Abstract
Chitosan is a natural polymer with applications in agriculture, which causes plasma membrane permeabilisation and induction of intracellular reactive oxygen species (ROS) in plants. Chitosan has been mostly applied in the phylloplane to control plant diseases and to enhance plant defences, but has also been considered for controlling root pests. However, the effect of chitosan on roots is virtually unknown. In this work, we show that chitosan interfered with auxin homeostasis in Arabidopsis roots, promoting a 2–3 fold accumulation of indole acetic acid (IAA). We observed chitosan dose-dependent alterations of auxin synthesis, transport and signalling in Arabidopsis roots. As a consequence, high doses of chitosan reduce WOX5 expression in the root apical meristem and arrest root growth. Chitosan also propitiates accumulation of salicylic (SA) and jasmonic (JA) acids in Arabidopsis roots by induction of genes involved in their biosynthesis and signalling. In addition, high-dose chitosan irrigation of tomato and barley plants also arrests root development. Tomato root apices treated with chitosan showed isodiametric cells respect to rectangular cells in the controls. We found that chitosan causes strong alterations in root cell morphology. Our results highlight the importance of considering chitosan dose during agronomical applications to the rhizosphere.
Introduction
Soil-borne pathogens cause some of the most serious diseases of cultivated crops and pose a serious threat to global food security1. Infections caused by soil borne organisms such as nematodes, fungi or bacteria can result in crop losses of more than $120 billion dollars per year in the USA only2. These losses together with the restriction in the use of fungicides, bactericides and nematicides justify the need to study and develop more sustainable methods of control. Chitosan has been used in agriculture to control plant damage by viruses and viroids, bacteria, fungi, nematodes and other pests3. Chitosan is a highly deacetylated form of chitin, with numerous applications in agriculture4. Chitosan is biodegradable, friendly to the environment and non-toxic to mammals and humans in particular5,6.
Chitosan is known to cause important physiological changes in plants7 such as growth stimulation of Eustoma grandiflorum seedlings (Raf.) Shinn8 and Robusta coffee (L.)9 or enhancing seed germination of Dendrobium spp.10. In addition, chitosan also promotes tomato11 and orchid production12, reduces flowering time of Dendrobium spp.13 and stimulates plant tissue differentiation14. Chitosan has been described as enhancer of photosynthetic rate in plants15 and modulator of their nutritional status. Chitosan acts as elicitor16 of plant secondary metabolites such as alkaloids17, withanolides18 and lignin19.
However, chitosan has mainly been applied to the phylloplane for controlling pests and diseases or modifying plant growth and defences. The effect of its aerial application has been also described as a stress-like response, which translates into the priming of plant defences20–22. Chitosan induces oxidative stress in tomato plants23 as well as in fungi6,24. Response to chitosan in plants is mediated by stomatal closure, which is independent of endogenous abscisic acid or jasmonates25. The manipulation of the plant immune system by using chitosan has recently been reviewed26. However, less is known about the effect on root development when chitosan is applied in the rhizosphere. Baque et al.27 found that chitosan applied to adventitious root cultures enhances secondary metabolite production and decreases root growth on Morindia plants. On the contrary, Khalil & Badawy (2012)28 found promotion of root growth by chitosan but applied only once and in plants inoculated with the root-knot nematode Meloidogyne incognita. Still the direct effect of chitosan on plant root at physiological and cellular levels is not known.
In this study we investigate the effect of chitosan on root development trying to establish the basis for its application in agriculture. We use the model plant Arabidopsis thaliana (Arabidopsis) as well as barley and tomato plants which are profitable crops and important for food security. We have investigated the activity of chitosan on plant growth and physiology, and in particular the effect of chitosan on root cell architecture and organisation. We study the production of plant hormones in roots treated with chitosan. We have also determined the activity of chitosan on gene expression regulating auxin, jasmonic and salicylic acids biosynthesis, signalling and regulation. In addition, we have also analysed how chitosan acts on quiescent centre (QC) organisation in Arabidopsis root meristem. In this paper, we have also applied chitosan in the irrigation system of tomato and barley plants. We have determined the effect of chitosan on plant growth and apical root meristem cell dynamics. This work provides a better understanding of the mode of action of chitosan on root development, focusing on hormone homeostasis.
Results
Chitosan reduces root growth and development in Arabidopsis
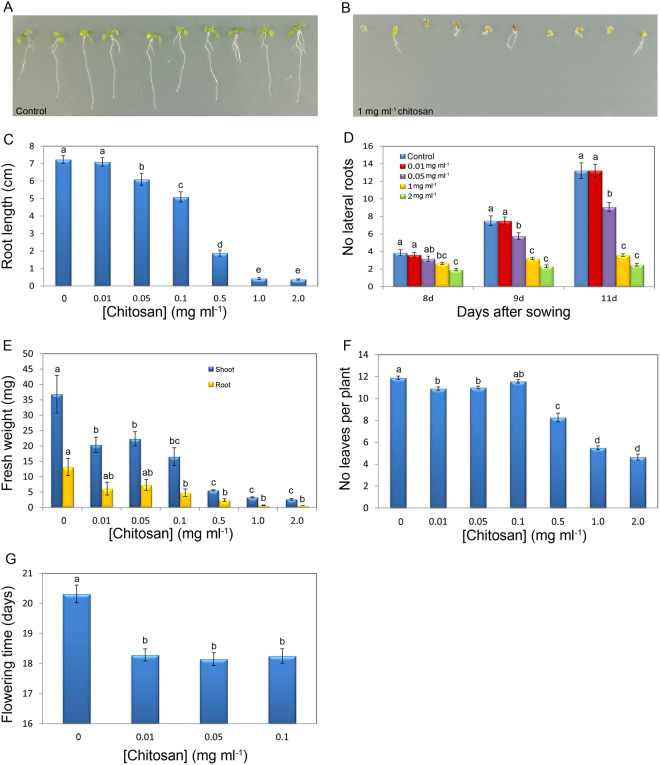
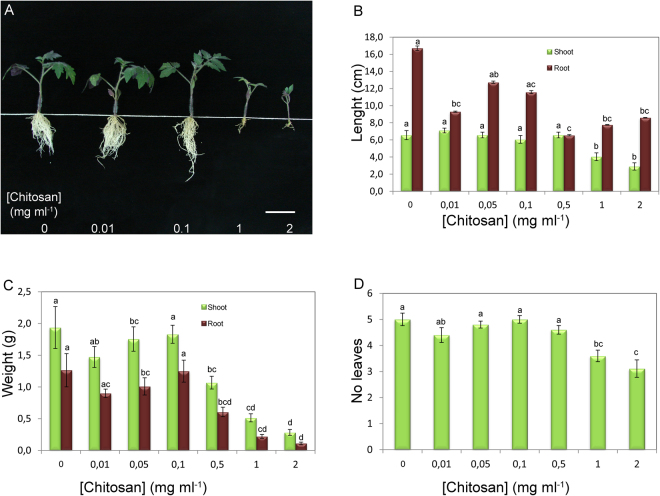
To investigate the possible effects on root growth of chitosan applied to the rhizosphere, we examined the phenotype of Arabidopsis plants (Col-0) treated with increasing doses of chitosan (Fig. 1A and B). After 21d, low doses of chitosan (0.01–0.1 mg ml−1) have little impact on Arabidopsis growth, however higher chitosan concentrations (>0.5 mg ml−1) severely arrested Arabidopsis root development (Fig. 1C). Plants in contact with 2 mg ml−1 chitosan for 11 days, displayed 80% reduction (p-value < 0.05) in the number of secondary roots respect to controls (Fig. 1D). However, lower doses of chitosan (0.05 and 0.1 mg ml−1) only caused a slight reduction of root biomass. Arabidopsis roots exposed to 1 mg ml−1 chitosan solution for 2 h showed an accumulation of reactive oxygen species (ROS) (Fig. S1). This suggests chitosan causes a systemic stress response.
Figure 1.
Chitosan alters Arabidopsis growth and development. (A) Untreated (control) plants (B) Plants treated with 1 mg ml−1 chitosan. (C) Effect of chitosan on root length. (D) Chitosan affecting secondary root formation. (E) Effect of chitosan on shoot and root weight. (F) Activity of chitosan on leaf production. (G) Evaluation of the activity of chitosan on flowering time.
Chitosan (0.5 mg ml−1 and upwards) also arrested shoot development (Fig. 1E). The number of leaves per plant also decreased significantly (p < 0.05) in plants grown with 0.5 mg ml−1 chitosan upwards (Fig. 1F). Besides, low chitosan doses (0.01–0.1 mg ml−1) significantly reduce flowering (bolting) time (Fig. 1G). Indicating that low chitosan doses cause slight stress which is translated in to flowering induction. However, plants treated with high chitosan doses (>0.5 mg ml−1), exposed to high stress, did not flower after 30d.
