Abstract
The Gram-negative bacterium Xanthomonas euvesicatoria (Xe) is the causal agent of bacterial spot disease of pepper and tomato. Xe delivers effector proteins into host cells through the type III secretion system to promote disease. Here, we show that the Xe effector XopAU, which is conserved in numerous Xanthomonas species, is a catalytically active protein kinase and contributes to the development of disease symptoms in pepper plants. Agrobacterium-mediated expression of XopAU in host and non-host plants activated typical defense responses, including MAP kinase phosphorylation, accumulation of pathogenesis-related (PR) proteins and elicitation of cell death, that were dependent on the kinase activity of the effector. XopAU-mediated cell death was not dependent on early signaling components of effector-triggered immunity and was also observed when the effector was delivered into pepper leaves by Xanthomonas campestris pv. campestris, but not by Xe. Protein-protein interaction studies in yeast and in planta revealed that XopAU physically interacts with components of plant immunity-associated MAP kinase cascades. Remarkably, XopAU directly phosphorylated MKK2 in vitro and enhanced its phosphorylation at multiple sites in planta. Consistent with the notion that MKK2 is a target of XopAU, silencing of the MKK2 homolog or overexpression of the catalytically inactive mutant MKK2K99R in N. benthamiana plants reduced XopAU-mediated cell death and MAPK phosphorylation. Furthermore, yeast co-expressing XopAU and MKK2 displayed reduced growth and this phenotype was dependent on the kinase activity of both proteins. Together, our results support the conclusion that XopAU contributes to Xe disease symptoms in pepper plants and manipulates host MAPK signaling through phosphorylation and activation of MKK2.
Author summary
Many bacterial pathogens inject effector proteins into their eukaryotic host cells through the type III secretion system to modulate host cellular processes. Elucidating the function of bacterial effectors and identification of their host targets is important for understanding the molecular mechanisms of pathogenicity and host immunity. In this study, we analyzed the mode of action of XopAU, a type III effector from the pepper and tomato pathogen Xanthomonas euvesicatoria. We found that XopAU is a catalytically active protein kinase providing the first report of an effector from a plant bacterial pathogen that displays such an enzymatic activity. We show that expression of XopAU activates immune responses and contributes to the development of disease symptoms. Interestingly, XopAU-mediated phenotypes are altered when the effector is expressed by different species of Xanthomonas, suggesting an interplay between this effector and other species-specific virulence determinants. Furthermore, we provide biochemical and genetic evidence that XopAU interferes with host immune signaling by activating the MAPKK MKK2. Together, our results provide new insights into the interaction between the plant immune system and bacterial type III effector proteins.
Introduction
Plant immunity against microbial pathogens relies on a complex detection and signaling network [1]. A first line of plant immune responses is activated by cell surface-exposed pattern recognition receptors (PRRs) that detect broadly conserved pathogen molecules (pathogen/microbe-associated molecular patterns, PAMP/MAMPs) [2]. Activation of PRRs initiates downstream signaling events that lead to the production of reactive oxygen species, stimulation of mitogen-activated protein kinase (MAPK) cascades, defense gene induction, release of ethylene, and callose deposition at the plant cell wall [3,4]. These host responses limit the growth of a large number of potential pathogens and are referred to as pattern-triggered immunity (PTI). Host-adapted pathogens overcome PTI through the activity of effector proteins that are targeted to the plant apoplast or delivered into the host cytoplasm [5]. To cope with these pathogens, plants have evolved other types of receptors known as resistance (R) proteins that specifically recognize effectors or their activity [6]. R proteins activate effector-triggered immunity (ETI) that consists of defense responses similar to PTI, but more robust and often accompanied by a localized cell death known as the hypersensitive response (HR) [7].
Mitogen-activated protein kinases (MAPKs) cascades play a fundamental role in plant immunity and are involved in both PTI and ETI signaling [8]. The tomato MAPKKK MAP3Kα and MAP3Kε were found to participate in signaling pathways that mediate elicitation of the ETI-associated HR in N. benthamiana plants, and to be required for disease resistance to bacterial pathogens in tomato [9,10]. The MEK2 MAPKK was identified as a central regulator of the HR elicited upon detection of effectors by several R proteins in N. benthamiana plants, and as required for tomato disease resistance to Pseudomonas and Xanthomonas bacteria [11]. Epistasis analysis revealed that MEK2 acts downstream of both MAP3Kα and MAP3Kε and upstream of the SIPK and WIPK MAP kinases [9,10,12]. Notably, SIPK and WIPK, and their respective Arabidopsis homologs MPK6 and MPK3, are also important regulators of PTI [13,14]. In line with these findings, the Arabidopsis MKK4 and MKK5, which are the MAPKKs upstream of MPK6 and MPK3, were also shown to participate in PTI signaling [13].
Many Gram-negative plant pathogenic bacteria utilize a type III secretion system to deliver effector proteins into the host cells [15]. Type III effector proteins contribute to bacterial virulence by subverting plant signaling pathways, suppressing immune responses, and modulating host metabolism and hormone signaling [16,17]. MAPK cascades have emerged as important targets of type III effectors of plant and mammalian bacterial pathogens [18]. For example, Yersinia pestis YopJ interferes with the activation of immune responses in mammalian cells by inhibiting phosphorylation of MAPKK6 through acetylation of Ser and Thr residues in the activation loop of the kinase [19]. The Salmonella phosphothreonine lyase effector SpvC irreversibly removes a phosphate from ERK1/2 MAPK to downregulate cytokine release from infected cells [20]. Several Pseudomonas syringae effectors were found to suppress immunity in Arabidopsis by interfering with the activity of components of MAPK cascades: HopAI1 encodes a phosphothreonine lyase that irreversibly removes a phosphate from MPK3 and MPK6 thereby suppressing PTI activation [21]. HopF2 inhibits PTI through inactivation of MKK5, the upstream MAPKK of MPK3 and MPK6, by ADP-ribosylation [22]. Finally, AvrB enhances plant susceptibility by promoting phosphorylation and activation of MPK4, which perturbs hormone signaling to the benefit of the bacterium [23].
The Gram-negative bacterium Xanthomonas euvesicatoria (Xe) is the causal agent of bacterial spot disease in pepper and tomato plants [24]. Xe bacteria penetrate into plant tissues through wounds and stomata, proliferate and colonize the apoplast of the aerial parts of the plants, and cause the appearance of water soaked lesions that develop into necrotic black spots. The ability of Xe to cause disease largely depends on the type III secretion system. To date, the pool of known Xe effectors includes approximately 35 proteins mostly identified in the 85–10 strain [25–27]. Biochemical activity and cellular targets have been elucidated only for a few Xe effectors. The XopD effector is a SUMO protease that alters host transcription to suppress hormone signaling [28]. The XopN and XopQ effectors target host 14-3-3 proteins to suppress PTI and ETI signaling, respectively [29,30]. The XopJ effector causes degradation of a proteasome subunit to suppress salicylic acid-mediated defense and protein secretion [31–33].
By a machine learning approach applied to the Xe strain 85–10, we have recently identified XopAU as a type III secreted effector and demonstrated its translocation into cells of pepper leaves [26]. XopAU is conserved in multiple Xanthomonas spp. and in a few Acidovorax spp., and encodes a putative serine/threonine protein kinase [26]. The xopAU gene from Xe 85–10 contains a plant-inducible promoter (PIP) box and expression of its homolog from Xanthomonas citri was found to be regulated by the HrpG/HrpX-regulon, which controls transcription of genes encoding structural components of the type III secretion system and some effector genes [34]. Here, we investigated XopAU molecular properties and virulence function. We found that XopAU is a catalytically active protein kinase that contributes to the development of disease symptoms in susceptible plants. In addition, we identified the MAPKK MKK2 as a binding partner and direct substrate of XopAU phosphorylation. Moreover, by genetic and functional analysis we provide evidence that MKK2 is required for the XopAU molecular function.
Results
The Xanthomonas type III effector XopAU encodes a catalytically active protein kinase
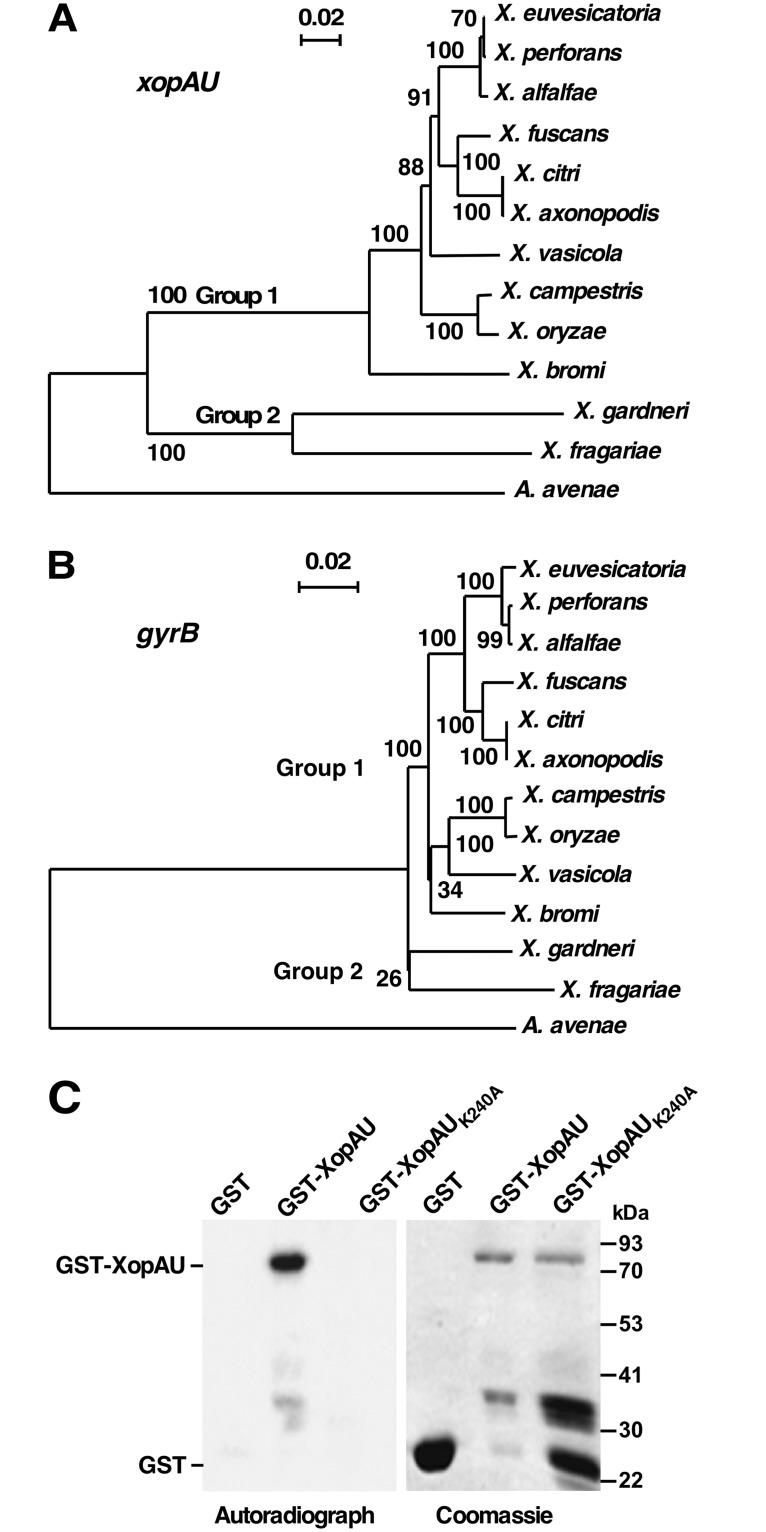
XopAU is a type III secreted effector originally identified in the Xanthomonas euvesicatoria strain 85–10 (Xe) [26]. Homologs of the effector are present in multiple species of the Xanthomonas genus and in Acidovorax spp. Promoter regions of all xopAU homologs contain a PIP box motif (S1 Table) indicating that their expression is controlled by the HrpG/HrpX regulon [35]. To determine evolutionary relationships between xopAU homologous genes, a representative of each Xanthomonas species encoding xopAU and an Acidovorax avenae homolog were used to construct a phylogenetic tree (Fig 1A, S1 Fig and S1 Table). xopAU homologs classified into two allelic groups and their phylogenetic relationships correlated to the relationships among the corresponding Xanthomonas species that were deduced by a sequence comparison of the gyrB phylogenetic marker gene [36] (Fig 1 and S1 Table). This correlation suggests that the two alleles were transmitted vertically after their acquisition in parental strains. xopAU homologs of group 1 share a low degree of sequence similarity to group 2 homologs, have a different GC content (group 1, 62.9%-64.3%; group 2, 55.2%-55.8%), and a distinct genomic location (S1 Table; S2 Fig) implying that the two xopAU alleles were independently acquired by Xanthomonas spp. The Xanthomonas species containing the group 1 xopAU allele correspond to a complete clade in the Xanthomonas genus (Fig 1A) [37]. Conversely, Xanthomonas species containing the group 2 xopAU allele (X. fragariae and X. gardneri) are members of a clade that also includes the X. arboricola species [37], which does not encode a xopAU allele. The borders of the xopAU deletion in the genome of a X. arboricola strain are shown in S2B Fig.
Fig 1. The xopAU gene of Xe strain 85–10 is conserved in multiple Xanthomonas species and encodes a protein kinase.
Phylogenetic tree of xopAU (A) and gyrB (B) homologs generated with Clustal X [68] using the standard neighbor joining phylogenetic tree definitions. NCBI xopAU and gyrB accession numbers and a sequence alignment of the xopAU genes included in the tree are reported in S1 Table and S1 Fig, respectively. Bootstrap values of 100 replications are shown on nodes. (C) In vitro kinase assay performed by incubating the indicated proteins in the presence of [γ-32P]ATP. Proteins were separated by SDS-PAGE and detected by autoradiography or Coomassie staining.
Analysis of the NCBI conserved domains database revealed that Xe XopAU contains a putative protein kinase domain at the C-terminus (203–493 amino acids), which includes kinase subdomains I-XI and the majority of the residues that are nearly invariant throughout the kinase superfamily [38] (S3 Fig). Notably, the kinase nearly invariant residues identified in Xe XopAU are also conserved in its homologs from other Xanthomonas strains. To test whether XopAU is a catalytically active protein kinase, it was expressed in E. coli as a glutathione S-transferase (GST) fusion, purified and assayed for kinase activity in the presence of [γ-32P]ATP. The GST-XopAU fusion was able to autophosphorylate in vitro and its activity was abolished by the introduction of an alanine substitution at the conserved lysine (K240) of the putative ATP binding site (Fig 1C). Together, this analysis revealed that the xopAU gene is conserved in the genome of numerous Xanthomonas species and XopAU of the Xe strain 85–10 is a catalytically active protein kinase.
