Abstract
The gastrointestinal microbiome is a diverse consortium of bacteria, archaea, fungi, protozoa, and viruses that inhabit the gut of all mammals. Studies in humans and other mammals have implicated the microbiome in a range of physiologic processes that are vital to host health including energy homeostasis, metabolism, gut epithelial health, immunologic activity, and neurobehavioral development. The microbial genome confers metabolic capabilities exceeding those of the host organism alone, making the gut microbiome an active participant in host physiology. Recent advances in DNA sequencing technology and computational biology have revolutionized the field of microbiomics, permitting mechanistic evaluation of the relationships between an animal and its microbial symbionts. Changes in the gastrointestinal microbiome are associated with diseases in humans and animals including inflammatory bowel disease, asthma, obesity, metabolic syndrome, cardiovascular disease, immune‐mediated conditions, and neurodevelopmental conditions such as autism spectrum disorder. While there remains a paucity of data regarding the intestinal microbiome in small animals, recent studies have helped to characterize its role in host animal health and associated disease states. This review is intended to familiarize small animal veterinarians with recent advances in the field of microbiomics and to prime them for a future in which diagnostic tests and therapies will incorporate these developments into clinical practice.
Keywords: Metagenomics, Microbiomics, Microbiota, Prebiotics, Probiotics
Abbreviations
- 16S rDNA
16S ribosomal subunit gene
- FISH
fluorescence in situ hybridization
- FMT
fecal microbial transplant
- FOS
fructooligosaccharide
- GALT
gut‐associated lymphoid tissue
- IBD
inflammatory bowel disease
- LPS
lipopolysaccharide
- MAMP
microbe‐associated molecular patterns
- NOD
nucleotide‐binding oligomerization domains
- PCR
polymerase chain reaction
- PRR
pattern recognition receptor
- RCDI
recurrent Clostridium difficile infection
- SCFA
short‐chain fatty acids
- SIBO
small intestinal bacterial overgrowth
- TLR
toll‐like receptors
- TRD
tylosin‐responsive diarrhea
- Treg
T regulatory cell
All mammals are inhabited by communities of microorganisms essential to the normal form and function of the host. In terms of cellular composition, genetic diversity, and metabolic capacity, the host animal should be regarded as a multispecies hybrid organism composed of host and microbial cells operating in dynamic and symbiotic equilibrium. The diverse consortium of bacteria, archaea, fungi, protozoa, viruses, and their collective genome found on and within the body comprises the microbiome.1, 2 The microbiome makes vital contributions to energy homeostasis, metabolism, gut epithelial health, immunologic activity, and neurodevelopment.1, 2, 3 The microbiome is dynamic and subject to important changes during the life of the host in response to a variety of factors including diet, environment, medical interventions, and disease states.
Microbiomics is an emerging investigative field seeking to identify the constituents of the microbiome, analyze the microbial genome, characterize the interactions between the microbiome and host, and determine its influence on the pathobiology of disease (Fig 1). The advent of high‐throughput DNA sequencing technologies, coupled to advances in bioinformatics, has revolutionized the field of microbiomics. Commonly, these methods involve amplification and sequencing of targeted microbial DNA regions followed by statistical analysis of microbial identity and diversity based on sequence similarity and comparisons to reference microbial genomic databases. For bacteria, these methods target the 16S ribosome gene (16S rDNA), a conserved bacterial gene region with hypervariable sequences that differ between species. More recently, investigators have employed whole‐genome shotgun sequencing which permits taxonomic analysis, investigation of microbial gene repertoires, and identification of nonbacterial microbes that are excluded from 16S rDNA sequencing. Differences in sample processing techniques, sequencing technologies, and statistical methods complicate any direct comparison between different sequencing studies. However, these methods have supplanted culture‐based approaches, permitting the analysis of increasingly complex characteristics of the microbiome. Most studies of the gastrointestinal microbiome in mammals have focused on the fecal microbiome, although it is unclear how well fecal samples reflect the microbiomes of other intestinal regions. One study in humans demonstrated a strong positive association between fecal and mucosa‐associated microbial communities for some genera (e.g, Bifidobacteria spp.), while other studies have revealed relevant differences between the microbial populations in feces and mucosal biopsy samples.4, 5, 6
Figure 1.
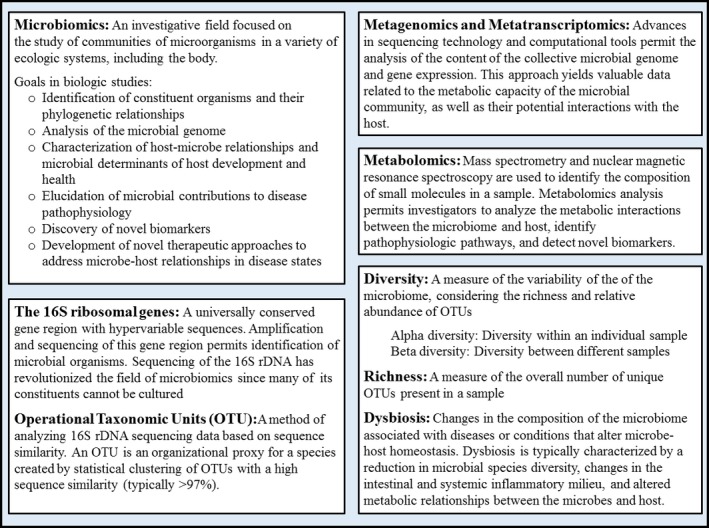
Definitions and methods of analysis of the intestinal microbiome. Terminology and technical aspects related to the analysis of microbiomic data can be a barrier to understanding the available literature. The information here is not intended to be a thorough technical overview of the field. Rather, these definitions are meant to familiarize the reader with the fundamental concepts and terminology present in the literature.
To date, there are limited comprehensive reviews of the intestinal microbiome that integrate recent developments from studies in humans into the existing literature regarding the importance of the intestinal microbiome in dogs and cats. Notably absent in existing reviews is a discussion of the acquisition of the neonatal microbiome and its impact on host health and development. Given the importance of the microbiome in normal host physiology and its role in association with a variety of diseases, a thorough review of this rapidly evolving topic is warranted. This review is intended to update the veterinary community on recent advances in knowledge from studies in humans and other mammals, and to summarize the current state of microbiomics in small animal medicine, with an emphasis on the bacterial inhabitants of the gastrointestinal tract.
Assembly and Development of the Intestinal Microbiome
Before birth, all mammals were thought to be sterile with inoculation with microbes occurring at the time of birth. There is some evidence supporting the vertical transfer of microbes before birth, but these findings are controversial and more research will be required to verify these claims.7, 8, 9 Most studies in human neonates suggest that the early colonizers of the infant intestine are acquired through contact with maternal and environmental microbes.10, 11 This concept is supported by studies demonstrating an association between the composition of the infant intestinal microbiome and the route of delivery in humans. Infants delivered vaginally harbor microbial communities, dominated by Lactobacillus spp. and Bifidobacterium spp., which are similar to their mother's vaginal canal.11, 12 Conversely, infants delivered by cesarean section are colonized by microbial communities composed of common skin microbes with Staphylococcus as the predominant genus.11 These studies suggest that neonatal acquisition of the intestinal microbiome is dependent on the organisms encountered in the first hours and days of life from the mother and environment. These early colonizers are enriched for genes governing the metabolism of sugars found in milk as well as those involved in the de novo synthesis of folate, representing critical metabolic functions in the developing host gut.13, 14 Partial restoration of the microbiome was accomplished in a pilot study of newborns delivered via C section that were exposed to the maternal vaginal fluids at birth.15 The gut, oral, and skin microbiomes in these infants were enriched with bacterial communities that resembled vaginally delivered babies.15 These, and other studies in humans, show that the early stages of colonization are characterized by profound interindividual and temporal variation and limited intra‐individual taxonomic diversity in the composition of the intestinal microbiome.10, 14, 16, 17, 18
In the third trimester of pregnancy, the human maternal intestinal microbiome undergoes a profound shift resulting in decreased compositional diversity with associated systemic inflammatory and metabolic consequences resembling metabolic syndrome.19 Interestingly, neonates do not tend to inherit these microbial signatures from their mothers. Instead, the neonatal gut microbiome more closely resembles that of its mother in the first trimester. This is evidence for the presence of a selective environment in the neonatal gut, permitting colonization by certain organisms while inhibiting others. In mice and humans, the formation of a microbial‐mucosal symbiotic unit is guided by complex interactions between bacterial and host immunologic signaling pathways that result in immunologic tolerance and niche colonization.20, 21 One study in human infants found that the mode of birth, antibiotics, and diet influenced the pace of microbial colonization, but not the pattern of maturation.22 These studies provide evidence that microbial colonization of the neonatal gut is initially influenced by the source and quality of the postparturient inoculum, but then follows a developmental maturation process guided by complex, reciprocal interactions between the host and microbiome.
Once the intestinal microbiome is established, an orderly developmental program unfolds during which its composition and functional capacity converge toward a mature configuration.10, 13, 18, 23, 24 This process is punctuated by dramatic shifts that correlate with events and circumstances in early childhood, especially the introduction of solid food during weaning.10, 13, 22, 25 These factors effect a transition whereby the gut microbiome evolves the ability to scavenge energy from complex carbohydrates, metabolize xenobiotics, and participate in vitamin biosynthesis (cobalamin, biotin, thiamine, among others); all features of a mature microbiome.26, 27, 28 Interestingly, bacterial communities involved with plant polysaccharide metabolism might be present before the introduction of solid food, preparing the infant gut for plant‐derived nutrients.14, 18, 29 This finding emphasizes the impact of the microbiome on the developmental trajectory of the host. The end state of this process is characterized by high interpersonal variation and is influenced by diet, geography, and culture. However, the developmental succession of the microbiome toward increasing compositional and functional diversity and complexity is conserved across diet and geography.13
In addition to the microbiome's role in the adaptation to complex nutrients, it is also involved in a variety of other critical developmental processes. Recent studies in laboratory animals point toward a window in which the intestinal microbiome directs the development of the immune system, gut epithelium, and brain, among other body systems.30, 31, 32, 33, 34 While compositional and functional changes in the intestinal microbiome are associated with normal developmental milestones, negative health events in childhood also have important impacts on the gut microbiome. Maternal health status and milk quality, antibiotic administration, premature birth, and malnutrition are associated with abnormal developmental patterns in the intestinal microbiome.32, 35, 36, 37, 38, 39 Malnutrition for instance has been associated with profound dysbiosis leading to persistent gut microbial immaturity that is refractory to nutritional treatment.38, 39 Additionally, asthma, atopy, childhood obesity, and autism spectrum disorder have all been linked to childhood antibiotic use in humans.32, 40 In 1 study, antibiotic administration before 6 months of age was associated with the development of asthma in children with no family history of asthma.41 Despite the evidence that abnormal development of the intestinal microbiome has long‐term implications on host health, the causal contributions of these abnormalities to disease states have yet to be elucidated.
