Abstract
The ecosystem of the human gastrointestinal (GI) tract traverses a number of environmental, chemical, and physical conditions because it runs from the oral cavity to the anus. These differences in conditions along with food or other ingested substrates affect the composition and density of the microbiota as well as their functional roles by selecting those that are the most suitable for that environment. Previous studies have mostly focused on Bacteria, with the number of studies conducted on Archaea, Eukarya, and Viruses being limited despite their important roles in this ecosystem. Furthermore, due to the challenges associated with collecting samples directly from the inside of humans, many studies are still exploratory, with a primary focus on the composition of microbiomes. Thus, mechanistic studies to investigate functions are conducted using animal models. However, differences in physiology and microbiomes need to be clarified in order to aid in the translation of animal model findings into the context of humans. This review will highlight Bacteria, Archaea, Fungi, and Viruses, discuss differences along the GI tract of healthy humans, and perform comparisons with three common animal models: rats, mice, and pigs.
Keywords: Microbiome, mycobiome, virome, human gastrointestinal (GI) tract, animal models, diet
Researchers have been investigating the ecology of the intestinal microbiota for decades (120, 165) in order to identify, characterize, and count their numbers. These extensive efforts are due to the important roles the intestinal microbiota play in digestion, the production of essential vitamins, and protection of the gastrointestinal (GI) tract from pathogen colonization (141). In the past few decades, molecular techniques targeting the 16S rRNA gene and other genetic markers have been developed to characterize and analyze bacterial communities. These methods have been used to reveal the important roles played by microbes in the GI tract (23, 180, 183, 184, 189, 212). In healthy individuals, the microbiome (microbial community) and host have a mutualistic relationship in which both partners benefit; however, pathogens may invade and cause disease under certain conditions. The initial aim of most studies was to elucidate the role of the microbiome in disease. More recently, surveys have been performed on healthy individuals in order to assess the contribution of the microbiota to health, particularly in response to dietary changes/supplementation with probiotics and/or prebiotics.
The human GI tract is a complex system that starts from the oral cavity, continues through the stomach and intestines, and finally ends at the anus (Fig. 1). The density and composition of the microbiome change along the GI tract, with major populations being selected by the functions performed at the various locations. Bacteria along the GI tract have several possible functions, many of which are beneficial for health including vitamin production, the absorption of ions (Ca, Mg, and Fe), protection against pathogens, histological development, enhancement of the immune system, and the fermentation of “non-digestible foods” to short chain fatty acids (SCFA) and other metabolites (19, 58, 63, 77, 138). The roles of fungi and viruses have not been examined in as much detail; however, they are known to play important roles in microbiota dynamics and host physiology/immunity related to health and disease (45, 94, 133).
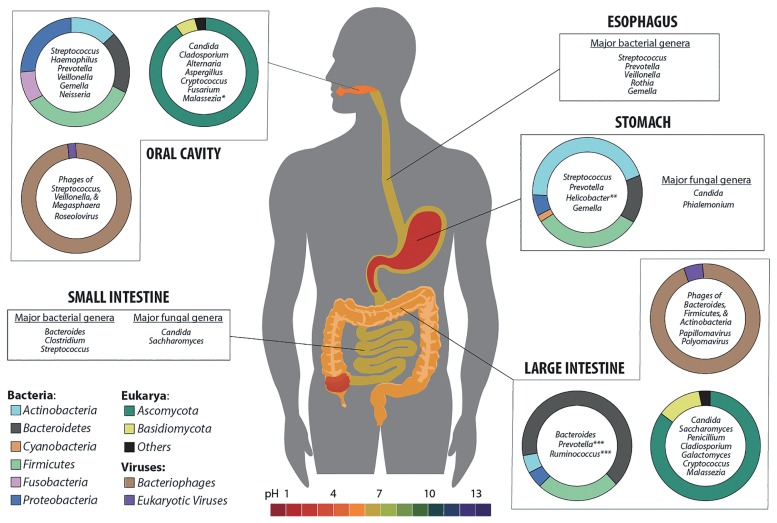
Fig. 1.
Microbiome composition of Bacteria (1, 5, 20, 21, 43, 147, 156, 223), Eukarya (52, 85, 114, 126, 182, 197), and Viruses (45, 134, 151, 215) among the physiological niches of the human gastrointestinal (GI) tract. Phylum level compositional data are presented where available along with the most common genera in each GI tract location. The colors on the doughnut plots correspond to the legend in the lower left corner; the GI tract is colored according to the pH scale shown at the bottom of Fig. 1. (* Malassezia was very abundant in one study and was not detected in another study. ** The abundance of Helicobacter may vary greatly between individuals. *** Proportions of these and other colon genera vary with age, diet, & geographical location.)
Food passes through the GI tract and the absorption rate of nutrients is largely dependent on the activities of various enzymes in the digestive system, such as amylase in saliva, pepsin in the stomach, and pancreatic enzymes in the small intestine. These mechanisms have been extensively examined (61, 62), particularly in the stomach. However, many food components cannot be digested in the upper GI tract and are passed into the lower intestinal tract, in which they are fermented by microbes. Functional studies commonly use animal models in order to obtain a better understanding of the processes in the GI tract that may lead to better health or decrease disease. However, information from animal models may not be directly translatable to humans. Therefore, researchers need to consider the limitations of the selected animal model when extrapolating findings to humans.
Although microbiome studies often include an ecological component, most of the research performed to date has focused on Bacteria and not all of the biota. This represents a logical approach because Bacteria comprise most of the microbiome. However, even biota representing a small proportion of the microbiome may play important roles in the ecosystem (133). Therefore, researchers need to start shifting their approach to include eukaryotic, prokaryotic, and viral (33, 133) interactions in efforts to elucidate the roles of all components of the microbiome.
In recent years, a number of reviews have summarized findings from the increasing number of studies being performed in this field (36, 73, 176, 188). While most studies have focused on disease, the microbiome is also important for maintaining health. We herein highlight differences in the microbiome (Bacteria, Archaea, Fungi, and Viruses) along the GI tract of healthy humans, and how it compares to those of typical animal models used in research. One finding that is consistent to most studies is that the microbiome of healthy individuals is unique; however, there are still some generalities that will be discussed in this review.
Microbiome diversity
Many factors contribute to the diversity of microbiomes, and most studies have demonstrated the individuality of microbiomes among subjects. Previous findings support microbial communities being more similar in subjects that are genetically related (191), of a similar age (135, 213), or with common diets (including the influences of ethnicity and geography) (63). Diseases will also have an impact on microbiome diversity, including autoimmune and neoplastic diseases, such as inflammatory bowel disease, diabetes, obesity, cardiovascular diseases, allergies, and cancer (37, 121). Treatments for diseases may also affect a patient’s gut microbiota, and the consequences of antibiotic use have been intensively investigated (22, 95).
The host genotype has been shown to influence the development of the gut microbiota, and the immune system has been identified as a contributing factor (188). Crosstalk between the microbiome and human immune system occurs in response to a number of environmental factors, such as diet, xenobiotics, and pathogens. Microbial host interactions occur in the gut, mainly in the epithelial cell layer, myeloid cells, and innate lymphoid cells, in which crosstalk and feedback loops contribute to the microbiome composition, host physiology, and disease susceptibility. These interactions contribute not only to the bacterial community along the GI tract, but also to the other microbiota (Fungi, Archaea, and Viruses). Our understanding of the immunology associated with Fungi (150) and Archaea is currently limited. Transkingdom commensal relationships among microbiota (including Viruses) are considered to form from infancy (29, 30, 106, 200) and several co-occurring relationships have been identified (35, 75, 76, 85, 214).
Bacteria
A more complete picture of human-associated bacterial communities obtained using molecular techniques has revealed that their diversity is greater than initially considered through cultivation (9, 20, 56, 90, 113). Using almost full-length 16S rRNA gene sequences, predicted taxa numbers range from 100–300 (20, 56), while pyrosequencing suggests there are 1000s of phylotypes (38, 49). Most of the gut bacteria identified by 16S rRNA gene sequencing belong to the five phyla originally identified by cultivation, namely, Bacteroidetes, Firmicutes, Actinobacteria, Proteobacteria, and Verrucomicrobia (90), and, at lower proportions, Fusobacteria, Tenericutes, Spirochaetes, Cyanobacteria, and TM7 (189). At lower levels of the taxonomic classification, microbiome compositions vary with each individual. Attempts have been made to identify a single core microbiome of Bacteria in the GI tract. Although this has not been possible in the lower GI tract (mainly using fecal samples) based on taxonomy, it appears there are core microbial functions (152, 189, 191). It is possible to identify some core microbiota in the oral cavity, esophagus, and stomach (148). Although extensive efforts have been made to cultivate representative gut microbiota in an attempt to gain a better understanding of the relationship between taxa and function (156), there are still many undescribed taxa with unknown functional roles in the gut.
As the price of sequencing decreases, it is becoming more common to use a metagenomic approach that provides information on all microbiota and potential functions (3, 70, 167, 189). This provides a means to go beyond Bacteria and obtain information on eukaryotic microbes (mainly fungi) and viruses. Although Fungi, Archaea, and Viruses in the microbiome are a part of the ‘rare biosphere’ (organisms that comprise <0.1% of the microbiome) (173), they still have a significant impact on host health.
Fungi
Fungi are considered to comprise approximately 0.03% of the fecal microbiome (143); making them approximately 3,300-fold less abundant than Bacteria. Fungal diversity in the human gut is also lower than that of Bacteria (143, 166), although more taxa are being found as the number of individuals being studied using next generation sequencing is increasing (44, 126, 166, 182). In 2015, a review of 36 fungal gut microbiome studies revealed that there have been at least 267 distinct fungi identified in the human gut (181), while another study reported 221 (72). Despite the number of taxa that have been reported, most fungi are highly variable among individuals, with few appearing to be common to all.
Cultivation-based analyses have typically identified Candida as the most common fungal genus (166), and it is also frequently identified using non-cultivation-based methods, whereas the other taxa identified have been variable, which may be because of the analytical method used and/or subject variability. For example, 66 genera of fungi were found using pyrosequencing when 98 individuals were examined, with the genera Saccharomyces, Candida, and Cladosporium being the most prevalent (85). Mucor was common in Spanish individuals (126) and the most common fungi in 16 vegetarians were Fusarium, Malassezia, Penicillium, and Aspergillus (182). These studies suggested that some taxa, e.g., Penicillium and Aspergillus, are not resident in the gut and enter through environmental sources, such as food and water, in which they are commonly found. This may account for some of the variability in taxa reported in various studies and for the increasing number of fungi being identified as more studies are being performed, even those based on cultivation (71). Under certain conditions, some fungi may flourish and become pathogenic including Candida, Aspergillus, Fusarium, and Cryptococcus (44, 84, 140, 143). More information on fungal interactions and diseases is available in a review by Wang et al. (204).
Despite their low abundance, fungi appear to have developed in mammalian guts along with the rest of the body from infancy (106, 169). Although there is no consensus of a core mycobiome, Candida, Saccharomyces, and Malassezia have been commonly reported (72). Most of the fungal species detected appear to be either transient or environmental fungi that cannot colonize the gut and are often found in a single study and/or one host only. A previous study indicated that the fungal community is unstable; only 20% of the initially identified fungi were detected again 4 months later (78). More studies on the stability of the mycobiome are needed in order to establish the ecological roles of the components of the mycobiome. Many non-bacterial organisms have been found in numerous mammalian systems, which indicates that they play an important role that has been largely overlooked and may lead to important discoveries and understanding in the coming years.
