Abstract
Variation in gene expression can provide insights into organismal responses to environmental stress and physiological mechanisms mediating adaptation to habitats with contrasting environmental conditions. We performed an RNA-sequencing experiment to quantify gene expression patterns in fish adapted to habitats with different combinations of environmental stressors, including the presence of toxic hydrogen sulphide (H2S) and the absence of light in caves. We specifically asked how gene expression varies among populations living in different habitats, whether population differences were consistent among organs, and whether there is evidence for shared expression responses in populations exposed to the same stressors. We analysed organ-specific transcriptome-wide data from four ecotypes of Poecilia mexicana (nonsulphidic surface, sulphidic surface, nonsulphidic cave and sulphidic cave). The majority of variation in gene expression was correlated with organ type, and the presence of specific environmental stressors elicited unique expression differences among organs. Shared patterns of gene expression between populations exposed to the same environmental stressors increased with levels of organismal organization (from transcript to gene to physiological pathway). In addition, shared patterns of gene expression were more common between populations from sulphidic than populations from cave habitats, potentially indicating that physiochemical stressors with clear biochemical consequences can constrain the diversity of adaptive solutions that mitigate their adverse effects. Overall, our analyses provided insights into transcriptional variation in a unique system, in which adaptation to H2S and darkness coincide. Functional annotations of differentially expressed genes provide a springboard for investigating physiological mechanisms putatively underlying adaptation to extreme environments.
Keywords: caves, extreme environments, hydrogen sulphide, local adaptation, poeciliidae, transcriptomes
1 | INTRODUCTION
High-throughput sequencing technologies have transformed the quantification of genome-wide gene expression patterns in natural systems (Wang, Gerstein, & Snyder, 2009). While studies of gene expression cannot identify causal loci underlying adaptation, transcriptional variation between organisms exposed to contrasting environmental conditions can provide important clues about genes and physiological pathways contributing to plastic or adaptive responses to novel environments (e.g., Cheviron, Whitehead, & Brumfield, 2008; Guo et al., 2016; López-Maury, Marguerat, & Bähler, 2008; Morris et al., 2014). Studies often focus on systems where evolutionarily replicated lineages are exposed to the same environmental conditions, which allows for the identification of shared and unique (i.e., lineage-specific) gene expression responses (Kelley et al., 2016; Whitehead, Pilcher, Champlin, & Nacci, 2012). Shared differentially expressed genes among lineages exposed to the same environmental conditions represent prime candidates for loci and physiological pathways that may be modulated during acclimation or adaptation (Whitehead, 2012). However, the degree to which gene expression patterns are shared among lineages exposed to the same environmental conditions may be structured hierarchically across levels of organismal organization, with shared responses increasing at higher levels of organization. Such patterns have been well established by studies that illuminate the genetic basis of phenotypic convergence, which may be caused by the same mutation, different mutations in the same gene, mutations in different genes of the same developmental pathway, or genes in different developmental pathways (see Manceau, Domingues, Linnen, Rosenblum, & Hoekstra, 2010; Elmer & Meyer, 2011; Rosenblum, Parent, & Brandt, 2014 for reviews). Analogously, organismal and evolutionary gene expression responses to a common environmental stressor may be mediated by the differential expression of the same transcript, different transcripts derived from the same gene, different genes in the same physiological pathway, or genes in different physiological pathways. It remains largely unknown what factors determine how shared expression responses are distributed across hierarchical levels, but the effects of specific sources of selection, phylogenetic history, demography, and genetic constraints have all been implicated as potential candidates (Rosenblum et al., 2014). Addressing this question requires integrative data analysis that explicitly considers the functional links among transcripts in genome-wide expression studies.
Another key challenge in the analysis of gene expression data from natural systems is that gene expression is notoriously plastic over short periods of time and in response to diverse endogenous and exogenous stimuli (Bajić & Poyatos, 2012; Lehner, 2010). At the same time, selective regimes even among apparently discrete habitat types are often multifarious (Holmstrup et al., 2010; Kaeuffer, Peichel, Bolnick, & Hendry, 2012). Nonetheless, we know relatively little about genome-wide expression responses in natural systems where closely related populations are exposed to multiple sources of selection that occur in different combinations. Organismal responses to multiple environmental cues may be additive such that their combined effects represent the sum of the effects of individual stressors (Folt, Chen, Moore, & Burnaford, 1999). Alternatively, multiple stressors may cause synergistic (responses greater than the sum of individual stressors) and antagonistic effects (responses smaller than the sum of individual stressors; Folt et al., 1999; Altshuler et al., 2011). Interactive effects and trade-offs that underlie nonadditive responses hamper our ability to identify shared gene expression patterns that may be indicative of adaptation and to predict organismal responses to environmental stress when multiple sources of selection coincide (Kopec et al., 2011; Shu, Kang, Yang, & Kaminski, 2003).
Understanding variation in gene expression in response to multiple stressors is also complicated by organ-specific responses. While single stressors with broad, systemic effects (e.g., thermal stress) may elicit relatively consistent expression responses across different organs (Akashi, Cádiz Díaz, Shigenobu, Makino, & Kawata, 2016), exposure to many physiochemical stressors—and especially combinations of different stressors—likely causes idiosyncratic patterns of gene expression that vary substantially among organs, tissues and even cell types (Bailey et al., 2013; Birnbaum et al., 2003; Bos, Pulliainen, Sundström, & Freitak, 2016). For example, expression responses to hypoxia exposure vary due to differences in the metabolic requirements among organs (Gracey, Troll, & Somero, 2001; Whitehead & Crawford, 2005). Disentangling the effects of multiple stressors on gene expression should therefore involve the analysis of multiple organs, which can also inform our understanding of how organ-specific expression changes relate to potential functional consequences and patterns of local adaptation (Oleksiak, Churchill, & Crawford, 2002).
Here, we characterized gene expression variation in a system of extremophile fish that includes populations exposed to different combinations of environmental stressors. We asked whether the analysis of gene expression patterns in multiple organs (gill, liver and brain) and at multiple levels of organization (transcript, gene and physiological pathway) affects inferences about the putative functional consequences of expression variation. Poecilia mexicana (Poeciliidae) is a small live-bearing fish that has colonized sulphide springs and caves in Mexico’s Cueva del Azufre system, giving rise to a unique group of closely related populations living in geographically proximate habitat types with vastly different environmental conditions (nonsulphidic surface streams, sulphidic surface streams, a nonsulphidic cave, and a sulphidic cave; see Figure 1; Parzefall, 2001; Tobler et al., 2008). Sulphidic habitats are characterized by high concentrations of hydrogen sulphide (H2S), a potent respiratory toxicant that binds to cytochrome c oxidase in the respiratory chain, effectively halting aerobic ATP production (Bagarinao, 1992; Cooper & Brown, 2008). Caves are characterized by the absence of light, which has direct effects on organismal function (e.g., sensory biology and circadian rhythms; Poulson & White, 1969; Langecker, 2000; Niemiller & Soares, 2015). Both H2S and permanent darkness also shape the biotic environment, affecting resource availability as well as competition and predation regimes (Hüppop, 2000; Roach, Tobler, & Winemiller, 2011). Previous studies have indicated that populations of P. mexicana exposed to specific combinations of H2S and darkness are locally adapted, exhibiting phenotypic differences in behavioural (Parzefall, 2001), sensory (Tobler, Coleman, Perkins, & Rosenthal, 2010), physiological (Passow, Greenway, Arias-Rodriguez, Jeyasingh, & Tobler, 2015), morphological (Tobler et al., 2008) and life-history (Riesch, Plath, Schlupp, & Marsh-Matthews, 2010) characteristics. In addition, microsatellite analyses have indicated significant genetic differentiation and low rates of gene flow among populations in different habitat types (Plath et al., 2010), and reproductive isolation is in part mediated by natural and sexual selection against nonadapted migrants (Riesch, Plath, & Schlupp, 2011; Tobler, 2009; Tobler, Riesch, Tobler, Schulz-Mirbach, & Plath, 2009).
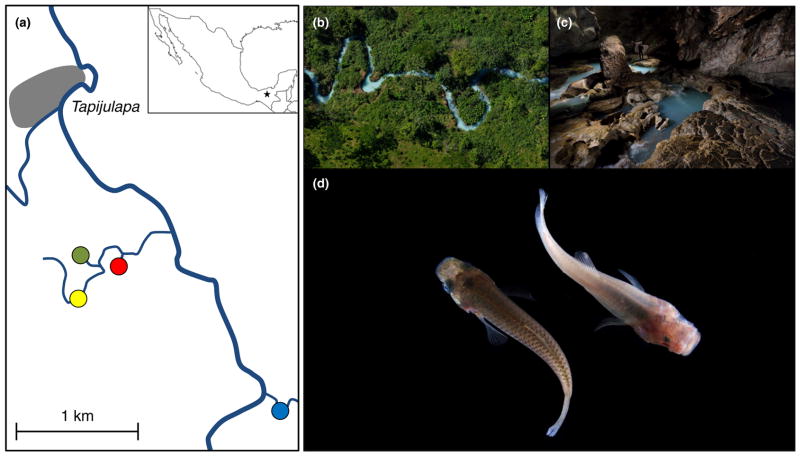
FIGURE 1.