Little is known about the acquisition and development of the intestinal microbiome in dogs and cats. Knowledge of the topic is limited to a handful of studies in kittens and puppies. To date, only 1 longitudinal study of developing kittens has been published.42 The results of this study confirmed that, as in humans, the early fecal microbiome is characterized by a high degree of interindividual variation and that intra‐individual diversity and compositional stability increase with age. Also similar to humans, the relative abundance of Lactobacillus and Bifidobacterium decreased with age, while Bacteriodes and bacterial genes associated with the ability to metabolize complex carbon sources increased with age. However, there was no major change in taxonomy or bacterial gene repertoires between weeks 30 and 42 in these kittens. Another study found that there was little change in the fecal microbiome of kittens between 8 and 16 weeks of age.43 These results suggest that, as in humans, the developing gut microbiome converges upon a temporally stable configuration as kittens mature. Only 1 study has employed DNA sequencing methods to investigate the gut microbiome in puppies, revealing that temporal instability and substantial interindividual variability are also features of the puppy fecal microbiome.44
Other studies designed to assess the impact of diet have revealed valuable information about the intestinal microbiome in young cats. In 1 such study, the fecal microbiomes of kittens fed high‐ or moderate‐protein diets after weaning were more similar between littermates at 8 weeks of age, but at 12 weeks of age, the effect of kinship was diminished and diet emerged as the primary factor driving interindividual similarities between the cats.45 This is evidence that the composition of the neonatal intestinal microbiome is strongly influenced by environment and kinship, whereas subsequent developmental shifts are driven by diet. These investigators also identified changes in the composition of the gut microbiome after weaning, a finding that is confirmed in several other studies of young cats, which also demonstrate age‐related changes in the microbial gene pool.43, 45, 46, 47 Interestingly, one of these studies found associations between diet‐induced changes in the microbiota and complex metabolic and hormonal effects in the host.46 In these kittens, fecal presence of Lactobacillaceae was negatively correlated with body weight and positively correlated with blood leptin concentrations. Bifidobacteraceae was positively correlated with blood ghrelin and negatively correlated with blood triglycerides. Kittens fed a high‐protein, low‐carbohydrate diet had a lower relative abundance of these organisms. The association between diet‐induced compositional differences in the fecal microbiome and hormones regulating satiety and lipid metabolism is intriguing and is suggestive of a large role for the intestinal microbiome in regulating developmental metabolism. However, the specific macronutrients, micronutrients, or both, responsible for these changes, and the mechanisms driving these host‐microbe interactions are unknown. Collectively, these findings are consistent with data in humans and might indicate a similar age‐related convergence of the developing microbiome toward an increasingly diverse and more stable mature configuration. It is also increasingly clear that the intestinal microbiome is an active participant in host developmental physiology.
Studies in humans and small animals suggest that the patterned maturation of the gut microbiome is a conserved feature of mammalian development. The findings summarized here are suggestive of a critical developmental window, during which long‐term host developmental trajectories are set in association with age, environment, illness, and diet‐related changes in the gastrointestinal microbiome. However, there is a paucity of longitudinal studies of the developing microbiome in small animals and fundamental questions regarding its maturation, and impact on host physiology, remain unanswered. Additional longitudinal studies are needed to characterize the origin, structure, and functional characteristics of the neonatal gut microbiome in veterinary patients. The compositional and temporal patterns by which these features evolve toward a mature configuration are unknown. The long‐term impact of events that alter maturation of the microbiome in small animals during the developmental window is also undescribed and deserves further investigation.
Maintenance and Stability of the Gastrointestinal Microbiome in Dogs and Cats
In contrast to the infant, the mature human intestinal microbiome is characterized by profound compositional and functional diversity, and long‐term temporal stability.48 These essential features of the adult human microbiome appear to be mirrored in adult dogs and cats. A variety of factors influence the gut microbiome's composition as well as its long‐term stability and resilience to change (Fig 2). Several studies describe the gastrointestinal microbiome in healthy, mature dogs and cats. Additionally, there are reports describing the impact of heredity, diet, and medical interventions on its compositional structure, diversity, and functional capacity. The predominant bacterial phyla in the intestinal tracts of adult dogs and cats are Firmicutes, Bacteroidetes, Proteobacteria, Fusobacteria, and Actinobacteria (Fig 3). The relative abundances of these phyla vary between the existing published reports, likely a result of differences in individual host genetics, diet, age, sample collection method and source, DNA extraction techniques, and analytic methodology. As in humans, the majority of studies in dogs reveal that the predominant bacterial phyla in the canine feces are Firmicutes and Bacteroidetes.26, 49, 50, 51, 52 Clostridium is the dominant genus with a relative abundance ranging from 7–22%.51, 52 In contrast with humans and other animals, Fusobacteria appears to be relatively abundant in canine feces, and some studies have found that Fusobacteria was either the most abundant phylum, or that it was codominant with Firmicutes and Bacteroidetes.53, 54, 55 The relevance of the increased proportion of Fusobacteria in dogs relative to humans is unclear, although members of the phylum have been associated with inflammatory bowel disease (IBD) and colorectal cancer in humans.56, 57 Conversely, dogs with IBD tend to have lower relative abundances of Fusobacteria, compared with healthy dogs.58 Although cats are obligate carnivores with no strict requirement for complex carbohydrates, their colonic microbiome is similar in composition to dogs and other mammals. Most studies indicate that Firmicutes are the predominant phylum in cats with a relative abundance ranging from 36 to 92%.52, 59, 60, 61 Within Firmicutes, Clostridiales is the most abundant order and Clostridium the most abundant genus.52, 59, 60 In contrast with dogs, Proteobacteria comprise a substantial proportion of the feline colonic microbiome, with a relative abundance of 7.9–14%.59, 60, 61
Figure 2.
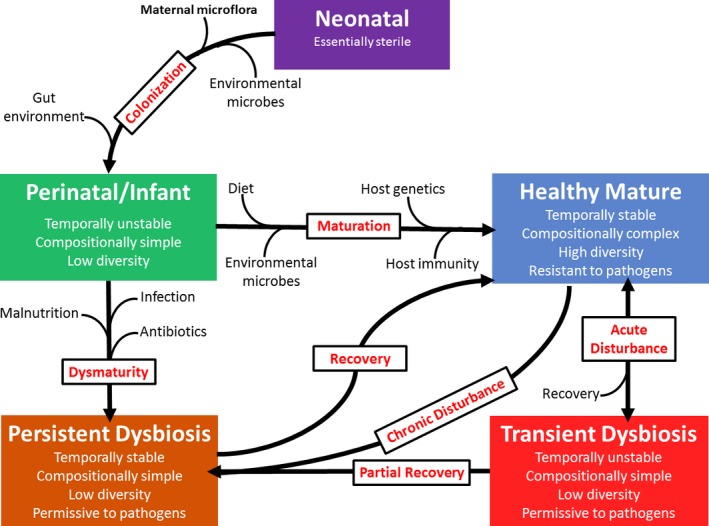
Factors influencing gut microbial development and steady states over time. The adult gastrointestinal microbiome is remarkably stable, although several factors influence gut microbial steady states starting at the time of birth. The infant microbiome is derived from maternal and environmental organisms and develops under selective pressure in the gut. Diet, drug administration, and disease states impact the intestinal microbiome, leading to transient or persistent dysbiosis, depending on the nature of the insult and its duration.
Figure 3.
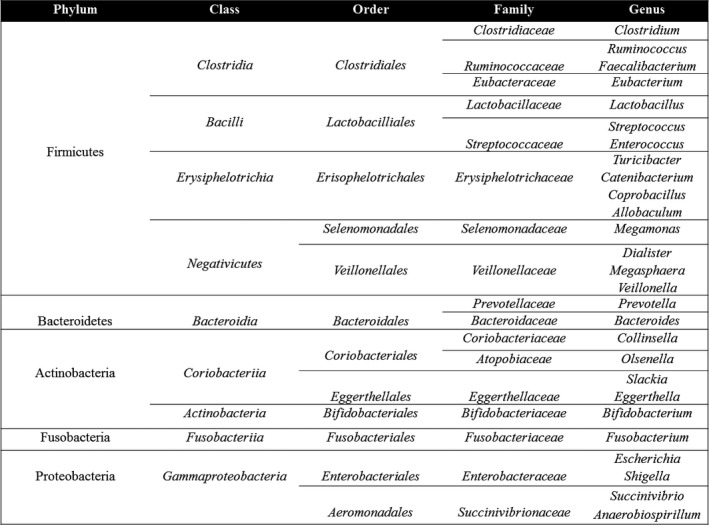
Taxonomy and phylogeny of common constituents of the gastrointestinal microbiome. Microbiomic studies rely on taxonomic classification of bacteria based on DNA sequencing analysis. This literature commonly features results in which organisms with similar names but different phylogenetic levels are listed side‐by‐side. This figure was prepared to provide readers with a reference containing the most common organisms found in the gut, along with their phylogenetic relationships. Taxonomic and phylogenetic information courtesy of the NCBI Taxonomy Browser (https://www.ncbi.nlm.nih.gov).
Although most studies in dogs and cats have focused on the characterization of the fecal microbiome, there are substantial differences in the composition of the microbiome in different segments of the gut in dogs and cats.59, 62 In dogs, Lactobacillus is distributed uniformly throughout the intestinal tract whereas the relative abundances of Enterobacteriales, Fusobacteriales, and Clostridiales varied along its length. Enterobacteriales had a higher relative abundance in the canine small intestine, and Clostridiales was the predominant order in the duodenum and jejunum (40% and 39%, respectively).62 This contrasts with the ileum and colon in which Fusobacteriales and Bacteroidales were the predominant bacterial orders. In cats, Lactobacillales is also distributed along the length of the gut, particularly in the jejunum and colon.59 The feline small intestinal microbiome is predominated by Firmicutes and Bacteroides, whereas Proteobacteria and Actinobacteria are the dominant phyla of the ileum, and the colonic microbiome contains a higher abundance of Firmicutes, Proteobacteria, and Fusobacteria.59
The influence of host genetics on the composition of the gut microbiome is unclear. Some 16S rDNA sequencing studies suggest that the intestinal microbiome of related humans is more similar in composition than that of unrelated individuals and that specific host gene elements are associated with the heritability of the gut microbiome.63, 64, 65 Similarly, the fecal microbiomes of genetically related dogs are more similar to each other than unrelated dogs.55 However, a study of several hundred humans, including mono and dizygotic twins, found that the gut microbiome of related individuals was no more similar than that of unrelated individuals within the same geographic and cultural region.13 This study, and others like it, reveals that humans from different geographic regions can be distinguished from one another based on the composition and genomic features of the intestinal microbiome.13, 48 These studies implicate diet and environment as the primary influences on long‐term steady states of intestinal microbial communities. Indeed, recent studies of humans and other mammalian species found that diet was the primary influence on the structural and functional characteristics of the fecal microbiome.66, 67, 68 Likewise, diet affects both the short‐term composition and long‐term composition in the canine and feline fecal microbiome.26, 43, 46, 49, 50, 54, 61, 69, 70, 71, 72, 73, 74, 75
The convergence of the intestinal microbiome around a stable, mature configuration has led to the emergence of the concept of a phylogenetic “core” microbiome.76 However, recent studies in dogs and humans refute the existence of a “core” microbiome. Metagenomic investigations have revealed that despite phylogenetic diversity, there is substantial interpersonal overlap in functional bacterial gene families.13, 65 These findings suggest that the “core” microbiome is defined by the contents of the collective bacterial genome rather than a conserved phylogeny. Interestingly, 1 study in dogs revealed that the relative abundance of Prevotella and Bacteroides is inversely proportional to that of Fusobacteria, a relationship that is also present in humans.55 Considering the broad functional overlap in microbial gene pools between individuals, the profound compositional variation in the intestinal microbiome of mammals likely precludes the existence of a “core” microbiome centered on a conserved phylogeny. It is more likely that gut microbial niches are occupied not by specific organisms, but by organisms with overlapping functional repertoires and that the “core” microbiome is defined by a relatively constant microbial gene pool despite variation in compositional structure.
Analysis of the metagenome of the canine intestinal microbiome has revealed functional similarity to mice and humans.50 These data show that the predominant bacterial gene categories in the canine gut include carbohydrate metabolism (12–13% of all sequences), protein and amino acid metabolism (8–9 and 7%, respectively), cell wall synthesis (7–8%), vitamin and cofactor synthesis (6%), and nucleic acid synthesis (7%).50 The feline gut microbiome is also involved in carbohydrate and protein metabolism, cell wall biosynthesis, nucleic acid synthesis, and cofactor and vitamin biosynthesis.77 Genes for virulence and antibiotic resistance are also common features of human, canine, and feline microbial gene pools.50, 77, 78 Interestingly, functional gene families do not differ between dogs fed high‐ and low‐fiber diets or between healthy dogs and those with intestinal dysbiosis.50, 79, 80 However, it is likely that current shotgun sequencing and statistical methods are not yet sensitive enough to detect small but potentially important changes in the metagenome under experimental and disease conditions.79 Future studies of the fecal transcriptome by quantitative PCR might permit the detection of biologically relevant changes in microbial gene expression from sequencing methods that are broader in nature.
