Abstract
Conditional knockout mice with targeted disruption of B-cell associated protein (BAP)31 in adult mouse liver were generated and challenged with a high-fat diet (HFD) for 36 or 96 days and markers of obesity, diabetes, and hepatic steatosis were determined. Mutant mice were indistinguishable from WT littermates, but exhibited increased HFD-induced obesity. BAP31-deletion in hepatocytes increased the expression of SREBP1C and the target genes, including acetyl-CoA carboxylase 1 and stearoyl-CoA desaturase-1, and increased hepatic lipid accumulation and HFD-induced liver steatosis. Immunoprecipitation assay showed that BAP31 interacts with SREBP1C and insulin-induced gene 1 (INSIG1), and BAP31-deletion reduces INSIG1 expression, suggesting that BAP31 may regulate SREBP1C activity by modulating INSIG1 protein levels. Additionally, BAP31-deletion induced glucose and insulin intolerance, decreased Akt and glycogen synthase kinase 3β phosphorylation, and enhanced hepatic glucose production in mice. Expression of endoplasmic reticulum (ER) stress markers was significantly induced in BAP31-mutant mice. HFD-induced inflammation was aggravated in mutant mice, along with increased c-Jun N-terminal kinase and nuclear factor-κB activation. These findings demonstrate that BAP31-deletion induces SREBP activation and promotes hepatic lipid accumulation, reduces insulin signaling, impairs glucose/insulin tolerance, and increases ER stress and hepatic inflammation, explaining the protective roles of BAP31 in the development of liver steatosis and insulin resistance in HFD-induced obesity in animal models.
Keywords: B-cell associated protein 31, liver steatosis, diabetes, insulin resistance, endoplasmic reticulum stress, sterol regulatory element-binding protein 1C, B-cell activated protein 31
Nonalcoholic fatty liver disease (NAFLD), one of the most common chronic liver disorders, affects up to one-third of adults in the USA (1). NAFLD begins with lipid deposition in the liver (liver steatosis) and may progress to become nonalcoholic steatohepatitis (NASH), a state in which steatosis is combined with inflammation and fibrosis, with approximately 20% of patients developing cirrhosis, terminal liver failure, or hepatocellular carcinoma (2). NAFLD is highly associated with the features of metabolic syndrome, including obesity, T2D, and dyslipidemia, and promotes the development of insulin resistance (IR), characterized by fasting hyperglycemia and enhanced hepatic glucose production (3).
SREBPs, which belong to the basic helix-loop-helix-leucine zipper family of transcription factors, contain three isoforms, namely, SREBP1A, SREBP1C, and SREBP2, and are key regulators of NAFLD (4). Newly synthesized SREBPs, or precursor SREBPs (pSREBPs), are anchored in the endoplasmic reticulum (ER) membranes and form a tight complex with SREBP cleavage-activating protein (SCAP). In sterol-depleted cells, the SREBP/SCAP complex is incorporated into COPII-coated vesicles. Subsequently, the vesicles migrate from the ER and escort SREBPs to the Golgi apparatus, where they are cleaved by two proteases. This process liberates the transcription factor portion into the cytosol, where it translocates into the nucleus [i.e., nuclear (n)SREBP] and activates the transcription of multiple genes involved in FA and cholesterol (Chol) synthesis, including FAS, acetyl-CoA carboxylase 1 (ACC1), stearoyl-CoA desaturase-1 (SCD1), HMG-CoA synthase (HMG-CoA Syn), and HMG-CoA reductase (HMG-CoA Red), thus leading to enhanced hepatic lipid accumulation (5, 6). Sterols promote the binding of insulin-induced gene 1 (INSIG1) to SCAP, causing the retention of the SREBP/SCAP complex in the ER, thereby limiting the transcription of SREBP target genes (7). When sterol is scarce, the SREBP/SCAP complex dissociates from INSIG1 and the dissociated INSIG1 is then degraded via the ubiquitin-proteasome system, thereby promoting SREBP proteolytic processing and inducing downstream signal transduction (8). Additionally, hypotonic or ER stress induces SREBP activation, which is highly associated with the degradation of INSIG1 (9), and deletion of INSIG1 results in constitutive activation of SREBP (10). In contrast, overexpression of INSIG1 in the liver inhibits SREBP processing and reduces insulin-induced lipogenesis, supporting a possible mechanism(s) of regulating nSREBP expression and signaling pathway activation by modulating the cellular protein levels of INSIG1 (11).
B-cell associated protein (BAP)31, which is one of the most abundant ER-associated proteins (12, 13), plays an integral role in the recognition of misfolded protein by triggering ER-associated degradation (ERAD) (14). Our previous study reported that BAP31 promoted the retrotranslocation of CFTRΔF508 into the cytosol and maintained the cellular protein-folding homeostasis (15). However, the function of BAP31 in glucose and lipid metabolism is uncertain. Patients with BAP31 and ABCD1 deletion presented with infantile death and liver dysfunction of intracanalicular and ductal cholestasis (16). In addition, patients exhibited hepatomegaly, mild to moderate fibrosis, vacuoles in hepatocytes (indicating excess hepatic lipid accumulation), and an increase of serum alanine transaminase (ALT) and aspartate aminotransferase (AST) levels, suggesting that the contiguous mutation in BAP31 and ABCD1 may result in fatty liver (17). The contiguous deletion of BAP31 and solute carrier family 6 member 8 (SLC6A8) also leads to liver dysfunction (18). However, Cacciagli et al. (19) reported that mutations in BAP31 caused deafness, dystonia, and disturbances of ER metabolism, but they failed to detect liver dysfunction, suggesting that BAP31-deletion would not induce liver disease, or even fatty liver. This inconsistency may be due to the cases that had reported the contiguous mutation of BAP31 with the adjacent genes of ABCD1 and SLC6A8, and not the single gene mutation of BAP31, which may explain the conflicting clinical presentation and complicate the interpretation of the data.
In this study, conditional knockout mice with targeted disruption of BAP31 in the adult mouse liver were generated. The mice were fed a high-fat diet (HFD) for 36 or 96 days and markers of obesity, liver steatosis, ER stress, and hepatic inflammation were determined. We demonstrate that BAP31-deletion in hepatocytes induces SREBP activation and increases lipogenic gene expression and hepatic lipid accumulation. Additionally, BAP31-deletion in hepatocytes impairs insulin signaling, increases hepatic gluconeogenesis, aggravates ER stress and hepatic inflammation, and eventually worsens IR in mice.
MATERIALS AND METHODS
Generation of mice for hepatocyte-specific deletion of BAP31
Details on targeting construct and targeting procedure can be found in the supplemental materials. Mice having the BAP31flox allele (BAP31flox/−) on the C57BL/6 background were mated with the transgenic mice expressing Cre recombinase under the control of rat albumin promoter (Alb-Cre mice) to obtain hemizygous Alb-Cre;BAP31flox/− offspring, which were then crossed with BAP31flox/flox to obtain the hemizygous Alb-Cre;BAP31flox/− of BAP31flox allele with Alb-Cre recombinase combined (MT) and their BAP31flox/− allele littermates (WT). Age-matched male WT and MT mice were used. Mice were housed in light- and humidity-controlled rooms with a 12 h light/dark cycle. All animal procedures were approved by the institutional review board of Northeastern University in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
HFD-induced obesity
Male WT and MT mice (4–6 weeks old) were fed a standard chow diet (SD) (LM 485, 10 kcal% fat; Harlan Teklad) or HFD (Research Diets 12492, 60 kcal% fat) for a short-term (36 days) or a long-term (96 days). Then the liver, heart, kidneys, and white adipose tissue (WAT) were collected and weighed.
Food intake measurement
Male mice of the four experimental groups were housed individually. Food intake for each mouse was measured every day for at least 1 week.
Glucose tolerance test, insulin tolerance test, and pyruvate tolerance test
Male WT and MT mice fasted 16, 5, or 16 h were injected (intraperitoneally) with glucose (2 g/kg), insulin (0.75 U/kg), or sodium pyruvate (2 g/kg) for glucose tolerance test, insulin tolerance test, and pyruvate tolerance test, respectively. Blood glucose was determined by measuring tail blood at 0, 15, 30, 60, and 120 min post glucose, insulin, or pyruvate solution administration.
Acute insulin treatment in vivo
Male WT and MT mice (8–10 weeks old) fasted overnight were anesthetized (ketamine, 150 mg/kg; xylazine, 5 mg/kg) and a maximal bolus of insulin (5 U/kg) was administered via the portal vein for 5 min. Then the liver tissues and gastrocnemius were collected and snap-frozen in liquid nitrogen for the next steps.
Fasting/refeeding treatment
Male WT and MT mice (16–18 weeks old) fasted 24 h were refed with the SD for 1 h or 6 h. Then mice were euthanized under anesthesia and the liver tissues were collected and snap-frozen in liquid nitrogen for the next steps.
Cell culture and primary hepatocyte glucose production
shRNA lentivirus was obtained from Novobio Scientific (Shanghai, China). HepG2 cells were infected with shRNA targeting BAP31 and the scrambled nontarget negative control based on the manufacturer’s instructions. Primary hepatocytes isolated from male WT and MT mice using two-step collagenase perfusion (Thermo Fisher Scientific) were seeded on collagen-coated 6-well plates, as described before (20). Hepatocytes were cultured in serum-free William’s E medium containing 1% ITS supplement (Thermo Fisher Scientific) and 100 nM dexamethasone (Sigma-Aldrich) for 16 h, then washed with sterile PBS and the glucose production was determined after an 8 h incubation in glucose-free William’s E medium containing lactate/sodium pyruvate (10:1 mM) and 100 nM dexamethasone alone or with 8-Br-cAMP (100 μM) (Sigma-Aldrich) (21).
