Abstract
The papillomavirus (PV) helicase protein E1 recruits components of the cellular DNA replication machinery to the PV replication fork, such as Replication Protein A (RPA), DNA polymerase α-primase (pol α) and topoisomerase I (topo I). Here we show that E1 binds to DNA polymerase ϵ (pol ϵ) and dramatically stimulates the DNA synthesis activity of pol ϵ. This stimulation of pol ϵ by E1 is highly specific and occurs even in the absence of the known pol ϵ cofactors Replication Factor C (RFC), Proliferating Cell Nuclear Antigen (PCNA) and RPA. This stimulation is due to an increase in the processivity of pol ϵ and occurs independently of pol ϵ’s replication cofactors. This increase in processivity is dependent on the ability of the E1 helicase to hydrolyze ATP, suggesting it is dependent on E1’s helicase action. In addition, RPA, thought to be vital for processive DNA synthesis by both pol ϵ and pol δ, was found to be dispensable for processive synthesis by pol ϵ in the presence of E1. Overall, E1 appears to be conferring processivity to pol ϵ by directly tethering pol ϵ to the DNA parental strand and towing ϵ behind the E1 helicase as the replication fork progresses; and thereby apparently obviating the need for RPA for leading strand synthesis. Thus far only pol α and pol δ have been implicated in the DNA replication of mammalian viruses; this is the first reported example of a virus recruiting pol ϵ. Furthermore, this demonstrates a unique capacity of a viral helicase having evolved to stimulate a cellular replicative DNA polymerase.
INTRODUCTION
Viruses that do not express their own polynucleotide synthetases have evolved to recruit cellular enzymes to carry out viral genome synthesis. While many DNA viruses encode their own viral DNA polymerases, many of the small DNA virus families do not, but recruit one or more of the cellular replicative DNA polymerases for viral genome replication. The three eukaryotic replicative DNA polymerases are: DNA polymerase α-primase (pol α, a non-processive DNA polymerase linked to a RNA primase enzyme that is a priming enzyme rather than a DNA polymerase that synthesizes much of the genome), DNA polymerase ϵ (pol ϵ, the putative leading strand DNA polymerase), and DNA polymerase δ (pol δ, the primary polymerase involved in lagging strand synthesis; although polymerase δ has also been shown to function as the leading strand polymerase if pol ϵ is dislodged from the leading strand, and appears to play the key synthetic role in many DNA repair pathways) (1–8). It is noteworthy that while the genes encoding all three of these DNA polymerases are essential, yeast can survive with a catalytically dead pol ϵ, possibly reflecting the ability of pol δ to compensate in pol ϵ’s absence (5). With the importance and flexibility of pol δ, it is perhaps not surprising that many small DNA tumor viruses have evolved to utilize this enzyme to replicate their viral genomes. The simplest case is the parvoviruses, which have been shown to use pol δ and its cofactors to initiate synthesis from the specific ssDNA break induced by the viral helicase/nuclease and to synthesize the entire parvovirus genome (9); and indeed, pol ϵ was shown to not be required for the DNA replication of this virus (9,10). Other small dsDNA mammalian viruses that do not encode their own DNA polymerases, specifically the polyomaviruses (PyVs) and papillomaviruses (PVs), have been shown to require pol α, to prime both leading strand synthesis and the lagging strand Okazaki fragments (11–14). Pol δ has also been shown to be the major synthetic DNA polymerase for PyV DNA replication (as shown in the major PyV model system, simian virus 40 (SV40)), replicating both leading and lagging strands (11–15). Conversely pol ϵ was not required for the reconstitution of SV40 DNA replication and when added had no effect on SV40 DNA replication (11,13,14,16). Additionally, introduction of pol ϵ-neutralizing antibodies into human cells containing SV40 replicons inhibited cellular, but not SV40 DNA replication, and unlike pol δ, pol ϵ was not cross linked to newly synthesized nascent SV40 DNA strands (17,18). Like PyVs, PVs have been shown to require both pol α and pol δ to carry out their viral DNA replication in vitro (19,20). However, unlike the case with SV40, extensive PV DNA synthesis could not be reconstituted with pol α, pol δ, and the other proteins required for the synthesis aspects of viral DNA replication (the viral DNA helicase, RFC, PCNA, RPA, and topoisomerase) (20). Hence, the question was still extant as to whether pol ϵ might be playing an important role in PV DNA replication.
A physical and biochemical link between DNA replicative helicases and their associated DNA polymerases are known to exist for several DNA replication systems, including bacterial, bacteriophage, viral and human (21–28). Recently such a link has been shown between the cellular replicative helicase, the Cdc45-MCM-GINS (CMG) complex and the putative cellular leading strand DNA polymerase, pol ϵ (21). This interaction had substantial effects on each complex's biochemical functions required for DNA replication; pol ϵ stimulated the DNA helicase function of CMG, and conversely, CMG stimulated the DNA polymerase function of pol ϵ (21). There have been cases of viral DNA helicases evolving to interact with cellular DNA polymerases not only physically, but also in a productive enzymatic manner, likely important for the overall replication of the viral DNA genomes (22–26,28,29). With this background in perspective, the experiments described in this study were designed to address whether the putative cellular leading strand DNA polymerase, pol ϵ, is recruited by the PV replicative DNA helicase, E1, to participate in PV DNA replication, and moreover, whether that interaction has substantive effects on either enzyme's known biochemical functions involved in the DNA replication process.
Here we demonstrate novel physical and functional interactions between pol ϵ and the PV E1 helicase. The DNA synthesis activity of pol ϵ is stimulated in the presence of E1, independently of pol ϵ’s DNA synthesis cofactors RFC, PCNA and RPA. In addition, we show that PV E1 confers processivity to pol ϵ. However, PCNA, RFC and RPA, which in combination are capable of stimulating the processivity of pol ϵ, are not required for E1’s stimulation of pol ϵ’s processivity. Inhibition of E1’s ATPase function was used to show that stimulation of pol ϵ’s processivity appears to be dependent upon E1’s hydrolysis of ATP, and presumably its DNA helicase function. Collectively, the results presented herein suggest a mechanism in which the E1 helicase tethers pol ϵ to the DNA through a direct interaction and tows it along the DNA leading strand template as the helicase drives the DNA replication fork.
MATERIALS AND METHODS
Purification of recombinant proteins
Pols δ and ϵ were expressed in the baculovirus expression system and purified as described (30).
HPV 11 EE-E1 was expressed in High Five insect cells using baculovirus expression. Briefly, ten T150 flasks were seeded with 2 × 107 High Five insect cells per flask and left to settle for 1 hour. Cells were infected at a multiplicity of infection of 3; virus stock was diluted into 10 ml of High Five Express medium per flask and incubated for 1 h at 27°C. Fifteen ml of additional media was then added to each flask and the incubation continued for 48 h at 27°C. Cells were combined and harvested by centrifugation at 1700 × g for 20 min at 4°C. The cells were washed with cold PBS and again subjected to centrifugation at 1700 × g. Twenty ml of ice cold lysis buffer (20 mM Hepes–NaOH (pH 7.5), 400 mM NaCl, 0.5 mM EDTA, 1 mM MgCl2, 1 mM DTT, 1 mM PMSF) was added to the cell pellet. The cells were resuspended and incubated on ice for 10 min, and then subjected to Dounce homogenization by 20 strokes with the loose pestle. Lysates were subjected to centrifugation (31 000 × g) for 10 minutes at 4°C. The supernatant was diluted with Buffer Q (20 mM Tris, (pH 7.0) and 10 mM beta mercaptoethanol) until the ionic strength was equivalent to Buffer Q with 50 mM NaCl. The diluted supernatant was applied to a Q Sepharose column (10 ml, 1.5 cm × 10cm). The column was washed with 50 ml Buffer Q containing 50 mM NaCl. Proteins were eluted with 10 ml Buffer Q containing 500 mM NaCl. Fractions were evaluated using the Bradford protein assay and the peak fractions were pooled and applied to a 1 ml affinity column (anti-EE monoclonal antibody conjugated to Protein A Sepharose 4B; (31)). The column was washed extensively with 20 mM Tris (pH 7.0) with 0.5 M NaCl. The column was eluted with 5 ml of 50 mM triethylamine (pH 11.5). Fractions of 0.5 ml were collected into tubes containing 50 μl Tris (pH 7.0). Fractions were analyzed using SDS-PAGE and fractions containing EE-E1 were pooled. Fractions were concentrated using Millipore Centricon™ (10 kDa MWCO) filters as per the manufacturers instructions.
MBP-E1 was purified from Escherichia coli using the pMal-boHPV11E1 expression vector, which encodes for the HPV11 E1 protein optimized for expression in E. coli (GenScript Inc.) fused to the E. coli maltose-binding protein (MBP) at the N-terminus. The coding sequence construct, provided by GenScript in pUC19, was directly sub-cloned out of the GenScript vector via PCR and into the pMalC2 vector's BamHI and EcoRI restriction sites (New England Biolabs, Inc.) as per the In-Fusion HD cloning kit (Takara Bio USA) instructions. For protein expression, transformed BL21 (DE3) E. coli cultures were grown to A595 = 0.4 at 37°C. The temperature was reduced to 18°C, further grown to A595 = 0.6 and isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to 0.4 mM to induce expression. Cells were grown for 2 h at 18°C. Cells were collected by centrifugation (5000 × g for 20 min at 4°C), washed with ice cold PBS and again collected by centrifugation (5000 × g for 20 min at 4°C). Cells were frozen using liquid nitrogen and stored at –80°C. Before use, cells were thawed on ice and resuspended in 25 ml MBP column buffer (20 mM Tris (pH 7.5), 200 mM NaCl, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, 5% glycerol, 0.01% NP40). The cell suspension was subjected to 1500 psi for 5 min in a cold Parr™ cell disruption chamber, and the suspension released slowly to lyse. Lysates were then sonicated for five 20 s pulses on ice, and subjected to centrifugation for 20 min at 20 000 × g at 4°C. Supernatants were collected, diluted 1:6 in MBP column buffer and applied to 1 ml of pre-equilibrated amylose resin (New England Biolabs, Inc.). Resin was washed with 20 ml MBP column buffer containing 1 M NaCl, and further washed with 20 ml MBP column buffer containing 0.2 M NaCl. MBP-E1 was eluted using 2 ml MBP column buffer containing 10 mM maltose. Fractions were evaluated using SDS-PAGE and MBP-E1 peak fractions were pooled.
