Summary
Physicochemical properties of storage starch largely determine rice grain quality and food characteristics. Therefore, modification of starch property is effective to fine‐tune cooked rice textures. To obtain new resources with modified starch property as breeding materials, we screened a mutant population of a japonica cultivar Nipponbare and found two independent mutant lines, altered gelatinization (age)1 and age2, with moderate changes in starch gelatinization property. A combination of conventional genetic analyses and the latest mapping method, MutMapPlus, revealed that both of these lines harbour novel independent mutant alleles of starch branching enzyme IIb (BEIIb) gene. In age1, amino acid substitution of Met‐723 to Lys completely abolished BEIIb enzyme activity without significant reduction in its protein level. A transposon insertion in an intron of BEIIb gene reduced BEIIb protein level and activity in age2. Production of a series of the mutant lines by combining age alleles and indica‐type starch synthase IIa allele established stepwise alteration of the physicochemical properties of starch including apparent amylose content, thermal property, digestibility by α‐amylase and branched structures of amylopectin. Consistent with the alteration of starch properties, the results of a sensory evaluation test demonstrated that warm cooked rice of the mutants showed a variety of textures without marked reduction in overall palatability. These results suggest that a series of the mutant lines are capable of manipulation of cooked rice textures.
Keywords: amylopectin, cooked rice texture, mutant allele, MutMapPlus, Oryza sativa, starch branching enzyme
Introduction
Starch constitutes most of the weight of cereal grains and is the most important carbohydrate glucose polymer for humans as a dietary source of energy. Starch consists of linear or slightly branched amylose and highly branched amylopectin. The amylose content and chain length distribution (CLD) of amylopectin branches are the predominant determinant of the physicochemical properties of starch and the cooking properties of grains.
Starch is synthesized by the orchestrated functional interactions of four classes of enzymes, ADP‐glucose pyrophosphorylase, starch synthase (SS), starch branching enzyme (BE) and starch debranching enzyme (DBE) (Ball and Morell, 2003; Nakamura, 2002; Smith et al., 1997; Zeeman et al., 2010). Of these, SS elongates the nonreducing end of α‐1,4‐glucan chains of amylose and amylopectin by the addition of glucose from ADP‐glucose, while BE introduces α‐1,6‐branched glucosidic linkages into α‐1,4‐glucan chains. These activities are essential for the synthesis of the branched structure of amylopectin.
In rice, SSI, SSIIa and SSIIIa are highly expressed in developing endosperm and elongate very short (degree of polymerization (DP) 6 and 7), short (DP 6–10) and long (DP > 33) amylopectin chains, respectively (Fujita et al., 2006, 2007; Nakamura et al., 2005; Ohdan et al., 2005). Two groups of rice varieties, indica and japonica, harbour different SSIIa alleles (referred to as SSIIa indica and SSIIa japonica, respectively), which produce active and inactive SSIIa, respectively (Umemoto et al., 2004). Such a difference in the SSIIa alleles alters the structure of amylopectin, of which intermediate chains (DP 12–24) are enriched, and short chains (DP < 11) are depleted in indica varieties compared to japonica varieties (Umemoto et al., 2002). Four amino acid substitutions were found between SSIIa japonica and SSIIa indica alleles, and two of them were identified as indispensable amino acids for full SS activity (Bao et al., 2006; Nakamura et al., 2005; Umemoto and Aoki, 2005; Waters et al., 2006).
Three BE isoforms, BEI, BEIIa and BEIIb, are encoded in the rice genome. In vitro biochemical analysis revealed that BEIIb exclusively transfers short chains (DP 6 and 7), and BEIIa has a more extensive transfer preference (DP 6–15), while BEI transfers intermediate chains (DP 26–39) in addition to short chains (DP 6–12) (Nakamura et al., 2010). Compared to BEI‐ and BEIIa‐deficient mutants, several BEIIb‐deficient mutants, referred to as amylose extender (ae), exhibit severe effects on grain appearance and starch properties (Kubo et al., 2010; Nishi et al., 2001; Satoh et al., 2003a,b; Tanaka et al., 2004). Starch of an ae mutant, EM10, showed about 13 °C higher peak gelatinization temperature (T p) compared to the original cultivar Kinmaze (Tanaka et al., 2004). Elevation of gelatinization temperature in EM10 is consistent with the structure of amylopectin, in which short chains (DP 6–17) and intermediate to long chains (DP > 18) were markedly decreased and increased, respectively. Complementation experiments of EM10 revealed that altered starch properties were restored by introducing functional BEIIb driven by its own promoter and that the degree of the restoration depended on the level of BEIIb activity (Tanaka et al., 2004). This suggests that control of BEIIb activity is effective to manipulate the starch properties.
Among recent breeding programmes of rice grain quality, variation of cooked rice texture without impairment of overall palatability is expected. For example, harder and less sticky rice cultivars can avoid excessive adhesion of cooked materials onto containers, and thus improve handling efficiency during packaging in food industry. To achieve this requirement, development of a series of lines showing a stepwise difference in starch property is demanded. A combination of mutant alleles for starch synthesizing enzymes that specifically function in endosperm, such as BEIIb and SSIIa, would be good candidates to modify characteristics of storage starch. As alleles with severe defects in the functions of the enzymes often lead to an excess in the effects on starch properties and profound reduction in palatability [e.g. assumed reasonable utilizations of EM10 are not cooked rice but food industrial materials (Takahashi et al., 2001)], the isolation of alleles with modest effects is desired.
Upon conventional genetic mapping procedure, repeated evaluation of a phenotype of interest in a large number of segregating populations until identification of the causal gene/mutation is a labour‐intensive and time‐consuming process. Recently developed novel genetic mapping procedures using high‐throughput next‐generation DNA sequencers (NGS) solved the bottleneck by greatly reducing the number of times of the phenotyping process and accelerated identification of the causal mutation governing the agronomic trait. MutMap is one of the procedures and is based on a crossing of the mutant to the original parental line used for mutagenesis, followed by resequencing of the genome DNA of a bulk of F2 segregants with mutant phenotypes obtained by self‐pollination of F1 (Abe et al., 2012). Takagi et al. (2015) applied MutMap to identify a mutation related to salt‐tolerance and verified the advantage of the procedure by production of a high salt‐tolerant rice cultivar within a few years. However, MutMap requires an original parental line to build the high‐quality reference genome and to eliminate background false‐positive SNPs, although in many mutant populations such parental line has not been available. Furthermore, MutMap cannot be applied to mutants that are not amenable for crossing. To address these fundamental issues of MutMap, an advanced method, MutMapPlus, in which whole‐genome sequences between two sibling bulks of mutant (M) and wild‐type (WT) segregants were compared to identify causal mutations, was developed (Fekih et al., 2013).
In this study, to provide novel breeding resources with linked DNA markers for fine‐tuning cooked rice textures, we screened the mutant lines exhibiting altered grain starch properties and identified two novel mutant alleles of BEIIb designated altered gelatinizations (ages) by a combination of the conventional mapping and MutMapPlus procedures. A point mutation of BEIIb gene in age1 allele caused BEIIb‐deficient phenotype, which is permissive for cooked rice even though there was a complete disappearance of its enzyme activity. In addition, a transposon insertion into the same gene yielded another age allele, named age2, resulting in milder modification of the activity. Both age alleles exhibited moderate effects on cooked rice textures as well as the physicochemical properties of their starch. Our work would not only provide breeding materials for fine‐tuning cooked rice textures and confirm the general versatility of MutMapPlus but also shed novel insights into the regulation of BEIIb functions in starch synthesis.
Results
Isolation of novel mutant lines with altered gelatinization properties
We screened a Nipponbare mutant population by evaluation of solubility of endosperm starch in 3.6 m urea solution, in which Nipponbare starch is gelatinized. Among the 3300 mutant lines examined, we found two recessive mutant lines whose starches were hardly gelatinized in the urea solution (Figure 1a). Starches from these lines were gelatinized by urea solution at different concentrations, suggesting that these lines harbour independent alleles affecting the gelatinization property of starch. Alleles with severe and moderate effects on starch gelatinization were referred to as age1 and age2, respectively. The grain appearance of age lines was almost normal, and a small proportion of white core grains was observed (Figure 1b).
Figure 1.
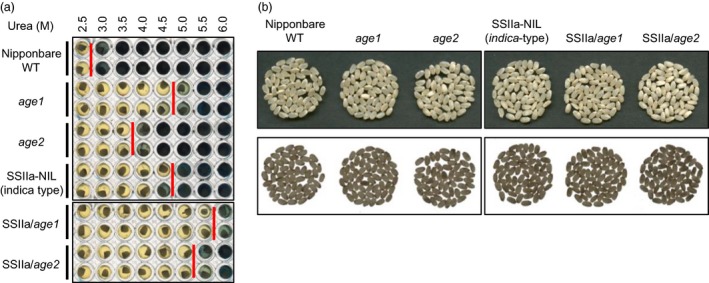
Gelatinization properties and grain appearance of age1, age2, SSIIa‐NIL and their double allele lines. (a) Gelatinization of endosperm starch at various concentrations of urea solution. Urea concentration of onset gelatinization was shown by red bars. (b) Grain appearance. Images by reflected light (upper panel) and transmitted light (lower panel) are shown. Parent plants of the seeds used in this experiment were grown in the paddy field of Hokuriku Research Station (Joetsu, Japan) in 2016.
