Abstract
During chloride salinity, the pH of the leaf apoplast (pHapo) transiently alkalizes. There is an ongoing debate about the physiological relevance of these stress-induced pHapo changes. Using proteomic analyses of expanding leaves of corn (Zea mays L.), we show that this transition in pHapo conveys functionality by (i) adjusting protein abundances and (ii) affecting the rheological properties of the cell wall. pHapo was monitored in planta via microscopy-based ratio imaging, and the leaf-proteomic response to the transient leaf apoplastic alkalinization was analyzed via ultra-high performance liquid chromatography–MS. This analysis identified 1459 proteins, of which 44 exhibited increased abundance specifically through the chloride-induced transient rise in pHapo. These elevated protein abundances did not directly arise from high tissue concentrations of Cl− or Na+ but were due to changes in the pHapo. Most of these proteins functioned in growth-relevant processes and in the synthesis of cell wall–building components such as arabinose. Measurements with a linear-variable differential transducer revealed that the transient alkalinization rigidified (i.e. stiffened) the cell wall during the onset of chloride salinity. A decrease in t-coumaric and t-ferulic acids indicates that the wall stiffening arises from cross-linkage to cell wall polymers. We conclude that the pH of the apoplast represents a dynamic factor that is mechanistically coupled to cellular responses to chloride stress. By hardening the wall, the increased pH abrogates wall loosening required for cell expansion and growth. We conclude that the transient alkalinization of the leaf apoplast is related to salinity-induced growth reduction.
Keywords: abscisic acid (ABA), chloride transport, extracellular matrix, proteomics, ribosome, sodium transport, tubulin
Introduction
Plants frequently experience unfavorable abiotic conditions such as salt stress. To deal with stressful conditions, plants developed mechanisms to acclimate with the aim to complete the life cycle. Systemic changes in the free apoplastic proton concentration are implicated in responses that are involved in acclimation to stress (1, 2). Currently, there is an ongoing scholarly debate over how exactly changes in the free apoplastic proton concentration can be of physiological relevance under stress. During pathogen–host plant interactions, apoplastic alkalinizations that propagate from root to shoot were related to the degree of resistance toward fungal pathogens (3). Drought-induced systemic increases in xylem pH were linked with the regulation of stomatal aperture via effects exerted on the stomata regulating phytohormone abscisic acid (ABA)2 (4, 5). As part of our previous studies on chloride salinity, we found that the leaf apoplastic pH (pHapo) transiently increased in response to the chloride component of NaCl stress (6). This transient alkalinization modulated the compartmental distribution of ABA, directed the hormone toward the guard cells, and induced them to close during the initiation of NaCl stress (7).
Besides effecting the mobility of hormones by changing their protonation state (8), it is well established that changes of the free apoplastic proton concentration are of great significance for transport processes of metabolites and nutrients across the plasma membrane (9, 10). Moreover, the pH of the apoplast facilitates “acid growth” (11) and integrates into models of cytosolic pH signaling (12).
Lager et al. (13) established that changes in the external pH alter patterns of global gene expression in roots and shoots of Arabidopsis thaliana being associated with cell wall modifications. Given the general importance of changing pH patterns, and inspired by the findings from Lager and colleagues (13), we aimed to test whether chloride-induced transient leaf apoplastic alkalinizations influence (i) the abundances of leaf proteins such as cell wall modifying enzymes and (ii) cell wall rheological properties during the initiation of chloride salinity. For this, a unique experimental work flow composed of life cell pHapo imaging in combination with a high throughput proteomics experiment was conducted to analyze the proteome of growing maize leaves that transiently alkalized their apoplast in response to chloride salinity. We reveal that protein abundances across different cellular compartments significantly differ, which was only correlated to the change in apoplastic pH, but not on high tissue concentrations of Cl− or Na+ ions. The majority of those proteins function in growth-relevant processes and the synthesis of cell wall components. This might explain why (i) certain components from cell wall matrix polysaccharides were affected in abundance and why (ii) the cell wall stiffened, as we show by measurements with a linear variable differential transducer. To our knowledge, this functional impact of the pH of the leaf apoplast on both the adjustment of protein abundances and cell wall rheological properties has never been reported.
Results
Leaf apoplast transiently alkalizes in response to the chloride component of NaCl stress
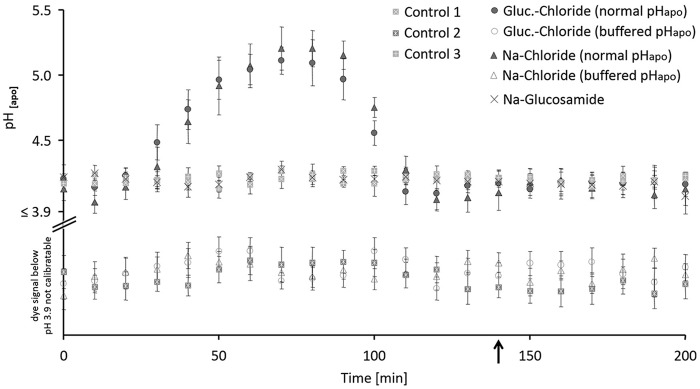
The real-time in planta pH monitoring showed that only the chloride component of NaCl salinity stress induced the leaf apoplast to alkalize transiently (Fig. 1, closed circle and triangle; Table 1). Plants that were salt-stressed without chloride, doing so by using glucosamide− as a sodium accompanying counteranion, did not show the alkalinization, even though 50 mm sodium was added (Fig. 1, cross). pHapo in the three control groups also remained stable (Fig. 1, gray squares filled with a white plus, cross, or asterisks). This reveals that the transient alkalinization is specific to chloride.
Figure 1.
In planta monitoring of leaf apoplastic pH shows that the formation of the transient alkalinization is inducible by the chloride component of NaCl stress. Plants were stressed with 50 mm of the salts, corresponding controls without salt. Chloride stress treatments are given as glucosamine (circle) or sodium chloride (triangle) under conditions when pHapo acted over its normal range (closed symbol) or when pHapo was clamped into the acid range at pH 3.7 (open symbol). Sodium glucosamide–treated plants are labeled by a cross and three different control groups (squares) guard the experiment against effects from the circadian rhythm or stress from the infiltration. Black arrow indicates the time point of taking leaf samples for the proteomics experiment, the cell wall extension assays, and the cell wall compound analysis. For pH measurement, 5 biological replicate samples were generated for each experimental group. Representative kinetic of one of the 5 biological replicates is shown. For this, pH quantitation was technically averaged with n = 6 regions of interest per ratio image and time point. Mean ± S.E.
Table 1.
Experimental setup shows which groups transiently alkalize the leaf apoplast
| Experimental group |
Alklinization | |
|---|---|---|
| Full name | Abbreviation | |
| Control-treated plants | Control 1 | No |
| Control-treated plants plus infiltration of pH buffer | Control 2 | No |
| Control-treated plants plus infiltration of water | Control 3 | No |
| Glucosamine chloride–treated plants plus infiltration of pH buffer | Gluc.-chloride (buffered pHapo) | No |
| Glucosamine chloride–treated plants without infiltration of pH buffer | Gluc.-chloride (normal pHapo) | Yes |
| Sodium glucosamide–treated plants | Na-glucosamide | No |
| Sodium chloride–treated plants plus infiltration of pH buffer | Na-chloride (buffered pHapo) | No |
| Sodium chloride–treated plants without infiltration of pH buffer | Na-chloride (normal pHapo) | Yes |
This chloride-induced rise in pH was blocked by the infiltration of a pH buffer (pH 3.7) into the leaf apoplast (Fig. 1, open circle and triangle). This enabled to have experimental groups that were exposed to chloride salinity and accumulated chloride and/or sodium in the leaves (Table 2) but did not transiently alkalize the leaf apoplast. A direct comparison of the groups “with pH buffer infiltration” (Fig. 1, open circle = glucosamine chloride; open triangle = sodium chloride) versus groups that consists of plants that were chloride-stressed and alkalized the apoplast (“no pH buffer infiltration,” Fig. 1, closed circle = glucosamine chloride; closed triangle = sodium chloride) is meant to (i) reveal proteins whose change in abundance is only correlated to the change of apoplastic pH and (ii) to determine whether the cell wall rheological properties changes specific to the pHapo.
Table 2.
Chloride and sodium leaf concentration
Lowercase letters indicate significant mean differences between chloride concentrations (p < 0.05). Uppercase letters indicate significant mean differences between sodium concentrations (p < 0.05).
| Experimental group | mg Cl− g−1 leaf DM | mg Na+ g−1 leaf DM |
|---|---|---|
| Control-treated plants | 6.16 ± 0.53a | 0.90 ± 0.09A |
| Control-treated plants plus infiltration of pH buffer | 6.50 ± 0.50a | 0.86 ± 0.13A |
| Control-treated plants plus infiltration of water | 6.32 ± 0.43a | 0.84 ± 0.12A |
| Glucosamine chloride–treated plants plus infiltration of pH buffer | 10.02 ± 1.00b | 1.50 ± 0.11B |
| Glucosamine chloride–treated plants without infiltration of pH buffer | 10.16 ± 0.94b | 0.90 ± 0.11A |
| Sodium glucosamide–treated plants | 6.38 ± 0.39a | 1.70 ± 0.24BC |
| Sodium chloride–treated plants plus infiltration of pH buffer | 11.22 ± 1.30b | 2.58 ± 0.14C |
| Sodium chloride–treated plants without infiltration of pH buffer | 11.36 ± 1.30b | 2.08 ± 0.14C |
Moreover, real-time pH imaging revealed the time point at which the leaf apoplast was re-acidified back to the pH range that prevailed in the apoplast before the alkalinization had started (time point indicated by black arrow in Fig. 1). At this time point, leaf material was sampled for (i) the proteomic study and (ii) the assays to assess the stiffening of the cell walls.
