Abstract
Epigenetic mechanisms that cause maternally and paternally inherited alleles to be expressed differently in offspring have the potential to radically change our understanding of the mechanisms that shape disease susceptibility, phenotypic variation, cell fate, and gene expression. However, the nature and prevalence of these effects in vivo have been unclear and are debated. Here, I consider major new studies of epigenetic allelic effects in cell lines and primary cells and in vivo. The emerging picture is that these effects take on diverse forms, and this review attempts to clarify the nature of the different forms that have been uncovered for genomic imprinting and random monoallelic expression (RME). I also discuss apparent discrepancies between in vitro and in vivo studies. Importantly, multiple studies suggest that allelic effects are prevalent and can be developmental stage- and cell type-specific. I propose some possible functions and consider roles for allelic effects within the broader context of gene regulatory networks, cellular diversity, and plasticity. Overall, the field is ripe for discovery and is in need of mechanistic and functional studies.
Keywords: non-genetic allelic effects/imprinting/gene regulatory networks/gene expression
Understanding gene regulatory networks at the allele level: recipes for cellular, anatomical, physiological, and behavioral phenotypes
Specific gene expression programs in the genome evolved to orchestrate different biological processes, including developmental processes, metabolic processes, and other cellular processes 1. The gene regulatory networks that govern gene expression programs are modular and hierarchically organized. They include highly conserved and essential subcircuits, called kernels, as well as various different logic gates and feedback loops to control gene expression in a precise temporal and spatial manner 1– 3. The explosion of interest in gene regulation over the past several years has been driven by the recognition that genetic and epigenetic variations in noncoding regulatory elements shape disease risk and phenotypic variation 4, 5, and the evolution of new phenotypes frequently involves changes to cis-regulation rather than changes to protein sequence 6, 7.
Defining the architecture and logic of the gene regulatory networks and gene expression programs that control different biological processes is challenging. A rare example of a relatively well-defined gene regulatory network in human cells is that controlling embryonic stem cell (ESC) pluripotency 8. However, in most cases, our understanding of the gene regulatory networks that control the development and function of the myriad of different cell types in the brain and body has just begun. Enticingly, beyond mechanisms for cell fate, studies of gene regulatory mechanisms in the nervous system have the potential to define gene regulatory networks and gene expression programs that control the development of specific features of behavior, such as particular social behavior traits, anxiety states, and different cognitive and sensorimotor abilities. However, while our understanding of and interest in gene regulatory networks is growing, most approaches assume that the maternal and paternal alleles for a given gene are expressed and regulated equally. Here, I discuss recent and growing evidence for diverse forms of non-genetic effects that cause alleles to be differentially expressed and consider some implications for understanding the regulatory mechanisms and gene expression programs governing cell fate and mammalian phenotypes.
Genomic imprinting and the differential expression of maternal and paternal alleles at the cellular level
Epigenetic allelic effects that cause maternal and paternal alleles to be expressed differently in vivo are best understood from studies of canonical genomic imprinting 9, random X-inactivation in females 10, 11, allelic exclusion of immunoglobulins 12, and RME of clustered protocadherins 13 and olfactory receptors 14. Many of these cases of established in vivo epigenetic allelic effects involve genes with a uniquely clustered organization in the genome. However, others have found evidence for a broader landscape of epigenetic allelic effects in the genome 15– 17, although this research area is new, rapidly evolving, and debated. Below, I discuss recent studies that have advanced our understanding of allelic effects and refer readers seeking a more comprehensive literature review to the aforementioned articles.
Genomic imprinting is an important phenomenon that causes maternal and paternal alleles to be differentially expressed in offspring. I and others previously described noncanonical imprinting (also referred to as parental allelic biases), which involves maternal or paternal allele expression biases at the tissue level 17– 20, in contrast to the allele-silencing effects exhibited by canonical imprinted genes 18 ( Figure 1). Some early studies overestimated 21, 22 or underestimated 23, 24 the prevalence of these effects in the mouse genome. Noncanonical imprinting is less robust and more variable between different individuals than canonical imprinting, and therefore sensitive methods and sufficient statistical power are required to detect these effects accurately 18– 20, 25. It is now clear that noncanonical imprinting is a bona fide, highly reproducible epigenetic allelic effect that is especially enriched in the brain and more prevalent than canonical imprinting in the mouse genome 18, 20. Furthermore, noncanonical imprinting can shape offspring phenotypes 17, 18 and therefore there is a strong motivation to learn more about it.
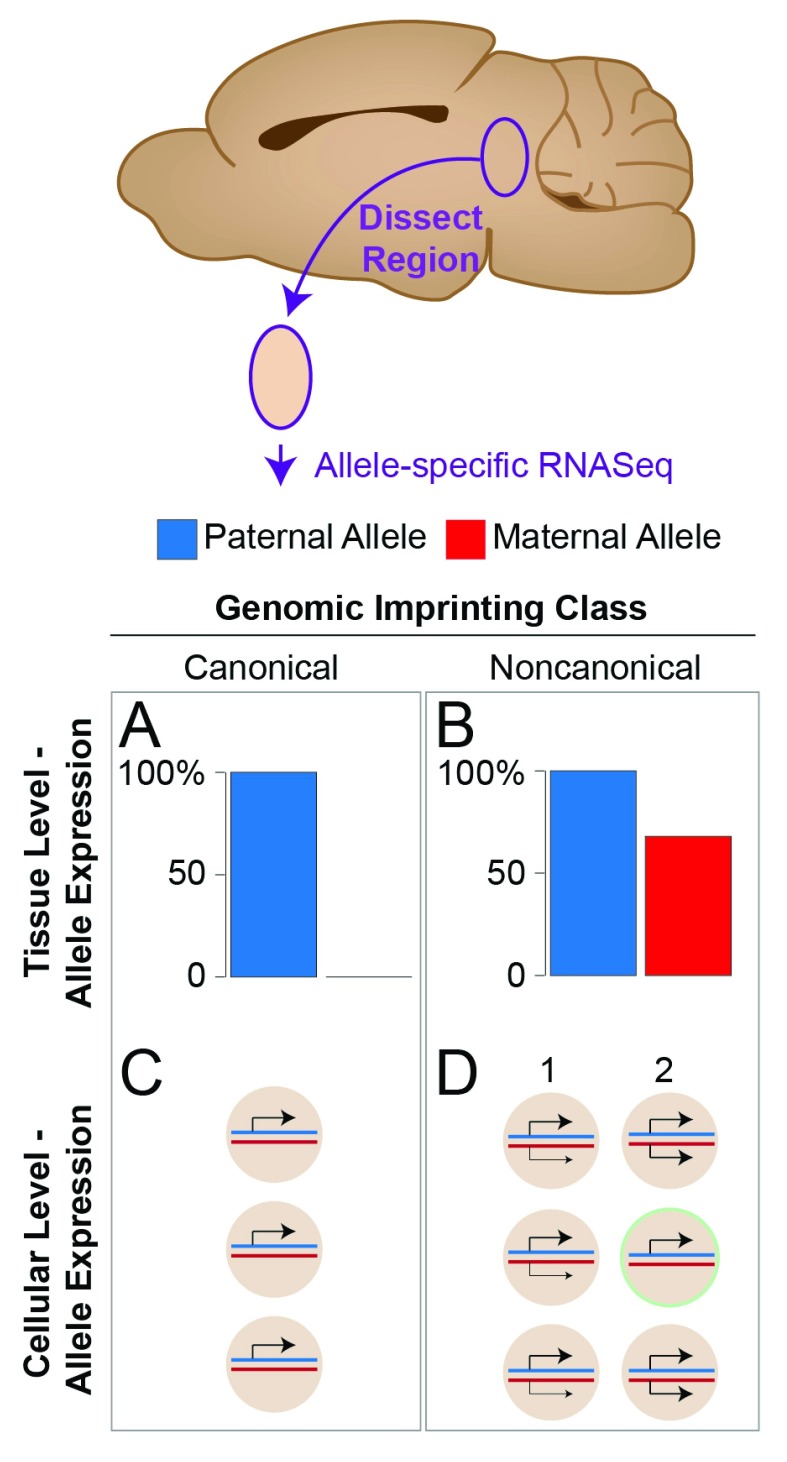
Figure 1. Schematic depiction of canonical versus noncanonical genomic imprinting identified in the mouse.
