Abstract
In the field of gene set enrichment analysis (GSEA), meta-analysis has been used to integrate information from multiple studies to present a reliable summarization of the expanding volume of individual biomedical research, as well as improve the power of detecting essential gene sets involved in complex human diseases. However, existing methods, Meta-Analysis for Pathway Enrichment (MAPE, [1]), may be subject to power loss because of (i) using gross summary statistics for combining end results from component studies and (ii) using enrichment scores whose distributions depend on the set sizes. In this paper, we adapt meta-analysis approaches recently developed for genome-wide association studies, which are based on fixed effect (FE) and random effects (RE) models, to integrate multiple GSEA studies. We further develop a mixed strategy via adaptive testing for choosing RE versus FE models to achieve greater statistical efficiency as well as flexibility. In addition, a size-adjusted enrichment score based on a one-sided Kolmogorov-Smirnov statistic is proposed to formally account for varying set sizes when testing multiple gene sets. Our methods tend to have much better performance than the MAPE methods, and can be applied to both discrete and continuous phenotypes. Specifically, the performance of the adaptive testing method seems to be the most stable in general situations.
Keywords: between-study heterogeneity, GSEA, generalized linear model, fixed effect, integrative GSEA, adjusted Kolmogorov-Smirnov statistic, MAPE, random effects
1. Introduction
In transcriptome studies, great attention has been drawn to identification of pathways, or more broadly, groups of biologically related genes involved in complex human diseases or other biological processes. A major type of such analysis is called Gene Set Enrichment Analysis (GSEA), which determines whether a gene set is over-represented by genes associated with a trait of interest. Gene sets can be pre-defined according to a variety of criteria, including genes/proteins participating in common pathways, sharing similar annotated functions or related sequence motifs, interacting with and co-regulating each other, and serving as oncogenic, immunologic or other disease signature genes. In general, GSEA is designed to detect coordinated expression changes in a group of related genes, and such changes are, in whole or in part, cellular reactions to changes related to disease phenotypes or therapeutic treatments. Thus, gene sets identified from GSEA can provide key insights into biological processes underlying disease pathogenesis or treatment effects.
Various statistical methods have been developed for GSEA using a single mRNA dataset. An early method for GSEA is to associate gene expression with phenotype changes to identify differentially expressed (DE) genes based on a statistic measuring the degree of differential expression, and then determine whether a gene set contains significantly more DE genes than would be expected by chance using Fisher’s exact test [2]. Subramanian et al. [3] proposed an improved GSEA method, which has become one of the most well-known and currently widely used GSEA algorithms. It makes use of the ranks of genes according to the degree of differential expression, to compute the enrichment score of a gene set based on a weighted Kolmogorov-Smirnov (KS) test. Then it estimates the statistical significance of the gene set using an empirical null distribution of the enrichment score obtained from a permutation procedure. Later, many other methods for GSEA were further developed. For example, [4] modified the GSEA algorithm by [3] using a max–mean statistic and a re-standardization procedure; and [5] proposed a random set approach. For a detailed review about the methodological development of GSEA, see [6, 7]. Due to mature statistical analytics, GSEA has been widely applied in biomedical fields, where GSEA plays critical roles in the innovation of disease prevention and intervention strategies, including revealing novel genes and key regulatory modules, detecting ensembles of diagnostic and prognostic markers, and discovering potential therapeutic targets [8–13].
In the past decades, enormous amounts of data have been generated from various biomedical experiments; and the volume continues to expand. Consortia have been recently formed and public databases have been constructed and regularly updated, making it increasingly feasible to access data from multiple research projects. Despite significant successes GSEA has achieved, it is striking that findings are often unstable and thus are inconsistent among independent studies targeting the same disease or biological problem. This is partly because of small sample sizes relative to an overwhelming number of genes, as is typical in individual genome-wide transcriptomic studies, making estimation and inference highly volatile. Thus, there is an increasingly urgent need to perform integrative GSEA, i.e., integrating multiple relevant GSEA studies, to turn individual data into collective knowledge.
Integrative GSEA (iGSEA), when performed properly, can effectively increase the sample size of the analysis, greatly facilitate information sharing, and improve the power of detecting truly interesting gene classes, as well as increasing the reproducibility and interpretability of research results. However, methods for iGSEA are rather scant. [1] systematically developed and evaluated three methods for Meta-Analysis of Pathway Enrichment (MAPE), including MAPE-P, MAPEG and MAPE-I. All these methods use the maximum, minimum or Fisher’s statistic to combine p-values from multiple studies, and so inevitably lose power by using such gross summaries. Further, when testing multiple pathways, the MAPE methods do not account for different set sizes in their permutation-based procedures. In addition, the lack of ability to formally handle between-study heterogeneity, which may exist in GSEA studies due to the varying quality of experiments and the noisy nature of genomic data, can affect the performance of the MAPE methods. More recently, a Bayesian method has been proposed for integrative GSEA by [14] to improve the detection of enriched gene sets, which simultaneously models gene set information and original gene expression data from all component studies. This method can only be applied to binary phenotypes. When the number of genes or gene sets or component studies gets large, it can become computationally formidable. In addition, detecting the convergence of Markov chains and selecting starting points may require great human efforts.
Motivated by the room for improvement of the existing methods, we focus on the development of new methods for iGSEA that are (i) statistically efficient; (2) computationally affordable and (3) applicable to both discrete and continuous phenotypes. Here, we adapt and extend meta-analysis approaches [15–17] newly developed for genome-wide association studies (GWAS), which are based on fixed effect (FE) and random effects (RE) models, to integrate multiple GSEA studies. Specifically, we propose a hybrid strategy for choosing RE versus FE models, with an attempt to achieve great statistical efficiency as well as stability in performance in various practical situations. In addition, unlike the MAPE methods, our proposed iGSEA methods formally account for different set sizes when testing a database of gene sets.
In the next section of this paper, we describe our modeling and testing strategy in an individual study, where a generalized linear model (GLM) is used to fit the relationship between the expression of an individual gene and the phenotype, and then gene-level statistics are constructed to quantify the strength of the association. In Section 3, we propose several meta-analysis methods to compute an overall gene-level statistic that integrates the gene-level statistics from individual studies. In Section 4, we focus on gene set analysis, where we propose size-adjusted set-level statistics via a one-sided KS test, estimate their significance based on permutation, and adjust for multiple testing when more than one gene set is tested. Sections 5 and 6 present results from simulation studies and an example using gene expression data from five lung cancer studies. Section 7 concludes the paper with a brief discussion. The algorithm for the proposed iGSEA methods is outlined in the appendix.
2. Modeling and testing in an individual study
We are interested in combining K independent GSEA studies that share a common phenotype Y. Suppose there are G genes in a genome that appear at least once in the K studies. Let Jk be the sample size in study k, where k = 1, … K; let Yjk be the phenotype of sample j in study k, where j = 1, …, Jk; and let Xjgk be the expression level of gene g for sample j in study k, where g = 1, …, G. We use βgk to denote the effect of gene g’s expression on the phenotype Y in study k. We assume that different studies may have different genes from the same genome (i.e., some genes’ information can be missing in one or more studies), which allows us to include more studies in our integrative analysis.