The leaf proteome changes under chloride stress in dependence of the pHapo
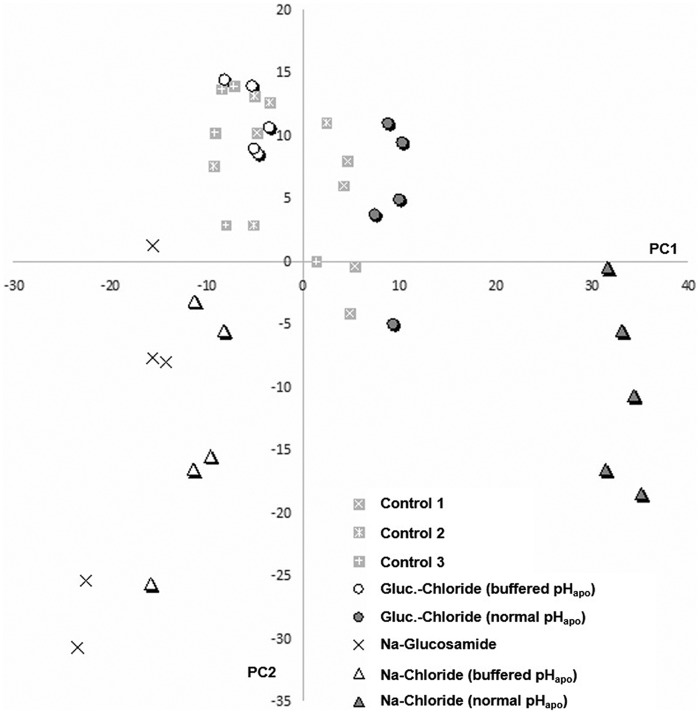
In total, 1459 leaf proteins were identified over the 8 experimental groups (supplemental Table S1) at a false discovery rate of 0.8%. The blind principle component analysis (PCA) resulted in a first principle component (PC) that explained 28.7% of the variation (Fig. 2). This first PC revealed remarkable differences in protein concentrations between (i) the samples from both chloride-stressed plants that alkalized the apoplast (Fig. 2, closed circle and triangle) and (ii) the chloride-stressed plants that did not alkalize the leaf apoplast because the apoplast was clamped in the acid range by the pH buffer (Fig. 2, open circle and triangle). However, the pH-buffered and non-pH-buffered plants all accumulated the same amount of Cl− and Na+ ions in the leaves when salt-stressed (Table 2). This shows that the chloride-inducible transient alkalinization of the leaf apoplast effects the leaf proteome during the initiation phase of salt stress.
Figure 2.
Leaf protein levels evaluated by PCA show distinct grouping of samples that undergo a transient leaf apoplastic alkalinization in dependence on chloride stress. Scores plot for leaf samples from chloride stress treatments given as glucosamine (circle) or sodium chloride (triangle) under conditions when pHapo acted over its normal range (closed symbol) or when pHapo was clamped into the acid range (open symbol). Sodium glucosamide–treated plants are labeled by a cross and three different control groups (squares) guard the experiment against effects from the circadian rhythm or stress from the infiltration. Plants were stressed with 50 mm of the salts, corresponding to controls without salt. PC1 explains 28.7% of the variation between samples. 5 biological replicate samples were generated for each group.
44 proteins increase abundance via effects exerted through the transient alkalinization of the apoplast
Among all proteins that increased abundance via effects exerted through the chloride-induced transient leaf apoplastic alkalinization, 189 proteins were selected for further statistical evaluation. This was done according to their loadings on the first PC that marked them as being most influential in terms of contributing to the separation on the first PC. The list of 189 proteins was subjected to a strict univariate analysis to limit false-positive candidates and to validate that their abundance did only increase in both chloride-stressed plants that alkalized the apoplast (“no pH buffer infiltration,” Fig. 1, closed circle and triangle) when compared with all 6 other experimental groups (2 chloride-stress groups that did not alkalize the apoplast due pH buffer treatment; 1 sodium-stress group, and 3 control groups). This resulted in a list with 44 significant proteins (Table 3).
Table 3.
List with 44 significant proteins that increased abundance specific to pHapo
| Protein | Annotation | ANOVA | Number of identified peptides | Best ion Score |
|---|---|---|---|---|
| A0A096PMM2 | Chaperonin 60 subunit α1, chloroplastic | 5.5E-08 | 10 | 105.2 |
| A0A096RK52 | β-Tubulin R2242-rice | 1.8E-15 | 9 | 88.1 |
| A0A096RTH2 | 5-Methyltetrahydropteroyltriglutamate-homocysteine methyltransferase 1 | 7.4E-14 | 14 | 74.74 |
| A0A096T909 | UDP-glucose 6-dehydrogenase 2 | 1.4E-06 | 6 | 61.41 |
| A0A096TTJ9 | Phenylalanine ammonia-lyase | 0.0E + 00 | 7 | 58.28 |
| A0A096UBL1 | Cinnamyl alcohol dehydrogenase 5 | 2.4E-14 | 3 | 64.17 |
| A0A096UCQ5 | Tubulin β-8 chain | 3.4E-15 | 2 | 41.83 |
| A0A097C0T5 | Sucrose synthase 1 | 2.2E-14 | 21 | 73.8 |
| A0A097C0X0 | Sucrose synthase 1 | 6.5E-13 | 11 | 63.29 |
| B4F946 | Tubulin β-8 chain | 1.4E-10 | 1 | 43.72 |
| B4FAG0 | UDP-glucuronic acid decarboxylase 6 | 0.0E + 00 | 9 | 78.03 |
| B4FIE9 | S-Adenosylmethionine synthase | 2.5E-09 | 2 | 34.2 |
| B4FL64 | 60S ribosomal protein L19 | 5.5E-05 | 3 | 29.9 |
| B4FNQ8 | Calmodulin | 1.7E-10 | 2 | 46.01 |
| B4FTP2 | Thioredoxin-like protein CDSP32, chloroplastic | 3.9E-06 | 3 | 40.35 |
| B4FUK7 | At2g39050/T7F6.22 | 0.0E + 00 | 1 | 48.41 |
| B4G0K5 | At2g39050/T7F6.22 | 2.6E-12 | 3 | 58.72 |
| B6SHD8 | Non-cyanogenic β-glucosidase | 1.9E-10 | 3 | 44.76 |
| B6SKK1 | d-3-Phosphoglycerate dehydrogenase | 3.3E-16 | 2 | 62.6 |
| B6SPX4 | Tubulin α-3 chain | 2.0E-15 | 17 | 102.16 |
| B6SS31 | At2g39050/T7F6.22 | 2.7E-12 | 7 | 85.32 |
| B6SX33 | 40S ribosomal protein S25–1 | 9.0E-08 | 2 | 38.7 |
| B6T681 | S-Adenosylmethionine synthase | 0.0E + 00 | 14 | 146.39 |
| B6T7J7 | Serine hydroxymethyltransferase | 1.3E-12 | 13 | 82.21 |
| B6T8K2 | 40S ribosomal protein S20 | 1.0E-11 | 4 | 68.18 |
| B6TB29 | Fructokinase-2 | 4.7E-14 | 1 | 91.39 |
| B6TDJ0 | 60S ribosomal protein L12 | 3.3E-07 | 3 | 60.77 |
| B6TP93 | Fructokinase-2 | 0.0E + 00 | 9 | 83.13 |
| B6TPJ8 | Fructokinase-2 | 0.0E + 00 | 9 | 84.63 |
| B6TSG5 | 40S ribosomal protein S27a | 3.1E-06 | 1 | 32.4 |
| B6U7D8 | Cinnamyl alcohol dehydrogenase | 1.3E-13 | 3 | 55.9 |
| B7ZYP5 | UDP-glucose-6-dehydrogenase | 0.0E + 00 | 10 | 91 |
| B7ZYX8 | UDP-glucose-6-dehydrogenase | 0.0E + 00 | 4 | 86.72 |
| B8A046 | Phenylalanine ammonia-lyase | 0.0E + 00 | 15 | 113.91 |
| B8A068 | S-Adenosylmethionine synthase | 0.0E + 00 | 4 | 71.56 |
| C0PCV2 | 40S ribosomal protein S8 | 1.5E-07 | 2 | 21.53 |
| C4J722 | At2g39050/T7F6.22 | 1.0E-15 | 3 | 72.2 |
| K7UNP2 | Tubulin β-6 chain | 5.5E-14 | 1 | 82.38 |
| K7V5Z8 | Sucrose synthase Shrunken1 | 5.3E-08 | 5 | 52.41 |
| K7VQR1 | Putative PrMC3 | 4.1E-08 | 1 | 81.74 |
| Q41785 | Tubulin β-8 chain | 6.6E-13 | 5 | 73.71 |
| Q43706 | Sucrose synthase 1 | 0.0E + 00 | 10 | 99.83 |
| Q6VWE6 | O-Methyltransferase | 0.0E + 00 | 6 | 64.74 |
| Q8VXG7 | Phenylalanine ammonia-lyase | 0.0E + 00 | 22 | 102.27 |
Annotations of the 44 identified proteins to cellular components
The cellular component ontology by Gene Ontology (GO) was annotated in the database for 17 of the 44 significant pHapo-responsive proteins (supplemental Table S2). The cellular component for 6 proteins was the ribosome (40S ribosomal proteins S25–1, -S20, -S27a, and -S8, 60S ribosomal proteins L12 and -L19; protein accessions in the above order: B6SX33, B6T8K2, B6TSG5, C0PCV2, B6TDJ0, and B4FL64), 5 were annotated to be located in the cytoplasm (phenylalanine ammonia-lyase, phenylalanine/tyrosine ammonia-lyase, tubulin β-8 chain, and two uncharacterized proteins: accession numbers B8A046, Q8VXG7, Q41785, A0A096PMM2, and A0A096TTJ9), 5 to the cytoplasm and microtubule (tubulin α-3 chain, and four uncharacterized proteins: accession numbers A0A096RK52, A0A096UCQ5, B4F946, B6SPX4, and K7UNP2), and 1 uncharacterized protein to the chloroplast envelope (B4FTP2).
