Abstract
The first major developmental transition in vertebrate embryos is the maternal-to-zygotic transition (MZT) when maternal mRNAs are degraded and zygotic transcription begins. During the MZT, the embryo takes charge of gene expression to control cell differentiation and further development. This spectacular organismal transition requires nuclear reprogramming and the initiation of RNAPII at thousands of promoters. Zygotic genome activation (ZGA) is mechanistically coordinated with other embryonic events including changes in the cell cycle, chromatin state, and nuclear-to-cytoplasmic component ratios. Here, we review progress in understanding vertebrate ZGA dynamics in frogs, fish, mice, and humans to explore differences and emphasize common features.
Introduction
“Life created without parents” was the front-page New York Times headline on Nov. 28, 1937. The article described experiments by Ethel Browne Harvey showing that sea urchin egg divisions could be triggered even in the absence of sperm or maternal DNA. Harvey had removed egg nuclei by centrifugation and then activated development with salt water. Surprisingly, cell division began and continued until these non-nucleated embryos contained about 500 cells (Harvey, 1936). The slightly less sensational subheading—“New view of cytoplasm”—perhaps more accurately captured the work’s significance: the experiments had conclusively demonstrated that crucial aspects of early development, such as the first rapid cell divisions, can be driven exclusively by maternal cytoplasm without any contribution from the maternal and paternal nuclei and genomes.
Eventually, however, the zygotic genome is required for further development. When embryonic RNA production is blocked with transcriptional inhibitors, development eventually stalls (Brachet et al., 1964; Golbus et al., 1973). Such experiments helped define the concept of the maternal-to-zygotic transition (MZT), which we consider not a single transition but as a series of events following fertilization up to the stage when the embryonic genome is fully transcriptionally active and maternal stores of many mRNA are depleted (Davidson, 1986; Lee et al., 2014; Tadros and Lipshitz, 2009).
The MZT is essential for development because it coordinates cell division and zygotic gene activation to prepare the embryo for cell differentiation and further development. In many cases, a primary role of early embryonic gene expression is to provide the molecular substrates to initiate gastrulation, which entails coordinated cell movements and germ layer specification. It is at this point in development that the animal begins to acquire different cell fates and specific morphological forms. While it is unknown what precise effect ZGA failure has on human development, it may contribute to pre-implantation pregnancy failure of abnormal embryos (Hertig et al., 1956; Niakan et al., 2012).
Two key features distinguish ZGA from transcription in other cellular transitions. First, ZGA takes the embryo from a state in which there is little if any transcription to a state where up to thousands of genes are transcribed. This contrasts with other developmental transitions wherein a cell’s global transcription profile remains largely unperturbed and a few transcription factors are sufficient to instruct cell fate along specific lineages by activating subsets of genes. Second, embryo cell division can proceed without cell growth because the massive oocytes are filled with maternal mRNA and protein. This results in drastic changes in the ratio between maternally deposited proteins and nuclear constituents such as the genome, a property exploited to direct molecular events.
Here, we review the mechanisms for how embryonic gene expression and genomic reprogramming are initiated and regulated during vertebrate embryogenesis. Although faster developing embryos are usually considered distinct from slower developing mammals, here, we review model species from both categories. We focus primarily on mechanistic models for zygotic genome activation (ZGA) and, where appropriate, refer readers to other reviews for discussions on other aspects of the broader MZT including the maternal contribution and degradation of maternal mRNAs (Svoboda et al., 2015; Yartseva and Giraldez, 2015).
Two broad categories of pre-gastrulation vertebrate embryo development
Vertebrates are divided into two classes: anamniotes, which lay eggs externally in water and include bony fish and amphibians, and amniotes, which lay fertilized eggs on land or retain them in the mother in a protective membrane impermeable to water, and include mammals, birds, and reptiles. Anamniote embryos contain everything needed to fuel development until feeding structures allow them to obtain external nutrients (O’Farrell, 2015). In addition, anamniote embryos typically have a period of rapid cell divisions following fertilization, whereas amniote embryos develop more slowly. This distinction in fast and slow developmental modes provides a useful dichotomy through which to explore differences and identify whether conserved principles of the vertebrate ZGA exist.
Here, as examples of fast-developing species, we review work on the model amphibian, the western clawed frog Xenopus (both X. laevis and X. tropicalis) and the model teleost fish, Danio rerio (zebrafish). As examples of slow-developing species, we primarily review work on the house mouse, Mus musculus, and, when data are available, humans. Within each vertebrate developmental mode, there are both broad similarities and important molecular differences, both of which we endeavor to highlight. We relate concepts from invertebrates such as Drosophila when they inform the vertebrate mechanism, but refer the reader to recent reviews for a more extensive treatment of invertebrate ZGA (Blythe and Wieschaus, 2015; Harrison and Eisen, 2015).
Fast-developing embryos
One of the most salient distinctions between fast- and slow-developing embryos is the duration of the first cell division cycles (Fig. 1). Fast-developing embryos often exhibit rapid synchronous cell cycles that rapidly amplify cell number before gastrulation. Early embryonic divisions do not require transcription and are driven by maternally supplied RNAs and proteins. Moreover, these early cell cycles lack G1 and G2 gap phases and instead alternate between S and M phases. Then, after a regulated number of divisions, the cell cycle becomes somatic-like prior to gastrulation.
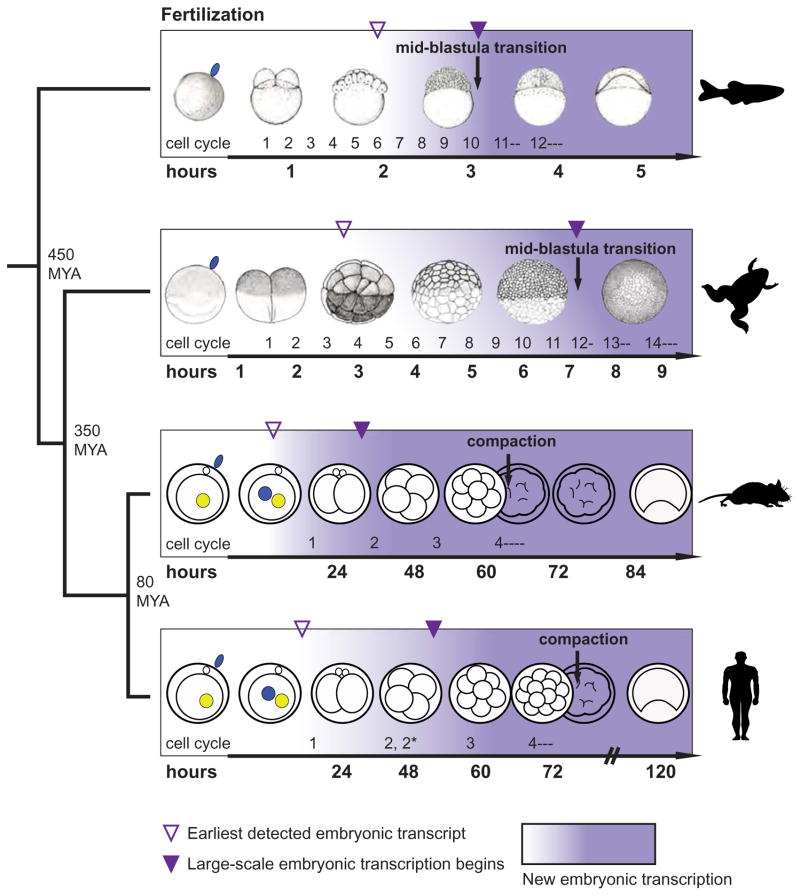
Figure 1. Timing of transcriptional onset and early embryonic events varies in vertebrates.
Evolutionary distance, embryo morphology, cell division timing and number in pre-gastrulation development for zebrafish (Danio rerio), frog (Xenopus laevis), mouse (Mus musculus), and human (Homo sapiens). X. laevis and X. tropicalis have broadly similar activation timing with respect to cell division number (Yanai et al., 2011). Purple shading density illustrates amount of zygotic transcription. In the fast developing species, fish and frog, cell divisions are rapid until the mid-blastula transition at the 10th and 12th cycles. In slow developing species, the initial divisions take place over days. Divisions ‘2’ and ‘2*’ refer to the two asynchronous divisions in the 2nd cleavage cycle. Xenopus embryo images adapted from (Nieuwkoop and Faber, 1994); Danio rerio embryo images were reproduced from (Kimmel et al., 1995) with permission (John Wiley and Sons).
Rapid cell cycles proceed up to the Mid-Blastula Transition (MBT), a series of cellular changes whose outlines are conserved in many fast-developing vertebrates. For example, in zebrafish, rapid maternally driven cell cycles occur until the 10th cycle, when S-phase lengthens, gap phases appear, and cell cycles become sensitive to DNA damage (Ikegami et al., 1999; Kane and Kimmel, 1993; Kane et al., 1996). In Xenopus laevis, the first cell division occurs ~90 minutes after fertilization, followed by 11 synchronous divisions every ~30 minutes, depending on temperature (Dettlaff and Rudneva, 1975; Satoh, 1977). At the 12th division, the cell cycle slows when S-phase lengthens, divisions of neighboring cells become asynchronous, and cells become motile (Iwao et al., 2005; Newport and Kirschner, 1982a). Concurrently, replication and DNA damage checkpoints are activated, and G1 and G2 phases emerge (Anderson et al., 1997; Kermi et al., 2015; Masui and Wang, 1998). In both species, the initial cell cycle lengthening coincides with a significant increase in zygotic transcription, but does not necessarily depend on zygotic transcription. Indeed, the timing of cell cycle lengthening in early Xenopus and zebrafish embryos is unchanged when zygotic RNA production is experimentally inhibited (Brachet et al., 1964; Kane and Kimmel, 1993; Kirschner et al., 1980; Newport and Kirschner, 1982a).
Slow-developing embryos
Early mammalian development is divided into pre- and post-implantation stages. The cleavage-stage of pre-implantation development is similar to rapid developing embryos in that it entails cell divisions without cell growth (Niakan et al., 2012). However, mammalian cleavage cycles are much slower: the first division occurs after 18–36 hours, and subsequent cleavage divisions take place every ~12–24 hours until blastocyst formation. Finally, several days after fertilization, the mammalian embryo hatches out of the zona pellucida, implants into the uterine wall, and grows rapidly while obtaining material and nutrients from the mother’s blood stream.
Transcription dynamics in fast-developing embryos
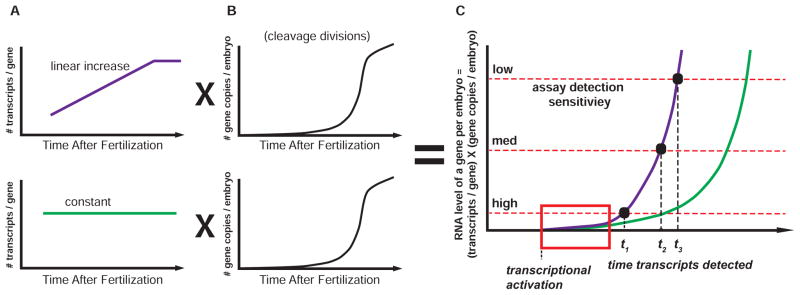
When is the zygotic genome first activated? While this question has been posed many times, obtaining an accurate answer has been difficult due to technical and conceptual challenges (see Box1). The interpretation of what constitutes zygotic transcriptional activation for a specific gene is complicated by the fact that a measured increase in transcript levels results from both an increase in the number of genomes per embryo and an increase in the transcription rate per gene (Fig. 2A–B). Even without an increase in the transcription rate per gene, the number of a given gene’s zygotic transcripts per whole embryo will double with each cell division as long as there is some basal rate of transcription. In addition, maternal RNA stores and a dynamic ‘total’ transcriptome can confound accurate transcript quantification. Thus, detection of a specific gene’s zygotic transcripts above an assay’s detection threshold may not accurately reflect when transcription of that gene was initiated (Fig. 2C).
Technology Box. Distinguishing Zygotic and Maternal transcripts.
-RNA-seq and differential expression
RNA-seq time courses are a straightforward way to define zygotic transcripts. Genes whose expression increases over several consecutive time points or genes whose expression rises above that of the unfertilized egg are likely activated in the embryo. This approach is sensitive enough to detect transcript-level changes above maternal background for many genes. However, one cannot definitively distinguish maternal and zygotic transcripts, as RNA level dynamics are a composite of increasing zygotic expression and maternal mRNA degradation. Best practices include normalizing transcript levels to spike-in abundance controls, and using sufficient replicates or high frequency time points for better variability assessment and statistical power to detect differentially expressed transcripts. In embryos, the commonly used RNA-seq preparation method of polyA selection may lead to erroneous conclusions, as maternal polyA lengths are often dynamic after fertilization, which could alter a gene’s perceived relative expression level. Approaches that avoid this issue include rRNA depletion and random priming during cDNA synthesis, or direct probe-based RNA counting methods (e.g., nanostring technology). In general, the sensitivity of RNA-seq based approaches depends upon read depth and the relative abundance of the transcript(s) of interest. For example, in their embryogenesis RNA-seq time course, Owens et al. estimated that, on average, ~1300 transcripts per embryo were required to generate a single read at a sequencing depth of ~35M reads per sample (Owens et al., 2016). This is approximately 1–2 transcripts per cell at the 1000 or 2000 cell stage, prior to the bulk of zygotic transcription. However, in practice, one often requires a higher minimum number of reads per transcript to consider that gene “expressed”.
