SUMMARY
Large datasets describing the quantitative effects of mutations on protein function are becoming increasingly available. Here, we leverage these datasets to develop Envision, which predicts the magnitude of a missense variant’s molecular effect. Envision combines 21,026 variant effect measurements from nine large-scale experimental mutagenesis datasets, a hitherto untapped training resource, with a supervised, stochastic gradient boosting learning algorithm. Envision outperforms other missense variant effect predictors both on large-scale mutagenesis data and on an independent test dataset comprising 2,312 TP53 variants whose effects were measured using a low-throughput approach. This data set was never used for hyperparameter tuning or model training, and thus serves as an independent validation set. Envision prediction accuracy is also more consistent across amino acids than other predictors. Finally, we demonstrate that Envision’s performance improves as more large-scale mutagenesis data is incorporated. We precompute Envision predictions for every possible single amino acid variant in human, mouse, frog, zebrafish, fruit fly, worm and yeast proteomes (https://envision.gs.washington.edu/).
eTOC BLURB
We present Envision, an accurate predictor of protein variant molecular effect trained using large-scale experimental mutagenesis data.
INTRODUCTION
Mutations have the power to reshape protein structure, stability or activity and can have drastic effects on evolutionary fitness, protein function and human health. For example, mutations were used to improve the pharmacokinetic and pharmacodynamic properties of insulin (Vigneri et al., 2010). Moreover, a recent survey of genetic variation in humans revealed that each individual harbors ~50 private missense variants, most of which are of unknown effect (Karczewski et al., 2017; Zou et al., 2016). This example highlights how DNA sequencing advances have facilitated detection of genetic variation. However, in both laboratory and clinical settings, determining the impact of a missense variant on a protein’s function remains a challenge (MacArthur et al., 2014).
Experiments can reveal a variant’s molecular effect, and recent advances in multiplex assays have enabled the assessment of large numbers of variants (Fowler and Fields, 2014; Gasperini et al., 2016). However, we are far from having a comprehensive atlas of missense variant effects in the human proteome, and such an atlas is a distant goal for model organisms. Thus, variant effect predictors such as PolyPhen2 (Adzhubei et al., 2010), SIFT (Sim et al., 2012), SNAP2 (Hecht et al., 2015), Evolutionary Action (Katsonis and Lichtarge, 2014), CADD (Kircher et al., 2014) and a host of others (Tang and Thomas, 2016) will continue to be widely used to predict missense variant effects. Some predictors are products of sophisticated supervised machine learning algorithms, and are developed using features and training data that make them suited for a particular type of prediction problem. For instance, the PolyPhen2 HumDiv model is a support vector machine trained on thousands of human Mendelian disease-associated and neutral variants, and is thus optimized to predict the clinical variant effects (Adzhubei et al., 2010). SNAP2, an ensemble of neural network models, is trained on human pathogenic and neutral variants as well as variants that impact molecular function (Hecht et al., 2015). Given the breadth of training data, SNAP2 predictions encompass both the clinical and molecular effects of missense variants. Conversely, SIFT and Evolutionary Action are not products of machine learning, but instead rely on evolutionary patterns to predict variant effects. Despite their simplicity, SIFT and Evolutionary Action perform similarly to PolyPhen2 and SNAP2 (Katsonis and Lichtarge, 2014), which highlights the importance of evolutionary information to successful variant effect prediction. A recently described unsupervised method, EVmutation, leverages evolutionary signatures of epistasis to predict variant effects, and has demonstrated enhanced accuracy over SIFT and PolyPhen2 for both molecular and clinical effect prediction (Hopf et al., 2017). These tools are all used to prioritize variants in clinical and laboratory settings.
Current predictors face two major limitations. First, most are optimized to predict categorical variant effects (e.g. damaging vs. benign), and cannot accurately predict effect magnitude. This limitation arises primarily from the structure of variant effect databases used to train predictors. For example, the Human Gene Mutation Database (Stenson et al., 2012), Online Mendelian Inheritance of Man (Amberger et al., 2015), and ClinVar (Landrum et al., 2013) all categorize variants as clinically deleterious or benign. Swiss-Prot and the Protein Mutant Database contain categorical measures of variant effects in laboratory assays. Second, most predictors focus on predicting the clinical effect of human variants rather than the molecular effects on protein function (Adzhubei et al., 2010; Sim et al., 2012). However, the relationship between molecular effect and clinical effect is complex, and most predictors do not deal well with this complexity. For example, both gain- and loss-of-function variants of BRAF can be pathogenic (Rodriguez-Viciana et al., 2006; Wan et al., 2004). Variants of PTEN variants can drive carcinogenesis when they occur somatically, or can cause autism or a tumor syndrome when they occur in the germline (Mester and Eng, 2013). Thus, we suggest that accurate clinical effect prediction should start with accurate, quantitative predictions of molecular effect whose subsequent interpretation is guided by specific knowledge about gene-disease associations.
Here we address the need for an accurate, quantitative predictor of molecular effect by leveraging deep mutational scanning data. In a deep mutational scan, selection for protein function among a library of nearly all possible single amino acid variants of a protein is coupled to high-throughput DNA sequencing (Fowler and Fields, 2014; Fowler et al., 2014). Sequencing reveals how each variant’s frequency changes during selection, yielding quantitative scores that describe the functional effect of each variant in the library. The resulting large-scale mutagenesis datasets have a distinct advantage over traditionally-used variant effect predictor training datasets like HumDiv/HumVar, HGMD and the Protein Mutant Database. Traditional datasets contain a large number of proteins, each with a median of four to six variant effect measurements. A large-scale mutagenesis dataset contains deep and unbiased information, capturing the effects of most variants at every position in a single protein. We hypothesize that large-scale mutagenesis datasets contain informative and generalizable patterns that can be used to predict variant effects in disparate proteins.
