Abstract
In Staphylococcus aureus, an important Gram-positive human pathogen, the SaeRS two-component system is essential for the virulence and a good target for the development of anti-virulence drugs. In this study, we screened 12,200 small molecules for Sae inhibitors and identified two anti-cancer drugs, streptozotocin (STZ) and floxuridine (FU), as lead candidates for anti-virulence drug development against staphylococcal infections. As compared with STZ, FU was more efficient in repressing Sae-regulated promoters and protecting human neutrophils from S. aureus-mediated killing. FU inhibited S. aureus growth effectively whereas STZ did not. Intriguingly, RNA-seq analysis suggests that both compounds inhibit other virulence-regulatory systems such as Agr, ArlRS, and SarA more efficiently than they inhibit the Sae system. Both compounds induced prophages from S. aureus, indicating that they cause DNA damages. Surprisingly, a single administration of the drugs was sufficient to protect mice from staphylococcal intraperitoneal infection. Both compounds showed in vivo efficacy in a murine model of blood infection too. Finally, at the experimental dosage, neither compound showed any noticeable side effects on blood glucose level or blood cell counts. Based on these results, we concluded that STZ and FU are promising candidates for anti-virulence drug development against S. aureus infection.
Introduction
The emergence of multi-drug resistant bacterial pathogens is a huge medical problem1. The problem is compounded by the fact that the number of new antibiotics entering markets is decreasing2. Also, the growth inhibition activity of antibiotics inevitably selects for resistant strains3,4. Therefore, to tackle the problem, along with the development of new class of antibiotics, we need to take a broad range of alternative approaches such as phage therapy and development of anti-quorum sensing drugs or anti-virulence drugs5,6.
S. aureus is a Gram-positive human pathogen colonizing skin, anterior nares and other mucosal surface and causes a variety of diseases ranging from skin and soft-tissue infections to life-threatening diseases such as endocarditis, toxic shock syndrome, and necrotizing pneumonia7,8. The success of S. aureus as a human pathogen is mainly due to the production of a large number of virulence factors. In S. aureus, the production of virulence factors is coordinately controlled by multiple regulators such as SarA, Agr, ArlRS, and the SaeRS two-component system (TCS)9–12. Of those, the SaeRS TCS is an excellent target for anti-virulence drug development13–15. Conserved in all clinical S. aureus strains, the SaeRS TCS controls the production of more than 20 important virulence factors including toxins (e.g., alpha-hemolysin, gamma hemolysin, and leukocidins), coagulases, adhesins, and enzymes (e.g., nucleases and proteases)12. More importantly, the SaeS’s kinase activity correlates with the bacterial virulence in mice16, suggesting that the SaeRS system is a viable target for the development of anti-virulence drugs against staphylococcal infections. Since no structural information is available for SaeS, however, a rational design of Sae inhibitors is not feasible yet.
In this study, by taking a high-throughput approach with a GFP-reporter system for the SaeRS TCS, we screened small molecule libraries for Sae-inhibitors and found that two anti-cancer drugs have excellent in vivo efficacy in a murine model of staphylococcal infection. To understand their in vivo efficacy, we further studied the effect of the compounds on S. aureus.
Results
Screening small compound libraries for Sae inhibitors
With two SaeR binding sites, the P1 promoter of the sae operon is an excellent reporter for the Sae activity17 (Fig. 1a). The P1-gfp fusion was cloned in the multi-copy plasmid pYJ335, and the resulting plasmid pYJ-P1-gfp was inserted into S. aureus strain USA300, the predominant CA-MRSA (community associated-methicillin resistant S. aureus) in the USA18. Using the reporter strain, we screened 12,200 small compounds for Sae-inhibition activity and identified 85 compounds that suppress the GFP expression by more than 50% at 10 μM. Subsequently, all 85 compounds were studied in a murine model of intraperitoneal infection, and, out of the initial tests, 10 compounds showed in vivo efficacy to some extent. When the in vivo experiment was repeated for these 10 compounds, the following three FDA-approved anti-cancer drugs consistently showed statistically significant in vivo efficacy: streptozotocin (STZ), floxuridine (FU) and doxorubicin (Fig. 1b and c). The structures of the compounds are different from other reported TCS-inhibitors19–22. Due to their excellent in vivo efficacy, STZ and FU were further studied.
Figure 1.
Identification of three anti-cancer agents with in vivo efficacy. (a) Overall procedure of the screening process. The number in parenthesis is the total number of compounds screened. The promoter sequences, −35 and −10, in the P1 promoter are indicated. SBS, the SaeR binding site. Parts of images were adapted from Motifolio Drawing Toolkits (www.motifolio.com). (b) Chemical structures and molecular weight of the identified compounds. (c) In vivo efficacy of the identified compounds. S. aureus (2 × 108 CFU) was i.p. injected into 18 mice. At 1 h post-infection, the corresponding compounds (100 μg, 5 mg/kg body weight) were i.p. injected once every day for 7 days. Statistical significance was assessed by Log-rank test. STZ, streptozotocin; FU, floxuridine.
Repression of the SaeRS system by STZ and FU
Along with the P1 promoter of the sae operon, the alpha-hemolysin promoter (Phla) is another well-characterized target of the SaeRS TCS17,23. In particular, the minimal Phla (Phlamin) has only the SaeR binding sites and the −35 and −10 promoter sequence of Phla. To compare their Sae-inhibitory activity, we measured IC50 (50% inhibitory concentration) of STZ and FU for P1 and Phlamin. As shown in Fig. 2, FU repressed the Sae-target promoters more efficiently than STZ did (0.5 μM vs. 13.5 μM for P1; 0.4 μM vs. 4.0 μM for Phlamin). Neither compound repressed the promoters of non-Sae-targets such as mecA and hrtB (Supplementary Fig. 1), indicating that the repression is target-specific.