-RNA-seq and Natural Sequence Variation
Paternal mRNAs are generally not present in sperm. This property can be exploited to detect zygotic RNAs from the paternally supplied genome by mating parental strains with enough sequence divergence to distinguish paternal and maternal transcripts, as performed in Drosophila and Zebrafish (Harvey et al., 2013; Lu et al., 2009). This method is straightforward, but requires two fully sequenced strains and sufficient SNP variation between transcripts to avoid mis-alignment of reads.
Introns as proxy for nascent transcripts
Although mRNAs are rapidly spliced during transcript elongation, they are initially unspliced as they dissociate from the DNA-RNAPII-transcript ternary complex. Thus, intronic read levels from RNA-seq likely correlate with nascent embryonic transcription (Lee et al., 2013), and can be used to define zygotic gene timing. However, the recently discovered circular RNAs–which can contain unspliced introns–expressed in human oocytes and embryos (Dang et al., 2016) and stable intronic sequences (sisRNAs) in early frog embryos can be confounding factors (Talhouarne and Gall, 2014).
-Blocking transcription and measuring differential expression
One can block transcription with small molecule RNAPII inhibitors such as α-amanitin or triptolide, and compare RNA levels to those of untreated embryos. This approach may be sensitive to α-amanitin levels, however, or the timing of treatment.
-Positive selection of zygotic transcripts using metabolic labeling
Metabolic labeling of newly transcribed RNA uses UTP analogs, such as BrdU, 4SU, EU or FU, which are injected into embryos or taken up in cell culture (Tani and Akimitsu, 2012). After modified nucleotides are incorporated into nascent RNAs, RNA is extracted and labeled with biotin or other chemical moieties used to isolate the RNA. The captured RNA is eluted and sequenced with standard RNA-seq preparations. This approach distinguishes zygotic from maternal transcripts (Heyn et al., 2014), as maternal transcripts should remain in the supernatant during the isolation procedure. There exists some potential for length bias, however if nucleotides are not efficiently incorporated into short transcripts. Embryos must also be carefully examined for viability, as some nucleoside analogs can be toxic to the organism at high doses (Burger et al., 2013).
-Single-molecule FISH (smFISH)
smFISH uses multiple dye-labeled probes that hybridize to a specific target mRNA to produce a punctate fluorescent signal for each mRNA. Although the sensitivity of smFISH has not been established in each model vertebrate, in cell culture one can reliably detect single mRNA molecules (Raj and Tyagi, 2010).
Figure 2. Initiation of embryonic transcription is difficult to detect.
Several factors influence the total transcription of a specific gene in embryos. (A) When a gene is activated after fertilization, the number of transcripts per gene (i.e., per copy of the genome) is likely linearly increasing (purple) or constant (green). (B) In addition, the number of copies of the genome per embryo is doubling with each cell division. Embryos exhibit an exponential increase in genome copies per unit time during cleavage divisions. (C) Total transcription—and total mRNA production—for a given gene is related to the product of the transcription per genome and the number of genomes. The total mRNA per embryo may thus increase exponentially or even hyper-exponentially in the first hours after activation. The more sensitive the method, the earlier one expects to detect zygotic transcription as indicated by times t1, t2, and t3. Yet, even with a dramatic increase in assay sensitivity (2-fold and 10-fold increases are shown), it may be difficult to detect the earliest transcription when assaying total embryo RNA (red box).
It is now clear that the view that zygotic transcription is ‘activated’ at a single moment is not accurate. Recent studies have elucidated the entire time-course of zygotic gene expression by examining differential expression of individual genes over consecutive time points (Collart et al., 2014; Mathavan et al., 2005; Paranjpe et al., 2013; Tan et al., 2013; Yanai et al., 2011; Yang et al., 2013). This showed that ZGA consists of different genes being activated at distinct developmental stages, many of which precede cell cycle lengthening.
Transcription dynamics are broadly similar in frogs and fish. In Xenopus, the first transcribed zygotic RNAs are detected in the 8-cell embryo and include the miRNA, pri-mir427, which likely regulates maternal mRNA degradation (Giraldez et al., 2006; Owens et al., 2016). The first detected phase of ZGA includes dozens of zygotic transcripts at the 128- and 256-cell stages. These early expressed genes include several transcription factors and the beta-catenin targets nodal5 and nodal6 (Blythe et al., 2010; Skirkanich et al., 2011), which likely function to prepare the embryo for later gene expression. Transcription continues to increase over several cell cycles. Several hundred genes initiate expression closer to the 10th and 11th cycle, with further increases as embryos traverse the MBT (Collart et al., 2014; Owens et al., 2016).
As in frogs, Zebrafish ZGA begins during the early cleavage divisions and is phased in across early development up through the MBT and gastrulation. Zygotic transcripts have been detected as early as the 64-cell stage and include the mir-430 transcript, which is crucial for clearing maternal transcripts (Giraldez et al., 2006; Heyn et al., 2014; Wei et al., 2012). Approximately 600 zygotic transcripts have been detected by the 512-cell stage. Many of these early transcripts are short, which suggests the brief interphase is insufficient to complete long transcripts before transcription is aborted in mitosis, as occurs in flies (Heyn et al., 2014; Shermoen and O’Farrell, 1991). However, this model remains to be tested in vertebrates. Finally, a larger-scale increase in zygotic transcription takes place around the 10th division, coinciding with the MBT (Aanes et al., 2011; Harvey et al., 2013; Mathavan et al., 2005; Yang et al., 2013). As in frogs, zebrafish ZGA expresses diverse transcript types including mRNA, miRNA, piRNA, and lncRNAs (Forouzmand et al., 2016; Houwing et al., 2007; Pauli et al., 2012; Wei et al., 2012). While much progress has been made in describing ZGA dynamics, we know much less about the functions in early development of the diverse set of zygotic transcripts.
Mammalian transcription dynamics
Mouse ZGA is characterized by phases of transcription often referred to as “waves, although this definition may be dependent on sampling frequency. Nonetheless, many genes appear to be transcribed only in a specific stage, while a minority of genes maintain active transcription throughout pre-implantation development (Hamatani et al., 2004). After fertilization in mouse, genetic material from the mouse egg and sperm remain in separate pronuclei in the zygote. The first embryonic transcripts are detected in the male pronucleus during the G2 phase of the first cell cycle (Aoki et al., 1997; Nothias et al., 1996). The pronuclei then fuse and commence the first embryonic division approximately 20 hours after fertilization. Transcription at the 2-cell stage is RNAPII-dependent and is required for further cell division (Bolton et al., 1984; Golbus et al., 1973; Hamatani et al., 2004; Li et al., 2010; Xue et al., 2013). A second phase of transcription initiates at the 4-to-8 cell transition and marks the beginning of dynamic morphological changes that lead to formation of the blastocyst (Hamatani et al., 2004; Wang and Davis, 2014). At the 8-cell stage, embryo blastomeres increase the surface area of their cell contacts in a process known as compaction. Around this time, the first cell fates are specified as inner cell mass (ICM) and trophectoderm (TE), which will form the embryonic and extraembryonic (placental) tissues, respectively, followed by cavitation wherein fluid accumulates between blastocysts and separates the ICM and TE. Because mammalian embryos need to implant to receive nutrients, a primary goal of mammalian ZGA may be to prepare for ICM-TE differentiation so that the TE can specify the extraembryonic tissue necessary for post-implantation nutrient acquisition.
The morphological features of human early embryo development resemble that of mice in that they undergo pronuclear fusion, several cleavage divisions, compaction, morula formation, and cavitation. However, the timing of ZGA and morphological changes are slightly different, as human ZGA accelerates at the 4–8 cell stage rather than the 2-cell stage as occurs in mouse. Two recent studies of human embryos found ~150 genes upregulated from the oocyte to 1-cell stage, and ~1000 genes upregulated from the 2-cell to the 4-cell stages (Xue et al., 2013; Yan et al., 2013). The first large cohort of RNAPII-mediated transcription of ~2500 genes then occurs at the 4–8 cell stage and is required for cell divisions past the 8-cell stage (Braude et al., 1988; Dobson et al., 2004; Taylor et al., 1997; Tesarík et al., 1987; Vassena et al., 2011; Xue et al., 2013; Yan et al., 2013), followed by ~2500 genes activated at the 8–16 cell stage (Yan et al., 2013). The timing of human ZGA at later cell stages is similar to other mammals, including cow, sheep, rabbit, and macaque, suggesting that human early development may be more representative of mammals than mice (Christians et al., 1994; Crosby et al., 1988; De Sousa et al., 1998; Frei et al., 1989; Schramm and Bavister, 1999).
Strikingly, early mammalian transcription includes several transposons and repetitive elements including endogenous retroviruses (Kigami et al., 2003). The synthesis of viral particles during ZGA was first discovered almost 40 years ago, when Intracisternal A-type viral particles (IAP) were found present in the 2-cell mouse embryo, but not gametes, indicating that IAP expression is likely of early embryonic origin (Biczysko et al., 1973; Calarco, 1975). In mouse, many of these elements are de-repressed early, then repressed again during the MZT to prevent mutations and maintain genome integrity in the germline or early embryo (Macfarlan et al., 2012). In humans, the endogenous retroviral (ERV) element, HERVK-HML-2, expresses at the 8-cell stage through the blastocyst stage before being repressed again (Grow et al., 2015). Unlike other HERVs with coding mutations preventing expression of their viral proteins, a HERVK-HML2 encoded protein was detected in pluripotent stem cells and affected ribosome occupancy on mRNA. This suggests a dedicated developmental function for some ERV gene products (Grow et al., 2015).
Do these elements express similarly early in frogs and fish? Currently, we do not know. In fish, the expression of piwiRNAs (piRNA)–many of which are derived from LTRs of transposable elements–increases across the blastula stage to mid-gastrulation (Houwing et al., 2007; Wei et al., 2012). Yet whether these piRNAs are required for embryo development, or are merely leftover maternal products involved in transposon silencing and genome integrity maintenance is unclear. Most vertebrate genomes comprise a large proportion of repeats and transposable elements, so it will be interesting to see if early frog and fish also express repetitive elements and retroviruses in early cleavage stages and what role this may have in their ZGA.
Reprogramming chromatin for embryonic transcription—a clean slate?
How is the genome prepared for zygotic transcription? In John Gurdon’s classic experiment, transferring the nucleus of a frog somatic cell into an enucleated frog oocyte ‘reprograms’ the somatic nucleus so that it supports the development of an adult frog (Gurdon, 1962). The maternal cytoplasm thus has the remarkable ability to reset the chromatin state of a terminally differentiated nucleus. An analogous process occurs after fertilization: The transition from specialized and transcriptionally silent gametes into a totipotent and transcriptionally active embryo requires comprehensive reorganization of zygotic chromatin (Biechele et al., 2015; Bultman et al., 2006; Burton and Torres-Padilla, 2014).
Vertebrate chromatin remodeling begins immediately after fertilization when protamines, positively charged proteins that tightly package the genome inside the sperm nucleus, are replaced by maternally supplied histones (Braun, 2001). The rapid protamine exchange is viewed as generating a somatic chromatin state that sets the stage for ZGA. Although most sperm DNA is compacted with protamines, some of the DNA is not—about 10–15% in humans, 5–10% in mouse, the entire zebrafish genome, and species-dependent amounts in amphibians is instead bound by paternal nucleosomes (Jung et al., 2017; Mann et al., 1982; Wu et al., 2011). This raises the possibility that stable nucleosome occupancy at particular genes could influence zygotic gene expression possibly by maintaining histone modifications at these loci (Carone et al., 2014; Hammoud et al., 2009; Jung et al., 2017; Royo et al., 2016; Teperek et al., 2016; van der Heijden et al., 2005). Some discrepancies exist among which loci are believed to retain nucleosomes, perhaps due to slight differences in experimental conditions (Saitou and Kurimoto, 2014). Nevertheless, histone retention differs between the sperm of fertile and infertile men (Hammoud et al., 2011) and, collectively, these studies suggest that paternal chromatin may influence embryonic gene activation.
Following rapid protamine-histone exchange, several maternal-specific histone variants are gradually replaced with somatic variants. Some maternal histone variants exchange through cessation of their embryonic synthesis and dilution through DNA replication. Others, such as the Xenopus H1 linker variant, H1M, persist in the chromatin until their somatic variants are synthesized at the MBT (Dimitrov and Wolffe, 1996; Dworkin-Rastl et al., 1994). Generally, histone variant exchange may promote structural changes in promoter nucleosomes to make local chromatin more or less accessible. For example, in frogs, H1M is less basic than somatic H1 and potentially generates less stable chromatin overall, which could be useful during the rapid early cell divisions and for initiating transcription (Freedman and Heald, 2010). In mice, deposition of the H3 variant H3.3 maintains open chromatin in pre-implantation embryos, consistent with the association of H3.3 with transcriptionally active loci in general (Lin et al., 2013; 2014; Torres-Padilla et al., 2006; van der Heijden et al., 2005). Altogether, these early chromatin exchanges help ‘reprogram’ the differentiated germ cell chromatin to prepare for embryonic transcription.
Models for MZT timing
Three broad classes of models help explain ZGA timing. The first model claims that MZT dynamics are triggered by the accumulation of maternally deposited activating transcription factors. A second model claims that MZT dynamics are triggered by reaching a threshold ratio of a nuclear component to the cytoplasmic volume, often referred to as the “N/C-ratio”. A third model posits that the de novo establishment of chromatin states permissive for transcription is critical for ZGA timing because embryos must start from a state with no transcription or active RNAPII. Importantly, these three timing mechanisms are not mutually exclusive and additional mechanisms may also exist.
