Abstract
The switch from skotomorphogenesis to photomorphogenesis is a key developmental transition in the life of seed plants. While much of the underpinning proteome remodeling is driven by light-induced changes in gene expression, the proteolytic removal of specific proteins by the ubiquitin-26S proteasome system is also likely paramount. Through mass spectrometric analysis of ubiquitylated proteins affinity-purified from etiolated Arabidopsis seedlings before and after red-light irradiation, we identified a number of influential proteins whose ubiquitylation status is modified during this switch. We observed a substantial enrichment for proteins involved in auxin, abscisic acid, ethylene, and brassinosteroid signaling, peroxisome function, disease resistance, protein phosphorylation and light perception, including the phytochrome (Phy) A and phototropin photoreceptors. Soon after red-light treatment, PhyA becomes the dominant ubiquitylated species, with ubiquitin attachment sites mapped to six lysines. A PhyA mutant protected from ubiquitin addition at these sites is substantially more stable in planta upon photoconversion to Pfr and is hyperactive in driving photomorphogenesis. However, light still stimulates ubiquitylation and degradation of this mutant, implying that other attachment sites and/or proteolytic pathways exist. Collectively, we expand the catalog of ubiquitylation targets in Arabidopsis and show that this post-translational modification is central to the rewiring of plants for photoautotrophic growth.
Keywords: Arabidopsis, ubiquitin, mass spectrometry, photomorphogenesis, phytochrome degradation
INTRODUCTION
The switch from growth in the dark (skotomorphogenesis) to growth in the light (photomorphogenesis or de-etiolation) is a critical developmental transition in plants. It enables young seedlings to become photosynthetically competent, and continues as plants mature to optimize light capture to the prevailing light environment and entrain the associated developmental programs to the seasonal cycles (Schäfer and Nagy, 2006; Kami et al., 2010). This switch is triggered by a collection of photoreceptors, including members of the red and far-red light-absorbing phytochrome (Phy) family (Franklin and Quail, 2010; Burgie and Vierstra, 2014), the blue light-absorbing phototropin (PHOT1), cryptochrome and Zeitlupe-type photoreceptors (Kim et al., 2007; Chaves et al., 2011; Liscum et al., 2014), and the UV-B light-absorbing UV-RESISTANT-8 protein (Rizzini et al., 2011).
Collectively, they facilitate perception of fluence rate (intensity), directionality, duration, day/night transitions, and spectral quality (wavelength) to provide detailed light information across a broad color spectrum. Ultimately, signals from these photoreceptors affect vital processes such as seed germination, chloroplast development from etioplasts, leaf expansion, photo-and gravitropism, circadian rhythm entrainment, shade avoidance, entry into the reproductive phase, and senescence (Schäfer and Nagy, 2006).
Not surprisingly, the skotomorphogenic to photomorphogenic transition is initiated by extensive remodeling of plant proteomes, much of which is orchestrated by massive alterations in transcription (Wu, 2014). In Arabidopsis for example, as much as one third of its mRNAs change in abundance after exposing dark-grown seedlings to light (Ma et al., 2001; Tepperman et al., 2001, 2006; Hu et al., 2009). These modifications are driven by whole-genome-scale adjustments of chromatin accessibility through changes in DNA methylation and histone acetylation, which are followed by sophisticated networks of signaling events that activate or repress the expression of extensive gene suites (Wu, 2014). Particularly influential are regulons controlled by specific cis elements (e.g., G-, PBF-, and CT-boxes) interacting with dedicated transcriptional regulators, including the LONG HYPOCOTYL (HY)-5, HY5 HOMOLOGY (HYH), LONG HYPOCOTYL IN FAR-RED (HFR)-1, and G-Box Binding Factor (GBF)-1 activators, and the Phy-Interacting Factor (PIF) repressors (Schindler et al., 1992; Fairchild et al., 2000; Zhang et al., 2011, 2013). Ultimately, these changes impinge on most, if not all, hormone signaling pathways that coordinate photomorphogenic development among the various tissues/ organs (de Wit et al., 2016).
In addition, changes in protein abundance and activity also occur during the photomorphogenic transition, which are driven by light-triggered effects on translation and various post-translational modifications (PTMs) (Wu, 2014). PTMs of note include protein phosphorylation, SUMOylation, and ubiquitylation, which affect, either singly or collectively, numerous targets upon exposing plants to light. In fact, PHOT1, and possibly the Phy photoreceptors, act as light-regulated protein kinases to directly initiate phosphorelays after photoexcitation (Yeh and Lagarias, 1998; Demarsy et al., 2012). This complexity is exemplified by the PhyA and PhyB isoforms that direct photomorphogenesis by the photointerconversion between their biologically inactive, red-light-absorbing Pr states and their biologically active, far-red-light-absorbing Pfr states (Franklin and Quail, 2010). While the mRNA abundance for PhyA declines rapidly upon exposing etiolated seedlings to light via autoregulated transcriptional repression of the PHYA gene, the PHYB mRNA remains at a constitutively low level (Somers and Quail, 1995; Tepperman et al., 2006). Following photoconversion to Pfr, both chromoproteins enter the nucleus where they activate gene expression. Concomitantly, the levels of both drop through selective ubiquitylation and subsequent proteasomal degradation of Pfr (Clough and Vierstra, 1997), which for PhyB is directed by a phosphorylation-mediated, mutually assured destruction pathway involving its PIF binding partners and the Light-Regulated BTB (LRB) 1-3 Ub ligases (Al-Sady et al., 2006; Christians et al., 2012; Ni et al., 2014). In addition, PhyB also becomes SUMOylated upon red-light exposure, which appears to repress its signaling potential (Sadanandom et al., 2015).
Ubiquitin (Ub) conjugation, typically to internal lysines, should be a particularly influential effector of proteome remodeling during photomorphogenesis given its roles in selective protein turnover through the Ub/26S proteasome system (UPS) and autophagy, and in directing protein localization via endosomal trafficking (Vierstra, 2009; Li and Vierstra, 2012). To further examine the effect of Ub, we present here an in-depth catalog of ubiquitylation targets (i.e., the ubiquitylome) from etiolated Arabidopsis seedlings, and describe how this catalog changes during the skotomorphogenic/photomorphogenic switch. Remarkably, large-scale changes in the profile and abundance of Ub conjugates (both up and down) were evident, with additional alterations seen upon exposing the seedlings to the proteasome inhibitor MG132. Notable substrates include those involved in light perception, synthesis, transport and signaling of the auxin, abscisic acid (ABA), ethylene and brassinosteroid hormones, disease resistance, peroxisome function, and protein phosphorylation.
PhyA was one of the most dramatically affected targets, with ubiquitylation sites mapped to six lysines in the polypeptide. Blocking Ub addition to these sites by lysine to arginine substitutions markedly slowed PhyA turnover and generated a hyperactive photoreceptor, indicating a role for Ub and these lysines in Pfr degradation. However, these substitutions did not completely inhibit PhyA breakdown as Pfr, nor prevent its ubiquitylation, implying that other amino acids can also be modified, and/or that alternative proteolytic pathways become engaged. Collectively, our results further expand the reach of ubiquitylation in plant development and show that this PTM is also critical for reconfiguring plant proteomes during seedling photomorphogenesis.
RESULTS
Mass Spectrometric Analysis of Ubiquitylated Proteins in Etiolated Seedlings
To explore the role(s) of Ub in the skotomorphogenic/photomorphogenic transition by mass spectrometry (MS), we developed a liquid medium growth regime capable of this switch and also amenable to control by proteasome inhibitors such as MG132 (Supplemental Figure 1B–1D). Ub conjugates were then isolated by a stringent purification strategy that exploits a transgenic Arabidopsis line accumulating high levels of 6His-Ub against a background of wild-type Ub (Saracco et al., 2009; Kim et al., 2013). As shown in Supplemental Figure 1E, both 6His-Ub monomers and oligomers accumulate in 7-day-old, dark-grown hexa(6His-Ub) seedlings, with the latter reflecting faithful entry of the tagged Ubs into the conjugation cascade.
6His-Ub adducts were purified by a two-step affinity protocol that first enriches for ubiquitylated species with beads coated with tandem Ub-binding entities (TUBEs; Hjerpe et al., 2009) fused to glutathione S-transferase (GST) (Supplemental Figure 1A). The TUBEs were created by concatenating four Ub-binding UBA domains from Arabidopsis RAD23 in tandem (Farmer et al., 2010), which confers high affinity for Ub, especially when assembled into poly-Ub chains (Kim et al., 2013). This step was followed by nickel-nitrilotriacetic acid (Ni-NTA) chelate chromatography, which enriches for Ub conjugates based on the 6His moiety. The initial extraction buffer contained a cocktail of protease inhibitors to block deubiquitylating enzymes (DUBs) from releasing the target-linked Ub moieties (Maor et al., 2007; Kim et al., 2013). Subsequent buffers also included high concentrations of denaturants (7 M guanidine-HCl or 8 M urea) to further suppress DUBs and to help avoid isolating proteins that would bind Ub or its adducts non-covalently. The cumulative effect of this protocol was to preferentially isolate Ub conjugates over free Ub monomers. As shown in Figure 1A, the protocol allowed us to efficiently isolate ubiquitylated proteins from etiolated hexa(6His-Ub) seedlings kept in the dark or exposed to a short pulse of red light, compared with wild-type seedlings that were subjected to GST-alone matrix and Ni-NTA chromatography steps. For a simple measure of conjugate abundance, we employed a label-free method that counts the peptide spectral matches (PSMs) for all peptides assigned to each protein from the full MS scans and averages them across the biological replicates.
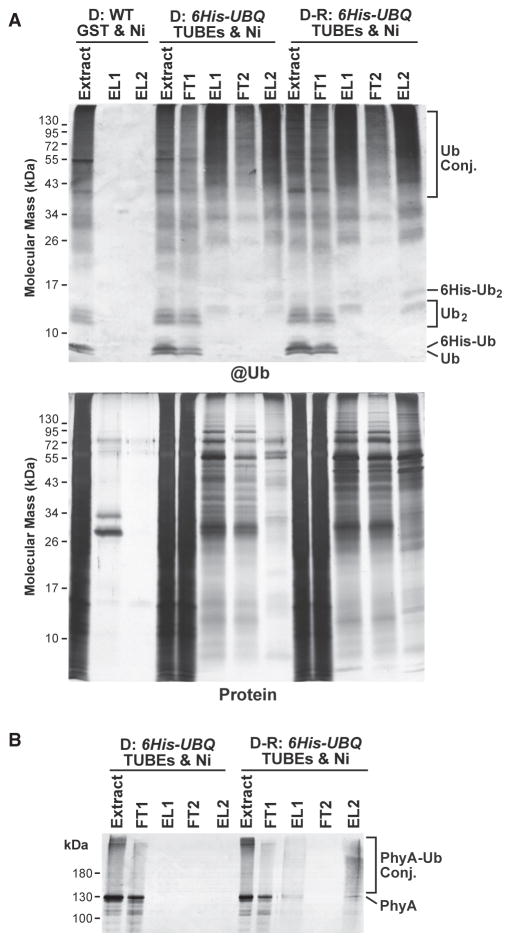
Figure 1. Two-Step Affinity Purification of Ubiquitylated Proteins from Etiolated Arabidopsis Seedlings Before and After Red-Light Irradiation.
Ub conjugates were affinity purified by sequential TUBEs (1) and Ni-NTA (Ni) chromatography (2) steps from 7-day-old 6His-UBQ seedlings either kept in the dark (D) or exposed to 1 h of red light (D-R). Samples from dark-grown wild-type (WT) seedlings, which were subjected to the same purification protocol using GST and Ni-NTA beads, were included for comparison.
(A) SDS–PAGE analysis of the various fractions during the purification. The gels were either subjected to immunoblotting with anti-Ub antibodies (@Ub) or stained for protein with silver (Protein). FT and EL represent flow through and elution fractions, respectively. Extract represents the initial clarified seedling extract. Ub conjugates and the Ub and 6His-Ub monomers and dimers are indicated.
(B) The two-step affinity purification also enriches for PhyA-Ub conjugates that accumulate in etiolated seedlings after R. The samples analyzed in (A) were immunoblotted with the monoclonal antibody 073D against PhyA.
Given our goal to characterize rapid changes in the ubiquitylome during photomorphogenesis, we examined samples prepared from dark-grown seedlings or following only a 1-h exposure to red light. Prior studies revealed that PhyA-Ub conjugates accumulate maximally during this period (Shanklin et al., 1987; Clough et al., 1999). Indeed, when we probed the affinity-purified samples subjected to SDS–PAGE with a monoclonal antibody that recognizes PhyA, a smear of high molecular mass species that likely represents Ub adducts of Pfr was detected in the red-light-irradiated samples but not in dark samples (Figure 1B).
The purified Ub conjugates were digested with trypsin, and the resulting peptides were analyzed by reverse-phase nanoflow liquid chromatography (LC) followed by electrospray ionization tandem MS (MS/MS) in the high-energy collision mode. The peptide spectra were then matched using SEQUEST against the Arabidopsis ecotype Col-0 protein database (IPI database, version 3.85; http://www.arabidopsis.org). A likely Ub conjugate was included in the ubiquitylome catalog based on the MS identification of two or more different matching peptides with a ≤0.01 false discovery rate (FDR), or if a single matching peptide of equivalent stringency was identified with a canonical Ub footprint. These footprints were detected as a missed trypsin cleavage at the lysine affected combined with an increased mass of 114 Da for this residue derived from the isopeptide-linked Gly-Gly Ub vestige that remains after trypsinization (Peng et al., 2003; Kim et al., 2013). To help eliminate contaminants, we also generated a database of background proteins from wild-type plants that were retained through the GST and Ni-NTA chromatography steps (Supplemental Dataset 1); these proteins were culled from the LC-MS/MS catalogs generated with the hexa(6His-Ub) seedlings.
In this study, we analyzed 7-day-old etiolated seedlings either kept in the dark, irradiated with red light for 1 h, and/or exposed to 50 μM MG132 for 4 h before harvesting, which we found to be effective in inhibiting proteasomes (Kim et al., 2013; Gladman et al., 2016). A 2-h LC-MS/MS run typically detected ~3000–5000 PSMs that could be assigned to ~750 proteins on average. Each condition (D, D + MG132, D-R, and D-R + MG132) analyzed in triplicate generated reasonably overlapping datasets based on the criteria above (Supplemental Figure 2), which were then combined to generate catalogs containing 564, 561, 736, and 670 Ub targets, respectively (Figure 2A, Supplemental Datasets 2 and 3). Approximately 17%–19% of the proteins were detected in all three biological replicates, while 40%–54% were detected in two of the three, which we considered to be high confidence targets (Supplemental Figure 2).