Canonical and noncanonical imprinting was characterized in different mouse tissues by using RNA-Seq in which allele expression was profiled in a piece of tissue dissected from the brain or in another tissue. ( A) In this chart, canonical imprinting manifests as complete silencing of one parent’s allele (silent maternal allele shown). ( B) In contrast, noncanonical imprinting manifests as a significant bias to express one parental allele at a higher level than the other parental allele (paternal allele bias shown). ( C) At the cellular level, canonical imprinting involves complete silencing of one allele in all cells expressing the gene. ( D) Noncanonical imprinting may involve either (1) an allelic bias in each cell or (2) allele silencing in a subpopulation of cells in the tissue. Distinguishing between these models (1 versus 2) is an active area of research.
For many genes, the relative strength of noncanonical imprinting changes between different brain regions and tissue types 18, 19. For example, two enzymes that synthesize catecholamines in the brain—tyrosine hydroxylase and dopa decarboxylase ( Ddc)—exhibit noncanonical imprinting involving a maternal allele expression bias in the arcuate nucleus (ARN), dorsal raphe nucleus (DRN), and locus coeruleus, but the imprinting effect is not observed in the ventral tegmental area (VTA) for either enzyme 18. For Ddc, the imprinting is especially strong in the ARN. Overall, these results suggest that noncanonical imprinting is likely influenced by the cellular composition of the target brain region (or tissue) and reflects highly cell type-specific allelic effects in the brain. In support of this interpretation, nascent RNA in situ hybridization revealed that brain regions in the mouse that have stronger noncanonical imprinting for Ddc are associated with more brain cells that exhibit monoallelic expression, while cells in the VTA, where the imprinting is absent, exhibit biallelic expression 18. Thus, an emerging picture is that at least some noncanonical imprinting cases shape maternal and paternal allele expression in a cell type-dependent manner ( Figure 1D).
Other new studies have begun to further clarify the complexities of imprinting at the cellular level in mice. Stelzer and colleagues recently developed a novel reporter of cellular genomic methylation effects that involves placing a differentially methylated region of interest in front of the minimal imprinted promoter region for the gene, SNRPN, driving the expression of a green fluorescent protein or tdTomato reporter 26. If the differentially methylated region is unmethylated, the reporter is expressed, and if it is methylated, the reporter is silent. With this technology, Stelzer and colleagues recently investigated DNA methylation dynamics at the cellular level in vivo for a differentially methylated region that controls imprinting at the Dlk1-Dio3 imprinted gene cluster 27. The study revealed highly cell type- and tissue-specific imprinting as well as imprinting changes during development. In the brain, mosaic methylation of the differentially methylated region was observed in dopaminergic neurons and Purkinje neurons and other cell populations; loss of parent-specific methylation was observed in neural stem cells, consistent with previous work 28, and variation between individuals was also found. Thus, the authors discovered that imprinted DNA methylation is more dynamic and varied at the cellular level than was previously known. The field is gaining a deeper appreciation for the complexity of imprinting at the cellular level and, although there is substantial precedence in the literature for such effects 17, the prevalence and function of cell type-specific imprinting remain unclear. New in situ hybridization-based strategies to resolve allele-specific expression at the cellular level have provided an expanded tool kit for the field to study these effects 18, 29– 31. In the brain, cell type-specific maternal and paternal imprinting may shape the development and function of specific brain cells and circuits to modulate particular aspects of offspring brain function and behavior. The identity of the cells, circuits, and brain functions that are impacted and the mechanisms involved are important areas for research.
Currently, less is known about cell type-specific imprinting and noncanonical imprinting in humans. Recent efforts to identify imprinted genes from Genotype-Tissue Expression (GTEx) consortium data were designed to uncover canonical imprinting that involves robust monoallelic expression that is consistent among individuals 32, 33. This strategy was necessary because parental genome information was not available to phase the RNA-Seq reads according to parental allele and to avoid various potential artifacts. Nonetheless, these studies have begun to define the landscape of imprinting in the human body and tissue-specific imprinting was found. One study reported relatively more imprinted genes in the human brain compared with other tissues 32. As in the mouse, future work analyzing human imprinting at the cellular level is also likely to reveal new information. Additionally, imprinting in the mouse brain is most prevalent and robust in the hypothalamus and in monoaminergic nuclei 18, 21, 34, but, other than the hypothalamus, few subcortical regions were included in the GTEx studies, indicating another important area for further study in humans.
New and diverse forms of epigenetic allelic effects uncovered in vitro and in vivo
Beyond imprinting, evidence exists for other forms of epigenetic allelic effects, although the nature and prevalence of these effects in vivo are debated 15, 16 and some new studies have improved our understanding. Widespread RME on the autosomes was first described in human lymphoblastoid cell lines by Gimelbrant and colleagues 35. The initial description of this phenomenon indicated similarities to random X-inactivation, such that the monoallelic effect was inherited by daughter cells derived from a single precursor and therefore is clonal. It was estimated that clonal RME impacts 5–15% of genes in human and mouse lymphoblastoid cell lines ( Figure 2A) 35, 36. Furthermore, a chromatin signature involving H3K36me3 (activating) and H3K27me3 (repressive) marks was found to distinguish RME genes from other autosomal genes in lymphoblastoid cell lines and then was applied to identify RME genes in human cells and tissues. These studies led the authors to estimate that 20–30% of human genes are subject to RME 37, 38. Interestingly, genes with this chromatin signature are more genetically variable in humans 39 and appear to be resistant to pathogenic variants impacting expression levels 40. This body of work suggests that clonal RME shapes the expression of a large but defined subset of autosomal genes with implications for understanding human genetic variation. However, some other studies suggest a different picture (see below).
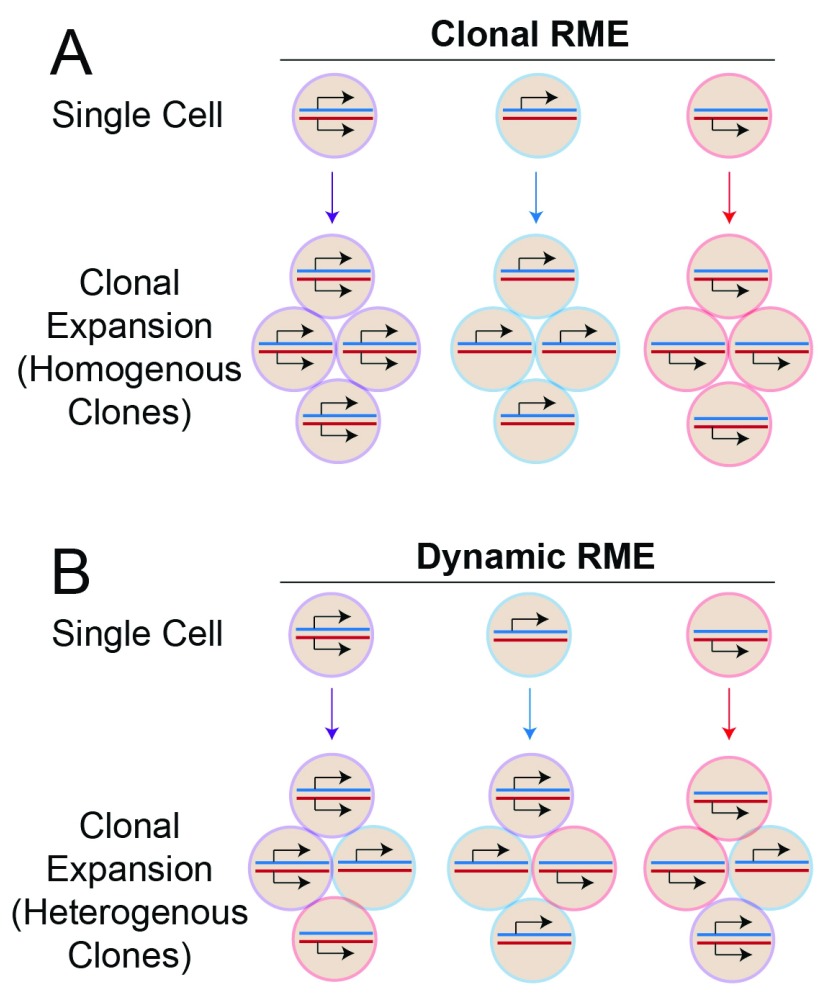
Figure 2. Schematic depiction of clonal versus dynamic random monoallelic expression (RME).