Nuclear-to-Cytoplasmic ratio mechanisms
One of the unique aspects of vertebrate oocytes is their massive size. Across metazoans, oocytes can be thousands of times the size of mitotically competent somatic cells such as epithelial or stem cells, despite containing a single haploid genome. Once fertilized, the enormous cell volumes in the early embryo are decreased two-fold with each division cycle leading to a doubling of the nuclear-to-cytoplasmic or DNA-to-cytoplasmic ratio (the “N/C-ratio”). The hypothesis that the exponential increase in N/C-ratio through early development can regulate embryonic events developed over many decades by observations in multiple species (Dettlaff, 1964; Gerhart, 1980; Kirschner et al., 1980). Observations in Triturus (newt) described a progressive increase in the N/C-ratio that stabilized during the sharp transition from synchronous to asynchronous cell cycles (Schönmann, 1938). This transition was later defined as a singular event termed the “midblastula transition” (MBT) from studies in Axolotl (Signoret and Lefresne, 1971). Irradiation experiments in loach fish linked a functional genome to changes in cell cycle timing, roughly at the stage now known as the MBT (Neyfakh, 1959). Later authors speculated that this event could be triggered by a particular “threshold ratio between quantity of nuclear DNA and other cell substances” (Rott and Sheveleva, 1968).
While the N/C ratio model broadly refers to the ratio of one or more nuclear components to the embryo cytoplasm, experiments manipulating zygotic ploidy helped establish DNA as the most likely nuclear numerator (Kirschner et al., 1980; Newport and Kirschner, 1982b). In animals as diverse as the loach, salamander, newt, frog, or zebrafish, experimentally generated haploid embryos or embryos with more cytoplasm underwent the MBT one cell cycle later than diploid embryos. Conversely, tetraploid embryos and embryos with reduced cytoplasmic volume undergo the MBT one cell cycle earlier (Chulitskaia, 1970; Kane and Kimmel, 1993; Kobayakawa and Kubota, 1981; Masui and Wang, 1998; Rott and Sheveleva, 1968).
A role for the DNA-to-cytoplasmic ratio in controlling the timing of multiple events in early Xenopus development was further supported by experimental manipulation of the two parts of this ratio (Newport and Kirschner, 1982a; Clute and Masui, 1995). A 1-cell Xenopus embryo was partially constricted into two equal sized lobes. The half of the embryo containing the nucleus divided two to three times before a division introduced a complement of chromosomes into the other half of the embryo. In one half, the MBT occurred 2 to 3 cycles before the other half. Thus, the MBT was triggered in both halves at the same DNA-to-cytoplasmic ratio, but after different numbers of cell divisions, and at different times since fertilization. Importantly, bulk RNA synthesis, measured using radiolabeling, was also controlled by the DNA-to-cytoplasm ratio at the MBT (Newport and Kirschner, 1982b). Taken together, these experiments show that one or more N/C ratios control the MBT in fast developing animals.
Molecular titration mechanisms sensing the N/C ratio
One proposal for how the DNA-to-cytoplasm ratio controls transcription is through titrated repressors. In this model, the DNA content doubling with each round of division titrates away maternally deposited inhibitory factors until zygotic gene activation begins at a critical DNA-to-cytoplasm ratio (Fig. 3A–D).
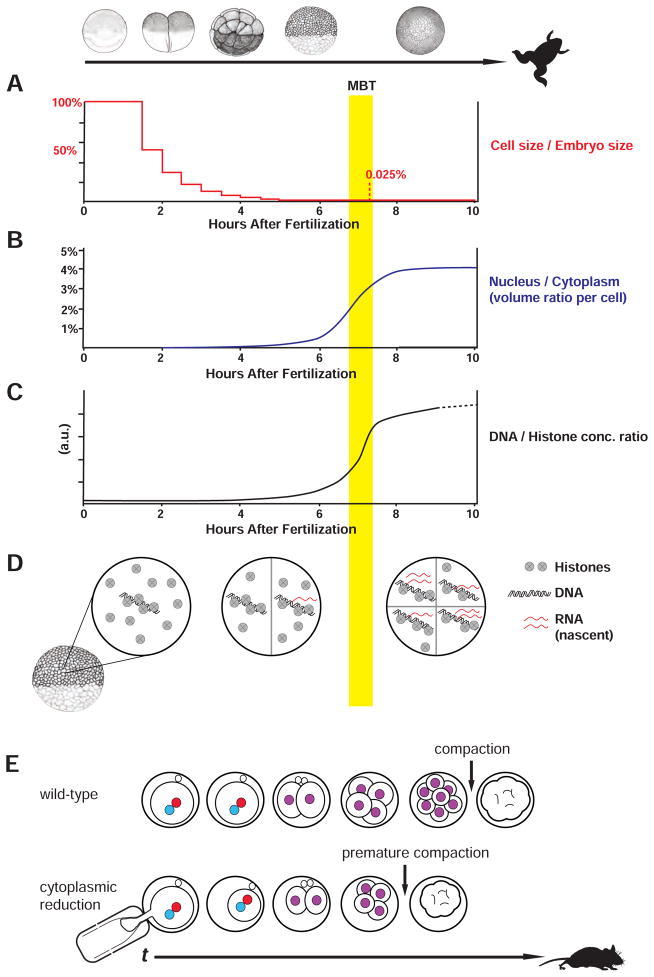
Figure 3. Nuclear-to-cytoplasmic (N/C) ratio-based models for determining the timing of early developmental events.
The numerator in the N/C model refers either to nuclear volume or some other nuclear component such as DNA. (A–C) Frog ZGA is regulated, in part, by the N/C ratio. (A) Cell size (i.e., volume) decreases by half with each division in an embryo of constant volume. (B) The ratio of nuclear volume to cytoplasmic volume increases through the mid-blastula transition and may control transcription through an unknown mechanism. Data re-plotted from (Jevtić and Levy, 2015). (C) The ratio of DNA-to-cytoplasm may be encoded by the ratio of DNA-to-histones as histone concentration is constant before the MBT. (D) Cartoon illustrates how a histone titration model may control transcription. (E) In mouse embryos, cell size decreases by half with each division before implantation (top row). N/C ratio may control compaction, where the embryo transitions from a loosely connected group of spherical cells to a tight mass without gaps between cells due to increased cell-to-cell contacts. (bottom row) Removal of ~50% of zygote cytoplasm induces compaction one cell cycle earlier (Lee et al., 2001). Zygote pronuclei are in red and blue; embryonic nuclei are in purple.
The first suspects for the elusive titrated inhibitor were chromatin components required to compact DNA and repress transcription (Almouzni and Wolffe, 1995; Prioleau et al., 1994). In a key set of experiments, transcription was measured following reporter plasmid injection. Plasmid reporter transcription is repressed from fertilization up to the MBT, at which time it reactivates (Prioleau et al., 1994). Moreover, pre-MBT repression correlated with plasmid chromatinization, which could be prevented by the injection of excess DNA. It was therefore proposed that an excess of histones could outcompete limiting transcription factors to create a repressive environment during early cleavages. Then, repression would be relieved by histone-DNA titration that allows transcription factors to bind (Almouzni and Wolffe, 1995; Pálfy et al., 2017; Prioleau et al., 1994; 1995). In addition, alleviation of repressive chromatin may also require the accumulation of rate-limiting general activators such as the TATA-binding protein, TBP (Veenstra et al., 1999).
An independent set of experiments provided evidence for titration of histones to regulate the MBT in vivo (Amodeo et al., 2015). The transcriptional inhibitory activity of Xenopus extract was exploited to reconstitute a DNA-to-cytoplasmic ratio-dependent transcriptional response in vitro. Through sequential biochemical purifications, the inhibitory activity was isolated as being due to histones H3 and H4. Crucially, manipulations of histone levels in embryos altered the timing of the MBT in the predicted fashion. Histone H3 morpholino injection at the one-cell stage reduced H3 levels by ~50% at the MBT. This H3 reduction advances the MBT by precisely one cell cycle as assayed by global transcription and cell cycle lengthening (Amodeo et al., 2015). In addition to these experiments, histones fulfill several criteria for a globally titrated inhibitor: 1) histones bind DNA across the genome; 2) histones are numerous in the early embryo (Woodland and Adamson, 1977); 3) histones inhibit transcription (Han and Grunstein, 1988); 4) histone depletion in somatic cells can increase the RNAPII transcriptional elongation rate (Jimeno-González et al., 2015; Prado et al., 2016); 5) total histone levels are roughly constant or only slowly increase from fertilization to the MBT, so that their ratio to DNA accurately reflects the DNA-to-cytoplasm (N/C) ratio throughout early development. Thus, histone titration is likely linked to chromatin assembly, cell cycle elongation, and transcription in the frog embryo.
How would histone titration work at the level of individual gene expression? One possibility is that transcriptional onset is determined by the competition of free histones and specific transcription factors at gene loci (Joseph et al., 2017). Consistent with this model, in zebrafish, addition of histones delays transcription, while addition of activating transcription factors advances transcription. Moreover, reducing the concentration of free histones advances transcription of several tested genes (Joseph et al., 2017; Pálfy et al., 2017). Thus, critical activation thresholds could be reached at different times for different genes depending on the concentrations and DNA binding kinetics of the specific activator. Whether such competition occurs during histone deposition after the replication fork, or during replication-independent nucleosome exchange, and whether this mechanism applies broadly or only to subsets of genes, remains to be determined.
In addition to histones, DNA replication factors have been implicated in Xenopus MBT regulation. S-phase is the first part of the cell cycle to elongate at the MBT (Iwao et al., 2005), and, in a key experiment, four conserved eukaryotic replication factors Cut5, RecQ4, Treslin, and Drf1, were overexpressed together in vivo to induce additional rapid cell cycles in a dose-dependent manner (Collart et al., 2013). Interestingly, these replication factors are specifically degraded near the MBT, suggesting that the effects on replication may be downstream of an earlier MBT trigger, which might target replication factors for degradation to elongate S-phase. However, the effects on zygotic transcription were moderate (Collart et al., 2013), consistent with the likely existence of multiple titrated molecules, with some having greater influence on replication and others—such as histones—perhaps having greater influence on transcription (Amodeo et al., 2015; Joseph et al., 2017). In addition to histones and replication factors, other candidate titrated molecules include the phosphatase PP2A-B55 (Murphy and Michael, 2013), maternal histone variants (Yue et al., 2013), the DNA methyl transferase xDnmt1 (Dunican et al., 2008), and dNTPs (Landström et al., 1975; Vastag et al., 2011).
In addition to DNA, other nuclear components could also contribute to the N/C-ratio, or affect nuclear concentrations of titrated factors. For example, nuclear size changes have been implicated in transcriptional activation in Xenopus embryos (Jevtić and Levy, 2015). The N/C volume ratio increases sharply during early embryogenesis, and this presumably affects nuclear import rates, and likely alters the nuclear concentration of transcription factors and potentially titrated molecules such as histones or replication factors. In addition, as ploidy changes correlate with nuclear size in many vertebrates (Fankhauser, 1945; Vuković et al., 2016), there exists the intriguing possibility that nuclear size changes could explain some aspects of ploidy phenotypes. Taken together, this recent body of work suggests that a unified molecular model for how the N/C ratio controls developmental events may need to incorporate a diverse set of factors. Importantly, such a model will also need to resolve the mechanism(s) for how N/C-ratio based changes regulate the transcription of specific loci.
Does the N/C ratio control early mammalian development?
While it has long been appreciated that N/C ratio mechanisms control aspects of early development in many fast-developing species, such mechanisms may also apply to slower-developing vertebrates. Just as in fast developing species, early pre-implantation mammalian development entails cell division without cell growth, which drives a dramatic increase in the N/C ratio (Tsichlaki and FitzHarris, 2016). For example, in pre-implantation mouse embryos, the amount of cytoplasm remains constant through the late-blastocyst 64-cell stage leading to an exponential increase in the N/C ratio with each division cycle (Aiken et al., 2004).
An N/C ratio mechanism may control the first major morphological event of mouse development, cell compaction (Kojima et al., 2014). In mouse embryos, compaction occurs at the 8–16 cell stage and denotes the transition from a loosely connected group of spherical cells to a tight mass without gaps between cells (White et al., 2016). Despite the clear intercellular nature of the event, compaction may depend on the N/C ratio within single cells rather than the total number of cells in the embryo. When 4-cell embryos are split into two 2-cell embryos, compaction begins when these half-sized embryos reach the 4–8 rather than 8–16 cell stage (Fernandez and Izquierdo, 1980; Smith and McLaren, 1977). Moreover, reducing embryo size 2-fold by extracting cytoplasm at the 1-cell stage advances compaction by a single cell cycle (Lee et al., 2001) (Fig. 3E). Inhibiting cell and nuclear division, but allowing DNA replication, results in compaction at a similar time since fertilization at the same DNA-to-cytoplasm ratio as control embryos (Pratt et al., 1981). Analysis of total transcription in cytoplasm-reduced embryos suggested it is largely unaffected. Therefore, it seems unlikely that titration of a global chromatin regulator, such as histones, would sense the N/C-ratio in mammals. However, in other model organisms, such as flies, the N/C ratio controls only a subset of genes (Lu et al., 2009), so it would be interesting to revisit these mouse experiments using modern single-cell sequencing technologies to test whether N/C-ratio changes regulate any genes in mammals.
