Abstract
Liddle’s Syndrome (LS) is considered a rare Mendelian hypertension. We have previously described three reportedly unrelated families, native of an Italian area around the Strait of Messina, carrying the same mutation (βP617L) of the epithelial sodium channel.
The aims of our study were: 1) to evaluate whether a close genomic relationship exists between the three families through the analysis of mitochondrial DNA (mtDNA) and Y chromosome; 2) to quantify the genomic relatedness between the LS patients belonging to the three families and assess the hypothesis of a mutation shared through Identity By Descent (IBD).
The hypervariable region HVRI of the mtDNA genome and the Y chromosome short tandem repeats profiles were analysed in individuals of the three families. Genotyping 542,585 genome-wide SNPs was performed in all the LS patients of the three families and some of their relatives. A panel of 780 healthy Italian adult samples typed for the same set of markers was used as controls.
Despite different lineages between the three families based on the analysis of mtDNA and Y chromosome, the three probands and their six affected relatives share the same ~5Mbp long haplotype which encompasses the mutant allele. Using an approach based on coalescent theory we estimate that the three families inherited the mutant allele from a common ancestor ~13 generations ago, and that such an ancestor may have left around 20 carriers alive today.
The prevalence of LS in the region of origin of the three families may be much higher than that estimated worldwide.
Keywords: Liddle’s syndrome, epithelial sodium channel, renin, aldosterone, genome-wide genotyping, coalescent theory, evolutionary medicine
Introduction
First described by Grant Liddle in 1963, LS is a hereditary condition, transmitted in an autosomal-dominant mode [1]. Its main clinical features include early onset of salt-sensitive hypertension, increased incidence of premature cardio- and cerebro-vascular events, hypokalaemic metabolic alkalosis, suppression of both renin and aldosterone secretion, and unresponsiveness to mineralocorticoid receptor (MR) antagonists, as opposed to the response to epithelial sodium channel (ENaC) inhibitors (i.e., amiloride and triamterene) [1–3]. Patients with LS are most often diagnosed between 10 to 30 years of age, but diagnosis is sometimes made earlier. LS occurs worldwide, with no apparent ethnic or sex predilection, and is classified by Orphanet as a “rare disease”, with an estimated prevalence of less than 1/106 within the global population (http://www.orpha.net/consor/cgi-bin/OC_Exp.php?Expert=526).
The mutations so far identified in LS cause constitutive activation of ENaC, which is composed of three homologous subunits, α, β and γ, sharing around 30-40% sequence identity. Each subunit is encoded by a specific gene (SCNN1A, SCNN1B, and SCNN1G, respectively) containing 13 exons. SCNN1A is located on 12p13.31, while SCNN1G and SCNN1B reside within a common 400-kb fragment on human chromosome 16p13-p12. Each ENaC subunit consists of two transmembrane domains, a large extracellular loop, and short cytoplasmic amino and carboxyl termini [4–6].
ENaC is expressed in the apical membrane of the principal cells of the aldosterone-sensitive-distal nephron (ASDN), which comprises the late distal convolute tubule, the connecting tubule, and the entire collecting duct. ENaC, which is highly selective for Na+ ions and inhibited by amiloride, mediates the rate-limiting step in Na+ absorption in the ASDN. The activity of ENaC depends on channel open probability (Po) and on the number of channels expressed at the apical membrane of ASDN principal cells. Aldosterone, which is the main regulator of ENaC, increases both the number of ENaCs expressed at the apical membrane and the channel Po [7–9].
To our knowledge, twenty-nine LS-causing alleles have been reported to date, occurring in familial or sporadic cases [10–11]. Apart from γENaC N530S and αENaC C479R, which increase the channel Po without changing the cell surface expression of ENaC [11, 12], all the other mutations cluster within exon 13 of either SCNN1B or SCNN1G genes, and are frameshift or nonsense or missense mutations that delete or alter a proline-rich PY motif, corresponding to the sequence PPPXY (where X is any aminoacid) in the cytoplasmic C-terminus of the β or γ subunit. Of the 22 β-subunit mutations, 12 cause deletion of the PY motif, and 10 alter the sequence of the PY motif [13–23].
The PY motif plays a fundamental role in the internalization of ENaCs. Indeed, PY motifs of all the three ENaC subunits are the recognition site for the ubiquitin-protein-ligase Nedd4-2, which interacts with these motifs and ubiquitinates ENaC. Ubiquitination targets the ENaC for internalization and degradation, thus decreasing the number of ENaC expressed in the apical membrane. Mutations causing deletion or alteration of the PY motif prevent Nedd4-2 binding to ENaC. As a consequence, internalization and degradation of ENaC fail to occur, resulting in increased density of ENaC at the apical membrane of the ASDN. In addition to affecting the number of channels at the membrane, LS mutations also increase channel Po [24].
The increased ENaC activity in the apical surface of ASDN leads to a greater Na+ reabsorption, plasma volume expansion, high blood pressure, and consequent decrease in renin and aldosterone secretion. In addition, the increased Na+ reabsorption through ENaC generates a transepithelial lumen-negative voltage that drives K+ secretion across the apical membrane, resulting in hypokalaemic metabolic alkalosis [8].
Here we analyze three previously reported families affected by LS caused by the same allele (βENaC P617L). The three families were native of a limited area of Italy, consisting of the two neighbouring Provinces of Messina (Sicily) and Reggio Calabria (Calabria) located on the shores of the Messina Strait, and were reportedly unrelated. Considering the rarity of LS, the lack of other reports of the βP617L mutation, and the restricted geographical area of origin shared by the three families, we checked the hypothesis that the three kindreds were carrying a mutation identical by descent. We therefore investigated, using genome-wide analyses, whether the LS-causing allele shared by the three probands and some of their relatives is a consequence of recurrent mutation or identity by descent (IBD) and, in the latter case, how many affected individuals should be expected, based on coalescent theory and forward simulation.