For each gene g included in study k, we use a GLM to model the relationship between Xjgk and Yjk:
| (1) |
where l(·) is the link function, and Yjk is assumed to follow an exponential family distribution.
To test the null hypothesis H0: βgk = 0, we can compute the score statistic Ugk and its corresponding variance Vgk based on the distribution of Y, whose probability distribution function can be written as
where ϕk is the dispersion parameter, and θjk is the natural parameter. Here, a(·), b(·), and c(·) are known functions, determined by the type of distribution of the phenotype Y. For example, if Y is binary, then the distribution is Bernoulli so that a (ϕ) = 1, b (θ) = log (1 + eθ) and c (Y, ϕ) = 0. We use b′ (·) and b″ (·) to denote the first and second derivatives of b(·). Since E(Yjk) = b′ (θjk), θjk is equal to b′ −1 ∘ l−1(αgk + βgkXjgk). We can therefore construct the likelihood function and then derive the score statistic and the corresponding estimated variance:
where ϕ̂gk and α̂gk are the maximum likelihood estimates, and θ̂jgk = b′−1 ∘ l−1(α̂gk + β̂gkXjgk). Note that under the null hypothesis of no association between Xgk and Yk, β̂gk ≡ 0; and asymptotically follows a chi-square distribution with one degree of freedom ( ).
3. Computing overall gene-level statistics
To combine multiple GSEA studies, we rely on meta-analysis to compute a statistic per gene, using the gene-level statistics (Ugk, Vgk) from individual studies, for measuring the overall strength of association between gene g’s expression and the phenotype. Below we consider three approaches: (1) testing based on a fixed-effect (FE) model; (2) testing based on a random-effects (RE) model; and (3) adaptive testing (AT). The first two adapt the recent FE and RE testing methods for GWAS meta-analysis [15–17] into iGSEA, respectively. The third aims to combine the strength of the first two and achieve robustness against model mis-specification.
3.1. FE testing
A fixed-effect model that assumes no heterogeneity among GSEA studies is specified as follows:
| (2) |
where μg stands for the common genetic effect of gene g among the different studies. Let Tgk indicate whether gene g is included in study k (1 if included; 0 otherwise). Motivated by [18] and [17], we use the following statistic to test the null hypothesis H0: μg = 0:
| (3) |
where follows an asymptotic distribution of under H0. Here, we do not need to calculate the P-value of and decide whether H0 is rejected. This is because in the latter sections, a gene set will be tested based on the ordering of s as larger values of s indicate more evidence to reject H0 and so imply smaller P-values no matter what the actual reference distribution of is.
Although meta-analysis is generally believed to be less statistically efficient than mega-analysis (i.e., joint analysis of individual-level raw data from all component studies), [18] proved that under the FE model, meta-analysis based on score statistics can achieve the same efficiency as mega-analysis. Thus, unlike using the coarse summary statistics in the MAPE methods, this model-based method has almost no information loss when testing the common effect in (2).
3.2. RE testing
To accommodate between-study heterogeneity, one can specify βgk as a random effect. The results from different studies for gene g are therefore combined based on a random effects model specified by
| (4) |
where μg stands for the mean genetic effect among studies, and εgk is the random effect representing the study-specific deviation of the effect from the mean effect μg. It is assumed that εgks are independent and follow a normal distribution with mean 0 and variance τg.
In GWAS, however, researchers prefer to using the FE approach to combine multiple genomic studies, even when between-study heterogeneity exists, due to a controversial phenomenon [19]. That is, the traditional RE approach that tests H0: μg = 0 usually provides less significant P-values than the corresponding FE approach so that RE does not give any new findings compared with FE in most cases. [16] investigated this conservative nature of the traditional RE approach and proposed an improved RE approach that tests the hypothesis H0: μg = 0 and τg = 0 in genomic settings. The new approach has been shown to achieve higher power than FE when there is heterogeneity. Here, we adapt this approach and test the null hypothesis H0: μg = 0 and τg = 0 rather than H0: μg = 0 under the RE model. The test statistic is specified as follows:
| (5) |
The first term is the statistic to test μg = 0 under the FE model (i.e., τg = 0) and the second term is to test τg = 0 given μg = 0. Again, we do not need to calculate the P-value because we will rely on the ordering of for testing a gene set.
3.3. Adaptive testing
The above FE and RE methods apply the same class of models to all genes. In practical situations, however, some genes, especially those “silent” with zero effect, tend to fit in the FE model while the others are likely to fit in the RE model. For instance, in lung cancer research, it is found that the effect size of gene “SLC35A5” seem to be quite stable, but that of gene “CYCS” differs greatly from study to study [20–22]. Thus, we propose a data-adaptive testing procedure that is robust to model mis-specification.
We begin with the more general RE model (4) and for each gene g, we first test the between-study heterogeneity . If is rejected, then no more testing is needed because H0: μg = 0 and τg = 0is also rejected, meaning that this gene is associated with the phenotype in at least one of the studies. If is not rejected, we switch to the FE model to test using . Note that if , we directly go to .
Let p1g and p2g be the P-value in stage 1 and 2, respectively. We can calculate p2g based on the asymptotic distribution of under , as mentioned in Section 3.1, or based on a standard permutation procedure. In Section 3.3.1, we explain how to compute p1g when testing . In Section 3.3.2, we compute an overall P-value of the two-stage test, denoted by , for each gene to combine p1g and p2g. When testing a gene set, the ordering of the genes will be produced based on the overall P-value from this adaptive testing method.
3.3.1. Testing the existence of between-study heterogeneity
Under the RE model, a classical approach to test the between-study heterogeneity τ is Cochran’s Q test [23, 24], where the Q statistic is computed by summing the squared deviation of each study’s estimated effect size from the estimated overall effect size, with the contribution of each study weighted by its inverse variance. More recently, three measures including the H, R, and I2 statistics have been proposed to assess the between-study heterogeneity in meta-analysis, each of which has its own characteristics as discussed in [25]. In this paper, we use the Q statistic to test the heterogeneity of gene g’s effect because its asymptotic distribution is relatively simple and the other three statistics are all computed based on the Q statistics. Under our context, the Q statistic of gene g can be defined by
| (6) |
where β̂gk is the estimator of βgk fit by the GLM with variance is the estimated precision of β̂gk within study k, and β̂g is a weighted average of the study estimates, using the estimated precisions as weights:
We can set p1g to be the P-value of Qg based on its asymptotic null distribution; that is, when holds, Qg asymptotically follows a chi-square distribution with degrees of freedom .
Alternatively, we can use a permutation-based method to test the heterogeneity τg in a meta-analysis. [26] summarized seven methods, which include the variance component type estimator (VC), the method of moments estimator (MM), the maximum likelihood estimator (ML), the restricted maximum likelihood estimator (REML), the empirical Bayes estimator (EB), the model error variance type estimator (MV), a variation of the MV estimator (MVvc), for estimating τg under the RE model; and among them, MVvc and EB are found to be the most accurate in general, particularly when τg is moderate to large. Below we describe a permutation procedure based on the MVvc estimator of τg because of its good performance as well as its computational ease based on a non-iterative procedure.