Agrobacterium-mediated expression of XopAU in planta promotes activation of immune responses
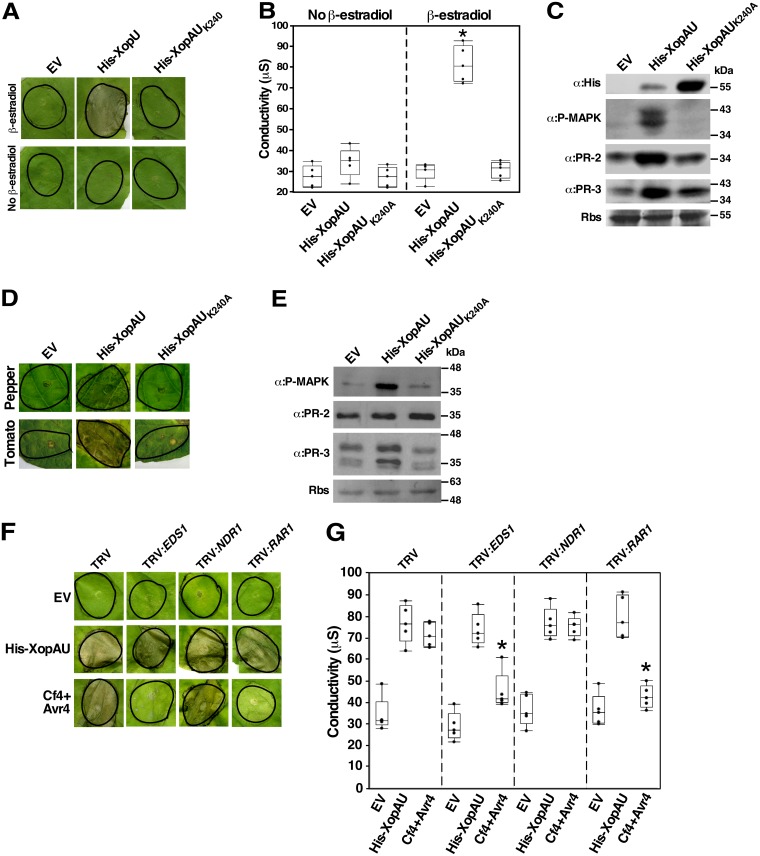
To examine whether XopAU from Xe causes detectable phenotypes in planta, the effector was fused to a His tag (His-XopAU) and transiently expressed via Agrobacterium under the control of an estradiol inducible system in leaves of the Xe non-host plant N. benthamiana. At 24–48 hours after estradiol application, cell death was visible in leaf areas expressing His-XopAU and confirmed by a higher ion leakage than in leaf areas infiltrated with Agrobacterium carrying an empty vector (Fig 2A and 2B). No cell death or ion leakage was observed without estradiol application. Furthermore, the His-XopAUK240A kinase deficient variant did not induce a visible cell death and ion leakage (Fig 2A and 2B) indicating that XopAU kinase activity was required for this phenotype. Expression of wild-type and kinase deficient His-XopAU variants in the infiltrated areas after estradiol application was confirmed by Western blot analysis (Fig 2C).
Fig 2. Agrobacterium-mediated expression of XopAU induces plant immune responses.
(A-E) N. benthamiana leaves were infiltrated with Agrobacterium strains for the expression of His-XopAU or His-XopAUK240A driven by an estradiol-inducible system, or carrying an empty vector (EV), and treated with 17β-estradiol 24 h later. (A) Photographs of inoculated areas at 48 h after 17β-estradiol application. (B) Electrolyte leakage at 24 h with or without 17β-estradiol application. The box plot displays 25th, 50th (middle line) and 75th percentiles (n = 5). An asterisk indicates a significant difference (Mann-Whitney U test, p value <0.05) compared to EV. (C) Total protein was extracted from N. benthamiana leaves 8 h after 17β-estradiol application and immunoblotted with the indicated antibodies. Rbs, Rubisco loading control stained by Ponceau S. (D) Leaves of the pepper line ECW30R or the tomato line Hawaii 7981 were infiltrated with Agrobacterium strains as in (A) and photographed at 96 h after 17β-estradiol application. (E) Total protein was extracted from pepper leaves at 8 h after 17β-estradiol application and immunoblotted with the indicated antibodies. (F and G) N. benthamiana plants were infected with TRV, TRV:EDS1, TRV:NDR1 and TRV:RAR1. Leaves of silenced plants were inoculated with Agrobacterium (OD600 = 0.02) carrying an empty vector (EV), a vector for expression of His-XopAU from an estradiol-inducible system or Cf4/Avr4 driven by the CaMV 35S promoter. Inoculated leaves were treated with 17β-estradiol 24 h later. (F) Photographs of inoculated areas at 36 h after 17β-estradiol application. (G) Electrolyte leakage at 24 h after 17β-estradiol application. The box plot displays 25th, 50th (middle line) and 75th percentiles (n = 5). An asterisk indicates a significant difference (Mann-Whitney U test, p value <0.05) compared to EV. (A-G) Experiments were repeated at least three times with similar results.
Because cell death is a typical immune response triggered by pathogens in resistant plants, we hypothesized that XopAU activates immune signaling. To test this hypothesis, we transiently expressed His-XopAU in N. benthamiana leaves and monitored phosphorylation of MAP kinases and accumulation of pathogenesis-related (PR) proteins, which are additional phenotypes associated with plant immunity [8,39]. Phosphorylation of MAP kinases was assessed by using antibodies against the phosphorylated form of mammalian MAP kinases of the ERK family that recognize also phosphorylated plant MAP kinases. Accumulation of PR proteins was monitored with antibodies against the tobacco PR-2 and PR-3 isoforms. As shown in Fig 2C, MAP kinase phosphorylation and PR protein accumulation were induced in leaves of N. benthamiana plants expressing His-XopAU, but no induction was observed in leaves expressing the kinase deficient variant His-XopAUK240A.
To examine whether XopAU induces immune responses in Xe host plants, wild-type and kinase deficient His-XopAU variants were transiently expressed via Agrobacterium in leaves of the pepper line ECW30R and tomato line Hawaii 7981. Similar as in N. benthamiana leaves, expression of wild-type but not kinase deficient His-XopAU in these plants induced a cell death at 48–72 hours after estradiol application (Fig 2D). In pepper leaves, cell death was accompanied by enhanced MAP kinase phosphorylation and higher accumulation of the PR-3 protein, but not of PR-2 (Fig 2E). Together, these results suggest that expression of XopAU via Agrobacterium activated immune signaling in Xe host and non-host plants.
To assess whether activation of immune signaling by XopAU is caused by recognition of the effector by an R protein, we tested if silencing of early components of ETI signaling affects XopAU-mediated cell death. The genes silenced in these experiments were EDS1 and NDR1, which are required for ETI mediated by multiple R genes of the TIR-NBS-LRR and CC-NBS-LRR class, respectively, and RAR1, which is required for ETI mediated by multiple R genes of different structural classes [40]. Virus-induced gene silencing (VIGS) techniques based on the tobacco rattle virus (TRV) vector were used to silence the genes in N. benthamiana plants [41]. Four weeks after infection of plants with the TRV vector carrying fragments of the genes to be silenced, transcript levels of NDR1, EDS1 and RAR1 were reduced by about 60% to 80% in silenced plants as compared to plants infected with the TRV empty vector (S4 Fig). At this time, Agrobacterium strains expressing His-XopAU were used to inoculate silenced and control plants, and cell death was monitored visually and quantified by measuring ion leakage. As a control, silenced leaves were also inoculated with Agrobacterium expressing the R gene/effector gene pair Cf4/avr4, which elicits a hypersensitive response in N. benthamiana leaves that is dependent on expression of EDS1 and RAR1, but not of NDR1 [42]. As expected, silencing of EDS1 and RAR1, but not of NDR1 severely reduced Cf4/avr4-mediated cell death and ion leakage (Fig 2F and 2G). Conversely, cell death mediated by His-XopAU was not affected by silencing of any of the tested ETI signaling components (Fig 2F and 2G). These results suggest that it is unlikely that the cell death observed upon XopAU expression in leaf tissues is triggered by recognition of the effector by an R protein.
XopAU promotes chlorosis and accumulation of PR proteins when delivered by Xe in pepper leaves
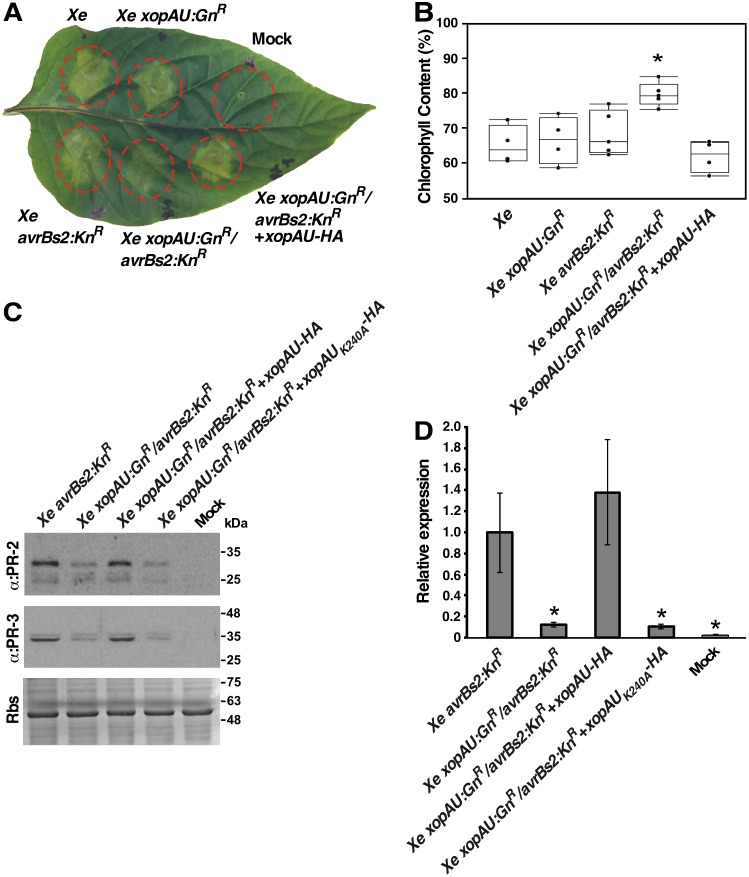
To assess the contribution of XopAU to bacterial virulence, the corresponding gene was inactivated in Xe bacteria by insertion mutagenesis. The mutant strain Xe xopAU:GnR and wild-type Xe were used to infect ECW30R pepper plants that were then monitored for bacterial growth and development of disease symptoms. Disease symptoms were estimated visually and quantified by measuring chlorophyll content and ion leakage, as parameters of chlorosis and necrosis that are typically observed in pepper leaves infected by Xe bacteria. Leaves infected with the mutant strain Xe xopAU:GnR displayed a similar chlorophyll content, ion leakage and bacterial growth as leaves infected with wild-type Xe bacteria (Fig 3 and S5A Fig).
Fig 3. Phenotypic analysis of xopAU gene inactivation.
Leaves of the pepper line ECW30R were syringe-infiltrated with a 10 mM MgCl2 mock solution or with suspensions (1 x 107 CFU/ml) of the indicated strains. (A) Photograph of an inoculated leaf at 5 days post-inoculation (dpi). (B) Chlorophyll content relative to mock-inoculated areas at 5 dpi. The box plot displays 25th, 50th (middle line) and 75th percentiles (n = 4 or 5 biological repeats). An asterisk indicates a significant difference (Mann-Whitney U test, p value <0.05) relative to Xe avrBs2:KnR. (C) Total protein was extracted from the infected leaves at 3 days post -inoculation (dpi) and immunoblotted with the indicated antibodies. Rbs, Rubisco loading control stained by Ponceau S. (D) mRNA abundance of the PR-1 gene in the inoculated areas was measured by qRT-PCR at 72 h post-inoculation and calculated relative to areas inoculated with the Xe avrBs2:KnR strain. Values are means ± SD of three biological repeats. Asterisks indicate a significant difference (Student’s t test, p value < 0.05) relative to Xe avrBs2:KnR.
We hypothesized that a weak contribution of XopAU to Xe pathogenicity is more likely to be revealed in an attenuated bacterial strain. To test this possibility, we generated the double mutant strain Xe xopAU:GnR/avrBs2:KnR, mutated in both the xopAU and avrBs2 effector genes. The AvrBs2 effector was chosen for this analysis because it was previously shown to contribute to Xe virulence activity and its deletion allowed to reveal the virulence activity of other effectors [43,44]. The double mutant along with wild-type Xe and the single mutants Xe xopAU:GnR and Xe avrBs2:KnR were used to infect ECW30R pepper plants that were then monitored for bacterial growth, chlorophyll content and ion leakage. In infected leaves, the Xe avrBs2:KnR mutant displayed reduced bacterial growth and ion leakage, but similar chlorophyll content, as compared to Xe wild-type and Xe xopAU:GnR (Fig 3 and S5A Fig). The Xe xopAU:GnR/avrBs2:KnR double mutant was similar to Xe avrBs2:GnR in bacterial growth and ion leakage, but caused less chlorosis as indicated by a higher chlorophyll content (Fig 3 and S5 Fig). To confirm that the reduction in chlorotic symptoms was the result of a mutation in xopAU, the gene was re-introduced into the Xe xopAU:GnR/avrBs2:KnR strain driven by its native promoter. When inoculated into pepper leaves the complemented Xe xopAU:GnR/avrBs2:KnR(xopAU) strain caused similar disease symptoms as the Xe avrBs2:KnR strain (Fig 3 and S5A Fig).
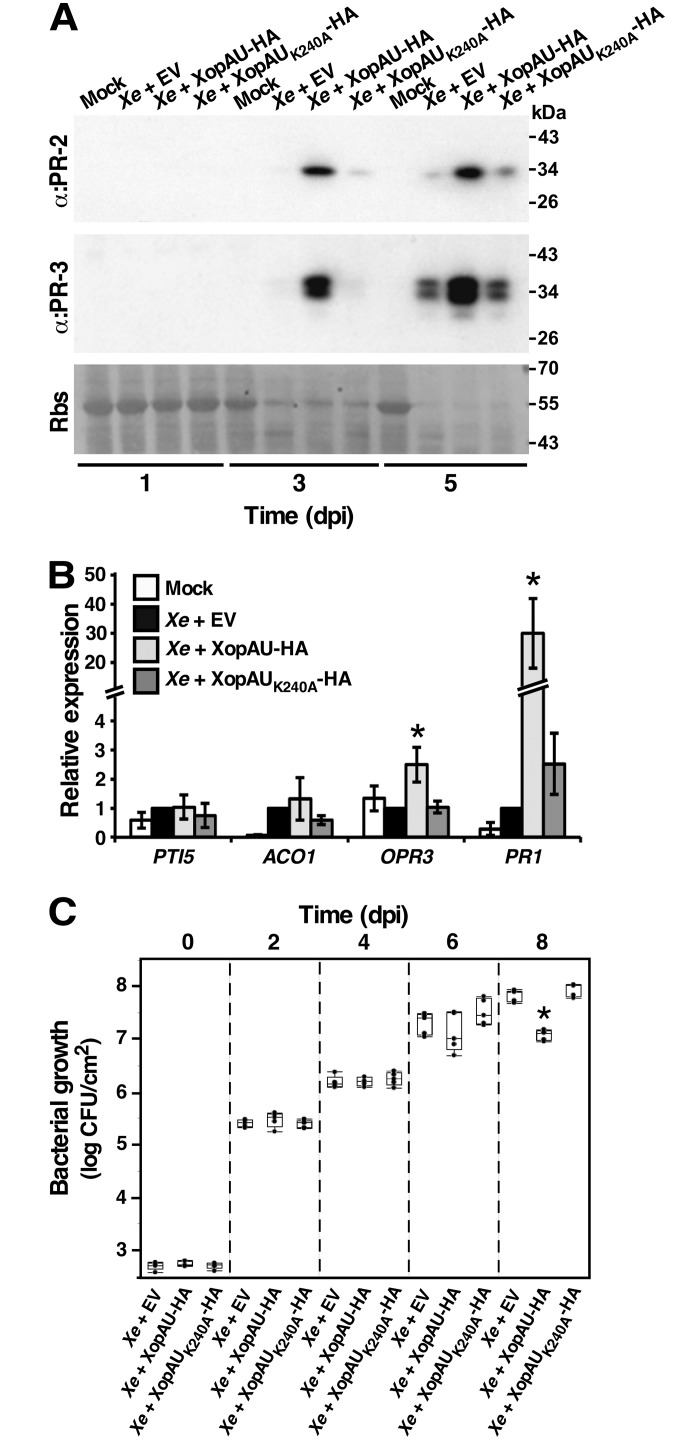
Because expression of His-XopAU via Agrobacterium activated immune signaling (see above), we tested whether deletion of the xopAU gene negatively affects the activation of defense responses. We monitored accumulation of the PR proteins PR-2 and PR-3 and mRNA levels of the PR-1 gene in infected pepper leaves at 3 dpi. Western blot analysis revealed that PR-2 and PR-3 accumulation was lower in leaves inoculated with the Xe xopAU:GnR/avrBs2:KnR double mutant strain than in leaves inoculated with the Xe avrBs2:KnR strain (Fig 3C). Leaves inoculated with the Xe xopAU:GnR/avrBs2:KnR double mutant complemented with XopAU-HA driven by its native promoter, but not with the kinase deficient XopAU-HAK240A, accumulated similar PR-2 and PR-3 protein levels as leaves infected with the Xe avrBs2:KnR strain. Similarly, qRT-PCR analysis revealed that transcript levels of the PR-1 gene were about 8.6-fold lower in leaves inoculated with the Xe xopAU:GnR/avrBs2:KnR double mutant or with this strain complemented with the kinase deficient XopAU-HAK240A than in leaves infected with the Xe avrBs2:KnR strain or with the double mutant strain complemented with XopAU-HA (Fig 3D). These results indicate that expression of a catalytically active XopAU in Xe strains promotes the activation of defense responses in infected pepper leaves.