In addition to diet and environment, medical interventions influence steady states in gut microbial communities. For instance, the human fecal microbiome is profoundly affected by the administration of ciprofloxacin which reduces taxonomic diversity and richness.81, 82 After discontinuation of the medication, the microbiome only partially recovers to its pre‐antibiotic configuration resulting in a new, long‐term compositional steady state that differs from the pre‐antibiotic state. In dogs, macrolide administration induces compositional shifts in the fecal microbiome characterized by a decrease in compositional diversity and richness.83 In this study, the response to tylosin administration varied between individual dogs, although treatment resulted in a persistent reduction in bacterial species diversity and richness in 40% of dogs. In addition to antibiotics, other medications can also affect bacterial populations in the canine gastrointestinal tract. The administration of a proton pump inhibitor (omeprazole) to dogs causes a reduction in Helicobacter and an increase in the proportion of Firmicutes and Fusobacteria in the stomach of dogs.84 Additionally, omeprazole administration was associated with an increase in the duodenal bacteria as well as changes in the fecal microbiome characterized by an increased relative abundance of Lactobacillus and a decrease in proportions of Faecalibacterium and Bacteroides.84
The studies reviewed here suggest that the adult intestinal microbiome of humans, dogs, and cats is characterized by long‐term stability and profound compositional and functional diversity. It is also clear that vital host physiologic processes are governed by interactions between the host and resident microbiome and that conditions which affect the structure of the microbiome may impact host health. The long‐term stability and susceptibility to change during pathologic and aberrant metabolic states make the gut microbiome a viable diagnostic and therapeutic target. The following sections of this review will explore host‐microbiome relationships in more detail, while reviewing diseases associated with alterations in the intestinal microbiome.
Microbe‐Host Interactions and Associations between Dysbiosis and Disease States
The ancient maxim, “All disease begins in the gut,” apparently stated by Hippocrates in the 3rd Century B.C.E, has, in principle, held up to modern scientific scrutiny. The microbiome participates in vital physiologic and immunologic processes including energy homeostasis and metabolism, the synthesis of vitamins and other nutrients, endocrine signaling, prevention of enteropathogen colonization, regulation of immune function, and metabolism of xenobiotic compounds.27, 28 Indeed, many gastrointestinal and systemic diseases have been associated with aberrant gut microbial communities. It is unclear whether the microbiome participates directly in the pathogenesis of these disease states. However, a growing body of evidence implicates the microbiome directly in pathogeneses of disease via complex interactions between the microbiome and host metabolic and immune systems. Here, we will review the current state of knowledge regarding the role of the intestinal microbiome in conditions affecting gastrointestinal and systemic health, with an emphasis on obesity and chronic enteropathies.
Interactions between the microbiome and host depend on local crosstalk networks with each intestinal niche harboring its own microbial community, adapted and responsive to the local environment. These communities differ along the length and width of the GI tract as well as within the different layers of mucus. The ileum and colon are much more active immunologically than the proximal small intestine.85 The architecture of the host‐cell‐microbe interaction is extremely complex and the interacting cells differ considerably. The microbiome in small intestinal crypts, for instance, regulates enterocyte proliferation by influencing DNA replication and gene expression, while the microbiome at the tips of the villi regulate the expression of genes involved in metabolic and immune function.34 Enterocytes and Paneth cells in turn regulate luminal bacteria through the production of mucus and antibacterial factors.86
The maintenance of mucosal immunologic homeostasis is an enormous task demanding discrimination between billions of harmless microbes and a rare, pathogenic invader. Both innate and adaptive immune responses function to prevent pathogen colonization and direct local and systemic inflammatory responses in the face of continuous exposure to foreign microbial and dietary antigens. These defense mechanisms consist of redundant systems to guard against rogue pathogens. The gut‐associated lymphoid tissue (GALT) provides the bulk of immune surveillance and defense. Lymphoid follicles containing T cells, B cells, and dendritic cells are poised to initiate either an inflammatory or an anti‐inflammatory response depending on the specific microbial signals. Specialized epithelial cells (M cells) sample luminal antigens and deliver them to dendritic cells for assessment.87 Innate lymphoid cells might also be important in the maintenance of intestinal barrier integrity and the development of tolerance to commensals.88 Additionally, microbes are essential in the development of GALT as evidenced by severely underdeveloped lymphoid follicles, Peyer's patches, and mesenteric lymph nodes in germ‐free animals.89 Colonization of germ‐free mice with segmented filamentous bacteria stimulates development of the host's immune system; these mice have relatively normal numbers of Th1, Th17, and Treg cells compared with conventional mice.90
Recognition of microbes begins with the 2 main pattern recognition receptor systems (PRRs): Toll‐like receptors (TLRs) and nucleotide‐binding oligomerization domain molecules (NODs).91 These are widely expressed in and on intestinal epithelial cells, as well as macrophages, and dendritic cells in the intestine. These PRRs recognize molecular patterns, called microbe‐associated molecular patterns (MAMPs), on pathogens and commensals.91 Once a microbe has been recognized, internalized, or has invaded the epithelial layer, it initiates an immunologic response appropriate for the microbe (Fig 4). Microbes can exert both pathogenic and protective effects depending on specific microbial signaling through PRRs and MAMPs and the subsequent immune response. Protective effects are mediated by the downregulation of pro‐inflammatory cytokines (IL‐8, IL‐12, IL‐23) and upregulation of anti‐inflammatory cytokines such as IL‐10 produced by T regulatory (Treg) cells.92, 93, 94 In the case of commensals, the dendritic cells present the antigen to naive T cells, which differentiate into Treg cells. Secretion of anti‐inflammatory cytokines ensues, propagating systemic and local tolerance.95, 96 Commensal organisms also decrease the migration of phagocytes, which traffic microbial antigens to local lymphoid tissues and promote B‐ and T‐cell activation.97 Additionally, goblet cell differentiation and production of the protective mucosal mucus layer are stimulated by commensal organisms.98 Conversely, pathogenic bacteria cause dendritic cells to secrete pro‐inflammatory cytokines that cause naïve T cells to differentiate into Th1 and Th17 cells leading to pro‐inflammatory immune responses.95, 96, 97 Additionally, TLRs 4 and 5, which are normally present on the basolateral side of epithelial cells, are expressed on the apical side in states of chronic inflammation, promoting a pro‐inflammatory steady state.22 Different gram‐negative bacteria have LPS modifications that vary in their potential to stimulate TLR so that not all bacteria will induce the same immunologic response.99
Figure 4.
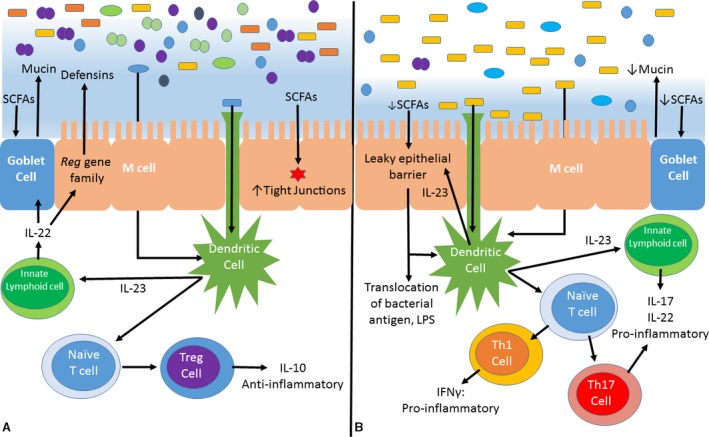
Balance between anti and pro‐inflammatory states in the intestinal mucosa. Mucosal‐microbial homeostasis is a complex and rapidly evolving subject. This simplified diagram is intended to provide examples of some of the most well‐described immunologic interactions between the microbiota and host mucosa. This image represents a schematic cross section of the gut epithelium, mucus layer, and lumen. The colorful rods, ovals, and circles at the top of the image represent luminal microbiota and the blue haze in which some reside is the mucus layer. A. In healthy individuals, pro‐ and anti‐inflammatory signals are balanced such that commensal organisms are recognized and tolerated, while pathogens are prevented from penetrating the mucus layer and underlying epithelium. SCFAs strengthen epithelial tight junctions and stimulate the production of the mucus layer. Commensal organisms are recognized by dendritic cells and specialized epithelial M cells, promoting the maturation of naïve T‐lymphocytes into Treg cells that secrete anti‐inflammatory and immunomodulatory cytokines. Innate lymphocytes stimulate the overlying epithelial cells to secrete antimicrobial defensins. B. States of dysbiosis are characterized by reduced diversity in the resident microbiota depicted here as an increased relative abundance of yellow rods. Recognition of a shifting antigenic milieu by dendritic cells and M cells results in the maturation of naïve T cells into Th1 and Th17 cells, both of which secrete pro‐inflammatory cytokines. Additionally, altered microbial metabolism (e.g, reduced SCFA production) promotes degradation of host protective factors, such as the mucus layer, that stabilize gut microbial communities and prevent colonization by pathogens.
While there is an intuitive link between the microbiome and local gut inflammation, recent metabolomic studies reveal that the microbiome is also a crucial participant in host systemic metabolism. Metabolomics is the study of small molecules present in biologic specimens whereas the metabolome is the collective pool of metabolites in an organism or individual sample. A recent study in mice reported significant differences in the plasma metabolomes of germ‐free and conventional mice, concluding that the microbiome interacted with approximately 10% of host metabolic pathways and that numerous plasma metabolites are derived exclusively from the microbiome.100 In 1 study, dogs with acute diarrhea had concurrent fecal dysbiosis and an altered systemic metabolic state, providing additional evidence for the metabolic links between microbe and host.80 Another study revealed changes in the fecal microbiomes and serum metabolomes of dogs with IBD compared to healthy controls.101 Interestingly, the IBD dogs improved clinically after treatment but this was not accompanied by changes in the microbiome or serum metabolite profiles.
In addition to their contribution to systemic metabolic states, microbial metabolites also modulate mucosal immune responses. Microbial fermentation of complex carbohydrates (e.g, resistant starch, inulin) produces short‐chain fatty acids (SCFA), including acetate, propionate, and butyrate. SCFAs are an essential energy source for colonocytes, maintain the epithelial barrier by strengthening tight junctions, help to regulate intestinal motility, and stimulate the production of anti‐inflammatory compounds.39 SCFAs also bind a G protein receptor on neutrophils to decrease migration, leading to downregulation of inflammation and have been shown to limit colitis in experimental animal models.39, 102, 103, 104 Alternatively, diminished SCFA production, the elaboration of toxic microbial metabolites, interruption in mucosal barrier function, and microbe‐mediated host immune dysregulation perpetuate the conditions which promote the establishment of a new long‐term, pro‐inflammatory steady state.94, 105 One study of dogs with acute diarrhea identified reductions in fecal propionate that was correlated with a decrease in fecal Faecalibacterium.80 Other classes of metabolites, such as bile acids have immunomodulatory effects.106, 107, 108 Primary bile acids are metabolized by gut microbes, generating secondary bile acids. Secondary bile acids bind and activate several host receptors to a greater extent than the primary bile acids of the host.109
Diversity of the intestinal microbiome is emerging as a critical determinant of host health, and a loss of diversity has been associated with a variety of gastrointestinal and systemic diseases in humans and other mammals.110, 111, 112, 113, 114, 115 Intestinal dysbiosis is a loosely defined concept referring to any change in the intestinal microbiome that adversely affects the health of the host organism. Dysbiosis is characterized by broad shifts in community microbial compositional structures, reduced species diversity, and changes in the relative proportion of particular organisms, whereby symbionts normally representing a small proportion of the microbiome develop pathogenic features.94, 113 Although the nature of dysbiosis varies according to the individual, as well as the pathologic condition, reductions in the relative proportion of obligate anaerobes and increases in facultative anaerobes including pathogens such as E. coli, Salmonella, Proteus, Klebsiella, and Shigella are common features of dysbiosis in humans and laboratory animals.105 Dysbiosis does not always involve pathogens as absence of important commensals can be detrimental without the presence of pathogens.