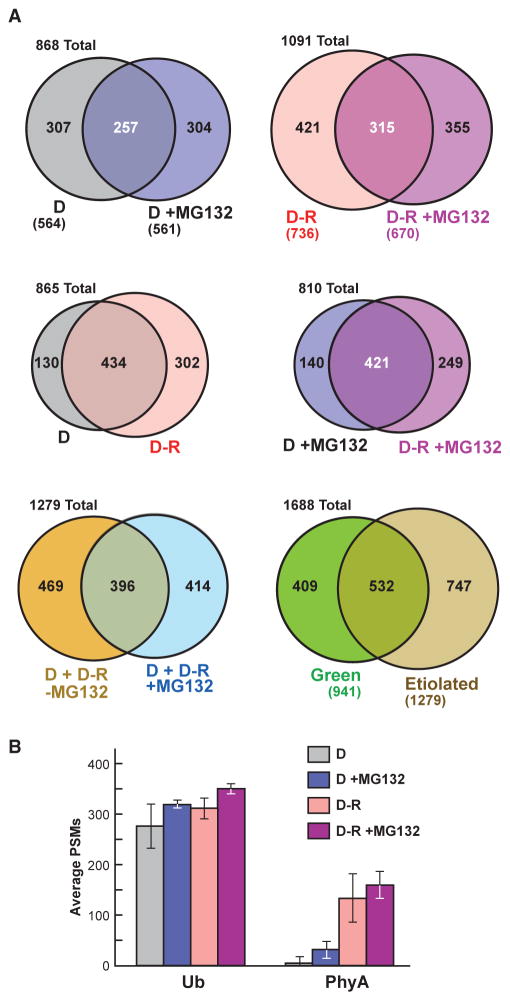
Figure 2. Comparisons of the Ubiquitylated Protein MS Datasets Obtained from Etiolated Arabidopsis Seedlings Before (D) and After Red-Light Irradiation (R) and/or MG132 Exposure.
(A) Venn diagrams showing overlap of the ubiquitylated protein data-sets. The number of proteins in each sector is indicated. The numbers in parentheses indicate the total number of proteins in each dataset. The etiolated dataset represents the combined catalog of proteins detected in dark-grown seedlings with or without exposure to 50 μM MG132 and/or red light. The green MS dataset was obtained from 14-day-old, light-grown seedlings and was described previously (Kim et al., 2013).
(B) Abundance of Ub and PhyA peptides in the MS datasets as quantified by peptide spectral matches (PSMs). Each bar represents the average of three biological replicates (±SD).
In total, 1279 different ubiquitylated proteins were present in the composite database. When this etiolated dataset was combined with that determined previously using green seedlings (Kim et al., 2013), at least 1688 distinct Ub targets are possible in Arabidopsis. As with green seedlings, the ubiquitylation status of many etiolated seedling targets rose upon MG132 treatment, further implicating Ub addition in directing proteasomal breakdown. For example, of the top 20 most abundant ubiquitylated proteins in D + MG132 samples, the PSM counts for 70% substantially increased in MG132-treated versus untreated tissues (Table 1). Surprisingly, only 42% of the etiolated seedling targets were also identified in green seedlings (532 of 1279), thus providing the first indications that a substantially different profile of ubiquitylated proteins accumulates in etiolated seedlings (Figure 2A; Supplemental Figure 3). Notable targets strongly enriched in the etiolated seedling ubiquitylome included the two plasma membrane ATPases (AHA1 and AHA2) involved in generating the electrochemical gradient needed for turgor-driven cell expansion, enzymes involved in secondary metabolism (e.g., PAL1/2, C4H, and clCDH), several proteasome subunits (RPN1a, RPT1a, and PBE1), RHM1 needed for rhamnose biosynthesis, several peroxisome enzymes (ICL, MDAR1, and ACX3), the COP1/2 coatomers required for non-clathrin-mediated retrograde transport from the ER to the Golgi, annexin-4 involved in Golgi-mediated secretion, and MLP28, which is a member of the major latex protein superfamily reported to bind hormones and secondary metabolites (Table 1).
Table 1.
| Locus | Protein Name | Description | Average PSMsb | |||
|---|---|---|---|---|---|---|
| D | D + MG | D-R | D-R + MG | |||
| Dark | ||||||
| At3g03250 | UDPG1 | UDP glucose pyrophosphorylase 1 | 31 | 7 | 20 | 8 |
| At3g21720 | ICL | isocitrate lyase | 30 | 12 | 38 | 10 |
| At4g30190 | AHA2 | H+-ATPase 2 | 30 | 39 | 34 | 51 |
| At1g70830 | MLP28 | MLP-like protein 28 | 28 | 28 | 24 | 33 |
| At2g20580 | RPN1a | 26S proteasome RPN subunit 1a | 26 | 25 | 40 | 20 |
| At2g37040 | PAL1 | phenylalanine ammonia lyase 1 | 24 | 17 | 33 | 12 |
| At2g38750 | ANNAT4 | annexin 4 | 24 | 26 | 20 | 35 |
| At5g39570 | – | transmembrane, unknown | 24 | 8 | 16 | 8 |
| At2g18960 | AHA1 | H+-ATPase 1 | 23 | 35 | 30 | 46 |
| At1g53750 | RPT1a | 26S proteasome RPT subunit 1a | 21 | 2 | 25 | 0 |
| At5g03300 | ADK2 | adenosine kinase 2 | 21 | 0 | 16 | 0 |
| At3g52880 | MDAR1 | monodehydroacorbate reductase 1 | 20 | 0 | 20 | 4 |
| At4g23670 | – | polyketide cyclase/dehydrase | 20 | 8 | 13 | 12 |
| At3g16460 | JAL34 | jacalin-related lectin 34 | 19 | 3 | 19 | 0 |
| At4g09000 | GRF1 | general regulatory factor 1 | 19 | 8 | 19 | 6 |
| At1g78300 | GRF2 | general regulatory factor 2 | 18 | 5 | 12 | 4 |
| At3g21370 | BGLU19 | b-glucosidase 19 | 18 | 0 | 18 | 0 |
| At5g16590 | LRR1 | leucine-rich protein kinase 1 | 17 | 7 | 11 | 10 |
| At5g38480 | GRF3 | general regulatory factor 3 | 17 | 5 | 17 | 2 |
| At5g40870 | UPRT1 | uracil phosphoribosyl transferase 1 | 17 | 20 | 16 | 21 |
| Dark-Red Light | ||||||
| At1g09570 | PhyA | phytochrome A | 9 | 33 | 134 | 160 |
| At2g20580 | RPN1a | 26S proteasome RPN subunit 1a | 26 | 25 | 40 | 20 |
| At3g21720 | ICL | isocitrate lyase | 30 | 12 | 38 | 10 |
| At4g30190 | AHA2 | H+-ATPase 2 | 30 | 39 | 34 | 51 |
| At2g37040 | PAL1 | phenylalanine ammonia lyase 1 | 24 | 17 | 33 | 12 |
| At2g18960 | AHA1 | H+-ATPase 1 | 23 | 35 | 30 | 46 |
| At1g13060 | PBE1 | 26S proteasome CP subunit b6 | 0 | 16 | 29 | 14 |
| At5g60790 | ABCF1 | ABC transporter F family 1 | 16 | 22 | 27 | 26 |
| At2g34470 | UREG | urease accessory protein G | 8 | 6 | 26 | 8 |
| At3g53260 | PAL2 | phenylalanine ammonia lyase 2 | 11 | 0 | 26 | 0 |
| At5g01410 | PDX1 | aldolase type TIM barrel | 14 | 4 | 26 | 8 |
| At1g48410 | AGO1 | argonaute RNA dicer 1 | 13 | 4 | 25 | 5 |
| At1g53750 | RPT1a | 26S proteasome RPT subunit 1a | 21 | 2 | 25 | 0 |
| At4g31500 | CYP83B1 | cytochrome P454 subfamily B, 1 | 4 | 6 | 25 | 7 |
| At1g70830 | MLP28 | MLP-like protein 28 | 28 | 28 | 24 | 33 |
| At5g42590 | CYP71A16 | cytochrome P454 subfamily A, 16 | 0 | 6 | 24 | 12 |
| At1g57720 | EF1B | translation elongation factor 1B | 0 | 8 | 21 | 14 |
| At2g43970 | LARP6B | LA-related protein 6B | 8 | 0 | 21 | 3 |
| At1g78570 | RNM1 | rhamnose biosynthesis 1 | 6 | 27 | 20 | 23 |
| At2g30490 | C4H | cinnamate-4-hydroxylase | 4 | 24 | 20 | 24 |
| Dark + MG132 | ||||||
| At4g30190 | AHA2 | H+-ATPase 2 | 30 | 39 | 34 | 51 |
| At2g18960 | AHA1 | H+-ATPase 1 | 23 | 35 | 30 | 46 |
| At1g09570 | PhyA | phytochrome A | 9 | 33 | 134 | 160 |
| At1g36160 | ACC1 | acetyl-CoA carboxylase 1 | 6 | 32 | 4 | 32 |
| At1g70830 | MLP28 | MLP-like protein 28 | 28 | 28 | 24 | 33 |
| At1g78570 | RHM1 | rhamnose biosynthesis 1 | 6 | 27 | 20 | 23 |
| At2g38750 | ANNAT4 | annexin 4 | 24 | 26 | 20 | 35 |
| At4g23650 | CPK3 | calcium-dependent protein kinase 3 | 6 | 26 | 9 | 29 |
| At1g06290 | ACX3 | acyl-CoA oxidase 3 | 8 | 25 | 4 | 21 |
| At2g20580 | RPN1a | 26S proteasome RPN subunit 1a | 26 | 25 | 40 | 20 |
| At5g11520 | ASP3 | Asp aminotransferase 3 | 13 | 25 | 10 | 23 |
| At1g20960 | EMB1507 | U5 SNR helicase | 6 | 24 | 11 | 20 |
| At2g30490 | C4H | cinnamate-4-hydroxylase | 4 | 24 | 20 | 24 |
| At3g43190 | SUS4 | sucrose synthase 4 | 4 | 24 | 3 | 13 |
| At1g10630 | ARFA1F | ADP ribosylation factor A1F | 13 | 22 | 18 | 16 |
| At5g60790 | ABCF1 | ABC transporter F family 1 | 16 | 22 | 27 | 26 |
| At1g53580 | GLX2-3 | glyoxalase II 3 | 5 | 21 | 4 | 16 |
| At4g13940 | EMB1395 | S-adenosyl-L-homo-Cys hydrolase | 4 | 21 | 12 | 17 |
| At3g19820 | DWF1 | DWARF1 | 9 | 20 | 17 | 29 |
| At5g40870 | UPRT1 | uracil phosphoribosyl transferase 1 | 17 | 20 | 16 | 21 |
| Dark-Red Light + MG132 | ||||||
| At1g09570 | PhyA | phytochrome A | 9 | 33 | 134 | 160 |
| At4g30190 | AHA2 | H+-ATPase 2 | 30 | 39 | 34 | 51 |
| At2g18960 | AHA1 | H+-ATPase 1 | 23 | 35 | 30 | 46 |
| At2g38750 | ANNAT4 | annexin 4 | 24 | 26 | 20 | 35 |
| At1g70830 | MLP28 | MLP-like protein 28 | 28 | 28 | 24 | 33 |
| At1g36160 | ACC1 | acetyl-CoA carboxylase 1 | 6 | 32 | 4 | 32 |
| At4g31490 | b-COP2 | coatomer b subunit 2 | 0 | 0 | 0 | 30 |
| At3g19820 | DWF1 | DWARF 1 | 9 | 20 | 17 | 29 |
| At4g23650 | CPK3 | calcium-dependent protein kinase 3 | 6 | 26 | 9 | 29 |
| At3g48990 | AAE3 | acyl-activating enzyme 3 | 13 | 18 | 9 | 28 |
| At3g16400 | NSP1 | nitrile specifier protein 1 | 0 | 8 | 5 | 27 |
| At1g19570 | DHAR1 | dehydroascorbate reductase 1 | 12 | 15 | 5 | 26 |
| At5g60790 | ABCF1 | ABC transporter F family 1 | 16 | 22 | 27 | 26 |
| At1g63660 | – | glutamine amidotransferase | 9 | 19 | 11 | 24 |
| At2g30490 | C4H | cinnamate-4-hydroxylase | 4 | 24 | 20 | 24 |
| At5g56030 | HSP81.2 | heat shock protein 81-2 | 16 | 0 | 17 | 24 |
| At1g65930 | clCDH | cytosol NADP-isocitrate dehydrogenase | 8 | 19 | 8 | 23 |
| At1g78570 | RPHM1 | rhamnose biosynthesis 1 | 6 | 27 | 20 | 23 |
| At4g31480 | b-COP1 | coatomer b subunit 1 | 11 | 20 | 10 | 23 |
| At5g11520 | ASP3 | Asp aminotransferase 3 | 13 | 25 | 10 | 23 |
PSM values represent the average of three biological replicates. PSM values for each of the four conditions are included for comparison. Total PSMs: dark, 3119; dark + MG132, 3634; dark-red, 4576; dark-red + MG132, 4897.
See Supplemental Datasets 1, 2, 3, and 4 for complete list.
The average number of PSMs for Ub in each sample was relatively consistent across the four datasets (Figure 2B). In contrast, the number of PSMs assigned to PhyA was markedly higher in the D-R versus D samples (13.3-fold up), and increased further in the D-R samples upon MG132 treatment (1.2-fold up) (Figure 2B), in agreement with the enhanced ubiquitylation and breakdown of PhyA after photoconversion to Pfr (Shanklin et al., 1987; Jabben et al., 1989; Clough and Vierstra, 1997). Altogether, ubiquitylated forms of PhyA went from comprising just 0.22% of the ubiquitylome (7 of 3119 total PSMs) in etiolated seedlings kept in the dark, to now representing 2.93% of the pool (134 of 4576 total PSMs) after just 1 h of red-light exposure, thus making PhyA the most abundant ubiquitylated protein detected in irradiated seedlings. We discovered that MG132 also increased by 4.1-fold the PSM count for PhyA in the D samples (7 of 3119 total PSMs in D samples versus 33 of 3634 total PSMs in D + MG132 samples), which is consistent with previous data showing that Pr is also degraded via the 26S proteasome, albeit more slowly compared with Pfr (Debrieux and Fankhauser, 2010).
