Abstract
Enzyme and metabolic engineering offer the potential to develop biocatalysts for converting natural resources into a wide range of chemicals. To broaden the scope of potential products beyond natural metabolites, methods of engineering enzymes to accept alternative substrates and/or perform novel chemistries must be developed. DNA synthesis can create large libraries of enzyme-coding sequences, but most biochemistries lack a simple assay to screen for promising enzyme variants. Our solution to this challenge is structure-guided mutagenesis in which optimization algorithms select the best sequences from libraries based on specified criteria (i.e. binding selectivity). Here, we demonstrate this approach by identifying medium-chain (C6-C12) acyl-ACP thioesterases through structure-guided mutagenesis. Medium-chain fatty acids, products of thioesterase-catalyzed hydrolysis, are limited in natural abundance compared to long-chain fatty acids; the limited supply leads to high costs of C6-C10 oleochemicals such as fatty alcohols, amines, and esters. Here, we applied computational tools to tune substrate binding to the highly-active ‘TesA thioesterase in Escherichia coli. We used the IPRO algorithm to design thioesterase variants with enhanced C12- or C8-specificity while maintaining high activity. After four rounds of structure-guided mutagenesis, we identified three thioesterases with enhanced production of dodecanoic acid (C12) and twenty-seven thioesterases with enhanced production of octanoic acid (C8). The top variants reached up to 49% C12 and 50% C8 while exceeding native levels of total free fatty acids. A comparably sized library created by random mutagenesis failed to identify promising mutants. The chain length-preference of ‘TesA and the best mutant were confirmed in vitro using acyl-CoA substrates. Molecular dynamics simulations, confirmed by resolved crystal structures, of ‘TesA variants suggest that hydrophobic forces govern ‘TesA substrate specificity. We expect that the design rules we uncovered and the thioesterase variants identified will be useful to metabolic engineering projects aimed at sustainable production of medium-chain oleochemicals.
Keywords: Molecular Dynamics, IPRO, Thioesterase, ‘TesA, Fatty Acid, Redesign, Dodecanoic Acid, Tetradecanoic Acid
Introduction
Free fatty acids (FFAs) are energy-rich precursors of membrane lipids, natural oils, liquid transportation fuels (a.k.a. biodiesel), and high-value oleochemicals (e.g. fatty alcohols, aldehydes, olefins, and waxes)1–4. Oleochemical properties such as energy content, melting point, and volatility are dictated by the chain length, degree of saturation, and branching pattern of the acyl-chain5. Fuels and oleochemicals derived from microbially produced FFAs could displace current, unsustainable plant feedstocks and reduce carbon footprints relative to petrochemical alternatives6–7. Unfortunately, natural sources of medium-chain length FFAs and lipids are significantly less abundant than longer chain compounds. The limited supply and costly petrochemical synthesis alternative leads to higher selling prices for medium-chain oleochemicals (e.g., 1-octanol costs approximately twice as much as 1-hexadecanol per pound)4. These economic drivers make bioproduction of medium-chain length FFA and oleochemicals an attractive opportunity if biosynthesis pathways with high yield and selectivity can be assembled and optimized.
While many oleochemical pathways have been demonstrated in model hosts, chain length selectivity remains an unsolved challenge. The product distribution of most metabolic engineering efforts has been restricted to the chain-length of the most abundant acyl-thioester in the cell or the distribution created by expression of a thioesterase. Acyl chains that comprise FFAs, lipids, and oleochemicals are made by an iterative series of elongation, keto-reduction, dehydration and enoyl-reduction reactions acting on acyl-coenzyme A (acyl-CoA) or acyl-acyl-carrier protein (acyl-ACP) thioesters4. An acyl-chain is elongated by two carbon atoms per cycle until it is trans-esterified (into phospholipids, waxes, or esters), reduced (to a fatty aldehyde or alcohol), or hydrolyzed (yielding a FFA)1. The chain length distribution of these terminal products is controlled by the relative kinetics of elongation, transesterification, reduction, and/or hydrolysis. In Escherichia coli, the activity of phospholipid synthases (PlsB, PlsC) and fatty acid synthases (FabB, FabF, FabH) constrain the lipid composition to mostly C16 or C18 acyl-chains with little FFA content8. In contrast, E. coli can produce high titers of FFA with a wide range of chain-length distributions by the upregulation of native thioesterases (TesA and TesB) or heterologous expression of plant and bacterial thioesterases (see Table S1). In the absence of pathways for catabolizing FFA and/or acyl-thioesters (i.e. beta-oxidation), the specificity of the acyl-ACP thioesterase controls the chain-length distribution and the chemical properties of downstream oleochemicals (see Figure 1). Collectively, thioesterases exhibit a wide range of substrate specificities2, 9–14 that has been further diversified through protein engineering and evolution15–17. Even with this known diversity, very few thioesterases are specific towards a single aliphatic chain length10. Worse, heterologous expression of thioesterases frequently begets unexpected product distributions4, 10, poor FFA yields18–19, or both. Of the many studied thioesterases, ‘TesA (a cytosolic E. coli TesA lacking the N-terminal signal peptide) has been used extensively in metabolic engineering studies and has a crystallographically resolved structure20. However, ‘TesA has broad substrate specificity with relatively low medium-chain content2, 9. For these reasons, ‘TesA is an attractive system for applying enzyme engineering to improve thioesterase selectivity towards medium chain lengths.
Figure 1. Overview of fatty acid biosynthesis and role of acyl-ACP thioesterases.
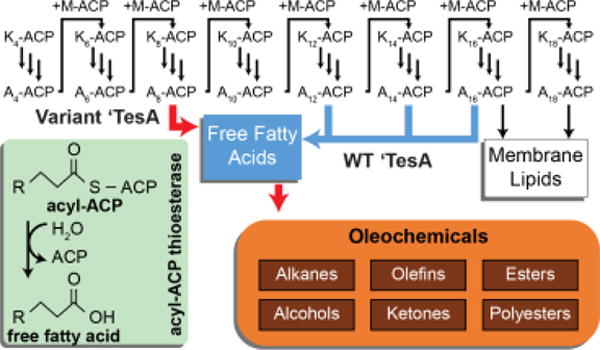
In each cycle of fatty acid biosynthesis, two carbons are added from a malonyl-ACP (M-ACP) yielding a β-ketoacyl-ACP (Kx-ACP where x is the number of carbons). Three reactions (three vertical arrows) reduce the Kx-ACP to a saturated acyl-ACP (Ax-ACP). In E. coli, A16-ACP and A18-ACP are incorporated into membrane lipids. Thioesterases produce free fatty acids by hydrolyzing the acyl-thioester bond (green inset). Tailoring the specificity of the acyl-ACP thioesterase (‘TesA) dictates free fatty acid and downstream oleochemical chain lengths (red arrows).
Enzyme engineering is generally pursued using directed evolution approaches that rely on high-throughput screening of large mutant libraries21. These large libraries are constructed using various mechanisms for diversifying the gene pool, including homologous (e.g., DNA shuffling) or non-homologous (e.g., overlap extension PCR) recombination, random mutagenesis (e.g., error-prone PCR), or combinations thereof (see Ref.22 for review). Large library sizes rely on high-throughput screening that takes advantage of optical properties, such as fluorescent or colorimetric assays. Currently, no high-throughput screen that can discriminate between different FFA chain lengths has been developed. When optical screens are unsuitable, more laborious experiments (e.g., mass spectrometry or NMR spectroscopy) can be used in low-throughput screens that mandate small, more focused libraries. Focused libraries can be generated through site saturation mutagenesis (e.g., degenerate oligonucleotide-primed PCR) but this approach can only include a small number of sites (three saturated sites would yield 8,000 variants) with a high percentage of inactive mutants. Smaller library sizes can be formed through site-directed mutagenesis23 (i.e., rational design) but so far it has been very difficult to forecast the effect of multiple mutations. When a protein structure is known, molecular modeling tools can suggest a handful of promising mutations while considering sequence-structure relationships as well as ther approximations.
Structure-based protein redesign procedures such as the Iterative Protein Redesign and Optimization (IPRO) method24 used here, offer several advantages over tools that simply suggest “hot spot” residues25–30, as they can capture the simultaneous effect of multiple mutations. Another class of computational protein redesign tools uses mostly sequence information to suggest crossover locations for generating combinatorial libraries31–37. However, these combinatorial libraries are limited by the parental sequence space and do not take full advantage of available structural information. De novo enzyme designs38–42 are usually less active than native ones (without the aid of directed evolution)38–39, 41. Thus, structure-based protein redesign can harness natural protein performance and suggest directed modifications using structural insight to meet a single or multiple design objectives. IPRO differs from other structure-based protein redesign procedures in that it employs a mixed-integer linear program to guarantee a global minimum for a given protein backbone structure, it can handle multiple decision criterion simultaneously, and distance restraints can be easily imposed to keep catalytic machinery intact24, 43–44. More extensive reviews of existing computational protein engineering procedures have been provided by Pantazes et al.45, Samish et al.46, and Huang et al.40
In this study, we applied the Iterative Protein Redesign and Optimization (IPRO) method24 to guide ‘TesA mutagenesis in search of variants that both improve medium-chain FFA specificity and maintain high thioesterase activity. Specifically, we engineered ‘TesA to yield additional dodecanoic acid (C12:0) and octanoic acid (C8:0) at the expense of the natively preferred tetradecanoic acid (C14:0). The employed redesign procedure involved recursively predicting in silico ‘TesA mutants with enhanced binding capabilities, analyzing the in vivo FFA composition, and modifying the computational explorations in a Design-Build-Test-Learn cycle47. We identified three separate ‘TesA mutants that exhibited a statistically significant (p<0.05) improvement in C12 composition over wild-type (WT) and twenty-two mutants with a statistically significant improvement in C8 composition. In comparison to previously studied thioesterases (see Table S1), our computationally predicted mutants include four of the ten most C12-specific and one of the ten most C8-specific thioesterases. While computational enzyme redesign that reaches industrially relevant performance metrics has so far remained elusive45, the results presented here demonstrate the potential of the adopted Design-Build-Test-Learn paradigm to pinpoint promising enzyme mutants45, 47–48.
Results and Discussion
Overview of the Design-Build-Test-Learn approach
In support of efforts to alter the product profile of ‘TesA towards medium chain length FFAs, we performed four rounds of site-directed mutagenesis structured around a Design-Build-Test-Learn cycle (Figure 2). In each round, mutagenesis targets and specific amino acid substitutions were selected after analyzing simulations of enzyme-substrate binding performed with IPRO24. Genes encoding the designed ‘TesA variants were constructed by Quikchange® or Gibson Assembly® of PCR products (Materials and Methods) and cloned into arabinose-inducible expression vectors. The impact of each mutation was assessed by quantifying the FFA content of a ‘TesA expressing E. coli culture (see Table S1). After each round, the product distributions for each mutant were analyzed and used to improve to the IPRO framework24 via changes in the scoring function parameters or refocusing the mutagenesis targets. The following sections describe the deployment of our computational enzyme Design phase, results of the Build-Test phase, and a discussion of what we have Learned about thioesterase selectivity after each round.