Archaea
The most commonly reported genus of Archaea that has been found in the GI tract is Methanobrevibacter (51, 55, 66, 85, 109). Other genera that have also been detected are Methanosphaera (51), Nitrososphaera, Thermogynomonas, and Thermoplasma (85) and the new candidate species, Methanomethylophilus alvus (27, 131). Although Archaea comprise a very small proportion of the microbiota, Methanobrevibacter species are important contributors to methanogenesis (66). Differences in Archaea in microbiome samples may be due to the method used (51) and/or complex relationships with other microbiota. For example, Methanobrevibacter and Nitrososphaera were previously shown to be mutually exclusive and potentially related to carbohydrate intake (85). More studies are needed in order to clarify the interaction between Archaea and other microbiota groups, which may contribute to our understanding of their fitness and function (beyond methanogenesis) in the microbiome.
Viruses
Viruses in the human microbiome have also been understudied and available information is limited (161); the majority of data are related primarily to disease and do not address the commensal virome (34, 40). The majority of viral reads in studies that have been performed cannot be assigned to a known group; this has contributed to the difficulties associated with assessing their roles in the GI tract (124, 160). A number of teams have made extensive efforts in order to advance human virome studies (157, 161). In the last ten years, the number of identified polyomaviruses has increased from 4 to 13 species (some that cause disease and some that do not) (47), and the accuracy of identification techniques has been improved to identify taxa at the genus level (199) and use metagenomic information for viral taxonomy (172). Viral communities are mainly comprised of bacteria-infecting phage families (~90%), while eukaryotic viruses (~10%) are in lower abundance (157, 161). Metagenomic analyses have suggested that the new bacteriophage, crAssphage associated with Bacteroides, is potentially common in humans (53). The greatest diversity of phages is considered to occur in infants and decreases with age, in contrast to increases in bacterial diversity (116, 117, 162). With the availability of methods to enrich viruses in samples (41), and with more metagenomic sequences and bioinformatics tools to identify viral sequences (53, 139), more information will be obtained on viral diversity and associated physiological factors in humans.
Similar to the microbiota, considerable variability appears to exist in the viral taxa found among subjects (133). Limited information is currently available on the functional roles of most viruses in the human GI tract. However, some possible functions are: to increase bacterial fitness as sources of genetic information (e.g., the source of antibiotic resistance genes), to increase the immunity of bacteria or the human host, and to protect against pathogens (40, 64, 157). The general consensus is that the presence of bacteria is beneficial for viruses that are increasingly trying to evade the immune system. This relationship may also be beneficial to bacteria as viruses may be sources of potentially advantageous genes (resistance or tolerance to stress environments). Researchers are now examining the ecological and evolutionary influences of phages on bacterial ecosystems (102), and the findings obtained may provide insights into the important roles played by phages in the gut microbiome.
The GI tract
Many challenges are associated with studying the microbial ecology of the GI tract because it is composed of chemically and physically diverse microhabitats stretching from the esophagus to the rectum, providing a surface area of 150–200 m2 for colonization or transient occupation by microbes (16). The adult GI tract was initially estimated to harbor 1014 bacteria, 10 times more cells than the human body (16, 120); however, a more recent calculation estimates there to be 1013 bacteria, which is equivalent to the number of human cells (170). Lower bacterial numbers (103 to 104 bacteria mL−1 of intestinal content) are found in the upper end of the GI tract, stomach, and small intestine, in which pH is low and the transit time is short (16). The highest biodiversity (richness and evenness) of bacteria (1010–1011 bacteria g−1 of intestinal content) is in the colon, in which cell turnover rate is low, redox potential is low, and the transit time is long. This section highlights the different functions and associated microbiota along the human GI tract starting from the oral cavity, then the esophagus, stomach, and intestines (Fig. 1).
The oral cavity
Activity in the mouth may have a large impact on the further digestion of food in the lower GI tract. Food is mechanically ground into small particles, typically 0.1 mm, which increases the surface area. The oral microbiome is composed of transient and commensal populations that often form biofilms on soft and hard surfaces in the mouth (8). The most up-to-date information on taxa of the oral microbiome may be found in the Human Oral Microbiome Database (HOMD, http://www.homd.org/) (50). Information in this database is limited to Bacteria and one Archaea. Cultivation-independent analyses indicate that the most common genus is Streptococcus, while other genera include Neisseria, Gemella, Granulicatella, and Veillonella, but not in all individuals examined (1, 91, 92, 107). The taxa present appear to be dependent on interactions between microbes within the community. For example, using a graph theory-based algorithm of an organism’s nutritional profile, the species Streptococcus oralis and S. gordonii have low metabolic complementarity and high metabolic competition, indicating they are antagonistic to each other (110). In contrast, Porphyromonas gingivalis was shown to have high metabolic complementarity, indicating its ability to grow symbiotically with diverse oral microbiota taxa. This computational method was tested and confirmed with growth assays, making it a viable means to assess the ability of species to inhabit the same environment. This has also been shown using an in situ spectral analysis of microbiota in biofilm plaques. Biofilms were shown to be composed of a number of taxa with Corynebacterium at the foundation (209). The other taxa are considered to play complementary roles driven by the environmental and chemical gradients formed in biofilms that control nutrient availability. These findings indicate that, despite the large number of taxa identified in oral microbiome studies, the core taxa of all microbiota may be identified in the future based on spatial locations and functional roles (10).
Similar to Bacteria, large variations have been noted in viruses found in the oral cavity among subjects (151). Most viruses are bacteriophages (approx. 99% of known sequences). Viral communities are reproducible across time points within a subject, suggesting that they are stable; however, the human and bacterial host significantly influence compositions (2, 151, 163). In addition to interactions among oral bacteria, many may associate with phages (57). Depending on the host range of the oral virome, this may make phages very common inhabitants of the oral cavity. Furthermore, in addition to survival within bacterial hosts, phages may also survive in the oral mucosa and contribute to host immunity (11). These are all new avenues of oral virome research that will likely be investigated in greater depth in the future.
In addition to the bacterial microbiome, two cultivation-independent studies have been conducted on oral fungi. Approximately 100 fungal species (20 genera) were detected in one study of the oral mycobiome of healthy individuals (68). Among the fungi detected, Candida species were the most common and abundant, while the other genera consisted of Cladosporium, Aureobasidium, Saccharomycetales, Aspergillus, Fusarium, and Cryptococcus. Most of these genera were also detected in a recent study on three subjects; however, Malassezia, a skin pathogen, accounted for the most sequence reads (52). Most of the other studies conducted on the oral mycobiome have focused on the role of fungi in disease (69, 136). Since the oral microbial community is directly exposed to the environment, the presence of a dynamic and transient community is expected, but warrants further study.
Esophagus
After swallowing, food is transported down the esophagus by peristalsis to the stomach. Limited information is available on microbes inhabiting the esophagus (5, 91, 147), and this may be due to the difficulties associated with obtaining samples because biopsies have typically been used. However, a less invasive method using an esophageal string has recently been demonstrated to be a feasible alternative and yields similar findings to non-cultivation-based analyses of biopsies (60). Similar to the oral cavity, the most common genus found in the esophagus is Streptococcus; however, an overall comparison of the two communities has indicated that the number of taxa significantly differ between the two locations (15, 60). Among the few studies conducted on the viral and fungal microbiota of the esophagus, the focus has been on association with disease (204) and none of the pathogenic taxa inhabit healthy individuals.
Stomach
The stomach is the first digestive organ in the body (89). It holds food and mechanically mixes it with proteolytic enzymes and gastric acids that aid in the breakdown and subsequent absorption of nutrients. The growth of many common bacteria is inhibited by these acidic conditions (pH<4), making this a unique community with the lowest number of microbes, ranging between 101 and 103 CFU g−1. In addition to digestion, the acidic conditions of the stomach are considered to have evolved as a means of protection from pathogens. This hypothesis is supported by the recent finding of a lower pH in the stomachs of scavengers and higher pH in herbivores, which are less likely to encounter pathogens in their food (13). Caution is needed when comparing the findings of various studies throughout the GI tract because gastric juice has a lower pH than the mucosal layer, resulting in differences in the microbiota present (89).
Despite the low pH, non-cultivation-based analyses on stomach biopsies revealed a more diverse microbiota than expected (5, 20, 115). Regardless of variations among subjects, there appears to be two major groups of individuals: those with and without Helicobacter pylori (20). There is a third subset in which H. pylori is present in lower proportions in some individuals that were negative using conventional testing. Microbiomes dominated by H. pylori had significantly greater proportions of the phylum Proteobacteria, of which it is a member, and lower alpha diversity (5, 20). Other common genera are Streptococcus and Prevotella, both of which are also found in the oral and esophageal communities; however, the communities at these locations appear to differ (5). Limited information is available on fungi analyzed in biopsy samples; although a cultivation study detected Candida species, this appeared to be associated more with disease (224). The major interaction currently studied in the stomach microbiota is with Helicobacter because of its association with gastritis, peptic ulcers, and gastric cancer. However, this taxon has been suggested to be beneficial for health, leading some to question whether the complete eradication of this microbe is the best option (67, 89).
In contrast, less information is available on the microbiome of stomach fluids; it appears to harbor fewer Helicobacter and an analysis of transcripts indicated that Actinobacteria are the most active phylum; however, the other major phyla, Firmicutes, Bacteroidetes, and Proteobacteria, are also present (197). In the same study, it also appeared to harbor novel fungi; 77.5% of the ITS reads were not identified at the phylum level or lower. Candida and Phialemonium were the only two identifiable fungal genera in all subjects tested, whereas an additional 66 genera were present in at least one of the nine subjects examined. Based on the infrequency and number of reads in this analysis, most of the taxa identified in stomach fluids appear to be transient, and those playing an active role are limited in this location.
Intestines
After mixing in the stomach, chime slowly passes through the pyloric sphincter and enters the intestines, in which the major digestion and absorption of nutrients begin (12). Humans have a small and large intestine. The small intestine, the main location in which food digestion and absorption occurs, is further divided into three parts, the duodenum, jejunum, and ileum. The duodenum, in which food chime enters from the stomach, is directly associated with digestion and is linked to the pancreas and gallbladder. Bile salts from the gallbladder and enzymes from the pancreas enter the duodenum and mix with stomach chime in order to start the digestion process. The epithelium in the jejunum and ileum is responsible for glucose absorption into the bloodstream via glucose transporters and sodium ions. The small intestine is followed by the large intestine (colon), which has a larger diameter, but shorter length and is divided into four sections: the ascending colon (cecum), transverse colon, descending colon, and sigmoid colon (123). Water and minerals are continuously absorbed along the colon before excretion. Furthermore, complex foods that cannot be digested by the host are used as growth substrates for the colonic microbiota (25, 178).
Spatial and temporal variabilities have been noted in the microbial composition among the different intestinal structures based on their functional roles and timing of food intake (18, 129, 186). Although spatial variability exists along the intestinal tract, the bacterial microbiome at the phylum level is considered to remain fairly stable over time (43, 155); however, many factors may affect its stability (119). Undigested food and most of the microbiota are found in the lumen, the central space surrounded by the mucosal layer of the tubular intestinal structure. The main absorption of growth substrates occurs through the epithelial cells of the mucosa, which also prevents the entry of the microbiota into host cells (174). A number of important host-microbe interactions occur within the mucosa. Energy from microbially produced metabolites, such as butyrate, contributes to epithelial metabolism (97). Most of the gut is anaerobic, but there is an oxygen gradient in the mucosa that provides a competitive advantage for facultative anaerobes (174). Recent studies have also shown the importance of metabolites produced by transkingdom microbiota to host physiology (185, 187, 188). Microbiota, such as Akkermansia mucinophila, are commonly found residing in the mucus layer and feed on mucin (39, 48). Therefore, the effects of host interactions with the gut microbiota, particularly those in the large intestine, have a prominent impact on overall human health, including energy reabsorption and immune system development.
