Abstract
After having revolutionized our understanding of the mechanisms of animal development, Drosophila melanogaster has more recently emerged as an equally valid genetic model in the field of animal metabolism. An increasing number of studies have revealed that many signaling pathways that control metabolism in mammals, including pathways controlled by nutrients (insulin, TOR), steroid hormone, glucagon, and hedgehog, are functionally conserved between mammals and Drosophila. In fact, genetic screens and analyses in Drosophila have identified new players and filled in gaps in the signaling networks that control metabolism. This review focuses on data that show how these networks control the formation and breakdown of triacylglycerol energy stores in the fat tissue of Drosophila.
Keywords: Lipid Metabolism, Drosophila, Obesity, Insulin, TOR, Ecdysone
1. Introduction
Organisms are highly ordered structures that can only assemble and maintain themselves through processes that require energy. How organisms harness sources of energy is one of the fundamental questions in biology. This question has gained additional importance by the growing obesity pandemic among human populations (Ng et al., 2014). The excessive accumulation of body fat in the obese is associated with metabolic dysregulation that manifests itself as type II diabetes and that can lead to cancer, cardiovascular disease and other health problems (Bastien et al., 2014; Gallagher and LeRoith, 2010; Seidell, 2000). Efforts to curb the obesity pandemic will be critically informed by basic research into the fundamental principles of fat metabolism. Much of this basic research is done in humans and mammalian models. However, in recent years it has become evident that the fruit fly Drosophila melanogaster, a simple genetic model organism, is uniquely situated to uncover basal causal relationships between adiposity and metabolic changes on a physiological level. Fruit flies will become obese under certain dietary conditions and obese flies show signs of diabetes, exhibit insulin resistance, and have impaired cardiac function. These studies have established Drosophila as a model for obesity studies and are discussed in several excellent recent reviews (Baker and Thummel, 2007; Diop and Bodmer, 2015; Graham and Pick, 2017; Kühnlein, 2011; Owusu-Ansah and Perrimon, 2014). Here, I will first briefly review anabolic and catabolic pathways that determine the size of storage fat depots in Drosophila (for an in-depth review, see Kühnlein, 2012). The main focus of this review, however, will be on pathways and mechanisms that control the formation and breakdown of fat stores in Drosophila.
2. Fat storage and transport in Drosophila
Animals use neutral fats or triacylglycerols (TAGs) to store chemical energy that is not immediately needed for the maintenance of normal cellular and organismal functions. As fatty acid esters of glycerol, TAGs are energy-dense and highly hydrophobic, forming compact energy stores in the form of cytoplasmic lipid droplets. In recent years it has become clear that lipid droplets not only serve as energy stores, but are dynamic cell organelles that have a variety of other cellular functions and roles in the etiology of metabolic diseases (for reviews, see Krahmer et al., 2013; Welte, 2015). The mobilization of fatty acids from lipid droplet TAGs for energy production becomes critical when energetic needs cannot be met by nutritional intake or other sources of chemical energy, in particular glycogen, which is stored in the liver of vertebrates and the fat body of insects. Thus, TAGs are essential for the survival of periods of starvation and of conditions that preclude the uptake of nutrients, which exist, for instance, in hibernating animals or insects that undergo metamorphosis, such as Drosophila.
As in other insects, in Drosophila TAG is primarily stored in a specialized tissue, the fat body. The fat body is functionally equivalent to the mammalian adipose tissue, but it also has liver-like functions, storing glycogen for instance. A second tissue with liver-like functions are the oenocytes, which store and process lipids during periods of fasting and, accordingly, express many lipid-metabolizing genes (Gutierrez et al., 2007). Neutral lipid droplets are not only found in the fat body and oenocytes, but also in many other tissues, including the brain, the imaginal discs and, especially, the midgut. Next to the fat body, the midgut is another major lipogenic organ in Drosophila. The midgut produces diacylglycerol (DAG) for distribution to other tissues using dietary or de novo-synthesized fatty acids (Palm et al., 2012). DAG is the major form in which fatty acids are transported in Drosophila and in other insects (Canavoso et al., 2001; Palm et al., 2012). In the hemolymph, DAG is bound to the lipoprotein Lipophorin (Lpp), which binds more than 95% of all hemolymph lipids (Palm et al., 2012). DAGs constitute 70% of those lipids. DAGs released from the gut are loaded onto Lpp by the Lipid Transfer Particle (LTP) protein. As in other insects, in Drosophila Lpp is produced and secreted into the hemolymph by the fat body, and the same is true for LTP (Canavoso et al., 2001; Palm et al., 2012). Drosophila has two Lpp receptor genes, lpr1 and lpr2, which give rise to multiple receptor isoforms. Somewhat surprisingly, Lpp receptors are required for the uptake of neutral lipids into oocytes and imaginal disc cells, but in the fat body the number and size of fat droplets does not change in the absence of Lpp receptors (Parra-Peralbo and Culi, 2011). In oocytes and imaginal discs, Lpp receptors in the plasma membrane are bound by LTP and this interaction is required for the efficient transfer of lipid from Lpp into the cells (Rodríguez-Vázquez et al., 2015). Notably, fat body TAG levels do not decrease when larvae are fed a lipid-free diet or if Lpp is reduced by RNAi (Palm et al., 2012). This suggests that the fat body can maintain constant TAG stores through de novo synthesis, independent of the delivery of external DAGs. In contrast, reduction of LTP by RNAi does reduce fat body TAGs by 30%. In addition, it strongly reduces medium-chain DAGs in the fat body that are predominantly produced in the gut. This suggests that the fat body indeed takes up DAGs from the hemolymph, a conclusion that is supported by studies in the tobacco hornworm, Manduca sexta, demonstrating uptake of radiolabeled DAG from Lpp into the fat body (Canavoso et al., 2004). Interestingly, fat body TAGs contain comparatively few medium-chain acyl residues, suggesting that medium-chain DAGs coming from the gut are not directly converted to TAGs (Palm et al., 2012).
3. The de novo synthesis of neutral fats
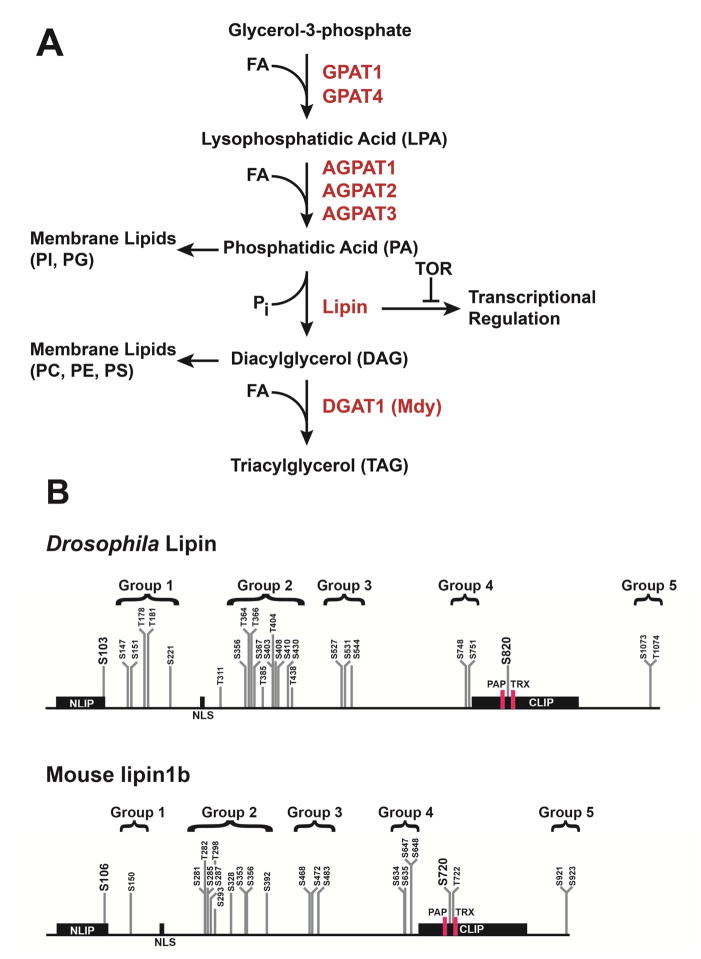
Neutral fats are derived from two basic building blocks, glycerol-3 phosphate and fatty acids. If not derived from nutritional sources or the breakdown of stored fat, fatty acids can be synthesized de novo from acetyl-CoA that is derived from the breakdown of carbohydrates and proteins. De novo synthesis of neutral fats occurs through the glycerol-3-phosphate pathway, which converts glycerol-3-phosphate into TAG by esterification with fatty acids in 4 enzymatic steps (Fig. 1A). The last step in the pathway, the conversion of DAG into TAG, is carried out by diacylglycerol acyltransferases (DGATs). In Drosophila, an enzyme with demonstrated in vitro DGAT activity is encoded by the midway (mdy) gene (Buszczak et al., 2002). TAG stores are reduced in mdy mutant and RNAi knockdown flies, confirming the role of the protein in TAG synthesis in vivo (Beller et al., 2010; Buszczak et al., 2002). A second potential DGAT homolog, encoded by CG1942, has been referred to as DGAT2 (Wilfling et al., 2013). However, the CG1942 product shows higher sequence identity to the vertebrate monoacylglycerol acyltransferase MGAT2 than to DGAT2 (39% vs 30.8% compared with the human proteins), leaving the question open which substrates are used by this enzyme. In S2 cells, reduction of the CG1942 product leads to a reduction in the number of large lipid droplets, but it does not reduce the total amount of TAG (Wilfling et al., 2013). A role of the CG1942 product in TAG synthesis in larvae and adults is suggested by expression in the larval midgut and the adult fat body and midgut (Chintapalli et al., 2007). Expression in the midgut is consistent with the known role of MGAT2 in intestinal triglyceride absorption in mammals (Cao et al., 2012). The CG1942 product belongs to a group of enzymes that includes GPAT4, AGPAT3 and Lipin, which are recruited to the lipid droplet surface during TAG synthesis. In contrast, the mdy product DGAT1 is not associated with lipid droplets during TAG synthesis, but remains localized within the ER (Wilfling et al., 2013).
Fig. 1.
The lipogenic glycerol-3-phosphate pathway in Drosophila. (A) Overview of pathway and enzymes; FA, fatty acid; PI, phosphatidylinositol; PG, phosphatidylglycerol; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PS, phosphatidylserine; (B) Comparison of mapped serine and threonine phosphorylation sites in Drosophila Lipin and mouse lipin 1b (Bodenmiller et al., 2008; Bridon et al., 2012; Harris et al., 2007). Sites are grouped in similar regions of the proteins. Two sites in the conserved NLIP and CLIP regions, which are highlighted by larger font, are homologous to one another. The locations of the catalytic motif, DIDGT (PAP), and of the transcriptional co-regulator motif, LGHIL (TRX), are indicated by red boxes; NLS, nuclear localization sequence.
Whereas DGATs are solely dedicated to the production of TAGs, the enzyme that produces DAG in the glycerol-3-phosphate pathway is involved in both TAG production and the production of membrane phospholipids. DAG is derived from phosphatidic acid (PA) by phosphatidic acid phosphatase (PAP), which is encoded in Drosophila by the single Lipin gene. In contrast, mice and humans possess three lipin genes, lipin 1, 2, and 3 (Csaki and Reue, 2010; Harris and Finck, 2011). Mutations in both mouse lipin 1 and Drosophila Lipin lead to a severe lipodystrophy phenotype (Peterfy et al., 2001; Ugrankar et al., 2011). In addition to TAG production, lipins provide DAG as a precursor for the synthesis of specific membrane phospholipids. The lipin substrate, PA, is used for the synthesis of membrane phospholipids as well, which puts lipins at the crossroads between storage and membrane lipid synthesis (Fig. 1A). Lipins are not only found in animals, but also in plants and fungi, and are characterized by highly conserved domains referred to as the NLIP and CLIP domains (Peterfy et al., 2001) (Fig. 1B).
