Abstract
Activation of the Hedgehog (Hh) pathway effector GLI1 is linked to tumorigenesis and invasiveness in a number of cancers, with targeting of GLI1 by small molecule antagonists shown to be effective. We profiled a collection of GLI antagonists possessing distinct mechanisms of action for efficacy in phenotypic models of inflammatory and non-inflammatory breast cancer (IBC and non-IBC) that we showed expressed varying levels of Hh pathway mediators. Compounds GANT61, HPI-1, and JK184 decreased cell proliferation, inhibited GLI1 mRNA expression and decreased the number of colonies formed in TN-IBC (SUM149) and TNBC (MDA-MB-231 and SUM159) cell lines. In addition, GANT61 and JK184 significantly down-regulated GLI1 targets that regulate cell cycle (cyclin D and E) and apoptosis (Bcl2). GANT61 reduced SUM149 spheroid growth and emboli formation, and in orthotopic SUM149 tumor models significantly decreased tumor growth. We successfully utilized phenotypic profiling to identify a subset of GLI1 antagonists that were prioritized for testing in in vivo models. Our results indicated that GLI1 activation in TN-IBC as in TNBC, plays a vital role in promoting cell proliferation, motility, tumor growth, and formation of tumor emboli.
Keywords: Hedgehog, GLI1, GANT61, JK184, SUM149, IBC
1. Introduction
Developmental signaling pathways such as Hedgehog (Hh) play an important role in the pathogenesis and progression of metastatic triple negative breast cancers (TNBC) [1, 2], with new therapeutic approaches targeting this pathway for inhibition currently being tested in advanced preclinical studies or early clinical trials [1, 3]. The Hh pathway is also an emerging target for breast cancer stem cells [4, 5]. Hh signaling is initiated by the binding of Hh ligand to its transmembrane receptor Patched (PTCH) which in turn relieves the inhibition of co-receptor Smoothened (SMO) allowing signaling to proceed. Hh pathway activation is mediated by the GLI family of transcription factors with GLI1 (GLIoma-associated oncogene homolog 1) being an essential mediator of Hh pathway activation [6–9]. Due to its role in a wide spectrum of cancers, pharmacological inhibitors of the Hh pathway have therapeutic value [10–13]. Hh pathway inhibitors that act on SMO have been approved [14], with SMO inhibitors currently in clinical trials for TNBC and metastatic solid breast tumors [4]. However, due to acquired resistance of tumors to SMO inhibitors [15–17], there is a need to identify inhibitors that target other parts of the pathway.
GLI1 acts as the terminal effector of Hh signaling and its status controls cellular proliferation, differentiation, and survival [6–9]. Activated GLI1 translocates into the nucleus, stimulating the transcription of Hh pathway target genes, including GLI1, PTCH1 and many pro-survival pathway genes [18]. Conditional expression of GLI1 in the mouse mammary gland results in the expansion of progenitor cell types and the development of mammary tumors that are aggressive, ER-negative and basal-like [19]. In addition, activation of GLI1, via gene amplification [20], has been implicated in the initiation, progression and invasiveness of multiple cancers [21–27], including basal-like breast cancers [28], with its expression correlating with unfavorable survival in human breast cancer [25].
Elevated GLI1 expression has also been observed in inflammatory breast cancer (IBC) [29], a highly aggressive and unique form of locally advanced breast cancer (LABC) [30], for which nearly 40% of patients exhibit the TN phenotype compared to ~20% for non-IBC [31]. The presence of tumor emboli is a hallmark of IBC [32], in which distinctive clusters of tumor cells migrate collectively, driving lymphatic invasion and metastasis [33]. Gene expression profiling of IBC tumors has identified Hh/GLI1 pathway overexpression as a predictor of the IBC phenotype [34]. We previously observed that the basal-like IBC cell line SUM149, a widely used model for IBC [35–39], exhibits elevated GLI1 expression and that knock down of GLI1 reduced cell proliferation and motility [29]. Furthermore, a Hh/SMO-independent non-canonical mechanism for GLI1 activation in SUM149 was implicated [29]. Due to these non-canonical mechanisms of GLI1 activation [40, 41], there is growing utility in targeting the Hh pathway downstream at the level of GLI [42–44]. Phenotypic screens have yielded inhibitors targeting the Hh pathway downstream of SMO [45–49], with some that target GLI activation (GANTs [46], HPI-1 [50]), having shown efficacy in in vivo cancer models. Furthermore, targeting at the level of GLI has been shown to overcome SMO inhibitor resistance [51].
In this study, we assessed a collection of small molecule GLI inhibitors with varying mechanisms of action for efficacy in in vitro and in vivo IBC and non-IBC models. Using a panel of phenotypic assays, we identified a subset of GLI antagonists with growth inhibitory effects. In particular, GANT61 displayed significant inhibitory activity in 3D models while also exhibiting efficacy in vivo in an IBC cell-line derived orthotropic mammary fat pad xenograft tumor model.
2. Materials and methods
2.1 Cell lines and reagents
The SUM149, SUM159 and SUM190 cell lines were obtained from Asterand Inc (Detroit, MI) and cultured per manufacturer’s instructions and as described previously [35]. MDA-MB-231, SKBR3 cells and C3H10T1/2 cell lines were from American Type Culture Collection (ATCC) (Manassas, VA) and cultured per their instructions. Human mammary epithelial cells (HMEC) were obtained from Lonza Group Ltd. (Allendale, NJ) and cultured per manufacturer’s instructions using their Mammary Epithelial Growth Medium Kit (CC-3150). GANT58 (Cat # 3889), GANT61 (Cat # 3191), JK184 (Cat # 3341) and cyclopamine were obtained from Tocris/R&D Systems (Minneapolis, MN). GANT61 in gram quantities was from DC Chemicals (Shanghai, China). HPI-1/2/3/4 were a kind gift from Dr. James K. Chen, Stanford School of Medicine. HPI-1/3/4 were also purchased from Sigma Aldrich (St. Louis, MO). Cytochalasin D was from MP Biomedicals, LLC (Solon, OH). Hoechst-33342 and YOYO-1 dyes were from Thermo Fisher Scientific
2.2 Real time quantitative RT-PCR
Total RNA was isolated using the RNeasy Mini Kit (Qiagen). cDNA was synthesized from 2 µg of RNA using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Gene expression primers (GLI1 (Hs00171790_m1), GLI2 (Hs00257977_m1), GLI3 (Hs00609233_m1), PTCH1 (Hs00181117_m1), Smo (Hs00170665 m1), Bcl2 (Hs00153350 m1), CCND1 (Hs00277039_m1), CCNE1 (Hs00233356_m1), β2-microglobulin (B2M) (Hs00984230_m1) and β-Actin (Hs99999903_m1) were obtained from Applied Biosystems. For mRNA quantification, quantitative real-time PCR was performed using an ABI 7500 Fast Real-Time PCR or QuantStudio 6 Flex Real-Time PCR (Thermo Fisher Scientific) system. β-actin and B2M were used as endogenous controls for expression normalization. All assays were performed in triplicate and the fold change in mRNA expression was determined according to the method of 2-∆∆Ct and represented changes were normalized to a reference HMEC cell line or DMSO control.
2.3 C3H10T1/2 cell-based assay for hedgehog activity
The C3H10T1/2 assay was performed essentially as previously described [52, 53]. C3H10T1/2 cells were maintained in DMEM medium containing 10% FBS and plated in 96-well plates at 5000 cells/well. For competition experiments, Sonic hedgehog (Shh) protein was added 24h later at a fixed concentration of 2 µg/ml along with various concentrations of inhibitors. Cells were incubated for a further 5 days, lyzed and assayed for alkaline phosphatase (AP) using pNPP with plates read at 405 nm. Inhibition constant (IC50) values were determined by nonlinear regression in GraphPad 6.0.
2.4 Dose-response plates
Compounds were purchased as dry powders, dissolved in Dimethyl Sulfoxide (DMSO) to give 10 mM stock concentrations and stored at −20°C. A Biomek®NX robotics workstation (Beckman Coulter Inc.), was used to serially dilute compounds with 100% DMSO in 384-well Greiner polypropylene plates (Greiner Bio-One) to generate 10-point 2-fold dose responses (5 µL final volume in each well).
2.5 High content cell imaging and cell proliferation assays
High content imaging was carried out essentially as we recently described [54], based on [55]. 384-well plates (black, clear bottom tissue culture treated) were seeded with SUM149 (700 cells per well), SUM159 (800 cells per well) or MDA-MB-231 (1000 cells per well) and cells allowed to attach overnight. Compounds or DMSO (50 nL) were then added from dose response plates to the cell plates (50 µL) using the Biomek NX equipped with Pintools and the cells incubated for a further 72 h. After washing, Hoechst 33342 dye (10 µg/ml) was added and after 45 min, cells fixed with 10% formalin for 15 min. Fluorescence quantification and localization were determined using a CellInsight NXT and 3-channel Cell Health Profiling protocol in HCS Screen software (ThermoFisher). Excitation wavelength was 386 nm for Hoechst 33342. Images were acquired using an Olympus UPlanFLN 10X/0.30 objective and 2×2 camera binning. A nuclear mask was established using channel 1 signaling (Hoechst 33342) and used to determine nuclear characteristics (nuclear count). All conditions were conducted in quintuplicate. Data analysis was performed retrieving data sets from HCS View software (ThermoFisher) and normalized to mean DMSO values. IC50 values were determined using a four-parameter dose-response (variable slope) equation in GraphPad Prism 6 (GraphPad Software, San Diego, CA).
For cell proliferation assays, compounds were incubated with cells for 72h. PrestoBlue™ Cell Viability Reagent (ThermoFisher Scientific) added to each well and cells incubated for a further 2 h at 37°C. Fluorescence intensity was read at 560 nm excitation/590 nm emission using a BMG PheraStar micro plate reader (BMG Labtech, Cary, NC). Heat maps using percent inhibition values were generated in Microsoft Excel using conditional formatting.
2.6 Colony formation assay
SUM149, SUM159 (250 cells/well) and MDA-MB-231 (350 cells/well) were seeded in triplicate in 6-well plates (Corning Inc. Tewksbury, MA), and cells allowed to adhere overnight. Briefly, after 24 h, cells were washed with PBS, fresh media containing compounds added and cells allowed to grow for 5–14 days. Cells were fixed with 10% formalin, colonies stained with 1% (w/v) crystal violet and colonies counted using an automated ColCount (Oxford Optronix, Abingdon, UK).
2.7 Wound closure assay
Cells were seeded in an interlock 96-well plate (Essen Bioscience, Ann Arbor, Michigan) and incubated overnight. The wound-maker module (Essen BioScience) was used to simultaneously create a vertical wound/scratch in all wells, after which cells were washed with PBS, and compounds added to the plate (n = 5). Cell motility was measured using the IncuCyte® ZOOM System (Essen Bioscience) Live Cell Analysis System with images taken at 0, 4, 12 18 and 24 h using a 10× objective. The relative wound density was determined using measures of both wound width and wound confluence to take into account any effects of proliferation (IncuCyte motility assay protocol). The relative wound density (%RWD) was calculated as follows: %RWD(t)=100×((w(t)-w(0))/(c(t)-w(0) )); where w(0) = initial wound density, w (t) = wound density at each successive time point, c (0) = initial cell region density, and c (t) = cell region density at each successive time point.
2.8 3D tumor spheroid high content imaging assay
The high throughput 3D tumor spheroid assay in 384-well format was carried out essentially as we recently described [54]. Cells were harvested from a sub-confluent flask and plated at 400 cells per well (in 50 µL of media) into ultra-low attachment 384-well spheroid microplates (Corning 3830) using a Multidrop bulk dispenser. Plates were then centrifuged at 200 rcf for 1 min followed by overnight incubation at 37°C. The following day, cells were treated with compounds (n = 5) using a D300E Digital Dispenser (Hewlett Packard. Palo Alto, CA). After a further 72 h incubation, spheroids were fixed, stained (with 10 µg/ml Hoechst 33342 and 100 nM YOYO-1) and imaged using a CellINsight NXT (ThermoFisher) as recently described in [56]. HCS Screen software (ThermoFisher) collected data using a modified “Cell health profiling” algorithm which was compiled in HCS View software, normalized against DMSO controls and plotted in ScreenAble (ScreenAble, Chapel Hill, NC) for analysis.
2.9 Tumor emboli assay
SUM149 cells were plated at 10,000 cells/well in 24-well ultra-low attachment plates (Corning Inc., Corning, NY) in serum-free minimum essential medium as recently described [56]. Compounds were added at time of seeding and cells allowed to grow for 7 d. For each well, emboli were identified (>20 cells) and imaged using a Motic AE2000 microscope (North America, Richmond, BC, Canada), Staurosporine was used as a positive control to inhibit tumor emboli formation in SUM149 cells [35, 57].