Measurement of serum metabolites and liver extracts
Serum insulin and TNFα were measured using a mouse insulin and TNFα ELISA kit (RayBiotech, Norcross, GA; eBioscience, San Diego, CA). Interleukin (IL)-1β in liver extracts was determined by ELISA kit from R&D Systems (Minneapolis, MN) and was normalized with protein content. Triglycerides (TGs), FFA, and Chol were determined with reagent kits (Pointe Scientific, Canton, MI; Wako Chemicals USA, North Chesterfield, VA). Glucose, β-hydroxybutyrate, HDL-Chol, LDL-Chol, AST, ALT, and total bile acid were determined with reagent kits from Nanjing Jiancheng Biomedical Company (Nanjing, China). Tissues (50 mg) or feces (100 mg) were homogenized with PBS and the lipids were extracted with a mixture of chloroform-methanol (2:1; v/v), as described before (22). The lipid content was normalized with tissue or feces weight.
Histopathology
Sections (5 μm) of paraffin-embedded livers were cut and stained with hematoxylin and eosin before histopathologic analysis. For Oil Red O staining, frozen sections were fixed and incubated with Oil Red O solution (six parts Oil Red O stock solution and four parts water; Oil Red O stock solution is 0.5% Oil Red O in 100% isopropanol) for 15 min, then counterstained with hematoxylin and mounted in glycerin jelly.
RNA isolation and real-time PCR
Total RNA was isolated using TRIzol reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. Two micrograms of total RNA were converted to cDNA and the relative mRNA levels were quantified using a CFX96 Touch™ real-time PCR detection system (Bio-Rad Laboratories). SYBR green chemistry was used. The sequences of primers used are listed in supplemental Table S1.
Immunoblot analysis
Nuclear proteins from liver tissues or HepG2 cells were isolated with an NE-PER kit following the manufacturer’s instructions (Pierce, Rockford, IL). Homogenates or nuclear extracts were resolved by SDS-PAGE and then transferred to PVDF membrane. The membrane was blocked with 5% nonfat dry milk in TBST followed by incubation with primary antibodies overnight. Bands were visualized with a Bio-Rad ChemiDoc™ imaging system using an ECL detection kit. The sources and dilutions of antibodies are listed in supplemental Table S2.
Immunoprecipitation
Liver extracts [50 mM HEPES buffer (pH 7.6), 100 mM NaCl, 1.5 mM MgCl2, 1% (w/v) CHAPS, and protease inhibitors] were incubated with a specific antibody against BAP31 at 4°C overnight. Protein G Sepharose beads were added next, followed by incubation for 4 h at 4°C. After washing three times with lysis buffer, the immune complexes were resolved on SDS-PAGE. Blots were incubated with anti-SREBP1, anti-INSIG1, or anti-SCAP antibodies.
Statistical analysis
Quantitative data are presented as mean ± SEM. Statistical differences were determined by a two-way ANOVA followed by Student-Newman-Keuls comparisons using SPSS 13.0 or by a two-tailed Student’s t-test for two independent samples. All statistical tests with P < 0.05 were considered to be significant.
RESULTS
BAP31-deletion in hepatocytes increases HFD-induced obesity
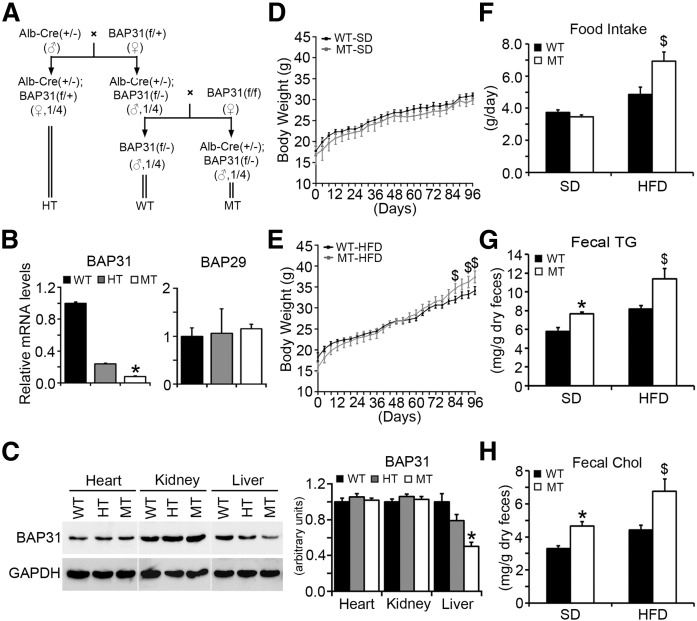
A conditional knockout mouse model containing a genetically engineered BAP31flox/+ allele in which exon 3 is flanked by loxP sites was generated. Detailed information about the construct design and the generation of BAP31 floxed allele mice can be found in the supplemental materials (supplemental Fig. S1) and the breeding strategy is described in the Materials and Methods. The offspring were born with Mendelian frequency (Fig. 1A, the frequency of female mice has not been shown). As the BAP31 gene is X-chromosome linked, one allele deletion results in 93% mRNA ablation in male hemizygous mice (MT) and 79% mRNA ablation in female heterozygous mice (HT) (Fig. 1B, left). BAP29, the homolog of BAP31, forming the complex of BAP29/BAP31 with BAP31, and is preferentially interacted with membrane protein of immunoglobulin D (12); and the mRNA levels were similar among these three types of mice, suggesting that BAP31-deletion in hepatocytes did not alter the homolog family expression of BAP29 (Fig. 1B, right). Immunoblot analysis confirmed that BAP31 was appropriately ablated and that the gene deletion was specifically targeted to the liver, with no deletion observed in the heart and kidney tissues (Fig. 1C).
Fig. 1.
BAP31-deletion in hepatocytes increases HFD-induced obesity. A: Breeding scheme for generating WT, HT, and MT mice. B: BAP31 and BAP29 relative mRNA levels from livers of WT, HT, and MT mice. C: Immunoblot analysis of BAP31 in heart, kidney, and liver tissues from WT, HT, and MT mice. Body weight of WT and MT mice fed the SD (D) or HFD (E) for 96 days. Body weight was measured every 4 days (n = 6–7 per group). F: Food intake from the four experimental groups. Fecal TGs (G) and Chol (H) are shown in WT and MT mice fed the SD or HFD for 96 days (n = 6–7 per group). *P < 0.05, MT compared with WT mice fed SD. $P < 0.05, MT compared with WT mice fed HFD.
In order to evaluate the function of BAP31 in lipid metabolism, WT and MT mice were subjected to HFD-feeding for multiple time-courses: 36 days of feeding for the short-term duration and 96 days for the long-term duration. No difference of body weight between the WT and MT mice over time on the SD was observed (Fig. 1D). MT mice were 10% heavier than WT littermates at the end of the treatment (Fig. 1E, Table 1). Liver, heart, and kidney weights were similar. Higher food intake may have contributed partly to the higher body weight in MT mice (Fig. 1F). Fecal TG and Chol were higher in MT mice than in WT mice (Fig. 1G, H). No difference in serum glucose, insulin, TG, β-hydroxybutyrate, Chol, HDL-Chol, or LDL-Chol was observed between WT and MT mice, except for serum FFA, which was increased in MT mice fed SD or HFD (P = 0.09) (Table 1). MT mice that were fed a HFD for 36 days had increased body weight gain compared to WT mice. Liver weight was increased by 13% in MT mice fed HFD for 36 days. No difference was observed in heart and kidney weights of these mice (supplemental Fig. S2).
TABLE 1.
Metabolic parameters of serum measured in WT and MT mice fed the SD or HFD
| Parameters | Unit | SD | HFD | ||
| WT | MT | WT | MT | ||
| Body weight | g | 30.63 ± 0.61 | 29.85 ± 0.41 | 34.15 ± 0.96 | 37.40 ± 1.40a |
| Liver weight | g | 1.58 ± 0.04 | 1.59 ± 0.04 | 1.65 ± 0.08 | 1.78 ± 0.07 |
| Heart weight | g | 0.13 ± 0.01 | 0.12 ± 0.01 | 0.14 ± 0.01 | 0.14 ± 0.01 |
| Kidney weight | g | 0.37 ± 0.01 | 0.36 ± 0.01 | 0.43 ± 0.02 | 0.43 ± 0.02 |
| Glucose | mg/dl | 179.52 ± 6.14 | 169.46 ± 18.69 | 168.92 ± 10.51 | 170.43 ± 10.66 |
| Insulin | μU/ml | 24.36 ± 3.25 | 25.11 ± 0.58 | 38.44 ± 7.87 | 33.88 ± 2.78 |
| TGs | mg/dl | 94.32 ± 7.10 | 101.47 ± 10.32 | 117.23 ± 14.89 | 106.39 ± 7.76 |
| FFA | mEq/l | 0.61 ± 0.05 | 0.71 ± 0.05b | 0.66 ± 0.08 | 0.84 ± 0.08 |
| β-Hydroxybutyrate | mmol/l | 0.15 ± 0.01 | 0.16 ± 0.02 | 0.14 ± 0.01 | 0.14 ± 0.01 |
| Chol | mg/dl | n.d. | n.d. | 169.93 ± 18.02 | 174.09 ± 8.93 |
| HDL-Chol | mg/dl | 12.72 ± 0.43 | 12.98 ± 0.51 | 32.39 ± 2.82 | 32.81 ± 1.86 |
| LDL-Chol | mg/dl | 16.98 ± 1.29 | 15.06 ± 1.18 | 72.80 ± 12.68 | 89.81 ± 3.59 |
Body weight, liver weight, heart weight, kidney weight, serum glucose, insulin, TGs, FFA, β-hydroxybutyrate, Chol, HDL-Chol, LDL-Chol were measured from WT and MT mice fed the SD or HFD for 96 days. Data are shown as mean ± SEM (n = 6–7 per group). n.d., not detected.
