Abstract
DNA methylation has been linked to gene silencing in cancer. Primary effusion lymphoma (PEL) and myeloma are lymphoid malignancies that arise from terminally differentiated B cells. Interestingly, PEL do not express immunoglobulins or most B lineage-specific genes. The B cell-specific B29 (Igβ/CD79b) gene is silenced in PEL and some myelomas but is expressed in other normal and malignant B cells. B29 expression was reactivated in PEL by demethylating and histone deacetylase inhibiting treatments. Bisulfite sequencing revealed two types of DNA methylation in silenced B29 promoters: at conventional CpG and at CC(A/T)GG B29 promoter sites. The pattern of methylated CpG (mCpG) and CmC(A/T)GG B29 promoter methylation observed was similar to that recently reported for epigenetic silencing of an integrated retrovirus. Methylation of CmC(A/T)GG sites in the B29 promoter significantly repressed in vivo transcriptional activity. Also, methylation of a central conserved CmCTGG B29 promoter site blocked the binding of early B cell factor. This methylated motif formed DNA–protein complexes with nuclear extracts from all cell types examined. Therefore, CmC(A/T)GG methylation may represent an important type of epigenetic marker on mammalian DNA that impacts transcription by altering DNA–protein complex formation.
Primary effusion lymphoma (PEL) is an infrequent neoplasia of severe immunodeficiency and is usually observed in those with longstanding AIDS (1, 2). PEL exhibit features both in common with and distinct from other B cell neoplasms. Like myeloma, PEL arise from postgerminal center, developmentally mature B lymphocytes (1–3). Unlike myeloma, PEL grow in body cavities as liquid effusions, are almost always infected with human herpesvirus-8 (HHV-8) and lack expression of B-lineage-specific genes (reviewed in ref. 2). The loss of B lineage gene expression in PEL could be caused by DNA methylation and epigenetic silencing. In accord with this idea, γ-herpesvirus genomes such as HHV-8, herpesvirus saimiri, and Epstein–Barr virus, are commonly methylated (4, 5). The same mechanism(s) responsible for viral DNA methylation may also be involved in methylation and silencing of B lineage-specific genes in PEL.
DNA methylation in mammalian cells largely occurs on cytosines in symmetric CpG dinucleotides and is associated with repressed gene transcription (6–10). Methyl-binding domain proteins engage methylated CpG (mCpG) and recruit histone deacetylase (HDAC) and transcriptional repressors to form stable repression complexes that induce local chromatin remodeling and gene silencing (11–16). Early mammalian embryos and germ cells, like plants and fungi, also methylate non-CpG cytosines (17–21). A recent report indicates that peripheral blood leukocytes may methylate the internal cytosine of symmetric CCTGG but not CCAGG motifs (22). However, little is known about the gene specificity, frequency, and functional significance of this latter type of symmetric non-CpG methylation.
The B29 (Igβ/CD79b) component of the B cell surface receptor is encoded by the B29 gene and is absent in all PEL lines and some myelomas (1, 23). The B cell-specific B29 promoter is well characterized and provides an ideal model to analyze DNA methylation in B lineage gene silencing in PEL and myeloma. Early B cell factor (EBF), in concert with Octamer, Ets, Sp1, and Ikaros transcription factors, regulates B29 promoter activity in early B cell development, whereas non-EBF factors control B29 gene expression at later stages of B cell differentiation (24). The human B29 promoter contains 20 CpG dinucleotides and 6 CCAGG or CCTGG motifs within a 450-bp span (25). CpG dinucleotides are present in single Sp1 and EBF consensus-binding sites, whereas the CCAGG and CCTGG motifs are concentrated in a central promoter control region containing essential EBF sites.
Here we report that the B29 promoter in PEL and nonexpressing myeloma cells is methylated at CpG and CC(A/T)GG sites. Because the methylation pattern observed at these B29 promoter sites is similar to that reported in epigenetic retroviral silencing, we propose that B cell gene extinction occurs through a similar mechanism. We also find that CC(A/T)GG methylation repressed B29 promoter activity and replaced transcription factors with new protein complexes.
Materials and Methods
DNA Demethylation and HDAC Inhibition.
BCBL-1 cultures were treated with 5-aza-2′-deoxycytidine (400 nM, 64 h, Sigma) and trichostatin A (TsA, 50 nM, 24 h, Sigma), alone or in combination, or with sodium butyrate (0.3 mM, 64 h, Aldrich). BCBL-1 PEL cells were stably infected with MSCV-GFP-IRES-PURO, a green fluorescent protein-expressing retrovirus, to monitor the effects of demethylating and HDAC-inhibiting treatments. Growth, viability, and general gene expression were assessed by cell counts, trypan blue exclusion, and green fluorescent protein intensity. Total RNA isolation and reverse transcription with random primers was followed by 40 cycles of PCR amplification with B29-specific primers (5′-GGAGCCTCGGACGTTGTCA-3′ and 5′-CGACCTGGCTCTCACTCCT-3′). Southern blot analysis was performed by using a random-primed αP32-dCTP-labeled B29 cDNA fragment (PrimeIt II, Stratagene).
Genomic Bisulfite Sequencing.