Figure 2. Overview of Predict-Design-Revise-Learn approach used to guide ‘TesA redesign.
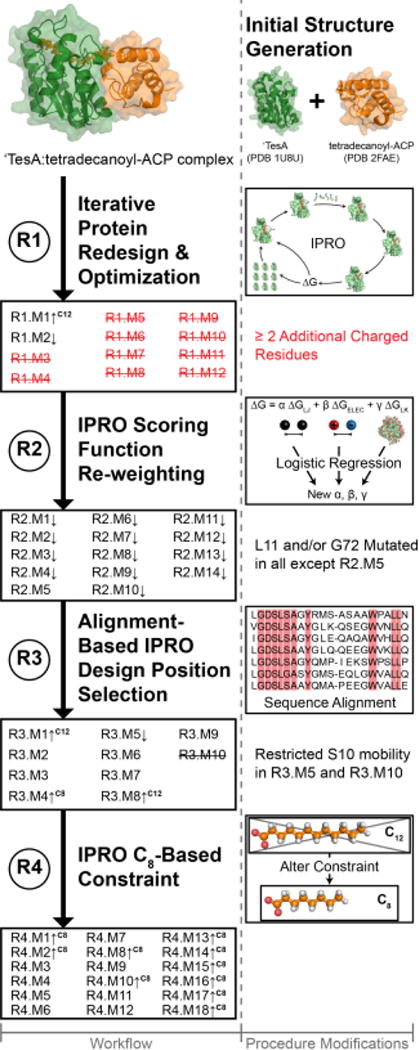
Pictorial illustration of the steps traversed (left column) and mutants identified with improved specificity towards C12- or C8- acyl-ACPs. The right column denotes the changes in the computational procedure modifications in response to the experimental results. Variants that produced significant improvements in the C12 (p<0.05) or C8 (p<0.005) fraction while maintaining WT production levels are indicated with an upward arrow followed by the FFA. Major improvements in C8 are indicated by p<0.005 rather than p<0.05 to highlight top designs Struck through variants indicate enzyme inactivity, and names followed by a downward arrow represent variants with reduced total production levels relative to WT.
IPRO constraints and implementation
IPRO enzyme redesign requires a three-dimensional model of the protein complex structure, a set of predetermined mutatable residues known as design positions, and a set of constraints that quantify the desired improvements that protein variants need to reach relative to wild-type. Examples of such constraints include imposing relations that require a (i) stronger interaction energy with the new substrate and (ii) weaker interaction energy with the native substrate compared to wild-type. Here, we constructed a structural model of ‘TesA bound to an acyl-ACP with acyl chains ranging from C8 to C14. The model was assembled from published structures of ‘TesA (PDB 1U8U)20 and decanoyl-ACP (PDB 2FAE)49. Docking between ‘TesA and octanoyl-ACP was modeled by systematically rotating the octanoyl-ACP structure about its phosphopantetheine linker until it aligned with bound octanoic acid in the ‘TesA crystal structure (PDB 1U8U, Materials and Methods). Other ‘TesA:acyl-ACP complexes were derived from the ‘TesA:octanoyl-ACP complex structure by adding atoms to the ω-1 carbon (i.e., the carbon furthest from ACP) of the octanoyl-ACP structure. The IPRO algorithm was used to search for mutations that led to improved binding between ‘TesA and the desired substrate while simultaneously discouraging binding of undesired substrate(s). Design positions for ‘TesA were selected based on proximity to the ω-1 carbon of bound tetradecanoyl-ACP while not considering positions vital for catalytic turnover (Materials and Methods). The fitness of each predicted variant was assessed using interaction energy as a proxy for binding energy (i.e., ΔG) thereby reducing force field dependence and requiring fewer calculations. Interaction energy is defined as GEnz:FFA,mm-GEnz-GFFA, where G is Gibbs free energy, “Enz” represents the enzyme (‘TesA or a variant thereof), and “min” indicates that the molecule(s) have undergone an energy minimization. In contrast, binding energy is defined as GEnz:FFA,min -GEnz,min -GFFA,min. The first constraint is aimed at worsening the interaction energy between ‘TesA and the native substrate, tetradecanoyl-ACP (e.g., C14). This first constraint safeguards against binding to even longer acyl-ACPs (e.g., C16, C18) because the repulsive interactions, which disfavor interactions with the long acyl-ACP (e.g., C14 and longer), are further exaggerated due to steric clashes or hydrophobicity. The second constraint requires interaction energy improvements between ‘TesA and the medium-chain acyl-ACP (e.g., C12). The imposition of the second constraint attempts to enhance binding with C12 or even shorter acyl-ACPs (e.g., C8, C10).
IPRO operates by successively performing redesign iterations for a preset number of cycles until a variant that simultaneously satisfies all constraints and optimizes the objective function is found. IPRO has been applied previously to modify E. coli β-glucuronidase substrate specificity50, alter the cofactor specificity of Candida boidnii xylose reductase51, graft a calciumbinding pocket into Thermoactinomyces vulgaris thermitase or a copper-binding pocket into E. coli thioredoxin52, and de novo design antibody variable regions that target influenza hemagglutinin, HIV gp120 and Ebola GP1-GP2 viruses42. Each IPRO iteration begins with a local backbone perturbation nearby a randomly selected design position from the requisite input set. The second step implements a mixed integer linear program (MILP) to identify the optimal set of amino acids for the new backbone conformation. The third step of IPRO executes a local, rigid-body docking (i.e., intramolecular movements are prohibited and only relative positioning between the enzyme and ligand is considered) to reorient the ligand (i.e., the acyl-ACP) within the binding site. The fourth and fifth steps perform an energy minimization of the entire enzyme complex. The sixth step and final steps evaluate the constraints set forth (i.e., (i) reducing binding to C14 and (ii) increasing binding to C12). The results of the iteration are retained or discarded based on the Metropolis criterion whereby worsening solutions are accepted with a decaying exponential probability as in simulated annealing (see Ref.24 for further details). The two constraints are imposed only at the ground state of the thioester hydrolysis with no additional calculations at the transition state. Instead, simple restraints on catalytic distances were imposed during all IPRO iterations so as to preserve, but not necessarily boost, catalytic activity50. Multiple IPRO trajectories (~10 independent trajectories) were simulated to discover alternative routes for improving specificity. For each variant, the difference of interaction energies for the short and long acyl-ACP with ‘TesA (i.e., ΔIE = IEC12 − IEC14) was calculated. The variants were prioritized based on the extent of the energy differences, and 10-20 variants were selected to build a focused library for experimental testing.
Method implementation for ‘TesA redesign
The first round (R1) of IPRO-guided mutagenesis helped tune the parameters of the scoring function to improve prediction accuracy. Our initial objective was a modest shift in substrate preference from C14 to C12, that makes up to 20% of the native ‘TesA product profile, in order to assess the efficacy of the redesign protocol. Of twelve tested variants, one improved C12 composition) and maintained WT production levels (i.e., ≥ WTTotal, p<0.05; R1.M1; Table S1). In contrast, ten were inactive (i.e., < ControlTotal) and included more than two charged substitutions (all except R1.M1 and R1.M2). The scoring function implemented within IPRO’s MILP rotamer-residue selection algorithm was identified as the source of the charged residue bias. The scoring function energy terms were re-weighted using logistic regression on a dataset of high-quality protein structures53. The updated scoring function roughly doubled native rotamer recovery relative to the existing scoring function (Materials and Methods). The former scoring function was adequate for earlier systems24, 42, 50–52, but the high hydrophobicity of ‘TesA (the energy term that was underemphasized in the former scoring function) made this enzyme especially susceptible to unsuccessful designs.
The modified scoring function was used to design a second round (R2) of variants with improved activity on C12 acyl chains. While all fourteen variants were active, thirteen mutants (all except R2.M5) produced less total FFA than WT and none improved the C12 fraction (p<0.05). All thirteen variants incorporated a mutation at either position L11 or G72. These positions were in hindsight deemed conserved (exhibiting 86.3% and 60.0% sequence conservation, respectively, across the L1-like lysophospholipase subgroup of the SGNH-hydrolase family) as they are immediately adjacent to S10 and N73 which are part of the oxyanion hole20. Therefore, both L11 and G72 were eliminated from the list of design positions in subsequent rounds.
The third round (R3) of mutagenesis, based on the updated set of design positions and revised scoring function, resulted in a higher fraction of variants that maintained WT activity (80.0%) compared to that of R1 and R2 (8.3% and 7.1%, respectively). Of the ten tested R3 designs, two improved the C12 product composition (R3.M1, R3.M8; p<0.05) and one led to a major increase in the C8 mole fraction (R3.M4; p<0.005). Notably, as the design constraints imposed by IPRO in rounds R1 through R3 did not preclude binding to acyl-ACPs smaller than C12 (i.e., C6, C8, C10), it led to the serendipitous isolation of an octanoyl-ACP dominant variant (R3.M4). The discovery of a C8 mutant R3.M4 and the higher commercial value of octanoic acid4 prompted a final round of computational predictions focusing on C8 composition only.
In the fourth round (R4), the IPRO imposed design constraints were similar to that of rounds R1 through R3 except that dodecanoyl-ACP (the “desired substrate”) was replaced with octanoyl-ACP, while tetradecanoyl-ACP (the “undesired substrate”) was replaced with dodecanoyl-ACP. These changes were made to drive more aggressive mutagenesis towards C8 preference. Of the eighteen R4 variants tested, all were active, sixteen maintained WT FFA production levels (all except R4.M11 and R4.M12), thirteen improved the C8 mole fraction, but none produced more octanoic acid than R3.M4. The reduced production levels for R4.M11 and R4.M12 could be explained by the rearrangement of aromatic side chains in the binding crevice (i.e., R108F, F139, Y145, and Y145F) relative to the active R4 variants, which may have disrupted the hydrophobicity of the binding crevice.
Through four rounds of the Design-Build-Test-Learn paradigm, 54 variants were tested, 43 were active, 25 maintained WT production levels (23 in R3-R4), 3 improved the C12 mole fraction, and 27 raised C8 composition (see Tables S1 and S2). All three C12-specific variants and all but six of the 27 C8-specific variants (R1.M2, R2.M2, R2.M3, R2.M4, R3.M3, and R4.M12) maintained or exceeded WT production levels. The success rate of computational enzyme design varies drastically based on the procedures employed, the system studied and the ambition of the (re)design (published values can range from as low as 7% to as high as 78%), and the frequency of favorable outcomes from IPRO in this study (24% for R1-R4, 43% for R3-R4) falls in line with these previously reported values47–48, 54–57. The variant with the highest C12 mole fraction (48±8%, a 1.8-fold improvement over WT), R3.M1, consisted of three mutations: S122K, Y145K, and L146K. Despite attempting to target octanoic acid production in R4, the variant with the highest C8 mole fraction was R3.M4 (50±3%, a 10-fold improvement over WT), containing mutations M141L, Y145K, and L146K. The FFA profiles for the top C12- and C8-specific variants are summarized in Figure 3.