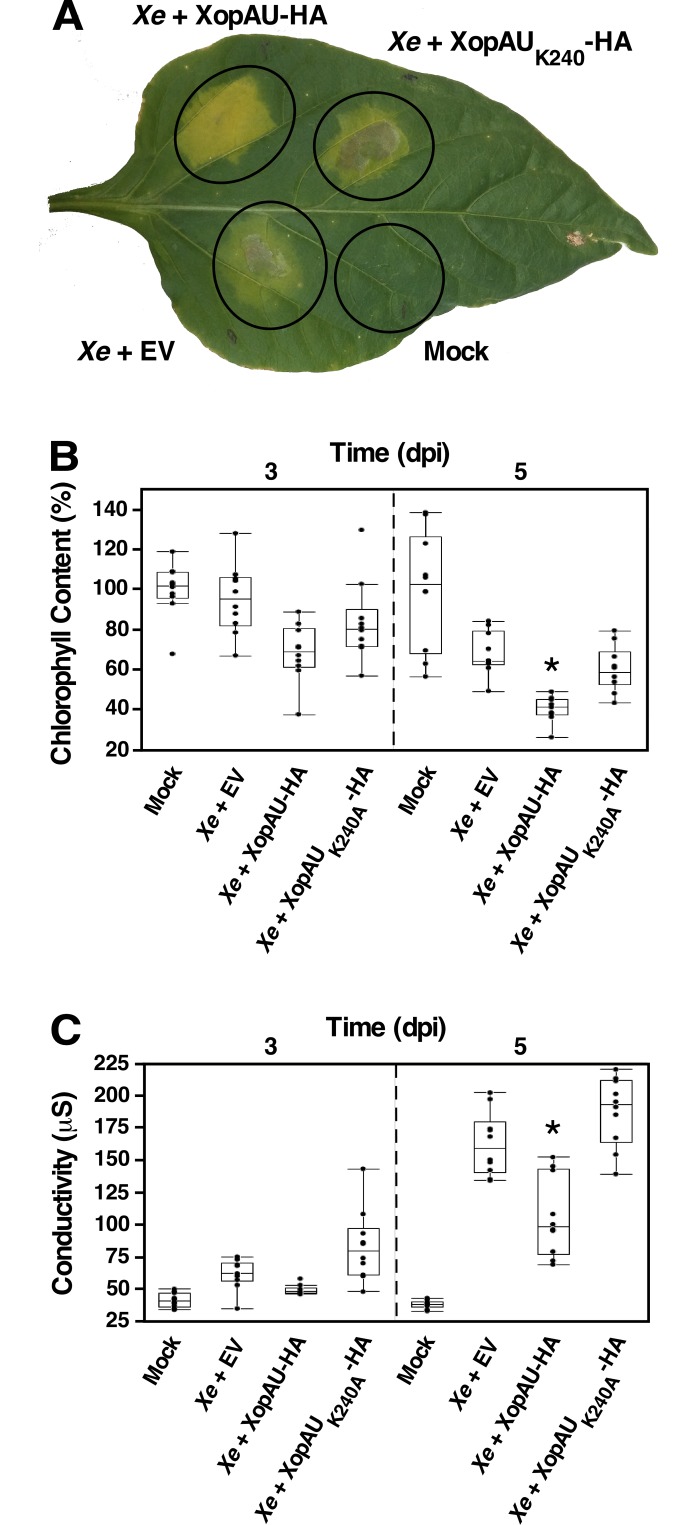
To further assess the contribution of XopAU to development of disease symptoms caused by Xe, we engineered a Xe strain carrying a HA-tagged XopAU variant (XopAU-HA) driven by a constitutive lac promoter in a broad-host plasmid. Overexpression of XopAU-HA in this strain was validated by Western blot analysis (S6A Fig). A mock solution, Xe bacteria overexpressing XopAU-HA or carrying an empty vector were infiltrated into ECW30R pepper leaves that were then monitored for the development of chlorosis and necrosis (i.e. chlorophyll content and ion leakage, respectively) at 1, 3 and 5 days post-inoculation (dpi). Xe overexpressing XopAU-HA displayed lower chlorophyll content and reduced ion leakage, compared to Xe containing an empty vector (Fig 4). To confirm that the observed phenotype is due to the biochemical activity of XopAU, we generated an Xe strain overexpressing the catalytically inactive XopAUK240A-HA variant. No difference was observed in chlorosis and ion leakage between pepper plants inoculated with Xe overexpressing XopAUK240A-HA and Xe carrying an empty vector (Fig 4). Together, observations obtained by using Xe xopAU:GnR/avrBs2:KnR double mutant and Xe bacteria overexpressing XopAU-HA suggest that the XopAU effector participates in the development of disease symptoms.
Fig 4. Phenotypic analysis of Xe strains overexpressing XopAU.
Leaves of the pepper line ECW30R were syringe-infiltrated with a 10 mM MgCl2 mock solution or with suspensions (1 x 107 CFU/ml) of Xe strains carrying a vector either empty (EV) or for expression of XopAU-HA or XopAUK240A-HA. (A) Photograph of an inoculated leaf at 5 days post-inoculation (dpi). (B) Chlorophyll content relative to mock-inoculated areas at 3 and 5 dpi. (C) Electrolyte leakage in the inoculated areas at 3 and 5 dpi. In B and C, box plots display 25th, 50th (middle line) and 75th percentiles (n = 10). Asterisks indicate a significant difference (Mann-Whitney U test, p value <0.05) relative to Xe containing an empty vector. Experiments were repeated at least three times with similar results.
Next, we tested whether infection of ECW30R pepper leaves with Xe overexpressing XopAU-HA caused activation of defense responses as observed when the effector was expressed via Agrobacterium. First, we monitored accumulation of PR proteins in infected leaf tissues by Western blot analysis. Both PR-2 and PR-3 accumulated at higher levels in plants inoculated with Xe overexpressing XopAU-HA than in plants inoculated with Xe carrying an empty vector or overexpressing the kinase deficient variant XopAUK240A-HA at 3 and 5 dpi (Fig 5A). We then assessed the mRNA levels of four genes (PTI5, ACO1, OPR3 and PR-1), whose expression reflects the activation of different defense and stress pathways, at the early stages of infection (16 hours after inoculation). qRT-PCR analysis revealed that transcript levels of the PR-1 gene, which is known to be induced by salicylic acid and pathogen attack [45], were about 30 fold higher in pepper leaves inoculated with Xe overexpressing XopAU-HA than in plants inoculated with Xe carrying an empty vector or overexpressing the kinase deficient variant XopAUK240A-HA (Fig 5B). The mRNA levels of the PTI5 and ACO1 genes, which are involved in ethylene signaling and biosynthesis, respectively [46], were not significantly altered by overexpression of XopAU-HA (Fig 5B). Finally, transcripts of the OPR3 gene, which encodes a component of the jasmonic acid biosynthesis pathway and is induced by wounding [47], displayed only a slight induction (about 2 fold) when leaves were infected with Xe overexpressing XopAU-HA (Fig 5B). This analysis suggests that XopAU overexpression activated plant defense signaling. Because accumulation of PR proteins might affect the ability of bacteria to colonize the plant, we examined whether overexpression of XopAU-HA affects bacterial growth in pepper leaves. As shown in Fig 5C, Xe bacteria overexpressing XopAU-HA displayed a similar growth as wild-type and bacteria overexpressing XopAUK240A-HA up to 6 dpi, and a reduced growth only at the late stages of infection (8 dpi), which may be ascribed to high accumulation of PR proteins.
Fig 5. Expression of defense and stress marker genes in leaves infected with Xe overexpressing XopAU.
Leaves of the pepper line ECW30R were syringe-infiltrated with a 10 mM MgCl2 mock solution (Mock) or with suspensions (1 x 107 CFU/ml) of Xe strains carrying a vector either empty (EV) or for expression of XopAU-HA or XopAUK240A-HA. (A) Total protein was extracted from the infected leaves at the indicated days post-inoculation (dpi) and immunoblotted with the indicated antibodies. Rbs, Rubisco loading control stained by Ponceau S. (B) mRNA abundance of the indicated genes in the inoculated areas was measured by qRT-PCR at 16 h post-inoculation and calculated relative to areas inoculated with the Xe strain carrying an empty vector. Values are means ± SE of three biological repeats. Asterisks indicate a significant difference (Student’s t test, p value < 0.05) relative to Xe containing an empty vector. (C) Leaves were infiltrated with bacterial suspensions (1 x 105 CFU/ml) of Xe strains carrying a vector either empty (EV) or for expression of XopAU-HA or XopAUK240A-HA, and bacterial growth was quantified at the indicated dpi. A box plot displays 25th, 50th (middle line) and 75th percentiles (n = 5). Asterisks indicate a significant difference (Mann-Whitney U test, p value <0.05) relative to Xe containing an empty vector. Experiments were repeated at least three times with similar results.
Delivery of XopAU in plant cells by Xcc causes cell death
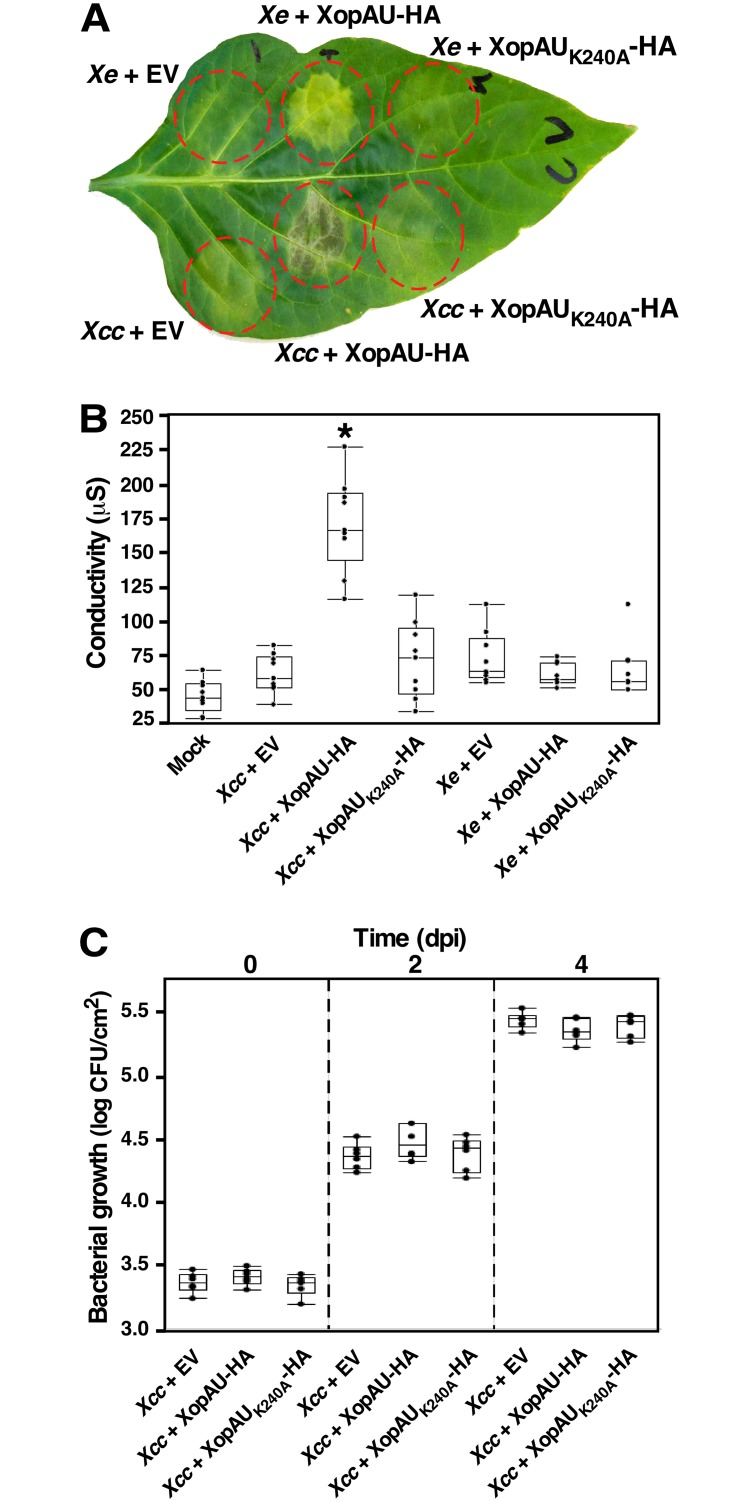
Activation of defense responses by XopAU is accompanied by cell death when XopAU is expressed through Agrobacterium but not when the effector is delivered in plant cells by Xe bacteria. This discrepancy may be related to the interplay between XopAU and other species-specific virulence determinants. To explore this possibility, a plasmid for overexpression of XopAU-HA or XopAUK240A-HA was mobilized into the crucifer pathogen Xanthomonas campestris pv. campestris strain 8004 (Xcc), which does not encode a XopAU homolog. ECW30R pepper leaves were inoculated with Xe or Xcc strains overexpressing XopAU-HA, the kinase deficient variant XopAUK240A-HA or an empty vector, and cell death was monitored visually and quantified by ion leakage. Leaf areas inoculated with Xcc overexpressing XopAU-HA displayed cell death and a concomitant increase in ion leakage at 2 to 3 dpi, while Xcc containing an empty vector or overexpressing XopAUK240A-HA did not induce any visible phenotype (Fig 6A and 6B). At the same time, overexpression of XopAU-HA through Xe induced strong chlorosis (Fig 6A). To examine if the cell death induced by the expression of XopAU from Xcc is host specific, Xcc bacteria overexpressing XopAU-HA, XopAUK240A-HA or an empty vector were infiltrated into the leaves of N. benthamiana. Leaves inoculated with Xcc overexpressing XopAU-HA, but not XopAUK240A-HA or an empty vector, displayed cell death, which was confirmed by increased ion leakage at 24–48 h after inoculation (S7A and S7B Fig), thus indicating that this phenotype was not host specific. Overexpression of XopAU in Xcc did not affect bacterial growth in infected pepper and N. benthamiana leaves (Fig 6C and S7C Fig). It should be pointed out that expression of XopAU-HA and XopAUK240A-HA was higher in Xcc as compared to Xe bacteria as detected by Western blot analysis (S6A Fig). Together, these observations suggest that the phenotype caused by XopAU when delivered by Xe may be tuned by bacterial determinants absent in Xcc strains. Alternatively, the different phenotypes may derive from the higher expression levels of XopAU-HA in Xcc compared to Xe.