2.10 Xenografts
The authors thank Dr. Matthias Lauth (Philipps University of Marburg) for his technical advice on GANT61 formulation and dosing. Animal experiments were conducted in accordance with accepted standards of humane animal care and approved by the Animal Care and Use Committees at the University of North Carolina-Chapel Hill and at North Carolina Central University. At 12 weeks of age, abdominal region mammary fat pads of female nude NU/J (Study 1) or NOD SCID (Study 2) mice were implanted with 1×106 SUM149 cells suspended in 50% reduced growth factor Matrigel® and PBS. Mice were randomized into treatment groups (n = 5 per treatment group).
Study 1
Once tumors reached 5 × 5 mm in size (approximately six weeks), animals were treated i.p. with either GANT61 3× per week 50 mg/kg (DC Chemicals) or DMSO vehicle control. Tumor response was assessed by weekly caliper measurements. Tumor-bearing mice were euthanized at 5 weeks for morbidity, tumor ulceration or tumor size of more than 2.0 cm in diameter. Growth curves were plotted as the mean relative treatment group tumor volume ± standard error. Differences in tumor volumes were assessed using Student t-test in GraphPad Prism 6.0. Comparisons in tumor growth were made using linear regression in GraphPad Prism 6.0. Tumors were snap frozen for protein extraction. MDA-MB-231 and SUM159 xenografts were carried out as described in study 1 (n = 6).
Study 2
When SUM149 tumor volumes became greater than 0.2 cm, mice were treated with either vehicle (corn oil:ethanol, 4:1 [46]) or GANT61 in vehicle (50 mg/kg) i.p. once per day. After 21 days, animals were euthanized, tumors were surgically removed and weighed. Tumors were divided into separate pieces for the following uses: in RNA Later for RNA extraction and snap frozen for protein extraction. Tumor weights were compared across groups using Student t-test in GraphPad Prism 6.0.
2.12 Western Immunoblotting
Protein lysates were prepared from the tumor tissues using T-PER Tissue Protein extraction reagent (Thermo Scientific). T-PER lysis buffer containing protease inhibitors (10 µg/ml) was added to the tumor tissue and lysis carried out using a Bullet Blender BBX24B (Next Advance, Inc. Averill Park, NY, USA). Samples were centrifuged at 10,000g for 5 min and supernatants collected. Protein concentration was determined using the Pierce BCA Protein Assay Kit (ThermoFisher). Lysates (30 µg) were electrophoresed on NU-PAGE Bis-Tris 4–12% gels (Life Technologies) and transferred to Immobilon-Fl membranes (Millipore, Billerica, MA) that had been pre-wet in methanol and transfer buffer. After blot transfer, membranes were stained for total protein quantification for normalization purposes using the REVERT Stain kit protocol (LI-COR, P/N 926–11010). Membranes were then blocked (5% non-fat milk) for 1 h at RT. Membranes were incubated with primary antibodies for GLI1: (rabbit Ab TA310536; Origene, Rockville, MD) at 1:5,000 and β-actin (mouse Ab Santa Cruz Biotechnology, Dallas, TX) at 1:10,000 overnight at 4°C. Purified recombin ant human GLI1 protein (Origene) was included as a positive control. Membranes were washed (PBST) and subsequently incubated with appropriate LI-COR (LI-COR Biosciences, Lincoln, NE) Odyssey secondary IRDye antibodies (1:15,000) for 1 h at RT. After washing, blots were visualized using LI-COR Odyssey and Image Studio software used to quantify bands. For each sample, GLI1 protein expression was normalized to β-actin or the total protein quantification and the mean for each treatment calculated according to Normalization Accuracy for Western Blotting guidelines (LI-COR Biosciences).
2.13 Statistical analysis
Unless stated, statistical analyses were conducted using GraphPad Prism version 6 (GraphPad Software, Inc) student’s 2-tailed t-test. *p < 0.05, **p < 0.01, ***p < 0.001 were considered statistically significant compared with controls.
3. Results
3.1 GLI1 and GLI2 are highly expressed in SUM149, SUM159 and MDA-MB-231 TNBC cell lines
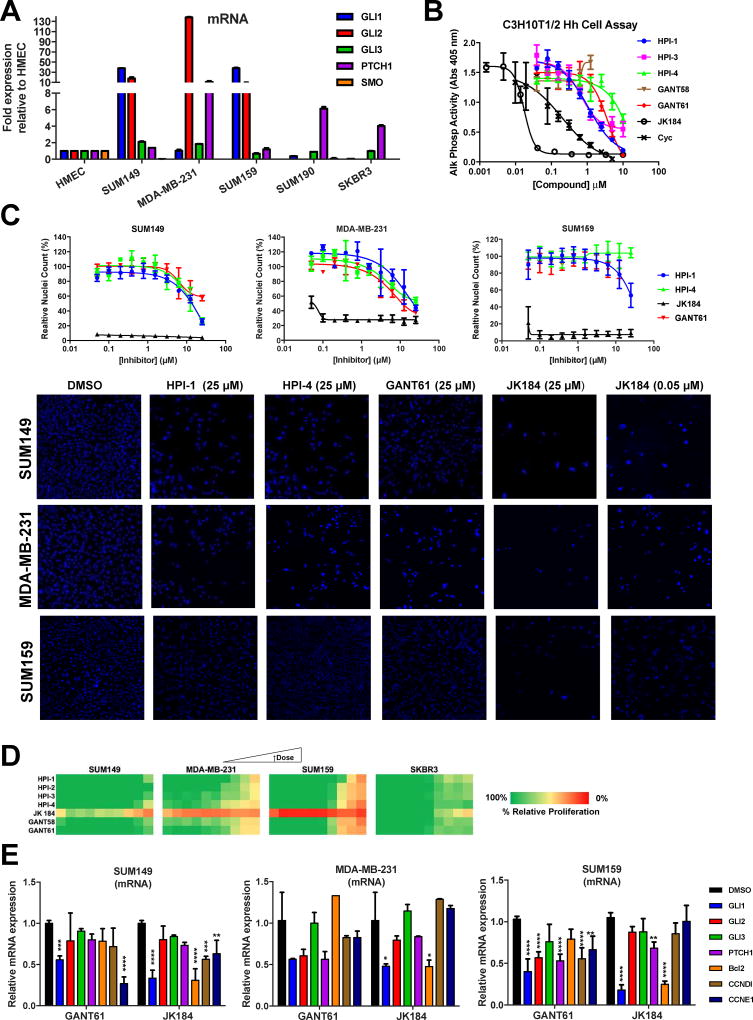
In this study, we assessed by qRT-PCR the expression of the major components of the Hh pathway, GLI1, GLI2, GLI3, SMO, and PTCH1, in a panel of breast cancer cell lines, including IBC cell lines SUM149 (TN, basal-like [58]) and SUM190 (HER2+), and the non-IBC cell lines MDA-MB-231 (TN), SUM159 (TN) and SKBR3 (HER2+). To ensure consistency, gene expression in cell lines was assessed at low passage numbers (<10 after obtained from vendor). The HER2+ cell lines exhibited consistently lower levels of GLI1 and GLI2, similar to expression levels in HMEC cells. The TN/basal-like cell lines were less consistent, with both the TN-IBC SUM149 and TN SUM159 demonstrating higher levels of GLI1 (~40-fold relative to HMEC for both) and GLI2 (~20 and ~10-fold relative to HMEC respectively), while the MDA-MB-231 had low GLI1 (comparable to HMEC) but significantly high GLI2 expression (>130-fold relative to HMEC) (Fig. 1A). The high expression of GLI2 and the low expression of GLI1 and GLI3 that we observed for MDA-MB-231 are consistent with previous reports [59]. Relative to HMEC, PTCH1 expression was highest in MDA-MB-231 cells and at comparable levels in the other cell lines tested. GLI3 and SMO levels were relatively low in all cell lines. With all genes tested there was no consistent pattern between the basal-like IBC and non-IBC models, however a similar expression pattern was observed in HER2+ cell lines.
Fig. 1.
Effect of GLI antagonists on breast cancer cell proliferation and Hh pathway activity. (A) mRNA levels of GLI1/2/3, PTCH1, and SMO in IBC and non-IBC cell lines. Data are expressed as mean ± SD. (B) Testing of GLI antagonists for Hh pathway inhibition in C3H10T1/2 hedgehog functional assay. C3H10T12 cells were stimulated with Shh protein (2 µg/ml) in the presence of GLI antagonists (0.001 – 10 µM) and alkaline phosphatase measured at 5 d. Dose response curves were generated using non-linear regression and IC50 values determined in GraphPad Prism 6. (C) SUM149, MDA-MB-231 and SUM159 cells were incubated with GLI antagonists (0.001 – 25 µM) for 72 h, stained with Hoechst-33342 nuclear stain and cell numbers determined by high content imaging. Upper: Dose response curves for relative nuclei count. For each concentration run in triplicate, percent inhibition values were calculated and data normalized to vehicle. Data is mean ± SD (n = 3). Dose response curves were generated using non-linear regression and IC50 values determined in GraphPad Prism 6. Lower: Representative images. (D) SUM149, MDA-MB-231, SUM159 and SKBR3 cells were incubated with GLI antagonists (0.001 – 25 µM) for 72 h, and proliferation assessed by Presto Blue. For each concentration, percent inhibition values were calculated and data normalized to vehicle. The heat map was generated in Excel using conditional formatting with red representing 100% inhibition through to green for 0% inhibition. (E) qRT-PCR comparative expression of Hh-GLI downstream targets in SUM149, MDA-MB-231 and SUM159 cells treated with vehicle (0.1% DMSO), GANT61 (20 µM) or JK184 (30 nM) for 72 h. Mean ± SD shown, n=4, two-way ANOVA with Tukey’s post-test (*p<0.05, **p<0.005, ***p<0.0005 and ****p<0.0001; significantly different from respective controls).
3.2 Assessment of a GLI antagonist panel reveals compounds with anti-hedgehog pathway activity in C3H10T1/2 cells
We have previously observed that siRNA-mediated downregulation of GLI1 decreases IBC cell proliferation and motility [29]. Furthermore, we went on to show that GLI1 activation occurred primarily through a non-canonical mechanism [29]. Hence, in this study we wanted to assess the effects of small molecule inhibitors that act downstream in the Hh pathway (reviewed in [60]) and as a result we assembled a set of GLI antagonists that have been identified with varying mechanisms and understanding of action, consisting of GANT58 and GANT61 [46], HPIs 1–4 [45], and JK184 [47] (compound structures shown in Supplementary Fig. 1A).
The Hh pathway activity in response to each GLI antagonist was assessed in a widely accepted Hh functional cell-based assay, the C3H10T1/2 cell line which we and others have used to assess Hh pathway inhibition [53, 61]. In this Hh-responsive cell line, addition of Hh to the cells induces alkaline phosphatase (AP) activity, a marker of Hh-mediated cell differentiation. For inhibition testing, C3H10T1/2 cells were incubated with Hh at its EC50 of 2 µg/ml and GLI antagonists titrated as 10-point, 2-fold dose responses with 10 µM as the highest concentration. The SMO-directed inhibitor cyclopamine (Cyc) [62] was included as a positive control for the C3H10T1/2 assay. Resulting dose response curves (Fig. 1B) and corresponding IC50 values for the GLI antagonist set are shown in Table 1. JK184 was the most potent inhibitor with an IC50 value of 20 nM, comparable to that previously reported for its activity in this assay [47]. HPI-1, HPI-3 and GANT61 were also effective inhibitors in this assay with IC50 values of 1.1, 0.7 and 3.7 µM, respectively.
Table 1.
IC50 values for GLI antagonists tested in C3H10T1/2 hedgehog functional assay and in Hoechst proliferation assay.
| IC50 (µM) (AP activity)a |
IC50 (µM) (Proliferation)b |
|||
|---|---|---|---|---|
| C3H10T1/2 | SUM149 | MDA-MB-231 | SUM159 | |
| HPI-1 | 1.1 | 29 | > 25 | 20.5 |
| HPI-2 | ||||
| HPI-3 | 0.7 | |||
| HPI-4 | 50% @ 10 µM | 15.9 | 6.2 | No curve |
| GANT58 | >10 | >25 | ||
| GANT61 | 3.7 | 6.3 | 5.6 | 6.1 |
| JK184 | 0.02 | 0.01 | 0.08 | 0.05 |
| Cyclopamine | 0.2 |
GLI antagonists were assayed in dose response in the C3H10T1/2 Hh functional assay in the presence of 2 µg/ml Hh protein, and AP activity measured after 5 days.
GLI antagonists were tested in dose response in Hoechst assay as described in Materials and Methods. IC50 values were determined from a minimum of three independent experiments.
