Abstract
Endogonales is a lineage of early diverging fungi within Mucoromycota. Many species in this order produce small sporophores (“sporocarps”) containing a large number of zygospores, and many species form symbioses with plants. However, due to limited collections, subtle morphological differentiation, difficulties in growing these organisms in vitro, and idiosyncrasies in their rDNA that make PCR amplification difficult, the systematics and character evolution of these fungi have been challenging to resolve. To overcome these challenges we generated a multigene phylogeny of Endogonales using sporophores collected over the past three decades from four continents. Our results show that Endogonales harbour significant undescribed diversity and form two deeply divergent and well-supported phylogenetic clades, which we delimit as the families Endogonaceae and Densosporaceae fam. nov. The family Densosporaceae consists of the genus Densospora, Sphaerocreas pubescens, and many diverse lineages known only from environmental DNA sequences of plant-endosymbiotic fungi. Within Endogonaceae there are two clades. One corresponds to Endogone and includes the type species, E. pisiformis. Species of Endogone are characterized by above- and below-ground sporophores, a hollow and infolded sporophore form, a loose zygosporangial hyphal mantle, homogeneous gametangia, and an enigmatic trophic mode with no evidence of ectomycorrhizal association for most species. For the other clade we introduce a new generic name, Jimgerdemannia gen. nov. Members of that genus (J. flammicorona and J. lactiflua species complexes, and an undescribed species) are characterized by hypogeous sporophores with a solid gleba, a well-developed zygosporangial hyphal mantle, heterogeneous gametangia, and an ectomycorrhizal trophic mode. Future studies on Densosporaceae and Endogonaceae will be important for understanding fungal innovations including evolution of macroscopic sporophores and symbioses with plants.
Keywords: Densosporaceae, Endogone, endophytes, Jimgerdemannia, multigene phylogeny
INTRODUCTION
Endogonales is an order of early diverging fungi that belong to the subphylum Mucoromycotina and is represented by the single family Endogonaceae. This family currently includes five genera: Endogone (Link 1809; with E. pisiformis as the type of the genus), Peridiospora (Wu & Lin 1997), Sclerogone (Warcup 1990), Youngiomyces (Yao et al. 1995) and the fossil genus Jimwhitea (Krings et al. 2012) reported from Middle Triassic formations. These fungi are rarely collected and phylogenetic affiliations of several taxa still need to be tested with molecular data. Some species of Endogone participate in ecto- or endomycorrhizal associations with diverse vascular and non-vascular plants ( Warcup 1990, Walker 1985, Field et al. 2015a, Yamamoto et al. 2017a). Similar to the obligately biotrophic arbuscular mycorrhizal fungi in Glomeromycotina (Spatafora et al. 2016), many lineages of Endogonales cannot be maintained in vitro. However, a few have been successfully isolated and maintained axenically in the laboratory with extensive efforts (Berch & Fortin 1982, Berch & Castellano 1986, Field et al. 2015b; Yamamoto et al. 2017b). Recent studies indicate that ectomycorrhizal symbioses and endosymbioses with several lineages of land plants have emerged independently more than once (Tedersoo & Smith 2013, Field et al. 2015a, Orchard et al. 2017a, Yamamoto et al. 2017a). Recently, Tedersoo & Smith (2017) considered four ectomycorrhizal lineages in Mucoromycotina: the “/densospora lineage” (Endogone group C sensu Yamamoto et al. 2017a) comprising members of the genus Densospora (McGee 1996) and uncultured ectomycorrhizal fungi associated with Eucalyptus and Nothofagus (Tedersoo et al. 2008); the “/endogone1 lineage” (Endogone group B sensu Yamamoto et al. 2015) which includes members of the Endogone flammicorona (Trappe & Gerdemann 1972) and E. lactiflua (Berkley & Broome 1846) complex; the “/endogone2 lineage” with Endogone tuberculosa (Lloyd 1918), Youngiomyces aggregatus (Yao et al. 1995) and, potentially, Sclerogone eucalypti (Warcup 1990). Endogone tuberculosa and S. eucalyptii were reported to grow axenically (Warcup 1990) but no sequence data are available for these taxa so there is no information about their phylogenetic position. The “/endogone3 lineage” is based on environmental DNA sequences putatively related to the saprotrophic E. pisiformis and generated from Quercus ectomycorrhizas (Yamamoto et al. 2017b). The ectomycorrhizal fungal lineages “/endogone2" and “/endogone3” were both within Endogone group A (sensu Yamamoto et al. 2015).
It has recently been hypothesized that mutualistic Mucoromycotina fungi related to Endogone played a crucial role during the colonization of terrestrial environments by early land plants (Bidartondo et al. 2011, Desirò et al. 2013, Field et al. 2015a). These studies highlight the ecology of Endogone-like Mucoromycotina fungi as plant-fungal symbionts, and challenge the paradigm of Glomeromycotina as the ancestral fungal mutualists of land plants (Field et al. 2015a). Similar to Glomeromycotina, Endogone can harbour Mollicutes-related endobacteria (MRE) in their mycelia and spores (Desirò et al. 2015). Even though the biology of these bacteria is still poorly understood, MRE might have played a role in the evolution of symbiotic interactions between plants and fungi (Bonfante & Desirò 2017). Given that Endogonales represent an early origin of a symbiotic nutrition mode by fungi, independent from arbuscular mycorrhizal Glomeromycotina and ectomycorrhizal Dikarya, there is now renewed interest in the diversity and evolutionary relationships of this early diverging group of fungi (Bidartondo et al. 2011, Desirò et al. 2013, Field et al. 2015b, Orchard et al. 2017b).
The genus Endogone comprises species that produce sporophores (“sporocarps”) containing a large number of zygospores. Endogone is among the earliest lineages in the fungal kingdom to produce macroscopic sporophores. The sporophores are the result of sexual reproduction by compatible apposed gametangia that lead to the production of zygospores. Sporophores of Endogone are sequestrate or enclosed and often hypogeous, although some epigeous species may produce sporophores within or upon heavily decayed wood or twigs, decaying polypore basidiomes, leaf litter, or amongst mosses and liverworts (Gerdemann & Trappe 1974, Tandy et al. 1975, Yamamoto et al. 2015). Given their range in diversity, morphology, and growth habits, the taxonomy and systematics of Endogone and related lineages have been in a state of flux over the past 200 years (Stürmer 2012).
As currently circumscribed, while Endogonales contains four extant genera (see above), there is poor resolution of the phylogenetic relationships of the taxa within the order, and still uncertainty whether Sphaerocreas pubescens (Saccardo 1882), members of Densospora (McGee 1996), and numerous Endogone-related Mucoromycotina associated with plants belong to Endogonales. Even though they are distributed across temperate and tropical habitats in the Northern and Southern Hemispheres, sporophores of most of these fungi are rarely collected and molecular data are limited or not available. Further, there are idiosyncratic challenges when working with Endogonales rDNA, because ITS rDNA does not amplify or sequence well, or is degenerate (Tedersoo et al. 2016). Consequently, Endogonales are often conspicuously underrepresented in environmental molecular surveys and databases (e.g. GenBank) that rely on rDNA markers (Větrovský et al. 2015). Moreover, when detected, these fungi are difficult to place within a phylogenetic and taxonomic framework.
