Abstract
Background
Domain of Unknown Function 231-containing proteins (DUF231) are plant specific and their function is largely unknown. Studies in the model plants Arabidopsis and rice suggested that some DUF231 proteins act in the process of O-acetyl substitution of hemicellulose and esterification of pectin. However, little is known about the function of DUF231 proteins in woody plant species.
Results
This study provides evidence supporting that one member of DUF231 family proteins in the woody perennial plant Populus deltoides (genotype WV94), PdDUF231A, has a role in the acetylation of xylan and affects cellulose biosynthesis. A total of 52 DUF231-containing proteins were identified in the Populus genome. In P. deltoides transgenic lines overexpressing PdDUF231A (OXPdDUF231A), glucose and cellulose contents were increased. Consistent with these results, the transcript levels of cellulose biosynthesis-related genes were increased in the OXPdDUF231A transgenic lines. Furthermore, the relative content of total acetylated xylan was increased in the OXPdDUF231A transgenic lines. Enzymatic saccharification assays revealed that the rate of glucose release increased in OXPdDUF231A transgenic lines. Plant biomass productivity was also increased in OXPdDUF231A transgenic lines.
Conclusions
These results suggest that PdDUF231A affects cellulose biosynthesis and plays a role in the acetylation of xylan. PdDUF231A is a promising target for genetic modification for biofuel production because biomass productivity and compositional quality can be simultaneously improved through overexpression.
Electronic supplementary material
The online version of this article (10.1186/s13068-017-0998-3) contains supplementary material, which is available to authorized users.
Keywords: Acetylation, Cell wall, Cellulose, DUF231, Populus, Sugar release, Xylan
Background
The plant cell wall is important for preventing pathogen attack and structural damage from environmental perturbations and mechanical stress. Recently, plant cell walls have been highlighted as important bioenergy sources through degrading structural polymer complexes of lignocellulosic products such as cellulose, hemicellulose, pectin and lignin. Among these, pectin, lignin and hemicellulose are regarded as substrates of O-acetylation that impact the industrial production of biofuel and inhibit the microbial fermentation for converting sugar to ethanol by released acetate [1–3]. In particular, the acetylation of hemicellulose has been studied to a greater extent due to its relevance to biomass recalcitrance. The acetylation of xyloglucan in dicots occurs mainly on the galactosyl residues in side chains [4, 5]. In contrast, the acetylation occurs at the glucosyl residue on xyloglucan backbone in the monocot such as Poaceae, though such an acetylation was also found in dicot plant Solanaceae [6–9]. In the woody plant, the glucoronoxylan and glucomannans are mainly acetylated at the O-2 position and/or the O-3 position in xylopyranosyl or mannopyranosyl residue [3]. The acetylation at O-2 position of xylan has been reported to be mediated by reduced wall acetylation (RWA) in hybrid aspen [10].
In Arabidopsis, three classes of proteins including reduced wall acetylation (AtRWA), altered xyloglucan (AtAXY), and trichome birefringence (AtTBR)/TBR-LIKE (AtTBL) have been reported as modifiers of acetylation of cell wall polysaccharides. Four AtRWA genes have been identified and loss of function of AtRWA resulted in alternation of acetylation of polysaccharides. The rwa2 single mutant reduced acetylation of pectin, xyloglucan, and xylan by up to 20% [11]. Acetylation in the quadruple loss-of-function mutant of AtRWA genes was reduced by 63% compared with wild type, indicating RWAs facilitate acetylation in cell wall polymers [12]. The other two protein classes of AtAXY and AtTBR/AtTBL share the conserved TBL domain and Domain of Unknown Function 231 (DUF231) [1] and are referred to as DUF231 family proteins. A total of 46 members of the DUF231 family proteins were found in the Arabidopsis genome [13]. The TBL domain has a conserved Gly-Asp-Ser (GDS) motif that can be found in esterases and lipases [14]. The DUF231 domain contains a conserved Asp-X-X-His (DXXH) motif localized toward the C-terminus following the TBL domain in most DUF231 proteins [14]. Loss of AXY4 in Arabidopsis abolished the acetylation of xyloglucan, indicating that AXY4 functions as a xyloglucan-specific O-acetyltransferase [4]. AtESK1/AtTBL29, a member of AtTBL family, has been shown to transfer the acetyl residue to the 2-O and 3-O positions on xylan in vitro, and loss-of-function mutation in ESK1/TBL29 rendered partial loss of 2-O and 3-O-acetylated xylan, implying that ESK1/TBL29 can function as a xylan acetyltransferase [15, 16]. It was reported that AtESK1 generates an even pattern of acetyl esters on xylan, thereby mediating the interaction of xylan with hydrophilic cellulose fibrils [17]. AtTBL3 and AtTBL31 were recently proposed to be compensators for the partial acetyltransferase activity of ESK1/TBL29 in xylan acetylation [18]. Additional AtTBL family proteins, including AtTBL32, 33, 34, and 35, have recently been reported as being mono-O-acetyltransferases in Arabidopsis [19, 20]. In vitro acetylation test showed that recombinant Arabidopsis TBL proteins acetylated either O-2 or O-3 mono position or 2,3-di-O-acetylation site [21]. In rice, a total of 66 TBL proteins were identified and, among them, OsTBL1 has been shown to function as a xylan mono-O-acetyltransferase [22]. Interestingly, the rice mutants of ostbl1 and ostbl2 were more sensitive to leaf blight pathogen, suggesting that xylan acetylation mediated by TBL plays a role in pathogen resistance [22]. Another rice GDSL motif-containing protein, brittle leaf sheath1 (BS1), was reported as the GDSL esterase for xylan deacetylation [23].
DUF231 family proteins’ activity is not limited to the acetyl transferase activity on hemicellulose. For example, the loss-of-function mutant of TBR and TBL3 had increased pectin content and reduced esterification of pectin [13]. Loss of powdery mildews resistance 5 (PMR5) in Arabidopsis resulted in reduction in pectin modification in cell walls together with a defect in cell expansion [24]. Through comparative genomics and amino acid sequence profiling, it was proposed that PMR5 may play a role in controlling the acylation levels of glycans via its predicted acyltransferase and esterase domain [25]. Interestingly, AtESK1 was also proposed to have similar functions as PMR5 [25]. The reduction of crystalline cellulose content was observed in the esk1/tbl29 Arabidopsis mutants [15]. In addition, microarray results showed that AtTBR and AtTBL3 were co-expressed with cellulose biosynthesis genes, indicating a close relationship between TBR and cellulose biosynthesis [13]. On the other hand, many Arabidopsis xylan backbone synthesis mutants have reduced cellulose content. For example, loss-of-function mutants of Arabidopsis irregular xylem (IRX) 15 and 15-L, members of DUF579 family that have been reported as biosynthetic genes related to xylan and cellulose formation have reduced cellulose content [5, 26]. Taken together, these findings suggest that DUF231 family proteins are important polysaccharide modifiers on various cell wall polymers in Arabidopsis.
So far, all functional characterizations of DUF231 family proteins have been limited to herbaceous plants, but bioinformatics analyses indicate that DUF231 proteins are also present in other species [1, 13]. In this study, we identified a total of 52 DUF231 family proteins in the woody perennial plant Populus. We provide characterization of one member of Populus DUF231 family proteins and propose that this gene is involved in both xylan O-acetylation and cellulose biosynthesis.
Results
Bioinformatics analysis of Populus trichocarpa DUF231 family proteins (PtDUF231)
To identify DUF231-containing proteins in Populus, we performed a protein homolog search in the Populus genome (Populus trichocarpa v3.0 annotation) at Phytozome v11.0 web site (https://phytozome.jgi.doe.gov/pz/portal.html) using the DUF231 domain of AtTBR as a template [13]. A total of 52 Populus proteins were identified as DUF231-containing proteins (Additional file 1). PtDUF231 protein family members had an amino acid sequence identity of > 30% with Arabidopsis DUF231 proteins. Forty-eight of the 52 PtDUF231 proteins shared each node with Arabidopsis DUF231 proteins in the phylogenetic tree (Fig. 1a). All PtDUF231 family proteins contain a plant-specific TBL domain and a DUF231 domain (Fig. 1b). One protein, Potri.001G010900, lacks an N-terminal region, but contains both the TBL domain and the DUF231 domain (Fig. 1b). A conserved GDSL motif was identified in the TBL domain which contains approximately 50 amino acids (Fig. 1c) [14]. The TBL domain is located in proximity to the DUF231 domain in PtDUF231 proteins, similar to what was reported for Arabidopsis DUF231 (AtDUF231) proteins (Fig. 1c) [13]. As expected, the RNQWESLxCxL conserved amino acid sequences aligned next to the GDSL motif (Fig. 1c). The signature DUF231 domain motifs, LLBITxLSxxRKDGHPSxY and DCxHWCLPGxPDTWNELLYAxL, were found at the C-terminus of the proteins (Fig. 1c).
Fig. 1.
