Here, Rogers et al. investigated how piRNA precursors are selected and channeled into the endonuclease Zucchini (Zuc)-dependent processing pathway in Drosophila germ cells. They engineered a modular system that can induce primary piRNA biogenesis at an arbitrary locus even in the absence of native piRNA precursors. They also established a subcellular compartmentalization as a key factor in RNA processing.
Keywords: piRNA biogenesis, nuage, Zucchini, tethering, Piwi, RNA sequestration
Abstract
The piRNA pathway represses transposable elements in the gonads and thereby plays a vital role in protecting the integrity of germline genomes of animals. Mature piRNAs are processed from longer transcripts, piRNA precursors (pre-piRNAs). In Drosophila, processing of pre-piRNAs is initiated by piRNA-guided Slicer cleavage or the endonuclease Zucchini (Zuc). As Zuc does not have any sequence or structure preferences in vitro, it is not known how piRNA precursors are selected and channeled into the Zuc-dependent processing pathway. We show that a heterologous RNA that lacks complementary piRNAs is processed into piRNAs upon recruitment of several piRNA pathway factors. This processing requires Zuc and the helicase Armitage (Armi). Aubergine (Aub), Argonaute 3 (Ago3), and components of the nuclear RDC complex, which are required for normal piRNA biogenesis in germ cells, are dispensable. Our approach allows discrimination of proteins involved in the transcription and export of piRNA precursors from components required for the cytoplasmic processing steps. piRNA processing correlates with localization of the substrate RNA to nuage, a distinct membraneless cytoplasmic compartment, which surrounds the nucleus of germ cells, suggesting that sequestration of RNA to this subcellular compartment is both necessary and sufficient for selecting piRNA biogenesis substrates.
Mature piRNAs are processed from longer transcripts, piRNA precursors (pre-piRNAs), most of which are derived from piRNA clusters—genomic regions with a high density of transposon sequences (Brennecke et al. 2007; Gunawardane et al. 2007). Insertion of a heterologous sequence in piRNA clusters leads to its processing into piRNAs, indicating that piRNAs can be processed from any sequence (Todeschini et al. 2010; Muerdter et al. 2012). piRNA biogenesis is a multistep process starting with transcription and early processing of precursor RNA in the nucleus, export, precursor cleavage followed by processing to mature piRNAs, and loading into piwi proteins. In the cytoplasm, piRNA precursors are processed by two mechanisms: the ping-pong cycle and Zucchini (Zuc)-dependent piRNA biogenesis (Brennecke et al. 2007; Gunawardane et al. 2007; Malone et al. 2009). In the ping-pong cycle, processing of pre-piRNAs is initiated by the endonucleolytic (slicer) activity of cytoplasmic piwi proteins Aubergine (Aub) or Argonaute3 (Ago3), loaded with complementary piRNAs (Brennecke et al. 2007; Gunawardane et al. 2007). The characteristic features of ping-pong biogenesis are the presence of complementary piRNA pairs whose 5′ ends are separated by 10 nucleotides (nt) and the generation of piRNAs with a bias for U at position 1 and/or A at position 10 of their sequence (Wang et al. 2014). Zuc-dependent biogenesis, in which endonucleolytic cleavage of pre-piRNAs by Zuc generates the 5′—and in part the 3′—ends of piRNAs, does not depend on either the slicer activity of piwi proteins or the presence of complementary piRNAs (Malone et al. 2009). piRNAs generated through Zuc-dependent biogenesis show a bias for U at their 5′ end but lack a bias for A at position 10. Many of the cytoplasmic piRNA biogenesis factors—including Aub and Ago3 as well as numerous factors that were genetically identified to act in the pathway, such as Vasa, Armitage (Armi), and Krimper—localize to membraneless perinuclear structures called nuage in germ cells and Yb bodies in the surrounding somatic follicular cells, leading to the assumption that these structures are the site of piRNA biogenesis (Hay et al. 1988; Lasko and Ashburner 1990; Lim and Kai 2007; Kirino et al. 2009; Malone et al. 2009; Qi et al. 2011; Ipsaro et al. 2012; Nishimasu et al. 2012; Ohtani et al. 2013; Webster et al. 2015).
How piRNA precursors are identified by the biogenesis machinery remains unresolved. Other RNA processing events, such as microRNA (miRNA) or CRISPR RNA biogenesis, splicing, or polyadenylation, require specific sequence and/or structure motifs in precursor RNA that are recognized by the processing machinery (Zeng et al. 2005; Park et al. 2011; Barrangou and Marraffini 2014; Li and Patel 2016; Tsai and Joung 2016). Common sequence or structural motifs that are shared by all piRNA precursors were not identified (Muerdter et al. 2012). In the absence of sequence motifs, two mutually nonexclusive models have been proposed to explain precursor selection in the germline, which we call the “persistent nuclear mark model” and the “selection by pre-existing piRNA model.”
The “persistent nuclear mark model” proposes that specific proteins associate with piRNA precursors in the nucleus and remain associated with the transcripts in the cytoplasm, where they interact with the processing machinery. In the germline, genomic regions that give rise to the majority of piRNAs, called piRNA clusters, are marked by the histone 3 Lys9 trimethylation (H3K9me3) mark and the RDC complex. RDC consists of the HP1 homolog Rhino and two additional proteins, Cutoff (Cuff) and Deadlock, and is required for transcription and early processing of cluster transcripts (Klattenhoff et al. 2009; Pane et al. 2011; Le Thomas et al. 2014; Mohn et al. 2014; Zhang et al. 2014; Chen et al. 2016). The well-conserved transcription export (TREX) complex is also enriched at cluster loci and binds piRNA precursors cotranscriptionally in an RDC-dependent fashion (Zhang et al. 2012; Hur and Chung 2016; Hur et al. 2016). Components of either the RDC or the TREX complex were proposed to constitute the mark that triggers processing in the cytoplasm. However, the nature of the mark and the mechanism by which it engages the processing machinery remain unclear. Evidence for RDC or TREX remaining associated with piRNA precursors after nuclear export is also lacking. Finally, tethering of Rhino to a single-stranded transgene (without its concomitant tethering to a complementary antisense transgene) does not trigger piRNA biogenesis (Zhang et al. 2014), arguing against the idea that binding of RDC by itself is sufficient to specify piRNA precursors.
The “selection by pre-existing piRNA model” suggests that precursors are specified in the cytoplasm by complementary piRNAs associated with the cytoplasmic piwi proteins. Existing piRNAs can target pre-piRNAs and induce their processing via the ping-pong cycle (Brennecke et al. 2007; Gunawardane et al. 2007). piRNA-dependent cleavage not only generates complementary piRNAs (ping-pong partners) but also leads to phased processing of the precursor downstream from the initial cleavage (Han et al. 2015; Homolka et al. 2015; Mohn et al. 2015; Senti et al. 2015; Wang et al. 2015). This process generates Piwi-loaded piRNAs. Mutation of the two piwi proteins involved in the ping-pong cycle, Aub and Ago3, greatly reduces Piwi-bound piRNAs in the germline, leading to the suggestion that Zuc-mediated piRNA biogenesis in germ cells is triggered by the ping-pong cycle (Han et al. 2015; Homolka et al. 2015; Mohn et al. 2015; Senti et al. 2015; Wang et al. 2015).
Follicular cells of the fly ovary lack ping-pong biogenesis factors as well as the RDC complex yet produce piRNAs, suggesting an alternative precursor selection mechanism. Some piRNA precursors specific to follicular cells, such as the mRNA of traffic-jam (tj) and transcripts from the flamenco piRNA cluster, harbor sequences that are bound by the RNA-binding protein Yb (Ishizu et al. 2015) and can trigger processing into piRNAs (Homolka et al. 2015; Ishizu et al. 2015). Whether other somatic piRNA precursors also harbor sequence motifs that are bound by Yb and whether binding of Yb leads to piRNA processing remain to be elucidated.
Genetically, many factors have been identified to be essential for piRNA biogenesis; however, a mechanistic dissection of which steps these factors are involved in (i.e., transcription, export, recognition, or cytoplasmic processing) is largely missing. Assigning many of the identified factors to specific steps of the biogenesis pathway is hindered by the absence of an in vitro system that allows independent analysis of different stages of the pathway. In vivo analysis is impaired because disrupting proteins acting in early steps of the pathway, such as transcription of pre-piRNAs, eliminates piRNA precursors and thereby the possibility of testing whether these factors also act in downstream steps of processing.