Due to the difficulties associated with collecting multiple samples along a healthy human GI tract in order to capture the spatial heterogeneity of microbes in this environment, most studies use fecal samples as a surrogate. However, this limits the availability of regio-specific community information on the GI tract, resulting in portions, such as the small intestine, remaining poorly characterized. The few studies conducted on the small intestine have limited subject numbers because they used biopsy samples (4, 201, 203) or ileotomy patients (108, 195, 222). The bacterial genera most commonly found among these studies were Clostridium, Streptococcus, and Bacteroides. The number of studies that include fungi are even more limited, with the genera Candida and Saccharomyces being the most frequently detected (108, 114). Caution is also needed when extrapolating these findings to all individuals because the health of some subjects was compromised when samples were obtained.
Bacteria in the colon account for approximately 70% of all bacteria in the human body because it is the main site for the bacterial fermentation of non-digestible food components such as soluble fiber. The small number of studies that have examined microbial communities directly in the colon suggests that the bacterial composition is similar to that found in feces (86). However, fecal communities do not represent a single colonic environment, such as the mucosa (223), but a mixture of indigenous and transient microbes from the entire GI tract. In studies in which a global view of the GI tract microbial community is of interest, fecal material represents a good surrogate and is easily obtained, allowing for multiple samples to be obtained over short and long time periods from healthy individuals. The majority of microbiome reviews have extensively covered colonic communities using feces (74, 92, 121, 148, 189); therefore, we will not describe its composition in detail. However, later in this review, the impact of diet on the microbiome composition will be discussed. Furthermore, the above sections on fungi and viruses provide information on the taxa of these groups in the intestines.
Summary of the GI tract
The use of non-cultivation-based methods to investigate the microbiota in the GI tract has increased our knowledge of their diversity. One group that we neglected to mention in this review was Protozoans/Protists; however, recent reviews are available (79, 145). Despite representing a smaller biomass than fungi, they also appear to be important to the ecological structure of the gut microbiome. The predator-prey relationship they have with other microbiota (145) may, in some cases, lead to disease prevention (7). Difficulties are associated with elucidating the functional roles played by these various taxa at different points along the GI tract. Therefore, it is still important to obtain cultivated representatives to investigate their role and ecological significance along the GI tract. This consideration is important for all microbiota; however, it represents a larger issue for low diversity groups, such as fungi, which may not be numerically abundant, but still play a significant role (17).
Use of animal models
Animal models have been widely adopted in human gut microbiome research (28, 98, 220) to reduce confounding experimental factors such as genetics, age, and diet, which may be more easily controlled in laboratory animals. Additionally, animal models with modified genetic backgrounds are available for investigating potential mechanisms (137). Ideally, animal models with relatively similar genetic information (217), gut structures, metabolism (142), and diets and behavior patterns (202) to humans need to be selected. Comprehensive comparisons of mice (137) and pigs (217) to humans were recently conducted in order to aid in translating information from animal models to humans. In this section, we will highlight some of their findings and compare GI tract structures and microbial community compositions. Furthermore, some advantages and limitations associated with the use of animal models in human microbiome research will be discussed.
Similarities exist in the anatomy of the GI tract between humans and most animal models (Table 1). However, differences in anatomical structures and pH at different locations along the GI tract may contribute to differences in the microbiota found in humans versus animal models (26). The human colon also has a thicker mucosal layer than those of mice and rats (137), which may have an effect on the diversity of the microbiota colonizing the colon. Human gut bacteria are dominated by two phyla: Firmicutes and Bacteroidetes (189), which also dominate the GI tract of commonly used model animals (112). However, at lower taxonomic levels, some differences have been reported in microbiome compositions in the gut between humans and animal models (Table 2). The dominant taxa reported have varied as the number of comparisons performed has increased (137, 152); therefore, the findings shown in Table 2 need to be used cautiously.
Table 1.
Comparison of the anatomy of the intestinal tract in humans and animal models
| Human | Mouse | Rat | Pig | |
|---|---|---|---|---|
| Stomach | Four regions: cardia, fundus, body, and pylorus | Three regions: forestomach, body, and pylorus | Three regions: forestomach, body, and pylorus | Four regions: esophagus, cardia, fundus, and pylorus |
| pH 1.5 to 3.5 | pH 3.0 to 4.0 | pH 3.0 to 4.0 | pH 1.5 to 2.5 | |
| Small intestine | 5.5–6.4 m in length | 350 mm in length | 1,485 mm in length | 1.2–2.1 m in length |
| pH 6.4 to 7.3 | pH 4.7 to 5.2 | pH 5.0 to 6.1 | pH 6.1 to 6.7 | |
| Cecum | Smaller than the colon | Larger than the colon | Larger than the colon | Smaller than the colon |
| No fermentation | Main fermentation | Main fermentation | Some fermentation | |
| pH 5.7 | pH 4.4 to 4.6 | pH 5.9 to 6.6 | pH 6.0 to 6.4 | |
| Appendix | Present | Absent | Absent | Absent |
| Colon | Divided into the ascending, transcending, and descending colon | Not divided | Not divided | Divided into the ascending, transcending, and descending colon |
| Main fermentation | No fermentation | No fermentation | Main fermentation | |
| Thick mucosa | Thinner mucosa | Thinner mucosa | Thick mucosa | |
| pH 6.7 | pH 4.4 to 5.0 | pH 5.5 to 6.2 | pH 6.1 to 6.6 |
Table 2.
Major taxa of the gut microbiota in humans and animal models
| Human | Mouse | Rat | Pig | |
|---|---|---|---|---|
| Bacteria | Firmicutes | Firmicutes | Firmicutes | Firmicutes |
| Bacteroidetes | Bacteroidetes | Bacteroidetes | Bacteroidetes | |
| Actinobacteria | ||||
| Proteobacteria | ||||
| Archaea | Methanobrevibacter | Methanobrevibacter | Methanobrevibacter | Methanomicrobia, |
| Nitrososphaera | Methanosphaera | |||
| Viruses | Herpesviridae | Variable | Variable | Picornaviridae |
| Papillomaviridae | Astroviridae | |||
| Polyomaviridae | Coronaviridae | |||
| Adenoviridae | Caliciviridae | |||
| Eukarya | Candida | Ascomycota | Ascomycota | Kazachstania |
| Malassezia | Basidiomycota | Basidiomycota | Candida | |
| Saccharomyces | Chytridiomycota | Chytridiomycota | Galactomyces | |
| Cladosporium | Zygomycota | Zygomycota | Issatchenkia |
A pig gut gene catalogue of metabolic function was recently developed and compared to catalogues available for humans and mice (217). They found that 96% of the KEGG orthologs in humans were also present in pigs, whereas the overlap at the gene level was markedly lower (9.46%). However, there was a greater overlap between humans and pigs than between humans and mice. Microbial activity also differs along the GI tract, with the most relevant being fermentation occurring in the ceca of most animal models, but not in humans (137). Strengths and weaknesses are associated with the major animal models being used, and these need to be taken into consideration when conducting translational research.
Rats
The use of rats as lab animals dates back to the 1850s. They were considered to be a good candidate for human microbiome research because the rat contains the same four dominant bacteria phyla in the GI tract (31), with Firmicutes (74%) and Bacteroidetes (23%) representing the largest proportions (221). The advantages of using rats in human microbiome research include quick reproduction, a fully sequenced genome, and easy handling and maintenance due to their relatively small size. The limitation of this model is that the diet used in rats differs from that for humans, and their behavior and living environment are also different, which will affect the gut microbiota. The diet used in rat studies is normal chow that is rich in fiber (205), and diet may rapidly alter gut microbiota diversity (46). Although most studies emphasize the impact of diet on the microbiota in the cecum and/or colon (feces), the oral cavity of rats has been used to clarify the impact of diet on the microbiome (93).
Mice
Many of the strengths and weaknesses associated with using rats are also applicable to mice. Similar to humans, the microbiota in the GI tract of mice is dominated by Firmicutes (74%) and Bacteroidetes (23%) at the phylum level (217). However, there are differences at the genus level, and this has led to the use of “humanized” mice. This is achieved by inoculating human gut microbiota into germ-free (GF) mice (192) or mice treated with antibiotics to eliminate their gut microbiome (83). The microbiome of these mice after fecal transplants may have a composition at the phyla level that is 100% similar to humans and 88% at the genus level (137). A recent study (175) used humanized mice to test microbiome diversity after feeding with poorly accessible carbohydrates, and found a similar reduction in OTU numbers to a human study (219). However, there are also some limitations to using these animals, including the diet and environmental living conditions. Furthermore, gnotobiotic mice may not reflect the human-microbe relationship due to their weaker immune system (6).
Approximately 10 years ago, Scupham (168) showed that all four major fungal phyla, Ascomycota, Basidiomycota, Chytridiomycota, and Zygomycota, were present in the murine gut. Additionally, many genera were identified, including Acremonium, Monilinia, Fusarium, Cryptococcus, Filobasidium, Scleroderma, Catenomyces, Spizellomyces, Neocallimastix, Powellomyces, Entophlyctis, Mortierella, and Smittium. When comparing these studies to the human gut, it is important to note that this study indicated a more diverse fungal community than those found in humans; the eukaryotic diversity of the human gut is low (143).
Pigs
Pigs have been used as surrogates for human microbiome research due to their highly similar genetics, physiological structures, behavior, metabolism, and immune functions to those of humans (81, 202). The greater similarities in the omnivorous diet and GI tract structure between pigs and humans are more advantageous than the murine model. The microbiome of pigs is dominated by two phyla: Firmicutes and Bacteroidetes (104); however, there are some notable differences at the genus level. The genus Prevotella was found to be common in two pig metagenomic studies (104, 118). Since the number of pigs used in most studies is less than humans, the pig core microbiome at the genus level may change as more pigs are studied. Another contributing factor to shaping the microbiome composition is diet. Most studies have found that the number of Bifidobacteria in pigs, even those on high fiber diets, is lower than that in humans (132, 218), while that of Lactobacillus is higher (149). In nutrition studies, humans and pigs are both dependent on the quality of the nutrient load; however, the pig cecum has a larger capacity to ferment indigestible compounds than the human cecum (54). The microbiota composition in pigs may differ from that in humans due in part to differences in diet (81). Similar to mice, humanized GF pigs have been developed and the microbiome after human fecal transplantation more closely resembles that of the donor than conventional pigs (144). However, the same disadvantages associated with using GF mice are also true for GF pigs.
The genome of pigs may be mutated to study human diseases; this is typically performed using miniature pigs such as those from the Ossabaw and Gottingen islands (146). Genetic mutations for metabolic syndrome and insulin resistance have successfully been performed using Ossabaw pigs to study human diseases such as type 2 diabetes (14, 177) and obesity (101). The ratio of Firmicutes to Bacteroidetes is higher in obese Ossabaw pigs than in lean pigs (146), similar to some obese humans (111, 190). This finding suggests that Ossabaw pigs are a good model for researching the role of the microbiota in human obesity. However, disadvantages are associated with using miniature pigs, mainly the higher cost for maintenance and longer reproductive period than rodents (146).
Although more extensive efforts have been made to investigate fungi in pigs than in other animal models, many of these studies were cultivation-based or for use as probiotics. Fungi in pigs have been recently studied using a non-cultivation approach and up to 17 species of yeast (belonging to the genera Kazachstania, Galactomyces, Candida, Issatchenkia, Pichia, Rhodotorula, and Trichosporon) were common in the gut (194). The number of studies on viruses is limited, but the composition appears to be highly variable among samples (164, 171) and affected by disease (24). These groups need to be examined in more detail in order to establish whether pigs are good models for use in understanding fungi and viruses in humans.
Animal model summary
The convenience and cost of using animal models for human research are appealing. However, researchers need be very careful when selecting animal models appropriate for their objectives, particularly when the objective is to directly extrapolate findings from animals to humans, due to the significant differences in GI tract physiology and microbiome composition (65, 137, 217).