Two micrograms of isolated genomic DNA was restriction digested with ApaI and subjected to sodium bisulfite treatment as described (26, 27). Two microliters of sodium bisulfite-treated DNA was PCR amplified for 47 cycles by using multiple sense-strand primer pair combinations followed by 30 cycles of nested PCR amplification. Primer pairs that amplified the largest fragment possible from sodium bisulfite treated genomic DNA were used (published as supplemental data on the PNAS web site, www.pnas.org). PCR products were subcloned into the pCR2.1-TOPO vector (Invitrogen) and cycle sequenced.
Transient Transfection Assay.
Cell line sources are listed in the supplemental data. Transient cotransfection of BL-41 B cells (1 × 107 cells/sample) with 10 μg of Renilla reporter constructs pRL-null, pRLB29, or pRLSV40 was performed with 5 μg of firefly reporter construct pGL3 control for transfection efficiency normalization by the DEAE-Dextran method (28) and analyzed with the Dual Luciferase Assay System (Promega). pRLB29 contains the genomic B29 promoter fragment from −193 to +47. Reporter constructs were passed through dcm+ DH5α or dcm− scs110 bacteria and bisulfite sequenced to confirm the methylation status of CmC(A/T)GG sites (data not shown).
Protein Preparation and Electrophoretic Mobility-Shift Assay (EMSA).
Human EBF protein was in vitro transcribed and translated (IVT) from pCDNA3hEBF, kindly provided by Mikael Sigvardsson (Lund University, Lund, Sweden) by using the T7 TNT Quick Coupled Transcription and Translation kit (Promega). Preparation of crude nuclear extracts was as described (29). EMSA was performed by using either 20 μg of nuclear extract or 2 μl of IVT hEBF, as described (28). EMSA probes and cold competitors were complementary double-stranded (ds)DNA oligonucleotides: 5′-CTGGTGCCTCCCCTGGGTCCCAATT-3′ (C−91); 5′-CTGGTGCCTCCCmCTGGGTCCCAATT-3′ (mC−91); 5′-CATGAATGGGGGTGGCAGAGA-3′ (Ikaros); 5′-GATCGATCGGGGCGGGGCGATC-3′ (Sp1); 5′-GATCTCGAGCAGGAAGTTCGA-3′ (Ets-1); and 5′-GAGAGAGACTCAAGGGAATTGTGGCCAGCC-3′ (EBF). BL-41 and Ramos genomic cell line DNA competitors were prepared by using the Wizard genomic DNA Prep kit (Promega). Saccharomyces cerevisiae, adult Drosophila melanogaster, and dcm+ Escherichia coli competitor genomic DNAs were kindly provided by Michael Grunstein, Utpal Banerjee, and Jeffrey Miller (University of California, Los Angeles), respectively.
Results
B29 Gene Expression in PEL Is Activated by Demethylation and HDAC Inhibition.
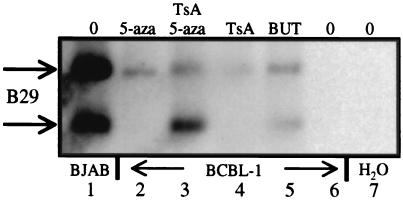
Twenty B cell lines from distinct developmental stages and one T cell line were examined for B29 gene expression. All four PEL lines, the myeloma line RPMI-8226, and Jurkat T cells lacked B29 mRNA by Northern blot and reverse transcription–PCR analyses (see supplemental data). Transient transfections of B29 promoter-luciferase reporter constructs were performed to determine the transcription competency of the B29 negative lines. B29-expressing cell transfectants resulted in a 4- to 20-fold luciferase activity, whereas PEL and RPMI-8226 cells exhibited lower but significant (P < 0.05) 3- to 5-fold activations over control background transfections (data not shown). These results are within experimental variability observed for B29 reporter constructs in other B and plasma cell lines (24). The data argue that factors needed for B29 expression are present in PEL and RPMI-8226 cells and that silencing of the endogenous locus may be because of DNA methylation. Therefore, B29-negative BCBL-1 PEL cells were treated with demethylating and HDAC-inhibiting agents to test whether these treatments could activate B29 gene expression. Treatments with 5-aza-2′-deoxycytidine, TsA, and sodium butyrate, alone or in combination, activated B29 gene expression (Fig. 1; refs. 5, 30). The extent of activation was similar to the levels of activation reported for other silenced genes with these drug treatments (31). These results show that in BCBL-1 PEL cells, the direct or indirect effects of DNA methylation and HDAC activity repress B29 gene transcription.
Figure 1.
DNA demethylation and HDAC inhibition activates endogenous B29 gene expression detected by reverse transcription–PCR. Products (750 and 438 bp) correspond to the normal and alternately spliced forms of B29 mRNA (48). Lanes: (1) BJAB untreated; (2) BCBL-1 + 5-aza-2′-deoxycytidine (5-aza); (3) BCBL-1 + 5-aza/TsA; (4) BCBL-1 + TsA; (5) BCBL-1 + sodium butyrate; (6) BCBL-1 untreated; and (7) water blank.
CpG and CC(A/T)GG Methylation of the B29 Minimal Promoter.