The core of the cytoplasmic processing machinery, including the endonuclease Zuc, is shared by nurse and follicular cells, raising the question of how the same processing machinery has evolved to recognize its targets by two distinct (sequence-specific in follicular cells and sequence-independent in nurse cells) mechanisms. We show that direct recruitment of a heterologous sequence to the cytoplasmic piRNA processing machinery can bypass the necessity for sequence-specific recognition, as well as the need for both complementary piRNAs and a unique chromatin architecture. Our work reveals that sequestration of RNA to the cytoplasmic processing machinery is both necessary and sufficient for selecting substrates for piRNA biogenesis, leading to a unified model of piRNA specification in the two cell types. The described experimental approach bypasses nuclear biogenesis steps, enabling identification and functional dissection of factors that act in Zuc-mediated processing downstream from nuclear export.
Results
Recruitment of Yb and Piwi to RNA in follicular cells leads to piRNA processing
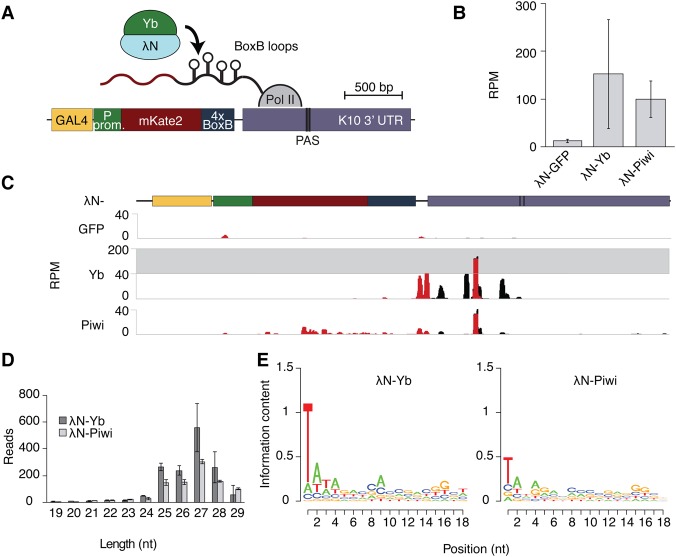
Two transcripts that are processed into piRNAs in follicular cells, the mRNA of tj and transcripts from the flamenco piRNA cluster, were shown to harbor sequences that associate with the Yb protein and trigger piRNA processing when transferred to heterologous transcripts (Homolka et al. 2015; Ishizu et al. 2015). To determine whether the interaction of a transcript with Yb is sufficient to induce processing, we tested whether we could trigger piRNA production in follicular cells by tethering a heterologous sequence that does not have sequence homology with natural piRNA substrates to Yb (Fig. 1A). The sequence of the reporter mRNA contains four BoxB sequences in its 3′ untranslated region (UTR) that are bound by the λN domain of anti-terminator protein N fused to Yb (Fig. 1A). We coexpressed λN-tagged Yb with the reporter in follicular cells of the fly ovary and sequenced ovarian small RNAs. Tethering of the transcript to Yb resulted in production of reporter-derived small RNAs, while small RNAs were absent upon recruitment of the control GFP protein (Fig. 1B,C). The majority of small RNAs produced from the reporter upon Yb tethering was 25–28 nt long and had a U bias at the first nucleotide (82.2% of all reporter-derived reads had a 5′U), consistent with these sequences being piRNAs (Fig. 1D,E). Thus, recruitment of Yb to a novel transcript is sufficient to trigger piRNA processing in follicular cells in the absence of any sequence or structure motifs in the RNA.
Figure 1.
Yb and Piwi recruitment to RNA in follicular cells triggers piRNA production from reporter mRNA. (A) Schematic diagram of the reporter construct used to study the effect of piRNA pathway component recruitment to RNA. (PAS) PolyA signal. (B) Recruitment of Yb and Piwi, but not GFP, to a reporter results in small RNA reads mapping to the reporter sequence. Shown are small RNA sequencing reads from 19- to 30-nt total ovarian RNA. RPM (reads per million mapped reads) was calculated for reads mapping exclusively to the reporter, normalized to total reads mapping to the DM3 genome. Error bars indicate standard deviation between replicates. n = 2. (C) The small RNA reads produced upon tethering Yb or Piwi map to the reporter. Shown are profiles for all reads (black) and reporter-specific reads (red) mapping to the reporter, normalized to all reads mapping to the DM3. (D) Tethering of Yb or Piwi results in production of piRNA-length reads from the reporter. Shown are size profiles of all reads mapping to the reporter. Error bars indicate standard deviation between replicates. n = 2. (E) Reporter-derived sequences upon Yb or Piwi tethering exhibit a 1U bias. The Weblogos were generated from all reads mapping to the reporter.
In follicular cells, piRNAs are loaded into Piwi, the only member of the piwi clade expressed in these cells. Therefore, we asked whether recruitment of Piwi itself to RNA is sufficient to trigger processing. Tethering of Piwi also induced generation of piRNAs from the reporter (Fig. 1B–E). We note that the piRNA profiles along the reporter differ depending on the factor recruited. Piwi recruitment leads to processing of the entire transcript, while Yb recruitment leads to piRNA biogenesis only downstream from the BoxB sites, to which the Yb fusion protein binds (Fig. 1C). This is similar to the piRNAs observed in the tj mRNA and when using the recognition sequence from flamenco, both of which lead to processing only downstream from the recognition sequence (Homolka et al. 2015; Ishizu et al. 2015). Our data indicate that Yb binding not only identifies piRNA precursors but also specifies the entry site for processing. Thus, in follicular cells, recruitment of either Yb or Piwi is sufficient to trigger piRNA processing from a novel substrate, and Yb might be responsible for specifying where processing initiates within the RNA.
Recruitment of the nuclear RDC and TREX complexes or Zuc is not sufficient to trigger piRNA generation in germ cells
In the germline, piRNA biogenesis is more complex than in somatic follicular cells and depends on numerous nuclear and cytoplasmic factors. Two nuclear complexes—RDC and TREX, which localize to chromatin of piRNA loci and nascent pre-piRNA, respectively—were proposed to play a role in selecting piRNA precursors and guiding them to the processing machinery in germ cells. To test whether the recruitment of RDC or TREX to the heterologous transcript is sufficient to trigger piRNA processing in germ cells, we tethered the RDC component Cuff and the TREX component Thoc5 to the reporter in the female germline and profiled ovarian small RNAs. Tethering of these factors did not trigger piRNA generation (Fig. 2A,B), suggesting that recruitment of RDC or TREX is not sufficient to trigger biogenesis from novel RNA substrates in germ cells.
Figure 2.