Ubiquitylation Affects Many Processes in Etiolated Seedlings
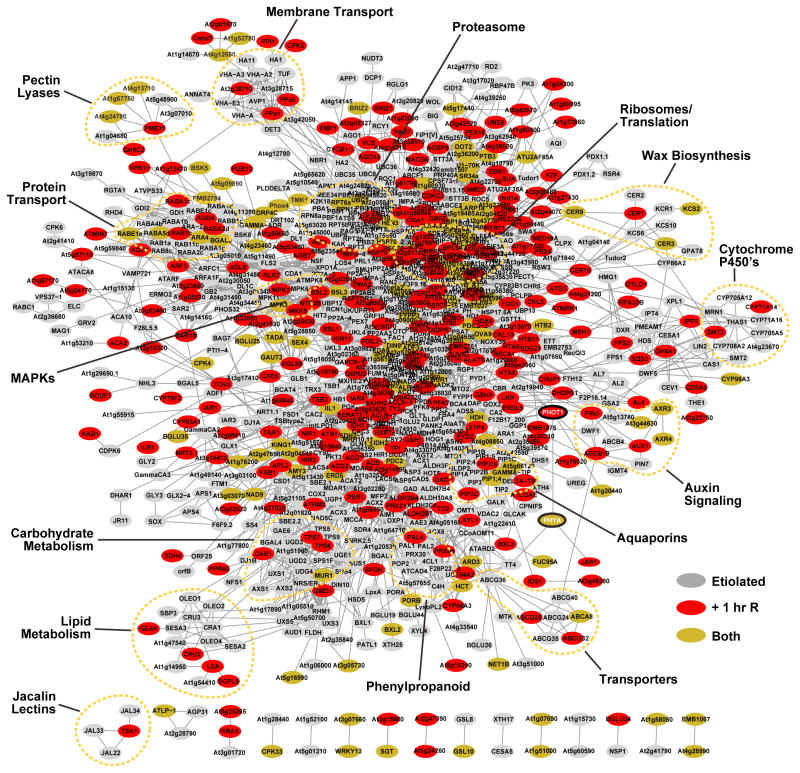
To probe how ubiquitylation globally affects etiolated seedling development and photomorphogenesis, we generated an interaction network using the Search Tool for Retrieval of Interacting Genes/Proteins database (Szklarczyk et al., 2015). Among our 1279 Ub targets, associations could be found for 1071 with a high confidence score (≥0.70). A number of sub-networks were identified, including many subunits of the proteasome and ribosome, peroxisomal proteins, enzymes involved in carbohydrate, lipid, wax and phenylpropanoid metabolism, cyto-chrome P450 enzymes, numerous water, ion, and protein transporters, cellulose synthesis, and components of MAP kinase cascades. Whereas most targets were detected in etiolated seedlings both before and after red-light irradiation (434 proteins), a surprisingly larger number appeared only after the short light treatment (302 proteins), compared with those that were detected only in the dark samples (130 proteins; Figure 2A), suggesting that light rapidly triggers the ubiquitylation of a number of proteins, possibly to induce their turnover or subcellular relocalization (Table 1, Supplemental Datasets 2 and 3). In addition, 414 new targets appeared in the D and D-R samples after MG132 exposure, many of which likely represent UPS targets. These striking changes are most evident when comparing the top 20 most abundant ubiquitylation targets in each dataset as judged by PSMs (Table 1). In addition to PhyA, which is the dominant target in the D-R and D-R + MG132 datasets (see above), many of the remaining top 20 most abundant targets were affected by the inhibitor.
Intriguing examples besides PhyA, whose ubiquitylation was up-regulated after only a 1-h red-light treatment, include the PHOT1 photoreceptor, CHUP1 that helps direct chloroplast photomovement, the auxin transcriptional regulator ARF3 and several auxin transporters (PIN3, AUX1, and ABCB1), the calmodulin-like protein TCH3, Cyclin B1-1, the CER1 protein involved in cuticular wax biosynthesis, the SUA RNA splicing factor, several RAB GTPases (RAB1c, RABA1c, and RABA2d), the RAPTOR regulator of TOR signaling, several kinases (KIN11, MPK2, and MKK5), and regulators of brassinosteroid signaling and biosynthesis (BSK5, BSK8, CAS1, and 7RED). SUMO1 was also detected only in the red-light-irradiated samples, which might reflect a role in directing subsequent ubiquitylation (Miller et al., 2010).
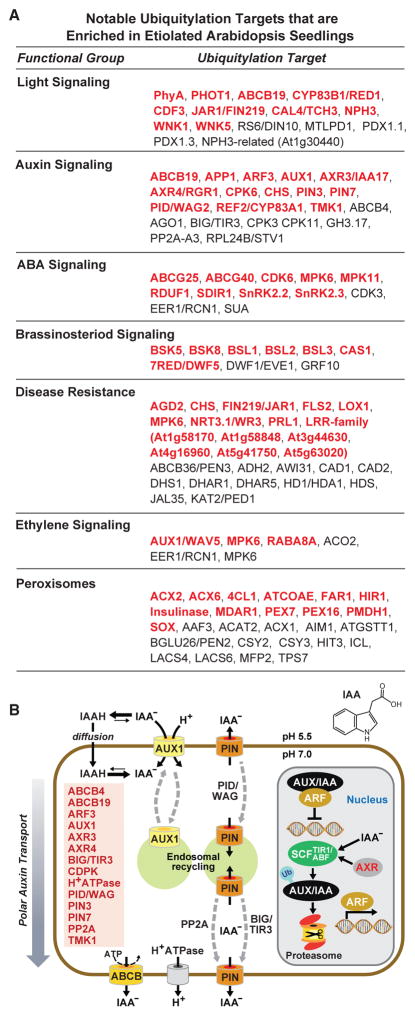
When we examined the combined etiolated datasets (D, D + MG132, D-R, and D-R + MG132), striking connections between Ub and a number of processes relevant to etiolated seedling development emerged. Particularly notable were involvements in light perception, auxin, ABA and brassinosteroid hormone signaling, disease resistance, protein kinases, and peroxisomes (Figures 3 and 4A; Supplemental Figure 3). For example, prior studies linked Ub to auxin signaling via a collection of SCFTIR1,ABF1–4 E3 complexes that direct the UPS-mediated turnover of AUX/IAA transcriptional repressors (Salehin et al., 2015). Here, we extended this involvement through identification of other Ub targets that run the gamut of auxin import/export and perception, including the PIN3/7, ABCB4/19, and AUX1 auxin transporters, the PP2a and PINOID/WAG proteins that promote PIN endocycling between the plasma membrane and endosomes, BIG/TIR3 that regulates auxin efflux, AXR4 that helps localize AUX1, the AXR3 target of the SCFTIR1,ABF1–4 E3s, and the TMK1 receptor kinase that promotes cell expansion in response to auxin (Figure 4B). This concentration of auxin regulators in our etiolated seedling datasets coincides with the prominent role for this hormone in helping etiolated seedlings reach the soil surface through rapid hypocotyl elongation, and subsequent functions in cotyledon/leaf expansion, meristem differentiation, and apical dominance (Leyser, 2010).
Figure 3. Protein Interaction Network of Ubiquitylated Proteins Identified in Etiolated Arabidopsis Seedlings Before and Soon After Red-Light (R) Irradiation.
Protein interaction networks were generated with the complete list of 955 (etiolated only) and 1071 (etiolated and following 1-h R irradiation) ubiquitylated proteins using the STRING database and visualized using the Cytoscape program. The node colors reflect proteins identified only in etiolated (D and D + MG132) (gray), R-irradiated (D-R and D-R + MG132) (red), or in both pairs of datasets. Important functional clusters are surrounded by dashed lines. The red circles locate PhyA and PHOT1. Either the Arabidopsis thaliana locus identifier or the protein name abbreviation (where known) is shown for each substrate.
Figure 4. Ubiquitylation Targets Influential in Etiolated Arabidopsis Seedlings.
(A) Notable ubiquitylation targets involved in various aspects of hormone and light signaling, peroxisome biology, and disease resistance. Targets shown in red were only detected in etiolated seedlings but not previously in green seedlings (Kim et al., 2013). See Supplemental Datasets 2 and 3 for the complete list.
(B) Schematic diagram of auxin transport and perception highlighting the key proteins that are ubiquitylated in etiolated seedlings. The proteins and their proposed position within the auxin system are indicated. The structure of IAA is shown at the top right.
Our etiolated ubiquitylome datasets also contained numerous peroxisome proteins (26 total) (Figure 4A), including three acyl-CoA oxidases (ACX1 ACX2, and ACX6), acyl-activating enzyme (AAE1), acetylaceto-CoA thiolase (ACAT2), fatty acid reductase (FAR1) and long-chain acyl-CoA synthase required for fatty acid β-oxidation, isocitrate lyase (ICL), peroxisomal citrate synthase (CSY2), and PEX7 and PEX16 required for peroxisomal biogenesis (Reumann and Bartel, 2016). Presumably, some of this ubiquitylation reflects peroxisome breakdown through Ub-mediated autophagy as germinating Arabidopsis cotyledons transition away from using stored lipids for nutrition. For brassinosteroid signaling (Wang et al., 2012), ubiquitylated forms of several enzymes required for brassinosteroid synthesis (CAS1 and 7RED), the BSK5 and BSK8 signaling kinases, and the BSL1-3 relatives of the BRI1 cell surface receptor kinase were seen (Figure 4A). ABA signaling components encompassed the aforementioned splicing factor SUA, the ABCG25 and ABCG40 transporters that catalyze ATP-dependent ABA efflux, and several proteins kinases connected to ABA perception, including the calcium-responsive CPK3 and CPK6 kinases, the SNF1-related protein kinases SnRK2.2 and SnRK2.3, and two in the MAP kinase cascade (MPK11 and MPK6) (Cutler et al., 2010). Some of the more intriguing Ub targets related to pathogen defense are FLS2 and other members of the leucine-rich-repeat, Toll-like receptor family (Figure 4A). FLS2 in particular detects a pathogen-associated molecular pattern peptide derived from bacterial flagellin as part of a general innate immune response (Boller and Felix, 2009). Previous studies showed that its abundance is controlled by ubiquitylation upon ligand binding (Lu et al., 2011). Interestingly, one of the more abundant Ub conjugates in MG132-treated etiolated seedlings was protochlorophyllide oxidoreductase (POR)-B, which catalyzes the conversion of protochlorophyllide to chlorophyllide (Supplemental Datasets 2 and 3). Presumably, PORB ubiquitylation and likely turnover by the UPS suppresses chlorophyll synthesis before young seedlings see light.
An Increased Database of Ubiquitylation Sites and Analysis of Poly-Ubiquitylation
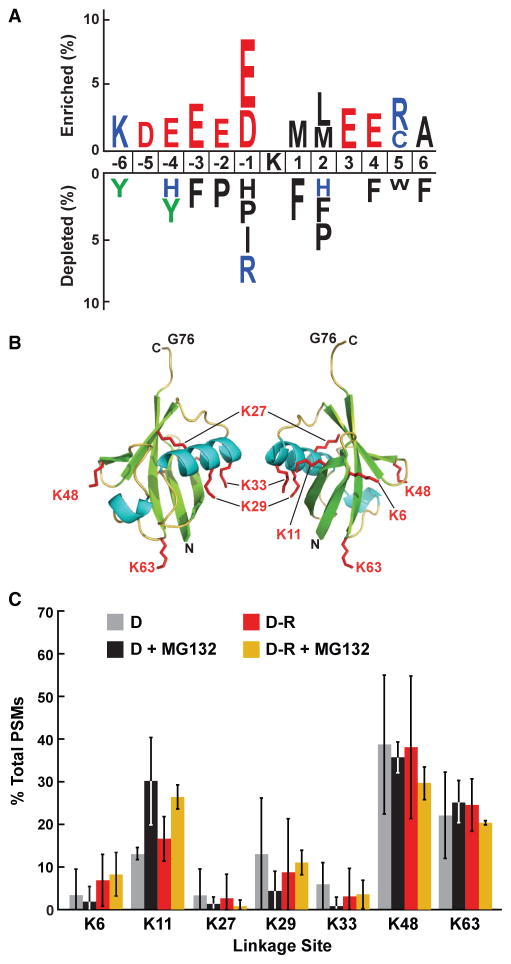
When we searched our etiolated seedling MS spectra for the canonical Ub footprint (KGG), we greatly increased the number of known attachment sites. In total, 227 ubiquitylation sites were assigned onto 199 Arabidopsis proteins (Supplemental Dataset 4). Combined with the previously mapped sites in green seedlings by Kim et al. (2013) and Maor et al. (2007), a database of 413 peptides bearing 478 predicted Ub-attachment sites is now available. When the total collection was aligned by IceLogo, using a window of six amino acids upstream and downstream of the affected lysine, we failed to find any significant consensus sequence(s) that might help predict ubiquitylation sites in other proteins (Figure 5A). The only preferences were an enrichment for charged amino acids both before and after the Ub-attachment site and a subtle exclusion of hydrophobic residues, implying a general, and not unexpected, requirement that the modified lysine be solvent exposed.
Figure 5. Analysis of Ub-Binding Sites within Various Targets and within Ub Itself from Etiolated Seedlings.
(A) Analysis of the amino acid sequences surrounding Ub attachment sites in the catalog of Ub targets from both etiolated and green seedlings (Kim et al., 2013). The analysis used a window of six residues N- and C-terminal to the modified lysine for 413 peptides with 478 Ub footprints to identify significant position-specific under- or over-representation of amino acids flanking the Ub-attachment lysine.
(B) 3D ribbon front and side models of plant Ub highlighting the lysines that could bind Ub covalently. α helices, β strands, and the side chains of lysines are shown in cyan, green, and red, respectively. N, N terminus. C, C terminus. G76, C-terminal active-site glycine. Adapted from PDB: 1UBQ (Vijay-Kumar et al., 1987).
(C) Percentage of Ub footprints identified by tandem MS that represents poly-Ub chains linked internally through the various Ub lysines. Ubiq-uitylated proteins were purified from dark-grown (D) seedlings before or after red-light (R) irradiation with or without pretreatment with 50 μM MG132. Each bar represents the average of three independent MS analyses (±SD).