Chitosan modifies root growth repressing WOX5
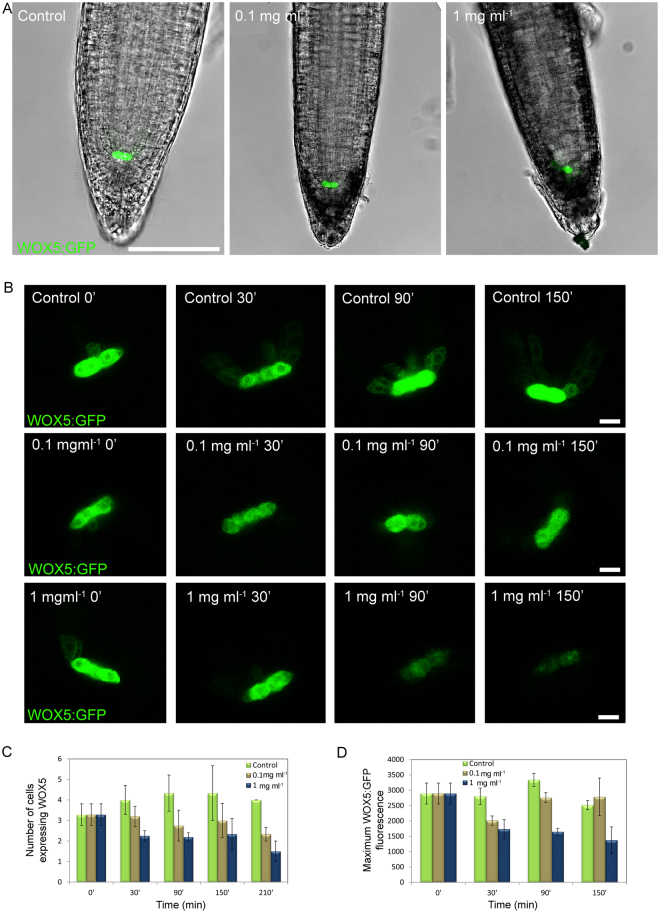
Chitosan altered root physiology and cellular activity. After 30 min with chitosan, roots did not display visual changes. However, after 90 min exposure to 1 mg ml−1 chitosan roots became dark and displayed auto-fluorescence in the root cap and epidermal cells when excited at 580–620 nm (Fig. 2A and Fig. S2). After 150 min, roots exposed to 0.1 mg ml−1 chitosan displayed these dark/auto-fluorescent compounds (likely phenols) only in root tips. Whereas at this time point (150 min) larger chitosan dose (1 mg ml−1) caused widespread accumulation of these compounds in the roots (Fig. 2A).
Figure 2.
Altered root development by chitosan correlates with the repression of the quiescent centre gene WOX5. (A) Tips from 5d-old-roots after 150 min in contact with MS medium (control), 0.1 and 1 mg ml−1 chitosan. (Scale bar 75 µm). (B) Time-course expression of WOX5 in roots exposed to MS medium amended with chitosan. Untreated controls were also included. (Scale bar 5 µm). (C) Number of cells expressing WOX5:GFP. (D) Maximum fluorescence emission from cells expressing WOX5:GFP.
Morphological changes observed in roots treated with chitosan led us to characterize the activity of the root meristematic quiescent centre (QC) (Fig. 2 and Fig. S2). Five-day-old plants placed in chitosan solutions displayed changes in the root apex that correlated with miss-expression of the WUSCHEL RELATED HOMEOBOX 5 (WOX5) gene. WOX5 encodes a transcription factor key to stem cell maintenance in the QC located using WOX5-GFP29 (Fig. 2A). In untreated roots, WOX5-GFP expression was observed in the two stem cells of the QC and to a lower extent in the neighbouring cells. Chitosan (0.1–1 mg ml−1) reduced both the expression level in these cells (Fig. 2B) and also the number of root cells expressing WOX5 (Fig. 2C). In roots exposed to high doses of chitosan (1 mg ml−1) expression of WOX5 in cells of the QC was clearly reduced after 90 min, and by 150 min expression was almost lost (Fig. 2B). The reduction in WOX5 expression was confirmed by fluorescence quantification in primary (Fig. 2D) and secondary roots (Fig. S3).
In view of the activity of chitosan on WOX5 expression and localisation, we also evaluated the activity of this polymer on ACTIVE QUIESCENT CENTRE (AQC1) gene expression (Fig. S4). We demonstrated that low doses of chitosan (0.1 mg ml−1) induced AQC1 gene expression more than 5-fold. However, high chitosan dose (1 mg ml−1) repressed this gene as happened with WOX5 in roots. This result is in agreement with the reduction of root growth in plants exposed to high chitosan doses.
Chitosan induces changes in hormone levels in Arabidopsis roots
In view of the changes in WOX5 and AQC1 gene expression caused by chitosan and since the expression of these genes is regulated at least by auxin30,31, we evaluated the hormonal status in response to the polymer. Roots from Arabidopsis plants grown 3d on MS solid medium amended with 1 mg ml−1 chitosan increased 3.6-fold (p < 0.05) auxin (IAA) content respect to controls. Chitosan also significantly increased (2–3 fold; p < 0.05) salicylic acid (SA) and jasmonic acid (JA) compared with plants not exposed to this polymer (Table 1). However, levels of abscisic acid (ABA) were slightly reduced in roots exposed to chitosan. We also observed that the levels of the active gibberellic acid (GA4) did not change in chitosan-treated roots.
Table 1.
Activity of chitosan on plant hormones in Arabidopsis roots. Chitosan induces accumulation of auxins measured by accumulation of IAA. Chitosan also propitiates accumulation of JA and SA hormones significantly. Asterisks indicate significant differences *(p < 0.05) respect to control (no chitosan).
| Control | 1 mg ml−1 chitosan | ||
|---|---|---|---|
| [Hormone] ng g−1 root | [Hormone] ng g−1 root | ||
| Hormones | GA4 | 0.08 ± 0.01 | 0.04 ± 0.001 |
| ABA | 2.39 ± 0.14 | 0.49 ± 0.07* | |
| JA | 13.88 ± 1.41 | 35.14 ± 2.27* | |
| SA | 28.82 ± 3.16 | 79.88 ± 4.98* | |
| IAA | 46.56 ± 5.30 | 167 ± 11.77* |
GA4: gibberellic acid; ABA: abscisic acid; JA: jasmonic acid; SA: salicylic acid; IAA: indole acetic acid.
Chitosan induces tryptophan-dependent pathway for IAA biosynthesis
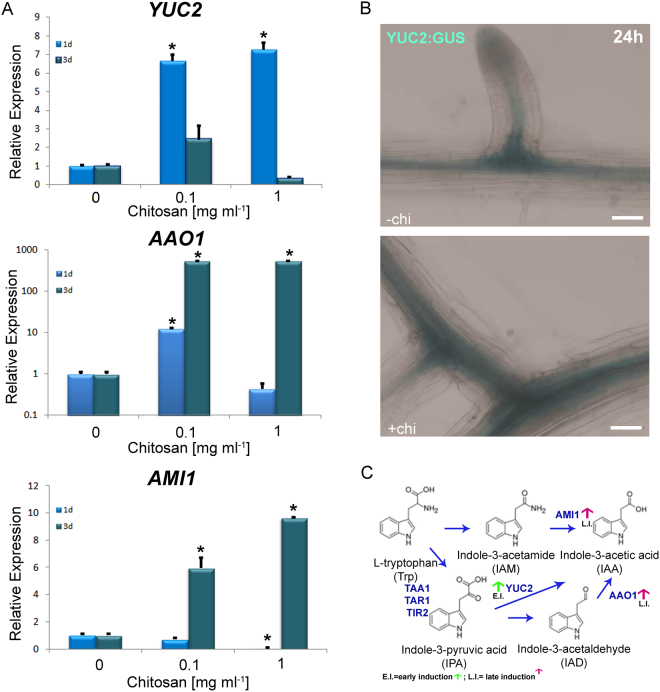
To determine if the increase in auxin levels is due to transcriptional changes in auxin biosynthetic genes, we tested the expression of genes that participate in the Trp-dependent pathway that is the major auxin biosynthetic pathway in Arabidopsis. Chitosan induced IAA accumulation in Arabidopsis roots via L-tryptophan (Trp) dependent pathway (Fig. 3). Chitosan (1 mg ml−1) in MS solid medium induced YUCCA2 (YUC2) gene expression more than 8 fold in Arabidopsis plants exposed to this polymer for 1d (Fig. 3A). We confirmed this result using YUC2:GUS plants exposed to 1 mg ml−1 chitosan solutions for 24 h. We showed YUC2 overexpression in the vascular tissue and in zones of secondary root differentiation (Fig. 3B). However, chitosan repressed this gene in plants exposed 3d to chitosan. Besides, transcriptional analysis by qRT-PCR also showed a large (ca. 100-fold) overexpression of ALDEHIDE OXIDASE1 (AAO1 encoding indole-3-acetaldeyde oxidase) in plants grown for 3d on MS plates amended with 1 mg ml−1 chitosan (Fig. 3A). Likewise, AMIDASE1 (AMI1 encoding indole-3-acetamide hydrolase) is overexpressed more than 9-fold in plants exposed for 3d to 1 mg ml−1 chitosan. These results suggest that late overexpression of AAO1 and AMI1 in response to chitosan induced auxin accumulation in roots. We also showed the same trend in the gene expression of those genes related with IAA synthesis at low chitosan doses (0.1 mg ml−1) (Fig. 3A). Chitosan induced YUC2 in plants exposed for 1d to 0.1 mg ml−1 chitosan and AAO1 and AMI1 in plants exposed for 3d to the same chitosan concentration. In conclusion, exposure of Arabidopsis plants to chitosan triggered IAA synthesis by a tryptophan-dependent pathway which results in an accumulation of IAA in roots (Fig. 3C).