To address these limitations, we generated a multigene phylogeny for Endogonales based on rDNA (18S; 28S) and protein coding genes (EF1-α; RPB2) from a global sampling of Endogone sporophores, and integrated available Endogonales and environmental DNA sequences into this phylogeny. We also employed the RPB2 gene for the first time as a marker to enhance the phylogenetic resolution of Endogonales. Our results provide a phylogenetic placement of two Endogone species that were previously unresolved and define two families within Endogonales: Endogonaceae and Densosporaceae fam. nov. Within Endogonaceae, we delimit two deeply divergent monophyletic lineages that differ in morphology, sporing habit, and potentially also ecology. We introduce Jimgerdemannia gen. nov. to accommodate one of these two lineages, and also synonymize Youngiomyces with Endogone s. str.
METHODS
Collections sampled
Dried fungarium specimens of Endogone flammicorona, E. incrassata (Thaxter 1922), Endogone lactiflua, E. oregonensis (Gerdmann & Trappe 1974), E. pisiformis (Link 1908), E. tuberculosa, and unidentified Endogone were obtained from private collections or the following institutions: University of Florida Herbarium (FLAS), Michigan State University Herbarium (MSC), National Herbarium of Victoria (MEL), Oregon State University Herbarium (OSC), and Western Australian Herbarium (PERTH). In total, 45 collections of Endogone from Australia, Italy, Mexico, the United Kingdom, and USA were analyzed (Table 1).
Table 1.
List of the Endogonaceae sporophores analyzed in this study and available relative information about voucher as herbarium and/or collector number (when both are available, collector numbers are in parentheses), site and date of collection.
| Species | Voucher/Collector No | Collection Site | Collection Date |
|---|---|---|---|
| Endogone incrassata | MEXU 26467 | Volcano Nevado de Colima National Park, Jalisco, Mexico | 25 Sep. 2009 |
| Endogone incrassata | T32417 | Cofre de Perote, Veracruz, Mexico | 17 Sep. 2007 |
| Endogone incrassata | T32492 | San José Teacalco, Tlaxcala, Mexico | 21 Sep. 2007 |
| Endogone oregonensis | OSC 130614 | Polk, Oregon, USA | 23 Feb. 2008 |
| Endogone oregonensis | T36235 | Benton County, Oregon, USA | 14 March 2013 |
| Endogone oregonensis | AD153 | Monmouth, Oregon, USA | 29 Dec. 2015 |
| Endogone pisiformis | AD152 | Corvallis, Oregon, USA | 11 March 2017 |
| Endogone pisiformis | FLAS F-59194 (MES1451) | Bartlett Experimental Forest, Carroll County, New Hampshire, USA | 11 Aug. 2015 |
| Endogone pisiformis | OSC 80931 (T28028) | Benton, Oregon, USA | 7 Feb. 2002 |
| Endogone pisiformis | OSC 112172 (T31477) | Washington, USA | 24 April 2006 |
| Endogone pisiformis | OSC 149839 (T37049) | White Mountain National Forest, Carroll County, New Hampshire, USA | 17 Aug. 2015 |
| Endogone pisiformis | T37093 | Lane County, Oregon, USA | 21 May 2013 |
| Endogone sp. | MEL 2024690 | Loftia Recreation Park, Adelaide Hills, Australia | 26 Aug. 1984 |
| Endogone sp. | FLAS F-59071 (MES866) | Ordway-Swisher Reserve, Melrose, Florida, USA | 23 Feb. 2015 |
| Endogone sp. | PERTH 7567251 | Atherton, Queensland, Australia | 7 May 1991 |
| Endogone sp. | PERTH 7591853 | Cape York, Australia | - |
| Endogone sp. | PERTH 7603037 | Bluewater Park, Queensland, Australia | 13 April 1989 |
| Endogone sp. | PERTH 7648049 | Dwellingup, Australia | 10 May 2002 |
| Endogone sp. | PERTH 7648847 | Dwellingup, Australia | 25 June 2002 |
| Endogone sp. | PERTH 7672527 | Mount Windsor Tableland, Queensland, Australia | 2 Feb. 1992 |
| Endogone sp. | PERTH 8092931 | Leeuwin-Naturaliste National Park, Australia | 17 May 2007 |
| Endogone sp. | PERTH 8127840 | Karakamia Sanctuary, Australia | 4 Oct. 2006 |
| Endogone sp. | PERTH 8473986 | Boorabbin National Park, Australia | 20 Aug. 2009 |
| Endogone sp. | T26631 | Bournda National Park, Australia | 22 Nov. 2000 |
| Endogone tuberculosa | OSC 146000 (T34145) | Australian Capital Territory, Australia | 14 May 2010 |
| Jimgerdemannia flammicorona | AD002 | Veglio, Piemonte, Italy | 7 Sep. 2013 |
| Jimgerdemannia flammicorona | MSC 0242545 (AD239) | Lake Lansing Park North, Haslett, Michigan, USA | 7 Oct. 2016 |
| Jimgerdemannia flammicorona | MSC 0242546 (AD244) | Lake Lansing Park North, Haslett, Michigan, USA | 7 Oct. 2016 |
| Jimgerdemannia flammicorona | AD245 | Lake Lansing Park North, Haslett, Michigan, USA | 7 Oct. 2016 |
| Jimgerdemannia flammicorona | GB716 | Lake Lansing Park North, Haslett, Michigan, USA | 13 Sep. 2015 |
| Jimgerdemannia flammicorona | MSC 0242548 (GB737) | Lake Lansing Park North, Haslett, Michigan, USA | 17 Sep. 2015 |
| Jimgerdemannia flammicorona | RH932 | Ledges State Park, Iowa, USA | 27 June 2009 |
| Jimgerdemannia flammicorona | T33849 | Bosque la Primavera, Jalisco, Mexico | 2 Oct. 2009 |
| Jimgerdemannia flammicorona | T33851 | Bosque la Primavera, Jalisco, Mexico | 2 Oct. 2009 |
| Jimgerdemannia lactiflua | AD001 | Veglio, Piemonte, Italy | 7 Sep. 2013 |
| Jimgerdemannia lactiflua | MSC 0242547 (AD251) | Mason, Michigan, USA | 13 Oct. 2016 |
| Jimgerdemannia lactiflua | AD256 | Mason, Michigan, USA | 13 Oct. 2016 |
| Jimgerdemannia lactiflua | AM2190 | Cavola, Emilia Romagna, Italy | 22 July 2000 |
| Jimgerdemannia lactiflua | CH9142 | Derbyshire, United Kingdom | 12 Nov. 2012 |
| Jimgerdemannia lactiflua | T32409 | Cofre de Perote, Veracruz, Mexico | 17 Sep. 2007 |
| Jimgerdemannia lactiflua | T32490 | San José Teacalco, Tlaxcala, Mexico | 21 Sep. 2007 |
| Jimgerdemannia lactiflua | T32544 | Huamantla, Tlaxcala, Mexico | 23 Sep. 2007 |
| Jimgerdemannia lactiflua | T32674 | Miquihuana, Tamaulipas, Mexico | 2 Aug. 2008 |
| Jimgerdemannia sp. | T34745-A | Main Ranges National Park, Queensland, Australia | 3 June 2010 |
| Jimgerdemannia sp. | T34745-B | Main Ranges National Park, Queensland, Australia | 3 June 2010 |
Molecular analyses
