Abstract
The epidermis is essential for animal survival, providing both a protective barrier and cellular sensor to external environments. The generally conserved embryonic origin of the epidermis, but the broad morphological and functional diversity of this organ across animals is puzzling. We define the transcriptional regulators underlying epidermal lineage differentiation in the planarian Schmidtea mediterranea, an invertebrate organism that, unlike fruitflies and nematodes, continuously replaces its epidermal cells. We find that Smed-p53, Sox and Pax transcription factors are essential regulators of epidermal homeostasis, and act cooperatively to regulate genes associated with early epidermal precursor cell differentiation, including a tandemly arrayed novel gene family (prog) of secreted proteins. Additionally, we report on the discovery of distinct and previously undescribed secreted organelles whose production is dependent on the transcriptional activity of soxP-3, and which we term Hyman vesicles.
Keywords: epidermis, stem cells, transcription, p53, zfp-1, pax-2/5/8, soxP-3
Introduction
The epidermis is a central innovation in the evolutionary history of animals. This outer cell layer not only serves as the first line of defense to environmental vicissitudes such as injury and infection, but also plays diverse roles including regulation of body temperature and water balance, respiration, sensing the surroundings, and producing signals related to behavior (Tyler, 1984). Because of its complexity and constant environmental interactions, epidermal cellular composition, architecture and homeostasis are remarkably diverse both morphologically and biochemically throughout the animal kingdom (Tyler, 1984). The planarian adult epidermis is a monostratified tissue that consists of a single layer of post-mitotic, differentiated, multi-ciliated and non-ciliated cell types (Rompolas et al., 2010). This non-cuticularized epidermis sits atop a basement membrane overlying the body wall musculature (Hyman, 1951). Several past andrecent studies have shown that the planarian epidermis is composed of a remarkable diversity of differentiated cell types (McGee et al., 1997; Tyler and Tyler, 1997; Liana et al., 2012; Tu et al., 2015). Moreover, these differentiated cells are continuously replaced by post-mitotic epithelial precursors derived from a pool of dividing pluripotent stem cells known as neoblasts that reside deep in the mesenchyme (Newmark and Sánchez Alvarado, 2000). Because planarians belong to the Lophotrochozoan/Spiralian superclade, an animal grouping distinct from the Ecdysozoa (e.g., flies and nematodes), yet share a common ancestor with the Deuterostomes (e.g., vertebrates), S. mediterranea provides new opportunities to explore and inform the molecular processes underpinning the evolution of extant functional attributes of the animal epidermis.
Unlike the epidermis of adult flies and nematodes, the adult planarian epidermis is made from post-mitotic stem cell progeny that migrate from the internal mesenchyme, intercalate and then differentiate into the many different cell types that compose this organ, an observation originally made by Hallez in 1887 (Hallez, 1887). These migratory cells form part of a cohort of precursor epidermal cells that undergo multiple transition states on their way to epithelial differentiation (Tu et al., 2015). Lineage experiments have uncovered two populations of epidermal precursors. The first, defined by Smed-prog-1+ expression in early differentiating progeny; and the second characterized by the expression of Smed-agat-1+ in later, differentiating progeny (Eisenhoffer et al., 2008; van Wolfswinkel et al., 2014; Tu et al., 2015). Intriguingly, in situ hybridization of early and late progeny markers show little co-localization, suggesting that their transcriptional activation is spatiotemporally regulated (Zhu et al., 2015). Moreover, single-cell transcriptional profiling partitions the early and late progeny cells into two distinct populations (Wurtzel et al., 2015) and indicates that epidermal progenitors respond to positional cues along the planarian dorsoventral axis (Wurtzel et al., 2017). Together, these observations support the hypothesis that discrete changes must be occurring in the transcriptional landscape supporting epidermal lineage progression in planarians.
The intricate spatiotemporal differentiation of precursors into epidermal cells is reflected by the transcriptional complexity recently uncovered in planarian neoblasts. These studies identified a sub-population of neoblasts expressing Smed-zfp-1 and Smed-p53 (i.e., the zeta-class) to be the origin of epidermal progenitors (van Wolfswinkel et al., 2014). RNAi knockdown of zfp-1 or p53 reduces the number of intermediate precursors for the epidermal lineage. Such precursor depletion ultimately leads to ventral curling of the animals, a phenotype attributed to loss of epidermal integrity preferentially in the ventral epidermis (Pearson and Sánchez Alvarado, 2010; Wagner et al., 2012). Even more strikingly, zfp-1 RNAi depletes the entire zeta-class population, yet leaves other classes of neoblasts (i.e., the sigma- and gamma-classes) unaffected (van Wolfswinkel et al., 2014), suggesting that zfp-1 functions specifically in the epidermal lineage. Smed-zfp-1 encodes a zinc-finger protein with a highly conserved C2H2 DNA binding domains (Wagner et al., 2012), while Smed-p53 shares a highly conserved DNA-binding domain with that of vertebrate TP63 (Pearson and Sánchez Alvarado, 2010).
In this study, we dissect the transcriptional regulatory network underlying the planarian epidermal lineage during homeostasis. We describe several conserved transcription regulators and demonstrate a fundamental, upstream role for p53 in planarians in the transcriptional cascade leading to epidermal lineage progression. Furthermore, we identify two transcription factors that together are not generally associated with the epidermal lineages of other animals: a sry-box homolog, Smed-soxP-3, and a pair-box homolog, Smed-pax-2/5/8 acting downstream of p53 and zfp-1 and functioning cooperatively to specify the identity of early progeny cells. We show that SoxP-3 and pax-2/5/8 are required for the expression of a novel family of secreted proteins, PROG, which are specific to the epidermal lineage and likely reflect a planarian-specific adaptation. Moreover, we describe for the first time novel PROG+ secretory vesicles whose production depends on SoxP-3 activity. Altogether, our work has uncovered a defined set of transcriptional modules integrated into a hierarchical regulatory program that likely drive the specification and homeostasis of the planarian epidermis, adding to our understanding of mechanisms underpinning epidermal cell diversity in animals.
Results
p53-and zfp-1(RNAi) yield similar transcriptional changes in planarian stem cells likely associated with epidermal differentiation
The zeta-class neoblasts associated with the epidermal lineage are characterized by their higher expression of zfp-1, fgfr-1, p53, soxP-3, and egr-1 (van Wolfswinkel et al., 2014). However, only zfp-1 and p53 have been reported to induce a ventral curling phenotype after RNAi (Pearson and Sánchez Alvarado, 2010; Wagner et al., 2012). We sought to take advantage of these data to identify regulators of epidermal differentiation. We first confirmed that zfp-1 and p53 RNAi each depleted early and late progeny cells with similar kinetics (Figure 1 Supplement 1A, B). Second, we determined the extent of co-expression of these transcription factors in vivo in neoblasts (piwi-1+ cells) via fluorescent in situ hybridization (FISH) and showed that zfp-1 and p53 co-localized extensively to the piwi-1+ compartment (Figure 1 Supplement 1C), suggesting that they function in the same stem cell sub-population. p53, however, was detected in a broader domain spanning both neoblasts and post-mitotic progeny cells (piwi-1neg prog-1+), indicating that p53 may have multiple, cell lineage-specific roles (Figure Supplement 1C).
We then profiled by RNA-Seq genome-wide transcriptional changes of neoblasts (“X1” population) isolated from p53- and zfp-1(RNAi) animals to systematically identify potential regulators involved in epidermal differentiation at the neoblast stage (Figure 1A). It was reported that RNAi of zfp-1 specifically eliminates the zeta-class neoblasts without affecting other progenitors that regenerate brain, muscle, gut and protonephridia (van Wolfswinkel et al., 2014). In contrast, RNAi of p53 induces hyper-proliferation of neoblasts followed by rapid decline of these cells (Figure 1 Supplement 1D, F) (Pearson and Sánchez Alvarado, 2010). Based on the onset of the zfp-1 and p53 phenotypes, we chose to isolate X1 cells at 6, 9 and 13 dpf to include the earliest observed absence of early progeny cells, prior to the depletion of neoblasts in p53(RNAi) animals (Figure 1A). We identified a total of 576 genes that showed changes in expression of at least 1.5 fold and an adjusted p-value < 0.01 for at least two time points in either p53- or zfp-1(RNAi) X1 populations (Figure 1B; Supplementary Table 1). The expression levels of these genes showed similar kinetics in p53- and zfp-1(RNAi) X1 cells by RNA-Seq (correlation coefficient r = 0.9, Figure 1B) indicating that p53 and zfp-1 regulate similar biological processes in neoblasts. We surveyed the expression levels of previously reported genes and confirmed that all known zeta-class markers are down-regulated in p53- and zfp-1(RNAi) (Figure 1 Supplement 1E). Markers of other lineages are either unchanged or are up-regulated. In particular, neuronal and protonephridial lineage markers showed increased expression along the time course suggesting an increase of other tissue progenitor cells at the expense of the epidermal lineage (Figure 1 Supplement 1E).
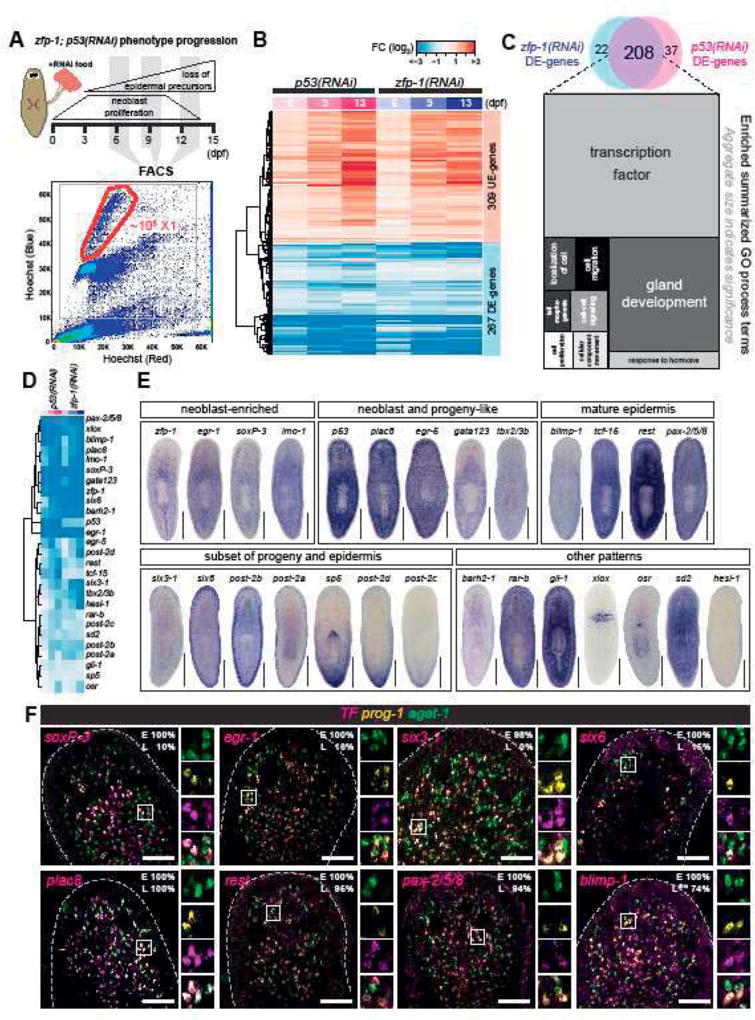
Figure 1. p53-and zfp-1 (RNAi) analyses define shared transcriptional changes in neoblasts and their differentiation progeny.
(A) Schematic of experimental strategy. Planarians fed with dsRNA food were dissociated into cell suspensions on 6, 9, and 13 dpf, and the neoblasts were FACS-sorted for RNA-Seq. A representative FACS plot (control 9 dpf) shows the gating of dividing neoblasts (X1). (B) RNA-Seq analysis of 576 genes shows highly correlated expression profiles in p53-and zfp-1(RNAi) X1 cells (selection criteria: FC > 1.5, p-adj < 0.01, ≥ 2 time points). Each column represents data of one time point. DE, down-regulated in expression. UE, up-regulated in expression. Colors represent log2 fold change (FC) to control. Color key is the same in Figure 1B and 1D. (C) Venn diagram of p53-and zfp-1(RNAi) DE-genes and the major GO terms enriched in the 208 common DE-genes. Related GO terms are merged in the same shade and the size indicates significance of a group of GO terms (Supek et al., 2011). (D) Expression profiles of the transcription factors (TFs) identified in the common DE-genes of p53-and zfp-1(RNAi) X1 cells. (E) WISH patterns of candidate TFs. Scale bar, 500µm. (F) Co-localization of TF, prog-1 and agat-1 analyzed by FISH. Box region is split into individual channels to the right. Percentage of early (E) or late (L) progeny cells detected positive for candidate TF are shown in the upper right corner. Scale bar, 100µm.