The expression level and enzyme activity of several starch biosynthetic enzymes in developing caryopses were examined by quantitative RT‐PCR (qRT‐PCR), immunoblot (IB) and activity staining analyses. ISA1 and BEIIb mRNA levels were significantly down‐regulated in age1 and age2, respectively (Figure 2a). Despite a slight reduction of BEIIb protein in age1 (76% of Nipponbare), its enzyme activity was completely abolished (Figure 2b,c). In age2, the BEIIb protein level (67% of Nipponbare) and its enzyme activity were lower than that of Nipponbare. An additional smaller molecular weight protein was detected in age2 (Figure 2b). We also analysed starch‐binding ability of starch biosynthetic enzymes. SSIIaindica showed strong binding to starch granules as reported by Umemoto and Aoki (2005) (Figure S1). Slight increase in SSIIa, BEI, BEIIa and BEIIb in age1 was also observed. These results suggest that the altered gelatinization property of age1 and age2 is related to the alteration of BEIIb activity.
Figure 2.
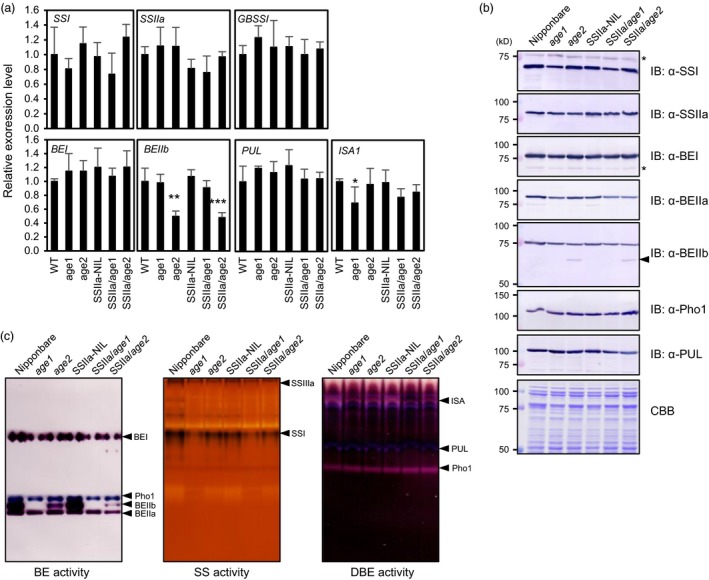
Expression level and enzyme activity of starch biosynthetic enzymes. (a) mRNA levels of starch biosynthetic enzyme genes in developing caryopses were analysed by qRT‐PCR. eEF‐1α was used as an internal control. The value for Nipponbare (WT) was set to 1, and relative values were shown. Asterisks indicate significant difference from background lines (Nipponbare or SSIIa‐NIL) of each line (Student's t test, *P < 0.05, **P < 0.01, ***P < 0.001). (b) Immunoblot (IB) analysis of soluble endosperm extract from developing seeds. Arrowhead on the right indicates bands corresponding to truncated BEIIb. Asterisks indicate nonspecific signals. (c) branching enzyme, starch synthase and debranching enzyme activities in soluble extract of developing endosperm were detected by activity staining after native‐PAGE. Enzymes corresponding to each activity band were indicated on the right in each panel.
Identification of causal mutation of age1 by a combination of conventional mapping and MutMapPlus
Rough mapping was performed by using F2 populations derived from the crosses of each age line and another japonica cultivar, Koshihikari. Illumina single‐nucleotide polymorphism (SNP) array analysis indicated that the mutated locus in both age1 and age2 was mapped within the region between 11 and 21 Mb on chromosome 2 (Figure 3a). However, as the region is common among Nipponbare and Koshihikari (Yamamoto et al., 2010) and no SNP was detected in the region, we could not narrow down the genomic region corresponding to age phenotype. Then, further mapping was conducted with F2 progenies generated by crosses of each age line and SL09, which have a chromosome segment of an indica cultivar, Kasalath, between 6.6 and 27.1 Mb of chromosome 2 on Nipponbare background genome, so that the effect of another gelatinization‐controlling allele, SSIIa indica, which is located on chromosome 6, was eliminated. The mapping using SSR markers focused both age mutations between 19.2 and 19.8 Mb (Figure 3b), presuming that the causal gene is BEIIb, which resides at 19.35 Mb of the chromosome.
Figure 3.
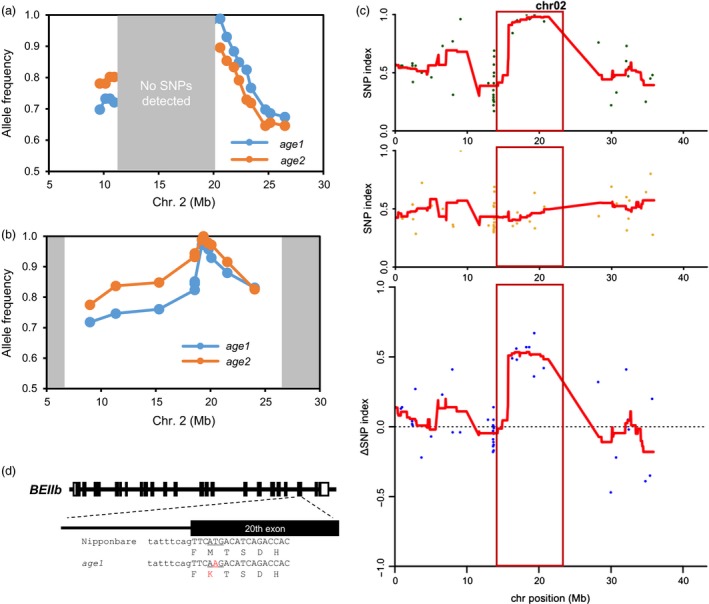
Genetic mapping of age1. (a) and (b) Nipponbare‐type allele frequency plot of chromosome 2 generated by rough mappings. The results of age mutants‐Koshihikari (a) and age mutants‐SL09 (b) F2 populations are shown. The regions in which no polymorphisms were detected are shaded with grey boxes. (c) SNP index and Δ(SNP index) plots of the chromosome 2 generated by MutMapPlus analysis. SNP index of the mutant (M) bulk (upper panel) and the wild‐type (WT) bulk (middle panel) and Δ(SNP index) calculated by subtraction of the index of WT bulk from that of M bulk (lower panel) is shown. Red lines indicate the sliding window average of 4 Mb intervals with 50 kb increment. Candidate region confined by the Fisher's P value of <0.05 is indicated with a red box. (d) Schematic diagrams of BEIIb gene in age1 allele. Open and black boxes indicate untranslated regions and exons, respectively. Uppercase and lowercase letters indicate DNA sequence of exon and intron, respectively. Translated amino acids are shown under the DNA sequence. Substituted nucleotide and amino acid are shown by red letter.
In order to exclude the possibility that the age phenotypes are attributed to mutations of closely linked neighbouring genes other than BEIIb, we next employed MutMapPlus for identification of a corresponding mutation. We applied this method to the F2 progenies of age1‐Nipponbare and age2‐Nipponbare crosses with a modification to detect all types of SNPs, as the original MutMapPlus algorithm extracted only transition‐type SNPs. For the analysis of age1, 54 802 734 and 54 196 761 aligned reads of M and WT bulks, corresponding to 21.1 and 20.9 times coverage of the rice genome, respectively, drew similar charts of SNP index, with an exception that a unique peak was detected only for the M bulk at 15–25 Mb on chromosome 2, and the Δ(SNP index) was also peaked at the same location (the expected Δ(SNP index) is 0.67) (Figure 3c, Figure S2). Within the candidate region confined by the Fisher's P values lower than 0.05, corresponding to 13.75–23.3 Mb, we extracted eight SNPs, that nearly agreed with the expected SNP indexes, 1 and 0.33 for M and WT bulks, respectively (Table 1). Among them, the most likely candidate SNP (19 357 116 base) was located at the 20th exon of BEIIb gene that leads to nonsynonymous substitution of Met‐723 (from translation start site) to Lys (M723K) (Figure 3d, Table 1). The other seven SNPs were located at intergenic regions, although one mutation of 18 242 584 base seemed to reside in a promoter region of a hypothetical gene.
Table 1.
Single‐nucleotide polymorphisms (SNPs) within the candidate genomic region identified by MutMapPlus analysis of age1
| Chromosome | Base | Nipponbare reference base | Mutant bulk | Wild‐type bulk | Fisher's P value | Annotation | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Base | Number of reads | SNP index | Base | Number of reads | SNP index | |||||
| chr02 | 16 289 640 | C | Y | 19 | 0.84 | Y | 17 | 0.35 | 0.005379 | Intergenic. |
| chr02 | 16 865 416 | A | G | 19 | 0.94 | R | 16 | 0.38 | 0.000556 | Intergenic. |
| chr02 | 16 929 731 | A | T | 23 | 0.95 | W | 32 | 0.47 | 0.000111 | Intergenic. |
| chr02 | 18 242 584 | C | T | 25 | 1.00 | Y | 23 | 0.43 | 0.000006 | 110 bp upstream of Hypothetical protein (Os02g0510400) gene. |
| chr02 | 18 648 446 | G | A | 26 | 0.96 | R | 23 | 0.39 | 0.000019 | Intergenic. |
| chr02 | 19 320 101 | G | C | 26 | 1.00 | S | 11 | 0.64 | 0.004997 | Intergenic. |
| chr02 | 19 357 116 | A | T | 18 | 1.00 | W | 27 | 0.33 | 0.000004 | Starch branching enzyme, BEIIb (Os02g0528200), M723K. |
| chr02 | 20 670 926 | T | A | 18 | 0.94 | W | 27 | 0.52 | 0.002882 | Intergenic. |
To confirm M723K substitution of BEIIb is the causal mutation of age1 phenotype, constructs consisting of a genomic fragment of WT or M723K mutant‐type BEIIb gene including the 2.4 kb upstream region from the translation start site were introduced into age1 (Figure 4a). Constructs containing four times‐repeated Myc epitope tags at the carboxy (C)‐terminal of BEIIb were also introduced. The endosperm starch of BEIIb‐ or BEIIb‐Myc‐expressing lines was gelatinized in 5.0 m urea solution, in which vector control seeds of Nipponbare background but not age1 background were gelatinized, indicating successful restoration of age1 phenotype (Figure 4b, Figure S3). In contrast, the expression of M723K or M723K‐Myc failed to restore the gelatinization property of age1 (Figure 4b). The enzyme activity of BEIIb was detected in BEIIb‐ or BEIIb‐Myc‐expressing lines but not in M723K‐ or M723K‐Myc‐expressing lines (Figure 4c). The difference in urea concentration for gelatinization in this experiment from that shown in Figure 1a would arise from the difference in ripening conditions between the paddy field and the greenhouse, as the ripening temperature affects the crystallinity of synthesized starch. These lines of evidence indicate that M723K mutation in BEIIb eliminates its enzyme activity and is the causal mutation of age1 phenotype.