3.3 GLI antagonists have differential effects on proliferation and cytotoxicity of GLI1/2 positive TNBC cell lines
To assess any role of the relative expression of GLI1 versus GLI2 levels on GLI antagonist sensitivity, we focused on the three TNBC cell lines; SUM149 and SUM159 with high GLI1/low GLI2 and MDA-MB-231 which has low GLI1 and high GLI2. To comprehensively assess the effects of the GLI antagonists on cell proliferation, viability, and cytotoxicity, we used live-cell high content automated microcopy in which cells were incubated with the GLI antagonists in dose response for 72h, stained with Hoechst 33342 (nuclear stain) and cell count determined (dose response curves and representative images in Fig. 1C). JK184 significantly reduced cell counts for all three cell lines with SUM149, MDA-MB-231 and SUM159 (Figs 1C) having IC50 values of 11 nM, 80 nM and 50 nM respectively (Table 1). SUM149 cells were relatively more sensitive to JK184 than MDA-MB-231 or SUM159 cells with SUM149 nuclei counts low at all concentrations tested (Figs 1C). The other GLI antagonists had modest micromolar activity in these cells with HPI-1, HPI-4, and GANT61 all showing decreased cell number as assessed by Hoechst staining (Figs 1C and Table 1). GANT61 had moderate effects on cell growth of SUM149, MDA-MB-231 and SUM159 with IC50 values in the range of 5–10 µM for all three cell lines (Table 1).
We also assessed the effects of the GLI antagonists in an alternative measure of proliferation, the Presto Blue assay. In this assay, JK184 treatment strongly reduced cell proliferation of SUM149, MDA-MB-231 and SUM159 cells at all concentrations tested, with the other GLI antagonists having modest micromolar potencies in these cell lines. Further, in SKBR3 cells that have very low levels of GLI1/2, we observed that the GLI antagonists including the potent JK184 were far less effective at reducing cell proliferation (Fig. 1D).
We next evaluated by qRT-PCR, those GLI antagonists that demonstrated anti-proliferative activity (JK184, GANT61 and HPI-1) for effects on GLI1 mRNA expression and the expression of GLI1 target genes. SUM149, MDA-MB-231 and SUM159 cells were treated with GANT61, JK184 (at their ~IC50 doses) or vehicle control (DMSO) for 72h. After RNA isolation and cDNA synthesis, qRT-PCR analysis was carried out using specific Taqman primer sets and mRNA expression levels were normalized to cells treated with vehicle control.
GLI1 mRNA expression in SUM149 and SUM159 cells was significantly reduced by GANT61 (20 µM; SUM149, adjusted p-value (adj p) = 0.0004; SUM159, adj p <0.0001) and by JK184 (30 nM; SUM149, adj p <0.0001; SUM159, adj p <0.0001) (Fig. 1E). GLI1 mRNA expression in SUM149 was also reduced by HPI-1 (Supplementary Fig. 1B). In MDA-MB-231 cells, JK184 (30 nM; adj p = 0.0198) but not GANT61 (20 µM) significantly reduced GLI1 expression. With the concentrations used, the effect of GANT61 and JK184 on reducing GLI1 expression in SUM149 cells was comparable and not significantly different. A similar finding was observed for SUM159 cells. In SUM149 cells and MDA-MB-231 cells, GLI2 and PTCH1 mRNA expression were not significantly affected by either treatment. In contrast, in SUM159 cells, GANT61 reduced both GLI2 (adj p <0.0001) and PTCH1 (adj p <0.0001) expression whereas JK184 only had a significant effect on PTCH1 (adj p = 0.0021).
Next, we looked at the effects of GLI antagonists on cell cycle progression and anti-apoptosis genes promoted by GLI activation [63, 64]. JK184 but not GANT61 significantly reduced Bcl2 expression in all three cell lines (SUM149, adj p <0.0001; MDA-MB-231, adj p =0.0185; SUM159, adj p <0.0001). CCNE1 expression was reduced by GANT61 (adj p <0.0001) and JK184 (adj p =0.0052) in SUM149, and by GANT61 in SUM159 cells (adj p =0.0023). CCND1 expression was significantly reduced by JK184 in SUM149 cells (adj p = 0.0005) and by GANT61 in SUM159 cells (adj p <0.0001). In SUM149 and SUM159 cells, there was a significant difference in the effect of JK184 compared to GANT61 on reducing the expression of Bcl2 (SUM149, adj p <0.0001; SUM159, adj p <0.0001) and CCNE1 (SUM149, adj p = 0.002; SUM159, adj p =0.002). These results indicated that GANT61 and JK184 have significant effects in SUM149 and SUM159 cells on targeting and inhibiting GLI1-mediated activity.
3.4 A subset of GLI antagonists decrease colony formation and cell motility of GLI1/2 positive TNBC cell lines
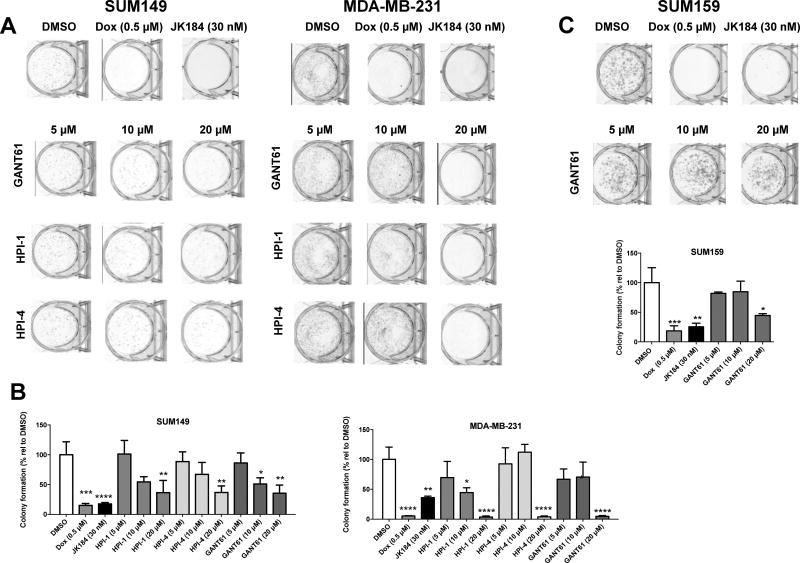
We further evaluated the subset of GLI antagonists that demonstrated anti-proliferative effects (JK184, GANT61, HPI-1 and HPI-4) for their ability to alter anchorage independent growth via colony formation assays (Fig. 2). For both SUM149 and MDA-MB-231 cells, JK184 (30 nM) was comparable to doxorubicin (Dox, 0.5 µM), our positive control for inhibition of colony formation, and significantly decreased the numbers of colonies formed compared to vehicle control (DMSO) (SUM149, adj p <0.0001; MDA-MB-231, adj p = 0.0028) (representative images Figs. 2A; quantification Fig. 2B). HPI-1, HPI-4 and GANT61 demonstrated comparable concentration dependent effects on colony formation for SUM149. Both cell lines showed fewer colonies after GANT61 (20 µM) treatment (SUM149, adj = 0.0022; MDA-MB-231, adj p <0.0001) and for SUM149 also with the lower 10 µM GANT61 treatment (adj p = 0.0337). At the higher 20 µM treatment, HPI-1 (SUM149, adj p = 0.0026; MDA-MB-231, adj p <0.0001) and HPI-4 (SUM149, adj p = 0.0028; MDA-MB-231, adj p <0.0001) had significant effects on colony formation. For SUM159, JK184 (adj p = 0.0023) and Dox adj p = 0.0003) had significant effects on colony formation (Fig 2C). Significant inhibition by GANT61 on SUM159 colony formation was observed at the highest concentration (20 µM) tested (adj p = 0.0207).
Fig. 2.
Effect of GLI antagonists JK184, HPI-1, HPI-4 and GANT61 on SUM149, MDA-MB-231 and SUM159 colony formation. For colony formation, cells were seeded in 6-well plates, treated with the indicated compounds for 5–14 days, fixed, colonies stained and counted. (A) Representative images for SUM149 and MDA-MB-231 colonies stained with crystal violet. A minimum of two independent experiments were carried out. (B) SUM149 and MDA-MB-231 percent colony formation relative to vehicle. (C) Representative images for SUM159 colonies and percent colony formation relative to vehicle. Data represents mean ± SD percentage of colonies relative to vehicle/untreated for a minimum of three wells. One-way ANOVA with Tukey’s post-test (*p<0.05, **p<0.005, ***p<0.0005 and ****p<0.0001; significantly different from respective controls).
Next, we determined whether GLI antagonists affect cell motility. For these studies, we used an automated in vitro wound healing assay in which confluent cells in 96 well plates are scratched simultaneously. This format allowed us to run multiple replicates for a number of compounds and follow scratch closure in real time with the effects of cell proliferation taken into account. Movement of cells into the scratch was imaged using a 10× objective every 2h. Image analysis with Incucyte (Essen BioScience, MI) software masks the unoccupied scratch area (pink) and the area of the scratch into which cells have moved (purple). For SUM149 cells, in the presence of vehicle control DMSO, the scratch was >90% closed at 24h, and less than 10% closed in the presence of cytochalasin D (0.5 µM), a positive control motility inhibitor [65] (representative images Supplementary Fig. 2A). Graphical results are shown as relative wound density (%RWD) which is calculated from measures of both wound width and wound confluence to factor in any effects of cell proliferation. RWD uses both the density of cell region (cell-occupied) and the density of wound region (cell-free regions) at a specific time to determine RWD. Hence, although GANT61 had modest effects on cell motility, this effect was not significant when cell proliferation was taken into account as determined as %RWD (Supplementary Fig 2B). MDA-MB-231 cells were less motile in this assay, with only 70–80% closure (DMSO) at 24h. While cytochalasin D significantly blocked motility in both cell lines (p-value <0.001), HPI-1, HPI-4 and GANT61 had no significant effect on decreasing SUM149 or MDA-MB-231 cell motility.
3.5 GANT 61 decreased 3D tumor spheroid and tumor emboli formation in SUM149
We next evaluated GLI antagonist effects in three dimensional (3D) in vitro models that have been shown to better mimic the tumor microenvironment and that take into account the spatial arrangement of cells and how they interact with each other [66, 67]. To comprehensively assess the effects of GLI antagonists on 3D tumor spheroid formation, we used an automated 3D spheroid assay that we have developed in which cells are plated in 384-well ultra-low attachment plates and spheroids allowed to form over four days [54]. We have previously observed that SUM149 cells form tight spheroids in this assay format [54]. For imaging, multi-parametric high-content analysis was used in which spheroids were simultaneously stained with Hoechst 33342 (nuclear stain) and YOYO-1 (plasma membrane permeability/cell death), and then imaged on a CellInsight high content screening platform. Compounds were added over a large concentration range (5 replicates) 24 h after cell plating and incubated for a further 72 h.
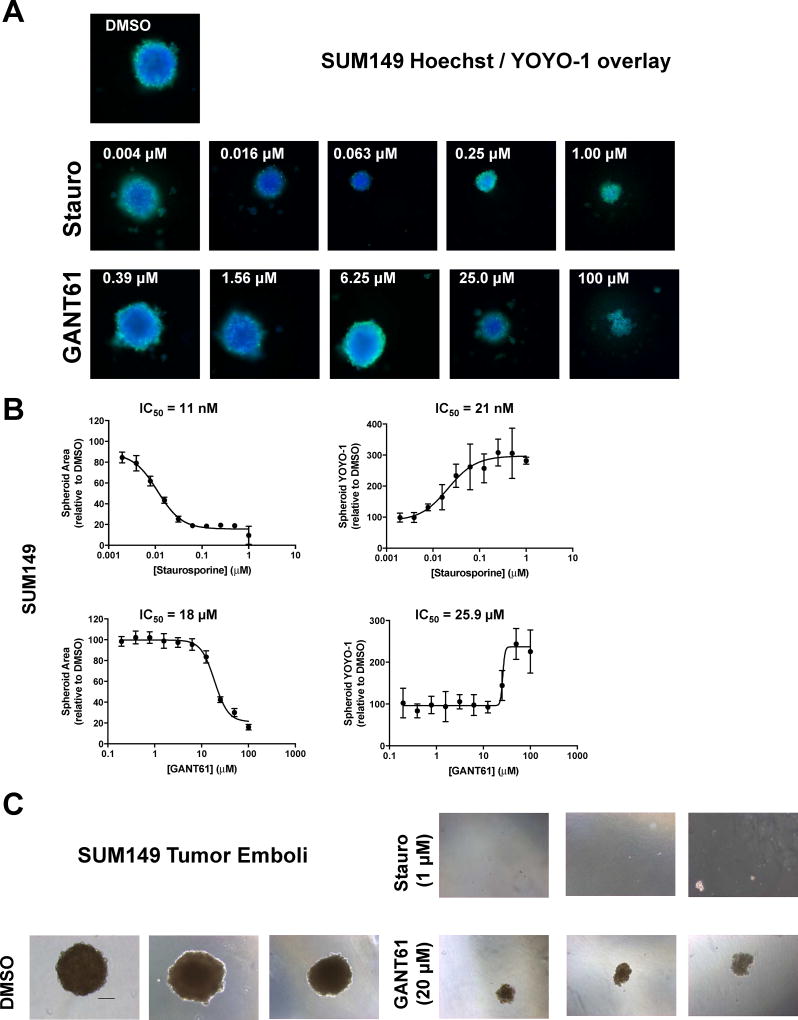
For SUM149 cells, high nuclear spheroid area (Hoechst) and minimal YOYO-1 were observed with DMSO indicating no observable cytotoxic effect of vehicle (representative image Fig. 3A). Staurosporine (Stauro) was included as a positive cytotoxic control and showed a dose dependent effect on spheroid area (IC50 value = 11 nM) with decreasing size with increasing concentration (Figs. 3A,B, Table 2). Staurosporine showed statistically significant decreases in spheroid area at all concentrations tested (1.9 nM to 1 µM; Supplementary Fig. 3A). Staurosporine had dose dependent effects on spheroid YOYO-1 (IC50 = 21 nM) with significant spheroid YOYO-1 staining observed at concentrations at and above the 7.8 nM treatment (Supplementary Fig. 3A) signifying cell membrane disruption. Although staurosporine decreased spheroid area, it did not appear to inhibit spheroid formation.