Diet in health
Many studies have found that diet is one of the main factors shaping the composition of gut microbial populations. Dietary approaches, such as the ingestion of non-digestible carbohydrates (prebiotics) and fermented food products containing live cultures (probiotics), have been suggested to confer health benefits by enhancing the growth of beneficial intestinal bacteria (100, 158). As described earlier, the microbiota may break down food components, such as non-digestible carbohydrates, which are indigestible by the host in order to aid in maximizing available nutrients (9) and produce metabolites that contribute to host health. Probiotics have been used as a means to replenish the gut with “beneficial” microbiota after antibiotic treatments or to treat diseases (82, 159). This section will highlight some studies that demonstrated the health benefits of prebiotics and probiotics and possible roles played by the microbiota.
Dietary prebiotics and probiotics
Non-digestible and fermentable food components are often consumed as prebiotics to selectively stimulate the growth and/or activity of endogenous colonic bacteria that may be beneficial to host health. The increased consumption of prebiotics often correlates with enhancements in certain bacterial genera (a common example is Bifidobacterium sp.); however, the reason they are beneficial remains unclear (208). Challenges are associated with elucidating the role being played by specific bacterial phylotypes because many of their processes are interactive (207). For example, SCFA produced by bacterial fermentation may lower intestinal pH, thereby increasing the solubility of essential minerals, such as calcium, iron, and magnesium, and consequently enhancing their absorption and improving health. Metabolites produced by microbes may also play an important role in cellular differentiation and proliferation in the colonic mucosa by inducing apoptosis and may confer protection against colitis and colorectal cancer by modulating oncogene expression. These functions do not appear to be performed by a single species; a number of different species may be acting independently or in combination. Research is leading to an understanding of microbial community structure and composition dynamics with respect to diet aids in establishing testable hypotheses for future research in health and beneficial microbes (32). Most research has been performed on the influence of beneficial intestinal bacteria such as Bifidobacterium spp. and Lactobacillus spp. on host health monitored using a cultivation approach. Cultivation-independent approaches have now become more popular, leading to the identification of new beneficial microbiota taxa and their potential functional roles in the gut as they relate to diet.
Dietary fibers and oligosaccharides are carbohydrate ingredients that vary in composition and structure, but are considered to be non-digestible because of the lack of appropriate intestinal enzymes to hydrolyze them or structural hindrances that prevent enzyme access in the gut. Although bacteria in the lower gut may ferment these carbohydrates, the rate and degree of fermentation vary with the polysaccharide (80). The range of fermentation in the colon for various fibers is broad, from approximately 5% for cellulose to nearly 100% for pectin (42). The resulting SCFA, including butyrate and propionate, are considered to reduce pH and solubilize minerals, thereby improving their absorption and subsequent utilization. Inulin, a long chain fructooligosaccharide (FOS) often obtained from chicory root, and FOS from other sources are the fibers that have been studied in the most detail (206). Several novel fibers have been tested in an in vitro large intestine model for their effects on the microbial stimulation and production of SCFA (122). All these novel fibers stimulated the growth of beneficial Bifidobacteria and some Lactobacillus species along with increases in SCFA production. Only a few studies have examined the effects of fibers and resistant starches on the human microbiome (87, 127, 198, 210, 211). A soluble corn fiber product has been demonstrated to increase Ca absorption in a number of different studies (210, 211). More benefits to human health may be attributed to the consumption of prebiotics and fermentation by the gut microbiome.
The number of studies that include diet effects on Archaea, Fungi, and/or Viruses are limited; however, some examples are included herein. Examinations of Archaea, Fungi, and Bacteria correlations in response to diet revealed a syntrophic model involving Candida, Prevotella, Ruminococcus, and Methanobrevibacter (85). Candida was considered to break down carbohydrates into metabolites used by Prevotella and Ruminococcus that produce CO2 for Methanobrevibacter (85). However, shifts in carbon sources or breaking down starches via amylases from the human mouth may alter this relationship because Prevotella may no longer be dependent on Candida. This is a good example of how Archaea, which represent a very small portion of the microbiome, are a key contributor to methanogenesis and waste decomposition. The absence of Archaea may have severe effects on the surrounding community as hydrogen, glucose metabolites, and other carbon sources accumulate. Other organisms will eventually fill this niche, but may diminish or accumulate new metabolites that ultimately shift the surrounding community based on their fitness for using these substrates.
A recent study investigated rapid changes in the microbiome composition when diets were either high in animal-based or plant-based fat and protein (46). The fungus Candida was found to increase in subjects placed on a plant-based diet, whereas Penicillium increased on animal-based diets. The most commonly found fungi in vegetarians were Fusarium, Malassezia, Penicillium, Aspergillus, and Candida (182). Caution is needed when interpreting findings because some of these fungi may be found on food prior to ingestion (46, 78, 182)
Phages assembled in the gut may also be modified by diet. A recent study examined changes in the fecal viral community over an 8-d period in six subjects supplied different diets (134). Shotgun sequencing of virus-like particles revealed that interpersonal differences in the virome were the largest source of variations in this study. However, the virome of subjects whose diets were changed differed more than in those who maintained their normal diet. Although this is only one study with a few human subjects, studies using a mouse model and different dietary fats support these findings (88, 99). Collectively, these findings indicate that diet plays a key role in shaping the gut virome, and further research is needed in order to investigate interactions between diet and the virome.
Summary
Advances have been made in the last decade in our understanding of the role of the GI tract microbiome in human health. This review has highlighted changes and differences in the microbiome along the GI tract that are due to changes in physical, chemical, and biological interactions. Although extensive research has been conducted on Bacteria in fecal samples, the main kingdom inhabiting the gut, our knowledge is still insufficient, particularly in other regions of the GI tract. Furthermore, other groups (Archaea, Fungi, and Viruses) have not yet been investigated in adequate detail, demonstrating a real void in knowledge. This highlights that the basic ecology of microbiomes is important for gaining a greater understanding to improve human health and decrease disease. In order to achieve this goal, it is important to include all microbiota in studies and remain cognizant of the limitations associated with understanding the entire GI tract of humans despite challenges in sampling and cultivation. Furthermore, the use of appropriate animal models in mechanistic studies requires careful consideration.
References
- 1.Aas J.A., Paster B.J., Stokes L.N., Olsen I., Dewhirst F.E. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abeles S.R., Robles-Sikisaka R., Ly M., Lum A.G., Salzman J., Boehm T.K., Pride D.T. Human oral viruses are personal, persistent and gender-consistent. ISME J. 2014;8:1753–1767. doi: 10.1038/ismej.2014.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abubucker S., Segata N., Goll J., et al. Metabolic reconstruction for metagenomic data and Its application to the human microbiome. PLoS Comput Biol. 2012;8:e1002358. doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmed S., Macfarlane G.T., Fite A., McBain A.J., Gilbert P., Macfarlane S. Mucosa-associated bacterial diversity in relation to human terminal ileum and colonic biopsy samples. Appl Environ Microbiol. 2007;73:7435–7442. doi: 10.1128/AEM.01143-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersson A.F., Lindberg M., Jakobsson H., Bäckhed F., Nyrén P., Engstrand L. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One. 2008;3:e2836. doi: 10.1371/journal.pone.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atarashi K., Tanoue T., Oshima K., et al. T-reg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 7.Audebert C., Even G., Cian A., Loywick A., Merlin S., Viscogliosi E., Chabe M. Colonization with the enteric protozoa Blastocystis is associated with increased diversity of human gut bacterial microbiota. Sci Rep. 2016;6:25255. doi: 10.1038/srep25255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Avila M., Ojcius D.M., Yilmaz Ö. The oral microbiota: Living with a permanent guest. DNA Cell Biol. 2009;28:405–411. doi: 10.1089/dna.2009.0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Backhed F., Ley R.E., Sonnenburg J.L., Peterson D.A., Gordon J.I. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 10.Baker J.L., Bor B., Agnello M., Shi W.Y., He X.S. Ecology of the oral microbiome: Beyond bacteria. Trends Microbiol. 2017;25:362–374. doi: 10.1016/j.tim.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barr J.J., Auro R., Furlan M., et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc Natl Acad Sci USA. 2013;110:10771–10776. doi: 10.1073/pnas.1305923110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrett K.E. Gastrointestinal Physiology. 2nd ed. Lange: Medical Books/McGraw-Hill; 2014. [Google Scholar]
- 13.Beasley D.E., Koltz A.M., Lambert J.E., Fierer N., Dunn R.R. The evolution of stomach acidity and is relevance to the human microbiome. PLoS One. 2015;10:e.0134116. doi: 10.1371/journal.pone.0134116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bellinger D.A., Merricks E.P., Nichols T.C. Swine models of type 2 diabetes mellitus: Insulin resistance, glucose tolerance, and cardiovascular complications. ILAR J. 2006;47:243–258. doi: 10.1093/ilar.47.3.243. [DOI] [PubMed] [Google Scholar]
- 15.Benitez A.J., Hoffmann C., Muir A.B., Dods K.K., Spergel J.M., Bushman F.D., Wang M.-L. Inflammation-associated microbiota in pediatric eosinophilic esophagitis. Microbiome. 2015;3:23. doi: 10.1186/s40168-015-0085-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berg R.D. The indigenous gastrointestinal microflora. Trends Microbiol. 1996;4:430–435. doi: 10.1016/0966-842x(96)10057-3. [DOI] [PubMed] [Google Scholar]
- 17.Berrilli F., Di Cave D., Cavallero S., D’Amelio S. Interactions between parasites and microbial communities in the human gut. Front Cell Infect Microbiol. 2012;2:141. doi: 10.3389/fcimb.2012.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biggs M.B., Medlock G.L., Moutinho T.J., Lees H.J., Swann J.R., Kolling G.L., Papin J.A. Systems-level metabolism of the altered Schaedler flora, a complete gut microbiota. ISME J. 2017;11:426–438. doi: 10.1038/ismej.2016.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bik E.M. Composition and function of the human-associated microbiota. Nutr Rev. 2009;67:S164–S171. doi: 10.1111/j.1753-4887.2009.00237.x. [DOI] [PubMed] [Google Scholar]