GST-E1 was purified from E. coli using the pGex6p (pGex-boHPV11E1) expression plasmid. The bacterially optimized HPV 11 E1 ORF described above was subcloned out of vector pUC19 and into pGex6p using the In-Fusion HD cloning kit (Takara Bio USA) into restriction sites BamHI and EcoRI. Briefly, transformed BL21 (DE3) E. coli was grown to A595 = 0.4 at 37°C. The temperature was reduced to 18°C, further grown to A595 = 0.6 and isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to 0.3 mM to induce expression. Cells were grown for 12 h at 18°C. Cells were collected by centrifugation (5000 × g for 20 min at 4°C), washed with cold PBS and again collected by centrifugation (5000 × g for 20 min at 4°C). Cells were resuspended on ice in 25 ml Buffer A (50 mM tris (pH 7.5), 100 mM NaCl, 10 mM EDTA, 2 mM DTT, 20% sucrose and 1 mM PMSF). The cell suspension was subjected to 1500 psi for 5 min in a cold Parr™ cell disruption chamber, and the suspension released slowly to lyse. Lysates were then sonicated for five 20-s pulses on ice. NP40 was added to 0.1% final concentration and the lysates were subjected to centrifugation for 20 min at 20 000 × g at 4°C. 1 ml of pre-equilibrated glutathione Sepharose (GE Healthcare Life Sciences) was added to the lysates and incubated for 3 h at 4°C. The resin was washed twice with 40 ml of Buffer A and poured into column format. The resin was washed with 5 ml Buffer A containing 0.1 mM PMSF, 5 ml of Buffer B (50 mM tris (pH 8.8), 1 mM EDTA, 2 mM DTT, 10% glycerol) with 1 M NaCl and 5 ml of Buffer B with 0.2 M NaCl. Protein was eluted with 6 ml of Buffer B containing 0.2 M NaCl and 10 mM glutathione. Fractions were evaluated using SDS-PAGE and fractions containing GST-E1 were pooled.
Recombinant bacterially expressed human RPA was expressed and purified as described (32). Bacterially expressed recombinant human PCNA was prepared as described (33). Recombinant human RFC was prepared using baculovirus expression system as described (34).
Binding studies
Enzyme-linked immunoassays (ELISAs) to evaluate protein–protein interactions were performed in polyvinyl 96-well plates essentially as previously described (35). Briefly, wells were coated for 1 hour at room temperature by immobilizing 100 ng of pol δ, pol ϵ or BSA in TBS. The wells were washed with TBS-T, blocked for 1 h at room temperature (with 5% nonfat milk and 2% calf serum in TBS-T) and washed again with TBS-T. Increasing concentrations (as indicated) of the second protein (MBP-E1 or MBP) were added to the wells and incubated for 1 h at room temperature with gentle rocking. The wells were washed and the primary anti-MBP polyclonal antibody raised in rabbit (36) was diluted 1:2000 in TBS-T and added to the wells for 1 h at room temperature. An anti-rabbit HRP-conjugated secondary antibody (Thermo Scientific, 31466), was diluted 1:4000 was applied to the wells for 1 h at room temperature. The wells were washed extensively with TBS-T and incubated with 0.05 ml of substrate solution (110 mM sodium acetate (pH 5.5)), containing the chromogenic substrate 3.3′,5,5′-tetramethylbenzidine (0.02 mg/ml) and hydrogen peroxide (0.0075% [vol/vol]). After 5 min, the reaction was stopped with the addition of 0.05 ml of 2 M sulfuric acid. These assays were quantified spectrophotometrically by absorbance at 450 nm. This assay was performed at least three times, each time in duplicate.
Far western blot analysis was performed by subjecting 10 μg of single-step affinity purified pol δ or pol ϵ to electrophoresis on a 4–12% SDS-PAGE tris-glycine gel (Thermofisher Scientific). (Note that the single-step affinity purified polymerases are highly enriched, but unlike the purified polymerases used in the biochemical studies (30), still contain a substantial number of contaminating insect cell protein bands.) The gel was transferred onto a nitrocellulose membrane using the Thermofisher Scientific iBlot 2 transfer device (Thermofisher Scientific). Denatured proteins on the membrane were renatured via subsequent incubations with decreasing concentrations of guanidine–HCl from 6 to 0 M (37). The membrane was blocked for 1 h at room temperature with 5% nonfat milk in TBS-T (20 mM Tris–HCl, 150 mM NaCl, 0.1% Triton X-100). The membrane was then incubated with 10 μg/ml of GST-E1 for 3 h at room temperature. GST was used to probe a second, identical membrane as a control. The membranes were washed extensively with TBS-T and then probed with an anti-GST primary antibody (Thermofisher Scientific), 1 μg/ml, diluted in 3% non-fat milk/TBS-T overnight at 4°C. The membrane was washed three times with TBS-T for 10 min each and probed with the horseradish peroxidase (HRP)-conjugated goat anti-mouse secondary antibody (Thermo Scientific) diluted 1:4000 in TBS-T for 1 h at room temperature. The membrane was washed and developed using chemiluminescent substrate (Thermo Scientific SuperSignal West) and imaged with the BioRad Chemidoc. Additionally, 1 μg of pol ϵ or pol δ was subjected to electrophoresis on a 4–12% SDS-PAGE tris-glycine gel (as described above) and stained with Coomassie brilliant blue. The gel was destained using a 15% acetic acid/15% methanol solution and imaged using a BioRad imager.
Primer extension assays
Primed M13 DNA substrate was prepared by annealing a 17 nucleotide oligonucleotide (5′-GTAAAACGACGGCCAGT-3′, 1 pmol) to 4.5 μg circular ssM13 in TE (10 mM Tris–HCl (pH 7.4), 1 mM EDTA) buffer with 100 mM NaCl. The 17 nucleotide oligonucleotide was combined with ssM13 and heated for 5 min at 100°C. The reaction was slow cooled by moving the heat block to 37°C incubation overnight. Each reaction was performed in a 10 μl reaction volume using 100 ng of primed M13 (except as indicated for processivity assays), 0.1 mg/ml acetylated BSA, DNA replication buffer (40 mM sodium creatine phosphate, 20 mM Tris (pH 7.5), 7 mM MgCl2, 4 mM ATP, 200 μM each of CTP, UTP and GTP, 25 μM dATP, 100 μM each of dATP, dCTP, dGTP and dTTP and 0.5 mM DTT) and 32P-dATP (∼3000 cpm/pmol, ∼1 μCi per reaction). RPA, RFC, PCNA, pol ϵ, pol δ and HPV EE-E1 were each individually titrated for optimal DNA synthesis activity. Replication Factors (‘RF’) were used at the following concentrations: RFC (3 ng/μl), PCNA (10 ng/μl), RPA (70 ng/μl). Pol ϵ was used at 1 ng/μl unless otherwise indicated. HPV E1 was used at 60 ng/μl unless indicated otherwise. Reactions were incubated at 37°C for 1.5 h unless indicated otherwise. Reactions were terminated with M13 stop buffer (2% SDS [wt/vol], 50 mM EDTA, 5% glycerol [vol/vol] and 0.2 mg/ml proteinase K for 15 min at 37°C). Reference size markers were created by end-labeling the GeneRuler 1kb ladder (Thermofisher Scientific) with γ-32P-ATP (3000 cpm/pmol) using T4 polynucleotide kinase as per instructions (Thermo Scientific). Free nucleotide label was removed using G50 gel filtration with TE buffer. DNA products were then denatured (by addition of an equal volume of: 0.5 M NaOH, 5 mM EDTA, 5% Ficoll [w/v]) and subjected to electrophoretic separation using a 25 cm × 20 cm 1% agarose gel in 50 mM NaOH and 1 mM Na2EDTA at 40 V for 16 h.
Gels were fixed using 20% methanol [vol/vol] and 15% acetic acid [vol/vol], dried and exposed to a phosphorimager screen. Screens were analyzed using a Typhoon phosphorimager. Synthesis of DNA in the lanes of the agarose gels were quantified using Bio Rad Quantity One software. Experiments were performed at least three times. Three different preparations of pol ϵ were used in these assays; overall levels of stimulation were comparable with all three preparations, although the comparative levels of synthesis in the consistent smear like pattern versus the pattern showing ‘hard stops’ varied with the different preparations. Each experiment was repeated at least three times per preparation. All replicates of each experiment were quantified and global background (a ‘lane’ where no sample was applied) was subtracted from the experimental values. Due to isotope variation, quantifications across multiple experiments were plotted as percent synthesis, with pol ϵ in the presence of replication factors set to 100% (unless otherwise indicated). Statistical analyses were performed using student's t-tests using GraphPad software. P values are indicated in the figure legends.