(a) Overview of the study area near the village of Tapijulapa, Tabasco, Mexico. Depicted are the four focal populations: nonsulphidic surface (blue), sulphidic surface (yellow), nonsulphidic cave (red) and sulphidic cave (green). The insert indicates the study region within Mexico. (b) The sulphidic surface stream and (c) the sulphidic cave in which fish were collected for this study. (d) Phenotypic differences are evident between fish from the sulphidic surface (left) and the sulphidic cave habitat (right). Photographs are courtesy of Robbie Shone (http://www.shonephotography.com) [Colour figure can be viewed at wileyonlinelibrary.com]
We performed transcriptome analyses on three different organs and populations exposed to all combinations of the presence and absence of H2S and light. We focused analyses on gills, liver and brain, because there are a priori expectations about the roles of these organs in adaptation to the environmental conditions encountered in the Cueva del Azufre system. Gills are an important organ involved in the maintenance of homeostasis in fish (Evans & Claiborne, 2006; Evans et al., 2011), and since they exhibit a high surface area directly suspended in the water, they provide an important access point for H2S exposure (Tobler, Passow, Greenway, Kelley, & Shaw, 2016). The liver plays roles in detoxification and the modulation of energy metabolism (Dorman et al., 2002; Green, Takahashi, & Bass, 2008), and both processes are relevant in the context of H2S and cave adaptation (Aspiras, Rohner, Martineau, Borowsky, & Clifford, 2015; Bagarinao & Vetter, 1990). Finally, the brain has previously been shown to play a role in adaptation to perpetual darkness (Langecker, 2000; Poulson, 2001), and H2S can have neurotoxic effects (Kombian, Warenyica, Mele, & Reiffenstein, 1988). In addition, brain morphology has been shown to vary among populations of Poecilia in the Cueva del Azufre system (Eifert et al., 2015). Analysing gene expression variation across habitats and organs, we specifically addressed the following questions: (i) How does gene expression vary among organs and populations living in different habitat types? We predicted significant differences in gene expression among fish from different habitat types, reflecting their exposure to starkly different environmental conditions. Considering that H2S impacts the function of mitochondria and can therefore have systemic effects (Li, Rose, & Moore, 2011), gene expression differences between sulphidic and nonsulphidic populations were expected to be relatively consistent across all organs. In contrast, we expected idiosyncratic, organ-specific gene expression differences between populations from cave and surface habitats. (ii) Is there evidence for shared expression responses in populations exposed to the same environmental conditions (presence of H2S or absence of light), and how do shared responses vary among hierarchical levels of organismal organization? Shared responses between populations exposed to the same environmental conditions were expected to increase with increasing level organization (from transcript to gene to physiological pathway). At each level of organization, we also expected a higher degree of shared responses between the sulphidic populations than between the cave populations, because physiochemical stressors with clear molecular targets have been hypothesized to limit the diversity of organismal coping strategies. (iii) What inferences can analyses of differential expression provide about physiological mechanisms that are modulated during exposure to specific environmental conditions? We expected that physiological mechanisms associated with living in sulphidic and cave environments largely correspond to the findings of previous analyses. Transcriptome studies on organisms living in H2S-rich environments have focused on gill tissues and documented differential expression of genes associated with enzymatic H2S detoxification and the processing of sulphur compounds, as well as aerobic and anaerobic ATP production and energy metabolism (Kelley et al., 2016; Liu et al., 2015; Wong et al., 2015). Analysis of transcriptomes of cave dwellers have focused on eyes or whole organisms and revealed differential expression of genes associated with eye development and function, circadian rhythms and energy metabolism (Gross, Furterer, Carlson, & Stahl, 2013; Meng et al., 2013; Strickler, Byerly, & Jeffery, 2007).
2 | MATERIALS AND METHODS
2.1 | Sample collections
Samples for transcriptome analyses were collected in the Cueva del Azufre system that is located in the Rio Tacotalpa drainage near the village of Tapijulapa, Tabasco, Mexico (Figure 1a; Tobler et al., 2008). Fish were caught at four sites: Arroyo Bonita (nonsulphidic surface stream), El Azufre II (sulphidic surface stream), Cueva Luna Azufre (nonsulphidic cave) and Cueva del Azufre (sulphidic cave) (see Tobler et al., 2008 for a detailed description of these sampling sites). Upon capture, four adult females per site were immediately euthanized, weighed and measured (Table S1). Gills, livers, brains and eyes were then extracted using sterilized scissors and forceps and separately preserved and stored in Invitrogen RNAlater. Procedures for all experiments were approved by the Institutional Animal Care and Use Committee at Kansas State University (Protocol #3418).
2.2 | RNA extraction, cDNA library preparation and sequencing
We adopted a general protocol for transcriptome analyses that was previously developed for P. mexicana (Kelley et al., 2012). In brief, 10–30 mg of tissue were placed into a Covaris TT1 tissue TUBE (designed for <1 g of tissue), frozen using liquid nitrogen and then pulverized with a Covaris Cryoprep at setting 3. Total RNA was extracted using the Qiagen RNeasy Plus Mini Kit and quantified using both the Thermo Fisher Scientific Qubit RNA Assay Kit and Agilent RNA 6000 Nano total RNA kit on an Agilent 2100 Bioanalyzer. We then purified mRNA by depleting rRNA from 1 μg of total RNA using the Epicentre Ribo-Zero Magnetic Gold Kit (human/mouse/rat). The remaining RNA was cleaned up twice using Beckman Coulter Life Sciences Agencourt RNAClean XP beads. The eluted RNA was then fragmented to 400 nucleotides using New England BioLabs (NEB) RNA fragmentation buffer, incubating at 94°C for 4 min. First-strand cDNA synthesis was performed by combining the fragmented mRNA with 1 μl of random hexamers: oligo-dT primers (3 μg:1 μg), 4 μl of Invitrogen 5× first-strand reaction buffer, 2 μl of Invitrogen 0.1 M DTT and 1 μl of NEB 10 mM dNTP mix. We then added 1 μl Invitrogen SuperScript III Reverse Transcriptase to the mixture. The solutions were incubated at 25°C for 50 min and immediately placed on ice to terminate the reaction. For second-strand synthesis, we added 5 μl of Invitrogen 5× first-strand buffer, 1 μl of Applied Biosystems DTT, 2 μl of 10 mM dNTP mix with dUTP, 15 μl of Invitrogen 5× second-strand reaction buffer and 3.75 μl of NEB second-strand enzyme mix. The reaction mix was incubated at 16°C for 2 hr. After second-strand synthesis, the reaction was cleaned up using Beckman Coulter Life Sciences Agen-court RNAclean XP beads and eluted into 50 μl of nuclease-free H2O. The double-stranded cDNA was used as an input for the KAPA Biosystems KAPA HTP Library Preparation Kit for end-repair, A-tailing, adapter ligation with Illumina TruSeq barcoded adapters and library amplification. For the library amplification reaction, we ran the initial denaturation at 98°C for 45 seconds (s), followed by 12 cycles of denaturation at 98°C for 15 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s and finishing with a final extension at 72°C for 60 s. RNA-sequencing libraries were quantified using Thermo Fisher Scientific Qubit dsDNA HS Assay Kit and Agilent High Sensitivity DNA Analysis Kit on the 2100 Bioanalyzer. Libraries were pooled based on nanomolar concentrations and sequenced on an Illumina HiSeq 2000 across two lanes with paired-end 101 base pair (bp) reads. Due to low coverage, some samples were rerun on an additional Illumina HiSeq 2000 lane, and reads were then concatenated together for each sample. Read mappings for eye samples were not sufficient for data analyses; thus, all analyses focused on gill, liver and brain samples.
2.3 | Reference transcriptome assembly and annotation
Reference transcriptome assembly and annotation generally followed previous published methods (Kelley et al., 2016). All raw RNA-seq reads were trimmed to remove primer dimers and low-quality bases using the program Trim Galore! (Krueger, 2014; Table S2). Trimmed reads were then mapped to the Poecilia mexicana reference genome (version 1.0, GenBank accession number: LMXC00000000.1) plus corresponding mitochondrial sequences (GenBank Accession Number: KC992991; Pfenninger et al., 2014) using BWA-mem (Li, 2013; Li & Durbin, 2009), which yielded the highest alignment percentage (results from other aligners not shown). We then used the cufflinks package (version 2.2.1) to extract expressed regions, merge the regions for all individuals and create a multifasta file containing the reference transcriptome for further analyses (Trapnell et al., 2010). Transcripts were annotated using a BLASTx search against the human SwissProt database (critical E-value = 0.001). This database provides more informative functional annotations than other databases (e.g., NCBI nonredundant protein database), which facilitates downstream analyses (see below). We retained the top BLAST hit for further analyses, and sequences with a BLAST match were also annotated with Gene Ontology (GO) IDs (Gene Ontology Consortium 2004) using Blast2GO (Conesa et al., 2005).
2.4 | Quantifying gene expression variation
To analyse variation in gene expression among populations and organs, we mapped all trimmed reads to the multifasta file (reference transcriptome) using BWA-mem (Li, 2013; Li & Durbin, 2009) and then estimated transcript abundance with eXpress (Roberts & Pachter, 2013). Transcripts with very low expression (less than two counts per million in at least three samples) were removed from further analyses. To describe multivariate variation in gene expression, we conducted a weighted gene coexpression network analysis on the top 10,000 expressed genes in the R package WGCNA (Langfelder & Horvath, 2008), as described in Oldham, Horvath, and Geschwind (2006). Before constructing the networks, we used the variance-stabilizing transformation function in the DESEQ2 package in R (Love, Huber, & Anders, 2014), which transforms data normalized for library size to a log2-scale (Anders & Huber, 2010), and set the soft threshold (i.e., power) to 3, which was the lowest value that optimized topology (Langfelder & Horvath, 2008). WGCNA quantifies correlations between the top 10,000 expressed genes using Pearson correlations, which are converted into measures of connection strength between the genes. Genes with similar connection strengths to other genes (i.e., high topological overlap) are identified and clustered together into modules. Once modules were identified, we constructed a consensus dendrogram based on the most significant modules and tested for associations between individual modules and predictor variables (organ, individual, population of origin, presence of H2S in natural habitat, absence of light in natural habitat) using Pearson correlation as implemented in WGCNA. Complementary to the weighted coexpression network analysis, we also conducted multidimensional scaling (MDS) of the top 10,000 expressed genes using plotMDS in the limma package in R. We analysed MDS scores with a multivariate analysis of variance (MANOVA) with “organ” and “population” as predictor variables. F-values were approximated using Wilks’ lambda and effect sizes by use of partial eta-squared ( ). To visualize potential differences in gene expression patterns among populations, we also conducted separate MDS for each organ.
To quantify differential gene expression among populations, we treated the ancestral, nonsulphidic surface population as a control and conducted pairwise comparisons with each of the populations inhabiting an extreme environment (sulphidic surface, nonsulphidic cave and sulphidic cave). Pairwise comparisons were conducted separately for each organ using the exactTest function in the Bioconductor package edgeR (Robinson, McCarthy, & Smyth, 2010; Robinson & Oshlack, 2010). The false discovery rate was set to .05. These analyses provided a list of up- and downregulated transcripts for each extremophile population and organ as compared to the ancestral nonsulphidic surface population.