Studies of mammalian and yeast lipins revealed functions of lipins not only in lipid synthesis, but also in gene regulation (for reviews, see Csaki and Reue, 2010; Harris and Finck, 2011; Siniossoglou, 2013). The CLIP domain contains an LXXIL transcriptional co-regulator motif that is similar to the LXXLL transcriptional co-regulator motif found in nuclear receptor co-regulators (Finck et al., 2006). Indeed, it has been shown that lipin 1 and 3 can physically interact with nuclear receptors and co-regulators (Finck et al., 2006). Gene-regulatory functions of Drosophila Lipin seem to be dispensable for survival under normal culture conditions in the laboratory (Rudolf, X.V., Hood, S.E., Lehmann, M.; unpublished data). However, data obtained for both Drosophila and vertebrates suggest that they are vital under starvation conditions (Peterson et al., 2011; Schmitt et al., 2015; Ugrankar et al., 2011). The full arrays of genes controlled by the co-regulator functions of lipins await identification. However, gene expression profiling after over-expression of lipin 1 in mouse liver suggests that lipin 1 activates genes involved in fatty acid utilization and oxidative phosphorylation, whereas it represses genes involved in de novo lipogenesis (Finck et al., 2006). Since lipin 1 also activates expression of the nuclear receptors PPARα and HNF4α, it is unclear which of these genes respond directly to lipin 1 and which are activated by these transcription factors. However, since lipin 1 can physically interact with the two receptors and also with the nuclear receptor co-regulator PGC-1α, many of the identified genes may be activated in a cooperative fashion. PPARα and PGC-1α are known key regulators of lipid and energy metabolism and HNF4 has been associated with the control of lipid metabolism in Drosophila (see below) (Kersten, 2014; Palanker et al., 2009; Sugden et al., 2010). The physical interaction data are consistent with the results of chromatin immunoprecipitation (ChIP) experiments in yeast and mice, which support a model in which lipins bind to specific gene loci as transcriptional co-regulators (Finck et al., 2006; Santos-Rosa et al., 2005). However, it is interesting to note that mouse lipin 1 may also control gene expression more indirectly by controlling nuclear localization of the transcription factor SREBP1 (Peterson et al., 2011).
In both mice and flies, nuclear translocation of lipins is regulated by the nutrient-sensitive TOR pathway (Peterson et al., 2011; Schmitt et al., 2015) (for the relationship of lipins to TOR and SREBP, see the discussion further below). Interestingly, the activity and intracellular localization of Drosophila Lipin has been shown to depend on Torsin, an evolutionarily conserved AAA+ ATPase family protein that localizes to the ER and nuclear envelope (Grillet et al., 2016). Null mutants of torsin show fat body defects that include elevated TAG levels in early larvae and fat body cell size reduction in late larvae. Fat body DAG is highly elevated in the early larvae as well, whereas PA is strongly reduced, suggesting that Torsin normally has a negative impact on the PAP activity of Lipin. Indeed, reduction of Lipin expression partially rescues Torsin mutant phenotypes. Lipin re-localizes to the nuclear area upon loss of Torsin, suggesting that Torsin exerts its effects by controlling where Lipin acts in the cell. However, whether this is achieved by direct physical interaction or by other means remains to be explored.
Enzymes catalyzing the first two steps of the glycerol-3-phospate pathway are not as well characterized in Drosophila as Mdy and Lipin. Glycerol-3-phosphate acyltransferases (GPATs) are responsible for the first step of the pathway, which converts glycerol-3-phosphate into lysophosphatidic acid (Fig. 1A). The Drosophila genome contains three genes that are predicted to encode GPATs, GPAT 1, GPAT 4 and CG15450 (Wilfling et al., 2013). While the biological function of CG15450 may be restricted to testis (Boutanaev et al., 2002), both GPAT 1 and 4 have been implicated with fat droplet formation in S2 cells. Similar to a reduction of DGAT1, reduction of GPAT4 in S2 cells leads to a significant decrease in TAG content (Wilfling et al., 2013). Surprisingly, a putative null mutant of GPAT4 does not exhibit reduced amounts of TAG in larval fat body cells, suggesting that GPAT4 acts with GPAT1 in a redundant manner (Tian et al., 2011; Yan et al., 2015). Consistent with this interpretation, GPAT1 transfection rescues total TAG levels in GPAT4-depleted S2 cells, although GPAT1 knockdown does not lead to changes in total TAG content or fat droplet size (Wilfling et al., 2013). Lysophosphatidic acid produced by GPATs is converted to PA by acylglycerol-3-phosphate acyltransferases (AGPATs). The Drosophila genome encodes four homologs of the mammalian AGPATs that have been analyzed to some extent in S2 cells (Wilfling et al., 2013). Whereas AGPAT3 is required for production of normal amounts of TAG in these cells, AGPAT1 and 2 are not. These and other proteins of the glycerol-3-phosphate pathway await a more detailed genetic characterization in whole animals.
4. The breakdown of neutral fats
The synthesis of fat is balanced by its breakdown, which is carried out by lipases in both mammals and Drosophila. Drosophila has two lipases that have been shown to contribute to lipolysis, the adipocyte triglyceride lipase (ATGL) homolog Brummer (BMM) and the hormone-sensitive lipase homolog HSL (Bi et al., 2012; Gronke et al., 2005). Additional putative lipases await characterization (Horne et al., 2009), among them lipases encoded by CG1882, CG5966 and the brummer-like dob gene. All three of these genes are expressed in fat body and the CG1882 product has been shown to co-purify with fat droplets (Birner-Gruenberger et al., 2012; Cermelli et al., 2006). Fat breakdown by lipases is controlled by proteins of the perilipin family, which associate with the surface of lipid droplets. Whereas mammals have five perilipins with different and partially redundant functions (Itabe et al., 2017), Drosophila has only two perilipins, Lsd-1 and Lsd-2 (also referred to as PLIN1 and PLIN2) (Bi et al., 2012; Miura et al., 2002; Teixeira et al., 2003). Similar to the mammalian proteins, Lsd-1 and Lsd-2 have both opposing and partially redundant functions. Lsd-1 promotes fat mobilization by recruitment of HSL to the lipid droplet surface (Bi et al., 2012). A role of Lsp-1 in the activation of lipolysis had initially been suggested by studies in another insect, Manduca sexta, which showed that increased phosphorylation of Lsd-1 correlates with increased lipolysis and increased activity of adipokinetic hormone, as discussed further below (Arrese et al., 2008a; Patel et al., 2005). Consistent with a role of Lsd-1 in promoting lipolysis, Lsd-1 mutant flies are obese and show a giant lipid droplet phenotype (Beller et al., 2010). Lsd-2 null mutants have significantly reduced TAG stores, suggesting a role in protecting lipid droplets from lipolysis (Gronke et al., 2003). However, the observation that Lsd-2 is required for the formation of lipid droplets in the cortical cytoplasm of fat body cells provides an alternative interpretation of this phenotype (Diaconeasa et al., 2013). Lsd-2 specifically associates with a subpopulation of small lipid droplets in the cortex. These droplets, but also larger cortical lipid droplets that do not bind Lsd-2, are eliminated when Lsd-2 is reduced by RNAi. These data suggest a role of Lsd-2 in the uptake of Lpp-delivered lipids from the hemolymph. Interestingly, Lsd-1 can partially replace Lsd-2 function in Lsd-2 mutants, suggesting some functional redundancy (Bi et al., 2012). Considering the important roles of perilipins in fat droplet dynamics, it comes as a surprise that flies that completely lack both Lsd-1 and Lsd-2 are viable and capable of regulating fat depots. This suggests that functions of perilipins are not essential in storage fat regulation in Drosophila under normal feeding conditions. However, they gain importance during times of reduced nutrient availability (Beller et al., 2010).
5. The hormonal and physiological regulation of fat stores
5.1 Insulin and TOR
5.1.1 Overview of insulin and TOR pathways
How does the availability of nutrients drive lipogenesis and lipolysis? Two major signaling pathways that mediate the effects of nutrients are the insulin and TOR (target of rapamycin) pathways. In both mammals and Drosophila, the effects of insulin or insulin-like peptides are mediated by an insulin receptor that activates a signaling axis consisting of the phosphatidyl-inositol 3-kinase PI3K and the protein kinase Akt (aka protein kinase B) (Manning and Toker, 2017). In mammals, the same axis can be activated by insulin-like growth factor 1 (IGF-1) and its receptor IGF-1R, which complicates the analysis of insulin signaling in mammalian systems (Kitamura et al., 2003; Oldham and Hafen, 2003). This complexity is reduced in Drosophila, which has only a single insulin receptor (InR) (Brogiolo et al., 2001). However, InR can potentially be activated by eight different insulin-like peptides (Dilps) (for reviews, see Nässel et al., 2013; Owusu-Ansah and Perrimon, 2014). Four of these Dilps, Dilp 1, 2, 3 and 5, are produced by two groups of insulin-producing neurosecretory cells in the brain, the IPCs (Brogiolo et al., 2001; Rulifson et al., 2002). Key downstream targets of the InR-PI3K-Akt pathway are FOXO transcription factors, which are phosphorylated by Akt and, thus, retained in the cytoplasm. Under fasting conditions that are associated with low insulin signaling, FOXOs translocate into the nucleus to control tissue-specific transcriptional programs that are important for cell survival, cell growth and metabolism, including lipid metabolism (S. Lee and Dong, 2017; Manning and Toker, 2017).
The TOR pathway can be directly activated by nutrients, in particular amino acids, or by insulin and other growth factors via crosstalk with the PI3K-Akt pathway (González and Hall, 2017; Wullschleger et al., 2006). Cross-activation of TOR by insulin, which is mediated by Akt phosphorylation of the TOR inhibitor tuberous sclerosis protein TSC2, can make it difficult to distinguish effects of insulin pathway activation that are independent of the TOR pathway. Mammalian TOR (mTOR) is a member of two functionally distinct complexes, mTORC1 and mTORC2. While an essential role of mTORC1 in protein synthesis and cellular growth has been known for quite some time, more recent data have established that both mTORC1 and mTORC2 also play important roles in adipogenesis and lipogenesis (Lamming and Sabatini, 2013). mTORC1 is not only required for normal adipogenesis, the development of adipose tissue, but it also stimulates lipogenesis by promoting the activation of lipogenic genes.
Critical for mediating the effects of both TORC1 and insulin on lipogenesis in the mammalian liver are the sterol regulatory element-binding proteins, SREBP1 and SREBP2. SREBPs are key regulators of lipid synthesis not only in vertebrates, but also in invertebrates (Kersten, 2001; Kunte et al., 2006; Shao and Espenshade, 2012). However, it is interesting to note that in mammalian adipose tissue, SREBP1 and SREBP2 may not have the same, critical role that they have in the liver, although essential functions may be obscured by redundancy between the two proteins (Shimano et al., 1997; Vergnes et al., 2016). Effects of mTORC1 on SREBP are mediated by both the ribosomal protein S6 kinase 1 (S6K1) and the lipin 1 protein (Düvel et al., 2010; Peterson et al., 2011). Controlling SREBP localization requires the PAP activity of lipin 1, suggesting the involvement of an intranuclear signaling pathway.
5.1.2 Insulin signaling and TOR in fat store regulation in Drosophila
Work in Drosophila has made important contributions to a better understanding of conserved aspects of the TOR and insulin regulatory networks (Hietakangas and Cohen, 2009; Oldham and Hafen, 2003). Here, I will discuss evidence indicating that this conservation extends to the control of lipogenesis and lipolysis. In Drosophila, insulin signaling promotes the accumulation of fat stores in a cell-autonomous manner. Up-regulation of the Drosophila InR specifically in the adult fat body increases TAG stores. Importantly, a similar increase is seen if the effect of InR on TAG levels is uncoupled from its effect on cell proliferation. This suggests that InR indeed promotes lipogenesis and/or attenuates lipolysis (DiAngelo and Birnbaum, 2009). A similar function in promoting fat storage is suggested for TOR by the phenotype of animals carrying hypomorphic alleles of TOR, which have decreased lipid levels and smaller fat droplets (Luong et al., 2006). However, some data seemingly contradict these results. For instance, TAGs are increased 4-fold in InR mutant flies and total lipids 2-fold in mutant flies lacking the insulin receptor substrate protein Chico (Böhni et al., 1999; Tatar et al., 2001). Consistent with these findings, ablation of the IPCs of the brain leads to a modest increase in whole-body TAGs (Broughton et al., 2005). Thus, interference with systemic release or systemic reception of the insulin signal results in increased lipid levels. These conflicting data can be reconciled by assuming that whole-organism effects mask cell-autonomous effects that altered insulin signaling has in the fat body. The same reasoning may explain the observation that feeding of the TOR inhibitor rapamycin results in increased fat levels in flies (Teleman et al., 2005).