We identified 208 genes with significantly decreased expression levels (DE-genes) in both p53- and zfp-1(RNAi) X1 cells (FC > 1.5, p-adj < 0.01, ≥ 2 time points) and analyzed the resulting gene set by assigning gene ontology (GO) terms (Figure 1C, Supplementary Table 2). We found that transcription factor activity (GO:0003700) and gland development (GO:0048732) are the most enriched terms. Among the identified 208 DE-genes, 105 genes were previously classified by single-cell sequencing (SCS) technology (Wurtzel et al., 2015), and 83/105 (79%) genes were assigned to the epidermal lineages (Supplementary Table 1). We hypothesized from these data that the transcriptionally identified gene set are likely enriched in regulators of epidermal differentiation.
We tested this hypothesis by selecting 27 transcription factors (TFs) from the DE-gene list, (Figure 1D), and performed whole-mount in situ hybridization (WISH) (Figure 1E). The majority of genes tested showed expression patterns characteristic of epidermal lineage (Figure 1E). For instance, like zfp-1, the genes egr-1, soxP-3, and lmo-1 appear to be highly expressed in neoblasts. Other genes like plac8, egr-5, gata123 and tbx2/3b are expressed in both neoblasts and discrete sub-epidermal cells in a manner that is not dissimilar to p53 expression. Additionally, blimp-1, tcf-15, rest, and pax-2/5/8 are expressed in both discrete sub-epidermal cells and in mature epidermis. We also found several TFs expressed in a subset of mature epidermal cells. For example, six3-1, six6, post-2b, and post-2a are all enriched in cells located at the dorsoventral boundary, possibly marking adhesion gland cells. Another subset of genes represented by sp5, post-2d and abdBa-2 exhibited graded expression along the anteroposterior axis. The rest of the genes examined showed strong expression in non-epidermal tissues.
We then used irradiation to rapidly and specifically eliminate neoblasts and, subsequently, epidermal progeny cells (Eisenhoffer et al., 2008), and checked the expression levels of the above TFs in an irradiation time-course by RNA-Seq (Figure 1 Supplement 2; Supplementary Table 3). We found that only those genes with expression enriched in neoblasts and progeny cells (zfp-1, egr-1, soxP-3, lmo-1, p53, egr-5, gata123, tcf-15 and six6) were significantly down-regulated upon irradiation (Figure 1 Supplement 2). We then performed double FISH of TFs with early (prog-1) and late (agat-1) progeny markers (summary in Supplementary Table 4). Among all TFs examined, expression of soxP-3, egr-1, six3-1, and six6 show substantial co-localization with the early progeny marker prog-1, but not late progeny marker agat-1. In contrast, plac8, rest, pax-2/5/8, and blimp-1 extend their expression to late progeny and mature epidermis (Figure 1F). These expression patterns reflect on a complex temporal and spatial regulation of transcription as cells transition from early to late progeny cell stages.
RNAi screen of p53-and zfp-1-modulated genes identifies soxP-3 and pax-2/5/8 as early transcriptional regulators in epidermal precursors
Based on their spatial distribution and association with progenitor cell markers (Figure 1F, Supplementary Table 4), we investigated 17 TFs for their possible roles in the differentiation of progeny cells. RNAi animals for each of the tested TFs were fixed and examined by WISH for prog-1, prog-2 and agat-1 expression (Figure 2A). Notably, of all tested TFs, only soxP-3- and pax-2/5/8(RNAi) animals showed significant reduction in expression of the early progeny markers prog-1 and prog-2, but not the late progeny marker agat-1. This result was unexpected, as we and others have hypothesized that the late progeny arise from the differentiation of early progeny. Two possibilities may explain this result. First, that the early progeny cells are being eliminated in soxP-3- and pax-2/5/8 (RNAi) animals with the late progeny cells originating from another lineage that is independent from the early progeny cells. Second, the early progeny cells in soxP-3-and pax-2/5/8 (RNAi) animals are not expressing prog-1 and prog-2, markers but are still capable of differentiating into late progeny cells. To better understand the effects of soxP-3- and pax-2/5/8(RNAi) on the early stage of epidermal differentiation, we examined the expression of additional markers specific to the early progeny cells.
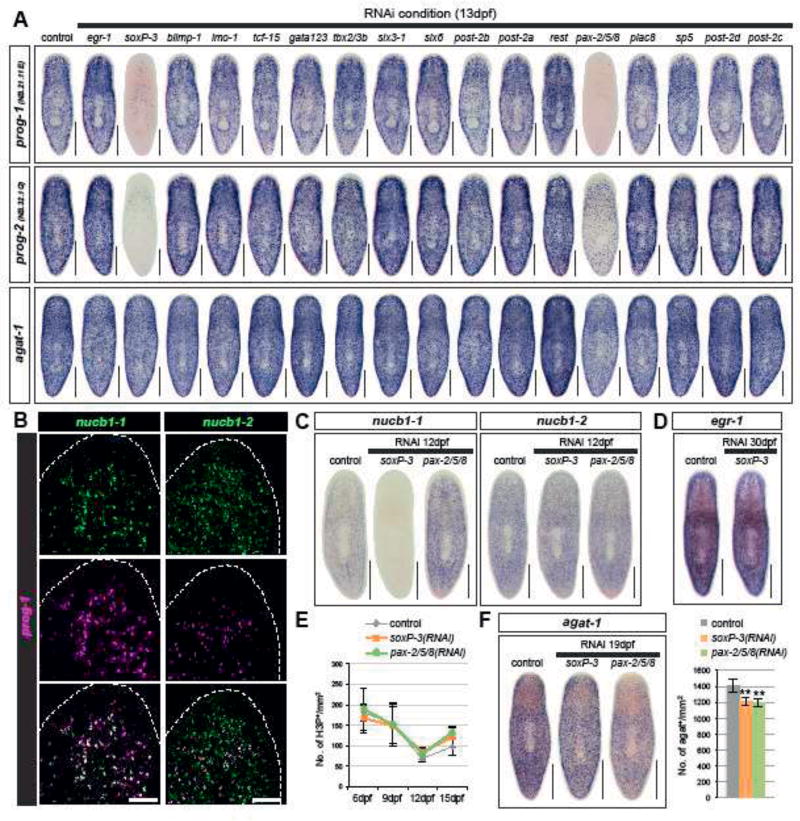
Figure 2. soxP-3 and pax-2/5/8 regulate early epidermal precursor trasncription.
(A) soxP-3-and pax-2/5/8(RNAi) animals show reduced WISH signals of early progeny markers (prog-1 and prog-2, 100% penetrance) but not late progeny marker agat-1 in the TF RNAi screen (n > 8 per RNAi, 2 independent experiments). Scale bar, 500µm. (B) nucb1-1 is 100% co-localized with prog-1 analyzed by FISH. nucb1-2 is partially overlapped with prog-1 and may label late progeny cells. Scale bar, 100µm. (C) Expression of nucb1-1 and nucb1-2 analyzed by WISH. nucb1-1 expression is eliminated in soxP-3(RNAi) animals but is up-regulated in pax-2/5/8(RNAi) animals (n > 6 per RNAi, 100% penetrance). nucb1-2 expression is unaffected in both soxP-3-and pax-2/5/8(RNAi) animals. Scale bar, 500µm. (D) egr-1 expression is unaffected by soxP-3 RNAi. Scale bar, 500µm. (E) Neoblast proliferation is unaffected by soxP-3-and pax-2/5/8 RNAi analyzed by H3P immunostaining (mean ± s.d., n > 8 per time point). (F) agat-1+ cells are slightly reduced in soxP-3-and pax-2/5/8(RNAi) animals. Scale bar, 500µm. Quantification of agat-1+ cells is shown in the bar graph (mean ± s.d., n > 8). Double asterisks, p<0.001. Unpaired Student’s t-test.
Two nucleobindin-1 (nucb1) homologs, nucb1-1 and nucb1-2 were previously shown to be expressed in the epidermal lineage (Tu et al., 2015; Zhu et al., 2015). nucb1-1 is expressed only in the early progeny cells whereas nucb1-2 is expressed in both early and likely late progeny cells (Figure 2B). Like other early progeny markers in the soxP-3(RNAi), nucb1-1 expression is also undetectable. Surprisingly, nucb1-1 expression level increases upon pax-2/5/8 RNAi knockdown (Figure 3C) suggesting that early progeny cells are indeed still present in these animals and capable of expressing other early markers. Because egr-1 expression is co-localized 100% with early progeny cells (Figure 2C), we looked at its expression in soxP-3 RNAi animals and found no difference from control animals (Figure 3D). These results indicate that abrogation of both soxP-3 and pax-2/5/8 by RNAi affects the transcriptional output of the early progeny genes rather than the viability of the cells.
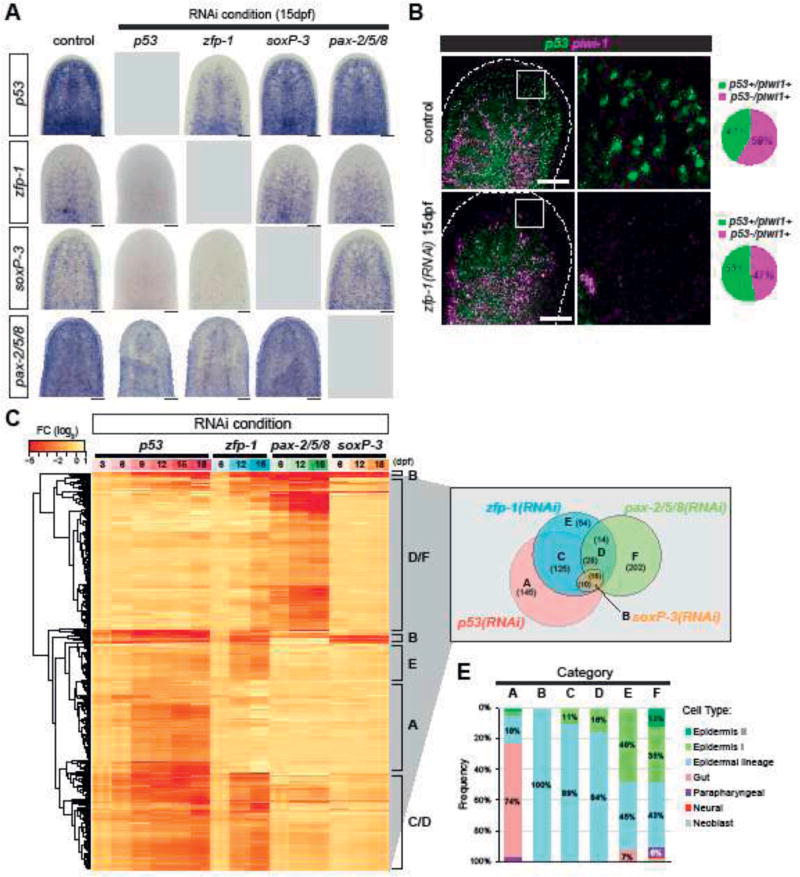
Figure 3. p53, zfp-1, soxP-3 and pax-2/5/8 regulate shared and distinct gene sets in a hierarchical transcriptional network.
(A) WISH of zfp-1, p53, soxP-3 and pax-2/5/8 in the corresponding RNAi. p53 RNAi eliminates the expression of all three TFs (n > 8, 100% penetrance). zfp-1 RNAi eliminates soxP-3 and pax-2/5/8 expression, and reduce p53 expression in subset of mesenchymal cells, likely due to the loss of early and late progeny cells. soxP-3 and pax-2/5/8 RNAi do not affect the expression of any of the other three TFs. Scale bar, 100µm. (B) p53 and piwi-1 double FISH shows that p53 expression in the neoblast is not reduced by zfp-1 RNAi. Scale bar, 100µm. Quantification of p53+ neoblasts of the representative images is shown in the pie charts (total cells counted: control = 1028; zfp-1(RNAi) = 648). (C) FISH of pooled riboprobes of sigma-class (soxP-1 and soxP-2), gamma-class (hnf-4 and gata456), and zeta-class (zfp-1, egr-1, soxP-3 and fgfr-1) is performed to identify neoblast sub-populations and their co-localization with p53. Boxed region is split into individual channels to the right. Arrowheads point to the triple positive cells. Scale bar, 100µm. (D) Expression profiles of 594 DE-genes identified in the RNA-Seq analysis of p53-, zfp-1-, pax-2/5/8-and soxP-3(RNAi) animals. Selection criteria of the gene list: FC > 2, p-adj < 0.001, ≥ 2 time points, except for p53(RNAi) in which FC > 2, p-adj < 0.001, ≥ 4 time points were used. DE-genes are classified into 7 major categories (A–F) and the numbers of each category are shown in the Venn diagram. (E) Cell type specific genes (Wurtzel et al., 2015) represented in the DE-gene Categories. Category A contains large numbers of gut markers. Categories B/C/D/E/F contain genes expressed along the epidermal lineage (including early progeny, late progeny and mature epidermis). More genes expressed in the mature epidermis are present in Category E and F.
