Abstract
Telomere homeostasis is controlled by both telomerase machinery and end protection. Telomere shortening induces DNA damage sensing kinases ATM/ATR for telomerase recruitment. Yet, whether telomere shortening also governs end protection is poorly understood. Here we discover that yeast ATM/ATR controls end protection. Rap1 is phosphorylated by Tel1 and Mec1 kinases at serine 731, and this regulation is stimulated by DNA damage and telomere shortening. Compromised Rap1 phosphorylation hampers the interaction between Rap1 and its interacting partner Rif1, which thereby disturbs the end protection. As expected, reduction of Rap1–Rif1 association impairs telomere length regulation and increases telomere–telomere recombination. These results indicate that ATM/ATR DNA damage checkpoint signal contributes to telomere protection by strengthening the Rap1–Rif1 interaction at short telomeres, and the checkpoint signal oversees both telomerase recruitment and end capping pathways to maintain telomere homeostasis.
INTRODUCTION
McClintock and Muller first speculated that the ends of chromosomes might play some protective roles (1,2), and without such protection, chromosome ends are recognized as DNA double-strand breaks (DSBs), resulting in detrimental chromosome rearrangements, genomic instability and the associated risk of cancer (3–8). Telomeres are dynamic DNA-protein complexes that protect the ends of linear chromosomes, which are composed of tandem repeats of short G-rich sequences and synthesized by the enzyme telomerase (9,10). The catalytic core of telomerase is composed of a reverse transcriptase and an RNA subunit. The reverse transcriptase utilizes the RNA subunit as a template to add the G-rich repeats onto the 3′ ends of the telomere (9–11).
Hayflick observed a cellular senescence phenomenon (12,13), which was explained by the end-replication problem. Most human somatic cells are devoid of telomerase activity and suffer replicative senescence due to their gradually shortened telomeres during consecutive cell divisions. When telomeres become extremely short, they gradually lose the ability to protect the ends of the chromosomes from being recognized as broken ends and being prone to nuclease attacking and recombinational repair. Successive telomere shortening in human fibroblasts results in chromosome fusions, crisis, and apoptosis (14). Some human cells can circumvent such complications either through telomerase reactivation or an alternative recombination pathway for telomere lengthening (15–17).
In budding yeast Saccharomyces cerevisiae, the double-stranded telomeric TG1–3 repeats are occupied by the DNA-binding protein Rap1. Rap1 is an essential protein that functions as a general transcriptional activator at about 300 genomic loci and serves as a repressor at two HM silent mating type loci and telomeres (18–20). At telomeres, Rap1 plays a vital role in maintaining telomere homeostasis by counting telomere length, inhibiting telomere resection, protecting telomere from telomere–telomere fusion and sheltering telomere from unregulated activation of the DNA damage checkpoint (7).
Several domains of Rap1 have been characterized, including the N-terminal BRCT domain, a central DNA-binding (DBD) domain with two Myb-like folds, a transcriptional activation (TA) domain, and the C-terminal protein–protein interaction (RCT) domain (21). The telomeric protective function of Rap1 is largely contributed by its RCT domain, which recruits shelterin proteins Rif1/Rif2 (22–25) and gene silencing Sir3/Sir4 complex (26). Budding yeast contains three protective telomere capping complexes: Rap1–Rif1–Rif2, Yku70–Yku80 and the Cdc13–Stn–Ten1 (CST) complexes. The CST complex binds 3′ single-stranded telomeric tails to prevent telomeric degradation during S phase (27) and is required for both telomere capping and telomerase recruitment (28–30). The Yku70-Yku80 complex plays a central role in preventing telomeres from exonucleolytic attack in non-dividing cells (31). The Rap1–Rif1–Rif2 complex builds a negative feedback loop (protein counting model) that controls telomerase action through the amount of bounded Rap1–Rif1–Rif2 complex to negatively regulate telomere length (25,32,33), but the molecular details of this feedback mechanism are not entirely understood. Rif1 and Rif2 execute nonoverlapping roles in masking telomeric ends and preventing G2/M checkpoint activation (34,35). They limit end resection through distinct mechanisms (31,36). Rif2 prevents telomere fusions and the association of Tel1/MRX complex to telomeres (36–38). On the other hand, Rap1 and Rif1 serve as the central mediator to maintain telomere length homeostasis (39,40). Rif1 becomes essential for cell viability when CST activity is compromised (41). These data indicate that Rif1 and Rif2 participate in distinct regulatory mechanisms.
Telomere healing assay revealed that DNA breaks occurring adjacent to long or short telomeric repeats induce different initial responses (42). Long telomeric tracts recruit less Mre11, Cdc13 and telomerase and this binding suppression requires Rap1. Conversely, short telomeres induce rapid Tel1- (43–46) and telomerase-dependent (47,48) elongation. Telomere length homeostasis is controlled by the balance between telomerase-mediated elongation and telomere capping, but the linkage between them is still not fully understood. Recent studies demonstrated that the Tel1-mediated phosphorylation of single-stranded capping protein Cdc13 is important for telomerase recruitment through strengthening the Cdc13–Est1 interaction (49–51). We were interested in whether phosphorylation of double-stranded capping protein Rap1 also plays a role in maintaining telomere length homeostasis. Here, we found that impairment of Rap1 S731 phosphorylation leads to telomere lengthening. Both DNA damage and telomere shortening boost the Tel1/Mec1-mediated Rap1 S731 phosphorylation. The phosphomimetic S731D mutation strengthens its interaction with Rif1, but not with Rif2 and silencing factor Sir3 and increases the Rif1 occupancy on telomeres. Interestingly, while both Cdc13 and Rap1 are phosphorylated by Tel1/Mec1, the telomere lengthening phenotype of Rap1 dephosphorylation is compromised by deprivation of the telomerase recruitment function of Cdc13 phosphorylation, suggesting that Rap1 phosphorylation-mediated telomere lengthening is still telomerase-dependent. All these findings provide Tel1/Mec1 a central role to coordinately modify two telomeric binding proteins to control end capping, telomerase recruitment and telomere length homeostasis.
MATERIALS AND METHODS
Strains and plasmids
All yeast manipulations were conducted by standard methods (52). Strains and plasmids used in this study are listed in Supplementary Tables S2 and 3. The yeast strains carrying yku80, tlc1, mec1 sml1, tel1 mec1 sml1, rif1, rif2, rif1 rif2, cdc13-S314A, Rap1-HA3 and Rap1-Myc13 are isogenic to YPH499 (MATa ura3–52 lys2–801_amber ade2–101_ochre trp1-Δ63 his3-Δ200 leu2-Δ1). The tel1 and pif1-m2 mutants are isogenic to YPH500 (MATα ura3–52 lys2–801_amber ade2–101_ochre trp1-Δ63 his3-Δ200 leu2-Δ1). MS179, MS206 (gifts from Dr Virginia Zakian) and Sir3-HA3 are isogenic to W303 (MATa leu2–3,112 trp1–1 can1–100 ura3–1 ade2–1 his3–11,15). UCC3505, UCC3515 and UCC4564 silencing reporter strains were kindly proved by Dr Daniel. E. Gottschling. pRS306RAP1 was constructed by polymerase chain reaction (PCR) amplifying a DNA fragment containing the full or partial RAP1 open reading frame and the downstream 300 nt from S. cerevisiae genomic DNA and ligating into the pRS306 vector. pRS304RAP1 was constructed by PCR amplifying a DNA fragment encoding residues 170–827 of Rap1 and the downstream 300 nt from genomic DNA and ligating into the pRS304 vector. pRS304-Rap1-ΔC (672–827) was constructed by one-step site-directed deletion mutagenesis PCR (53) using primer sets RAP1-del2014–2481 and RAP1-del2014–2481 antisense to delete the Rap1 C-terminal (RCT) 672–827 amino acid region on pRS304RAP1. All point mutations were introduced into RAP1 using QuickChange site-directed mutagenesis (Stratagene). To generate rap1 mutants, the pRS306rap1 point-mutation plasmids were linearized by BlpI or SphI (New England Biolabs) and transformed into yeast cells. The URA3 pop-out mutants were selected from the 5-fluoroorotic acid (5-FOA) resistant transformants. The rap1 mutations were confirmed by PCR and sequencing. To generate strains for testing silencing effects, the pRS304RAP1 and pRS304-Rap1-ΔC (672–827) were linearized by BlpI and transformed into UCC3505, UCC3515 and UCC4564 reporter cells. These mutants were selected from the synthetic complete plates without tryptophan (SC-Trp) and further confirmed by PCR and sequencing. The Rap1-HA3 and Rap1-Myc13 strains were constructed by transforming the RAP1-HA3::KanMX6, RAP1-Myc13::TRP1 and RAP1-Myc13::KanMX6 PCR fragments, respectively, and selected. The Sir3-HA3 strain was constructed by transforming the SIR3-HA3::HIS3 PCR fragments and selected. The yku80::HIS3 and sir3::HIS3 mutants were constructed by transforming yku80::HIS3 and sir3::HIS3 PCR fragments, respectively, and selected. Plasmid pGEX-4T-Rif1 (1709–1916) was constructed by ligating PCR products containing amino acids 1709–1916 of Rif1 into EcoRI- and XhoI-digested pGEX-4T-1. pGEX-4T-Rif2 (1–395) was constructed by ligating PCR products containing full-length Rif2 into EcoRI- and XhoI-digested pGEX-4T-1. pGEX-4T-Rap1 (353–827) was constructed by ligating PCR products containing amino acids 353–827 of Rap1 into EcoRI- and XhoI-digested pGEX-4T-1. pGEX-4T-Rap1 (716–746) was constructed by ligating PCR products containing amino acids 716–746 of Rap1 into BamHI- and XhoI-digested pGEX-4T-1. To generate YEpFAT7-SIR3-HA3, PCR products containing SIR3 open reading frame and HA3 tag were amplified from the Sir3-HA3 strain and subcloned into pGEM-T easy (Promega). The SIR3-HA3 fragments were then obtained by NotI digestion and further ligated into the YEpFAT7 vector. All primer sequences for PCR and mutagenesis are provided in Supplementary Table S4.