Fig 6. Cell death caused by delivery of XopAU in pepper cells by Xanthomonas campestris pv. campestris.
Leaves of the pepper line ECW30R were syringe-infiltrated with a 10 mM MgCl2 mock solution (Mock) or with suspensions (1 x 107 CFU/ml) of Xe or Xcc strains containing a vector for expression of XopAU-HA and XopAUK240A-HA, or an empty vector (EV). (A) Photograph of an inoculated leaf at 3 days post-inoculation (dpi). (B) Electrolyte leakage at 2 dpi. (C) Leaves were syringe-infiltrated with bacterial suspensions (1 x 105) of Xe and Xcc strains as in (A) and bacterial growth was quantified at the indicated dpi. In (B) and (C), box plots display 25th, 50th (middle line) and 75th percentiles (in A, n = 7 or 9; in C, n = 6). An asterisk indicates a significant difference (Mann-Whitney U test, p value <0.05) relative to Xe containing an empty vector. Experiments were repeated three times with similar results.
MEK2 is required for elicitation of cell death caused by XopAU in N. benthamiana leaves
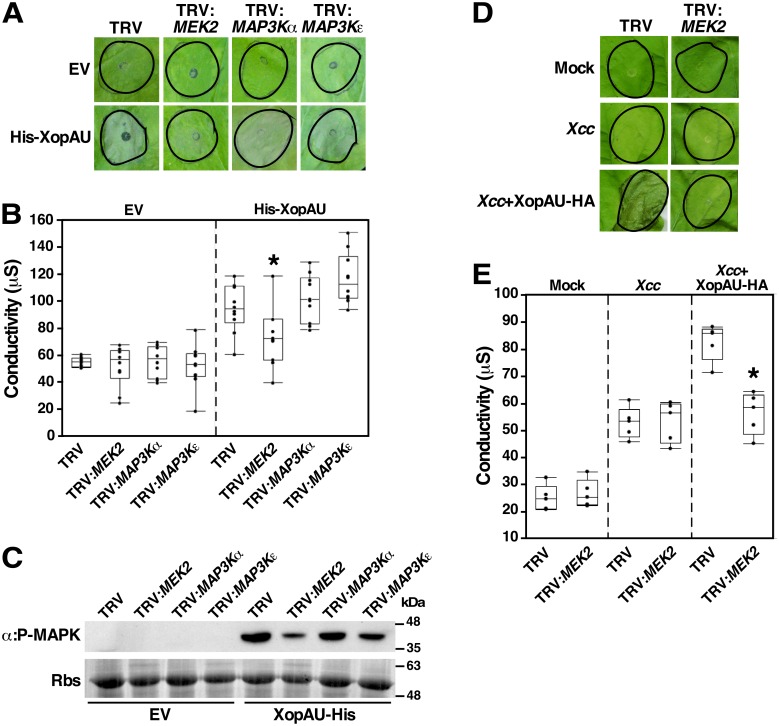
MAP kinase cascades were previously shown to be involved in cell death signaling associated with plant immunity [12]. We hypothesized that XopAU induces immune responses by manipulating and activating components of MAP kinase cascades. To test this hypothesis, we examined the ability of His-XopAU to elicit cell death in N. benthamiana plants that were silenced either for the MEK2 gene, which encodes a positive regulator of cell death, or for the MAP3Kα and MAP3Kε genes, which encode MAPKKKs acting upstream of MEK2 [9,10]. For gene silencing by VIGS, N. benthamiana plants were inoculated with Agrobacterium strains containing plasmids for the expression of TRV either empty or carrying a fragment of the gene to be silenced. Four weeks later, MEK2, MAP3Kα and MAP3Kε transcript levels were reduced by at least 75% in silenced plants as compared to TRV-infected plants (S4 Fig). At this time, Agrobacterium strains expressing His-XopAU were used to inoculate silenced and control plants, and cell death was monitored visually and quantified by measuring ion leakage. Cell death and ion leakage induced by His-XopAU were significantly reduced in MEK2-silenced plants compared to MAP3Kα- and MAP3Kε-silenced plants, and to TRV-infected plants (Fig 7A and 7B). In addition, Western blot analysis revealed that phosphorylation of MAPKs induced by His-XopAU was reduced in the MEK2-silenced plants (Fig 7C). Similarly, a reduction in cell death and ion leakage was also observed in MEK2-silenced plants challenged with Xcc overexpressing XopAU (Fig 7D and 7E).
Fig 7. Silencing of MEK2 in N. benthamiana reduces XopAU-induced cell death.
N. benthamiana plants were infected with TRV, TRV:MEK2, TRV:MAP3Kα and TRV:MAP3Kε. In (A), (B), and (C), leaves of silenced plants were inoculated with Agrobacterium (OD600 = 0.02) carrying a vector either empty (EV) or for expression of His-XopAU from an estradiol-inducible system, and treated with 17β-estradiol 24 h later. (A) Photographs of inoculated areas at 36 h after 17β-estradiol application. (B) Electrolyte leakage at 24 h after 17β-estradiol application. (C) Total proteins were extracted at 12 h after 17β-estradiol application and samples were immunoblotted with α:P-MAPK antibodies. Rbs, Rubisco loading control stained by Ponceau S. In (D) and (E), leaves of silenced plants were inoculated with mock or suspensions (5 x 107 CFU/ml) of Xcc carrying a vector either empty (EV) or for expression of XopAU-HA. (D) Photographs of inoculated areas at 48 h after Xcc inoculation. (E) Electrolyte leakage at 24 h after Xcc inoculation. In (B) and (E), box plots display 25th, 50th (middle line) and 75th percentiles. (in B, n = 10; in E, n = 5 or 7). Asterisks indicate a significant difference (Mann-Whitney U test, p value <0.05) relative to TRV empty control. Experiments were repeated three times with similar results.
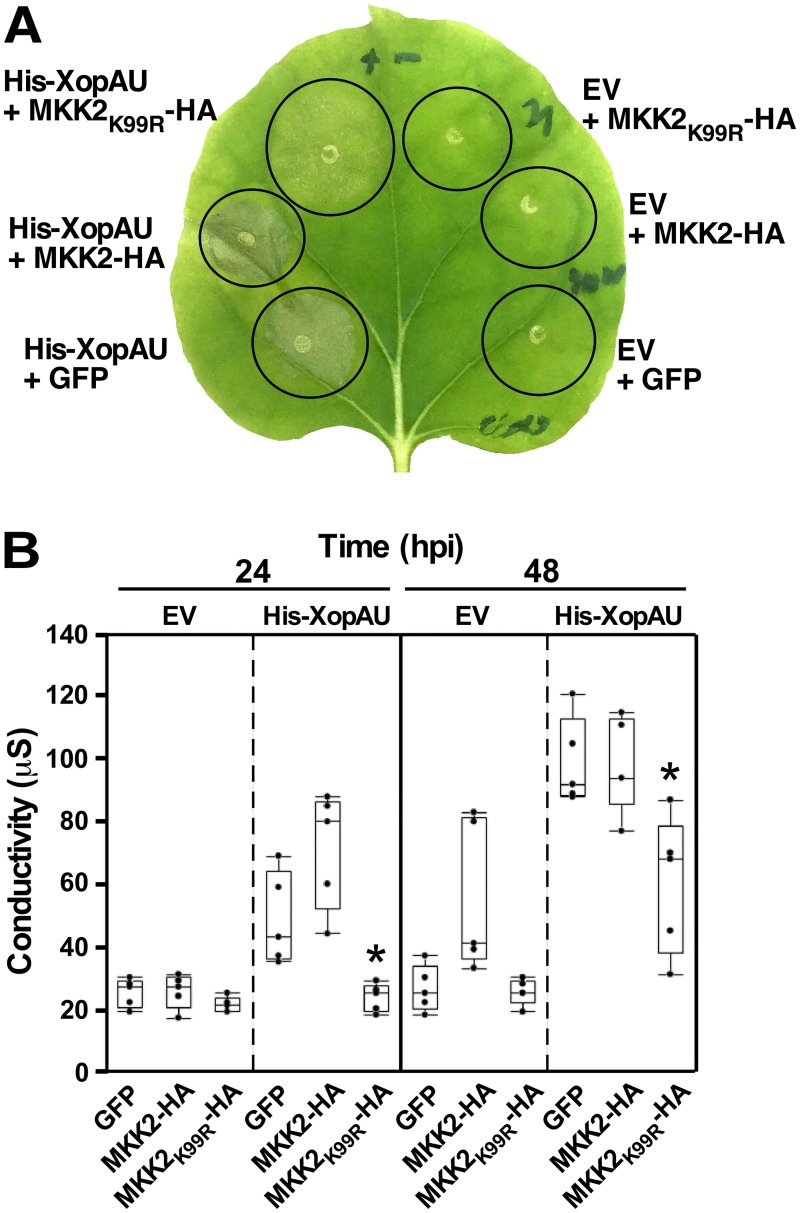
To provide additional evidence that cell death induced by XopAU requires a functional MEK2, we examined whether expression of the catalytically inactive variant of the tomato MEK2 homolog MKK2 (MKK2K99R) causes a dominant negative effect on XopAU-mediated cell death. His-XopAU was co-expressed via Agrobacterium in N. benthamiana leaves with MKK2-HA, MKK2K99R-HA, or an unrelated protein (GFP) driven by an estradiol inducible system. Cell death was visually monitored in the inoculated leaves and quantified by measuring ion leakage at 48 h after estradiol application. Expression of MKK2K99R-HA, but not that of MKK2-HA or GFP, significantly reduced the cell death and ion leakage induced by His-XopAU (Fig 8) indicating that a catalytically active MEK2/MKK2 is required for XopAU-mediated cell death.
Fig 8. Expression of MKK2K99R suppresses cell death mediated by XopAU.
N. benthamiana leaves were inoculated with Agrobacterium to co-express His-XopAU or an empty vector (EV) with MKK2-HA, MKK2K99R-HA or GFP. Expression was driven by an estradiol-inducible system and 17β-estradiol was applied 24 h after agro-infiltration. (A) Photograph taken at 48 h after 17β-estradiol application. (B) Electrolyte leakage at 24 h and 48 h after 17β-estradiol application. A box plot displays 25th, 50th (middle line) and 75th percentiles. (n = 5). Asterisks indicate a significant difference (Mann-Whitney U test, p value <0.05) relative to co-expression of His-XopAU with a GFP control. The experiment was repeated three times with similar results.
Co-expression of XopAU and MKK2 causes a growth inhibition phenotype in yeast
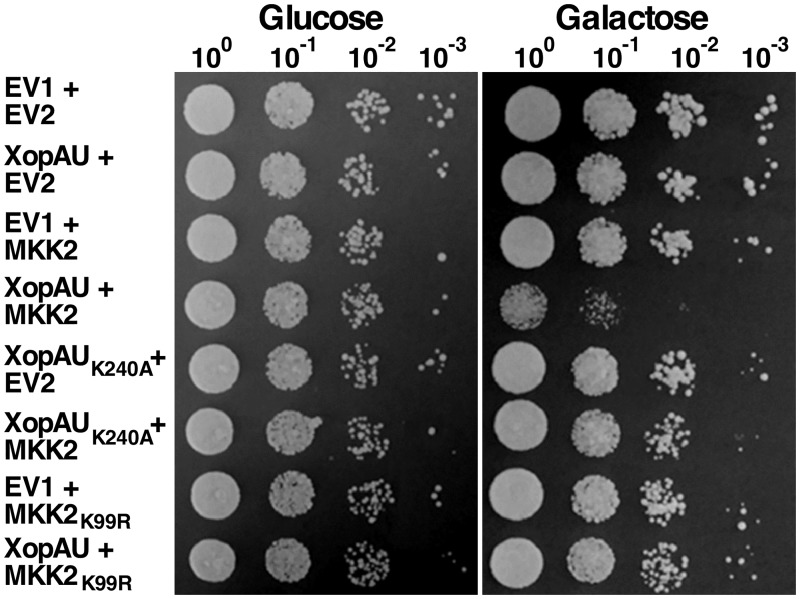
Expression of certain type III effectors in yeast has been shown to cause phenotypes that can be exploited to elucidate effector function, biochemical activity and host targets [48]. To test whether XopAU causes a detectable phenotype in the yeast Saccharomyces cerevisiae, the effector was fused to a c-myc tag, expressed in the yeast strain W303 driven by the GAL1 promoter, and protein accumulation was confirmed by Western blot analysis (S6B Fig). The effect of XopAU on yeast growth was examined by serially diluting yeast cultures that carry a vector either empty or for expression of the effector and plating them onto repressing (glucose) or inducing (galactose) media. The strain expressing XopAU exhibited similar growth as the control strain containing an empty vector both in repressing and inducing media (Fig 9).
Fig 9. Co-expression of XopAU and MKK2 inhibits yeast growth.
Yeast cultures containing the pGML10 vector either empty (EV1) or for expression of XopAU and XopAUK240A, and the pGMU10 vector either empty (EV2) or for expression of MKK2 and MKK2K99R were normalized to OD600 = 0.1, and serial dilutions were spotted onto selective media containing 2% glucose or 2% galactose and 1% raffinose. Plates were incubated at 30°C for 72 h (2% glucose) or 96 h (2% galactose and 1% raffinose) and photographed. The experiment was repeated three times with similar results.
Because MEK2/MKK2 was required for XopAU-mediated phenotypes in planta, we hypothesized a similar requirement in yeast. To test this hypothesis, yeast were engineered to express under the control of the GAL1 promoter either XopAU, XopAU kinase deficient (XopAUK240A), MKK2 and MKK2 kinase deficient (MKK2K99R) with an empty vector or the following protein combinations: XopAU with MKK2, XopAU with MKK2K99R, and XopAUK240A with MKK2. All the proteins were fused to a c-myc tag and their expression was validated by Western blot analysis (S6B Fig). Yeast co-expressing XopAU with MKK2 displayed a significant reduced growth when plated on inducing medium, but not on repressing medium, as compared to yeast strains expressing each protein alone or protein combinations that included a kinase deficient variant of either XopAU or MKK2 (Fig 9). These results indicate that XopAU required MKK2 to cause growth inhibition in yeast and this phenotype was dependent on the kinase activity of both proteins.
