Abstract
The secretion of virulence factors by parasitic protists into the host environment plays a fundamental role in multifactorial host–parasite interactions. Several effector proteins are known to be secreted by Trichomonas vaginalis, a human parasite of the urogenital tract. However, a comprehensive profiling of the T. vaginalis secretome remains elusive, as do the mechanisms of protein secretion. In this study, we used high-resolution label-free quantitative MS to analyze the T. vaginalis secretome, considering that secretion is a time- and temperature-dependent process, to define the cutoff for secreted proteins. In total, we identified 2 072 extracellular proteins, 89 of which displayed significant quantitative increases over time at 37 °C. These 89 bona fide secreted proteins were sorted into 13 functional categories. Approximately half of the secreted proteins were predicted to possess transmembrane helixes. These proteins mainly include putative adhesins and leishmaniolysin-like metallopeptidases. The other half of the soluble proteins include several novel potential virulence factors, such as DNaseII, pore-forming proteins, and β-amylases. Interestingly, current bioinformatic tools predicted the secretory signal in only 18% of the identified T. vaginalis-secreted proteins. Therefore, we used β-amylases as a model to investigate the T. vaginalis secretory pathway. We demonstrated that two β-amylases (BA1 and BA2) are transported via the classical endoplasmic reticulum-to-Golgi pathways, and in the case of BA1, we showed that the protein is glycosylated with multiple N-linked glycans of Hex5HexNAc2 structure. The secretion was inhibited by brefeldin A but not by FLI-06. Another two β-amylases (BA3 and BA4), which are encoded in the T. vaginalis genome but absent from the secretome, were targeted to the lysosomal compartment. Collectively, under defined in vitro conditions, our analysis provides a comprehensive set of constitutively secreted proteins that can serve as a reference for future comparative studies, and it provides the first information about the classical secretory pathway in this parasite.
Trichomonas vaginalis is an anaerobic, aerotolerant pathogen that causes trichomoniasis, the most widespread nonviral sexually transmitted disease in humans. Although the majority of infections are asymptomatic, approximately one-third of infected women develop symptoms such as vaginitis and urethritis (1). In addition, trichomonad infection has been associated with poor birth outcomes and increased risk of Human Immunodeficiency Virus (HIV) acquisition (2). In men, the infection is rarely symptomatic; however, the parasite can damage sperm cells (3, 4), and chronic infection has been associated with prostate cancer (5, 6).
In the female urogenital tract, T. vaginalis is challenged by factors such as nutrient limitation, the host immune response, physiological changes during the menstrual cycle, the continual flow of vaginal fluid, and coexistence with other members of the vaginal microbiota (7). Upon transmission to men, the parasite must adapt to the different environmental conditions within the male urogenital tract, including increased concentrations of zinc in the prostatic fluid that may kill the parasite (8, 9). Thus, the establishment of trichomonad infection within such hostile environments is dependent on multifactorial host–parasite interactions that involve both contact-dependent and contact-independent mechanisms (10). The former include the adherence of the parasite to vaginal epithelial cells, the contact-dependent extracellular killing of host cells (11–14), and active phagocytosis of host cells and bacteria (15, 16). The contact-independent mechanisms include the secretion of soluble biologically active molecules, particularly proteases with diverse effects (10, 17, 18). Finally, T. vaginalis has been shown to pack specific sets of macromolecules into microvesicles (exosomes) that are secreted and that influence the parasite's binding to the host cell (19).
With regard to nutrients, the energy metabolism of T. vaginalis is dependent on glucose to generate ATP via anaerobic fermentation in the cytosol and via the extended glycolytic pathway in hydrogenosomes, an anaerobic form of mitochondria (20, 21). The main source of glucose in the vaginal fluid is likely free glycogen derived from vaginal epithelial cells (VECs) (22–26). To be utilized by T. vaginalis, glycogen and glucose-containing polymers must be extracellularly digested to monomeric glucose, which is then transported into the cells. Glycogen-hydrolyzing enzymes include endo-acting α-amylases (EC 3.2.1.1) that randomly hydrolyze α-1,4-linkages of glycogen, exo-acting β-amylases (EC 3.2.1.2) that hydrolyze α-1,4-linkages of glycogen at the nonreducing end to liberate β-maltose, and α-glucosidases (EC 3.2.1.20) that act on α-1,4-linkages of oligosaccharides to liberate d-glucose. Early studies suggested that T. vaginalis secretes α-glucosidase to hydrolyze maltose to glucose (27). More recently, enzymes with α-amylase and β-amylase activities that utilize glycogen as a substrate were found to be released by T. vaginalis (28, 29).
High-resolution mass-spectrometry-based proteomic studies have been used to analyze the T. vaginalis surface proteome (30) and the exosome proteome (19), which have revealed a number of new candidate proteins with potential roles in T. vaginalis–host interactions and in the parasite's pathogenicity. More recent quantitative proteomic analyses have identified surface membrane proteins that are released to the T. vaginalis environment upon cleavage by rhomboid protease (31). The best-studied group of secreted proteins is the proteases, including cysteine proteases and metalloproteases (17, 32, 33). Kucknoor et al. (34) identified 32 various secreted proteins, including a putative adhesin, AP65, via 2-D SDS-PAGE followed by MALDI-TOF (34). In addition, Twu et al. showed that the parasite secretes a macrophage migration inhibitory factor (5). However, information about the T. vaginalis secretome remains rather incomplete.
The major challenge for studies of the secretome using high-resolution MS is to identify bona fide secreted proteins and avoid artifacts caused by protein contamination. Here, we used quantitative MS and considered the fact that secretion is a time- and temperature-dependent process in defining the cutoff for T. vaginalis-secreted proteins. After bioinformatic sorting of the secreted proteins, we focused on β-amylases as model secreted proteins to investigate the T. vaginalis secretory pathway. Moreover, β-amylases are absent from humans and animals and may provide a suitable target for the development of novel antiparasitic strategies.
EXPERIMENTAL PROCEDURES
Cell Cultivation
T. vaginalis strain Tv17–48 was isolated from a symptomatic patient, and the axenic culture was immediately stored in liquid nitrogen (35). The strain was cultivated in tryptone-yeast extract-maltose medium (TYM) supplemented with 10% inactivated horse serum (36).
Cell Incubation and Sample Preparation
T. vaginalis cells in the logarithmic phase of growth were harvested by centrifugation and washed twice in isotonic Doran's medium (37) with 15 mm maltose (Doran's medium with maltose). The cells were then resuspended at a concentration of 1 × 106 cells/ml, and 15 ml of suspension was incubated in 15 ml tubes for 10, 30, 60, and 120 min at 37 °C. Control cells were incubated for 60 and 120 min on ice. After incubation, the cells were removed by centrifugation at 1,000 × g for 5 min at 4 °C, and then the supernatant was centrifuged at 10,000 × g for 10 min to remove cell debris, filtered through a 0.22 μm filter, and centrifuged at 100,000 × g for 75 min to remove microvesicles (5). The proteins in the final supernatant were precipitated with TCA for 10 min at 4 °C (one volume of TCA to four volumes of supernatant). The precipitated proteins were pelleted at 12,000 × g for 20 min at 4 °C, washed with cold acetone, dried, and stored at −80 °C.
Cell Integrity Assays
During the incubation described above, the cell integrity was monitored under a light microscope using the trypan blue exclusion test (38). In parallel, at each time point, we determined the free activity of the cytosolic enzyme NADH oxidase in the cell suspension (39). In addition, aliquots of trichomonad suspensions taken at each time point were processed for transmission electron microscopy. The cell samples were centrifuged at 3 000 × g for 10 min and fixed in 2.5% glutaraldehyde and 5 mm CaCl2 in 0.1 m cacodylate buffer, pH 7.2, overnight at 4 °C. The cells were then postfixed in 0.1 m cacodylate buffer containing 1.6% ferricyanide, 10 mm CaCl2, and 2% OsO4 at 4 °C for 15 min, dehydrated in acetone, and embedded in the epoxy resin EMBed 812 (Electron Microscopy Sciences, Hatfield, PA, USA). Ultrathin sections were stained with uranyl acetate and observed using a JEOL JEM-1011.
Protein Preparation
The cell-free samples of TCA-precipitated proteins were dissolved in 100 mm triethylammonium bicarbonate buffer with 2% sodium deoxycholate, reduced with 5 mm tris(2-carboxyethyl)phosphine for 30 min at 60 °C, and alkylated with 10 mm S-methyl methanethiosulfonate for 10 min at room temperature. Total protein concentrations were measured via the bicinchoninic acid assay (Sigma-Aldrich, St. Louis, MO, USA). Next, 100 μg of proteins were digested with trypsin (trypsin:protein ratio 1:50) overnight at 37 °C. After digestion, 1% trifluoroacetic acid (TFA) was added. Sodium deoxycholate was removed by extraction to ethyl acetate as previously described (40). The remaining ethyl acetate was removed using vacuum centrifugation at 45 °C for 10 min, and then 1% TFA was added. The samples were desalted using C18 sorbent (Supelco 66883-U, supplied by Sigma-Aldrich). The eluents were dried and resuspended in 20 μl of 1% TFA.
Mass Spectrometry Data Acquisition
A nano reversed-phase column (EASY-Spray column, 50 cm × 75 μm inner diameter, PepMap C18, 2 μm particles, 100 Å pore size) was used for nanoLC-MS analysis. Mobile phase buffer A consisted of water, 2% acetonitrile, and 0.1% formic acid. Mobile phase B consisted of 80% acetonitrile and 0.1% formic acid. Two micrograms of each sample were loaded onto the trap column (Acclaim PepMap300, C18, 5 μm, 300 Å Wide Pore, 300 μm × 5 mm) at a flow rate of 15 μl/min. The loading buffer consisted of water, 2% acetonitrile, and 0.1% TFA. Peptides were eluted with a gradient from 2% to 40% B over 60 min at a flow rate of 300 nl/min. The peptide cations eluted were converted to gas-phase ions via electrospray ionization and analyzed on a Thermo Orbitrap Fusion (Q-OT- qIT, Thermo Fisher Scientific, Waltham, MA, USA). Spectra were acquired with a 2 s duty cycle. Full MS spectra were acquired in the Orbitrap within a mass range of 350–1,400 m/z, at a resolution of 120,000 at 200 m/z and with a maximum injection time of 50 ms. The most intense precursors were isolated by quadrupole ion trapping with a 1.6 m/z isolation window and fragmented via higher-energy collisional dissociation with the collision energy set to 30%. Fragment ions were detected in the ion trap with the scan range mode set to normal and the scan rate set to rapid with a maximum injection time of 35 ms. The fragmented precursors were excluded from fragmentation for 60 s.
Analysis of Mass Spectrometry Data
For label-free quantification (LFQ), the data were processed in MaxQuant LFQ version 1.5.8.3 (41). Searches were performed using the latest version of the T. vaginalis database from UniProt (release 2017_4, 60,330 entries) and a common contaminant database. Trypsin was used to generate the peptides, and two missed cleavages were allowed. The protein modifications were set as follows: cysteine (unimod nr: 39) as static and methionine oxidation (unimod: 1384) and protein N terminus acetylation (unimod: 1) as variable. The precursor ion mass tolerance in the initial search was 20 ppm, the tolerance in the main search was 4.5 ppm, and the fragment ion mass tolerance was 0.5 Da. The false discovery rates for peptides and for proteins were set to 1%.
For each identified protein, the cell localization and secretory pathway signal were predicted using the SignalP 4.1 server (http://www.cbs.dtu.dk/services/TMHMM/), TargetP 1.1 server (http://www.cbs.dtu.dk/services/TargetP/), and SecretomeP 2.0 server (http://www.cbs.dtu.dk/services/SecretomeP/). Transmembrane helixes and topology were predicted using the TMHMM server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/). Conserved domains were predicted using Pfam 31.0 (http://pfam.xfam.org/), and distant homologies were detected using the HHpred search against the CD database https://toolkit.tuebingen.mpg.de and Evolutionary Classification of Protein Domains (42) Molecular function gene ontology (http://geneontology.org/page/molecular-function-ontology-guidelines) and manual curation were used to sort the identified proteins.
Experimental Design and Statistical Rationale
Three independent biological experiments were performed, and each biological sample was analyzed in three technical replicates using LFQ mass spectrometry. Proteins with LFQ values determined in at least two biological replicates with two valid values within the technical replicates were used for further processing. Changes in the LFQ values between two consecutive time points were calculated for each biological replicate as the difference in the LFQ binary logarithm between the means of technical replicates. The significance of each change was estimated using Student's t test. To distinguish actively released proteins from contaminants, we used two criteria: (i) the secreted protein displayed more significant increases than decreases in LFQ values over time, and (ii) the difference between the LFQ values for a given protein at 37 °C and 4 °C was greater than 1 LFQ unit. The secretion score (SecS) was then calculated as the sum of all decreases and significant increases (p value < 0.05). Hierarchical clustering was performed using the standard UPGMA hierarchical clustering method with the Scipy package (https://www.scipy.org/). Secreted proteins were ordered according to the means of LFQ values of technical replicates at 37 °C. Boxplot analysis of dominant clusters was performed based on the ratio [sum LFQ values (10 min, 30 min)+1]/[sum LFQ values (60 min, 120 min)+1]. The median, 25th, and 75th percentiles were computed for each cluster using the Scipy package (https://www.scipy.org/).