Southern blot analysis of telomere length
Individual colonies were inoculated into 2 ml yeast extract peptone adenine dextrose (YPAD) medium at 30°C. Spore colonies from tetrad plates were serially restreaked onto YPAD plates. Genomic DNAs were digested with KpnI (New England Biolabs) and separated by 1% agarose gel electrophoresis and transferred to Genescreen Plus membrane (PerkinElmer). The blot was probed by a 550-bp 32P-labeled (Invitrogen) EcoRI fragment of the TG1–3 sequence. For survivor type analysis, individual colonies from dissected spores were repeatedly restreaked on YPAD plates at 30°C. The genomic DNAs of survivors were digested with a mixture of four-base cutters (AluI, HaeIII, HinfI and MspI) or XhoI. The telomere length images were quantified and analyzed by ImageQuant TL software (GE healthcare).
Telomeric and mating-type locus silencing assays
Silencing at telomeres was assayed in UCC3505 strain, which harbors a URA3 reporter cassette at VII-L subtelomeric region (54). Silencing at the HML and HMR mating-type loci were determined in UCC3515 and UCC4564 stains, containing a URA3 reporter cassette in HML and HMR loci, respectively (55). To create the Rap1 point mutations, the pRS304rap1 mutants were BlpI-digested and transformed into the reporter strains. Cells from overnight culture were spotted in 10-fold serial dilutions (starting with 107 cells) on YPAD, the synthetic complete medium lacking uracil (SC-Ura) and synthetic complete medium with 5-FOA (SC 5-FOA) plates. Expression of URA3 was examined on plates containing 5-FOA, which is lethal to cells expressing URA3 (inability of growth indicates a loss of silencing).
Immunoprecipitation, generation of phospho-specific antibodies and λ phosphatase assays
Cells were grown at 30°C in YPAD medium to log phase. Cell lysates were prepared in lysis buffer (140 mM NaCl, 50 mM HEPES-KOH, pH 7.5, 1 mM ethylenediaminetetraacetic acid (EDTA), 1 mM EGTA, 1% Nonidet P-40, 0.1% deoxycholate, 2 mM imidazole, 2 mM sodium orthovanadate, 100 mM sodium fluoride, 60 mM β-glycerophosphate, 30 mM sodium pyrophosphate and Roche protease inhibitors cocktail). Rap1-Myc13 was immunoprecipitated by 2 μg anti-Myc monoclonal antibodies (9E10, Roche) and coupled with Pierce Protein G agarose beads (ThermoFisher) at 4°C for 2 h. The beads were washed three times with lysis buffer and boiled in Laemmli sample buffer for 5 min. Proteins were resolved by 7% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE). Rap1-Myc13 was detected by the anti-Myc monoclonal antibody (9E10, Roche). Affinity-purified rabbit anti-Rap1 pS731 phospho-specific antibodies, in-house customized via GeneTex, were raised against CEVISGDYEPpSQAEK phosphopeptides to detect phosphorylation of Rap1 S731. A cysteine residue was added to the N-terminus to facilitate conjugation with a carrier protein for greater immunogenicity. The specificity of antibodies was verified by peptide dot blot analysis and λ phosphatase assay (Supplementary Figure S1). For λ phosphatase assay, immunoprecipitated Rap1-Myc13 was incubated with λ phosphatase (New England Biolabs) at 30°C for 30 min and further examined by western blotting.
Dot blot assay
Dot blot assay was conducted as previously described (56). A quantity of 50, 5 or 0.5 ng of the phosphorylated or unphosphorylated peptides was spotted onto nitrocellulose membranes and probed with the anti-Rap1 pS731 phospho-specific antibody.
Cell cycle analysis
Overnight culture was refreshed to log phase in YPAD medium at 30°C. Cells were arrested at G1 by α factor for 2 h at 30°C, and released into cell cycle at 24°C. Samples were collected at 20-min intervals and processed for fluorescence-activated cell sorting and western blot analysis.
Co-immunoprecipitation (Co-IP)
YEpFAT7 and YEpFAT7-SIR3-HA3 was transformed into 303–1A and sir3 mutant cells, respectively. Cells were grown at 30°C in SC-Ura medium. Log phase cell lysates prepared in lysis buffer (50 mM NaCl, 50 mM HEPES-KOH, pH 7.5, 1 mM EDTA, 10% glycerol, 0.1% Nonidet P-40 and Roche protease inhibitors cocktail) were immunoprecipitated with 3 μg anti-Rap1 polyclonal antibody (sc6662, Santa Cruz Biotechnology) coupled to Pierce Protein G agarose beads (ThermoFisher) at 4°C for 3 h. The beads were washed three times with lysis buffer and boiled in Laemmli sample buffer for 5 min. Proteins were resolved by 10% SDS-PAGE. Endogenous Rap1 and overexpressed Sir3-HA3 proteins were detected by the anti-Rap1 polyclonal antibody (sc6662, Santa Cruz Biotechnology) and anti-HA monoclonal antibody (12CA5, Roche), respectively.
Pull-down assay
GST, GST-Rif1 (1709–1916) or GST-Rif2 (1–395) proteins were prepared from E. coli BL21 (DE3) pLys (Invitrogen) carrying pGEX-4T-1, pGEX-4T-Rif1 (1709–1916) or pGEX-4T-Rif2 (1–395), respectively. After induction with 1 mM IPTG for 3 h at 30°C, GST proteins were purified by glutathione sepharose beads according to the instructions of the manufacturer (GE Healthcare). Log phase cells were lysed in pulldown buffer (100 mM NaCl, 20 mM Tris, pH 8.0, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, 0.5% Triton X-100 and Roche protease inhibitor cocktail). Aliquots of glutathione sepharose beads coupled with purified GST, GST-Rif1 (amino acids 1709–1916) or GST-Rif2 (amino acids 1–395) were incubated with the yeast lysates at 4°C for 3 h. The beads were washed three times with pulldown buffer. Bound proteins were released by boiling in Laemmli sample buffer and analyzed by immunoblotting.
Chromatin immunoprecipitation (ChIP) assay
Chromatin immunoprecipitation (ChIP) analysis was performed as described (35,57). In brief, cells were harvested at log phase and crosslinked in 1% formaldehyde at room temperature for 15 min, followed by adding final 125 mM glycine to quench the formaldehyde. Cell pellets were washed twice with ice-cold HBS buffer (50 mM HEPES-KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, pH 8.0) and stored at −80°C. The anti-Myc monoclonal (9E10, Roche) antibody, anti-Rap1 polyclonal antibody (sc6662, Santa Cruz Biotechnology) and anti-HA monoclonal antibody (12CA5, Roche) were utilized. The anti-normal mouse IgG (sc2025, Santa Cruz Biotechnology) and anti-normal goat IgG (sc2028, Santa Cruz Biotechnology) were served as negative controls. For all ChIP experiments, samples were amplified in duplicate to obtain an average value for each sample. Primers used in this study are listed in Supplementary Table S4. The amount of DNA in ChIP and input samples was quantitated using real-time PCR (BioRad CFX Connect). All real-time experiments were conducted according to MIQE Guidelines (58) and checklist (Supplementary Table S5). Each real-time PCR reaction (20 μl) contained sample DNA, 1× KAPA SYBR® FAST qPCR Master Mix (KAPA BIOSYSTEMS) and 0.2 μM primers. PCR amplification was performed at 95°C 3 min, then 40 cycles of 95°C 3 s, 55°C 1 min, followed by melt curve 65–95°C (increment 0.5°C per 5 s). The data were analyzed by Bio-Rad CFX Manager 3.1 software (Bio-Rad) and presented as fold enrichment of telomeric sequence over the non-telomeric ARO1 sequence in the same samples. Data were shown as the mean plus or minus one standard deviation (error bars). A two-tailed Student's t-test was used to determine statistical significance.
In vitro kinase assay
In vitro kinase assay was conducted as previously described (59). Recombinant GST-Rap1 (amino acids 716–746) and GST-Rap1-S731A (amino acids 716–746) substrates were purified from E. coli BL21 (DE3) pLys (Invitrogen). Myc18-Mec1 and the kinase dead Myc18-Mec1KD were purified from strains LSS90 and LSS93, kindly provided by Dr Akira Matsuura. Tel1-HA and the kinase dead Tel1KD-HA were purified from strains CWY148 and CWY150, carrying plasmid pKR5 (TEL1-HA) and pJM8 (tel1-HA(KD)), respectively. The plasmids pKR5 and pJM8 were kindly provided by Dr Thomas D. Petes.
In brief, protein extracts were prepared by lysis buffer (50 mM HEPES-KOH, pH 7.5, 100 mM KCl, 0.1 mM EDTA, 0.2% Tween 20, 1 mM DTT, 200 μM β-glycerophosphate, 200 μM sodium orthovanadate, 40 μM sodium fluoride, 1 mM PMSF and Roche protease inhibitors cocktail). Cell lysates were incubated at 4°C for 2 h with Pierce Protein G agarose beads (ThermoFisher) and 2 μg anti-Myc monoclonal (9E10, Roche) or 3 μg anti-HA monoclonal antibody (12CA5, Roche). The beads were washed twice with 0.5 M LiCl, three times with lysis buffer, and once with kinase buffer (20 mM HEPES-KOH, pH 7.5, 4 mM MgCl2, 4 mM MnCl2), and separated into equal portions for immunoblotting and kinase reaction. The kinase reactions were started in 25 μl of kinase buffer by addition of the buffer to the final of 20 μM ATP, 10 μCi [γ-32P]ATP and 4 μg recombinant GST-Rap1 proteins, and incubated at 30°C for 30 min. The reaction was stopped by addition of 5× sample buffer and the eluted proteins were analyzed by 12% SDS-PAGE. The gels were stained with Coomassiae blue and dried for autoradiography.
Electrophoretic mobility shift assay (EMSA)
pGEX-4T-Rap1 (353–827) was transformed into E. coli BL21 (DE3) pLys (Invitrogen) to express a GST-Rap1 fusion protein with 1 mM IPTG at 16°C overnight induction. Electrophoretic mobility shift assay (EMSA) was performed as described (60). In brief, an aliquot of 4 pmol 32P-labeled double-stranded synthetic oligonucleotides (Scer19: 5′-TGTGGTGTGTGGGTGTGTG-3′, which contains the wild-type telomere sequence) was mixed in binding buffer (20 mM Tris, pH 7.5, 75 mM KCl, 5 mM MgCl2, 0.5 M EDTA, 1 mM DTT) with 2 μg non-specific competitor poly(dI-dC). An aliquot of the GST-Rap1 fusion protein was added to react at 25°C for 15 min and analyzed by 6% non-denaturing polyacrylamide gels.