P < 0.05, MT compared with WT mice fed HFD.
P < 0.05, MT compared with WT mice fed the SD.
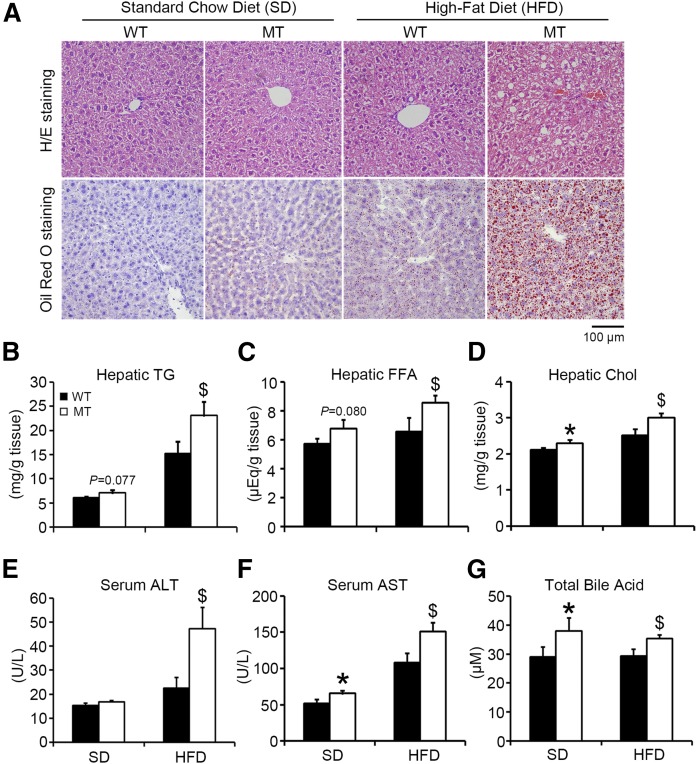
BAP31-deletion in hepatocytes increases hepatic lipid accumulation and HFD-induced liver steatosis
There were higher neutral TG and lipid staining in MT mice than in WT mice fed HFD (Fig. 2A). Compared with WT mice, hepatic TG, FFA, and Chol were increased by 16% (P = 0.077), 19% (P = 0.080), and 8% in MT mice fed SD, respectively, and were increased by 52, 31, and 20% in MT mice fed HFD, respectively (Fig. 2B–D). Additionally, hepatic TG was increased in MT mice fed HFD for 36 days; however, no difference was observed in hepatic FFA and Chol (supplemental Fig. S3). Serum ALT was increased by 110% in MT mice fed HFD compared to WT mice (Fig. 2E). AST was increased by 27% in MT compared with WT mice fed SD, and increased by 40% compared with those fed HFD (Fig. 2F). Total bile acid was increased by 31% in MT mice compared with WT mice, and increased by 20% compared with those fed HFD (Fig. 2G).
Fig. 2.
BAP31-deletion in hepatocytes increases hepatic lipid accumulation and HFD-induced liver steatosis. A: Representative hematoxylin and eosin (H/E) staining (top) and Oil Red O staining (bottom) of livers from WT and MT mice fed the SD or HFD for 96 days (200× magnification, scale bar = 100 µm). Hepatic TGs (B), FFA (C), and Chol(D) from WT and MT mice fed the SD or HFD for 96 days. Serum ALT (E), AST (F), and total bile acid (G) from mice fed the SD or HFD for 96 days (n = 6–7 per group). *P < 0.05, MT compared with WT mice fed SD. $P < 0.05, MT compared with WT mice fed HFD.
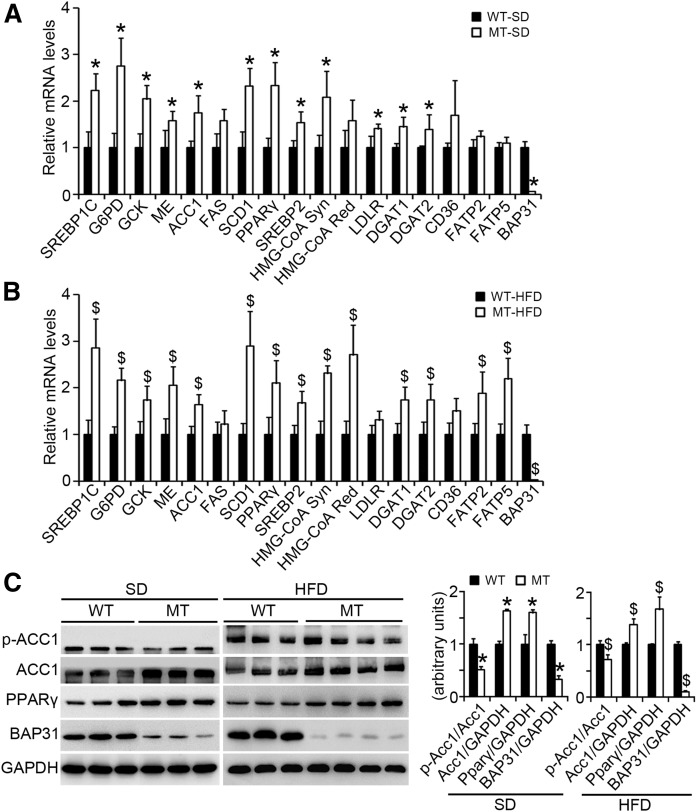
BAP31-deletion in hepatocytes induces lipogenic gene expression in mouse livers
BAP31 was ablated in MT mice. SREBP1C and the target genes, including glucose-6-phosphate dehydrogenase (G6PD), glucokinase (GCK), malic enzyme (ME), ACC1, and SCD1, were significantly induced in MT mice fed SD or HFD. SREBP2 and the target genes of HMG-CoA Syn and LDL receptor (LDLR) were also induced. Diacylglycerol acyltransferases (DGATs) (DGAT1 and DGAT2), which catalyze the formation of TG from diacylglycerol and acyl-CoA, considered as the terminal and only committed step in TG synthesis (23), were also increased in MT mice. No change in FA translocase [cluster of differentiation 36 (CD36)] expression was determined. FA transport proteins (FATPs) (FATP2 and FATP5) were increased in MT mice fed HFD (Fig. 3A, B). Total ACC1 protein levels were increased and the phosphorylation levels (p-ACC1) were decreased in MT mice. PPARγ was elevated in MT mice (Fig. 3C). In the 36 day HFD-feeding, PPARγ, ACC1, and FAS were increased, but no change in CD36, DGAT1, and DGAT2 expression between WT and MT mice was observed (supplemental Fig. S4).
Fig. 3.
BAP31-deletion in hepatocytes induces lipogenic gene expression in mouse liver. A, B: Total RNA was extracted from livers of WT and MT mice fed the SD or HFD for 96 days. The relative mRNA levels were determined by real-time PCR and normalized with 18S rRNA levels (n = 6–7 per group). C: Immunoblot analysis of proteins associated with lipogenesis in WT and MT mice fed the SD or HFD for 96 days. *P < 0.05, MT compared with WT mice fed SD. $P < 0.05, MT compared with WT mice fed HFD.
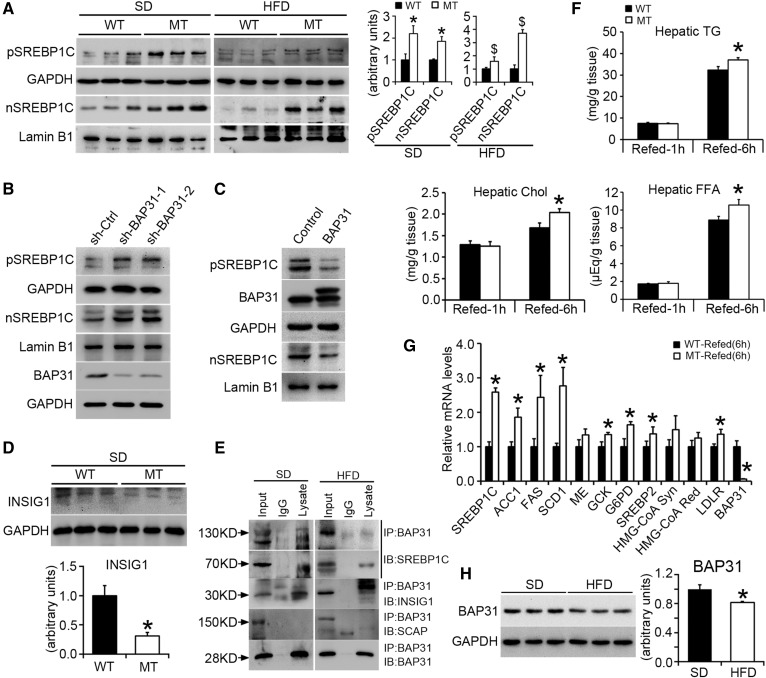
BAP31-deletion in hepatocytes promotes SREBP signaling activation and increases the response to fasting/refeeding treatment
SREBPs function as the master regulators of FA and Chol biosynthesis (24). The mature form of nSREBP1C was more abundant in MT mice, indicating that BAP31-deletion may enhance SREBP1C proteolytic cleavage (Fig. 4A). The mRNA and the precursor form of SREBP1C were also robustly induced, suggesting that the enhanced proteolytic cleavage of the precursor is compensated for by the induced SREBP1C transcription, which may be due to a feed-forward mechanism, as the SREBP1C promoter is the target of mature SREBP1C itself (Fig. 3A, B). HepG2 cells stably infected with lentivirus targeting BAP31 exhibited enhanced nSREBP1C and pSREBP1C expression (Fig. 4B), and overexpression of BAP31 reduced nSREBP1C and pSREBP1C expression (Fig. 4C). INSIG1, which mediates the export of the SREBP/SCAP complex from the ER to the Golgi apparatus, was significantly reduced in MT mice, thereby facilitating the export of SREBP1C to the Golgi apparatus and inducing the activation of downstream genes (Fig. 4D). Data from the immunoprecipitation assay further demonstrated that BAP31 interacted with SREBP1C and INSIG1. However, no interaction between BAP31 and SCAP was observed, suggesting that BAP31 may form a complex with SREBP1C and INSIG1, and that modulate SREBP1C activation by modulating INSIG1 cellular content in mouse livers (Fig. 4E).