We also searched the MS/MS spectra for Ub footprints on Ub itself, which would reflect the types of poly-Ub chains employed in etiolated Arabidopsis seedlings. Linkages to all of the seven Ub lysines were detected, even to K27, which was not detected by us previously in green seedlings (Kim et al., 2013). A strong preference was seen for K11, K48, and K63 linkages, and a moderate preference for K29 linkages (Figure 5B and 5C). Prior studies with a range of eukaryotes connected K11- and K48-linked poly-Ub chains to 26S proteasome-mediated turnover, whereas K63-linked poly-Ub chains are often associated with endosomal sorting (Komander and Rape, 2012). Only K11-linked chains were significantly upregulated in both D and D-R samples treated with MG132 (Figure 5C), strongly suggesting that K11-linked poly-Ub chains be considered as a signal for UPS-mediated proteolysis in plants in addition to the K48-linked versions.
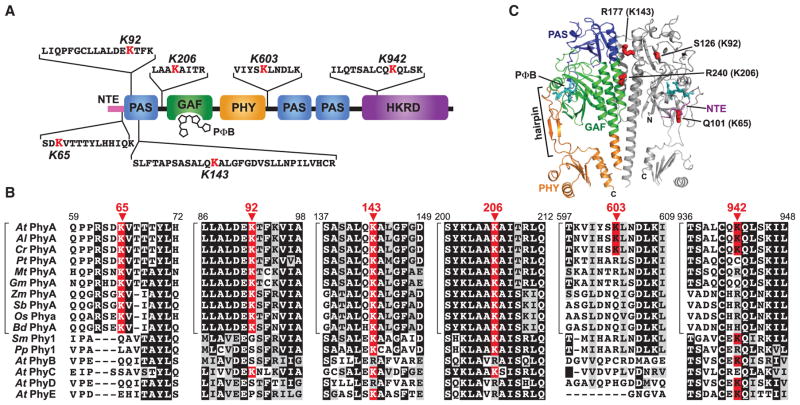
PhyA Ubiquitylation as Pfr Could Be Detected on Six Lysines
MaxQuant searches of the Ub-attachment site library using a site localization probability filter of ≥0.8 uncovered six high probability conjugation sites on PhyA that could only be detected in red-light-irradiated samples (D-R and D-R + MG132) (Supplemental Dataset 4). These sites (K65, K92, K143, K206, K603, and K942) were dispersed throughout the polypeptide, and where 3-D structural information could be predicted (Burgie et al., 2014), were located on solvent-exposed regions (Figure 6A–6C). Under the assumption that PhyA ubiquitylation and turnover as Pfr is pervasive among spermatophytes, we assessed the conservation of these sites in an alignment containing PhyA representatives from eudicots, monocots, and seedless plants, along with the four other Phy isoforms (PhyB-E) in Arabidopsis. Only the 65, 92, 143, and 206 positions contained a lysine in all PhyA proteins, with positions 143 and 206 also containing lysines in seedless plants (Figure 6B; Supplemental Figure 4), suggesting that these invariant residues are important for PhyA turnover. By contrast, K603 and K942 appear weakly conserved among PhyA types, being mainly restricted to the three Brassicaceae species examined. The four lysines within the N-terminal half were also intriguing given previous data examining chimeras between the potato and Arabidopsis PhyA and their more light-stable PhyB isoforms, which implicated the N-terminal half of PhyA, especially the GAF domain, in Pfr ubiquitylation and rapid turnover (Clough et al., 1999; Oka et al., 2012).
Figure 6. Location and Conservation of the Ubiquitylation Sites in Arabidopsis PhyA.
(A) Diagram of the PhyA locating the identified ubiquitylation sites relative to the PAS, GAF, PHY, and HKRD domains. The MS-identified peptide sequence bearing each Ub footprint is shown with the modified lysine colored in red. NTE, N-terminal extension. PΦB, phytochromobilin.
(B) Amino acid sequence conservation surrounding the ubiquitylation sites. The alignment windows include sequences from representative PhyA isoforms from A. thaliana (At), A. lyrata (Al), Capsella rubella (Cr), Populus trichocarpa (Pt), Medicago truncatula (Mt), Glycine max (Gm), Zea mays (Zm), Sorghum bicolor (Sb), Oryza sativa (Os) and Brachypodium distachyon (Bd), PhyA proteins from the bryophytes Selaginella moellendorffii (Sm) and Physcomitrella patens (Pp), and the PhyB, PhyC, PhyD, and PhyE isoforms from A. thaliana. Identical and similar amino acids are highlighted in black and grey boxes, respectively. Dashes denote gaps. The modified lysines are highlighted by the red boxes with the residue number indicated above the arrowheads. Brackets locate members of the PhyA subfamily. See Supplemental Figure 4 for the complete sequence alignment and protein identifier numbers.
(C) Predicted 3D structure of the PhyA photosensory module showing the expected positions of the four ubiquitylated lysines in this region. The model was generated by SWISS-MODEL (http://swissmodel.expasy.org) using the equivalent region in Arabidopsis PhyB as the template (PDB: 4OUR; Burgie et al., 2014). N, amino terminus. C, carboxyl terminus.
To further clarify the importance of these ubiquitylation sites, we generated an Arabidopsis line expressing FLAG-tagged PhyA in the phyA-119 null background and used these seedlings to isolate PhyA-Ub conjugates for MS analysis. After irradiating the seedlings with red light for 1 h, PhyA species were first enriched from total protein extracts with anti-FLAG antibody beads, the bound proteins were released with the FLAG epitope peptide, and the eluate was then subjected to a second round of purification with GST-TUBE beads to enrich for the PhyA-Ub species. As shown in Supplemental Figure 5A, this protocol robustly enriched for PhyA-Ub conjugates from PHYA-FLAG phyA-119 seedling compared with phyA-119 seedlings. The final preparation was subjected to SDS–PAGE, the lane was sliced into apparent mass regions, and each slice was in-gel digested with trypsin followed by LC-MS/MS analysis.
PhyA peptides were readily identified in most, if not all, slices, with the largest number of PSMs detected in the high molecular mass region (slices 1 and 2), as expected for the slower SDS–PAGE migration of PhyA-Ub conjugates versus unmodified PhyA (Supplemental Figure 5B). However, despite extensive MS/MS sequence coverage of PhyA (68 different peptides covering 71% of the 1122 amino acid polypeptide), we failed to detect PhyA peptides with Ub footprints. However, we did find Ub peptides in these slices, including peptides containing a Ub footprint at K48, suggesting that PhyA becomes decorated with K48 poly-Ub chains (Supplemental Figure 5B). Interestingly, we also detected several proteins that co-purified with ubiquitylated PhyA. The most notable were SIZ1, a SUMO E3 ligase that might direct PhyA SUMOylation (Sadanandom et al., 2015), PHOT1, the SKP15 subunit of SCF-type E3 ligases, and the HSP81-2 chaperone.
Roles of Ubiquitylation Sites in PhyA Turnover
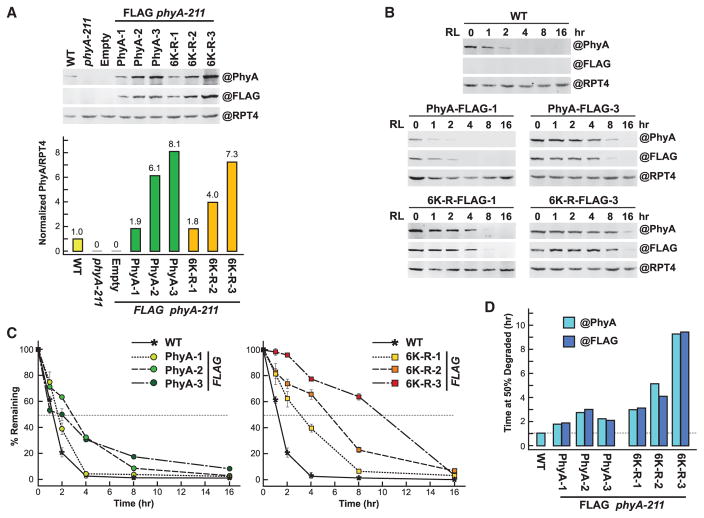
With the expectation that one or more of the six mapped ubiquitylation sites participate in the UPS-mediated turnover of PhyA, we expressed in the phyA-211 null mutant FLAG-tagged versions of either unmodified PhyA or a mutant version in which all six lysines were replaced with arginines (6K-R), and compared their red-light-induced turnover in etiolated seedlings. In preliminary studies examining the chromoproteins immunopurified from etiolated PHYA-FLAG phyA-211 or 6K-R-FLAG phyA-211 seedlings with anti-FLAG antibody-decorated beads, we found that the 6K-R-FLAG variant retained normal photochemistry despite the substitutions. Like PhyA-FLAG, the 6K-R-FLAG protein readily assembled with its bilin chromophore, phytochromobilin (PΦB), to generate a photoactive photoreceptor, as shown by zinc-induced fluorescence of the bound chromophore following SDS–PAGE, and by difference absorption spectra obtained after sequential red- and far-red-light irradiations, which revealed normal absorption maxima of 666 nm and 728 nm for the Pr and Pfr states, respectively (Supplemental Figure 6A and 6B). The rate of photoconversion of Pr to Pfr was also unaltered for 6K-R-FLAG compared with PhyA-FLAG (rate constants of 1.13 ± 0.04 min−1 versus 0.95 ± 0.02 min−1 [±SE] at 25°C under 12 μmol m−2 s−1 of red light), but thermal reversion of Pfr back to Pr in the dark was slightly faster (0.0111 ± 0.0007 min−1 versus 0.0061 ± 0.0003 min−1 [±SE] at 25°C) (Supplemental Figure 6C and 6D). However, this slower reversion rate should be inconsequential relative to the photoconversion rate in affecting Pfr degradation under the conditions used here, in which we exposed seedlings continuously to strong red light (50 μmol m−2 s−1) to saturate Pfr levels.
Based on our preliminary findings that the rate of Pfr degradation is profoundly sensitive to PhyA levels (see below), we selected three independent homozygous lines that express the 6K-R-FLAG or PhyA-FLAG polypeptides at levels close to each other and to unmodified PhyA in wild-type plants. As judged by the immunoblot signals obtained from dark-grown seedlings using anti-PhyA and anti-FLAG antibodies, measured with a near-infrared fluorescence imaging system and then normalized to the signals generated with anti-RPT4 control antibodies, the three 6K-R-FLAG lines expressed 1.9, 6.1, and 8.1 times more PhyA than wild-type, whereas the unmodified PhyA-FLAG lines expressed 1.8, 4.0, and 7.3 times more (Figure 7A).
Figure 7. Preventing Ubiquitylation at the Six Known Sites in Arabidopsis PhyA Slows but Does Not Block Degradation as Pfr.
Either the normal PhyA sequence (PhyA) or one in which the six ubiquitylated lysines were substituted for arginines (6K-R) were expressed with a FLAG tag in the phyA-211 background.
(A) Upper panel: Levels of PhyA in 4-day-old etiolated seedlings before irradiation. Clarified extracts from WT, phyA-211, or phyA-211 seedlings transformed with an empty vector or the PHYA-FLAG and 6K-R-FLAG transgenes were subjected to SDS–PAGE and immunoblotting with either the 073D anti-PhyA or anti-FLAG monoclonal antibody. Three independent transformants were analyzed for PhyA and 6K-R. Near equal protein loading was confirmed by immunoblotting with anti-RPT4 antibodies. Lower panel: Quantification of PhyA levels by densitometric scans of the anti-PhyA antibody immunoblots shown in (A). The values were normalized to the PhyA level in WT.
(B) Degradation of PhyA in R. Four-day-old etiolated seedlings were irradiated with red light (R) for the indicated times and harvested. Clarified extracts were subjected to immunoblotting as in (A).
(C) Quantification of PhyA degradation rates by densitometric scans of the anti-PhyA antibody immunoblots shown in (B). Each point represents the average of three biological replicates (±SD), which was normalized to the value for PhyA at t = 0. Dashed lines highlight the time when 50% of PhyA was degraded. Each bar represents the average of three independent experiments.
(D) Estimated time needed to degrade 50% of PhyA or 6K-R as Pfr based on the degradation kinetics shown in (C) using either the anti-PhyA or FLAG monoclonal antibodies for detection. Dashed line highlights the value for WT PhyA.
The PhyA turnover rate in these etiolated seedlings during continuous red-light irradiation was then measured in triplicate by the same quantitative immunoblotting. Compared with wild-type PhyA and the PhyA-FLAG protein expressed at similar levels, the in vivo degradation rate of 6K-R-FLAG was substantially slower (Figure 7B–7D). For example, whereas the time required to degrade 50% of the PhyA pool was calculated to be 1.2 h for wild-type plants, it was 1.6, 1.9 and 2.7 h, respectively, for the PhyA-FLAG-1, PhyA-FLAG-2, and PhyA-FLAG-3 lines, but was extended to 3.2, 5.2, and 9.1 h, respectively for the 6K-R-FLAG-1, 6K-R-FLAG-2, and 6K-R-FLAG-3 lines. In fact, some of the initial PhyA pool in the 6K-R-FLAG-3 line was still detectable after 16 h of continuous red light, but not for the PhyA-FLAG-3 line (Figure 7B). On average, the 6K-R-FLAG chromoprotein was degraded 2.5-fold slower as Pfr than PhyA-FLAG, indicating that one or more of the six lysines are important to turnover.
Following the completion of this analysis, Rattanapisit et al. (2016) reported, using a candidate approach based on amino acid sequence alignments followed by turnover assays of lysine to arginine substitutions, that K206 is critical for PhyA ubiquitylation and degradation as Pfr, but unfortunately, this study did not account for the strong effects of initial PhyA abundance on turnover. Here, we repeated this degradation kinetic using three independent homozygous lines expressing the FLAG-tagged K206-R chromoprotein at levels similar to those of wild-type PhyA and PhyA-FLAG (Supplemental Figure 7A). As seen above, the rate of degradation for K206-R-FLAG was dependent on chromoprotein levels. However, while the turnover rate for K206-R-FLAG was slower than that for PhyA-FLAG when comparing lines with similar expression levels, it was faster than that for 6K-R-FLAG (Supplemental Figure 7B and 7C), indicating that ubiquitylation at lysines other than K206 also affects Pfr breakdown.