Figure 3. Fatty acid production profiles for the most C12-specific and C8-specific computationally designed variants.
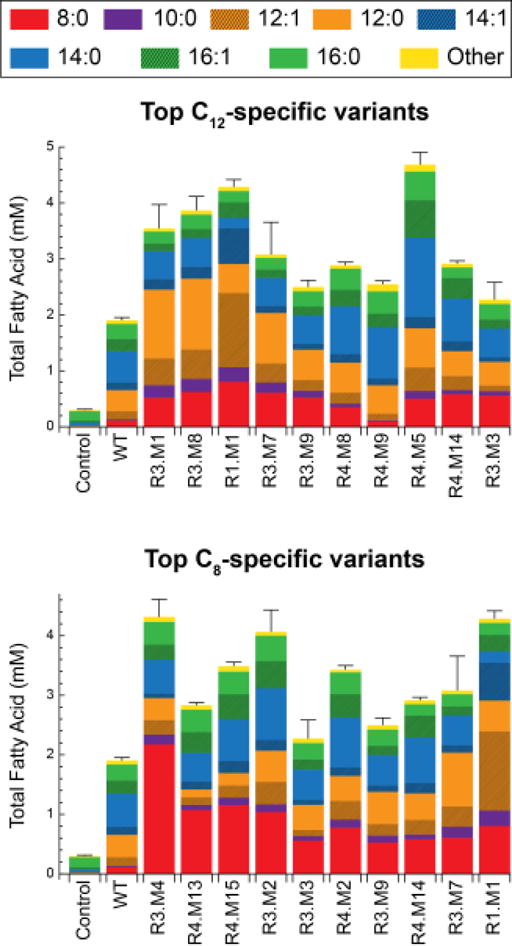
FFA titers are shown as bars, where error bars indicate total FFA standard deviation. Profiles of uninduced cells (Control) and wild-type ‘TesA (WT) are provided for reference. All profiles are listed in Table S1.
Computation-guided design outperforms random mutagenesis
Several of our best performing variants have a small number of mutations that may have been recovered from classical random mutagenesis approaches. Therefore, we created a small library of randomly mutated ‘TesA variants by error-prone PCR mutagenesis and screened for changes in product profile. The purpose of this library was to provide a negative control for testing that a library not directed by IPRO would not achieve the same level of success. The FFA profile of 61 E. coli cultures harboring expression vectors for unique ‘TesA variants was measured (N=1). Of the 61 random mutants (RMs) screened, 46 were active (i.e., ≥ 240 μM), and 20 maintained WT FFA production levels (i.e., ≥ 1750 μM). The best dodecanoic acid producing RM (RM.M39, 44±6%) demonstrated a comparable C12 composition to the top computationally predicted variant (R3.M1, 48±8%) but at the expense of a substantial reduction in total FFA titer (p<0.05, 80% of R3.M1). The best octanoic acid producing RM (RM.M29, 21.9%) produced a lower fraction of C8 than R3.M4 (50±3%, 44% reduction) and displayed only 58% of the total activity of R3.M4 (see Table S3). A comparison of these results shows that IPRO-guided mutagenesis generated more hits, more active mutants, and better leads than a library of similar size made through random mutagenesis.
Analysis of successful ‘TesA redesigns
The best C12 producing variants were dominated by three mutations: S122K, Y145K, and L146K. Mutation S122K (R3.RD3, Table S1) alone was sufficient to shift the C12 fraction to 35% of total FFAs, equal to the best C12-producing mutant R3.M1. A non-polar mutation at the same position (S122L – R3.RD4) had a similar but less pronounced shift towards C12 at the expense of C14, indicating that S122 is an important residue in the active site (see Figure S1). The additional mutations in R3.M1 (Y145K and L146K – equivalent to the R3.M7 mutation) also reduced the long-chain composition but produced a higher fraction of C8 and lower total activity. Nearly all of the top C8 producing variants contained a mutation at Y145 with lysine or phenylalanine as the dominant substituents. The best C8 producing mutant (R3.M4) contained Y145K and L146K mutations as well as a M141L, which by itself (R3.M2) was able to dramatically increase the C8 composition. The Y145K mutation drastically increased the fraction of unsaturated products in the C12 and C14 chain lengths.
Crystal structures and simulations show that hydrophobic interactions govern specificity
Crystal structures of WT ‘TesA and R3.M4 bound to octanoic acid were solved to confirm the structures predicted by IPRO and facilitate analysis of helpful mutations. The WT crystal structure was very similar to the previously published structure20 with an all-atom root-mean-square deviation (RMSD) of 1.1 Å. The X-ray resolved structures corroborated the structures predicted by our computational methods, quantified by an all-atom RMSD of 1.6 Å for R3.M4:octanoyl-ACP (Figure 4). The structural differences in loop111-120 may have affected IPRO’s ability to accept or reject mutations. However, the strong structural similarity between the crystallized and modeled structure at positions M141L, Y145K and L146K indicate that the conformation of these side chains is favorable despite the movement of loop111-120. Comparison of the WT and R3.M4 structures reveal that the largest differences (RMSD ≥ 2.5 Å) occur at (i) the mutated positions (M141L, Y145K, L146K), (ii) the flexible regions as suggested by MD-derived B factors (L11-A19, Q32-S33, G44-D45, N73, I107-R115 and D153-I156), (iii) the solvent-exposed residues adjacent to these flexible loops (i.e., H157 and R160), and (iv) the C-terminus (i.e., L177, Figure 5). Whereas structural differences in the flexible regions are possibly artifacts due to high residue mobility, the structural differences at the mutated positions provide insight to enzyme specificity determinants. The M141L side chain extends laterally towards the ω-1 carbon of the FFA yielding an attractive dispersion force (Figure 6A). Y145K partially occludes the binding crevice by forming a barrier between the ω-1 carbon of the FFA and solvent. L146K forms a salt bridge with E143 that may stabilize the conformation of Y145K but does not directly influence binding. Because mutant R3.M4’s (and all other active variants’) catalytic machinery includes an oxyanion hole that necessitates a negative charge for binding, R3.M4 in complex with octanoic acid (pKa 4.9) was also crystallized at a higher pH to ensure binding site occupancy. The additional experiments included crystallization at pH 7.5 and crystallization at pH 5.0 followed by additional octanoic acid buffered at pH 7.5. These additional experiments revealed substantial differences (RMSD ≥ 2.5 Å) at R16 (2.534 Å), K34 (3.024 Å), A111 (4.362 Å), Y113 (9.311 Å), G114 (3.536 Å), and L177 (3.723 Å, Figure S2). These highly flexible regions indicate that their movement is highly sensitive to changes in hydrophobicity and their motion may be essential for catalytic turnover. The structure of N112 was not solved for R3.M4 at pH 5.0 because of low-resolution electron density maps.
Figure 4. Comparison of computationally-predicted and crystallized structure of R3.M4.
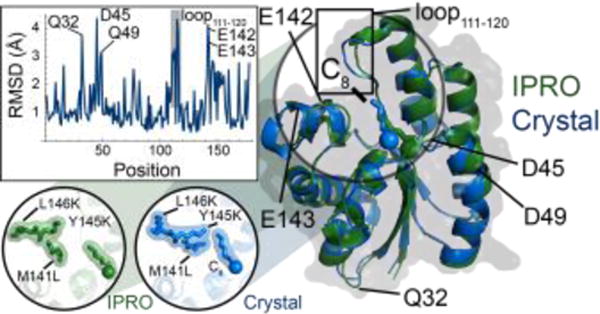
The IPRO-derived structure (green) and crystallized structure at pH 5.0 (blue) of R3.M4 are shown as cartoons bound to octanoyl-ACP (truncated at the thioester bond) and octanoic acid (C8), respectively. The carbonyl oxygen of each FFA structure is shown as a sphere. Regions of relatively large structural differences (RMSD ≥ 3.0 Å) are shown in a plot of RMSD versus position and annotated. Design positions (141, 145, and 146) are shown in the bottom left inset, where each residue is shown by sticks and a transparent surface. Hydrogen atoms are excluded from the IPRO predicted structure.
Figure 5. Comparison of WT and R3.M4 crystal structures.
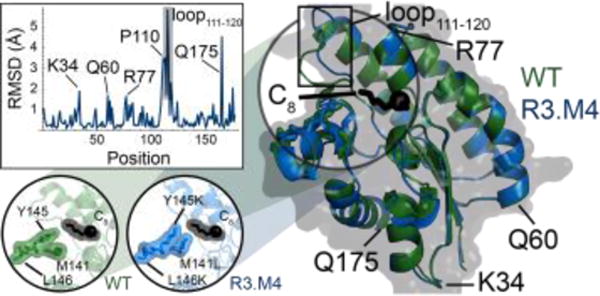
Both WT (green) and R3.M4 (blue, pH 5.0) are shown as cartoons to the right of the figure bound to octanoic acid (C8). The carbonyl oxygen of each FFA structure is shown as a sphere. Regions with relatively large changes (RMSD ≥ 1.5 Å) are annotated. Quantification of the structural differences is shown with the inset plot of RMSD versus sequence position. The structural differences between the residues at positions 141, 145, and 146 (the mutated positions in R3.M4) are depicted in the bottom left inset. Here, each of the three residues, as well as octanoic acid, is represented as sticks with a transparent surface.
Figure 6. Hydrophobicity drives substrate specificity as demonstrated by (A) R3.M4 crystal structure and (B) a two-atom model for the Y145K mutation.
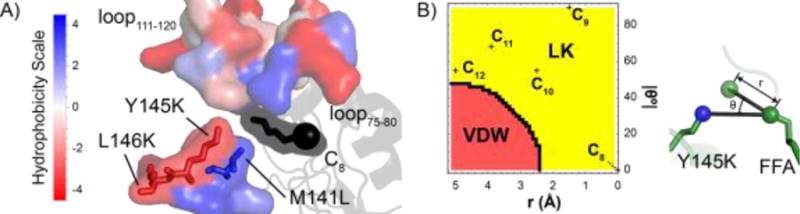
In (A), crystallized R3.M4 (pH 5.0) is bound to octanoic acid (C8). The two critical hydrophobic loops (loop75-80 and loop111-120) as well as the residues nearby the design positions are shown as molecular surfaces and colored according to their hydrophobicity score. The FFA is represented as sticks (with its carbonyl oxygen shown as a sphere) and a black molecular surface, as its hydrophobicity score is unknown. In (B), a two-atom model is shown to examine the ability of Y145K to eliminate binding to tetradecanoyl-ACP. The ε-amino nitrogen of Y145K (blue sphere) and the ω-1 carbon of FFA (green sphere) constitute the two-atom model. The model shows the effect of extending the acyl chain, which changes the hypothetical position of the ω-1 atom (transparent green sphere). r represents the interatomic distance between the C8 ω-1 atom and the hypothetical ω-1 atom. θ represents the angle between the ε-amino nitrogen, the C8 ω-1 atom and the hypothetical ω-1 atom. At (r=0 Å, 0=0°), the ω-1 atom occupies the position of the crystallized ω-1 C8 atom. The energy term with the largest contribution towards the interaction energy between the ε-amino nitrogen and the hypothetical ω-1 atom at a given r and 0 is provided in the contour plot (red=van der Waals energy, yellow=Lazaridis-Karplus solvation energy). The positions of the hypothetical ω-1 atoms using ideal FFA geometry (C-C bond length of 1.54 Å, 109.5° angle, 180° dihedral angle) are also labeled in the contour plot. C13 (r=6.4 Å, 0=62.8°) and C14 (r=7.5 Å, θ=54.9°) are beyond the boundaries of the contour plot. Molecular structures were generated using PyMOL.