2.5 | Testing for organ-specific responses
We assessed whether gene expression responses to the same sources of selection were quantitatively and qualitatively consistent across organs or whether there are idiosyncratic, organ-specific responses. To do so, we first tested whether the number of differentially expressed transcripts varied among organs using analysis of variance (ANOVA). We included direction of differential expression (i.e., up- or downregulated), population and organ as independent variables. Furthermore, we tested whether the biological processes putatively associated with differentially expressed genes in each habitat were consistent among organs. To do so, we first performed a pathway enrichment analysis for each organ and population using the enrichPathway function from the ReactomePA package in R (Yu & He, 2016) to identify the enriched pathways (p-value cut-off .05). We then implemented the compareCluster function in the CLUSTERPRO-FILER package in R (Yu, Wang, Han, & He, 2012), which calculates and compares enriched functional categories of each gene cluster among organs of the same population (p-value cut-off .05). Using gene ratios (proportion of genes enriched in each category) and adjusted p-values, we constructed a dot plot in R to visualize variation among organs of the same population. Finally, we implemented the rcorr function in the HMISC package in R to compute a matrix of Pearson’s r rank correlation coefficients (r) and significance levels (p-value) based on the gene ratios in the enriched pathways to evaluate the consistency of responses among all organ pairs (gill vs. liver, gill vs. brain and brain vs. liver).
2.6 | Identifying shared expression responses in H2S and cave environments
We tested for shared expression responses to H2S and perpetual darkness by comparing differentially expressed genes between the sulphidic surface and sulphidic cave populations (shared responses to H2S), as well as between the nonsulphidic and the sulphidic cave populations (shared responses to darkness). Because theory predicts that the degree of shared responses should be a function of the level of biological organization (see Introduction), we quantified shared responses at the level of the transcript (transcript ID based on our reference transcriptome), gene (same BLAST hit in SwissProt) and function (same GO terms in biological processes associated with the BLAST hit). The number of shared and unique up- and downregulated transcripts, genes and functions was quantified by generating Venn diagrams with the vennCounts and vennDiagram functions in the package LIMMA in R (see Fig. S1 for the cave populations and Fig. S2 for the populations from sulphidic habitats). The distribution of values in these Venn diagrams was then summarized for quantitative analysis using the Jaccard index, which was calculated as the size of the intersection between two samples (number of responses in the intersection of the Venn diagrams) divided by size of the union between two samples (total number of responses, unique and shared, in the Venn diagrams) (Krebs, 1999; Pfenninger et al., 2015). Jaccard indices (log10-square-root-transformed) were analysed using ANOVA. We included expression direction (i.e., up- and downregulated), environmental factor (i.e., H2S and darkness), organization level and organ as independent variables.
2.7 | Biological functions associated with differentially expressed genes
We assessed the biological functions of differentially expressed genes to explore potential functional consequences associated with adaptation to H2S and perpetual darkness. To do so, we conducted a GO term enrichment analysis (p-value cut-off .001) as implemented in GOrilla (Eden, Navon, Steinfeld, Lipson, & Yakhini, 2009). Enrichment analyses were conducted separately for each population and organ.
3 | RESULTS
3.1 | Reference transcriptome
We obtained 133,785,284 reads for gill tissues (14 individuals in final data set), 95,374,496 reads for brain tissues (11 individuals in final data set) and 136,779,442 reads for liver tissues (16 individuals in final data set; see Table S2 for details). Due to low coverage, we excluded five brain and two gill samples. Mapping against the reference genome resulted in a total of 63,590 unique transcripts from 38,764 unique loci (see Table S3 for additional summary statistics). The total length of the merged transcriptome was 225,876,000 bp, with an N50 of 5,290 bp. BLAST against the human SwissProt database resulted in matches for 48,242 transcripts (75.9% of transcripts; see Table S4) corresponding to 14,005 unique SwissProt entries, 13,504 of which were associated with Gene Ontology (GO) terms.
3.2 | Organ-specific gene expression
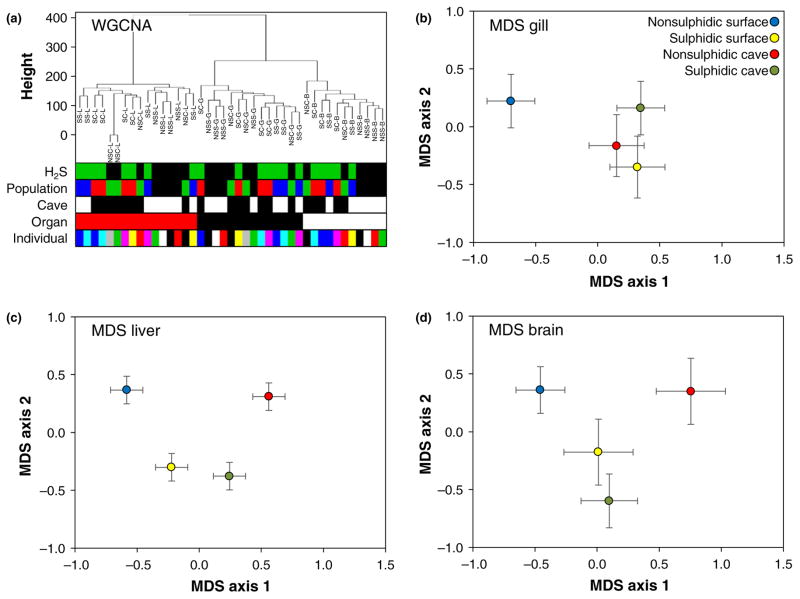
Weighted gene coexpression network analysis revealed five modules of coexpressed transcripts (Fig. S3). Organ identity was significantly correlated with all modules, while none of the other predictor variables (presence or absence of H2S, presence or absence of light, population and individual) exhibited any significant associations (Figure 2a; Table 1). This general pattern was also evident by visualizing the MDS results on the full data set, which indicated a clear separation of different organs in multivariate space (Fig. S4). Multivariate analyses of variance using MDS scores as the dependent variables confirmed the strong effect of organ on gene expression patterns (F,56 = 3516.07, p < .001, ) and also indicated a significant interaction between organ type and population of origin (F12,56 = 2.01, p = .040, ). Population as a main factor had no significant effect (F6,56 = 1.021, p = .421, ). Gene expression consequently varied substantially among organs, while effects of environmental conditions were comparatively subtle. Hence, we analysed among population variation in gene expression separately for each organ. Visualization of MDS on each organ indicated significant differences in gene expression patterns among populations from different habitat types (Figure 2b–d).
FIGURE 2.
(a) Results of the weighted gene coexpression network analysis (WGCNA) indicated that gene expression variation among samples was primarily driven by organ type (see Table 1). (b–d) MDS on each organ separately revealed significant differences in gene expression patterns among fish from different habitat types. MDS plots depict mean scores along axes 1 and 2 (± standard error). Data from different habitat types are colour-coded as indicated in the legend in panel (b) [Colour figure can be viewed at wileyonlinelibrary.com]
TABLE 1.
Results of the weighted gene coexpression network analysis. Correlation between module eigenvalues and predictor variables indicated that organ type was the best predictor of coexpressed genes
| Cluster ID | Individual | Organ | Population | Cave | H2S |
|---|---|---|---|---|---|
| 1 | 0.110 (0.5) | 0.820 (6e-11) | 0.098 (0.5) | −0.008 (1.0) | 0.037 (0.8) |
| 2 | −0.045 (0.8) | −0.540 (3e-4) | −0.043 (0.8) | −0.059 (0.7) | −0.054 (0.7) |
| 3 | −0.069 (0.7) | 0.410 (0.008) | −0.058 (0.7) | 0.110 (0.5) | −0.014 (0.9) |
| 4 | −0.120 (0.4) | −0.990 (5e-39) | −0.110 (0.5) | −0.052 (0.7) | −0.071 (0.7) |
| 5 | −0.092 (0.6) | −0.520 (5e-4) | −0.080 (0.6) | −0.002 (1.0) | −0.033 (0.8) |
Each row corresponds to a module of coexpressed genes (see Fig. S3 for visualization), and we report Pearson’s correction coefficients and p-values (in parentheses) between each module and predictor variables. Significant effects (p < .05) are highlighted in bold font.
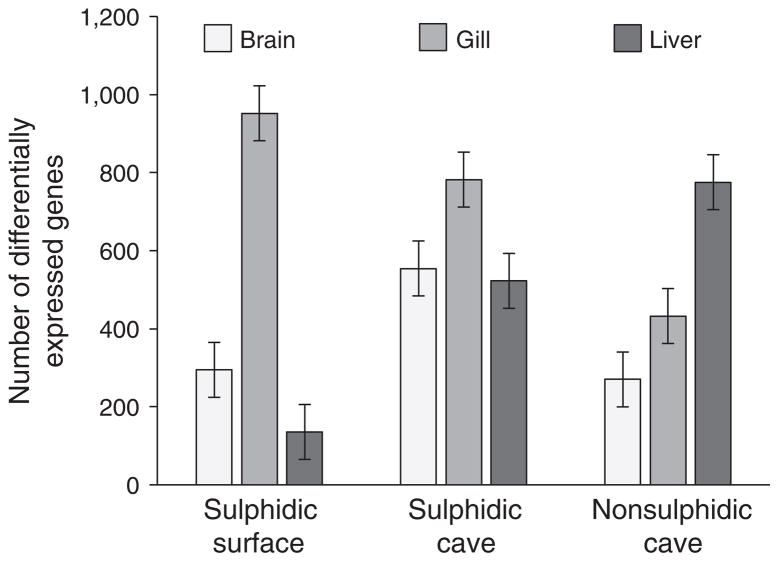
Comparing the number of differentially expressed genes among populations from the different extreme habitats (relative to the ancestral, nonsulphidic surface population) indicated that organ and the interaction between organ and habitat type explained the majority of variation (Table 2). In the two sulphidic habitats, the number of differentially expressed genes was the highest in the gills (Figure 3). In the nonsulphidic cave, however, the number of differentially expressed genes was greatest in the liver and lower in the brain and gills (Figure 3).
TABLE 2.
