Significance
Repeated sequences make up approximately two-thirds of the human genome, which become fully accountable when very large DNA molecules are analyzed. Long, single DNA molecules are problematic using common experimental techniques and fluidic devices because of mechanical considerations that include breakage, dealing with the massive size of these coils, or the huge length of stretched DNAs. Accordingly, we harness analyte “issues” as exploitable advantages by invention and characterization of the “molecular gate,” which controls and synchronizes formation of stretched molecules as DNA dumbbells within nanoslit geometries that may also offer new routes to separation. This was accomplished by theoretical studies and experiments leveraging a series of electrical forces acting on DNA molecules, device walls, and the fluid flows within our devices.
Keywords: single DNA molecules, nanofluidics, genomics, devices
Abstract
Very large DNA molecules enable comprehensive analysis of complex genomes, such as human, cancer, and plants because they span across sequence repeats and complex somatic events. When physically manipulated, or analyzed as single molecules, long polyelectrolytes are problematic because of mechanical considerations that include shear-mediated breakage, dealing with the massive size of these coils, or the length of stretched DNAs using common experimental techniques and fluidic devices. Accordingly, we harness analyte “issues” as exploitable advantages by our invention and characterization of the “molecular gate,” which controls and synchronizes formation of stretched DNA molecules as DNA dumbbells within nanoslit geometries. Molecular gate geometries comprise micro- and nanoscale features designed to synergize very low ionic strength conditions in ways we show effectively create an “electrostatic bottle.” This effect greatly enhances molecular confinement within large slit geometries and supports facile, synchronized electrokinetic loading of nanoslits, even without dumbbell formation. Device geometries were considered at the molecular and continuum scales through computer simulations, which also guided our efforts to optimize design and functionalities. In addition, we show that the molecular gate may govern DNA separations because DNA molecules can be electrokinetically triggered, by varying applied voltage, to enter slits in a size-dependent manner. Lastly, mapping the Mesoplasma florum genome, via synchronized dumbbell formation, validates our nascent approach as a viable starting point for advanced development that will build an integrated system capable of large-scale genome analysis.
The Precision Medicine Initiative is pressing for the development of new approaches for knowing the molecular underpinnings of disease through the detailed measurement of individuals, which may ramp up to a large cohort of one million participants (1). Meeting this challenge means that genome analysis approaches must advance to become more informative across the entire human genome, by leveraging large DNA molecules that can span across repeated sequences to foster their elucidation. Accordingly, systems employing single-molecule analytes have emerged, but not without much teething pain. Early single-molecule sequencing systems (2–5) have pointed the way forward to meeting these challenges, but despite costly commercialization efforts by Pacific Biosciences and Oxford Nanopore, issues still remain to be solved for moving industrialized versions of these systems into widespread use within biomedical settings.
Single-molecule approaches to human genome mapping provide a counterpart to sequencing efforts through discernment of structural variation (SV), in ways that elude sequence analysis (6, 7). The invention of optical mapping (8, 9) and its advanced version—nanocoding (10, 11), now being commercialized by BioNanoGenomics (12), is offering insights into structural variation present in normal human (6) and cancer genomes (7, 13). Such variants are difficult to fully characterize by sequencing because the human genome comprises vast stretches of complex, repeat-ridden regions harboring SVs that were comprehensively functionalized by the Encyclopedia of DNA Elements (ENCODE) Project (14). The new insights provided by ENCODE are substantiating the biological importance of these previously neglected portions within the human genome, and this new knowledge is also motivating development of new technologies that readily reveal complex variants.
As such, our previous work (10, 15, 16) dealt with these issues through development of a robust DNA labeling and presentation approach, “nanocoding,” which barcodes molecules with nicking restriction enzymes whose cleavage sites are then marked by nick translation using fluorochrome-labeled nucleotides. Thus formed punctates are imaged by fluorescence resonance energy transfer (FRET) microscopy along stretched molecules using nanoconfinement regimes leveraging low ionic strength () conditions. Because the DNA persistence length increases with lowered solution ionic strength (17, 18), these conditions synergized DNA stretching within relatively large slits. Other groups later built upon these developments (12, 19). More specifically, our first nanoslit devices were fabricated from poly(dimethylsiloxane) (PDMS) using soft lithography techniques that featured high aspect ratio slits (100 nm × 1,000 nm). Although much smaller slit dimensions are required for stretching DNA molecules, confinement conditions were greatly enhanced by using electrostatic effects mediated by very low ionic strength conditions (∼0.2 mM). Later work (16) modified slit geometries (250 nm × 400 nm) and ionic strength conditions, which further enhanced DNA stretch (, where L is the molecule contour length), but loading molecules into the nanoslits became more difficult. This is a common problem affecting most nanofluidic devices, since the entropic cost is substantial when threading large random-coil molecules into slit geometries comparable to the DNA persistence length.
Our early investigations revealed that large DNA molecules, under low ionic strength conditions, would sometimes partially load into nanoslits (100 nm × 1,000 nm), but were bracketed outside of the slits by random-coil portions that formed “DNA dumbbells” (11). Importantly, DNA molecules in a dumbbell conformation showed enhanced stretching (), which we previously reported (11) cannot be the result of the vanishingly small entropic forces exerted by the “molecular lobes.” Instead, our theoretical treatments and simulations identified the combined effects of electrostatic and hydrodynamic interactions as the dominant factors mediating enhanced stretching.