To test whether the epidermal lineage of soxP-3- and pax-2/5/8(RNAi) animals can still differentiate properly, we examined the expression of the late progeny markers nucb1-2 and found no difference as compared to control (Figure 3C). Similarly, other previously identified late differentiation markers (Tu et al., 2015; Zhu et al., 2015) are expressed normally in soxP-3(RNAi) animals (Figure 2 Supplement 1) confirming that these early precursor cells are still capable of differentiating into later stages despite of their failure to express some of the early progeny markers. Additionally, neither soxP-3- nor pax-2/5/8(RNAi) induce neoblast proliferation over the time course in treated animals (Figure 2E), suggesting that the integrity of the epidermis is not severely compromised and that a wound response does not appear to be triggered as in the case of other downstream lineage effectors such as egr-5(RNAi) (Tu et al., 2015). Nevertheless, we did observe a small but significant difference in the density of agat-1+ cells in soxP-3- and pax-2/5/8(RNAi) animals (Figure 2F) suggesting that subtle phenotypes associated with cell fate allocation may exist under these conditions.
Hierarchically deployed transcriptional events drive epidermal fate decisions
We sought to define the most likely role soxP-3 and pax-2/5/8 may play in a transcriptional network controlling planarian epidermal differentiation. Therefore, we performed reciprocal RNAi followed by WISH of each TF to determine their most likely epistatic relationships to p53 and zfp-1 in the epidermal lineage (Figure 3A). As expected, soxP-3 and pax-2/5/8 expression in the mesenchyme were undetectable in the p53- and zfp-1(RNAi) animals. Conversely, p53 and zfp-1 expression were not affected by RNAi of soxP-3 and pax-2/5/8. Furthermore, the expression of soxP-3 was not dependent on pax-2/5/8 and vice versa. These observations strongly indicated that soxP-3 and pax-2/5/8 act in parallel to each other and that both are downstream of p53 and zfp-1, likely initiating the expression of early progeny genes cooperatively in the epidermal lineage. Moreover, the epistatic interactions uncovered by our work appear to be epidermal-lineage specific. We noted that pax-2/5/8 expression is also detected in a subset of neuronal cells (Figure 3 Supplement 1), and that such expression is not immediately affected by p53- and zfp-1(RNAi), suggesting that pax-2/5/8 expression in those tissues may be independent of p53 and zfp-1 functions.
Interestingly, while zfp-1 expression is undetectable in the p53(RNAi) animals, p53 expression is only partially eliminated by zfp-1 RNAi (Figure 4A, B). For instance, p53+piwi-1neg cells in front of the photoreceptors (e.g., differentiating early and late progeny cells) completely disappear in the zfp-1(RNAi) animals. However, p53+piwi-1+ cells are still present in the mesenchyme in the zfp-1(RNAi) animals although with a slight increase in their proportion (53% versus 41% in control, Figure 3B). These data suggest that p53 expression in the neoblast compartment may be independent of zfp-1 function, and that there may be increased proliferation in the p53+ neoblast pool in response to the loss of the epidermal lineage.
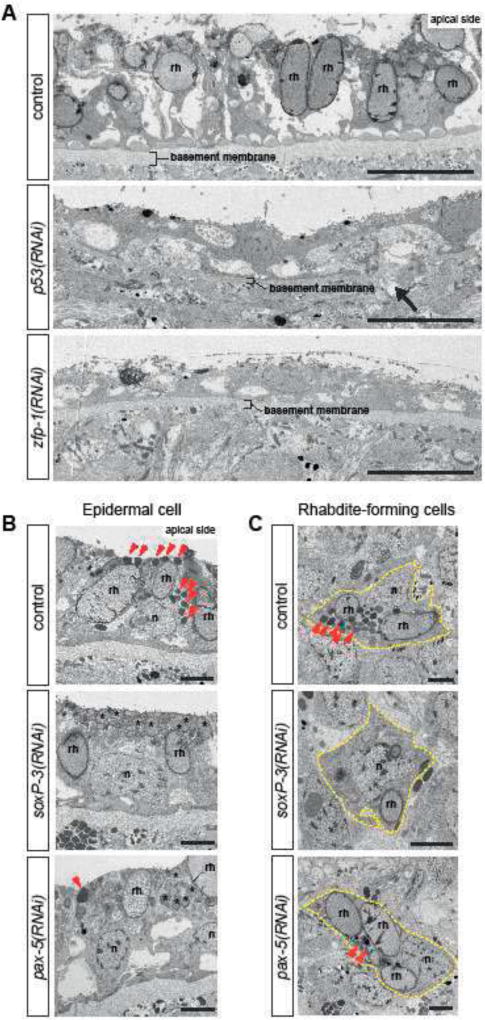
Figure 4. Ultrastructural analysis show distinct epidermal defects inp53-, zfp-1-, soxP-3- and pax-2/5/8 (RNAi) animals.
(A) TEM micrographs of ventral epidermis. p53-and zfp-1(RNAi) epidermis appear to be stretched, contain reduced number of rhabdites (rh) and sit on thinner basement membrane (arrow points to a rupture). n = 3, 100% penetrance. Scale bar, 10µm. (B) TEM micrographs of dorsal epidermis. The numbers of electron-dense granules (arrowheads) in the epidermal cells are greatly reduced by soxP-3-and pax-2/5/8 RNAi. Instead, the apical side of soxP-3 and pax-2/5/8(RNAi) epidermis are filled with rhabdite-like vesicles (asterisks). n, nucleus; rh, rhabdite. Scale bar, 2µm. (C) TEM micrographs of rhabdite-forming cells. Electron-dense granules (arrowheads) observed in the control cell are greatly reduced in soxP-3-and pax-2/5/8(RNAi) cells. Scale bar, 2µm.
p53, zfp-1, soxP-3 and pax-2/5/8 regulate shared and distinct gene sets in the epidermal lineage
To investigate the functions that each of the identified TFs may play in the postmitotic differentiation of the epidermal lineage cells, we generated RNA-Seq datasets of zfp-1-, soxP-3- and pax-2/5/8(RNAi) animals on 6, 12, 18 dpf. Together with a previous p53(RNAi) dataset (Tu et al., 2015), we selected for further analysis 594 DE-genes significantly down-regulated by 2-fold with an adjusted p-value < 0.001 in at least 2 time points in the zfp-1-, soxP-3- or pax-2/5/8(RNAi) time courses, and at least 4 time points of the p53(RNAi) time course (Venn diagram in Figure 3C; Supplementary Table 5). Based on the expression profiles across different RNAi conditions, we catalogued these DE-genes into 6 major categories (A–F) (Figure 3C). 145 genes in Category A are specifically down-regulated in p53(RNAi) but not in other TF RNAi conditions. Category B (the smallest gene set) is composed of genes that are down-regulated in soxP-3(RNAi). Categories C/D/E contain zfp-1(RNAi) DE-genes but may or may not be down-regulated in other RNAi conditions. And Category F encompasses genes specifically down-regulated in pax-2/5/8(RNAi).
We analyzed the data for the presence of cell types influenced by each TF by correlating the DE-genes of each category to a previously published list of cell-type specific markers (Wurtzel et al., 2015) (Figure 3D). We found that DE-genes in Categories B/C/D/E/F contain markers specifically expressed in the epidermal lineage, with more mature epidermal markers in Category E and F. Interestingly, Category A, the p53(RNAi) DE-genes, showed enrichment with gut markers suggesting a role for p53 in the gut or gut progenitors. We tested this prediction by analyzing gut morphology via TEM and observed severe defects in this tissue in p53(RNAi) animals but not in zfp-1(RNAi) specimens (arrowhead, Figure 3 Supplement 2).
Taken together, the data indicate that the RNAi-defined epistasis (Figure 3A) and the corresponding analyses of global transcriptomic changes (Figure 3C) define an epidermal lineage progression that includes a neoblast stage in which the most upstream regulator is p53, which besides targeting genes important for the gut lineage (Figure 3 Supplement 2), also robustly drives the expression of zfp-1 (Figure 3A). In turn, zfp-1 appears to act cooperatively with p53 to induce the expression of multiple TFs, including soxP-3 and pax-2/5/8 (Figure 3A and C), likely responsible for the maturation and diversification of the epidermal lineage (Figure 3D).
Ultrastructural analyses implicate Rhabdite-forming cells are migratory epidermal precursors
In contrast to the readily detectable consequences of abrogating p53-and zfp-1, soxP-3-and pax-2/5/8(RNAi) failed to produce overt epidermal deficiencies. We were struck by the range of phenotypic severity caused by the RNAi of these 4 transcription factors on the planarian epidermis. To better understand these observations, we sought to take advantage of the precise spatial and temporal transcriptional regulation of the epidermal lineage to define ultrastructural characteristics associated with the specification and differentiation of epidermal cells. To guide our analyses, we focused our efforts on rhabdite-forming cells. Rhabdites are specialized, rod-shaped structures found in the epidermis of all flatworms and some nemertene worms (Walker and Anderson, 2001). Rhabdites are discharged in mucous secretions and have been associated with both defensive mechanisms (Hyman, 1951) and adhesion (Martin, 1978).
Rhabdites have distinct morphology and are relatively easy to locate and identify by TEM. We examined tiled TEM images of animal cross sections for the presence of both epidermal rhabdites and rhabdite-forming cells residing in the internal mesenchyme. Rhabdites were readily identified in both epidermal and mesenchymal cells in control animals (rh in Figure 4A, B), but were undetectable in either of these cell types in p53-and zfp-1(RNAi) (Figure 4A and Figure 4 Supplement 1). Moreover, the epidermis of p53-and zfp-1(RNAi) animals appeared stretched and deformed, and the basement membranes noticeably thinner (Figure 4A). Interestingly, we also noticed that while p53(RNAi) affected the presence of rhabdites in both dorsal and ventral epithelia, zfp-1(RNAi) appeared to primarily affect ventral rhabdites (Figure 4 Supplement 1) supporting recent observations that epidermal progenitors respond to positional cues along the planarian dorsoventral axis (Wurtzel et al., 2017). These observations suggest that rhabdite-forming cells may be epidermal precursors and that in their absence epidermis homeostasis is severely compromised. In contrast, ultrastructural analyses of pax-2/5/8-and soxP-3(RNAi) animals revealed a marked reduction or a complete absence, respectively, of a cohort of dark granules normally found in both epidermal and rhabdite forming cells without noticeably altering the rhabdites (Figure 4B and C). Additionally, we found that the space at the apical side of the epidermal cell is instead filled with rhabdite-looking granules in the soxP-3-and pax-2/5/8animals. Altogether, we conclude from these data that planarian epidermal differentiation is likely a multi-step process in which neoblasts produce post-mitotic intermediate cells that progressively acquire features of mature epidermal cells (e.g., rhabdites and other secreted granules) as they integrate into the epidermis.