Figure 4.
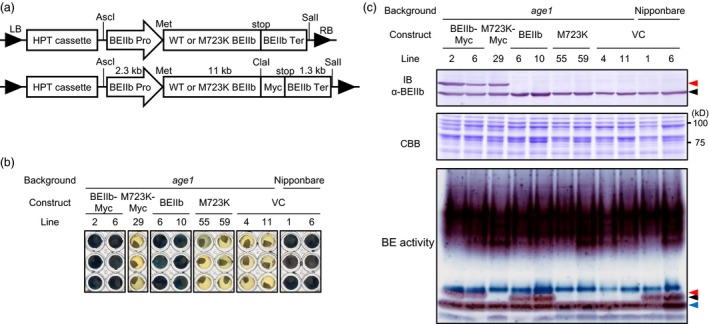
Complementation of age1 by genomic fragment of BEIIb gene. (a) Schematic diagrams of constructs used for complementation. LB, left border; RB, right border; HPT, hygromycin phosphotransferase; Pro, promoter; Ter, terminator; Met, translation start site; stop, translation termination site. Restriction sites were indicated above. (b) Gelatinization of endosperm starch of complementation lines in 5.0 m urea solution. VC, vector control. (c) IB analysis using anti‐BEIIb antibody (upper panel), CBB staining (middle panel) and branching enzyme activity staining (lower panel) of soluble extract of developing endosperm. Black, red and blue arrowheads indicate bands corresponding to BEIIb, Myc‐tagged BEIIb and BEIIa, respectively. The transgenic plants were grown in a greenhouse.
Tos17 insertion in BEIIb gene in age2
To analyse allelism between age1 and age2, they were crossed, and gelatinization of the endosperm starch of F1 grains by urea was determined by the volume of sediment (Figure 5a, Figure S4). Starches obtained by crossing age1 and age2 were not gelatinized in 5.0 m urea solution, while those obtained by crossing Nipponbare and age1 or age2 were gelatinized, indicating that age1 and age2 are allelic. Sequence analysis of BEIIb gene in age2 found an insertion of a retrotransposon, Tos17, in the 17th intron of BEIIb gene (Figure 5b). Application of MutMapPlus to age2 failed to detect functional SNPs, but the survey of short reads mapped around BEIIb indicated that insertion of an interrupting sequence at the position described above was found in M bulk (data not shown). Furthermore, whole‐genome resequencing data confirmed that no SNPs introducing amino acid substitutions were found within the region from 10 Mb to the end of chromosome 2 in age2 (Table S1).
Figure 5.
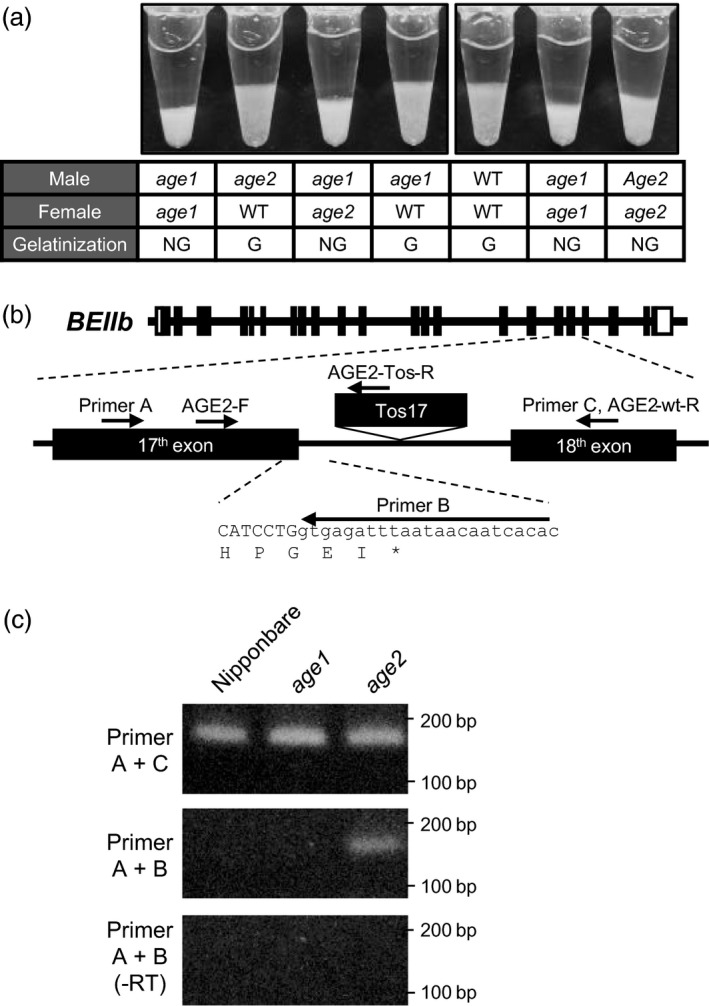
Insertion of Tos17 in BEIIb gene in age2. (a) Gelatinization of endosperm starch of F1 seeds in 5.0 m urea solution. Lines used for each cross and evaluation of gelatinization were shown under the image. WT, G and NG indicate Nipponbare, gelatinized and not gelatinized, respectively. (b) Schematic diagrams of BEIIb gene in age2. Primers used in (c) and Figure 6 were shown by arrows. Open and black boxes indicate untranslated regions and exons, respectively. Location of Tos17 insertion is indicated above the gene. Uppercase and lowercase letters indicate DNA sequence of exon and intron, respectively. Translated amino acids were shown under the DNA sequence. (c) Alternatively spliced BEIIb transcript in age2 was detected by RT‐PCR analysis. Total RNA without reverse transcription (‐RT) was used as a template for negative control (lower panel). DNA size markers are indicated on the right.
The smaller molecular weight‐immunoreacted protein in age2 endosperm extract in Figure 2b is estimated to be a truncated form of BEIIb, because the Tos17 insertion likely leads to aberrant splicing in the 17th exon–intron junction and introduces a premature stop codon in the BEIIb transcripts (Figure 5b). Indeed, a 17th intron‐containing BEIIb transcript as well as a correctly spliced one was produced in age2 (Figure 5c). Migration of the truncated BEIIb corresponds to its estimated molecular weight (approximately 67 kD) in comparison with that of intact mature BEIIb (approximately 86 kD) (Figure 2c). Therefore, the reduction of full‐length BEIIb transcript in age2 by occasional mis‐splicing caused by a Tos17 insertion leads to a weak BEIIb‐deficient phenotype. These results clearly illustrate the different severities of the altered gelatinization phenotype between age1 and age2.
Development of DNA markers for detection of age1 and age2 alleles
We developed a PCR‐based SNP marker for detection of age1 allele. Two forward primers whose 3′ ends locate to the position of SNP (one is specific to WT allele and the other is specific to age1 allele) were designed with an additional mismatch introduced in the third nucleotide from the 3′ end, while a reverse primer is common (Figure 6a). These specific primer sets exclusively amplified WT or age1 alleles, respectively (Figure 6b).
Figure 6.
