Abstract
The intestinal microbiota has been identified as an environmental factor that markedly impacts energy storage and body fat accumulation, yet the underlying mechanisms remain unclear. Here we show that the microbiota regulates body composition through the circadian transcription factor NFIL3. Nfil3 transcription oscillates diurnally in intestinal epithelial cells and the amplitude of the circadian oscillation is controlled by the microbiota through group 3 innate lymphoid cells (ILC3), STAT3, and the epithelial cell circadian clock. NFIL3 controls expression of a circadian lipid metabolic program and regulates lipid absorption and export in intestinal epithelial cells. These findings provide mechanistic insight into how the intestinal microbiota regulates body composition and establish NFIL3 as an essential molecular link among the microbiota, the circadian clock, and host metabolism.
The worldwide obesity epidemic presents a pressing public health crisis. More than 2.1 billion individuals are overweight or obese throughout the world, and ~3.4 million deaths are caused each year by obesity-related disease (1). Consequently, there is an urgent need to identify host and environmental factors that regulate human metabolism and energy homeostasis.
The intestinal microbiota is an environmental factor that markedly impacts mammalian body composition. The microbiota promotes energy storage in adipose tissue, and thus microbiologically sterile (germ-free) mice have less body fat relative to conventionally-raised mice (2). This is because in part the microbiota enhance energy harvest from the host diet (3) and promote storage of that energy in adipose tissue (2). Less is known about how microbial regulation of host metabolic pathways might also impact energy storage and body composition.
Many host metabolic pathways are synchronized with day-night light cycles through the circadian clock. The mammalian circadian clock is a network of transcription factors, present in all cells of the body, that drives rhythmic ~24-hour oscillations in gene expression. Synchronization of metabolism with the clock couples energetically expensive metabolic pathways to the availability of dietary substrates, optimizing energy utilization. Emerging evidence indicates that the microbiota interacts with the circadian clock in ways that profoundly impact host metabolism, and that disrupting these interactions can lead to obesity and other metabolic diseases (4, 5). However, little is known about the mechanisms that govern microbiota interactions with the circadian clock and how these interactions alter host metabolism.
NFIL3, also known as E4BP4, is a basic leucine zipper transcription factor that is expressed in a variety of immune cells and controls immune functions that vary by cell type (6, 7). We found that small intestinal epithelial cells also expressed NFIL3, and that expression was markedly reduced in germ-free mice (Fig. 1A). This accorded with prior findings in antibiotic-treated mice (8), and suggested that epithelial NFIL3 might regulate a physiological activity that is responsive to the intestinal microbiota.
Figure 1. Nfil3ΔIEC mice are resistant to high fat diet (HFD)-induced obesity.
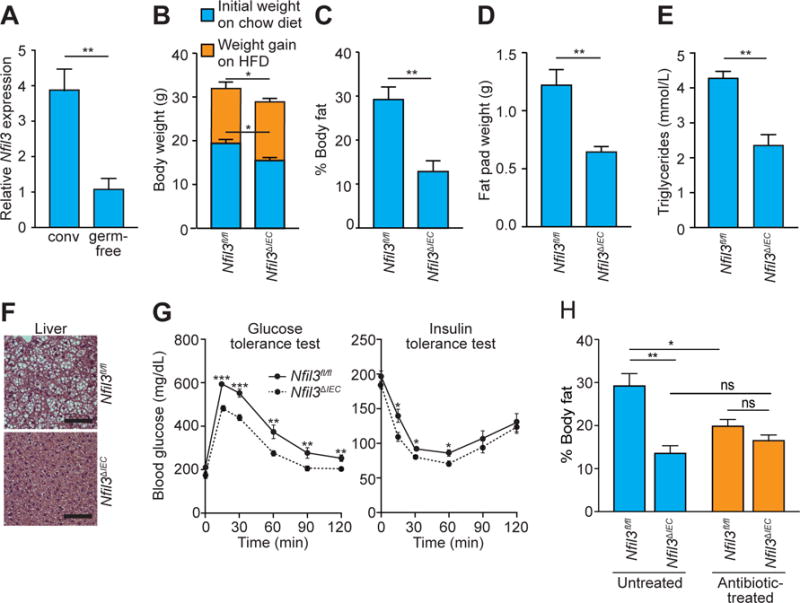
(A) qRT-PCR analysis of Nfil3 transcript abundance in small intestinal epithelial cells recovered by laser capture microdissection from conventional (conv) and germ-free mice. (B) Age-matched Nfil3fl/fl and Nfil3ΔIEC mice were co-housed and placed on a high fat diet (HFD) for 10 weeks. Body weight was measured before and after diet switching. (C) Body fat percentages of mice in (B). (D) Epididymal fat pad weight, (E) serum triglyceride concentration, (F) hematoxylin & eosin (H&E) staining of liver (scale bar=100 μm), and (G) glucose tolerance and insulin tolerance tests. (H) Body fat percentage of mice treated with or without antibiotics after switching to HFD. All data represent two independent experiments with 4–8 mice per group. Male mice were used in all experiments. Means±SEM are plotted; statistics were performed with Student’s t-test or one-way ANOVA. *p<0.05; **p<0.01; ***p<0.001; ns, not significant.