ATPase assays
ATPase assays were assembled as 10 μl reactions with a reaction buffer containing 30mM Tris–HCl (pH 7.8), 7mM MgCl2, 0.1mM DTT, 50 μg/ml BSA and 1 μCi γ-32P-ATP (3000 cpm/pmol). 75 ng/μl of EE-E1 was preincubated with or without the nonhydrolyzable ATP analogs for 15 min at 37°C (either Adenosine 5′-(3-thiotriphosphate) tetralithium salt (ATPγS), Sigma) or Adenosine 5-(β,γ-imido) triphosphate lithium salt hydrate (AMP-PNP, Sigma), ranging in concentration from 0.1 mM to 20 mM. Reactions were then incubated for 45 minutes at 37°C, and stopped by adding 0.2 μl of 0.5 M EDTA (pH 8.0) to each reaction. One μl of each reaction was spotted on a PEI-cellulose plate (Sigma). The plate was developed in 0.5 M lithium chloride/1 M formic acid until the liquid phase migrated ∼75% of the way up the plate. Plates were dried and exposed to phosphorimaging screens and imaged with a Typhoon phosphoimager. ATPase activity was quantified using Bio Rad Quanitity One software. Experiments were performed three times. All replicates were quantified and background was subtracted from experimental values. E1 without inhibitor was set to 100% and samples with increasing concentrations of inhibitor were plotted relative to E1 without inhibitor.
RESULTS
HPV E1 physically interacts with pol ϵ
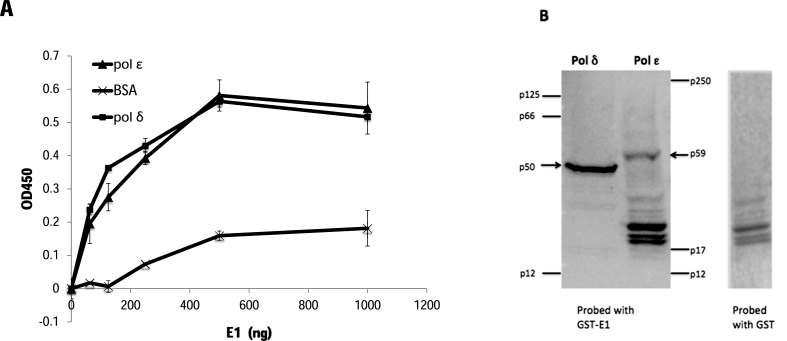
The PV E1 helicase interacts with several host DNA replication proteins, including pol α, RPA and topo I (36,38–42), and some of those interactions have been shown to modulate functions of these proteins critical for DNA replication (41,42). To determine whether the HPV E1 protein physically interacts with pol ϵ or pol δ enzyme-linked immuno-sorbent assay (ELISA)-based protein interaction analyses were performed as described previously (42). Purified pol ϵ or pol δ was immobilized in ELISA plate wells, washed, and after blocking MBP-E1 (HPV type 11) was added in increasing amounts. When MBP-E1 was added, binding to both pol ϵ and pol δ, substantially above background (BSA), was clearly detected (Figure 1A). The MBP tag did not interact with either DNA polymerase.
Figure 1.
HPV E1 physically interacts with DNA polymerases ϵ and δ. (A) Enzyme-linked Immunoassays (ELISAs) were performed using 100 ng of pol ϵ, pol δ or BSA as the immobilized protein. After washing, increasing amounts of MBP-E1 (as indicated) were added to the wells. Binding was detected using a polyclonal anti-MBP antibody followed by a secondary HRP-conjugated antibody. After washing, and addition of HRP substrate, absorbance was measured at 450 nm. Error bars are defined as Standard Deviation (s.d.); n = 6. (B) Far western blot analysis was performed by subjecting 10 μg of single-step affinity purified pol ϵ or pol δ to electrophoresis, transferring and then probing the membrane with 10 μg/ml of either GST-E1 (first panel) or GST (second panel) (100 μg total), according to the Materials and Methods section. A monoclonal anti-GST antibody was used as the primary antibody, and detected using HRP-linked secondary antibody as described in the Materials and Methods.
Pol ϵ is a large, multi-subunit protein that is conserved throughout all eukaryotes and has been implicated in a variety of important cellular processes (2). The p59 subunit of pol ϵ has been shown to physically interact with pol ϵ’s known processivity factor, PCNA (2,6,43). Partially purified, highly enriched preparations of pol δ and pol ϵ complexes were subjected to far western analysis probed with HPV E1. The results indicates that E1 interacts directly with the p59 subunit of pol ϵ (Figure 1B; note that all bands in the pol ϵ lane other than p59 also appear in the pol ϵ lane probed with GST alone, indicating they are not E1-specific. These bands represent contaminating insect cell proteins still present in the single-step affinity-purified pol ϵ complex. It is also noteworthy that none of the other pol ϵ subunits are bound by either E1 or GST.) E1 also interacts with the p50 subunit of pol δ, which has previously been shown to be required for HPV DNA replication (19,20).
HPV E1 stimulates DNA synthesis by pol ϵ
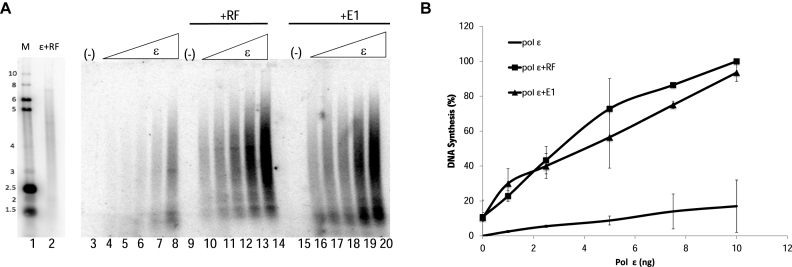
To determine whether the physical interaction between E1 and pol ϵ modulates pol ϵ activity, DNA synthesis by pol ϵ was analyzed on a primed circular ssM13 template in the presence and absence of E1, as well as known polymerase cofactors, PCNA, RFC and RPA (RF; Figure 2A). Increasing levels of pol ϵ alone produced a basal level of DNA synthesis (lanes 4–8); addition of RF to pol ϵ resulted in stimulation of synthesis by pol ϵ (lanes 10–14), consistent with those previously reported in the literature. When E1 was added to pol ϵ, E1 was observed to stimulate DNA synthesis by pol ϵ (lanes 16–20); this stimulation occurred even in the absence of pol ϵ’s RF. Quantification of this experiment showed an approximate six- to fifteen-fold stimulation of pol ϵ (varying depending on the levels of pol ϵ); and at all levels of pol ϵ showed a similar degree of stimulation by either RF or E1 (Figure 2B). The variation in degree of stimulation, as well as the pattern of products produced (which particularly varies for stimulated synthesis by RF), is dependent primarily on the pol ϵ preparation utilized in each experiment; however the overall level of stimulation of synthesis seen with each pol ϵ preparation is always very similar for E1 and RF, even for a number of different E1 preparations. Time course experiments show that the stimulation of pol ϵ by E1 exhibits a longer lag time than the stimulation seen with RF (Supplementary Figure S1, panel A). At 90 min, stimulation of pol ϵ by E1 is approximately two-fold greater than stimulation of pol ϵ by RF (Supplementary Figure S1, panel B). Little additional synthesis is seen with the addition of both E1 and RF, and addition of increasing levels of E1 into a reaction with fixed levels of pol ϵ and RF just results in an increasing shift from the pol ϵ plus RF synthesis pattern to a pol ϵ plus E1 pattern (a more continuous range of sized products, rather than a product range showing preferred stop sites) (Supplementary Figure S2).
Figure 2.
HPV E1 stimulates pol ϵ. (A) Pol ϵ was added to reactions containing primed M13 template in increasing amounts from 1, 2.5, 5, 7.5 and 10 ng (as indicated, panel B). Pol ϵ was incubated with template alone (lanes 3–8). Pol ϵ was also added in increasing concentrations to reactions containing replication cofactors (RF; RFC (3 ng/μl), PCNA (10 ng/μl), RPA (70 ng/μl; lanes 10–14), or HPV E1 (60 ng/μl; lanes 16–20). RF or E1 alone with no pol ϵ showed no synthesis (lanes 9 and 15, respectively). Lanes 1 and 2 represent a reference panel containing an end-labeled molecular weight marker (M, lane 1) and pol ϵ with RF (lane 2). (B) Synthesis was quantified as in the Methods and graphed, and includes the averages of six experiments. Pol ϵ (10 ng) with RF was set to 100% and all other values were plotted relative to that value. Error bars are defined as s.d. An unpaired student's t-test compared pol ϵ + RF to pol ϵ + E1 at 10 ng of pol ϵ and was not found to be statistically different (lanes 14 and 20; P > 0.05). Compared to pol ϵ alone at 10 ng, results were found to be statistically significant (lanes 8 and 14; P = 0.009).