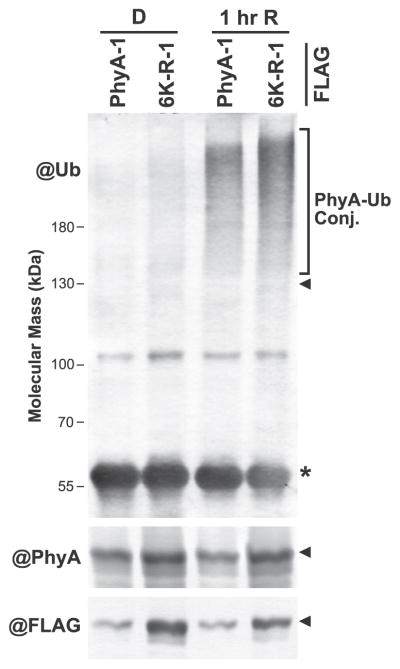
Given that the 6K-R-FLAG chromoprotein was still degraded as Pfr (albeit more slowly), we considered it likely that other residues are also ubiquitylated. To examine this possibility, we immunopurified PhyA with anti-FLAG antibodies from the PhyA-FLAG-1 and 6K-R-FLAG-1 lines and compared their ubiquitylation levels before and after a 1-hr exposure to red light. As expected based on the MS data (Table 1), the ubiquitylation state of the PhyA-FLAG chromoprotein rose substantially during the light treatment. Remarkably, the 6K-R-FLAG mutant retained ubiquitylation as Pfr in planta, as judged by the smear of high molecular mass species immunodetectable with the anti-Ub antibody (Figure 8). Thus, other ubiquitylation sites in PhyA likely exist, or alternative residues can participate once one or more of the six sites studied here (K65, K92, K143, K206, K603, and K942) are eliminated.
Figure 8. Substitution of the Six Known Ubiquitylation Sites in PhyA for Arginines Does Not Block Pfr Ubiquitylation.
PhyA-FLAG was immunoprecipitated from PhyA-1 phyA-211 and 6K-R-1 phyA-211 plants (see Figure 7A) before or after a 1-h irradiation with red light (R), and immunoblotted with antibodies against Ub, PhyA, or FLAG. PhyA-Ub conjugates are located by the bracket. The arrowheads and asterisk indicate the position of PhyA-FLAG and the anti-FLAG IgG heavy chain used for immunoprecipitation, respectively.
Selective and Non-selective Pathways for PhyA Turnover?
Prior studies broadly implicated the SCF-type Cullin-RING Ub ligases (CRLs) in PhyA degradation by the ability of weak Cullin (CUL)-1 mutants to slow Pfr turnover (Quint et al., 2005; Moon et al., 2007). Here, we extended these studies to CRL E3s in general (Hua and Vierstra, 2011) given the fact that another CRL subtype (BTB E3s) assembled with the substrate adaptor proteins LRB1–3 affects PhyB ubiquitylation and degradation (Christians et al., 2012; Ni et al., 2014). The mutant panel included weak knockdown alleles compromising the CUL1 subunit of the SCF E3s (axr6-3, cul1-6, and icu13; Quint et al., 2005; Moon et al., 2007; Esteve-Bruna et al., 2013), the CUL3a/b subunits of the BTB E3s (cul3a-3 cul3b-1; Gingerich et al., 2005; Thomann et al., 2009), the CUL4 subunit of the DWD E3s (ascul4; Bernhardt et al., 2006), and the CAND1 factor necessary for recycling all CRL complexes (eta2-1; Chuang et al., 2004). (All three Cullins and CAND1 are essential in Arabidopsis, thus precluding the use of null alleles; Shen et al., 2002; Gingerich et al., 2005; Chen et al., 2006.) As shown in Supplemental Figure 8, PhyA turnover was not detectably slowed in either cul3a-3 cul3b-1 or ascul4 seedlings, thus eliminating BTB and DWD E3 as participating. Conversely, degradation was noticeably slower in the mutant panel affecting SCF complex assembly (axr6-3, cul1-6, icu13, and eta2-1), further supporting the proposals that an SCF-type E3 is involved in at least some portion of PhyA turnover (Quint et al., 2005; Moon et al., 2007).
It was also conceivable that other proteolytic pathways participate in PhyA turnover, especially given its dynamic relocalization during signaling and/or turnover, including nuclear import and concentration into cytoplasmic and nuclear puncta, variously called photobodies and sequestering (Chen and Chory, 2011). Autophagy was especially intriguing because of its role in clearing protein aggregates that morphologically resemble photobodies. However, when we tested PhyA degradation in null mutants that either block ATG8-mediated autophagy (atg7-2 and atg5-1), its upstream regulatory kinase cascade (atg11-1 and atg13a-2 atg13b-2), or eliminate the NBR1 autophagic receptor that recognizes ubiquitylated proteins (nbr1-2), none slowed PhyA turnover in red light, thus eliminating autophagy as a major route for Pfr breakdown (Supplemental Figure 8C and 8D).
The 6K-R Mutant Is Hyperactive in Signaling
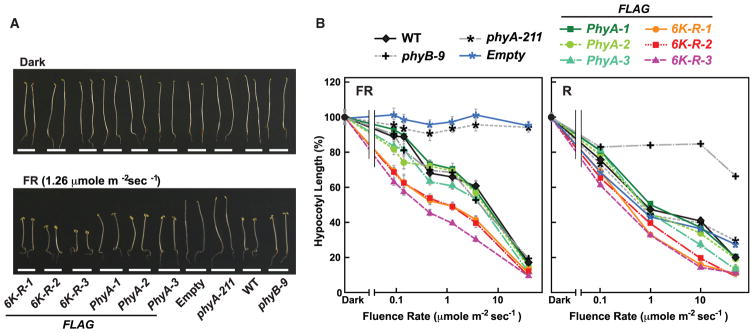
One outcome of dampened Pfr turnover should be greater PhyA signaling caused by increased retention of the bioactive state. From analysis of the suppressive effects of red and far-red light on hypocotyl growth for the wild-type, phyA-211 PhyA-FLAG, and the 6K-R-FLAG lines, we indeed found that the 6K-R-FLAG chromoprotein is hyperactive. Here, we exposed etiolated seedlings to various fluence rates beginning at germination and measured their hypocotyl lengths after 4 days. Whereas the phyA-211 seedlings were markedly insensitive to far-red light and the complemented PhyA-FLAG phyA-211 lines behaved like wild-type, the 6K-R-FLAG phyA-211 lines more effectively responded to the light across a range of far-red light fluences (Figure 9). A modest increase in 6K-R-FLAG signaling was also observed using red light compared with that from PhyA-FLAG (Figure 9).
Figure 9. PhyA Missing the Ubiquitylated Lysines Are Hyperactive in Seedling Photomorphogenesis.
Plants expressing FLAG-tagged PhyA without (PhyA) or with the known ubiquitylated lysines converted to arginines (6K-R) were germinated in the dark or under various fluences of continuous far-red (FR) or red light (R). After 7 days, hypocotyl lengths were measured.
(A) Images of representative seedlings either kept in the dark or irradiated with FR. The three independent transformants of phyA-211 with the FLAG-tagged versions of either PhyA or 6K-R as described in Figure 7 were compared. WT, phyB-9, and phyA-211 seedlings either alone or transformed with an empty vector were included as controls.
(B) Fluence responses of the mutant collection in (A) under continuous R or FR. Each point (±SE) represents the average of three independent experiments, each containing at least 30 seedlings.
DISCUSSION
The transcriptional events that underpin reprogramming of plant proteomes as they transition from skotomorphogenic to photomorphogenic growth are well documented in numerous studies (e.g., Ma et al., 2001; Tepperman et al., 2001, 2006; Hu et al., 2009). More recent MS studies with Arabidopsis, rice, and maize have in turn provided a preliminary glimpse into how the corresponding proteomes are affected (Wang et al., 2006; Shen et al., 2009; Hamamoto et al., 2012). As expected, the dominant feature is the rapid accumulation of proteins involved in photosynthetic light capture and carbon assimilation as etioplasts convert to chloroplasts during the acquisition of photoautotrophic metabolism. Because this developmental transition is also likely influenced by removing proteins contradictory to, or less essential for green plants, understanding how proteolysis affects etiolated seedling proteomes is also needed to provide a full picture of photomorphogenesis. Certainly, one strategy would be to globally compare the half-lives of proteins during this process, but the technical challenges posed by such comprehensive measurements remain insurmountable in plants, many due to inherent difficulties in stable isotope labeling of proteins (Schulze and Usadel, 2010). Here, we assayed Ub conjugation, with or without the proteasome inhibitor MG132, as a surrogate for turnover given the roles of this PTM in driving selective degradation of cytoplasmic and nuclear proteins and membrane proteins that face these compartments, either by the 26S proteasome or autophagy (Vierstra, 2009; Li and Vierstra, 2012).
Altogether, our Ub-conjugate datasets prepared from etiolated Arabidopsis seedlings either before or after a short red-light irradiation, combined with our prior datasets from green seedlings (Saracco et al., 2009; Kim et al., 2013), demonstrate that rapid and dramatic sculpting of seedling proteomes driven by Ub occurs during photomorphogenesis. While some of these differences in Ub-conjugate profiles between green and etiolated seedlings certainly reflect the morphological differences between the two developmental stages sampled (mainly hypocotyls and cotyledons versus leaves), we propose that most reflect the distinctions between the skotomorphogenic and photomorphogenic programs. For example, the predominance of proteins involved in signaling by various hormones can be explained by the need of hypocotyl cells to expand rapidly to help the germinating seedling reach the soil surface, while a repression of cotyledon and leaf expansion is encouraged to limit stored food consumption. Once the seedling reaches the soil surface, hypocotyl expansion is dramatically repressed and rapid cotyledon and leaf expansion commences as the plant becomes photosynthetic. Given the importance of auxin, ABA, and brassiosteroids to hypocotyl elongation (Wang et al., 2012; de Wit et al., 2016), it is not surprising that signaling components for these hormones are enriched in our Ub-conjugate datasets from etiolated versus green seedlings (Figure 4A). In particular, transporters facilitating the movement of both auxin (AUX1/LAX3, PIN1, PIN3, PIN4, PIN7, and ABCB19; Wu et al., 2010; Zadnikova et al., 2010) and ABA (ABCG25 and ABCG40; Kang et al., 2015) were detected. It is also worth noting that two plasma membrane H+-ATPase isoforms are also enriched (AHA1 and AHA2; Table 1). As the electrochemical gradient generated by these proton pumps is essential for ion uptake and subsequent turgor pressure and is strongly influenced by auxins (Takahashi et al., 2012; Spartz et al., 2014) and light (Hohm et al., 2014), we expect that controlling their activities would be essential for the rapid cell expansion of hypocotyls and cotyledons, and in directing photo- and gravitropism.
Surprisingly, our ubiquitylome datasets generated here and by Kim et al. (2013) are mostly devoid of known/predicted transcription factors (19 and nine in etiolated and green seedlings, respectively, with one overlap; Supplemental Table 2), despite the likelihood that the UPS regulates their abundance (Vierstra, 2009). A fuller view of Ub in regulating transcription and other nuclear functions would certainly benefit from a focused MS analysis of nucleus-enriched preparations given the low cellular levels for many of these factors.
The Ub conjugate in etiolated seedlings whose abundance was most dramatically influenced by red light was PhyA, which rose from just one of many Ub adducts in dark-grown seedlings to easily become the most abundant conjugate after just 1 h of red light. This rise is likely underpinned by an efficient conjugation system directed at the Pfr conformer, with the end result being a dramatic drop in PhyA levels through UPS-mediated proteolysis as etiolated seedlings reach the soil surface and immediately transition to become green seedlings (Clough and Vierstra, 1997). Our MS data also potentially connected K48-linked poly-Ub chains to PhyA, which is a strong signal for proteasome recognition (Komander and Rape, 2012). A number of other factors important to light perception in general, and Phy signaling in particular, were also detected here, including PHOT1 and its signaling partner NPH3 (Figure 4A), and CDF3 linked to circadian rhythm entrainment. Notably, we did not detect PhyB or any of its PIF-binding partners in our ubiquitylome datasets. The absence of PhyB likely reflects the low levels of this isoform in etiolated seedlings compared with PhyA, and possibly, lower steady-state amounts of conjugates given the slower turnover of PhyB in red light (Christians et al., 2012).
While numerous Ub conjugates in our etiolated seedling datasets are likely targets of the UPS based on their detection only in MG132-treated samples, or by having significantly increased PSM counts (2-fold) upon inhibitor treatment (68% [868 of 1279] of our etiolated seedling datasets), other functions are also possible. As examples, Ub conjugation could commit proteins to autophagy or correctly traffic proteins to their intracellular destinations via endosomal sorting (Mukhopadhyay and Riezman, 2007; Li and Vierstra, 2012). In fact, the detection of ubiquitylated 26S proteasome subunits here and elsewhere (Book et al., 2010; Kim et al., 2013) likely reflects the autophagic removal of this proteolytic machine (Marshall et al., 2015). Non-proteolytic roles of Ub addition have also connected to chromatin structure and transcription through modifying histones and other core components (Weake and Workman, 2008). In fact, several histones (H1, H2A, and H2B) and enzymes predicted to ubiquitylate/deubiquitylate them (e.g., BRE1-like and HUB2) are present in our etiolated and green Ub target catalogs (Kim et al., 2013; this report).
The accumulating library of canonical Ub-binding sites detected via the KGG Ub footprint now offers a strategy to examine the roles of Ub in protein function and half-life via lysine to arginine substitutions. Here, we tested this notion by examining the red-light-induced turnover of PhyA in which the six identified ubiquitylation sites were replaced. Surprisingly, while we could slow Pfr turnover by ~2.5-fold, we were unable to completely stabilize the photoreceptor or block its ubiquitylation. Retained ubiquitylation and turnover could reflect: (1) our failure to identify all the lysine acceptor sites, (2) the ability of the corresponding conjugation machinery to attack other lysines in their absence, or, interestingly, (3) the use of other residues for adduct formation. While our MS evidence for the six ligation sites appeared statistically strong, we cannot rule out other sites whose tryptic peptides are either less amenable to MS/MS detection or too large for our MS parameters due to the missed trypsin cleavage at the modified lysine. We also note that several of our mapped sites have neighboring lysines that could become alternatives (e.g., K95, K202, and K608, and K946; Figure 6B). Rattanapisit et al. (2016) investigated the K202 and K206 pair and found that the single K202 chromoprotein was degraded like wild-type PhyA and that the double mutant was no more stable than the K206 variant, suggesting that K202 is not a relevant substitute. They and Clough et al. (1999) also tested several other conserved lysines in PhyA (e.g., K165, K284, K286, K400, K739, K744, K784, and K796); none were found to be effective in stabilizing Pfr.