soxP-3 and pax-2/5/8 function cooperatively to regulate the expression of novel planarian genes
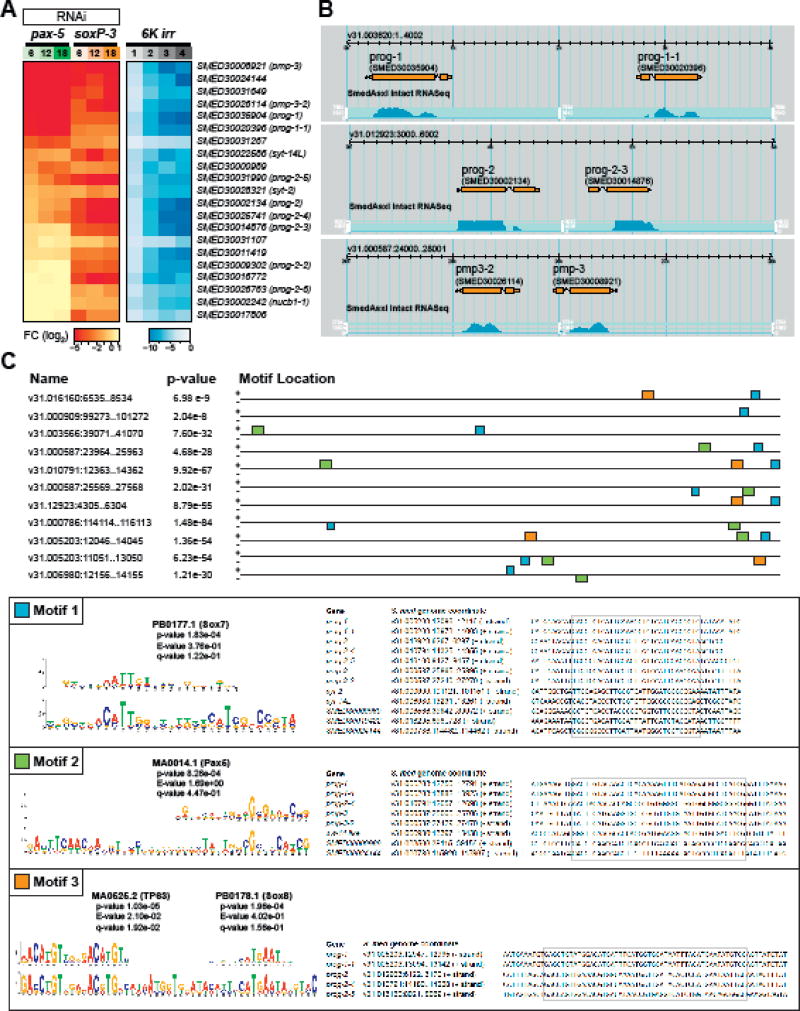
We noted that soxP-3(RNAi) specifically down-regulated a small number of genes (Figure 3C). Except for the homologs of nucleobindin (nucb1-1) and synaptotagmins (syt-2 and syt-14L), all soxP-3-specific transcriptional changes are associated with novel genes devoid of clear homologs in other animal species. Some of these genes were previously identified by irradiation or FACS isolation of X1, X2 and Xin cell populations and named after prog (progeny) or pmp (post-mitotic progeny) (Eisenhoffer et al., 2008; Zhou et al., 2009) We examined the expression profiles of 21 soxP-3(RNAi) DE-genes identified via RNA-Seq from an irradiation time-course and found that they exhibit similar expression kinetics to prog-1 and prog-2 (Figure 5A) suggesting that they are expressed in the irradiation-sensitive early progeny cells. Indeed, double FISH labeling showed 100% co-localization between prog-1 and the tested novel prog genes (Figure 5 Supplement 1A; (Zhu et al., 2015)). Interestingly, only 13 out of 21 soxP-3(RNAi) DE-genes were also down-regulated by pax-2/5/8 RNAi (Figure 5A). We confirmed this by WISH and observed that while prog-1, prog-1-1, prog-2 and prog-2-5 expression levels are markedly reduced in pax-2/5/8(RNAi) animals, prog-2-4 is only reduced moderately and prog-2-2 expression is not noticeably affected when compared to controls (Figure 5 Supplement 1B). Since soxP-3 and pax-2/5/8 are expressed in all early progeny cells, we suspected that the organization of the soxP-3(RNAi) DE-genes may share common genomic features such as nearby DNA sequences that may correlate with their responsiveness to soxP-3 and pax-2/5/8 activities.
Figure 5. soxP-3 and pax-2/5/8 regulate the expression of a novel gene family uniquely arranged in the S. mediterranea genome.
(A) Expression profiles of Category B DE-genes (from Figure 4D) in the pax-2/5/8(RNAi), soxP-3 (RNAi) and irradiation time course RNA-Seq analysis. (B) Screenshot of SmedGD browser showing paired prog and pmp gene loci in the S. mediterranea genome V3.1 (Robb et al., 2015). (C) Top three motifs over-represented in the upstream 2kb sequences of DE-genes common in soxP-3-and pax-2/5/8(RNAi). The consensus matrix of vertebrate Sox7, Pax5, TP63 and Sox8 binding sites derived from TRANSFAC database and the genome coordinates of the motif sites are shown in the box.
We mapped the 13 DE-genes that are down-regulated in both soxP-3-and pax-2/5/8(RNAi) to the S. mediterranea genome V3.1 and obtained 2kb upstream of coding sequences for further analysis. Surprisingly, we found that some of these DE-genes are located in close proximity to each other and form opposing oriented “pairs” on the same genomic contig suggesting that they may share regulatory elements for transcription (Figure 5B Supplement 1). We performed de novo motif analysis using the MEME suite (Bailey et al., 2015) on 2kb upstream sequences from 64,028 mapable genes in the whole genome as background (Figure 5C). After we retrieved the top-enriched motifs, we compare the sequences to the JASPER database and identified at least three highly conserved TF binding sites (Gupta et al., 2007). Motif 1 is shared by 11 out of 13 DE-genes and contains a vertebrate Sox7 binding site. Motif 2 is identified in the eight DE-genes with higher fold-change in the pax-2/5/8(RNAi) animals including prog-1, prog-1-1, pmp-3, pmp-3-2, SMED30000989 and SMED30024144 and contains a vertebrate Pax-2/5/8 binding site. Motif 3 is found in the most highly expressed five DE-genes including prog-1, prog-1-1, prog-2, prog-2-4 and prog-2-5 (mean RPKM = 372 as compared to mean RPKM = 8.9 for the 8 remaining genes). Remarkably, Motif-3 contains not only a vertebrate Sox8 binding site, but also a highly conserved vertebrate TP63 binding site. We also find putative TP63 binding sites in the promoter region of zfp-1 (Figure 5 Supplemental 2) Based on the highly homologous DNA-binding domains of planarian p53 and TP63, their binding sites may also be conserved. We conclude from these data that planarian p53 may bind to the identified putative regulatory elements and potentially modulate the subsequent binding of SoxP-3 and the resulting expression of prog genes.
A novel gene family likely encodes epidermal secreted molecules in planarians
To further investigate the properties and functions of the novel prog genes, we performed SMART domain analysis (Schultz et al., 1998). We identified signal peptides at the N-terminus suggesting that the gene products may be secreted (Figure 6 Supplement 1A). In addition, the predicted proteins from these novel genes share similar architecture (coiled-coil and internal repeats) and nucleotide sequences. For example, prog-2-2, prog-2-3, prog-2-4 and prog-2-5 share higher than 50% identity at the nucleotide level (Zhu et al., 2015) suggesting that gene duplication events may have been involved in the evolution of this planarian-specific gene family.
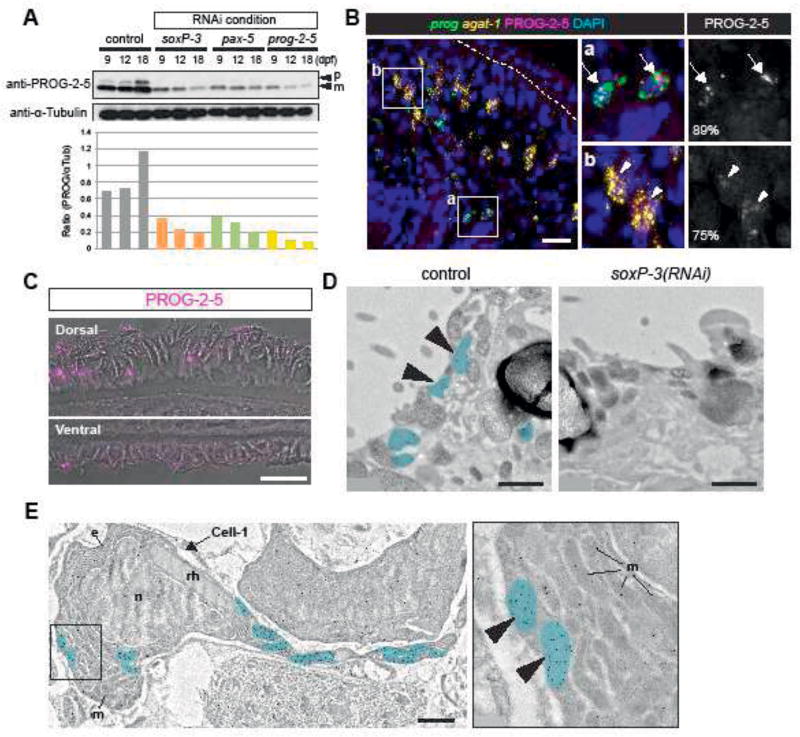
We sought to investigate the cellular distribution of the novel PROG proteins by raising polyclonal antibodies against the protein product of prog-2-5. Purified polyclonal antibodies from two rabbits were tested for specificity on Western blots (Figure 6 Supplement 1B). The polyclonal obtained from Rabbit-2 not only displayed robust and specific reactivity, but also reproducibly detected two protein bands near 15 kDa standard. The two sizes likely correspond to the predicted full-length peptide (16.8 kDa) and the mature form of secreted products lacking the signal peptide (13.6 kDa). To verify specificity, we tested the antibodies on proteins obtained from soxP-3-, pax-2/5/8-, and prog-2-5(RNAi) animals and observed that the two different sizes of PROG-2-5 were both decreased in relation to control (Figure 6A). We conclude from these data that the obtained polyclonal antibodies are likely specific to prog-2-5 protein products.
Figure 6. PROG-2-5 proteins are found associated with granules in the planarian epidermis.
(A) Western blot analysis of PROG-2-5 in soxP-3-, pax-2/5/8-and prog-2-5(RNAi) whole worm lysates. Two bands corresponding to the precursor form (p) and mature form (m) of PROG-2-5 proteins are both significantly down-regulated in the RNAi lysates. Relative densities of PROG-2-5 bands to α-Tubulin are shown at the bottom. (B) Immunostaining of anti-PROG-2-5 combined with double FISH of prog (prog-1 + prog-2 mix) and agat-1. Higher magnification of the boxed regions (a and b) is shown in the right panel. Large PROG-2-5+ puncta are observed in 89% of early progeny (prog+) cells (a, arrows). Smaller PROG-2-5+ puncta are also detected in 75% of late progeny (agat-1+) cells (b, arrowheads). Total 341 cells sampled from 5 animals are analyzed. Scale bar, 20µm. (C) Confocal/DIC images of PROG-2-5 immunostaining in the epidermis. Anti-PROG-2-5 detects puncta on both dorsal and ventral epidermis. Scale bar, 20µm. (D) IEM images showing the apical side of epidermal cells. In the control epidermal cell, immuno-gold labeled anti-PROG-2-5 antibodies detect signals in the oval-shaped granules (pseudo-colored in blue). PROG-2-5+ granules are undetectable in soxP-3(RNAi) epidermal cells. n = 2. Scale bar, 1µm. (E) IEM image of a mesenchymal cell labeled with anti-PROG-2-5 immuno-gold. Large rhod-shaped granules resembling rhabdites are found in the same cell labeled with PROG-2-5+ oval-shaped granules (pseudo-colored in blue) (also see Figure 7 - figure supplement 3). Higher magnification of the boxed region is shown in the right panel. e, endoplasmic reticulum; m, mitochondria; n, nucleus; rh, rhabdite. Scale bar, 1µm.
The newly developed polyclonal antibodies revealed that the mature form of PROG-2-5 (13.6 kDa) was detectable in p53(RNAi) samples even after 20 dpf when all prog-2-5 transcript expressing early progeny cells are gone (Figure 6 Supplement 1C). We interpreted this result by hypothesizing that the protein may have a longer half-life than the mRNA that codifies it. As such, we would expect mature PROG-2-5 proteins to be distributed in places other than the early progeny cells. We tested this hypothesis by developing immunofluorescence (IF) assays for anti-PROG-2-5 in combination with FISH of epidermal lineage markers. We observed a 89% of co-localization of PROG-2-5 puncta with early progeny cells confirming the specificity of the IF staining (Figure 6B, Figure 6 Supplement 2). In addition to the bright puncta observed in the early progeny cells, we also detected less well defined PROG-2-5 IF signal in the majority of the late progeny cells (Figure 6B), and in the mature epidermis on both the dorsal and ventral surface of the animals (Figure 6C, Figure 6 Supplement 3). We conclude from these data that PROG-2-5 proteins are first produced in the epidermal progenitor cells as they differentiate and mature.
PROG-2-5 localizes to discrete vesicles in both precursor and mature epidermal cells
The punctate staining pattern of PROG-2-5 suggested that this protein may be associated with specific organelles such as the rhabdites or the many vesicles found in the cells of the epidermal lineage (Figure 4B and C). In order to examine the subcellular localization of PROG-2-5, we developed reagents and protocols for immune-electron-microscopy (IEM) detection of proteins on fixed planarian sections. The immuno-gold-labeled anti-PROG-2-5 antibodies detected 0.5–1µm-size granules not only on the epidermis (Figure 6D), but also in a cohort of mesenchymal cells with quite distinctive ultrastructural morphology, namely, elongated cell processes, an abundance of mitochondria and secretory granules, densely packed granular ER around the nucleus, and large rod-shape granules (rhabdite precursors) (Figure 6E, Figure 6 Supplement 3). These data clearly indicated that the vesicles containing PROG-2-5 immunogold label are distinct from rhabdites in both precursor and mature cells. Importantly, we find that soxP-3(RNAi) abolishes the production of the PROG-2-5 containing vesicles (Figure 6D, Figure 4B and C). Therefore, we conclude from these data that PROG-2-5 localizes to a distinct and previously undescribed secreted organelle whose production is dependent on the transcriptional activity of soxP-3. We refer to this organelle as the Hyman vesicle (see discussion for details).