The challenge we address here is to further understand and then harness electrostatic effects and DNA polymer dynamics within nanofluidic systems in ways that would readily load and present very large DNA molecules as dumbbells, important considerations since DNA dumbbells, when formed by random loading events, would be difficult to produce en masse. Accordingly, our work presented here enables the synchronized formation of DNA dumbbells by a multiscale approach to device geometries. Given these insights, we report advances for greatly enhancing DNA presentation that leverage electrostatic properties (20), inherent to both nanofluidic devices and highly charged DNA molecules, through specifically engineered geometries in place of just confinement effects commonly used by many nanofluidic devices. Such advances in DNA presentation are tested here for genome analysis and shown to successfully map the Mesoplasma florum (M. florum) genome, and, after system hardening, may support high-throughput operation for enabling analysis of complex genomes.
Results
Multiscale Theoretical Approach Toward Device Design and Functionality.
A comprehensive theoretical study was performed, using multiple length scales, which informed the design and functionalities of the nanofluidic device featuring microchannel and nanoslit geometries (Fig. 1A). Electrostatic conditions, posed by device features and ionic strength conditions, affect both electroosmotic (electrically driven fluid flows) and electrophoretic forces, controlling DNA migration. These forces were studied using Brownian dynamics (BD), continuum finite-element (FE) simulations, and arguments from polymer physics, thereby engineering device features that leverage both molecular confinement and electrostatic effects (Methods). The FE calculations were performed on two levels: a detailed electrostatic study (Figs. S1 and S2), complemented by full momentum and mass balance simulations (Nernst–Planck/Stokes flow) that explored the microchannel/cup/nanoslit geometry (Fig. 1B). The electrostatic simulations guided electrode locations through calculation of resulting electric field lines and electrostatic potentials within the device immersed in the surrounding buffer medium. The BD simulations provided insights into enabling electrokinetic effects within the device for moving, parking, and loading DNA molecules.
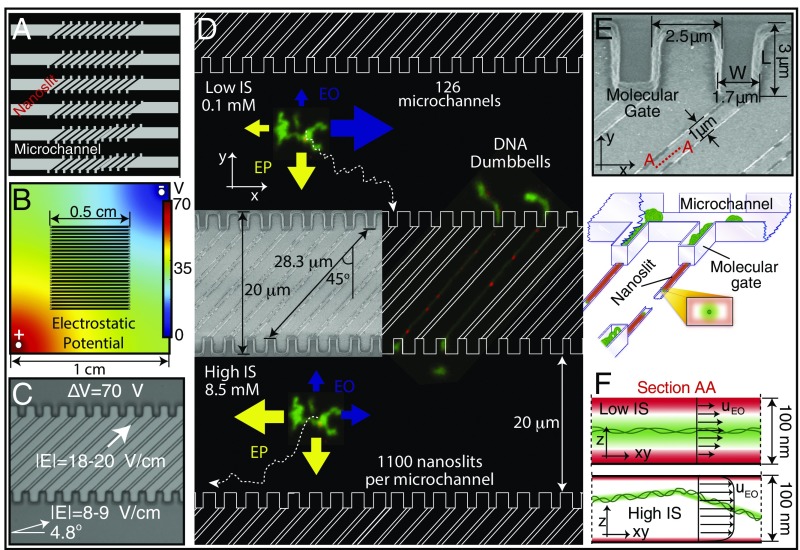
Fig. 1.
Electrostatic confinement and manipulation of DNA: device considerations. (A) Microchannel/nanoslit device schematic (top view): 1.6-μm height × 20-μm width microchannels (molecule bus) connecting 100-nm-high × 1-μm-wide × 28.3-μm-long nanoslits. Entire device, 1.0 cm × 1.0 cm square, comprises 126 microchannels, each one harboring 1,100 nanoslits bounded by molecular gates. (B) Electrostatic potential determined by FE simulation of the entire device within the buffer chamber. Such simulations guided electrode locations for producing the appropriate field lines within the microchannel/nanoslit device. (C) Microchannel/nanoslit device (imaged by DIC microscopy) is superimposed with arrows showing the direction and magnitude of field lines within device microchannel and nanoslit features (70 V applied). (D) Cartoon (top view) shows the direction and magnitude of the electrokinetic forces for low and high ionic strength conditions. (Inset) SEM micrograph (top view) of a patterned silicon master detailing nanoslits and molecular gates. Micrographs of DNA dumbbells bearing nanocoded labels (red punctates) are shown placed within the device. At low I, EO (blue arrows) guides molecules along the microchannel, while EP (yellow arrows) drives them toward the molecular gates. At high I, both directions are dominated by EP. Molecular trajectories (dotted line) are also drawn. (E) SEM of cup-like molecular gate features and dimensions (top view) of a silicon master. Illustration below shows DNA molecules (green) within a microchannel (1.6 μm high). Several molecular gates are shown bearing DNAs threaded into nanoslits (100 nm high), which pass through to the other side to form dumbbells. Note small 1-μm × 100-nm slit openings at the bottom of molecular gates. Cross-sectional view (Inset) depicts intersecting ion distributions (green) surrounding DNA and the nanoslit walls (red). (F) Perspective drawing showing DNA molecules (green balls/threads) within a microchannel. Ion clouds surround DNA and device walls. Lateral cross-sectional view within a nanoslit (see E; view A–A), showing ion clouds, under low and high ionic strength surrounding a DNA molecule (green) and nanoslit (red). At low I, an “electrostatic bottle” is created because ion clouds overlap, electrostatically confining the now-stiffened (increased persistence length) DNA molecule. In contrast, high I engenders a short Debye length, allowing the molecule to more freely diffuse throughout the entire height of the nanoslit. Furthermore, ionic strength conditions collectively affect the profile of the EO flow fields, illustrated by arrows, where the maximum velocity depends directly on the ratio between confinement dimensions and Debye length.
