Supplemental Digital Content is Available in the Text.
Keywords: microRNA, Cancer pain mechanisms, R-type calcium channels, miRNA target prediction
Abstract
Pathophysiological mechanisms underlying pain associated with cancer are poorly understood. microRNAs (miRNAs) are a class of noncoding RNAs with emerging functional importance in chronic pain. In a genome-wide screen for miRNAs regulated in dorsal root ganglia (DRG) neurons in a mouse model of bone metastatic pain, we identified miR-34c-5p as a functionally important pronociceptive miRNA. Despite these functional insights and therapeutic potential for miR-34c-5p, its molecular mechanism of action in peripheral sensory neurons remains unknown. Here, we report the identification and validation of key target transcripts of miRNA-34c-5p. In-depth bioinformatics analyses revealed Cav2.3, P2rx6, Oprd1, and Oprm1 as high confidence putative targets for miRNA-34c-5p. Of these, canonical and reciprocal regulation of miR-34c-5p and Cav2.3 was observed in cultured sensory neurons as well as in DRG in vivo in mice with cancer pain. Coexpression of miR-34c-5p and Cav2.3 was observed in peptidergic and nonpeptidergic nociceptors, and luciferase reporter assays confirmed functional binding of miR-34c-5p to the 3′ UTR of Cav2.3 transcripts. Importantly, knocking down the expression of Cav2.3 specifically in DRG neurons led to hypersensitivity in mice. In summary, these results show that Cav2.3 is a novel mechanistic target for a key pronociceptive miRNA, miR-34c-5p, in the context of cancer pain and indicate an antinociceptive role for Cav2.3 in peripheral sensory neurons. The current study facilitates a deeper understanding of molecular mechanisms underlying cancer pain and suggests a potential for novel therapeutic strategies targeting miR-34c-5p and Cav2.3 in cancer pain.
1. Introduction
According to a recent report by the World Health Organization (WHO), incidence and mortality of cancer have increased globally to alarming levels and predicted approximately 35% increase in following years worldwide.56 On the other hand, the 5-year survival rate of cancer patients has been greatly improved owing to improvements in diagnosis and treatment options available to the majority of the world population. It has been reported that up to 90% of cancer survivors suffer from varying degrees of pain, and 30% of them experience severe pain throughout their lifespan.60 Bone is the most vulnerable metastases target in a variety of cancers which ultimately leads to bone degeneration, surrounding soft tissue malformation, and structural changes in the nerve endings penetrating bone tissue. All these changes occurring in bone metastatic conditions lead to excruciating ongoing pain in late-stage cancer patients and in cancer survivors. Despite such a severity of this problem, underlying pathophysiological mechanisms are poorly understood. Clinical options available for the treatment of cancer pain are either nonsteroidal anti-inflammatory analgesics or opioid therapy which have their own severe side effects on major organ systems,10 underscoring an immediate need for investigating the causal mechanisms behind cancer pain development.
microRNAs (miRNAs) are approximately 21 nucleotides in length and well-studied noncoding RNA species in terms of their biogenesis and functions. The emerging literature on miRNA-mediated control of distinct pathologies reiterates their regulatory role in the genome.5,6 Although there are several studies reporting miRNA dysregulation in different pain conditions, eg, neuropathic pain, there has been only one study to address the role of miRNAs in cancer pain modulation7 so far. On the other hand, even within well-studied pain models in terms of miRNAs, there are only isolated studies addressing miRNA-mediated mechanisms in cancer pain modulation.6,41 These observations point to an enormous potential for miRNA-mediated regulation of pain pathology and an immediate need to investigate miRNA-mediated mechanisms in pain modulation in general and in cancer pain states in particular. miR-34c-5p is one of the widely investigated miRNAs in cancer conditions1,21,22,42,63 as well as in neurological conditions30,37,66 and development.4,32,48 We recently reported that bone metastatic tumor leads to massive changes in the miRNA expression repertoire in peripheral sensory neurons, and these changes are more pronounced in the hyperalgesia maintenance phase than in the establishing phase.7 miR-34c-5p is one of the highly upregulated miRNAs in sensory neurons at 8d but not at 4d post-tumor implantation, and inhibition of such tumor-mediated upregulation alleviates tumor-mediated hyperalgesia. However, the mechanistic details of such a pronociceptive role of miR-34c-5p have not been studied. In the current study, we comprehensively investigated mRNA targets of miR-34c-5p in the context of cancer pain. By employing extensive in silico analyses together with advanced molecular, genetic, and behavioral experiments, we identified miR-34c-5p and Cav2.3 as a novel functional pair in the context of cancer pain and Cav2.3 as an antinociceptive Ca2+ channel in the peripheral sensory neurons.
2. Methods
2.1. Animal model of tumor-evoked pain
All animal usage procedures were in accordance with ethical guidelines laid down by the local governing body (Regierungspräsidium Karlsruhe). All behavioral measurements were done in awake, unrestrained, age‐matched adult (more than 2 month old) C3H/HeNCrl mice. The model of bone metastases–associated pain was implemented as described previously.11,52 Briefly, National Collection of Type Cultures (NCTC) clone 2472 fibrosarcoma cells (ATCC, Manassas, VA) were cultured and injected into and around the calcaneus bone of wild-type C3H/HeNCrl mice as described previously.
2.2. Sensory neuronal cultures and transfections
Adult dorsal root ganglia (DRG) neuronal cultures were prepared following the protocol explained previously.52 Briefly, neuronal cells isolated from adult wild-type mice were seeded on Poly-L-Lysine–coated 24-well plates and maintained in DMEM Media (Gibco, Darmstadt, Germany) supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen), 1% penicillin/streptomycin (Gibco), and 0.5% L-Glutamine (Gibco). After culturing for 4 days, cells were transfected with miR-34c-5p mimic (Thermoscientific custom meridian: C-120849-00-600, Darmstadt, Germany) or with nontargeting negative control mimic (CN-120848-00-600) using Lipofectamine RNAimax reagent (13778100, Thermofischer Scientific). Total RNA was isolated 48 hours after transfection and used for quantitative real-time polymerase chain reaction (qRTPCR) analysis.
2.3. Gene ontology and pathway enrichment analysis
Gene ontology enrichment analyses were performed using the bioCompendium (http://biocompendium.embl.de) web portal developed at the European Molecular Biology Laboratory, Heidelberg, Germany. Pathway enrichment analysis was performed by uploading the list of 1533 genes, which were commonly predicted as targets for miR-34c-5p by 6 independent target prediction algorithms, to the WebGestalt (WEB-based GEne SeT AnaLysis Toolkit) online server and following all default parameters.62,65
2.4. RNA isolation from DRGs
Mice were killed using CO2, spinal column isolated, and rinsed in cold 1× phosphate-buffered saline (PBS), and Lumbar level 3, 4 DRGs were quickly isolated into a microcentrifuge tube and flash frozen in liquid nitrogen until RNA isolation was performed. Total RNA was isolated using mirVana miRNA Isolation Kit (AM 1561; Ambion) following manufacturer's instructions to enrich miRNA fraction by adding 1.25 times of absolute ethanol to the upper phase isolated from DRG lysate + chloroform: Phenol mixture. RNA was dissolved in nuclease-free water. Concentration was determined using the NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE).
2.5. qRTPCR analysis of miRNAs and mRNAs
For the generation of miR-34c-5p specific first strand cDNA, 20 ng of total RNA was reverse transcribed by miRNA-specific RT primer using TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, 4366597) following manufacturer's instructions. cDNA was synthesized from 20 ng of total RNA using random primers from the High Capacity cDNA Reverse Transcription Kit (4368814; Applied Biosystems, Darmstadt, Germany) following manufacturer's instructions for mRNA amplification. cDNA was PCR-amplified in each reaction using the corresponding miRNA‐ or mRNA‐specific primers using TaqMan Universal Master Mix II, (Applied Biosystems, 4440040) following manufacturer's instructions on Roche LC 96 system. The expression level of the target miRNA was normalized to expression of small nucleolar RNA 202 (sno202) and that of target mRNA was normalized to the expression of GAPDH. Each miRNA or mRNA was amplified in triplicates, and Ct values were recorded. Fold change in the miRNA or mRNA expression in DRGS isolated from tumor-bearing mice over corresponding Sham samples in triplicate samples was calculated using DDCT method,19 which measures the relative change in expression of a miRNA (or gene) from treatment to control compared with the reference small RNA (or gene). All miRNA and mRNA assays were purchased from Applied Biosystems, and Assay IDs are as follows: snoRNA202: 001232; miR‐34c‐5p: 000428; GAPDH: 4352932E; Oprd1 Mm00443063_m1; Oprm1: Mm01188089_m1; Cacna1e: Mm0049444_m1; and Prx6: mm00440591_m1.
2.6. Cloning of Cacna1e 3′UTR into luciferase reporter vector
A primer set (5′-GCTAGCGACACGGAAGAAGACGATAAGT-3′ and 5′-TTATTAGGAGGCAGTAGGAAAC-3′) was designed to amplify partial 3′UTR of Cacna1e (NM_009782.3). The first strand cDNA was prepared from total RNA isolated from mouse (C57BL/6j) DRG (High-Capacity cDNA Reverse Transcription kit, Applied Biosystems, 436881), and PCR was conducted with 0.5 µL of cDNA and 2× Phusion Flash high fidelity master mix (Thermo Fisher, Darmstadt, Germany) with following PCR conditions: 98°C for 1 minute; 95°C for 15 seconds, 61°C for 30 seconds, 72°C for 1 minute for 35 cycles; and 72°C for 3 minutes. Amplicons were resolved on 1% agarose gel in TAE buffer, and an expected single band of the size 2.76 kb was observed. Amplified DNA was purified (QIAquick PCR Purification Kit; QIAGEN, Hilden, Germany), ligated into the cloning vector (Zero Blunt TOPO PCR Cloning Kit; Invitrogen, Darmstadt, Germany), and further used to transform chemical competent cells (One Shot TOP10 E. coli; Invitrogen). Colonies were picked randomly and inoculated into 5 mL LB medium for miniprep. Clones were sequenced for verifying the identity (GATC Biotech, Konstanz, Germany). Correct recombinant vector was double-digested with restriction enzymes XbaI and Sac I to release the sequence of interest, and the released DNA was further collected by gel purification. In the meantime, the empty luciferase reporting vector (pmirGLO Dual-Luciferase miRNA Target Expression Vector, Promega, Mannheim, Germany) was treated with XbaI I and Sac I enzymes to generate appropriate ends for directional cloning. The ligation reaction was prepared with partial cacna1e 3′UTR and the linearized reporter vector, and the reaction product was used to transform chemical competent cells. Positive clones were verified using restriction digestion with ECORI. For the generation of a reporter construct containing a mutated binding site for miR-34c-5p in the 3′ UTR of Cacna1e, the above-cloned reporter vector was PCR amplified by using following forward and reverse primer sets in which mutated miRNA-binding site was incorporated. In the following primer sequence, the gray highlighted region represents the miR-34c-binding site, and the bold text represents mutated nucleotides.