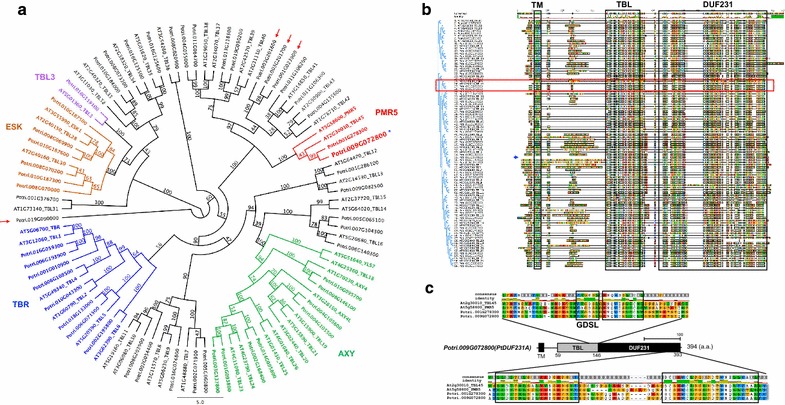
Bioinformatics analysis of DUF231-containing proteins in Arabidopsis and Populus. a Maximum likelihood phylogenetic tree of Arabidopsis and Populus DUF231-containing proteins. The numbers at the branches denote bootstrap confidence values. Note that 48 of the 52 PtDUF231 proteins share each node with Arabidopsis DUF231 proteins (red arrows mark four PtDUF231 proteins that are not shared a node with Arabidopsis DUF231 proteins). The location of PtDUF231A (Potri.009G072800) is indicated in the PMR5 clade by a blue asterisk. b Amino acid sequence alignment by MUSCLE. Note that PtDUF231A (Potri.009G072800) shows 52.5% amino acid identity with Arabidopsis PMR5 and 55.9% identity with TBL45. The closest paralog for PtDUF231A is PtDUF231B (Potri.001G278300) with 89.4% identity at the amino acid level. Blue lined brackets shown in the left illustrate the phylogenic tree as shown in a. Red horizontal box marks the amino acid alignment of PtDUF231A-containing node. Blue arrow points Potri.001G010900 which contains both TBL and DUF231 domains but without N-terminal sequences. Three well-conserved protein domains including TM, TBL, and DUF231 are indicated in boxes. c Diagram of amino acid sequence alignment of TBL and DUF231 domains among PtDUF231A, its Populus paralog, its Arabidopsis ortholog (PMR5) and TBL45, in the node shown in b. Consensus sequence was defined by 50% threshold of amino acid sequence identity. The upper panel shows sequence identity using different colors (yellow: over 50%, red: 100% conserved). All conserved regions including GDSL are indicated by the black box. Note that the TBL and DUF231 domains are highly conserved in the PdDUF231A protein
To examine how many PtDUF231 family proteins can be assigned as membrane binding proteins, as reported in Arabidopsis [1], we examined the presence of transmembrane domains (TM) in PtDUF231 proteins. Among 52 PtDUF231 proteins, 39 proteins were predicted to possess at least one TM domain at the N-terminal region (Fig. 1b; Additional file 1). Potri.010G187600 and Potri.006G140300 (with 530 and 512 amino acids, respectively; 100 more amino acids than others) were predicted to contain two TM domains (Additional file 1). In contrast, 13 PtDUF231 family proteins were predicted to not contain a TM domain (Additional file 1). The signal peptide, an indicator for transferring the protein to the endoplasmic reticulum (ER) or Golgi, was also found in nine PtDUF231 proteins, with six predicted to not contain a TM domain and three predicted to contain a single TM domain (Additional file 1).
Expression pattern of PdDUF231A in different organs/tissues
No functional characterization has been reported for any member of DUF231 family proteins in Populus. In this study, we reported the characterization of one member of PtDUF231 family proteins, Potri.009G072800, designated as PtDUF231A. PtDUF231A clustered with the PMR5 subfamily (Fig. 1a) [24], together with its paralog encoded by Potri.001G278300 (PtDUF231B) (sharing 89.4% amino acid sequence identity with PtDUF231A). The PMR5 subfamily has been poorly characterized in plants with indications that it may function in carbohydrate modification [24, 25]. Both PtDUF231A and PtDUF231B were predicted to contain a TM domain at the N-terminus (Additional file 1).
As the first step toward investigating the function of PtDUF231A, we examined its expression pattern across various tissues and organs. We isolated RNA from various tissues and organs of Populus deltoides clone ‘WV94’. The full-length open reading frame of DUF231A gene in P. deltoides was designated as PdDUF231A. This was also the gene used for the transgenic study in the P. deltoides clone ‘WV94’ background described below. We designed gene-specific primers to distinguish PdDUF231A and PdDUF231B and performed a quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis. As shown in Fig. 2, PdDUF231A was ubiquitously expressed in all tested tissues and organs, with relatively high expression in young leaf, phloem and stem. PdDUF231B was similarly detected in all tested tissues and organs (Fig. 2). The only difference was that the transcript of PdDUF231A was higher than that of PdDUF231B in root (Fig. 2).
Fig. 2.
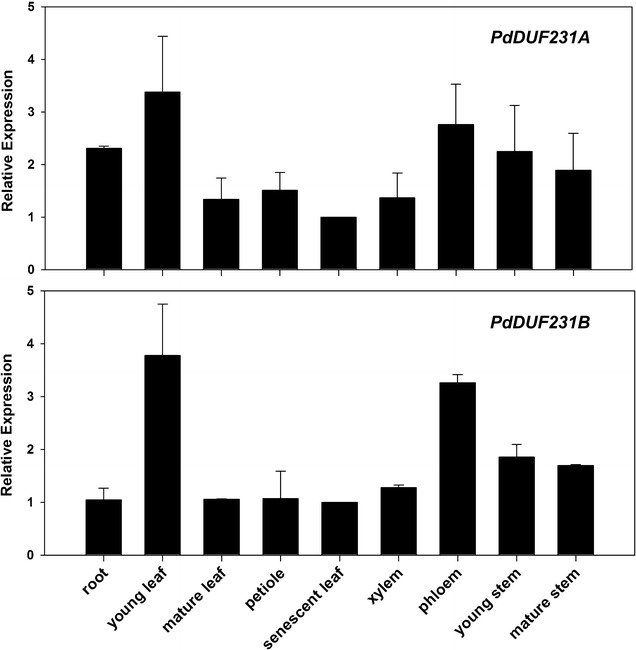
Expression pattern of PdDUF231A across various tissues and organs. Shown are qRT-PCR analysis using gene-specific primers for PdDUF231A (Potri.009G072800) and its paralog PdDUF231B (Potri.001G278300). The PdUBCc (Populus UBIQUITIN C) was used as an internal control. The relative expression range in each tissue/organ was determined by comparing expression level of senescent leaf (set as 1). Shown are mean values ± standard deviation (SD) of three technical replicates
Generation of Populus transgenic plants overexpressing PdDUF231A
To further investigate the function of PdDUF231A, we generated transgenic plants overexpressing PdDUF231A in the P. deltoides (genotype ‘WV94’) background. The expression of PdDUF231A was driven by a constitutive UBIQUITIN3 promoter (Fig. 3a). A total of ten independent transgenic lines were generated (Additional file 2). RT-PCR analysis indicated that five among those ten transgenic lines had higher expression of PdDUF231A (Additional file 2). We selected two independent transgenic lines with higher PdDUF231A expression for further characterization and these two lines were designated as OXPdDUF231A-1 and OXPdDUF231A-2. PCR analysis indicated that the copy number of the transgene was 1.8 ± 0.2 and 2.2 ± 0.4 for OXPdDUF231A-1 and OXPdDUF231A-2, respectively (Additional file 3).
Fig. 3.
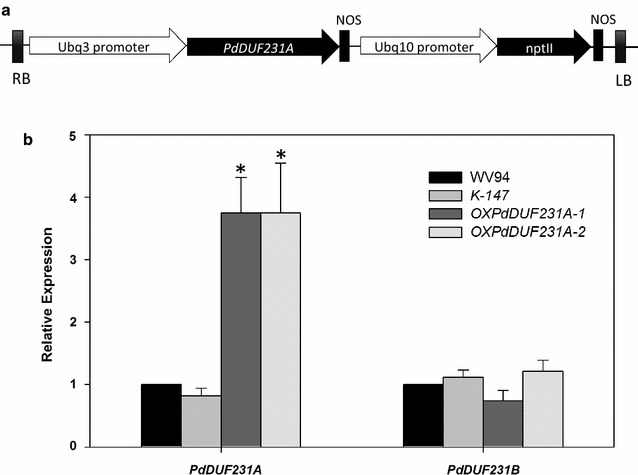
Quantitative RT-PCR analysis of the expression of PdDUF231A in Populus transgenic lines. a Diagram illustrating the gene construct used to generate OXPdDUF231A transgenic lines. b PdDUF231A expression in OXPdDUF231A transgenic lines. The cDNA generated from total RNA of scrapped xylem tissue was used for qRT-PCR. The PdUBCc (Populus UBIQUITIN C) was used as an internal control. Two independent PdDUF231A overexpression lines were examined, together with the wild-type WV94 and the empty vector control K-147. Statistical analysis was performed with three replicates in two different plants per individual transgenic line (n = 6). Asterisk indicates statistical significance compared to WV94 (p < 0.01)
To quantitatively assess the PdDUF231A transcript level in transgenic lines, we performed qRT-PCR analysis using gene-specific primers for PdDUF231A and compared the transcript level of PdDUF231A in the transgenic plants with that in the wild-type WV94 and vector only transgenic plants (K-147). PdDUF231B expression was also assessed to validate that PdDUF231A, but not PdDUF231B, was overexpressed in OXPdDUF231A. As shown in Fig. 3, the PdDUF231A transcript was about fourfold higher in two transgenic lines than in WV94 and K-147, whereas the transcript level of PdDUF231B did not differ.
Cellulose and glucose contents were higher in the OXPdDUF231A transgenic plants
To examine whether carbohydrate content was altered in the OXPdDUF231A transgenic lines, we measured monosaccharide content from stem by the NREL method [27]. The content of glucose was significantly higher in both transgenic lines than that in the control plants, whereas the contents of arabinose, galactose, xylose, and mannose were similar in all tested plants (Fig. 4a). The glucose content in OXPdDUF231A-1 and -2 was increased by 8.5 ± 4.0 and 11.4 ± 2.7% compared to WV94, respectively (Fig. 4a). To examine whether the higher glucose content observed in the OXPdDUF231A transgenic lines was due to higher amount of cellulose, we performed an in vitro anthrone assay to estimate the content of cellulose [28]. Both OXPdDUF231A transgenic lines contained significantly higher cellulose content (increased by 8–21%) than the control plants (Fig. 4b), suggesting that the higher glucose content observed in the OXPdDUF231A transgenic lines is likely due to higher cellulose content in the cell walls.
Fig. 4.