Glycopeptide MS Analysis
The BA1 coding gene was subcloned into the modified TagVag vector (43) to allow for the expression of BA1 in T. vaginalis with a streptavidin tag at the C terminus. The logarithmic cell culture (2.5 liter) was harvested, the cells were broken by sonication, and the cell lysate was spun down by ultracentrifugation (100,000 × g for 25 min at 4 °C). Tagged BA1 was isolated from the supernatant using the Strep-Tactin system (IBA GmbH, Göttingen, Germany). The purity of the isolated protein was checked by SDS-PAGE.
Recombinant BA1 protein was transferred to 50 mm ammonium bicarbonate buffer, pH 7.8, with Amicon Ultra 0.5 ml centrifugal filters MWCO 3 kDa (Merck, Darmstadt, Germany). Twenty micrograms of BA1 protein were reduced by dithiothreitol (Sigma-Aldrich) and alkylated by iodoacetamide (Sigma-Aldrich). The protein was digested by trypsin (Promega, Madison, WI, USA) overnight at 37 °C. To reduce the size of certain tryptic peptides, the endoproteinase Glu-C (Roche, Basel, Switzerland) was added to the sample and incubated overnight at 25 °C. The peptide mixture was separated by a reversed-phase HPLC connected to a 15 T solariX XR mass spectrometer (Bruker Daltonics, Billerica, MA USA) operating in data-dependent mode. Data were processed by the DataAnalysis 4.2 software (Bruker Daltonics), and glycopeptides were identified by manual data curation based upon measured mass values. Annotated, mass-labeled spectra for all glycopeptides identified are presented in Fig. S1A–S1G.
β-Amylase Phylogeny
The T. vaginalis BA1 protein sequence was used as a query for a BLAST search in the NCBI RefSeq protein database (GenBank release 220.0), and 1,312 homologues were used to build a preliminary phylogeny using Fast Tree (44). Then, 166 representative sequences were manually selected and aligned using MAFFT (45), and the alignment was trimmed with BMGE (354 sites) (46). The phylogenetic tree was inferred using PhyloBayes (47) v. 4.1 under the CAT + GTR model with a burn-in of 1,000 generations and a postburn-in sampling of ∼30,000 generations. The maximum likelihood bootstrap support was calculated using PHYML (48) with the best-fit model (LG+I+G4) and 200 bootstrap replicates.
Quantitative Real-Time PCR
Trichomonads were grown in TYM medium lacking maltose with 5% inactivated dialyzed fetal bovine serum (Sigma-Aldrich) and supplemented with either maltose or oyster glycogen at a final concentration of 5%. The cells were subcultured twice under these conditions (48 h) prior to RNA isolation. The iron-depleted cells were subcultured daily in TYM medium supplemented with 5% inactivated dialyzed fetal bovine serum, 5% maltose, and 60 μm 2,2-dipyridyl (Sigma-Aldrich) for 5 days prior to RNA isolation.
An RNA Isolation Kit (Pharmacia, Whitehouse Station, NJ, USA) was used to extract the total RNA from the cells, and the complementary DNA was synthesized using a SuperScript VILO™ cDNA Synthesis Kit (Thermo Fisher Scientific). The RNA was isolated in quadruplicate. The DNATopII, α-tubulin, and actin genes were selected as reference genes for quantitative RT-PCR (qRT-PCR) analysis (49). The specific primers for the amplification of β-amylases are listed in Table S1. Gene expression levels were evaluated using a LightCycler 480 instrument (Roche). Each amplification reaction (10 μl) contained specific primers (400 nm final concentration), 20 ng of cDNA template, iQ SYBR Green Supermix (Bio-Rad, Hercules, CA, USA) and RNase-free water. PCR thermal cycling conditions: 95 °C for 3 min followed by 50 cycles of 95 °C for 10 s, 63 °C for 20 s, and 72 °C for 30 s. The specificity of the amplified PCR product was assessed by performing a melting curve analysis. The PCR data were analyzed using the GenEx software (MultiD Analyses). Relative gene expression levels were normalized to the expression levels of reference genes selected by the Normfinder software (https://moma.dk/normfinder-software).
Cellular Localization of Biotinylated β-Amylases
A partial sequence of T. vaginalis protein disulfide isomerase (TVAG_267400) encoding the first 75 amino acids (AAs) was amplified by PCR from genomic DNA and fused at the C terminus with Escherichia coli gene for biotin ligase (BirA, WP_023308552) amplified from pET21a-BirA (50). Protein disulfide isomerase-BirA was subcloned to modified pTagVag (43), in which the C-terminal hemagglutinin tag was replaced with the 2xV5 epitope tag and the KQEL sequence, and the neomycin phosphotransferase cassette was replaced with the puromycin-N-acetyltransferase gene (pTagVag-V5-Pur). The vector was introduced into T. vaginalis by electroporation as previously described (43), and transformants were selected in the presence of 40 μg/ml puromycin (strain Tv17–48-BirA).
The β-amylase-coding genes BA1–4 were amplified by PCR and subcloned into the modified pTagVag, into which we introduced a sequence encoding the biotin acceptor peptide (AP, GLNDIFEAQKIEWHE), which allowed for the production of recombinant proteins with a C-terminal AP tag. The BA1–4 genes were expressed under the control of the native promotor (ca. 500 bp of upstream noncoding sequences). The constructs were electroporated into the Tv17–48-BirA strain, and the double transformants were selected in the presence of 200 μg/ml geneticin and 40 μg/ml puromycin. All primers used for gene amplification and cloning are listed in Supplemental Table S1.
Immunofluorescence Microscopy
For biotin labeling, the double transformants were incubated in TYM medium with 1 mm biotin for 30 min at 37 °C. Cells were then fixed in 1% formaldehyde for 30 min, washed in PEM (100 mM PIPES, 1 mM EGTA, 0.1 mM MgSO4) buffer (51) and placed on high-performance cover glasses (Carl Zeiss, Oberkochen, Germany). The attached cells were treated in 1% Triton X-100 for 20 min in PEM and stained using rabbit polyclonal α-V5 Ab1 (Sigma-Aldrich) and the secondary antibody Alexa Fluor 594 donkey α-rabbit Ab (Life Technologies, Carlsbad, CA, USA). Biotinylated proteins were detected using streptavidin Alexa Fluor 488 conjugate (Life Technologies). Structured illumination microscopy (SIM) was performed on a 3D N-SIM microscope (Nikon Eclipse Ti-E, Nikon, Japan) equipped with a Nikon CFI SR Apo TIRF objective (100x oil, NA 1.49). The structured illumination pattern projected into the sample plane was created on a diffraction grating block (100 EX V-R 3D-SIM) at laser wavelengths of 488 and 561 nm. The excitation and emission light were separated by filter sets SIM488 (excitation 470–490, emission 500–545) and SIM561 (excitation 556–566 nm, emission 570–640 nm). The emission light was projected through a 2.5x relay lens onto the chip of an EM CCD camera (AndoriXon Ultra DU897, 10 MHz at 14-bit, 512 × 512 pixels, Andor Technology, Belfast, U.K.). Three-color z-stacks (z-step: 125 nm) were acquired in the NIS-Elements AR software (Laboratory Imaging, Prague, Czech Republic). The laser intensity, EM gain, and camera exposure time were set independently for each excitation wavelength. The intensity of the fluorescence signal was maintained within the linear range of the camera. Fifteen images (three rotations and five phase shifts) were recorded for every plane and color. The SIM data were processed in the NIS-Elements AR software. Before sample measurement, the symmetry of the point spread function was checked with 100 nm red fluorescent beads (580/605, Carboxylate-Modified Microspheres, Life Technologies) mounted in Prolong Diamond Antifade Mountant (Life Technologies) and optimized by adjusting the objective correction collar. The signal for 4,6-diamidine-2-phenylindole dihydrochloride (DAPI) was observed in wide-field mode.
Effect of Inhibitors on β-Amylase Glycosylation and Secretion
Trichomonads episomally expressing HA-tagged BA1–4 were inoculated into TYM medium (1 × 105 cells/ml) supplemented with 5 μg/ml tunicamycin (Sigma-Aldrich) and grown for 24 h. The control cells were grown without tunicamycin. After 24 h, the cells were harvested by centrifugation, and the cell lysate was examined by Western blot analysis with mouse monoclonal α-HA Ab (Sigma-Aldrich). Brefeldin A (Sigma-Aldrich) and FLI-06 (Sigma-Aldrich) were tested using T. vaginalis episomally expressing HA-tagged BA1–3. The cells (1 × 106 cells/ml of TYM without serum) were preincubated for 30 min at 24 °C with brefeldin A (50 μg/ml) or FLI-06 (50 μm), and then the cells were washed with the medium and incubated with the inhibitors for 5 and 60 min at 37 °C. The cells were then removed by centrifugation, and the proteins in the supernatant were precipitated by TCA as described above and examined by Western blot analysis with mouse monoclonal α-HA Ab. Western blots were quantified using ImageJ v1.47 (National Institute of Health, Bethesda, MD, USA), and statistical analysis was performed using the Mann-Whitney nonparametric test (GraphPad Software, Inc., La Jolla, CA, USA).
Treatment of β-Amylases with N-glycosidase F (PNGase F)
T. vaginalis strains expressing HA-tagged BA1–4 were harvested by centrifugation (∼2 × 106 cells), washed twice with 50 mm sodium phosphate buffer, pH 7.5, resuspended in 100 μl of the same buffer with 0.2% SDS and 100 mm β-mercaptoethanol, and denatured at 100 °C for 10 min. Then, Triton X-100 was added to a final concentration of 1%, and a 50 μl aliquot was treated with 1 U PNGase F (Sigma-Aldrich) at 37 °C for 120 min. After incubation, the sample was incubated at 100 °C for 5 min and examined by Western blot analysis with mouse monoclonal α-HA Ab.
Localization of BA4 in Lactoferrin-Labeled Vesicles
After preliminary experiments to optimize the conditions, trichomonads episomally expressing HA-tagged BA1–4 were incubated at 20 °C for 10 min in TYM with 450 μg/ml lactoferrin (Sigma-Aldrich) labeled with FITC as previously described (http://www.ridgeviewinstruments.com). The cells were then washed twice in TYM and processed for immunofluorescence microscopy as described above. BA1–4 were labeled using mouse monoclonal α-HA Ab (Sigma-Aldrich) and the secondary Alexa Fluor 594 donkey α-mouse Ab (Life Technologies).
RESULTS
Analysis of Secreted Proteins Using Quantitative Mass Spectrometry
To study the secretome of T. vaginalis, we developed a protocol for analyzing the time-dependent accumulation of extracellular proteins via high-resolution nano LC coupled with ESI-linear-ion trap and MS/MS LFQ. The cells were incubated in Doran's medium with maltose for 10, 30, 60, and 120 min at 37 °C, and the amounts of secreted proteins were analyzed at each time point. The control cells were incubated for 60 and 120 min on ice. The viability of the cells during incubation was monitored via a trypan blue exclusion test that revealed >99% (n = 12) viable cells during all experiments. The intactness of the cells was further monitored via an enzyme assay using NADH oxidase as a cytosolic marker enzyme (Table S2). In addition, the integrity of most cells was confirmed by transmission electron microscopy, which revealed that >95% (n = 6) of cells were intact. In some cells, we observed budding of extracellular microvesicles (ectosomes), and occasionally we found destroyed cells with hydrogenosomes (Fig. S2).
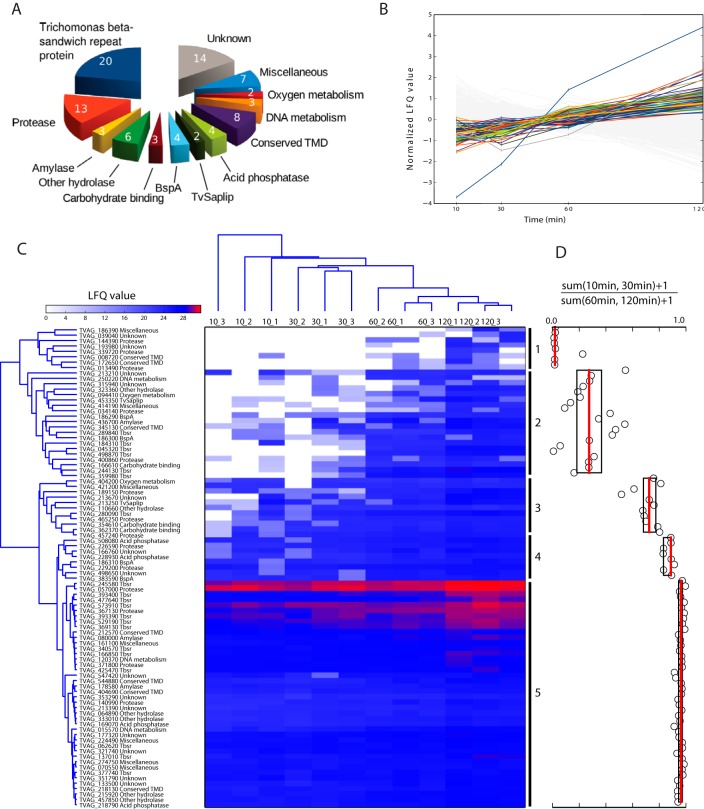
LFQ values were calculated for individual proteins at each time point. Altogether, we identified 2,072 proteins (Table S3). To distinguish bona fide secreted proteins from contaminants, we applied two main criteria. First, the amount of the secreted protein significantly increased over time (Fig. 1), and second, the LFQ values for a given protein at 37 °C were greater than LFQ values at 4 °C for more than 1 LFQ unit. Then, for each protein, we calculated SecS as the sum of the differences in LFQ values determined for each time point. The application of these criteria resulted in a list of 89 actively released proteins (Table I) that were manually sorted into 13 functional categories considering TrichoDB annotations, HHpred searches, Pfam motif identifications, Evolutionary Classification of Protein Domains classification, and molecular function gene ontology (GOMF) names (Fig. 1, Table I, and Table S3). Most categories contain both soluble proteins and proteins with predicted transmembrane domains (TMD) (Table I, Table S3).
