Abstract
Proteasome inhibition is an effective therapy for multiple myeloma (MM) patients; however, the emergence of drug resistance is common. Novel therapeutic strategies to overcome proteasome inhibitor resistance are needed. In this study, we examined whether targeting deubiquitylating (DUB) enzymes upstream of 20S proteasome overcomes proteasome inhibitor resistance. Gene expression analysis, immunohistochemical studies of MM patient bone marrow, reverse transcription–PCR and protein analysis show that Rpn11/POH1, a DUB enzyme upstream of 20S proteasome, is more highly expressed in patient MM cells than in normal plasma cells. Importantly, Rpn11 expression directly correlates with poor patient survival. Loss-of-function studies show that Rpn11-siRNA knockdown decreases MM cell viability. Pharmacological inhibition of Rpn11 with O-phenanthroline (OPA) blocks cellular proteasome function, induces apoptosis in MM cells and overcomes resistance to proteasome inhibitor bortezomib. Mechanistically, Rpn11 inhibition in MM cells activates caspase cascade and endoplasmic stress response signaling. Human MM xenograft model studies demonstrate that OPA treatment reduces progression of tumor growth and prolongs survival in mice. Finally, blockade of Rpn11 increases the cytotoxic activity of anti-MM agents lenalidomide, pomalidomide or dexamethasone. Overall, our preclinical data provide the rationale for targeting DUB enzyme Rpn11 upstream of 20S proteasome to enhance cytotoxicity and overcome proteasome inhibitor resistance in MM.
INTRODUCTION
Protein ubiquitylation and deubiquitylation is regulated by various enzymes.1–3 Specifically, ubiquitin ligases link ubiquitin moiety to proteins, thereby facilitating their degradation via the 20S proteasome. In contrast, deubiquitylating enzymes (DUBs) remove ubiquitin from proteins and prevent their degradation.4,5 DUBs play a pivotal role in maintaining cellular protein homeostasis by regulating protein activation, turnover rate, recycling and localization.6–9 Many human diseases are linked to dysfunction of DUBs, suggesting that inhibitors of DUB enzymes represent a potential novel therapeutic strategy.10–12
DUBs are categorized into cysteine proteases (USP, UCH, OTU, MJD) and metalloproteases (JAMM/Jab1/Mov34 metalloenzyme).13 Among these, Rpn11/POH1, UCHL5 and USP14 are primarily responsible for mediating 19S proteasome DUB activity;1–3,14–16 therefore, modulating their function may affect the uptake of protein substrate for degradation through the downstream 20S proteasome subunit. Our previous study showed that inhibition of USP14 and UCHL5 overcomes proteasome inhibitor resistance in multiple myeloma (MM).11,17 To date, the role of Rpn11 in MM biology remains undefined.
Rpn11 DUB removes the ubiquitin chain from the proteins at the 19S proteasome entry gate, allowing for the translocation of proteins from 19S proteasome lid to the 20S proteasome subunit for degradation.18,19 Mutations in the JAMM domain of Rpn11 cause lethality in yeast as well as in mammalian cells.18–22 Other studies showed that knockdown of Rpn11 using small interfering RNA (siRNA) inhibits the proliferation of Hela cells.21,22 These studies suggest that Rpn11 mediates deubiquitylation and protein degradation. In this study, we utilized our in vitro and in vivo models of MM to examine (1) the prognostic relevance and functional significance of Rpn11 and (2) whether blockade of Rpn11 DUB activity triggers cytotoxicity and overcomes bortezomib resistance.
RESULTS
Prognostic relevance and functional significance of Rpn11/POH1 in MM
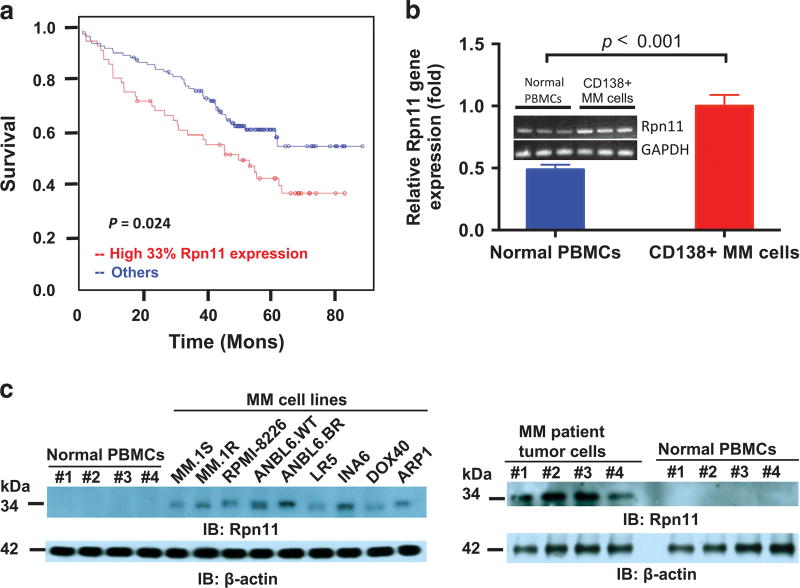
We first analyzed the correlation between baseline Rpn11 gene expression and overall survival of MM patients using gene expression data sets from 170 and 550 newly diagnosed uniformly treated MM patients. High Rpn11 levels at diagnosis correlated with poor overall survival in MM (P = 0.024; Figure 1a and Supplementary Figure 1). Rpn11 expression was analyzed in samples from normal individuals and patients with monoclonal gammopathy of undetermined significance or smoldering multiple myeloma as well as active MM. Rpn11 expression correlates with the progression of the disease (Supplementary Figure 2). Similarly, Rpn11 expression was elevated in tumor cells from patients versus normal plasma cells (Supplementary Figure 3). Reverse transcription–PCR (RT–PCR) analysis confirmed higher Rpn11 levels in patient MM cells compared with normal peripheral blood mononuclear cells (Figure 1b). Consistent with gene expression studies, we found elevated Rpn11 protein levels in MM cells (Figure 1c). Immunohistochemistry analysis of bone marrow (BM) biopsies from MM patients and normal donors showed higher Rpn11 expression in MM cells than normal cells (Supplementary Figure 4). Importantly, transfection of Rpn11-siRNA, but not scr-siRNA, reduced viability of MM cells in a time-dependent manner (Figure 2a, P < 0.0001). Together, our findings imply that Rpn11 may contribute in MM pathogenesis.
