Abstract
DNA methylation in the germline is among the most important factors influencing the evolution of mammalian genomes. Yet little is known about its evolutionary rate or the fraction of the methylome that has undergone change. We compared whole-genome, single-CpG DNA methylation profiles in sperm of seven species—human, chimpanzee, gorilla, rhesus macaque, mouse, rat, and dog—to investigate epigenomic evolution. We developed a phylo-epigenetic model for DNA methylation that accommodates the correlation of states at neighboring sites and allows for inference of ancestral states. Applying this model to the sperm methylomes, we uncovered an overall evolutionary expansion of the hypomethylated fraction of the genome, driven both by the birth of new hypomethylated regions and by extensive widening of hypomethylated intervals in ancestral species. This expansion shows strong lineage-specific aspects, most notably that hypomethylated intervals around transcription start sites have evolved to be considerably wider in primates and dog than in rodents, whereas rodents show evidence of a greater trend toward birth of new hypomethylated regions. Lineage-specific hypomethylated regions are enriched near sets of genes with common developmental functions and significant overlap across lineages. Rodent-specific and primate-specific hypomethylated regions are enriched for binding sites of similar transcription factors, suggesting that the plasticity accommodated by certain regulatory factors is conserved, despite substantial change in the specific sites of regulation. Overall our results reveal substantial global epigenomic change in mammalian sperm methylomes and point to a divergence in trans-epigenetic mechanisms that govern the organization of epigenetic states at gene promoters.
Mammalian DNA methylation is an epigenetic modification that occurs primarily on cytosines in the context of CpG dinucleotide and has wide-ranging connections to mammalian development (Reik et al. 2001; Smith and Meissner 2013). A typical mammalian genome contains 20–30 million CpG sites with an average methylation level between 70% and 80% in most cell types (Lister et al. 2013; Kundaje et al. 2015). Hypomethylated regions punctuate this globally high methylation profile and often coincide with gene regulatory elements that comprise gene promoters, enhancers, and insulators (Jones 2012). Programmed changes to these methylome features, for example altering the methylation state through a promoter, correlate with gene expression changes during development and differentiation (Meissner et al. 2008; Hodges et al. 2011). These dynamics can exhibit high precision, for example, when boundaries of hypomethylated regions shift to uncover CpG island “shores” and allow additional regulatory elements to receive signals (Irizarry et al. 2009).
DNA methylation has substantially impacted the large-scale evolution of mammalian genomes. Spontaneous deamination elevates the mutation rate at methylated cytosines (Bird 1980). Consequently, germline DNA methylation profiles have shaped the CpG landscape of mammalian genomes (Cohen et al. 2011), resulting in the CpG island phenomenon (Gardiner-Garden and Frommer 1987). In germ cells, DNA methylation is necessary for suppressing germline retroelement proliferation (Walsh et al. 1998; Bourc'his and Bestor 2004). Mounting evidence supports a contribution of germ cell DNA methylation to the epigenomic state of embryonic cells and the early activation of developmental genes (Brykczynska et al. 2010; Carrell and Hammoud 2010; Smith et al. 2012; Atsem et al. 2016; Branco et al. 2016). Examination of germline DNA methylation is therefore of particular interest in understanding how the requirements of epigenomic regulatory programming are balanced against the increased mutational burden of DNA methylation.
Taking a broader view, patterns of DNA methylation and the way they vary between species afford us a glimpse into the evolution of gene regulation. As highlighted by the seminal work of King and Wilson (1975) and validated in the current genomics area, species with high genetic similarity in their protein coding regions can show substantial phenotypic differences that may be related to differences in gene expression and regulation (Karaman et al. 2003; Odom et al. 2007; Bauernfeind et al. 2015). Epigenetic divergence reflects altered protein–DNA interactions, which may associate with changes in both the sequence of cis-regulatory elements (Heijmans et al. 2007; Lienert et al. 2011) and the binding preference of trans-acting factors (Cheng et al. 2014). Thus, characterizing epigenetic divergence is a first step toward understanding the evolution of regulatory systems in different species.
Existing comparative analyses of mammalian methylomes have increased our understanding of DNA methylation divergence. Methylation states across multiple primate species recapitulate their phylogenetic relationship (Martin et al. 2011), and greater variation has been observed between tissues than between species (Molaro et al. 2011; Pai et al. 2011). Regions with species-specific DNA methylation have been reported to harbor excessive species-specific substitutions (Hernando-Herraez et al. 2015). However, such studies have been limited to species most closely related to human and have focused on species-specific differentially methylated regions, instead of systematically studying methylation changes in all lineages of the phylogenetic tree without bias.
It remains unclear how and why DNA methylation patterns have evolved in mammalian species: Where in the genome, and when during evolutionary history, have changes occurred? Addressing these questions involves modeling an evolutionary process that may be viewed as analogous to sequence evolution. In homogeneous healthy cell populations, and over nearly the entire genome, continuous methylation levels at CpG sites can be functionally understood as having either high or low methylation, and a methylome can be modeled as a sequence of binary states. Phylogenetic modeling for molecular sequence evolution has a rich history (Felsenstein 1981; Yang 1994; Siepel and Haussler 2004), and approaches have been applied to DNA methylation changes in tumor growth and development (Siegmund et al. 2009; Capra and Kostka 2014). However, modeling methylome evolution at individual CpG sites presents a unique challenge: The majority of those sites have mutated between distant species due to their hypermutability. Focusing on CpG sites that are fully conserved in the relevant species dramatically reduces the portion of the orthologous genome that can be analyzed. Solutions that aggregate data, for example in predetermined windows along the genome, lower resolution while only partially solving the problem. At the same time, it is well known that methylation states of neighboring CpG sites are highly interdependent and a phenotypically relevant epigenomic change tends to involve multiple consecutive CpG sites. Horizontal dependence provides information that may be leveraged to overcome this challenge.
In this study, we examined whole-genome bisulfite sequencing data from the sperm genomes of seven mammalian species—human, chimpanzee, gorilla, rhesus macaque, mouse, rat, and dog—to investigate the evolution of germline DNA methylation. We designed a phylo-epigenetic model that accounts for loss of CpG sites between species by incorporating the interdependence of methylation states at neighboring CpG sites to infer the history of methylation divergence on each branch of the phylogenetic tree. We were able to detect genome-wide trends and lineage-specific features of methylome evolution, identify evolutionary events at a high resolution, and investigate the relationship with divergence of genomic DNA, the association with other epigenetic modifications, and the functional relevance of recently evolved methylome features.
Results
Comparison of sperm methylomes reveals lineage-specific features
We generated whole-genome bisulfite sequencing (WGBS) data for sperm cells from gorilla, dog, and rat. Sperm methylomes of human, chimpanzee, rhesus macaque, and mouse were produced in previous WGBS studies (Molaro et al. 2011; Hammoud et al. 2014; Lu et al. 2015). For each species, the sequencing had a total depth greater than 10× and covered the entire mappable fraction of the corresponding reference genome (Supplemental Table S1). In parts of our study, we used methylomes of embryonic stem cells (ESC) and somatic cells (B-cell, whole blood, peripheral blood mononuclear cells, and left ventricle) from different species for comparison with sperm methylomes (Methods). We used Ensembl gene annotations (Release 75) (Flicek et al. 2014) and defined gene promoter as the 2-kbp region surrounding the transcription start site (TSS). We identified hypomethylated regions (HMRs) from individual methylomes using a hidden Markov model as previously described (Molaro et al. 2011).
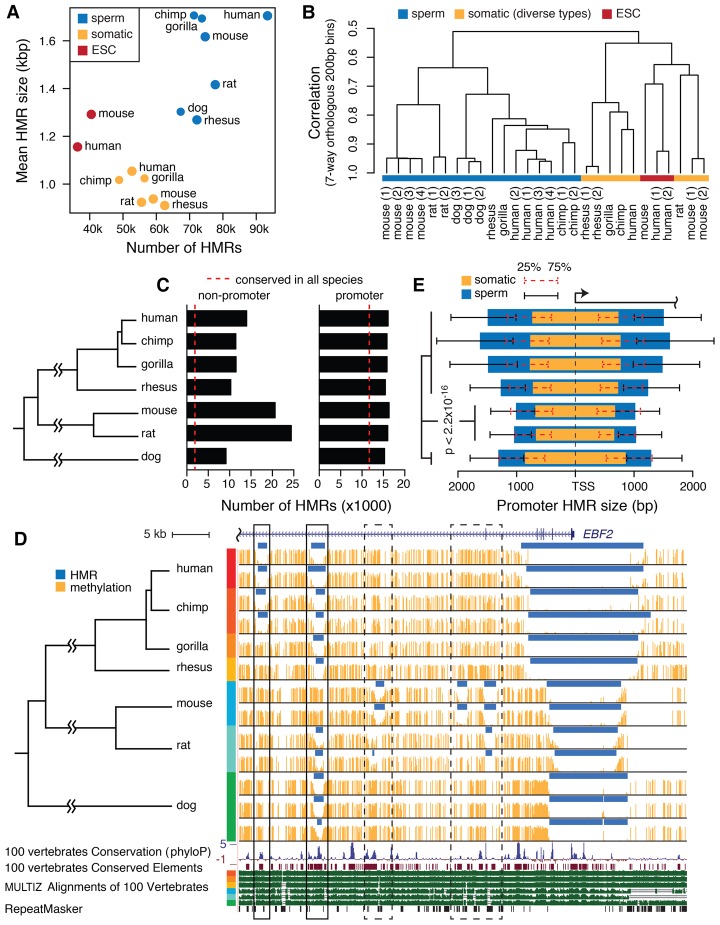
All species exhibited global methylation in sperm, with genome-wide average DNA methylation levels ranging between 0.68 and 0.79 (Supplemental Table S2). The methylation levels at single CpG sites formed a bimodal distribution (Supplemental Fig. S1A). Sperm methylomes contained more and wider HMRs than those in the examined ESC and somatic methylomes (Fig. 1A; Supplemental Table S2). Sperm HMRs overlapped retrotransposons and pericentromeric satellites more frequently than ESC and somatic HMRs (Supplemental Table S3; Supplemental Fig. S1B). These observations are consistent with patterns observed in human, chimpanzee, and mouse (Molaro et al. 2011, 2014).