Here, we use the molecular effects of 21,026 variants of eight proteins, determined through deep mutational scans, to train Envision, a decision tree ensemble-based quantitative variant effect predictor. Envision uses a stochastic gradient boosting learning algorithm, which excels at analyzing nonlinear interactions between features and has performed well in a myriad of regression tasks (Friedman, 2002). To maximize Envision’s generalizability, proteins in the Envision training set have disparate structures and functions, and are drawn from diverse organisms. We demonstrate the generality of Envision’s predictions by iteratively training models that exclude a single protein dataset and then comparing the resulting model’s predictions to the observed variant effects for the excluded protein. We also assess performance using independent variant effect data that was not generated by deep mutational scanning nor included in Envision’s training. Envision’s predictions are generally more accurate than other state-of-the-art predictors. Envision’s prediction accuracy is also consistent across different amino acids, unlike other predictors that perform well on some amino acids and poorly on others. We pre-computed Envision predictions for all possible single amino acid variants of proteins in the human, mouse, fruit fly, clawed frog, zebrafish, worm, and yeast proteomes. We provide a web-based tool allowing users to visualize and explore predicted protein sequence-function maps, which can be used to prioritize variants. Envision is available at https://envision.gs.washington.edu.
RESULTS
Data collection and curation
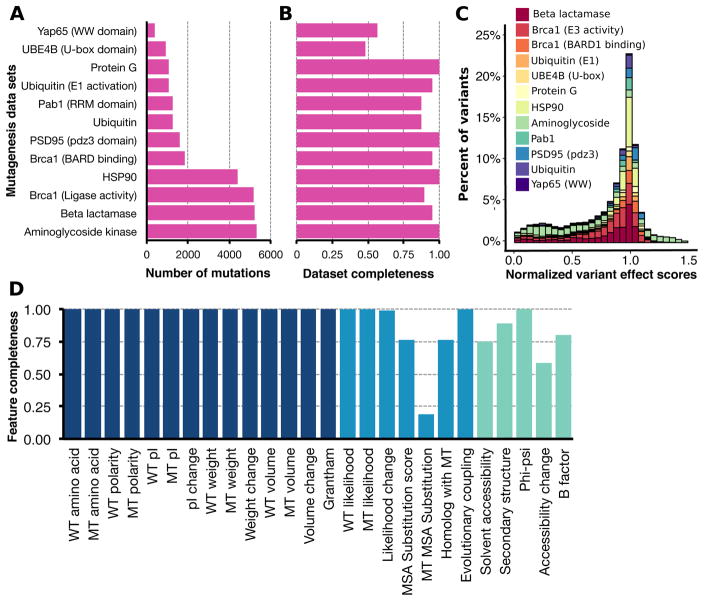
We collected previously published, large-scale mutagenesis datasets with quantitative measures of variant effect on protein function. Exploratory analysis led to the following inclusion criteria: 1) the experiment must have measured single amino acid variant effects, rather than averaging across different genetic backgrounds; 2) the experiment must have been on a natural protein instead of a designed protein; and 3) the experiment must have quantitated effects for at least ~50% of all possible variants of the mutagenized region. Ultimately, deep mutational scans of ten proteins from twelve studies comprising 28,545 single amino acid variant effects met these criteria (Figure 1A, Supplementary Table 1). Variant coverage ranged from ~50% for the Ube4b domain of murine E3 ligase to 100% for the IgG-binding domain of influenza protein G and the PDZ domain of human PSD-95 (Figure 1B). Variant coverage depended on experimental details like the protocol used for library generation (e.g., doped oligomer (Matteucci and Heyneker, 1983) vs. site saturation mutagenesis (Jain and Varadarajan, 2014)), the number of clones generated and the sequencing depth. The proteins in the dataset were distinct, coming from different organisms, having different structures and having functions ranging from catalysis to peptide binding (Supplementary Table 1). To make datasets comparable, we normalized variant effect scores in each dataset such that variants that were more active than wild-type had a variant effect score greater than one, wild-type-like variants had a score of one and variants that were less active than wild-type had a score less than one (Supplementary Figure 1, Figure 1C).
Figure 1. Large-scale mutagenesis data and descriptive features used to train Envision.
The number of single mutants (A) collected from different protein or protein domain large-scale mutagenesis datasets and the mutational completeness of each dataset (B) are shown. Mutational completeness was calculated by dividing the number of observed single mutants by the number possible single mutants. (C) The distribution of variant effect scores for each large-scale mutagenesis dataset is shown. For each dataset, variant effect scores were normalized such that a score of one is wild type-like and a score of zero is inactivating (see Supplementary Figure 1 for unnormalized score distribution). Each collected variant was annotated with 27 features, which describe physicochemical (dark blue), evolutionary (blue) or structural (green) variant attributes (Supplementary Table 2). (D) The proportion of variants in the collected large-scale mutagenesis datasets having each feature is shown (WT = wild type, MT = mutant).
Next, we annotated each variant with 27 biological, structural and physicochemical features. The biological features captured evolutionary constraints using both site-specific and co-varying conservation metrics. The structural features included local density and solvent accessibility, while the physicochemical features describe properties of amino acids, such as polarity and size. Physicochemical and biological features were available for nearly all variants, but structural features were not (Figure 1D, Supplementary Table 2).
Predicting quantitative variant effects
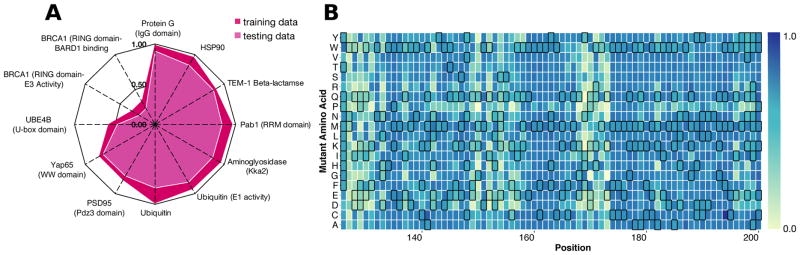
We first tested whether a stochastic gradient boosting regression algorithm could model the relationships between our 27 features and quantitative variant effect scores for each protein, individually. To train each single-protein model, hyperparameters, such as the number of decision trees in the ensemble and tree depth were tuned using tenfold cross-validation. After hyperparameter tuning, we reserved 20% of mutations for testing, allowing us to estimate the generality of each model to unseen variants. Nine of the twelve models performed well (median Pearson’s R = 0.83, Spearman’s ρ = 0.80, Figure 2A), while three, the BRCA1 RING domain BARD binding, BRCA1 RING domain E3 activity and E4B ubiquitin ligase models, performed poorly (median R = 0.22, ρ = 0.35).