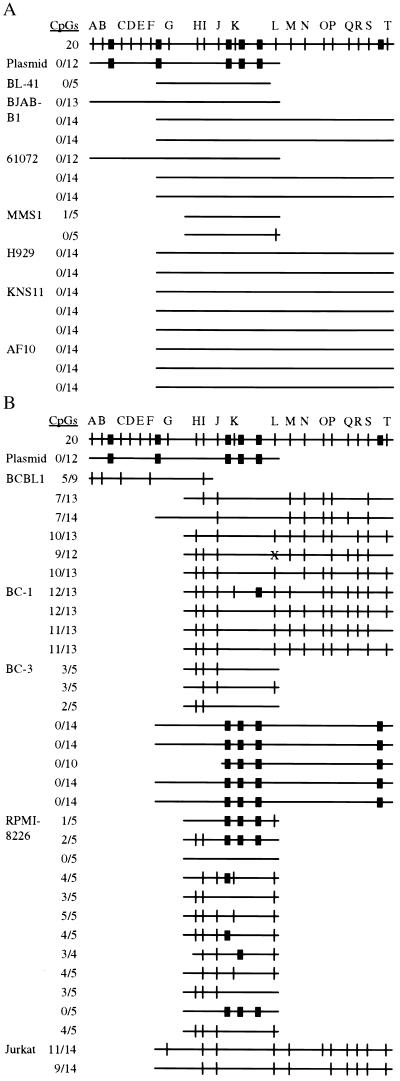
Multiple B29-expressing and nonexpressing lines were surveyed by genomic bisulfite sequencing to determine whether silencing is because of DNA methylation of the B29 promoter (Fig. 2 A and B and supplemental data; ref. 32). Results from 17 independent DNA clones derived from seven B29-expressing cell lines indicate that B29 expression is correlated with promoter region hypomethylation (Fig. 2A). In contrast, 13 clones from three different B29 silenced PEL lines exhibited dense CpG methylation that ranged from 40 to 92% of sites (Fig. 2B). Interestingly, several clones from B29 nonexpressing BC-3 PEL and RPMI-8226 myeloma lines had minimal or no CpG methylation. Instead, these clones showed specific methylation of three or four non-CpG cytosines at positions −119, −91, −60, and +58 relative to the major transcriptional start site, all localizing to the internal cytosine within the consensus sequence CC(A/T)GG (Fig. 2B and supplemental data). One clone from RPMI-8226 myeloma cells was not methylated; however, the only PCR primer set that worked for this bisulfite-converted DNA amplified a short span of the B29 promoter in these cells. We cannot exclude the influence of DNA methylation outside of the examined region in this clone. Strikingly, CC(A/T)GG forms the target sequence for dcm methyltransferase activity in bacteria (33). These bisulfite sequencing results indicate that CmC(A/T)GG also occurs in mammalian cells, as recently reported in studies of genomic Southern blotting of the myogenic Myf-3 gene (22). We did not detect B29 promoter methylation at additional symmetric and nonsymmetric non-CpG sites, including 22 C(A/T)G and 5 CpCpG sites. mCpG and CmC(A/T)GG are strongly correlated with extinguished gene activity, suggesting that both forms of methylation may be involved in B29 gene silencing.
Figure 2.
Genomic bisulfite sequencing of the endogenous human B29 promoter. Compiled results of bisulfite sequencing from B29-expressing (A) and nonexpressing (B) cell lines. The CpG positions analyzed are labeled A-T in alphabetic order (see supplemental data). The cell lines (Left) list the B29 promoter clones examined. Data for BC-3 cells are from two independent bisulfite conversions on distinct days. The span sequenced for each DNA clone is indicated by the extent of the horizontal line. A vertical slash indicates a mCpG at that position; a darkened box indicates a methylated CmC(A/T)GG at that position. X = C to G polymorphism. The plasmid is the B29 minimal promoter that was bisulfite sequenced after passage through dcm+ DH5α bacteria and demonstrates positions of CmC(A/T)GG sites.
CmC(A/T)GG Represses B29 Promoter Activity in B Cells.
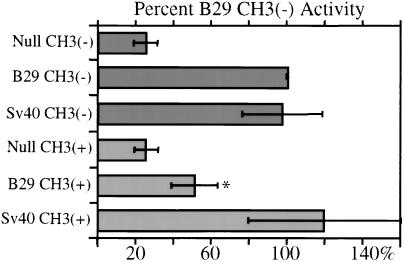
To determine whether CmC(A/T)GG sites repressed B29 promoter activity in vivo, we tested CC(A/T)GG site-specific methylated and unmethylated reporter constructs by transient transfections of BL-41 B cells. B29 reporter constructs were propagated in dcm+ DH5α or dcm− scs110 E. coli and were bisulfite sequence confirmed. The B29 promoter in DH5α-propagated constructs was methylated on three distinct CmC(A/T)GG sites, whereas scs110-propagated constructs were unmethylated (data not shown). The dcm-methylated B29 promoter constructs reproducibly showed statistically significant reductions of 50% in reporter gene activity compared with dcm-unmethylated constructs in B29-expressing BL-41 B-cells (P < 0.05; Fig. 3). Importantly, control simian virus 40 promoter driven constructs that do not contain CC(A/T)GG sites showed no statistically significant difference in activity between DH5α- and scs110-propagated constructs (P > 0.50).
Figure 3.