To identify the physiological functions of NFIL3 in intestinal epithelial cells, we generated an epithelial cell-specific Nfil3 knockout mouse (Nfil3ΔIEC). Nfil3ΔIEC mice raised on a chow diet weighed less than their Nfil3fl/fl littermates (Fig. 1B, fig. S1A) and had reduced body fat and increased lean body mass relative to Nfil3fl/fl littermates (fig. S1B,C). The body composition differences were not due to off-target effects of CRE expression (fig. S1D,E), differences in food intake (fig. S2A), physical activity (fig. S2B), or energy utilization (fig. S33A–C), which were similar between the two groups. We also did not detect differences in the response to intestinal injury (fig. S4A–D) or in the expression of key inflammatory cytokines and antimicrobial proteins (fig. S5A–E).
When placed on a high fat, Western-style diet (HFD) for 10 weeks, both Nfil3fl/fl and Nfil3ΔIEC mice gained weight (Fig. 1B). However, the Nfil3ΔIEC mice maintained lower body weights (Fig. 1B) and had lower body fat percentages (Fig. 1C) and higher lean body mass percentages (fig. S6A). The Nfil3ΔIEC mice also had lower epididymal fat pad weights (Fig. 1D, fig. S6B), and were protected from elevated blood triglycerides (Fig. 1E), liver fat accumulation (Fig. 1F), lowered glucose tolerance (Fig. 1G), and increased insulin resistance (Fig. 1G). 16S ribosomal RNA sequencing of fecal microbiotas indicated that the metabolic differences between Nfil3fl/fl and Nfil3ΔIEC mice were not due to differences in microbiota taxonomic composition (fig. S7). Thus, epithelial NFIL3 regulates lipid storage and body composition in mice.
Because epithelial NFIL3 expression is microbiota-dependent we sought to determine if NFIL3-dependent body fat accumulation also depends on the microbiota. Depletion of the microbiota through antibiotic treatment produced lower fat and higher lean body mass in the Nfil3fl/fl mice, resulting in body compositions that were not significantly different from those of Nfil3ΔIEC mice (Fig. 1H, fig. S8). Thus, high fat diet-induced body fat accumulation requires both NFIL3 and a microbiota.
We next studied the mechanism by which the microbiota regulates NFIL3 expression. Nfil3 transcription is controlled by the circadian clock (9) and accordingly, Nfil3 transcript abundance oscillated diurnally in an enriched population of small intestinal epithelial cells acquired by laser capture microdissection (Fig. 2A). Comparison of Nfil3 expression levels across a circadian cycle in wild-type germ-free and conventional mice revealed that the microbiota is required for maximal Nfil3 expression and thus governs the amplitude of Nfil3 transcriptional rhythms (Fig. 2A). This was reflected in protein expression levels (Fig. 2B) and accords with prior findings in antibiotic-treated mice (8).
Figure 2. The microbiota induces epithelial NFIL3 expression through the circadian clock factor REV-ERBα and a DC-ILC3 signaling relay.
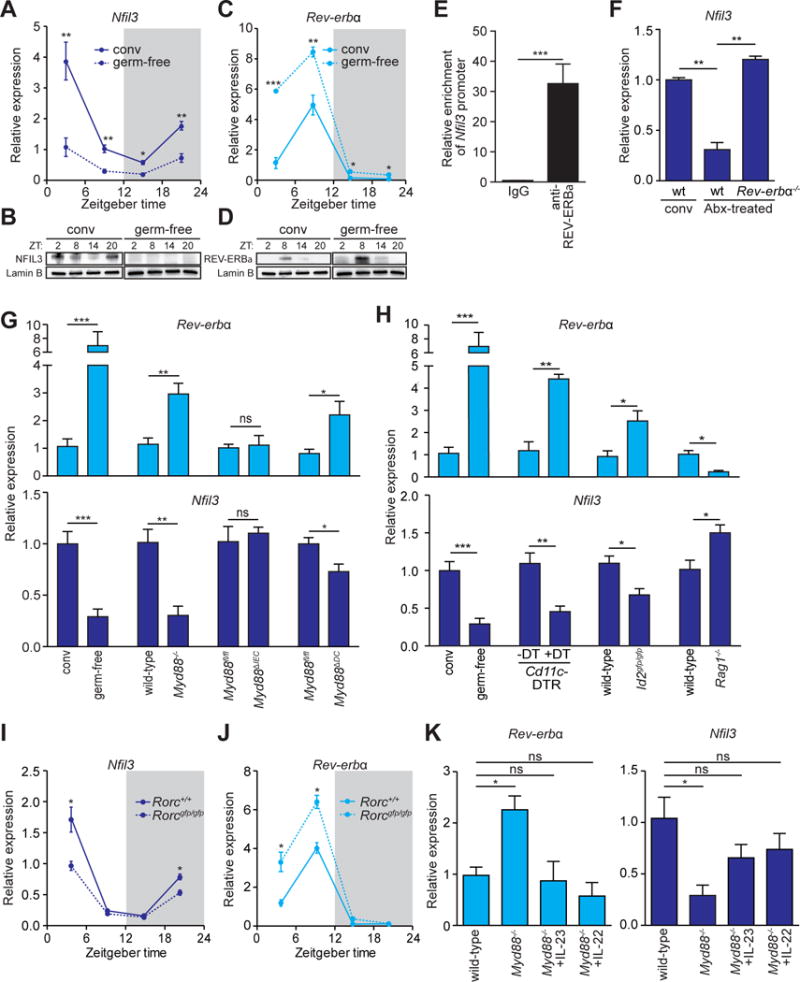
(A–D) qRT-PCR analysis of Nfil3 (A) and Rev-erbα (C) transcript abundance in small intestinal epithelial cells from germ-free (dotted line) and conventional mice (solid line) across a 24-hour day-night light cycle. Western blot analysis of NFIL3 (B) and REV-ERBα (D) was performed on small intestinal epithelial cells isolated from conventional or germ-free mice, Lamin B is the loading control. (E) Chromatin immunoprecipitation assay on intestinal epithelial cells using immunoglobulin G (IgG) or anti-REV-ERBα antibody. Precipitated fragments of the Nfil3 promoter were detected by qRT-PCR. (F) qRT-PCR analysis of epithelial Nfil3 expression in conventional wild-type, antibiotic (Abx)-treated wild-type