Indeed, estimating the number of generations that separate reportedly unrelated patients, has important consequences on the prediction of the number of additional affected individuals yet to be diagnosed in a given population source. Previously, successful strategies were deployed to apply evolutionary inference to tackle this problem [25, 26]. Particularly, approaches based on coalescent theory were adopted, where the convergence of the alleles of a given locus to their common ancestor from an earlier time is used to make inferences on the evolutionary history of the examined DNA sequence and, hence, on the genetic relationships of the screened probands.
Methods
The data that support the findings of this study are available from the corresponding author upon reasonable request. Scripts from Y.D. are available here: https://github.com/ydiekmann/Pagani_Hypertension_2017
Probands discovery and sampling
Some of us (E.R., E.F., B.C., F.M.) have previously identified a novel allele of the β subunit of ENaC (βP617L) causing LS in a proband whose paternal line was strongly suspected of carrying the allele. Expression of this ENaC variant in Xenopus oocytes caused a three-fold increase in amiloride-sensitive Na+ current compared with wild-type channels [15]. Later, the same group of authors found a second family carrying the same allele [27]. Lastly, a third family carrying the βP617L allele was recently identified by some other members of our current group (M.R., A.C., F.N., O.Z.) [28].
All subjects participating in the study provided their informed consent. The study was approved by the Ethics Committees of Santa Maria Nuova Hospital, Reggio Emilia, and MultiMedica Hospital, Sesto San Giovanni, Milan, respectively.
Genome-wide genotyping
Genomic DNA of the following samples (A IV-1, A III-2, A II-3, B V-1, B IV-2, B IV-3, B IV-4, B III-3, C IV-1, C III-2, C III-3, C II-2) (see Figure 1 for their family trees) was quantified with the Quant-iT dsDNA Broad-Range Assay Kit (Invitrogen Life Technologies, Carlsbad, CA, USA) and 200 ng of DNA were used to genotype 542,585 genome-wide SNPs with the CoreExomeChip v.1.1 Array (Illumina, San Diego, CA, USA). A panel of 780 healthy Italian adult samples typed for the same set of markers was used as controls and was already described elsewhere [29]. Control samples showing more than 1% missing genotypes, as well as those presenting discordant genetic/ascertained sex (as revealed by comparison of each individual’s mean homozygosity rate across X-chromosome markers with the expected rate) were removed from subsequent analyses. Moreover, autosomal heterozygosity rate per individual was assessed and subjects with excessive or reduced proportion of heterozygote genotypes (i.e. with values exceeding ± 3 standard deviations from the mean) were filtered out. Further quality checks (PLINK commands –geno 0.1 –irem 0.08) and phasing were then performed on the entire dataset (i.e. including both healthy and disease individuals) to avoid batch effects.
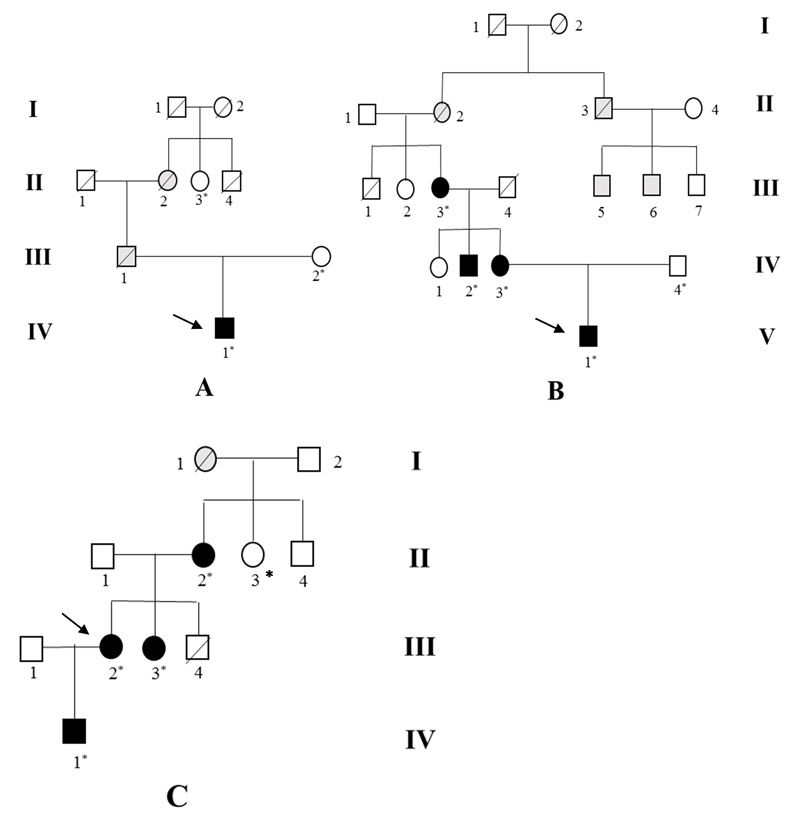
Figure 1.
Pedigrees of the three families (A, B, C) carrying the βENaC P617L mutation. Squares indicate males and circles females. Deceased members are indicated with diagonal lines. The individuals submitted to genetic testing are marked with an asterisk. Individuals with proven βENaC P617L mutation in the genetic analysis are indicated as filled black symbols. Individuals with either early hypertension or premature cardiovascular events suggestive of Liddle syndrome are indicated as filled grey symbols. The probands are marked by arrows.
mtDNA and Y chromosome analyses
The hypervariable region HVRI of the mtDNA genome and the 17 Y chromosome short tandem repeats profiles were analysed in individuals from the three families with LS. Moreover, the complete mitochondrial genomes of two probands (A IV-1 and C IV-1) were sequenced to confirm the haplogroup inferred from the HVSI sequence. The mitochondrial genomes were sequenced with the Ion PGM™ System (Life Technologies, GrandIsland, NY, USA) taking advantage of an optimized protocol [30].