Effect of CmC(A/T)GG methylation on B29 promoter activity in B29 expressing BL-41 B cells. Renilla luciferase B29 promoter, SV40 promoter and promoter-less (null) reporter constructs were propagated in dcm(+) DH5α [CH3(+)] or dcm(−) scs110 [CH3(−)] E. coli. Renilla luciferase activities are SV40 firefly luciferase normalized (pGL3 control) with the ±SD of at least three transfections by using three preparations of DNA. The data are presented as percent activity relative to the scs110-propagated B29 Renilla luciferase construct, [B29 CH3(−)]. The activity of SV40 constructs, which contain no CC(A/T)GG sites, is statistically equivalent whether passed through dcm+ or dcm− bacteria (Student's two-sided t test, P > 0.50). *, statistical significance, P < 0.05.
CmCTGG Methylation Yields a Unique DNA–Protein Complex in Multiple Cell Types.
Because B29 promoter activity was repressed by CmC(A/T)GG methylation, we analyzed the effect of this methylation on transcription factor binding in EMSA. The human and mouse B29 core promoters contain three and two CC(A/T)GG motifs, respectively. One CCTGG site is highly conserved between species in a central critical region of the promoters, with the methylated internal cytosine located at position −91 (C−91) relative to the major transcriptional start site in the human sequence (25, 28). Prior DNase I footprint and EMSA have shown that purified Sp1, Pu.1, Ets-1, Ikaros, and EBF transcription factors bind the mouse B29 promoter in this region (24, 28). The human B29 promoter contains these same consensus sequences, although binding of specific transcription factors has only been inferred by use of these consensus motifs as cold competitors in EMSA (34).
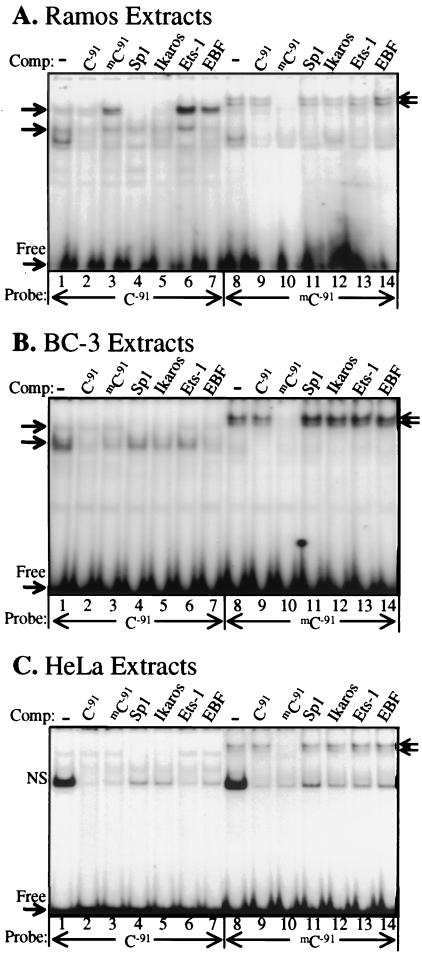
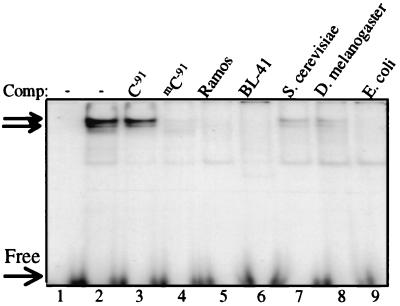
Two dsDNA oligonucleotides were synthesized with and without symmetrical methylation of the target C−91 residue. Nuclear extracts were prepared from multiple cell lines that either express (Ramos, Raji, 729, and BL-41) or do not express (BC-1, BC-3, KS-1, BCBL-1, Jurkat, and HeLa) the human B29 gene. Several specific protein complexes were observed in EMSA by using the unmethylated human B29 dsDNA oligonucleotide as a probe with Ramos nuclear extracts (Fig. 4A). Similar complexes were also seen with nuclear extracts from all of the B29-expressing cell lines and from the nonexpressing PEL cell lines (BC-1, BC-3, BCBL-1, and KS-1; Fig. 4 and data not shown). This observation confirms that PEL contain transcription factor complexes similar to those seen in B29-expressing cell lines. Sp1, Ikaros, and EBF cold competitor oligonucleotides inhibited several DNA–protein complexes with the unmethylated target probe, consistent with results previously obtained with the corresponding mouse B29 promoter region (24, 28). Importantly, cold competitor dsDNA oligonucleotides with symmetric methylation of C−91 (mC−91) residues failed to inhibit formation of these protein complexes. Moreover, when the mC−91 probe was used as the target, the specific DNA–protein complexes seen with both Ramos and BC-3 nuclear extracts were not observed (Fig. 4 A and B). Instead, mC−91 showed new specific bands of reduced mobility, indicating alterations in DNA–protein complex formation. Neither unmethylated cold competitor probe (C−91) nor transcription factor-specific cold competitors inhibited this banding pattern. Fig. 4C shows that this reduced mobility banding pattern was not unique to B cell lines. These complexes were also seen with non-B lineage HeLa and Jurkat nuclear extracts incubated with the mC−91 probe (Fig. 4C and data not shown). In fact, all 10 cell line extracts tested displayed an identical banding pattern with the mC−91 probe (data not shown). This banding pattern was not inhibited by a symmetrically CpG-methylated probe, indicating that the mC−91 complexes were distinct from CpG methylation-specific complexes (see supplemental data). Also, 1 μg of genomic DNA from organisms that do not or perhaps minimally methylate their DNA, such as S. cerevisiae and adult D. melanogaster, could not compete for this DNA–protein complex, whereas organisms that methylate their DNA, such as humans and E. coli, completely inhibited complex formation (Fig. 5; ref. 35). B29-expressing Ramos and BL-41 genomic DNAs also abrogated complex formation, predicting that genes other than B29 contain CmC(A/T)GG methylated sites in these lines. These observations indicate that CmCTGG site-specific methylation in the B29 promoter replaced normal factor binding with distinct methylation-dependent DNA–protein complexes of ubiquitous protein factors.