Figure 2.
Repression of the SaeRS system by the anti-cancer agents. S. aureus USA300 carrying either pYJ-P1-gfp or pCL-Phlamin-gfp was grown to exponential growth phase in TSB; then a varying concentration of the anti-cancer agents was added. At 3 h post-incubation, GFP expression was measured and normalized by OD600.
Protection of neutrophils from S. aureus-mediated killing
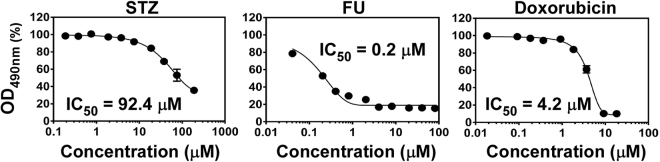
Neutrophils are the most abundant white blood cells in human blood and critical for the defense against staphylococcal infection24. However, it is also well established that S. aureus can kill human neutrophils25. To understand the protective effect of STZ and FU on the host, we assessed whether the compounds could protect human neutrophils from killing by S. aureus. As with the Sae inhibition assay, FU protected human neutrophils from S. aureus-mediated killing much more efficiently than STZ did (IC50, 0.2 μM vs. 92.4 μM) (Fig. 3). Since STZ and FU showed similar in vivo efficacy (Fig. 1), these results might indicate that the neutrophil protection activity of a compound is not a good indicator for its in vivo efficacy. To examine this notion further, we measured IC50 for doxorubicin, which showed the least in vivo efficacy among them (Fig. 1). Again, doxorubicin protected human neutrophil more efficiently than STZ did (IC50, 4.2 μM vs. 92.4 μM) (Fig. 3), showing that the neutrophil-protection activity of a compound does not correlate well with its in vivo efficacy in a murine model of intraperitoneal infection.
Figure 3.
Protection of human neutrophils by the anti-cancer drugs. S. aureus USA300 (106 CFU) and human neutrophils (105 cells) were mixed, and the test compounds were added to the concentration indicated for 4 h. The viability of human neutrophils was measured by CellTiter assay (Promega). In the graph, the OD490 in the absence of compound was set to 100%.
Bacterial growth inhibition by STZ and FU
STZ and FU are known to have not only anti-cancer activity but also antibacterial activity26,27. Therefore, it is possible that the excellent in vivo efficacy of the compounds is due to their antibacterial activity. To examine this possibility, we determined the MIC (minimum inhibitory concentration) of the compounds using S. aureus USA300 in two different growth media: Mueller Hinton broth and tryptic soy broth. In the condition employed, STZ showed almost negligible antibacterial activity whereas FU inhibited staphylococcal growth efficiently (Table 1). As compared with STZ, even doxorubicin showed more than 10 times higher activity of growth inhibition. Therefore, it is possible that the in vivo efficacy of FU might be, in part, due to its growth inhibitory activity, whereas the in vivo efficacy of STZ cannot be explained by its antibacterial activity.
Table 1.
MIC of the anti-cancer agents against S. aureus.
| Compounds | Mueller Hinton Broth | Tryptic Soy Broth |
|---|---|---|
| Streptozotocin | >256 μg/mL (965 μM) | >256 μg/mL (965 μM) |
| Floxuridine | 0.0625 μg/mL (0.25 μM) | 0.2 μg/mL (0.81 μM) |
| Doxorubicin | 32 μg/mL (58.9 μM) | 10 μg/mL (18.4 μM) |
Transcriptional alteration in S. aureus by STZ and FU
To understand the molecular basis of the in vivo efficacy of STZ and FU, we analyzed genome-wide transcriptional changes caused by the compounds. S. aureus USA300 at exponential growth phase was treated with 1 μg/mL (~4 μM) of either compound at 37 °C for 3 h (Supplementary Fig. 2); then total RNA was purified and subjected to RNA-seq. Both STZ and FU caused significant transcriptional changes in S. aureus (STZ, 818 genes; FU 1180 genes, Fig. 4a, and Supplementary Table 1–4). The majority of STZ-affected genes (76% up-regulated and 85% of down-regulated) were also similarly affected by FU, indicating that most anti-virulence mechanisms of STZ are shared by FU. At 4 μM, STZ is not expected to inhibit the SaeRS TCS (Fig. 2). Indeed, in the qRT-PCR analysis, STZ did not reduce the transcription of saeQ and saeS (Fig. 4b), although it did reduce the transcription of saeP. On the other hand, FU reduced the transcription of saeP and saeQ, but it increased the transcription of saeS (Fig. 4b). The molecular mechanism of the differential sae-repression is not clear. Intriguingly, both compounds appeared to inhibit the transcription of other virulence-regulatory systems such as Agr, ArlRS, and SarA more effectively than they inhibit the Sae system (Fig. 4b). The repression also appears to be specific because the transcription of the control gene gyrB and most other staphylococcal two-component systems including the SrrAB TCS was not significantly affected (gyrB and srrB in Fig. 4b and Supplementary Tables 1–4). Taken altogether, these data suggest that the highly effective in vivo efficacy of STZ and FU could be due to their repression of multiple regulatory systems.