Dysbiosis and mucosal inflammation are interrelated and a plethora of data points to inflammation as either a cause of dysbiosis, a consequence of it, or some combination of the 2. Research in murine and rat models of IBD suggests the microbiome is essential to the initiation and progression of mucosal inflammation, which is absent or attenuated in these animals when raised in germ‐free conditions.116, 117, 118, 119, 120 In humans, many of the genes that are associated with IBD are involved in the regulation of host‐microbe interactions.121 For instance, variation in genes (NOD2 and ATG16L1) involved with the intracellular processing of bacterial components has been associated with Crohn's disease in humans.121 Additionally, research in animal models has demonstrated that transfer of the microbiome from animals with IBD to healthy animals increased the susceptibility of the healthy animal to the development of IBD.16, 122 Dysbiosis is a consistent feature of IBD in humans and, while the clinical characteristics of IBD differ between humans and animals, researchers have identified states of dysbiosis in dogs and cats with IBD and other gastrointestinal disorders.58, 74, 123
Dysbiosis in dogs is also characterized by decreased diversity and a reduction in the relative proportion of species that are known to produce SCFAs.58, 74, 113 Historically, the term small intestinal bacterial overgrowth (SIBO) had been used to describe quantitative and qualitative changes in the intestinal microbiome, based on cultures of duodenal and jejunal juice, associated with exocrine pancreatic insufficiency and antibiotic‐responsive enteropathies in dogs.124 More recently, the term dysbiosis has been used to describe alterations in the gut microbiome associated with disease states or other conditions (e.g., antibiotic administration) in order to reflect the more discriminating findings of modern genomic studies. Several microbial community compositional abnormalities have been identified in dogs with various chronic and acute enteropathies.72, 74, 125, 126, 127 It is clear from these data that intestinal dysbiosis is a common feature of a variety of enteropathies in dogs. The number of studies is small and, as in humans, no universal pattern of dysbiosis has emerged from the data. A majority of sequencing‐based studies in dogs with dysbiosis have identified a lower relative abundance of Bacteroidetes in addition to increased proportions of Clostridia and Proteobacteria, as well as expansion of other organisms that are minor constituents of the microbiome of healthy animals.58, 101 There were substantial differences between samples from dogs with IBD and healthy control dogs, and dogs with IBD had significantly lower alpha diversity compared with healthy controls. However, there was no correlation with diet, body condition, or several other factors with the microbiome analysis.58 A dysbiosis index created by these authors was negatively correlated with alpha diversity in this study.58 Recently, a fecal qPCR panel provided rapid diagnosis of dysbiosis based on the dysbiosis index score.a In this study, a negative dysbiosis index score indicated eubiosis and as the value became greater, it indicated a stronger deviation from eubiosis such that a value of 0 had 84% sensitivity and 86% specificity for differentiation of dysbiosis from eubiosis. To date, no 16S rDNA or shotgun sequencing studies have been published in cats with intestinal dysbiosis; however, 2 studies employing fluorescence in situ hybridization (FISH) suggest that cats with chronic enteropathies have significant dysbiosis compared with healthy cats. One study identified decreased Bifidobacterium spp and Bacteroides spp. as well as increased Desulfovibrio, a producer of toxic sulfur‐containing compounds, in the feces of cats with IBD.123 Another FISH study identified increased mucosa‐associated Enterobacteriaceae associated with inflammatory infiltrates in cats with IBD.128 The clinical importance of these findings is unclear, particularly when attempting to compare small intestinal biopsies and fecal analysis. Future research efforts should be aimed at elucidating the pathophysiologic mechanisms underlying intestinal dysbiosis in veterinary patients with enteropathies.
There is particularly strong evidence for an etiologic link between obesity and metabolic syndrome and certain forms of intestinal dysbiosis. Obesity is a major problem in human and veterinary medicine, and diagnostic and therapeutic tools to prevent and treat obesity are in critical demand. Recent studies have revealed compelling associations between obesity and intestinal microbiome in humans, laboratory animals, and pets. Proposed mechanisms for these associations include increased dietary energy harvest, microbe‐induced changes in host glucose and lipid metabolism, microbial signaling through host endocrine systems, and chronic low‐grade inflammation leading to insulin resistance.129 A recent metagenomics study found that the fecal microbiomes of obese humans are enriched for genes that govern the harvesting of dietary energy though the production of SCFAs.110 SCFAs are a direct energy source but are also effective signaling molecules, acting through receptors (GPR41) in enteroendocrine cells to decrease gut transit time, increase SCFA recovery, and promote adiposity.130 One study conducted in germ‐free mice inoculated with cecal contents from conventional mice suggests a direct, causal link between the intestinal microbiome and increased adiposity.131 Weight gain in these mice occurred despite calorie restriction via increased intestinal monosaccharide absorption and increased hepatic lipogenesis. Furthermore, the microbiome in these mice suppressed a host gene (Fiaf) coding a circulating lipoprotein lipase inhibitor (Angptl4), causing an increase in triglyceride deposition in adipose tissue.132 Lipopolysaccharide (LPS), a component of gram‐negative bacterial cell walls, has been associated with low‐grade systemic inflammation in mouse models of obesity. LPS levels are also positively correlated with total energy intake and are higher in patients with type 2 diabetes.129 High‐fat diets induce a reduction in the relative proportion of Bifidobacteria, resulting in increased intestinal permeability and higher plasma LPS levels.133 Conversely, probiotic supplementation with Bifidobacteria lowers serum LPS levels, improves glucose tolerance, and reduces inflammation.129
Dysbiosis characterized by an increased Firmicutes:Bacteroidetes ratio has been identified in obese mice compared with lean mice as well as in high‐fat diet‐induced obese mice.65, 110, 111 Recent 16S rDNA sequencing studies of fecal samples from obese humans, including 1 study of 154 mothers and twins with obese and lean phenotypes, reveal that human obesity is also associated with decreased diversity and lower proportions of fecal Bacteroidetes and that weight loss is associated with a proportional increase in Bacteroidetes.110, 111, 134, 135 Similarly, obese cats have lower proportions of Bacteroidetes than healthy cats, a feature of moderately and severely obese cats, suggesting that changes in the microbiome occur before clinically relevant obesity is apparent.136 A single study comparing lean and obese pet dogs did not identify large shifts in the microbiome associated with obesity.127 However, this study did identify changes in the fecal microbiome under different feeding conditions in a research colony of beagles. In these dogs, Clostridiales increased with ad lib feeding while Gammaproteobacteria and Alphaproteobacteria decreased in dogs fed a calorie‐restricted diet. These results should be interpreted with caution given the influence of the different environmental factors on the microbiomes of dogs from research colonies compared with pets. Additional studies will be required to clarify the role of the microbiome in obesity in dogs and cats.
In humans and laboratory animals, a diverse array of other systemic diseases including immune‐mediated conditions (rheumatoid arthritis, atopy, and asthma) and neurodevelopmental disorders (autism spectrum disorder) have been linked to intestinal dysbiosis.32, 33 These associations have not been identified in small animals and are beyond the scope of this review. Despite the paucity of data, and lack of clarity regarding the microbiome's role in the pathogenesis of the conditions reviewed here, there are clear links between the intestinal microbiome and systemic health. Therefore, the intestinal microbiome should be considered a viable diagnostic and therapeutic target in future studies designed to assess the clinical impact of these findings.
Therapeutic Manipulation of the Gastrointestinal Microbiome
Therapeutic manipulation of the intestinal microbiome is generally achieved through modification of the diet, administration of prebiotics, probiotics, or antibiotics, and more recently, fecal microbiome transplantation (FMT).b , c , d The therapeutic targets of these interventions and their efficacy are not well established and have only recently been described in the literature. These therapies are meant to shift microbial community steady states associated with dysbiosis to those associated with health. Several studies have reported amelioration of clinical signs after treatment with various microbial concoctions, as will be discussed in this section. Interpretation of these findings is challenging as changes in the microbiome might not necessarily translate into clinical improvements, clinical improvements can occur without detectable changes in the microbiome, and clinical signs might be unaffected, or transiently affected by alterations in the microbiome.