Figure 4.
Methylation of C−91 in CmCTGG dramatically alters transcription factor binding and results in a unique DNA–protein complex. Symmetrically mC−91-methylated or unmethylated dsDNA oligonucleotide probes were analyzed by EMSA with 20 μg of nuclear extracts from (A) Ramos, a B29-expressing B cell line, (B) BC-3, a B29 nonexpressing PEL cell line, and (C) HeLa, a B29 nonexpressing carcinoma cell line. Oligonucleotide cold competitor (500-fold) was added as indicated. EMSA banding patterns were similar in all the B cell extracts tested (data not shown). Unlabeled arrows denote specific complexes. NS, nonspecific complex.
Figure 5.
Methylation negative genomic DNA does not compete for the mC−91 DNA–protein complex. Symmetrically mC−91-methylated dsDNA oligonucleotide probe was analyzed by EMSA with 20 μg of Ramos B cell nuclear extracts. One microgram of genomic DNA cold competitor was added, as indicated. Unlabeled arrows denote specific complexes.
CmCTGG Methylation Blocks EBF Transcription Factor Binding.
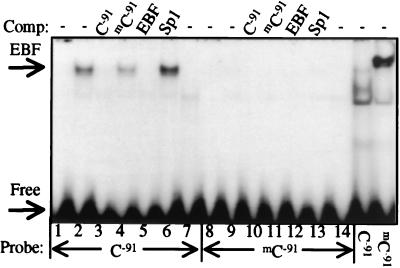
The conserved mC−91 region contains a consensus EBF-binding site that binds purified EBF in EMSA studies of the mouse B29 promoter (24). This site has not been investigated directly for EBF binding in the human promoter sequence, except as a cold competitor in EMSA (34). Therefore, we performed EMSA to determine whether an IVT EBF binds the C−91 site in the human sequence and whether mC−91 alters this binding. Fig. 6 demonstrates that IVT EBF bound the unmethylated C−91 probe and binding was completely inhibited by competition with itself but not by competition with cold mC−91 probe or a cold Sp1 consensus oligonucleotide. In contrast, when the mC−91 sequence was used as a probe, IVT EBF binding was completely abolished (Fig. 6). These results indicate that CmCTGG methylation is sufficient to block known transcription factor binding.
Figure 6.
Methylation of C−91 in CmCTGG blocks EBF binding. Symmetrically mC−91-methylated or unmethylated dsDNA oligonucleotide probes were analyzed by EMSA with human IVT EBF (lanes 2–6 and 9–13), control IVT luciferase (lanes 7 and 14), or 20 μg Ramos nuclear extracts (lanes 15 and 16). Oligonucleotide cold competitor (250-fold) was added, as indicated.
Discussion
PEL and RPMI-8226 myeloma cells must have expressed B29 during earlier times in normal development. Multiple results strongly argue that B29 silencing in PEL and RPMI-8226 cells is directly attributable to promoter region methylation. First, B29 expression is reactivated by DNA demethylation and HDAC inhibition. Second, the B29 promoter is active in transfections of both B29 producing and silenced B cells. Third, symmetric methylation at both mCpG and CmC(A/T)GG is associated with B29 gene silencing in PEL and RPMI-8226 lines. The pattern of methylation at these two motifs in the B29 promoter is similar to that reported for epigenetic silencing of an integrated retrovirus, where the density of CmC(A/T)GG methylation is negatively correlated with the density of mCpG methylation (36).
This is, to our knowledge, the first clear example of CmC(A/T)GG methylation in endogenous gene silencing detected by direct DNA sequencing in mammalian cells. Previously, symmetric CmC(A/T)GG methylation was detected and implicated in retroviral gene silencing after viral integration into mammalian DNA (36). Also, methylation was detected in the Myf-3 gene by genomic Southern analysis at CmCTGG sequences and not at CmCAGG sites (22). Although our study used tumor cell lines, CmCTGG methylation of the Myf-3 gene was found in peripheral blood lymphocytes, indicating that this type of symmetric methylation occurs in normal and malignant lymphoid cells. However, this latter study did not address the functional significance of CmCTGG methylation, and the exclusion of CmCAGG methylation is distinct from results reported here and by Lorincz et al. (36). Our study provides evidence of methylation on CmC(A/T)GG in endogenous genes in mammalian cells, indicating that this pattern is not merely a cellular response to invading nucleic acids (18).