Results of analyses of variance (ANOVA) comparing the number of differentially expressed genes among habitat types (sulphidic surface, nonsulphidic cave, sulphidic cave), organs (gills, liver, brain) and expression direction (up vs. downregulated)
| Term | df | F | p |
|
|
|---|---|---|---|---|---|
| Habitat type | 2 | 4.3 | .101 | .682 | |
| Organ | 2 | 19.5 | .009 | .907 | |
| Expression direction | 1 | 0.1 | .724 | .035 | |
| Habitat × Expression direction | 2 | 0.8 | .501 | .292 | |
| Organ × Expression direction | 2 | 1.6 | .313 | .441 | |
| Habitat × Organ | 4 | 18.0 | .008 | .947 |
The number of differentially expressed genes varied among organs, but in a habitat-specific manner (see Figure 3). Effect sizes were estimated with partial eta-squared ( ), and significant effects (p < .05) are high-lighted in bold font.
FIGURE 3.
The number (estimated marginal means ± standard error) of differentially expressed genes (up and downregulated) among populations and organs. Estimated marginal means were derived from the ANOVA model in Table 2
We also found significant differences in the enrichment of physiological pathways associated with differentially expressed genes across organs (Fig. S5). Overall, positive correlations in the enrichment of functional classes across organs were relatively rare, indicating that the nature of differentially expressed genes is organ-specific. Among the few exceptions were similar functional responses in upregulated transcripts between brain and liver in the nonsulphidic cave, upregulated transcripts between brain and gill in the sulphidic surface habitat, upregulated transcripts between liver and gills in the sulphidic cave and downregulated transcripts between brain and liver in the sulphidic cave. When detected, correlations in the functional responses between organs were primarily driven by genes associated with metabolism and metabolism of lipids and lipoproteins (Fig. S5).
3.3 | Shared expression responses in H2S and cave environments
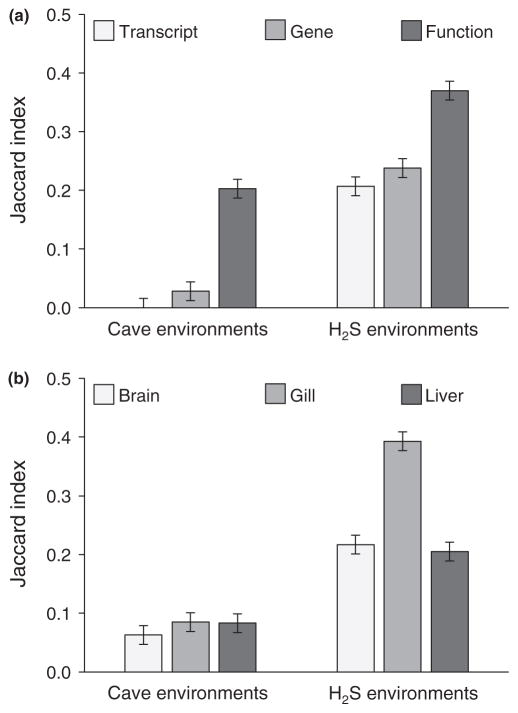
By comparing differentially expressed transcripts between populations, we identified shared responses to H2S-rich and cave environments. The extent of shared responses was a function of interactive effects between environmental factors, organs and the level of biological organization (Table 3). As predicted, the number of shared responses generally increased with level of biological organization (Figure 4a). However, the amount of shared responses at each level of organization and the magnitude of increase from one level to the next significantly differed between environmental factors. In cave environments, there were very few shared differentially expressed transcripts and only about 20% of GO terms associated with differentially expressed genes were shared between populations (Fig. S1). In contrast, shared responses were much higher between sulphidic environments at each level of organization, with about 40% of GO terms associated with differentially expressed genes appearing in both sulphidic populations (Fig. S2). Population differences in the sulphidic and cave-adapted fish were also evident among organs (Figure 4b). While there were no differences in the degree of shared responses among organs in the two cave populations, shared responses were significantly more prevalent in gills between the two sulphidic populations as compared to the brain and liver.
TABLE 3.
Results of analyses of variance (ANOVA) comparing the proportions of shared responses (Jaccard index) among environmental factors (sulphidic vs. cave environments), organs (gills, liver, brain), expression direction (up vs. downregulated) and level of organismal organization (transcript, gene, gene function)
| Term | df | F | p |
|
|
|---|---|---|---|---|---|
| Expression direction | 1 | 0.3 | .619 | .016 | |
| Environmental factor | 1 | 450.4 | <.001 | .966 | |
| Level of organization | 2 | 152.7 | <.001 | .950 | |
| Organ | 2 | 24.4 | <.001 | .753 | |
| Expression direction × Environmental factor | 1 | 2.0 | .178 | .111 | |
| Expression direction × Level of organization | 2 | 0.7 | .489 | .086 | |
| Expression direction × Organ | 2 | 2.3 | .128 | .227 | |
| Environmental factor × Level of organization | 2 | 27.7 | <.001 | .776 | |
| Environmental factor × Organ | 2 | 15.2 | <.001 | .655 | |
| Level of organization × Organ | 4 | 0.5 | .711 | .118 |
The amount of shared responses increased with level of organization, but shared responses were higher for sulphidic than for cave populations (Figure 4a). In addition, the amount of shared responses also varied between sulphidic and cave populations in an organ-specific manner (Figure 4b). Effect sizes were estimated with partial eta-squared ( ), and significant effects (p < .05) are highlighted in bold font.
FIGURE 4.
The proportion (estimated marginal mean of Jaccard index ± standard error) of shared differences (a) across populations and levels of biological organization and (b) across populations and organs. Estimated marginal means were derived from the ANOVA model in Table 3
3.4 | Biological functions associated with differentially expressed genes
To identify functional changes potentially associated with adaptation to the different environmental conditions, we conducted a GO term enrichment analysis for up- and downregulated genes for each tissue and population (Table S5). For brevity, we only discuss enriched terms that are shared between both sulphidic or both cave habitats.
While no GO terms were enriched across all organs in the two sulphidic habitats (Table S6), several genes associated with H2S detoxification and sulphur metabolism were consistently upregulated in the gills (Table S6A). This included terms associated with sulphur (GO:0044272, GO:0006790) and glutathione metabolism (GO:0006749). In addition, upregulated genes in the gills of sulphidic populations were enriched in terms associated with oxidative stress responses (GO:0006979). Downregulated genes in the gills were predominately associated with the transport of ions and other molecules (GO:0030001, GO:0006814, GO:0006811 and associated terms; Table S6B). In the livers of sulphidic populations, downregulated genes were disproportionally associated with metabolism of lipids and fatty acids (GO:0035337, GO:0006631, GO:003583 and associated terms; Table S6B). In the brains of sulphidic populations, upregulated genes were enriched for processes associated with energy metabolism and ATP production (GO:0006096, GO:00016051, GO:00006757; Table S6A). Downregulated genes in the brain were associated with the organization of the extracellular matrix and DNA integration (GO:0030198, GO:0015074).
No GO terms were significantly enriched in all organs of the two cave populations. In the gills, up- and downregulated genes were primarily involved in ion transport and the regulation of pH (Table S6C, D). In the livers of the two cave populations, we found no shared enrichment in upregulated genes, and downregulated genes were associated with responses to chemicals and the regulation of biological quality and protein secretion (GO:0042221, GO:0065008 and associated terms; Table S6D). There was no consistent enrichment in any GO term for the brains of the two cave populations (Table S6C,D).
4 | DISCUSSION
Transcriptome analyses among closely related fish populations in proximate but environmentally distinct habitat types uncovered significant variation in gene expression. Gene expression variation was primarily correlated with organ type, and the number and function of differentially expressed genes in each habitat type was also organ-specific. As predicted by theory, shared responses between the two sulphidic and the two cave habitats increased with the level of biological organization, from relatively few shared patterns at the level of differentially expressed transcripts to higher levels of overlap in physiological pathways associated with differentially expressed transcripts. However, shared responses at any level of organization were much more pronounced in the H2S-rich habitats than the cave habitats, highlighting that H2S impacts gene regulation in a more predictable manner than the absence of light. Our analyses provided insights into transcriptional variation in a unique system with coinciding sources of selection, and functional annotation of differentially expressed genes provides a springboard for investigating physiological mechanisms putatively underlying adaptation to extreme environments.
4.1 | Expression variation among organs and environments
Considering different organs are fulfilling vastly different functions despite being composed of cells with the same genome, it was not surprising that the majority of variation in gene expression was observed among organ types. However, the gene expression responses to the presence of H2S and to the absence of light in caves also significantly varied among organs, both in terms of the number of genes that were differentially expressed and their function (Figure 3 and Fig. S5). So, even though H2S-rich and cave environments are often assumed to exert strong selection (Niemiller & Soares, 2015; Tobler et al., 2016), and H2S has specific and well-documented biochemical and physiological effects (Olson, 2012), the consequences of exposure to these environments are not systemic and uniform but specific to particular organs. In the nonsulphidic cave for example, the majority of differentially expressed genes occurred in the liver (Figure 3), and the enrichment in Reactome pathways differed among organs (especially gills vs. liver and brain; Fig. S5). In both sulphidic habitats, differentially expressed genes were mostly concentrated in the gills (Figure 3), which are directly exposed to environmental H2S (Tobler et al., 2016). Furthermore, genes associated with H2S detoxification and the metabolic processing of sulphur were primarily differentially expressed in the gills but not the brain or the liver. This may suggest that the bulk of H2S is detoxified peripherally in the gills (rather than the liver as in other vertebrates; Dorman et al., 2002), potentially shielding internal organs from the toxic effects. Some organisms exposed to H2S in marine environments have also been hypothesized to sequester sulphide peripherally in external mucosa in order to minimize toxic effects (Goffredi, Jones, Erhlich, Springer, & Vrijenhoek, 2008; Oeschger & Vetter, 1992). In addition, modification of the integumentary system may be common during exposure to environmental stressors in general (Ao et al., 2015; Micallef et al., 2012).
Our results indicate that the choice of tissue substantially affects inferences about gene expression variation in nature. Many studies still focus on the analysis of single organs (Narum & Campbell, 2015; Wang et al., 2014), whole organisms (Gross et al., 2013) or lack formal analyses of how gene expression varies among organs (Hinaux et al., 2013; Uyhelji, Cheng, & Besansky, 2016). This is not necessarily problematic when a priori hypotheses are being tested, but a focus on single organs may also lead to skewed or misleading results that affect our inferences about organismal responses to environmental variation. The advent of single-cell sequencing (Gawad, Koh, & Quake, 2016)—which is now commonly applied in the biomedical sciences—will provide increasing resolution to understand the nature of organismal responses to environmental stressors in the future.
