Abstract
The study of phenomes or phenomics has been a central part of biology. The field of automatic phenotype acquisition technologies based on images has seen an important advance in the last years. As with other high-throughput technologies, it addresses a common set of problems, including data acquisition and analysis. In this review, we give an overview of the main systems developed to acquire images. We give an in-depth analysis of image processing with its major issues and the algorithms that are being used or emerging as useful to obtain data out of images in an automatic fashion.
Keywords: Algorithms, artificial vision, deep learning, hyperspectral cameras, machine learning, segmentation
Background
The development of systems to monitor large fields using the Normalized Difference Vegetation Index (NDVI) started more than 25 years ago when NDVI was used in the so-called remote sensing field [1]. It was an important milestone in the advance of automatic methods for analysing plant growth and biomass [2]. Ever since, new technologies have increased our capacity to obtain data from biological systems. The ability to measure chlorophyll status from satellite images allowed plant health to be measured in large fields and predict crops and productivity in very large areas such as the Canadian prairies, Burkina Faso, or the Indian Basin in Pakistan [3–6]. Thus, the field of remote sensing is an important basis where knowledge about data acquisition and analysis started. The development of phenotyping devices using local cameras for crops took off using an array of technologies including Infrared thermography to measure stomatal opening or osmotic stress [7–9]. Extraction of quantitative data from images has been developed to study root development [10–12] and has found a niche to identify germplasm resistant to abiotic stresses in plants such as cereals [13], Arabidopsis [14], and large-scale field phenotyping [15]. There are several recent reviews addressing the different types of growing setups [16–22], and we will not cover them in the current review.
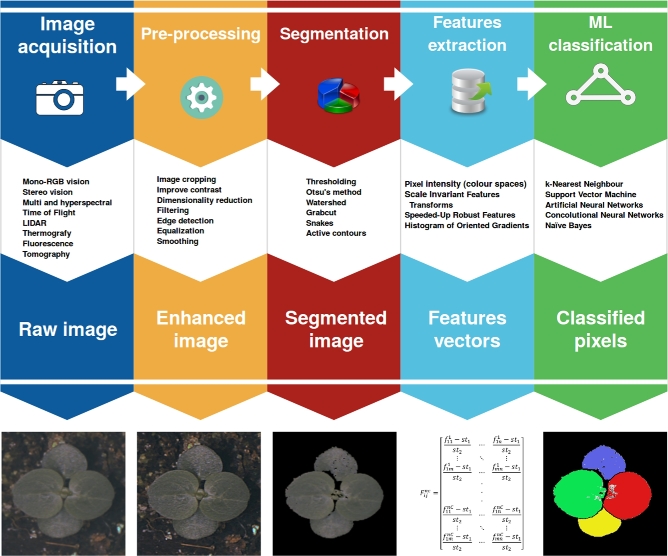
Two main aspects to consider are the type of image acquired and how to process it. There are a number of recent reviews on phenomics and high-throughput image data acquisition [15, 23–26]. In contrast, the majority of the literature concerning image processing and analysis is found in books where methods are described in detail [27–31]. There are some very good reviews on aspects of data acquisition and analysis, i.e., imaging techniques [32], machine learning (ML) for high-throughput phenotyping [33], or software for image analysis [34], but a detailed review on the different types of data analysis is lacking. In this review, we cover the current and emerging methods of image acquisition and processing that allow image-based phenomics (Fig. 1).
Figure 1:
Basic workflow in computer vision–based plant phenotyping.
Review
Image acquisition
Image acquisition is the process through which we obtain a digital representation of a scene. This representation is known as an image, and its elements are called pixels (picture elements). The electronic device used to capture a scene is known as an imaging sensor. A charge-coupled device (CCD) and complementary metal oxide semiconductor (CMOS) are the most broadly used technologies in image sensors. A light wavelength is captured by small analogic sensors, which will acquire major or minor charge depending on the amount of incident light. These signals are amplified, filtered, transported, and enhanced by means of specific hardware. A suitable output interface and a lens in the same housing are all that is needed to perform image acquisition. The elements enumerated above comprise the main element of computer vision systems, the camera. Time delay and integration (TDI) is an imaging acquisition mode that can be implemented over CCD [35] or CMOS [36]. It improves the features of the image acquisition system considerably. TDI is used in applications that require the ability to operate in extreme lighting conditions, requiring both high speed and high sensitivity, e.g., inline monitoring, inspection, sorting, and remote sensing (for weather or vegetation observation) [36].
The aforementioned technologies, CCD, CMOS, and TDI, confer unique characteristics, which define the type of data a camera can provide with a degree of robustness. There are fundamental differences in the types of performance the different sensors offer. In recent years, CMOS technology has outperformed CCDs in most visible imaging applications. When selecting an imaging sensor (a camera), CCD technology causes less noise and produces higher-quality images, mainly in scenes with bad illumination. It has a better depth of colour due to the higher dynamic range. On the other hand, the CMOS sensors are faster at processing images. Due to the hardware architecture for pixel extraction, they need less electrical power to operate, they allow a region of interest to be processed on the device, and they are cheaper than CCDs. Furthermore, TDI mode with CCD or CMOS imaging sensors is used for high-speed and low–light level applications [37]. The latest technological developments in cameras show that the trend of the manufacturers such as IMEC, world-leader in nanoelectronics, is to fuse TDI technology with CCD and CMOS characteristics in the same device [38]. TDI technology is expected to be applied to high-throughput phenotyping processes in the near future.
The field of image acquisition is extremely developed in the literature, but image acquisition systems can be classified into 7 groups that are suitable for phenotyping.
Mono-RGB vision
Mono-RGB vision systems are composed of a set comprising a lens, imaging sensor, specific hardware, and input/output (IO) interface. Depending on if they use a line or matrix of pixels, they are classified as line cameras (or scanners) or matrix cameras. Most computer vision phenotyping devices are based on mono-RGB vision systems. Examples of mono-RGB vision devices include Smart tools for the Prediction and Improvement of Crop Yield, an automated phenotyping prototype of large pepper plants in the greenhouse. The system uses multiple RGB cameras to extract 2 types of features: features from a 3D reconstruction of the plant canopy and statistical features derived directly from RGB images [39]. A different approach has been used with 2 cameras inside a growth chamber to measure circadian growth features of Petunia, Antirrhinum, and Opuntia [40]. Two cameras with low and high magnifications were used to carry out phenotype studies of Arabidopsis thaliana seeds. The system is mounted on a 3-axis gantry, and the rotation of the samples allows the gravitropic bending response to be determined in the roots, as well as its posterior quantification [41]. Recently a high-throughput RGB system has been developed to identify quantitative trait loci (QTL) involved in yield in large recombinant inbred lines in maize [42], demonstrating the increasing impact of this approach in phenomics.
These devices have excellent spatial and temporal resolution; i.e., they can produce a very large number of images in very short periods and at a very low cost. They are portable, and there are many software tools to perform image processing (Table 1). Systems based on mono-RGB vision allow a quantification of the plant canopy [43], as well as sufficient computation of vegetation indices for most purposes. The main disadvantages are caused by the overlap of plant organs during growth and nutation phases and the relative position of the organs with respect to the device that makes the precise quantification difficult. In addition, these devices are affected by variations in illumination when used outdoors. The trend in outdoor plant phenotyping is to combine mono-RGB systems with other systems such as light detection and ranging (LIDAR) devices (see below) or thermal imaging, or adding new bands or filters to the camera that allow the segmenting of specific regions of the spectrum [44, 45].
Table 1:
List of software tools for image processing
| Vision libraries | Source | Language |
|---|---|---|
| OpenCV | http://opencv.org | C++, Python, Java, C# |
| EmguCV | http://www.emgu.com/ | |
| PlantCV | http://plantcv.danforthcenter.org | Python |
| Scikit-image | http://scikit-image.org | |
| Bioimagetools, bayesimages, edci, DRIP, dpmixsim, raster, … | https://cran.r-project.org/ | R |
| Cimg | http://cimg.eu | C++ |
| Simplecv | http://simplecv.org | |
| Fastcv | https://developer.qualcomm.com/software/fastcv-sdk | |
| Ccv | http://libccv.org | |
| Vxl | http://vxl.sourceforge.net | |
| BoofCV | http://boofcv.org | Java |
| OpenIMAJ | http://openimaj.org | |
| JavaCV | https://github.com/bytedeco/javacv |
Stereo vision
Stereo vision systems try to correct a drawback of mono-RGB vision systems for distance measurement. The architecture of stereo vision systems emulates the behaviour of human vision using 2 mono vision systems. Basically, after locating a point in 2 mono vision systems, it is possible to compute the distance from the point to the system. Images produced are known as depth maps [46]. A stereo vision system has been used by Biskup and colleagues [47] to obtain structural features of plant canopies. The 3D reconstruction has been successfully employed to obtain 3D models of plants, thus demonstrating the power of this approach [48]. Simple depth reconstructions help to define stems, leaves, and grapes, showing the potential of this technology [49]. An RGB camera mounted on a mobile robot is used as an automated 3D phenotyping of vineyards under field conditions. Sequentially, the system captures a set of images, which are used to reconstruct a textured 3D point cloud of the whole grapevine row [50]. Stereo vision has been developed to perform high-throughput analysis of rapeseed leaf traits. The system uses 2 identical RGB cameras to obtain stereo images for canopy and 3D reconstruction [51]. Developing a 3D-mesh segmentation has allowed cotton growth to be analysed [52], showing the further possibilities of 3D imaging.
The main advantage of 3D systems is their simplicity; 2 cameras are enough to obtain depth maps. Stereo vision has evolved into multi-view stereo (MSV) and has found a place in plant phenotyping [53]. Furthermore, MSV is a low-cost 3D image acquisition system compared with other technologies such as LIDAR or tomography imaging [54]. Stereo vision systems have important weaknesses. They are affected by changes of the scene illumination, they need a high-performance computational system to carry out stereo-matching algorithms, and they have poor depth resolution [55]. These limitations are increased in outdoor environments, as image segmentation becomes more challenging.
Multi- and hyperspectral cameras
Multispectral and hyperspectral cameras have been used in numerous fields of science and in industrial applications [56–61]. The spectral resolution is the main factor that distinguishes multispectral imagery from hyperspectral imagery [62]. Multispectral cameras are devices able to capture images from a number of discrete spectral bands. The number of bands has increased in the last decade as technology has improved. Currently, the main camera manufacturers offer multispectral cameras acquiring between 3 and 25 bands, including visible RGB channels, near infrared (NIR), or a set of custom bands, with a tendency to provide increasing number of bands [63]. The spectral bands may not be continuous; thus for 1 pixel we obtain a vector of information comprising the number of elements corresponding to the number of bands registered. Hyperspectral systems may reach resolutions of a few nanometers in wavelength, obtaining for each pixel a digital signature that may contain several hundreds of continuous bands within a specific range of wavelengths [64]. Traditionally, both multispectral and hyperspectral imaging have been used for remote sensing and have an increased number of applications in phenomics. A multispectral system has been developed to improve the original colour of images for fruit recognition [65]. The authors fused the original colour image with an infrared image using nonlinear Daubechies wavelet transform (DWT). Thus, the additional information from the second image allows the original to be improved.