CmC(A/T)GG methylation does not depend on the presence of γ-herpesviruses, because B29-silenced human herpesvirus-8-negative RPMI-8226 cells exhibit this type of methylation. Also, multiple B29-expressing Epstein–Barr virus-positive Burkitt lymphomas do not have CmC(A/T)GG. This finding predicts that an enzyme with bacteria-like dcm methyltransferase activity is present in mammalian cells. A potential candidate would be Dnmt2, a mammalian structural homologue of the Schizosaccharomyces pombe pmt1, which can specifically bind CmC(A/T)GG sequences in vitro (37). However, methyltransferase activity for Dnmt2 has not been demonstrated (38). We can exclude contamination with dcm+ bacteria as the source of these results by noting the co-occurrence of both mCpG and CmC(A/T)GG methylation in the same DNA clones from RPMI-8226 cells (39). The detection of abundant mCpG methylation in BCBL-1 DNA clones with no unconverted CC(A/T)GG sites confirms complete bisulfite treatment conditions despite nearby methylated cytosines (40, 41). Also, the 5′ cytosine in CC(A/T)GG sites was always converted with bisulfite treatment, indicating that structural changes caused by the methylation of an adjacent internal cytosine did not impede the reaction (40). Furthermore, we show that different types of symmetric methylation may be present singly or concurrently in distinct DNA sequences. Overall, these observations suggest that the enzymatic activities responsible for mCpG and CmC(A/T)GG are separable, perhaps independently regulated and active within a single cell.
Among the B29-silenced cell lines, only BC-3 and RPMI-8226 had abundant CmC(A/T)GG methylation in distinct sequences. Interestingly, for both cell types, there is an apparent negative correlation between mCpG and CmC(A/T)GG methylation in that sequences with abundant methylation of one type tend to lack methylation of the other type and vice versa (Fig. 2B). This pattern of methylation has been reported previously for silencing of a retroviral gene in mouse erythroleukemia cells (36). To determine whether the patterns are likely to be equivalent, we applied two statistical tools to the data derived for methylation of 12 RPMI-8226 sequences in Fig. 2B and 24 sequences from clone 18 in figure 4 of Lorincz et al. The standard Pearson correlation coefficient (r) between mCpG and CmC(A/T)GG methylation density was r = −0.82 for the RPMI-8226 sequences (excluding the one sequence that was not methylated at all) and r = −0.63 for data derived from clone 18. Each correlation coefficient is significantly different from zero (1-sided P < 0.005), suggesting that each data set represents a negative correlation between mCpG and CmC(A/T)GG methylation density. Furthermore, Kendall's rank correlation coefficient (τ) corrected for ties allows a direct comparison of the two data sets (42). The estimated τ (and asymptotic standard error) for RPMI-8226 and clone 18 sequences are −0.532 (0.200) and −0.374 (0.176), respectively. Each τ is significantly different from zero (1-sided P = 0.02 and 0.01, respectively, by exact permutation test), indicating it is likely they represent the same degree of negative correlation because this hypothesis cannot be rejected. If the unmethylated sequence is included in the RPMI-8226 data, the estimated coefficients are somewhat reduced in magnitude (r = −0.55 and τ = −0.352) but are even closer to the corresponding estimates for clone 18 sequences. These results suggest that the differential methylation mechanism operating on an integrated retrovirus may be similar to that operating on the endogenous B29 gene in RPMI-8226 cells.
Most strikingly, our results show that the EBF transcription factor is displaced and that a unique methylation-dependent DNA–protein complex forms instead on a symmetrically methylated CmCTGG site. mCpG competitors did not inhibit complex formation, indicating that the protein(s) bound to this CmCTGG site are distinct from those that bind mCpG. Our data further demonstrate that EBF, along with other transcription factors (on the basis of EMSA competition assays with nuclear extracts) was displaced by a symmetrically methylated CmCTGG site, similar to descriptions of transcription factor displacement by mCpG sites (reviewed in ref. 9). Although some transcription factors, such as Sp1, MTF-1, and Krox-20, are still able to bind mCpG methylated consensus sequences, other transcription factors, such as ATF-like and RBF1, are blocked by mCpG methylation in their consensus sequences (43, 44). Apparently, some factors are insensitive to mCpG, although data indicate that Sp1 is blocked from DNA binding if methylation occurs on two consecutive cytosine residues in the consensus-binding region (45). Interestingly, inhibition of ATF-like and RBF1 binding by mCpG methylation strongly correlates with repression of Rb1 promoter activity in vivo (44). Similarly, we demonstrate that B29 promoter activity in vivo is ≈50% inhibited by methylation of three CmC(A/T)GG sites. In both cases, this repression is likely because of recruitment of HDAC and chromatin remodeling complexes to the methylated, in vivo chromatinized plasmids, as has been shown directly for Ikaros-mediated repression of reporter gene transcription (46). Our B29 expression results with methylated CmC(A/T)GG sites are also comparable to the reductions produced by site-directed mutations that block EBF binding to the mouse B29 promoter. These site-directed mutations caused a 30–40% reduction in B29 promoter activity in mature B cells (24). Additionally, site-directed mutations within EBF consensus sites in both the CD19 and λ5 promoters resulted in a similar 40–65% reduction in promoter activity (34, 47). Furthermore, previous data have shown that multiple transcription factors are capable of binding the critical EBF-consensus region of the mouse B29 promoter, indicating that mutations or DNA methylation in this region may effect more than just EBF factor binding (24, 28). Taken together, promoter inhibition and blockage of EBF binding by CmC(A/T)GG methylation in our studies are consistent with data generated from EBF-binding site mutation studies. Our study is, to our knowledge, the first to demonstrate a link between site-specific non-CpG methylation, methylation-specific DNA–protein complex formation, and cellular gene repression. These findings indicate that CmC(A/T)GG methylation contributes to promoter activity control in mammalian cells and may have a companion role with CpG methylation in epigenetic gene regulation.