Nonetheless, our data agree with Rattanapisit et al. (2016) that K206 is an influential residue, but also provide a cautionary example that PhyA levels prior to photoexcitation significantly influence the rate of Pfr clearance. We hypothesize that the effect of initial PhyA levels on degradation rate stems from limited capacity in the machinery needed to recognize and ubiquitylate PhyA and is further accentuated by our experimental conditions that used a sharp dark to red-light transition to provoke a rapid influx of Pfr. Under more physiological conditions where the dark/light transition is more gradual, such dependency might be less dominant.
With respect to alternative amino acids, evidence is rapidly emerging in animals that residues besides lysine can form adducts with Ub, including internal serines, threonines, and cysteines, and the N-terminal amino group (Komander and Rape, 2012; Kravtsova-Ivantsiv and Ciechanover, 2012; Elliott, 2016). Such non-canonical linkages have not yet been chemically confirmed in plants; in fact, designated searches of our MS datasets for these linkages failed to uncover any, suggesting that if they exist, they are likely rare in Arabidopsis. However, Gilkerson et al. (2015) recently provided indirect support for serine-/threonine-based Ub linkages from turnover studies with a lysine-less versions of the AUX/IAA protein IAA1. Clearly, future MS analysis of the ubiquitylated forms of 6K-R is needed to address this possibility. Whatever the outcome, our experiences with PhyA and those with IAA1 complicate the use of lysine to arginine substitutions to confirm ubiquitylation sites for plant targets.
Given the complex dynamics undertaken by Phys after photo-conversion as they are transported from the cytoplasm to nucleus, bind to chromatin-bound PIFs to negate their transcriptional suppression, and concomitantly aggregate into photobodies, it is likely that multiple proteolytic pathways in at least two compartments (cytoplasm and nucleus) affect Pfr removal, some of which could be Ub and proteasome independent. Previous phenotypic studies of weak SCF mutants implicated an SCF-type E3 in at least some portion of PhyA turnover as Pfr (Quint et al., 2005; Moon et al., 2007; Esteve-Bruna et al., 2013). We confirmed this importance from the analysis of the axr6-3, cul1-6, icu13, and eta2-1 mutants, but failed to detect contributions for other CRL subtypes (BTB and DWD ligases). For further support regarding SCF E3(s), we detected another component of the SCF complex (SKP15) during the MS analysis of PhyA-Ub conjugates purified from Arabidopsis. Given that PhyA coalesces into nuclear photobodies after Pfr formation (Chen and Chory, 2011), it was also possible that Ub-driven autophagy participates in Pfr turnover. However, this route does not appear to be influential given the normal Pfr turnover rates seen for seedlings missing a number of components within the autophagy system. Whatever the reasons, the mechanism(s) devoted to PhyA turnover upon Pfr formation remains enigmatic.
In conclusion, our MS analysis of ubiquitylation in etiolated seedlings before and soon after the transition to photomorphogenic development provides a deep catalog of targets from which to better understand how plant proteomes are reconfigured during seedling greening. In particular, the list of targets associated with light and hormone signaling, peroxisome function, and disease resistance can now be used to test the role(s) of Ub addition associated with these events.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis thaliana ecotype Columbia (Col)-0 was used as wild-type and was grown in a growth chamber on solid half-strength Murashige and Skoog (MS) medium containing 0.5 g/L MES (pH 5.7) and 0.7% agar at 22°C and a 16-h light/8-h dark cycle unless indicated otherwise. After stratification for 3 days in the dark at 4°C, seeds were illuminated with white light for 2 h to induce germination. The hexa(6His-UBQ) transgenic line was as described previously (Saracco et al., 2009). The phyA-211 and phyB-9 mutants (Reed et al., 1993, 1994) were obtained from the Arabidopsis Biological Resource Center (ABRC) at Ohio State University (https://abrc.osu.edu/). The PHYA T-DNA insertion mutant phyA-119 conferring Basta resistance was obtained from the SAIL insertion collection (ID: CS802119) available at ABRC (Sessions et al., 2002). The cul1-6, axr6-3 (CUL1), icu-13 (CUL1), cul3a-3 cul3b-1, ascul4, and eta2-1 (CAND1) seedlings were studied previously (Chuang et al., 2004; Quint et al., 2005; Bernhardt et al., 2006; Moon et al., 2007; Thomann et al., 2009; Esteve-Bruna et al., 2013). The atg5-1, atg7-2, atg11-1, atg13a-2 atg13b-2, and nbr1-2 mutants were as described (Thompson et al., 2005; Suttangkakul et al., 2011; Li et al., 2014; Marshall et al., 2015).
The PHYA-FLAG complemented phyA-211 and phyA-119 lines, which express either unmodified FLAG-tagged PhyA or versions in which one or more of the six ubiquitylated lysines (K65, K92, K143, K206, K603, and K942) were replaced with arginines (K-R), were generated with a full-length PHYA cDNA sequence (Sharrock and Quail, 1989), which was appended by PCR using the oligonucleotide GGA GGA GGA GAT TAC AAA GAT GAT GAT GAT AAA TAG to terminate in a FLAG tag (DYKDDDDK) preceded by a GGG sequence. The PHYA promoter with 5′ UTR (2454 bp) terminating just proximal to the ATG codon and PHYA 3′ UTR (242 bp) were PCR amplified from genomic DNA using the primer pairs AAT ACG GAA GTT TTG GTG TAA AGA ACC and TTT TTT CCT GAC ACA GAG ACA AGA CAA C, and TCC CCA AAA GAA AAG GGG TCT GG and AGC TCA TTT CTC CTA TAT TAC TTA TTT TG, respectively. The final pPHYA:PHYA-FLAG construction for complementing the phyA-211 lines was cloned through the Gateway BP reaction system into the pDONOR221 vector and then transferred via the Gateway LR reaction into the pMDC99 T-DNA destination vector, which confers hygromycin resistance. For the complemented phyA-119 lines, the pPHYA:PHYA-FLAG construction was cloned into the kanamycin-resistant pMDC100 plasmid by the Gateway reactions.
The K-R substitutions were introduced into the pPHYA:PHYA-FLAG template by PCR with mutagenic primer pairs (Supplemental Table 1). Multiple K-R substitutions were added iteratively by subsequent PCR amplifications. All mutations were verified as correct by full-sequence analysis of the transgenes. Wild-type pPHYA:PHYA and the library of K-R mutants were transformed into the Agrobacterium tumefaciens strain GV3101 and then introduced into homozygous phyA-211 or phyA-119 plants by the floral-dip method (Saracco et al., 2009). Expression of a functional PhyA-FLAG protein in the phyA-211 and phyA-119 backgrounds was confirmed by restoration of far-red light sensitivity of hypocotyl growth. Three independent T4 homozygous lines, with similar PhyA expression levels as determined by immunoblotting (see below), were analyzed for PhyA degradation and ubiquitylation, and for phenotypic effects on Arabidopsis development.
Affinity Enrichment of Ubiquitylated Proteins
Wild-type and hexa(6His-UBQ) seedlings (starting from ~200 μg of seeds) were grown in 50 mL of liquid Gamborg’s B5 minimal medium (Sigma-Aldrich) supplemented with 2% Suc and 0.5 g/L MES (pH 5.7) for 7 days, either in darkness, or in darkness followed by 1 h of red light (50 μmol m−2 s−1; 660 nm maximum) provided by a bank of light-emitting diodes (LEDs). For the MG132 (N-benzyloxycarbonyl-Leu-Leu-Leu-al; Selleckchem) treatments, the cultures were made either 50 μM in inhibitor or an equivalent volume of DMSO, and incubated for 4 h before harvesting for both dark- and red-light-treated seedlings. Seedlings were collected, blotted dry, rapidly frozen at liquid nitrogen temperatures, and then stored at −80°C until use. Unless indicated, all manipulations of the seedlings and subsequent purifications were performed under a dim green safe light.
Ubiquitylated proteins were enriched from hexa(6His-UBQ) and wild-type (control) seedlings by the two-step TUBEs/Ni2+-NTA affinity chromatography method as described (Kim et al., 2013). Frozen tissue was powdered at liquid nitrogen temperatures and then mixed with 2 mL/g tissue of extraction buffer (EB; 50 mM Tris-HCl [pH 7.2], 200 mM NaCl, and 0.25% Triton X-100) containing 1 × protease inhibitor cocktail (Roche), 2 mM phenylmethanesulfonyl fluoride, 10 mM 2-chloroacetamide, 10 mM sodium metabisulfite, and 1 mM N-ethylmaleimide added just before use. The homogenate was filtered through two layers of Miracloth and one layer of cheesecloth and clarified at 12 000 g for 20 min. Clarified extracts equivalent to 40 g fresh weight of tissue were incubated for 6 h at 4°C with gentle shaking with 1 mL of GST-TUBEs or GST alone (control) coupled to AffiGel-15 beads (Bio-Rad). The beads were washed three times with EB supplemented with 2 M NaCl, and then bound proteins were eluted at room temperature with 10 mL of 7 M guanidine-HCl, 100 mM NaH2PO4, and 10 mM Tris-HCl (pH 8.0). The eluates were made 20 mM imidazole and 10 mM 2-chloroacetamide and incubated for 12 h at 4°C with Ni-NTA agarose (Qiagen) pre-equilibrated in the same buffer. Afterward, the beads were washed once in 6 M guanidine-HCl, 0.1% SDS, 100 mM NaH2PO4, and 10 mM Tris-HCl (pH 8.0), once in urea buffer (URB; 8 M urea, 100 mM NaH2PO4, and 10 mM Tris-HCl [pH 8.0]) plus 0.1% Triton X-100, twice in URB plus 20 mM imidazole, and three times in URB alone. Bound proteins were eluted with URB containing 400 mM imidazole and concentrated by centrifugation with an Ultracel-10K filter (Millipore). The purity of the preparations was assessed after SDS–PAGE by staining for total protein with silver and by immunoblot analysis with anti-Ub antibodies (Shanklin et al., 1989).
Mass Spectrometry
Purified Ub conjugate (100 μL) or control samples were reduced in 10 mM dithiothreitol for 1 h, carbamidomethylated with 50 mM iodoacetamide for 1 h followed by quenching the reaction with 20 μL of 200 mM dithiothreitol for 10 min, and then alkylated in 30 mM 2-chloroacetamide for 1 h (Kim et al., 2013). Samples were diluted 10-fold with 25 ammonium bicarbonate and digested for 12 h at 37°C with 2 μg of sequencing-grade trypsin (Promega), followed by a second digestion for 6 h with an additional 2 μg of trypsin. Digested samples were desalted using a C18 solid-phase extraction pipette tip (SPEC PT C18, Varian), vacuum dried, and reconstituted in 10 μL of 5% acetonitrile and 0.1% formic acid in water for MS analysis.
LC-MS/MS was performed with a nanoflow liquid chromatography system (nanoAcquity; Waters) coupled to an electrospray ionization FT/ion-trap mass spectrophotometer (LTQ Orbitrap Velos; Thermo Fisher Scientific). The LC system utilized a 15 cm × 365 μm fused silica microcapillary column packed with 3-μm-diameter, 100-Å pore size, C18 beads (Magic C18; Bruker), with the emitter tip pulled to ~2 μm using a laser puller (Sutter Instruments). Peptides were loaded onto the column for 30 min at 500 μL/min, and eluted by a 120-min, 2%–30% acetonitrile gradient in 0.1% formic acid at a 200 nL/min flow rate. MS spectra were acquired between 300 and 1500 mass-to-charge ratio (m/z) at a resolution of 60 000, followed by 10 MS/MS scans of the 10 highest intensity parent ions at 42% relative collision energy and 7500 resolution, with a mass range starting at 100 mass-to-charge ratio. Dynamic exclusion was enabled for 30 s and an exclusion window for 120 s with a repeat count of 2.
For further peptide mapping of PhyA-Ub conjugates, the TUBEs eluates prepared with PHYA-FLAG phyA-119 seedlings exposed to red light for 1 h were affinity enriched using anti-FLAG M2 antibody agarose beads (Sigma-Aldrich) and separated by SDS–PAGE. The gel lanes were sliced into various apparent molecular mass fractions, each of which was de-stained twice for 5 min with 800 μL of a freshly prepared 1:1 solution of 100 mM sodium thiosulfate and 30 mM potassium ferricyanide, followed by two washes with 1 mL of water. Gel pieces were dehydrated for 5 min with 800 μL of 25 mM NH4HCO3 and 50% acetonitrile, then once more for 30 s with 100% acetonitrile followed by vacuum drying. Dried gel pieces were reduced in 25 mM dithiothreitol for 20 min at 65°C, carbamidomethylated with 55 mM iodoacetamide for 1 h in the dark, and followed by a wash with 20 volumes of water. Dried and rehydrated gel pieces were digested for 16 h at 37°C with 2 μg of sequencing-grade trypsin (Promega), followed by a second digestion for 8 h with an additional 2 μg of trypsin. Digested peptides were extracted first with 0.1% trifluoroacetic acid (TFA) and then with 5% TFA and 70% acetonitrile. Elutes were pooled, vacuum dried, rehydrated in 1.9% acetonitrile and 0.1% formic acid, desalted using a C18 solid-phase extraction pipette tip (SPEC PT C18, Varian), vacuum dried, and reconstituted in 10 μL of 5% acetonitrile and 0.1% formic acid for MS analysis.
Mass Spectrometry Data Analysis
The MS/MS data were searched using SEQUEST version 1.2 (Thermo Fisher Scientific) against the A. thaliana ecotype Col-0 protein database (IPI database, version 3.85 containing 39 677 entries available at The Arabidopsis Information Resource [TAIR], http://www.arabidopsis.org). Masses for both precursor and fragment ions were treated as mono-isotopic. Met oxidation (+15.994915 Da), Cys carbamidomethylation (+57.021464 Da), the Gly-Gly Ub footprint reflecting Ub addition ± the lysine directly affected (114.043 Da and 242.1 Da, respectively) were set as variable modifications. The database search allowed for up to two missed trypsin cleavages, and the mass tolerances were set to 10 ppm for precursor and 0.1 Da for product ions. The data were filtered using a 1% FDR (Rohrbough et al., 2006). Inclusion in the ubiquitylation list required a minimum of two peptide matches or at least one peptide if it included a GGK Ub footprint. In total, the MS analyses identified 8408 peptides that could be assigned to 1279 individual ubiquitylated proteins following subtraction of contaminants.