Fig. 4.
BAP31-deletion in hepatocytes promotes SREBP signaling activation and increases the response to fasting/refeeding treatment. A: Immunoblot analysis of SREBP1C precursor and nuclear form in WT and MT mice fed the SD or HFD for 96 days. *P < 0.05, MT compared with WT mice fed SD. $P < 0.05, MT compared with WT mice fed HFD. B, C: Immunoblot analysis of SREBP1C precursor and nuclear form in HepG2 cell lines with stable knockdown of the BAP31 gene or with overexpression of the BAP31 gene. D: Immunoblot analysis of INSIG1 in WT and MT mice fed the SD. Statistical differences were determined by a two-tailed Student’s t-test. *P < 0.05, MT compared with WT mice fed SD. E: Liver lysates (SD and HFD mouse livers) were subjected to immunoprecipitation assay with polyclonal anti-BAP31 antibody. Immunoprecipitated proteins were probed by immunoblot with anti-SREBP1, anti-INSIG1, anti-SCAP, and anti-BAP31 antibodies. F: Male WT and MT mice were fasted 24 h and then refed the SD for 1 h or 6 h, respectively. Hepatic TGs, FFA, and Chol were extracted and quantified spectrophotometrically (n = 6 per group). G: Total RNA was extracted from the livers of WT and MT mice fasted 24 h followed by refeeding the SD for 6 h. The relative mRNA levels were determined by real-time PCR and normalized with 18S rRNA levels (n = 6 per group). *P < 0.05, MT compared with WT mice refed for 6 h. H: Immunoblot analysis of BAP31 in mice fed the SD or HFD for 96 days. GAPDH and lamin B1 were used as a loading control. Statistical differences were determined by a two-tailed Student’s t-test. *P < 0.05, mice fed HFD compared with SD.
It has been widely recognized that SREBP1C plays key roles in response to fasting and refeeding treatment, and is involved in nutrition induction of lipogenic enzyme genes (25). In order to confirm that BAP31-deletion in hepatocytes induced SREBP1C activation in the liver, WT and MT mice were fasted for 24 h and then refed with the SD for 1 or 6 h. The hepatic lipid accumulation did not change much between these two genotypes of mice following the 1 h refeeding, but was increased significantly in MT mice upon the 6 h refeeding (Fig. 4F). A quantitative real-time PCR assay demonstrated that BAP31-deletion in hepatocytes increased the induction of lipogenic genes, including ACC1, FAS, and SCD1, as well as other genes, such as ME, GCK, G6PD, and LDLR in mouse livers, suggesting that there is an increase of SREBP1C activation in MT mice following fasting/refeeding treatment (Fig. 4G). It was noted that HFD-treatment for 96 days reduced BAP31 protein levels in mouse livers (Fig. 4H).
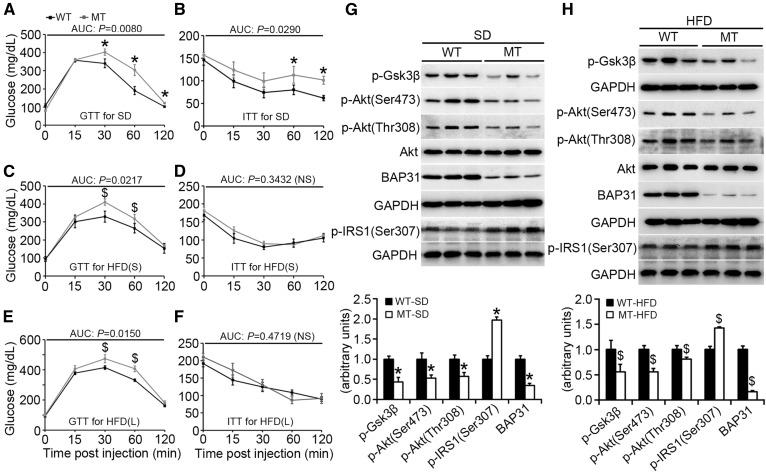
BAP31-deletion in hepatocytes impairs glucose and insulin tolerance in mice
MT mice exhibited glucose and insulin intolerance compared with WT mice fed SD (Fig. 5A, B) and continuously exhibited glucose intolerance following HFD-feeding, both short- and long-term (Fig. 5C, E), but did not feature more insulin intolerance (Fig. 5D, F). The relative protein levels of p-Akt (Ser473) and p-Akt (Thr308) were decreased in MT mice, with no change in Akt expression. The phosphorylation levels of the target gene of glycogen synthase kinase 3β (Gsk3β) [phosphorylated Gsk3β (p-Gsk3β)] were also decreased in WT mice, suggesting that BAP31-deletion in hepatocytes reduces Akt activation, thus resulting in glucose and insulin intolerance. BAP31-deletion in hepatocytes increased insulin receptor substrate 1 (IRS1) phosphorylation at serine (307) in mouse livers, when fed either SD or HFD (Fig. 5G, H). BAP31-deletion in the liver did not change p-Akt or p-Gsk3β levels in WAT, which had normal expression levels of BAP31 (supplemental Fig. S5), indicating that the reduced Akt activation in MT mice was due specifically to BAP31-deletion in hepatocytes.
Fig. 5.
BAP31-deletion in hepatocytes impairs glucose and insulin tolerance in mice. Blood glucose concentration during glucose tolerance test (GTT) and insulin tolerance test (ITT) assay from WT and MT mice fed the SD (n = 5–6 per group) (A, B), the HFD for short-term (36 days) [HFD(S)] (n = 5–6 per group) (C, D), or the HFD for long-term (96 days) [HFD(L)] (n = 6–7 per group) (E, F). AUC (area under the curve) was calculated for each assay. *P < 0.05, MT compared with WT mice fed SD. $P < 0.05, MT compared with WT mice fed HFD. G, H: Immunoblot analysis of Akt, p-Akt (Ser473), p-Akt (Thr308), p-Gsk3β, p-IRS1(Ser307), and BAP31 in livers of mice fed the SD or HFD (long-term). GAPDH was used as a loading control.
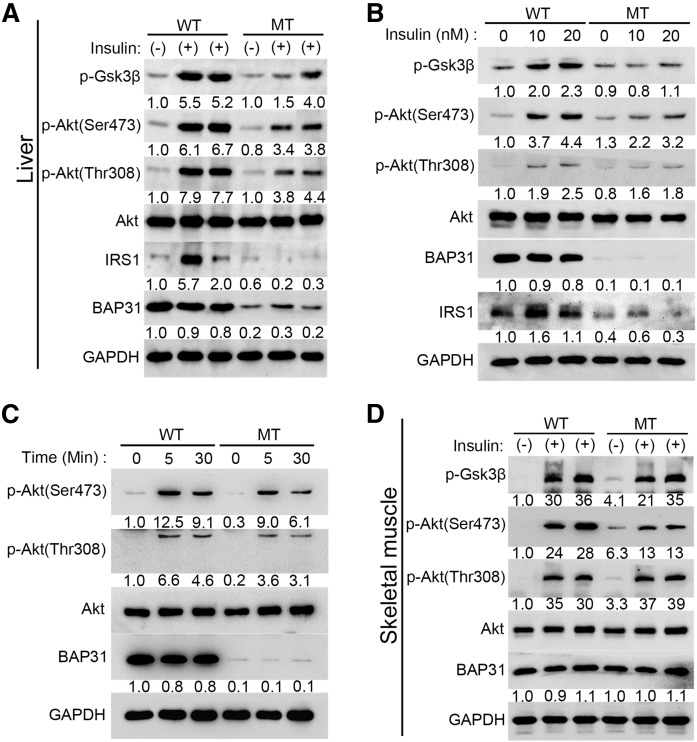
BAP31-deletion in hepatocytes impairs insulin signal transduction in vivo and in vitro
To confirm BAP31 effects on insulin signal transduction, mice fasted overnight were perfused with insulin for 5 min, and then the phosphorylation levels of Akt were determined. Insulin administration induced IRS1 expression and increased p-Akt (Ser473 and Thr308) and p-Gsk3β levels in WT mice, but the induction was dampened in MT mice (Fig. 6A). Primary hepatocytes were isolated and the insulin response ability was evaluated in vitro. Insulin significantly induced IRS1 expression and promoted p-Akt (Ser473), p-Akt (Thr308), and p-Gsk3β levels in WT hepatocytes, but these effects were completely blocked in MT hepatocytes via a dosage-dependent mechanism (Fig. 6B). Hepatocytes exhibited the peak of the phosphorylation levels of Akt (Ser473 and Thr308) at 5 min post insulin administration and then decreased at the 30 min time point. This induction was significantly reduced in MT hepatocytes in a time-course treatment (Fig. 6C), demonstrating that BAP31-deletion impairs insulin signal transduction in hepatocytes, which is consistent with the in vivo observations. In skeletal muscles, there was no difference in p-Akt and p-Gsk3β levels in response to insulin administration between WT and MT mice, which exhibited similar BAP31 expression, further suggesting that the impairment of insulin signal transduction is liver specific (Fig. 6D).