Figure 1.
Mammalian sperm methylome characteristics. (A) The number and average size of HMRs in native assemblies. (B) Hierarchical clustering of aligned seven-way orthologous methylomes of multiple species and cell types. (C) The number of promoter HMRs and nonpromoter HMRs in seven-way orthologous sperm methylomes. Dashed lines indicate the average number of conserved HMRs across species. (D) Sperm DNA methylation of seven species in an example orthologous region. Methylome alignment is shown along with conservation tracks from MULTIZ alignment of 100 vertebrates and human repeat elements by RepeatMasker (Smit et al. 2013–2015). Regions in solid boxes show divergent methylation states at well-conserved elements. See zoomed-in browser image for dashed boxes in Supplemental Figure S2A. (E) The median size of hypomethylated regions upstream of and downstream from TSS in somatic (orange) and sperm (blue) methylomes of different species. HMR sizes are measured in their respective native genomes. Whiskers mark the 25th and 75th percentiles of HMR sizes upstream of or downstream from TSS. Wilcoxon rank-sum tests for all pairs of species between rodent species and nonrodent species showed significantly narrower HMRs in rodents.
To study epigenomic evolution across species, we aligned nonhuman methylomes and HMRs to the human reference genome at single-CpG resolution (Methods). The genomic composition of the seven-way orthologous genome is shown in Supplemental Figure S1C. The orthologous genome contains, on average, 17% (4.3 million) of the total CpG sites in individual species. The majority of the CpG sites have diverged between species, and only 0.4 million CpG sites are conserved across all species, the majority (59.7%) of which reside in codons. We therefore do not focus on fully conserved CpG sites, because they are limited in number and may be confounded by evolutionary pressures associated with protein coding regions. The number of HMRs located in the orthologous genome ranges between 24,000 and 40,000, and the average size ranges between 1.1 and 1.5 kbp for individual species (Supplemental Table S4; Supplemental Fig. S1D).
For a global view of the relationships between sperm methylomes, we clustered the sperm methylomes along with ESC and somatic methylomes based on pairwise correlation of average methylation levels in 200-bp bins (Fig. 1B; Supplemental Fig. S1E). There was a clear separation of sperm from ESC and somatic methylomes, consistent with tissue-specific characteristics of methylation (Molaro et al. 2011; Pai et al. 2011). The hierarchy in the sperm methylome cluster was largely consistent with the known phylogeny, except that dog and primates were more similar to each other than either was to rodents, despite a more recent common ancestor for rodents and primates (dos Reis et al. 2012). This may not be unexpected. Comparative genomic studies have reported a threefold higher neutral mutation rate in rodents compared with primates (Rat Genome Sequencing Project Consortium 2004; Lindblad-Toh et al. 2011).
Methylation features are conserved at promoters and divergent distally
All species have comparable numbers of promoter HMRs, although rodents have many more nonpromoter HMRs than other species (Fig. 1C). The intersection of HMRs across all species contained a total of 15,000 regions, with an average size of 1.0 kbp. If a HMR contains a region that is hypomethylated in all species, we call it a conserved HMR. Conserved HMRs were located predominantly at gene promoters and constitute on average 74% of the hypomethylated promoters in each species (Fig. 1C). Nonpromoter HMRs were far less conserved across species, with conserved HMRs constituting 8%–22% of the nonpromoter HMRs in each species. The example interval in Figure 1D (Supplemental Fig. S2B) shows the conserved presence of an HMR at the promoter in each species. The gene within this interval, EBF2 in human, is expressed in a variety of somatic cells, but has low expression in testis (Uhlén et al. 2015). This promoter HMR size varied between species, and there were lineage-specific HMR gain and loss events more distal to TSS. These observations are consistent with slower change at promoters, and more rapid evolution of enhancers, as has been observed in a comparative study of histone modifications focused on mammalian liver (Villar et al. 2015).
Taking advantage of the precision afforded by WGBS, we compared the sizes of promoter-associated HMRs across species. HMRs at orthologous gene promoters had similar sizes in ESC and somatic cells across species. In sperm, in contrast, HMRs showed substantial variation (Fig. 1E; Supplemental Fig. S1F,G). Rodent sperm promoter HMRs are significantly shorter compared with primates and dog (Wilcoxon rank-sum test P < 2.2 × 10−16). Within primates, rhesus macaque had significantly shorter HMRs than other species examined (Wilcoxon rank-sum test P < 2.2 × 10−16). Similar levels of variation were also evident at the replicate level between species (Supplemental Table S2). These sperm promoter HMR size differences indicate a global divergence in the organization of the sperm epigenome.
The cross-species HMR size differences in orthologous regions indicate that an ancestral HMR has evolved to have different boundary locations in different species. For a conserved HMR in human, on average, the core region of hypomethylation shared in all species only constituted 53% of the HMR. This proportion was 64% for rodents. Boundary variation was also common for HMRs specific to a lineage. Altogether, a considerable amount of cross-species epigenomic variation took the form of adjustments in boundaries of these putative regulatory intervals—in addition to complete gain or loss. Subsequently, we implement a formal model to quantify such boundary movements.
Despite prevalent HMR boundary variation, a subset of orthologous HMRs had relatively conserved boundary locations across species. We identified 250 such “ultraconserved” HMRs (Supplemental Methods), all of which overlapped phastCons-conserved elements for placental mammals (Siepel et al. 2005), and 92% overlapped gene transcription start sites (Supplemental Table S5). Relative to genes with promoter HMRs in all seven species, those with ultraconserved promoter HMRs were enriched in multiple biological processes, including developmental process, signaling, and regulation of multicellular organismal process (Supplemental Table S6).
Methylation loss exceeds methylation gain across species
We compared sperm HMRs across the seven species and identified species-specific HMRs and species-specific methylated regions. For each species, there was more species-specific hypomethylation than methylation (Supplemental Fig. S3A; Supplemental Table S7). This hints at a nonstationary process and prompted us to ask whether global trends and rates of epigenomic change can be extracted from these methylomes.
We designed a model that combines two processes: (1) the “horizontal” process that induces autocorrelation of methylation states along the genome within a species, and (2) the “vertical” inheritance of methylation states from the ancestral methylome. The vertical process is modeled with a continuous-time Markov process in the usual way (Felsenstein 1981). To model the horizontal dependency between the methylation states of two neighboring sites in the same methylome, we introduce a transition probability to model the conditional distribution of a site's methylation state given the state at the site's neighbors (Supplemental Fig. S3B). Combining these two processes, our model is a Bayesian network that describes the probability of a site having a particular methylation state as a function of its ancestral state and the contemporaneous state of its previous neighbor (Methods). The inclusion of the horizontal process allows us to treat nonconserved CpG sites as sites with latent data to be modeled using a computationally tractable set of dependencies (Supplemental Fig. S3B).
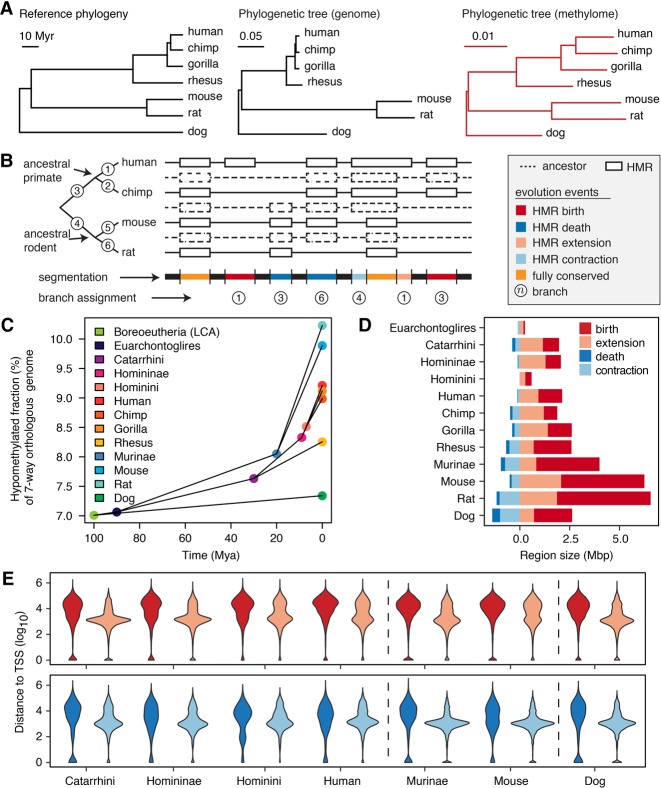
We estimated the sperm methylome evolutionary tree with the proposed model (Fig. 2A; Supplemental Table S8). Branches within rodents were significantly longer than the primate lineage (Wilcoxon signed-rank test P = 6.6 × 10−9) (Supplemental Methods), indicating faster epigenomic evolution in sperm of rodents than primates. The rat sperm methylome exhibited faster evolution than mouse (Wilcoxon signed-rank test P = 1.6 × 10−8), which coincided with a higher nucleotide substitution rate in rat (Rat Genome Sequencing Project Consortium 2004). However, the methylome evolution rates were much higher at the terminal branches than internal branches, suggesting relatively fast turnover of sperm HMRs in mammals. With an exponential decay model, we estimated the half-life of HMRs to be 117 million yr (Myr) overall, 439 Myr at gene promoters and 39 Myr for distal HMRs (Supplemental Fig. S3C).