Figure 1. Expression and prognostic relevance of deubiquitylating enzyme Rpn11 in MM.
(a) Kaplan–Meier plots of Rpn11 expression versus overall survival of newly diagnosed, uniformly treated, MM patients (n = 170). Red line points to the patient group with elevated Rpn11 expression and shorter survival, whereas blue line represents patient cohort with lower Rpn11 and longer survival. The raw data for expression profiling and the CEL files can be found at the website Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number: GSE39754. (b) RNA from MM patient BM (CD138+) and (n = 3) or normal peripheral blood mononuclear cells (PBMCs; n = 3) were analyzed for Rpn11 expression using RT–PCR. (c) Analyses of Rpn11 expression in MM cell lines, patient tumor cells and normal PBMCs using immunoblotting with indicated antibodies. A representative blot from three independent experiments is shown.
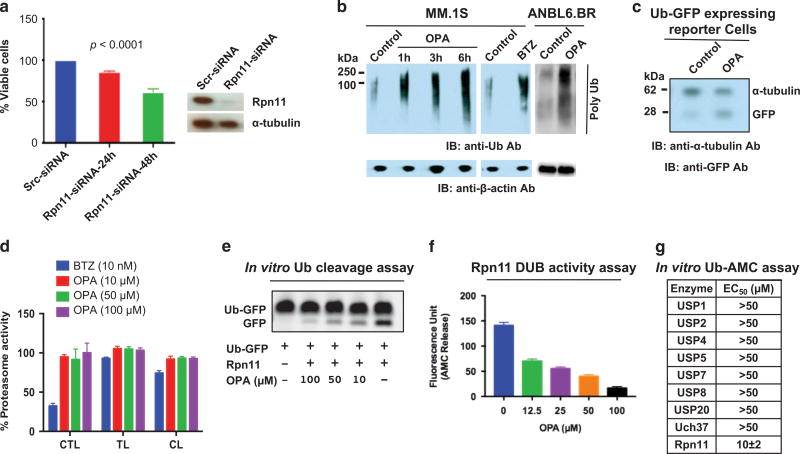
Figure 2. Functional significance of Rpn11.
(a) Effect of Rpn11 knockdown on MM.1S cell viability was assessed using scr-siRNA or Rpn11-siRNA with WST assay at 24 and 48 h post transfection (mean ± s.d.; n = 3; P < 0.0001). Immunoblotting confirmed knockdown of Rpn11 at 24 h post transfection. (b) Effect of OPA (10 µm) versus bortezomib (BTZ; 5 nm) on intracellular polyubiquitylation in MM.1S or ANBL6.BR cells. Immunoblotting was performed with anti-ubiquitin or anti-β-actin antibodies (Abs). (c) Effect of OPA on the accumulation of Ub-GFP in GFPu-1 reporter cell line. GFPu-1 cells were treated with OPA for 15 h, and total cell extracts were analyzed with immunoblotting using anti-GFP, or anti-tubulin Abs. (d) Analysis of proteasomal activities in OPA-or bortezomib-treated MM.1S cells. Cells were treated with indicated drugs for 3 h, and cell extracts were analyzed for proteasome activities (C-L, caspase-like; CT-L, chymotrypsin-like; T-L, trypsin-like proteasome activity) (mean ± s.d.; n = 3). (e) Recombinant human Rpn11 (1 µg) was incubated with OPA for 30 min at room temperature; 1 µg Ub-GFP was then added to the reaction for 1 h, followed by analysis of cleaved-GFP using SDS–polyacrylamide gel electrophoresis (SDS–PAGE) and western blotting with anti-GFP Abs. (f) Recombinant human Rpn11 was incubated with OPA for 30 min at 37 °C, followed by the assessment of DUB activity using Ub-AMC assay (mean ±s.d.; P < 0.005, n=3). (g) Recombinant proteins USP1, USP2, USP4, USP5, USP7, USP8, USP20, UCH37 or Rpn11 were incubated with dimethyl sulfoxide (DMSO) control or OPA for 30 min at 37 °C, followed by assessment of DUB activity using Ub-AMC assay.
OPA targets Rpn11 and blocks protein degradation
A prior study showed Rpn11 metalloprotease mediates deubiquitylation and facilitates protein degradation via proteasome.19 To determine whether Rpn11 blockade affects Rpn11 DUB activity and/or protein degradation in MM cells, we utilized a metallopeptidase inhibitor O-phenanthroline (OPA) that inhibits Rpn11 activity.19 Bortezomib was utilized as an inducer of polyubiquitylation. OPA treatment increased the levels of polyubiquitylated proteins in both bortezomib-sensitive MM.1S and bortezomib-resistant ANBL6.BR cells (Figure 2b). An increase in ubiquitylated proteins is a hallmark of proteasome inhibition,23 and we here observed a similar increase in ubiquitylated protein in OPA-treated MM cells. Furthermore, OPA-treated GFPu-1 cells showed a marked accumulation of Ub-GFP, indicating impaired proteasome-mediated protein degradation (Figure 2c). No significant inhibition of chymotrypsin-like, trypsin-like or caspase-like proteasome activities was observed in OPA-treated MM cells (Figure 2d). Our data are in concert with other studies reporting a similar role for Rpn11 in proteasome-mediated protein degradation.2,3
We next examined whether OPA-triggered blockade in proteasome function is due to inhibition of Rpn11 DUB activity. For these studies, we utilized two distinct assays. First, recombinant human Rpn11 was incubated with dimethyl sulfoxide control or OPA for 30 min; Ub-GFP substrate was then added for 1 h, followed by analysis for cleaved green fluorescent protein (GFP) using SDS–polyacrylamide gel electrophoresis and western blotting with anti-GFP antibody. Results showed that OPA inhibits Rpn11-mediated Ub cleavage from Ub-GFP in a concentration-dependent manner (Figure 2e). Second, we utilized Ub-AMC (ubiquitin 7-amino-4-methycoumar) assays to assess the effect of OPA on Rpn11 versus other DUBs using recombinant DUB proteins. OPA inhibits Rpn11 DUB activity (half-maximal effective concentration range: 10–12 µm) (Figure 2f). Similar analyses confirmed that OPA did not significantly affect the activity of other recombinant DUBs (USP1/USP2/USP4/USP5/USP7/USP8/USP20/UCH37) (see Table in Figure 2g). Taken together, these findings provide evidence for the specificity of OPA against Rpn11.