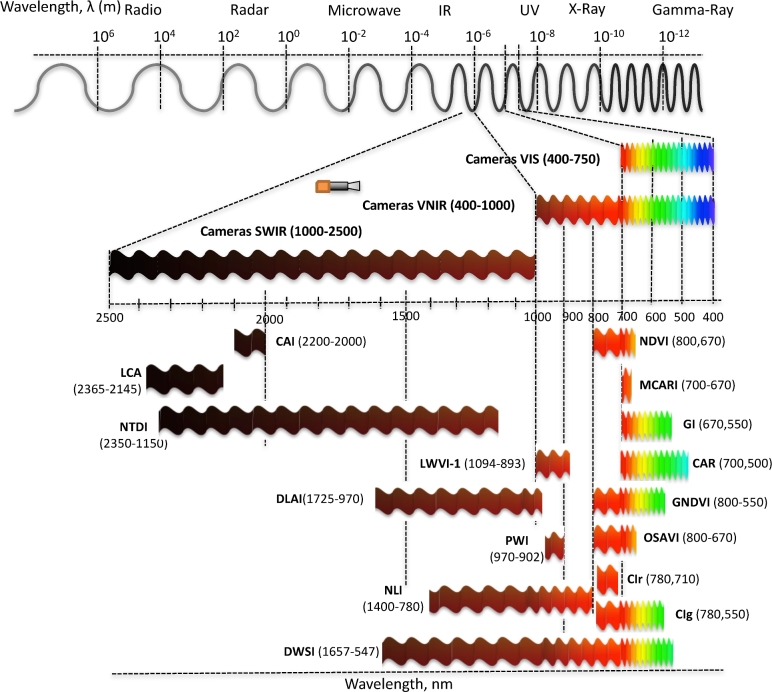
The use of hyperspectral cameras is increasing in phenotyping experiments as they allow the identification of physiological responses, pathologies, or pests in a noninvasive way. Using hyperspectral images, a system has been developed to identify pathogens in barley leaves using probabilistic topic models [66]. A hyperspectral microscope was used to determine spectral changes on the leaf and cellular level of barley (Hordeum vulgare) during resistance reactions against powdery mildew (Blumeria graminis f.sp. hordei, isolate K1) [67]. A detailed description of the different wavelengths and combinations used in multispectral and hyperspectral cameras can be seen in Fig. 2, and their uses in Table 2. We expect to see an increase in phenomic setups using multispectral and hyperspectral cameras in the future. An emerging issue will be the data analysis as the number of pictures doubles with each additional spectrum used for analysis (see below).
Figure 2:
An overview of different spectra used for phenotyping and the associated cameras. The names of different indexes are found in Table 2.
Table 2:
A list of indexes, the corresponding wavelength ranges, and their use to analyse plant material
| Index | Range, nm | Applications |
|---|---|---|
| CAI—Cellulose Absorption Index | 2200–2000 | Quantification of mixed soil–plant litter scenes [178], estimation of non-photosynthetic biomass [179] |
| LCA—Lignin-Cellulose Absorption Index | 2365–2145 | Measure of the effects of soil composition and mineralogy of crop residue cover [180] |
| NTDI—Normalized Difference Tillage Index | 2359–1150 | Used for identifying crop residue cover in conventional and conservation tillage systems [181] |
| LWVI-1 – Normalized Difference Leaf water VI 2 | 1094–893 | Discrimination of sugarcane varieties, allowed to detect large amounts of non-photosynthetically active constituents within the canopy [182] |
| DLAI—Difference Leaf Area Index | 1725–970 | Used for estimating leaf area index based on the radiation measurements in the visible and near-infrared [183] |
| PWI—Plant Water Index | 970–902 | Water content estimation and study of the characteristics of canopy spectrum and growth status [184, 185] |
| NLI—Nonlinear Vegetation Index | 1400–780 | Measurement of plant leaf water content; in combination with others, indexes can detect interaction of biochemicals such as protein, nitrogen, lignin, cellulose, sugar, and starch [186] |
| DWSI—Disease Water Stress Index | 1657–547 | To predict larval mosquito presence in wetland [187] and detect sugarcane “orange rust” disease [188] |
| NDVI—Normalized Difference Vegetation Index | 800–670 | Measurement of significant variations in photosynthetic activity and growing season length at different latitudes [189] |
| MCARI—Modified Chlorophyll Absorption Ratio Index | 700–670 | Study of vegetation biophysical parameters, as well as external factors affecting canopy reflectance [190] |
| GI—Greenness Index | 670–550 | Characterization of corn nitrogen status [191] |
| CAR—Chlorophyll Absorption Ratio | 700–500 | Estimating the concentration of individual photosynthetic pigments within vegetation [192] |
| GNDVI—Green Normalized Difference Vegetation Index | 800–550 | Providing important information for site-specific agricultural decision-making [193] and for identification of chlorophyll content and tissue nitrogen [194] |
| OSAVI—Optimized Soil Adjusted Vegetation Index | 800–670 | Measurement of highly sensitive chlorophyll content variations that are very resistant to the variations of LAI and solar zenith angle [195] |
| CI r—Coloration Index red | 780–710 | Mapping of coastal dune and salt marsh ecosystems [196] |
| CI g—Coloration Index green | 780–550 | Characterization of the state of soil degradation by erosion [197] |
ToF cameras
The Time of Flight cameras (ToF cameras) have been one of the last imaging devices to be incorporated into automatic plant phenotyping [68]. ToF has as a general principle the measurement of the distance between the objective of the camera and each pixel. This is achieved by measuring the time it takes for a signal emitted in NIR to come back, reflected by the object. This allows a precision 3D reconstruction. Stereo vision coupled with ToF images has been implemented to increase the performance of methods of image segmentation to obtain leaf areas [69]. Beyond the tedious hand work required for manual analysis, sampling is done in a non-destructive way. Depth maps obtained by a ToF camera, together with colour images, are used to carry out the 3D modelling of leaves. The system is mounted on a robotic arm, which allows image acquisition to be automated [70]. A ToF has been successfully used to identify QTL regulating shoot architectures of Sorghum by means of 3D reconstruction [71].
Microsoft Kinect is a low-cost image acquisition system designed for video gaming that can be used for characterization and for tracking of phenological parameters [72]. The device is composed of an infrared projector and a camera that generates a grid from which the location of a nearby object in 3D can be ascertained [73]. Kinect has been used to measure plant structure and size for 2 species growing in California grasslands [74]. The quantitative 3D measurements of the architecture of the shoot and structure of the leaves can be performed when proper segmentation algorithms are developed, suggesting some potential for ToF systems [75].
The main disadvantages of this acquisition system are the low resolution, a reduced distance range of a few meters, and the high dependence on the reflecting surface for imaging. As a result, it cannot operate under strong sunlight and is more appropriate for indoor conditions. Its reduced cost and the possibility of obtaining 3D structures of entire plants, as well as of individual organs, make this system very attractive for indoor phenotyping.
LIDAR technology
LIDAR is a remote sensing technology developed at the beginning of the 70s to monitor the Earth's surface [76]. LIDAR uses a laser pulse light to measure the distance between the light source and the object by calculating the time of emission and the time of reflected light detection. It allows the creation of a cloud of points that reconstruct the 3D structure of an object [77, 78]. LIDAR has been used in image acquisition from distances of thousands of kilometres to centimetres, demonstrating the great potential of these types of devices. Satellite-based LIDAR systems are used for the measurements of vegetation canopy height, area, volume or biomass, etc. [79–81]. Recent development using both manned and unmanned flights has allowed the estimation of biomass dynamics of a coniferous forest using Landsat satellite images, together with ground and airborne LIDAR measurements [82]. Terrestrial LIDAR sensors are applied to detect and discriminate maize plants and weeds from soil surface [83]. Short-range LIDAR can be deployed for high-throughput phenotyping systems for cotton plant phenotyping in the field [84] or tomato leaf area by 3D laser reconstruction [85]. Fully automated crop monitoring is feasible using centimetre ranges from robotized or gantry systems [43]. An autonomous robotic system has allowed 3D mapping of plant structures to be performed with millimetric precision [86]. A LASER SCAN mounted on an XYZ gantry system was used to estimate the growth measures and structural information of plants through laser triangulation techniques [87]. Thus, using different devices, LIDAR has an impressive range of possibilities for plant phenomics.
Some shortcomings of LIDAR devices for plant phenotyping are the absence of colour in the measurement, excessive time to compute the cloud points, low precision for massive phenotyping, scanning noises caused by wind, rain, insects, and small particles in the air, and the requirement of calibration. Recent advantages suggest that the use of LIDAR technologies could overcome some of the challenges for the next-generation phenotyping technologies [88]. Developments in multispectral LIDAR instruments show novel systems that are capable of measuring multiple wavelengths and obtaining vegetation indexes (see below) [89, 90] or measuring arboreal parameters [91]. The massive adoption of LASER technologies by autonomous car manufactures has fostered the development of 3D high-definition LIDAR (HDL) with real-time (RT) capacities. The new 3D HDLs are capable of generating 1.3 million points per second with a precision of 2 cm and distances of up to 120 meters [92]. These new devices open the door to the RT massive phenotyping in outdoor and indoor crops.
Thermography and fluorescence imaging
Thermography is a widely used technology in remote sensing and plant phenotyping [93–96]. Thermographic cameras are able to acquire images at wavelengths ranging from 300 to 14 000 nm [97], thus allowing the conversion of the irradiated energy into temperature values once the environmental temperature is assessed. Plants open stomata in response to environmental cues and circadian clock depending on the type of photosynthetic metabolism they have [98, 99]. The evapotranspiration can be assessed with thermography [100], and quantification can be made at different scales, such as a leaf, a tree, a field, or a complete region. Water stress and irrigation management are 2 fields of application of thermography imaging [101–104]. Thermography imaging can detect local changes of temperature produced due to pathogen infection or defence mechanisms [105]. Oerke et al. used a digital infrared thermography to correlate the maximum temperature difference (MTD) of apple leaves with all stages of scab development [106].
Fluorescence imaging has been used in a large number of experimental setups, as ultraviolet (UV) light in the range of 340–360 nm is reflected by different plant components as discrete wavelengths [32]. The corresponding wavelengths emitted are cinnamic acids in the range of green-blue (440–520 nm). Early experiments using reflected fluorescence allowed the identification of phenylpropanoid synthesis mutants in Arabidopsis [107]. Chlorophyll fluorescence emits in red and far-red (690–740 nm). It is an important parameter that has been studied as a proxy for different biological processes such as circadian clock or plant health [8, 108, 109]. A system based on a UV light lamp and a conventional camera with a UV filter to avoid RGB and infrared (IR) images has been used to identify changes in UV absorbance related to pollination [110]. Multicolour fluorescence detection uses the combination of chlorophyll and secondary metabolite–emitted fluorescence to determine plant health in leaf tissues [111].
Thermography imaging results in an estimable tool for the monitoring of genotypes and detection of plant diseases [112] where all the specimens are located under strict control conditions: Temperature, wind velocity, irradiance, leaf angle, and canopy leaf structures are potential issues for quality image acquisition. The next generation of thermography imaging for phenotyping will have to resolve drawbacks related to temporal variations of environment conditions, aspects relating to angles of view, distance, sensitivity, and the reproducibility of the measurements [104]. Both thermographic and fluorescent images capture a single component, and images are in principle easy to analyse as segmentation based on thresholds can be applied to the acquired images. Combining thermographic and fluorescent imaging requires sophisticated data analysis methods based on neural networks to obtain quality data, but it is an emerging solution [111].
Tomography imaging
Magnetic resonance imaging (MRI) is a non-invasive imaging technique that uses radio frequency (RF) magnetic fields to construct tomographic images [113]. Commonly, MRI has been used to investigate the anatomical structure of the body (especially the brain) in both health and disease [114]. In plant phenomics, MRI is used to visualize internal structures and metabolites. This method poses a great potential to monitor physiological processes occurring in vivo [115]. MRI has allowed the development of root systems over time in the bean to be mapped [116], moisture distribution to be visualized during development in rice [117], and water presence to be analysed during the maturity process of barley grains [118].
Positron emission tomography (PET) is a nuclear medicine imaging modality that allows the assessment of biochemical processes in vivo, to diagnose and stage diseases and monitor their treatment [119]. Karve et al. [120] presented a study about C-allocation (carbon allocation from CO2 through photosysthesis) in large grasses such as Sorghum bicolor. The study concluded that the commercial PET scanners can be used reliably, not only to measure C-allocation in plants but also to study dynamics in photoassimilate transport.
X-ray computed tomography (x-ray CT) employs x-rays to produce tomographic images of specific areas of the scanned object. The process of attenuation of rays together with a rotation and axial movement over objects produces 3D images [32]. A high-throughput phenotyping system based on x-ray CT is 10 times more efficient than human operators, being capable of detecting a single tiller mutant among thousands of rice plants [121]. The remarkable penetration of x-rays has made this technology a great ally of phenotyping carried out below ground. The study of root systems and their quantification has been a field of habitual application of x-ray CT [122–126]. New developments address the reduction of penetrability and the increase of the image resolution of x-ray CT in plant tissue using phosphotungstate as a contrast agent, due to its capacity of increasing the contrast and penetrability of thick samples [127].