XopAU interacts with tomato MKK2 and MAP kinases
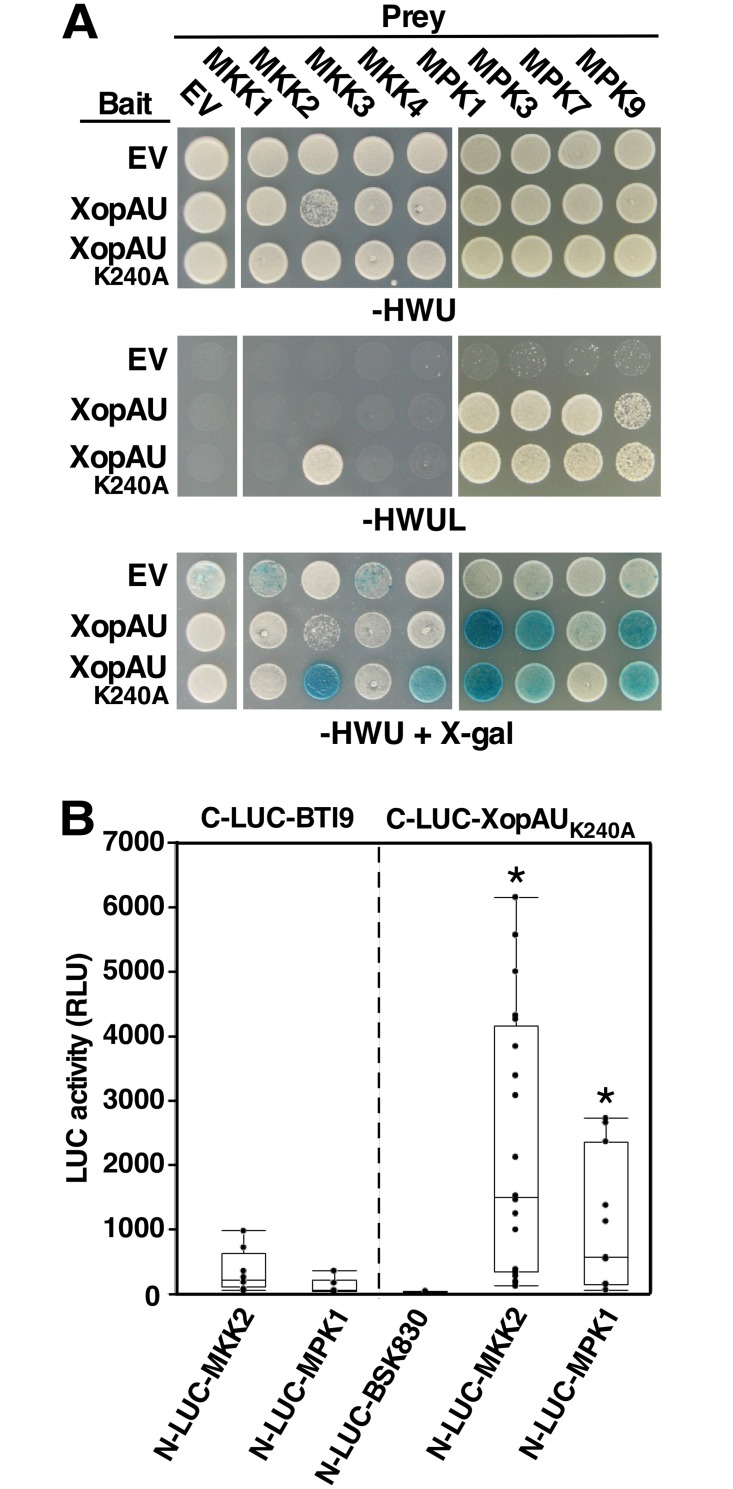
Because XopAU-mediated phenotypes were dependent on MEK2/MKK2 in planta and yeast, we hypothesized that MKK2 is a direct plant target manipulated by XopAU virulence activity. To explore this hypothesis, we tested whether XopAU physically interacts with MKK2 in a yeast two-hybrid system. To this aim, XopAU and its catalytically deficient form XopAUK240A were used as bait in yeast cells that expressed the tomato MAPKKs MKK1, MKK2, MKK3 or MKK4 as preys. All bait and prey proteins were expressed in the yeast cells as confirmed by Western blot analysis (S6C and S6D Fig). While XopAU did not interact with any of the tomato MAPKKs, XopAUK240A specifically interacted with MKK2, as evident by activation of the reporter genes LEU2 and lacZ (Fig 10A). XopAUK240A also interacted with N. benthamiana MEK2 and pepper MKK2 (S8 Fig). The lack of interaction between the catalytically active XopAU and MKK2 could be the consequence of the growth inhibition phenotype observed when both proteins were co-expressed in yeast (Fig 9).
Fig 10. Physical interaction of XopAU with components of plant MAPK cascades in yeast and in planta.
(A) Yeast expressing the indicated combinations of bait and prey were spotted on either selective medium (-HWUL) or non-selective medium (-HWU) with or without the addition of X-gal. (B) The indicated combinations of fusions proteins were co-expressed in epidermal cells of N. benthamiana leaves via Agrobacterium, and luciferase activity was quantified as relative luciferase units (RLU) at 48 h post-infiltration. C-LUC and N-LUC indicate the luciferase N-terminal (N-LUC) or C-terminal (C-LUC) region, respectively. A box plot displays 25th, 50th (middle line) and 75th percentiles. (n = at least 6). Asterisks indicate a significant difference (Mann-Whitney U test, p value <0.05) relative to C-LUC empty and C-LUC-BTI9. The experiment was repeated three times with similar results.
In parallel investigation aimed at the identification of additional candidate plant targets of the effector, XopAU was used as bait in a yeast two-hybrid screen of a tomato cDNA library [49]. This screen identified three MAPKs (MPK1, MPK3, and MPK9) that consistently interacted with the kinase active and inactive forms of XopAU resulting in the activation of both reporter genes (Fig 10A and S6D Fig).
Next, we used split luciferase complementation assays to validate in planta protein-protein interactions that were observed in yeast. Wild-type XopAU could not be used in these experiments because it caused cell death when fused to the C-terminus of the firefly luciferase protein (C-LUC) and failed to accumulate in leaves. Instead, we used XopAUK240A that was fused to C-LUC and co-expressed in N. benthamiana leaves through Agrobacterium along with MPK1 (representative of the XopAU-interacting MAP kinases) or MKK2 fused to the N-terminus of the firefly luciferase (N-LUC). As negative controls, C-LUC-XopAUK240A was co-expressed with N-LUC fused to the tomato receptor-like cytoplasmic kinase BSK830, while N-LUC-MPK1 and N-LUC-MKK2 were co-expressed with C-LUC fused to the kinase domain of the tomato receptor-like kinase BTI9. Expression of all the fusion proteins was validated by Western blot analysis (S6E Fig). Protein-protein interactions in planta were quantified by measurements of luminescence at 48 h after agro-infiltration. Co-expression of C-LUC-XopAUK240A and N-LUC-MPK1 or N-LUC-MKK2 resulted in emission of significantly higher luminescence compared to the negative controls indicating a physical interaction in planta between these two pairs of fusion proteins (Fig 10B). Together, these results indicate that XopAU physically interacts with multiple components of MAP kinase cascades at the MAPK and MAPKK levels.
XopAU phosphorylates MKK2 in vitro and promotes phosphorylation of MKK2 at multiple sites in planta
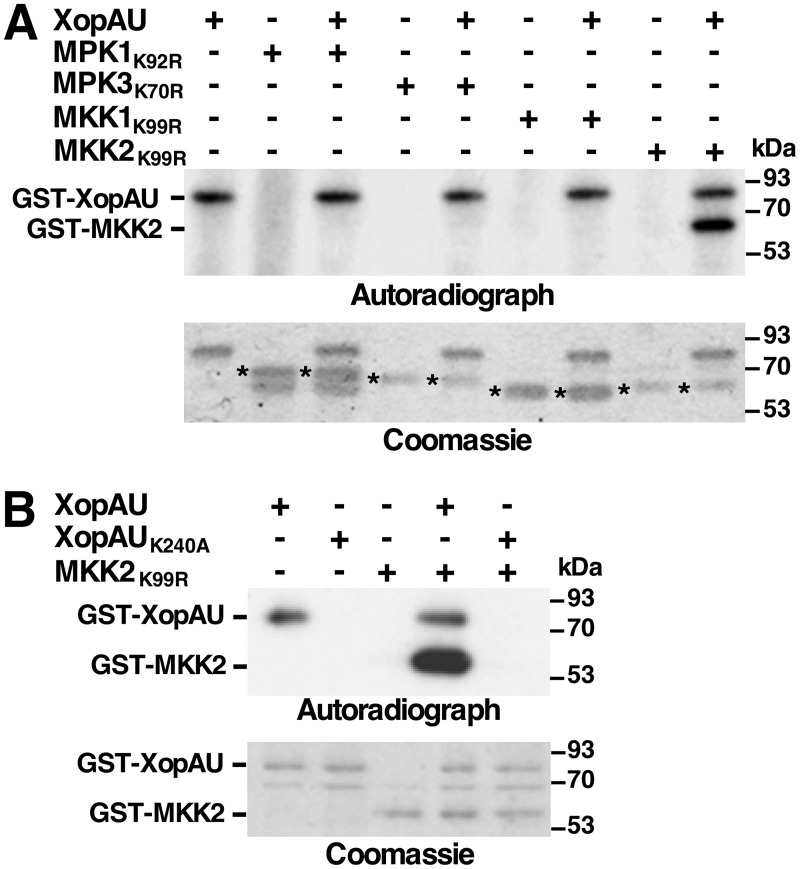
In vitro kinase assays were performed to test whether proteins that physically interacted with XopAU are substrates of XopAU phosphorylation. Kinase deficient variants of MPK1 (MPK1K92R), MPK3 (MPK3K70R), MKK2 (MKK2K99R) and MKK1 (MKK1K99R), which did not interact in yeast with XopAU and thus served as a negative control, were expressed as GST fusions in E. coli, purified and incubated with GST-XopAU in the presence of [γ-32P]ATP. As shown in Fig 11A, GST-XopAU phosphorylated GST-MKK2K99R, but not GST-MPK1K92R, GST-MPK3K70R or GST-MKK1K99R. The kinase deficient GST-XopAUK240A was not able to phosphorylate GST-MKK2K99R confirming that labeling of GST-MKK2K99R was dependent on the GST-XopAU catalytic activity (Fig 11B).
Fig 11. XopAU phosphorylates MKK2.
In vitro kinase assays testing phosphorylation of XopAU-interacting proteins by XopAU (A), and MKK2K99R by XopAUK240A (B). The indicated proteins were incubated in a kinase assay in the presence of [γ-32P]ATP, fractionated by SDS-PAGE, and exposed to autoradiography (upper panel) or stained by Coomassie (lower panel). Asterisks in (A) mark bands corresponding to proteins that were tested as XopAU substrates. The experiments were repeated three times with similar results.
The effect of XopAU on the phosphorylation state of MKK2 was then examined in planta. To this aim, MKK2 tagged with a HA epitope tag (MKK2-HA) was co-expressed via Agrobacterium in leaves of N. benthamiana plants along with His-XopAU in the wild-type or the kinase deficient form (His-XopAUK240A). Expression of MKK2-HA, His-XopAU and His-XopAUK240A was driven by an estradiol inducible system. MKK2-HA was immunoprecipitated from leaf samples, which were collected at 12 h after estradiol application, digested with trypsin, and analyzed by quantitative mass spectrometry of phosphopeptides. This analysis identified six residues (Thr33, Ser73, Tyr176, Thr215, Ser221 and Ser269) that were differentially phosphorylated in the presence of His-XopAU compared to His-XopAUK240A (Table 1 and S4 Table). Remarkably, expression of His-XopAU resulted in an average increase of 200 fold in the phosphorylation of both Thr215 and Ser221 (Table 1 and S4 Table), which are part of the S/TxxxS/T activation motif of MKK2 [12]. Phosphorylation of Thr33, Ser73, Tyr176 and Ser269 was also enhanced upon expression of XopAU by about 36, 28, 51 and 73 folds, respectively (Table 1). Together, these observations demonstrate that XopAU phosphorylated MKK2 in vitro and either directly or indirectly promoted phosphorylation of multiple MKK2 sites in planta, possibly resulting in its activation.
Table 1. XopAU-mediated phosphorylation of MKK2 in planta.
| Peptide | Site | Fold change* |
|---|---|---|
| RTDLTLPLPQR | T33 | 42 ± 5 |
| TDLTLPLPQR | T33 | 30 ± 7 |
| IGSGTGGTVYK | S73 | 28 ± 6 |
| QVLSGLYYLHR | Y176 | 51 ± 14 |
| VLAQTMDPCNSSVGTIAYMSPER | T215 | 43 ± 4 |
| VLAQTMDPCNSSVGTIAYMSPER | T215, S221 | 215 ± 118 |
| FPFSVGR | S269 | 73 ± 46 |
| DVDNPNVVR | - | 0.6 ± 0.3 |
| Total ion current | - | 1 ± 0.2 |
Phosphorylated residues are underlined and bold. The ratio of the DVDNPNVVR peptide of MKK2, which does not contain any phosphorylation site, and total ion current between samples expressing His-XopAU and His-XopAUK240A were determined to confirm that similar protein amounts were analyzed for the compared samples.
*Change in the amount of MKK2 phosphorylated peptides between samples expressing His-XopAU and His-XopAUK240A ± SE (n = 3).
Discussion
In this study, we uncovered biochemical properties of the Xanthomonas euvesicatoria type III effector XopAU that encodes a protein kinase and contributes to the development of disease symptoms in pepper plants. In addition, we show that XopAU manipulates MAP kinase signaling by activating the immunity-associated MAPKK MKK2.
This is the first report of a type III effector of phytopathogenic bacteria that encodes a catalytically active serine/threonine protein kinase representing a novel enzymatic activity for type III effectors acting within plant cells. Type III effectors with protein kinase activity were previously identified in bacterial pathogens that infect mammalian cells and they include the YpkA/YopO effector from Yersinia and OspG from Shigella [50]. YpkA/YopO phosphorylates the heterotrimeric G-protein Gαq in the GTP binding loop inhibiting Gαq activation and signal transduction [51]. While OspG substrates are yet to be identified, its kinase activity is required to inhibit degradation of phosphorylated IkBα and NF-κB activation induced by TNF-α stimulation, resulting in the interference of host innate immune responses [52]. Interestingly, kinase activity of both YpkA/YopO and OspG requires binding of a host factor for activation (i.e. actin and ubiquitin, respectively) possibly to prevent undesired activity while in the bacterium [53–55]. It will be interesting to test whether XopAU is a constitutively active kinase or it is activated in the plant cell by binding of a host factor or by posttranslational modifications.
Protein-protein interaction studies revealed that XopAU interacts in yeast with multiple tomato MAPKs and with the immunity-associated MAPKK MKK2. The interactions of XopAU with the MPK1 MAP kinase and MKK2 were also confirmed in planta. Moreover, MKK2, but not the MAPKs, was a substrate of XopAU phosphorylation in vitro. In several instances components of MAP kinase cascades were found to be targeted by type III effectors. With the exception of activation of the MAP kinase MPK4 by the P. syringae effector AvrB [23], other effector-MAPK interactions results in inactivation of the host MAPKs and interference with MAPK signaling. For example, members of the HopAI1 effector family of phosphothreonine lyases from plant and animal bacterial pathogens interact with MAP kinases and suppress their activities by irreversibly removing a phosphate to inhibit host immune responses [56]. Similarly, Yersinia YopJ and P. syringae HopF2 inhibit the signaling ability of MAPKKs by acetylation and ADP-ribosylation, respectively [19,22]. Our results demonstrate that this is not the case for XopAU. In fact, expression of XopAU in planta promoted phosphorylation of MAP kinases and MKK2 in their activation domains, and induced plant defense responses that are typically observed upon MKK2 activation [12]. The possibility that XopAU-mediated activation of defense responses is the result of recognition of the effector by a plant R protein is unlikely because silencing of early components of ETI signaling in N. benthamiana plants did not affect XopAU-mediated cell death.