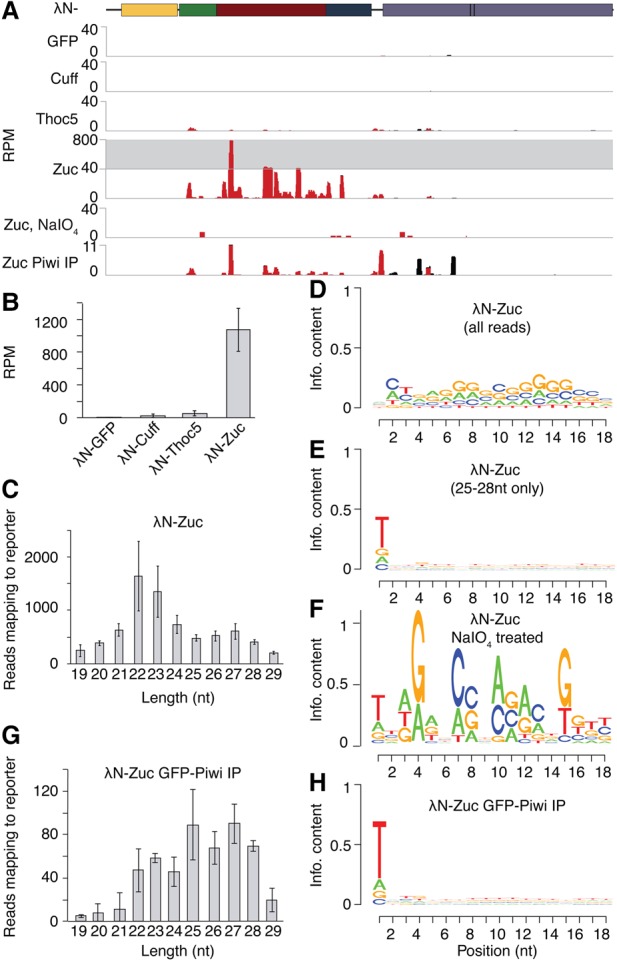
Recruitment of the nuclear RDC and TREX complexes or Zuc is not sufficient to trigger piRNA processing in germ cells. (A) Recruitment of Zuc—but not Cuff, Thoc5, or GFP—to the reporter results in small RNA production. However, upon NaIO4 treatment, reporter-derived small RNAs generated upon Zuc tethering are lost. Shown are profiles for all reads (black) and reporter-specific reads (red) mapping to the reporter, normalized to all reads mapping to the DM3. (B) Normalized small RNA-seq reads (cumulative RPM) upon germline tethering of GFP, Cuff, Thoc5, or Zuc. RPM was calculated for 19- to 30-nt total ovarian RNA reads mapping exclusively to the reporter, normalized to total reads mapping to the DM3 genome. Error bars indicate standard deviation between replicates. n = 2. (C) Zuc tethering leads to production of two classes of small RNAs. Shown is the size profile of all reads mapping to the reporter. Error bars indicate standard deviation between replicates. n = 2. (D) Reporter-derived sequences upon Zuc tethering do not show any nucleotide bias. The Weblogo was generated from all reads mapping to the reporter. (E) piRNA-sized (25- to 28-nt) reporter sequences produced upon Zuc tethering exhibit a slight 1U bias. The Weblogo was generated from all 25- to 28-nt reads mapping to the reporter. (F) The Weblogo of NaIO4-treated reporter-derived small RNAs upon Zuc tethering indicates a slight 1U bias but very few reads. (G) Piwi-loaded small RNAs upon Zuc tethering are piRNA length. Shown is the size profile of all reads mapping to the reporter. Error bars indicate standard deviation between replicates. n = 2. (H) Reporter-derived sequences upon Zuc tethering that are loaded into Piwi show a 1U bias, indicating that they are bona fide piRNAs. The Weblogo was generated from all reads mapping to the reporter.
Next we tested whether recruitment of Zuc—the endonuclease responsible for generating the 5′ end and, to some extent, the 3′ end of piRNAs (Malone et al. 2009; Ipsaro et al. 2012; Nishimasu et al. 2012; Han et al. 2015; Mohn et al. 2015)—is sufficient to trigger piRNA biogenesis. Small RNA sequencing (RNA-seq) upon Zuc tethering revealed the presence of abundant small RNAs arising from the reporter transcript (Fig. 2A,B). However, small RNA sequences were predominantly 22–23 nt in length (mostly from the 5′ region of the reporter) (Supplemental Fig. S1) and were depleted of 5′U reads (13.9% of all reporter-mapped reads had a 5′U; standard deviation = 1.3; note that the reporter is depleted of uracils, with an overall U content of 21.3%) (Fig. 2C–F). Small RNAs produced upon Zuc tethering were sensitive to NaIO4 treatment (Fig. 2A,F), further confirming that the majority of reporter-derived reads upon tethering Zuc is not bona fide piRNAs. Thus, Zuc tethering induces processing of the transcript but not the efficient production of piRNAs. A closer look at the size profile revealed a smaller peak (27.4% of all reporter-derived reads; standard deviation = 0.01) around the 25- to 28-nt piRNA size range (Fig. 2C). This small RNA population had slightly more 5′U RNAs (28.4% had 5′U; standard deviation = 0.03) and was derived from the entire length of the reporter (Fig. 2E; Supplemental Fig. S1).
To test whether any genuine piRNAs are formed upon recruitment of Zuc, we immunoprecipitated Piwi/piRNA complexes and cloned the isolated piRNAs. Analysis of Piwi-associated RNAs yielded reporter-derived small RNAs with a size profile and 5′U bias characteristic of piRNAs (41.8% had 5′U) (Fig. 2A,G,H). Thus, tethering of Zuc leads to reporter processing into small RNAs; however, only a small fraction of the formed small RNAs is genuine piRNAs.
Recruitment of Piwi and cytoplasmic piRNA pathway components triggers piRNA biogenesis in germ cells
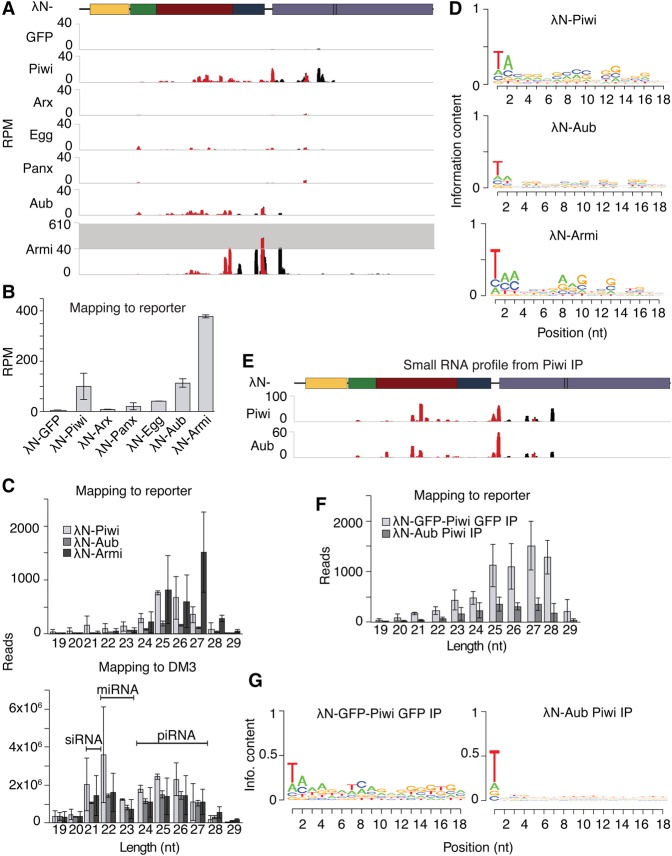
Recent studies suggested that in germ cells, both slicer-dependent processing and Zuc-mediated processing are triggered by initial Aub- or Ago3-mediated cleavage guided by complementary piRNAs (Han et al. 2015; Mohn et al. 2015; Senti et al. 2015; Wang et al. 2015). Our reporter lacks complementary piRNAs, based on both bioinformatics analysis (Supplemental Fig. S2) and the fact that no processing of the reporter was detected in control flies upon recruitment of GFP. Thus, our inability to trigger piRNA biogenesis in germ cells might be due to the absence of piRNAs that are complementary to the reporter. We tested whether the recruitment of Piwi—which, unlike Yb, is expressed in both cell types (Cox et al. 2000)—can trigger piRNA generation in the absence of cognate piRNAs in germ cells. Small RNA profiling revealed that, like in follicular cells, recruitment of Piwi in germ cells triggers small RNA processing from the reporter (Fig. 3A,B). Similar results were obtained with a reporter inserted in a different genomic location and containing a different sequence (except for the BoxB sites) (Supplemental Fig. S3; Sienski et al. 2015), indicating that neither the genomic location of the target nor its sequence was crucial for processing.
Figure 3.