Our analyses also indicated that gene expression responses in populations exposed to multiple stressors are not the mere sum of the responses to individual stressors. Although the sulphidic cave population (exposed to H2S and darkness) had the highest total number of differentially expressed genes, the number of differentially expressed genes in the gills was higher in the sulphidic surface population, and the number of differentially expressed genes in the liver was higher in the nonsulphidic cave population (Figure 3). In addition, only a subset of responses in the sulphidic cave were actually shared with the sulphidic surface and the nonsulphidic cave population (see below) such that the functional overlap of responses was lower than what a comparison of mere numbers implied. This result highlights that there are likely interactive, nonadditive effects between H2S exposure and living in a cave environment. Such interactions have also been documented in experimental studies that manipulated exposure to multiple physiochemical stressors (Garcia-Reyero et al., 2012; Maes et al., 2013; Yang et al., 2007) and will complicate our ability to predict organismal responses along complex environmental gradients.
4.2 | Shared responses and the predictability of gene expression in different environments
The number of shared responses between the two sulphidic and the two cave populations increased with levels of biological organization (from transcript to gene to function; Figure 4a). This was particularly evident in the two cave populations that did not share a single differentially expressed transcript in some organs, yet differentially expressed genes shared about 20% of their functional attributes (GO terms). The increase in shared responses with level of organization is predicted by theory (Elmer & Meyer, 2011; Manceau et al., 2010; Rosenblum et al., 2014), and it has been documented in a variety of studies that investigated patterns of molecular evolution or the genetic basis of convergent trait evolution (Linnen, Kingsley, Jensen, & Hoekstra, 2009; Rosenblum, Römpler, Schöneberg, & Hoekstra, 2010; Shapiro et al., 2004). Hierarchical structuring in the context of gene expression is less explored both conceptually and empirically (but see Fong, Joyce, & Palsson, 2005; Mallarino, Linden, Linnen, & Hoekstra, 2017). However, expression changes of different transcripts or genes may lead to equivalent organismal performance (see Arnold, 1983; Losos, 2011; Wainwright, Alfaro, Bolnick, & Hulsey, 2005 for analogous concepts involving different levels of organismal integration), just like different mutations in the same gene or mutations in different genes can lead to equivalent phenotypic changes (e.g., Chen, DeVries, & Cheng, 1997; Hoekstra, Hirschmann, Bundey, Insel, & Crossland, 2006; Wilkens & Strecker, 2003). The key—but largely unexplored—difference is that differential gene expression may occur plastically over short periods of time, opening the possibility that individuals can embark on different solutions to cope with an environmental stressor (Koolhaas, 2008; Schulte, 2014). From a transcriptome perspective, future research needs to address how plastic and evolved changes in gene regulation affect our inferences about the hierarchical structuring of responses to specific environmental stressors.
The proportion of shared responses was also significantly higher between the two sulphidic than the two cave habitats. This was particularly evident at the levels of transcripts and genes (>20% of responses shared between sulphidic habitats, <3% of responses shared between cave habitats), but substantial differences were also observed at a functional level (37% vs. 20%). There are three non-mutually exclusive hypotheses about the mechanisms that could be driving this pattern. (i) The number of shared differentially expressed genes may be a function of phylogenetic relatedness; that is, shared responses may have been higher between the two sulphidic populations, because they are more closely related (Rohlfs, Harrigan, & Nielsen, 2013). To test this, we identified single nucleotide polymorphisms (SNPs) in our data set and analysed the relationship among populations (see supplementary material for details). The results indicated that the three extremophile populations were closely related, and unlike analyses based on microsatellites (e.g., Plath et al., 2010), there was no evidence for significant population structure in the coding portion of the genome (Fig. S6). Nonetheless, inference of population splits indicated that the two cave populations were more closely related to each other than to the sulphidic surface population (Fig. S7). Hence, phylogenetic history alone is unlikely to explain the low level of shared responses between the two cave populations. (ii) Our classification of habitats into discrete types may be an oversimplification of complex ecological gradients (see Kaeuffer et al., 2012). Besides the presence of H2S and the absence of light, there are likely additional sources of selection in the nonsulphidic cave that we have not considered and may have shaped the distinct patterns of gene expression. For example, despite their proximity, the two caves differ in ambient temperature, the ionic composition of the water (other than the presence of H2S) and the availability of trophic resources (see Tobler & Plath, 2011 for a review). (iii) Some sources of selection may elicit more predictable evolutionary responses than others. At least in the organs investigated here, the presence of H2S—unlike the absence of light—has clear-cut molecular targets (e.g., cytochrome c oxidase in the respiratory chain and haemoglobin) that affect specific physiological functions (e.g., energy metabolism, oxygen transport and oxidative stress; Li et al., 2011; Olson, 2012; Tobler et al., 2016). H2S’s direct interaction with specific proteins could constrain the diversity of adaptive solutions that mitigate the toxic effects, ultimately leading to more predictable outcomes in gene expression variation in replicated populations. Indeed, previous studies have found that sulphide spring populations exhibit positive selection on and differential expression of genes associated with highly conserved pathways involved in H2S toxicity and detoxification (Kelley et al., 2016; Pfenninger et al., 2014, 2015). Furthermore, the notion that H2S elicits more predictable organismal responses is supported by the level of shared responses between populations being disproportionally high in the gills, which are directly exposed to environmental H2S (Tobler et al., 2016). Physiochemical stressors with clear molecular targets eliciting predictable patterns of genetic evolution have also been documented in other organisms, such as killifish adapted to anthropogenic pollutants (Reid et al., 2016), insects adapted to secondary plant metabolites (Dobler, Dalla, Wagschal, & Agrawal, 2012) and poison frogs adapted to their own skin toxins (Tarvin, Santos, O’Connell, Zakon, & Cannatella, 2016).
4.3 | Potential mechanisms mediating adaptation to extreme environments
Functional annotations of differentially expressed genes provided insights about potential regulatory and physiological mechanisms mediating adaptation to sulphidic and cave environments. Populations in sulphidic environments upregulated key enzymes involved in enzymatic H2S detoxification, including multiple components of the sulphide: quinone oxidoreductase and the glutathione pathways (Hildebrandt & Grieshaber, 2008; Jackson, Melideo, & Jorns, 2012). In addition, we found evidence for upregulation of genes associated with the toxic effects of H2S. Specifically, H2S negatively affects aerobic ATP production and creates oxidative stress through the interruption of the mitochondrial respiratory chain (Cooper & Brown, 2008; Eghbal, Pennefather, & O’Brien, 2004), and genes associated with energy metabolism, anaerobic ATP production and oxidative stress responses were consistently upregulated. Enrichment analysis also indicated downregulation of ion transporters in the gills, genes associated with lipid and fatty acid metabolism in the liver and extra-cellular matrix components in the brain. Since the sulphidic surface and cave habitats also differ from normal surface streams in a variety other aspects of the environment (e.g., salinity, pH and oxygen concentrations; Tobler & Plath, 2011), some of the documented gene expression changes may have arisen in response to these correlated environmental variables. Future studies will need to address the potential functional consequences of these expression changes and test how they actually affect organismal performance in sulphidic environments.
There were only a few shared enriched GO terms in the two cave populations. Changes in the expression of ion transporters and the regulation of pH were likely driven by variation in water chemistry and not sources of selection that are typically associated with cave environments, although some ion channels have been associated with the development of pigmentation (Bellono, Escobar, Lefkovith, Marks, & Oancea, 2014), and both cave populations exhibit significant reductions of pigmentation in the skin (Joachim, Riesch, Jeffery, & Schlupp, 2013). Despite the lack of shared responses, it is important to note that there was evidence for significant enrichment in each cave population for several functions typically associated with cave adaptation. This includes genes associated with energy and lipid metabolism that could be related to resource scarcity in caves (Hüppop, 2000), with regulation of circadian rhythms that are likely related to the absence of light (Beale et al., 2013; Koilraj, Sharma, Marimuthu, & Chandrashekaran, 2000) and with neuronal development and axon guidance that may be related to changes in the brain anatomy of cavefish (Eifert et al., 2015). While the high proportion of shared responses between the two sulphidic habitats facilitated the identification of potential physiological pathways contributing to adaptation to H2S, the largely unique expression patterns in the two caves make inferences about the molecular underpinnings of adaptation to perpetually dark habitats much less straightforward. This is somewhat surprising because evolution in caves is typically associated with strong patterns of convergent phenotypic evolution (Jeffery, 2009; Porter & Crandall, 2003), and regressive evolution of pigmentation in cavefish is even caused by convergent molecular modifications (Protas et al., 2006). Perhaps, other organs not investigated here (e.g., eyes and skin) exhibit a higher degree of shared gene expression differences between the two cave populations.
5 | CONCLUSIONS
Overall, this study has documented significant variation in gene expression among spatially proximate and genetically closely related populations. Gene expression varied among populations in an organ-specific manner, suggesting that regulatory changes are likely important in coping with the environmental stressors present in this system (see Fay & Wittkopp, 2008; Romero, Ruvinsky, & Gilad, 2012). In addition, shared gene expression responses in H2S-rich and cave habitats increase significantly with level of organismal organization, which indicates that alternative transcriptional responses—albeit often in the same physiological pathways—mediate organismal responses to life in different environments. A key question remaining is whether the transcriptional variation among populations is a consequence of population-specific, plastic induction of gene expression in response to ambient environmental conditions, or whether there are evolved changes in gene regulation. Laboratory experiments using two of the extremophile populations studied here (nonsulphidic and sulphidic surface) have indicated that both plastic and evolved changes in gene regulation contribute to gene expression variation in natural habitats (Passow et al., unpublished data). Future studies will need to address how plasticity and evolution shape gene expression in the cave populations and identify potential regulatory mutations that could contribute to transcriptional variation among populations. Considering the rampant variation in functional traits among closely related populations, the fish of the Cueva del Azufre system provide a unique opportunity to study the genetic basis of complex phenotypic variation in nature, including patterns of gene expression. Accordingly, future studies should focus on identifying mechanisms linking genomic variation to phenotypes and organismal function (Barrett & Hoekstra, 2011; Storz & Wheat, 2010).