Limitations of N/C ratio models and the complexity of MBT regulation
In its simplest iteration, the concept of a singular N/C-ratio mechanism that coordinates both transcription and cell cycle timing in multiple fast developing species has been challenged (Yasuda and Schubiger, 1992; Yuan et al., 2016). For example, the MBT in frogs was defined by the near coincident onset of transcription, cell motility, and cell cycle lengthening. However, zygotic transcription initiates prior to cell cycle lengthening in fish and frogs, and it is unknown if the timing of these early transcripts depends on an N/C ratio. At the very least, different genes would have to be activated in response to very different N/C ratios at different cleavage divisions. Moreover, while some bulk transcription appears sensitive to changes in N/C ratio in fast developing vertebrates, it remains to be shown whether this is true for most genes. Thus, models based only on the alleviation of transcriptional repression are likely insufficient to describe sequential gene activation during ZGA.
Transcriptional Activation by Activators
Transcription requires DNA binding by sequence-specific transcriptional activators, which must therefore be central components of ZGA timing models. While activator models are typically discussed in general terms, it is useful to consider how they might work quantitatively. First, the translation of maternal mRNA of a transcriptional activator is initiated by fertilization. This activator then accumulates to a critical concentration at which transcription is initiated. The timing of the activation of specific target genes is then determined by the amount of time it takes to reach the critical concentration. For example, in pre-implantation mouse embryos, the general transcriptional activator TBP begins to accumulate at the two-cell stage so that its concentration increases through early development (Gazdag et al., 2007). Importantly, the activator accumulation model does not require cell division and depends strongly on time, translation rate, and mRNA concentration. The relatively long duration of the early mammalian embryo cell cycles allows the activator mechanism sufficient time to reach critical concentrations during the course of a single cell cycle. Additional cell cycles (i.e., time) or increased maternal mRNA levels may be required for critical concentrations to be reached in the fast developing frogs and fish.
A related model is that timing results from a sequence of biochemical events. These may include regulated and dynamic polyadenylation of mRNAs, translation of a maternal transcription factor mRNA, transcription factor import into the nucleus, remodeling of nucleosomes, and, finally, recruitment and elongation of RNAPII. The minimum time to expression would be constrained by biochemical activities of expressing and translating a cascade of one or several TFs (Fig. 4). This activation cascade could account for the timing of a gene’s onset at a particular stage. For example, in Xenopus, polyA lengthening of many maternal transcripts occurs in the first 2 hours after fertilization (Collart et al., 2014), which, presumably, stabilizes the mRNA for increased translation. Maternal transcription factors such as VegT are then required, directly or indirectly, for zygotic expression of the mesoderm-promoting genes such as the T-box transcription factor Brachyury (Kofron et al., 1999). Brachyury expression initiates at the MBT and peaks several hours later when Brachyury protein activates several direct binding targets to propagate zygotic gene expression (Collart et al., 2014; Gentsch et al., 2013). Importantly, both the cascade and transcriptional activator models can be tested by measuring and manipulating the concentrations of key predicted regulatory factors through developmental time-courses.
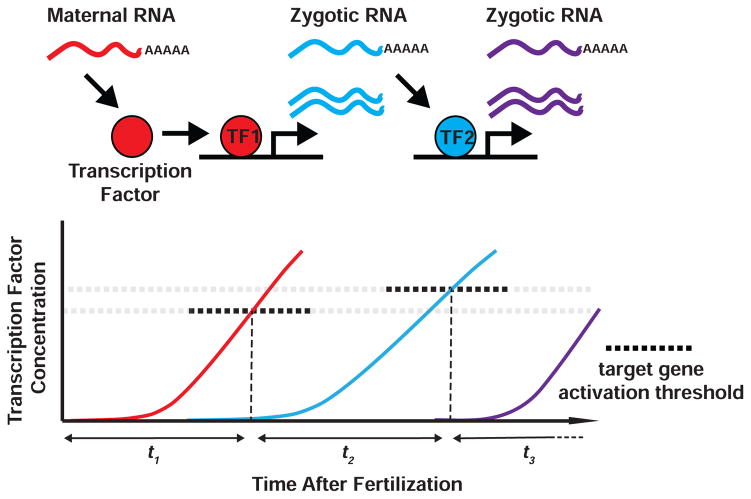
Figure 4. Activator-accumulation model for determining the timing of zygotic transcription.
Upon fertilization, maternal proteins are deposited and RNA translation is initiated, which triggers the accumulation of transcriptional activators (e.g., TF1). Once this transcription factor reaches a critical concentration (dotted line in graph), specific zygotic target genes initiate transcription. Differential timing of transcriptional activation can arise when the product of an early target gene affects subsequent transcriptional events (e.g., TF2). Transcriptional activation may be negatively regulated by site-specific repressors, competing chromatin components, or rate-limiting cofactors. Importantly, if transcription initiation depends solely on transcription factor concentration, the timing of the earliest transcription events may be independent of ploidy.
While the qualitative aspects of activator models remain to be determined, progress has been made in identifying specific ZGA activators. The canonical example of a “master” ZGA regulator comes from studies in Drosophila embryos, where the site-specific transcription factor Zelda binds the promoters of hundreds of genes and is required for their activation (Bosch et al., 2006; Harrison et al., 2011; Liang et al., 2008; Nien et al., 2011). Zelda may function as a “pioneer” TF that can bind to nucleosomes to promote chromatin opening (Sun et al., 2015). Until recently, it was unclear if such wide-binding activating factors existed in vertebrates because Zelda has no clear ortholog in fish, amphibians, or mammals. However, two complementary zebrafish studies have now shown that the transcription factors Nanog, SoxB1, and Pou5f3 (Oct4) are required to initiate ~75% of the first major phase of zygotic gene activation (Lee et al., 2013; Leichsenring et al., 2013). One target of Nanog is miR-430, the highly expressed miRNA responsible for clearing maternal transcripts (Giraldez et al., 2006; Lee et al., 2013). This demonstrates that Nanog, SoxB1, and Pouf51 are important early MZT transcription factors in vertebrates that may function analogously to Zelda in flies.
Although frogs are more closely related to mammals than zebrafish (last common ancestor ~350 MYA compared to ~430 MYA) their Oct4/Pou5f1 orthologs, Oct91 and Oct25, and Nanog-related Ventx proteins are not expressed until the MBT, after ZGA has initiated (Owens et al., 2016). Thus, these transcription factors are unlikely to fulfill an early ZGA activator role in frogs. It remains to be seen whether Sox proteins or a third Pou5f1 ortholog, Oct60, that is maternally provided (Hinkley et al., 1992), play a role in frog ZGA.
Nanog, SoxB1, and Oct4 (Pouf1) activate transcription in mammalian pre-implantation embryos (Boyer et al., 2005), and may play a role in mouse ZGA. However, these factors do not function identically in these two species. In fish embryos, Nanog, SoxB1, and Oct4 promote differentiation by driving towards gastrulation, while, in mammals, these factors are best known for their role in maintaining the pluripotency of embryonic stem cells to prevent differentiation. Nevertheless, there are enough parallels to raise the question—do these pluripotency factors also function during mammalian ZGA? Oct4 is highly expressed in mouse oocytes, which makes it a likely candidate to regulate ZGA (Foygel et al., 2008). However, knockout experiments show that loss of Oct4 alone does not prevent the accumulation of critical zygotic transcripts in the first ZGA cohort (Frum et al., 2013; Wu et al., 2013). Another pluripotency factor, Sox2, is expressed in oocytes and translocates to the nuclei at the 2-cell stage, and Sox2 overexpression arrests development between the 2- and 8-cell stage (Pan and Schultz, 2011). However, it is unclear how this phenotype relates to ZGA.
Other TFs, however, have been directly implicated in mouse ZGA by oocyte-specific deletion experiments (Li et al., 2010). General transcription regulators, like TIF1α, which translocates to transcription foci of pronuclei, are often required for progression past the 2-cell stage (Torres-Padilla et al., 2006). Mouse ZGA is also controlled by Yap1, the Hippo pathway transcriptional regulator best known for its role in organ size control. Yap1 is highly expressed in mammalian oocytes, but remains inactive due to cytoplasmic sequestration until after fertilization (Abbassi et al., 2016) when it gradually translocates to the nucleus. Maternal deletion of Yap1 results in failure to form a blastocyst, downregulation of ~3000 ZGA transcripts, and upregulation of ~1300 otherwise degraded transcripts. Other activators include the Zscan transcription factors, which are expressed specifically in the 2 cell embryo (Falco et al., 2007; Ko, 2016), NFYa, which likely directly regulates some genes activated in the 2-cell embryo (Lu et al., 2016), and the DUX (mouse) and DUX4 (human) homeobox transcription factors (De Iaco et al., 2017; Hendrickson et al., 2017; Whiddon et al., 2017). Intriguingly, DUX4 directly induces several hundred early human ZGA target genes, including HERVL retrotransposons, at the 4–8 cell stage. DUX4/DUX family genes are found in telomeric and pericentromeric regions as multicopy loci and appear specific to placental mammals, suggesting that DUX transcription factors could be critical drivers of embryonic genome activation in the mammalian lineage (De Iaco et al., 2017; Hendrickson et al., 2017; Whiddon et al., 2017). DUX itself is zygotically expressed by unknown mechanisms, indicating that key maternally deposited regulators of human and mouse ZGA remain to be identified.
Chromatin dynamics and transcriptional timing
Histone tails are extensively modified by maternal proteins during the MZT and may play a role in determining gene expression timing (Hontelez et al., 2015; Vastenhouw et al., 2010). If this is the case, a specific set of modifications should exist marking genes for early expression before activation. Indeed, in zebrafish, promoter histone modifications associated with gene expression (e.g., H3K4me3) are observed prior to gene expression, leading to a “pre-patterning” model consistent with a causal relationship between a modification and subsequent transcription. In Xenopus, histone modifications at specific promoters exist before the MBT (Blythe et al., 2010), but the majority appear to arise during the MBT and the first major zygotic transcription phase (Akkers et al., 2009). In fish, a transcriptionally permissive chromatin state (H3K4me3) and well-positioned nucleosomes appear to be established both prior to and during gene activation, and can even be found in the absence of RNAPII, the elongation mark H3K36me3, or detectable gene expression (Lindeman et al., 2011; Pálfy et al., 2017; Vastenhouw et al., 2010; Zhang et al., 2014). These experiments suggest that some active loci can be marked by histone modifications prior to transcription, consistent with influencing initiation, although the mechanisms by which these marks are targeted to specific embryonic loci remain unknown.
Intriguingly, some loci in fish exhibit both active- (H3K4me3) and inactive- (H3K27me3) promoter associated marks, reminiscent of the “bivalent” chromatin domains in pluripotent mouse ES cells (Bernstein et al., 2006; Vastenhouw et al., 2010). Although their functional relevance remains unclear, bivalent states are presumed to exist on genes poised for expression, i.e., they are repressed, but may need to be rapidly activated. However, accurate quantification of the fraction of histones modified at specific loci is difficult because in pooled cells it is not known what fraction of cells have a particular histone mark at a given locus. Thus, a locus could be falsely characterized as bivalent even if an overwhelming fraction of cells have only one of the two modifications at that locus. Isolating the chromatin associated with both marks using sequential ChIP analysis on a single sample can circumvent this problem (Vastenhouw et al., 2010). In addition, new approaches show promise in resolving this issue quantitatively by using recombinant and semisynthetic histones as quantitative spike-in standards for normalization (Grzybowski et al., 2015) or single-molecule imaging of individual nucleosomes (Shema et al., 2016).
Histone modifications in mammalian early embryos have also recently been measured genome-wide (Dahl et al., 2016; Liu et al., 2016; Zhang et al., 2016). In mouse oocytes, prior to the ZGA, the “active” mark, H3K4me3, which is normally found in sharp peaks at transcription start sites (TSS), was present in broad 5–10kb domains over loci (Fig. 5). These “non-canonical” H3K4me3 domains became restricted to the TSS only at the 2-cell stage, concurrent with mouse ZGA. Since ZGA is required for removal of these broad domains, they may be remnants of prior transcription, or even exist as a “repressive” mark to keep these loci silenced before the ZGA (Dahl et al., 2016). Notably, loss of H3K4me3 results in loss of paternal pronuclear ZGA in mouse (Aoshima et al., 2015). Just as in fish and frogs, H3K27me3 tends to accumulate after H3K4me3 in mouse, suggesting that repressive domains may increase later during mouse ZGA as the transcriptionally competent chromatin state is restricted to promoters and enhancers (Akkers et al., 2009; Lindeman et al., 2011; Liu et al., 2016; van Heeringen et al., 2014).
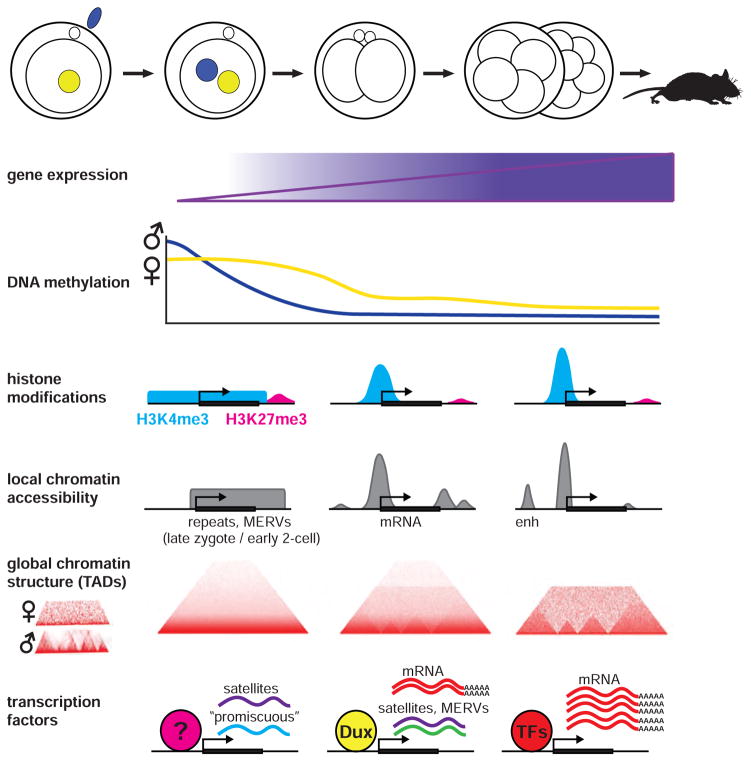
Figure 5. Chromatin state dynamics in early mouse embryos.