Figure 2. Protein-specific gradient boosting models can accurately predict variant effect scores.
We trained a model for each protein using a randomly selected 80% of data, with 20% reserved for testing. (A) A radar plot of Pearson’s correlation coefficients between observed and predicted variant effect scores illustrates protein-specific model performance on both training (dark red) and testing data (light red). The PAB1 RRM domain-specific model predicts the effects of variants withheld from training well (Pearson’s R > 0.75), and was used to predict the 197 missing variant effect scores. (B) The completed Pab1 RRM domain sequence-function map is shown for positions 126–200. Each mutagenized position is a column, and each amino acid substitution is a row. Wild type-like variants are colored dark blue and inactive variants are colored light blue. Predicted effects are denoted by black borders.
Experimental noise cannot account for these models’ poor performance, since the correlation of model predictions with the training and testing data is much lower than the correlation between replicate experiments (Supplementary Table 1). We hypothesized that poor performance arose because correlations between the features and variant effect scores were low (Supplementary Figure 2). Low correlation might occur because the assays did not test every function of these proteins. For instance, BCRA1 RING domain variants were assayed for E3 ligase activity and BARD binding. However, BRCA1 has many functions and interacts with >25 other proteins (Deng and Brodie, 2000; Kerrien et al., 2012). Another possibility is that these two datasets were missing some structural features. However, the YAP65 WW domain dataset, missing the same features, resulted in an accurate model. Thus, we could not identify the cause of poor performance in the BRCA1 RING domain and E4B ubiquitin ligase models. We excluded these three datasets from subsequent analyses.
For most proteins, our feature set and learning procedure generated accurate models of variant effect. Beyond validating our approach, these single protein models enabled us to complete each large-scale mutagenesis dataset by predicting missing variant effect scores (Supplementary Table 3). For example, we used the Pab1 model (R = 0.86; ρ = 0.79) to predict the ~20% of scores that were missing, completing the Pab1 dataset (Figure 2B).
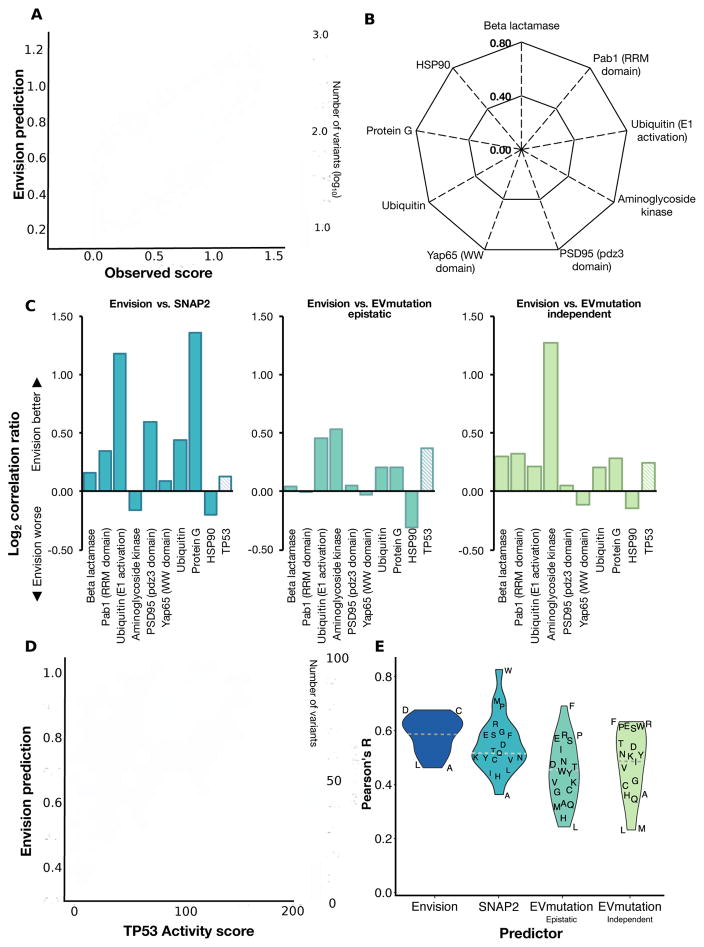
Next, we trained a global model with the 21,026 empirically-derived variant effect scores in the nine large-scale mutagenesis datasets. We tuned hyperparameters using a leave-one-protein-out approach designed to avoid protein-specific overtraining (Supplementary Figure 3, Supplementary Table 4). Once hyperparameters were tuned, we trained Envision with all available data, except for a random 5% of variant effect scores that we withheld for testing and to assess overfitting. Training and testing data root mean squared errors were similar at each model training iteration, indicating that the model is not overfitted to the training data (Supplementary Figure 4). Envision predicted the training data well (R = 0.79, ρ = 0.76; Figure 3A).
Figure 3. Envision outperforms other quantitative variant effect predictors.
(A) A hexagonal bin plot shows the correlation between predicted and observed variant effect scores for all the large-scale mutagenesis data used to train Envision (Pearson’s R = 0.79). To evaluate performance on data not used in training, models were retrained excluding each one of the nine proteins (see Supplementary Figure 3–4 for cross-validation scheme and training performance). (B) A radar plot shows the correlation (Pearson’s R) between predicted and observed variant effect scores when the indicated protein was left out (see Supplementary Figure 5 for scatter plots). (C) We also compared the leave-one-protein-out models to SNAP2 (left panel), EVmutation-epistatic (middle panel) and EVmutation-independent (right panel). The log2 ratio of each leave-one-protein-out model’s Pearson’s R to another predictor Pearson’s R on the left-out data is shown. Hashed bars indicate relative performance on a set of 2,312 TP53 transactivation activity scores measured in a low-throughput assay and not used in training (see Supplementary Figure 7 for raw comparision). (D) A hexagonal bin plot shows the correlation between Envision predictions and TP53 activity scores (Pearson’s R = 0.58). (E) A violin plot illustrates the distribution of Pearson’s correlation coefficients for variant effect scores and Envision, SNAP2 and EVmutation predictions for different mutant amino acids. The dashed horizontal line indicates the median Pearson’s correlation coefficients for each predictor (see Supplementary Figure 9A–B for heatmap of correlations).
