be attributed to more relaxed constraints on A+T content at 3rd position than at 1st and 2nd position due to the phenomena of degeneracy in genetic code.
Nucleotide skewness was determined to measure the relative number of Gs to Cs and As to Ts. The AT skewness in A. assamensis mitogenome was found to be negative (-0.02) which indicates the occurrence of more Ts than As and lies within the range of Saturniid moths [25–26]. This value differed from the selected Bombycidae species having positive AT skew values e.g. 0.06 in B. mori, B. mandarina, etc. [30–31]. Similarly, mitogenome GC skewness also exhibited negative value (-0.22) as observed in many Bombycoids such as M. sexta (-0.18) and S. ricini (-0.23). The GC skewness varied with major and minor strands of PCGs, however, no significant change was observed in AT skewness. Minor strand was G-skewed i.e. rich in G nucleotides while more ‘C’s were encoded by major strand (S5 Table). The strand asymmetry is common in insects and has been reported in all the lepidopterans which occur due to asymmetrical mutation pressure on mitogenome. These skewness patterns were also observed in rRNAs, tRNAs, control region and are similar to the other sequenced lepidopteran insects.
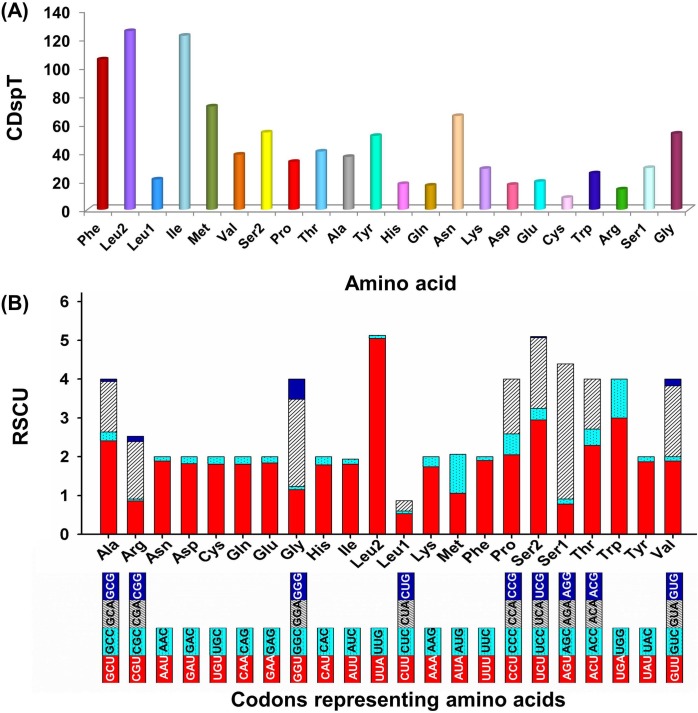
Nucleotide biasness was also reflected in the codon usage of PCGs that determines the frequency of synonymous codon usage (SCU) in various genomes at inter and intra levels. Based on the codon usage and relative SCU results, codons with A or T nucleotides at the 3rd codon position were highly used in comparison with G or C nucleotides as discussed above. The five most frequently used amino acids were Leu, Ile, Phe, Met and Asn (Fig 6) and the most prevalent codons for these corresponding amino acids were UUA, AUU, UUU, AUA and AAU with a usage frequency of 45.53% to 48.95% (S2 Fig). Cys was found to be the least used amino acid as detected in other selected Bombycoids. Biasness in SCU has been observed in many other lepidopterans to avoid errors due to mis-incorporation of amino acids which depends upon various factors like natural selection (gene length and function), mutation bias (base mutation and GC content), etc. [24, 35]. This A+T biasness may result in the change in amino acid composition similar to the other lepidopterans. Therefore, the codon usage analysis will have great significance in studying gene expression and evolutionary studies in silkworms.
Fig 6. Codon usage in A. assamensis mitogenome.
(A) Codon distribution where CDspT = codons per thousand codons (B) Relative synonymous codon usage (RSCU). Termination codons were excluded in the study.