Fig. 1.
Proteins secreted by T. vaginalis. (A) Functional categories. (B) Normalized LFQ values (binary logarithm) determined for proteins released between 10 and 120 min of incubation of T. vaginalis in Doran's medium with maltose at 37 °C. Secreted proteins are color-coded as in (A), and proteins in gray are under the defined cutoff. Accession numbers of color-coded proteins are given in Fig. S3. (C) Hierarchical clustering. The means of LFQ values of three technical replicates for three biological replicates were clustered over 10–120 min cell incubation at 37 °C. X_Y; X, time point in min; Y, code of biological experiment according to Table S3. (D) Five dominant clusters were tested by boxplot analysis as the ratio [sum LFQ values (10 min, 30 min)+1]/[sum LFQ values (60 min, 120 min)+1]. The median (red line) and 25th and 75th percentiles were computed for each cluster and are shown as boxplots. Circles indicate [sum LFQ values (10 min, 30 min)+1]/[sum LFQ values (60 min, 120 min)+1] calculated for each protein.
Table I. The list of T. vaginalis secreted proteins.
| Category | Accession No. | Annotation | Length | TMD | TMD-C | SignalP | SecretomeP | SecS |
|---|---|---|---|---|---|---|---|---|
| Acid phosphatase | TVAG_218790 | acid phosphatase, putative | 379 | 0 | 0.401 | 0.433 | 6.34 | |
| TVAG_508080 | acid phosphatase, putative | 391 | 0 | 0.304 | 0.470 | 4.99 | ||
| TVAG_169070 | acid phosphatase, putative | 394 | 0 | 0.356 | 0.406 | 3.45 | ||
| TVAG_228930 | acid phosphatase, putative | 390 | 0 | 0.272 | 0.486 | 3.42 | ||
| Amylase | TVAG_178580 | alpha-amylase, putative | 494 | 0 | 0.374 | 0.724 | 1.86 | |
| TVAG_080000 | beta-amylase, putative | 377 | 0 | 0.106 | 0.568 | 3.97 | ||
| TVAG_436700 | beta-amylase, putative | 428 | 0 | 0.335 | 0.654 | 0.67 | ||
| BspA | TVAG_186310 | leucine-rich repeat protein, BspA family | 1,363 | 0 | 0.109 | 0.644 | 5.85 | |
| TVAG_383590 | leucine-rich repeat protein, BspA family | 927 | 1 | 23 | 0.182 | 0.260 | 3.91 | |
| TVAG_186290 | leucine-rich repeat protein, BspA family | 1,474 | 0 | 0.133 | 0.697 | 1.43 | ||
| TVAG_186300 | leucine-rich repeat protein, BspA family | 1,434 | 0 | 0.554 | 0.710 | 1.37 | ||
| Carbohydrate binding | TVAG_362370 | carbohydrate-binding domain, FA58C | 224 | 0 | 0.416 | 0.813 | 4.76 | |
| TVAG_354610 | carbohydrate-binding domain, FA58C | 218 | 0 | 0.178 | 0.487 | 2.25 | ||
| TVAG_166610 | carbohydrate-binding module, cd14489 | 354 | 0 | 0.325 | 0.755 | 2.57 | ||
| Conserved TMD | TVAG_544880 | conserved protein | 290 | 1 | 23 | 0.109 | 0.256 | 7.33 |
| TVAG_345130 | conserved protein | 383 | 1 | 26 | 0.373 | 0.257 | 5.60 | |
| TVAG_218130 | conserved protein | 1,085 | 1 | 24 | 0.570 | 0.572 | 5.51 | |
| TVAG_404690 | conserved protein | 593 | 1 | 79 | 0.100 | 0.19 | 2.53 | |
| TVAG_212570 | conserved protein | 624 | 1 | 71 | 0.443 | 0.345 | 0.90 | |
| TVAG_172650 | conserved protein | 356 | 1 | 71 | 0.106 | 0.121 | 0.90 | |
| TVAG_008720 | conserved protein | 1,300 | 1 | 24 | 0.190 | 0.457 | 0.47 | |
| TVAG_140990 | conserved protein | 276 | 1 | 12 | 0.540 | 0.427 | 6.86 | |
| DNA metabolism | TVAG_015570 | deoxyribonuclease II precursor, putative | 337 | 0 | 0.272 | 0.658 | 10.30 | |
| TVAG_250220 | DNA replication licensing factor MCM4, putative | 752 | 0 | 0.102 | 0.462 | 0.70 | ||
| TVAG_120370 | extracellular ribonuclease precursor, putative | 233 | 0 | 0.279 | 0.742 | 4.29 | ||
| Miscellaneous | TVAG_421200 | APC10 subunit of the anaphase-promoting complex | 217 | 0 | 0.117 | 0.239 | 1.20 | |
| TVAG_224490 | cystatin-like domain, cd00042 | 188 | 0 | 0.382 | 0.475 | 4.19 | ||
| TVAG_274750 | GTP-binding protein alpha subunit, gna, putative | 363 | 0 | 0.107 | 0.617 | 0.95 | ||
| TVAG_186390 | guanylate cyclase, putative | 1,540 | 11 | 374 | 0.105 | 0.745 | 0.30 | |
| TVAG_161100 | heat shock protein 70kD, putative | 659 | 0 | 0.112 | 0.272 | 1.29 | ||
| TVAG_414190 | pqiA integral membrane protein, TIGR00155 | 1,074 | 9 | 29 | 0.505 | 0.656 | 1.29 | |
| TVAG_070550 | syntaxin, putative | 298 | 1 | 4 | 0.106 | 0.395 | 0.70 | |
| Other hydrolase | TVAG_064890 | alpha-l-fucosidase, putative | 1,021 | 1 | 27 | 0.212 | 0.448 | 1.81 |
| TVAG_110660 | beta-hexosaminidase, putative | 553 | 1 | 521 | 0.198 | 0.654 | 1.68 | |
| TVAG_323360 | GH36 glycosyl hydrolase family 36, cd14791 | 2,104 | 1 | 19 | 0.145 | 0.366 | 1.36 | |
| TVAG_215920 | phospholipase B | 621 | 1 | 19 | 0.183 | 0.48 | 5.93 | |
| TVAG_333010 | phospholipase B | 533 | 0 | 0.501 | 0.676 | 4.47 | ||
| TVAG_457850 | N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase | 1,000 | 1 | 35 | 0.600 | 0.606 | 6.53 | |
| Oxygen metabolism | TVAG_404200 | superoxide dismutase, putative | 217 | 0 | 0.154 | 0.474 | 3.04 | |
| TVAG_094410 | thioredoxin-like | 197 | 0 | 0.296 | 0.908 | 1.01 | ||
| Protease | TVAG_339720 | M60 peptidase | 1,148 | 1 | 11 | 0.483 | 0.456 | 0.20 |
| TVAG_229200 | cathepsin C-like protein | 433 | 0 | 0.215 | 0.751 | 2.15 | ||
| TVAG_057000 | clan CA, family C1, papain-like cysteine peptidase | 314 | 0 | 0.181 | 0.399 | 0.51 | ||
| TVAG_034140 | clan CA, family C1, papain-like cysteine peptidase | 317 | 0 | 0.257 | 0.525 | 0.24 | ||
| TVAG_457240 | clan CA, family C40, NlpC/P60 superfamily cysteine peptidase | 275 | 0 | 0.208 | 0.57 | 2.13 | ||
| TVAG_400860 | clan MA, family M8, protease GP63 | 589 | 0 | 0.101 | 0.337 | 1.79 | ||
| TVAG_226590 | clan MA, family M8, surface protease GP63 | 637 | 1 | 22 | 0.105 | 0.111 | 4.66 | |
| TVAG_367130 | clan MA, family M8, surface protease GP63 | 630 | 1 | 21 | 0.519 | 0.248 | 3.40 | |
| TVAG_371800 | clan MA, family M8, surface protease GP63 | 704 | 1 | 14 | 0.252 | 0.047 | 3.02 | |
| TVAG_013490 | clan MG, family M24, aminopeptidase P-like metallopeptidase | 383 | 0 | 0.100 | 0.443 | 0.30 | ||
| TVAG_465250 | clan SB, family S8, subtilisin-like serine peptidase | 768 | 1 | 2 | 0.311 | 0.500 | 2.93 | |
| TVAG_144390 | peptidase C1A subfamily | 573 | 1 | 53 | 0.118 | 0.616 | 0.30 | |
| TVAG_189150 | peptidase M60 domain | 1,247 | 1 | 21 | 0.304 | 0.437 | 8.44 | |
| TBSR protein | TVAG_244130 | conserved protein | 751 | 1 | 53 | 0.323 | 0.405 | 12.31 |
| TVAG_045320 | conserved protein | 581 | 1 | 7 | 0.244 | 0.194 | 9.80 | |
| TVAG_137010 | conserved protein | 1,080 | 1 | 62 | 0.475 | 0.417 | 7.22 | |
| TVAG_289840 | conserved protein | 972 | 1 | 69 | 0.656 | 0.364 | 6.88 | |
| TVAG_280090 | conserved protein | 763 | 1 | 62 | 0.122 | 0.369 | 6.60 | |
| TVAG_498870 | conserved protein | 734 | 1 | 6 | 0.175 | 0.210 | 6.35 | |
| TVAG_393400 | conserved protein | 563 | 1 | 2 | 0.377 | 0.500 | 5.75 | |
| TVAG_245580 | conserved protein | 2,365 | 1 | 66 | 0.184 | 0.388 | 4.58 | |
| TVAG_166850 | conserved protein | 748 | 1 | 61 | 0.132 | 0.444 | 4.35 | |
| TVAG_062620 | conserved protein | 1,365 | 1 | 8 | 0.533 | 0.561 | 3.61 | |
| TVAG_184310 | conserved protein | 1,239 | 1 | 7 | 0.173 | 0.423 | 3.47 | |
| TVAG_377740 | conserved protein | 391 | 0 | 0.101 | 0.408 | 3.37 | ||
| TVAG_573910 | conserved protein | 1,225 | 1 | 63 | 0.113 | 0.330 | 3.33 | |
| TVAG_477640 | conserved protein | 621 | 0 | 0.280 | 0.520 | 3.24 | ||
| TVAG_425470 | conserved protein | 2,061 | 1 | 66 | 0.172 | 0.407 | 2.92 | |
| TVAG_529190 | conserved protein | 1,088 | 0 | 0.524 | 0.507 | 2.82 | ||
| TVAG_340570 | conserved protein | 752 | 1 | 6 | 0.243 | 0.250 | 2.61 | |
| TVAG_393390 | conserved protein | 563 | 1 | 2 | 0.238 | 0.437 | 2.29 | |
| TVAG_369130 | conserved protein | 523 | 1 | 3 | 0.097 | 0.318 | 1.86 | |
| TVAG_359980 | conserved protein | 1,592 | 1 | 43 | 0.171 | 0.383 | 1.63 | |
| TvSaplip | TVAG_213250 | TvSaplip8 | 126 | 0 | 0.653 | 0.944 | 3.53 | |
| TVAG_453350 | TvSaplip6 | 131 | 0 | 0.382 | 0.673 | 1.03 | ||
| Unknown | TVAG_213670 | conserved protein | 733 | 1 | 29 | 0.149 | 0.314 | 8.74 |
| TVAG_133500 | conserved protein | 255 | 0 | 0.133 | 0.636 | 6.80 | ||
| TVAG_213390 | conserved protein | 285 | 0 | 0.227 | 0.125 | 5.34 | ||
| TVAG_177320 | conserved protein | 963 | 1 | 44 | 0.154 | 0.610 | 4.93 | |
| TVAG_498650 | conserved protein | 988 | 2 | 14 | 0.217 | 0.509 | 4.10 | |
| TVAG_351790 | conserved protein | 991 | 2 | 15 | 0.115 | 0.594 | 2.50 | |
| TVAG_353290 | conserved protein | 252 | 0 | 0.276 | 0.455 | 2.24 | ||
| TVAG_547420 | conserved protein | 158 | 0 | 0.430 | 0.663 | 1.66 | ||
| TVAG_321740 | conserved protein | 803 | 1 | 19 | 0.202 | 0.617 | 1.52 | |
| TVAG_213210 | conserved protein | 1,023 | 1 | 55 | 0.269 | 0.421 | 0.76 | |
| TVAG_193980 | conserved protein | 511 | 0 | 0.122 | 0.409 | 0.30 | ||
| TVAG_315940 | conserved protein | 296 | 1 | 11 | 0.327 | 0.309 | 0.20 | |
| TVAG_039040 | conserved protein | 554 | 1 | 49 | 0.109 | 0.253 | 0.20 | |
| TVAG_166760 | unknown protein | 927 | 1 | 44 | 0.179 | 0.485 | 5.26 |
Length, number of amino acids; TMD, number of predicted transmembrane helixes; TMD-C, number of amino acids from the last TMD to the C-terminus; SignalP prediction of signal peptide (default cutoff value 0.450); SecretomeP, prediction of nonclassical protein secretion (default cutoff value 0.600); SecS, secretion score indicating time-dependent protein secretion.