Assessing Envision’s performance
To evaluate Envision’s performance, we employed a jack-knife leave-one-protein-out (LOPO) approach. Here, we repeated the training procedure described above, leaving one protein completely out of the hyperparameter tuning and model training process. Then, we used the resulting model to predict variant effect scores for the left-out protein and determined performance. We repeated this procedure for all nine proteins. Variant effect scores for left-out proteins were predicted with Pearson’s R ranging from 0.38 to 0.69 and Spearman’s ρ ranging from 0.30 to 0.74 (Figure 3B; Supplementary Figure 5). To determine the effect of our variant effect score normalization scheme on model training and performance, we compared LOPO models trained using either normalized or non-normalized variant effect scores. Models trained using normalized data predicted variant effect scores for the left-out protein better than models trained using non-normalized data (median R = 0.56 vs 0.39, median ρ = 0.51 vs 0.35; Supplementary Figure 6). This result highlights the utility of our normalization scheme.
Next, we compared our LOPO models’ performance to other predictors. PolyPhen2 is trained to predict the categorical clinical effect of variants, but also generates a numerical score. This score is the naïve Bayes posterior probability that a variant is damaging, and, although quantitative, it is not designed to predict the magnitude of a variant’s molecular effect. As expected, for the only human protein in our dataset, YAP65 WW domain, our LOPO model outperformed PolyPhen2 when predicting WW domain variant effect scores (R = 0.46 vs. 0.17; ρ = 0.36 vs. 0.19). Like PolyPhen2, SIFT also generates categorical predictions and scores for human proteins. SIFT scores represent the scaled probability of a missense variant being tolerated, and are also not expected to capture the magnitude of variant molecular effects. The WW domain LOPO model also outperformed SIFT scores (R 0.46 vs. 0.03; ρ = 0.36 vs. 0.04). PolyPhen2 and SIFT were not designed to predict variant effect magnitude, and our results confirm that they should not be used to do so.
SNAP2, EVmutation and Evolutionary Action were developed to predict variant effect magnitude (Hecht et al., 2015; Hopf et al., 2017; Katsonis and Lichtarge, 2014). Evolutionary Action scores could not be obtained by batch query, preventing us from including them in our analysis. SNAP2 predicted variant effect scores much better than PolyPhen2 or SIFT, but not as well as our LOPO models, which outperformed SNAP2 on seven of nine datasets (median R 0.56 vs. 0.44; Figure 3C). EVmutation predicts variant effect magnitude using either an epistatic or an independent conservation-based unsupervised statistical model. Our LOPO models outperformed EVmutation’s epistatic model on six out of nine datasets (median R 0.56 vs. 0.47) and EVmutation’s independent model on seven of nine (mean R 0.56 vs. 0.48; Figure 3C). An equivalent analysis using Spearman’s ρ revealed similar results (Supplementary Figure 7). Across all datasets, our LOPO models’ predictions are 4%, 14% and 21% more correlated with the observed variant effect scores than predictions from EVmutation epistatic, EVmutation independent and SNAP2, respectively.
Next, we analyzed what factors led to our improved performance. Envision’s features are similar to those used by SNAP2 and PolyPhen2, so the improvement we observed is not likely due to feature choice. Instead, we hypothesized that our use of deep mutational scanning data and our cross-validation approach, designed to yield a generalizable model, are the two attributes that led to improved performance. The lack of a large database of quantitative variant effects measured by means other than deep mutational scanning made it impossible to evaluate the performance advantage conferred by using deep mutational scanning data. However, we quantified the impact of our cross-validation approach by comparing the performance of models trained using standard tenfold cross-validation to models trained using our LOPO scheme. We found our LOPO approach improved performance by ~10–20% over all protein datasets compared to tenfold cross-validation (median R = 0.56 vs. 0.45, ρ = 0.50 vs. 0.45; Supplementary Figure 8). We suggest that our LOPO approach, designed to yield generalizable models, was especially important given that our training data set contained relatively few proteins.
Our leave-one-protein-out analysis demonstrated that Envision provided improved quantitative predictions of variant effects measured using deep mutational scanning. However, Envision’s performance advantage might have arisen because it learned deep mutational scanning-specific patterns in the data. To ensure that Envision was not overfitted to deep mutational scanning data, we obtained a TP53 tumor suppressor mutagenesis dataset where the effects of 2,312 variants on TP53 transactivation were measured individually using a fluorescent reporter (Kato et al., 2003). We predicted these TP53 variant effect scores using Envision, which was trained on all nine large-scale mutagenesis data sets. The TP53 data were never used, directly or indirectly, in the training procedure. Despite the fact that the TP53 dataset was not acquired using deep mutational scanning, Envision predicted the TP53 variant effect scores well (R = 0.58, ρ = 0.53; Figure 3C, D). Importantly, Envision outperformed SNAP2 (R = 0.53; ρ = 0.50), whose training dataset included the effects of ~400 human TP53 mutations, and EVmutation (epistatic R = 0.45, ρ = 0.49; independent R = 0.49, ρ = 0.52; Figure 3C). Thus, Envision learned patterns of the molecular effects of variants that do not depend on the measurement method.
Next, we sought to determine whether Envision performance depended on the identity of either the mutant or wild type amino acid. We evaluated performance on the TP53 dataset to enable comparison to EVmutation and SNAP2. We found that Envision prediction performance did not depend much on the identity of the mutant amino acid (Figure 3E, Supplementary Figure 9A). However, EVmutation and SNAP2 showed large biases in performance. For instance, EVmutation predicted mutations to phenylalanine with high accuracy (R = 0.69, ρ = 0.70), but predicted mutations to leucine with low accuracy (R = 0.24, ρ = 0.33). SNAP2 performance was also biased in favor of mutations to tryptophan and methionine and against mutations to alanine. These biases are also apparent for the wild-type amino acid, where EVmutation predicted mutations from wild type cysteine well (R = 0.82, ρ = 0.71) and wild type aspartic acid poorly (R = 0.02, ρ = 0.05; Supplementary Figure 9B). Consequently, in addition to greater overall accuracy, Envision performance was more consistent.