Electrical Effects: Electrophoretic vs. Electroosmotic.
We employ electrostatic considerations for controlling the Debye lengths of both DNA molecules and device features for efficient electrokinetic loading into nanoslits. The Debye length, defined as (where is the Boltzmann constant, is the temperature, is Avogadro’s number, is the elementary charge, ε0 is the vacuum permittivity, is the dielectric constant, and is the ionic strength), determines the length of the electrical double layer (ion cloud) near charged walls and DNA molecules. The unusual design theme here is to foster, rather than hinder, electroosmotic flows. Our thinking is that purely electrophoretic forces may be insufficient for efficient loading of molecules into nanoslits decorated by micropillars (21), which suffer entanglement and are not suitable for dealing with very large DNA molecules. Fig. 1 A–D shows the overall layout of the microfluidic/nanofluidic device. Fig. 1D details the design and functionalities of the device for DNA manipulations using ionic strength regimes engendering electroosmotic flows. The device uses a series of parallel microchannels (a molecule bus) for transporting DNA molecules to the molecular gate features (cup-like structures), which abut each diagonally oriented nanoslit. We expected electroosmotic perturbation of DNA migration due to low ionic strength ( < 0.75 mM) buffer conditions and the presence of negatively charged walls of the device (PDMS walls are O2 plasma treated) (10). By increasing the ionic strength ( > 2 mM), we see the net direction of migrating DNA molecules reverse, relative to electrode polarity, indicating that the dominating force transitions from electroosmotic to electrophoretic; independent evidence of electroosmotic flow is presented in Fig. S2.
FE calculations informed device geometrical design and placements of electrodes within the microscope-mounted buffer tank; multiple systems were simulated for optimization of the effective electric field that would enhance molecular manipulations (parking and loading; see Molecular Gates: Parking, Loading, and Synchronized Dumbbell Formation). Electrode positions control the field magnitude and direction within the microchannels, thereby guiding molecules to gates according to ionic strength (as we will describe below). Fig. 1C presents the magnitude and direction of the electric field, calculated by FE, for the electrode configuration in Fig. 1B under 70 V applied at the electrodes. Within the microchannels, the field has an ∼5o angle while in the nanoslit it follows the 45° geometrical direction. Importantly, the small cross-sectional area of the nanoslits, compared with microchannels, increases electrical resistance, which increases the electric field strength within that device feature (18–20 vs. 8–9 V/cm).
We then define DNA migration direction relative to nanoslit features and microchannels as being “acute” (electroosmotic flow), or “obtuse” (electrophoretic) under low ( = 0.44–0.89 mM), or high ( = 9.0 and 17 mM) ionic strength conditions. Fig. 2A shows time-lapse imaging, rendered as one composite image that reveals migrational trajectories of adeno DNA molecules (35.9 kb) within the device under low and high ionic strength conditions. Remarkably, low ionic strength conditions enable adeno DNA molecules to readily load into the nanoslit features of the device and then exit, as evidenced by sparse occupancies within all device features. In contrast, under high ionic strength conditions, molecules migrate by skirting along the molecular gate/microchannel interface, and consequently do not appreciably load, or pass through the nanoslits. Fig. 2B echoes these findings over a range of ionic strength conditions evaluated at two applied voltages (50 and 70 V) and gauged by loading efficiency into the nanoslits . At very low ionic strength ( = 0.44–0.89 mM) adeno DNA molecules quantitatively load in nanoslits at 50 and 70 V but then loading dramatically decreases, dropping to nearly zero at the highest ionic strength conditions ( = 9.0 and 17 mM; 50 V).
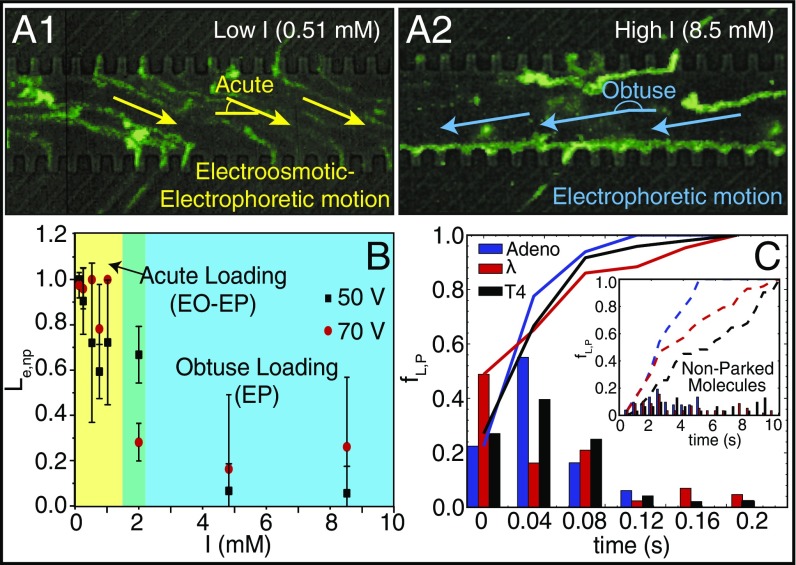
Fig. 2.