Expression of XopAU along with MKK2 in planta enhanced phosphorylation of MKK2 at six residues, including Thr215 and Ser221 that are part of the S/TxxxS/T MKK2 activation motif. It remains to be established whether XopAU directly phosphorylates these residues in planta or activates a mechanism resulting in their phosphorylation by another kinase(s). Additional host proteins, such as the XopAU-interacting protein MPK1, MPK3 and MPK9 may be involved in the activation of MKK2 by XopAU. MPK1 might modulate XopAU activity and substrate affinity by phosphorylation, and promote phosphorylation and activation of MKK2 by XopAU. Alternatively, XopAU might enhance MPK1 phosphorylation by MKK2 by acting as a scaffold protein that interacts and bridges between MKK2 and MPK1.
Functional evidence provides further support to the biochemical data for a role of MKK2 as a target of XopAU. In planta, silencing of NbMEK2, the N. benthamiana ortholog of MKK2, or overexpression of a kinase dead variant of MKK2 suppressed XopAU-mediated cell death and MAPK phosphorylation indicating that MKK2 is required for XopAU molecular function. In yeast, expression of both XopAU and MKK2 in a catalytic active form, but not that of each protein alone, resulted in growth inhibition suggesting molecular cooperation between the two proteins. MKK2 does not have a closely related homolog in yeast, while its downstream MAPKs, MPK1 and MPK3, share 53% and 48% identity to FUS3 (NCBI acc. num. AAA34613.1) and HOG1 (NCBI acc. num. AJV50684.1), respectively. Interestingly, activation of either FUS3 or HOG1 pathways was reported to promote cell cycle arrest [57,58]. Based on these observations and on the finding that MKK2 is a substrate of XopAU phosphorylation in vitro, we hypothesize that the growth inhibition phenotype caused by co-expression of MKK2 and XopAU in yeast is a result of MKK2 activation by XopAU and subsequent MKK2 initiation of yeast MAPK cascades involved in cell cycle arrest.
Gene inactivation analysis in an attenuated Xe strain revealed that XopAU contributes to the appearance of disease symptoms in susceptible pepper plants, but not to bacterial growth. In addition, molecular analysis demonstrated that XopAU overexpression activates plant defense responses in Xe host and non-host plants. Accumulation of defense proteins reaches high levels at late stages of infection in pepper leaves infected with Xe overexpressing XopAU and may be the source of the decreased bacterial growth observed for this strain at 8 dpi. Remarkably, defense responses induced by XopAU were accompanied by the appearance of chlorosis when the effector was expressed by Xe, or by cell death when XopAU was delivered/expressed by Agrobacterium and Xcc. These different phenotypes may be related to different XopAU expression levels in the various experimental systems, as observed in Xe and Xcc bacteria overexpressing XopAU. Alternatively, the differential response observed when XopAU is delivered/expressed by different bacteria may result from inhibition of XopAU-mediated cell death by other Xe determinants. Possible XopAU antagonists are Xe 85–10 type III effectors, such as XopE1 and XopM, that are absent in Xcc 8004 and were found to suppress cell death induced by a constitutively active form of the immunity-associated MAPKK MEK2 [59], the tobacco ortholog of MKK2, which we report here to be activated by XopAU. Additional candidates are effectors that were shown to suppress ETI-dependent or independent cell death. For example, XopB inhibits ETI-related cell death triggered by recognition of the AvrBsT effector in pepper plants, as well as cell death induced by several other effectors in tobacco [60]. AvrBsT suppresses the ETI-related cell death induced by AvrBs1 in pepper [61], while XopJ delays the appearance of necrotic disease symptoms interfering with host salicylic acid signaling [32].
Despite the fact that MKK2 was identified as a target of XopAU, it is yet to be established how activation of this immunity-associated MKK by XopAU may contribute to Xe pathogenicity. MAP kinase cascades activated by tomato MKK2 and its orthologs in other plant species have been implicated as key signaling modules not only in plant immunity [8], but also in other physiological processes, such as the response to abiotic stress (e.g. wounding, osmotic and oxidative stress), stomata development and floral senescence [62–66]. It is possible that XopAU-mediated activation of MKK2 selectively induces a subset of cellular responses that are beneficial to the pathogen. Alternatively, activation of defense responses through MKK2 could be connected to the contribution of XopAU to the development of disease symptoms. In support of this hypothesis, a correlation was observed between the appearance of chlorotic symptoms and accumulation of PR proteins in pepper leaves infected with Xe strains expressing catalytically active and inactive variants of XopAU.
In summary, we provide evidence that XopAU is a functional protein kinase that manipulates host MAPK signaling by activating the immunity-associated MAPKK MKK2. In addition, based on the different phenotypes observed when the effector is expressed by different bacteria, we propose a functional interaction between XopAU and other bacterial determinant(s). This study provides new insights about a possible role for activation of host immunity-associated MAPK cascades in disease development.
Materials and methods
Phylogenetic analysis
The genomic region of Xe 85–10 (NZ_CP017190.1) from position 4,861,200 to 4,862,753 (base pairs), which contains the ORF of the xopAU gene [67], was used to search homologous sequences in bacterial genomes of the non-redundant NCBI database. The xopAU and gyrB genes from a representative strain for each Xanthomonas species were selected for the phylogenetic analysis (S1 Table). Phylogenetic analysis was performed by using the neighbor joining method based on the xopAU and gyrB sequence alignments that were obtained by using Clustal X [68]. The bootstrap consensus tree inferred from 100 replicates is taken to represent the evolutionary history of the taxa analyzed.
Plant material, bacterial and yeast strains
Bacterial and yeast strains used in this study are listed in S2 Table and were grown as follows: Escherichia coli in Lysogeny Broth (LB) medium at 37°C; Xanthomonas euvesicatoria (Xe), Xanthomonas campestris pv. campestris (Xcc), and Agrobacterium tumefaciens in LB medium at 28°C; yeast (Saccharomyces cerevisiae) at 30°C in selective synthetic complete medium supplemented with 2% glucose, or 2% galactose and 1% raffinose [69].
Plant cultivars used in this study are: pepper (Capsicum annuum) ECW20R [43] and ECW30R [70], Nicotiana benthamiana [71], and tomato (Solanum lycopersicum) Hawaii 7981 [72].
DNA manipulation
Plasmid constructs used in this study are described in S3 Table. For cloning, DNA fragments were amplified from the Xanthomonas genome or cDNA of pepper, tomato or N. benthamiana plants, using Phusion DNA Polymerases (Thermo Fisher Scientific, Inc. Waltham MA, USA) or PrimeSTAR HS DNA Polymerase (Clontech Laboratories, Inc. Mountain View CA, USA). Site-directed mutagenesis was carried out using the QuikChange II kit (Agilent technologies, Inc. Santa Clara CA, USA). Sequences of oligonucleotides used in this study are available upon request.
Mutagenesis and overexpression in Xanthomonas bacteria
To generate an Xe insertion mutant in the avrBs2 gene (XCV0052) by single crossover, an avrBs2 DNA fragment (187–827 bp) was cloned into the pVIK165 plasmid. The obtained plasmid was mobilized into the Xe or Xe xopAU:GnR [67] strains and bacteria were plated on LB media with kanamycin selection. Gene disruption was verified by PCR and loss of Xe avirulence in resistant ECW20R pepper plants. For overexpression of the xopAU gene, the xopAU coding region was fused to a HA epitope tag and cloned into the pBBR1MCS2 broad host vector driven by the lac promoter. For complementation of the Xe xopAU:GnR/avrBs2:KnR strain, the xopAU gene was cloned with its native promoter (646 bp upstream to the start codon) into the pBBR1MCS-3 plasmid in reverse orientation to the lac promoter. Plasmids were mobilized into Xanthomonas strains by triparental mating [73].
Agrobacterium-mediated transient expression
Binary vectors were transformed into Agrobacterium GV2260 by electroporation. For transient expression, Agrobacterium overnight cultures were pelleted, resuspended in induction medium (10 mM MgCl2, 10 mM MES pH 5.6, 200 mM acetosyringone), and incubated at 25°C with shaking for 4 h. Bacterial cultures were diluted to OD600 = 0.1 and infiltrated into leaves of N. benthamiana, pepper or tomato plants using a needleless syringe. When using the XVE estradiol-inducible system [74], plants were sprayed with an induction solution (5 μM 17β-estradiol, 1% Tween-20) at 24 h after agro-infiltration.
Plant inoculations, measurement of bacterial growth, chlorophyll content and ion leakage
For inoculation, 7-week-old pepper or 4-weeks-old N. benthamiana and tomato plants were infiltrated with bacterial suspensions (105 CFU/mL when monitoring bacterial growth; 107 CFU/mL when measuring ion leakage and chlorophyll content) in 10 mM MgCl2 by using a needleless syringe.
For measurement of bacterial growth, three 1-cm-diameter leaf discs were sampled from at least three plants and ground in 1 mL of 10 mM MgCl2. Bacterial numbers were determined by plating 10 μL from 10-fold serial dilutions and counting the resulting colonies.
For measurements of chlorophyll content, 10–20 1-cm2 leaf disks were sampled for each treatment, placed in a tube containing 2 ml of acetone, and incubated overnight in the dark. Absorption was determined at OD660 and OD642. Total chlorophyll content was quantified with the equation: 7.12 × OD660 + 16.8 × OD642 [75]. Chlorophyll content was calculated for each inoculated leaf area relative to a mock-infiltrated area of the same leaf.
For the measurements of ion leakage, two 1.5-cm-diameter leaf disks were sampled from inoculated areas of at least five plants, and floated in 10-mL tubes containing 5 mL of double-distilled water for 4 h at 25 °C with shaking. Conductivity was measured using a DDS-12DW conductivity meter (BANTE Instruments, Shanghai, China).
Yeast two-hybrid interactions
Yeast two-hybrid (Y2H) interactions and library screening were conducted as described [76]. To enable the use of 17β-estradiol to activate the GAL1 promoter, the yeast EGY48 strain was integrated with the Gal4-ER-VP16 transactivator [77] and renamed EGY48ES. The xopAU and xopAUK240A genes were cloned into the bait plasmid pEG202 and plasmids were transformed into EGY48ES by lithium acetate transformation. Baits were tested for interactions with either a tomato cDNA library [49] or tomato proteins (S3 Table) that were fused to the pB42 transcriptional activation domain in the prey plasmid pJG4-5. Expression of prey constructs was induced by growing yeast on media supplemented with 2% glucose and 0.5 μM of 17β-estradiol. Y2H interactions were tested using the LEU2 and lacZ reporter genes by plating yeast on selective media plates lacking leucine or containing x-gal, respectively.
Yeast growth inhibition assays
The xopAU and MKK2 genes were cloned into the yeast galactose inducible expression vectors pGML10 and pGMU10, respectively. Plasmids were co-transformed into the yeast strain W303 by lithium acetate transformation. For monitoring growth, yeast cultures were grown overnight at 30°C in liquid selective media containing 2% glucose, washed twice in 10 mM MgCl2, and normalized to OD600 = 0.1. Ten-fold serial dilutions were spotted (10 μl) onto repressing (2% glucose) or inducing (2% galactose and 1% raffinose) solid selective media, plates were incubated in 30°C for 72–96 h, and monitored visually for yeast growth inhibition.
Split luciferase complementation assay
The xopAU, xopAUK240A, MKK2, MPK1, BSK830 (GenBank acc. num. XP_004252882.1) and BTI9 [78] genes were cloned in frame to firefly luciferase fragments in the binary vectors pCAMBIA:N-LUC and pCAMBIA:C-LUC [79]. The obtained vectors were transformed into Agrobacterium and co-expressed in N. benthamiana leaves. Luciferase activity was measured at 48 h after infiltration: 3 mm diameter leaf disks were harvested and floated in 100 μL water in a white 96-well plate. Samples were supplemented with 0.5 mM D-luciferin (Sigma-Aldrich, St. Louis MO, USA) and incubated in the dark for 10 min. Luminescence was measured using a Veritas Microplate Luminometer (Promega Corporation, Madison WI, USA).
Expression and purification of GST fusion proteins in E. coli
MPK1K92R, MPK3K70R, MKK2, MKK2K99R, MKK1K99R, xopAU and xopAUK240A were cloned into the pGEX-4T-1 GST fusion expression vector (GE Healthcare, Little Chalfont, UK). Plasmids were transformed into E. coli Rosetta strain (MERCK, Kenilworth NJ, USA). Bacterial cultures were grown at 37°C while shaking to OD600 = 0.4–0.6, supplemented with 0.1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG), and incubated overnight at 16°C with shaking. Bacteria were pelleted, resuspended in binding buffer (Tris pH 7.4 containing 1 mM PMSF, 5 μg/mL leupeptin and 5 μg/mL aprotinin), lysed using a French press and centrifuged. Supernatants were incubated with glutathione agarose (Sigma-Aldrich) and proteins were purified according to manufacturer’s instructions.
In vitro kinase assay
GST fusion proteins (0.1–0.5 μg) were incubated in a kinase assay solution [50 mM Tris-HCl, pH 7.0, 1 mM dithiothreitol, 10 mM MgCl2, 20 μM ATP, 10 μCi [γ-32P]ATP (3,000 Ci/mmol; PerkinElmer, Inc. Waltham MA, USA)] at 25°C for 30 min. Reactions were stopped by the addition of SDS-sample buffer. Half of the reaction volume was fractionated on SDS-PAGE and stained with Coomassie blue. The second half was fractionated on SDS-PAGE, transferred onto a PVDF membrane, and the membrane was exposed to autoradiography.
Virus-induced gene silencing
For TRV infection, cotyledons of one-week-old N. benthamiana plants were co-infiltrated with Agrobacterium containing pTRV1 and pTRV2 in 1:1 ratio as described [10]. TRV2 plasmids used for silencing are described in S3 Table. Plants were grown in a growth chamber at 20°C in long day conditions (16 h light, 8 h dark).
RNA isolation and quantitative RT-PCR
Total RNA was isolated from leaves (50 mg) using the SV total RNA isolation system (Promega Corporation). RNA samples (2 μg) were reverse-transcribed using qScript cDNA Synthesis Kit (Quanta BioSciences, Inc. Gaithersburg MD, USA) and subjected to quantitative RT-PCR using gene specific primers (available upon request). cDNAs were amplified using the SYBR Premix Ex Taq II (Clontech Laboratories) and the Mx3000P qPCR System (Agilent technologies, Inc. Santa Clara CA, USA). The GAPDH gene was used for normalization, and gene expression was calculated by the comparative Ct method [80].
Protein extraction
For protein extraction from yeast and bacteria, overnight cultures were pelleted, resuspended in lysis buffer (4% SDS, 100 mM Tris/HCl pH 7.6, 0.1 M dithiothreitol) and incubated in 95°C with SDS sample buffer for 10 min. For protein extraction from plant tissues, 3–6 leaf disks of 1 cm diameter were frozen in liquid nitrogen, homogenized in extraction buffer (100 mM Tris pH 7.4, 1% Triton X-100, 1 mM PMSF, 5 μg/mL leupeptin, 5 μg/mL aprotinin, 50 mM NaF and 1 mM Na3VO4), and centrifuged.
Immunoprecipitation
MKK2-HA was transiently co-expressed in N. benthamiana leaves with either His-XopAU or His-XopAUK240A driven by the XVE estradiol inducible system [74]. Ten gram of leaf tissues was harvested at12 h after estradiol application and ground in liquid nitrogen. Protein extraction buffer was added to the powder and samples were centrifuged. The supernatant was collected, centrifuged again, filtered through Miracloth and incubated overnight at 4°C with Monoclonal α:HA-agarose (Sigma-Aldrich) on a tube roller. HA-agarose beads were washed twice in Tris pH 7.4 and submitted to phosphopeptide mass-spectrometry analysis.