A small fragment of gleba tissue from each sporophore was sampled and genomic DNA extracted with a CTAB-based method (Doyle 1991). All PCR reactions were carried out with DreamTaq Green PCR Master Mix (Thermo Fisher Scientific, Waltham, MA). A fragment of the 18S rRNA gene was amplified with primers EndAD1f (Desirò et al. 2013) and EF3 (Smit et al. 1999). For samples that failed to amplify, PCR products were diluted 1:10 in sterile water and used as template for a semi-nested PCR with the reverse primer EndAD2r (Desirò et al. 2013). The PCR conditions followed those of Desirò et al. (2013). The ITS2 region and a partial fragment of the 28S rRNA gene were amplified with a semi-nested PCR approach. The first PCR was carried out with the new primers EndAD7f (5’-CTGCTAAATAGYTAKGCCAAC-3’, designed on the 18S rRNA gene) and EndAD28Sr (5’- CATTAMGYCAGCGACCYAAG-3’, designed on the 28S rRNA gene). The cycling conditions were: an initial step at 95 °C for 5 min, 30 cycles at 95 °C for 30 s, 55 °C for 30 s, 72 °C for 1 min 15 s and a final extension step at 72 °C for 7 min. The PCR amplicons were then diluted and used as template for the second PCR with the forward primers ITS3 (White et al. 1990), fITS9 (Ihrmark et al. 2012) or LR0R (Vilgalys & Hester 1990) in combination with EndAD28Sr. The cycling conditions for the second step of the semi-nested PCR were: an initial step at 95 °C for 5 min, 27 cycles at 95 °C for 30 s, 55 °C for 30 s, 72 °C for 1 min and a final extension step at 72 °C for 7 min. A partial fragment of the elongation factor 1 alpha (EF1-α) gene was amplified with the primers 983F and 2218R (Rehner & Buckley 2005). The cycling conditions in this case were: an initial step at 95 °C for 5 min, 35 cycles at 95 °C for 30 s, 55 °C for 30 s, 72 °C for 1 min and a final extension step at 72 °C for 7 min. When amplification was not successful, PCR products were diluted and used as template for a semi-nested PCR with the new forward primer EndADef1f (5’-TWCACVCTYGGYGTGCGTC-3’) and PCR conditions as detailed above with 27 cycles. A partial fragment of the second largest subunit of RNA polymerase II (RPB2) gene was amplified with the new primers RPB2AD3f (5’-GAAGGTCARGCKTGYGGTC-3’) or RPB2AD2f (5’-ATTCATCCSAGTATGATTC-3’) and RPB2AD1r (5’- AASGGTGTRGCRTCACCTTC-3’). The cycling conditions were: an initial step at 95 °C for 5 min, 35 cycles at 95 °C for 30 s, 55 °C for 30 s, 72 °C for 1 min and a final extension step at 72 °C for 7 min. When amplification with these two primer combinations was unsuccessful, a semi-nested PCR was attempted with forward primers RPB2AD2f or RPB2AD1f (5’- ATGGARGARTTTGARAAGCC-3’). The semi-nested PCR conditions were as detailed above with 27 cycles. All PCR amplicons were purified and either sequenced directly or cloned with the TOPO-TA cloning kit (Thermo Fisher Scientific). The PCR amplicons or cloned fragments were sequenced with an ABI 3730XL DNA Analyzer (Applied Biosystems, Foster City, CA). The DNA sequences generated in this study are deposited in GenBank (MF478989-MF479111).
Phylogenetic analyses
Sequences were assembled and curated in Geneious v.8.1.7 (Kearse et al. 2012), and were used as queries for conducting BLAST searches (Altschul et al. 1990). Four single-locus datasets were created (i.e. 18S, 28S, EF1-α, RPB2). The datasets included 123 sequences generated in this study and 187 obtained from the NCBI and UNITE databases. In particular, we used sequences generated from the recently described Bifiguratus adelaidae (Torres-Cruz et al. 2017), Calcarisporiella thermophila, Densospora nuda and D. solicarpa, several Endogone spp., Sphaerocreas pubescens, Youngiomyces aggregatus (syn. E. aggregata, see below) and other undescribed Mucoromycotina spp., Mortierella verticillata (NRRL 6337) (Mortierellomycotina) was included as outgroup in single-locus analyses. The 18S, 28S, EF1-α and RPB2 datasets included 140, 67, 65 and 34 taxa, respectively (Supplementary Table 1). Datasets were aligned with MAFFT (Katoh & Standley 2013) or MUSCLE (Edgar 2004) and then manually edited. The ITS2 region and introns of EF1-α and RPB2 genes were excluded from subsequent analyses. Each alignment was then trimmed with GBlocks v.0.91b (Castresana 2000) using the least stringent conditions. The single-locus alignments had a total of 1 435 (18S), 877 (28S), 839 (EF1-α), and 904 (RPB2) nucleotide positions. We also created a concatenated dataset that included, when available, all four loci for 56 taxa (Supplementary Table 1). Densospora solicarpa (DAR 69421; DAR 74956; Tedersoo et al. 2016), Sphaerocreas pubescens (NBRC 109377) and an unidentified Mucoromycotina sp. (MES1534; Truong et al. 2017) were used as outgroups in the multi-locus analysis. Missing loci were treated as missing data. Because the four single-locus trees appeared largely congruent, they were combined after aligning and trimming steps described above. The concatenated alignment had a total of 3568 nucleotide positions. Trimmed single-locus and concatenated alignments were deposited in TreeBase (http://purl.org/phylo/treebase/phylows/study/TB2:S21409).
Prior to phylogenetic inferences, best-fit nucleotide substitution models were estimated for each dataset by using jModelTest v.2.1.9 (Darriba et al. 2012). Phylogenetic analyses were carried out using MrBayes v.3.2.6 (Ronquist et al. 2012) and RAxML v.8.2.4 (Stamatakis 2014). A Markov chain Monte Carlo was run for five million generations under the TIM3+I+G (18S, 28S), TrN+G (EF1-α) and TrNef+I (RPB2) nucleotide substitution models. A partitioned analysis was carried out for the concatenated dataset and run for five million generations using the models as above for the 28S and RPB2 partitions; the TrN+I+G and TIM2ef+G nucleotide substitution models were applied for the 18S and EF1-α partitions. Maximum likelihood analyses were carried out under the GTRCAT nucleotide substitution model with the “autoMR” option for bootstrap replicates (Pattengale et al. 2010).
RESULTS
Specimens examined and phylogenetic analyses
In total, 45 Endogone specimens were analyzed in this study. Thirty-three specimens were identified morphologically and 12 were unidentified (Table 1). Previously used primer pairs (Bidartondo et al. 2011, Desirò et al. 2013) together with new primer combinations designed in this study allowed us to amplify all of the four target genes from 19 out of 45 specimens. For the other 26 specimens, three (12 specimens), two (13 specimens) or one (1 specimen) gene sequences were successfully generated (Supplementary Table 1). Most missing gene sequence data were of the ITS-28S rRNA region, which is known to be problematic for Endogonales (Tedersoo et al. 2016). The RPB2 gene also proved challenging to amplify and sequence. Although primer bias cannot be excluded, multiple factors such as age and preservation mode of the samples may affect DNA integrity and therefore PCR success (Osmundson et al. 2013). Indeed, most amplification failures were from specimens collected in the 1980s and 1990s whereas three or four target genes were amplified for most of the specimens collected within the last 10 years.