Figure 2.
Methylation loss exceeds methylation gain during evolution. (A) Phylogenetic tree representing consensus divergence time (Hedges et al. 2015) genome evolution and sperm methylome evolution. Unit branch length represents 1 million yr, one substitution/site, and one methylation state change/CpG site, respectively. (B) Schematic segmentation of the orthologous genome according to the history of methylation evolution and annotation of methylation mutation events. (C) Fraction of orthologous genome hypomethylated in extant and ancestral species, estimated by the interdependent-site phylo-epigenetic model. (D) Total size of different types of methylation evolution events on individual branches. (E) The distribution of distances from methylome evolution events to closest TSS in the seven-way orthologous genome. The color legend is as shown in D.
The inferred methylation states in ancestral species indicate an increasing proportion of hypomethylation in the orthologous genome along each branch of the phylogenetic tree (Fig. 2C). We estimated that HMRs made up 7.0% of the orthologous genome in the last common ancestor of the seven species; 7.6% in the primate common ancestor and 8.1% in the rodent common ancestor; and 8.3%–9.2% in extant primates and 9.9%–10.2% in extant rodents (Fig. 2C). The overall increase in the hypomethylated fraction of the orthologous genome was a result of DNA methylation loss exceeding gain along all branches of the tree, with the dog coming closest to balancing the two (Supplemental Fig. S3D). To examine whether the methylome evolutionary process was stationary, we applied independent-site phylogenetic models on discretized methylation states in 200-bp bins (Methods). We compared a reversible model assuming stationary evolutionary process with an unrestricted model (i.e., lacking the reversibility assumption). The unrestricted model fit the observed data better and estimated an evolutionary process with increasing hypomethylation fraction (Supplemental Fig. S3E,F).
To examine whether our preceding observations are robust to the technical aspect of using human-referenced alignment, we performed the same analyses using mouse-referenced alignment (Supplemental Methods). In addition, to investigate whether sperm methylomes have similar evolutionary features at well-conserved sequence elements as in the entire orthologous genome, we restricted analyses to CpG sites located within placental mammal conserved elements (Siepel et al. 2005). The genomic context composition and distribution of CpG methylation levels vary slightly for the mouse-referenced seven-way orthologous genome (Supplemental Fig. S4A,B). However, they vary substantially for well-conserved elements that are mostly protein coding sequences and important regulatory elements under strict evolutionary constraints (Supplemental Fig. S5A,B). Nevertheless, the results in both analyses are consistent with results presented above. Rodents display faster evolution rates (Supplemental Figs. S4C,D, S5C,D). Sperm methylomes have experienced expansion of hypomethylation in the overall orthologous genome (Supplemental Fig. S4E,F) and in well-conserved elements (Supplemental Fig. S5E,F). Methylation patterns are more conserved at promoter-proximal elements than distal elements (Supplemental Fig. S5G).
Widening of hypomethylated regions has contributed significantly to methylation loss
Loss of DNA methylation during evolution reflects an increase in either the size or the number of HMRs. We classified methylation loss (HMR gain) events into HMR birth events or HMR extension events; similarly, we classified HMR loss events into HMR deaths or HMR contractions (Fig. 2B,D; Methods). The HMR birth and death events showed different genomic distribution from the extensions and contractions (Fig. 2E). Distal (more than 10 kbp away from TSS) events constitute 56% of HMR births and 40% of deaths, in contrast to 30% of extensions and 16% of contractions.
Only a small proportion (<10%) of the orthologous genome was hypomethylated. However, HMR extensions comprise 21%–74% of total HMR gains (Fig. 2D; Supplemental Table S9), which correspond to >20-fold more than expected if HMR gains were to randomly occur in the genome (Supplemental Fig. S6; Methods). Among HMR loss events, deaths occurred more frequently than expected (Supplemental Fig. S6). These observations suggest that existing HMRs may facilitate the evolutionary formation of a new HMR in the adjacent region; meanwhile, loss of HMR is more likely to occur as methylation through an entire ancestral HMR rather than only a fraction of it.
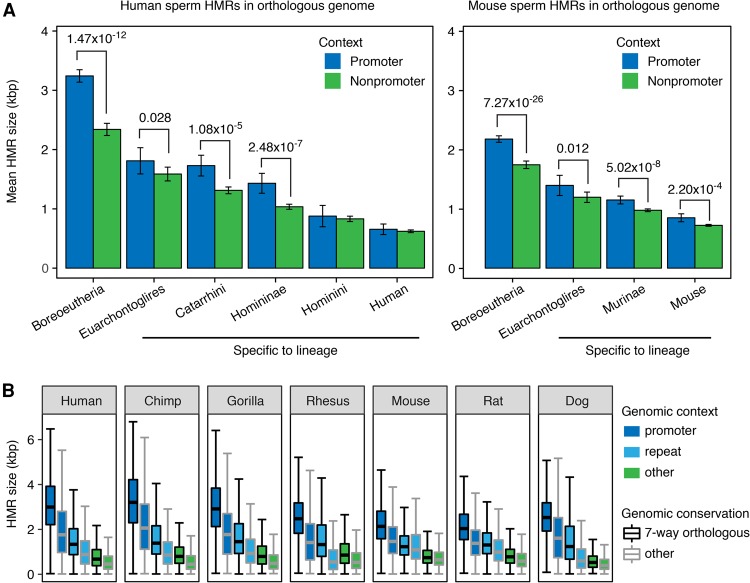
We found two forms of evidence in line with our hypothesis that once established, a typical HMR can gradually evolve to span a larger genomic region. First, we compared the HMR sizes between older and younger groups, and found positive correlation between the age and the size of HMRs (Fig. 3A), with faster expansion at promoter HMRs than nonpromoter HMRs. Second, we found that HMRs in the orthologous genome were consistently larger in size within each genomic context group than those in species-specific genomic regions (Fig. 3B). An alternative hypothesis may state that HMRs randomly arise in the genome with substantial variation in size, but wider HMRs are more likely to survive through evolution. Although this alternative hypothesis may also explain the size–age correlation of HMRs, it offers no explanation to the substantial cross-species size variation of conserved HMRs. Therefore, we find progressive extension a more likely hypothesis. Together, these observations suggest that recently appearing epigenomic features encompass smaller genomic intervals than more ancient features. Of note, such a global trend would have progressed independently and may have different rates along parallel evolutionary lineages.
Figure 3.
Older and more conserved sperm HMRs are wider. (A) Mean sizes of promoter and nonpromoter HMRs in the seven-way orthologous genome of human and mouse sperm, grouped by estimated HMR emergence time in mammalian evolution. Error bars indicate standard errors of the mean. Significant Wilcoxon rank-sum test P-values are shown. The Boreoeutheria group contains HMRs conserved in all seven species, representing HMRs formed before and during the speciation of Boreoeutheria. Other groups represent HMRs formed during the speciation of respective lineages. (B) Distribution of HMR sizes in native genome assemblies, grouped by genomic context and conservation level.
Sequence signatures driven by sperm methylome divergence
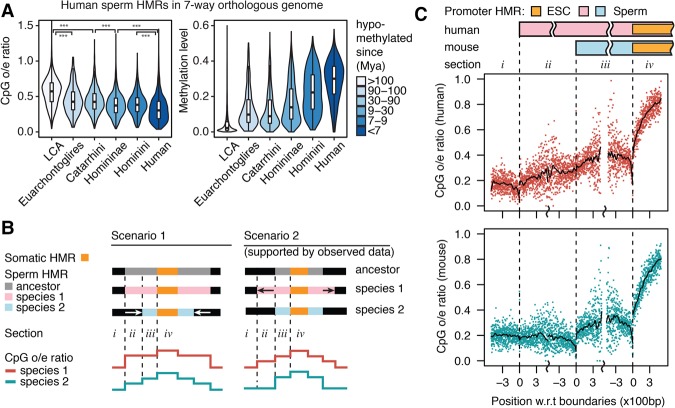
Because methylated CpGs experience elevated mutation rates driven by deamination, we predicted that observed/expected (o/e) ratio of CpG frequency in a region is related to the length of time a region has been methylated. To test this prediction, we measured CpG frequency in human HMRs separated into six different age groups based on estimated ancestral methylation states. As predicted, older HMRs had higher CpG o/e ratios compared to younger HMRs (Fig. 4A). In addition, older HMRs displayed lower levels of DNA methylation than younger HMRs, suggesting that conserved elements are subject to more stringent epigenetic regulation and maintenance, whereas younger elements may be involved in more dynamic interactions. These findings remained unchanged after we controlled for DNA methylation level (Supplemental Fig. S7A).
Figure 4.