Stimulation of pol ϵ by HPV E1 is specific for pol ϵ
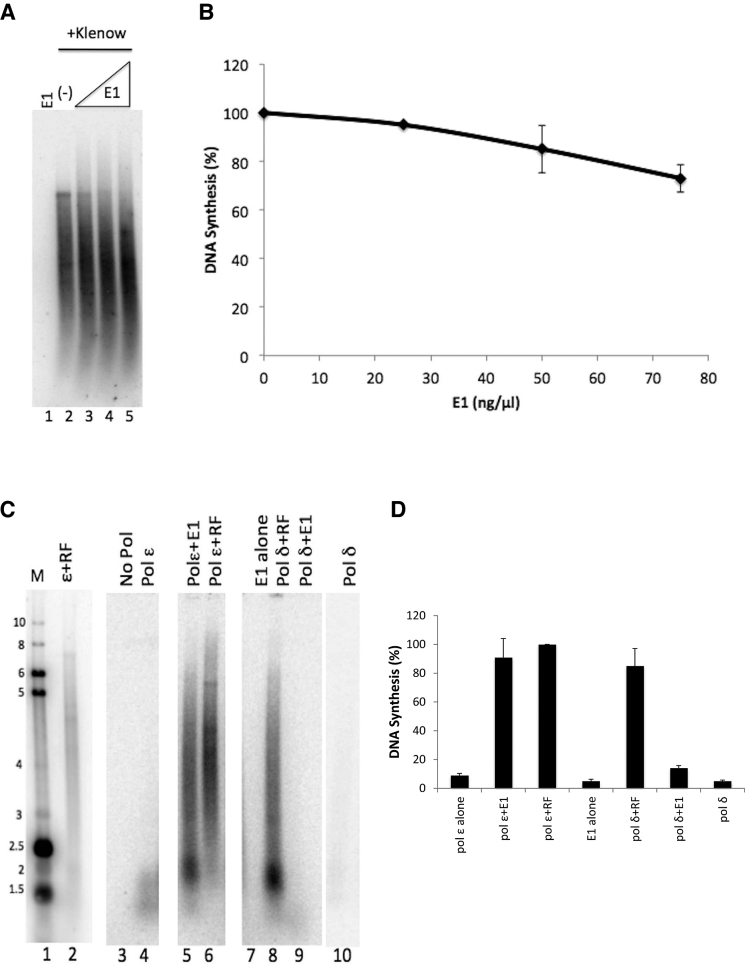
To address whether the E1 stimulation of pol ϵ was specific, or whether it could be a general effect on DNA polymerases, we evaluated whether E1 could stimulate synthesis by other DNA polymerases. First we evaluated whether an unrelated DNA polymerase, E. coli DNA polymerase I, could be stimulated by E1. Increasing levels of E1 (consistent with levels used in Figure 2) were added to primed M13 template reactions with a fixed level of E. coli DNA polymerase I (Klenow fragment). No overall increase in DNA polymerase I activity was detected (although addition of E1 created a slight shift in the length of the reaction products so that the average size was slightly shorter) (Figure 3A and B). To address whether E1 was capable of stimulating a much more closely related DNA polymerase, and since we previously demonstrated that HPV E1 interacts with pol δ, we also evaluated whether E1 was capable of stimulating pol δ. Figure 3 (panels C and D) demonstrates that whereas both pol ϵ and pol δ were clearly stimulated by the addition of RF, 11-fold and 9-fold respectively, (lane 6 compared to lane 4, and lane 8 compared to lane 10), and that E1 was capable of stimulating pol ϵ (lane 4 compared to lane 5); E1 did not stimulate pol δ in these conditions (lane 10 compared to lane 9). Hence, we concluded that the stimulation of pol ϵ by E1 is highly specific to the pol ϵ DNA polymerase.
Figure 3.
HPV E1 did not stimulate E. coli DNA Polymerase I or human pol δ. (A) Primed M13 template assays were used to analyze DNA synthesis by the E. coli DNA polymerase I (Klenow fragment) in the presence and absence of HPV E1. 1 unit of Klenow was incubated with template with increasing concentrations of EE-E1 (25, 50 and 75 ng/μl; lanes 3–5, as indicated in panel B) for 1 h at 37°C. E1 alone produced no detectable synthesis (lane 1). The basal synthesis observed with this level of DNA polymerase I with no E1 is shown in lane 2. (B) Synthesis was quantified as in the Methods and graphed, and includes the averages of three experiments. Klenow alone (lane 2) was set to 100% and all other values were plotted relative to that. Error bars are defined as s.d. (C) Primed M13 template assays were used to analyze DNA synthesis by pol ϵ or pol δ in the presence of HPV E1 or RF (RFC, PCNA, RPA). Lanes 1 and 2 represent reference size markers, as described in Materials and Methods. Template was incubated with 1 ng/μl of either pol ϵ or pol δ for 1 h at 37°C (lanes 4 and 10, respectively). RFC, PCNA and RPA at 3, 10 and 70 ng/μl, respectively, were added to reactions containing 1 ng/μl of pol ϵ or pol δ (lanes 6 and 8, respectively). EE-E1 (60 ng/μl) was also added to reactions containing 1 ng/μl of either pol ϵ or δ (lanes 5 and 9, respectively). EE-E1 (60 ng/μl) was also incubated alone with template (lane 7). Lanes 3–10 represent results from a single experiment on a single day, run on the same gel and exposed for the same time; they were reassembled in this order for more logical presentation. (D) Synthesis from the experiment in figure 3C and the same experiment carried out on two other occasions were quantified as in the Materials and Methods the values for each experiment were normalized setting pol ϵ plus RF to 100% for each set of values, and graphed. Error bars are defined as s.d. An unpaired Student's t-test indicated significant differences between pol ϵ+E1 and pol δ+E1 (lanes 5 and 8; P = 0.0005), and between pol ϵ+E1 and pol ϵ alone or E1 alone (lanes 5, 4 and 7; P = 0.0003 and 0.0004, respectively). Differences were not found to be significant between pol ϵ + E1 and pol ϵ + RF (P = 0.2967) and pol ϵ + E1 and pol δ + RF (P = 0.60), as well as pol ϵ + RF and pol δ + RF (P = 0.30).
The stimulation of pol ϵ by PV E1 is helicase-specific
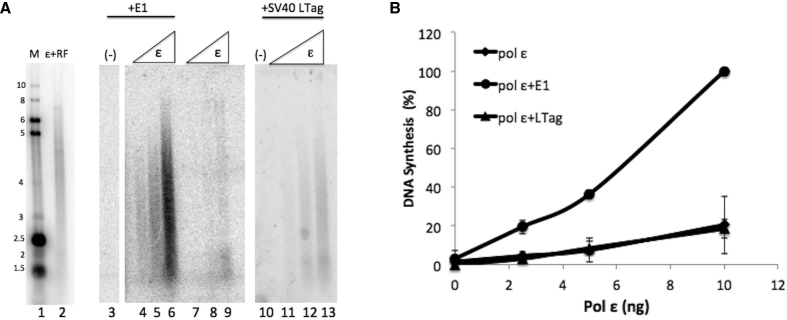
We also sought to determine whether the stimulation of pol ϵ by E1 is specific for the DNA helicase used. SV40 Large T-antigen (LTag) is also a SF3 family helicase that is related to E1 structurally, that shares some protein sequence motifs to a greater degree than other SF3 AAA+ helicases, and certainly shares many DNA replication strategies with E1 (44). To determine whether the activity of pol ϵ could be stimulated by this related DNA helicase, we added increasing amounts of pol ϵ to reactions containing primed ssM13 and a known working concentration of LTag (comparable levels of protein to the levels of E1 used, and sufficient levels of LTag to support in vitro SV40 DNA replication, data not shown). Unlike PV E1, which showed a clear ∼5- to 8-fold stimulation of pol ϵ (Figure 4, panel A, compare lanes 4–6 with lanes 7–9, and see panel B), SV40 LTag did not stimulate DNA synthesis activity of pol ϵ (Figure 4, panel A, lanes 7–9 compared to lanes 11–13, and Figure 4B). These results demonstrate that the stimulation of pol ϵ by E1 is specific with regard to the viral DNA helicase.
Figure 4.
Pol ϵ is not stimulated by the SV40 Large T-antigen helicase. (A) Lanes 1 and 2 represent reference size markers, as described in Materials and Methods. Pol ϵ was added in increasing amounts (2.5, 5 and 10 ng, as indicated in panel B) to reactions containing primed M13 template. Pol ϵ was incubated alone (lanes 7–9), with HPV E1 at 60 ng/μl (lanes 4–6) or SV40 Large T-antigen at 60 ng/μl (lanes 11–13). E1 alone had no effect on the template (lane 3) and neither did Large T-antigen (lane 10) (both at 60 ng/μl). (B) Synthesis was quantified for three separate experiments as in the Materials and Methods and graphed. All values were normalized relative to the pol ϵ (10 ng) + RF value for that experiment which was set to 100%. Error bars are defined as s.d. An unpaired Student's t-test indicated statistical significance between pol ϵ + E1 compared to pol ϵ + LTag at 10 ng (lanes 6 and 13; P < 0.0001). No statistical significance was indicated when comparing pol ϵ + LTag and pol ϵ alone (lanes 9 and 13; P = 0.82).