OPA inhibits MM cell growth and overcomes drug resistance
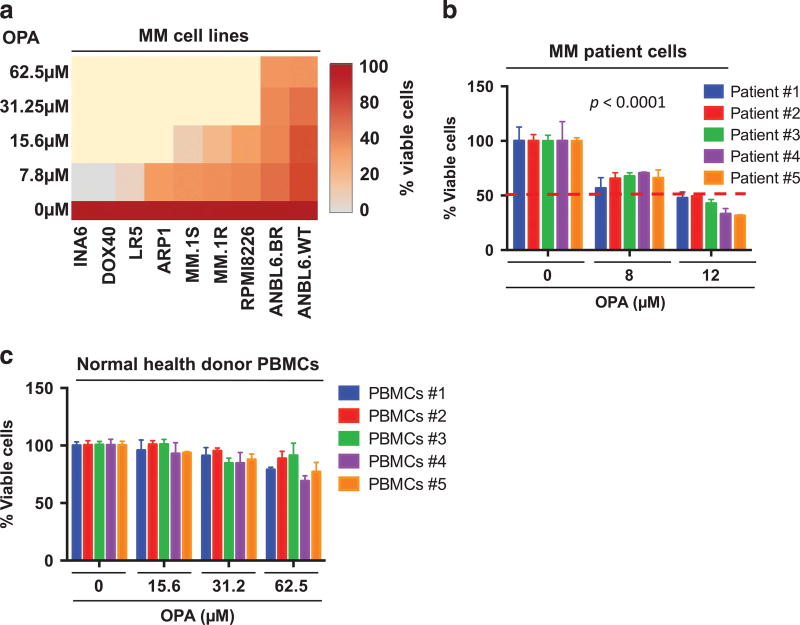
The cytotoxic effect of OPA was examined using MM cell lines as well as tumor cells from MM patients resistant to various therapies (bortezomib, lenalidomide, dexamethasone (Dex)). OPA decreases the viability of various MM cell lines in a concentration-dependent manner (Figure 3a). As for MM cell lines, OPA also reduced the viability of all patient MM cells, including tumor cells from patients refractory to bortezomib, lenalidomide and Dex therapies (Figure 3b). In contrast, OPA at the half-maximal inhibitory concentration (10–12 µm) for patient MM cells showed no significant toxicity against normal peripheral blood mononuclear cells (Figure 3c), suggesting a favorable therapeutic index for OPA in MM.
Figure 3. Rpn11 inhibitor OPA triggers MM cell death.
(a) Various MM cell lines were treated with OPA for 48 h, and cell viability was measured using WST assay (P < 0.05; n = 3). Data are shown in a Heatmap format. (b) Patient MM cells were treated with OPA for 48 h, and cell viability was measured using CellTiter-Glo assay (mean ± s.d. of triplicate cultures; P < 0.0001 for all patient samples). (c) Normal peripheral blood mononuclear cells (PBMCs) were treated with OPA for 48 h, and cell viability was measured using CellTiter-Glo assay (mean ± s.d. of quadruplicate cultures).
OPA inhibits tumor-promoting activity of MM BM microenvironment
Bone marrow stromal cells (BMSCs) regulate MM cell growth and protect against drug cytotoxicity by both tumor cell–BMSC adhesion and cytokine secretion.24 Similarly, plasmacytoid dendritic cells (pDCs) are key components of MM BM milieu, and enhance tumor growth and survival.11,25 Using in vitro BMSC-MM or pDC-MM cell co-culture assays, we show that OPA inhibited both BMSC- or pDC-induced MM.1S growth (Supplementary Figure 5), without affecting the viability of BMSCS or pDCs (data not shown).
Cytotoxic activity of OPA against MM cells
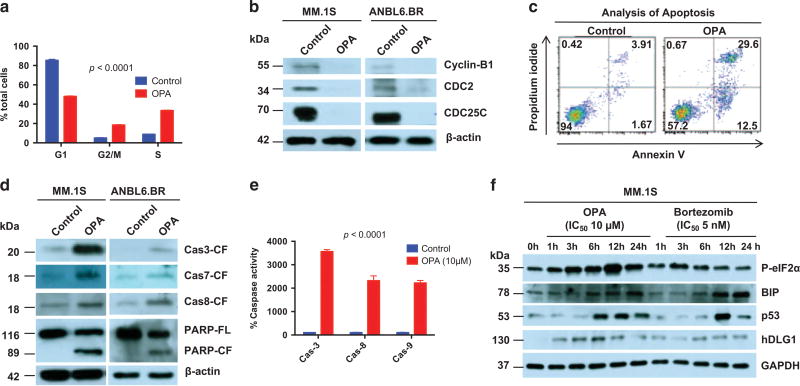
Cell-cycle analysis showed that OPA treatment is associated with increase in S phase and growth arrest in G2/M phase (Figure 4a). OPA treatment decreased growth arrest-related protein CDC25C and its downstream protein CDC2, as well as cyclin B1, in both MM.1S and ANBL6.BR cell lines (Figure 4b). Furthermore, OPA induced apoptosis in MM cells (Figure 4c), evidenced by Annexin V/propidium iodide staining, activation of caspase-3, caspase-7, caspase-8 and caspase-9 as well as poly (ADP-ribose) polymerase cleavage (Figures 4d and e). OPA-triggered apoptosis was markedly inhibited in the presence of a pan-caspase inhibitor (Supplementary Figure 6).
Figure 4. Mechanism of action of OPA in MM cells.