Estimating tMRCA and number of carriers
Firstly, we estimated the time to the most recent common ancestor (tMRCA) of the 4.5Mbp haplotype encompassing the causal mutation and shared by the three probands using an analytical approach based on coalescent theory. The linkage map positions [31] corresponding to the physical chromosomal positions were converted into a recombination rate r per generation using Haldane’s map function
where d is the genetic distance between the markers delimiting the conserved haplotype in Morgan. We set the effective size Ne of the population that the carriers are from to of the census population size for the Italian provinces of Reggio Calabria (census population 2014: 557,993, http://www.tuttitalia.it/calabria/provincia-di-reggio-calabria/statistiche/censimenti-popolazione/) and Messina (census population 2014: 645,296, http://www.tuttitalia.it/sicilia/provincia-di-messina/statistiche/censimenti-popolazione/), however, the tMRCA estimates remain largely constant for all values of Ne > 400 (data not shown). Under the assumptions of coalescent theory—most importantly neutrality—the probability distribution of time to the most recent common ancestor t given the haplotype data D, here |D| = 3, can be expressed as the distribution of the sum (and therefore the convolution) of independent exponential random variables with parameters [32].
Secondly, we estimated the number of carriers today by forward simulations of a Wright-Fisher population. In order to model population growth over the last thousand years, either an exponential f(t; N0, r) = N0rt+1 or a hyper-exponential model f(t; N0, b, r) = N0bbrt−1 was fitted to the historical demographic data of Italy (https://en.wikipedia.org/wiki/Demographics_of_Italy) by least squares minimisation (Figure S1). For fitting, N0 was set to the current census population size of Italy (https://en.wikipedia.org/wiki/Demographics_of_Italy) (time t therefore taking negative values backwards in time) and later scaled down to the current joint census population of Reggio Calabria and Messina. We performed 5 × 105 simulations starting with a single allele at a time point in the past sampled from the distribution of tMRCA established above, multiplied with a generation time sampled uniformly between 25 and 31 years. Finally, we conditioned the resulting distribution of number of carriers to values greater or equal than 3, accounting for the prior knowledge from direct observation of three carriers.
Results
Sequencing analysis had previously detected a heterozygous C to T allele at codon 617 in exon 13 of SCNN1B in A IV-1, B V-1, B IV-2, B IV-3, B III-3, C IV-1, C III-2, C III-3, C II-2. Therefore, screening the three probands’ relatives led to the identification of six more carriers of the same allele. The mutant allele is predicted to replace the second proline with leucine in the sequence 616PPPXY620 (PY motif) of the β subunit (β P617L mutation). Each of the three families was characterized by an early onset of hypertension and premature cardiovascular events in several members across generations, which is consistent with an autosomal-dominant pattern of inheritance (Figure 1).
Age at presentation of hypertension ranged from 11 to 31 years. The time lapse between the ascertainment of hypertension and diagnosis of LS was remarkably variable among our LS patients, from a few years to decades, confirming that LS may be misdiagnosed and undertreated. Severity of hypertension among our LS patients ranged from mild to severe. Plasma potassium levels were low in the probands of family A and C, but completely normal in the proband’s sister of family C and in family B. The proband’s sister of family C (C III-3) was normotensive at the beginning of our research, but developed hypertension during the preparation of our manuscript. Low plasma level of both renin and aldosterone was the only laboratory feature shared by the LS patients belonging to the three families. Treatment with amiloride at a daily dose ranging from 5 to 20 mg resulted in normalization of blood pressure, correction of hypokalaemia or maintenance of normal potassium levels in those with pre-treatment normokalaemia, and increase in plasma levels of renin and aldosterone (Table 1).
Table 1. Clinical characteristics of the examined subjects of the three pedigrees carrying the βP617L mutation.
| Subjects with proven βP617L mutation | A IV-1 | B V-1 | B IV-2 | B IV-3 | B III-3 | C IV-1 | C III-2 | C III-3 | C II-2 |
| Sex | M | M | M | F | F | M | F | F | F |
| Age at genetic analysis, ys | 19 | 21 | 41 | 48 | 71 | 0.1 | 24 | 22 | 44 |
| Age at diagnosis of HT, ys | 16 | 17 | 31 | 18 | 22 | - | 11 | 25 | 19 |
| SBP/DBP before treatment, mmHg | 164/94 | 168/105 | 145/100 | 150/94 | 156/96 | - | 180/134 | 140/100 | 190/130 |
| Post-Tx SBP/DBP, mmHg | 127/70 | 128/84 | - | 122/80 | 130/76 | - | 125/80 | - | 148/94 |
| [K+], mmol/L | 2.8 | 4.6 | 4.7 | 4.3 | 3.9 | 4.5 | 3.0 | 3.8 | - |
| Post-Tx [K+] | 4.3 | 5.2 | - | 4.2 | 4.4 | - | 4.1 | - | - |
| Pre-Tx PRA, ng/ml/h | 0.1 | 0.3 | 0.3 | 0.8 | 0.8 | - | 0.1 | 0.2 | - |
| Post-Tx PRA, ng/ml/h | 0.8 | 2.5 | - | 6.4 | 2.7 | 3.0 | - | - | |
| Pre-Tx p[Aldo], ng/dL | <1.0 | 3.0 | 1.5 | 1.5 | 3.0 | - | <1.0 | 3.4 | - |
| Post-Tx p[Aldo], ng/dL | 1.5 | 10.0 | - | 7.5 | 6.8 | - | 5.9 | - | - |
HT, hypertension; SBP, systolic blood pressure; DBP, diastolic blood pressure; Tx, treatment with amiloride; PRA, plasma renin activity in sitting position; p[Aldo], plasma aldosterone in sitting position. The patients are indicated as in the Figure 1. B IV-2 declined treatment and follow-up. C IV-1 was newly born at the time of genetic analysis. C III-3 developed hypertension 3 years after the genetic analysis
Comparison of mtDNA and Y chromosome showed different lineages between the lines (either maternal or paternal) carrying the β-P617L mutation (Table 2). However, the probands’ mtDNA haplotypes in families A and C are typical of people of Sicilian-Calabrian descent, suggesting a localized geographic origin in the provinces of Messina (Sicily, Italy) and Reggio Calabria (Calabria, Italy) based also on surname localization (data not shown to protect the anonymity of the probands). We therefore compared the total proportion of shared genome (using the PLINK –genome function) between pairs of affected samples and between affected and control samples to uncover relatedness at the genomic level. The proportion of genome shared by IBD in pairs of affected samples each belonging to a different family was comparable to the proportion of genome shared between these and the healthy controls (no comparison was above the top 1% of the empirical distribution). This confirms that the three LS families are not closely related at a genomic level, consistently with the mtDNA and Y chromosome results.