Let be the VC estimator of τ, where
and . Let be the plug-in estimator for the ratio of within-study vs. between-study heterogeneity, i.e., ; and v̂gk ≡ r̂gk + 1. Then according to [27], the MVvc estimator of τg can be calculated by
| (7) |
with
In case that , we replace it with a small value (e.g., 0.01) to compute r̂gk. We permute sample labels over different studies to obtain the empirical null distribution of and then calculate the P-value of the observed statistic.
3.3.2. Combining P-values
We first discuss how to combine P-values from individual stages for a two-stage test defined by a set of general decision rules (the subscript g is dropped whenever there is no ambiguity). Let α be the overall size of the two-stage test, and αi be the size of the ith-stage test, satisfying 0 < αi < α for i = 1, 2. Further, let α0 be a predetermined upper limit such that 0 < α < α0 ≤ 1. Typically, the test uses the following decision rules: (1) if p1 ≤ α1, reject ; if p1 > α0, fail to reject ; and in either case, the test stops. (2) If α1 < p1 ≤ α0, the test proceeds to the second stage: is rejected if and only if F(p1, p2), a predetermined function, is less than or equal to f, and f is determined by the following equality
where I(·) is the indicator function. Then according to [28], the overall P-value of the two-stage test can be given by
Many existing methods for computing the overall P-value use the framework above, such as the Fisher’s weighted product test [29] and the weighted inverse normal method [30].
For our adaptive testing procedure, it is obvious that α0 is set to 1. We further set F(p1, p2) = p2, proposed by [31]. Thus, the overall P-value of our test is given by
If the tests in the two stages are independent, then the following relationship holds:
| (8) |
In our context, it might be plausible to argue that the result of the second stage is not related to that of the first stage as they involve testing the mean and variance of the effect sizes, respectively, which are two distinctive characteristics of data. As mentioned in [31], the above method to combine P-values is easy to implement and only depends on one parameter α1, which lies in (0, α) and can be determined by prior information about the existence of between-study heterogeneity or the common effect size. When prior information is not available, we could simply set α1 = α2 and then solve the equality (8), yielding ; or alternatively, we could set α1 based on some exploratory data analysis for assessing the heterogeneity.
4. Gene set analysis with size-adjusted enrichment scores
If there is only one gene set (say set s) to test, a straightforward approach to enrichment analysis is to choose some reasonable set-level statistic as the enrichment score vs and compute its significance through a permutation procedure. In detail, we randomly shuffle the gene labels B times (so that a different set of genes randomly selected from the larger pool of G genes is included in set s each time) and compute the permuted enrichment scores, say , 1 ≤ b ≤ B. The P-value of the observed vs can be approximated by
When more than one gene set is tested, the Q-value is computed to account for multiplicity ([1, 7, 32]), which is defined as the minimum false discovery rate (FDR) at which a set is claimed to be statistically significant. The Q-value of the observed vs is evaluated by
| (9) |
where π̂0 is a rough estimate of the proportion of non-enriched sets and S is the number of gene sets being tested. We calculate π̂0 using the method described in [32], which is implemented in an R package called “qvalue” [33]. Note that for the MAPE methods, π̂0 is always set to 1 ([1]), a conservative choice. Our preliminary simulation has found that using qvalue with MAPEs leads to worse results in FDR control. Gene sets with a Q-value < δ are claimed to be enriched. Throughout this paper, δ is set to the default value 0.05.
As to choosing an enrichment score, we consider a one-sided KS test, which is to determine whether the distribution of the overall gene-level statistic (say ug) for genes in set s is stochastically larger/smaller than the distribution of the same statistic for genes out of the set. To keep the direction of the one sided KS test the same over the different meta-analysis approaches, we set for the FE method, for the RE method, and for the AT method. Suppose set s contains Gs genes. We order the total G genes according to one of the three overall gene-level statistics. For example, let A and B denote the statistic for genes in and out of set s, respectively. The order statistics are A(1),A(2), …,A(Gs) and B(1),B(2), …,B(G−s), where G−s ≡ G − Gs. Let FA and FB denote the underlying cumulative distribution functions (CDF) for A and B, respectively. Then the null and alternative hypotheses are H0: FA = FB for all x, and Ha: FA ≤ FB for all x, FA < FB for some x.
The one-sided two sample KS test statistic for set s is given by
where F̂A(x) is the empirical CDF of A, defined by
and F̂B(x) is defined similarly.
We mention that the KS-type statistics have been commonly used as enrichment scores in the literature. For example, the popular GSEA algorithm by [3] used a weighted version of the two-sided KS statistic; and the existing MAPE methods for integrative GSEA by [1] used the one-sided KS statistic as well, but for testing the opposite direction. However, an important fact about the KS-type statistics is often ignored: for gene sets of different sizes, their KS statistics follow different distributions. While enrichment analysis is commonly applied to a database of gene sets, whose sizes vary in a wide range, none of the existing methods adjust the KS-type statistics to formally account for varying set sizes when computing the Q-value to control the FDR. [3] obtained normalized enrichment scores from separately rescaling the positive and negative scores by dividing by the mean of the permuted scores. However, there is no theoretical ground provided for their adjustment and so it is ad hoc.
Below we propose a size-adjusted KS statistic as our enrichment score:
| (10) |
According to [34] and [35], vs has an asymptotic distribution whose CDF is given by
| (11) |
which is independent of the size of the gene set. A comparison of the empirical CDF of 1000 replicates and the asymptotic CDF is given in Section 1 of Supplementary Material. We find that they are very close, especially when Gs ≥ 30. Therefore, the size-adjusted KS statistics for gene sets of varying sizes approximately follow the same distribution, making the permutation-based computation of Q-value considerably improved.
5. Simulation
We designed two simulation studies, one for binary phenotypes and the other for continuous phenotypes, to assess the performance of the proposed iGSEA methods and compare them with the existing methods under default settings. Our methods are labeled by iGSEA-FE, iGSEA-RE and iGSEA-AT, respectively, according to the meta-analysis strategies used, as discussed in Section 3. In each study, we first compared the power in identifying enrichment via a one-gene-set simulation model as in [1]; and we further examined the sensitivity and specificity of the methods via a multiple-gene-set simulation model. Throughout this section, we fixed the significance level at 0.05 for every test conducted; and we set B = 500 for the one-gene-set model and B = 200 for the multiple-gene-set model. For iGSEA-AT, we set the first-stage significant level α1 ∈ {0.02, 0.03, 0.04} in our simulation and find that its performance was not sensitive to the change of α1. Thus we report the results based on α1 = 0.02. We also note that due to the mixed strategy of using the FE and RE models, as discussed in Section 3.3, it is unrealistic to expect that iGSEA-AT outperforms iGSEA-FE and iGSEA-RE uniformly; instead, we anticipate that its performance can mimic the better of the two closely in most cases.
5.1. Binary phenotypes
Power comparison
Suppose there are G = 500 genes in a genome and the first 100 genes belong to the gene set of interest. For DE genes, we simulated both down-regulated (DR) and up-regulated (UR) genes. We generated a random variable dg to indicate whether gene g is an UR, DR, or equally expressed (EE) gene, which is represented by dg = 1, −1, and 0, respectively. There are DE genes out of the first 100 genes that belong to the gene set and DE genes out of the rest 400 genes. We fixed ω0 = 0.2 in the simulation, and so the gene set is enriched if ω > 0.2. We assume there are (ω − 10%) UR and 10% DR genes in the gene set, and 10% UR and 10% DR genes out of the gene set. We set ω ∈ {0.2, 0.3, 0.4, 0.5} to represent zero, weak, medium and strong enrichment signals, respectively.