Fig. 6.
BAP31-deletion in hepatocytes impairs insulin signal transduction in vivo and in vitro. Mice (8–10 weeks old) fasted overnight were anesthetized with ketamine (150 mg/kg) and xylazine (5 mg/kg) and then perfused with insulin for 5 min (5 U/kg body weight), the liver tissues (A) and gastrocnemius (D) were excised and frozen immediately. The protein levels of Akt, p-Akt (Ser473), p-Akt (Thr308), p-Gsk3β, and IRS1 were determined by immunoblot analysis. Primary hepatocytes were isolated from 2- to 3-month-old male WT and MT mice and cultured in serum-free William’s E medium containing 1% ITS supplement and 100 nM dexamethasone for 16 h. The next day, cells were switched to medium without insulin for 8 h and then treated with various doses of insulin (10 nM or 20 nM) for 30 min (B) or treated with 10 nM insulin for 5 min or 30 min (C). At the end of treatment, cells were harvested in RIPA buffer and the protein levels of Akt, p-Akt (Ser473), p-Akt (Thr308), p-Gsk3β, and IRS1 were determined by immunoblot analysis. GAPDH was used as a loading control.
BAP31-deletion in hepatocytes enhances glucose production and increases ER stress and HFD-induced inflammation
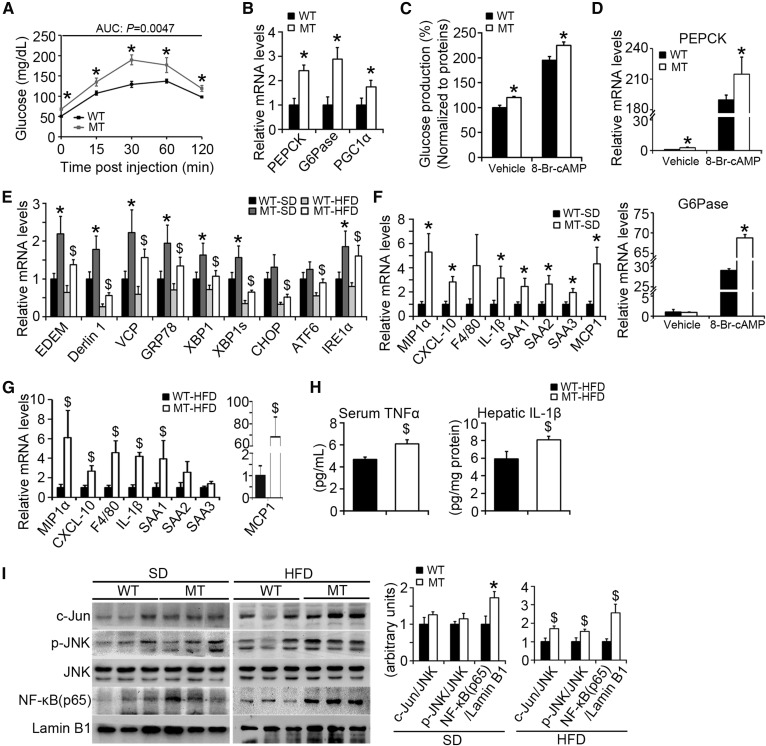
Patients with diabetes exhibit fasting hyperglycemia, which may be due to the failure of insulin-induced suppression of gluconeogenesis (26). During prolonged fasting, gluconeogenesis is stimulated to maintain glucose homeostasis. The mitochondrial matrix is thought to be the majority of gluconeogenic carbon flux and pyruvate is the possible major substrate (27). The change of blood glucose in response to a bolus of pyruvate is a reflection of hepatic gluconeogenesis (28). It is simplified, as pyruvate is also able to be utilized by many tissues and can possibly affect glucose levels by competition oxidative processes in extrahepatic tissues, including skeletal muscles. MT mice (16–18 weeks old) exhibited elevated basal levels of blood glucose and continuously exhibited elevated blood glucose for the remaining time points (Fig. 7A). Phosphoenolpyruvate carboxykinase (PEPCK) and glucose-6-phosphatase (G6Pase), the two rate-limited enzymes catalyzing gluconeogenesis, were increased by 140% and 190% in MT mice, respectively. PPARγ coactivator-1α (PGC1α) activation, which is required for fasting induction (29), was also significantly increased in MT mice compared to WT mice (by 80%) (Fig. 7B). The effects of BAP31 on gluconeogenesis were also evaluated in primary hepatocytes. BAP31-deletion increased the cumulative glucose production in primary hepatocytes stimulated with cAMP analog (8-Br-cAMP) or without (Fig. 7C), and increased the mRNA levels of the key gluconeogenic enzyme genes (PEPCK and G6Pase), which is consistent with the in vivo findings (Fig. 7D).
Fig. 7.
BAP31-deletion in hepatocytes enhances glucose production and increases ER stress and HFD-induced inflammation. A: Pyruvate tolerance test (PTT) was performed on WT and MT mice (16–18 weeks old). The blood glucose levels were determined via tail bleeding post pyruvate administration of 0, 15, 30, 60, and 120 min. AUCs (areas under the curve) were calculated (n = 6 per group). B: BAP31-deletion in hepatocytes induced PEPCK, G6Pase, and PGC1α mRNA levels in mouse livers (n = 6–7 per group). *P < 0.05, MT compared with WT mice fed SD. C: BAP31-deletion increased glucose production in the primary hepatocytes. The glucose concentrations have been normalized to cellular proteins. D: At the end of the incubation period, total RNA was extracted by using TRIzol reagent. The relative mRNA levels of PEPCK and G6Pase were determined by real-time PCR and normalized with 18S rRNA levels. *P < 0.05, MT compared with WT primary hepatocytes. E: The transcription levels of ER stress markers in WT and MT mice fed with SD or HFD. *P < 0.05, MT compared with WT mice fed SD. $P < 0.05, MT compared with WT mice fed HFD. F, G: Expression of markers of macrophages and pre-inflammatory genes were measured by real-time PCR in the livers from mice fed the SD or HFD for 96 days (n = 6–7 per group). H: BAP31-deletion in hepatocytes increased inflammatory cytokines. Serum TNFα (n = 6 per group) and hepatic IL-1β (n = 6 per group) in liver extracts were determined by ELISA. I: Immunoblot analysis of c-Jun, p-JNK, JNK, and NF-κB (p65) in the nuclei from the livers of mice fed the SD or HFD for 96 days. *P < 0.05, MT compared with WT mice fed SD. $P < 0.05, MT compared with WT mice fed HFD.
ER stress has been linked with NAFLD and other metabolic disorders (30). Genes of ER degradation enhancing mannosidase-like proteins (EDEMs), Derlin1, valosin-containing protein (VCP), glucose regulated protein 78 (GRP78), X-box binding protein 1 (XBP1), spliced XBP1 (XBP1s), C/EBP homologous protein (CHOP), activating transcription factor-6 (ATF6), and inositol-requiring enzyme 1α (IRE1α), related to ER stress, were significantly induced in MT compared with WT mice, either fed with SD or HFD, suggesting that BAP31-deletion in hepatocytes increased ER stress in mouse livers (Fig. 7E).
It has been increasingly recognized that obesity is highly associated with inflammation (31). BAP31-deletion significantly induced inflammatory markers in the livers of SD-fed mice, including macrophage inflammatory protein 1α (MIP1α), C-X-C motif chemokine ligand 10 (CXCL-10), IL-1β, serum amyloid A (SAA)1, SAA2, SAA3, and monocyte chemoattractant protein-1 (MCP1), with an insignificant increase for F4/80 expression (Fig. 7F), and induced MIP1α, CXCL-10, F4/80, IL-1β, SAA1, and MCP1 expression in mice fed HFD, with an insignificant increase for SAA2 and SAA3 expression (Fig. 7G). Serum TNFα (4.68 ± 0.20 pg/ml vs. 6.06 ± 0.38 pg/ml) and hepatic IL-1β (5.90 ± 0.87 pg/mg protein vs. 8.08 ± 0.42 pg/mg protein) were both significantly induced, suggesting that enhanced hepatic inflammation occurred in MT mice (Fig. 7H). c-Jun N-terminal kinase (JNK) and nuclear factor-κB (NF-κB) pathways have been reported playing key roles in the metabolic response to obesity and liver steatosis (32, 33). BAP31-deletion in hepatocytes increased the protein levels of c-Jun and p-JNK in livers of HFD-fed mice, but no differences were observed in the livers of mice fed SD. Nuclear translocation of transcription factor NF-κB was increased in MT mice, when fed either SD or HFD (Fig. 7I).
Abdominal and overall adiposity are closely associated with NAFLD (34). WAT mass was increased slightly in mice fed SD and was increased significantly following the HFD feeding (supplemental Fig. S6, S7). Compared with WT mice, WAT TG and FFA were increased by 140% and 74% in MT mice fed SD, respectively, and were increased by 18% and 25% in MT mice fed HFD, respectively (supplemental Fig. S7). CD36 and LPL, associated with FA transport to WAT (35), were increased in MT mice. Adipose TG lipase (ATGL), associated with lipolysis in WAT (36), was reduced in the mice fed HFD. PPARγ, ACC1, FAS, and DGAT1 were reduced in MT mice fed HFD, which may be regulated by a negative feedback mechanism due to the high lipid accumulation in WAT (supplemental Fig. S8).
