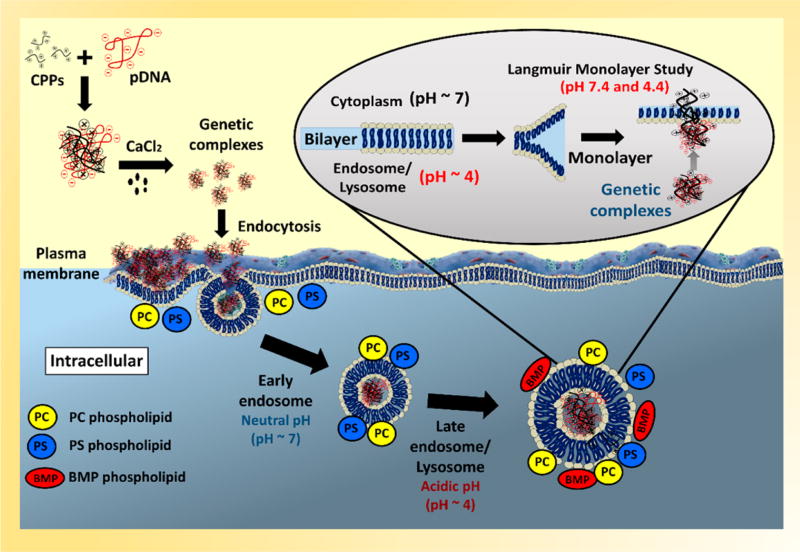
Abstract
Noncovalently condensed complexes of genetic material, cell penetrating peptides (CPPs), and calcium chloride present a nonviral route to improve transfection efficiency of nucleic acids (e.g., pDNA and siRNA). However, the exact mechanisms of membrane insertion and delivery of macromolecule complexes to intracellular locations as well as their stability in the intracellular environment are not understood. We show that calcium condensed gene complexes containing different hydrophilic (i.e., dTAT, K9, R9, and RH9) and amphiphilic (i.e., RA9, RL9, and RW9) CPPs formed stable cationic complexes of hydrodynamic radii 100 nm at neutral pH. However, increasing the acidity caused the complexes to become neutral or anionic and increase in size. Using zwitterionic and anionic phospholipid monolayers as models that mimic the membrane composition of the outer leaflet of cell membranes and intracellular vesicles and pHs that mimic the intracellular environment, we study the membrane insertion potential of these seven gene complexes (CPP/pDNA/Ca2+ complexes) into model membranes. At neutral pH, all gene complexes demonstrated the highest insertion potential into anionic phospholipid membranes, with complexes containing amphiphilic peptides showing the maximum insertion. However, at acidic pH, the gene complexes demonstrated maximum monolayer insertion into zwitterionic lipids, irrespective of the chemical composition of the CPP in the complexes. Our results suggest that in the neutral environment the complexes are unable to penetrate the zwitterionic lipid membranes but can penetrate through the anionic lipid membranes. However, the acidic pH mimicking the local environment in the late endosomes leads to a significant increase in adsorption of the complexes to zwitterionic lipid headgroups and decreases for anionic headgroups. These membrane–gene complex interactions may be responsible for the ability of the complexes to efficiently enter the intracellular environment through endocytosis and escape from the endosomes to effectively deliver their genetic payload.
Graphical abstract
1. INTRODUCTION
Efficient intracellular delivery of genetic material (DNA and RNAi) suffers from several bottlenecks. Unlike micromolecules that can enter the cell by passive diffusion or through specific channels, the larger size and anionic nature of the macromolecules prevent them from penetrating through the plasma membrane. As a result, most macromolecules enter the cell through the endocytosis mechanism.1,2 However, this process suffers from bottlenecks of its own, including effective cellular uptake as well as timely escape from the endosomal pathway to prevent enzymatic degradation of the nucleic acids in lysosomes.3–6 The plasma membranes as well as the membranes in the intracellular compartment serve as barriers that prevent penetration or escape of macromolecules and are the principal deterrents to efficient intracellular delivery.7,8 Subsequently, tremendous efforts have been made to develop gene vectors (both viral and nonviral) that can easily penetrate the plasma membrane as well as effectively avail of the cell’s intracellular transport mechanism to enable intracellular delivery of genetic material. Vector unpacking at the intracellular site is also a necessary step in this process.9 Viral vectors are highly efficient in this process, but safety concerns (e.g., immunogenicity) still persist.10–12 Nonviral vectors, on the other hand, offer several advantages over viral vectors, such as ease of synthesis, low cost, and low degree of immunogenicity. 12–14 Therefore, efficient design of nonviral vectors for intracellular gene delivery necessitates an understanding of the mechanisms through which the nonviral vectors can effectively trigger cellular uptake as well as escape from late endosomes to deliver their payload.
The main structural component of the plasma membrane as well as the various compartments in the intracellular trafficking pathway is phospholipids. While the zwitterionic phosphatidylcholine (PC) headgroups make up the major phospholipid content of all eukaryotic cell membranes, anionic phospholipids such as phosphotidylserine (PS) are present in the inner leaflet of the plasma membrane as well as the lipid membranes that make the endosomes.7,15 Cell membranes also contain very small amounts of anionic phosphotidylglycerol (PG), while late endosomes are unique in their composition of bis-(monoacylglycero)phosphate (BMP), which is a structural isomer of PG. In fact, compared to the outer leaflet of the cell membrane, the endosomes are significantly enriched in anionic lipids such as PS and BMP.16 In addition to the change in the lipid headgroup moieties, the intracellular pH also decreases along the endocytic pathway, from the early endosome to the lysosomes.15 Therefore, understanding the mechanisms of nonviral gene delivery vectors requires an understanding of their interactions with these biological barriers.
Noncovalent complexation of genetic material (e.g., DNA and siRNA) using polycations has received attention as potential nonviral gene vectors that can deliver larger genetic payloads at the required delivery site.12 Unfortunately, the most efficient polycations, such as polyethylenimine (PEI), are also the most toxic. Further, while high molecular weight polycations are often essential to condense larger genetic material such as plasma DNA (pDNA) into small particles, the increase in molecular weight often increases cytotoxicity as well.17 We and others have shown that the use of synthetic cell penetrating peptides (CPPs) is a potentially interesting method for condensation of genetic material as well as their delivery across the cell membrane.6,11,12,18–20 CPPs consist of low molecular weight cationic or amphiphilic sequences of about 30 residues.12,17,18,21 Since their discovery in the early 1990s, they have attracted a lot of attention as potential candidates for delivery of biomolecules to cells.12,22,23 In general, CPPs can be classified into three types, depending on their origin: natural CPPs (protein-derived) (e.g., TAT peptide), synthetic CPPs (e.g., polyarginine), and chimeric CPPs (e.g., transportan).12 Simple polyarginine or polylysine molecules influenced by a highly basic minimal transduction domain of the protein-derived TAT protein, typically consisting of nine amino acids (RKKRRQRRR), were shown to cross the plasma membrane even more readily than the TAT peptide.2,24 Because of the faith in “arginine’s magic”, several groups, including us, have previously reported on the ability of using these polypeptides as CPPs.25–27 It has also been shown that the membrane insertion potential of synthetic peptides is dependent on the positive charge on the CPPs interacting with anionic phospholipids within the membrane.28 Particularly, synthetic CPPs with 7–9 amino acids have demonstrated the best cell penetration efficiency, and this efficiency has been found to go down as the residue sequence length is either increased or decreased.17,29,30 Further, we and others have shown that the presence of hydrophobic moieties, such as tryptophan, can also enhance phospholipid penetration.2,31 Particularly, the properties of the guanidyl group were extremely efficient in increasing the membrane penetration behavior of arginine polypeptides.25
Numerous studies have also demonstrated that adding calcium chloride (CaCl2) to CPP/siRNA or pDNA complexes produced stable and small nanoparticles that improved the transfection efficiency of these gene complexes in vitro and in vivo.6,11,14,20,32 It was also found that adding 100 mM CaCl2 to the CPP/pDNA samples yielded the highest gene expression compared to 50, 150, and 300 mM,17 possibly by enhancing their endosomal release before degradation by lysosomes.33,34 However, how does complexing CPPs with genetic material in the presence of CaCl2 impact their cell insertion potential has not been previously reported. While the calcium condensed CPP/pDNA complex uptake may be aided by endocytosis or cell penetration, which in turn involves interactions with specific cell membrane lipids or possibly interactions with membrane associated proteoglycans, the desirable goal of endosomal escape is typically destabilization of the endosomal membrane, to allow timely escape of genetic material.22,35–37 Therefore, understanding the mechanism of cell penetration (or cellular uptake) of these CPP-induced condensed complexes as well as the mechanisms of escape of these nonviral gene vectors from endosomal compartments is essential for designing efficient CPP/pDNA vectors.2,38,39
In order to better understand the stability of gene complexes and their interactions with phospholipid membranes in the endocytic pathway (plasma, endosome, and lysosome membranes), the mechanisms of interactions between the condensed gene complexes with phospholipid membranes forms the focus of this study. In particular, we report on the interactions of seven different gene complexes with model PC, PS, and PG phospholipid monolayers at two pHs (7.4 and 4.4). Based on previous studies, our choice of CPPs consists of four hydrophilic CPPs (i.e., dTAT, K9, R9, and RH9) and three amphiphilic arginine-rich peptides (RW9, RL9, and RA9) (Table 1). Positively charged synthetic polypeptides—polyarginine R9 and polylysine K9—were used to study the effect of specific amino acids on gene complex formation and interaction with the plasma membrane. Arginine-rich CPPs where the arginine residues at positions 3, 4, and 7 are replaced with tryptophan, leucine, alanine, and histidine (RW9, RL9, RA9, and RH9) were used to study the effect of a combination of positively charged and hydrophobic residues on the insertion efficiency of the CPP/gene complexes.2 dTAT (double-TAT) is a natural HIV protein derived CPP that was used as a control. In our studies, unsaturated fatty acids with mixed alkyl chains were used as simple models of the phospholipid barriers. POPC monolayers were used as model zwitterionic monolayers, while POPS and POPG phospholipids were used as model anionic monolayers (Table 2) to study the effect of lipid headgroup charge density on the phospholipid insertion potential of gene complexes. One of the advantages of using model membranes is that their packing can be easily controlled and varied. Additionally, model membranes have previously been used to study the individual interactions of DNA as well as CPPs to better understand their mechanisms of interaction. The two pHs represent the extreme limits in the acidic nature of the environment in the endocytic pathway. Finally, since several previous in vitro and in vivo studies have shown that calcium improves the transfection efficiency of gene complexes, 6,11,14,17,20,32,40,41 here we also present the effect of the addition of calcium on the stability and membrane insertion potential of our gene complexes.