MD simulations were used to elucidate the mechanisms by which FFA composition is controlled. Seven total trajectories were analyzed using MD. These included WT (bound to C14, C14, or C8 acyl-ACPs), R3.M1 (bound to C14 or C12), and R3.M4 (bound to C12 or C8). The MD results revealed an alternate binding mode nearby T46, S47, N73, and R77 that is only assumed for preferred chain lengths in the WT and R3.M4 trajectories. This binding mode is likely not observed for R3.M1:dodecanoyl-ACP because of insufficient conformational sampling. In addition to the alternate binding mode, MD analysis revealed that enzymes bound to acyl-ACPs beyond their preferred chain lengths have a deformed loop between G75 and Q80. With the exception of R3.M4:octanoyl-ACP and WT:octanoyl-ACP, all other enzymes bound to an acyl-ACP equal to or just below (≤ 2 carbon atoms) its preferred chain length maintain a constant conformation of this loop (Figure S3). Finally, B factors for each of the seven simulated enzymes were estimated and exhibited six regions with increased mobility. These regions were found from L11-A19, Q32-S33, G44-D45, N73, I107-R115, and D153-I156 .
Combining the MD and crystallography results with previously published investigations of ‘TesA acyl-ACP specificity20 support the theory that hydrophobic interactions formed between loop75-80 (i.e., residues forming the loop between G75 and Q80), the acyl chain of acyl-ACP, and loop111-120 govern substrate selectivity. Lo et al.20 previously postulated that loop75-80, described therein as the “switch loop” that is in one conformation (i.e., “on”) when the acyl-ACP is bound and in another conformation (i.e., “off’) when the substrate is not bound. The switch loop is dependent on the acyl chain length and stabilizes the enzyme:substrate complex during hydrolysis. Our MD simulations are consistent with these findings, as the switch loop is in the “on” position for variants bound to an acyl-ACP equal to or just less than (≤ 2 carbon atoms; i.e., WT bound to C12) its preferred chain length. Prior studies20, 58 revealed that rigidity of the switch loop or the lack of a bound acyl-ACP force the switch loop into the “off” conformation. MD simulations for WT:octanoyl-ACP, R3.M1:tetradecanoyl-ACP, and R3.M4:dodecanoyl-ACP occupy intermediate states between the “on” and “off” conformations. The switch loop is in a completely different conformation for R3.M4:octanoyl-ACP, which is due to the increased hydrophobicity in the binding crevice towards the C-1 terminus caused by M141L. For R3.M4:octanoyl-ACP, the movement of the switch loop towards loop111-120 allows octanoyl-ACP to slide underneath the switch loop into the alternate binding cavity, which could be important for octanoic acid release. The pH-dependent conformation of loop111-120 for R3.M4 from the crystallography experiments suggests that a change in protonation state of a titratable residue (i.e., an acidic or basic amino acid) causes a major conformational change, demonstrating the sensitivity of the hydrophobic region to electrostatics. At pH 7.5, the hydrophobic loop111-120 extends away from the typically hydrophobic binding crevice towards the highly hydrophilic bulk solvent (Figure S2), which is unexpected since nonpolar substances typically aggregate (i.e., the hydrophobic effect). This movement of loop111-120 is postulated to be due to a newly charged residue in the binding crevice, weakening the binding crevice’s hydrophobic environment. This observation is consistent with octanoic acid serving as the titratable residue. While M141L increases the hydrophobicity near the C-8 of R3.M4, Y145K limits hydrolysis of longer acyl-ACPs by reducing binding crevice hydrophobicity near the ω-1 atom (i.e., C-10, C-12, C-14) thereby disrupting the conformation of the switch loop. A simple two-atom model involving the FFA ω-1 carbon and the ε-amino nitrogen (i.e., the side chain nitrogen) atom had pairwise energies calculated and demonstrates that the Lazaridis-Karplus solvation term, a computationally accessible proxy for hydrophobicity, governs unfavorable interactions with the ω-1 carbon (Figure 6B). The two-atom model illustrates that solvation energy dominates the phase space with the exception of the van der Waals region. The ω-1 carbon would not be expected to occupy the van der Waals region because the energy is much more unfavorable (i.e., positive) at these close distances (E~r−12 for van der Waals, E~exp(-r2) for Lazaridis-Karplus solvation). Longer FFA chain lengths extend closer to the ε-amino nitrogen, exacerbating the repulsive solvation energy. Adding additional carbon atoms to the point where the ω-1 is further from the ε-amino nitrogen will only add to the total repulsive energy, although the marginal cost will decrease with each additional carbon. Finally, the large degree of switch loop and loop111-120 mobility could explain the broad substrate specificity that is typically observed for ‘TesA.
ΔΔG (ΔGVariant−ΔGwt) values estimated from the computations exhibited good agreement with experimentally derived values (Supporting Information). A Pearson correlation coefficient of 0.6 ± 0.2 was found for R3 and 0.43 ± 0.07 for R4. These correlation coefficients are in line with earlier computational studies24, 59–60. Notably, the regions with elevated B factors from the MD trajectories (loop11-19, loop32-33, loop44-45, loop73, loop107-115, and loop153-156) overlap with the regions with elevated B factors from the crystallography experiments (loop30-35, loop59-62, loop98-100, loop111-115, and loop153) and other published20, 58 crystallographic structures (loop31-35, loop75-80, and loop111-120).
In vitro assays of ‘TesA WT and R3.M4 C8-specific confirms in vivo results
‘TesA can catalyze hydrolysis of both acyl-CoA and acyl-ACP substrates. Given the relative availability of these substrates, we compared the kinetic activity of WT ‘TesA and R3.M4 on acyl-CoAs ranging from 6-16 carbons in length. We monitored reaction progress by tracking the abundance of free CoA released by hydrolysis. The highest in vitro ‘TesA WT activity (Figure 7A) was observed for C12-CoA, C14-CoA and C16-CoA, consistent with the in vivo data for release of FFAs from acyl-ACPs (Figure 3). In contrast, R3.M4 showed a significant increase in activity on C8-CoA compared to WT, consistent with the observed in vivo production of octanoic acid, and a modest decrease in activity on C12-CoA and C14-CoA activity. For both enzymes, we observed an unexpected drop in activity on hexadecanoyl-CoA (C16-CoA) beyond a threshold concentration. Interestingly, in the R3.M4 mutant the inhibitory effect of C16-CoA is exacerbated and C14-CoA also shows inhibition (not seen on ‘TesA WT). Given the linear reaction progress curves we observed, we suspected that the enzymes were substrate inhibited. Therefore, we performed assays with both C8-CoA and the inhibitory CoA species (Figure 7B–E). Competitive binding assays were performed at a constant concentration of C8-CoA (50 μM) and variable concentrations of C14-CoA (Figure 7B for WT and 7C for R3.M4) and C16-CoA (Figure 7D for WT and 7E for R3.M4). In all cases, production of free CoA was inhibited by C14-CoA and C16-CoA in a concentration dependent manner consistent with the original assay in Figure 7A.
Figure 7. Enzymatic assays of ‘TesA WT and R3.M4 (A) confirm the mutant’s increase in specificity for C8 species, and competitive binding assays (B-E) show ‘TesA WT and R3.M4 activity on C8-CoA as a function of C14-CoA (B and C) and C16-CoA (D and E) concentration.
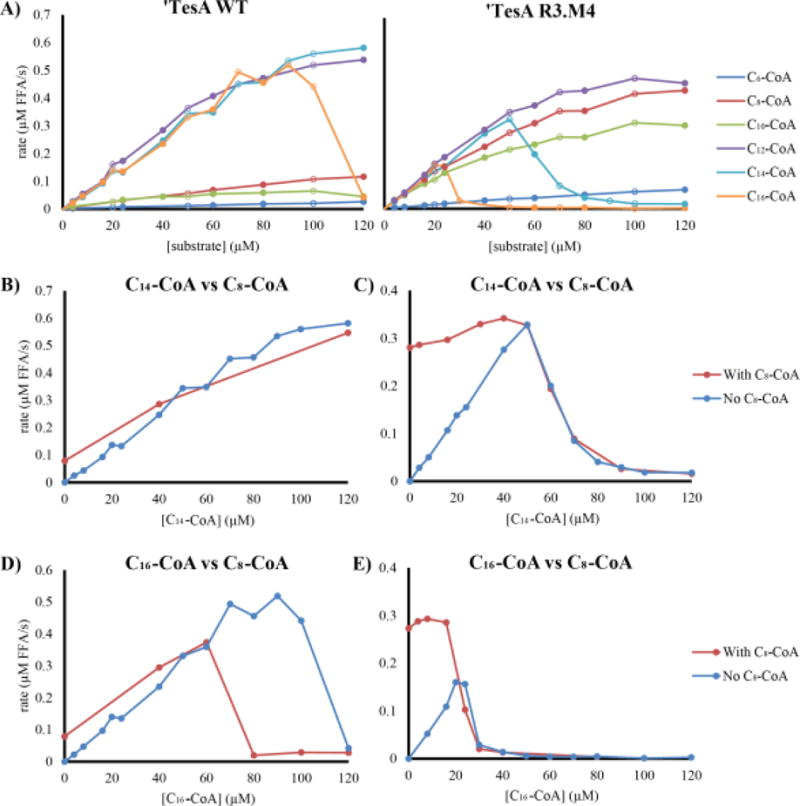
In (A), the activity of ‘TesA WT (left) and R3.M4 (right) as a function of substrate concentration for six different acyl-CoA substrates. Open and closed circles indicate measurements taken in separate days. Competitive binding assays (B-E) were done to see the effect that an increase in C14-CoA and C16-CoA would have on the activity of the enzymes on C8-CoA. Competitive binding assays were done at a constant C8-CoA concentration of 50 μM and variable concentrations of C14-CoA ((B) for WT and (C) for R3.M4) and C16-CoA ((D) for WT and (E) for R3.M4). In all cases the activity of ‘TesA was impacted by the C14-CoA and C16-CoA in a concentration dependent manner consistent with the original assay in (A).
Conclusions
The potential of the IPRO algorithm to aid in protein engineering efforts was demonstrated using a Design-Build-Test-Learn approach to alter the substrate preference of ‘TesA. Our approach leverages computational protein design procedures to achieve successful experimental redesign beyond what has been achievable so far15–17, yielding two top variants. One, R3.M1, produced 48±8% C12 composition, a 1.8-fold improvement over WT, while maintaining native production levels. Despite a preference for C14 production in WT ‘TesA, three amino acid substitutions constitute R3.M1, which is the third most C12-specific thioesterase known to date (see Table 1). Similarly, R3.M4 produced 50±3% C8 composition, a 10-fold improvement, while maintaining WT production levels. R3.M4 is the tenth most C8-specific thioesterase described to date. In total, the Design-Build-Test-Learn paradigm yielded three and twenty-one variants with significant (p<0.05) improvements in C12 and C8 mole fraction, respectively, while maintaining WT production levels.