The actively released proteins were then ordered by hierarchical clustering according to LFQ values over 10–120 min at 37 °C (Fig. 1C). This analysis revealed five main clusters with different dynamics of protein release. The most striking difference appeared between 10 to 30 min versus 60 to 120 min, which is supported by boxplot analysis (Fig. 1D). As indicated by the heat map, proteins in cluster 1 (eight proteins) were entirely absent (seven proteins) or present in a low quantity (one protein) during 10–30 min incubation (late secreted proteins). Two of the proteins were detected only after 120 min of incubation (TVAG_186390, and TVAG_193980). On the other side of the spectrum, proteins of Group 5 (52 proteins) were present in relatively high quantity beginning 10 min after incubation (early secreted proteins) and included a subcluster of the most abundant proteins, such as TVAG_057000. We suspected that differences in the dynamics of protein secretion may reflect the presence or absence TMD; however, we did not find such a correlation.
Transmembrane Proteins
Predictions of TMD using the TMHMM software detected 51 secreted proteins with putative TMDs, among which 46 proteins possess a single-spanning TMD and 5 proteins possess 2–11 TMDs (Table I). The peptides determined in the secretome corresponded exclusively to the soluble protein domains, whereas the TMD domains were predicted based on the corresponding AA sequences in TrichDB. The largest group of single spanning proteins (17 entries) is a heterogeneous group of unknown proteins that we named Trichomonas beta-sandwich repeat (TBSR) protein for the presence of a glycosyl hydrolase domain-like of a beta-sandwich structure at the N terminus followed by 2–5 immunoglobulin-like beta-sandwich domains and a conserved transmembrane domain close to the C terminus. Most of the TBSR proteins are grouped into two clusters. Cluster 2 contains seven TBSR proteins, including TVAG_244130, TVAG_045320, and TVAG_289840, that are among the 10 top proteins with the highest SecS (Table I, Table S3). Nine TBSR proteins with TMD and three TBRS proteins without TMD are grouped in cluster 5 (Fig. 1C). These TBSR proteins are among the most abundant proteins in the secretome with the top TBSR protein TVAG_245580. A single TBSR protein is present in cluster 3 (Fig. 1C). The topology of TMD at the C terminus of TBSR proteins and the high score for secretory signal prediction at the N terminus (Table I, and Table S3) correspond to type I membrane proteins with the N-terminal part outside the cell and the C-terminal part facing the cytosol (52). Analysis of the C termini revealed that 11 TBSR proteins possess an NPXY-type signal for endocytic internalization, which further supports their type I topology. We also found the NPXY motif at the C termini of two proteins in the conserved TMD category (Fig. S4). Interestingly, most of the C-terminal domains of TBSR proteins and two conserved TMD proteins possess the DDPFA motif that was previously noted at the C terminus of 21 surface TvBspA proteins (53). Four TBSR proteins scattered among cluster 2, 3, and 5 possess the motif for parasite rhomboid protease within TMD (Fig. S4) (31). The other transmembrane proteins consist of seven proteases, including three GP63-like membrane proteases, a single TvBspA protein, four putative hydrolases, 10 unknown proteins, and three proteins of miscellaneous category.
Soluble Proteins
The second half of the T. vaginalis secretome consisted of soluble proteins of various categories. The highest SecS of the soluble proteins belonged to a putative deoxyribonuclease II family protein (TVAG_015570, TvDNaseII). The other soluble proteins include four acid phosphatases of the histidine phosphatase family; three soluble TvBspA proteins; three putative carbohydrate-binding proteins; two other proteins involved in DNA metabolism; two oxygen-metabolizing enzymes; six proteases, including papain-like cysteine peptidase TvCP2 of cluster 5 (TVAG_057000), which appeared to be the most abundant soluble protein; three TBSR proteins; two proteins with saposin-like domains; and seven hydrolases (Table I). The largest number of soluble proteins (12 proteins) was in the miscellaneous category of proteins with known functional domains but with currently unclear functions in the T. vaginalis secretome. Finally, eight soluble proteins belong to the category of unknown proteins.
Glycoside Hydrolases
For further investigation, we selected soluble glycoside hydrolases as model secreted proteins. These enzymes have the potential to metabolize external glycogen, a key substrate for the energy metabolism of the parasite. Searches in our dataset revealed the presence of a single α-amylase (TVAG_178580, glycoside hydrolases family 13) and two β-amylases (glycoside hydrolases family 14) (http://www.cazy.org/). In the T. vaginalis genome, proteins with α-amylase domains are encoded by at least 19 genes that form three distinct clades (Fig. S5). The secreted α-amylase TVAG_178580 belongs to clade I together with two other paralogues in the genome. Another α-amylase of clade II (TVAG_112500) was identified among the proteins under the SecS cutoff.
Four β-amylase paralogues are encoded in the T. vaginalis genome, among which we identified TVAG_436700 (BA1) grouped in cluster 2 and TVAG_080000 (BA2) grouped in cluster 5 (Fig. 1C). Two other paralogues were named BA3 (TVAG_175670) and BA4 (TVAG_236600). Comparing the AA sequences of BA1–4 revealed that BA1–3 are highly similar, with a high AA sequence identity of 72.9–88.6%, whereas BA4 displayed only 35.2–36.5% sequence similarity to the other paralogues. In addition, BA4 possesses a 19 AA N-terminal extension and a predicted N-terminal transmembrane helix (17–33 AA). BA3 and BA4 were not identified in the secretome and were not present among the proteins below the SecS cutoff.
Peculiar Distribution of β-Amylases in Eukaryotes
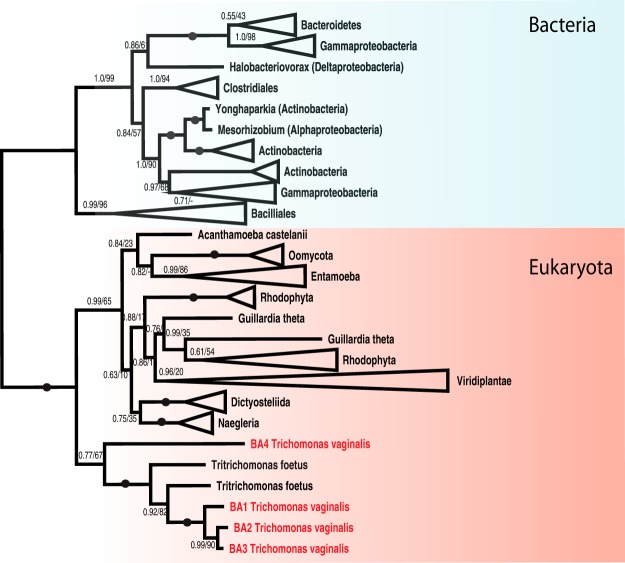
The secretion of β-amylase by T. vaginalis is noteworthy, as the production of this enzyme has been observed in only a few eukaryotic lineages, including land plants and Entamoeba histolytica (54). Thus, to gain further insight into β-amylase distribution, we searched for β-amylase coding genes across eukaryotic supergroups (Fig. 2 and Fig. S6). In addition to T. vaginalis, we identified β-amylase genes in the related bovine pathogen Tritrichomonas fetus. Naegleria gruberi, and N. fowleri appeared to be the only other members of the Excavata supergroup with putative β-amylase-coding genes. As expected, we found β-amylases in land plants, but we also found orthologues in members of the Chlorophyta and Rhodophyta groups. Moreover, we identified β-amylase genes in several members of Oomycota and Cryptophyta. Whereas patchy distribution is apparent in the other eukaryotic groups, β-amylases appear to be common in Amoebozoa. However, animals and fungi appear to be devoid of β-amylases.
Fig. 2.
Phylogeny of β-amylases. The phylogeny of beta-amylase homologues inferred by PhyloBayes (CAT+GTR model) and PhyML (LG+I+G model). For each branch, the posterior probability and ML bootstrap support are given. Branches with maximum support (1.0/100) are indicated by black circles. Supports with posterior probability below 0.5 are omitted.
Expression of β-Amylases and Influence of Environmental Conditions
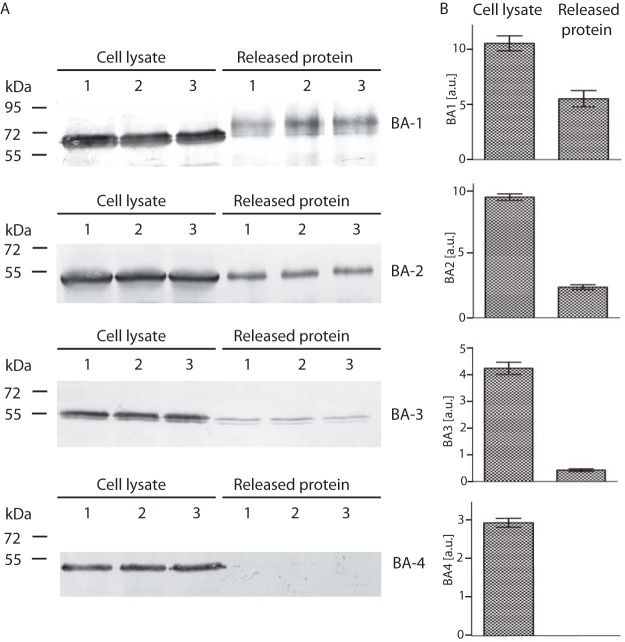
The presence of only two of four β-amylase paralogues in the T. vaginalis secretome may reflect differences in gene expression among BA1–4 under certain environmental conditions. First, using qRT-PCR, we compared BA1–4 expression in trichomonads cultivated in standard medium with maltose and detected transcripts for all four paralogues (Fig. 3A). The gene for BA2 showed the highest expression level, which corresponded to the highest LFQ values determined for BA2 in the secretome. The expression of ba1 was lower by two orders of magnitude, and ba3 displayed the lowest expression. Interestingly, the expression of ba4 was higher than that of ba1. When maltose was replaced by glycogen, the expression level significantly increased for ba1–3, whereas ba4 expression showed no significant effect (Fig. 3B). We observed a similar trend when trichomonads were cultivated under iron-limited conditions, which resulted in increased transcription of ba1–3 with no effect on ba4 (Fig. 3B). Next, we expressed recombinant BA3 and BA4 under the control of a strong promotor of succinyl CoA synthase with a C-terminal HA tag in T. vaginalis (55). BA1 and BA2 were expressed as a positive control. The transformed cells were incubated for 60 min in TYM. BA1, BA2, and a low level of BA3 were detected in cell-free TYM, but BA4 was not (Fig. 4). All four proteins were detected in the cell lysates. These experiments confirmed our suspicion that BA4 is not secreted, whereas the secretion of BA1–3 depends on the expression level.
Fig. 3.
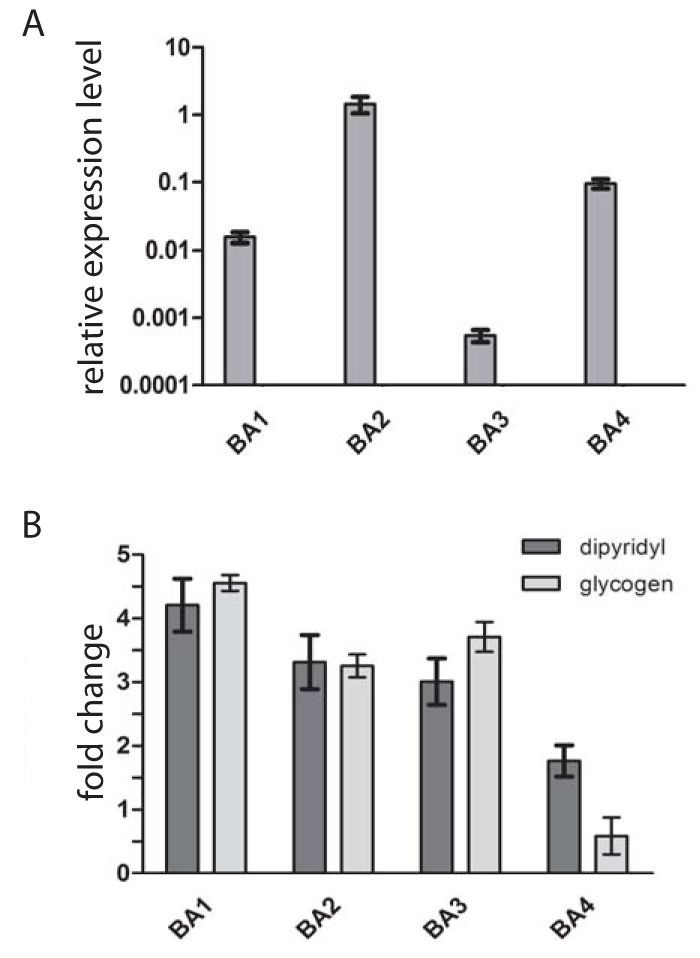
Effect of environmental factors on expression of β-amylases analyzed by qRT-PCR. (A) Relative mRNA expression levels of BA1–4 in T. vaginalis cultivated in standard TYM medium. Expression levels of β-amylases were normalized using the DNATopII reference gene. (B) Relative mRNA expression level of BA1–4 in T. vaginalis cultivated in TYM when maltose is replaced with glycogen or in standard TYM medium with a restricted level of iron. Data are expressed as -fold increases relative to the expression of the corresponding gene in standard TYM medium. Error bars represent the standard deviation of the mean.