Schematic of changes in chromatin state after fertilization in mouse. Time points shown are the 1-cell zygote, 2-cell stage, and 4- to 8-cell stage. Gene expression begins in pronuclei of the 1-cell zygote and increases through the 2-, 4-, and 8-cell stages. DNA methylation (5mC) decreases sharply on paternal alleles by both active and passive mechanisms, and decreases gradually on maternal alleles largely by passive mechanisms such as dilution by replication. H3K4me3 is present in broad (>10kb) non-canonical domains in the zygote and restricts to promoters during ZGA. H3K27me3 is present distal to promoters and at low levels throughout early embryogenesis, and only becomes present near bivalent promoters in the later blastocyst stages. At the 2-cell stage, local chromatin is accessible in broad domains across repetitive genomic elements, and at gene promoters and transcription end sites. At the 4–8 cell stage, accessible chromatin more resembles that of somatic cells. Topologically associating domains (TADs) are removed after fertilization and gradually reform starting at the 4–8 cell stage. TAD images were reproduced with minor modifications from (Ke et al., 2017) with permission (Elsevier). The sequence of major gene expression phases is driven by different transcription factors and include different types of RNA.
Profiling accessible chromatin in early mouse embryos also revealed broad open domains over repetitive loci that are transcriptionally active in late 1-cell and early 2-cell embryos, including ERVs (Wu et al., 2016). Sharp open chromatin peaks over promoters gradually appeared from the 2-cell stage to ICM blastocysts as genes become expressed during the ZGA. This is consistent with experiments showing that transcription from a plasmid reporter requires an enhancer in the 2-cell stage, but not the 1-cell stage, in mouse embryos (Majumder and DePamphilis, 1995). An emerging perspective is that repeat elements and “promiscuous” transcripts that lack complete splicing or polyA-tail lengthening are among the first transcribed RNAs, which may help establish the chromatin states influencing later transcription (Abe et al., 2015; Probst et al., 2010).
Recent work has also revealed extensive post-fertilization remodeling of global chromatin in mouse. In somatic cells, chromatin is organized into topologically associated domains (TADs) on the scale of several hundred kb to ~1Mb that play important roles in regulating transcription and DNA replication (Hi-C Review). DNA within a TAD interacts much more frequently with itself than with DNA outside the TAD. The compact genomes of mature sperm have a TAD structure similar to that of somatic cells, with even more long-range interchromosomal interactions, whereas mature oocytes exhibit a weak TAD structure and have a smaller proportion of distal interactions (Du et al., 2017; Flyamer et al., 2017; Ke et al., 2017). Strikingly, both maternal and paternal alleles of the zygote and the 2-cell embryo appear to lack well-defined TADs. Instead, TADs gradually organize as the embryo proceeds through its cleavage stages, with clear TAD structure arising only at the 8-cell stage, well after the major ZGA phases (Du et al., 2017; Ke et al., 2017). Given the strong correlation between TADs and transcription in somatic cells, it is likely that the substantial global remodeling of parental genomes upon fertilization strongly influences the ZGA.
Finally, another post-fertilization chromatin modification is DNA methylation of cysteine residues (5mC), which is thought to generally inhibit transcription. Conversely, de-methylation removes 5mC, largely by replication-mediated dilution or by oxidation to several intermediate methylation states (5hmC, 5fC, 5caC and others) that could each have their own effects on gene expression (Fraser and Lin, 2016). DNA methylome dynamics have been extensively studied in frog (Bogdanovic et al., 2011; Stancheva et al., 2002), fish (Jiang et al., 2013; Potok et al., 2013), mouse (Shen et al., 2014), and human (Guo et al., 2014), with notable species differences. One tacit model is that de-methylation of the parental genomes may be a pre-requisite for timing of some zygotic genes’ expression. For example, during embryogenesis in mammals, both parental genomes appear to shift towards a hypo-methylated state. However, Xenopus embryonic genomes remain hyper-methylated through the early cleavage cycles (Bogdanovic et al., 2011; Veenstra and Wolffe, 2001) and in zebrafish, the oocyte DNA is hypo-methylated compared to sperm, but embryonic methylation levels then increase to match that of sperm by the MBT (Jiang et al., 2013; Potok et al., 2013). Finally, in human embryos broad de-methylation precedes the major ZGA by several days (Guo et al., 2014). Taken together, these studies indicate that global methylation dynamics are not conserved across species. Moreover, the precise effects of methylation on transcription of individual genes, and intervening events between (de-)methylation and altered gene expression are unclear (Biechele et al., 2015; Fraser and Lin, 2016).
Histone modifications—correlation or causation?
Despite the enormous progress in understanding chromatin state dynamics during ZGA, it will be crucial to move from cataloging temporal dynamics towards understanding the causality between chromatin dynamics and gene expression. Manipulation of histone modification enzymes often results in gene expression changes, indicating their importance for transcription. But, at the level of specific loci, whether such modifications determine activation timing or are mere accompaniments of the normal gene expression program remains an active area of investigation (Henikoff and Shilatifard, 2011). Several key questions include: 1) Do histone marks help recruit transcription factors? Or 2) do site-specific transcription factors bind first to recruit histone modification enzymes? 3) Which chromatin changes instruct transcription, and which are maintenance mechanisms? Ideally, we should eventually be able to understand the precise sequence of chromatin-associated events at a single locus leading to transcription and to elucidate the causative mechanisms.
Timing models are not mutually exclusive
For fast developing vertebrates, the combination of transcriptional activators and N/C-ratio mechanisms can explain most aspects of ZGA timing. In these species, N/C-ratio increases may provide a permissive context upon which transcriptional activators control specific loci. For instance, sudden or even gradual histone depletion may allow greater DNA access to maternally supplied transcription factors, and thus histone vs. transcription factor competition models could explain precise ZGA timing preceding the MBT (Joseph et al., 2017; Pálfy et al., 2017; Prioleau et al., 1995). Other cellular remodeling events at the MBT—including lamina exchange and a much longer S-phase—may reinforce a transcriptionally permissive state. In mammals, activator models seem to explain most ZGA timing, as there is currently no evidence that the N/C-ratio regulates early transcription in mouse or human. Additionally, in both fast and slow developing vertebrates, the various chromatin and methylation state changes also likely influence transcriptional competence globally and at specific genes. Of course, other timing models exist, and the ZGA can be orchestrated by diverse mechanisms.
Conclusions
In summary, the direct comparison of genome activation in different vertebrates reveals several common events. However, these events may be largely regulated by different molecular mechanisms and regulatory pathways based on the distinct needs of fast or slow dividing species. Broad similarities across the vertebrate lineage—and, in fact, most metazoans—include large oocytes, a maternal stockpile of RNA and proteins to initiate transcription and drive the early divisions, chromatin remodeling, and transcriptional activation cascades. Some molecular features may also be conserved, as evident from the common role of Oct4, Nanog, and Sox2 transcription factors in pluripotent stem cell maintenance in mammals and activation of zygotic genes in the pluripotent blastomeres of zebrafish. While N/C-ratio based timing mechanisms are better appreciated and perhaps more obviously applicable to fast developing species such as frogs and fish, it remains possible that they are used in mammalian development because all fertilized embryos undergo cytoplasmic subdivision during early cleavage divisions. These broad similarities in early development among diverse vertebrates suggests studies of MZT control in model organisms will have a wide impact through identifying regulatory principles also present in the early embryos of our own species.
In the coming decade we expect technical advances to be harnessed to answer long-standing questions in embryo gene activation. First, we anticipate a better understanding of the spatial organization of zygotic transcripts and how they control spatial aspects of differentiation (Satija et al., 2015). Second, with a few exceptions, current studies are limited by our lack of knowledge of activating transcription factors in frogs, fish and mice. It remains unknown if these transcription factors are generally conserved, as is the case for many developmental transcriptional regulators such as the Hox genes and the germ layer specification genes. Third, what insights will arise from studies in other vertebrate species such as birds or reptiles? Fourth, as emphasized in this review, we need a mechanistic understanding of how N/C-ratio and activator models work at the level of individual promoters. This should provide insight into how these timing models and chromatin state changes are interconnected to determine genome activation timing and differentiation in the early embryo. Finally, we expect the application of single cell technologies that circumvent the major constraint for studying human embryos—the limited amount of material—to lead to exciting breakthroughs in understanding of how gene activation drives development through the first days of human life.
Acknowledgments
We thank Eric Siggia, Nadine Vastenhouw, Amanda Amodeo, Julia Arand, Aaron Straight, Pranidhi Sood, and the Skotheim lab for critical discussions or comments on the manuscript. We thank the following funding sources for support: NIH NRSA F32GM108295 (DJ) and NIH RO1 HD085135 and HHMI-Simons Faculty Scholar Award (JMS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aanes H, Winata CL, Lin CH, Chen JP, Srinivasan KG, Lee SGP, Lim AYM, Hajan HS, Collas P, Bourque G, et al. Zebrafish mRNA sequencing deciphers novelties in transcriptome dynamics during maternal to zygotic transition. Genome Res. 2011;21:1328–1338. doi: 10.1101/gr.116012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbassi L, Malki S, Cockburn K, Macaulay A, Robert C, Rossant J, Clarke HJ. Multiple Mechanisms Cooperate to Constitutively Exclude the Transcriptional Co-Activator YAP from the Nucleus During Murine Oogenesis. Biol Reprod. 2016;94:102. doi: 10.1095/biolreprod.115.137968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe K-I, Yamamoto R, Franke V, Cao M, Suzuki Y, Suzuki MG, Vlahovicek K, Svoboda P, Schultz RM, Aoki F. The first murine zygotic transcription is promiscuous and uncoupled from splicing and 3′ processing. Embo J. 2015;34:1523–1537. doi: 10.15252/embj.201490648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiken CEM, Swoboda PPL, Skepper JN, Johnson MH. The direct measurement of embryogenic volume and nucleo-cytoplasmic ratio during mouse pre-implantation development. Reproduction. 2004;128:527–535. doi: 10.1530/rep.1.00281. [DOI] [PubMed] [Google Scholar]
- Akkers RC, van Heeringen SJ, Jacobi UG, Janssen-Megens EM, Françoijs KJ, Stunnenberg HG, Veenstra GJC. A hierarchy of H3K4me3 and H3K27me3 acquisition in spatial gene regulation in Xenopus embryos. Dev Cell. 2009;17:425–434. doi: 10.1016/j.devcel.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almouzni G, Wolffe AP. Constraints on transcriptional activator function contribute to transcriptional quiescence during early Xenopus embryogenesis. Embo J. 1995;14:1752–1765. doi: 10.1002/j.1460-2075.1995.tb07164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amodeo AA, Jukam D, Straight AF, Skotheim JM. Histone titration against the genome sets the DNA-to-cytoplasm threshold for the Xenopus midblastula transition. Proc Natl Acad Sci USa. 2015;112:E1086–E1095. doi: 10.1073/pnas.1413990112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JA, Lewellyn AL, Maller JL. Ionizing radiation induces apoptosis and elevates cyclin A1-Cdk2 activity before but not after the midblastula transition in Xenopus. Mol Biol Cell. 1997;8:1195–1206. doi: 10.1091/mbc.8.7.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki F, Worrad DM, Schultz RM. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev Biol. 1997;181:296–307. doi: 10.1006/dbio.1996.8466. [DOI] [PubMed] [Google Scholar]