Table 1. Top ten C12-specific and C8-specific thioesterases published to date.
Data was collected from Table S1 and sorted by dodecanoic acid (either saturated or unsaturated) and octanoic acid mole fractions in the product pool. WT compositions were provided as a reference. “Composition” refers to mole fraction of the corresponding FFA. Names match those provided in Table S1. The most C12-specific and C8-specific ‘TesA variants found in R1-R4 correspond to the third most C12-specific and tenth most C8-specific thioesterases identified to date respectively.
| Dodecanoic Acid (C12) | Octanoic Acid (C8) | |||||
|---|---|---|---|---|---|---|
| Rank | Name | Composition | Source | Name | Composition | Source |
| – | WT | 28±1 | This study | WT | 5.9±0.3 | This study |
| 1 | Q41635 | 76.9a,b | 13 | AAC49179 | 97.5±0.2 | 10 |
| 2 | rTE15 | 49.0a | 16 | AAC72882 | 93.5a | 16 |
| 3 | R3.M1 | 49±8 | This study | EEI82564 | 87±2 | 10 |
| 4 | R3.RD3 | 48±10 | This study | ABG82470 | 70±4 | 10 |
| 5 | R3.M8 | 46±4 | This study | CAD63310 | 68.0±0.8 | 10 |
| 6 | RM.M39 | 44±6 | This study | CvB2MT40 | 61.4a | 16 |
| 7 | R1.M1 | 43±2 | This study | ABJ63754 | 55.5±0.7 | 10 |
| 8 | AEM72521 | 40±2 | 10 | AEM72522 | 52±6 | 10 |
| 9 | R3.M7 | 40±11 | This study | TEGm258 | 50.4a | 16 |
| 10 | R4.RD11 | 37.7±0.6 | This study | R3.M4 | 50±3 | This study |
Standard deviation not provided
Data approximated using image processing software
Despite sampling a similar library size, random mutagenesis yielded fewer active mutants than the computationally guided library. In addition, the random library produced two fewer C12-specific variants with native production levels and twenty fewer C8-specific variants with at least WT productivity. Unlike random mutagenesis, variants from R3 and R4 achieved high levels of activity by directly enforcing catalytic contacts through imposed restraints and preserving conserved amino acids. Furthermore, R3 and R4 variants systematically tailored the binding crevice environment to generate hydrophobic clusters between loop111-120, the acyl-ACP, and the switch loop (loop75-80). MD and crystallography results from this work and elsewhere20, 58 suggest that this hydrophobic packing is essential for enzyme functionality. Whereas the in silico method directly accounts for hydrophobicity (albeit only approximately) through the Lazaridis-Karplus solvation energy term, random mutagenesis techniques can only improve binding site hydrophobicity by chance alone.
The results from this work not only establish the potential of computational methods in enzyme redesign, but the lessons learned from earlier rounds of design (i.e., R1 and R2) may inform redesign work with other systems. Computational procedures are especially valuable when high throughput screening is impractical. We found that the number of inactive designs was reduced by applying a modified scoring function that alleviated bias towards senseless mutations and avoiding design positions that are highly conserved in family sequence alignments. These initial rounds thus established the essentiality of working with a correctly calibrated scoring function and carefully selecting design positions with the aid of sequence alignments. The integrated deployment of computations with experiments in a sequential manner allows for the “early on” identification of deficiencies in molecular modeling and erroneously targeted design positions, providing a tractable workflow for engineering enzymes for higher specificity and activity.
Materials and methods
‘TesA model construction
The structure of ‘TesA was derived from PDB 1U8U, where it is in complex with octanoic acid20. The acyl-ACP structures were derived from PDB 2FAE, where decanoyl-ACP is held in an internal binding cavity49. Other acyl-ACP structures include hexanoyl-ACP from PDB 2FAC49 and heptanoyl-ACP from PDB 2FAD49 are similar to decanoyl-ACP as demonstrated by respective all-atom RMSDs of 1.1 Å and 1.1 Å. In order to dock octanoyl-ACP with ‘TesA, the acyl chain was systematically rotated about the phosphopantetheine linker and superimposed with the bound octanoic acid in 1U8U. The rotation that led to the lowest root-mean-square deviation was energy-minimized within CHARMM3461. Acyl-ACPs with different chain lengths were adapted from this initial complex by either deleting atoms or adding atoms using CHARMM’s internal coordinate system. Lazaridis-Karplus solvation files and CHARMM input files were constructed using published parameters for lipids and proteins. The constructed topology and parameter files were in close agreement with CGenFF-derived parameters62–63.
Structure-based redesign and analysis
All computationally-predicted mutants were identified using multiple IPRO trajectories that each ran for 1000 iterations without ensemble structure refinements24. The primary constraint of each trajectory was to eliminate binding to a larger acyl-ACPs (C14 for Rounds 1-3, C12 for Round 4) with a secondary constraint to improve binding to the shorter acyl-ACPs (C12 for Rounds 1-3, C8 for Round 4). Round 4 C8/C14 and C8/C12 fraction ratios correlate with r = 0.88 ± 0.02, alluding to a similar repulsive force for C12 and C14 binding (Figure S4). Design position selection is described in the “Design Position Selection” subsection. Restraints were imposed to ensure that the intermolecular catalytic distances (± 0.2 Å) were maintained (i.e., S10, G44, N73, D154 and H157). All other IPRO parameters were set to their standard values, and calculations were run on the Lion-XF system at Penn State University. Error propagation was performed manually and replicated using the Python uncertainties module64. Statistical differences were calculated using Welch’s t-test between the WT and mutant FFA profiles. For a given enzyme-FFA complex, the interaction energy is found using IE=GEnz-FFA,min−GEnz−GFFA. Mutants were sorted by the interaction energy difference between the short-chain FFA and C14 (ΔIE=IEC12,C8−IEC14,C12). Therefore, for a given round, Mutant 1 (smallest ΔIE) would be expected to show the biggest change in specificity.
Design Position Selection
For R1, ‘TesA residues were sorted by distance to carbon atoms in the acyl group of the ‘TesA:tetradecanoyl-ACP complex (i.e., C-1 through C-14). The minimum interatomic distance between the residue’s heavy atoms (i.e., not hydrogen atoms) and the acyl carbon atoms was used for sorting. In R1, residues constituting the catalytic triad (i.e., S10, D154, H157) and the oxyanion hole (i.e., S10, G44, N73) were not considered during design position selection. At the time design positions for R1 were designated, IPRO was unable to handle mutations from proline so these residues were not considered during the selection of design positions24. L109 was considered to be important for ‘TesA functionality and was also removed from consideration as a design position20. Residues that were near the undesired end of the acyl group (i.e., near C-1 instead of ω-1) were no longer regarded as potential design positions. Residues considered “near C-1” were those that contained a heavy atom within 4.5 Å of the thioester sulfur atom. A final set of residues that were strongly oriented away from the ω-1 terminus of the acyl moiety were also no longer considered as potential design positions. Residues “strongly oriented away from the ω-1 terminus” were those whose Cα atom was over 0.75 Å closer to the ω-1 atom than the Cβ atom (glycines not considered). The sorted residues are provided in Table S4 with any exceptions annotated. The eight nearest residues that were not filtered out were selected as design positions. The set of design positions used for R1 were L11, G72, L76, I107, R108, A111, F139, and Y145.
For R2, ‘TesA residues were once again sorted by distance to carbon atoms in the acyl group of the ‘TesA:tetradecanoyl-ACP complex. Residues constituting the catalytic triad and oxyanionic hole, proline residues, residues near the C-1 terminus of the acyl moiety, and L109 were once again removed from consideration, as described for the R1 design position selection procedure. In addition, residues belonging to the three flexible loops (i.e., loop31-35, loop75-80, loop111-120) were no longer recognized as prospective design positions for the same reason that L109 was removed from consideration20. Finally, I107 and R108 were removed as possible design positions because these positions invariably mutated to lysines in R1 and all instances of I107K and R108K abolished catalytic activity (see Figure 2, Table S2). The set of design positions (i.e., the eight nearest residues that were not filtered out) employed for R2 was L11, G72, F139, M141, E142, Y145, G155, and I156.
For R3 and R4, a very different approach was used when compared to R1 and R2. Instead of sorting residues by distance to carbon atoms in the acyl group of ‘TesA:tetradecanoyl-ACP, residues were sorted by distance between the residue’s Cβ atom and the ω-1 atom of dodecanoyl-ACP from the ‘TesA: dodecanoyl-ACP complex. In lieu of the Cβ atom, Cα was used to calculate interatomic distances for glycine residues, and Cγ was used for H180 because Cα and Cβ were not part of the solved crystal structure20. Unlike R1 and R2, only residues that were aligned to gaps or had ≥40% sequence conservation were not considered as candidate design positions. Sequence alignment was performed using the conserved domain database, where 81 members (including ‘TesA) of the lysophospholipase L1-like subgroup from the SGNH-hydrolase superfamily were found65. The sorted residues are provided in Table S4. Residues aligned to gaps or conserved residues are also noted in Table S4. The final set of design positions (i.e., the eight nearest residues that were not filtered out) used for R3 and R4 was I107, R108, L109, S122, M141, E142, Y145, and L146.
Scoring function re-weighting
A new set of weights for the IPRO scoring function was found that approximately doubles native rotamer recovery relative to the unmodified scoring function. The scoring function was modified using a symmetric logistic regression within Weka, a collection of machine-learning algorithms66. A dataset of native and non-native rotamers was collected from the top8000 database, which is a dataset of 8000 high-resolution (< 2 Å), quality-filtered (< 2.0 MolProbity score67), nonhomologous (<70% identity) protein structures53. Of these structures, 50 were randomly selected for use with the machine-learning training set. A separate set of 80 structures was randomly selected to validate the results. From these 130 structures, the native rotamer was found by finding the rotamer (of the same amino acid type) with the lowest root-mean-square deviation to the crystallized side chain. Then the van der Waals, electrostatic, and Lazaridis-Karplus solvation energies were calculated for each rotamer (regardless of amino acid type) as well as a binary indicator as to whether this was the native rotamer at the position or not. The data was separated for residues at the protein surface (≤ 20 Cβ atoms within 10 Å) and within the core of the protein (> 20 Cβ atoms within 10 Å) using a distance-based metric developed by Kuhlman and Baker68. Finally, since the number of non-native rotamers heavily outweighed the number of native rotamers, non-native rotamers were randomly removed until there was approximately a 60:40 split of non-native:native rotamers. The rotamer data was used to determine the set of weights that can optimally classify a rotamer as native or non-native. The updated energy equation for the core residues is E = 0.04 EVDW + 0.02 EElec + 0.16 ELK. For the surface residues, the updated energy equation is E = 0.03 EVDW + 0.01 EElec + 0.09 ELK. The inaccuracy of the original scoring function that used equally-weighted energy terms (i.e., E = 0.01 EVDW + 0.01 EElec + 0.01 ELK) stemmed mostly from the Lazaridis-Karplus implicit solvation energy term.