To identify consensus Ub-attachment site(s) in the collection of 431 Ub footprints identified here (279) and elsewhere (Maor et al., 2007; Saracco et al., 2009; Book et al., 2010; Kim et al., 2013), the region flanking the modified lysine was compared by IceLogo (Colaert et al., 2009). Levels of specific ubiquitylation targets were roughly quantified by the number of PSMs for assigned peptides. GO annotations were assigned using the GO database in TAIR (http://.www.arabidopsis.org/). Protein interaction networks were generated by the STRING database version 10.0 (Szklarczyk et al., 2015) and visualized by Cytoscape version 3.4.0 (Shannon et al., 2003) using the preferred layout to highlight subclusters.
PhyA Protein Sequence Alignments and 3D Visualization
PhyA amino acid sequences from representative land plants were identified by BLASTP (https://blast.ncbi.nlm.nih.gov/) using the Arabidopsis PhyA sequence (At1g09570) as the query. Protein sequence alignments and domain identifications were generated with the Clustal Omega server (http://www.ebi.ac.uk/Tools/msa/clustalo/) and Pfam (http://pfam.xfam.org), respectively. The 3D structure of the photosensory module from Arabidopsis PhyA was predicted by SWISS-MODEL (http://swissmodel.expasy.org) using the equivalent region in Arabidopsis PhyB as the template (PDB: 4OUR; http://www.wwpdg.org; Burgie et al., 2014), and visualized using PyMOL v1.7.03 (http://www.pymol.org).
Spectroscopic Analyses of PhyA
Absorption spectra, Pr→Pfr photoconversion, and Pfr→Pr thermal reversion of PhyA-FLAG and the 6K-R-FLAG mutant were measured in vitro following affinity enrichment of the tagged chromoproteins. Four-day-old seedlings, grown on solid medium in the dark at 22°C, were either kept in the dark or irradiated with red light for 1 h, blotted dry, and frozen in liquid nitrogen. Frozen tissue was pulverized at liquid nitrogen temperatures, and homogenized into 2 mL/g fresh weight of Phy EB (PEB; 45% ethylene glycol, 90 mM Tris-HCl [pH 8.0]), containing 1×protease inhibitor cocktail (Roche), 18 mM sodium metabisulfite, and 4 mM phenylmethane-sulfonyl fluoride (Clough et al., 1999). The homogenate was filtered through two layers of Miracloth and one layer of cheesecloth, clarified at 20 000 g for 10 min, and then incubated for 1 h at 4°C with anti-FLAG M2 agarose beads pre-equilibrated with same buffer. The beads were washed three times with PEB, and the remaining bound PhyA-FLAG was eluted with FLAG EB (150 mM KCl, 50 mM HEPES-KOH [pH 7.8]) and 500 ng/μL of FLAG peptide (Sigma-Aldrich) (Burgie et al., 2014). Absorption spectra were recorded using a Cary60 spectrophotometer (Agilent Technologies, Santa Clara, CA). Red light and far-red light were provided by 660-nm and 730-nm peak output LEDs (Digi-Key, Thief River Falls, MN), respectively.
PhyA Degradation Assays
For PhyA degradation assays, seedlings were grown for 4 days as above and either kept in darkness or subjected to continuous red-light irradiation (50 μmol m−2 s−1) for various times before harvest and immediate freezing. Frozen tissue was pulverized at liquid nitrogen temperatures, and homogenized directly in SDS–PAGE sample buffer (125 mM Tris-HCl [pH 6.8], 4% [w/v] SDS, 20% [v/v] glycerol, 0.001% [w/v] bromophenol blue, and 10% [v/v] 2-mercaptoethanol). The homogenates were heat denatured and clarified at 13 000 g for 10 min, and the resulting supernatants were subjected to SDS–PAGE followed by transfer onto Immobilon-FL-PVDF (EMD Millipore) membranes. The membranes were blocked in 5% (w/v) nonfat milk in PBS and then probed with either a mouse anti-PhyA monoclonal antibody (073D; Shanklin et al., 1989), an anti-FLAG-M2 monoclonal antibody (Sigma-Aldrich), or rabbit anti-RPT4 polyclonal antibodies (Marshall et al., 2015). Goat anti-rabbit IRDye 800 or donkey anti-mouse IRDye 800 (Li-Cor Biosciences) antibodies were used as the secondary antibody. Fluorescent signals from the membranes were detected with an Odyssey-Fc imaging system (Li-Cor Bioscience) and quantified using Image Studio Lite 5.2 according to the manufacturer’s recommendations. The anti-PhyA or anti-FLAG antibody fluorescence signals for a dilution of each sample were normalized to that generated with the anti-RPT4 antibody, and then adjusted using a dilution series to that generated with samples at t = 0, which was assigned as 100% remaining.
Immunoblot Detection of Ubiquitylated PhyA
Ubiquitylated PhyA-FLAG was enriched from 4-day-old dark-grown or red-light irradiated seedlings grown as above following homogenization with PEB containing 1× protease inhibitor cocktail (Roche), 2 mM phenyl-methanesulfonyl fluoride, 10 mM 2-chloroacetamide, 10 mM sodium metabisulfite, and 1 mM N-ethylmaleimide added just before use. The homogenate was filtered through two layers of Miracloth and one layer of cheesecloth, clarified at 12 000 g for 20 min, and incubated for 2 h at 4°C with anti-FLAG M2 agarose beads pre-equilibrated with PEB. Unbound proteins were released by three washes with PEB, and the remaining bound PhyA-FLAG was eluted with an equal volume of twice-strength SDS–PAGE sample buffer, followed by SDS–PAGE and immunoblot analyses with anti-FLAG (Sigma-Aldrich) or anti-PhyA (073D, Shanklin et al., 1989) monoclonal antibodies, or polyclonal antibodies against Ub.
Phenotypic Analysis of Lines Expressing Wild-Type PhyA and the K-R Mutants
The effect of light on hypocotyl elongation was measured for the wild-type, phyA-211, phyB-9, PHYA-FLAG phyA-211, and 6K-R-FLAG phyA-211 seedlings grown on half-strength MS medium containing 0.5 g/L MES (pH 5.7) and 0.7% agar as above. After seed stratification, the plates were either kept in darkness or exposed continuously to various fluence rates of red or far-red light provided by a bank of LEDs (E-30LED-controlled environment chamber; Percival). Hypocotyl lengths after 3 days were measured using ImageJ software (http://rsb.info.nih.gov.ij (Abramoff et al., 2004)).
Supplementary Material
Acknowledgments
FUNDING
This work was supported by an NSF grant (MCB-1329956) to R.D.V., grants from NIH/NHGRI (1P50HG004952) and NIH-NIGMS (P01GM081629) to M.S. and L.M.S., the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2015R1D1A1A01059372) to D.-Y.K., and a postdoctoral fellowship from the Mexican National Council for Science and Technology (CONACYT) (208247 and 250963) to V.A.-H.
We thank Drs. David Gemperline, E. Sethe Burgie, and Fionn McLoughlin for technical advice. Various mutants were graciously provided by Drs. William Gray, Mark Estelle, Hanjo Helleman, Jose Luis Micol, Allison Philips, Pascal Genschik, and Scott Sarraco. Dr. Robert Sharrock kindly provided the PHYA cDNA. No conflict of interest declared.
Footnotes
ACCESSION NUMBERS
Protein identifiers and/or corresponding gene accession numbers for the catalog of Arabidopsis ubiquitylation targets identified here can be found in the various tables and Supplemental Datasets.
Supplemental Information is available at Molecular Plant Online.
AUTHORS CONTRIBUTIONS
D.-Y.K. developed the affinity purification strategy and generated the PhyA and Ub-conjugate preparations for MS analysis. D.-Y.K. and V.A.-H. created and organized the K-R substitution transgenic lines. V.A.-H. analyzed PhyA degradation, characterized the 6K-R protein, and conducted the phenotypic analysis of the PHYA transgenic lines. R.J.S. created the PHYA-FLAG phyA-119 line. M.S. and L.M.S. generated the ubiquitylome datasets by LC-ESI-MS/MS. V.A.-H., D.-Y.K., and R.D.V proposed the research, analyzed the data, and wrote the paper with advice from M.S. and L.M.S.
References
- Abramoff M, Magalhaes P, Ram S. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- Al-Sady B, Ni W, Kircher S, Schafer E, Quail PH. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell. 2006;23:439–446. doi: 10.1016/j.molcel.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Bernhardt A, Lechner E, Hano P, Schade V, Dieterle M, Anders M, Dubin MJ, Benvenuto G, Bowler C, Genschik P, et al. CUL4 associates with DDB1 and DET1 and its downregulation affects diverse aspects of development in Arabidopsis thaliana. Plant J. 2006;47:591–603. doi: 10.1111/j.1365-313X.2006.02810.x. [DOI] [PubMed] [Google Scholar]
- Boller T, Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- Book AJ, Gladman NP, Lee SS, Scalf M, Smith LM, Vierstra RD. Affinity purification of the Arabidopsis 26S proteasome reveals a diverse array of plant proteolytic complexes. J Biol Chem. 2010;285:25554–25569. doi: 10.1074/jbc.M110.136622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgie ES, Vierstra RD. Phytochromes: an atomic perspective on photoactivation and signaling. Plant Cell. 2014;26:4568–4583. doi: 10.1105/tpc.114.131623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgie ES, Bussell AN, Walker JM, Dubiel K, Vierstra RD. Crystal structure of the photosensing module from a red/farred light-absorbing plant phytochrome. Proc Natl Acad Sci USA. 2014;111:10179–10184. doi: 10.1073/pnas.1403096111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, Essen LO, van der Horst GTJ, Batschauer A, Ahmad M. The cryptochromes: blue light photoreceptors in plants and animals. Annu Rev Plant Biol. 2011;62:335–364. doi: 10.1146/annurev-arplant-042110-103759. [DOI] [PubMed] [Google Scholar]
- Chen M, Chory J. Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol. 2011;21:664–671. doi: 10.1016/j.tcb.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Shen Y, Tang X, Yu L, Wang J, Guo L, Zhang Y, Zhang H, Feng S, Strickland E, et al. Arabidopsis CULLIN4 forms an E3 ubiquitin ligase with RBX1 and the CDD complex in mediating light control of development. Plant Cell. 2006;18:1991–2004. doi: 10.1105/tpc.106.043224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christians MJ, Gingerich DJ, Hua Z, Lauer TD, Vierstra RD. The light-response BTB1 and BTB2 proteins assemble nuclear ubiquitin ligases that modify phytochrome B and D signaling in Arabidopsis. Plant Physiol. 2012;160:118–134. doi: 10.1104/pp.112.199109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang HW, Zhang W, Gray WM. Arabidopsis ETA2, an apparent ortholog of the human cullin-interacting protein CAND1, is required for auxin responses mediated by the SCF(TIR1) ubiquitin ligase. Plant Cell. 2004;16:1883–1897. doi: 10.1105/tpc.021923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough RC, Vierstra RD. Phytochrome degradation. Plant Cell Environ. 1997;20:713–721. [Google Scholar]
- Clough RC, Jordan-Beebe ET, Lohman KN, Marita JM, Walker JM, Gatz C, Vierstra RD. Sequences within both the N- and C-terminal domains of phytochrome A are required for Pfr ubiquitination and degradation. Plant J. 1999;17:155–167. doi: 10.1046/j.1365-313x.1999.00360.x. [DOI] [PubMed] [Google Scholar]
- Colaert N, Helsens K, Martens L, Vandekerckhove J, Gevaert K. Improved visualization of protein consensus sequences by IceLogo. Nat Methods. 2009;6:786–787. doi: 10.1038/nmeth1109-786. [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- de Wit M, Galvao VC, Fankhauser C. Light-mediated hormonal regulation of plant growth and development. Annu Rev Plant Biol. 2016;67:513–537. doi: 10.1146/annurev-arplant-043015-112252. [DOI] [PubMed] [Google Scholar]
- Debrieux D, Fankhauser C. Light-induced degradation of phyA is promoted by transfer of the photoreceptor into the nucleus. Plant Mol Biol. 2010;73:687–695. doi: 10.1007/s11103-010-9649-9. [DOI] [PubMed] [Google Scholar]
- Demarsy E, Schepens I, Okajima K, Hersch M, Bergmann S, Christie J, Shimazaki K, Tokutomi S, Fankhauser C. Phytochrome kinase substrate 4 is phosphorylated by the phototropin 1 photoreceptor. EMBO J. 2012;31:3457–3467. doi: 10.1038/emboj.2012.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott PR. Molecular basis for specificity of the Met1-linked polyubiquitin signal. Biochem Soc Trans. 2016;44:1581–1602. doi: 10.1042/BST20160227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve-Bruna D, Perez-Perez JM, Ponce MR, Micol JL. incurvata13, a novel allele of AUXIN RESISTANT6, reveals a specific role for auxin and the SCF complex in Arabidopsis embryogenesis, vascular specification, and leaf flatness. Plant Physiol. 2013;161:1303–1320. doi: 10.1104/pp.112.207779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairchild CD, Schumaker MA, Quail PH. HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev. 2000;14:2377–2391. [PMC free article] [PubMed] [Google Scholar]
- Farmer LM, Book AJ, Lee KH, Lin YL, Fu H, Vierstra RD. The RAD23 family provides an essential connection between the 26S proteasome and ubiquitylated proteins in Arabidopsis. Plant Cell. 2010;22:124–142. doi: 10.1105/tpc.109.072660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Quail PH. Phytochrome functions in Arabidopsis development. J Exp Bot. 2010;61:11–24. doi: 10.1093/jxb/erp304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkerson J, Kelley DR, Tam R, Estelle M, Callis J. Lysine residues are not required for proteasome-mediated proteolysis of the auxin/indoleacetic acid protein IAA1. Plant Physiol. 2015;168:708–720. doi: 10.1104/pp.15.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingerich DJ, Gagne JM, Salter DW, Hellmann H, Estelle M, Ma L, Vierstra RD. Cullins 3a and 3b assemble with members of the broad complex/tramtrack/bric-a-brac (BTB) protein family to form essential ubiquitin-protein ligases (E3s) in Arabidopsis. J Biol Chem. 2005;280:18810–188821. doi: 10.1074/jbc.M413247200. [DOI] [PubMed] [Google Scholar]