or Abx-treated Rev-erbα−/− mice. (G) qRT-PCR analysis of epithelial Rev-erbα and Nfil3 expression in germ-free and conventional wild-type mice and conventional Myd88fl/fl, Myd88−/−, Myd88ΔIEC (epithelial cell-specific knockout) and Myd88ΔDC (DC-specific knockout) mice. (H) qRT-PCR analysis of epithelial Rev-erbα and Nfil3 expression in germ-free and conventional wild-type mice, conventional Cd11c-DTR mice that were untreated or treated with Diphtheria toxin (DT), Id2gfp/gfp and Rag1−/− mice. (I,J) qRT-PCR analysis of epithelial Nfil3 (I) and Rev-erbα (J) expression in Rorc+/+ (solid line) and Rorcgfp/gfp (dotted line) mice. (K) qRT-PCR analysis of epithelial Rev-erbα and Nfil3 expression in Myd88−/− mice treated with recombinant IL-23, IL-22 or vehicle. Data in E,F,G,H, and K were collected at ZT4. N=3–8 mice per group. Means±SEM are plotted; statistics were performed with Student’s t-test or one-way ANOVA. *p<0.05; **p<0.01; ***p<0.001; ns, not significant; conv, conventional; ZT, Zeitgeber time.
Nfil3 expression is directly regulated by the core circadian clock transcriptional repressor REV-ERBα. In T cells and liver cells, REV-ERBα binds to a consensus sequence in the Nfil3 gene locus and represses transcription, resulting in a rhythmic diurnal Nfil3 expression pattern (6, 10). Rev-erbα transcript and protein abundance also oscillated diurnally in intestinal epithelial cells but was higher in germ-free than in conventional mice (Fig. 2C,D). This suggested that REV-ERBα governs circadian rhythmicity in Nfil3 expression, and that the microbiota might induce Nfil3 expression by repressing Rev-erbα expression. Indeed, REV-ERBα bound directly to the Nfil3 promoter in intestinal epithelial cells as assessed by chromatin immunoprecipitation (ChIP) assay (Fig. 2E). Further, Nfil3 expression in antibiotic-treated Rev-erbα−/− mice was similar to that in conventional wild-type mice (Fig. 2F). Thus, microbiota regulation of Nfil3 expression is REV-ERBα-dependent, and the microbiota elevates Nfil3 expression by repressing Rev-erbα expression.
We next sought to determine how bacterial signals are relayed to the epithelial circadian clock to regulate Rev-erbα and Nfil3 expression. Intestinal epithelial cells sense the microbiota through Toll-like receptors (TLR) and their common signaling adaptor MyD88 to regulate expression of key genes (11). We therefore tested if MyD88 is required for microbiota regulation of Rev-erbα and Nfil3 expression in epithelial cells. We quantified Rev-erbα and Nfil3 transcripts in epithelial cells from Myd88−/− mice and wild-type littermates at Zeitgeber time (ZT4), which is when Nfil3 expression is near-peak in conventional mice (Fig. 2A). Epithelial Rev-erbα expression was increased and Nfil3 expression was consequently reduced to germ-free levels in Myd88−/− mice (Fig. 2G), indicating that microbiota regulation of the Rev-erbα-Nfil3 cascade requires MyD88. While epithelial MyD88 was dispensable for microbiota-induced alterations in Rev-erbα and Nfil3 expression, Myd88 expression in a CD11c+ cell population (which includes some dendritic cells) was required for maximal repression of Rev-erbα and induction of Nfil3 (Fig. 2G), suggesting a requirement for dendritic cells (DCs).
To further test for DC involvement, we used a mouse model of DC depletion in which Diphtheria toxin receptor (DTR) is expressed under the control of the Cd11c promoter (12). On administration of Diphtheria toxin (DT), CD11c+ cells are selectively killed (12). On CD11c+ cell depletion, Rev-erbα expression was increased and Nfil3 expression was decreased (Fig. 2H), supporting a requirement for DCs in microbiota induction of Nfil3.