Table 1.
Identification of the Seven CPPs and Their Properties (Peptide Sequence, Number of Residues, Classification, Net Charge at pH 7, and Molecular Weight)
| name | interpreted sequence | no. of residues | classification | charge | mol wt (g/mol) | |
|---|---|---|---|---|---|---|
|
| ||||||
| pH 7.4 | pH 4.4 | |||||
| dTAT | RKKRRQRRRHRRKKR | 15 | hydrophilic | +13 | +14 | 2200.75 |
| K9 | KKKKKKKKK | 9 | hydrophilic | +9 | +9 | 1170.65 |
| R9 | RRRRRRRRR | 9 | hydrophilic | +9 | +9 | 1422.74 |
| RH9 | RRHHRRHRR | 9 | hydrophilic | +6 | +9 | 1365.62 |
| RA9 | RRAARRARR | 9 | amphiphilic | +6 | +6 | 1167.41 |
| RL9 | RRLLRRLRR | 9 | amphiphilic | +6 | +6 | 1293.68 |
Table 2.
Name, Synonyms, Classification, and Structure of the POPC, POPS, and POPG Membrane Models
| Name | Synonyms | Classification | Relative Charge * | pKa * | Structure | |
|---|---|---|---|---|---|---|
| pH 7.4 | pH 4.4 | |||||
|
| ||||||
| POPC | 1-hexadecanoyl-2-(9Z-octadecenoyl)-sn-glycero-3 phosphocholine | Zwitterionic Phospholipid (Unsaturated) | 0 | 0 | pKa ~ 1 |
|
|
| ||||||
| POPS | 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (sodium salt) | Anionic Phospholipid (Unsaturated) | ~ − 1.0 | ~ − 1.0 | pKa ~ 5.5 |
|
| pKa ~ 2.6 | ||||||
| pKa~ 11.55 | ||||||
|
| ||||||
| POPG | 1-hexadecanoyl-2-(9Z-octadecenoyl)-sn-glycero-3-phospho-(1'-rac-glycerol) | Anionic Phospholipid (Unsaturated) | ~ − 0.9 | ~ − 0.9 | pKa ~ 3.5 |
|
Marsh, D. CRC Handbook of Lipid Bilayers, 1990.
2. MATERIALS AND METHODS
2.1. Materials
Plasmid DNA (pDNA) encoding firefly luciferase (pGL3, 4818 bp) was obtained from Promega (Madison, WI). The pDNA purity level was determined by UV-spectroscopy and agarose gel electrophoresis. dTAT (RKKRRQRRRHRRKKR; Mw = 2201.7 Da) peptide, K9 (KKKKKKKKK; Mw = 1170.65 Da) peptide, R9 (RRRRRRRRR; Mw = 1422.74 Da) peptide, RH9 (RRHHRRHRR; Mw = 1365.62 Da) peptide, RA9 (RRA ARRARR; Mw = 1167.41 Da) peptide, RL9 (RRLLRRLRR; Mw = 1293.68 Da) peptide, and RW9 (RRWWRRWRR; Mw = 1512.83 Da) peptide were purchased from Biomatik Corporation (Cambridge, Ontario, Canada) (purity >95%). 1-Hexadecanoyl-2-(9Z-octadecenoyl)-sn-glycero-3 phosphocholine (sodium salt) (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine (sodium salt) (POPS), and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (sodium salt) (POPG) were purchased from Avanti Polar Lipids (Alabaster, AL) as organic mixtures in chloroform. Agarose (medium-EEO/protein electrophoresis grade) was purchased from Fisher Scientific. Bench Top DNA Ladder was purchased from Promega (Madison, WI). SYBR Green I Nucleic Acid Gel Stain was obtained from Invitrogen (Carlsbad, CA). Calcium chloride dehydrate (CaCl2·2H2O) was purchased from Fisher Scientific. Other organic chemicals used for this work was purchased from Fisher Scientific. Petri dishes (Falcon 1008, Becton-Dickinson Labware, Franklin Lakes, NJ) were purchased from Fisher Scientific. All samples and lipid mixtures were stored at −20 °C when not in use.
2.2. Methods
2.2.1. Preparation of CPP/PDNA/Ca2+ Complexes
CPP/pDNA complexes were prepared by adding 15 µL of CPPs solution (0.28 µg/µL) to 10 µL (0.1 µg/µL) of pDNA (TAE Buffer (1×) was used as a solution for DNA storage), followed by fast pipetting for 20 s. At that point, 15 µL of CaCl2 of desired molarity (100 mM in most cases) was added and mixed by fast pipetting for a further 20 s. This process resulted in the formation of condensed complexes containing pDNA and the CPP. After preparing the gene complexes, they were stored at 4 °C for 15–20 min and then used for further analysis. The peptide-to-lipid (P/L) molar ratio at an initial SP of 20 and 30 mN/m was 0.25.
2.2.2. Agarose Gel Electrophoresis
4 µL (1×) of Tris-acetate-EDTA (TAE) buffer was added to the solution containing the gene complexes of interest. EDTA interacts with the divalent metal ions in solution and thus inhibits metal-dependent nucleases. 4 µL of SYBR Green 1 (a highly sensitive DNA gel stain used for visualization of DNA in agarose or acrylamide gels) was mixed with the gene complexes. The mixture was well-mixed and stored at 4 °C for 15–20 min. After the storage, 7 µL of 6× DNA loading dye (dye used to prepare DNA markers and samples for loading on agarose gels) was added to allow visual tracking of DNA migration during electrophoresis. A 1 kb DNA ladder was used. The presence of glycerol ensures that the DNA in the ladder and sample forms a layer at the bottom of the well. The approximate mass of DNA in each of the bands is provided for approximating the mass of DNA in comparatively concentrated samples of similar size. The mixture solutions were loaded onto a 1% agarose gel and electrophoresed for 30 min at 110 V.
2.2.3. Particle Size
The particle size (effective diameter (nm)) of CPP/pDNA complexes with calcium chloride was determined by dynamic light scattering (Brookhaven Instruments, Holtsville, NY). All samples intended for particle size measurements were prepared using PBS at pH 7.4 and 4.4.
2.2.4. Zeta Potential
The zeta potentials of the complexes were measured by Zeta PALS dynamic light scattering (Brookhaven Instrument, Holtsville, NY). All samples intended for particle size measurements were prepared using PBS, Nuclease Free Water (NFW), and Serum Free Media (SFM). All samples intended for zeta-potential measurements were prepared using PBS at pH 7.4 and pH 4.4.
2.2.5. Langmuir Trough Experiments
The insertion potential of the different synthetic complexes and CPPs was measured using model phospholipid monolayers (PMs) containing different amounts of a zwitterionic (POPC) or negatively charged phospholipid (POPS, and POPG) at the air–PBS buffer interface. SP changes were recorded by a Wilhelmy plate sensor, which is part of the KSV-NIMA Langmuir trough purchased from Biolin Scientific, while Petri dishes (volume 4 mL, 35 × 10) were used as “mini-troughs” for the experiments. Insertion of the gene complexes and CPPs into pre-formed monoalyers was recorded in the presence of phospholipid membranes. A wet calibrated filter paper flag was dipped into this buffer solution and used as a probe to monitor changes in the surface pressure due to adsorption of surface active material. The phospholipid membranes were spread from chloroform solutions, on PBS subphase, at pH 7.4 and 4.4, using a Hamilton microsyringe (Hamilton Co., Reno, NV). The spreading phospholipid solvents were allowed to evaporate for 20 min prior to adding of the gene complexes (or free CPPs). The gene complexes (and free CPPs) aqueous solutions were injected underneath the surface of lipid membranes, and the changes in surface pressure with respect to time were measured immediately. Precaution was taken to ensure that the injection did not disrupt the phospholipid monolayer. Adsorption to a bare air/buffer interface was recorded for all the gene complexes (and free CPPs) studied here to ensure that the changes in the SP of the monolayers were due to interaction of the gene complexes (or free CPPs) with the cell membrane. Each experiment was run for 60 min at 22 ± 2 °C. All figures were normalized by subtraction of the control [the phospholipids (POPC, POPS, and POPG) without complexes or free CPPs] at pH 7.4 and 4.4 to ensure that the differences in the SP of the monolayers were due to interaction of complexes and peptides.
2.2.6. Statistical Analysis
Data were analyzed by using the GraphPad software. Statistical evaluation between the means of the data was performed using an unpaired t-test. One-way ANOVA, Tukey post test was used to analyze the differences when more than two data sets were compared. Each experiment was repeated three times (n = 3).