DISCUSSION
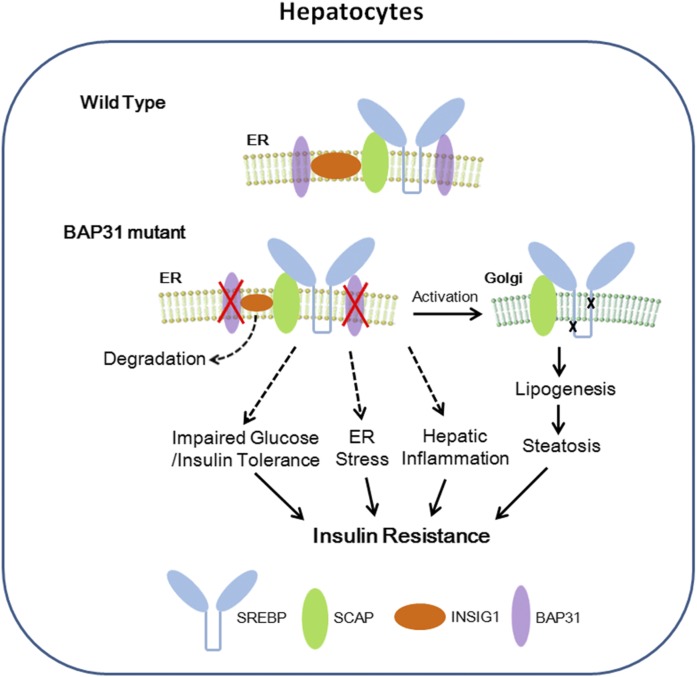
In the current study, conditional knockout mice with targeted disruption of BAP31 in adult mouse liver were generated and challenged with a Western diet and the effects of BAP31 on glucose and lipid metabolism were evaluated. In normal physiological conditions, it is possible that BAP31 forms a complex with INSIG1 and SREBP1C proteins. Hepatocyte-depletion of BAP31 may destroy the integrity of the complex, and induced the degradation of INSIG1, which reduced INSIG1 expression and led to SREBP1C activation, thus increased lipogenic gene expression and hepatic lipid accumulation. Also, BAP31-deletion in hepatocytes impaired glucose and insulin tolerance, reduced Akt phosphorylation and insulin signal transduction, and increased hepatic glucose production. In addition, BAP31-deletion increased ER stress and enhanced JNK and NF-κB signaling pathway activation, thus promoting hepatic inflammation. Therefore, we propose here that BAP31-deletion in hepatocytes induces SREBP signaling activation and promotes hepatic lipid accumulation and HFD-induced liver steatosis. Additionally, we propose that BAP31-deletion induces hyperglycemia and glucose/insulin intolerance, increases ER stress and hepatic inflammation, and eventually induces IR in a mouse model of HFD-induced obesity (summarized in Fig. 8). To the best of our knowledge, this is the first report elucidating BAP31 function in glucose and lipid metabolism in HFD-induced obesity in animal models, pointing to the potential roles of BAP31 against obesity and diabetes mellitus.
Fig. 8.
Model depicts that BAP31 interacts with SREBP/SCAP complex and INSIG1 protein in normal physiological conditions. BAP31-deletion in hepatocytes linked SREBP activation, increased lipogenesis, and induced liver steatosis. Additionally, BAP31-deletion in hepatocytes impaired glucose and insulin tolerance, increased ER stress and hepatic inflammation, and eventually induced IR in mouse livers.
HFD-induced fatty liver is a gradual process. We used multiple durations of HFD-feeding with the mice, aiming to demonstrate whether BAP31 plays the same roles in the development of HFD-induced fatty liver. The current study demonstrates that BAP31-deletion increased lipogenic gene expression and promoted HFD-induced liver steatosis, either short- or long-term, indicating the negative role of BAP31 in the regulation of lipid metabolism. SREBP1C is one of the key transcription factors in the regulation of de novo lipogenesis and modulates the transcriptional levels of genes related to lipid synthesis. Forced SREBP1C expression caused a mild hepatomegaly and a moderate increase in hepatic TG accumulation, mediated by increased ACC1, FAS, and SCD1 expression (6). INSIG1 binds to the SREBP/SCAP complex and leads to the ER retention of SCAP, thereby limiting the delivery of the complex to the Golgi for proteolytic release. Overexpression of INSIG1 inhibited SREBP processing and reduced lipogenesis in rodent animal models (11, 37). BAP31-deletion reduced INSIG1 expression, which may promote SREBP/SCAP complex export from the ER to the Golgi apparatus for SREBP proteolytic processing, and then increases SREBP activation and the lipogenesis process, thus promotes hepatic lipid accumulation and results in liver steatosis. Immunoprecipitation assays further reported that BAP31 interacted with the endogenous SREBP1C and INSIG1 proteins, suggesting that BAP31 may correlate with SREBP1C/INSIG1 physiologically, and forming a complex of BAP31/SREBP1C/INSIG1, thus regulated the downstream genes of lipogenesis process. However, whether and/or how BAP31 modulates ER export of SREBP/SCAP complex is still unknown and whether BAP31 regulates INSIG1 expression directly or modulates the cellular protein amount of INSIG1 via modulating the proteasomal degradation system is uncertain and warrants future studies.
BAP31-deletion impaired glucose and insulin tolerance, reduced insulin signal transduction, and induced IR. NAFLD and hepatic IR are highly correlated and increase the risk of T2D and atherosclerosis (38, 39). Increased hepatic FFA accumulation triggers the activation of serine/threonine kinase that directly reduces insulin receptor/insulin receptor substrate-dependent signaling cascades, which impairs insulin-stimulated Akt-Gsk3β activation (40). Akt is the most important factor that mediates insulin-induced glucose and lipid metabolism. Insulin binds to its receptor, which results in the autophosphorylation of tyrosine residues, allowing PI3K and Akt phosphorylation and activation, deactivates Gsk3β signaling, leading to glycogen synthesis from glucose and glycogen accumulation. The activation of JNK signaling induced the phosphorylation levels of IRS1 at Ser307 and weakened insulin signal transduction (41). BAP31-deletion increased p-IRS1 (Ser307) levels, resulted in reduced phosphorylation levels of Akt and Gsk3β, impaired glucose and insulin tolerance, thus induced IR in BAP31-mutant mice. Kim et al. (42) demonstrated that BAP31-depletion decreased cyclin D1 and cyclin E expression, reduced PI3K/Akt signaling, and induced cell death in human embryonic stem cells, which is consistent with the current study. However, we failed to determine any difference of insulin tolerance between WT and BAP31-mutant mice when fed HFD for either a short- or long-term duration. A possible reason for this observation is that HFD-feeding have impaired insulin tolerance and induced IR in these two types of mice already, thereby overwhelmed the difference observed from the normal chow diet, thus made no difference of insulin tolerance between WT and BAP31-mutant mice fed HFD.
Under normal conditions, insulin inhibited glucose production from glycogenolysis and gluconeogenesis in the liver, promoting glucose storage in the form of glycogen and helping to control postprandial glucose levels. However, the ability of insulin to shut down glucose production from the liver is diminished under a state of hepatic IR, in turn leads to the manifestation of hyperglycemia in diabetic states (43), which due to increased expression of gluconeogenic genes, including PEPCK and G6Pase (44). Deletion of BAP31 in hepatocytes induced IR, increased glucose production, accompanied by increased PEPCK, G6Pase and PGC1α expression, pointing to the protective roles of BAP31 against hyperglycemia or enhanced hepatic gluconeogenesis. A high level of glucose has been reported to induce ER stress in various cellular models (45, 46). Chronic ER stress perturbs the ER homeostasis and promotes the development of NAFLD and diabetes. The protein levels of BAP31 were decreased in mouse livers after HFD-feeding, suggesting that excess intake of nutrients reduces BAP31 expression and, thus, decreases the protective roles of BAP31 against the adverse effects resulting from ER stress. So it is possible that BAP31-deletion or BAP31-reduction (in HFD-induced obesity) in hepatocytes increases hepatic glucose production and hepatic lipid accumulation, and the increased cellular glucose and lipid content lead to ER stress occurring, thus aggravating the development of NAFLD and diabetes mellitus, which reduces BAP31 expression in the liver. Also, the reduced BAP31 expression possibly worsens the characters of NAFLD and diabetes, and increases hepatic glucose and lipid accumulation more severely, which is in a vicious circle.
BAP31-deletion promoted HFD-induced inflammation in the liver and activated NF-κB and JNK signaling pathways, indicating that BAP31 may have some roles in preventing the initiation of hepatic inflammation and is involved in the progression from liver steatosis to NASH. Sustained activation of JNK and NF-κB signaling is linked to the inflammatory pathway, initiates the inflammatory cascade, and induces the release of various chemokines and cytokines, such as TNFα, IL-6, and IL-1β (47). The “two hit” hypothesis for the development of steatosis to NASH has been widely recommended. The “first hit” constitutes the massive accumulation of TGs in the cytoplasm of hepatocytes. BAP31-deletion increased lipid accumulation in the liver, which may promote the “first hit” and accelerate the development to steatohepatitis. Additional cellular events, including hepatic inflammation, promote cell death, apoptosis, and fibrosis, which are the characteristic features of NASH (48). Activated JNK can also lead to liver injury and hepatocyte apoptosis, enhancing the progression from steatosis to NASH (49). We failed to detect any obvious increase of c-Jun and JNK activation in BAP31-mutant mice fed SD. One of the possible reasons is that, under normal conditions, the basal levels of hepatic inflammation response are relatively low and the protein levels of c-Jun and p-JNK in the liver are relatively low; thus, it might be difficult to detect any differences by immunoblot assay due to the low levels of gene expression. HFD-feeding facilitates the accumulation of excess lipids in the liver, which induces oxidative stress and consequently results in JNK signaling activation (50). Contiguous mutation of BAP31 and ABCD1 is associated with increased serum ALT and AST levels (17), which are the enzymes located in hepatocytes that are leaked into the blood circulation when the hepatocytes are injured, and have been considered as the indicators of liver inflammation. The current study reported that BAP31-depletion in the adult mouse liver increased serum ALT and AST content, further indicating the existence of liver injury and inflammation in the mutant mice.