Yeast two-hybrid assay
The yeast two-hybrid assay was conducted as described (61). The L40 reporter strain (Invitrogen) was used for the studies. Point mutations were introduced into RAP1 using QuickChange site-directed mutagenesis (Stratagene). LexA, LexA/Rap1 (679–827), LexA/Rap1-S731A (679–827) and LexA/Rap1-S731D 679–827) were expressed from pBTM116 and pBTM116-derived plasmids. GAD-fusion proteins were expressed from pACT2 and pACT2-derived plasmids. LexA, LexA/Rap1 (679–827), GAD/Rif1 (1614–1916), GAD/Rif2 (14–395) and GAD-Sir3 (356–978) were kindly provided by Dr Elizabeth A. Feeser. The interactions among proteins were quantitated by the yeast β-galactosidase assay (56,62). In each assay, a minimum of four independent colonies of each mutant were analyzed and a two-tailed Student's t-test was used to determine statistical significance.
Amplification of the telomere–telomere fusions by PCR
The single individual colony was inoculated in the YEPD and grown at 30°C for 6 days to reach saturation. Genomic DNAs were extracted with phenol–chloroform, precipitated with ethanol and resuspended in ddH2O. Fusion PCR procedure and PCR primers between X and Y’ telomeres were performed as previously described (63). In brief, a PCR reaction (30 μl) contained ∼10 ng genomic DNA, 1× HF Phusion reaction buffer, 200 μM each dNTP, 0.5 μM primers and 2 units of DNA Phusion polymerase. PCR amplification was performed at 98°C 30 s, then 26 or 34 cycles of 98°C 10 s, 65°C 20 s, 72°C 1 min, followed by 72°C 5 min. The PCR products (10 μl) were separated on 1% Tris/Borate/EDTA (TBE) gel containing 0.1% GelRed. The rap1-(Δ) mutant was used as a positive control.
Statistical analysis
All of the continuous variables are expressed as the mean ± standard deviation (s.d.). Differences between groups were tested using two-tailed Student's t-test. The P-values < 0.05 was considered as statistically significant.
RESULTS
Phosphorylation of Rap1 serine 731 regulates telomere length, but not silencing effect
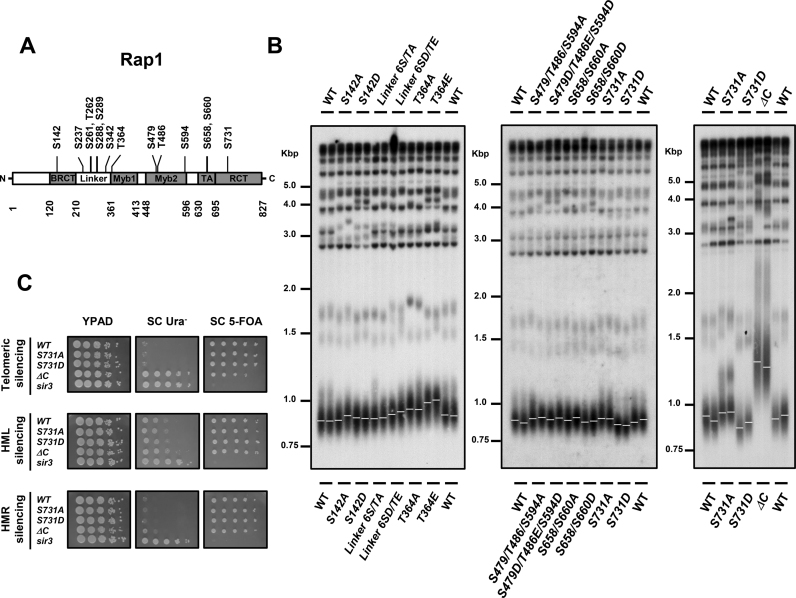
To understand whether cells control telomere homeostasis through Rap1 modification, chromosomally-tagged Rap1 was subjected to immunoprecipitation and liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyses to detect potential phosphorylation sites. Additionally, global analyses of all yeast phosphorylation atlas have previously identified several Rap1 phosphorylation sites (Figure 1A and Table 1). To examine the functions of these phosphorylations, we clustered these fourteen phosphorylation sites into six known domains (regions) according to their distributions on Rap1: BRCT (S142), Linker (S237, S261, T262, S288, S289 and S342), Myb1 (T364), Myb2 (S479, T486 and S594), TA (S658 and S660) and RCT (S731) domains. We first mutated these sites in the YPH499 strain from serine or threonine to alanine that mimics non-phosphorylation and serine or threonine to aspartic acid or glutamic acid that mimics phosphorylation. As shown in Figure 1B, none of the mutants at BRCT, Linker, Myb2 and TA domains exhibited apparent telomere length variations. Strangely, both rap1-T364A and rap1-T364E cells displayed telomere lengthening (Figure 1B). Since threonine 364 is located on the DNA binding domain of Rap1, we speculated that the amino acid substitutions might change the binding ability of Rap1 and further alter the telomere length homeostasis. Interestingly, the telomeres in rap1-S731A cells were slightly longer than those in wild-type (WT) cells, while the telomeres in rap1-S731D cells were shorter (Figure 1B). To exclude the possibility of a strain-specific effect, we also examined telomere length in the rap1 mutants in the W303–1A strain. The telomere lengthening and shortening phenotypes were also observed in the rap1-S731 mutants in the W303–1A background (Supplementary Figure S1), demonstrating that S731 phosphorylation-mediated telomere regulation is not a strain-specific phenotype.
Figure 1.
Phosphorylation of Rap1 serine 731 modulates telomere length regulation. (A) Schematic diagram of Rap1 illustrates its domain structure and putative phosphorylation sites. The BRCT, linker, DNA-binding (Myb1 and Myb2), transactivation (TA) and regulatory C-terminal (RCT) domains are indicated. (B) Telomere analysis of rap1 mutants in YPH499 background. Genomic DNA from each colony was digested with KpnI, separated in a 1% agarose gel, transferred to a nylon membrane and hybridized with a TG1–3 probe. The maximum integrity of the autoradiographic signal was determined by ImageQuant software and is indicated as a white line (n = 2). (C) Deficiency of Rap1 S731 phosphorylation does not attenuate telomere position effect and mating type locus silencing. Mutation of RAP1 S731 was introduced into the URA3 reporter strains, UCC3505 (telomeric silencing), UCC3515 (HML silencing) and UCC4564 (HMR silencing). 10-fold serial dilution of cells was spotted onto YPAD, SC Ura− and SC 5-FOA plates. Inability to grow on 5-FOA plates indicates a loss of silencing effect. The isogenic rap1-ΔC and sir3 mutants were used as controls (n = 2).
Table 1. Putative phosphorylation sites of Rap1.
| Phosphorylation site | Sequence (N’→C’) | Predicted Kinasea | Scorea | References |
|---|---|---|---|---|
| S142 | 139 AHDSLND | Cak1, 2 group | 0.09 | (89) |
| S237 | 234 NSNSDNK | Sky1 | 0.14 | (89) |
| S261 | 258 TEDSTSE | Rad53 | 0.09 | (89) |
| T262 | 259 EDSTSEK | Pho85, Cdc28 group | 0.13 | (89) |
| S288 | 285 QHVSSTA | Gcn2 | 0.12 | (90) |
| S289 | 286 HVSSTAS | Rim15 | 0.14 | (90) |
| S342 | 339 GNASFQA | Kin3 | 0.08 | (89) |
| T364 | 361 ASFTDEE | Cak1, 2 group | 0.24 | This study |
| S479 | 476 TGRSLIT | Atg1 | 0.14 | (91) |
| T486 | 483 DEDTPTA | Pho85, Cdc28 group | 0.15 | This Study, (66,90,92) |
| S594 | 591 NYNSAAK | Pho85, Cdc28 group | 0.17 | (89) |
| S658 | 655 SNISNSL | Rim15 | 0.12 | (93) |
| S660 | 657 ISNSLPF | Sky1 | 0.15 | (66) |
| S731 | 728 YEPSQAE | Mec1, Tel1 | 0.38, 0.28 | (66) |
aKinases were predicted and scored by NetPhorest 2.0 (http://www.netphorest.info/) (94).
The silent information regulator proteins Sir2–4 govern silencing at mating type loci and telomeres (64,65). Loss of the C-terminus of Rap1 reduces the telomere position effect at telomeres (20). To test whether S731 phosphorylation plays a role in telomere silencing, a mutation of S731 was introduced into the strains containing a URA3 reporter at either telomeric, HMR or HML loci. None of these S731 mutations moderated the silencing effect (Figure 1C). The isogenic sir3 mutant, which displayed increased growth on SC-Ura and repressed growth on 5-FOA plates, was used as a positive control. Deletion of the C-terminal 672–827 amino acids of Rap1 (rap1-ΔC) elongated telomeres (Figure 1B) and reduced the telomeric silencing effect (Figure 1C).