Using the modified scoring function weights, the classifier was able to correctly classify 78.4% of the core rotamers and 68.1% of the surface rotamers as either native or non-native from the validation set. Revisiting the complete validation set (before modifying to the 60:40 ratio), the full set of rotamers for each position was sorted from lowest energy to highest. The native rotamer was found to be the lowest-energy rotamer for 8.48% of the total dataset for the modified scoring function, up from 3.46%. Furthermore, the native rotamer was in the top 1.5%, 3.0%, and 6.0% of the energy-sorted rotamers in 19.09%, 31.64%, and 45.89% of the time. This was up from 9.09%, 16.86%, and 30.36%, respectively, for the original scoring function. Similar improvements were demonstrated on two different validation sets: one used to train the Rosetta scoring function69 (native: 8.38% (3.57%), 1.5%: 18.56% (8.79%), 3.0%: 13.01% (16.30%), 6.0%: 45.07% (29.20%)) and one on high-quality antibody structures (native: 8.02% (4.00%), 1.5%: 16.68% (9.05%), 3.0%: 28.31% (16.46%), 6.0%: 43.10% (30.19%)). The success of the modified scoring function on diverse sets of protein structures that do not include ‘TesA implies that the new scoring function is equally accurate across most (if not all) systems.
DNA synthesis and ‘TesA variant construction
All mutants were created starting with WT ‘tesA gene cloned into a pBAD18 plasmid70 to link ‘TesA expression to the presence of L-arabinose. Round 1 mutants were constructed using Agilent technologies QuickChange II site directed mutagenesis kit following the given protocol. For Rounds 2-4, all mutants were constructed using Gibson assembly strategies using primers containing the desired mutations in the 5′ tails. All cloning was performed in E. coli DH5α strain.
‘TesA in pBad18 was randomly mutagenized using Gene Morph II random mutagenesis kit following the kit protocol to make a library of 61 mutants with a mutation rate of 1.8 amino acids per gene (Table S2). Primers were designed to include the start and stop codons to ensure keeping those positions of the mutants invariant.
Bacterial culturing and fatty acid production
FFA production was assayed from small batch cultures (5-50 mL) of E. coli strain RL08ara (K-12 MG1655 ΔfadD ΔaraBAD ΔaraFGH Φ(ΔaraEp PCP18-araE)3) harboring each thioesterase expression vector. Three single colonies of each mutant were grown overnight on LB media containing 100 mg/L of ampicillin. Overnight cultures of each strain were diluted 1:100 into 25 mL of LB media containing 100 mg/L of ampicillin and 0.4% w/v glycerol in a 250 mL baffled shake flask and grown at 37°C and 250 rpm. When the OD600 reached 0.2-0.3, cultures were induced with 0.2% w/v L-arabinose and shaken for 24 h. All mutants were tested in triplicate and error bars represent the standard error of the measurements.
Lipid extraction and quantification
After 24 h post induction, 2.5 mL culture samples were collected in 10 mL glass centrifuge tubes, and 5 μL of 10 g/L heptadecanoic acid in ethanol solution was added as an internal standard. For fatty acid extraction into a chloroform layer, 100 mL of glacial acetic acid was added, followed by 5 mL of a 1:1 v/v solution of chloroform and methanol. Samples were vortexed and centrifuged for 10 min at 1000g to separate the layers. The chloroform extract was dried using a SpeedVac SC250EXP concentrator at no heat setting for 75 minutes and 1.0 torr. Samples were further dried for 30 min in a lyophilizer to remove any residual liquid. To methylate the dried extract, 0.5 mL of 1.25 M HCl in methanol was added and left overnight at 50°C. Finally, 5 mL of a 100 g/L sodium bicarbonate solution was added and fatty acid methyl esters were then extracted twice with 0.5 mL hexane for GC-FID quantification. Samples were collected and analyzed using a GC-FID model Shimadzu GC-2010 equipped with an AOC-20i auto-injector and a 30 m, 0.25 mm ID RTX-5 column. The GC temperature protocol was 100°C for 2 min, ramp to 150°C (at 80°C/min), hold for 4 min, ramp to 218°C (at 4°C/min), ramp to 250°C (at 8°C/min), and hold for 2.5 min.
Protein expression and purification of WT ‘TesA
The WT ‘tesA gene was cloned into pET28t, a vector previously modified to contain a TEV protease site rather than a thrombin cleavage site between ‘tesA and a N-terminal polyhistidine tag71. When translated, the modified enzyme was fused to the following peptide: MGSSHHHHHHSSENLYFQGGGG. The pET28t-tesA plasmid was used to transform E. coli Rosetta2(DE3) cells (Novagen). Cultures were grown at 37°C with shaking in lysogeny broth supplemented with 50 mg/L kanamycin and 50 mg/L chloramphenicol until the OD600 reached 0.8. Flasks were cooled in an ice bath, induced with 1 mM isopropyl-β-D-thiogalactopyranoside, and incubated overnight at 21°C. Cells were harvested by centrifugation and frozen as pellets in liquid nitrogen. The frozen cell pellets were sonicated on ice in a lysis buffer composed of 50 mM sodium phosphate, 20 mM imidazole, 10% glycerol, and 300 mM NaCl (pH 8.0). The lysate was cleared by centrifugation, and ‘TesA was purified at 4°C utilizing Ni-nitrilotriacetic acid resin (Qiagen) according to the manufacturer’s instructions. TEV protease was added in a 1:20 molar ratio to the pooled protein solution and subsequently dialyzed against 50 mM sodium phosphate, 300 mM NaCl, and 20 imidazole (pH 8.0) at 4°C for 36 h. Both the TEV protease and the uncleaved protein were removed by passage over a Ni-nitrilotriacetic acid resin. The cleaved protein was collected and dialyzed against 10 mM Tris-HCl (pH 8.0) and 200 mM NaCl and concentrated to ~35 g/L based on an extinction coefficient of 0.62 (g/L)−1cm−1.
Crystallization and structural analysis of WT ‘TesA
Crystallization conditions for ‘TesA were surveyed by the hanging drop method of vapor diffusion using a laboratory-based sparse matrix screen. The enzyme was initially tested either in the presence or absence of 3 mM C8 FFA. Crystals were subsequently grown from 22-26% poly(ethylene glycol) (PEG) 5000 with 100 mM Homo-PIPES buffer (pH 5.0). The protein solution used contained 3 mM C8 FFA. Crystals belonged to the monoclinic space group P21 with unit cell dimensions of a = 40.9 Å, b = 82.1 Å, c = 53.9 Å, and P = 90.4°. The asymmetric unit was comprised of two monomers. Prior to X-ray data collection at 100K, the crystals were transferred to a cryoprotectant solution composed of 30% PEG 5000, 250 mM NaCl, 3 mM C8 FFA, and 13% ethylene glycol with Homo-PIPES buffer (pH 5.0). An X-ray data set was collected with a Bruker AXS Platinum-135 CCD detector using the PROTEUM software suite (Bruker AXS Inc.) The X-ray source was Cu Ka radiation from a Rigaku RU200 X-ray generator equipped with Montel optics and operated at 50 kV and 90 mA. Data were processed with SAINT and scaled with SADABS (Bruker AXS Inc.). X-ray data collection statistics are listed in Table S5. The structure of WT ‘TesA was solved via molecular replacement using the software package PHASER72 and the PDB 1U8U as the search model20. Model refinement with REFMAC73 and manual model building with COOT74–75 reduced the overall R-factor to 18.8% at 1.65 A resolution. Refinement statistics are presented in Table S6.
Crystallization and structural analysis of R3.M4
The R3.M4 ‘tesA gene was subcloned, expressed, and purified as described for WT. Crystals were obtained at both pH 5.0 and pH 7.5. Those obtained at pH 5.0 were grown from 20-25% PEG 5000 with 100 mM Homo-PIPES buffer (protein solution contained 3 mM C8 FFA). The crystals were cryoprotected as described for WT ‘TesA. Crystals belonged to the monoclinic space group P21 with unit cell dimensions of a = 40.7 Å, b = 55.2 Å, c = 42.3 Å, and P = 105.2° with a single monomer in the asymmetric unit. R3.M4 crystals obtained at pH 7.5 were grown from 24-28% PEG with 100 mM HEPES buffer (pH 7.5). Again the protein solution contained 3 mM C8 FFA. These crystals were isomorphous to those obtained at pH 5.0 and were cryoprotected with a solution composed of 32% PEG, 250 mM NaCl, 3 mM C8 FFA, and 13% ethylene glycol with 100 mM HEPES buffer (pH 7.5). Given the concern that at pH 5.0 the C8 FFA would most likely not bind at full occupancy, these crystals were subsequently moved in a final experiment to solutions buffered at pH 7.5 that contained an additional 3 mM C8 FFA. These “soaked” crystals were cryoprotected in a similar manner to those grown at pH 7.5. X-ray data from R3.M4 crystals obtained at pH 5.0, pH 7.5, and from the “soaked” crystals were collected as described for WT. The structure R3.M4 at pH 5.0 was solved by molecular replacement using the WT model as the search probe whereas the structures either grown or soaked at pH 7.5 were solved via Fourier difference analyses. X-ray data collection statistics and model refinement statistics are provided in Tables S5 and S6, respectively.
Purification for enzymatic assays of WT ‘TesA and R3.M4
The ‘tesA gene was subcloned as described for WT. Expression followed the same procedure, except the construct was transformed into BL21(DE3) cells. The frozen cell pellets were sonicated on ice in a lysis buffer composed of 50 mM sodium phosphate, 10 mM imidazole, and 300 mM NaCl (pH 8.0). The lysate was cleared by centrifugation, and ‘TesA was purified at 20°C utilizing Ni-nitrilotriacetic acid resin (Qiagen) according to the manufacturer’s instructions. TEV protease was added in a 1:20 molar ratio to the pooled protein solution and subsequently dialyzed against 10 mM Tris-HCl (pH 7.5) at 4°C for 18 h. Both the TEV protease and the uncleaved protein were removed by passage over a Ni-nitrilotriacetic acid resin. The cleaved protein was concentrated to 2.5 mL to be solvent exchanged into 50mM potassium phosphate (pH 7.0), and 30% glycerol using a PD-10 desalting column (GE) according to the manufacturer’s instructions.