Previous studies have identified a subepithelial cellular signaling relay in the small intestine that captures microbiota signals and passes them to epithelial cells to alter expression of key epithelial cell genes (13, 14). In this circuit, bacteria activate TLR-MyD88 signaling in DCs, and the bacterial signals are relayed from DC to group 3 innate lymphoid cells (ILC3) through the cytokine interleukin-23 (IL-23). ILC3 then signal to the epithelium through the production of IL-22 (15).
To determine if ILCs were required for microbiota-induced Nfil3 expression we studied ID2-deficient mice (Id2gfp/gfp), which lack all known ILC subsets (16, 17). Id2gfp/gfp mice showed increased Rev-erbα expression and consequent decreased Nfil3 expression in the small intestinal epithelium (Fig. 2H), consistent with a requirement for ILCs in regulating Rev-erbα expression. Rag1−/− mice, which lack T and B cells, showed decreased epithelial Rev-erbα expression and increased Nfil3 expression (Fig. 2H). The fact that Nfil3 expression was higher in Rag1−/− mice than in conventional wild-type mice is likely due to the elevated bacterial loads as well as aberrant expansion of ILC3 in the small intestines of these mice (18), and indicates that T and B cells are not required for induction of Nfil3 expression. RORγt-deficient (Rorcgfp/gfp) mice (19), which lack both TH17 cells and ILC3, showed circadian Rev-erbα and Nfil3 expression patterns (Fig. 2I,J) that were similar to those of germ-free mice (Fig. 2A,B). This establishes that ILC3 are required for microbiota induction of Nfil3 expression through Rev-erbα. Treatment of Myd88−/− mice with recombinant IL-23 or IL-22 restored epithelial Rev-erbα and Nfil3 expression to wild-type conventional levels (Fig. 2K), further supporting the idea that the subepithelial DC-ILC3 circuit relays microbiota signals to epithelial cells to regulate Nfil3 expression.
The intestinal DC-ILC3 circuit can be triggered by flagellin or lipopolysaccharide (LPS), which is present in the outer membranes of Gram-negative bacteria (20,21). Accordingly, treatment of germ-free mice with flagellin and LPS decreased Rev-erbα expression and increased Nfil3 expression (fig. S9A,B). Further, Rev-erbα expression decreased and Nfil3 expression increased when we monoassociated germ-free mice with Gram-negative, flagellated bacterial species, including Salmonella typhimurium and Escherichia coli (fig. S9A,B). Rev-erbα and Nfil3 expression were not markedly altered by monoassociation with the Gram-positive species Enterococcus faecalis or with the Gram-negative non-flagellated species Bacteroides thetaiotaomicron, suggesting that Nfil3 expression is selectively activated by Gram-negative, motile bacteria. This is likely because such bacteria produce both flagellin and LPS, and because they can readily penetrate the intestinal epithelial barrier and contact lamina propria DCs.
We next sought to identify epithelial cell-intrinsic pathways downstream of IL-22 that regulate Rev-erbα and Nfil3 expression. The transcription factor STAT3 is a key response element downstream of the IL-22 receptor (IL-22R)(14). Activation of the IL-22R leads to phosphorylation of STAT3, which then binds to the promoters of its target genes and either activates or inhibits their transcription. Indeed, ChIP analysis of intestinal epithelial cells showed STAT3 binding to the Rev-erbα promoter (Fig. 3A,B). STAT3 binding was markedly reduced in epithelial cells from germ-free mice (Fig. 3A,B). Co-transfection of STAT3-encoding vectors with luciferase reporters fused to the Rev-erbα promoter resulted in decreased luciferase activity in HEK-293T cells (Fig. 3C), and expression of a dominant active form of STAT3 further inhibited the luciferase activity (Fig. 3C). Thus, STAT3 binds to the Rev-erbα promoter and inhibits its transcription.
Figure 3. STAT3 represses Rev-erbα transcription by binding directly to its promoter.