Fig. 4.
Test of BA-3 and BA4 secretion in Tv17–48 strain. (A) HA-tagged versions of BA1, BA2, BA3, and BA4 were expressed under a strong promotor using the TagVag vector. BA1 and BA2, which were detected in the T. vaginalis secretome, were used as a positive control. Cells were incubated for 60 min at 37 °C in TYM, and BA proteins were then detected using an antitag antibody by immunoblotting of the cell lysate (L) and of the cell-free TYM (S). Each experiment was performed in three biological replicates. (B) Bar graph shows the relative amount of BA proteins associated with cell and released to the media that were quantified from three independent experiments. Error bar indicates standard deviation. a.u., arbitrary unit.
Secretion of β-Amylases via Classical Secretory Pathway
The secretory pathway in T. vaginalis has not yet been studied. Thus, we were interested in whether T. vaginalis β-amylases are secreted via the classical endoplasmic reticulum (ER)-to-Golgi secretory pathway or a nonclassical secretory pathway that is independent of the ER (56). The predictions of classical and nonclassical secretion using the SignalP/TargetP and SecretomeP servers, respectively, were not conclusive (Table I, Table S3). Thus, we prepared T. vaginalis strain Tv17–48-BirA expressing biotin ligase (BirA) fused at the N terminus with the first 75 AA of protein disulfide isomerase to target BirA to the ER and with the V5 tag at the C terminus for BirA visualization. This cell line allows for the specific biotinylation of proteins containing the AP tag within the ER and their subsequent visualization using fluorochrome-conjugated avidin. When we co-expressed BA1, BA2, and BA4 in this cell line, all β-amylases were biotinylated and detected in the ER (Fig. 5). We observed a strongly labeled ring of ER around the nucleus and a rich ER network within the T. vaginalis cell. Moreover, BA1 and BA2 appeared in rod-like structures close to the nucleus whose appearance corresponds to that of the Golgi apparatus, whereas no Golgi labeling was observed in the cells expressing BA4. Multiple attempts to express BA3 in the Tv17–48-BirA strain under the control of a native promotor were not successful. Therefore, we expressed BA3 in Tv17–48 strain without BirA (Fig. S7). The BA3 was observed in ER structures, whereas its presence in the Golgi apparatus was not conclusive. These data indicate that recombinant BA1 and BA2 are biotinylated within the ER and transported to the Golgi apparatus.
Fig. 5.
Visualization of β-amylases in the secretory pathway using super-resolution fluorescence microscopy (SFM). BA1, BA2, and BA4 with AP tag were biotinylated by BirA in ER and visualized using Alexa Fluor 488 avidin conjugate (green). BirA targeted to ER was visualized using rabbit polyclonal α-V5 Ab and the secondary Alexa Fluor 594 donkey α-rabbit Ab (red). The nucleus is labeled with 4′,6-diamidino-2-phenylindole (DAPI). Arrows indicate Golgi apparatus. Bar = 2.5 μm.
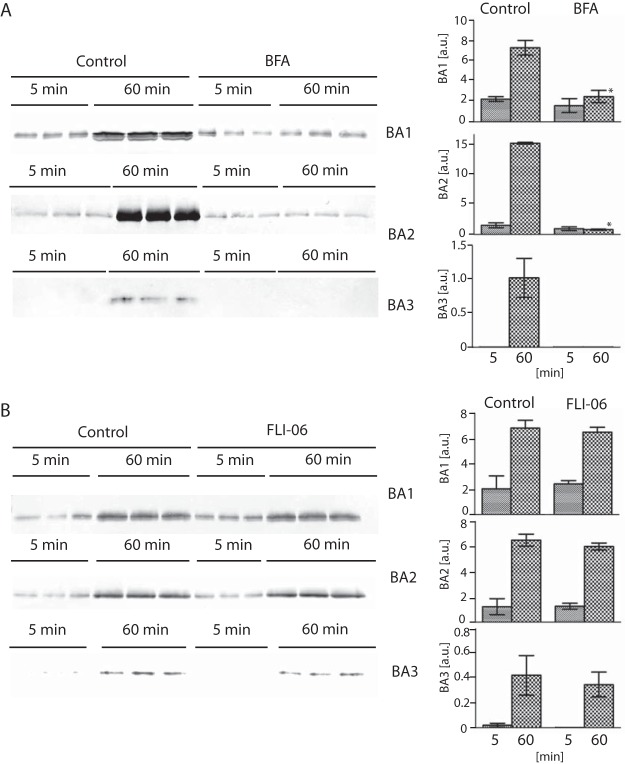
To gain further insight into the secretion mechanisms, we tested the effect of two secretion inhibitors, brefeldin A and FLI-6, with different modes of action. Brefeldin A causes a collapse of the Golgi apparatus via stimulation of retrograde transport from the Golgi to the ER (57), whereas FLI-06 inhibits the recruitment of cargo to the ER and trans-Golgi network exit sites (58). The treatment of T. vaginalis with brefeldin A clearly inhibited BA1, BA2, and BA3 secretion, as expected for the ER–Golgi secretory pathway. Interestingly, we observed no effect of FLI-06 on BA1–3 secretion despite the use of a range of concentrations up to 50 μm (Fig. 6).
Fig. 6.
Effect of brefeldin A and FLI-06 on BA1–3 secretion. T. vaginalis strains producing HA-tagged BA1–3 were incubated for 5 and 60 min in TYM supplemented with brefeldin A or FLI-06, and secreted BA proteins were detected in conditioned cell-free TYM medium via Western blot analysis with mouse monoclonal α-HA Ab. Control cells were incubated without inhibitors. Bar graph shows the relative amount of secreted BA proteins quantified from three independent experiments. Error bar indicates standard deviation. Star (*) indicates significant difference between relative quantity of protein secreted after 60 min incubation of control cells and cells treated with inhibitor (p < 0.05).
N-Linked Glycosylation of T. vaginalis β-Amylases
N-linked glycosylation is a major modification of secretory proteins upon their translocation to the ER. Searches for the conserved sequon NX[ST], which serves as a glycan acceptor, predicted seven glycosylation sites in BA1, none in BA2, one in BA3, and two in BA4 (Table S4). To investigate the predicted N-glycosylation, we expressed HA-tagged BA1–4 in T. vaginalis and treated the cell lysate from each strain with N-glycosidase F (PNGase F), which cleaves N-linked oligosaccharides. Western blot analysis of the treated and untreated lysates showed the most prominent shift in molecular weight in BA1 (∼11 kDa), which is consistent with the prediction of the largest number of glycosylation sites in this protein (Fig. 7A). Untreated BA3 appeared as a double band that was shifted by ∼1.5 kDa by PNGase treatment. Although two glycosylation sites were predicted for BA4, we did not observe any shift in this case.
Fig. 7.
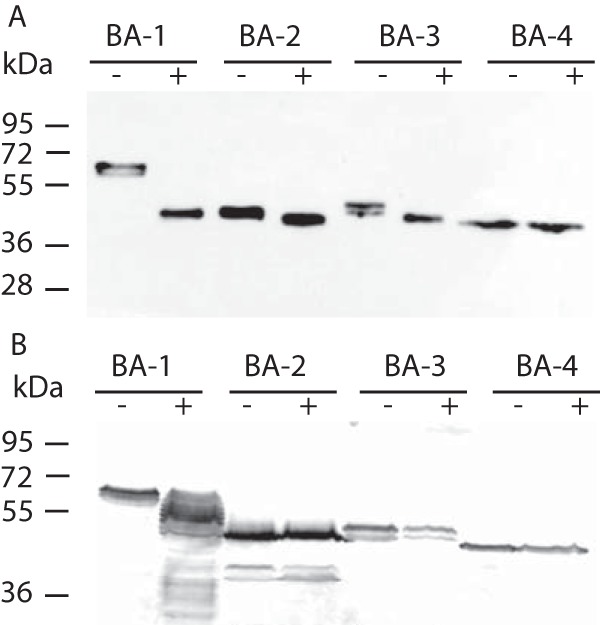
Effect of PNGase and tunicamycin on β-amylase glycosylation. (A) Representative Western blot analysis (n = 3) of HA-tagged BA1–4 produced by T. vaginalis cells that were treated with PNGase F (+) or untreated (-). (B) Representative Western blot analysis (n = 3) of T. vaginalis cell lysate producing HA-tagged BA1–4 in the presence (+) or absence (-) of tunicamycin.
Next, we treated the T. vaginalis strains expressing HA-tagged BA1–4 with tunicamycin, a nucleoside antibiotic that inhibits N-linked glycan biosynthesis. The immunoblot analysis of T. vaginalis incubated for 24 h with tunicamycin revealed a series of BA1 forms of 51–63 kDa that differ by ∼1 kDa (Fig. 7B), indicating impaired N-glycosylation. However, BA3 formed a double band under both conditions. Next, we expressed streptavidin-tagged BA1 in the Tv17–48 strain, purified the protein by affinity chromatography, and subjected the purified protein to MS analysis. The analysis confirmed that all seven predicted sites in BA1 were glycosylated. Moreover, all seven glycosylated peptides contained glycans with the same composition of Hex5HexAc2 (Table S5).
BA3 and BA4 Are Present in Lactoferrin-Labeled Vesicles
BA4 protein is apparently not secreted into the T. vaginalis environment, and only small amount of BA3 is released when it is expressed under a control of a strong promotor. Thus, we were interested in whether these proteins are recruited to the lysosomal compartment. To label the endosomal/lysosomal compartment, we incubated trichomonads in medium containing FITC-labeled lactoferrin, which is known to be internalized via receptor-mediated endocytosis and to release lactoferrin-associated iron upon reaching the acidified compartment (59, 60). After incubation, the cells were processed for immunofluorescence microscopy by labeling the HA-tagged β-amylases. For comparison, we performed the same experiment with BA1–2. As described above, BA4 appeared in the ER structure surrounding the nucleus and in numerous vesicles scattered within the cell. FITC-labeled lactoferrin appeared in tiny endosomes and in large vesicles corresponding to lysosomes, in which it was co-localized with BA4 (Fig. 8). Lactoferrin-labeled vesicles were co-stained also with BA3. In contrast, labeling of BA2 did not co-localize with lactoferrin. BA1-labeled structures were mostly distinct from lysosomes, although 18% (n = 3) of lactoferrin-labeled vesicles also displayed a BA1 signal. These results indicate that BA4 and BA3 are targeted to the T. vaginalis lysosomal compartment, whereas secretion of BA2 and that of most of BA1 are independent of lysosomes.
Fig. 8.
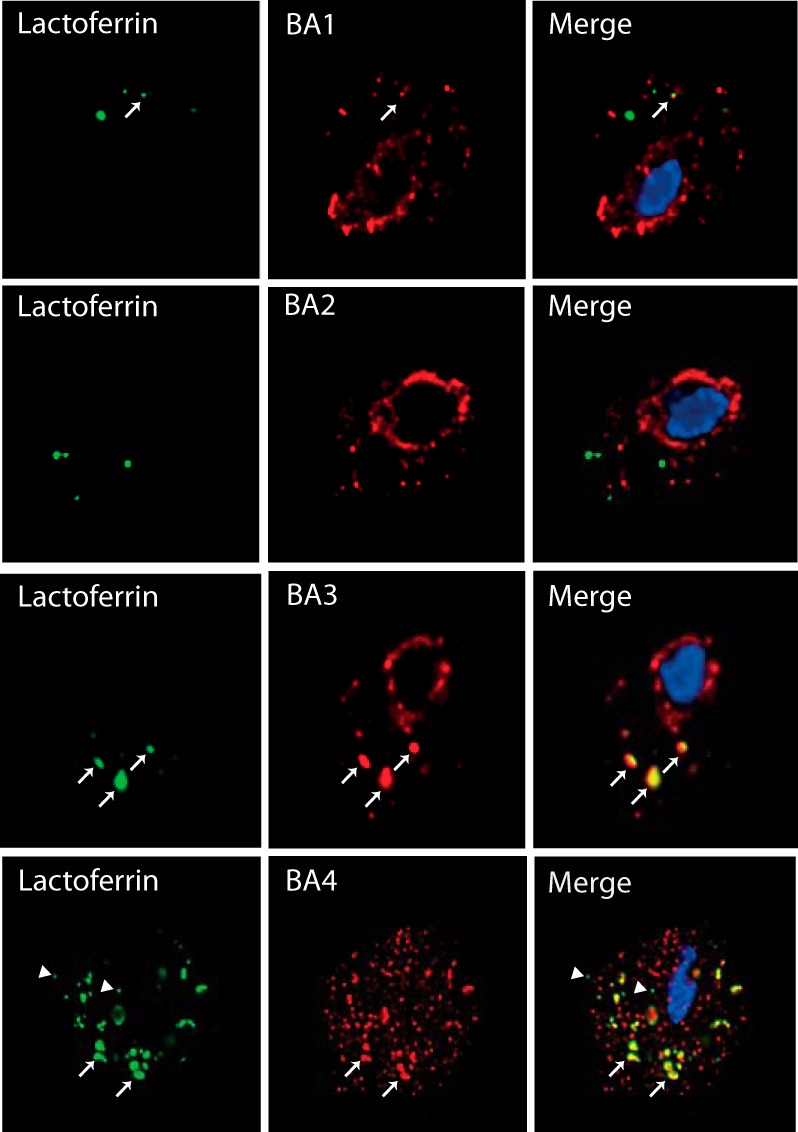
Visualization of BA1–4 and lactoferrin-labeled endosomal/lysosomal compartment. T. vaginalis strains expressing HA-tagged BA1–4 were incubated for 10 min with FITC-labeled lactoferrin (green) and then processed for SIM microscopy. BA proteins were visualized using mouse monoclonal α-HA Ab and the secondary Alexa Fluor 594 donkey α-mouse Ab (red). Arrow indicates co-localization of lactoferrin and BA proteins in lysosomes; arrow head indicates small endosomes with lactoferrin that do not contain BA.