Fig. 3.
GANT61 disrupts formation of SUM149 tumor spheroids and tumor emboli. For the tumor spheroid assay, cells were seeded in ultralow attachment 384-well plates and treated 24 h later with either vehicle control (DMSO), staurosporine, or GANT61 at the indicated concentrations. Cells were grown for a further 72 h, spheroids stained with Hoechst/YOYO-1 and high content 3D images obtained using the CellInsight HCS platform. (A) representative images are shown for each treatment. Three independent experiments were carried out. (B) Data represent mean ± SD percent normalized to DMSO vehicle control for spheroid area (Hoechst) and spheroid YOYO-1 comprising a minimum of five replicate wells. Dose response curves were generated using non-linear regression and IC50 values determined in GraphPad Prism 6. (C) Tumor emboli: Representative images (10 × – scale bar = 100 microns) of SUM149 cells grown under tumor emboli conditions. Cells were treated at the time of plating with vehicle, staurosporine or GANT61 and cells grown for 7d. Images from at least two independent experiments comprising a minimum of three replicate wells.
Table 2.
IC50 values for GLI antagonists tested in 3D tumor spheroid assay.
| IC50 (µM)a | ||||
|---|---|---|---|---|
| SUM149 | MDA-MB-231 | SUM159 | ||
| Stauropsorine | Spheroid Area | 0.011 | No curve | 0.007 |
| Spheroid YOYO-1 | 0.021 | No curve | 0.065 | |
| GANT61 | Spheroid Area | 18 | 16.7 | 22.6 |
| Spheroid YOYO-1 | 25.9 | No curve | 37.8 | |
GLI antagonists were tested in dose response in 3D tumor spheroid assay, stained with Hoechst and YOYO-1 and imaged as described in Materials and Methods. IC50 values were determined by non-linear regression in Graphpad Prism 6. A minimum of three independent experiments were conducted.
Treatment of SUM149 spheroids with GANT61 altered spheroid formation and decreased spheroid area with an IC50 value of 18 µM (Fig. 3A, B, Table 2), with GANT61 at 12.5 µM or above having significant effects on spheroid area (Supplementary Fig 3A). Significant YOYO-1 staining was observed at and above the 25 µM dose (Supplementary Fig 3A) with cytotoxic effects increasing as the GANT61 dose increased (IC50 = 26 µM; Fig 3B, Table 2). Loss of spheroid shape and increased spheroid fragmentation was observed at GANT61 concentrations at and above 25 µM (Fig. 3A, B, Supplementary Fig. 3A). Staurosporine and GANT61 exhibited differences in their effects on spheroid formation suggesting differing mechanisms of actions.
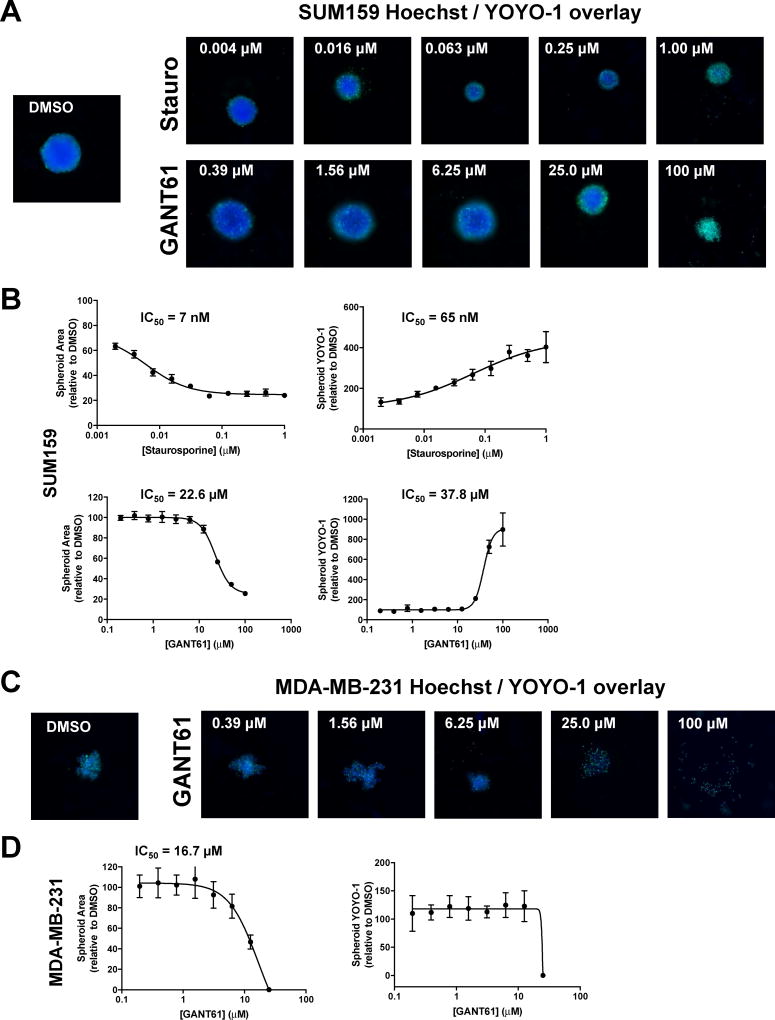
The effects of GANT61 on spheroid formation were also assessed in the two other TNBC lines MDA-MB-231 and SUM159 (Fig. 4). SUM159 like SUM149, were also observed to form tight spheroids in this assay system. For SUM159, GANT61 and staurosporine had effects on spheroid area and spheroid YOYO-1 (Fig. 4A and B, Table 2, Supplementary Fig. 3B) comparable to that observed on SUM149 cells. In contrast, in this format in which cells are plated and allowed to form spheroids for 24 h before treatment, we observed that MDA-MB- 231 cells routinely formed clumped aggregates as opposed to the more spherical tight spheroids formed by SUM149 and SUM159 cells. Indeed, MDA-MB-231 have been classified as forming cell aggregates when using these ultra-low attachment plates (Corning spheroid technical document). Further, it has been reported that MDA-MB-231 cells form only loose spheroids [68] making their use challenging when assessing drug effects in 3D culture systems. Hence, the effect we observed for GANT61 treatment of MDA-MB-231 appeared to be more of a disaggregation effect (a loosening of cell aggregates) at concentrations at or above 6.25 µM (Fig. 4C and D, Supplementary Fig. 3C rather than an effect on spheroid fragmentation. Indeed, staursporine had little effect on these MDA-MB-231 cell aggregates (Supplementary Fig. 3D).
Fig. 4.
Effect of GANT61 on formation of SUM159 and MDA-MB-231 tumor spheroids. The tumor spheroid assay was carried out as described in Fig. 3. Representative images (A) and dose response curves (B) are shown for SUM159 treated with staurosporine and GANT61. Representative images (C) and dose response curves (D) are shown for MDA-MB-231 treated with GANT61. A minimum of two independent experiments were carried out. Data represent mean ± SD percent normalized to DMSO vehicle control for spheroid area (Hoechst) and spheroid YOYO-1 comprising a minimum of five replicate wells. Dose response curves were generated using non-linear regression and IC50 values determined in GraphPad Prism 6.
An in vitro IBC tumor emboli model has been recently developed with the resulting emboli shown to have similar size and shape to those from IBC patients [69]. We have recently adapted this model to develop a high content assay to identify compounds that can inhibit the formation of IBC tumor clusters/embolic structures [56]. Cells are plated in a media containing PEG and rocked to mimic the viscosity and sheer flow dynamics of the lymphatic system with compounds added at the time of seeding. In this current study, the vehicle treated SUM149 emboli showed very tight cell-cell contacts (Fig. 3C) comparable to those observed by Lehman [69]. No tumor emboli were observed with staurosporine (1 µM), a cytotoxic agent. GANT61 (20 µM) significantly inhibited SUM149 tumor emboli (Fig. 3C). These results indicate that GANT61 is effective in targeting IBC spheroids and tumor clusters in SUM149.
3.6 GANT61 decreased GLI1 positive IBC tumor growth in vivo
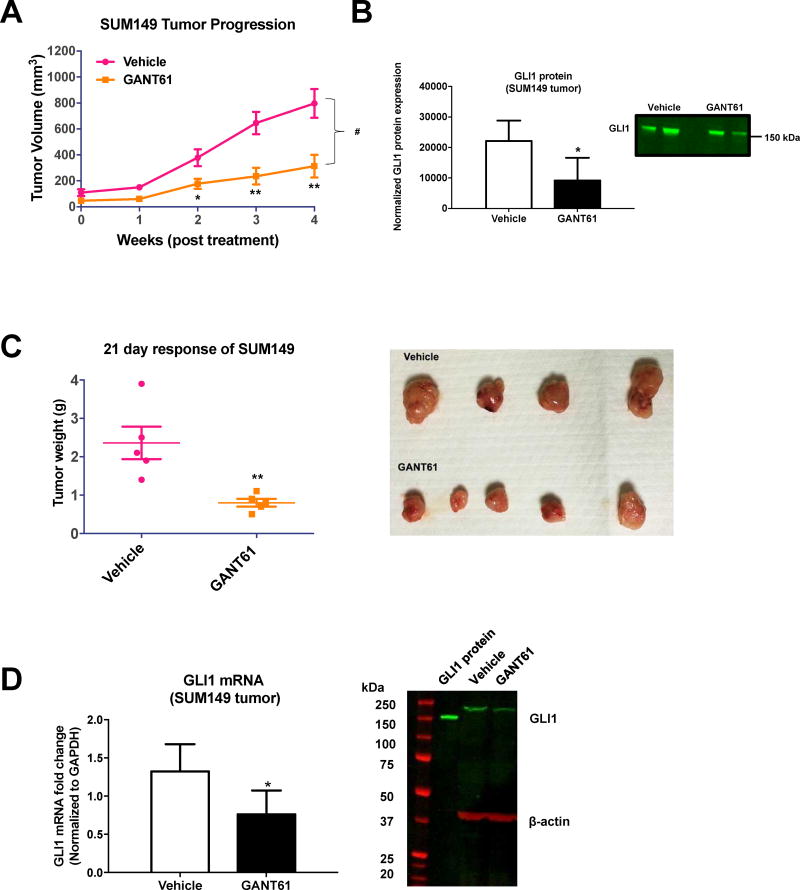
Based on our data that GANT61 significantly reduced spheroid formation and tumor emboli growth in vitro, we next assessed its effect in vivo, in the GLI1-positive SUM149 tumor model. Two SUM149 xenograft studies were carried out using different mouse strains. As the mammary fat pad is the tissue of origin for breast epithelial tumors and provides the proper host environment for tumor growth over time, in both studies, mice were orthotopically injected with cells into the abdominal mammary fat pad. In the first study, the effect of GANT61 on SUM149 tumor growth was assessed in nude NU/J mice. For the GANT61 (DC Chemicals) used for this study, compound structure was confirmed by NMR (Supplementary Fig. 4A) and potency confirmed in vitro using the C3H10T1/2 Hh functional assay (IC50 = 11 µM; Supplementary Fig. 4B). After tumors had reached 125 mm3, GANT61 was administered (50 mg/kg) i.p. 3 days per week. This 50 mg/kg dose was chosen as it has been utilized in the majority of studies testing GANT61 in vivo [46, 70–73]. GANT61 significantly reduced tumor growth (Fig. 5A). After four weeks of treatment, the average tumor volumes for the vehicle and GANT61 groups were 796.1 mm3 and 313.5 mm3 respectively. Statistically significant differences in tumor volumes were seen between vehicle and GANT61 treatment beginning at 2 weeks (Fig. 5A), with a significant inhibition of tumor growth observed (#p <0.025 for GANT61 compared to vehicle). GANT61 decreased the expression of GLI1 protein in the tumors (*p <0.05 for GANT61 compared to vehicle, Fig. 5B). No adverse side effects of GANT61 were observed with the mice gaining weight during the study. In the same nude NU/J mice mouse model, we also assessed the effect of GANT61 in MDA-MB-231 and SUM159 xenografts and observed a statistically significant effect of GANT61 (50 mg/kg) on reducing SUM159 tumor growth ((#p = 0.0012 for GANT61 compared to vehicle) (Supplementary Fig. 4D) but not on MDA-MB-231 tumors (Supplementary Fig. 4C).