( A) Clonal RME is identified when a single cell is expanded to form a colony of daughter cells and each daughter cell has the same allele expression pattern as the original parent cell. Typically, studies of this phenomenon use cell lines and expand them clonally and then profile allelic expression from the entire batch of cells in the clone. They find that some clones exclusively express one allele, others are biallelic, and others express only the other allele (homogenous clones). ( B) Dynamic RME occurs when a single cell is expanded to form a colony but the individual cells in the colony have different allelic expression patterns (heterogeneous clones). This phenomenon is detectable only by using single-cell transcriptome analysis and cannot be identified from profiles of the whole batch of cells in the clone, as was done in previous cell line studies reporting widespread clonal RME.
Two studies of RME using a similar strategy in mouse ESC lines uncovered a related but more dynamic picture 41, 42. In these studies, RME impacted relatively few genes in ESCs but became more prevalent following differentiation into neural progenitor cells, ultimately impacting hundreds of genes. Once established, the monoallelism for a given gene is stable in the neural progenitor cell lines, indicating clonal RME. Thus, the findings from these two major studies suggest that RME is not fixed for specific genes but can change developmentally.
While studies of RME in cell lines yielded a provocative new picture of widespread clonal RME on the autosomes, others have challenged these conclusions and the prevalence of clonal RMEs 16. Furthermore, large consortiums studying human allele-specific expression effects in vivo concluded that most allele expression differences are explained by genetic variants (expression quantitative trait loci) rather than epigenetic effects 43, 44. Similarly, while allelic differences in DNA methylation and chromatin composition are widespread in vivo 45, 46 and in vitro 5, 47, these effects also have frequently been attributed to genetic variation 48– 50. Thus, in vitro versus in vivo studies yielded an apparent discrepancy regarding the nature and prevalence of epigenetic allelic effects. While a number of possible explanations exist, recent studies have added some new information.
A new study of RME using single-cell transcriptome profiling in primary cells has challenged previous findings in cell lines regarding the prevalence of clonal RME for mouse and human autosomal genes 51. Earlier studies using single-cell transcriptome profiling found RME effects, but few effects were inherited by daughter cells 16. A point of note is that over 80% of stochastic allelic expression in single-cell RNA-Seq is potentially due to technical noise 52. Thus, this approach is arguably best suited for ruling out clonal RME rather than discovering new RME effects in single cells. Nonetheless, the approach is suitable for the main conclusions drawn in this new study 51. Rather than analyzing cell lines, Reinius and colleagues isolated primary mouse fibroblasts and human T cells and expanded single cells to form clones in vitro 51. By performing single-cell RNA-Seq profiling for individual cells within a clone, they were able to show that clonal RME is very rare, impacting less than 1% of genes in mouse fibroblasts or human T cells, and is associated with genes expressed at very low levels. The identity of the genes impacted in different clones frequently differed. On the other hand, RME that differs between individual cells within a clone, referred to as dynamic RME ( Figure 2B), is frequent and impacts about 13% of genes in fibroblasts and 60–85% of genes in human T cells. The results of this study indicate that clonal RME is very rare in vivo, in agreement with the authors’ previous assessment of the field 16, but reveals that dynamic RME is frequent and more prevalent in human T cells than mouse fibroblasts. The authors also provide evidence that dynamic RME is related to the transcriptional activity within a cell.
In a recent study, my colleagues and I also sought to gain a deeper understanding of the different forms of non-genetic allelic effects that exist in vivo 53. We devised a robust genomics and statistical methodology that tests the null hypothesis that the maternal and paternal alleles for a given gene are equally co-expressed (or correlated) across different RNA-Seq biological replicates from a particular brain region or tissue type ( Figure 3A). This screening approach has the potential to uncover a wide range of different allelic effects in vivo, including imprinting, clonal RME, dynamic RME, and other possible effects ( Figure 3B). Genes with clonal RME in vivo will typically have negatively correlated allelic expression ( Figure 3B), and, as expected, many randomly inactivated X-linked genes in females are detected with this signature. Genes with biallelic expression have positively correlated allele expression, and, finally, genes with canonical imprinting, cell-specific imprinting, cell-specific RME, or other possible allelic effects will manifest with low or no allele correlation, which we refer to broadly as differential allele expression effects ( Figure 3B). The screen was performed in the DRN in postnatal day 5 (P5) and P15 and adult female mice and in the ARN in the hypothalamus, the liver, and skeletal muscle as well as in the juvenile DRN of female cynomolgus macaques.
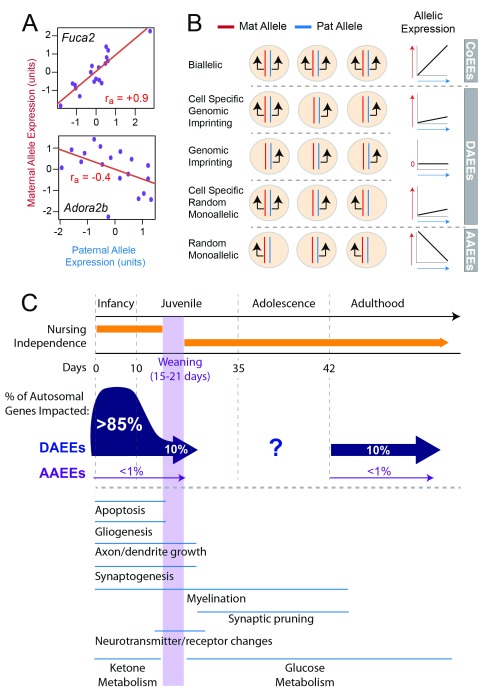
Figure 3. RNA-Seq approach to screen for high-confidence, non-genetic differential allele expression effects in vivo.
( A) The approach involves RNA-Seq profiling of maternal and paternal allele expression levels across a population of individuals and examining the correlated expression of the two alleles across the population. For example, Fuca2 exhibits highly correlated allelic expression, while the maternal and paternal alleles for Adora2b are negatively correlated. Statistical modeling is performed to estimate the effects of technical noise, biological variation, and genetic variation on the data. The resulting statistic identifies high-confidence, non-genetic allelic effects in a genome-wide manner for any tissue. ( B) This in vivo screening approach can detect diverse forms of allelic effects. Biallelic expression at the cellular level is expected to manifest as highly correlated allelic expression. Clonal random monoallelic expression (RME) that is similar to X-inactivation will manifest as a negative allele correlation (antagonistic allele expression effects, or AAEEs), since more maternal allele-expressing cells arise at the expense of paternal allele-expressing cells and vice versa. Genome imprinting and cell-specific imprinting or RME will manifest as a weak correlation or no correlation between the alleles; we refer to these cases more generally as differential allele expression effects (DAEEs). ( C) Profiling of non-genetic allelic effects in the postnatal day 5 (P5) and P15 and adult mouse DRN revealed major developmental differences. Most genes exhibit evidence for high-confidence DAEEs in the P5 DRN, but these effects are reduced by P15 and in adults such that only 10% of autosomal genes exhibit DAEEs at these older ages. AAEEs are rare in vivo and impact less than 1% of all autosomal genes expressed. The in vivo developmental shift in non-genetic allelic effects is presented relative to other major developmental milestones and processes in the mouse brain. We applied a similar approach to study DAEEs in the primate brain. CoEE, co-expression effect.