Prebiotics
Although fewer studies exist, the potential benefits of prebiotic supplementation, especially by the addition of plant‐derived polysaccharides, is a mainstay of treatment directed at modification of the intestinal microbiome. Referred to as prebiotics, these include inulin, nonstarch polysaccharides found in some cereal grains and seaweed or algae, disaccharides (lactulose), and polysaccharides including fructooligosaccharides (FOS) and galactooligosaccharides. Prebiotics are fermented by colonic bacteria, generating end‐products such as SCFAs that provide essential nutrients for the enteric epithelium. They also induce anti‐inflammatory Tregs, and lower luminal pH.137, 138, 139 Induction of Tregs maintains an anti‐inflammatory milieu and the acidic pH prevents specific pathogens from gaining a foothold. Prebiotics are also speculated to protect the gut epithelium by increasing the mucus layer, elongating the microvilli, increasing numbers of epithelial cells, and by preventing adherence of pathogenic strains to the epithelial cells.140, 141 Other SCFAs such as propionate appear to induce de novo generation of Tregs in the peripheral immune system.142 A newly published study in mice revealed that when deprived of fiber, commensal bacteria will degrade the protective mucosal mucus layer, permitting invasion by commensals and pathogens alike.143
Several studies have evaluated the effects of prebiotics in dogs. In 1 study, chicory root, a source of inulin, improved fecal scores, increased Bifidobacteria, and decreased C. perfringens in the feces of healthy dogs.144 Meta‐analysis of 15 studies including 65 different treatment conditions showed that fecal SCFAs concentrations increase linearly with prebiotic dose.145 This analysis also revealed that fecal Bifidobacteria and Lactobacillus increase with prebiotic dose, although no changes were observed for pathogenic C. perfingens or E. coli. The impacts of prebiotics were not related to the composition of the dog's diet, suggesting that prebiotic therapies can provide benefits independent of diet. The potential benefits of prebiotic supplementation are also evident in cats. In 1 study, pectin administration increased levels of Lactobacilli and fecal SCFA concentrations, while supplementation with FOS led to increased Bifidobacteria and decreased E. coli as well as increased concentrations of fecal butyrate.146
Probiotics
Probiotics are formulations of live organisms that confer beneficial effects on the recipient when delivered in adequate amounts. Proposed mechanisms through which probiotics improve host health include reducing intestinal permeability by upregulation of tight junction proteins, increasing mucin secretion by goblet cells, increasing secretion of defensins which prevent pathogen colonization, production of SCFAs, stimulation of IgA secretion, decreasing luminal pH, and enhancing and directing immune cells to promote tolerance to commensals while maintaining protection against pathogens.147 Even nonviable organisms might confer health benefits by adhering to the mucus layer and stimulating immune function in dogs.148, 149 Capsular polysaccharide from Bacteroides fragilis, for instance, activates Tregs and decreases severity of colitis in mice.150 The most commonly used probiotic organisms include Lactobacillus and Bifidobacterium. Additional bacteria used include Bacillus and Streptococcus as well as the yeast, Saccharomyces boulardii. It is essential to remember that probiotic organisms are often strain specific.b In order to remain viable, probiotics must survive the acidic environment of the stomach and bile acid to colonize the intestines. One study detected probiotic species in 11 of 12 dogs during probiotic administration but was not able to detect the probiotic species before or after completion of the trial.151 However, probiotics appear to confer benefits without changing the microbiome permanently and transient colonization is still associated with beneficial effects in the host.152
Evidence of a therapeutic effect of probiotics dates to the early 20th century. A study describing improvement in autoimmune arthritis after supplementation with live cultures of Streptococcus lacticus and Bacillus bulgaricus was reported in 1909.153 More recent studies have shown that Bifidobacterium longum subspecies longum (JCM 1217) has been shown to protect against E. coli O157:H7‐induced enteropathogenic infection in mice.154 A large body of research has focused on a probiotic mixture of lyophilized bacteria representing Lactobacillus (L. casei, L. plantarum, L. acidophilus, L. bulgaricus), Bifidobacterium (B. longum, B. breve, B. infantis), and Streptococcus salivaris termed VSL#3. In a meta‐analysis of VSL#3 in humans with ulcerative colitis, the supplement was associated with longer remission times compared with placebos.155 In 29 children with ulcerative colitis, VSL#3 was associated with higher remission rates (92.8%) compared with placebo (36.4%) as well as decreased relapse (21.4%) compared with placebo (73.3%) at 1 year.156 A systematic review of 23 randomized controlled trials (RCTs) in 1917 patients reported that probiotics were associated with shortened duration of acute gastroenteritis compared with placebo in humans.157 Another review of 3 RCTs revealed improvement in clinical signs associated with rotavirus diarrhea in the probiotic group compared with a placebo in infants.158 In yet another meta‐analysis including 2963 patients, the probiotic Lactobacillus rhamnosus was associated with decreased duration of diarrhea compared with a placebo.159 A systematic review of probiotics for prevention of antibiotic‐associated diarrhea reported that probiotics were associated with a reduction in diarrhea in humans receiving antibiotics.160
Probiotic supplementation benefited small animals in several clinical trials. A small clinical trial with a probiotic strain of Saccharomyces boulardii improved clinical signs in dogs with IBD and protein losing enteropathy.a In dogs with food‐responsive diarrhea treated with lyophilized Lactobacillus for 21 days along with diet change, there were increased Lactobacilli and decreased Enterobacteria in the feces accompanied by improved clinical signs.161 In another study of 36 dogs with acute gastroenteritis, a probiotic combination improved clinical signs compared to a placebo.162 Enterococcus faecium SF68 is a lactic acid‐producing strain that has been shown to increase antibody secretion in mice with Giardiasis as well as decrease fecal shedding of Giardia.163 When administered to puppies, E. faecium SF68 was associated with increased serum IgA levels.163 In a shelter‐based study, this probiotic, administered with metronidazole, improved fecal scores compared to dogs treated with metronidazole alone.164 In kittens from a shelter, E. faecium SF68 significantly decreased the incidence of diarrhea.165 Only 9.5% of the probiotic group developed diarrhea compared with 60% of kittens that were not given probiotics. There was also a decrease in fecal C. perfringens and an increase in Bifidobacteria associated with probiotic administration. In another large trial of 217 shelter cats, E. faecium decreased episodes of diarrhea compared with a placebo.166 A multispecies synbiotic containing Enterococcus faecium, Bacillus coagulans, and Lactobacillus acidophilus, along with prebiotics, improved fecal scores and reduced the incidence of diarrhea in healthy Alaskan sled dogs.167 In dogs with IBD, VSL#3 improved clinical signs and increased immune‐histochemical staining for FoxP3+ cells, a marker for Tregs, compared with a control group that was treated with metronidazole and prednisolone.168
Antibiotic Administration
Therapeutic manipulation of the microbiome can also be achieved through the administration of select antibiotics. In several human studies, metronidazole was associated with disease remission in patients with Crohn's disease.169, 170, 171 Metronidazole is a nitroimidazole antibiotic and antiprotozoal that is commonly used in small animals with acute and chronic gastrointestinal disorders. A recent study in healthy dogs revealed that metronidazole decreases fecal bacterial diversity but increases the proportions of putatively beneficial bacteria such as Bifidobacterium.172 Tylosin is a macrolide antibiotic that is typically used to treat chronic enteropathies in dogs, especially a variant of antibiotic‐responsive diarrhea termed tylosin‐responsive diarrhea (TRD). One study evaluating its effect in healthy dogs revealed that it causes a reduction in jejunal bacterial diversity and increased the proportion of putatively beneficial Enterococcus species.83 Interestingly, it also caused an increase in potentially pathogenic bacteria such as C. perfingens, E. coli, and Pasteurella, although no dogs in this study developed any new clinical signs of gastrointestinal disease during treatment. Clinical signs in dogs with TRD resolve within the first 3 days of therapy. Additionally, tylosin administration is associated with increases in Enterococcus species in dogs with TRD.173 Rifaximin, a semisynthetic rifamycin with broad‐spectrum activity which undergoes minimal systemic absorption, was found to significantly improve clinical signs in a cohort of dogs with IBD, suggesting potential manipulation of either pathogenic strains (decreased), beneficial commensals (increased), or a combination of the 2.174, 175
Fecal Microbiome Transplantation (FMT)
Fecal microbiome transplantation entails the transfer of feces from a healthy donor into the intestinal tract of a diseased recipient. The first modern record of FMT, for treatment of humans with pseudomembranous colitis, was reported in 1958 and all 4 patients that were treated survived.176 The route of delivery varies between studies and includes oral capsules, nasogastric, nasoduodenal and colonoscopic administration, and via enema.177 It is unclear whether the benefits of FMT are derived from the transfer of viable commensals, or via the delivery of viruses, proteins, bile acids, vitamins, SCFAs, and the myriad of other substances not yet identified in feces, or some combination of these. The first condition where the FDA has granted investigational new drug status for FMT in the United States was Clostridium difficile infection. Patients with recurrent C. difficile (RCDI) have decreased proportions of fecal Bacteroidetes and Firmicutes, and an ~90% cure rate is expected after FMT.178, 179 In patients with RCDI after FMT, the recipient's fecal microbiome mirrors that of the donor, up to 24 weeks after FMT.180, 181, 182 There are also reports of clinical improvement after FMT in humans with IBD, multiple sclerosis, myoclonus dystonia, and refractory ulcerative colitis.23, 183 A double‐blind, randomized controlled trial of FMT in 18 men with metabolic syndrome used autologous feces as the control and donor feces from lean men for the treatment arm.184 The treatment group had improvement in insulin sensitivity and increases in butyrate‐producing bacteria after 6 weeks of weekly infusion compared with the control group.
Two recent abstracts provide information from case reports of FMT in dogs. One dog with eosinophilic IBD improved substantially after an FMT enema, and remained free of clinical signs for 3 months afterward.c Sequencing of 16S rDNA revealed that 2 days after FMT, the dog's fecal microbiome was more similar to the donor that its own pretreatment sample. Additionally, the post‐FMT fecal sample had higher microbial diversity than the pre‐FMT sample. Another report documented successful FMT in 8 dogs with refractory C. perfingens‐associated diarrhea.d In this report, FMT was also administered by enema and diarrhea resolved in all dogs after treatment. These results, as well as those from studies in humans, indicate that FMT is a highly successful treatment for Clostridium‐associated diarrhea in humans and dogs, although the efficacy in the treatment for other gastrointestinal diseases such as IBD and systemic diseases associated with dysbiosis is unclear. Although reports suggest that FMT is a safe therapeutic approach, some humans with ulcerative colitis developed a fever and increased C‐reactive protein after FMT.185 Given the paucity of data demonstrating its safety and efficacy, indications for its use, and uncertainty in regard to selection of appropriate donors, preparation of the donor fecal samples, and delivery methods, the authors do not recommend FMT except for cases of Clostridium‐associated diarrhea and other chronic enteropathies that are refractory to standard therapies. A recent commentary provides a thorough review of FMT in veterinary medicine.186
Conclusion: Unanswered Questions and Future Directions for Research
The gastrointestinal microbiome is an essential contributor to mammalian health that participates in vital physiologic processes and guides host development. Abnormalities in gut microbial populations have been associated with a variety of gastrointestinal and systemic diseases, making the intestinal microbiome a viable diagnostic and therapeutic target. While recent advances in DNA sequencing and computational technology have revolutionized the field of microbiomics, many fundamental questions remain unanswered. Future directions for research should aim to elucidate the mechanisms underlying interactions between the microbiome and host, describe the process of microbiome maturation during host development and its impact on early‐life and adult health outcomes, clarify its role in the pathogenesis of disease states, and assess the viability of diagnostic tests and therapies designed to assess and treat conditions associated with intestinal dysbiosis.
Supporting information
Table S1. Adapted with permission from P. Deng and K. S. Swanson, British Journal of Nutrition (2015), 113, S6–S17.
Table S2. Adapted with permission from P. Deng and K. S. Swanson, British Journal of Nutrition (2015), 113, S6–S17.
Table S3. Adapted with permission from P. Deng and K. S. Swanson, British Journal of Nutrition (2015), 113, S6–S17.
Acknowledgments
Conflict of Interest Declaration
All of the authors have received research funding from corporations that manufacture prebiotic and probiotic supplements and prescription veterinary diets.
Off‐label Antimicrobial Declaration
Authors declare no off‐label use of antimicrobials.
This review was authored at the University of Illinois College of Veterinary Medicine in Urbana, IL
This review article was not supported by any grants or gifts.
This review was not presented at any scientific or professional meetings.