Figure 4.
The effect of streptozotocin (STZ) and floxuridine (FU) on the transcription in S. aureus USA300. (a) Summary of RNA-seq analysis. (b) Confirmation of the repression of other virulence regulatory systems by qRT-PCR. Y-axis indicates the mRNA level relative to that of gyrB, whose transcription was not significantly affected by the compounds. -, no treatment. Statistical significance was measured by unpaired, two-tailed Student’s t-test. *p < 0.05; **p < 0.01; ***p < 0.001; ns, not significant.
Prophage induction by STZ and FU
Most clinical isolates of S. aureus carry prophages. Since induction of prophages kills the S. aureus host, it can be an effective antibacterial mechanism. The genome of the strain USA300 contains two prophages (ΦSa2usa and ΦSa3usa), of which ΦSa2usa is replication-defective28. The RNA-seq analysis showed that the treatment with the anti-cancer drugs increased the transcription of ΦSa3usa genes (Supplementary Tables S1 and S3), indicating induction of the prophage. To confirm the results, we grew S. aureus USA300 in TSB containing either STZ or FU (0.2 μg/mL) for 18 h and analyzed the culture supernatant by the soft-agar method. As shown, the culture supernatants produced a large number of plaques whereas the non-treated culture supernatant did not (Fig. 5), confirming the prophage induction by the compounds. These results also indicate that both compounds probably cause damages in the staphylococcal chromosome and that the prophage induction likely contributes to in vivo efficacy of the compounds.
Figure 5.
Prophage induction by streptozotocin (STZ) and floxuridine (FU). NC, No compound.
Dose-dependent in vivo efficacy of STZ and FU
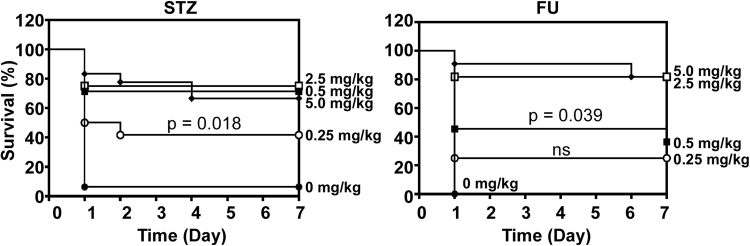
Next, we tested the dose-dependency of STZ and FU in their protective effect on the mice. A daily dose of 0.25 mg/kg (STZ) or 0.5 mg/kg (FU) was sufficient to show statistically significant protection against S. aureus infection (Fig. 6).
Figure 6.
Dose-dependent efficacy of streptozotocin (STZ) and floxuridine (FU). Mice (11–17 e.a.) were i.p. injected with S. aureus USA300 (2 × 108 CFU). At 1 h post-infection, a various amount of the compounds was i.p. injected. The compounds were administered once per day for 7 days. ns, not significant. The statistical significance was measured by Log-rank test.
Effect of administration frequency on in vivo efficacy of STZ and FU
The biological half-life of STZ is 35–40 min29. The half-life of FU is also rather short (t1/2β = 2 h in human)30. Although in the previous experiments, the compounds were administered every day, due to their short half-lives, it is likely that S. aureus cells were exposed to the compounds for a brief period. Therefore, we hypothesized that a brief exposure to the compounds is sufficient to give significant in vivo efficacy. To examine the hypothesis, at 1 h post-infection with S. aureus, we administered the compounds only once (1.25 mg/kg body weight for STZ and FU; 2.5 mg/kg body weight for doxorubicin) and watched the infected mice for 7 days. As shown in Fig. 7, the single administration of the compounds showed in vivo efficacy similar to that of daily administration.
Figure 7.
Effect of single administration of the compounds on murine survival. Fifteen mice were used for streptozotocin (STZ, 50 μg) and floxuridine (FU, 50 μg) while ten mice were used for doxorubicin (50 μg). For a comparison purpose, the daily administration results were superimposed. Statistical significance was measured against the no compound control (black circle) by Log-rank test. ****p < 0.0001; *p < 0.05.
In vivo efficacy of STZ and FU in a murine model of blood infection
In our drug screening and subsequent animal studies, we used a murine model of intraperitoneal infection. However, the peritoneum is not a typical site of S. aureus infection. Therefore, STZ and FU were further tested in a murine model of blood infection, a common type of staphylococcal infections. S. aureus USA300 was administered into mice via retro-orbital injection; then, at 1 h post-infection, the compounds (2.5 mg/kg body weight) were injected into the mice via the intraperitoneal route. The drugs were administered once every day, and the mice were watched for 2 weeks. As shown in Fig. 8, both STZ and FU showed a statistically significant protective effect on murine survival whereas doxorubicin did not, confirming in vivo efficacy of STZ and FU in a clinically relevant animal model.
Figure 8.

Efficacy of the anti-cancer drugs (50 μg) on staphylococcal blood infection. Ten mice were used for each condition. STZ, streptozotocin; FU, floxuridine; Dox, doxorubicin. Statistical significance was measured by Log-rank test against no drug control. ****p < 0.0001; *p < 0.05; ns, not significant. The survival difference between the STZ-treated mice and the FU-treated mice is not statistically significant (p = 0.105).