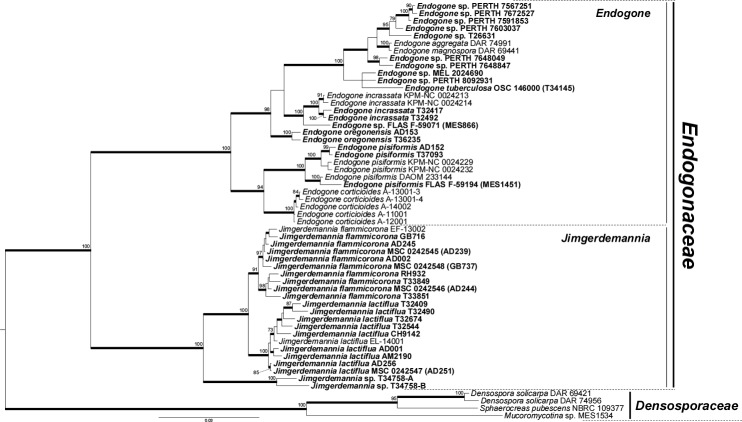
Single- and multi-locus phylogenetic reconstructions showed two deeply divergent and well-supported monophyletic groups within Endogonaceae. The first group corresponded to the genus Endogone whereas the second represents the new genus Jimgerdemannia. The Endogone clade included the type species, E. pisiformis, together with Youngiomyces aggregatus (syn. E. aggregata; see below), E. corticioides, E. incrassata, E. magnospora, E. oregonensis, E. tuberculosa, all the unidentified Endogone specimens investigated, and several environmental sequences. The Endogone group was divided into two clades. The first clade encompassed E. pisiformis and its sister lineage E. corticioides (Figs 1–2, Supplementary Fig. 1–2). Interestingly, putative ectomycorrhizal Endogone sequences (Yamamoto et al. 2017a) were nested within the pisiformis-corticiodes clade together with a clade constituted by environmental DNA sequences generated from fungal symbionts of liverworts (Bidartondo et al. 2011) (Fig. 2, Supplementary Fig. 2). The second clade included E. aggregata, E. incrassata, E. magnospora, E. oregonensis, E. tuberculosa, and unidentified Endogone specimens. The position of the taxa within this clade was not clearly resolved by single-locus analyses, however, our multigene phylogeny supported the placement of E. oregonensis as the most basal taxon within this clade and E. incrassata as sister group to the Endogone species complex that contains E. aggregata, E. magnospora, and E. tuberculosa (Fig. 1).
Fig. 1.
Phylogenetic reconstruction of Endogonaceae based on a concatenated dataset of 18S, 28S, EF1-α and RPB2 sequences. The family Densosporaceae, represented by Densospora solicarpa, Sphaerocreas pubescens and an unidentified Mucoromycotina sp., was used as outgroup. The tree shows the topology obtained with the Bayesian method; branches with Bayesian posterior probabilities ≥0.95 are thickened and ML bootstrap support values ≥70 are shown. Specimens analyzed in this study are in bold.
Fig. 2.


Phylogenetic reconstruction of Endogonales based on 18S rDNA sequences. The node “D” defines the family Densosporaceae. Mortierella verticillata was used as outgroup. The tree shows the topology obtained with the Bayesian method; branches with Bayesian posterior probabilities ≥0.95 are thickened and ML bootstrap support values ≥70 are shown. Sequences generated in this study are in bold.
The Jimgerdemannia clade included J. flammicorona and J. lactiflua, and an undescribed Jimgerdemannia sp. (T34758-A and T34758-B). In contrast to the Endogone clade, the relationships within Jimgerdemannia were well resolved; J. flammicorona and J. lactiflua are sister groups, while the undescribed Jimgerdemannia sp. (T34758-A and T34758-B) is sister of the flammicorona-lactiflua clade (Fig. 1, Supplementary Fig. 1, 2, 3). Curiously, phylogenetic reconstructions showed two distinct J. flammicorona clades. Furthermore, several environmental fungal sequences retrieved from hornworts and liverworts clustered together with Jimgerdemannia sp. (T34758-A and T34758-B) or were closely related to it (“group E” sensu Desirò et al. 2013) (Fig. 2, Supplementary Fig. 1–2). A third group (“groups D and H“ sensu Desirò et al. 2013) comprised Mucoromycotina spp. associated with bryophytes and ferns (Bidartondo et al. 2011; Desirò et al. 2013; Rimington et al. 2015; Field et al. 2016) and a fine root endophyte (Orchard et al. 2017a) nested between the Endogone and Jimgerdemannia clades (Fig. 2, Supplementary Fig. 2). Finally, Jimgerdemannia was sister of a clade of hornwort- and liverwort-associated fungi (“group G” sensu Desirò et al. 2013), whereas a second clade encompassing DNA sequences retrieved from the liverwort Neohodgsonia mirabilis was the first diverging clade of Endogonaceae (Fig. 2).
All the sporophore and environmental DNA sequences described above were included within a monophyletic clade referred to here as Endogonaceae. A more distantly related monophyletic sister group mostly consisted of environmental Mucoromycotina sequences generated from undescribed fungal symbionts of early diverging land plants. Several well-supported phylogroups were present in this clade. Fine root endophytes (Orchard et al. 2017a) clustered within two phylogroups whereas a third very diverse phylogroup encompassed numerous environmental Mucoromycotina sequences, Densospora nuda, D. solicarpa, Sphaerocreas pubescens, an unidentified Mucoromycotina sp. (MES1534) (Truong et al. 2017), and uncultured ectomycorrhizal Mucoromycotina (Tedersoo et al. 2008). Below we circumscribe this monophyletic sister lineage that has been referred to previously as the Sphaerocreas-Densospora clade (Yamamoto et al. 2015, Truong et al. 2017) as the new family Densosporaceae.
TAXONOMY
Densosporaceae Desirò, M.E. Sm., Bidartondo, Trappe & Bonito, fam. nov.
MycoBank MB821851
Type genus: Densospora McGee 1996.
Diagnosis: The family Densosporaceae is erected here to apply to all descendants of the node “D” defined in the phylogeny (Fig. 2) as the terminal Densosporaceae clade. We define Densosporaceae as the least inclusive clade containing the genus Densospora and Sphaerocreas pubescens (sensu Hirose et al. 2014), but also environmental DNA sequences generated from fungal symbionts of non-vascular and vascular plants (JF414222, JF414224, KC708392, KC708404, KC708409, KC708417, KC708436, KJ952212, KJ952213, UDB002714).
Discussion: Most of the phylogenetic diversity within Densosporaceae is known from environmental DNA sequences generated from fungal symbionts of bryophytes, rather than from sporophore collections. Consequently, we have used these environmental DNA sequences to help define this family. Taxa within Densosporaceae have previously been classified as belonging to Endogonales and Glomerales, and some are currently classified as incertae sedis (http://www.indexfungorum.org/). However, single-locus and multigene phylogenetic reconstructions resolve Densosporaceae as a distant sister clade of Endogonaceae within the order Endogonales. Densosporaceae encompasses multiple divergent monophyletic clades that mostly comprise undescribed Mucoromycotina lineages, but also species such as Densospora nuda, D. solicarpa and Sphaerocreas pubescens that produce sporophores. Like Endogonaceae, fungi in Densosporaceae are frequently associated with liverworts, hornworts (Bidartondo et al. 2011, Desirò et al. 2013, Hirose et al. 2014), lycopods and ferns (Rimington et al. 2015). Some short NGS sequences from fine root endophytes (Glomus tenue s. lat.) associated with Trifolium roots (Orchard et al. 2017a) also cluster within Densosporaceae. Furthermore, sequences from ectomycorrhizal fungi associated with Eucalyptus and Nothofagus (Tedersoo et al. 2008) and an unidentified Mucoromycotina sp. (MES1534; Truong et al. 2017) are placed within this clade.
Jimgerdemannia Trappe, Desirò, M.E. Sm., Bonito & Bidartondo, gen. nov.