Annotations of the 44 identified proteins to biological processes
Biological process ontology assigned 7 of the 44 pHapo-responsive maize leaf proteins to protein metabolism and protein processing such as protein refolding, protein polymerization, or translation (supplemental Table S2) (40S ribosomal proteins S20, S27a, and S8; 60S ribosomal proteins L12 and L19, tubulin β-8 chain and tubulin α-3 chain; protein accessions in the above order: B6T8K2, B6TSG5, C0PCV2, B6TDJ0, B4FL64, Q41785 and B6SPX4). 6 of the 44 proteins were annotated to a microtubule-based process (tubulin α-3 chain, tubulin β-8 chain, and 4 uncharacterized proteins: accession numbers B6SPX4, Q41785, A0A096RK52, A0A096UCQ5, B4F946, and K7UNP2). The proteins phenylalanine ammonia-lyase (PAL; B8A046) and phenylalanine/tyrosine ammonia-lyase (Q8VXG7) were annotated to a phenylpropenoic acid biosynthetic process and a further uncharacterized protein (A0A096TTJ9) that shared this biological process ontology had an 87.5% sequence similarity to a PAL from Oryza sativa subsp. japonica (P14717) (supplemental Table S2). Some of the pH-responsive proteins were assigned to amino acid metabolism. Among them are S-adenosylmethionine synthase (B4FIE9, B6T681, and B8A068), d-3-phosphoglycerate dehydrogenase 2 (B6SKK1), serine hydroxymethyltransferase (B6T7J7), and an uncharacterized protein (A0A096RTH2) that is similar (87.1%) to 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase 1 (O50008) of Arabidopsis. Other proteins shared biological process ontology to sucrose- or carbohydrate metabolic processes including 3 sucrose synthase entries (SUS; A0A097C0T5, A0A097C0X0, and Q43706), a sucrose synthase SH-1 (K7V5Z8), and a non-cyanogenic β-glucosidase (B6SHD8).
Next, a singular enrichment analysis (SEA) overrepresentation test was conducted to hierarchically sort the overrepresented GO terms that were enriched in the group of pHapo-responsive proteins. The hierarchical tree graph of overrepresented GO terms in the biological process category (supplemental Fig. S1) displays that annotations to the “cellular amino acid metabolic process” (GO:0006520), “translation” (GO:0006412), “protein polymerization” (GO:0051258), “lignin biosynthetic process” (GO:0009809), “microtubule-based movement” (GO:0007018), and “response to salt stress” (GO:0009651) enriched in the set of the 44 pHapo-responsive proteins. The latter was based on increased abundances of tubulin β-6 chains (P29514 and P29511), tubulin β-8 chain (P29516), 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase (O50008), and S-adenosylmethionine synthetase 1 (P23686).
Increase in arabinose from cell wall polysaccharides
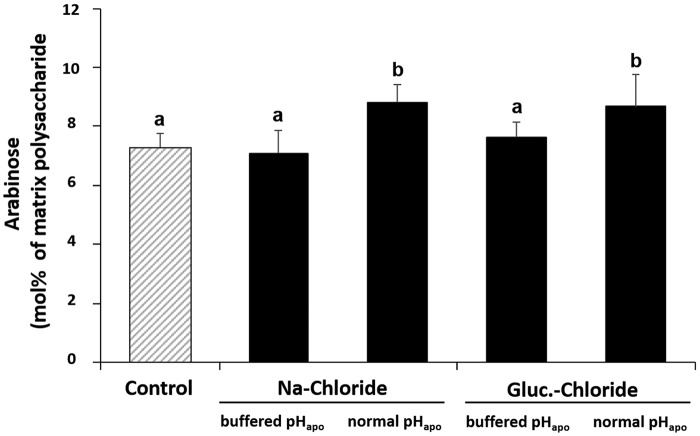
The formation of the chloride stress-specific alkalinization resulted in an increased amount of arabinose in cell wall–bound hemicellulose (Fig. 3).
Figure 3.
The chloride-inducible transient leaf apoplastic alkalinization increases abundance of arabinose in hemicellulose matrix polysaccharides in the leaf cell wall. Plants were either stressed with 50 mm sodium chloride or glucosamine chloride. pHapo was clamped into the acid range at pH 3.7 (buffered pHapo) or acted over its normal range (normal pHapo). Leaf material was harvested for arabinose analysis when the pHapo, which prevailed in the apoplast before the alkalinization had started, was restored (see arrow in Fig. 1). Corresponding controls are without salt. Mean ± S.D. of 5 biological replicates. Statistically significant mean differences (p ≤ 0.05) are indicated by letters. Multiple t-tests were adjusted according to Bonferroni-Holm.
The leaf cell wall stiffens under chloride stress in dependence of the pHapo
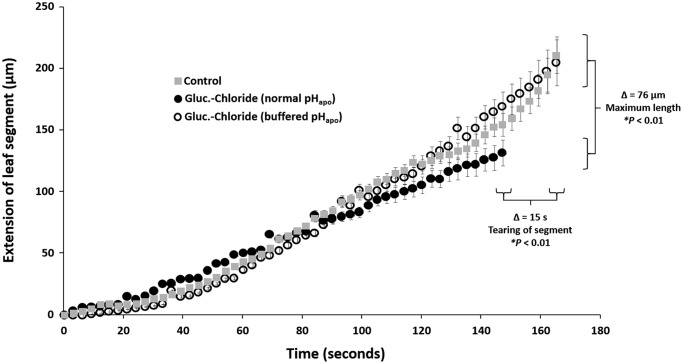
To assess the influence of the chloride-inducible leaf apoplastic alkalinization on the capacity of the leaf cell wall to extend, an in vitro assay was conducted using a linear variable differential transducer. Resulting in leaf segments that underwent a chloride-inducible transient alkalinization of the apoplast torn earlier when the exerted force was increased, viz. were stiffer (Fig. 4, closed circles), compared with leaf segments derived from pH-buffered (closed circle) or control (gray squares) plants that both did not show this pH event. Consequently, the capacity to extend was about 76 μm higher in both, the control and the pH-buffered leaf segments (total size of all segments was 2.5 cm length × 0.75 cm width). In other words, the transient alkalinization of the leaf apoplast is making the cell wall more rigid by stiffening it, consequently, more resistant to the expanding forces of turgor pressure that drives wall extension during expansion growth.
Figure 4.
The chloride-inducible transient leaf apoplastic alkalinization stiffens the leaf cell wall. In vitro assay to assess the capacity of the leaf cell walls to extend or stiffen. Extension of frozen-thawed leaf sections (2.5 cm length × 0.75 cm width) in response to a force applied to leaf segments that either underwent a chloride-induced transient pHapo alkalinization (closed circles) or were pH-buffered before chloride was added (open circles). Non-stressed controls are indicated (gray square). Extension is in micrometers, as measured with a linear variable differential transducer until leaf segments tore. 5 biological replicate samples were generated for each experimental group. Representative kinetic of one of the 5 biological replicates is shown. For this, assay was technically averaged using a total of three leaf segments taken from the identical leaf. Mean ± S.E. Statistically significant mean difference (p ≤ 0.01) are indicated by asterisk. Multiple t-tests were adjusted according to Bonferroni-Holm.
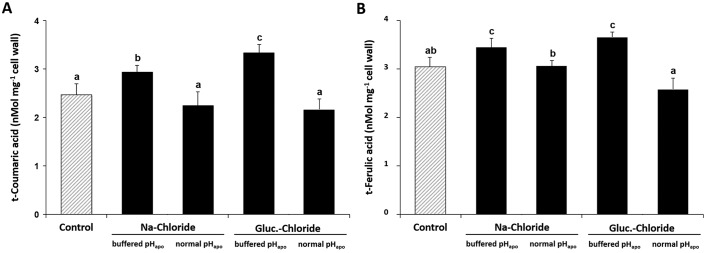
pHapo-specific decrease in free coumaric and free ferulic acid
The chloride-inducible transient leaf apoplastic alkalinization led to a decreased extractability of the two major free monophenols t-coumaric acid and t-ferulic acid from the cell wall. These phenols are relevant for cell wall extensibility because they can get cross-linked via esterification to cell wall polymers, restricting locally further extension of the cell wall (Fig. 5).
Figure 5.
The chloride-inducible transient leaf apoplastic alkalinization decreases abundance of free t-coumaric acid and t-ferulic acid that were extracted from the cell wall. Plants were either stressed with 50 mm sodium chloride or glucosamine chloride. pHapo was clamped into the acid range at pH 3.7 (buffered pHapo) or acted over its normal range (normal pHapo). Leaf material was sampled for analysis when the pHapo, which prevailed in the apoplast before the alkalinization had started, was restored (see black arrow in Fig. 1). Corresponding controls are without salt. A, t-coumaric acid; B, t-ferulic acid. Mean ± S.D. of 5 biological replicates. Statistically significant mean differences (p ≤ 0.05) are indicated by letters. Multiple t-tests were adjusted according to Bonferroni-Holm.
Discussion
Chloride stress-specific transient alkalinization of the leaf apoplast affects protein abundances
As alkalinizations of the leaf apoplast are discussed to be involved in reactions to coping with stress (3, 4), by e.g. (i) modulating the expression of stress-related genes (13), (ii) effecting on the compartmental distribution of plant hormones such as ABA (7), or (iii) inducing growth-related cell wall modifications (11, 13), we decided to investigate in maize whether a chloride salinity-induced transient alkalinization of the leaf apoplast effects: 1) abundances of stress-related proteins or cell wall modifying enzymes and 2) whether this is an event that impacts cell wall rheological properties. The presented results show that this is clearly the case, finally highlighting the importance of integrating leaf apoplastic pH dynamics into models of stress responses that ensue in the cell during the establishment of chloride stress.
First we analyzed if the leaf proteomic pattern changed in response to the formation of the chloride-specific transient alkalinization of the leaf apoplast. For this, 5 biological replicates were generated for each of the 8 experimental groups (Table 1). Real-time in planta imaging of the pH of the leaf apoplast revealed that the pHapo transiently alkalized specific to the chloride-component of the NaCl stress (Fig. 1). The import of chloride from the apoplast into the cell that is energetically coupled to the symport of 2 H+ from the apoplastic site across the plasma membrane into the cytosol (2, 3) is likely to account for this alkalizing effect (6).