Fig. 5.
GANT61 treatment inhibits TN-IBC SUM149-derived xenograft tumor growth. (A) Nude NU/J mic were injected orthotopically with SUM149 cells into the mammary fat pad. Once tumors reached 5 × 5 mm (6 weeks) they were treated with vehicle or GANT61 (50 mg/kg i.p. 3× per week for n = five animals). Tumor response was assessed by weekly caliper measurements. Tumor volumes (Mean ± SEM) are shown for each time point. Statistical significance relative to respective control *P<0.05, **P<0.005, ***P<0.001 (Student t-test). Comparisons in tumor growth were also made by linear regression in GraphPad Prism 6.0. Statistical significance relative to control #P<0.05. (B) Quantification of the study shown in (A) for GLI1 protein expression by immunoblot analysis. Mean band intensity normalized to total protein (± SD) for vehicle compared to GANT61 treated tumor lysates (left panel). *P<0.05 (Student t-test). Representative immunoblot for GLI1 protein expression from three independent analyses of tumor cell lysates (B, right panel). (C) NOD SCID mice were injected orthotopically with SUM149 cells into the mammary fat pad. Once tumors were palpable, mice were treated with vehicle or GANT61 in vehicle (50 mg/kg i.p. once per day for n = five animals). After 21 days, tumors were excised and weighed. Weights are shown as Mean ± SEM. *P<0.05, **P<0.005, ***P<0.001 (Student t-test). (D) Quantification from study shown in (C) of GLI1 mRNA in the SUM149-derived tumors (left panel). *P<0.05 (Student t-test). Representative immunoblot from three independent experiments of tumor tissue lysates for GLI1 protein expression (D, right panel). Recombinant human GLI1 protein (GLI1 protein) was included as a positive control for the Western.
In a second study at a different animal facility, SUM149 cells were injected orthotopically into the mammary fat pad of NOD SCID mice, and after tumors had reached a palpable size, GANT61 was administered (50 mg/kg) i.p. three days per week (Fig. 5C). After a 21-day treatment, GANT61 again significantly reduced tumor growth in this TN-IBC model (Fig 5C, *p = 0.0072). In addition, GANT61 significantly decreased GLI1 mRNA expression at 21 days after GANT61 dosing (Fig. 5D, p = 0.04), a result consistent with our in vitro qPCR findings (Fig 1E). Further, GANT61 treatment dampened GLI1 protein levels in tumors (19 ± 5% decrease in GLI1 protein compared to vehicle) (Fig. 5D).
Hence, in two in vivo studies for IBC, GANT61 was able to reduce tumor volume and slow growth over time but not fully eradicate the tumors.
4. Discussion
There is overwhelming evidence that GLI1, the downstream effector of Hh signaling, has a critical role in the progression and invasiveness of multiple cancers [21–25], including basal-like breast cancers [28]. We have previously reported that the highly invasive basal-like TN-IBC cell line SUM149 had elevated GLI1 expression and that siRNA mediated knockdown of GLI1 reduced cell proliferation and motility [29]. The current study describes how small molecule antagonists targeting GLI were identified for testing for in vivo activity through the use of a series of 2D and 3D assays with varying phenotypic endpoints, a number of which we recently developed as models for IBC [54, 56, 74]. First, herein, we demonstrated that TN-IBC (SUM149) and TNBC (SUM159 and MDA-MB-231) cell lines expressed elevated levels of GLI1 and/or GLI2 compared to the HER2+ cell lines tested. Several of the GLI antagonists had anti-proliferative activity in the GLI+ but not GLI- cell lines and were able to downregulate the expression of GLI1 and associated target genes. Second, in assays that are proposed to be more predictive of in vivo efficacy [66, 67, 75–77] including colony forming, 3D spheroid and tumor emboli models, we showed that a subset of GLI antagonists had significant activity in reducing colony formation and spheroid growth. Lastly, we discovered that one of the subset, GANT61, previously shown to be a direct binder of GLI1 [78, 79], had significant in vivo efficacy in orthotropic xenograft models for IBC. Most importantly, this is the first demonstration of activity of a hedgehog pathway inhibitor in a model of inflammatory breast cancer.
In this study, we first wanted to assess the expression of Hh pathway components in breast cancer cell lines of differing subtypes, as some variation in findings for relative GLI levels had been previously reported [25, 26, 29, 59, 80–82]. For example, in the TNBC MDA-MB-231 cells, both high [25, 82] and low expression [59] of GLI1 relative to HMEC has been observed. The latter study [59] along with recent studies [83, 84] observed high GLI2/low GLI1 in MDA-MB-231, findings consistent with our observation of low GLI1 and high GLI2 in these cells. Further, microarray data presented in heat map format indicated that GLI1 mRNA expression was higher in SUM149 compared to MDA-MB-231 [59]. We have consistently observed high levels of GLI1 in SUM149 (~30–40 fold relative to HMEC) and we observed very comparable levels of GLI1 and GLI2 for SUM149 and SUM159. A recent study showed GLI1 protein levels to be comparable in SUM149 and SUM159 cells and low in MDA-MB-231 [85].
As there is growing evidence that GLI1 activation in a number of cancers is primarily through non-canonical mechanisms [40, 41], including for breast cancer [29, 81], we decided to assess the effects of inhibitors that act downstream of the Hh pathway [60]. Indeed, the majority of breast cancer cell lines do not appear to possess intact canonical Hh signaling [81]. A number of small molecules that act at the level of the GLI transcription factors have been identified with varying mechanisms and understanding of action and so we assembled a GLI antagonist panel consisting of GANT58 and GANT61 [46], HPIs 1–4 [45], and JK184 [47] that were previously identified from GLI-reporter cell-based screens. In a Hh functional cell based assay, C3H10T1/2, that we and others have used to assess the activity of Hh ligands and Hh inhibitors [52, 53], we found that these GLI antagonists showed varying effects at blocking canonical Hh signaling with JK184 being the most potent. JK184 was previously shown to reduce GLI1 mRNA expression in this cell-based assay [47]. The HPIs have been shown to have inhibitory effects on Hh pathway activity in C3H10T1/2 cells [45], and in the same assay we observed comparable activity with the following IC50 values for HPI-1 (1 µM), HPI-3 (0.7 µM), and HPI-4 (10 µM).
We demonstrated that the cell lines possessing high GLI1/lowGLI2 (TN-IBC SUM149; TNBC non-IBC SUM159) or high GLI2/low GLI1 (MDA-MB-231) expression, were sensitive to a subset of these GLI antagonists (GANT61, HPI-1, HPI-4 and JK184) when tested in a 2D proliferation assay. While GANT58 and GANT61 have been shown to decrease cell growth in numerous types of cancer cell lines, GANT61 is typically the more potent [43, 46, 60]. Likewise, we observed GANT61 to be more effective than GANT58 on reducing proliferation of both SUM149 and MDA-MB-231 cells. JK184 treatment of Claudin-low breast cancer cell lines that express elevated GLI2 compared to GLI1 has been shown to result in increased cell death [59].
To further explore the effects of the GLI antagonists, we assessed their effects on transcriptional targets. We found that GANT61, HPI-1 and JK184 all reduced GLI1 mRNA expression in SUM149 cells, with GANT61 reducing both GLI1 and GLI2 levels in SUM159. JK184, a Hh pathway inhibitor [47] and microtubule destablizer [86] was able to reduce GLI1 in all three cell lines. Both GANT61 and JK184 reduced PTCH1 expression in SUM159 cells. GANT61 has been shown to bind directly to zinc fingers 2 and 3 of GLI1 thereby blocking GLI1-mediated transcriptional activity [78]. In addition, GANT61 has been shown to downregulate GLI1 and GLI2 levels [46] and to block binding of GLI1 and GLI2 to GLI target gene promoters [87]. The effect of GANT61 on GLI2 may be indirect as a consequence of GLI1 inhibition and requires further investigation.
Hh-GLI signaling promotes G1 cell cycle progression which enhances the expression of cyclins (cyclin D1, cyclin D2, and cyclin E) [64]. Bcl-2 (B-cell lymphoma 2), an anti-apoptosis oncogene, has also been shown to be regulated by GLI activation in cancer [63]. Our findings that GANT61 in SUM149 reduced genes such as cyclin E1 that promote cell cycle progression is consistent with observations of GANT61’s effect in other cancers including colon [87, 88] and rhabdomyosarcoma [72]. We also demonstrated that GANT61 decreased CCND1 and CCNE1 in SUM159. Inhibition by GANT61 of GLI1 in SUM149 and GLI1/2 in SUM159 leads to reduced expression of these GLI targets. In other models, GANT61 inhibition of GLI transcription has been shown to inhibit several cell activities including apoptosis (Bcl2) and cell cycle progression (cyclin D1 & E1) [43], and we are currently investigating further GANT61’s mechanism in these IBC models. Our findings here add to the idea that GANT61 targeting of GLI1 impairs cell cycle progression leading to reduced tumor growth.
In addition to 2D cell proliferation assays, we utilized assay models that are purported to be capable of predicting effects on tumor growth in vivo [76, 89], including colony formation, 3D spheroid and tumor emboli assays. GANT61 significantly decreased the proliferative potential of SUM149, MDA-MB-231 and SUM159 cells as assessed by colony formation assay. The 3D spheroid assays are thought to more accurately reflect cell interactions within tumors as they grow, whereas cells grown in 2D on plastic lack these cell connections and 3D organization. In the 3D spheroid model, we observed that GANT61 at low micromolar concentrations altered spheroid formation, decreased spheroid cross-sectional area and at higher concentrations promoted spheroid fragmentation for SUM149 and SUM159 cells. For SUM149, GANT61 was ~3–4 more potent in 2D culture compared to the 3D format which may reflect differences in spheroid biology. Differential drug effects in 2D versus 3D culture have been reported with cells in tight spheroids being more resistant to drug effects [68, 90, 91] due to increases in cell survival pathways [68, 90], transporter proteins [90] and potential rewiring of signaling pathways [91]. The effect on MDA-MB-231 was difficult to assess in our assay format as these cells formed cell aggregates rather than tight spheroids although GANT61 has been reported to decrease mammosphere numbers of MDA-MB-231 [83]. While 3D spheroid formation is characteristic of most breast cancer cells, the formation of tumor cell emboli are a distinct feature of IBC cells with dermal lymphatic emboli promoting the rapid spread of IBC throughout the body [69, 92, 93]. In the tumor emboli model, when tested at its IC50 concentration, GANT61 was effective at disrupting tumor emboli formation.
We demonstrated that GANT61 was also effective in vivo in reducing tumor growth in orthotopic xenograft models for IBC. Further, we showed that GANT61 treatment reduced GLI1 expression in the tumors, a finding also seen in vivo with GANT61 treatment in rhabdomyosarcoma [72], prostate [70] and pancreatic [94] xenograft models. Our findings demonstrating the efficacy of GANT61 in in vitro and in vivo IBC models also builds upon recent studies showing GANT61 to be effective in breast cancer xenografts when combined with a second Hh pathway inhibitor itraconazole [95], and when tested in a mouse mammary cancer xenograft model [84]. We also found that GANT61 significantly reduced tumor growth in SUM159 but not in MDA-MB-231 xenografts (Supplementary Fig. 4C) suggesting GANT61 has increased efficacy in GLI1+ compared to GLI2+ in vivo models. GANT61 has shown efficacy in vivo in a number of different xenograft models including prostate [46, 70], pancreatic [94], lung [71], neuroblastoma [73], and rhabdomyosarcoma [72]. JK184 demonstrated toxicity at relatively low concentrations in our cell models and so prior to testing it in xenograft models we are assessing its maximum tolerated dose to identify any potential toxicity effects in vivo (Darr and Williams, unpublished data). The GLI antagonists used in our study have differing mechanisms of action and so it would be of interest to assess whether combinations would have an additive or synergistic effect.
To our knowledge, a comprehensive profiling of the GLI antagonists has not been previously undertaken in a cancer model. Ridzewski et al compared four SMO inhibitors in rhabdomyosarcoma cell lines and observed cell line dependent effects [96]. In some cases a paradoxical increase in GLI1 levels by SMO inhibitors was observed in some cell lines [96], which has also been seen with cyclopamine on breast cancer cell lines [81]. Benvenuto et al [84] compared a SMO inhibitor with GANT61 in breast cancer cell models and found that GANT61 was more effective than the SMO inhibitor in reducing cell survival and Hh pathway activity, presumably as we (herein) and others [81] have shown that the majority of breast cancer cell lines possess very low levels of SMO, with GLI1 typically activated by Hh/SMO-independent non-canonical mechanisms as we previously found for IBC [29].