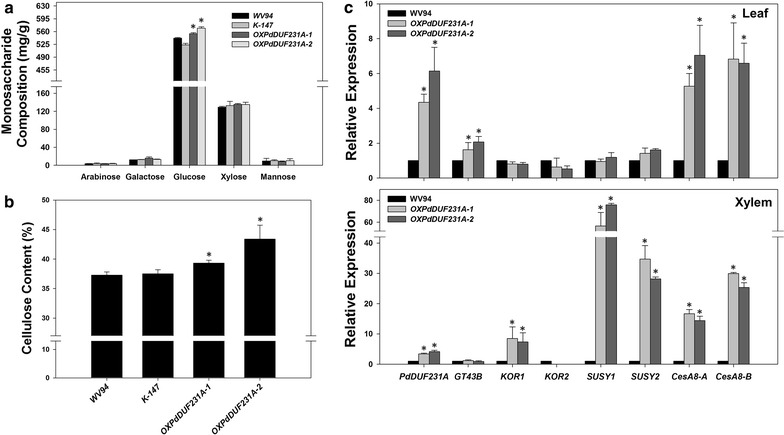
Carbohydrate and gene expression analyses in OXPdDUF231A transgenic lines. Two independent OXPdDUF231A transgenic lines (OXPdDUF231A-1 and OXPdDUF231A-2), WV94 (wild type) and K-147 (empty vector control) were grown under greenhouse conditions. a Monosaccharide composition analysis. The monosaccharide composition was determined by ion chromatography after a two-step acid treatment. b Cellulose content analysis by anthrone dye staining. c Relative gene expression of cellulose biosynthesis-related genes in leaf and xylem. Shown are the mean value ± SD of three technical repeats of three biological replicates of leaf or xylem (n = 9). Asterisks indicate statistical significance compared to WV94 (p < 0.01)
To seek further evidence supporting the involvement of PdDUF231A in cellulose biosynthesis, we examined the expression of several genes in the cellulose and hemicellulose biosynthesis pathways. qRT-PCR was performed using gene-specific primers for genes encoding Populus cellulose synthases (CesA), sucrose synthases (SUSY), and KORRIGAN (KOR) in leaf and xylem [29–31]. We also included a gene proposed to be involved in hemicellulose biosynthesis, GT43B [5, 32]. Among all tested genes in leaf, the most drastic changes were found for cellulose biosynthesis genes CesA8, whose transcript levels were four- to sixfold higher in both OXPdDUF231A transgenic lines than wild type (Fig. 4c). The transcript of GT43B, a gene encoding xylan backbone elongation factor, was also increased by approximately twofold in both OXPdDUF231A transgenic lines (Fig. 4c). On the other hand, the expression levels of SUSY and KOR were not significantly altered in the OXPdDUF231A transgenic lines (Fig. 4c). In xylem, the expression of SUSY family was most dramatically increased (30- to 80-fold) in both OXPdDUF231A transgenic plants (Fig. 4c). CesA8 and KOR1 were also expressed at higher levels in both OXPdDUF231A transgenic plants than WV94 control plant (Fig. 4c). The expression of KOR2 and GT43B was not significantly altered (Fig. 4c). Collectively, we observed increased expression of genes associated with cellulose biosynthesis in OXPdDUF231A transgenic plants, though gene expression differences were observed between leaf and xylem tissues. These results supported that PdDUF231A affects cellulose biosynthesis.
Saccharification efficiency of OXPdDUF231A transgenic lines
Because PdDUF231A appeared to affect cellulose biosynthesis and contained higher content of cellulose (Fig. 4), we wanted to examine whether lignin content was altered in the OXPdDUF231A transgenic plants. As shown in Fig. 5a, the lignin content was reduced by 6.4–7.4% in the OXPdDUF231A lines compared with that in the wild type. Because both cellulose and lignin affect sugar release, subsequently we wanted to assess the enzymatic saccharification efficiency in OXPdDUF231A transgenic lines. We measured the amount of glucose released from enzymatic saccharification and calculated it against the total glucose content in each line. Significantly higher glucose yield in both OXPdDUF231A transgenic lines was observed after 48 h enzyme treatment, compared with wild-type control (Fig. 5b). At 72 h duration of enzyme digestion, the glucose yield was approximately 4% higher in OXPdDUF231A transgenic plants than the wild type.
Fig. 5.
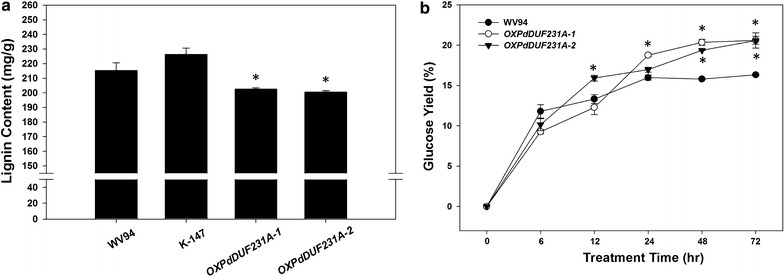
Lignin content and enzymatic saccharification assay of OXPdDUF231A transgenic plants. Dried Populus stem after debarking was subjected for lignin content measurement and saccharification analysis. a Lignin content of dried stem, b glucose yield based on total glucose content in each plant. X axis denotes enzymatic hydrolysis time. Each data point represents average value of two biological replicates ± SD. Asterisks indicate statistical significance compared to WV94 (p < 0.05)
Xylan acetylation in OXPdDUF231A transgenic lines
The acetyl substitution of hemicellulose, such as xyloglucan and xylan, was previously observed in the loss-of-function mutant of Arabidopsis DUF231 genes [4, 15, 16, 18–20]. Therefore, we investigated whether acetyl groups in xylan were also affected in the PdDUF231A overexpression lines. We performed 2D 1H-13C HSQC NMR analysis [33] to calculate the relative acetylation levels in xylan molecules in Populus stems. As shown in Fig. 6a, five different types of xylan structures including 2-O-acetylated (2-O-β-d-AcXylp), 3-O-acetylated (3-O-β-d-AcXylp), 2,3-di-O-acetylated (2,3,di-O-Ac-β-d-Xylp) xylosyl residues, 4-O-methyl-α-d-glucuronic acid (4-O-Me-GlcA), and xylan backbone [(1-4)-β-d-Xylp] were observed in the NMR spectra of OXPdDUF231A transgenic lines and wild-type plants (Fig. 6a, b). The internal anomeric xylan correlation peak ((1-4)-β-d-Xylp) appeared at 101.68/4.35 ppm, while 2-O-Ac-β-d-Xylp, 3-O-Ac-β-d-Xylp and 2,3-di-O-Ac-β-d-Xylp were observed at 99.41/4.55, 101.60/4.50, and 99.26/4.71 ppm, respectively (Fig. 6b). These peaks partially overlapped, and thus the acetylated xylans were quantified with 2-O-Ac-β-d-Xylp (C2/H2) at 73.20/4.54 ppm, 3-O-Ac-β-d-Xylp (C3/H3) at 74.76/4.83 ppm, and 2,3-di-O-Ac-β-d-Xylp (C2/H2) at 71.08/4.61 ppm, and compared to the xylan backbone ((1-4)-β-d-Xylp) peak to obtain the relative abundance of each type of acetylated xylan. The acetyl group in each Populus stem was compared in two different ways. First, the total acetyl group at ~ 20.7/1.97 ppm in the cell wall samples was quantified with total xylan content based on the aforementioned assigned peaks. Since hemicellulose acetylation mostly occurs on xylan in plant cell walls [34], the observed results indirectly indicate the abundance of acetylated xylan. In addition, the relative abundance of acetyl group in OXPdOXDUF231A transgenic lines was confirmed by an alternate comparison using the acetylated and non-acetylated xylan peaks. The relative abundance of 2-O-Ac-β-d-Xylp (C2/H2) was nearly the same in wild-type and OXPdDUF231A lines, whereas those of 3-O-Ac-β-d-Xylp (C3/H3) increased from 7.9% in wild-type to 10.0–11.5% in OXPdDUF231A transgenic lines (Fig. 6c). The 2,3-di-O-Ac-β-d-Xylp (C2/H2) was increased from 9.4% in the wild-type to 12.4–13.1% in the transgenic plants (Fig. 6c). The total acetylated xylan was increased from 65.7% in wild-type to 70–71.5% in OXPdDUF231A transgenic plants (Fig. 6c). The 4-O-methyl-α-d-glucuronic acid (MeGlcA) substitution reported in a previous study [35] was only barely observed in this NMR analysis. These results indicated that acetylation of xylan was influenced by overexpression of PdDUF231A.
Fig. 6.
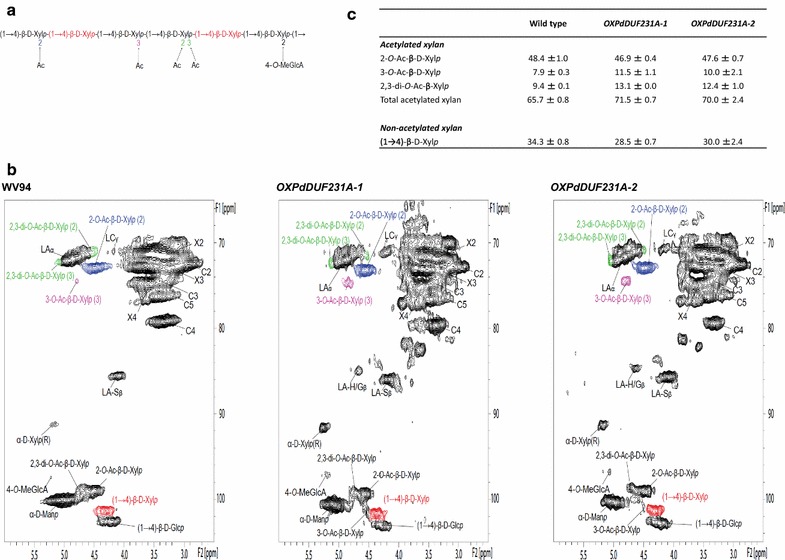
Integration analysis of xylan acetylation in OXPdDUF231A transgenic lines. a Target chemical structure detected by 13C-1H 2D HSQC NMR. b The NMR spectra from cell wall gels. Five different spectral types including 2-O-acetylated (2-O-AcXyl), 3-O-acetylated (3-O-AcXyl), 2,3-di-O-acetylated (2,3,-di-AcXyl), 4-O-methyl-alpha-d-glucuronic acid (4-O-Me-GlcA) and xylan backbone ((1-4)-β-d-Xylp) were detected in this NMR analysis. The resonance peaks of lignin were also assigned together here; LA β-aryl ether (β-O-4), LA-H/Gβ β-aryl ether (β-O-4-H/G), LA-Sβ β-aryl ether (β-O-4-S), LC resinol (β-β). The acetylated and non-acetylated xylan resonance peaks were used to perform integration analysis. c The relative integration result of acetylated groups and non-acetylated xylan. Note that 3-O-AcXyl and 2,3-di-O-AcXyl were increased in OXPdDUF231A transgenic lines. Shown are the mean values of two biological replicates each line ± SD
Biomass production in OXPdDUF231A transgenic lines
We observed that OXPdDUF231A transgenic lines were larger than control plants under our greenhouse conditions. Therefore, we measured the diameter and height and used the stem volume to estimate the biomass amount of OXPdDUF231A plants and compared it with the WV94 control plants. As shown in Fig. 7, the stem volumes of both OXPdDUF231A transgenic plants were significantly higher than those of the control plant, suggesting that overexpression of PdDUF231A increases biomass production.