The subsequent comparison of the 8 proteomes acquired from the 8 different experimental groups clearly demonstrated that this chloride-specific pHapo event changes the leaf proteome (Fig. 2). Univariate statistical analysis revealed that 44 proteins increased abundance in leaves of chloride-stressed plants via effects exerted through the transient alkalinization of the apoplast that is formed specific to the chloride component of salt stress (Table 3). The accumulation of Cl− and Na+ ions in leaves can be excluded as a factor that directly effects abundance of those 44 proteins. This is stated because tissue concentrations of those ions also increased in those chloride-stressed plants that, however, did not show the increase in the 44 proteins because the apoplast was pH-buffered in the acid range (Table 2). This shows that during the initiation of chloride salinity in maize, the higher abundances of those 44 proteins does not directly rely on high tissue concentrations of Cl− or Na+ ions, but must be related to changes in the free apoplastic proton concentration or associated downstream events, which are as yet unresolved, with the exception of pHapo-based effects on the tissue distribution of ABA (7).
Remote effect on the abundances of symplastic proteins
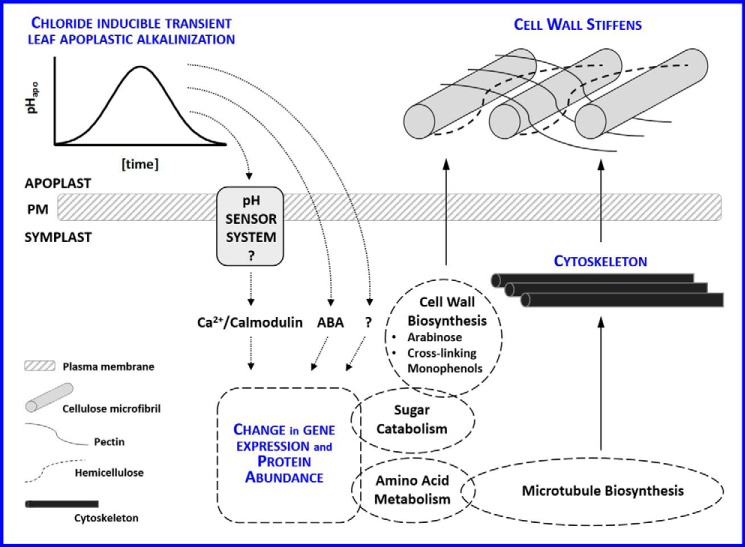
Following Gene Ontology annotations, 17 of the 44 pHapo-effected proteins were located to symplastic compartments (6 protein accessions to ribosomes; 5 to cytoplasm; 5 to microtubule and cytoplasm; and 1 to chloroplast envelope; supplemental Table S2). Cross-species annotations gathered for the non-annotated proteins revealed as well symplastic locations (membrane systems, plasmodesmata, cytosol, or other subcellular compartments such as peroxisome, Golgi apparatus, chloroplast, or nucleus; supplemental Table S2). It is striking that the vast majority of the pHapo-responsive proteins was exclusively annotated to the symplast, because, after all, the apoplast was the compartment that undergoes the transition in pH. As presented in the working model illustrated in Fig. 6, we speculate that the change in the apoplastic free proton concentration is perceived via a yet unknown mechanism that is somehow linked to changes in protein abundances. The elucidation as to whether these proteomic changes convey any molecular or physiological functions is of paramount importance for assessing the importance of this phenomena.
Figure 6.
Working model that links the chloride-induced transient alkalinization of the leaf apoplast with cell wall rheological changes via effects exerted on protein abundances. The change in the apoplastic free proton concentration is perceived via a yet unknown mechanism. The downstream response is the modulation of various symplastic protein abundances. Abscisic acid, which was previously shown to accumulate in the symplast in a pHapo-dependent manner (7) and other as yet unknown factors such as, e.g. calmodulin, which increases abundance in response to the chloride-induced alkalinization (Table 3), are postulated to control adjustments of protein abundances. In the presented study, enzymes increased abundance that allocate carbon flux to cell wall polysaccharide precursor synthesis. Arabinose in cell wall–bound hemicellulose increased in the leaf cell wall. Other proteins that are building blocks of the cytoskeleton also increase in a pHapo-specific manner. Because (i) the organization of the cytoskeleton is relevant for controlling growth and for depositing polysaccharides into the wall and considering that (ii) several other proteins that produce cell wall precursors are under control of the pHapo, we postulate that this chloride-induced change of the apoplastic pH influences growth-related attributes of the wall via the adjustment of metabolic pathways that are catalyzed by the proteins. In good agreement, the cell wall stiffens in response to the alkalinization and free monophenols that were extracted from the cell wall decrease in abundance, which indicates that these compounds get cross-linked to cell wall polymers that decrease cell wall extensibility. In conclusion, the pH of the leaf apoplast is instrumental in hardening the cell wall during the onset of chloride salinity.
Interpretation of the proteomic changes
The most pressing question to arise from these results concerns the meaning of those pHapo-induced proteomic changes. We used a bioinformatics approach, based on Gene Ontology, to acquire information about the biological functions of those 44 proteins. This revealed that the pHapo-responsive proteins function preeminently in (i) amino acid synthesis, translation, and protein polymerization, (ii) microtubule synthesis, or (iii) lignin synthesis (supplemental Fig. S1). All these anabolic processes have in common that they contribute to the control of growth, which is why we anticipate an effect of the chloride-induced change in pHapo on the rheological properties of the cell wall (Fig. 6).
pHapo links chloride stress to enzymes related to lignin and hemicellulose synthesis
The working model in Fig. 6 outlines an effect of the pHapo on the cell wall, because we found that: (i) enzymes from the phenylpropanoid pathway that synthesize precursors of cell wall building blocks, as well as (ii) enzymes from the lignin and hemicellulose synthesis were more abundant in leaves that underwent a transient alkalinization of the pHapo (Table 3). The increase of 2 phenylalanine ammonia-lyase (PAL) members (B8A046; Q8VXG7) proved to be specifically induced by the transient change in pHapo (Table 3). PAL catalyzes the deamination of phenylalanine (or tyrosine; TAL activity) to yield trans-cinnamic acid and NH4 (14). This is the first step in the general phenylpropanoid pathway enabling the production of lignin and other cell wall compounds.
Another enzyme that participates in a more specific branch of the phenylpropanoid pathway is the cinnamyl alcohol dehydrogenase, which catalyzes the last step in the lignin precursor synthesis (15). Cinnamyl alcohol dehydrogenase (B6U7D8) increased in abundance specific to the pHapo (Table 3).
We further identified pHapo-responsive enzymes that play a role in the biosynthesis of hemicellulose. The enzyme UDP-glucuronic acid decarboxylase uses UDP-d-glucuronic acid to synthesize UDP-xylose, which is a precursor for cell wall–bound hemicellulose (16). An uncharacterized maize protein (B4FAG0; Table 3) that has 86.8% sequence similarity to the UDP-glucuronic acid decarboxylase 2 from Arabidopsis (Q9ZV36; supplemental Table S2), as well as three UDP-glucose 6-dehydrogenase (UDPGDH) entries (B7ZYP5, B7ZYX8, A0A096T909; Table 3) increased specifically in response to the alkalinization. Both enzymes are annotated to the cell wall. The UDP-glucose dehydrogenases of maize oxidize UDP-d-glucose to UDP-d-glucuronic acid, which is then decarboxylated, by the UDP-glucuronic acid decarboxylase for UDP-d-xylose production as described above. By the action of an epimerase, the UDP-d-xylose will be converted to UDP-β-l-arabinose, which is a component of the hemicellulose matrix polysaccharides (17). Lager et al. (13) discussed that such changes in the abundance of cell wall modifying enzymes that are mediated by shifts in external pH might be relevant for the acclimation of the wall structure to a changed growth environment. Following this, and taking into account that hemicellulose is a fundamental component that determines the rigidity of the cell wall, we postulate that the chloride-inducible transient leaf apoplastic alkalinization functionally impacts the rheological property of the cell wall. This assumption may require that certain carbohydrates from the fine structure of the cell-bound hemicellulose are affected in abundance by the formation of the transient leaf apoplastic alkalinization. To investigate whether such a response was mediated, we isolated hemicellulose from the cell wall and further hydrolyzed it with acid into its monosaccharides, of which arabinose is one dominant form. Fig. 3 shows that the proposed assumption is indeed the case, because both chloride treatments significantly increased the abundance of arabinose in hemicellulose only when the transient alkalinization was formed.
Transient apoplastic alkalinization stiffens the cell walls of maize leaves
The data show that the pHapo is instrumental by increasing the abundance of (i) the cell wall component arabinose and by acting on (ii) the abundance of enzymes that synthesize cell wall building blocks. However, from this we do not know as to whether these components are either needed to support the increasing demand of cell wall compounds to enable growth (either via cell wall expansion or cell division) or if these compounds are incorporated into the existing cell wall to stiffen it. Increased transfer of hemicellulose and other building blocks in a cell wall that stopped turgor-driven expansion growth will thicken and rigidify it (18). After all, hardening of cell walls of young growing maize roots is known to be a growth inhibitory response to NaCl stress (19).
To test the functional impact of the changing apoplastic pH on the wall rheological properties, an in vitro assay was conducted. These rheological measurements clearly demonstrate for the first time that the chloride-inducible transient leaf apoplastic alkalinization stiffens the cell wall of young leaves that are stressed by salt treatment (Fig. 4). Although it is known that cell wall hardening is an important component of a growth inhibition under salinity (19), the nature of the mechanisms that convey cell wall stiffening was not understood and the elucidation of the exact mechanisms remained an elusive target. With respect to this, our data add novel mechanistic information: the apoplastic pH is a factor that conveys such a stiffening effect under salt stress, possibly via increasing the abundances of enzymes that synthesize cell wall compounds.
Decrease in free monophenols suggests cell wall rigidifications by cross-linking of matrix polysaccharides
To further substantiate our finding that cell wall rigidification is, at least in part, mediated by the formation of the transient alkalinization of the leaf apoplast, we reasoned that plants that underwent the chloride-dependent pHapo change have a lower amount of free extractable coumaric and ferulic acids in the cell wall. This is hypothesized because during cell wall stiffening, the two free monophenols coumaric acid and ferulic acid will be cross-linked to hemicellulose matrix polysaccharides, resulting locally in a decreased wall extensibility. This coupling will also decrease the amount of free form of both acids that can be extracted from the cell wall because they get covalently linked to cell wall polysaccharides, which prevents them from being extracted with the protocol that was used (see “Experimental procedures” and citations therein). The data in Fig. 5 show decreasing amounts of free coumaric and ferulic acids in the cell wall when the pH was transiently alkalized in chloride-stressed leaves, which confirms our metabolic hypothesis for the stiffening of the leaf cell wall.