In conclusion, we propose that GANT61 was effective in IBC models as SUM149 has high GLI1 and GANT61 is GLI1-directed. Our findings indicate that GANT61 inhibition of GLI1 in IBC results in reduced cell proliferation and impaired cell cycle progression leading to reduced tumor growth. Hence, our studies highlight the importance of targeting at the level of GLI and add more evidence to the potential of GLI1-directed binders such as GANT61 [78] as potential therapeutics for those cancers including IBC where GLI activation is driven primarily by non-canonical mechanisms. Our studies have demonstrated that the use of GLI inhibitors can be a valid therapeutic approach for treating GLI-dependent TNBC cancers such as IBC for which there are few therapeutic options.
Supplementary Material
Supplementary Fig. 1. Structures of GLI antagonists and HPI-1 effect on gene expression in SUM149 cells. (A) Structures of GLI antagonists. (B) qRT-PCR comparative expression of Hh-GLI downstream targets in SUM149 cells treated with vehicle (0.1% DMSO) or HPI-1 (10 µM) for 72 h. Mean ± SD shown. One-way ANOVA with Tukey’s post-test (*p<0.05, **p<0.005, ***p<0.0005 and ****p<0.0001). Significantly different from respective controls.
Supplementary Fig. 2. Effect of GLI antagonists GANT61 and HPI-1 on SUM149 and MDA-MB-231 cell motility. For the wound healing/scratch assay, cells were seeded in 96-well plates, incubated overnight and the wound-maker module used to simultaneously create a vertical wound/scratch in all wells, cells were washed with PBS and DMSO, GANT61, HPI-1 and cytochalasin D (Cyto D) added to the plate (n = 8). Cell motility was measured using the IncuCyte ZOOM System and images taken at 0, 4, 12 18, 24 h with 10x objective. (A) Representative images from wound healing assay for SUM149 and MDA-MB-231. Initial wound shown as pink mask with cell occupied area shown with purple mask. (B) Percent relative wound density (RWD) was calculated as described in methods using measures of wound width and wound confluence for compound effects on SUM149 and MDA-MB-231motility. Statistical significance relative to DMSO control *P<0.05, (Student t-test).
Supplementary Fig. 3. Data for spheroid area and spheroid YOYO-1 raw values for GANT61 and staurosporine treatment on SUM149, MDA-MB-231 and SUM159 tumor spheroids. Shown is data from Figures 3 and 4 replotted using the raw values for spheroid area and spheroid YOYO-1 for DMSO, GANT61 and staurosporine treatment of SUM149 (A), SUM159 (B) and MDA-MB-231 (C, GANT61 only) spheroids. Statistical significance relative to DMSO control *P<0.05, **P<0.005, ***P<0.001 (Student t-test). (D) Representative images and dose response curves are shown for MDA-MB-231 spheroids treated with staurosporine. A minimum of two independent experiments were carried out. Dose response curves were generated using nonlinear regression and IC50 values determined in GraphPad Prism 6.
Supplementary Fig. 4. GANT61 structural confirmation, testing in C3H10T1/2 assay and MDA-MB-231 and SUM159 xenograft models. (A) NMR spectra of GANT61 used for in vivo studies determined using 500 MHz Varian instrument. (B) Testing of GANT61 used for in vivo study for inhibition in C3H10T1/2 Hh functional assay. C3H10T12 cells were stimulated with Shh protein (2 µg/ml) in the presence of GANT61 (0.001 – 10 µM) and alkaline phosphatase measured at 5 d. Dose response curve were generated using non-linear regression and IC50 values determined in GraphPad Prism 6. Nude NU/J mic were injected orthotopically with MDA-MB-231 (C) or SUM 159 (D) cells into the mammary fat pad. Once tumors reached 5×5 mm (6 weeks) they were treated with vehicle or GANT61 (50 mg/kg i.p. 3x per week for n = six animals). Tumor response was assessed by weekly caliper measurements. Tumor volume (percent change over 21 days) and tumor progression (tumor volume over 28 days, mean ± SEM) are shown for MDA-MB-231 and SUM159 models respectively. Statistical significance relative to respective control; ns = not significant *P<0.05, **P<0.005, ***P<0.001 (Student t-test). Comparisons in tumor growth were also made by linear regression in GraphPad Prism 6.0. Statistical significance relative to control #P<0.05.
Highlights.
TN-IBC (SUM149) and TNBC (SUM159) cell lines expressed elevated levels of GLI1.
GANT61 and JK184 inhibited proliferation and colony formation of SUM149/159 and MDA-MB-231.
GANT61 and JK184 reduced expression of GLI1 and genes involved in cell cycle progression.
In 3D models of SUM149 and SUM159, GANT61 reduced tumor spheroid size and tumor emboli formation.
GANT61 had significant in vivo efficacy in orthotropic xenograft models for IBC.
Acknowledgments
This study was supported in part by Department of Defense award W81WXH-13-1-0141 (KPW and GRD), NIH awards SC2CA137844 (KPW), U54CA156735 (KPW and DMD), P20CA202924 (KPW and GRD), T32CA009111 (SJS), and Komen Graduate Training in Disparities Research award GTDR16377604 (KPW, HOO and JMF), School of Medicine Dean’s Office supported Duke Consortium for IBC funds (GRD), with additional funding from the Golden LEAF Foundation and the BIOIMPACT Initiative of the State of North Carolina. Co-senior authors (KPW and GRD).
The authors would like to thank Dr. Ned Sharpless for preliminary discussions; Chris Laudeman, Zainab Thomas and Jennai Pettis for technical assistance; David (Sam) Lamson and Donna Crabtree for their critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- 1.O'Toole SA, Beith JM, Millar EK, West R, McLean A, Cazet A, Swarbrick A, Oakes SR. Therapeutic targets in triple negative breast cancer. J. Clin. Pathol. 2013;66:530–542. doi: 10.1136/jclinpath-2012-201361. [DOI] [PubMed] [Google Scholar]
- 2.Tao Y, Mao J, Zhang Q, Li L. Overexpression of Hedgehog signaling molecules and its involvement in triple-negative breast cancer. Oncol Lett. 2011;2:995–1001. doi: 10.3892/ol.2011.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Habib JG, O'Shaughnessy JA. The hedgehog pathway in triple-negative breast cancer. Cancer Med. 2016;5:2989–3006. doi: 10.1002/cam4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zardavas D, Baselga J, Piccart M. Emerging targeted agents in metastatic breast cancer. Nature Reviews Clinical Oncology. 2013;10:191–210. doi: 10.1038/nrclinonc.2013.29. [DOI] [PubMed] [Google Scholar]
- 5.Takebe N, Miele L, Harris PJ, Jeong W, Bando H, Kahn M, Yang SX, Ivy SP. Targeting Notch Hedgehog and Wnt pathways in cancer stem cells: clinical update. Nature reviews Clinical oncology. 2015;12:445–464. doi: 10.1038/nrclinonc.2015.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kasper M, Regl G, Frischauf AM, Aberger F. GLI transcription factors: mediators of oncogenic Hedgehog signalling. Eur J Cancer. 2006;42:437–445. doi: 10.1016/j.ejca.2005.08.039. [DOI] [PubMed] [Google Scholar]
- 7.Kinzler KW, Ruppert JM, Bigner SH, Vogelstein B. The GLI gene is a member of the Kruppel family of zinc finger proteins. Nature. 1988;332:371–374. doi: 10.1038/332371a0. [DOI] [PubMed] [Google Scholar]
- 8.Pasca di Magliano M, Hebrok M. Hedgehog signalling in cancer formation and maintenance. Nat Rev Cancer. 2003;3:903–911. doi: 10.1038/nrc1229. [DOI] [PubMed] [Google Scholar]
- 9.Zhu H, Lo HW. The Human Glioma-Associated Oncogene Homolog 1 (GLI1) Family of Transcription Factors in Gene Regulation and Diseases. Current Genomics. 2010;11:238. doi: 10.2174/138920210791233108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lauth M, Toftgard R. The Hedgehog pathway as a drug target in cancer therapy. Curr Opin Investig Drugs. 2007;8:457–461. [PubMed] [Google Scholar]
- 11.Mas C, Ruiz i Altaba A. Small molecule modulation of HH-GLI signaling: Current leads trials and tribulations. Biochemical Pharmacology. 2010;80:712–723. doi: 10.1016/j.bcp.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 12.Stanton B, Peng L. Small-molecule modulators of the Sonic Hedgehog signaling pathway. Molecular BioSystems. 2010;6:44–54. doi: 10.1039/b910196a. [DOI] [PubMed] [Google Scholar]
- 13.Scales S, de Sauvage F. Mechanisms of Hedgehog pathway activation in cancer and implications for therapy. Trends in pharmacological sciences. 2008;30:303–312. doi: 10.1016/j.tips.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 14.Burness CB. Sonidegib: First Global Approval. Drugs. 2015;75:1559–1566. doi: 10.1007/s40265-015-0458-y. [DOI] [PubMed] [Google Scholar]
- 15.Yauch R, Dijkgraaf G, Alicke B, Januario T, Ahn C, Holcomb T, Pujara K, Stinson J, Callahan C, Tang T. Smoothened mutation confers resistance to a hedgehog pathway inhibitor in medulloblastoma. Science. 2009;326:572. doi: 10.1126/science.1179386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dijkgraaf GJP, Alicke B, Weinmann L, Januario T, West K, Modrusan Z, Burdick D, Goldsmith R, Robarge K, Sutherlin D. Small molecule inhibition of GDC-0449 refractory smoothened mutants and downstream mechanisms of drug resistance. Cancer Research. 2011;71:435. doi: 10.1158/0008-5472.CAN-10-2876. [DOI] [PubMed] [Google Scholar]
- 17.Metcalfe C, de Sauvage FJ. Hedgehog Fights Back: Mechanisms of Acquired Resistance against Smoothened Antagonists. Cancer Research. 2011;71:5057. doi: 10.1158/0008-5472.CAN-11-0923. [DOI] [PubMed] [Google Scholar]
- 18.Jiang J, Hui C-c. Hedgehog signaling in development and cancer. Developmental Cell. 2008;15:801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fiaschi M, Rozell B, Bergstrom A, Toftgard R. Development of mammary tumors by conditional expression of GLI1. Cancer Res. 2009;69:4810–4817. doi: 10.1158/0008-5472.CAN-08-3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katoh Y, Katoh M. Hedgehog Target Genes: Mechanisms of Carcinogenesis Induced by Aberrant Hedgehog Signaling Activation. Current Molecular Medicine. 2009;9:873–886. doi: 10.2174/156652409789105570. [DOI] [PubMed] [Google Scholar]
- 21.Das S, Harris L, Metge B, Liu S, Riker A, Samant R, Shevde L. The Hedgehog pathway transcription factor GLI1 promotes malignant behavior of cancer cells by up-regulating Osteopontin. Journal of Biological Chemistry. 2009;284:22888–22897. doi: 10.1074/jbc.M109.021949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inaguma S, Kasai K, Ikeda H. GLI1 facilitates the migration and invasion of pancreatic cancer cells through MUC5AC-mediated attenuation of E-cadherin. Oncogene. 2011;30:714–723. doi: 10.1038/onc.2010.459. [DOI] [PubMed] [Google Scholar]
- 23.Mori Y, Okumura T, Tsunoda S, Sakai Y, Shimada Y. Gli-1 expression is associated with lymph node metastasis and tumor progression in esophageal squamous cell carcinoma. Oncology. 2006;70:378–389. doi: 10.1159/000098111. [DOI] [PubMed] [Google Scholar]
- 24.Nagai S, Nakamura M, Yanai K, Wada J, Akiyoshi T, Nakashima H, Ohuchida K, Sato N, Tanaka M, Katano M. Gli1 contributes to the invasiveness of pancreatic cancer through matrix metalloproteinase 9 activation. Cancer Science. 2008;99:1377–1384. doi: 10.1111/j.1349-7006.2008.00822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.ten Haaf A, Bektas N, von Serenyi S, Losen I, Arweiler EC, Hartmann A, Knuchel R, Dahl E. Expression of the glioma-associated oncogene homolog (GLI) 1 in human breast cancer is associated with unfavourable overall survival. Bmc Cancer. 2009;9 doi: 10.1186/1471-2407-9-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu L, Kwon Y, Frolova N, Steg A, Yuan K, Johnson M, Grizzle W, Desmond R, Frost A. Gli1 promotes cell survival and is predictive of a poor outcome in ER -negative breast cancer. Breast cancer research and treatment. 2010;123:59–71. doi: 10.1007/s10549-009-0617-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang K, Pan L, Che X, Cui D, Li C. Gli1 inhibition induces cell-cycle arrest and enhanced apoptosis in brain glioma cell lines. Journal of neuro-oncology. 2010;98:319–327. doi: 10.1007/s11060-009-0082-3. [DOI] [PubMed] [Google Scholar]