Supplementary Material
Acknowledgments
Funding information
Society of Integrative and Comparative Biology; American Livebearer Association; U.S. Army Research Office, Grant/Award Number: W911NF-15-1-0175; National Science Foundation, Grant/Award Number: IOS-1121832, IOS-1463720, IOS-1557860, IOS-1557795; National Institutes of Health, Grant/Award Number: 2R24OD011198-04A1
We would like to thank N. Franssen for assistance collecting field samples and the Centro de Investigación e Innovación para la Enseñanza y Aprendizaje (CIIEA) for providing logistical support. We also thank the Oklahoma State University High Performance Computing Center (OSUHPCC) for providing access, support and resources, especially D. Brunson and J. Schafer for all their guidance. Permits were provided by the Mexican Government (DGOPA.00093.120110.-0018). Research was supported by a Fellowship of Graduate Student Travel from the Society of Integrative and Comparative Biology and a Vern Parish Award from the American Livebearer Association to CNP, as well as grants from the National Science Foundation (IOS-1121832, IOS-1463720, IOS-1557860 and IOS-1557795) and the U.S. Army Research Office (W911NF-15-1-0175) to MT and JLK. Genome reference work was supported by a grant to WCW (NIH: 2R24OD011198-04A1).
Footnotes
DATA ACCESSIBILITY
Raw reads of gill (N = 14), liver (N = 16) and brain (N = 11) transcriptomes of P. mexicana are archived in the NCBI Short Read Archive (SRA; study accession ID: PRJNA369434). The P. mexicana reference genome was obtained from NCBI GenBank (Accession no: LMXC00000000.1) along with the mitochondrial reference sequences (Accession no: KC992991). Scripts for all analyses performed are located at https://github.com/jokelley/cave-molly-transcriptomes.
AUTHOR CONTRIBUTIONS
C.N.P. and M.T. designed the study C.N.P., L.A.R. and M.T. conducted fieldwork. C.N.P., M.-C.Y., A.S. and J.L.K. conducted laboratory work. C.N.P., A.P.B, J.L.K. and M.T. conducted the analysis. M.S., W.C.W. and C.B. provided some reagents and analytical tools. C.N.P. and M.T. wrote the manuscript, and all authors contributed substantially to revisions.
Additional Supporting Information may be found online in the supporting information tab for this article.
References
- Akashi HD, Cádiz Díaz A, Shigenobu S, Makino T, Kawata M. Differentially expressed genes associated with adaptation to different thermal environments in three sympatric Cuban Anolis lizards. Molecular Ecology. 2016;25:2273–2285. doi: 10.1111/mec.13625. [DOI] [PubMed] [Google Scholar]
- Altshuler I, Demiri B, Xu S, Constantin A, Yan ND, Cristescu ME. An integrated multi-disciplinary approach for studying multiple stressors in freshwater ecosystems: Daphnia as a model organism. Integrative and Comparative Biology. 2011;51:623–633. doi: 10.1093/icb/icr103. [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biology. 2010;11:1. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao J, Mu Y, Xiang LX, Fan D, Feng M, Zhang S, … Chen X. Genome sequencing of the Perciform fish Larimichthys crocea provides insights into molecular and genetic mechanisms of stress adaptation. PLoS Genetics. 2015;11:e1005118. doi: 10.1371/journal.pgen.1005118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold SJ. Morphology, performance and fitness. American Zoologist. 1983;23:347–361. [Google Scholar]
- Aspiras AC, Rohner N, Martineau B, Borowsky RL, Clifford CJ. Melanocortin 4 receptor mutations contribute to the adaptation of cavefish to nutrient-poor conditions. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:9968–9973. doi: 10.1073/pnas.1510802112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagarinao T. Sulfide as an environmental factor and toxicant: Tolerance and adaptations in aquatic organisms. Aquatic Toxicology. 1992;24:21–62. [Google Scholar]
- Bagarinao T, Vetter R. Oxidative detoxification of sulfide by mitochondria of the California killifish Fundulus parvipinnis and the speckled sanddap Citharichthys stignaeus. Journal of Comparative Physiology B. 1990;160:519–527. [Google Scholar]
- Bailey NW, Veltsos P, Tan YF, Millar AH, Ritchie MG, Simmons LW. Tissue-specific transcriptomics in the field cricket Teleogryllus oceanicus. G3: Genes, Genomes, Genetics. 2013;3:225–230. doi: 10.1534/g3.112.004341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajić D, Poyatos JF. Balancing noise and plasticity in eukaryotic gene expression. BMC Genomics. 2012;13:343. doi: 10.1186/1471-2164-13-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett RDH, Hoekstra HE. Molecular spandrels: Tests of adaptation at the genetic level. Nature Reviews Genetics. 2011;12:767–780. doi: 10.1038/nrg3015. [DOI] [PubMed] [Google Scholar]
- Beale A, Guibal C, Tamai TK, Klotz L, Cowen S, Peyric E, … Whitmore D. Circadian rhythms in Mexican blind cavefish Astyanax mexicanus in the lab and in the field. Nature Communications. 2013;4:2769. doi: 10.1038/ncomms3769. [DOI] [PubMed] [Google Scholar]
- Bellono NW, Escobar IE, Lefkovith AJ, Marks MS, Oancea E. An intracellular anion channel critical for pigmentation. eLife. 2014;3:e04543. doi: 10.7554/eLife.04543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN. A gene expression map of the Arabidopsis root. Science. 2003;302:1956–1960. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- Bos N, Pulliainen U, Sundström L, Freitak D. Starvation resistance and tissue-specific gene expression of stress-related genes in a naturally inbred ant population. Royal Society Open Science. 2016;3:160062. doi: 10.1098/rsos.160062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, DeVries AL, Cheng CH. Convergent evolution of antifreeze glycoproteins in Antarctic notothenioid fish and Arctic cod. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:3817–3822. doi: 10.1073/pnas.94.8.3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheviron ZA, Whitehead A, Brumfield RT. Transcriptomic variation and plasticity in rufous-collared sparrows (Zonotrichia capensis) along an altitudinal gradient. Molecular Ecology. 2008;17:4556–4569. doi: 10.1111/j.1365-294X.2008.03942.x. [DOI] [PubMed] [Google Scholar]
- Conesa A, Goetz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Cooper CE, Brown GC. The inhibition of mitochondrial cytochrome oxidase by the gases carbon monoxide, nitric oxide, hydrogen cyanide and hydrogen sulfide: Chemical mechanism and physiological significance. Journal of Bioenergy and Biomembranes. 2008;40:533–539. doi: 10.1007/s10863-008-9166-6. [DOI] [PubMed] [Google Scholar]
- Dobler S, Dalla S, Wagschal V, Agrawal AA. Community-wide convergent evolution in insect adaptation to toxic cardenolides by substitutions in the Na, K-ATPase. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:13040–13045. doi: 10.1073/pnas.1202111109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman DC, Moulin FJ, McManus BE, Mahle KC, James RA, Struve MF. Cytochrome oxidase inhibition induced by acute hydrogen sulfide inhalation: Correlation with tissue sulfide concentrations in the rat brain, liver, lung, and nasal epithelium. Toxicological Sciences. 2002;65:18–25. doi: 10.1093/toxsci/65.1.18. [DOI] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eghbal MA, Pennefather PS, O’Brien PJ. H2S cytotoxicity mechanism involves reactive oxygen species formation and mitochondrial depolarisation. Toxicology. 2004;203:69–76. doi: 10.1016/j.tox.2004.05.020. [DOI] [PubMed] [Google Scholar]
- Eifert C, Farnworth M, Schulz-Mirbach T, Riesch R, Bierbach D, Klaus S, … Plath M. Brain size variation in extremophile fish: Local adaptation vs. phenotypic plasticity. Journal of Zoology. 2015;295:143–153. [Google Scholar]
- Elmer KR, Meyer A. Adaptation in the age of genomics: Insights from parallelism and convergence. Trends in Ecology & Evolution. 2011;26:298–306. doi: 10.1016/j.tree.2011.02.008. [DOI] [PubMed] [Google Scholar]
- Evans DH, Claiborne JB. The physiology of fishes. Boca Raton, FL: Taylor & Francis; 2006. [Google Scholar]
- Evans TG, Hammill E, Kaukinen K, Schulze AD, Patterson DA, English KK, … Miller KM. Transcriptomics of environmental acclimatization and survival in wild adult Pacific sockeye salmon (Oncorhynchus nerka) during spawning migration. Molecular Ecology. 2011;20:4472–4489. doi: 10.1111/j.1365-294X.2011.05276.x. [DOI] [PubMed] [Google Scholar]
- Fay JC, Wittkopp PJ. Evaluating the role of natural selection in the evolution of gene regulation. Heredity. 2008;100:191–199. doi: 10.1038/sj.hdy.6801000. [DOI] [PubMed] [Google Scholar]
- Folt CL, Chen CY, Moore MV, Burnaford J. Synergism and antagonism among multiple stressors. Limnology and Oceanography. 1999;44:864–877. [Google Scholar]
- Fong SS, Joyce AR, Palsson BØ. Parallel adaptive evolution cultures of Escherichia coli lead to convergent growth phenotypes with different gene expression states. Genome Research. 2005;15:1365–1372. doi: 10.1101/gr.3832305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Reyero N, Escalon BL, Loh PR, Laird JG, Kennedy AJ, Berger B, Perkins EJ. Assessment of chemical mixtures and groundwater effects on Daphnia magna transcriptomics. Environmental Science & Technology. 2012;46:42–50. doi: 10.1021/es201245b. [DOI] [PubMed] [Google Scholar]
- Gawad C, Koh W, Quake SR. Single genome sequencing: Current state of the science. Nature Reviews Genetics. 2016;17:175–188. doi: 10.1038/nrg.2015.16. [DOI] [PubMed] [Google Scholar]
- Gene Ontology Consortium. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Research. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffredi SK, Jones WJ, Erhlich H, Springer A, Vrijenhoek RC. Epibiotic bacteria associated with the recently discovered Yeti crab, Kiwa hirsuta. Environmental Microbiology. 2008;10:2623–2634. doi: 10.1111/j.1462-2920.2008.01684.x. [DOI] [PubMed] [Google Scholar]