DISCUSSION
Products secreted by parasitic protists play a fundamental role in the host–parasite relationship. Here, we analyzed the T. vaginalis secretome using high-resolution LFQ MS. Altogether, we identified 2,072 extracellular proteins, which is the largest protein set of T. vaginalis origin identified to date. However, our filter for distinguishing bona fide secreted proteins from contaminants resulted in only 89 proteins that fulfilled our strict criteria. Most experimental designs for studying the secretome of parasitic protists are based on a single time period of incubation under optimal conditions to minimize cell lysis, followed by MS analysis of the proteins released to the medium (conditioned medium) (31, 34, 61–64). Although this approach has identified important secreted proteins, eliminating contaminant proteins is problematic. More precise identification could be achieved by quantitative MS analysis of metabolically labeled cells, which makes it possible to define a cutoff based on the ratio of protein levels in the conditioned medium to the corresponding protein levels associated with the cells (65). Our approach was based on the LFQ MS analysis of time- and temperature-dependent secretion. This strategy makes it possible to define a simple cutoff based on the increasing concentration of a given protein over time. We also removed all proteins with similar levels at 37 °C and 4 °C, as exocytosis is a temperature-dependent process.
Analysis of the T. vaginalis secretome revealed surprisingly high participation of proteins (over 50%) that contain TMD. Most of these proteins possess a single C-terminal transmembrane helix (47 proteins), and of this majority, 40 proteins were previously found in the T. vaginalis surface proteome (30). Most likely, these proteins are embedded in the cellular membrane via the C-terminal TMD, and their N-terminal domains are subsequently released to the environment upon proteolytic cleavage. Indeed, Riestra et al. (31) found that T. vaginalis possesses an active rhomboid secretory protease, TvROM1, that is localized in the plasma membrane. This enzyme has been shown to catalyze the cleavage within the TMD of two proteins, TVAG_166850 and TVAG_280090. Moreover, the treatment of TvROM1-transfected cells with the serine protease inhibitor 3,4-dichloroisocoumarin caused a statistically significant decrease in the secretion of four additional proteins with predicted TMDs (31). These proteins were not cleaved by TvROM1, but they all contain a bacterial-like rhomboid substrate motif. All these proteins belong to the family of 17 TBSR proteins that we identified in the secretome. Interestingly, all TBSR proteins with C-terminal TMDs and two conserved TMD proteins possess the endosomal signal NPX[YFW] at the C terminus facing the cytosol. The same signal was previously observed at the C termini of 15 putative surface TvBspA proteins (53). The NPXY sorting signal is known to mediate the rapid internalization of membrane proteins such as receptors in humans. It would be interesting to test the role of this sorting signal in T. vaginalis. The signal may facilitate the internalization of complete proteins, such as receptors with cargo, or it may clear the C-terminal domains after the release of the N-terminal part. Altogether, our results indicate that the proteolytic cleavage of membrane proteins greatly contributes to the protein spectrum in the T. vaginalis secretome and that, in addition to TvROM1, other proteases are likely involved in the membrane protein secretion. In contrast to the membrane proteome, there is very low overlap (eight proteins) between our dataset of secreted proteins and the set obtained in a previous proteomic analysis of T. vaginalis exosomes (19). This finding is consistent with the concept that secretory proteins and exosomal proteins represent two distinct sets of extracellular proteins released by two different mechanisms.
The function of most of these secreted membrane proteins is unknown; however, members of at least two protein groups, TBSR and TvBspA proteins, are likely involved in the adhesion of the parasite to the host cell. Exogenous expression of the TBSR protein TVAG_166850 leads to increased attachment of trichomonads to the host ectocervical cells (31). In the secretome, we found six TBSR protein paralogous to TVAG_166850 that may have similar properties. TvBspA proteins are known as membrane adhesins, originally described in Bacteroides forsythus, that mediate the co-aggregation of bacteria as well as their adhesion to host cells (53, 66). In the secretome, we identified a single TvBspA protein with C-terminal TMD, TVAG_383590, and three soluble TvBspA proteins, TVAG_186300, TVAG_186290, and TVAG_186310. The latter three proteins have also been found in the surface proteome of two different T. vaginalis strains (30). Notably, the T. vaginalis genome encodes an extraordinarily large set of TvBspA-like proteins, numbering 911 genes, and there is evidence of the transcription of more than 30% of these genes (53). Thus, the identification of a rather limited number of identical TvBspA proteins exposed to the cell environment in different T. vaginalis strains suggests that they have a specific function that should be elucidated in future studies. Interestingly, six hydrogenosomal enzymes involved in energy metabolism have been previously reported to serve as adhesins in addition to their metabolic functions. These enzymes include three paralogues of malic enzyme (AP65 1–3), the α- and β-subunits of succinyl CoA synthetase (AP51, AP33), and pyruvate:ferredoxin oxidoreductase (AP120) (67, 68). We found over 30 hydrogenosomal proteins, including all putative adhesins, in our dataset under the SecS cutoff. These proteins most likely represent hydrogenosomal contaminants resulting from the presence of hydrogenosomes released from some broken cells, which we observed in our samples by electron microscopy. β-Amylases are part of the second half of the T. vaginalis secretome, which consists of soluble proteins. These enzymes have been found in several pathogenic protists but are absent in human and animal proteomes. For example, E. histolytica expresses eight β-amylases that are proposed to be involved in the utilization of host mucus glycans as a carbon source for energy metabolism and to contribute to the mucosa invasion (69). Our genome searches and phylogenetic analysis revealed that β-amylases are also present in other amoebozoans, in the bovine pathogen T. fetus, and in N. fowleri, the causative agent of human primary meningoencephalitis. However, nothing is currently known about the role of β-amylase in these pathogens. In T. vaginalis, β-amylase likely contributes to the utilization of free glycogen and oligosaccharides in the vaginal fluids by breaking down the substrate into maltose. Indeed, Smith et al. 2016 (29) showed that recombinant TVAG_080000 produces a detectable amount of maltose from glycogen. Maltose is then extracellularly hydrolyzed by α-glucosidase, and glucose is taken up via a putative glucose transporter into the cell (27). The proposed production of maltose from glycogen is supported by our finding of two β-amylases (BA1 and BA2) that are actively released to the secretome in a time-dependent manner, although with different dynamics. Whereas BA2 clusters with the most abundant proteins after 10 min of incubation and increases markedly over time (high SecS value), BA1 clusters with proteins of low abundance between 10 and 30 min after incubation and slowly increases (low SecS value). The involvement of BA1 and BA2 in the glycogen metabolism is further supported by the observed up-regulation of corresponding gene transcription when glycogen was added to the cultivation media instead of maltose, as determined by qRT-PCR. Moreover, Smith et al. (29) purified proteins with glucosidase activity from culture medium in which T. vaginalis strain G3 was grown. In contrast, the conversion of maltose to glucose remains unclear. It has been shown that α-glucosidase (maltase) activity is associated with the T. vaginalis outer membrane (27); however, no candidate for α-glucosidase activity has been found either among the list of T. vaginalis membrane proteins (30) or in the secretome (this study).
T. vaginalis is currently the only eukaryote that has been shown to actively secrete β-amylases into its environment. In E. histolytica, β-amylases have been identified only in the proteome of the lysosomes (54) or associated with the cellular surface (69); however, none of the multiple β-amylase paralogues have been found in the E. histolytica secretome (64). Although the secretory pathway in T. vaginalis has not been studied, trichomonads possess an ER and a well-developed Golgi apparatus with a specific structure. The Golgi apparatus is organized along two striated filaments forming a “V”-shaped body. To trace protein transport through the ER–Golgi pathway in T. vaginalis, we established a system using specific in vivo biotinylation of the target protein by BirA (70). This tool allowed us to demonstrate that both BA1 and BA2 proceed through the ER, where they are labeled by ER-targeted BirA and are subsequently transported to the Golgi apparatus and exported from the cell. As expected, the export was inhibited by brefeldin A, which prevents ER-Golgi transport; however, the export was resistant to FLI-06, which interferes with the function of ER and trans-Golgi network exit sites. These results indicate that BA1 and BA2 are secreted by the classical secretory pathway, but the resistance to FLI-06 suggests that there are likely subtleties in the mechanisms of T. vaginalis secretion, in which this parasite may differ from classical models.
Interestingly, BA1 appeared to be remarkably N-glycosylated. According to MS analysis of the purified protein, BA1 possesses seven glycosylation sites that are all glycosylated. In the majority of eukaryotes, N-glycans consist of two N-acetyl glucosamines, nine mannose residues, and three glucose residues that are linked to the sequon NX[ST] in the nascent protein in the ER and then modified in the Golgi apparatus. Based on the reduced set of glycosyltransferases responsible for sequential glycan synthesis, Samuelson et al. (71) predicted a simplified glycan structure consisting of GlcNAc2Man5 in T. vaginalis and E. histolytica. This finding is consistent with the Hex5HexNAc2 type of glycosylation that we detected in BA1.
Two other β-amylase paralogues encoded in the genome (BA3 and BA4) were not identified either in the secretome or in the membrane proteome. The absence of BA3 in the secretome is consistent with the very low transcription level of the corresponding gene. When BA3 was episomally expressed under the control of a strong promotor, still only low secretion of BA3 was observed, while a majority was associated with the cell. BA4 was not secreted under any conditions, although native ba4 transcription is comparable with that of ba1 and ba2. This protein appeared to be exclusively associated with the cell. The partial co-localization of BA3 and BA4 with lactoferrin in the endosomal/lysosomal compartment suggests that these amylases are targeted preferentially to lysosomes, similarly to β-amylase EAL51020 in E. histolytica. Surprisingly, the transcription of ba4 was downregulated in the presence of glycogen, which argues against glycogen as a preferential substrate. T. vaginalis is a mucin-dwelling parasite that binds and degrades mucin with mucinases (72) and various glycosidases (73). In the T. vaginalis secretome, we identified β-hexosaminidase (TVAG_110660), which is also present in E. histolytica lysosomes. Thus, oligosaccharides released from sources other than glycogen might serve as an alternative substrate for BA4.
In addition to β-amylases, we identified several other families of soluble proteins in the T. vaginalis secretome that may play an important role in the parasite's virulence. As expected, the most prominent family appeared to be proteases. Currently, five proteases (CP2, TVAG_057000; CP3, TVAG_090100; CP4, TVAG_467970; CPT, TVAG_298080; and CP65, TVAG_096740) have been shown to be secreted by T. vaginalis (17, 32). Altogether, we identified seven proteases, including soluble GP63-like protease (TVAG_400860), two papain-like cysteine proteases (TVAG_057000; TVAG_034140), NlpC/P60 superfamily cysteine protease (TVAG_457240), cathepsin C protease (TVAG_229200), and two metalloproteases of the M24 (TVAG_013490) and M60 (TVAG_339720) superfamilies. Thus, in our dataset, only CP2, which appeared to be the most abundant soluble protein in the secretome, was previously identified. CP3, CP4, and CPT were found together with 58 other proteases under the SecS cutoff, whereas we did not detect CP65 (Table S3). The observed differences in the secretion of soluble CPs are not surprising. The T. vaginalis genome encodes 220 genes for CPs, and the differential expression of CPs between various strains (74) as well as in response to various external conditions has been reported (75).
TvDNaseII displayed the second highest SecS and in hierarchical clustering appeared among abundant early secreted proteins. It has been shown that neutrophils form an extracellular DNA trap (NET) as a form of innate response that degrades virulence factors and kills microorganisms (76, 77). To defend against NET, various pathogens secrete NET-degrading DNases (78, 79). Moreover, in the roundworm Trichinella spiralis, members of the secreted DNase II subfamily have been found to counteract host innate immune responses (80). In this context, the high secretion of TvDNaseII by T. vaginalis is a noteworthy and attractive topic for further studies.
The TvSaplip category of secreted proteins includes two putative pore-forming proteins, TvSaplip-6 (TVAG_453350) and TvSaplip-8 (TVAG 213250). These proteins belong to the saponin-like protein family, which includes the amoebapore and naegleriapore pore-forming proteins, which are secreted by E. histolytica (81) and N. fowleri, (82), respectively. TvSaplip-6 and -8 possess a single SAPLIP domain, and the latter contains a predicted signal peptide for the classical secretory pathway. In the T. vaginalis genome, there are 12 predicted genes for TvSaplips encoding proteins with 1–7 SAPLIP domains, 11 of which have been found to be transcribed (83). However, TvSaplip-6 and -8 are the first SAPLIP proteins observed to be actively secreted by T. vaginalis. In addition, four other SAPLIP proteins (TvSaplip1–4) were found under the SecS cutoff. Further studies are necessary to investigate whether T. vaginalis utilizes this potential weapon against host cells or bacteria via the formation of destructive pores within the target membrane.