Footnotes
Mustafa A, Wajid B, Guard M, Minamoto Y, Lidbury J, Steiner J, Serpedin E, Suchodolski J. A Dysbiosis Index to Assess Microbial Changes in fecal samples of dogs with chronic enteropathy. Journal of Veterinary Internal Medicine 2016;17
Bresciani F, D'Angelo S, Fracassi F, Galuppi R, Diana A, Linta N, et al. Efficacy of Saccharomyces boulardii in the treatment of dogs with chronic enteropathies – randomized double‐blind placebo controlled study. European College of Veterinary Internal Medicine – Companion Animal Proceedings. 2014
Weese J.S. Preliminary clinical and microbiome assessment of stool transplantation in the dog and cat. Journal of Veterinary Internal Medicine 2013;27, 604–756
Murphy T., Chaitman J. & Han E. Use of fecal transplant in eight dogs with refractory Clostridium perfringens associated diarrhea. Journal of Veterinary Internal Medicine 2014; 28, 976–1134
References
- 1. Shreiner AB, Kao JY, Young VB. The gut microbiome in health and in disease. Curr Opin Gastroenterol 2015;31:69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Turnbaugh PJ, Ley RE, Hamady M, et al. The human microbiome project. Nature 2007;449:804–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cho I, Blaser MJ. The human microbiome: At the interface of health and disease. Nat Rev Genet 2012;13:260–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ouwehand AC, Salminen S, Arvola T, et al. Microbiota composition of the intestinal mucosa: Association with fecal microbiota? Microbiol Immunol 2004;48:497–500. [DOI] [PubMed] [Google Scholar]
- 5. Green GL, Brostoff J, Hudspith B, et al. Molecular characterization of the bacteria adherent to human colorectal mucosa. J Appl Microbiol 2006;100:460–469. [DOI] [PubMed] [Google Scholar]
- 6. Zoetendal EG, von Wright A, Vilpponen‐Salmela T, et al. Mucosa‐associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol 2002;68:3401–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aagaard K, Ma J, Antony KM, et al. The placenta harbors a unique microbiome. Sci Transl Med 2014;6:237–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wassenaar TM, Panigrahi P. Is a foetus developing in a sterile environment? Lett Appl Microbiol 2014;59:572–579. [DOI] [PubMed] [Google Scholar]
- 9. Ardissone AN, de la Cruz DM, Davis‐Richardson AG, et al. Meconium microbiome analysis identifies bacteria correlated with premature birth. PLoS ONE 2014;9:e90784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Palmer C, Bik EM, DiGiulio DB, et al. Development of the human infant intestinal microbiota. PLoS Biol 2007;5:e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dominguez‐Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A 2010;107:11971–11975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Biasucci G, Rubini M, Riboni S, et al. Mode of delivery affects the bacterial community in the newborn gut. Early Hum Dev 2010;86(Suppl 1):13–15. [DOI] [PubMed] [Google Scholar]
- 13. Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature 2012;486:222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kurokawa K, Itoh T, Kuwahara T, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res 2007;14:169–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dominguez‐Bello MG, De Jesus‐Laboy KM, Shen N, et al. Partial restoration of the microbiota of cesarean‐born infants via vaginal microbial transfer. Nat Med 2016;22:250–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Garrett WS, Gallini CA, Yatsunenko T, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe 2010;8:292–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eggesbo M, Moen B, Peddada S, et al. Development of gut microbiota in infants not exposed to medical interventions. APMIS 2011;119:17–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Koenig JE, Spor A, Scalfone N, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A 2011;108(Suppl 1):4578–4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Koren O, Goodrich JK, Cullender TC, et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell 2012;150:470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Round JL, Lee SM, Li J, et al. The Toll‐like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 2011;332:974–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stockinger S, Hornef MW, Chassin C. Establishment of intestinal homeostasis during the neonatal period. Cell Mol Life Sci 2011;68:3699–3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. La Rosa PS, Warner BB, Zhou Y, et al. Patterned progression of bacterial populations in the premature infant gut. Proc Natl Acad Sci U S A 2014;111:12522–12527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Borody TJ, Warren EF, Leis S, et al. Treatment of ulcerative colitis using fecal bacteriotherapy. J Clin Gastroenterol 2003;37:42–47. [DOI] [PubMed] [Google Scholar]
- 24. Azad MB, Konya T, Maughan H, et al. Gut microbiota of healthy Canadian infants: Profiles by mode of delivery and infant diet at 4 months. CMAJ 2013;185:385–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fallani M, Amarri S, Uusijarvi A, et al. Determinants of the human infant intestinal microbiota after the introduction of first complementary foods in infant samples from five European centres. Microbiology 2011;157:1385–1392. [DOI] [PubMed] [Google Scholar]
- 26. Middelbos IS. Implications of Dietary Fiber and Fermentable Carbohydrates on Gut Health and Intestinal Microbial Ecology of the Dog. Urbana‐Champaign, Illinois: University of Illinois at Urbana‐Champaign; 2008:330. [Google Scholar]
- 27. Nicholson JK, Holmes E, Kinross J, et al. Host‐gut microbiota metabolic interactions. Science 2012;336:1262–1267. [DOI] [PubMed] [Google Scholar]
- 28. Wilson ID, Nicholson JK. The Modulation of Drug Efficacy and Toxicity by the Gut Microbiome In: Kochhar S, Martin FP, ed. Metabonomics and Gut Microbiota in Nutrition and Disease, Molecular and Integrative Toxicology. London: Springer‐Verlag; 2015:323. [Google Scholar]
- 29. Vaishampayan PA, Kuehl JV, Froula JL, et al. Comparative metagenomics and population dynamics of the gut microbiota in mother and infant. Genome Biol Evol 2010;2:53–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Martinez FD. The human microbiome. Early life determinant of health outcomes. Ann Am Thorac Soc 2014;11(Suppl 1):S7–S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sommer F, Backhed F. The gut microbiota–masters of host development and physiology. Nat Rev Microbiol 2013;11:227–238. [DOI] [PubMed] [Google Scholar]
- 32. Arrieta MC, Stiemsma LT, Amenyogbe N, et al. The intestinal microbiome in early life: Health and disease. Front Immunol 2014;5:427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yang I, Corwin EJ, Brennan PA, et al. The infant microbiome: Implications for infant health and neurocognitive development. Nurs Res 2016;65:76–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Olszak T, An D, Zeissig S, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 2012;336:489–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Penders J, Thijs C, Vink C, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 2006;118:511–521. [DOI] [PubMed] [Google Scholar]
- 36. Lemas DJ, Young BE, Baker PR 2nd, et al. Alterations in human milk leptin and insulin are associated with early changes in the infant intestinal microbiome. Am J Clin Nutr 2016;103:1291–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mueller NT, Bakacs E, Combellick J, et al. The infant microbiome development: Mom matters. Trends Mol Med 2015;21:109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Subramanian S, Huq S, Yatsunenko T, et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 2014;510:417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Smith PM, Howitt MR, Panikov N, et al. The microbial metabolites, short‐chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013;341:569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kozyrskyj AL, Bahreinian S, Azad MB. Early life exposures: Impact on asthma and allergic disease. Curr Opin Allergy Clin Immunol 2011;11:400–406. [DOI] [PubMed] [Google Scholar]
- 41. Risnes KR, Belanger K, Murk W, et al. Antibiotic exposure by 6 months and asthma and allergy at 6 years: Findings in a cohort of 1,401 US children. Am J Epidemiol 2011;173:310–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Deusch O, O'Flynn C, Colyer A, et al. A longitudinal study of the feline faecal microbiome identifies changes into early adulthood irrespective of sexual development. PLoS ONE 2015;10:e0144881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Deusch O, O'Flynn C, Colyer A, et al. Deep Illumina‐based shotgun sequencing reveals dietary effects on the structure and function of the fecal microbiome of growing kittens. PLoS ONE 2014;9:e101021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Burton EN, O'Connor E, Ericsson AC, et al. Evaluation of fecal microbiota transfer as treatment for postweaning diarrhea in research‐colony puppies. J Am Assoc Lab Anim Sci 2016;55:582–587. [PMC free article] [PubMed] [Google Scholar]
- 45. Vester BMDBL, Middelbos IS, Apanavicius CJ, et al. Faecal microbial populations of growing kittens fed high‐ or moderate‐ protein diets. Arch Animal Nutrit 2009;63:254–265. [Google Scholar]
- 46. Hooda S, Vester Boler BM, Kerr KR, et al. The gut microbiome of kittens is affected by dietary protein:carbohydrate ratio and associated with blood metabolite and hormone concentrations. Br J Nutr 2013;109:1637–1646. [DOI] [PubMed] [Google Scholar]
- 47. Bermingham EN, Kittelmann S, Young W, et al. Post‐weaning diet affects faecal microbial composition but not selected adipose gene expression in the cat (Felis catus). PLoS ONE 2013;8:e80992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lozupone CA, Stombaugh JI, Gordon JI, et al. Diversity, stability and resilience of the human gut microbiota. Nature 2012;489:220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kerr KR, Forster G, Dowd SE, et al. Effects of dietary cooked navy bean on the fecal microbiome of healthy companion dogs. PLoS ONE 2013;8:e74998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Swanson KS, Dowd SE, Suchodolski JS, et al. Phylogenetic and gene‐centric metagenomics of the canine intestinal microbiome reveals similarities with humans and mice. ISME J 2011;5:639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Garcia‐Mazcorro JF, Dowd SE, Poulsen J, et al. Abundance and short‐term temporal variability of fecal microbiota in healthy dogs. Microbiologyopen 2012;1:340–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Handl S, Dowd SE, Garcia‐Mazcorro JF, et al. Massive parallel 16S rRNA gene pyrosequencing reveals highly diverse fecal bacterial and fungal communities in healthy dogs and cats. FEMS Microbiol Ecol 2011;76:301–310. [DOI] [PubMed] [Google Scholar]
- 53. Middelbos IS, Vester Boler BM, Qu A, et al. Phylogenetic characterization of fecal microbial communities of dogs fed diets with or without supplemental dietary fiber using 454 pyrosequencing. PLoS ONE 2010;5:e9768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Panasevich MR, Kerr KR, Dilger RN, et al. Modulation of the faecal microbiome of healthy adult dogs by inclusion of potato fibre in the diet. Br J Nutr 2015;113:125–133. [DOI] [PubMed] [Google Scholar]
- 55. Hand D, Wallis C, Colyer A, et al. Pyrosequencing the canine faecal microbiota: Breadth and depth of biodiversity. PLoS ONE 2013;8:e53115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gevers D, Kugathasan S, Denson LA, et al. The treatment‐naive microbiome in new‐onset Crohn's disease. Cell Host Microbe 2014;15:382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chen W, Liu F, Ling Z, et al. Human intestinal lumen and mucosa‐associated microbiota in patients with colorectal cancer. PLoS ONE 2012;7:e39743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vazquez‐Baeza Y, Hyde ER, Suchodolski JS, et al. Dog and human inflammatory bowel disease rely on overlapping yet distinct dysbiosis networks. Nat Microbiol 2016;1:16177. [DOI] [PubMed] [Google Scholar]
- 59. Ritchie LE, Steiner JM, Suchodolski JS. Assessment of microbial diversity along the feline intestinal tract using 16S rRNA gene analysis. FEMS Microbiol Ecol 2008;66:590–598. [DOI] [PubMed] [Google Scholar]
- 60. Ritchie LE, Burke KF, Garcia‐Mazcorro JF, et al. Characterization of fecal microbiota in cats using universal 16S rRNA gene and group‐specific primers for Lactobacillus and Bifidobacterium spp. Vet Microbiol 2010;144:140–146. [DOI] [PubMed] [Google Scholar]
