Abstract
In this issue of Blood, Kataoka et al1 leverage their landscape of adult T-cell leukemia/lymphoma (ATL) to define associations between outcome and specific genetic alterations.2 They identify gene alterations in both high-grade and low-grade disease that correlate with outcome. Future studies are needed to refine and validate this clinicogenetic classification of ATL. Once validated, prospective studies should assess whether patients with poor-risk disease benefit from alternative strategies and whether those with low-risk disease can achieve long-term survival with less intensive treatment.
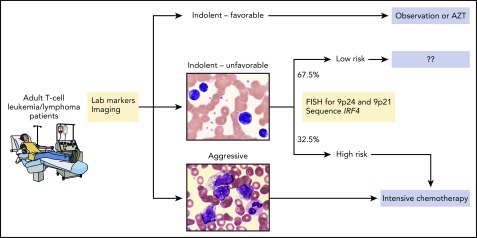
Shown is one possible approach to treating patients that is based on known clinical and laboratory factors as well as genetic alterations identified by Kataoka et al. Specifically, patients with chronic type, unfavorable disease who harbor 1 or more of the 3 genetic alterations would receive intensive therapy for aggressive disease whereas those who do not harbor any of those alterations may not require intensive therapy. Future analyses and prospective trials are needed to address whether this type of approach can refine patient selection and maximize therapeutic benefit. AZT, zidovudine; FISH, fluorescent in situ hybridization. The microscopic images were kindly provided by Peter Maslak, Memorial Sloan Kettering Cancer Center. Professional illustration by Patrick Lane, ScEYEnce Studios.
Human T-cell leukemia virus type-1 infection is endemic within parts of Japan, the Caribbean, Latin America, Africa, Iran, and Romania. Among infected persons, the lifetime risk of developing ATL is approximately 5%. Laboratory and imaging findings can classify patients with ATL into 4 subtypes: acute, lymphoma, chronic, and smoldering.3 The chronic subtype can be further divided into favorable and unfavorable groups on the basis of the presence of high serum lactate dehydrogenase, high blood urea nitrogen, and low albumin levels.4
The acute, lymphoma, and unfavorable chronic subtypes are considered aggressive disease, whereas smoldering and favorable chronic subtypes are considered indolent.3 This distinction is pivotal for selecting therapy. Patients with aggressive disease typically receive highly intensive chemotherapy with or without allogeneic stem cell transplantation. The anti-CCR4 antibody mogamulizumab is also widely used in countries where it is available. Overall outcomes, even with this intensive approach, remain poor. In stark contrast, watchful waiting or combined interferon-α and zidovudine therapy is standard treatment for indolent disease.4
Kataoka and colleagues recently published a landmark description of ATL genetics.2 Like other T-cell malignancies, the ATL exomes are dominated by recurrent alterations in factors involved in T-cell receptor and cytokine signaling, immune escape, and transcriptional regulation.2,5-7 In the Kataoka study in this issue, the authors used Cox proportional hazards regression modeling to identify statistically robust associations between genetic alterations and survival in patients with ATL. There were several notable findings. First, almost all single-nucleotide and insertion/deletion mutations recurrently identified in ATL are subclonal. Thus, even though genetic alterations can be targeted with available agents (eg, mutations in CSNK1A1 with immunomodulatory imide drugs8), those agents may only be active against subclones.
The second notable finding comes as no surprise: mutations in aggressive disease differ from those in indolent disease. In particular, mutations of IRF4 (also known as MUM1), which is a central transcription factor in both B- and T-cell maturation, were enriched nearly fivefold in high-risk disease. These mutations are believed to be gain-of-function and were essentially all subclonal; therefore, they may indicate a dependence on IRF4 that has yet to be exploited therapeutically. Strategies for targeting IRF4 with proteolysis-targeting chimeras or other novel approaches are therefore a high priority in this disease.
Finally, Kataoka et al show that gene mutations can add prognostic value to the traditional classification of ATL. They identify age ≥70 years, mutations of PRKCB, and amplifications of chr.9p24 (including CD274, which encodes PD-L1) as independent risk factors for poor outcome among patients with aggressive disease. These factors were particularly impactful in patients with moderate-risk disease (according to the Japan Clinical Oncology Group prognostic index) because none of the 12 with 2 or more risk factors had long-term survival.