Finally, we assessed the utility of Envision scores for clinical effect prediction, evaluating performance by constructing ROC curves using variants annotated as either pathogenic or benign in ClinVar. Envision predictions were better than random guessing (AUROC = 0.72), but not as good as PolyPhen2, CADD and SIFT (AUROCs = 0.86, 0.85, 0.84; Supplementary Figure 10). This result is not surprising because Envision was not designed or optimized for this task, and because comparison of predictor performance on clinical data is difficult given that many predictors are trained on or optimized to predict these data (Grimm et al., 2015). Furthermore, the relationship between the magnitude of a variant’s molecular effect and disease phenotype is likely to be different for each disease-associated protein. For example, a weakly damaging variant in some proteins may be sufficient to cause disease, whereas only strongly damaging variants lead to disease in other proteins. Finally, we note that the rate at which training datasets grow in the coming years may be much greater for deep mutational scans than for clinical variants.
Feature importance and future improvements
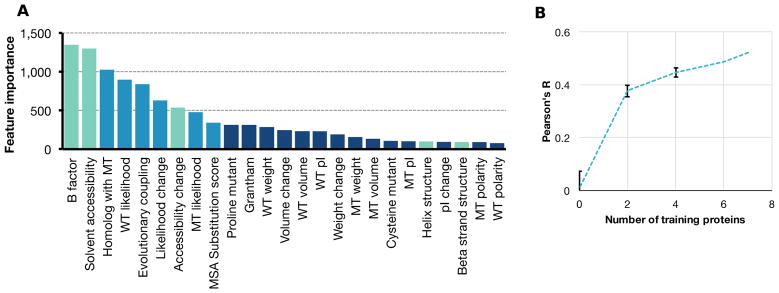
Features known to be predictive of variant effects, including solvent accessibility and evolutionary conservation, were the most highly represented in the Envision decision tree ensemble (Figure 4A; Supplementary Table 5) (Kumar et al., 2009; Saunders and Baker, 2002). However, unlike for other feature-driven predictors (Adzhubei et al., 2010; Hecht et al., 2015), we found that the mutant amino acid identity was informative. This amino acid identity effect was largely driven by proline. Proline variants are generally disruptive of protein function and, indeed, proline variants were the most damaging substitutions in the large-scale mutagenesis datasets (proline mean effect score = 0.60 vs. all AA mean = 0.81; paired t-test P ≪ 0.001, n = 8; Supplementary Figure 11). Envision predicted the effects of proline variants about as accurately as the effects of other variants (Supplementary Figure 12). Thus, rather than simply predicting that all proline variants were strongly damaging, Envision predicted the degree to which proline variants maintain or disrupt function.
Figure 4. Envision is an interpretable model that will improve with more training data.
The number of times each feature is used in Envision’s decision tree ensemble is a measure of feature importance. (A) Feature importance for every physicochemical (dark blue), biological (blue) and structural (green) feature is shown (WT = wild type, MT = mutant). See Supplementary Figure 11–12 for proline feature analysis. (B) To assess the impact of adding more training data to Envision, we conducted a downsampling analysis. Models were trained with increasing numbers of randomly selected protein datasets, and tested on mutations from proteins withheld from training. The mean Pearson’s correlation coefficient between predicted and observed variant effects across testing datasets are shown, organized by the number of proteins included in the training set. Error bars indicate the standard deviation of correlation coeffcients obtained from ten random samplings of proteins to include in the training set. A naïve model (i.e. number of training proteins = 0) was also generated by randomizing feature values for all proteins and repeating the training procedure. The error bars for the naïve model indicate the standard deviation of correlation coefficients obtained from ten different feature randomizations. See Supplementary Figure 13 for left-out feature analysis.
Structural and evolutionary features are important for Envision’s predictions but are not always available. Thus, we quantified predictive performance when these features were missing by masking them for each of the nine LOPO models when they predicted the left-out protein’s variant effect scores. As expected, models performed worse without structural features. For example, the β-lactamase LOPO model predicted β-lactamase variant effect scores 15% worse when structural features were masked (R = 0.69 vs 0.59; Supplementary Figure 13). Similarly, the β-lactamase LOPO model predicted β-lactamase variant effect scores 13% worse when evolutionary features were masked (R = 0.69 vs 0.60). Across the nine LOPO models, we found that masking structural features degraded performance by 39% and masking evolutionary features degraded performance by 18%. Thus, we strongly encourage users to consider feature completeness when using Envision’s predictions. Feature information is available, along with predictions, on the Envision website. We note that all feature-driven predictors suffer when key features are unavailable.
Finally, we determined how the number of proteins in our training dataset affected Envision performance. We trained versions of Envision with different numbers of proteins and tested on the left-out proteins. We found that model performance increased as more proteins were used in training, suggesting that accumulation of more data will improve Envision’s predictive performance (Figure 4B).
Availability of Envision predictions
Envision predictions are available for proteins from seven commonly studied organisms: human (N = 20,130), mouse (N = 16,836), fruit fly (N = 3,375), clawed frog (N = 1,704), zebrafish (N = 2,982), worm (N = 3,802), and yeast (N = 8,322). We provide predictions for all 19 alternative amino acids at each position, with batch query and download options available. Along with predictions, features are also available for download.
DISCUSSION
We developed Envision, the first variant molecular effect predictor trained on large-scale mutagenesis data. Envision accurately predicts variant effects in large-scale mutagenesis data withheld from training as well as variant effects from low-throughput experiments. Overall, Envision outperforms other quantitative predictors like SNAP2 and EVmutation in predicting experimentally measured molecular effects. In particular, the quality of Envision predictions is relatively uniform across different amino acid substitutions, whereas other predictors’ accuracy is driven by high performance on some substitutions and poor performance on others. The promise of using large-scale mutagenesis data to develop variant effect predictors is highlighted by the fact that Envision was trained from deep mutational data on only nine proteins, but can outperform established methods that are trained using sparse mutational data on thousands of proteins. As more large-scale mutagenesis data becomes available, Envision will continue to improve.