Table 2. Mitochondrial haplogroups and Y chromosome STR profile in subjects belonging to the three families with LS (see Figure 1).
| Subjects* | mtDNA HVRI haplotype/lineage | Y STRs** haplotype/lineage |
|---|---|---|
| A IV-1 | 16129, 16183, 16189, 16193insC, 16223, 16249, 16311/M1a | 14, 12, 22, 28, 15, 14, 12/14, 13, 10, 11, 22, 11, 11, 16, 10, 20/I1 |
| A III-2 | 16130, 16183, 16189, 16193insC, 16223, 16249, 16311/M1a | |
| A II-3 | 16223, 16270, 16292/W | |
| B V-1 | 16147, 16183, 16189, 16193insC, 16240, 16270/U5b | 15, 13, 23, 30, 17.2, 13, 12/17, 12, 10, 12, 20, 11, 11, 14, 10, 20/J1 |
| B IV-3 | 16148, 16183, 16189, 16193insC, 16240, 16270/U5b | |
| B IV-2 | 16149, 16183, 16189, 16193insC, 16240, 16270/U5b | |
| B III-3 | 16150, 16183, 16189, 16193insC, 16240, 16270/U5b | |
| B IV-4 | rCRs/H | 15, 13, 23, 30, 17.2 13, 12/17, 12, 10, 12, 20, 11, 11, 14, 10, 20/J1 |
| C IV-1 | 146, 152, 247, 489, 769, 813, 825, 1018, 2758, 2885, 3594, 4104, 4312, 6446, 6473, 6671, 6680, 7146, 7256, 7521, 8468, 8655, 10400, 10664, 10688, 10810, 10915, 11257, 11914, 12403, 12950, 13105, 13276, 13506, 13637, 13650, 14110, 14783, 15043, 15301, 16187, 16223, 16230, 16249, 16278/M1a3a | 16, 13, 23, 29, 16, 13, 13-17, 13, 10, 10, 23, 11, 11, 15, 9, 19 |
| C III-2 | 146, 152, 247, 489, 769, 813, 825, 1018, 2758, 2885, 3594, 4104, 4312, 6446, 6473, 6671, 6680, 7146, 7256, 7521, 8468, 8655, 10400, 10664, 10688, 10810, 10915, 11257, 11914, 12403, 12950, 13105, 13276, 13506, 13637, 13650, 14110, 14783, 15043, 15301, 16187, 16223, 16230, 16249, 16278/M1a3a | |
| C III-3 | 146, 152, 247, 489, 769, 813, 825, 1018, 2758, 2885, 3594, 4104, 4312, 6446, 6473, 6671, 6680, 7146, 7256, 7521, 8468, 8655, 10400, 10664, 10688, 10810, 10915, 11257, 11914, 12403, 12950, 13105, 13276, 13506, 13637, 13650, 14110, 14783, 15043, 15301, 16187, 16223, 16230, 16249, 16278/M1a3a | |
| C II-2 | 146, 152, 247, 489, 769, 813, 825, 1018, 2758, 2885, 3594, 4104, 4312, 6446, 6473, 6671, 6680, 7146, 7256, 7521, 8468, 8655, 10400, 10664, 10688, 10810, 10915, 11257, 11914, 12403, 12950, 13105, 13276, 13506, 13637, 13650, 14110, 14783, 15043, 15301, 16187, 16223, 16230, 16249, 16278/M1a3a | |
| C II-1 | 14, 13, 24, 29, 16, 14, 11-14, 12, 10, 12, 25, 13, 11, 15, 12, 19 |
indicated as in the Figure 1
n° of alleles for the following STRs loci: DYS456, DYS389I, DYS390, DYS389II, DYS458, DYS19, DYS385a/b, DYS393, DYS391, DYS439, DYS635, DYS392, Y GATA H4, DYS437, DYS438, DYS448. mtDNA, mithocondrial DNA; STR, short tandem repeat; rCRS, revised Cambridge Reference Sequence
However, on closer inspection, the shared LS-causing allele tagged a genomic haplotype shared by all the nine carriers, which spans 4.5 Mbps on chromosome 16 and encompasses a smaller “core” haplotype also detected among the healthy control samples (Figure S2). Given its length, such a haplotype is unlikely to have arisen by chance in all three families and is therefore best explained as the three families sharing the LS-causing allele by common descent. We therefore chose to estimate the tMRCA of this haplotype, and in turn used this date to estimate how many un-sampled carriers to expect in the same population.