- 28.Li Y, Yang W, Yang Q, Zhou S. Nuclear localization of GLI1 and elevated expression of FOXC2 in breast cancer is associated with the basal-like phenotype. Histology and Histopathology. 2012;27:475. doi: 10.14670/HH-27.475. [DOI] [PubMed] [Google Scholar]
- 29.Thomas ZI, Gibson W, Sexton JZ, Aird KM, Ingram SM, Aldrich A, Lyerly HK, Devi GR, Williams KP. Targeting GLI1 expression in human inflammatory breast cancer cells enhances apoptosis and attenuates migration. British Journal of Cancer. 2011;104:1575–1586. doi: 10.1038/bjc.2011.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Houchens NW, Merajver SD. Molecular determinants of the inflammatory breast cancer phenotype. Oncology. 2008;22:1. [PubMed] [Google Scholar]
- 31.Masuda H, Baggerly KA, Wang Y, Iwamoto T, Brewer T, Pusztai L, Kai K, Kogawa T, Finetti P, Birnbaum D, Dirix L, Woodward WA, Reuben JM, Krishnamurthy S, Symmans W, Van Laere SJ, Bertucci F, Hortobagyi GN, Ueno NT. Comparison of molecular subtype distribution in triple-negative inflammatory and non-inflammatory breast cancers. Breast Cancer Res. 2013;15:R112. doi: 10.1186/bcr3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charafe-Jauffret E, Tarpin C, Viens P, Bertucci F. Defining the molecular biology of inflammatory breast cancer. Seminars in oncology. 2008;35:41–50. doi: 10.1053/j.seminoncol.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 33.Robertson FM, Bondy M, Yang W, Yamauchi H, Wiggins S, Kamrudin S, Krishnamurthy S, Le-Petross H, Bidaut L, Player AN, Barsky SH, Woodward WA, Buchholz T, Lucci A, Ueno NT, Cristofanilli M. Inflammatory breast cancer: the disease the biology the treatment. CA Cancer J Clin. 2010;60:351–375. doi: 10.3322/caac.20082. [DOI] [PubMed] [Google Scholar]
- 34.Van Laere S, Limame R, Van Marck E, Vermeulen P, Dirix L. Is there a role for mammary stem cells in inflammatory breast carcinoma? Cancer. 2010;116:2794–2805. doi: 10.1002/cncr.25180. [DOI] [PubMed] [Google Scholar]
- 35.Aird KM, Ghanayem RB, Peplinski S, Lyerly HK, Devi GR. X-linked inhibitor of apoptosis protein inhibits apoptosis in inflammatory breast cancer cells with acquired resistance to an ErbB1/2 tyrosine kinase inhibitor. Mol Cancer Ther. 2010;9:1432–1442. doi: 10.1158/1535-7163.MCT-10-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dong HM, Liu G, Hou YF, Wu J, Lu JS, Luo JM, Shen ZZ, Shao ZM. Dominant-negative E-cadherin inhibits the invasiveness of inflammatory breast cancer cells in vitro. J Cancer Res Clin Oncol. 2007;133:83–92. doi: 10.1007/s00432-006-0140-6. [DOI] [PubMed] [Google Scholar]
- 37.Forozan F, Veldman R, Ammerman CA, Parsa NZ, Kallioniemi A, Kallioniemi O-P, Ethier SP. Molecular cytogenetic analysis of 11 new breast cancer cell lines. Br J Cancer. 1999;81:1328–1334. doi: 10.1038/sj.bjc.6695007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoffmeyer MR, Wall KM, Dharmawardhane SF. In vitro analysis of the invasive phenotype of SUM 149 an inflammatory breast cancer cell line. Cancer Cell Int. 2005;5:11. doi: 10.1186/1475-2867-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu M, Wu Z, Rosenthal D, Rhee E, Merajver S. Characterization of the roles of RHOC RHOA GTPases in invasion, motilityand matrix adhesion in inflammatory and aggressive breast cancers. Cancer. 2010;116:2768–2782. doi: 10.1002/cncr.25181. [DOI] [PubMed] [Google Scholar]
- 40.Jenkins D. Hedgehog signalling: emerging evidence for non-canonical pathways. Cellular signalling. 2009;21:1023–1034. doi: 10.1016/j.cellsig.2009.01.033. [DOI] [PubMed] [Google Scholar]
- 41.Lauth M, Toftgard R. Non-canonical activation of GLI transcription factors: implications for targeted anti-cancer therapy. Cell Cycle. 2007;6:2458–2463. doi: 10.4161/cc.6.20.4808. [DOI] [PubMed] [Google Scholar]
- 42.Di Magno L, Coni S, Di Marcotullio L, Canettieri G. Digging a hole under Hedgehog: downstream inhibition as an emerging anticancer strategy. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer. 2015;1856:62–72. doi: 10.1016/j.bbcan.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 43.Gonnissen A, Isebaert S, Haustermans K. Targeting the Hedgehog signaling pathway in cancer: beyond Smoothened. Oncotarget. 2015;6:13899–13913. doi: 10.18632/oncotarget.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Infante P, Mori M, Alfonsi R, Ghirga F, Aiello F, Toscano S, Ingallina C, Siler M, Cucchi D, Po A, Miele E, D'Amico D, Canettieri G, De Smaele E, Ferretti E, Screpanti I, Barretta GU, Botta M, Botta B, Gulino A, Di Marcotullio L. Gli1/DNA interaction is a druggable target for Hedgehog-dependent tumors. Embo Journal. 2015;34:200–217. doi: 10.15252/embj.201489213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hyman JM, Firestone AJ, Heine VM, Zhao Y, Ocasio CA, Han K, Sun M, Rack PG, Sinha S, Wu JJ, Solow-Cordero DE, Jiang J, Rowitch DH, Chen JK. Small-molecule inhibitors reveal multiple strategies for Hedgehog pathway blockade. Proc Natl Acad Sci U S A. 2009;106:14132–14137. doi: 10.1073/pnas.0907134106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lauth M, Bergstrom A, Shimokawa T, Toftgard R. Inhibition of GLI-mediated transcription and tumor cell growth by small-molecule antagonists. Proc Natl Acad Sci U S A. 2007;104:8455–8460. doi: 10.1073/pnas.0609699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee J, Wu X, Pasca di Magliano M, Peters EC, Wang Y, Hong J, Hebrok M, Ding S, Cho CY, Schultz PG. A small-molecule antagonist of the hedgehog signaling pathway. Chembiochem. 2007;8:1916–1919. doi: 10.1002/cbic.200700403. [DOI] [PubMed] [Google Scholar]
- 48.Mahindroo N, Connelly M, Punchihewa C, Kimura H, Smeltzer M, Wu S, Fujii N. Structure Activity Relationships and Cancer-Cell Selective Toxicity of Novel Inhibitors of Glioma-Associated Oncogene Homologue 1 (Gli1) Mediated Transcription. Journal of Medicinal Chemistry. 2009;52:361–372. doi: 10.1021/jm900106f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mahindroo N, Punchihewa C, Fujii N. Hedgehog-Gli Signaling Pathway Inhibitors as Anticancer Agents. Journal of Medicinal Chemistry. 2009;52:3829–3845. doi: 10.1021/jm801420y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu Y, Chenna V, Hu C, Sun HX, Khan M, Bai H, Yang XR, Zhu QF, Sun YF, Maitra A. Polymeric Nanoparticle-Encapsulated Hedgehog Pathway Inhibitor HPI-1 (NanoHHI) Inhibits Systemic Metastases in an Orthotopic Model of Human Hepatocellular Carcinoma. Clinical Cancer Research. 2012;18:1291–1302. doi: 10.1158/1078-0432.CCR-11-0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim J, Aftab BT, Tang JY, Kim D, Lee AH, Rezaee M, Kim J, Chen B, King EM, Borodovsky A. Itraconazole and arsenic trioxide inhibit Hedgehog pathway activation and tumor growth associated with acquired resistance to smoothened antagonists. Cancer cell. 2013;23:23–34. doi: 10.1016/j.ccr.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.House AJ, Daye LR, Tarpley M, Addo K, Lamson DS, Parker MK, Bealer WE, Williams KP. Design and characterization of a photo-activatable hedgehog probe that mimics the natural lipidated form. Archives of Biochemistry and Biophysics. 2015;567:66–74. doi: 10.1016/j.abb.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williams KP, Rayhorn P, Chi-Rosso G, Garber EA, Strauch KL, Horan GS, Reilly JO, Baker DP, Taylor FR, Koteliansky V, Pepinsky RB. Functional antagonists of sonic hedgehog reveal the importance of the N terminus for activity. J Cell Sci. 1999;112(Pt 23):4405–4414. doi: 10.1242/jcs.112.23.4405. [DOI] [PubMed] [Google Scholar]
- 54.Sauer SJ, Tarpley M, Shah I, Save AV, Lyerly HK, Patierno SR, Williams KP, Devi GR. Bisphenol Aactivates EGFR ERK promoting proliferation tumor spheroid formation and resistance to EGFR pathway inhibition in estrogen receptor-negative inflammatory breast cancer cells. Carcinogenesis. 2017;38:252–260. doi: 10.1093/carcin/bgx003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abraham VC, Towne DL, Waring JF, Warrior U, Burns DJ. Application of a high-content multiparameter cytotoxicity assay to prioritize compounds based on toxicity potential in humans. J Biomol Screen. 2008;13:527–537. doi: 10.1177/1087057108318428. [DOI] [PubMed] [Google Scholar]
- 56.Arora J, Sauer SJ, Tarpley M, Vermeulen P, Rypens C, Van Laere S, Williams KP, Devi GR, Dewhirst MW. Inflammatory breast cancer tumor emboli express high levels of anti-apoptotic proteins: use of a quantitative high content and high-throughput 3D IBC spheroid assay to identify targeting strategies. Oncotarget. 2017;5 doi: 10.18632/oncotarget.15667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aird KM, Allensworth JL, Batinic-Haberle I, Lyerly HK, Dewhirst MW, Devi GR. ErbB1/2 tyrosine kinase inhibitor mediates oxidative stress-induced apoptosis in inflammatory breast cancer cells. Breast Cancer Research and Treatment. 2012;132:109–119. doi: 10.1007/s10549-011-1568-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. The Journal of clinical investigation. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Colavito SA, Zou MR, Yan Q, Nguyen DX, Stern DF. Significance of glioma-associated oncogene homolog 1 (GLI1) expression in claudin-low breast cancer and crosstalk with the nuclear factor kappa-light-chain-enhancer of activated B cells (NF inverted question markB) pathway. Breast Cancer Res. 2014;16:444. doi: 10.1186/s13058-014-0444-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rimkus TK, Carpenter RL, Qasem S, Chan M, Lo HW. Targeting the Sonic Hedgehog Signaling Pathway: Review of Smoothened and GLI Inhibitors. Cancers (Basel) 2016;8 doi: 10.3390/cancers8020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang L, Liu Z, Gambardella L, Delacour A, Shapiro R, Yang J, Sizing I, Rayhorn P, Garber E, Benjamin C. Conditional disruption of hedgehog signaling pathway defines its critical role in hair development and regeneration. Journal of Investigative Dermatology. 2000;114:901–908. doi: 10.1046/j.1523-1747.2000.00951.x. [DOI] [PubMed] [Google Scholar]
- 62.Chen JK, Taipale J, Cooper MK, Beachy PA. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 2002;16:2743–2748. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bigelow RL, Chari NS, Unden AB, Spurgers KB, Lee S, Roop DR, Toftgard R, McDonnell TJ. Transcriptional regulation of bcl-2 mediated by the sonic hedgehog signaling pathway through gli-1. J Biol Chem. 2004;279:1197–1205. doi: 10.1074/jbc.M310589200. [DOI] [PubMed] [Google Scholar]
- 64.Kenney AM, Rowitch DH. Sonic hedgehog promotes G1 cyclin expression and sustained cell cycle progression in mammalian neuronal precursors. Molecular and cellular biology. 2000;20:9055–9067. doi: 10.1128/mcb.20.23.9055-9067.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hayot C, Debeir O, Van Ham P, Van Damme M, Kiss R, Decaestecker C. Characterization of the activities of actin-affecting drugs on tumor cell migration. Toxicol Appl Pharmacol. 2006;211:30–40. doi: 10.1016/j.taap.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 66.Xu X, Farach-Carson MC, Jia X. Three-dimensional in vitro tumor models for cancer research and drug evaluation. Biotechnology advances. 2014;32:1256–1268. doi: 10.1016/j.biotechadv.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ryan S-L, Baird A-M, Vaz G, Urquhart AJ, Senge h, Richard DJ, O'Byrne KJ, Davies AM. Drug discovery approaches utilizing three-dimensional cell culture. Assay and drug development technologies. 2016;14:19–28. doi: 10.1089/adt.2015.670. [DOI] [PubMed] [Google Scholar]
- 68.Imamura Y, Mukohara T, Shimono Y, Funakoshi Y, Chayahara N, Toyoda M, Kiyota N, Takao S, Kono S, Nakatsura T. Comparison of 2D–and 3D–culture models as drug-testing platforms in breast cancer. Oncology reports. 2015;33:1837–1843. doi: 10.3892/or.2015.3767. [DOI] [PubMed] [Google Scholar]