The stiffening of the cell wall is thought to help plants to survive conditions of salt stress–induced water shortages. Salinity reduces the osmotic potential of the root solution, making it harder for the plant to take up water (20). Kutschera and Schopfer (21) proposed that a cell wall rigidification that counteracts turgor-driven cell wall expansion might be favorable during water stress, where wall loosening would greatly augment further water loss and wilting.
Sucrose catabolism may fuel cell wall matrix polysaccharide biosynthesis
Despite enzymes from the lignin or hemicellulose synthesis, seven proteins from the sucrose metabolism have a higher abundance in response to the transient rise in pHapo. Here we report the increased abundances of sucrose synthases (SUS1, A0A097C0T5; SUS1, A0A097C0X0; SUS1, Q43706; and Shrunken1, K7V5Z8) and fructokinases (B6TB29, B6TP93, and B6TPJ8) (Table 3), which is indicative for a role in the production of UDP-glucose for cellulose biosynthesis. According to the results from Mellerowicz and Sundberg (22) such a concomitant increase in SUS and fructokinase abundances strongly indicates that carbon is allocated to cellulose. SUS produces the nucleotide sugar uridine diphosphate glucose (UDPG) by transferring a uridine diphosphate to the glucose monomer of sucrose, releasing fructose and UDPG (23). The activated glucose monomer UDPG can be shunted into starch or cellulose biosynthesis (24). At the latter, UDPG is polymerized to cellulose by the activity of the transmembrane cellulose synthase enzyme complex that deposits cellulose to the wall.
By silencing the expression of fructokinases 2 (FRK2) in Populus, Roach et al. (25) found that the activity of the fructokinase is also required for carbon partitioning to cellulose. The fructokinase transfers a phosphate group from adenosine triphosphate to fructose yielding fructose 6-phosphate and adenosine diphosphate. The phosphate binding energy from this phosphorylated hexose is then used to fuel the dimerization with glucose to form sucrose. The RNAi-mediated reduction of FRK2 activity resulted in an accumulation of glucose and fructose and in a decrease in phosphorylated hexoses (glucose 6-phosphate and fructose 6-phosphate) and UDPG, showing that carbon flux to cell wall polysaccharide precursors decreased. It was also reported that a reduced FRK2 activity led to thinner cell walls with a reduction in the proportion of cellulose (25). Although it was not tested here, the pHapo-dependent increase of the abundances of the sucrose synthases and fructokinases could mean that carbon is allocated to form cell wall polysaccharide building blocks during the onset of chloride salinity (see model in Fig. 6).
Proteins from the cytoskeleton increase in abundance
A further group of proteins with implications in cellular growth has proven to be under control of the chloride-inducible change in pHapo. β-Tubulins are basic cytoplasmic proteins that form together with α-tubulins the microtubules, which are a major component of the cytoskeleton (26). The dynamic microtubule cytoskeleton network coordinates cellular transport, cell shape, cell division, and cell growth (27, 28). In dependence to the alkalinization, we observe an increased abundance of a number of tubulins (Table 3). They are tubulin α-3 chain (B6SPX4), tubulin β-8 chain (Q41785), and 3 uncharacterized maize proteins that share homology with the Arabidopsis tubulin proteins β-6 chain (P29514; 96.4% sequence identify) and β-8 chain (P29516; 95.1%) and the β-tubulin R2242-rice protein (D7LH43; 95.9%). Microtubules are sensitive to exogenous supply of high concentrations of NaCl because salt stress effects the helical cell growth via disrupting the organization of the microtubules (29). Under salt stress, the dynamic microtubules array is known to be remodeled (30). This reorientation from transverse to parallel to the longitudinal axis was discussed to be responsible for the inhibition of cell elongation in maize roots under chloride toxicity (31). This is because microtubules bind to the plasma membrane where they assist cellulose microfibril deposition into the cell wall by facilitating the movement of the transmembrane cellulose synthase complex. This enzyme complex polymerizes glucose to form cellulose fibrils that are deposited to the wall. A reorientation of the microtubules from transverse to parallel to the longitudinal axis is diminishing cell growth because deposition of cellulose fibrils in the cell wall will also reorient, thus ultimately affecting cell expansion (30, 32–34). Given the relevance of tubulins for cell wall composition and growth inhibition, the pHapo-mediated effect on tubulins could impact on cell wall stiffness and growth during the initiation phase of chloride salinity (see model in Fig. 6).
Ribosomal proteins
Six ribosomal subunits increased in abundance in response to the pHapo (Table 3). They are the 40S ribosomal proteins S25-1 (B6SX33), 40S (B6T8K2), S27a (B6TSG5), S8 (C0PCV2), the 60S ribosomal protein L12 (B6TDJ0), and the ribosomal protein L19 (B4FL64). It was reported that the de novo synthesis of ribosomes closely correlates to (i) changing growth rates and (ii) to an increased ability to synthesize proteins (35–38). Fig. 2 in combination with Table 3 clearly show that the plants that underwent alkalinization synthesized the new proteins.
Proteins responsive to ABA
The presented study revealed that the apoplastic located and ABA-inducible (39) enzyme non-cyanogenic β-glucosidase (40) increased in abundance specific to the alkalinization (B6SHD8; Table 3). During the phototropic response in maize coleoptiles, blue light illumination is well known to up-regulate the β-glucosidase causing a growth inhibition that leads to a phototropic curvature (41). We observe that this growth-inhibiting enzyme increases in abundance the leaves that stiffen their cell walls.
Proposed mode of action
From these results, the question arises how the chloride-inducible transient leaf apoplastic alkalinization stiffens the cell wall and adjusts abundances of cell wall compounds and symplastic proteins that are overall annotated to different biological processes and cellular components. There must be an unknown factor that links the pHapo mechanistically with the observed changes. ABA could be such a node because transcriptomic studies show that ABA controls the expression of a myriad of genes that contribute to aspects of induced tolerance toward salinity and water stress (42, 43). Based on this, it might be a realistic scenario that pHapo orchestrates ABA tissue concentrations, resulting in the modulation of ABA-inducible gene expression. This notion is further substantiated by our recent data for Vicia faba L. that shows the chloride salinity inducible alkalinization of the leaf apoplast is indeed instrumental for the compartmentation of ABA between (guard cell) symplast and the surrounding apoplast (7). Taking into account that, for Arabidopsis (i) 1–10% of the genome has been identified as ABA-regulated, that (ii) 25–50% of the genes regulated by ABA are also regulated by drought or salinity, and that (iii) ABA-repressible genes are often cell wall–associated proteins required for growth (44), it is likely that ABA is a hormonal factor that acts downstream of the alkalinization linking the changes of the pHapo with changes in symplastic proteins abundances (Fig. 6).
Conclusion
We analyzed maize leaves and observed increases in 44 protein abundances that were mechanistically linked to the transient alkalinization of the leaf apoplastic pH and this pH event was a specific response to the chloride component of NaCl salinity. For the first time, proteomic data in combination with live apoplast pH imaging revealed that the transient leaf apoplastic alkalinization is a factor that functions on the modulation of protein abundances during chloride stress. The majority of those proteins is implicated in the synthesis of cell wall building blocks that could be relevant for either facilitating or inhibiting cell wall extension growth. It was found that arabinose in wall-bound hemicellulose increased in abundance. Measurements with a linear-variable differential transducer revealed that the formation of the alkalinization is instrumental for rigidifying (stiffening) the cell wall during the onset of salinity. Decreasing concentrations of the two free monophenols t-coumaric and t-ferulic acids in the cell wall indicates that the cell wall stiffens via a coupling reaction of cell wall polymers to those phenols.
It is well known that crops such as maize show impaired growth under salt stress due to impaired extensibility of the cell wall, whereas many aspects of how extensibility is reduced are not understood. Here, we provide evidence for a new and unexpected role of the pH of the leaf apoplast for the rheological properties of the cell wall and in doing so identify that the transient alkalinization hardens the wall during the initiation of chloride salinity (Fig. 6).
Experimental procedures
Experimental design
Chloride stress is known to alkalize the leaf apoplast whereby this event is transient. It is formed 10–20 min after stressing the roots with chloride containing salts and lasts over a period of ∼1–2 h, which depends on factors such as stress intensity, transpiration rate, or distance between root and leaf (45). Here it was aimed to test whether the formation of this chloride salinity–induced apoplastic pH peak (i) induces proteomic changes and (ii) modulates cell wall rheological properties of expanding maize leaves.
For this, leaf material was sampled that underwent a full chloride-inducible transient leaf apoplastic alkalinization. This was required to monitor the formation of the pH increase and the subsequent re-acidification in real-time and in planta via microcopy-based pH imaging. Leaf material was harvested when the pH that prevailed in the apoplast before the alkalinization had started was restored (at ∼20 min after the leaf apoplast re-acidified). Expanding leave blades from leaf number 4 were investigated that emerged from the sheath 2 days before treatment was started. According to the leaf length and leaf elongation measurements from Neves-Piestun and Bernstein (46), these blades were in the phase of extension growth. 5 biological replicate samples were generated for each group.