For the purpose of meta-analysis, we simulated four independent studies in each generated dataset. The chance of each gene to be included in study k is determined by a universal sampling rate λ, where we set λ ∈ {0.5, 0.6, 0.7, 0.8, 0.9, 1}. Each study has J = 40 samples, including 20 normal samples with Yj = 0 and 20 tumor samples with Yj = 1. For a DE gene, we generated a random binary variable rg to indicate whether the effects of this DE gene across different studies are random or fixed. If rg = 1, this DE gene is called a RE gene, otherwise rg = 0. The proportion of the RE genes out of the DE genes is represented by γ, where we set γ ∈ {0, 0.2, 0.4, 0.6, 0.8, 1}.
Since Y is binary, we used a logistic regression model: for sample j in study k, logit(E(Ykj = 1|Xgkj = x)) = αgk + βgkx. According to the Bayes theorem, we can generate the expression levels Xgkj from N(βgk, 1) given the value of Ykj ([36]), where βgk = dgμ if Ykj = 1 and rg = 0 (i.e., DE genes from the FE model); βgk ~ N(dgμ, τ2) if Ykj = 1, rg = 1 (i.e., DE genes from the RE model); and βgk = 0 otherwise (i.e., EE genes or Ykj = 0).
We mainly consider those situations where a wise choice about which method to use can make a difference in identifying an enriched gene set, so we set the mean effect size of the DE genes μ ∈ {0.3, 0.45, 0.6} to make the signal-to-noise ratio not too high (otherwise, all the methods perform well). We further set τ ∈ {0.52, 1}. A total number of 500 (1000 for ω = 0.2) independent replicate datasets were simulated for each combination of the design parameters (ω, λ, γ, μ, τ).
We first examined the test size for all the methods compared. For the cases with ω = 0.2 where the null hypothesis of no enrichment holds for the gene set, we computed type I errors (i.e., test sizes) and then compared them with the nominal significance level 0.05. We report the results of simulated test sizes in Section 2.1 of Supplementary Material. We find that under the null, our iGSEA methods and MAPE-G seem to be a bit conservative and so tend to reject the null less than expected; MAPE-P seems to be aggressive and so tend to reject the null more than expected, especially for large γ; and MAPE-I is often somewhere between MAPE-G and MAPE-P. Thus, for a fair comparison in power, we need to match the type 1 errors of all the methods. To do so, for each non-null setting (i.e., ω ≠ 0.2), we used 1000 replicates under ω = 0.2 to simulate the critical value from the empirical reference distribution of the enrichment score; and we computed the power based on the simulated critical value so that the type I error of each method was controlled at 0.05.
We examined the power for all the combinations of (ω, λ, γ, μ, τ); and in Section 2.2 of Supplementary Material, we report the results for all the non-null settings except for those in which all the three proposed iGSEA methods worked well and have nearly 100% power. In our simulation, we observe that among the three existing methods, their power typically follows the order MAPE-P>MAPE-I>MAPE-G. Thus, to reduce the number of lines in the figures, we only plot the maximum power of the three MAPE methods in each setting, labeled by maxMAPE, instead of each individual power. As we expect, the increase of the enrichment signal ω or the mean effect size μ would boost the power of all the involved tests. The increase of the sampling rate λ also has a positive impact on the power. Among all the methods, either iGSEA-AT or iGSEA-RE appears to be the top performer in most of the settings; and maxMAPE has lower power than the above two methods except for only a few settings where ω = 0.3.
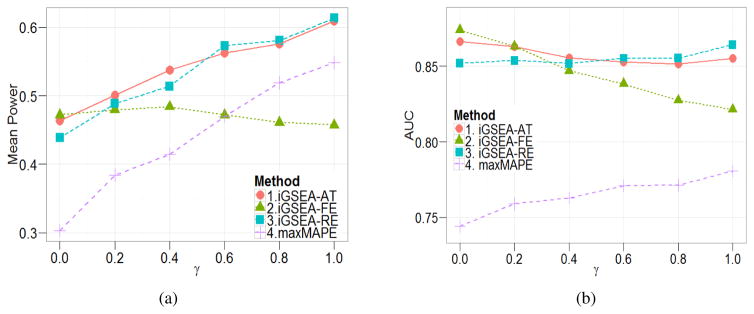
Figure 1(a) displays the mean power over the different settings stratified by the proportion of the RE genes γ. It seems that γ plays an important role in the relative performance of the three proposed iGSEA methods. When γ = 0 (i.e., all genes follow the FE model), it is not surprising to observe that iGSEA-FE has the highest mean power; and iGSEA-AT has mean power quite close to iGSEA-FE. When γ is small, iGSEA-AT outperforms both iGSEA-FE and iGSEA-RE. As γ is moving to 1, iGSEA-RE tends to outperform the other methods; however, the performance of iGSEA-AT is very close to that of iGSEA-RE. Overall, in terms of the mean power, iGSEA-AT is better than iGSEA-RE when there is no or a small proportion of RE genes; and it is much better than iGSEA-FE when there exist RE genes. In addition, iGSEA-AT is better than maxMAPE for all γ. Thus, in realistic situations where γ is unknown, we recommend iGSEA-AT as a safe choice for its stable performance.
Figure 1.
Simulation results for binary phenotypes: (a) the mean power (b) the mean AUC stratified by the proportion of RE genes γ.
Sensitivity vs. specificity
We proceed to compare the sensitivity and specificity of the methods via ROC curves by generating multiple gene sets. We assume that each generated dataset contains four independent studies, each having 20 normal samples and 20 tumor samples as before; and there are 1000 genes in the genome of interest, of which the first 100 are UR genes, the last 100 are DR genes, and the rest are EE genes. We generated 100 gene sets of varying sizes, of which the first 30% are enriched by UR genes, the next 30% are enriched by DR genes and the last 40% are non-enriched. For each of these gene sets, its size was independently generated from N(100, 302) and then left-truncated at 25; and UR, DR and EE genes were randomly chosen from the corresponding populations. We set ω = 0.3, μ = 0.45, τ = 0.52, and λ = 0.7. The detail about constructing the different types of gene sets and generating expression levels for the different types of genes can be found in Section 2.3 of Supplementary Material.
We present an example of ROC curves in each setting of γ using one randomly generated dataset in Section 2.4 of Supplementary Material. The curves show that all the three iGSEA methods have better performance than the MAPE methods. Among them, iGSEA-FE seems to be the best for small γ but the poorest for large γ while iGSEA-RE shows an opposite pattern; and iGSEA-AT is the best for medium γ, and otherwise, it is somewhere between the other two. We further examine the average AUC (area under the ROC curve) of each method by simulating 200 datasets under each setting considered. As clearly shown in Figure 1(b), the three iGSEA methods have much higher AUC than the MAPE methods. Further, as γ increases, the AUC of iGSEA-FE tends to decrease and the AUC of iGSEA-RE tends to increase while that of iGSEA-AT is steadier. Overall, iGSEA-AT has the best performance in terms of AUC as it is close to iGSEA-FE for small γ, close to iGSEA-RE for large γ and it is the best in the middle. This pattern is similar to what we observed from power results in Figure 1(a), which leads to the same conclusion that iGSEA-AT should be chosen in situations when γ is not known.