Telomere shortening stimulates Tel1 and Mec1 to directly phosphorylate Rap1 S731
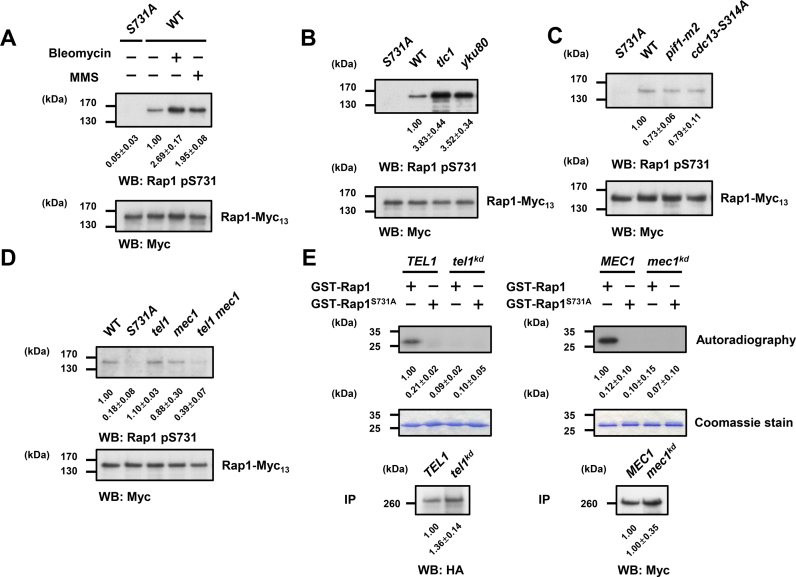
To ask whether Rap1 serine 731 is indeed phosphorylated in vivo, we generated an S731 phospho-specific antibody (Supplementary Figure S2A and B). Previous large-scale phosphoproteomic studies indicated that phosphorylation of Rap1 S731 may be induced upon methyl methanesulfonate (MMS) treatment (66). Therefore, we asked whether DNA damage could boost S731 phosphorylation. As shown in Figure 2A, an enhancement of phosphorylation on Rap1 S731 was observed under bleomycin or MMS treatment. Previous studies revealed that short telomeres exert similar biological and mechanistic traits of DNA damages (67–71). To characterize whether telomere shortening enhances the phosphorylation level of Rap1, S731 phosphorylation was detected in yku80 cells and in senescing tlc1 cells (about 75 population doublings after sporulation), which contain shortened and continuously shortening telomeres, respectively. The phosphorylation levels of Rap1 S731 were augmented in both strains (Figure 2B), suggesting that DNA damage and telomere shortening stimulate S731 phosphorylation. We then asked whether telomere lengthening could repress phosphorylation of Rap1 S731. Pif1 helicase is responsible for inhibiting both telomerase-mediated telomere elongation (72–74) and de novo telomere addition at double strand breaks (74). The pif1-m2 cells show defects in telomere maintenance and have lengthened telomeres (74). Aurora kinase-mediated Cdc13 S314 phosphorylation helps telomerase removal through alleviation of the Est1–TLC1 interaction at the M phase, and blunting this phosphorylation causes telomere lengthening (49,50). The telomere lengthening was detected in pif1-m2 and cdc13-S314A mutants (Supplementary Figure S3). Interestingly, both pif1-m2 and cdc13-S314A cells displayed reduced phosphorylation of Rap1 (Figure 2C). Thus, the phosphorylation level of Rap1 S731 is associated with the length of telomeres.
Figure 2.
Tel1/Mec1-mediated Rap1 phosphorylation on S731 is associated with telomere length variation. (A) DNA damage increases Rap1-S731 phosphorylation. Log phase cells were grown at 30°C in YPAD containing 50 mU/ml bleomycin or 0.05% Methyl methanesulfonate (MMS) for 3 h. Endogenous Rap1-Myc13 proteins were immunoprecipitated and analyzed by western blotting. The Rap1 pS731 phospho-specific antibody detected the Rap1-S731 phosphorylation upon bleomycin and MMS treatment in WT. The levels of signal compared with that of the WT were shown below and expressed as the mean ± s.d. (n = 3). The total Rap1 protein level was detected by the anti-Myc antibodies. (B) Telomere shortening increases Rap1-S731 phosphorylation. Rap1 S731 phosphorylation was examined for the individual colony from dissected tlc1 spore, which had senesced for about 75 generations (population doubling, PD75) and the yku80 spores. Endogenous Rap1-Myc13 proteins were immunoprecipitated and analyzed by western blotting as in (A). The levels of signal compared with that of the WT were shown below and expressed as the mean ± s.d. (n = 3). (C) Telomere lengthening decreases Rap1 S731 phosphorylation. The pif1-m2 and cdc13-S314A mutants showed the declined level of Rap1-S731 phosphorylation compared with that of the WT. Endogenous Rap1-Myc13 proteins were immunoprecipitated and analyzed by western blotting as in (A). The levels of signal compared with that of the WT were shown below and expressed as the mean ± s.d. (n = 3). (D) Rap1 S731 phosphorylation in vivo is Tel1- and Mec1-dependent. The Rap1 S731 phosphorylation levels were decreased in tel1 mec1 double mutants. Endogenous Rap1-Myc13 proteins were immunoprecipitated and analyzed by western blotting as in (A). The levels of signal compared with that of the WT were shown below and expressed as the mean ± s.d. (n = 3). (E) Tel1 and Mec1 phosphorylate Rap1 S731 in vitro. Left panel, in vitro Tel1 kinase assay was conducted using immunoprecipitants of HA-tagged Tel1 or kinase-dead mutant on recombinant GST-Rap1 (716–746) or GST-Rap1-S731A (716–746) substrates. Samples were loaded onto the 12% SDS-PAGE, and the phosphorylated proteins were detected by autoradiography (shown at the top, n = 3). The same gel was subsequently stained with Coomassie blue to confirm that all proteins were equally loaded (shown at the middle). The Tel1 kinases were resolved by 5% SDS-PAGE and western blotted with anti-HA antibodies (shown at the bottom, n = 3). Right panel, in vitro Mec1 kinase assay was performed as the left panel using Myc18-tagged Mec1 or kinase-dead mutants as kinases (n = 3). The Mec1 kinases were resolved by 5% SDS-PAGE and western blotted with anti-Myc antibodies (shown at the bottom, n = 3). The levels of signal compared with that of the WT were shown below and expressed as the mean ± s.d.
We next examined which kinase is responsible for the phosphorylation of Rap1. Bioinformatic analysis identified that S731 fits into a putative consensus sequence (SQ/TQ motif) of ATM/ATR family kinase substrates (Table 1). In budding yeast, TEL1 and MEC1 are homologous to the human ATM and ATR, respectively (75). Tel1 and Mec1 play functionally redundant roles in regulating checkpoint and telomere length (76,77). Tel1 preferentially associated with short telomere to promote telomere addition (44–46,48,78). Phosphorylation of Rap1 S731 was significantly reduced only in the tel1 mec1 double mutants, indicating that both Tel1 and Mec1 are redundantly responsive for phosphorylating S731 (Figure 2D). To determine whether Tel1 and Mec1 can directly phosphorylate Rap1, IP-kinase assay was conducted using immunoprecipitated Tel1 or Mec1 kinases and recombinant Rap1 substrates. As shown in Figure 2E, Tel1 and Mec1 both phosphorylated recombinant Rap1, but not Rap1-S731A, in vitro. These data indicate that telomere length shortening triggers Tel1/Mec1 to directly phosphorylate Rap1. To identify at which cell cycle stage Rap1 is phosphorylated, WT cells were synchronized with α factor and released. The phosphorylation of Rap1 S731 was observed throughout the cell cycle without obvious perturbation (Supplementary Figure S4A and B), suggesting that phosphorylation of Rap1 S731 is not regulated by cell cycle.
Phosphorylation of Rap1 S731 promotes the interaction with Rif1, but not Rif2
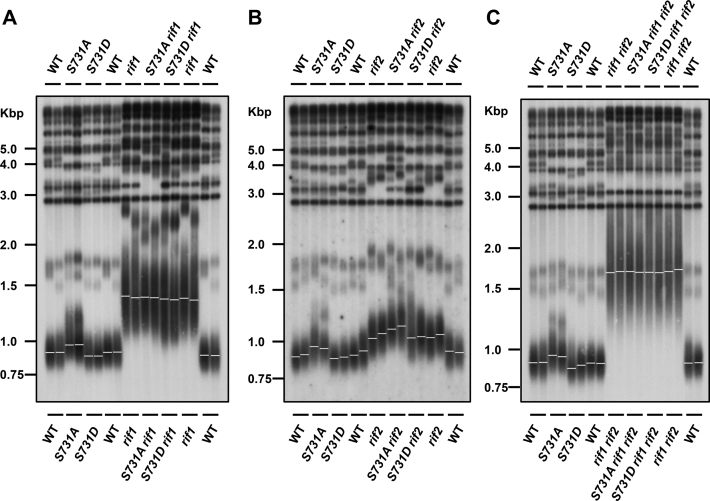
Given that S731 is located at the RCT domain, which is responsible for protein–protein interaction, we hypothesized that the function of phosphorylation at S731 might alter the interaction between Rap1 and its interacting proteins. Previous studies demonstrated that Rif1 and Rif2 are recruited to telomeres through the RCT domain (22–25). The Rap1–Rif1–Rif2 complex exerts a negative feedback mechanism to repress telomere elongation (25,32,33). We examined the telomere length in rif1, rif2 and rif1 rif2 strains in combination with the rap1-S731 mutation. Southern blot analysis showed that the telomere length in rap1-S731A rif1 and rap1-S731D rif1 cells retained similar length comparing to that of rif1 mutation alone (Figure 3A). These results suggest that Rif1 may play a role in Rap1 S731-mediated telomere regulation. However, mutation of rif1 results in very long telomeres that may mask the S731D phenotype (shortened telomeres). To differentiate between long telomeres and Rif1-dependent effects, the rap1-S731D rif1 cells were backcrossed into a strain carrying only the rap1-S731D mutation. Although the telomeres of mated rif1/RIF1 hybrids remained elongated initially at 25 PDs (Supplementary Figure S5B, labeled as the A strain), rap1-S731D/rap1-S731D rif1/RIF1 cells displayed shorter telomeres (C strain). The rap1-S731D/rap1-S731A rif1/RIF1 (D strain) served as a control. Interestingly, when these diploid cells were passaged for additional generations, the telomere phenotypes of S731A (elongated, B strain) and S731D (shortened, C strain) were observed in the haploid cells (Supplementary Figure S5C), suggesting that the reoccurrence of telomere phenotype was due to recovery of the Rif1 capping function. These genetic results indicate that Rif1 participates in the pathway of Rap1 S731 phosphorylation to regulate telomere length.
Figure 3.
Telomere length variations of rap1-S731 mutants in the rif1, rif2 and rif1 rif2 backgrounds. (A) Telomere analysis of WT, rap1-S731A and rap1-S731D mutations in the rif1 background (n = 2). (B) Telomere analysis of WT, rap1-S731A and rap1-S731D mutations in the rif2 background (n = 2). (C) Telomere analysis of WT, rap1-S731A and rap1-S731D mutations in the rif1 rif2 background. The genomic DNA was processed and probed with the same approach as described in Figure 1B (n = 2).