How are the effects of Dilps and TOR on fat storage mediated in fat body cells? Evidence indicates that both lipogenic and lipolytic activities are the targets. TOR regulates intracellular localization of Lipin and insulin signaling seems to positively affect the role of Lipin in fat droplet formation (Schmitt et al., 2015). When nutrients are scarce and TOR signaling is low, Drosophila Lipin translocates into the cell nucleus, a property that it has in common with mammalian lipin 1 (Peterson et al., 2011). However, unlike down-regulation of TOR, down-regulation of the insulin pathway does not lead to nuclear translocation of Lipin (Schmitt et al., 2015). Instead, reduced InR activity strongly enhances the small lipid droplet phenotype observed after reduction of Lipin. These data contrast with data obtained in mice, which have, so far, not provided evidence that insulin has regulatory inputs on lipin 1 that are independent of TOR (Harris et al., 2007; Huffman et al., 2002; Péterfy et al., 2010). It will be interesting to determine in more detail how the InR-PI3K-Akt axis controls Lipin function independently of TOR in Drosophila and to further explore independent regulation in mammalian systems. Lipin 1 contains multiple phosphorylation sites that are sensitive to the TOR inhibitor rapamycin and that undergo phosphorylation in response to insulin signaling (Harris et al., 2007; Huffman et al., 2002; Peterson et al., 2011; Péterfy et al., 2010). In both lipin 1 and Drosophila Lipin these sites are clustered in similar regions of the proteins, suggesting functional conservation (Fig. 1B). Dephosphorylation leads to translocation into the nucleus and is catalyzed in mammals by the CTDNEP1(aka Dullard)/NEP1-R1 phosphatase complex (Han et al., 2012; Y. Kim et al., 2007). Homologs of this complex in yeast have been shown to dephosphorylate the yeast lipin homolog, Pah1p (Karanasios et al., 2010). Although the evolutionary conservation of this process suggests that the CTDNEP1 and NEP1-R1 homologs of Drosophila have the same function, this remains to be shown experimentally. It is also unknown if Drosophila Lipin targets that same genes that have been identified as putative direct targets of mouse lipin 1 (Finck et al., 2006).
In mammalian cells, nuclear translocation of lipin 1 leads to the exclusion of SREBP1 from the nucleus (Peterson et al., 2011), which raises the possibility that Drosophila Lipin acts in a similar way. In contrast to mammals, Drosophila has only one SREBP gene and Drosophila SREBP is activated by fatty acids and not by cholesterol (Seegmiller et al., 2002). However, like the mammalian SREBPs, Drosophila SREBP plays a key role in the activation of lipogenic genes (Kunte et al., 2006; Porstmann et al., 2008). SREBP mutants are not viable, but can be rescued by SREBP expression exclusively in two tissues, the midgut and the fat body, indicating that SREBP has an essential role in the fat body (Kunte et al., 2006).
An interesting observation worth noting is that Drosophila Lipin is required for the normal growth of fat body cells, as shown by genetic mosaic studies. This suggests insensitivity of cells lacking Lipin to growth factor stimulation (Schmitt et al., 2015). Indeed, insulin pathway activity is cell-autonomously down-regulated in the cells and, on the organismal level, hemolymph sugar levels are elevated. This suggests that tissues lacking Lipin become insulin resistant, an observation that is consistent with the severe insulin resistance observed in fld mice that lack lipin 1 (Reue et al., 2000). Reduction of two other enzymes of the glycerol-3-phosphate pathway, GPAT4 and AGPAT3, has effects on insulin pathway activity in the fat body that are similar to those of reduced Lipin (Schmitt et al., 2015). Consistent with this observation, a GPAT4 loss-of-function mutant exhibits elevated mRNA levels of Dilp 2 and 3 and decreased insulin responsiveness (Yan et al., 2015). These observations link the activity of the whole glycerol-3-phosphate pathway to cellular insulin responsiveness. It will be interesting to unravel the underlying molecular mechanism that connects the two pathways.
Work in Drosophila shows that not only lipogenesis, but also lipolysis, is under the control of insulin signaling and TOR. Expression of the BMM lipase depends on FOXO, the downstream effector of the insulin pathway, which promotes the synthesis of bmm mRNA under fasting conditions when insulin signaling is low (Wang et al., 2011). FOXO is negatively regulated through phosphorylation by the insulin pathway kinase Akt. However, FOXO can also be regulated by reversible acetylation (Brunet et al., 2004; Daitoku et al., 2004). Enzymes that deacetylate FOXO and, thereby, modulate its transcriptional functions, are the sirtuins (discussed below) and the class IIa histone deacetylase HDAC4 (Wang et al., 2011). HDAC4 is under negative control by InR and Akt that is mediated by the AMP-activated protein kinase SIK3. Loss of SIK3 leads to elevated expression of the BMM lipase and decreased fat stores (Choi et al., 2015; Wang et al., 2011). Interestingly, adipokinetic hormone (AKH) (to be discussed below) has a regulatory input on the SIK3-HDAC4-FOXO signaling axis by inhibiting LKB1, a kinase that directly activates SIK3 (Choi et al., 2015). Thus, feeding (insulin) and fasting (AKH) signals converge on SIK3 to control fat mobilization via HDAC4 and FOXO. Bmm mRNA is also up-regulated in animals carrying hypomorphic alleles of TOR (Luong et al., 2006). Thus, bmm appears to be regulated on a transcriptional level by insulin, TOR, and AKH to balance fat levels. It will be interesting to learn more about how regulation of Drosophila HSL contributes to this balance and if it is controlled by hormone-induced posttranslational modification similar to its mammalian counterpart.
It will also be interesting to determine in more detail which of the eight Dilps of Drosophila that potentially act through the insulin pathway have specific functions in the regulation of fat stores. So far, evidence suggests that the Drosophila InR can be activated by Dilp2, one of four Dilps that are produced by the IPCs in the brain (Brogiolo et al., 2001; Rulifson et al., 2002). Another of these Dilps, Dilp3, has been show to activate TOR activity in the fat body (J. Kim and Neufeld, 2015). These observations link both of Dilp 2 and Dilp 3 to the regulation of fat stores. Another Dilp that has been implicated with the regulation of fat stores is Dilp 6. Dilp 6 expression is strongly induced upon starvation in a FOXO-dependent manner in the larval fat body (Slaidina et al., 2009). The loss of body mass that is normally observed during starvation is enhanced in Dilp 6 mutants, but it has not been determined if this is associated with changes in TAG stores. Under fed conditions, loss of Dilp 6 does not affect fat stores in the larval fat body (Slaidina et al., 2009). However, in freshly eclosed adult flies, TAGs are increased after RNAi knockdown of Dilp 6 during the pupal stage, suggesting a defect in lipid processing. Consistent with this observation, it has been shown that, when adult flies are starved, fat body-produced Dilp 6 signals to the oenocytes and is required for lipid uptake and processing by the oenocytes (Chatterjee et al., 2014).
5.2 Adipokinetic Hormone and Octopamine
Fat stores in mammals are not only controlled by insulin and TOR, but also by glucagon, which antagonizes insulin signaling, and β-adrenergic signaling, which controls the rapid mobilization of fat stores during stress responses. Glucagon and adrenaline/noradrenaline have in common that they act through activation of intracellular signaling by the second messenger cAMP. cAMP-activated protein kinase A (PKA) promotes fat mobilization by phosphorylation of mammalian HSL and perilipins, with the result that HSL is recruited to the lipid droplet surface (Holm, 2003).
In Drosophila and other insects, adipokinetic hormone (AKH) takes on the role that glucagon has in mammals, and octopamine is the insect equivalent of noradrenaline/adrenaline in β-adrenergic signaling (Arrese and Soulages, 2010; Roeder, 2005). AKH is produced and released by the corpora cardiaca (CC). In Drosophila, these endocrine glands are fused with two other hormone-producing glands, the prothoracic glands (PG) and the corpora allata, to form the composite ring gland. Studies in a number of insect species, including pioneering studies in locusts and moths (Arrese et al., 1999; Lum and Chino, 1990; Z. Wang et al., 1990), have established a shared function of AKH in the mobilization of fat and glycogen from the fat body that is mediated by the second messengers cAMP and Ca2+ (reviewed in Arrese and Soulages, 2010). In Drosophila, mutations of the AKH gene or the AKH receptor (AKHR) gene lead to obesity in adult flies, whereas over-expression of AKH or AKHR leads to a dramatic reduction of fat stores (Baumbach et al., 2014b; Gáliková et al., 2015; Gronke et al., 2007). The obesity of AKHR mutants is strongly enhanced by loss of bmm, leading to extreme fat accumulation and suggesting that AKH acts on fat stores mostly through activating a different lipase. Considering the vertebrate data, a promising candidate is Drosophila HSL. Indeed, HSL translocates to lipid droplets during starvation, suggesting that an AKH-dependent phosphorylation of Lsd-1 and/or HSL by PKA is responsible for the translocation (Bi et al., 2012). However, although Drosophila Lsd-1 is susceptible to phosphorylation by PKA (Arrese et al., 2008b), mutation of potential PKA phosphorylation sites in Lsd-1 did not provide evidence in support of this hypothesis (Beller et al., 2010), and phosphorylation sites of mammalian HSL are not conserved in Drosophila HSL (Bi et al., 2012). Clearly, further studies will be necessary to gain a comprehensive understanding of the mechanism by which AKH activates lipolysis in Drosophila. With the recent discovery that AKH influences gene expression through the same signal transduction pathway used by mammalian glucagon, the door has opened for a genetic dissection of how the AKH signaling pathway influences the expression of lipogenic and lipolytic genes (Song et al., 2017). It seems unlikely that this occurs exclusively through convergence with the PI3K-Akt-FOXO axis, which, as discussed earlier, leads to the activation of lipolytic genes by FOXO.
In contrast to the adult stage, Drosophila larvae that cannot produce AKH have normal TAG and glycogen stores, and changes in TAG and carbohydrates that are characteristic for metamorphosis are undisturbed (Gáliková et al., 2015). Similarly, ablation of the AKH-producing cells of the CC does not affect larval TAG stores (G. Lee and Park, 2004). These observations serve as an important reminder that results obtained with adult fat body do not necessarily translate to larval fat body and vice versa. Environmental and feeding conditions under which larvae and adults live are quite different and this entails significant differences in fat storage regulation.
Interestingly, both cell ablation and AKH mutation suppress hyperactivity that flies normally display when starved, suggesting that AKH is responsible for behavioral changes during starvation. These changes are possibly mediated by the activation of octopaminergic neurons, which is required for starvation-induced hyperactivity in Drosophila (Yang et al., 2015). However, although it has been shown that AKH can activate octopaminergic signaling in Drosophila (Metaxakis et al., 2014), this is a hypothesis that remains to be tested. Flies lacking octopamine have considerably increased TAG depots despite lower food intake (Li et al., 2016). Besides the reduced locomotion, a reduced metabolic resting state was identified as the likely cause of this increase. At least part of the effects of the lack of octopamine can be explained by increased release of Dilp 2 by the IPCs.
5.3 Cytokine Signaling
The larval fat body of Drosophila operates as a nutrient sensor that coordinates the availability of nutrients with organismal growth (Colombani et al., 2003). A factor released by the fat body that has been shown to control systemic growth is the cytokine Unpaired 2 (Upd2) (Rajan and Perrimon, 2012). Both larvae and adults that cannot express upd2, or in which upd2 is specifically down-regulated in the fat body, have considerably reduced TAG levels, and larvae show growth defects. Starvation leads to a dramatic reduction of usd2 transcripts in adult flies that accompanies the reduction of fat stores in these flies. Importantly, starvation does not lead to fat breakdown in the fat body when upd2 expression is artificially maintained in the tissue. Further, it has been shown that Upd2 maintains fat stores in the fed state non-cell autonomously by signaling to the brain and stimulating Dilp 2 and Dilp 5 secretion by the IPCs. Knockdown of STAT92E, a component of the Upd2-controlled JAK/STAT pathway, in neurons projecting onto the IPCs, but not in the fat body, blocks Dilp release and leads to the depletion of fat stores. Intriguingly, human Leptin can replace Upd2 in Drosophila and rescue upd2 mutant phenotypes. This suggests that Upd2 is a functional homolog of Leptin in the fly, although reduction of Upd2 does not alter feeding behavior (Rajan and Perrimon, 2012).
5.4 Steroid Hormone Signaling
Vertebrate steroid hormones, in particular glucocorticoids, are known to have important roles in the control of lipid metabolism (Peckett et al., 2011). In contrast to vertebrates, insects produce only one class of steroid hormones that are referred to as ecdysteroids. Ecdysteroids act through two members of the nuclear receptor family, the Ecdysone Receptor (EcR) and Ultraspiracle (Usp), which form the heterodimeric ecdysteroid receptor EcR/Usp (for a review, see King-Jones and Thummel, 2005). 20-hydoxyecdysone (20E) is an ecdysteroid widely used as a hormone by many insect species, including Drosophila melanogaster, but the major ecdysteroid with biological activity found in Drosophila is makisterone A (Lavrynenko et al., 2015) (henceforth, and for the sake of simplicity, I will refer to the active compound as 20E). 20E is produced in peripheral tissues, including the fat body, from the ecdysteroid ecdysone, which is produced and secreted into the hemolymph by the PG portion of the ring gland. 20E acts as a key regulator of development and metabolism in Drosophila and other insects (for reviews, see Tennessen and Thummel, 2011; Yamanaka et al., 2013).