Figure 3.
Chitosan induces the Trp-dependent pathway for auxin synthesis. (A) Exposition of Arabidopsis plants to chitosan for 1d induces relative expression of YUC2. AAO1is overexpressed after 3d in contact with 1 mg ml−1. However, AMI1 is repressed after 1d in contact with chitosan. Higher doses of chitosan (1 mg ml−1) allows overexpression of AMI1 after 3d in contact with chitosan. Besides, low doses of chitosan (0.1 mg ml−1) induces Trp-dependent pathway for auxin synthesis. Arabidopsis plants exposed for 1d to low chitosan doses propitiates overexpression of YUC2. However, Arabidopsis plants exposed for 3d to low chitosan doses propitiates overexpression of AAO1 and AMI1. Asterisks indicate significant differences *(p < 0.05) respect to control (B) Histochemical analysis shows chitosan induces YUC2:GUS transgene expression in Arabidopsis roots. (Scale bar 50 µm). (C) Simplified view of IAA and its modulation by chitosan.
Chitosan alters IAA transport and signalling
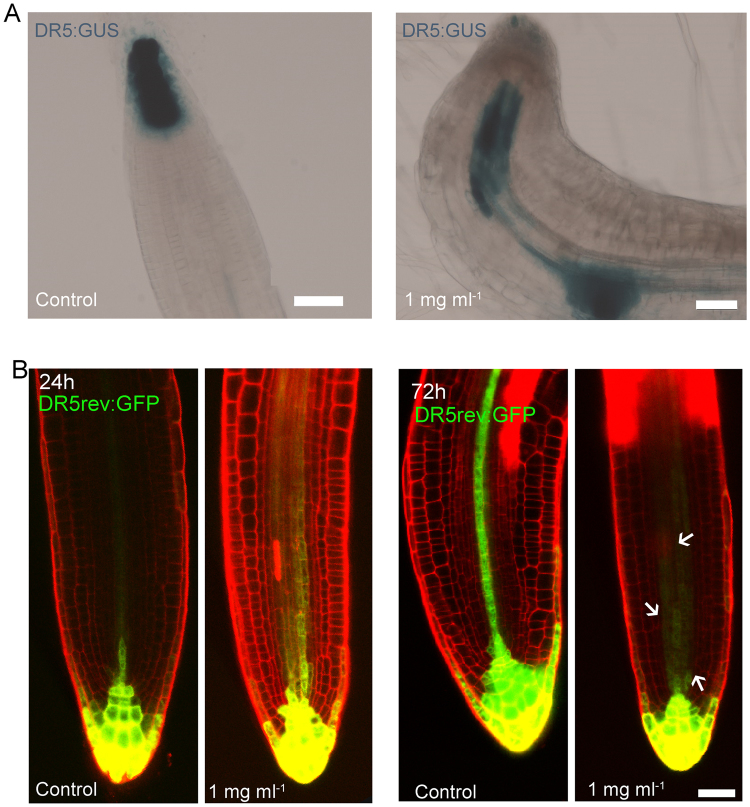
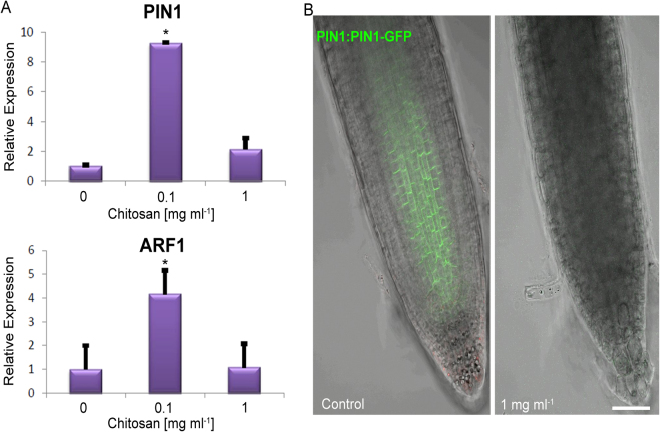
In agreement with the increase in IAA levels, a 2 h treatment with chitosan (1 mg ml−1) enhanced auxin signaling in Arabidopsis primary roots as detected by the DR5:GUS reporter (Fig. 4A). Interestingly, the activity of the reporter extended to the newly emerged secondary roots (Fig. 4B). Despite the fact that this reporter is transcriptional and does not respond directly to auxin levels, it is reasonable to think that the increase in its activity is due, at least partly, to chitosan-induced accumulation of IAA. We also showed that AUXIN RESPONSE FACTOR1 (ARF1), known to repress gene expression in response to auxin32, was induced ca. 5-fold at low chitosan dose (0.1 mg ml−1) (Fig. 5A), suggesting it is part of the signalling pathway triggered by chitosan. A higher dose of this polymer (1 mg ml−1), however, caused no changes in its expression.
Figure 4.
Chitosan causes auxin accumulation in Arabidopsis roots. (A) Two hours exposition to liquid chitosan provoke changes in DR5:GUS localisation in primary roots exposed this polymer. Untreated plants shows DR5:GUS expression located in the meristematic zone and stem cell niche. Plants exposed to chitosan showed accumulation of DR5:GUS expression in the transition zone and meristematic zone. (Scale bar 50 µm). (B) Chitosan causes mislocalisation of DR5rev:GFP in secondary roots. DR5rev:GFP localise in the meristematic zone and vascular tissue. Arrows indicate expression of DR5 in the meristematic zone. (Scale bar 50 µm).
Figure 5.
Chitosan modify auxin transport and auxin signalling gene expression in Arabidopsis roots. (A) Relative expression of PIN1 and ARF1 at low chitosan doses (0.1 mg ml−1). High chitosan doses (1 mg ml−1) repress expression of both genes. Asterisks indicate significant differences *(p < 0.05) respect to control. (B) Chitosan (1 mg ml−1) abolishes PIN1-GFP fusion protein visualization in Arabidopsis roots in comparison with untreated Arabidopsis roots. (Scale bar 10 µm).
We next tested if chitosan was also acting on the transport of auxin. Low doses of chitosan (0.1 mg ml−1) induced expression of the auxin efflux carrier gene PIN1 (Fig. 5A). On the contrary, higher chitosan dose (1 mg ml−1) abolished PIN1-GFP signal in Arabidopsis roots (Fig. 5B), suggesting it could potentially affect auxin transport propitiating auxin accumulation in roots. In summary, chitosan can impair root growth and morphogenesis modifying auxin synthesis and delivery.
Chitosan induces the expression of SA- and JA-related genes
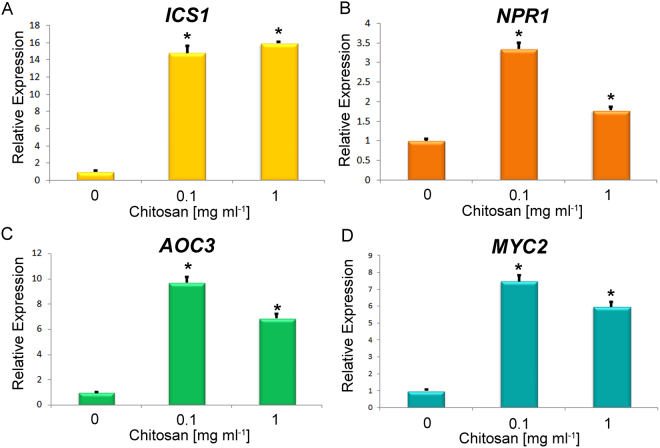
Similar to the effect on auxin-related genes, chitosan induced the expression of genes involved in the synthesis and perception of SA and JA (Figs 6 and S5). This supports the increase in JA and SA found in roots of Arabidopsis plants exposed to chitosan (Table 1). More specifically, the expression of genes involved in the isochorismate pathway (ICS1 and ICS2) for SA biosynthesis was induced by chitosan between 6 and 15-fold (Figs 6A and S5A), in agreement with the larger accumulation of SA (Table 1). Chitosan induction (ca. 3.5–1.5 fold) of NON-INDUCIBLE IMMUNITY PROTEIN (NPR1) gene, required for normal systemic acquired resistance (SAR), also supports a possible enhancement of SA mediated signalling in Arabidopsis exposed to chitosan (Fig. 6B). With respect to JA biosynthesis, ALLENE OXIDE CYCLASE3 (AOC3) was induced 9-fold with low chitosan dose (0.1 mg ml−1) (Fig. 6C). The same trend was also found with higher doses of chitosan (1 mg ml−1). LIPOXIGENASE3 (LOX3) was only slightly induced by 1 mg ml−1chitosan (Fig. S5B). Likewise, CYTOCHROME P450 94B1 (CYP94B) which mediates jasmonoyl-isoleucine hormone oxidation in the last stages of the biosynthesis of the active hormone was also induced (ca. 5-fold) by chitosan (Fig. S5C). An equivalent induction (6–7 fold) was found for MYC2, encoding a JA-dependent transcription factor (Fig. 6D). In summary, induction of SA and JA by chitosan could mediate balancing on plant immunity.