- Aoshima K, Inoue E, Sawa H, Okada Y. Paternal H3K4 methylation is required for minor zygotic gene activation and early mouse embryonic development. EMBO Rep. 2015;16:803–812. doi: 10.15252/embr.201439700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Biczysko W, Pienkowski M, Solter D, Koprowski H. Virus particles in early mouse embryos. J Natl Cancer Inst. 1973;51:1041–1050. doi: 10.1093/jnci/51.3.1041. [DOI] [PubMed] [Google Scholar]
- Biechele S, Lin C-J, Rinaudo PF, Ramalho-Santos M. Unwind and transcribe: chromatin reprogramming in the early mammalian embryo. Curr Opin Genet Dev. 2015;34:17–23. doi: 10.1016/j.gde.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blythe SA, Wieschaus EF. Coordinating Cell Cycle Remodeling with Transcriptional Activation at the Drosophila MBT. Curr Top Dev Biol. 2015;113:113–148. doi: 10.1016/bs.ctdb.2015.06.002. [DOI] [PubMed] [Google Scholar]
- Blythe SA, Cha SW, Tadjuidje E, Heasman J, Klein PS. beta-Catenin primes organizer gene expression by recruiting a histone H3 arginine 8 methyltransferase, Prmt2. Dev Cell. 2010;19:220–231. doi: 10.1016/j.devcel.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanovic O, Long SW, van Heeringen SJ, Brinkman AB, Gómez-Skarmeta JL, Stunnenberg HG, Jones PL, Veenstra GJC. Temporal uncoupling of the DNA methylome and transcriptional repression during embryogenesis. Genome Res. 2011;21:1313–1327. doi: 10.1101/gr.114843.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton VN, Oades PJ, Johnson MH. The relationship between cleavage, DNA replication, and gene expression in the mouse 2-cell embryo. J Embryol Exp Morphol. 1984;79:139–163. [PubMed] [Google Scholar]
- Bosch ten JR, Benavides JA, Cline TW. The TAGteam DNA motif controls the timing of Drosophila pre-blastoderm transcription. Development. 2006;133:1967–1977. doi: 10.1242/dev.02373. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachet J, Denis H, Devitry F. The effects of actinomycin D and puromycin on morphogenesis in amphibian eggs and acetabularia mediterranea. Dev Biol. 1964;9:398–434. doi: 10.1016/0012-1606(64)90033-8. [DOI] [PubMed] [Google Scholar]
- Braude P, Bolton V, Moore S. Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature (London) 1988;332:459–461. doi: 10.1038/332459a0. [DOI] [PubMed] [Google Scholar]
- Braun RE. Packaging paternal chromosomes with protamine. Nat Genet. 2001;28:10–12. doi: 10.1038/ng0501-10. [DOI] [PubMed] [Google Scholar]
- Bultman SJ, Gebuhr TC, Pan H, Svoboda P, Schultz RM, Magnuson T. Maternal BRG1 regulates zygotic genome activation in the mouse. Genes Dev. 2006;20:1744–1754. doi: 10.1101/gad.1435106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger K, Mühl B, Kellner M, Rohrmoser M, Gruber-Eber A, Windhager L, Friedel CC, Dölken L, Eick D. 4-thiouridine inhibits rRNA synthesis and causes a nucleolar stress response. RNA Biol. 2013;10:1623–1630. doi: 10.4161/rna.26214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton A, Torres-Padilla M-E. Chromatin dynamics in the regulation of cell fate allocation during early embryogenesis. Nat Rev Mol Cell Biol. 2014;15:723–734. doi: 10.1038/nrm3885. [DOI] [PubMed] [Google Scholar]
- Calarco PG. Intracisternal A particle formation and inhibition in preimplantation mouse embryos. Biol Reprod. 1975;12:448–454. doi: 10.1095/biolreprod12.4.448. [DOI] [PubMed] [Google Scholar]
- Carone BR, Hung JH, Hainer SJ, Chou MT, Carone DM, Weng Z, Fazzio TG, Rando OJ. High-resolution mapping of chromatin packaging in mouse embryonic stem cells and sperm. Dev Cell. 2014;30:11–22. doi: 10.1016/j.devcel.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christians E, Rao VH, Renard JP. Sequential Acquisition of Transcriptional Control during Early Embryonic Development in the Rabbit. Dev Biol. 1994;164:160–172. doi: 10.1006/dbio.1994.1188. [DOI] [PubMed] [Google Scholar]
- Chulitskaia EV. Desynchronization of cell divisions in the course of egg cleavage and an attempt at experimental shift of its onset. J Embryol Exp Morphol. 1970;23:359–374. [PubMed] [Google Scholar]
- Clute P, Masui Y. Regulation of the appearance of division asynchrony and microtubule-dependent chromosome cycles in Xenopus laevis embryos. Dev Biol. 1995;171:273–285. doi: 10.1006/dbio.1995.1280. [DOI] [PubMed] [Google Scholar]
- Collart C, Allen GE, Bradshaw CR, Smith JC, Zegerman P. Titration of four replication factors is essential for the Xenopus laevis midblastula transition. Science. 2013;341:893–896. doi: 10.1126/science.1241530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart C, Owens NDL, Bhaw-Rosun L, Cooper B, De Domenico E, Patrushev I, Sesay AK, Smith JN, Smith JC, Gilchrist MJ. High-resolution analysis of gene activity during the Xenopus mid-blastula transition. Development. 2014;141:1927–1939. doi: 10.1242/dev.102012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby IM, Gandolfi F, Moor RM. Control of protein synthesis during early cleavage of sheep embryos. J Reprod Fertil. 1988;82:769–775. doi: 10.1530/jrf.0.0820769. [DOI] [PubMed] [Google Scholar]
- Dahl JA, Jung I, Aanes H, Greggains GD, Manaf A, Lerdrup M, Li G, Kuan S, Li B, Lee AY, et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature. 2016;537:548–552. doi: 10.1038/nature19360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Yan L, Hu B, Fan X, Ren Y, Li R, Lian Y, Yan J, Li Q, Zhang Y, et al. Tracing the expression of circular RNAs in human pre-implantation embryos. Genome Biol. 2016;17:130. doi: 10.1186/s13059-016-0991-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH. Gene Activity in Early Development. Academic Press; 1986. [Google Scholar]
- Dekker J, Mirny L. The 3D Genome as Moderator of Chromosomal Communication. Cell. 2016;164:1110–1121. doi: 10.1016/j.cell.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Iaco A, Planet E, Coluccio A, Verp S, Duc J, Trono D. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat Genet. 2017;487:57. doi: 10.1038/ng.3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sousa PA, Watson AJ, Schultz RM. Transient expression of a translation initiation factor is conservatively associated with embryonic gene activation in murine and bovine embryos. Biol Reprod. 1998;59:969–977. doi: 10.1095/biolreprod59.4.969. [DOI] [PubMed] [Google Scholar]
- Dettlaff TA. Cell divisions, duration of interkinetic states and differentiation in early stages of embryonic development. Adv Morphog. 1964;4:323–362. doi: 10.1016/b978-1-4831-9950-4.50011-4. [DOI] [PubMed] [Google Scholar]
- Dettlaff TA, Rudneva TB. The clawed frog Xenopus laevis Daudin. Objects of Developmental Biology 1975 [Google Scholar]
- Dimitrov S, Wolffe AP. Remodeling somatic nuclei in Xenopus laevis egg extracts: molecular mechanisms for the selective release of histones H1 and H1(0) from chromatin and the acquisition of transcriptional competence. Embo J. 1996;15:5897–5906. [PMC free article] [PubMed] [Google Scholar]
- Dobson AT, Raja R, Abeyta MJ, Taylor T, Shen S, Haqq C, Pera RAR. The unique transcriptome through day 3 of human preimplantation development. Hum Mol Genet. 2004;13:1461–1470. doi: 10.1093/hmg/ddh157. [DOI] [PubMed] [Google Scholar]
- Du Z, Zheng H, Huang B, Ma R, Wu J, Zhang X, He J, Xiang Y, Wang Q, Li Y, et al. Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature. 2017;547:232–235. doi: 10.1038/nature23263. [DOI] [PubMed] [Google Scholar]
- Dunican DS, Ruzov A, Hackett JA, Meehan RR. xDnmt1 regulates transcriptional silencing in pre-MBT Xenopus embryos independently of its catalytic function. Development. 2008;135:1295–1302. doi: 10.1242/dev.016402. [DOI] [PubMed] [Google Scholar]
- Dworkin-Rastl E, Kandolf H, Smith RC. The maternal histone H1 variant, H1M (B4 protein), is the predominant H1 histone in Xenopus pregastrula embryos. Dev Biol. 1994;161:425–439. doi: 10.1006/dbio.1994.1042. [DOI] [PubMed] [Google Scholar]
- Falco G, Lee S-L, Stanghellini I, Bassey UC, Hamatani T, Ko MSH. Zscan4: a novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev Biol. 2007;307:539–550. doi: 10.1016/j.ydbio.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser G. Maintenance of normal structure in heteroploid salamander larvae, through compensation of changes in cell size by adjustment of cell number and cell shape. J Exp Zool. 1945;100:445–455. doi: 10.1002/jez.1401000310. [DOI] [PubMed] [Google Scholar]
- Fernandez MS, Izquierdo L. Blastocoel formation in half and double mouse embryos. Anat Embryol. 1980;160:77–81. doi: 10.1007/BF00315650. [DOI] [PubMed] [Google Scholar]
- Flyamer IM, Gassler J, Imakaev M, Brandão HB, Ulianov SV, Abdennur N, Razin SV, Mirny LA, Tachibana-Konwalski K. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature. 2017;544:110–114. doi: 10.1038/nature21711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forouzmand E, Owens NDL, Blitz IL, Paraiso KD, Khokha MK, Gilchrist MJ, Xie X, Cho KWY. Developmentally regulated long non-coding RNAs in Xenopus tropicalis. Dev Biol. 2016 doi: 10.1016/j.ydbio.2016.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foygel K, Choi B, Jun S, Leong DE, Lee A, Wong CC, Zuo E, Eckart M, Reijo Pera RA, Wong WH, et al. A novel and critical role for Oct4 as a regulator of the maternal-embryonic transition. PLoS ONE. 2008;3:e4109. doi: 10.1371/journal.pone.0004109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser R, Lin CJ. Epigenetic reprogramming of the zygote in mice and men: on your marks, get set, go! Reproduction. 2016;152:R211–R222. doi: 10.1530/REP-16-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman BS, Heald R. Functional comparison of H1 histones in Xenopus reveals isoform-specific regulation by Cdk1 and RanGTP. Curr Biol. 2010;20:1048–1052. doi: 10.1016/j.cub.2010.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei RE, Schultz GA, Church RB. Qualitative and quantitative changes in protein synthesis occur at the 8–16 cell stage of embryogenesis in the cow. J Reprod Fertil. 1989;86:637–641. doi: 10.1530/jrf.0.0860637. [DOI] [PubMed] [Google Scholar]
- Frum T, Halbisen MA, Wang C, Amiri H, Robson P, Ralston A. Oct4 cell-autonomously promotes primitive endoderm development in the mouse blastocyst. Dev Cell. 2013;25:610–622. doi: 10.1016/j.devcel.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazdag E, Rajkovic A, Torres-Padilla ME, Tora L. Analysis of TATA-binding protein 2 (TBP2) and TBP expression suggests different roles for the two proteins in regulation of gene expression during oogenesis and early mouse development. Reproduction. 2007;134:51–62. doi: 10.1530/REP-06-0337. [DOI] [PubMed] [Google Scholar]
- Gentsch GE, Owens NDL, Martin SR, Piccinelli P, Faial T, Trotter MWB, Gilchrist MJ, Smith JC. In vivo T-box transcription factor profiling reveals joint regulation of embryonic neuromesodermal bipotency. Cell Rep. 2013;4:1185–1196. doi: 10.1016/j.celrep.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart JC. Biological Regulation and Development. Boston, MA: Springer US; 1980. Mechanisms Regulating Pattern Formation in the Amphibian Egg and Early Embryo; pp. 133–316. [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Golbus MS, Calarco PG, Epstein CJ. The effects of inhibitors of RNA synthesis (α-amanitin and actinomycin D) on preimplantation mouse embryogenesis. Journal of Experimental Zoology. 1973;186:207–216. doi: 10.1002/jez.1401860211. [DOI] [PubMed] [Google Scholar]
- Grow EJ, Flynn RA, Chavez SL, Bayless NL, Wossidlo M, Wesche DJ, Martin L, Ware CB, Blish CA, Chang HY, et al. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature. 2015;522:221–225. doi: 10.1038/nature14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzybowski AT, Chen Z, Ruthenburg AJ. Calibrating ChIP-Seq with Nucleosomal Internal Standards to Measure Histone Modification Density Genome Wide. Mol Cell. 2015;58:886–899. doi: 10.1016/j.molcel.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Zhu P, Yan L, Li R, Hu B, Lian Y, Yan J, Ren X, Lin S, Li J, et al. The DNA methylation landscape of human early embryos. Nature. 2014;511:606–610. doi: 10.1038/nature13544. [DOI] [PubMed] [Google Scholar]
- Gurdon JB. The developmental capacity of nuclei taken from intestinal epithelium cells of feeding tadpoles. J Embryol Exp Morphol. 1962;10:622–640. [PubMed] [Google Scholar]
- Hamatani T, Carter MG, Sharov AA, Ko MSH. Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell. 2004;6:117–131. doi: 10.1016/s1534-5807(03)00373-3. [DOI] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Hammoud AO, Gibson M, Cairns BR, Carrell DT. Genome-wide analysis identifies changes in histone retention and epigenetic modifications at developmental and imprinted gene loci in the sperm of infertile men. Hum Reprod. 2011;26:2558–2569. doi: 10.1093/humrep/der192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M, Grunstein M. Nucleosome loss activates yeast downstream promoters in vivo. Cell. 1988;55:1137–1145. doi: 10.1016/0092-8674(88)90258-9. [DOI] [PubMed] [Google Scholar]
- Harrison MM, Eisen MB. Transcriptional Activation of the Zygotic Genome in Drosophila. Curr Top Dev Biol. 2015;113:85–112. doi: 10.1016/bs.ctdb.2015.07.028. [DOI] [PubMed] [Google Scholar]