Conversely, the telomere length of rap1-S731A rif2 cells remained longer than that of rif2 cells (Figure 3B). We did not observe any noticeable telomere length variation between rap1-S731A rif1 rif2 and rif1 rif2 mutants (Figure 3C). The impaired S731 phosphorylation also did not perturb telomere length in isogenic yku80 strains (Supplementary Figure S6). Moreover, the total Rap1 protein levels in these strains were similar (Supplementary Figure S7A–C), indicating that loss of phosphorylation of S731 does not change Rap1 stability in rif1, rif2 and rif1 rif2 cells. These results demonstrate that Rif1 plays a key role in S731 phosphorylation-mediated telomere length regulation.
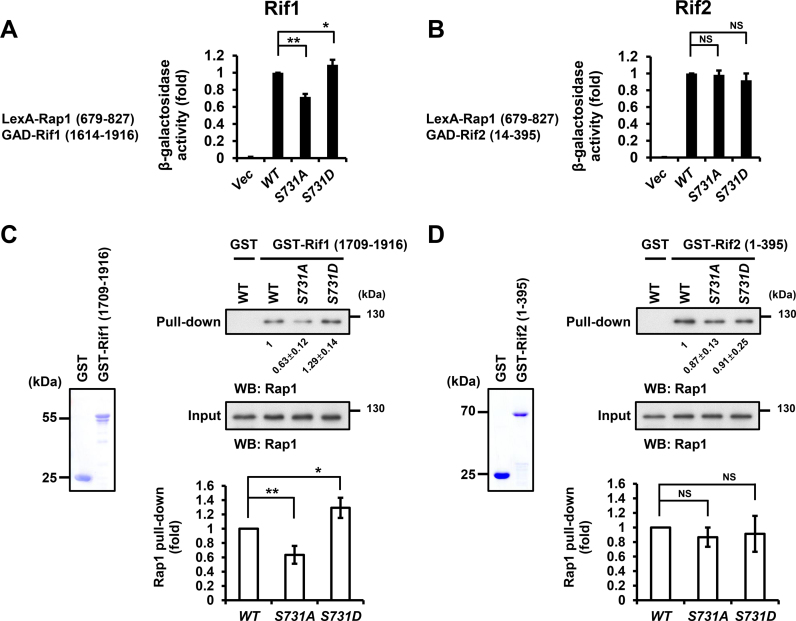
To dissect whether Rap1–Rif1 protein–protein interaction is specifically influenced by the Rap1 S731 phosphorylation, we first performed yeast two-hybrid assay to examine the interactions of Rap1–Rif1 and Rap1–Rif2. LexA/Rap1-S731A fusion protein decreased 28% of its interaction with Rif1, while LexA/Rap1-S731D increased 9% of the interaction (Figure 4A). Both LexA/Rap1 mutants did not exhibit significant change on the interactions with Rif2 (Figure 4B). To further confirm the yeast two-hybrid interaction data, we purified GST-fused Rif1 and Rif2 recombinant proteins to pull-down the endogenous Rap1 from yeast extracts. While the GST alone could not pull-down any endogenous Rap1 (Figure 4C, top-right panel lane 1), mutation of S731 to A decreased the interaction between Rap1 and Rif1 (Figure 4C, top-right panel lane 3), but a mutation to aspartic acid (Figure 4C, top-right panel lane 4) enhanced the interaction. Conversely, mutation of S731 to alanine or aspartic acid did not change their interactions with Rif2 (Figure 4D, top-right panel). These data indicate that phosphorylation of S731 stimulates the interaction between Rap1 and Rif1.
Figure 4.
Rap1 S731A mutation reduces its interaction with Rif1, but not Rif2. (A) Yeast two-hybrid assay indicated that rap1-S731A mutation significantly reduces, whereas rap1-S731D increases, the Rap1–Rif1 interaction. The Y axis shows the relative folds of β-galactosidase activity. Error bars indicate the s.d. (n = 4, *P-values < 0.05, **P-values < 0.001, Student's t-test, two-tailed). (B) Yeast two-hybrid assay indicated that WT and rap1-S mutants display similar Rap1-Rif2 interaction (n = 4, NS, non-significant, Student's t-test, two-tailed,). Bars, s.d. (C) Left panel, the aliquot of GST and GST-Rif1 (1709–1916) fusion proteins was resolved on SDS-PAGE and stained with Coomassie blue. Top-right panel, GST pulldown assay indicated that GST-Rif1 (1709–1916) significantly reduce its interaction with Rap1-S731A, whereas increase that with Rap1-S731D. Lower panel, the quantitative data of GST-Rif1 pulldown (n = 4, *P-values < 0.05, **P-values < 0.001, Student's t-test, two-tailed). Bars, s.d. (D) Left panel, the aliquot of GST and GST-Rif2 (1–395) fusion proteins was resolved on SDS-PAGE and stained with Coomassie blue. Top-right panel, GST pulldown assay demonstrated that the level of GST-Rif2 (1–395) interacting with Rap1 is not significantly different between WT and Rap1-S731A or Rap1-S731D, respectively. Lower panel, the quantitative data of GST-Rif2 pulldown (n = 4, NS, non-significant, Student's t-test, two-tailed). Bars, s.d.
Rap1 S731 phosphorylation facilities the Rif1 loading onto telomeres
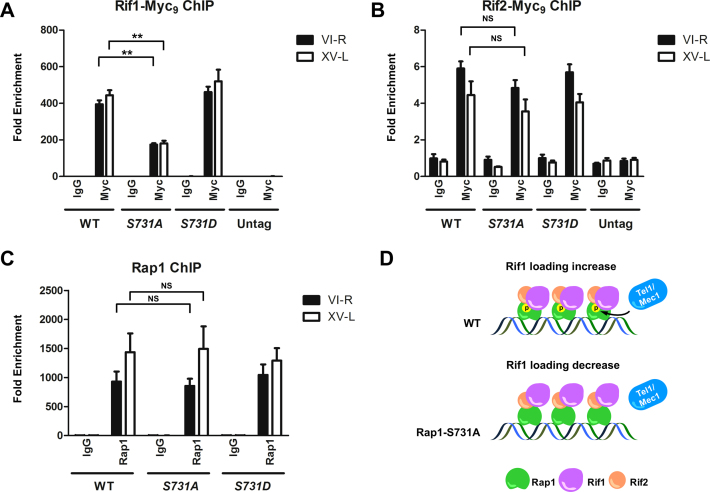
We next asked whether Rap1 S731 phosphorylation could alter the Rif1 and Rif2 occupancy on telomeres. We measured the amounts of Myc9-tagged Rif1 and Rif2 bound to VI-R and XV-L telomeres in WT, rap1-S731A and rap1-S731D cells by ChIP assay. The amounts of Rif1 on VI-R and XV-L telomeres were reduced in S731A cells (Figure 5A), whereas the loading of Rif2 on VI-R and XV-L telomeres was unaffected by Rap1 S731 mutations (Figure 5B). To dissect whether the reduction of Rif1 was caused by losing the phosphorylation of Rap1 or by changing the amounts of Rap1 on telomeres, we performed Rap1 ChIP in WT, rap1-S731A and rap1-S731D cells. The ChIP data indicate that the Rap1 protein on VI-R and XV-L telomeres displayed similar amounts as those in WT, rap1-S731A and rap1-S731D cells (Figure 5C), suggesting that mutation of S731 does not influence the Rap1 occupancy on telomeres. Furthermore, EMSA confirmed that mutation of S731 did not change the Rap1 binding affinity to the telomere sequence (Supplementary Figure S8A–C). These data imply that absence of Rap1 S731 phosphorylation decreases Rif1 binding to telomeres, alleviates the telomere capping ability and thereby extends the telomere length.
Figure 5.
Rif1, but not Rif2, occupancy at telomeres is reduced in rap1-S731A cells. (A and B). ChIP assay demonstrated that Rif1, but not Rif2, occupancy is lower at rap1-S731A versus WT telomeres. (A). The telomere binding level of Rif1 is significantly reduced in rap1-S731A cells (n = 4, **P-values < 0.001, Student's t-test, two-tailed). Strains expressing Myc-tagged proteins or untagged strain were immunoprecipitated with anti-Myc or anti-normal mouse IgG antibodies. Eluted DNA was analyzed by quantitative PCR to measure the occupancy of binding proteins on VI-R and XV-L telomeres. The data were presented as fold enrichment of telomeric sequence over the non-telomeric ARO1 sequence in the same samples. Bars, s.d. (B). The telomere binding level of Rif2 is indistinguishable between WT and rap1-S731A cells (n = 4, NS, non-significant, Student's t-test, two-tailed). The experiment manipulations and data presentations were same as in (A). Bars, s.d. (C). ChIP assay showed that Rap1 content on VI-R and XV-L telomeres is not significantly different between WT and rap1-S731A cells (n = 5, NS, non-significant, Student's t-test, two-tailed). Endogenous Rap1 was immunoprecipitated with anti-Rap1 antibodies. The data were presented as in (A). Bars, s.d. (D). Schematic model of Tel1 promoted Rap1–Rif1 interaction. Unphosphorylated Rap1-S731A causes diminished Rif1 binding to telomeres.
Rap1 S731 phosphorylation does not modulate the Sir3 occupancy on telomeres
Sir proteins compete with Rif proteins for binding to Rap1 (25,79). Moretti et al. previously demonstrated that Rif1 and Sir3 compete for binding to the carboxyl domain of Rap1 (26), and the Sir complex contributes to telomere position effect (64). Our data revealed that Rif1 binding was affected by S731 mutation. Hence, we were interested in whether S731 phosphorylation could also change the Rap1–Sir3 interaction. To examine this possibility, yeast two-hybrid assay was performed between LexA/Rap1 and GAD/Sir3 fusion proteins. Mutation of S731 did not alter the Rap1–Sir3 interaction (Supplementary Figure S9A). To further confirm this finding, HA-tagged Sir3 was expressed in sir3 mutant cells. Endogenous Rap1 was precipitated by anti-Rap1 antibodies and co-precipitated Sir3-HA3 was examined. Mutation of S731 to alanine did not enforce the interaction between Rap1 and Sir3 (Supplementary Figure S9B and C). Next, we used ChIP to examine whether mutation of S731 on Rap1 would promote Sir3 binding to telomeres. As shown in Supplementary Figure S9D, Sir3 occupancies on VI-R and XV-L telomeres were maintained at the similar level between WT and rap1-S731 mutants. Taken together, our data suggest that S731 mutation does not modulate the Rap1–Sir3 interaction and the silencing function of Sir3 on telomere.