Figure 6.
Chitosan induces expression of genes involved in SA and JA biosynthetic and signalling pathways. (A) Chitosan significantly (p < 0.05) overexpress SA biosynthetic genes isochorismate synthase 1 (ICS1) under low (0.1 mg ml−1) and high (1 mg ml−1) chitosan doses in Arabidopsis plants (Col-0). (B) Chitosan doses (0.1 and 1 mg ml−1) induce NPR1 gene related with SA signalling in Arabidopsis plants. Chitosan also significantly (p < 0.05) induces expression of JA related genes (C) AOC3 (JA biosynthesis) and (D) MYC2 (JA signalling) respect to untreated controls.
Hormone involvement on root growth defects caused by chitosan
To evaluate if only the changes in hormone levels could explain the reduction of root growth caused by chitosan, wild-type seedlings were grown in the presence of IAA, SA and JA levels equivalent to the concentrations found after chitosan treatment (Table 1). When IAA, SA and JA were applied together, root length was significantly reduced respect to control (Fig. S6A). This reduction was lower than that found in chitosan treated plants. However, the three hormones combined increased root weight (Fig. S6B). This suggests hormone homeostasis of IAA, JA and SA together with other factors such as induction of ROS is involved in the growth reduction caused by chitosan.
We evaluated the role of hormone perception on root growth reduction by chitosan. For this purpose, we tested Arabidopsis mutants with reduced sensitivity to either SA or JA (SA: npr1–1; JA: coi1–40 and myc2/3/4 triple mutant). After 15d on MS medium amended with chitosan (0.1 mg ml−1) JA insensitive mutants (coi1–40 and myc2/3/4) had larger (p < 0.05) root biomass (ca. 2-fold) than these mutants growing on MS only (Table 2). The same phenotype was shown in Col-0 plants amended with IAA and SA (with lack in JA) (Fig. S5). These results suggest that JA could play a relevant role on shortening root by chitosan. Conversely, the mutant with reduced sensitivity to SA (npr1–1) did not show significant differences on plant biomass. However, npr1-1 plants showed a significant rise in number of leaves per plant (ca. 10%; Table 2). On the contrary, higher chitosan dose (1 mg ml−1) severely reduced growth of both JA and SA mutants.
Table 2.
Effect of chitosan on Arabidopsis mutants with reduced JA/SA sensitivity.
| Chitosan (mg ml−1): | MRL (cm) | RFW (mg) | SFW (mg) | No leaves | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.1 | 1 | 0 | 0.1 | 1 | 0 | 0.1 | 1 | 0 | 0.1 | 1 | |
| Col-0 | 8.56 ± 0.28a | 8.96 ± 0.34a | 1.12 ± 0.06b | 6.45 ± 0.35a | 6.74 ± 0.29a | 0.2 ± 0.02b | 20.18 ± 0.62a | 20.48 ± 0.58a | 1.16 ± 0.09b | 9.68 ± 0.19a | 10.13 ± 0.18a | 3.67 ± 0.19b |
| npr1–1 (SA insensitive) | 7.08 ± 0.21a | 7.39 ± 0.45a | 0.59 ± 0.04b | 2.15 ± 0.12a | 3.64 ± 0.27a | 0.12 ± 0.02b | 10.7 ± 0.24a | 13.9 ± 0.49a | 1.67 ± 0.03b | 6.53 ± 0.24a | 7.4 ± 0.16 b | 4.06 ± 0.12c |
| coi1–40 (JA insensitive) | 8.19 ± 0.38a | 8.09 ± 0.59a | 0.71 ± 0.07b | 3.18 ± 0.25a | 5.77 ± 0.33 b | 0.15 ± 0.01c | 12.28 ± 0.63a | 14.97 ± 0.48a | 1.47 ± 0.04b | 8.67 ± 0.25a | 8.67 ± 0.27a | 4.34 ± 0.16b |
| myc2/3/4 (JA insensitive) | 7.63 ± 0.26a | 8.1 ± 0.41a | 0.99 ± 0.09b | 1.97 ± 0.13a | 4.6 ± 0.03 b | 0.17 ± 0.01c | 12.44 ± 0.64a | 16.74 ± 0.2 b | 1.55 ± 0.04c | 8.9 ± 0.2a | 9.55 ± 0.23a | 4.2 ± 0.11b |
Letters indicate significant differences (p < 0.05) between treatments. Numbers in bold show growth promotion in 0.1 mg ml−1 chitosan treatment vs. MS untreated controls. MRL: Maximum root length, RFW: Root fresh weight, SFW: Shoot fresh weight. SA: Salicylic acid, JA: Jasmonic acid.
Chitosan reduces root growth and development of tomato and barley plantlets
To test the effect of chitosan on root development in other plants, we investigated the behaviour of two phylogenetically distant plants from Arabidopsis, tomato (Solanum lycopersicum Mill cv. Marglobe; eudicot) and barley (Hordeum vulgare L. var. Disticum; monocot). We showed that at concentrations higher than 0.1 mg ml−1 chitosan also impaired development of tomato (Figs 7 and S7) and barley plants (Fig. S8). High doses of chitosan (0.5, 1 and 2 mg ml−1) applied in the irrigation system for 21d blocked root elongation and caused a strong decrease (more than 2.5-fold) in tomato plant biomass (Fig. 7B and C) and reduced the number of leaves per plant (Fig. 7D). Application of chitosan also induced accumulation of violaceous compounds (likely anthocyanins) on the abaxial side of tomato leaves (Fig. S7). This is perhaps an indication of the stress suffered by plants irrigated with chitosan. However, low doses (0.01–0.1 mg ml−1) did not affect tomato plant growth and development.
Figure 7.
Irrigation with chitosan inhibits tomato root and shoot development. (A) Overview of the inhibitory effect of chitosan on development of tomato plantlets after 21 days of treatment. (Scale bar 5 cm). (B–D) Effect of chitosan on tomato (B) shoot and root length, (C) biomass of shoot and roots and (D) the number of leaves per plant. Different letters indicate significant differences (p < 0.05) between treatments.
High chitosan doses also reduced growth of barley plants (Fig. S8). Plants irrigated with 1 and 2 mg ml−1 chitosan for 21d showed roots 2-fold shorter than those of controls (Fig. S8A and S8B). However, shoot length remained unaffected. On the contrary, barley plants irrigated with low doses of chitosan (0.01 and 0.1 mg ml−1, Fig. S8) exhibited a slight induction of root and shoot growth (Fig. S8B).
Chitosan blocks cell differentiation and alters root architecture in tomato plants
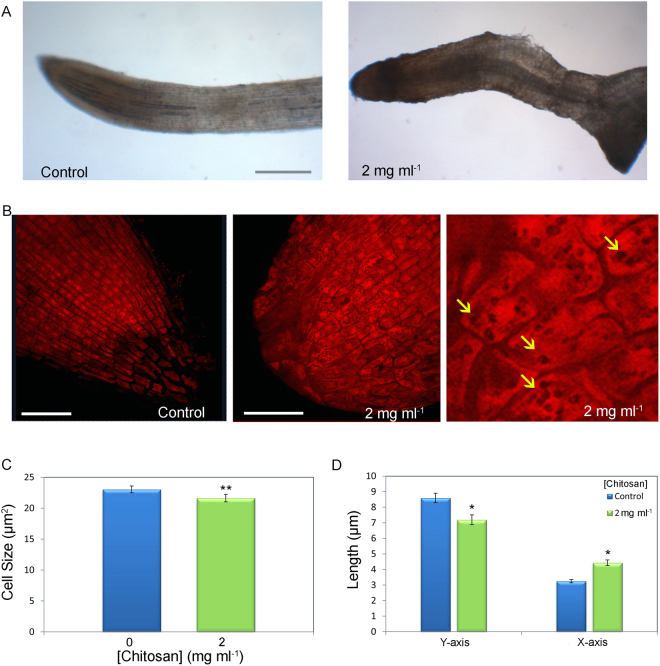
Chitosan changed tomato root development and root cell architecture (Fig. 8). Irrigation with 2 mg ml−1 chitosan for 21d altered morphology of tomato roots (Fig. 8A). It caused deformation of the root apex and caused accumulation of dark compounds (like phenols) as described in Arabidopsis, which might be associated with the response to stress. Besides, root growth lost polarity and root apices displayed a round shape, unlike sharp apices from control plants (Fig. 8). Cells from untreated control plants were rectangular and formed strips in the direction of root elongation (Fig. 8B). Instead, root apex cells from chitosan-irrigated plants were isodiametric, and not organised in a given direction (Fig. 8B). Chitosan exposed roots accumulated dark vesicles in their cytoplasm likely related to stress response. Cell imaging analysis showed that chitosan caused a significant reduction (p < 0.01) in the size of root cells (Fig. 8C) as well as changes in their shape, being both shorter and wider (Fig. 8D).