MRI, PET, and x-ray imaging techniques are available for screening 3D objects. MRI and PET are 2 non-destructive and non-invasive scanning technologies that have been applied in plant sciences to acquire 3D structural information [128]. MRI and PET data acquisition is time consuming, and software tools need to be further developed to analyse data and obtain physiologically interpretable results [97]. High-resolution x-ray computed tomography (HRXCT) promises to be the broadest non-destructive imaging method used in plant sciences. HRXCT will provide 3D data at a resolution suited for detailed analysis of morphological traits of in vivo plant samples and at a cellular resolution for ex vivo samples [128]. In terms of the development of devices, the trend will be to increase the resolution of images, the size of the fields of view, and increase the devices’ portability [129].
Image analysis
Extracting information from images is performed through the process of segmentation. The aim of a segmentation procedure is to extract the components of an image that are of interest, i.e., object or region of interest from the rest of the image, i.e., background of the image or irrelevant components. Thus, we end up with a partitioned image with significant regions. The significant regions may be defined as foreground vs background or by selecting a number of individual components from an image. The construction of the selected regions is based on the image characteristics such as colour (colour spaces), spectral radiance (vegetation indexes), edge detection, neighbour similarity [130], or combinations that are integrated via a machine learning process [131]. In some cases, preprocessing is required in order to obtain a meaningful segmentation.
Image preprocessing
Image preprocessing is an important aspect of image analysis. The aim of image preprocessing is to improve contrast and eliminate noise in order to enhance the objects of interest in a given image [132]. This process can be extremely helpful to enhance the feature extraction quality and the downstream image analysis [133]. Preprocessing can include simple operations such as image cropping, contrast improvement, or other significantly more complex operations such as dimensionality reduction via principal component analysis or clustering [33]. One preprocessing pipeline has been proposed for plant phenotyping based on converting the image to grayscale, application of a median filter, binarization, and edge detection [134]. A similar preprocessing method has been developed to identify plant species under varying illumination conditions [135]. It comprises conversion to grayscale, image binarization, smoothing, and application of a filter to detect edges. In a comparative study to analyze leaf diseases, histogram equalization was found to be the best way to obtain preprocessing of color images converted to grayscale [136]. However, RGB images have been found to perform better than grayscale conversions when identifying leaf pathogens [137].
We cannot conclude that a single preprocessing method will outperform other methods. The quality and type of image are fundamental to selecting a type of preprocessing procedure. Nevertheless, preprocessing is a basic step that can improve image analysis, and sometimes make it possible. It should be described in the materials and methods of image procedures to make data comply to the new standards—Findability, Accessibility, Interoperability, and Reusability (FAIR) [138].
Image segmentation
As we mentioned above, image segmentation is the core of image processing for artificial vision-based plant phenotyping. Segmentation allows the isolation and identification of objects of interest from an image, and it aims to discriminate background or irrelevant objects [139]. The objects of interest are defined by the internal similarity of pixels in parameters such as texture, colour, statistic [133], etc. (See a list of Open software libraries for image segmentation in Table 1.)
One of the simplest algorithms used is threshold segmentation, based on creating groups of pixels on a grayscale according to the level of intensity, thus separating the background from targets. Such an approach has been used with Android OS (ApLeaf) in order to identify plant leaves [140].
The Otsu's method [141] is a segmentation algorithm that searches for a threshold that minimizes the weighted within-class variance [132]. This method has been used for background subtraction in a system that records and performs automatic plant recognition [142] and can give high-contrast segmented images in an automatic fashion [143]. Under certain circumstances, it can underestimate the signal, causing under segmentation, and is significantly slower than other thresholding methods [132].
The Watershed [144] transformation is a popular algorithm for segmentation. It treats an image as a topological surface that is flooded, and seed regions are included, usually by the user. This generates an image with gradients of magnitudes, where crests appear in places where borders are apparent (strong edges) and causes segmentation to stop at those points [130]. It has been used to identify growth rate [145], recognition of partially occluded leaves [56], individual tree crown delineation [146], and leaf segmentation [147].
Grabcut [148] is a segmentation algorithm based on graph cut [149]. It is created on graph theory to tackle the problem of separating an object or foreground from the background. The user should mark a rectangle (bounding box) surrounding the object of interest, thus defining the outrebound of the box as background [150]. This algorithm has been tested to extract trees from a figure, but it has been successful only with very simple backgrounds [151]. More recently, Grabcut has been deployed as a segmentation algorithm in a pipeline for plant recognition with multimodal information, i.e., leaf contour, flower contour, etc. [152]. Grabcut loses precision or even fails when pictures have complex backgrounds but is highly precise with simple backgrounds [151, 142].
Snakes are a special type of active contour [153] and are used as methods to fit lines (splines) either to open or close edges and lines in an image. These methods have been used for face recognition, iris segmentation, and medical image analysis. Within the field of plant phenotyping, there are procedures where active contours are used inside a protocol constructing a vector of features with data of colour intensity, local texture, and a previous knowledge of the plant incorporated via Gaussian mixture models, previously segmented [154]. These steps give an initial rough segmentation, upon which active contours can operate with a much higher precision.
Active contours have used images of flowers for plant recognition [155], based on a combination of the algorithm proposed by Yonggang and Karl [156] and the model of active contours without edges [157]. Whilst the work proposed by Minervini et al. [154] appears to give significantly better results compared with the results of Suta et al. [155], the usage of images with a natural background may be related to the apparent differences in segmentation. Thus, a current problem concerning the comparison of algorithms and procedures lies with the different backgrounds used for image acquisition.
Features extraction
Features extraction constitutes one of the pillars of the identification and classification of objects based on computer vision. Beyond the raw image, a feature is information that is used to resolve a specific computer vision problem. The features extracted from an image are disposed in the so-called “feature vectors.” The construction of feature vectors uses a wide set of methods to identify the objects in an image. The main features are edges, intensity of image pixels [39], geometries [158], textures [154, 159], image transformations, e.g., Fourier [160] or Wavelet [65, 161] or combinations of pixels of different colour spaces [131]. The end goal of feature extraction is to feed up a set of classifiers and machine learning algorithms (see below).
One system proposed uses a feature vector composed of a combination of RGB and CIE L*a*b* colour spaces to segment the images captured during the day [131]. The night-time image segmentation computed a vector composed of statistical features over 2 decomposition levels of the wavelet transform using IR images.
Iyer-Pascuzzi et al. presented an imaging and analysis platform for automatic phenotyping to identify genes underlying root system architecture. The authors employed a set of 16 statistical, geometric, and shape features obtained from 2297 images from 118 individuals such as median and maximum number of roots, the total root length, perimeter, and depth, among others [162].
There are a number of algorithms to identify invariant feature detectors and descriptors. This type of image analysis ensures the detection of points of interest in a scale- and rotation-independent manner. This is crucial for camera calibration and for matching to produce a set of corresponding image points in 3D image reconstruction. Furthermore, it allows the identification of points of interest even when they change scale and/or position or situations of uncontrolled illumination, a common issue when phenotyping plants. The Scale Invariant Features Transforms (SIFT) [163], Speeded-Up Robust Features (SURF) [164], and the Histograms of Oriented Gradients (HoG) [165] are algorithms used to extract characteristics in computer vision, and they have been extended to plant phenotyping. Wei et al. [166] presented an image-based method that automatically detects the flowering of paddy rice. The method uses a scale-invariant feature transform descriptor, bag of visual words, and a machine learning method. The SIFT algorithm has been used to combine stereo and ToF images with automatic plant phenotyping. It can create dense depth maps to identify pepper leaf in glasshouses [69]. SIFT and SURF algorithms have been tested for detecting local invariant features for obtaining a 3D plant model from multi-view stereo images [167]. A HoG framework allows the extraction of a reliable quantity of phenotypic data of grapevine berry using a feature vector composed of colour information [168].
So far, feature extraction has been an arduous and difficult task, requiring the testing of hundreds of feature extraction algorithms and a greater number of combinations between them. This task demands expert skills in different subjects. The success in the identification does not depend on the robustness of the classification methods, but on the robustness of the data.
Machine learning in plant image analysis
The amount of data generated in current and future phenomic setups with high-throughput imaging technologies has brought the use of machine learning (ML) statistical approaches. Machine learning is applied in many fields of research [169–171]. As phenotyping can generate terabytes of information, ML tools provide a good framework for data analysis. A list of ML libraries can be found in Table 3. A major advantage of ML is the possibility of exploring large datasets to identify patterns using combinations of factors instead of performing independent analysis [33].
Table 3:
List of machine learning software libraries and their languages
| Libraries ML/DL | Source | Language |
|---|---|---|
| MICE, rpart, Party, CARET, randomForest, nnet, e1071, KernLab, igraph, glmnet, ROCR, tree, Rweka, earth, klaR, | https://cran.r-project.org/ | R |
| Scikit-learn | http://scikit-learn.org/stable/ | Python |
| Tensorflow | https://www.tensorflow.org/ | |
| Theano | http://deeplearning.net/software/theano | |
| Pylearn2, | http://deeplearning.net/software/pylearn2 | |
| NuPIC | http://numenta.org/ | |
| Caffe | http://caffe.berkeleyvision.org/ | |
| PyBrain | http://pybrain.org/ | |
| Weka | http://www.cs.waikato.ac.nz/ml/weka/ | Java |
| Spark | http://spark.apache.org/ | |
| Mallet | http://mallet.cs.umass.edu/ | |
| JSAT | https://github.com/EdwardRaff/JSAT | |
| ELKI | http://elki.dbs.ifi.lmu.de/ | |
| Java-ML | http://java-ml.sourceforge.net/ | |
| Accord | http://accord-framework.net/ | C#, C++, C |
| Multiboost | http://www.multiboost.org/ | |
| Shogun | http://shogun-toolbox.org/ | |
| LibSVM | http://www.csie.ntu.edu.tw/∼cjlin/libsvm/ | |
| mlpack | http://mlpack.org/ | |
| Shark | http://image.diku.dk/shark/ | |
| MLC++ | http://www.sgi.com/tech/mlc/source.html |
Among the ML algorithms, a predictive model of regression has been used to phenotype Arabidopsis leaves, based on geometric features as a training dataset [158]. Three different algorithms were tested, k Nearest Neighbour (kNN), Support Vector Machine (SVM), and Naïve Bayes, to segment Antirrhinum majus leaves. Colour images have a characteristic vector intensity in the RBG and CIE L*a*b*, while the NIR vector is obtained with the wavelet transform. The best results were obtained with kNN for colour images and SVM for NIR. This shows that segmentation has several components, as mentioned before, including the wavelength of image acquisition [131].
As the specific wavelength used for image acquisition plays a key role in the type of data obtained, hyperspectral cameras are becoming important tools; however, hyper images can be in the order of gigabites of size, making ML a necessity. Examples of coupling hyperspectral and thermal imaging with ML have allowed the early detection of stress caused by Alternaria in Brassica [172]. The best image classification was obtained doing a second derivative transformation of the hyperspectral images together with a back propagation of neural networks, allowing the identification of fungi on leaves days after infection [172].
A current concept derived from ML is deep learning (DL), comprising a set of algorithms aimed to model with a high level of abstraction. This allows the development of complex concepts starting from simpler ones, thus getting closer to the idea of artificial intelligence (AI) [173]. Convolutional neural networks (CNN) are an example of DL derived from artificial neural networks (ANN). These multi-layered networks are formed by a layer of neurons that work in a convolutional way, reducing the sampling process to end with a layer of perception neurons for final classification [174]. Recently DL has been implemented using a CNN to automatically classify and identify different plant parts [175], thus obtaining both classification and localization that significantly improve the current methods. A CNN has been used to detect plant pathogen attacks [176]. Although the training period is computationally heavy, requiring several hours of CPU clusters, classification was performed in less than 1 second [176]. Nevertheless, DL is a step forward in ML and has great potential to allow the management and analysis of the data produced in phenomic experiments.
Although direct testing maybe the best way to determine the superior algorithm in each case, there are a number of examples that may guide initial approaches [33, 177, 178]. As a general rule, discriminating methods such as SVM, ANN, and kNN give better results in large datasets that are labelled [33]. Generative methods such as Naive Bayes, Gaussian mixture models, and Hide Markov models give better results with smaller datasets, both labelled and unlabelled. The use of unsupervised algorithms, i.e., k-means, may help identify unexpected characteristics of a dataset. As mentioned above, preprocessing plays a fundamental role in increasing the ML output. A summary of the complete pipeline of image analysis, including sensors, preprocessing, segmentation procedures, feature extractions, and machine learning algorithms, can be found in Table 4.