In germ cells, recruitment of Piwi and cytoplasmic piRNA pathway components triggers piRNA production from the reporter. (A) Tethering of Piwi, Aub, or Armi in the germline produces piRNA from the reporter, whereas tethering of the chromatin factors Arx, Egg, and Panx does not. Shown are profiles for all reads (black) and reporter-specific reads (red) mapping to the reporter, normalized to all reads mapping to the DM3. (B) Shown is cumulative RPM from 19- to 30-nt total ovarian RNA. RPM was calculated for reads mapping exclusively to the reporter, normalized to total reads mapping to the DM3 genome. Error bars indicate standard deviation between replicates. n = 2. (C) Tethering of Piwi, Aub, or Armi leads to piRNA-length reads from the reporter. Shown are size profiles of all reads mapping to the reporter (top) and, as a size selection control, the DM3 genome (bottom) upon tethering Piwi, Aub, or Armi. Error bars indicate standard deviation between replicates. n = 2. (D) Upon recruitment of Piwi, Aub, or Armi, reporter-derived sequences show a 1U bias. The Weblogos were generated from all reads mapping to the reporter. (E) Upon tethering of Piwi or Aub, small RNA sequences derived from the reporter are loaded into Piwi. Shown are profiles for all reads (black) and reporter-specific reads (red) mapping to the reporter, normalized to all reads mapping to the DM3. (F) Piwi-loaded reporter small RNA sequences obtained upon Piwi or Aub tethering are piRNA length. Shown are size profiles of all reads mapping to the reporter. Error bars indicate standard deviation between replicates. n = 2. (G) Piwi-loaded reporter small RNA sequences show a 1U bias. The Weblogos were generated from all reads mapping to the reporter.
We tested whether other components of the piRNA pathway can trigger processing in germ cells upon their recruitment to the reporter. Recruitment of nuclear proteins that work together with or downstream from Piwi to repress Piwi targets such as Asterix (Arx), Panoramix (Panx), and the histone methyltransferase SetDB1/Egg (Donertas et al. 2013; Muerdter et al. 2013; Sienski et al. 2015; Yu et al. 2015) did not trigger piRNA biogenesis (Fig. 3A,B). Recruitment of the cytoplasmic components Aub and Armi, two nuage components involved in pong-pong and Zuc-dependent processing, respectively, resulted in small RNA biogenesis (Fig. 3A,B). Small RNAs generated upon Piwi, Aub, or Armi tethering were resistant to NaIO4 treatment, confirming that they are generated by the canonical piRNA biogenesis machinery (Supplemental Fig. S4A).
The bulk of the reporter-mapped small RNAs upon Piwi, Aub, and Armi tethering was 25–28 nt in length (Fig. 3C), and the majority (60.4% [standard deviation = 1.8], 55.9%, and 78.4% [standard deviation = 0.06], respectively) had a strong bias for U at the first position (Fig. 3D; Supplemental Fig. S4B), both features of piRNAs. Cloning of small RNAs isolated from immunoprecipitated Piwi complex upon Piwi or Aub tethering confirmed that they are indeed bona fide piRNAs (Fig. 3E–G). Reporter piRNAs are derived only from the sense strand of the reporter (99.4% [standard deviation = 0.001], 99.4% and 99.8% [standard deviation = 0.001], respectively) and lack a bias for A at position 10 (a feature of ping-pong piRNAs) (Fig. 3D,G). Together with the lack of complementary piRNAs, this suggests that these piRNAs are formed through a ping-pong-independent mechanism. We note that neither of our tethering experiments resulted in phased small RNAs from the reporter, although we did detect phasing in these libraries when we analyzed cluster 42AB (Supplemental Fig. S4C). We conclude that tethering of several different factors, such as Piwi, Aub, and Armi, to the reporter in germ cells triggers piRNA biogenesis. The mechanism by which reporter piRNAs are generated does not require complementary piRNAs and ping-pong processing and thus seems similar, if not identical, to the mechanism observed in follicular cells.
Identification of factors required for piRNA processing in germ cells
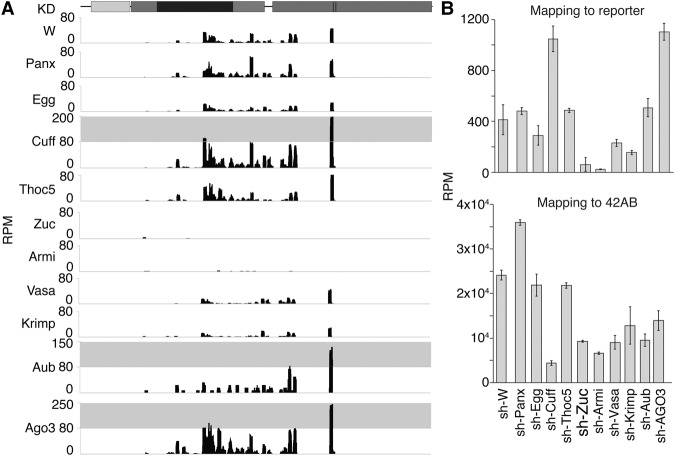
Having shown that tethering of Piwi, Aub, or Armi is sufficient for piRNA biogenesis, we set out to identify factors that are required for piRNA processing of the reporter mRNA. We combined tethering of Piwi to the reporter with germline knockdown (Supplemental Fig. S5) of the respective gene followed by small RNA cloning.
Factors that work with Piwi to induce transcriptional repression (Panx and Egg) (Donertas et al. 2013; Muerdter et al. 2013; Sienski et al. 2015; Yu et al. 2015) are not required for piRNA biogenesis upon Piwi tethering (Fig. 4A,B). Similarly, Cuff and Thoc5, components of the RDC and TREX complexes, respectively, were dispensable for generation of reporter piRNAs (Fig. 4A,B). These results indicate that nuclear Piwi partners as well as RDC and TREX are not required for piRNA biogenesis triggered by Piwi recruitment.
Figure 4.
Chromatin factors are dispensable, but primary biogenesis factors are required for piRNA generation upon Piwi tethering. (A) Knockdown of piRNA factors concomitantly to Piwi tethering leads to loss of piRNA only if Zuc-mediated cytoplasmic biogenesis factors are affected. Shown are small RNA profiles along the reporter for 25- to 28-nt reads mapping exclusively to the reporter, normalized to 25- to 28-nt reads mapping to the DM3. (B) Shown is the cumulative RPM of small RNA from 25- to 28-nt total ovarian RNA calculated for all reads mapping to the reporter (top) and total reads uniquely mapping to cluster 42AB (bottom), normalized to total reads mapping to the DM3 genome. Knockdown of White was used as a control.
In contrast, reporter piRNAs were strongly (sevenfold) reduced upon knockdown of Zuc (Fig. 4A,B). Similarly, strong (14-fold) reduction of piRNA biogenesis was observed upon knockdown of Armi (a putative RNA helicase). These data confirm that reporter-derived small RNAs are genuine piRNAs formed through the canonical Zuc-dependent biogenesis pathway. A somewhat weaker effect on piRNA biogenesis was seen upon knockdown of other cytoplasmic piRNA processing proteins, such as the nuage component Vasa, an RNA helicase, and the Tudor domain protein Krimper, which reduced reporter piRNA levels approximately twofold (Fig. 4A,B). The cytoplasmic piwi proteins Aub and Ago3 were dispensable for piRNA generation from the reporter upon Piwi tethering. This result further confirms that reporter piRNAs are generated independently of the ping-pong cycle (Fig. 4A,B). Knockdown of Cuff and Ago3 resulted in an apparent increase in reporter-derived small RNAs when depicted as RPM of all mapping reads (Fig. 4A,B), likely due to the massive loss of endogenous piRNAs mapping to the DM3 genome. Overall, our results indicate that reporter RNA is processed by the canonical piRNA processing machinery, which includes Zuc and Armi, and that tethering bypasses the requirement of nuclear proteins.
Tethering to Piwi leads to transcript accumulation in nuage
We used RNA-FISH to test whether piRNA biogenesis triggered by tethering of Piwi leads to a change in the subcellular localization of the reporter RNA. Reporter RNA was uniformly distributed in the cytoplasm in control cells upon GFP tethering. Accumulation of the transcript in distinct perinuclear foci, which also show vasa enrichment, was observed upon Piwi tethering, indicating that Piwi recruits the bound RNA to nuage (Fig. 5A). Thus, induction of piRNA biogenesis from a heterologous reporter correlates with sequestration of the reporter mRNA to nuage.
Figure 5.