Figure 8.
Chitosan affects architecture of tomato roots altering cell division. (A) Root apex of plant irrigated with water (control) and chitosan (2 mg ml−1) for 21d under visual light. (Scale bar 250 µm). (B) Microscopically evaluation of root apices morphology from control plants (left) and plants irrigated with chitosan (centre, right) for 21d. Close-up (right) shows dark intracellular vesicles (arrows) in cells from chitosan treated roots. (Scale bar 50 µm). (C) Measurement of tomato root cell size irrigated with chitosan during 21d. (D) Effect of chitosan on root cell shape. Cells Y-axis (in the root’s growth direction) and X-axis (perpendicular to growth direction). Asterisks indicate significant differences *(p < 0.05) and **(p < 0.01).
Discussion
Chitosan has wide applications in agriculture4. It has been used as a plant defence inducer33 and against infections caused by viruses, bacteria, fungi and nematodes in plants3. Most chitosan applications have been performed on the phylloplane as modulator of defences and innate immune responses in plants34. However, recent studies have analysed the effect of chitosan applied in the rhizosphere in order to improve plant yield and to enhance virulence of biocontrol fungi such as P. chlamydosporia 24,35,36. In our study we use Arabidopsis to understand the mechanisms underlying the activity of chitosan on root growth. Chitosan causes important changes on Arabidopsis development, reducing root growth and delaying plant development. The effect of chitosan on root development is concentration dependent; 0.01–0.1 mg ml−1 chitosan cause small changes on plant growth, while 0.5–2 mg ml−1 chitosan arrest Arabidopsis root elongation. Chitosan also reduces the rate of secondary roots emerged in Arabidopsis. Secondary root emergence plays an essential role in water and nutrient uptake37. Their inhibition could explain the distortion caused by chitosan on plant development. Besides, chitosan causes accumulation of auxin in Arabidopsis roots. This is due to increased IAA biosynthesis since YUC2, AMI1 and especially AAO1 are overexpressed in chitosan treated roots. Chitosan also causes auxin accumulation by blocking PIN1 gene expression involved in IAA transport. Decreases in primary root length and reduction in secondary root emergence caused by chitosan could be explained by auxin overproduction in roots38,39.
Auxin accumulation is known to repress the homeodomain transcription factor WOX5 mediated by ARF1, ARF6 and ARF10 30. Chitosan causes WOX5 repression, reducing its localisation in the neighbouring QC cells. WOX5 is a major regulator of root stem cell activity in the quiescent centre which controls cell division. This could explain why chitosan alters root cell dynamics (root cell shape and size changes) and reduces root elongation. These changes would be directly related to the alteration in the rate of cell division in roots and the morphological changes caused by chitosan. Chitosan also represses AQC1 gene expression suggesting regulation of cell division in the root meristem. To this respect the effect of chitosan on cell cycle regulators such as cyclins mediated by SHORT-ROOT/SCARECROW pathway, up-stream of WOX5 40, should be considered in future studies.
The systemic effect of chitosan in plant development could also be explained by changes in homeostasis of hormones other than auxins. Previous authors described chitosan as pathogen/microbe-associated molecular patter (PAMP/MAMP)41, which modulates stress-related hormones. In this work we show that chitosan reduces the presence of ABA in roots. This may be related to ABA translocation from root to aerial parts under stress conditions42. However, chitosan causes accumulation of JA and SA43,44 which is reflected in an induction of biosynthetic and signalling of SA and JA genes by this polymer. This indicates that the increase in these stress-related hormones is translated in their perception. Similar results were observed in plants exposed to abiotic stresses such as heat, salt or drought45. ICS1 and ICS2 (SA biosynthetic genes) and NPR1 (SA signalling genes) are overexpressed by chitosan. Induction of NPR1 and SA signalling could trigger plant immunity mainly PTI (PAMP/MAMP Triggered Immunity)46. Previous studies described that increased levels of SA induce synthesis and accumulation of phenols47 as we found in chitosan-treated roots. Genes related to JA biosynthesis (AOC3 and CYP94B) and signalling (MYC2) were also overexpressed by chitosan. In view of these results, we also confirmed plant perception of chitosan induced stress is reflected in the growth increase of JA insensitive mutants. This effect is chitosan concentration dependent since growth promotion in the mutants was abolished under high chitosan dose. This suggests factors in addition to hormone perception such as ROS levels or phenols accumulation are involved in the activity of chitosan during Arabidopsis root growth. Both, JA and SA hormones have been described in the plant response to stress caused by chitosan when is is applied in leaves42,44,48. An induction of stress-related hormones under low doses of chitosan could be associated with a moderate stress which is described that causes an induction of flowering49 as we observe in our experiments.
However, the application of chitosan on plant roots and its effect on plant growth and plant defence activation has not been fully investigated. Therefore, we set the present study applying chitosan in the root system of tomato and barley to provide evidences about the activity of chitosan on roots. Chitosan overexposure also arrests development of tomato and barley roots, ultimately altering cell size and shape in root tips. These structural modifications of cells could be related to lignification, cytoplasmic acidification, membrane depolarisation and generation of reactive oxygen species caused by chitosan in plant cells26. This phenotype has been also observed when a triazine-base synthetic small compound (rootin) is applied to Arabidopsis roots50. We also describe the formation of intracellular dark vesicles in cells of tomato roots treated with chitosan. Their autofluorescence suggests that they maybe phenolic compounds accumulated in response to chitosan-induced cell stress. Similar vesicles have also been previously described in Mimosa pudica cells treated with chitosan51. High doses of this polymer induce peroxidase and polyphenol oxidases, which in turn generate phenolic compounds3 and calose depositions52 in plant cells. Chitosan also triggers induction of programed-cell death (PCD) in plants53. The involvement of PCD in the inhibitory effect of chitosan on root development should be investigated in future studies. Our work suggests that chitosan, a pathogen/molecular-associated molecular pattern (PAMP/MAMP)41, can affect plant development via hormone homeostasis modification. Since plant immunity and growth converge46 chitosan could induce both PTI (PAMP triggered immunity) and the hormone changes described in this paper.
Our mechanistic study about the effect of chitosan on plant root development opens up new possibilities to implement this polymer in field applications. Doses, frequency and formulation of chitosan should be adjusted to prevent negative effects on plant development as reported in this study. Besides, we have also proven that chitosan is an experimental tool to manipulate root development, physiology, gene expression and hormone homeostasis. Our study contributes to the understanding of chitosan in the rhizosphere. This will help to develop chitosan application to control important pest and diseases of economically important crops by balancing plant immunity and yield.
Material and Methods
Plant material
Arabidopsis thaliana (L. Heynh), ecotype Columbia (Col-0) reporter lines, transformants DR5rev:GFP and DR5:GUS54, WOX5:GFP29, PIN1:PIN1-GFP55, YUC2:GUS56, mutants npr1–1 57, coi1–40 58 and the triple mutant myc2/3/4 59, were tested with chitosan. Arabidopsis seeds were surface-sterilised using 1% NaClO for 2 min and then washed 3 times with sterile distilled water. Surface sterilised seed were stratified at 4 °C for 48 h and then grown on MS (Murashige and Skoog medium; Sigma) plates as in Ripoll et al.60.
Tomato (Solanum lycopersicum Mill cv. Marglobe) and barley (Hordeum vulgare L. var. Disticum) seeds were also surface-sterilised using 50 ml 4% sodium hypochlorite with 3 drops of Tween-20 (Sigma-Aldrich) for 20 min and 1 h, respectively. They were then washed three times (5 min each) with sterile distilled water. Surface-sterilised seeds were plated on 9 cm petri dishes with a germinating medium (1.2% agar supplemented with glucose (10 g l−1), peptone (0.1 g l−1) and yeast extract (0.1 g l−1)61. Seeds were stratified for two days at 4 °C and then incubated at 25 °C in the dark for 5 days and finally 4 days under a 16 h/8 h (light/darkness) photoperiod.