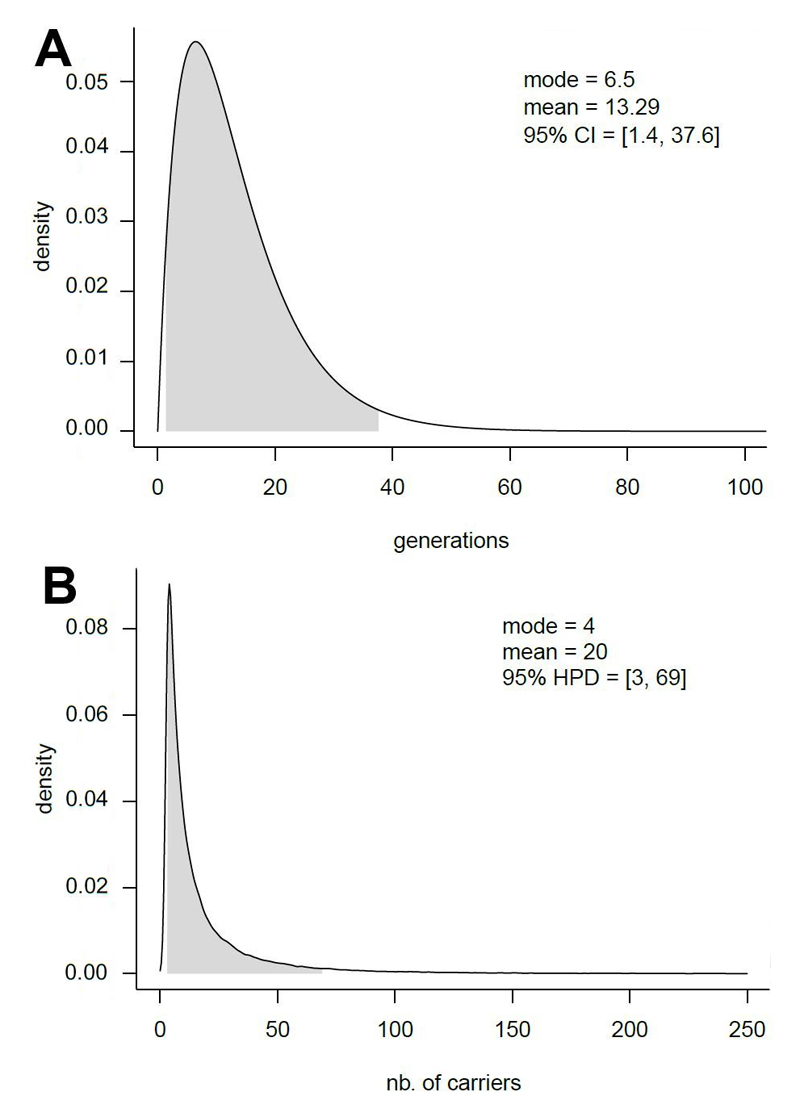
As a starting point we selected the population of Messina and Reggio Calabria (shown to be a continuum from a biodemographic viewpoint [33], total census size ~1.2 Million) as the source population of the three cases. The tMRCA for the disease allele-associated haplotype in these 3 families was estimated to have lived 13 (95% CI: 1.4 – 37.6) generations ago (Figure 2a). By performing forward simulations conditioned on at least 3 carriers being present in today’s generation we estimate that 20 (95% highest posterior density (HPD): 3 - 69) carriers are present in the region of origin of the affected individuals (Figure 2b), 17 in addition to the already sampled probands and their relatives. This number is even larger (99, 95% HPD: 3 - 193) when using an exponential rather than a hyper-exponential function to model population growth in Southern Italy. Therefore, we conservatively estimate the prevalence of the Liddle syndrome due to β-P617L mutation to be 20/1,200,000=1.67 x 10-5 (95% HPD: 2.5 x 10-6 - 5.75 x 10-5), which is at least 17 times higher than the estimated prevalence of <1 x 10-6 worldwide.
Figure 2.
Demographic simulations. Estimated tMRCA for the three carriers of the LS haplotype (A). Estimated number of carriers of the LS haplotype in the population of Reggio Calabria and Messina, under the hyper-exponential model and assumptions described in the Methods section (B).
Discussion
Our findings show that, when ascertaining reportedly unrelated individuals belonging to a circumscribed population for the presence of a rare disease, the chances of discovering cryptic relatedness in the form of a long shared haplotype at the disease-causing locus are high. In the present study we report that the three probands shared a common ancestor 13 generations ago.
From a clinical viewpoint, our study confirms the variable expressivity of LS with regard to age at presentation, degree of hypertension, and plasma potassium concentration, despite sharing the same causative allele. Both blood pressure and plasma potassium values can vary greatly among patients within the same pedigree, or unrelated patients with the same allele, presumably as a result of both allelic variants of other genes and different sodium and potassium intakes, which may modify the effects of the mutant alleles [3, 22, 27]. On the contrary, reduced plasma levels of renin associated with low- or normal values of plasma aldosterone are a common finding in LS patients. One carrier of the β P617L allele (C IV-1) did not exhibit the LS phenotype. However, he was newly born (now 4 years old). Since LS usually manifests itself between the ages of 10 and 30 years, he could develop LS in the future. Alternatively, he could be spared from developing LS, since LS alleles are incompletely penetrant, even within the same kin [3]. Since this subject is actually at risk of LS, we have planned periodic monitoring of his blood pressure, plasma potassium values, and urinary sodium excretion (in order to check his intake of sodium).
Though LS is considered a very rare condition, its prevalence in very large cohorts of hypertensives has not been investigated to date. However, in two different cohorts of 330 and 766 Chinese patients with early-onset hypertension, the prevalence of LS genetically confirmed turned out to be 1.5% and 0.9%, respectively. Moreover, genetic screening of the probands’ relatives identified many more cases with LS, as expected from the Mendelian dominant nature of the disease [34, 35].
These findings suggest that the prevalence of LS may not be as low as commonly reported. Moreover, one of the above studies [34] may have failed to identify some LS patients, since only the hypokalaemic subjects of the cohort were selected for genetic testing. Indeed, plasma potassium concentration may range from severe hypokalaemia to normal values both among LS patients belonging to the same kin and among reportedly unrelated kin carrying the same allele [3, 27], as shown in the present study. This variability of plasma potassium concentration is reminiscent of other conditions characterized by increased activity of ENaC, in primis primary aldosteronism. Indeed, screening of hypertensives for primary aldosteronism regardless of plasma potassium values has considerably increased the prevalence of this condition among hypertensives from less than 1% to more than 10%, and showed that the majority of the patients are actually normokalaemic [36]. By contrast, LS is currently considered exclusively in hypokalaemic hypertensives, while normokalaemic patients with LS are identified only following the genetic screening of the relatives of hypokalaemic probands. Thus, existing approaches may fail to diagnose and appropriately treat many normokalaemic patients with LS. Therefore, LS should be taken into account in all patients with early-onset hypertension, regardless of the plasma potassium concentration. Moreover, LS should be considered even in young hypertensives without a family history of early-onset hypertension, since de novo mutations may be responsible for sporadic cases of LS [20, 37, 38].