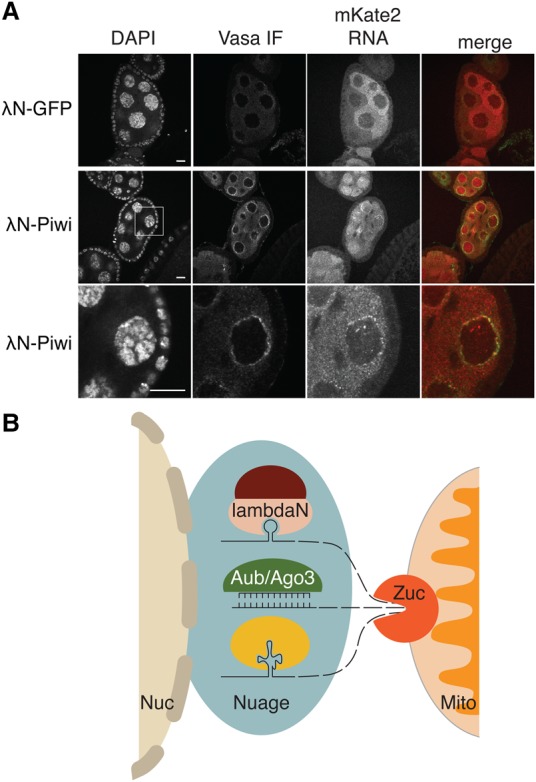
Tethering to Piwi leads to transcript sequestration in nuage. (A) Artificial tethering of RNA to Piwi leads to its accumulation in perinuclear granules. RNA-FISH experiments detected dispersed mKate-4BoxB reporter mRNA upon tethering to GFP (control) and perinuclear colocalization of the RNA with Vasa upon Piwi tethering. Higher magnification of the boxed section is shown in the third row. Bars, 10 µm. (B) An overarching model of precursor selection for primary piRNA biogenesis.
Discussion
Compared with the processing of other classes of small RNAs, piRNA biogenesis is very complex and requires numerous proteins with, to date, unspecified roles in piRNA processing. How the precursor RNAs are discriminated from other cellular RNA and directed to the processing machinery is also not clear. As standard genetic and biochemical approaches failed to provide clear answers to these questions, we used an alternative approach, which is based on tethering of several piRNA pathway components to a heterologous reporter, to gain a better understanding of piRNA biogenesis. We found that tethering of several proteins (Piwi, Aub, and Armi) to an RNA that lacks homology with other piRNA precursors triggers its processing into piRNAs (Fig. 3A,B). The piRNAs produced by this approach have all of the features of canonical piRNAs: They have the correct size and a bias for U at the first nucleotide position and are loaded into Piwi (Fig. 3C–G). Biogenesis of these piRNAs is mediated by the canonical processing machinery (Fig. 4).
Experimental approach to dissect the piRNA biogenesis pathway
Most of the numerous proteins required for piRNA biogenesis were identified through genetic screens (Handler et al. 2011, 2013; Muerdter et al. 2013). Mutations in these genes diminish or eliminate mature piRNAs; however, it was difficult to assign specific molecular functions to these proteins, largely due to the fact that we do not know intermediate steps of piRNA processing that could be analyzed in these mutants. For example, the nuclear RDC and TREX complexes are required for piRNA biogenesis and were proposed to direct pre-piRNAs to the processing machinery (Klattenhoff et al. 2009; Pane et al. 2011; Le Thomas et al. 2014; Mohn et al. 2014; Zhang et al. 2014; Chen et al. 2016; Hur and Chung 2016; Hur et al. 2016); however, the molecular mechanism by which these complexes might bring precursors to the cytoplasmic machinery remains unresolved.
The approach used in this study, tethering combined with knockdown of piRNA biogenesis factors, provides a valuable tool to dissect steps of the piRNA biogenesis pathway and discriminate proteins involved in processing and precursor localization per se from factors involved in upstream steps of biogenesis, such as transcription and nuclear export of pre-piRNAs. As expected, processing absolutely requires Zuc, the endonuclease that mediates the formation of both the 5′ and 3′ ends of piRNAs (Fig. 4; Malone et al. 2009; Ipsaro et al. 2012; Nishimasu et al. 2012; Han et al. 2015; Mohn et al. 2015). While recruitment of Zuc to RNA leads to processing of the transcript, the majority of the obtained small RNAs is not piRNAs, indicating that Zuc by itself is not sufficient for efficient piRNA processing (Fig. 2). We show that another factor that is required for piRNA biogenesis upon tethering is the putative RNA helicase Armi (Fig. 4). Thus, efficient piRNA processing requires the interaction between Zuc and other proteins such as Armi.
We found that components of the nuclear RDC and TREX complexes, which associate with chromatin of piRNA clusters and nascent pre-piRNAs, respectively, are not required for piRNA processing upon tethering (Fig. 4), suggesting that their requirement for piRNA biogenesis can be bypassed. Previous studies have shown that both RDC and TREX are required for transcription of pre-piRNAs (Klattenhoff et al. 2009; Pane et al. 2011; Le Thomas et al. 2014; Mohn et al. 2014; Zhang et al. 2014; Chen et al. 2016; Hur and Chung 2016; Hur et al. 2016). TREX is likely also involved in nuclear export of pre-piRNAs. It is possible that RDC and TREX are only necessary in the nucleus for transcription and export of pre-piRNAs and do not interact with the cytoplasmic processing machinery. Alternatively, these complexes might be required to channel natural precursors to the processing machinery, but this function can be effectively bypassed by recruitment of several different proteins to the RNA. Either way, RDC and TREX are not part of the core processing machinery. The same approach can be used to assign roles to other (including yet unidentified) piRNA biogenesis factors.
Zuc-dependent, ping-pong-independent piRNA processing exists in germ cells
Tethering of several proteins to an RNA that has no complementarity to mature piRNAs triggers its processing to piRNAs. This processing is observed in both follicular and germ cells (Figs. 1,3) and is independent of pre-existing piRNAs (Supplemental Fig. S2) and ping-pong. Historically, two types of piRNA biogenesis were proposed: the ping-pong cycle, which relies on the catalytic (slicer) activity of Aub and Ago3 to generate the 5′ end of new piRNAs (Brennecke et al. 2007; Gunawardane et al. 2007), and the so-called primary processing, which was thought to be independent of ping-pong (Malone et al. 2009). Recent studies suggested that in germ cells, “primary” biogenesis is triggered by initial ping-pong processing (Han et al. 2015; Mohn et al. 2015; Senti et al. 2015; Wang et al. 2015). These results suggested that “primary” piRNA biogenesis is actually downstream from ping-pong (or “secondary” biogenesis), making these titles obsolete. Our data show that piRNA biogenesis that is independent of the ping-pong machinery can be achieved—at least by the artificial tethering approach—in germ cells. Therefore, both germ cells and somatic follicular cells (which lack the ping-pong machinery) (Malone et al. 2009) possess the same machinery, which allows them to process piRNAs in a ping-pong-independent fashion. We propose that Aub and Ago3 might promote Zuc-dependent processing of piRNA precursors through sequestration of these RNA to a specific cellular compartment rather than through its slicing activity (see below).
The sequestration to nuage might explain the selection of substrates for piRNA processing
How piRNA precursors are recognized by the piRNA processing machinery is still unresolved. The core piRNA processing machinery, composed of Zuc and Armi, is similar in both cell types, suggesting that a common principle for precursor selection should exist. piRNAs could be generated from artificial sequences inserted into piRNA clusters (Muerdter et al. 2012). piRNA processing from genic and tRNA transcripts was also observed (correlating with transcript abundance) in Kc167 cells. Both of these observations argue against the need for specific sequence/structure motifs in precursor transcripts (Vrettos et al. 2017). We propose that sequestration of RNA into a distinct cellular compartment might be the central principle for precursor selection that is shared by germline and follicular cells. Based on our proposed model, any RNA that is localized to the processing compartment will be processed to piRNAs in a sequence-independent fashion (Fig. 5B).