Fig. 7.
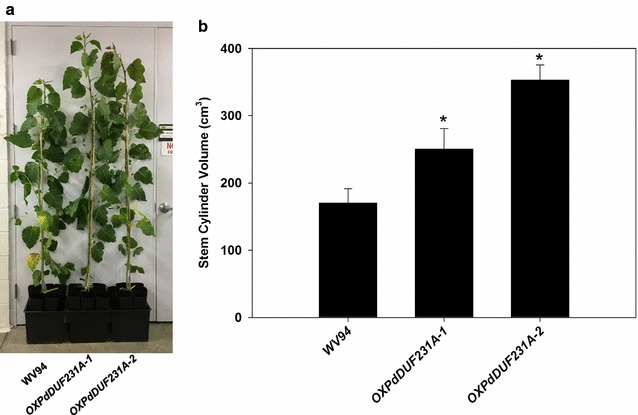
Stem volume of OXPdDUF231A transgenic plants. a Whole plant images of 6-month-old OXPdDUF231A plants grown under greenhouse conditions (bar = 30 cm). b Comparison of estimated stem volume between OXPdDUF231A transgenic plants and WV94 control. The stem volume was estimated by using the πr 2 h equation with height and diameter of primary stem. Shown are the average values of calculated stem cylinder volumes ± SD (n = 3)
Metabolite profiles of OXPdDUF231A transgenic lines
To seek further evidence supporting a role of PdDUF231A in modifying cell wall chemistry, we analyzed the metabolite profiles of OXPdDUF231A transgenic lines. We found that overexpression of PdDUF231 had greatly altered the leaf metabolite profiles relative to that observed for the empty vector control plants (Tables 1, 2; Additional file 4). The greatest upregulated metabolite that was statistically significant (p ≤ 0.05) was a 30.75-fold increase for a partially identified metabolite (13.27 235 xylopyranoside) that is likely an aromatic glycoside. With the two major m/z being 204 and 235 and the metabolite eluting earlier than known glucosides, the metabolite was tentatively identified to be a xylopyranoside conjugated to an aromatic moiety, possibly coniferyl alcohol, which would generate the observed m/z 235 if conjugated on the alcohol rather than on the aromatic ring as it is for coniferin. Additionally, other major upregulated aromatic metabolites included 1,5-dicaffeoyl-shikimate (25.92-fold), 1,2,3-benzenetriol (pyrogallol) (5.42-fold) and salicyl alcohol (3.38-fold). Several organic acid and fatty acid metabolites were also upregulated, including maleic acid (3.49-fold), erythronic acid (3.46-fold), phytol (2.31-fold), digalactosylglycerol (2.12-fold) and linoleic acid (1.27-fold). In contrast to the relatively small number of upregulated metabolites, there was a large number of downregulated metabolites including amino acids, organic acids, and flavonoids. Significantly downregulated amino acids included lysine, asparagine, ornithine (includes that generated from arginine breakdown), glutamine, 5-oxo-proline, threonine, alanine, phenylalanine, glutamic acid, aspartic acid, and serine, which ranged from 0.03- to 0.40-fold of that observed in control plants. Downregulated organic acids included oxalomalic acid, α-keto-glutaric acid, citraconic acid, citric acid, and succinic acid, which were reduced from 0.08- to 0.63-fold of the controls. Flavonoids that were reduced included rutin, luteolin, unknown 17.80 501 559 471 flavonoid, quercetin, and kaempferol that were reduced to 0.04- to 0.12-fold of that of controls. Although most higher-order salicylates were unchanged, those that are conjugated to hydroxycinnamates, including populosides B and C, were reduced to 0.21- and 0.41-fold, respectively. Similarly, many partially identified aromatics conjugated to hydroxycinnamates, including p-coumaric acid, caffeic acid, and ferulic acid were also greatly reduced in leaves of plants overexpressing PdDUF231A. These metabolites have characteristic m/z of 219, 307, and 249, respectively, when conjugated as esters. An exception was 1,5-dicaffeoyl-shikimate which was elevated almost 26-fold, but coupled with a decline in shikimic acid by 0.34-fold. Together, these results indicate major shifts in major aromatic pathways with restricted production of flavonoids and most hydroxycinnamate conjugates.
Table 1.
List of metabolites with increased content (µg/g FW sorbitol equivalents) in leaf tissues of 6-month-old greenhouse-grown OXPdDUF231A versus K-147 plants
| Metabolite [retention time (min); key m/z] | OXPdDUF231A/K-147 | OXPdDUF231A | K-147 | |||
|---|---|---|---|---|---|---|
| Fold change | p value | Avg | sem | Avg | sem | |
| 19.63 171 Feruloyl-caffeoyl glycoside | #DIV/0! | 0.282 | 0.4 | 0.3 | 0 | 0 |
| 17.04 354 Guaiacyl lignan | #DIV/0! | 0.094 | 1.0 | 0.3 | 0 | 0 |
| 17.33 354 Syringyl lignan glycoside | #DIV/0! | 0.081 | 1.2 | 0.4 | 0 | 0 |
| 18.48 Feruloyl-caffeoyl-shikimate glycoside | 59.16 | 0.083 | 15.2 | 5.0 | 0.3 | 0.0 |
| 13.27 235 Xylopyranoside | 30.75 | 0.001 | 2685.9 | 330.7 | 87.3 | 15.0 |
| 1,5-Dicaffeoyl-shikimate | 25.92 | 0.008 | 99.0 | 17.9 | 3.8 | 0.3 |
| 21.01 557 193 647 Glycoside | 15.61 | 0.252 | 4.5 | 2.3 | 0.3 | 0.0 |
| 13.63 278 309 227 | 11.67 | 0.002 | 7.6 | 1.0 | 0.7 | 0.0 |
| 18.43 616 386 Caffeoyl-shikimate glycoside conjugate | 9.61 | 0.243 | 0.8 | 0.4 | 0.1 | 0.0 |
| Syringin | 9.06 | 0.199 | 13.7 | 5.9 | 1.5 | 0.1 |
| 1,2,3-Benzenetriol | 5.42 | 0.002 | 15.2 | 1.8 | 2.8 | 0.5 |
| Raffinose | 4.75 | 0.066 | 86.6 | 21.4 | 18.2 | 4.0 |
| Maleic acid | 3.49 | 0.028 | 406.5 | 69.4 | 116.5 | 38.4 |
| Erythronic acid | 3.46 | 0.046 | 42.4 | 8.5 | 12.2 | 1.0 |
| 18.11 235 Phenolic glycoside | 3.46 | 0.263 | 0.6 | 0.2 | 0.2 | 0.0 |
| Salicyl alcohol | 3.38 | 0.020 | 113.5 | 18.1 | 33.5 | 5.3 |
| Galactinol | 3.25 | 0.160 | 74.7 | 22.4 | 23 | 4.1 |
| 12.81 539 359 320 | 3.23 | 0.026 | 15.1 | 2.5 | 4.7 | 0.3 |
Metabolites were analyzed as trimethylsilyl derivatives by gas chromatography–mass spectrometry. Data are the average (avg) and standard error of the mean (sem) of six OXPdDUF231A plants (three plants from each of two independent transgenic lines) and three K-147 control plants. Unknown and partially identified metabolites are designated by retention time, key mass-to-charge (m/z) ratios, and identified moieties. #DIV/0! represents an integration area divided by zero (absence)
Table 2.
List of metabolites with decreased content (µg/g FW sorbitol equivalents) in leaf tissues of 6-month-old greenhouse-grown OXPdDUF231A versus K-147 plants
| Metabolite [retention time (min); key m/z] | OXPdDUF231A/K-147 | OXPdDUF231A | K-147 | |||
|---|---|---|---|---|---|---|
| Fold change | p value | Avg | sem | Avg | sem | |
| Iminodiacetic acid | 0.02 | 0.003 | 3.3 | 0.9 | 188.2 | 63.6 |
| Asparagine | 0.03 | 0.000 | 2.4 | 1.0 | 68.5 | 14.7 |
| 10.84 158 200 302 | 0.03 | 0.000 | 0.6 | 0.1 | 21.2 | 2.2 |
| Lysine | 0.03 | 0.003 | 2.3 | 1.0 | 90.7 | 29.9 |
| Ornithine | 0.04 | 0.000 | 50.1 | 35.3 | 1290.1 | 170.1 |
| Rutin | 0.04 | 0.000 | 0.5 | 0.4 | 12.8 | 2.6 |
| Luteolin | 0.05 | 0.004 | 0.6 | 0.1 | 11.4 | 3.9 |
| 17.80 501 559 471 Flavonoid | 0.06 | 0.005 | 1.4 | 0.3 | 21.2 | 7.4 |
| 7.92 218 231 | 0.06 | 0.000 | 0.3 | 0.1 | 4.7 | 0.6 |
| 13.75 219 Coumaroyl glycoside | 0.07 | 0.001 | 1.3 | 0.1 | 17.6 | 4.3 |
| 9.49 302 316 288 242 208 | 0.07 | 0.000 | 4.5 | 1.7 | 68.5 | 11.6 |
| α-Keto-glutaric acid | 0.08 | 0.000 | 9.7 | 2.3 | 114.7 | 14.5 |
| 1,6-Anhydroglucose | 0.08 | 0.000 | 16.7 | 3.0 | 209.7 | 37.7 |
| Oxalomalic acid | 0.08 | 0.000 | 7.5 | 1.3 | 96.6 | 17.6 |
| Glutamine | 0.08 | 0.001 | 6.8 | 1.8 | 90.3 | 24.4 |
| 14.11 521 171 289 Glycoside | 0.09 | 0.013 | 1.9 | 0.7 | 20.0 | 8.2 |
| 3-Amino-2-piperidone | 0.09 | 0.007 | 3.7 | 3.1 | 39.8 | 12.9 |
| 10.58 315 330 200 172 | 0.09 | 0.000 | 1.3 | 0.9 | 14.3 | 2.0 |
| 10.15 368 353 271 242 184 | 0.09 | 0.000 | 6.4 | 2.4 | 71.1 | 10.9 |
| 8.99 242 257 99 | 0.10 | 0.003 | 2.2 | 0.7 | 22.0 | 6.7 |
Metabolites were analyzed as trimethylsilyl derivatives by gas chromatography–mass spectrometry. Data are the average (avg) and standard error of the mean (sem) of six OXPdDUF231A plants (three plants from each of two independent transgenic lines) and three K-147 control plants. Unknown and partially identified metabolites are designated by retention time, key mass-to-charge (m/z) ratios, and identified moieties
Discussion
In this study, we identified a total of 52 DUF231-containing proteins in Populus (Fig. 1) and characterized one member of this protein family, PdDUF231A. PtDUF231A was clustered with the PMR5 subfamily in the phylogenetic tree (Fig. 1a). The PMR5 subfamily has been poorly characterized in plants with indications that it may function in carbohydrate modification [24, 25]. Overexpression of PdDUF231A resulted in increases in cellulose content, sugar release, and 3-O-acetylated xylan and 2,3-O-acetylated xylan (Figs. 4, 5, 6), suggesting that PdDUF231A plays a role in both xylan acetylation and cellulose biosynthesis. The phenotype of increased 3-O-acetylation on xylan in Populus transgenic plants overexpressing PdDUF231A is opposite to that reported in loss-of-function mutants of DUF231 genes in Arabidopsis [17, 18], suggesting that a common function of xylan acetylation by DUF231 genes may exist in both herbaceous and woody species. The increased xylan acetylation which would limit xylan chain elongation may have driven the large accumulation of the partially identified xylopyranoside aromatic metabolite eluting at 13.27 min with a key m/z 235 (Table 1).