More extensive searching for, and earlier diagnosis of LS patients is justified for several reasons. Firstly, patients with misdiagnosed LS are at high risk of both cerebrovascular and cardiovascular events, including sudden death, and are unresponsive to both conventional antihypertensive drugs and MR-antagonists. Secondly, in many patients diagnosis of LS is delayed by many years, and many more patients remain undiagnosed in their lifetime. Thirdly, screening of the probands’ relatives often leads to the identification of additional LS patients, previously misdiagnosed and improperly treated. Lastly, ENaC blockers (amiloride and triamterene) together with dietary salt restriction are impressively efficient in both reversing the clinical manifestations and preventing the severe complications of LS.
Conclusions
With the present study, we infer the evolutionary history of the disease-causing allele detected in all three affected families, building on an approach developed for another successful prediction of under-diagnosed cases of a rare disease [25, 26].
Perspectives
We predict that a considerable number of undiagnosed cases may reside in the area of origin of these three families, which constitutes between a 17- and 82-fold increase in the prevalence (depending on the assumed underlying demographic model). Our results suggest that evolutionary approaches combined with clinical data may increase the accuracy of epidemiological prediction, at least in original populations of circumscribed geographical areas, and contribute to the understanding of the prevalence and hopefully the ways in which we can improve the treatment of an otherwise under-diagnosed disease.
Supplementary Material
Novelty and Significance.
What is new?
Genome-wide analysis of three kindreds from southern Italy with the same mutation responsible for Liddle syndrome (LS) and without a family relationship between each other showed a common haplotype encompassing the pathogenic allele. An evolutionary approach based on coalescent analysis showed that the three families inherited the mutant allele from a common ancestor who lived about 13 generations earlier and that several undiagnosed patients with the same LS mutation live in the same area of origin.
What is relevant?
Hypertension due to Liddle syndrome may be more common in southern Italy than elsewhere.
Summary
Genome-wide analysis supplemented by coalescent analysis allowed us to infer the evolutionary history of a LS mutation shared by three kindreds and predict the approximate number of undiagnosed cases living in the area of origin.
Acknowledgements
The authors would like to thank the sample donors and their families, who made this research possible.
Sources of Funding
L.P. is supported by the European Union through the European Regional Development Fund (Project No. 2014-2020.4.01.16-0030, No. 2014-2020.4.01.15-0012, and No. 2014-2020.4.01.16-0024, MOBTT53). M.G.T. and Y.D. are supported by a Wellcome Trust Senior Research Fellowship awarded to M.G.T. D.L is supported by the Italian Ministry of Education, University and Research (PRIN2010EL8TXP_006).
Footnotes
Disclosures
All the Authors declare no conflicts of interest.
Contributor Information
Luca Pagani, Department of Biology, University of Padova, Padova, Italy; Estonian Biocentre, Tartu, Estonia.
Yoan Diekmann, Research Department of Genetics, Evolution and Environment, University College London, UK.
Marco Sazzini, Department of Biological Geological and Environmental Sciences, University of Bologna, Bologna, Italy.
Sara De Fanti, Department of Biological Geological and Environmental Sciences, University of Bologna, Bologna, Italy.
Maurizio Rondinelli, IRCCS Centro Cardiologico Monzino, Milano, Italy.
Enrico Farnetti, Laboratory of Molecular Biology, Department of Oncology and advanced Technologies, IRCCS Santa Maria Nuova Hospital, Reggio Emilia, Italy.
Bruno Casali, Laboratory of Molecular Biology, Department of Oncology and advanced Technologies, IRCCS Santa Maria Nuova Hospital, Reggio Emilia, Italy.
Amelia Caretto, Department of Endocrinology and Metabolic Diseases, San Raffaele Scientific Institute, Milano, Italy.
Francesca Novara, Department of Molecular Medicine, University of Pavia, Pavia, Italy.
Orsetta Zuffardi, Department of Molecular Medicine, University of Pavia, Pavia, Italy.
Paolo Garagnani, Department of Experimental, Diagnostic and Specialty Medicine, University of Bologna, Bologna, Italy.
Franco Mantero, Endocrinology Unit, Department of Medicine, University of Padova, Padova, Italy.
Mark G. Thomas, Research Department of Genetics, Evolution and Environment, University College London, UK
Donata Luiselli, Department of Biological Geological and Environmental Sciences, University of Bologna, Bologna, Italy.
Ermanno Rossi, Department of Internal Medicine, IRCCS Santa Maria Nuova Hospital, Reggio Emilia, Italy.