Comparative phylogenetic analysis
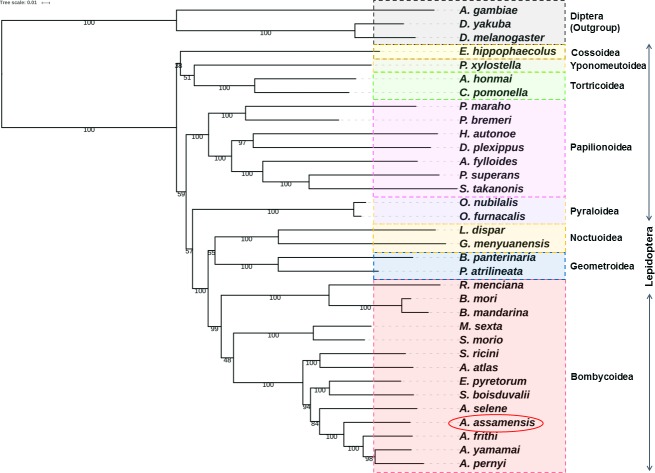
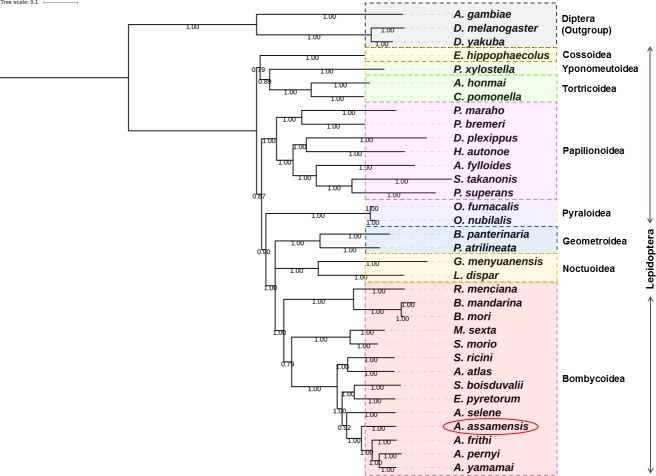
Maximum likelihood (ML) and Bayesian Inference (BI) phylogenetic trees were constructed based on nucleotide sequences of 13 PCGs and 13 PCGs+2 rRNAs datasets, respectively (Figs 7 and 8, S7 and S8 Figs). The phylogenetic trees obtained using both the methods were found to be of similar topologies. The tree clustering showed that A. assamensis belongs to Saturniidae family which is clustered in the same branch with other families (Bombycidae+Sphingidae) of Bombycoidea superfamily within the order Lepidoptera. Within the Saturniidae family, A. assamensis has clustered around with other Antheraea species and was also close to A. selene. Further, our study supported the triplet superfamily (Bombycoidea+Geometroidea+Noctuoidea) relationship under Macroheterocera which along with Pyraloidea and Papilionoidea formed Obtectomera, a group within Apoditrysia [8–9]. These results were consistent with the previously reported studies [26, 32, 35, 75]. More mitogenome sequence information on Bombycoidea superfamily is needed to gain deep insights into the families/superfamilies relationships within Lepidoptera. The present study supports the fact that intra and inter family level relationships are well resolved within the superfamily Bombycoidea providing a better evolutionary relationship among the silk producing insects.
Fig 7. Phylogenetic tree inferred from nucleotide sequences of 13 PCGs of 34 organisms using maximum likelihood (ML) method in RaxmlGUI v1.3 (1000 bootstrap replicates).
The tree is drawn to scale with bootstrap values indicated along with the branches.
Fig 8. Phylogenetic tree inferred from nucleotide sequences of 13 PCGs of 34 organisms using bayesian inference (BI) method in MrBayes v3.2.6.
The tree is drawn to scale with bayesian posterior probability values indicated along with the branches.