Table 4:
A list of current procedures for image analysis based on the type of sensor used
| Data type/source | Preprocessing | Segmentation | Feature extraction | Machine learning |
|---|---|---|---|---|
| Mono—RGB | *Homomorphic filtering to minimize illumination issues in outdoor images [198] | *Many vegetation indexes apply to segmentation in [132] | *Fourier descriptors and Zernike moments [200] | *ANN to detect Phalaenopsis seedling diseases [202] |
| *Filtering and histogram equalization in plant disease detection [136] | *NDVI index to discriminate background and foreground [199] | *Statistical parameters and Wavelet transform with geometric characteristics [131] | *SVM to detect tomato leaf viruses [203] | |
| *Cellular neural networks edge detection [200] | *SIFT and SURF in 3D reconstruction images from multiple RGB cameras with basil specimen [167] | *Gaussian mixture model to detect biotic stress in wheat [204] | ||
| *HSV algorithm [205] | *Histogram to color features and Fast Fourier Transform + Discrete Wavelet Transform to texture features extraction [145] | *k-NN to identify leaf disease [206] | ||
| *Probabilistic Neural Networks and Genetic Algorithm [200] | ||||
| *Random forest to QTL analysis [207] | ||||
| StereoVision | *Complete and general preprocessing pipeline [208] | *Otsu's method & growing region [209] | *Graph-cut and local correlation [210] | *SVM to identify diseased pixels in leaves [211] |
| *Rectification of image based on SIFT and epipolar transformation, in vitis vinifera segmentation [212] | *SVM to remove background [211] | *SURF to stereo-match images based on their feature vectors [209] | *SVM & Gaussian Processes Classifier to detect soil moisture deficit [213] | |
| *Camera stereo calibration, leaf quantifying Brassica napus [214] | *Combined with thermal images (global and local features (temperature, depth, color) using PCA and analysis of variance [211] | |||
| *RGB2GrayScale [209] | *Simple statistical and intensity values [213] | |||
| *Align and depth estimation [213, 211] | ||||
| Multi-Hyper spectral | *Savitzky-Golay filter: remove noise and smooth the image [215] | *NDVI (750–705/750+705) nm with threshold of 0.20 [216] | *Pixels averaged to obtain average reflectance [216] | *Cascade of data mining techniques to detect foliar disease in barley leaves [217] |
| *Gaussian filter to remove noise: detection of disease in banana leaves [218] | *Bayes, logistic, random forest, and decision trees to detect biotic stress in Alternaria genus [172] | |||
| *Savitzky-Golay filter: detection of disease in plants [219] | *k-NN to identify leaf disease [206] | |||
| *PCA and partial least squares regression to predict water, macronutrient, and micronutrient concentrations [216] | ||||
| ToF | *Correction of the distance error caused by the extra contribution of electrons from sunlight using an offset parameter [68] | *Combine hierarchical color segmentation with quadratic surface fitting using ToF depth data [70] | *SIFT, Hough Transform, and RANSAC algorithms to extract relevant features [220] | |
| * Carry out a calibration stage before fusing the depth data and color information [69, 70] | * The maximally stable extremal regions algorithm for the segmentation of single object over background in gray level images [221] | |||
| *Removal of spurious individual points (outliers) using statistical filter [74] | *Removal of background by simple thresholding pixel values greater than a certain threshold [220] | |||
| *Removal of lens distortion [220] | *Segmentation inspired from the maximally stable extremal regions algorithm [221] | |||
| LIDAR | *RANSAC algorithm to detect ground plane [222] | *Clustering to detect individual plants [222] | *Statistical features from reflectace and geometry [222] | *ANN for Wheat Green Area Index measurement [223] |
| *Reduction of noise, filtering point clouds based on deviation [224] | *Surface feature histograms to characterize the grapevine and wheat organs [225] | *ANN, SVM, logistic regression for plant identification (the best results) [222] | ||
| *Generalized linear model (the best) to model plant richness [226] | ||||
| *SVM obtained a highly reliable classification of about 96% [225] | ||||
| Thermography/Flourescence | *Align with stereo images (in combination with stereo images) [213, 211] | *Semi-automated segmentation through a geometric algorithm implemented in Python-based software ChopIt [227] | *Combined with thermal images (global and local features: temperature, depth, color) using PCA and analysis of variance [211] | *SVM to identify diseased pixels in leaves [211] |
| *Normalize thermal information with thermal indexes [228] | *Manual thresholding comparing conventional color images with fluorescent images (Fv/Fm) [229] | *SVM and Gaussian Processes Classifier to detect soil moisture deficit [213] | ||
| *Trimming extraneous images from image stack [227] | *Analysis of variance (not ML) to analyze different water status [228] | |||
| *ANN and SVM to detect zinc deficiency stress using fluorescence imaging [230] | ||||
| MRI/Tomography | *2D and 3D Fourier transformations (MRI) [231] | *Yang 2011: watershed segmentation [232] | *Intensity features, Haralick textural features, intensity local binary pattern features, contrast features, and Gabor intensity textural features [233] | *Supervised learning with ANN, Mahalanobis distance, linear discriminant analysis, and quadratic discriminant analysis to determine boundary lines [233] |
| *Median filter, binaryzation, fill holes, remove small particles, and morphological filter (erosion) [232] | *Histogram thresholding method to binaryze the image [233] | |||
| *Re-slicing, cropping, and contrast enhancement [233] |
Conclusions and Future Prospects
The implementation of phenomic technologies is a welcome change toward reproducibility and unbiased data acquisition in basic and applied research. A successful approach requires integrating sensors with wavelength and image acquisitions that will allow the proper identification of the items under analysis. A lot of work has been conducted in indoor setups, where reasonable conditions can be created to obtain high-quality images amenable to further processing. The difficulty with outdoor setups increases as a result of limitations in the actual image acquisition devices and the uncontrolled conditions that directly affect image quality. The new technologies such as the high-definition LIDAR or the multi-hyperspectral cameras have great potential to improve in the near future, especially in outdoor environments.
Preprocessing and segmentation data are 2 aspects of data treatment and acquisition that require careful design in order to avoid distortions and reproducibility [138]. As images are machine-produced data, but image types and processing procedures may be very different, the standardization of image capture, preprocessing, and segmentation may play an important role. Furthermore, a single procedure for image analysis cannot be considered a better choice, and it is the researcher that needs to assess the different algorithms to come up with an optimized procedure for their specific setup. It is a matter of time until databases with raw images become part of the standard in phenomics; using images very much like NCBI or Uniprot plays a key role in genomic and proteomic projects. With the decrease in the price of hyperspectral devices, new experiments may be performed that produce even larger datasets, and these datasets will have to go through artificial intelligence–based data analysis in order to give the researchers results interpretable by humans. We guess that, like in other omic approaches, there will be a confluence of standard procedures that are not currently common ground, making the current literature look intimidatingly diverse. Nevertheless, most of the basic processes described here are shared by the different experimental setups and data analysis pipes.
Abbreviations
AI: artificial intelligence; ANN: artificial neural networks; CAI: Cellulose Absorption Index; CAR: chlorophyll absorption ratio; CCD: charge coupled device; Cig: Coloration Index green; Cir: Coloration Index red; CMOS: complementary metal oxide semiconductor; CNN: convolutional neural networks; CPU: central processing unit; DL: deep learning; DLAI: Difference Leaf Area Index; DSWI: disease water stress index; DWT: Daubechies wavelet transform; EVI: Enhanced Vegetation Index; FAIR: Findability, Accessibility, Interoperability, and Reusability; GI: Greenness Index; GMM: Gaussian mixture model; GNDVI: Green Normalized Difference Vegetation Index; HoG: Histograms of Oriented Gradients; KNN: K nearest neighbour; LAI: Leaf Area Index; LCA: Lignin-Cellulose Absorption Index; LIDAR: light detection and ranging; LWVI-1: Normalized Difference Leaf water VI 1; MCARI: Modified Chlorophyll Absorption Ratio Index; MCFI: multicolour fluorescence imaging; ML: machine learning; NDVI: Normalized Difference Vegetation Index; NIR: near infrared; NLI: Nonlinear Vegetation Index; NTDI: Normalized Tillage Difference Index; OSAVI: Optimized Soil Adjusted Vegetation Index; PCA: principal component analysis; PET: positron emission tomography; PWI: Plant Water Index; QTL: quantitative trait locus; RF: radiofrequency; RGB: red, green, blue; RT: real-time; SIFT: Scale Invariant Features Transforms; SURF: Speeded-Up Robust Features; SVM: Support Vector Machine; TDI: time delay and integration; ToF: time of flight; UV: ultraviolet.
Competing interests
The authors declare they have no competing interests.
Funding
This work was funded by grants FEDER BFU-2013–45 148-R, Fundación Séneca 19 398/PI/14 to MEC and FEDER ViSelTR (TIN2012–39 279) to PJN.
Author contributions
F.P.S., M.E.C., and P.J.N. defined the scope of the manuscript. F.P.S., M.E.C., and P.J.N. wrote and corrected the manuscript. M.E.C. and P.J.N. wrote the grant applications.
Supplementary Material
Acknowledgments
We would like to thank Leanne Rebecca Miller for the edition of the manuscript and Victoria Ruiz-Hernández and Julia Weiss for comments on the manuscript.