DNA parking synchronizes nanoslit loading controlled by ionic strength conditions. (A1 and A2) Green traces show trajectories of adeno DNA molecules traveling through a microchannel, without parking, loading into nanoslits captured by superimposition of 174 image frames (0.03-s interval); device is detailed in Fig. 1. (A1) Yellow arrows indicate overall direction of DNA migration (low ionic strength: 0.51 mM) under EO and EP forces. Accumulation of intense fluorescence along the molecular gate/microchannel interface (A2) indicates lack of passage through nanoslits. (A2) Same conditions as A1, except blue arrows indicate DNA migration dominated by EP forces under high ionic strength (8.5 mM). (B) Plot shows how loading efficiency Le,nP or the yield of adeno DNA molecules as imaged being present at a molecular gate that then goes on to load into nanoslits, without a parking step, varies with ionic strength and applied voltage (square-wave signals: 0 to 70 V, or 0 to 50 V; 0.1 Hz). Error bars are SDs on the means; sample size for the experiments ranged from 18 to 94 molecules. Colors highlight DNA loading regimes: [yellow: acute loading (EO-EP), green: transition, and blue: obtuse (EP)]. (C) Histogram showing the frequency of loading, after parking, fL,P, over time, across three DNA sizes: adeno (35.9 kb), lambda (48.5 kb), and T4 (165.6 kb). (Inset) Loading frequencies fL,nP for molecules without a parking step; lines represent cumulative frequency for each DNA sample (23–67 measurements).
Electroosmosis produces a flow-driven force that transports charged molecules toward the similarly charged electrode. Here, the flow field drags DNA molecules; therefore, the electroosmotic force depends on the “Zimm” frictional coefficient [, where , , and are the molecule radius of gyration, effective width, and persistence length, respectively], the electroosmotic (EO) mobility (), and the applied electric field (): . In contrast, during DNA electrophoresis, molecules move toward the electrode with opposite charge. Because polyelectrolytes (i.e., DNA) are free-draining during electrophoresis, meaning no hydrodynamic shielding, the electrophoretic force is now a function of the “Rouse” frictional coefficient . Consequently, the electrophoretic (EP) mobility () and the applied electric field scale as (): .
Intuitively, obtuse migration of DNA molecules should enhance loading, but we observe a noticeable difference in the loading rate between low and high ionic strength conditions. We attribute this difference to EO flows within the nanoslits; under high ionic strength conditions “push” molecules away from the molecular gates. Within a nanoslit, the EO velocity field can be calculated, as a first approximation, from Stokes equations providing an estimate for the characteristic velocity , where is the magnitude of the EO velocity, is the slit height, and is the Debye length. At high ionic strength, the Debye length is small compared with the nanoslit height, allowing a fully developed EO flow within the nanoslit. However, under low ionic strength conditions the flow field will be attenuated by an enlarged Debye length, now comparable to the nanoslit height (Fig. 1E), thereby removing this flow, which prevents loading.
Molecular Gates: Parking, Loading, and Synchronized Dumbbell Formation.
The molecular gate dimensions (Fig. 1E)––comparable to the Rg of the DNA coils––are designed for placing and holding an individual molecule at the entrance of each nanoslit. As such, these device features, differing vastly in scale, present support-controlled and synchronous loading of DNA molecules into nanoslits. We reasoned that molecules under low applied voltage () would “park” within the molecular gates, and could then be triggered at high voltage () to synchronously “load” within the nanoslits. We first tested this concept by measuring the loading times for a population of individual molecules sized 35.9, 48.5, and 165.6 kb (Fig. 2C). This plot shows a relatively tight distribution of loading into nanoslits that completes at ∼80 ms (80–90%). In contrast (Fig. 2C, Inset), nonparked molecules demonstrate a rather broad distribution of loading times that now span seconds, because molecules directly enter nanoslits from the microchannels by passing through the molecular gates without parking. Although these loading times (no parking) do not foster synchronous loading across multiple nanoslits, this experiment shows that molecular gates support efficient loading of large DNA molecules into nanoslits, even without the parking step.
Given that parked molecules load within a short period of time (Fig. 2C), we then evaluated this effect for the synchronous formation of dumbbells within multiple slits. Fig. 3 shows T4 DNA molecules moving through microchannels, with , becoming stably parked within molecular gates. Application of a short pulse () synchronously loads parked molecules into nanoslits and traps them as dumbbells when . During parking, is carefully selected so that molecules within molecular gates are compressed, as visually judged, but do not load into adjoining nanoslits. Once parked, the loading voltage triggers passage into nanoslits through a nondiffusive and fast translocation, fostering synchronized loading. This transition from parked to loaded is sharp for a population of molecules, indicating a kinetic energy barrier in the process (Fig. 2C). The detailed dynamics of loading into nanoslits is complex and will be developed through simulations in another publication. Here, we develop scaling arguments that were used for the design and operation of the device.