The ER regulates protein folding and the biosynthesis of macromolecules, such as steroids, lipids, and carbohydrates. Enhanced ER stress has been implicated in the development of NAFLD, which was highly associated with IR. Forced expression of GRP78 reduced ER stress markers and SREBP1C cleavage, and decreased SREBP1C and the target genes expression, which prevented hepatic lipid accumulation in leptin-mutant mice, suggesting the possibility of that enhanced ER stress increased SREBP1C activation and induced lipid synthesis (51). BAP31 is largely localized to the ER and plays key roles in maintaining ER homeostasis (19, 52). Mutation of BAP31 in the liver may destroy this homeostasis and then induce ER stress, which has been confirmed in the current study. SREBP1C and the target genes were significantly induced in BAP31-mutant mice. It is possible that BAP31-deletion in hepatocytes induced ER stress and then enhanced SREBP1C cleavage, which increased SREBP signaling activation, leading to increased expression of lipogenic genes and promotion of lipogenesis. It was noted that serum bile acid was increased in BAP31-mutant mice. Bile acids are derivatives of Chol synthesized in the hepatocytes. After the meal, bile acids are released into the intestinal tract and facilitate fat digestion and absorption, and are efficiently reabsorbed in the ileum and transported back to the liver. Elevated serum bile acid content hints that there may be a disturbance of bile acid metabolism in BAP31-mutant mice, and this disturbance influences lipid digestion and absorption. Also, residual TG and Chol were increased in the feces, meaning that there is a high amount of lipid remaining in the feces, further suggesting that lipid absorption was impaired in BAP31-mutant mice. Maybe this is the reason why we found higher food intake in BAP31-mutant mice fed with HFD, but failed to find any pronounced body weight change between the two types of mice, suggesting that there is an energy imbalance in the defective mice. All of these factors will affect the glucose and lipid metabolism integrally, thus inducing IR in animal models.
In summary, we report the novel roles of BAP31 in regulating glucose and lipid metabolism. Specific deletion of the BAP31 gene in hepatocytes increased SREBP1C activation and excessively enhanced lipogenesis and hepatic lipid accumulation. BAP31-deletion in hepatocytes impaired insulin signaling, increased ER stress and liver inflammation, and eventually induced IR in mice. This study raises the possibility that the pharmacological activation of BAP31 could provide a useful therapeutic approach for combating the effects of the disturbance of glucose or lipid metabolism in the process of diabetes and metabolic syndrome.
Supplementary Material
Footnotes
Abbreviations:
- ACC1
- acetyl-CoA carboxylase 1
- ALT
- alanine transaminase
- AST
- aspartate aminotransferase
- BAP
- B-cell associated protein
- CD36
- cluster of differentiation 36
- Chol
- cholesterol
- CXCL-10
- C-X-C motif chemokine ligand 10
- DGAT
- diacylglycerol acyltransferase
- ER
- endoplasmic reticulum
- FATP
- FA transport protein
- GCK
- glucokinase
- G6Pase
- glucose-6-phosphatase
- G6PD
- glucose-6-phosphate dehydrogenase
- GRP78
- glucose regulated proteins 78
- Gsk3β
- glycogen synthase kinase 3β
- HFD
- high-fat diet
- HMG-CoA Red
- HMG-CoA reductase
- HMG-CoA Syn
- HMG-CoA synthase
- IL
- interleukin
- INSIG1
- insulin-induced gene 1
- IR
- insulin resistance
- IRS1
- insulin receptor substrate 1
- JNK
- c-Jun N-terminal kinase
- LDLR
- LDL receptor
- MCP1
- monocyte chemoattractant protein-1
- ME
- malic enzyme
- MIP1α
- macrophage inflammatory protein 1α
- NAFLD
- nonalcoholic fatty liver disease
- NASH
- nonalcoholic steatohepatitis
- NF-κB
- nuclear factor-κB
- nSREBP
- nuclear SREBP
- PEPCK
- phosphoenolpyruvate carboxykinase
- PGC1α
- PPARγ coactivator-1α
- p-Gsk3β
- phosphorylated glycogen synthase kinase 3β
- pSREBP
- precursor SREBP
- SAA
- serum amyloid A
- SCAP
- SREBP cleavage-activating protein
- SCD1
- stearoyl-CoA desaturase-1
- SD
- standard chow diet
- SLC6A8
- solute carrier family 6 member 8
- TG
- triglyceride
- XBP1
- X-box binding protein 1
- WAT
- white adipose tissue
This work was supported by National Natural Science Foundation of China Grants 81570788, 31401473, 81341102, and 31370784 and Fundamental Research Funds for Northeastern University (N162004005, N162004004, N130220001, N120820002, N141008001/8, and N130120001).
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Bellentani S., Scaglioni F., Marino M., and Bedogni G.. 2010. Epidemiology of non-alcoholic fatty liver disease. Dig. Dis. 28: 155–161. [DOI] [PubMed] [Google Scholar]
- 2.McCullough A. J. 2004. The clinical features, diagnosis and natural history of nonalcoholic fatty liver disease. Clin. Liver Dis. 8: 521–533. [DOI] [PubMed] [Google Scholar]
- 3.Marchesini G., Brizi M., Bianchi G., Tomassetti S., Bugianesi E., Lenzi M., McCullough A. J., Natale S., Forlani G., and Melchionda N.. 2001. Nonalcoholic fatty liver disease: a feature of the metabolic syndrome. Diabetes. 50: 1844–1850. [DOI] [PubMed] [Google Scholar]
- 4.Horton J. D., Goldstein J. L., and Brown M. S.. 2002. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109: 1125–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strable M. S., and Ntambi J. M.. 2010. Genetic control of de novo lipogenesis: role in diet-induced obesity. Crit. Rev. Biochem. Mol. Biol. 45: 199–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shimano H., Horton J. D., Shimomura I., Hammer R. E., Brown M. S., and Goldstein J. L.. 1997. Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. J. Clin. Invest. 99: 846–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang T., Espenshade P. J., Wright M. E., Yabe D., Gong Y., Aebersold R., Goldstein J. L., and Brown M. S.. 2002. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 110: 489–500. [DOI] [PubMed] [Google Scholar]
- 8.Gong Y., Lee J. N., Lee P. C., Goldstein J. L., Brown M. S., and Ye J.. 2006. Sterol-regulated ubiquitination and degradation of Insig-1 creates a convergent mechanism for feedback control of cholesterol synthesis and uptake. Cell Metab. 3: 15–24. [DOI] [PubMed] [Google Scholar]
- 9.Lee J. N., and Ye J.. 2004. Proteolytic activation of sterol regulatory element-binding protein induced by cellular stress through depletion of Insig-1. J. Biol. Chem. 279: 45257–45265. [DOI] [PubMed] [Google Scholar]
- 10.McFarlane M. R., Liang G., and Engelking L. J.. 2014. Insig proteins mediate feedback inhibition of cholesterol synthesis in the intestine. J. Biol. Chem. 289: 2148–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engelking L. J., Kuriyama H., Hammer R. E., Horton J. D., Brown M. S., Goldstein J. L., and Liang G.. 2004. Overexpression of Insig-1 in the livers of transgenic mice inhibits SREBP processing and reduces insulin-stimulated lipogenesis. J. Clin. Invest. 113: 1168–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim K. M., Adachi T., Nielsen P. J., Terashima M., Lamers M. C., Kohler G., and Reth M.. 1994. Two new proteins preferentially associated with membrane immunoglobulin D. EMBO J. 13: 3793–3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adachi T., Schamel W. W., Kim K. M., Watanabe T., Becker B., Nielsen P. J., and Reth M.. 1996. The specificity of association of the IgD molecule with the accessory proteins BAP31/BAP29 lies in the IgD transmembrane sequence. EMBO J. 15: 1534–1541. [PMC free article] [PubMed] [Google Scholar]
- 14.Geiger R., Andritschke D., Friebe S., Herzog F., Luisoni S., Heger T., and Helenius A.. 2011. BAP31 and BiP are essential for dislocation of SV40 from the endoplasmic reticulum to the cytosol. Nat. Cell Biol. 13: 1305–1314. [DOI] [PubMed] [Google Scholar]
- 15.Wang B., Heath-Engel H., Zhang D., Nguyen N., Thomas D. Y., Hanrahan J. W., and Shore G. C.. 2008. BAP31 interacts with Sec61 translocons and promotes retrotranslocation of CFTRDeltaF508 via the derlin-1 complex. Cell. 133: 1080–1092. [DOI] [PubMed] [Google Scholar]
- 16.Corzo D., Gibson W., Johnson K., Mitchell G., LePage G., Cox G. F., Casey R., Zeiss C., Tyson H., Cutting G. R., et al. . 2002. Contiguous deletion of the X-linked adrenoleukodystrophy gene (ABCD1) and DXS1357E: a novel neonatal phenotype similar to peroxisomal biogenesis disorders. Am. J. Hum. Genet. 70: 1520–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwasa M., Yamagata T., Mizuguchi M., Itoh M., Matsumoto A., Hironaka M., Honda A., Momoi M. Y., and Shimozawa N.. 2013. Contiguous ABCD1 DXS1357E deletion syndrome: report of an autopsy case. Neuropathology. 33: 292–298. [DOI] [PubMed] [Google Scholar]