References
- 1. Tucker C. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens Environ 1979. Available at: http://www.sciencedirect.com/science/article/pii/0034425779900130. Accessed 11 October 2016. [Google Scholar]
- 2. Myneni RB, Keeling CD, Tucker CJ et al. Increased plant growth in the northern high latitudes from 1981 to 1991. Nature 1997;386:698–702. [Google Scholar]
- 3. DeFries R, Townshend J. NDVI-derived land cover classifications at a global scale. Int J Remote 1994. Available at: http://www.tandfonline.com/doi/abs/10.1080/01431169408954345. Accessed 11 October 2016. [Google Scholar]
- 4. Pettorelli N, Vik J, Mysterud A et al. Using the satellite-derived NDVI to assess ecological responses to environmental change. Trends Ecol 2005. Available at: http://www.sciencedirect.com/science/article/pii/S016953470500162X. Accessed 11 October 2016. [DOI] [PubMed] [Google Scholar]
- 5. Mkhabela MS, Bullock P, Raj S et al. Crop yield forecasting on the Canadian Prairies using MODIS NDVI data. Agric For Meteorol 2011;151:385–93. [Google Scholar]
- 6. GROTEN SME. NDVI—crop monitoring and early yield assessment of Burkina Faso. Int J Remote Sens 1993;14:1495–515. [Google Scholar]
- 7. Jones HG, Stoll M, Santos T et al. Use of infrared thermography for monitoring stomatal closure in the field: application to grapevine. J Exp Bot 2002;53:2249–60. [DOI] [PubMed] [Google Scholar]
- 8. Chaerle L, Van der Straeten D. Seeing is believing: imaging techniques to monitor plant health. Biochim Biophys Acta-Gene Struct Expr 2001;1519:153–66. [DOI] [PubMed] [Google Scholar]
- 9. Sirault XRR, James RA, Furbank RT et al. A new screening method for osmotic component of salinity tolerance in cereals using infrared thermography. Funct Plant Biol 2009;36:970. [DOI] [PubMed] [Google Scholar]
- 10. Lobet G, Pagès L, Draye X. A novel image analysis toolbox enabling quantitative analysis of root system architecture. Plant Physiol 2011;157:29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Galkovskyi T, Mileyko Y, Bucksch A et al. GiA Roots: software for the high throughput analysis of plant root system architecture. BMC Plant Biol 2012;12:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. French A, Ubeda-Tomas S, Holman TJ et al. High-throughput quantification of root growth using a novel image-analysis tool. Plant Physiol 2009;150:1784–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Golzarian MR, Frick RA, Rajendran K et al. Accurate inference of shoot biomass from high-throughput images of cereal plants. Plant Methods 2011;7:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fabre J, Dauzat M, Negre V et al. PHENOPSIS DB: an information system for Arabidopsis thaliana phenotypic data in an environmental context. BMC Plant Biol 2011;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Araus JL, Cairns JE. Field high-throughput phenotyping: the new crop breeding frontier. Trends Plant Sci 2014;19:52–61. [DOI] [PubMed] [Google Scholar]
- 16. Furbank RT. Plant phenomics: from gene to form and function. Funct Plant Biol 2009;36:V–Vi. [DOI] [PubMed] [Google Scholar]
- 17. Poorter H, Fiorani F, Pieruschka R et al. Tansley review: Pampered inside, pestered outside? Differences and similarities between plants growing in controlled conditions and in the field. New Phytol 2016;838–55. [DOI] [PubMed] [Google Scholar]
- 18. Yang W, Duan L, Chen G et al. Plant phenomics and high-throughput phenotyping: accelerating rice functional genomics using multidisciplinary technologies. Curr Opin Plant Biol 2013;16:180–7. [DOI] [PubMed] [Google Scholar]
- 19. White J, Andrade-Sanchez P, Gore M. Field-based phenomics for plant genetics research. F Crop 2012. Available at: http://www.sciencedirect.com/science/article/pii/S037842901200130X. Accessed 31 August 2016. [Google Scholar]
- 20. Fahlgren N, Gehan MA, Baxter I. Lights, camera, action: high-throughput plant phenotyping is ready for a close-up. Curr Opin Plant Biol 2015:93–9. [DOI] [PubMed] [Google Scholar]
- 21. Furbank RT, Tester M. Phenomics - technologies to relieve the phenotyping bottleneck. Trends Plant Sci 2011;16:635–44. [DOI] [PubMed] [Google Scholar]
- 22. Granier C, Vile D. Phenotyping and beyond: modelling the relationships between traits. Curr Opin Plant Biol 2014;18:96–102. [DOI] [PubMed] [Google Scholar]
- 23. White J, Andrade-Sanchez P, Gore M et al. Field-based phenomics for plant genetics research. F Crop 2012;133:101-12. [Google Scholar]
- 24. Furbank RT, Tester M. Phenomics - technologies to relieve the phenotyping bottleneck. Trends Plant Sci 2011;16:635–44. [DOI] [PubMed] [Google Scholar]
- 25. Simko I, Jimenez-Berni JA, Sirault XRR. Phenomic approaches and tools for phytopathologists. Phytopathology 2016. Available at: http://apsjournals.apsnet.org/doi/10.1094/PHYTO-02-16-0082-RVW. Accessed 28 August 2017. [DOI] [PubMed] [Google Scholar]
- 26. da Silva Marques J. Monitoring photosynthesis by in vivo chlorophyll fluorescence: application to high-throughput plant phenotyping. Appl Photosynth-New Prog 2016;Intech:3–22. [Google Scholar]
- 27. Gonzalez RC, Woods RE. Digital Image Processing. Upper Saddle River, New Jersey, USA: Prentice Hall Press; 2002. [Google Scholar]
- 28. Russ J, Woods R. The image processing handbook. 1995. Available at: http://journals.lww.com/jcat/Citation/1995/11000/The_Image_Processing_Handbook_2nd_Ed.26.aspx. Accessed 25 April 2017.
- 29. Jain A. Fundamentals of digital image processing. 1989. Available at: http://dl.acm.org/citation.cfm?id=59921. Accessed 25 April 2017.
- 30. Sonka M, Hlavac V, Boyle R. Image Processing, Analysis, and Machine Vision. 4th ed CL Engineering. Berlin, Germany: Springer; 2014. Available at: https://books.google.es/books?hl=en&lr=&id=QePKAgAAQBAJ&oi=fnd&pg=PR11&dq=image+analysis+a+review&ots=95qB21F9B-&sig=kSGTMS9GfxkddVJUHnxnBzU2VL8. Accessed 24 April 2017. [Google Scholar]
- 31. Soille P. Morphological Image Analysis: Principles and Applications. Berlin, Germany: Springer; 2013. Available at: https://books.google.es/books?hl=en&lr=&id=ZFzxCAAAQBAJ&oi=fnd&pg=PA1&dq=image+analysis+a+review&ots=-oc-0SEZ6g&sig=wLoRbdNSusr-5UtgD_RvtMHVqjQ. Accessed 25 April 2017. [Google Scholar]
- 32. Li L, Zhang Q, Huang D. A review of imaging techniques for plant phenotyping. Sensors 2014;14:20078–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Singh A, Ganapathysubramanian B, Singh AK et al. Machine learning for high-throughput stress phenotyping in plants. Trends Plant Sci 2016;110–24. [DOI] [PubMed] [Google Scholar]
- 34. Fiorani F, Schurr U. Future scenarios for plant phenotyping. Annu Rev Plant Biol 2013;64:267–91. [DOI] [PubMed] [Google Scholar]
- 35. Lepage G, Bogaerts J, Meynants G. Time-delay-integration architectures in CMOS image sensors. IEEE Trans Electron Devices 2009;56:2524–33. [Google Scholar]
- 36. Yu C, Nie K, Xu J et al. A low power digital accumulation technique for digital-domain CMOS TDI image sensor. Sensors 2016;16:1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Teledyne Dalsa. https://www.teledynedalsa.com/corp/. Accessed 4 April 2017.
- 38. IMEC. Imec launches TDI, multispectral and hyperspectral sensors. Available at: http://optics.org/news/8/2/8. Accessed 24 April 2017.
- 39. Van Der Heijden G, Song Y, Horgan G et al. SPICY: towards automated phenotyping of large pepper plants in the greenhouse. Funct Plant Biol 2012;39:870–7. [DOI] [PubMed] [Google Scholar]
- 40. Navarro PJ, Fernández C, Weiss J et al. Development of a configurable growth chamber with a computer vision system to study circadian rhythm in plants. Sensors (Basel) 2012;12:15356–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Subramanian R, Spalding EP, Ferrier NJ. A high throughput robot system for machine vision based plant phenotype studies. Mach Vis Appl 2013;24:619–36. [Google Scholar]
- 42. Zhang X, Huang C, Wu D et al. High-throughput phenotyping and QTL mapping reveals the genetic architecture of maize plant growth. Plant Physiol 2017;173(3):1554–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Virlet N, Sabermanesh K, Sadeghi-Tehran P et al. Field Scanalyzer: an automated robotic field phenotyping platform for detailed crop monitoring. Funct Plant Biol 2017;44:143. [DOI] [PubMed] [Google Scholar]
- 44. Deery D, Jimenez-Berni J, Jones H et al. Proximal remote sensing buggies and potential applications for field-based phenotyping. Agronomy 2014;4:349–79. [Google Scholar]
- 45. Comar A, Burger P, de Solan B et al. A semi-automatic system for high throughput phenotyping wheat cultivars in-field conditions: description and first results. Funct Plant Biol 2012;39:914. [DOI] [PubMed] [Google Scholar]
- 46. Brown MZ, Burschka D, Hager GD. Advances in computational stereo. IEEE Trans Pattern Anal Mach Intell 2003;25:993–1008. [Google Scholar]
- 47. Biskup B, Scharr H, Schurr U et al. A stereo imaging system for measuring structural parameters of plant canopies. Plant Cell Environ 2007;30:1299–308. [DOI] [PubMed] [Google Scholar]
- 48. Nguyen TT, Slaughter DC, Max N et al. Structured light-based 3D reconstruction system for plants. Sensors (Switzerland) 2015;15:18587–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Klodt M, Herzog K, Töpfer R et al. Field phenotyping of grapevine growth using dense stereo reconstruction. BMC Bioinformatics 2015;16:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rose J, Kicherer A, Wieland M et al. Towards automated large-scale 3D phenotyping of vineyards under field conditions. Sensors 2016;16:2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xiong X, Yu L, Yang W et al. A high-throughput stereo-imaging system for quantifying rape leaf traits during the seedling stage. Plant Methods 2017;13:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Paproki A, Sirault XRR, Berry S et al. A novel mesh processing based technique for 3D plant analysis. BMC Plant Biol 2012;12:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nguyen TT, Slaughter DC, Maloof JN et al. Plant phenotyping using multi-view stereo vision with structured lights. In: Valasek J, Thomasson JA, eds. International Society for Optics and Photonics. Baltimore, Maryland, USA: SPIE Commercial + Scientific Sensing and Imaging; 2016:986608 Available at: http://proceedings.spiedigitallibrary.org/proceeding.aspx?doi=10.1117/12.2229513. Accessed 19 May 2017. [Google Scholar]
- 54. Rose JC, Paulus S, Kuhlmann H. Accuracy analysis of a multi-view stereo approach for phenotyping of tomato plants at the organ level. Sensors (Basel) 2015;15:9651–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schwartz S. An overview of 3D plant phenotyping methods. Phenospex Smart Plant Anal 2015. Available at: https://phenospex.com/blog/an-overview-of-3d-plant-phenotyping-methods/#ref. Accessed 19 June 2017. [Google Scholar]
- 56. Lee K-S, Cohen WB, Kennedy RE et al. Hyperspectral versus multispectral data for estimating leaf area index in four different biomes. Remote Sens Environ 2004;91:508–20. [Google Scholar]
- 57. Dozier J, Painter TH. Multispectral and hyperspectral remote sensing of alpine snow properties. Annu Rev Earth Planet Sci 2004;32:465–94. [Google Scholar]
- 58. Adam E, Mutanga O, Rugege D. Multispectral and hyperspectral remote sensing for identification and mapping of wetland vegetation: a review. Wetl Ecol Manag 2010;18:281–96. [Google Scholar]
- 59. Qin J, Chao K, Kim MS et al. Hyperspectral and multispectral imaging for evaluating food safety and quality. J Food Eng 2013;118:157–71. [Google Scholar]
- 60. van der Meer FD, van der Werff HMA, van Ruitenbeek FJA et al. Multi- and hyperspectral geologic remote sensing: a review. Int J Appl Earth Obs Geoinf 2012;14:112–28. [Google Scholar]
- 61. Mehl PM, Chao K, Kim M et al. Detection of defects on selected apple cultivars using hyperspectral and multispectral image analysis. Appl Eng Agric 2002;18:219. [Google Scholar]
- 62. Ferrato L-J. Comparing hyperspectral and multispectral imagery for land classification of the lower don river, Toronto. MsC Thesis. Ryerson University. Available at: http://www.geography.ryerson.ca/wayne/MSA/LisaJenFerratoMRP2012.pdf. Accessed 27 April 2017. [Google Scholar]
- 63. Cubert S. 137 - ButterflEYE NIR - Cubert-GmbH. Available at: http://cubert-gmbh.com/product/s-137-butterfleye-nir/. Accessed 4 June 2017.
- 64. Kise M, Park B, Heitschmidt GW et al. Multispectral imaging system with interchangeable filter design. Comput Electron Agric 2010;72:61–8. [Google Scholar]
- 65. Li P, Lee S-H, Hsu H-Y et al. Nonlinear fusion of multispectral citrus fruit image data with information contents. Sensors 2017;17:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wahabzada M, Mahlein A-K, Bauckhage C et al. Plant phenotyping using probabilistic topic models: uncovering the hyperspectral language of plants. Sci Rep 2016;6:22482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kuska M, Wahabzada M, Leucker M et al. Hyperspectral phenotyping on the microscopic scale: towards automated characterization of plant-pathogen interactions. Plant Methods 2015;11:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Klose R, Penlington J. Usability study of 3D time-of-flight cameras for automatic plant phenotyping. Bornimer 2009. Available at: https://www.hs-osnabrueck.de/fileadmin/HSOS/Homepages/COALA/Veroeffentlichungen/2009-CBA-3DToF.pdf. Accessed 2 May 2017. [Google Scholar]
- 69. Song Y, Glasbey CA, van der Heijden GWAM et al. Combining Stereo and Time-of-Flight Images with Application to Automatic Plant Phenotyping. Berlin: Springer; 2011:467–78. [Google Scholar]
- 70. Alenyà G, Dellen B, Torras C. 3D modelling of leaves from color and ToF data for robotized plant measuring. Robot Autom (ICRA) 2011. Available at: http://ieeexplore.ieee.org/abstract/document/5980092/. Accessed 2 May 2017. [Google Scholar]
- 71. McCormick RF, Truong SK, Mullet JE. 3D sorghum reconstructions from depth images identify QTL regulating shoot architecture. Plant Physiol 2016;172:823–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Paulus S, Behmann J, Mahlein A et al. Low-cost 3D systems: suitable tools for plant phenotyping. Sensors 2014. Available at: http://www.mdpi.com/1424-8220/14/2/3001/htm. Accessed 2 May 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Microsoft. Kinect for Windows sensor components and specifications. 2010. Available at: https://msdn.microsoft.com/en-us/library/jj131033.aspx. Accessed 7 May 2017.