Our broad analysis of the secretome using high-resolution proteomics and rigorous analyses of the obtained data provides valuable information about new trichomonad virulence factors. Our set of 89 secreted proteins highlights several new protein targets that are highly attractive for future investigations. However, this set of proteins is likely not complete, as we used strict criteria for the data evaluation based on proteins that were constitutively secreted in a time- and temperature-dependent manner. Previous transcriptomic studies have revealed dramatic changes in T. vaginalis gene expression in response to various environmental stimuli such as iron level (84), glucose availability (85), interaction with fibronectin (86), and contact with vaginal epithelial cells (87), which may affect various cellular functions, including protein secretion. Therefore, the presented T. vaginalis secretome provides a solid set of proteins that are constitutively secreted under defined in vitro conditions that can serve as a reference for future comparative studies.
DATA AVAILABILITY
MaxQuant results were uploaded to the PRIDE partner repository (https://www.ebi.ac.uk/pride/archive/login) with the data set identifier PXD007034 (88) and to MS-Viewer with the search key ajo748srzk (http://msviewer.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msviewer) (89). MaxQuant LFQ version 1.5.8.3 software was deposited at https://drive.google.com/file/d/0B_13YRpRFPM5c0pwVDAzZEVLc2s/view.
Supplementary Material
Acknowledgments
We would like to thank Michaela Marcincikova for the excellent technical support and the proteomic core facility BIOCEV.
Footnotes
* This work was supported by the programs KONTAKT II (LH15254), NPU II (LQ1604), and the Biotechnology and Biomedicine Center of the Academy of Sciences and Charles University CZ.1.05/1.1.00/02.0109 provided by the Ministry of Education, Youth and Sport of the Czech Republic. We acknowledge the Imaging Methods Core Facility at BIOCEV supported by the Ministry of Education, Youth (the Czech-BioImaging RI project LM2015062 and CZ.02.1.01/0.0/0.0/16_013/0001775) and the project Operational Program Prague Competitiveness CZ.2.16/3.1.00/21515 funded by the European Regional Development Fund, and the Center of Molecular Structure (supported by the CIISB research infrastructure (LM2015043 funded by the Ministry of Education, Youth and Sport of the Czech Republic).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- Ab
- antibody
- AP
- acceptor peptide
- DAPI
- 4′,6-eiamidino-2-phenylindole
- FITC
- fluorescein isothiocyanate
- HA
- hemagglutinin
- qRT-PCR
- quantitative RT-PCR
- PEM
- 100 mM PIPES, 1 mM EGTA, 0.1 mM MgSO4
- PNGase F
- N-glycosidase F
- SecS
- secretion score
- SIM
- structured illumination microscopy
- TMD
- transmembrane domain
- TBSR
- trichomonas beta-sandwich repeat protein
- TYM
- tryptone-yeast extract-maltose medium.
REFERENCES
- 1. Petrin D., Delgaty K., Bhatt R., and Garber G. (1998) Clinical and microbiological aspects of Trichomonas vaginalis. Clin. Microbiol. Rev. 11, 300–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kissinger P., and Adamski A. (2013) Trichomoniasis and HIV interactions: A review. Sex Transm. Infect. 89, 426–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tuttle J. P. Jr, Holbrook T. W., and Derrick F. C. (1977) Interference of human spermatozoal motility by Trichomonas vaginalis. J. Urol. 118, 1024–1025 [DOI] [PubMed] [Google Scholar]
- 4. Benchimol M., d Andrade R. I, da Silva F. R., and Burla Dias A. J. (2008) Trichomonas adhere and phagocytose sperm cells: Adhesion seems to be a prominent stage during interaction. Parasitol. Res. 102, 597–604 [DOI] [PubMed] [Google Scholar]
- 5. Twu O., Dessí D., Vu A., Mercer F., Stevens G. C., de M. N., Rappelli P., Cocco A. R., Clubb R. T., Fiori P. L., and Johnson P. J. (2014) Trichomonas vaginalis homolog of macrophage migration inhibitory factor induces prostate cell growth, invasiveness, and inflammatory responses. Proc. Natl. Acad. Sci. U.S.A. 111, 8179–8184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stark J. R., Judson G., Alderete J. F., Mundodi V., Kucknoor A. S., Giovannucci E. L., Platz E. A., Sutcliffe S., Fall K., Kurth T., Ma J., Stampfer M. J., and Mucci L. A. (2009) Prospective study of Trichomonas vaginalis infection and prostate cancer incidence and mortality: Physicians' Health Study. J. Natl. Cancer Inst. 101, 1406–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Farage M. A., and Maibach H. I. (2011) Morphology and physiological changes of genital skin and mucosa. Curr. Probl. Dermatol. 40, 9–19 [DOI] [PubMed] [Google Scholar]
- 8. Langley J. G., Goldsmid J. M., and Davies N. (1987) Venereal trichomoniasis: Role of men. Genitourin. Med. 63, 264–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gardner W. A. Jr, O'Hara C., Bailey J., and Bennett B. D. (1981) In vitro susceptibility of Trichomonas vaginalis to zinc. Prostate 2, 323–325 [DOI] [PubMed] [Google Scholar]
- 10. Ryan C. M., de Miguel N., Johnson P. J. (2011) Trichomonas vaginalis: Current understanding of host-parasite interactions. Essays Biochem. 51, 161–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Krieger J. N., Ravdin J. I., and Rein M. F. (1985) Contact-dependent cytopathogenic mechanisms of Trichomonas vaginalis. Infect. Immun. 50, 778–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lin W. C., Chang W. T., Chang T. Y., and Shin J. W. (2015) The pathogenesis of human cervical epithelium cells induced by interacting with Trichomonas vaginalis. PLoS One. 10, e0124087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fiori P. L., Rappelli P., Addis M. F., Mannu F., and Cappuccinelli P. (1997) Contact-dependent disruption of the host cell membrane skeleton induced by Trichomonas vaginalis. Infect. Immun. 65, 5142–5148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lustig G., Ryan C. M., Secor W. E., and Johnson P. J. (2013) Trichomonas vaginalis contact-dependent cytolysis of epithelial cells. Infect. Immun. 81, 1411–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Midlej V., and Benchimol M. (2010) Trichomonas vaginalis kills and eats—Evidence for phagocytic activity as a cytopathic effect. Parasitol. 137, 65–76 [DOI] [PubMed] [Google Scholar]
- 16. Juliano C., Cappuccinelli P., and Mattana A. (1991) In vitro phagocytic interaction between Trichomonas vaginalis isolates and bacteria. Eur. J. Clin. Microbiol. Infect. Dis. 10, 497–502 [DOI] [PubMed] [Google Scholar]
- 17. Sommer U., Costello C. E., Hayes G. R., Beach D. H., Gilbert R. O., Lucas J. J., and Singh B. N. (2005) Identification of Trichomonas vaginalis cysteine proteases that induce apoptosis in human vaginal epithelial cells. J. Biol. Chem. 280, 23853–23860 [DOI] [PubMed] [Google Scholar]
- 18. Kusdian G., Woehle C., Martin W. F., and Gould S. B. (2013) The actin-based machinery of Trichomonas vaginalis mediates flagellate-amoeboid transition and migration across host tissue. Cell Microbiol. 15, 1707–1721 [DOI] [PubMed] [Google Scholar]
- 19. Twu O., de Miguel N., Lustig G., Stevens G. C., Vashisht A. A., Wohlschlegel J. A., and Johnson P. J. (2013) Trichomonas vaginalis exosomes deliver cargo to host cells and mediate hostratioparasite interactions. PLoS Pathog. 9, e1003482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Müller M., Mentel M., van Hellemond J. J., Henze K., Woehle C., Gould S. B., Yu R. Y., van der Giezen M., Tielens A. G., and Martin W. F. (2012) Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol. Mol. Biol. Rev. 76, 444–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hrdý I., Tachezy J., and Müller M. (2008) Metabolism of trichomonad hydrogenosomes, In: Tachezy J. (ed), Hydrogenosomes and Mitosomes:Mitochondria of Anaerobic Euakryotes, pp. 114–145, Springer-Verlag, Berlin, Heidelberg [Google Scholar]
- 22. Mirmonsef P., Hotton A. L., Gilbert D., Gioia C. J., Maric D., Hope T. J., Landay A. L., and Spear G. T. (2016) Glycogen levels in undiluted genital fluid and their relationship to vaginal pH, estrogen, and progesterone. PLoS One. 11, e0153553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mirmonsef P., Hotton A. L., Gilbert D., Burgad D., Landay A., Weber K. M., Cohen M., Ravel J., and Spear G. T. (2014) Free glycogen in vaginal fluids is associated with Lactobacillus colonization and low vaginal pH. PLoS One. 9, e102467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rajan N., Cao Q., Anderson B. E., Pruden D. L., Sensibar J., Duncan J. L., and Schaeffer A. J. (1999) Roles of glycoproteins and oligosaccharides found in human vaginal fluid in bacterial adherence. Infect. Immun. 67, 5027–5032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gregoire A. T. (1963) Carbohydrates of human vaginal tissue. Nature 198, 996. [DOI] [PubMed] [Google Scholar]
- 26. Sumawong V., Gregoire A. T., Johnson W. D., and Rakoff A. E. (1962) Identification of carbohydrates in the vaginal fluid of normal females. Fertil. Steril. 13, 270–280 [DOI] [PubMed] [Google Scholar]
- 27. ter Kuile B. H., and Müller M. (1995) Maltose utilization by extracellular hydrolysis followed by glucose transport in Trichomonas vaginalis. Parasitol. 110, 37–44 [DOI] [PubMed] [Google Scholar]
- 28. Huffman R. D., Nawrocki L. D., Wilson W. A., and Brittingham A. (2015) Digestion of glycogen by a glucosidase released by Trichomonas vaginalis. Exp. Parasitol. 159, 151–159 [DOI] [PubMed] [Google Scholar]
- 29. Smith R. W., Brittingham A., and Wilson W. A. (2016) Purification and identification of amylases released by the human pathogen Trichomonas vaginalis that are active towards glycogen. Mol. Biochem. Parasitol. 210, 22–31 [DOI] [PubMed] [Google Scholar]
- 30. de Miguel N., Lustig G., Twu O., Chattopadhyay A., Wohlschlegel J. A., and Johnson P. J. (2010) Proteome analysis of the surface of Trichomonas vaginalis reveals novel proteins and strain-dependent differential expression. Mol. Cell. Proteomics 9, 1554–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Riestra A. M., Gandhi S., Sweredoski M. J., Moradian A., Hess S., Urban S., and Johnson P. J. (2015) A Trichomonas vaginalis rhomboid protease and its substrate modulate parasite attachment and cytolysis of host cells. PLoS Pathog. 11, e1005294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hernández H. M., Sariego I., Alvarez A. B., Marcet R., Vancol E., Alvarez A., Figueredo M., and Sarracent J. (2011) Trichomonas vaginalis 62 kDa proteinase as a possible virulence factor. Parasitol. Res. 108, 241–245 [DOI] [PubMed] [Google Scholar]
- 33. Arroyo R., Cárdenas-Guerra R. E., Figueroa-Angulo E. E., Puente-Rivera J., Zamudio-Prieto O., and Ortega-López J. (2015) Trichomonas vaginalis cysteine proteinases: Iron response in gene expression and proteolytic activity. Biomed. Res. Int. 2015, 946787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kucknoor A. S., Mundodi V., and Alderete J. F. (2007) The proteins secreted by Trichomonas vaginalis and vaginal epithelial cell response to secreted and episomally expressed AP65. Cell Microbiol. 9, 2586–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kulda J., Vojtěchovská M., Tachezy J., Demes P., and Kunzová E. (1982) Metronidazole resistance of Trichomonas vaginalis as a cause of treatment failure in trichomoniasis—A case report. Br. J. Vener. Dis. 58, 394–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Diamond L. S. (1957) The establishment of various trichomonads of animals and man in axenic cultures. J. Parasitol. 43, 488–490 [PubMed] [Google Scholar]
- 37. Doran D. J. (1959) Studies on trichomonads: III. Inhibitors, acid production, and substrate utilization by 4 strains of Tritrichomonas foetus. J. Protozool. 6, 177–182 [Google Scholar]
- 38. Strober W. (2001) Trypan blue exclusion test of cell viability. Curr. Protoc. Immunol. Appendix 3 [DOI] [PubMed] [Google Scholar]
- 39. Linstead D. J., and Bradley S. (1988) The purification and properties of two soluble reduced nicotinamide: Acceptor oxidoreductases from Trichomonas vaginalis. Mol. Biochem. Parasitol. 27, 125–133 [DOI] [PubMed] [Google Scholar]
- 40. Masuda T., Tomita M., and Ishihama Y. (2008) Phase transfer surfactant-aided trypsin digestion for membrane proteome analysis. J. Proteome. Res. 7, 731–740 [DOI] [PubMed] [Google Scholar]