- 61. Barry KA, Middelbos IS, Vester Boler BM, et al. Effects of dietary fiber on the feline gastrointestinal metagenome. J Proteome Res 2012;11:5924–5933. [DOI] [PubMed] [Google Scholar]
- 62. Suchodolski JS, Camacho J, Steiner JM. Analysis of bacterial diversity in the canine duodenum, jejunum, ileum, and colon by comparative 16S rRNA gene analysis. FEMS Microbiol Ecol 2008;66:567–578. [DOI] [PubMed] [Google Scholar]
- 63. Goodrich JK, Davenport ER, Beaumont M, et al. Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe 2016;19:731–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Khachatryan ZA, Ktsoyan ZA, Manukyan GP, et al. Predominant role of host genetics in controlling the composition of gut microbiota. PLoS ONE 2008;3:e3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature 2009;457:480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Muegge BD, Kuczynski J, Knights D, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 2011;332:970–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wu GD, Chen J, Hoffmann C, et al. Linking long‐term dietary patterns with gut microbial enterotypes. Science 2011;334:105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. De Filippo C, Cavalieri D, Di Paola M, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A 2010;107:14691–14696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hang I, Rinttila T, Zentek J, et al. Effect of high contents of dietary animal‐derived protein or carbohydrates on canine faecal microbiota. BMC Vet Res 2012;8:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bermingham EN, Young W, Kittelmann S, et al. Dietary format alters fecal bacterial populations in the domestic cat (Felis catus). Microbiologyopen 2013;2:173–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Beloshapka AN, Dowd SE, Suchodolski JS, et al. Fecal microbial communities of healthy adult dogs fed raw meat‐based diets with or without inulin or yeast cell wall extracts as assessed by 454 pyrosequencing. FEMS Microbiol Ecol 2013;84:532–541. [DOI] [PubMed] [Google Scholar]
- 72. Allenspach K, House A, Smith K, et al. Evaluation of mucosal bacteria and histopathology, clinical disease activity and expression of Toll‐like receptors in German shepherd dogs with chronic enteropathies. Vet Microbiol 2010;146:326–335. [DOI] [PubMed] [Google Scholar]
- 73. Suchodolski JS, Markel ME, Garcia‐Mazcorro JF, et al. The fecal microbiome in dogs with acute diarrhea and idiopathic inflammatory bowel disease. PLoS ONE 2012;7:e51907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Suchodolski JS, Xenoulis PG, Paddock CG, et al. Molecular analysis of the bacterial microbiota in duodenal biopsies from dogs with idiopathic inflammatory bowel disease. Vet Microbiol 2010;142:394–400. [DOI] [PubMed] [Google Scholar]
- 75. Williams DA, Batt RM, McLean L. Bacterial overgrowth in the duodenum of dogs with exocrine pancreatic insufficiency. J Am Vet Med Assoc 1987;191:201–206. [PubMed] [Google Scholar]
- 76. Tap J, Mondot S, Levenez F, et al. Towards the human intestinal microbiota phylogenetic core. Environ Microbiol 2009;11:2574–2584. [DOI] [PubMed] [Google Scholar]
- 77. Tun HM, Brar MS, Khin N, et al. Gene‐centric metagenomics analysis of feline intestinal microbiome using 454 junior pyrosequencing. J Microbiol Methods 2012;88:369–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Forslund K, Sunagawa S, Coelho LP, et al. Metagenomic insights into the human gut resistome and the forces that shape it. BioEssays 2014;36:316–329. [DOI] [PubMed] [Google Scholar]
- 79. Guard BC, Suchodolski JS. HORSE SPECIES SYMPOSIUM: Canine intestinal microbiology and metagenomics: From phylogeny to function. J Anim Sci 2016;94:2247–2261. [DOI] [PubMed] [Google Scholar]
- 80. Guard BC, Barr JW, Reddivari L, et al. Characterization of microbial dysbiosis and metabolomic changes in dogs with acute diarrhea. PLoS ONE 2015;10:e0127259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Dethlefsen L, Huse S, Sogin ML, et al. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 2008;6:e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A 2011;108:4554–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Suchodolski JS, Dowd SE, Westermarck E, et al. The effect of the macrolide antibiotic tylosin on microbial diversity in the canine small intestine as demonstrated by massive parallel 16S rRNA gene sequencing. BMC Microbiol 2009;9:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Garcia‐Mazcorro JF, Suchodolski JS, Jones KR, et al. Effect of the proton pump inhibitor omeprazole on the gastrointestinal bacterial microbiota of healthy dogs. FEMS Microbiol Ecol 2012;80:624–636. [DOI] [PubMed] [Google Scholar]
- 85. Sommer F, Backhed F. The gut microbiota engages different signaling pathways to induce Duox2 expression in the ileum and colon epithelium. Mucosal Immunol 2015;8:372–379. [DOI] [PubMed] [Google Scholar]
- 86. Pelaseyed T, Bergström JH, Gustafsson JK, et al. The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol Rev 2014;260:8–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Klose CS, Artis D. Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nat Immunol 2016;17:765–774. [DOI] [PubMed] [Google Scholar]
- 88. Hepworth MR, Monticelli LA, Fung TC, et al. Innate lymphoid cells regulate CD4+ T‐cell responses to intestinal commensal bacteria. Nature 2013;498:113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Stepankova R, Kovaru F, Kruml J. Lymphatic tissue of the intestinal tract of germfree and conventional rabbits. Folia Microbiol (Praha) 1980;25:491–495. [DOI] [PubMed] [Google Scholar]
- 90. Ivanov II, Atarashi K, Manel N, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009;139:485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Moresco EMY, LaVine D, Beutler B. Toll‐like receptors. Curr Biol 2011;21:R488–R493. [DOI] [PubMed] [Google Scholar]
- 92. Underwood MA. Intestinal dysbiosis: Novel mechanisms by which gut microbes trigger and prevent disease. Prev Med 2014;65:133–137. [DOI] [PubMed] [Google Scholar]
- 93. Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti‐inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A 2008;105:16731–16736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Kamada N, Seo SU, Chen GY, et al. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol 2013;13:321–335. [DOI] [PubMed] [Google Scholar]
- 95. Pabst O, Herbrand H, Friedrichsen M, et al. Adaptation of solitary intestinal lymphoid tissue in response to microbiota and chemokine receptor CCR7 signaling. J Immunol 2006;177:6824–6832. [DOI] [PubMed] [Google Scholar]
- 96. Lorenz RG, Chaplin DD, McDonald KG, et al. Isolated lymphoid follicle formation is inducible and dependent upon lymphotoxin‐sufficient B lymphocytes, lymphotoxin beta receptor, and TNF receptor I function. J Immunol 2003;170:5475–5482. [DOI] [PubMed] [Google Scholar]
- 97. Diehl GE, Longman RS, Zhang JX, et al. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX(3)CR1(hi) cells. Nature 2013;494:116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Wrzosek L, Miquel S, Noordine ML, et al. Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol 2013;11:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Pulendran B, Kumar P, Cutler CW, et al. Lipopolysaccharides from distinct pathogens induce different classes of immune responses in vivo. J Immunol 2001;167:5067–5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Wikoff WR, Anfora AT, Liu J, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci 2009;106:3698–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Minamoto Y, Otoni CC, Steelman SM, et al. Alteration of the fecal microbiota and serum metabolite profiles in dogs with idiopathic inflammatory bowel disease. Gut Microbes 2015;6:33–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Maslowski KM, Vieira AT, Ng A, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 2009;461:1282–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Arpaia N, Campbell C, Fan X, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T‐cell generation. Nature 2013;504:451–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Furusawa Y, Obata Y, Fukuda S, et al. Commensal microbe‐derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013;504:446–450. [DOI] [PubMed] [Google Scholar]
- 105. Walker AW, Lawley TD. Therapeutic modulation of intestinal dysbiosis. Pharmacol Res 2013;69:75–86. [DOI] [PubMed] [Google Scholar]
- 106. Ridlon JM, Kang DJ, Hylemon PB, et al. Bile acids and the gut microbiome. Curr Opin Gastroenterol 2014;30:332–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Begley M, Gahan CG, Hill C. The interaction between bacteria and bile. FEMS Microbiol Rev 2005;29:625–651. [DOI] [PubMed] [Google Scholar]
- 108. Islam KB, Fukiya S, Hagio M, et al. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology 2011;141:1773–1781. [DOI] [PubMed] [Google Scholar]
- 109. Kawamata Y, Fujii R, Hosoya M, et al. A G protein‐coupled receptor responsive to bile acids. J Biol Chem 2003;278:9435–9440. [DOI] [PubMed] [Google Scholar]
- 110. Turnbaugh PJ, Ley RE, Mahowald MA, et al. An obesity‐associated gut microbiome with increased capacity for energy harvest. Nature 2006;444:1027–1031. [DOI] [PubMed] [Google Scholar]
- 111. Ley RE, Backhed F, Turnbaugh P, et al. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A 2005;102:11070–11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Ianiro G, Tilg H, Gasbarrini A. Antibiotics as deep modulators of gut microbiota: Between good and evil. Gut 2016;65:1906–1915. [DOI] [PubMed] [Google Scholar]
- 113. Frank DN, St Amand AL, Feldman RA, et al. Molecular‐phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A 2007;104:13780–13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Tong M, Li X, Wegener Parfrey L, et al. A modular organization of the human intestinal mucosal microbiota and its association with inflammatory bowel disease. PLoS ONE 2013;8:e80702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Scher JU, Sczesnak A, Longman RS, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife 2013;2:e01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Schultz M, Tonkonogy SL, Sellon RK, et al. IL‐2‐deficient mice raised under germfree conditions develop delayed mild focal intestinal inflammation. Am J Physiol 1999;276:G1461–G1472. [DOI] [PubMed] [Google Scholar]
- 117. Bamias G, Okazawa A, Rivera‐Nieves J, et al. Commensal bacteria exacerbate intestinal inflammation but are not essential for the development of murine ileitis. J Immunol 2007;178:1809–1818. [DOI] [PubMed] [Google Scholar]
- 118. Sellon RK, Tonkonogy S, Schultz M, et al. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin‐10‐deficient mice. Infect Immun 1998;66:5224–5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Dianda L, Hanby AM, Wright NA, et al. T cell receptor‐alpha beta‐deficient mice fail to develop colitis in the absence of a microbial environment. Am J Pathol 1997;150:91–97. [PMC free article] [PubMed] [Google Scholar]
- 120. Rath HC, Wilson KH, Sartor RB. Differential induction of colitis and gastritis in HLA‐B27 transgenic rats selectively colonized with Bacteroides vulgatus or Escherichia coli. Infect Immun 1999;67:2969–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Cho JH. The genetics and immunopathogenesis of inflammatory bowel disease. Nat Rev Immunol 2008;8:458–466. [DOI] [PubMed] [Google Scholar]
- 122. Elinav E, Strowig T, Kau AL, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 2011;145:745–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Inness VL, McCartney AL, Khoo C, et al. Molecular characterisation of the gut microflora of healthy and inflammatory bowel disease cats using fluorescence in situ hybridisation with special reference to Desulfovibrio spp. J Anim Physiol Anim Nutr (Berl) 2007;91:48–53. [DOI] [PubMed] [Google Scholar]
- 124. Rutgers HC, Batt RM, Elwood CM, et al. Small intestinal bacterial overgrowth in dogs with chronic intestinal disease. J Am Vet Med Assoc 1995;206:187–193. [PubMed] [Google Scholar]
- 125. Chaban B, Links MG, Hill JE. A molecular enrichment strategy based on cpn60 for detection of epsilon‐proteobacteria in the dog fecal microbiome. Microb Ecol 2012;63:348–357. [DOI] [PubMed] [Google Scholar]
- 126. Xenoulis PG, Palculict B, Allenspach K, et al. Molecular‐phylogenetic characterization of microbial communities imbalances in the small intestine of dogs with inflammatory bowel disease. FEMS Microbiol Ecol 2008;66:579–589. [DOI] [PubMed] [Google Scholar]
- 127. Handl S, German AJ, Holden SL, et al. Faecal microbiota in lean and obese dogs. FEMS Microbiol Ecol 2013;84:332–343. [DOI] [PubMed] [Google Scholar]