- 18.Osaka H., Takagi A., Tsuyusaki Y., Wada T., Iai M., Yamashita S., Shimbo H., Saitsu H., Salomons G. S., Jakobs C., et al. . 2012. Contiguous deletion of SLC6A8 and BAP31 in a patient with severe dystonia and sensorineural deafness. Mol. Genet. Metab. 106: 43–47. [DOI] [PubMed] [Google Scholar]
- 19.Cacciagli P., Sutera-Sardo J., Borges-Correia A., Roux J. C., Dorboz I., Desvignes J. P., Badens C., Delepine M., Lathrop M., Cau P., et al. . 2013. Mutations in BCAP31 cause a severe X-linked phenotype with deafness, dystonia, and central hypomyelination and disorganize the Golgi apparatus. Am. J. Hum. Genet. 93: 579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu J., Kulkarni S. R., Li L., and Slitt A. L.. 2012. UDP-glucuronosyltransferase expression in mouse liver is increased in obesity- and fasting-induced steatosis. Drug Metab. Dispos. 40: 259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foretz M., Hebrard S., Leclerc J., Zarrinpashneh E., Soty M., Mithieux G., Sakamoto K., Andreelli F., and Viollet B.. 2010. Metformin inhibits hepatic gluconeogenesis in mice independently of the LKB1/AMPK pathway via a decrease in hepatic energy state. J. Clin. Invest. 120: 2355–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu J., Kulkarni S. R., Donepudi A. C., More V. R., and Slitt A. L.. 2012. Enhanced Nrf2 activity worsens insulin resistance, impairs lipid accumulation in adipose tissue, and increases hepatic steatosis in leptin-deficient mice. Diabetes. 61: 3208–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cases S., Smith S. J., Zheng Y. W., Myers H. M., Lear S. R., Sande E., Novak S., Collins C., Welch C. B., Lusis A. J., et al. . 1998. Identification of a gene encoding an acyl CoA:diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc. Natl. Acad. Sci. USA. 95: 13018–13023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eberlé D., Hegarty B., Bossard P., Ferré P., and Foufelle F.. 2004. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie. 86: 839–848. [DOI] [PubMed] [Google Scholar]
- 25.Stoeckman A. K., and Towle H. C.. 2002. The role of SREBP-1c in nutritional regulation of lipogenic enzyme gene expression. J. Biol. Chem. 277: 27029–27035. [DOI] [PubMed] [Google Scholar]
- 26.Basu R., Chandramouli V., Dicke B., Landau B., and Rizza R.. 2005. Obesity and type 2 diabetes impair insulin-induced suppression of glycogenolysis as well as gluconeogenesis. Diabetes. 54: 1942–1948. [DOI] [PubMed] [Google Scholar]
- 27.Katz J., and Tayek J. A.. 1999. Recycling of glucose and determination of the Cori cycle and gluconeogenesis. Am. J. Physiol. 277: E401–E407. [DOI] [PubMed] [Google Scholar]
- 28.Hughey C. C., Wasserman D. H., Lee-Young R. S., and Lantier L.. 2014. Approach to assessing determinants of glucose homeostasis in the conscious mouse. Mamm. Genome. 25: 522–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoon J. C., Puigserver P., Chen G., Donovan J., Wu Z., Rhee J., Adelmant G., Stafford J., Kahn C. R., Granner D. K., et al. . 2001. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 413: 131–138. [DOI] [PubMed] [Google Scholar]
- 30.Jiang S., Yan C., Fang Q. C., Shao M. L., Zhang Y. L., Liu Y., Deng Y. P., Shan B., Liu J. Q., Li H. T., et al. . 2014. Fibroblast growth factor 21 is regulated by the IRE1alpha-XBP1 branch of the unfolded protein response and counteracts endoplasmic reticulum stress-induced hepatic steatosis. J. Biol. Chem. 289: 29751–29765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wellen K. E., and Hotamisligil G. S.. 2005. Inflammation, stress, and diabetes. J. Clin. Invest. 115: 1111–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han M. S., Jung D. Y., Morel C., Lakhani S. A., Kim J. K., Flavell R. A., and Davis R. J.. 2013. JNK expression by macrophages promotes obesity-induced insulin resistance and inflammation. Science. 339: 218–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lawrence T. 2009. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb. Perspect. Biol. 1: a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sung K. C., Ryan M. C., Kim B. S., Cho Y. K., Kim B. I., and Reaven G. M.. 2007. Relationships between estimates of adiposity, insulin resistance, and nonalcoholic fatty liver disease in a large group of nondiabetic Korean adults. Diabetes Care. 30: 2113–2118. [DOI] [PubMed] [Google Scholar]
- 35.Goldberg I. J., Eckel R. H., and Abumrad N. A.. 2009. Regulation of fatty acid uptake into tissues: lipoprotein lipase- and CD36-mediated pathways. J. Lipid Res. 50 (Suppl.): S86–S90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lass A., Zimmermann R., Haemmerle G., Riederer M., Schoiswohl G., Schweiger M., Kienesberger P., Strauss J. G., Gorkiewicz G., and Zechner R.. 2006. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman syndrome. Cell Metab. 3: 309–319. [DOI] [PubMed] [Google Scholar]
- 37.Takaishi K., Duplomb L., Wang M. Y., Li J., and Unger R. H.. 2004. Hepatic insig-1 or -2 overexpression reduces lipogenesis in obese Zucker diabetic fatty rats and in fasted/refed normal rats. Proc. Natl. Acad. Sci. USA. 101: 7106–7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bugianesi E., Moscatiello S., Ciaravella M. F., and Marchesini G.. 2010. Insulin resistance in nonalcoholic fatty liver disease. Curr. Pharm. Des. 16: 1941–1951. [DOI] [PubMed] [Google Scholar]
- 39.Birkenfeld A. L., and Shulman G. I.. 2014. Nonalcoholic fatty liver disease, hepatic insulin resistance, and type 2 diabetes. Hepatology. 59: 713–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chattopadhyay M., Selinger E. S., Ballou L. M., and Lin R. Z.. 2011. Ablation of PI3K p110-alpha prevents high-fat diet-induced liver steatosis. Diabetes. 60: 1483–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Draznin B. 2006. Molecular mechanisms of insulin resistance: serine phosphorylation of insulin receptor substrate-1 and increased expression of p85alpha: the two sides of a coin. Diabetes. 55: 2392–2397. [DOI] [PubMed] [Google Scholar]
- 42.Kim W. T., Seo Choi H., Min Lee H., Jang Y. J., and Ryu C. J.. 2014. B-cell receptor-associated protein 31 regulates human embryonic stem cell adhesion, stemness, and survival via control of epithelial cell adhesion molecule. Stem Cells. 32: 2626–2641. [DOI] [PubMed] [Google Scholar]
- 43.Rizza R. A. 2010. Pathogenesis of fasting and postprandial hyperglycemia in type 2 diabetes: implications for therapy. Diabetes. 59: 2697–2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dong B., Saha P. K., Huang W., Chen W., Abu-Elheiga L. A., Wakil S. J., Stevens R. D., Ilkayeva O., Newgard C. B., Chan L., et al. . 2009. Activation of nuclear receptor CAR ameliorates diabetes and fatty liver disease. Proc. Natl. Acad. Sci. USA. 106: 18831–18836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mooradian A. D., and Haas M. J.. 2011. Glucose-induced endoplasmic reticulum stress is independent of oxidative stress: a mechanistic explanation for the failure of antioxidant therapy in diabetes. Free Radic. Biol. Med. 50: 1140–1143. [DOI] [PubMed] [Google Scholar]
- 46.Zhong Y., Li J., Chen Y., Wang J. J., Ratan R., and Zhang S. X.. 2012. Activation of endoplasmic reticulum stress by hyperglycemia is essential for Muller cell-derived inflammatory cytokine production in diabetes. Diabetes. 61: 492–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baker R. G., Hayden M. S., and Ghosh S.. 2011. NF-kappaB, inflammation, and metabolic disease. Cell Metab. 13: 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Browning J. D., and Horton J. D.. 2004. Molecular mediators of hepatic steatosis and liver injury. J. Clin. Invest. 114: 147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malhi H., Bronk S. F., Werneburg N. W., and Gores G. J.. 2006. Free fatty acids induce JNK-dependent hepatocyte lipoapoptosis. J. Biol. Chem. 281: 12093–12101. [DOI] [PubMed] [Google Scholar]
- 50.Pereira S., Park E., Mori Y., Haber C. A., Han P., Uchida T., Stavar L., Oprescu A. I., Koulajian K., Ivovic A., et al. . 2014. FFA-induced hepatic insulin resistance in vivo is mediated by PKCdelta, NADPH oxidase, and oxidative stress. Am. J. Physiol. Endocrinol. Metab. 307: E34–E46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kammoun H. L., Chabanon H., Hainault I., Luquet S., Magnan C., Koike T., Ferre P., and Foufelle F.. 2009. GRP78 expression inhibits insulin and ER stress-induced SREBP-1c activation and reduces hepatic steatosis in mice. J. Clin. Invest. 119: 1201–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Namba T., Tian F., Chu K., Hwang S. Y., Yoon K. W., Byun S., Hiraki M., Mandinova A., and Lee S. W.. 2013. CDIP1–BAP31 complex transduces apoptotic signals from endoplasmic reticulum to mitochondria under endoplasmic reticulum stress. Cell Rep. 5: 331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.