Our study revealed that over 85% of genes exhibit high-confidence differential allele expression in the developing P5 DRN, indicating profound differences in the expression patterns of maternal and paternal alleles at this stage, and very few cases involved imprinting ( Figure 3C). In P15 juveniles and adults, only about 10% of genes are impacted and most genes shift toward allele co-expression at these later developmental stages ( Figure 3C). We further show that genes with allele co-expression predominantly exhibit biallelic expression at the cellular level but that genes with differential allele expression predominantly exhibit monoallelic expression. These results reveal a developmental shift in allelic effects in mice, which is associated with the progression of neuronal and glial cell differentiation, cell and synaptic pruning, and circuit formation and maturation in the brain ( Figure 3C). Together with previous in vitro studies of RME 41, 42, an emerging picture is that non-genetic allelic effects are relatively infrequent in ESCs, increase in frequency in neural progenitors, are highly prevalent in the neonatal brain, and decrease in the mature brain 41, 42, 53. These observations suggest that many allelic effects are more than just stochastic transcriptional noise in genes expressed at a low level. In fact, genes with in vivo differential allelic expression are not expressed at lower levels than genes without these effects 53 and they appear to be a major feature of developmental gene expression programs.
While we found that differential allelic expression is frequent in vivo, we also found that very few autosomal genes exhibit evidence for clonal RME effects that are similar to random X-inactivation, which manifests as a negative allele correlation with our approach. Many X-linked genes in females exhibit a negative allele correlation, but fewer than 15 autosomal genes exhibit evidence for these types of effects in the mouse in any of the tissues examined 53. These findings may be consistent with the results of Reinius and colleagues 16, 51, since they indicate that strict clonal RME is indeed rare in vivo. Some of the differential allelic expression effects we observe in vivo could involve clonal RME in a subpopulation of cells in vivo, as was observed in cell lines 35, 36, 41, 42, or dynamic RME 16, 51; we currently do not know the underlying mechanisms involved. Importantly, dynamic RME simply refers to allelic effects that differ between cells in the same clone 51 and the temporal stability of these effects is not known. Overall, further studies are needed to investigate clonal versus dynamic RME in vivo and in specific cell lineages and the temporal stability of these effects for different genes.
Potential functional roles for allelic effects in shaping cellular diversity and gene regulatory network plasticity
Studies of stochastic gene expression effects across otherwise identical prokaryotic and eukaryotic cell populations have shown that cellular gene expression variability can function to diversify an otherwise homogeneous cell population 54– 57. Others have argued that stochastic epigenetic variation in humans and mice promotes phenotypic variation 58– 60, providing a powerful evolutionary strategy to cope with changing and unpredictable environments by diversifying a population of organisms. Similarly, clonal and dynamic RME may be an important source of variation, placing otherwise similar cells into different states and thereby introducing diversity and plasticity into the population ( Figure 4A and B). By increasing the diversity of gene expression programs in a cell population, some cells may respond differently to the same signal, but at least some will respond appropriately ( Figure 4B). Thus, the population is prepared for cues arising in a changing and unpredictable cellular environment.
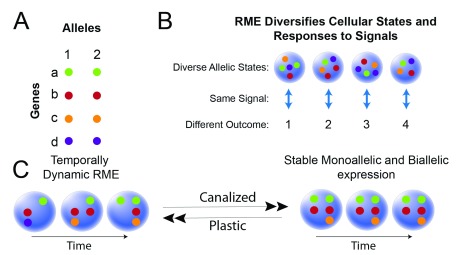
Figure 4. Possible functions for different forms of random monoallelic expression (RME) in promoting cellular diversity and plasticity in mammalian cells.
( A) Schematic of two alleles (1 and 2) for four genes (a–d). ( B) A population of four cells is diversified by clonal or dynamic RME such that each cell expresses a different allelic combination. Exposure of the population to a signal in the environment is predicted to result in different outcomes for each cell (1, 2, 3, 4). Some cells will be in a state that responds better (more quickly or correctly) to the signal than other cells. ( C) Cellular and gene transitions from a temporally dynamic RME state to a stable biallelic or monoallelic (clonal RME or imprinting) state are predicted to shape plasticity versus canalization for gene networks within a cell. The temporally dynamic RME state for different genes is a predicted state of increased plasticity because the cell has access to different allelic combinations that are maintained in a poised state, but these combinations are no longer available once a gene and cell commit to a stable allelic expression state.
At the level of individual cells, commitment to stable biallelic or monoallelic expression states (imprinting or clonal RME) is expected to promote the canalization of gene regulatory networks, thereby committing cells to a particular fate ( Figure 4C). In contrast, if dynamic RME effects can change temporally for different genes in the same cell, such that different allelic combinations are maintained in a poised state and available to be expressed, this mechanism could function to increase plasticity and expand the landscape of gene regulatory networks and programs available to the cell ( Figure 4C). Interestingly, transitions between different cellular states during development have recently been shown to involve a destabilization of gene expression, such that the cell can respond to diverse environmental cues during the transition from one state into a new state 61. I speculate that the presence versus absence of dynamic RME could reflect this type of destabilization, promoting cellular plasticity and permitting the formation of new epigenetic states within a cell 62. Seemingly consistent with this prediction is the finding that about 60–85% of genes in human T cells exhibit dynamic RME 51, since T cells are a highly plastic cell type that must respond to unpredictable environmental cues 63. In contrast, only 13% of genes exhibit dynamic RME in mouse fibroblasts, which presumably are less plastic than T cells 51. Interestingly, several examples of RME have been described in immune cells over the years 15; however, the function of effects other than allelic exclusion is currently unknown.
Enticingly, a model for allelic effects in regulating gene expression plasticity and diversity predicts that allelic effects could have roles in shaping cell fate decisions in the developing brain and how environmental factors, such as stress, diet, drugs, infection, and disease, impact cells in the brain and body. However, in apparent opposition to this model is the observation that ESCs have a relatively low frequency of RME 41, 42 yet they are a highly plastic cell type. It is possible that the pluripotent state does not benefit from allelic diversity in the same way as more committed cell types. Alternatively, allelic effects may serve other functions that remain to be uncovered and are not related to promoting gene regulatory network plasticity and cellular diversity.
Interactions between allelic effects and genetic variation at the cellular level
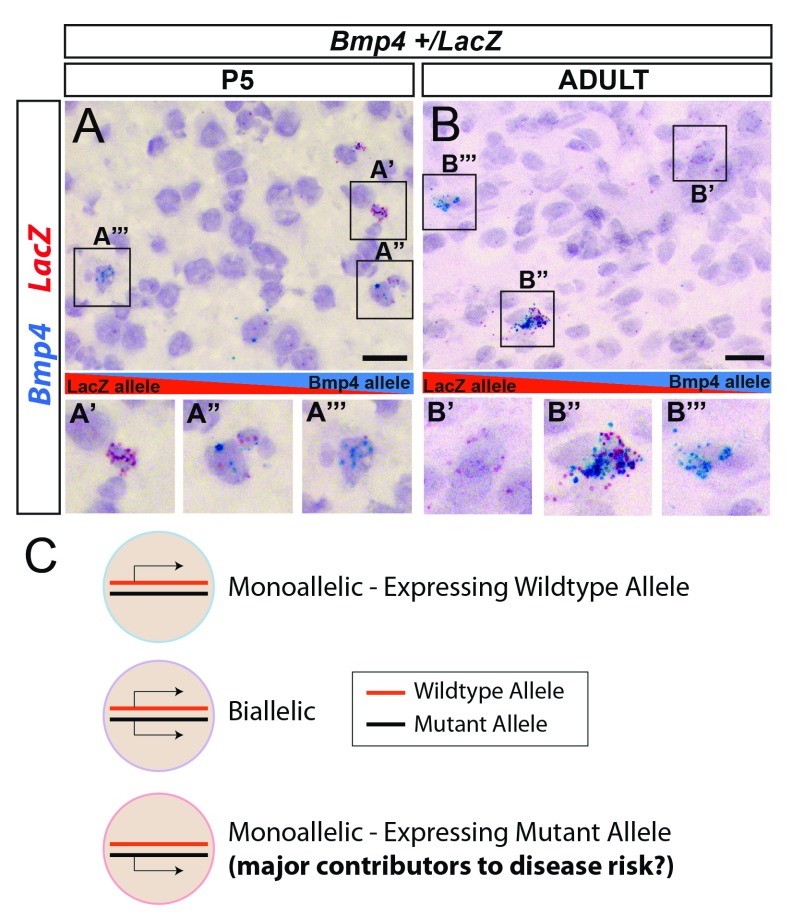
When considered in the context of genetic variation, cells exhibiting RME for a gene are diversified by the potential to express not only different combinations of alleles but also different combinations of heterozygous variants. The effect of such genetic allelic diversity across a population of cells is predicted to further contribute to diversity in cellular responses to the environment and physiological state of the organism. We recently demonstrated that a heterozygous mutation in a gene with differential allelic expression, such as Bmp4, results in mosaics of cells in the mouse brain, such that some brain cells express the mutated allele, some express the wild-type allele, and some express both alleles ( Figure 5) 53. This mosaic pattern was found to differ according to cell type for some genes. However, while our study showed how these effects can interact with genetic variants, we focused on the RNA level and it is unclear whether such effects also manifest at the protein level, which is important to determine in order to fully understand the impact on genetic architecture.