3. RESULTS AND DISCUSSION
3.1. Physical Characterization of the CPP/PDNA/Ca2+ Complexes
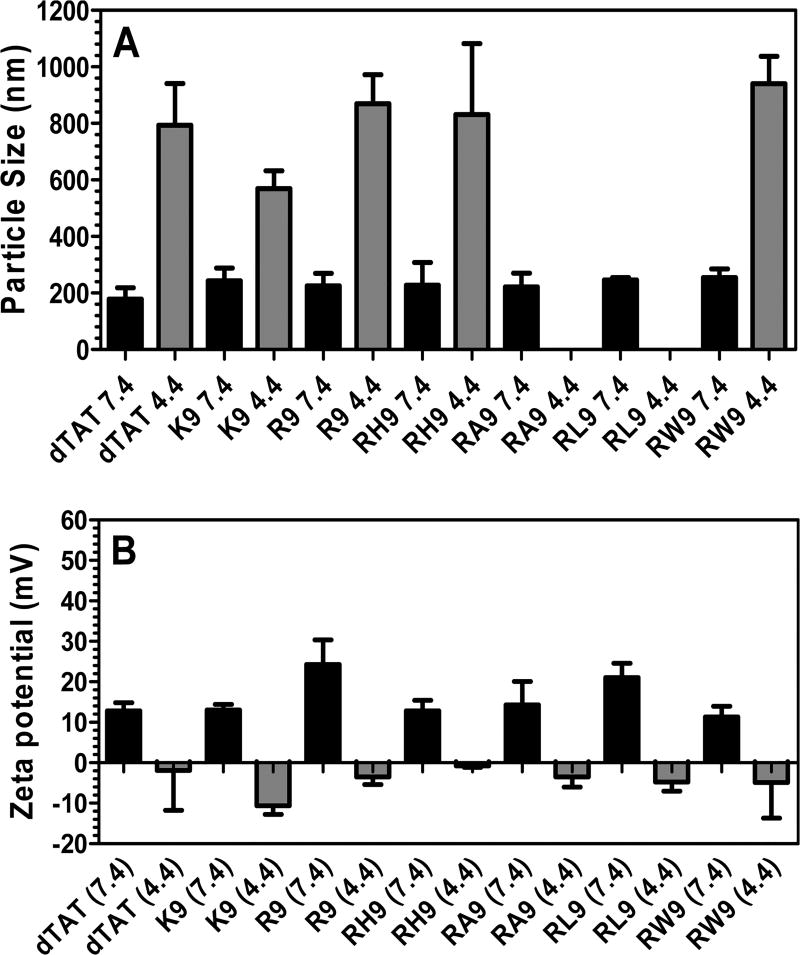
Two important characteristics of CPP/pDNA/Ca2+ complexes are their particle size and overall charge. Figure 1A illustrates the particle size of the seven CPP (dTAT, K9, R9, RH9, RA9, RL9, and RW9)/pDNA/Ca2+ complexes in PBS (ionic strength, 185 mM) at pH 7.4 and 4.4. Overall, the particle size of the complexes was around 250 nm at pH 7.4 (with relatively narrow polydispersity (<0.2)), irrespective of the CPP used, suggesting that the choice of CPP has little effect on the particle size at normal pH. Further, an increase in particle size was noted at low pH (4.4), compared to the physiological pH (7.4), suggesting that an acidic environment reverses the condensing effect of Ca2+. Moreover, a reduction in the photon counting rate (kilocounts per second; kcps), at the more acidic pH also supports the argument that these gene complexed may be partially dissociated or are no longer condensed at pHs that are relevant to the late endosomal or lysosomal environment. Such acidic pH-induced dissociation has previously been reported for DNA/calcium complexes.17,42 Figure 1B presents the zeta-potential of these gene complexes at normal and acidic pHs. We find that at the neutral pH the zeta-potential values are positive, although there are small differences in the individual values. This is possibly due to the charge on the CPPs making up the condensed gene complexes. The net positive zeta-potential of our condensed complexes verifies the cationic nature of the complexes and confirms that while there is a core of pDNA, the CPP, being in excess form the shell of these gene complexes. Figure 1B also shows that the net charge on the particles is switched when the pH is lowered, and the gene complexes become anionic or neutral. These observations confirm that a lowering of the environmental pH destabilized the noncovalently condensed pDNA complexes, allowing the pDNA to be exposed or released from the complex.
Figure 1.
(A) Particle size of the seven CPPs (dTAT, K9, R9, RH9, RA9, RL9, and RW9)/pDNA/Ca2+ complexes, which was measured in phosphate buffer saline and at pH 7.4 and 4.4. For missing data points, the diameter was undetectable. (B) Zeta-potentials of CPPs (dTAT, K9, R9, RH9, RA9, RL9, and RW9)/pDNA/Ca2+ complexes in phosphate buffer saline and at pH 7.4 and 4.4. Results are presented as mean ± SD (n = 3).
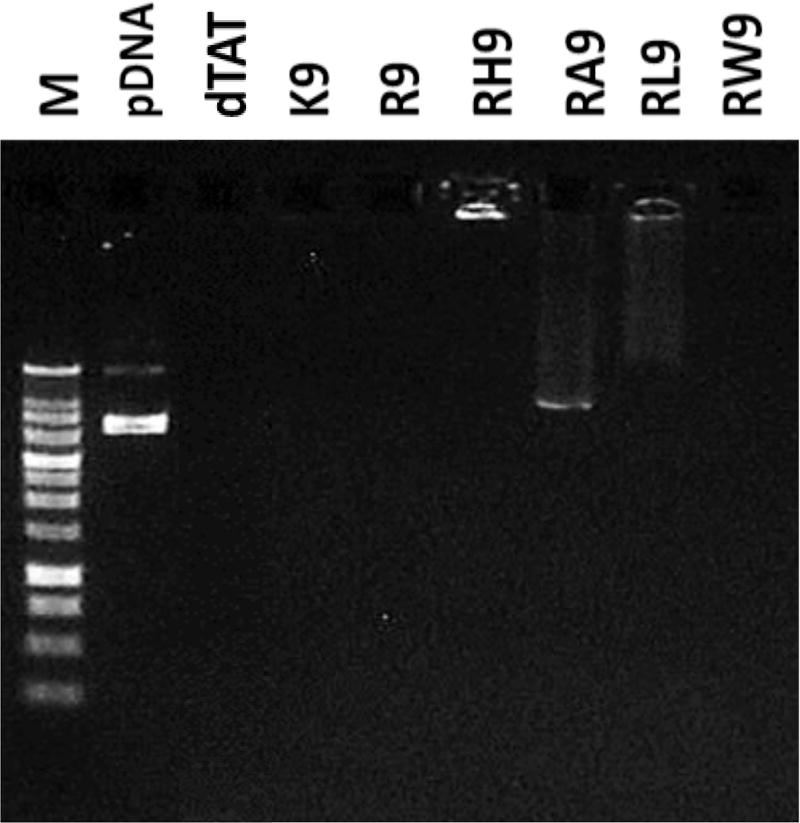
To further prove that at pH ~ 7.4 the pDNA is immobilized with the CPP to form complexes, we ran agarose gel electrophoresis assay for these complexes. Figure 2 illustrates the results of this assay for these complexes. Free pDNA was used as a control, while the leftmost column shows the molecular weight marker. When an electric field is applied across the cells, the appearance of different bands along the column corresponds to the migration of any free DNA. Our results show that most of the gene complexes completely immobilized the pDNA (no bands were observed during electrophoresis), except RA9 containing complexes, which looked slightly unstable.
Figure 2.
Agarose gel electrophoresis study of the seven CPPs (dTAT, K9, R9, RH9, RA9, RL9, and RW9)/pDNA/Ca2+ complexes. “M” refers to DNA molecular weight marker.
3.2. Insertion of the CPP/PDNA/Ca2+ Complexes into Model Membranes
As cell membranes are complex systems, many model membranes have become commonly used for studying the membrane activity of various natural and synthetic compounds (e.g., peptides, drugs, and surfactants). Lipid monolayers are the most common biomimetic systems used to study the insertion of biomolecules into cell membranes. These monolayers can be considered as models for one leaflet of the cell membrane. These two-dimensional models show many advantages compared to other models (e.g., lipid vesicles). Some parameters (e.g., the composition of the subphase (pH and ionic strength), temperature, and the packing of the molecules) can be easily varied in a controlled manner allowing several biophysical studies of the phospholipid–gene complex interactions. However, one of the limitations of the monolayer system is that they represent only one leaflet of the cell membranes without any other components such as membrane proteins Therefore, they do not reflect the complexity of cell structures.2,43 Yet, these simple models are often used to understand the biophysical interactions governing molecular interactions in phospholipid membranes. For example, it is well-known that if the area of the monolayer is kept constant (constant area assay), adsorption followed by insertion of molecules into the hydrophobic region of phospholipid monolayers (tails) can cause a significant increase in the surface pressure. On the other hand, particles that interact with the hydrophilic region (head groups) typically induce only slight changes in surface pressure.2,44 In order to understand the physical mechanisms of interactions of calcium condensed gene complexes with cell membranes in the endosomal environment, the membrane insertion potential of seven different complexes was recorded using two model phospholipid monolayers. Zwitterionic phospholipid (POPC) and anionic phospholipids (POPS and POPG) were used as simple mimics of the cell membranes. PC is the most common phospholipid in the cell membrane of eukaryotic cells,45 whereas anionic PS are normally found in the inner leaflet of the plasma membrane as well as endosomal membranes.7,46,47 Even though PG headgroups are present at 1–2% in the plasma membrane, they have often been used as model anionic lipids to study the function of CPP.7,46 More relevant to this study, BMP, an isomer of PG with the same headgroup charge,48,49 but differences in the position of the glycerol units on the phosphate moiety, is a unique anionic lipid found in significant fractions in the late endosomes.7,49,50 Since BMP was not stable as a monolayer in our experimental setup because of its high solubility and tendency to form vesicles,51–53 we chose to use PG headgroups as model lipid system that remains anionic at the pHs relevant to this study, in order to understand the interactions of the CPPs with anionic headgroups that are enriched in the late endosomes.48–50
The effect of hydrophobic interactions vs electrostatic interactions on the phospholipid membrane insertion potential of gene complexes was monitored using gene complexes that contained both hydrophilic and amphiphilic peptides. Additionally, we have studied the effect of pH of the cytoplasm/early endosome vs late endosome/lysosome on this insertion process by monitoring the changes in the surface pressures at two different pHs, 7.4 and 4.4. Therefore, by studying the insertion potential of these different cationic gene complexes into phospholipids at different pHs, we expect to develop a mechanistic understanding of interactions of the gene complexes with phospholipids in the endosomal pathway. While the insertion potential of the gene complexes was initially measured for different monolayer surface pressures (20–40 mN/m), our previous2 and current work indicate that the maximum difference in the results between the samples was monitored at a surface pressure of 20 mN/m (Supporting Information Figure S1). Therefore, in this work, we report the insertion of our gene complexes into monolayers held at an initial surface pressure of 20 mN/m.