Enzymatic assays of WT ‘TesA and R3.M4
Enzymatic thioesterase assay was performed with WT and R3.M4 ‘TesA to compare their activities on various chain lengths. The reaction conditions follow as per Shin et al. (63), except the enzyme concentration used was 40 nM and substrate concentrations ranged 0-120 μM for six saturated acyl-CoA substrates of 6-16 carbons (hexanoyl-CoA, octanoyl-CoA, decanoyl-CoA, dodecanoyl-CoA, tetradecanoyl-CoA and hexadecanoyl-CoA). The assay tracks generation of free-CoA as ‘TesA hydrolyses the thiol bond in the acyl-CoA. This hydrolysis is tracked by the increase of absorbance at 412 nm due to the free-CoA dependent reduction of 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) present in the reaction mixture (63). Absorbance at 412 nm was followed using a NanoDrop 2000c (Thermo Scientific) using a path length of 10 mm and measurements were taken for 2 minutes in 10 second intervals.
Molecular dynamics
VMD was used to solvate enzyme-FFA complexes within a 12.0 Å water box with 0.17 M NaCl and contained ≈49,000 atoms76. Each complex was minimized and slowly heated to 310K and 1 atm over 7 ns using Langevin dynamics. Force field parameters were identical to those used for the IPRO trajectories. Periodic boundary conditions were applied, and long-range electrostatic forces were considered using the particle mesh Ewald method. 40 ns production simulations were performed using NAMD over 30 nodes on the Lion-XF cluster at Penn State University using the NVE ensemble77.
Supplementary Material
Acknowledgments
Support for this work was provided by the National Science Foundation Award CBET-0967062, CBET-1149678, the Advanced Research Projects Agency-Energy Award DE-AR0000431, and the National Institutes of Health (NIH GM115921). Nestor Hernandez-Lozada is the recipient of a NIH Chemistry-Biology Interface Training Program fellowship (T32 GM008505) and a Graduate Engineering Research Scholars fellowship from the UW-Madison College of Engineering.
References
- 1.Lennen RM, Pfleger BF. Microbial production of fatty acid-derived fuels and chemicals. Curr Opin Biotech. 2013;24(6):1044–1053. doi: 10.1016/j.copbio.2013.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steen EJ, Kang YS, Bokinsky G, Hu ZH, Schirmer A, McClure A, del Cardayre SB, Keasling JD. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature. 2010;463(7280):559–U182. doi: 10.1038/nature08721. [DOI] [PubMed] [Google Scholar]
- 3.Lennen RM, Braden DJ, West RM, Dumesic JA, Pfleger BF. A Process for Microbial Hydrocarbon Synthesis: Overproduction of Fatty Acids in Escherichia coli and Catalytic Conversion to Alkanes. Biotechnol Bioeng. 2010;106(2):193–202. doi: 10.1002/bit.22660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pfleger BF, Gossing M, Nielsen J. Metabolic engineering strategies for microbial synthesis of oleochemicals. Metabolic engineering. 2015;29:1–11. doi: 10.1016/j.ymben.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 5.Knothe G. “Designer” biodiesel: Optimizing fatty ester (composition to improve fuel properties. Energ Fuel. 2008;22(2):1358–1364. [Google Scholar]
- 6.Howard TP, Middelhaufe S, Moore K, Edner C, Kolak DM, Taylor GN, Parker DA, Lee R, Smirnoff N, Aves SJ, Love J. Synthesis of customized petroleum-replica fuel molecules by targeted modification of free fatty acid pools in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(19):7636–7641. doi: 10.1073/pnas.1215966110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liao JC, Mi L, Pontrelli S, Luo S. Fuelling the future: microbial engineering for the production of sustainable biofuels. Nat Rev Microbiol. 2016;14(5):288–304. doi: 10.1038/nrmicro.2016.32. [DOI] [PubMed] [Google Scholar]
- 8.Magnuson K, Jackowski S, Rock CO, Cronan JE., Jr Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol Rev. 1993;57(3):522–42. doi: 10.1128/mr.57.3.522-542.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi YJ, Lee SY. Microbial production of short-chain alkanes. Nature. 2013;502(7472):571–+. doi: 10.1038/nature12536. [DOI] [PubMed] [Google Scholar]
- 10.Jing FY, Cantu DC, Tvaruzkova J, Chipman JP, Nikolau BJ, Yandeau-Nelson MD, Reilly PJ. Phylogenetic and experimental characterization of an acyl-ACP thioesterase family reveals significant diversity in enzymatic specificity and activity. Bmc Biochem. 2011;12 doi: 10.1186/1471-2091-12-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, Li M, Agrawal A, San KY. Efficient free fatty acid production in Escherichia coli using plant acyl-ACP thioesterases. Metabolic engineering. 2011;13(6):713–22. doi: 10.1016/j.ymben.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 12.Lu X, Vora H, Khosla C. Overproduction of free fatty acids in E. coli: implications for biodiesel production. Metabolic engineering. 2008;10(6):333–9. doi: 10.1016/j.ymben.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 13.Voelker TA, Davies HM. Alteration of the specificity and regulation of fatty acid synthesis of Escherichia coli by expression of a plant medium-chain acyl-acyl carrier protein thioesterase. J Bacteriol. 1994;176(23):7320–7. doi: 10.1128/jb.176.23.7320-7327.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dormann P, Voelker TA, Ohlrogge JB. Cloning and Expression in Escherichia-Coli of a Novel Thioesterase from Arabidopsis-Thaliana Specific for Long-Chain Acyl-Acyl Carrier Proteins. Arch Biochem Biophys. 1995;316(1):612–618. doi: 10.1006/abbi.1995.1081. [DOI] [PubMed] [Google Scholar]
- 15.Hom L, Trinh N, Alibhai M. Methods and compositions related to thioesterase enzymes. 2010 [Google Scholar]
- 16.Jing F. ProQuest Dissertations. 2013. Characterization of acyl-ACP thioesterases for the purpose of diversifying fatty acid synthesis pathway. Publishing. [Google Scholar]
- 17.Mayer KM, Shanklin J. Identification of amino acid residues involved in substrate specificity of plant acyl-ACP thioesterases using a bioinformatics-guided approach. Bmc Plant Biol. 2007;7 doi: 10.1186/1471-2229-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang FZ, Carothers JM, Keasling JD. Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nature biotechnology. 2012;30(4):354–U166. doi: 10.1038/nbt.2149. [DOI] [PubMed] [Google Scholar]
- 19.Liu T, Vora H, Khosla C. Quantitative analysis and engineering of fatty acid biosynthesis in E. coli. Metabolic engineering. 2010;12(4):378–86. doi: 10.1016/j.ymben.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 20.Lo Y-C, Lin S-C, Shaw J-F, Liaw Y-C. Substrate Specificities of Escherichia coli Thioesterase I/Protease I/Lysophospholipase L1 Are Governed by Its Switch Loop Movement†. Biochemistry. 2005;44(6):1971–1979. doi: 10.1021/bi048109x. [DOI] [PubMed] [Google Scholar]
- 21.Bloom JD, Meyer MM, Meinhold P, Otey CR, MacMillan D, Arnold FH. Evolving strategies for enzyme engineering. Curr Opin Struc Biol. 2005;15(4):447–452. doi: 10.1016/j.sbi.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 22.Packer MS, Liu DR. Methods for the directed evolution of proteins. Nat Rev Genet. 2015;16(7):379–394. doi: 10.1038/nrg3927. [DOI] [PubMed] [Google Scholar]
- 23.Gajewski J, Pavlovic R, Fischer M, Boles E, Grininger M. Engineering fungal de novo fatty acid synthesis for short chain fatty acid production. Nat Commun. 2017;8:14650. doi: 10.1038/ncomms14650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pantazes RJ, Grisewood MJ, Li T, Gifford NP, Maranas CD. The Iterative Protein Redesign and Optimization (IPRO) suite of programs. J Comput Chem. 2014 doi: 10.1002/jcc.23796. [DOI] [PubMed] [Google Scholar]
- 25.Pavelka A, Chovancova E, Damborsky J. HotSpot Wizard: a web server for identification of hot spots in protein engineering. Nucleic Acids Res. 2009;37:W376–83. doi: 10.1093/nar/gkp410. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grosdidier S, Fernandez-Recio J. Identification of hot-spot residues in protein-protein interactions by computational docking. BMC bioinformatics. 2008;9:447. doi: 10.1186/1471-2105-9-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng H, Reetz MT. Manipulating the stereoselectivity of limonene epoxide hydrolase by directed evolution based on iterative saturation mutagenesis. J Am Chem Soc. 2010;132(44):15744–51. doi: 10.1021/ja1067542. [DOI] [PubMed] [Google Scholar]
- 28.Kortemme T, Baker D. A simple physical model for binding energy hot spots in protein-protein complexes. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(22):14116–21. doi: 10.1073/pnas.202485799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kortemme T, Kim DE, Baker D. Computational alanine scanning of protein-protein interfaces. Sci STKE. 2004;2004(219):pl2. doi: 10.1126/stke.2192004pl2. [DOI] [PubMed] [Google Scholar]
- 30.Darnell SJ, Page D, Mitchell JC. An automated decision-tree approach to predicting protein interaction hot spots. Proteins. 2007;68(4):813–23. doi: 10.1002/prot.21474. [DOI] [PubMed] [Google Scholar]
- 31.Kawarasaki Y, Griswold KE, Stevenson JD, Selzer T, Benkovic SJ, Iverson BL, Georgiou G. Enhanced crossover SCRATCHY: construction and high-throughput screening of a combinatorial library containing multiple non-homologous crossovers. Nucleic Acids Res. 2003;31(21):e126. doi: 10.1093/nar/gng126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer MM, Hochrein L, Arnold FH. Structure-guided SCHEMA recombination of distantly related beta-lactamases. Protein Eng Des Sel. 2006;19(12):563–70. doi: 10.1093/protein/gzl045. [DOI] [PubMed] [Google Scholar]
- 33.Voigt CA, Martinez C, Wang ZG, Mayo SL, Arnold FH. Protein building blocks preserved by recombination. Nature structural biology. 2002;9(7):553–8. doi: 10.1038/nsb805. [DOI] [PubMed] [Google Scholar]
- 34.Pantazes RJ, Saraf MC, Maranas CD. Optimal protein library design using recombination or point mutations based on sequence-based scoring functions. Protein Eng Des Sel. 2007;20(8):361–73. doi: 10.1093/protein/gzm030. [DOI] [PubMed] [Google Scholar]
- 35.Moore GL, Maranas CD. Computational challenges in combinatorial library design for protein engineering. AIChE Journal. 2004;50(2):262–272. [Google Scholar]
- 36.Saraf MC, Maranas CD. Using a residue clash map to functionally characterize protein recombination hybrids. Protein engineering. 2003;16(12):1025–34. doi: 10.1093/protein/gzg129. [DOI] [PubMed] [Google Scholar]
- 37.Moore GL, Maranas CD. Identifying residue-residue clashes in protein hybrids by using a second-order mean-field approach. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(9):5091–6. doi: 10.1073/pnas.0831190100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rothlisberger D, Khersonsky O, Wollacott AM, Jiang L, DeChancie J, Betker J, Gallaher JL, Althoff EA, Zanghellini A, Dym O, Albeck S, Houk KN, Tawfik DS, Baker D. Kemp elimination catalysts by computational enzyme design. Nature. 2008;453(7192):190–U4. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- 39.Jiang L, Althoff EA, Clemente FR, Doyle L, Rothlisberger D, Zanghellini A, Gallaher JL, Betker JL, Tanaka F, Barbas CF, Hilvert D, Houk KN, Stoddard BL, Baker D. De novo computational design of retro-aldol enzymes. Science. 2008;319(5868):1387–1391. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang PS, Boyken SE, Baker D. The coming of age of de novo protein design. Nature. 2016;537(7620):320–7. doi: 10.1038/nature19946. [DOI] [PubMed] [Google Scholar]