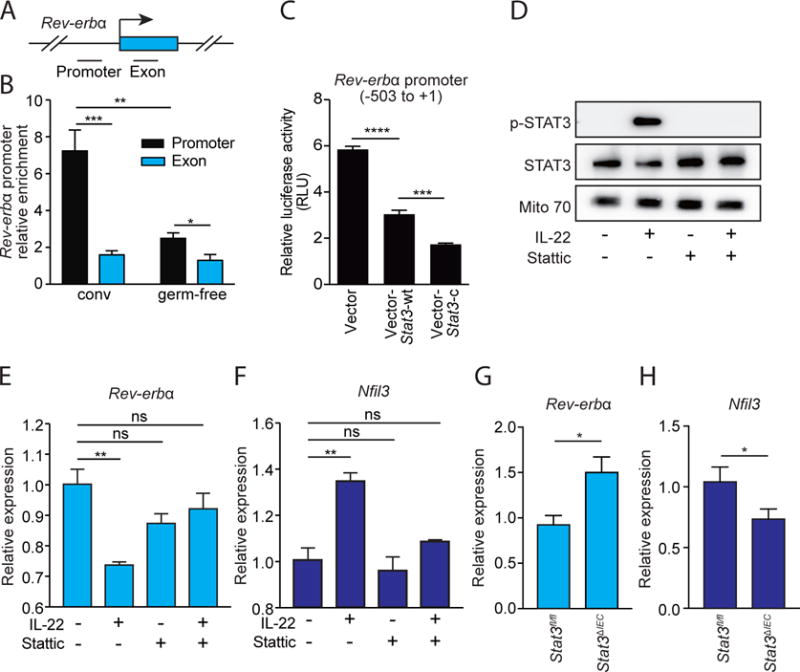
(A) Schematic of the Rev-erbα gene promoter. (B) ChIP analysis of intestinal epithelial cells from conventional (conv) or germ-free mice using immunoglobulin G (IgG) or anti-STAT3 antibody. Precipitated fragments of the Rev-erbα promoter or control exon were detected by qRT-PCR. (C) Luciferase reporter assay. A 504 bp fragment of Rev-erbα promoter was fused to a firefly luciferase reporter. HEK-293T cells were transfected with reporters and either empty vector, a wild-type STAT3-encoding vector (Stat3-wt), or a dominant active STAT3-encoding vector (Stat3-c). (D) Western-blot of total STAT3, phosphorylated STAT3 (p-STAT3) in small intestinal organoids treated with IL-22 and/or the STAT3 inhibitor Stattic. Mito 70 is the loading control. (E,F) qRT-PCR analysis of Rev-erbα (E) and Nfil3 (F) expression in small intestinal organoids treated with IL-22 and/or Stattic. (G,H) qRT-PCR analysis of epithelial Rev-erbα (G) and Nfil3 (H) expression in Stat3fl/fl and Stat3ΔIEC mice at ZT4. N=3–8 samples per group. Means±SEM are plotted; statistics were performed with Student’s t-test or one-way ANOVA. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001; ns, not significant.
We further tested the role of STAT3 in repressing Rev-erbα transcription using cultured intestinal organoids. Addition of recombinant IL-22 to organoid cultures resulted in STAT3 phosphorylation (Fig. 3D), supporting prior findings that IL-22 activates STAT3 in intestinal epithelial cells (14). At the same time, expression of Rev-erbα was decreased and expression of Nfil3 was increased, compared to controls (Fig. 3E,F). In contrast, when we added a STAT3 phosphorylation inhibitor (Stattic) (22) to organoids together with recombinant IL-22, STAT3 phosphorylation was inhibited (Fig. 3D), and Rev-erbα and Nfil3 showed expression levels similar to those of the controls (Fig. 3E,F). These findings further support the idea that STAT3 is a transcriptional repressor of Rev-erbα.
We next investigated whether STAT3 is required for microbiota repression of epithelial Rev-erbα expression in vivo. We generated a mouse with an epithelial cell-specific deletion of Stat3 (Stat3ΔIEC) and assayed for Rev-erbα and Nfil3 expression in intestinal epithelial cells. Consistent with our in vitro findings, the Stat3ΔIEC mice showed increased expression of epithelial Rev-erbα and decreased expression of Nfil3 as compared to their Stat3fl/fl littermates (Fig. 3G,H). The expression of Stat3 and the activation of STAT3 in intestinal epithelial cells did not exhibit diurnal rhythms (fig. S10A,B), indicating that STAT3 does not generate the rhythmicity in Rev-erbα and Nfil3 expression. Instead, our findings suggest that diurnal rhythms in NFIL3 expression are generated by the circadian clock through REV-ERBα, while the amplitude of these rhythms is fine-tuned by the microbiota through STAT3.
To understand the mechanism by which epithelial NFIL3 regulates fat storage and body composition, we compared the transcriptomes of epithelial cells from Nfil3fl/fl and Nfil3ΔIEC mice. We performed an RNAseq analysis on intestinal epithelial cells at multiple time points across the 24-hour day-night light cycle. We identified 33 transcripts having differential abundances in Nfil3fl/fl and Nfil3ΔIEC mice, noting that expression of a number of these genes was diurnally rhythmic in Nfil3fl/fl mice (Fig. 4A). Rhythmic expression of the clock genes Bmal1 (Arnt1), Per2, and Nr1d1 (Rev-erbα) was maintained in Nfil3ΔIEC mice (Fig. 4A), indicating that the core clock mechanism remains intact. Seventeen of the transcripts encoded proteins that are known to function in lipid uptake and metabolism (Fig. 4A). These included Cd36, encoding a transporter that imports dietary fatty acids into cells (23); Scd1, encoding a stearoyl-CoA-desaturase 1 (24); Cyp2e1, encoding a fatty acid hydroxylase (25); and Fabp4, encoding a fatty acid binding protein (26). Deletion of each of these genes protects against HFD-induced obesity and/or insulin-resistance (24–27), suggesting that their lowered expression in Nfil3ΔIEC mice could account in part for the metabolic phenotypes of these mice.