Conclusion
Our study deciphered the complete mitogenome of A. assamensis which shares similarity with majority of the lepidopterans, particularly Saturniids, in several characteristics such as genome organization and content, PCG size and structure, AT/GC skewness, tRNA structure and anti-codons, OS and IGS regions. Our analysis indicates that A. assamensis PCGs evolved under strong purifying selection and tRNAs genes showed high base substitution/mismatches. Transition-transversion ratio suggests that nad4l gene may be the significant contributor for evolution to form A. assamensis within Antheraea genus. A six-bp indel (deletion) and two highly conserved sequence blocks in control region suggest its potential application as a marker for delineating other closely related Antheraea sp. and subsequent usage in phylogenetic studies at lower taxonomic levels. The largest tRNAGln-nad2 spacer found in the A. assamensis mitogenome may serve as another origin of replication. Our study assigns the taxonomic status of A. assamensis using an optimal model based phylogenetic tree construction. This is the 12th representative organism of Saturniidae family with a completely sequenced mitogenome. As A. assamensis is the sole producer of unique golden Muga silk which is the backbone of Assam’s sericulture industry, our study on its mitogenomic landscape is an important addition to the existing genome informatics resources on silkworms.
Supporting information
(TIF)
(A) G+C content versus Ka/Ks ratios of 13 concatenated PCGs in A. assamensis mitogenome (B) The usage rate of five frequently used codons in Bombycoidea members.
(TIF)
trnY lacked TΨC loop. tRNA here are represented as trn. AA denotes amino acyl arm and AC denotes anticodon arm.
(TIF)
trnS1 lacked DHU loop. tRNA here are represented as trn. AA denotes amino acyl arm and AC denotes anticodon arm.
(TIF)
AA denotes Aminoacyl arm and AC denotes Anticodon arm. S-L denotes Stem-Loop of tRNA gene. tRNA is represented as trn.
(TIF)
AA denotes Aminoacyl arm and AC denotes Anticodon arm. S-L denotes Stem-Loop of tRNA gene. tRNA is represented as trn.
(TIF)
The tree is drawn to scale with bootstrap values indicated along with the branches.
(TIF)
The tree is drawn to scale with bayesian posterior probability values indicated along with the branches.
(TIF)
(PDF)
C here denotes conserved sites, I denotes phylogenetically informative sites, V denotes variable sites, N denotes nucleotide level, A denotes amino acid level. The number of sites is represented in terms of percentage values.
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability
All relevant data are within the paper and its Supporting Information files. The full annotated mitogenome sequence and SRA data of A. assamensis submitted to NCBI GenBank are available under the accession numbers KU379695 and SRR3948351, respectively.
Funding Statement
The authors thank the Department of Biotechnology, Govt. of India, New Delhi for supporting the research through the UXCEL project (Sanction Order No: BT/411/NE/U-Excel/2013 dated 06.02.2014). The funding agency had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Andersson SG, Zomorodipour A, Andersson JO, Sicheritz-Pontén T, Alsmark UC, Podowski RM, et al. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 1998;396: 133–140. doi: 10.1038/24094 [DOI] [PubMed] [Google Scholar]
- 2.Salvioli S, Bonafè M, Capri M, Monti D, Franceschi C. Mitochondria, aging and longevity–a new perspective. FEBS Lett. 2001;492: 9–13. doi: 10.1016/S0014-5793(01)02199-8 [DOI] [PubMed] [Google Scholar]