- 69.Lehman HL, Dashner EJ, Lucey M, Vermeulen P, Dirix L, Van Laere S, van Golen KL. Modeling and characterization of inflammatory breast cancer emboli grown in vitro. Int J Cancer. 2013;132:2283–2294. doi: 10.1002/ijc.27928. [DOI] [PubMed] [Google Scholar]
- 70.Gonnissen A, Isebaert S, McKee CM, Dok R, Haustermans K, Muschel RJ. The hedgehog inhibitor GANT61 sensitizes prostate cancer cells to ionizing radiation both in vitro and in vivo. Oncotarget. 2016;7:84286–84298. doi: 10.18632/oncotarget.12483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang L, Walter V, Hayes DN, Onaitis M. Hedgehog-GLI signaling inhibition suppresses tumor growth in squamous lung cancer. Clin Cancer Res. 2014;20:1566–1575. doi: 10.1158/1078-0432.CCR-13-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Srivastava RK, Kaylani SZ, Edrees N, Li C, Talwelkar SS, Xu J, Palle K, Pressey JG, Athar M. GLI inhibitor GANT-61 diminishes embryonal and alveolar rhabdomyosarcoma growth by inhibiting Shh/AKT-mTOR axis. Oncotarget. 2014;5:12151. doi: 10.18632/oncotarget.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wickstrom M, Dyberg C, Shimokawa T, Milosevic J, Baryawno N, Fuskevag OM, Larsson R, Kogner P, Zaphiropoulos PG, Johnsen JI. Targeting the hedgehog signal transduction pathway at the level of GLI inhibits neuroblastoma cell growth in vitro and in vivo. Int J Cancer. 2013;132:1516–1524. doi: 10.1002/ijc.27820. [DOI] [PubMed] [Google Scholar]
- 74.Williams KP, Allensworth JL, Ingram SM, Smith GR, Aldrich AJ, Sexton JZ, Devi GR. Quantitative high-throughput efficacy profiling of approved oncology drugs in inflammatory breast cancer models of acquired drug resistance and re-sensitization. Cancer Lett. 2013;337:77–89. doi: 10.1016/j.canlet.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Benton G, DeGray G, Kleinman HK, George J, Arnaoutova I. In vitro microtumors provide a physiologically predictive tool for breast cancer therapeutic screening. PloS one. 2015;10:e0123312. doi: 10.1371/journal.pone.0123312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Edmondson R, Broglie JJ, Adcock AF, Yang L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev Technol. 2014;12:207–218. doi: 10.1089/adt.2014.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kenny PA, Lee GY, Myers CA, Neve RM, Semeiks JR, Spellman PT, Lorenz K, Lee EH, Barcellos-Hoff MH, Petersen OW. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Molecular oncology. 2007;1:84–96. doi: 10.1016/j.molonc.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Agyeman A, Jha BK, Mazumdar T, Houghton JA. Mode and specificity of binding of the small molecule GANT61 to GLI determines inhibition of GLI-DNA binding. Oncotarget. 2014;5:4492–4503. doi: 10.18632/oncotarget.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang R, Wu J, Ferrandon S, Glowacki KJ, Houghton JA. Targeting GLI by GANT61 involves mechanisms dependent on inhibition of both transcription and DNA licensing. Oncotarget. 2016;7:80190. doi: 10.18632/oncotarget.13376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sun Y, Wang YS, Fan C, Gao P, Wang XW, Wei GW, Wei JM. Estrogen promotes stemness and invasiveness of ER-positive breast cancer cells through Gli1 activation. Molecular Cancer. 2014;13 doi: 10.1186/1476-4598-13-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang X, Harrington N, Moraes RC, Wu M-F, Hilsenbeck SG, Lewis MT. Cyclopamine inhibition of human breast cancer cell growth independent of Smoothened (Smo) Breast cancer research and treatment. 2009;115:505–521. doi: 10.1007/s10549-008-0093-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhao J, Chen G, Cao D, Li Y, Diao F, Cai H, Jin Y, Lu J. Expression of Gli1 correlates with the transition of breast cancer cells to estrogen-independent growth. Breast Cancer Res Treat. 2009;119:39–51. doi: 10.1007/s10549-009-0323-3. [DOI] [PubMed] [Google Scholar]
- 83.Koike Y, Ohta Y, Saitoh W, Yamashita T, Kanomata N, Moriya T, Kurebayashi J. Anti-cell growth and anti-cancer stem cell activities of the non-canonical hedgehog inhibitor GANT61 in triple-negative breast cancer cells. Breast Cancer. 2017;24:683–693. doi: 10.1007/s12282-017-0757-0. [DOI] [PubMed] [Google Scholar]
- 84.Benvenuto M, Masuelli L, De Smaele E, Fantini M, Mattera R, Cucchi D, Bonanno E, Di Stefano E, Frajese GV, Orlandi A, Screpanti I, Gulino A, Modesti A, Bei R. In vitro and in vivo inhibition of breast cancer cell growth by targeting the Hedgehog/GLI pathway with SMO (GDC-0449) or GLI (GANT-61) inhibitors. Oncotarget. 2016;7:9250–9270. doi: 10.18632/oncotarget.7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Di Mauro C, Rosa R, D'Amato V, Ciciola P, Servetto A, Marciano R, Orsini RC, Formisano L, De Falco S, Cicatiello V. Hedgehog signalling pathway orchestrates angiogenesis in triple-negative breast cancers. British Journal of Cancer. 2017;116:1425. doi: 10.1038/bjc.2017.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cupido T, Rack PG, Firestone AJ, Hyman JM, Han K, Sinha S, Ocasio CA, Chen JK. The imidazopyridine derivative JK184 reveals dual roles for microtubules in Hedgehog signaling. Angew Chem Int Ed Engl. 2009;48:2321–2324. doi: 10.1002/anie.200805666. [DOI] [PubMed] [Google Scholar]
- 87.Mazumdar T, Devecchio J, Agyeman A, Shi T, Houghton JA. Blocking Hedgehog survival signaling at the level of the GLI genes induces DNA damage and extensive cell death in human colon carcinoma cells. Cancer Res. 2011;71:5904–5914. doi: 10.1158/0008-5472.CAN-10-4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shi T, Mazumdar T, DeVecchio J, Duan Z-H, Agyeman A, Aziz M, Houghton JA. cDNA microarray gene expression profiling of hedgehog signaling pathway inhibition in human colon cancer cells. PloS one. 2010;5:e13054. doi: 10.1371/journal.pone.0013054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Reid BG, Jerjian T, Patel P, Zhou Q, Yoo BH, Kabos P, Sartorius CA, LaBarbera DV. Live multicellular tumor spheroid models for high-content imaging and screening in cancer drug discovery. Current chemical genomics and translational medicine. 2014;8 doi: 10.2174/2213988501408010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Breslin S, O'Driscoll L. The relevance of using 3D cell cultures in addition to 2D monolayer cultures when evaluating breast cancer drug sensitivity and resistance. Oncotarget. 2016;7:45745. doi: 10.18632/oncotarget.9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Riedl A, Schlederer M, Pudelko K, Stadler M, Walter S, Unterleuthner D, Unger C, Kramer N, Hengstschläger M, Kenner L. Comparison of cancer cells in 2D vs 3D culture reveals differences in AKT-mTOR-S6K signaling and drug responses. J Cell Sci. 2017;130:203–218. doi: 10.1242/jcs.188102. [DOI] [PubMed] [Google Scholar]
- 92.Alpaugh ML, Tomlinson JS, Kasraeian S, Barsky SH. Cooperative role of E-cadherin and sialyl-Lewis X/A-deficient MUC1 in the passive dissemination of tumor emboli in inflammatory breast carcinoma. Oncogene. 2002;21:3631–3643. doi: 10.1038/sj.onc.1205389. [DOI] [PubMed] [Google Scholar]
- 93.Kleer CG, van Golen KL, Merajver SD. Molecular biology of breast cancer metastasis. Inflammatory breast cancer: clinical syndrome and molecular determinants. Breast Cancer Res. 2000;2:423–429. doi: 10.1186/bcr89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fu J, Rodova M, Roy SK, Sharma J, Singh KP, Srivastava RK, Shankar S. GANT-61 inhibits pancreatic cancer stem cell growth in vitro and in NOD/SCID/IL2R gamma null mice xenograft. Cancer Lett. 2013;330:22–32. doi: 10.1016/j.canlet.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang X, Wei S, Zhao Y, Shi C, Liu P, Zhang C, Lei Y, Zhang B, Bai B, Huang Y. Anti-proliferation of breast cancer cells with itraconazole: Hedgehog pathway inhibition induces apoptosis and autophagic cell death. Cancer Letters. 2017;385:128–136. doi: 10.1016/j.canlet.2016.10.034. [DOI] [PubMed] [Google Scholar]
- 96.Ridzewski R, Rettberg D, Dittmann K, Cuvelier N, Fulda S, Hahn H. Hedgehog inhibitors in rhabdomyosarcoma: a comparison of four compounds and responsiveness of four cell lines. Frontiers in oncology. 2015;5 doi: 10.3389/fonc.2015.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. Structures of GLI antagonists and HPI-1 effect on gene expression in SUM149 cells. (A) Structures of GLI antagonists. (B) qRT-PCR comparative expression of Hh-GLI downstream targets in SUM149 cells treated with vehicle (0.1% DMSO) or HPI-1 (10 µM) for 72 h. Mean ± SD shown. One-way ANOVA with Tukey’s post-test (*p<0.05, **p<0.005, ***p<0.0005 and ****p<0.0001). Significantly different from respective controls.
Supplementary Fig. 2. Effect of GLI antagonists GANT61 and HPI-1 on SUM149 and MDA-MB-231 cell motility. For the wound healing/scratch assay, cells were seeded in 96-well plates, incubated overnight and the wound-maker module used to simultaneously create a vertical wound/scratch in all wells, cells were washed with PBS and DMSO, GANT61, HPI-1 and cytochalasin D (Cyto D) added to the plate (n = 8). Cell motility was measured using the IncuCyte ZOOM System and images taken at 0, 4, 12 18, 24 h with 10x objective. (A) Representative images from wound healing assay for SUM149 and MDA-MB-231. Initial wound shown as pink mask with cell occupied area shown with purple mask. (B) Percent relative wound density (RWD) was calculated as described in methods using measures of wound width and wound confluence for compound effects on SUM149 and MDA-MB-231motility. Statistical significance relative to DMSO control *P<0.05, (Student t-test).
Supplementary Fig. 3. Data for spheroid area and spheroid YOYO-1 raw values for GANT61 and staurosporine treatment on SUM149, MDA-MB-231 and SUM159 tumor spheroids. Shown is data from Figures 3 and 4 replotted using the raw values for spheroid area and spheroid YOYO-1 for DMSO, GANT61 and staurosporine treatment of SUM149 (A), SUM159 (B) and MDA-MB-231 (C, GANT61 only) spheroids. Statistical significance relative to DMSO control *P<0.05, **P<0.005, ***P<0.001 (Student t-test). (D) Representative images and dose response curves are shown for MDA-MB-231 spheroids treated with staurosporine. A minimum of two independent experiments were carried out. Dose response curves were generated using nonlinear regression and IC50 values determined in GraphPad Prism 6.
Supplementary Fig. 4. GANT61 structural confirmation, testing in C3H10T1/2 assay and MDA-MB-231 and SUM159 xenograft models. (A) NMR spectra of GANT61 used for in vivo studies determined using 500 MHz Varian instrument. (B) Testing of GANT61 used for in vivo study for inhibition in C3H10T1/2 Hh functional assay. C3H10T12 cells were stimulated with Shh protein (2 µg/ml) in the presence of GANT61 (0.001 – 10 µM) and alkaline phosphatase measured at 5 d. Dose response curve were generated using non-linear regression and IC50 values determined in GraphPad Prism 6. Nude NU/J mic were injected orthotopically with MDA-MB-231 (C) or SUM 159 (D) cells into the mammary fat pad. Once tumors reached 5×5 mm (6 weeks) they were treated with vehicle or GANT61 (50 mg/kg i.p. 3x per week for n = six animals). Tumor response was assessed by weekly caliper measurements. Tumor volume (percent change over 21 days) and tumor progression (tumor volume over 28 days, mean ± SEM) are shown for MDA-MB-231 and SUM159 models respectively. Statistical significance relative to respective control; ns = not significant *P<0.05, **P<0.005, ***P<0.001 (Student t-test). Comparisons in tumor growth were also made by linear regression in GraphPad Prism 6.0. Statistical significance relative to control #P<0.05.