Sequence signatures driven by sperm methylome evolution. (A) CpG enrichment (observed/expected ratio of CpG occurrences) and regional average DNA methylation level in human HMRs in seven-way orthologous genome grouped by estimated hypomethylation age. (B) Two opposite scenarios explaining the evolution path to different promoter HMR sizes in two extant species and the expected CpG enrichment schematic profile in the extant species as a result of methylation-induced CpG decay. (C) Human and mouse CpG enrichment profile at hypomethylated orthologous gene promoters, where human sperm HMRs are wider than mouse. Profiles show CpG enrichment in 2-bp windows by distance to indicated HMR boundaries. Smoothed lines were generated with local polynomial regression fitting (LOESS).
The relationship between germline methylation history and CpG decay through a genomic interval suggests we might use CpG o/e ratios to polarize the promoter HMR size divergence observed in Figure 1D. The substantial number of promoter HMRs that are much wider in human than in mouse may have arisen from a general widening along the human lineage or a narrowing along the mouse lineage. Figure 4B illustrates a method for distinguishing these possibilities. We selected gene promoters that were hypomethylated in sperm and ESC of both human and mouse, with a wider sperm HMR than ESC HMR in both species and with a wider sperm HMR in human than in mouse. The mouse CpG o/e ratios on opposite sides of positions orthologous to human sperm HMR boundaries (sections i versus ii) showed no significant difference (Fig. 4C), suggesting that both sides in the mouse genome shared a common methylation history during evolution. Meanwhile, regions corresponding to rodent-lineage-specific sperm HMR loss showed higher CpG o/e ratios than the methylated genomic background level (Wilcoxon rank-sum test P = 1.3 × 10−4) (Supplemental Fig. S7B), which suggests that ancestral CpGs have not had enough evolutionary time to decay following recent increases in methylation. In contrast, there was a significant drop of human CpG o/e ratios (Wilcoxon rank-sum test P = 1.3 × 10−31) crossing the mouse sperm HMR boundaries from section iii to ii. Although regions on both sides were hypomethylated in present-day human sperm, the outer side (section ii) must have become hypomethylated later than the inner side (section iii). Dog sperm methylome also showed wider promoter HMRs than mouse (Fig. 1D), and we observed similar profiles of CpG o/e ratios in the dog–mouse comparison (Supplemental Fig. S7C). These observations suggest that although sperm promoter HMR sizes may have increased on parallel lineages, the changes likely occurred at different rates, and that ancestral promoter HMR size in sperm was closer to that in rodents, with both dog and primates evolving wider intervals of hypomethylation around promoters in sperm.
Methylation divergence is associated with histone modification and sequence divergence
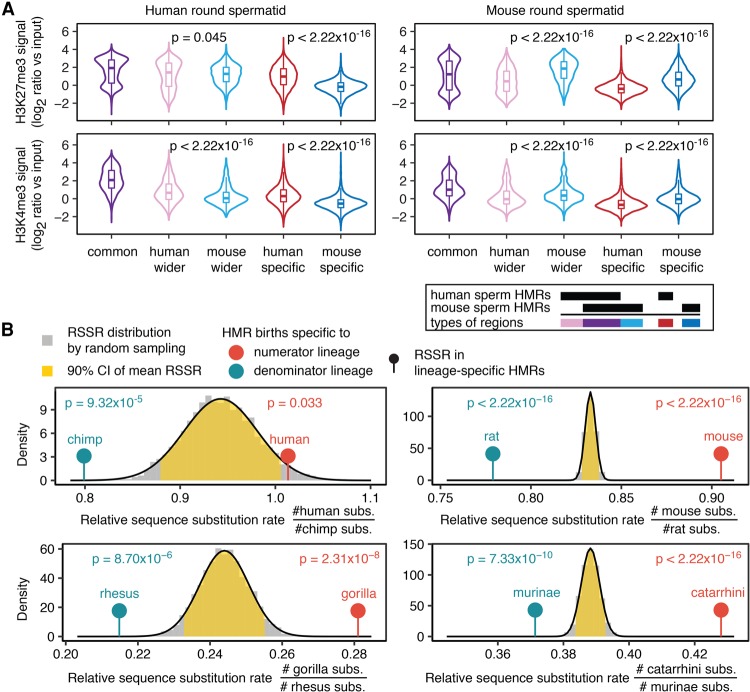
To examine whether sperm DNA methylation divergence is accompanied by histone modification divergence, we analyzed H3K4me3 and H3K27me3 ChIP-seq data from round spermatids of human and mouse (Lesch et al. 2016) and H3K4me1 ChIP-seq data from human and mouse sperm (Hammoud et al. 2014; Jung et al. 2017). Lineage-specific sperm HMR births and extensions in human and mouse were associated with lineage-specific enrichment of histone modifications (Fig. 5A; Supplemental Fig. S8A,B). In both human and mouse, promoter HMRs showing species-specific widening in sperm, especially those showing the same widening in ESC, were enriched with bivalent histone marks in round spermatid (Supplemental Table S10). The association between divergent DNA methylation and divergent histone modification in sperm indicates that sperm DNA methylation is an integral part of the germline epigenome evolution.
Figure 5.
Sperm DNA methylation divergence is accompanied by sperm histone modification and DNA sequence divergence. (A) Mouse- and human-specific sperm HMRs and HMR extensions show species-specific H3K4me3 and H3K27me3 enrichment. (B) Lineage-specific HMRs are associated with relatively more sequence substitutions. For each pair of parallel lineages, expected distribution of RSSR is shown as a gray histogram, and fit is shown with normal distribution (black curve). The yellow area marks 90% confidence interval for mean RSSR. Upper (lower) tail of the distribution indicates significantly more substitutions in the numerator (denominator) lineage. Each lollipop marks the observed RSSR in HMRs specific to the indicated lineage, with one-sided P-value shown on top.
The sequence conservation levels (Pollard et al. 2010) were substantially higher in lineage-specific HMRs than their flanking regions, suggesting that regulatory elements may be conserved at the sequence level, but diverged at the epigenetic level. Moreover, all groups of lineage-specific sperm HMRs showed a sizable (>30%) overlap with human Roadmap somatic HMRs, which are potentially active regulatory elements in human somatic tissues (Supplemental Table S11; Kundaje et al. 2015). Overlapping with human somatic HMRs considerably increased the sequence conservation level in lineage-specific sperm HMRs (Supplemental Fig. S7D). Rodent lineage-specific HMRs not overlapping human somatic HMRs remained at higher sequence conservation level than neighboring regions (Supplemental Fig. S7D), suggesting conserved regulatory regions between human and mouse yet to be observed. These observations are consistent with exaptation of existing elements as a major source for species to evolve new regulatory interactions (Villar et al. 2015).
To examine whether lineage-specific HMRs are associated with lineage-specific substitutions, we computed the relative sequence substitution rate (RSSR) in orthologous regions between two parallel lineages and compared the RSSR in lineage-specific HMRs against randomly sampled regions from the background orthologous genome (Methods). We studied four pairs of sister or parallel lineages and observed that lineage-specific HMR births were associated with significantly elevated sequence substitution rates on the same lineage (Fig. 5B). After excluding CpG sites, the differences in relative substitution rates remained significant (Supplemental Table S12). These observations, further extending previous results, reveal a local link between sequence substitutions and evolution of new epigenetic features.
Lineage-specific changes involve common transcription factors and developmental genes
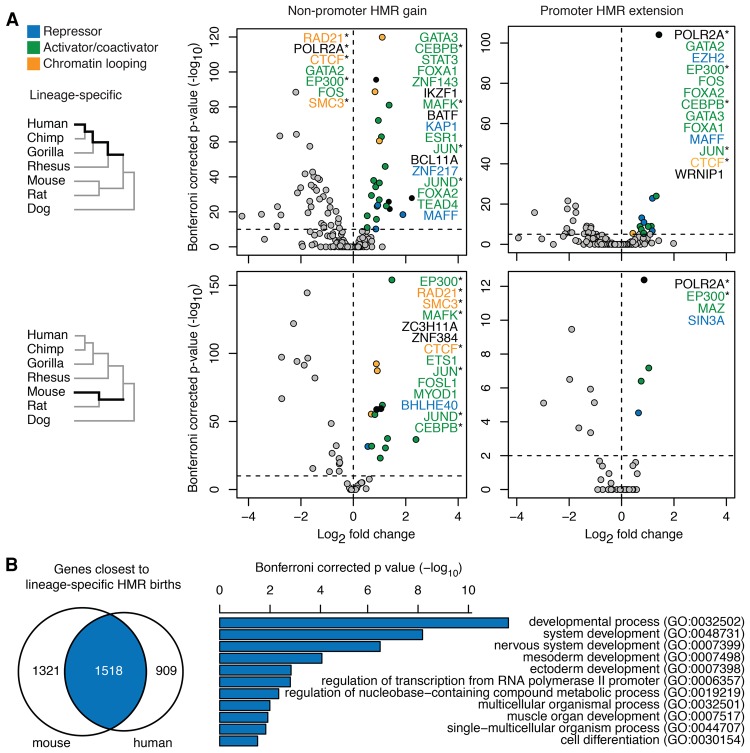
DNA hypomethylation reflects cis-regulatory interactions (Bird 2002; Hodges et al. 2011; dos Santos et al. 2015). We tested whether certain transcription factors were enriched in lineage-specific promoter HMR extensions and lineage-specific nonpromoter HMR gains in human and mouse (Fig. 6A; Methods). At lineage-specific nonpromoter HMRs, a number of transcriptional activators were enriched on both lineages, such as EP300; activator protein 1 (AP-1) subunits FOS, FOSL1, and JUN; and AP-1 interacting factor MAFK. In addition, chromatin-looping factors including CTCF and cohesin components RAD21 and SMC3 were overrepresented on both lineages. Of the CTCF binding sites found in recently evolved lineage-specific nonpromoter sperm HMRs, 33% and 43% of the sites were also lineage-specific hypomethylated in the ESCs of human and mouse. These observations suggest a degree of evolutionary plasticity for these factors in their roles associated with 3D organization of the genome and long-range promoter-enhancer interaction; considering results presented above, this plasticity seems to be increasing the number of regions involved in these functions.