- 3.Cameron SL. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 2014;59: 95–117. doi: 10.1146/annurev-ento-011613-162007 [DOI] [PubMed] [Google Scholar]
- 4.Boore JL. The use of genome-level characters for phylogenetic reconstruction. Trends Ecol Evol. 2006;21: 439–446. doi: 10.1016/j.tree.2006.05.009 [DOI] [PubMed] [Google Scholar]
- 5.Mandal SD, Chhakchhuak L, Gurusubramanian G, Kumar NS. Mitochondrial markers for identification and phylogenetic studies in insects–A Review. DNA Barcodes 2014;2: 1–9. doi: 10.2478/dna-2014-0001 [Google Scholar]
- 6.Arunkumar KP, Metta M, Nagaraju J. Molecular phylogeny of silkmoths reveals the origin of domesticated silkmoth, Bombyx mori from Chinese Bombyx mandarina and paternal inheritance of Antheraea proylei mitochondrial DNA. Mol Phylogenet Evol. 2006;40: 419–427. doi: 10.1016/j.ympev.2006.02.023 [DOI] [PubMed] [Google Scholar]
- 7.Liu YQ, Li YP, Wang H, Xia RX, Chai CL, Pan MH, et al. The complete mitochondrial genome of the wild type of Antheraea pernyi (Lepidoptera: Saturniidae). Ann Entomol Soc Am. 2012;105: 498–505. doi: 10.1603/AN11156 [Google Scholar]
- 8.Timmermans MJTN, Lees DC, Simonsen TJ. Towards a mitogenomic phylogeny of Lepidoptera. Mol Phylogenet Evol. 2014;79: 169–178. doi: 10.1016/j.ympev.2014.05.031 [DOI] [PubMed] [Google Scholar]
- 9.Mitter C, Davis DR, Cummings MP. Phylogeny and Evolution of Lepidoptera. Annu Rev Entomol. 2017,62: 265–283. doi: 10.1146/annurev-ento-031616-035125 [DOI] [PubMed] [Google Scholar]
- 10.Nässig WA, Lampe REJ, Kager S. Heterocera Sumatrana: The Saturniidae of Sumatra (Lepidoptera). Vol. 10 Gottingen: Heterocera Sumatrana Society; 1996. [Google Scholar]
- 11.Tikader A, Vijayan K, Saratchandra B. Muga silkworm, Antheraea assamensis (Lepidoptera: Saturniidae)-an overview of distribution, biology and breeding. Eur J Entomol. 2013;110: 293 doi: 10.14411/eje.2013.096 [Google Scholar]
- 12.Kasoju N, Bhonde RR, Bora U. Fabrication of a novel micro–nano fibrous nonwoven scaffold with Antheraea assama silk fibroin for use in tissue engineering. Mater Lett. 2009;63: 2466–2469. doi: 10.1016/j.matlet.2009.08.037 [Google Scholar]
- 13.Chetia H, Kabiraj D, Singh D, Mosahari PV, Das S, Sharma P, et al. De novo transcriptome of the muga silkworm, Antheraea assamensis (Helfer). Gene 2017;611: 54–65. doi: 10.1016/j.gene.2017.02.021 [DOI] [PubMed] [Google Scholar]
- 14.Chakravorty R, Barah A, Neog K, Rahman SAS, Ghose J. Package of practices for muga culture Package of practices of Muga, Eri and Mulberry Sericulture for North Eastern Region of India, Central Muga Eri Research & Training Institute, Central Silk Board, Lahdoigarh, Jorhat: 2005: 1–23. [Google Scholar]
- 15.Andrews S. FastQC: A quality control tool for high throughput sequence data. 2010. Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 16.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19: 455–477. doi: 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9: 868–877. doi: 10.1101/gr.9.9.868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 2013;69: 313–319. doi: 10.1016/j.ympev.2012.08.023 [DOI] [PubMed] [Google Scholar]
- 19.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41: 95–98. doi: 10.1021/bk-1999-0734.ch008 [Google Scholar]
- 20.Thompson JD, Gibson TJ, Higgins DG. Multiple Sequence Alignment Using ClustalW and ClustalX. Curr Protoc Bioinformatics 2002; 2.3.1–2.3.22. doi: 10.1002/0471250953.bi0203s00 [DOI] [PubMed] [Google Scholar]