Discussion
The homeostatic dynamics and cellular diversity of the planarian epidermis provides an experimentally accessible model to study fundamental aspects of stem cell fate regulation and developmental biology. Here, we uncovered striking molecular similarities underpinning the epidermal lineage progression programs of planarians and mammals. Further, we identified novel components unique to planarians both at the regulatory network and gene level. Together with the phylogenetic position of planarians in relation to both vertebrates and traditional invertebrate model systems (e.g., flies and nematodes), the planarian epidermis provides unique opportunities to inform our understanding of how conserved and derived mechanisms may have evolved to produce, maintain and diversify the metazoan epidermis.
Epidermal lineage progression is mediated by a hierarchical transcriptional program
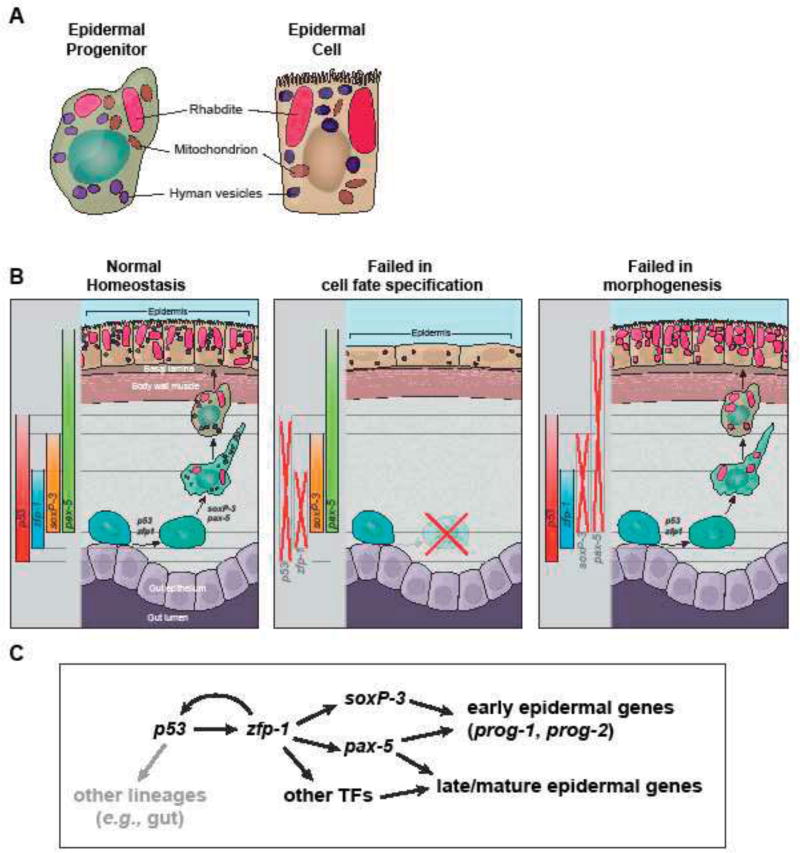
The planarian epidermal lineage is thought to transition through several morphologically and molecularly distinct stages before reaching the body surface (van Wolfswinkel et al., 2014; Tu et al., 2015). Here, we demonstrate that P53 likely initiates a hierarchical transcriptional program mediated by a defined cohort of TFs that sequentially activate gene expression as neoblast progressively differentiate into epidermal cells (Figure 7C). Our RNAi data support a model in which zfp-1 is a transcriptional target of P53. Indeed, we found putative, highly conserved, vertebrate-like TP63 binding sites within a 5kb DNA region upstream the likely transcription start site of the zfp-1 locus (Figure 5 Supplement 2; Supplementary Table 7). ZFP-1, in turn, likely acts cooperatively with P53 to control the expression of other downstream TFs associated with epidermal functions (Figure 7C). This feed-forward, hierarchical arrangement of epidermal-associated TFs in planarians is characteristic of gene regulatory networks that mediate developmental processes, and often induce the sequential expression of gene repertoires that effect the functional and morphological changes of a cell (Erwin and Davidson, 2009).
Figure 7. Model of epidermal lineage progression in planarian.
(A) Schematic of ultrastructures of epidermal precursor and mature epidermal cell. Epidermal precursors contain features of the mature epidermal cells including large rod-shape rhabdites and small oval-shape granules. They are also abundant in mitochondria. (B) Schematic of the planarian epidermal lineage progression and the phenotypic differences in p53-, zfp-1-, soxP-3 and pax-2/5/8 (RNAi) animals. (C) Proposed transcription network in the epidermal lineage progression. Gray arrow represents aspects of the network not studied in depth.
The observed spatiotemporal transcriptional organization of the planarian epidermal lineage may be a consequence of transcriptional priming, an epigenetic mechanism thought to act at enhancers to bookmark loci for transcriptional activity at later stages of differentiation (Vernimmen and Bickmore, 2015). In fact, TP63 is thought to function as a pioneer factor, and binding of TP63 to chromatin is associated with the deposition of transcriptional activation marks that provide instructive cues that facilitate the binding of other TFs (Kouwenhoven et al., 2015). Given the recent demonstration that p63 depletion in mice results in significant changes in the expression of genes involved in chromatin organization, remodeling and epigenetic modification (Boxer et al., 2014; Sun et al., 2015), it should be fruitful to determine whether SMEDP53 functions as a pioneer factor or not. In addition to zfp-1, the identification of predicted and highly conserved TP63 binding sites in some of the early progeny genes suggests that planarian P53 may facilitate SOXP-3 and PAX-2/5/8 binding. We have recently established an experimental approach for quantitatively assessing epigenetic landscapes in planarian neoblasts (Duncan et al., 2015) that could be extended to measuring transcription factor occupancy via chromatin immunoprecipitation (ChIP). Efforts to develop a ChIP-grade antibody against planarian P53 are currently underway as this will be an essential reagent in understanding not only how p53 influences the activity of downstream TFs, but also whether it too plays a role in regulating the global epigenetic landscape of the epidermal progenitor cells.
The planarian epidermis as an invertebrate model to study dynamic epidermal organs
The regulatory association of Smed-p53 and zfp-1 is reminiscent of what is presently known about mammalian epidermis homeostasis, where one of the key targets of P63 is also a zinc finger protein, i.e., ZNF750 (Sen et al., 2012; Zarnegar et al., 2012). Even though, zfp-1 and znf750 are both members of the large family of zinc-finger proteins, their functional homology remains to be established. However, we find it curious that a network architecture involving members of two distinct family of proteins (TP53/63 and ZNF) share similar gene regulatory architecture and hierarchy in both planarian and mammalian epidermal development. The P63/ZNF750 network in the epidermis has been a focus of intense research for its relevance to many human pathologies including atopic dermatitis, psoriasis, ichthyosis vulgaris, and skin cancer (Lopez-Pajares et al., 2013). However, because of the complexity of p63 isoforms and the crosstalk between p53/p73 and p63 due to their highly conserved DNA-binding domains, it has been a challenge to dissect p63 mediated downstream pathways responsible for its diverse functions (Vanbokhoven et al., 2011). Additionally, it is not known whether deletion of ZNF750 will cause developmental defects in epidermal tissue other than psoriasis, which is caused by point mutations that lead to hypomorph phenotype of this gene in human patients, as there are presently no published reports on the targeted elimination of this gene in mice. Nevertheless, based on the data presented here, the fact that the only member of the P53 family of proteins in S. mediterranea is P53, and that the p53 locus only encodes a single mRNA species (Pearson and Sánchez Alvarado, 2010), we propose that planarians offer a unique opportunity to resolve the spatiotemporal transcriptional organization of dynamic epithelia. If in fact such network architecture shares a common evolutionary origin, a clear and testable prediction would be that abrogation of ZNF750 in the mouse epidermis should result in early lineage epidermal deficiencies similar to those observed in zfp-1(RNAi) treated planarians.
Altogether, our data establish an intriguing parallel between mammals and planarians of key molecular mechanisms driving the self-renewing capacity and commitment of progenitor cells in the respective epidermal tissues of these organisms.
p53 family may play an ancient role in generating ectodermally derived tissues
In vertebrates, the p53 gene family of transcription factors is composed of three members (tp53, tp63 and tp73) and is known to play essential roles in tumor suppression, development and germ-line protection (Dötsch et al., 2010). Phylogenetic analyses of vertebrate p53 family members suggested that they originated from a tp63-like common ancestor (Joerger et al., 2014). Our study supports this finding. We show that Smedp53 is essential for maintaining the homeostasis of the planarian epidermis, a role played in the mammalian epidermis by the vertebrate TP63. This observation suggests that an ancient role of a P53 family member in epidermal development may have predated the split of deuterostomes and protostomes. How this genetic program may have been lost in flies and C. elegans is unknown. One explanation might be that the mature adult epidermis of both organisms does not turn over as rapidly as the vertebrate stratified epidermis or the planarian epidermis. Therefore, flies and nematodes likely evolved distinct developmental strategies by deploying alternative genetic programs. This is supported by recent studies of adult epidermal wound healing and repair in the fly D. melanogaster. In this organism, repair of damaged abdominal epidermis does not involve the recruitment and/or activation of epithelial progenitors, but rather the polyploidization of the surviving epidermal cells surrounding the wound via activation of Hippo and JNK signaling, resulting in efficient covering and healing of the damaged epidermis (Losick et al., 2013; Losick et al., 2016). Studies of more clades of animals that renew and regenerate their epidermis will be necessary to further investigate this hypothesis.
A conserved Sox/Pax transcription factor pair is deployed to drive specific aspects of planarian epidermal morphogenesis
The Pax and Sox families of transcription factors are known for their diverse functions in development and disease. In the vertebrate eye development, Sox2 partners with Pax6 to activate the expression of δ-crystallin, a water-soluble structural protein found in the lens and the cornea (Kamachi et al., 2001). Interestingly, Pax-6 alone binds poorly to the lens-specific enhancer DC5 and requires the binding of Sox-2 to DC5 to facilitate the formation of a ternary complex of Pax6/Sox2/DC5 (Narasimhan et al., 2015). Remarkably, the reporter carrying vertebrate DC5 enhancer is activated by Drosophila Pax and Sox homologs, D-Pax2 and SoxN, in the Crystallin secreting cone cells of the compound eyes suggesting that Pax/Sox cooperative binding is evolutionary conserved.
In the epidermis, SOX and PAX proteins appear to act independently of each other. The SOX family of transcription factors, for example, has been extensively implicated in the modulation of fate lineages in the mammalian epidermis, particularly in driving the differentiation of epidermal fibroblasts into dermal papillae and keratinocytes (Driskell et al., 2009; Driskell et al., 2013; Lesko et al., 2013; Perdigoto et al., 2014). In addition, the highly specialized mechanoreceptor of the vertebrate skin develop thorough an entirely postmitotic and progressive maturation process that is also specified by SOX2 (Perdigoto et al., 2014). Interestingly, a role for PAX6 proteins in the maturation of Merkel cells has been also identified and appears to be the only reported function of this transcription factor in epidermal development and maintenance thus far (Parisi and Collinson, 2012).
Our study, however, found a similar role for a member of the PAX family of transcription factors in modulating the fate of epidermal progenitors (Figures 2A and 3A). To our knowledge, a partnership of Sox/Pax proteins in the epidermis has not yet been described. Our data indicate that planarian soxP-3 and pax-2/5/8 function cooperatively in the expression of a subset of prog genes during epidermal differentiation. Given their genomic architecture (Figure 5B), the newly identified prog genes are likely the result of gene duplication in the S. mediterranea genome, and may be a recently evolved gene family for an adaptation specific to planarians. Irrespective of the evolutionary origin of the PROG family of proteins, it remains intriguing that their regulation appears to be under the control PAX and SOX proteins, which themselves belong to gene families that can be dated back to the appearance of multicellularity (Degnan et al., 2009). Therefore, the SoxP-3/Pax-2/5/8 pair controlling the expression of prog genes may be a re-deployment of another more ancient network, which was integrated into the P53-mediated epidermal program. Given the roles that the TP53, SOX and PAX families of proteins play in the maintenance, diversification and specialization of the mammalian and planarian epidermis, S. mediterranea provides us with an opportunity to readily explore how conserved gene regulatory networks may be co-opted to drive the emergence of new specialized structures in evolutionarily conserved tissues.