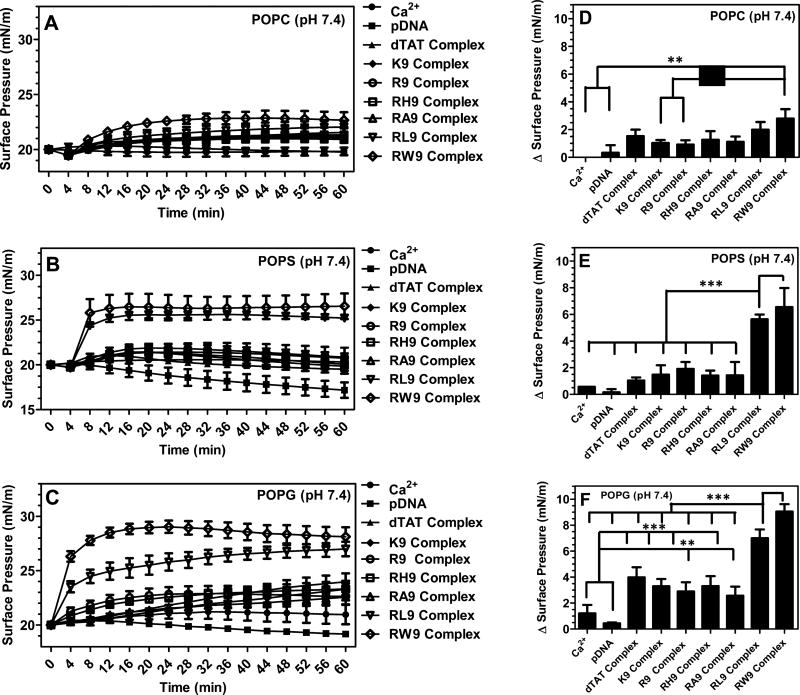
Figure 3 shows the changes in surface pressure of the three phospholipid monolayers as a function of time, following interactions with the seven complexes at pH 7.4. Figure 3A presents the change in surface pressure with time while Figure 3D presents the maximum change in surface pressure [Δsurface pressure, the maximum absolute value of the surface pressure recorded during the 60 min period] for each of the seven complexes, when injected below the POPC monolayer. The change in surface pressure with time is significantly higher for gene complexes condensed using the CPPs and calcium when compared with the control (pDNA alone), indicating that the gene complexes have significantly higher insertion efficiency into the model zwitterionic phospholipid monolayer, when compared with an equal amount of free pDNA. However, no significant difference in the surface pressure was observed between the different cationic gene complexes. Since the sizes of these condensed complexes are comparable, our results suggest that at physiological pH the gene complexes do not show significant insertion into the zwitterionic phospholipids. However, the significant difference in the insertion potential of the complexes when compared to the change in surface pressure of the PC monolayer in the presence of free pDNA suggests that condensing the complex with CPPs enabled increased interactions of the genetic material with the plasma membrane. Further, this observation also confirms that the increased interactions in the presence of gene complexes cannot be associated with pDNA phospholipid monolayer interactions that occur in lipoplexes.54 To further verify that the increase in surface pressure was not due to the presence of cations, we also performed control experiments in the presence of 100 mM calcium chloride without any complexes, even though we do not expect such a high concentration of free ions (the calcium is added during the complexation process and is expected to associate with the pDNA/CPP complex). As shown in Figure 3A, the increase in surface pressure in the presence of gene complexes was highly significant, when compared with calcium chloride alone, verifying that the change in the surface pressure is due to phospholipid–gene complex interactions and not due to the presence of calcium ions.
Figure 3.
Changes in surface pressure as a function of time following injection of the seven CPPs (dTAT, K9, R9, RH9, RA9, RL9, and RW9)/pDNA/Ca2+ complexes below POPC (A), POPS (B), and POPG (C) monolayers held at an initial surface pressure of 20 mN/m and at pH 7.4. The maximum change in surface pressure (plateau values) of the seven complexes when inserted below (D) POPC, (E) POPS, and (F) POPG monolayers. Results are presented as mean SD (n = 3) (***p < 0.0001, **p < 0.001, and *p < 0.05, one-way ANOVA, Tukey post test).
Figures 3B and 3E show the recorded changes in the surface pressure for the seven complexes, when injected below the anionic POPS monolayer. Similarly, changes in the surface pressure with time and the maximum change in surface pressure, of a POPG monolayer, due to insertion of the seven complexes, are shown in Figure 3C,F. Our results demonstrated that for both anionic lipids studied here the increase in surface pressure with time was significantly higher for all the complexes, when compared with the free pDNA. Moreover, the amphiphilic RL9 and RW9 containing complexes demonstrated a significant increase in the surface pressure of the anionic phospholipid monolayer, when compared with the hydrophilic complexes. Therefore, Figure 3 provides conclusive evidence that gene complexes containing amphiphilic RL9 and RW9 peptides show enhanced insertion into anionic lipid monolayers at neutral pH.
We and others have previously shown that electrostatic interactions between CPP and phospholipids are enhanced by hydrophobic interactions, if the peptide is also able to penetrate the phospholipid membrane.2,55 To elucidate if this cooperative interaction is also true for our gene complexes, Figure 4 presents a comparison of the maximum change in surface pressure of each of the seven complexes with the three monolayers (POPC, POPS, and POPG) at pH 7.4. The hydrophilic, positively charged dTAT, K9, R9, and RH9 containing gene complexes as well as the amphiphilic RA9 containing complex showed a slight increase in the maximum surface pressure change in the case of POPS monolayers, when compared to the POPC monolayers. However, unlike the gene complexes containing hydrophilic CPPS, the complexes composed of amphiphilic RW9 and RL9 peptides demonstrated a significantly higher increase in surface pressure, when they were injected below a POPS monolayer, compared to a POPC monolayer. Further, POPG showed the maximum increase in surface pressure for all the gene complexes. The overall increase in the insertion potential of gene complexes into anionic POPS and POPG phospholipids can be attributed to electrostatic interactions between the anionic phospholipid monolayers and the cationic gene complexes. However, the significant increase in surface pressure for anionic lipid monolayers in the presence of gene complexes containing RL9 and RW9 supports the idea that addition of hydrophobic amino acids potentially enhances the insertion potential of these complexes. On the basis of our results, we conclude that for the cationic gene complexes studied here the initial surface activity of the complexes in the presence of anionic lipids is probably driven by electrostatic interactions between the charged amino acids and the anionic lipid headgroups, while the insertion is controlled by the hydrophobic nature of the tryptophan and leucine residues. Complexes containing RW9 present the highest insertion into the anionic lipids, possibly because of the strong hydrophobicity or the aromatic ring of tryptophan amino acids in RW9. Complexes with amphiphilic RA9 peptides do not follow this trend, possibly because of the weak hydrophobicity and the small size of the alanine amino acid, which prevents the complexes from penetrating deeper into the monolayer. Therefore, we conclude that calcium condensed gene complexes containing hydrophilic cationic peptides do not disrupt the phospholipid monolayers, but the presence of large hydrophobic amino acid moieties can increase the insertion potential of condensed gene complexes.
Figure 4.
Maximum change in the surface pressure (plateau values) following injection of the seven CPPs ((A) dTAT, (B) K9, (C) R9, (D) RH9, (E) RA9, (F) RL9, and (G) RW9)/pDNA/Ca2+ complexes below POPC, POPS, and POPG monolayers held at an initial pressure of 20 mN/m and at pH 7.4. Results are presented as mean SD (n = 3) (***p < 0.0001 and **p < 0.001, one-way ANOVA, Tukey post test).
However, the difference in the maximum surface pressure of the anionic POPS and POPG lipids in the presence of the gene complexes cannot be explained by electrostatic or hydrophobic interactions. To understand the difference in the insertion into POPS and POPG monolayers, differences in the organization of the phospholipid headgroups should also be considered. Even though both POPS and POPG are anionic phospholipids, the dipole moment P−–N+ for the PS and PG headgroups are different. The polar PS headgroup has both a positive charge due to the NH3+ and a negative charge due to the presence of COO− groups. As a result, the cationic gene complexes can interact with the phospholipid headgroup without significant insertion into the membrane. On the other hand, the polar PG headgroup is itself uncharged. The negative charge on PG is due to the phosphate moiety that binds the polar headgroup with the glycerol backbone. This implies that the gene complexes would have to insert deeper into the PG membranes to interact with the negatively charged phosphate moeity, which was indeed found to be the case.