For conducting this study, the leaf apoplast of hydroponically grown maize was induced to transiently alkalized, as described by Geilfus and Mühling (6), by adding 50 mm chloride containing salts, in this case sodium chloride or glucosamine hydrochloride, into the nutrient solution. To investigate responses that ensue in response to the alkalinization, experimental groups were introduced consisting of plants whose leaf apoplast was buffered (clamped) in the acid range of 3.7 (i.e. did not show the transient apoplastic alkalinization although chloride stress was applied). This was achieved by using a 10 mm citric acid/sodium citrate buffer as described by Geilfus et al. (7). Plants from these groups were then stressed with chloride salinity and compared with chloride-stressed plants whose leaf apoplastic pH was not buffered but acted over its normal pH range (i.e. transiently alkalized the apoplast after chloride stress was applied). This direct comparison between chloride stress conditions where pHapo was clamped versus chloride stress conditions where pHapo acted over its normal pH range allowed to identify proteins that exclusively changed abundance via pHapo-based effects in chloride salinity–stressed plants. To achieve high reliability, this comparison was carried out under two different chloride-stressed conditions. For this, plants were either stressed with NaCl or glucosamine hydrochloride. Only proteins that were up-regulated under both chloride-stressed conditions in a pHapo-specific manner were considered in this study. A further experimental group was introduced with the aim to identify and reject proteins that were up-regulated in response to the sodium component of NaCl stress. Plants from this group were treated with 50 mm sodium glucosamide. A comparison with this group also enabled to exclude proteins that were up-regulated by the osmotic stress component of the salts because addition of sodium glucosamide decreases the osmotic potential of the nutrient solution (47). Importantly, we used a further 3 different control groups to guard the experiment against changes that are likely the result of the circadian rhythm or the result from the infiltration procedure itself that was necessary to infiltrate the pH buffer into the leaf apoplast; and for these reasons unrelated to the rise in pHapo. Leaves of the 1st control group received no stress treatment. Leaves of the 2nd control group received no stress treatment but were infiltrated with the pH buffer that was used to clamp the apoplastic pH in the acid range. Last, the 3rd control group received no stress treatment but was infiltrated with water to mimic the infiltration.
To keep experimental frameworks identical, experimental groups that did not show the alkalinization in the leaf apoplast, e.g. (i) the three control groups, (ii) the sodium glucosamide group, and (iii) the two groups that were stressed with chloride-containing salts but were infiltrated with a pH buffer to block the formation of the transient leaf apoplastic alkalinization (see Table 1), were also suspected in the pH monitoring procedure and were also sampled at the same time of the day. Whole leaf tissue was sampled between 16:00 and 17:00 h and was either frozen at −20 °C for cell wall extension assays or in liquid nitrogen before it was freeze-dried before the proteome was analyzed.
Plant cultivation
Zea mays (cultivar Susann, Nordsaat Saatzucht GmbH, Langenstein, Germany) was grown in hydroponic culture in a controlled environment chamber. Seeds were embedded in aerated water for 1 day and placed in moistened quartz sand for a period of 6 days for germination. Seedlings were transferred in 7.5-liter plastic pots containing one-quarter strength nutrient solution. After 2 days of cultivation, the nutrient concentration was increased to half-strength and, after 4 days of cultivation to full strength. The solution was changed every 3.5 days to avoid nutrient depletion. The nutrient solution had the following composition: 2.5 mm Ca(NO3)2, 1.0 mm K2SO4, 0.2 mm KH2PO4, 0.6 mm MgSO4, 2.5 mm CaCl2, 0.5 mm NaCl, 1.0 μm H3BO4, 2.0 μm MnSO4, 0.5 μm ZnSO4, 0.3 μm CuSO4, 0.005 μm (NH4)6Mo7O24, 200 μm Fe-EDTA. Plants grew under a 12 h (18 °C):12 h (20 °C) dark:light cycle (photoperiod 08:00–20:00 h) with an atmospheric water vapor pressure deficit of 0.58 kilopascal (75% RH) during the photoperiod. Light intensity was 230–250 μmol s−1 m−2 above leaf canopy. Plants grew 11 days in full strength nutrient solution before being subjected to different treatments for ∼3 h. Expanding leave number 4 was harvested and analyzed. 5 biological replicates were generated for each experimental group.
Ratiometric pH quantification in intact leaves
pH of the leaf apoplast was monitored in real-time using the technique of ratio imaging. For in vivo quantification of leaf apoplastic pH, 25 μm solution of the fluorescent pH indicator dye Oregon Green 488-dextran (Invitrogen GmbH, Darmstadt, Germany) was infiltrated into the leaf apoplast of intact maize using a needleless syringe. Fluorescence images were collected as a time series with a Leica inverted microscope (DMI6000B; Leica Microsystems, Wetzlar, Germany) at 440/20 and 490/10 nm, whereas emission was collected at 535/25 nm for both channels (48). The fluorescence ratio F490/F440 was obtained as a measurement of pH on a pixel-by-pixel basis. Fluorescence ratio data were converted into apoplastic pH values by using an in vivo calibration and the Boltzmann fit was used to fit sigmoidal curves to the calibration data (49).
Protein extraction and digestion
Proteins were extracted following the phenol extraction/ammonium acetate precipitation method described by Carpentier et al. (50). 80 mg of mortared freeze-dried maize leaf tissue was suspended in 750 μl of ice-cold extraction buffer (0.1 m Tris-HCl, pH 8.3, 5 mm EDTA, 0.1 m KCl, 1% dithiothreitol, 30% sucrose, 1 complete Mini EDTA-free protease inhibitor mixture tablet (purchased by F. Hoffmann-La Roche Ltd., Vilvoorde, Belgium)) and vortexed for 30 s. 750 μl of ice-cold Tris-buffered phenol (pH 8.0) was added and vortexed for 10 min at 4 °C. After 10 min of centrifugation (12,000 rpm, 4 °C) the phenolic phase was collected and re-extracted by adding 750 μl of extraction buffer and gently vortexed. The mixture was centrifuged again for 5 min (12,000 rpm, 4 °C), the phenolic phase was collected and left overnight for precipitation with 5 volumes of 0.1 m ammonium acetate in methanol at −20 °C. The samples were centrifuged for 60 min at 13,000 rpm at 4 °C, and after removal of the supernatant the pellet was rinsed with 2 ml of cold acetone, 0.2% DTT and incubated at −20 °C for 1 h. After centrifugation (13,000 rpm, 4 °C) the rinsing solution was discarded and samples were rinsed a second time with 2 ml of the same acetone/DTT mixture and centrifuged for 30 min (13,000 rpm, 4 °C). The pellet was briefly air-dried and re-suspended in 75 μl of buffer (8 m urea, 5 mm DTT, 30 mm Tris base). Protein concentration was determined by a 2-D Quant Kit assay (GE Healthcare, Diegem, Belgium) using BSA as standard and measuring absorbance at 480 nm. For tryptic digestion, DTT was added to 20 μg of protein-containing sample up to a final concentration of 20 mm, gently vortexed, and incubated for 15 min. Iodoacetamide was added to the samples up to a final concentration of 50 mm and vortexed before it was incubated for 30 min in the dark. Samples were diluted 4 times with a stock solution of 150 mm ammonium bicarbonate to reduce urea concentration from 8 to 2 m. Trypsin was added in a ratio of 0.2 μg of trypsin, 20 μg of extracted protein and incubated overnight at 37 °C. Samples were acidified with trifluoroacetic acid (0.1% final concentration) and de-salted using C18 solid phase extraction according to the manufacturer (PierceTM C18 Spin Columns, Thermo Fisher Scientific, Gent, Belgium). Peptides were eluted with 40 μl of 70% ACN, after which solvents were evaporated using a Speed-Vac and dissolved in 5% ACN, 0.1% formic acid.
Peptide separation and MS analysis
The UPLC-MS/MS analysis was performed on a Q Exactive Orbitrap mass spectrometer (Thermo Scientific) as described by Vanhove et al. (51). The samples (5 μl containing 1 μg of peptides) were injected and separated on an Ultimate 3000 UPLC system (Dionex, Thermo Scientific) equipped with a C18 PepMap100 precolumn (5 μm, 300 μm × 5 mm, Thermo Scientific) and an EasySpray C18 column (3 μm, 75 μm × 15 cm, Thermo Scientific) using a gradient of 5–20% ACN in 0.1% formic acid (FA) in 10 min followed by a gradient of 10–35% ACN in 0.1% FA in 4 min, and then a final gradient from 35 to 95% ACN in 0.1% FA in 2.5 min. The flow-rate was set at 250 μl/min. The Q Exactive was operated in a positive ion mode with a nanospray voltage of 1.5 kV and a source temperature of 250 °C. ProteoMass LTQ/FT-Hybrid ESI Pos. Mode CalMix (MSCAL5–1EA SUPELCO, Sigma) was used as an external calibrant and the lock mass 445.12003 as an internal calibrant. The instrument was operated in a data-dependent acquisition mode with a survey MS scan at a resolution of 70,000 (FWHM at m/z 200) for the mass range of m/z 350–1800 for precursor ions, followed by MS/MS scans of the top 10 most intense peaks with +2, +3, and +4 charged ions above a threshold ion count of 16,000 at a 35,000 resolution using a normalized collision energy of 28 eV with an isolation window of 3.0 m/z and dynamic exclusion of 10 s. All data were acquired with Xcalibur 2.2 software (Thermo Scientific).
Protein identification
All raw data were converted into mgf files using Progenesis v4.1 (Nonlinear Dynamics, UK). The spectra were searched using Mascot (version 2.2.06; Matrix Science, London, United Kingdom) against an in-house database containing all UniProt Z. mays accessions plus contaminants of keratin and trypsin (85176 sequences). The following settings were used: the enzyme was trypsin and two miscleavages were allowed, cysteine carbamidomethylation was chosen as a fixed modification and methionine oxidation as a variable one. Precursor peptide charge state was +2 and +3, error window on the experimental peptide mass values was 10 ppm, and 20 absolute millimass units were chosen for fragment ion mass tolerance. Peptides assigned to keratin or trypsin were not taken into account. For data validation, the false discovery rate was calculated using Scaffold (version Scaffold 3.6.3; Proteome Software Inc., Portland, OR). An integrated version of X! Tandem (version CYCLONE, 2010.12.01.1) in Scaffold was used for an additional database searching. For false discovery rate calculation, the searching results from both Mascot and X! Tandem were combined automatically by Scaffold with the following settings: a peptide confidence level of 95%, a protein confidence level of 80%, a minimum peptide number of 1, and the thresholds of each search engines separately. For functional annotations of the identified proteins by GO, annotations were retrieved from UniProt database using our in-house software tool (labtrop.shinyapps.io/Uniprot2Go/).3 When the protein of interest was uncharacterized in maize or poorly annotated, the sequence was aligned by UniProtKB blast to Arabidopsis or O. sativa subsp. japonica and cross-species annotations were engaged. The concern with cross-species annotations is that putative homologous genes from different species must not necessarily be identical in function. Accordingly, data based on cross-species annotations are interpreted with care throughout the presented work. A hierarchical SEA was conducted to identify enriched GO terms in the sets of proteins that increased abundance specific to the pHapo using the agriGO SEA tool with the Arabidopsis background according to Du et al. (52), a significance level was 0.05. Because the identified Z. mays sequences were not completely annotated, the enrichment analysis was done based on A. thaliana cross-species annotations.