Chitosan
Medium molecular weight chitosan (T8: 70 kDa and 85% deacetylation degree) was from Marine BioProducts GmbH (Bremerhaven, Germany). Chitosan was prepared as described in Palma-Guerrero et al.62. The resulting solution was dialyzed against distilled water and then autoclaved. It was stored at 4 °C until used and never kept longer than 5 weeks.
Characterisation of Arabidopsis growth and development with chitosan in solid media
Arabidopsis seeds (Col-0, DR5rev:GFP, DR5:GUS, WOX5:GFP, YUC2:GUS, PIN1:PIN1-GFP, coi1–40, myc2/3/4 and npr1–1) were surface sterilised. Seeds were then plated on MS medium (Sigma) supplemented with 0.05% 2-(N-morpholino) ethanesulfonic acid (MES, Sigma), 1% sucrose and 1% technical agar and amended with chitosan. Plates with no chitosan were used as controls. Seed were placed at 4 °C for 48 h in the dark to synchronise germination. They were then incubated at 21 °C and 65% relative humidity (RH) with continuous light for 21d upright to allow root elongation. Every two days plants were checked for secondary root and bolting emergence in order to identify flowering time. At harvest time, total plant weight, root weight and length and number of leaves in the rosette per plant and treatment were scored. This experiment was performed in triplicate.
Intracellular hydroxyl, peroxyl and other reactive oxygen species were detected using 2′-7′ dichlorodihydrofluorescein diacetate (H2DCFDA; Sigma, St. Louis, MO, USA). Arabidopsis roots were exposed to 1 mg ml−1 chitosan for 2 h and then were dipped in H2DCFDA. Finally, we observed ROS in roots by confocal microscopy with a 490 nm excitation and a 535 nm emission filters. Untreated control roots were incorporated to evaluate the ROS levels in standard conditions.
Characterisation of the physiological and cellular effect of chitosan on Arabidopsis roots
Five-day-old Arabidopsis Col-0, DR5rev:GFP, PIN1:PIN1-GFP and WOX5:GFP seedlings obtained as above were transferred to liquid MS medium amended with 0.1 and 1 mg ml−1 chitosan. Liquid MS without chitosan was used as control. After 0–150 min (primary roots) or 24–72 h (secondary roots) in contact with chitosan, seedlings were sampled to determine root reaction to chitosan. Roots were stained with a drop 50 µg ml−1 propidium iodide (PI) for root cell wall labelling. PI and GFP fluorescence were detected in root cells using respectively 488-nm and 580–620 nm (autof.) or 505–530 nm (GFP) as excitation and detection wavelengths in the laser confocal microscope. GFP expression in micrographs was quantified using MetaMorph (Molecular devices, Sunnyvale, Ca, USA). These experiments were performed per triplicate.
Plant hormone quantification in Arabidopsis roots treated with chitosan
Arabidopsis Col-0 wild-type plants were grown under sterile conditions for 4d on upright plates containing 0.5x MS agar. Plants were then transferred to plates with MS (controls) or MS amended with 1 mg ml−1 chitosan and incubated upright for 3d. Roots from ca. 50 plants per treatment were collected and ground in liquid nitrogen. Approximately 100 mg of ground tissue (n = 4 per treatment) per sample and treatment (control/chitosan) were used for hormone extraction. Root tissue was suspended in 80% methanol-1% acetic acid containing internal standards and mixed by 1 h shaking at 4 °C. The extract was centrifuged at −20 °C and the supernatant dried in a vacuum evaporator overnight. Dry residue was dissolved in 1% acetic acid and passed through an Oasis HLB (reverse phase) column as described in Seo et al.63. For gibberellic acid (GAs), indole acetic acid (IAA), abscisic acid (ABA), salicylic acid (SA) and jasmonate (JA) quantification, dried eluate was dissolved in 5% acetonitrile and 1% acetic acid. Hormones were separated using reverse phase UHPLC chromatography (2.6 µm Accucore RP-MS column, 50 mm length × 2.1 mm i.d.; ThermoFisher Scientific) with a 5 to 50% acetonitrile gradient containing 0.05% acetic acid, at 400 µL/min for 14 min. Internal standards for quantification were deuterium-labelled hormones except for JA where dhJA was used. Hormones were identified with a Q-Exactive mass spectrometer (Orbitrap detector; ThermoFisher Scientific) targeted Selected Ion Monitoring (SIM). Hormone concentrations in the extracts were determined using embedded calibration curves as well as program Xcalibur 2.2 SP1 build 48 and TraceFinder.
Hormone Treatments
To evaluate the relevance of hormone balance in the response of Arabidopsis to chitosan, we tested the importance of IAA, SA and JA including these hormones in the MS media. After IAA, SA and JA quantification in Arabidopsis roots, we prepared MS medium amended with these hormones alone and in combination in the same amount (+10%) quantified in Arabidopsis roots exposed to chitosan (Table 1). We also included MS medium (negative control) and MS amended with 1 mg ml−1 chitosan (positive control). Arabidopsis (Col-0) seeds were then plated on MS solid media, stratified, grown and plant scored as described above for 15d.
Histochemical Analysis
For histochemical analysis β-glucuronidase (GUS) activity, DR5:GUS and YUC2:GUS64 A. thaliana seedlings (15 day-old) were incubated for 24 h in MS plates amended with 1 mg ml−1 chitosan. MS plates without chitosan were used for controls. Roots were then incubated overnight at 37 °C in a GUS reaction buffer (0.5 mg ml−1 5-bromo-4-chloro-indolyl-β-D-glucoronide in 100 ml sodium phosphate, pH = 7). Stained seedlings were rinsed in sodium phosphate for 5 min, and then analysed by microscopy65. For each marker line, at least 5–7 transgenic plants were analysed. A representative plant was chosen and photographed, using a digital camera connected to a binocular microscope Nikon SMZ1500 and analysed using ACT-1 2.70 software.
Quantification of gene expression by qRT-PCR
RNA was obtained from eighty roots of 15day-old Col-0 plants exposed for 24 h to chitosan (0.1 or 1 mg ml−1) in MS plates. MS plates without chitosan were used as controls. Roots were frozen and ground and then RNA isolated using Trizol reagent (Thermo Fisher Scientific) following manufacturer’s instructions. Isolated RNA was treated with TurboDNA free (Ambion) for removing DNA remains. cDNA was then synthetized with a retro-transcriptase RevertAid (Thermo Fisher Scientific) using oligo dT (Thermo Fisher Scientific). Gene expression was quantified using real-time reverse transcription PCR (qRT-PCR). SYBR Green with ROX (Roche) was used following the manufacturer instructions. Gene quantifications were performed in a Step One Plus real-time PCR system (Applied Biosystems). Relative gene expression was estimated with the ∆∆Ct methodology66 with three technical replicates per condition. Primers used to quantify the expression of genes related to Arabidopsis response to chitosan are shown in Table S1. ACTIN2 (ACT2) and ORNITHINE CARBAMOYLTRANSFERASE (OTC) genes were used as endogenous controls for all experiments, since their expression showed Ct stability for all conditions tested.
Application of chitosan to the irrigation system of tomato and barley plantlets
Tomato and barley seedlings were planted individually in 150 ml sterile cylindrical containers (Deltalab) each containing 70 cm3 sterilised sand. Twenty-tree ml of 1/10 Gamborg’s B-5 basal medium (Sigma) amended with chitosan (0.01; 0.05; 0.1; 0.5; 1 and 2 mg ml−1) were used to irrigate each seedling. Seedlings watered with 1/10 Gamborg’s only were used as controls. Seedlings were incubated at 25 °C, 65% relative humidity and under a photoperiod (16 h light/8 h dark) in a growth chamber (Fitoclima 10000EHVP). Twenty-one days after planting, 10 plants per treatment were sampled. Fresh shoot weights (FSW), maximum shoot length (MSL), fresh root weight (FRW) and maximum root length (MRL) were measured per plant67. For tomato plants, the number of leaves per plant was also scored. These experiments were replicated 4 times.
Evaluation of cell morphology in tomato plants exposed to chitosan
To assess the effect of chitosan on tomato roots we evaluated root apex cell morphology. The primary root apex from 20 day-old plants was observed in a Leica TCS-SP2 laser-scanning confocal microscope using 488-nm and 580–620 nm as excitation and emission wavelengths68. We scored at least 75 cells per root apex from 3 plants both irrigated with chitosan and controls. We measured Y-axis (root growth direction), X-axis (perpendicular to Y-axis) and surface for each root cell in a 200 × 200 pixels area using Image Analysis Software MetaMorph.
Statistical analyses
Analysis of variance (ANOVA) was used to test for significant differences in experiments69. Homogeneity of variance was checked using Bartlett’s test70. When significant heterogeneity was found, data were 1/x transformed and then ANOVA F-test (p-value < 0.05) was used. When significant heterogeneity was not removed by transformations, analyses were performed on the untransformed data, but F-test α-value was set at 0.0169. Post-hoc analyses were conducted using Tukey HSD comparisons. Analyses were performed using R version 3.0.2. (The R Foundation for Statistical Computing, 2015). All data were reported as mean ± standard error (SE) and statistical tests were conducted with significance level α = 0.05.