(a) MM.1S cells were treated with OPA (10 µm) for 24 h, followed by analysis of DNA content using propidium iodide (PI) staining (mean±s.d.; n=3; P < 0.001). (b) MM.1S cells were treated with OPA for 12 h, and cell extracts were then analyzed for indicated molecules using western blotting. (c) MM.1S cells were treated with OPA (10 µm) for 24 h, followed by analysis of apoptosis using Annexin V/PI staining (mean ±s.d.; n =3; P < 0.002). (d) MM.1S and ANBL6.BR cells were treated with OPA for 12 h; cell extracts were then analyzed for the indicated molecules using immunoblotting. (e) MM.1S cells were treated with OPA (10 µm) for 12 h, and enzymatic activity of indicated caspases was then analyzed (mean ± s.d.; n = 3; P < 0.0001). (f) MM.1S cells were treated with OPA or bortezomib, and cell extracts were then analyzed for indicated molecules using immunoblotting.
Blockade of Rpn11 DUB activity OPA triggers ER stress response
As shown in Figure 2b, OPA induces accumulation of ubiquitylated proteins. Earlier reports showed that increased intracellular ubiquitylated protein initiates the endoplasmic reticulum (ER) response signaling and cell death,23 and we therefore examined whether OPA induces ER stress response signaling via p-eIf2α and BIP. Parallel experiments were performed using proteasome inhibitor bortezomib as a positive control for ER stress induction. A more robust upregulation of p-eIf2α in MM cells was observed in response to treatment with OPA than bortezomib (Figure 4f). Although similar levels of BIP were induced in OPA- or bortezomib-treated MM cells, OPA triggered an earlier induction of p53 compared with bortezomib (Figure 4f). In addition, OPA robustly induced another tumor suppressor protein hDLG1,26,27 whereas only modest increases in hDLG1 levels were observed in bortezomib-treated cells. Of note, OPA triggered similar induction of ER stress response signaling and tumor suppressor proteins p53 and hDLG1 in bortezomib-resistant ABL6.BR cells (Supplementary Figure 7).
Cytotoxic activity of OPA in in vivo model of MM
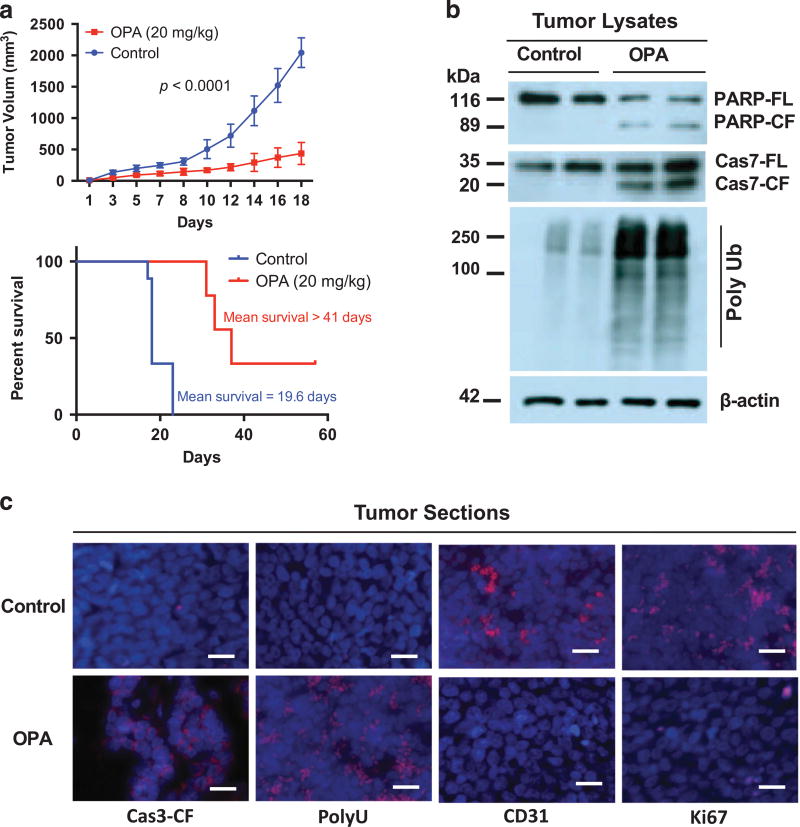
Having defined the anti-MM activity of Rpn11 DUB inhibitor in vitro, we next examined whether OPA similarly affects MM cell growth in vivo using a murine xenograft model of human MM.11,28,29 OPA reduced tumor progression and extended survival time in mice (Figure 5a). In vivo anti-MM activity of OPA is further evident from analysis of mice tumors for the markers of growth inhibition, apoptosis, angiogenesis and polyubiquitylation (Figures 5b and c).
Figure 5. In vivo anti-MM activity of Rpn11 inhibitor OPA.
(a) SCID mice with MM.1S tumors were treated with vehicle control or OPA (20 mg/kg; intraperitoneal (i.p.), 3 × each week) for 18 days. Tumor volume was measured at the indicated times (mean tumor volume ± s.d., 10 mice/group). Mice survival was assessed using Kaplan–Meier plots. (b) Cellular extracts from mice tumors were analyzed for the indicated molecules using immunoblotting. (c) Mice tumors were analyzed for the indicated molecules by immunostaining. All images were obtained with a Nikon inverted TE2000 microscope (40 × magnification; scale bar, 10 µm).
Combining Rpn11 DUB inhibitor OPA with immunomodulatory drugs lenalidomide, pomalidomide or conventional agent Dex triggers synergistic anti-MM activity
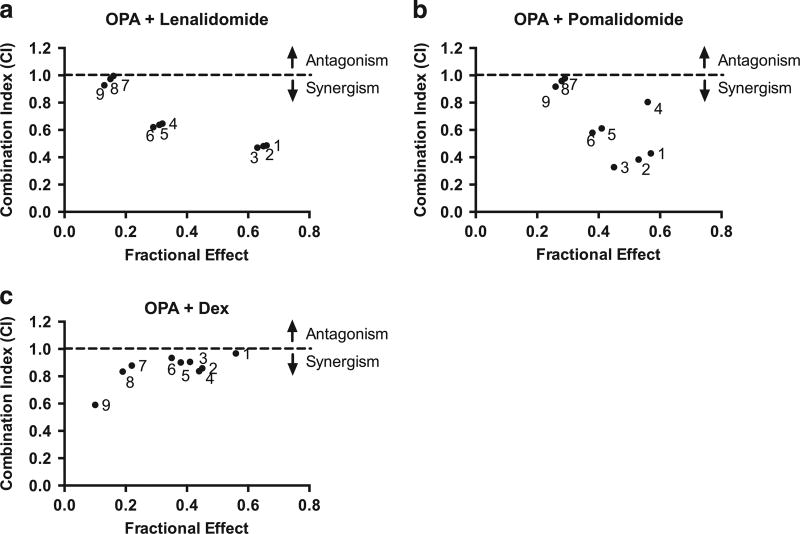
Bortezomib combination therapy is now widely used in the treatment of MM patients. Similar to the mechanism of action of bortezomib, Rpn11 DUB blockade also inhibits protein degradation. Based on these findings, we examined the efficacy of combinations of OPA against MM cells. Results showed that OPA adds to the anti-MM activity of immunomodulatory drugs (pomalidomide and lenalidomide) and conventional agent (Dex) (combination index < 1.0) (Figures 6a–c and Supplementary Figures 8–10).