Figure 5. Interactions between non-genetic allelic effects and heterozygous variants can shape genetic architecture at the cellular level.
( A, B) Single-molecule mRNA in situ hybridization for the mutant ( LacZ, red) and wild-type ( Bmp4, blue) alleles in a heterozygous knockout reporter Bmp4 LacZ/+ mouse line. Images of the postnatal day 5 (P5) and adult mouse brain are shown and reveal a mosaic of cells that preferentially express the mutant allele (red; A’ and B’), wild-type allele (blue; A’’ and B’’), and biallelic (red and blue co-expressed; A’’’ and B’’’) cells. ( C) Monoallelic mutant allele-expressing cells might be more dysfunctional than biallelic or monoallelic wild-type allele-expressing cells for some mutations.
Interactions between epigenetic allelic effects and heterozygous genetic variation could improve our understanding of the factors that drive phenotypic variation in different disorders, such as mental illnesses 15, 16, 64, 65. Indeed, we found that differential allelic expression effects exist in vivo in the primate brain and impact genes linked to mental illness in the macaque and human brain, including autism-linked genes and huntingtin 53. RME for disease-linked genes has also been observed in cell lines 35, 41, 42, 66 and recently for the autism-linked gene FOXP2 in humans 67. Allelic effects in the autistic brain have also been identified, although it is unclear whether the effects are genetic or epigenetic in origin 68. As others have proposed 15, 64, 65, we speculate that cells that preferentially express mutated alleles, or particular combinations of mutated alleles, due to epigenetic allelic effects may play important roles in contributing to disease risk and phenotypic variance ( Figure 5C). However, these ideas remain to be tested.
Possible links between imprinting and random monoallelic expression
The prevalence of RME in vivo provides opportunities for discovery and reflection. In an insightful review by Ohlsson and colleagues in 2001, it was proposed that genomic imprinting and random X-inactivation evolved from stochastic allele expression effects in the genome, perhaps to better coordinate the expression of groups of genes 69. With the expanded landscape of RME effects that have been uncovered in vivo and in vitro, and the characterization of noncanonical imprinting in vivo, it is worth revisiting the relationship between RME and imprinting. If some forms of RME function to promote plasticity within cellular gene regulatory networks or cellular diversity or both, then imprinting is expected to constrain these effects, as noted above ( Figure 4C). Thus, noncanonical and canonical imprinting may function to reduce plasticity and cellular diversity, stabilizing particular gene regulatory networks and promoting specific cellular states and phenotypic traits in offspring. The strength of the imprinting effect (for example, canonical versus noncanonical) may be related to the strength of the imprinting constraint placed on the RME effect for some genes. Indeed, preliminary data in our lab suggest that some noncanonical imprinted genes also exhibit RME at the cellular level in the brain (Paul J. Bonthuis and C. Gregg, unpublished observations).
Uncovering the mechanisms governing non-genetic allelic effects
Our review of the literature above indicates that non-genetic allelic effects are prevalent and take on many different forms. We summarize the current in vivo landscape of these effects in Figure 6. In most cases, the mechanisms involved are not known. For instance, the mechanisms involved in causing rare clonal RME versus dynamic RME versus biallelic expression are not known. Furthermore, for dynamic RME, we do not know how stable these effects are over time; some may be stable during specific developmental stages and others may be more transient. Our study revealed 335 genes that exhibit differential allelic expression in the mouse DRN across all developmental stages examined 53. We also found 69 genes that exhibit differential allelic expression across all tissue types examined (brain, liver, and muscle). However, most in vivo effects appear to be developmental stage-, tissue-, and cell type-specific. Chromatin structure and transcription factor dosage are likely to have important roles in regulating these effects. Indeed, in the brain, profound epigenetic changes are known to occur over the course of development and chromatin is highly cell type-specific 70– 73.
Figure 6. The current landscape of in vivo non-genetic allelic effects in mammals.
( A) Non-genetic allelic effects that impact X-linked genes in females are shown. ( B) Effects impacting autosomal genes are shown. Tissue- and age-specific allele co-expression effects for X-linked genes were observed by Huang and Ferris and colleagues 53 (2017), but the underlying cause is not yet known. Additionally, the frequency of clonal random monoallelic expression (RME) effects in vivo is debated and requires further investigation.
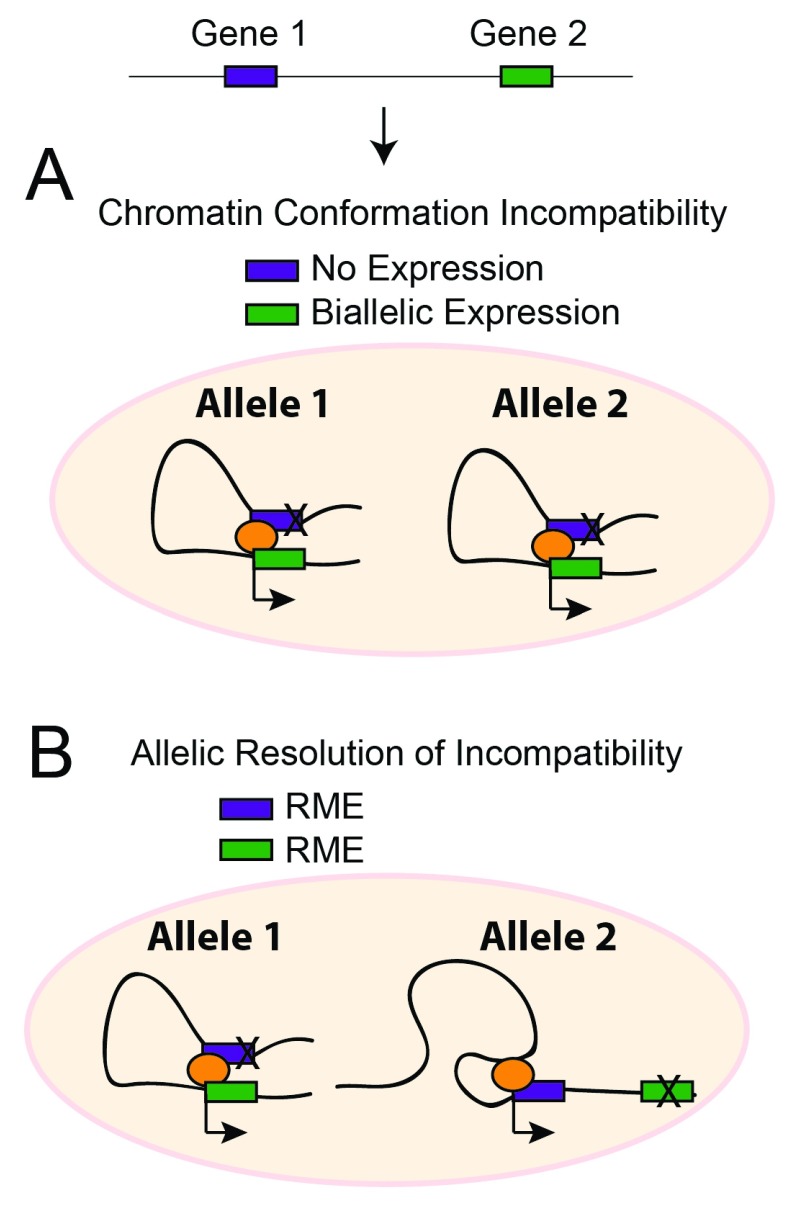
In one model of RME, chromatin conformation incompatibilities may prevent some genes from being expressed simultaneously from the same allele in the same cell, in which case RME effects would resolve these incompatibilities and permit particular combinations of genes to be expressed simultaneously in the same cell ( Figure 7). Indeed, such allelic incompatibility is known for the imprinted genes Igf2 and H19, which form allele-specific chromatin loops that provide both genes access to the same upstream enhancers 74, 75. Allelic competition for enhancers also contributes to singular allelic expression for olfactory receptors in olfactory neurons 76 and is a plausible mechanism contributing to other autosomal clonal and dynamic RME effects. RME might also arise due to transcriptional interference if the enhancer or transcriptional start site for one gene is located in a position that disrupts the expression of another gene and the interference is resolved in an allele-specific manner. Indeed, many genes in the genome overlap 77, which can result in transcriptional interference leading to allele-specific expression effects, as was shown for some imprinted genes 78. Furthermore, about 51% of enhancers reside within the introns or exons of coding genes 79 and can regulate the expression of neighboring genes 80, 81. Thus, RME might resolve various potential regulatory incompatibilities in the genome, allowing diverse gene regulatory networks to be differentially active or poised in different cells within a cell population.