Rhabdite-forming cells are epidermal precursors in planarians
Rhabdites are large rod-shaped granules found in the epidermis of most Turbellarians (Hyman, 1951). They are embedded in the epidermis and are discharged when the animals are irritated. Planarian rhabdites are thought to contain mucopolysaccharides that form a mucous cuticle protecting the animal from environmental chemicals and pathogens. They are also deposited in the slimy tracks left behind by the animals, and therefore are also suggested to facilitate cilia-based gliding (Martin, 1978; Rompolas et al., 2010). Although the function of rhabdites have not been formally tested, this epidermal specialization is likely to play important roles in the interaction between planarians and their environment.
Previous studies have documented that rhabdite-forming cells in the mesenchyme may be epidermal precursors in some triclad species. Skaer observed that rhabdite-forming cells develop at the later stage of the embryogenesis of Polycelis tenuis embryos, and become the animals’ mature epidermis before they hatch (Skaer, 1965). Hori observed that rhabdite-forming cells participate in the regeneration of epidermis in the amputated Dugesia japonica (Hori, 1978). Here we made polyclonal antibodies that successfully detect the expression of an early progeny marker, PROG-2-5, and observed co-localization of PROG-2-5 with rhabdite-forming cells in the planarian S. mediterranea. Moreover, p53 and zfp-1 RNAi eliminate rhabdite-forming cells in the mesenchyme supporting that they are epidermal precursors (Figure 4 Supplement 1). Our molecular and genetic studies, therefore, unambiguously validate previous ultrastructural and morphological observations, and confirm that these otherwise mysterious mesenchymal cells with distinct organelles are indeed planarian epidermal precursors. We postulate that the combined activities of p53 and zfp-1 are likely responsible for cell fate specification, while soxP-3 and pax-2/5/8 are more likely involved in driving aspects of epidermal lineage morphogenesis.
PROG-2-5 is a novel secreted protein that defines the Hyman vesicles, specialized organelles specific to the planarian epidermis
The molecular properties and functions of the non-rhabdite epidermal granules, generally termed rhabdoids (Hyman, 1951), are completely unknown. Their likely secretory properties make them prime candidates in modulating the chemical microenvironment at the epidermal surface of planarians. Several hypothesis have been proposed regarding the nature of these secretory vesicles. Coward and Piedelato suggested that the rhabdoids may be responsible for generating the noxious taste characteristic of Turbellarians, thus repulsing potential predators (Coward and Piedilato, 1972). Additionally, Tyler suggested that rhabdoids may contain materials that can be added to the apical surface of the epidermal cells to strengthen their barrier function, or adhesive functions (Tyler, 1976). Here we show that a member of the PROG family of small, secreted proteins (PROG-2-5) is a specific component of these vesicles. Because the term rhabdoid is currently used in the literature to describe either pediatric malignant tumors (Geller et al., 2015) or the rod-shaped protoplasmic body found in the sensitive cells of insectivorous plant leaves (Darwin, 1875), we thought it appropriate to honor the memory and meticulous work of Libbie Hyman (Hyman, 1951) by naming these PROG-2-5+ organelles Hyman vesicles.
Still, the function of these vesicles remains to be determined. Their elimination by chronic soxP-3 dsRNA treatment (3 months) did not result in an observable defect under standard laboratory conditions, suggesting that these vesicles may have highly specialized functions associated with environmental selective pressures we have yet to recapitulate in captivity. The fact that prog genes exist in pairs and are all exquisitely regulated by soxP-3 suggests that these genes have been subjected to selective pressure. Gene duplications are a molecular mechanism known to mediate the generation of functional novelty. As such, it is thought that selective pressures acting on the gene copies immediately after the duplication event shape the maintenance and functional divergence of duplicated genes (Hittinger and Carroll, 2007; Des Marais and Rausher, 2008). It still remains a distinct possibility, therefore, that prog genes may exist in other organisms, as they may represent fast evolving orthologs whose homology by sequence alone cannot be easily ascertained. This possibility can only be tested by first carrying out extensive taxon sampling to overcome long evolutionary distances, followed by functional studies of the identified candidates to establish orthologies. Such an approach, if systematically executed, should help further determine the extent of conservation that may or may not exist between the different strategies deployed by animals with homeostatically dynamic epidermal tissues to diversify and specialize their respective epithelial cells.
Conclusion
Our work demonstrates that both conserved and unique transcriptional modules are integrated into a hierarchical regulatory program (Figure 7B) driving the specification and differentiation of the planarian epidermis (Figure 7C). We have shown that planarians, like mammals, require TP53, zinc-finger, and SOX proteins to modulate the progressive, postmitotic differentiation of epidermal cells. This is in contrast to other invertebrate model systems (e.g., flies and nematodes) in which P53 is dispensable for adult epidermal development. We also uncovered a shared role between mammals and planarians for PAX proteins in modulating the fate of epidermal progenitors into specialized cells. In planarians, however, pax-2/5/8 cooperates with soxP-3 and togwtther promote the morphological differentiation of epidermal progenitors through the activation of a likely recently expanded family of novel and/or fast-evolving PROG proteins. Our study sheds light on the genetic and molecular programs that likely underpin the evolution of complex, dynamic epidermal tissues. It also puts forward the planarian epidermis as a useful invertebrate model system to study the conserved and derived mechanisms coordinating cell proliferation and differentiation in epidermal tissues undergoing constant homeostasis.
Material and Method
Animal care and irradiation
The CIW4 clonal line of S. mediterranea was maintained at 20°C in 1X Montjuic salts (Cebrià and Newmark, 2005) supplemented with 50 ug/mL gentamicin sulfate and fed homogenized calf liver paste once per week. Planarians 2–3 mm in length were starved for at least 1 week before use. Irradiation was done with a GammaCell 40 Exactor irradiator at a dose rate of 85 Rad/minute.
Gene cloning
Genes in this study were cloned from a CIW4 regeneration time-course cDNA library into the pPR-T4P vector as previously described (Gurley et al., 2008).
Primers used in this study:
| Smednr ID | Name | Sequence | |
|---|---|---|---|
| SMED30026364 | p53 | forward | CATTACCATCCCGAACTAGACATTATCGATGGCCC |
| reverse | CCAATTCTACCCGGCACATAGCACACATGACAAAG | ||
| SMED30023708 | zfp-1 | forward | CATTACCATCCCGCAGCCAGTTTTATTTCACCAGG |
| reverse | CCAATTCTACCCGTCTACGTCCATGGAGTCAGTTG | ||
| SMED30012332 | egr-1 | forward | CATTACCATCCCGCATCGTTTGAGGTTCATTCG |
| reverse | CCAATTCTACCCGTGATATTTCCGTTGATGGGG | ||
| SMED30008505 | soxP-3 | forward | CATTACCATCCCGTTACCTGTGCAGACAATTCG |
| reverse | CCAATTCTACCCGATCAAGGAACTGACGGATGG | ||
| SMED30006353 | pax-2/5/8 | forward | CATTACCATCCCGGACCTCTCCCTAACAGCGTG |
| reverse | CCAATTCTACCCGTATAGAGGCAACCCCACTGC | ||
| SMED30010878 | six3-1 | forward | CATTACCATCCCGGGTTTGTGAAACGCTAGAAG |
| reverse | CCAATTCTACCCGTAGACCGTAGGAAACGAATG | ||
| SMED30000938 | six6 | forward | CATTACCATCCCGACGATGAGACTATGACTACGC |
| reverse | CCAATTCTACCCGATGTCGGTGTTTTTGGTCTC | ||
| SMED30006464 | tcf-15 | forward | CATTACCATCCCGGGAATTAGTAAACGATCGGC |
| reverse | CCAATTCTACCCGAGCACTCTTGAAATCTGTCG | ||
| SMED30023969 | lmo-1 | forward | CATTACCATCCCGCGAATGAATAGTAGCAACGC |
| reverse | CCAATTCTACCCGGGTCAGAACTTCAGAAAACG | ||
| SMED30011950 | gata123 | forward | CATTACCATCCCGCCCTATCAAGCCTCACTATG |
| reverse | CCAATTCTACCCGGCTGGTTTGAGGTATTGAAG | ||
| SMED30022628 | plac8 | forward | CATTACCATCCCGTGTGGTGAATTTTGTTTCC |
| reverse | CCAATTCTACCCGTATTTGTAATCCGGTTGTCC | ||
| SMED30032499 | rest | forward | CATTACCATCCCGCCATCCAGATTTACCATTCG |
| reverse | CCAATTCTACCCGGCTAGAGATCTTCTGCTGCG | ||
| SMED30000485 | blimp-1 | forward | CATTACCATCCCGTCGACAAGTTGCTCAATGAC |
| reverse | CCAATTCTACCCGCCAATGATATTTGAGTCCGC | ||
| SMED30010086 | barh2-1 | forward | CATTACCATCCCGGGTTCGATTGGTTCATGTC |
| reverse | CCAATTCTACCCGGTTGTTTCTAGCTGGGATTG | ||
| SMED30015704 | post-2d | forward | CATTACCATCCCGATTGGACCGACAATTCACTC |
| reverse | CCAATTCTACCCGTGGCCAGTGGATGTTATTTC | ||
| SMED30000303 | post-2c | forward | CATTACCATCCCGGGATTTTGAAGCTGTGGAGC |
| reverse | CCAATTCTACCCGTTGGGTTTGCTGTAATTGGC | ||
| SMED30013674 | post-2b | forward | CATTACCATCCCGGCAATAAAATCACTGGCTGG |
| reverse | CCAATTCTACCCGAATCAATGAGTCCTGGCAAC | ||
| SMED30021036 | post-2a | forward | CATTACCATCCCGGAACCACTTCCTCCTATTTGTG |
| reverse | CCAATTCTACCCGGAGGTCATTCTTTATCCGGTC | ||
| SMED30019046 | sd2 | forward | CATTACCATCCCGGTCAAAAGAAGCTGAAGGTG |
| reverse | CCAATTCTACCCGATACATCGGATGGACAACTG | ||
| SMED30033839 | sp5 | forward | CATTACCATCCCGACGTCGATTCACGTTTTACC |
| reverse | CCAATTCTACCCGGCGCTGTTTGAGACAGAAAG | ||
| SMED30006011 | tbx2/3b | forward | CATTACCATCCCGTGTAAGCATTGTCAGTGGTGTC |
| reverse | CCAATTCTACCCGGATCTGGTGTCTCTCGTTTTTC | ||
| SMED30015041 | rar-b | forward | CATTACCATCCCGGTGAAGTATCGACTCGACGG |
| reverse | CCAATTCTACCCGAGCGCGTAGATCTCACAGTC | ||
| SMED30011047 | gli-1 | forward | CATTACCATCCCGTAGGTGTGTACCGGTGTGTG |
| reverse | CCAATTCTACCCGCCGAATCCATTTGTAACGAG | ||
| SMED30022043 | osr | forward | CATTACCATCCCGTCGTTGCAGTCCATTCAATC |
| reverse | CCAATTCTACCCGACTTCTTGGCTGGCAGAATC |
RNAi
RNAi was performed by feeding as previously described (Gurley et al., 2008) with modifications. Briefly, cloned gene vectors were transformed into bacterial strain HT115, cultured in 2XYT to O.D.= 0.8 – 1, induced to express dsRNA for 2 hours with IPTG (final 1mM) at 37°C with vigorous shaking. Bacterial pellets were rinsed once with Milli-Q water, and mixed with an equal weight of calf liver paste. dsRNA food was given to the animals every 3 days for three feedings.
FACS
FACS sorting of X1s was performed as previously described (Reddien et al., 2005; Hayashi et al., 2006) with modifications. Briefly, animals were minced and dissociated in ice-cold CMF supplemented with 1% BSA. Cell suspension was filtered through 20 µm nylon mesh, pelleted, resuspended in room temperature IPM (Schürmann and Peter, 2001), supplemented with Hoechst 33342 (final = 50 µg/mL), and incubated at R.T. in the dark for 40 minutes. Propidium iodide was added (final = 1 µg/mL) for the last 5 minutes to label dead cells. Viable cells were then sorted using Influx (BD Biosciences).