Figure 4. Epithelial NFIL3 controls expression of a circadian lipid metabolic program and regulates lipid absorption in intestinal epithelial cells.
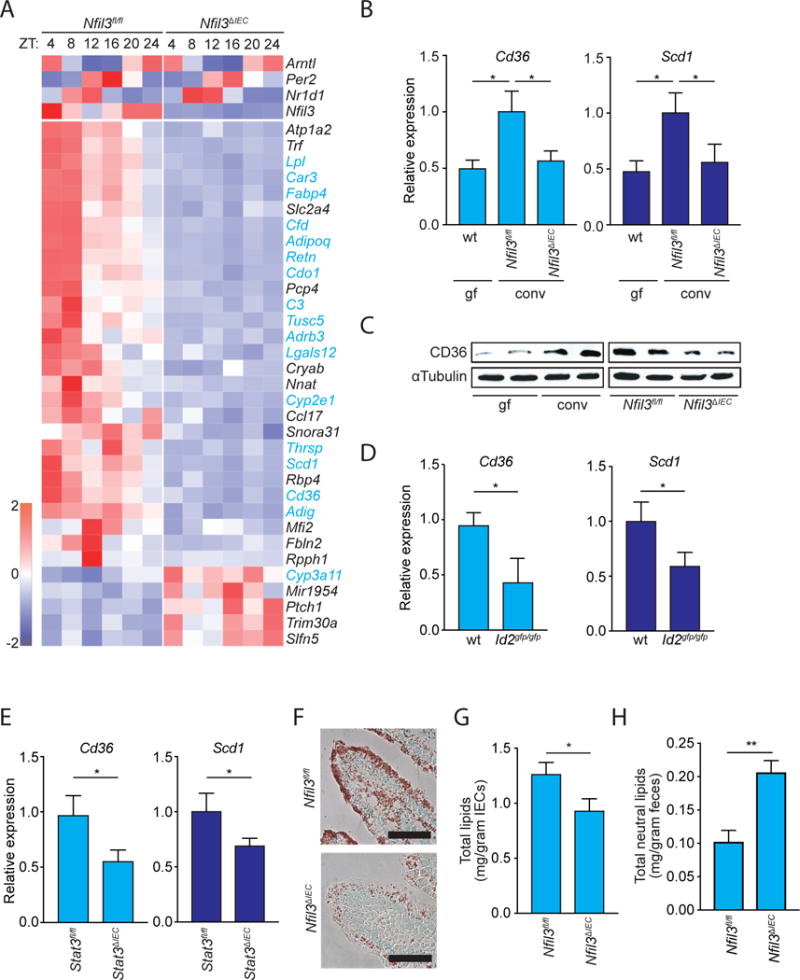
(A) RNAseq analysis of epithelial cell transcripts in Nfil3fl/fl and Nfil3ΔIEC mice across a circadian cycle. The heatmap visualizes expression levels of the 33 genes that have altered expression in Nfil3ΔIEC mice as compared to Nfil3fl/fl mice. Genes encoding proteins that function in lipid metabolism are highlighted in blue. Top panels show sustained circadian expression of the core clock genes Bmal1 (Arnt1), Per2, and Nr1d1 (Rev-erbα) in Nfil3ΔIEC mice. (B) qRT-PCR analysis of epithelial Cd36 and Scd1 expression in germ-free wild-type (wt) and conventional Nfil3fl/fl and Nfil3ΔIEC mice at ZT4. (C) Western blot of epithelial CD36 in germ-free (gf) and conventional wild-type (conv) mice, and in conventional Nfil3fl/fl and Nfil3ΔIEC mice. All mice were fed a HFD. Mice were sacrificed at ZT4. α-tubulin is the loading control. (D,E) qRT-PCR analysis of epithelial Cd36 and Scd1 expression in conventional wild-type (wt) and ID2-deficient (Id2gfp/gfp) mice (D) and Stat3fl/fl and Stat3ΔIEC mice (E) at ZT4. (F) Oil red O detection of lipids in the small intestines of Nfil3fl/fl and Nfil3ΔIEC mice fed on HFD. Nuclei were stained with Methyl Green. Scale bar=40 μm. (G) Total lipid concentrations in isolated small intestinal epithelial cells from Nfil3fl/fl and Nfil3ΔIEC mice fed on HFD. (H) Total neutral lipid concentrations in feces of Nfil3fl/fl and Nfil3ΔIEC mice fed on HFD. Data in B,D,E,G,H have N=5-12 mice per group. Means±SEM are plotted; statistics were performed with Student’s t-test or one-way ANOVA. *p<0.05; **p<0.01; ns, not significant; ZT, Zeitgeber time
We further analyzed Cd36 and Scd1 transcripts by qRT-PCR, confirming that expression of both genes required epithelial NFIL3 and the microbiota (Fig. 4B). Western blot analysis confirmed that CD36 protein levels were reduced in germ-free and Nfil3ΔIEC mice (Fig. 4C). Additionally, expression of Cd36 and Scd1 was reduced in Id2gfp/gfp and Stat3ΔIEC mice (Fig. 4D,E), consistent with microbiota regulation of Nfil3 expression through ILC and epithelial STAT3. Thus, the microbiota regulates an NFIL3-dependent lipid metabolic program that is intrinsic to intestinal epithelial cells.