HPV E1 confers processivity to pol ϵ
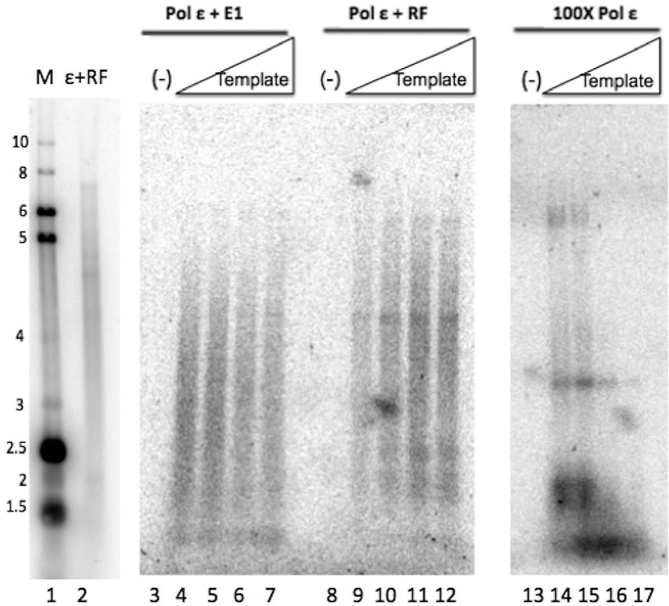
The fairly long products produced when E1 is stimulating pol ϵ, as well as the similar degree to which pol ϵ is stimulated by either E1 or pol ϵ’s known processivity factors, RF, led us to investigate whether the stimulation of pol ϵ by E1 is due to an increase in pol ϵ’s processivity. To determine whether E1 is conferring processivity to pol ϵ, singly-primed ssM13 template was added in increasing concentrations (with the template in excess of polymerase). Under these conditions, with increasing template levels non-processive polymerase action would result in the polymerase repeatedly transferring to a ‘new’ template, resulting in overall shorter products; with processive polymerase action the polymerase would stay associated with the initial template, resulting in product length that remains consistent even in the presence of the increasing template levels (45,46). When pol ϵ was combined with RF, long DNA product lengths (up to full-length, ∼7 kb) were observed under all concentrations of template, indicating pol ϵ synthesis in the presence of RF is processive (Figure 5, lanes 9–12). Conversely, synthesis by pol ϵ without any co-factors shows a dramatic decrease in nascent strand length, as well as overall inhibition, with increasing levels of template, indicative of non-processive DNA synthesis (Figure 5, lanes 14–18, note that 100-fold higher levels of pol ϵ had to be used in these reactions to obtain detectable levels of synthesis). In the presence of E1, pol ϵ synthesized nascent strands on average only slightly shorter than those produced in the presence of RF, but clearly longer than the majority of the nascent strands synthesized by pol ϵ alone. Moreover, addition of increasing levels of template resulted in little change in synthesis levels, and no appreciable change in nascent strand length (Figure 5, lanes 4–7). This clearly indicates that addition of E1 induces pol ϵ to synthesize DNA processively. In the presence of the combination of both RF and E1, pol ϵ also synthesized DNA processively, with no appreciable synergy nor interference (data not shown). These results indicate that E1 confers processivity to pol ϵ, similar to the effect of pol ϵ’s known processivity factor, PCNA.
Figure 5.
HPV E1 confers processivity to pol ϵ. Pol ϵ processivity was examined by performing primed M13 reactions (essentially as in Figure 2) under conditions in which template is in great excess over polymerase (45). Pol ϵ (2.9 fmol) was incubated with E1 (60 ng/μl) increasing concentrations of template (5, 10, 20, and 40 fmoles, lanes 3–7), or with RF with the same increasing concentrations of template (lanes 9–12). A high concentration of pol ϵ (290 fmol) was similarly incubated with the same increasing concentrations of the primed M13 template (lanes 14–17). No template was present in lanes 3, 8 and 13. Lanes 1 and 2 represent a reference marker, as described in Materials and Methods.
Stimulation of pol ϵ by E1 is dependent on ATP hydrolysis
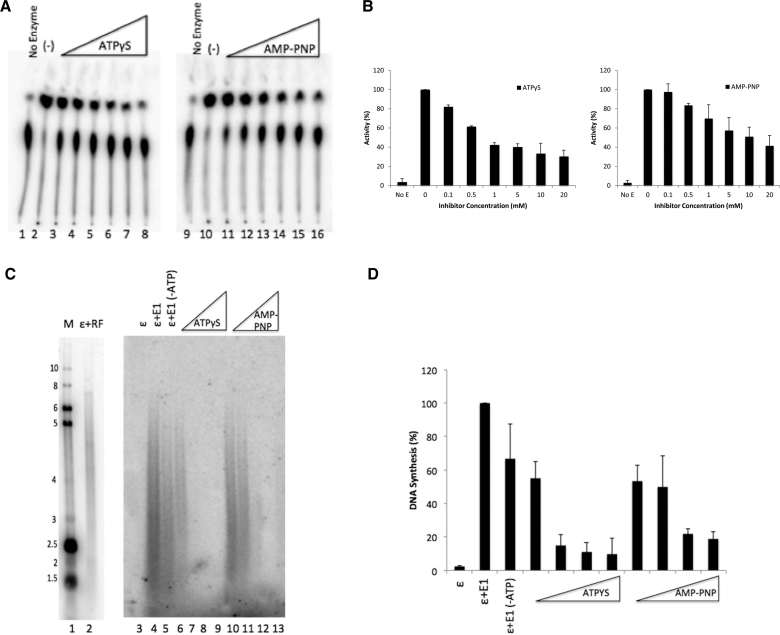
Since E1 physically interacts with pol ϵ and confers processivity to pol ϵ, one could envision a model in which pol ϵ is linked to E1, being towed behind the helicase as it unwinds duplex DNA (see Figure 8). In order to track along DNA, helicase proteins must hydrolyze ATP (44,47). In Figure 6, we demonstrate that in the presence of limiting ATP, E1’s ATPase activity can be inhibited by addition of nonhydrolyzable ATP analogs. Lanes 2 and 10 represent E1’s ATPase activity in the absence of inhibitor (Figure 6A). We compared two different analogs, adenosine 5′-[gamma–thio] triphosphate (ATPγS) (Figure 6A, lanes 3–8) and adenosine 5′-[beta,gamma–imido]]triphosphate (AMP-PNP) (Figure 6A, lanes 11–16) and saw up to 70% inhibition at the highest concentration of inhibitor for ATPγS and up to 60% inhibition for AMP-PNP (Figure 6B). Under similar low ATP levels, we were still able to see substantial stimulation of pol ϵ synthesis by E1 in the primed M13 assay (Figure 6C, compare lane 5 with lane 4 and the no E1 lane, 3). Addition of the ATPase inhibitors resulted in inhibition the E1 stimulation of pol ϵ synthesis at levels consistent with those shown to inhibit E1’s ATPase activity (compare Figure 6D with 6B). The ATPase inhibitors were not observed to inhibit the intrinsic activity of pol ϵ alone (Supplementary Figure S4). Together, these results indicate that E1’s ability to utilize ATP is critical for stimulating pol ϵ.
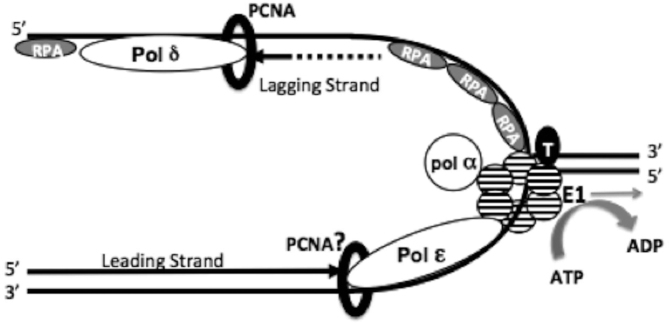
Figure 8.
Interactions at the HPV replication fork. The replicative helicase, E1, is depicted as a hexamer at the core of the replisome (striped circles). E1 interacts with various cellular replication proteins including topo I (black circle, T), RPA (grey ovals, RPA), and pol α-primase (white circle, pol α). We have previously demonstrated that E1 physically interacts with pol δ, but have not detected stimulation of activities in these studies. It is reasonable to surmise that pol δ may be playing a primary role in HPV lagging strand synthesis, with the RPA ssDNA binding complex, RFC clamp loader complex and the PCNA processivity clamp (black ring) aiding in pol δ’s processivity when synthesizing Okazaki fragments, as with host cell DNA replication. We have recently demonstrated a novel physical and functional interaction between E1 and pol ϵ. E1 stimulates pol ϵ processivity by producing a leading strand synthesis complex that is independent of the usual polymerase cofactors (RFC and PCNA, as well as RPA) and is capable of synthesizing kilobases of DNA before dissociating. This stimulation of pol ϵ by E1 is dependent on E1’s ability to hydrolyze ATP (gray curved arrow). This suggests a mechanism in which the stimulation of pol ϵ processivity may be the result of the physical interaction between E1 and pol ϵ, in which pol ϵ is tethered to E1 and is being pulled behind the helicase as it tracks along the leading strand DNA template.
Figure 6.
ATP hydrolysis is required for E1 stimulation of pol ϵ. (A) ATPase assays were performed by incubating HPV E1 in the presence and absence of nonhydrolyzable analogs ATPγS and AMP-PNP. E1 was incubated alone (lanes 2 and 10) or with increasing amounts (as indicated in panel B) of ATPγS (0.1 mM to 20 mM; lanes 3–8) or AMP-PNP (0.1 mM to 20 mM; lanes 11–16). The no enzyme control (lanes 1 and 9) contained only reaction buffer and [γ-32P] ATP. (B) Quantification of panel A. ATPase assays were quantified as in Methods and graphed. E1 without inhibitor was set to 100%. All other values were plotted relative to that value. Error bars are defined as s.d.; n = 3. (C) Primed M13 template assays were performed in the presence and absence of the nonhydrolyzable analogs ATPγS and AMP-PNP. Pol ϵ was combined with HPV E1 with ATP (4 mM; lane 4) or without ATP in the reaction buffer (lanes 5–13). Increasing amounts of either ATPγS (4, 8, 16 and 32 mM; lanes 6–9) or AMP-PNP (4, 8, 16 and 32; lanes 10–13) were titrated into the reactions. Lanes 1 and 2 represent reference size markers, as described in Materials and Methods. (D) Quantification of panel C. Assays were quantified as in Methods and graphed. E1 with ATP, but without inhibitor was set to 100%. All other values were plotted relative to that value. Error bars are defined as s.d.; n = 3. An unpaired Student's t-test determined that pol ϵ + E1 in the presence or absence of ATP in the reaction buffer (lanes 5 and 6) were not statistically different (P > 0.5). Statistical significance was observed when comparing lanes 5 to lanes 7–9 and to lanes 12 and 13 (P < 0.001).