Figure 6. OPA adds to the anti-MM activity of immunomodulatory agents and Dex.
(a) MM.1S cells were treated for 48 h with OPA, lenalidomide or OPA plus lenalidomide and then analyzed for cell viability. Raw data were subjected to isobologram analysis to measure synergistic activity. The graphs were obtained from the values given in the Supplementary Figure 8. The combination index (CI) of < 1.0 indicates synergy. (b) MM.1S cells were treated for 48 h with OPA, pomalidomide or OPA plus pomalidomide and then analyzed for cell viability. Raw data were subjected to synergy analysis as in (a). The graphs were obtained from the values given in the Supplementary Figure 9. (c) MM.1S cells were treated for 48 h with OPA, Dex or OPA plus Dex and then analyzed for cell viability. Raw data were subjected to synergy analysis as in (a). The graphs were obtained from the values given in the Supplementary Figure 10.
DISCUSSION
In this study we demonstrate that targeting 19S proteasome-associated DUB enzyme Rpn11 triggers apoptosis in MM cells, including those that are resistant to conventional (Dex, melphalan or doxorubicin) and novel agents (bortezomib). Our study highlights for the first time the prognostic and functional relevance of DUB enzyme Rpn11 in MM. Analysis of Gene Expression Profiles (GEP) data set from 170 newly diagnosed uniformly treated MM patients showed that Rpn11 is a poor prognostic factor in MM. RT–PCR and protein expression analysis showed elevated Rpn11 levels in MM versus normal cells. Our data are consistent with the tumor-promoting role of Rpn11 observed in hepatocellular carcinoma models,30 and indicate that Rpn11 contributes in MM pathogenesis.
The role of Rpn11 in MM was further examined using siRNA and pharmacological inhibition strategies. Rpn11-siRNA reduced MM cell viability, as was observed in prior studies of Rpn11 knockdown in yeast and mammalian cells.19,22 Furthermore, Rpn11 knock-down triggered a time-dependent significant decrease in MM.1S cell viability. Rpn11 DUB activity is linked to protein degradation via the proteasome, and we therefore next asked whether Rpn11 inhibition blocks protein degradation in MM cells using Rpn11 inhibitor OPA.19 OPA increased levels of ubiquitylated proteins, reflecting inhibition of cellular protein degradation via the proteasome. Consistent with this finding, results showed accumulation of Ub-GFP, indicating impaired proteasome degradation. Rpn11 is localized in the 19S proteasome subunit, whereas the proteasomal catalytic activities (CT-L, C-L and T-L) mediating protein degradation reside within the 20S proteasome. As expected, OPA blocked cellular proteasome function without inhibiting 20S proteasome proteolytic activities. We next confirmed the specificity of OPA against Rpn11 in two ways: first, we showed that OPA blocks the Rpn11 DUB activity in an in vitro assay using Ub-GFP; and second, we utilized DUB enzymatic assays with recombinant DUBs to show that OPA selectively inhibits Rpn11 activity, without affecting other DUBs (USP1/USP2/USP4/USP5/USP7/USP8/USP20/UCH37). These data show that OPA inhibits both Rpn11 DUB activity and proteasome function, without altering 20S proteolytic activities or other DUBs. The therapeutic strategy to inhibit Rpn11 may be therefore clinically relevant, especially for the MM patients who are refractory to bortezomib therapy. For example, mutations or defects in the 20S core proteolytic activities can confer bortezomib resistance; and importantly, proteasome blockade by targeting Rpn11 will occur irrespective of defects in the proteasomal activities.
Prior studies showed the requirement of Rpn11 DUB activity for survival of mammalian cells.22,30 OPA inhibits this activity and causes MM cell death. Anti-MM activity of OPA was observed against cells exhibiting varied cytogenetic abnormalities as well as those that are resistant to current therapies.31–35 Furthermore, OPA decreased MM cell viability even in the presence of the MM-host BM microenvironment. Our findings therefore demonstrate that blockade of Rpn11 DUB activity in MM cells using OPA overcomes the tumor-protective activity of the MM-host BM milieu.
Delineation of OPA-induced MM cell death signaling pathways showed involvement of apoptotic caspases. Moreover, OPA induced p53 and ER stress signaling cascade. Our finding that OPA decreases viability of p53-null ARP1 MM cells supports the therapeutic potential of Rpn11 blockade in patients with p53 mutations. Of note, the anti-MM activity of OPA was observed even against bortezomib-resistant MM cells. Other studies have reported a role of Rpn11 in signaling pathways mediating cell differentiation, DNA repair pathways and transcription, as well as in multidrug resistance and mitochondrial function.36–40 However, it remains to be determined whether all these functions of Rpn11 are linked to DUB activity of Rpn11; and importantly, whether Rpn11 inhibition will modulate these biological processes in MM cells.
We also confirmed the in vitro anti-MM activity of OPA in using in vivo xenograft models of human MM. OPA blocks tumor growth progression in vivo, and extend host survival. Moreover, tumor cell apoptosis is associated with anti-angiogenic activity in vivo.