Several lines of evidence support this hypothesis. First, efficient piRNA biogenesis can be triggered by recruitment of several different proteins to RNA, which have little in common except for their localization to nuage. Some of these factors, such as Aub, localize to nuage in steady state, while others localize transiently, such as Piwi on its way to the nucleus (Lim and Kai 2007; Le Thomas et al. 2013; Huang et al. 2014; Webster et al. 2015). The only compartment where both Aub and Piwi are present together is nuage (Malone et al. 2009). Recruitment of Zuc—the central player in processing, according to both previous results and our data—does not lead to efficient processing into piRNAs (Fig. 2). Zuc associates with mitochondria and therefore is localized throughout the cytoplasm and not exclusively to nuage (Handler et al. 2013). Second, we found that even though Zuc-dependent piRNA biogenesis does not require ping-pong, biogenesis is reduced upon knockdown of Vasa and Krimp, proteins that localize to nuage and were implicated previously in ping-pong (Fig. 4; Xiol et al. 2014; Webster et al. 2015). Knockdown of Vasa disrupts the overall structure of nuage (Liang et al. 1994); this might be true for knockdown of Krimp as well, making biogenesis that relies on physical integrity of this compartment less efficient. Finally, we observed that reporter RNA normally distributed throughout the cytoplasm is localized to nuage upon tethering (Fig. 5A). A study published during the revision of this report showed that artificial reporter tethering to Armi in follicular cells, where Armi localizes to Yb bodies, leads to reporter processing into piRNAs. Mutating either the helicase or the ATP-binding domain of Armi delocalizes it from Yb bodies, and, consistent with our results, reporter tethering to these mutant (and thus delocalized) versions of Armi greatly reduced reporter processing into piRNAs (Pandey et al. 2017). Thus, it seems that processing directly correlates with localization of the substrate to nuage.
How are pre-piRNAs recruited to nuage in germ cells and to Yb bodies in follicular cells? In our experiments, artificial tethering of the heterologous RNA to Piwi was sufficient to sequester it to nuage. Natural piRNA substrates might be recruited to Yb bodies through RNA-binding proteins that recognize specific motifs in RNA sequences, such as the sequences identified in tj or Flamenco (Homolka et al. 2015; Ishizu et al. 2015). In germ cells, pre-piRNAs might be recruited to nuage through interaction with Aub and Ago3 loaded with complementary piRNAs. Piwi-loaded piRNAs are substantially reduced in the Aub/Ago3 double mutant, which was interpreted as a requirement for slicer cleavage to trigger Zuc-dependent processing (Han et al. 2015; Mohn et al. 2015; Senti et al. 2015; Wang et al. 2015). We propose that these results can be explained by failure to sequester pre-piRNAs to nuage in the Aub/Ago3 double mutant. Expression of catalytically impaired Ago3 and Aub, which cannot cleave precursors but are likely able to bind and recruit them to nuage, can partially rescue Zuc-dependent processing of Piwi-associated piRNAs (Wang et al. 2015).
Compartmentalization is important for many RNA processing pathways, such as splicing, rRNA maturation, etc. (Zhang et al. 1994; Henras et al. 2015). However, our hypothesized model of piRNA precursor selection goes beyond stating that compartmentalization is important for piRNA processing: While RNA sequence motifs are still required for processing in all other pathways (Zeng et al. 2005; Park et al. 2011; Barrangou and Marraffini 2014; Li and Patel 2016; Tsai and Joung 2016), indicating that localization of substrates to the processing compartments is necessary but not sufficient to trigger processing, we propose that for the piRNA pathway, localization of RNA into nuage/Yb granules is both necessary and sufficient to initiate piRNA processing.
Our model of piRNA biogenesis allows unification of two seemingly different mechanisms for selection of piRNA precursors operating in germ and follicular cells. Processing induced by artificial recruitment enables separation of proteins involved in upstream steps of piRNA biogenesis from factors involved in Zuc-mediated processing. Finally, selection of piRNA precursors through recruitment to the processing machinery enables the design of artificial sequences that can be processed efficiently into piRNAs and thus opens possibilities of using the piRNA pathway as a tool for both transcriptional and post-transcriptional gene regulation.
Materials and methods
Fly stocks
The sh-White, sh-AGO3, sh-Armi, sh-Aub, sh-Cuff, sh-Krimper, sh-Vasa, and sh-Zuc lines were obtained from the Bloomington Drosophila Stock Center (BDSC; nos. 33623, 35232, 34789, 33728, 35182, 35230, 32434, and 35227). The GFP-Piwi line was a generous gift from the Hannon laboratory. The sh-SetDB1, sh-Panoramix, λN-Flag-Piwi, λN-Flag, and Tubulin-GFP-BoxB lines were generous gifts from the Brennecke laboratory. The TJ-Gal4 line was obtained from the Kyoto Stock Center (DGGR; no. 104055). To obtain the sh-Thoc5 line, the short hairpin sequences were ligated into the pValium20 vector (Ni et al. 2011) using T4 DNA ligase from New England Biolabs (M0202), according to the manual, and then integrated into the attP2 landing site (BDSC, 25710). Hairpin sequences are listed in Supplemental Table S2. UASp-λN-eGFP-Piwi, UASp-λN-eGFP-Aub, UASp-Armi-HA-λN, UASp-Zuc-HA-λN, UASp-λN-eGFP-GFP, UASp-λN-HA-Yb, UASp-λN-eGFP-Thoc5, UASp-λN-eGFP-Cuff, UASp-λN-eGFP-Asterix, UASp-λN-eGFP-Panoramix, UASp-λN-eGFP-Egg, and UASp-mKate2-4xBoxB-K10polyA were generated in the laboratory as described previously (Chen et al. 2016). These constructs were generated by P-element integration and driven by maternal α-tubulin67C-Gal4 (BDSC, 7063).
Flies carrying the λN transgenes and the α-tubulin67C-Gal4 driver were crossed to flies carrying the UASp-mKate2-4xBoxB-K10polyA reporter and, where applicable, the respective hairpin to obtain flies that express all components of the system in the female germline. For expression in follicular cells, flies carrying the λN transgenes and the TJ-gal4 driver were crossed to flies carrying the UASp-mKate2-4xBoxB-K10polyA reporter. Flies were put on yeast for 3–4 d prior to dissection and were 5–8 d old at the time of dissection.
RT-qPCR
Total RNA was isolated from 20 ovaries with Ribozol (Amresco, N580) followed by DNase treatment with amplification-grade DNase I (Invitrogen, 18068-015) according to the manual. Reverse transcription was carried out using SuperScript III (Invitrogen) with oligo d(T)15. qPCR was performed on a Mastercycler ep Realplex PCR thermal cycler machine (Eppendorf). RT-qPCR data target expression was normalized to the rp49 locus expression. PCR primers are listed in Supplemental Table S2.
Small RNA-seq
Ovaries were dissected, and total RNA was isolated with Ribozol (Amresco, N580). Small RNAs within a 19- to 29-nt window were isolated from 15% polyacrylamide gels from 4 µg of ovarian total RNA. For samples that were NaIO4-treated, 5× borate buffer (pH 8.6; 150 mM borax, 150 mM boric acid) and 200 mM sodium periodate were added to the size-selected small RNA, and the samples were incubated for 30 min at 25°C. The NaIO4-treated small RNA was then ethanol-precipitated before proceeding to library construction. The small RNA libraries were constructed using the NEBNext small RNA library preparation set for Illumina (no. E7330S), according to the protocol, using NEBNext multiplex oligos for Illumina (no. E7335S). Libraries were sequenced on the Illumina HiSeq 2500 (SE 50-bp reads) platform. The resulting reads were mapped against the reporter sequence and the DM3 genome using Bowtie 0.12.7 (Langmead et al. 2009) with the settings “ -v 0 -m 10000 --best -strata,” retaining reads that map with zero mismatches. The reads were also mapped to RepBase using Bowtie 0.12.7 (Langmead et al. 2009) with the settings “-v 3 --best -strata.” Reads that specifically mapped to the reporter sequence and not to the DM3 genome with zero mismatches were extracted from the libraries. Where applicable, 22- to 23-nt reads or 25- to 28-nt reads were extracted from the reads mapping exclusively to the reporter. All four sets of reads (all reads, all reads-specific, 22- to 23-nt reads-specific, and 25- to 28-nt reads-specific) were plotted along the reporter sequence normalized to the number of total reads or 25- to 28-nt reads mapping to the DM3 genome, respectively, using Integrative Genomics Viewer 2.3.68 (Robinson et al. 2011). Reads that mapped specifically to the 42AB locus (chromosome 2R: 2,144,349–2,386,719) and not to the rest of the DM3 genome were extracted from the libraries. Weblogos were generated using an R script that uses the seqLogo library (https://rdrr.io/bioc/seqLogo) to display the nucleotide frequency occurring at each position of the input reads calculated within a homemade bash script. Sequencing data are summarized in Supplemental Table S1.