We report the mean AUC values for each proposed iGSEA method before and after our size adjustment in Table 1. It is clear that the use of the size-adjusted KS statistic in (10) consistently improves the mean AUC of all the three iGSEA methods. In addition, the performance of all the six methods in FDR control is reported in Section 2.5 of Supplementary Material, where we find that the iGSEA methods outperform the MAPE methods, regardless of the γ value.
Table 1.
Mean AUC values of the proposed iGSEA methods before and after our size adjustment.
| Method | Adjustment | γ = 0 | γ = 0.2 | γ = 0.4 | γ = 0.6 | γ = 0.8 | γ = 1 |
|---|---|---|---|---|---|---|---|
| iGSEA-FE | Before | 0.862 | 0.848 | 0.832 | 0.824 | 0.810 | 0.805 |
| After | 0.874 | 0.863 | 0.847 | 0.838 | 0.827 | 0.821 | |
|
| |||||||
| iGSEA-RE | Before | 0.837 | 0.839 | 0.840 | 0.842 | 0.841 | 0.851 |
| After | 0.852 | 0.854 | 0.852 | 0.855 | 0.855 | 0.864 | |
|
| |||||||
| iGSEA-AT | Before | 0.852 | 0.848 | 0.842 | 0.840 | 0.838 | 0.839 |
| After | 0.866 | 0.863 | 0.856 | 0.853 | 0.852 | 0.855 | |
5.2. Continuous phenotypes
In practice, many continuous response data can be approximated closely by normal distributions, especially after appropriate transformation. As a typical example of continuous phenotypes, we assume that the response Y follows a normal distribution, where the GLM (1) becomes a linear regression model: E(Ykj|Xjgk = x) = αgk + βgkx.
Power comparison
We used the same settings for the total number of genes in the genome (G), the size of the generated gene set (Gs), the number of GSEA studies (K), the numbers of the UR, DR, EE genes, and the proportion of RE genes (γ ∈ {0, 0.2, 0.4, 0.6, 0.8, 1}) as in the binary case. We set λ = 0.7, ω0 = 0.2, and ω ∈ {0.2, 0.3}. We assume that Xjgk and Yjk follow a bivariate normal distribution BV N(μx, μy, σx, σy, ρgk) for j = 1, ⋯, 20, where μx = μy = 0, and σx = σy = 1, and ρgk = βgk. So we simulated Yjk from N(0, 1) and then simulated the expression levels based on the conditional distribution . For a RE gene, we set βgk ~ N(μg, 0.252), where μg ~ N(0.3dg, 0.12); otherwise, we set βgk = μg, where μg ~ N(0.3dg, 0.12) for a DE gene and μg = 0for an EE gene. Note that βgk ∈ (−1, 1), we truncated its value at -0.9 and 0.9 if βgk > 0.9 and βgk < −0.9.
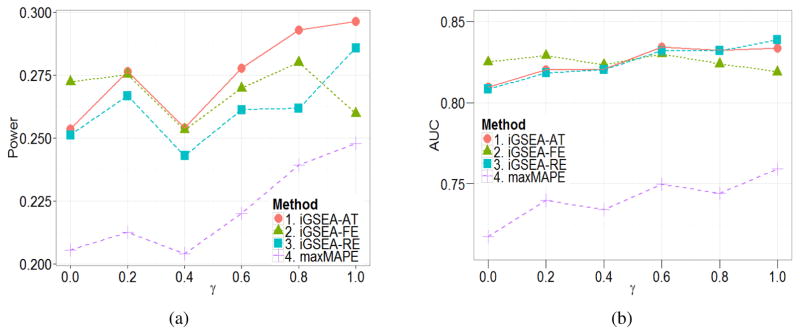
By examining the type I errors of the methods under the settings with the enrichment signal ω = 0.2, we find that the three iGSEA methods are relatively conservative, which is similar to what we find in the case of binary phenotypes. Thus, we used simulated critical values for each method to control the type I error at 0.05 and compared the power in Figure 2(a). Again, all the three proposed methods work better than the three MAPE methods for all γ. Unlike the binary case, iGSEA-FE seems to outperform iGSEA-RE except for γ = 1; and iGSEA-AT seems to outperform iGSEA-FE for medium or large γ. Overall, iGSEA-AT seems to be the best in terms of power.
Figure 2.
Simulation results for continuous phenotypes: (a) the power; (b) the mean AUC stratified by the proportion of RE genes γ.
Sensitivity vs. specificity
The way we generated the different types of genes and gene sets is the same as in the multiple-gene-set model for the binary case; and we generated μg, βgk, Yjk and Xjgk as in the single-gene-set model for the normal case. The average AUC over 200 datasets for each γ value is shown in Figure 2(b). It is clear that the three iGSEA methods outperform the MAPE methods. For small γ, iGSEA-AT is slightly better than iGSEA-RE; and it is better than iGSEA-FE for large γ.
6. Data example
Here, we illustrate the proposed methods using real expression data and real gene sets. To identify pivotal gene sets involved in lung cancer, we conducted integrative GSEA of five studies using pathways in Kyoto Encyclopedia of Genes and Genomes (KEGG), which is a comprehensive public database containing a large collection of human curated pathways [37]. The data contain four microarray mRNA datasets, including three from [21] and the other from [20], and one RNA-seq dataset [22]. Each of the five expression datasets contains both case and control samples. The detail of the datasets, including the source, the type of experiment and the sample size, is given in Section 3 of Supplementary Material. All expression data were log2-transformed and then standardized.
6.1. Performance Evaluation
To draw ROC curves, we constructed 60 benchmark pathways, including 30 positive controls (PC) and 30 negative controls (NC). A PC pathway includes 25 “essential” genes and 25 “non-essential” genes while a NC pathway includes 50 “nonessential” genes. To randomly generate PCs and NCs, we used the list of “essential” genes given in [14], which contains genes that are believed to be highly related to lung cancer according to the literature, while the list of “non-essential” genes contains those excluded from the list of “essential” genes and any KEGG pathway.