Effects of STZ and FU on murine blood glucose and bone marrow
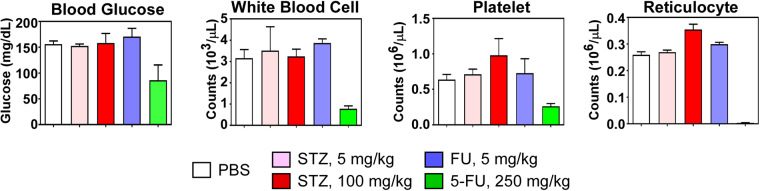
At high doses, STZ is known to induce diabetes in rodents by killing β cells in the pancreas31,32. Besides, a high dose of 5-fluorouracil (5-FU), a metabolite of FU, is known to cause bone-marrow depression33. Therefore, we examined whether the dosage used in our animal experiment can cause those side effects. As expected, the 5-FU-treated mice showed a lower level of blood cell counts along with a sign of hypoglycemia, whereas the mice treated with either STZ or FU showed no such changes (Fig. 9). Even at 100 mg/kg, 20 times higher dose than the highest dose we used, STZ did not cause any significant changes in either blood glucose level or blood cell counts. These results indicate that both STZ and FU can reduce staphylococcal virulence at a safe dosage.
Figure 9.
Effect of streptozotocin (STZ) and floxuridine (FU) on murine blood glucose and bone marrow depression. STZ and FU of the indicated amount were i.p. injected into three female mice. Seven days later, mice were sacrificed, and blood glucose concentration and blood cell counts were measured. PBS, phosphate buffered saline; 5-FU, 5′-fluorouracil. PBS and 5-FU were used as a negative and positive control, respectively. The normal blood glucose level was reported to be 153–171 mg/dL49. The normal values of cell counts are as follows: WBC, 3.2–12.7 (103/μL); Platelet, 0.766–1.657 (106/μL); Reticulocyte, 0.1258–0.469 (106/μL).
Discussion
In this study, through a small molecule screening with a reporter for the SaeRS TCS, we identified two anti-cancer drugs, STZ and FU, as attractive lead candidates for novel anti-virulence drugs against staphylococcal infections. For lead candidates, these two compounds have the following promising properties: (1) simple molecular structure with low molecular weight (Fig. 1); (2) excellent in vivo efficacy even at low dosage and with single administration (Figs 1 and 6); (3) repression of multiple virulence regulators (Fig. 4); and (4) no overt adverse effect at the administration dosage (Fig. 9). In particular, with its excellent anti-growth and anti-virulence activities against S. aureus, FU has a potential to be developed into a dual agent, which inhibits not only the growth but also the virulence of S. aureus.
Although three compounds identified in the screening are all anti-cancer drugs, not all anti-cancer drugs have anti-staphylococcal activity. In the 12,200 small compounds screened in this study, 89 anti-cancer drugs were included. However, only three drugs, STZ, FU, and doxorubicin, repressed the Sae system and showed in vivo efficacy against S. aureus infection (Fig. 1c), suggesting that the anti-cancer activity itself is not sufficient to inhibit the SaeRS system and to protect the host from S. aureus-mediated killing. Intriguingly, of the 85 compounds showing anti-Sae activity in the initial screening (Fig. 1a), only three compounds showed significant in vivo activity, demonstrating that the in vitro Sae-inhibition activity does not guarantee in vivo efficacy of the compound. Since the sae-deletion mutant completely lost its virulence at the bacterial dosage used in the experiment16, these results imply that a partial repression of the Sae system is not sufficient to protect the host from staphylococcal infections. The repression of multiple regulatory systems (Fig. 4) and phage induction (Fig. 5) likely contribute to the in vivo efficacy of STZ and FU. Also, other pharmacological characteristics including in vivo half-life, plasma protein binding, and toxicity will also affect the overall in vivo efficacy of a compound. Therefore, in a drug development process, it might be prudent to employ an in vivo efficacy assay at an early stage as possible.
Both STZ and FU are known to have antibacterial activity. In fact, STZ was initially identified as an antibiotic that inhibits the growth of both Gram-negative and Gram-positive bacteria including S. aureus26. In the original study, STZ was reported to inhibit S. aureus growth by 50% at 0.75 μg/mL26. FU was also reported to be a very potent inhibitor of staphylococcal growth (MIC, 0.025–0.00313 μM)34. However, in our study, STZ showed a negligible anti-growth effect on S. aureus USA300 (Table 1). Although much more potent than STZ, the anti-growth effect of FU was about 10 times lower than that reported previously (Table 1). These discrepancies might be due to the differences in the test strain and the assay conditions employed. Interestingly, despite its low growth-inhibition activity, STZ showed a robust efficacy in vivo similar to that of FU (Figs 1, 6–8), signifying the importance of the anti-virulence activity of the compounds. Since 76–85% of the transcriptional effect of STZ are also observed in S. aureus treated with FU (Fig. 4), it is likely that most of the anti-virulence effects of STZ are also shared by FU.
Quinolone antibiotics such as ciprofloxacin are known to induce prophages via SOS-response35–38. Indeed, the 2 h-treatment of S. aureus 8325 with ciprofloxacin induced 16 prophage genes39. Analysis of non-prophage genes affected by STZ, FU, or ciprofloxacin showed that only a total of 22 genes (11 up-regulated, and 11 down-regulated) were commonly regulated by the three compounds (Supplementary Fig. 3 and Supplementary Table 5), suggesting that the effect of ciprofloxacin on S. aureus is largely different from that of STZ or FU. Of the 11 up-regulated genes, eight are involved in DNA repair, showing the induction of SOS-response is the primary effect shared by the three compounds. Intriguingly, along with alpha-hemolysin (hla), the virulence regulatory systems, Agr and SarA, were also commonly repressed by all three compounds. Although the molecular mechanism of the repression is not known, it is tempting to hypothesize that the induction of SOS-response gives negative impact on the virulence gene expression. Also, since STZ and FU appear to inhibit Agr more effectively than they inhibit the Sae system (Fig. 4b), it is possible that the repression of hla is through the repression of Agr, not the Sae system40.