- 20.Bik E.M., Eckburg P.B., Gill S.R., Nelson K.E., Purdom E.A., Francois F., Perez-Perez G., Blaser M.J., Relman D.A. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci USA. 2006;103:732–737. doi: 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bik E.M., Long C.D., Armitage G.C., et al. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 2010;4:962–974. doi: 10.1038/ismej.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blaser M.J. Antibiotic use and its consequences for the normal microbiome. Science. 2016;352:544–545. doi: 10.1126/science.aad9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blaut M., Collins M.D., Welling G.W., Dore J., van Loo J., de Vos W. Molecular biological methods for studying the gut microbiota: the EU human gut flora project. Brit J Nutr. 2002;87:S203–S211. doi: 10.1079/BJNBJN/2002539. [DOI] [PubMed] [Google Scholar]
- 24.Blomstrom A.L., Fossum C., Wallgren P., Berg M. Viral metagenomic analysis displays the co-infection situation in healthy and PMWS affected pigs. PLoS One. 2016;11:e0166863. doi: 10.1371/journal.pone.0166863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bolam D.N., Sonnenburg J.L. Mechanistic insight into polysaccharide use within the intestinal microbiota. Gut microbes. 2011;2:86–90. doi: 10.4161/gmic.2.2.15232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Booijink C., Zoetendal E.G., Kleerebezem M., de Vos W.M. Microbial communities in the human small intestine: coupling diversity to metagenomics. Fut Microbiol. 2007;2:285–295. doi: 10.2217/17460913.2.3.285. [DOI] [PubMed] [Google Scholar]
- 27.Borrel G., Harris H.M., Tottey W., et al. Genome sequence of “Candidatus Methanomethylophilus alvus” Mx1201, a methanogenic archaeon from the human gut belonging to a seventh order of methanogens. J Bacteriol. 2012;194:6944–6945. doi: 10.1128/JB.01867-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowey E., Adlercreutz H., Rowland I. Metabolism of isoflavones and lignans by the gut microflora: a study in germ-free and human flora associated rats. Food Chem Toxicol. 2003;41:631–636. doi: 10.1016/s0278-6915(02)00324-1. [DOI] [PubMed] [Google Scholar]
- 29.Breitbart M., Hewson I., Felts B., Mahaffy J.M., Nulton J., Salamon P., Rohwer F. Metagenomic analyses of an uncultured viral community from human feces. J Bacteriol. 2003;185:6220–6223. doi: 10.1128/JB.185.20.6220-6223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Breitbart M., Haynes M., Kelley S., et al. Viral diversity and dynamics in an infant gut. Res Microbiol. 2008;159:367–373. doi: 10.1016/j.resmic.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 31.Brooks S.P.J., McAllister M., Sandoz M., Kalmokoff M.L. Culture-independent phylogenetic analysis of the faecal flora of the rat. Can J Microbiol. 2003;49:589–601. doi: 10.1139/w03-075. [DOI] [PubMed] [Google Scholar]
- 32.Brownawell A.M., Caers W., Gibson G.R., Kendall C.W.C., Lewis K.D., Ringel Y., Slavin J.L. Prebiotics and the health benefits of fiber: Current regulatory status, future research, and goals. J Nutr. 2012;142:962–974. doi: 10.3945/jn.112.158147. [DOI] [PubMed] [Google Scholar]
- 33.Cadwell K. Expanding the role of the virome: Commensalism in the gut. J Virol. 2015;89:1951–1953. doi: 10.1128/JVI.02966-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cadwell K. The virome in host health and disease. Immunity. 2015;42:805–813. doi: 10.1016/j.immuni.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cavalcanti I.M.G., Nobbs A.H., Ricomini A.P., Jenkinson H.F., Cury A.A.D. Interkingdom cooperation between Candida albicans, Streptococcus oralis and Actinomyces oris modulates early biofilm development on denture material. Pathog Dis. 2016;74 doi: 10.1093/femspd/ftw002. ftw002. [DOI] [PubMed] [Google Scholar]
- 36.Charbonneau M.R., Blanton L.V., DiGiulio D.B., Relman D.A., Lebrilla C.B., Mills D.A., Gordon J.I. A microbial perspective of human developmental biology. Nature. 2016;535:48–55. doi: 10.1038/nature18845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cho I., Blaser M.J. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13 doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Claesson M.J., O’Sullivan O., Wang Q., Nikkilä J., Marchesi J.R., Smidt H., de Vos W.M., Ross R.P., O’Toole P.W. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS ONE. 2009;4:e6669. doi: 10.1371/journal.pone.0006669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Collado M.C., Derrien M., Isolauri E., de Vos W.M., Salminen S. Intestinal Integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl Environ Microbiol. 2007;73:7767–7770. doi: 10.1128/AEM.01477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Columpsi P., Sacchi P., Zuccaro V., Cima S., Sarda C., Mariani M., Gori A., Bruno R. Beyond the gut bacterial microbiota: The gut virome. J Med Virol. 2016;88:1467–1472. doi: 10.1002/jmv.24508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Conceição-Neto N., Zeller M., Lefrère H., et al. Modular approach to customise sample preparation procedures for viral metagenomics: a reproducible protocol for virome analysis. Sci Rep. 2015;5:16532. doi: 10.1038/srep16532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cook S.I., Sellin J.H. Review article: short chain fatty acids in health and disease. Ailment Pharmacol Ther. 1998;12:499–507. doi: 10.1046/j.1365-2036.1998.00337.x. [DOI] [PubMed] [Google Scholar]
- 43.Costello E.K., Lauber C.L., Hamady M., Fierer N., Gordon J.I., Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cui L., Morris A., Ghedin E. The human mycobiome in health and disease. Genome Med. 2013;5:63. doi: 10.1186/gm467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dalmasso M., Hill C., Ross R.P. Exploiting gut bacteriophages for human health. Trends Microbiol. 2014;22:399–405. doi: 10.1016/j.tim.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 46.David L.A., Maurice C.F., Carmody R.N., et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DeCaprio J.A., Garcea R.L. A cornucopia of human polyomaviruses. Nat Rev Microbiol. 2013;11:264–276. doi: 10.1038/nrmicro2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Derrien M., Vaughan E.E., Plugge C.M., de Vos W.M. Akkermansia muciniphila gen. nov., sp nov., a human intestinal mucin-degrading bacterium. Internat J Syst Evol Microbiol. 2004;54:1469–1476. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- 49.Dethlefsen L., Huse S., Sogin M.L., Relman D.A. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dewhirst F.E., Chen T., Izard J., Paster B.J., Tanner A.C.R., Yu W.-H., Lakshmanan A., Wade W.G. The human oral microbiome. J Bacteriol. 2010;192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dridi B., Raoult D., Drancourt M. Archaea as emerging organisms in complex human microbiomes. Anaerobe. 2011;17:56–63. doi: 10.1016/j.anaerobe.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 52.Dupuy A.K., David M.S., Li L., Heider T.N., Peterson J.D., Montano E.A., Dongari-Bagtzoglou A., Diaz P.I., Strausbaugh L.D. Redefining the human oral mycobiome with improved practices in amplicon-based taxonomy: Discovery of Malassezia as a prominent commensal. PLoS One. 2014;9:e90899. doi: 10.1371/journal.pone.0090899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dutilh B.E., Noriko C.W., McNair K., et al. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat Commun. 2014;5:4498. doi: 10.1038/ncomms5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eberhard M., Hennig U., Kuhla S., Brunner R.M., Kleessen B., Metges C.C. Effect of inulin supplementation on selected gastric, duodenal, and caecal microbiota and short chain fatty acid pattern in growing piglets. Arch Animal Nutr. 2007;61:235–246. doi: 10.1080/17450390701431631. [DOI] [PubMed] [Google Scholar]
- 55.Eckburg P.B., Lepp P.W., Relman D.A. Archaea and their potential role in human disease. Infect Immun. 2003;71:591–596. doi: 10.1128/IAI.71.2.591-596.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eckburg P.B., Bik E.M., Bernstein C.N., Purdom E., Dethlefsen L., Sargent M., Gill S.R., Nelson K.E., Relman D.A. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Edlund A., Santiago-Rodriguez T.M., Boehm T.K., Pride D.T. Bacteriophage and their potential roles in the human oral cavity. J Oral Microbiol. 2015;7:27423. doi: 10.3402/jom.v7.27423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Egert M., de Graaf A.A., Smidt H., de Vos W.M., Venema K. Beyond diversity: functional microbiomics of the human colon. Trends Microbiol. 2006;14:86. doi: 10.1016/j.tim.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 59.Fallingborg J., Christensen L.A., Ingeman-Nielsen M., Jacobsen B.A., Abildgaard K., Rasmussen H.H. pH-profile and regional transit times of the normal gut measured by a radiotelemetry device. Aliment Pharmacol Ther. 1989;3:605–613. doi: 10.1111/j.1365-2036.1989.tb00254.x. [DOI] [PubMed] [Google Scholar]
- 60.Fillon S.A., Harris J.K., Wagner B.D., et al. Novel device to sample the esophageal microbiome—The esophageal string test. PLoS One. 2012;7:e42938. doi: 10.1371/journal.pone.0042938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fleet J.C. Molecular regulation of calcium metabolism. In: Weaver C.M., Heaney R.P., editors. Calcium in Human Health. Humana Press Inc; Totowa, NJ: 2006. pp. 163–189. [Google Scholar]
- 62.Fleet J.C., Schoch R.D. Molecular mechanisms for regulation of intestinal calcium absorption by vitamin D and other factors. Crit Rev Clinl Lab Sci. 2010;47:181–195. doi: 10.3109/10408363.2010.536429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Flint H.J., Duncan S.H., Scott K.P., Louis P. Interactions and competition within the microbial community of the human colon: links between diet and health. Environ Microbiol. 2007;9:1101–1111. doi: 10.1111/j.1462-2920.2007.01281.x. [DOI] [PubMed] [Google Scholar]
- 64.Foca A., Liberto M.C., Quirino A., Marascio N., Zicca E., Pavia G. Gut inflammation and immunity: What is the role of the human gut virome? Mediat Inflamm. 2015 doi: 10.1155/2015/326032. Article ID 326032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fritz J.V., Desai M.S., Shah P., Schneider J.G., Wilmes P. From meta-omics to causality: experimental models for human microbiome research. Microbiome. 2013;1:14. doi: 10.1186/2049-2618-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gaci N., Borrel G., Tottey W., O’Toole P.W., Brugère J.-F. Archaea and the human gut: New beginning of an old story. World J Gastroenterol. 2014;20:16062–16078. doi: 10.3748/wjg.v20.i43.16062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gagnaire A., Nadel B., Raoult D., Neefjes J., Gorvel J.-P. Collateral damage: insights into bacterial mechanisms that predispose host cells to cancer. Nat Rev Microbiol. 2017;15:109–128. doi: 10.1038/nrmicro.2016.171. [DOI] [PubMed] [Google Scholar]
- 68.Ghannoum M.A., Jurevic R.J., Mukherjee P.K., Cui F., Sikaroodi M., Naqvi A., Gillevet P.M. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 2010;6:e1000713. doi: 10.1371/journal.ppat.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gholizadeh P., Eslami H., Yousefi M., Asgharzadeh M., Aghazadeh M., Kafil H.S. Role of oral microbiome on oral cancers, a review. Biomed Pharmacother. 2016;84:552–558. doi: 10.1016/j.biopha.2016.09.082. [DOI] [PubMed] [Google Scholar]