Supplementary Material
Acknowledgments
We thank Stephen Smale, Arnie Berk, Michael Grunstein, and York Marahrens for valuable advice. Special thanks to Elliot Landaw and Stephen Teitell for statistical analyses, to Lisa Patrone for human nuclear extracts (University of California, Los Angeles), and to Larry Souza for continued support and encouragement. This work was supported by grants from the National Institutes of Health (T32CA09056, CA74929, and CA85841), the Amgen/U.C. BioSTAR Project (S98–35), the Jonsson Comprehensive Cancer Foundation, and the Lymphoma Research Foundation of America.
Abbreviations
- PEL
primary effusion lymphoma
- HDAC
histone deacetylase
- EBF
early B cell factor
- TsA
trichostatin A
- IVT
in vitro translated
- dsDNA
double-stranded DNA
- mCpG
methylated CpG
- EMSA
electrophoretic mobility-shift assay
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 10034.
References
- 1.Drexler H G, Uphoff C C, Gaidano G, Carbone A. Leukemia. 1998;12:1507–1517. doi: 10.1038/sj.leu.2401160. [DOI] [PubMed] [Google Scholar]
- 2.Gaidano G, Carbone A. Adv Cancer Res. 2001;80:115–146. doi: 10.1016/s0065-230x(01)80014-2. [DOI] [PubMed] [Google Scholar]
- 3.Matolcsy A, Nador R G, Cesarman E, Knowles D M. Am J Pathol. 1998;153:1609–1614. doi: 10.1016/S0002-9440(10)65749-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karlin S, Mocarski E S, Schachtel G A. J Virol. 1994;68:1886–1902. doi: 10.1128/jvi.68.3.1886-1902.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J, Ueda K, Sakakibara S, Okuno T, Parravicini C, Corbellino M, Yamanishi K. Proc Natl Acad Sci USA. 2001;98:4119–4124. doi: 10.1073/pnas.051004198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Razin A, Cedar H. Microbiol Rev. 1991;55:451–458. doi: 10.1128/mr.55.3.451-458.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tate P H, Bird A P. Curr Opin Genet Dev. 1993;3:226–231. doi: 10.1016/0959-437x(93)90027-m. [DOI] [PubMed] [Google Scholar]
- 8.Kass S U, Pruss D, Wolffe A P. Trends Genet. 1997;13:444–449. doi: 10.1016/s0168-9525(97)01268-7. [DOI] [PubMed] [Google Scholar]
- 9.Jost J P, Bruhat A. Prog Nucleic Acid Res Mol Biol. 1997;57:217–248. doi: 10.1016/s0079-6603(08)60282-2. [DOI] [PubMed] [Google Scholar]
- 10.Chan M F, Liang G, Jones P A. Curr Top Microbiol Immunol. 2000;249:75–86. doi: 10.1007/978-3-642-59696-4_5. [DOI] [PubMed] [Google Scholar]
- 11.Nan X, Ng H H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Nature (London) 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 12.Jones P L, Veenstra G J, Wade P A, Vermaak D, Kass S U, Landsberger N, Strouboulis J, Wolffe A P. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 13.Wade P A, Gegonne A, Jones P L, Ballestar E, Aubry F, Wolffe A P. Nat Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 14.Fuks F, Burgers W A, Brehm A, Hughes-Davies L, Kouzarides T. Nat Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- 15.Ohki I, Shimotake N, Fujita N, Jee J, Ikegami T, Nakao M, Shirakawa M. Cell. 2001;105:487–497. doi: 10.1016/s0092-8674(01)00324-5. [DOI] [PubMed] [Google Scholar]
- 16.Ballestar E, Wolffe A P. Eur J Biochem. 2001;268:1–6. doi: 10.1046/j.1432-1327.2001.01869.x. [DOI] [PubMed] [Google Scholar]
- 17.Woodcock D M, Crowther P J, Diver W P. Biochem Biophys Res Commun. 1987;145:888–894. doi: 10.1016/0006-291x(87)91048-5. [DOI] [PubMed] [Google Scholar]
- 18.Toth M, Muller U, Doerfler W. J Mol Biol. 1990;214:673–683. doi: 10.1016/0022-2836(90)90285-T. [DOI] [PubMed] [Google Scholar]
- 19.Clark S J, Harrison J, Frommer M. Nat Genet. 1995;10:20–27. doi: 10.1038/ng0595-20. [DOI] [PubMed] [Google Scholar]
- 20.Woodcock D M, Lawler C B, Linsenmeyer M E, Doherty J P, Warren W D. J Biol Chem. 1997;272:7810–7816. doi: 10.1074/jbc.272.12.7810. [DOI] [PubMed] [Google Scholar]