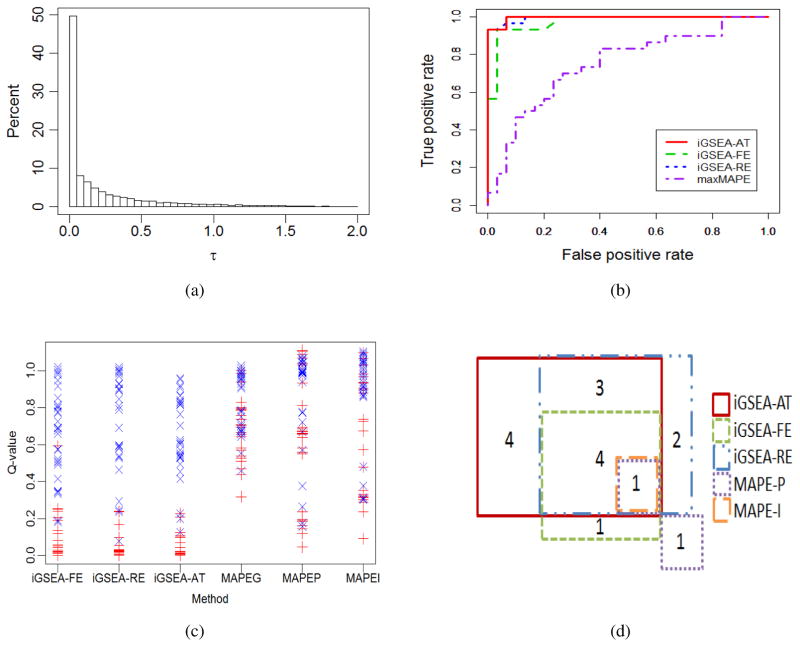
Through an exploratory analysis of the data, we find that although the estimated between-study heterogeneity is close to zero for 50% of the genes, it varied largely among potentially DE genes and 16% of the genes have estimated values greater than 0.5, as seen in Figure 3(a). This obviously indicates neither the FE nor RE model holds for all the genes considered. Due to the conservative nature of iGSEA-AT, we set α1 = 0.04, making it a bit easier to reject than the default value 0.0253. Figure 3(b) shows the ROC curves of the three iGSEA methods and MAPEI, since MAPEI is slightly better than MAPEG and MAPEP in this example; and Table 2 presents the AUC value for each of the six methods. The three iGSEA methods clearly have better performance than the three MAPE methods. As seen from the AUC table, the performance of iGSEA-AT and iGSEA-RE is quite comparable, and both have greater AUC than iGSEA-FE. Recall that in our simulation for the binary case, iGSEA-AT and iGSEA-RE often performed better than iGSEA-FE when γ is large. Thus, the above AUC results might hint that the between-study heterogeneity cannot be ignored for a large portion of the DE genes in this example. Figure 3(c) further shows the estimated Q-values of the benchmark pathways computed from the six methods. The three iGSEA methods separate the PC pathways (red “×”s) from the NC pathways (blue “+”s) very well, while the three MAPE methods yield a much poorer distinction.
Figure 3.
Data example: (a) the histogram of estimated between-study heterogeneity ; (b) ROC curves of the three iGSEA methods and MAPEI using 60 constructed benchmark pathways; (c) Estimated Q-values of benchmark pathways from the six methods, where red “×”s and blue “+”s represent positive and negative controls, respectively; (d) Venn diagram of enriched KEGG pathways identified by at least one of the methods.
Table 2.
Data example: area under the ROC curve of each method considered.
| FE | RE | AT | MAPE-G | MAPE-P | MAPE-I | |
|---|---|---|---|---|---|---|
| AUC | 0.972 | 0.994 | 0.996 | 0.707 | 0.709 | 0.748 |
6.2. Results
We tested KEGG pathways and report the estimated Q-values of those identified by any of the methods in Section 3 of Supplementary Material. In total, iGSEA-FE, iGSEA-RE and iGSEA-AT reported 6, 10 and 12 enriched pathways, respectively. By contrast, MAPE-P, MAPE-G and MAPE-I only reported 2, 0 and 1, respectively, even with π̂0 = 0.5 in (9). Figure 3(d) shows the Venn diagram for the methods. There are four pathways can be detected by all the three iGSEA methods but none of the MAPE methods. For example, “glyoxylate and dicarboxylate metabolism” is a pathway that has been found to be significantly correlated with loss of tumor differentiation [38]. Also, there are four pathways that were detected only by iGSEA-AT. Among them, “primary immunodeficiency” is a complex series of diseases, and may be associated with adenocarcinoma [39]. This pathway has been reported by [40] to be associated with early-stage lung adenocarcinoma. Also, for the pathway “glycosaminoglycan degradation”, it is known that the structural characteristics of glycosaminoglycans and enzymes involved in their degradation are involved in cancer progression [41]. Thus, these findings are consistent with recent studies in lung cancer while none of the other methods identified them.
7. Discussion
We have shown that the proposed iGSEA methods typically outperformed the MAPE methods through simulation and a data example. In particular, iGSEA-AT has good overall performance; and unlike iGSEA-FE and iGSEA-RE, it seems not to be sensitive to model specification in meta-analysis, due to a data-adaptive strategy of choosing FE vs. RE models. Thus, we recommend iGSEA-AT for combining multiple GSEA studies in practical situations where there is typically no one-size-fits-all model.
We mention that in our numerical studies, for iGSEA-AT, we used Cochran’s Q test to estimate the first-stage P-value p1g and the asymptotic test of to estimate the second-stage P-value p2g. In our preliminary simulation, we find that using permutation-based methods led to similar results. This is because whether the permutation or asymptotic methods are used may not affect the ordering of much. However, the permutation procedures were much slower when the number of genes is large.
Computational efficiency is critical in practice given the increasingly large numbers of genes, gene sets, samples, and available datasets. The three iGSEA methods are fairly fast to conduct and numerically stable. To illustrate the relative efficiency in computing, we report the time to run each method with B = 500 for a randomly generated dataset of four studies with λ = 1 under the one-gene-set model for the binary case in Section 5.1: it takes iGSEA-FE and iGSEA-RE less than 1 second to finish, iGSEA-AT about 4 seconds, and the three MAPE methods 83–86 seconds, using a machine with Windows 8.1 64-bit Operating System, Intel(R) Core(TM) i7-4700MQ CPU @2.40GHz and 8 GB of memory.
In some applications, it would be desirable to adjust for individual-level confounding covariates/factors such as age, race, environmental exposures, etc. Using the GLM setup described in Section 2, the proposed iGSEA methods can easily provide covariate-adjusted estimates as well as covariate-adjusted score statistics and associated variances within each study, and then they can be combined in the same way as we discussed in Section 3. [18] mentioned that using meta-analysis methods based on score statistics, the numbers and types of covariates even need not be the same among the component studies.
Although it covers a wide range of models and distributions, the GLM is not the best way to model censored survival outcomes. Instead, a standard approach is to use Cox proportional hazards models [42]. We note that the extension of our iGSEA methods to survival outcomes is straightforward. Here, we define Yjk as the observed time (either censoring or event time) for sample j in study k. Using the partial likelihood function under the Cox model, Ugks and Vgks can be constructed accordingly, and all the subsequent steps in our iGSEA methods remain unchanged.
The proposed methods are applicable to situations when expression data are from both microarray and NGS experiments, as shown in our data example. To enhance comparability among studies and ensure estimation of the same parameter, we should carefully review inclusion criteria and adjustments of covariates, and conduct appropriate data preprocessing including annotation and alignment across all different platforms and versions, background correction and normalization of expression data, removal of batch effects whenever possible. For highly complex datasets where the above could fail, blindly applying the proposed methods would be inappropriate; and we suggest to develop robust iGSEA methods based on aggregation of ranked lists from component studies. We further note that although presented in the context of gene expression studies, the proposed iGSEA methods seem to be equally applicable to meta-analysis of other omics data, e.g., epigenomics/methylation studies in large consortia.
Finally, software for the proposed methods is available as an R package named “iGSEA” and is freely distributed on CRAN after testing.
Supplementary Material
Figure 1: Comparison of the empirical CDF of vs with the asymptotic CDF.