MycoBank MB821846
Etymology: In honour of James (“Jim”) W. Gerdemann, researcher and professor, who was instrumental in bringing Endogonales and Glomerales into modern taxonomy before the advent of DNA and phylogenetic analyses.
Type species: Jimgerdemannia flammicorona (Trappe & Gerd.) Trappe et al. 2017.
Diagnosis: Differs from Endogone by pairing a large gametangium with a small one, the tip of the small one fusing to the side of the large one.
Discussion: The zygospore typically buds from the tip of the large gametangium or occasionally from the junction of the two gametangia. Spores typically becoming enveloped in tightly appressed hyphae that fuse at maturity with the spore wall to form a surface ornamentation. Spores are distributed randomly among the glebal hyphae, and not clustered. Phylogenetic analyses confirm that Jimgerdemannia and Endogone s. str. represent separate lineages within Endogonaceae. All known Jimgerdemannia species putatively form ectomycorrhizas with various species of Pinaceae and the sporophores of taxa of this group are usually hypogeous among host rootlets.
Jimgerdemannia flammicorona (Trappe & Gerd.) Trappe, Desirò, M.E. Sm., Bonito, Bidartondo, comb. nov.
MycoBank MB821847
Basionym: Endogone flammicorona Trappe & Gerd., Trans. Brit. Mycol. Soc. 59: 405 (1972).
Jimgerdemannia lactiflua (Berk. & Broome) Trappe, Desirò, M.E. Sm., Bonito & Bidartondo, comb. nov.
MycoBank MB 821853
Basionym: Endogone lactiflua Berk. & Broome, Ann. Mag. Nat. Hist., ser. 1, 18: 81 (1846).
Discussion: Jimgerdemannia flammicorona and J. lactiflua have been compared in detail by Trappe & Gerdemann (1972), and demonstrated to form ectomycorrhizas with Pinus contorta, P. lambertiana, P. monticola, P. peuce, P. radiata, P. strobus and Pseudotsuga menziesii. These taxa are common and sporulate in beds of Pinaceae seedlings in Europe and North America (Fassi & Palenzona 1969, Fassi et al. 1969, Chu-Chou & Grace 1979, 1984). Jimgerdemannia lactiflua was reported to form mycorrhizas and produce sporophores on three-month old Pinus contorta nursery seedlings (Walker 1985). These species are widely distributed in Pinus radiata plantations in Australia and New Zealand, where they were likely accidentally introduced with imported seedlings in the early 20th century (Chu-Chou & Grace 1984, Trappe, unpubl.). Both species commonly appear in faeces (scats) of bush rats (Rattus fuscipes) in Pinus radiata plantations in Australia (Trappe, unpubl) and are also found in small mammal droppings in North America (Maser et al. 1978).
Endogone Link, Ges. Naturf. Freunde Berlin Mag. Neuesten Entdeckt. Gesammten Naturk. 3: 33 (1809).
Type species: Endogone pisiformis Link 1809.
Synonym: Youngiomyces Y.-J. Yao, Kew Bull. 50: 350 (1995).
Diagnosis: Differs from Jimgerdemannia by pairing two gametangia of similar size that fuse near their tips and the zygospores mostly then budding from the junction of the two.
Discussion: Spores mostly not enveloped in appressed hyphae, or, if so then the hyphae not fused with the spore wall to form a surface ornamentation at maturity. Spores either distributed randomly among the glebal hyphae or clustered in discrete aggregations separated by hyphal tissue. Phylogenetic analyses reveal that Endogone s. str. (as circumscribed here) and Jimgerdemannia represent separate lineages within Endogonaceae. In contrast to the ectomycorrhizal Jimgerdemannia, most Endogone s. str. species are either putatively saprotrophic or perhaps fungicolous and may be either hypogeous or epigeous, the latter sporing on various substrates. Ectomycorrhizas putatively identified as related to E. pisiformis were detected in oak forest in Japan (Yamamoto et al. 2017a). Even though their functional role is unknown, some taxa within the Endogone clade have been documented as symbionts of liverworts.
Endogone pisiformis, undoubtedly the most widely distributed species of the genus, has been reported from Asia, Europe, and North America and proliferates on diverse substrates, including forest litter, brown-cubical rotted wood, decaying polypore basidiomes, and among mosses. It sometimes produces sporophores at the edge of melting snowbanks or immersed in meltwater on saturated organic matter. It can grow in pure culture (Jabaji-Hare & Charest 1987) and produce zygospores in vitro in the absence of mycorrhiza formation (Berch & Castellano 1986). Endogone tuberculosa and Youngiomyces aggregatus (i.e. E. aggregata) were reported to grow on agar media (Warcup 1990). However, molecular tools should be applied to validate these findings. Yamamoto et al. (2017b) showed a limited vegetative growth of E. corticioides in pure culture. Warcup (1990) inoculated various Eucalyptus spp. with Y. aggregatus (i.e. E. aggregata) and E. tuberculosa. Although ectomycorrhizas formed, the methods used did not preclude contamination by other fungi, so the results were inconclusive.
Endogone carolinensis (Y.-J. Yao) Desirò, M.E. Sm., Bonito, Bidartondo & Trappe, comb. nov.
MycoBank MB821852
Basionym: Youngiomyces carolinensis Y.-J. Yao, Kew Bull. 50: 351 (1995).
Discussion: The key feature used to distinguish Youngiomyces from Endogone was that in Youngiomyces the zygosporangium has two, three, or four openings (Yao et al. 1995). However, E. pisiformis, and rarely E. incrassata, have been reported to have zygosporangia with two openings (Yamamoto et al. 2015). Although the type of the genus Youngiomyces (Y. caroliniensis) was not included in our phylogeny, the phylogenetic position of Y. aggregatus is nested deep within the Endogone clade (Fig. 1), close to E. magnospora and other Endogone species devoid of multiple openings in their zygosporangia. For those reasons, and in line with Yamamoto et al. (2015), we synonymize Youngiomyces with Endogone s. str. This requires recombining the type species Y. carolinensis into Endogone s. str. and returning the other species assigned to Youngiomyces to their original status as Endogone species: E. aggregata, E. multiplex, and E. stratosa.
DISCUSSION
Single-locus and multigene phylogenetic analyses using rDNA and single-copy protein-coding genes resolved the phylogeny of Endogonales into two monophyletic clades showing a deep divergence with significant support. Based on these results, we revised the taxonomy of Endogonales and introduced the new family Densosporaceae. We also introduced the new genus Jimgerdemannia within Endogonaceae and synonymized the genus Youngiomyces with Endogone s. str. The new genus Jimgerdemannia includes two species, J. flammicorona and J. lactiflua, and an undescribed Jimgerdemannia lineage, which is sister to the flammicorona-lactiflua clade. Furthermore, we also provided a more detailed phylogenetic placement for E. oregonensis and placed E. tuberculosa within the genus Endogone: the single-locus and multigene phylogeny placed E. oregonensis and E. turberculosa together with E. aggregata, E. incrassata, E. magnospora and several unidentified Endogone spp. within a clade that is sister to the corticioides-pisiformis clade.
Jimgerdemannia appears sister to Endogone. However, when environmental Mucoromycotina sequences were included in the phylogenetic reconstructions, several additional novel clades were detected within Endogonaceae. Furthermore, Jimgerdemannia was sister to a clade consisting of DNA sequences belonging to fungi associated with liverworts and hornworts (“group G” sensu Desirò et al. 2013). Similarly, Endogone is nested within a monophyletic group that also included two clades of fungal symbionts of liverworts, hornworts and ferns (“groups D and H” sensu Desirò et al. 2013). Sequences from a taxon of fine root endophytes (Glomus tenue) associated with roots of Trifolium (Orchard et al. 2017a) clusters within one of these two clades (“group D” sensu Desirò et al. 2013). However, the fine root endophyte sequences are short NGS reads (ca. 200 bp) so their placement will need to be revisited based on longer sequences for better resolution. Lastly, all other Endogonaceae sequences are sister to a clade of fungal sequences retrieved from the thallus of the liverwort Neohodgsonia mirabilis (Field et al. 2016).