3.3. Insertion of the CPP/PDNA/Ca2+ Complexes into POPC, POPS, and POPG PMs at Acidic pH
Cytoplasmic delivery of genetic material to subcellular targets can be achieved by initiating endosomal escape of the macromolecules and thus preventing their enzymatic degradation in the lysosome.56 One possible route of endosomal escape is via disruption of the endosomal membranes, which would result in leakage of the genetic payload.12,25 Since the transmembrane pH has been proposed to play a significant role in endosomal escape of peptides, we hypothesized that changes in the subphase pH (from a neutral pH in the plasma membrane to an acidic pH in endosomes) can also effect the interactions of our gene complexes with model phospholipid membranes.25 Since the pH decreases along the endocytic pathway from the early endosome (pH 6) via the late endosome (pH 5.0) to the lysosome (pH 4.5), we have chosen to study the effect of the most acidic pH on the penetration potential and stability of our CPP/pDNA complexes.15
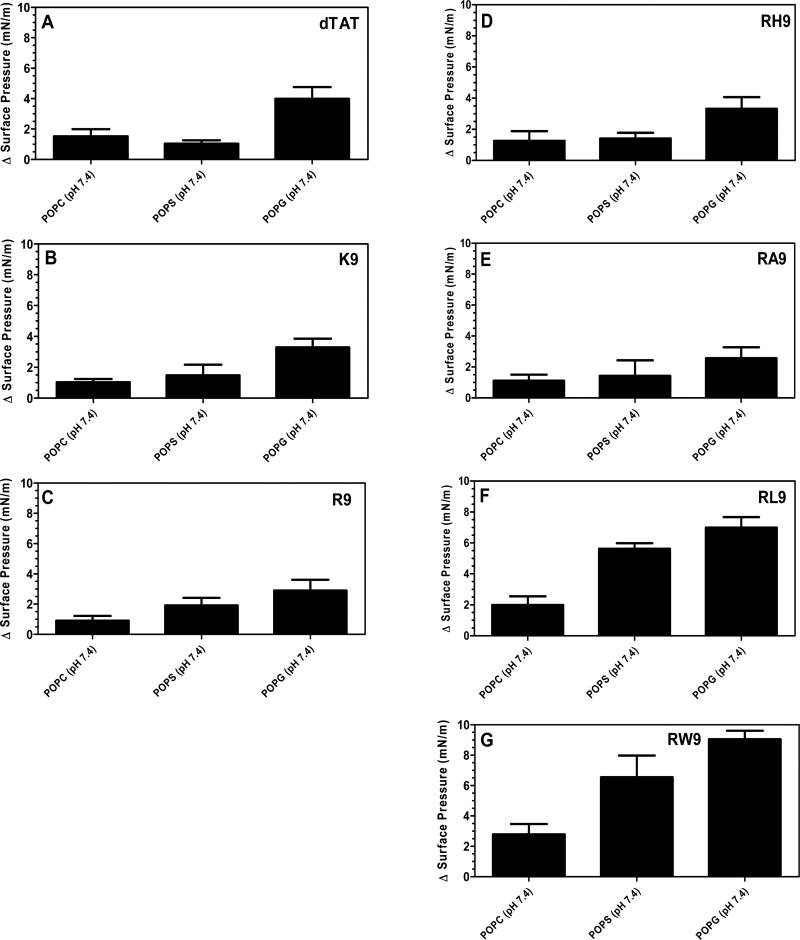
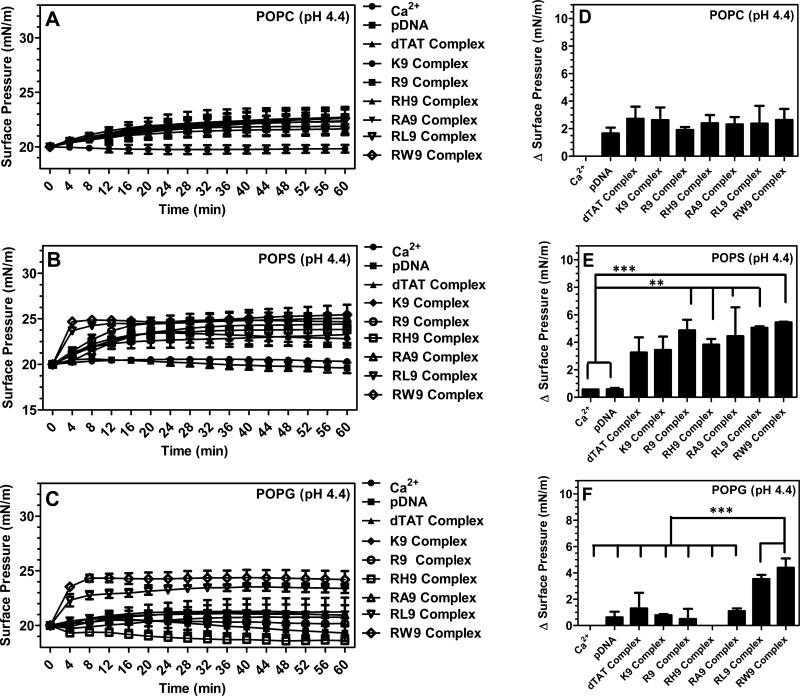
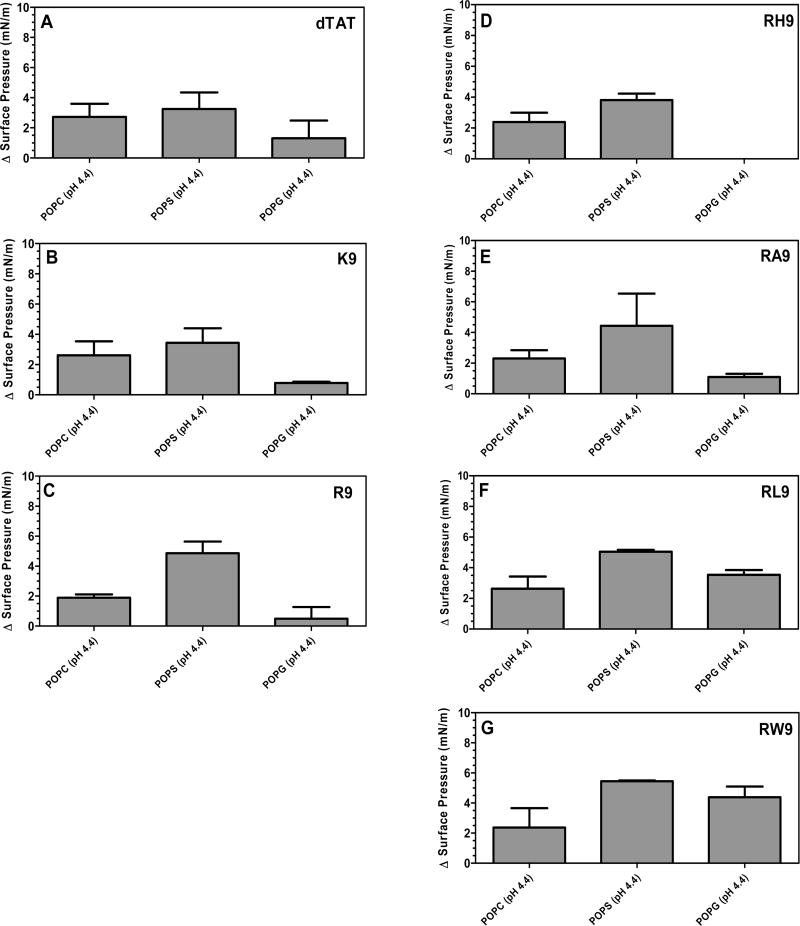
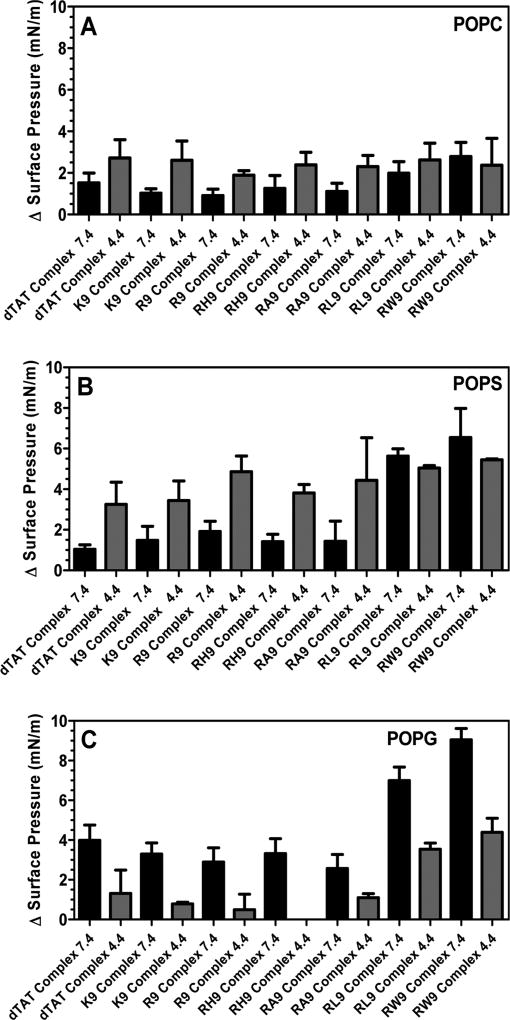
Figure 5 shows the changes in surface pressure of a phospholipid monolayer as a function of time, following interactions with the seven complexes at a subphase pH of 4.4. Figure 5A represents the measured change in surface pressure with time while Figure 5D shows the maximum change in surface pressure that was measured for each of the complexes, when injected below a POPC monolayer. We found that there was no statistically significant difference in the insertion potential of the gene complexes compared with pDNA. On the other hand, Figures 5B and 5E show that both the measured change in surface pressure with time and the maximum change in surface pressure are significantly higher for the gene complexes containing arginine based CPPs compared to the free pDNA, when injected below the POPS monolayers. The other complexes also exhibit higher surface activities compared to the free pDNA. Similarly, Figures 5C and 5F show that both the measured change in surface pressure with time and the maximum change in surface pressure of POPG monolayers is significantly higher for the gene complexes compared to the free pDNA, although the change is lower than for the POPS monolayers. In order to explore the role of electrostatics on the insertion potential of each gene complex into different phospholipid membranes at acidic subphase pH, Figure 6 presents a comparison of the maximum change in surface pressure of each of the seven complexes, when injected below POPC, POPS, or POPG monolayers at a subphase pH of 4.4. Further, Figure 7 presents a comparison of the maximum change in surface pressure of the seven complexes, when injected below POPC, POPS, or POPG monolayers at the two pHs, to develop a fundamental understanding of the effect of pH on the change in the surface pressure of different phospholipid monolayers. Complexes containing hydrophilic peptides show an increase in membrane insertion in a more acidic pH compared to neutral pH. However, as seen in Figure 7C, the gene complexes showed a reduced tendency to change the surface pressure when injected below POPG monolayers at a subphase pH of 4.4.
Figure 5.