20E has been shown to have a suppressive effect on InR-PI3K pathway activity in the larval fat body that promotes nuclear translocation of FOXO (Colombani et al., 2005). However, it is unclear how this is achieved mechanistically, although it may involve 20E repression of microRNA mir-8 (Jin et al., 2012) (see below). 20E-induced attenuation of signaling through InR-PI3K should lead to increased lipolysis in the fat body given the known role of FOXO in the regulation of this process. Indeed, RNAi knockdown of EcR in the larval fat body leads to increased TAG levels (Kamoshida et al., 2012). Interestingly, when signaling through PI3K is reduced in the PG, basal titers of circulating ecdysteroids decrease (Colombani et al., 2005). This observation supports a model in which insufficient nutrient availability lowers basal ecdysteroid titers which, in turn, promotes fat storage. If the same scenario applies to adult flies, this may explain why systemic reduction of insulin signaling has the net effect of increasing fat stores despite its cell-autonomous role in the fat body to promote fat storage. However, at least in female flies, measurements of ecdysteroid titers indicate that these rather increase than decrease upon starvation (Terashima et al., 2005). Interestingly, decreased activity of TOR in the PG leads to a developmental delay that is associated with an inability of the animals to generate peak levels of ecdysteroids (Layalle et al., 2008). Such a delay is not seen after reduction of PI3K (Colombani et al., 2005). Thus, it seems that PI3K and TOR regulate ecdysone production independently to modulate basal titers and peak titers, the former leading to metabolic changes and the latter inducing developmental transitions. Interestingly, on the transcriptional level, increased 20E signaling leads to increased expression of the adipose (adp) gene in larval fat body, which may explain part of the effect that lowering of the ecdysteroid titer has on fat stores (Kamoshida et al., 2012). Mutations in adp, which has long been known to control body fat depots in Drosophila, lead to obese flies (Doane, 1960). Importantly, the function of adp in fat store regulation is highly conserved between invertebrates and mammals (Suh et al., 2007). In mammals, Adp binds to histones and the histone deacetylase HDAC3. Liver-specific knockout of HDAC3 in the mouse leads to de novo lipogenesis and increased expression of lipogenic genes, and HDAC3 has been shown to play important roles in the circadian control of lipid metabolism genes (Sun et al., 2011). This, and the known role of circadian pathways in the control of ecdysteroid titers (Di Cara and King-Jones, 2013), raises the intriguing possibility that an ecdysteroid-Adp axis controls circadian fluctuations in lipid metabolism in Drosophila.
5.5 Juvenile Hormone
Juvenile hormone (JH) is an important hormone in insects that cooperates with 20E to control the nature of developmental transitions. In addition, it plays an important role in insect reproduction (Jindra et al., 2013). JH is produced by the corpora allata, which are part of the ring gland in Drosophila. Genetic ablation of the corpora allata in Drosophila leads to stunted larval growth, the formation of undersized pupae, and impaired adult fat body development (Mirth et al., 2014; Yamamoto et al., 2013). Reduced growth can be rescued by reducing the level of FOXO activity, suggesting that it is primarily caused by changes in insulin signaling (Mirth et al., 2014). This suggests that JH acts on lipid metabolism, at least in part, indirectly via crosstalk with the insulin pathway. However, measurements of TAG levels are required to demonstrate that JH has, in fact, an effect on fat stores in Drosophila. Animals with ablated corpora allata show changes in the expression of several proteins in the larval fat body that are involved in energy metabolism such as hexokinase and isocitrate dehydrogenase (Liu et al., 2009). Gene expression profiling of adult females with reduced JH titers revealed that genes responding to JH are enriched in the Gene Ontology category “oxidation reduction” (Yamamoto et al., 2013). These data suggest participation of JH in the regulation of energy metabolism in Drosophila. This conclusion is supported by data from other insect species. For instance, in the cricket Gryllus firmus, treatment with the JH analog methoprene leads to decreased synthesis of total lipid and TAG, which correlates with the reduced activity of lipogenic enzymes in the fat body, and to increased oxidation of fatty acids (Zera and Z. Zhao, 2004). Consistent with these findings, in the mosquito Aedes aegypti, reduction of the JH receptor Met by RNAi leads to an up-regulation of genes involved in lipid and carbohydrate metabolism in the fat body (Zou et al., 2013).
In many insects, including Drosophila, JH is required for vitellogenesis, the incorporation of nutrients into maturing oocytes (Bownes, 1982). It has been demonstrated for a number of insects that JH is required for the synthesis of yolk protein precursors (vitellogenins) by the fat body and the uptake of these proteins into oocytes (Tufail and Takeda, 2008). However, little is known about the mechanism by which JH controls the import of lipids, which constitute 30–40% of the dry weight of insect oocytes, and are derived mostly from the fat body (Ziegler and Van Antwerpen, 2006). Clearly, the demonstrated importance of JH for the formation of lipid stores in oocytes warrants a closer investigation of how JH controls TAG synthesis in different tissues, the transport of lipids between tissues, and the tissue uptake of lipids.
5.6 Hedgehog Signaling
One of the main attractions of Drosophila is the ease with which unbiased genetic screens can be carried out to identify genes of functional importance. At least three such screens have been conducted that identified new players in the control of body fat depots. Pospisilik et al. (2010) carried out an in vivo RNAi screen for altered TAG levels in adult flies targeting over 10,000 genes. This screen confirmed known and identified a treasure trove of new genes that have an effect on TAG stores. Interestingly, in particular alteration of expression of genes of the Hedgehog (Hh) pathway led to robust changes in adult fat depots. Whereas reduction of Hh signaling in the fat body stimulated fat deposition, increases in Hh signaling decreased fat depots. The same responses to manipulation of the Hh pathway were observed in larval fat body (Suh et al., 2006). These results inspired experiments in transgenic mice, which showed that activation of the Hh pathway in adipose tissue leads to a dramatic reduction of white, but not brown, fat tissue (Pospisilik et al., 2010). Consistent with these results, Sonic Hh protein has been shown to inhibit adipocyte differentiation in mouse cell culture (Suh et al., 2006). A role of the Hh pathway in mammalian adipogenesis is further supported by gene expression studies showing that Hh represses adipogenic genes (Pospisilik et al., 2010). An involvement of Hh in adipogenesis has also been demonstrated in Drosophila, although here Hh seems to play a positive rather than a negative role in fat body development during embryogenesis (Riechmann et al., 1998). Later, during larval development, Hh produced by the midgut acts systemically as a hormone promoting fat store mobilization in the larval fat body under starvation conditions (Rodenfels et al., 2014). Block of Hh production by the midgut completely blocks lipolysis in the fat body. Levels of Hh protein found in the fat body correlate with the amount of Hh produced by the gut and an increase or decrease of Hh signaling in the fat body leads to reduced or enhanced organismal growth, respectively (Rodenfels et al., 2014). In the liver of mice, selective down-regulation of Hh signaling leads to activation of lipogenic genes, including SREBP1 and GPAT, and fat droplet accumulation in hepatocytes (steatosis) (Matz-Soja et al., 2016). Together, these studies establish roles of Hh in adipogenesis and lipid metabolism with a conserved role as a negative regulator of TAG stores. Interestingly, in Drosophila larvae, Hh produced by the midgut also signals to the prothoracic gland to inhibit expression of genes of the ecdysone biosynthetic pathway and to delay pupariation (Rodenfels et al., 2014). This suggests that Hh signaling coordinately controls both energy homeostasis and developmental timing in response to nutritional cues.
5.7 Sirtuins and HNF4
A second genetic screen for obesity genes employed a buoyancy assay examining mutant fly stocks for obese larvae (Reis et al., 2010). Examining about 500 genes, this screen identified 66 candidate genes for the control of fat stores, among them the Drosophila homolog of mammalian SIRT1, Sir2. SIRT1/Sir2 belongs to the sirtuin family of NAD+-dependent deacetylases that have conserved and evolutionarily old functions in metabolic regulation (Schwer and Verdin, 2008). Down-regulation of Sir2 specifically in the fat body leads to increased fat stores, and up-regulation in fat body cell clones to depletion of cellular fat stores. Loss of Sir2 correlates with changes in metabolic gene expression that favor fat accumulation (Reis et al., 2010), including the down-regulation of eleven genes annotated as lipases (Palu and Thummel, 2016). Together, these results show that Sir2 cell-autonomously controls fat stores in the fat body and add Drosophila to the list of organisms with conserved roles of sirtuins in the regulation of lipid metabolism (Schwer and Verdin, 2008).
At least one downstream target that mediates the effects of Sir2 on lipid metabolism in Drosophila is the nuclear receptor HNF4 (Palu and Thummel, 2016). Sir2 deacetylates and physically interacts with HNF4 to increase its activity and stability. HNF4, in turn, has been shown to be required for TAG breakdown under starvation conditions and to activate genes that act in lipolysis and fatty acid β-oxidation (Palanker et al., 2009). Interestingly, target genes regulated by HNF4 in Drosophila are similar to target genes of the mammalian peroxisome proliferator-activated receptor PPARα. PPARα is a key regulator of lipid metabolism in mammals that maintains energy homeostasis under starvation conditions (Kersten, 2014). Since orthologs of PPARs do not exist in Drosophila, it has been proposed that ancestral functions of HNF4 in lipid metabolism have been adopted by PPARα in the vertebrate lineage (Palanker et al., 2009).
5.8 Calcium
In a third genetic screen for obesity and anti-obesity genes, Baumbach et al. (2014a) targeted 6,796 genes (~50% of the protein-encoding genes of Drosophila) by conditional RNAi in adult flies. Again, aside from confirming known genes involved in lipid storage, this screen identified 58 previously unknown obesity-related genes, 79% of which have human orthologs. Several of the identified genes encode components of the store-operated calcium entry machinery that controls cytosolic Ca2+ (iCa2+) levels by regulating entry of ER-sequestered and extracellular Ca2+. Genetic manipulations that increased iCa2+ levels in the fat body led to reduced fat stores, whereas manipulations that decreased iCa2+ levels led to increased fat stores. Interestingly, the effects of iCa2+ on fat storage appear to be caused, at least in part, by a non-cell autonomous mechanism that involves activation of production of sNPF, a functional homolog of mammalian orexigenic neuropeptide F, in cells of the brain. Elevated levels of sNPF, produced when levels of iCa2+ in the fat body are low, cause increased food intake and, in turn, increased TAG accumulation in adult flies. Changes in iCa2+ in adult fat body cells have also been linked to AKH signaling, which acts through both cAMP and Ca2+ as second messengers to mobilize fats (Baumbach et al., 2014b). While these data establish a role of Ca2+ signaling in the fat body of adults, manipulation of iCa2+ levels in the larval fat body did not have a significant effect on whole-body TAG levels, consistent with the observation that AKH does not have an effect on larval fat stores (Baumbach et al., 2014a; Gáliková et al., 2015).
5.9 DHR96
As discussed above, the nuclear receptors HNF4 and EcR/Usp have important roles in the regulation of fat stores in Drosophila. Another nuclear receptor with a key role in lipid metabolism in Drosophila is DHR96. DHR96 is expressed in the midgut where it is responsible for transcriptional activation of the margo gene, which encodes gastric lipase (Sieber and Thummel, 2012; 2009). This homolog of mammalian gastric lipase is secreted into the gut lumen to digest dietary TAGs for fatty acid absorption. Consistent with this function, DHR96 mutants have depleted fat body TAG stores (Sieber and Thummel, 2009). Interestingly, DHR96 also controls cholesterol homeostasis and mediates the transcriptional response to cholesterol (Bujold et al., 2010). Cholesterol, which is a vitamin in insects, can directly bind to DHR96, although it is unclear if cholesterol or a different ligand controls transcriptional activity of the receptor (Horner et al., 2009). Based on its primary structure, DHR96 is most closely related to vertebrate receptors PXR (pregnane X receptor) and CAR (androstane receptor). However, functionally, it seems to be more similar to the liver X receptor LXRα, which has similar roles in the regulation of cholesterol homeostasis in mammals (C. Zhao and Dahlman-Wright, 2010).
5.10 microRNAs
How microRNAs are tied into the signaling networks discussed above, including the insulin, ecdysteroid, and Hh pathways, has been extensively covered in a recent review article (Carthew et al., 2017). Therefore, I will confine myself here to a few examples where the impact of microRNAs on fat stores has been directly measured. A recent proteomics study in Drosophila has identified the mir-310 family of microRNAs as important regulators of lipid metabolism (Çiçek et al., 2016). The total TAG content of female flies lacking the mir-310 microRNA genes is reduced to one half and, strikingly, 20% of the genes that respond to mir-310s encode proteins that are lipid droplet associated. Mir-310s respond to nutritional changes and regulate stem cell maintenance in the ovarian stem cell niche via a control of Hh signaling. However, it is currently unknown if mir-310s have a similar role in Hh signaling in other tissues. It would be interesting to determine if, for instance, they are involved in the control of Hh signaling in the larval or adult fat body or in Hh release from the larval midgut.