RNA-Seq and gene expression analysis
Whole worms or sorted cells were homogenized in TRIzol reagent (Sigma) and RNA was isolated as described in the manufacturer’s manual. RNA from whole worms was treated with RNase-free DNase on QIAGEN RNAeasy columns and eluted in nuclease-free water. For each replicate, 1 µg RNA from five worms or 100 ng RNA from 105 sorted cells was used to generate RNA-Seq libraries using the Illumina TruSeq Stranded kit. Libraries were sequenced in 50bp single reads (whole worms) or in 150bp paired reads (sorted cells) using the Illumina HiSeq 2500 sequencer. RNA-Seq analysis was carried out by mapping reads to the transcriptome as previously described (version: smed_20140614) (Robb et al., 2015; Tu et al., 2015). Reads were aligned using Bowtie with the following parameters: --best --strata -v 2 -m 5, and read counts to genes were tallied from the SAM files with a custom script. Differential gene expression was evaluated using R and the edgeR library (Robinson et al., 2010). P-values were adjusted, as previously described (Benjamini and Hochberg, 2000). Analyses were based on the mean expression values of four biological replicates, except for the p53(RNAi) whole worm dataset in which 3 replicates were used (Tu et al., 2015). Fold changes were normalized and compared to controls of the same time-point (unc-22 dsRNA treated animals). The RNA-Seq data and the transcriptome against which it was quantified are archived at GEO (Edgar et al., 2002) and are available under accession number: GSE80562.
GO-enrichment analysis
S. mediterranea transcripts were aligned with NCBI BLASTX to Swiss-Prot protein database (Consortium, 2015). The top BLAST hits with a minimum e-value of 0.001 were used to annotate transcripts. GO functional categories were assigned and the enrichment was generated using DAVID (version 6.7) (Huang et al., 2009). Terms were clustered using ReVigo script (Supek et al., 2011) at a similarity cutoff of 0.7 (Medium). A representative term from each cluster was plotted based on GO term enrichment p-value.
Transcription factors homology predictions
Possible transcription factors were identified by domain annotation as well as manual curation. PFAM domains were identified in target sequences using HMMER search (Eddy, 1998; Finn et al., 2016) with an e-value cutoff of 0.01. The list was then compared to another list created from those annotated by the Gene Ontology Consortium with GO term “regulation of transcription” (GO: 0006355) (Ashburner et al., 2000).
In situ hybridization, immunostaining, and TUNEL assay
Anti-sense riboprobes were made by in vitro transcription with T7 polymerase from genes in the pPR-T4P vector as previously described (Pearson et al., 2009). Whole mount in situ hybridization (WISH) and fluorescent in situ hybridization (FISH) were performed as previously described (King and Newmark, 2013).
Immunostaining of anti-H3P (dilution 1:2000, Millipore, 63-1C-8) was performed after whole-mount FISH, and was detected with Alexa Fluor-conjugated antibodies (dilution 1:1000, Invitrogen). Both antibody incubations were performed for overnight at 4°C.
Immunostaining of anti-PROG-2-5 was performed on cryosections following FISH. Briefly, 4–6 mm planarians were rinsed with ice-cold 1X Montjuic salt, fixed in ice-cold 4% formaldehyde/PBSTx (0.3% Triton-X) for overnight, rinsed with PBSTx, infiltrated with 30% sucrose/PBS, embedded in O.C.T, frozen on dry ice and cut into 10 µm sections. After cryo-sectioning, samples were rinsed in PBS, bleached with 3% H2O2/PBS for 30 minutes under light, rinsed with PBS, equilibrated with 50% hybridization buffer, and hybridized with appropriate riboprobes (0.1–0.2 µg/mL) at 60°C for overnight on a humidified horizontal tray. Excess riboprobes were then removed by stringent washes in the 15-mL slide mailer (1X with 50% formamide/5X SSC, 3X with 2X SSC, 3X with 0.2X SSC, all done in 60°C with 20 minutes intervals). Samples were rinsed with PBS, blocked with 5% hose serum (Sigma) and 0.5% Western blocking reagent (Roche) in PBSTw (0.1% Tween-20), incubated with appropriate HRP-conjugated antibodies diluted in PBSTw at R.T. for 2 hours (antibody dilutions: anti-DIG, 1:500; anti-DNP, 1:200, anti-FITC, 1:500). Antibodies were washed with PBSTw and developed with tyramide-conjugated fluorophores diluted in 0.006% H2O2/borate buffer. After desired signals were developed, HRP activity was quenched with 200mM sodium azide. Immunostaining of anti-PROG-2-5 was performed at 4°C for overnight (1:5000 diluted in PBSTx), washed, and incubated with Alexa Fluor-conjugated secondary antibodies (1:1000 diluted in PBST) for 2 hours at R.T.
TUNEL assay was performed as previously described (Pellettieri et al., 2010; Tu et al., 2015).
Antibody production
Full length prog-2-5 was cloned into pET21a vector for recombinant protein induction in E. coli. Rabbit polyclonal antibodies were generated against purified recombinant PROG-2-5 and affinity purified (Proteintech).
Western blotting
Planarian total lysate was extracted in ice-cold 2X lysis buffer (4% SDS, 200mM DTT, 250mM Tris-HCl pH6.8, supplemented with 2X cOmplete ULTRA (Roche) prior use) and centrifuged at full speed (>10,000 g) for 15 minutes at 4°C. The supernatant was mixed with an equal volume of 2X sample buffer, boiled for 5 minutes, loaded on 4–20% Mini-Protean TGX gels (BioRad), and transferred to 0.45 µm PVDF membranes. The membranes were blocked in 5% non-fat milk/TBSTw (0.1% Tween-20), incubated with appropriate primary antibodies (diluted in blocking reagent) overnight at 4°C with shaking, washed with TBSTw, incubated with appropriate HRP-conjugated secondary antibodies (diluted in TBSTw) for 1 hour at R.T., washed and developed using Amersham ECL Western Blotting Detection Reagent or SuperSignal West Dura Extended Duration Substrate (ThermoFisher). Antibody dilutions as follow: Rabbit anti-PROG-2-5 polyclonal IgG 1:10,000; Mouse anti-αTubulin monoclonal IgG (Sigma, T5168) 1:10,000.
Electron microscopy
For ultrastructural analysis, worms were fixed in cold fixative containing 2.5% glutaraldehyde and 2% paraformaldehyde in cacodylate buffer (pH 7.35) and stored at 4°C in fixative until processing. After rinse in the same buffer, samples were fixed in 1% Osmium tetroxide 2 hours and En Bloc staining overnight in 0.5% aqueous uranyl acetate. Samples were then dehydrated in gradient acetone at 4°C, infiltrated at room temperature with Spurr’s resin (Electron Microscopy Sciences Kit and recipe), and polymerized at 60°C for 48 hours. Thin sections were cut at 50 nm on a Leica UC6 ultramicrotome,collected on slot grids, and post-stained in Sato’s lead and 4% uranyl acetate.
For immunogold labeling, planarians were fixed in 4% paraformaldehyde and 0.1% glutaraldehyde in 0.1 M PBS (pH 7.4) for overnight, dehydrated in gradient ethanol, and infiltrated and embedded in LR White resin. Sections of about 70nm were stained with rabbit anti-PROG-2-5 primary antibody (1:200) followed by 12 nm colloidal gold goat-anti-rabbit secondary antibody (Jackson Immunoresearch Labs; 1:25 dilution). All images were taken on a FEI Technai BioTwin microscope equipped with a Gatan Ultrascan 1000 digital camera.
De novo motif analysis
The S. mediterranea transcriptome (version: smed_20140614) were aligned to the S. mediterranea sexual genome assembly version 3.1 (SmedSxl_v3.1) using map2assembly, a script included in the MAKER package (v2.31.8) (Holt and Yandell, 2011). The alignment for each gene of interest with the best map2assembly score was used to pinpoint the genomic sequence regions for which to extract promoters, 2kb or 5kb upstream of the coding sequence.
To identify putative regulatory regions, the MEME Suite (4.11.1) (Bailey et al., 2015) was used. Two sets of upstream sequences (2kb), the experimental set (13 sequences from genes down-regulated in both soxP-3-and pax-2/5/8(RNAi) animals) and the background set (32,104 sequences from all S. mediterranea genes mapped) were first processed with psp-gen to generate a position-specific prior that was then used as input to MEME. TOMTOM was run with the JASPER motif database to identify consensus to known motifs (Gupta et al., 2007; Mathelier et al., 2016).
To determine the p63 motifs, p63scan program was downloaded from http://131.174.198.125/bioinfo/p53scan/ and supplied a FASTA file with the promoter sequences (5kb) for 19 TFs (of which expression levels are down-regulated in both p53-and zfp-1(RNAi) X1s) using optimal threshold as described previously (Kouwenhoven et al., 2010).
Supplementary Material
Highlights.
We identify a role in early epidermal progenitor specification for two transcription factors, SOXP-3 and PAX-2/5/8
We put together a framework for a transcriptional hierarchy/network with analyses of previously established epidermal factors P53 and ZFP-1, and with the identification of putative transcription factor binding sites for P53/SOX/PAX factors near the loci of epidermal specification genes
We developed an antibody specific for a family member of the strongly expressed PROG family of proteins
We uncover the presence of PROG-2-5 protein in epidermal cells and solidify early progeny cells as epidermal progenitors
We expand on epidermal lineage with ultrastructural analyses of epidermis and progenitors and discover a new epidermal vesicle we termed Hyman vesicles in honor of the late zoologist.
Acknowledgments
We would like to thank members of the Sánchez Alvarado lab for critical reading of the manuscript, especially Sarah Elliot for the editing of our first draft. We thank Hanh Vu and Erin Davies for kindly sharing their plasmids and RNA probes. We thank Eric Ross and Kirsten Gotting for bioinformatics assistance, Sean McKinney for image processing assistance, Melania McClain and Kexi Yi for EM assistance, Allison Peak for RNA-seq library assistance, Mark Miller for illustration assistance, and all members of the Cytometry, Histology, Molecular Biology and Planaria core facilities at the Stowers Institute for their technical support. This work was supported by NIH R37GM057260 to ASA. ASA is an investigator of the Howard Hughes Medical Institute and the Stowers Institute for Medical Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Additional Information
Original data underlying this manuscript can be accessed from the Stowers Original Data Repository at http://www.stowers.org/research/publications/LIBPB-1174
Author contributions
LCC, ASA, conception and design, drafting and revising the article; LCC, KTU, CWS, SMR, FLG, acquisition of data, and analysis; LCC, KTU, CWS, SMR, FLG, ASA interpretation of data.