Table 1: Type I errors of each method when λ = 0.5, μ = 0.3 and τ = 1.
Table 2: Type I errors of each method when λ = 0.5, μ = 0.45 and τ = 1.
Table 3: Type I errors of each method when λ = 0.5, μ = 0.6 and τ = 1.
Figure 2: Power comparison for the settings with ω = 0.3, μ = 0.3 and τ = 0.52.
Figure 3: Power comparison for the settings with ω = 0.3, μ = 0.3 and τ = 1.
Figure 4: Power comparison for the settings with ω = 0.3, μ = 0.45 and τ = 0.52.
Figure 5: Power comparison for the settings with ω = 0.3, μ = 0.45 and τ = 1.
Figure 6: Power comparison for the settings with ω = 0.3, μ = 0.6 and τ = 0.52.
Figure 7: Power comparison for the settings with ω = 0.3, μ = 0.6 and τ = 1.
Figure 8: Power comparison for the settings with ω = 0.4, μ = 0.3 and τ = 0.52.
Figure 9: Power comparison for the settings with ω = 0.4, μ = 0.3 and τ = 1.
Figure 10: Power comparison for the settings with ω = 0.4, μ = 0.45 and τ = 0.52.
Figure 11: Power comparison for the settings with ω = 0.4, μ = 0.45 and τ = 1.
Figure 12: Power comparison for the settings with ω = 0.5, μ = 0.3 and τ = 0.52.
Figure 13: Power comparison for the settings with ω = 0.5, μ = 0.3 and τ = 1.
Table 4: Design detail in constructing gene sets; 30% of the gene sets are enriched by UR genes, another 30% are enriched by DR genes, and the remaining 40% are non-enriched.
Table 5: Design detail in generating expression levels for different types of genes. Genes 1–100 are UR genes, Genes 101–900 are EE genes, and Genes 901- 1000 are DR genes.
Figure 14: ROC curves for detecting multiple enriched gene sets using iGSEA-FE, iGSEA-RE, iGSEA-AT, and maxMAPE. The purple curve on the plots actually represents the performance of MAPE-P because it is the best of its kind.
Table 6: Empirical FDRs of each method in the multiple-gene-set simulation for binary phenotypes.
Table 7: Lung adenocarcinoma datasets involved in data analysis
Table 8: The estimated Q-values of identified KEGG pathways
Acknowledgments
This work was supported by the NIH grant R15GM113157 (PI: Xinlei Wang).
Appendix: Algorithm
I. Computing gene-level statistics
For each study k, compute the estimated effect β̂gk, the estimated precision wgk, the score statistic Ugk, and the corresponding variance estimate Vgk for the genes involved in study k, where g = 1, …, G and k = 1, …, K.
II. Meta-analysis
For each gene g, compute the overall gene-level statistic ug, where for the FE method, for the RE method, and for the AT method.
III. Gene set analysis
For each gene set s, order the genes in and out of the set according to the values of ug (from small to large), and then compute the enrichment score using the size-adjusted one-sided KS statistic vs.
Randomly assign genes to set s B times and compute the permuted statistics, , 1 ≤ b ≤ B, 1 ≤ s ≤ S.
- Estimate the P-value of set s by
Estimate the Q-value of gene set s by (9).
Report those gene sets with Q-value< δ as enriched.
References
- 1.Shen K, Tseng GC. Meta-analysis for pathway enrichment analysis when combining multiple genomic studies. Bioinformatics. 2010;26:1316–1323. doi: 10.1093/bioinformatics/btq148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with ease. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15 545–15 550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Efron B, Tibshirani R. On testing the significance of sets of genes. The Annals of Applied Statistics. 2007;1:107–129. [Google Scholar]
- 5.Newton MA, Quintana FA, den Boon Srikumar Sengupta JA, Ahlquist P. Random-set methods identify distinct aspects of the enrichment signal in gene-set analysis. Annals of Applied Statistics. 2007;1:85–106. [Google Scholar]
- 6.Ackermann M, Strimmer K. A general modular framework for gene set enrichment analysis. BMC Bioinformatics. 2009;10:47. doi: 10.1186/1471-2105-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hung JH, Yang TH, Hu Z, Weng Z, DeLisi C. Gene set enrichment analysis: performance evaluation and usage guidelines. Brief Bioinform. 2012;13:281–291. doi: 10.1093/bib/bbr049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Downward J. Cancer biology: signatures guide drug choice. Nature. 2006;439:274–275. doi: 10.1038/439274a. [DOI] [PubMed] [Google Scholar]
- 9.Wang X. Identification of common tumor signatures based on gene set enrichment analysis. In Silico Biol. 2011;11:1–10. doi: 10.3233/ISB-2012-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ullah U, Tripathi P, Lahesmaa R, Rao KVS. Gene set enrichment analysis identifies lif as a negative regulator of human th2 cell differentiation. Sci Rep. 2012;2:464. doi: 10.1038/srep00464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng B, Liao Z, Locascio JJ, Lesniak KA, Roderick SS, Watt ML, Eklund AC, Zhang-James Y, Kim PD, Hauser MA, et al. Pgc-1α, a potential therapeutic target for early intervention in parkinson’s disease. Sci Transl Med. 2010;2:52–73. doi: 10.1126/scitranslmed.3001059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4:682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 13.Farkas IJ, Korcsmáros T, Kovács IA, Mihalik A, Palotai R, Simkó GI, Szalay KZ, Szalay-Beko M, Vellai T, Wang S, et al. Network-based tools for the identification of novel drug targets. Sci Signal. 2011;4:pt3. doi: 10.1126/scisignal.2001950. [DOI] [PubMed] [Google Scholar]
- 14.Chen M, Zang M, Wang X, Xiao G. A powerful bayesian meta-analysis method to integrate multiple gene set enrichment studies. Bioinformatics. 2013;29:862–869. doi: 10.1093/bioinformatics/btt068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu YJ, Berndt SI, Gustafsson S, Ganna A, Hirschhorn J, North KE, Ingelsson E, Lin DY Consortium GIANT. Meta-analysis of gene-level associations for rare variants based on singlevariant statistics. Am J Hum Genet. 2013 Aug;93(2):236–248. doi: 10.1016/j.ajhg.2013.06.011. URL http://dx.doi.org/10.1016/j.ajhg.2013.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han B, Eskin E. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. The American Journal of Human Genetics. 2011;88:586–598. doi: 10.1016/j.ajhg.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang ZZ, Lin DY. Meta-analysis of sequencing studies with heterogeneous genetic associations. Genetic Epidemiology. 2014;38:389–401. doi: 10.1002/gepi.21798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin DY, Zeng D. Meta-analysis of genome-wide association studies: no efficiency gain in using individual participant data. Genetic Epidemiology. 2010;34:60–66. doi: 10.1002/gepi.20435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang ZZ, Lin DY. Mass: meta-analysis of score statistics for sequencing studies. Bioinformatics. 2013;29:1803–1805. doi: 10.1093/bioinformatics/btt280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu CQ, Ding K, Strumpf D, Weir BA, Meyerson M, Pennell N, Thomas RK, Naoki K, Ladd-Acosta C, Liu N, et al. Prognostic and predictive gene signature for adjuvant chemotherapy in resected non-small-cell lung cancer. Journal of Clinical Oncology. 2010;28:4417–4424. doi: 10.1200/JCO.2009.26.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shedden K, Taylor JMG, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, Eschrich S, Jurisica I, Giordano TJ, Misek DE, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nature Medicine. 2008;14:822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim S, Jung Y, Park J, Cho S, Seo C, Kim J, Kim P, Park J, Seo J, Kim J, et al. A high-dimensional, deep-sequencing study of lung adenocarcinoma in female never-smokers. PLoS ONE. 2013;8:e55–596. doi: 10.1371/journal.pone.0055596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cochran WG. The combination of estimates from different experiments. Biometrics. 1954;10:101–129. [Google Scholar]