- Harrison MM, Li X-Y, Kaplan T, Botchan MR, Eisen MB. Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet. 2011;7:e1002266. doi: 10.1371/journal.pgen.1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey EB. Parthenogenetic Merogony or Cleavage, without Nuclei in Arbacia punctulata. Biological Bulletin. 1936;71:101. [Google Scholar]
- Harvey SA, Sealy I, Kettleborough R, Fenyes F, White R, Stemple D, Smith JC. Identification of the zebrafish maternal and paternal transcriptomes. Development. 2013;140:2703–2710. doi: 10.1242/dev.095091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson PG, Doráis JA, Grow EJ, Whiddon JL, Lim J-W, Wike CL, Weaver BD, Pflueger C, Emery BR, Wilcox AL, et al. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat Genet. 2017;487:57. doi: 10.1038/ng.3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Shilatifard A. Histone modification: cause or cog? Trends Genet. 2011;27:389–396. doi: 10.1016/j.tig.2011.06.006. [DOI] [PubMed] [Google Scholar]
- Hertig AT, Rock J, Adams EC. A description of 34 human ova within the first 17 days of development. Am J Anat. 1956;98:435–493. doi: 10.1002/aja.1000980306. [DOI] [PubMed] [Google Scholar]
- Heyn P, Kircher M, Dahl A, Kelso J, Tomancak P, Kalinka AT, Neugebauer KM. The earliest transcribed zygotic genes are short, newly evolved, and different across species. Cell Rep. 2014;6:285–292. doi: 10.1016/j.celrep.2013.12.030. [DOI] [PubMed] [Google Scholar]
- Hinkley CS, Martin JF, Leibham D, Perry M. Sequential expression of multiple POU proteins during amphibian early development. Mol Cell Biol. 1992;12:638–649. doi: 10.1128/mcb.12.2.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hontelez S, van Kruijsbergen I, Georgiou G, van Heeringen SJ, Bogdanovic O, Lister R, Veenstra GJC. Embryonic transcription is controlled by maternally defined chromatin state. Nat Commun. 2015;6:10148. doi: 10.1038/ncomms10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Ikegami R, Hunter P, Yager TD. Developmental activation of the capability to undergo checkpoint-induced apoptosis in the early zebrafish embryo. Dev Biol. 1999;209:409–433. doi: 10.1006/dbio.1999.9243. [DOI] [PubMed] [Google Scholar]
- Iwao Y, Uchida Y, Ueno S, Yoshizaki N, Masui Y. Midblastula transition (MBT) of the cell cycles in the yolk and pigment granule-free translucent blastomeres obtained from centrifuged Xenopus embryos. Dev Growth Differ. 2005;47:283–294. doi: 10.1111/j.1440-169X.2005.00802.x. [DOI] [PubMed] [Google Scholar]
- Jevtić P, Levy DL. Nuclear size scaling during Xenopus early development contributes to midblastula transition timing. Curr Biol. 2015;25:45–52. doi: 10.1016/j.cub.2014.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Zhang J, Wang JJ, Wang L, Zhang L, Li G, Yang X, Ma X, Sun X, Cai J, et al. Sperm, but not oocyte, DNA methylome is inherited by zebrafish early embryos. Cell. 2013;153:773–784. doi: 10.1016/j.cell.2013.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimeno-González S, Payán-Bravo L, Muñoz-Cabello AM, Guijo M, Gutierrez G, Prado F, Reyes JC. Defective histone supply causes changes in RNA polymerase II elongation rate and cotranscriptional pre-mRNA splicing. Proc Natl Acad Sci USa. 2015;112:14840–14845. doi: 10.1073/pnas.1506760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph SR, Pálfy M, Hilbert L, Kumar M, Karschau J, Zaburdaev V, Shevchenko A, Vastenhouw NL. Competition between histone and transcription factor binding regulates the onset of transcription in zebrafish embryos. Elife. 2017;6:e23326. doi: 10.7554/eLife.23326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung YH, Sauria MEG, Lyu X, Cheema MS, Ausio J, Taylor J, Corces VG. Chromatin States in Mouse Sperm Correlate with Embryonic and Adult Regulatory Landscapes. Cell Rep. 2017;18:1366–1382. doi: 10.1016/j.celrep.2017.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane DA, Kimmel CB. The zebrafish midblastula transition. Development. 1993;119:447–456. doi: 10.1242/dev.119.2.447. [DOI] [PubMed] [Google Scholar]
- Kane DA, Maischein HM, Brand M, van Eeden FJ, Furutani-Seiki M, Granato M, Haffter P, Hammerschmidt M, Heisenberg CP, Jiang YJ, et al. The zebrafish early arrest mutants. Development. 1996;123:57–66. doi: 10.1242/dev.123.1.57. [DOI] [PubMed] [Google Scholar]
- Ke Y, Xu Y, Chen X, Feng S, Liu Z, Sun Y, Yao X, Li F, Zhu W, Gao L, et al. 3D Chromatin Structures of Mature Gametes and Structural Reprogramming during Mammalian Embryogenesis. Cell. 2017;170:367–381. e20. doi: 10.1016/j.cell.2017.06.029. [DOI] [PubMed] [Google Scholar]
- Kermi C, Prieto S, van der Laan S, Tsanov N, Recolin B, Uro-Coste E, Delisle MB, Maiorano D. RAD18 Is a Maternal Limiting Factor Silencing the UV-Dependent DNA Damage Checkpoint in Xenopus Embryos. Dev Cell. 2015;34:364–372. doi: 10.1016/j.devcel.2015.06.002. [DOI] [PubMed] [Google Scholar]
- Kigami D, Minami N, Takayama H, Imai H. MuERV-L is one of the earliest transcribed genes in mouse one-cell embryos. Biol Reprod. 2003;68:651–654. doi: 10.1095/biolreprod.102.007906. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kirschner MW, Butner KA, Newport JW, Black SD, Scharf SR, Gerhart JC. Spatial and Temporal Changes in Early Amphibian Development. Netherlands Journal of Zoology. 1980;31:50–77. [Google Scholar]
- Ko MSH. Zygotic Genome Activation Revisited: Looking Through the Expression and Function of Zscan4. Curr Top Dev Biol. 2016;120:103–124. doi: 10.1016/bs.ctdb.2016.04.004. [DOI] [PubMed] [Google Scholar]
- Kobayakawa Y, Kubota HY. Temporal pattern of cleavage and the onset of gastrulation in amphibian embryos developed from eggs with the reduced cytoplasm. J Embryol Exp Morphol. 1981;62:83–94. [PubMed] [Google Scholar]
- Kofron M, Demel T, Xanthos J, Lohr J, Sun B, Sive H, Osada S, Wright C, Wylie C, Heasman J. Mesoderm induction in Xenopus is a zygotic event regulated by maternal VegT via TGFbeta growth factors. Development. 1999;126:5759–5770. doi: 10.1242/dev.126.24.5759. [DOI] [PubMed] [Google Scholar]
- Kojima Y, Tam OH, Tam PPL. Timing of developmental events in the early mouse embryo. Semin Cell Dev Biol. 2014;34:65–75. doi: 10.1016/j.semcdb.2014.06.010. [DOI] [PubMed] [Google Scholar]
- Landström U, Løvtrup-Rein H, Løvtrup S. Control of cell division and cell differentiation by deoxynucleotides in the early embryo of Xenopus laevis. Cell Differentiation. 1975;4:313–325. doi: 10.1016/0045-6039(75)90016-0. [DOI] [PubMed] [Google Scholar]
- Lee DR, Lee JE, Yoon HS, Roh SI, Kim MK. Compaction in preimplantation mouse embryos is regulated by a cytoplasmic regulatory factor that alters between 1- and 2-cell stages in a concentration-dependent manner. J Exp Zool. 2001;290:61–71. doi: 10.1002/jez.1036. [DOI] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, Giraldez AJ. Zygotic genome activation during the maternal-to-zygotic transition. Annu Rev Cell Dev Biol. 2014;30:581–613. doi: 10.1146/annurev-cellbio-100913-013027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, Takacs CM, Bazzini AA, DiVito KR, Fleming ES, Giraldez AJ. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature. 2013;503:360–364. doi: 10.1038/nature12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leichsenring M, Maes J, Mössner R, Driever W, Onichtchouk D. Pou5f1 transcription factor controls zygotic gene activation in vertebrates. Science. 2013;341:1005–1009. doi: 10.1126/science.1242527. [DOI] [PubMed] [Google Scholar]
- Li L, Zheng P, Dean J. Maternal control of early mouse development. Development. 2010;137:859–870. doi: 10.1242/dev.039487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, Rushlow C. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008;456:400–403. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CJ, Conti M, Ramalho-Santos M. Histone variant H3.3 maintains a decondensed chromatin state essential for mouse preimplantation development. Development. 2013;140:3624–3634. doi: 10.1242/dev.095513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CJ, Koh FM, Wong P, Conti M, Ramalho-Santos M. Hira-mediated H3.3 incorporation is required for DNA replication and ribosomal RNA transcription in the mouse zygote. Dev Cell. 2014;30:268–279. doi: 10.1016/j.devcel.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeman LC, Andersen IS, Reiner AH, Li N, Aanes H, Østrup O, Winata C, Mathavan S, Müller F, Aleström P, et al. Prepatterning of developmental gene expression by modified histones before zygotic genome activation. Dev Cell. 2011;21:993–1004. doi: 10.1016/j.devcel.2011.10.008. [DOI] [PubMed] [Google Scholar]
- Liu X, Wang C, Liu W, Li J, Li C, Kou X, Chen J, Zhao Y, Gao H, Wang H, et al. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature. 2016;537:558–562. doi: 10.1038/nature19362. [DOI] [PubMed] [Google Scholar]
- Lu F, Liu Y, Inoue A, Suzuki T, Zhao K, Zhang Y. Establishing Chromatin Regulatory Landscape during Mouse Preimplantation Development. Cell. 2016;165:1375–1388. doi: 10.1016/j.cell.2016.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Li JM, Elemento O, Tavazoie S, Wieschaus EF. Coupling of zygotic transcription to mitotic control at the Drosophila mid-blastula transition. Development. 2009;136:2101–2110. doi: 10.1242/dev.034421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder S, DePamphilis ML. A unique role for enhancers is revealed during early mouse development. Bioessays. 1995;17:879–889. doi: 10.1002/bies.950171010. [DOI] [PubMed] [Google Scholar]
- Mann M, Risley MS, Eckhardt RA, Kasinsky HE. Characterization of spermatid/sperm basic chromosomal proteins in the genus Xenopus (Anura, Pipidae) J Exp Zool. 1982;222:173–186. doi: 10.1002/jez.1402220209. [DOI] [PubMed] [Google Scholar]
- Masui Y, Wang P. Cell cycle transition in early embryonic development of Xenopus laevis. Biol Cell. 1998;90:537–548. [PubMed] [Google Scholar]
- Mathavan S, Lee SGP, Mak A, Miller LD, Murthy KRK, Govindarajan KR, Tong Y, Wu YL, Lam SH, Yang H, et al. Transcriptome analysis of zebrafish embryogenesis using microarrays. PLoS Genet. 2005;1:260–276. doi: 10.1371/journal.pgen.0010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CM, Michael WM. Control of DNA replication by the nucleus/cytoplasm ratio in Xenopus. J Biol Chem. 2013;288:29382–29393. doi: 10.1074/jbc.M113.499012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell. 1982a;30:675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell. 1982b;30:687–696. doi: 10.1016/0092-8674(82)90273-2. [DOI] [PubMed] [Google Scholar]
- Neyfakh AA. X-ray inactivation of nuclei as method for studying their function in the early development of fishes. J Embryol Exp Morphol. 1959;7:173–192. [PubMed] [Google Scholar]
- Niakan KK, Han J, Pedersen RA, Simon C, Pera RAR. Human pre-implantation embryo development. Development. 2012;139:829–841. doi: 10.1242/dev.060426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nien C-Y, Liang H-L, Butcher S, Sun Y, Fu S, Gocha T, Kirov N, Manak JR, Rushlow C. Temporal coordination of gene networks by Zelda in the early Drosophila embryo. PLoS Genet. 2011;7:e1002339. doi: 10.1371/journal.pgen.1002339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (Daudin): A Systematical and Chronological Survey of the Development from the Fertilized Egg till the end of …. New York: Garland Pub; 1994. [Google Scholar]
- Nothias JY, Miranda M, DePamphilis ML. Uncoupling of transcription and translation during zygotic gene activation in the mouse. Embo J. 1996;15:5715–5725. [PMC free article] [PubMed] [Google Scholar]
- O’Farrell PH. Growing an Embryo from a Single Cell: A Hurdle in Animal Life. Cold Spring Harb Perspect Biol. 2015;7:a019042. doi: 10.1101/cshperspect.a019042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens NDL, Blitz IL, Lane MA, Patrushev I, Overton JD, Gilchrist MJ, Cho KWY, Khokha MK. Measuring Absolute RNA Copy Numbers at High Temporal Resolution Reveals Transcriptome Kinetics in Development. Cell Rep. 2016;14:632–647. doi: 10.1016/j.celrep.2015.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H, Schultz RM. Sox2 modulates reprogramming of gene expression in two-cell mouse embryos. Biol Reprod. 2011;85:409–416. doi: 10.1095/biolreprod.111.090886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paranjpe SS, Jacobi UG, van Heeringen SJ, Veenstra GJC. A genome-wide survey of maternal and embryonic transcripts during Xenopus tropicalis development. BMC Genomics. 2013;14:762. doi: 10.1186/1471-2164-14-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ, Fan L, Sandelin A, Rinn JL, Regev A, et al. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 2012;22:577–591. doi: 10.1101/gr.133009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pálfy M, Joseph SR, Vastenhouw NL. The timing of zygotic genome activation. Curr Opin Genet Dev. 2017;43:53–60. doi: 10.1016/j.gde.2016.12.001. [DOI] [PubMed] [Google Scholar]
- Potok ME, Nix DA, Parnell TJ, Cairns BR. Reprogramming the maternal zebrafish genome after fertilization to match the paternal methylation pattern. Cell. 2013;153:759–772. doi: 10.1016/j.cell.2013.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado F, Jimeno-González S, Reyes JC. Histone availability as a strategy to control gene expression. RNA Biol. 2016:1–6. doi: 10.1080/15476286.2016.1189071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt HP, Chakraborty J, Surani MA. Molecular and morphological differentiation of the mouse blastocyst after manipulations of compaction with cytochalasin D. Cell. 1981;26:279–292. doi: 10.1016/0092-8674(81)90310-x. [DOI] [PubMed] [Google Scholar]