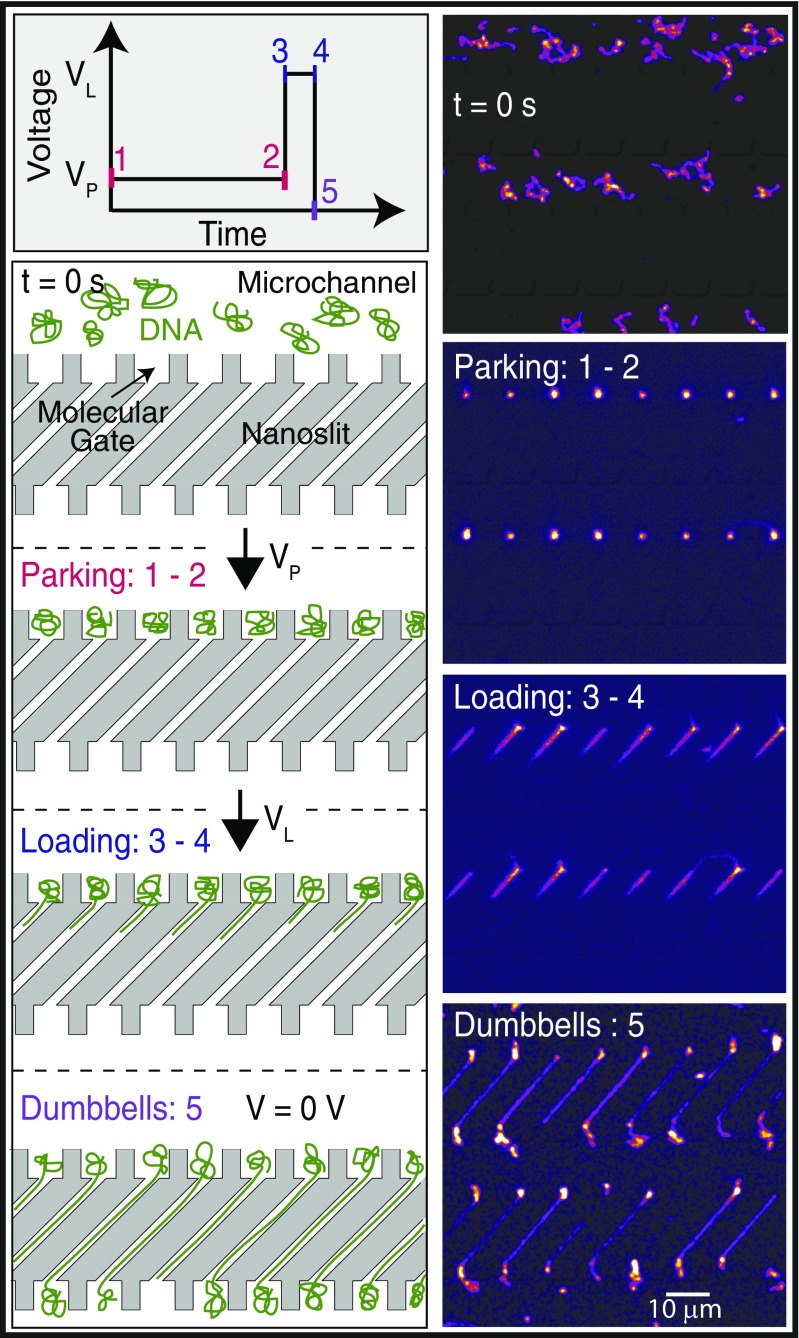
Fig. 3.
Parking, loading, and synchronized formation of T4 DNA dumbbells. Schematic of electrical signal triggering synchronized loading and dumbbell formation of parked DNA molecules into nanoslits. Micrographs (superimposed fluorescence and DIC), accompanied by cartoons, show T4 (165.6 kb) DNA molecules within several microchannels (t = 0 s) migrating toward the molecular gates (Vp = 20 V) for parking. (Parking: 1–2) A portion of these DNA molecules now resides (Vp = 20 V; t = 70.10 s) within molecular gates, and are now parked. (Loading: 3–4) Parked molecules are triggered (t = 70.32 s) to synchronously load into the adjoining slits by a short, higher-voltage pulse (1.0 s; VL = 70 V) to form an array of dumbbells (Dumbbells: 5) (t = 74.01 s).
As argued in Supporting Information, when there is a barrier we adopt a transition-state approximation. The DNA segments in both channels are effectively in equilibrium because the rate of leaking of the DNA molecule into the nanoslit is comparatively slow. The electrokinetic phenomena discussed above result in a front factor within our expression for the rate. The other factor is of the Boltzmann type exp(−Δ F/kBT), where ΔF(E) is the free energy of the barrier. For electric fields E much smaller than the cutoff , the DNA coils are parked in the molecular gates, or cups. At a larger field , the leaking into the nanoslit becomes immediate, since the barrier disappears []. Within this scenario, the loading voltage VL refers to the associated electric field that should be close to .
Applied Voltage Differentially Loads DNA Molecules as a Function of Size.
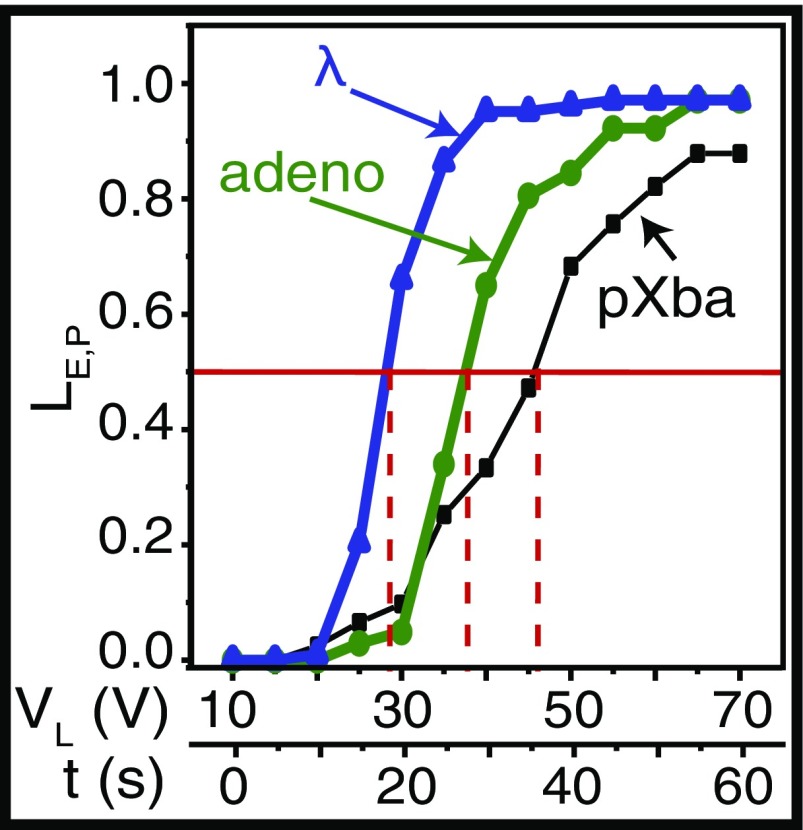
Our scaling arguments indicate that loading into nanoslits, after parking, at a given applied voltage should show a pronounced dependence on the size of a DNA coil, with larger molecules triggered to loading before smaller ones. We explored this concept by increasing the applied voltage (10–70 V; 5 V per 5-s interval) in a stepwise manner and assessed the loading efficiency for different DNA molecule sizes: pXba (22.6 kb), adeno (35.9 kb), and λ (48.5 kb). We define as the number of molecules that load after parking and as the voltage at which 50% of the parked molecules load into the nanoslits. Fig. 4 plots vs. , showing steep transitions from the parked to loaded state for the larger molecules, λ and adeno, less so for pXba. Differential loading effects are apparent under this voltage stepping scheme: Consider that 66% of the parked λ DNA loads at 30 V compared with only 5% of the adeno DNA molecules. Analysis of this plot also reveals an inverse linear relationship for size-dependent loading, ( in kbp), confirming that larger molecules load before smaller ones (22). Although direct separation of molecules was not attempted, this plot suggests that excellent size-dependent separations may be achievable.