Combining therapeutic agents targeting protein degradation with immunomodulatory drugs has demonstrated synergistic preclinical MM cytotoxicity.41–44 Furthermore, combination of proteasome inhibitor bortezomib and immunomodulatory agents lenalidomide or pomalidomide are effective therapies for newly diagnosed or relapsed/refractory MM. As OPA blocks protein degradation, we hypothesized that it may similarly induce additive/synergistic anti-MM activity in combination with immunomodulatory drugs. Indeed, we found that combining OPA with lenalidomide, pomalidomide or Dex induces synergistic anti-MM activity. Future clinical combinations strategies will determine whether Rpn11 inhibitors can overcome proteasome inhibitor resistance with a favorable therapeutic index.
Collectively, our data provide the rationale for the development of novel therapeutics targeting 19S proteasome-associated DUB enzyme Rpn11 in MM.
MATERIALS AND METHODS
Cell culture
MM cell lines (MM.1S, MM.1R, RPMI8226, INA6, Dox40, LR5, ARP1) and peripheral blood mononuclear cells from normal healthy donors were cultured in RPMI-1640 medium containing 10% fetal bovine serum and antibiotics. ANBL6 cells were kindly provided by Dr Robert Orlowski (Houston, TX, USA).45 Tumor cells, BMSCs and pDCs were cultured as described previously.28,29 Helsinki protocol was followed for obtaining informed consent from all patients. Bortezomib, dexamethasone, lenalidomide or pomalidomide were obtained from Selleck Chemicals LLC (Houston, TX, USA), and OPA was obtained from Calbiochem (San Diego, CA, USA). Recombinant DUBs were obtained from Creative BioMart (Rpn11/ PSMD14, Shirley, NY, USA), Boston Biochem (USP1, USP7, Cambridge, MA, USA) and LifeSensors Inc. (USP2, USP4, USP5, USP8, USP20, Uch37, Malvern, PA, USA).
Plasma cell isolation
Plasma cells were isolated from patient BM samples using CD138 microbeads from Miltenyi Biotec Inc. (San Diego, CA, USA) following the manufacturer’s instructions. The purity of CD138+ cells is over 99%.
Cell viability, apoptosis and immunoblot analysis
Cell viability was assessed by WST (Clontech, Mountain View, CA, USA)/CellTiter-Glo Luminescent assays (Promega, Madison, WI, USA) as described previously.11 Apoptosis was assessed with Annexin/propidium iodide staining.46 Caspase activity was measured using caspase-8 and caspase-9 Fluorometric Assay Kit (Enzo Life Sciences, Farmingdale, NY, USA) as per the manufacturer’s instructions. Cell cycle and immunoblot analysis was performed as described previously.47
Transient transfections
Genome control siRNA or Rpn11 ON-TARGET plus SMART pool siRNA (Dharmacon, Inc., Lafayette, CO, USA; target sequences: 5′-GAACAAGUCUA-UAUCUCUU-3′; 5′-GGCAUUAAUUCAUGGACUA-3′; 5′-AGAGUUGGAUGG-AAGGUUU-3′; and 5′-GAUGGUUGUUGGUUGGUAU-3′) were transfected using the cell line Nucleofector Kit V (Amaxa Biosystems, Cologne, Nordrhein-Westfalen, Germany). Cells were harvested 24 h post transfection, followed by analysis using both immunoblotting and cell viability assays.
Ubiquitin-AMC assays
Of each recombinant DUB, 20 nm was incubated with dimethyl sulfoxide control or OPA in assay buffer (40 mm Tris/HCl pH 7.4; 5% glycerol; 0.005% Tween-20; 1 mm dithiothreitol; 0.05 mg/ml ovalbumin) for 30 min at 37 °C, and Ub-AMC (500 nm) was then added for 20 min, followed by measurement of fluorescence intensity.
Ubiquitin-GFP cleavage assay
Recombinant human Rpn11 was incubated with dimethyl sulfoxide control or OPA for 30 min at room temperature; Ub-GFP substrate was then added to the reaction mixture for 1 h, followed by analysis of cleaved GFP using SDS–polyacrylamide gel electrophoresis and western blotting with anti-GFP antibodies.
Reverse transcription–PCR
Total cellular RNA was extracted from purified CD138+ MM cells or normal peripheral blood mononuclear cells using Trizol reagent and subjected to reverse transcription reaction with iScript cDNA synthesis kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA), followed by RT–PCR analysis.
Analysis of Rpn11
The Affymetrix Human Exon 1.0 ST Array data for 170 newly diagnosed MM patients were quality controlled and normalized with aroma.affymetrix package (aroma.affymetrix_3.1.0.tar.gz). Gene expression was estimated with a PLM model. The survival analysis was carried out using the R-package ‘Survival’. High and low expression groups were compared with log-rank test. Raw data are available at http://www.ncbi.nlm.nih.gov/geo/ (accession number: GSE39754).
Animal model study
All studies were performed with prior approval from the institutional animal care and use committee (DFCI). MM xenografts were established as previously described.11 Tumor-bearing SCID mice (10 mice/group) received vehicle control or OPA (20 mg/kg) 3 times/week for 18 days. Tumor volume was measured every third day. Mice survival is shown in the Kaplan–Meier plot (P < 0.005). Tumors from mice were analyzed for apoptosis, ubiquitylation and angiogenesis with immunostaining, as previously described.11,28 Immunostained tissues were imaged by microscopy (Nikon inverted TE2000 microscope; Tokyo, Japan). Protein extracts from mice tumors were also assessed for apoptotic markers using immunoblotting.
Statistics
Student’s t-test was applied to derive statistical significance. Mice survival was calculated with GraphPad Prism software (GraphPad Software, Inc., La Jolla, CA, USA). Cytotoxic activity of drug combinations was calculated using isobologram analysis and CalcuSyn software program (Biosoft, Great Shelford, Cambridge, UK).
Supplementary Material
Acknowledgments
This investigation was supported by National Institutes of Health Specialized Programs of Research Excellence (SPORE) Grants P50100707, R01CA207237 and RO1CA050947. KCA is an American Cancer Society Clinical Research Professor.
Footnotes
CONFLICT OF INTEREST
KCA is on advisory board of Celgene, Millenium, Gilead and Bristol Myers Squibb, and is a Scientific Founder of Acetylon, Oncopep and C4 Therapeutics. The other authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
DC conceived the project, designed research, analyzed data and wrote the manuscript; YS designed and performed the experiments and interpreted data; SL helped in RT–PCR and animal studies; AR and DSD helped in cytotoxicity assays; JQ helped in western studies; MKS performed the GEP data set analyses; NM and Y-TT provided patient samples; RDC helped in immunohistochemistry analysis; and KCA reviewed the manuscript.