Rap1 S731 phosphorylation inhibits type II telomere–telomere recombination
The senescing telomerase-minus cells gradually lose the viability, and a population of cells can abruptly bypass the senescence and survive through recombination (80). These cells elongate telomeres through either Y’-Y’ (type I) or telomere–telomere (type II) recombination (81). To understand whether S731 phosphorylation participates in telomere–telomere recombination in telomerase-minus tlc1 cells, we obtained individual tlc1, tlc1 rap1-S731A and tlc1 rap1-S731D spores from heterozygous tlc1/TLC1 rap1-S731A/RAP1 and tlc1/TLC1 rap1-S731D/RAP1 diploid strains, respectively. These colonies were serially streaked on solid YEPD plates and grown at 30°C. The telomeric patterns of survivors were analyzed by Southern blotting. Loss of S731 phosphorylation in tlc1 strains increased the ratio of type II survivor formation (Supplementary Table S1), which is similar to the percentage of type II survivors in tlc1 rif1 background (82). These results imply that the Rap1 S731 phosphorylation may contribute to the inhibition of type II telomere–telomere recombination pathway in telomerase-deficient cells.
Rap1 also plays a major role in blocking telomere–telomere fusion through a cis-inhibition of non-homologous end-joining (NHEJ) (83). Rap1 inhibits NHEJ through Rif2 and Sir4, but not Rif1 (38). To address whether the S731 phosphorylation contributes to Rap1-mediated suppression of end-to-end fusion, we detected the telomeric fusions by PCR amplification. Specific primers in X and Y’ elements were used to amplify the end-to-end fusions (Supplementary Figure S10A) in stationary phase cells. The rap1-(Δ) mutant served as a positive control (63,83). As shown in Supplementary Figure S10B, telomere–telomere fusions were not detected in rap1-S731A and rap1-S731D cells, indicating that NHEJ is still strongly inhibited at telomeres in these cells (Stéphane Marcand’s personal communication) as expected for mutations only impacting on Rif1 recruitment.
Rap1 phosphorylation-mediated telomere length regulation depends on Cdc13 phosphorylation-mediated telomerase recruitment
Previous studies indicate that Tel1 and Mec1 phosphorylate Cdc13 S249 and S255 to promote telomerase recruitment through enhancing Cdc13 and Est1 interaction (49–51). Since Rap1 S731 is also phosphorylated by Tel1 and Mec1 (Figure 2D and E), we were curious whether both Tel1/Mec1-mediated phosphorylations on Cdc13 and Rap1 could exhibit some interplay to regulate telomere length homeostasis. We generated mutation of Rap1 S731 on heterozygous diploid cdc13-S249S255A/CDC13 cells, and the freshly dissected combinational mutant spores were restreaked on YPAD solid plates serially for grown at 30°C. The cdc13-S249/S255A cells displayed gradual telomere shortening and cellular senescence phenotypes as previously reported (49,50). The telomere length of the rap1-S731 mutants in the cdc13-S249S255A background, which was measured after ∼75 population doublings, exhibited a comparable telomere shortening pattern as that in cdc13-S249S255A cells (Supplementary Figure S11). These data suggest that the Cdc13-S249S255 phosphorylation-mediated telomerase recruitment controls the final telomere length regulated by Tel1/Mec1-mediated Rap1 S731 phosphorylation.
DISCUSSION
About half century ago, Hayflick and Moorhead found that normal human fibroblasts cannot proliferate endlessly in laboratory culture condition (13). The telomere length is the main issue to determine the replicative lifespan of cells. Dysfunctional telomeres, which arise by either progressive telomere shortening or loss of shelterin complex, elicit a strong DNA damage response and genomic instability. In metazoan, telomere shortening and telomere uncapping activate ATM/ATR kinases to phosphorylate downstream kinases CHK1 and CHK2, which initiates p53-dependent replicative senescence and apoptosis pathways to suppress tumorigenesis (84) (Supplementary Figure S12).
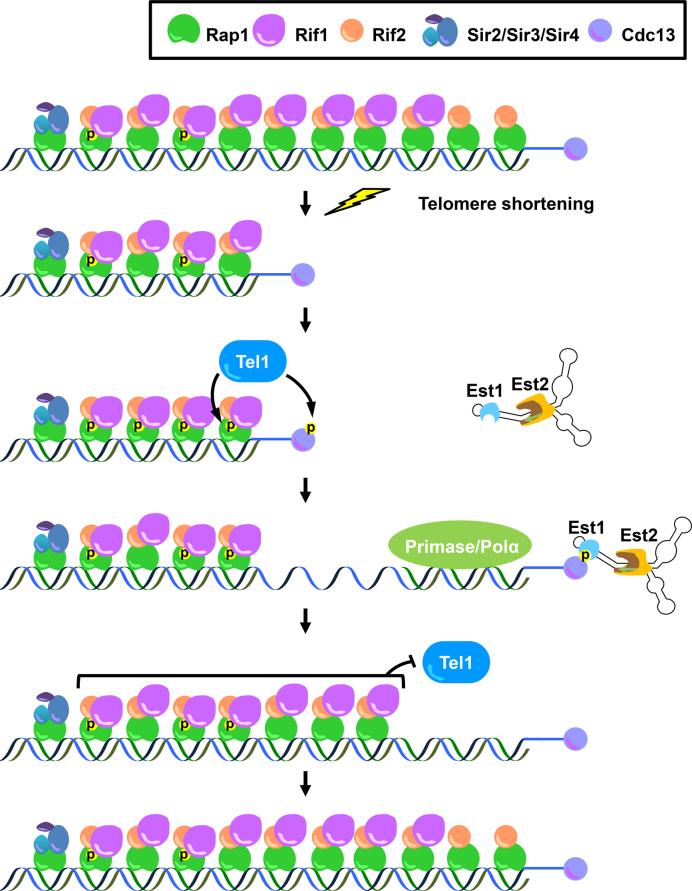
We previously demonstrated that Tel1 and Mec1 phosphorylate Cdc13 at S249 and S255 in vitro and in vivo, which facilitates the Cdc13–Est1 interaction (49–51). Other laboratories revealed genetic and physical evidence that Tel1 is recruited to short telomeres (44,46,48). In addition to the telomerase recruitment function, here we discover that Tel1 and Mec1 also participate in promoting end protection, especially in telomere-shortened cells. The major double strand telomere-binding protein Rap1 is phosphorylated by Tel1 and Mec1 at its protein–protein interaction domain. This phosphorylation is enhanced by DNA damage and telomere shortening. Compromised Rap1 phosphorylation impedes the interaction between Rap1 and Rif1, which relieving the Rif1 binding at telomeres, and increases the risk of telomere–telomere recombination. The previous study indicate that short telomeres generated by incomplete replication bear a relatively low level of Rif2 but maintaining similar amounts of Rif1 compared with long telomeres, which hints that shortened telomeres prefer to lose Rif2 before Rif1 during telomere shortening (35,46). Interestingly, we found that the interaction between Rif1 and Rap1 is regulated by Rap1 S731 phosphorylation, which is enhanced during telomere shortening. Taken together, we propose a model that once telomeres become shortened, progressive Rif2 loss weakens the capping ability to inhibit Tel1 recruitment. Tel1 localizes to shortened telomeres and phosphorylates Rap1 S731. This reinforces the interaction between Rap1 and Rif1 and/or increases Rif1 recruitment to compensate for the partial loss of end protection ability by a low number of Rap1 complexes. Furthermore, Tel1 also phosphorylates Cdc13 S249 and S255 to recruit telomerase for telomere elongation. After telomerase extends the 3′ ends of the G-rich strand, RNA-primed DNA replication by primase/DNA polyermase α fills in the complementary C strand. The elongated telomeres then recruit more Rap1 complex. They gradually reach the threshold of end protection to inhibit Tel1 loading and prevent telomerase, recombinational machinery and exonucleases from attacking the Rap1 complex-occupied DNA ends (Figure 6). Therefore, the ATM/ATR DNA damage sensing kinases play a critical role in balancing the telomere capping and telomere elongation at shortened telomeres.
Figure 6.
Tel1 orchestrates the telomere capping and telomerase recruitment pathways in telomere-shortened cells. The schematic model describes how telomere shortening enhances Rap1 phosphorylation and triggers a feedback loop to promote telomere end protection.
We demonstrated that the loss of Rap1 S731 phosphorylation causes telomere lengthening, which is diminished by RIF1 deletion but not by RIF2 deletion (Figure 3A and B). The physical interaction and ChIP data indicate that impaired Rap1 S731 phosphorylation decreases the interaction between Rap1 and Rif1, which results in low occupancy of Rif1 at telomeres (Figures 4 and 5A). Telomere lengthening in rap1-S731A cells (Figure 1B) may be due to relieved Rif1 capping ability that partially derepresses telomerase-based telomere extension, which is consistent with the ‘protein-counting’ model for telomere length regulation (25,32,33). We further rule out the possibility that S731 phosphorylation regulates the Rap1–Sir3 interaction (Supplementary Figure S9). Taken together, we think that S731 phosphorylation might directly and specifically enhance the interaction between Rap1 and Rif1. Alternatively, it is possible that S731 phosphorylation might increase Rif1 abundance and its recruitment onto telomeres.