Another microRNA with a link to Hh signaling is mir-14, which is one of the first Drosophila microRNAs shown to have a role in fat store regulation (Xu et al., 2003). Mir-14 was identified as a microRNA that can modulate Hh signaling by reducing Hh expression in the wing imaginal disc (Kim et al., 2014). Flies that cannot express mir-14 have an about 2-fold increase in body stores of TAG and adult fat body cells contain enlarged fat droplets. DAG is increased as well. While a role of mir-14 in regulating fat stores through effects on Hh signaling remains unexplored, it has been shown that mir-14 exerts at least part of its effect on fat stores by acting on systemic insulin signaling. Specific down-regulation of mir-14 in the Dilp-producing IPCs leads to increased fat storage, whereas down-regulation in the fat body has no effect on fat stores (Varghese et al., 2010). The direct target of mir-14 in the IPCs is the mRNA of the transcription factor Sugarbabe, which negatively regulates expression of Dilps 3 and 5. Thus, mir-14 exerts its effect on fat storage systemically through inhibition of an inhibitor of Dilp expression.
Loss of the mir-278 micoRNA has the opposite effect on fat stores. Mir-278 mutants are lean and show increased Dilp expression in the IPCs. However, the reduction of fat stores in these animals is not caused by a direct action of mir-278 in the IPCs. Instead, loss of mir-278 in the fat body causes insulin resistance of the tissue, which in turn leads to elevated hemolymph sugar levels and increases Dilp expression (Teleman et al., 2006).
Another microRNA that is required for normal insulin sensitivity of the fat body is mir-8 (Hyun et al., 2009). Interestingly, mir-8 is transcriptionally repressed in the larval fat body by ecdysteroid signaling (Jin et al., 2012), which may explain, at least in part, the repressive effects of ecdysteroid on InR-PI3K activity in the fat body (Colombani et al., 2005). Mir-8 represses expression of the zinc finger protein U-shaped (USH) in the fat body. The vertebrate homolog of USH has been shown to directly suppress PI3K signaling by binding to the regulatory subunit of PI3K, suggesting that this is the mechanism by which USH acts in Drosophila as well (Hyun et al., 2009).
Mir-8 and other microRNAs, including mir-305, are down-regulated by starvation. This suggests that lack of nutrients affects the entire microRNA-processing pathway in the fat body. In support of this model, it has been shown that reduction of dicer 1 expression in the fat body increases survival of flies under starvation conditions, although it was not determined if this is accompanied by altered fat stores (Barrio et al., 2014). Reduced expression of mir-305 under starvation conditions depends on reduced TOR signaling and leads to enhanced expression of p53, which promotes survival under starvation conditions by controlling the rate at which glycogen and TAG stores are depleted. The genes targeted by p53 are unknown, but several target genes of mammalian p53 are known to act in lipid metabolism and Drosophila homologs of these genes should be good candidates (Parrales and Iwakuma, 2016). These examples illustrate that microRNAs are intricately integrated into the fine-tuning of signaling pathways that respond to nutrition and control organismal fat stores.
6. Concluding Remarks
The power of the established toolset of genetic analysis in Drosophila together with newly emerging techniques such as CRISPR/Cas9 mutagenesis ensures that Drosophila will continue to make seminal contributions to our understanding of metabolic regulation. We have only begun to appreciate the complexity of regulatory networks that govern the control of body fat depots. For instance, it has been estimated that more than 60% of all protein-encoding genes in humans are subject to control by microRNAs (Friedman et al., 2009). Thus, we have probably only scratched the surface in understanding the complex roles of microRNAs in metabolic control. Similarly, both the brain and the gut of Drosophila produce numerous secreted peptides with potential roles in the control of feeding behavior and adiposity (Nässel and Winther, 2010; Reiher et al., 2011). Some of these, such as neuropeptide F, allatostatin A, and tachykinin have already been shown to impact adiposity and others are likely to follow (Baumbach et al., 2014a; Hentze et al., 2015; Song et al., 2014). The genetic screens discussed here have provided numerous candidate genes that will keep the comparatively small number of Drosophila laboratories working in the field of metabolic regulation busy for years to come. As discussed in this review, many homologs of known mammalian enzymes and regulators still await genetic characterization in Drosophila. These studies may not only confirm known functions but also bring roles to light that were previously unknown.
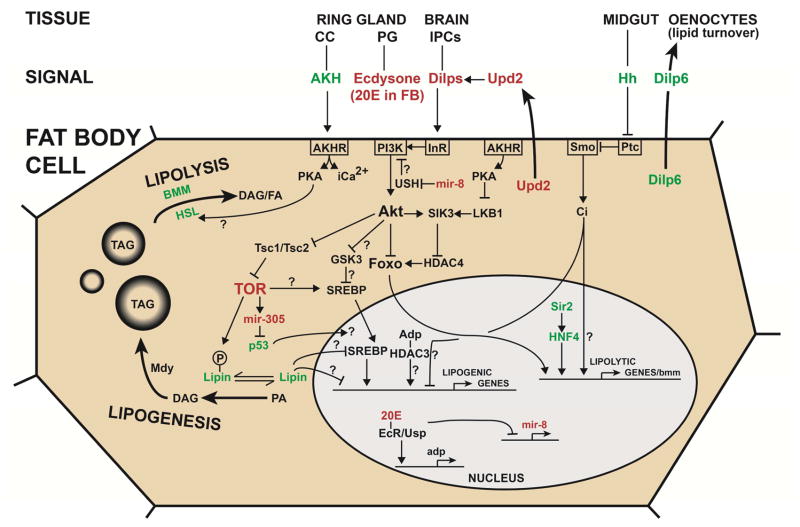
Fig. 2.
Model of the regulatory network that controls lipogenesis and lipolysis in the fat body of Drosophila. Only signals that control adiposity by directly acting on the fat body, or by being released from the tissue, are shown. The color RED indicates down-regulation upon starvation and GREEN up-regulation upon starvation. Question marks indicate connections that have been shown to exist in mammalian systems, but not yet in Drosophila, or that need clarification of mechanistic details. The model combines data derived from studies in larvae and adults and it is important to emphasize that regulatory mechanisms are not necessary the same in larval and adult fat body. For instance, a role for AKH and iCa2+ has so far only been described for adult fat body; the effects of mir-8 and of 20E on adp and of mir-8 have only been demonstrated for larvae, although Adp controls adiposity in both larvae and adults; Sir2 has been shown to control fat depots in both larvae and adults, but interaction with HNF4 has only been described for adults; Hh controls adiposity in both larvae and adults, but secretion by the midgut has only been demonstrated for larvae; the LKB1-SIK3-HDAC4 signaling axis has been studied in larval, but not in adult, fat body; Dilp 6 plays a role in both larval and adult fat body, but signaling to oenocytes has only been demonstrated for adults; finally, the effects of PI3K and TOR down-regulation in the PG suggest that ecdysone is down-related in larvae in response to starvation, but this is not the case in female adult flies that show an up-regulation of ecdysteroids.
Highlights.
Similar signaling processes govern fat store regulation in flies and mammals.
Genetic screens in Drosophila discover new regulators of body fat depots.
Drosophila genetics gauges the biological importance of fat store regulators.
Acknowledgments
I am grateful to an anonymous reviewer for making excellent suggestions for improving this review, and I am grateful to Stephanie E. Hood for critical reading of the manuscript. Work in the author’s laboratory is supported by the National Institutes of Health [grant number 1R15DK114748-01] and the Arkansas Biosciences Institute.
Abbreviations
- AGPAT
acylglycerol-3-phosphate acyltransferase
- AKH
adipokinetic hormone
- BMM
Brummer
- CC
corpora cardiac
- DAG
diacylglycerol
- Dilp
Drosophila insulin-like peptide
- DGAT
diacylglycerol acyltransferase
- EcR
ecdysone receptor
- GPAT
glycerol-3-phosphate acyltransferase
- HDAC
histone deacetylase
- Hh
hedgehog
- HSL
hormone-sensitive lipase
- InR
insulin receptor
- IPC
insulin-producing cell
- JH
juvenile hormone
- Mdy
Midway
- PA
phosphatidic acid
- PAP
phosphatidic acid phosphatase
- PG
prothoracic gland
- PI3K
phospha-tidylinositol 3-kinase
- PPAR
peroxisome proliferator-activated receptor
- SREBP
sterol regulatory element-binding protein
- TAG
triacylglycerol
- TOR
target of rapamycin
- Upd2
Unpaired 2
- USH
U-shaped
- Usp
Ultraspiracle
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arrese EL, Flowers MT, Gazard JL, Wells MA. Calcium and cAMP are second messengers in the adipokinetic hormone-induced lipolysis of triacylglycerols in Manduca sexta fat body. J Lipid Res. 1999;40:556–564. [PubMed] [Google Scholar]
- Arrese EL, Mirza S, Rivera L, Howard AD, Chetty PS, Soulages JL. Expression of lipid storage droplet protein-1 may define the role of AKH as a lipid mobilizing hormone in Manduca sexta. Insect Biochem Mol Biol. 2008a;38:993–1000. doi: 10.1016/j.ibmb.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrese EL, Rivera L, Hamada M, Mirza S, Hartson SD, Weintraub S, Soulages JL. Function and structure of lipid storage droplet protein 1 studied in lipoprotein complexes. Arch Biochem Biophys. 2008b;473:42–47. doi: 10.1016/j.abb.2008.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrese EL, Soulages JL. Insect fat body: energy, metabolism, and regulation. Annu Rev Entomol. 2010;55:207–225. doi: 10.1146/annurev-ento-112408-085356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker KD, Thummel CS. Diabetic larvae and obese flies-emerging studies of metabolism in Drosophila. Cell Metab. 2007;6:257–266. doi: 10.1016/j.cmet.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrio L, Dekanty A, Milán M. MicroRNA-mediated regulation of Dp53 in the Drosophila fat body contributes to metabolic adaptation to nutrient deprivation. Cell Rep. 2014;8:528–541. doi: 10.1016/j.celrep.2014.06.020. [DOI] [PubMed] [Google Scholar]
- Bastien M, Poirier P, Lemieux I, Després J-P. Overview of epidemiology and contribution of obesity to cardiovascular disease. Prog Cardiovasc Dis. 2014;56:369–381. doi: 10.1016/j.pcad.2013.10.016. [DOI] [PubMed] [Google Scholar]
- Baumbach J, Hummel P, Bickmeyer I, Kowalczyk KM, Frank M, Knorr K, Hildebrandt A, Riedel D, Jäckle H, Kühnlein RP. A Drosophila in vivo screen identifies store-operated calcium entry as a key regulator of adiposity. Cell Metab. 2014a;19:331–343. doi: 10.1016/j.cmet.2013.12.004. [DOI] [PubMed] [Google Scholar]
- Baumbach J, Xu Y, Hehlert P, Kühnlein RP. Gαq, Gγ1 and Plc21C control Drosophila body fat storage. J. Genet. Genomics. 2014b;41:283–292. doi: 10.1016/j.jgg.2014.03.005. [DOI] [PubMed] [Google Scholar]
- Beller M, Bulankina AV, Hsiao H-H, Urlaub H, Jäckle H, Kühnlein RP. PERILIPIN-dependent control of lipid droplet structure and fat storage in Drosophila. Cell Metab. 2010;12:521–532. doi: 10.1016/j.cmet.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Bi J, Xiang Y, Chen H, Liu Z, Gronke S, Kühnlein RP, Huang X. Opposite and redundant roles of the two Drosophila perilipins in lipid mobilization. J Cell Sci. 2012;125:3568–3577. doi: 10.1242/jcs.101329. [DOI] [PubMed] [Google Scholar]
- Birner-Gruenberger R, Bickmeyer I, Lange J, Hehlert P, Hermetter A, Kollroser M, Rechberger GN, Kühnlein RP. Functional fat body proteomics and gene targeting reveal in vivo functions of Drosophila melanogaster α-Esterase-7. Insect Biochem Mol Biol. 2012;42:220–229. doi: 10.1016/j.ibmb.2011.12.004. [DOI] [PubMed] [Google Scholar]
- Bodenmiller B, Campbell D, Gerrits B, Lam H, Jovanovic M, Picotti P, Schlapbach R, Aebersold R. PhosphoPep—a database of protein phosphorylation sites in model organisms. Nat Biotechnol. 2008;26:1339–1340. doi: 10.1038/nbt1208-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutanaev AM, Kalmykova AI, Shevelyov YY, Nurminsky DI. Large clusters of co-expressed genes in the Drosophila genome. Nature. 2002;420:666–669. doi: 10.1038/nature01216. [DOI] [PubMed] [Google Scholar]