PdDUF231A and cellulose biosynthesis
Cellulose forms the largest portion of secondary cell walls. For biofuel conversion and production using plant biomass, the availability and utilization of cellulose is critical. Increases in cellulose and glucose contents were observed in two independent Populus transgenic lines overexpressing PdDUF231A (Fig. 4). In Arabidopsis, reduction in cellulose content has been observed in loss-of-function mutants of DUF231 genes, such as esk1 and tbr [13, 16], suggesting that involvement in cellulose biosynthesis may be another common feature of DUF231 genes in herbaceous and woody species. Given that the other major cell wall monosaccharides were not negatively impacted, the bulk of increased carbon partitioning to glucose and cellulose in plants overexpressing PdDUF231A likely occurred at the expense of soluble flavonoids and hydroxycinnamate conjugates as indicated in the metabolite profiles (Tables 1, 2).
Co-expression analysis based on microarray results showed that TBR (At5G06700) and TBL3 (At5G01360) are co-expressed with cellulose biosynthesis genes, although whether the expression of cellulose synthase genes is affected by the modification of DUF231 gene expression has not been tested. Here, we showed that overexpression of PdDUF231A resulted in increase in the expression of SUSY and CesA8 and increase of cellulose and glucose contents (Fig. 4), reinforcing the view of close relationship between DUF231 proteins and cellulose biosynthesis. To date, CesA4, CesA7, and CesA8 have been reported to be involved in the assembly of the CesA complex responsible for secondary cell wall formation [36, 37]. SUSY also participates in cellulose biosynthesis by producing UDP-glucose to elongate cellulose fibril. Populus transgenic plants heterologously expressing the cotton (Gossypium hirsutum L.) SUSY gene had elevated cellulose content [38]. Reduction of hybrid aspen (Populus tremula L. × tremuloides Michx.) SUSY resulted in a decrease in wood density together with reduced contents of lignin, hemicellulose, and cellulose [39]. In addition, the transgenic tobacco expressing P. simonii × P. nigra SUSY2, a protein highly similar to Populus trichocarpa SUSY2, showed increased cellulose content and fiber length [40]. SUSY gene expression was most drastically elevated in the xylem of OXPdDUF231A transgenic plants, supporting the view that PdDUF231A is involved in cellulose biosynthesis (Fig. 4). Given that PdDUF231A does not appear to be a transcription factor (i.e., without a DNA binding motif), its influence on SUSY and CesA8 expression is likely an indirect effect.
PdDUF231A and xylan biosynthesis
It should be noted that the expression of GT43B, a putative marker gene for xylan biosynthesis, was slightly upregulated in the leaf, but was not altered in the xylem of OXPdDUF231A transgenic plants (Fig. 4). Carbohydrate composition analysis did not indicate alteration in xylose content in the stem samples (Fig. 4). Characterization of xylan-deficient mutants irx9, irx10, and irx10-like suggested that GT43 and GT47 are involved in xylan elongation, and their xylan synthase activity has been demonstrated experimentally [32, 41, 42]. Although we cannot rule out a possible role of PdDUF231A in xylan biosynthesis, given the slight increase of GT43B expression in the leaf of OXPdDUF231A transgenic plants, such a role may not be major since the expression of GT43B in the xylem was not altered in PdDUF231A overexpression lines (Fig. 4).
PdDUF231A and xylan acetylation
Although PdDUF231A may have a minor role in xylan biosynthesis, it potentially has an important role in the modification of xylan. The 2D-HSQC NMR analysis showed an increase of O-acetylated xylan in OXPdDUF231A transgenic lines (Fig. 6), suggesting that PdDUF231A is involved in the acetylation of xylan. More specifically, acetylations at 3-O- and 2,3,-di-O-xylosyl residues on xylan were increased in both OXPdDUF231A transgenic lines (Fig. 6), indicating that PdDUF231A may specifically regulate these two types of acetylation. Acetylation at 3-O-xylosyl residue by PdDUF231A is consistent with the studies on DUF231 proteins in Arabidopsis [18–20]. It should be noted that the acetylation at 2,3-di-O-xylan was also increased in the OXPdDUF231A transgenic lines, but we could not specify whether this increase was induced by another acetylation of either mono-acetylated xylan or by simultaneous acetylation at 2- and 3-xylosyl residues on xylan. As a xylan-specific acetyltransferase among AtDUF231 family protein, AtESK1 mutant has a drastic reduction of 2-O-acetylated xylan [16]. However, mono 2-O-acetylated xylan was not drastically altered in OXPdDUF231A (Fig. 6), implying the acetylation at 3-O-xylosyl residue on xylan was not compensated by reduction of 2-O-AcXylp in P. deltoides. Additionally, because AtESK1 has recently been shown to be necessary for generating the even pattern of acetyl esters on xylan which is required for normal interaction with cellulose fibrils [17] and OXPdDUF231A transgenic lines showed increased glucose release (Fig. 5), it remains unknown whether excess xylan acetylations (i.e., via PdDUF231A overexpression in the present study) may have made cellulose fibrils more accessible for digestion by enzymes.
Although in the present study, we present evidence supporting the association of PdDUF231A with cellulose biosynthesis and xylan acetylation, the biochemical activity of PdUDF231A remains to be determined. We cannot rule out the possibility that PdDUF231A may also have a role in the modification of other cell wall polysaccharides. The specific mechanism underlying increased acetylation of xylan and increased cellulose content in PdDUF231A overexpression lines remains unknown. However, because reduced xylan acetylation and reduced cellulose content were observed in loss-of-function mutants of AtDUF231 in Arabidopsis, the association of xylan acetylation and cellulose biosynthesis may represent a general feature of action of DUF231 proteins. A precise mechanism of such correlations is worth further investigation and may have profound impact on the conversion of plant biomass for biofuel production. In addition, reduced lignin content was observed in the Populus transgenic lines overexpressing PdDUF231A. It is unknown whether this is an indirect effect due to increased cellulose biosynthesis. Finally, increased sugar release was observed in PdDUF231A overexpression lines. How increased cellulose content, reduced lignin content, and increased xylan acetylation were playing out in the process of enzymatic saccharification is an interesting topic that is worth further investigation.
Conclusions
PdDUF231A enhances both cellulose biosynthesis and xylan acetylation, coupled with large-scale shifts in carbon partitioning away from flavonoids and many hydroxycinnamate conjugates. One important feature of PdDUF231A overexpression lines is that both the saccharification efficiency and biomass production were increased. This makes PdDUF231A an attractive target for genetic modification through overexpression for biofuel conversion and production.
Methods
Protein amino acid sequence analysis and phylogenetic analysis
To identify DUF231-containing proteins encoded by the Populus genome, we used the amino acid sequence of the DUF231 domain (from amino acid 429 to amino acid 590) of the AtTBR (AT5G06700) protein as a query to search the Populus trichocarpa v3.0 genome annotation database through a BLAST search by TBLASTN (v. 2.2.26) using the BLOSUM62 database integrated in Phytozome v11.0 (https://phytozome.jgi.doe.gov). In a second search, we used the full-length amino acid sequence of Potri.001G010900, the PtDUF231 family protein showing the highest amino acid sequence identity (61.7%) with AtTBR, as a query. The proteins with short amino acid length (< 300 A.A.) or low amino acid sequence identity (< 30%) with the DUF231 domain of AtTBR were filtered out of the protein alignment and phylogenetic analyses.
The Arabidopsis DUF231-containing proteins were adopted from the published study [13]. Complied full-length PtDUF231 and AtDUF231 proteins were aligned using MUSCLE [43] integrated in Geneious software (v8.1.2; Biomatters Ltd., New Zealand). For phylogenetic analysis, amino acid alignments were subjected to the PhyML 3.0 [44]. The phylogenetic tree was constructed by LG matrix for protein substitution modeling with bootstrap resampling using 1000 replicates. To predict the TM domain, the full-length amino acid sequences of PtDUF231 proteins were subjected to the TMHMM Web-based software (v2.0) (www.cbs.dtu.dk/servies/TMHMM) [45]. Significant TM predictions were determined by selecting the probability score over 0.8. To assess the probability of signal peptides, the same amino acid sequences were subjected to SignalP v4.1 server (http://www.cbs.dtu.dk/services/SignalP) under a valuable signal sequence selection score over 0.5 [46].
Plant materials and biomass measurement
The full-length open reading frame of PdDUF231A was amplified from P. deltoides clone ‘WV94’, cloned into the binary vector and used in Agrobacterium-mediated transformation at ArborGen LLC, Ridgeville (SC), as described previously [47, 48]. A total of ten independent transgenic lines were generated. Transgenic plants including the empty vector transformed control plants and wild type (WV94) were grown in the greenhouse at Oak Ridge National Laboratory at constant 25 °C and 16-h day length.