Forward: 5′-CGCGTACATAGTCCTGCCTCTTTGCTGGGGAAA-3′
Reverse: 5′-GTACGCGCCCATGTTGCAAAGGGAAATAATCCA-3′
2.7. HEK293 cell culture and luciferase assay
HEK293 cells were cultured in Dulbecco's modified Eagle's medium (21969, Gibco) containing 10% fetal bovine serum (FBS) (Gibco, 10270), 200 units/mL of Penicillin and 200 μg/mL of Streptomycin (15140, Gibco). Approximately 2.5 × 104 cells were plated into each well of 96 wells and one day later cotransfected with 1, 5, 10 nM of either miR-34c-mimic or nontargeting control Mirdian mimic together with 300 ng of Cav2.3 reporter or mutant Cav2.3 reporter vectors into each well of 96-well plate using Lipofectamine 2000 (0.5 μL/well, Invitrogen) following manufacturer's instructions. Forty-eight hours later, luciferase activities were quantified using Dual-Glo Luciferase Assay kit (Promega E2920) and normalized to the respective control experiment. Firefly luminescence signals were normalized to Renilla Luciferase signals.
2.8. Western blotting analysis
Western blots were performed by following standard immunoblotting protocols on the protein lysates isolated from either mouse lumbar 3 and 4 DRGs isolated from the mice injected with adeno-associated viral (AAV) or lentivirus (LV) or the sensory neuronal cultures treated with miR-34c-5p specific or nontargeting mimic. Following polyacrylamide gel electrophoresis and protein transfer onto the nitrocellulose membrane, the blots were probed with anti-cav2.3 at 1:500 dilution (Alomone Labs, Jerusalem, Israel) or monoclonal Anti-β-Tubulin Isotype III antibody (1: 2500 dilution, 5076, Sigma, Taufkirschen, Germany) as loading control and anti-Rabbit HRP (1:2500, sigma A0545) secondary antibody, and signals were developed using Amersham ECL (GE Healthcare, Freiburg, Germany) and Hyperfilm MP (Amersham, GE Healthcare).
2.9. miRNA fluorescent in situ hybridization for miRNA combined with immunofluorescence for protein marker
LNA-based 5′ and 3′ DIG-labeled miR-34c-5p specific (38542-15) or scrambled (99004-15) probes were purchased from Exiqon, Denmark. All the reagents were prepared in RNase-free buffers, and experimental areas and tools were maintained RNAse-free. Mice were transcardially perfused with ice-cold PBS and 4% cold paraformaldehyde (PFA) and lumbar 3 and 4 DRGs were extracted, kept for 24 hours each in 4% PFA and 0.5 M sucrose before cryosectioning at 13 μm thickness. Slides with DRG sections were dried for 1 hour before proceeding with the ISH protocol. The slides were then washed with following reagents: 10 minutes with 4% PFA, 3× 5 minutes in 1× PBS, and 10 minutes in acetylation buffer (containing 2.33 mL triethanolamine and 500 μL acetic anhydride and rest DEPC water for 200 mL of acetylation buffer, freshly prepared before use). During the acetylation step, a hybridization buffer (Exiqon) containing miR-34c-5p specific or negative control ISH probe (25 pmol) was heated at 65°C for 5 minutes and immediately chilled on ice. The hybridization buffer was added to the slides and covered with hybrislips (GBL714022 Sigma) placed in humidified chamber, and the chamber was placed in an incubator overnight at 53°C. Next day, hybrislips were removed by adding 5× SSC buffer and washed 2× 30 minutes with 50% formamide in 1× SSC containing 0.1% tween-20 and incubated at the same temperature as the hybridization temperature. The slides were then washed for 15 minutes with 0.2× SSC and 2× 15 minutes with 1× PBS at room temperature (RT). Blocking solution (containing 0.5% blocking reagent [Roche # 1096176], 10% goat serum heat-inactivated at 70°C for 30 minutes and 0.1% Tween in 1× PBS) was added to each slide and incubated for 1 hour at RT. Anti-DIG-POD (11207733910; Roche Diagnostics, Mannheim, Germany) antibody 1:100 in blocking solution was added to each slide and incubated overnight at 4°C. For combining immunofluorescence (IF) with fluorescence in situ hybridization (FISH) protocol, required primary antibody was added into the same blocking solution. Slides were washed 3× 10 minutes with 1× PBS at RT and incubated with required secondary antibody in blocking buffer for 1 hour at RT. Following secondary antibody incubation, slides were washed 2× 10 minutes with phosphate buffer saline with Tween20 (PBST). For amplification and visualization of FISH signals, Cy3.5 standard from Cy3.5-TSA kit (NEL763001KT, Perkin Elmer, Germany) was diluted 1:100 in the provided diluent buffer, added to the slides and incubated at RT for 10 minutes. Slides were washed for 2× 10 minutes with PBST, followed by a wash with PBST containing DAPI (1:10,000 dilution). Slides were then washed for 10 minutes with PBST and 2× 10 minutes with PBS before mounting with Mowiol. Primary antibodies used for IF are Guinea pig anti-PGP9.5 (1:100 dilution, 14104, Neuromics, Edina, MN), Guinea pig anti-HCN1 (1:100, Alomone Labs, AGP203) rabbit anti-Cav2.3 antibody (1:80, Alomone Labs, ACC-006), Biotinylated-Isolectin B4 (1:100; B-1205, Vector, Burlingame, CA), Guinea pig Substance P (1:150; Neuromics GP14103), Anti-GFAP (1:500; NeuroMab clone N206A/8, UC Davis, Davis, CA) and Chicken anti-NF200 (1:500; Neuromics CH23015). In the experiments to investigate the specificity of Cav2.3 antibody, the Cav2.3 antibody was incubated with its blocking peptide at 1:10 v/v ratio in the blocking buffer for 30 min at 37° C before adding to the slide. Secondary antibodies used were 1:200 Streptavidin, coupled with Alexa 647 (S21374 Invitrogen), 1:500 anti-chicken coupled with Alexa 647 (A-21449; Thermoscientific), 1:500 anti-rabbit coupled with Alexa 488 (Thermoscientific 11034), 1:500 anti-mouse coupled with Alexa 488 (Thermoscientific, R37114) and 1:500 anti-guinea pig coupled with Alexa 647 (Thermoscientific 21450). Images were acquired using a confocal laser-scanning microscope (Leica TCS SP8 AOBS, Wetzlar, Germany) and analyzed with Fiji-ImageJ software.
2.10. Cav2.3 shRNA cloning and selection of potent shRNA
A set of 3 mission shRNAs were obtained against the Cav2.3 (Gene ID: 12290) coding region (Cat. No. TR500244, Sigma-Aldrich), with each shRNA sequence cloned by standard methods into the AAV backbone peptidyl glycine alpha amidating monooxygenase (PAM) vector along with the U6 promoter. The expression of the native green fluorescent protein (GFP) reporter cassette in the final clones was confirmed by the strong GFP signal following transfection into HEK293 cells. shRNA sequences were as following (the highlighted sequence represents the target binding site):
In order to identify the shRNA with best knockdown efficiency among 3 shRNAs, all 3 Cav2.3-shRNAs or Scr-shRNA were cloned into AAV backbone vector together with the GFP reporter gene. shRNA vectors were transfected into DRG cultures using the DC100 program with the Lonza 4D-Nucleofector X-unit system. At 72 hours after transfection, transfection efficiency was identified to be approximately 25% with the help of GFP expression. Thirty GFP-positive cells were isolated from each transfection with the help of a patch pipette and used for the qRTPCR analysis of Cav2.3 expressions. Cells were lysed with Taqman gene expression cells to Ct kit (AM1729; Ambion), and cDNA synthesis and qRTPCR was performed according to manufacturer's instructions. Following qRTPCR analyses, Cav2.3-shRNA-2 was identified to have approximately 100% knockdown efficiency. Because we analyzed only those neurons into which the shRNA-vector was introduced, it is not surprising to see virtually no Cav2.3 signal from those cells.
2.11. Recombinant adeno-associated virus carrying shRNA against Cav2.3 and lentivirus carrying miR-34c-5p specific mimic
The recombinant adeno-associated virus serotype 2/8 particles carrying shRNA against Cav2.3 (AAV-Cav2.3-shRNA) or a scrambled shRNA (AAV-Scr-shRNA) were generated in-house by co-transfecting the cloned recombinant adeno-associated virus backbone plasmid, and the helper 2/8 plasmids into HEK293 cells following standard protocols as previously described.18 The AAVs carrying scrambled control shRNA or Cav2.3-shRNA-2 are referred as AAV-Scr-shRNA and AAV-Cav2.3-shRNA, respectively, in the article.
Lentivirus carrying nontargeting mimic (S05-005000-01) or miR-34c pre-miRNA (VSM6215-213639165) were purchased from GE Healthcare Europe GmbH, Freiburg, Germany.
2.12. Intra ganglionic injections
Injection of adeno-associated virus (AAV) or LV was performed by adapting the protocol reported previously.36,52 Briefly, mice were deeply anesthetized by intraperitoneal injection of Fentanyl/domitor/dormicum mix in 4:6:16 (vol/vol/vol) ratio at 0.7 mL/g b.w. Unilateral lumbar 3 and 4 DRGs were exposed following a laminectomy and 500 nL of AAV or LV, mixed with Fast Green (Sigma-Aldrich, F7258) at < 1% concentration, was injected into each DRG using a 35 G glass needle at a rate of 16.6 nL/min. The opening of the bone was filled with an absorbable haemostatic gelatin sponge (Curaspon, CS-010, Curamedical BV, Assendelft, the Netherlands), and the skin opening was sutured. Mice were maintained at 37°C temperature until they recovered from anesthesia. At the end of the surgery, analgesic (Rimadyl, Pfizer) diluted in saline at 1:1000 dilution was injected intraperitoneally at a dose of 10 mL/Kg body weight for 3 days. Following postoperative recovery, the mice were housed at standard conditions for at least 3 W before performing behavior experiments. At the end of behavioral experiments, mice were killed with CO2, DRGs injected with AAVs were quickly collected, flash-frozen in liquid nitrogen and stored at −80°C until further experiments.