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
References
- 1.Lee MJ, Lee BH, Hanna J, King RW, Finley D. Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes. Mol Cell Proteomics. 2011;10:003871. doi: 10.1074/mcp.R110.003871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lander GC, Estrin E, Matyskiela ME, Bashore C, Nogales E, Martin A. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lasker K, Forster F, Bohn S, Walzthoeni T, Villa E, Unverdorben P, et al. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc Natl Acad Sci USA. 2012;109:1380–1387. doi: 10.1073/pnas.1120559109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 5.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 6.Adams J. The proteasome: a suitable antineoplastic target. Nat Rev Cancer. 2004;4:349–360. doi: 10.1038/nrc1361. [DOI] [PubMed] [Google Scholar]
- 7.Chauhan D, Hideshima T, Anderson KC. Proteasome inhibition in multiple myeloma: therapeutic implication. Annu Rev Pharmacol Toxicol. 2005;45:465–476. doi: 10.1146/annurev.pharmtox.45.120403.100037. [DOI] [PubMed] [Google Scholar]
- 8.Hershko A. The ubiquitin system for protein degradation and some of its roles in the control of the cell-division cycle (Nobel lecture) Angew Chem Int Ed Engl. 2005;44:5932–5943. doi: 10.1002/anie.200501724. [DOI] [PubMed] [Google Scholar]
- 9.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hussain S, Zhang Y, Galardy PJ. DUBs and cancer: the role of deubiquitinating enzymes as oncogenes, non-oncogenes and tumor suppressors. Cell Cycle. 2009;8:1688–1697. doi: 10.4161/cc.8.11.8739. [DOI] [PubMed] [Google Scholar]
- 11.Chauhan D, Tian Z, Nicholson B, Kumar KG, Zhou B, Carrasco R, et al. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell. 2012;22:345–358. doi: 10.1016/j.ccr.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim KH, Baek KH. Deubiquitinating enzymes as therapeutic targets in cancer. Curr Pharm Des. 2013;19:4039–4052. doi: 10.2174/1381612811319220013. [DOI] [PubMed] [Google Scholar]
- 13.Liu CW, Jacobson AD. Functions of the 19S complex in proteasomal degradation. Trends Biochem Sci. 2013;38:103–110. doi: 10.1016/j.tibs.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhattacharyya S, Yu H, Mim C, Matouschek A. Regulated protein turnover: snapshots of the proteasome in action. Nat Rev Mol Cell Biol. 2014;15:122–133. doi: 10.1038/nrm3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D'Arcy P, Brnjic S, Olofsson MH, Fryknas M, Lindsten K, De Cesare M, et al. Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat Med. 2011;17:1636–1640. doi: 10.1038/nm.2536. [DOI] [PubMed] [Google Scholar]
- 16.Borodovsky A, Kessler BM, Casagrande R, Overkleeft HS, Wilkinson KD, Ploegh HL. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 2001;20:5187–5196. doi: 10.1093/emboj/20.18.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian Z, D'Arcy P, Wang X, Ray A, Tai YT, Hu Y, et al. A novel small molecule inhibitor of deubiquitylating enzyme USP14 and UCHL5 induces apoptosis in multiple myeloma and overcomes bortezomib resistance. Blood. 2014;123:706–716. doi: 10.1182/blood-2013-05-500033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 19.Verma R, Aravind L, Oania R, McDonald WH, Yates JR, 3rd, Koonin EV, et al. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 20.Maytal-Kivity V, Reis N, Hofmann K, Glickman MH. MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 2002;3:28. doi: 10.1186/1471-2091-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koulich E, Li X, DeMartino GN. Relative structural and functional roles of multiple deubiquitylating proteins associated with mammalian 26S proteasome. Mol Biol Cell. 2008;19:1072–1082. doi: 10.1091/mbc.E07-10-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallery M, Blank JL, Lin Y, Gutierrez JA, Pulido JC, Rappoli D, et al. The JAMM motif of human deubiquitinase Poh1 is essential for cell viability. Mol Cancer Ther. 2007;6:262–268. doi: 10.1158/1535-7163.MCT-06-0542. [DOI] [PubMed] [Google Scholar]
- 23.Menendez-Benito V, Verhoef LG, Masucci MG, Dantuma NP. Endoplasmic reticulum stress compromises the ubiquitin-proteasome system. Hum Mol Genet. 2005;14:2787–2799. doi: 10.1093/hmg/ddi312. [DOI] [PubMed] [Google Scholar]
- 24.Chauhan D, Uchiyama H, Akbarali Y, Urashima M, Yamamoto K, Libermann TA, et al. Multiple myeloma cell adhesion-induced interleukin-6 expression in bone marrow stromal cells involves activation of NF-kappa B. Blood. 1996;87:1104–1112. [PubMed] [Google Scholar]
- 25.Ray A, Das DS, Song Y, Richardson P, Munshi NC, Chauhan D, et al. Targeting PD1-PDL1 immune checkpoint in plasmacytoid dendritic cell interactions with T cells, natural killer cells and multiple myeloma cells. Leukemia. 2015;29:1441–1444. doi: 10.1038/leu.2015.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bassal S, Nomura N, Venter D, Brand K, McKay MJ, van der Spek PJ. Characterization of a novel human cell-cycle-regulated homologue of Drosophila dlg1. Genomics. 2001;77:5–7. doi: 10.1006/geno.2001.6570. [DOI] [PubMed] [Google Scholar]
- 27.Watson RA, Rollason TP, Reynolds GM, Murray PG, Banks L, Roberts S. Changes in expression of the human homologue of the Drosophila discs large tumour suppressor protein in high-grade premalignant cervical neoplasias. Carcinogenesis. 2002;23:1791–1796. doi: 10.1093/carcin/23.11.1791. [DOI] [PubMed] [Google Scholar]