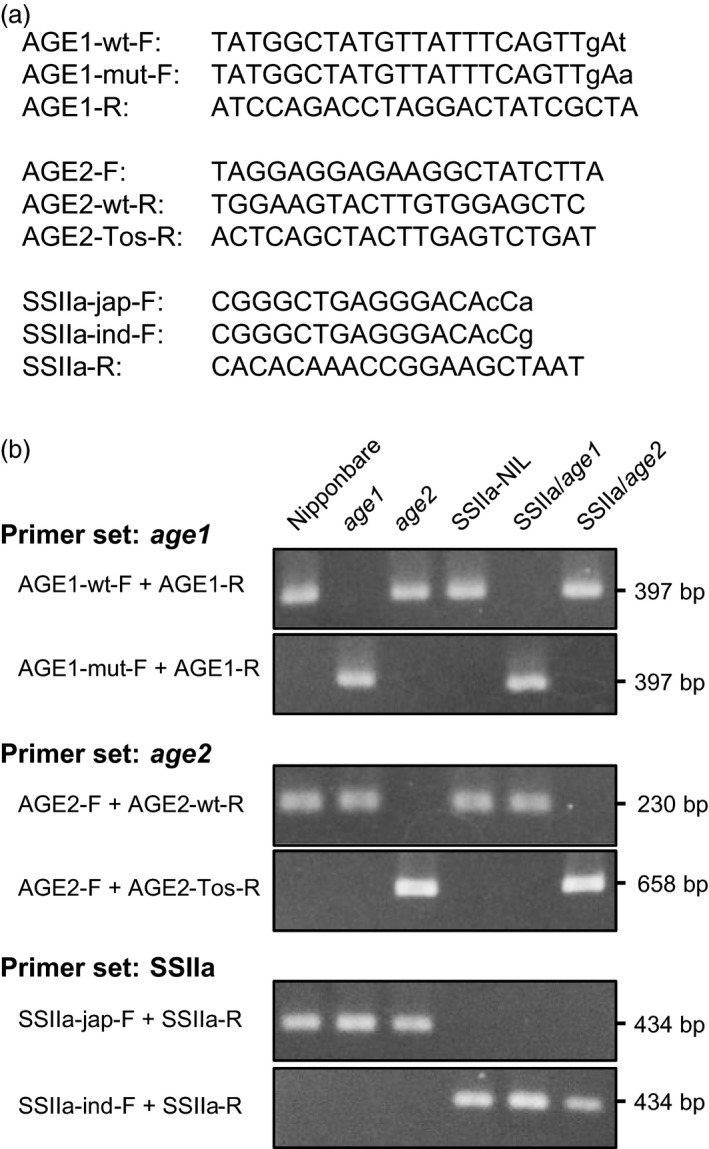
DNA markers for detection of age1 and age2 alleles and SSIIa indica allele. (a) Primers for DNA markers. DNA sequence shown is 5′ to 3′. Allele‐specific and additional mismatch nucleotides are shown in lowercase letters. (b) Band patterns of amplified DNA. Amplicon lengths are indicated on the right.
For age2, a common forward primer and WT‐specific reverse primer were designed on the 17th exon and the 18th exon, respectively, for detection of WT allele (Figures 5b and 6a). Another reverse primer was designed on Tos17. A combination of the reverse primer and common forward primer specifically amplified a DNA fragment composed of a part of the BEIIb genomic region and Tos17 flanking region.
These DNA markers established strict discrimination of WT and age alleles of BEIIb gene.
Introgression of SSIIa indica allele into age1 and age2
To obtain further variation in the starch gelatinization property, we crossed age1 and age2 with Nipponbare near‐isogenic line (NIL) harbouring SSIIa indica allele (Umemoto et al., 2004). A PCR‐based SNP marker to distinguish SSIIa indica and SSIIa japonica alleles was designed based on the primer set for SNP3 (Met/Val‐737 in japonica/indica) (Hiratsuka et al., 2010) with slight modification to optimize to our PCR condition (Figure 6). Plants homozygous for age1 or age2 allele and SSIIa indica allele were obtained by the DNA marker‐assisted selection and designated as SSIIa/age1 or SSIIa/age2, respectively. Starches of the double allele lines were gelatinized in urea solution at a higher concentration compared to their parental age lines (Figure 1a). The expression level and activity of BEIIb in the double allele lines were comparable to those in each parental age line (Figure 2). These results indicate that the increased severity of the gelatinization property in the double allele lines arises from a combination of impairment of BEIIb and active SSIIa. We could not identify the SSIIa activity band in SSIIa‐NIL extract by SS activity staining (Figure 2c) because SSIIa activity is weak and hardly detected by this staining method as observed previously (Umemoto et al., 2004).
Grain size and starch parameters
The grain weight of age1, SSIIa/age1 and SSIIa/age2 was reduced (Table 2). It is considered that reduction of grain weight in age1 and SSIIa/age2 originates from reduced thickness of the grains, while SSIIa/age1 reduced grain weight without a significant change of grain size. The effect of age alleles on the grain weight was quite weak compared to EM10, whose grain weight was about 65% of that of Kinmaze (Nishi et al., 2001).
Table 2.
Grain size, apparent amylose content and thermal properties of starch
| Weight (mg)a | Length (mm)b | Width (mm)b | Thickness (mm)b | Amylose content (%)c | Gelatinization temperature (°C)d | |||
|---|---|---|---|---|---|---|---|---|
| T o | T p | T c | ||||||
| Nipponbare | 22.14 ± 0.35 (100) | 5.16 ± 0.03 | 2.83 ± 0.09 | 2.07 ± 0.02 | 21.5 ± 0.9 | 63.4 ± 0.2 | 75.3 ± 0.2 | 69.8 ± 0.1 |
| age1 | 19.49 ± 1.50* (88) | 5.18 ± 0.09 | 2.84 ± 0.03 | 1.96 ± 0.05* | 29.4 ± 0.4*** | 71.5 ± 0.6*** | 85.3 ± 0.4*** | 79.5 ± 0.5*** |
| age2 | 22.37 ± 0.55 (101) | 5.38 ± 0.00*** | 2.87 ± 0.03 | 2.07 ± 0.02 | 22.9 ± 0.2* | 68.4 ± 0.3*** | 81.3 ± 0.4*** | 74.2 ± 0.3*** |
| SSIIa‐NIL | 21.50 ± 0.57 (97) | 5.33 ± 0.08* | 2.88 ± 0.03 | 2.09 ± 0.02 | 17.7 ± 0.5** | 72.2 ± 0.0*** | 80.5 ± 0.2*** | 76.5 ± 0.1*** |
| SSIIa/age1 | 19.78 ± 0.37** (89) | 5.28 ± 0.01** | 2.77 ± 0.04 | 2.05 ± 0.03 | 28.8 ± 0.5*** | 79.2 ± 0.5*** | 89.4 ± 1.2*** | 84.7 ± 0.2*** |
| SSIIa/age2 | 20.41 ± 0.24** (92) | 5.32 ± 0.13 | 2.82 ± 0.05 | 2.01 ± 0.01* | 24.4 ± 0.2** | 76.4 ± 0.4*** | 89.2 ± 0.7*** | 83.7 ± 0.5*** |
Weight is mean ± SD of at least three independent sets of measurement of 40–70 mature brown rice. Percentages of the weight relative to Nipponbare weight were shown in parentheses.
Length, width and thickness are mean ± SD of three independent sets of measurement of five mature brown rice.
Values are the means ± SD (n = 3).
T o, T p and T c indicate onset, peak and conclusion temperatures, respectively.
Asterisks indicate significant difference from Nipponbare (Student's t test, *P < 0.05, **P < 0.01, ***P < 0.001).
Compared to Nipponbare (21.5%), age1 and age2 lines showed increased apparent amylose content to around 30% and 23%, respectively (Table 2). This is consistent with the results of ae mutants (Nakamura et al., 2011; Nishi et al., 2001). Although the apparent amylose content of SSIIa‐NIL was significantly lower than Nipponbare, SSIIa/age1 and SSIIa/age2 showed almost the same apparent amylose content as age1 and age2, respectively. The increase of apparent amylose content in the mutant lines is, at least in part, likely to be due to the altered CLD of amylopectin as described below (Figure 7).
Figure 7.
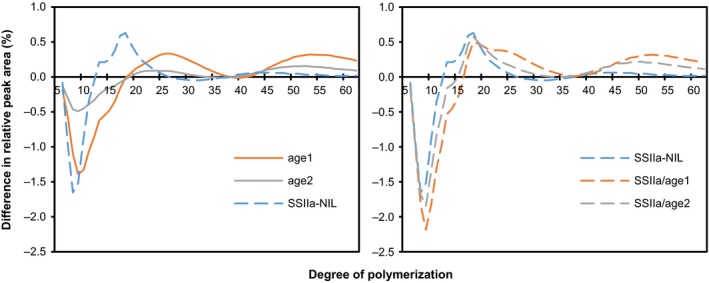
Chain length distribution (CLD) patterns of amylopectin. CLD of amylopectin was examined by HPAEC‐PAD, and then the difference in the profiles between each line and Nipponbare was calculated by subtracting the ratio of a chain of given length of the sample with that of the mean value of Nipponbare. Values are the means of three replicates. The grains used for this experiment were ripened in the paddy field.
The thermal properties of purified starch were analysed by differential scanning calorimetry. T p in age1 and age2 lines was 10 °C and 6 °C higher than that of Nipponbare, respectively (Table 2). SSIIa‐NIL also exhibited a significantly (around 5 °C) higher T p value than Nipponbare, and SSIIa/age1 and SSIIa/age2 showed a further increase in it. Particularly, T p of SSIIa/age2 was comparable to that of SSIIa/age1 (14 °C higher than Nipponbare), suggesting that a combination of SSIIa indica and age2 alleles provided an additive or even transcending effect on the elevation of T p.
Fine structure of amylopectin
The CLD of amylopectin was examined by high‐performance anion‐exchange chromatography‐pulsed amperometric detection (HPAEC‐PAD) (Figure 7). Amylopectin in age1 and age2 showed similar CLD patterns, in which the proportion of short chains (DP 6–17) was significantly reduced, and that of intermediate (DP 19–33) and long chains (DP > 41) was increased. Consistent with BEIIb activity (Figure 2c), the extent of the alteration was markedly severer in age1 than in age2. In SSIIa‐NIL, short chains (DP 6–12) and intermediate chains (DP 13–25) were decreased and increased, respectively, as reported previously (Umemoto et al., 2004). A combination of age and SSIIa indica alleles further increased the severity of alteration of CLD of amylopectin. The detailed effects of each allele on the profiles of CLD in the combination lines are considered in the Discussion section.