- Bownes M. Hormonal and genetic regulation of vitellogenesis in Drosophila. Q Rev Biol. 1982;57:247–274. doi: 10.1086/412802. [DOI] [PubMed] [Google Scholar]
- Böhni R, Riesgo-Escovar J, Oldham S, Brogiolo W, Stocker H, Andruss BF, Beckingham K, Hafen E. Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1-4. Cell. 1999;97:865–875. doi: 10.1016/s0092-8674(00)80799-0. [DOI] [PubMed] [Google Scholar]
- Bridon G, Bonneil E, Muratore-Schroeder T, Caron-Lizotte O, Thibault P. Improvement of phosphoproteome analyses using FAIMS and decision tree fragmentation. application to the insulin signaling pathway in Drosophila melanogaster S2 cells. J Proteome Res. 2012;11:927–940. doi: 10.1021/pr200722s. [DOI] [PubMed] [Google Scholar]
- Brogiolo W, Stocker H, Ikeya T, Rintelen F, Fernandez R, Hafen E. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol. 2001;11:213–221. doi: 10.1016/s0960-9822(01)00068-9. [DOI] [PubMed] [Google Scholar]
- Broughton SJ, Piper MDW, Ikeya T, Bass TM, Jacobson J, Driege Y, Martinez P, Hafen E, Withers DJ, Leevers SJ, Partridge L. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc Natl Acad Sci USA. 2005;102:3105–3110. doi: 10.1073/pnas.0405775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Bujold M, Gopalakrishnan A, Nally E, King-Jones K. Nuclear receptor DHR96 acts as a sentinel for low cholesterol concentrations in Drosophila melanogaster. Mol Cell Biol. 2010;30:793–805. doi: 10.1128/MCB.01327-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buszczak M, Lu X, Segraves WA, Chang TY, Cooley L. Mutations in the midway gene disrupt a Drosophila acyl coenzyme A: diacylglycerol acyltransferase. Genetics. 2002;160:1511–1518. doi: 10.1093/genetics/160.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canavoso LE, Jouni ZE, Karnas KJ, Pennington JE, Wells MA. Fat metabolism in insects. Annu Rev Nutr. 2001;21:23–46. doi: 10.1146/annurev.nutr.21.1.23. [DOI] [PubMed] [Google Scholar]
- Canavoso LE, Yun HK, Jouni ZE, Wells MA. Lipid transfer particle mediates the delivery of diacylglycerol from lipophorin to fat body in larval Manduca sexta. J Lipid Res. 2004;45:456–465. doi: 10.1194/jlr.M300242-JLR200. [DOI] [PubMed] [Google Scholar]
- Cao G, Konrad RJ, Li SD, Hammond C. Glycerolipid acyltransferases in triglyceride metabolism and energy homeostasis-potential as drug targets. Endocr Metab Immune Disord Drug Targets. 2012;12:197–206. doi: 10.2174/187153012800493459. [DOI] [PubMed] [Google Scholar]
- Carthew RW, Agbu P, Giri R. MicroRNA function in Drosophila melanogaster. Sem Cell Dev Biol. 2017;65:29–37. doi: 10.1016/j.semcdb.2016.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermelli S, Guo Y, Gross SP, Welte MA. The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr Biol. 2006;16:1783–1795. doi: 10.1016/j.cub.2006.07.062. [DOI] [PubMed] [Google Scholar]
- Chatterjee D, Katewa SD, Qi Y, Jackson SA, Kapahi P, Jasper H. Control of metabolic adaptation to fasting by dILP6-induced insulin signaling in Drosophila oenocytes. Proc Natl Acad Sci USA. 2014;111:17959–17964. doi: 10.1073/pnas.1409241111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow JAT. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- Choi S, Lim D-S, Chung J. Feeding and Fasting Signals Converge on the LKB1-SIK3 Pathway to Regulate Lipid Metabolism in Drosophila. PLoS Genet. 2015;11:e1005263. doi: 10.1371/journal.pgen.1005263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombani J, Bianchini L, Layalle S, Pondeville E, Dauphin-Villemant C, Antoniewski C, Carré C, Noselli S, Leopold P. Antagonistic actions of ecdysone and insulins determine final size in Drosophila. Science. 2005;310:667–670. doi: 10.1126/science.1119432. [DOI] [PubMed] [Google Scholar]
- Colombani J, Raisin S, Pantalacci S, Radimerski T, Montagne J, Leopold P. A nutrient sensor mechanism controls Drosophila growth. Cell. 2003;114:739–749. doi: 10.1016/s0092-8674(03)00713-x. [DOI] [PubMed] [Google Scholar]
- Csaki LS, Reue K. Lipins: multifunctional lipid metabolism proteins. Annu Rev Nutr. 2010;30:257–272. doi: 10.1146/annurev.nutr.012809.104729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çiçek IÖ, Karaca S, Brankatschk M, Eaton S, Urlaub H, Shcherbata HR. Hedgehog Signaling Strength Is Orchestrated by the mir-310 Cluster of MicroRNAs in Response to Diet. Genetics. 2016;202:1167–1183. doi: 10.1534/genetics.115.185371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daitoku H, Hatta M, Matsuzaki H, Aratani S, Ohshima T, Miyagishi M, Nakajima T, Fukamizu A. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc Natl Acad Sci USA. 2004;101:10042–10047. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cara F, King-Jones K. How clocks and hormones act in concert to control the timing of insect development. Curr Top Dev Biol. 2013;105:1–36. doi: 10.1016/B978-0-12-396968-2.00001-4. [DOI] [PubMed] [Google Scholar]
- Diaconeasa B, Mazock GH, Mahowald AP, Dubreuil RR. Genetic studies of spectrin in the larval fat body of Drosophila melanogaster: evidence for a novel lipid uptake apparatus. Genetics. 2013;195:871–881. doi: 10.1534/genetics.113.155192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiAngelo JR, Birnbaum MJ. Regulation of fat cell mass by insulin in Drosophila melanogaster. Mol Cell Biol. 2009;29:6341–6352. doi: 10.1128/MCB.00675-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diop SB, Bodmer R. Gaining Insights into Diabetic Cardiomyopathy from Drosophila. Trends Endocrinol Metab. 2015;26:618–627. doi: 10.1016/j.tem.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doane WW. Developmental physiology of the mutant female sterile(2)adipose of Drosophila melanogaster. I. Adult morphology, longevity, egg production, and egg lethality. J Exp Zool. 1960;145:1–21. doi: 10.1002/jez.1401450102. [DOI] [PubMed] [Google Scholar]
- Düvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, Vander Heiden MG, MacKeigan JP, Finan PM, Clish CB, Murphy LO, Manning BD. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finck BN, Gropler MC, Chen Z, Leone TC, Croce MA, Harris TE, Lawrence JC, Kelly DP. Lipin 1 is an inducible amplifier of the hepatic PGC-1alpha/PPARalpha regulatory pathway. Cell Metab. 2006;4:199–210. doi: 10.1016/j.cmet.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK-H, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher EJ, LeRoith D. Insulin, insulin resistance, obesity, and cancer. Curr Diab Rep. 2010;10:93–100. doi: 10.1007/s11892-010-0101-y. [DOI] [PubMed] [Google Scholar]
- Gáliková M, Diesner M, Klepsatel P, Hehlert P, Xu Y, Bickmeyer I, Predel R, Kühnlein RP. Energy Homeostasis Control in Drosophila Adipokinetic Hormone Mutants. Genetics. 2015;201:665–683. doi: 10.1534/genetics.115.178897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González A, Hall MN. Nutrient sensing and TOR signaling in yeast and mammals. EMBO J. 2017;36:397–408. doi: 10.15252/embj.201696010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham P, Pick L. Drosophila as a Model for Diabetes and Diseases of Insulin Resistance. Curr Top Dev Biol. 2017;121:397–419. doi: 10.1016/bs.ctdb.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillet M, Dominguez Gonzalez B, Sicart A, Pöttler M, Cascalho A, Billion K, Hernandez Diaz S, Swerts J, Naismith TV, Gounko NV, Verstreken P, Hanson PI, Goodchild RE. Torsins Are Essential Regulators of Cellular Lipid Metabolism. Dev Cell. 2016 doi: 10.1016/j.devcel.2016.06.017. [DOI] [PubMed] [Google Scholar]
- Gronke S, Beller M, Fellert S, Ramakrishnan H, Jäckle H, Kühnlein RP. Control of fat storage by a Drosophila PAT domain protein. Curr Biol. 2003;13:603–606. doi: 10.1016/s0960-9822(03)00175-1. [DOI] [PubMed] [Google Scholar]
- Gronke S, Mildner A, Fellert S, Tennagels N, Petry S, Müller G, Jäckle H, Kühnlein RP. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab. 2005;1:323–330. doi: 10.1016/j.cmet.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Gronke S, Müller G, Hirsch J, Fellert S, Andreou A, Haase T, Jäckle H, Kühnlein RP. Dual lipolytic control of body fat storage and mobilization in Drosophila. PLoS Biol. 2007;5:e137. doi: 10.1371/journal.pbio.0050137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez E, Wiggins D, Fielding B, Gould AP. Specialized hepatocyte-like cells regulate Drosophila lipid metabolism. Nature. 2007;445:275–280. doi: 10.1038/nature05382. [DOI] [PubMed] [Google Scholar]
- Han S, Bahmanyar S, Zhang P, Grishin N, Oegema K, Crooke R, Graham M, Reue K, Dixon JE, Goodman JM. Nuclear envelope phosphatase 1-regulatory subunit 1 (formerly TMEM188) is the metazoan Spo7p ortholog and functions in the lipin activation pathway. J Biol Chem. 2012;287:3123–3137. doi: 10.1074/jbc.M111.324350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TE, Finck BN. Dual function lipin proteins and glycerolipid metabolism. Trends Endocrinol Metab. 2011;22:226–233. doi: 10.1016/j.tem.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TE, Huffman TA, Chi A, Shabanowitz J, Hunt DF, Kumar A, Lawrence JC. Insulin controls subcellular localization and multisite phosphorylation of the phosphatidic acid phosphatase, lipin 1. J Biol Chem. 2007;282:277–286. doi: 10.1074/jbc.M609537200. [DOI] [PubMed] [Google Scholar]
- Hentze JL, Carlsson MA, Kondo S, Nässel DR, Rewitz KF. The Neuropeptide Allatostatin A Regulates Metabolism and Feeding Decisions in Drosophila. Sci Rep. 2015;5:11680. doi: 10.1038/srep11680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietakangas V, Cohen SM. Regulation of tissue growth through nutrient sensing. Annu Rev Genet. 2009;43:389–410. doi: 10.1146/annurev-genet-102108-134815. [DOI] [PubMed] [Google Scholar]
- Holm C. Molecular mechanisms regulating hormone-sensitive lipase and lipolysis. Biochem Soc Trans. 2003;31:1120–1124. doi: 10.1042/bst0311120. [DOI] [PubMed] [Google Scholar]
- Horne I, Haritos VS, Oakeshott JG. Comparative and functional genomics of lipases in holometabolous insects. Insect Biochem Mol Biol. 2009;39:547–567. doi: 10.1016/j.ibmb.2009.06.002. [DOI] [PubMed] [Google Scholar]
- Horner MA, Pardee K, Liu S, King-Jones K, Lajoie G, Edwards A, Krause HM, Thummel CS. The Drosophila DHR96 nuclear receptor binds cholesterol and regulates cholesterol homeostasis. Genes Dev. 2009;23:2711–2716. doi: 10.1101/gad.1833609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffman TA, Mothe-Satney I, Lawrence JC. Insulin-stimulated phosphorylation of lipin mediated by the mammalian target of rapamycin. Proc Natl Acad Sci USA. 2002;99:1047–1052. doi: 10.1073/pnas.022634399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun S, Lee JH, Jin H, Nam J, Namkoong B, Lee G, Chung J, Kim VN. Conserved MicroRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell. 2009;139:1096–1108. doi: 10.1016/j.cell.2009.11.020. [DOI] [PubMed] [Google Scholar]
- Itabe H, Yamaguchi T, Nimura S, Sasabe N. Perilipins: a diversity of intracellular lipid droplet proteins. Lipids Health Dis. 2017;16:83. doi: 10.1186/s12944-017-0473-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Kim VN, Hyun S. Conserved microRNA miR-8 controls body size in response to steroid signaling in Drosophila. Genes Dev. 2012;26:1427–1432. doi: 10.1101/gad.192872.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jindra M, Palli SR, Riddiford LM. The juvenile hormone signaling pathway in insect development. Annu Rev Entomol. 2013;58:181–204. doi: 10.1146/annurev-ento-120811-153700. [DOI] [PubMed] [Google Scholar]