Telomere length homeostasis is particularly important for cells under stress condition because DNA damage agents can also cause telomere breakage. The fact that DNA damage causing an upward level of Rap1 phosphorylation is consistent with the recent report that Rap1/Rif1 complex serves as the central mediator to regulate telomere length homeostasis for cells under environmental stress (40). Interestingly, Rap1, Rif1 and Rif2 complexes modulate DNA damage-induced de novo telomere addition (37,42,85). They repress the action of telomerase at telomeric DSBs (86). Nevertheless, the absence of Rif1 or Rif2 causes different resection effects (34,36). In cdc13–1 mutants, which possess an altered cap, Rif1, but not Rif2, is important for preventing end resection (41). These data imply that Rif1 and Rif2 prevent ends from resection by different pathways. We propose that abnormal telomeres induce Tel1/Mec1 to phosphorylate Rap1 S731, thereby increasing Rif1 recruitment. Interestly, the ribonucleotide reductase 3 (RNR3) gene, which is required for DNA repair and telomere expansion, is activated at its promoter by a Mec1-dependent Rap1 recruitment and the C-terminal domain of Rap1 is required for RNR3 activation (87). Upon telomere shortening, Rap1 relocalizes to the upstream promoters of hundreds of new Rap1 targets at senescence, which are preferentially activated at cell senescence stage, not simply due to loss of Rap1 telomere-binding site, but rather through an active Mec1-mediated mechanism (88). Here we identified that Tel1/Mec1-mediated Rap1 S731 phosphorylation is boosted during DNA damage and cellular senescence. It will be of interest to know whether this Rap1 phosphorylation participates in connecting between DNA damage responses at telomeres and global gene expression changes during senescence. These Rap1-dependent mechanisms for maintaining genomic integrity under environmental stress require further investigation.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ginger Zakian, Daniel Gottschling, Akira Matsuura, Thomas D. Petes, Elizabeth A. Feeser and Jing-Jer Lin for the gifts of plasmids and yeast strains, and Meng-Hsun Hsieh, Hsuan-Ming Chen and Stéphane Marcand for their critical reading of the manuscript.
Author contributions: S.-C.T. conceived and designed the experiments; C.-W.Y., S.-F.T. and C.-J.Y. performed the experiments with the assistance of C.-Y.C., C.-Y.C. and S.P.; and S.-C.T. wrote the paper with the help of C.-W.Y.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology of Taiwan [MoST-105–2311-B-002–015-MY3]; National Taiwan University [NTU-ERP-106R8805A1 to S.C.T.]. Funding for open access charge: Ministry of Science and Technology of Taiwan; National Taiwan University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Muller H.J. The remaking of chromosomes. Collecting Net. 1938; 13:181–195. [Google Scholar]
- 2. McClintock B. The behavior in successive nuclear divisions of a chromosome broken at meiosis. Proc. Natl. Acad. Sci. U.S.A. 1939; 25:405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zakian V.A. Structure, function, and replication of Saccharomyces cerevisiae telomeres. Annu. Rev. Genet. 1996; 30:141–172. [DOI] [PubMed] [Google Scholar]
- 4. McEachern M.J., Krauskopf A., Blackburn E.H.. Telomeres and their control. Annu. Rev. Genet. 2000; 34:331–358. [DOI] [PubMed] [Google Scholar]
- 5. Shay J.W., Zou Y., Hiyama E., Wright W.E.. Telomerase and cancer. Hum. Mol. Genet. 2001; 10:677–685. [DOI] [PubMed] [Google Scholar]
- 6. Shore D., Bianchi A.. Telomere length regulation: coupling DNA end processing to feedback regulation of telomerase. EMBO J. 2009; 28:2309–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wellinger R.J., Zakian V.A.. Everything you ever wanted to know about Saccharomyces cerevisiae telomeres: beginning to end. Genetics. 2012; 191:1073–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xu L., Li S., Stohr B.A.. The role of telomere biology in cancer. Annu. Rev. Pathol. 2013; 8:49–78. [DOI] [PubMed] [Google Scholar]
- 9. Greider C.W., Blackburn E.H.. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985; 43:405–413. [DOI] [PubMed] [Google Scholar]
- 10. Vega L.R., Mateyak M.K., Zakian V.A.. Getting to the end: telomerase access in yeast and humans. Nat. Rev. Mol. Cell Biol. 2003; 4:948–959. [DOI] [PubMed] [Google Scholar]
- 11. Palm W., de Lange T.. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008; 42:301–334. [DOI] [PubMed] [Google Scholar]
- 12. Hayflick L. The cell biology of aging. J. Invest. Dermatol. 1979; 73:8–14. [DOI] [PubMed] [Google Scholar]
- 13. Hayflick L., Moorhead P.S.. The serial cultivation of human diploid cell strains. Exp. Cell Res. 1961; 25:585–621. [DOI] [PubMed] [Google Scholar]
- 14. Artandi S.E., DePinho R.A.. A critical role for telomeres in suppressing and facilitating carcinogenesis. Curr. Opin. Genet. Dev. 2000; 10:39–46. [DOI] [PubMed] [Google Scholar]
- 15. Bryan T.M., Englezou A., Dalla-Pozza L., Dunham M.A., Reddel R.R.. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 1997; 3:1271–1274. [DOI] [PubMed] [Google Scholar]
- 16. Dunham M.A., Neumann A.A., Fasching C.L., Reddel R.R.. Telomere maintenance by recombination in human cells. Nat. Genet. 2000; 26:447–450. [DOI] [PubMed] [Google Scholar]
- 17. Reddel R.R., Bryan T.M., Colgin L.M., Perrem K.T., Yeager T.R.. Alternative lengthening of telomeres in human cells. Radiat. Res. 2001; 155:194–200. [DOI] [PubMed] [Google Scholar]
- 18. Kurtz S., Shore D.. Rap1 protein activates and silences transcription of mating-type genes in yeast. Genes Dev. 1991; 5:616–628. [DOI] [PubMed] [Google Scholar]
- 19. Sussel L., Shore D.. Separation of transcriptional activation and silencing functions of the RAP1-encoded repressor/activator protein 1: isolation of viable mutants affecting both silencing and telomere length. Proc. Natl. Acad. Sci. U.S.A. 1991; 88:7749–7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kyrion G., Liu K., Liu C., Lustig A.J.. Rap1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes Dev. 1993; 7:1146–1159. [DOI] [PubMed] [Google Scholar]
- 21. Shore D. Rap1: a protean regulator in yeast. Trends Genet. 1994; 10:408–412. [DOI] [PubMed] [Google Scholar]
- 22. Conrad M.N., Wright J.H., Wolf A.J., Zakian V.A.. Rap1 protein interacts with yeast telomeres in vivo: overproduction alters telomere structure and decreases chromosome stability. Cell. 1990; 63:739–750. [DOI] [PubMed] [Google Scholar]
- 23. Hardy C.F., Sussel L., Shore D.. A Rap1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 1992; 6:801–814. [DOI] [PubMed] [Google Scholar]
- 24. Lustig A.J., Kurtz S., Shore D.. Involvement of the silencer and UAS binding protein Rap1 in regulation of telomere length. Science. 1990; 250:549–553. [DOI] [PubMed] [Google Scholar]
- 25. Wotton D., Shore D.. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 1997; 11:748–760. [DOI] [PubMed] [Google Scholar]
- 26. Moretti P., Freeman K., Coodly L., Shore D.. Evidence that a complex of Sir proteins interacts with the silencer and telomere-binding protein Rap1. Genes Dev. 1994; 8:2257–2269. [DOI] [PubMed] [Google Scholar]
- 27. Vodenicharov M.D., Wellinger R.J.. DNA degradation at unprotected telomeres in yeast is regulated by the CDK1 (Cdc28/Clb) cell-cycle kinase. Mol. Cell. 2006; 24:127–137. [DOI] [PubMed] [Google Scholar]
- 28. Chandra A., Hughes T.R., Nugent C.I., Lundblad V.. Cdc13 both positively and negatively regulates telomere replication. Genes Dev. 2001; 15:404–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lin J.J., Zakian V.A.. The Saccharomyces Cdc13 protein is a single-strand TG1-3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:13760–13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nugent C.I., Hughes T.R., Lue N.F., Lundblad V.. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996; 274:249–252. [DOI] [PubMed] [Google Scholar]
- 31. Vodenicharov M.D., Laterreur N., Wellinger R.J.. Telomere capping in non-dividing yeast cells requires Yku and Rap1. EMBO J. 2010; 29:3007–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Levy D.L., Blackburn E.H.. Counting of Rif1p and Rif2p on Saccharomyces cerevisiae telomeres regulates telomere length. Mol. Cell. Biol. 2004; 24:10857–10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Marcand S., Gilson E., Shore D.. A protein-counting mechanism for telomere length regulation in yeast. Science. 1997; 275:986–990. [DOI] [PubMed] [Google Scholar]
- 34. Ribeyre C., Shore D.. Anticheckpoint pathways at telomeres in yeast. Nat. Struct. Mol. Biol. 2012; 19:307–313. [DOI] [PubMed] [Google Scholar]
- 35. McGee J.S., Phillips J.A., Chan A., Sabourin M., Paeschke K., Zakian V.A.. Reduced Rif2 and lack of Mec1 target short telomeres for elongation rather than double-strand break repair. Nat. Struct. Mol. Biol. 2010; 17:1438–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bonetti D., Clerici M., Anbalagan S., Martina M., Lucchini G., Longhese M.P.. Shelterin-like proteins and Yku inhibit nucleolytic processing of Saccharomyces cerevisiae telomeres. PLoS Genet. 2010; 6:e1000966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hirano Y., Fukunaga K., Sugimoto K.. Rif1 and Rif2 inhibit localization of Tel1 to DNA ends. Mol. Cell. 2009; 33:312–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Marcand S., Pardo B., Gratias A., Cahun S., Callebaut I.. Multiple pathways inhibit NHEJ at telomeres. Genes Dev. 2008; 22:1153–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Harari Y., Romano G.H., Ungar L., Kupiec M.. Nature vs nurture: interplay between the genetic control of telomere length and environmental factors. Cell Cycle. 2013; 12:3465–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Romano G.H., Harari Y., Yehuda T., Podhorzer A., Rubinstein L., Shamir R., Gottlieb A., Silberberg Y., Pe’er D., Ruppin E. et al. Environmental stresses disrupt telomere length homeostasis. PLoS Genet. 2013; 9:e1003721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Anbalagan S., Bonetti D., Lucchini G., Longhese M.P.. Rif1 supports the function of the CST complex in yeast telomere capping. PLoS Genet. 2011; 7:e1002024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Negrini S., Ribaud V., Bianchi A., Shore D.. DNA breaks are masked by multiple Rap1 binding in yeast: implications for telomere capping and telomerase regulation. Genes Dev. 2007; 21:292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nakada D., Matsumoto K., Sugimoto K.. ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev. 2003; 17:1957–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bianchi A., Shore D.. Increased association of telomerase with short telomeres in yeast. Genes Dev. 2007; 21:1726–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hector R.E., Shtofman R.L., Ray A., Chen B.R., Nyun T., Berkner K.L., Runge K.W.. Tel1p preferentially associates with short telomeres to stimulate their elongation. Mol. Cell. 2007; 27:851–858. [DOI] [PubMed] [Google Scholar]