As with cancer cells, bacteria can replicate indefinitely as long as a permissible growth condition is provided. Indeed, similarities between cancer cells and bacterial pathogens have been noted: high replication rates, damages to the host, spreading and dissemination within the host, and rapid development of resistance against therapeutic agents41. Even the immune response against bacterial pathogens has been used to treat certain types of tumor42–44. Although those similarities might be purely superficial without any biological connections, it is surprising to see that, of the 12,200 compounds screened, three compounds with in vivo efficacy are all anti-cancer agents. It is likely that the rapidly replicating nature of both types of cells makes them vulnerable to the anti-replication activity of the anti-cancer agents. Nonetheless, it remains to be seen whether there are other similarities between the pathogenic bacteria and cancer cells that can be targeted by the same therapeutic agent.
Methods
Ethics Statement
The human subject experiment (i.e., purification of human neutrophils) was approved by the Indiana University Institutional Review Board (Study number: 1010002390). Before taking blood, informed written consent was obtained from each human subject. All experiments were performed in accordance with relevant guidelines and regulations. The animal protocol was approved by the Committee on the Ethics of Animal Experiments of the Indiana University School of Medicine-Northwest (Protocol Number: NW-43). The animal experiment was performed by following the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. Every effort was made to minimize the suffering of the animals.
Bacterial strains and growth conditions
The S. aureus strains and plasmids used in this study are described in Supplementary Tables 6. S. aureus was grown in tryptic soy broth (TSB). When necessary, antibiotics were added to the growth media at the following concentrations: erythromycin, 10 μg/mL; and chloramphenicol, 5 μg/mL.
Construction of a GFP-promoter fusion
The reporter plasmids, pYJ-P1-gfp, pCL-Phlamin-gfp, pCL-PmecA-gfp, and pCL-PhrtB-gfp were generated by a ligation independent cloning method45. First, vectors were PCR-amplified from pYJ-gfp16 with the primers P1969/P1747 or from pCL55-gfp14 with the primers P1990/P1991 (for pCL-Phlamin-gfp), P3016/P3017 (for pCL-PmecA-gfp), or P3286/P3287 (for pCL-PhrtB-gfp) (Supplementary Table 7). The insert DNA fragment containing the promoter sequence was amplified with primer pairs P1971/P1972 for P1, P1992/P1993 (for Phlamin), P3014/P3015 (for PmecA), or P3284/P3285 (for PhrtB) (Supplementary Table 7). The PCR products were treated with T4 DNA polymerase in the presence of dCTP (vector) or dGTP (insert DNA) and mixed. DNA mixture was used to transform E. coli DH5α. Once verified, all plasmids were electroporated into S. aureus strain RN4220 and subsequently transduced into S. aureus strain USA300 with ϕ85.
High-throughput screening of small molecule libraries
S. aureus USA300 harboring pYJ-P1-gfp was used to screen 12,200 compounds (3,000 compounds from Spectrum Library [MicroSource Discovery Systems], 6,000 from the discovery collection library [DCL, MicroSource Discovery Systems], and 3,200 natural compounds [Indiana University]) in DMSO (stock concentration 10 mM). The compounds were dry transferred into 384 plates using an Echo 550 Acoustic Liquid Transfer System. S. aureus USA300 (pYJ-P1-gfp) was grown in TSB overnight. The overnight culture was diluted 1:100 into fresh TSB and grown until OD600 = 0.5. Then 30 μL of the culture was transferred into the plate, resulting in 10 μM of the compounds. A negative control (DMSO blank) was included in each plate. Initial cell density and GFP fluorescence were measured after filling the plates. Plates were then sealed and incubated on a plate shaker-incubator at 37 °C for 3 h. The GFP signal (excitation 485 nm, emission 515 nm) and OD600 were measured with a Tecan Infinite M1000 Microplate Reader. The GFP signal was normalized by OD600, and the normalized value was used to identify the Sae inhibitor candidates.
Animal test
An overnight culture of S. aureus was 100-fold diluted into fresh TSB and further incubated at 37 °C for 4 h. Cells were collected by centrifugation, washed with sterile PBS, and suspended in sterile PBS to OD600 = 4 (i.e., 1 × 109 CFU mL−1). Sex-matched 8-week-old C57BL/6 mice were i.p. injected with S. aureus (2 × 108 CFU); then, at 1 h post-infection, a varying amount of compound was administered through i.p. injection. DMSO was used as a negative control. For daily dose experiment, the compound was i.p. injected every 24 h for seven days.
For retro-orbital studies for the compounds, the bacterial suspension (107 CFU in 100 μL) was administered into 10 sex-matched 8-week-old C57BL/6 mice via retro-orbital injection. At 1 h post-infection, test compounds (50 μg, 2.5 mg/kg) was administered by i.p. injection. The compounds were i.p. injected every 24 h for 14 days.
Statistical significance in murine survival was measured by Log-rank (Mantel-Cox) test with Prism 6 (GraphPad).