LC-MS/MS analysis
Mass spectrometry analysis was performed at The Nancy & Stephen Grand Israel National Center for Personalized Medicine, Weizmann Institute of Science. For the identification of MKK2 sites phosphorylated in planta in the presence XopAU or XopAUK240A, immunoprecipitated samples were analyzed by LC-MS/MS as described [81]. Briefly, samples were subjected to in-solution, on-bead, trypsin digestion, and separated by using Split-less Nano Ultra Performance Liquid Chromatography (nanoUPLC; 10 kpsi NanoAcquity, Waters). The nanoUPLC was coupled online through a nanoESI emitter (10 μm tip; New Objective; Woburn, MA, USA) to a quadrupole orbitrap mass spectrometer (Q Exactive Plus, Thermo Scientific) using a FlexIon nanospray apparatus (Proxeon). For “Discovery” analysis and calculation of total ion current data was acquired in Data Dependent Acquisition (DDA) mode, using a Top 12 method [82]. For “Targeted” analysis, data was acquired in parallel reaction monitoring (PRM) mode [83], using an inclusion list containing all relevant peptides in the phosphorylated or un-modified form, as well as MKK2 peptide DVDNPNVVR. For DDA data analysis, raw data was imported into the Expressionist software (GeneData). The software was used for retention time alignment and peak detection of precursor peptides. A master peak list was generated from all MS/MS events and sent for database searching using Mascot v2.5 (Matrix Sciences). Data was searched against a protein database containing all available Nicotianoideae protein sequences from UniprotKB [84], Agrobacterium tumefaciens protein sequences, tagged MKK2 and XopAU, and 125 common laboratory contaminant proteins. Peptide identifications were filtered such that the global false discovery rate was maximum of 1%, and were then imported back to Expressionist to annotate identified peaks and calculate peptide intensities. For “Targeted” analysis [85] of MKK2 peptides, raw data were imported into the Skyline software [86]. Each peptide was manually curated to select the three most intense and reliable transitions, as well as to determine exact peak boundaries. Spectral libraries from the DDA experiments were also constructed and used in Skyline to evaluate the confidence in peak peptide assignment. Peak areas were then exported to a Microsoft Excel file, where the ratio of phosphorylated versus unmodified species of the peptide was calculated for each experiment. This ratio was then used to determine differential phosphorylation of MKK2 between samples that expressed XopAU or XopAUK240A (S4 Table). The intensity of the DVDNPNVVR peptide of MKK2, which does not contain any phosphorylation site, was determined in each sample and along with total ion current of the analyzed samples was used to compare MKK2 and total protein levels.
Supporting information
(XLSX)
(DOCX)
(DOCX)
(XLSX)
Nucleic acid sequences of xopAU homologs were aligned with ClustalX multiple sequence alignment tool. Sequences and corresponding NCBI accession numbers are reported in S1 Table. A dashed line represents a gap in the alignment. Asterisks indicate nucleotide conserved in all the homologs. Nucleotides are color-coded.
(PDF)
(A) Genomic location of the xopAU allelic variant of group 1 Xanthomonas strains in X. euvesicatoria (acc. num. NC_007508.1) and X. oryzae (acc. num. CP003057.2). (B) Genomic location of the xopAU group 2 allelic variant in X. fragariae (acc. num. CP016830.1) and X. gardneri (acc. num. CP018728.1), and corresponding genomic region in the X. arboricola strain (acc. num. CP012251.1), which does not contain the xopAU allele. Numbers and arrows represent genomic location and open reading frames (ORF), respectively. Locus tags are indicated below each ORF, which are colored based on DNA sequence homology.
(PDF)
Protein sequences of the XopAU homologs (S1 Table) were aligned with the COBALT multiple sequence alignment tool (https://www.ncbi.nlm.nih.gov/tools/cobalt/cobalt.cgi?) using default parameters. Blue fonts represent identical amino acids; yellow fonts represent nearly invariant residues in the protein kinase superfamily [38]. Roman numerals above the sequence indicate conserved kinase subdomains [38].
(PDF)
N. benthamiana plants were infected with TRV, TRV:MEK2, TRV:MAP3Kα, TRV:MAP3Kε, TRV:EDS1, TRV:NDR1, and TRV:RAR1. Four weeks after infection, qRT-PCR was used to assess the expression of the targeted gene in the silenced plants relative to plants infected with empty TRV. Values are means ± SE of three biological repeats.
(TIF)
Leaves of the pepper line ECW30R were syringe-infiltrated with a 10 mM MgCl2 mock solution or with suspensions (1 x 107 CFU/ml) of the following Xe strains: Xe wild-type, Xe xopAU:GnR, Xe avrBs2:KnR, Xe xopAU:GnR/avrBs2:KnR, and Xe xopAU:GnR/avrBs2:KnR complemented with xopAU. Electrolyte leakage (A) and bacterial growth (B) in the inoculated areas were quantified at the indicated days post-inoculation (dpi). The box plots display 25th, 50th (middle line) and 75th percentiles (in A, n = 4; in B, n = 5). An asterisk indicates a significant difference (Mann-Whitney U test, p value <0.05) compared to Xe avrBs2:KnR.
(TIF)
Total protein was extracted from Xe or Xcc bacteria (A), yeast (B, C and D), and N. benthamiana plants (E), separated by SDS-PAGE and immunoblotted with the indicated antibodies. In (E) asterisks indicate bands corresponding to the full-length proteins.
(TIF)
N. benthamiana leaves were syringe-infiltrated with a 10 mM MgCl2 mock solution (Mock) or with suspensions (5 x 107 CFU/ml) of Xcc strains containing a vector for expression of XopAU-HA and XopAUK240A-HA, or an empty vector (EV). (A) Photograph of an inoculated leaf at two days post-inoculation (dpi). Electrolyte leakage (B) and bacterial growth (C) in the inoculated areas was quantified at the indicated hours (hpi) and days post-inoculation (dpi), respectively. The box plots display 25th, 50th (middle line) and 75th percentiles (n = 5). An asterisk indicates a significant difference (Mann-Whitney U test, p value <0.05) compared to Xcc containing an EV.
(TIF)
(A) Yeast expressing the indicated combinations of bait and prey were spotted on either selective medium (-HWUL) or non-selective medium (-HWU) with or without the addition of X-gal. (B) Western blot analysis to assess expression of NbMEK2 and CaMKK2 in yeast. Total protein was extracted from yeast, separated by SDS-PAGE and immunoblotted with α:HA antibodies.
(TIF)
Acknowledgments
We thank Dor Salomon for critically reading of the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was funded by the United States-Israel Binational Science Foundation (http://www.bsf.org.il/BSFPublic/Default.aspx; grant no. 2015062; GS) and is based upon work from COST Actions FA1208 SUSTAIN (https://www.cost-sustain.org) and CA16107 EuroXanth (https://euroxanth.eu), supported by COST (European Cooperation in Science and Technology). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jones JD, Dangl JL. The plant immune system. Nature 2006; 444: 323–329. doi: 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- 2.Zipfel C. Plant pattern-recognition receptors. Trends Immunol 2014; 35: 345–351. [DOI] [PubMed] [Google Scholar]
- 3.Boller T, Felix G. A Renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 2009; 60: 379–406. doi: 10.1146/annurev.arplant.57.032905.105346 [DOI] [PubMed] [Google Scholar]
- 4.Macho AP, Zipfel C. Plant PRRs and the activation of innate immune signaling. Mol Cell 2014; 54: 263–272. doi: 10.1016/j.molcel.2014.03.028 [DOI] [PubMed] [Google Scholar]
- 5.Asai S, Shirasu K. Plant cells under siege: plant immune system versus pathogen effectors. Curr Opin Plant Biol 2015; 28: 1–8. doi: 10.1016/j.pbi.2015.08.008 [DOI] [PubMed] [Google Scholar]
- 6.Cui H, Tsuda K, Parker JE. Effector-triggered immunity: from pathogen perception to robust defense. Annu Rev Plant Biol 2015; 66: 487–511. doi: 10.1146/annurev-arplant-050213-040012 [DOI] [PubMed] [Google Scholar]
- 7.Mur LA, Kenton P, Lloyd AJ, Ougham H, Prats E. The hypersensitive response; the centenary is upon us but how much do we know? J Exper Bot 2008; 59: 501–520. [DOI] [PubMed] [Google Scholar]
- 8.Meng X, Zhang S. MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol 2013; 51: 245–266. doi: 10.1146/annurev-phyto-082712-102314 [DOI] [PubMed] [Google Scholar]
- 9.del Pozo O, Pedley KF, Martin GB. MAPKKKα is a positive regulator of cell death associated with both plant immunity and disease. The EMBO J 2004; 23: 3072–3082. doi: 10.1038/sj.emboj.7600283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Melech‐Bonfil S, Sessa G. Tomato MAPKKKε is a positive regulator of cell‐death signaling networks associated with plant immunity. The Plant J 2010; 64: 379–391. [DOI] [PubMed] [Google Scholar]
- 11.Oh C-S, Martin GB. Tomato 14-3-3 protein TFT7 interacts with a MAP kinase kinase to regulate immunity-associated programmed cell death mediated by diverse disease resistance proteins. J Biol Chem 2011; 286: 14129–14136. doi: 10.1074/jbc.M111.225086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang K-Y, Liu Y, Zhang S. Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc Natl Acad Sci USA 2001; 98: 741–746. doi: 10.1073/pnas.98.2.741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asai T, Tena G, Plotnikova J, Willmann MR, Chiu W-L, et al. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 2002; 415: 977–983. doi: 10.1038/415977a [DOI] [PubMed] [Google Scholar]
- 14.Segonzac C, Feike D, Gimenez-Ibanez S, Hann DR, Zipfel C, et al. Hierarchy and roles of pathogen-associated molecular pattern-induced responses in Nicotiana benthamiana. Plant Physiol 2011; 156: 687–699. doi: 10.1104/pp.110.171249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galan JE, Lara-Tejero M, Marlovits TC, Wagner S. Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annu Rev Microbiol 2014; 68: 415–438. doi: 10.1146/annurev-micro-092412-155725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng F, Zhou JM. Plant-bacterial pathogen interactions mediated by type III effectors. Curr Opin Plant Biol 2012; 15: 469–476. doi: 10.1016/j.pbi.2012.03.004 [DOI] [PubMed] [Google Scholar]
- 17.Macho AP. Subversion of plant cellular functions by bacterial type III effectors: beyond suppression of immunity. New Phytol 2016; 210: 51–57. doi: 10.1111/nph.13605 [DOI] [PubMed] [Google Scholar]
- 18.Shan L, He P, Sheen J. Intercepting host MAPK signaling cascades by bacterial type III effectors. Cell Host Microbe 2007; 1: 167–174. doi: 10.1016/j.chom.2007.04.008 [DOI] [PubMed] [Google Scholar]
- 19.Mukherjee S, Keitany G, Li Y, Wang Y, Ball HL, et al. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science 2006; 312: 1211–1214. doi: 10.1126/science.1126867 [DOI] [PubMed] [Google Scholar]
- 20.Haneda T, Ishii Y, Shimizu H, Ohshima K, Iida N, et al. Salmonella type III effector SpvC, a phosphothreonine lyase, contributes to reduction in inflammatory response during intestinal phase of infection. Cell Microbiol 2012; 14: 485–499. doi: 10.1111/j.1462-5822.2011.01733.x [DOI] [PubMed] [Google Scholar]
- 21.Zhang J, Shao F, Li Y, Cui H, Chen L, et al. A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 2007; 1: 175–185. doi: 10.1016/j.chom.2007.03.006 [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Li J, Hou S, Wang X, Li Y, et al. A Pseudomonas syringae ADP-ribosyltransferase inhibits Arabidopsis mitogen-activated protein kinase kinases. Plant Cell 2010; 22: 2033–2044. doi: 10.1105/tpc.110.075697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cui H, Wang Y, Xue L, Chu J, Yan C, et al. Pseudomonas syringae effector protein AvrB perturbs Arabidopsis hormone signaling by activating MAP kinase 4. Cell Host Microbe 2010; 7: 164–175. doi: 10.1016/j.chom.2010.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones JB, Stall RE, Bouzar H. Diversity among xanthomonads pathogenic on pepper and tomato. Annu Rev Phytopathol 1998; 36: 41–58. doi: 10.1146/annurev.phyto.36.1.41 [DOI] [PubMed] [Google Scholar]
- 25.Roden JA, Belt B, Ross JB, Tachibana T, Vargas J, et al. A genetic screen to isolate type III effectors translocated into pepper cells during Xanthomonas infection. Proc Natl Acad Sci USA 2004; 101: 16624–16629. doi: 10.1073/pnas.0407383101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teper D, Burstein D, Salomon D, Gershovitz M, Pupko T, et al. Identification of novel Xanthomonas euvesicatoria type III effector proteins by a machine‐learning approach. Mol Plant Pathol 2016; 17: 398–411. doi: 10.1111/mpp.12288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thieme F, Koebnik R, Bekel T, Berger C, Boch J, et al. Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. J Bacteriol 2005; 187: 7254–7266. doi: 10.1128/JB.187.21.7254-7266.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim J-G, Stork W, Mudgett MB. Xanthomonas type III effector XopD desumoylates tomato transcription factor SlERF4 to suppress ethylene responses and promote pathogen growth. Cell Host Microbe 2013; 13: 143–154. doi: 10.1016/j.chom.2013.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor KW, Kim JG, Su XB, Aakre CD, Roden JA, et al. Tomato TFT1 is required for PAMP-triggered immunity and mutations that prevent T3S effector XopN from binding to TFT1 attenuate Xanthomonas virulence. PLoS Pathog 2012; 8: e1002768 doi: 10.1371/journal.ppat.1002768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teper D, Salomon D, Sunitha S, Kim JG, Mudgett MB, et al. Xanthomonas euvesicatoria type III effector XopQ interacts with tomato and pepper 14-3-3 isoforms to suppress effector‐triggered immunity. Plant J 2014; 77: 297–309. doi: 10.1111/tpj.12391 [DOI] [PubMed] [Google Scholar]
- 31.Bartetzko V, Sonnewald S, Vogel F, Hartner K, Stadler R, et al. The Xanthomonas campestris pv. vesicatoria type III effector protein XopJ inhibits protein secretion: evidence for interference with cell wall-associated defense responses. Mol Plant Microbe Interact 2009; 22: 655–664. doi: 10.1094/MPMI-22-6-0655 [DOI] [PubMed] [Google Scholar]
- 32.Üstün S, Bartetzko V, Börnke F. The Xanthomonas campestris type III effector XopJ targets the host cell proteasome to suppress salicylic-acid mediated plant defence. PLoS Pathog 2013; 9: e1003427 doi: 10.1371/journal.ppat.1003427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Üstun S, Bornke F. The Xanthomonas campestris type III effector XopJ proteolytically degrades proteasome subunit RPT6. Plant Physiol 2015; 168: 107–119. doi: 10.1104/pp.15.00132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo Y, Figueiredo F, Jones J, Wang N. HrpG and HrpX play global roles in coordinating different virulence traits of Xanthomonas axonopodis pv. citri. Mol Plant Microbe Interact 2011; 24: 649–661. doi: 10.1094/MPMI-09-10-0209 [DOI] [PubMed] [Google Scholar]