RPA may be dispensable for HPV leading strand DNA replication
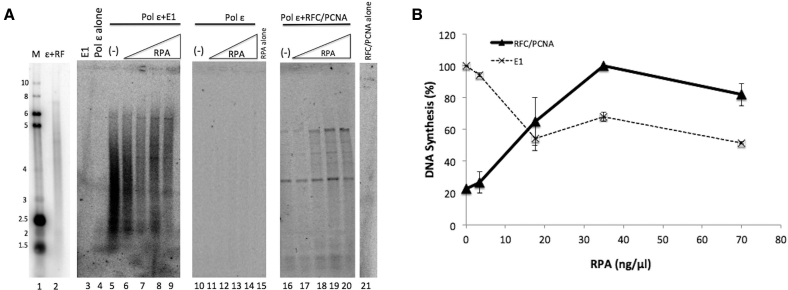
The single-stranded DNA binding complex, RPA, is a critical part of the eukaryotic replication fork and has been shown to stimulate polymerase processivity and to be important for pol ϵ-dependent leading strand synthesis (21,46). In addition, RPA is essential for PV DNA replication and physically interacts with E1 (19,20,36,42). Here we show that RPA is not required for efficient synthesis or pol ϵ stimulation in the presence of E1, as synthesis is robust even in RPA’s absence (Figure 7A, see also Figures 2–5). In the absence of RPA, pol ϵ was strongly stimulated by E1 (Figure 7A, lane 5). However, only a small portion of the DNA products were full length. Increasing the concentration of RPA resulted in some shift to a larger proportion of fully extended primers (Figure 7A, lanes 6–9), and a substantial (∼40–50%) inhibition of overall synthesis with the levels of RPA used (Figure 7B, dashed line). This inhibition is likely due to RPA’s strong binding to ssDNA impeding E1 progression along the primed ssDNA template in this reaction (note, this would not occur when E1 acts at a replication fork, where the dsDNA template ahead of E1 would provide a poor template for RPA binding). Pol ϵ alone was not detectably stimulated by RPA (Figure 7A, lanes 11–14). When pol ϵ was combined with RFC and PCNA, and RPA was added at varying levels, RPA both substantially stimulated synthesis by the pol ϵ/RFC/PCNA combination (Figure 7B solid line, by over 4-fold in these experiments), as well as resulted in longer products (Figure 7A, lanes 16–20), both aspects consistent with previous publications (21,46). These results indicate that unlike the pol ϵ stimulation by RFC and PCNA, RPA is not vital for E1’s stimulation of pol ϵ.
Figure 7.
RPA may be dispensable for synthesis by pol ϵ in the presence of E1. (A) Primed M13 template assays were performed with increasing amounts of RPA (3.5, 17.5, 35 and 70 ng/μl) in the presence of pol ϵ (lanes 11–14), pol ϵ and E1 (lanes 6–9) and pol ϵ and RFC/PCNA (lanes 17–20). Pol ϵ and E1 with no RPA serves as the positive control standard (lane 5). E1 alone (lane 3), pol ϵ alone (lane 4), RPA alone (lane 15) and RFC/PCNA alone (lane 21) were incubated with template and serve as controls, in which little (pol ϵ, lane 4) or no synthesis was observed. Lanes 1 and 2 represent reference size markers, as described in Materials and Methods. (B) Synthesis was quantified as in the Materials and Methods and graphed for three experiments. Results were quantified as in the Methods. Values for pol ϵ + E1 (dashed line) were plotted with pol ϵ + E1 with 0 ng/μl of RPA set to 100%. Values for pol ϵ + RFC/PCNA (solid line) were plotted setting pol ϵ + RFC/PCNA with 35 ng/μl of RPA to 100%, as this is the RPA level used for 100% synthesis of pol ϵ + RFC/PCNA throughout this manuscript and consistent with other published studies (11,20,21). Error bars are defined as s.d.
DISCUSSION
Several families of small DNA viruses do not encode their own DNA polymerases and instead rely on recruiting cellular DNA polymerases to replicate their viral genomes. These viruses require polymerases with both processivity and fidelity and so have evolved to recruit processive proofreading DNA polymerases involved in cellular DNA replication. The polyomaviruses, exemplified by SV40, have been shown to utilize pol δ to synthesize the majority of their DNA during DNA replication (15). The fractionation and reconstitution of the cellular factors required to support SV40 DNA replication showed a requirement for pol δ, but never showed any requirement for pol ϵ (11–14). In addition, pol ϵ was found to be dispensable in the DNA replication process for SV40 both by UV-crosslinking of polymerases to replicating DNA (18) and through the use of pol ϵ-blocking antibodies that inhibited cellular but not SV40 DNA replication (17). Our demonstration that unlike PV E1, SV40 LTag was unable to stimulate pol ϵ in primed M13 assays (Figure 4), is consistent with the lack of a role for pol ϵ in SV40 DNA replication. Parvoviruses also do not encode their own DNA polymerases, and have been shown to require DNA pol δ for AAV DNA synthesis; it was further shown that pol ϵ cannot replace pol δ in this reaction (9,10). It is noteworthy that both of these viral families have evolved to utilize pol δ and not pol ϵ, possibly reflecting the greater versatility of pol δ and the essential nature of its enzymatic function, in contrast to pol ϵ (1,48–50). Pol δ has also been shown to be required for PV DNA replication (19,20); however, pol α and pol δ were not shown to be sufficient to carry out PV DNA replication in vitro, possibly suggesting a role for pol ϵ in PV DNA replication (20). The results shown herein provide a compelling case implicating pol ϵ in HPV DNA replication. This obviously does not preclude a role for pol δ in HPV DNA replication, as a requirement for pol δ in PV DNA replication has been shown (19,20). To our knowledge this is the first known instance of a virus utilizing pol ϵ to replicate its viral genome. One could imagine that PV DNA replication either utilizes both pol δ and pol ϵ for DNA replication, much like cellular DNA replication, with one polymerase synthesizing the leading strand and the other polymerase extending the lagging strand products (2–4,15). It is unlikely that PV DNA replication uses the two polymerases on an either/or basis, since pol δ has been shown to be required for PV DNA replication in a partially purified soluble system that utilized pol ϵ-containing fractions (20).
Several examples of functional coupling between helicases and polymerases have been documented (21,22,26,51). The T4 bacteriophage helicase, gp41, was reported to stimulate the activity of the T4 polymerase, gp43, and reciprocal stimulation of gp41 by gp43 was also demonstrated (22,51). It was reported that efficient DNA synthesis in the T7 bacteriophage system is only achieved with the combined action of the helicase and polymerase (26). Coupling of helicases and polymerases also occur for eukaryotic DNA replication; recently the human CMG helicase complex was shown to interact with pol ϵ and stimulate its activity. Pol ϵ was also shown to increase the processivity of the CMG helicase complex (21). These findings suggest that strand displacement and DNA synthesis are linked. However, our studies did not reveal any effect of pol ϵ on either the ATPase or helicase functions of E1 (Supplementary Figure S3). It may be that this viral system does not exhibit the complementary effect on the DNA helicase seen with host cell replication factors, or it may be that additional factors and/or different reaction conditions will be required to observe such an effect. However, this is the first known case of a viral DNA helicase acting to stimulate a cellular DNA polymerase acting as the major synthetic DNA polymerase for viral genome synthesis (DNA pol α has been shown to be stimulated by SV40 LTag, but pol α is essentially a RNA-DNA primer synthetase, and the LTag stimulation of pol α function is consistent with this role rather than that of a replicative DNA polymerase (29)).
Functional coupling between helicases and polymerases have also been shown to result in an increase in polymerase processivity (21,22,28,52,53). This is true in T7 bacteriophage DNA replication, in which there are two modes of replication that enhance polymerase processivity (28). In one mode, the T7 DNA polymerase utilizes the bacterially produced thioredoxin as a processivity factor to facilitate the polymerase sliding along the DNA without dissociation. In the second mode, interactions between the polymerase and the C-terminal tail of the helicase ensure a high local concentration of the DNA polymerase-thioredoxin complex in the event that the replicating polymerases dissociates. Furthermore, tethering relationships have also been characterized in the T4 bacteriophage replication system, as well as in the E. coli DNA replication system between pol III and DnaB, through the pol III tau subunit (22,23,53).
Our results may suggest HPV DNA replication may use a helicase-polymerase coupling mechanism similar to the E. coli DNA replication model or perhaps a simplified version of the T7 helicase-polymerase coupling mechanism. In this system the HPV E1 helicase would act as the processivity factor as well as a tether to the DNA template. The fact that E1 confers processivity to pol ϵ synthesis (Figure 5) provides strong support for a tethering mechanism, in which E1 holds pol ϵ to the DNA, preventing dissociation. Further, the demonstration that E1’s ability to hydrolyze ATP, and presumably track along the DNA, is critical for stimulation of pol ϵ (Figure 6), supports a combined tethering/tracking model for how E1 acts to stimulate pol ϵ. The 3′ to 5′ directional translocation of the E1 helicase places it tracking along the leading strand template, which is an ideal position to tether a leading strand DNA polymerase, presumably pol ϵ (see Figure 8, E1, striped circles). This is unlike the case for E. coli, where the dnaB helicase tracks along the lagging strand template, creating a more complicated helicase-leading strand polymerase coupling model (23,54).