- 74. Azzari G, Goulden M, Rusu R. Rapid characterization of vegetation structure with a Microsoft Kinect sensor. Sensors 2013;13:2384–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chéné Y, Rousseau D, Lucidarme P et al. On the use of depth camera for 3D phenotyping of entire plants. Comput Electron Agric 2012;82:122–7. [Google Scholar]
- 76. Wang G, Weng Q. Remote Sensing of Natural Resources. Available at: https://books.google.es/books?id=wIDNBQAAQBAJ&pg=PA9&dq=Light+Detection+and+Ranging+(LIDAR)+1970s&hl=es&sa=X&ved=0ahUKEwi0mbSksePTAhVJDxoKHaKxC6UQ6AEIJjAA#v=onepage&q=LightDetectionandRanging (LIDAR)1970s&f=false. Accessed 9 May 2017.
- 77. Lin Y. LiDAR: an important tool for next-generation phenotyping technology of high potential for plant phenomics? Comput Electron Agric 2015;119:61–73. [Google Scholar]
- 78. Vázquez-Arellano M, Griepentrog HW, Reiser D et al. 3-D imaging systems for agricultural applications-a review. Sensors (Basel) 2016;16(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Chen JM, Cihlar J. Retrieving Leaf Area Index of boreal conifer forests using Landsat TM images. Remote Sens Environ 1996;55:155–62. [Google Scholar]
- 80. Gwenzi D, Helmer E, Zhu X et al. Predictions of tropical forest biomass and biomass growth based on stand height or canopy area are improved by Landsat-scale phenology across Puerto Rico and the U.S. Virgin Islands. Remote Sens 2017;9:123. [Google Scholar]
- 81. Kellndorfer JM, Walker WS, LaPoint E et al. Statistical fusion of lidar, InSAR, and optical remote sensing data for forest stand height characterization: a regional-scale method based on LVIS, SRTM, Landsat ETM+, and ancillary data sets. J Geophys Res Biogeosciences 2010;115. [Google Scholar]
- 82. Badreldin N, Sanchez-Azofeifa A. Estimating forest biomass dynamics by integrating multi-temporal Landsat satellite images with ground and airborne LiDAR data in the Coal Valley Mine, Alberta, Canada. Remote Sens 2015;7:2832–49. [Google Scholar]
- 83. Andújar D, Rueda-Ayala V, Moreno H et al. Discriminating crop, weeds and soil surface with a terrestrial LIDAR sensor. Sensors (Switzerland) 2013;13:14662–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Sun S, Li C, Paterson A. In-field high-throughput phenotyping of cotton plant height using LiDAR. Remote Sens 2017;9:377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hosoi F, Nakabayashi K, Omasa K. 3-D modeling of tomato canopies using a high-resolution portable scanning lidar for extracting structural information. Sensors (Basel) 2011;11:2166–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Chaudhury A, Ward C, Talasaz A et al. Computer vision based autonomous robotic system for 3D plant growth measurement. In: 12th Conference on Computer and Robot Vision. Halifax, NS, Canada: IEEE; 2015; DOI: 10.1109/CRV.2015.45 Accessed 10 May 2017. [DOI] [Google Scholar]
- 87. Kjaer KH, Ottosen C-O. 3D laser triangulation for plant phenotyping in challenging environments. Sensors (Basel) 2015;15:13533–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Lin Y. LiDAR: An important tool for next-generation phenotyping technology of high potential for plant phenomics? Comput Electron Agric 2015;119:61–73. [Google Scholar]
- 89. Wallace A, Nichol C, Woodhouse I. Recovery of forest canopy parameters by inversion of multispectral LiDAR data. Remote Sens 2012;4:509–31. [Google Scholar]
- 90. Morsy S, Shaker A, El-Rabbany A. Multispectral LiDAR data for land cover classification of urban areas. Sensors 2017;17:958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wallace AM, McCarthy A, Nichol CJ et al. Design and evaluation of multispectral LiDAR for the recovery of arboreal parameters. IEEE Trans Geosci Remote Sens 2014;52:4942–54. [Google Scholar]
- 92. Navarro P, Fernández C, Borraz R et al. A machine learning approach to pedestrian detection for autonomous vehicles using high-definition 3D range data. Sensors 2016;17:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Padhi J, Misra RK, Payero JO. Estimation of soil water deficit in an irrigated cotton field with infrared thermography. F Crop Res 2012;126:45–55. [Google Scholar]
- 94. Guilioni L, Jones HG, Leinonen I et al. On the relationships between stomatal resistance and leaf temperatures in thermography. Agric For Meteorol 2008;148:1908–12. [Google Scholar]
- 95. Kranner I, Kastberger G, Hartbauer M et al. Noninvasive diagnosis of seed viability using infrared thermography. Proc Natl Acad Sci U S A 2010;107:3912–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Jones HG. Use of infrared thermography for monitoring stomatal closure in the field: application to grapevine. J Exp Bot 2002;53:2249–60. [DOI] [PubMed] [Google Scholar]
- 97. Fiorani F, Rascher U, Jahnke S et al. Imaging plants dynamics in heterogenic environments. Curr Opin Biotechnol 2012;23:227–35. [DOI] [PubMed] [Google Scholar]
- 98. Mallona I, Egea-Cortines M, Weiss J. Conserved and divergent rhythms of CAM-related and core clock gene expression in the cactus Opuntia ficus-indica. Plant Physiol 2011;156:1978–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Somers DE, Webb A A, Pearson M et al. The short-period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development 1998;125:485–94. [DOI] [PubMed] [Google Scholar]
- 100. Costa JM, Grant OM, Chaves MM et al. Thermography to explore plant-environment interactions. J Exp Bot 2013;64:3937–49. [DOI] [PubMed] [Google Scholar]
- 101. Zia S, Romano G, Spreer W et al. Infrared thermal imaging as a rapid tool for identifying water-stress tolerant maize genotypes of different phenology. J Agron Crop Sci 2013;199:75–84. [Google Scholar]
- 102. Jones HG, Serraj R, Loveys BR et al. Thermal infrared imaging of crop canopies for the remote diagnosis and quantification of plant responses to water stress in the field. Funct Plant Biol 2009;36:978. [DOI] [PubMed] [Google Scholar]
- 103. Jones HG, Stoll M, Santos T et al. Use of infrared thermography for monitoring stomatal closure in the field: application to grapevine. J Exp Bot 2002;53:2249–60. [DOI] [PubMed] [Google Scholar]
- 104. Prashar A, Jones H. Infra-Red thermography as a high-throughput tool for field phenotyping. Agronomy 2014;4:397–417. [Google Scholar]
- 105. Mahlein A-K. Precision agriculture and plant phenotyping are information-and technology-based domains with specific demands and challenges for. Plant Dis 2016;100:241–51. [DOI] [PubMed] [Google Scholar]
- 106. Oerke E-C, Fröhling P, Steiner U. Thermographic assessment of scab disease on apple leaves. Precis Agric 2011;12:699–715. [Google Scholar]
- 107. Chapple CC, Vogt T, Ellis BE et al. An Arabidopsis mutant defective in the general phenylpropanoid pathway. Plant Cell 1992;4:1413–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Gould PD, Diaz P, Hogben C et al. Delayed fluorescence as a universal tool for the measurement of circadian rhythms in higher plants. Plant J 2009;58:893–901. [DOI] [PubMed] [Google Scholar]
- 109. Sweeney BM, Prezelin BB, Wong D et al. In vivo chlorophyll-a fluorescence transients and the circadian-rhythm of photosynthesis in gonyaulax-polyedra. Photochem Photobiol 1979;30:309–11. [DOI] [PubMed] [Google Scholar]
- 110. Sheehan H, Moser M, Klahre U et al. MYB-FL controls gain and loss of floral UV absorbance, a key trait affecting pollinator preference and reproductive isolation. Nat Genet 2015. Available at: http://www.nature.com/doifinder/10.1038/ng.3462. Accessed 30 June 2017. [DOI] [PubMed] [Google Scholar]
- 111. Pérez-Bueno ML, Pineda M, Cabeza FM et al. Multicolor fluorescence imaging as a candidate for disease detection in plant phenotyping. Front Plant Sci 2016;7:1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Fang Y, Ramasamy R. Current and prospective methods for plant disease detection. Biosensors 2015;5:537–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Boviki A. Handbook of Image am Video Processing. Austin, Texas, USA: Academic Press; 2005. [Google Scholar]
- 114. Zhang Y, Wang S, Sun P. Pathological brain detection based on wavelet entropy and Hu moment invariants. Bio-medical Mater 2015. at: http://content.iospress.com/articles/bio-medical-materials-and-engineering/bme1426. Accessed 18 October 2016. [DOI] [PubMed] [Google Scholar]
- 115. Borisjuk L, Rolletschek H, Neuberger T. Surveying the plant's world by magnetic resonance imaging. Plant J 2012;129–46. Available at: http://doi.wiley.com/10.1111/j.1365-313X.2012.04927.x. Accessed 15 May 2017. [DOI] [PubMed] [Google Scholar]
- 116. Rascher U, Blossfeld S, Fiorani F et al. Non-invasive approaches for phenotyping of enhanced performance traits in bean. Funct Plant Biol 2011;38:968–83. [DOI] [PubMed] [Google Scholar]
- 117. Horigane A, Engelaar W, Maruyama S. Visualisation of moisture distribution during development of rice caryopses (Oryza sativa L.) by nuclear magnetic resonance microimaging. J Cereal 2001. Available at: http://www.sciencedirect.com/science/article/pii/S0733521000903485. Accessed 15 May 2017. [Google Scholar]
- 118. Glidewell S. NMR imaging of developing barley grains. J Cereal Sci 2006. Available at: http://www.sciencedirect.com/science/article/pii/S0733521005000913. Accessed 15 May 2017. [Google Scholar]
- 119. Converse A, Ahlers E, Bryan T. Positron emission tomography (PET) of radiotracer uptake and distribution in living plants: methodological aspects. J Radioanal Nucl Chem 2013;297(2):241–6. [Google Scholar]
- 120. Karve AA, Alexoff D, Kim D et al. In vivo quantitative imaging of photoassimilate transport dynamics and allocation in large plants using a commercial positron emission tomography (PET) scanner. BMC Plant Biol 2015;15:273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Yang W, Xu X, Duan L et al. High-throughput measurement of rice tillers using a conveyor equipped with x-ray computed tomography. Rev Sci Instrum 2011;82:25102. [DOI] [PubMed] [Google Scholar]
- 122. Tracy SR, Roberts JA, Black CR et al. The X-factor: visualizing undisturbed root architecture in soils using X-ray computed tomography. J Exp Bot 2010;61:311–3. [DOI] [PubMed] [Google Scholar]