- 70.Gill S.R., Pop M., DeBoy R.T., et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gouba N., Raoult D., Drancourt M. Eukaryote culturomics of the gut reveals new species. PLoS One. 2014;9:e106994. doi: 10.1371/journal.pone.0106994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gouba N., Drancourt M. Digestive tract mycobiota: A source of infection. Med Mal Infect. 2015;45:9–16. doi: 10.1016/j.medmal.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 73.Greenhalgh K., Meyer K.M., Aagaard K.M., Wilmes P. The human gut microbiome in health: establishment and resilience of microbiota over a lifetime. Environ Microbiol. 2016;18:2103–2116. doi: 10.1111/1462-2920.13318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grice E.A., Segre J.A. The human microbiome: Our second genome. Chakravarti A., Green E., editors. Ann Rev Genom Hum Genet. 2012;13:151–170. doi: 10.1146/annurev-genom-090711-163814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grimaudo N.J., Nesbitt W.E., Clark W.B. Coaggregation of Candida albicans with oral Actinomyces species. Oral Microbiol Immunol. 1996;11:59–61. doi: 10.1111/j.1399-302x.1996.tb00337.x. [DOI] [PubMed] [Google Scholar]
- 76.Grimaudo N.J., Nesbitt W.E. Coaggregation of Candida albicans with oral Fusobacterium species. Oral Microbiol Immunol. 1997;12:168–173. doi: 10.1111/j.1399-302x.1997.tb00374.x. [DOI] [PubMed] [Google Scholar]
- 77.Guarner F., Malagelada J.-R. Gut flora in health and disease. The Lancet. 2003;361:512–519. doi: 10.1016/S0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- 78.Hallen-Adams H.E., Kachman S.D., Kim J., Legge R.M., Martínez I. Fungi inhabiting the healthy human gastrointestinal tract: A diverse and dynamic community. Fung Ecol. 2015;15:9–17. [Google Scholar]
- 79.Hamad I., Raoult D., Bittar F. Repertory of eukaryotes (eukaryome) in the human gastrointestinal tract: Taxonomy and detection methods. Parasite Immunol. 2016;38:12–36. doi: 10.1111/pim.12284. [DOI] [PubMed] [Google Scholar]
- 80.Hamaker B.R., Tuncil Y.E. A perspective on the complexity of dietary fiber structures and their potential effect on the gut microbiota. J Mol Biol. 2014;426:3838–3850. doi: 10.1016/j.jmb.2014.07.028. [DOI] [PubMed] [Google Scholar]
- 81.Heinritz S.N., Mosenthin R., Weiss E. Use of pigs as a potential model for research into dietary modulation of the human gut microbiota. Nutr Res Rev. 2013;26:191–209. doi: 10.1017/S0954422413000152. [DOI] [PubMed] [Google Scholar]
- 82.Hemarajata P., Versalovic J. Effects of probiotics on gut microbiota: mechanisms of intestinal immunomodulation and neuromodulation. Therapeu Adv Gastroenterol. 2013;6:39–51. doi: 10.1177/1756283X12459294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hintze K.J., Cox J.E., Rompato G., Benninghoff A.D., Ward R.E., Broadbent J., Lefevre M. Broad scope method for creating humanized animal models for animal health and disease research through antibiotic treatment and human fecal transfer. Gut Microbes. 2014;5:183–191. doi: 10.4161/gmic.28403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hoarau G., Mukherjee P.K., Gower-Rousseau C., et al. Bacteriome and mycobiome interactions underscore microbial dysbiosis in familial Crohn’s disease. mBio. 2016;7:e01250–16. doi: 10.1128/mBio.01250-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hoffmann C., Dollive S., Grunberg S., Chen J., Li H., Wu G.D., Lewis J.D., Bushman F.D. Archaea and fungi of the human gut microbiome: Correlations with diet and bacterial residents. PLoS One. 2013;8:e66019. doi: 10.1371/journal.pone.0066019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hold G.L., Pryde S.E., Russell V.J., Furrie E., Flint H.J. Assessment of microbial diversity in human colonic samples by 16S rDNA sequence analysis. FEMS Microbiol Ecol. 2002;39:33–39. doi: 10.1111/j.1574-6941.2002.tb00904.x. [DOI] [PubMed] [Google Scholar]
- 87.Hooda S., Boler B.M.V., Serao M.C.R., Brulc J.M., Staeger M.A., Boileau T.W., Dowd S.E., Fahey G.C., Swanson K.S. 454 Pyrosequencing reveals a shift in fecal microbiota of healthy adult men consuming polydextrose or soluble corn fiber. The J Nutr. 2012;142:1259–1265. doi: 10.3945/jn.112.158766. [DOI] [PubMed] [Google Scholar]
- 88.Howe A., Ringus D.L., Williams R.J., Choo Z.N., Greenwald S.M., Owens S.M., Coleman M.L., Meyer F., Chang E.B. Divergent responses of viral and bacterial communities in the gut microbiome to dietary disturbances in mice. ISME J. 2016;10:1217–1227. doi: 10.1038/ismej.2015.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hunt R.H., Camilleri M., Crowe S.E., et al. The stomach in health and disease. Gut. 2015;64:1650–1668. doi: 10.1136/gutjnl-2014-307595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huse S.M., Dethlefsen L., Huber J.A., Mark Welch D., Relman D.A., Sogin M.L. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 2008;4:e1000255. doi: 10.1371/journal.pgen.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huse S.M., Ye Y., Zhou Y., Fodor A.A. A core human microbiome as viewed through 16S rRNA sequence clusters. PLoS ONE. 2012;7:e34242. doi: 10.1371/journal.pone.0034242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huttenhower C., Gevers D., Knight R., et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hyde E.R., Luk B., Cron S., et al. Characterization of the rat oral microbiome and the effects of dietary nitrate. Free Radic Biol Med. 2014;77:249–257. doi: 10.1016/j.freeradbiomed.2014.09.017. [DOI] [PubMed] [Google Scholar]
- 94.Iliev I.D., Funari V.A., Taylor K.D., et al. Interactions between commensal fungi and the C-type lectin receptor dectin-1 Influence colitis. Science. 2012;336:1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jernberg C., Lofmark S., Edlund C., Jansson J.K. Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiol. 2010;156:3216–3223. doi: 10.1099/mic.0.040618-0. [DOI] [PubMed] [Google Scholar]
- 96.Kararli T.T. Comparison of the gastrointestinal anatomy, physiology, and biochemistry of humans and commonly used laboratory animals. Biopharm Drug Dispos. 1995;16:351–380. doi: 10.1002/bdd.2510160502. [DOI] [PubMed] [Google Scholar]
- 97.Kelly C.J., Zheng L., Campbell E.L., et al. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe. 2015;17:662–671. doi: 10.1016/j.chom.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kibe R., Sakamoto M., Yokota H., Ishikawa H., Aiba Y., Koga Y., Benno Y. Movement and fixation of intestinal microbiota after administration of human feces to germfree mice. Appl Environ Microbiol. 2005;71:3171–3178. doi: 10.1128/AEM.71.6.3171-3178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kim M.-S., Bae J.-W. Spatial disturbances in altered mucosal and luminal gut viromes of diet-induced obese mice. Environ Microbiol. 2016;18:1498–1510. doi: 10.1111/1462-2920.13182. [DOI] [PubMed] [Google Scholar]
- 100.Kleerebezem M., Vaughan E.E. Probiotic and gut Lactobacilli and Bifidobacteria: Molecular approaches to study diversity and activity. Ann Rev Microbiol. 2009;63:269–290. doi: 10.1146/annurev.micro.091208.073341. [DOI] [PubMed] [Google Scholar]
- 101.Koopmans S.J., Schuurman T. Considerations on pig models for appetite, metabolic syndrome and obese type 2 diabetes: From food intake to metabolic disease. Eur J Pharmacol. 2015;759:231–239. doi: 10.1016/j.ejphar.2015.03.044. [DOI] [PubMed] [Google Scholar]
- 102.Koskella B., Brockhurst M.A. Bacteria–phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol Rev. 2014;38:916–931. doi: 10.1111/1574-6976.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lam V., Su J.D., Koprowski S., Hsu A.N., Tweddell J.S., Rafiee P., Gross G.J., Salzman N.H., Baker J.E. Intestinal microbiota determine severity of myocardial infarction in rats. FASEB J. 2012;26:1727–1735. doi: 10.1096/fj.11-197921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lamendella R., Domingo J.W.S., Ghosh S., Martinson J., Oerther D.B. Comparative fecal metagenomics unveils unique functional capacity of the swine gut. BMC Microbiol. 2011;11:103. doi: 10.1186/1471-2180-11-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lamendella R., Santo Domingo J.W., Ghosh S., Martinson J., Oerther D.B. Comparative fecal metagenomics unveils unique functional capacity of the swine gut. BMC Microbiol. 2011;11:103. doi: 10.1186/1471-2180-11-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.LaTuga M.S., Ellis J.C., Cotton C.M., Goldberg R.N., Wynn J.L., Jackson R.B., Seed P.C. Beyond bacteria: A study of the enteric microbial consortium in extremely low birth weight infants. PLoS ONE. 2011;6:e27858. doi: 10.1371/journal.pone.0027858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lazarevic V., Whiteson K., Huse S., Hernandez D., Farinelli L., Osteras M., Schrenzel J., Francois P. Metagenomic study of the oral microbiota by Illumina high-throughput sequencing. J Microbiol Meth. 2009;79:266–271. doi: 10.1016/j.mimet.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Leimena M.M., Ramiro-Garcia J., Davids M., et al. A comprehensive metatranscriptome analysis pipeline and its validation using human small intestine microbiota datasets. BMC Genom. 2013;14:530. doi: 10.1186/1471-2164-14-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lepp P.W., Brinig M.M., Ouverney C.C., Palm K., Armitage G.C., Relman D.A. Methanogenic Archaea and human periodontal disease. Proc Natl Acad Sci USA. 2004;101:6176–6181. doi: 10.1073/pnas.0308766101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Levy R., Borenstein E. Metabolic modeling of species interaction in the human microbiome elucidates community-level assembly rules. Proc Natl Acad Sci USA. 2013;110:12804–12809. doi: 10.1073/pnas.1300926110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ley R.E., Turnbaugh P.J., Klein S., Gordon J.I. Microbial ecology—Human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 112.Ley R.E., Hamady M., Lozupone C., et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li M., Wang B., Zhang M., et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci USA. 2008;105:2117–2122. doi: 10.1073/pnas.0712038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li Q., Wang C., Zhang Q., Tang C., Li N., Ruan B., Li J. Use of 18S ribosomal DNA polymerase chain reaction–denaturing gradient gel electrophoresis to study composition of fungal community in 2 patients with intestinal transplants. Hum Pathol. 2012;43:1273–1281. doi: 10.1016/j.humpath.2011.09.017. [DOI] [PubMed] [Google Scholar]
- 115.Li X.-X., Wong G.L.-H., To K.-F., Wong V.W.-S., Lai L.H., Chow D.K.-L., Lau J.Y.-W., Sung J.J.-Y., Ding C. Bacterial microbiota profiling in gastritis without Helicobacter pylori infection or non-steroidal anti-inflammatory drug use. PLoS One. 2009;4:e7985. doi: 10.1371/journal.pone.0007985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lim E.S., Zhou Y.J., Zhao G.Y., et al. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat Med. 2015;21:1228–1234. doi: 10.1038/nm.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lim E.S., Wang D., Holtz L.R. The bacterial microbiome and virome milestones of infant development. Trends Microbiol. 2016;24:801–810. doi: 10.1016/j.tim.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 118.Looft T., Johnson T.A., Allen H.K., et al. In-feed antibiotic effects on the swine intestinal microbiome. Proc Natl Acad Sci USA. 2012;109:1691–1696. doi: 10.1073/pnas.1120238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lozupone C.A., Stombaugh J.I., Gordon J.I., Jansson J.K., Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Luckey T.D. Introduction to intestinal microecology Amer. J Clin Nutr. 1972;25:1292–1294. doi: 10.1093/ajcn/25.12.1292. [DOI] [PubMed] [Google Scholar]
- 121.Lynch S.V., Pedersen O. The human intestinal microbiome in health and disease. New Engl J Med. 2016;375:2369–2379. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 122.Maathuis A., Hoffman A., Evans A., Sanders L., Venema K. The effect of the undigested fraction of maize products on the activity and composition of the microbiota determined in a dynamic in vitro model of the human proximal large intestine. J Am Coll Nutr. 2009;28:657–666. doi: 10.1080/07315724.2009.10719798. [DOI] [PubMed] [Google Scholar]