Figure 6.
HMRs gained on parallel lineages are associated with similar transcription factors and developmental genes. (A) Enrichment of transcription factor binding sites in HMR birth and extension regions on the human lineage and mouse lineage. (*) Factors enriched in both lineages. (B) Lineage-specific HMRs are associated with gene sets with significant overlap and that are enriched in developmental processes.
Lineage-specific promoter HMR extensions on both lineages showed enriched binding sites of EP300 and RNA polymerase II, whereas extensions on the human lineage showed enrichment of many transcriptional activators that are also enriched in human lineage-specific nonpromoter HMR gains. Genes with primate lineage-specific promoter HMR extension showed enrichment in developmental process and nervous system process (Supplemental Fig. S9A; Supplemental Table S13). About 12% of primate lineage-specific HMR gains in human sperm were also hypomethylated in human ESC and as much as 82.5% overlap with ENCODE transcription factor binding sites, DNase I hypersensitive sites (Thurman et al. 2012), or Roadmap somatic HMRs. This suggests that recently evolved sperm HMRs likely can take on roles of regulatory elements in developmental stages beyond germ cell development.
Despite being lineage specific, HMR births on these two parallel lineages sometimes occurred in each other's neighborhood in the orthologous genome (Supplemental Fig. S9B), and they shared a sizeable list of common associated genes (Jaccard index = 0.41, Fisher's exact test P < 2.2 × 10−16) (Methods). These common genes were enriched for functions in embryonic developmental processes, including mesoderm and ectoderm development and nervous system development (Fig. 6B). Meanwhile, analysis of associated genes that were specific to one species did not yield any enrichment of biological processes. Together, these results suggest that independently evolving features of the sperm epigenome are involved in regulating common genes that are important for embryonic development. This may be viewed as a form of convergent evolution: Regulatory features are emerging independently for the same genes.
Discussion
We used high-resolution whole-genome DNA methylation profiles and quantitatively measured the divergence among the sperm methylomes of seven mammals with a newly developed phylo-epigenetic model. Several key findings emerged from our analysis. There is a global trend toward expansion of the hypomethylated fraction of the genome on all phylogenetic lineages. Hypomethylated intervals around transcription start sites have evolved to be substantially wider in primates and dog than in rodents. Genomic intervals with lineage-specific hypomethylation are associated with common developmental processes and are often close to sets of genes with significant overlap. Analysis of transcription factor binding data indicates that regions of rodent-specific and primate-specific hypomethylation are enriched for binding sites of orthologous transcription factors.
DNA hypomethylation reveals cell-type- and species-specific distribution of active or poised regulatory elements (Lister et al. 2009; Stadler et al. 2011; The ENCODE Project Consortium 2012). Evolution and divergence of DNA methylation patterns reflect the evolution of regulatory elements. We found that the fraction of DNA hypomethylation in the orthologous genome has been expanding, with frequent HMR births and HMR extensions on all lineages of the phylogenetic tree, and provided evidence for progressive widening of HMRs at both promoters and potential distal enhancers. These results suggest a mode by which new regulatory elements may evolve to increase specificity of transcriptional regulation and are in line with the findings from a recent theoretical study of transcription factor binding site evolution (Tuğrul et al. 2015). Specifically, in promoters and enhancers, which harbor clusters of binding sites, the additive effect of transcription factor binding on gene expression and cooperative binding of transcription factors can accelerate emergence of new transcription factor binding sites. In contrast, evolution of an isolated binding site would typically be much slower. Considering models of regulatory elements, such as the “enhanceosome” and “billboard” models for transcriptional enhancers (Arnosti and Kulkarni 2005), our phylo-epigenetic model provides a phylogenetic tool for elements that lack the degree of sequence conservation often required to infer function.
Our analyses of DNA sequence and methylation divergence revealed principal features of the relationship between genomic and epigenomic evolution. We observed methylome evolution rate differences between lineages, similar to those observed in genomic evolution, reflecting a genetic component underlying epigenetic divergence. Sequence divergence has been associated with gain and loss of some regulatory elements (Schmidt et al. 2010). Linking DNA sequence divergence with DNA methylation changes, Hernando-Herraez et al. (2015) have shown within multiple primate species that species-specific methylation is associated with elevated species-specific nucleotide substitutions. We add to this picture by showing that lineage-specific hypomethylation is associated with increased sequence substitutions compared with the parallel sister lineage in the phylogenetic tree. Interestingly, in the seven-way orthologous genome, sequences in lineage-specific sperm HMRs are usually more conserved than flanking regions, perhaps reflecting a functional exaptation of ancestral sequence and repurposing of ancestral regulatory elements in species-specific and cell-type–specific ways.
Germline DNA methylation patterns have shaped the mammalian genomic distribution of CpG dinucleotides. In human germline cells across many generations, the promoter HMRs alternate between a wider size in sperm cells and a narrower size in ESCs, which has left distinctive signatures of deamination-induced CpG depletion in the human genome (Molaro et al. 2011). In this study, we add further support to this theory by delineating the occurrence time during evolution of human sperm HMRs and showing that CpG enrichment in HMRs is strongly correlated with HMR age. In addition, CpG enrichment around promoter HMR boundaries in primates, rodents, and dog offers additional support to our inference of parallel promoter HMR extension in primates and dog by phylo-epigenetic modeling. Moorjani et al. (2016) reported a more clock-like behavior of CpG transitions compared with other substitutions. Understanding germline methylation patterns as well as their evolutionary history in different species would enable better calibration of divergence time by such a CpG-based molecular clock.
Several factors in the mature sperm of human and mouse, such as histone retention and modification, CTCF binding, and microRNAs, have been proposed to have regulatory importance in embryonic development (Carone et al. 2014; Hammoud et al. 2014; Rodgers et al. 2015; Teperek et al. 2016). For example, Lesch et al. (2016) reported that epigenetically poising developmental regulator genes with bivalent histone modifications (H3K4me3 and H3K27me3) at gene promoters was a conserved phenomenon in mammalian sperm, and differences in gene poising in sperm were correlated with differential developmental expression between species. However, of all epigenetic modifications, DNA methylation is the only epigenetic mark with known mechanisms of stable transmission across cell divisions (Henikoff and Greally 2016). Although most DNA methylation marks are removed during two waves of epigenetic reprogramming, regulatory elements that can escape demethylation have been observed in both zygotic and primordial germ cell reprogramming (Borgel et al. 2010; Hackett et al. 2013; Wang et al. 2014). In mouse, oocyte-derived methylation is involved in regulating trophoblast development (Branco et al. 2016), and paternal age-related sperm DNA methylation changes are associated with transcriptional dysregulation of developmental genes in the offspring (Milekic et al. 2015). We have observed lineage-specific HMRs associated with conserved developmental processes and even proximal to the same genes. Meanwhile, these lineage-specific regions are enriched with binding sites of chromatin-looping factors and enhancer binding factors. These observations suggest that the plasticity accommodated by certain regulatory factors is conserved, despite substantial turnover in the specific sites of regulation. Such plasticity may reflect or permit greater divergence between species in how the associated genes are regulated even in the context of developmental programs that are deeply conserved within mammals.
In parallel with emergence of species-specific regulatory elements, divergence in the organization of gene promoters may be of equal significance in mammalian evolution. The striking differences in sperm promoter HMR sizes between species indicate a global divergence in epigenetic regulation and chromatin organization of orthologous genes including key developmental genes. We observed binding sites of repressive factors EZH2 (a subunit of polycomb repressive complex 2, PRC2) and SIN3A to be enriched respectively in human and mouse lineage promoter HMR extension regions. Although the data from somatic cells does not necessarily imply binding at the same sites in male germ cells, those two repressor complexes have been found indispensable for proper repression of soma-specific genes through chromatin remodeling during germ cell development (Gallagher et al. 2013; Mu et al. 2014). In mouse ESC, it has been observed that both complexes interact with TET1 but at distinct gene promoters (Williams et al. 2011; Neri et al. 2013). The binding profile of TET1 spreads wider at bivalent gene promoters (Neri et al. 2013), possibly due to association with PRC2, which has a proposed mechanism for spreading at bivalent promoters (Margueron et al. 2009). In contrast, TET1 binding is much narrower when it co-binds with SIN3A, which has a narrower binding profile as well (Neri et al. 2013). We also observed lineage-specific enrichment of histone modifications in lineage-specific HMR extensions, indicating that the breadth of sperm histone marks differs between species. Broad H3K4me3 domains at gene promoters have been connected with increased RNA polymerase II pausing, transcriptional consistency, and cell identity (Benayoun et al. 2014). Although mature sperm are usually assumed to be transcriptionally inactive, the presence of RNA polymerase II has been detected in the nuclei of mature mouse sperm (Lin et al. 2013). Considering these results, it is possible that divergent recruitment of trans-acting factors during germ cell development contributes to different sizes of promoter HMRs between species and has further influence in zygotic gene activation and embryo development. It remains to be examined whether the occupation of these chromatin-modifying and transcription complexes differ between species in the context of germ cell development and how they are associated with promoter methylation patterns in different species.