Protein quantification
Quantitative analysis was performed using Progenesis LC-MS version 4.1 (Nonlinear Dynamics) as described by Buts et al. (53) with some modifications. For alignment, a reference run was selected, after which the files were aligned automatically, manual landmark vectors were not necessary. The sensitivity of the peak picking limits was put to four. With these settings, Progenesis LC-MS generates an aggregate run that contains all ions from the analyzed runs. Peptides with charges 2–5 were retained in the filter step, and then the data were normalized by calculating abundance ratios to a reference run. The accuracy of ion alignment was double-checked manually for those ions that belong to proteins that are discussed here. Feature tables of the different fractions were combined to a complete overview of the peptide quantification of the samples. Peptides with p value <0.05 (Progenesis) were retained for further analysis and abundances of all corresponding significant peptides were summed per protein accession number. By this, protein abundances were calculated based on peptide abundances.
Statistics of the proteomic data
The software STATISTICA (version 8.0; Tulsa, OK) was used for data analysis. Statistical evaluation started with a PCA, see for example, Carpentier et al. (54). Based on this PCA, the most relevant variables (calculated proteins in this case), were selected via their loadings for univariate analysis of variance and post-hoc analysis. The Fisher's LSD test was used to evaluate the homogeneity of the different experimental groups (p = 0.05; n = 5 biological replicates).
Ion analysis
The analysis of Na+ and Cl− was performed with 7.5 mg of dried leaf material (n = 5 biological replicates for each experimental group) that were boiled for 5 min in 0.8 ml of deionized water. After cooling, samples were centrifuged and proteins were precipitated in the supernatant by washes in chloroform. Thereafter, samples were cleaned by passage through a C-18 column. Na+ and Cl− concentrations were analyzed using ion chromatography (Dionex ICS-5000+, Life Technologies GmbH).
Linear variable differential transducer measurements
The influence of the transient leaf apoplastic alkalinization on the capacity of the cell wall to extend or stiffen was assayedin vitro by using a linear variable differential transducer as described by Geilfus et al. (55). In brief, frozen-thawed leaf segments (2.5 cm length × 0.75 cm width) were connected to the linear variable differential transducer by a thread (via glue) that was pulled over the wheel of the potentiometer. The free end of the threat was connected to a container that represented the weight that was used to pull, viz. to exert force on, the segment. The free end of the leaf segment (opposite to the end that was connected to the thread) needed to be fixed during measurement and this was done by using a clamp to position it to a perforated plate. Extension was induced by continuously increasing the counterweight by means of adding water into the container with a flow rate of 650 μl/s. Readings were taken from the transducer at 3-s by 3-s intervals. Data were recorded by a data logger (Delta-T Device, Cambridge, UK). Measurements were run until the leaf segment tore (point of breakage). Kinetics from 5 independent plants were recorded for each experimental group.
Cell wall compound analysis
Maize cell walls were prepared and analyzed essentially as described (56). Samples (1 mg of lyophilized grinded powder) were hydrolyzed in diluted H2SO4 for 1 h at 121 °C. After cooling, the samples were diluted 1:20 in water and directly analyzed by high performance anion exchange chromatography with pulsed amperometric detection (HPAEC-PAD) on an ICS300 system (Thermo Fisher Scientific). A Carbopac PA20 column was used for separation (57).
Cell wall phenolics esterified to cell wall polymers were extracted from 5 mg of cell wall material by incubation overnight (16 h) in 0.5 ml of NaOH (0.1 m) with rotation similar as described (58). The next day the samples were acidified with 35 μl of 2 m trifluoroacetic acid and extracted twice with 400 μl of ethyl acetate. The organic phases were combined and dried under reduced pressure. The resulting pellet was redissolved in 200 μl of 50% methanol, cleared by centrifugation, and analyzed on a Gravity RP18 column (150 × 2 mm) on an U3000 HPLC (Thermo Fisher Scientific). Solvent A was 0.1% formic acid; solvent B was acetonitrile with 0.1% formic acid. The gradient t0 = 5% B; t2 = 5% B; t12 = 35% B; t15 = 90% B; t20 = 90% B; t21 = 5% B; t30 = 5% B was used to separate phenolic compounds. Quantification was performed at 310 nm using authentic standards (Sigma).
Author contributions
C. M. G., S. S. C., and R. T. designed and performed the study, reviewed the results, wrote the paper, and approved the final version of the manuscript.
Supplementary Material
Acknowledgment
We are grateful to Kusay Arat for excellent technical assistance.
This work was supported by a Coopération européenne dans le domaine de la recherche scientifique et technique STSM scholarship COST Action FA1306 (to C.-M. G.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Tables S1 and S2 and Fig. S1.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- ABA
- abscisic acid
- pHapo
- apoplastic pH
- FA
- formic acid
- PCA
- principal component analysis
- PC
- principal component
- PAL
- phenylalanine/tyrosine ammonia-lyase
- SUS
- sucrose synthase
- PM
- plasma membrane
- UDPG
- uridine diphosphate glucose
- SEA
- singular enrichment analysis
- UPLC
- ultra-performance liquid chromatography.
References
- 1. Wilkinson S. (1999) pH as a stress signal. Plant Growth Regul. 29, 87–99 [Google Scholar]
- 2. Felle H. H. (2001) pH: signal and messenger in plant cells. Plant Biol. 3, 577–591 [Google Scholar]
- 3. Felle H. H., Herrmann A., Schäfer P., Hückelhoven R., and Kogel K. H. (2008) Interactive signal transfer between host and pathogen during successful infection of barley leaves by Blumeria graminis and Bipolaris sorokiniana. J. Plant Physiol. 165, 52–59 [DOI] [PubMed] [Google Scholar]
- 4. Wilkinson S., and Davies W. J. (1997) Xylem sap pH increase: a drought signal received at the apoplastic face of the guard cell that involves the suppression of saturable abscisic acid uptake by the epidermal symplast. Plant Physiol. 113, 559–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Davies W. J., Kudoyarova G., and Hartung W. (2005) Long-distance ABA signaling and its relation to other signaling pathways in the detection of soil drying and the mediation of the plant's response to drought. J. Plant Growth Regul. 24, 285–295 [Google Scholar]
- 6. Geilfus C. M., and Mühling K. H. (2013) Ratiometric monitoring of transient apoplastic alkalinizations in the leaf apoplast of living Vicia faba plants: chloride primes and PM-H+-ATPase shapes NaCl-induced systemic alkalinizations. New Phytol. 197, 1117–1129 [DOI] [PubMed] [Google Scholar]
- 7. Geilfus C. M., Mithöfer A., Ludwig-Müller J., Zörb C., and Muehling K. H. (2015) Chloride-inducible transient apoplastic alkalinizations induce stomata closure by controlling abscisic acid distribution between leaf apoplast and guard cells in salt-stressed Vicia faba. New Phytol. 208, 803–816 [DOI] [PubMed] [Google Scholar]
- 8. Wilkinson S., and Davies W. J. (2002) ABA-based chemical signalling: the co-ordination of responses to stress in plants. Plant Cell Environ. 25, 195–210 [DOI] [PubMed] [Google Scholar]
- 9. Blatt M. R. (1992) K+ channels of stomatal guard cells: characteristics of the inward rectifier and its control by pH. J. Gen. Physiol. 99, 615–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Maathuis F. J., and Sanders D. (1999) Plasma membrane transport in context: making sense out of complexity. Curr. Opin. Plant Biol. 2, 236–243 [DOI] [PubMed] [Google Scholar]
- 11. Kutschera U. (1994) The current status of the acid-growth hypothesis. New Phytol. 126, 549–569 [Google Scholar]
- 12. Monshausen G. B., Bibikova T. N., Messerli M. A., Shi C., and Gilroy S. (2007) Oscillations in extracellular pH and reactive oxygen species modulate tip growth of Arabidopsis root hairs. Proc. Natl. Acad. Sci. U.S.A. 104, 20996–21001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lager I., Andréasson O., Dunbar T. L., Andreasson E., Escobar M. A., and Rasmusson A. G. (2010) Changes in external pH rapidly alter plant gene expression and modulate auxin and elicitor responses. Plant Cell Environ. 33, 1513–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rösler J., Krekel F., Amrhein N., and Schmid J. (1997) Maize phenylalanine ammonia-lyase has tyrosine ammonia-lyase activity. Plant Physiol. 113, 175–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baucher M., Chabbert B., Pilate G., Van Doorsselaere J., Tollier M. T., Petit-Conil M., Cornu D., Monties B., Van Montagu M., Inze D., Jouanin L., and Boerjan W. (1996) Red xylem and higher lignin extractability by down-regulating a cinnamyl alcohol dehydrogenase in poplar. Plant Physiol. 112, 1479–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bindschedler L. V., Tuerck J., Maunders M., Ruel K., Petit-Conil M., Danoun S., Boudet A. M., Joseleau J. P., and Bolwell G. P. (2007) Modification of hemicellulose content by antisense down-regulation of UDP-glucuronate decarboxylase in tobacco and its consequences for cellulose extractability. Phytochemistry 68, 2635–2648 [DOI] [PubMed] [Google Scholar]
- 17. Kärkönen A., Murigneux A., Martinant J. P., Pepey E., Tatout C., Dudley B. J., and Fry S. C. (2005) UDP-glucose dehydrogenases of maize: a role in cell wall pentose biosynthesis. Biochem. J. 391, 409–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alberts B., Johnson A., Lewis J., Walter P., Raff M., and Roberts K. (2002) Molecular Biology of the Cell, 4th Edition, International Student Edition, Garland Science, New York [Google Scholar]