References
- 1.Liddle GW, Bledsoe T, Coppage WS., Jr A familial renal disorder simulating primary aldosteronism but with negligible aldosterone secretion. Trans Assoc Am Phys. 1963;76:199–213. [Google Scholar]
- 2.Warnock DG. Liddle syndrome: genetics and mechanisms of Na+ channel defects. Am J Med Sci. 2001;322:302–307. doi: 10.1097/00000441-200112000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Botero-Velez M, Curtis JJ, Warnock DG. Brief report: Liddle's syndrome revisited--a disorder of sodium reabsorption in the distal tubule. N Engl J Med. 1994;330:178–181. doi: 10.1056/NEJM199401203300305. [DOI] [PubMed] [Google Scholar]
- 4.Canessa CM, Horisberger JD, Rossier BC. Epithelial sodium channel related to proteins involved in neurodegeneration. Nature. 1993;361:467–470. doi: 10.1038/361467a0. [DOI] [PubMed] [Google Scholar]
- 5.Canessa CM, Schild L, Buell G, Thorens B, Gautschi I, Horisberger JD, Rossier BC. Amiloride-sensitive epithelial Na+ channel is made of three homologous subunits. Nature. 1994;367:463–467. doi: 10.1038/367463a0. [DOI] [PubMed] [Google Scholar]
- 6.Canessa CM, Merillat AM, Rossier BC. Membrane topology of the epithelial sodium channel in intact cells. Am J Physiol. 1994;267:C1682–C1690. doi: 10.1152/ajpcell.1994.267.6.C1682. [DOI] [PubMed] [Google Scholar]
- 7.Kellenberger S, Schild L. International Union of Basic and Clinical Pharmacology. XCI. Structure, Function, and Pharmacology of Acid-Sensing Ion Channels and the Epithelial Na+ Channel. Pharmacol Rev. 2015;67:1–35. doi: 10.1124/pr.114.009225. [DOI] [PubMed] [Google Scholar]
- 8.Rossier BC, Staub O, Hummler E. Genetic dissection of sodium and potassium transport along the aldosterone-sensitive distal nephron: Importance in the control of blood pressure and hypertension. FEBS Letters. 2013;587:1929–1941. doi: 10.1016/j.febslet.2013.05.013. [DOI] [PubMed] [Google Scholar]
- 9.Palmer LG, Patel A, Frindt G. Regulation and dysregulation of epithelial Na+ channels. Clin Exp Nephrol. 2012;16:35–43. doi: 10.1007/s10157-011-0496-z. [DOI] [PubMed] [Google Scholar]
- 10.Cui Y, Tong A, Jiang J, Wang F, Li C. Liddle syndrome: clinical and genetic profiles. J Clin Hypertens. 2017;19:524–29. doi: 10.1111/jch.12949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salih M, Gautschi I, van Bemmelen MX, Di Benedetto M, Brooks AS, Lugtenberg D, Schild L, Hoorn E. A missense mutation in the extracellular domain of αENaC causes Liddle syndrome. JASN. 2017 doi: 10.1681/ASN.2016111163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiltunen TP, Hannila-Handelberg T, Petajaniemi N, Kantola I, Tikkanen I, Virtamo J, Gautschi I, Schild L, Kontula K. Liddle’s syndrome associated with a point mutation in the extracellular domain of the epithelial sodium channel γ subunit. J Hypertens. 2002;20:2383–2390. doi: 10.1097/00004872-200212000-00017. [DOI] [PubMed] [Google Scholar]
- 13.Shimkets RA, Warnock DG, Bositis CM, Nelson-Williams C, Hansson JH, Schambelan M, Gill JR, Ulick S, Milora RV, Findling JW, Canessa CM, et al. Liddle's syndrome: heritable human hypertension caused by mutations in the beta subunit of the epithelial sodium channel. Cell. 1994;79:407–414. doi: 10.1016/0092-8674(94)90250-x. [DOI] [PubMed] [Google Scholar]
- 14.Gao L, Wang L, Liu Y, Zhou X, Hui R, Hu A. A Family with Liddle Syndrome Caused by a Novel Missense Mutation in the PY Motif of the Beta-Subunit of the Epithelial Sodium Channel. J Pediatr. 2013;162:166–70. doi: 10.1016/j.jpeds.2012.06.017. [DOI] [PubMed] [Google Scholar]
- 15.Rossi E, Farnetti E, Debonneville A, Nicoli D, Grasselli C, Regolisti G, Negro A, Perazzoli F, Casali B, Mantero F, Staub O. Liddle's syndrome caused by a novel missense mutation (P617L) of the epithelial sodium channel beta subunit. J Hypertens. 2008;26:921–927. doi: 10.1097/HJH.0b013e3282f85dfe. [DOI] [PubMed] [Google Scholar]
- 16.Inoue J, Iwaoka T, Tokunaga H, Takamune K, Naomi S, Araki M, Takahama K, Yamaguchi K, Tomita K. A family with Liddle’s syndrome caused by a new missense mutation in the β subunit of the epithelial sodium channel. J Clin Endocrinol Metab. 1998;83:2210–2213. doi: 10.1210/jcem.83.6.5030. [DOI] [PubMed] [Google Scholar]
- 17.Sawathiparnich P, Sumboonnanonda A, Weerakulwattana P, Limwongse C. A novel mutation in the β-subunit of the epithelial sodium channel gene (SCNN1B) in a Thai family with Liddle’s syndrome. J Pediatr Endocrinol Metab. 2009;22:85–89. doi: 10.1515/jpem.2009.22.1.85. [DOI] [PubMed] [Google Scholar]
- 18.Hansson JH, Schild L, Lu Y, Wilson TA, Gautschi I, Shimkets R, Nelson-Williams C, Rossier BC, Lifton RP. A de novo missense mutation of the β subunit of the epithelial sodium channel causes hypertension and Liddle syndrome, identifying a proline-rich segment critical for regulation of channel activity. Proc Natl Acad Sci USA. 1995;92:11495–11499. doi: 10.1073/pnas.92.25.11495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freundlich M, Ludwig M. A novel epithelial sodium channel β-subunit mutation associated with hypertensive Liddle syndrome. Pediatr Nephrol. 2005;20:512–515. doi: 10.1007/s00467-004-1751-2. [DOI] [PubMed] [Google Scholar]
- 20.Uehara Y, Sasaguri M, Kinoshita A, Tsuji E, Kiyose H, Taniguchi H, Noda K, Ideishi M, Inoue J, Tomita K, Arakawa K. Genetic analysis of the epithelial sodium channel in Liddle’s syndrome. J Hypertens. 1998;16:1131–1135. doi: 10.1097/00004872-199816080-00008. [DOI] [PubMed] [Google Scholar]