In vitro digestion profile of starch by α‐amylase
To evaluate the digestive property of purified starches, raw, gelatinized and retrograded starches were treated with α‐amylase and generated reducing sugars were determined. Raw starches of age1, age2 and SSIIa‐NIL were digested slower than those of Nipponbare (Figure 8). The digestibility of starches in the double allele lines was further slowed down. Interestingly, although age2 starch was digested faster than age1 starch, SSIIa/age2 showed slower digestibility than SSIIa/age1. When starches were gelatinized, the digestive property of the starches of all mutants was comparable to that of Nipponbare (Figure 8). When gelatinized starches were retrograded, only SSIIa/age1 starch exhibited slower digestibility (Figure 8).
Figure 8.
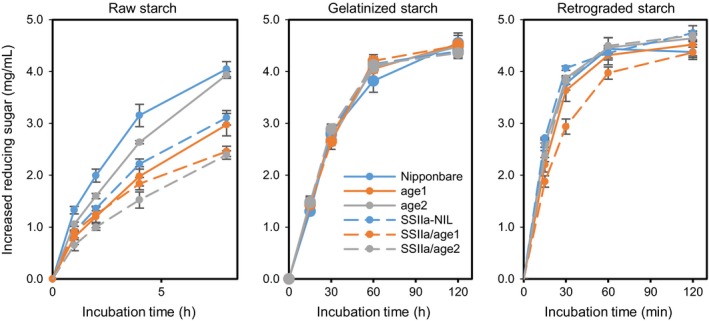
α‐amylase digestion properties of purified starch. Raw, gelatinized or retrograded starches were treated with α‐amylase for indicated time. The amount of reducing sugar produced by the reaction was determined. Values are the means ± SD of three replicates.
Eating quality of the cooked rice of the mutant lines
The eating quality of cooked rice of the mutant lines was evaluated by a sensory evaluation test. A series of the mutants demonstrated stepwise changes of scores related to rice textures including stickiness and hardness. The result with warm rice (10 m after cooking) showed a significant reduction of stickiness in SSIIa/age1, leading to the loss of its overall eating quality, as our sensory test grades sticky rice as superior (Figure 9, Table S2). However, the other lines presented various degrees of stickiness and hardness. When the cooked rice was cooled down for 2 h, the variations in stickiness and hardness were more pronounced (Figure 9, Table S2). These parameters of age1, SSIIa/age1 and SSIIa/age2 were significantly decreased. Especially, SSIIa/age1 exhibited the least sticky and hard texture. Although SSIIa‐NIL rice exhibited a hard and less sticky texture after prolonged cooling at a lower temperature (e.g. 5 °C for 16 h) (Umemoto et al., 2008), it was evaluated as a slightly higher eating quality rice compared to Nipponbare in both tested conditions. However, a combination of alleles with reduced BEIIb activity and SSIIa indica allele drastically reduced the scores of most features of eating quality especially in cooled rice. Taken together, a series of materials with a variety of the textures of cooked rice were produced.
Figure 9.
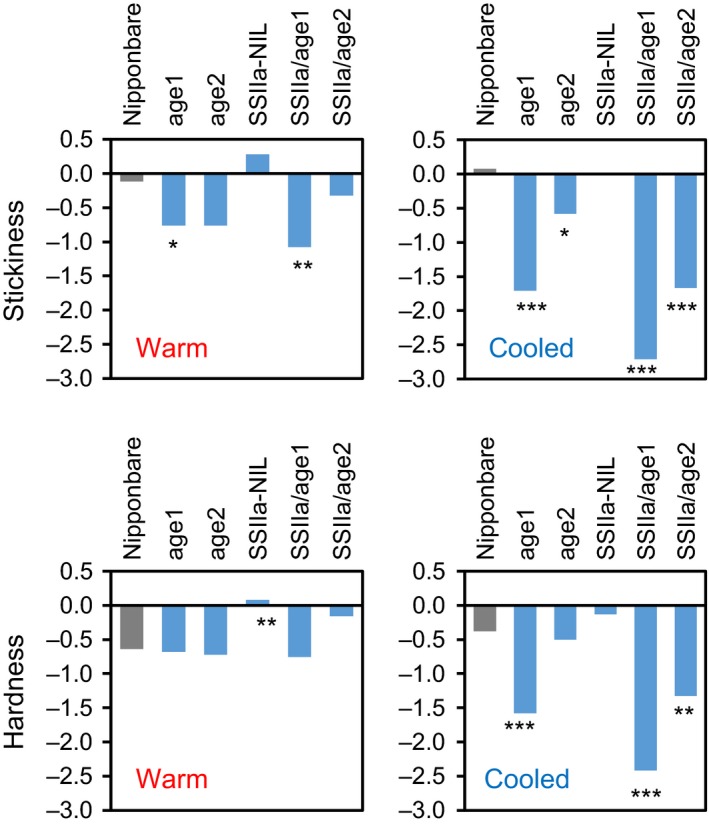
Stickiness and hardness of the cooked rice of the mutant lines as measured by a sensory evaluation test. Each sample was evaluated by 24‐trained panellists under two conditions, in which cooked rice was cooled for 10 m (warm) or 2 h (cooled) at room temperature. For hardness, lower scores are harder texture. Asterisks denote significant differences from Nipponbare (Student's t test, *P < 0.05, **P < 0.01, ***P < 0.001).
Discussion
Advantages of application of MutMapPlus
In this study, we isolated age mutant lines with moderate changes of grain starch properties and identified them as novel mutants of BEIIb by a combination of conventional mapping and MutMapPlus. Before application of MutMapPlus, we tried to identify the causal mutation of age1 by MutMap. However, the Nipponbare mutant population, from which we found age mutants, was generated two decades ago, and the original parental plant subjected to the mutagenesis was not available. Furthermore, as the Nipponbare individuals, which were used for the crossing to establish the F2 progenies were not preserved, we sequenced different individuals to build the reference sequence for the mapping analysis. Under these situations, a trial of MutMap analysis gave multiple candidate regions, where SNP indexes approached 1, throughout the genome due to background SNPs among Nipponbare individuals used for mutagenesis, crossing and reference sequencing, and failed to pick the age1 causal mutation (Figure S5). So, we employed the MutMapPlus procedure, where the sequences of F2 siblings showing M and WT phenotypes were compared. Although MutMapPlus was originally intended to analyse mutants that are not amenable for crossing, such as sterility, we applied the procedure for age1 mutant without the available original parental line and successfully narrowed down the candidate SNPs in a single region of the genome. Our result indicated an effective utilization of the MutMapPlus procedure for the mapping of mutants without an available original parental line.
Although we tried to identify the causal mutation in age2 by MutMapPlus, the analysis alone hardly detected the Tos17 insertion in BEIIb gene because it is difficult to find an indel of a large DNA fragment by this method. However, we could find gaps in every short read aligned to the Tos17 insertion site when we paid attention to the genomic region around BEIIb gene. This fact suggests that, as we conducted in this study, a combination of MutMapPlus with other genetic analyses makes it easy to identify causal mutations including indels.
As BEIIb is well‐studied and characterized for affecting starch properties among the genes in the genomic region identified by rough mapping, it is speculated that if we did not employ MutMapPlus to our analysis, we could focus on BEIIb gene and finally identify M723K mutation as the causal mutation corresponding to age1 phenotype. Application of MutMapPlus, however, markedly shortened the period for identification of the causal mutation and provided solid information about other candidate SNPs around the genomic region. In case of the mapping of agronomic traits with poor information under the situation that the exact reference genome sequence of the original mutagenized plant is not available, MutMapPlus analysis should fully exert its advantages for identification of causal genes/mutations governing the traits. In addition, identification of the causal mutation directly linked with establishment of the DNA marker, which never segregates with the phenotype, to detect the useful allele.
Biochemical characteristics of BEIIb produced by age alleles
Branching enzyme proteins consist of three common characteristic domains, carbohydrate‐binding module 48, catalytic α‐amylase domain and α‐amylase C‐terminal domain (Pfister and Zeeman, 2016). Deletion analysis using chimeric protein of maize BEIIb and BEI revealed that the C‐terminal portion of the C‐terminal domain, which corresponds to a region between Leu‐747 and Glu‐825 of full‐length rice BEIIb, is required for enzyme activity (Hong and Preiss, 2000). In age2, insertion of Tos17 located in a position corresponding to the intermediate region of the C‐terminal domain of BEIIb gene would result in the production of inactive BEIIb protein lacking the C‐terminal portion of the C‐terminal domain. This is consistent with the result in which BE activity corresponding to the truncated BEIIb protein was not detected (Figure 2, Figure S6).
Met‐723 residue in rice BEIIb is conserved among BEIIa and BEIIb in maize, wheat and rice and is located within the C‐terminal domain but not in the region required for the enzyme activity described above (Hong and Preiss, 2000). M723K mutation abolished enzyme activity of BEIIb and affected starch properties without a marked decrease in BEIIb protein (Figures 2 and 7, Table 2). However, the degree of the effects on grain quality and amylopectin structure in age1 was significantly weaker than those in EM10 (Figure S7). One possible explanation of the difference between age1 and EM10 is the presence of inactive BEIIb protein in age1. Starch biosynthetic enzymes in endosperm form large protein complexes (Ahmed et al., 2015; Crofts et al., 2015; Hennen‐Bierwagen et al., 2008; Liu et al., 2009, 2012; Tetlow et al., 2004, 2008). In maize, ae1.1, lacking BEIIb protein, and ae1.2, expressing inactive BEIIb, alleles exhibited a different organization and assembly of protein complexes of starch biosynthetic enzymes (Liu et al., 2009, 2012). This implies that deficiency of a specific isozyme involved in starch synthesis is compensated by other isozymes through modification of assembly of the protein complexes (Abe et al., 2014; Fujita et al., 2011; Li and Gilbert, 2016; Tetlow and Emes, 2014). We detected an additional slower migration band only in age1 extract by IB analysis after native‐PAGE (Figure S6). The protein complex corresponding to the band is considered to be exclusively organized by M723K‐mutated BEIIb. In addition, composition of starch‐bound proteins was altered in age1 (Figure S1) as observed in maize inactive BEIIb mutant ae1.2 (Liu et al., 2012). M723K mutation slightly up‐regulated starch‐binding ability of BEIIb (Figure S1). These results indicate that physical protein–protein interactions between starch biosynthetic enzymes and their starch‐binding states are rearranged in age1 by the expression of M723K‐mutated BEIIb. These alterations may lead to partial compensation of BEIIb function in age1. Further detailed researches concerning the contribution of starch‐bound and unbound respective enzymes to the altered starch properties are necessary.