- Kamoshida Y, Fujiyama-Nakamura S, Kimura S, Suzuki E, Lim J, Shiozaki-Sato Y, Kato S, Takeyama K-I. Ecdysone receptor (EcR) suppresses lipid accumulation in the Drosophila fat body via transcription control. Biochem Biophys Res Commun. 2012;421:203–207. doi: 10.1016/j.bbrc.2012.03.135. [DOI] [PubMed] [Google Scholar]
- Karanasios E, Han G-S, Xu Z, Carman GM, Siniossoglou S. A phosphorylation-regulated amphipathic helix controls the membrane translocation and function of the yeast phosphatidate phosphatase. Proc Natl Acad Sci USA. 2010;107:17539–17544. doi: 10.1073/pnas.1007974107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten S. Integrated physiology and systems biology of PPARα. Mol Metab. 2014;3:354–371. doi: 10.1016/j.molmet.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten S. Mechanisms of nutritional and hormonal regulation of lipogenesis. EMBO Rep. 2001;2:282–286. doi: 10.1093/embo-reports/kve071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Neufeld TP. Dietary sugar promotes systemic TOR activation in Drosophila through AKH-dependent selective secretion of Dilp3. Nat Commun. 2015;6:6846. doi: 10.1038/ncomms7846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Vinayagam A, Perrimon N. A rapid genome-wide microRNA screen identifies miR-14 as a modulator of Hedgehog signaling. Cell Rep. 2014;7:2066–2077. doi: 10.1016/j.celrep.2014.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Gentry MS, Harris TE, Wiley SE, Lawrence JC, Dixon JE. A conserved phosphatase cascade that regulates nuclear membrane biogenesis. Proc Natl Acad Sci USA. 2007;104:6596–6601. doi: 10.1073/pnas.0702099104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King-Jones K, Thummel CS. Nuclear Receptors - A Perspective from Drosophila. Nat Rev Genet. 2005;6:311–323. doi: 10.1038/nrg1581. [DOI] [PubMed] [Google Scholar]
- Kitamura T, Kahn CR, Accili D. Insulin receptor knockout mice. Annu Rev Physiol. 2003;65:313–332. doi: 10.1146/annurev.physiol.65.092101.142540. [DOI] [PubMed] [Google Scholar]
- Krahmer N, Farese RV, Walther TC. Balancing the fat: lipid droplets and human disease. EMBO Mol Med. 2013;5:973–983. doi: 10.1002/emmm.201100671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunte AS, Matthews KA, Rawson RB. Fatty acid auxotrophy in Drosophila larvae lacking SREBP. Cell Metab. 2006;3:439–448. doi: 10.1016/j.cmet.2006.04.011. [DOI] [PubMed] [Google Scholar]
- Kühnlein RP. Thematic review series: Lipid droplet synthesis and metabolism: from yeast to man. Lipid droplet-based storage fat metabolism in Drosophila. J Lipid Res. 2012;53:1430–1436. doi: 10.1194/jlr.R024299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühnlein RP. The contribution of the Drosophila model to lipid droplet research. Prog Lipid Res. 2011;50:348–356. doi: 10.1016/j.plipres.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Lamming DW, Sabatini DM. A Central role for mTOR in lipid homeostasis. Cell Metab. 2013;18:465–469. doi: 10.1016/j.cmet.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavrynenko O, Rodenfels J, Carvalho M, Dye NA, Lafont R, Eaton S, Shevchenko A. The ecdysteroidome of Drosophila: influence of diet and development. Development. 2015;142:3758–3768. doi: 10.1242/dev.124982. [DOI] [PubMed] [Google Scholar]
- Layalle S, Arquier N, Leopold P. The TOR pathway couples nutrition and developmental timing in Drosophila. Dev. Cell. 2008;15:568–577. doi: 10.1016/j.devcel.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Lee G, Park JH. Hemolymph sugar homeostasis and starvation-induced hyperactivity affected by genetic manipulations of the adipokinetic hormone-encoding gene in Drosophila melanogaster. Genetics. 2004;167:311–323. doi: 10.1534/genetics.167.1.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Dong HH. FoxO integration of insulin signaling with glucose and lipid metabolism. J Endocrinol. 2017;233:R67–R79. doi: 10.1530/JOE-17-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Hoffmann J, Li Y, Stephano F, Bruchhaus I, Fink C, Roeder T. Octopamine controls starvation resistance, life span and metabolic traits in Drosophila. Sci Rep. 2016;6:35359. doi: 10.1038/srep35359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Sheng Z, Liu H, Wen D, He Q, Wang S, Shao W, Jiang RJ, An S, Sun Y, Bendena WG, Wang J, Gilbert LI, Wilson TG, Song Q, Li S. Juvenile hormone counteracts the bHLH-PAS transcription factors MET and GCE to prevent caspase-dependent programmed cell death in Drosophila. Development. 2009;136:2015–2025. doi: 10.1242/dev.033712. [DOI] [PubMed] [Google Scholar]
- Lum PY, Chino H. Primary role of adipokinetic hormone in the formation of low density lipophorin in locusts. J Lipid Res. 1990;31:2039–2044. [PubMed] [Google Scholar]
- Luong N, Davies CR, Wessells RJ, Graham SM, King MT, Veech R, Bodmer R, Oldham SM. Activated FOXO-mediated insulin resistance is blocked by reduction of TOR activity. Cell Metab. 2006;4:133–142. doi: 10.1016/j.cmet.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Manning BD, Toker A. AKT/PKB Signaling: Navigating the Network. Cell. 2017;169:381–405. doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz-Soja M, Rennert C, Schönefeld K, Aleithe S, Boettger J, Schmidt-Heck W, Weiss TS, Hovhannisyan A, Zellmer S, Klöting N, Schulz A, Kratzsch J, Guthke R, Gebhardt R. Hedgehog signaling is a potent regulator of liver lipid metabolism and reveals a GLI-code associated with steatosis. Elife. 2016;5:2062. doi: 10.7554/eLife.13308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metaxakis A, Tain LS, Gronke S, Hendrich O, Hinze Y, Birras U, Partridge L. Lowered insulin signalling ameliorates age-related sleep fragmentation in Drosophila. PLoS Biol. 2014;12:e1001824. doi: 10.1371/journal.pbio.1001824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirth CK, Tang HY, Makohon-Moore SC, Salhadar S, Gokhale RH, Warner RD, Koyama T, Riddiford LM, Shingleton AW. Juvenile hormone regulates body size and perturbs insulin signaling in Drosophila. Proc Natl Acad Sci USA. 2014;111:7018–7023. doi: 10.1073/pnas.1313058111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura S, Gan J-W, Brzostowski J, Parisi MJ, Schultz CJ, Londos C, Oliver B, Kimmel AR. Functional conservation for lipid storage droplet association among Perilipin, ADRP, and TIP47 (PAT)-related proteins in mammals, Drosophila, and Dictyostelium. J Biol Chem. 2002;277:32253–32257. doi: 10.1074/jbc.M204410200. [DOI] [PubMed] [Google Scholar]
- Nässel DR, Kubrak OI, Liu Y, Luo J, Lushchak OV. Factors that regulate insulin producing cells and their output in Drosophila. Front Physiol. 2013;4:252. doi: 10.3389/fphys.2013.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nässel DR, Winther AME. Drosophila neuropeptides in regulation of physiology and behavior. Prog Neurobiol. 2010;92:42–104. doi: 10.1016/j.pneurobio.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Ng M, Fleming T, Robinson M, Thomson B, Graetz N, Margono C, Mullany EC, Biryukov S, Abbafati C, Abera SF, Abraham JP, Abu-Rmeileh NME, Achoki T, AlBuhairan FS, Alemu ZA, Alfonso R, Ali MK, Ali R, Guzman NA, Ammar W, Anwari P, Banerjee A, Barquera S, Basu S, Bennett DA, Bhutta Z, Blore J, Cabral N, Nonato IC, Chang JC, Chowdhury R, Courville KJ, Criqui MH, Cundiff DK, Dabhadkar KC, Dandona L, Davis A, Dayama A, Dharmaratne SD, Ding EL, Durrani AM, Esteghamati A, Farzadfar F, Fay DFJ, Feigin VL, Flaxman A, Forouzanfar MH, Goto A, Green MA, Gupta R, Hafezi-Nejad N, Hankey GJ, Harewood HC, Havmoeller R, Hay S, Hernandez L, Husseini A, Idrisov BT, Ikeda N, Islami F, Jahangir E, Jassal SK, Jee SH, Jeffreys M, Jonas JB, Kabagambe EK, Khalifa SEAH, Kengne AP, Khader YS, Khang YH, Kim D, Kimokoti RW, Kinge JM, Kokubo Y, Kosen S, Kwan G, Lai T, Leinsalu M, Li Y, Liang X, Liu S, Logroscino G, Lotufo PA, Lu Y, Ma J, Mainoo NK, Mensah GA, Merriman TR, Mokdad AH, Moschandreas J, Naghavi M, Naheed A, Nand D, Narayan KMV, Nelson EL, Neuhouser ML, Nisar MI, Ohkubo T, Oti SO, Pedroza A, Prabhakaran D, Roy N, Sampson U, Seo H, Sepanlou SG, Shibuya K, Shiri R, Shiue I, Singh GM, Singh JA, Skirbekk V, Stapelberg NJC, Sturua L, Sykes BL, Tobias M, Tran BX, Trasande L, Toyoshima H, van de Vijver S, Vasankari TJ, Veerman JL, Velasquez-Melendez G, Vlassov VV, Vollset SE, Vos T, Wang C, Wang X, Weiderpass E, Werdecker A, Wright JL, Yang YC, Yatsuya H, Yoon J, Yoon SJ, Zhao Y, Zhou M, Zhu S, Lopez AD, Murray CJL, Gakidou E. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2014;384:766–781. doi: 10.1016/S0140-6736(14)60460-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham S, Hafen E. Insulin/IGF and target of rapamycin signaling: a TOR de force in growth control. Trends Cell Biol. 2003;13:79–85. doi: 10.1016/s0962-8924(02)00042-9. [DOI] [PubMed] [Google Scholar]
- Owusu-Ansah E, Perrimon N. Modeling metabolic homeostasis and nutrient sensing in Drosophila: implications for aging and metabolic diseases. Dis Model Mech. 2014;7:343–350. doi: 10.1242/dmm.012989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanker L, Tennessen JM, Lam G, Thummel CS. Drosophila HNF4 regulates lipid mobilization and beta-oxidation. Cell Metab. 2009;9:228–239. doi: 10.1016/j.cmet.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, Sampaio JL, Brankatschk M, Carvalho M, Mahmoud A, Shevchenko A, Eaton S. Lipoproteins in Drosophila melanogaster - Assembly, function, and influence on tissue lipid composition. PLoS Genet. 2012;8:e1002828. doi: 10.1371/journal.pgen.1002828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palu RAS, Thummel CS. Sir2 Acts through Hepatocyte Nuclear Factor 4 to maintain insulin Signaling and Metabolic Homeostasis in Drosophila. PLoS Genet. 2016;12:e1005978. doi: 10.1371/journal.pgen.1005978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra-Peralbo E, Culi J. Drosophila lipophorin receptors mediate the uptake of neutral lipids in oocytes and imaginal disc cells by an endocytosis-independent mechanism. PLoS Genet. 2011;7:e1001297. doi: 10.1371/journal.pgen.1001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrales A, Iwakuma T. p53 as a Regulator of Lipid Metabolism in Cancer. Int J Mol Sci. 2016;17:2074. doi: 10.3390/ijms17122074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel RT, Soulages JL, Hariharasundaram B, Arrese EL. Activation of the lipid droplet controls the rate of lipolysis of triglycerides in the insect fat body. J Biol Chem. 2005;280:22624–22631. doi: 10.1074/jbc.M413128200. [DOI] [PubMed] [Google Scholar]
- Peckett AJ, Wright DC, Riddell MC. The effects of glucocorticoids on adipose tissue lipid metabolism. Metab Clin Exp. 2011;60:1500–1510. doi: 10.1016/j.metabol.2011.06.012. [DOI] [PubMed] [Google Scholar]
- Peterfy M, Phan J, Xu P, Reue K. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat Genet. 2001;27:121–124. doi: 10.1038/83685. [DOI] [PubMed] [Google Scholar]
- Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, Sabatini DM. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–420. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péterfy M, Harris TE, Fujita N, Reue K. Insulin-stimulated interaction with 14-3-3 promotes cytoplasmic localization of lipin-1 in adipocytes. J Biol Chem. 2010;285:3857–3864. doi: 10.1074/jbc.M109.072488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, Griffiths JR, Chung Y-L, Schulze A. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8:224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospisilik JA, Schramek D, Schnidar H, Cronin SJF, Nehme NT, Zhang X, Knauf C, Cani PD, Aumayr K, Todoric J, Bayer M, Haschemi A, Puviindran V, Tar K, Orthofer M, Neely GG, Dietzl G, Manoukian A, Funovics M, Prager G, Wagner O, Ferrandon D, Aberger F, Hui CC, Esterbauer H, Penninger JM. Drosophila genome-wide obesity screen reveals hedgehog as a determinant of brown versus white adipose cell fate. Cell. 2010;140:148–160. doi: 10.1016/j.cell.2009.12.027. [DOI] [PubMed] [Google Scholar]