In conclusion, we characterized evolutionary changes in mammalian sperm methylomes at high spatial and temporal resolution. Our analyses reveal the global trend and lineage-specific features of expanding DNA hypomethylation during evolution and provide insights to how evolution of the epigenome may contribute to species-specific fine-tuning of conserved mammalian developmental programs.
Methods
Whole-genome bisulfite sequencing data sets
We collected sperm samples from one gorilla individual, two rat individuals, and three dog individuals and generated whole-genome bisulfite sequencing (WGBS) data sets. See Supplemental Methods for details of sample collection, library preparation, and sequencing.
Public WGBS data used in this study include human and chimpanzee sperm (Gene Expression Omnibus accession GSE49624 and GSE30340) (Hammoud et al. 2014; Molaro et al. 2011), human H1 ESC (GSE16256) (Lister et al. 2013), human H9 ESC (GSE19418) (Laurent et al. 2010), human and chimpanzee B-cell (GSE31971) (Hodges et al. 2011), gorilla whole blood (NCBI SRA accession SRP059313) (Hernando-Herraez et al. 2015), rhesus macaque sperm (GSE60166) (Lu et al. 2015), rhesus macaque PBMC (GSE34129) (Tung et al. 2012), rat left ventricle (European Nucleotide Archive accession number ERP002215) (Johnson et al. 2014), mouse sperm (GSE49624) (Hammoud et al. 2014), mouse B-cell (SRP029721) (Kieffer-Kwon et al. 2013), mouse ESC (GSE30206) (Stadler et al. 2011), and the dog kidney epithelial cell line MDCK (GSE48527) (Carmona et al. 2014).
Mapping and annotation
Reads from whole-genome bisulfite sequencing were mapped with the RMAP software (Smith et al. 2009) to respective reference genome of each species: hg19, panTro4, gorGor3, rheMac3, mm10, rn5, and canFam3. Subsequent analyses on duplicate removal, bisulfite conversion rates, methylation levels, and HMRs were carried out with tools from the MethPipe package (Song et al. 2013). Mapping statistics are provided in Supplemental Table S1. We did not observe significant non-CpG methylation in any species and focused our analyses on CpG methylation. We used Ensembl gene annotations (Release 75) (Flicek et al. 2014) for species involved in this study. Coordinates of rhesus macaque gene annotation were converted to assembly rheMac3 from rheMac2 with liftOver from the Genome Browser tool suite (Kent et al. 2002).
Methylome alignment
We produced multiple methylome alignment by mapping nonhuman CpG positions and methylation levels to the human genome assembly hg19, using the 100 vertebrate MULTIZ alignment (hg19) downloaded from UCSC Genome Browser (Rosenbloom et al. 2015). We used UCSC Genome Browser binary utility mafSpeciesSubset to filter out sequence records from species other than the seven species of interest. Next, we fused together adjacent alignments blocks that were separated by species not included in this study, using Galaxy script maf_thread_for_species.py (https://github.com/bxlab/bx-python/blob/master/scripts/maf_thread_for_species.py) (Giardine et al. 2005; Blankenberg et al. 2010; Goecks et al. 2010). Then we parsed the locations of CpG sites in each species and the corresponding coordinates in the human reference genome from alignment blocks that contain sequences from all seven species. We assessed whether using the latest human genome assembly GRCh38 would significantly impact the methylome alignment. Although GRCh38 has improved coverage at centromere sequences and increased diversity with alternate loci, sequences in genomic regions conserved with other mammalian species are highly consistent with the older assembly GRCh37/hg19. Only 4596 (0.067%) CpGs in the seven-species alignment blocks under the new assembly (alternate loci and unplaced sequences excluded) were absent from the old assembly, 1308 (0.019%) were non-CpG sites in the old assembly, and 78 (0.001%) had insertion/deletion or nonunique mapping, whereas the rest of CpG sites had one-to-one mapping between the two versions of human reference genome assemblies.
To define the seven-way orthologous genome, we excluded any regions that are >1 kbp in size and contain no CpGs or have zero read coverage in at least one species; we considered the resulting set of genomic regions, covering 370 million bases, as the regions conserved across all seven species. We chose 1 kbp as the minimum CpG desert size because the correlation of methylation levels at neighboring CpG sites drops to background level at distance >1 kbp.
To map HMRs from the native genome assembly to the human reference genome, we kept track of HMR membership of individual CpGs and then merged consecutive CpGs that belonged to the same HMR in the original native genome and were not separated by CpG deserts into a HMR. We filtered out HMRs with fewer than five CpG sites.
Phylo-epigenetic model considering CpG-site interdependence
We constructed a phylo-epigenetic evolution model Θ = {τ, π, Q, F, G}, where τ is a phylogenetic tree with known phylogeny and unknown branch lengths; π is the distribution of methylation states at the first site in the last common ancestor (LCA; root of the phylogenetic tree); F is the transition probability matrix for methylation states between neighboring CpG sites in the LCA; Q is a continuous-time Markov chain transition rate matrix for the evolution of methylation state at a single site; G is a transition probability matrix for neighboring CpG sites in descendant species. The methylome of the LCA is modeled by a two-state discrete-time Markov chain with model parameters (π,F). Evolution of the first site in the genome is modeled with continuous-time Markov process Q along the phylogenetic tree τ, as in the standard independent-site phylogenetic model. The main component of our phylo-epigenetic model is the combination of the horizontal inter-CpG dependence process described by G and the vertical inheritance process described by Q and divergence time. Consider a parent–child pair of species (u,v) with branch length ℓuv, and let the methylation state at the site i of v be vi. Suppose the methylation states of vi’s parent site and its previous site are given. We define the conditional distribution of vi given vi − 1 and ui as
This modeling strategy achieves a balance between computational tractability and incorporation of dependence relations between neighboring CpG within and across species (for discussion of alternative models, see Supplemental Methods). The input data can be binary methylation states, continuous methylation probabilities at individual CpG sites in extant species, and missing data at sites in an extant species when its orthologous sites have observed data in other species. See Supplemental Methods for details about the model and parameter estimation methods. In brief, we used a Monte Carlo Expectation Maximization (MCEM) algorithm (Wei and Tanner 1990), where the posterior distribution of methylation states at unobserved CpG sites is approximated by Gibbs sampling in the E-step. In-house software can be downloaded from https://github.com/smithlabcode/Epiphyte.
Model selection between reversible and unrestricted evolution processes
We used Akaike information criterion (AIC) to compare two independent-site phylogenetic models, with and without the assumption of reversible evolutionary process. We discretized average methylation probabilities (cutoff 0.5) to determine the methylation states in 200-bp bins in the seven-way orthologous genome. Only bins with nonzero CpG coverage in all species are included to form complete observation at extant species. The reversible model and the unrestricted model were estimated with R package RPHAST (Supplemental Table S8; Hubisz et al. 2011).
Types of methylome evolution events
We inferred HMRs in ancestral species using the proposed interdependent-site phylo-epigenetic model. Methylome evolution events—HMR gain in the form of birth and extension and HMR loss in the form of death and contraction—along each branch of the phylogenetic tree were determined by comparison of HMRs in the corresponding parent and child species. We filtered out events of size <50 bp or located at the boundaries of seven-way orthologous genome fragments.
To examine the enrichment of de novo (birth or death) and secondary (extension or contraction) methylome evolution events on each phylogenetic tree branch, we computed the distances between HMR gains to the closest ancestral HMRs, and between HMR losses to the closest descendant HMRs. We formed the expected distance distribution by randomly shuffling HMR gain events within all methylated regions in the ancestral methylome and HMR loss events within ancestral HMRs, using BEDTools (Quinlan and Hall 2010). The enrichment of different types of HMR gain and loss events was measured with the ratio between the observed and expected distance distribution (Supplemental Fig. S6).
CpG enrichment at promoter HMRs
In the human-referenced seven-way orthologous methylome alignment, we filtered for human and mouse sperm HMRs such that both their intersection and union contain exactly one gene transcription start site (TSS). We obtained human promoter HMR extension regions relative to the mouse promoter HMRs with minimum length of 200 bp. The two boundaries of such an extension region correspond to a human HMR boundary and a mouse HMR boundary. We masked out regions that are not alignable between human and mouse according to their pairwise sequence alignment. We used a sliding 2-bp window with 1-bp step size around the human HMR boundary of the extended HMR, the mouse HMR boundary contained within a wider human HMR, and around TSS along the human genome to measure GC content and CpG occurrences, and produced a meta plot for CpG enrichment relative to the boundaries and TSS. To make these measurements in the mouse genome, we mapped human methylomes to mouse mm10 genome assembly through the mouse-referenced MULTIZ alignment of 60 species and followed the same analysis procedures. We performed the same analysis for dog–mouse comparison using mouse-referenced and dog-referenced methylome alignments.