- 123.Macfarlane G.T., Cummings J.H. The colonic flora, fermentation and large bowel digestive function. Raven Press; New York, N.Y.: 1991. [Google Scholar]
- 124.Manrique P., Bolduc B., Walk S.T., van der Oost J., de Vos W.M., Young M.J. Healthy human gut phageome. Proc Natl Acad Sci USA. 2016;113:10400–10405. doi: 10.1073/pnas.1601060113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mao S.Y., Yang C.F., Zhu W.Y. Phylogenetic analysis of methanogens in the pig feces. Curr Microbiol. 2011;62:1386–1389. doi: 10.1007/s00284-011-9873-9. [DOI] [PubMed] [Google Scholar]
- 126.Mar Rodríguez M., Pérez D., Javier Chaves F., et al. Obesity changes the human gut mycobiome. Sci Rep. 2015;5:14600. doi: 10.1038/srep14600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Martinez I., Kim J., Duffy P.R., Schlegel V.L., Walter J. Resistant starches types 2 and 4 have differential effects on the composition of the fecal microbiota in human subjects. PLoS One. 2010;5:e15046. doi: 10.1371/journal.pone.0015046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.McConnell E.L., Basit A.W., Murdan S. Measurements of rat and mouse gastrointestinal pH, fluid and lymphoid tissue, and implications for in-vivo experiments. J Pharm Pharmacol. 2008;60:63–70. doi: 10.1211/jpp.60.1.0008. [DOI] [PubMed] [Google Scholar]
- 129.McHardy I.H., Goudarzi M., Tong M., et al. Integrative analysis of the microbiome and metabolome of the human intestinal mucosal surface reveals exquisite inter-relationships. Microbiome. 2013;1:17. doi: 10.1186/2049-2618-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Merchant H.A., McConnell E.L., Liu F., Ramaswamy C., Kulkarni R.P., Basit A.W., Murdan S. Assessment of gastrointestinal pH, fluid and lymphoid tissue in the guinea pig, rabbit and pig, and implications for their use in drug development. Eur J Pharm Sci. 2011;42:3–10. doi: 10.1016/j.ejps.2010.09.019. [DOI] [PubMed] [Google Scholar]
- 131.Mihajlovski A., Alric M., Brugère J.-F. A putative new order of methanogenic Archaea inhabiting the human gut, as revealed by molecular analyses of the mcrA gene. Res Microbiol. 2008;159:516–521. doi: 10.1016/j.resmic.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 132.Mikkelsen L.L., Bendixen C., Jakobsen M., Jensen B.B. Enumeration of bifidobacteria in gastrointestinal samples from piglets. Appl Environ Microbiol. 2003;69:654–658. doi: 10.1128/AEM.69.1.654-658.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mills S., Shanahan F., Stanton C., Hill C., Coffey A., Ross R.P. Movers and shakers: Influence of bacteriophages in shaping the mammalian gut microbiota. Gut Microbes. 2013;4:4–16. doi: 10.4161/gmic.22371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Minot S., Sinha R., Chen J., Li H.Z., Keilbaugh S.A., Wu G.D., Lewis J.D., Bushman F.D. The human gut virome: Inter-individual variation and dynamic response to diet. Genome Res. 2011;21:1616–1625. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mitsuoka T. Intestinal flora and aging. Nutr Rev. 1992;50:438–446. doi: 10.1111/j.1753-4887.1992.tb02499.x. [DOI] [PubMed] [Google Scholar]
- 136.Mukherjee P.K., Chandra J., Retuerto M., et al. Oral mycobiome analysis of HIV-infected patients: Identification of Pichia as an antagonist of opportunistic fungi. PLoS Pathog. 2014;10:e1003996. doi: 10.1371/journal.ppat.1003996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Nguyen T.L.A., Vieira-Silva S., Liston A., Raes J. How informative is the mouse for human gut microbiota research? Dis Mod Mech. 2015;8:1–16. doi: 10.1242/dmm.017400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Nicholson J.K., Holmes E., Wilson I.D. Gut microorganisms, mammalian metabolism and personalized health care. Nat Rev Microbiol. 2005;3:431–438. doi: 10.1038/nrmicro1152. [DOI] [PubMed] [Google Scholar]
- 139.Nielsen H.B., Almeida M., Juncker A.S., et al. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat Biotechnol. 2014;32:822–828. doi: 10.1038/nbt.2939. [DOI] [PubMed] [Google Scholar]
- 140.Noble S.M., Gianetti B.A., Witchley J.N. Candida albicans cell-type switching and functional plasticity in the mammalian host. Nat Rev Microbiol. 2017;15:96–108. doi: 10.1038/nrmicro.2016.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.O’Hara A.M., Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Odle J., Lin X., Jacobi S.K., Kim S.W., Stahl C.H. The suckling piglet as an agrimedical model for the study of pediatric nutrition and metabolism. Ann Rev Anim Biosci. 2014;2:419–444. doi: 10.1146/annurev-animal-022513-114158. [DOI] [PubMed] [Google Scholar]
- 143.Ott S.J., Kuhbacher T., Musfeldt M., et al. Fungi and inflammatory bowel diseases: Alterations of composition and diversity. Scand J Gastroenterol. 2008;43:831–841. doi: 10.1080/00365520801935434. [DOI] [PubMed] [Google Scholar]
- 144.Pang X.Y., Hua X.G., Yang Q., Ding D.H., Che C.Y., Cui L., Jia W., Bucheli P., Zhao L.P. Inter-species transplantation of gut microbiota from human to pigs. ISME J. 2007;1:156–162. doi: 10.1038/ismej.2007.23. [DOI] [PubMed] [Google Scholar]
- 145.Parfrey L.W., Walters W.A., Knight R. Microbial eukaryotes in the human microbiome: ecology, evolution, and future directions. Front Microbiol. 2011;2:153. doi: 10.3389/fmicb.2011.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pedersen R., Ingerslev H.C., Sturek M., Alloosh M., Cirera S., Christoffersen B.O., Moesgaard S.G., Larsen N., Boye M. Characterisation of gut microbiota in Ossabaw and Gottingen minipigs as models of obesity and metabolic syndrome. PLoS One. 2013;8:e 0056612. doi: 10.1371/journal.pone.0056612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Pei Z.H., Bini E.J., Yang L.Y., Zhou M.S., Francois F., Blaser M.J. Bacterial biota in the human distal esophagus. Proc Natl Acad Sci USA. 2004;101:4250–4255. doi: 10.1073/pnas.0306398101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Pflughoeft K.J., Versalovic J. Human Microbiome in Health and Disease. Ann Revi Pathol. 2012;7:99–122. doi: 10.1146/annurev-pathol-011811-132421. [DOI] [PubMed] [Google Scholar]
- 149.Pieper R., Janczyk P., Zeyner A., Smidt H., Guiard V., Souffrant W.B. Ecophysiology of the developing total bacterial and Lactobacillus communities in the terminal small intestine of weaning piglets. Microb Ecol. 2008;56:474–483. doi: 10.1007/s00248-008-9366-y. [DOI] [PubMed] [Google Scholar]
- 150.Pothoulakis C. Review article: anti-inflammatory mechanisms of action of Saccharomyces boulardii. Alimen Pharmacol Therapeut. 2009;30:826–833. doi: 10.1111/j.1365-2036.2009.04102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Pride D.T., Salzman J., Haynes M., Rohwer F., Davis-Long C., White R.A., III, Loomer P., Armitage G.C., Relman D.A. Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome. ISME J. 2012;6:915–926. doi: 10.1038/ismej.2011.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Qin J., Li R., Raes J., et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Qiu X.Y., Zhang F., Yang X., Wu N., Jiang W.W., Li X., Li X.X., Liu Y.L. Changes in the composition of intestinal fungi and their role in mice with dextran sulfate sodium-induced colitis. Sci Rep. 2015;5:10416. doi: 10.1038/srep10416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Queipo-Ortuno M.I., Seoane L.M., Murri M., Pardo M., Gomez-Zumaquero J.M., Cardona F., Casanueva F., Tinahones F.J. Gut microbiota composition in male rat models under different nutritional status and physical activity and its association with serum leptin and ghrelin levels. PLoS One. 2013;8:e65465. doi: 10.1371/journal.pone.0065465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Rajilic-Stojanovic M., Heilig H.G.H.J., Tims S., Zoetendal E.G., de Vos W.M. Long-term monitoring of the human intestinal microbiota composition. Environ Microbiol. 2013;15:1146–1159. doi: 10.1111/1462-2920.12023. [DOI] [PubMed] [Google Scholar]
- 156.Rajilic-Stojanovic M., de Vos W.M. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev. 2014;38:996–1047. doi: 10.1111/1574-6976.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Rascovan N., Duraisamy R., Desnues C. Metagenomics and the human virome in asymptomatic individuals. Gottesman S., editor. Ann Rev Microbiol. 2016;70:125–141. 125–141. doi: 10.1146/annurev-micro-102215-095431. [DOI] [PubMed] [Google Scholar]
- 158.Rastall R.A., Gibson G.R., Gill H.S., Guarner F., Klaenhammer T.R., Pot B., Reid G., Rowland I.R., Sanders M.E. Modulation of the microbial ecology of the human colon by probiotics, prebiotics and synbiotics to enhance human health: An overview of enabling science and potential applications. FEMS Microbiol Ecol. 2005;52:145. doi: 10.1016/j.femsec.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 159.Rauch M., Lynch S.V. The potential for probiotic manipulation of the gastrointestinal microbiome. Curr Opin Biotechnol. 2012;23:192–201. doi: 10.1016/j.copbio.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 160.Reyes A., Haynes M., Hanson N., Angly F.E., Heath A.C., Rohwer F., Gordon J.I. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Reyes A., Semenkovich N.P., Whiteson K., Rohwer F., Gordon J.I. Going viral: next-generation sequencing applied to phage populations in the human gut. Nat Rev Microbiol. 2012;10:607–617. doi: 10.1038/nrmicro2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Reyes A., Blanton L.V., Cao S., et al. Gut DNA viromes of Malawian twins discordant for severe acute malnutrition. Proc Natl Acad Sci USA. 2015;112:11941–11946. doi: 10.1073/pnas.1514285112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Robles-Sikisaka R., Ly M., Boehm T., Naidu M., Salzman J., Pride D.T. Association between living environment and human oral viral ecology. ISME J. 2013;7:1710–1724. doi: 10.1038/ismej.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sachsenroder J., Twardziok S.O., Scheuch M., Johne R. The general composition of the faecal virome of pigs depends on age, but not on feeding with a probiotic bacterium. PLoS One. 2014;9:e88888. doi: 10.1371/journal.pone.0088888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Savage D.C. Microbial ecology of gastrointestinal-tract. Ann Revi Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 166.Scanlan P.D., Shanahan F., O’Mahony C., Marchesi J.R. Culture-independent analyses of temporal variation of the dominant fecal microbiota and targeted bacterial subgroups in Crohn’s Disease. J Clin Microbiol. 2006;44:3980–3988. doi: 10.1128/JCM.00312-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Schwartz S., Friedberg I., Ivanov I.V., et al. A metagenomic study of diet-dependent interaction between gut microbiota and host in infants reveals differences in immune response. Genome Biol. 2012;13:r32. doi: 10.1186/gb-2012-13-4-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Scupham A.J., Presley L.L., Wei B., et al. Abundant and diverse fungal microbiota in the murine intestine. Appl Environ Microbiol. 2006;72:793–801. doi: 10.1128/AEM.72.1.793-801.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Seed P.C. The Human Mycobiome. Cold Spring Harbor Perspectives in Medicine. 2015;5 doi: 10.1101/cshperspect.a019810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sender R., Fuchs S., Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14:e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Shan T.L., Li L.L., Simmonds P., Wang C.L., Moeser A., Delwart E. The fecal virome of pigs on a high-density farm. J Virol. 2011;85:11697–11708. doi: 10.1128/JVI.05217-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Simmonds P., Adams M.J., Benko M., et al. Virus taxonomy in the age of metagenomics. Nat Rev Microbiol. 2017;15:161–168. doi: 10.1038/nrmicro.2016.177. [DOI] [PubMed] [Google Scholar]