- Rajan A, Perrimon N. Drosophila cytokine unpaired 2 regulates physiological homeostasis by remotely controlling insulin secretion. Cell. 2012;151:123–137. doi: 10.1016/j.cell.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiher W, Shirras C, Kahnt J, Baumeister S, Isaac RE, Wegener C. Peptidomics and peptide hormone processing in the Drosophila midgut. J Proteome Res. 2011;10:1881–1892. doi: 10.1021/pr101116g. [DOI] [PubMed] [Google Scholar]
- Reis T, Van Gilst MR, Hariharan IK. A buoyancy-based screen of Drosophila larvae for fat-storage mutants reveals a role for Sir2 in coupling fat storage to nutrient availability. PLoS Genet. 2010;6:e1001206. doi: 10.1371/journal.pgen.1001206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reue K, Xu P, Wang XP, Slavin BG. Adipose tissue deficiency, glucose intolerance, and increased atherosclerosis result from mutation in the mouse fatty liver dystrophy (fld) gene. J Lipid Res. 2000;41:1067–1076. [PubMed] [Google Scholar]
- Riechmann V, Rehorn KP, Reuter R, Leptin M. The genetic control of the distinction between fat body and gonadal mesoderm in Drosophila. Development. 1998;125:713–723. doi: 10.1242/dev.125.4.713. [DOI] [PubMed] [Google Scholar]
- Rodenfels J, Lavrynenko O, Ayciriex S, Sampaio JL, Carvalho M, Shevchenko A, Eaton S. Production of systemically circulating Hedgehog by the intestine couples nutrition to growth and development. Genes Dev. 2014;28:2636–2651. doi: 10.1101/gad.249763.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Vázquez M, Vaquero D, Parra-Peralbo E, Mejía-Morales JE, Culi J. Drosophila Lipophorin Receptors Recruit the Lipoprotein LTP to the Plasma Membrane to Mediate Lipid Uptake. PLoS Genet. 2015;11:e1005356. doi: 10.1371/journal.pgen.1005356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder T. Tyramine and octopamine: ruling behavior and metabolism. Annu Rev Entomol. 2005;50:447–477. doi: 10.1146/annurev.ento.50.071803.130404. [DOI] [PubMed] [Google Scholar]
- Rulifson EJ, Kim SK, Nusse R. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science. 2002;296:1118–1120. doi: 10.1126/science.1070058. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Leung J, Grimsey N, Peak-Chew S, Siniossoglou S. The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 2005;24:1941. doi: 10.1038/sj.emboj.7600672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt S, Ugrankar R, Greene SE, Prajapati M, Lehmann M. Drosophila Lipin interacts with insulin and TOR signaling pathways in the control of growth and lipid metabolism. J Cell Sci. 2015;128:4395–4406. doi: 10.1242/jcs.173740. [DOI] [PubMed] [Google Scholar]
- Schwer B, Verdin E. Conserved metabolic regulatory functions of sirtuins. Cell Metab. 2008;7:104–112. doi: 10.1016/j.cmet.2007.11.006. [DOI] [PubMed] [Google Scholar]
- Seegmiller AC, Dobrosotskaya I, Goldstein JL, Ho YK, Brown MS, Rawson RB. The SREBP pathway in Drosophila: regulation by palmitate, not sterols. Dev. Cell. 2002;2:229–238. doi: 10.1016/s1534-5807(01)00119-8. [DOI] [PubMed] [Google Scholar]
- Seidell JC. Obesity, insulin resistance and diabetes - a worldwide epidemic. Br J Nutr. 2000;83(Suppl 1):8. doi: 10.1017/s000711450000088x. [DOI] [PubMed] [Google Scholar]
- Shao W, Espenshade PJ. Expanding roles for SREBP in metabolism. Cell Metab. 2012;16:414–419. doi: 10.1016/j.cmet.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimano H, Shimomura I, Hammer RE, Herz J, Goldstein JL, Brown MS, Horton JD. Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J Clin Invest. 1997;100:2115–2124. doi: 10.1172/JCI119746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber MH, Thummel CS. Coordination of triacylglycerol and cholesterol homeostasis by DHR96 and the Drosophila LipA homolog magro. Cell Metab. 2012;15:122–127. doi: 10.1016/j.cmet.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber MH, Thummel CS. The DHR96 nuclear receptor controls triacylglycerol homeostasis in Drosophila. Cell Metab. 2009;10:481–490. doi: 10.1016/j.cmet.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S. Phospholipid metabolism and nuclear function: roles of the lipin family of phosphatidic acid phosphatases. Biochim Biophys Acta. 2013;1831:575–581. doi: 10.1016/j.bbalip.2012.09.014. [DOI] [PubMed] [Google Scholar]
- Slaidina M, Delanoue R, Gronke S, Partridge L, Leopold P. A Drosophila insulin-like peptide promotes growth during nonfeeding states. Dev. Cell. 2009;17:874–884. doi: 10.1016/j.devcel.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Cheng D, Hong S, Sappe B, Hu Y, Wei N, Zhu C, O’Connor MB, Pissios P, Perrimon N. Midgut-Derived Activin Regulates Glucagon-like Action in the Fat Body and Glycemic Control. Cell Metab. 2017;25:386–399. doi: 10.1016/j.cmet.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Veenstra JA, Perrimon N. Control of lipid metabolism by tachykinin in Drosophila. Cell Rep. 2014;9:40–47. doi: 10.1016/j.celrep.2014.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden MC, Caton PW, Holness MJ. PPAR control: it’s SIRTainly as easy as PGC. J Endocrinol. 2010;204:93–104. doi: 10.1677/JOE-09-0359. [DOI] [PubMed] [Google Scholar]
- Suh JM, Gao X, McKay J, McKay R, Salo Z, Graff JM. Hedgehog signaling plays a conserved role in inhibiting fat formation. Cell Metab. 2006;3:25–34. doi: 10.1016/j.cmet.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Suh JM, Zeve D, McKay R, Seo J, Salo Z, Li R, Wang M, Graff JM. Adipose is a conserved dosage-sensitive antiobesity gene. Cell Metab. 2007;6:195–207. doi: 10.1016/j.cmet.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Feng D, Everett LJ, Bugge A, Lazar MA. Circadian epigenomic remodeling and hepatic lipogenesis: lessons from HDAC3. Cold Spring Harb Symp Quant Biol. 2011;76:49–55. doi: 10.1101/sqb.2011.76.011494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatar M, Kopelman A, Epstein D, Tu MP, Yin CM, Garofalo RS. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science. 2001;292:107–110. doi: 10.1126/science.1057987. [DOI] [PubMed] [Google Scholar]
- Teixeira L, Rabouille C, Rørth P, Ephrussi A, Vanzo NF. Drosophila Perilipin/ADRP homologue Lsd2 regulates lipid metabolism. Mech Dev. 2003;120:1071–1081. doi: 10.1016/s0925-4773(03)00158-8. [DOI] [PubMed] [Google Scholar]
- Teleman AA, Chen Y-W, Cohen SM. 4E-BP functions as a metabolic brake used under stress conditions but not during normal growth. Genes Dev. 2005;19:1844–1848. doi: 10.1101/gad.341505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teleman AA, Maitra S, Cohen SM. Drosophila lacking microRNA miR-278 are defective in energy homeostasis. Genes Dev. 2006;20:417–422. doi: 10.1101/gad.374406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JM, Thummel CS. Coordinating growth and maturation - insights from Drosophila. Curr Biol. 2011;21:R750–7. doi: 10.1016/j.cub.2011.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima J, Takaki K, Sakurai S, Bownes M. Nutritional status affects 20-hydroxyecdysone concentration and progression of oogenesis in Drosophila melanogaster. J Endocrinol. 2005;187:69–79. doi: 10.1677/joe.1.06220. [DOI] [PubMed] [Google Scholar]
- Tian Y, Bi J, Shui G, Liu Z, Xiang Y, Liu Y, Wenk MR, Yang H, Huang X. Tissue-autonomous function of Drosophila seipin in preventing ectopic lipid droplet formation. PLoS Genet. 2011;7:e1001364. doi: 10.1371/journal.pgen.1001364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufail M, Takeda M. Molecular characteristics of insect vitellogenins. J Insect Physiol. 2008;54:1447–1458. doi: 10.1016/j.jinsphys.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Ugrankar R, Liu Y, Provaznik J, Schmitt S, Lehmann M. Lipin is a central regulator of adipose tissue development and function in Drosophila melanogaster. Mol Cell Biol. 2011;31:1656. doi: 10.1128/MCB.01335-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese J, Lim SF, Cohen SM. Drosophila miR-14 regulates insulin production and metabolism through its target, sugarbabe. Genes Dev. 2010;24:2748–2753. doi: 10.1101/gad.1995910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergnes L, Chin RG, de Aguiar Vallim T, Fong LG, Osborne TF, Young SG, Reue K. SREBP-2-deficient and hypomorphic mice reveal roles for SREBP-2 in embryonic development and SREBP-1c expression. J Lipid Res. 2016;57:410–421. doi: 10.1194/jlr.M064022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Moya N, Niessen S, Hoover H, Mihaylova MM, Shaw RJ, Yates JR, Fischer WH, Thomas JB, Montminy M. A hormone-dependent module regulating energy balance. Cell. 2011;145:596–606. doi: 10.1016/j.cell.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Hayakawa Y, Downer RGH. Factors influencing cyclic AMP and diacylglycerol levels in fat body of Locusta migratoria. Insect Biochem. 1990;20:325–330. doi: 10.1016/0020-1790(90)90051-u. [DOI] [Google Scholar]
- Welte MA. Expanding roles for lipid droplets. Curr Biol. 2015;25:R470–81. doi: 10.1016/j.cub.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfling F, Wang H, Haas JT, Krahmer N, Gould TJ, Uchida A, Cheng JX, Graham M, Christiano R, Fröhlich F, Liu X, Buhman KK, Coleman RA, Bewersdorf J, Farese RV, Walther TC. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev Cell. 2013;24:384–399. doi: 10.1016/j.devcel.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13:790–795. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- Yamamoto R, Bai H, Dolezal AG, Amdam G, Tatar M. Juvenile hormone regulation of Drosophila aging. BMC Biol. 2013;11:85. doi: 10.1186/1741-7007-11-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka N, Rewitz KF, O’Connor MB. Ecdysone control of developmental transitions: lessons from Drosophila research. Annu Rev Entomol. 2013;58:497–516. doi: 10.1146/annurev-ento-120811-153608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Wang H, Chen H, Lindström-Battle A, Jiao R. Ecdysone and insulin signaling play essential roles in readjusting the altered body size caused by the dGPAT4 mutation in Drosophila. J. Genet. Genomics. 2015;42:487–494. doi: 10.1016/j.jgg.2015.06.008. [DOI] [PubMed] [Google Scholar]
- Yang Z, Yu Y, Zhang V, Tian Y, Qi W, Wang L. Octopamine mediates starvation-induced hyperactivity in adult Drosophila. Proc Natl Acad Sci USA. 2015;112:5219–5224. doi: 10.1073/pnas.1417838112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zera AJ, Zhao Z. Effect of a juvenile hormone analogue on lipid metabolism in a wing-polymorphic cricket: implications for the endocrine-biochemical bases of life-history tradeoffs. Physiol Biochem Zool. 2004;77:255–266. doi: 10.1086/383500. [DOI] [PubMed] [Google Scholar]
- Zhao C, Dahlman-Wright K. Liver X receptor in cholesterol metabolism. J Endocrinol. 2010;204:233–240. doi: 10.1677/JOE-09-0271. [DOI] [PubMed] [Google Scholar]
- Ziegler R, Van Antwerpen R. Lipid uptake by insect oocytes. Insect Biochem Mol Biol. 2006;36:264–272. doi: 10.1016/j.ibmb.2006.01.014. [DOI] [PubMed] [Google Scholar]
- Zou Z, Saha TT, Roy S, Shin SW, Backman TWH, Girke T, White KP, Raikhel AS. Juvenile hormone and its receptor, methoprene-tolerant, control the dynamics of mosquito gene expression. Proc Natl Acad Sci USA. 2013;110:E2173–81. doi: 10.1073/pnas.1305293110. [DOI] [PMC free article] [PubMed] [Google Scholar]