Compared to single age lines, SSIIa/age double allele lines showed a severe effect on starch properties (Figures 1a and 8, Table 2). The decline in BEIIb activity by age alleles restrains generation of new short branches by cleaving longer chains, and active SSIIa indica allele increases longer chains by elongation of short branches during amylopectin synthesis, resulting in production of amylopectin composed of less‐branched longer glucan chains in crystalline lamellae (Figure 7). Longer glucan chains in amylopectin also increase apparent amylose content (Table 2). As the proportion of short chains (DP < 13) of amylopectin is correlated with the digestibility and gelatinization temperature of starch positively and negatively, respectively (Tanaka et al., 2004; Zhang et al., 2008), profound effects on starch properties in the double allele lines are explained by alteration of CLD of amylopectin.
We analysed the contribution of each allele in the double allele lines to CLD of amylopectin (Figure S8). The contribution of age alleles in the single and double allele lines to the distribution of intermediate to long glucan chains (DP > 14) was almost the same, whereas those to short chains (DP 6–12) were different. A similar trend was observed in the effect of SSIIa indica allele. The effect of age1 and SSIIa indica on starch properties was severely attenuated in SSIIaindica/age1 compared to that of age2 and SSIIa indica in SSIIaindica/age2 (Figure S8), resulting in a similar degree of effects of age1 and age2 in the double allele lines on starch properties (Figure 8, Table 2). We hypothesized that BEIIb and SSIIa act independently for synthesizing longer chains, whereas they have functional interaction, which is probably originated from physical interactions of these enzymes (Crofts et al., 2015), to some extent, for making short chains. According to the hypothesis, CLD of amylopectin in ss3a ss4b double mutant showed a different pattern from that in ss3a and ss4b single mutants (Toyosawa et al., 2016). In contrast, two double mutants, ss1 be1 and ss1 be2b, displayed additive patterns of their respective single mutants in CLD of amylopectin (Abe et al., 2014). Accumulation of understandings from these approaches should contribute to interpret physiological functions of the organization of a specific protein complex by starch biosynthesis‐related proteins during starch biosynthesis and uncover the missing links in the mechanisms of starch biosynthesis in future studies.
Availability and advantages of age alleles for breeding
BEIIb‐deficient mutants have been reported to produce rice grain with a chalky appearance, which reflects the morphology of starch granules (Kubo et al., 2010). Rice grains with chalky appearance reduce the yield of head rice after polishing and lower their market value (Fitzgerald et al., 2009). In contrast, production of almost normal grains in age1 and age2 may be involved in moderate alteration of their starch properties (Figure 1b). Although occasional production of white core grains in age1 and age2 was observed regardless of their growth conditions in the paddy field, greenhouse and plant incubator, the ratios of white core grain and severity of chalkiness in age grains are significantly lower than those in EM10 (Figure S7b). In addition, the effects of age alleles on grain size and weight were weak (Table 2). The less negative effect of age alleles on grain appearance is not presumed to reduce the market value of the rice, being a great advantage in the utilization of age alleles for breeding.
In conclusion, comprehensive assessment of biochemical and food chemical characteristics demonstrated that age alleles produce starch with moderately changed properties and are capable of utilization for practical breeding. Development of DNA markers linked with the alleles can accelerate utilization of these alleles. Recently, consumers’ preferences became diversified owing to the variety of food cultures. Markets require a wide range of cooked rice textures, not only traditionally preferred soft rice but also less sticky rice. Rice breeders can select one or several lines produced in this study for breeding depending on the objective of breeding. We hope that our work facilitates rice breeding towards new cultivars with novel features of cooked rice.
Experimental procedures
Plant growth, screening of age mutants, mapping of mutant genes, plasmid construction, generation of transgenic plants, measurements of starch properties, RT‐PCR and protein analyses were performed according to previous reports with slight modification (Asai et al., 2014; Fujita et al., 1999; Hirochika, 2001; Kosugi et al., 2013; Kuroda et al., 2010; Li and Durbin, 2009; Li et al., 2009; Miyao et al., 2003; Nakagawa et al., 2007; Nelson, 1944; Toki, 1997; Yamakawa et al., 2007). Details of the Supplementary Experimental Procedures were provided in the Appendix S1. Primers used in this study and antigens for raising specific polyclonal antibodies are listed in Tables S3 and S4, respectively.
Supporting information
Figure S1 Distribution of starch biosynthetic enzymes in developing endosperm. Immunoblot (IB) analysis of soluble protein (SP) and proteins loosely (LBP) or tightly (TBP) bound to starch granule in developing seeds. Protein fractions were prepared as reported by Asai et al. (2014). Antibodies used were indicated below the images. Positions of molecular markers were shown on the left of the images.
Figure S2 SNP index and Δ(SNP index) plots of rice chromosomes generated by MutMapPlus analysis of age1. SNP index and Δ(SNP index) plots were generated by MutMapPlus analysis of bulked F2 mutant progeny obtained by a crossing between age1 and Nipponbare. (a) SNP index of the mutant (M) bulk. (b) SNP index of the wild‐type (WT) bulk. (c) Δ(SNP index) calculated by subtraction of the index of WT bulk from that of M bulk. Red lines indicate average value of SNP index or Δ(SNP index) obtained by the sliding window analysis of 4 Mb intervals with 50 kb increment. Candidate region confined by the Fisher's P value of <0.05 is indicated with a red box.
Figure S3 Investigation of urea concentration for gelatinization of starch of vector control lines of Nipponbare and age1. Halved rice grains were incubated with various concentrations of urea solution overnight and gelatinization of starch was evaluated by iodine staining. Transgenic plants were grown in a greenhouse. Temperatures after flowering were set at 27 °C/22 °C during 15 h light/9 h dark periods, respectively.
Figure S4 Investigation of urea concentration for gelatinization of endosperm starch of F1 seed in allelism test. Endosperm powder of F1 seeds (2.5 mg) was incubated with various concentrations of urea solution (150 μL) and gelatinization of starch was evaluated by the volumes of sediments. Grains used for this experiment were ripened in a warm and closed room in summer (see Experimental procedures).
Figure S5 Trial of mapping age1 mutation by MutMap analysis. SNP index plot was generated by MutMap analysis of bulked F2 mutant progeny obtained by a crossing between age1 and Nipponbare. Red lines indicate average value of SNP index obtained by the sliding window analysis of 4 Mb interval with 50 kb increment. Green lines represent the 95% statistical confidence limit.
Figure S6 BE staining and immunoblot analyses after native‐PAGE. Ten micrograms of soluble endosperm extract was separated by native‐PAGE. After electrophoresis, the gels were subjected to BE activity staining or immunoblot analyses with anti‐BEIIb antibody. The slight delay in the migration of the immunoreactive band of age1 occurred probably due to the change in electrical charge by the M723K substitution. An additional band observed in age1 was indicated by a red arrowhead.
Figure S7 Comparison of EM10 and age1. (a) The distribution of the length of amylopectin side chains was examined by HPAEC‐PAD, and then the difference in the profiles between EM10 and Kinmaze or age1 and Nipponbare was calculated by subtracting the ratio of a chain of given length of the sample with that of the mean value of Kinmaze or Nipponbare, respectively. Values are the means of three biological replicates. (b) Grain appearance of age1 and EM10. Images by reflected light (left panel) and transmitted light (right panel) are shown. Grains used in this experiment were ripened in a plant incubator.
Figure S8 Comparison of chain length distribution between single and double allele lines. The difference in the chain length profiles of amylopectin between the double allele lines and their parental single allele lines was shown. The values were calculated by subtracting the ratio of a chain of given length of the double allele line with that of the single allele line shown in Figure 7.
Table S1 SNPs identified in age2 by BWA mapping and Coval extraction between 10 Mb and 3′ end of chromosome 2.
Table S2 Eating quality of the cooked rice of the mutant lines as measured by a sensory evaluation test.
Table S3 Primers used in this study.
Table S4 Peptide sequences of antigens for raising polyclonal antibodies.
Appendix S1 Experimental procedures.
Acknowledgements
We thank Drs. Shibaya Taeko and Masahiro Yano in Institute of Crop Science, NARO for supporting Illumina Golden Gate genotyping, Ms. Naomi Sugiura for genotyping with SSR markers and Rice Genome Resource Center of National Institute of Agrobiological Sciences, Dr. Toshihiro Kumamaru in Kyushu University, Dr. Naoko Fujita in Akita Prefectural University and Dr. Tsuyoshi Nakagawa in Shimane University for providing SL09 seed and Nipponbare Tos17 insertion mutant library, EM10 seed, anti‐GBSSI antibody and pUGW16 vector, respectively. Computations were partially performed on the supercomputers at ROIS National Institute of Genetics and Advanced Analysis Center, NARO. This work was supported by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan [Genomics‐based Technology for Agricultural Improvement, IVG3001] and a Japan Society for the Promotion of Science Grant‐in‐Aid for Scientific Research (C) [15K07279] from the Ministry of Education, Culture, Sports, Science and Technology of Japan to H.Y. The authors declare no conflict of interest.