Electronic supplementary material
Acknowledgements
This work was supported by AGL 2015 66833-R Grant from the Spanish Ministry of Economy and Competitiveness Grant AGL 2015. We would like to thank Drs Isabel Lopez-Diaz and Esther Carrera for plant hormone quantitation (IBMCP, Valencia, Spain). Part of this work was filed for a patent (P201431399) by L.V. Lopez-Llorca, F. Lopez-Moya and N. Escudero as inventors. We would like to thank Dr Michael Kershaw (University of Exeter) for his English revision and critical comments of the manuscript. We also thank Ms Marta Suarez-Fernandez (University of Alicante) and Mr Alfonso Prieto for their technical support. All the authors reviewed and approved the manuscript.
Author Contributions
F.L.-M.: design and performance of the research, data collection and data analysis, writing the manuscript. N.E.: performance of the research and technical support and writing the manuscript. E.A.Z.-G.: performance of the research and technical support and data interpretation. D.E.-B.: performance of the research and technical support and data interpretation. M.A.B. and D.A.: hormone analysis, data interpretation, manuscript revision. L.V.L.-L.: design of the research, data interpretation, writing the manuscript. All authors have read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-16874-5.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Savary S, Ficke A, Aubertot J-N, Hollier C. Crop losses due to diseases and their implications for global food production losses and food security. Food Sec. 2012;4(4):519–37. doi: 10.1007/s12571-012-0200-5. [DOI] [Google Scholar]
- 2.Pimentel D, Zuniga R, Morrison D. Update on the environmental and economic costs associated with alien-invasive species in the United States. Ecol. Econ. 2005;52(3):273–88. doi: 10.1016/j.ecolecon.2004.10.002. [DOI] [Google Scholar]
- 3.El-Hadrami A, Adam LR. Hadrami El, I. & Daayf, F. Chitosan in plant protection. Mar. Drugs. 2010;8(4):968–987. doi: 10.3390/md8040968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar RMNV. A review of chitin and chitosan applications. React. Funct. Poly. 2000;46(1):1–27. doi: 10.1016/S1381-5148(00)00038-9. [DOI] [Google Scholar]
- 5.Mayakrishnan V, Kannappan P, Abdullah N, Ali AAB. Cardioprotective activity of polysaccharides derived from marine algae: an overview. Trends Food Sci. Technol. 2013;30:98–104. doi: 10.1016/j.tifs.2013.01.007. [DOI] [Google Scholar]
- 6.Lopez-Moya F, et al. Carbon and nitrogen limitation increase chitosan antifungal activity in Neurospora crassa and fungal human pathogens. Fun. Biol. 2015;119(2-3):154–69. doi: 10.1016/j.funbio.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Benhamou N, Theriault G. Treatment with chitosan enhances resistance of tomato plants to the crown and root rot pathogen Fusarium oxysporum f. sp. radicis-lycopersici. Physiol Mol Plant Path. 1992;44(1):33–52. doi: 10.1016/0885-5765(92)90047-Y. [DOI] [Google Scholar]
- 8.Ohta K, Taniguchi A, Konishi N, Hosoki T. Chitosan treatment affects plant growth and flower quality in Eustoma grandiflorum. HortScience. 1999;34(2):233–234. [Google Scholar]
- 9.Van SN, Minh HD, Anh DN. Study on chitosan nanoparticles on biophysical characteristics and growth of Robusta coffee in green house. Biocatal Agric Biotechnol. 2013;2(4):289–294. [Google Scholar]
- 10.Kananont N, Pichvangkura R, Chanprame S, Chadchawan S, Limpanavech P. Chitosan specificity for the in vitro seed germination of two Dendrobium orchids (Asparagales: Orchidaceae) Sci. Hortic. 2010;124(2):239–247. doi: 10.1016/j.scienta.2009.11.019. [DOI] [Google Scholar]
- 11.Malerba M, Cerana R. Chitosan Effects on Plant Systems. Int. J. Mol. Sci. 2016;17(7):pii: E996. doi: 10.3390/ijms17070996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uthairatanakij A, da Silva JAT, Obsuwan K. Chitosan for improving orchid production and quality. Orchid Sci. Biotechnol. 2007;1(1):1–5. [Google Scholar]
- 13.Limpanavech P, et al. Chitosan effects on floral production, gene expression, and anatomical changes in the Dendrobium orchid. Sci. Hortic. 2008;116(1):65–72. doi: 10.1016/j.scienta.2007.10.034. [DOI] [Google Scholar]
- 14.Nge KL, New N, Chandrkrachang S, Stevens WF. Chitosan as a growth stimulator in orchid tissue culture. Plant Sci. 2006;170(6):1185–90. doi: 10.1016/j.plantsci.2006.02.006. [DOI] [Google Scholar]
- 15.Khan TA, Peh KK, Ch’ng HS. Reporting degree of deacetylation values of chitosan: the influence of analytical methods. J. Pharma. Sci. 2002;5(3):205–12. [PubMed] [Google Scholar]
- 16.Iriti M, Faoro F. Bioactivity of grape chemicals for human health. Nat. Prod. Commun. 2009;4(5):611–34. [PubMed] [Google Scholar]
- 17.Pitta-Alvarez SI, Giulietti AM. Influence of chitosan, acetic acid and citric acid on growth and tropane alkaloid production in transformed roots of Brugmansia candida effect of medium pH and growth phase. Plant Cell Tissue Organ Cult. 1999;59(1):31–38. doi: 10.1023/A:1006359429830. [DOI] [Google Scholar]
- 18.Sivanandhan G, et al. Chitosan enhances with anolides production in adventitious root cultures of Withania somnifera (L.) Dunal. Ind. Crops Prod. 2012;37:124–129. doi: 10.1016/j.indcrop.2011.11.022. [DOI] [Google Scholar]
- 19.Chatelain PG, Pintado ME, Vasconcelos MW. Evaluation of chitooligosaccharide application on mineral accumulation and plant growth in Phaseolus vulgaris. Plant Sci. 2014;214(15):134–40. doi: 10.1016/j.plantsci.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 20.El-Tantawi EM. Behaviour of tomato plants as affected by spraying with chitosan and aminofort as natural stimulator substances under application of soil organic amendments. Pak. J. Biol. Sci. 2009;12(17):1164–73. doi: 10.3923/pjbs.2009.1164.1173. [DOI] [PubMed] [Google Scholar]
- 21.Sharp RG. 2013. A review of the applications of chitin and its derivatives in agriculture to modify plant-microbial interactions and improve crop yields. Agronomy. 2013;3:757–793. doi: 10.3390/agronomy3040757. [DOI] [Google Scholar]
- 22.Aranega-Bou P, de la O Leyva M, Finiti I, García-Agustín P, González-Bosch C. Priming of plant resistance by natural compounds. Hexanoic acid as a model. Front. Plant Sci. 2014;1(5):488. doi: 10.3389/fpls.2014.00488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwak JM, Nguyen V, Schroeder JI. The role of reactive oxygen species in hormonal responses. Plant Physiol. 2006;141(2):323–9. doi: 10.1104/pp.106.079004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez-Moya F, Lopez-Llorca LV. Omics for investigating chitosan as an antifungal and gene modulator. J. Fungi. 2016;2(1):1–11. doi: 10.3390/jof2010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Issak M, et al. Neither endogenous abscisic acid nor endogenous jasmonate is involved in salicylic acid-, yeast elicitor-, orchitosan-induced stomatal closure in Arabidopsis thaliana. Biosc. Biotechnol. Biochem. 2013;77(5):1111–3. doi: 10.1271/bbb.120980. [DOI] [PubMed] [Google Scholar]
- 26.Vidhyasekaran, P. Switching on Plant Innate Immunity Signaling Systems: Bioengineering and Molecular Manipulation Of PAMP PIMP PRR Signaling Complex. Signaling and Communication In Plants. Chapt. 3. Switching on Plant Immune Signaling Systems using Microbe-Associated Molecular Patterns/section 3.6 Manipulation of Plant Immune System using Chitosan 144–148, (Springer, 2016).