- 21.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27: 573 doi: 10.1093/nar/27.2.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alikhan NF, Petty NK, Zakour NLB, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 2011;12: 402 doi: 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen MM, Li Y, Chen M, Wang H, Li Q, Xia RX, et al. Complete mitochondrial genome of the atlas moth, Attacus atlas (Lepidoptera: Saturniidae) and the phylogenetic relationship of Saturniidae species. Gene 2014;545: 95–101. doi: 10.1016/j.gene.2014.05.002 [DOI] [PubMed] [Google Scholar]
- 24.Kim JS, Park JS, Kim MJ, Kang PD, Kim SG, Jin BR, et al. Complete nucleotide sequence and organization of the mitochondrial genome of eri-silkworm, Samia cynthia ricini (Lepidoptera: Saturniidae). J Asia Pac Entomol. 2012;15: 162–173. doi: 10.1016/j.aspen.2011.10.002 [Google Scholar]
- 25.Liu Y, Li Y, Pan M, Dai F, Zhu X, Lu C, et al. The complete mitochondrial genome of the Chinese oak silkmoth, Antheraea pernyi (Lepidoptera: Saturniidae). Acta Biochim Biophys Sin (Shanghai) 2008;40: 693–703. doi: 10.1111/j.1745-7270.2008.00449.x [PubMed] [Google Scholar]
- 26.Kim SR, Kim MI, Hong MY, Kim KY, Kang PD, Hwang JS, et al. The complete mitogenome sequence of the Japanese oak silkmoth, Antheraea yamamai (Lepidoptera: Saturniidae). Mol Biol Rep. 2009;36: 1871–1880. doi: 10.1007/s11033-008-9393-2 [DOI] [PubMed] [Google Scholar]
- 27.Liu QN, Zhu BJ, Dai LS, Wei GQ, Liu CL. The complete mitochondrial genome of the wild silkworm moth, Actias selene. Gene 2012;505: 291–299. doi: 10.1016/j.gene.2012.06.003 [DOI] [PubMed] [Google Scholar]
- 28.Hong MY, Lee EM, Jo YH, Park HC, Kim SR, Hwang JS, et al. Complete nucleotide sequence and organization of the mitogenome of the silk moth Caligula boisduvalii (Lepidoptera: Saturniidae) and comparison with other lepidopteran insects. Gene 2008;413: 49–57. doi: 10.1016/j.gene.2008.01.019 [DOI] [PubMed] [Google Scholar]
- 29.Jiang ST, Hong GY, Yu M, Li N, Yang Y, Liu YQ, et al. Characterization of the complete mitochondrial genome of the giant silkworm moth, Eriogyna pyretorum (Lepidoptera: Saturniidae). Int J Biol Sci. 2009;5: 351–365. doi: 10.7150/ijbs.5.351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan M, Yu Q, Xia Y, Dai F, Liu Y, Lu C, et al. Characterization of mitochondrial genome of Chinese wild mulberry silkworm, Bomybx mandarina (Lepidoptera: Bombycidae). Sci China C Life Sci. 2008;51: 693–701. doi: 10.1007/s11427-008-0097-6 [DOI] [PubMed] [Google Scholar]
- 31.Yukuhiro K, Sezutsu H, Itoh M, Shimizu K, Banno Y. Significant levels of sequence divergence and gene rearrangements have occurred between the mitochondrial genomes of the wild mulberry silkmoth, Bombyx mandarina, and its close relative, the domesticated silkmoth, Bombyx mori. Mol Biol Evol. 2002;19: 1385–1389. doi: 10.1093/oxfordjournals.molbev.a004200 [DOI] [PubMed] [Google Scholar]
- 32.Kong W, Yang J. The complete mitochondrial genome of Rondotia menciana (Lepidoptera: Bombycidae). J Insect Sci. 2015;15: 48 doi: 10.1093/jisesa/iev032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cameron SL, Whiting MF. The complete mitochondrial genome of the tobacco hornworm, Manduca sexta, (Insecta: Lepidoptera: Sphingidae), and an examination of mitochondrial gene variability within butterflies and moths. Gene 2008;408: 112–123. doi: 10.1016/j.gene.2007.10.023 [DOI] [PubMed] [Google Scholar]
- 34.Kim MJ, Choi SW, Kim I. Complete mitochondrial genome of the larch hawk moth, Sphinx morio (Lepidoptera: Sphingidae). Mitochondrial DNA 2013;24: 622–624. doi: 10.3109/19401736.2013.772155 [DOI] [PubMed] [Google Scholar]