Fig. 4.
DNA loading kinetics, after parking, is governed by size and applied voltage. For each DNA sample, 103–123 molecules were measured and I was fixed at 0.62 mM; molecules were stably parked for 20 s at 10 V before incrementally stepping applied voltage at 5-s intervals, 5 V, from 10 to 70 V (lower x axis). Plot shows LE,P vs. VL(V); VL is the voltage at which 50% of parked molecules are observed loading into nanoslits (horizontal red line); dashed red lines indicate respective VL values at LE,P = 50% for pXba (22.6 kb), adeno (35.9 kb), and lambda (48.5 kb) DNA molecules.
At present, we do not have a detailed theory of the full curves displayed in Fig. 4. Nevertheless, our scaling analysis in Supporting Information does present a qualitative explanation for what is happening at . In that case, the voltage times the length of the DNA (VL) should be a constant within a zero-order approximation. This seems to be borne out reasonably well; the products are 1,040 (pXba), 1,364 (adeno), and 1,358 (lambda) in units volts per kilobase. However, Eq. S7 underestimates this product by an order of magnitude although the numerical coefficient on the right-hand side is unknown.
Genome Mapping via DNA Dumbbells.
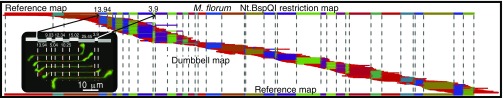
We evaluated the effectiveness of the parking and loading scheme for mapping genomes using M. florum (793 kb) genomic DNA labeled for nanocoding (ref. 10 and Methods). Briefly, nick translation places fluorescently labeled nucleotides at nick sites created by Nt.BspQI that are imaged as FRET pairs formed by YOYO-1 (green donor) staining and the covalently incorporated Alexa Fluor-647 (red acceptor) moieties. This labeling step effectively barcodes individual DNA molecules through later measurement of punctate spacing, using image processing, to create one restriction map per molecule—termed, “Nmap.” Such distance measurements (pixels, nanometers) are converted into fragment sizes as kilobase pairs by using DNA stretch estimations, determined by alignment using SOMA software (refs. 6, 23, and 24 and Methods), which are mediated by ionic strength (10) and the amount of YOYO-1 bound to DNA molecules (25). Accordingly, the pairwise alignment rate of the entire Nmap dataset (906 Nmaps) against the M. florum reference map maximized at 86% (781 aligned/906 total) using a stretch of 0.85; Fig. 5 shows these alignments spanning across the entire genome. Briefly, SOMA uses a series of error models, reflecting labeling rates (false and missing) and sizing errors to score and then optimally place Nmaps onto a reference genome. The reference genome is simply an ordered restriction map created in the computer from available sequence. We generate confidence scores (P values) using an approach similar to that used by Vingron and Waterman for sequence alignments (26).
Fig. 5.
Genomic M. florum molecules, labeled by nanocoding, mapped as dumbbells and then aligned to a reference map. Track (Top) shows a linear Nt.BspQI in silico restriction map for the M. florum genome (793,224 bp) comprising 39 restriction fragments presented as arbitrarily colored bars. Below, 781 SOMA aligned Nmaps are shown spanning across the entire genome, color-keyed to the reference; vertical red lines demarcate restriction fragments. (Inset) Montage of five nanocode-labeled dumbbells aligned to the indicated portion of the M. florum reference detailed as gray, undulating bars with their respective restriction fragment sizes (kb). Micrographs of dumbbell molecules overlaid by red punctates, label Nt.BspQI nick sites.
Discussion
We have created an electrostatically inspired approach for genome analysis through design of a nanofluidic device embracing a series of synergistic functionalities exhibited by both DNA molecules and the device itself. Here, very low ionic strength conditions augment stretching and strategically combine for effective transport and temporal control of molecules loading into nanoslits. These advances empower DNA dumbbells through parking and loading, which greatly enhance DNA stretching, to be synchronously or consistently formed and analyzed for mapping M. florum using genomic DNA molecules, which were potentiated by low rates of DNA breakage within the device. We accomplished this through the elucidation and harnessing of two major electrostatic effects: (i) Enhanced confinement of DNA molecules within relatively large, easily fabricated nanoslits. (ii) Electrokinetic actions using both EO and EP forces, which greatly facilitate and synchronize loading DNA molecules into nanoslits via molecular gates. Collectively, the coordination of these somewhat antagonistic effects posed special challenges for enabling DNA dumbbells for genome analysis. We have addressed these challenges by a unique design and device strategy: Our multiscale theoretical approach allowed direct engineering of device features that exploit electrostatic and fluctuating hydrodynamic forces for controlling the concerted and parallel manipulation of large DNA molecules. Such advancements fostered invention of the molecular gate and discovery of the “parking and loading effect,” which now enfranchises dumbbell molecules as practical analytes.