To estimate stem volume, we measured stem diameter at a position that was 1 cm above the base of the primary stem and measured the total height from the base of the primary stem to the apical top. By using these parameters, we estimated the stem volume using the v = πr 2 h equation (v: volume, r: diameter, h: height).
RT-PCR and qRT-PCR analyses
For the expression analysis of PdDUF231A expression in different tissues/organs, total RNA was prepared from root, young leaf, mature leaf, young stem (internodes 1–3), mature stem (internodes 6–8), petiole of mature leaf, phloem (bark of mature stem), and xylem (scrapped stem under bark of mature stem) [49]. Total RNA extraction and qRT-PCR were performed by the same method as described previously [48].
For RT-PCR analysis for transgenic line selection, the PCR was performed with dreamTaq enzyme solution with 1 µL of two times diluted cDNA (Thermo Fisher Scientific). PCR were performed as follows: denaturation at 95 °C for 2 min followed by 30 cycles of 95 °C for 30 s, 56 °C for 30 s and 72 °C for 20 s. The final extension reaction was performed at 72 °C for 7 min. As an internal control, we used PdUBCc gene in the same manner as above, but replaced the 28 cycles with an annealing temperature of 57 °C in the PCR. The gene-specific primers used and their sequences are listed in Additional file 5.
Gene copy number quantification in transgenic plants
To determine the copy number of PdDUF231A transgene in the transgenic lines compared to WV94, genomic DNA of PdDUF231A gene was quantified by quantitative PCR [50]. Genomic DNA was extracted from mature leaf using a DNeasy Plant Mini kit (Qiagen, Heiden, Germany). One hundred ng of genomic DNA was amplified with PdDUF231A-specific primes as described in “RT-PCR and qRT-PCR analyses”. PdUBCc was used for internal control. The relative transgene quantification was determined by the 2−ΔΔCt equation [51].
Cell wall chemical composition analysis
Two-step sulfuric acid (H2SO4) hydrolysis with the extractives-free biomass to analyze carbohydrate contents in the air-dried stem was performed as described previously [48]. The extractive-free stem was prepared by ethanol/toluene (1:2, v/v) extraction followed by hydrolyzing with 72% H2SO4 at 30 °C for 1 h. The mixture was diluted to 4% concentration of H2SO4, and then more hydroxylation performed at 121 °C using an autoclave for 1 h. The hydrolysate and residual solids after two-step acid hydrolysis were separated by filtration. The filtered liquid fraction was used for sugar composition analysis using a Dionex ICS-3000 ion chromatography system.
To measure lignin content, we collected separately acid-soluble and -insoluble fraction from hydrolysate and solid residue. Acid-soluble lignin content was measured at 240 nm with UV/Vis spectroscopy. The lignin content in the acid-insoluble fraction was determined using solid pellet after filtration by the NREL protocol [27].
Anthrone assay
To determine glucose content using colorimetric measurement with anthrone dye, we used a total of 15 mg of milled dried stems of 6-month-old Populus plants. Sample preparation and anthrone binding assay have been described previously [48]. A total of 15 mg of milled dried stem of Populus transgenic plants and WV94 control plants (6-month-old grown in greenhouse) were dissolved in 500 µL of acetic nitric acid reagent [1:8:2 (v/v) of nitric acid:acetic acid:water] (Sigma-Aldrich, St. Louis, MO) followed by heating at 98 °C for 30 min. The undissolved pellet was collected by centrifugation for 10 min at 14,000 rpm. The pellet was dissolved in 600 μL of 67% sulfuric acid for 1 h at room temperature. The dissolved solvent phase was separated from the pellet by centrifugation for 5 min at 14,000 rpm. Twenty μL of solution was diluted to ten times with deionized water. The diluted solution was diluted again to five times and then mixed with freshly prepared anthrone solution (0.5 mg of anthrone/mL of concentrated sulfuric acid) (Sigma-Aldrich, St. Louis, MO). The anthrone and sample mixture was boiled at 96 °C for 10 min and cooled down at 4 °C. The glucose content was determined by measuring the absorbance at 630 nm wavelength compared to glucose standard solution. Based on the measurement of glucose content, the cellulose content (%) was converted by applying the equation of [(glucose quantity × 600 (dilution factor)]/[15(initial sample amount) × 1000)] × 100.
Two-dimensional heteronuclear single quantum coherence nuclear magnetic resonance (2D-HSQC NMR) analysis
Two biological replicates of each line were used for 2D-HSQC NMR analysis. Populus stems were ground with a Wiley mill and extracted with ethanol:toluene (1:2, v:v) for 24 h. The extractives-free samples were air dried at ambient temperature and ground using a planetary ball mill (Retsch PM 100) spinning at 580 rpm with zirconium dioxide (ZrO2) vessels (50 mL) containing ZrO2 ball bearings (10 mm × 10) for 2 h and 30 min (5 min grinding and 5 min break) for whole cell wall NMR analysis [33]. The ball-milled, whole cell wall sample (100–130 mg) was loaded in a 5 mm NMR tube with DMSO-d 6/HMPA-d 18 (4:1, v:v, ~ 0.5 mL). NMR spectra were acquired at 298 K using a Bruker Advance III 400-MHz spectroscopy equipped with a 5-mm Broadband Observe probe (5-mm BBO 400 MHz W1 with Z-gradient probe, Bruker). Two-dimensional (2D) 1H-13C heteronuclear single quantum coherence (HSQC) experiment was performed using a Bruker standard pulse sequence (‘hsqcetgpsi2’) with the following parameters: spectral width of 11 ppm in F2 (1H) with 2048 data points and 190 ppm in F1 (13C) with 256 data points; 128 scans (NS) and 1 s interscan delay (D1). Volume integration of contours in HSQC spectra was carried out using Bruker’s TopSpin 2.1 software. Assignments of peaks from NMR spectra were based on previous publications [52, 53]. For comparing the relative content of acetyl group in xylan, non-acetylated (1 → 4)-β-d-Xylp and acetylated ones including 2-O-acetylated (2-O-Ac-β-d-Xylp), 3-O-acetylated (3-O-Ac-β-d-Xylp), and 2,3,-di-O-acetylated (2,3-di-O-Ac-β-d-Xylp) xylosyl residues in 2D 1H-13C HSQC NMR spectra were integrated.
Enzymatic saccharification assay
Air-dried Populus stem of 6-month-old after peeling was Wiley-milled with 40 mesh. The methods for enzyme treatment and sugar detection have been described previously [48]. The enzymatic saccharification assay was performed without any pretreatment process (i.e., without strong acid solution treatment). For each sample, 250 mg of dried sample was dissolved in 50 mM citrate buffer (pH 4.8) complemented with Novozymes CTec2 (70 mg of enzyme/gram of biomass) and then incubated at 50°C with 200 rpm shaking. The time course samples were collected at 0, 6, 12, 24, 48, and 72 h after incubation. The enzyme was deactivated by boiling water before carbohydrate measurement. Ion chromatography was performed to measure the released sugar with Dionex ICS-3000 ion chromatography system. The measurement value displayed the average value of two biological replicates.
Metabolite profiling by gas chromatography–mass spectrometry
Leaves (LPI 5) of ~ 9-month-old transgenic OXPdDUF231A (DUF231A) (n = 6; 3 plants from each of two independent transgenic lines) and empty vector control (K-147) P. deltoides ‘WV94’ plants (n = 3) growing in the greenhouse were fast frozen in liquid nitrogen and stored at − 80 °C. The leaf tissues were ground with liquid nitrogen in a chilled mortar and pestle with ~ 50 mg FW of leaf tissue, and were subsequently twice extracted with 2.5 mL 80% ethanol overnight and then combined prior to drying a 1.0 mL aliquot in a nitrogen stream. Sorbitol was added before extraction as an internal standard to correct for differences in extraction efficiency, subsequent differences in derivatization efficiency, and changes in sample volume during heating. Dried extracts were dissolved in 500 μL of silylation–grade acetonitrile, followed by the addition of 500 μL N-methyl-N-trimethylsilyltrifluoroacetamide (MSTFA) with 1% trimethylchlorosilane (TMCS) (Thermo Scientific, Bellefonte, PA), and samples then heated for 1-h at 70 °C to generate trimethylsilyl (TMS) derivative [54, 55]. After 2 days, 1-μL aliquots were injected into an Agilent Technologies Inc. (Santa Clara, CA) 5975C inert XL gas chromatograph-mass spectrometer (GC–MS), fitted with an Rtx-5MS with Integra-guard (5% diphenyl/95% dimethyl polysiloxane) 30 m × 250 µm × 0.25 µm film thickness capillary column. The standard quadrupole GC–MS was operated in the electron impact (70 eV) ionization mode, targeting 2.5 full-spectrum (50–650 Da) scans per second, as described previously [55]. Metabolite peaks were extracted using a key selected ion, characteristic m/z fragment, rather than the total ion chromatogram, to minimize integrating co-eluting metabolites. The extracted peaks of known metabolites were scaled back up to the total ion current using predetermined scaling factors. Peaks were quantified by area integration and concentrations normalized to the quantity of the internal standard (sorbitol) recovered, amount of sample extracted, derivatized, and injected. A large user-created database (> 2400 spectra) of mass spectral electron impact ionization (EI) fragmentation patterns of TMS-derivatized compounds, as well as the Wiley Registry 10th Edition combined with NIST 2014 mass spectral database, were used to identify the metabolites of interest to be quantified. Unidentified metabolites were denoted by their retention time as well as key mass-to-charge (m/z) ratios and partial naming given the typical identity of specific m/z.
Statistical analysis
Statistical analysis to determine statistical significance was performed by Student’s t tests of paired samples (against WV94). We used the t test function integrated in Excel software with p < 0.01 (Microsoft, Redmond, WA). The asterisk in each figure indicates significant difference compared to WV94 or control samples (p < 0.01 or < 0.05).
Additional files
Additional file 1. Protein domain structure prediction of PtDUF231 family proteins.
Additional file 2. PdDUF231A expression in OXPdDUF231A transgenic Populus. RT-PCR was performed using cDNA generated from total RNA isolated from petiole of mature leaves. PdUBCc was used as an internal control. Red asterisks indicate the two transgenic lines selected for subsequent analyses.