These findings accord with the key functions of intestinal epithelial cells in fatty acid uptake and metabolism. Following absorption by enterocytes, fatty acids are processed and packaged into chylomicrons for export into the circulation. These processes show circadian rhythmicity, resulting in diurnal variations in circulating lipids (28), and disruption of rhythms in chylomicron export, as seen in mice lacking Nocturnin, is also associated with a lean phenotype (29). Thus, we hypothesized that the metabolic phenotypes of Nfil3ΔIEC mice might arise from reduced epithelial cell uptake and processing of dietary lipids with consequent lowered export of lipids to the circulation for storage in adipose tissue.
We tested this idea by visualizing lipid stores in small intestinal tissues from HFD-fed Nfil3ΔIEC and Nfil3fl/fl mice. Lipids were detected with oil red O staining of small intestinal sections, revealing that the intestinal epithelial cells of Nfil3fl/fl mice harbor abundant lipids (Fig. 4F). By contrast, there was lowered detection of lipids in intestinal epithelial cells from Nfil3ΔIEC mice (Fig. 4F). Nfil3ΔIEC mice also showed reduced oil red O staining in subepithelial intestinal tissues, suggesting reduced export of lipids into the lymphatic capillaries that transport packaged lipids to the bloodstream (fig. S11). Consistent with these findings, lipid concentrations were also lower in intestinal epithelial cells (Fig. 4G) but higher in feces (Fig. 4H) from Nfil3ΔIEC mice as compared to Nfil3fl/fl mice. Thus, epithelial NFIL3 regulates lipid absorption and export in intestinal epithelial cells, potentially explaining why Nfil3ΔIEC mice have a limited body fat gain on a high fat diet.
Here we have shown that NFIL3 is an essential molecular link among the microbiota, the circadian clock, and host metabolism. Our findings show that the microbiota acts through NFIL3 to regulate lipid uptake and storage, thus providing new insight into how the intestinal microbiota regulates host metabolism and body composition. Further, we have shown that the ILC3-STAT3 signaling relay forms an essential conduit between the microbiota and the epithelial circadian clock, thus identifying key molecular circuitry through which the microbiota interacts with the clock (fig. S12). These results potentially provide a deeper understanding of why perturbing microbiota-clock interactions can lead to metabolic disease (4, 5). Our studies could also help to explain why circadian clock disruptions in humans, arising from shift work or international travel, are associated with an increased occurrence of metabolic diseases including obesity, diabetes, and cardiovascular disease (30, 31). Ultimately, our findings could lead to new strategies for treating metabolic disease by targeting NFIL3, STAT3, the microbiota, or the circadian clock.
Supplementary Material
One sentence summary.
The intestinal microbiota regulates body composition through the transcription factor NFIL3, which controls a circadian lipid metabolic pathway in the intestine.
Acknowledgments
We thank C.L. Behrendt-Boyd, Tess Leal, and Brian Hassell for assistance with mouse experiments and other members of the Hooper laboratory for fruitful discussions and critical reading of the manuscript. This work was supported by NIH R01 DK070855 (L.V.H.), a Burroughs Wellcome Foundation Investigators in the Pathogenesis of Infectious Diseases Award (L.V.H.), the Welch Foundation (L.V.H.) and the Howard Hughes Medical Institute (L.V.H.). All data and code to understand and assess the conclusions of this research are available in the main text, supplementary materials and via the GEO repository with accession number GSE100339.
Footnotes
References
- 1.Ng M, et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2014;384:766–781. doi: 10.1016/S0140-6736(14)60460-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bäckhed F, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–131. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 4.Thaiss CA, et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 2014;159:514–529. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
- 5.Leone V, et al. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe. 2015;17:681–689. doi: 10.1016/j.chom.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu X, et al. TH17 cell differentiation is regulated by the circadian clock. Science. 2013;342:727–730. doi: 10.1126/science.1243884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu X, et al. The basic leucine zipper transcription factor NFIL3 directs the development of a common innate lymphoid cell precursor. eLife. 2014;3:e04406. doi: 10.7554/eLife.04406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mukherji A, Kobiita A, Ye T, Chambon P. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell. 2013;153:812–827. doi: 10.1016/j.cell.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 9.Mitsui S, Yamaguchi S, Matsuo T, Ishida Y, Okamura H. Antagonistic role of E4BP4 and PAR proteins in the circadian oscillatory mechanism. Genes Dev. 2001;15:995–1006. doi: 10.1101/gad.873501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duez H, et al. Regulation of bile acid synthesis by the nuclear receptor Rev-erbα. Gastroenterology. 2008;135:689–698. doi: 10.1053/j.gastro.2008.05.035. [DOI] [PubMed] [Google Scholar]