2.13. Data analysis and statistical analyses
All data are expressed as SEM. Two‐way repeated measures analysis of variance (ANOVA) followed by Bonferroni post hoc test was used to assess statistical significance in behavioral experiments. Analysis of variance followed by post hoc Fischer test was used to assess statistical significance in all other experiments. Changes with P ≤ 0.05 were considered to be statistically significant.
3. Results
3.1. Validated and predicted targets for miR-34c-5p
In order to understand the nature of the genes which are already validated as targets for miR-34c-5p, we started our analysis with the complete list of miR-34c-5p validated targets. We retrieved the list of all validated targets for miR-34c-5p from the latest version of miRTarBase, an online repository for archiving validated mRNA targets for all known miRNAs.12 To verify the extent of miR-34c-5p targets across different species, we first compiled the complete list without applying any species filter. This list resulted in a total of 49 unique validated targets, out of which 2 are from the human system and the rest from mouse (Suppl. Table 1, available online at http://links.lww.com/PAIN/A430). Out of 47 validated targets from mouse, 12 were disregarded after consulting the original publications because of incorrect annotation in the miRTarBase repository (highlighted in red in Suppl. Table 1, available online at http://links.lww.com/PAIN/A430). Out of 35 remaining validated targets for miR-34c-5p, 34 were validated in either cardiac, pulmonary, gonadal, or developmental models, making them less likely to be relevant in pain modulation in sensory neurons. However, one target namely phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 (Prex2) is validated in the mouse brain glioblastoma model via HITS-CLIP analyses, marking it as a potential target for miR-34c-5p in the context of sensory modulation. To test this hypothesis, we checked for the change in Prex2 expression in DRGs following tumor cell implantation in the calcaneus bone. qRTPCR analysis revealed that the expression of Prex2 was increased by 2-fold in DRGs isolated from tumor-bearing mice at 8 days following tumor induction as compared to that of the sham control group (Fig. 1, panel A, P > 0.05 as compared to sham group, ANOVA followed by post hoc Fischer's test), suggesting Prex2 as a pronociceptive gene. Furthermore, Prex2 expression was not changed in sensory neurons in the presence of miR-34c-5p specific inhibitor as compared to mismatch inhibitor transfected controls (Fig. 1, panel A, P ≤ 0.05 as compared to the sham group, ANOVA followed by post hoc Fischer test). Taken these observations together, it is evident that there was no canonical pairing between miR-34c-5p and Prex2 in sensory neurons in the context of cancer pain.
Figure 1.
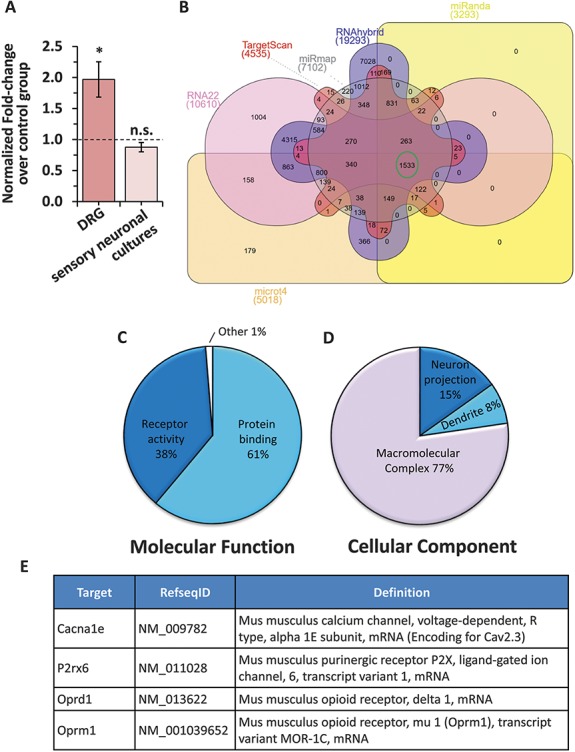
Analyses of previously validated and predicted targets for miR-34c-5p. (A) Change in the expression of Prex2, one of the previously validated targets for miR-34c-5p in DRGs isolated from tumor-bearing mice as compared to sham controls (dark colored bar) or in sensory neurons transfected with a miR-3c-5p specific inhibitor as compared to scramble-inhibitor transfected controls (light colored bar). (B) Venn diagram representation of predicted targets for miR-34c-5p by 6 different target prediction algorithms. The number of genes consistently predicted as targets for miR-34c-5p by all programs is highlighted in a green circle. (C, D) Pie-chart representation of significantly enriched (Adj. P ≤ 0.05 calculated by the multiple test adjustment method as compared to number of reference genes in the category genome-wide) molecular function (C) and cellular component (D) Gene ontology terms by unique genes from the list of 1533 genes commonly predicted as targets for miR-34c-5p by 6 independent algorithms. (E) Annotation details for 4 genes prioritized as putative targets for miR-34c-5p in sensory neurons.
3.2. Predicted targets for miR-34c-5p
Thus, after excluding validated targets of miR-34c-5p as a potential functional pair of miR-34C-5p in sensory neurons, we set out to identify potential mRNA targets for miR-34c-5p by taking a comprehensive approach. We started with identifying putative mRNA targets for miR-34c-5p by using standard prediction algorithms available. As each prediction algorithm resulted in a huge list of mRNAs to be predicted as targets for miR-34c-5p (Fig. 1, panel B and Suppl. Table 2, available online at http://links.lww.com/PAIN/A430), we concentrated our further analyses on 1533 genes, which were consistently predicted as putative mRNA targets for miR-34c-5p by 6 widely used miRNA target prediction algorithms namely TargetScan,3 Miranda,8 miRmap,24 RNA22,34 RNAhybrid,47 and microT446 (Fig. 1, panel B, highlighted in green circle). In order to identify the biological relevance of the predicted targets, we performed system-level bioinformatics analyses by taking those 1533 enriched predictions as a template. Performing Gene ontology enrichment analysis associated with each GO term by taking unique genes from the template list revealed that the majority of predicted targets of miR-34c-5p belong to the protein binding (61%) and the receptor activity (38%) category (Fig. 1, panel C). In the same lines, the cellular component analysis revealed that the majority out of the enriched list of miR-34c-5p predicted targets belong to the macromolecular complex. Interestingly, significantly enriched cellular component terms identified from the same gene list included 'dendrite' and 'neuronal projection,' suggesting an intimate association of miR-34c-5p with neuronal functions (Fig. 1, panel D).
Pathway enrichment analysis of the enriched list of 1533 predicted targets for miR-34c-5p revealed that components of several biological pathways were significantly enriched (Suppl. Table 3, available online at http://links.lww.com/PAIN/A430). Interestingly, the majority of them belong to cancer-relevant pathways supporting the well-studied tumor-suppressive role of miR-34c-5p. However, other pathways such as calcium, MAPK, chemokine, wnt, and VEGF signaling pathways which are well known to be closely involved in pain modulatory mechanisms,20,31,53–55,58,59 or neuronal pathways such as amyotrophic lateral sclerosis, axon guidance, long-term potentiation, neuroactive ligand–receptor interaction were also significantly enriched, suggesting involvement of miR-34c-5p in pain modulation, potentially via these pathways. On one hand, miR-34c-5p is known to be a pronociceptive miRNA7 and on the other hand, it is well documented that miRNAs predominantly function by reducing the target gene expression.61 In this light, we hypothesize that miR-34c-5p is exerting its pronociceptive functions via an antinociceptive target. Therefore, in the next step, we investigated potential pathways that would explain this canonical pairing between miR-34c-5p and its target. Upon careful investigation, it was observed that the calcium signaling pathway is one of the significantly enriched (Suppl. Table 3, http://links.lww.com/PAIN/A430, adj. P = 0.0028 calculated by the multiple test adjustment method as compared to a number of reference genes in the category genome-wide) pathways among the enriched list of miR-34c-5p putative targets. Interestingly, 21 of the calcium signaling pathway components happened to be within the enriched list of miR-34c-5p putative targets (Suppl. Fig. 1, available online at http://links.lww.com/PAIN/A431) and suggest a potential involvement of miR-34c-5p in regulating the calcium signaling pathway. Another such pathway significantly enriched in the list of enriched miR-34c-5p targets was the 'Neuroactive ligand-receptor interaction' pathway (Suppl. Table 3 [available online at http://links.lww.com/PAIN/A430], adj. P < 0.0043 calculated by the multiple test adjustment method as compared to a number of reference genes in the category genome- and Suppl. Fig. 2, available online at http://links.lww.com/PAIN/A431). We then identified key members of these 2 pathways, namely: calcium channel, voltage-dependent, R type, alpha 1E subunit (Cacna1e, encoding Cav2.3), purinergic receptor P2X, ligand-gated ion channel, 6, transcript variant 1 (P2rx6), opioid receptor, delta 1 (oprd1) and opioid receptor, mu 1, transcript variant MOR-1C (Oprm1) (Fig. 1, panel E), which might explain canonical pairing and pronociceptive mechanisms of miR-34c-5p. Thus, by starting with a huge list of predicted targets and by implementing constructive exclusion criteria and combining with available biological knowledge, we prioritized 4 candidate mRNAs to be potential targets for miR-34c-5p.
3.3. miR-34c-5p and Cacna1e are a functional pair
In the next steps, we sought to investigate the functional relation between miR-34c-5p and each of 4 prioritized targets. In cultured sensory neurons, we transfected either specific miR-34c-5p mimic or nontargeting mimic. Expression analysis with quantitative real-time PCR (qRTPCR) revealed that miR-34c-5p expression was increased by more than 20-fold following miR-34c-5p mimic transfection as compared to nontargeting mimic transfected neurons (Fig. 2, panel A, P ≤ 0.05 as compared to control group, ANOVA followed by post hoc Fischer test). In the same experiment, we then checked for the change in the expression of 4 prioritized targets of miR-34c-5p. qRTPCR analyses revealed that the expression of Cav2.3 and Oprm1 was significantly reduced, expression of P2rx6 was significantly increased (Fig. 2, panel A, P ≤ 0.05 as compared to control group, ANOVA followed by post hoc Fischer's test), and that of oprd1 was unchanged in the presence of miR-34c-5p. In the next step, we performed a complementary experiment where miR-34c-5p expression was reduced in sensory neuronal cultures by transfecting with miR-34c-5p specific inhibitor. As shown in Figure 2 panel B, expression of miR-34c-5p was significantly reduced to 0.2-fold following miR-34c-5p inhibitor transfection as compared to scrambled miR-35c-5p inhibitor (Fig. 2, panel B, P ≤ 0.05 as compared to the sham group, ANOVA followed by post hoc Fischer's test). Analysing the expression of 4 putative targets in the presence of miR-34-5p inhibition revealed that the expression of Cav2.3 and P2rx6 was significantly increased, expression of oprd1 was significantly reduced (Fig. 2, panel B, P ≤ 0.05 as compared to control group, ANOVA followed by post hoc Fischer's test), and that of oprm1 was unchanged. Taken the results from these 2 complementary experiments together, it is evident that the expression of P2rx6, oprd1, or oprm1 was not in classical reciprocal relation with the expression of miR-34c-5p. However, expression of Cav2.3 was significantly and inversely changed as compared to the expression of miR-34c-5p in sensory neuronal cultures providing the first level of evidence for canonical pairing between miR-34c-5p and Cav2.3 in sensory neurons.