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry MJ, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. Nature Genetics. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Johnson J, Grant CE, Noble WS. The MEME Suite. Nucleic Acids Research. 2015;43(W1) doi: 10.1093/nar/gkv416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. On the Adaptive Control of the False Discovery Rate in Multiple Testing with Independent Statistics. Journal of Educational and Behavioral Statistics. 2000;25(1):60. [Google Scholar]
- Boxer LD, Barajas B, Tao S, Zhang J, Khavari PA. ZNF750 interacts with KLF4 and RCOR1, KDM1A, and CTBP1/2 chromatin regulators to repress epidermal progenitor genes and induce differentiation genes. Genes & Development. 2014;28(18):2013–2026. doi: 10.1101/gad.246579.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebrià F, Newmark PA. Planarian homologs of netrin and netrin receptor are required for proper regeneration of the central nervous system and the maintenance of nervous system architecture. Development. 2005;132(16):3691–3703. doi: 10.1242/dev.01941. [DOI] [PubMed] [Google Scholar]
- Consortium T. UniProt: a hub for protein information. Nucleic Acids Research. 2015;43(D1) doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coward SJ, Piedilato JW. The behavioral significance of the rhabdite cells in planarians. Journal of Biological Psychology 1972 [Google Scholar]
- Darwin C. Insectivorous Plants. Albemarle Street, London: John Murray; 1875. [Google Scholar]
- Degnan BM, Vervoort M, Larroux C, Richards GS. Early evolution of metazoan transcription factors. Current Opinion in Genetics & Development. 2009;19(6):591–599. doi: 10.1016/j.gde.2009.09.008. [DOI] [PubMed] [Google Scholar]
- Des Marais DL, Rausher MD. Escape from adaptive conflict after duplication in an anthocyanin pathway gene. Nature. 2008;454(7205):762–5. doi: 10.1038/nature07092. [DOI] [PubMed] [Google Scholar]
- Dötsch V, Bernassola F, Coutandin D, Candi E, Melino G. p63 and p73, the ancestors of p53. Cold Spring Harbor Perspectives in Biology. 2010;2(9) doi: 10.1101/cshperspect.a004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driskell RR, Giangreco A, Jensen KB, Mulder KW, Watt FM. Sox2-positive dermal papilla cells specify hair follicle type in mammalian epidermis. Development. 2009;136(16):2815–23. doi: 10.1242/dev.038620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driskell RR, Lichtenberger BM, Hoste E, Kretzschmar K, Simons BD, Charalambous M, Ferron SR, Herault Y, Pavlovic G, Ferguson-Smith AC, et al. Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature. 2013;504(7479):277–81. doi: 10.1038/nature12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan EM, Chitsazan AD, Seidel CW, Sánchez Alvarado A. Set1 and MLL1/2 Target Distinct Sets of Functionally Different Genomic Loci In Vivo. Cell Reports. 2015;13(12):2741–2755. doi: 10.1016/j.celrep.2015.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14(9):755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhoffer GT, Kang H, Sánchez Alvarado A. Molecular Analysis of Stem Cells and Their Descendants during Cell Turnover and Regeneration in the Planarian Schmidtea mediterranea. Cell stem cell. 2008;3(3):327–339. doi: 10.1016/j.stem.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin DH, Davidson EH. The evolution of hierarchical gene regulatory networks. Nature reviews. Genetics. 2009;10(2):141–148. doi: 10.1038/nrg2499. [DOI] [PubMed] [Google Scholar]
- Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Research. 2016;44(D1) doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller JI, Roth JJ, Biegel JA. Biology and Treatment of Rhabdoid Tumor. Crit Rev Oncog. 2015;20(3–4):199–216. doi: 10.1615/critrevoncog.2015013566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble W. Quantifying similarity between motifs. Genome Biology. 2007;8(2) doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley KA, Rink JC, Sánchez Alvarado A. β-Catenin Defines Head Versus Tail Identity During Planarian Regeneration and Homeostasis. Science. 2008;319(5861):323–327. doi: 10.1126/science.1150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallez P. Embryogénie des Dendrocoeles d'eau douce, par le Dr Paul Hallez: O. Doin 1887 [Google Scholar]
- Hayashi T, Asami M, Higuchi S, Shibata N, Agata K. Isolation of planarian X - ray - sensitive stem cells by fluorescence - activated cell sorting. Development, Growth & Differentiation. 2006;48(6):371–380. doi: 10.1111/j.1440-169X.2006.00876.x. [DOI] [PubMed] [Google Scholar]
- Hittinger CT, Carroll SB. Gene duplication and the adaptive evolution of a classic genetic switch. Nature. 2007;449(7163):677–81. doi: 10.1038/nature06151. [DOI] [PubMed] [Google Scholar]
- Holt C, Yandell M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics. 2011;12(1):491. doi: 10.1186/1471-2105-12-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori I. Possible role of rhabdite-forming cells in cellular succession of the planarian epidermis. Journal of Electron Microscopy. 1978;27(2):89–102. [PubMed] [Google Scholar]
- Huang D, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman LH. The invertebrates. The invertebrates 1951 [Google Scholar]
- Joerger AC, Wilcken R, Andreeva A. Tracing the Evolution of the p53 Tetramerization Domain. Structure. 2014;22(9):1301–1310. doi: 10.1016/j.str.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamachi Y, Uchikawa M, Tanouchi A, Sekido R, Kondoh H. Pax6 and SOX2 form a co-DNA-binding partner complex that regulates initiation of lens development. Genes & Development. 2001;15(10):1272–1286. doi: 10.1101/gad.887101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RS, Newmark PA. In situ hybridization protocol for enhanced detection of gene expression in the planarian Schmidtea mediterranea. BMC Developmental Biology. 2013;13(1):8. doi: 10.1186/1471-213X-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouwenhoven EN, Oti M, Niehues H, van Heeringen SJ, Schalkwijk J, Stunnenberg HG, Bokhoven H, Zhou H. Transcription factor p63 bookmarks and regulates dynamic enhancers during epidermal differentiation. EMBO reports. 2015;16(7):863–878. doi: 10.15252/embr.201439941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouwenhoven EN, van Heeringen SJ, Tena JJ, Oti M, Dutilh BE, Alonso ME, de la Calle-Mustienes E, Smeenk L, Rinne T, Parsaulian L, et al. Genome-wide profiling of p63 DNA-binding sites identifies an element that regulates gene expression during limb development in the 7q21 SHFM1 locus. PLoS Genetics. 2010;6(8) doi: 10.1371/journal.pgen.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesko MH, Driskell RR, Kretzschmar K, Goldie SJ, Watt FM. Sox2 modulates the function of two distinct cell lineages in mouse skin. Developmental Biology. 2013;382(1):15–26. doi: 10.1016/j.ydbio.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liana MK, Quiroga SY, Bolaños MD, Litvaitis MK. Comparative morphology of the epidermis of seven species of polyclad flatworms (Platyhelminthes: Rhabditophora) Zoologischer Anzeiger - A Journal of Comparative Zoology. 2012;251(3):206–214. [Google Scholar]
- Lopez-Pajares V, Yan K, Zarnegar BJ, Jameson KL, Khavari PA. Genetic pathways in disorders of epidermal differentiation. Trends in genetics : TIG. 2013;29(1):31–40. doi: 10.1016/j.tig.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick VP, Fox DT, Spradling AC. Polyploidization and cell fusion contribute to wound healing in the adult Drosophila epithelium. Current Biology. 2013;23(22):2224–32. doi: 10.1016/j.cub.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick VP, Jun AS, Spradling AC. Wound-Induced Polyploidization: Regulation by Hippo and JNK Signaling and Conservation in Mammals. PLoS ONE. 2016;11(3):e0151251. doi: 10.1371/journal.pone.0151251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GG. A new function of rhabdites: Mucus production for ciliary gliding. Zoomorphologie. 1978;91(3):235–248. [Google Scholar]
- Mathelier A, Fornes O, Arenillas DJ, Chen C-y, Denay G, Lee J, Shi W, Shyr C, Tan G, Worsley-Hunt R, et al. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Research. 2016;44(D1) doi: 10.1093/nar/gkv1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee C, Fairweather I, Blackshaw RP. Ultrastructural features of the epidermis of the planarian Artioposthia triangulata (Dendy) Hydrobiologia. 1997;347(1/3):15–24. [Google Scholar]
- Narasimhan K, Pillay S, Huang Y-H, Jayabal S, Udayasuryan B, Veerapandian V, Kolatkar P, Cojocaru V, Pervushin K, Jauch R. DNA-mediated cooperativity facilitates the co-selection of cryptic enhancer sequences by SOX2 and PAX6 transcription factors. Nucleic Acids Research. 2015;43(3):1513–1528. doi: 10.1093/nar/gku1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmark PA, Sánchez Alvarado A. Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Developmental Biology. 2000;220(2):142–153. doi: 10.1006/dbio.2000.9645. [DOI] [PubMed] [Google Scholar]
- Parisi I, Collinson JM. Regulation of Merkel cell development by Pax6. International Journal of Developmental Biology. 2012;56(5):341–50. doi: 10.1387/ijdb.113406ip. [DOI] [PubMed] [Google Scholar]
- Pearson BJ, Eisenhoffer GT, Gurley KA, Rink JC, Miller DE, Sánchez Alvarado A. Formaldehyde - based whole - mount in situ hybridization method for planarians. Developmental Dynamics. 2009;238(2):443–450. doi: 10.1002/dvdy.21849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson BJ, Sánchez Alvarado A. A planarian p53 homolog regulates proliferation and self-renewal in adult stem cell lineages. Development. 2010;137(2):213–221. doi: 10.1242/dev.044297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellettieri J, Fitzgerald P, Watanabe S, Mancuso J, Green DR, Sánchez Alvarado A. Cell death and tissue remodeling in planarian regeneration. Developmental Biology. 2010;338(1):76–85. doi: 10.1016/j.ydbio.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdigoto CN, Bardot ES, Valdes VJ, Santoriello FJ, Ezhkova E. Embryonic maturation of epidermal Merkel cells is controlled by a redundant transcription factor network. Development. 2014;141(24):4690–4696. doi: 10.1242/dev.112169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sánchez Alvarado A. SMEDWI-2 Is a PIWI-Like Protein That Regulates Planarian Stem Cells. Science. 2005;310(5752):1327–1330. doi: 10.1126/science.1116110. [DOI] [PubMed] [Google Scholar]
- Robb S, Gotting K, Ross E, Sánchez Alvarado A. SmedGD 2.0: The Schmidtea mediterranea genome database. Genesis. 2015;53(8):535–546. doi: 10.1002/dvg.22872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompolas P, Patel-King RS, King SM. An Outer Arm Dynein Conformational Switch Is Required for Metachronal Synchrony of Motile Cilia in Planaria. Molecular Biology of the Cell. 2010;21(21):3669–3679. doi: 10.1091/mbc.E10-04-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(11):5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürmann W, Peter R. Planarian cell culture: a comparative review of methods and an improved protocol for primary cultures of neoblasts. Belgian Journal of Zoology. 2001;131(s1) [Google Scholar]
- Sen GL, Boxer LD, Webster DE, Bussat RT, Qu K, Zarnegar BJ, Johnston D, Siprashvili Z, Khavari PA. ZNF750 Is a p63 Target Gene that Induces KLF4 to Drive Terminal Epidermal Differentiation. Developmental Cell. 2012;22(3):669–677. doi: 10.1016/j.devcel.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaer RJ. The origin and continuous replacement of epidermal cells in the planarian Polycelis tenuis (IIJIMA) Journal of embryology and experimental morphology. 1965;13:129–139. [PubMed] [Google Scholar]
- Sun BK, Boxer LD, Ransohoff JD, Siprashvili Z, Qu K, Lopez-Pajares V, Hollmig TS, Khavari PA. CALML5 is a ZNF750-and TINCR-induced protein that binds stratifin to regulate epidermal differentiation. Genes & Development. 2015;29(21):2225–2230. doi: 10.1101/gad.267708.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE. 2011;6(7) doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu KC, Cheng L-C, Vu HTK, Lange JJ, McKinney SA, Seidel CW, Sánchez Alvarado A. Egr-5 is a post-mitotic regulator of planarian epidermal differentiation. Elife. 2015;4 doi: 10.7554/eLife.10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler S. Comparative ultrastructure of adhesive systems in the turbellaria. Zoomorphologie. 1976;84(1):1–76. [Google Scholar]
- Tyler S. Turbellarian Platyhelminths. In: Bereiter-Hahn J, Matoltsy GA, Richards SK, Tyler S, editors. Biology of the Integument. Springer; 1984. [Google Scholar]
- Tyler S, Tyler MS. Origin of the epidermis in parasitic platyhelminths. International Journal for Parasitology. 1997;27(6):715–738. doi: 10.1016/s0020-7519(97)00013-1. [DOI] [PubMed] [Google Scholar]
- van Wolfswinkel JC, Wagner DE, Reddien PW. Single-cell analysis reveals functionally distinct classes within the planarian stem cell compartment. Cell stem cell. 2014;15(3):326–339. doi: 10.1016/j.stem.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanbokhoven H, Melino G, Candi E, Declercq W. p63, a story of mice and men. The Journal of investigative dermatology. 2011;131(6):1196–1207. doi: 10.1038/jid.2011.84. [DOI] [PubMed] [Google Scholar]
- Vernimmen D, Bickmore WA. The Hierarchy of Transcriptional Activation: From Enhancer to Promoter. Trends in Genetics. 2015;31(12):696–708. doi: 10.1016/j.tig.2015.10.004. [DOI] [PubMed] [Google Scholar]
- Wagner DE, Ho JJ, Reddien PW. Genetic Regulators of a Pluripotent Adult Stem Cell System in Planarians Identified by RNAi and Clonal Analysis. Cell stem cell. 2012;10(3):299–311. doi: 10.1016/j.stem.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JC, Anderson DT. The Platyhelminthes, Nemertea, Entoprocta, and Gnathostomulida. Invertebrate Zoology. 2001:59–85. [Google Scholar]
- Wurtzel O, Cote LE, Poirier A, Satija R, Regev A, Reddien PW. A Generic and Cell-Type-Specific Wound Response Precedes Regeneration in Planarians. Developmental Cell. 2015;35(5):632–645. doi: 10.1016/j.devcel.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzel O, Oderberg IM, Reddien PW. Planarian Epidermal Stem Cells Respond to Positional Cues to Promote Cell-Type Diversity. Developmental Cell. 2017;40(5):491–504. doi: 10.1016/j.devcel.2017.02.008. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnegar BJ, Webster DE, Lopez-Pajares V, Vander Stoep Hunt B, Qu K, Yan KJ, Berk DR, Sen GL, Khavari PA. Genomic profiling of a human organotypic model of AEC syndrome reveals ZNF750 as an essential downstream target of mutant TP63. American Journal of Human Genetics. 2012;91(3):435–443. doi: 10.1016/j.ajhg.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zheng H, Chen Y, Zhang L, Wang K, Guo J, Huang Z, Zhang B, Huang W, Jin K, et al. The Schistosoma japonicum genome reveals features of host-parasite interplay. Nature. 2009;460(7253):345–351. doi: 10.1038/nature08140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Hallows SE, Currie KW, Xu C, Pearson BJ. A mex3 homolog is required for differentiation during planarian stem cell lineage development. Elife. 2015:4. doi: 10.7554/eLife.07025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.