Accession numbers: The FASTQ sequence data are available from DDBJ Sequence Read Archive, DRA005449.
References
- Abe, A. , Kosugi, S. , Yoshida, K. , Natsume, S. , Takagi, H. , Kanzaki, H. , Matsumura, H. et al (2012) Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 30, 174–178. [DOI] [PubMed] [Google Scholar]
- Abe, N. , Asai, H. , Yago, H. , Oitome, N.F. , Itoh, R. , Crofts, N. , Nakamura, Y. et al (2014) Relationships between starch synthase I and branching enzyme isozymes determined using double mutant rice lines. BMC Plant Biol. 14, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed, Z. , Tetlow, I.J. , Ahmed, R. , Morell, M.K. and Emes, M.J. (2015) Protein‐protein interactions among enzymes of starch biosynthesis in high‐amylose barley genotypes reveal differential roles of heteromeric enzyme complexes in the synthesis of A and B granules. Plant Sci. 233, 95–106. [DOI] [PubMed] [Google Scholar]
- Asai, H. , Abe, N. , Matsushima, R. , Crofts, N. , Oitome, N.F. , Nakamura, Y. and Fujita, N. (2014) Deficiencies in both starch synthase IIIa and branching enzyme IIb lead to a significant increase in amylose in SSIIa‐inactive japonica rice seeds. J. Exp. Bot. 65, 5497–5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball, S.G. and Morell, M.K. (2003) From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Annu. Rev. Plant Biol. 54, 207–233. [DOI] [PubMed] [Google Scholar]
- Bao, J.S. , Corke, H. and Sun, M. (2006) Nucleotide diversity in starch synthase IIa and validation of single nucleotide polymorphisms in relation to starch gelatinization temperature and other physicochemical properties in rice (Oryza sativa L.). Theor. Appl. Genet. 113, 1171–1183. [DOI] [PubMed] [Google Scholar]
- Crofts, N. , Abe, N. , Oitome, N.F. , Matsushima, R. , Hayashi, M. , Tetlow, I.J. , Emes, M.J. et al (2015) Amylopectin biosynthetic enzymes from developing rice seed form enzymatically active protein complexes. J. Exp. Bot. 66, 4469–4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekih, R. , Takagi, H. , Tamiru, M. , Abe, A. , Natsume, S. , Yaegashi, H. , Sharma, S. et al (2013) MutMap plus: genetic mapping and mutant identification without crossing in rice. PLoS ONE, 8, e68529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald, M.A. , McCouch, S.R. and Hall, R.D. (2009) Not just a grain of rice: the quest for quality. Trends Plant Sci. 14, 133–139. [DOI] [PubMed] [Google Scholar]
- Fujita, N. , Kubo, A. , Francisco, P.B. , Nakakita, M. , Harada, K. , Minaka, N. and Nakamura, Y. (1999) Purification, characterization, and cDNA structure of isoamylase from developing endosperm of rice. Planta, 208, 283–293. [DOI] [PubMed] [Google Scholar]
- Fujita, N. , Yoshida, M. , Asakura, N. , Ohdan, T. , Miyao, A. , Hirochika, H. and Nakamura, Y. (2006) Function and characterization of starch synthase I using mutants in rice. Plant Physiol. 140, 1070–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, N. , Yoshida, M. , Kondo, T. , Saito, K. , Utsumi, Y. , Tokunaga, T. , Nishi, A. et al (2007) Characterization of SSIIIa‐deficient mutants of rice: the function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm. Plant Physiol. 144, 2009–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, N. , Satoh, R. , Hayashi, A. , Kodama, M. , Itoh, R. , Aihara, S. and Nakamura, Y. (2011) Starch biosynthesis in rice endosperm requires the presence of either starch synthase I or IIIa. J. Exp. Bot. 62, 4819–4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennen‐Bierwagen, T.A. , Liu, F. , Marsh, R.S. , Kim, S. , Gan, Q. , Tetlow, I.J. , Emes, M.J. et al (2008) Starch biosynthetic enzymes from developing maize endosperm associate in multisubunit complexes. Plant Physiol. 146, 1892–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsuka, M. , Umemoto, T. , Aoki, N. and Katsuta, M. (2010) Development of SNP markers of Starch synthase IIa (alk) and haplotype distribution in Rice Core Collections. Rice Genet. Newsl. 25, 80–82. [Google Scholar]
- Hirochika, H. (2001) Contribution of the Tos17 retrotransposon to rice functional genomics. Curr. Opin. Plant Biol. 4, 118–122. [DOI] [PubMed] [Google Scholar]
- Hong, S.J. and Preiss, J. (2000) Localization of C‐terminal domains required for the maximal activity or for determination of substrate preference of maize branching enzymes. Arch. Biochem. Biophys. 378, 349–355. [DOI] [PubMed] [Google Scholar]
- Kosugi, S. , Natsume, S. , Yoshida, K. , MacLean, D. , Cano, L. , Kamoun, S. and Terauchi, R. (2013) Coval: improving alignment quality and variant calling accuracy for next‐generation sequencing data. PLoS ONE, 8, e75402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo, A. , Akdogan, G. , Nakaya, M. , Shojo, A. , Suzuki, S. , Satoh, H. and Kitamura, S. (2010) Structure, physical, and digestive properties of starch from wx ae double‐mutant rice. J. Agric. Food Chem. 58, 4463–4469. [DOI] [PubMed] [Google Scholar]
- Kuroda, M. , Kimizu, M. and Mikami, C. (2010) A simple set of plasmids for the production of transgenic plants. Biosci. Biotechnol. Biochem. 74, 2348–2351. [DOI] [PubMed] [Google Scholar]
- Li, H. and Durbin, R. (2009) Fast and accurate short read alignment with Burrows‐Wheeler transform. Bioinformatics, 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C. and Gilbert, R.G. (2016) Progress in controlling starch structure by modifying starch‐branching enzymes. Planta, 243, 13–22. [DOI] [PubMed] [Google Scholar]
- Li, H. , Handsaker, B. , Wysoker, A. , Fennell, T. , Ruan, J. , Homer, N. , Marth, G. et al and 1000 Genome Project Data Processing Subgroup (2009) The sequence alignment/map format and SAMtools. Bioinformatics, 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, F.S. , Makhmoudova, A. , Lee, E.A. , Wait, R. , Emes, M.J. and Tetlow, I.J. (2009) The amylose extender mutant of maize conditions novel protein‐protein interactions between starch biosynthetic enzymes in amyloplasts. J. Exp. Bot. 60, 4423–4440. [DOI] [PubMed] [Google Scholar]
- Liu, F.S. , Ahmed, Z. , Lee, E.A. , Donner, E. , Liu, Q. , Ahmed, R. , Morell, M.K. et al (2012) Allelic variants of the amylose extender mutation of maize demonstrate phenotypic variation in starch structure resulting from modified protein‐protein interactions. J. Exp. Bot. 63, 1167–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyao, A. , Tanaka, K. , Murata, K. , Sawaki, H. , Takeda, S. , Abe, K. , Shinozuka, Y. et al (2003) Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon‐rich regions of the genome. Plant Cell, 15, 1771–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, T. , Kurose, T. , Hino, T. , Tanaka, K. , Kawamukai, M. , Niwa, Y. , Toyooka, K. et al (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104, 34–41. [DOI] [PubMed] [Google Scholar]
- Nakamura, Y. (2002) Towards a better understanding of the metabolic system for amylopectin biosynthesis in plants: rice endosperm as a model tissue. Plant Cell Physiol. 43, 718–725. [DOI] [PubMed] [Google Scholar]
- Nakamura, Y. , Francisco, P.B. , Hosaka, Y. , Sato, A. , Sawada, T. , Kubo, A. and Fujita, N. (2005) Essential amino acids of starch synthase IIa differentiate amylopectin structure and starch quality between japonica and indica rice varieties. Plant Mol. Biol. 58, 213–227. [DOI] [PubMed] [Google Scholar]
- Nakamura, Y. , Utsumi, Y. , Sawada, T. , Aihara, S. , Utsumi, C. , Yoshida, M. and Kitamura, S. (2010) Characterization of the reactions of starch branching enzymes from rice endosperm. Plant Cell Physiol. 51, 776–794. [DOI] [PubMed] [Google Scholar]
- Nakamura, S. , Satoh, H. and Ohtsubo, K. (2011) Characteristics of pregelatinized ae mutant rice flours prepared by boiling after preroasting. J. Agric. Food Chem. 59, 10665–10676. [DOI] [PubMed] [Google Scholar]
- Nelson, N. (1944) A photometric adaptation of the Somogyi method for the determination of glucose. J. Biol. Chem. 153, 375–380. [Google Scholar]
- Nishi, A. , Nakamura, Y. , Tanaka, N. and Satoh, H. (2001) Biochemical and genetic analysis of the effects of amylose‐extender mutation in rice endosperm. Plant Physiol. 127, 459–472. [PMC free article] [PubMed] [Google Scholar]
- Ohdan, T. , Francisco, P.B. , Sawada, T. , Hirose, T. , Terao, T. , Satoh, H. and Nakamura, Y. (2005) Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J. Exp. Bot. 56, 3229–3244. [DOI] [PubMed] [Google Scholar]
- Pfister, B. and Zeeman, S.C. (2016) Formation of starch in plant cells. Cell. Mol. Life Sci. 73, 2781–2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh, H. , Nishi, A. , Fujita, N. , Kubo, A. , Nakamura, Y. , Kawasaki, T. and Okita, T.W. (2003a) Isolation and characterization of starch mutants in rice. J. Appl. Glycosci. 50, 225–230. [Google Scholar]