Current models of eukaryotic DNA replication often include the presence of the ssDNA DNA binding protein, RPA, between the helicase and the leading strand DNA polymerase complex (15). This is likely due to the substantial positive effect of RPA on synthesis by both pol ϵ and pol δ (46). The human CMG helicase complex was shown to interact with pol ϵ and increase its processivity; however, RPA was still found to be an important part of this DNA synthesis complex, and acted to enhance both helicase and polymerase processivity (21). Results presented here indicate that RPA is not required for processive DNA synthesis by pol ϵ in the presence of E1 (Figure 7); indeed the addition of RPA produces dramatically differing effects on synthesis by the two pol ϵ synthesis complexes (a 50% decrease for the E1–pol ϵ complex versus a 300% increase for the RFC–PCNA–pol ϵ complex). The role of RPA in synthesis by the pol ϵ and pol δ processive complexes (with RFC and PCNA) can be replaced by other ssDNA binding proteins, such as E. coli SSB (21). Since RPA, and other ssDNA binding proteins, when added at sufficient levels, can eliminate secondary structure on the template strand that could otherwise block DNA polymerase progression, we speculate that the critical role of RPA in the pol ϵ and pol δ processive complexes is just that—prevention of secondary structure and/or annealing of the ssDNA template. However, since the E1 helicase is leading the PV DNA replication fork, creating the ssDNA template for synthesis of the leading strand, having pol ϵ tethered to the E1 helicase could ensure that no secondary structures/ssDNA annealing would occur on the template strand as it exits E1 and encounters pol ϵ. Furthermore, such tight spacing might even preclude RPA from gaining access to the newly exposed ssDNA. This model is indeed consistent with our data. In the presence of E1, pol ϵ is able to synthesize long chains of DNA, similar in length to those observed when pol ϵ synthesizes DNA in the presence of RFC, PCNA and RPA (Figure 7A). Furthermore, the highly continuous nature of the nascent strands synthesized by the E1-pol ϵ complex (as opposed to the strands synthesized by the RFC-PCNA-RPA-pol ϵ complex, which although producing long strands, still show the presence of strong stop sites) is also consistent with a decreased effect of ssDNA secondary structure on E1-pol ϵ DNA synthesis (Figure 7A). Although RPA isn’t required for synthesis by this leading strand E1–pol ϵ complex, we still observed a shift to more fully extended DNA products in the presence of RPA (Figure 7A). It is possible that RPA could affect synthesis by the E1–pol ϵ complex by preventing slippage (22,26); alternately, this could be due to RPA binding inhibiting the well-established tendency of M13 ssDNA to anneal into very stable secondary structures that might otherwise slow helicase progression. However, the overall finding, that RPA is not required for efficient synthesis and the production of long DNA strands by pol ϵ, leads us to hypothesize that at the HPV DNA replication fork pol ϵ is synthesizing the leading strand directly attached to the E1 helicase in the absence of RPA on the leading template (Figure 8). Although this model indicates that RPA would be dispensable for leading strand synthesis, RPA would of course play an integral role in lagging strand DNA synthesis, consistent with its known importance in synthesis by pol δ, and consistent with our view of the role of RPA in synthesis by pol ϵ and pol δ processive complexes – prevention of secondary structure/annealing of a ssDNA template in front of the complex.
The findings reported here have led us to change our current view of the HPV DNA replication fork (Figure 8). Briefly, following origin recognition and unwinding by E1, pol α synthesizes the first RNA-DNA primers. Primer synthesis (both during initiation and Okazaki fragment priming) occurs through interactions between E1, RPA and pol α, in an analogous fashion to that of SV40 (29,55,56). As E1 splits apart duplex DNA, E1 actively loads RPA onto the lagging strand template (42). Topo I functionally interacts with E1, after possibly aiding in origin recognition, but also to associate with E1 in its helicase conformation to relieve torsional stress ahead of the progressing replication fork (41,57). After each RNA-DNA primer is synthesized, pol α is prevented from associating with and extending this primer through the action of RFC and PCNA and the RFC-PCNA complex is recognized by pol δ in a process known as polymerase switching (11,14,58). On the leading strand, pol ϵ physically interacts with E1, which stimulates DNA synthesis by pol ϵ and increases the processivity of pol ϵ. This process is dependent on E1 hydrolyzing ATP (grey curved arrow) providing the motive force for E1 to track 3′ to 5′ along the template DNA (grey straight arrow) for leading strand synthesis, pulling pol ϵ behind it. This physical tether also apparently obviates the need for RPA to prevent secondary structure/annealing of ssDNA, which would otherwise inhibit pol ϵ progression. Additionally, the conferral of processivity by E1, and the lack of a synergistic effect on pol ϵ by the combination of E1 and RFC/PCNA, may suggest that PCNA may not be required for synthesis by the E1-pol ϵ leading strand complex. Of course PCNA and RPA would both continue to play a vital role in lagging strand DNA synthesis.
Overall these studies have provided evidence for a new view of the PV DNA replication fork, incorporating the cellular DNA replication leading strand polymerase as a likely PV leading stand DNA polymerase; a polymerase which has until now not been shown to play a role in any other viral DNA replication system. Further, the novel finding of a eukaryotic virus DNA helicase that has evolved to act as a motor and processivity factor for a cellular DNA polymerase is surprising and will provide many interesting avenues for further investigation.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jerard Hurwitz and Inger Tappin for their generous gifts of the human DNA polymerases δ and ϵ baculoviruses, as well as purified human replication proteins.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
National Institute of Health F31 fellowship [5 F31 CA189383 to M.C.] as well as by National Institute of Health grant [R01 Al095632 to T.M.]. Funding for open access charge: National Institutes of Health, NIAID.
Conflict of interest statement. None declared.
REFERENCES
- 1. Flood C.L., Rodriguez G.P., Bao G., Shockley A.H., Kow Y.W., Crouse G.F.. Replicative DNA polymerase delta but not epsilon proofreads errors in Cis and in Trans. PLoS Genet. 2015; 11:e1005049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kunkel T.A., Burgers P.M.. Dividing the workload at a eukaryotic replication fork. Trends Cell Biol. 2008; 18:521–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lujan S.A., Williams J.S., Kunkel T.A.. DNA Polymerases Divide the Labor of Genome Replication. Trends Cell Biol. 2016; 26:640–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Miyabe I., Mizuno K., Keszthelyi A., Daigaku Y., Skouteri M., Mohebi S., Kunkel T.A., Murray J.M., Carr A.M.. Polymerase delta replicates both strands after homologous recombination-dependent fork restart. Nat. Struct. Mol. Biol. 2015; 22:932–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ohya T., Kawasaki Y., Hiraga S., Kanbara S., Nakajo K., Nakashima N., Suzuki A., Sugino A.. The DNA polymerase domain of pol(epsilon) is required for rapid, efficient, and highly accurate chromosomal DNA replication, telomere length maintenance, and normal cell senescence in Saccharomyces cerevisiae. J. Biol. Chem. 2002; 277:28099–28108. [DOI] [PubMed] [Google Scholar]
- 6. Pursell Z.F., Kunkel T.A.. DNA polymerase epsilon: a polymerase of unusual size (and complexity). Prog. Nucleic Acid Res. Mol. Biol. 2008; 82:101–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stillman B. DNA polymerases at the replication fork in eukaryotes. Mol. Cell. 2008; 30:259–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sun J., Shi Y., Georgescu R.E., Yuan Z., Chait B.T., Li H., O’Donnell M.E.. The architecture of a eukaryotic replisome. Nat. Struct. Mol. Biol. 2015; 22:976–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nash K., Chen W., McDonald W.F., Zhou X., Muzyczka N.. Purification of host cell enzymes involved in adeno-associated virus DNA replication. J. Virol. 2007; 81:5777–5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ni T.H., McDonald W.F., Zolotukhin I., Melendy T., Waga S., Stillman B., Muzyczka N.. Cellular proteins required for adeno-associated virus DNA replication in the absence of adenovirus coinfection. J. Virol. 1998; 72:2777–2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tsurimoto T., Melendy T., Stillman B.. Sequential initiation of lagging and leading strand synthesis by two different polymerase complexes at the SV40 DNA replication origin. Nature. 1990; 346:534–539. [DOI] [PubMed] [Google Scholar]
- 12. Lee S.H., Pan Z.Q., Kwong A.D., Burgers P.M., Hurwitz J. Synthesis of DNA by DNA polymerase epsilon in vitro. J. Biol. Chem. 1991; 266:22707–22717. [PubMed] [Google Scholar]
- 13. Weinberg D.H., Collins K.L., Simancek P., Russo A., Wold M.S., Virshup D.M., Kelly T.J.. Reconstitution of simian virus 40 DNA replication with purified proteins. Proc. Natl. Acad. Sci. U.S.A. 1990; 87:8692–8696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Waga S., Stillman B.. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature. 1994; 369:207–212. [DOI] [PubMed] [Google Scholar]