Envision also has limitations. Envision’s predictions are provided as quantitative scores that range from ~0 to ~1, where scores less than one are damaging as compared to wild-type. Envision can predict the scores of strongly damaging and wild type-like mutations well, but predicts mutations of intermediate effect less well (Supplementary Figure 5). Envision also relies on structural and evolutionary features that are not available for every protein, and predictive performance degraded when these features were missing. Thus, while Envision predictions are available for millions of variants, we recommend caution when key features are missing. The Envision web tool reveals missing features for each prediction.
To train Envision, we employed large-scale mutagenesis data from two types of deep mutational scans. One type is based on a generalized selection for protein function whereas the other type is based on selection for a specific protein function. Specific selections could fail to capture the effect of variants on other functions of the protein like binding to a different substrate or catalysis. Envision’s was trained using data from both generalized and specific deep mutational scans, and did not distinguish between them. Therefore, Envision predicts generalized variant effects, and does not distinguish between specific molecular effects like enzymatic activity for one substrate or another, or binding versus catalysis. Collection of more large-scale mutagenesis data for the specific molecular effects of variants may enable the development of predictors that capture these specific functional effects.
We anticipate that Envision will be useful for identifying candidate variants that tune protein activity levels. Envision’s predictions of molecular effect may also be useful when the relationship between protein function and disease is clear. Furthermore, Envision will continue to improve as new datasets become available.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Douglas Fowler (dfowler@uw.edu).
METHOD DETAILS
Training data collection
Published large-scale mutagenesis datasets were used as training data if they met several criteria. First, we required that at least 50% of all possible single amino acids were substituted at each mutagenized position. Thus, alanine and proline scans did not qualify for this study. Second, we only accepted mutational scans of native proteins assayed for native biological function. Third, we excluded scans in which the complete variant sequence was unknown. We also removed variants with more than one mutation. In total, we accepted twelve datasets comprising ~30,000 missense mutations. These scans were performed on proteins from different organisms: human, mouse, rat, S. cerevisiae, and bacteria (Supplementary Table 1).
Normalization
Each large-scale mutagenesis dataset was generated using a distinct experimental assay, which resulted in different variant effect score distributions. To enable meaningful comparison between datasets, we normalized them. For each dataset, every variant effect score was normalized to the wild type score and then log2 transformed. Next, we subtract the median effect of synonymous variants, if available. Synonymous variants were unavailable for the PSD95 (Pdz3 domain), Protein G (IgG domain), UBE4B (U-box domain) and BRCA1 datasets, so we instead subtracted 0 from each score in those datasets. Lastly, we divided each score by the negative median score of the bottom 1% of mutations of each dataset and added one. Our normalization scheme is expressed as Snormalized = (Sreported i − Smedian synonymous)/( −Smedian bottom 1%)+1, where S signifies score. This normalization scheme results in variants that are more active than wild type having scores of greater than one, wild type-like variants having scores of one, and damaging variants having scores of less than one.
Variant annotation
Mutations were annotated with three general types of descriptive annotations: evolutionary, biochemical and structural (Supplementary Table 2). Several evolutionary features used in our model were obtained using the PolyPhen2 annotation pipeline (Sunyaev et al., 1999). We also derived a measure of average mutational covariance between a given position and all other positions in a multiple sequence alignment from EVfold (Hopf et al., 2017). To obtain structural information, we use DSSP (http://www.cmbi.ru.nl/dssp.html) (Kabsch and Sander, 1983) and PDB files from the Protein Data Bank (http://www.rcsb.org/pdb/home/home.do) (Rose et al., 2011). Our biochemical annotations include measures of amino acid size, weight, volume, isoelectric point, and Grantham scores (Grantham, 1974).
Machine learning
Stochastic gradient boosting is a method of machine learning that uses an ensemble of weak prediction models (e.g., decision trees) for classification or regression problems (Friedman, 2002). We constructed stochastic gradient boosting tree regression models using the GraphLab Create framework from Turi (https://turi.com/products/create/). Hyperparameters were optimized using a grid search. For each predictive model, we tuned six parameters in a stepwise fashion. First, we optimized for the number of decision trees in the ensemble. Next, we tuned the maximum depth of a decision tree and the minimum number of observances allowed in a terminal node of a tree. Then, we determined the value that the squared-loss must be reduced by in order to add an additional node to a tree. Finally, we identified the optimal proportion of variant effect scores and features used to train each tree. Once hyperparameters were tuned, we reduced the learning rate from 0.1 to 0.01 and increased the number of decision trees by fivefold. All tuned and trained models treat missing feature data as such, i.e., no imputing procedures were performed. Instead, during training, the algorithm uses variants with missing features to determine how feature missingness should handled by the model at each tree node (Chen and Guestrin, 2016).
Single protein models
To filter out datasets that are noisy or contain variant effects that cannot be explained by our evolutionary, structural or physicochemical features, we performed gradient boosting machine learning on a randomly selected 80% of variant effect scores from each protein dataset. This resulted in a model for each protein, which we used to predict the 20% of variant effect scores withheld from model training. Proteins whose specific models performed poorly on withheld data (Pearson’s R < 0.5) were excluded from the LOPO and global models.
Training Envision
Envision was trained using the same approach as our single protein models with an added leave-one-protein-out cross-validation procedure, where, at each round, a different protein was removed from the training set and used for validation (Supplementary Figure 3). Thus, after each round of training, a model’s generality was tested on variant effect scores from a protein not used to train the model. This cross-validation procedure allowed us to test an array of hyperparameters to see which parameter sets yielded the most generalizable models. Here, model generality was determined by measuring the root mean squared error between model predictions and variant effect scores from a left-out protein. Once all hyperparameters were optimized (Supplementary Table 4), we trained Envision with all available data except for a randomly selected 5% of which we excluded to evaluate model generality and ensure that the model was not overfitted. The resulting model was used to make all the Envision predictions available on our website.