- Satoh, H. , Nishi, A. , Yamashita, K. , Takemoto, Y. , Tanaka, Y. , Hosaka, Y. , Sakurai, A. et al (2003b) Starch‐branching enzyme I‐deficient mutation specifically affects the structure and properties of starch in rice endosperm. Plant Physiol. 133, 1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, A.M. , Denyer, K. and Martin, C. (1997) The synthesis of the starch granule. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 65–87. [DOI] [PubMed] [Google Scholar]
- Takagi, H. , Tamiru, M. , Abe, A. , Yoshida, K. , Uemura, A. , Yaegashi, H. , Obara, T. et al (2015) MutMap accelerates breeding of a salt‐tolerant rice cultivar. Nat. Biotechnol. 33, 445–449. [DOI] [PubMed] [Google Scholar]
- Takahashi, H. , Ohtsubo, K. , Romero, M. , Toyoshima, H. , Okadome, H. , Nishi, A. and Sato, H. (2001) Evaluation of basic physical and chemical properties of amylose extender mutants of rice for the presumption of the suitable utilizations (in Japanese). Nihon Shokuhin Kagaku Kōgakkai, 48, 617–621. [Google Scholar]
- Tanaka, N. , Fujita, N. , Nishi, A. , Satoh, H. , Hosaka, Y. , Ugaki, M. , Kawasaki, S. et al (2004) The structure of starch can be manipulated by changing the expression levels of starch branching enzyme llb in rice endosperm. Plant Biotechnol. J. 2, 507–516. [DOI] [PubMed] [Google Scholar]
- Tetlow, I.J. and Emes, M.J. (2014) A review of starch‐branching enzymes and their role in amylopectin biosynthesis. IUBMB Life, 66, 546–558. [DOI] [PubMed] [Google Scholar]
- Tetlow, I.J. , Wait, R. , Lu, Z.X. , Akkasaeng, R. , Bowsher, C.G. , Esposito, S. , Kosar‐Hashemi, B. et al (2004) Protein phosphorylation in amyloplasts regulates starch branching enzyme activity and protein‐protein interactions. Plant Cell, 16, 694–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetlow, I.J. , Beisel, K.G. , Cameron, S. , Makhmoudova, A. , Liu, F. , Bresolin, N.S. , Wait, R. et al (2008) Analysis of protein complexes in wheat amyloplasts reveals functional interactions among starch biosynthetic enzymes. Plant Physiol. 146, 1878–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toki, S. (1997) Rapid and efficient Agrobacterium‐mediated transformation in rice. Plant Mol. Biol. Rep. 15, 16–21. [Google Scholar]
- Toyosawa, Y. , Kawagoe, Y. , Matsushima, R. , Crofts, N. , Ogawa, M. , Fukuda, M. , Kumamaru, T. et al (2016) Deficiency of starch synthase IIIa and IVb alters starch granule morphology from polyhedral to spherical in rice endosperm. Plant Physiol. 170, 1255–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemoto, T. and Aoki, N. (2005) Single‐nucleotide polymorphisms in rice starch synthase IIa that alter starch gelatinisation and starch association of the enzyme. Funct. Plant Biol. 32, 763–768. [DOI] [PubMed] [Google Scholar]
- Umemoto, T. , Yano, M. , Satoh, H. , Shomura, A. and Nakamura, Y. (2002) Mapping of a gene responsible for the difference in amylopectin structure between japonica‐type and indica‐type rice varieties. Theor. Appl. Genet. 104, 1–8. [DOI] [PubMed] [Google Scholar]
- Umemoto, T. , Aoki, N. , Lin, H.X. , Nakamura, Y. , Inouchi, N. , Sato, Y. , Yano, M. et al (2004) Natural variation in rice starch synthase IIa affects enzyme and starch properties. Funct. Plant Biol. 31, 671–684. [DOI] [PubMed] [Google Scholar]
- Umemoto, T. , Horibata, T. , Aoki, N. , Hiratsuka, M. , Yano, M. and Inouchi, N. (2008) Effects of variations in starch synthase on starch properties and eating quality of rice. Plant Prod. Sci. 11, 472–480. [Google Scholar]
- Waters, D.L.E. , Henry, R.J. , Reinke, R.F. and Fitzgerald, M.A. (2006) Gelatinization temperature of rice explained by polymorphisms in starch synthase. Plant Biotechnol. J. 4, 115–122. [DOI] [PubMed] [Google Scholar]
- Yamakawa, H. , Hirose, T. , Kuroda, M. and Yamaguchi, T. (2007) Comprehensive expression profiling of rice grain filling‐related genes under high temperature using DNA microarray. Plant Physiol. 144, 258–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, T. , Nagasaki, H. , Yonemaru, J. , Ebana, K. , Nakajima, M. , Shibaya, T. and Yano, M. (2010) Fine definition of the pedigree haplotypes of closely related rice cultivars by means of genome‐wide discovery of single‐nucleotide polymorphisms. BMC Genom. 11, 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeman, S.C. , Kossmann, J. and Smith, A.M. (2010) Starch: its metabolism, evolution, and biotechnological modification in plants. Annu. Rev. Plant Biol. 61, 209–234. [DOI] [PubMed] [Google Scholar]
- Zhang, G. , Ao, Z. and Hamaker, B.R. (2008) Nutritional property of endosperm starches from maize mutants: a parabolic relationship between slowly digestible starch and amylopectin fine structure. J. Agric. Food Chem. 56, 4686–4694. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Distribution of starch biosynthetic enzymes in developing endosperm. Immunoblot (IB) analysis of soluble protein (SP) and proteins loosely (LBP) or tightly (TBP) bound to starch granule in developing seeds. Protein fractions were prepared as reported by Asai et al. (2014). Antibodies used were indicated below the images. Positions of molecular markers were shown on the left of the images.
Figure S2 SNP index and Δ(SNP index) plots of rice chromosomes generated by MutMapPlus analysis of age1. SNP index and Δ(SNP index) plots were generated by MutMapPlus analysis of bulked F2 mutant progeny obtained by a crossing between age1 and Nipponbare. (a) SNP index of the mutant (M) bulk. (b) SNP index of the wild‐type (WT) bulk. (c) Δ(SNP index) calculated by subtraction of the index of WT bulk from that of M bulk. Red lines indicate average value of SNP index or Δ(SNP index) obtained by the sliding window analysis of 4 Mb intervals with 50 kb increment. Candidate region confined by the Fisher's P value of <0.05 is indicated with a red box.
Figure S3 Investigation of urea concentration for gelatinization of starch of vector control lines of Nipponbare and age1. Halved rice grains were incubated with various concentrations of urea solution overnight and gelatinization of starch was evaluated by iodine staining. Transgenic plants were grown in a greenhouse. Temperatures after flowering were set at 27 °C/22 °C during 15 h light/9 h dark periods, respectively.
Figure S4 Investigation of urea concentration for gelatinization of endosperm starch of F1 seed in allelism test. Endosperm powder of F1 seeds (2.5 mg) was incubated with various concentrations of urea solution (150 μL) and gelatinization of starch was evaluated by the volumes of sediments. Grains used for this experiment were ripened in a warm and closed room in summer (see Experimental procedures).
Figure S5 Trial of mapping age1 mutation by MutMap analysis. SNP index plot was generated by MutMap analysis of bulked F2 mutant progeny obtained by a crossing between age1 and Nipponbare. Red lines indicate average value of SNP index obtained by the sliding window analysis of 4 Mb interval with 50 kb increment. Green lines represent the 95% statistical confidence limit.
Figure S6 BE staining and immunoblot analyses after native‐PAGE. Ten micrograms of soluble endosperm extract was separated by native‐PAGE. After electrophoresis, the gels were subjected to BE activity staining or immunoblot analyses with anti‐BEIIb antibody. The slight delay in the migration of the immunoreactive band of age1 occurred probably due to the change in electrical charge by the M723K substitution. An additional band observed in age1 was indicated by a red arrowhead.
Figure S7 Comparison of EM10 and age1. (a) The distribution of the length of amylopectin side chains was examined by HPAEC‐PAD, and then the difference in the profiles between EM10 and Kinmaze or age1 and Nipponbare was calculated by subtracting the ratio of a chain of given length of the sample with that of the mean value of Kinmaze or Nipponbare, respectively. Values are the means of three biological replicates. (b) Grain appearance of age1 and EM10. Images by reflected light (left panel) and transmitted light (right panel) are shown. Grains used in this experiment were ripened in a plant incubator.
Figure S8 Comparison of chain length distribution between single and double allele lines. The difference in the chain length profiles of amylopectin between the double allele lines and their parental single allele lines was shown. The values were calculated by subtracting the ratio of a chain of given length of the double allele line with that of the single allele line shown in Figure 7.
Table S1 SNPs identified in age2 by BWA mapping and Coval extraction between 10 Mb and 3′ end of chromosome 2.
Table S2 Eating quality of the cooked rice of the mutant lines as measured by a sensory evaluation test.
Table S3 Primers used in this study.
Table S4 Peptide sequences of antigens for raising polyclonal antibodies.
Appendix S1 Experimental procedures.