Additional file 3. The gene copy number of PdDUF231A in OXPdDUF231A transgenic plants.
Additional file 4. Fold change and relative metabolite concentrations (µg/g FW sorbitol equivalents) of OXPdDUF231A transgenic plants versus empty vector control (K-147) Populus deltoides ‘WV94’ leaves (LPI 5) of 6-month-old greenhouse-grown plants. Metabolites were analyzed as trimethylsilyl derivatives by gas chromatography-mass spectrometry. Data are the average (Avg) and standard error of the mean (sem) of six OXPdDUF231A plants (three plants from each of two independent transgenic lines) and three K-147 control plants. Unknown and partially identified metabolites are designated by retention time, key mass-to-charge (m/z) ratios, and identified moieties.
Additional file 5. Primers used for RT-PCR and quantitative RT-PCR analyses in this study.
Authors’ contributions
YY performed bioinformatics, gene copy determination, RT-PCR and qRT-PCR analyses, and measured cellulose content. CGY, YP, and AJR performed chemical composition analysis and measured sugar release. KAW, CMC, and MAW generated Populus transgenic lines. LEG and SSJ measured biomass production. NLE and TJT analyzed metabolite profiles. XY designed the gene construct of PdDUF231A for Populus transformation. GAT and JGC conceived the study, coordinated research, and contributed to experimental design and data interpretation. All authors read and approved the final manuscript.
Acknowledgements
Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the U.S. Department of Energy under Contract Number DE-AC05-00OR22725. The US Government retains, and the publisher, by accepting the article for publication acknowledges that the US Government retains, a non-exclusive, paid-up, irrevocable, worldwide license to publish or reproduce the published form of this manuscript, or allow others to do so, for US Government purposes. The Department of Energy will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (http://energy.gov/downloads/doe-public-access-plan).
Competing interests
The strategy to produce improved biomass feedstock described in this paper has been included in a patent application.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its additional files.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This research was supported by the U.S. Department of Energy BioEnergy Science Center project. The BioEnergy Science Center is a U.S. Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research in the U.S. Department of Energy Office of Science. The funding body has no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- DUF231
Domain of Unknown Function 231
- GPC
gel permeation chromatography
- NMR
nuclear magnetic resonance
- TM
transmembrane domain
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13068-017-0998-3) contains supplementary material, which is available to authorized users.
Contributor Information
Yongil Yang, Email: yyang98@utk.edu.
Chang Geun Yoo, Email: yooc@ornl.gov.
Kimberly A. Winkeler, Email: kwinkeler1@gmail.com
Cassandra M. Collins, Email: cmcolli@arborgen.com
Maud A. W. Hinchee, Email: mhinchee@agricensci.com
Sara S. Jawdy, Email: jawdys@ornl.gov
Lee E. Gunter, Email: gunterle@ornl.gov
Nancy L. Engle, Email: englenl@ornl.gov
Yunqiao Pu, Email: puy1@ornl.gov.
Xiaohan Yang, Email: yangx@ornl.gov.
Timothy J. Tschaplinski, Email: tschaplinstj@ornl.gov
Arthur J. Ragauskas, Email: ragauskasaj@ornl.gov
Gerald A. Tuskan, Email: tuskanga@ornl.gov
Jin-Gui Chen, Phone: (865) 574-9094, Email: chenj@ornl.gov.
References
- 1.Gille S, Pauly M. O-acetylation of plant cell wall polysaccharides. Front Plant Sci. 2012;3:12. doi: 10.3389/fpls.2012.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klein-Marcuschamer D, Oleskowicz-Popiel P, Simmons BA, Blanch HW. The challenge of enzyme cost in the production of lignocellulosic biofuels. Biotechnol Bioeng. 2012;109:1083–1087. doi: 10.1002/bit.24370. [DOI] [PubMed] [Google Scholar]
- 3.Pawar PM, Koutaniemi S, Tenkanen M, Mellerowicz EJ. Acetylation of woody lignocellulose: significance and regulation. Front Plant Sci. 2013;4:118. doi: 10.3389/fpls.2013.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gille S, de Souza A, Xiong G, Benz M, Cheng K, Schultink A, Reca IB, Pauly M. O-acetylation of Arabidopsis hemicellulose xyloglucan requires AXY4 or AXY4L, proteins with a TBL and DUF231 domain. Plant Cell. 2011;23:4041–4053. doi: 10.1105/tpc.111.091728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scheller HV, Ulvskov P. Hemicelluloses. Annu Rev Plant Biol. 2010;61(263–28):9. doi: 10.1146/annurev-arplant-042809-112315. [DOI] [PubMed] [Google Scholar]
- 6.Gibeaut DM, Pauly M, Bacic A, Fincher GB. Changes in cell wall polysaccharides in developing barley (Hordeum vulgare) coleoptiles. Planta. 2005;221:729–738. doi: 10.1007/s00425-005-1481-0. [DOI] [PubMed] [Google Scholar]
- 7.Hayashi T, Kaida R. Functions of xyloglucan in plant cells. Mol Plant. 2011;4:17–24. doi: 10.1093/mp/ssq063. [DOI] [PubMed] [Google Scholar]
- 8.Hoffman M, Jia Z, Pena MJ, Cash M, Harper A, Blackburn AR, Darvill A, York WS. Structural analysis of xyloglucans in the primary cell walls of plants in the subclass Asteridae. Carbohydr Res. 2005;340:1826–1840. doi: 10.1016/j.carres.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 9.Jia Z, Cash M, Darvill AG, York WS. NMR characterization of endogenously O-acetylated oligosaccharides isolated from tomato (Lycopersicon esculentum) xyloglucan. Carbohydr Res. 2005;340:1818–1825. doi: 10.1016/j.carres.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 10.Pawar PM, Patke C, Balasubramanian VK, Chong S, Grandla ML, Adriasola M, Sparrman T, Hedenstrom M, Szwaj K, Debra-Marceluch M, Gaertner C, Mouille G, Ezcurra I, Tenkanen M, Johnson LJ, Mellerowicz EJ. Down-regulation of RWA genes in hybrid aspen affects xylan acetylation and wood saccharification. New Phytol. 2017;214:1491. doi: 10.1111/nph.14489. [DOI] [PubMed] [Google Scholar]
- 11.Manabe Y, Nafisi M, Verhertbruggen Y, Orfila C, Gille S, Rautengarten C, Cherk C, Marcus SE, Somerville S, Pauly M, Knox JP, Sakuragi Y, Scheller HV. Loss-of-function mutation of REDUCED WALL ACETYLATION2 in Arabidopsis leads to reduced cell wall acetylation and increased resistance to Botrytis cinerea. Plant Physiol. 2011;155:1068–1078. doi: 10.1104/pp.110.168989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manabe Y, Verhertbruggen Y, Gille S, Harholt J, Chong SL, Pawar PM, Mellerowicz EJ, Tenkanen M, Cheng K, Pauly M, Scheller HV. Reduced wall acetylation proteins play vital and distinct roles in cell wall O-acetylation in Arabidopsis. Plant Physiol. 2013;163:1107–1117. doi: 10.1104/pp.113.225193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bischoff V, Nita S, Neumetzler L, Schindelasch D, Urbain A, Eshed R, Persson S, Delmer D, Scheible WR. TRICHOME BIREFRINGENCE and its homolog AT5G01360 encode plant-specific DUF231 proteins required for cellulose biosynthesis in Arabidopsis. Plant Physiol. 2010;153:590–602. doi: 10.1104/pp.110.153320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bischoff V, Selbig J, Scheible WR. Involvement of TBL/DUF231 proteins into cell wall biology. Plant Signal Behav. 2010;5:1057–1059. doi: 10.4161/psb.5.8.12414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiong G, Cheng K, Pauly M. Xylan O-acetylation impacts xylem development and enzymatic recalcitrance as indicated by the Arabidopsis mutant tbl29. Mol Plant. 2013;6:1373–1375. doi: 10.1093/mp/sst014. [DOI] [PubMed] [Google Scholar]
- 16.Yuan Y, Teng Q, Zhong R, Ye ZH. The Arabidopsis DUF231 domain-containing protein ESK1 mediates 2-O- and 3-O-acetylation of xylosyl residues in xylan. Plant Cell Physiol. 2013;54:1186–1199. doi: 10.1093/pcp/pct070. [DOI] [PubMed] [Google Scholar]
- 17.Grantham NJ, Wurman-Rodrich J, Terrett OM, Lyczakowski JJ, Stott K, Iuga D, Simmons TJ, Durand-Tardif M, Brown SP, Dupree R, Busse-Wicher M, Dupree P. An even pattern of xylan substitution is critical for interaction with cellulose in plant cell walls. Nat Plants. 2017 doi: 10.1038/s41477-017-0030-8. [DOI] [PubMed] [Google Scholar]
- 18.Yuan Y, Teng Q, Zhong R, Ye ZH. TBL3 and TBL31, two Arabidopsis DUF231 domain proteins, are required for 3-O-monoacetylation of xylan. Plant Cell Physiol. 2016;57:35–45. doi: 10.1093/pcp/pcv172. [DOI] [PubMed] [Google Scholar]
- 19.Yuan Y, Teng Q, Zhong R, Ye ZH. Roles of Arabidopsis TBL34 and TBL35 in xylan acetylation and plant growth. Plant Sci. 2016;243:120–130. doi: 10.1016/j.plantsci.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 20.Yuan Y, Teng Q, Zhong R, Haghighat M, Richardson EA, Ye ZH. Mutations of Arabidopsis TBL32 and TBL33 affect xylan acetylation and secondary wall deposition. PLoS ONE. 2016;11:e0146460. doi: 10.1371/journal.pone.0146460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhong R, Cui D, Ye Z. Regiospecific acetylation of xylan is mediated by a group of DUF231-containing O-acetyltransferases. Plant Cell Physiol. 2017 doi: 10.1093/pcp/pcx147. [DOI] [PubMed] [Google Scholar]
- 22.Gao Y, He C, Zhang D, Liu X, Xu Z, Tian Y, Liu X, Zhang S, Pauly M, Zhou Y, Zhang B. Two trichome birefringence-like proteins mediate xylan acetylation, which is essential for leaf blight resistance in rice. Plant Physiol. 2017;173:470–481. doi: 10.1104/pp.16.01618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang B, Zhang L, Li F, Zhang D, Liu X, Wang H, Xu Z, Chu C, Zhou Y. Control of secondary cell wall patterning involves xylan deacetylation by a GDSL esterase. Nat Plants. 2017;3:17017. doi: 10.1038/nplants.2017.17. [DOI] [PubMed] [Google Scholar]
- 24.Vogel JP, Raab TK, Somerville CR, Somerville SC. Mutations in PMR5 result in powdery mildew resistance and altered cell wall composition. Plant J. 2004;40:968–978. doi: 10.1111/j.1365-313X.2004.02264.x. [DOI] [PubMed] [Google Scholar]
- 25.Anantharaman V, Aravind L. Novel eukaryotic enzymes modifying cell-surface biopolymers. Biol Direct. 2010;5:1. doi: 10.1186/1745-6150-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown D, Wightman R, Zhang Z, Gomez LD, Atanassov I, Bukowski JP, Tryfona T, McQueen-Mason SJ, Dupree P, Turner S. Arabidopsis genes IRREGULAR XYLEM (IRX15) and IRX15L encode DUF579-containing proteins that are essential for normal xylan deposition in the secondary cell wall. Plant J. 2011;66:401–413. doi: 10.1111/j.1365-313X.2011.04501.x. [DOI] [PubMed] [Google Scholar]
- 27.Sluiter A, Hames B, Ruiz R, Scarlata C, Sluiter J, Templeton D, Crocker D. Determination of structural carbohydrates and lignin in biomass. In: Laboratory analytical procedure (LAP). National Renewable Energy Laboratory; 2012. p. 3–13. https://www.nrel.gov/docs/gen/fy13/42618.pdf.