Figure 2.
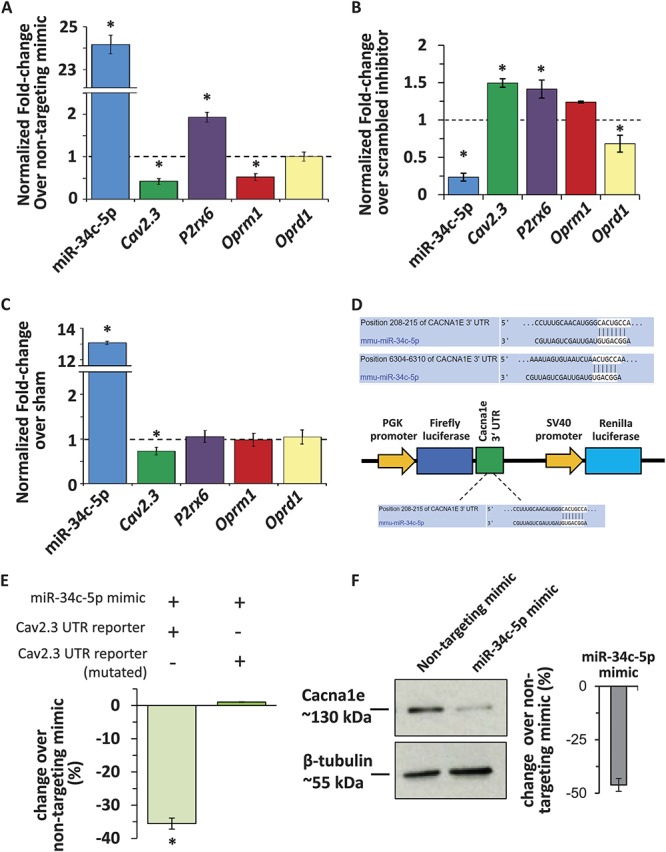
Analyses of prioritized putative targets for miR-3c-5p in sensory neurons isolated from tumor-bearing mice. (A) qRTPCR analyses demonstrating the change in the expression of miR-34c-5p and 4 of its putative targets in cultured sensory neurons in the presence of miR-34c-5p specific mimic as compared to nontargeting mimic transfected controls. (B) qRTPCR analyses demonstrating the change in the expression of miR-34c-5p and 4 of its putative targets in cultured sensory neurons in the presence of miR-34c-5p specific inhibitor as compared to scrambled nontargeting inhibitor-transfected controls. (C) qRTPCR analyses demonstrating the change in the expression of miR-34c-5p and 4 of its putative targets in the DRGs isolated from tumor-bearing mice as compared to sham controls. In panels A-C, *denotes P ≤ 0.05 as compared to control group, analysis of variance followed by post hoc Fischer test from at least 3 biological replicate experiments. The dotted line represents expression levels in the control group, bars above the line represent upregulation and below the line represent downregulation of tested transcripts. (D) Representation of miR-34c-5p binding sites in the 3′UTR of Cav2.3 and strategy for the cloning of 3′UTR sequence into the dual luciferase vector. (E) Luciferase reporter assay in HEK293 cells demonstrating changes in the translation of the Cav2.3 gene in the presence of intact or mutated binding sites for miR-34c-5p and following induction of miR‐34c‐5p expression via specific mimic. (F) Representative western blot analysis images and their quantification for Cacna1e or β-tubulin protein expression in the lysates of cultured DRG neuronal cells following transfection with control (non-targeting mimic) or miR-34c-5p specific mimic. *Denotes P ≤ 0.05 as compared to nontargeting mimic group, analysis of variance followed by post hoc Fischer test, n = 3 independent experiments.
In the next step, to investigate the functional relationship between miR-34c-5p and its 4 prioritized targets in cancer states, we tested for the change in the expression of 4 prioritized predicted targets in sensory neurons in tumor conditions in vivo. qRTPCR analysis revealed that the expression of miR-34c-5p was more than 10-fold higher in the DRGs isolated from tumor-bearing mice as compared to sham mice (Fig. 2, panel C, P ≤ 0.05 as compared to sham group, ANOVA followed by post hoc Fischer test), which is in accordance with our previous report.7 In the same tissue samples, analysis of 4 prioritized putative targets revealed that the expression of Cav2.3 was significantly reduced (Fig. 2, panel C, P ≤ 0.05 as compared to the sham group, ANOVA followed by post hoc Fischer test), while that of P2rx6, oprm1, or oprd1 was unchanged. This observed upregulation of miR-34c-5p and downregulation of Cav2.3 in sensory neurons in vivo, when tumor-induced hyperalgesia is significantly present,7 provided a second level of evidence for canonical pairing between miR-34c-5p and Cav2.3 and their potential involvement in bone metastasis–induced pain.
In the next set of experiments, we focused on Cav2.3 and asked whether miR-34c-5p is able to directly bind the 3′ UTR of Cav2.3 and regulate the translation. Analysis of the 3′ UTR of Cav2.3 revealed that it is 7681 bp in length and has 2 potential binding sites for the seed region of miR-34c-5p (Fig. 2, panel D). Owing to its size and envisioned difficulties in cloning, we chose to clone first 2500 bp of the 3′UTR, which harbors the classical 7 bp binding site or mutated binding site for miR-34c-5p, into the dual Luciferase reporter construct under the Luciferase promoter (Fig. 2, panel D). The Cav2.3 UTR reporter constructs were transfected into HEK293 cells together with the miR-34c-5p specific mimic or nontargeting mimic, and change in the translated Luciferase protein levels was measured 48 hours after transfection in the form of luminescence signals in an enzymatic assay. Analysis of the results revealed that the luminescence signals from the HEK293 cells transfected with the UTR reporter construct were significantly less as compared to nontargeting mimic transfected cells, and there was no effect of miR-34c-5p mimic on the luminescence signals from the HEK293 cells transfected with the reporter construct containing mutated seed regions (Fig. 2, panel E). We then tested the impact of miR-34c-5p overexpression on the expression of Cacna1e protein in the cultured sensory neurons by following the same protocol explained above for Figure 2, panel A. Immunoblot analyses in the presence of miR-34c-5p specific or control mimic revealed that the expression of Cav2.3 protein is significantly less in the sensory neurons expressing miR-34c-5p specific mimic as compared to the cells transfected with nontargeting mimic (Fig. 2, panel F). These observations provided a third level and conclusive proof for functional binding between miR-34c-5p and Cav2.3.
3.4. mir-34c-5p and Cacne1e are coexpressed in nociceptive neurons
Having thus confirmed inverse regulation and functional relation between miR-34c-5p and Cav2.3, we studied the cellular localization of these 2 entities in sensory neurons in vivo by FISH-IF protocol. Analysis using miR-34c-5p specific or scrambled ISH probes on DRGs isolated from wild-type mice revealed that miR-34c-5p specific signals could be detected in DRG cells, while there was no detectable signal from the negative control probe (Fig. 3, panel A). In order to investigate Cav2.3 expression, we first confirmed the specificity of previously used anti-Cav2.3 antibody49 in the IF staining by preincubating the tissue sections with blocking peptide and by performing secondary antibody only control staining (Suppl. Fig. 3, available online at http://links.lww.com/PAIN/A431). Analysis revealed that Cav2.3 specific signals were observed in the cytoplasm and cell membrane of DRG cells of all sizes, suggesting its ubiquitous expression profile in peripheral sensory neurons in vivo (Fig. 3, panel A). Furthermore, the signals from both miR-34c-5p and Cav2.3 are colocalized in the DRG cells (Fig. 3, panel A), supporting their functional association observed in above-explained experiments. Further colocalization experiments for miR-34c-5p using previously characterized IB4-binding,7 antisubstance P antibody36 or neurofilament 200 antibodies25,33 revealed that 17.8% of miR-34c-5p expressing neurons were isolectin-B4-binding nonpeptidergic nociceptors, 24% were substance-p positive peptidergic nociceptors, and 19% were NF200-positive large diameter sensory neurons (Fig. 3, panels B-E). In next experiments, we investigated the Cav2.3 expression in neuronal and nonneuronal cells using previously characterized anti-PGP9.5 and anti-GFAP antibodies, respectively, and observed that Cav2.3 is expressed in majority of neurons (88%) and few GFAP-positive satellite cells (11%) in DRG (Fig. 4, panels A-C). We further analyzed membrane localization of Cav2.3 by colabeling with a previously characterized antibody against HCN1 (hyperpolarization-activated cyclic nucleotide-gated)44 protein which is expressed in DRGs and localized to the membrane of sensory neuronal cells.2 Analyses of single plane confocal images revealed colocalization of HCN1 and Cav2.3 specific signals suggesting membrane expression of Cav2.3 (Fig. 4, panel C). We then performed more labeling studies to identify the extent of Cav2.3 expression in neuronal subpopulations in DRGs isolated from WT mice. Confocal analyses of colocalization and quantification revealed that 18% of Cav2.3 expressing neurons were isolectin-B4-binding nonpeptidergic nociceptors (Fig. 5, panels A and D), 11% were substance-p positive peptidergic nociceptors (Fig. 5, panels B and D), and 17% were NF200-positive large diameter sensory neurons (Fig. 5, panels C and D).
Figure 3.