Figure 7. Random monoallelic expression (RME) effects may resolve chromatin conformation incompatibilities in the genome, allowing the activation of diverse gene regulatory networks across a cell population.
A schematic representation of a chromatin incompatibility that influences the cellular expression of two genes is shown. Gene 1 and gene 2 compete for a particular chromatin state for their expression. ( A) In the absence of RME effects, only one gene can win and is expressed in a biallelic manner in a cell. ( B) However, if RME effects are present, then different chromatin structures can form on each allele to resolve the incompatibility and permit the simultaneous expression of both genes in the same cell. This model of RME effects implies that these effects may function to allow cells in a population access to diverse potential gene regulatory networks, thereby further contributing to cellular plasticity.
A new study by Xu and colleagues used ATAC-Seq to uncover the nature of random monoallelic chromatin architecture and DNA accessibility in mouse ESC lines differentiated into neural progenitors 82. The results reveal that allele-specific DNA accessibility increases in prevalence following the differentiation of ESCs into neural progenitors and that most allele-specific open chromatin sites occur in promoter regions rather than distal regulatory elements. These effects are stable once they are established at specific genomic sites in clonal neural progenitor cell lines. It may be feasible to adapt this powerful strategy to perform in vivo profiling of differential allelic DNA accessibility in tissues and purified cell populations using the statistical methods we recently developed 53. The new findings by Xu and colleagues are a major first step toward understanding the mechanisms involved in clonal autosomal RME.
Opportunities for the discovery of novel allelic effects at the chromatin and cellular levels are likely to be plentiful. Early studies revealed that chromosomes occupy specific territories in the nucleus, and elegant studies using fluorescent in situ hybridization and chromatin confirmation capture found that genome topology changes in response to cellular differentiation and can vary at the cellular level and between cell types 83, 84. At the time, however, little attention appears to have been paid to the relative locations of the maternal and paternal chromosomes in these studies, yet it is clear from the reported data that the two chromosomes frequently occupy different relative positions in the nucleus 85, 86. The same appears to be true for the relative positioning of alleles in healthy and diseased cells 87, 88. Therefore, at the cellular level, the topological organization, globular structure, and chromatin architecture of maternal and paternal chromosomes may differ frequently, which could reflect differences in gene regulation at the cis or trans level or both. Indeed, recent work in macrophages uncovered dynamic changes from monoallelic to biallelic expression for TNFalpha in response to pro-inflammatory cues, which was associated with changes to the relative location of the alleles in the nucleus 89.
Conclusions
Throughout this article, I have highlighted many exciting opportunities for discovery in this expanding field. I suggest that a deeper understanding of noncanonical imprinting and RME at the cellular level in vivo is needed, and I propose that some allelic effects might function to regulate the plasticity versus canalization of gene regulatory networks within a cell and shape cellular diversity within a population. Overall, new studies in the field have uncovered diverse forms of non-genetic allelic effects in vitro and in vivo and, when heterozygous mutations are present, these effects can shape the expression of mutated versus wild-type alleles at the cellular level, at least at the RNA level. Although the temporal stability of RME has not been established, it appears that many genes have the capacity to move in and out of an RME state. Therefore, unlike imprinting, which impacts a defined subset of genes in the genome, RME appears to be a more general and dynamic property of gene expression, particularly during development. The prevalence of clonal RME in vivo appears to be less than was initially thought from cell lines, but more studies are warranted. The prevalence of noncanonical imprinting has now been clarified in mice and shown to impact offspring phenotypes, but the mechanisms and function are undefined and little is known in humans. Overall, many opportunities for new mechanistic and functional investigations exist. The field is poised to improve our understanding of gene regulation and genetic architecture in development, neurobiology, and disease.
Abbreviations
ARN, arcuate nucleus; Ddc, dopa decarboxylase; DRN, dorsal raphe nucleus; ESC, embryonic stem cell; GTEx, Genotype-Tissue Expression; P, postnatal day; RME, random monoallelic expression; VTA, ventral tegmental area.
Acknowledgments
The author wishes to thank members of his lab for feedback on the manuscript. This work has been supported by NIH R01MH109577, a Simons Foundation Autism Research Initiative Explorer Award, and a University of Utah Seed Grant. The author is a New York Stem Cell Foundation–Robertson Investigator. This research was supported by the New York Stem Cell Foundation.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Rebecca Oakey, King's College London, London, UK
Catherine Dulac, Department of Molecular and Cellular Biology, Harvard University, Cambridge, MA, 02138, USA
Funding Statement
This work has been supported by NIH R01MH109577, a Simons Foundation Autism Research Initiative Explorer Award, and a University of Utah Seed Grant. The author is a New York Stem Cell Foundation–Robertson Investigator. This research was supported by the New York Stem Cell Foundation.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; referees: 2 approved]
References
- 1. Thompson D, Regev A, Roy S: Comparative analysis of gene regulatory networks: from network reconstruction to evolution. Annu Rev Cell Dev Biol. 2015;31:399–428. 10.1146/annurev-cellbio-100913-012908 [DOI] [PubMed] [Google Scholar]
- 2. Davidson EH: Emerging properties of animal gene regulatory networks. Nature. 2010;468(7326):911–20. 10.1038/nature09645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Erwin DH, Davidson EH: The evolution of hierarchical gene regulatory networks. Nat Rev Genet. 2009;10(2):141–8. 10.1038/nrg2499 [DOI] [PubMed] [Google Scholar]
- 4. Vockley CM, Barrera A, Reddy TE: Decoding the role of regulatory element polymorphisms in complex disease. Curr Opin Genet Dev. 2017;43:38–45. 10.1016/j.gde.2016.10.007 [DOI] [PubMed] [Google Scholar]
- 5. Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, et al. : Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–30. 10.1038/nature14248 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 6. Wray GA: The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8(3):206–16. 10.1038/nrg2063 [DOI] [PubMed] [Google Scholar]
- 7. Carroll SB: Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134(1):25–36. 10.1016/j.cell.2008.06.030 [DOI] [PubMed] [Google Scholar]
- 8. Boyer LA, Lee TI, Cole MF, et al. : Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122(6):947–56. 10.1016/j.cell.2005.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 9. Bartolomei MS, Ferguson-Smith AC: Mammalian genomic imprinting. Cold Spring Harb Perspect Biol. 2011;3(7): pii: a002592. 10.1101/cshperspect.a002592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee JT: Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nat Rev Mol Cell Biol. 2011;12(12):815–26. 10.1038/nrm3231 [DOI] [PubMed] [Google Scholar]
- 11. Deng X, Berletch JB, Nguyen DK, et al. : X chromosome regulation: diverse patterns in development, tissues and disease. Nat Rev Genet. 2014;15(6):367–78. 10.1038/nrg3687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vettermann C, Schlissel MS: Allelic exclusion of immunoglobulin genes: models and mechanisms. Immunol Rev. 2010;237(1):22–42. 10.1111/j.1600-065X.2010.00935.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen WV, Maniatis T: Clustered protocadherins. Development. 2013;140(16):3297–302. 10.1242/dev.090621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Monahan K, Lomvardas S: Monoallelic expression of olfactory receptors. Annu Rev Cell Dev Biol. 2015;31:721–40. 10.1146/annurev-cellbio-100814-125308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chess A: Monoallelic Gene Expression in Mammals. Annu Rev Genet. 2016;50:317–27. 10.1146/annurev-genet-120215-035120 [DOI] [PubMed] [Google Scholar]