- 41.Garrabou X, Wicky BI, Hilvert D. Fast Knoevenagel Condensations Catalyzed by an Artificial Schiff-Base-Forming Enzyme. J Am Chem Soc. 2016;138(22):6972–4. doi: 10.1021/jacs.6b00816. [DOI] [PubMed] [Google Scholar]
- 42.Li T, Pantazes RJ, Maranas CD. OptMAVEn–a new framework for the de novo design of antibody variable region models targeting specific antigen epitopes. PLoS One. 2014;9(8):e105954. doi: 10.1371/journal.pone.0105954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fazelinia H, Cirino PC, Maranas CD. Extending Iterative Protein Redesign and Optimization (IPRO) in protein library design for ligand specificity. Biophys J. 2007;92(6):2120–30. doi: 10.1529/biophysj.106.096016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saraf MC, Moore GL, Goodey NM, Cao VY, Benkovic SJ, Maranas CD. IPRO: an iterative computational protein library redesign and optimization procedure. Biophys J. 2006;90(11):4167–80. doi: 10.1529/biophysj.105.079277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pantazes RJ, Grisewood MJ, Maranas CD. Recent advances in computational protein design. Curr Opin Struct Biol. 2011;21(4):467–72. doi: 10.1016/j.sbi.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 46.Samish I, MacDermaid CM, Perez-Aguilar JM, Saven JG. Theoretical and computational protein design. Annual review of physical chemistry. 2011;62:129–49. doi: 10.1146/annurev-physchem-032210-103509. [DOI] [PubMed] [Google Scholar]
- 47.Privett HK, Kiss G, Lee TM, Blomberg R, Chica RA, Thomas LM, Hilvert D, Houk KN, Mayo SL. Iterative approach to computational enzyme design. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(10):3790–5. doi: 10.1073/pnas.1118082108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baker D. An exciting but challenging road ahead for computational enzyme design. Protein Sci. 2010;19(10):1817–1819. doi: 10.1002/pro.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roujeinikova A, Simon WJ, Gilroy J, Rice DW, Rafferty JB, Slabas AR. Structural studies of fatty acyl-(acyl carrier protein) thioesters reveal a hydrophobic binding cavity that can expand to fit longer substrates. J Mol Biol. 2007;365(1):135–45. doi: 10.1016/j.jmb.2006.09.049. [DOI] [PubMed] [Google Scholar]
- 50.Grisewood MJ, Gifford NP, Pantazes RJ, Li Y, Cirino PC, Janik MJ. OptZyme: Computational Enzyme Redesign Using Transition State Analogues. PLoS ONE. 2013;8(10):e75358. doi: 10.1371/journal.pone.0075358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khoury GA, Fazelinia H, Chin JW, Pantazes RJ, Cirino PC, Maranas CD. Computational design of Candida boidinii xylose reductase for altered cofactor specificity. Protein Sci. 2009;18(10):2125–38. doi: 10.1002/pro.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fazelinia H, Cirino PC, Maranas CD. OptGraft: A computational procedure for transferring a binding site onto an existing protein scaffold. Protein Sci. 2009;18(1):180–95. doi: 10.1002/pro.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Richardson JS, Keedy DA, Richardson DC. “the Plot” Thickens: More Data, More Dimensions, More Uses. Biomolecular Forms and Functions: A Celebration of 50 Years of the Ramachandran Map. 2013:46–61. [Google Scholar]
- 54.Feldmeier K, Hocker B. Computational protein design of ligand binding and catalysis. Curr Opin Chem Biol. 2013;17(6):929–33. doi: 10.1016/j.cbpa.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 55.Khare SD, Kipnis Y, Greisen PJr, Takeuchi R, Ashani Y, Goldsmith M, Song Y, Gallaher JL, Silman I, Leader H, Sussman JL, Stoddard BL, Tawfik DS, Baker D. Computational redesign of a mononuclear zinc metalloenzyme for organophosphate hydrolysis. Nat Chem Biol. 2012;8(3):294–300. doi: 10.1038/nchembio.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gordon SR, Stanley EJ, Wolf S, Toland A, Wu SJ, Hadidi D, Mills JH, Baker D, Pultz IS, Siegel JB. Computational design of an alpha-gliadin peptidase. J Am Chem Soc. 2012;134(50):20513–20. doi: 10.1021/ja3094795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen CY, Georgiev I, Anderson AC, Donald BR. Computational structure-based redesign of enzyme activity. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(10):3764–9. doi: 10.1073/pnas.0900266106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lo YC, Lin SC, Shaw JF, Liaw YC. Crystal structure of Escherichia coli thioesterase I/protease I/lysophospholipase L-1: Consensus sequence blocks constitute the catalytic center of SGNH-hydrolases through a conserved hydrogen bond network. J Mol Biol. 2003;330(3):539–551. doi: 10.1016/s0022-2836(03)00637-5. [DOI] [PubMed] [Google Scholar]
- 59.Morozov AV, Havranek JJ, Baker D, Siggia ED. Protein-DNA binding specificity predictions with structural models. Nucleic Acids Res. 2005;33(18):5781–98. doi: 10.1093/nar/gki875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Meiler J, Baker D. ROSETTALIGAND: protein-small molecule docking with full side-chain flexibility. Proteins. 2006;65(3):538–48. doi: 10.1002/prot.21086. [DOI] [PubMed] [Google Scholar]
- 61.Brooks BR, Brooks CL, 3rd, Mackerell AD, Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30(10):1545–614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vanommeslaeghe K, MacKerell AD. Automation of the CHARMM General Force Field (CGenFF) I: Bond Perception and Atom Typing. J Chem Inf Model. 2012;52(12):3144–3154. doi: 10.1021/ci300363c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vanommeslaeghe K, Raman EP, MacKerell AD. Automation of the CHARMM General Force Field (CGenFF) II: Assignment of Bonded Parameters and Partial Atomic Charges. J Chem Inf Model. 2012;52(12):3155–3168. doi: 10.1021/ci3003649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lebigot EO. Uncertainties: a Python package for calculations with uncertainties. http://pythonhosted.org/uncertainties/ (accessed February 23)
- 65.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu SN, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang ZX, Yamashita RA, Zhang DC, Zheng CJ, Bryant SH. Cdd: Ncbi’s Conserved Domain Database. Nucleic Acids Res. 2015;43(D1):D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. The WEKA data mining software: an update. SIGKDD Explor Newsl. 2009;11(1):10–18. [Google Scholar]
- 67.Davis IW, Murray LW, Richardson JS, Richardson DC. MolProbity: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kuhlman B, Baker D. Native protein sequences are close to optimal for their structures. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(19):10383–10388. doi: 10.1073/pnas.97.19.10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang YM, Bystroff C. Expanded Explorations into the Optimization of an Energy Function for Protein Design. Ieee Acm T Comput Bi. 2013;10(5):1176–1187. doi: 10.1109/TCBB.2013.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight Regulation, Modulation, and High-Level Expression by Vectors Containing the Arabinose P-Bad Promoter. J Bacteriol. 1995;177(14):4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thoden JB, Holden HM. The molecular architecture of human N-acetylgalactosamine kinase. The Journal of biological chemistry. 2005;280(38):32784–91. doi: 10.1074/jbc.M505730200. [DOI] [PubMed] [Google Scholar]
- 72.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 74.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 75.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. Journal of molecular graphics. 1996;14(1):33–8. 27–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 77.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ghosh SK, Bhattacharjee A, Jha JK, Mondal AK, Maiti MK, Basu A, Ghosh D, Ghosh S, Sen SK. Characterization and cloning of a stearoyl/oleoyl specific fatty acyl-acyl carrier protein thioesterase from the seeds of Madhuca longifolia (latifolia) Plant Physiol Bioch. 2007;45(12):887–897. doi: 10.1016/j.plaphy.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 79.Jha JK, Maiti MK, Bhattacharjee A, Basu A, Sen PC, Sen SK. Cloning and functional expression of an acyl-ACP thioesterase FatB type from Diploknema (Madhuca) butyracea seeds in Escherichia coli. Plant Physiol Bioch. 2006;44(11-12):645–655. doi: 10.1016/j.plaphy.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 80.Dong SB, Huang JC, Li YN, Zhang J, Lin SZ, Zhang ZX. Cloning, characterization, and expression analysis of acyl-acyl carrier protein (ACP)-thioesterase B from seeds of Chinese Spicehush (Lindera communis) Gene. 2014;542(1):16–22. doi: 10.1016/j.gene.2014.03.028. [DOI] [PubMed] [Google Scholar]
- 81.Chen G, Peng ZY, Shan L, Xuan N, Tang GY, Zhang Y, Li L, He QF, Bi YP. Cloning of acyl-ACP thioesterase FatA from Arachis hypogaea L. and its expression in Escherichia coli. Journal of biomedicine & biotechnology. 2012;2012:652579. doi: 10.1155/2012/652579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zheng Y, Li L, Liu Q, Qin W, Yang J, Cao Y, Jiang X, Zhao G, Xian M. Boosting the free fatty acid synthesis of Escherichia coli by expression of a cytosolic Acinetobacter baylyi thioesterase. Biotechnology for biofuels. 2012;5(1):76. doi: 10.1186/1754-6834-5-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jha JK, Sinha S, Maiti MK, Basu A, Mukhopadhyay UK, Sen SK. Functional expression of an acyl carrier protein (ACP) from Azospirillum brasilense alters fatty acid profiles in Escherichia coli and Brassica juncea. Plant physiology and biochemistry : PPB / Societe francaise de physiologie vegetale. 2007;45(6-7):490–500. doi: 10.1016/j.plaphy.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 84.Gong Y, Guo X, Wan X, Liang Z, Jiang M. Characterization of a novel thioesterase (PtTE) from Phaeodactylum tricornutum. Journal of basic microbiology. 2011;51(6):666–72. doi: 10.1002/jobm.201000520. [DOI] [PubMed] [Google Scholar]
- 85.Serrano-Vega MJ, Garces R, Martinez-Force E. Cloning, characterization and structural model of a FatA-type thioesterase from sunflower seeds (Helianthus annuus L. Planta. 2005;221(6):868–80. doi: 10.1007/s00425-005-1502-z. [DOI] [PubMed] [Google Scholar]
- 86.Lee S, Park S, Park C, Pack SP, Lee J. Enhanced free fatty acid production by codon-optimized Lactococcus lactis acyl-ACP thioesterase gene expression in Escherichia coli using crude glycerol. Enzyme and microbial technology. 2014;67:8–16. doi: 10.1016/j.enzmictec.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 87.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a Program to Check the Stereochemical Quality of Protein Structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