Relative sequence substitution rate
We used 15-way eutherian mammals Enredo-Pecan-Ortheus (EPO) multiple alignments corresponding to the Release 75 of Ensembl (Herrero et al. 2016), and we extracted the alignment of the seven species in this study and the ancestral species in the corresponding phylogeny. Sequence substitutions were annotated to individual lineages by comparing the ancestral sequence and the descendant sequence. We only considered alignment columns that have no insertion or deletion in all seven species. Relative sequence substitution rate (RSSR) in orthologous regions between two sister lineages is defined as the ratio of the total number of substitutions on one lineage to the total number of substitutions on the other lineage. To form the background distribution of RSSR, we randomly sampled regions from the seven-way orthologous genome. The sampled region size was chosen to match the smaller size of the total aligned bases in lineage-specific HMRs between the two lineages. We independently sampled 5000 times and created the sampling distribution of RSSR for each pairs of sister lineages. The significance (one-sided P-value) of observed RSSRs in lineage-specific HMRs was calculated using a normal distribution fit to the empirical sampling results.
Enrichment of transcription factor binding sites
We used ENCODE transcription factor binding sites for human and mouse (Gerstein et al. 2012; The ENCODE Project Consortium 2012; Wang et al. 2013) and mouse EZH2 binding sites from studies by Ku et al. (2008) and Peng et al. (2009). To evaluate enrichment of the binding sites of each TF in, for example, human lineage-specific HMRs, we used Fisher's exact test on the contingency table of binding site counts: (factor, other factors) × (lineage-specific HMRs, other human HMRs). See Supplemental Methods for more details.
Gene ontology analyses
For genes with promoter HMR extensions in the primate lineage, we used GOrilla (Eden et al. 2009) to identify enriched biological processes at a false discovery rate of 0.05 and removed ontology term redundancy with REVIGO (Supek et al. 2011). For genes closest to lineage-specific HMR gains that are common between parallel lineages, we used the PANTHER overrepresentation test (Mi et al. 2016) to find overrepresented biological processes with <0.05 Bonferroni corrected P-value. More details can be found in Supplemental Methods.
Data access
The whole-genome bisulfite sequencing data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE79566. Methylome alignment, inference of ancestral methylation states, and HMRs are provided in UCSC Genome Browser Track Data Hubs (Raney et al. 2014) as part of the public hub DNA Methylation (Song et al. 2013). Custom analysis scripts are available as Supplemental Scripts.
Supplementary Material
Acknowledgments
We thank Dr. Tara Stoinski, Zoo Atlanta, for the gorilla sample, and Dr. Beckie Williams, Yorba Regional Animal Hospital, for dog samples. We thank Michael Kessler for sample collection; Brent Young for dog sperm DNA extraction. We thank Dr. Joseph G. Hacia, Dr. Peter Calabrese, and Giorgia Battistoni for helpful discussions and comments. We thank Vanderbilt Technologies for Advanced Genomics (VANTAGE) and BGI (formerly Beijing Genomics Institute) for sequencing services. We thank the University of Southern California High Performance Computing Centers for computational support. This research was partially supported by National Institutes of Health grant R01 HG005238 to A.D.S.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.225896.117.
References
- Arnosti DN, Kulkarni MM. 2005. Transcriptional enhancers: intelligent enhanceosomes or flexible billboards? J Cell Biochem 94: 890–898. [DOI] [PubMed] [Google Scholar]
- Atsem S, Reichenbach J, Potabattula R, Dittrich M, Nava C, Depienne C, Böhm L, Rost S, Hahn T, Schorsch M, et al. 2016. Paternal age effects on sperm FOXK1 and KCNA7 methylation and transmission into the next generation. Hum Mol Genet 25: 4996–5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauernfeind AL, Soderblom EJ, Turner ME, Moseley MA, Ely JJ, Hof PR, Sherwood CC, Wray GA, Babbitt CC. 2015. Evolutionary divergence of gene and protein expression in the brains of humans and chimpanzees. Genome Biol Evol 7: 2276–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benayoun BA, Pollina EA, Ucar D, Mahmoudi S, Karra K, Wong ED, Devarajan K, Daugherty AC, Kundaje AB, Mancini E, et al. 2014. H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell 158: 673–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AP. 1980. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res 8: 1499–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev 16: 6–21. [DOI] [PubMed] [Google Scholar]
- Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. 2010. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol Chapter 19: Unit 19.10.1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgel J, Guibert S, Li Y, Chiba H, Schübeler D, Sasaki H, Forné T, Weber M. 2010. Targets and dynamics of promoter DNA methylation during early mouse development. Nat Genet 42: 1093–1100. [DOI] [PubMed] [Google Scholar]
- Bourc'his D, Bestor TH. 2004. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431: 96–99. [DOI] [PubMed] [Google Scholar]
- Branco MR, King M, Perez-Garcia V, Bogutz AB, Caley M, Fineberg E, Lefebvre L, Cook SJ, Dean W, Hemberger M, et al. 2016. Maternal DNA methylation regulates early trophoblast development. Dev Cell 36: 152–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, Roloff TC, Beisel C, Schübeler D, Stadler MB, Peters AH. 2010. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol 17: 679–687. [DOI] [PubMed] [Google Scholar]
- Capra JA, Kostka D. 2014. Modeling DNA methylation dynamics with approaches from phylogenetics. Bioinformatics 30: i408–i414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona FJ, Davalos V, Vidal E, Gomez A, Heyn H, Hashimoto Y, Vizoso M, Martinez-Cardus A, Sayols S, Ferreira HJ, et al. 2014. A comprehensive DNA methylation profile of epithelial-to-mesenchymal transition. Cancer Res 74: 5608–5619. [DOI] [PubMed] [Google Scholar]
- Carone BR, Hung JH, Hainer SJ, Chou MT, Carone DM, Weng Z, Fazzio TG, Rando OJ. 2014. High-resolution mapping of chromatin packaging in mouse embryonic stem cells and sperm. Dev Cell 30: 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrell DT, Hammoud SS. 2010. The human sperm epigenome and its potential role in embryonic development. Mol Hum Reprod 16: 37–47. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Ma Z, Kim BH, Wu W, Cayting P, Boyle AP, Sundaram V, Xing X, Dogan N, Li J, et al. 2014. Principles of regulatory information conservation between mouse and human. Nature 515: 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen NM, Kenigsberg E, Tanay A. 2011. Primate CpG islands are maintained by heterogeneous evolutionary regimes involving minimal selection. Cell 145: 773–786. [DOI] [PubMed] [Google Scholar]
- dos Reis M, Inoue J, Hasegawa M, Asher RJ, Donoghue PC, Yang Z. 2012. Phylogenomic datasets provide both precision and accuracy in estimating the timescale of placental mammal phylogeny. Proc R Soc Lond B Biol 279: 3491–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos CO, Dolzhenko E, Hodges E, Smith AD, Hannon GJ. 2015. An epigenetic memory of pregnancy in the mouse mammary gland. Cell Rep 11: 1102–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17: 368–376. [DOI] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, et al. 2014. Ensembl 2014. Nucleic Acids Res 42: D749–D755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher SJ, Kofman AE, Huszar JM, Dannenberg JH, DePinho RA, Braun RE, Payne CJ. 2013. Distinct requirements for Sin3a in perinatal male gonocytes and differentiating spermatogonia. Dev Biol 373: 83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M. 1987. CpG islands in vertebrate genomes. J Mol Biol 196: 261–282. [DOI] [PubMed] [Google Scholar]
- Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et al. 2012. Architecture of the human regulatory network derived from ENCODE data. Nature 489: 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, et al. 2005. Galaxy: a platform for interactive large-scale genome analysis. Genome Res 15: 1451–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J; The Galaxy Team. 2010. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11: R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, Surani MA. 2013. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science 339: 448–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud SS, Low DHP, Yi C, Carrell DT, Guccione E, Cairns BR. 2014. Chromatin and transcription transitions of mammalian adult germline stem cells and spermatogenesis. Cell Stem Cell 15: 239–253. [DOI] [PubMed] [Google Scholar]
- Hedges SB, Marin J, Suleski M, Paymer M, Kumar S. 2015. Tree of life reveals clock-like speciation and diversification. Mol Biol Evol 32: 835–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijmans BT, Kremer D, Tobi EW, Boomsma DI, Slagboom PE. 2007. Heritable rather than age-related environmental and stochastic factors dominate variation in DNA methylation of the human IGF2/H19 locus. Hum Mol Genet 16: 547–554. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Greally JM. 2016. Epigenetics, cellular memory and gene regulation. Curr Biol 26: R644–R648. [DOI] [PubMed] [Google Scholar]
- Hernando-Herraez I, Heyn H, Fernandez-Callejo M, Vidal E, Fernandez-Bellon H, Prado-Martinez J, Sharp AJ, Esteller M, Marques-Bonet T. 2015. The interplay between DNA methylation and sequence divergence in recent human evolution. Nucleic Acids Res 43: 8204–8214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero J, Muffato M, Beal K, Fitzgerald S, Gordon L, Pignatelli M, Vilella AJ, Searle SMJ, Amode R, Brent S, et al. 2016. Ensembl comparative genomics resources. Database 2016: bav096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges E, Molaro A, Dos Santos CO, Thekkat P, Song Q, Uren PJ, Park J, Butler J, Rafii S, McCombie WR, et al. 2011. Directional DNA methylation changes and complex intermediate states accompany lineage specificity in the adult hematopoietic compartment. Mol Cell 44: 17–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubisz MJ, Pollard KS, Siepel A. 2011. PHAST and RPHAST: phylogenetic analysis with space/time models. Brief Bioinform 12: 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. 2009. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet 41: 178–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MD, Mueller M, Adamowicz-Brice M, Collins MJ, Gellert P, Maratou K, Srivastava PK, Rotival M, Butt S, Game L, et al. 2014. Genetic analysis of the cardiac methylome at single nucleotide resolution in a model of human cardiovascular disease. PLoS Genet 10: e1004813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA. 2012. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13: 484–492. [DOI] [PubMed] [Google Scholar]