Determination of MIC
Minimum inhibitory concentration (MIC) for streptozotocin, floxuridine, and doxorubicin was determined according to the recommendations proposed by the Clinical and Laboratory Standards Institute (CLSI) using microdilution method46.
RNA-seq analysis
An overnight culture of S. aureus in TSB was diluted 100 times in a fresh TSB (2 mL in a 15 mL test tube) and incubated in a shaking incubator at 37 °C for 2 h. Then 1 μg/mL of streptozotocin or floxuridine were added to the culture and further incubated for 3 h. After immediate stabilization of RNA in all samples by RNAprotect Bacteria Reagent (Qiagen), total RNA was isolated with RNeasy Mini Kit (Qiagen) according to the manufacturer’s recommendations. For each condition, three RNA samples were isolated from three independent bacterial cultures. The isolated RNAs were sent to the Center for Genomics and Bioinformatics at Indiana University. Sequencing libraries were constructed using the ScriptSeq Complete Kit for Bacteria (Epicentre). The statistical analysis of the RNA-seq results was done with DeSeq. 2 as described before47.
qRT-PCR analysis
The total RNA used for RNA-seq was also used for qRT-PCR analysis. cDNA was synthesized using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Quantification of transcripts was carried out by real-time PCR using SYBR Green PCR Master Mix (Applied Biosystems) in a QuantStudio 7 Flex Real-Time PCR System (Applied Biosystems). Primers used to detect specific transcripts are shown in Supplementary Table 6. The relative amount of cDNA was determined using a standard curve obtained from PCR with serially diluted genomic DNA, and results were normalized to the levels of gyrB RNA. Statistical significance was measured by unpaired, two-tailed Student’s t-test with Prism 6 (GraphPad).
GFP assay
An overnight culture of S. aureus harboring pYJ-P1-gfp, pCL-Phla min-gfp, pCL-PmecA-gfp, or pCL-PhrtB-gfp was diluted 100 times in a fresh TSB and incubated in a shaking incubator at 37 °C for 2 h. Then various concentrations of streptozotocin and floxuridine were added to cells, and the cells were incubated for 3 h. The GFP signal (excitation 485 nm, emission 515 nm) and OD600 were measured with Enspire Plate Reader (Perkin Elmer). All GFP expressions were normalized by OD600. The half maximal inhibitory concentration (IC50) was calculated from at least three independent experiments via nonlinear regression analysis (sigmoidal dose-response with variable slope) using Prism 6 (GraphPad).
Neutrophil killing assay
Human neutrophils were purified from healthy adult blood donors by Ficoll-Paque gradient method48. The purified neutrophils were added to a 96-well tissue culture plate (2 × 105/well) in 100 μL RPMI (Gibco) supplemented with 10 mM HEPES and 10% human serum. S. aureus cells (1 × 106 CFU) and various concentrations of streptozotocin, floxuridine, and doxorubicin were added to each well. Then, 10 μL of CellTiter 96® AQueous One Solution (Promega) was added to each well and incubated at 37 °C in 5% CO2 for 3–5 h. Neutrophil viability was assessed with the Enspire plate reader (Perkin Elmer).
Prophage induction by streptozotocin and floxuridine
S. aureus USA300 was grown in TSB containing 0.2 μg/mL of the compounds (STZ or FU) or no compound (a negative control) at 37 °C for 18 h; then the supernatant was collected by filtration (0.22 μm). The supernatant (100 μL) was mixed with the overnight culture (100 μL) of S. aureus RN4220 suspended in HIB (heart infusion broth) containing 5 mM of CaCl2. The mixture was incubated at 37 °C for 10 min, mixed with soft TSA (0.8%), spread on TSA, and incubated at 37 °C for 18 h.
Blood glucose and bone marrow suppression tests in mice
Streptozotocin (0.1 mg [=5 mg/kg] and 2 mg [=100 mg/kg]) and floxuridine (0.1 mg, 5 mg/kg) were i.p. injected into 8-week-old C57BL/6 mice. PBS and 5-fluorouracil (5 mg, 250 mg/kg) were used a negative and positive control, respectively. Seven days later, mice were sacrificed, and blood was collected. The collected blood was sent to Biologic Resources Laboratory at the University of Illinois at Chicago, where blood cells were counted. Blood glucose concentration was measured by Contour blood glucose monitoring system (Asensia).
Accession number
The RNA-seq data were deposited in GEO (Gene Expression Omnibus) with the accession number GSE104069.
Data Availability
All processed RNA-seq results were included in Supplementary Tables 1–4. The raw data deposited to GEO (the accession number GSE104069) will be released on September 19, 2019.
Electronic supplementary material
Acknowledgements
This study was supported by NIH (AI121664) to TB, and National Research Foundation of Korea to RA (2015R1C1A2A01056004) and KKK (2017M3A9E4078553). The funders had no role in study design, data collection, and interpretation, or the decision to submit the work for publication.