Changes in the surface pressure as a function of time following injection of the seven CPPs (dTAT, K9, R9, RH9, RA9, RL9, and RW9)/pDNA/Ca2+ complexes below POPC (A), POPS (B), and POPG (C) monolayers held at an initial surface pressure of 20 mN/m and at pH 4.4. The maximum change in the surface pressure (plateau values) of the seven complexes upon interaction with (D) POPC, (E) POPS, and (F) POPG monolayers. Results are presented as mean SD (n = 3) (***p < 0.0001, **p < 0.001, and *p < 0.05, one-way ANOVA, Tukey post test).
Figure 6.
Maximum change in surface pressure (plateau values) following injection of the seven CPPs (((A) dTAT, (B) K9, (C) R9, (D) RH9, (E) RA9, (F) RL9, and (G) RW9)/pDNA/Ca2+ complexes below POPC, POPS, and POPG PMs held at an initial SP of 20 mN/m and at pH 4.4. Results are presented as mean SD (n = 3) (***p < 0.0001 and **p < 0.001, one-way ANOVA, Tukey post test).
Figure 7.
Maximum change in surface pressure (plateau values) of (A) POPC, (B) POPS, and (C) POPG held at an initial SP of 20 mN/m and at pH 7.4 and 4.4. in the presence of the seven CPPs (dTAT, K9, R9, RH9, RA9, RL9, and RW9)/pDNA/Ca2+ complexes. Results are presented as mean SD (n = 3).
To interpret the differences in the pH-induced changes in the membrane insertion potential of our condensed CPP/pDNA complexes, we first discuss the effects of pH on the phospholipid headgroup charge. While the zwitterionic PC headgroups are stable at both pHs, the acidic environment can lower the relative charge on the anionic lipids. The POPS phospholipid has three pKas ~ 11.55, 5.5, and 2.6 (R–NH3, R–COOH, and R–H2PO, respectively), whereas the POPG phospholipid has one pKa ~ 3.5 (R2-HPO4) at an ionic strength of 0.1 M NaCl.57 Therefore, both PS and PG lipids undergo a decrease in the relative headgroup charge at lower pHs, although this effect is more pronounced for the PS monolayers. For the PS and PC monolayers, the net headgroup charge is zero (i.e., the lipid headgroups are zwitterionic) at a pH of 4.4, while the PG headgroup has a negative relative charge. Since the charge on the zwitterionic PC headgroup remains unaltered by a drop in the subphase pH, the increase in phospholipid insertion potential of the complexes at pH 4.4 is unclear at this time. It can be attributed to the physical characteristics of the gene complexes at pH 4.4. Particle size analysis shows that the gene complexes become larger at the lower pH, while zeta-potential measurements demonstrate that the particles become neutral or anionic. Previous results with gold nanoparticles show that positively charged nanoparticles penetrate into a zwitterionic bilayer at a neutral pH, while negatively charged particles do not.58 Applying this knowledge to our system would imply that the gene complexes demonstrate a lowering of the surface pressure change, contrary to results presented here. Therefore, we conclude that the increase in the environmental acidity helps the ability of the gene complexes to penetrate the zwitterionic phospholipid monolayer by increasing the size of the complex. In the case of the PS phospholipid, a pure electrostatic interaction based argument would imply a decrease in the maxium surface pressure in the acidic pH, which is contrary to the observed results. Therefore, on the basis of our results, we conclude that even for the PS lipids, the increased size of the gene complexes helps with their insertion potential. In fact, even for complexes that seem to dissociate at lower pHs (RA9 and RL9), a significant increase in the insertion was noted, further validating our conclusion that for phospholipid with a net neutral headgroup charge, unstable complexes or complexes with large size increase the insertion potential of the complex into phospholipids when compared with condensed complexes. However, the behavior of the complexes when interacting with the POPG lipids is possibly controlled by the electrostatic repulsion between the anionic gene complexes and the anionic lipid headgroup, which results in a decrease in the insertion potential into the POPG monolayers. It must be noted that while PG headgroups are not common in the late endosome membranes, late endosomes are characterized by an increase in the composition of anionic lipids including BMP, which is a structural isomer of PG. Our results imply that for the calcium condensed gene complexes studied here the enrichment of anionic lipids in the late endosome would prevent the insertion and escape of the genetic payload, but early endosomes rich in anionic lipids can enable endosomal escape and delivery of the genetic material in the cytosol.
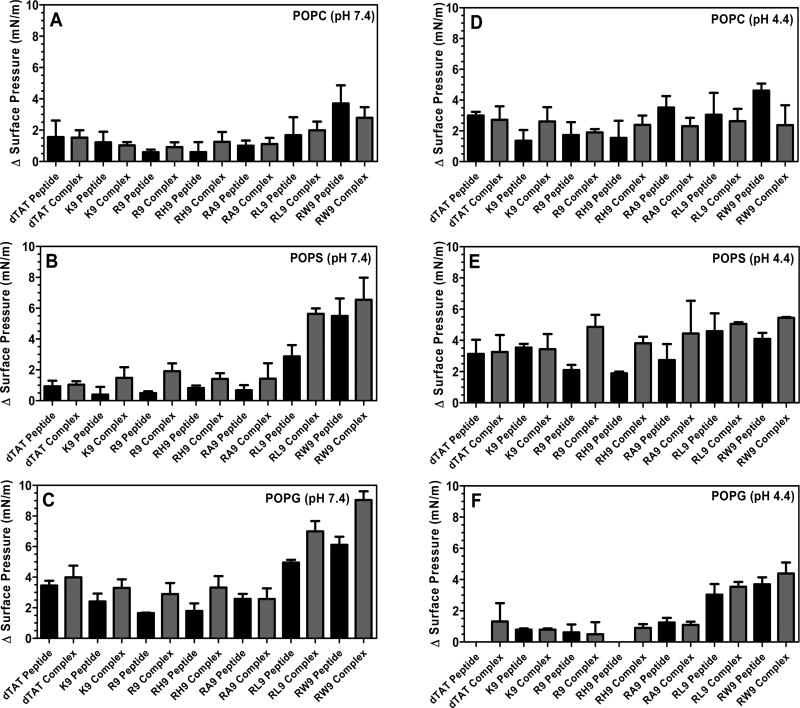
To further understand if the insertion is due to the pDNA, the CPPs or both, we have also measured the changes in the surface pressure of the seven CPPs alone at pH 7.4 and 4.4, when injected below the POPC, POPS, and POPG monolayers (Supporting Information Figures S2 and S3). Our results are summarized in Figure 8, which presents a comparison of the maximum change in the SP of the seven complexes and the seven CPPs at pH 7.4 and 4.4. Generally, the maximum change in surface pressure of the phospholipids was found to be higher for the gene complexes compared to the free CPPs when they were injected below the anionic phospholipids (i.e., POPS and POPG) (Figure 8B,C,E,F), particularly for the complexes containing amphiphilic peptides RW9 and RL9. On the other hand, no difference was noted when the different complexes were injected below the zwitterionic lipid (POPC) (Figure 8A,D). Since the total CPP concentration was kept constant in both the experiments (free CPPs vs condensed complexes), and we expect most of the CPP to exist as a condensed complex with the pDNA, the increase in the surface pressure of the phospholipids in the presence of gene complexes can be attributed to the presence of CPPs on the complexes.
Figure 8.
Maximum change in surface pressure (plateau values) of (A) POPC at pH 7.4, (B) POPS at pH 7.4, (C) POPG at pH 7.4, (D) POPC at pH 4.4, (E) POPS at pH 4.4, and (F) POPG at pH 4.4 due to interactions with the seven CPPs (dTAT, K9, R9, RH9, RA9, RL9, and RW9) and the seven CPPs (dTAT, K9, R9, RH9, RA9, RL9, and RW9)/pDNA/Ca2+ complexes. The initial surface pressure of the monolayers was 20 mN/m. Results are presented as mean SD (n = 3).
Therefore, our results suggest that the mechanisms of interactions of gene complexes with phospholipid membranes during cellular uptake and intracellular delivery are determined by both the changes in the physical characteristics of the gene complexes and the phospholipid headgroup charge. Further, our results also confirm that among all the polypeptides used as CPPs, condensed complexes of pDNA with amphiphilic CPPs show the maximum membrane insertions into anionic phospholipids at normal pH. However, no difference is noted in the insertion potential of the seven peptides into zwitterionic POPC monolayers. A schematic of our proposed mechanism of gene complex action is shown in Figure 9. On the basis of our results, we propose that the CPP/pDNA complexes initially interact with the zwitterionic PC headgroups and trigger cellular uptake, regardless of the composition of the CPP. Once endocytosed, the complexes undergo a pH induced partial disassembly of the complex. This uncondensed gene complex induces changes in the membranes, possibly by interacting with the CPP, allowing the pDNA to escape into the cytosol. Surprisingly, we found that at the more acidic pH the insertion potential of the gene complexes did not depend on the composition of the CPPs. Analysis of kinetics of adsorption of the complexes into different phospholipid systems will provide additional understanding of the nature of the adsorption and insertion of the complexes into different monolayer systems at neutral and acidic pHs and is the topic for a future publication. The findings described here make the noncovalently condensed gene complexes interesting candidates as nonviral gene delivery vectors. Future studies will focus on in vitro and in vivo performance of these gene delivery vectors.
Figure 9.
Schematic of our proposed mechanism of gene complex action at neutral pH (cytoplasm) and acidic pH (late endosome and lysosome).