Immunoprecipitation small RNA-seq
Ovaries (∼100 per immunoprecipitation) from flies expressing UASp-λN-eGFP-Piwi, UASp-λN-eGFP-Aub, or UASp-Zuc-HA-λN with UASp-GFP-Piwi, and the UASp-mKate2-4xBoxB-K10 polyA reporter line driven by maternal α-tubulin67C-Gal4 were dissected and lysed on ice in 250 µL of lysis buffer [30 mM Hepes-KOH at pH7.4, 100 mM KOAc, 2 mM Mg(OAc)2, 5 mM DTT, 0.5% (v/v) NP40, proteinase inhibitor (Roche, 11836170001), RNasin Plus (Promega, N2611)]. Lysate was dounced and clarified by centrifugation at maximum speed at 4°C. The supernatant was incubated with protein A/G agarose beads (Thermo Fisher, 20421) preconjugated with rabbit polyclonal anti-GFP (Covance, affinity-purified in our laboratory) or anti-Piwi (Brennecke et al. 2007) for 2 h at 4°C. The immunoprecipitation and RNA isolation were carried out as described previously (Vagin et al. 2006). A fifth of the RNA was CIP-treated (New England Biolabs, M0290S) in NEB buffer #3 (New England Biolabs, B7003S) for 30 min at 37°C and then ethanol-precipitated after phenol:chloroform and chloroform extraction. The CIP-treated RNA was then PNK-treated with 1 µL of 10× T4 polynucleotide kinase buffer (New England Biolabs, B0201S), 2 µL of [γ-P32]ATP (PerkinElmer, BLU502A250UC), and 1 µL of T4 polynucleotide kinase (New England Biolabs, M0201S) for 45 min at 37°C. The CIP- and PNK-treated RNA was added back to the remainder of the RNA isolated from the immunoprecipitation. Size selection, library preparation, and analysis were performed as described above, except that fragments were gel-extracted based on labeled immunoprecipitation material.
RNA-FISH and immunofluorescence
Probes specific to the mKate2 transcript were designed and ordered from Stellaris (Biosearch Technologies) and resuspended in 200 µL of DEPC-treated TE buffer (10 mM Tris-HCl, 1 mM EDTA at pH 8.0) and stored at −20°C as stock. FISH was performed on 30–50 pairs of ovaries that were dissected and washed with ice-cold DEPC-treated PBS. Prior to fixation, the ovaries were exposed to blue light for 15 min to quench GFP fluorescence from the tethering construct (UASp-λN-eGFP-Piwi and UASp-λN-eGFP-GFP). The ovaries were fixed with 300 µL of fixation solution (4% paraformaldehyde, 0.15% Triton X-100 in DEPC-treated PBS) with shaking at 300 rpm at room temperature and washed three times with PBX (DEPC-treated PBS, 0.3% Triton X-100) for 10 min each at room temperature. Samples were dehydrated in 500 µL of 70% ethanol (in DEPC-treated water) and permeabilized overnight at 4°C on a nutator. Stock probes were diluted 100 times in hybridization buffer (DEPC-treated 2× SSC, 10% [w/v] dextran sulfate, 10% [v/v] formamide). Following rehydration of ovaries in 500 µL of wash buffer (DEPC-treated 2× SSC, 10% [v/v] formamide) for 5 min, the diluted probe was added to the samples for overnight incubation at 37°C in a hybridization chamber. Samples were washed twice with wash buffer and incubated in wash buffer twice for 30 min at 37°C. Samples were blocked with SBX buffer (DEPC-treated 2× SSC, 1% [w/v] BSA, 0.3% Triton X-100) for 2 h at room temperature and incubated with 1:100 diluted anti-Vasa (Developmental Studies Hybridoma Band, A.C. Spradling and D. Williams) overnight at 4°C followed by five washes with SBX buffer for 10 min at room temperature and incubation with 1:400 diluted chicken anti-rat IgG secondary antibody and Alexa fluor 647 (Thermo Fisher, A21472) in SBX buffer for 4 h at room temperature in the dark without agitation. Ovaries were washed five times for 10 min with SSCT buffer (DEPC-treated 2× SSC, 0.1% Triton X-100) at room temperature and incubated with 1:5000 diluted DAPI in 2× SSC buffer for 10 min at room temperature on a nutator. Ovaries were washed twice with 2× SSC buffer and then mounted on a glass slide using Prolong Gold anti-fade reagent (Thermo Fisher, P36934).
Accession numbers
High-throughput sequencing data for small RNA-seq experiments are available through Gene Expression Omnibus (GSE102961).
Supplementary Material
Acknowledgments
We are very grateful to Alexei Aravin for discussions and critical reading and editing of the manuscript. We thank members of the Fejes Toth and Aravin laboratoriess for discussions and suggestions, and Maria Ninova for assistance with data analysis. We thank Howard Lipshitz for critical reading and comments on the manuscript. We are grateful to the Brennecke laboratory, the Bloomington Stock Center, and the Kyoto Stock Center for fly stocks. Stocks obtained from the Bloomington Drosophila Stock Center (National Institutes of Health [NIH] P400D018537) and the Drosophila Genomics Resource Center (NIH 2P40OD010949) were used in this study. We are grateful to Igor Antoschechkin at the Millard and Muriel Jacobs Genetics and Genomics Laboratory at California Institute of Technology for sequencing, and the California Institute of Technology Biological Imaging Facility for support with confocal imaging. The Vasa antibody developed by A.C. Spradling and D. Williams was obtained from the Developmental Studies Hybridoma Bank, created by the National Institute of Child Health and Human Development of the NIH and maintained at The University of Iowa Department of Biology. This work was supported by a grant from the NIH (R01GM110217), and the Ellison Medical Foundation award to K.F.T. A.K.R. is a National Science Foundation (NSF) Graduate Research Fellowship Program (GRFP) fellow. This material is based on work supported by the NSF GRFP (DGE-1144469). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation. A.K.R. and K.F.T. designed and executed the experiments. Flies were generated by A.K.R., E.M.P., and K.S. The manuscript was written by K.F.T. and edited by Alexei Aravin, A.K.R., and K.S.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.303214.117.