- 21.Furuhashi M, Kitamura K, Adachi M, Miyoshi T, Wakida N, Ura N, Shikano Y, Shinshi Y, Sakamoto K, Hayashi M, Satoh N, et al. Liddle’s syndrome caused by a novel mutation in the proline-rich PY motif of the epithelial sodium channel β-subunit. J Clin Endocrinol Metab. 2005;90:340–344. doi: 10.1210/jc.2004-1027. [DOI] [PubMed] [Google Scholar]
- 22.Tamura H, Schild L, Enomoto N, Matsui N, Marumo F, Rossier BC, Sasaki S. Liddle disease caused by a missense mutation of β subunit of the epithelial sodium channel gene. J Clin Invest. 1996;97:1780–1784. doi: 10.1172/JCI118606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang K-Q, Lu C-X, Xiao Y, Liu Y-X, Jiang XJ, Zhang X, Zhou XL. A novel frameshift mutation of epithelial sodium channel β-subunit leads to Liddle syndrome in an isolated case. Clinical Endocrinology. 2015;82:611–614. doi: 10.1111/cen.12650. [DOI] [PubMed] [Google Scholar]
- 24.Knight KK, Olson DR, Zhou R, Snyder PM. Liddle’s syndrome mutations increase Na+ transport through dual effects on epithelial Na+ channel surface expression and proteolytic cleavage. Proc Natl Acad Sci USA. 2006;103:2805–2808. doi: 10.1073/pnas.0511184103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chahal HS, Stals K, Unterlander M, et al. AIP mutation in pituitary adenomas in the 18th century and today. N Engl J Med. 2011;364:43–50. doi: 10.1056/NEJMoa1008020. [DOI] [PubMed] [Google Scholar]
- 26.Radian S, Diekmann Y, Gabrovska P, et al. Increased Population Risk of AIP-Related Acromegaly and Gigantism in Ireland. Hum Mutat. 2017;38:78–85. doi: 10.1002/humu.23121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rossi E, Farnetti E, Nicoli D, Sazzini M, Perazzoli F, Regolisti G, Grasselli C, Santi R, Negro A, Mazzeo V, Mantero F, et al. A clinical phenotype mimicking essential hypertension in a newly discovered family with Liddle's syndrome. Am J Hypertens. 2011;24:930–935. doi: 10.1038/ajh.2011.76. [DOI] [PubMed] [Google Scholar]
- 28.Caretto A, Primerano L, Novara F, Zuffardi O, Genovese S, Rondinelli M. A Therapeutic Challenge: Liddle's Syndrome Managed with Amiloride during Pregnancy. Case reports in obstetrics and gynecology. 2014;2014:156250. doi: 10.1155/2014/156250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sazzini M, Gnecchi Ruscone GA, Giuliani C, et al. Complex interplay between neutral and adaptive evolution shaped differential genomic background and disease susceptibility along the Italian peninsula. Scientific reports. 2016;6:32513. doi: 10.1038/srep32513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Fanti S, Vianello D, Giuliani C, Quagliariello A, Cherubini A, Sevini F, Iaquilano N, Franceschi C, Sazzini M, Luiselli D. Massive parallel sequencing of human whole mitochondrial genomes with Ion Torrent technology: an optimized workflow for Anthropological and Population Genetics studies. Mitochondrial DNA Part A, DNA Map Seq Anal. 2016:1–8. doi: 10.1080/24701394.2016.1197218. [DOI] [PubMed] [Google Scholar]
- 31.Matise TC, Chen F, Chen W, De La Vega FM, Hansen M, He C, Hyland FC, Kennedy GC, Kong X, Murray SS, Ziegle JS, et al. A second-generation combined linkage physical map of the human genome. Genome Res. 2007;17:1783–1786. doi: 10.1101/gr.7156307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Donnelly P, Tavare S, Balding DJ, Griffiths RC. Estimating the age of the common ancestor of men from the ZFY intron. Science. 1996;272:1357–1359. doi: 10.1126/science.272.5266.1357. author reply 1361-1352. [DOI] [PubMed] [Google Scholar]
- 33.Boattini A, Lisa A, Fiorani O, Zei G, Pettener D, Manni F. General method to unravel ancient population structures through surnames, final validation on Italian data. Hum Biol. 2012;84:235–270. doi: 10.3378/027.084.0302. [DOI] [PubMed] [Google Scholar]
- 34.Wang LP, Yang KQ, Jiang XJ, Wu HY, Zhang HM, Zou YB, Song L, Bian J, Hui RT, Liu YX, Zhou XL. Prevalence of Liddle Syndrome Among Young Hypertension Patients of Undetermined Cause in a Chinese Population. J Clin Hypertens (Greenwich) 2015;17:902–907. doi: 10.1111/jch.12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu K, Quin F, Sun X, et al. Analysis of the genes involved in Mendelian forms of low-renin hypertension in Chinese early-onset hypertensive patients. J Hypertens. 2017;35 doi: 10.1097/HJH.0000000000001556. 000-000. [DOI] [PubMed] [Google Scholar]
- 36.Rossi GP, Bernini G, Caliumi C, et al. A prospective study of the prevalence of primary aldosteronism in 1,125 hypertensive patients. J Am Coll Cardiol. 2006;48:2293–2300. doi: 10.1016/j.jacc.2006.07.059. [DOI] [PubMed] [Google Scholar]
- 37.Nakano Y, Ishida T, Ozono R, Matsuura H, Yamamoto Y, Kambe M, Chayama K, Oshima T. A frameshift mutation of beta subunit of epithelial sodium channel in a case of isolated Liddle syndrome. J Hypertens. 2002;20:2379–2382. doi: 10.1097/00004872-200212000-00016. [DOI] [PubMed] [Google Scholar]
- 38.Yamashita Y, Koga M, Takeda Y, Enomoto N, Uchida S, Hashimoto K, Yamano S, Dohi K, Marumo F, Sasaki S. Two sporadic cases of Liddle's syndrome caused by de novo ENaC mutations. Am J Kidney Dis. 2001;37:499–504. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.