These device effects and functionalities hinge on controlling the Debye lengths () inherent to DNA molecules (polyelectrolyte) and the charged device features by varying buffer ionic strength conditions. Here, high ionic strength solutions (∼8 mM) produce compact ion clouds (3.4 nm), whereas low ionic strength solutions (∼0.1 mM) generate expansive ion clouds (∼30 nm). Accordingly, at low ionic strength, device nanoslit (100 nm high) and DNA (60-nm Debye diameter) Debye layers intersect (Fig. 1F) to enhance DNA confinement (11) and consequent stretching (16). Low ionic strength conditions also increase the “stiffness,” or persistence length of DNA molecules, which is yet another effect that further enhances DNA stretch within the device. This stiffness follows Odijk–Skolnick–Fixman theory (17, 18) , where is the persistence length excluding electrostatic considerations, which governs the average dimension of a DNA random coil, explicitly described by the radius of gyration: (. Accordingly, increased ionic strength decreases coil dimensions; for example, we see that as I increases (0.1 vs. 8.5 mM), shrinks (358 vs. 53 nm), thereby reducing (1.9 vs. 0.7 μm). Importantly, our previous work (10) had shown that increasing DNA persistence length under nanoconfinement greatly increases its stretch: , where is the measured molecule length, is the polymer contour length, and and are the slit height and width, respectively.
Electrostatic considerations allowed us to engineer a device modality that synchronously loads DNA molecules into nanoslit geometries in ways that portend its broad application. The overall utility of the molecular gate geometry, complemented by low ionic strength conditions, showed usefulness for genome analysis via synchronous dumbbell formation within nanoslits parking/loading, and an almost “digital-like” separation ability (Fig. 4), where under certain conditions, closely sized DNA molecules exhibit either great mobility, or effectively none. In addition, we showed facile entry of large DNA molecules into nanoslits, even without using the parking and loading routine (Fig. 2C). Nascent systems for genome analysis gain credibility when they demonstrate the potential for high-throughput operation. Although a limited portion of our device was sampled for the complete mapping of the M. florum genome, the device harbors 138,600 nanoslits, each 28 μm in length. With a total length of almost 4 m, the device can hold DNA molecules corresponding to ∼4 haploid human genome equivalents. Given such capacity, automated data acquisition schemes are easily envisioned where serial dumbbell formation and concerted imaging over occupied portions of the device would enable high-throughput operation.
Lastly, we envision future applications that may leverage the potential of the molecular gate to aptly manipulate arrays of DNA molecules for the cell-free synthesis of entire chromosomes.
Methods
Device Design and Fabrication.
Device fabrication was multistep via standard photolithography and electron-beam lithography techniques: (i) Fiduciary marks, UVIII were spin coated (∼600 nm) onto a silicon wafer, then exposed using a JBX5DII electron-beam lithography system (JEOL; CNTech). Oxygen descum process removed organic deposits or residual resist before evaporating metal. An ∼20-nm layer of platinum was placed by electron-beam evaporation (CNTech) to promote adhesion between the silicon wafer; a gold layer (∼60 nm thick) was then deposited for a high-contrast mark for alignment between multiple layers. Sonication (acetone) facilitated liftoff of the excess metal, followed by isopropyl alcohol (IPA) rinse, water rinse, and air-drying. (ii) SU8 2000.5 photoresist (∼250 nm; MicroChem) was applied and exposed as boxes over the alignment marks, protecting marks against subsequent etching steps. (iii) SU8 2000.5 was spin coated (∼250 nm) onto a wafer and the nanoslits were exposed, developed using SU8 remover, and IPA. SU8 nanoslits were etched into the silicon wafer with CF4 (8 min, 10 mTorr; Unaxis 790, Unaxis Wafer Processing), placed in a piranha bath (80% H2SO4 and 20% H2O2) for 5 min to remove the residual SU8, and rinsed with water to remove the acid. Finally, the microchannel with molecular gates, aligned with the slits by global and chip fiduciary marks, were exposed by electron-beam lithography. SI Methods details silicone replica creation.
DNA Sample Preparation and Mapping.
Previously published sample preparation and labeling techniques were used as detailed in SI Methods.
Microscopy and Data Acquisition for Elongated DNAs with Nanoslits.
Previously published microscopy and data acquisition were used as detailed in SI Methods.
Device Setup, Parking, and Loading.
Acid-cleaned glass coverslips (9) with a PDMS device adhered were affixed to a Plexiglas holder using paraffin wax. Capillary action loaded device microchannels using a 3-μL solution containing final concentrations of DNA (0.615 ng/μL), YOYO-1 (in water; 0.38 μM), beta-mercaptoethanol (3.65%), and POP6 (0.091%; ThermoFisher Scientific). Next, devices were immersed in 2 mL of 0.05 × TE buffer (10 mM Tris⋅HCl, 1 mM EDTA; pH = 7.9; solution dilutions checked by conductivity) for 20 min before electrokinetic loading of DNA molecules into nanoslits using platinum electrodes inserted into the reservoirs. DNA was loaded into nanoslits via parking and loading using an electrical signal [∼20 s: square waveform (20–70 V; 0.025 Hz)] with electrodes 2.5 cm apart. Thus, parked molecules were then synchronously loaded into adjoining nanoslits (70 V using a square-wave signal; ∼1-s duration).