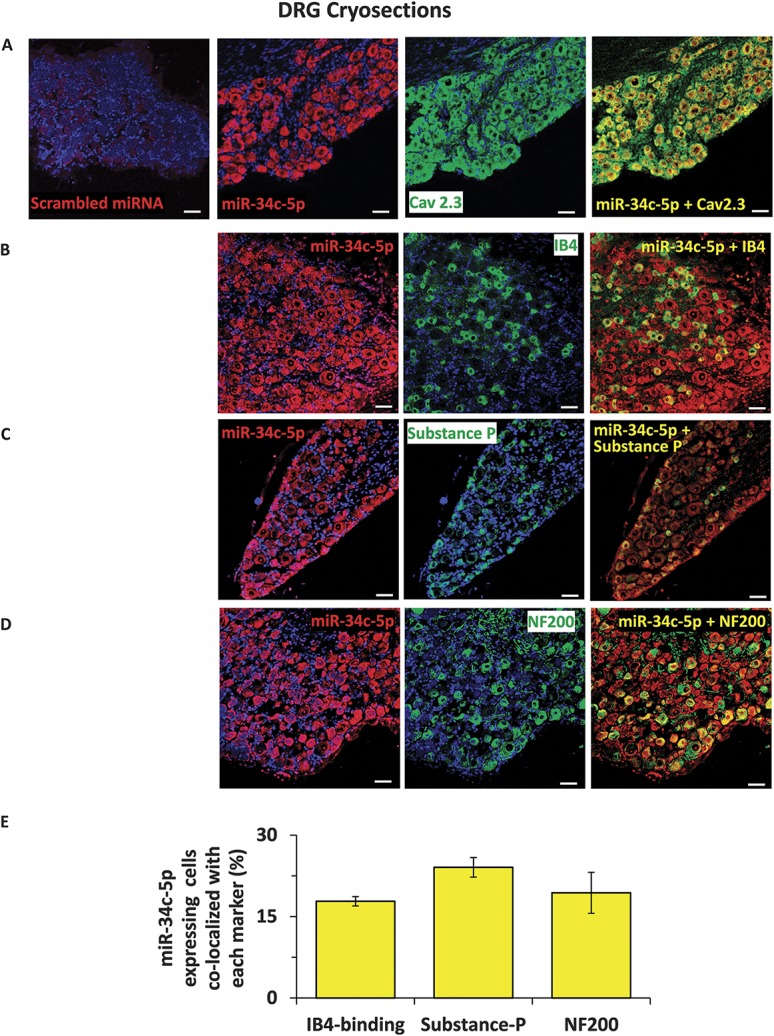
Analyses of cellular expression of miR-34c-5p in dorsal root ganglia in vivo. Representative images to demonstrate fluorescent in situ hybridization (FISH) analysis of miR-34c-5p expression in mouse DRG with specific or negative control probes (A) and immunofluorescence analysis of colabeling with its mRNA target Cav2.3 (A), isolectin‐B4‐binding (IB4) nonpeptidergic nociceptors (B), substance P‐positive peptidergic nociceptors (C) and NF200-positive large diameter sensory neurons (D) in mouse DRG. Cell nuclei were counterstained with DAPI. Quantification of coexpression of each neuronal subtype with the miR-34c-5p expressing neurons is shown in panel E. Scale bars represent 50 µm in all panels. Tissue samples from 3 independent mice were analyzed.
Figure 4.
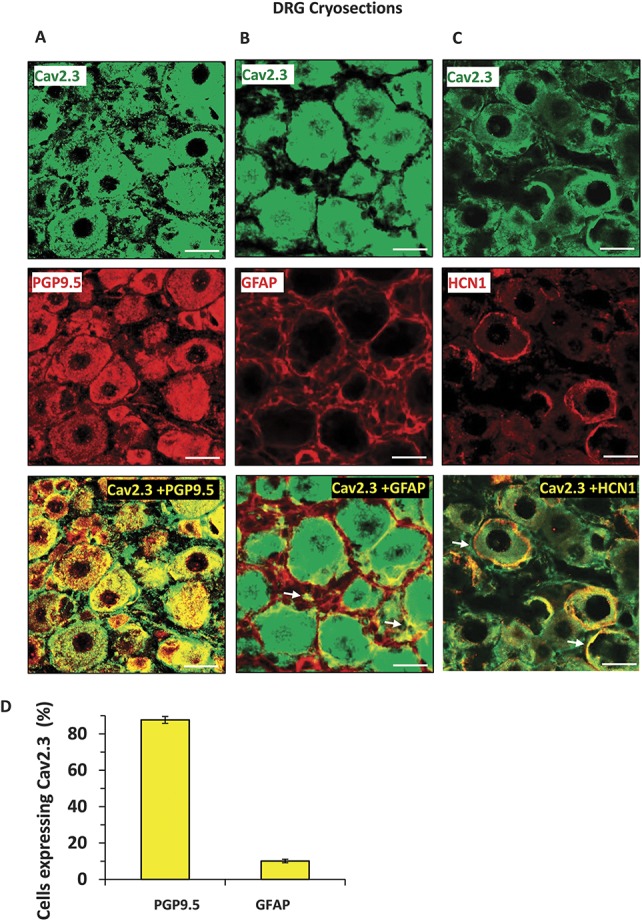
Analyses of expression of the Cav2.3 protein in neuronal and nonneuronal cells or in cell membrane of the cells in dorsal root ganglia in vivo. Representative images to demonstrate expression of Cav2.3 in mouse DRG and colabeling with PGP9.5-positive neuronal cells (A), GFAP-positive satellite cells (B) and with HCN1 (hyperpolarization-activated cyclic nucleotide-gated) channel in the cell membrane (C). Observed colocalization is highlighted with white arrows. Quantification of coexpression of each neuronal subtype with the Cav2.3 expressing neurons is shown in panel D. Scale bars represent 50 µm in all panels. Tissue samples from 3 independent mice were analyzed.
Figure 5.
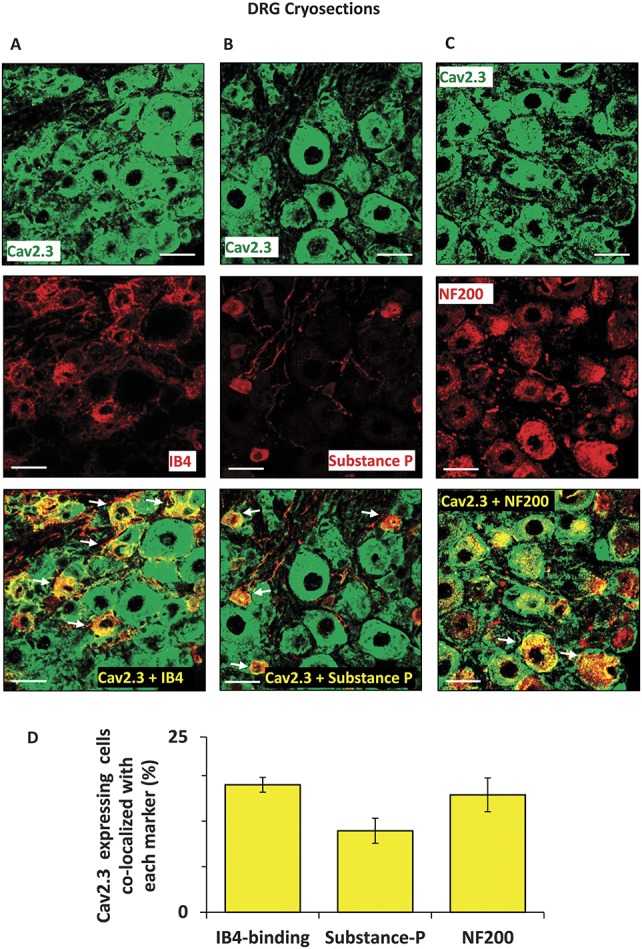
Analyses of cellular expression of the Cav2.3 protein in dorsal root ganglia in vivo. Representative images to demonstrate expression of Cav2.3 in mouse DRG and colabeling with isolectin‐B4‐binding (IB4) nonpeptidergic nociceptors (A), substance P‐positive peptidergic nociceptors (B) and NF200-positive large diameter sensory neurons (C) in mouse DRG. Observed colocalization is highlighted with white arrows. Quantification of coexpression of each neuronal subtype with the Cav2.3 expressing neurons is shown in panel D. Scale bars represent 50 μm in all panels. Tissue samples from 3 independent mice were analyzed.
3.5. Cav2.3 functions as an antinociceptive transcript
After conclusively establishing the synchronous and inverse change in the expression levels of miR-34c-5p and Cav2.3 in sensory neurons, we next asked whether Cav2.3 or miR-34c-5p alone is sufficient to modulate sensitivity in basal conditions. To test the impact of Cav2.3 on the mediation of mechanical sensitivity, we designed 3 independent shRNA sequences targeting different regions of the coding sequence of Cav2.3 and cloned into a dual-promoter AAV expression vector in which expression of shRNA and GFP was driven by U6 and CBA promoters, respectively. After selecting the shRNA sequence with best knockdown efficiency against Cav2.3 (Suppl. Fig. 4, available online at http://links.lww.com/PAIN/A431), AAVs carrying either shRNA against Cav2.3 (AAV-Cav2.3-shRNA) or scrambled shRNA (AAV-Scr-shRNA) were generated. The AAVs were then injected into lumbar 3 and 4 DRGs of 2 groups of WT mice by following intraganglionic injection procedure previously described.36 Behavioral analyses revealed that the withdrawal frequency to a range of calibrated Von Frey filaments was significantly higher in the group of mice injected with AAV-Cav2.3-shRNA at 3W following viral injection as compared to the sensitivity observed before viral injection (basal) or to the group of mice injected with control AAV-Scr-shRNA virus (Fig. 6, panel A, P ≤ 0.05 as compared to basal readings, 2-way ANOVA of repeated measures followed by the Bonferroni multiple comparisons post hoc test, n = 8 mice per group). Analyses of mechanical response threshold (Fig. 6, panel B) also revealed the same results. There was no change in the hypersensitivity in the paws contralateral to DRGs injected with either AAV-Scr-shRNA or AAV-Cav2.3-shRNA (Suppl. Fig. 6, panel A, available online at http://links.lww.com/PAIN/A431). At the end of the behavioral experiments, lumbar DRGs were collected to analyze the change in Cav2.3 protein expression following intraganglionic injections of AAV-shRNA and to confirm knockdown of Cav2.3 using previously characterized antibody against Cav2.3 protein.49 Western blot analyses revealed that there were 2 specific bands for Cav2.3 protein corresponding to approximately 252 and 130 kDa, and both of them were almost undetectable in the AAV-Cav2.3-shRNA injected group as compared to that in the AAV-Scr-shRNA injected group (Fig. 6, panel C). The quantification Cav2.3 protein–specific signals revealed the reduction of 95% in the Cav2.3-shRNA injected group as compared to the AAV-Scr-shRNA injected group (Fig. 6, panel C), confirming shRNA-mediated potent knockdown of Cav2.3 in vivo in DRGs.