- 41. Cox J., Hein M. Y., Luber C. A., Paron I., Nagaraj N., and Mann M. (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vizcaíno J. A., Csordas A., Del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, 11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hrdy I., Hirt R. P., Dolezal P., Bardonová L., Foster P. G., Tachezy J., and Embley T. M. (2004) Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I. Nature 432, 618–622 [DOI] [PubMed] [Google Scholar]
- 44. Price M. N., Dehal P. S., and Arkin A. P. (2010) FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS One. 5, e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Katoh K., and Standley D. M. (2013) MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol Evol. 30, 772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Criscuolo A., and Gribaldo S. (2010) BMGE (block mapping and gathering with entropy): A new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol 10, 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lartillot N., Lepage T., and Blanquart S. (2009) PhyloBayes 3: A Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 25, 2286–2288 [DOI] [PubMed] [Google Scholar]
- 48. Guindon S., Delsuc F., Dufayard J. F., and Gascuel O. (2009) Estimating maximum likelihood phylogenies with PhyML. Methods Mol. Biol 537, 113–137 [DOI] [PubMed] [Google Scholar]
- 49. Dos S. O., de Vargas R. G., Frasson A. P., Macedo A. J., and Tasca T. (2015) Optimal reference genes for gene expression normalization in Trichomonas vaginalis. PLoS One. 10, e0138331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Howarth M., and Ting A. Y. (2008) Imaging proteins in live mammalian cells with biotin ligase and monovalent streptavidin. Nat. Protoc. 3, 534–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dawson S. C., Sagolla M. S., Mancuso J. J., Woessner D. J., House S. A., Fritz-Laylin L., and Cande W. Z. (2007) Kinesin-13 regulates flagellar, interphase, and mitotic microtubule dynamics in Giardia intestinalis. Eukaryot. Cell 6, 2354–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Goder V., and Spiess M. (2001) Topogenesis of membrane proteins: Determinants and dynamics. FEBS Lett. 504, 87–93 [DOI] [PubMed] [Google Scholar]
- 53. Noël C. J., Diaz N., Sicheritz-Ponten T., Safarikova L., Tachezy J., Tang P., Fiori P. L., and Hirt R. P. (2010) Trichomonas vaginalis vast BspA-like gene family: Evidence for functional diversity from structural organisation and transcriptomics. BMC Genomics 11, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Okada M., Huston C. D., Oue M., Mann B. J., Petri W. A. Jr, Kita K., and Nozaki T. (2006) Kinetics and strain variation of phagosome proteins of Entamoeba histolytica by proteomic analysis. Mol. Biochem. Parasitol. 145, 171–183 [DOI] [PubMed] [Google Scholar]
- 55. Rada P., Makki A. R., Zimorski V., Garg S., Hampl V., Hrdý I., Gould S. B., and Tachezy J. (2015) N-Terminal presequence-independent import of phosphofructokinase into hydrogenosomes of Trichomonas vaginalis. Eukaryot. Cell 14, 1264–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nickel W. (2003) The mystery of nonclassical protein secretion. A current view on cargo proteins and potential export routes. Eur. J. Biochem. 270, 2109–2119 [DOI] [PubMed] [Google Scholar]
- 57. Fujiwara T., Oda K., Yokota S., Takatsuki A., and Ikehara Y. (1988) Brefeldin A causes disassembly of the Golgi complex and accumulation of secretory proteins in the endoplasmic reticulum. J. Biol. Chem. 263, 18545–18552 [PubMed] [Google Scholar]
- 58. Yonemura Y., Li X., Müller K., Krämer A., Atigbire P., Mentrup T., Feuerhake T., Kroll T., Shomron O., Nohl R., Arndt H. D., Hoischen C., Hemmerich P., Hirschberg K., and Kaether C. (2016) Inhibition of cargo export at ER exit sites and the trans-Golgi network by the secretion inhibitor FLI-06. J. Cell Sci. 129, 3868–3877 [DOI] [PubMed] [Google Scholar]
- 59. Peterson K. M., and Alderete J. F. (1984) Iron uptake and increased intracellular enzyme activity follow host lactoferrin binding by Trichomonas vaginalis receptors. J. Exp. Med. 160, 398–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tachezy J., Kulda J., Bahníkova I., Suchan P., Rázga J., and Schrével J. (1996) Tritrichomonas foetus: Iron acquisition from lactoferrin and transferrin. Exp. Parasitol. 83, 216–228 [DOI] [PubMed] [Google Scholar]
- 61. Pollo-Oliveira L., Post H., Acencio M. L., Lemke N., van den Toorn H., Tragante V., Heck A. J., Altelaar A. F., and Yatsuda A. P. (2013) Unravelling the Neospora caninum secretome through the secreted fraction (ESA) and quantification of the discharged tachyzoite using high-resolution mass spectrometry-based proteomics. Parasit. Vectors 6, 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Geiger A., Hirtz C., Bécue T., Bellard E., Centeno D., Gargani D., Rossignol M., Cuny G., and Peltier J. B. (2010) Exocytosis and protein secretion in Trypanosoma. BMC Microbiol. 10, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Queiroz R. M., Ricart C. A., Machado M. O., Bastos I. M., de Santana J. M., de Sousa M. V., Roepstorff P., and Charneau S. (2016) Insight into the exoproteome of the tissue-derived trypomastigote form of Trypanosoma cruzi. Front. Chem. 4, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ujang J. A., Kwan S. H., Ismail M. N., Lim B. H., Noordin R., and Othman N. (2016) Proteome analysis of excretory-secretory proteins of Entamoeba histolytica HM1:IMSS via LC-ESI-MS/MS and LC-MALDI-TOF/TOF. Clin. Proteomics 13, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Silverman J. M., Chan S. K., Robinson D. P., Dwyer D. M., Nandan D., Foster L. J., and Reiner N. E. (2008) Proteomic analysis of the secretome of Leishmania donovani. Genome Biol. 9, R35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sharma A., Sojar H. T., Glurich I., Honma K., Kuramitsu H. K., and Genco R. J. (1998) Cloning, expression, and sequencing of a cell surface antigen containing a leucine-rich repeat motif from Bacteroides forsythus ATCC 43037. Infect. Immun. 66, 5703–5710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Engbring J., O'Brien J. L., and Alderete J. F. (1996) Trichomonas vaginalis adhesin proteins display molecular mimicry to metabolic enzymes, In: Kahane, and Ofek (eds), Toward Anti-Adhesion Therapy for Microbial Diseases, pp. 207–223, Plenum Press, New York: [DOI] [PubMed] [Google Scholar]
- 68. Meza-Cervantez P., González-Robles A., Cárdenas-Guerra R. E., Ortega-López J., Saavedra E., Pineda E., and Arroyo R. (2011) Pyruvate:ferredoxin oxidoreductase (PFO) is a surface-associated cell-binding protein in Trichomonas vaginalis and is involved in trichomonal adherence to host cells. Microbiology 157, 3469–3482 [DOI] [PubMed] [Google Scholar]
- 69. Thibeaux R., Weber C., Hon C. C., Dillies M. A., Avé P., Coppée J. Y., Labruyère E., and Guillén N. (2013) Identification of the virulence landscape essential for Entamoeba histolytica invasion of the human colon. PLoS Pathog. 9, e1003824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Martincová E., Voleman L., Pyrih J.,Žárský V., Vondráčková P., Kolísko M., Tachezy J., and Doležal P. (2015) Probing the biology of Giardia intestinalis mitosomes using in vivo enzymatic tagging. Mol. Cell. Biol. 35, 2864–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Samuelson J., Banerjee S., Magnelli P., Cui J., Kelleher D. J., Gilmore R., and Robbins P. W. (2005) The diversity of dolichol-linked precursors to Asn-linked glycans likely results from secondary loss of sets of glycosyltransferases. Proc. Natl. Acad. Sci. U.S.A. 102, 1548–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lehker M. W., and Sweeney D. (1999) Trichomonad invasion of the mucous layer requires adhesins, mucinases, and motility. Sex Transm. Infect. 75, 231–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Connaris S., and Greenwell P. (1997) Glycosidases in mucin-dwelling protozoans. Glycoconj. J. 14, 879–882 [DOI] [PubMed] [Google Scholar]
- 74. Cuervo P., Cupolillo E., Britto C., González L. J., FC E. S.-F., Lopes L. C., Domont G. B., and De Jesus J. B. (2008) Differential soluble protein expression between Trichomonas vaginalis isolates exhibiting low and high virulence phenotypes. J. Proteomics 71, 109–122 [DOI] [PubMed] [Google Scholar]
- 75. Figueroa-Angulo E. E., Rendón-Gandarilla F. J., Puente-Rivera J., Calla-Choque J. S., Cárdenas-Guerra R. E., Ortega-López J., Quintas-Granados L. I., Alvarez-Sánchez M. E., and Arroyo R. (2012) The effects of environmental factors on the virulence of Trichomonas vaginalis. Microbes. Infect. 14, 1411–1427 [DOI] [PubMed] [Google Scholar]
- 76. Brinkmann V., Reichard U., Goosmann C., Fauler B., Uhlemann Y., Weiss D. S., Weinrauch Y., and Zychlinsky A. (2004) Neutrophil extracellular traps kill bacteria. Science 303, 1532–1535 [DOI] [PubMed] [Google Scholar]
- 77. Urban C. F., Reichard U., Brinkmann V., and Zychlinsky A. (2006) Neutrophil extracellular traps capture and kill Candida albicans yeast and hyphal forms. Cell Microbiol. 8, 668–676 [DOI] [PubMed] [Google Scholar]
- 78. Buchanan J. T., Simpson A. J., Aziz R. K., Liu G. Y., Kristian S. A., Kotb M., Feramisco J., and Nizet V. (2006) DNase expression allows the pathogen group A Streptococcus to escape killing in neutrophil extracellular traps. Curr. Biol. 16, 396–400 [DOI] [PubMed] [Google Scholar]
- 79. Zhang X., Zhao S., Sun L., Li W., Wei Q., Ashman R. B., and Hu Y. (2017) Different virulence of Candida albicans is attributed to the ability of escape from neutrophil extracellular traps by secretion of DNase. Am. J. Transl. Res. 9, 50–62 [PMC free article] [PubMed] [Google Scholar]
- 80. Liu M. F., Wu X. P., Wang X. L., Yu Y. L., Wang W. F., Chen Q. J., Boireau P., and Liu M. Y. (2008) The functions of deoxyribonuclease II in immunity and development. DNA Cell Biol. 27, 223–228 [DOI] [PubMed] [Google Scholar]
- 81. Leippe M., Andrä J., Nickel R., Tannich E., and Müller-Eberhard H. J. (1994) Amoebapores, a family of membranolytic peptides from cytoplasmic granules of Entamoeba histolytica: Isolation, primary structure, and pore formation in bacterial cytoplasmic membranes. Mol. Microbiol. 14, 895–904 [DOI] [PubMed] [Google Scholar]
- 82. Herbst R., Marciano-Cabral F., and Leippe M. (2004) Antimicrobial and pore-forming peptides of free-living and potentially highly pathogenic Naegleria fowleri are released from the same precursor molecule. J. Biol. Chem. 279, 25955–25958 [DOI] [PubMed] [Google Scholar]
- 83. Hirt R. P., de Miguel N., Nakjang S., Dessi D., Liu Y. C., Diaz N., Rapelli P., Acosta-Serano A., Fiori P. L., and Mottram J. C. (2011) Trichomonas vaginalis Pathobiology: New insights from the Genome Sequence, 1st Ed., pp. 87–130, Elsevier, Amsterdam: [DOI] [PubMed] [Google Scholar]
- 84. Horváthová L., Šafariková L., Basler M., Hrdy I., Campo N. B., Shin J. W., Huang K. Y., Huang P. J., Lin R., Tang P., and Tachezy J. (2012) Transcriptomic identification of iron-regulated and iron-independent gene copies within the heavily duplicated Trichomonas vaginalis genome. Genome Biol. Evol. 4, 1017–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Huang K. Y., Chen Y. Y., Fang Y. K., Cheng W. H., Cheng C. C., Chen Y. C., Wu T. E., Ku F. M., Chen S. C., Lin R., and Tang P. (2014) Adaptive responses to glucose restriction enhance cell survival, antioxidant capability, and autophagy of the protozoan parasite Trichomonas vaginalis. Biochim. Biophys. Acta 1840, 53–64 [DOI] [PubMed] [Google Scholar]
- 86. Huang K. Y., Huang P. J., Ku F. M., Lin R., Alderete J. F., and Tang P. (2012) Comparative transcriptomic and proteomic analyses of Trichomonas vaginalis following adherence to fibronectin. Infect. Immun. 80, 3900–3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Gould S. B., Woehle C., Kusdian G., Landan G., Tachezy J., Zimorski V., and Martin W. F. (2013) Deep sequencing of Trichomonas vaginalis during the early infection of vaginal epithelial cells and amoeboid transition. Int. J. Parasitol. 43, 707–719 [DOI] [PubMed] [Google Scholar]
- 88. Vizcaíno J. A., Csordas A., Del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, 11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Baker P. R., and Chalkley R. J. (2014) MS-viewer: A web-based spectral viewer for proteomics results. Mol. Cell. Proteomics 13, 1392–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
MaxQuant results were uploaded to the PRIDE partner repository (https://www.ebi.ac.uk/pride/archive/login) with the data set identifier PXD007034 (88) and to MS-Viewer with the search key ajo748srzk (http://msviewer.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msviewer) (89). MaxQuant LFQ version 1.5.8.3 software was deposited at https://drive.google.com/file/d/0B_13YRpRFPM5c0pwVDAzZEVLc2s/view.