- 19. Neumann P. M., Azaizeh H., and Leon D. (1994) Hardening of root cell walls: a growth inhibitory response to salinity stress. Plant Cell Environ. 17, 303–309 [Google Scholar]
- 20. Munns R. (2002) Comparative physiology of salt and water stress. Plant Cell Environ. 25, 239–250 [DOI] [PubMed] [Google Scholar]
- 21. Kutschera U., and Schopfer P. (1986) Effect of auxin and abscisic acid on cell wall extensibility in maize coleoptiles. Planta 167, 527–535 [DOI] [PubMed] [Google Scholar]
- 22. Mellerowicz E. J., and Sundberg B. (2008) Wood cell walls: biosynthesis, developmental dynamics and their implications for wood properties. Curr. Opin. Plant Biol. 11, 293–300 [DOI] [PubMed] [Google Scholar]
- 23. Kleczkowski L. A., Kunz S., and Wilczynska M. (2010) Mechanisms of UDP-glucose synthesis in plants. CRC Crit. Rev. Plant Sci. 29, 191–203 [Google Scholar]
- 24. Baroja-Fernández E., Muñoz F. J., Li J., Bahaji A., Almagro G., Montero M., Etxeberria E., Hidalgo M., Sesma M. T., and Pozueta-Romero J. (2012) Sucrose synthase activity in the sus1/sus2/sus3/sus4 Arabidopsis mutant is sufficient to support normal cellulose and starch production. Proc. Natl. Acad. Sci. U.S.A. 109, 321–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Roach M., Gerber L., Sandquist D., Gorzsás A., Hedenström M., Kumar M., Steinhauser M. C., Feil R., Daniel G., Stitt M., Sundberg B., and Niittylä T. (2012) Fructokinase is required for carbon partitioning to cellulose in aspen wood. Plant J. 70, 967–977 [DOI] [PubMed] [Google Scholar]
- 26. Snustad D. P., Haas N. A., Kopczak S. D., and Silflow C. D. (1992) The small genome of Arabidopsis contains at least nine expressed β-tubulin genes. Plant Cell 4, 549–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kopczak S. D., Haas N. A., Hussey P. J., Silflow C. D., and Snustad D. P. (1992) The small genome of Arabidopsis contains at least six expressed α-tubulin genes. Plant Cell 4, 539–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bürstenbinder K., Möller B., Plötner R., Stamm G., Hause G., Mitra D., and Abel S. (2017) The IQD family of calmodulin-binding proteins links calmodulin signaling to microtubules, membrane microdomains, and the nucleus. Plant Physiol. 10.1104/pp.16.01743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shoji T., Suzuki K., Abe T., Kaneko Y., Shi H., Zhu J. K., Rus A., Hasegawa P. M., and Hashimoto T. (2006) Salt stress affects cortical microtubule organization and helical growth in Arabidopsis. Plant Cell Physiol. 47, 1158–1168 [DOI] [PubMed] [Google Scholar]
- 30. Dhonukshe P., Laxalt A. M., Goedhart J., Gadella T. W., and Munnik T. (2003) Phospholipase D activation correlates with microtubule reorganization in living plant cells. Plant Cell 15, 2666–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Blancaflor E. B., and Hasenstein K. H. (1997) The organization of the actin cytoskeleton in vertical and graviresponding primary roots of maize. Plant Physiol. 113, 1447–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Green P. B. (1962) Mechanism for plant cellular morphogenesis. Science 138, 1404–1405 [DOI] [PubMed] [Google Scholar]
- 33. Mueller S. C., and Brown R. M. Jr. (1982) The control of cellulose microfibril deposition in the cell wall of higher plants. I. Can directed membrane flow orient cellulose microfibrils? indirect evidence from freeze-fractured plasma membranes of maize and pine seedlings. Planta 154, 489–500 [DOI] [PubMed] [Google Scholar]
- 34. Baskin T. I. (2001) On the alignment of cellulose microfibrils by cortical microtubules: a review and a model. Protoplasma 215, 150–171 [DOI] [PubMed] [Google Scholar]
- 35. Fromont-Racine M., Senger B., Saveanu C., and Fasiolo F. (2003) Ribosome assembly in eukaryotes. Gene 313, 17–42 [DOI] [PubMed] [Google Scholar]
- 36. Brinker M., Brosché M., Vinocur B., Abo-Ogiala A., Fayyaz P., Janz D., Ottow E. A., Cullmann A. D., Saborowski J., Kangasjärvi J., Altman A., and Polle A. (2010) Linking the salt transcriptome with physiological responses of a salt-resistant Populus species as a strategy to identify genes important for stress acclimation. Plant Physiol. 154, 1697–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kosová K., Vítámvás P., Prášil I. T., and Renaut J. (2011) Plant proteome changes under abiotic stress-contribution of proteomics studies to understanding plant stress response. J. Proteomics 74, 1301–1322 [DOI] [PubMed] [Google Scholar]
- 38. Omidbakhshfard M. A., Omranian N., Ahmadi F. S., Nikoloski Z., and Mueller-Roeber B. (2012) Effect of salt stress on genes encoding translation-associated proteins in Arabidopsis thaliana. Plant Signal. Behav. 7, 1095–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Seki M., Narusaka M., Ishida J., Nanjo T., Fujita M., Oono Y., Kamiya A., Nakajima M., Enju A., Sakurai T., Satou M., Akiyama K., Taji T., Yamaguchi-Shinozaki K., Carninci P., Kawai J., Hayashizaki Y., and Shinozaki K. (2002) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 31, 279–292 [DOI] [PubMed] [Google Scholar]
- 40. Kakes P. (1985) Linamarase and other β-glucosidases are present in the cell walls of Trifolium repens L. leaves. Planta 166, 156–160 [DOI] [PubMed] [Google Scholar]
- 41. Jabeen R., Yamada K., Shigemori H., Hasegawa T., Hara M., Kuboi T., and Hasegawa K. (2006) Induction of β-glucosidase activity in maize coleoptiles by blue light illumination. J. Plant Physiol. 163, 538–545 [DOI] [PubMed] [Google Scholar]
- 42. Ingram J., and Bartels D. (1996) The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 377–403 [DOI] [PubMed] [Google Scholar]
- 43. Peleg Z., Apse M. P., and Blumwald E. (2011) Engineering salinity and water-stress tolerance in crop plants. Adv. Bot. Res. 57, 405–443 [Google Scholar]
- 44. Finkelstein R. (2013) Abscisic acid synthesis and response. The Arabidopsis Book, 11, e0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Geilfus C. M., and Mühling K. H. (2012) Transient alkalinization in the leaf apoplast of Vicia faba L. depends on NaCl stress intensity: an in situ ratio imaging study. Plant Cell Environ. 35, 578–587 [DOI] [PubMed] [Google Scholar]
- 46. Neves-Piestun B. G., and Bernstein N. (2001) Salinity-induced inhibition of leaf elongation in maize is not mediated by changes in cell wall acidification capacity. Plant Physiol. 125, 1419–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sümer A., Zörb C., Feng Y., and Schubert S. (2004) Evidence of sodium toxicity for the vegetative growth of maize (Zea mays L.) during the first phase of salt stress. J. Appl. Bot. Food Qual. 78, 135–139 [Google Scholar]
- 48. Geilfus C. M., and Mühling K. H. (2011) Real-time imaging of leaf apoplastic pH dynamics in response to NaCl stress. Front. Plant Sci. 2, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Geilfus C. M., Mühling K. H., Kaiser H., and Plieth C. (2014) Bacterially produced Pt-GFP as ratiometric dual-excitation sensor for in planta mapping of leaf apoplastic pH in intact Avena sativa and Vicia faba. Plant Methods 10, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Carpentier S. C., Witters E., Laukens K., Deckers P., Swennen R., and Panis B. (2005) Preparation of protein extracts from recalcitrant plant tissues: an evaluation of different methods for two-dimensional gel electrophoresis analysis. Proteomics 5, 2497–2507 [DOI] [PubMed] [Google Scholar]
- 51. Vanhove A. C., Vermaelen W., Swennen R., and Carpentier S. C. (2015) A look behind the screens: characterization of the HSP70 family during osmotic stress in a non-model crop. J. Proteomics 119, 10–20 [DOI] [PubMed] [Google Scholar]
- 52. Du Z., Zhou X., Ling Y., Zhang Z., and Su Z. (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38, W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Buts K., Michielssens S., Hertog M. L., Hayakawa E., Cordewener J., America A. H., Nicolai B. M., and Carpentier S. C. (2014) Improving the identification rate of data independent label-free quantitative proteomics experiments on non-model crops: a case study on apple fruit. J. Proteomics 105, 31–45 [DOI] [PubMed] [Google Scholar]
- 54. Carpentier S. C., Panis B., Swennen R., and Lammetryn J. (2008) Finding the significant markers. in Clinical Proteomics, pp. 327—347, Humana Press, New York: [DOI] [PubMed] [Google Scholar]
- 55. Geilfus C. M., Ober D., Eichacker L. A., Mühling K. H., and Zörb C. (2015) Down-regulation of ZmEXPB6 (Zea mays β-expansin 6) protein is correlated with salt-mediated growth reduction in the leaves of Z. mays L. J. Biol. Chem. 290, 11235–11245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yeats T. H., Sorek H., Wemmer D. E., and Somerville C. R. (2016) Cellulose deficiency is enhanced on hyper accumulation of sucrose by a H+-coupled sucrose symporter. Plant Physiol. 171, 110–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Reboul R., Geserick C., Pabst M., Frey B., Wittmann D., Lütz-Meindl U., Léonard R., and Tenhaken R. (2011) Down-regulation of UDP-glucuronic acid biosynthesis leads to swollen plant cell walls and severe developmental defects associated with changes in pectic polysaccharides. J. Biol. Chem. 286, 39982–39992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Waldron K. W., Parr A. J., Ng A., and Ralph J. (1996) Cell wall esterified phenolic dimers: identification and quantification by reverse phase high performance liquid chromatography and diode array detection. Phytochem. Anal. 7, 305–312 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.