Figure 6.
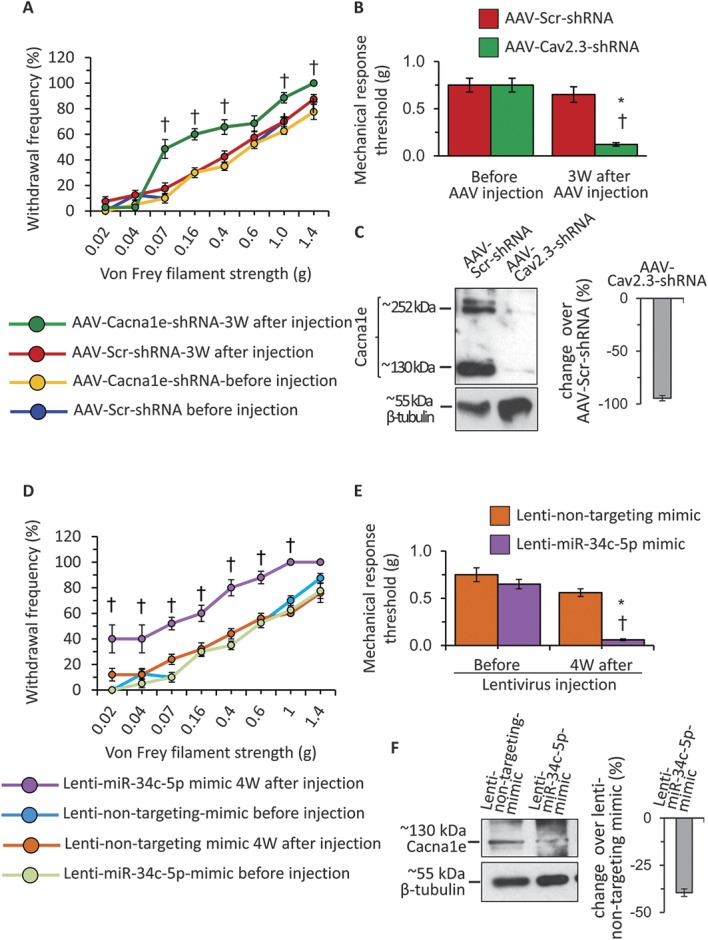
Analyses of functional contribution of Cav2.3 and miR-34c-5p in the mediation of mechanical sensitivity in wild-type mice. (A) Change in the frequency of paw withdrawal to the plantar application of graded von Frey filament forces of different strength in ipsilateral paws following intraganglionic injection of AAVs carrying either scrambled shRNA (AAV-Scr-shRNA) or shRNA directed against coding sequence of Cav2.3 (AAV-Cav2.3-shRNA), measured before and at 3W after viral injections. (B) Mechanical response thresholds calculated as von Frey filament strength required to achieve 60% withdrawal frequency in ipsilateral paws following intraganglionic injection of AAV-Scr-shRNA or AAV-Cav2.3-shRNA, measured before and at 3W following viral injection. (C) Representative western blot analysis images and their quantification for Cacna1e or β-tubulin protein expression in the DRG tissue lysates following intraganglionic injection of AAV-Scr-shRNA or AAV-Cav2.3-shRNA. (D) Change in the frequency of paw withdrawal to the plantar application of graded von Frey filament forces of different strength in ipsilateral paws following intraganglionic injection of lentivirus carrying either nontargeting miRNA mimic (Lenti-nontargeting-mimic) miR-34c-5p specific mimic (Lenti-miR-34c-mimic), measured before and at 4W after viral injections. (E) Mechanical response thresholds calculated as von Frey filament strength required to achieve 60% withdrawal frequency in ipsilateral paws following intraganglionic injection of Lenti-nontargeting-mimic or Lenti-miR-34c-mimic, measured before and at 3W following viral injection. (F) Representative western blot analysis images and their quantification for Cacna1e or β-tubulin protein expression in the DRG tissue lysates following intraganglionic injection of Lenti-nontargeting-mimic or Lenti-miR-34c-mimic. In panels A, B, D, and E, *P ≤ 0.05 as compared to basal readings and †denotes P ≤ 0.05 as compared to a corresponding data point in other 3 groups, 2-way analysis of variance of repeated measures followed by Bonferroni multiple comparisons post hoc test, n = 8 mice per group. In panels C and F, *P ≤ 0.05 as compared to control group, analysis of variance followed by post hoc Fischer test, n = 5 independent experiments.
In order to test the impact of miR-34c-5p overexpression on the basal hypersensitivity in WT mice, we procured LV expressing modified pre-miRNA sequences for the miR-34c, which will facilitate preferential incorporation of the miR-34c-5p by the RNA-induced silencing complex (RISC) and subsequent degradation of miR-34c-3p, and corresponding control nontargeting mimic. After measuring the basal mechanical sensitivity, lentivirions were injected directly into the lumbar 3 and 4 DRGs of WT mice, and the mechanical sensitivity was measured at 4W following viral injections. Behavioral analyses revealed that the withdrawal frequency to a range of calibrated Von Frey filaments was significantly higher in the group of mice injected with Lenti-miR34c-5p-mimic at 4W following viral injection as compared to the sensitivity observed before viral injection (basal) or to the group of mice injected with control Lenti-nontargeting-mimic (Fig. 6, panel D, P ≤ 0.05 as compared to basal readings, 2-way ANOVA of repeated measures followed by Bonferroni multiple comparisons post hoc test, n = 8 mice per group). Analyses of mechanical response threshold revealed the same results (Fig. 6, panel E), and analyses of behavioral data from the paw contralateral to the LV injection revealed no difference among groups (Suppl. Fig. 6, panel B, available online at http://links.lww.com/PAIN/A431). At the end of behavioral analyses, DRGs injected with LV were collected and analyzed for the change in Cav2.3 protein expression by immunoblotting. Quantification of Cav2.3 specific expression after normalizing to β-tubulin loading control revealed that there was significant and 40% reduction in the Cav2.3 isoform corresponding to ∼130 kDa size as compared to the lenti-nontargeting-mimic injected group (Fig. 6 panel F, P ≤ 0.05 as compared to control group, ANOVA followed by post hoc Fischer test, n = 5 mice per group).
4. Discussion
Our understanding of miRNA-mediated mechanisms underlying cancer pain is still in its infancy. Most of the studies addressing the role of miRNAs in chronic pain conditions are confined to reporting expression or their altered regulation but fail to investigate their target level mechanisms. The current study aims at demonstrating an extended experimental pipeline to identify a functional miRNA-mRNA pair for a key pronociceptive miRNA in the context of cancer pain. In this study, we, therefore, adopted a comprehensive approach combining in silico, in vitro, and in vivo analyses to tackle this issue and identified (1) miR-34c-5p and Cav2.3 as a novel functional pair in cancer pain modulation and (2) an antinociceptive role for Cav2.3 containing Ca2+ channels in peripheral sensory neurons.
4.1. mRNA targets for miR-34c-5p
One of the current biggest challenges in miRNA research is to interpret biological relevance from “putative target predictions” usually containing tens of candidate genes. We previously reported a successful strategy for miR-1a-3p, in which expression change of all 62 enriched predictions was investigated by qRTPCR to identify Clcn3 as a novel functional pair for miR-1a-3p.7 However, this strategy is difficult to implement in the cases where even the prioritized list of predicted targets contains several hundreds of candidate genes. For miR-34c-5p, 6 independent target prediction algorithms consistently predicted 1533 genes as putative targets. Therefore, here, we analyzed in silico observations in the light of the literature on key genes associated with pain to further narrow down the list of enriched predictions for functional analyses. By performing complementary in silico experiments, we identified that components of calcium, MAPK, chemokine, Wnt, VEGF signaling, and neuroactive ligand-receptor interaction pathways, which have a well-established role in pain, are enriched among those 1533 predicted targets.7,20,31,53–55,59 However, owing to well characterized pronociceptive properties for key components of those signaling pathways, it is less likely that they constitute direct canonical targets for miR-34c-5p, which itself is a pronociceptive miRNA. On close observation, it is identified that the majority members of calcium signaling and neuroactive ligand-receptor interaction pathways have binding sites for miR-34c-5p in their 3′ UTR (Suppl. Table 3, available online at http://links.lww.com/PAIN/A430). Of 21 such members of the calcium signaling pathway, Cacna1e has 2 conserved binding sites for the seed region of miR-34c-5p, whereas others have one binding site (Fig. 2), suggesting a potentially stronger functional association between Cav2.3 and miR-34c-5p. Therefore, we prioritized Cav2.3 as a potential target for miR-34c-5p. Adapting the same strategy, we prioritized P2rx6, oprd1, and Oprm1 as potential binding partners for miR-34c-5p from the neuroactive ligand-receptor interaction pathway. Expression analyses of 4 prioritized targets together with miR-34c-5p in DRGs isolated from tumor-bearing mice or in sensory neuronal cultures in the presence of a miR-34c-5p specific inhibitor or its mimic consistently confirmed reciprocal regulation of miR-34c-5p and Cav2.3 but not of the other 3 candidates. Luciferase reporter assay further confirmed functional binding between miR-34c-5p and Cav2.3. It is interesting that despite highly stringent bioinformatics predictions, 3 of 4 prioritized mRNAs were not regulated at mRNA level in sensory neurons in cancer conditions. Each gene can be regulated by more than one miRNA, and several miRNAs are regulated in sensory neurons in cancer pain conditions.7 Therefore, it is possible that even though P2rx6, oprd1, and Oprm1 are targets of miR-34c-5p, regulation by other endogenous miRNAs would have resulted in the neutralization of miR-34c-5p–mediated effects on these targets. The potential impact of miR-34c-5p on those targets via translational repression will have to be addressed in future studies. Our results indicate the importance of context-dependent changes in a miRNA and its targets when analyzing miRNA-mRNA functional interactions. Thus, by implementing state-of-the-art in silico analyses and stringent expression analyses combined with constructive exclusion criteria in each step, starting with 21,801 of unique genes as putative targets for miR-34c-5p, the current study identified Cav2.3 as a novel and bona fide functional target for miR-34c-5p in sensory neurons.