A number of points can be made regarding the morphology, sporing habit and ecology of Jimgerdemannia. In particular, species in this genus have a developed zygosporangial hyphal mantle and heterogeneous gametangia. They usually produce below ground sporophores and are considered ectomycorrhizal with Pinaceae (Fassi et al. 1969, Fassi & Palenzona 1969, Chu-Chou & Grace 1979, Walker 1985; Warcup, 1990). Interestingly, our results show that sequences from fungal symbionts of hornworts and liverworts are closely related to an undescribed Jimgerdemannia sp. This indicates that this Jimgerdemannia species might engage in symbiotic interactions with early diverging land plant lineages. We cannot rule out the possibility that members of this genus can form ectomycorrhizas on some hosts and be endophytic or form other biotrophic interactions with other hosts.
In contrast, taxa in Endogone have a loose zygosporangial hyphal mantle and homogeneous gametangia. They may have hypogeous or epigeous sporophores. Most Endogone species are saprotrophic or perhaps fungicolous and have generally been considered non-ectomycorrhizal. However, Warcup (1990) reported mycorrhiza formation on Eucalyptus spp. inoculated with E. aggregata (syn. Youngiomyces aggregatus) and E. tuberculosa. Although ectomycorrhizas formed, molecular confirmation is needed to verify the fungal identity and exclude potential contamination by other fungi. Curiously, Yamamoto et al. (2017a) recently reported ectomycorrhizas on two oak root tips putatively formed by a novel lineage related to E. corticioides and E. pisiformis. It is interesting to note that phylogenetic reconstructions based on the EF1-α gene placed sequences from fungal symbionts associated with the liverworts Treubia lacunosa and T. pygmaea (Bidartondo et al. 2011) close to the ones retrieved from oak roots, suggesting the possibility that some Endogone species might associate with multiple plant partners.
Phylogenetic reconstructions placed Youngiomyces aggregatus (i.e. E. aggregata), the only representative of the genus Youngiomyces in our study, within Endogone. It has recently been shown that the key morphological character used to distinguish Youngiomyces from Endogone (i.e. a zygosporangium with two to four openings) can also be observed in E. pisiformis and rarely in E. incrassata (Yamamoto et al. 2015). Based on this rationale, we synonymize Youngiomyces with Endogone s. str.
Densospora nuda, D. solicarpa, and Sphaerocreas pubescens, short DNA sequences from environmental sequencing of fine root endophytes (Glomus tenue), and numerous environmental Mucoromycotina sequences clustered together within a diverse and well supported monophyletic clade that we name Densosporaceae here. This group has previously been referred to as the Sphaerocreas-Densospora clade (Yamamoto et al. 2015, Truong et al. 2017). Due to limited taxon and/or gene sampling in previous (Lin et al. 2014; Spatafora et al. 2016) and present studies, there is still uncertainty regarding the diversity and relationships among Densosporaceae and the phylogenetic position of Endogonales within Mucoromycotina. However, in our analyses the monophyletic Densosporaceae is sister to the monophyletic Endogonaceae. Most of the DNA sequences included in Densosporaceae were undescribed Mucoromycotina. Densospora, Sphaerocreas pubescens, an unidentified Mucoromycotina sp. (MES1534; Truong et al. 2017), and uncultured ectomycorrhizal Mucoromycotina spp. (Tedersoo et al. 2008) clustered within a monophyletic clade together with several liverwort- and hornwort-associated lineages. The remaining clades were formed by fungal DNA sequences retrieved from liverworts, hornworts, lycopods, and ferns, whose identity is unknown. Some of these apparently undescribed taxa are also similar to short DNA reads of fine root endophytes in the Glomus tenue species complex (Orchard et al. 2017a).
The presence of several clades constituted only by environmental DNA sequences indicates that there are several undescribed genera and species in Endogonales, so additional phylogenetic studies are needed based on additional fresh specimens. In particular, sequence data are needed for several described but rare species of Endogone, Peridiospora, and Sclerogone to determine whether or not these fungi really belong to Endogonaceae. Species of Peridiospora and Sclerogone were not sampled in this study and sequences for these two fungal lineages were not available in public databases. However, preliminary data indicate that Peridiospora might be phylogenetically related to Glomeromycotina, not Mucoromycotina (C Walker & MI Bidartondo, unpubl.). Additional molecular data from putatively related taxa could help in identifying and providing a taxonomic placement for some of these diverse and enigmatic fungi, and further clarify the taxonomy of Endogonales and Mucoromycotina.
Similar to Endogone and Jimgerdemannia, Densospora and Sphaerocreas pubescens produce sporophores. Densospora tubiformis can form ectomycorrhizas (Warcup 1985; McGee 1996) and Densospora sporophores are often found on the soil surface. However, S. pubescens sporulates on decaying wood or twigs, and also leaf litter or rotten basidiomes of Polyporaceae. This suggests a possible fungicolous behaviour but little information is available on this fungus (Hirose et al. 2014). Endogone species also sporulate on gametophores of mosses as well as rotten wood or twigs, and rarely on old polypore basidiomes (Yamamoto et al. 2015). Sphaerocreas pubescens was also considered to be a saprotroph on the basis of failed mycorrhizal synthesis experiments (McGee & Trappe 2002). However, sequences retrieved from fungal symbionts associated with liverworts and hornworts suggest that these fungi are biotrophic (Hirose et al. 2014).
Notwithstanding that their ecology remains poorly understood, our results support the hypothesis that lineages of Mucoromycotina co-evolved independently with different lineages of vascular and non-vascular plants, among them the early diverging bryophytes whose ancestors were involved in the colonization of terrestrial environments. As such, Endogonales provides essential context for studying the origin, evolution and biology of plant-fungal symbioses. However, many challenges remain regarding Endogonales, including difficulties in collecting sporophores and detecting these fungi in environmental surveys, and the integration of environmental data with collection-based datasets.
Further sampling and research is needed to provide a more comprehensive investigation of Endogonales. This group of fungi requires urgent attention to: (1) provide a formal name to the clades of environmental DNA sequences related to Endogone, Jimgerdemannia, and Densosporaceae; (2) provide taxonomic descriptions of the potentially novel species of Endogone and Jimgerdemannia used in this study; (3) generate molecular data in order to place species of Peridiospora and Sclerogone within a phylogenetic framework; (4) investigate the fine root endophytes of vascular plants and the undescribed Mucoromycotina spp. of early diverging land plants for taxonomic treatment; (5) shed light on the ecophysiology and trophic status of the various different lineages within Endogonales; and (6) clarify taxonomic inconsistencies between Sphaerocreas pubescens (sensu Hirose et al. 2014) and Sclerocystis in Glomeromycotina.