- 46. Sabourin M., Tuzon C.T., Zakian V.A.. Telomerase and Tel1p preferentially associate with short telomeres in S. cerevisiae. Mol. Cell. 2007; 27:550–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Goudsouzian L.K., Tuzon C.T., Zakian V.A.. S. cerevisiae Tel1p and Mre11p are required for normal levels of Est1p and Est2p telomere association. Mol. Cell. 2006; 24:603–610. [DOI] [PubMed] [Google Scholar]
- 48. Chang M., Arneric M., Lingner J.. Telomerase repeat addition processivity is increased at critically short telomeres in a Tel1-dependent manner in Saccharomyces cerevisiae. Genes Dev. 2007; 21:2485–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shen Z.J., Hsu P.H., Su Y.T., Yang C.W., Kao L., Tseng S.F., Tsai M.D., Teng S.C.. PP2A and Aurora differentially modify Cdc13 to promote telomerase release from telomeres at G2/M phase. Nat. Commun. 2014; 5:5312–5322. [DOI] [PubMed] [Google Scholar]
- 50. Shen Z.J., Hsu P.H., Su Y.T., Yang C.W., Kao L., Tseng S.F., Tsai M.D., Teng S.C.. Corrigendum: PP2A and Aurora differentially modify Cdc13 to promote telomerase release from telomeres at G2/M phase. Nat. Commun. 2015; 6:7819–7829. [DOI] [PubMed] [Google Scholar]
- 51. Tseng S.F., Lin J.J., Teng S.C.. The telomerase-recruitment domain of the telomere binding protein Cdc13 is regulated by Mec1p/Tel1p-dependent phosphorylation. Nucleic Acids Res. 2006; 34:6327–6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Amberg D.C., Burke D.J., Strathern J.N.. Methods in Yeast Genetics: a Cold Spring Harbor Laboratory Course Manual, 2005 Edition. 2005; NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 53. Liu H., Naismith J.H.. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 2008; 8:91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Singer M.S., Gottschling D.E.. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994; 266:404–409. [DOI] [PubMed] [Google Scholar]
- 55. Singer M.S., Kahana A., Wolf A.J., Meisinger L.L., Peterson S.E., Goggin C., Mahowald M., Gottschling D.E.. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics. 1998; 150:613–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kao L., Wang Y.T., Chen Y.C., Tseng S.F., Jhang J.C., Chen Y.J., Teng S.C.. Global analysis of Cdc14 dephosphorylation sites reveals essential regulatory role in mitosis and cytokinesis. Mol. Cell Proteomics. 2014; 13:594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fisher T.S., Taggart A.K., Zakian V.A.. Cell cycle-dependent regulation of yeast telomerase by Ku. Nat. Struct. Mol. Biol. 2004; 11:1198–1205. [DOI] [PubMed] [Google Scholar]
- 58. Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009; 55:611–622. [DOI] [PubMed] [Google Scholar]
- 59. Wakayama T., Kondo T., Ando S., Matsumoto K., Sugimoto K.. Pie1, a protein interacting with Mec1, controls cell growth and checkpoint responses in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001; 21:755–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wahlin J., Cohn M.. Saccharomyces cerevisiae Rap1 binds to telomeric sequences with spatial flexibility. Nucleic Acids Res. 2000; 28:2292–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Feeser E.A., Wolberger C.. Structural and functional studies of the Rap1 C-terminus reveal novel separation-of-function mutants. J. Mol. Biol. 2008; 380:520–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Guarente L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983; 101:181–191. [DOI] [PubMed] [Google Scholar]
- 63. Lescasse R., Pobiega S., Callebaut I., Marcand S.. End-joining inhibition at telomeres requires the translocase and polySUMO-dependent ubiquitin ligase Uls1. EMBO J. 2013; 32:805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Aparicio O.M., Billington B.L., Gottschling D.E.. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991; 66:1279–1287. [DOI] [PubMed] [Google Scholar]
- 65. Rine J., Herskowitz I.. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987; 116:9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Albuquerque C.P., Smolka M.B., Payne S.H., Bafna V., Eng J., Zhou H.. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol. Cell Proteomics. 2008; 7:1389–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. AS I.J, Greider C.W.. Short telomeres induce a DNA damage response in Saccharomyces cerevisiae. Mol. Biol. Cell. 2003; 14:987–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Enomoto S., Glowczewski L., Berman J.. MEC3, MEC1, and DDC2 are essential components of a telomere checkpoint pathway required for cell cycle arrest during senescence in Saccharomyces cerevisiae. Mol. Biol. Cell. 2002; 13:2626–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Karlseder J., Broccoli D., Dai Y., Hardy S., de Lange T.. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science. 1999; 283:1321–1325. [DOI] [PubMed] [Google Scholar]
- 70. Karlseder J., Smogorzewska A., de Lange T.. Senescence induced by altered telomere state, not telomere loss. Science. 2002; 295:2446–2449. [DOI] [PubMed] [Google Scholar]
- 71. Lin Y.H., Chang C.C., Wong C.W., Teng S.C.. Recruitment of Rad51 and Rad52 to short telomeres triggers a Mec1-mediated hypersensitivity to double-stranded DNA breaks in senescent budding yeast. PLoS One. 2009; 4:e8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhou J., Monson E.K., Teng S.C., Schulz V.P., Zakian V.A.. Pif1p helicase, a catalytic inhibitor of telomerase in yeast. Science. 2000; 289:771–774. [DOI] [PubMed] [Google Scholar]
- 73. Boule J.B., Vega L.R., Zakian V.A.. The yeast Pif1p helicase removes telomerase from telomeric DNA. Nature. 2005; 438:57–61. [DOI] [PubMed] [Google Scholar]
- 74. Schulz V.P., Zakian V.A.. The Saccharomyces Pif1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell. 1994; 76:145–155. [DOI] [PubMed] [Google Scholar]
- 75. Abraham R.T. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001; 15:2177–2196. [DOI] [PubMed] [Google Scholar]
- 76. Craven R.J., Greenwell P.W., Dominska M., Petes T.D.. Regulation of genome stability by TEL1 and MEC1, yeast homologs of the mammalian ATM and ATR genes. Genetics. 2002; 161:493–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ritchie K.B., Mallory J.C., Petes T.D.. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1999; 19:6065–6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Viscardi V., Bonetti D., Cartagena-Lirola H., Lucchini G., Longhese M.P.. MRX-dependent DNA damage response to short telomeres. Mol. Biol. Cell. 2007; 18:3047–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Palladino F., Laroche T., Gilson E., Axelrod A., Pillus L., Gasser S.M.. Sir3 and Sir4 proteins are required for the positioning and integrity of yeast telomeres. Cell. 1993; 75:543–555. [DOI] [PubMed] [Google Scholar]
- 80. Lundblad V., Blackburn E.H.. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell. 1993; 73:347–360. [DOI] [PubMed] [Google Scholar]
- 81. Teng S.C., Zakian V.A.. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999; 19:8083–8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Teng S.C., Chang J., McCowan B., Zakian V.A.. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol. Cell. 2000; 6:947–952. [DOI] [PubMed] [Google Scholar]
- 83. Pardo B., Marcand S.. Rap1 prevents telomere fusions by nonhomologous end joining. EMBO J. 2005; 24:3117–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Deng Y., Chan S.S., Chang S.. Telomere dysfunction and tumour suppression: the senescence connection. Nat. Rev. Cancer. 2008; 8:450–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Frank C.J., Hyde M., Greider C.W.. Regulation of telomere elongation by the cyclin-dependent kinase CDK1. Mol. Cell. 2006; 24:423–432. [DOI] [PubMed] [Google Scholar]
- 86. Gallardo F., Laterreur N., Cusanelli E., Ouenzar F., Querido E., Wellinger R.J., Chartrand P.. Live cell imaging of telomerase RNA dynamics reveals cell cycle-dependent clustering of telomerase at elongating telomeres. Mol. Cell. 2011; 44:819–827. [DOI] [PubMed] [Google Scholar]
- 87. Tomar R.S., Zheng S., Brunke-Reese D., Wolcott H.N., Reese J.C.. Yeast Rap1 contributes to genomic integrity by activating DNA damage repair genes. EMBO J. 2008; 27:1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Platt J.M., Ryvkin P., Wanat J.J., Donahue G., Ricketts M.D., Barrett S.P., Waters H.J., Song S., Chavez A., Abdallah K.O. et al. Rap1 relocalization contributes to the chromatin-mediated gene expression profile and pace of cell senescence. Genes Dev. 2013; 27:1406–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Swaney D.L., Beltrao P., Starita L., Guo A., Rush J., Fields S., Krogan N.J., Villen J.. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat. Methods. 2013; 10:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Holt L.J., Tuch B.B., Villen J., Johnson A.D., Gygi S.P., Morgan D.O.. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009; 325:1682–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Helbig A.O., Rosati S., Pijnappel P.W., van Breukelen B., Timmers M.H., Mohammed S., Slijper M., Heck A.J.. Perturbation of the yeast N-acetyltransferase NatB induces elevation of protein phosphorylation levels. BMC Genomics. 2010; 11:685–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Li X., Gerber S.A., Rudner A.D., Beausoleil S.A., Haas W., Villen J., Elias J.E., Gygi S.P.. Large-scale phosphorylation analysis of alpha-factor-arrested Saccharomyces cerevisiae. J. Proteome Res. 2007; 6:1190–1197. [DOI] [PubMed] [Google Scholar]
- 93. Smolka M.B., Albuquerque C.P., Chen S.H., Zhou H.. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:10364–10369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Horn H., Schoof E.M., Kim J., Robin X., Miller M.L., Diella F., Palma A., Cesareni G., Jensen L.J., Linding R.. KinomeXplorer: an integrated platform for kinome biology studies. Nat. Methods. 2014; 11:603–604. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.