- 35.Yang X, Xue D, Han H. The complete mitochondrial genome of Biston panterinaria (Lepidoptera: Geometridae), with phylogenetic utility of mitochondrial genome in the Lepidoptera. Gene 2013;515: 349–358. doi: 10.1016/j.gene.2012.11.031 [DOI] [PubMed] [Google Scholar]
- 36.Yang L, Wei ZJ, Hong GY, Jiang ST, Wen LP. The complete nucleotide sequence of the mitochondrial genome of Phthonandria atrilineata (Lepidoptera: Geometridae). Mol Biol Rep. 2009;36: 1441–1449. doi: 10.1007/s11033-008-9334-0 [DOI] [PubMed] [Google Scholar]
- 37.Gong YJ, Wu QL, Wei SJ. The first complete mitogenome for the superfamily Cossoidea of Lepidoptera: The seabuckthorn carpenter moth Eogystia hippophaecolus. Mitochondrial DNA 2014;25: 288–289. doi: 10.3109/19401736.2013.792071 [DOI] [PubMed] [Google Scholar]
- 38.Zhao F, Huang DY, Sun XY, Shi QH, Hao JS, Zhang LL, et al. The first mitochondrial genome for the butterfly family Riodinidae (Abisara fylloides) and its systematic implications. Dongwuxue Yanjiu 2013;34: E109 doi: 10.11813/j.issn.0254-5853.2013.E4-5.E109 [PubMed] [Google Scholar]
- 39.Kim MJ, Kang AR, Jeong HC, Kim KG, Kim I. Reconstructing intraordinal relationships in Lepidoptera using mitochondrial genome data with the description of two newly sequenced lycaenids, Spindasis takanonis and Protantigius superans (Lepidoptera: Lycaenidae). Mol Phylogenet Evol. 2011;61: 436–445. doi: 10.1016/j.ympev.2011.07.013 [DOI] [PubMed] [Google Scholar]
- 40.Wu LW, Lees DC, Yen SH, Hsu YF. The complete mitochondrial genome of the near-threatened swallowtail, Agehana maraho (Lepidoptera: Papilionidae): evaluating sequence variability and suitable markers for conservation genetic studies. Entomol News 2010;121: 267–280. doi: 10.3157/021.121.0308 [Google Scholar]
- 41.Kim MI, Baek JY, Kim MJ, Jeong HC, Kim KG, Bae CH, et al. Complete nucleotide sequence and organization of the mitogenome of the red-spotted apollo butterfly, Parnassius bremeri (Lepidoptera: Papilionidae) and comparison with other lepidopteran insects. Mol Cells 2009;28: 347–363. doi: 10.1007/s10059-009-0129-5 [DOI] [PubMed] [Google Scholar]
- 42.Kim MJ, Wan X, Kim K, Hwang JS, Kim I. Complete nucleotide sequence and organization of the mitogenome of endangered Eumenis autonoe (Lepidoptera: Nymphalidae). Afr J Biotechnol. 2010; 9 doi: 10.5897/AJB09.1486 [Google Scholar]
- 43.Yuan ML, Zhang QL. The complete mitochondrial genome of Gynaephora menyuanensis (Lepidoptera: Lymantriidae) from the Qinghai-Tibetan Plateau. Mitochondrial DNA 2013;24: 328–330. doi: 10.3109/19401736.2012.760077 [DOI] [PubMed] [Google Scholar]
- 44.Dai LS, Zhu BJ, Qian C, Zhang CF, Li J, Wang L, et al. The complete mitochondrial genome of the diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae). Mitochondrial DNA Part A 2016;27: 1512–1513. doi: 10.3109/19401736.2014.953116 [DOI] [PubMed] [Google Scholar]
- 45.Lee ES, Shin KS, Kim MS, Park H, Cho S, Kim CB. The mitochondrial genome of the smaller tea tortrix Adoxophyes honmai (Lepidoptera: Tortricidae). Gene 2006;373: 52–57. doi: 10.1016/j.gene.2006.01.003 [DOI] [PubMed] [Google Scholar]
- 46.Shi BC, Liu W, Wei SJ. The complete mitochondrial genome of the codling moth Cydia pomonella (Lepidoptera: Tortricidae). Mitochondrial DNA 2013;24: 37–39. doi: 10.3109/19401736.2012.716054 [DOI] [PubMed] [Google Scholar]
- 47.Coates BS, Sumerford DV, Hellmich RL, Lewis LC. Partial mitochondrial genome sequences of Ostrinia nubilalis and Ostrinia furnicalis. Int J Biol Sci. 2005;1: 13 doi: 10.7150/ijbs.1.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Bruijn MH. Drosophila melanogaster mitochondrial DNA, a novel organization and genetic code. Nature 1983;304: 234–241. doi: 10.1038/304234a0 [DOI] [PubMed] [Google Scholar]