Among patients with more indolent disease, those with IRF4 mutations, amplifications of chr.9p24, or deletions of chr.9p21 had a worse prognosis (median overall survival, <2 years) compared with those with none of these abnormalities (median overall survival, not reached). From a treatment standpoint, this genetic information may be most important for patients with unfavorable chronic subtype; among the two-thirds of these patients who lacked any of the genetic risk factors, median overall survival was approximately 5 years compared with approximately 1 year for those with 1 or more genetic risk factors.
These findings suggest but do not prove that genetic testing can help guide treatment selection among patients with ATL (see figure). However, several outstanding issues must first be addressed. The analysis by Kataoka et al must be validated using an independent cohort, preferably from a region outside Japan. Additional alterations known to occur in ATL but not analyzed by Kataoka et al should also be considered. These include structural variations within the 3′ untranslated region of CD274 that significantly increase transcript levels, rearrangements involving CD28, and high levels of soluble interleukin-2 receptor.9 From a statistical standpoint, it may or may not be preferable to create a risk score that sums predictor values weighted by Lasso coefficients for each clinical or genetic factor10 instead of using the approach of Kataoka et al.
Once all of that is done, it remains to be seen whether outcomes for patients with poor-risk disease can be improved with new therapeutic strategies and whether de-intensification of therapy for patients with good-risk disease (especially those with the unfavorable chronic subtype) will compromise their good outcomes. Inhibitors of both PD-1/PD-L1 and CTLA4 are likely to have activity in patients with ATL who harbor specific genetic lesions, but the appropriate timing for these agents may also depend on their risk with current therapies. In other words, we now seem to have multiple agents that can target vulnerabilities in ATL. If we do the right trials and the right analyses of patients on those trials over the next decade, patients with ATL are likely to benefit forever after.
Footnotes
Conflict-of-interest disclosure: The authors declare no competing financial interests.
REFERENCES
- 1.Kataoka K, Iwanaga M, Yasunaga J-i, et al. . Prognostic relevance of integrated genetic profiling in adult T-cell leukemia/lymphoma. Blood. 2018;131(2):215-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kataoka K, Nagata Y, Kitanaka A, et al. . Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat Genet. 2015;47(11):1304-1315. [DOI] [PubMed] [Google Scholar]
- 3.Shimoyama M. Diagnostic criteria and classification of clinical subtypes of adult T-cell leukaemia-lymphoma. A report from the Lymphoma Study Group (1984-87). Br J Haematol. 1991;79(3):428-437. [DOI] [PubMed] [Google Scholar]
- 4.Ishitsuka K, Tamura K. Human T-cell leukaemia virus type I and adult T-cell leukaemia-lymphoma. Lancet Oncol. 2014;15(11):e517-e526. [DOI] [PubMed] [Google Scholar]
- 5.Vallois D, Dobay MP, Morin RD, et al. . Activating mutations in genes related to TCR signaling in angioimmunoblastic and other follicular helper T-cell-derived lymphomas. Blood. 2016;128(11):1490-1502. [DOI] [PubMed] [Google Scholar]
- 6.Schatz JH, Horwitz SM, Teruya-Feldstein J, et al. . Targeted mutational profiling of peripheral T-cell lymphoma not otherwise specified highlights new mechanisms in a heterogeneous pathogenesis. Leukemia. 2015;29(1):237-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiel MJ, Velusamy T, Rolland D, et al. . Integrated genomic sequencing reveals mutational landscape of T-cell prolymphocytic leukemia. Blood. 2014;124(9):1460-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krönke J, Fink EC, Hollenbach PW, et al. . Lenalidomide induces ubiquitination and degradation of CK1α in del(5q) MDS. Nature. 2015;523(7559):183-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katsuya H, Shimokawa M, Ishitsuka K, et al. . Prognostic index for chronic- and smoldering-type adult T-cell leukemia-lymphoma. Blood. 2017;130(1):39-47. [DOI] [PubMed] [Google Scholar]
- 10.Jurinovic V, Kridel R, Staiger AM, et al. . Clinicogenetic risk models predict early progression of follicular lymphoma after first-line immunochemotherapy. Blood. 2016;128(8):1112-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]