- 128. Janeczko S, Atwater D, Bogel E, et al. The relationship of mucosal bacteria to duodenal histopathology, cytokine mRNA, and clinical disease activity in cats with inflammatory bowel disease. Vet Microbiol 2008;128:178–193. [DOI] [PubMed] [Google Scholar]
- 129. Ley RE. Obesity and the human microbiome. Curr Opin Gastroenterol 2010;26:5–11. [DOI] [PubMed] [Google Scholar]
- 130. Samuel BS, Shaito A, Motoike T, et al. Effects of the gut microbiota on host adiposity are modulated by the short‐chain fatty‐acid binding G protein‐coupled receptor, Gpr41. Proc Natl Acad Sci U S A 2008;105:16767–16772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Backhed F, Ding H, Wang T, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A 2004;101:15718–15723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Backhed F, Manchester JK, Semenkovich CF, et al. Mechanisms underlying the resistance to diet‐induced obesity in germ‐free mice. Proc Natl Acad Sci U S A 2007;104:979–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Cani PD, Delzenne NM. The role of the gut microbiota in energy metabolism and metabolic disease. Curr Pharm Des 2009;15:1546–1558. [DOI] [PubMed] [Google Scholar]
- 134. Nadal I, Santacruz A, Marcos A, et al. Shifts in clostridia, bacteroides and immunoglobulin‐coating fecal bacteria associated with weight loss in obese adolescents. Int J Obes (Lond) 2009;33:758–767. [DOI] [PubMed] [Google Scholar]
- 135. Santacruz A, Marcos A, Warnberg J, et al. Interplay between weight loss and gut microbiota composition in overweight adolescents. Obesity (Silver Spring) 2009;17:1906–1915. [DOI] [PubMed] [Google Scholar]
- 136. Kieler IN, Molbak L, Hansen LL, et al. Overweight and the feline gut microbiome – a pilot study. J Anim Physiol Anim Nutr (Berl) 2016;100:478–484. [DOI] [PubMed] [Google Scholar]
- 137. Gibson PR, Rosella O, Wilson AJ, et al. Colonic epithelial cell activation and the paradoxical effects of butyrate. Carcinogenesis 1999;20:539–544. [DOI] [PubMed] [Google Scholar]
- 138. Wang HB, Wang PY, Wang X, et al. Butyrate enhances intestinal epithelial barrier function via up‐regulation of tight junction protein Claudin‐1 transcription. Dig Dis Sci 2012;57:3126–3135. [DOI] [PubMed] [Google Scholar]
- 139. Wong JM, de Souza R, Kendall CW, et al. Colonic health: Fermentation and short chain fatty acids. J Clin Gastroenterol 2006;40:235–243. [DOI] [PubMed] [Google Scholar]
- 140. Lennon G, Balfe A, Earley H, et al. Influences of the colonic microbiome on the mucous gel layer in ulcerative colitis. Gut Microbes 2014;5:277–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Mendis M, Leclerc E, Simsek S. Arabinoxylans, gut microbiota and immunity. Carbohydr Polym 2016;139:159–166. [DOI] [PubMed] [Google Scholar]
- 142. Ahmed I, Roy BC, Khan SA, et al. Microbiome, metabolome and inflammatory bowel disease. Microorganisms 2016;4:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Desai MS, Seekatz AM, Koropatkin NM, et al. A dietary fiber‐deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 2016;167:1339–1353 e1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Zentek J, Marquart B, Pietrzak T, et al. Dietary effects on bifidobacteria and Clostridium perfringens in the canine intestinal tract. J Anim Physiol Anim Nutr (Berl) 2003;87:397–407. [DOI] [PubMed] [Google Scholar]
- 145. Patra AK. Responses of feeding prebiotics on nutrient digestibility, faecal microbiota composition and short‐chain fatty acid concentrations in dogs: A meta‐analysis. Animal 2011;5:1743–1750. [DOI] [PubMed] [Google Scholar]
- 146. Barry KA, Wojcicki BJ, Middelbos IS, et al. Dietary cellulose, fructooligosaccharides, and pectin modify fecal protein catabolites and microbial populations in adult cats. J Anim Sci 2010;88:2978–2987. [DOI] [PubMed] [Google Scholar]
- 147. Gallo A, Passaro G, Gasbarrini A, et al. Modulation of microbiota as treatment for intestinal inflammatory disorders: An uptodate. World J Gastroenterol 2016;22:7186–7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Kanasugi H, Hasegawa T, Goto Y, et al. Single administration of enterococcal preparation (FK‐23) augments non‐specific immune responses in healthy dogs. Int J Immunopharmacol 1997;19:655–659. [DOI] [PubMed] [Google Scholar]
- 149. Grzeskowiak L, Endo A, Beasley S, et al. Microbiota and probiotics in canine and feline welfare. Anaerobe 2015;34:14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Shen Y, Giardino Torchia ML, Lawson GW, et al. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe 2012;12:509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Garcia‐Mazcorro JF, Lanerie DJ, Dowd SE, et al. Effect of a multi‐species synbiotic formulation on fecal bacterial microbiota of healthy cats and dogs as evaluated by pyrosequencing. FEMS Microbiol Ecol 2011;78:542–554. [DOI] [PubMed] [Google Scholar]
- 152. Ceapa C, Wopereis H, Rezaiki L, et al. Influence of fermented milk products, prebiotics and probiotics on microbiota composition and health. Best Pract Res Clin Gastroenterol 2013;27:139–155. [DOI] [PubMed] [Google Scholar]
- 153. Warden CC. The toxemic factor in rheumatoid arthritis. Cal State J Med 1909;7:299–301. [PMC free article] [PubMed] [Google Scholar]
- 154. Fukuda S, Toh H, Hase K, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 2011;469:543–547. [DOI] [PubMed] [Google Scholar]
- 155. Mardini HE, Grigorian AY. Probiotic mix VSL#3 is effective adjunctive therapy for mild to moderately active ulcerative colitis: A meta‐analysis. Inflamm Bowel Dis 2014;20:1562–1567. [DOI] [PubMed] [Google Scholar]
- 156. Miele E, Pascarella F, Giannetti E, et al. Effect of a probiotic preparation (VSL#3) on induction and maintenance of remission in children with ulcerative colitis. Am J Gastroenterol 2009;104:437–443. [DOI] [PubMed] [Google Scholar]
- 157. Allen SJ, Okoko B, Martinez E, et al. Probiotics for treating infectious diarrhoea. Cochrane Database Syst Rev 2004;2:CD003048. [DOI] [PubMed] [Google Scholar]
- 158. Szajewska H, Wanke M, Patro B. Meta‐analysis: The effects of Lactobacillus rhamnosus GG supplementation for the prevention of healthcare‐associated diarrhoea in children. Aliment Pharmacol Ther 2011;34:1079–1087. [DOI] [PubMed] [Google Scholar]
- 159. Szajewska H, Skorka A, Ruszczynski M, et al. Meta‐analysis: Lactobacillus GG for treating acute gastroenteritis in children–updated analysis of randomised controlled trials. Aliment Pharmacol Ther 2013;38:467–476. [DOI] [PubMed] [Google Scholar]
- 160. Hempel S, Newberry SJ, Maher AR, et al. Probiotics for the prevention and treatment of antibiotic‐associated diarrhea: A systematic review and meta‐analysis. JAMA 2012;307:1959–1969. [DOI] [PubMed] [Google Scholar]
- 161. Sauter SN, Benyacoub J, Allenspach K, et al. Effects of probiotic bacteria in dogs with food responsive diarrhoea treated with an elimination diet. J Anim Physiol Anim Nutr (Berl) 2006;90:269–277. [DOI] [PubMed] [Google Scholar]
- 162. Herstad HK, Nesheim BB, L'Abee‐Lund T, et al. Effects of a probiotic intervention in acute canine gastroenteritis–a controlled clinical trial. J Small Anim Pract 2010;51:34–38. [DOI] [PubMed] [Google Scholar]
- 163. Benyacoub J, Perez PF, Rochat F, et al. Enterococcus faecium SF68 enhances the immune response to Giardia intestinalis in mice. J Nutr 2005;135:1171–1176. [DOI] [PubMed] [Google Scholar]
- 164. Fenimore A, Groshong L, Scorza V, Lappin MR. Evaluation of enterococcus faecium SF68 supplementation with metronidazole for the treatment of non‐specific diarrhea in dogs houses in animal shelters. J Vet Intern Med 2012;26:793. [Google Scholar]
- 165. Czarnecki‐Maulden GCC, Lawler D, Benyacoub J. Incidence of naturally occurring diarrhea in kittens fed enterococcus faecium SF68. Compend Contin Educ Vet 2007;29:37. [Google Scholar]
- 166. Bybee SNSAV, Lappin MR. Effect of the probiotic enterococcus faecium SF68 on the presences of diarrhea in cats and dogs housed in an animal shelter. J Vet Intern Med 2011;25:856–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Gagne JW, Wakshlag JJ, Simpson KW, et al. Effects of a synbiotic on fecal quality, short‐chain fatty acid concentrations, and the microbiome of healthy sled dogs. BMC Vet Res 2013;9:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Rossi G, Pengo G, Caldin M, et al. Comparison of microbiological, histological, and immunomodulatory parameters in response to treatment with either combination therapy with prednisone and metronidazole or probiotic VSL#3 strains in dogs with idiopathic inflammatory bowel disease. PLoS ONE 2014;9:e94699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Blichfeldt P, Blomhoff JP, Myhre E, et al. Metronidazole in Crohn's disease. A double blind cross‐over clinical trial. Scand J Gastroenterol 1978;13:123–127. [DOI] [PubMed] [Google Scholar]
- 170. Ambrose NS, Allan RN, Keighley MR, et al. Antibiotic therapy for treatment in relapse of intestinal Crohn's disease. A prospective randomized study. Dis Colon Rectum 1985;28:81–85. [DOI] [PubMed] [Google Scholar]
- 171. Scribano ML, Prantera C. Antibiotics and inflammatory bowel diseases. Dig Dis 2013;31:379–384. [DOI] [PubMed] [Google Scholar]
- 172. Igarashi H, Maeda S, Ohno K, et al. Effect of oral administration of metronidazole or prednisolone on fecal microbiota in dogs. PLoS ONE 2014;9:e107909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Kilpinen S, Rantala M, Spillmann T, et al. Oral tylosin administration is associated with an increase of faecal enterococci and lactic acid bacteria in dogs with tylosin‐responsive diarrhoea. Vet J 2015;205:369–374. [DOI] [PubMed] [Google Scholar]
- 174. Menozzi A, Dall'Aglio M, Quintavalla F, et al. Rifaximin is an effective alternative to metronidazole for the treatment of chronic enteropathy in dogs: A randomised trial. BMC Vet Res 2016;12:217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Gerard L, Garey KW, DuPont HL. Rifaximin: A nonabsorbable rifamycin antibiotic for use in nonsystemic gastrointestinal infections. Expert Rev Anti Infect Ther 2005;3:201–211. [DOI] [PubMed] [Google Scholar]
- 176. Eiseman B, Silen W, Bascom GS, et al. Fecal enema as an adjunct in the treatment of pseudomembranous enterocolitis. Surgery 1958;44:854–859. [PubMed] [Google Scholar]
- 177. Choi HH, Cho YS. Fecal microbiota transplantation: Current applications, effectiveness, and future perspectives. Clin Endosc 2016;49:257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Brandt LJ, Aroniadis OC, Mellow M, et al. Long‐term follow‐up of colonoscopic fecal microbiota transplant for recurrent Clostridium difficile infection. Am J Gastroenterol 2012;107:1079–1087. [DOI] [PubMed] [Google Scholar]
- 179. Gupta S, Allen‐Vercoe E, Petrof EO. Fecal microbiota transplantation: In perspective. Therap Adv Gastroenterol 2016;9:229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Li SS, Zhu A, Benes V, et al. Durable coexistence of donor and recipient strains after fecal microbiota transplantation. Science 2016;352:586–589. [DOI] [PubMed] [Google Scholar]
- 181. Khoruts A, Dicksved J, Jansson JK, et al. Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent Clostridium difficile‐associated diarrhea. J Clin Gastroenterol 2010;44:354–360. [DOI] [PubMed] [Google Scholar]
- 182. Grehan MJ, Borody TJ, Leis SM, et al. Durable alteration of the colonic microbiota by the administration of donor fecal flora. J Clin Gastroenterol 2010;44:551–561. [DOI] [PubMed] [Google Scholar]
- 183. Borody TJ, Leis S, Campbell J, et al. Fecal microbiota transplantation (FMT) in multiple sclerosis (MS). Am J Gastroenterol 2011;106:S352. [Google Scholar]
- 184. Vrieze A, Van Nood E, Holleman F, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 2012;143:913–916 e917. [DOI] [PubMed] [Google Scholar]
- 185. Angelberger S, Reinisch W, Makristathis A, et al. Temporal bacterial community dynamics vary among ulcerative colitis patients after fecal microbiota transplantation. Am J Gastroenterol 2013;108:1620–1630. [DOI] [PubMed] [Google Scholar]
- 186. Chaitman J, Jergens AE, Gaschen F, et al. Commentary on key aspects of fecal microbiota transplantation in small animal practice. Vet Med‐Res Rep 2016;7:71–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Deng P, Swanson KS. Gut microbiota of humans, dogs and cats: current knowledge and future opportunities and challenges. Brit J Nutr 2015;113:S6–S17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Adapted with permission from P. Deng and K. S. Swanson, British Journal of Nutrition (2015), 113, S6–S17.
Table S2. Adapted with permission from P. Deng and K. S. Swanson, British Journal of Nutrition (2015), 113, S6–S17.
Table S3. Adapted with permission from P. Deng and K. S. Swanson, British Journal of Nutrition (2015), 113, S6–S17.