- 16. Reinius B, Sandberg R: Random monoallelic expression of autosomal genes: stochastic transcription and allele-level regulation. Nat Rev Genet. 2015;16(11):653–64. 10.1038/nrg3888 [DOI] [PubMed] [Google Scholar]
- 17. Perez JD, Rubinstein ND, Dulac C: New Perspectives on Genomic Imprinting, an Essential and Multifaceted Mode of Epigenetic Control in the Developing and Adult Brain. Annu Rev Neurosci. 2016;39:347–84. 10.1146/annurev-neuro-061010-113708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bonthuis PJ, Huang WC, Stacher Hörndli CN, et al. : Noncanonical Genomic Imprinting Effects in Offspring. Cell Rep. 2015;12(6):979–91. 10.1016/j.celrep.2015.07.017 [DOI] [PubMed] [Google Scholar]
- 19. Perez JD, Rubinstein ND, Fernandez DE, et al. : Quantitative and functional interrogation of parent-of-origin allelic expression biases in the brain. eLife. 2015;4:e07860. 10.7554/eLife.07860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Crowley JJ, Zhabotynsky V, Sun W, et al. : Analyses of allele-specific gene expression in highly divergent mouse crosses identifies pervasive allelic imbalance. Nat Genet. 2015;47(4):353–60. 10.1038/ng.3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gregg C, Zhang J, Weissbourd B, et al. : High-resolution analysis of parent-of-origin allelic expression in the mouse brain. Science. 2010;329(5992):643–8. 10.1126/science.1190830 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 22. Gregg C, Zhang J, Butler JE, et al. : Sex-specific parent-of-origin allelic expression in the mouse brain. Science. 2010;329(5992):682–5. 10.1126/science.1190831 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 23. Wang X, Sun Q, McGrath SD, et al. : Transcriptome-wide identification of novel imprinted genes in neonatal mouse brain. PLoS One. 2008;3(12):e3839. 10.1371/journal.pone.0003839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Babak T, Deveale B, Armour C, et al. : Global survey of genomic imprinting by transcriptome sequencing. Curr Biol. 2008;18(22):1735–41. 10.1016/j.cub.2008.09.044 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 25. Deveale B, van der Kooy D, Babak T: Critical evaluation of imprinted gene expression by RNA-Seq: a new perspective. PLoS Genet. 2012;8(3):e1002600. 10.1371/journal.pgen.1002600 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 26. Stelzer Y, Shivalila CS, Soldner F, et al. : Tracing dynamic changes of DNA methylation at single-cell resolution. Cell. 2015;163(1):218–29. 10.1016/j.cell.2015.08.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stelzer Y, Wu H, Song Y, et al. : Parent-of-Origin DNA Methylation Dynamics during Mouse Development. Cell Rep. 2016;16(12):3167–80. 10.1016/j.celrep.2016.08.066 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 28. Ferrón SR, Charalambous M, Radford E, et al. : Postnatal loss of Dlk1 imprinting in stem cells and niche astrocytes regulates neurogenesis. Nature. 2011;475(7356):381–5. 10.1038/nature10229 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 29. Levesque MJ, Ginart P, Wei Y, et al. : Visualizing SNVs to quantify allele-specific expression in single cells. Nat Methods. 2013;10(9):865–7. 10.1038/nmeth.2589 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 30. Ginart P, Kalish JM, Jiang CL, et al. : Visualizing allele-specific expression in single cells reveals epigenetic mosaicism in an H19 loss-of-imprinting mutant. Genes Dev. 2016;30(5):567–78. 10.1101/gad.275958.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hansen CH, van Oudenaarden A: Allele-specific detection of single mRNA molecules in situ. Nat Methods. 2013;10(9):869–71. 10.1038/nmeth.2601 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 32. Babak T, Deveale B, Tsang EK, et al. : Genetic conflict reflected in tissue-specific maps of genomic imprinting in human and mouse. Nat Genet. 2015;47(5):544–9. 10.1038/ng.3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Baran Y, Subramaniam M, Biton A, et al. : The landscape of genomic imprinting across diverse adult human tissues. Genome Res. 2015;25(7):927–36. 10.1101/gr.192278.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ivanova E, Kelsey G: Imprinted genes and hypothalamic function. J Mol Endocrinol. 2011;47(2):R67–74. 10.1530/JME-11-0065 [DOI] [PubMed] [Google Scholar]
- 35. Gimelbrant A, Hutchinson JN, Thompson BR, et al. : Widespread monoallelic expression on human autosomes. Science. 2007;318(5853):1136–40. 10.1126/science.1148910 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 36. Zwemer LM, Zak A, Thompson BR, et al. : Autosomal monoallelic expression in the mouse. Genome Biol. 2012;13(2):R10. 10.1186/gb-2012-13-2-r10 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 37. Nag A, Savova V, Fung HL, et al. : Chromatin signature of widespread monoallelic expression. eLife. 2013;2:e01256. 10.7554/eLife.01256 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 38. Nag A, Vigneau S, Savova V, et al. : Chromatin Signature Identifies Monoallelic Gene Expression Across Mammalian Cell Types. G3 (Bethesda). 2015;5(8):1713–20. 10.1534/g3.115.018853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Savova V, Chun S, Sohail M, et al. : Genes with monoallelic expression contribute disproportionately to genetic diversity in humans. Nat Genet. 2016;48(3):231–7. 10.1038/ng.3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Savova V, Vinogradova S, Pruss D, et al. : Risk alleles of genes with monoallelic expression are enriched in gain-of-function variants and depleted in loss-of-function variants for neurodevelopmental disorders. Mol Psychiatry. 2017;22(12):1785–1794. 10.1038/mp.2017.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gendrel AV, Attia M, Chen CJ, et al. : Developmental dynamics and disease potential of random monoallelic gene expression. Dev Cell. 2014;28(4):366–80. 10.1016/j.devcel.2014.01.016 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 42. Eckersley-Maslin MA, Thybert D, Bergmann JH, et al. : Random monoallelic gene expression increases upon embryonic stem cell differentiation. Dev Cell. 2014;28(4):351–65. 10.1016/j.devcel.2014.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 43. GTEx Consortium: Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648–60. 10.1126/science.1262110 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 44. Chen L, Ge B, Casale FP, et al. : Genetic Drivers of Epigenetic and Transcriptional Variation in Human Immune Cells. Cell. 2016;167(5):1398–1414.e24. 10.1016/j.cell.2016.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schultz MD, He Y, Whitaker JW, et al. : Human body epigenome maps reveal noncanonical DNA methylation variation. Nature. 2015;523(7559):212–6. 10.1038/nature14465 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 46. Leung D, Jung I, Rajagopal N, et al. : Integrative analysis of haplotype-resolved epigenomes across human tissues. Nature. 2015;518(7539):350–4. 10.1038/nature14217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dixon JR, Jung I, Selvaraj S, et al. : Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518(7539):331–6. 10.1038/nature14222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kilpinen H, Waszak SM, Gschwind AR, et al. : Coordinated effects of sequence variation on DNA binding, chromatin structure, and transcription. Science. 2013;342(6159):744–7. 10.1126/science.1242463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Heinz S, Romanoski CE, Benner C, et al. : Effect of natural genetic variation on enhancer selection and function. Nature. 2013;503(7477):487–92. 10.1038/nature12615 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 50. Kasowski M, Kyriazopoulou-Panagiotopoulou S, Grubert F, et al. : Extensive variation in chromatin states across humans. Science. 2013;342(6159):750–2. 10.1126/science.1242510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Reinius B, Mold JE, Ramsköld D, et al. : Analysis of allelic expression patterns in clonal somatic cells by single-cell RNA-seq. Nat Genet. 2016;48(11):1430–5. 10.1038/ng.3678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kim JK, Kolodziejczyk AA, Ilicic T, et al. : Characterizing noise structure in single-cell RNA-seq distinguishes genuine from technical stochastic allelic expression. Nat Commun. 2015;6:8687. 10.1038/ncomms9687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Huang WC, Ferris E, Cheng T, et al. : Diverse Non-genetic, Allele-Specific Expression Effects Shape Genetic Architecture at the Cellular Level in the Mammalian Brain. Neuron. 2017;93(5):1094–1109.e7. 10.1016/j.neuron.2017.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kaern M, Elston TC, Blake WJ, et al. : Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet. 2005;6(6):451–64. 10.1038/nrg1615 [DOI] [PubMed] [Google Scholar]