- Gladman NP, Marshall RS, Lee KH, Vierstra RD. The proteasome stress regulon is controlled by a pair of NAC transcription factors in Arabidopsis. Plant Cell. 2016;28:1279–1296. doi: 10.1105/tpc.15.01022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamoto K, Aki T, Shigyo M, Sato S, Ishida T, Yano K, Yoneyama T, Yanagisawa S. Proteomic characterization of the greening process in rice seedlings using the MS spectral intensity-based label free method. J Proteome Res. 2012;11:331–347. doi: 10.1021/pr200852q. [DOI] [PubMed] [Google Scholar]
- Hjerpe R, Aillet F, Lopitz-Otsoa F, Lang V, England P, Rodriguez MS. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 2009;10:1250–1258. doi: 10.1038/embor.2009.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohm T, Demarsy E, Quan C, Allenbach Petrolati L, Preuten T, Vernoux T, Bergmann S, Fankhauser C. Plasma membrane H(+) -ATPase regulation is required for auxin gradient formation preceding phototropic growth. Mol Syst Biol. 2014;10:751. doi: 10.15252/msb.20145247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Su YS, Lagarias JC. A light-independent allele of phytochrome B faithfully recapitulates photomorphogenic transcriptional networks. Mol Plant. 2009;2:166–182. doi: 10.1093/mp/ssn086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z, Vierstra RD. The cullin-RING ubiquitin-protein ligases. Annu Rev Plant Biol. 2011;62:299–334. doi: 10.1146/annurev-arplant-042809-112256. [DOI] [PubMed] [Google Scholar]
- Jabben M, Shanklin J, Vierstra RD. Ubiquitin-phytochrome conjugates - pool dynamics during in-vivo phytochrome degradation. J Biol Chem. 1989;264:4998–5005. [PubMed] [Google Scholar]
- Kami C, Lorrain S, Hornitschek P, Fankhauser C. Light-regulated plant growth and development. Curr Top Dev Biol. 2010;91:29–66. doi: 10.1016/S0070-2153(10)91002-8. [DOI] [PubMed] [Google Scholar]
- Kang J, Yim S, Choi H, Kim A, Lee KP, Lopez-Molina L, Martinoia E, Lee Y. Abscisic acid transporters cooperate to control seed germination. Nat Commun. 2015;6:8113. doi: 10.1038/ncomms9113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Fujiwara S, Suh SS, Kim J, Kim Y, Han LQ, David K, Putterill J, Nam HG, Somers DE. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature. 2007;449:356–360. doi: 10.1038/nature06132. [DOI] [PubMed] [Google Scholar]
- Kim DY, Scalf M, Smith LM, Vierstra RD. Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. Plant Cell. 2013;25:1523–1540. doi: 10.1105/tpc.112.108613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- Kravtsova-Ivantsiv Y, Ciechanover A. Non-canonical ubiquitin-based signals for proteasomal degradation. J Cell Sci. 2012;125:539–548. doi: 10.1242/jcs.093567. [DOI] [PubMed] [Google Scholar]
- Leyser O. The power of auxin in plants. Plant Physiol. 2010;154: 501–505. doi: 10.1104/pp.110.161323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Vierstra RD. Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 2012;17:526–537. doi: 10.1016/j.tplants.2012.05.006. [DOI] [PubMed] [Google Scholar]
- Li F, Chung T, Vierstra RD. AUTOPHAGY-RELATED11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis. Plant Cell. 2014;26:788–807. doi: 10.1105/tpc.113.120014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Askinosie SK, Leuchtman DL, Morrow J, Willenburg KT, Coats DR. Phototropism: growing towards an understanding of plant movement. Plant Cell. 2014;26:38–55. doi: 10.1105/tpc.113.119727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Lin W, Gao X, Wu S, Cheng C, Avila J, Heese A, Devarenne TP, He P, Shan L. Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science. 2011;332:1439–1442. doi: 10.1126/science.1204903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Li J, Qu L, Hager J, Chen Z, Zhao H, Deng XW. Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell. 2001;13:2589–2607. doi: 10.1105/tpc.010229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maor R, Jones A, Nühse TS, Studholme DJ, Peck SC, Shirasu K. Multidimensional protein identification technology (MudPIT) analysis of ubiquitinated proteins in plants. Mol Cell Proteomics. 2007;6:601–610. doi: 10.1074/mcp.M600408-MCP200. [DOI] [PubMed] [Google Scholar]
- Marshall RS, Li F, Gemperline DC, Book AJ, Vierstra RD. Autophagic degradation of the 26S proteasome is mediated by the dual ATG8/ubiquitin receptor RPN10 in Arabidopsis. Mol Cell. 2015;58:1053–1066. doi: 10.1016/j.molcel.2015.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MJ, Barrett-Wilt GA, Hua Z, Vierstra RD. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc Natl Acad Sci USA. 2010;107:16512–16517. doi: 10.1073/pnas.1004181107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Zhao Y, Dai X, Zhang W, Gray WM, Huq E, Estelle M. A new CULLIN 1 mutant has altered responses to hormones and light in Arabidopsis. Plant Physiol. 2007;143:684–696. doi: 10.1104/pp.106.091439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- Ni W, Xu SL, Tepperman JM, Stanley DJ, Maltby DA, Gross JD, Burlingame AL, Wang ZY, Quail PH. A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science. 2014;344:1160–1164. doi: 10.1126/science.1250778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka Y, Ono Y, Toledo-Ortiz G, Kokajo K, Matsui M, Mochizuki H, Nagatani A. Arabidopsis phytochrome A is modularly structured to integrate the multiple features that are required for a highly sensitized phytochrome. Plant Cell. 2012;24:2949–2962. doi: 10.1105/tpc.111.094201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- Quint M, Ito H, Zhang W, Gray WM. Characterization of a novel temperature-sensitive allele of the CUL1/AXR6 subunit of SCF ubiquitin-ligases. Plant J. 2005;43:371–383. doi: 10.1111/j.1365-313X.2005.02449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattanapisit K, Cho MH, Bhoo SH. Lysine 206 in Arabidopsis phytochrome A is the major site for ubiquitin-dependent protein degradation. J Biochem. 2016;159:161–169. doi: 10.1093/jb/mvv085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J. Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell. 1993;5:147–157. doi: 10.1105/tpc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagatani A, Elich TD, Fagan M, Chory J. Phytochrome-A and phytochrome-B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 1994;104:1139–1149. doi: 10.1104/pp.104.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Bartel B. Plant peroxisomes: recent discoveries in functional complexity, organelle homeostasis, and morphological dynamics. Curr Opin Plant Biol. 2016;34:17–26. doi: 10.1016/j.pbi.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzini L, Favory JJ, Cloix C, Faggionato D, O’Hara A, Kaiserli E, Baumeister R, Schafer E, Nagy F, Jenkins GI, et al. Perception of UV-B by the Arabidopsis UVR8 protein. Science. 2011;332:103–106. doi: 10.1126/science.1200660. [DOI] [PubMed] [Google Scholar]
- Rohrbough JG, Breci L, Merchant N, Miller S, Haynes PA. Verification of single-peptide protein identifications by the application of complementary database search algorithms. J Biomol Tech. 2006;17:327–332. [PMC free article] [PubMed] [Google Scholar]
- Sadanandom A, Adam E, Orosa B, Viczian A, Klose C, Zhang C, Josse EM, Kozma-Bognar L, Nagy F. SUMOylation of phytochrome-B negatively regulates light-induced signaling in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2015;112:11108–11113. doi: 10.1073/pnas.1415260112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehin M, Bagchi R, Estelle M. SCFTIR1/AFB-based auxin perception: mechanism and role in plant growth and development. Plant Cell. 2015;27:9–19. doi: 10.1105/tpc.114.133744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saracco SA, Hansson M, Scalf M, Walker JM, Smith LM, Vierstra RD. Tandem affinity purification and mass spectrometric analysis of ubiquitylated proteins in Arabidopsis. Plant J. 2009;59:344–358. doi: 10.1111/j.1365-313X.2009.03862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer E, Nagy F. Photomorphogenesis in Plants and Bacteria. Dordrecht: Springer; 2006. [Google Scholar]
- Schindler U, Menkens AE, Beckmann H, Ecker JR, Cashmore AR. Heterodimerization between light-regulated and ubiquitously expressed Arabidopsis GBF bZIP proteins. EMBO J. 1992;11:1261–1273. doi: 10.1002/j.1460-2075.1992.tb05170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze WX, Usadel B. Quantitation in mass-spectrometry-based proteomics. Annu Rev Plant Biol. 2010;61:491–516. doi: 10.1146/annurev-arplant-042809-112132. [DOI] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al. A high-throughput Arabidopsis reverse genetics system. Plant Cell. 2002;14:2985–2994. doi: 10.1105/tpc.004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanklin J, Jabben M, Vierstra RD. Red light-induced formation of ubiquitin-phytochrome conjugates - identification of possible intermediates of phytochrome degradation. Proc Natl Acad Sci USA. 1987;84:359–363. doi: 10.1073/pnas.84.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanklin J, Jabben M, Vierstra RD. Partial purification and peptide mapping of ubiquitin-phytochrome conjugates from oat. Biochemistry. 1989;28:6028–6034. [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock RA, Quail PH. Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev. 1989;3:1745–1757. doi: 10.1101/gad.3.11.1745. [DOI] [PubMed] [Google Scholar]
- Shen WH, Parmentier Y, Hellmann H, Lechner E, Dong A, Masson J, Granier F, Lepiniec L, Estelle M, Genschik P. Null mutation of AtCUL1 causes arrest in early embryogenesis in Arabidopsis. Mol Biol Cell. 2002;13:1916–1928. doi: 10.1091/mbc.E02-02-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z, Li P, Ni RJ, Ritchie M, Yang CP, Liu GF, Ma W, Liu GJ, Ma L, Li SJ, et al. Label-free quantitative proteomics analysis of etiolated maize seedling leaves during greening. Mol Cell Proteomics. 2009;8:2443–2460. doi: 10.1074/mcp.M900187-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Quail PH. Phytochrome-mediated light regulation of PHYA- and PHYB-GUS transgenes in Arabidopsis thaliana seedlings. Plant Physiol. 1995;107:523–534. doi: 10.1104/pp.107.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz AK, Ren H, Park MY, Grandt KN, Lee SH, Murphy AS, Sussman MR, Overvoorde PJ, Gray WM. SAUR inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. Plant Cell. 2014;26:2129–2142. doi: 10.1105/tpc.114.126037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttangkakul A, Li F, Chung T, Vierstra RD. The ATG1/ ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell. 2011;23:3761–3779. doi: 10.1105/tpc.111.090993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Hayashi K, Kinoshita T. Auxin activates the plasma membrane H+-ATPase by phosphorylation during hypocotyl elongation in Arabidopsis. Plant Physiol. 2012;159:632–641. doi: 10.1104/pp.112.196428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperman JM, Zhu T, Chang HS, Wang X, Quail PH. Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc Natl Acad Sci USA. 2001;98:9437–9442. doi: 10.1073/pnas.161300998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperman JM, Hwang YS, Quail PH. phyA dominates in transduction of red-light signals to rapidly responding genes at the initiation of Arabidopsis seedling de-etiolation. Plant J. 2006;48:728–742. doi: 10.1111/j.1365-313X.2006.02914.x. [DOI] [PubMed] [Google Scholar]
- Thomann A, Lechner E, Hansen M, Dumbliauskas E, Parmentier Y, Kieber J, Scheres B, Genschik P. Arabidopsis CULLIN3 genes regulate primary root growth and patterning by ethylene-dependent and -independent mechanisms. PLoS Genet. 2009;5:e1000328. doi: 10.1371/journal.pgen.1000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 2005;138:2097–2110. doi: 10.1104/pp.105.060673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol. 2009;10:385–397. doi: 10.1038/nrm2688. [DOI] [PubMed] [Google Scholar]
- Vijay-Kumar S, Bugg CE, Wilkinson KD, Vierstra RD, Hatfield PM, Cook WJ. Comparison of the three-dimensional structures of human, yeast, and oat ubiquitin. J Biol Chem. 1987;262: 6396–6399. [PubMed] [Google Scholar]
- Wang BC, Pan YH, Meng DZ, Zhu YX. Identification and quantitative analysis of significantly accumulated proteins during the Arabidopsis seedling de-etiolation process. J Integr Plant Biol. 2006;48:104–113. [Google Scholar]
- Wang ZY, Bai MY, Oh E, Zhu JY. Brassinosteroid signaling network and regulation of photomorphogenesis. Annu Rev Genet. 2012;46:701–724. doi: 10.1146/annurev-genet-102209-163450. [DOI] [PubMed] [Google Scholar]
- Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Wu SH. Gene expression regulation in photomorphogenesis from the perspective of the central dogma. Annu Rev Plant Biol. 2014;65:311–333. doi: 10.1146/annurev-arplant-050213-040337. [DOI] [PubMed] [Google Scholar]
- Wu GS, Cameron JN, Ljung K, Spalding EP. A role for ABCB19-mediated polar auxin transport in seedling photomorphogenesis mediated by cryptochrome 1 and phytochrome B. Plant J. 2010;62:179–191. doi: 10.1111/j.1365-313X.2010.04137.x. [DOI] [PubMed] [Google Scholar]
- Yeh KC, Lagarias JC. Eukaryotic phytochromes: light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc Natl Acad Sci USA. 1998;95:13976–13981. doi: 10.1073/pnas.95.23.13976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadnikova P, Petrasek J, Marhavy P, Raz V, Vandenbussche F, Ding ZJ, Schwarzerova K, Morita MT, Tasaka M, Hejatko J, et al. Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development. 2010;137:607–617. doi: 10.1242/dev.041277. [DOI] [PubMed] [Google Scholar]
- Zhang H, He H, Wang X, Wang X, Yang X, Li L, Deng XW. Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 2011;65:346–358. doi: 10.1111/j.1365-313X.2010.04426.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Mayba O, Pfeiffer A, Shi H, Tepperman JM, Speed TP, Quail PH. A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 2013;9:e1003244. doi: 10.1371/journal.pgen.1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.