4. CONCLUSION
Understanding the interactions of gene complexes with phospholipid membranes can be an important first step in predicting the membrane insertion efficiency of calcium condensed gene complexes containing CPPs and efficiently delivering genetic material to the cytosolic environment. In this study, phospholipid membranes POPC, POPS, and POPG have been used to study the membrane insertion of the seven CPP/pDNA/Ca2+ complexes, containing hydrophilic (i.e., dTAT, K9, R9, and RH9) and amphiphilic (i.e., RA9, RL9, and RW9) CPPs, at different pHs (pH 7.4 and 4.4). Our results showed that using CPPs with differences in the net charge and hydrophobicity did not alter their ability to form complexes with pDNA. In fact, the complexes had the same size at neutral pH. Further, the complexes were found to be cationic due to an excess of the CPP during the synthesis process. However, an increase in the acidity of the environment did cause an increase in the particle size, possibly because of partial dissociation of the gene complex, which also caused a decrease in the surface charge on the particles. Using Langmuir monolayers as our model phospholipid membranes, we also showed that there was no difference in the insertion potential of gene complexes of different composition, when interacting with a zwitterionic phospholipid at a neutral pH. However, when interacting with anionic phospholipid membranes, all complexes showed an increase in their insertion potential, although there was a significant difference in the membrane insertion potential of complexes containing amphiphilic CPPs. This suggests that while the initial interactions of our calcium condensed gene complexes with phospholipid membranes are facilitated by electrostatic interactions, the hydrophobic amino acids further facility membrane insertion. However, in more acidic environments such as those in the intracellular vesicles (endosomes/lysosomes) all gene complexes show a greater insertion into the POPC and POPS membranes, irrespective of the composition. Since both POPC and POPS are zwitterionic at the acidic pH, we conclude that the acidic pH induced an increase in the size of the gene complexes possibly due to a reversal of the condensing effect of Ca2+. This increase in the complex size may trigger the disturbance and even disruption of endosomal membranes, leading to escape of the genetic payload into the cytosol. In conclusion, the Langmuir monolayer technique can help to determine the type and degree of interaction of gene complexes with membrane systems. We believe that measuring the insertion potential of gene complexes into model membranes will serve as a screening method to test the potential insertion ability of gene complexes, before they are tested in cells or in more involved therapeutic situations. Our results also suggest that, overall, designing gene complexes with amphiphilic peptides may not necessarily demonstrate more transfection efficiency because of the differences in the insertion potential of the amphiphilic peptides at neutral and acidic pH, even though the amphiphilic peptides have the potential to disrupt anionic membranes at neutral pH. However, additional experiments with in vitro cell assays are essential to obtain a complete understanding on the mechanism of action of the full transfection mechanism. Ultimately, our results will aid in the design of the next generation of CPP-based nonviral gene complexes and is a first step in our overall goal of using short synthetic CPPs for efficient intracellular delivery of nucleic acids. Further, our results also propose the potential of synthetic CPPs with hydrophobic amino acid sequences for use in designing antibacterial membranes, where the CPPs will interact with anionic PG headgroups and may enable membrane disruptions.
Supplementary Material
Acknowledgments
We acknowledge financial support from the following sources: Higuchi Biosciences Center, the NIH (P20 GM103638) and Faculty of Pharmacy, King Abdulaziz University, Jeddah, Saudi Arabia.
Footnotes
ASSOCIATED CONTENT
Figure S1 showing the maximum change in surface pressure of the seven CPPs (dTAT, K9, R9, RH9, RA9, RL9, and RW9) below POPG phospholipid monolayers held at an initial surface pressure of 20 mN/m and at pH 7.4; Figure S2 displaying the changes in surface pressure as a function of time and the maximum change in surface pressure following injection of the seven CPPs (dTAT, K9, R9, RH9, RA9, RL9, and RW9) below POPC, POPS, and POPG phospholipid monolayers held at an initial surface pressure of 20 mN/m and at pH 7.4; Figure S3 showing the changes in surface pressure as a function of time and the maximum change in surface pressure following injection of the seven CPPs (dTAT, K9, R9, RH9, RA9, RL9, and RW9) below POPC, POPS, and POPG phospholipid monolayers held at an initial surface pressure of 20 mN/m and at pH 4.4. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
References
- 1.Doherty GJ, McMahon HT. Mechanisms of endocytosis. Annu. Rev. Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 2.Alhakamy NA, Kaviratna A, Berkland CJ, Dhar P. Dynamic measurements of membrane insertion potential of synthetic cell penetrating peptides. Langmuir. 2013;29(49):15336–15349. doi: 10.1021/la403370p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Figueiredo IR, Freire JM, Flores L, Veiga AS, Castanho MA. Cell-penetrating peptides: A tool for effective delivery in gene-targeted therapies. IUBMB Life. 2014;66(3):182–194. doi: 10.1002/iub.1257. [DOI] [PubMed] [Google Scholar]
- 4.Margus H, Padari K, Pooga M. Cell-penetrating peptides as versatile vehicles for oligonucleotide delivery. Mol. Ther. 2012;20(3):525–533. doi: 10.1038/mt.2011.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakamura Y, Kogure K, Futaki S, Harashima H. Octaarginine-modified multifunctional envelope-type nano device for siRNA. J. Controlled Release. 2007;119(3):360–367. doi: 10.1016/j.jconrel.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 6.Baoum AA, Berkland C. Calcium condensation of DNA complexed with cell-penetrating peptides offers efficient, noncytotoxic gene delivery. J. Pharm. Sci. 2011;100(5):1637–1642. doi: 10.1002/jps.22407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang S-T, Zaitseva E, Chernomordik LV, Melikov K. Cell-penetrating peptide induces leaky fusion of liposomes containing late endosome-specific anionic lipid. Biophys. J. 2010;99(8):2525–2533. doi: 10.1016/j.bpj.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belting M, Wittrup A. Macromolecular drug delivery: basic principles and therapeutic applications. Mol. Biotechnol. 2009;43(1):89–94. doi: 10.1007/s12033-009-9185-5. [DOI] [PubMed] [Google Scholar]
- 9.Chen HH, Ho Y-P, Jiang X, Mao H-Q, Wang T-H, Leong KW. Quantitative comparison of intracellular unpacking kinetics of polyplexes by a model constructed from quantum dot-FRET. Mol. Ther. 2008;16(2):324–332. doi: 10.1038/sj.mt.6300392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karagiannis ED, Alabi CA, Anderson DG. Rationally designed tumor-penetrating nanocomplexes. ACS Nano. 2012;6(10):8484–8487. doi: 10.1021/nn304707b. [DOI] [PubMed] [Google Scholar]
- 11.Khondee S, Baoum A, Siahaan TJ, Berkland C. Calcium condensed LABL-TAT complexes effectively target gene delivery to ICAM-1 expressing cells. Mol. Pharmaceutics. 2011;8(3):788–798. doi: 10.1021/mp100393j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alhakamy NA, Nigatu AS, Berkland CJ, Ramsey JD. Noncovalently associated cell-penetrating peptides for gene delivery applications. Ther. Delivery. 2013;4(6):741–757. doi: 10.4155/tde.13.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen S, Han K, Yang J, Lei Q, Zhuo R-X, Zhang X-Z. Bioreducible polypeptide containing cell-penetrating sequence for efficient gene delivery. Pharm. Res. 2013;30(8):1968–1978. doi: 10.1007/s11095-013-1040-5. [DOI] [PubMed] [Google Scholar]
- 14.Baoum A, Xie S-X, Fakhari A, Berkland C. “Soft” calcium crosslinks enable highly efficient gene transfection using TAT peptide. Pharm. Res. 2009;26(12):2619–2629. doi: 10.1007/s11095-009-9976-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hullin-Matsuda F, Taguchi T, Greimel P, Kobayashi T. Seminars in Cell & Developmental Biology. Elsevier; Amsterdam: 2014. Lipid compartmentalization in the endosome system. [DOI] [PubMed] [Google Scholar]
- 16.Zaitseva E, Yang S-T, Melikov K, Pourmal S, Chernomordik LV. Dengue virus ensures its fusion in late endosomes using compartment-specific lipids. PLoS Pathog. 2010;6(10):e1001131. doi: 10.1371/journal.ppat.1001131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alhakamy NA, Berkland CJ. Polyarginine molecular weight determines transfection efficiency of calcium condensed complexes. Mol. Pharmaceutics. 2013;10(5):1940–1948. doi: 10.1021/mp3007117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ziegler A, Li Blatter X, Seelig A, Seelig J. Protein transduction domains of HIV-1 and SIV TAT interact with charged lipid vesicles. Binding mechanism and thermodynamic analysis. Biochemistry. 2003;42(30):9185–9194. doi: 10.1021/bi0346805. [DOI] [PubMed] [Google Scholar]
- 19.Lindgren M, Hällbrink M, Prochiantz A, Langel Ü. Cell-penetrating peptides. Trends Pharmacol. Sci. 2000;21(3):99–103. doi: 10.1016/s0165-6147(00)01447-4. [DOI] [PubMed] [Google Scholar]