- 15. Stillman B. Reconsidering DNA Polymerases at the Replication Fork in Eukaryotes. Mol Cell. 2015; 59:139–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li J.J., Kelly T.J.. Simian virus 40 DNA replication in vitro. Proc. Natl. Acad. Sci. U.S.A. 1984; 81:6973–6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pospiech H., Kursula I., Abdel-Aziz W., Malkas L., Uitto L., Kastelli M., Vihinen-Ranta M., Eskelinen S., Syvaoja J.E.. A neutralizing antibody against human DNA polymerase epsilon inhibits cellular but not SV40 DNA replication. Nucleic Acids Res. 1999; 27:3799–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zlotkin T., Kaufmann G., Jiang Y., Lee M.Y., Uitto L., Syvaoja J., Dornreiter I., Fanning E., Nethanel T.. DNA polymerase epsilon may be dispensable for SV40- but not cellular-DNA replication. EMBO J. 1996; 15:2298–2305. [PMC free article] [PubMed] [Google Scholar]
- 19. Muller F., Seo Y.S., Hurwitz J.. Replication of bovine papillomavirus type 1 origin-containing DNA in crude extracts and with purified proteins. J. Biol. Chem. 1994; 269:17086–17094. [PubMed] [Google Scholar]
- 20. Melendy T., Sedman J., Stenlund A.. Cellular factors required for papillomavirus DNA replication. J. Virol. 1995; 69:7857–7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kang Y.H., Galal W.C., Farina A., Tappin I., Hurwitz J.. Properties of the human Cdc45/Mcm2-7/GINS helicase complex and its action with DNA polymerase epsilon in rolling circle DNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:6042–6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Delagoutte E., von Hippel P.H.. Molecular mechanisms of the functional coupling of the helicase (gp41) and polymerase (gp43) of bacteriophage T4 within the DNA replication fork. Biochemistry. 2001; 40:4459–4477. [DOI] [PubMed] [Google Scholar]
- 23. Kim S., Dallmann H.G., McHenry C.S., Marians K.J.. tau couples the leading- and lagging-strand polymerases at the Escherichia coli DNA replication fork. J. Biol. Chem. 1996; 271:21406–21412. [DOI] [PubMed] [Google Scholar]
- 24. Nakai H., Richardson C.C.. Interactions of the DNA polymerase and gene 4 protein of bacteriophage T7. Protein-protein and protein-DNA interactions involved in RNA-primed DNA synthesis. J. Biol. Chem. 1986; 261:15208–15216. [PubMed] [Google Scholar]
- 25. Notarnicola S.M., Mulcahy H.L., Lee J., Richardson C.C.. The acidic carboxyl terminus of the bacteriophage T7 gene 4 helicase/primase interacts with T7 DNA polymerase. J. Biol. Chem. 1997; 272:18425–18433. [DOI] [PubMed] [Google Scholar]
- 26. Stano N.M., Jeong Y.J., Donmez I., Tummalapalli P., Levin M.K., Patel S.S.. DNA synthesis provides the driving force to accelerate DNA unwinding by a helicase. Nature. 2005; 435:370–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gambus A., van Deursen F., Polychronopoulos D., Foltman M., Jones R.C., Edmondson R.D., Calzada A., Labib K.. A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase alpha within the eukaryotic replisome. EMBO J. 2009; 28:2992–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee S.J., Richardson C.C.. Choreography of bacteriophage T7 DNA replication. Curr. Opin. Chem. Biol. 2011; 15:580–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Collins K.L., Kelly T.J.. Effects of T antigen and replication protein A on the initiation of DNA synthesis by DNA polymerase alpha-primase. Mol. Cell Biol. 1991; 11:2108–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bermudez V.P., Farina A., Raghavan V., Tappin I., Hurwitz J.. Studies on human DNA polymerase epsilon and GINS complex and their role in DNA replication. J. Biol. Chem. 2011; 286:28963–28977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lane D., Harlow E.. Antibodies: A Laboratory Manual. 1988; CSHL Press, Cold Spring Harbor Laboratory. [Google Scholar]
- 32. Binz S.K., Dickson A.M., Haring S.J., Wold M.S.. Functional assays for replication protein A (RPA). Methods Enzymol. 2006; 409:11–38. [DOI] [PubMed] [Google Scholar]
- 33. Fien K., Stillman B.. Identification of replication factor C from Saccharomyces cerevisiae: a component of the leading-strand DNA replication complex. Mol. Cell. Biol. 1992; 12:155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cai J., Uhlmann F., Gibbs E., Flores-Rozas H., Lee C.G., Phillips B., Finkelstein J., Yao N., O’Donnell M., Hurwitz J.. Reconstitution of human replication factor C from its five subunits in baculovirus-infected insect cells. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:12896–12901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Clower R.V., Hu Y., Melendy T.. Papillomavirus E2 protein interacts with and stimulates human topoisomerase I. Virology. 2006; 348:13–18. [DOI] [PubMed] [Google Scholar]
- 36. Han Y., Loo Y.M., Militello K.T., Melendy T.. Interactions of the papovavirus DNA replication initiator proteins, bovine papillomavirus type 1 E1 and simian virus 40 large T antigen, with human replication protein A. J. Virol. 1999; 73:4899–4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wu Y., Li Q., Chen X.Z.. Detecting protein-protein interactions by Far western blotting. Nat. Protoc. 2007; 2:3278–3284. [DOI] [PubMed] [Google Scholar]
- 38. Park P., Copeland W., Yang L., Wang T., Botchan M.R., Mohr I.J.. The cellular DNA polymerase alpha-primase is required for papillomavirus DNA replication and associates with the viral E1 helicase. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:8700–8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Conger K.L., Liu J.S., Kuo S.R., Chow L.T., Wang T.S.. Human papillomavirus DNA replication. Interactions between the viral E1 protein and two subunits of human dna polymerase alpha/primase. J. Biol. Chem. 1999; 274:2696–2705. [DOI] [PubMed] [Google Scholar]
- 40. Bonne-Andrea C., Santucci S., Clertant P., Tillier F.. Bovine papillomavirus E1 protein binds specifically DNA polymerase alpha but not replication protein A. J. Virol. 1995; 69:2341–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Clower R.V., Fisk J.C., Melendy T.. Papillomavirus E1 protein binds to and stimulates human topoisomerase I. J. Virol. 2006; 80:1584–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Loo Y.M., Melendy T.. Recruitment of replication protein A by the papillomavirus E1 protein and modulation by single-stranded DNA. J. Virol. 2004; 78:1605–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li Y. 1998; Ph.D. University of California, Berkeley, Berkeley, CA. [Google Scholar]
- 44. Koonin E.V. A common set of conserved motifs in a vast variety of putative nucleic acid-dependent ATPases including MCM proteins involved in the initiation of eukaryotic DNA replication. Nucleic Acids Res. 1993; 21:2541–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bambara R.A., Fay P.J., Mallaber L.M.. Methods of analyzing processivity. Methods Enzymol. 1995; 262:270–280. [DOI] [PubMed] [Google Scholar]
- 46. Chilkova O., Stenlund P., Isoz I., Stith C.M., Grabowski P., Lundstrom E.B., Burgers P.M., Johansson E.. The eukaryotic leading and lagging strand DNA polymerases are loaded onto primer-ends via separate mechanisms but have comparable processivity in the presence of PCNA. Nucleic Acids Res. 2007; 35:6588–6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Enemark E.J., Joshua-Tor L.. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006; 442:270–275. [DOI] [PubMed] [Google Scholar]
- 48. Takemura M. Poxviruses and the origin of the eukaryotic nucleus. J. Mol. Evol. 2001; 52:419–425. [DOI] [PubMed] [Google Scholar]
- 49. Tahirov T.H., Makarova K.S., Rogozin I.B., Pavlov Y.I., Koonin E.V.. Evolution of DNA polymerases: an inactivated polymerase-exonuclease module in Pol epsilon and a chimeric origin of eukaryotic polymerases from two classes of archaeal ancestors. Biol. Direct. 2009; 4:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kesti T., Flick K., Keranen S., Syvaoja J.E., Wittenberg C.. DNA polymerase epsilon catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol. Cell. 1999; 3:679–685. [DOI] [PubMed] [Google Scholar]
- 51. Dong F., Weitzel S.E., von Hippel P.H.. A coupled complex of T4 DNA replication helicase (gp41) and polymerase (gp43) can perform rapid and processive DNA strand-displacement synthesis. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:14456–14461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Manosas M., Spiering M.M., Ding F., Croquette V., Benkovic S.J.. Collaborative coupling between polymerase and helicase for leading-strand synthesis. Nucleic Acids Res. 2012; 40:6187–6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. McInerney P., Johnson A., Katz F., O’Donnell M.. Characterization of a triple DNA polymerase replisome. Mol. Cell. 2007; 27:527–538. [DOI] [PubMed] [Google Scholar]
- 54. Schauer G.D., O’Donnell M.E.. Quality control mechanisms exclude incorrect polymerases from the eukaryotic replication fork. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:675–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Melendy T., Stillman B.. An interaction between replication protein A and SV40 T antigen appears essential for primosome assembly during SV40 DNA replication. J. Biol. Chem. 1993; 268:3389–3395. [PubMed] [Google Scholar]
- 56. Ott R.D., Rehfuess C., Podust V.N., Clark J.E., Fanning E.. Role of the p68 subunit of human DNA polymerase alpha-primase in simian virus 40 DNA replication. Mol. Cell Biol. 2002; 22:5669–5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hu Y., Clower R.V., Melendy T.. Cellular topoisomerase I modulates origin binding by bovine papillomavirus type 1 E1. J. Virol. 2006; 80:4363–4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tsurimoto T., Stillman B.. Multiple replication factors augment DNA synthesis by the two eukaryotic DNA polymerases, alpha and delta. EMBO J. 1989; 8:3883–3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.