- 27.Baque MdA, Shiragi MdHK, Lee EJ, Paek K-Y. Elicitor effect of chitosan and pectin on the biosynthesis of anthraquinones, phenolics and flavonoids in adventitious root suspension cultures of Morinda citrifolia (L.) Aust. J. Crop Sci. 2012;6(9):1349–1355. [Google Scholar]
- 28.Khalil MS, Badawy MEI. Nematicidal activity of a biopolymer chitosan at different molecular weights against root-knot nematode. Meloidogyne incognita. Plant Prot. Sci. 2012;48(4):170–178. [Google Scholar]
- 29.Sarkar AK, et al. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature. 2007;446(7137):811–4. doi: 10.1038/nature05703. [DOI] [PubMed] [Google Scholar]
- 30.Ding Z, Friml J. Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proc. Natl. Acad. Sci. USA. 2010;107(26):12046–12051. doi: 10.1073/pnas.1000672107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian H, et al. WOX5-IAA17 feedback circuit-mediated cellular auxin response is crucial for the patterning of root stem cell niches in Arabidopsis. Mol. Plant. 2014;7(2):277–89. doi: 10.1093/mp/sst118. [DOI] [PubMed] [Google Scholar]
- 32.Tiwari SB, Hagen G, Guilfoyle T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell. 2003;15(2):533–43. doi: 10.1105/tpc.008417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doares SH, Syrovets T, Weiler EW, Ryan CA. Oligogalacturonides and chitosan activate plant defensive genes through the octadecanoid pathway. Proc. Natl. Acad. Sci. USA. 1995;92(10):4095–4098. doi: 10.1073/pnas.92.10.4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chandra S, et al. Chitosan nanoparticles: A positive modulator of innate immune responses in plants. Sci. Rep. 2015;5:15195. doi: 10.1038/srep15195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Escudero N, et al. Chitosan enhances parasitism of Meloidogyne javanica eggs by the nematophagous fungus Pochonia chlamydosporia. Fun. Biol. 2016;120(4):572–85. doi: 10.1016/j.funbio.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 36.Escudero N, et al. Chitosan Increases Tomato Root Colonization by Pochonia chlamydosporia and Their Combination Reduces Root-Knot Nematode Damage. Front. Plant Sci. 2017;8:1415. doi: 10.3389/fpls.2017.01415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Atkinson NJ, Lilley CJ, Urwin PE. Identification of Genes Involved in the Response of Arabidopsis to Simultaneous Biotic and Abiotic Stresses. Plant Physiol. 2013;162(4):2028–2041. doi: 10.1104/pp.113.222372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kazan K. Auxin and the integration of environmental signals into plant root development. Ann Bot. 2013;112(9):1655–1665. doi: 10.1093/aob/mct229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Overvoorde P, Fukaki H, Beeckman T. Auxin control of root development. Cold Spring Harb. Perspect. Biol. 2010;2(6):a001537. doi: 10.1101/cshperspect.a001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petricka JJ, Winter CM, Benfey PN. Control of Arabidopsis Root Development. Ann. Rev. Plant Biol. 2012;63:563–590. doi: 10.1146/annurev-arplant-042811-105501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iriti M, Faoro F. Chitosan as a MAMP, searching for a PRR. Plant Signal Behav. 2009;4(1):66–68. doi: 10.4161/psb.4.1.7408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones AM. A new look at stress: abscisic acid patterns and dynamics at high-resolution. New Phytol. 2015;210(1):38–44. doi: 10.1111/nph.13552. [DOI] [PubMed] [Google Scholar]
- 43.Wasternack C. Jasmonates: An Update on Biosynthesis, Signal Transduction and Action in Plant Stress Response, Growth and Development. Ann. Bot. 2007;100(4):681–697. doi: 10.1093/aob/mcm079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wasternack C, Hause B. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. Ann. Bot. 2013;111(6):1021–1058. doi: 10.1093/aob/mct067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kazan K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 2015;20(4):219–29. doi: 10.1016/j.tplants.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 46.Ning Y, Liu W, Wang GL. Balancing Immunity and Yield in Crop Plants. Trends Plant Sci. 2017;, 1385(17):30206–6. doi: 10.1016/j.tplants.2017.09.010. [DOI] [PubMed] [Google Scholar]
- 47.Zhang RQ, Zhu HH, Zhao HQ, Yao Q. Arbuscular mycorrhizal fungal inoculation increases phenolic synthesis in clover roots via hydrogen peroxide, salicylic acid and nitric oxide signaling pathways. J. Plant Physiol. 2013;170(1):74–9. doi: 10.1016/j.jplph.2012.08.022. [DOI] [PubMed] [Google Scholar]
- 48.Dar TA, Uddin M, Masroor AM, Hakeem KR, Jaleel H. 2015. Jasmonates counter plant stress: A Review. Environ. Exp. Bot. 2015;115:49–57. doi: 10.1016/j.envexpbot.2015.02.010. [DOI] [Google Scholar]
- 49.Kazan K, Lyons R. 2016. The link between flowering time and stress tolerance. J Exp Bot. 2016;67(1):47–60. doi: 10.1093/jxb/erv441. [DOI] [PubMed] [Google Scholar]
- 50.Jeong S, et al. Rootin, a compound that inhibits root development through modulating PIN-mediated auxin distribution. Plant Sci. 2015;233:116–26. doi: 10.1016/j.plantsci.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 51.Amborabé BE, Bonmort J, Fleurat-Lessard P, Roblin G. Early events induced by chitosan on plant cells. J. Exp. Bot. 2008;59(9):2317–2324. doi: 10.1093/jxb/ern096. [DOI] [PubMed] [Google Scholar]
- 52.Chen XY, Kim JY. Callose synthesis in higher plants. Plant Signal Behav. 2009;4(6):489–492. doi: 10.4161/psb.4.6.8359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vasil’ev LA, et al. Chitosan-induced programmed cell death in plants. Biochem (Mosc). 2009;74(9):1035–43. doi: 10.1134/S0006297909090120. [DOI] [PubMed] [Google Scholar]
- 54.Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, et al. Hamann, et al. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature. 2003;426(6963):147–153. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- 55.Xu J, et al. A molecular framework for plant regeneration. Science. 2006;311(5759):385–8. doi: 10.1126/science.1121790. [DOI] [PubMed] [Google Scholar]
- 56.Cheng Y, Dai X, Zhao Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006;20(13):1790–1799. doi: 10.1101/gad.1415106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cao H, Bowling SA, Gordon AS, Dong X. Characterization of an Arabidopsis mutant that is nonresponsive to inducers of Systemic Acquired Resistance. Plant Cell. 1994;6(11):1583–92. doi: 10.1105/tpc.6.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dobón A, Wulff BB, Canet JV, Fort P, Tornero P. An allele of Arabidopsis COI1 with hypo- and hypermorphic phenotypes in plant growth, defence and fertility. Plos One. 2013;8(1):e55115. doi: 10.1371/journal.pone.0055115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fernández-Calvo P, et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell. 2011;23(2):701–15. doi: 10.1105/tpc.110.080788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ripoll JJ, Ferrándiz C, Martínez-Laborda A, Vera A. PEPPER, a novel K-homology domain gene, regulates vegetative and gynoecium development in Arabidopsis. Dev. Biol. 2006;289(2):346–59. doi: 10.1016/j.ydbio.2005.10.037. [DOI] [PubMed] [Google Scholar]
- 61.Bordallo JJ, et al. Colonization of plant roots by egg-parasitic and nematode-trapping fungi. New Phytol. 2002;154(2):491–499. doi: 10.1046/j.1469-8137.2002.00399.x. [DOI] [PubMed] [Google Scholar]
- 62.Palma-Guerrero J, Jansson HB, Salinas J, Lopez-Llorca LV. Effect of chitosan on hyphal growth and spore germination of plant pathogenic and biocontrol fungi. J. App. Microbiol. 2008;104(2):541–53. doi: 10.1111/j.1365-2672.2007.03567.x. [DOI] [PubMed] [Google Scholar]
- 63.Seo M, Jikumaru Y, Kamiya Y. Profiling of hormones and related metabolites in seed dormancy and germination studies. Methods Mol. Biol. 2011;773:99–111. doi: 10.1007/978-1-61779-231-1_7. [DOI] [PubMed] [Google Scholar]
- 64.Sabatini S, et al. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell. 1999;99:463–472. doi: 10.1016/S0092-8674(00)81535-4. [DOI] [PubMed] [Google Scholar]
- 65.Weigel, D. & Glazebrook, J. Arabidopsis: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 2002).
- 66.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 67.Escudero N, Lopez-Llorca LV. Effects on plant growth and root-knot nematode infection of an endophytic GFP transformant of the nematophagous fungus Pochonia chlamydosporia. Symbiosis. 2012;57(1):33–42. doi: 10.1007/s13199-012-0173-3. [DOI] [Google Scholar]
- 68.Maciá-Vicente JG, Jansson HB, Talbot NJ, Lopez-Llorca LV. Real-time PCR quantification and live-cell imaging of endophytic colonization of barley (Hordeum ytvulgare) roots by Fusarium equiseti and Pochonia chlamydosporia. New Phytolo. 2009;182(1):213–28. doi: 10.1111/j.1469-8137.2008.02743.x. [DOI] [PubMed] [Google Scholar]
- 69.Underwood, A. J. Experiments in ecology: their logical design and interpretation using analysis of variance. (Cambridge University Press, Cambridge, 1997).
- 70.Bartlett MS. 1937. Properties of sufficiency and statistical tests. Proc. Royal. Soci. London Math. 1937;160:268–282. doi: 10.1098/rspa.1937.0109. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.