Author Contributions
W.Y., K.K.K., H.J., and T.B. conceived and designed the experiments. W.Y., R.A., and K.H.C. performed the experiments. W.Y., K.K.K., H.J., and T.B. analyzed and interpreted the data. W.Y. and T.B. prepared figures and tables. W.Y. and T.B. co-wrote the first draft of the manuscript. All authors reviewed and contributed to the manuscript, and approved its final version.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-20617-5.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Medina E, Pieper DH. Tackling Threats and Future Problems of Multidrug-Resistant Bacteria. Curr Top Microbiol Immunol. 2016;398:3–33. doi: 10.1007/82_2016_492. [DOI] [PubMed] [Google Scholar]
- 2.Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. P T. 2015;40:277–283. [PMC free article] [PubMed] [Google Scholar]
- 3.Lewis JS, II., et al. Emergence of daptomycin resistance in Enterococcus faecium during daptomycin therapy. Antimicrob Agents Chemother. 2005;49:1664–1665. doi: 10.1128/AAC.49.3.945-951.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsiodras S, et al. Linezolid resistance in a clinical isolate of Staphylococcus aureus. Lancet. 2001;358:207–208. doi: 10.1016/S0140-6736(01)05410-1. [DOI] [PubMed] [Google Scholar]
- 5.Janardhanan J, et al. In Vitro and In Vivo Synergy of the Oxadiazole Class of Antibacterials with beta-Lactams. Antimicrob Agents Chemother. 2016;60:5581–5588. doi: 10.1128/AAC.00787-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Daniel PI, et al. Discovery of a new class of non-beta-lactam inhibitors of penicillin-binding proteins with Gram-positive antibacterial activity. J Am Chem Soc. 2014;136:3664–3672. doi: 10.1021/ja500053x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Archer GL. Staphylococcus aureus: a well-armed pathogen. Clin Infect Dis. 1998;26:1179–1181. doi: 10.1086/520289. [DOI] [PubMed] [Google Scholar]
- 8.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 9.Cheung AL, Nishina KA, Trotonda MP, Tamber S. The SarA protein family of Staphylococcus aureus. Int J Biochem Cell Biol. 2008;40:355–361. doi: 10.1016/j.biocel.2007.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu Rev Genet. 2008;42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 11.Fournier B, Klier A, Rapoport G. The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol Microbiol. 2001;41:247–261. doi: 10.1046/j.1365-2958.2001.02515.x. [DOI] [PubMed] [Google Scholar]
- 12.Liu, Q., Yeo, W. S. & Bae, T. The SaeRS Two-Component System of Staphylococcus aureus. Genes (Basel) 7 (2016).
- 13.Giraudo AT, Raspanti CG, Calzolari A, Nagel R. Characterization of a Tn551-mutant of Staphylococcus aureus defective in the production of several exoproteins. Can J Microbiol. 1994;40:677–681. doi: 10.1139/m94-107. [DOI] [PubMed] [Google Scholar]
- 14.Cho H, et al. Calprotectin Increases the Activity of the SaeRS Two Component System and Murine Mortality during Staphylococcus aureus Infections. PLoS Pathog. 2015;11:e1005026. doi: 10.1371/journal.ppat.1005026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nygaard TK, et al. SaeR Binds a Consensus Sequence within Virulence Gene Promoters to Advance USA300 Pathogenesis. J Infect Dis. 2010;201:241–254. doi: 10.1086/649570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Q, Cho H, Yeo WS, Bae T. The Extracytoplasmic Linker Peptide of the Sensor Protein SaeS Tunes the Kinase Activity Required for Staphylococcal Virulence in Response to Host Signals. PLoS Pathog. 2015;11:e1004799. doi: 10.1371/journal.ppat.1004799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho H, Jeong DW, Li C, Bae T. Organizational requirements of the SaeR binding sites for a functional P1 promoter of the sae operon in Staphylococcus aureus. J Bacteriol. 2012;194:2865–2876. doi: 10.1128/JB.06771-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diep BA, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 19.Watanabe T, Okada A, Gotoh Y, Utsumi R. Inhibitors targeting two-component signal transduction. Adv Exp Med Biol. 2008;631:229–236. doi: 10.1007/978-0-387-78885-2_16. [DOI] [PubMed] [Google Scholar]
- 20.Stephenson K, Yamaguchi Y, Hoch JA. The mechanism of action of inhibitors of bacterial two-component signal transduction systems. J Biol Chem. 2000;275:38900–38904. doi: 10.1074/jbc.M006633200. [DOI] [PubMed] [Google Scholar]
- 21.Macielag MJ, Goldschmidt R. Inhibitors of bacterial two-component signalling systems. Expert Opin Investig Drugs. 2000;9:2351–2369. doi: 10.1517/13543784.9.10.2351. [DOI] [PubMed] [Google Scholar]
- 22.Barrett JF, et al. Antibacterial agents that inhibit two-component signal transduction systems. Proc Natl Acad Sci USA. 1998;95:5317–5322. doi: 10.1073/pnas.95.9.5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mainiero M, et al. Differential target gene activation by the Staphylococcus aureus two-component system saeRS. J. Bacteriol. 2010;192:613–623. doi: 10.1128/JB.01242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spickett GP. Immune deficiency disorders involving neutrophils. J Clin Pathol. 2008;61:1001–1005. doi: 10.1136/jcp.2007.051185. [DOI] [PubMed] [Google Scholar]
- 25.DuMont AL, et al. Staphylococcus aureus LukAB cytotoxin kills human neutrophils by targeting the CD11b subunit of the integrin Mac-1. Proc Natl Acad Sci USA. 2013;110:10794–10799. doi: 10.1073/pnas.1305121110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vavra JJ, Deboer C, Dietz A, Hanka LJ, Sokolski WT. Streptozotocin, a new antibacterial antibiotic. Antibiot Annu. 1959;7:230–235. [PubMed] [Google Scholar]