Leave-one-protein-out (LOPO) models
To estimate Envision’s performance on proteins not used in model training, we generated nine LOPO models. These models were trained using the same protocol as Envision, except that in each case a different protein was left completely out of the hyperparameter tuning and final model training procedures. These LOPO models were used to estimate Envision’s performance on proteins not included in the training set.
Downsampling analysis
To evaluate the effect of additional training data on model performance, we trained models with 2, 4, 6 or 7 of the available nine protein datasets. Model training was performed as described in the Training Envision section above. Each model was used to predict variant effects in proteins that were not used during the training phase. Confidence intervals were generated by repeated rounds of randomly selecting proteins to use in the training phase (n = 8).
QUANTIFICATION AND STATISTICAL ANALYSIS
The details of the statistical test we conduct, as well as definitions of center and correlation can be found in the main text. Criteria for inclusion of deep mutational scanning data sets are described in the METHOD DETAILS section of the STAR Methods.
DATA AND SOFTWARE AVAILABILITY
All data and software in this study are freely available. The training data set and all code used to train the models and generate the figures presented in this manuscript are available at https://github.com/FowlerLab/Envision2017. Envision predictions, along with feature annotations, are available at https://envision.gs.washington.edu/.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Critical Commercial Assays | ||
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| Oligonucleotides | ||
| Recombinant DNA | ||
| Software and Algorithms | ||
| Python/2.7.3 | Python | https://www.python.org/ |
| Numpy/1.13.1 | Walt et al. (2011) | http://www.numpy.org/ |
| GraphLab/2.1 | Low et al. (2012) | https://turi.com/ |
| Scipy/0.19.1 | Jones et al. (2014) | https://www.scipy.org/ |
| scikit-learn/0.17.0 | Pedregosa et al. (2011) | http://scikit-learn.org/stable/ |
| R version 3.2.3 | R | https://cran.r-project.org/ |
| ggplot2/2.2.1 | Wickham (2016) | http://ggplot2.org/ |
| reshape2/1.4.2 | Wickham (2016) | https://cran.r-project.org/web/packages/reshape2/index.html |
| DSSP | Kabsch and Sander(1983) | http://swift.cmbi.ru.nl/gv/dssp/ |
| Polyphen2 (annotations and predictions) | Adzhubei et al. (2010) | http://genetics.bwh.harvard.edu/pph2/dokuwiki/downloads |
| SIFT predictions | Ng and Henikoff (2002) | http://sift.jcvi.org/ |
| EVmutation predictions | Hopf et al. (2017) | https://marks.hms.harvard.edu/evmutation/ |
| SNAP2 predictions | Hecht M, Bromberg Y & Rost B (2015) | https://rostlab.org/services/snap2web/ |
| Envision | This study | https://github.com/FowlerLab/Envision2017 |
| Other | ||
| TEM1 β-lactamase | Firnberg | PubmedID: 24567513 |
| Yap65 (WW domain) | Fowler | 20711194 |
| PSD95 (Pdz3 domain) | McLaughlin | 23041932 |
| Brca1 (RING domain)- E3 ligase activity | Starita | 25823446 |
| Brca1 (RING domain)- Bard1 binding | Starita | 25823446 |
| Aminoglycoside kinase | Melnikov | 24914046 |
| E4B (U-box domain) | Starita | 23509263 |
| Hsp90 | Mishra | 27068472 |
| Ubiquitin | Roscoe | 23376099 |
| Pab1 (RRM domain) | Melamed | 25671604 |
| Ubiquitin - E1 activity | Roscoe | 24862281 |
| Protein G (IgG domain) | Olson | 25455030 |
Supplementary Material
Single protein model predictions and datasets
HIGHLIGHTS.
Large-scale, quantitative mutagenesis data offers a novel source of training data
Envision outperforms other missense variant effect predictors on independent data
More mutagenesis data will improve Envision’s predictive performance
Envision predictions are available for download: https://envision.gs.washington.edu
Acknowledgments
We thank Bill Noble and Christine Queitsch for insightful comments. This research was supported by research grants from the National Science Foundation to V.E.G. [DGE-1256082], the National Institutes of Health to J.S. [DP1HG007811], and D.M.F. [R01GM109110]. R.J.H. is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation [DRG-2224-15].