- 24.Whitehead A, Whitehead J. A general parametric approach to the meta-analysis of randomized clinical trials. Statistics in Medicine. 1991;10:1665–1677. doi: 10.1002/sim.4780101105. [DOI] [PubMed] [Google Scholar]
- 25.Higgins, Thompson Quantifying heterogeneity in a meta-analysis. Statistics in medicine. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 26.Sidik K, Jonkman JN. A comparison of heterogeneity variance estimators in combining results of studies. Statistics in Medicine. 2007;26:1964–1981. doi: 10.1002/sim.2688. [DOI] [PubMed] [Google Scholar]
- 27.Sidik K, Jonkman JN. Simple heterogeneity variance estimation for meta-analysis. Journal of the Royal Statistical Society, Series C: Applied Statistics. 2005;54:367–384. [Google Scholar]
- 28.Tsiatis AA, Rosner GL, Mehta CR. Exact confidence intervals following a group sequential test. Biometrics. 1984;40:797–803. [PubMed] [Google Scholar]
- 29.Fisher RA. Statistical methods for research workers. London: Oliver & Boyd; 1932. [Google Scholar]
- 30.Lehmacher W, Wassmer G. Adaptive sample size calculations in group sequential trials. Biometrics. 1999;55:1286–1290. doi: 10.1111/j.0006-341x.1999.01286.x. [DOI] [PubMed] [Google Scholar]
- 31.Sheng J, Qiu P. On p-value calculation for multi-stage additive tests. Journal of Statistical Computation and Simulation. 2007;77:1057–1064. [Google Scholar]
- 32.Storey JD. A direct approach to false discovery rates. Journal of the Royal Statistical Society: Series B. 2002;64:479–498. [Google Scholar]
- 33.Bass J, Dabney A, Robinson D. r package version 2.8.0 2015. qvalue: Q-value estimation for false discovery rate control. [Google Scholar]
- 34.Smirnov N. On the derivations of the empirical distribution curve. Matematicheskii Sbornilt. 1939;6:2–26. [Google Scholar]
- 35.Gail MH, Green SB. A generalization of the one-sided two-sample kolmogorov-smirnov statistic for evaluating diagnostic tests. Biometrics. 1976;32:561–570. [PubMed] [Google Scholar]
- 36.Cornfield J. Joint dependence of risk of coronary heart disease on serum cholesterol and systolic blood pressure: a discriminant function analysis. Federation proceedings. 1962;21:58–61. [PubMed] [Google Scholar]
- 37.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. Kegg for integration and interpretation of large-scale molecular data sets. Nucleic Acids Research. 2012;40:D104–D109. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Creighton C, Hanash S, Beer D. Gene expression patterns define pathways correlated with loss of differentiation in lung adenocarcinomas. FEBS Letters. 2003;570:167–170. doi: 10.1016/s0014-5793(03)00259-x. [DOI] [PubMed] [Google Scholar]
- 39.Milner JD, Holland SM. The cup runneth over: lessons from the ever-expanding pool of primary immunodeficiency diseases. Nat Rev Immunol. 2013;13:635–648. doi: 10.1038/nri3493. [DOI] [PubMed] [Google Scholar]
- 40.Saji H, Tsuboi M, Shimada Y, Kato Y, Hamanaka W, Kudo Y, Yoshida K, Matsubayashi J, Usuda J, Ohira T, et al. Gene expression profiling and molecular pathway analysis for the identification of early-stage lung adenocarcinoma patients at risk for early recurrence. Oncology Reports. 2013;29:1902–1906. doi: 10.3892/or.2013.2332. [DOI] [PubMed] [Google Scholar]
- 41.Afratis N, Gialeli C, Nikitovic D, Tsegenidis T, Karousou E, Theocharis AD, Pavao MS, Tzanakakis GN, Karamanos NK. Glycosaminoglycans: key players in cancer cell biology and treatment. FEBS Journal. 2012;279:1177–1197. doi: 10.1111/j.1742-4658.2012.08529.x. [DOI] [PubMed] [Google Scholar]
- 42.Cox DR. Regression models and life-tables. Journal of the Royal Statistical Society. Series B (Methodological) 1972:187–220. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure 1: Comparison of the empirical CDF of vs with the asymptotic CDF.
Table 1: Type I errors of each method when λ = 0.5, μ = 0.3 and τ = 1.
Table 2: Type I errors of each method when λ = 0.5, μ = 0.45 and τ = 1.
Table 3: Type I errors of each method when λ = 0.5, μ = 0.6 and τ = 1.
Figure 2: Power comparison for the settings with ω = 0.3, μ = 0.3 and τ = 0.52.
Figure 3: Power comparison for the settings with ω = 0.3, μ = 0.3 and τ = 1.
Figure 4: Power comparison for the settings with ω = 0.3, μ = 0.45 and τ = 0.52.
Figure 5: Power comparison for the settings with ω = 0.3, μ = 0.45 and τ = 1.
Figure 6: Power comparison for the settings with ω = 0.3, μ = 0.6 and τ = 0.52.
Figure 7: Power comparison for the settings with ω = 0.3, μ = 0.6 and τ = 1.
Figure 8: Power comparison for the settings with ω = 0.4, μ = 0.3 and τ = 0.52.
Figure 9: Power comparison for the settings with ω = 0.4, μ = 0.3 and τ = 1.
Figure 10: Power comparison for the settings with ω = 0.4, μ = 0.45 and τ = 0.52.
Figure 11: Power comparison for the settings with ω = 0.4, μ = 0.45 and τ = 1.
Figure 12: Power comparison for the settings with ω = 0.5, μ = 0.3 and τ = 0.52.
Figure 13: Power comparison for the settings with ω = 0.5, μ = 0.3 and τ = 1.
Table 4: Design detail in constructing gene sets; 30% of the gene sets are enriched by UR genes, another 30% are enriched by DR genes, and the remaining 40% are non-enriched.
Table 5: Design detail in generating expression levels for different types of genes. Genes 1–100 are UR genes, Genes 101–900 are EE genes, and Genes 901- 1000 are DR genes.
Figure 14: ROC curves for detecting multiple enriched gene sets using iGSEA-FE, iGSEA-RE, iGSEA-AT, and maxMAPE. The purple curve on the plots actually represents the performance of MAPE-P because it is the best of its kind.
Table 6: Empirical FDRs of each method in the multiple-gene-set simulation for binary phenotypes.
Table 7: Lung adenocarcinoma datasets involved in data analysis
Table 8: The estimated Q-values of identified KEGG pathways