- 28.Chauhan D, Singh AV, Brahmandam M, Carrasco R, Bandi M, Hideshima T, et al. Functional interaction of plasmacytoid dendritic cells with multiple myeloma cells: a therapeutic target. Cancer Cell. 2009;16:309–323. doi: 10.1016/j.ccr.2009.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song Y, Ray A, Li S, Das DS, Tai YT, Carrasco RD, et al. Targeting proteasome ubiquitin receptor Rpn13 in multiple myeloma. Leukemia. 2016;30:1877–1886. doi: 10.1038/leu.2016.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang B, Ma A, Zhang L, Jin WL, Qian Y, Xu G, et al. POH1 deubiquitylates and stabilizes E2F1 to promote tumour formation. Nat Commun. 2015;6:8704. doi: 10.1038/ncomms9704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bergsagel PL, Kuehl WM. Molecular pathogenesis and a consequent classification of multiple myeloma. J Clin Oncol. 2005;23:6333–6338. doi: 10.1200/JCO.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 32.Bergsagel PL, Chesi M, Nardini E, Brents LA, Kirby SL, Kuehl WM. Promiscuous translocations into immunoglobulin heavy chain switch regions in multiple myeloma. Proc Natl Acad Sci USA. 1996;93:13931–13936. doi: 10.1073/pnas.93.24.13931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greenstein S, Krett NL, Kurosawa Y, Ma C, Chauhan D, Hideshima T, et al. Characterization of the MM.1 human multiple myeloma (MM) cell lines: a model system to elucidate the characteristics, behavior, and signaling of steroid-sensitive and -resistant MM cells. Exp Hematol. 2003;31:271–282. doi: 10.1016/s0301-472x(03)00023-7. [DOI] [PubMed] [Google Scholar]
- 34.Avet-Loiseau H, Attal M, Moreau P, Charbonnel C, Garban F, Hulin C, et al. Genetic abnormalities and survival in multiple myeloma: the experience of the Intergroupe Francophone du Myelome. Blood. 2007;109:3489–3495. doi: 10.1182/blood-2006-08-040410. [DOI] [PubMed] [Google Scholar]
- 35.Burger R, Guenther A, Bakker F, Schmalzing M, Bernand S, Baum W, et al. Gp130 and ras mediated signaling in human plasma cell line INA-6: a cytokine-regulated tumor model for plasmacytoma. Hematol J. 2001;2:42–53. doi: 10.1038/sj.thj.6200075. [DOI] [PubMed] [Google Scholar]
- 36.Spataro V, Toda T, Craig R, Seeger M, Dubiel W, Harris AL, et al. Resistance to diverse drugs and ultraviolet light conferred by overexpression of a novel human 26S proteasome subunit. J Biol Chem. 1997;272:30470–30475. doi: 10.1074/jbc.272.48.30470. [DOI] [PubMed] [Google Scholar]
- 37.Spataro V, Simmen K, Realini CA. The essential 26S proteasome subunit Rpn11 confers multidrug resistance to mammalian cells. Anticancer Res. 2002;22:3905–3909. [PubMed] [Google Scholar]
- 38.Rinaldi T, Ricci C, Porro D, Bolotin-Fukuhara M, Frontali L. A mutation in a novel yeast proteasomal gene, RPN11/MPR1, produces a cell cycle arrest, over-replication of nuclear and mitochondrial DNA, and an altered mitochondrial morphology. Mol Biol Cell. 1998;9:2917–2931. doi: 10.1091/mbc.9.10.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rinaldi T, Ricordy R, Bolotin-Fukuhara M, Frontali L. Mitochondrial effects of the pleiotropic proteasomal mutation mpr1/rpn11: uncoupling from cell cycle defects in extragenic revertants. Gene. 2002;286:43–51. doi: 10.1016/s0378-1119(01)00799-5. [DOI] [PubMed] [Google Scholar]
- 40.Rinaldi T, Pick E, Gambadoro A, Zilli S, Maytal-Kivity V, Frontali L, et al. Participation of the proteasomal lid subunit Rpn11 in mitochondrial morphology and function is mapped to a distinct C-terminal domain. Biochem J. 2004;381:275–285. doi: 10.1042/BJ20040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richardson PG, Xie W, Jagannath S, Jakubowiak A, Lonial S, Raje NS, et al. A phase 2 trial of lenalidomide, bortezomib, and dexamethasone in patients with relapsed and relapsed/refractory myeloma. Blood. 2014;123:1461–1469. doi: 10.1182/blood-2013-07-517276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Richardson PG, Weller E, Lonial S, Jakubowiak AJ, Jagannath S, Raje NS, et al. Lenalidomide, bortezomib, and dexamethasone combination therapy in patients with newly diagnosed multiple myeloma. Blood. 2010;116:679–686. doi: 10.1182/blood-2010-02-268862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richardson PG, Weller E, Jagannath S, Avigan DE, Alsina M, Schlossman RL, et al. Multicenter, phase I, dose-escalation trial of lenalidomide plus bortezomib for relapsed and relapsed/refractory multiple myeloma. J Clin Oncol. 2009;27:5713–5719. doi: 10.1200/JCO.2009.22.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richardson PG, Siegel DS, Vij R, Hofmeister CC, Baz R, Jagannath S, et al. Pomalidomide alone or in combination with low-dose dexamethasone in relapsed and refractory multiple myeloma: a randomized phase 2 study. Blood. 2014;123:1826–1832. doi: 10.1182/blood-2013-11-538835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuhn DJ, Chen Q, Voorhees PM, Strader JS, Shenk KD, Sun CM, et al. Potent activity of carfilzomib, a novel, irreversible inhibitor of the ubiquitin-proteasome pathway, against preclinical models of multiple myeloma. Blood. 2007;110:3281–3290. doi: 10.1182/blood-2007-01-065888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chauhan D, Singh A, Brahmandam M, Podar K, Hideshima T, Richardson P, et al. Combination of proteasome inhibitors bortezomib and NPI-0052 trigger in vivo synergistic cytotoxicity in multiple myeloma. Blood. 2008;111:1654–1664. doi: 10.1182/blood-2007-08-105601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chauhan D, Ray A, Viktorsson K, Spira J, Paba-Prada C, Munshi N, et al. In vitro and in vivo antitumor activity of a novel alkylating agent, melphalan-flufenamide, against multiple myeloma cells. Clin Cancer Res. 2013;19:3019–3031. doi: 10.1158/1078-0432.CCR-12-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.