Supplementary Material
Acknowledgments
We thank Tom Knight for providing M. florum. This work was supported by Grants NIH R01-HG-000225 and National Cancer Institute (NCI) CA182360 (to D.C.S.). G.T. was supported by National Human Genome Research Institute Grant T32 HG002760. The development and validation of the codes used here to simulate DNA, which couple particle-based simulations to hydrodynamics, was supported by the Department of Energy, Basic Energy Sciences, Materials Science and Engineering, through the Midwest Integrated Center for Computational Materials. The engineering of processes for charged-driven assembly of polyelectrolytes is supported by the National Institute of Standards and Technology through the Center for Hierarchical Materials Design.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711069114/-/DCSupplemental.
References
- 1.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ramanathan A, et al. An integrative approach for the optical sequencing of single DNA molecules. Anal Biochem. 2004;330:227–241. doi: 10.1016/j.ab.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 3.Braslavsky I, Hebert B, Kartalov E, Quake SR. Sequence information can be obtained from single DNA molecules. Proc Natl Acad Sci USA. 2003;100:3960–3964. doi: 10.1073/pnas.0230489100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levene MJ, et al. Zero-mode waveguides for single-molecule analysis at high concentrations. Science. 2003;299:682–686. doi: 10.1126/science.1079700. [DOI] [PubMed] [Google Scholar]
- 5.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci USA. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teague B, et al. High-resolution human genome structure by single-molecule analysis. Proc Natl Acad Sci USA. 2010;107:10848–10853. doi: 10.1073/pnas.0914638107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta A, et al. Single-molecule analysis reveals widespread structural variation in multiple myeloma. Proc Natl Acad Sci USA. 2015;112:7689–7694. doi: 10.1073/pnas.1418577112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwartz DC, et al. Ordered restriction maps of Saccharomyces cerevisiae chromosomes constructed by optical mapping. Science. 1993;262:110–114. doi: 10.1126/science.8211116. [DOI] [PubMed] [Google Scholar]
- 9.Dimalanta ET, et al. A microfluidic system for large DNA molecule arrays. Anal Chem. 2004;76:5293–5301. doi: 10.1021/ac0496401. [DOI] [PubMed] [Google Scholar]
- 10.Jo K, et al. A single-molecule barcoding system using nanoslits for DNA analysis. Proc Natl Acad Sci USA. 2007;104:2673–2678. doi: 10.1073/pnas.0611151104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kounovsky-Shafer KL, et al. Presentation of large DNA molecules for analysis as nanoconfined dumbbells. Macromolecules. 2013;46:8356–8368. doi: 10.1021/ma400926h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lam ET, et al. Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nat Biotechnol. 2012;30:771–776. doi: 10.1038/nbt.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ray M, et al. Discovery of structural alterations in solid tumor oligodendroglioma by single molecule analysis. BMC Genomics. 2013;14:505. doi: 10.1186/1471-2164-14-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Consortium EP. ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jo K, Schramm TM, Schwartz DC. A single-molecule barcoding system using nanoslits for DNA analysis: Nanocoding. Methods Mol Biol. 2009;544:29–42. doi: 10.1007/978-1-59745-483-4_3. [DOI] [PubMed] [Google Scholar]
- 16.Kim Y, et al. Nanochannel confinement: DNA stretch approaching full contour length. Lab Chip. 2011;11:1721–1729. doi: 10.1039/c0lc00680g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skolnick J, Fixman M. Electrostatic persistence length of a wormlike polyelectrolyte. Macromolecules. 1977;10:944–948. [Google Scholar]
- 18.Odijk T. Polyelectrolytes near rod limit. J Polym Sci Pol Phys. 1977;15:477–483. [Google Scholar]
- 19.Hastie AR, et al. Rapid genome mapping in nanochannel arrays for highly complete and accurate de novo sequence assembly of the complex Aegilops tauschii genome. PLoS One. 2013;8:e55864. doi: 10.1371/journal.pone.0055864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zwanikken JW, Olvera de la Cruz M. Tunable soft structure in charged fluids confined by dielectric interfaces. Proc Natl Acad Sci USA. 2013;110:5301–5308. doi: 10.1073/pnas.1302406110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao H, Tegenfeldt JO, Austin RH, Chou SY. Gradient nanostructures for interfacing microfluidics and nanofluidics. Appl Phys Lett. 2002;81:3058–3060. [Google Scholar]
- 22.Mannion JT, Reccius CH, Cross JD, Craighead HG. Conformational analysis of single DNA molecules undergoing entropically induced motion in nanochannels. Biophys J. 2006;90:4538–4545. doi: 10.1529/biophysj.105.074732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valouev A, et al. Alignment of optical maps. J Comput Biol. 2006;13:442–462. doi: 10.1089/cmb.2006.13.442. [DOI] [PubMed] [Google Scholar]
- 24.Sarkar D, Goldstein S, Schwartz DC, Newton MA. Statistical significance of optical map alignments. J Comput Biol. 2012;19:478–492. doi: 10.1089/cmb.2011.0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Günther K, Mertig M, Seidel R. Mechanical and structural properties of YOYO-1 complexed DNA. Nucleic Acids Res. 2010;38:6526–6532. doi: 10.1093/nar/gkq434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vingron M, Waterman MS. Sequence alignment and penalty choice. Review of concepts, case studies and implications. J Mol Biol. 1994;235:1–12. doi: 10.1016/s0022-2836(05)80006-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.