- Jung YH, Sauria ME, Lyu X, Cheema MS, Ausio J, Taylor J, Corces VG. 2017. Chromatin states in mouse sperm correlate with embryonic and adult regulatory landscapes. Cell Rep 18: 1366–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaman MW, Houck ML, Chemnick LG, Nagpal S, Chawannakul D, Sudano D, Pike BL, Ho VV, Ryder OA, Hacia JG. 2003. Comparative analysis of gene-expression patterns in human and African great ape cultured fibroblasts. Genome Res 13: 1619–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. 2002. The human genome browser at UCSC. Genome Res 12: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer-Kwon KR, Tang Z, Mathe E, Qian J, Sung MH, Li G, Resch W, Baek S, Pruett N, Grøntved L, et al. 2013. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell 155: 1507–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Wilson AC. 1975. Evolution at two levels in humans and chimpanzees. Science 188: 107–116. [DOI] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. 2008. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4: e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, et al. 2015. Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Sung KW, Rigoutsos I, Loring J, et al. 2010. Dynamic changes in the human methylome during differentiation. Genome Res 20: 320–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch BJ, Silber SJ, McCarrey JR, Page DC. 2016. Parallel evolution of male germline epigenetic poising and somatic development in animals. Nat Genet 48: 888–894. [DOI] [PubMed] [Google Scholar]
- Lienert F, Wirbelauer C, Som I, Dean A, Mohn F, Schübeler D. 2011. Identification of genetic elements that autonomously determine DNA methylation states. Nat Genet 43: 1091–1097. [DOI] [PubMed] [Google Scholar]
- Lin C, Garruss AS, Luo Z, Guo F, Shilatifard A. 2013. The RNA Pol II elongation factor Ell3 marks enhancers in ES cells and primes future gene activation. Cell 152: 144–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, Washietl S, Kheradpour P, Ernst J, Jordan G, Mauceli E, et al. 2011. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478: 476–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, et al. 2013. Global epigenomic reconfiguration during mammalian brain development. Science 341: 1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Yuan Z, Tan T, Wang J, Zhang J, Luo HJ, Xia Y, Ji W, Gao F. 2015. Improved tagmentation-based whole-genome bisulfite sequencing for input DNA from less than 100 mammalian cells. Epigenomics 7: 47–56. [DOI] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ III, Voigt P, Martin SR, Taylor WR, De Marco V, et al. 2009. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461: 762–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DI, Singer M, Dhahbi J, Mao G, Zhang L, Schroth GP, Pachter L, Boffelli D. 2011. Phyloepigenomic comparison of great apes reveals a correlation between somatic and germline methylation states. Genome Res 21: 2049–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454: 766–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD. 2016. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res 44: D336–D342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milekic MH, Xin Y, O'Donnell A, Kumar KK, Bradley-Moore M, Malaspina D, Moore H, Brunner D, Ge Y, Edwards J, et al. 2015. Age-related sperm DNA methylation changes are transmitted to offspring and associated with abnormal behavior and dysregulated gene expression. Mol Psychiatry 20: 995. [DOI] [PubMed] [Google Scholar]
- Molaro A, Hodges E, Fang F, Song Q, McCombie WR, Hannon GJ, Smith AD. 2011. Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates. Cell 146: 1029–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molaro A, Falciatori I, Hodges E, Aravin AA, Marran K, Rafii S, McCombie WR, Smith AD, Hannon GJ. 2014. Two waves of de novo methylation during mouse germ cell development. Genes Dev 28: 1544–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorjani P, Amorim CEG, Arndt PF, Przeworski M. 2016. Variation in the molecular clock of primates. Proc Natl Acad Sci 113: 10607–10612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu W, Starmer J, Fedoriw AM, Yee D, Magnuson T. 2014. Repression of the soma-specific transcriptome by Polycomb-repressive complex 2 promotes male germ cell development. Genes Dev 28: 2056–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri F, Incarnato D, Krepelova A, Rapelli S, Pagnani A, Zecchina R, Parlato C, Oliviero S. 2013. Genome-wide analysis identifies a functional association of Tet1 and Polycomb repressive complex 2 in mouse embryonic stem cells. Genome Biol 14: R91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom DT, Dowell RD, Jacobsen ES, Gordon W, Danford TW, MacIsaac KD, Rolfe PA, Conboy CM, Gifford DK, Fraenkel E. 2007. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat Genet 39: 730–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai AA, Bell JT, Marioni JC, Pritchard JK, Gilad Y. 2011. A genome-wide study of DNA methylation patterns and gene expression levels in multiple human and chimpanzee tissues. PLoS Genet 7: e1001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J. 2009. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell 139: 1290–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. 2010. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res 20: 110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raney BJ, Dreszer TR, Barber GP, Clawson H, Fujita PA, Wang T, Nguyen N, Paten B, Zweig AS, Karolchik D, et al. 2014. Track data hubs enable visualization of user-defined genome-wide annotations on the UCSC Genome Browser. Bioinformatics 30: 1003–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rat Genome Sequencing Project Consortium. 2004. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428: 493–521. [DOI] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J. 2001. Epigenetic reprogramming in mammalian development. Science 293: 1089–1093. [DOI] [PubMed] [Google Scholar]
- Rodgers AB, Morgan CP, Leu NA, Bale TL. 2015. Transgenerational epigenetic programming via sperm microRNA recapitulates effects of paternal stress. Proc Natl Acad Sci 112: 13699–13704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom KR, Armstrong J, Barber GP, Casper J, Clawson H, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, et al. 2015. The UCSC genome browser database: 2015 update. Nucleic Acids Res 43: D670–D681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, Kutter C, Watt S, Martinez-Jimenez CP, Mackay S, et al. 2010. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 328: 1036–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegmund KD, Marjoram P, Woo YJ, Tavaré S, Shibata D. 2009. Inferring clonal expansion and cancer stem cell dynamics from DNA methylation patterns in colorectal cancers. Proc Natl Acad Sci 106: 4828–4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Haussler D. 2004. Phylogenetic estimation of context-dependent substitution rates by maximum likelihood. Mol Biol Evol 21: 468–488. [DOI] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit A, Hubley R, Green P. 2013–2015. RepeatMasker Open-4.0. http://www.repeatmasker.org. [Google Scholar]
- Smith ZD, Meissner A. 2013. DNA methylation: roles in mammalian development. Nat Rev Genet 14: 204–220. [DOI] [PubMed] [Google Scholar]
- Smith AD, Chung WY, Hodges E, Kendall J, Hannon G, Hicks J, Xuan Z, Zhang MQ. 2009. Updates to the RMAP short-read mapping software. Bioinformatics 25: 2841–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A. 2012. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 484: 339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q, Decato B, Hong EE, Zhou M, Fang F, Qu J, Garvin T, Kessler M, Zhou J, Smith AD. 2013. A reference methylome database and analysis pipeline to facilitate integrative and comparative epigenomics. PLoS One 8: e81148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Schöler A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. 2011. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 480: 490–495. [DOI] [PubMed] [Google Scholar]
- Supek F, Bosnjak M, Skunca N, Smuc T. 2011. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6: e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teperek M, Simeone A, Gaggioli V, Miyamoto K, Allen GE, Erkek S, Kwon T, Marcotte EM, Zegerman P, Bradshaw CR, et al. 2016. Sperm is epigenetically programmed to regulate gene transcription in embryos. Genome Res 26: 1034–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. 2012. The accessible chromatin landscape of the human genome. Nature 489: 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuğrul M, Paixão T, Barton NH, Tkačik G. 2015. Dynamics of transcription factor binding site evolution. PLoS Genet 11: e1005639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung J, Barreiro LB, Johnson ZP, Hansen KD, Michopoulos V, Toufexis D, Michelini K, Wilson ME, Gilad Y. 2012. Social environment is associated with gene regulatory variation in the rhesus macaque immune system. Proc Natl Acad Sci 109: 6490–6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al. 2015. Tissue-based map of the human proteome. Science 347: 1260419. [DOI] [PubMed] [Google Scholar]
- Villar D, Berthelot C, Aldridge S, Rayner TF, Lukk M, Pignatelli M, Park TJ, Deaville R, Erichsen JT, Jasinska AJ, et al. 2015. Enhancer evolution across 20 mammalian species. Cell 160: 554–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CP, Chaillet JR, Bestor TH. 1998. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet 20: 116–117. [DOI] [PubMed] [Google Scholar]
- Wang J, Zhuang J, Iyer S, Lin XY, Greven MC, Kim BH, Moore J, Pierce BG, Dong X, Virgil D, et al. 2013. Factorbook.org: a Wiki-based database for transcription factor-binding data generated by the ENCODE consortium. Nucleic Acids Res 41: D171–D176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhang J, Duan J, Gao X, Zhu W, Lu X, Yang L, Zhang J, Li G, Ci W, et al. 2014. Programming and inheritance of parental DNA methylomes in mammals. Cell 157: 979–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei GCG, Tanner MA. 1990. A Monte Carlo implementation of the EM algorithm and the poor man's data augmentation algorithms. J Am Stat Assoc 85: 699–704. [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PAC, Rappsilber J, Helin K. 2011. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473: 343–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 1994. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J Mol Evol 39: 306–314. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.