- Gracey AY, Troll JV, Somero GN. Hypoxia-induced gene expression profiling in the euryoxic fish Gillichthys mirabilis. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:1993–1998. doi: 10.1073/pnas.98.4.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JB, Furterer A, Carlson BM, Stahl BA. An integrated transcriptome-wide analysis of cave and surface Astyanax mexicanus. PLoS ONE. 2013;8:e55659. doi: 10.1371/journal.pone.0055659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Liu R, Huang L, Zheng XM, Liu PL, Du YS, … Ge S. Widespread and adaptive alterations in genome-wide gene expression associated with ecological divergence of two Oryza species. Molecular Biology and Evolution. 2016;33:62–78. doi: 10.1093/molbev/msv196. [DOI] [PubMed] [Google Scholar]
- Hildebrandt TM, Grieshaber M. Three enzymatic activities catalyze the oxidation of sulfide to thiosulfate in mammalian and invertebrate mitochondria. FEBS Journal. 2008;275:3352–3361. doi: 10.1111/j.1742-4658.2008.06482.x. [DOI] [PubMed] [Google Scholar]
- Hinaux H, Poulain J, Da Silva C, Noirot C, Jeffery WR, Casane D, Rétaux S. De novo sequencing of Astyanax mexicanus surface fish and Pachón cavefish transcriptomes reveals enrichment of mutations in cavefish putative eye genes. PLoS ONE. 2013;8:e53553. doi: 10.1371/journal.pone.0053553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra H, Hirschmann RJ, Bundey RA, Insel PA, Crossland JP. A single amino acid mutation contributes to adaptive beach mouse color pattern. Science. 2006;313:101–104. doi: 10.1126/science.1126121. [DOI] [PubMed] [Google Scholar]
- Holmstrup M, Bindesbol AM, Janneke Oostingh G, Duschl A, Scheil V, Kohler HR, … Spurgeon DJ. Interactions between effects of environmental chemicals and natural stressors: A review. Science of the Total Environment. 2010;408:3746–3762. doi: 10.1016/j.scitotenv.2009.10.067. [DOI] [PubMed] [Google Scholar]
- Hüppop K. How do cave animals cope with the food scarcity in caves? In: Wilkens H, Culver DC, Humphries WF, editors. Ecosystems of the world 30: Subterranean ecosystems. Amsterdam: Elsevier Science; 2000. pp. 159–188. [Google Scholar]
- Jackson MR, Melideo SL, Jorns M. Human sulfide: Qui-none oxidoreductase catalyzes the first step in hydrogen sulfide metabolism and produces a sulfane sulfur metabolite. Biochemistry. 2012;51:6804–6815. doi: 10.1021/bi300778t. [DOI] [PubMed] [Google Scholar]
- Jeffery WR. Regressive evolution in Astyanax cavefish. Annual Review of Genetics. 2009;43:25–47. doi: 10.1146/annurev-genet-102108-134216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joachim BL, Riesch R, Jeffery WR, Schlupp I. Pigment cell retention in cavernicolous populations of Poecilia mexicana (Poecili-idae) Bulletin of Fish Biology. 2013;14:61–73. [Google Scholar]
- Kaeuffer R, Peichel CL, Bolnick DI, Hendry AP. Parallel and nonparallel aspects of ecological, phenotypic, and genetic divergence across replicate population pairs of lake and stream stickleback. Evolution. 2012;66:402–418. doi: 10.1111/j.1558-5646.2011.01440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley JL, Arias-Rodriguez L, Patacsil Martin D, Yee MC, Bustamante C, Tobler M. Mechanisms underlying adaptation to life in hydrogen sulfide rich environments. Molecular Biology and Evolution. 2016;33:1419–1434. doi: 10.1093/molbev/msw020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley JL, Passow C, Plath M, Arias-Rodriguez L, Yee MC, Tobler M. Genomic resources for a model in adaptation and speciation research: The characterization of the Poecilia mexicana transcriptome. BMC Genomics. 2012;13:652. doi: 10.1186/1471-2164-13-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koilraj AJ, Sharma VK, Marimuthu G, Chandrashekaran MK. Presence of circadian rhythms in the locomotor activity of a cave-dwelling millipede Glyphiulus cavernicolus sulu (Cambalidae, Spirostreptida) Chronobiology International. 2000;17:757–765. doi: 10.1081/cbi-100102111. [DOI] [PubMed] [Google Scholar]
- Kombian SB, Warenyica MW, Mele F, Reiffenstein RJ. Effects of acute intoxication with hydrogen sulfide on central amino acid transmitter systems. Neurotoxicology. 1988;9:587–596. [PubMed] [Google Scholar]
- Koolhaas JM. Coping style and immunity in animals: Making sense of individual variation. Brain, Behavior, and Immunity. 2008;22:662–667. doi: 10.1016/j.bbi.2007.11.006. [DOI] [PubMed] [Google Scholar]
- Kopec AK, D’Souza ML, Mets BD, Burgoon LD, Reese SE, Archer KJ, … Harkema J. Non-additive hepatic gene expression elicited by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) and 2,2′,4,4′,5,5′-hexachlorobiphenyl (PCB153) co-treatment in C57BL/6 mice. Toxicology and Applied Pharmacology. 2011;256:154–167. doi: 10.1016/j.taap.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs CJ. Ecological methodology. 2. Menlo Park, CA: Addison-Wesley Publishers; 1999. [Google Scholar]
- Krueger F. Trim Galore! version 0.3.7. 2014 http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/
- Langecker TG. The effect of continuous darkness on cave ecology and cavernicolous evolution. In: Wilkens H, Culver DC, Humphreys WF, editors. Ecosystems of the world 30: Subterranean ecosystems. Amsterdam: Elsevier Science; 2000. pp. 135–157. [Google Scholar]
- Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B. Conflict between noise and plasticity in yeast. PLoS Genetics. 2010;6:e1001185. doi: 10.1371/journal.pgen.1001185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. 2013 arXiv Preprint. [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Rose P, Moore PK. Hydrogen sulfide and cell signaling. Annual Review of Pharmacology and Toxicology. 2011;51:169–187. doi: 10.1146/annurev-pharmtox-010510-100505. [DOI] [PubMed] [Google Scholar]
- Linnen CR, Kingsley EP, Jensen JD, Hoekstra HE. On the origin and spread of an adaptive allele in deer mice. Science. 2009;325:1095–1098. doi: 10.1126/science.1175826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhang Z, Ma X, Li X, Zhou D, Gao B, Bai Y. Sulfide exposure results in enhanced sqr transcription through upregulating the expression and activation of HSF1 in echiuran worm Urechis unicinctus. Aquatic Toxicology. 2015;170:229–239. doi: 10.1016/j.aquatox.2015.11.021. [DOI] [PubMed] [Google Scholar]
- López-Maury L, Marguerat S, Bähler J. Tuning gene expression to changing environments: From rapid responses to evolutionary adaptation. Nature Reviews Genetics. 2008;9:583–593. doi: 10.1038/nrg2398. [DOI] [PubMed] [Google Scholar]
- Losos JB. Convergence, adaptation, and constraint. Evolution. 2011;65:1827–1840. doi: 10.1111/j.1558-5646.2011.01289.x. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes GE, Raeymaekers JA, Hellemans B, Geeraerts C, Parmentier K, De Temmerman L, … Belpaire C. Gene transcription reflects poor health status of resident European eel chronically exposed to environmental pollutants. Aquatic Toxicology. 2013;126:242–255. doi: 10.1016/j.aquatox.2012.11.006. [DOI] [PubMed] [Google Scholar]
- Mallarino R, Linden TA, Linnen CR, Hoekstra H. The role of isoforms in the evolution of cryptic coloration in Peromyscus mice. Molecular Ecology. 2017;26:245–258. doi: 10.1111/mec.13663. [DOI] [PubMed] [Google Scholar]
- Manceau M, Domingues VS, Linnen CR, Rosenblum EB, Hoekstra H. Convergence in pigmentation at multiple levels: Mutation, genes, and function. Philosophical Transactions of the Royal Society B. 2010;365:2439–2450. doi: 10.1098/rstb.2010.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F, Braasch I, Phillips JB, Lin XW, Titus T, Zhang C, Postlethwait JH. Evolution of the eye transcriptome under constant darkness in Sinocyclocheilus cavefish. Molecular Biology and Evolution. 2013;30:1527–1543. doi: 10.1093/molbev/mst079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micallef G, Bickerdike R, Reiff C, Fernandes JM, Bowman AS, Martin SA. Exploring the transcriptome of Atlantic salmon (Salmo salar) skin, a major defense organ. Marine Biotechnology. 2012;14:559–569. doi: 10.1007/s10126-012-9447-2. [DOI] [PubMed] [Google Scholar]
- Morris MR, Richard R, Leder EH, Barrett RD, Aubin-Horth N, Rogers SM. Gene expression plasticity evolves in response to colonization of freshwater lakes in threespine stickleback. Molecular Ecology. 2014;23:3226–3240. doi: 10.1111/mec.12820. [DOI] [PubMed] [Google Scholar]
- Narum SR, Campbell NR. Transcriptomic response to heat stress among ecologically divergent populations of redband trout. BMC Genomics. 2015;16:103. doi: 10.1186/s12864-015-1246-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemiller ML, Soares D. Cave environments. In: Riesch R, Tobler M, Plath M, editors. Extremophile fishes: Ecology, evolution, and physiology of teleosts in extreme environments. Heidelberg, Germany: Springer; 2015. pp. 161–191. [Google Scholar]
- Oeschger R, Vetter RD. Sulfide detoxification and tolerance in Halicryptus spinulosus: A multiple strategy. Marine Ecology Progress Series. 1992;86:167–179. [Google Scholar]
- Oldham MC, Horvath S, Geschwind DH. Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:17973–17978. doi: 10.1073/pnas.0605938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleksiak MF, Churchill GA, Crawford DL. Variation in gene expression within and among natural populations. Nature Genetics. 2002;32:261–266. doi: 10.1038/ng983. [DOI] [PubMed] [Google Scholar]
- Olson KR. A practical look at the chemistry and biology of hydrogen sulfide. Antioxidants & Redox Signaling. 2012;17:32–44. doi: 10.1089/ars.2011.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parzefall J. A review of morphological and behavioural changes in the cave molly, Poecilia mexicana, from Tabasco, Mexico. Environmental Biology of Fishes. 2001;62:263–275. [Google Scholar]