Interestingly, none of the previously validated targets for miR-34c-5p were confirmed in sensory neurons, which was also true for miR-1a-3p,7 underscoring context-specific miRNA actions in sensory neurons. Here, we identified Prex2 to be upregulated in sensory neurons innervating tumor-affected areas. Prex2 is a Rho guanine exchange factor for Rac14 and is expressed in neurons.15 While the expression of Prex2 has not been reported in DRG previously, its effector Rac1 is expressed in DRG neurons and regulates axonal growth dynamics.23,50 The structural reorganization is one of the important hallmarks in cancer pain states,28,51 and neuronal Rac GEFs thus hold immense potential in modulating this process.35
4.2. Cav2.3 functions as an antinociceptive calcium channel in peripheral sensory neurons
The most important finding of the current study was the identification of mRNA encoding Cav2.3 as a key target for miR-34c and the observations of antinociceptive properties of Cav2.3 in sensory neurons. Cav2.3 is encoded by Cacna1e, expressed in neuronal and endocrine tissues27,29,45,43,64 and constitutes the principle pore-forming subunit of R-type Ca2+ currents.13 Expression of 2 of 6 Cav2.3 splice variants has been reported in the DRG via RTPCR,16,17 and here we demonstrate expression of the Cav2.3 protein in the DRG. Interestingly, although, previous studies reported that Cav2.3 expression is unaffected in DRGs following chronic constriction injury, axotomy-induced neuropathy,26 or in the neuropathic phase of diabetes,64 we observed a significant reduction in the expression of Cav2.3 in sensory neurons corresponding to tumor-affected areas. Because neuropathic pain is an integral component of cancer pain,9,38,39 these observations imply a context-dependent modulation of Cav2.3 and suggest a selective involvement in specific mechanisms involving tumor-nerve interactions.
The mode of action of Cav2.3-containing Ca2+ channels is not well understood, and its potential functional role in modulation of pain has been discussed controversially. For instance, one study reported that intrathecal application of specific R-type calcium channel blocker SNX-482 increased formalin-induced pain behavior in the early phase but reduced it in the late phase,43 while another study reported attenuation of behavior in both phases in the same experimental paradigm.57 Furthermore, spinally expressed SNX-482 sensitive channels were suggested to function as antinociceptive channels in neuropathic pain conditions.40 Mice lacking Cav2.3 globally showed an unaltered response to distinct pain stimuli in basal conditions but develop reduced pain behavior in the second phase of formalin test.49 In this study, with conditional deletion of Cav2.3 specifically in DRGs via shRNA targeting or miR-34c-5p overexpression, we observed that mice showed exaggerated responses to mechanical stimuli. In the light of the previous literature on Cav2.3 functions, it is intriguing that our experiments suggest an antinociceptive role for Ca2+ channels containing Cav2.3, which is contrary to the lack of phenotype reported in global Cav2.3 KO mice under basal conditions.49 A likely explanation for this apparent discrepancy is that Cav2.3 is absent throughout the somatosensory axis in global knockout mice, which may have neutralized site-specific functions of Cav2.3 in peripheral sensory neurons. Our results suggest a specific antinociceptive role for Ca2+ channels containing Cav2.3 located on peripheral afferents, and future studies should reveal more functional insights into the underlying mechanisms.
Thus, this study demonstrates how functions of miRNAs identified in open-ended genome-wide screens can be elucidated in the context of a specific pain disorder. Starting with stringent bioinformatics, we demonstrate how the excessively long lists of potential mRNA targets of individual miRNAs can be narrowed down by incorporating expression analyses, promoter function analyses to discover true miRNA-mRNA interactions, and finally by functionally validating in the specific site in the nociceptive pathway in vivo. We identify Cav2.3 as a target of miRNA regulation in cancer pain, and our studies imply the therapeutic potential for this antinociceptive channel.
Conflict of interest statement
The authors have no conflict of interest to declare.
K. K. Bali is supported by a fellowship from the Medical Faculty of Heidelberg University. R. Kuner is a principal investigator in the Excellence Cluster CellNetworks, Heidelberg University. The research leading to these results has received funding from an ERC Advanced Investigator Grant (294293) from the European Research Council, by funding from the European Seventh Framework Programme (FP7/2007-2013) under grant agreement number 602133 and by funding from the Baden-Württemberg Stiftung under the project number BWST_NCRNA-037 to RK. J. Gandla was partially supported by a fellowship from the Hartmut Hoffmann-Berling International Graduate School for Molecular and Cellular Biology.
Supplementary Material
Acknowledgements
The authors thank Rose LeFaucheur for secretarial help and Dunja Baumgartl-Ahlert and Karin Meyer for technical assistance. The authors acknowledge support from the Interdisciplinary Neurobehavioral Core (INBC) for the behavioral experiments performed in this study.
Author contributions: J. Gandla performed the major portion of experiments and analyzed the data. S. K. Lomada and J. Lu contributed to experiments and performed data analysis. R. Kuner designed and supervised the study. K. K. Bali designed, performed some experiments, analyzed data and wrote the manuscript.
Appendix A. Supplemental Digital Content
Supplemental digital content associated with this article can be found online at http://links.lww.com/PAIN/A430 and http://links.lww.com/PAIN/A431.
Footnotes
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Web site (www.painjournalonline.com).
References
- [1].Achari C, Winslow S, Ceder Y, Larsson C. Expression of miR-34c induces G2/M cell cycle arrest in breast cancer cells. BMC Cancer 2014;14:538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Acosta C, McMullan S, Djouhri L, Gao L, Watkins R, Berry C, Dempsey K, Lawson SN. HCN1 and HCN2 in Rat DRG neurons: levels in nociceptors and non-nociceptors, NT3-dependence and influence of CFA-induced skin inflammation on HCN2 and NT3 expression. PLoS One 2012;7:e50442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015;4:e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Bae Y, Yang T, Zeng HC, Campeau PM, Chen Y, Bertin T, Dawson BC, Munivez E, Tao J, Lee BH. miRNA-34c regulates Notch signaling during bone development. Hum Mol Genet 2012;21:2991–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bali KK, Hackenberg M, Lubin A, Kuner R, Devor M. Sources of individual variability: miRNAs that predispose to neuropathic pain identified using genome-wide sequencing. Mol Pain 2014;10:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bali KK, Kuner R. Noncoding RNAs: key molecules in understanding and treating pain. Trends Mol Med 2014;20:437–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bali KK, Selvaraj D, Satagopam VP, Lu J, Schneider R, Kuner R. Genome-wide identification and functional analyses of microRNA signatures associated with cancer pain. EMBO Mol Med 2013;5:1740–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol 2010;11:R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Bloom AP, Jimenez-Andrade JM, Taylor RN, Castaneda-Corral G, Kaczmarska MJ, Freeman KT, Coughlin KA, Ghilardi JR, Kuskowski MA, Mantyh PW. Breast cancer-induced bone remodeling, skeletal pain, and sprouting of sensory nerve fibers. J Pain 2011;12:698–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Borda AP, Charnay-Sonnek F, Fonteyne V, Papaioannou EG. Guide-lines on pain management and palliative care. Arnhem, Netherlands: European Association of Urology (EAU), 2013. [Google Scholar]
- [11].Cain DM, Wacnik PW, Eikmeier L, Beitz A, Wilcox GL, Simone DA. Functional interactions between tumor and peripheral nerve in a model of cancer pain in the mouse. Pain Med 2001;2:15–23. [DOI] [PubMed] [Google Scholar]
- [12].Chou CH, Chang NW, Shrestha S, Hsu SD, Lin YL, Lee WH, Yang CD, Hong HC, Wei TY, Tu SJ, Tsai TR, Ho SY, Jian TY, Wu HY, Chen PR, Lin NC, Huang HT, Yang TL, Pai CY, Tai CS, Chen WL, Huang CY, Liu CC, Weng SL, Liao KW, Hsu WL, Huang HD. miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res 2016;44:D239–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Dietrich D, Kirschstein T, Kukley M, Pereverzev A, von der Brelie C, Schneider T, Beck H. Functional specialization of presynaptic Cav2.3 Ca2+ channels. Neuron 2003;39:483–96. [DOI] [PubMed] [Google Scholar]
- [14].Donald S, Hill K, Lecureuil C, Barnouin R, Krugmann S, John coadwell W, Andrews SR, Walker SA, Hawkins PT, Stephens LR, Welch HC. P-Rex2, a new guanine-nucleotide exchange factor for rac. FEBS Lett 2004;572:172–6. [DOI] [PubMed] [Google Scholar]
- [15].Donald S, Humby T, Fyfe I, Segonds-Pichon A, Walker SA, Andrews SR, Coadwell WJ, Emson P, Wilkinson LS, Welch HC. P-Rex2 regulates Purkinje cell dendrite morphology and motor coordination. Proc Natl Acad Sci USA 2008;105:4483–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Fang Z, Hwang JH, Kim JS, Jung SJ, Oh SB. R-type calcium channel isoform in rat dorsal root ganglion neurons. Korean J Physiol Pharmacol 2010;14:45–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Fang Z, Park CK, Li HY, Kim HY, Park SH, Jung SJ, Kim JS, Monteil A, Oh SB, Miller RJ. Molecular basis of Cav2.3 calcium channels in rat nociceptive neurons. J Biol Chem 2007;282:4757–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Florez-Paz D, Bali KK, Kuner R, Gomis A. A critical role for Piezo2 channels in the mechanotransduction of mouse proprioceptive neurons. Scientific Rep 2016;6:25923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Fu WJ, Hu J, Spencer T, Carroll R, Wu G. Statistical models in assessing fold change of gene expression in real-time RT-PCR experiments. Comput Biol Chem 2006;30:21–6. [DOI] [PubMed] [Google Scholar]
- [20].Hagenston AM, Simonetti M. Neuronal calcium signaling in chronic pain. Cell Tissue Res 2014;357:407–26. [DOI] [PubMed] [Google Scholar]