- 11.Vaishnava S, et al. The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jung S, et al. In vivo depletion of CD11c+ dendritic cells abrogates priming of CD8+ T cells by exogenous cell-associated antigens. Immunity. 2002;17:211–220. doi: 10.1016/s1074-7613(02)00365-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanos SL, et al. RORγt and commensal microflora are required for the differentiation of mucosal interleukin 22–producing NKp46+ cells. Nat Immunol. 2009;10:83–91. doi: 10.1038/ni.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sano T, et al. An IL-23R/IL-22 circuit regulates epithelial serum amyloid A to promote local effector Th17 responses. Cell. 2015;163:381–393. doi: 10.1016/j.cell.2015.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sonnenberg GF, Fouser LA, Artis D. Functional biology of the IL-22-IL-22R pathway in regulating immunity and inflammation at barrier surfaces. Adv Immunol. 2010;107:1–29. doi: 10.1016/B978-0-12-381300-8.00001-0. [DOI] [PubMed] [Google Scholar]
- 16.Boos MD, Yokota Y, Eberl G, Kee BL. Mature natural killer cell and lymphoid tissue-inducing cell development requires Id2-mediated suppression of E protein activity. J Exp Med. 2007;204:1119–1130. doi: 10.1084/jem.20061959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Savage AK, et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sawa S, et al. RORγt+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nat Immunol. 2011;12:320–326. doi: 10.1038/ni.2002. [DOI] [PubMed] [Google Scholar]
- 19.Eberl G, et al. An essential function for the nuclear receptor RORγt in the generation of fetal lymphoid tissue inducer cells. Nat Immunol. 2004;5:64–73. doi: 10.1038/ni1022. [DOI] [PubMed] [Google Scholar]
- 20.Kinnebrew MA, et al. Interleukin 23 Production by Intestinal CD103+CD11b+ Dendritic Cells in Response to Bacterial Flagellin Enhances Mucosal Innate Immune Defense. Immunity. 2012;36:276–287. doi: 10.1016/j.immuni.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pickard JM, et al. Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature. 2014;514:638–641. doi: 10.1038/nature13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindemans CA, et al. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature. 2015;528:560–564. doi: 10.1038/nature16460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coburn CT, et al. Defective uptake and utilization of long chain fatty acids in muscle and adipose tissues of CD36 knockout mice. J Biol Chem. 2000;275:32523–32529. doi: 10.1074/jbc.M003826200. [DOI] [PubMed] [Google Scholar]
- 24.Cohen P, et al. Role for stearoyl-CoA desaturase-1 in leptin-mediated weight loss. Science. 2002;297:240–243. doi: 10.1126/science.1071527. [DOI] [PubMed] [Google Scholar]
- 25.Zong H, Armoni M, Harel C, Karnieli E, Pessin JE. Cytochrome P-450 CYP2E1 knockout mice are protected against high-fat diet-induced obesity and insulin resistance. AJP: Endocrinol Metabol. 2012;302:E532–9. doi: 10.1152/ajpendo.00258.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hotamisligil GS, et al. Uncoupling of obesity from insulin resistance through a targeted mutation in aP2, the adipocyte fatty acid binding protein. Science. 1996;274:1377–1379. doi: 10.1126/science.274.5291.1377. [DOI] [PubMed] [Google Scholar]
- 27.Cai L, Wang Z, Ji A, Meyer JM, van der Westhuyzen DR. Scavenger receptor CD36 expression contributes to adipose tissue inflammation and cell death in diet-induced obesity. PLoS ONE. 2012;7:e36785. doi: 10.1371/journal.pone.0036785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pan X, Hussain MM. Clock is important for food and circadian regulation of macronutrient absorption in mice. J Lipid Res. 2009;50:1800–1813. doi: 10.1194/jlr.M900085-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Douris N, et al. Nocturnin regulates circadian trafficking of dietary lipid in intestinal enterocytes. Curr Biol. 2011;21:1347–1355. doi: 10.1016/j.cub.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buxton OM, et al. Adverse metabolic consequences in humans of prolonged sleep restriction combined with circadian disruption. Sci Transl Med. 2012;4:129ra43–129ra43. doi: 10.1126/scitranslmed.3003200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scheer FAJL, Hilton MF, Mantzoros CS, Shea SA. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci USA. 2009;106:4453–4458. doi: 10.1073/pnas.0808180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Motomura Y, et al. The transcription factor E4BP4 regulates the production of IL-10 and IL-13 in CD4+ T cells. Nat Immunol. 2011;12:450–459. doi: 10.1038/ni.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madison BB, et al. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J Biol Chem. 2002;277:33275–33283. doi: 10.1074/jbc.M204935200. [DOI] [PubMed] [Google Scholar]
- 34.Moh A, et al. Role of STAT3 in liver regeneration: survival, DNA synthesis, inflammatory reaction and liver mass recovery. Lab Invest. 2007;87:1018–1028. doi: 10.1038/labinvest.3700630. [DOI] [PubMed] [Google Scholar]
- 35.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stappenbeck TS, Hooper LV, Manchester JK, Wong MH, Gordon JI. Laser capture microdissection of mouse intestine: characterizing mRNA and protein expression, and profiling intermediary metabolism in specified cell populations. Meth Enzymol. 2002;356:167–196. doi: 10.1016/s0076-6879(02)56932-9. [DOI] [PubMed] [Google Scholar]
- 37.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