As we enter the “-omics” age it will be possible to use both fungal genomes and plant microbiomes to facilitate future studies of Endogonales, which are rapidly emerging as a key group of fungi to study. It will be critical to understand more about the evolution and trophic modes of this group of fungi in order to elucidate the origin and evolutionary history of plant-fungal symbioses. The ecology of many species of Densosporaceae and Endogonaceae is likely to be mycorrhizal (or mycorrhiza-like in rootless plants) involving carbon and nutrient transfer between fungus and plant host. This has only been tested thus far for a few liverwort species (Field et al. 2015b, 2016). Genomic data have the potential to determine the physiological abilities and differences among symbiotic and free-living species, and to clarify the position of Endogonales within the kingdom Fungi (i.e. their relationships to other Mucoromycota and particularly to the arbuscular mycorrhizal Glomeromycotina). Genome data will also enable functional metabolomic and transcriptomic studies of these fungi in symbiosis with their hosts in order to decipher their intriguing plant-fungal interactions.
Acknowledgments
We wish to thank Amer Montecchi, Begoña Aguirre-Hudson, Carolina Piña-Páez, Caroline Hobart, Christopher Walker, Efren Cazares, Kris Jacobson, Michael Castellano, Peter McGee, Roberto Garibay-Orijel, Rosanne Healy, R. Stephens, S. Loring, Michigan State University Herbarium, National Herbarium of Victoria, New Zealand Fungal and Plant Disease Collection, Oregon State University Herbarium, Royal Botanic Gardens Kew, Royal Botanical Garden of Madrid, University of Florida Herbarium, and Western Australian Herbarium that generously provided fungal specimens. Christopher Walker is also thanked for valuable comments on the manuscript, and Joey Spatafora, Mario Palenzona and Gian Maria Niccolò Benucci for their help in hunting Endogone and Jimgerdemannia sporophores in the field. AD and GB were supported through the US National Science Foundation (NSF) DEB 1737898 and Michigan State University AgBioResearch NIFA project MICL02416. MIB and AJ received support from a NERC standard grant (NE/N00941X/1) and WRR received support from a NERC studentship (DTP SSCP). MES received support from NSF grants DEB 1354802 (Symbiotic ectomycorrhizal fungi in southern South America) and DEB 1441677 (The Zygomycetes Genealogy of Life (ZyGoLife)- the conundrum of Kingdom Fungi) as well as the University of Florida’s Institute for Food and Agricultural Sciences (IFAS).
Supplementary Material
REFERENCES
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berch SM, Castellano MA. (1986) Sporulation of Endogone pisiformis in axenic and monoxenic culture. Mycologia 78: 292–295. [Google Scholar]
- Berch SM, Fortin JA. (1983) Endogone pisiformis: axenic culture and associations with Sphagnum, Pinus sylvestris, Allium cepa, and Allium porrum. Canadian Journal of Botany 61: 899–905. [Google Scholar]
- Berch SM, Fortin JA. (1982) Germination of zygospores of Endogone incrassata. Mycologia 74: 861–864. [Google Scholar]
- Berkeley MJ, Broome CE. (1874) Enumeration of the fungi of Ceylon. Part II. Botanical Journal of the Linnean Society 14: 29-141. [Google Scholar]
- Berkeley MJ, Broome CE. (1846) Notices of British hypogaeous fungi. Annals and Magazine of Natural History 18: 73–82. [Google Scholar]
- Bidartondo MI, Read DJ, Trappe JM, Merckx V, Ligrone R, et al. (2011) The dawn of symbiosis between plants and fungi. Biology Letters 7: 574–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfante P, Desirò A. (2017) Who lives in a fungus? The diversity, origins and functions of fungal endobacteria living in Mucoromycota. The ISME Journal 11: 1727–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu-Chou M, Grace LJ. (1984) Endogone flammicorona and Tuber sp. as mycorrhizal fungi of Pinus radiata in New Zealand. New Zealand Journal of Botany 22: 525–531. [Google Scholar]
- Chu-Chou M, Grace LJ. (1979) Endogone flammicorona as a mycorrhizal symbiont of Douglas fir in New Zealand. New Zealand Journal of Forest Science 9: 344–347. [Google Scholar]
- Castresana J. (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution 17: 540–552. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. (2012) jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desirò A, Faccio A, Kaech A, Bidartondo MI, Bonfante P. (2015) Endogone, one of the oldest plant-associated fungi, host unique Mollicutes-related endobacteria. New Phytologist 205: 1464–1472. [DOI] [PubMed] [Google Scholar]
- Desirò A, Duckett JG, Pressel S, Villarreal JC, Bidartondo MI. (2013) Fungal symbioses in hornworts: a chequered history. Proceedings of the Royal Society of London, B: Biological Sciences 280: 20130207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J. (1991) DNA protocols for plants. In: Molecular techniques in taxonomy (Hewitt GM, Johnston AWB. & Young JPW, eds): 283–293. Berlin: Springer. [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassi B, Fontana A, Trappe JM. (1969) Ectomycorrhizae formed by Endogone lactiflua with species of Pinus and Pseudotsuga. Mycologia 5: 412–414. [Google Scholar]
- Fassi B, Palenzona M. (1969) Mycorrhizal synthesis between Pinus strobus, Pseudotsuga menziesii and Endogone lactiflua. Allionia 15: 105–114. [Google Scholar]
- Fassi B. (1965) Ectotrophic mycorrhizae produced by Endogone lactiflua Berk, on Pinus strobus L. Allionia 11: 7–15. [Google Scholar]
- Field KJ, Rimington WR, Bidartondo MI, Allinson KE, Beerling DJ, et al. (2016) Functional analysis of liverworts in dual symbiosis with Glomeromycota and Mucoromycotina fungi under a simulated Palaeozoic CO2 decline. The ISME Journal 10: 1514–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field KJ, Pressel S, Duckett JG, Rimington WR, Bidartondo MI. (2015a) Symbiotic options for the conquest of land. Trends in Ecology and Evolution 30: 477–486. [DOI] [PubMed] [Google Scholar]
- Field KJ, Rimington WR, Bidartondo MI, Allinson KE, Beerling DJ, et al. (2015b) First evidence of mutualism between ancient plant lineages (Haplomitriopsida liverworts) and Mucoromycotina fungi and its response to simulated Palaeozoic changes in atmospheric CO2. New Phytologist 205: 743–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdemann JW, Trappe JM. (1974) Endogonaceae in the Pacific Northwest. Mycological Memoirs 5: 1–76. [Google Scholar]
- Hirose D, Degawa Y, Yamamoto K, Yamada A. (2014) Sphaerocreas pubescens is a member of the Mucoromycotina closely related to fungi associated with liverworts and hornworts. Mycoscience 55: 221–226. [Google Scholar]
- Ihrmark K, Bödeker IT, Cruz-Martinez K, Friberg H, Kubartova A, et al. (2012) New primers to amplify the fungal ITS2 region–evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiology Ecology 82: 666–677. [DOI] [PubMed] [Google Scholar]
- Jabaji-Hare SH, Charest PM. (1987) Ultrastructural and cytochemical observations on the somatic phase of Endogone pisiformis (Endogonaceae). Mycologia 79: 433–444. [Google Scholar]
- James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, et al. (2006) Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 443: 818–822. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, et al. (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krings M, Taylor TN, Dotzler N, Persichini G. (2012) Fossil fungi with suggested affinities to the Endogonaceae from the Middle Triassic of Antarctica. Mycologia 104: 835–844. [DOI] [PubMed] [Google Scholar]