- 123. Mooney SJ, Pridmore TP, Helliwell J et al. Developing X-ray computed tomography to non-invasively image 3-D root systems architecture in soil. Plant Soil 2012;352:1–22. [Google Scholar]
- 124. Metzner R, Eggert A, van Dusschoten D et al. Direct comparison of MRI and X-ray CT technologies for 3D imaging of root systems in soil: potential and challenges for root trait quantification. Plant Methods 2015;11:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Lontoc-Roy M, Dutilleul P, Prasher SO et al. Advances in the acquisition and analysis of CT scan data to isolate a crop root system from the soil medium and quantify root system complexity in 3-D space. Geoderma 2006;137:231–41. [Google Scholar]
- 126. Perret JS, Al-Belushi ME, Deadman M. Non-destructive visualization and quantification of roots using computed tomography. Soil Biol Biochem 2007;39:391–9. [Google Scholar]
- 127. Staedler YM, Masson D, Schönenberger J et al. Plant tissues in 3D via X-Ray tomography: simple contrasting methods allow high resolution imaging. PLoS One 2013;8:e75295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Dhondt S, Vanhaeren H, Van Loo D et al. Plant structure visualization by high-resolution X-ray computed tomography. Trends Plant Sci 2010;15:419–22. [DOI] [PubMed] [Google Scholar]
- 129. Brodersen CR, Roddy AB. New frontiers in the three-dimensional visualization of plant structure and function. Am J Bot 2016;103:184–8. [DOI] [PubMed] [Google Scholar]
- 130. Solem JE Programming computer vision with python. In: Andy Oram, Mike Hendrikosn, eds. Programming Computer Vision with Python. 1st ed Sebastopol, CA: O'Reilly Media; 2012;264 Available at: http://programmingcomputervision.com/. Accessed 15 July 2017. [Google Scholar]
- 131. Navarro PJ, Pérez F, Weiss J et al. Machine learning and computer vision system for phenotype data acquisition and analysis in plants. Sensors (Switzerland) 2016;16:641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Hamuda E, Glavin M, Jones E. A survey of image processing techniques for plant extraction and segmentation in the field. Comput Electron Agric 2016;184–99. Available at: http://www.sciencedirect.com/science/article/pii/S0168169916301557. Accessed 10 May 2017. [Google Scholar]
- 133. Krig S. Computer vision metrics: survey, taxonomy, and analysis. In: Scott K, ed. Apress; 2014. Available at: http://www.apress.com/us/book/9781430259299. Accessed September 2017. [Google Scholar]
- 134. Wang Z, Li H, Zhu Y et al. Review of plant identification based on image processing. Arch Comput Methods Eng 2016. Available at: http://link.springer.com/10.1007/s11831-016-9181-4. Accessed 10 May 2017. [Google Scholar]
- 135. Bhagwat R, Dandawate Y. Indian plant species identification under varying illumination and viewpoint conditions. In: 2016 Conference on Advances in Signal Processing. IEEE; 2016;469–73. Available at: http://ieeexplore.ieee.org/document/7746217/. Accessed 9 May 2017. [Google Scholar]
- 136. Thangadurai K, Padmavathi K. Computer visionimage enhancement for plant leaves disease detection. In: 2014 World Congress on Computing and Communication Technologies; 2014. IEEE; 2014;173–5. Available at: http://ieeexplore.ieee.org/document/6755131/. Accessed 10 May 2017. [Google Scholar]
- 137. Padmavathi K, Thangadurai K. Implementation of RGB and grayscale images in plant leaves disease detection - comparative study. Indian J Sci Technol 2016;9 Available at: http://www.indjst.org/index.php/indjst/article/view/77739. Accessed 20 May 2017. [Google Scholar]
- 138. Wilkinson MD, Dumontier M, Aalbersberg IJJ et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 2016;3:160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Singh V, Misra AK. Detection of plant leaf diseases using image segmentation and soft computing techniques. Inf Process Agric 2017;4:41–9. [Google Scholar]
- 140. Zhao ZQ, Ma LH, Cheung Y ming et al. ApLeaf: an efficient android-based plant leaf identification system. Neurocomputing 2015;151:1112–9. [Google Scholar]
- 141. Otsu N. A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern 1979;9:62–6. [Google Scholar]
- 142. Liu J-C, Lin T-M. Location and image-based plant recognition and recording system. J Inform Hiding and Multimedia Signal Processing 2015;6(5):898–910. Available at: http://www.jihmsp.org/∼jihmsp/2015/vol6/JIH-MSP-2015-05-007.pdf. Accessed 30 May 2017. [Google Scholar]
- 143. Chéné Y, Rousseau D, Belin É tienn et al. Shape descriptors to characterize the shoot of entire plant from multiple side views of a motorized depth sensor. Mach Vis Appl 2016;1–15. Available at: http://link.springer.com/10.1007/s00138-016-0762-x. Accessed 19 May 2017. [Google Scholar]
- 144. Vincent L, Vincent L, Soille P. Watersheds in digital spaces: an efficient algorithm based on immersion simulations. IEEE Trans Pattern Anal Mach Intell 1991;13:583–98. [Google Scholar]
- 145. Patil S, Soma S, Nandyal S. Identification of growth rate of plant based on leaf features using digital image processing techniques. Int J Emerg Technol Adv Eng 2013;3. [Google Scholar]
- 146. Barnes C, Balzter H, Barrett K et al. Individual tree crown delineation from airborne laser scanning for diseased larch forest stands. Remote Sens 2017;9:231. [Google Scholar]
- 147. Vukadinovic D, Polder G. Watershed and supervised classification based fully automated method for separate leaf segmentation. The Netherland Congress on Computer Vision; 2015;1–2. Available at: http://edepot.wur.nl/385470. Accessed 20 August 2017. [Google Scholar]
- 148. Rother C, Kolmogorov V, Blake A. GrabCut—interactive foreground extraction using iterated graph cuts. ACM Trans Graph 2004. Available at: https://www.microsoft.com/en-us/research/publication/grabcut-interactive-foreground-extraction-using-iterated-graph-cuts/. Accessed 10 May 2017. [Google Scholar]
- 149. Boykov YY, Jolly M-P. Interactive graph cuts for optimal boundary & region segmentation of objects in N-D images. In: Proceedings of “Internation Conference on Computer Vision”; Vancouver, Canada, July 2001. p. 105–12. IEEE; 2001. Available at: http://ieeexplore.ieee.org/document/937505/. Accessed 3 November 2016. [Google Scholar]
- 150. Sonka M, Hlavac V, Boyle R. Image Processing, Analysis, and Machine Vision. 3rd ed Thomson, ed. Toronto: Thomson; 2008. [Google Scholar]
- 151. Wang X. The grabcut segmentation technique as used in the study of tree image extraction. In: Zhu FG, X, eds. In: Proceedings of the 2009 International Workshop on Information Security and Application (IWISA 2009); Qingdao, China, November 2009. Qingdao, China: Academy Publisher; 2009. Available at: https://pdfs.semanticscholar.org/538c/ff4cf1f660e1b5ab8ed115f272b6122035e3.pdf. Accessed 10 September 2017. [Google Scholar]
- 152. Liu J-C, Chiang C-Y, Chen S. Image-Based plant recognition by fusion of multimodal information. In: 2016 10th International Conference on Innovative Mobile and Internet Services in Ubiquitous Computing (IMIS). IEEE; 2016;5–11. Available at: http://ieeexplore.ieee.org/document/7794433/. Accessed 30 May 2017. [Google Scholar]
- 153. Kass M, Witkin A, Terzopoulos D. Snakes: active contour models. Int J Comput Vis 1988;1:321–31. [Google Scholar]
- 154. Minervini M, Abdelsamea MM, Tsaftaris SA. Image-based plant phenotyping with incremental learning and active contours. Ecol Inform 2014;23:35–48. [Google Scholar]
- 155. Suta L, Bessy F, Veja C et al. Active contours: application to plant recognition. In: 2012 IEEE International Conference on Intelligent Computer Communication and Processing (ICCP). IEEE; 2012;181–7. Available at: http://ieeexplore.ieee.org/document/6356183/. Accessed 10 May 2017. [Google Scholar]
- 156. Shi Y, Karl WC. A real-time algorithm for the approximation of level-set-based curve evolution. IEEE Trans Image Process 2008;17:645–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Chan TF, Vese LA. Active contours without edges. IEEE Trans Image Process 2001;10:266–77. [DOI] [PubMed] [Google Scholar]
- 158. Pape J-M, Klukas C. Utilizing machine learning approaches to improve the prediction of leaf counts and individual leaf segmentation of rosette plant images. In: Computer Vision Problems in Plant Phenotyping (CVPPP 2015). British Machine Vision Association; 2015;1–12. Available at: http://www.bmva.org/bmvc/2015/cvppp/papers/paper003/index.html. Accessed 21 September 2016. [Google Scholar]
- 159. Arivazhagan S, Shebiah RN, Ananthi S et al. Detection of unhealthy region of plant leaves and classification of plant leaf diseases using texture features. Agric Eng Int CIGR J 2013;15:211–7. [Google Scholar]
- 160. Mouille G, Robin S, Lecomte M et al. Classification and identification of Arabidopsis cell wall mutants using Fourier-Transform InfraRed (FT-IR) microspectroscopy. Plant J 2003;35:393–404. [DOI] [PubMed] [Google Scholar]
- 161. Guijarro M, Riomoros I, Pajares G et al. Discrete wavelets transform for improving greenness image segmentation in agricultural images. Comput Electron Agric 2015;118:396–407. [Google Scholar]
- 162. Iyer-Pascuzzi AS, Symonova O, Mileyko Y et al. Imaging and analysis platform for automatic phenotyping and trait ranking of plant root systems. Plant Physiol 2010;152:1148–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Lowe DG. Distinctive image features from scale-invariant keypoints. Int J Comput Vis 2004;60:91–110. [Google Scholar]
- 164. Bay H, Ess A, Tuytelaars T et al. Speeded-Up Robust Features (SURF). Comput Vis Image Underst 2008;110:346–59. [Google Scholar]
- 165. Dalal N, Triggs B. Histograms of oriented gradients for human detection. In: International Conference on Computer Vision & Pattern Recognition (CVPR '05). IEEE; 2005;886–93. Available at: http://ieeexplore.ieee.org/document/1467360/. Accessed 9 December 2016. [Google Scholar]
- 166. Guo W, Fukatsu T, Ninomiya S. Automated characterization of flowering dynamics in rice using field-acquired time-series RGB images. Plant Methods 2015;11:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Santos T, Oliveira A. Image-based 3D digitizing for plant architecture analysis and phenotyping. In: SIBGRAPI 2012 XXV Conf. 2012. Available at: http://www.cnptia.embrapa.br/∼thiago/pool/2012-08-24_sibgrapi.pdf. Accessed 9 December 2016.