- 173.Sogin M.L., Morrison H.G., Huber J.A., Welch D.M., Huse S.M., Neal P.R., Arrieta J.M., Herndl G.J. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci USA. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Sommer F., Bäckhed F. Know your neighbor: Microbiota and host epithelial cells interact locally to control intestinal function and physiology. BioEssays. 2016;38:455–464. doi: 10.1002/bies.201500151. [DOI] [PubMed] [Google Scholar]
- 175.Sonnenburg E.D., Smits S.A., Tikhonov M., Higginbottom S.K., Wingreen N.S., Sonnenburg J.L. Diet-induced extinctions in the gut microbiota compound over generations. Nature. 2016;529:212–215. doi: 10.1038/nature16504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Sonnenburg J.L., Backhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. 2016;535:56–64. doi: 10.1038/nature18846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Spurlock M.E., Gabler N.K. The development of porcine models of obesity and the metabolic syndrome. J Nutr. 2008;138:397–402. doi: 10.1093/jn/138.2.397. [DOI] [PubMed] [Google Scholar]
- 178.Stephen A.M., Cummings J.H. Mechanism of action of dietary fibre in the human colon. Nature. 1980;284:283–284. doi: 10.1038/284283a0. [DOI] [PubMed] [Google Scholar]
- 179.Su Y., Bian G.R., Zhu Z.G., Smidt H., Zhu W.Y. Early methanogenic colonisation in the faeces of Meishan and Yorkshire piglets as determined by pyrosequencing analysis. Archaea. 2014;2014:547908. doi: 10.1155/2014/547908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Suau A., Bonnet R., Sutren M., Godon J.J., Gibson G.R., Collins M.D., Dore J. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol. 1999;65:4799–4807. doi: 10.1128/aem.65.11.4799-4807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Suhr M.J., Hallen-Adams H.E. The human gut mycobiome: pitfalls and potentials—a mycologist’s perspective. Mycologia. 2015;107:1057–1073. doi: 10.3852/15-147. [DOI] [PubMed] [Google Scholar]
- 182.Suhr M.J., Banjara N., Hallen-Adams H.E. Sequence-based methods for detecting and evaluating the human gut mycobiome. Lett Appl Microbiol. 2016;62:209–215. doi: 10.1111/lam.12539. [DOI] [PubMed] [Google Scholar]
- 183.Tannock G.W. Exploring the relationships between intestinal microflora and inflammatory conditions of the human bowel and spine. Anton Leeuw Int J G. 2002;81:529–535. doi: 10.1023/a:1020517603993. [DOI] [PubMed] [Google Scholar]
- 184.Tannock G.W. Molecular analysis of the intestinal microflora in IBD. Mucosal Immunol. 2008;1:S15–S18. doi: 10.1038/mi.2008.54. [DOI] [PubMed] [Google Scholar]
- 185.Thaiss C.A., Zeevi D., Levy M., et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 2014;159:514–529. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
- 186.Thaiss C.A., Zeevi D., Levy M., Segal E., Elinav E. A day in the life of the meta-organism: diurnal rhythms of the intestinal microbiome and its host. Gut Microbes. 2015;6:137–142. doi: 10.1080/19490976.2015.1016690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Thaiss C.A., Levy M., Korem T., et al. Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell. 2016;167:1495–1510.e12. doi: 10.1016/j.cell.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 188.Thaiss C.A., Zmora N., Levy M., Elinav E. The microbiome and innate immunity. Nature. 2016;535:65–74. doi: 10.1038/nature18847. [DOI] [PubMed] [Google Scholar]
- 189.The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Turnbaugh P., Ley R., Mahowald M.A., Magrini V., Mardis E.R., Gordon J. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1131. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 191.Turnbaugh P.J., Gordon J.I. The core gut microbiome, energy balance and obesity. J Physiol. 2009;587:4153–4158. doi: 10.1113/jphysiol.2009.174136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Turnbaugh P.J., Ridaura V.K., Faith J.J., Rey F.E., Knight R., Gordon J.I. The effect of diet on the human gut microbiome: A metagenomic analysis in humanized gnotobiotic mice. Sci Trans Med. 2009;1:6ra14–6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Underhill D.M., Lliev L.D. The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol. 2014;14:405–416. doi: 10.1038/nri3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Urubschurov V., Janczyk P., Souffrant W.B., Freyer G., Zeyner A. Establishment of intestinal microbiota with focus on yeasts of unweaned and weaned piglets kept under different farm conditions. FEMS Microbiol Ecol. 2011;77:493–502. doi: 10.1111/j.1574-6941.2011.01129.x. [DOI] [PubMed] [Google Scholar]
- 195.van den Bogert B., Erkus O., Boekhorst J., de Goffau M., Smid E.J., Zoetendal E.G., Kleerebezem M. Diversity of human small intestinal Streptococcus and Veillonella populations. FEMS Microbiol Ecol. 2013;85:376–388. doi: 10.1111/1574-6941.12127. [DOI] [PubMed] [Google Scholar]
- 196.Vdoviaková K., Petrovová E., Maloveská M., Krešáková L., Teleky J., Elias M.Z.J., Petrášová D. Surgical anatomy of the gastrointestinal tract and its vasculature in the laboratory rat. Gastroenterol Res Pract. 20162016 doi: 10.1155/2016/2632368. Article ID 2632368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.von Rosenvinge E.C., Song Y., White J.R., Maddox C., Blanchard T., Fricke W.F. Immune status, antibiotic medication and pH are associated with changes in the stomach fluid microbiota. ISME J. 2013;7:1354–1366. doi: 10.1038/ismej.2013.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Walker A.W., Ince J., Duncan S.H., et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011;5:220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Waller A.S., Yamada T., Kristensen D.M., Kultima J.R., Sunagawa S., Koonin E.V., Bork P. Classification and quantification of bacteriophage taxa in human gut metagenomes. ISME J. 2014;8:1391–1402. doi: 10.1038/ismej.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wampach L., Heintz-Buschart A., Hogan A., et al. Colonization and succession within the human gut microbiome by Archaea, Bacteria, and Microeukaryotes during the first year of life. Front Microbiol. 2017;8:738. doi: 10.3389/fmicb.2017.00738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wang M., Ahrn S., Jeppsson B., Molin G. Comparison of bacterial diversity along the human intestinal tract by direct cloning and sequencing of 16S rRNA genes. FEMS Microbiol Ecol. 2005;54:219. doi: 10.1016/j.femsec.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 202.Wang M., Donovan S.M. Human microbiota-associated swine: Current progress and future opportunities. ILAR J. 2015;56:63–73. doi: 10.1093/ilar/ilv006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wang X., Heazlewood S.P., Krause D.O., Florin T.H.J. Molecular characterization of the microbial species that colonize human ileal and colonic mucosa by using 16S rDNA sequence analysis. J Appl Microbiol. 2003;95:508–520. doi: 10.1046/j.1365-2672.2003.02005.x. [DOI] [PubMed] [Google Scholar]
- 204.Wang Z.K., Yang Y.S., Stefka A.T., Sun G., Peng L.H. Review article: fungal microbiota and digestive diseases. Alimen Pharmacol Therapeu. 2014;39:751–766. doi: 10.1111/apt.12665. [DOI] [PubMed] [Google Scholar]
- 205.Warden C.H., Fisler J.S. Comparisons of diets used in animal models of high fat feeding. Cell Metabol. 2008;7:277. doi: 10.1016/j.cmet.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Weaver C.M., McCabe L.D., McCabe G.M., Novotny R., Van Loan M.D., Going S.B., Boushey C., Savaiano D.A., Matkovic V. Bone mineral and predictors of whole body, total hip, and lumbar spine for 740 early pubertal white, Hispanic, and Asian girls. J Bone Miner Res. 2005;20:S314–S314. [Google Scholar]
- 207.Weaver C.M., Martin B.R., Story J.A., Hutchinson I., Sanders L. Novel fibers Increase bone calcium content and strength beyond efficiency of large intestine fermentation. J Ag Food Chem. 2010;58:8952–8957. doi: 10.1021/jf904086d. [DOI] [PubMed] [Google Scholar]
- 208.Weaver C.M., Martin B.R., Nakatsu C.H., et al. Galactooligosaccharides improve mineral absorption and bone properties in growing rats through gut fermentation. J Ag Food Chem. 2011;59:6501–6510. doi: 10.1021/jf2009777. [DOI] [PubMed] [Google Scholar]
- 209.Welch J.L.M., Rossetti B.J., Rieken C.W., Dewhirst F.E., Borisy G.G. Biogeography of a human oral microbiome at the micron scale. Proc Natl Acad Sci USA. 2016;113:E791–E800. doi: 10.1073/pnas.1522149113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Whisner C.M., Martin B.R., Nakatsu C.H., McCabe G.P., McCabe L.D., Peacock M., Weaver C.M. Soluble maize fibre affects short-term calcium absorption in adolescent boys and girls: a randomised controlled trial using dual stable isotopic tracers. Brit J Nutr. 2014;112:446–456. doi: 10.1017/S0007114514000981. [DOI] [PubMed] [Google Scholar]
- 211.Whisner C.M., Martin B.R., Nakatsu C.H., Story J.A., MacDonald-Clarke C.J., McCabe L.D., McCabe G.P., Weaver C.M. Soluble corn fiber increases calcium absorption associated with shifts in the gut microbiome: A randomized dose-response trial in free-living pubertal girls. Nutr J. 2016;146:1298–1306. doi: 10.3945/jn.115.227256. [DOI] [PubMed] [Google Scholar]
- 212.Wilson K., Blitchington R. Human colonic biota studied by ribosomal DNA sequence analysis. Appl Environ Microbiol. 1996;62:2273–2278. doi: 10.1128/aem.62.7.2273-2278.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Woodmansey E.J. Intestinal bacteria and ageing. J Appl Microbiol. 2007;102:1178–1186. doi: 10.1111/j.1365-2672.2007.03400.x. [DOI] [PubMed] [Google Scholar]
- 214.Wright C.J., Burns L.H., Jack A.A., Back C.R., Dutton L.C., Nobbs A.H., Lamont R.J., Jenkinson H.F. Microbial interactions in building of communities. Mol Oral Microbiol. 2013;28:83–101. doi: 10.1111/omi.12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Wylie K.M., Mihindukulasuriya K.A., Zhou Y., Sodergren E., Storch G.A., Weinstock G.M. Metagenomic analysis of double-stranded DNA viruses in healthy adults. BMC Biol. 2014;12:71. doi: 10.1186/s12915-014-0071-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Xiao L., Feng Q., Liang S., et al. A catalog of the mouse gut metagenome. Nat Biotechnol. 2015;33:1103–1108. doi: 10.1038/nbt.3353. [DOI] [PubMed] [Google Scholar]
- 217.Xiao L., Estellé J., Kiilerich P., et al. A reference gene catalogue of the pig gut microbiome. Nat Microbiol. 2016;1:16161. doi: 10.1038/nmicrobiol.2016.161. [DOI] [PubMed] [Google Scholar]
- 218.Yan H., Potu R., Lu H., et al. Dietary fat content and fiber type modulate hind gut microbial community and metabolic markers in the pig. PLoS One. 2013;8:e59581. doi: 10.1371/journal.pone.0059581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Yatsunenko T., Rey F.E., Manary M.J., et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Zhang Q., Widmer G., Tzipori S. A pig model of the human gastrointestinal tract. Gut Microbes. 2013;4:193–200. doi: 10.4161/gmic.23867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Zhu Y.Y., Li H., Xu X.L., Li C.B., Zhou G.H. The gut microbiota in young and middle-aged rats showed different responses to chicken protein in their diet. BMC Microbiol. 2016;16:281. doi: 10.1186/s12866-016-0895-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Zoetendal E.G., von Wright A., Vilpponen-Salmela T., Ben-Amor K., Akkermans A.D.L., de Vos W.M. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol. 2002;68:3401–3407. doi: 10.1128/AEM.68.7.3401-3407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Zoetendal E.G., Raes J., van den Bogert B., et al. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J. 2012;6:1415–1426. doi: 10.1038/ismej.2011.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Zwolinska-Wcislo M., Budak A., Trojanowska D., Bogdal J., Stachura J. Fungal colonization of the stomach and its clinical relevance. Mycoses. 1998;41:327–334. doi: 10.1111/j.1439-0507.1998.tb00346.x. [DOI] [PubMed] [Google Scholar]