- 20.Baoum A, Ovcharenko D, Berkland C. Calcium condensed cell penetrating peptide complexes offer highly efficient, low toxicity gene silencing. Int. J. Pharm. (Amsterdam, Neth.) 2012;427(1):134–142. doi: 10.1016/j.ijpharm.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 21.Wagstaff KM, Jans DA. Protein transduction: cell penetrating peptides and their therapeutic applications. Curr. Med. Chem. 2006;13(12):1371–1387. doi: 10.2174/092986706776872871. [DOI] [PubMed] [Google Scholar]
- 22.Walrant A, Vogel A, Correia I, Lequin O, Olausson BE, Desbat B, Sagan S, Alves ID. Membrane interactions of two arginine-rich peptides with different cell internalization capacities. Biochim. Biophys. Acta, Biomembr. 2012;1818(7):1755–1763. doi: 10.1016/j.bbamem.2012.02.024. [DOI] [PubMed] [Google Scholar]
- 23.Järver P, Langel Ü. Cell-penetrating peptides—a brief introduction. Biochim. Biophys. Acta, Biomembr. 2006;1758(3):260–263. doi: 10.1016/j.bbamem.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 24.Deshayes S, Morris M, Divita G, Heitz F. Cell-penetrating peptides: tools for intracellular delivery of therapeutics. Cell. Mol. Life Sci. 2005;62(16):1839–1849. doi: 10.1007/s00018-005-5109-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwieger C, Blume A. Interaction of poly (l-arginine) with negatively charged DPPG membranes: calorimetric and monolayer studies. Biomacromolecules. 2009;10(8):2152–2161. doi: 10.1021/bm9003207. [DOI] [PubMed] [Google Scholar]
- 26.Rothbard JB, Jessop TC, Lewis RS, Murray BA, Wender PA. Role of membrane potential and hydrogen bonding in the mechanism of translocation of guanidinium-rich peptides into cells. J. Am. Chem. Soc. 2004;126(31):9506–9507. doi: 10.1021/ja0482536. [DOI] [PubMed] [Google Scholar]
- 27.Futaki S. Oligoarginine vectors for intracellular delivery: Design and cellular-uptake mechanisms. Pept. Sci. 2006;84(3):241–249. doi: 10.1002/bip.20421. [DOI] [PubMed] [Google Scholar]
- 28.Dennison SR, Phoenix AJ, Phoenix DA. Effect of salt on the interaction of Hal18 with lipid membranes. Eur. Biophys. J. 2012;41(9):769–776. doi: 10.1007/s00249-012-0840-6. [DOI] [PubMed] [Google Scholar]
- 29.Futaki S, Goto S, Sugiura Y. Membrane permeability commonly shared among arginine-rich peptides. J. Mol. Recognit. 2003;16(5):260–264. doi: 10.1002/jmr.635. [DOI] [PubMed] [Google Scholar]
- 30.Futaki S, Nakase I, Tadokoro A, Takeuchi T, Jones A. Arginine-rich peptides and their internalization mechanisms. Biochem. Soc. Trans. 2007;35(4):784. doi: 10.1042/BST0350784. [DOI] [PubMed] [Google Scholar]
- 31.Witte K, Olausson BE, Walrant A, Alves ID, Vogel A. Structure and dynamics of the two amphipathic arginine-rich peptides RW9 and RL9 in a lipid environment investigated by solid-state NMR and MD simulations. Biochim. Biophys. Acta, Biomembr. 2013;1828(2):824–833. doi: 10.1016/j.bbamem.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 32.Kawabata A, Baoum A, Ohta N, Jacquez S, Seo G-M, Berkland C, Tamura M. Intratracheal administration of a nanoparticle-based therapy with the angiotensin II type 2 receptor gene attenuates lung cancer growth. Cancer Res. 2012;72(8):2057–2067. doi: 10.1158/0008-5472.CAN-11-3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zaitsev S, Haberland A, Otto A, Vorob’ev V, Haller H, Böttger M. H1 and HMG17 extracted from calf thymus nuclei are efficient DNA carriers in gene transfer. Gene Ther. 1997;4(6) doi: 10.1038/sj.gt.3300433. [DOI] [PubMed] [Google Scholar]
- 34.Khosravi-Darani K, Mozafari MR, Rashidi L, Mohammadi M. Calcium based non-viral gene delivery: an overview of methodology and applications. Acta Med. Iran. 2010;48(3):133–141. [PubMed] [Google Scholar]
- 35.Jiao C-Y, Delaroche D, Burlina F, Alves ID, Chassaing G, Sagan S. Translocation and endocytosis for cell-penetrating peptide internalization. J. Biol. Chem. 2009;284(49):33957–33965. doi: 10.1074/jbc.M109.056309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blume A. A comparative study of the phase transitions of phospholipid bilayers and monolayers. Biochim. Biophys. Acta, Biomembr. 1979;557(1):32–44. doi: 10.1016/0005-2736(79)90087-7. [DOI] [PubMed] [Google Scholar]
- 37.Joanne P, Galanth C, Goasdoué N, Nicolas P, Sagan S, Lavielle S, Chassaing G, El Amri C, Alves ID. Lipid reorganization induced by membrane-active peptides probed using differential scanning calorimetry. Biochim. Biophys. Acta, Biomembr. 2009;1788(9):1772–1781. doi: 10.1016/j.bbamem.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 38.Bally MB, Harvie P, Wong FM, Kong S, Wasan EK, Reimer DL. Biological barriers to cellular delivery of lipid-based DNA carriers. Adv. Drug Delivery Rev. 1999;38(3):291–315. doi: 10.1016/s0169-409x(99)00034-4. [DOI] [PubMed] [Google Scholar]
- 39.Peetla C, Jin S, Weimer J, Elegbede A, Labhasetwar V. Biomechanics and thermodynamics of nanoparticle interactions with plasma and endosomal membrane lipids in cellular uptake and endosomal escape. Langmuir. 2014;30(25):7522–7532. doi: 10.1021/la5015219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loyter A, Scangos GA, Ruddle FH. Mechanisms of DNA uptake by mammalian cells: fate of exogenously added DNA monitored by the use of fluorescent dyes. Proc. Natl. Acad. Sci. U. S. A. 1982;79(2):422–426. doi: 10.1073/pnas.79.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haberland A, Knaus T, Zaitsev SV, Stahn R, Mistry AR, Coutelle C, Haller H, Böttger M. Calcium ions as efficient cofactor of polycation-mediated gene transfer. Biochim. Biophys. Acta, Gene Struct. Expression. 1999;1445(1):21–30. doi: 10.1016/s0167-4781(99)00017-2. [DOI] [PubMed] [Google Scholar]
- 42.Bisht S, Bhakta G, Mitra S, Maitra A. pDNA loaded calcium phosphate nanoparticles: highly efficient non-viral vector for gene delivery. Int. J. Pharm. (Amsterdam, Neth.) 2005;288(1):157–168. doi: 10.1016/j.ijpharm.2004.07.035. [DOI] [PubMed] [Google Scholar]
- 43.Eeman M, Deleu M. From biological membranes to biomimetic model membranes. Base. 2010;14:719–736. [Google Scholar]
- 44.Zhang L, Rozek A, Hancock RE. Interaction of cationic antimicrobial peptides with model membranes. J. Biol. Chem. 2001;276(38):35714–35722. doi: 10.1074/jbc.M104925200. [DOI] [PubMed] [Google Scholar]
- 45.Song C, Nerdal W. Olanzapine interaction with dipalmitoyl phosphatidylcholine (DPPC) and 1-palmitoyl-2-oleoyl phosphatidylserine (POPS) bilayer: A 13C and 31P solid-state NMR study. Biophys. Chem. 2008;134(1):47–55. doi: 10.1016/j.bpc.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 46.Van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9(2):112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leventis PA, Grinstein S. The distribution and function of phosphatidylserine in cellular membranes. Ann. Rev. Biophys. 2010;39:407–427. doi: 10.1146/annurev.biophys.093008.131234. [DOI] [PubMed] [Google Scholar]
- 48.Bouvier J, Berry KAZ, Hullin-Matsuda F, Makino A, Michaud S, Geloën A, Murphy RC, Kobayashi T, Lagarde M, Delton-Vandenbroucke I. Selective decrease of bis (monoacylglycero) phosphate content in macrophages by high supplementation with docosahexaenoic acid. J. Lipid Res. 2009;50(2):243–255. doi: 10.1194/jlr.M800300-JLR200. [DOI] [PubMed] [Google Scholar]
- 49.Meikle P, Duplock S, Blacklock D, Whitfield P, Macintosh G, Hopwood J, Fuller M. Effect of lysosomal storage on bis (monoacylglycero) phosphate. Biochem. J. 2008;411:71–78. doi: 10.1042/BJ20071043. [DOI] [PubMed] [Google Scholar]
- 50.Hullin-Matsuda F, Kawasaki K, Delton-Vandenbroucke I, Xu Y, Nishijima M, Lagarde M, Schlame M, Kobayashi T. De novo biosynthesis of the late endosome lipid, bis(monoacylglycero) phosphate. J. Lipid Res. 2007;48(9):1997–2008. doi: 10.1194/jlr.M700154-JLR200. [DOI] [PubMed] [Google Scholar]
- 51.Frederick TE, Goff PC, Mair CE, Farver RS, Long JR, Fanucci GE. Effects of the endosomal lipid bis (monoacylglycero) phosphate on the thermotropic properties of DPPC: A 2H NMR and spin label EPR study. Chem. Phys. Lipids. 2010;163(7):703–711. doi: 10.1016/j.chemphyslip.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frederick TE, Chebukati JN, Mair CE, Goff PC, Fanucci GE. Bis(monoacylglycero) phosphate forms stable small lamellar vesicle structures: Insights into vesicular body formation in endosomes. Biophys. J. 2009;96(5):1847. doi: 10.1016/j.bpj.2008.12.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Urbina P, Flores-Díaz M, Alape-Girón A, Alonso A, Goñi FM. Effects of bilayer composition and physical properties on the phospholipase C and sphingomyelinase activities of Clostridium perf ringens α-toxin. Biochim. Biophys. Acta, Biomembr. 2011;1808(1):279–286. doi: 10.1016/j.bbamem.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 54.Gromelski S, Brezesinski G. DNA condensation and interaction with zwitterionic phospholipids mediated by divalent cations. Langmuir. 2006;22(14):6293–6301. doi: 10.1021/la0531796. [DOI] [PubMed] [Google Scholar]
- 55.Travkova OG, Brezesinski G. Adsorption of the antimicrobial peptide arenicin and its linear derivative to model membranes–A maximum insertion pressure study. Chem. Phys. Lipids. 2013;167:43–50. doi: 10.1016/j.chemphyslip.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 56.Zhang S, Nelson A, Coldrick Z, Chen R. The effects of substituent grafting on the interaction of pH-responsive polymers with phospholipid monolayers. Langmuir. 2011;27(13):8530–8539. doi: 10.1021/la105125d. [DOI] [PubMed] [Google Scholar]
- 57.Marsh D. CRC Handbook of Lipid Bilayers. CRC Press; Boca Raton, FL: 1990. [Google Scholar]
- 58.Tatur S, Maccarini M, Barker R, Nelson A, Fragneto G. Effect of functionalized gold nanoparticles on floating lipid bilayers. Langmuir. 2013;29(22):6606–6614. doi: 10.1021/la401074y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.