- Prioleau MN, Buckle RS, Méchali M. Programming of a repressed but committed chromatin structure during early development. Embo J. 1995;14:5073–5084. doi: 10.1002/j.1460-2075.1995.tb00189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prioleau MN, Huet J, Sentenac A, Méchali M. Competition between chromatin and transcription complex assembly regulates gene expression during early development. Cell. 1994;77:439–449. doi: 10.1016/0092-8674(94)90158-9. [DOI] [PubMed] [Google Scholar]
- Probst AV, Okamoto I, Casanova M, El Marjou F, Le Baccon P, Almouzni G. A strand-specific burst in transcription of pericentric satellites is required for chromocenter formation and early mouse development. Dev Cell. 2010;19:625–638. doi: 10.1016/j.devcel.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Raj A, Tyagi S. Detection of individual endogenous RNA transcripts in situ using multiple singly labeled probes. Meth Enzymol. 2010;472:365–386. doi: 10.1016/S0076-6879(10)72004-8. [DOI] [PubMed] [Google Scholar]
- Rott NN, Sheveleva GA. Changes in the rate of cell divisions in the course of early development of diploid and haploid loach embryos. J Embryol Exp Morphol. 1968;20:141–150. [PubMed] [Google Scholar]
- Royo H, Stadler MB, Peters AHFM. Alternative Computational Analysis Shows No Evidence for Nucleosome Enrichment at Repetitive Sequences in Mammalian Spermatozoa. Dev Cell. 2016;37:98–104. doi: 10.1016/j.devcel.2016.03.010. [DOI] [PubMed] [Google Scholar]
- Saitou M, Kurimoto K. Paternal nucleosomes: are they retained in developmental promoters or gene deserts? Dev. Cell. 2014;30:6–8. doi: 10.1016/j.devcel.2014.06.025. [DOI] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh N. “Metachronous” cleavage and initiation of gastrulation in amphibian embryos. Development, Growth & Differentiation. 1977;19:111–117. doi: 10.1111/j.1440-169X.1977.00111.x. [DOI] [PubMed] [Google Scholar]
- Schönmann W. Der diploide Bastard Triton palmatus♀ x Salamandra♂. Development Genes and Evolution. 1938 doi: 10.1007/BF00573809. [DOI] [PubMed] [Google Scholar]
- Schramm RD, Bavister BD. Onset of nucleolar and extranucleolar transcription and expression of fibrillarin in macaque embryos developing in vitro. Biol Reprod. 1999;60:721–728. doi: 10.1095/biolreprod60.3.721. [DOI] [PubMed] [Google Scholar]
- Shema E, Jones D, Shoresh N, Donohue L, Ram O, Bernstein BE. Single-molecule decoding of combinatorially modified nucleosomes. Science. 2016;352:717–721. doi: 10.1126/science.aad7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Inoue A, He J, Liu Y, Lu F, Zhang Y. Tet3 and DNA replication mediate demethylation of both the maternal and paternal genomes in mouse zygotes. Cell Stem Cell. 2014;15:459–470. doi: 10.1016/j.stem.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shermoen AW, O’Farrell PH. Progression of the cell cycle through mitosis leads to abortion of nascent transcripts. Cell. 1991;67:303–310. doi: 10.1016/0092-8674(91)90182-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signoret J, Lefresne J. Contribution à l’étude de la segmentation de l‘œuf d’axolotl. I. Définition de la transition blastuléenne. Ann Embryol Morphogen 1971 [Google Scholar]
- Skirkanich J, Luxardi G, Yang J, Kodjabachian L, Klein PS. An essential role for transcription before the MBT in Xenopus laevis. Dev Biol. 2011;357:478–491. doi: 10.1016/j.ydbio.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R, McLaren A. Factors affecting the time of formation of the mouse blastocoele. J Embryol Exp Morphol. 1977;41:79–92. [PubMed] [Google Scholar]
- Stancheva I, El-Maarri O, Walter J, Niveleau A, Meehan RR. DNA methylation at promoter regions regulates the timing of gene activation in Xenopus laevis embryos. Dev Biol. 2002;243:155–165. doi: 10.1006/dbio.2001.0560. [DOI] [PubMed] [Google Scholar]
- Sun Y, Nien C-Y, Chen K, Liu H-Y, Johnston J, Zeitlinger J, Rushlow C. Zelda overcomes the high intrinsic nucleosome barrier at enhancers during Drosophila zygotic genome activation. Genome Res. 2015;25:1703–1714. doi: 10.1101/gr.192542.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda P, Franke V, Schultz RM. Sculpting the Transcriptome During the Oocyte-to-Embryo Transition in Mouse. Curr Top Dev Biol. 2015;113:305–349. doi: 10.1016/bs.ctdb.2015.06.004. [DOI] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009;136:3033–3042. doi: 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- Talhouarne GJS, Gall JG. Lariat intronic RNAs in the cytoplasm of Xenopus tropicalis oocytes. Rna. 2014;20:1476–1487. doi: 10.1261/rna.045781.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan MH, Au KF, Yablonovitch AL, Wills AE, Chuang J, Baker JC, Wong WH, Li JB. RNA sequencing reveals a diverse and dynamic repertoire of the Xenopus tropicalis transcriptome over development. Genome Res. 2013;23:201–216. doi: 10.1101/gr.141424.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani H, Akimitsu N. Genome-wide technology for determining RNA stability in mammalian cells: historical perspective and recent advantages based on modified nucleotide labeling. RNA Biol. 2012;9:1233–1238. doi: 10.4161/rna.22036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DM, Ray PF, Ao A, Winston RM, Handyside AH. Paternal transcripts for glucose-6-phosphate dehydrogenase and adenosine deaminase are first detectable in the human preimplantation embryo at the three- to four-cell stage. Mol Reprod Dev. 1997;48:442–448. doi: 10.1002/(SICI)1098-2795(199712)48:4<442::AID-MRD4>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Teperek M, Simeone A, Gaggioli V, Miyamoto K, Allen GE, Erkek S, Kwon T, Marcotte EM, Zegerman P, Bradshaw CR, et al. Sperm is epigenetically programmed to regulate gene transcription in embryos. Genome Res. 2016;26:1034–1046. doi: 10.1101/gr.201541.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesarík J, Kopecný V, Plachot M, Mandelbaum J. Ultrastructural and autoradiographic observations on multinucleated blastomeres of human cleaving embryos obtained by in-vitro fertilization. Hum Reprod. 1987;2:127–136. doi: 10.1093/oxfordjournals.humrep.a136496. [DOI] [PubMed] [Google Scholar]
- Torres-Padilla M-E, Bannister AJ, Hurd PJ, Kouzarides T, Zernicka-Goetz M. Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and preimplantation embryos. Int J Dev Biol. 2006;50:455–461. doi: 10.1387/ijdb.052073mt. [DOI] [PubMed] [Google Scholar]
- Tsichlaki E, FitzHarris G. Nucleus downscaling in mouse embryos is regulated by cooperative developmental and geometric programs. Sci Rep. 2016;6:28040. doi: 10.1038/srep28040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heijden GW, Dieker JW, Derijck AAHA, Muller S, Berden JHM, Braat DDM, van der Vlag J, de Boer P. Asymmetry in histone H3 variants and lysine methylation between paternal and maternal chromatin of the early mouse zygote. Mech Dev. 2005;122:1008–1022. doi: 10.1016/j.mod.2005.04.009. [DOI] [PubMed] [Google Scholar]
- van Heeringen SJ, Akkers RC, van Kruijsbergen I, Arif MA, Hanssen LLP, Sharifi N, Veenstra GJC. Principles of nucleation of H3K27 methylation during embryonic development. Genome Res. 2014;24:401–410. doi: 10.1101/gr.159608.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassena R, Boué S, González-Roca E, Aran B, Auer H, Veiga A, Izpisua Belmonte JC. Waves of early transcriptional activation and pluripotency program initiation during human preimplantation development. Development. 2011;138:3699–3709. doi: 10.1242/dev.064741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastag L, Jorgensen P, Peshkin L, Wei R, Rabinowitz JD, Kirschner MW. Remodeling of the metabolome during early frog development. PLoS ONE. 2011;6:e16881. doi: 10.1371/journal.pone.0016881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw NL, Zhang Y, Woods IG, Imam F, Regev A, Liu XS, Rinn J, Schier AF. Chromatin signature of embryonic pluripotency is established during genome activation. Nature. 2010;464:922–926. doi: 10.1038/nature08866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenstra GJ, Wolffe AP. Constitutive genomic methylation during embryonic development of Xenopus. Biochim Biophys Acta. 2001;1521:39–44. doi: 10.1016/s0167-4781(01)00280-9. [DOI] [PubMed] [Google Scholar]
- Veenstra GJ, Destrée OH, Wolffe AP. Translation of maternal TATA-binding protein mRNA potentiates basal but not activated transcription in Xenopus embryos at the midblastula transition. Mol Cell Biol. 1999;19:7972–7982. doi: 10.1128/mcb.19.12.7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuković LD, Jevtić P, Edens LJ, Levy DL. New Insights into Mechanisms and Functions of Nuclear Size Regulation. Int Rev Cell Mol Biol. 2016;322:1–59. doi: 10.1016/bs.ircmb.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Wang J, Davis RE. Contribution of transcription to animal early development. Transcription. 2014;5:e967602. doi: 10.4161/21541264.2014.967602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C, Salichos L, Wittgrove CM, Rokas A, Patton JG. Transcriptome-wide analysis of small RNA expression in early zebrafish development. Rna. 2012;18:915–929. doi: 10.1261/rna.029090.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiddon JL, Langford AT, Wong C-J, Zhong JW, Tapscott SJ. Conservation and innovation in the DUX4-family gene network. Nat Genet. 2017;25:4577. doi: 10.1038/ng.3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MD, Bissiere S, Alvarez YD, Plachta N. Mouse Embryo Compaction. Curr Top Dev Biol. 2016;120:235–258. doi: 10.1016/bs.ctdb.2016.04.005. [DOI] [PubMed] [Google Scholar]
- Woodland HR, Adamson ED. The synthesis and storage of histones during the oogenesis of Xenopus laevis. Dev Biol. 1977 doi: 10.1016/0012-1606(77)90359-1. [DOI] [PubMed] [Google Scholar]
- Wu G, Han D, Gong Y, Sebastiano V, Gentile L, Singhal N, Adachi K, Fischedick G, Ortmeier C, Sinn M, et al. Establishment of totipotency does not depend on Oct4A. Nat Cell Biol. 2013;15:1089–1097. doi: 10.1038/ncb2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Huang B, Chen H, Yin Q, Liu Y, Xiang Y, Zhang B, Liu B, Wang Q, Xia W, et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature. 2016;534:652–657. doi: 10.1038/nature18606. [DOI] [PubMed] [Google Scholar]
- Wu S-F, Zhang H, Cairns BR. Genes for embryo development are packaged in blocks of multivalent chromatin in zebrafish sperm. Genome Res. 2011;21:578–589. doi: 10.1101/gr.113167.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L, Cai JY, Ma J, Huang Z, Guo MX, Fu LZ, Shi YB, Li WX. Global expression profiling reveals genetic programs underlying the developmental divergence between mouse and human embryogenesis. BMC Genomics. 2013;14:568. doi: 10.1186/1471-2164-14-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Yang M, Guo H, Yang L, Wu J, Li R, Liu P, Lian Y, Zheng X, Yan J, et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol. 2013;20:1131–1139. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- Yanai I, Peshkin L, Jorgensen P, Kirschner MW. Mapping gene expression in two Xenopus species: evolutionary constraints and developmental flexibility. Dev Cell. 2011;20:483–496. doi: 10.1016/j.devcel.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Zhou Y, Gu J, Xie S, Xu Y, Zhu G, Wang L, Huang J, Ma H, Yao J. Deep mRNA sequencing analysis to capture the transcriptome landscape of zebrafish embryos and larvae. PLoS ONE. 2013;8:e64058. doi: 10.1371/journal.pone.0064058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yartseva V, Giraldez AJ. The Maternal-to-Zygotic Transition During Vertebrate Development: A Model for Reprogramming. Curr Top Dev Biol. 2015;113:191–232. doi: 10.1016/bs.ctdb.2015.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda GK, Schubiger G. Temporal regulation in the early embryo: is MBT too good to be true? Trends Genet. 1992;8:124–127. doi: 10.1016/0168-9525(92)90369-F. [DOI] [PubMed] [Google Scholar]
- Yuan K, Seller CA, Shermoen AW, O’Farrell PH. Timing the Drosophila Mid-Blastula Transition: A Cell Cycle-Centered View. Trends Genet. 2016;32:496–507. doi: 10.1016/j.tig.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue H-M, Li Z, Wu N, Liu Z, Wang Y, Gui J-F. Oocyte-specific H2A variant H2af1o is required for cell synchrony before midblastula transition in early zebrafish embryos. Biol Reprod. 2013;89:82. doi: 10.1095/biolreprod.113.108043. [DOI] [PubMed] [Google Scholar]
- Zhang B, Zheng H, Huang B, Li W, Xiang Y, Peng X, Ming J, Wu X, Zhang Y, Xu Q, et al. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature. 2016;537:553–557. doi: 10.1038/nature19361. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Vastenhouw NL, Feng J, Fu K, Wang C, Ge Y, Pauli A, van Hummelen P, Schier AF, Liu XS. Canonical nucleosome organization at promoters forms during genome activation. Genome Res. 2014;24:260–266. doi: 10.1101/gr.157750.113. [DOI] [PMC free article] [PubMed] [Google Scholar]