- 55. Balázsi G, van Oudenaarden A, Collins JJ: Cellular decision making and biological noise: from microbes to mammals. Cell. 2011;144(6):910–25. 10.1016/j.cell.2011.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Raj A, van Oudenaarden A: Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135(2):216–26. 10.1016/j.cell.2008.09.050 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 57. Raj A, Rifkin SA, Andersen E, et al. : Variability in gene expression underlies incomplete penetrance. Nature. 2010;463(7283):913–8. 10.1038/nature08781 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 58. Feinberg AP, Irizarry RA: Evolution in health and medicine Sackler colloquium: Stochastic epigenetic variation as a driving force of development, evolutionary adaptation, and disease. Proc Natl Acad Sci U S A. 2010;107 Suppl 1:1757–64. 10.1073/pnas.0906183107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Alter MD, Hen R: Is there a genomic tone? Implications for understanding development, adaptation and treatment. Dev Neurosci. 2009;31(4):351–7. 10.1159/000216546 [DOI] [PubMed] [Google Scholar]
- 60. Alter MD, Rubin DB, Ramsey K, et al. : Variation in the large-scale organization of gene expression levels in the hippocampus relates to stable epigenetic variability in behavior. PLoS One. 2008;3(10):e3344. 10.1371/journal.pone.0003344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mojtahedi M, Skupin A, Zhou J, et al. : Cell Fate Decision as High-Dimensional Critical State Transition. PLoS Biol. 2016;14(12):e2000640. 10.1371/journal.pbio.2000640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Acar M, Becskei A, van Oudenaarden A: Enhancement of cellular memory by reducing stochastic transitions. Nature. 2005;435(7039):228–32. 10.1038/nature03524 [DOI] [PubMed] [Google Scholar]
- 63. DuPage M, Bluestone JA: Harnessing the plasticity of CD4 + T cells to treat immune-mediated disease. Nat Rev Immunol. 2016;16(3):149–63. 10.1038/nri.2015.18 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 64. Gendrel AV, Marion-Poll L, Katoh K, et al. : Random monoallelic expression of genes on autosomes: Parallels with X-chromosome inactivation. Semin Cell Dev Biol. 2016;56:100–10. 10.1016/j.semcdb.2016.04.007 [DOI] [PubMed] [Google Scholar]
- 65. Eckersley-Maslin MA, Spector DL: Random monoallelic expression: regulating gene expression one allele at a time. Trends Genet. 2014;30(6):237–44. 10.1016/j.tig.2014.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Jeffries AR, Collier DA, Vassos E, et al. : Random or stochastic monoallelic expressed genes are enriched for neurodevelopmental disorder candidate genes. PLoS One. 2013;8(12):e85093. 10.1371/journal.pone.0085093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Adegbola AA, Cox GF, Bradshaw EM, et al. : Monoallelic expression of the human FOXP2 speech gene. Proc Natl Acad Sci U S A. 2015;112(22):6848–54. 10.1073/pnas.1411270111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ben-David E, Shohat S, Shifman S: Allelic expression analysis in the brain suggests a role for heterogeneous insults affecting epigenetic processes in autism spectrum disorders. Hum Mol Genet. 2014;23:4111–24. 10.1093/hmg/ddu128 [DOI] [PubMed] [Google Scholar]
- 69. Ohlsson R, Paldi A, Graves JA: Did genomic imprinting and X chromosome inactivation arise from stochastic expression? Trends Genet. 2001;17(3):136–41. 10.1016/S0168-9525(00)02211-3 [DOI] [PubMed] [Google Scholar]
- 70. Mo A, Mukamel EA, Davis FP, et al. : Epigenomic Signatures of Neuronal Diversity in the Mammalian Brain. Neuron. 2015;86(6):1369–84. 10.1016/j.neuron.2015.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lister R, Mukamel EA, Nery JR, et al. : Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341(6146):1237905. 10.1126/science.1237905 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 72. Jaffe AE, Gao Y, Deep-Soboslay A, et al. : Mapping DNA methylation across development, genotype and schizophrenia in the human frontal cortex. Nat Neurosci. 2016;19(1):40–7. 10.1038/nn.4181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Szulwach KE, Li X, Li Y, et al. : 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011;14(12):1607–16. 10.1038/nn.2959 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 74. Kurukuti S, Tiwari VK, Tavoosidana G, et al. : CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci U S A. 2006;103(28):10684–9. 10.1073/pnas.0600326103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Leighton PA, Saam JR, Ingram RS, et al. : An enhancer deletion affects both H19 and Igf2 expression. Genes Dev. 1995;9(17):2079–89. 10.1101/gad.9.17.2079 [DOI] [PubMed] [Google Scholar]
- 76. Markenscoff-Papadimitriou E, Allen WE, Colquitt BM, et al. : Enhancer interaction networks as a means for singular olfactory receptor expression. Cell. 2014;159(3):543–57. 10.1016/j.cell.2014.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sanna CR, Li WH, Zhang L: Overlapping genes in the human and mouse genomes. BMC Genomics. 2008;9:169. 10.1186/1471-2164-9-169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. McCole RB, Oakey RJ: Unwitting hosts fall victim to imprinting. Epigenetics. 2008;3(5):258–60. 10.4161/epi.3.5.7052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Heintzman ND, Stuart RK, Hon G, et al. : Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39(3):311–8. 10.1038/ng1966 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 80. Birnbaum RY, Clowney EJ, Agamy O, et al. : Coding exons function as tissue-specific enhancers of nearby genes. Genome Res. 2012;22(6):1059–68. 10.1101/gr.133546.111 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 81. Harmston N, Lenhard B: Chromatin and epigenetic features of long-range gene regulation. Nucleic Acids Res. 2013;41(15):7185–99. 10.1093/nar/gkt499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Xu J, Carter AC, Gendrel AV, et al. : Landscape of monoallelic DNA accessibility in mouse embryonic stem cells and neural progenitor cells. Nat Genet. 2017;49(3):377–86. 10.1038/ng.3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Cavalli G, Misteli T: Functional implications of genome topology. Nat Struct Mol Biol. 2013;20(3):290–9. 10.1038/nsmb.2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Misteli T: Beyond the sequence: cellular organization of genome function. Cell. 2007;128(4):787–800. 10.1016/j.cell.2007.01.028 [DOI] [PubMed] [Google Scholar]
- 85. Parada LA, McQueen PG, Misteli T: Tissue-specific spatial organization of genomes. Genome Biol. 2004;5(7):R44. 10.1186/gb-2004-5-7-r44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Foster HA, Bridger JM: The genome and the nucleus: a marriage made by evolution. Genome organisation and nuclear architecture. Chromosoma. 2005;114(4):212–29. 10.1007/s00412-005-0016-6 [DOI] [PubMed] [Google Scholar]
- 87. Meaburn KJ, Agunloye O, Devine M, et al. : Tissue-of-origin-specific gene repositioning in breast and prostate cancer. Histochem Cell Biol. 2016;145(4):433–46. 10.1007/s00418-015-1401-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Meaburn KJ, Gudla PR, Khan S, et al. : Disease-specific gene repositioning in breast cancer. J Cell Biol. 2009;187(6):801–12. 10.1083/jcb.200909127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Stratigi K, Kapsetaki M, Aivaliotis M, et al. : Spatial proximity of homologous alleles and long noncoding RNAs regulate a switch in allelic gene expression. Proc Natl Acad Sci U S A. 2015;112(13):E1577–86. 10.1073/pnas.1502182112 [DOI] [PMC free article] [PubMed] [Google Scholar]