- 49.Clary DO, Wolstenholme DR. The mitochondrial DNA molecule of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol. 1985;22: 252–271. doi: 10.1007/BF02099755 [DOI] [PubMed] [Google Scholar]
- 50.Beard CB, Hamm D, Collins FH. The mitochondrial genome of the mosquito Anopheles gambiae: DNA sequence, genome organization, and comparisons with mitochondrial sequences of other insects. Insect Mol Biol. 1993;2: 103–124. doi: 10.1111/j.1365-2583.1993.tb00131.x [DOI] [PubMed] [Google Scholar]
- 51.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30: 2725–2729. doi: 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith C, Heyne S, Richter AS, Will S, Backofen R. Freiburg RNA Tools: a web server integrating INTARNA, EXPARNA and LOCARNA. Nucleic Acids Res. 2010;38: W373–W377. doi: 10.1093/nar/gkq316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Junqueira AC, Lessinger AC, Torres TT, da Silva FR, Vettore AL, Arruda P, et al. The mitochondrial genome of the blowfly Chrysomya chloropyga (Diptera: Calliphoridae). Gene 2004;339: 7–15. doi: 10.1016/j.gene.2004.06.031 [DOI] [PubMed] [Google Scholar]
- 54.Abascal F, Zardoya R, Telford MJ. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010;38: W7–13. doi: 10.1093/nar/gkq291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guindon S, Gascuel O. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Syst Biol. 2003;52: 696–704. doi: 10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- 56.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 2012;9: 772–772. doi: 10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Silvestro D, Michalak I. RaxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 2012;12: 335–337. doi: 10.1007/s13127-011-0056-0 [Google Scholar]
- 58.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001;17: 754–755. doi: 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- 59.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61: 539–42. doi: 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rambaut A, Suchard MA, Xie D, Drummond AJ. Tracer v1.6. 2014. Available from: http://tree.bio.ed.ac.uk/software/tracer/.
- 61.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44: W242–245. doi: 10.1093/nar/gkw290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park JS, Kim MJ, Kim I. The complete mitochondrial genome of the moon moth, Actias aliena (Lepidoptera: Saturniidae). Mitochondrial DNA A DNA Mapp Seq Anal. 2016;27: 149–50. doi: 10.3109/19401736.2013.878918 [DOI] [PubMed] [Google Scholar]
- 63.Cao YQ, Ma C, Chen JY, Yang DR. The complete mitochondrial genomes of two ghost moths, Thitarodes renzhiensis and Thitarodes yunnanensis: the ancestral gene arrangement in Lepidoptera. BMC Genomics 2012;13: 276 doi: 10.1186/1471-2164-13-276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dowton M, Austin AD. Evolutionary dynamics of a mitochondrial rearrangement" hot spot" in the Hymenoptera. Mol Biol Evol. 1999;16: 298–309. doi: 10.1093/oxfordjournals.molbev.a026111 [DOI] [PubMed] [Google Scholar]
- 65.Park JS, Kim MJ, Jeong SY, Kim SS, Kim I. Complete mitochondrial genomes of two gelechioids, Mesophleps albilinella and Dichomeris ustalella (Lepidoptera: Gelechiidae), with a description of gene rearrangement in Lepidoptera. Curr Genet. 2016;62: 809–826. doi: 10.1007/s00294-016-0585-3 [DOI] [PubMed] [Google Scholar]