- Passow C, Greenway R, Arias-Rodriguez L, Jeyasingh PD, Tobler M. Reduction of energetic demands through modification of body size and metabolic rates in locally adapted extremophile fishes. Comparative Biochemistry and Physiology. 2015;88:371–383. doi: 10.1086/681053. [DOI] [PubMed] [Google Scholar]
- Pfenninger M, Lerp H, Tobler M, Passow C, Kelley JL, Funke E, … Plath M. Parallel evolution of cox genes in H2S-tolerant fish as key adaptation to a toxic environment. Nature Communications. 2014;5:3873. doi: 10.1038/ncomms4873. [DOI] [PubMed] [Google Scholar]
- Pfenninger M, Patel S, Arias-Rodriguez L, Feldmeyer B, Riesch R, Plath M. Unique evolutionary trajectories in repeated adaptation to hydrogen sulphide-toxic habitats of a neotropical fish (Poecilia mexicana) Molecular Ecology. 2015;24:5446–5459. doi: 10.1111/mec.13397. [DOI] [PubMed] [Google Scholar]
- Plath M, Hermann C, Schröder R, Riesch R, Tobler M, Garcia de Leon FJ, Tiedemann R. Locally adapted fish populations maintain small-scale genetic differentiation despite perturbation by a catastrophic flood event. BMC Evolutionary Biology. 2010;10:256. doi: 10.1186/1471-2148-10-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter M, Crandall K. Lost along the way: The significance of evolution in reverse. Trends in Ecology & Evolution. 2003;18:541–547. [Google Scholar]
- Poulson TL. Adaptations of cave fishes with some comparisons to deep-sea fishes. Environmental Biology of Fishes. 2001;62:345–364. [Google Scholar]
- Poulson TL, White WB. The cave environment. Science. 1969;165:971–981. doi: 10.1126/science.165.3897.971. [DOI] [PubMed] [Google Scholar]
- Protas ME, Hersey C, Kochanek D, Zhou Y, Wilkens H, Jeffery WR, … Tabin CJ. Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nature Genetics. 2006;38:107–111. doi: 10.1038/ng1700. [DOI] [PubMed] [Google Scholar]
- Reid NM, Prostou DA, Clark BW, Warren WC, Colbourne JK, Shaw JR, … Whitehead A. The genomic landscape of rapid repeated evolutionary adaptation to toxic pollution in wild fish. Science. 2016;354:1305–1308. doi: 10.1126/science.aah4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesch R, Plath M, Schlupp I. Speciation in caves: Experimental evidence that permanent darkness promotes reproductive isolation. Biology Letters. 2011;7:909–912. doi: 10.1098/rsbl.2011.0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesch R, Plath M, Schlupp I, Marsh-Matthews E. Matrotrophy in the cave molly: An unexpected provisioning strategy in an extreme environment. Evolutionary Ecology. 2010;24:789–801. [Google Scholar]
- Roach K, Tobler M, Winemiller KO. Hydrogen sulfide, bacteria, and fish: A unique, subterranean food chain. Ecology. 2011;92:2056–2062. doi: 10.1890/11-0276.1. [DOI] [PubMed] [Google Scholar]
- Roberts A, Pachter L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nature Methods. 2013;10:71–73. doi: 10.1038/nmeth.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bio-conductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biology. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlfs R, Harrigan P, Nielsen R. Modeling gene expression evolution with an extended Ornstein-Uhlenbeck process accounting for within-species variation. Molecular Biology and Evolution. 2013;31:201–211. doi: 10.1093/molbev/mst190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero IG, Ruvinsky I, Gilad Y. Comparative studies of gene expression and the evolution of gene regulation. Nature Reviews Genetics. 2012;13:505–516. doi: 10.1038/nrg3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblum EB, Parent CE, Brandt EE. The molecular basis of convergent evolution. Annual Review of Ecology Evolution and Systematics. 2014;45:203–226. [Google Scholar]
- Rosenblum EB, Römpler H, Schöneberg T, Hoekstra HE. Molecular and functional basis of phenotypic convergence in white lizards at White Sands. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:2113–2117. doi: 10.1073/pnas.0911042107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte PM. What is environmental stress? Insights from fish living in variable environments. Journal of Experimental Biology. 2014;217:23–34. doi: 10.1242/jeb.089722. [DOI] [PubMed] [Google Scholar]
- Shapiro MD, Marks ME, Peichel CL, Blackman BK, Nereng KS, Jónsson B, … Kingsley DM. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature. 2004;428:717–723. doi: 10.1038/nature02415. [DOI] [PubMed] [Google Scholar]
- Shu J, Kang JS, Yang KH, Kaminski NE. Antagonism of aryl hydrocarbon receptor-dependent induction of CYP1A1 and inhibition of IgM expression by di-ortho-substituted polychlorinated biphenyls. Toxicology and Applied Pharmacology. 2003;187:11–21. doi: 10.1016/s0041-008x(02)00040-6. [DOI] [PubMed] [Google Scholar]
- Storz JF, Wheat CW. Integrating evolutionary and functional approaches to infer adaptation at specific loci. Evolution. 2010;64:2489–2509. doi: 10.1111/j.1558-5646.2010.01044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickler AG, Byerly MS, Jeffery WR. Lens gene expression analysis reveals downregulation of the anti-apoptotic chaperone alphaA-crystallin during cavefish eye degeneration. Development Genes and Evolution. 2007;217:771–782. doi: 10.1007/s00427-007-0190-z. [DOI] [PubMed] [Google Scholar]
- Tarvin RD, Santos JC, O’Connell LA, Zakon HH, Cannatella DC. Convergent substitutions in a sodium channel suggests multiple origins of toxin resistance in poison frogs. Molecular Biology and Evolution. 2016;33:1068–1081. doi: 10.1093/molbev/msv350. [DOI] [PubMed] [Google Scholar]
- Tobler M. Does a predatory insect contribute to the divergence between cave- and surface adapted fish populations? Biology Letters. 2009;5:506–509. doi: 10.1098/rsbl.2009.0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobler M, Coleman SW, Perkins BD, Rosenthal GG. Reduced opsin gene expression in a cave-dwelling fish. Biology Letters. 2010;6:98–101. doi: 10.1098/rsbl.2009.0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobler M, DeWitt TJ, Schlupp I, Garcia de Leon FJ, Herrmann R, Feulner P, … Plath M. Toxic hydrogen sulfide and dark caves: Phenotypic and genetic divergence across two abiotic environmental gradients in Poecilia mexicana. Evolution. 2008;62:2643–2649. doi: 10.1111/j.1558-5646.2008.00466.x. [DOI] [PubMed] [Google Scholar]
- Tobler M, Passow CN, Greenway R, Kelley JL, Shaw JH. The evolutionary ecology of animals inhabiting hydrogen sulfide-rich environments. Annual Review of Ecology, Evolution and Systematics. 2016;47:239–262. [Google Scholar]
- Tobler M, Plath M. Living in extreme habitats. In: Evans J, Pilastro A, Schlupp I, editors. Ecology and evolution of poeciliid fishes. Chicago: University of Chicago Press; 2011. pp. 120–127. [Google Scholar]
- Tobler M, Riesch R, Tobler CM, Schulz-Mirbach T, Plath M. Natural and sexual selection against immigrants maintains differentiation among micro-allopatric populations. Journal of Evolutionary Biology. 2009;22:2298–2304. doi: 10.1111/j.1420-9101.2009.01844.x. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, … Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyhelji HA, Cheng C, Besansky NJ. Transcriptomic differences between euryhaline and stenohaline malaria vector sibling species in response to salinity stress. Molecular Ecology. 2016;25:2210–2225. doi: 10.1111/mec.13609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainwright PC, Alfaro ME, Bolnick DI, Hulsey CD. Many-to-one mapping of form to function: A general principle of organismal design. Integrative and Comparative Biology. 2005;45:256–262. doi: 10.1093/icb/45.2.256. [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-seq: A revolutionary tool for transcriptomics. Nature Reviews Genetics. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Yang E, Smith KJ, Zeng Y, Ji G, Connon R, … Cai JJ. Gene expression responses of threespine stickleback to salinity: Implications for salt-sensitive hypertension. Frontiers in Genetics. 2014;5:312. doi: 10.3389/fgene.2014.00312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead A. Comparative genomics in ecological physiology: Toward a more nuanced understanding of acclimation and adaptation. Journal of Experimental Biology. 2012;215:884–891. doi: 10.1242/jeb.058735. [DOI] [PubMed] [Google Scholar]
- Whitehead A, Crawford DL. Variation in tissue-specific gene expression among natural populations. Genome Biology. 2005;6:R13. doi: 10.1186/gb-2005-6-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead A, Pilcher W, Champlin D, Nacci D. Common mechanism underlies repeated evolution of extreme pollution tolerance. Proceedings of the Royal Society B. 2012;279:427–433. doi: 10.1098/rspb.2011.0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkens H, Strecker U. Convergent evolution of the cavefish Astyanax (Characidae, Teleostei): Genetic evidence from reduced eye-size and pigmentation. Biological Journal of the Linnean Society. 2003;80:545–554. [Google Scholar]
- Wong YH, Sun J, He LS, Chen LG, Qiu JW, Qian PY. High-throughput transcriptome sequencing of the cold seep mussel Bathymodiolus platifrons. Scientific Reports. 2015;5:16597. doi: 10.1038/srep16597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Kemadjou JR, Zinsmeister C, Bauer M, Legradi J, Müller F, … Strähle U. Transcriptional profiling reveals barcode-like toxicogenomic responses in the zebrafish embryo. Genome Biology. 2007;8:R227. doi: 10.1186/gb-2007-8-10-r227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, He QY. ReactomePA: An R/Bioconductor package for reactome pathway analysis and visualization. Molecular BioSystems. 2016;12:477–479. doi: 10.1039/c5mb00663e. [DOI] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS: A Journal of Integrative Biology. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.