- 28.Updegraff DM. Semimicro determination of cellulose in biological materials. Anal Biochem. 1969;32:420–424. doi: 10.1016/S0003-2697(69)80009-6. [DOI] [PubMed] [Google Scholar]
- 29.Kumar M, Thammannagowda S, Bulone V, Chiang V, Han K, Joshi CP, Mansfield SD, Mellerowicz E, Sundberg B, Teeri T, Ellis BE. An update on the nomenclature for the cellulose synthase genes in Populus. Trends Plant Sci. 2009;14:248–254. doi: 10.1016/j.tplants.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 30.Zhang D, Xu B, Yang X, Zhang Z, Li B. The sucrose synthase gene family in Populus: structure, expression, and evolution. Tree Genet Genomes. 2011;7:443–456. doi: 10.1007/s11295-010-0346-2. [DOI] [Google Scholar]
- 31.Kalluri UC, Payyavula RS, Labbé JL, Engle N, Bali G, Jawdy SS, Sykes RW, Davis M, Ragauskas A, Tuskan GA, Tschaplinski TJ. Down-regulation of KORRIGAN-Like endo-β-1,4-glucanase genes impacts carbon partitioning, Mycorrhizal colonization and biomass production in Populus. Front Plant Sci. 2016;7. article 1455. [DOI] [PMC free article] [PubMed]
- 32.Lee C, Teng Q, Zhong R, Ye ZH. Molecular dissection of xylan biosynthesis during wood formation in poplar. Mol Plant. 2011;4:730–747. doi: 10.1093/mp/ssr035. [DOI] [PubMed] [Google Scholar]
- 33.Yoo CG, Pu Y, Li M, Ragauskas AJ. Elucidating structural characteristics of biomass using solution-state 2 D NMR with a mixture of deuterated dimethylsulfoxide and hexamethylphosphoramide. Chemsuschem. 2016;9:1090–1095. doi: 10.1002/cssc.201600135. [DOI] [PubMed] [Google Scholar]
- 34.Kim H, Ralph J. A gel-state 2D-NMR method for plant cell wall profiling and analysis: a model study with the amorphous cellulose and xylan from ball-milled cotton linters. RSC Adv. 2014;4:7549–7560. doi: 10.1039/C3RA46338A. [DOI] [Google Scholar]
- 35.Zhong R, Teng Q, Lee C, Ye ZH. Identification of a disaccharide side chain 2-O-α-d-galactopyranosyl-α-d-glucuronic acid in Arabidopsis xylan. Plant Signal Behav. 2014;9:e27933. doi: 10.4161/psb.27933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Endler A, Persson S. Cellulose synthases and synthesis in Arabidopsis. Mol Plant. 2011;4:199–211. doi: 10.1093/mp/ssq079. [DOI] [PubMed] [Google Scholar]
- 37.Taylor NG. Cellulose biosynthesis and deposition in higher plants. New Phytol. 2008;178:239–252. doi: 10.1111/j.1469-8137.2008.02385.x. [DOI] [PubMed] [Google Scholar]
- 38.Coleman HD, Yan J, Mansfiedl SD. Sucrose synthase affects carbon partitioning to increase cellulose production and altered cell wall ultrastructure. Proc Natl Acad Sci USA. 2009;106:13118–13123. doi: 10.1073/pnas.0900188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gerber L, Zhang B, Roach M, Rende U, Gorzsás A, Kumar M, Burgert I, Niittylä T, Sundberg B. Deficient sucrose synthase activity in developing wood does not specifically affect cellulose biosynthesis, but causes an overall decrease in cell wall polymers. New Phytol. 2014;203:1220–1230. doi: 10.1111/nph.12888. [DOI] [PubMed] [Google Scholar]
- 40.Wei Z, Qu Z, Zhang L, Zhao S, Bi Z, Ji X, Wang X, Wei H. Overexpression of poplar xylem sucrose synthase in tobacco leads to a thickened cell wall and increased height. PLoS ONE. 2015;10:e0120669. doi: 10.1371/journal.pone.0120669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jensen JK, Johnson NR, Wilkerson CG. Arabidopsis thaliana IRX10 and two related proteins from psyllium and Physcomitrella patens are xylan xylosyltransferases. Plant J. 2014;80:207–215. doi: 10.1111/tpj.12641. [DOI] [PubMed] [Google Scholar]
- 42.Urbanowicz BR, Peña MJ, Moniz HA, Moremen KW, York WS. Two Arabidopsis proteins synthesize acetylated xylan in vitro. Plant J. 2014;80:197–206. doi: 10.1111/tpj.12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 45.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 46.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 47.Bryan AC, Jawdy S, Gunter L, Gjersing E, Sykes R, Hinchee MA, Winkeler KA, Collins CM, Engle N, Tschaplinski TJ, Yang X, Tuskan GA, Muchero W, Chen JG. Knockdown of a laccase in Populus deltoides confers altered cell wall chemistry and increased sugar release. Plant Biotechnol J. 2016;14:2010–2020. doi: 10.1111/pbi.12560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Y, Yoo CG, Guo H, Rottmann W, Winkeler KA, Collins CM, Gunter LE, Jawdy SS, Yang X, Guo H, Pu Y, Ragauskas AJ, Tuskan GA, Chen J. Overexpression of a Domain of Unknown Function 266-containing protein results in high cellulose content, reduced recalcitrance, and enhanced plant growth in the bioenergy crop Populus. Biotechnol Biofuels. 2017;10:74. doi: 10.1186/s13068-017-0760-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Payyavula RS, Tschaplinski TJ, Jawdy S, Skyes RW, Tuskan GA, Kalluri UC. Metabolic profiling reveals altered sugar and secondary metabolism in response to UGPase overexpression in Populus. BMC Plant Biol. 2014;14:265–278. doi: 10.1186/s12870-014-0265-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma L, Chung WK. Quantitative analysis of copy number variants based on real-time lightcycler PCR. Curr Protoc Hum Genet. 2014;80:7.21.1–7.21.8. doi: 10.1002/0471142905.hg0721s80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−(ΔΔCt) method. Methods. 2011;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 52.Busse-Wicher M, Gomes TC, Tryfona T, Nikolovski N, Stott K, Grantham NJ, Bolam DN, Skaf MS, Dupree P. The pattern of xylan acetylation suggests xylan may interact with cellulose microfibrils as a twofold helical screw in the secondary plant cell wall of Arabidopsis thaliana. Plant J. 2014;79:492–506. doi: 10.1111/tpj.12575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim H, Ralph J. Solution-state 2D NMR of ball-milled plant cell wall gels in DMSO-d6/pyridine-d5. Org Biomol Chem. 2010;8:576–591. doi: 10.1039/B916070A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abraham P, Yin H, Borland AM, Weighill D, Lim SD, De Paoli HC, Engle NL, Agh R, Weston DJ, Wullschleger SD, Tschaplinski T, Jacobson D, Cushman JC, Hettich RL, Tuskan GA, Yang X. Transcript, protein and metabolite temporal dynamics in the CAM plant Agave. Nat Plant. 2016;2. Article number 16178. [DOI] [PubMed]
- 55.Tschaplinski TJ, Standaert RF, Engle NL, Martin MZ, Sangha AK, Parks JM, Smith JC, Samuel R, Jiang N, Pu Y, Ragauskas AJ, Hamilton CY, Fu C, Wang Z-Y, Davison BH, Dixon RA, Mielenz JR. Down-regulation of the caffeic acid O-methyltransferase gene in switchgrass reveals a novel monolignol analog. Biotechnol Biofuels. 2012;5:71. doi: 10.1186/1754-6834-5-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Protein domain structure prediction of PtDUF231 family proteins.
Additional file 2. PdDUF231A expression in OXPdDUF231A transgenic Populus. RT-PCR was performed using cDNA generated from total RNA isolated from petiole of mature leaves. PdUBCc was used as an internal control. Red asterisks indicate the two transgenic lines selected for subsequent analyses.
Additional file 3. The gene copy number of PdDUF231A in OXPdDUF231A transgenic plants.
Additional file 4. Fold change and relative metabolite concentrations (µg/g FW sorbitol equivalents) of OXPdDUF231A transgenic plants versus empty vector control (K-147) Populus deltoides ‘WV94’ leaves (LPI 5) of 6-month-old greenhouse-grown plants. Metabolites were analyzed as trimethylsilyl derivatives by gas chromatography-mass spectrometry. Data are the average (Avg) and standard error of the mean (sem) of six OXPdDUF231A plants (three plants from each of two independent transgenic lines) and three K-147 control plants. Unknown and partially identified metabolites are designated by retention time, key mass-to-charge (m/z) ratios, and identified moieties.
Additional file 5. Primers used for RT-PCR and quantitative RT-PCR analyses in this study.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its additional files.