References
- Barrangou R, Marraffini LA. 2014. CRISPR–Cas systems: prokaryotes upgrade to adaptive immunity. Mol Cell 54: 234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103. [DOI] [PubMed] [Google Scholar]
- Chen YC, Stuwe E, Luo Y, Ninova M, Le Thomas A, Rozhavskaya E, Li S, Vempati S, Laver JD, Patel DJ, et al. 2016. Cutoff suppresses RNA polymerase II termination to ensure expression of piRNA precursors. Mol Cell 63: 97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Lin H. 2000. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development 127: 503–514. [DOI] [PubMed] [Google Scholar]
- Donertas D, Sienski G, Brennecke J. 2013. Drosophila Gtsf1 is an essential component of the Piwi-mediated transcriptional silencing complex. Genes Dev 27: 1693–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. 2007. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1590. [DOI] [PubMed] [Google Scholar]
- Han BW, Wang W, Li CJ, Weng ZP, Zamore PD. 2015. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science 348: 817–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler D, Olivieri D, Novatchkova M, Gruber FS, Meixner K, Mechtler K, Stark A, Sachidanandam R, Brennecke J. 2011. A systematic analysis of Drosophila TUDOR domain-containing proteins identifies Vreteno and the Tdrd12 family as essential primary piRNA pathway factors. EMBO J 30: 3977–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler D, Meixner K, Pizka M, Lauss K, Schmied C, Gruber FS, Brennecke J. 2013. The genetic makeup of the Drosophila piRNA pathway. Mol Cell 50: 762–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay B, Jan LY, Jan YN. 1988. A protein component of Drosophila polar granules is encoded by vasa and has extensive sequence similarity to ATP-dependent helicases. Cell 55: 577–587. [DOI] [PubMed] [Google Scholar]
- Henras AK, Plisson-Chastang C, O'Donohue MF, Chakraborty A, Gleizes PE. 2015. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip Rev RNA 6: 225–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homolka D, Pandey RR, Goriaux C, Brasset E, Vaury C, Sachidanandam R, Fauvarque MO, Pillai RS. 2015. PIWI slicing and RNA elements in precursors instruct directional primary piRNA biogenesis. Cell Rep 12: 418–428. [DOI] [PubMed] [Google Scholar]
- Huang H, Li Y, Szulwach KE, Zhang G, Jin P, Chen D. 2014. AGO3 Slicer activity regulates mitochondria-nuage localization of Armitage and piRNA amplification. J Cell Biol 206: 217–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur JK, Chung YD. 2016. A novel model of THO/TREX loading onto target RNAs in metazoan gene expression. BMB Rep 49: 355–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur JK, Luo Y, Moon S, Ninova M, Marinov GK, Chung YD, Aravin AA. 2016. Splicing-independent loading of TREX on nascent RNA is required for efficient expression of dual-strand piRNA clusters in Drosophila. Genes Dev 30: 840–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ipsaro JJ, Haase AD, Knott SR, Joshua-Tor L, Hannon GJ. 2012. The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature 491: 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizu H, Iwasaki YW, Hirakata S, Ozaki H, Iwasaki W, Siomi H, Siomi MC. 2015. Somatic primary piRNA biogenesis driven by cis-acting RNA elements and trans-acting Yb. Cell Rep 12: 429–440. [DOI] [PubMed] [Google Scholar]
- Kirino Y, Kim N, de Planell-Saguer M, Khandros E, Chiorean S, Klein PS, Rigoutsos I, Jongens TA, Mourelatos Z. 2009. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat Cell Biol 11: 652–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C, Xi H, Li C, Lee S, Xu J, Khurana JS, Zhang F, Schultz N, Koppetsch BS, Nowosielska A, et al. 2009. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell 138: 1137–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasko PF, Ashburner M. 1990. Posterior localization of vasa protein correlates with, but is not sufficient for, pole cell development. Genes Dev 4: 905–921. [DOI] [PubMed] [Google Scholar]
- Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM, Hur JK, Aravin AA, Toth KF. 2013. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev 27: 390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Thomas A, Stuwe E, Li S, Du J, Marinov G, Rozhkov N, Chen YC, Luo Y, Sachidanandam R, Toth KF, et al. 2014. Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing. Genes Dev 28: 1667–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SS, Patel DJ. 2016. Drosha and Dicer: slicers cut from the same cloth. Cell Res 26: 511–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Diehl-Jones W, Lasko P. 1994. Localization of vasa protein to the Drosophila pole plasm is independent of its RNA-binding and helicase activities. Development 120: 1201–1211. [DOI] [PubMed] [Google Scholar]
- Lim AK, Kai T. 2007. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci 104: 6714–6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. 2009. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137: 522–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Sienski G, Handler D, Brennecke J. 2014. The rhino–deadlock–cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell 157: 1364–1379. [DOI] [PubMed] [Google Scholar]
- Mohn F, Handler D, Brennecke J. 2015. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science 348: 812–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muerdter F, Olovnikov I, Molaro A, Rozhkov NV, Czech B, Gordon A, Hannon GJ, Aravin AA. 2012. Production of artificial piRNAs in flies and mice. RNA 18: 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muerdter F, Guzzardo PM, Gillis J, Luo Y, Yu Y, Chen C, Fekete R, Hannon GJ. 2013. A genome-wide RNAi screen draws a genetic framework for transposon control and primary piRNA biogenesis in Drosophila. Mol Cell 50: 736–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JQ, Zhou R, Czech B, Liu LP, Holderbaum L, Yang-Zhou D, Shim HS, Tao R, Handler D, Karpowicz P, et al. 2011. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods 8: 405–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Ishizu H, Saito K, Fukuhara S, Kamatani MK, Bonnefond L, Matsumoto N, Nishizawa T, Nakanaga K, Aoki J, et al. 2012. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature 491: 284–287. [DOI] [PubMed] [Google Scholar]
- Ohtani H, Iwasaki YW, Shibuya A, Siomi H, Siomi MC, Saito K. 2013. DmGTSF1 is necessary for Piwi-piRISC-mediated transcriptional transposon silencing in the Drosophila ovary. Genes Dev 27: 1656–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey RR, Homolka D, Chen KM, Sachidanandam R, Fauvarque MO, Pillai RS. 2017. Recruitment of Armitage and Yb to a transcript triggers its phased processing into primary piRNAs in Drosophila ovaries. PLoS Genet 13: e1006956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A, Jiang P, Zhao DY, Singh M, Schupbach T. 2011. The Cutoff protein regulates piRNA cluster expression and piRNA production in the Drosophila germline. EMBO J 30: 4601–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN. 2011. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature 475: 201–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H, Watanabe T, Ku HY, Liu N, Zhong M, Lin H. 2011. The Yb body, a major site for Piwi-associated RNA biogenesis and a gateway for Piwi expression and transport to the nucleus in somatic cells. J Biol Chem 286: 3789–3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative Genomics Viewer. Nat Biotechnol 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senti KA, Jurczak D, Sachidanandam R, Brennecke J. 2015. piRNA-guided slicing of transposon transcripts enforces their transcriptional silencing via specifying the nuclear piRNA repertoire. Genes Dev 29: 1747–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienski G, Batki J, Senti KA, Donertas D, Tirian L, Meixner K, Brennecke J. 2015. Silencio/CG9754 connects the Piwi–piRNA complex to the cellular heterochromatin machinery. Genes Dev 29: 2258–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todeschini AL, Teysset L, Delmarre V, Ronsseray S. 2010. The epigenetic trans-silencing effect in Drosophila involves maternally-transmitted small RNAs whose production depends on the piRNA pathway and HP1. PLoS One 5: e11032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Joung JK. 2016. Defining and improving the genome-wide specificities of CRISPR–Cas9 nucleases. Nat Rev Genet 17: 300–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. 2006. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324. [DOI] [PubMed] [Google Scholar]
- Vrettos N, Maragkakis M, Alexiou P, Mourelatos Z. 2017. Kc167, a widely used Drosophila cell line, contains an active primary piRNA pathway. RNA 23: 108–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yoshikawa M, Han BW, Izumi N, Tomari Y, Weng Z, Zamore PD. 2014. The initial uridine of primary piRNAs does not create the tenth adenine that is the hallmark of secondary piRNAs. Mol Cell 56: 708–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Han BW, Tipping C, Ge DT, Zhang Z, Weng ZP, Zamore PD. 2015. Slicing and binding by Ago3 or Aub trigger Piwi-bound piRNA production by distinct mechanisms. Mol Cell 59: 819–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster A, Li S, Hur JK, Wachsmuth M, Bois JS, Perkins EM, Patel DJ, Aravin AA. 2015. Aub and Ago3 are recruited to nuage through two mechanisms to form a ping-pong complex assembled by Krimper. Mol Cell 59: 564–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiol J, Spinelli P, Laussmann MA, Homolka D, Yang Z, Cora E, Coute Y, Conn S, Kadlec J, Sachidanandam R, et al. 2014. RNA clamping by Vasa assembles a piRNA amplifier complex on transposon transcripts. Cell 157: 1698–1711. [DOI] [PubMed] [Google Scholar]
- Yu Y, Gu J, Jin Y, Luo Y, Preall JB, Ma J, Czech B, Hannon GJ. 2015. Panoramix enforces piRNA-dependent cotranscriptional silencing. Science 350: 339–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Yi R, Cullen BR. 2005. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J 24: 138–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Taneja KL, Singer RH, Green MR. 1994. Localization of pre-mRNA splicing in mammalian nuclei. Nature 372: 809–812. [DOI] [PubMed] [Google Scholar]
- Zhang F, Wang J, Xu J, Zhang Z, Koppetsch BS, Schultz N, Vreven T, Meignin C, Davis I, Zamore PD, et al. 2012. UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell 151: 871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wang J, Schultz N, Zhang F, Parhad SS, Tu S, Vreven T, Zamore PD, Weng Z, Theurkauf WE. 2014. The HP1 homolog rhino anchors a nuclear complex that suppresses piRNA precursor splicing. Cell 157: 1353–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.