- 21.Ramsahoye B H, Biniszkiewicz D, Lyko F, Clark V, Bird A P, Jaenisch R. Proc Natl Acad Sci USA. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franchina M, Kay P H. DNA Cell Biol. 2000;19:521–526. doi: 10.1089/104454900439755. [DOI] [PubMed] [Google Scholar]
- 23.Verschuren M C, Comans-Bitter W M, Kapteijn C A, Mason D Y, Brouns G S, Borst J, Drexler H G, van Dongen J J. Leukemia. 1993;7:1939–1947. [PubMed] [Google Scholar]
- 24.Akerblad P, Rosberg M, Leanderson T, Sigvardsson M. Mol Cell Biol. 1999;19:392–401. doi: 10.1128/mcb.19.1.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson A A, Wood W J, Jr, Gilly M J, Damore M A, Omori S A, Wall R. Blood. 1996;87:666–673. [PubMed] [Google Scholar]
- 26.Clark S J, Harrison J, Paul C L, Frommer M. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobsen S E, Sakai H, Finnegan E J, Cao X, Meyerowitz E M. Curr Biol. 2000;10:179–186. doi: 10.1016/s0960-9822(00)00324-9. [DOI] [PubMed] [Google Scholar]
- 28.Omori S A, Wall R. Proc Natl Acad Sci USA. 1993;90:11723–11727. doi: 10.1073/pnas.90.24.11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lo K, Landau N R, Smale S T. Mol Cell Biol. 1991;11:5229–5243. doi: 10.1128/mcb.11.10.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu Y, Black J B, Goldsmith C S, Browning P J, Bhalla K, Offermann M K. J Gen Virol. 1999;80:83–90. doi: 10.1099/0022-1317-80-1-83. [DOI] [PubMed] [Google Scholar]
- 31.Cameron E E, Bachman K E, Myohanen S, Herman J G, Baylin S B. Nat Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 32.Herman J G, Baylin S B. Curr Top Microbiol Immunol. 2000;249:35–54. doi: 10.1007/978-3-642-59696-4_3. [DOI] [PubMed] [Google Scholar]
- 33.Palmer B R, Marinus M G. Gene. 1994;143:1–12. doi: 10.1016/0378-1119(94)90597-5. [DOI] [PubMed] [Google Scholar]
- 34.Gisler R, Jacobsen S E, Sigvardsson M. Blood. 2000;96:1457–1464. [PubMed] [Google Scholar]
- 35.Meehan R R, Lewis J D, McKay S, Kleiner E L, Bird A P. Cell. 1989;58:499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- 36.Lorincz M C, Schubeler D, Goeke S C, Walters M, Groudine M, Martin D I. Mol Cell Biol. 2000;20:842–850. doi: 10.1128/mcb.20.3.842-850.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pinarbasi E, Elliott J, Hornby D P. J Mol Biol. 1996;257:804–813. doi: 10.1006/jmbi.1996.0203. [DOI] [PubMed] [Google Scholar]
- 38.Dong A, Yoder J A, Zhang X, Zhou L, Bestor T H, Cheng X. Nucleic Acids Res. 2001;29:439–448. doi: 10.1093/nar/29.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lyko F, Ramsahoye B H, Jaenisch R. Nature (London) 2000;408:538–540. doi: 10.1038/35046205. [DOI] [PubMed] [Google Scholar]
- 40.Harrison J, Stirzaker C, Clark S J. Anal Biochem. 1998;264:129–132. doi: 10.1006/abio.1998.2833. [DOI] [PubMed] [Google Scholar]
- 41.Rother K I, Silke J, Georgiev O, Schaffner W, Matsuo K. Anal Biochem. 1995;231:263–265. doi: 10.1006/abio.1995.1530. [DOI] [PubMed] [Google Scholar]
- 42.Gibbons J D. Nonparametric Statistical Inference. New York: Dekker; 1985. [Google Scholar]
- 43.Radtke F, Hug M, Georgiev O, Matsuo K, Schaffner W. Biol Chem Hoppe–Seyler. 1996;377:47–56. doi: 10.1515/bchm3.1996.377.1.47. [DOI] [PubMed] [Google Scholar]
- 44.Ohtani-Fujita N, Fujita T, Aoike A, Osifchin N E, Robbins P D, Sakai T. Oncogene. 1993;8:1063–1067. [PubMed] [Google Scholar]
- 45.Clark S J, Harrison J, Molloy P L. Gene. 1997;195:67–71. doi: 10.1016/s0378-1119(97)00164-9. [DOI] [PubMed] [Google Scholar]
- 46.Koipally J, Renold A, Kim J, Georgopoulos K. EMBO J. 1999;18:3090–3100. doi: 10.1093/emboj/18.11.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sigvardsson M. Mol Cell Biol. 2000;20:3640–3654. doi: 10.1128/mcb.20.10.3640-3654.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alfarano A, Indraccolo S, Circosta P, Minuzzo S, Vallario A, Zamarchi R, Fregonese A, Calderazzo F, Faldella A, Aragno M, et al. Blood. 1999;93:2327–2335. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.