- 27.Bean B, Tomasz A. 5-Fluoropyrimidine-resistant mutants of pneumococcus. J Bacteriol. 1973;113:1348–1355. doi: 10.1128/jb.113.3.1348-1355.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wirtz C, Witte W, Wolz C, Goerke C. Transcription of the phage-encoded Panton-Valentine leukocidin of Staphylococcus aureus is dependent on the phage life-cycle and on the host background. Microbiology. 2009;155:3491–3499. doi: 10.1099/mic.0.032466-0. [DOI] [PubMed] [Google Scholar]
- 29.Adolphe AB, Glasofer ED, Troetel WM, Weiss AJ, Manthei RW. Preliminary pharmacokinetics of streptozotocin, an antineoplastic antibiotic. J Clin Pharmacol. 1977;17:379–388. doi: 10.1002/j.1552-4604.1977.tb04620.x. [DOI] [PubMed] [Google Scholar]
- 30.Creaven PJ, et al. Phase I and pharmacokinetic evaluation of floxuridine/leucovorin given on the Roswell Park weekly regimen. Cancer Chemother Pharmacol. 1994;34:261–265. doi: 10.1007/BF00685087. [DOI] [PubMed] [Google Scholar]
- 31.Rakieten N, Rakieten ML, Nadkarni MV. Studies on the diabetogenic action of streptozotocin (NSC-37917) Cancer Chemother Rep. 1963;29:91–98. [PubMed] [Google Scholar]
- 32.Graham ML, Janecek JL, Kittredge JA, Hering BJ, Schuurman HJ. The streptozotocin-induced diabetic nude mouse model: differences between animals from different sources. Comp Med. 2011;61:356–360. [PMC free article] [PubMed] [Google Scholar]
- 33.Lu J, Sun W, Mao W. Establishment of mouse bone marrow depression and regeneration model and dynamic observations of morphological changes. J. Shanghai Jiaotong U. 2010;30:758–762. [Google Scholar]
- 34.Lau QY, et al. An FDA-Drug Library Screen for Compounds with Bioactivities against Meticillin-Resistant Staphylococcus aureus (MRSA) Antibiotics (Basel) 2015;4:424–434. doi: 10.3390/antibiotics4040424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goerke C, Koller J, Wolz C. Ciprofloxacin and trimethoprim cause phage induction and virulence modulation in Staphylococcus aureus. Antimicrob Agents Chemother. 2006;50:171–177. doi: 10.1128/AAC.50.1.171-177.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X, et al. Quinolone antibiotics induce Shiga toxin-encoding bacteriophages, toxin production, and death in mice. J Infect Dis. 2000;181:664–670. doi: 10.1086/315239. [DOI] [PubMed] [Google Scholar]
- 37.Ubeda C, et al. Antibiotic-induced SOS response promotes horizontal dissemination of pathogenicity island-encoded virulence factors in staphylococci. Mol Microbiol. 2005;56:836–844. doi: 10.1111/j.1365-2958.2005.04584.x. [DOI] [PubMed] [Google Scholar]
- 38.Lopez E, et al. Induction of prophages by fluoroquinolones in Streptococcus pneumoniae: implications for emergence of resistance in genetically-related clones. PLoS One. 2014;9:e94358. doi: 10.1371/journal.pone.0094358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cirz RT, et al. Complete and SOS-mediated response of Staphylococcus aureus to the antibiotic ciprofloxacin. J Bacteriol. 2007;189:531–539. doi: 10.1128/JB.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Recsei P, et al. Regulation of exoprotein gene expression in Staphylococcus aureus by agr. Mol Gen Genet. 1986;202:58–61. doi: 10.1007/BF00330517. [DOI] [PubMed] [Google Scholar]
- 41.Benharroch D, Osyntsov L. Infectious diseases are analogous with cancer. Hypothesis and implications. J Cancer. 2012;3:117–121. doi: 10.7150/jca.3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coley WB., II. Contribution to the Knowledge of Sarcoma. Ann Surg. 1891;14:199–220. doi: 10.1097/00000658-189112000-00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Linnebacher M, Maletzki C, Klier U, Klar E. Bacterial immunotherapy of gastrointestinal tumors. Langenbecks Arch Surg. 2012;397:557–568. doi: 10.1007/s00423-011-0892-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts NJ, et al. Intratumoral injection of Clostridium novyi-NT spores induces antitumor responses. Sci Transl Med. 2014;6:249ra111. doi: 10.1126/scitranslmed.3008982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Donnelly MI, et al. An expression vector tailored for large-scale, high-throughput purification of recombinant proteins. Protein Expr Purif. 2006;47:446–454. doi: 10.1016/j.pep.2005.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.CLSI. Performance standards for antimicrobial suspceptability testing. CLSI approved standard M100-S15. (Clinical and Laboratory Standards Institute, Wayne, PA, 2005).
- 47.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. 2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Voyich JM, et al. Insights into Mechanisms Used by Staphylococcus aureus to Avoid Destruction by Human Neutrophils. J Immunol. 2005;175:3907–3919. doi: 10.4049/jimmunol.175.6.3907. [DOI] [PubMed] [Google Scholar]
- 49.Togashi Y, et al. Evaluation of the appropriateness of using glucometers for measuring the blood glucose levels in mice. Sci Rep. 2016;6:25465. doi: 10.1038/srep25465. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All processed RNA-seq results were included in Supplementary Tables 1–4. The raw data deposited to GEO (the accession number GSE104069) will be released on September 19, 2019.