- 66.Liu QN, Xin ZZ, Zhu XY, Chai XY, Zhao XM, Zhou CL, et al. A transfer RNA gene rearrangement in the lepidopteran mitochondrial genome. Biochem Biophys Res Commun. 2017;489: 149–154. doi: 10.1016/j.bbrc.2017.05.115 [DOI] [PubMed] [Google Scholar]
- 67.Margam VM, Coates BS, Hellmich RL, Agunbiade T, Seufferheld MJ, Sun W, et al. Mitochondrial genome sequence and expression profiling for the legume pod borer Maruca vitrata (Lepidoptera: Crambidae). PloS One 2011;6: e16444 doi: 10.1371/journal.pone.0016444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stewart JB, Beckenbach AT. Characterization of mature mitochondrial transcripts in Drosophila, and the implications for the tRNA punctuation model in arthropods. Gene 2009;445: 49–57. doi: 10.1016/j.gene.2009.06.006 [DOI] [PubMed] [Google Scholar]
- 69.DeSalle R, Freedman T, Prager EM, Wilson AC. Tempo and mode of sequence evolution in mitochondrial DNA of Hawaiian Drosophila. J Mol Evol. 1987;26: 157–64. doi: 10.1007/BF02111289 [DOI] [PubMed] [Google Scholar]
- 70.Sarmah MC. Eri pupa: a delectable dish of North East India. Curr Sci. 2011;100: 279. [Google Scholar]
- 71.Meiklejohn CD, Montooth KL, Rand DM. Positive and negative selection on the mitochondrial genome. Trends Genet. 2007;23: 259–263. doi: 10.1016/j.tig.2007.03.008 [DOI] [PubMed] [Google Scholar]
- 72.Lavrov DV, Brown WM, Boore JL. A novel type of RNA editing occurs in the mitochondrial tRNAs of the centipede Lithobius forficatus. Proc Natl Acad Sci USA 2000;97: 13738–13742. doi: 10.1073/pnas.250402997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang DX, Hewitt GM. Insect mitochondrial control region: a review of its structure, evolution and usefulness in evolutionary studies. Biochem Syst Ecol. 1997;25: 99–120. doi: 10.1016/S0305-1978(96)00042-7 [Google Scholar]
- 74.Stolle E, Kidner JH, Moritz RFA. Patterns of Evolutionary Conservation of Microsatellites (SSRs) Suggest a Faster Rate of Genome Evolution in Hymenoptera Than in Diptera. Genome Biol Evol. 2013;5: 151–162. doi: 10.1093/gbe/evs133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu QN, Xin ZZ, Bian DD, Chai XY, Zhou CL, Tang BP. The first complete mitochondrial genome for the subfamily Limacodidae and implications for the higher phylogeny of Lepidoptera. Sci Rep. 2016;6: 35878 doi: 10.1038/srep35878 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(A) G+C content versus Ka/Ks ratios of 13 concatenated PCGs in A. assamensis mitogenome (B) The usage rate of five frequently used codons in Bombycoidea members.
(TIF)
trnY lacked TΨC loop. tRNA here are represented as trn. AA denotes amino acyl arm and AC denotes anticodon arm.
(TIF)
trnS1 lacked DHU loop. tRNA here are represented as trn. AA denotes amino acyl arm and AC denotes anticodon arm.
(TIF)
AA denotes Aminoacyl arm and AC denotes Anticodon arm. S-L denotes Stem-Loop of tRNA gene. tRNA is represented as trn.
(TIF)
AA denotes Aminoacyl arm and AC denotes Anticodon arm. S-L denotes Stem-Loop of tRNA gene. tRNA is represented as trn.
(TIF)
The tree is drawn to scale with bootstrap values indicated along with the branches.
(TIF)
The tree is drawn to scale with bayesian posterior probability values indicated along with the branches.
(TIF)
(PDF)
C here denotes conserved sites, I denotes phylogenetically informative sites, V denotes variable sites, N denotes nucleotide level, A denotes amino acid level. The number of sites is represented in terms of percentage values.
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. The full annotated mitogenome sequence and SRA data of A. assamensis submitted to NCBI GenBank are available under the accession numbers KU379695 and SRR3948351, respectively.