- Lin K, Limpens E, Zhang Z, Ivanov S, Saunders DG, et al. (2014) Single nucleus genome sequencing reveals high similarity among nuclei of an endomycorrhizal fungus. PLoS Genetics 10: p.e1004078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link HF. (1809) Observationes in ordines plantarum naturales. Dissertatio I. Magazin der Gesellschaft Naturforschenden Freunde Berlin 3: 3–42. [Google Scholar]
- Lloyd CG. (1918) Mycological Notes 56. Mycological Writings 5: 797–812. [Google Scholar]
- Maser C, Trappe JM, Nussbaum RA. (1978) Fungal-small mammal interrelationships with emphasis on Oregon coniferous forests. Ecology 59: 799–809. [Google Scholar]
- McGee PA, Trappe JM. (2002) The Australian zygomycetous mycorrhizal fungi. II. Further Australian sporocarpic Glomaceae. Australian Systematic Botany 15: 115–124. [Google Scholar]
- McGee PA. (1996) The Australian zygomycetous mycorrhizal fungi: the genus Densospora gen. nov. Australian Systematic Botany 9: 329–336. [Google Scholar]
- Orchard S, Hilton S, Bending GD, Dickie IA, Standish RJ, et al. (2017a) Fine endophytes (Glomus tenue) are related to Mucoromycotina, not Glomeromycota. New Phytologist 213: 481–486. [DOI] [PubMed] [Google Scholar]
- Orchard S, Standish RJ, Dickie IA, Renton M, Walker C, et al. (2017b) Fine root endophytes under scrutiny: a review of the literature on arbuscule-producing fungi recently suggested to belong to the Mucoromycotina. Mycorrhiza 27: 619–638. [DOI] [PubMed] [Google Scholar]
- Osmundson TW, Robert VA, Schoch CL, Baker LJ, Smith A, et al. (2013) Filling gaps in biodiversity knowledge for macrofungi: contributions and assessment of an herbarium collection DNA barcode sequencing project. PLoS One 8: E62419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattengale ND, Alipour M, Bininda-Emonds OR, Moret BM, Stamatakis A. (2010) How many bootstrap replicates are necessary? Journal of Computational Biology 17: 337–354. [DOI] [PubMed] [Google Scholar]
- Rehner SA, Buckley E. (2005) A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97: 84–98. [DOI] [PubMed] [Google Scholar]
- Rimington WR, Pressel S, Duckett JG, Bidartondo MI. (2015) Fungal associations of basal vascular plants: reopening a closed book? New Phytologist 205: 1394–1398. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, et al. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccardo PA. (1882) Fungi boreali-Americani. Michelia 2: 564–582. [Google Scholar]
- Smit E, Leeflang P, Glandorf B, Van Elsas JD, Wernars K. (1999) Analysis of fungal diversity in the wheat rhizosphere by sequencing of cloned PCR-amplified genes encoding 18S rRNA and temperature gradient gel electrophoresis. Applied and Environmental Microbiology 65: 2614–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spatafora JW, Chang Y, Benny GL, Lazarus K, Smith ME, et al. (2016) Zygomycete genealogy of life (ZyGoLife): a phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 108: 1028–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2014) RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stürmer SL. (2012) A history of the taxonomy and systematics of arbuscular mycorrhizal fungi belonging to the phylum Glomeromycota. Mycorrhiza 22: 247–258. [DOI] [PubMed] [Google Scholar]
- Tandy PA. (1975) Sporocarpic species of Endogonaceae in Australia. Australian Journal of Botany 23: 849–866. [Google Scholar]
- Tedersoo L, Smith ME. (2017) Ectomycorrhizal fungal lineages: detection of four new groups and notes on consistent recognition of ectomycorrhizal taxa in high-throughput sequencing studies. In: Biogeography of Mycorrhizal Symbiosis (Tedersoo L, ed.): 125–142. Springer International Publishing. [Google Scholar]
- Tedersoo L, Liiv I, Kivistik PA, Anslan S, Kõljalg U, et al. (2016) Genomics and metagenomics technologies to recover ribosomal DNA and single-copy genes from old fruit-body and ectomycorrhiza specimens. MycoKeys 13: 1. [Google Scholar]
- Tedersoo L, Smith ME. (2013) Lineages of ectomycorrhizal fungi revisited: foraging strategies and novel lineages revealed by sequences from belowground. Fungal Biology Reviews 27: 83–99. [Google Scholar]
- Tedersoo L, Jairus T, Horton BM, Abarenkov K, Suvi T, et al. (2008) Strong host preference of ectomycorrhizal fungi in a Tasmanian wet sclerophyll forest as revealed by DNA barcoding and taxon-specific primers. New Phytologist 180: 479–490. [DOI] [PubMed] [Google Scholar]
- Thaxter R. (1922) A revision of the Endogoneae. Proceedings of the American Academy of Arts and Sciences 57: 289–350. [Google Scholar]
- Torres-Cruz TJ, Tobias TL, Almatruk M, Hesse CN, Kuske CR. et al. (2017) Bifiguratus adelaidae gen. et sp. nov., a new member of Mucoromycotina in endophytic and soil-dwelling habitats. Mycologia 109: 363–378. [DOI] [PubMed] [Google Scholar]
- Trappe JM, Gerdemann JW. (1972) Endogone flammicorona sp. nov., a distinctive segregate from Endogone lactiflua. Transactions of the British Mycological Society 59: 403–407. [Google Scholar]
- Truong C, Mujic AB, Healy R, Kuhar F, Furci G, et al. (2017) How to know the fungi: combining field inventories and DNA-barcoding to document fungal diversity. New Phytologist 214: 913–919. [DOI] [PubMed] [Google Scholar]
- Větrovský T, Kolařík M, Žifčáková L, Zelenka T, Baldrian P. (2015) The rpb2 gene represents a viable alternative molecular marker for the analysis of environmental fungal communities. Molecular Ecology Resources 16: 388–401. [DOI] [PubMed] [Google Scholar]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker C. (1985) Endogone lactiflua forming ectomycorrhizas with Pinus contorta. Transactions of the British Mycological Society 84: 353–355. [Google Scholar]
- Warcup JH. (1990) Taxonomy, culture and mycorrhizal associations of zygosporic Endogonaceae. Mycological Research 94: 173–178. [Google Scholar]
- Warcup JH. (1985) Ectomycorrhizal formation by Glomus tubiforme. New Phytologist 99: 267–272. [Google Scholar]
- Warcup JH. (1975) A culturable Endogone associated with eucalyptus. In: Endomycorrhizas (Sanders FE, Mosse B, Tinker PB, eds): 53–63. London: Academic Press. [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor JW. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR protocols: a guide to methods and applications (Innis MA, Gelfand DH, Sninsky JJ. & White TJ, eds): 315–322. San Diego: Academic Press. [Google Scholar]
- Wu CG, Lin SJ. (1997) Endogonales in Taiwan: a new genus with unizygosporic sporocarps and a hyphal mantle. Mycotaxon 64: 179–188. [Google Scholar]
- Yamamoto K, Endo N, Degawa Y, Fukuda M, Yamada A. (2017a) First detection of Endogone ectomycorrhizas in natural oak forests. Mycorrhiza 27: 295–301. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Degawa Y, Takashima Y, Fukuda M, Yamada A. (2017b) Endogone corticioides sp. nov. from subalpine conifer forests in Japan and China, and its multi-locus phylogeny. Mycoscience 58: 23–29. [Google Scholar]
- Yamamoto K, Degawa Y, Hirose D, Fukuda M, Yamada A. (2015) Morphology and phylogeny of four Endogone species and Sphaerocreas pubescens collected in Japan. Mycological Progress 14: 86. [Google Scholar]
- Yao YJ, Pegler DN, Young TWK. (1995) Youngiomyces, a new genus in Endogonales (Zygomycotina). Kew Bulletin 50: 349–357. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.