- [21].Hagman Z, Haflidadottir BS, Ansari M, Persson M, Bjartell A, Edsjo A, Ceder Y. The tumour suppressor miR-34c targets MET in prostate cancer cells. Br J Cancer 2013;109:1271–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hagman Z, Larne O, Edsjo A, Bjartell A, Ehrnstrom RA, Ulmert D, Lilja H, Ceder Y. miR-34c is downregulated in prostate cancer and exerts tumor suppressive functions. Int J Cancer 2010;127:2768–76. [DOI] [PubMed] [Google Scholar]
- [23].Hansel DE, Quinones ME, Ronnett GV, Eipper BA. Kalirin, a GDP/GTP exchange factor of the Dbl family, is localized to nerve, muscle, and endocrine tissue during embryonic rat development. J Histochem Cytochem 2001;49:833–44. [DOI] [PubMed] [Google Scholar]
- [24].Hsu SD, Chu CH, Tsou AP, Chen SJ, Chen HC, Hsu PW, Wong YH, Chen YH, Chen GH, Huang HD. miRNAMap 2.0: genomic maps of microRNAs in metazoan genomes. Nucleic Acids Res 2008;36(Database Issue):D165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Johnson-Kerner BL, Ahmad FS, Diaz AG, Greene JP, Gray SJ, Samulski RJ, Chung WK, Van Coster R, Maertens P, Noggle SA, Henderson CE, Wichterle H. Intermediate filament protein accumulation in motor neurons derived from giant axonal neuropathy iPSCs rescued by restoration of gigaxonin. Hum Mol Genet 2015;24:1420–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kim DS, Choi JO, Rim HD, Cho HJ. Downregulation of voltage-gated potassium channel alpha gene expression in dorsal root ganglia following chronic constriction injury of the rat sciatic nerve. Brain Res Mol Brain Res 2002;105:146–52. [DOI] [PubMed] [Google Scholar]
- [27].Kubota M, Murakoshi T, Saegusa H, Kazuno A, Zong S, Hu Q, Noda T, Tanabe T. Intact LTP and fear memory but impaired spatial memory in mice lacking Ca(v)2.3 (alpha(IE)) channel. Biochem Biophys Res Commun 2001;282:242–8. [DOI] [PubMed] [Google Scholar]
- [28].Kuner R. Central mechanisms of pathological pain. Nat Med 2010;16:1258–66. [DOI] [PubMed] [Google Scholar]
- [29].Lee SC, Choi S, Lee T, Kim HL, Chin H, Shin HS. Molecular basis of R-type calcium channels in central amygdala neurons of the mouse. Proc Natl Acad Sci USA 2002;99:3276–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Li Y, Jia Z, Zhang L, Wang J, Yin G. Caspase-2 and microRNA34a/c regulate lidocaine-induced dorsal root ganglia apoptosis in vitro. Eur J Pharmacol 2015;767:61–6. [DOI] [PubMed] [Google Scholar]
- [31].Lin X, Wang M, Zhang J, Xu R. p38 MAPK: a potential target of chronic pain. Curr Med Chem 2014;21:4405–18. [DOI] [PubMed] [Google Scholar]
- [32].Liu WM, Pang RT, Chiu PC, Wong BP, Lao K, Lee KF, Yeung WS. Sperm-borne microRNA-34c is required for the first cleavage division in mouse. Proc Natl Acad Sci USA 2012;109:490–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Liu Y, Kelamangalath L, Kim H, Han SB, Tang X, Zhai J, Hong JW, Lin S, Son YJ, Smith GM. NT-3 promotes proprioceptive axon regeneration when combined with activation of the mTor intrinsic growth pathway but not with reduction of myelin extrinsic inhibitors. Exp Neurol 2016;283:73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Loher P, Rigoutsos I. Interactive exploration of RNA22 microRNA target predictions. Bioinformatics 2012;28:3322–3. [DOI] [PubMed] [Google Scholar]
- [35].Lu J, Luo C, Bali KK, Xie RG, Mains RE, Eipper BA, Kuner R. A role for Kalirin-7 in nociceptive sensitization via activity-dependent modulation of spinal synapses. Nat Commun 2015;6:6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Luo C, Gangadharan V, Bali KK, Xie RG, Agarwal N, Kurejova M, Tappe-Theodor A, Tegeder I, Feil S, Lewin G, Polgar E, Todd AJ, Schlossmann J, Hofmann F, Liu DL, Hu SJ, Feil R, Kuner T, Kuner R. Presynaptically localized cyclic GMP-dependent protein kinase 1 is a key determinant of spinal synaptic potentiation and pain hypersensitivity. PLoS Biol 2012;10:e1001283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Malmevik J, Petri R, Knauff P, Brattas PL, Akerblom M, Jakobsson J. Distinct cognitive effects and underlying transcriptome changes upon inhibition of individual miRNAs in hippocampal neurons. Scientific Rep 2016;6:19879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mantyh P. Bone cancer pain: causes, consequences, and therapeutic opportunities. PAIN 2013;154(suppl 1):S54–62. [DOI] [PubMed] [Google Scholar]
- [39].Mantyh PW. Cancer pain and its impact on diagnosis, survival and quality of life. Nat Rev Neurosci 2006;7:797–809. [DOI] [PubMed] [Google Scholar]
- [40].Matthews EA, Bee LA, Stephens GJ, Dickenson AH. The Cav2.3 calcium channel antagonist SNX-482 reduces dorsal horn neuronal responses in a rat model of chronic neuropathic pain. Eur J Neurosci 2007;25:3561–9. [DOI] [PubMed] [Google Scholar]
- [41].McDonald MK, Ajit SK. MicroRNA biology and pain. Prog Mol Biol Transl Sci 2015;131:215–49. [DOI] [PubMed] [Google Scholar]
- [42].Misso G, Di Martino MT, De Rosa G, Farooqi AA, Lombardi A, Campani V, Zarone MR, Gulla A, Tagliaferri P, Tassone P, Caraglia M. Mir-34: a new weapon against cancer? Mol Ther Nucleic Acids 2014;3:e194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Murakami M, Suzuki T, Nakagawasai O, Murakami H, Murakami S, Esashi A, Taniguchi R, Yanagisawa T, Tan-No K, Miyoshi I, Sasano H, Tadano T. Distribution of various calcium channel alpha(1) subunits in murine DRG neurons and antinociceptive effect of omega-conotoxin SVIB in mice. Brain Res 2001;903:231–6. [DOI] [PubMed] [Google Scholar]
- [44].Notomi T, Shigemoto R. Immunohistochemical localization of Ih channel subunits, HCN1-4, in the rat brain. J Comp Neurol 2004;471:241–76. [DOI] [PubMed] [Google Scholar]
- [45].Osanai M, Saegusa H, Kazuno AA, Nagayama S, Hu Q, Zong S, Murakoshi T, Tanabe T. Altered cerebellar function in mice lacking CaV2.3 Ca2+ channel. Biochem Biophys Res Commun 2006;344:920–5. [DOI] [PubMed] [Google Scholar]
- [46].Paraskevopoulou MD, Georgakilas G, Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, Filippidis C, Dalamagas T, Hatzigeorgiou AG. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res 2013;41(Web Server Issue):W169–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA 2004;10:1507–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Rokavec M, Li H, Jiang L, Hermeking H. The p53/miR-34 axis in development and disease. J Mol Cell Biol 2014;6:214–30. [DOI] [PubMed] [Google Scholar]
- [49].Saegusa H, Kurihara T, Zong S, Minowa O, Kazuno A, Han W, Matsuda Y, Yamanaka H, Osanai M, Noda T, Tanabe T. Altered pain responses in mice lacking alpha 1E subunit of the voltage-dependent Ca2+ channel. Proc Natl Acad Sci USA 2000;97:6132–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Sayyad WA, Fabris P, Torre V. The role of Rac1 in the growth cone dynamics and force generation of DRG neurons. PLoS One 2016;11:e0146842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Schmidt BL, Hamamoto DT, Simone DA, Wilcox GL. Mechanism of cancer pain. Mol Interv 2010;10:164–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Schweizerhof M, Stosser S, Kurejova M, Njoo C, Gangadharan V, Agarwal N, Schmelz M, Bali KK, Michalski CW, Brugger S, Dickenson A, Simone DA, Kuner R. Hematopoietic colony-stimulating factors mediate tumor-nerve interactions and bone cancer pain. Nat Med 2009;15:802–7. [DOI] [PubMed] [Google Scholar]
- [53].Selvaraj D, Gangadharan V, Michalski CW, Kurejova M, Stosser S, Srivastava K, Schweizerhof M, Waltenberger J, Ferrara N, Heppenstall P, Shibuya M, Augustin HG, Kuner R. A functional role for VEGFR1 expressed in peripheral sensory neurons in cancer pain. Cancer Cell 2015;27:780–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Simonetti M, Agarwal N, Stosser S, Bali KK, Karaulanov E, Kamble R, Pospisilova B, Kurejova M, Birchmeier W, Niehrs C, Heppenstall P, Kuner R. Wnt-Fzd signaling sensitizes peripheral sensory neurons via distinct noncanonical pathways. Neuron 2014;83:104–21. [DOI] [PubMed] [Google Scholar]
- [55].Simonetti M, Hagenston AM, Vardeh D, Freitag HE, Mauceri D, Lu J, Satagopam VP, Schneider R, Costigan M, Bading H, Kuner R. Nuclear calcium signaling in spinal neurons drives a genomic program required for persistent inflammatory pain. Neuron 2013;77:43–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Stewart BW, Wild CP. World Cancer Report 2014. France: International Agency for Research on Cancer 2014 and WHO, 2014. [PubMed] [Google Scholar]
- [57].Terashima T, Xu Q, Yamaguchi S, Yaksh TL. Intrathecal P/Q- and R-type calcium channel blockade of spinal substance P release and c-Fos expression. Neuropharmacology 2013;75:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Tsuda M. Microglia in the spinal cord and neuropathic pain. J Diabetes Investig 2016;7:17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Tsuda M, Inoue K. Neuron-microglia interaction by purinergic signaling in neuropathic pain following neurodegeneration. Neuropharmacology 2016;104:76–81. [DOI] [PubMed] [Google Scholar]
- [60].Turabi A, Plunkett AR. The application of genomic and molecular data in the treatment of chronic cancer pain. J Surg Oncol 2012;105:494–501. [DOI] [PubMed] [Google Scholar]
- [61].Vidigal JA, Ventura A. The biological functions of miRNAs: lessons from in vivo studies. Trends Cell Biol 2015;25:137–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res 2013;41(Web Server Issue):W77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Wang R, Ma J, Wu Q, Xia J, Miele L, Sarkar FH, Wang Z. Functional role of miR-34 family in human cancer. Curr Drug Targets 2013;14:1185–91. [DOI] [PubMed] [Google Scholar]
- [64].Yusaf SP, Goodman J, Gonzalez IM, Bramwell S, Pinnock RD, Dixon AK, Lee K. Streptozocin-induced neuropathy is associated with altered expression of voltage-gated calcium channel subunit mRNAs in rat dorsal root ganglion neurones. Biochem Biophys Res Commun 2001;289:402–6. [DOI] [PubMed] [Google Scholar]
- [65].Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res 2005;33(Web Server Issue):W741–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Zovoilis A, Agbemenyah HY, Agis-Balboa RC, Stilling RM, Edbauer D, Rao P, Farinelli L, Delalle I, Schmitt A, Falkai P, Bahari-Javan S, Burkhardt S, Sananbenesi F, Fischer A. microRNA-34c is a novel target to treat dementias. EMBO J 2011;30:4299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