Footnotes
AUTHOR CONTRIBUTIONS
The project was conceived and designed by all authors. V.E.G. and R.J.H. performed data analysis and generated figures. All authors wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, Hamosh A. OMIM. org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucl Acids Res. 2015;43:789–798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Guestrin C. XGBoost: A Scalable Tree Boosting System, the 22nd ACM SIGKDD International Conference; New York, New York, USA: ACM; 2016. [DOI] [Google Scholar]
- Deng CX, Brodie SG. Roles of BRCA1 and its interacting proteins. Bioessays. 2000;22:728–737. doi: 10.1002/1521-1878(200008)22:8<728::AID-BIES6>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Fowler DM, Fields S. Deep mutational scanning: a new style of protein science. Nat Methods. 2014;11:801–807. doi: 10.1038/nmeth.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler DM, Stephany JJ, Fields S. Measuring the activity of protein variants on a large scale using deep mutational scanning. Nat Protoc. 2014;9:2267–2284. doi: 10.1038/nprot.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JH. Stochastic gradient boosting. Comput Stat Data Anal. 2002;38:367–378. doi: 10.1016/S0167-9473(01)00065-2. [DOI] [Google Scholar]
- Gasperini M, Starita L, Shendure J. The power of multiplexed functional analysis of genetic variants. Nat Protoc. 2016;11:1782–1787. doi: 10.1038/nprot.2016.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- Grimm DG, Azencott CA, Aicheler F, Gieraths U, MacArthur DG, Samocha KE, Cooper DN, Stenson PD, Daly MJ, Smoller JW, Duncan LE, Borgwardt KM. The evaluation of tools used to predict the impact of missense variants is hindered by two types of circularity. Hum Mutat. 2015;36:513–523. doi: 10.1002/humu.22768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht M, Bromberg Y, Rost B. Better prediction of functional effects for sequence variants. BMC Genomics. 2015;16:1–12. doi: 10.1186/1471-2164-16-S8-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopf TA, Ingraham JB, Poelwijk FJ, Schärfe CPI, Springer M, Sander C, Marks DS. Mutation effects predicted from sequence co-variation. Nat Biotechnol. 2017;35:128–135. doi: 10.1038/nbt.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain PC, Varadarajan R. A rapid, efficient, and economical inverse polymerase chain reaction-based method for generating a site saturation mutant library. Anal Biochem. 2014;449:90–98. doi: 10.1016/j.ab.2013.12.002. [DOI] [PubMed] [Google Scholar]
- Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Karczewski KJ, Weisburd B, Thomas B, Solomonson M, Ruderfer DM, Kavanagh D, Hamamsy T, Lek M, Samocha KE, Cummings BB, Birnbaum D, Daly MJ, MacArthur DG The Exome Aggregation Consortium. The ExAC browser: displaying reference data information from over 60 000 exomes. Nucl Acids Res. 2017;45:D840–D845. doi: 10.1093/nar/gkw971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S, Han SY, Liu W, Otsuka K, Shibata H, Kanamaru R, Ishioka C. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc Natl Acad Sci. 2003;100:8424–8429. doi: 10.1073/pnas.1431692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsonis P, Lichtarge O. A formal perturbation equation between genotype and phenotype determines the Evolutionary Action of protein-coding variations on fitness. Genome Res. 2014;24:2050–2058. doi: 10.1101/gr.176214.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrien S, Aranda B, Breuza L, Bridge A, Broackes-Carter F, Chen C, Duesbury M, Dumousseau M, Feuermann M, Hinz U, Jandrasits C, Jimenez RC, Khadake J, Mahadevan U, Masson P, Pedruzzi I, Pfeiffenberger E, Porras P, Raghunath A, Roechert B, Orchard S, Hermjakob H. The IntAct molecular interaction database in 2012. Nucl Acids Res. 2012;40:841–846. doi: 10.1093/nar/gkr1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Suleski MP, Markov GJ, Lawrence S, Marco A, Filipski AJ. Positional conservation and amino acids shape the correct diagnosis and population frequencies of benign and damaging personal amino acid mutations. Genome Res. 2009;19:1562–1569. doi: 10.1101/gr.091991.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, Maglott DR. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucl Acids Res. 2013;44:862–868. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur DG, Manolio TA, Dimmock DP, Rehm HL, Shendure J, Abecasis GR, Adams DR, Altman RB, Antonarakis SE, Ashley EA, Barrett JC, Biesecker LG, Conrad DF, Cooper GM, Cox NJ, Daly MJ, Gerstein MB, Goldstein DB, Hirschhorn JN, Leal SM, Pennacchio LA, Stamatoyannopoulos JA, Sunyaev SR, Valle D, Voight BF, Winckler W, Gunter C. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matteucci MD, Heyneker HL. Targeted random mutagenesis: the use of ambiguously synthesized oligonucleotides to mutagenize sequences immediately 5′ of an ATG initiation codon. Nucl Acids Res. 1983;11:3113–3121. doi: 10.1093/nar/11.10.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mester J, Eng C. When overgrowth bumps into cancer: the PTEN-opathies. Am J Med Genet. 2013;163:114–121. doi: 10.1002/ajmg.c.31364. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Tetsu O, Tidyman WE, Estep AL, Conger BA, Cruz MS, McCormick F, Rauen KA. Germline mutations in genes within the MAPK pathway cause cardio-facio-cutaneous syndrome. Science. 2006;311:1287–1290. doi: 10.1126/science.1124642. [DOI] [PubMed] [Google Scholar]
- Rose PW, Beran B, Bi C, Bluhm WF, Dimitropoulos D, Goodsell DS, Prlić A, Quesada M, Quinn GB, Westbrook JD, Young J, Yukich B, Zardecki C, Berman HM, Bourne PE. The RCSB Protein Data Bank: redesigned web site and web services. Nucl Acids Res. 2011;39:392–401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders CT, Baker D. Evaluation of Structural and Evolutionary Contributions to Deleterious Mutation Prediction. J Mol Biol. 2002;322:891–901. doi: 10.1016/S0022-2836(02)00813-6. [DOI] [PubMed] [Google Scholar]
- Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucl Acids Res. 2012;40:452–457. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenson PD, Ball EV, Mort M, Phillips AD, Shaw K, Cooper DN. The Human Gene Mutation Database (HGMD) and its exploitation in the fields of personalized genomics and molecular evolution. Hum Genet. 2012;133:1–9. doi: 10.1002/0471250953.bi0113s39. [DOI] [PubMed] [Google Scholar]
- Sunyaev SR, Eisenhaber F, Rodchenkov IV, Eisenhaber B, Tumanyan VG, Kuznetsov EN. PSIC: profile extraction from sequence alignments with position-specific counts of independent observations. Protein Eng. 1999;12:387–394. doi: 10.1093/protein/12.5.387. [DOI] [PubMed] [Google Scholar]
- Tang H, Thomas PD. Tools for Predicting the Functional Impact of Nonsynonymous Genetic Variation. Genetics. 2016;203:635–647. doi: 10.1534/genetics.116.190033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigneri R, Squatrito S, Sciacca L. Insulin and its analogs: actions via insulin and IGF receptors. Acta Diabetol. 2010;47:271–278. doi: 10.1007/s00592-010-0215-3. [DOI] [PubMed] [Google Scholar]
- Wan PTC, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, Jones CM, Marshall CJ, Springer CJ, Barford D, Marais R Cancer Genome Project. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1038/nrc1347. [DOI] [PubMed] [Google Scholar]
- Zou J, Valiant G, Valiant P, Karczewski K, Chan SO, Samocha K, Lek M, Sunyaev S, Daly M, MacArthur DG. Quantifying unobserved protein-coding variants in human populations provides a roadmap for large-scale sequencing projects. Nat Commun. 2016;7:13293. doi: 10.1038/ncomms13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Single protein model predictions and datasets