- 35.Koebnik R, Kruger A, Thieme F, Urban A, Bonas U. Specific binding of the Xanthomonas campestris pv. vesicatoria AraC-type transcriptional activator HrpX to plant-inducible promoter boxes. J Bacteriol 2006; 188: 7652–7660. doi: 10.1128/JB.00795-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parkinson N, Cowie C, Heeney J, Stead D. Phylogenetic structure of Xanthomonas determined by comparison of gyrB sequences. Int J Syst Evol Microbiol 2009; 59: 264–274. doi: 10.1099/ijs.0.65825-0 [DOI] [PubMed] [Google Scholar]
- 37.Midha S, Patil PB. Genomic insights into the evolutionary origin of Xanthomonas axonopodis pv. citri and its ecological relatives. Appl Environ Microbiol 2014; 80: 6266–6279. doi: 10.1128/AEM.01654-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J 1995; 9: 576–596. [PubMed] [Google Scholar]
- 39.Stintzi A, Heitz T, Prasad V, Wiedemann-Merdinoglu S, Kauffmann S, et al. Plant ‘pathogenesis-related’ proteins and their role in defense against pathogens. Biochimie 1993; 75: 687–706. [DOI] [PubMed] [Google Scholar]
- 40.Martin GB, Bogdanove AJ, Sessa G. Understanding the functions of plant disease resistance proteins. Annu Rev Plant Biol 2003; 54: 23–61. doi: 10.1146/annurev.arplant.54.031902.135035 [DOI] [PubMed] [Google Scholar]
- 41.Lu R, Martin-Hernandez AM, Peart JR, Malcuit I, Baulcombe DC. Virus-induced gene silencing in plants. Methods 2003; 30: 296–303. [DOI] [PubMed] [Google Scholar]
- 42.Gabriels SH, Vossen JH, Ekengren SK, van Ooijen G, Abd-El-Haliem AM, et al. An NB-LRR protein required for HR signalling mediated by both extra- and intracellular resistance proteins. Plant J 2007; 50: 14–28. doi: 10.1111/j.1365-313X.2007.03027.x [DOI] [PubMed] [Google Scholar]
- 43.Kearney B, Staskawicz BJ. Widespread distribution and fitness contribution of Xanthomonas campestris avirulence gene avrBs2. Nature 1990; 346: 385–386. doi: 10.1038/346385a0 [DOI] [PubMed] [Google Scholar]
- 44.Wichmann G, Bergelson J. Effector genes of Xanthamonas axonopodis pv. vesicatoria promote transmission and enhance other fitness traits in the field. Genetics 2004; 166: 693–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lebel E, Heifetz P, Thorne L, Uknes S, Ryals J, et al. Functional analysis of regulatory sequences controlling PR-1 gene expression in Arabidopsis. Plant J 1998; 16: 223–233. [DOI] [PubMed] [Google Scholar]
- 46.Merchante C, Alonso JM, Stepanova AN. Ethylene signaling: simple ligand, complex regulation. Curr Opin Plant Biol 2013; 16: 554–560. doi: 10.1016/j.pbi.2013.08.001 [DOI] [PubMed] [Google Scholar]
- 47.Stintzi A. The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl Acad Sci USA 2000; 97: 10625–10630. doi: 10.1073/pnas.190264497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Popa C, Coll NS, Valls M, Sessa G. Yeast as a heterologous model system to uncover type III effector function. PLoS Pathog 2016; 12: e1005360 doi: 10.1371/journal.ppat.1005360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou J, Loh Y-T, Bressan RA, Martin GB. The tomato gene Pti1 encodes a serine/threonine kinase that is phosphorylated by Pto and is involved in the hypersensitive response. Cell 1995; 83: 925–935. [DOI] [PubMed] [Google Scholar]
- 50.Salomon D, Orth K. What pathogens have taught us about posttranslational modifications. Cell Host Microbe 2013; 14: 269–279. doi: 10.1016/j.chom.2013.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Navarro L, Koller A, Nordfelth R, Wolf-Watz H, Taylor S, et al. Identification of a molecular target for the Yersinia protein kinase A. Mol Cell 2007; 26: 465–477. doi: 10.1016/j.molcel.2007.04.025 [DOI] [PubMed] [Google Scholar]
- 52.Kim DW, Lenzen G, Page AL, Legrain P, Sansonetti PJ, et al. The Shigella flexneri effector OspG interferes with innate immune responses by targeting ubiquitin-conjugating enzymes. Proc Natl Acad Sci USA 2005; 102: 14046–14051. doi: 10.1073/pnas.0504466102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Juris SJ, Rudolph AE, Huddler D, Orth K, Dixon JE. A distinctive role for the Yersinia protein kinase: actin binding, kinase activation, and cytoskeleton disruption. Proc Natl Acad Sci USA 2000; 97: 9431–9436. doi: 10.1073/pnas.170281997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trasak C, Zenner G, Vogel A, Yuksekdag G, Rost R, et al. Yersinia protein kinase YopO is activated by a novel G-actin binding process. J Biol Chem 2007; 282: 2268–2277. doi: 10.1074/jbc.M610071200 [DOI] [PubMed] [Google Scholar]
- 55.Zhou Y, Dong N, Hu LY, Shao F. The Shigella type three secretion system effector OspG directly and specifically binds to host ubiquitin for activation. Plos One 2013; 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang J, Shao F, Li Y, Cui H, Chen L, et al. A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 2007; 1: 175–185. doi: 10.1016/j.chom.2007.03.006 [DOI] [PubMed] [Google Scholar]
- 57.Escote X, Zapater M, Clotet J, Posas F. Hog1 mediates cell-cycle arrest in G1 phase by the dual targeting of Sic1. Nat Cell Biol 2004; 6: 997–1002. doi: 10.1038/ncb1174 [DOI] [PubMed] [Google Scholar]
- 58.Fujimura HA. Yeast homolog of mammalian mitogen-activated protein kinase, FUS3/DAC2 kinase, is required both for cell fusion and for G1 arrest of the cell cycle and morphological changes by the cdc37 mutation. J Cell Sci 1994; 107: 2617–2622. [DOI] [PubMed] [Google Scholar]
- 59.Teper D, Sunitha S, Martin GB, Sessa G. Five Xanthomonas type III effectors suppress cell death induced by components of immunity-associated MAP kinase cascades. Plant Signal Behav 2015; 10: e1064573 doi: 10.1080/15592324.2015.1064573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schulze S, Kay S, Büttner D, Egler M, Eschen-Lippold L, et al. Analysis of new type III effectors from Xanthomonas uncovers XopB and XopS as suppressors of plant immunity. New Phytologist 2012; 195: 894–911. doi: 10.1111/j.1469-8137.2012.04210.x [DOI] [PubMed] [Google Scholar]
- 61.Szczesny R, Büttner D, Escolar L, Schulze S, Seiferth A, et al. Suppression of the AvrBs1-specific hypersensitive response by the YopJ effector homolog AvrBsT from Xanthomonas depends on a SNF1-related kinase. New Phytologist 2010; 187: 1058–1074. doi: 10.1111/j.1469-8137.2010.03346.x [DOI] [PubMed] [Google Scholar]
- 62.Cho SK, Larue CT, Chevalier D, Wang H, Jinn T-L, et al. Regulation of floral organ abscission in Arabidopsis thaliana. Proc Natl Acad Sci USA 2008; 105: 15629–15634. doi: 10.1073/pnas.0805539105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heinrich M, Baldwin IT, Wu J. Two mitogen-activated protein kinase kinases, MKK1 and MEK2, are involved in wounding and specialist lepidopteran herbivore Manduca sexta-induced responses in Nicotiana attenuata. J Exp Bot 2011: 62: 4355–4365. doi: 10.1093/jxb/err162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim S-H, Woo D-H, Kim J-M, Lee S-Y, Chung WS, et al. Arabidopsis MKK4 mediates osmotic-stress response via its regulation of MPK3 activity. Biochem Biophys Res Commun 2011; 412: 150–154. doi: 10.1016/j.bbrc.2011.07.064 [DOI] [PubMed] [Google Scholar]
- 65.Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S. Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 2007; 19: 63–73. doi: 10.1105/tpc.106.048298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu J, Yang KY, Yoo SJ, Liu Y, Ren D, et al. Reactive oxygen species in signalling the transcriptional activation of WIPK expression in tobacco. Plant Cell & Environ 2014; 37: 1614–1625. [DOI] [PubMed] [Google Scholar]
- 67.Teper D, Burstein D, Salomon D, Gershovitz M, Pupko T, et al. Identification of novel Xanthomonas euvesicatoria type III effector proteins by a machine-learning approach. Mol Plant Pathol 2015; 17: 398–411. doi: 10.1111/mpp.12288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007; 23: 2947–2948. doi: 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 69.Salomon D, Sessa G. Identification of growth inhibition phenotypes induced by expression of bacterial type III effectors in yeast. J Vis Exp 2010: doi: 10.3791/1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Minsavage G, Dahlbeck D, Whalen M, Kearney B, Bonas U, et al. Gene-for-gene relationships specifying disease resistance in Xanthomonas campestris pv. vesicatoria-pepper interactions. Mol Plant Microbe Interact 1990; 3: 41–47. [Google Scholar]
- 71.Goodin MM, Zaitlin D, Naidu RA, Lommel SA. Nicotiana benthamiana: its history and future as a model for plant-pathogen interactions. Mol Plant Microbe Interact 2008; 21: 1015–1026. doi: 10.1094/MPMI-21-8-1015 [DOI] [PubMed] [Google Scholar]
- 72.Scott JW, Jones JB, Somodi GC, Stall RE. Screening tomato accessions for resistance to Xanthomonas campestris pv. vesicatoria, race T3. Hortscience 1995; 30: 579–581. [Google Scholar]
- 73.Rott PC, Costet L, Davis MJ, Frutos R, Gabriel DW. At least two separate gene clusters are involved in albicidin production by Xanthomonas albilineans. J Bacteriol 1996; 178: 4590–4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zuo J, Niu QW, Chua NH. An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 2000; 24: 265–273. [DOI] [PubMed] [Google Scholar]
- 75.Porra RJ. The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynth Res 2002; 73: 149–156. doi: 10.1023/A:1020470224740 [DOI] [PubMed] [Google Scholar]
- 76.Golemis EA, Serebriiskii I, Finley RL Jr, Kolonin MG, Gyuris J, et al. Interaction trap/two-hybrid system to identify interacting proteins. Curr Protoc Cell Biol 2011; Chapter 17: Unit 17 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Quintero MJ, Maya D, Arévalo-Rodríguez M, Cebolla Á, Chávez S. An improved system for estradiol-dependent regulation of gene expression in yeast. Microb Cell Fact 2007; 6: 10 doi: 10.1186/1475-2859-6-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zeng L, Velásquez AC, Munkvold KR, Zhang J, Martin GB. A tomato LysM receptor-like kinase promotes immunity and its kinase activity is inhibited by AvrPtoB. Plant J 2012; 69: 92–103. doi: 10.1111/j.1365-313X.2011.04773.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen H, Zou Y, Shang Y, Lin H, Wang Y, et al. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 2008; 146: 368–376. doi: 10.1104/pp.107.111740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pfaffl MW. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res 2001; 29: e45–e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zigdon H, Savidor A, Levin Y, Meshcheriakova A, Schiffmann R, et al. Identification of a biomarker in cerebrospinal fluid for neuronopathic forms of Gaucher disease. PLoS One 2015; 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kelstrup CD, Young C, Lavallee R, Nielsen ML, Olsen JV. Optimized fast and sensitive acquisition methods for shotgun proteomics on a quadrupole orbitrap mass spectrometer. J Proteome Res 2012; 11: 3487–3497. doi: 10.1021/pr3000249 [DOI] [PubMed] [Google Scholar]
- 83.Rauniyar N. Parallel Reaction Monitoring: A targeted experiment performed using high resolution and high mass accuracy mass spectrometry. Int J Mol Sci 2015; 16: 28566–28581. doi: 10.3390/ijms161226120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.UniProt C. UniProt: a hub for protein information. Nucleic Acids Res 2015; 43: D204–212. doi: 10.1093/nar/gku989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gillette MA, Carr SA. Quantitative analysis of peptides and proteins in biomedicine by targeted mass spectrometry. Nature Meth 2013; 10: 28–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010; 26: 966–968. doi: 10.1093/bioinformatics/btq054 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(DOCX)
(DOCX)
(XLSX)
Nucleic acid sequences of xopAU homologs were aligned with ClustalX multiple sequence alignment tool. Sequences and corresponding NCBI accession numbers are reported in S1 Table. A dashed line represents a gap in the alignment. Asterisks indicate nucleotide conserved in all the homologs. Nucleotides are color-coded.
(PDF)
(A) Genomic location of the xopAU allelic variant of group 1 Xanthomonas strains in X. euvesicatoria (acc. num. NC_007508.1) and X. oryzae (acc. num. CP003057.2). (B) Genomic location of the xopAU group 2 allelic variant in X. fragariae (acc. num. CP016830.1) and X. gardneri (acc. num. CP018728.1), and corresponding genomic region in the X. arboricola strain (acc. num. CP012251.1), which does not contain the xopAU allele. Numbers and arrows represent genomic location and open reading frames (ORF), respectively. Locus tags are indicated below each ORF, which are colored based on DNA sequence homology.
(PDF)
Protein sequences of the XopAU homologs (S1 Table) were aligned with the COBALT multiple sequence alignment tool (https://www.ncbi.nlm.nih.gov/tools/cobalt/cobalt.cgi?) using default parameters. Blue fonts represent identical amino acids; yellow fonts represent nearly invariant residues in the protein kinase superfamily [38]. Roman numerals above the sequence indicate conserved kinase subdomains [38].
(PDF)
N. benthamiana plants were infected with TRV, TRV:MEK2, TRV:MAP3Kα, TRV:MAP3Kε, TRV:EDS1, TRV:NDR1, and TRV:RAR1. Four weeks after infection, qRT-PCR was used to assess the expression of the targeted gene in the silenced plants relative to plants infected with empty TRV. Values are means ± SE of three biological repeats.
(TIF)
Leaves of the pepper line ECW30R were syringe-infiltrated with a 10 mM MgCl2 mock solution or with suspensions (1 x 107 CFU/ml) of the following Xe strains: Xe wild-type, Xe xopAU:GnR, Xe avrBs2:KnR, Xe xopAU:GnR/avrBs2:KnR, and Xe xopAU:GnR/avrBs2:KnR complemented with xopAU. Electrolyte leakage (A) and bacterial growth (B) in the inoculated areas were quantified at the indicated days post-inoculation (dpi). The box plots display 25th, 50th (middle line) and 75th percentiles (in A, n = 4; in B, n = 5). An asterisk indicates a significant difference (Mann-Whitney U test, p value <0.05) compared to Xe avrBs2:KnR.
(TIF)
Total protein was extracted from Xe or Xcc bacteria (A), yeast (B, C and D), and N. benthamiana plants (E), separated by SDS-PAGE and immunoblotted with the indicated antibodies. In (E) asterisks indicate bands corresponding to the full-length proteins.
(TIF)
N. benthamiana leaves were syringe-infiltrated with a 10 mM MgCl2 mock solution (Mock) or with suspensions (5 x 107 CFU/ml) of Xcc strains containing a vector for expression of XopAU-HA and XopAUK240A-HA, or an empty vector (EV). (A) Photograph of an inoculated leaf at two days post-inoculation (dpi). Electrolyte leakage (B) and bacterial growth (C) in the inoculated areas was quantified at the indicated hours (hpi) and days post-inoculation (dpi), respectively. The box plots display 25th, 50th (middle line) and 75th percentiles (n = 5). An asterisk indicates a significant difference (Mann-Whitney U test, p value <0.05) compared to Xcc containing an EV.
(TIF)
(A) Yeast expressing the indicated combinations of bait and prey were spotted on either selective medium (-HWUL) or non-selective medium (-HWU) with or without the addition of X-gal. (B) Western blot analysis to assess expression of NbMEK2 and CaMKK2 in yeast. Total protein was extracted from yeast, separated by SDS-PAGE and immunoblotted with α:HA antibodies.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.