- 168. Roscher R, Herzog K, Kunkel A et al. Automated image analysis framework for high-throughput determination of grapevine berry sizes using conditional random fields. Comput Electron Agric 2014;100:148–58. Available at: http://www.sciencedirect.com/science/article/pii/S0168169913002780. Accessed 11 March 2017. [Google Scholar]
- 169. Lantz B. Machine Learning With R. 1st ed Jones J, Sheikh A, eds. Birmingham: Packt Publishing; 2013. [Google Scholar]
- 170. Müller A, Guido S. Introduction to Machine Learning With Python. 1st ed Schanafelt D, editor. Sebastopol: O’Reilly Media; 2016. [Google Scholar]
- 171. Smola A, Vishwanathan SV. Introduction to Machine Learning. 1st ed Cambridge: Cambridge University Press; 2008. [Google Scholar]
- 172. Baranowski P, Jedryczka M, Mazurek W et al. Hyperspectral and thermal imaging of oilseed rape (Brassica napus) response to fungal species of the genus Alternaria. PLoS One 2015;10:e0122913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Goodfellow I, Bengio Y, Courville A. Deep Learning. Boston, USA: MIT Press; Available at: www.deeplearningbook.org. Accessed 21 August 2017. [Google Scholar]
- 174. Fukushima K. Neocognitron: a self organizing neural network model for a mechanism of pattern recognition unaffected by shift in position. Biol Cybern 1980;36:193–202. [DOI] [PubMed] [Google Scholar]
- 175. Pound MP, Burgess AJ, Wilson MH et al. Deep machine learning provides state-of-the-art performance in image-based plant phenotyping. BioRxiv 2016;53033 Available at: http://biorxiv.org/lookup/doi/10.1101/053033. Accessed 21 September 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Mohanty SP, Hughes D, Salathé M. Using deep learning for image-based plant disease detection. 2016;1–7. [DOI] [PMC free article] [PubMed]
- 177. Tsaftaris SA, Minervini M, Scharr H. Machine learning for plant phenotyping needs image processing. Trends Plant Sci 2016;989–91. Available at: http://linkinghub.elsevier.com/retrieve/pii/S1360138516301613. Accessed 27 May 2017. [DOI] [PubMed] [Google Scholar]
- 178. Naik HS, Zhang J, Lofquist A et al. A real-time phenotyping framework using machine learning for plant stress severity rating in soybean. Plant Methods 2017;13:23 Available at: http://plantmethods.biomedcentral.com/articles/10.1186/s13007-017-0173-7. Accessed 21 August 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Nagler PL, Inoue Y, Glenn E. et al. Cellulose absorption index (CAI) to quantify mixed soil–plant litter scenes. Remote Sens Environ 2003;87:310–25. [Google Scholar]
- 180. Ren H, Zhou G, Zhang F et al. Evaluating cellulose absorption index (CAI) for non-photosynthetic biomass estimation in the desert steppe of Inner Mongolia. Chinese Sci Bull 2012;57:1716–22. [Google Scholar]
- 181. Serbin G, Daughtry CST, Hunt ER et al. Effects of soil composition and mineralogy on remote sensing of crop residue cover. Remote Sens Environ 2009;113:224–38. [Google Scholar]
- 182. Eskandari I, Navid H, Rangzan K. Evaluating spectral indices for determining conservation and conventional tillage systems in a vetch-wheat rotation. Int Soil Water Conserv Res 2016;4:93–8. [Google Scholar]
- 183. Galvão LS, Formaggio AR, Tisot DA. Discrimination of sugarcane varieties in Southeastern Brazil with EO-1 Hyperion data. Remote Sens Environ 2005;94:523–34. [Google Scholar]
- 184. Price J. Leaf area index estimation from visible and near-infrared reflectance data. Remote Sens Environ 1995;52:55–65. [Google Scholar]
- 185. Zarco-Tejada P, Berjón A, Miller J. Stress detection in crops with hyperspectral remote sensing and physical simulation models. Proc Airborne 2004. [Google Scholar]
- 186. Cai J, Golzarian MR, Miklavcic SJ. Novel image segmentation based on machine learning and its application to plant analysis. Int J Inf Electron Eng 2011;1. [Google Scholar]
- 187. Gong P, Pu R, Biging G. Estimation of forest leaf area index using vegetation indices derived from Hyperion hyperspectral data. IEEE Trans 2003. [Google Scholar]
- 188. Brown HE, Diuk-Wasser MA, Guan Y et al. Comparison of three satellite sensors at three spatial scales to predict larval mosquito presence in Connecticut wetlands. Remote Sens Environ 2008;112:2301–8. [Google Scholar]
- 189. Apan A, Held A, Phinn S et al. Detecting sugarcane “orange rust” disease using EO-1 Hyperion hyperspectral imagery. Int J Remote Sens 2004;25:489–98. [Google Scholar]
- 190. Tucker CJ, Slayback DA, Pinzon JE et al. Higher northern latitude normalized difference vegetation index and growing season trends from 1982 to 1999. Int J Biometeorol 2001;45:184–90. [DOI] [PubMed] [Google Scholar]
- 191. Haboudane D, Miller JR, Pattey E et al. Hyperspectral vegetation indices and novel algorithms for predicting green LAI of crop canopies: modeling and validation in the context of precision agriculture. Remote Sens Environ 2004;90:337–52. [Google Scholar]
- 192. Pagani A, Echeverría HE, Andrade FH et al. Characterization of corn nitrogen status with a greenness index under different availability of sulfur. Agron J Am Soc Agron 2009;101:315. [Google Scholar]
- 193. Blackburn GA. Spectral indices for estimating photosynthetic pigment concentrations: a test using senescent tree leaves. Int J Remote Sens 1998;19:657–75. [Google Scholar]
- 194. Hunt ER, Dean Hively W, Fujikawa SJ et al. Acquisition of NIR-green-blue digital photographs from unmanned aircraft for crop monitoring. Remote Sens 2010;2:290–305. [Google Scholar]
- 195. Bell GE, Howell BM, Johnson GV et al. Optical sensing of turfgrass chlorophyll content and tissue nitrogen. HortScience 2004;39:1130–2. [Google Scholar]
- 196. Haboudane D, Miller JR, Tremblay N et al. Integrated narrow-band vegetation indices for prediction of crop chlorophyll content for application to precision agriculture. Remote Sens Environ 2002;81:416–26. [Google Scholar]
- 197. Timm BC, McGarigal K. Fine-scale remotely-sensed cover mapping of coastal dune and salt marsh ecosystems at Cape Cod National Seashore using Random Forests. Remote Sens Environ 2012;127:106–17. [Google Scholar]
- 198. Parenteau MP, Bannari A, El-Harti A et al. Characterization of the state of soil degradation by erosion using the hue and coloration indices. IGARSS 2003. In: Geoscience and Remote Sensing Symposium, 2003. IGARSS '03. Proceedings. IEEE; 2003;2284–6. [Google Scholar]
- 199. Pajares G, Ruz JJ, de la Cruz JM. Performance analysis of homomorphic systems for image change detection. In: Pattern Recognition. Image Anal. Pt 1.Berlin: Springer; 2005;563–70. Available at: http://link.springer.com/10.1007/11492429_68. Accessed 25 July 2017. [Google Scholar]
- 200. Oluleye B, Leisa A, Jinsong L et al. On the application of genetic probabilistic neural networks and cellular neural networks in precision agriculture. Asian J Comput Inf Syst 2014;2:90–101. [Google Scholar]
- 201. Huang K-Y. Application of artificial neural network for detecting Phalaenopsis seedling diseases using color and texture features. Comput Electron Agric 2007;57:3–11. [Google Scholar]
- 202. Perez AJ, Lopez F, Benlloch J V et al. Colour and shape analysis techniques for weed detection in cereal fields. Comput Electron Agric 2000;25:197–212. [Google Scholar]
- 203. Mokhtar U, Ali MAS, Hassanien AE et al. Identifying Two of Tomatoes Leaf Viruses Using Support Vector Machine. New Delhi: Springer; 2015;771–82. Available at: http://link.springer.com/10.1007/978-81-322-2250-7_77. Accessed 11 September 2017. [Google Scholar]
- 204. Casanova J, O'Shaughnessy S, Evett S et al. Development of a wireless computer vision instrument to detect biotic stress in wheat. Sensors 2014;14:17753–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Knoll FJ, Holtorf T, Hussmann S. Modified 3D time-of-flight camera for object separation in organic farming. In: Kress BC, Osten W, Urbach HP, eds. International Society for Optics and Photonics; 2017;103351R Available at: http://proceedings.spiedigitallibrary.org/proceeding.aspx?doi=10.1117/12.2270276. Accessed 9 August 2017. [Google Scholar]
- 206. Bauer SD, Korč F, Förstner W. The potential of automatic methods of classification to identify leaf diseases from multispectral images. Precis Agric 2011;12:361–77. [Google Scholar]
- 207. Atkinson JA, Lobet G, Noll M et al. Combining semi-automated image analysis techniques with machine learning algorithms to accelerate large scale genetic studies. BioRxiv 2017. Available at: http://www.biorxiv.org/content/early/2017/06/20/152702. Accessed 8 August 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Ballabeni A, Apollonio FI, Gaiani M et al. Advances in image pre-processing to improve automated 3d reconstruction. Int Arch Photogramm Remote Sens Spat Inf Sci ISPRS Arch 2015;315–23. Available at: http://www.int-arch-photogramm-remote-sens-spatial-inf-sci.net/XL-5-W4/315/2015/. Accessed 20 July 2017. [Google Scholar]
- 209. Tsung-Shiang H. An improvement stereo vision images processing for object distance measurement. Int J Autom Smart Technol 2015;5:85–90. [Google Scholar]
- 210. Kazmi W, Foix S, Alenyà G et al. Indoor and outdoor depth imaging of leaves with time-of-flight and stereo vision sensors: analysis and comparison. ISPRS J Photogramm Remote Sens 2014;88:128–46. [Google Scholar]
- 211. Raza SEA, Prince G, Clarkson JP et al. Automatic detection of diseased tomato plants using thermal and stereo visible light images. PLoS One 2015;10:e0123262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Klodt M, Herzog K, Töpfer R et al. Field phenotyping of grapevine growth using dense stereo reconstruction. BMC Bioinformatics 2015;16:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Raza SEA, Smith HK, Clarkson GJJ et al. Automatic detection of regions in spinach canopies responding to soil moisture deficit using combined visible and thermal imagery. PLoS One 2014;9:e97612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Zhang X, Huang C, Wu D et al. High-throughput phenotyping and QTL mapping reveals the genetic architecture of maize plant growth. Plant Physiol 2017;01516 Available at: http://www.plantphysiol.org/lookup/doi/10.1104/pp.16.01516. Accessed 10 May 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Wahabzada M, Mahlein A-K, Bauckhage C et al. Plant phenotyping using probabilistic topic models: uncovering the hyperspectral language of plants. Sci Rep 2016;6:22482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Pandey P, Ge Y, Stoerger V et al. High throughput in vivo analysis of plant leaf chemical properties using hyperspectral imaging. Front Plant Sci 2017;8:1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Wahabzada M, Mahlein AK, Bauckhage C et al. Metro maps of plant disease dynamics-automated mining of differences using hyperspectral images. PLoS One 2015;10:e0116902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Ochoa D, Cevallos J, Vargas G et al. Hyperspectral imaging system for disease scanning on banana plants. In: Proceedings Volume 9864, Sensing for Agriculture and Food Quality and Safety VIII. 2016;4:98640M. [Google Scholar]
- 219. Mahlein A-K, Kuska MT, Thomas S et al. Plant disease detection by hyperspectral imaging: from the lab to the field. Adv Anim Biosci 2017;8:238–43. [Google Scholar]
- 220. Chattopadhyay S, Akbar SA, Elfiky NM et al. Measuring and modeling apple trees using time-of-flight data for automation of dormant pruning applications. In: 2016 IEEE Winter Conference on Applications of Computer Vision (WACV); 2016IEEE; 2016;1–9. Available at: http://ieeexplore.ieee.org/document/7477596/. Accessed 4 September 2017. [Google Scholar]
- 221. Chéné Y, Rousseau D, Lucidarme P et al. On the use of depth camera for 3D phenotyping of entire plants. Comput Electron Agric 2012;82:122–7. [Google Scholar]
- 222. Weiss U, Biber P, Laible S et al. Plant species classification using a 3D LIDAR sensor and machine learning. In: Ninth International Conference on Machine Learning and Applications; 2010. IEEE; 2010;339–45. Available at: http://ieeexplore.ieee.org/document/5708854/. Accessed 8 August 2017. [Google Scholar]
- 223. Liu S, Baret F, Abichou M et al. Estimating wheat green area index from ground-based LiDAR measurement using a 3D canopy structure model. Agric For Meteorol 2017;247:12–20. [Google Scholar]
- 224. Greaves HE, Vierling LA, Eitel JUH et al. Applying terrestrial lidar for evaluation and calibration of airborne lidar-derived shrub biomass estimates in Arctic tundra. Remote Sens Lett 2017;8:175–84. [Google Scholar]
- 225. Paulus S, Dupuis J, Mahlein A-K et al. Surface feature based classification of plant organs from 3D laser scanned point clouds for plant phenotyping. BMC Bioinformatics 2013;14:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Lopatin J, Dolos K, Hernández HJ et al. Comparing generalized linear models and random forest to model vascular plant species richness using LiDAR data in a natural forest in central Chile. Remote Sens Environ 2016;173:200–10. [Google Scholar]
- 227. Deery DM, Rebetzke GJ, Jimenez-Berni JA et al. Methodology for high-throughput field phenotyping of canopy temperature using airborne thermography. Front Plant Sci 2016;7:1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. García-Tejero IF, Hernández A, Padilla-Díaz CM et al. Assessing plant water status in a hedgerow olive orchard from thermography at plant level. Agric Water Manag 2017;188:50–60. [Google Scholar]
- 229. Rousseau C, Belin E, Bove E et al. High throughput quantitative phenotyping of plant resistance using chlorophyll fluorescence image analysis. Plant Methods 2013;9:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Wetterich CB, Felipe de Oliveira Neves R, Belasque J et al. Detection of Huanglongbing in Florida using fluorescence imaging spectroscopy and machine-learning methods. Appl Opt 2017;56:15. [DOI] [PubMed] [Google Scholar]
- 231. Hillnhutter C, Sikora RA, Oerke E-C et al. Nuclear magnetic resonance: a tool for imaging belowground damage caused by Heterodera schachtii and Rhizoctonia solani on sugar beet. J Exp Bot 2012;63:319–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Yang W, Xu X, Duan L et al. High-throughput measurement of rice tillers using a conveyor equipped with x-ray computed tomography. Rev Sci Instrum 2011;82:25102. [DOI] [PubMed] [Google Scholar]
- 233. Donis-González IR, Guyer DE, Fulbright DW et al. Postharvest noninvasive assessment of fresh chestnut (Castanea spp.) internal decay using computer tomography images. Postharvest Biol Technol 2014;94:14–25. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.